
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 02 March 2022
Sec. Infectious Agents and Disease
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.841286
 Linwei Wei1,2,3,4
Linwei Wei1,2,3,4 Jin Shao1,2,3,4
Jin Shao1,2,3,4 Yinggai Song1,2,3,4
Yinggai Song1,2,3,4 Zhe Wan1,2,3,4
Zhe Wan1,2,3,4 Limin Yao1,2,3,4
Limin Yao1,2,3,4 Hong Wang1,2,3,4
Hong Wang1,2,3,4 Jin Yu1,2,3,4*
Jin Yu1,2,3,4*
Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) has emerged as a powerful microorganism identification tool. Research on MALDI-TOF MS identification of rare filamentous fungi is still lacking. This study aimed to evaluate the performance of MALDI-TOF MS in the identification of Scedosporium, Acremonium-like, Scopulariopsis, and Microascus species. Sabouraud broth cultivation and formic acid/acetonitrile protein extraction were used for MALDI-TOF MS identification by a Bruker Biotyper system. An in-house database containing 29 isolates of Scedosporium, Acremonium-like, Scopulariopsis, and Microascus spp. was constructed. A total of 52 clinical isolates were identified using the Bruker Filamentous Fungi Library v1.0 (FFL v1.0) alone, and Filamentous Fungi Library v1.0 plus the in-house library, respectively. The mass spectrum profile (MSP) dendrograms of the 28 Scedosporium isolates, 26 Acremonium-like isolates, and 27 Scopulariopsis and Microascus isolates were constructed by MALDI Biotyper OC 4.0 software, respectively. The correct species identification rate significantly improved when using the combined databases compared with that when using FFL v1.0 alone (Scedosporium spp., 75% versus 0%; Acremonium-like spp., 100% versus 0%; Scopulariopsis and Microascus spp., 100% versus 62.5%). The MSP dendrograms differentiated Acremonium-like species, Scopulariopsis and Microascus species clearly, but cannot distinguish species in the Scedosporium apiospermum complex. In conclusion, with an expanded database, MALDI-TOF MS is an effective tool for the identification of Scedosporium, Acremonium-like, Scopulariopsis, and Microascus species.
In recent years, with the development of organ transplantation and the widespread use of immunosuppressants and antibiotics, the number of cases of invasive infections caused by filamentous fungi have increased (Brown et al., 2012). In addition to that of infection caused by the most common filamentous fungus, Aspergillus, the incidence of infections caused by non-aspergillus filamentous fungi such as Fusarium, Mucorales, Scedosporium, and other rare fungi is also increasing (Miceli and Lee, 2011). Scedosporium, Acremonium-like, Scopulariopsis, and Microascus spp. are saprobic fungi commonly found in the environment, some species have been reported as pathogens of humans, and most are opportunistic (Ramirez-Garcia et al., 2018; Perez-Cantero and Guarro, 2020a,b). Within the genus Scedosporium, the Scedosporium apiospermum species complex and Scedosporium aurantiacum are most related to human diseases; the former currently contains Scedosporium apiospermum, Scedosporium boydii, Scedosporium ellipsoideum, Scedosporium angustum, and Scedosporium fusarium (Ramirez-Garcia et al., 2018). Aremonium-like spp. comprise a high diversity of morphologically and genetically related fungi, among which Aremonium egyptiacum and Sarocladium kiliense are the most commonly involved in human diseases (Perez-Cantero and Guarro, 2020b). Scopulariopsis-like spp. include a group of hyaline and dematiaceous fungi, and most of the clinically relevant species belong to the genera Scopulariopsis and Microascus (Perez-Cantero and Guarro, 2020a). The above pathogenic fungi have raised concern in the field of medical mycology, as they show intrinsic resistance to multiple antifungal drugs (Ramirez-Garcia et al., 2018; Perez-Cantero and Guarro, 2020a,b). Additionally, differences in in vitro susceptibility have been reported among species within Scedosporium and Scopulariopsis-like spp. (Lackner et al., 2012; Yao et al., 2015). Therefore, rapid and accurate identification of these fungi is important for timely treatment, as well as for the supplementation of epidemiological data and drug susceptibility studies of a large range of species. However, due to the taxonomic complexity of these genera and the interspecies morphological similarities, morphological methods are often unable to identify these fungi at the species level (Perdomo et al., 2011; Ramirez-Garcia et al., 2018; Perez-Cantero and Guarro, 2020a). DNA sequencing allows accurate identification but is expensive, labor intensive, time consuming, and not suitable for routine laboratory testing.
Over the last few years, matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) has been increasingly used for the laboratory identification of yeast and filamentous fungi because of its high accuracy, simple operation and low cost (Wilkendorf et al., 2020). The main obstacles to the application of this technique to the identification of filamentous fungi are the lack of a sufficient database and of a rapid and effective protein extraction method (Wilkendorf et al., 2020). Few studies have been conducted on the identification of Scedosporium (Del Chierico et al., 2012; Normand et al., 2013; Ranque et al., 2014; Sitterle et al., 2014; Sleiman et al., 2016; Zvezdanova et al., 2019), Acremonium-like (Becker et al., 2014; Rychert et al., 2018), Scopulariopsis and Microascus species (Lau et al., 2013; Becker et al., 2014; Schulthess et al., 2014; Levesque et al., 2015; Riat et al., 2015; Stein et al., 2018; Sacheli et al., 2020) by MALDI-TOF MS. In this study, we developed an in-house database containing 29 strains covering 21 species of Scedosporium, Acremonium-like, Scopulariopsis, and Microascus and challenged the system with 52 clinical isolates to evaluate the identification performance of the Bruker Filamentous Fungi Library v1.0 (FFL v1.0) (Bruker Daltonics, Bremen, Germany) alone and FFL v1.0 plus the in-house database.
A total of 81 clinical isolates preserved at the Research Center for Medical Mycology of Peking University were included in this study. The sources and antifungal susceptibility data of these isolates were shown in the previous studies (Wang et al., 2015; Jagielski et al., 2016; Yao et al., 2019). Twenty out of 28 Scedosporium isolates (four species) were identified by DNA sequencing of the partial β-tubulin (BT2, exons 2–4), calmodulin (CAL, exons 3, 4), second large subunit of RNA polymerase II (RPB2), superoxide dismutase (SOD), and actin (ACT) loci in a previous study (Wang et al., 2015). The remaining eight Scedosporium isolates were identified by BT2 locus sequencing using the same method and the GenBank accession numbers of them were MW528357 to MW5283564. Twenty-six Acremonium-like isolates (seven species) were identified by sequencing the internal transcribed spacer (ITS), ribosomal large subunit (LSU), and transcription elongation factor 1-α (EF1-α) loci in a previous study (Yao et al., 2019). Twenty-seven Scopulariopsis and Microascus isolates (10 species) were identified by sequencing the ITS, LSU, TUB, and EF1-α loci as previously reported (Jagielski et al., 2016). Twenty-seven of the 81 isolates were selected as reference strains to build the novel Beijing Medical University (BMU) database (Table 1). Challenge isolates comprising 20 Scedosporium, 16 Acremonium-like, and 16 Scopulariopsis and Microascus isolates (Table 1) were used to assess the performance of FFL v1.0 alone versus that of the combined databases for species identification.
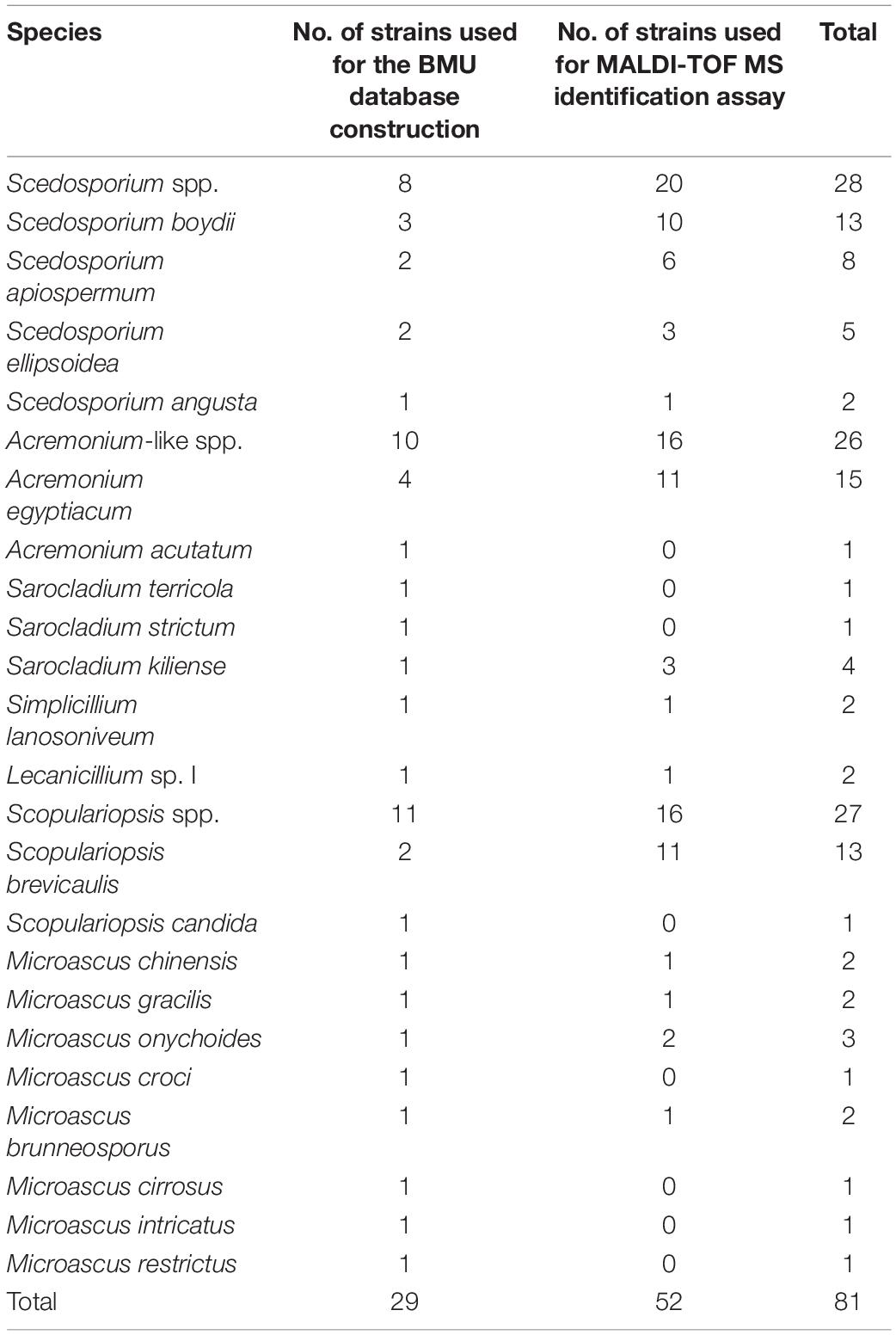
Table 1. Clinical isolates and species used in this study for Beijing Medical University (BMU) database construction and matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) identification assay.
All strains were cultivated on potato dextrose agar (PDA) slants at 28°C for 5–10 days until the colonies reached a size of approximately 1 cm in diameter. The spores and hyphae were scraped by a long, sterile, wet cotton swab; reinoculated in 5 mL of Sabouraud dextrose broth (SDB) (Becton Dickinson, Franklin Lakes, NJ, United States) at 28°C on a Loopster digital rotator (IKA, Staufen, Germany); and incubated for 24–48 h to yield a high quantity of small mycelial balls.
The ethanol/formic acid extraction procedure was performed according to the manufacturer’s instructions. Briefly, 1.5 mL of mycelium was transferred into a 1.5-mL tube (Eppendorf, Hamburg, Germany) after settling and centrifuged at 15,870 × g for 2 min. Then, the pellet was washed twice with 1 mL of deionized water. The supernatant was discarded, and then, 300 μL of deionized water and 900 μL of anhydrous ethanol (Sigma-Aldrich, St. Louis, MO, United States) were added and vortexed in sequence. After 10 min of incubation, the materials were centrifuged at 15,870 × g for 2 min, and the supernatant was discarded. After drying at 37°C, the pellet was thoroughly mixed in 25–100 μL of 70% formic acid (the volume of 70% formic acid depended on the size of the pellet), incubated for 10 min at ambient temperature, mixed with an equal volume of acetonitrile (Sigma-Aldrich, St. Louis, MO, United States) and incubated for another 10 min. This mixture was centrifuged at 15,870 × g for 2 min, and then 1 μL of the supernatant was transferred onto an MTP 384 polished steel MALDI target plate (Bruker Daltonik GmbH, Bremen, Germany), air dried and overlaid with 1 μL of saturated α-cyano-4 hydroxy-cinnamic acid (HCCA) matrix solution (Bruker Daltonics, Bremen, Germany). Finally, the MALDI target was placed into an autoflex speed TOF instrument (Bruker Daltonik, Bremen, Germany).
The reference spectra on Scedosporium, Acremonium-like, Scopulariopsis, and Microascus species in FFL v1.0 only include 4 S. apiospermum [anamorph] P. boydii [teleomorph], 1 A. strictum (now named S. strictum), 7 S. brevicaulis, 1 S. acremonium, and 1 S. brumptii. The isolates used for in-house database construction were deposited in eight target spots. The acquisition settings were as follows: ion source 1 at 19.50 kV, ion source 2 at 18.22 kV, lens at 7.01 kV, and mass range from 2,000 to 20,000 Da. The other parameters were kept the default settings. The process of in-house database building was in accordance with Bruker’s procedures. Three fingerprints from each target spot were manually obtained by Bruker Daltonics FlexControl 3.4 software (summing to a signal strength > 10,000); MALDI Biotyper OC 4.0 software was used to evaluate the quality of the 24 fingerprints, poorly quality and poor repeated mass spectra were removed in GelView, and then, 20–24 mass spectra were used to generate a reference main spectrum profile (MSP).
The challenge isolates were deposited in four target spots. Spectrum acquisition was performed automatically using MALDI Biotyper RTC 4.0 software (Bruker Daltonik, Bremen, Germany). MALDI Biotyper 3.1 (Bruker Daltonics, Bremen, Germany) was applied for spectral acquisition and comparison with reference spectra from FFL v1.0 and FFL v1.0 plus the BMU database. Identification scores ≥ 2.0 and 1.7–1.99 indicated species- and genus-level identification, respectively, and scores of <1.7 indicated no reliable identification. If the best matches in at least three of the four spots tested were consistent, the MALDI-TOF MS identification was considered interpretable. Correct identification indicates that the results were consistent with the DNA sequencing.
We used the representative composite correlation index (CCI) tool by MALDI Biotyper 4.0 software to analyze the relationships between spectra. CCI value around 1 represents a high conformance of spectra, while that near 0 indicates a clear diversity of the spectra. The CCI matrix translated into a heat map in which closely related spectra are represented in hot colors and unrelated spectra in cold colors.
Phylogenetic tree of 28 Scedosporium isolates was constructed by the BT2 sequence using the Kimura two-parameter model and the neighbor-joining (NJ) method. Multilocus phylogenetic tree of 26 Acremonium-like isolates was constructed by combining the ITS-LSU and EF1-α sequences using the Tamura-Nei model and the NJ method. Multilocus phylogenetic tree of 27 Scopulariopsis and Microascus isolates was constructed by combining the ITS, LSU, TUB, and EF1-α sequences using the Tamura-Nei model and the NJ method. The bootstrap method (1,000 replicates) was applied to test for phylogeny. The MSP dendrograms of 28 Scedosporium isolates, 26 Acremonium-like isolates, and 27 Scopulariopsis and Microascus isolates were constructed by MALDI Biotyper 4.0 software.
Statistical analysis was performed by SPSS 26. McNemar’s test was used to compare the species identification rates using FFL v1.0 alone and the combined databases, and a p-value < 0.05 was considered statistically significant.
The mass spectra of Scedosporium, Acremonium-like, Scopulariopsis, and Microascus species are shown in Figures 1–3, respectively. The results of MALDI-TOF MS identification of the 52 Scedosporium, Acremonium-like, Scopulariopsis, and Microascus isolates by FFL v1.0 alone and by the combined databases are shown in Table 2.
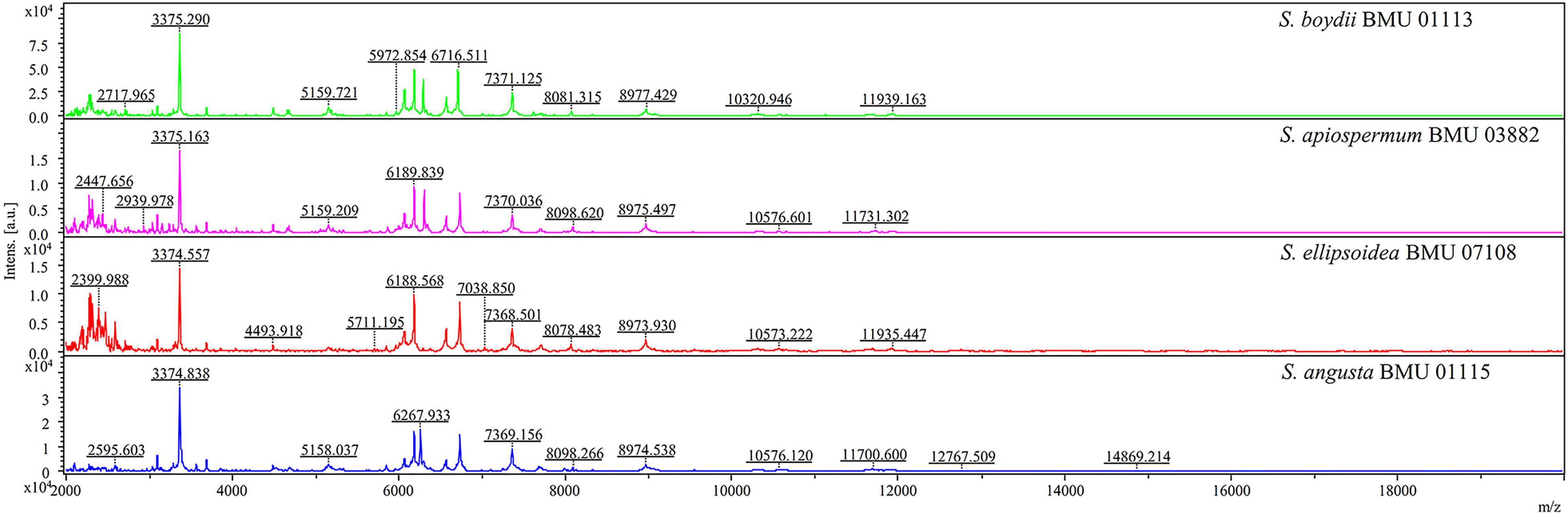
Figure 1. Matrix-assisted laser desorption ionization-time-of-flight mass spectra (m/z 2,000 to 20,000) of Scedosporium species.
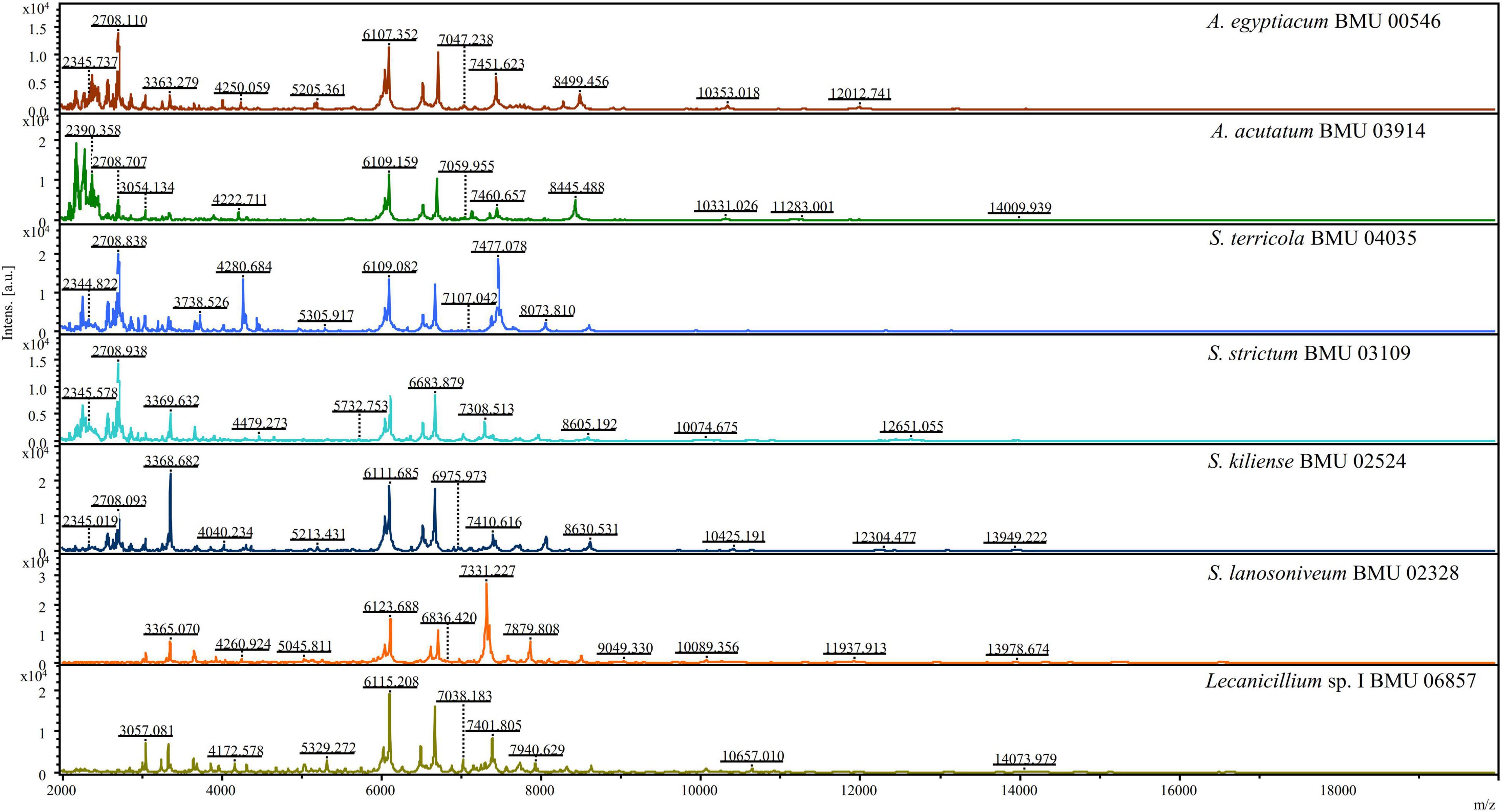
Figure 2. Matrix-assisted laser desorption ionization-time-of-flight mass spectra (m/z 2,000 to 20,000) of Acremonium-like species.
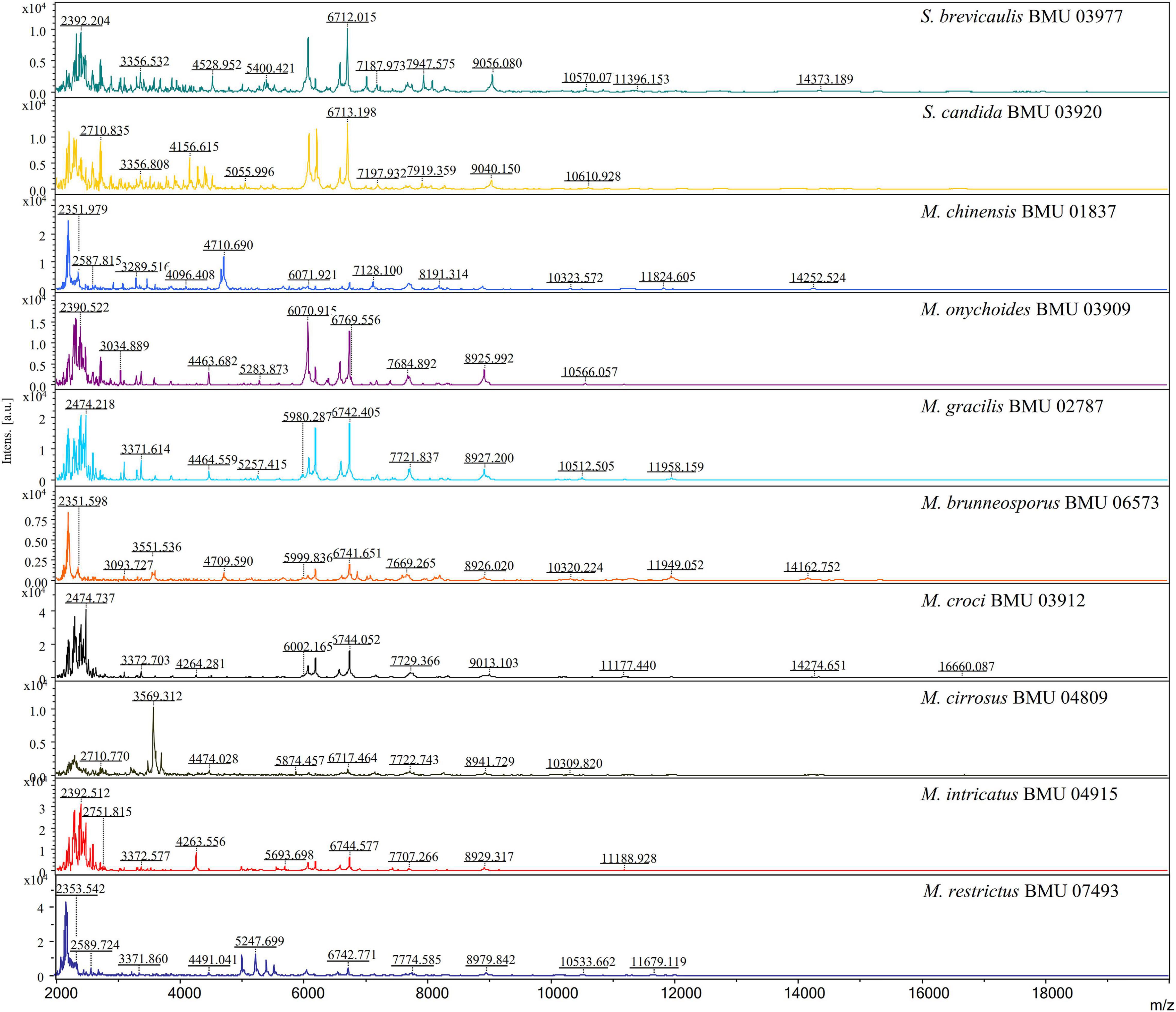
Figure 3. Matrix-assisted laser desorption ionization-time-of-flight mass spectra (m/z 2,000 to 20,000) of Scopulariopsis and Microascus species.
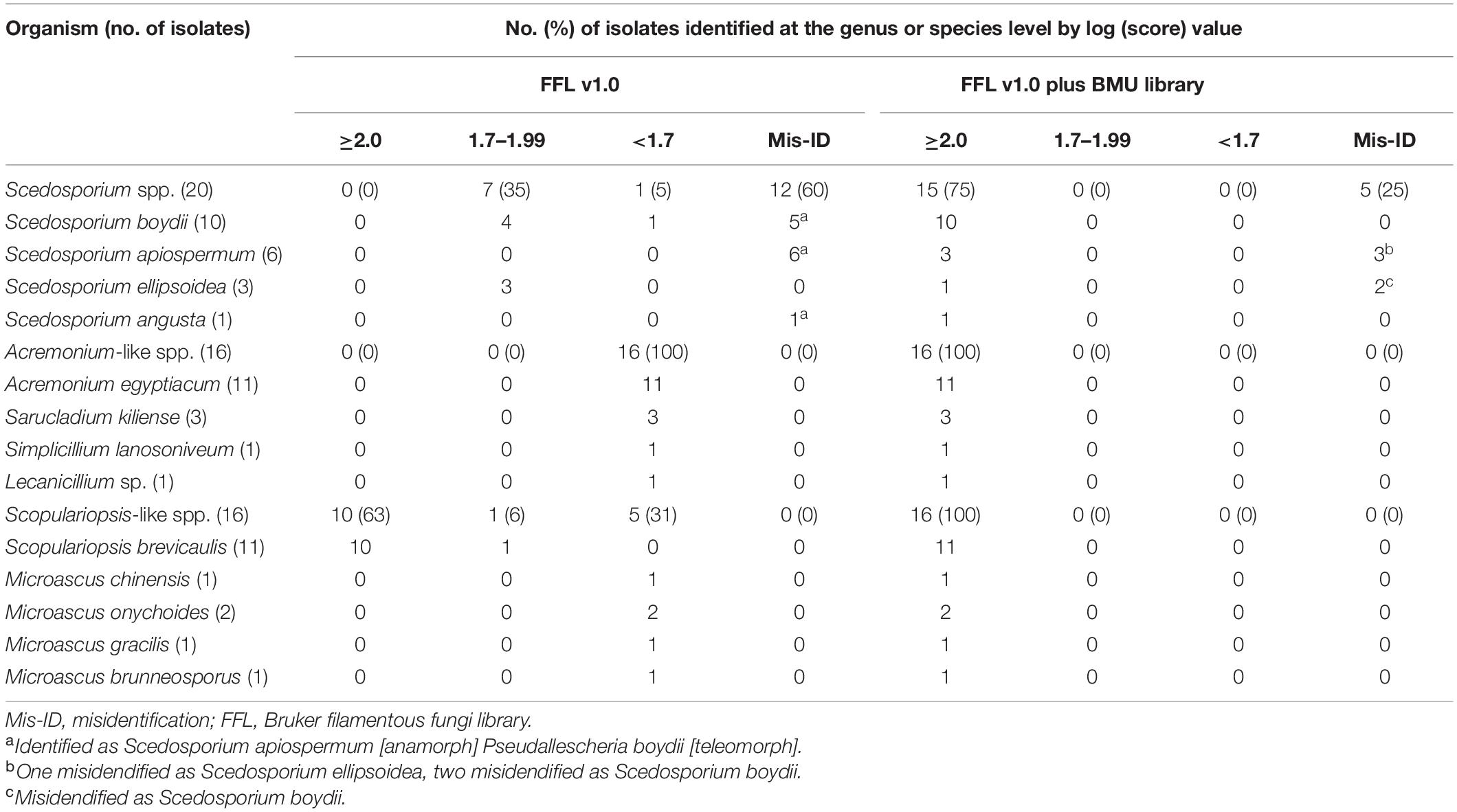
Table 2. Identification of 52 clinical isolates by the Bruker library and the Bruker library plus BMU database.
Using FFL v1.0 alone, 60% (12/20) of Scedosporium isolates were identified with scores ≥ 2.0, while all of them were identified as Scedosporium apiospermum [anamorph] Pseudallescheria boydii [teleomorph], which was the only Scedosporium species represented in FFL v1.0, so clear species identification was not available. Seven isolates (35%) were identified to the genus level and also identified as S. apiospermum [ana] P. boydii [teleo]. Using the combination of FFL v1.0 and the BMU database, the correct species-level identification rates increased significantly to 75% (15/20) (p < 0.001), and all 10 S. boydii isolates were correctly identified. The remaining five isolates were misidentified as species within the S. apiospermum species complex (Table 2).
Using FFL v1.0 alone, none of the Acremonium-like isolates were reliably identified. When supplemented with the in-house database, all isolates were correctly identified to the species level. Even though FFL v1.0 contains one strain of Acremonium strictum (now named Sarocladium strictum), there was no consistent identification of the quadruplicate tests when challenged with one isolate of S. strictum used for BMU database construction.
When challenged with Scopulariopsis and Microascus isolates, FFL v1.0, which contains seven Scopulariopsis brevicaulis, one Scopulariopsis acremonium and one Scopulariopsis brumptii MSPs, identified 62.5% (10/16) and 6.3% (1/16) of the isolates at the species and genus levels, respectively. All of the identified isolates were S. brevicaulis. With the combined databases, the species-level identification rate increased significantly to 100% (16/16) (p = 0.031).
The species- and genus-levels identification rates of Scedosporium, Acremonium-like, Scopulariopsis, and Microascus isolates using the BMU database alone were the same as that using the combined databases.
The CCI matrix of test species indicated that mass spectra within the S. apiospermum complex were highly similar, while those within Acremonium-like, Scopulariopsis, and Microascus spp. were more diverse (Figure 4).
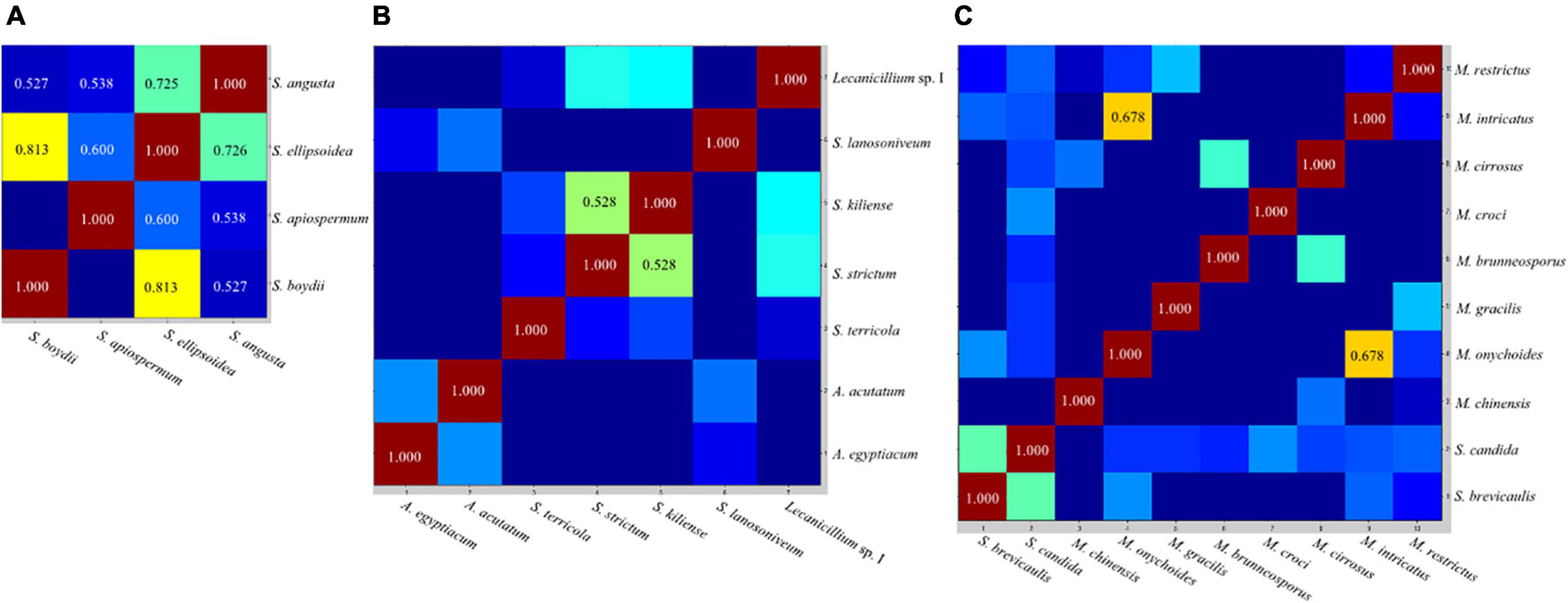
Figure 4. Representative composite correlation index (CCI) matrix derived from Scedosporium (A), Acremonium-like (B), Scopulariopsis and Microascus species (C). By comparing the spectra of different species, numerical correlation index is obtained to form the CCI matrix and translated into a heat map. Closely related spectra were represented in hot colors and with higher CCI value, and unrelated spectra in cold colors with lower CCI value. CCI value ≥ 0.5 were shown in the view.
The MSP dendrogram of the Scedosporium isolates cannot differentiate species in the S. apiospermum complex (Figure 5). As for the Acremonium-like isolates, the MSP dendrogram showed clear separation of different species, and the topology of the dendrogram appeared to be similar to that of the multilocus phylogenetic tree (Figure 6). The MSP dendrogram of Scopulariopsis and Microascus spp. clustered 27 isolates to the Scopulariopsis clade and Microascus clade (Figure 7). Isolates of M. gracilis and M. croci clustered in one subclade, whereas isolates of M. onychoides and M. intricatus clustered in another subclade, and the above two pairs of species were also close in the phylogenetic tree. The other isolates belonging to the same species clustered together into respective subclades. However, the topology of the MSP dendrogram and that of the multilocus phylogenetic tree of Microascus isolates differed.
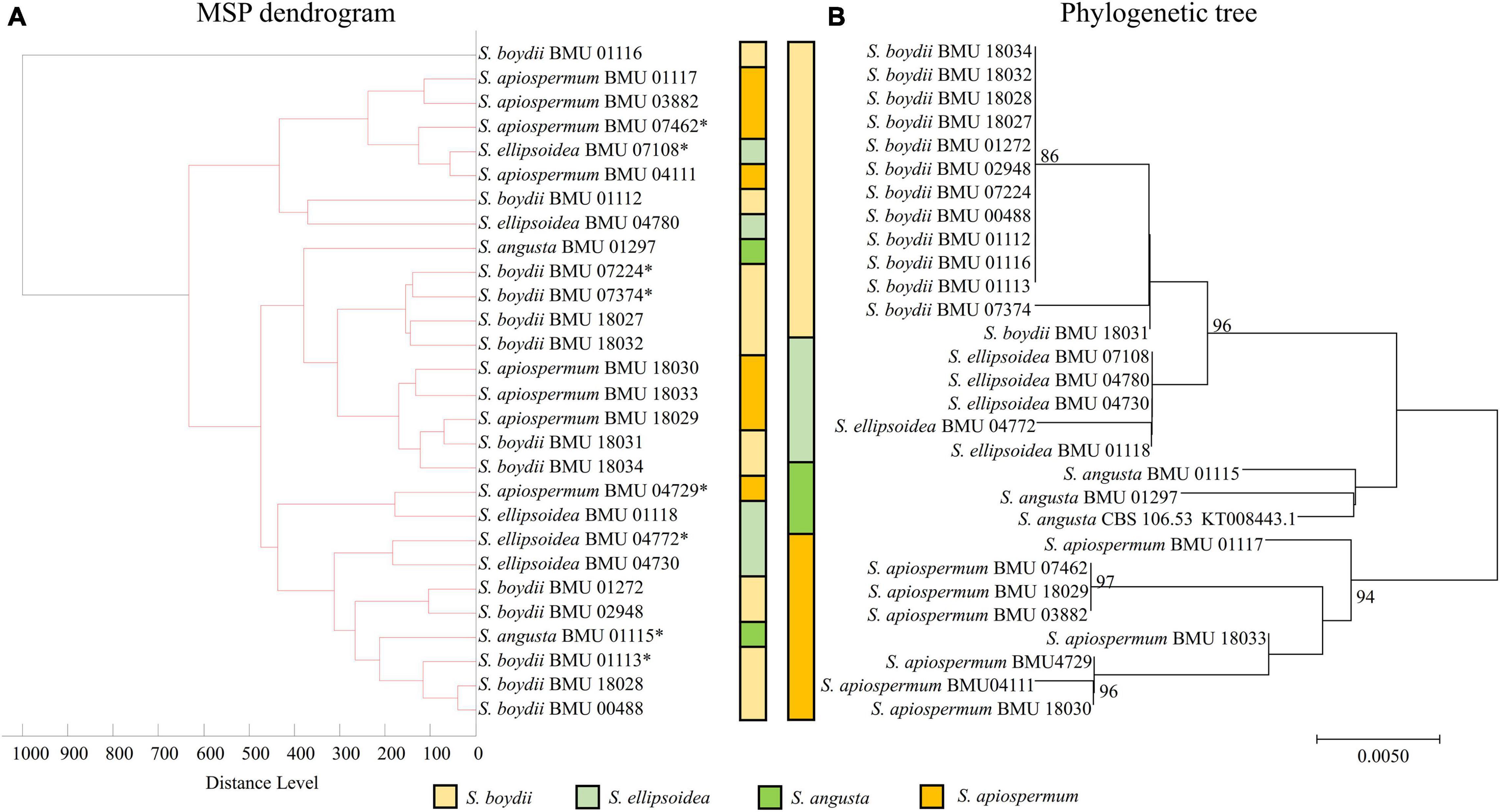
Figure 5. (A) Main spectrum profile (MSP) dendrogram of 28 Scedosporium isolates based on MALDI-TOF MS analysis; (B) molecular phylogenetic tree generated by the Neighbor Joining (NJ) method from the BT2 sequences of 28 Scedosporium isolates.
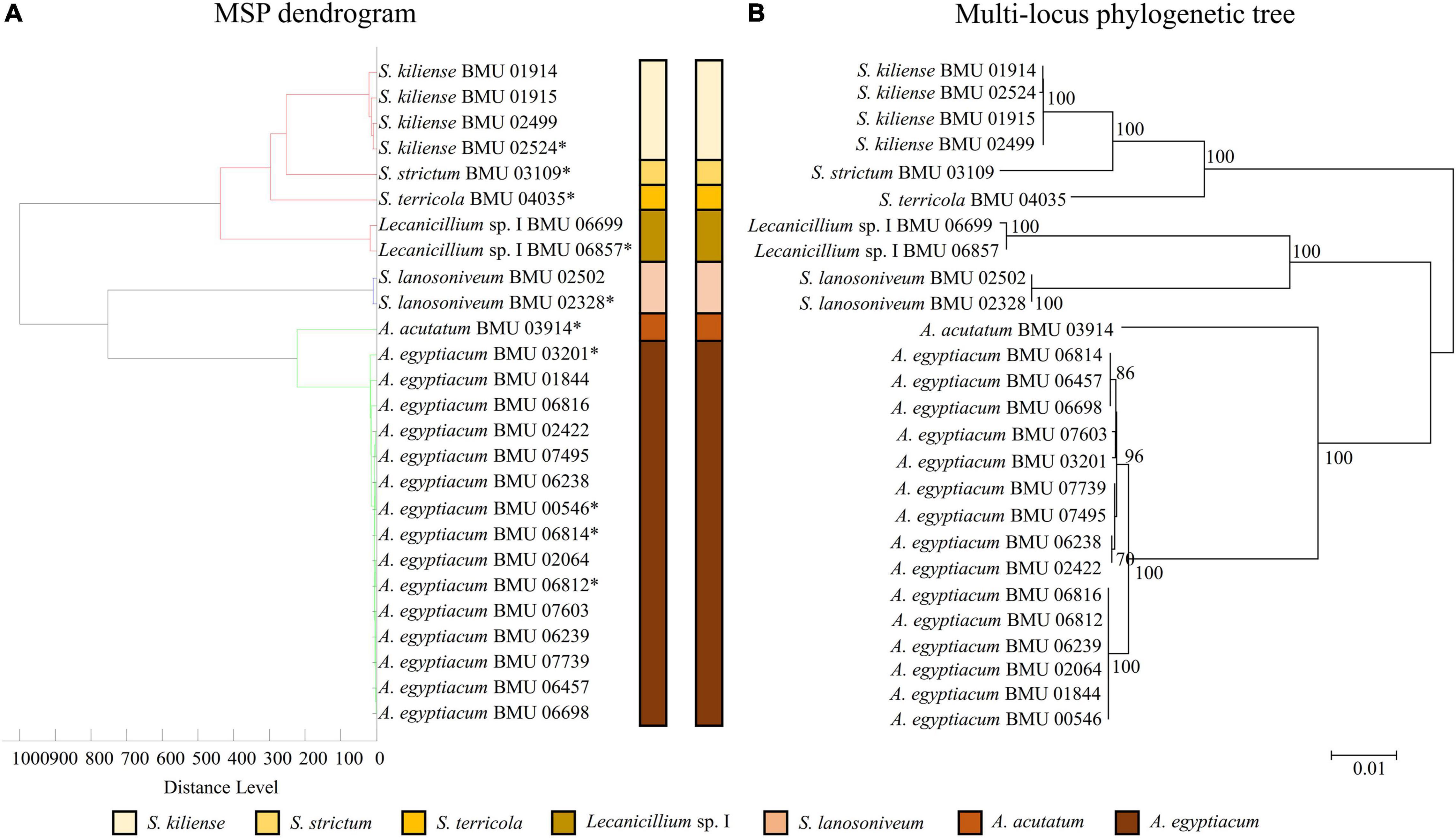
Figure 6. (A) Main spectrum profile dendrogram of 26 Acremonium-like isolates based on MALDI-TOF MS analysis; (B) molecular phylogenetic tree generated by the NJ method from the combined ITS-LSU and EF1-α sequences of 26 Acremonium-like isolates.
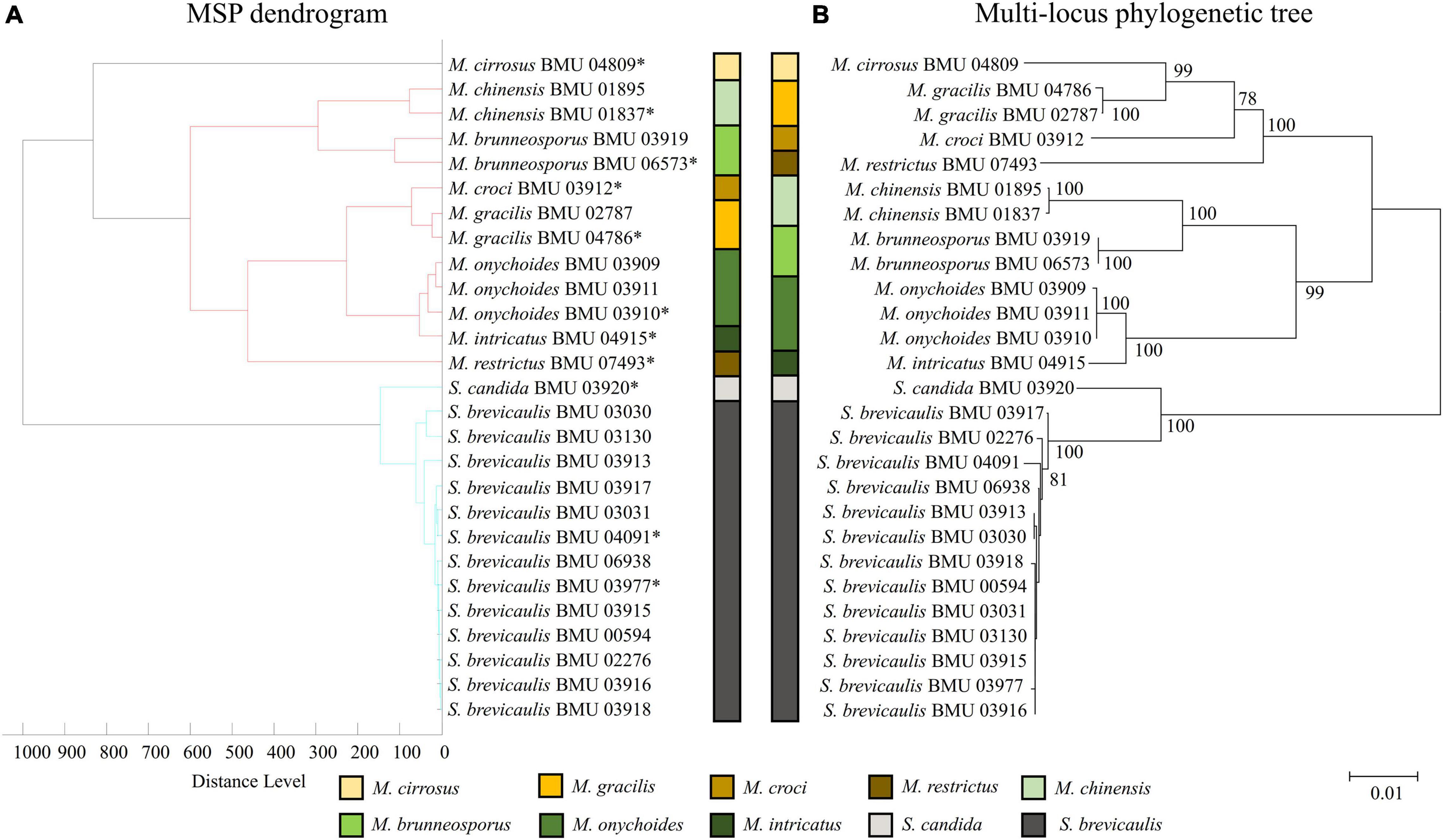
Figure 7. (A) Main spectrum profile dendrogram of 27 Scopulariopsis and Microascus isolates based on MALDI-TOF MS analysis; (B) molecular phylogenetic tree generated by the NJ method from the combined ITS, LSU, TUB, and EF1-α sequences of 27 Scopulariopsis and Microascus isolates.
The identification of filamentous fungi by MALDI-TOF MS relies mainly on an available database and an efficient protein extraction method. Liquid culture can improve the accuracy of MALDI-TOF MS identification and is usually used for research purpose. In clinical practice, liquid culture can be added when the identification of solid culture is unsatisfactory. The main obstacle of MALDI-TOF MS identification of Scedosporium, Acremonium-like, and Scopulariopsis-like species is a lack of reference spectral data in commercial databases. Our study indicated that MALDI-TOF MS is a powerful technique for rapid and accurate identification of the above species when using the Bruker database complimented with an in-house database and liquid culture.
Due to the non-updated nomenclature and a lack of reference species of Scedosporium in FFL v1.0, clear species identification is unavailable. With a supplementary database, the species identification rate of Scedosporium spp. ranged between 76 and 100% (Del Chierico et al., 2012; Normand et al., 2013; Ranque et al., 2014; Sleiman et al., 2016; Zvezdanova et al., 2019), and our study also found a correct identification rate of 75% using the combined databases. However, we found that the mass spectra within the S. apiospermum complex were highly similar, indicated by the high CCI values between species, so the strains in the complex could not be accurately differentiated, which was also reported by Bernhard et al. (2016). Zvezdanova et al. (2019) also found that even though 9 strains of S. apiospermum were included in an in-house database, three strains of S. apiospermum were misidentified at the species level (misidentified as S. boydii [n = 1] or Lomentospora prolificans [n = 2]). However, Sleiman et al. (2016) found that 17 strains of Scedosporium spp. and L. prolificans were correctly identified with a supplementary database, but the strains in the S. apiospermum complex included only five S. apiospermum and one S. boydii isolates. Sitterle et al. (2014) found with an in-house database containing 47 reference strains that the Andromas system identified 64 strains of Scedosporium spp. (including S. boydii, S. apiospermum, S. aurantiacum, S. minutispora, and S. dehoogii) and L. prolificans to the species level using the direct smearing method (without protein extraction). This indicated that the ability of different MALDI-TOF MS systems to identify Scedosporium spp. may be different, and a larger number of strains for database construction may improve the ability of species differentiation. Due to the lack of species, we were unable to clarify whether MALDI-TOF MS can distinguish the S. apiospermum complex from other species in the genus. In addition, by comparing the MSP dendrogram with the drug sensitivity data (Wang et al., 2015), we didn’t find a relation between the spectra difference and the drug sensitivity in the test strains. In conclusion, our results showed that MALDI-TOF MS identification for Scedosporium spp. by the Bruker Biotyper was relatively reliable after protein extraction from liquid cultivation and database combination.
To date, there have been no MALDI-TOF MS identification studies for a wide range of Acremonium-like species. Becker et al. (2014) correctly identified five out of six isolates of S. strictum to the species level with a supplemented database by the Bruker Biotyper system. Rychert et al. (2018) correctly identified 30 Acremonium sclerotigenum (now named A. egyptiacum) (Summerbell et al., 2018) isolates to the species level using the Vitek MS v3.0 database. Due to the limited species in these studies, the identification ability of MALDI-TOF MS for Acremonium-like species could not be defined. We conducted the first MALDI-TOF MS identification study on multiple species and genera of Acremonium-like spp. Due to a lack of reference MSPs, FFL v1.0 could not identify all of the Acremonium-like isolates. Although one S. strictum MSP was included in the commercial database, no identification results were found when tested with one S. strictum isolate. However, three of the four isolates of S. kiliense were identified as S. strictum, though with low scores (between 1.464 and 1.671, data not shown), suggesting that the reference strain of S. strictum in FFL v1.0 might actually be S. kiliense. Some isolates previously reported as S. strictum in the literature were also reconfirmed as S. kiliense (Perdomo et al., 2011; Summerbell et al., 2018), indicating the possibility of confusion. After supplementation of the BMU database, the identification accuracy by MALDI-TOF MS of Acremonium-like species was similar to that of multilocus sequencing, and the former method was faster, simpler and less expensive.
Only a few species and strains of Scopulariopsis-like fungi have yet been involved in MALDI-TOF MS identification studies. Using FFL v1.0, S. brevicaulis was correctly identified in some cases (Schulthess et al., 2014; Levesque et al., 2015; Riat et al., 2015; Stein et al., 2018; Sacheli et al., 2020), while S. candida, S. cinerea, S. brumptii, and M. cirrosus could not be identified (Schulthess et al., 2014; Levesque et al., 2015; Stein et al., 2018). The identification of Scopulariopsis-like fungi can be improved by constructing an in-house database (Lau et al., 2013; Becker et al., 2014). Lau et al. (2013) correctly identified three strains of S. brevicaulis to the species level using the Bruker Biotyper system plus the self-built NIH database. Becker et al. (2014) also correctly identified 12 strains of S. brevicaulis and one strain of S. candida with an in-house database. Our study found that FFL v1.0 performed well in the identification of S. brevicaulis but was unable to identify other species due to a lack of reference MSPs. After adding the reference MSPs of ten species, the Bruker Biotyper system was able to accurately identify Scopulariopsis and Microascus species.
The MSP dendrogram obtained by MALDI-TOF MS analysis indicates the relationship between the strains based on protein fingerprint differences and can be applied to the classification of microorganisms. This study found that the MSP dendrogram of Acremonium-like isolates clearly distinguished 7 different species, with similar topology to the multilocus phylogenetic tree. The MSP dendrogram of Scopulariopsis and Microascus spp. clustered isolates of the same species or close species to a branch, while the topology of Microascus spp. was inconsistent with that of the multilocus phylogenetic tree. These results indicated that the MSP dendrogram is efficient for species differentiation; however, there was a difference between molecular taxonomy at the nucleic acid and protein levels, which may be due to the varied protein expression related to the growth condition and life cycle of the fungi (Putignani et al., 2011). Shao et al. (2018) also found that the interspecific discrimination of MSP dendrograms and ITS-based trees of Rhizopus and Mucor species was consistent, while there were inconsistences at the intergeneric and intraspecific levels. Research on Lichtheimia species by Schrodl et al. (2012) showed that the MSP dendrogram clearly discriminated different species, but the topology was different from that of the phylogenetic tree. Therefore, MSP dendrogram is an excellent tool for interspecific differentiation but is limited in phylogenetic analysis (Shao et al., 2018). In addition, we found the MSP dendrogram cannot differentiate species in the S. apiospermum complex. Shao et al. (2020) also found that the MSP dendrogram was difficult to distinguish species in the Trichophyton mentagrophytes series. These results indicated discrimination of close species in some species complex by MSP dendrogram is difficult.
This study showed that with the supplementary database, MALDI-TOF MS represents a powerful tool to identify Scedosporium, Acremonium-like, Scopulariopsis, and Microascus species. As the test strains were all previously identified storage strains, and the species were rare and difficult to collect in a short time, blind test was not conducted. In the future, the identification effect will be clearer on the basis of continuous database expansion and amplification of test species and strains.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, MW528357 to MW5283564.
JY designed the study. ZW collected the samples. LW, JS, YS, LY, and HW participated in the performance of the research. LW participated in the writing of the manuscript and data analysis. All authors contributed to the article and approved the submitted version.
This research was supported by Major Infections Diseases such as AIDS and Viral Hepatitis Prevention and Control Technology Major Projects (Grant 2018ZX10712-001).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Becker, P. T., de Bel, A., Martiny, D., Ranque, S., Piarroux, R., Cassagne, C., et al. (2014). Identification of filamentous fungi isolates by MALDI-TOF mass spectrometry: clinical evaluation of an extended reference spectra library. Med. Mycol. 52, 826–834. doi: 10.1093/mmy/myu064
Bernhard, M., Zautner, A. E., Steinmann, J., Weig, M., Gross, U., and Bader, O. (2016). Towards proteomic species barcoding of fungi - An example using Scedosporium/Pseudallescheria complex isolates. Fungal Biol. 120, 162–165. doi: 10.1016/j.funbio.2015.07.001
Brown, G. D., Denning, D. W., Gow, N. A., Levitz, S. M., Netea, M. G., and White, T. C. (2012). Hidden killers: human fungal infections. Sci. Transl. Med. 4:165rv113. doi: 10.1126/scitranslmed.3004404
Del Chierico, F., Masotti, A., Onori, M., Fiscarelli, E., Mancinelli, L., Ricciotti, G., et al. (2012). MALDI-TOF MS proteomic phenotyping of filamentous and other fungi from clinical origin. J. Proteomics 75, 3314–3330. doi: 10.1016/j.jprot.2012.03.048
Jagielski, T., Sandoval-Denis, M., Yu, J., Yao, L., Bakula, Z., Kalita, J., et al. (2016). Molecular taxonomy of scopulariopsis-like fungi with description of new clinical and environmental species. Fungal Biol. 120, 586–602. doi: 10.1016/j.funbio.2016.01.014
Lackner, M., de Hoog, G. S., Verweij, P. E., Najafzadeh, M. J., Curfs-Breuker, I., Klaassen, C. H., et al. (2012). Species-specific antifungal susceptibility patterns of Scedosporium and Pseudallescheria species. Antimicrob. Agents Chemother. 56, 2635–2642. doi: 10.1128/AAC.05910-11
Lau, A. F., Drake, S. K., Calhoun, L. B., Henderson, C. M., and Zelazny, A. M. (2013). Development of a clinically comprehensive database and a simple procedure for identification of molds from solid media by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 51, 828–834. doi: 10.1128/JCM.02852-12
Levesque, S., Dufresne, P. J., Soualhine, H., Domingo, M. C., Bekal, S., Lefebvre, B., et al. (2015). A side by side comparison of bruker biotyper and VITEK MS: utility of MALDI-TOF MS technology for microorganism identification in a public health reference laboratory. PLoS One 10:e0144878. doi: 10.1371/journal.pone.0144878
Miceli, M. H., and Lee, S. A. (2011). Emerging moulds: epidemiological trends and antifungal resistance. Mycoses 54, e666–e678. doi: 10.1111/j.1439-0507.2011.02032.x
Normand, A. C., Cassagne, C., Ranque, S., L’Ollivier, C., Fourquet, P., Roesems, S., et al. (2013). Assessment of various parameters to improve MALDI-TOF MS reference spectra libraries constructed for the routine identification of filamentous fungi. BMC Microbiol. 13:76. doi: 10.1186/1471-2180-13-76
Perdomo, H., Sutton, D. A., Garcia, D., Fothergill, A. W., Cano, J., Gene, J., et al. (2011). Spectrum of clinically relevant Acremonium species in the United States. J. Clin. Microbiol. 49, 243–256. doi: 10.1128/JCM.00793-10
Perez-Cantero, A., and Guarro, J. (2020a). Current knowledge on the etiology and epidemiology of Scopulariopsis infections. Med. Mycol. 58, 145–155. doi: 10.1093/mmy/myz036
Perez-Cantero, A., and Guarro, J. (2020b). Sarocladium and Acremonium infections: new faces of an old opportunistic fungus. Mycoses 63, 1203–1214. doi: 10.1111/myc.13169
Putignani, L., Del Chierico, F., Onori, M., Mancinelli, L., Argentieri, M., Bernaschi, P., et al. (2011). MALDI-TOF mass spectrometry proteomic phenotyping of clinically relevant fungi. Mol. Biosyst. 7, 620–629. doi: 10.1039/c0mb00138d
Ramirez-Garcia, A., Pellon, A., Rementeria, A., Buldain, I., Barreto-Bergter, E., Rollin-Pinheiro, R., et al. (2018). Scedosporium and Lomentospora: an updated overview of underrated opportunists. Med. Mycol. 56(Suppl._1), 102–125. doi: 10.1093/mmy/myx113
Ranque, S., Normand, A. C., Cassagne, C., Murat, J. B., Bourgeois, N., Dalle, F., et al. (2014). MALDI-TOF mass spectrometry identification of filamentous fungi in the clinical laboratory. Mycoses 57, 135–140. doi: 10.1111/myc.12115
Riat, A., Hinrikson, H., Barras, V., Fernandez, J., and Schrenzel, J. (2015). Confident identification of filamentous fungi by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry without subculture-based sample preparation. Int. J. Infect. Dis. 35, 43–45. doi: 10.1016/j.ijid.2015.04.013
Rychert, J., Slechta, E. S., Barker, A. P., Miranda, E., Babady, N. E., Tang, Y. W., et al. (2018). Multicenter evaluation of the vitek MS v3.0 system for the identification of filamentous fungi. J. Clin. Microbiol. 56:e01353-17. doi: 10.1128/JCM.01353-17
Sacheli, R., Henri, A. S., Seidel, L., Ernst, M., Darfouf, R., Adjetey, C., et al. (2020). Evaluation of the new Id-Fungi plates from Conidia for MALDI-TOF MS identification of filamentous fungi and comparison with conventional methods as identification tool for dermatophytes from nails, hair and skin samples. Mycoses 63, 1115–1127. doi: 10.1111/myc.13156
Schrodl, W., Heydel, T., Schwartze, V. U., Hoffmann, K., Grosse-Herrenthey, A., Walther, G., et al. (2012). Direct analysis and identification of pathogenic Lichtheimia species by matrix-assisted laser desorption ionization-time of flight analyzer-mediated mass spectrometry. J. Clin. Microbiol. 50, 419–427. doi: 10.1128/JCM.01070-11
Schulthess, B., Ledermann, R., Mouttet, F., Zbinden, A., Bloemberg, G. V., Bottger, E. C., et al. (2014). Use of the Bruker MALDI Biotyper for identification of molds in the clinical mycology laboratory. J. Clin. Microbiol. 52, 2797–2803. doi: 10.1128/JCM.00049-14
Shao, J., Wan, Z., Li, R., and Yu, J. (2018). Species identification and delineation of pathogenic mucorales by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 56:e01886-17. doi: 10.1128/JCM.01886-17
Shao, J., Wan, Z., Li, R., and Yu, J. (2020). Species identification of dermatophytes isolated in China by matrix-assisted laser desorption ionisation-time-of-flight mass spectrometry. Mycoses 63, 1352–1361. doi: 10.1111/myc.13175
Sitterle, E., Giraud, S., Leto, J., Bouchara, J. P., Rougeron, A., Morio, F., et al. (2014). Matrix-assisted laser desorption ionization-time of flight mass spectrometry for fast and accurate identification of Pseudallescheria/Scedosporium species. Clin. Microbiol. Infect. 20, 929–935. doi: 10.1111/1469-0691.12574
Sleiman, S., Halliday, C. L., Chapman, B., Brown, M., Nitschke, J., Lau, A. F., et al. (2016). Performance of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of Aspergillus, Scedosporium, and Fusarium spp. in the Australian clinical setting. J. Clin. Microbiol. 54, 2182–2186. doi: 10.1128/JCM.00906-16
Stein, M., Tran, V., Nichol, K. A., Lagace-Wiens, P., Pieroni, P., Adam, H. J., et al. (2018). Evaluation of three MALDI-TOF mass spectrometry libraries for the identification of filamentous fungi in three clinical microbiology laboratories in Manitoba, Canada. Mycoses 61, 743–753. doi: 10.1111/myc.12800
Summerbell, R. C., Gueidan, C., Guarro, J., Eskalen, A., Crous, P. W., Gupta, A. K., et al. (2018). The protean Acremonium. A. sclerotigenum/egyptiacum: revision, food contaminant, and human disease. Microorganisms 6:88. doi: 10.3390/microorganisms6030088
Wang, H., Wan, Z., Li, R., Lu, Q., and Yu, J. (2015). Molecular identification and susceptibility of clinically relevant Scedosporium spp. in China. Biomed. Res. Int. 2015:109656. doi: 10.1155/2015/109656
Wilkendorf, L. S., Bowles, E., Buil, J. B., van der Lee, H. A. L., Posteraro, B., Sanguinetti, M., et al. (2020). Update on matrix-assisted laser desorption ionization-time of flight mass spectrometry identification of filamentous fungi. J. Clin. Microbiol. 58:e01263-20. doi: 10.1128/JCM.01263-20
Yao, L., Wan, Z., Li, R., and Yu, J. (2015). In vitro triple combination of antifungal drugs against clinical Scopulariopsis and Microascus species. Antimicrob. Agents Chemother. 59, 5040–5043. doi: 10.1128/AAC.00145-15
Yao, L., Wang, H., Wan, Z., Li, R., and Yu, J. (2019). The high diversity and variable susceptibility of clinically relevant acremonium-like species in China. Mycopathologia 184, 759–773. doi: 10.1007/s11046-019-00399-8
Keywords: MALDI-TOS MS, identification, Scedosporium, Acremonium, Scopulariopsis, Microascus
Citation: Wei L, Shao J, Song Y, Wan Z, Yao L, Wang H and Yu J (2022) Performance of Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry for Identification of Scedosporium, Acremonium-Like, Scopulariopsis, and Microascus Species. Front. Microbiol. 13:841286. doi: 10.3389/fmicb.2022.841286
Received: 22 December 2021; Accepted: 09 February 2022;
Published: 02 March 2022.
Edited by:
Jens Andre Hammerl, Bundesinstitut für Risikobewertung, GermanyReviewed by:
Jayaseelan Murugaiyan, SRM University, IndiaCopyright © 2022 Wei, Shao, Song, Wan, Yao, Wang and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jin Yu, eXVqaW42NzZAMTI2LmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.