- 1Department of Plant Pathology and Environmental Microbiology, Pennsylvania State University, University Park, PA, United States
- 2Department of Entomology and Plant Pathology, Auburn University, Auburn, AL, United States
Bacterial leaf spot (BLS) of lettuce caused by Xanthomonas hortorum pv. vitians (Xhv) was first described over 100 years ago and remains a significant threat to lettuce cultivation today. This study investigated the genetic relatedness of the Xhv strains and the possible genetic sources of this race-specific pathogenicity. Whole genome sequences of eighteen Xhv strains representing the three races, along with eight related Xanthomonas strains, were included in the analysis. A maximum likelihood phylogeny based on concatenated whole genome SNPs confirmed previous results describing two major lineages of Xhv strains. Gene clusters encoding secretion systems, secondary metabolites, and bacteriocins were assessed to identify putative virulence factors that distinguish the Xhv races. Genome sequences were mined for effector genes, which have been shown to be involved in race specificity in other systems. Two effectors identified in this study, xopAQ and the novel variant xopAF2, were revealed as possible mediators of a gene-for-gene interaction between Xhv race 1 and 3 strains and wild lettuce Lactuca serriola ARM-09-161-10-1. Transposase sequence identified downstream of xopAF2 and prophage sequence found nearby within Xhv race 1 and 3 insertion sequences suggest that this gene may have been acquired through phage-mediated gene transfer. No other factors were identified from these analyses that distinguish the Xhv races.
Introduction
Bacterial leaf spot (BLS) of lettuce is caused by Xanthomonas hortorum pv. vitians (Xhv) Morinière et al. (2020) (formally X. campestris pv. vitians), which was first described in 1918 following an outbreak in South Carolina lettuce fields (Brown, 1918). BLS is a sporadic, yet significant threat to lettuce production worldwide, and several subsequent outbreaks in the last century have prompted research to better understand this plant-pathogen interaction. Investigation into Xhv strain diversity has been vital to the development and application of effective strategies for the management of BLS. In 2015, Bull et al. (2015) demonstrated that some, but not all, of the 120 Xhv strains evaluated resulted in a hypersensitive response (HR) to limit the spread of infection upon inoculation into Lactuca sativa cvs. Little Gem, Pavane, and La Brillante, and that these strains did not cause HR in the other lettuce cultivars that were tested. This subset of Xhv strains was designated race 1 to correspond to the HR elicited in Little Gem, Pavane, and La Brillante and the hypothesized interaction with the resistance gene (R-gene) Xar1 mapped to chromosome 2 in these cultivars (Hayes et al., 2014). Another identified source of resistance to Xhv race 1 strains isolated from Florida lettuce fields was the Xcvr R-gene from L. serriola PI358001-1 (Wang et al., 2016). The presence of these R-genes led to the hypothesis that Xhv race 1 strains engage in a gene-for-gene interaction with these lettuce cultivars. Additional races of the pathogen were designated among those Xhv strains that did not result in HR in the Xar1- or Xcvr-encoding cultivars, but instead triggered HR in either L. serriola PI491114 (designated race 2) or L. serriola ARM-09-161-10-1 (designated race 3). Recent HR screening also suggests that Xhv race 1 strains might also induce HR in ARM-09-161-10-1 (Rosenthal et al., unpublished).
Bull et al. (2016) found that the diversity they observed in the resistance phenotypes of the lettuce cultivars corresponded to the genetic diversity of the pathogen. They demonstrated the presence of five distinct sequetypes among 120 Xhv strains using a multi-locus sequence analysis (MLSA) scheme originally designed for distinguishing Xanthomonas spp. (Maiden et al., 1998; Young et al., 2008). The results of this study corroborated previous work on Xhv diversity (Sahin et al., 2003; Fayette et al., 2016), and they found that the strains that induce HR upon injection into the Xar1- or Xcvr-encoding cultivars all belonged to sequetypes B, D, or E. The strains which triggered HR in PI491114 all belonged to sequetype A, and those that triggered HR in ARM-09-161-10-1 all belonged to sequetype C. These results supported the hypothesis that genotypic differences underly the differences in resistance phenotype. The relationship between sequetype and race also supported the finding that Xhv populations are predominantly clonal, though with some sequence variation due to recombination (Fayette et al., 2016).
Studies of plant-pathogen interactions have suggested a constant cycle in which plants and their pathogens evolve to outperform each other, in defense strategy and pathogenicity, respectively (Dangl and Jones, 2001; Zipfel and Felix, 2005). Pathogen presence can trigger basal plant defenses to prevent further colonization, which can be then interrupted by the delivery of bacterial effector proteins through the type three-secretion system. In turn, effector activity can be thwarted by plant host recognition of those effectors and activation of resistance (R-)genes, such as those that activate HR to kill off invaded tissues and limit the spread of infection. The presence of the R-genes Xar1 and Xcvr in lettuce cultivars capable of HR exclusively to Xhv race 1 (Hayes et al., 2014; Wang et al., 2016) suggested the possibility that Xhv race 1 might produce an effector that is recognized by those R-genes. Other gene-for-gene interactions might also be at play for Xhv races 2 and 3 and their respective HR-inducible lettuce cultivars, though R-genes have not yet been demonstrated for PI491114 and ARM-09-161-10-1.
In other bacterial plant pathogens, the proximity of many type III effectors (T3Es) to mobile genetic elements suggests that they were acquired or lost through horizontal gene transfer with other bacteria in their local microbiome (McCann and Guttman, 2008; Merda et al., 2017). Following effector gene acquisition or loss, the selective environment created by the host’s defenses may have caused the new genotype to be maintained and to be host-specific (Sarkar et al., 2006; Hajri et al., 2009) or even race-specific. Such a relationship between race-specific genotype and a tomato cultivar has already been demonstrated for X. euvesicatoria pv. perforans (Astua-Monge et al., 2000).
Other gene products, such as secondary metabolites and bacteriocins, have been shown to play a role in virulence. The Xanthomonas pigment Xanthomonadin and siderophore Xanthoferrin contribute to bacterial fitness by protecting bacterial cells from UV damage and sequestering iron in low-iron conditions, respectively; functional knockouts of these gene clusters resulted in reduced virulence according to Rajagopal et al. (1997) and Pandey et al. (2017). A variety of bacteriocins identified in Xanthomonas spp. enhance bacterial competitiveness through antimicrobial activity (Holtsmark et al., 2008). Although a role in host-specific pathogenicity for these components has not yet been demonstrated, it is possible that race-specific inclusion of these genes may be used to further distinguish the Xhv races.
This study was designed to investigate variation in effector repertoires between the Xhv races as a possible genetic source for race-specific HR elicitation. Several important virulence factors were also studied to look for genotypic differences that could be used to distinguish the Xhv races. Eighteen geographically diverse isolates of Xhv and seven related plant pathogenic Xanthomonas strains were sequenced and assembled. A genome assembly for X. hortorum pv. carotae M081 was also retrieved and included for comparison. Genome statistics and predicted features, including plasmids and secretions systems, were determined for each strain. A maximum likelihood phylogeny based on SNP data corroborated the taxonomic relationships predicted by MLSA (Bull et al., 2016; Fayette et al., 2016). Mining the genomes for effector sequences and other potential virulence factors revealed insights into the race-specificity of the BLS pathogen. A preliminary report of this data has been given (Rosenthal et al., 2018).
Methods
Bacterial Strains and Culturing Methods
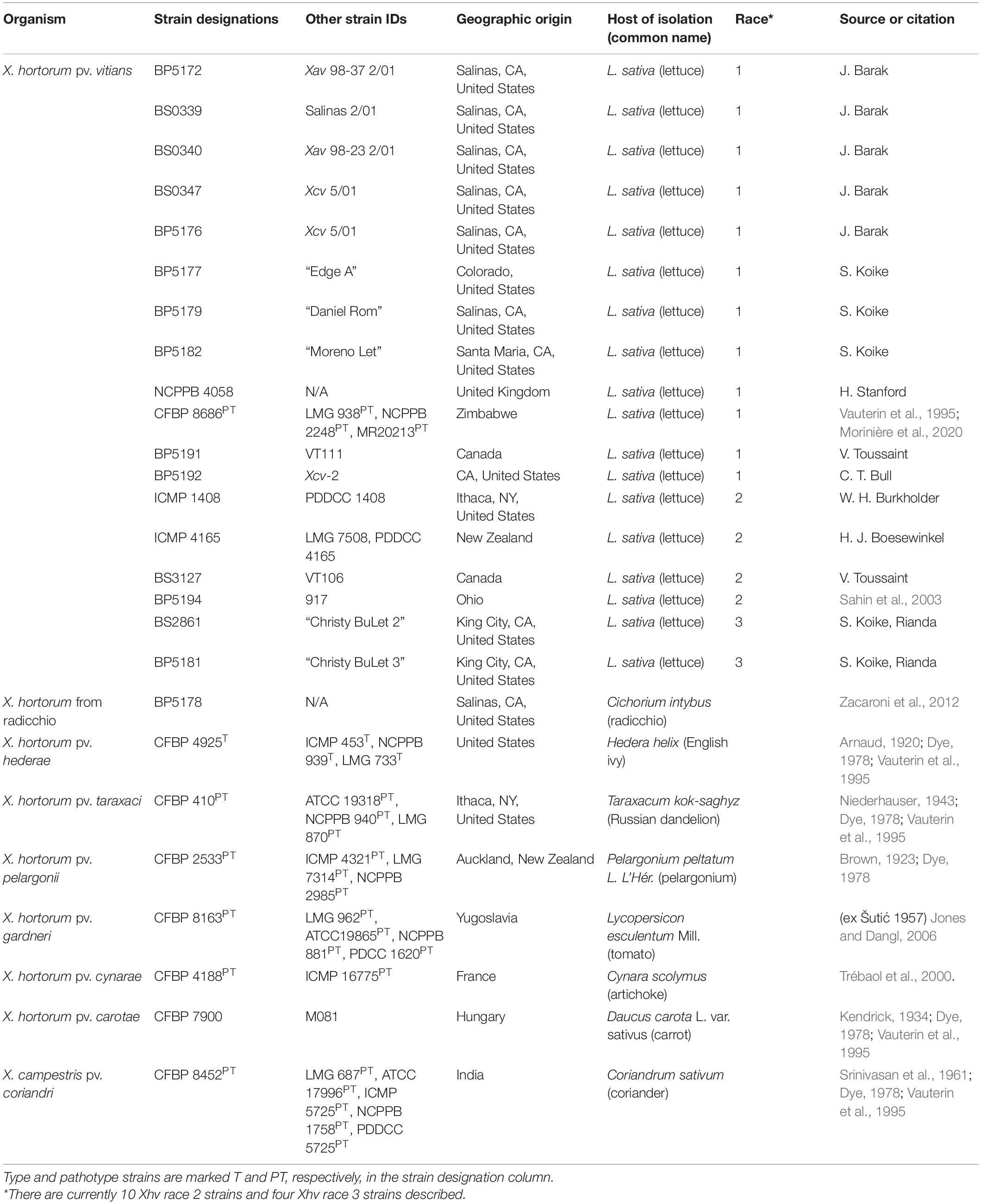
All strains used in this study and their sources are shown in Table 1. Eighteen strains of Xhv were selected as representatives of the three pathogenic races designated in previous studies: race 1 (12 strains), race 2 (4 strains), and race 3 (2 strains). The type and pathotype strains of X. hortorum pv. hederae, X. hortorum pv. taraxaci, X. hortorum pv. pelargonii, X. hortorum pv. gardneri, and X. hortorum pv. cynarae were included for comparison to the Xhv strains, as well as one Xanthomonas hortorum strain from radicchio. X. campestris pv. coriandri was included as an outgroup. Bacteria were routinely cultured on nutrient agar (NA) and in nutrient broth (NB), both at room temperature (20–28°C).
DNA Extraction and Whole Genome Sequencing
DNA extraction was performed using the Qiagen DNeasy UltraClean Microbial Kit (cat. no. 12224; Valencia, CA, United States) according to the manufacturer’s instructions, with the following change: five replicates were completed for each strain and after eluting each first replicate, that elute was used to elute the next replicate, and so on. This resulted in >2.0 μg of total DNA per strain, as measured using the Thermo Fischer Qubit Fluorometer 3.0 and the Invitrogen dsDNA Broad Range Assay Kit (cat nos. Q33216 and Q32850; Waltham, MA, United States). Library preparation and 250 × 250 paired-end whole genome sequencing was performed at the Pennsylvania State University Genomics Facility using the TruSeq DNA PCR Free Library Construction Kit and the Illumina MiSeq System (cat. nos. FC-121-3001 and SY-410-1003; San Diego, CA, United States).
Quality Control and Whole Genome and Plasmid Assembly
Trimmomatic PE v.0.39 (Bolger et al., 2014) was used to remove Illumina adapters and poor-quality sequence, defined as four consecutive bases with phred scores lower than 20 and DNA fragments shorter than 50 bases. Sequence quality reports were generated using FastQC v0.11.9 (Andrews, 2010) before and after using Trimmomatic PE to verify an improvement in sequence quality which would improve the accuracy of the genome assembly. This procedure was completed in a loop using trimming.sh, spades.sh, and spades_loop.txt (Supplementary Data 1–3).
Trimmed reads were assembled de novo using SPAdes v3.12.0 (Bankevich et al., 2012) and Quast v5.0.0 (Gurevich et al., 2013) was used to provide descriptive statistics about the assemblies (Table 2). Plasmid counts were predicted using the Recycler pipeline (Rozov et al., 2017) and plasmid sequences were predicted using plasmidSPAdes (Antipov et al., 2019); using both programs for plasmid analysis has been recommended by the creators of plasmidSPAdes due to the differing predictive strengths of each tool. Raw reads and whole genome sequence assemblies are available at BioProject PRJNA790934. Completeness of the whole genome assemblies was assessed using BUSCO v5.2.2 (lineage = xanthomonadales_odb10 and mode = genome; Manni et al., 2021) to compare these assemblies against several publicly available genome assemblies for related Xanthomonas strains obtained from NCBI’s genome database, including X. campestris pv. campestris (ATCC 33913T), X. populi (CFBP 1817T), X. arboricola pv. juglandis (CFBP 2528T), X. hortorum pv. pelargonii (CFBP 2533PT), X. hortorum pv. taraxaci (CFBP 410PT), X. hortorum pv. cynarae (CFBP 4188PT), X. hortorum pv. carotae (CFBP 7900), X. hortorum pv. gardneri (CFBP 8163PT), X. hortorum pv. vitians (CFBP 8686PT), X. campestris pv. coriandri (ICMP 5725PT), X. fragariae (PD 885T), and X. hortorum pv. hederae (WHRI 7744T).
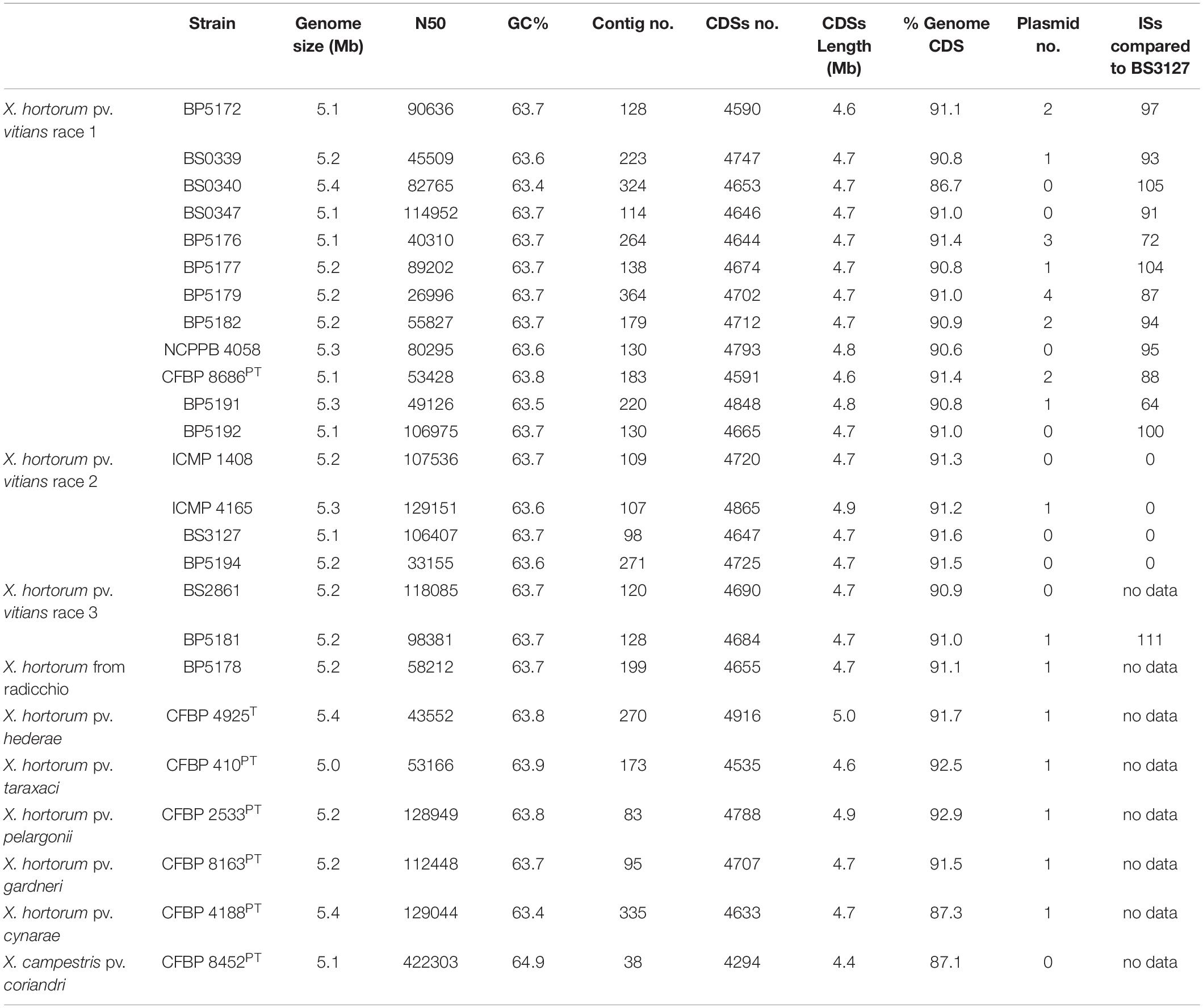
Table 2. Genome characteristics for Xanthomonas hortorum pv. vitians strains and related type and pathotype strains.
Variant-Based Phylogeny Using Whole Genome Sequences
To investigate the genetic relatedness of the Xhv strains and related strains used in this study, a phylogenetic tree was constructed based on a core alignment of single nucleotide polymorphisms (SNPs). The whole genome assembly of Xanthomonas hortorum pv. carotae (Xhc) M081 was downloaded from NCBI (GenBank assembly accession GCA_000505565.1) to be used for comparison. The snippy-multi pipeline v4.3.6 (Seemann, 2015) was used to map trimmed Illumina sequencing reads to the Xhc M081 reference and to identify and align a set of core SNPs. In the resulting core alignment, all non-[AGTCN-] characters were replaced using snippy-clean_full_aln and then loci with high levels of base substitution, indicating possible recombination, were removed using run_gubbins.py v2.4.1 (Croucher et al., 2015). SNPs were extracted from the filtered multi-fasta alignment using SNP-sites with the option “-c” to only output AGTC into a final alignment file. This file was imported into CLC Genomics Workbench and General Time Reversible with rate variation (four categories) and estimated topology was selected as the appropriate substitution model following model testing with the neighbor-joining clustering method. This model was used to produce a maximum-likelihood phylogeny with 1,000 bootstraps and branch lengths that represent the expected number of nucleotide substitutions per sequence site.
Effector Gene Mining
BLAST databases were constructed from each whole genome assembly scaffold file or plasmid scaffold file. A FASTA file containing known bacterial effector protein sequences, including those previously published for plant pathogenic Xanthomonas and Pseudomonas species, was provided (White et al., 2009). Translated nucleotide BLAST was used to align each entry of this effector sequence catalog to the whole genome sequence databases. Matches were filtered to include those with greater than 60% identity, greater than 40% query coverage per high scoring sequence pair, and less than 0.0001 expect value. Other potential matches were found using a filter of greater than 45% identity, greater than 40% qcovhsp, and less than 0.00001 e-value; this allowed for inclusion of some other effectors which may be present but with moderate sequence variation compared to those in our database. The same searches were performed using the predicted plasmid sequences to determine which effectors may be carried on plasmids. The filtered matches constitute the hypothesized effector repertoires of that strain. Mining for these effector repertoires was completed using the script effectorgene_mining.sh (Supplementary Data 4).
Gene Alignments and Trees
Nucleotide sequences for two genes of interest, xopAQ and xopAF, were extracted from our assembly files using the program samtools v1.9 faidx and the gene positions found from the effector gene mining procedure (Li et al., 2009). These sequences were used as input for the ORFfinder program available online at https://www.ncbi.nlm.nih.gov/orffinder/, and the longest ORFs were selected for protein alignment using MEGA11: Molecular Evolutionary Genetics Analysis (Tamura et al., 2021), along with the original protein sequences published for the homologs XopAQ, HopAQ1, Rip6, and Rip11 (Guttman et al., 2002; Mukaihara et al., 2010; Potnis et al., 2011); or XopAF (AvrXv3) and HopAF1 (Astua-Monge et al., 2000; Petnicki-Ocwieja et al., 2002). Overhanging sequences were trimmed. Maximum-likelihood phylogenies were generated from these alignments to show the relatedness of the extracted genes. The Jones-Taylor-Thornton (JTT) amino acid substitution model, an assumption of uniform substitution rates among sites, and the nearest-neighbor-interchange (NNI) method were used to generate the phylogenies. The phylogenies were tested using 1,000 bootstraps and branch lengths represented the number of amino acid substitutions per site.
Mobile Genetic Elements
Mobile genetic elements were assessed for their effector content, especially for xopAQ and xopAF presence, to find evidence of recent acquisition by a common ancestor of Xhv race 1 and 3 strains. Large insertion sequences in Xhv race 1 and 3 strains were identified by aligning their whole genome sequencing reads to the assembled sequence for Xhv race 2 strain BS3127, identifying the sequence regions present in their genomes that were not present in BS3127, and aligning the regions flanking the putative insertion to their whole genome assemblies to find the complete insertion sequences. These steps were completed following the MGEfinder pipeline (Durrant et al., 2020). Xhv race 3 strain BS2861 was excluded from this analysis because its inclusion caused the program to fail. The identified insertion sequences were then mined for effector genes using the method described above, as well as for transposase sequences included in the ISEScan package (Xie and Tang, 2017) using the same method and phage elements using PHASTER’s online platform (Arndt et al., 2016) with the default settings.
xopAF and xopAQ Gene Regions in Xhv Race 2 Strains
Whether there were other known genes present at the sites in Xhv race 2 strains where xopAF and xopAQ were absent was analyzed. Sequences flanking the xopAF and xopAQ genes from BP5172 (Xhv race 1) were extracted using samtools faidx, including 1,000 nucleotides on either side. Xhv race 2 strains were then parsed for these sequences using BLAST to identify the same regions of their genome sequence. Alignments of these regions were completed for a subset of Xhv race 1 and 2 strains, and both the Xhv race 3 strains. These alignments revealed the sequence within the Xhv race 2 genomes that were present instead of the xopAF and xopAQ genes. These sequences were copied into NCBI’s ORFfinder to look for coding sequence and NCBI’s Conserved Domain Search to determine the possible function of that coding sequence. One protein-coding gene of interest which was present in all Xhv race 2 strains instead of xopAF was used as a query in a BLAST search of Xhv race 1 and 3 genomes to determine whether this gene was specific to Xhv race 2 strains or present in all Xhv strains tested.
Secretion System Gene Mining
Genes associated with or encoding secretion system components were compiled into a database by downloading their sequences from representative species from NCBI (Pena et al., in preparation). Nucleotide BLAST using the script SSgene_mining.sh (Supplementary Data 5) revealed genes from our database which aligned to the whole genome sequence assemblies. Plasmid sequences were also queried for secretion system genes. Matches were filtered to retain only those that had 65% identity and 65% query coverage per high scoring sequence pair or higher and an e-value less than 0.00001. To confirm these results, additional global alignments of extracted secretion system gene clusters were completed in CLC Genomics Workbench, and aligned genes were again filtered for 65% identity. Reported results represent the sum of these two methods.
Secondary Metabolite/Bacteriocin Gene Mining
Secondary metabolite gene clusters were predicted for each strain using the antiSMASH v5.0 online server using the “relaxed strictness” parameter and the “KnownClusterBlast” feature (Blin et al., 2019). Individual genes within these clusters were manually filtered to include only hits with greater than 60% identity and 40% query coverage.
To verify the antiSMASH results, each whole genome assembly was also uploaded to the BAGEL4 webserver and queried against a database of known bacteriocin genes and motifs using default settings (van Heel et al., 2018). Areas of interest that were scored as significant by the program (where the sum of weight factors is greater than 175) are reported here along with their identified class.
Results and Discussion
Comparative Genomic Analysis
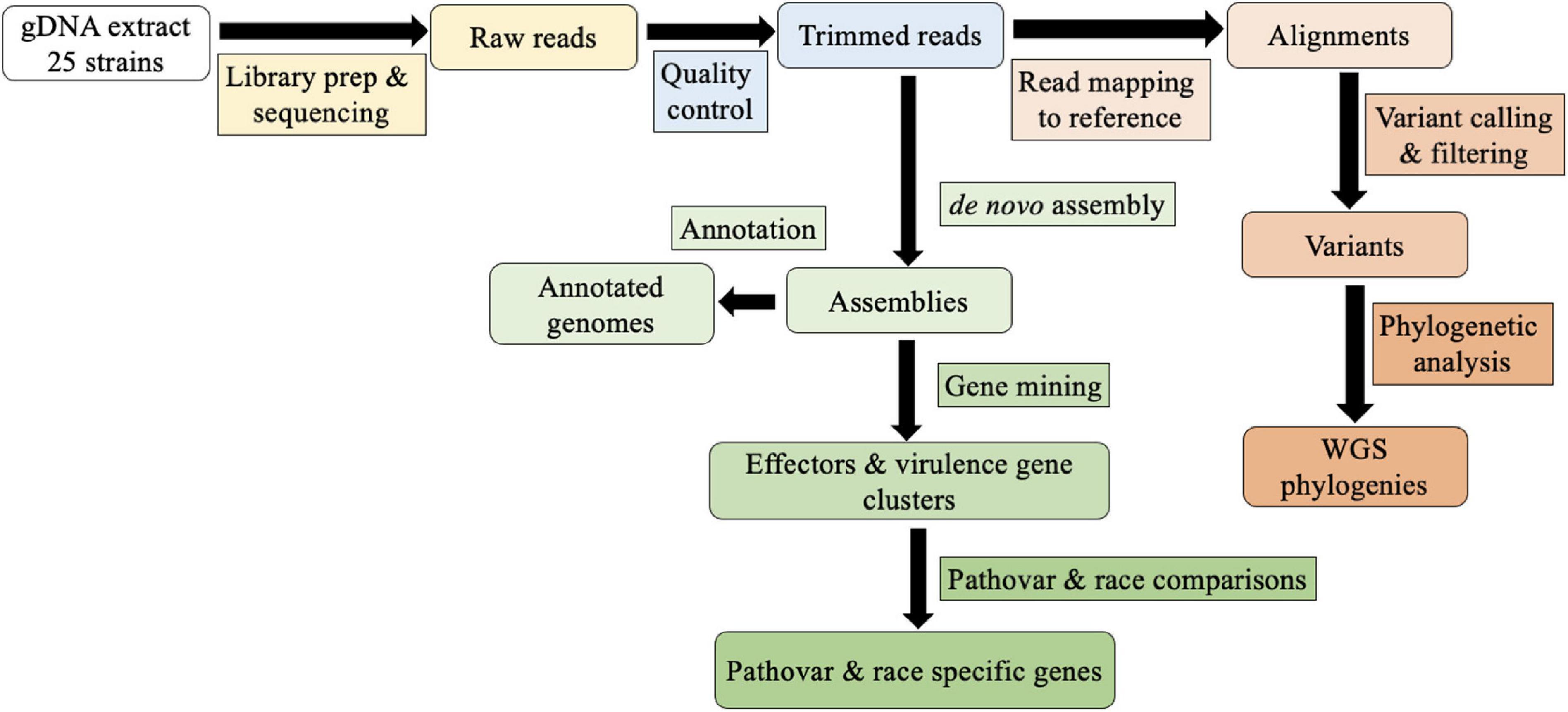
Eighteen assembled genomes representing the three pathogenic races of Xhv, as well as those of seven related Xanthomonas strains, are available at BioProject PRJNA790934. The bioinformatics pipeline used in this study is shown in Figure 1. Genome-associated statistics are reported in Table 2. The average Xhv genome size was 5.2 Mbp and in this study the number of contigs ranged from 98 for the Xhv race 2 strain BS3127 to 364 for the Xhv race 1 strain BP5179, with an average of 180 contigs per strain. The N50 for Xhv strains varied from 27,000 to 129,000 bp. Whole genome annotation showed that each Xhv strain contained about 4,700 coding sequences (CDS), with an average total length of CDS per genome being 4.7 Mb and accounting for 91% of the total genome size. The number of plasmids predicted for each Xhv strain varied from zero to four, with no pattern by race observed.
Compared to the Xhv strains, the related Xanthomonas strains had approximately the same genome sizes, fewer gaps in the sequence with an average of 170 contigs per strain, and similar variation in N50. Annotations for these reference strains revealed similar statistics for coding sequences, with 4,650 CDSs at an average length of 4.7 Mb and representing 91% of the total genome size. A single plasmid is predicted for each of the reference strains except for the pathotype of X. campestris pv. coriandri, which did not have any predicted plasmid sequence. BUSCO analysis for measuring the completeness of all of these sequence assemblies showed that they are complete and similar to other published Xanthomonas genomes (Figure 2).
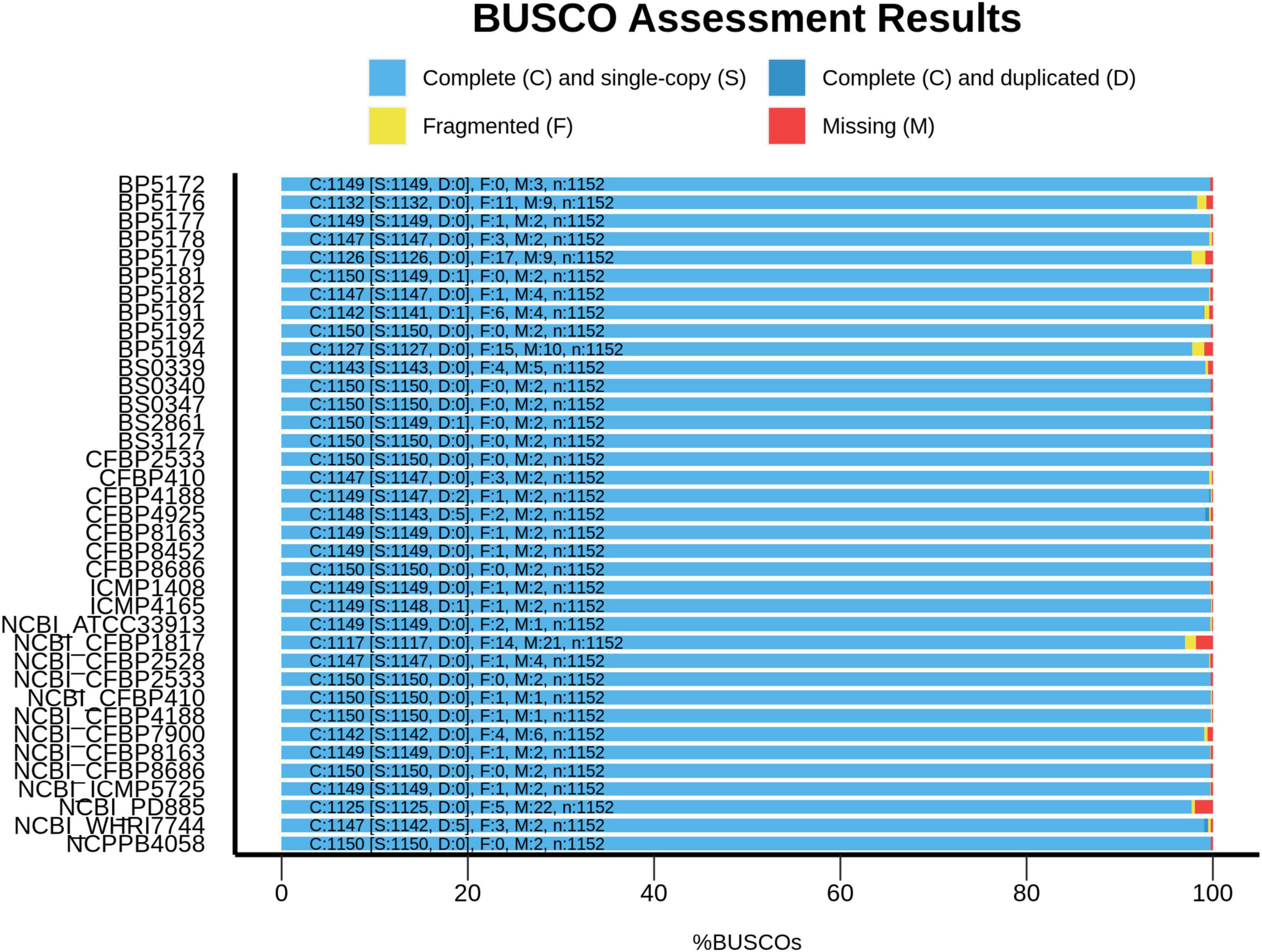
Figure 2. Genome assembly completeness assessment for Xhv and related strains. The completeness of the genome assemblies produced in this study was evaluated using the benchmarking universal single copy orthologs method (BUSCO; Manni et al., 2021). The proportions of complete and single-copy orthologs in these assemblies were compared to those of previously published Xanthomonas assemblies retrieved from NCBI’s genome database, including X. campestris pv. campestris (ATCC 33913T; GenBank assembly accession GCA_000007145.1), X. populi (CFBP 1817T; GCA_002940065.1), X. arboricola pv. juglandis (CFBP 2528T; GCA_001013475.1), X. hortorum pv. pelargonii (CFBP 2533PT; GCA_012922215.1), X. hortorum pv. taraxaci (CFBP 410PT; GCA_012922225.1), X. hortorum pv. cynarae (CFBP 4188PT; GCA_002939985.1), X. hortorum pv. carotae (CFBP 7900; GCA_000505565.1), X. hortorum pv. gardneri (CFBP 8163PT; GCA_012922265.1), X. hortorum pv. vitians (CFBP 8686PT; GCA_012922135.1), X. campestris pv. coriandri (ICMP 5725PT; GCA_019201305.1), X. fragariae (PD 885T; GCA_900183975.1), and X. hortorum pv. hederae (WHRI 7744T; GCA_003064105.1).
Phylogenetic Analysis
The maximum likelihood phylogeny using whole genome SNP data showed a distinct clade to which all the Xhv strains belong (Figure 3). Within this larger clade there were three groups that corresponded to the three Xhv races, originally distinguished by their differing host reactions on various lettuce plant introduction lines (Bull et al., 2016). Xhv race 1 and 2 appear to have diverged from a common ancestor that itself diverged from strains of X. hortorum pv. gardneri and X. hortorum pv. cynarae, which are pathogenic on tomato and artichoke, respectively. The Xhv race 3 strains appeared to be more recently evolved, stemming from Xhv race 1 strains. This corroborated the finding of Fayette et al. (2016) in which Xhv race 3 strains made up a single MLST. The relationship between the Xhv strains and the other strains of Xanthomonas hortorum corroborated results presented in Morinière et al. (2020), which were based on a predicted core proteome rather than the whole genome SNP data presented here.
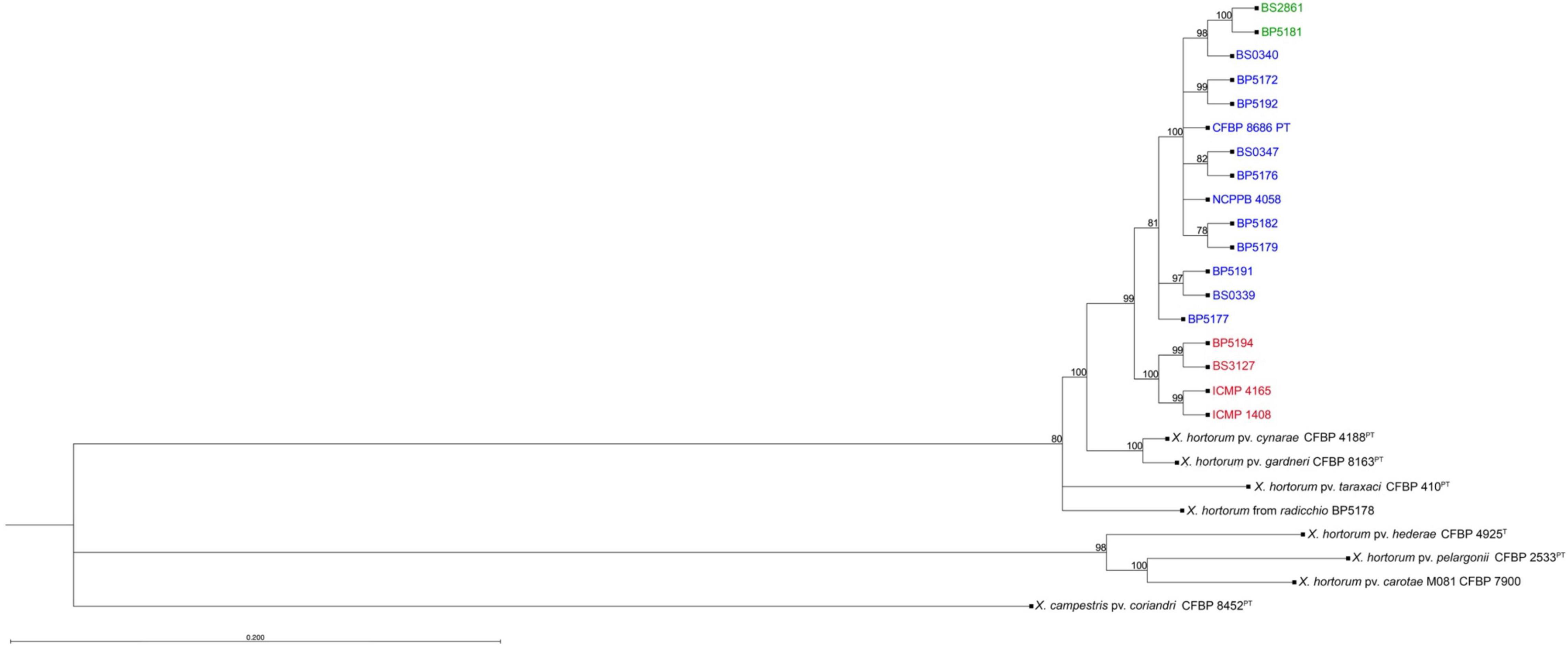
Figure 3. Phylogenetic tree constructed from whole genome SNP data. Maximum-likelihood phylogeny of Xanthomonas hortorum pv. vitians strains of race 1 (blue), race 2 (red), and race 3 (green) and related Xanthomonas type and pathotype strains (black) used in this study. SNP data was extracted from trimmed Illumina sequence reads and core aligned using the snippy-multi pipeline (Seemann, 2015) and the tree was built from sequence alignment using CLC Genomics Workbench. This phylogeny was constructed using the neighbor-joining clustering method and the general time-reversible substitution method with rate variation (4 categories) and estimated topology. Bootstrap values are shown above branches and branch length represents the expected number of nucleotide substitutions per site. Branches shorter than 0.0100 are shown as having a length of 0.0100.
Effector Repertoire Variations
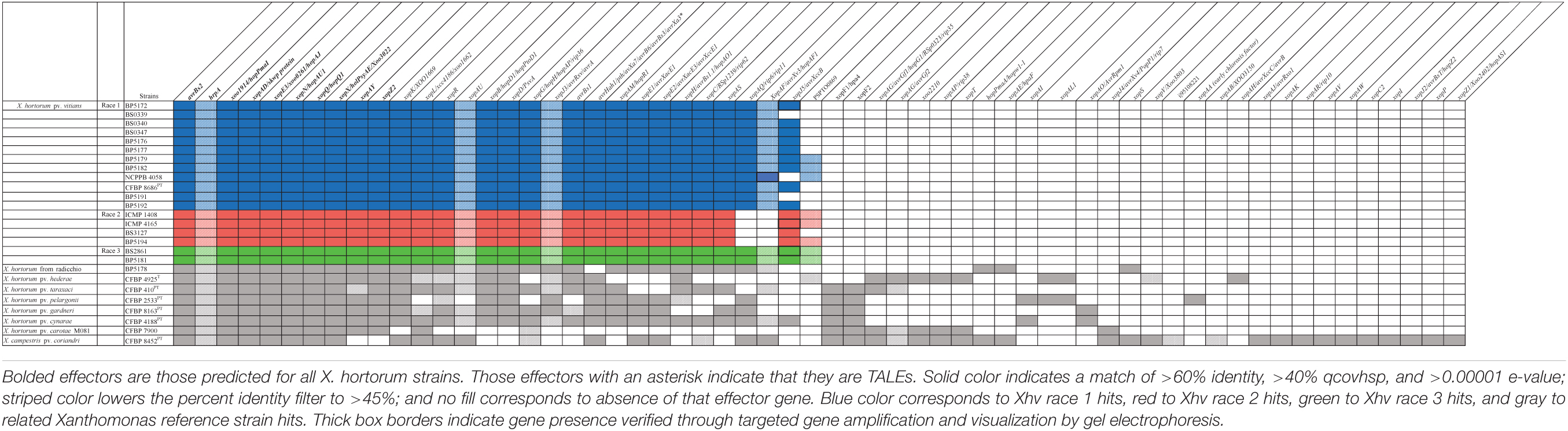
The focus of this study was to identify race-specific avirulence genes among the Xhv strains that could explain the race-specific HR phenotypes observed upon inoculation into various lettuce-cultivars. Of the total sixty-seven effector homologs predicted, fifteen were identified in all X. hortorum strain sequences tested here and constitute the core effector repertoire of the species (Table 3). Regarding the Xhv strains, thirty-four of the predicted effector homologs were present in all Xhv strains tested, representing the core repertoire of the pathovar. Homologs for the transcription-activator like effector (TALE) avrHah1 were identified in all Xhv strains tested here, as well as in the X. hortorum pv. hederae, X. hortorum pv. taraxaci, X. hortorum pv. gardneri, X. hortorum pv. cynarae, and X. campestris pv. coriandri sequences evaluated. This class of effectors is significant for its ability to enter the host cell nucleus and alter transcription in such a way that makes the host susceptible to disease (Bogdanove et al., 2010). Although race-specific variation in avrHah1 sequence was not observed in our strains, this could be a limitation in the ability of our short-read sequencing method to resolve the repeat sequences characteristic of TALEs. Further research is needed using a long-read sequencing platform that can demonstrate whether there are race-specific differences as well as pathovar-specific differences in the avrHah1 gene.
Two additional effectors, xopAF and xopAQ, were predicted in the genomes of all Xhv strains belonging to races 1 and 3 but were absent from those of Xhv race 2 strains. The presence of xopAF was also predicted in the genomes of X. hortorum pv. taraxaci, X. hortorum pv. cynarae, and X. campestris pv. coriandri but not in the genomes of X. hortorum pv. pelargonii, X. hortorum pv. gardneri, X. hortorum pv. carotae M081, or the X. hortorum strain from radicchio. Further, the presence of xopAQ was predicted in the genomes of X. hortorum from radicchio, X. hortorum pv. pelargonii, X. hortorum pv. cynarae, and X. campestris pv. coriandri but not of X. hortorum pv. hederae, X. hortorum pv. taraxaci, or X. hortorum pv. carotae M081.
The gene xopAF was first identified as avrXv3 in X. euvesicatoria pv. perforans strains of tomato race 3 (T3) and was demonstrated to be responsible for race-specific HR elicitation in tomato cultivar Hawaii 7981 (Astua-Monge et al., 2000). Acting intracellularly, xopAF causes the upregulation of defense related genes (Balaji et al., 2007). Since its original identification, homologs have been identified in strains of X. citri pv. citri (Xcc) isolated only from Mexican lime and X. vasicola pv. vasculorum (Xvv) isolated only from sugarcane. Experiments with xopAF mutants showed that the gene was not responsible for limiting the host range of Xcc, but did contribute to virulence (Jalan et al., 2013). Cultivar-specificity for Xvv strains does not appear to have been tested.
The other gene, xopAQ, was first identified as rip6/rip11 in a screen for Ralstonia solanacearum proteins that are injected into host plant cells (Mukaihara et al., 2010). The Xanthomonas homolog xopAQ was first described in X. hortorum pv. gardneri and hypothesized to be part of a group of pathotype-specific genes responsible for its aggressive disease compatibility on tomato and pepper (Potnis et al., 2011). Homologs have since been found in X. arboricola, X. citri, and X. euvesicatoria and have a proposed function in lipid modification that may be involved in host recognition and hypersensitive response (Thieme et al., 2007; Barak et al., 2016).
Five of the effectors predicted for all Xhv strains were found within their plasmid sequences: avrBs1, avrHah1, xopE1, xopE2, and xopH (Supplementary Data 6). Several other effector genes were predicted among various Xhv strains, including avrBs2, xopL, xopJ5, xopAQ, xopB, xopC, xopD, xopG, and xopZ2, but with no pattern that could underly the race-specific differences in HR elicitation in lettuce cultivars. Each of the reference strains had a different predicted repertoire of plasmid-borne effectors, and there were no plasmid-borne effectors that were predicted for all X. hortorum pathovars. The genes avrBs1, avrHah1, xopH, xopAQ, avrBsT (predicted only in X. campestris pv. coriandri), and xopAO (predicted only in X. hortorum pv. gardneri and X. hortorum pv. cynarae) are known to be plasmid-borne in other Xanthomonas species (Swanson et al., 1988; Minsavage et al., 1990; Schornack et al., 2008; Potnis et al., 2011; Newberry et al., 2019; Roach et al., 2019), and so were likely acquired through plasmid transfer in the strains tested here. All other predicted plasmid-borne effectors (see Supplementary Data 6 for the complete list) are not yet demonstrated to be plasmid-borne in other species but may still have been acquired through plasmid transfer to a strain ancestral to X. hortorum pv. vitians. Further study with completed Xhv genomes produced using long-read sequencing may be able to address these questions regarding the acquisition of these effectors. All effectors not predicted on plasmid sequence here are assumed to be chromosomal genes.
From these data we hypothesize that the differences in effector repertoire composition between Xhv races 1 and 3, and Xhv race 2, is responsible for the race-specific elicitation of HR in lettuce. It is possible that xopAQ and/or xopAF interact with an R-gene in ARM-09-161-10-1 to trigger resistance to Xhv race 1 and 3 strains, and that the absence of these genes in Xhv race 2 strains allows them to go undetected. Further work is needed to confirm the avirulence function of these xopAQ and xopAF homologs in ARM-09-161-10-1, and that the resistance in these cultivars is governed by a single dominant R gene.
XopAQ Homolog and New XopAF2 Variant
Alignment of the XopAQ protein sequences revealed that they were present among X. hortorum strains (Table 3), it was identical to XopAQ protein sequence from X. hortorum pv. gardneri. In the gene tree, all XopAQ protein sequences from X. hortorum strains clustered together and were separated from the HopAQ1 protein sequences from Pst DC3000 (Figure 4A).
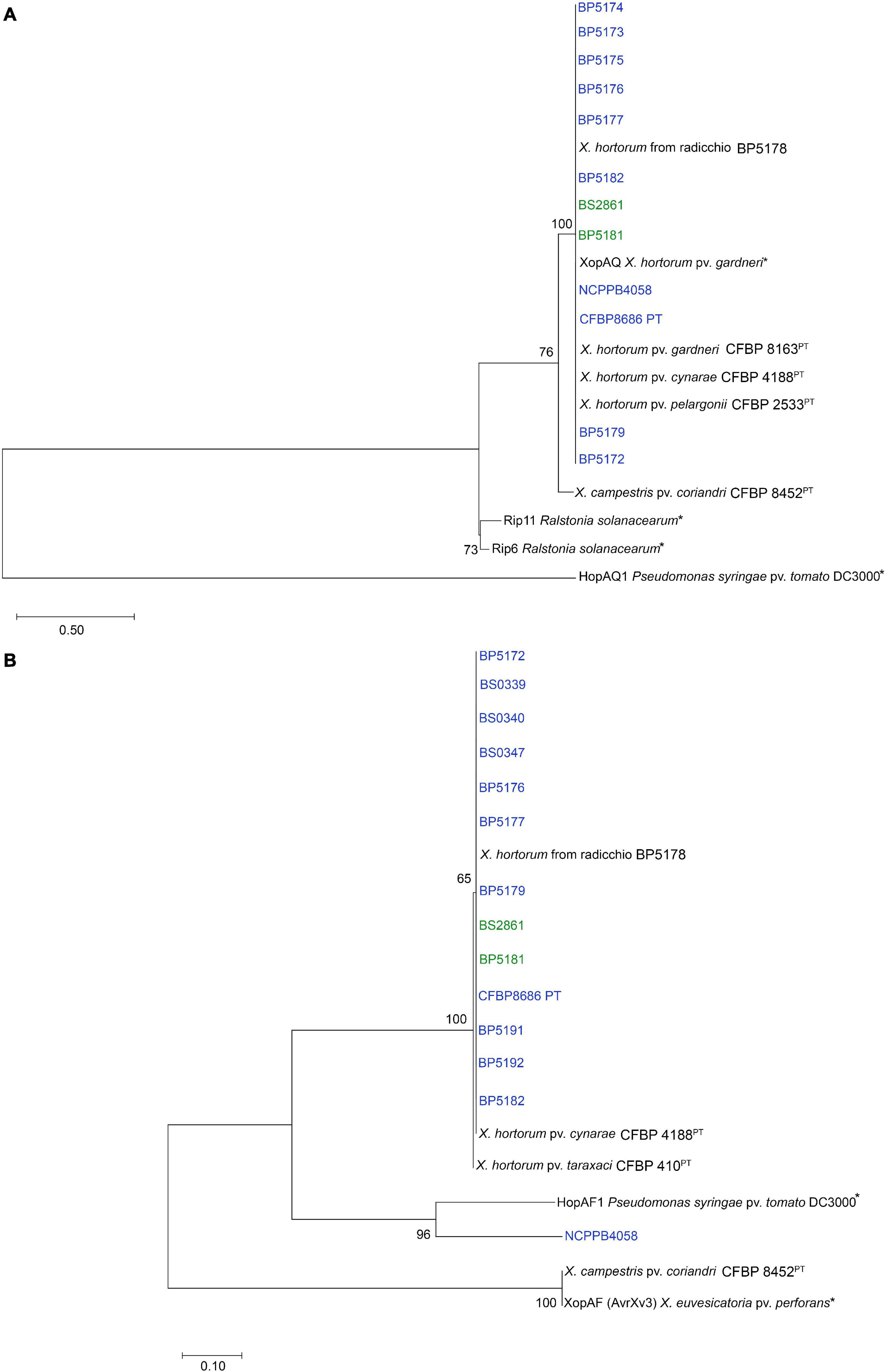
Figure 4. Phylogenetic trees constructed from (A) XopAQ amino acid alignment and (B) XopAF amino acid alignment. Maximum-likelihood phylogenies were constructed for the two genes present in Xhv race 1 and race 3 strains (blue and green) but missing from Xhv race 2 strains. Nucleotide sequences for these genes were extracted from the whole genome sequence assemblies and converted to amino acid sequences using OrthoFinder. Alignments and trimming were completed using MEGA11, as well as the phylogeny building using the Jones-Taylor-Thornton (JTT) substitution model, uniform substitution rates among sites, and nearest-neighbor-interchange (NNI) method for tree inference. Bootstrap values are shown beside branches and branch length represents the expected number of amino acid substitutions per site. Asterisks indicate published sequences used as references for proteins HopAF1 and XopAF (Astua-Monge et al., 2000; Petnicki-Ocwieja et al., 2002) and XopAQ, HopAQ1, Rip6, and Rip11 (Guttman et al., 2002; Mukaihara et al., 2010; Potnis et al., 2011).
The XopAF protein sequence alignment showed that all Xhv race 1 and 3 homologs were identical except for that of NCPPB 4058, which had significant variation. Phylogenetic analysis revealed that all XopAF protein sequences from X. hortorum strains clustered together and were more closely related to the HopAF1 protein sequences from Pseudomonas syringae pv. tomato (Pst) DC3000 than the XopAF/AvrXv3 from Xanthomonas euvesicatoria pv. perforans (Figure 4B). It is possible this effector gene was originally acquired by the ancestor of X. hortorum complex from a Pseudomonas species. Here, we designate this new effector variant XopAF2 for X. hortorum and propose to distinguish it from protein XopAF1 identified in X. euvesicatoria.
Determining the regions flanking xopAQ and xopAF2 in Xhv race 1 and 3 strains allowed for the identification of those same regions in Xhv race 2 strains, and alignments revealed the gene sequence present in place of those effector genes. Where xopAQ would have been present, the Xhv race 2 strains vary in the sequence at this position: conserved domain search revealed that ICMP 1408 encoded for a DNA breaking-rejoining enzyme, while ICMP 4165 encoded for an IS3 family transposase and BS3127 did not encode any conserved domain. In contrast, where xopAF would have been present, alignments showed that the Xhv race 2 strains all had the same sequence which was predicted to encode a glycosyl hydrolase family 3 C-terminal domain. Mining the Xhv race 1 and 3 genomes showed that this sequence could be found elsewhere in those genomes, and so its presence alone is not likely a distinguishing feature of a particular Xhv race.
In comparison to Xhv race 2 strain BS3127, the number of insertion sequences present in Xhv race 1 and 3 strains ranged from 64 to 111, with an average of 92 (Table 2). Searching these insertion sequences for effector genes yielded no matches using the same effector gene mining method used for the whole genome sequences. Searches for the transposase genes included in the ISEScan package reveals a negative sense transposase sequence on the same node as xopAF and 953 bp downstream. This transposase sequence belongs to insertion sequence family IS5, and the sequence itself is 371 bp in length. Further, searching the Xhv race 1 and 3 insertion sequences for phage elements using PHASTER reveals two intact prophage regions. These results indicate that the effector gene xopAF may have been acquired by a strain ancestral to Xhv race 1 and 3 strains through phage-mediated gene transfer. As expected, no insertion sequences were found in other Xhv race 2 strains when compared to BS3127, which supports the assertion that these strains are more related to one another than to the Xhv race 1 and 3 strains.
Secondary Metabolites
Though the main interest in this study was to identify effector repertoires as possible genetic sources of host-specificity, several other virulence factors were investigated for genetic differences among the strains tested that could be useful for characterization of the Xhv races. The secondary metabolites Xanthomonadin and Xanthoferrin are encoded by gene clusters that are conserved among Xanthomonas spp. Xanthomonadin was first described in X. oryzae pv. oryzae (Xoo) and is a yellow pigment that protects bacteria from the increase in lipid-degrading free radicals caused by UV exposure (Rajagopal et al., 1997; Ayala et al., 2014). Xanthoferrin is a siderophore that enhances bacterial survival in low-iron conditions, such as the plant epidermis, due to its iron search and uptake function. Knock-out mutants for these metabolites resulted in decreased virulence for X. campestris pv. campestris, the causal agent of black rot on cabbage. For Xanthomonadin, lipid peroxidation is higher in knock-out mutants than in WT strains (Rajagopal et al., 1997). For Xanthoferrin, the knock-out results in smaller leaf lesions and less migratory capacity through the tissue (Pandey et al., 2017).
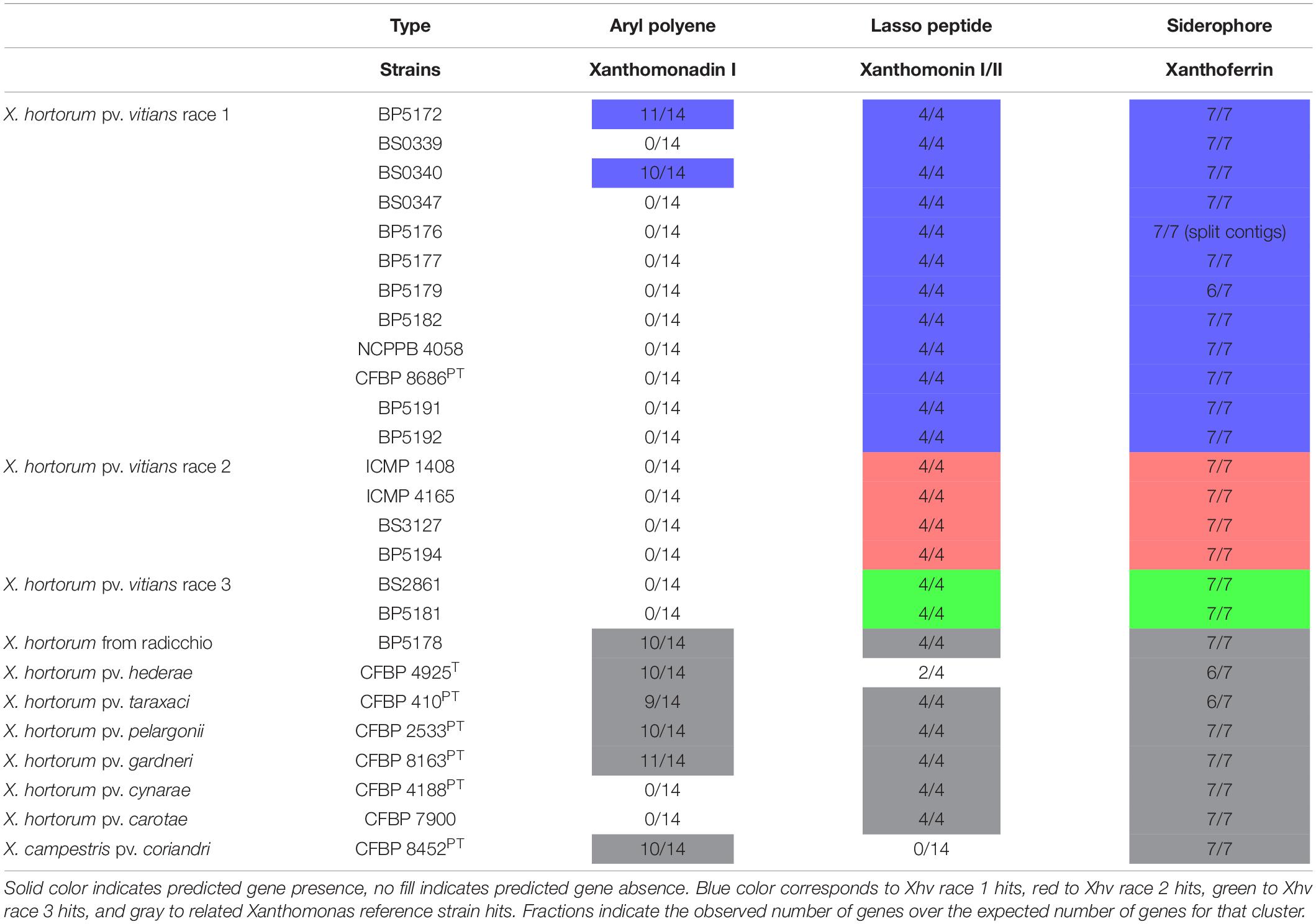
Secondary metabolite gene cluster mining of the whole genome sequences using antiSMASH revealed the possible presence of two clusters (Table 4). Nine to eleven genes of the 14-gene cluster encoding Xanthomonadin I, as described for Xathomonas oryzae pv. oryzae (Xoo), were found only in the genome sequences of Xhv race 1 strains BP5172 and BS0340, and those of the reference strains of X. hortorum from radicchio and X. hortorum pv. hederae. If functional, the production of Xanthomonadin I would afford these strains a fitness advantage in their ability to resist UV damage. The entire 7-gene cluster for Xanthoferrin production, as described for Xoo, was found in all genome sequences tested except for those of Xhv race 1 strain BP5179, the X. hortorum pv. hederae pathotype, and the X. hortorum pv. taraxaci pathotype, which were missing one gene, XOO1360. The strains that do produce Xanthoferrin would be better equipped to survive in low-iron conditions and may be able to spread further through host tissues than strains that do not produce Xanthoferrin. Further research is necessary to demonstrate the production and activity of these metabolites empirically.
Bacteriocins
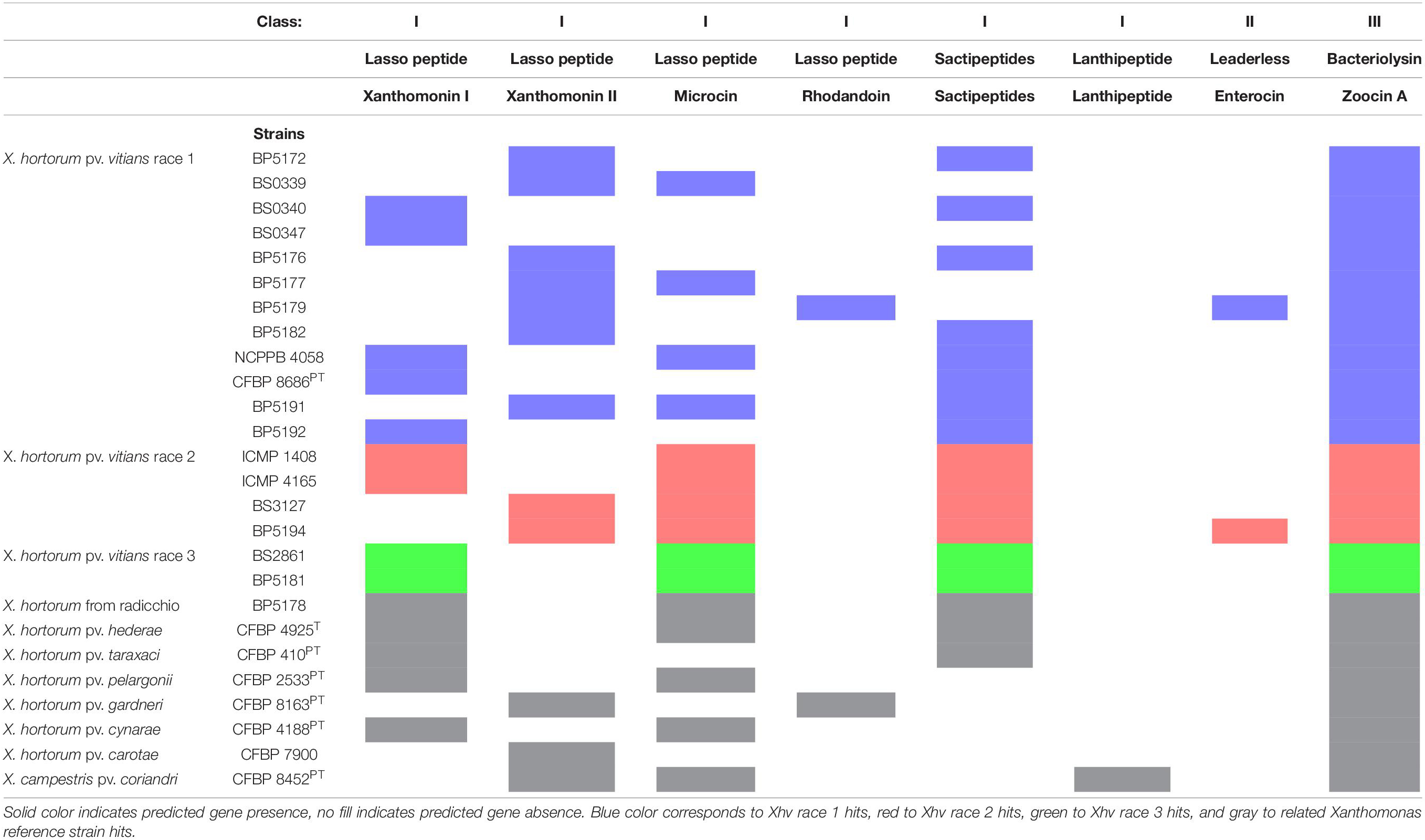
Bacteriocins are also potential virulence factors. Three classes of bacteriocins have been proposed: class I which are heat-stable and ribosomally produced, post-translationally modified peptides (RiPPs); class II which are heat-stable and unmodified; and class III which are thermolabile and unmodified. Xanthomonin I/II, microcin, and rhodanodin are all class I bacteriocins called lasso peptides, so named for their 3-D looped structure (Hegemann et al., 2013). Sactipeptides and lanthipeptides also belong to class I, and all class I peptides have a variety of functions and may aid in bacterial fitness via antibacterial or antiviral activity (Maksimov et al., 2012). Zoocin A is a class III bacteriocin known as a bacteriolysin, which also serves as an antimicrobial peptide but differs from the others in its ability to degrade target bacterial cell walls (Simmonds et al., 1996).
Searches for bacteriocin gene clusters using antiSMASH revealed the presence of one bacteriocin among the strains tested (Table 4), and a parallel analysis using BAGEL4 produced more detailed as well as some contradictory results (Table 5). The entire 4-gene cluster encoding Xanthomonin I/II, as previously described for Xanthomonas hortorum pv. gardneri (ATCC 19865), was found in all strains except the X. hortorum pv. hederae pathotype which was missing half of the cluster, and Xanthomonas campestris pv. coriandri, which was missing the entire cluster. This might suggest that all strains except X. hortorum pv. hederae have an advantage in bacterial fitness due to their production of Xanthomonin I/II. However, searches with BAGEL4 revealed that five of the Xhv race 1, two of the Xhv race 2, and both Xhv race 3 strains encoded for the Xanthomonin I cluster, and the remaining seven Xhv race 1 and two of the Xhv race 2 strains encoded for the Xanthomonin II cluster. Among the non-Xhv strains, X. hortorum from radicchio, X. hortorum pv. hederae, X. hortorum pv. taraxaci, X. hortorum pv. pelargonii, and X. hortorum pv. cynarae all encoded for the Xanthomonin I cluster; X. hortorum pv. gardneri and X. hortorum pv. carotae M081 encoded for the Xanthomonin II cluster. X. campestris pv. coriandri did not encode for either Xanthomonin cluster. These results suggest that all strains tested except X. campestris pv. coriandri might lack the fitness advantage afforded by Xanthomonin I/II production, however further study would be necessary to determine whether Xanthomonin I or II is expressed by these strains and confer a fitness advantage.
Several other bacteriocins were found to be encoded in the strains tested using BAGEL4. A cluster encoding for microcin production was found in 5 of the 12 Xhv race 1 strains, all the Xhv race 2 and 3 strains, and all the related Xanthomonas strains except for X. hortorum pv. hederae and X. hortorum pv. taraxaci. X. hortorum pv. hederaePT and X. hortorum pv. carotae M081 were the only strains encoding for rhodandoin and lanthipeptide, respectively. Nine Xhv race 1 strains, all the Xhv race 2 and 3 strains, X. hortorum from radicchio, X. hortorum pv. hederae, and X. hortorum pv. taraxaci encoded for sactipeptide. For each strain in which they are produced, these class I bacteriocins likely enhance that strain’s ability to compete for resources through antibacterial and antiviral activity. All strains tested encoded for zoocin A production, a type of class III bacteriocin known as a bacteriolysin. The presence of this cluster in all strains suggests that they have a fitness advantage due to the ability to degrade the cell walls of competitor bacteria.
Type II Secretion System
The type II, type III, type IV, and type VI bacterial secretion systems are important delivery systems for pathogenicity and virulence factors, and so the whole genome sequence data was also used to assess the possible presence and composition of these secretions system gene clusters. Type II secretion systems (T2SSs) enhance the virulence of plant pathogens by releasing cell wall degrading enzymes and toxins (Jha et al., 2005). The secretion complex is composed of an inner membrane platform, a pseudopilin extending from that platform into the periplasm, and a secretin forming a pore in the outer membrane. Its substrates are transported into the periplasm by the Sec or Tat pathways and into the path of the type II pseudopilin, which then pushes the substrate out through the outer membrane pore. There are two variations of the type II secretion system, known as Xps and Xcs; although only the former contributes to virulence, the latter has some homology that allows for it to complement the former. Both variations can be present in the same strain, or the Xps system may be present alone, and they are both encoded in gene clusters (xpsE-N, xpsD; xcsC-N, respectively).
T2SS gene cluster mining of each of the 25 whole genome sequences included in this study revealed that all encoded for the complete Xcs-T2SS (Figure 5A). However, the xcs gene cluster has not been implicated in the virulence of a plant pathogen to date, except in its ability to complement the xpsD-H genes of the Xps system, and so it is unlikely to play a role in virulence for the strains in included in this study. Regarding the xps gene cluster, X. hortorum pv. taraxaci and X. hortorum pv. gardneri genomes encoded for its entirety, and all the other genomes encoded for the entire cluster minus the xpsD gene (Figure 5A). One exception was the Xhv race 1 strain BP5179 genome sequence, which lacked xpsI and parts of xpsJ and xpsD—all of which may be explained by gaps between nodes at their expected positions in the sequence. The incomplete or absent xpsD genes may be complemented by the presence of xcsD due to significant homology. No T2SS genes were identified within predicted plasmid sequences, suggesting that these genes are chromosomal. Overall, these results suggest that all strains studied here contained a functional Xps-T2SS that may contribute to virulence through the delivery of toxins and cell-wall degrading enzymes, though empirical study is necessary to confirm this hypothesis.
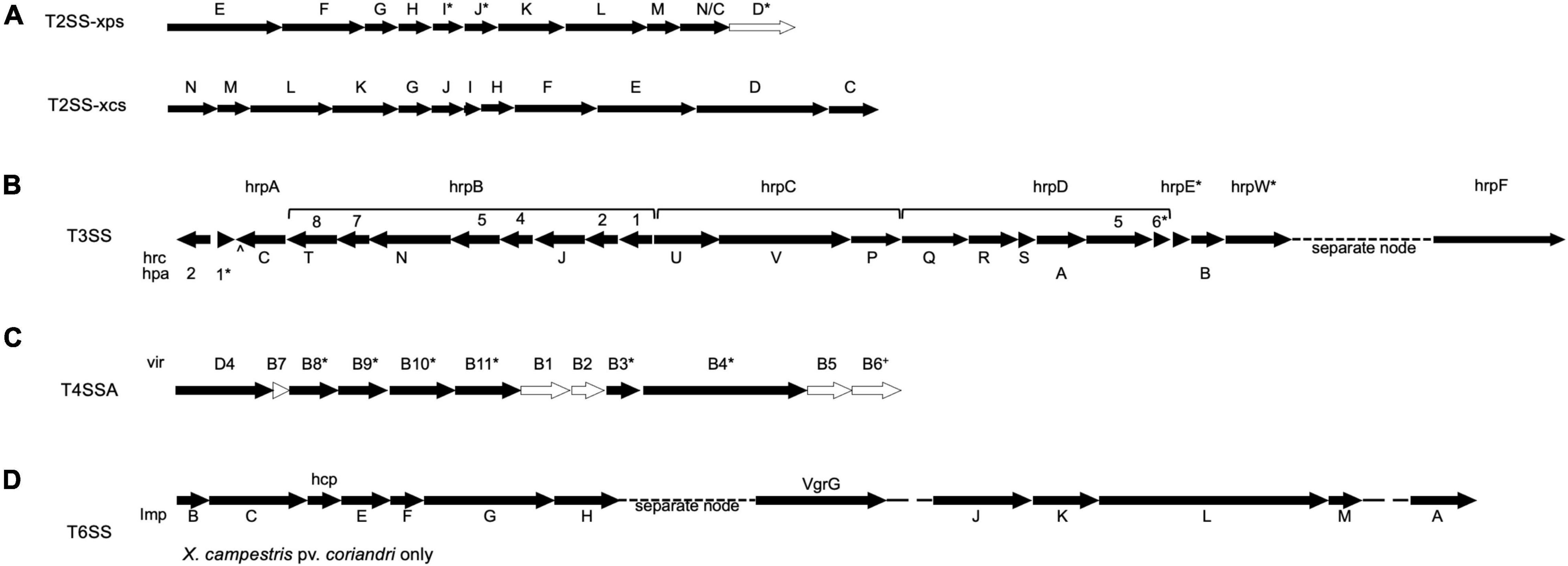
Figure 5. Predicted Secretion system gene clusters for Xhv and related Xanthomonas strains. Presence of each labeled gene in the majority of Xhv and related strains is indicated by a black arrow, and absence from all strains is indicated by a white arrow. Asterisks, carets, and crosses indicate exceptions listed here. (A) T2SSs xps and xcs—exceptions include X. hortorum pv. taraxaci and X. hortorum pv. gardneri, for which the entire xps gene cluster is predicted, and Xhv race 1 strain BP5179, for which the genes xpsI, xpsJ, and xpsD were truncated; (B) T3SS—exceptions include X. hortorum pv. hederae, which had a large insertion between hpa1 and hrcC and lacked hrpE, X. campestris pv. coriandri, which lacked hpa1 and hrpW, and X. hortorum pv. pelargonii, which lacked hrpE and hrpD6; (C) T4SSA—exceptions include Xhv race 1 strains BS0541 and BS3044, which encoded the additional gene virB6, X. hortorum pv. gardneri, which encoded the additional virB6 gene and lacked virB11, X. hortorum from radicchio, which only encoded for virD4, and X. campestris pv. coriandri, which only encoded for virB4, virB8, virB9, and virD4; and (D) T6SS—which was only predicted for the X. campestris pv. coriandri strain and not found in any other strain tested.
Type III Secretion System
Type III secretion systems (T3SSs) play a major role in the virulence and pathogenicity of bacterial plant pathogens (Büttner, 2016). The transmembrane complex includes a needle-like structure that extends into the extracellular space and can pierce the host plant cell wall to directly deliver effector proteins into the host cell cytoplasm. Once inside the host cell, the effectors can halt the basal immune response, tag R-proteins for degradation, disrupt protective phytohormones, or enter the nucleus and alter gene transcription—all of these are strategies to enhance virulence or enable pathogenicity. A cluster of approximately 20 genes encodes the T3SS (Rossier et al., 1999). The effector genes that they translocate are usually dispersed throughout the genome and near to mobile genetic elements, which facilitates their transfer and potential for gain or loss (Darrasse et al., 2013; Cesbron et al., 2015).
Twenty-four genes contributing to the T3SS structure and function were queried against the 25 Xanthomonas strains included in this study, and matches are demonstrated in Figure 5B. The genome sequences of the X. hortorum pv. vitians strains, X. hortorum from radicchio, X. hortorum pv. taraxaci, X. hortorum pv. gardneri, and X. hortorum pv. cynarae all encoded for the complete hrp gene cluster and likely have functional systems (Hueck, 1998). The X. campestris pv. coriandri genome sequence tested here lacked hpa1 and hrpW. The hpa1 gene product transports effectors across the T3SS apparatus in X. oryzae pv. oryzae (Wang et al., 2018) and the hrpW gene is a type III secreted protein that resembles both a harpin and pectate lyase (Charkowski et al., 1998). Either of these two genes may contribute to virulence, but they are not required for a functional T3SS; therefore, this X. campestris pv. coriandri strain likely has a functional T3SS. On the other hand, the genome sequences of X. hortorum pv. hederae and X. hortorum pv. pelargonii both lacked hrpE, and that of X. hortorum pv. pelargonii also lacked hrpD6. The hrpE gene encoded the type 3 pilus in X. vesicatoria (Weber et al., 2005) and the hrpD6 gene was responsible for regulating the expression of other T3SS structural genes and type three secreted effectors in X. oryzae pv. oryzicola (Li et al., 2011); without these genes the T3SS is likely not functional in these strains. Further research would be necessary to demonstrate the translocation activity of these secretion systems empirically, especially for X. hortorum pv. hederae and X. hortorum pv. pelargonii, which both encode T3Es but seem to lack key T3SS structural gene sequences in their genomes. No T3SS genes were found to be encoded by predicted plasmid sequences, indicating that they are chromosomal.
Type IV Secretion System
There are two classes of type IV secretion systems (T4SSs); class A has 12 core genes (virB1-11 and virD4; Tzfira and Citovsky, 2006) and class B has 5 core genes (Dot/IcmCDFG[H/K]) and up to 27 total genes (Hilbi et al., 2017; Qiu and Luo, 2017). Class A substrates have antimicrobial activity to reduce competition for resources; Class B substrates interfere with plant host cell signaling. Class B has not yet been found in Xanthomonas spp. The database used here for type IV secretion system gene mining only included the 10 class A core genes for which DNA sequences are available.
All genome sequences tested here encoded for incomplete T4SSs of class A (Figure 5C). The Xhv genome sequences and those of X. hortorum pv. hederae, X. hortorum pv. pelargonii, and X. hortorum pv. taraxaci, all encoded for the genes virB3, virB4, virB8, virB9, virB10, virB11, and virD4, and those of two Xhv race 1 strains also encoded for virB6 (BP5177 and NCPPB 4058). For three of the Xhv race 2 strains (ICMP 4165, BS3127, and BP5194), virD4 was encoded for on predicted plasmid sequence and all other predicted T4SS genes were predicted on chromosomal sequence (Supplementary Data 6). In other Xanthomonas species, T4SS the genes virB1, virB2, virB4, virB9, virB10, virD2, and virD4 have been shown to be plasmid-borne, and among those all but virD2 can be found duplicated in a larger T4SS gene cluster located on the chromosome (Potnis et al., 2011). The genomic sequence of X. hortorum pv. gardneri encoded for nearly the same T4SS composition as the Xhv strains but was missing virB11 and had the additional virB6 gene. The genome sequence of X. hortorum isolated from radicchio encoded for only virD4. X. campestris pv. coriandri genome sequence encoded for only virB4, virB8, virB9, and virD4. All chromosomal and plasmid sequences assessed here lacked genes that encoded the core complex and pilus of this system (Sgro et al., 2019). Without core structural genes, it is likely that none of these strains have functional T4SSs. More research is necessary to confirm this hypothesis.
Type VI Secretion System
The type VI secretion system (T6SS) in plant pathogenic bacteria has antimicrobial ability; they are used to directly inject toxins into competitor bacteria cells to reduce competition for resources (Bernal et al., 2018). The contribution of T6SS in plant host interactions, however, has not yet been investigated (Liyanapathiranage et al., 2021). A cluster of thirteen type six secretion genes, tssA-tssM, encode the system and were formerly referred to as imp (impaired in nitrogen fixation) genes. The injection structure they form resembles a bacteriophage and toxin delivery relies on its direct contact with bacterial cell walls. The genes tssD and tssI, also known as hcp and vgrG, respectively, are thought to be both structural components and substrates of this system (Russell et al., 2014).
The only whole genome sequence included in this study that encoded for a T6SS was X. campestris pv. coriandri (Figure 5D). The gene cluster was split between two nodes; node 6 contained genes tssB, tssC, tssD, tssE-H and node 15 contained genes tssI-M and tssA. None of these genes were found among predicted plasmid sequences, suggesting that they are all encoded chromosomally. The presence of all core genes suggests that the T6SS is functional in this strain of X. campestris pv. coriandri and may contribute to its fitness against competitor bacteria.
Secretion system mining revealed that the X. hortorum strains, including all Xhv strains tested here, likely have functional type II and III secretion systems and do not have type IV or type VI secretion systems. X. campestris pv. coriandri also likely has functional type II, III, and VI secretion systems but likely lacks the type IV secretion system. The presence of type II and type III secretion system gene clusters suggests that these strains can deliver type II secreted toxins and enzymes and type III secreted effectors that may enhance virulence. In X. campestris pv. coriandri, the type VI secretion system may offer a fitness advantage in the ability to secrete type VI effectors that target competitor bacteria.
Conclusion
This study revealed insights into the race structure of X. hortorum pv. vitians. Phylogenetic analysis showed that strains of Xhv races 1 and 3 were more closely related to each other than to strains of Xhv race 2. HR induction in ARM-09-161-10-1 upon inoculation with Xhv race 1 and 3 strains may have been due to a gene-for-gene interaction, in which a race-specific effector is recognized by a resistance protein expressed by this cultivar. Such a race-specific interaction has been demonstrated for tomato cultivars and three races of X. euvesicatoria pv. perforans (Astua-Monge et al., 2000). The one TALE identified among the genome sequences tested, avrHah1, did not show race-specific variation here, but long-read sequencing to resolve the repeat regions may yet reveal such variation. Two other putative effectors identified in this study are possible candidate genes responsible for race-specificity because they were present only in Xhv races 1 and 3 genome sequences and not in Xhv race 2 genome sequences. These genes are a xopAQ homolog and the novel effector variant xopAF2. Antisense transposase sequence downstream from xopAF2 and the prophage sequence identified among nearby Xhv race 1 and 3 insertion sequences suggest that an ancestor of Xhv race 1 and 3 strains may have acquired xopAF2 in a phage-mediated gene transfer. Our ongoing research seeks to demonstrate whether these effectors are responsible for HR induction in ARM-09-161-10-1. Additionally, more study is necessary to identify other possible gene products involved in the incompatible interactions of Xhv races 1 and 2 with Xar1-containing lettuce cultivars, such as Little Gem, and PI491114, respectively. Furthermore, the close relatedness of the Xhv race 1 and 3 strains may make it difficult to identify genotypic differences that explain their different disease phenotypes on lettuce cultivar Little Gem, as none were identified in this study.
This study also provided several insights into Xanthomonas genetic diversity. The genome statistics for our assemblies are consistent with what is expected for Xanthomonas spp. Secondary metabolite and bacteriocin production varied by strain, and no pattern was found that would be useful for characterizing the Xhv races. All strains tested here encoded for type II and type III secretion systems, and X. campestris pv. coriandri also encoded for a type VI secretion system. These systems all appear to be encoded chromosomally, as none of the genes were identified among predicted plasmid sequences.
Understanding the genetic variation among strains of Xhv is crucial for developing effective disease management strategies, especially the breeding of lettuce cultivars with durable resistance. Chemical control is most effective when applied prophylactically, but the sporadic nature of the disease can render this approach unnecessary and costly in years that the pathogen does not appear (Bull et al., 2007). A more efficient strategy is the use of resistant cultivars, especially those with multiple genetic sources of resistance against the different races of Xhv (Sandoya et al., 2019). Lettuce cultivars resistant to Xhv race 1 strains have already been found (Hayes et al., 2014; Wang et al., 2016) and additional germplasm is being sought (Sandoya et al., submitted manuscript). These resistant cultivars encode R-genes Xar1 and Xcvr, which have been mapped to lettuce chromosome two, but the precise sequence has not yet been determined. The presence of these R-genes suggests a possible gene-for-gene interaction between these cultivars and Xhv race 1, and the race-specific interactions of Xhv race 2 and 3 with lettuce cultivars PI491114 and ARM-09-161-10-1, respectively, may also be due to gene-for-gene interactions with other yet unidentified R-genes. With knowledge of the specific genes interacting with lettuce hosts to induce HR, research could proceed for the precise identification of R-gene interaction partners and subsequent engineering of lettuce cultivars expressing those R-genes.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
ER co-authored grants funding research, conducted the analyses, and wrote the manuscript. NP provided bioinformatic and phytobacterial expertise to direct the research. CB co-wrote grants funding research, designed research hypotheses, mentored Ph.D. candidate ER, and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by NSF GRFP Grant No. 2018265841 and USDA SCMP Grant No. 134853. This work was supported, in part, by the USDA National Institute of Food and Federal Appropriations.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Istvan Albert, Aswathy Sebastian, and Juan Francisco Iturralde Martinez for sharing their expertise in bioinformatics and genomics. We also thank Laura Ramos Sepulveda for her early contributions to this work and Samuel Martins for his unwavering support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.840311/full#supplementary-material
References
Andrews, S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed January 22, 2019).
Antipov, D., Raiko, M., Lapidus, A., and Pevzner, P. A. (2019). Plasmid detection and assembly in genomic and metagenomic data sets. Genome Res. 29, 961–968. doi: 10.1101/gr.241299.118
Arnaud, M. G. (1920). Une maladie bactcriennc du lierre (I fe-dera helix L.). C. R. Lebdomadaires Seances Acad. Sci. 171, 121–122.
Arndt, D., Grant, J. R., Marcu, A., Sajed, T., Pon, A., Liang, Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21. doi: 10.1093/nar/gkw387
Astua-Monge, G., Minsavage, G. V., Stall, R. E., Davis, M. J., Bonas, U., and Jones, J. B. (2000). Resistance of tomato and pepper to T3 strains of Xanthomonas campestris pv. vesicatoria is specified by a plant-inducible avirulence gene. Mol. Plant Microbe Interact. 13, 911–921. doi: 10.1094/MPMI.2000.13.9.911
Ayala, A., Muñoz, M. F., and Argüelles, S. (2014). Lipid peroxidation: production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid. Med. Cell. Longev. 2014:360438. doi: 10.1155/2014/360438
Balaji, V., Gibly, A., Debbie, P., and Sessa, G. (2007). Transcriptional analysis of the tomato resistance response triggered by recognition of the Xanthomonas type III effector AvrXv3. Funct. Integr. Genom. 7, 305–316. doi: 10.1007/s10142-007-0050-y
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Barak, J. D., Vancheva, T., Lefeuvre, P., Jones, J. B., Timilsina, S., Minsavage, G. V., et al. (2016). Whole-genome sequences of Xanthomonas euvesicatoria strains clarify taxonomy and reveal a stepwise erosion of type 3 effectors. Front. Plant Sci. 7:1805. doi: 10.3389/fpls.2016.01805
Bernal, P., Llamas, M. A., and Filloux, A. (2018). Type VI secretion systems in plant-associated bacteria. Environ. Microbiol. 20, 1–15. doi: 10.1111/1462-2920.13956
Blin, K., Shaw, S., Steinke, K., Villebro, R., Ziemert, N., Lee, S. Y., et al. (2019). antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47, W81–W87. doi: 10.1093/nar/gkz310
Bogdanove, A. J., Schornack, S., and Lahaye, T. (2010). TAL effectors: finding plant genes for disease and defense. Curr. Opin. Plant Biol. 13, 394–401. doi: 10.1016/j.pbi.2010.04.010
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Brown, N. A. (1923). Bacterial leaf spot of geranium in the eastern United States. J. Agric. Res. 23, 361–372.
Bull, C., Trent, M., and Hayes, R. (2016). Three races of Xanthomonas campestris pv. vitians causing bacterial leaf spot on lettuce identified. Phytopathol. S 4:100.
Bull, C. T., Gebben, S. J., Goldman, P. H., Trent, M. A., and Hayes, R. J. (2015). Host genotype and hypersensitive reaction influence population levels of Xanthomonas campestris pv. vitians in lettuce. Phytopathology 105, 316–324. doi: 10.1094/PHYTO-07-14-0185-R
Bull, C. T., Goldman, P. H., Hayes, R., Madden, L. V., Koike, S. T., and Ryder, E. (2007). Genetic diversity of lettuce for resistance to bacterial leaf spot caused by Xanthomonas campestris pv. vitians. Plant Health Prog. 8:15. doi: 10.1094/PHP-2007-0917-02-RS
Büttner, D. (2016). Behind the lines-actions of bacterial type III effector proteins in plant cells. FEMS Microbiol. Reviews 40, 894–937. doi: 10.1093/femsre/fuw026
Cesbron, S., Briand, M., Essakhi, S., Gironde, S., Boureau, T., Manceau, C., et al. (2015). Comparative genomics of pathogenic and nonpathogenic strains of Xanthomonas arboricola unveil molecular and evolutionary events linked to pathoadaptation. Front. Plant Sci. 6:1126. doi: 10.3389/fpls.2015.01126
Charkowski, A. O., Alfano, J. R., Preston, G., Yuan, J., He, S. Y., and Collmer, A. (1998). The Pseudomonas syringae pv. tomato HrpW protein has domains similar to harpins and pectate lyases and can elicit the plant hypersensitive response and bind to pectate. J. Bacteriol. 180, 5211–5217. doi: 10.1128/JB.180.19.5211-5217.1998
Croucher, N. J., Page, A. J., Connor, T. R., Delaney, A. J., Keane, J. A., Bentley, S. D., et al. (2015). Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 43:e15. doi: 10.1093/nar/gku1196
Dangl, J. L., and Jones, J. D. (2001). Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. doi: 10.1038/35081161
Darrasse, A., Carrère, S., Barbe, V., Boureau, T., Arrieta-Ortiz, M. L., Bonneau, S., et al. (2013). Genome sequence of Xanthomonas fuscans subsp. fuscans strain 4834-R reveals that flagellar motility is not a general feature of xanthomonads. BMC Genomics 14:761. doi: 10.1186/1471-2164-14-761
Durrant, M. G., Li, M. M., Siranosian, B. A., Montgomery, S. B., and Bhatt, A. S. (2020). A bioinformatic analysis of integrative mobile genetic elements highlights their role in bacterial adaptation. Cell Host Microbe 27, 140.e9–153.e9. doi: 10.1016/j.chom.2019.10.022
Dye, D. W. (1978). Genus IX Xanthomonas. In a proposed nomenclature and classification for plant pathogenic bacteria. N. Z. J. Agric. Res. 21, 153–177. doi: 10.1080/00288233.1978.10427397
Fayette, J., Raid, R., Roberts, P. D., Jones, J. B., Pernezny, K., Bull, C. T., et al. (2016). Multilocus sequence typing of strains of bacterial spot of lettuce collected in the United States. Phytopathology 106, 1262–1269. doi: 10.1094/PHYTO-11-15-0302-R
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Guttman, D. S., Vinatzer, B. A., Sarkar, S. F., Ranall, M. V., Kettler, G., and Greenberg, J. T. (2002). A functional screen for the type III (Hrp) secretome of the plant pathogen Pseudomonas syringae. Science 295, 1722–1726. doi: 10.1126/science.295.5560.1722
Hajri, A., Brin, C., Hunault, G., Lardeux, F., Lemaire, C., Manceau, C., et al. (2009). A “repertoire for repertoire” hypothesis: repertoires of type three effectors are candidate determinants of host specificity in Xanthomonas. PLoS One 4:e6632. doi: 10.1371/journal.pone.0006632
Hayes, R. J., Trent, M. A., Truco, M. J., Antonise, R., Michelmore, R. W., and Bull, C. T. (2014). The inheritance of resistance to bacterial leaf spot of lettuce caused by Xanthomonas campestris pv. vitians in three lettuce cultivars. Hortic. Res. 66:14066. doi: 10.1038/hortres.2014.66
Hegemann, J. D., Zimmermann, M., Zhu, S., Klug, D., and Marahiel, M. A. (2013). Lasso peptides from Proteobacteria: genome mining employing heterologous expression and mass spectrometry. Biopolymers 100, 527–542. doi: 10.1002/bip.22326
Hilbi, H., Nagai, H., Kubori, T., and Roy, C. R. (2017). Subversion of host membrane dynamics by the legionella Dot/Icm type IV secretion system. Curr. Top. Microbiol. Immunol. 413, 221–242. doi: 10.1007/978-3-319-75241-9_9
Holtsmark, I., Eijsink, V. G., and Brurberg, M. B. (2008). Bacteriocins from plant pathogenic bacteria. FEMS Microbiol. Lett. 280, 1–7. doi: 10.1111/j.1574-6968.2007.01010.x
Hueck, C. J. (1998). Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62, 379–433. doi: 10.1128/MMBR.62.2.379-433.1998
Jalan, N., Kumar, D., Andrade, M. O., Yu, F., Jones, J. B., Graham, J. H., et al. (2013). Comparative genomic and transcriptome analyses of pathotypes of Xanthomonas citri subsp. citri provide insights into mechanisms of bacterial virulence and host range. BMC Genomics 14:551. doi: 10.1186/1471-2164-14-551
Jha, G., Rajeshwari, R., and Sonti, R. V. (2005). Bacterial type two secretion system secreted proteins: double-edged swords for plant pathogens. Mol. Plant Microbe Interact. 18, 891–898. doi: 10.1094/MPMI-18-0891
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Li, Y. R., Zou, H. S., Che, Y. Z., Cui, Y. P., Guo, W., Zou, L. F., et al. (2011). A novel regulatory role of HrpD6 in regulating hrp-hrc-hpa genes in Xanthomonas oryzae pv. oryzicola. Mol. Plant Microbe Interact. 24, 1086–1101. doi: 10.1094/MPMI-09-10-0205
Liyanapathiranage, P., Jones, J. B., and Potnis, N. (2021). A mutation of a single core gene, tssM, of Type VI secretion system of Xanthomonas perforans influences virulence, epiphytic survival and transmission during pathogenesis on tomato. Phytopathology [Epub ahead of print]. doi: 10.1094/PHYTO-02-21-0069-R
Maiden, M. C., Bygraves, J. A., Feil, E., Morelli, G., Russell, J. E., Urwin, R., et al. (1998). Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U.S.A. 95, 3140–3145. doi: 10.1073/pnas.95.6.3140
Maksimov, M. O., Pan, S. J., and James Link, A. (2012). Lasso peptides: structure, function, biosynthesis, and engineering. Nat. Product Rep. 29, 996–1006. doi: 10.1039/c2np20070h
Manni, M., Berkeley, M. R., Seppey, M., Simão, F. A., and Zdobnov, E. M. (2021). BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol. Biol. Evol. 38, 4647–4654. doi: 10.1093/molbev/msab199
McCann, H. C., and Guttman, D. S. (2008). Evolution of the type III secretion system and its effectors in plant-microbe interactions. New Phytol. 177, 33–47. doi: 10.1111/j.1469-8137.2007.02293.x
Merda, D., Briand, M., Bosis, E., Rousseau, C., Portier, P., Barret, M., et al. (2017). Ancestral acquisitions, gene flow and multiple evolutionary trajectories of the type three secretion system and effectors in Xanthomonas plant pathogens. Mol. Ecol. 26, 5939–5952. doi: 10.1111/mec.14343
Minsavage, G. V., Dahlbeck, D., Whalen, M. C., Kearney, B., Bonas, U., Staskawicz, B. J., et al. (1990). Gene-for-gene relationships specifying disease resistance in Xanthomonas campestris pv. vesictoria-Pepper interactions. Mol. Plant Microbe Interact. 3, 41–47. doi: 10.1094/MPMI-3-041
Morinière, L., Burlet, A., Rosenthal, E. R., Nesme, X., Portier, P., Bull, C. T., et al. (2020). Clarifying the taxonomy of the causal agent of bacterial leaf spot of lettuce through a polyphasic approach reveals that Xanthomonas cynara Trébol et al. 2000 emend. Timilsina et al. 2019 is a later heterotypic synonym of Xanthomonas hortorum Vauterin et al. (1995). Syst. Appl. Microbiol. 43:126087. doi: 10.1016/j.syapm.2020.126087
Mukaihara, T., Tamura, N., and Iwabuchi, M. (2010). Genome-wide identification of a large repertoire of Ralstonia solanacearum type III effector proteins by a new functional screen. Mol. Plant Microbe Interact. 23, 251–262. doi: 10.1094/MPMI-23-3-0251
Newberry, E. A., Bhandari, R., Minsavage, G. V., Timilsina, S., Jibrin, M. O., Kemble, J., et al. (2019). Independent evolution with the gene flux originating from multiple Xanthomonas species explains genomic heterogeneity in Xanthomonas perforans. Appl. Environ. Microbiol. 85:e00885-19. doi: 10.1128/AEM.00885-19
Niederhauser, J. S. (1943). A bacterial leaf spot and blight of the Russian da∼delion. Phytopathology 33, 959–961.
Pandey, P., Irulappan, V., Bagavathiannan, M. V., and Senthil-Kumar, M. (2017). Impact of combined abiotic and biotic stresses on plant growth and avenues for crop improvement by exploiting physio-morphological traits. Front. Plant Sci. 8:537. doi: 10.3389/fpls.2017.00537
Petnicki-Ocwieja, T., Schneider, D. J., Tam, V. C., Chancey, S. T., Shan, L., Jamir, Y., et al. (2002). Genomewide identification of proteins secreted by the Hrp type III protein secretion system of Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. U.S.A. 99, 7652–7657. doi: 10.1073/pnas.112183899
Potnis, N., Krasileva, K., Chow, V., Almeida, N. F., Patil, P. B., Ryan, R. P., et al. (2011). Comparative genomics reveals diversity among xanthomonads infecting tomato and pepper. BMC Genom. 12:146. doi: 10.1186/1471-2164-12-146
Qiu, J., and Luo, Z. Q. (2017). Legionella and Coxiella effectors: strength in diversity and activity. Nat. Rev. Microbiol. 15, 591–605. doi: 10.1038/nrmicro.2017.67
Rajagopal, L., Sundari, C. S., Balasubramanian, D., and Sonti, R. V. (1997). The bacterial pigment xanthomonadin offers protection against photodamage. FEBS Lett. 415, 125–128. doi: 10.1016/s0014-5793(97)01109-5
Roach, R., Mann, R., Gambley, C. G., Chapman, T., Shivas, R. G., and Rodoni, B. (2019). Genomic sequence analysis reveals diversity of Australian Xanthomonas species associated with bacterial leaf spot of tomato, capsicum and chilli. BMC Genom. 20:310. doi: 10.1186/s12864-019-5600-x
Rosenthal, E. R., Sebastian, A., Potnis, N., Albert, I., and Bull, C. T. (2018). Investigating Effector Diversity as a Source of Race-Specific Pathogenicity for the Lettuce Bacterial Leaf Spot Pathogen. Boston, MA: The American Phytopathological Society.
Rossier, O., Wengelnik, K., Hahn, K., and Bonas, U. (1999). The Xanthomonas Hrp type III system secretes proteins from plant and mammalian bacterial pathogens. Proc. Natl. Acad. Sci. U.S.A. 96, 9368–9373. doi: 10.1073/pnas.96.16.9368
Rozov, R. Brown Kav, A., Bogumil, D., Shterzer, N., Halperin, E., Mizrahi, I., et al. (2017). Recycler: an algorithm for detecting plasmids from de novo assembly graphs. Bioinformatics 33, 475–482. doi: 10.1093/bioinformatics/btw651
Russell, A. B., Peterson, S. B., and Mougous, J. D. (2014). Type VI secretion system effectors: poisons with a purpose. Nat. Rev. Microbiol. 12, 137–148. doi: 10.1038/nrmicro3185
Sahin, F., Abbasi, P. A., Lewis Ivey, M. L., Zhang, J., and Miller, S. A. (2003). Diversity among strains of Xanthomonas campestris pv. vitians from lettuce. Phytopathology 93, 64–70. doi: 10.1094/PHYTO.2003.93.1.64
Sandoya, G. V., Maisonneuve, B., Truco, M. J., Bull, C. T., Simko, I., Trent, M., et al. (2019). Genetic analysis of resistance to bacterial leaf spot in the heirloom lettuce cultivar Reine des Glaces. Mol. Breed. 39:160. doi: 10.1007/s11032-019-1072-6
Sarkar, S. F., Gordon, J. S., Martin, G. B., and Guttman, D. S. (2006). Comparative genomics of host-specific virulence in Pseudomonas syringae. Genetics 174, 1041–1056. doi: 10.1534/genetics.106.060996
Schornack, S., Minsavage, G. V., Stall, R. E., Jones, J. B., and Lahaye, T. (2008). Characterization of AvrHah1, a novel AvrBs3-like effector from Xanthomonas gardneri with virulence and avirulence activity. New Phytol. 179, 546–556. doi: 10.1111/j.1469-8137.2008.02487.x
Seemann, T. (2015). Snippy: Fast Bacterial Variant Calling from NGS Reads. Available online at: https://github.com/tseemann/snippy (accessed November 23, 2020).
Sgro, G. G., Oka, G. U., Souza, D. P., Cenens, W., Bayer-Santos, E., Matsuyama, B. Y., et al. (2019). Bacteria-killing type IV secretion systems. Front. Microbiol. 10:1078. doi: 10.3389/fmicb.2019.01078
Srinivasan, M. C., Patel, M. K., and Thirumalachar, M. J. (1961). A bacterial blight disease of coriander. Proc. Indian Acad. Sci. U.S.A. 53B, 298–301.
Simmonds, R. S., Pearson, L., Kennedy, R. C., and Tagg, J. R. (1996). Mode of action of a lysostaphin-like bacteriolytic agent produced by Streptococcus zooepidemicus 4881. Appl. Environ. Microbiol. 62, 4536–4541. doi: 10.1128/aem.62.12.4536-4541.1996
Swanson, J., Kearney, B., Dahlbeck, D., and Staskawicz, B. J. (1988). Cloned avirulence gene of Xanthomonas campestris pv. vesicatoria complements spontaneous race-change mutants. Mol. Plant Microbe Interact. 1, 5–9.
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Thieme, F., Szczesny, R., Urban, A., Kirchner, O., Hause, G., and Bonas, U. (2007). New type III effectors from Xanthomonas campestris pv. vesicatoria trigger plant reactions dependent on a conserved N-myristoylation motif. Mol. Plant Microbe Interact. 20, 1250–1261. doi: 10.1094/MPMI-20-10-1250
Trébaol, G., Gardan, L., Manceau, C., Tanguy, J. L., Tirilly, Y., and Boury, S. (2000). Genomic and phenotypic characterization of Xanthomonas cynarae sp. nov., a new species that causes bacterial bract spot of artichoke (Cynara scolymus L.). Int. J. Syst. Evol. Microbiol. 50, 1471–1478. doi: 10.1099/00207713-50-4-1471
Tzfira, T., and Citovsky, V. (2006). Agrobacterium-mediated genetic transformation of plants: biology and biotechnology. Curr. Opin. Biotechnol. 17, 147–154. doi: 10.1016/j.copbio.2006.01.009
van Heel, A. J. de Jong, A., Song, C., Viel, J. H., Kok, J., and Kuipers, O. P. (2018). BAGEL4: a user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 46, W278–W281. doi: 10.1093/nar/gky383
Vauterin, L., Hoste, B., Kersters, K., and Swings, J. (1995). Reclassification of Xanthomonas. Int. J. Syst. Biol. 45, 472–489. doi: 10.1099/00207713-45-3-472
Wang, X., Zhang, L., Ji, H., Mo, X., Li, P., Wang, J., et al. (2018). Hpa1 is a type III translocator in Xanthomonas oryzae pv. oryzae. BMC Microbiol. 18:105. doi: 10.1186/s12866-018-1251-3
Wang, Y., Lu, H., and Hu, J. (2016). Molecular mapping of high resistance to bacterial leaf spot in lettuce PI 358001-1. Phytopathology 106, 1319–1325. doi: 10.1094/PHYTO-09-15-0238-R
Weber, E., Ojanen-Reuhs, T., Huguet, E., Hause, G., Romantschuk, M., Korhonen, T. K., et al. (2005). The type III-dependent Hrp pilus is required for productive interaction of Xanthomonas campestris pv. vesicatoria with pepper host plants. J. Bacteriol. 187, 2458–2468. doi: 10.1128/JB.187.7.2458-2468.2005
White, F. F., Potnis, N., Jones, J. B., and Koebnik, R. (2009). The type III effectors of Xanthomonas. Mol. Plant Pathol. 10, 749–766. doi: 10.1111/j.1364-3703.2009.00590.x
Xie, Z., and Tang, H. (2017). ISEScan: automated identification of insertion sequence elements in prokaryotic genomes. Bioinformatics 33, 3340–3347. doi: 10.1093/bioinformatics/btx433
Young, J. M., Park, D. C., Shearman, H. M., and Fargier, E. (2008). A multilocus sequence analysis of the genus Xanthomonas. Syst. Appl. Microbiol. 31, 366–377. doi: 10.1016/j.syapm.2008.06.004
Zacaroni, A. B., Koike, S. T., de Souza, R. M., and Bull, C. T. (2012). Bacterial leaf spot of radicchio (Cichorium intybus) is caused by Xanthomonas hortorum. Plant Dis. 96:1820. doi: 10.1094/PDIS-07-12-0672-PDN
Keywords: bacterial plant pathogens, plant-microbe interactions, comparative genomics, Xanthomonas, effectors
Citation: Rosenthal E, Potnis N and Bull CT (2022) Comparative Genomic Analysis of the Lettuce Bacterial Leaf Spot Pathogen, Xanthomonas hortorum pv. vitians, to Investigate Race Specificity. Front. Microbiol. 13:840311. doi: 10.3389/fmicb.2022.840311
Received: 21 December 2021; Accepted: 09 February 2022;
Published: 18 April 2022.
Edited by:
Chih-Horng Kuo, Academia Sinica, TaiwanReviewed by:
Joana G. Vicente, Fera Science Ltd., United KingdomNemanja Kuzmanovic, Julius Kühn-Institut, Germany
Copyright © 2022 Rosenthal, Potnis and Bull. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carolee T. Bull, Y2Fyb2xlZWJ1bGxAcHN1LmVkdQ==
†ORCID: Emma Rosenthal, orcid.org/0000-0002-6397-9696; Neha Potnis, orcid.org/0000-0002-5418-5894; Carolee T. Bull, orcid.org/0000-0002-2592-407X
 Emma Rosenthal
Emma Rosenthal Neha Potnis
Neha Potnis Carolee T. Bull
Carolee T. Bull