- 1Institute of Marine Science and Technology, Shandong University, Qingdao, China
- 2Research Centre for Indian Ocean Ecosystem, Tianjin University of Science and Technology, Tianjin, China
- 3College of Marine Science and Technology, China University of Geosciences, Wuhan, China
- 4State Key Laboratory of Biogeology and Environmental Geology, China University of Geosciences, Wuhan, China
Bacterial transformation and processing of phytoplankton-derived organic matter are extremely important for the formation of ubiquitous organic matter (OM) in aquatic ecosystems. Heterotrophic bacteria convert OM into biomass and recycle inorganic components, contributing to the production of microbial food webs. While phytoplankton-derived organic matter is commonly studied, the transformation and processing of dissolved OM (DOM) and lysate OM (LOM) by culturable epiphytic bacteria remains poorly understood. In this study, cultivable epiphytic bacteria from the marine diatom, Skeletonema dohrnii, were isolated, purified, and identified. Three bacteria, Roseobacteria sp., Marinobacter sp., and Bacillus sp., were selected to study the transformation and processing of S. dohrnii-derived DOM and LOM using excitation-emission matrix (EEM) fluorescence methods, and bacterial abundance, dissolved organic carbon (DOC) concentration, and transparent exopolymer particle (TEP) content were measured. Meanwhile, the bacterial transformation of DOM and LOM was further evaluated by the fluorescence index, biological index, β/α, and humification index. The primary fluorophores, peak A (humic-like), peak C (humic-like), peak M (humic-like), peak B (protein-like), and peak T (tryptophan-like), were present in the sample. The fluorescence of DOM and LOM was dominated by protein-like signal that became increasingly humic-like over time, suggesting that more complex molecules (e.g., recalcitrant OM) are being produced. The fluorescence of DOM and LOM was dominated by a protein-like signal that became increasingly humic-like over time, suggesting that epiphytic bacteria produced more complex molecules. Results showed that the bacteria utilized LOM more rapidly than DOM. While the three bacteria transformed OM to different degrees, all were able to facilitate microbial reprocessing of OM into refractory OM.
Introduction
Phytoplankton-derived dissolved organic matter (DOM) is the dominant form of organic matter (OM) in the aquatic environment. DOM contributes to the largest carbon stock on Earth (662 Pg C), of which more than 90% of dissolved organic carbon (DOC) is produced at a rate of 43 Tg per year (Hansell et al., 2012), and plays an important role in the global carbon cycle (Hansell et al., 2009; Thornton, 2014). The majority of DOC is modified into recalcitrant DOC and exported to the deep sea (Hansell et al., 2012). Chromophoric dissolved organic matter (CDOM) is the optically active fraction of DOM and is an essential part of the microbial process, controlling the attenuation of light and photochemical reactions, and influencing primary productivity (Blough and Siegel, 2002; Mopper et al., 2015). In comparison, lysate organic matter (LOM) from phytoplankton forms a pool of OM that is created extracellularly through metabolic excretion and adsorption (McIntyre and Guéguen, 2013). LOM can significantly protect algal cells from adverse environmental conditions (Pereira et al., 2009). Cell wall polymers of marine phytoplankton are considered one of the major components of recalcitrant dissolved organic matter (RDOM) (Tanoue et al., 1995; Benner and Kaiser, 2003). Recent studies show that carotenoid degradation products may become biological RDOM in the ocean (Arakawa et al., 2017). Fluorescent dissolved organic matter (FDOM) produced by microorganisms has also attracted wide attention due to its extended degradation time (Yamashita and Tanoue, 2008; Catalá et al., 2015). As a result, the microbial carbon pump (MCP) has been proposed to further elucidate the mechanisms by which carbon is sequestered in the ocean (Jiao et al., 2010).
Phytoplankton and bacteria have co-existed for over 200 million years (Falkowski et al., 2004), and epiphytic bacteria are essential for phytoplankton-derived DOM production (Liu et al., 2021a). Bacteria can use and transform 10–50% of the photosynthetic products of phytoplankton and together, bacteria and phytoplankton are important regulators of the structure and function of aquatic ecosystems (Seymour et al., 2017). Diatoms (Bacillariophyceae) are widespread in aquatic ecosystems and account for approximately 20% of fixed carbon in oceans (Nelson et al., 1995). Bacteria-diatom interactions typically take place in the phycosphere, a diffusive boundary layer surrounding diatom cells, or within free-floating or surface-attached cell aggregates (Amin et al., 2012). There are bacterial communities in the phycosphere that are distinct from the surrounding environment and the abundance and diversity of bacteria change during different phases of algal growth (Liu et al., 2021a). There is also specificity in the epiphytic bacteria associated with each algae species. Previous studies show that the epiphytic bacteria, Alexandrium tamarense, primarily consists of the genus Roseobacter sp. and Bacteroidetes (Jasti et al., 2005). Fandino et al. (2001) showed that free-living bacteria mainly belong to α-Proteobacteria and γ-Proteobacteria, and attached bacteria are primarily dominated by Cytophaga–Flexibacter–Bacteroides (CFB) during the Lingulodinium polyedrum dinoflagellate bloom. However, different selective pressures can impact the composition of bacterial communities in different survival environments (i.e., free-living or attached) (Grossart et al., 2005; Rooney-Varga et al., 2005). In general, epiphytic bacteria are responsible for significant uptake of monomeric carbohydrates and amino acids (Middelboe et al., 1995).
Transparent exopolymer particles (TEP) are acid-rich polysaccharides that form biologically or abiotically through condensation or exudation of extracellular polymeric substances (EPS) and can be stained by the specific dye, Alcian Blue (Passow and Alldredge, 1994; Passow, 2002). While phytoplankton are the main producers of TEP, bacteria can also release these particles (Ortega-Retuerta et al., 2010), however, the components of TEP production differ between the two. TEP produced by bacteria has a higher content of uronic acids which makes the surrounding environment more reactive with other surfaces (Bhaskar and Bhosle, 2005). As a result, bacteria are particularly important for carbon fixation by phytoplankton in aquatic ecosystems (Simon et al., 2002).
The Roseobacter sp. clade is one of the major groups of marine Proteobacteria, and is widely distributed in a variety of aquatic environments. Members of this clade share > 89% identity of the 16S rRNA genes (Buchan et al., 2005). Marinobacter sp., including halotolerant and halophilic microorganisms, is an ecologically important genus of Gammaproteobacteria found in diverse marine habitats, many species of which can degrade hydrocarbons. Bacillus also has the potential for degrading lignocellulosic biomass (Chen et al., 2018).
Excitation-emission matrix (EEM), also known as three-dimensional fluorescence (3D-EEM), is used commonly in fluorescence spectroscopy because it can produce a large amount of data, visual maps, and multidimensional information. EEM methods are frequently used to characterize DOM due to their remarkable sensitivity and selectivity (Coble, 1996). Protein- and humic-like fluorophores have been identified by EEM and their peak positions make them easy to discriminate.
In this study, epiphytic bacteria from the diatom S. dohrnii were isolated, cultured, and characterized. Given the potential importance of OM processing into biogeochemical cycles, the role and preferences of the epiphytic bacteria, Roseobacter sp., Marinobacter sp., and Bacillus sp., in DOM and LOM transformation by S. dohrnii were explored. This research builds upon laboratory-based studies lasting 90 days. The characteristics of fluorescent organic matter were examined using EEM methods and different parameters, including bacteria abundance, DOC, TEP, fluorescence index (FI), the biological index (BIX), the humification index (HIX), and β/α to further characterize how OM processing may differ by type of bacteria. It was hypothesized that the major bacterial taxa would correlate significantly with OM concentration and composition and would exhibit different patterns in these linkages.
Materials and Methods
Skeletonema dohrnii Cultures
Marine diatoms, Skeletonema spp., are widely distributed in offshore China. Skeletonema dohrnii was isolated from coastal seawaters around the Yellow Sea. The S. dohrnii was incubated at 25°C with 100 μmol photons m–2 s–1 irradiation in artificial seawater (ASW) liquid medium and the photoperiod was on a light: dark (14 h: 10 h) cycle. The conical flasks used for the ASW medium are pre-combusted (450°C, 5 h) to minimize the input of background organic carbon (Lechtenfeld et al., 2015). To regularly monitor algal growth, a 100 μL of culture solution was mounted onto a blood cell counting slide and algae cells were enumerated with a microscope (Olympus BX51, Japan).
Isolation, Culture, and Identification of Bacterial Strains
Culturable bacteria were isolated from the degradation growth stage of S. dohrnii using the gradient dilution method, plated on 2216E agar plates, and cultured in transparent conical flasks (500 mL) in a shaking incubator (26°C, 160 rpm). To identify the culturable bacteria, genomic DNA was extracted using the TIANamp Bacteria DNA Kit (Tiangen-Biotech, Beijing, China). For polymerase chain reaction (PCR), amplification of the 16S rDNA V3 region, a universal bacterial primer, was used. Bar-coded fragments of the 16S rDNA were amplified using the primer 27F (5′-AGAGTTTGATCCTGGCAG-3′) and 1492R (5′-TACGGTTACCTTGTTACGACTT-3′) (Bosshard et al., 2000). A phylogenetic tree (Neighbor-Joining tree, N-J tree) was constructed using the bacterial sequences and closest related sequences from GenBank, and the genetic distances were calculated. Three isolated bacterial strains, Roseobacter sp., Marinobacter sp., and Bacillus sp., shared 64–100% sequence identity with the valid species from GenBank (Supplementary Figure 1).
Flow Cytometry of Culturable Bacteria
The three culturable bacterial strains, Roseobacter sp., Marinobacter sp., and Bacillus sp., were cultured in 500 mL pre-combusted (450°C, 5 h) transparent conical flasks. The abundance of bacterial cells was measured using an Accuri C6 flow cytometer (BD Biosciences, Erembodegem, Belgium), as described previously (Moens et al., 2016). The sample was stained with 0.01% SYBR Green I for 30 min in the dark at 37°C (Marie et al., 1997). As an internal standard, 1 μm fluorescent beads (Polyscience, Warrington, PA, United States) were injected into each sample. The samples were measured at a flow rate of 0.25 μL s–1 for 1 min.
Measurement of Dissolved Organic Carbon and Absorption Spectroscopy
The dissolved organic carbon (DOC) was determined using a total organic carbon analyzer (TOC-3100, Germany). All samples were defrosted and acidified with phosphoric acid to a pH of 2 before being analyzed. Potassium hydrogen phthalate standards were used to quantify the DOC. Samples were gravity filtered using pre-combusted (450°C for 6 h) GF/F glass fiber filters (0.7 μm pore size, 47 mm diameter, Whatman, Maidstone, United Kingdom). While GF/F glass fiber filters do not retain as many bacterial cells as 0.2 μm polycarbonate filters, GF/F glass fiber filters can be cleaned by high-temperature combustion and can filter sufficient volumes without clogging, reducing potential sources of contamination. Therefore, the advantages of GF/F glass fiber filters prove that its application in this study is reasonable.
Transparent Exopolymer Particle Measurements
Lysate OM samples (100 mL) were filtered through duplicate 0.4 μm polycarbonate membranes (Millipore, Billerica, MA, United States) at low and constant pressure (<100 mm Hg) and particles retained on the filters were stained for < 5 s with 0.5 mL Alcian Blue solution (0.02% w/w at pH 2.5, Alcian Blue 8GX, Sigma) that had been prefiltered through 0.2 μm polycarbonate membranes (Millipore, United States) to fully remove reaggregated dye particles. Stained filters were rinsed twice with distilled water to remove excess dye that failed to bond with substrates. Alcian Blue-stained particles were extracted from filters by soaking in 6 ml of 80% H2SO4 for 2 h with gentle agitation at least three times during this period. The extracted TEP was then measured using spectrophotometry (referring to Passow and Alldredge, 1995) with a maximum absorption of 787 nm. Calibrations by the Gum Xanthan (GX, Sigma) were carried out for the recorded TEP absorbance values and the final results were expressed in micrograms of GX equivalents per liter (μg Xeq L–1) using the following equation:
where E787 is the sample absorption, B787 is the blank (distilled water) absorption, Vf is the filtered volume in liters, and fx is the calibration factor (Bittar et al., 2018).
Excitation-Emission Matrix Fluorescence Spectroscopy
Three dimensional (3D)-EEM measurements were made using a fluorescence spectrophotometer (Hitachi F-7100, Tokyo, Japan). The voltage of the photomultiplier tube was set to 700 V. Fluorescence spectra detected subsequent scanning of excitation (Ex) from 200 to 450 nm and emission (Em) from 250 to 550 nm. Ex and Em slits were maintained at 5 nm and the scanning speed was set at 12000 nm min–1. Instrument corrections were performed based on the procedure recommended by the Hitachi F-7100 instruction manual. To eliminate most of the Raman scatter, each DOM spectrum was subjected to blank subtraction using ultrapure water (Milli-Q). The correction was followed by Raman calibration as described previously (Lawaetz and Stedmon, 2009). A number of fluorescent peaks were used in this study, namely peak A, peak C, peak M, peak B and peak T. The components of these peaks include: humic-like (peaks A, C, and M) and protein-like (peak T). All peak characteristics mentioned are referenced in Table 1. Fluorescence intensity arbitrary units (a.u.) were utilized to analyze all data. Four indexes [including FI (McKnight et al., 2001), BIX (Huguet et al., 2009), β/α (Parlanti et al., 2000), and HIX (Ohno, 2002)] were calculated using the formula below:
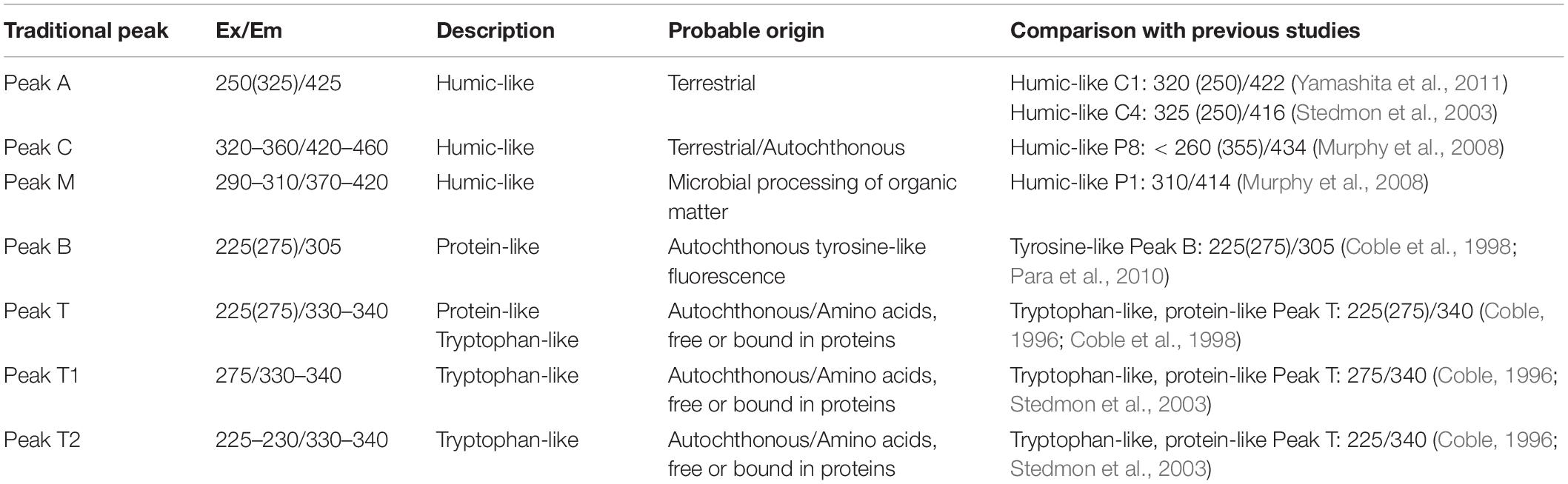
Table 1. Central regions of EEM fluorescence attributed to different sources of organic matter compared with previous studies.
where I is the fluorescence intensity at each wavelength. Ex and Em are represented as excitation wavelength and emission wavelength, respectively.
The FI greater than 1.8 indicate fluorescence component of OM was mainly produced by microorganisms (Loginova et al., 2016). The BIX can be used as indicators of DOM traceability in aquatic ecosystems (Huguet et al., 2009). The β/α (a ratio of two known fluorescing components, where β represents more recently derived DOM and α represents highly decomposed DOM) (Parlanti et al., 2000). The HIX is a fluorescence index of organic matter degradation (Ohno, 2002).
Experimental Set-Up
Experiments were conducted in transparent conical flasks that were pre-acid washed and rinsed with ultrapure water. S. dohrnii cells were cultivated in ASW medium, and after reaching the degradation growth phase, in which the algal concentration was approximately (3.84 ± 0.01) × 107 cells L–1, microalgae liquid was filtered through a 0.2 μm polycarbonate membrane (Millipore, United States) to remove the particles. To prevent carbon contamination, the filter system was pre-cleaned with Milli-Q water, and the filtrate was regarded as the DOM fraction. To isolate the LOM fraction, algal samples were centrifuged at 5000 rpm for 5 min. The supernatant was removed, resuspended with 0.05% NaCl solution, and heated at 60°C for 30 min. The extracted solutions were then centrifuged at 7000 rpm for 20 min and the supernatant was regarded as the LOM fraction (Xu and Jiang, 2013). LOM samples were filtered through a 0.2 μm polycarbonate membrane (Millipore, United States) to prevent the introduction of particulate matter. We collected, concentrated, centrifuged and washed the bacterial cells to prevent the effect of the substrate on the growth of the bacteria. To evaluate the ability of epiphytic bacteria derived from S. dohrnii to transform DOM and LOM, the following treatment groups were established: (1) Roseobacter sp. grown in DOM (Roseobacter sp. + DOM), (2) Marinobacter sp. grown in DOM (Marinobacter sp. + DOM), (3) Bacillus sp. grown in DOM (Bacillus sp. + DOM), (4) Roseobacter sp. grown in LOM (Roseobacter sp. + LOM), (5) Marinobacter sp. grown in LOM (Marinobacter sp. + LOM), and (6) Bacillus sp. grown in LOM (Bacillus sp. + LOM). Two blanks, (1) DOM filtrate (DOM Blank), and (2) LOM filtrate (LOM Blank), were also used. A total of eighteen 1.8 L borosilicate bottles per treatment were inoculated with an initial abundance of 7.25–7.27 × 105 bacteria cells mL–1, and the room temperature was maintained at 26°C. Three independent replicates were analyzed per experimental group (i.e., 24 conical flasks for 6 treatment group and 2 control group). In brief, all experiments were carried out in triplicate and under dark conditions.
Statistical Analysis
In this study, data were checked for normality and homogeneity of variance before analysis. A one-way analysis of variance (ANOVA) and t-tests via SPSS software (version 23.0) were used to analyze the data. If an ANOVA was significant, differences between DOM and LOM treatment groups (excluding DOM and LOM blank groups) were compared with a Tukey’s post hoc test. The p < 0.05 was considered statistically significant. The correlation analysis and DOM associated parameters were obtained using the “ggplot2” and “corrplot” package in RStudio software. Pearson correlation coefficients were used to evaluate the relationships between fluorescence index and fluorescence peak intensity. The measured values were expressed as the mean ± standard deviation (SD).
Results
Bacterial Abundance, Dissolved Organic Carbon, and Transparent Exopolymer Particle Concentrations
Growth of the three culturable bacterial strains, Roseobacter sp., Marinobacter sp., and Bacillus sp., in different treatment groups, is shown in Figure 1A. In all groups, the initial abundance of bacteria was around (7.26 ± 0.01) × 105 cells mL–1, and this increased over the course of the 90 days incubation period. While Bacillus sp. + DOM achieved the highest abundance of up to (12.86 ± 0.08) × 105 cells mL–1, Marinobacter sp. + DOM had a moderate abundance of (12.42 ± 0.03) × 105 cells mL–1 and the abundance of Roseobacter sp. + DOM slowly increased to (11.84 ± 0.09) × 105 cells mL–1. In contrast, bacterial abundance increased markedly in all the LOM groups, especially in the Bacillus sp. + LOM group, which increased to (20.25 ± 0.11) × 105 cells mL–1. The significant increase in bacterial abundance seen in all treatment groups was likely caused by the uptake of organic matter.
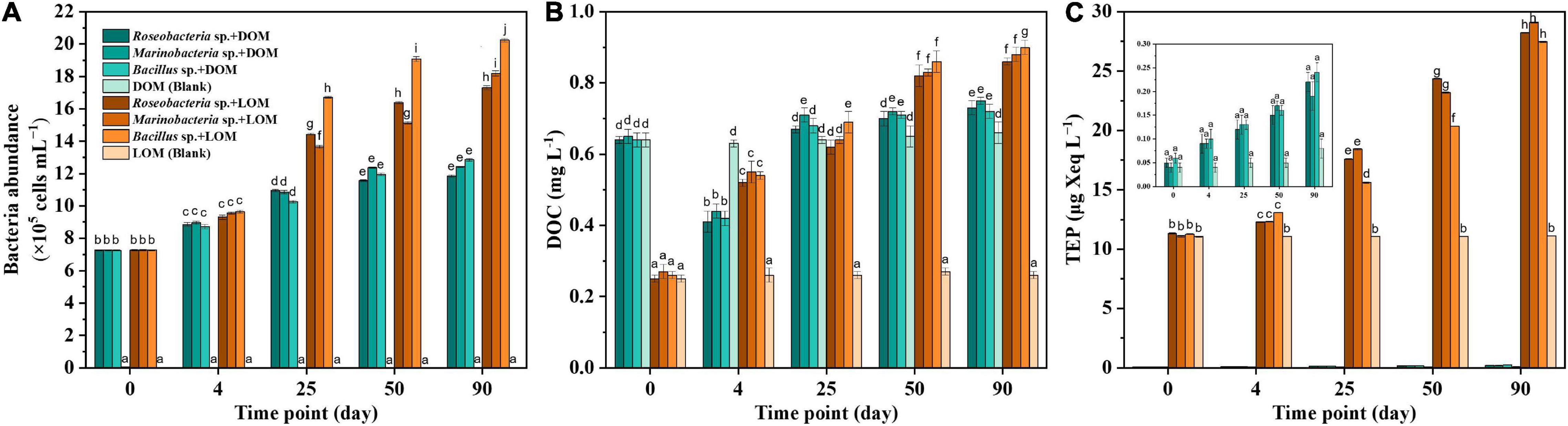
Figure 1. Bacteria abundance (A), DOC (dissolved organic carbon, B) and TEP (transparent exopolymer particles, C) for the different treatment groups and associated blanks. Error bars represent the standard error for triplicates cultures. Comparison between all DOM and LOM treatments for all experimental days. Different lowercase letters represented significant difference (p < 0.05).
As shown in Figure 1, the initial DOC concentration was 0.64 ± 0.01 mg L–1 for the DOM treatment groups and decreased over the four days of culture. The DOC concentration increased slightly on the remaining days but this difference was not significant. In contrast, the DOC concentration of the LOM treatment groups maintained an increasing trend. The largest DOC increase occurred in the Bacillus sp. + LOM group, which rose by 0.64 mg L–1 DOC over the 90-day period.
Figure 1C shows the TEP trend in the DOM and LOM treatment groups. It was visually apparent that while TEP concentrations increased in the DOM groups, they remained low. For example, Marinobacter sp. + DOM only increased from 0.04 ± 0.01 to 0.19 ± 0.03 μg Xeq L–1. TEP concentrations in the LOM treatment groups increased significantly over time.
Fluorescence Characteristics
Fluorescence patterns were similar between all the DOM and LOM treatment groups, with peak T (including T1 and T2, protein-like) being the most prominent (Figure 2, Supplementary Figures 2, S3, and Table 1). The peak T (T1 and T2) of the S. dohrnii-derived DOM was more pronounced until the overall peak pattern shifted to longer wavelengths. Roseobacter sp. + DOM maintained the two-peak pattern during the first 25 days (Supplementary Figures 2A–C). On day 50, a weaker peak M (Ex/Em, 335/400, humic-like) appeared (Supplementary Figure 2D). This phenomenon was also seen in the Marinobacter sp. + DOM and Bacillus sp. + DOM cultures (Supplementary Figures 2I,N), but was not significant. Marinobacter sp. + DOM showed a clear peak B (Ex/Em, 225/300, protein-like) on day four (Supplementary Figure 2G), after which the fluorescence characteristics weakened and disappeared. The overall fluorescence characteristics of Bacillus sp. + DOM had fluorescence characteristics that were similar to Roseobacter sp. + DOM, with peak M (humic-like) being relatively weak. Higher fluorescence intensities generally occurred on days 4 and 25 for all DOM treatment groups.
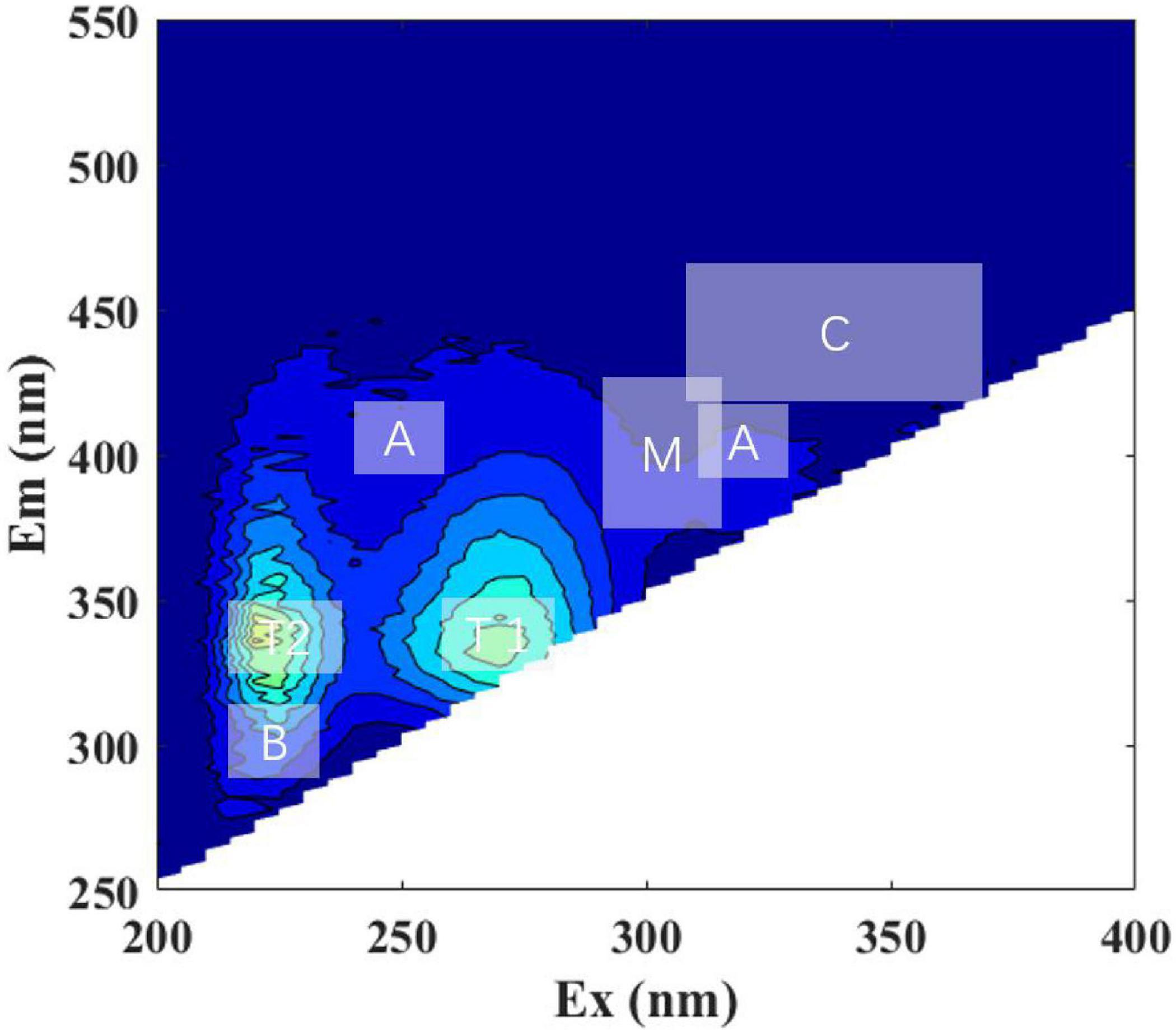
Figure 2. Representative excitation-emission matrices (EEMs) of Roseobacteria sp. + DOM treatment at initial time point (day 0). Major peak regions of EEMs are indicated in the picture. The ‘A,’ ‘C,’ and ‘M’ stands for humic-like; ‘B’ and ‘T’ stands for protein-like; ‘T1’ and ‘T2’ stands for tryptophan-like.
The overall peak pattern of LOM was similar to that of DOM (Supplementary Figures 2, 3), but these peaks were broader and less distinct. In all LOM treatment groups, there was an initial low-intensity fluorescence (a.u. < 800) in the region of peak T2 (Ex/Em, 275/340) (Supplementary Figures 3A,F,K). Peak T1 was relatively weak compared to the DOM treatment groups. On day four, the LOM treatment groups essentially formed a three-peak pattern (Supplementary Figure 3B,G,I), and unstructured fluorescence with slightly enhanced fluorescence appeared around peaks T1 and T2. The maximum fluorescence intensity gradually decreased over time. On day 90, the fluorophore changes in Marinobacter sp. + DOM were not as obvious as those seen in Roseobacter sp. + LOM and Bacillus sp. + LOM (Supplementary Figures 3C,J,O).
Fluorescence Indices and Peaks
Peaks A and C (humic-like) had similar fluorescence intensities in all treatment groups, which increased over time (Figures 3A,B). Peak M (humic-like) declined for the first two to three days and then increased slowly in all treatment groups (Figure 3C). For peak B, within-group samples were similar for the DOM and LOM treatment groups, except for Bacillus sp. + LOM which had the highest value at day 20 and then decreased (Figure 3D). For peak T, DOM treatment groups had the highest value on day four, while LOM treatment groups peaked on different days (Figure 3E).
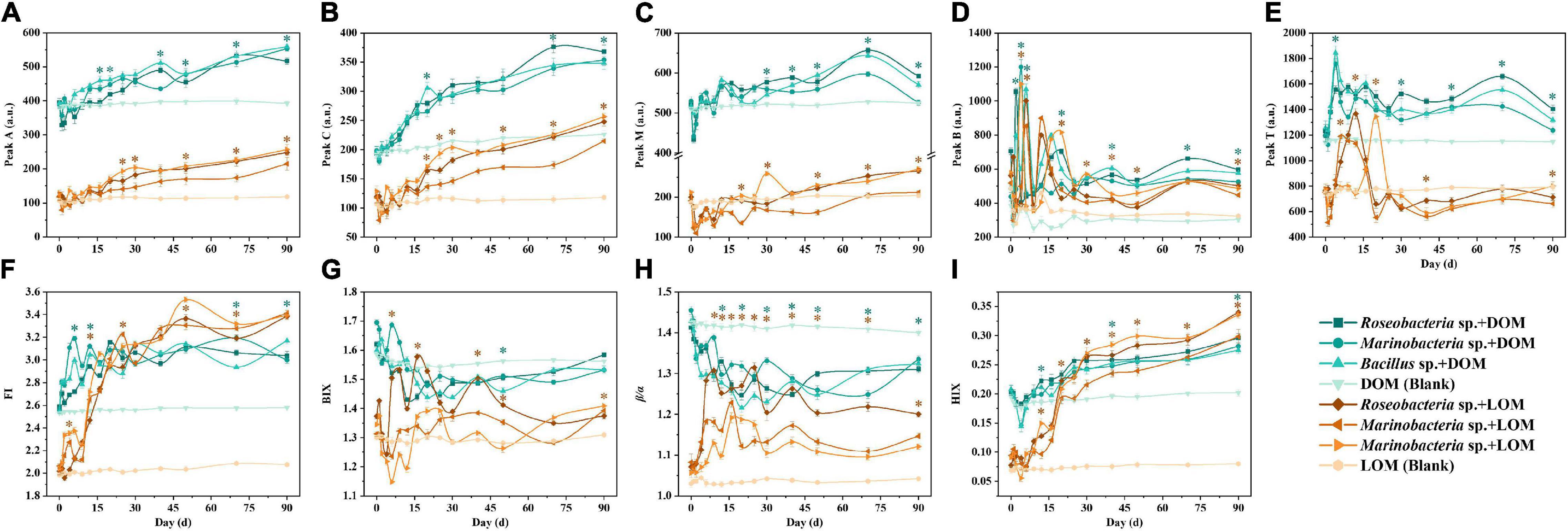
Figure 3. Changes in fluorescence peaks (A–E) and indices (F–I) of three culturable epiphytic bacteria, Roseobacteria sp., Marinobacter sp., and Bacillus sp. of S. dohrnii in the different OM treatments over the 90-day time period. Error bars represent the standard error for triplicates cultures. Note error bars are smaller than the symbol sizes in some cases. Tukey’s post hoc test were conducted between the DOM and LOM treatments, excluding the blank groups. Asterisk (*) represents statistical significance at the p < 0.05. Green and orange asterisk indicate statistical analysis for each experimental day for the DOM and LOM groups, respectively.
The fluorescence index was also used to indicate changes in fluorescence characteristics. Changes in the fluorescence indices, FI, BIX, β/α, and HIX, are shown in Figure 3. In general, LOM values were lower than DOM values across all fluorescent indices, with similar trends for FI and HIX (Figures 3F,I), and for BIX and β/α (Figures 3G,H). Compared to the DOM treatment groups, the LOM treatment groups showed a clear increase in FI and HIX. In addition, BIX and β/α decreased over time in the DOM treatment groups (Figure 3G).
Discussion
Most previous studies have investigated the dynamics of phytoplankton and bacterial CDOM with carbon and nutrient additions (Romera-Castillo et al., 2010; Goto et al., 2017, 2020). It has also been shown that phytoplankton produce DOM with different characteristics and fluorescence intensities under different growth conditions (Liu et al., 2021a,b). To better investigate the transformation of OM by culturable marine bacteria, diatom-derived DOM and LOM were used as the sole carbon sources.
In this study, culturable epiphytic bacteria were isolated from S. dohrnii, and identified. The 90-day transformation of S. dohrnii DOM and LOM by three bacterial strains, Roseobacter sp., Marinobacter sp., and Bacillus sp., was assessed. S. dohrnii-derived DOM was collected during degradation because it had multiple fluorescence characteristics and high fluorescence intensity. To ensure consistency of the experimental materials, S. dohrnii-derived LOM was also extracted during degradation. It was helpful to observe the transformation of DOM and LOM by epiphytic bacteria. Previously published data along with results from this study demonstrate that the characteristics of OM produced by diatoms are relatively similar (Liu et al., 2021a). However, while the fluorescence characteristics of S. dohrnii-derived DOM and LOM were similar, the fluorescence intensity of LOM was generally low (≤1200 a.u.).
Spectral Properties of Skeletonema dohrnii-Derived Dissolved Organic Matter and Lysate Organic Matter
All treatment groups produced a two- or three-peak patter dominated by discrete S. dohrnii-derived DOM and LOM fluorophores, indicating that different compounds, such as those resembling tryptophan, are present in algae CDOM. In addition, not all fluorophores were created from contamination during incubation or by the background ASW medium. Several studies have shown that protein peaks (e.g., tryptophan-like) are dominant during phytoplankton growth (Stedmon and Markager, 2005; Jørgensen et al., 2011; Liu et al., 2021a,b). In addition, peaks A and C (e.g., flavins and phenols) are thought to be humic substances from the earth (Wünsch et al., 2015). As a result, the experiments were conducted with an inorganic medium synthesized from artificial seawater, which lacks substances of terrestrial origin. Thus, the observed changes in unstructured fluorescence with slightly enhanced fluorescence, the formation of peaks A and C, and the subsequent red-shift were due to bacterial-transformed DOM and LOM rather than terrestrially derived fluorescence. DOM and LOM fluorescence had similar peak regions and DOM usually had a broader emission wavelength. It is likely that once algae are degraded, the algal-derived substances are released into the surrounding aquatic environment. In previous studies, DOM produced in phytoplankton and bacterial cultures showed similar fluorescence patterns as those presented here (Kinsey et al., 2018). Fukuzaki et al. (2014) also studied the DOM spectra of algal cultures (e.g., axenic diatoms, dinoflagellates, and chlorophytes) and found that even when DOM fluorophores are different, the DOM patterns produced are generally similar. In the current study, the fluorescence of peaks A, C, and T increased, while peak B increased significantly in a short period of time and then decreased to near its initial value. This suggests that epiphytic bacteria utilize DOM and LOM over a short period which leads to a brief increase in bacterial abundance before the OM is transformed into recalcitrant OM and the epiphytic bacteria are gradually accumulated. The incubations contained the protein-like fluorophore (e.g., tryptophan-like components), which increased by the fourth day likely because the production of bacteria exceeded consumption (Goto et al., 2017, 2020). Importantly, these changes occurred in all treatment groups. Specifically, there was a significant increase in peak B and T fluorescence intensity in the short term, which then decreased. This suggests that large polymers were still released but that some of these exopolymers were degraded into small peptides (Goto et al., 2017). The decrease in the tryptophan-like fluorophore was predominant, but stayed relatively stable for the remaining time, suggesting that small peptides produced by the degradation of large exopolymers correlated with reutilization of tryptophan-like fluorophores by the bacteria.
Previous studies of natural microbial community cultures have shown that a wide range of heterotrophic bacteria primarily produce humic-like substances during growth (Shimotori et al., 2009; Arai et al., 2018). It is noteworthy that these humus-like substances are recalcitrant during deep ocean circulation and only a small fraction of fluorescent dissolved organic matter are biodegradable (Catalá et al., 2015). In addition, the generally low fluorescence in the open ocean suggests that the degradation experiments in this study were conducted over a longer time period than those performed in other studies (Gruber et al., 2006; Fukuzaki et al., 2014). One study demonstrated that a single bacterial species cannot completely degrade large molecular weight OM compounds, implying that multiple species are required for degradation to occur (Horemans et al., 2013). The utilization and transformation of OM by a single bacterium takes considerably longer. The current study shows that the transformation and utilization of DOM by a single strain of bacteria requires more time than the transformation of LOM (Liu et al., 2022).
The FI provided a clear insight into the process of OM-related substance changes. The values were all greater than 1.8, indicating that the fluorescence component of OM was mainly produced by microorganisms (Loginova et al., 2016). The FI values of the LOM treatments rose significantly, surpassing the DOM treatments by day 40. The ratio of two known fluorescing components (β/α, where β and α represent more recently derived OM and highly decomposed OM, respectively) and BIX, where higher values indicate greater degradation of DOM. The significant changes in β/α and BIX values over a short period of time indicate that endogenous carbon products are most likely produced through bacterial processing of DOM and LOM. The higher values of HIX indicating higher molecular weight aromatic compounds (i.e., HIX is directly proportional to the humic content of DOM) (Huguet et al., 2009). The degree of humification gradually intensified over time in all treatment groups, especially the LOM groups (e.g., Bacillus sp. + LOM). These findings indicated that the alteration of the OM pool by bacteria leads to a tendency for the fluorescent component to become humified. Taken together, these results indicate that epiphytic bacteria used in this study have a strong ability to transform S. dohrnii-derived LOM, while only weakly transforming S. dohrnii-derived DOM.
Organic Matter Transformation
Transparent exopolymer particle is a class of exopolymers rich in acidic polysaccharides that form biotically or abiotically by exudation or coagulation of exopolymeric precursors (i.e., exopolymeric substances) (Alldredge et al., 1993). TEP is widely distributed in diatoms, coccolithophores, and cyanobacteria (Van Oostende et al., 2013; Chen and Thornton, 2015; Pannard et al., 2016). The current study compared epiphytic bacteria TEP production needed to utilize S. dohrnii-derived DOM and LOM. The DOM-treated groups showed a small amount of TEP production that increased slowly over time, indicating that epiphytic bacteria can produce and release TEP, as shown previously (Bhaskar and Bhosle, 2005). In contrast, the TEP concentration increased significantly in the LOM-treated groups indicating that the epiphytic bacteria were able to better utilize LOM and synthesize their own biomass.
In the present study, the significant positive correlation (p < 0.05) between bacterial abundance and different carbon pools (i.e., DOM and LOM) suggested a strong link between bacterial growth and activity, with epiphytic bacteria actively transforming organic compounds. The DOC concentration showed a distinct increase (net increases of 0.11–0.55 mg L–1), suggesting that OM components were transformed by bacteria to produce new DOC (i.e., production was higher than consumption). This is similar to DOC concentrations (0.41–0.96 mg L–1) observed in the natural environment (Hansell et al., 2009). The OM can be used as a substrate for remineralization of carbon by heterotrophic microorganisms. Microorganisms use enzymes to catalyze OM into smaller compounds that can be transported to the environment across bacterial cell membranes (Arnosti, 2011). Organic carbon is then incorporated into the biomass or excreted as DOC in the form of metabolic products (Arnosti, 2011). This provides a new perspective for observing the transformation process of diatoms-derived DOM and LOM by epiphytic bacteria.
Correlation Between Fluorescence Indices and Peak Parameters
By analyzing the relevant parameters of fluorescence indices and peaks, it was found that most parameters were correlated significantly, either positively or negatively (Figure 4). However, there were differences between each treatment group. Indeed, significant correlations were found between HIX and other parameters. In the DOM treatment groups (Figures 4A–C), the HIX was positively correlated with the FI, peak A, peak C, and peak M, but negatively correlated with the peak B, peak T, β/α, and BIX. Since HIX is a fluorescence index measuring the degradation of organic matter (aromatic compounds), with higher values indicating higher molecular weight, the results demonstrate that DOM is gradually converted to recalcitrant high molecular weight compounds by epiphytic bacteria. In the LOM treatment groups (Figures 4A–C), peak B was negatively correlated with most parameters (e.g., FI, HIX, peaks A, C, and M) and showed different correlations in each DOM treatment group. This observation suggests that epiphytic bacteria have different transforming abilities for DOM and LOM.
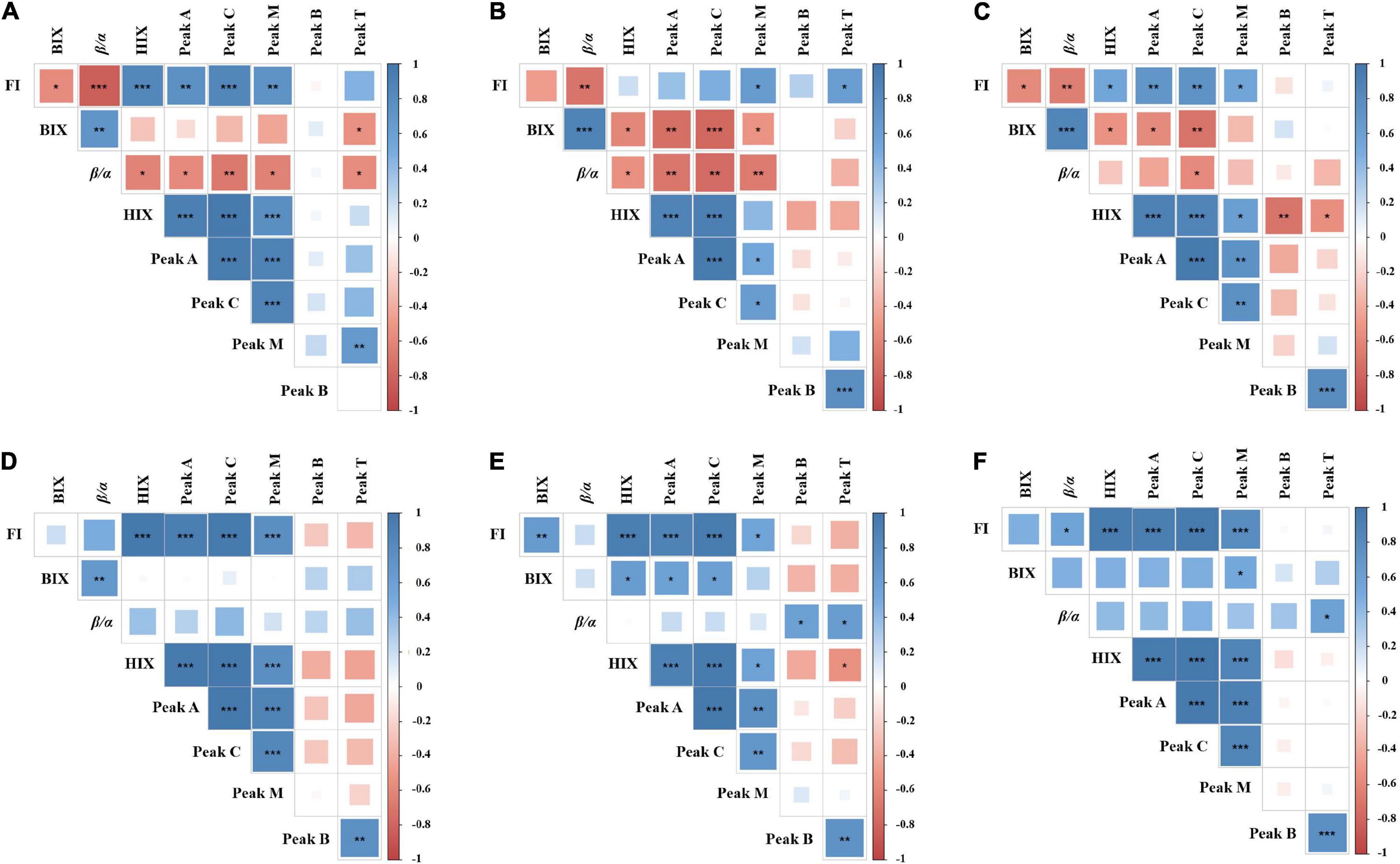
Figure 4. Correlation matrix among different fluorescence peaks and indices parameters. (A–C) Represent Roseobacter sp. + DOM, Marinobacter sp. + DOM, and Bacillus sp. + DOM, respectively. (D–F) Represent Roseobacter sp. + LOM, Marinobacter sp. + LOM, and Bacillus sp. + LOM, respectively. The color and size of the square circle indicate the Pearson’s correlation coefficient. *, **, and *** indicate significance levels at p < 0.05, p < 0.01, and p < 0.001, respectively. The blue color indicates a positive correlation and the red color indicates a negative correlation.
Conclusion
This study demonstrates that epiphytic bacteria are involved in the formation of unstructured “humus-like” fluorescence. In short, epiphytic bacteria are essential for the transformation and processing of diatom-derived OM. Previous studies have observed red-shift of CDOM peaks and these have been described as the result of microbial degradation (Burdige et al., 2004; Kinsey et al., 2018). This supports the conclusion that the altered fluorescence peaks and indices observed in this study are due to bacterial processing of DOM and LOM, and reprocessing into more complex molecules like RDOM (Lechtenfeld et al., 2015). The presence of molecules that are harder to degrade can facilitate charge transfer interactions between electron acceptors and donors. Indeed, the interaction between heterotrophic bacteria and OM is also an important source of deep-sea RDOM.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
JS and YL: conceptualization. JS: methodology, resources, supervision, project administration, and funding acquisition. YL: writing—original draft preparation and visualization. YL, XW, and JS: writing—review and editing. All authors: read and agreed to the published version of the manuscript.
Funding
This research was financially supported by the National Key Research and Development Project of China (2019YFC1407805), the National Natural Science Foundation of China (41876134), the Changjiang Scholar Program of Chinese Ministry of Education (T2014253), and supported by State Key Laboratory of Biogeology and Environmental Geology, China University of Geosciences (No. GKZ21Y645) to JS.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.840564/full#supplementary-material
References
Alldredge, A. L., Passow, U., and Logan, B. E. (1993). The abundance and significance of a class of large, transparent organic particles in the ocean. Deep. Res. Part I Oceanogr. Res. Pap. 40, 1131–1140. doi: 10.1016/0967-0637(93)90129-q
Amin, S. A., Parker, M. S., and Armbrust, E. V. (2012). Interactions between diatoms and bacteria. Microbiol. Mol. Biol. Rev. 76, 667–684. doi: 10.1128/MMBR.00007-12
Arai, K., Wada, S., Shimotori, K., Omori, Y., and Hama, T. (2018). Production and degradation of fluorescent dissolved organic matter derived from bacteria. J. Oceanogr. 74, 39–52. doi: 10.1007/s10872-017-0436-y
Arakawa, N., Aluwihare, L. I., Simpson, A. J., Soong, R., Stephens, B. M., and Lane-Coplen, D. (2017). Carotenoids are the likely precursor of a significant fraction of marine dissolved organic matter. Sci. Adv. 3:e1602976. doi: 10.1126/sciadv.1602976
Arnosti, C. (2011). Microbial extracellular enzymes and the marine carbon cycle. Annu. Rev. Mar. Sci. 3, 401–425. doi: 10.1146/annurev-marine-120709-142731
Benner, R., and Kaiser, K. (2003). Abundance of amino sugars and peptidoglycan in marine particulate and dissolved organic matter. Limnol. Oceanogr. 48, 118–128. doi: 10.4319/lo.2003.48.1.0118
Bhaskar, P. V., and Bhosle, N. B. (2005). Microbial extracellular polymeric substances in marine biogeochemical processes. Curr. Sci. 88, 45–53.
Bittar, T. B., Passow, U., Hamaraty, L., Bidle, K. D., and Harvey, E. L. (2018). An updated method for the calibration of transparent exopolymer particle measurements. Limnol. Oceanogr. Methods 16, 621–628. doi: 10.1002/lom3.10268
Blough, N. V., and Siegel, D. A. (2002). “Chromophoric DOM in the coastal environment,” in Biogeochemistry Of Marine Dissolved Organic Matter, eds D. A. Hansell and C. A. Carlson (San Diego, CA: Academic Press), 509–546. doi: 10.1016/b978-012323841-2/50012-9
Bosshard, P. P., Santini, Y., Grüter, D., Stettler, R., and Bachofen, R. (2000). Bacterial diversity and community composition in the chemocline of the meromictic alpine Lake Cadagno as revealed by 16S rDNA analysis. FEMS Microbiol. Ecol. 31, 173–182. doi: 10.1111/j.1574-6941.2000.tb00682.x
Buchan, A., González, J. M., and Moran, M. A. (2005). Overview of the marine Roseobacter lineage. Appl. Environ. Microbiol. 71, 5665–5677. doi: 10.1128/AEM.71.10.5665-5677.2005
Burdige, D. J., Kline, S. W., and Chen, W. (2004). Fluorescent dissolved organic matter in marine sediment pore waters. Mar. Chem. 89, 289–311. doi: 10.1016/j.marchem.2004.02.015
Catalá, T. S., Reche, I., Fuentes-Lema, A., Romera-Castillo, C., Nieto-Cid, M., Ortega-Retuerta, E., et al. (2015). Turnover time of fluorescent dissolved organic matter in the dark global ocean. Nat. Commun. 6:5986. doi: 10.1038/ncomms6986
Chen, J., and Thornton, D. C. (2015). Transparent exopolymer particle production and aggregation by a marine planktonic diatom (Thalassiosira weissflogii) at different growth rates. J. Phycol. 51, 381–393. doi: 10.1111/jpy.12285
Chen, L., Gu, W., Xu, H., Yang, G., Shan, X., Chen, G., et al. (2018). Comparative genome analysis of Bacillus velezensis reveals a potential for degrading lignocellulosic biomass. 3 Biotech 8:253. doi: 10.1007/s13205-018-1270-7
Coble, P. G. (1996). Characterization of marine and terrestrial DOM in seawater using excitation-emission matrix spectroscopy. Mar. Chem. 51, 325–346. doi: 10.1016/0304-4203(95)00062-3
Coble, P. G., Del Castillo, C. E., and Avril, B. (1998). Distribution and optical properties of CDOM in the Arabian Sea during the 1995 Southwest Monsoon. Deep Sea Res. Part II 45, 2195–2223. doi: 10.1016/S0967-0645(98)00068-X
Falkowski, P. G., Katz, M. E., Knoll, A. H., Quigg, A., Raven, J. A., Schofield, O., et al. (2004). The evolution of modern eukaryotic phytoplankton. Science 305, 354–360. doi: 10.1126/science.1095964
Fandino, L. B., Riemann, L., Steward, G. F., Long, R. A., and Azam, F. (2001). Variations in bacterial community structure during a dinoflagellate bloom analyzed by DGGE and 16S rDNA sequencing. Aquat. Microb. Ecol. 23, 119–130. doi: 10.3354/ame023119
Fukuzaki, K., Imai, I., Fukushima, K., Ishii, K. I., Sawayama, S., and Yoshioka, T. (2014). Fluorescent characteristics of dissolved organic matter produced by bloom-forming coastal phytoplankton. J. Plank. Res. 36, 685–694. doi: 10.1093/plankt/fbu015
Goto, S., Tada, Y., Suzuki, K., and Yamashita, Y. (2017). Production and reutilization of fluorescent dissolved organic matter by a marine bacterial strain, Alteromonas macleodii. Front. Microbiol. 8:507. doi: 10.3389/fmicb.2017.00507
Goto, S., Tada, Y., Suzuki, K., and Yamashita, Y. (2020). Evaluation of the production of dissolved organic matter by three marine bacterial strains. Front. Microbiol. 11:584419. doi: 10.3389/fmicb.2020.584419
Grossart, H. P., Levold, F., Allgaier, M., Simon, M., and Brinkhoff, T. (2005). Marine diatom species harbour distinct bacterial communities. Environ. Microbiol. 7, 860–873. doi: 10.1111/j.1462-2920.2005.00759.x
Gruber, D. F., Simjouw, J. P., Seitzinger, S. P., and Taghon, G. L. (2006). Dynamics and characterization of refractory dissolved organic matter produced by a pure bacterial culture in an experimental predator-prey system. Appl. Environ. Microbiol. 72, 4184–4191. doi: 10.1128/AEM.02882-05
Hansell, D. A., Carlson, C. A., Repeta, D. J., and Schlitzer, R. (2009). Dissolved organic matter in the ocean: a controversy stimulates new insights. Oceanography 22, 202–211. doi: 10.5670/oceanog.2009.109
Hansell, D. A., Carlson, C. A., and Schlitzer, R. (2012). Net removal of major marine dissolved organic carbon fractions in the subsurface ocean. Global Biogeochem. Cycles 26:1. doi: 10.1029/2011GB004069
Horemans, B., Vandermaesen, J., Smolders, E., and Springael, D. (2013). Cooperative dissolved organic carbon assimilation by a linuron-degrading bacterial consortium. FEMS Microbiol. Ecol. 84, 35–46. doi: 10.1111/1574-6941.12036
Huguet, A., Vacher, L., Relexans, S., Saubusse, S., Froidefond, J. M., and Parlanti, E. (2009). Properties of fluorescent dissolved organic matter in the Gironde Estuary. Org. Geochem. 40, 706–719. doi: 10.1016/j.orggeochem.2009.03.002
Jasti, S., Sieracki, M. E., Poulton, N. J., Giewat, M. W., and Rooney-Varga, J. N. (2005). Phylogenetic diversity and specificity of bacteria closely associated with Alexandrium spp. and other phytoplankton. Appl. Environ. Microbiol. 71, 3483–3494. doi: 10.1128/AEM.71.7.3483-3494.2005
Jiao, N., Herndl, G. J., Hansell, D. A., Benner, R., Kattner, G., Wilhelm, S. W., et al. (2010). Microbial production of recalcitrant dissolved organic matter: long-term carbon storage in the global ocean. Nat. Rev. Microbiol. 8, 593–599. doi: 10.1038/nrmicro2386
Jørgensen, L., Stedmon, C. A., Kragh, T., Markager, S., Middelboe, M., and Søndergaard, M. (2011). Global trends in the fluorescence characteristics and distribution of marine dissolved organic matter. Mar. Chem. 126, 139–148. doi: 10.1016/j.marchem.2011.05.002
Kinsey, J. D., Corradino, G., Ziervogel, K., Schnetzer, A., and Osburn, C. L. (2018). Formation of chromophoric dissolved organic matter by bacterial degradation of phytoplankton-derived aggregates. Front. Mar. Sci. 4:430. doi: 10.3389/fmars.2017.00430
Lawaetz, A. J., and Stedmon, C. A. (2009). Fluorescence intensity calibration using the Raman scatter peak of water. Appl. Spectrosc. 63, 936–940. doi: 10.1366/000370209788964548
Lechtenfeld, O. J., Hertkorn, N., Shen, Y., Witt, M., and Benner, R. (2015). Marine sequestration of carbon in bacterial metabolites. Nat. Commun. 6, 1–8. doi: 10.1038/ncomms7711
Liu, Y., Kan, J., He, C., Shi, Q., Liu, Y. X., Fan, Z. C., et al. (2021a). Epiphytic bacteria are essential for the production and transformation of algae-derived carboxyl-rich alicyclic Molecule (CRAM)-like DOM. Microbiol. Spectr. 9, e1531–e1521. doi: 10.1128/Spectrum.01531-21
Liu, Y., Kan, J., Yang, J., Noman, M. A., and Sun, J. (2021b). Bacterial community composition and chromophoric dissolved organic matter differs with culture time of Skeletonema dohrnii. Diversity 13:150. doi: 10.3390/d13040150
Liu, Y., Wang, X., and Sun, J. (2022). Transformations of diatom-derived dissolved organic matter by bacillus pumilus under warming and acidification conditions. Front. Microbiol. 84:833670. doi: 10.3389/fmicb.2022.833670
Loginova, A. N., Thomsen, S., and Engel, A. (2016). Chromophoric and fluorescent dissolved organic matter in and above the oxygen minimum zone off Peru. J. Geophys. Res. Oceans 121, 7973–7990. doi: 10.1002/2016JC011906
Marie, D., Partensky, F., Jacquet, S., and Vaulot, D. (1997). Enumeration and cell cycle analysis of natural populations of marine picoplankton by flow cytometry using the nucleic acid stain SYBR Green I. Appl. Environ. Microbiol. 63, 186–193. doi: 10.1128/aem.63.1.186-193.1997
McIntyre, A. M., and Guéguen, C. (2013). Binding interactions of algal-derived dissolved organic matter with metal ions. Chemosphere 90, 620–626. doi: 10.1016/j.chemosphere.2012.08.057
McKnight, D., Boyer, E., Westerhoff, P. K., Doran, P. T., Kulbe, T., and Andersen, D. (2001). Spectrofluorometric characterization of dissolved organic matter for indication of precursor organic material and aromaticity. Limnol. Oceanogr. 46, 38–48. doi: 10.4319/lo.2001.46.1.0038
Middelboe, M., Søndergaard, M., Letarte, Y., and Borch, N. H. (1995). Attached and free-living bacteria: production and polymer hydrolysis during a diatom bloom. Microb. Ecol. 29, 231–248. doi: 10.1007/BF00164887
Moens, F., Weckx, S., and De Vuyst, L. (2016). Bifidobacterial inulin-type fructan degradation capacity determines cross-feeding interactions between bifidobacteria and Faecalibacterium prausnitzii. Int. J. Food Microbiol. 231, 76–85. doi: 10.1016/j.ijfoodmicro.2016.05.015
Mopper, K., Kieber, D. J., and Stubbins, A. (2015). “Marine photochemistry of organic matter: processes and impacts,” in Biogeochemistry of Marine Dissolved Organic Matter, eds C. A. Carlson and D. A. Hansell (Amsterdam: Elsevier), 389–450. doi: 10.1016/b978-0-12-405940-5.00008-x
Murphy, K. R., Stedmon, C. A., Waite, T. D., and Ruiz, G. M. (2008). Distinguishing between terrestrial and autochthonous organic matter sources in marine environments using fluorescence spectroscopy. Mar. Chem. 108, 40–58. doi: 10.1016/j.marchem.2007.10.003
Nelson, D. M., Tréguer, P., Brzezinski, M. A., Leynaert, A., and Quéguiner, B. (1995). Production and dissolution of biogenic silica in the ocean: revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Global Biogeochem. Cycles 9, 359–372. doi: 10.1029/95GB01070
Ohno, T. (2002). Fluorescence inner-filtering correction for determining the humification index of dissolved organic matter. Environ. Sci. Technol. 36, 742–746. doi: 10.1021/es0155276
Ortega-Retuerta, E., Duarte, C. M., and Reche, I. (2010). Significance of bacterial activity for the distribution and dynamics of transparent exopolymer particles in the Mediterranean Sea. Microb. Ecol. 59, 808–818. doi: 10.1007/s00248-010-9640-7
Pannard, A., Pedrono, J., Bormans, M., Briand, E., Claquin, P., and Lagadeuc, Y. (2016). Production of exopolymers (EPS) by cyanobacteria: impact on the carbon-to-nutrient ratio of the particulate organic matter. Aquat. Ecol. 50, 29–44. doi: 10.1007/s10452-015-9550-3
Para, J., Coble, P. G., Charrìère, B., Tedetti, M., Fontana, C., and Sempere, R. (2010). Fluorescence and absorption properties of chromophoric dissolved organic matter (CDOM) in coastal surface waters of the northwestern Mediterranean Sea, influence of the Rhône River. Biogeosciences 7, 4083–4103. doi: 10.5194/bg-7-4083-2010
Parlanti, E., Wörz, K., Geoffroy, L., and Lamotte, M. (2000). Dissolved organic matter fluorescence spectroscopy as a tool to estimate biological activity in a coastal zone submitted to anthropogenic inputs. Org. Geochem. 31, 1765–1781. doi: 10.1016/S0146-6380(00)00124-8
Passow, U. (2002). Transparent exopolymer particles (TEP) in aquatic environments. Prog. Oceanogr. 55, 287–333. doi: 10.1016/S0079-6611(02)00138-6
Passow, U., and Alldredge, A. L. (1994). Distribution, size and bacterial colonization of transparent exopolymer particles (TEP) in the ocean. Mar. Ecol. Prog. Ser. 113, 185–198. doi: 10.3354/meps113185
Passow, U., and Alldredge, A. L. (1995). A dye-binding assay for the spectrophotometric measurement of transparent exopolymer particles (TEP). Limnol. Oceanogr. 40, 1326–1335. doi: 10.4319/lo.1995.40.7.1326
Pereira, S., Zille, A., Micheletti, E., Moradas-Ferreira, P., De Philippis, R., and Tamagnini, P. (2009). Complexity of cyanobacterial exopolysaccharides: composition, structures, inducing factors and putative genes involved in their biosynthesis and assembly. FEMS Microbiol. Rev. 33, 917–941. doi: 10.1111/j.1574-6976.2009.00183.x
Romera-Castillo, C., Sarmento, H., Álvarez-Salgado, X. A., Gasol, J. M., and Marrasé, C. (2010). Production of chromophoric dissolved organic matter by marine phytoplankton. Limnol. Oceanogr. 55, 446–454. doi: 10.4319/lo.2010.55.1.0446
Rooney-Varga, J. N., Giewat, M. W., Savin, M. C., Sood, S., LeGresley, M., and Martin, J. L. (2005). Links between phytoplankton and bacterial community dynamics in a coastal marine environment. Microb. Ecol. 49, 163–175. doi: 10.1007/s00248-003-1057-0
Seymour, J. R., Amin, S. A., Raina, J. B., and Stocker, R. (2017). Zooming in on the phycosphere: the ecological interface for phytoplankton-bacteria relationships. Nat. Microbiol. 2, 1–12. doi: 10.1038/nmicrobiol.2017.65
Shimotori, K., Omori, Y., and Hama, T. (2009). Bacterial production of marine humuc-like fluorescent dissolver organic matter and its biogeochemical importance. Aquat. Microb. Ecol. 58, 55–66. doi: 10.3354/ame01350
Simon, M., Grossart, H. P., Schweitzer, B., and Ploug, H. (2002). Microbial ecology of organic aggregates in aquatic ecosystems. Aquat. Microb. Ecol. 28, 175–211. doi: 10.3354/ame028175
Stedmon, C. A., and Markager, S. (2005). Tracing the production and degradation of autochthonous fractions of dissolved organic matter by fluorescence analysis. Limnol. Oceanogr. 50, 1415–1426. doi: 10.4319/lo.2005.50.5.1415
Stedmon, C. A., Markager, S., and Bro, R. (2003). Tracing dissolved organic matter in aquatic environments using a new approach to fluorescence spectroscopy. Mar. Chem. 82, 239–254. doi: 10.1016/S0304-4203(03)00072-0
Tanoue, E., Nishiyama, S., Kamo, M., and Tsugita, A. (1995). Bacterial membranes: possible source of a major dissolved protein in seawater. Geochim. Cosmochim. Acta 59, 2643–2648. doi: 10.1016/0016-7037(95)00134-4
Thornton, D. C. (2014). Dissolved organic matter (DOM) release by phytoplankton in the contemporary and future ocean. Eur. J. Phycol. 49, 20–46. doi: 10.1080/09670262.2013.875596
Van Oostende, N., Moerdijk-Poortvliet, T. C., Boschker, H. T., Vyverman, W., and Sabbe, K. (2013). Release of dissolved carbohydrates by Emiliania huxleyi and formation of transparent exopolymer particles depend on algal life cycle and bacterial activity. Environ. Microbiol. 15, 1514–1531. doi: 10.1111/j.1462-2920.2012.02873.x
Wünsch, U. J., Murphy, K. R., and Stedmon, C. A. (2015). Fluorescence quantum yields of natural organic matter and organic compounds: implications for the fluorescence-based interpretation of organic matter composition. Front. Mar. Sci. 2:98. doi: 10.3389/fmars.2015.00098
Xu, H., and Jiang, H. (2013). UV-induced photochemical heterogeneity of dissolved and attached organic matter associated with cyanobacterial blooms in a eutrophic freshwater lake. Water Res. 47, 6506–6515.
Yamashita, Y., Panton, A., Mahaffey, C., and Jaffé, R. (2011). Assessing the spatial and temporal variability of dissolved organic matter in Liverpool Bay using excitation-emission matrix fluorescence and parallel factor analysis. Ocean Dyn. 61, 569–579. doi: 10.1007/s10236-010-0365-4
Keywords: organic matter (OM), dissolved OM (DOM), lysate OM (LOM), culturable epiphytic bacteria, Skeletonema dohrnii, dissolved organic carbon, transparent exopolymer particles
Citation: Liu Y, Wang X and Sun J (2022) Bacterial Transformation and Processing of Diatom-Derived Organic Matter: A Case Study for Skeletonema dohrnii. Front. Microbiol. 13:840564. doi: 10.3389/fmicb.2022.840564
Received: 21 December 2021; Accepted: 25 March 2022;
Published: 28 April 2022.
Edited by:
Wei Qin, University of Oklahoma, United StatesReviewed by:
Michael Thomas Montgomery, Texas A&M University-Corpus Christi, United StatesEric Fouilland, UMR 9190 Centre Pour la Biodiversité Marine, l’Exploitation et la Conservation (MARBEC), France
Yu Wang, Xiamen University, China
Copyright © 2022 Liu, Wang and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Sun, cGh5dG9wbGFua3RvbkAxNjMuY29t
 Yang Liu
Yang Liu Xueru Wang2
Xueru Wang2 Jun Sun
Jun Sun