- 1Ministry of Agriculture Laboratory of Quality and Safety Risk Assessment for Dairy Products (Beijing), Institute of Animal Science, Chinese Academy of Agricultural Sciences, Beijing, China
- 2Ministry of Agriculture—Milk and Dairy Product Inspection Center (Beijing), Beijing, China
- 3Institute of Quality Standard and Testing Technology, Xinjiang Academy of Agricultural Sciences, Urumqi, China
Staphylococcus aureus (S. aureus) is one of the main pathogens in different raw milk and dairy products, which may lead to economic losses. Staphylococcus aureus is a significant and costly public health concern because it may enter the human food chain and contaminate milk causing foodborne illness. This study aimed to investigate the prevalence, antimicrobial susceptibility and virulence genes of S. aureus in raw milks. In total, 125 raw milk samples collected from goat (n = 50), buffalo (n = 25), camel (n = 25), and yak (n = 25) were collected from 5 provinces in China in 2016. Out of 125 samples, 36 (28.8%) S. aureus were isolated (16 from goat, 9 from buffalo, 6 from camel, and 5 from yak). Out of 36 S. aureus, 26 strains (26/36, 72.2%) showed antibiotics resistance, and 6 strains isolated from goats were identified as methicillin-resistant S. aureus (MRSA). The antimicrobial resistance against Penicillin G, tetracycline and gentamicin was 50% (18/36), 41.7% (15/36), and 36.1% (13/36), respectively. 19 S. aureus (52.8%) were considered as multidrug resistant. The highest prevalence of resistant S. aureus was observed in goat milk (13/36, 36.1%). Among the 36 strains, 16 isolates harbored three or more resistant genes. The resistance genes were detected in 25 S. aureus, including 13 strains in goat, 5 strains in buffalo, 4 strains in camel, and 3 strains in yak. Among the 26 resistant strains, 61.5% of isolates harbored three or more resistant genes. The resistance genes were detected in 25 S. aureus, including 13 strains in goat milk, 5 strains in buffalo milk, 4 strains in camel milk, and 3 strains in yak milk. The most predominant resistance genes were blaZ (18/26, 69.2%), aac6′-aph2″ (13/26, 50.0%), and tet(M) (10/26, 38.5%). The mecA, ant(6)-Ia and fexA gene were only detected in S. aureus from goat milk. The most predominant toxin gene were sec (8/26, 30.8%). The majority of S. aureus were multidrug resistant and carried multiple virulence genes, which may pose potential risk to public health. Our findings indicated that the prevalence and antimicrobial resistance of S. aureus was a serious concern in different raw milks in China, especially goat milks.
Introduction
Staphylococcus aureus (S. aureus) is one of the main pathogens in mastitis of involved in intramammary infections in cows, goats, and sheep, which may lead to economic losses due to reduced milk production and poor milk quality (Aires-de-Sousa et al., 2007; Stapels et al., 2014). Contamination of raw milk with S. aureus has a highly potentially hazardous in China (Chao et al., 2015; Lan et al., 2017; Liu et al., 2017). Staphylococcus aureus can cause specific toxin-mediated conditions, such as scalded skin syndrome, staphylococcal food poisoning and toxic shock syndrome (Schmidt et al., 2017). Risk assessment for S. aureus in different raw milk samples should been conducted (Cortimiglia et al., 2015; Xu et al., 2015; Ding et al., 2016).
Staphylococcus aureus infections are related to the expression of virulence factors. Many potential virulence factors, such as enzymes and exotoxins, contribute to cause S. aureus diseases. Staphylococcus aureus can harbor different virulence genes encoding for enterotoxins, enterotoxin-like exfoliative toxin, toxic shock syndrome toxin-1 (TSST-1), and Panton-Valentine leukocidin (PVL) (Kot et al., 2016; Wang et al., 2016). Thermostable staphylococcal enterotoxins generally retain their biological activity after pasteurization treatment (Guimaraes et al., 2013). Staphylococcus aureus with toxin genes were present in about 60.0% of goat, ovine, caprine, and bubaline origin (Mork et al., 2010; Carfora et al., 2015). An efficient screening to detect the virulence genes in different raw milks is necessary.
The use of antibiotics in veterinary practice could cause the selection of antibiotic resistant S. aureus, which is a public health problem (Gomes and Henriques, 2016). Staphylococcus aureus is frequently resistant to antibiotics because it produces exopolysaccharide which forms a barrier to bacterial cell penetration by several antimicrobial agents (Johnston, 1998). Moreover, S. aureus can also develop acquired resistance to many other antimicrobial agents by carrying various resistance traits on plasmids or transposons (Chajecka-Wierzchowska et al., 2015). Some studies have reported that S. aureus which originated from goats, buffaloes, camels, and yak milks, showed high antimicrobial resistance patterns (Pamuk et al., 2012; Biswas et al., 2014; Cortimiglia et al., 2015; Redwan et al., 2016). Therefore, monitoring antimicrobial resistance in S. aureus from different raw milks is necessary to predict the risk on antimicrobial resistance of S. aureus, and to make decisions regarding antibiotic treatments on animals from a pre-harvest food safety standpoint.
The antimicrobial resistance and virulence characteristics of S. aureus from cow milk samples are well described. However, data regarding S. aureus from different raw milk samples in China are relatively limited. Therefore, the present work aimed to determine the prevalence, antimicrobial susceptibility and key virulence genes of S. aureus isolated from goat, buffalo, camel, and yak milk samples in China.
Materials and Methods
Collection of Samples
A total of 125 raw milk samples were collected from 125 dairy farms located in China, including 25 goat milk samples from Shaanxi province, 25 goat milk samples from Shandong province, 25 camel milk samples from Xinjiang Uygur Autonomous Region, 25 buffalo milk samples from Guangxi Zhuang Autonomous Region, and 25 yak milk samples from Sichuan province, from April to June 2016 (Figure 1). All samples were collected from healthy animals by mechanical milking. The milking transferred into sterile bottles and transported immediately to laboratory at 4°C, for bacteriological analysis.
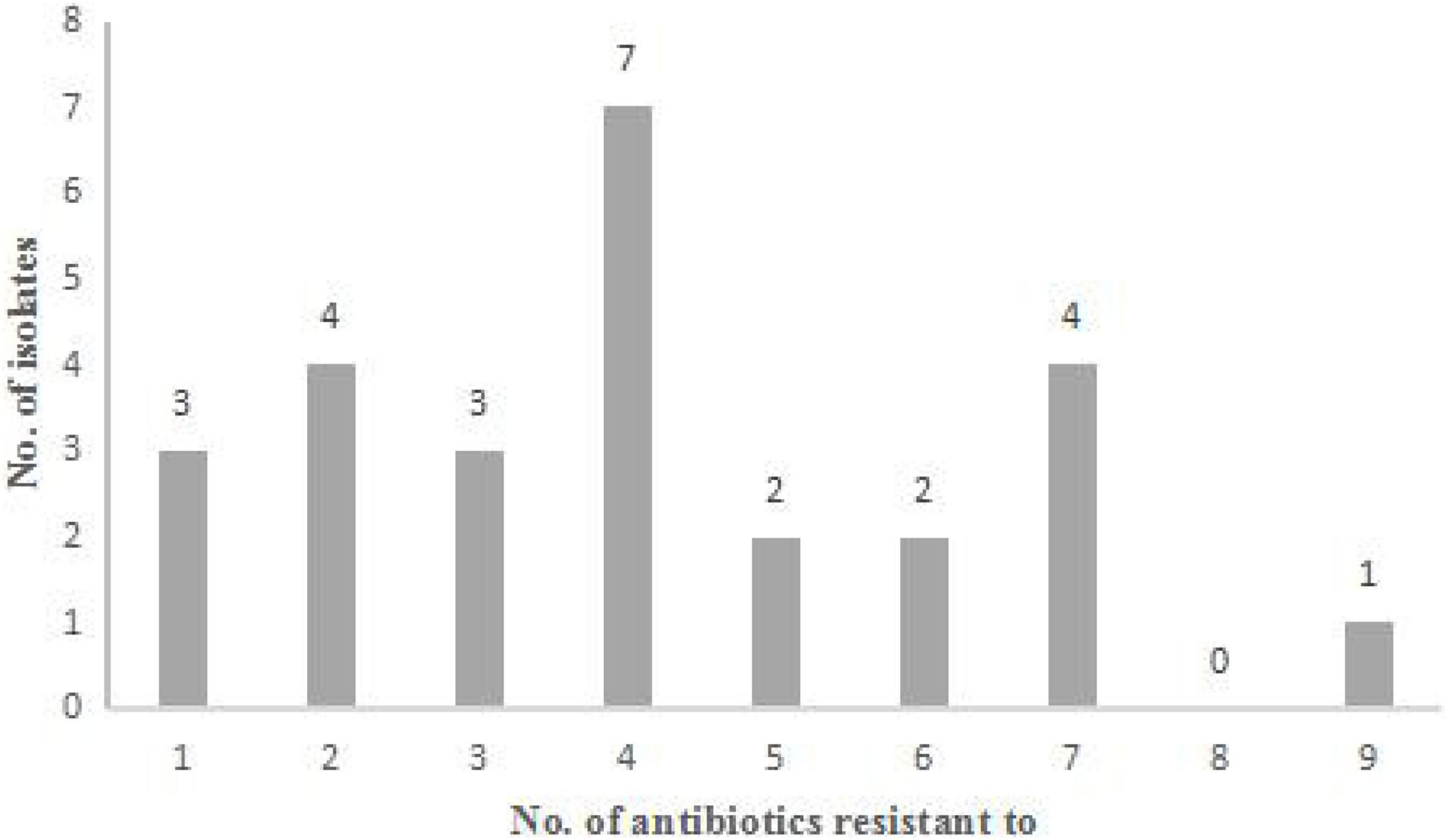
Figure 1. Antibiotic resistance characteristics of 26 Staphylococcus aureus strains. Distribution of the number of antibiotics that the strains exhibit resistance.
Isolation and Identification of Staphylococcus aureus
To isolate and detect the S. aureus, each sample was diluted 10-fold in sterile peptone water and homogenized. Aliquots (1 mL) were placed onto Baird-Parker agar supplemented with 5% egg yolk and tellurite (Beijing Land Bridge Technology Ltd., Beijing, China). The plates were incubated for 24–48 h at 37°C. Colonies with typical black appearance and surrounded by a clear zone were enumerated as S. aureus.
Presumptive colonies were confirmed by polymerase chain reaction (PCR) (Bio-Rad S1000, United States) detection of the thermonuclease gene (nuc, S. aureus specific) (Supplementary Table 1). DNA of the strains was extracted using the InstaGene Matrix DNA extraction kit (Bio-Rad Laboratories, Hercules, California, United States) following the manufacturer’s instructions. The amplification conditions and reagents for the PCR assays were those described by Liu et al. (2017). Negative control (without DNA template) and positive control (S. aureus ATCC 6538) templates were included in all PCR assays. After identification, one to three colonies per sample were randomly selected for subsequent analysis. All strains were stored with sterile magnetic beads at −80°C until further analysis.
Antimicrobial Resistance Screening
Antimicrobial resistance of S. aureus were determined by the agar disc diffusion method according to Clinical and Laboratory Standards Institute [CLSI] (2018a,b). According to the frequency of antibiotical therapy for dairy mastitis in China, the antimicrobials used for susceptibility testing included 13 antimicrobial agents: Penicillin G (10U), amoxicillin-clavulanic acid (20/10 μg), tetracycline (30 μg), gentamicin (10 μg), oxacillin (1 μg), streptomycin (10 μg), cefoxitin (30 μg), erythromycin (15 μg), ampicillin (10 μg), chloramphenicol (30 μg), kanamycin (30 μg), clindamycin (2μg), and trimethoprim-sulfamethoxazole (1.25/23.75 μg) were used as antimicrobial agents (Oxoid, Basingstoke, United Kingdom). Staphylococcus aureus ATCC 6538 and E. coli ATCC 25922 were used as quality controls. The antimicrobial resistance experiment of S. aureus was repeated twice.
Antimicrobial Resistance Genes
All resistant strains were screened for antimicrobial resistance genes by PCR amplification. The information of all primers is shown in Supplementary Table 1. The primers were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). The genes encoding penicillin (blaZ and mecA), cefoxitin (cfxA and mecA), aminoglycoside [aac6′-aph2″, ant(6)-Ia, ant(4′)-Ia], chloramphenicol (fexA and catA), tetracycline [tet(K), tet(L), tet(M) and tet(O)], erythromycin [erm(A), erm(B), erm(C), msr(A) and msr(B)], streptomycin [ant(6)-Ia], and oxacillin (mecA) resistance genes were detected by PCR. PCR products were electrophoresed in a 1.5% (m/v) agarose gels (Hydragene) with SYBR Safe DNA Stain (Invitrogen).
Detection of Virulence-Associated Genes
The detection of genes encoding staphylococcal enterotoxins (sea, seb, sec, sed, see, seg, seh, sei, sek, sel, ser, ses sej, and set), enterotoxin-like (selm, selo, selp, and selu), toxic-shock syndrome toxin (tst-1), exfoliative toxin genes (eta and etb), and Panton-Valentine leukocidin (pvl) was performed by PCR according to Wang et al. (2016). The target genes, primer sequences, and target fragment of PCR products are given in Supplementary Table 1.
Statistical Analysis
The data of prevalence and gene distribution between different categories of raw milks were analyzed using χ2-test and Fisher’s exact test with P < 0.05 considered as statistically significant in SPSS software (ver. 19.0, IBM Corp., Armonk, NY, United States).
Results
Prevalence of Staphylococcus aureus
Out of 125 samples, 36 strains of S. aureus were isolated, including 10 strains (40%) of 25 goat milk samples from Shaanxi province, 6 strains (24%) of 25 goat milk from Shandong province, 9 strains (36%) of 25 buffalo milk samples from Guangxi Zhuang Autonomous Region, and 6 strains (24%) of 25 camel milk samples from Xinjiang Uygur Autonomous Region and 5 strains (20%) of 25 yak milk samples from Sichuan province.
Antimicrobial Susceptibility Testing
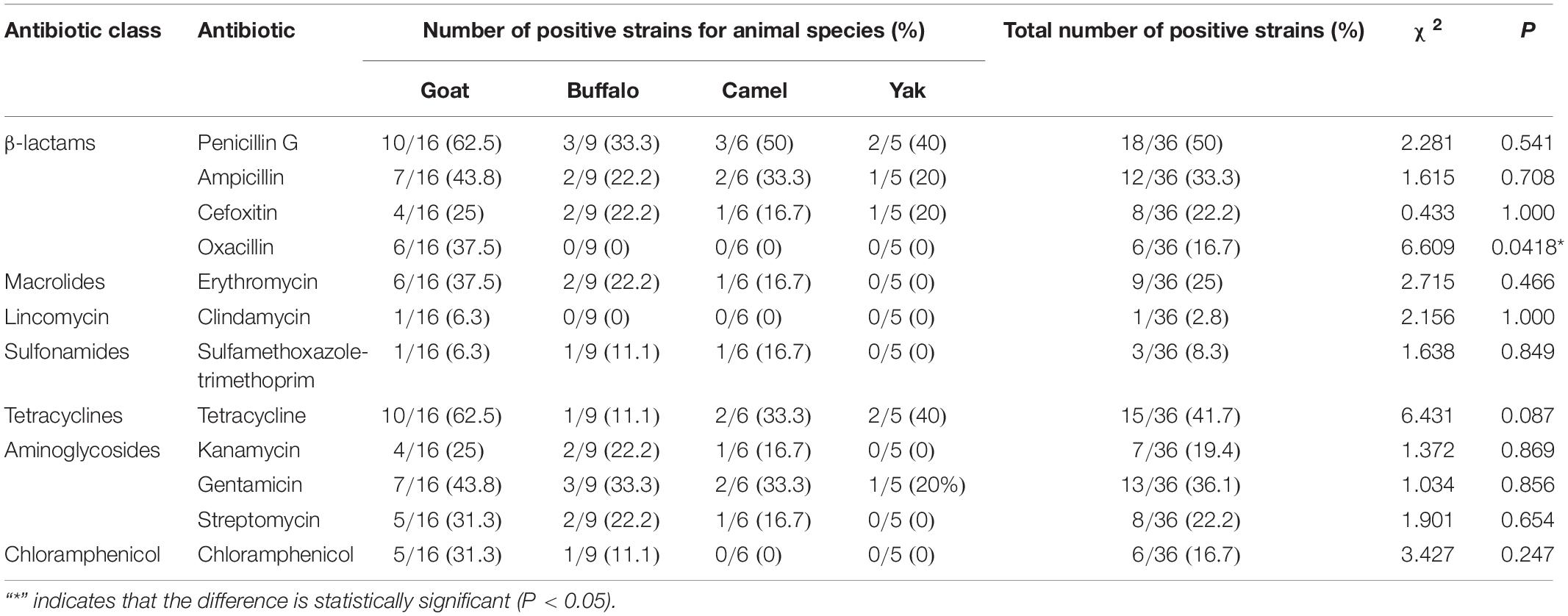
The 36 strains of S. aureus were tested for susceptibility against 13 antimicrobial agents by the disc diffusion method. A total of 26 strains (26/36, 72.2%) showed antibiotics resistance. 19 strains (19/36, 52.8%) showed resistance to at least three antimicrobial classes. Moreover, 4 strains (4/36, 11.1%) were resistant to 7 antibiotics tested, and 1 strain (1/36, 2.8%) was resistant to 9 antibiotics tested (Figure 1). Antimicrobial resistance was most frequently observed to penicillin G (18/36, 50%), followed by to tetracycline (15/36, 41.7%), gentamicin (13/36, 36.1%), ampicillin (12/36, 33.3%), erythromycin (9/36, 25%), cefoxitin (8/36, 22.2%), streptomycin (8/36, 22.2%), kanamycin (7/36, 19.4%), chloramphenicol (6/36, 16.7%), oxacillin (6/36, 16.7%), sulfamethoxazole-trimethoprim (3/36, 8.3%), clindamycin (1/36, 2.8%), and amoxicillin-clavulanic acid (0/36, 0) (Table 1). Among the 36 S. aureus, the antimicrobial resistance was frequently observed in S. aureus from goat milks (13/36, 36.1%), followed by buffalo milks (6/36, 16.7%), camel milks (4/36, 11.1%), and yak milks (3/36, 8.3%).
Moreover, six strains isolated from goats were identified as methicillin-resistant S. aureus (MRSA). Only one S. aureus, which was isolated from goat milk, showed resistance to clindamycin. There was no sulfamethoxazole-trimethoprim-resistant, kanamycin-resistant, and streptomycin-resistant strains isolated from yak milk samples, and there was no chloramphenicol-resistant strains isolated from yak and camel milk samples.
Screening of Antimicrobial Resistance Genes
The results of antimicrobial resistance genes detection are presented in Figure 2 and Tables 2, 3. Among the 26 resistant strains, 96.2% of strains (25/26) harbored at least one resistant gene (Figure 2).
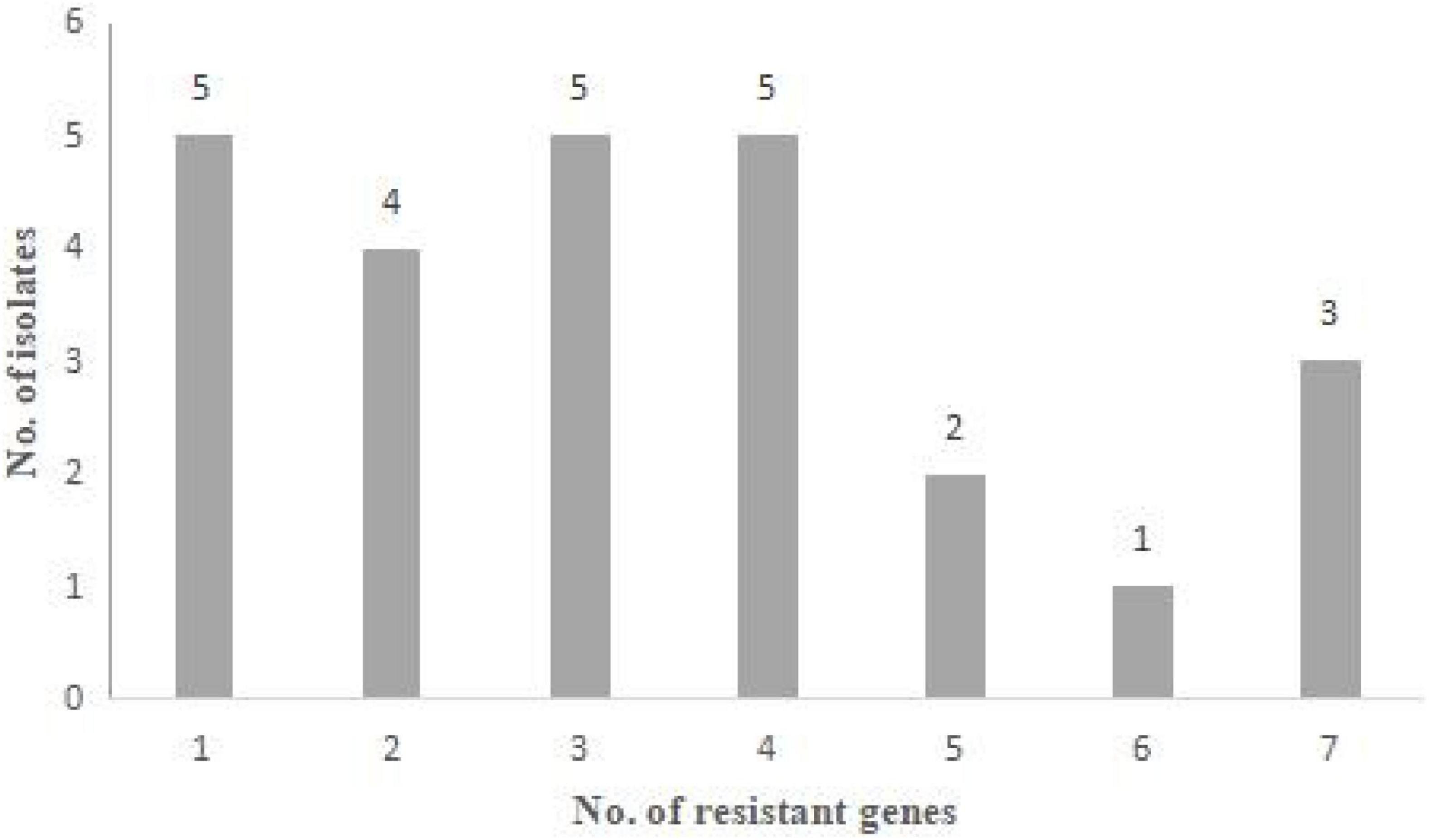
Figure 2. Antibiotic resistance genes of 26 Staphylococcus aureus strains. Distribution of the number of antibiotic resistance genes that the strains harbored.
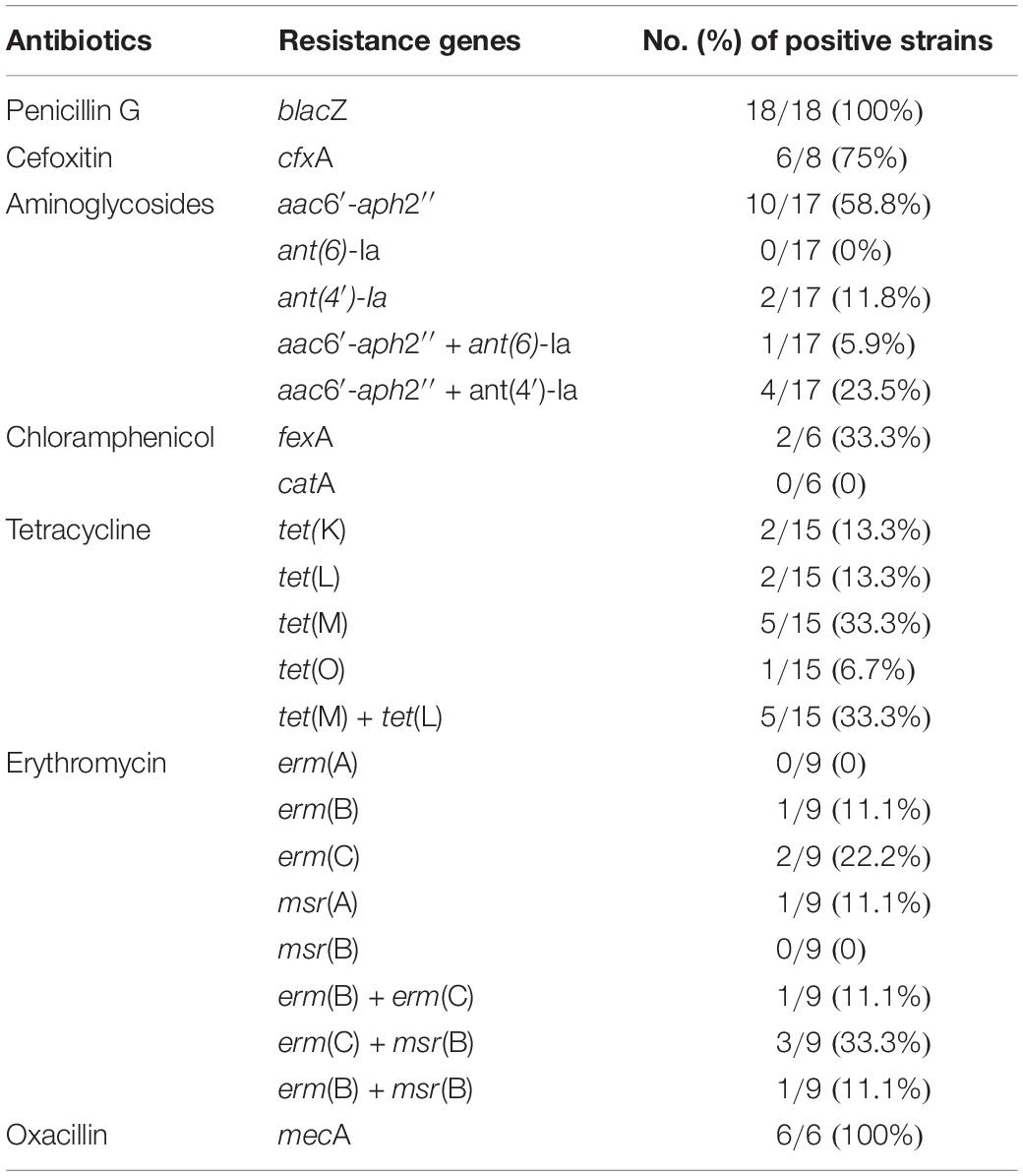
As shown in Supplementary Table 2, the antimicrobial resistance genes were frequently detected in S. aureus from goat milks (13/13, 100%), camel milks (4/4, 100%) and yak milks (3/3, 100%). The most predominant resistance genes were blaZ (18/26, 69.2%), aac6′-aph2″(13/26, 50.0%), tet(M) (10/26, 38.5%), tet(L) (7/26, 26.9%), mecA (6/26, 23.1%), erm(C) (6/26, 23.1%), ant(4′)-Ia (6/26, 23.1%), cfxA (6/26, 23.1%), msr(B) (4/26, 15.4%), erm(B) (3/26, 11.5%), tet(K) (2/26, 7.7%), fexA (2/26, 7.7%), tet(O) (1/26, 3.9%), and ant(6)-Ia (1/26, 3.9%). The catA, erm(A) and msr(A) genes were not detected in S. aureus (Table 2). Moreover, the mecA, ant(6)-Ia and fexA gene were only detected in the S. aureus from goat milk. The tet(K) and tet(O) genes were only detected in camel and yak milks, respectively.
Moreover, all the penicillin G-resistant S. aureus carried blaZ gene, and all the oxacillin-resistant S. aureus carried mecA gene. The aminoglycosides-resistant strains (n = 17) harbored one or more resistant genes, including aac6′-aph2″ (10/17, 58.8%), ant(4′)-Ia (2/17, 11.8%), aac6′-aph2″ + ant(4′)-Ia (4/17, 23.5%), aac6′-aph2″ and ant(6)-Ia (1/17, 5.9%). The tetracycline-resistant strains (n = 15) harbored one or more resistant genes, including tet(M) + tet(L) (5/15, 33.3%), tet(M) (5/15, 33.3%), tet(K) (2/15, 13.3%), tet(L) (2/15, 13.3%), and tet(O) (1/15, 6.7%). The erythromycin-resistant strains (n = 9) harbored one or more genes, including erm(B), erm(C), msr(A), and msr(B). The erm(A) genes were not detected in erythromycin-resistant strains. The msr(B) gene was detected together with erm(B) and erm(C) in the same strains.
Distribution of Virulence Genes
As shown in Table 4, the predominant toxin genes were sec (8/26, 30.8%), followed by sea (3/26, 11.5%), seg (3/26, 11.5%), tst (3/26, 11.5%), sel (2/26, 7.7%), sed (1/26, 3.9%), sei (1/26, 3.9%), sej (1/26, 3.9%), and pvl (1/26, 3.9%). Thirteen virulence genes, including seb, see, seh, sek, ser, ses, set, selm, selo, selp, selu, eta, and etb, were not detected in S. aureus. Moreover, 42.3% of the strains (11/26) harbored at least one virulence gene. Among the 26 of S. aureus, 6 strains from goat milk samples harbored 17 virulence genes, 1 strain from buffalo milk samples harbored 1 virulence gene, 2 strains from camel milk samples harbored 3 virulence genes, and 2 strains from yak milk samples harbored 2 virulence genes.
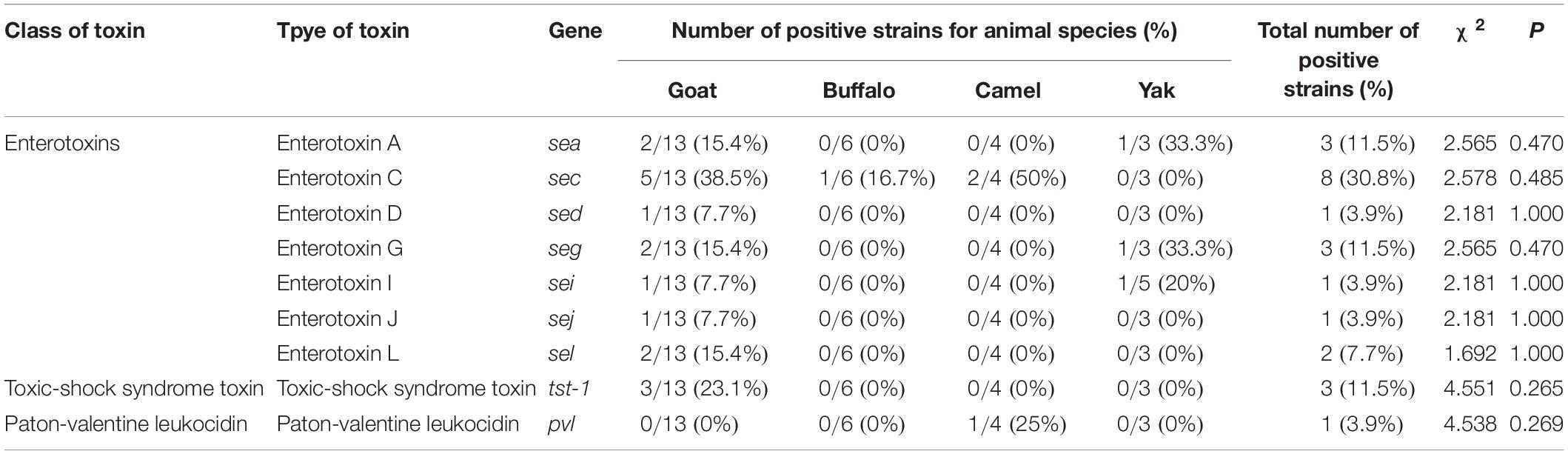
Table 4. Virulence genes identified in Staphylococcus aureus strains isolated from different raw milk samples (n = 26).
Discussion
Staphylococcus aureus is one of the main pathogens in raw milk and dairy products, which may lead to economic losses (Aires-de-Sousa et al., 2007; Stapels et al., 2014). Staphylococcus aureus has a strong capability to produce a wide variety of enterotoxins (Liu et al., 2017). The enterotoxins produced by S. aureus are common causes of bovine mastitis pathogenesis and food-borne illnesses (Wang et al., 2016). However, little is known about the presence of S. aureus in goat, buffalo, camel, and yak milk in China. In the present study, the prevalence, antimicrobial susceptibility, and key virulence genes of S. aureus isolated from goat, buffalo, camel, and yak milk samples in China has been assessed.
The prevalence of S. aureus in different raw milk samples varies depending on the country. In our study, raw buffalo milk samples showed the highest prevalence of S. aureus (9/25, 36%), followed by goat milk (16/50, 32%), camel milk (6/25, 24%), and yak milk (5/25, 20%). These results are significantly higher than that in previous reports which have shown that the occurrence rate of S. aureus in buffalo milk was 17.5%, followed by goat milk (7.5%) and camel milk (3.4%) in Iran (Ebrahim and Forough, 2013), 31.43% in goat milk in Egypt (Elbagory, 2017), and 15% in camel milk in Sudan (Shuiep et al., 2009). In contrast, much higher occurrence rate of S. aureus was 43.1% in goat milk in Northern Italy (Cortimiglia et al., 2015), 54.02% in camel milk in Pakistan (Aqib et al., 2017), 47.36% in yak milk in India (Biswas et al., 2014), and 32% in camel milk in Kenya (Njage et al., 2013). Overall, the results indicate that S. aureus is common in different raw milks, including goat, buffalo, camel and yak milk in China. This suggests that more attention should be paid to explore the method of controlling S. aureus occurrence in goat, buffalo, camel, and yak milk.
The antibiotics resistance has correlation to many factors, including excessive use of antibiotics, spread of resistance, and the background antibiotic resistance levels (Hu et al., 2014; Low et al., 2016; Crofts et al., 2017). The poorly regulated use of antibiotics on dairy animals in the different raw milk production systems examined may be an important reason for the high antimicrobial resistance. In this study, the antimicrobial resistance was the frequently observed in S. aureus from goat milk (36.1%) and buffalo milks (16.7%). The results indicated that the antibiotics resistance of S. aureus isolated from goat milk in Shandong and Shaanxi province, buffalo milk in Guangxi Zhuang Autonomous Region were very serious in China, although less than raw cow milk (Liu et al., 2017). A high emission densities of the antibiotics in China were reported in Shandong province, Shaanxi province, and Guangxi Zhuang Autonomous Region (Zhang et al., 2015). It implied that the high antimicrobial resistance of S. aureus from goat and buffalo milk may be related to it.
In the current study, S. aureus showed a high percentage of antimicrobial resistance to penicillin G, tetracycline, gentamicin, and ampicillin. Similar findings were reported by Oliveira et al. (2011), who found that 100% of S. aureus from buffalo milks were resistant to penicillin G and ampicillin, and by Rola et al. (2015), who found that coagulase-positive staphylococcus isolated from goat milk were resistant to penicillin G and tetracycline, and by Njage et al. (2013), who found that S. aureus isolated from camel milk from Kenya showed a high percentage of antimicrobial resistance to ampicillin, gentamicin, and tetracycline. Moreover, a total of 26 strains (26/36, 72.2%) showed antibiotics resistance, and 19 strains (19/26, 52.8%) showed resistance to at least three antimicrobial classes in our study. Based on the high percentage of resistance, these antibiotics should be used with caution for mastitis caused by S. aureus in goat, buffalo, and camel milk in China.
MRSA has been identified as an emerging pathogen in livestock animals that is readily transferable to humans in contact with livestock (Liu et al., 2017). Numerous reports have described the prevalence of MRSA in bovine milk (Caruso et al., 2016; Basanisi et al., 2017), and the transmission of MRSA between people working on farms and dairy cattle (Antoci et al., 2013; Mole, 2013). On the contrary, little is known about the prevalence of MRSA in other ruminants (e.g., goats, camels, buffalo, yaks), although the consumption of milk from different animals species is more and more common. In the present study, 16.7% of S. aureus (6/36) isolated from goat milk were identified as methicillin-resistant strains (MRSA). The results showed that there is a high prevalence in goat milks. The prevalence of MRSA in Southern Italy was 1.23% in goat milk and sheep bulk tank milk (Caruso et al., 2016), 8.3% in cow milk samples and dairy products (Basanisi et al., 2017). Moreover, there was no MRSA strains detected in buffalo, yak, and camel milk samples in our study. There was difference in data regarding the prevalence of MRSA strains in buffalo milk samples. Pamuk et al. (2012) reported that 2.5% of the buffalo samples were contaminated with MRSA in Turkey. El-Ashker et al. (2015) found that 24.5% of S. aureus in buffalo milk were identified as MRSA in Dakahlia Governorate, Egypt.
In the present study, 42.3% of the strains (11/26) harbored at least one virulence genes, which was much lower than reported in other studies. Liu et al. (2017) found that 60.3% of S. aureus from cow milk harbored one or more virulence genes. Martins et al. (2017) found that 62.5% of S. aureus from sheep milk samples showed some toxin-encoding genes. There are also reports showing that 67% of the detected S. aureus exhibited at least one enterotoxin gene, many caprine, or ovine raw milk products may be contaminated with low levels of enterotoxigenic S. aureus (Merz et al., 2016). Liu et al. (2019) reported that 62 S. aureus (72.5%) carried virulence genes, including set, hlb, hld, lukED, ebp, clfA, and clfB. Moreover, the largest percentage of sec (31.8%) genes was found in our study. Several authors have also reported that enterotoxin genes sea and sec are the most common in S. aureus isolated from raw milk (Carfora et al., 2015; Liu et al., 2017). Staphylococcus aureus from different kinds of milk samples exhibited different virulence gene patterns. For goat milk, sec gene was the most frequent in the study, and also reported as the most common virulence genes in Brazil (Lyra et al., 2013; Cavicchioli et al., 2015; Salaberry et al., 2015). Other researchers have reported that the most prevalent enterotoxin genes were sec and sel from goat milk samples in Switzerland (Merz et al., 2016). For camel milk, the sec and pvl genes have been detected in the study. Njage et al. (2013) found that pvl gene was detected in S. aureus from raw and fermented camel milk from Kenya and Somali. However, three S. aureus from camel milk were positive for sec, seg, sei, sem, sen, and seo, respectively (Shuiep et al., 2009). Majority of S. aureus were co-carried many virulence genes in goat, yak, camle, and bufflo milk, and it may pose great potential risk to public health. In order to estimate the actual impact on public health, it was necessary to the detection of virulence genes in goat, buffalo, and camel milk samples.
Conclusion
In conclusion, the antimicrobial resistance and virulence genes of S. aureus in goat, buffalo, camel, and yak milk in China were assessed. Our data indicated that S. aureus, which exhibit a high percentage of antimicrobial resistant and carried multiple virulence genes, were commonly found in different raw milks, especially goat milk. Although the prevalence of MRSA in raw milk from china was low, MRSA may pose a potential threat for consumers of raw milk or dairy products made with raw milk and for people working or living in close contact with animals. Therefore, it was very important to monitor the usage of antimicrobial agents and the potential transfer of antimicrobial resistance genes in goat milk.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
HL, LD, and YZ designed and performed the researches. LM helped with the data analysis. CW and JW gave advices to the researchers. NZ gave the opinions on the researches design. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the China Agriculture Research System of MOF and MARA, the Agricultural Science and Technology Innovation Program (ASTIP-IAS12), the State Key Laboratory of Animal Nutrition (2004DA125184G2108), and the Major Science and Technology Project of Xinjiang (2020A01001-3-1).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.840670/full#supplementary-material
References
Aires-de-Sousa, M., Parente, C. E. S. R., Vieira-da-Motta, O., Bonna, I. C. F., Silva, D. A., and de Lencastre, H. (2007). Characterization of Staphylococcus aureus isolates from buffalo, bovine, ovine, and caprine milk samples collected in Rio de Janeiro State, Brazil. Appl. Environ. Microb. 73, 3845–3849. doi: 10.1128/AEM.00019-07
Antoci, E., Pinzone, M. R., Nunnari, G., Stefani, S., and Cacopardo, B. (2013). Prevalence and molecular characteristics of methicillin-resistant Staphylococcus aureus (MRSA) among subjects working on bovine dairy farms. Infez. Med. 21, 125–129. doi: 10.1111/zph.12076
Aqib, A. I., Ijaz, M., Hussain, R., Durrani, A. Z., Anjum, A. A., Rizwan, A., et al. (2017). Identification of coagulase gene in Staphylococcus aureus isolates recovered from subclinical mastitis in camels. Pak. Vet. J. 37, 160–164.
Basanisi, M. G., Bella, G. L., Nobili, G., Franconieri, I., and La-Salandra, G. (2017). Genotyping of methicillin-resistant Staphylococcus aureus (MRSA) isolated from milk and dairy products in South Italy. Food. Microbiol. 62, 141–146. doi: 10.1016/j.fm.2016.10.020
Biswas, S., Mukherjee, R., Mahto, R. P., Chakravarti, U. K., De, S., Bera, A. K., et al. (2014). Antibacterial sensitivity and resistance pattern of yak intramammary infection from Arunachal Pradesh, India. Asian J. Anim. Vet. Adv. 9, 683–689. doi: 10.3923/ajava.2014.683.689
Carfora, V., Caprioli, A., Marri, N., Sagrafoli, D., Boselli, C., Giacinti, G., et al. (2015). Enterotoxin genes, enterotoxin production, and methicillin resistance in Staphylococcus aureus isolated from milk and dairy products in Central Italy. Int. Dairy. J. 42, 12–15. doi: 10.1016/j.idairyj.2014.10.009
Caruso, M., Latorre, L., Santagada, G., Fraccalvieri, R., Miccolupo, A., Sottili, R., et al. (2016). Methicillin-resistant Staphylococcus aureus (MRSA) in sheep and goat bulk milk from Southern Italy. Small Ruminant. Res. 135, 26–31. doi: 10.1016/j.smallrumres.2015.12.023
Cavicchioli, V. Q., Scatamburlo, T. M., Yamazi, A. K., Pieri, F. A., and Nero, L. A. (2015). Occurrence of Salmonella, Listeria monocytogenes and enteroxoxigenic Staphylococcus in goat milk from small and medium-sized farms located in Minas Gerais State, Brazil. J. Dairy. Sci. 98, 8386–8390. doi: 10.3168/jds.2015-9733
Chajecka-Wierzchowska, W., Zadernowska, A., Nalepa, B., Sierpinska, M., and Laniewska-Trokenheim, L. (2015). Coagulase-negative staphylococci (CoNS) isolated from ready-to-eat food of animal origin-phenotypic and genotypic antibiotic resistance. Food. Microbiol. 46, 222–226. doi: 10.1016/j.fm.2014.08.001
Chao, G. X., Bao, G. Y., Cao, Y. Z., Yan, W. G., Wang, Y., Zhang, X. R., et al. (2015). Prelalence and diversity of enterotoxin genes with genetic background of Staphylococcus aureus isolates from different origins in China. Int. J. Food. Microbiol. 211, 142–147. doi: 10.1016/j.ijfoodmicro.2015.07.018
Clinical and Laboratory Standards Institute [CLSI] (2018b). Performance Standards for Antimicrobial Susceptibility Testing 2018 Twenty-Second Informational Supplement. Document M100-S22. Wayne, PA: Clinical and Laboratory Standards Institute (CLSI).
Clinical and Laboratory Standards Institute [CLSI] (2018a). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals. CLSI supplement VET08. 4th edn. Wayne, PA: Clinical and Laboratory Standards Institute.
Cortimiglia, C., Bianchini, V., Franco, A., Caprioli, A., Battisti, A., Colombo, L., et al. (2015). Short communication: prevalence of Staphylococcus aureus and methicillin-resistant S. aureus in Northern Italy. J. Dairy Sci. 98, 2307–2311. doi: 10.3168/jds.2014-8923
Crofts, T., Gasparrini, A. J., and Dantas, G. (2017). Next-generation and approaches to understand and combat the antibiotic resistome. Nat. Rev. Microbiol. 15, 422–434. doi: 10.1038/nrmicro.2017.28
Ding, T., Yu, Y. Y., Schaffner, D. W., Chen, S. G., Ye, X. Q., and Liu, D. H. (2016). Farm to consumption risk assessmrent for Staphylococcus aureus and Staphylococcus enterotoxins in fluid milk in China. Food. Control. 59, 636–643. doi: 10.1016/j.foodcont.2015.06.049
Ebrahim, R., and Forough, A. (2013). Presence of enterotoxigenic Staphylococcus aureus in cow, camel, sheep, goat, and buffalo bulk tank milk. Vet. Arhiv. 83, 23–30.
El-Ashker, M., Gwida, M., Tomaso, H., Monecke, S., Ehricht, R., Ei-Gohary, F., et al. (2015). Staphylococci in cattle and buffaloes with mastitis in Dakahlia Governorate, Egypt. J. Dairy. Sci. 98, 7450–7459. doi: 10.3168/jds.2015-9432
Elbagory, A. (2017). Bacteriological quality and occurrence of some microbial pathogens in goat and ewe milk in Egypt. Int. Food. Res. J. 24, 847–851.
Gomes, F., and Henriques, M. (2016). Control of bovine mastitis: old and recent therapeutic approaches. Curr. Microbiol. 72, 377–382. doi: 10.1007/s00284-015-0958-8
Guimaraes, F. D. F., Nobrega, D. B., Richini-Pereira, V. B., Marson, P. M., and Langoni, H. (2013). Enterotoxin genes in coagulase-negative and coagulase-positive staphylococcus isolated from bovine milk. J. Dairy. Sci. 96, 2866–2872. doi: 10.3168/jds.2012-5864
Hu, Y. F., Yang, X., Lu, N., and Zhu, B. L. (2014). The abundance of antibiotic resistance genes in human guts has correlation to the consumption of antibiotics in animal. Gut. Microbiol. 5, 245–249. doi: 10.4161/gmic.27916
Johnston, A. M. (1998). Use of antimicrobial drugs in veterinary practice. BMJ 317, 665–667. doi: 10.1136/bmj.317.7159.665
Kot, B., Szweda, P., Frankowska-Maciejewska, A., Piechota, M., and Wolska, K. (2016). Virulence gene profiles in Staphylococcus aureus isolated from cows with subclinical mastitis in eastern Poland. J. Dairy. Res. 83, 228–235. doi: 10.1017/S002202991600008X
Lan, X. Y., Zhao, S. G., Zheng, N., Li, S. S., Zhang, Y. D., Liu, H. M., et al. (2017). Short communication: microbiological quality of raw cow milk and its association with herd management practices in Northern China. J. Dairy. Sci. 100, 4294–4299. doi: 10.3168/jds.2016-11631
Liu, B. G., Cai, T., Li, X. S., Liu, Y. Y., He, D. D., Kuang, X. H., et al. (2019). Serotyping, virulence genes and pulsed field gel electrophoresis (PFGE) typing of Staphylococcus aureus isolated from fresh milk. Food. Sci. 40, 281–286.
Liu, H. M., Li, S. L., Meng, L., Dong, L., Zhao, S. G., Lan, X. Y., et al. (2017). Prevalence, antimicrobial susceptibility, and molecular characterization of Staphylococcus aureus isolated from dairy herds in Northern China. J. Dairy. Sci. 100, 8796–8803. doi: 10.3168/jds.2017-13370
Low, A., Ng, C., and He, J. Z. (2016). Identification of antibiotic resistant bacteria community and a Geochip based study of resistome in urban watersheds. Water. Res. 106, 330–338. doi: 10.1016/j.watres.2016.09.032
Lyra, D. G., Sousa, F. G. C., Borges, M. F., Givisiez, P. E. N., Queiroga, R. C. R. E., Souza, E. L., et al. (2013). Enterotoxin-encoding genes in Staphylococcus spp. from bulk goat milk. Foodborne. Pathog. Dis. 10, 126–130. doi: 10.1089/fpd.2012.1256
Martins, K., Faccioli, P., Bonesso, M. F., Fernandes, S., Oliveira, A., Dantas, A., et al. (2017). Characteristics of resistance and virulence factors in different species of coagulase-negative staphylococci isolated from milk of healthy sheep and animals with subclinical mastitis. J. Dairy Sci. 3, 2184–2195. doi: 10.3168/jds.2016-11583
Merz, A., Stephan, R., and Johler, S. (2016). Staphylococcus aureus isolates from goat and sheep milk seem to be closely related and differ from isolates detected from bovine milk. Front. Microbiol. 7:319. doi: 10.3389/fmicb.2016.00319
Mole, B. (2013). Farming up trouble: microbiologists are trying to work out whether use of antibiotics on farms is fuelling the human epidemic of drug-resistant bacteria. Nature 499, 398–400.
Mork, T., Kvitle, B., Mathisen, T., and Jorgensen, H. J. (2010). Bacteriological and molecular investigations of Staphylococcus aureus in dairy goats. Vet. Microbiol. 141, 134–141. doi: 10.1016/j.vetmic.2009.08.019
Njage, P. M. K., Dolci, S., Jans, C., Wangoh, J., Lacroix, C., and Meile, L. (2013). Phenotypic and genotypic antibiotic resistance patterns of Staphylococcus aureus from raw and spontaneously fermented camel milk. Eur. J. Nutr. Food Saf. 3, 87–98. doi: 10.9734/EJNFS/2013/2853
Oliveira, A. A. F., Pinheiro, J. W., Mota, R. A., Cunha, M. L. R. S., Lopes, C. A. M., and Rocha, N. S. (2011). Phenotype characterization of Staphylococcus species strains isolated from buffalo (Bubalus bubalis) milk. J. Vet. Diagn. Invest. 23, 1208–1211. doi: 10.1177/1040638711428946
Pamuk, S., Yildirim, Y., Seker, E., Gurler, Z., and Kapa, R. (2012). A survey of the occurrence and properties of methicillin-resistant Staphylococcus aureus and methicillin-resistant Staphylococcus intermedius in water buffalo milk and dairy products in Turkey. Int. J. Dairy. Technol. 65, 416–422. doi: 10.1111/j.1471-0307.2012.00832.x
Redwan, E. M., Ei-Baky, N. A., Ai-Hejin, A. M., Baeshen, M. M., Almehdar, H. A., Elsaway, A., et al. (2016). Significant antibacterial activity and synergistic effects of camel lactoferrin with antibiotics against methicillin-resistant Staphylococcus aureus (MRSA). Res. Microbiol. 167, 480–491. doi: 10.1016/j.resmic.2016.04.006
Rola, J. G., Sosnowski, M., Ostrowska, M., and Osek, J. (2015). Prevalence and antimicrobial resistance of coagulase-positive staphylococci isolated from raw goat milk. Small Rumin. Res. 123, 124–128. doi: 10.1016/j.smallrumres.2014.11.010
Salaberry, S. R. S., Saidenberg, A. B. S., Zuniga, E., Melville, P. A., Santos, F. G. B., Guimaraes, E. C., et al. (2015). Virulence factors genes of Staphylococcus spp. Isolated from caprine subclinical mastitis. Microbial. Pathog. 85, 35–39. doi: 10.1016/j.micpath.2015.05.007
Schmidt, T., Kock, M., and Ehlers, M. M. (2017). Molecular Characterization of Staphylococcus aureus isolated from bovine mastitis and close human contacts in South African dairy herds: genetic diversity and inter-species host transmission. Front. Microbiol. 8:511. doi: 10.3389/fmicb.2017.00511
Shuiep, E. S., Kanbar, T., Eissa, N., Alber, J., Lammler, C., Zschock, M., et al. (2009). Phenotypic and genotypic characterization of Staphylococcus aureus isolated from raw camel milk samples. Res. Vet. Sci. 86, 211–215. doi: 10.1016/j.rvsc.2008.07.011
Stapels, D. A. C., Ramyar, K. X., Bischoff, M., van Kockritz-Blickwede, M., Milder, F. J., Ruyken, M., et al. (2014). Staphylococcus aureus secretes a unique class of neutrophil serine protease inhibitors. Proc. Natl. Acad. Sci. U.S.A. 111, 13187–13192. doi: 10.1073/pnas.1407616111
Wang, D., Zhang, L. M., Zhou, X. Z., He, Y. L., Yong, C. F., Shen, M. L., et al. (2016). Antimicrobial susceptibility, virulence genes, and randomly amplified polymorphic DNA analysis of Staphylococcus aureus recovered from bovine mastitis in Ningxia, China. J. Dairy. Sci. 99, 9560–9569. doi: 10.3168/jds.2016-11625
Xu, J., Tan, X., Zhang, X. Y., Xia, X. L., and Sun, H. C. (2015). The diversities of Staphylococcal species, virulence and antibiotic resistance genes in the subclinical mastitis milk from a single Chinese cow herb. Microb. Pathogenes 88, 29–38. doi: 10.1016/j.micpath.2015.08.004
Zhang, Q. Q., Ying, G. G., Pan, C. G., Liu, Y. S., and Zhao, J. L. (2015). Comprehensive evaluation of antibiotics emission and fate in the river basins of China: source analysis, multimedia modeling, and linkage to bacterial resistance. Environ. Sci. Technol. 49, 6772–6782. doi: 10.1021/acs.est.5b00729
Keywords: antimicrobial resistance, virulence genes, Staphylococcus aureus, public health, raw milk
Citation: Liu H, Dong L, Zhao Y, Meng L, Wang J, Wang C and Zheng N (2022) Antimicrobial Susceptibility, and Molecular Characterization of Staphylococcus aureus Isolated From Different Raw Milk Samples in China. Front. Microbiol. 13:840670. doi: 10.3389/fmicb.2022.840670
Received: 21 December 2021; Accepted: 28 March 2022;
Published: 13 May 2022.
Edited by:
Hosny El-Adawy, Institut für bakterielle Infektionen und Zoonosen, Friedrich Loeffler Institut, GermanyReviewed by:
Mohammed Elbediwi, Zhejiang University, ChinaTaurai Tasara, University of Zurich, Switzerland
Copyright © 2022 Liu, Dong, Zhao, Meng, Wang, Wang and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nan Zheng, emhlbmduYW5fMTk4MEAxMjYuY29t
†These authors have contributed equally to this work
 Huimin Liu
Huimin Liu Lei Dong
Lei Dong Yankun Zhao
Yankun Zhao Lu Meng
Lu Meng Jiaqi Wang
Jiaqi Wang Cheng Wang3
Cheng Wang3 Nan Zheng
Nan Zheng