
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 06 April 2022
Sec. Virology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.838382
Hepatitis Delta Virus (HDV) is the smallest mammalian single-stranded RNA virus. It requires host cells and hepatitis B virus (HBV) to complete its unique life cycle. The present review summarizes the specific regions on hepatitis D antigen (HDAg) and hepatitis B surface antigen (HBsAg) that drive HDV to utilize host cell machinery system to produce three types of RNA and two forms of HDAg, and hijack HBsAg for its secretion and de novo entry. Previously, interferon-α was the only recommended therapy for HDV infection. In recent years, some new therapies targeting these regions, such as Bulevirtide, Lonafarnib, Nucleic acid polymers have appeared, with better curative effects and fewer adverse reactions.
Hepatitis delta virus is a defective virus that requires HBV to proliferate in human hepatocytes. A recent meta-analysis estimated that 292.0 (251.5–341.1) million individuals were HBsAg positive, corresponding to a global prevalence of 3.9% (3.4–4.6) (Polaris Observatory Collaborators, 2018); another meta-analysis showed that 0.16% (0.11–0.25) of global population, totaling 12.0 (8.7–18.7) million people, were estimated to be anti-HDV positive (Stockdale et al., 2020). HDV prevalence varies according to geography (Table 1).
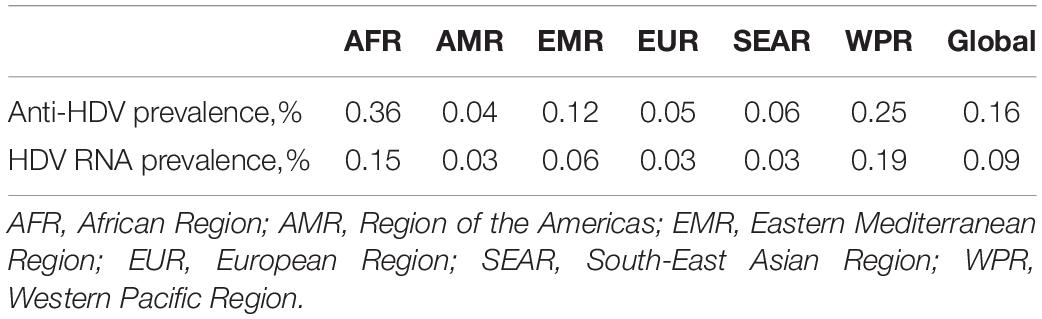
Table 1. Estimates of HDV prevalence in the general population (95% CI) [Source: Stockdale et al. (2020)].
Hepatitis delta virus infection occurs via two ways: coinfection and super-infection. HBV and HDV simultaneously co-infect healthy people, causing extensive hepatic necrosis and severe fulminant hepatitis with a high fatality rate. Most adults can eliminate both viruses and recover from coinfection, and < 5% of cases develop chronic hepatitis D (CHD) (Botelho-Souza et al., 2017). Compared with HBV mono-infection and two viruses’ coinfection, chronic hepatitis B patients with HDV super-infection face an accelerated disease progression to cirrhosis and an increased risk of developing hepatocellular carcinoma. Usually, such patients cannot clear HDV, and 90% of cases develop CHD (Fattovich et al., 2000).
Hepatitis B virus vaccine protects healthy individuals from HBV and HDV infections. However, there is currently no HDV vaccine available protecting chronic hepatitis B patients from HDV infection. Before the European Medicines Agency approved the conditional marketing authorization of Bulevirtide in July 2020, interferon (IFN)-α was the only therapy for CHD recommended by professional and societal guidelines. However, IFN-α can lead to common and sometimes severe adverse reactions, with a high recurrence rate (Abbas et al., 2011).
We summarize the specific regions on HDAg and HBsAg that drive HDV proliferation, and the curative effect of some therapies targeting these regions. We further list some mechanisms underlying the HDV life cycle that are yet ill-understood, and some regions that are potential therapeutic targets. Investigating HDV proliferation mechanisms and the functions of specific regions on HDAg and HBsAg will help us to develop new treatment strategies associated with better therapeutic effects, fewer adverse reactions, and lower recurrence rates.
Hepatitis delta virus is 35–37 nm in diameter and composed of HDV ribonucleoprotein (RNP) complex and HBsAg-containing envelope (Bonino et al., 1981). HDV RNP complex consists of HDV genome RNA (gRNA) and HDAg (Ryu et al., 1993).
There are currently eight HDV genotypes (HDV-1–8), the ribonucleotide number of HDV gRNA is between 1,672 and 1,697 (genotype-dependent) (Wang et al., 2021). HDV gRNA is a circular single-stranded RNA of negative polarity, and about 74% of ribonucleotides form dozens of consecutive base-paired regions (9–15 bp), interspersed with small internal loops or bulges formed by the unpaired ribonucleotides. All ribonucleotides fold into a quasi-double-stranded (ds), unbranched, rod-like structure (Casey, 2012; Lempp et al., 2016; Hsu et al., 2019; Figure 1A).
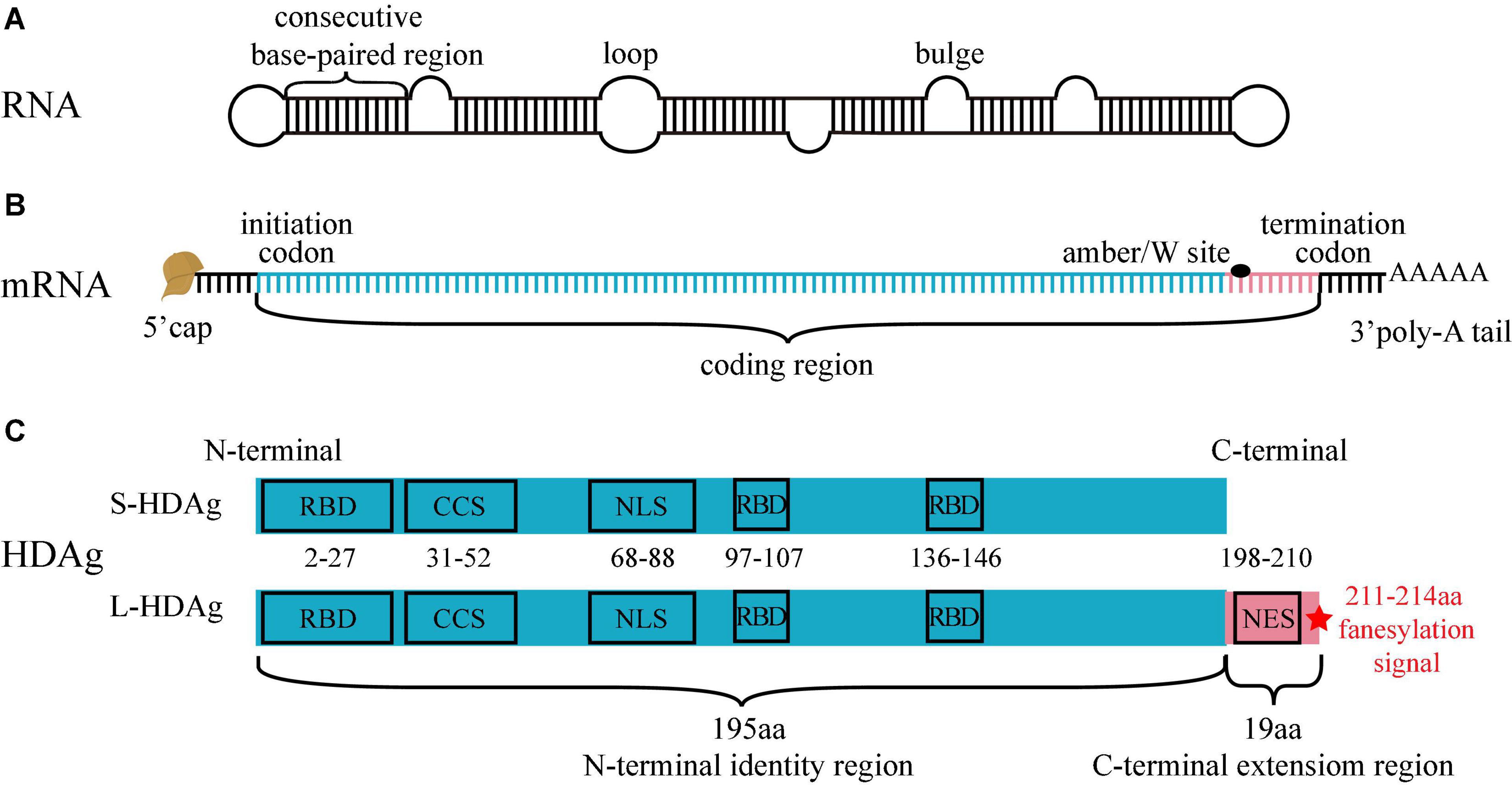
Figure 1. Structure of HDV RNA, mRNA and HDAg. (A) HDV RNA is a circular single-stranded RNA, 74% ribonucleotides form consecutive base-paired regions, interspersed by small loops or bulges formed by the unpaired ribonucleotides. All ribonucleotides fold into a quasi-double-stranded, unbranched rod-like structure. (B) There are a 5′ cap, a 3′ poly-A tail, and a coding region in HDV mRNA. The ribonucleotides from initiation codon to amber/W site (blue segment) encodes S-HDAg and L-HDAg N-terminal identity region, and the ribonucleotides from amber/W site to termination codon (pink segment) encodes L-HDAg C-terminal extension region. (C) S-HDAg and L-HDAg are characterized by three RNA-binding domains (RBD), a coiled-coil sequence (CCS), a nuclear localization signal (NLS) in the N-terminal identity region (blue box), and a nuclear export signal (NES), a farnesylation signal (red star) in the C-terminal extension region (pink box).
Genomic strand ribozyme and antigenome strand ribozyme are the only viral enzymes possessed by HDV, display related sequences and are similar in the predicted secondary structure, and both are crucial for HDV RNA replication (Riccitelli and Lupták, 2013). HDV gRNA only encodes two proteins in the same open reading frame (Xia et al., 1990).
Two forms of HDAg that are translated from HDV mRNA have been identified, including small-HDAg [S-HDAg with 195 amino acids (aa), except HDV-3, -4, and -6 with 194 aa] and large-HDAg (L-HDAg with 214 aa, except HDV-4, -6 with 213 aa). HDV mRNA structure is similar to that of the host mRNA, containing a 5′ cap, a 3′ poly-A tail, and a coding region (Taylor, 2015). A variable amber/W site on the coding region regulates the production of HDAg. The ribonucleotides from initiation codon to amber/W site encode S-HDAg and L-HDAg N-terminal identity region (195 or 194 aa), and the ribonucleotides from amber/W site to termination codon encode L-HDAg C-terminal extension region (19 or 20 aa). Genotypes of HDV except HDV-3, 4, and 6 are described in the following text (Lempp et al., 2016; Figure 1B).
The three critical regions on the S-HDAg and L-HDAg N-terminal identity region (1–195 aa) are as follows: (i) RNA-binding domain (RBD, 2–27 aa, 97–107 aa, and 136–146 aa), binding to HDV RNA to form HDV RNP (Lee et al., 1993; Poisson et al., 1993); (ii) Coiled-coil sequence (CCS, 31–52 aa), mediating the polymerization of HDAg (Xia and Lai, 1992); (iii) Nuclear localization signal (NLS, 68–88 aa), driving the translocation of HDAg from cytoplasm to nucleus (Xia et al., 1992). The two critical regions on the L-HDAg C-terminal extension region (196–214 aa) are as follows: (i) Nuclear export signal (NES, 198–210 aa), driving the translocation of HDV RNP from nucleus to cytoplasm (Lee et al., 2001); (ii) Farnesylation signal (211–214 aa), located at the last four amino acids of the N-terminal and covalently linked to farnesyl lipid group, which inhibits HDV RNA amplification and promotes progeny viruses assembly (Glenn and White, 1991; Hwang and Lai, 1993; Figure 1C).
Hepatitis delta virus RNP complex is 18.7 ± 2.5 nm in diameter and composed of HDV gRNA and 70–200 HDAg monomers (Ryu et al., 1993; Gudima et al., 2002). Before binding to RNA, HDAg monomers are subjected to antiparallel coiled-coil mediated dimerization via CCS (Zuccola et al., 1998). These HDAg dimers continue to polymerize into HDAg octamers that exist as either S-HDAg/L-HDAg homomultimers or S-HDAg and L-HDAg heteromultimers (Xia and Lai, 1992; Figure 2). The number of S-HDAg and L-HDAg monomers is equivalent in circulating HDV (Bergmann and Gerin, 1986).
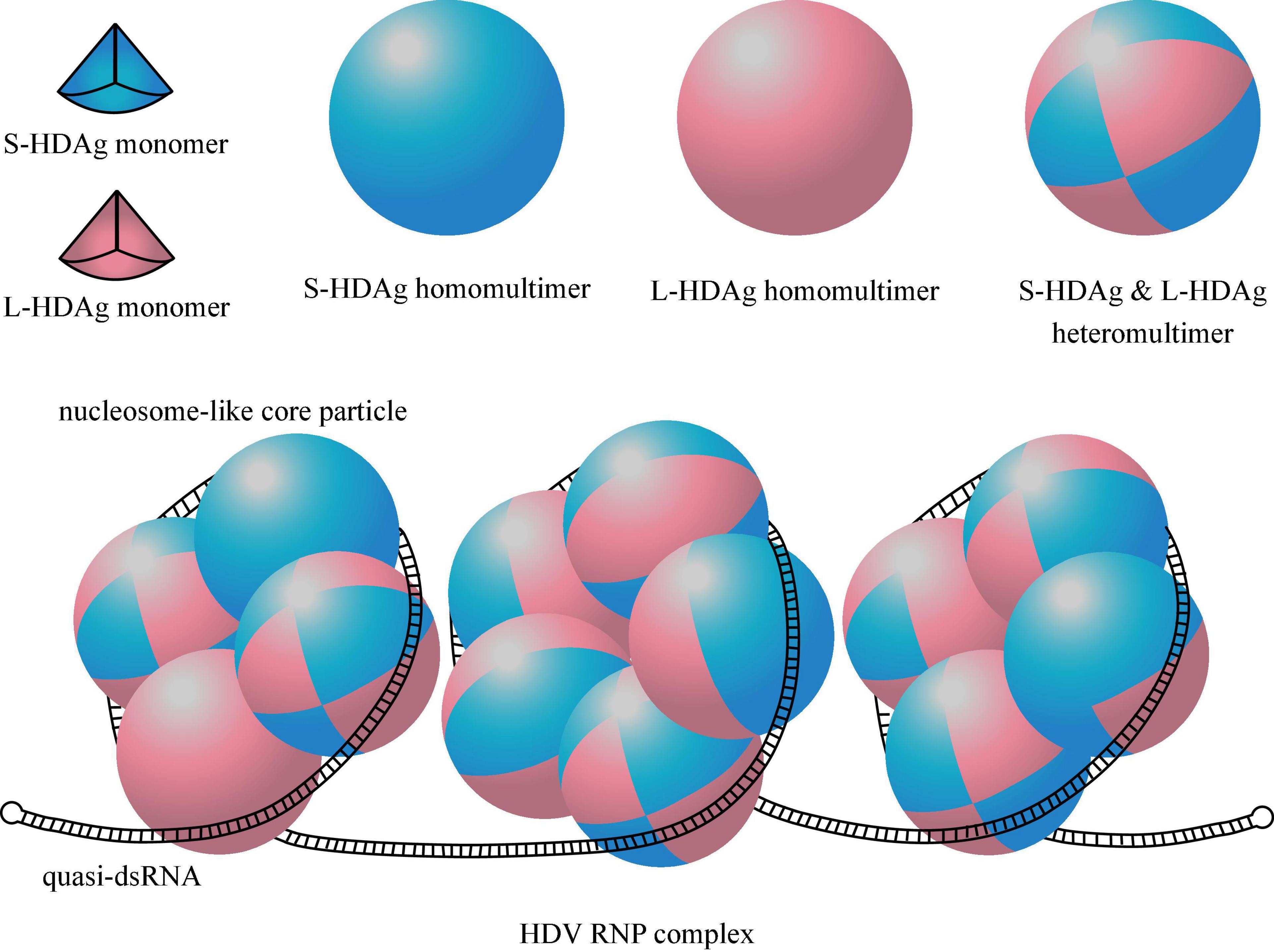
Figure 2. Structure of HDAg multimers and HDV ribonucleoprotein (RNP) complex. One eighth of the sphere represents a HDAg monomer, the blue one represents S-HDAg, the pink one represents L-HDAg. A HDAg octamer containing 8 S-HDAg or 8 L-HDAg is called homomultimer, and is called heteromultimer when it contains a mix of S-HDAg and L-HDAg. Quasi-double-stranded RNA (quasi-dsRNA) wrap around four or five HDAg octamers as a nucleosome-like core particle, and the whole HDV RNP contain 2–6 such core particles.
The loops and bulges contribute to the flexibility of the quasi-dsRNA to wrap around HDAg multimers via arginine-rich, positively charged RBD for optimal compression. Although the exact assembly structure of dsRNA and HDAg is not yet clear, it is speculated that quasi-dsRNA wraps around four or five HDAg octamers to form a nucleosome-like core particle (Abeywickrama-Samarakoon et al., 2020). The presence of about 70–200 HDAg monomers per HDV implies 2–6 such nucleosome-like core particles per HDV RNP (Griffin et al., 2014; Figure 2).
The HBsAg-containing envelope is a host-derived outer surface lipid coat containing three forms of HBsAgs (Steiner et al., 1974), including small-HBsAg (S-HBsAg, 226 aa), middle-HBsAg (M-HBsAg, 281 aa), and large-HBsAg (L-HBsAg, 388–400 aa, genotype-dependent). All these antigens are encoded by a single open reading frame divided by three in frame standing initiation codons, resulting in preS1 (107, 118, or 119 aa, genotype-dependent), preS2 (55 aa), and S (226 aa) translation regions respectively. S-HBsAg contains the S region, M-HBsAg contains the preS2 and S region, and L-HBsAg contains the preS1, preS2, and S region (Jiang and Hildt, 2020). In the N-terminal of L-HBsAg, a myristoyl group linked to Gly-2 (Bruss et al., 1996; Figure 3). Two promoter sequences regulate the production of HBsAg. The upstream preS1 promoter gives rise to 2.4 kb transcripts that are translated mainly into the L-HBsAg, while the downstream preS2/S promoter promotes the transcription of 2.1 kb mRNAs that are translated into the M-HBsAg and S-HBsAg (Siddiqui et al., 1986). The preS1 promoter is negatively regulated by preS2/S promoter, M-HBsAg and S-HBsAg are produced in excess relative to the L-HBsAg (Bulla and Siddiqui, 1989). The HBsAg-containing envelope of HDV comprises of approximately 95% S-HBsAg, 5% M-HBsAg, and 1% L-HBsAg (Bonino et al., 1986).
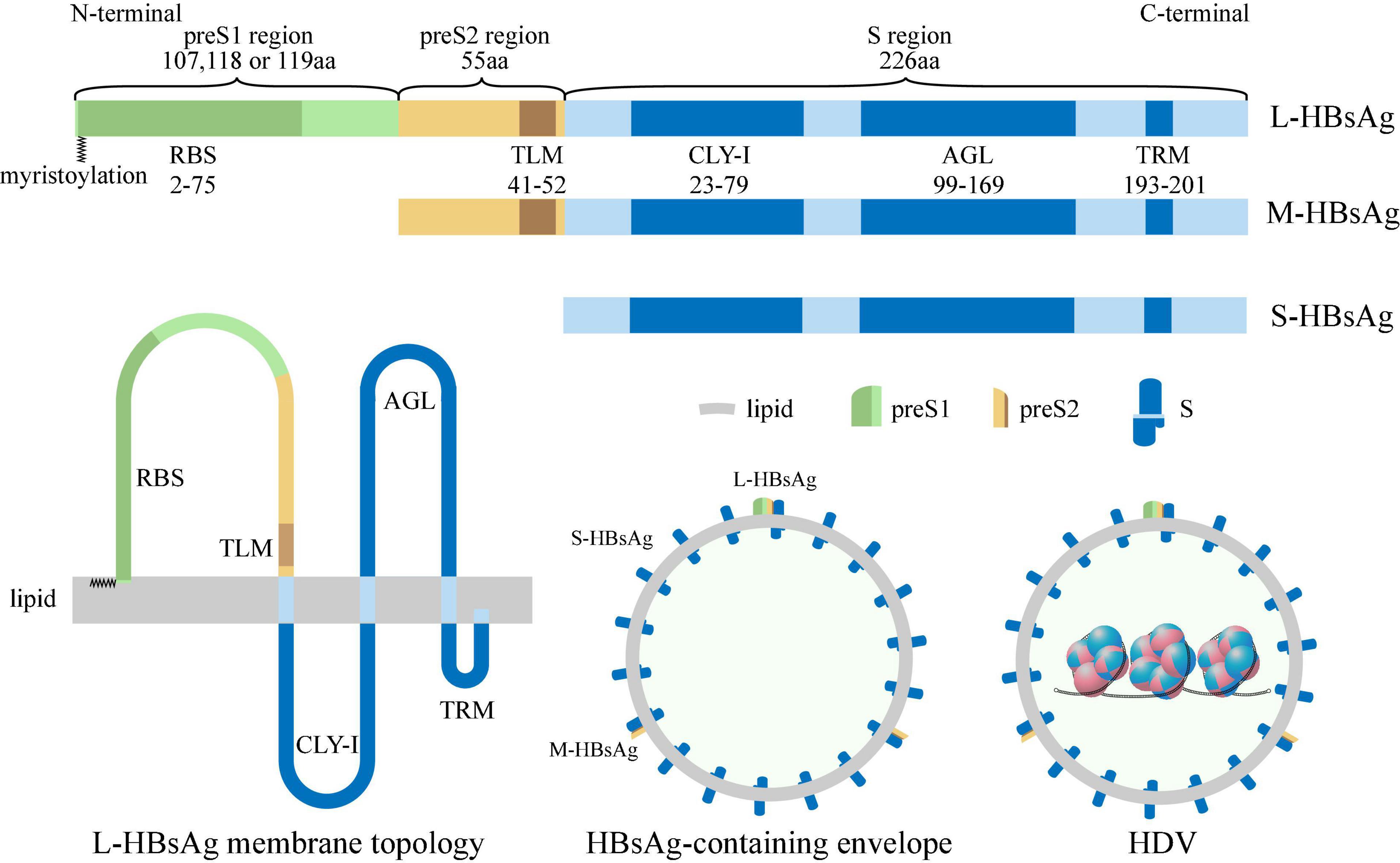
Figure 3. Structure of HBsAg, HBsAg-containing envelope and HDV. S-HBsAg contains S region (blue box), M-HBsAg contains preS2 (yellow box) and S region, and L-HBsAg contains preS1 (green box), preS2 and S region, and the N-terminal of L-HBsAg is myristoylated (zigzag line). Receptor-binding site (RBS) located at preS1 region; Translocation motif (TLM) located at preS2 region; Cytosolic loop I (CYL-I), antigenic loop (AGL), and try-rich motif (TRM) located at S region. The HBsAg-containing envelope of HDV is a host-derived outer surface lipid coat containing three forms of HBV surface antigens.
Also, several critical regions have been identified in HBsAg (Yang et al., 2021), including (i) receptor-binding site (RBS, 2–75 aa of the myristoylated preS1 region), binding to specific receptors during de novo entry (Le Duff et al., 2009); (ii) translocation motif (TLM, 41–52 aa of the preS2 region), driving translocation of HDV RNP across the HBsAg-containing envelope into the cytoplasm during de novo entry (Stoeckl et al., 2006); (iii) cytosolic loop I (CYL-I, 23–79 aa of the S region), which is crucial for envelopment (Löffler-Mary et al., 2000); (iv) antigenic loop (AGL, 99–169 aa of the S region), determining the antigenicity and attaching to the cell membrane during de novo entry (Sureau and Salisse, 2013); (v) Try-rich motif (TRM, 193–201 aa of the S region), binding to HDV RNP during HDV assembly (Komla-Soukha and Sureau, 2006; Figure 3).
The de novo entry of HDV relies on the HBsAg-containing envelope, and the mechanism underlying de novo entry is also similar to that in HBV infection. HDV primarily targets human hepatocytes, and no evidence has shown that HDV naturally infects other cells (Negro et al., 1989). HDV can be experimentally delivered into, and spontaneously replicate in a wide range of non-hepatic cells, such as mouse brain, kidney, testis, skeletal muscle cells (Polo et al., 1995), and Hela cells (Greco-Stewart et al., 2009). This proves that the hepatotropism of HDV is the tropism of de novo entry.
The de novo entry of HDV is mainly divided into three steps, including attachment, recognition, and entry into cells, and these processes rely on S region, preS1 region, preS2 region of HBsAg, respectively. L-HBsAg is necessary for de novo entry as it provides the only source of preS1 region for recognition.
At the beginning of the de novo entry, the AGL on the HBsAg S region, which is exposed on the surface of the HBsAg-containing envelope, attaches to the heparan sulfate proteoglycans (HSPG), which is expressed on the outer face of the hepatocyte membrane (Sureau and Salisse, 2013). One particular HSPG, glypican-5, is an entry factor, but it is not necessary for de novo entry, as its abrogation is insufficient to completely prevent infection (Verrier et al., 2016).
Attaching to low-affinity HSPG is important but not sufficient for HDV to enter into hepatocytes, as it should be further recognized by high-affinity specific receptor [human sodium taurocholate coreceptor peptide (hNTCP)] (Yan et al., 2012). Through interacting with E-cadherin, hNTCP is distributed on the basolateral membrane of differentiated hepatocytes and responsible for bile acid uptake (Hu et al., 2020). The bile acid-binding region of hNTCP interacts with the RBS on the myristoylated N-terminal of L-HBsAg, allowing HDV to enter into hepatocytes (Meier et al., 2013). Unlike dispensable glypican-5, hNTCP is essential for the de novo entry. Knockdown of hNTCP in HepaRG cells via small hairpin RNA prevents them from HDV infection (Ni et al., 2014). Primary hepatocytes of mouse, rat, dog, pig, rhesus, and cynomolgus macaque are resistant to HDV infection, but after being transduced with adeno-associated viral vectors encoding hNTCP, these cells are susceptible to HDV infection (Lempp et al., 2017).
The specific entry mechanism is not yet clear. However, considering that HDV has the same envelope as HBV, it might follow an entry process that is identical or very similar to that used by HBV. After binding with L-HBsAg, hNTCP is markedly oligomerized (Fukano et al., 2018). Then, triggered by epidermal growth factor receptor (Iwamoto et al., 2019), the cell membrane fuses and internalizes virus-hNTCP complex via clathrin-mediated endocytosis pathway (Herrscher et al., 2020). Epidermal growth factor receptor is also internalized in the virus-carried vesicle, triggering the relocation of the vesicle to the Rab5-containing early endosome and then to the Rab7-containing late endosome (Macovei et al., 2013; Iwamoto et al., 2020). Endosomal proteases expose TLM on HBsAg preS2, and the unmasking TLM drives translocation of HBV core particles or HDV RNP across the endosomal membrane into the cytoplasm (Stoeckl et al., 2006), but deleting TLM does not interfere with the infection (Blanchet and Sureau, 2007; Gudima et al., 2007).
After being released into the cytoplasm, HBV core particles seem to be delivered to the nucleus via microtubule-mediated transportation process (Rabe et al., 2006). HDV RNP is also transported to the nucleus, a process driven by NLS on HDAg (Tavanez et al., 2002). Supposedly, entry of a single RNP into the nucleus is sufficient for HDV infection (Macnaughton and Lai, 2006).
Hepatitis delta virus RNA is amplified in the nucleus and the amplification does not require the presence of HBV (Giersch et al., 2019). HDV does not express its RNA-dependent RNA polymerase or use HBV polymerase, rather uses the host’s DNA-dependent RNA polymerase for RNA amplification. Accumulating evidences have indicated that RNA polymerase I (Pol-I) and RNA polymerase II (Pol-II) are associated with the extremities of both polarities of HDV RNA and involved in amplification. Although RNA polymerase III (Pol-III) is also associated with HDV RNA and seems to be involved in amplification, its exact function is not yet clear (Greco-Stewart et al., 2009).
Hepatitis delta virus RNA amplification involves three types of HDV RNA, including genomic RNA (gRNA, ∼1,700 nt), antigenomic RNA (agRNA, ∼1,700 nt), and mRNA (∼800 nt). The copy number of these RNA is approximately 300,000, 60,000, and 600/cell, respectively (Taylor, 2015). Moreover, gRNA and agRNA bind with HDAg to form gRNP or agRNP, respectively. The three steps of RNA amplification are as follows: (i) In the nucleoplasm, Pol-II uses gRNP as a template to transcribe mRNA; (ii) In the nucleolus, Pol-I uses gRNP as a template to replicate agRNA; (iii) In the nucleoplasm, Pol-II uses agRNP as a template to replicate gRNA (Macnaughton and Lai, 2006; Abeywickrama-Samarakoon et al., 2020). In addition, the process of HDV RNA amplification is inseparable from the production of S-HDAg.
In nuclear, the parental gRNP migrates to Pol-II-containing speckles (Chang et al., 2008). Pol-II uses gRNP as a template to transcribe mRNA, which encodes the S-HDAg. HDV mRNA has a 5′ cap and a 3′ poly-A tail and migrates to the cytoplasm through a cell region maintenance 1-independent pathway (Macnaughton and Lai, 2002). Then mRNA is translated to S-HDAg in cytoplasm. Mediated by CCS and NLS, S-HDAg oligomerizes and re-enters the nucleus through the nuclear pore complex (Xia et al., 1992).
After enough mRNA are transcribed, parental gRNP migrates to the nucleolus for agRNA replication. Although the migration machinery is not yet clear, it seems to be related to the newly synthesized S-HDAg (Macnaughton and Lai, 2006). Subsequently, Pol-I in the nucleolus uses the parental gRNP as a template, through the rolling-circle mechanism (Negro et al., 1989), to replicate linear agRNA polymers that are complementary to the gRNA. HDV antigenome strand ribozyme cleaves linear agRNA monomers from the linear agRNA polymers (Wadkins et al., 1999). Then, linear agRNA monomers are self-ligated into circular agRNA molecules. The self-ligation might be catalyzed by HDV RNA ribozyme or cell ligase (Reid and Lazinski, 2000).
The structure of circular agRNA is similar to gRNA, it wraps around the newly synthesized HDAg multimers to form agRNP, and migrates to Pol-II-containing nuclear speckles. Pol-II uses agRNP as a template, through the rolling-circle mechanism, to replicate linear gRNA polymers. HDV genome strand ribozyme cleaves linear gRNA monomers from linear gRNA polymers. The linear gRNA monomers are self-ligated into circular gRNA molecules and wrap around the HDAg multimers to become the progeny gRNP.
According to the amplification process of HDV RNA, the progeny gRNP works as parental gRNP to amplify more mRNA or agRNA, or leaves the nucleus to form progeny virus.
As mentioned above, the amplification of HDV RNA is accompanied by the production of S-HDAg. Notably, after translation from mRNA, S-HDAg requires posttranslational modification, which is critical for its function. These critical modifications include: (i) Arginine methyltransferase catalyzes methylation at Arg-13, which is essential for RNA replication, affecting HDAg subcellular localization (Li et al., 2004); (ii) Acetyltransferase p300 catalyzes acetylation at Lys-72, which modulates HDAg subcellular localization and may be involved in viral RNA nucleocytoplasmic shuttling and replication (Mu et al., 2004); (iii) Extracellular signal-related kinases 1 and 2 catalyze phosphorylation at Ser-177, which mediates RNA replication and RNA edition (mention in 3.5.1) (Mu et al., 2001; Chen et al., 2008); (iv) Sumoylation at multiple lysine residues, enhancing RNA replication (Tseng et al., 2010).
After binding to HDV RNA, S-HDAg further interacts with several components of the host cell: (i) Pol-II. S-HDAg interacts with 9/12 subunits of Pol-II (Cao et al., 2009), the combination and loosening between S-HDAg and Pol-II clamp accelerate RNA elongation speed at the cost of fidelity (Yamaguchi et al., 2007). Except that, S-HDAg displaces negative elongation factor to promote elongation (Yamaguchi et al., 2001); (ii) Histone. S-HDAg N-terminal 2–67 aa bind to histone H1e central globular domain, and H1e plays a significant role in HDV replication (Lee and Sheu, 2008); (iii) Transcription factor. S-HDAg RBD and a GA/GK-rich region interact with transcription factor Yin Yang 1, and further bind to Yin Yang 1 associated acetyltransferases CBP and p300, these factor and transferases enhance HDV RNA replication (Huang et al., 2008); (iv) Nucleolar phosphoprotein. N-terminal domain of both forms of HDAg interacts with nucleolar phosphoprotein B23 (Huang et al., 2001) and nucleolin (Lee et al., 1998) to target nucleolus and modulate HDV replication; (v) Chromatin remodeling factor. S-HDAg contains a short linear interacting motif KacXXR, which binds with a chromatin remodeling factor, bromodomain adjacent to zinc finger domain 2B protein, to activate Pol-II-dependent HDV RNA amplification and sustain HDV replication (Abeywickrama-Samarakoon et al., 2020).
In the Pol-II-dependent mRNA and gRNA amplification, S-HDAg mainly activates RNA amplification and promotes RNA elongation. In the Pol-I dependent agRNA replication, S-HDAg stabilizes the agRNA, protects it from nuclease decomposition, and promotes its cleavage and self-ligation (Harichandran et al., 2019).
Hepatitis delta virus RNA amplification requires S-HDAg, while L-HDAg prevents HDV RNA amplification and is essential for virus assembly. After accumulating some S-HDAg in cells, HDV RNA will be edited to produce L-HDAg. The newly synthesized L-HDAg is also posttranslational modified.
A family of proteins called adenosine deaminases acting on RNA (ADAR) catalyzes the adenosine (A) to inosine (I) editing reaction on dsRNA (Bass, 2002). HDV RNA editing is catalyzed by Adenosine deaminases acting on RNA 1 (ADAR1), which converts UAG triplet of agRNA into UIG triplet, and the original agRNA is converted into edited-agRNA (Figure 4). Before editing, the adenosine (A) mismatch with a cytosine (C), the editing converts the A-C mismatch pair into I-C match pair, which is a stabler structure. Currently, there is no known method to reverse this editing during infection (Casey, 2012). Presumably, the newly synthesized agRNA is available to be edited only before it binds to HDAg (Sato et al., 2004).
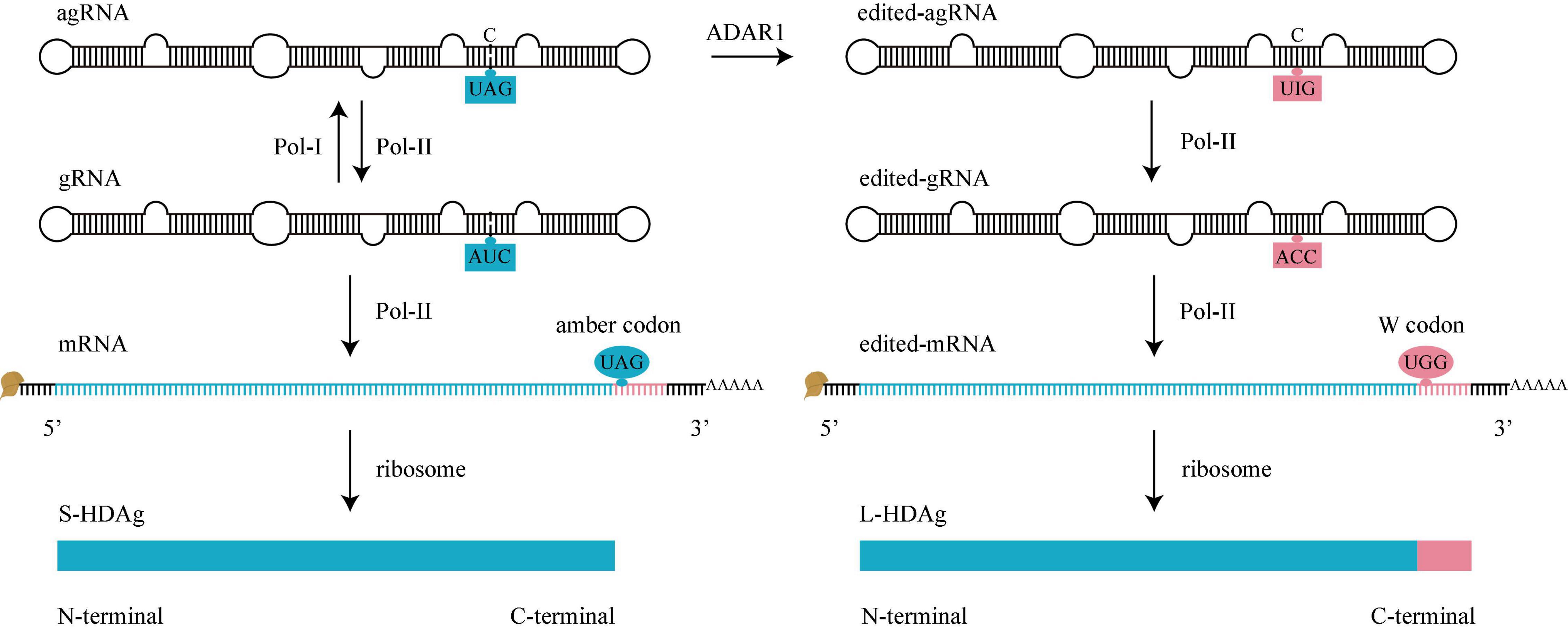
Figure 4. HDV RNA editing. Before editing (left), there is an amber codon (UAG, blue oval) at amber/W site of mRNA, translation will stop here to produce S-HDAg. During editing, adenosine deaminases acting on RNA 1 (ADAR1) converts UAG of agRNA to UIG (right). When Pol-II use these edited-agRNA as a template to replicate edited-gRNA, and use edited-gRNA as a template to transcribe edited-mRNA, there is a W codon (UGG, pink oval) at amber/W site of edited-mRNA, the translation won’t stop until next termination codon is met, producing an L-HDAg. gRNA, genomic RNA. agRNA, antigenomic RNA.
When Pol-II uses the edited-agRNA as a template, it replicates an ACC triplet on edited-gRNA. Pol-II further uses the edited-gRNA as a template to transcribe a W codon (UGG) on edited-mRNA. The edited-gRNA is defective for edited-agRNA replication (Sheu, 2002; Figure 4).
As mentioned above, there is a variable amber/W site in mRNA. For original mRNA, the site contains an amber codon (UAG, termination codon) and the translation stops here to produce an S-HDAg; For edited-mRNA, the site contains a W codon (UGG, encodes tryptophan), the translation doesn’t stop here, keeps adding the C-terminal 19 amino acids of L-HDAg until the next termination codon (Casey, 2012).
Adenosine deaminases acting on RNA 1 regulates the HDV life cycle by converting the early stage of HDV RNA amplification (mediated by S-HDAg) into the later stage of virus assembly (mediated by L-HDAg). Some genotypes of HDV (for example, HDV-6 and 8) do not display an A-C mismatch pair at the editing site, thereby use alternative transient conformation to increase the binding efficiency of ADAR1 (Dziri et al., 2021).
Phosphorylation, acetylation, methylation, and sumoylation are posttranslational modifications of S-HDAg that also occur in the N-terminal identity region (1–195 aa) of L-HDAg. Additionally, the C-terminal extension region (196–214 aa) of L-HDAg requires farnesylation. The last four amino acids at the C-terminal (211–214 aa) of L-HDAg are Cys-Arg/Thr-Pro/Gln-Gln (C-R/T-P/Q-Q), which constitute a farnesylation signal (Huang et al., 2006). The farnesyl lipid group is added covalently to Cys-211 by the host’s farnesyl transferase, generating farnesylated L-HDAg.
Nuclear localization signal drives both farnesylated and non-farnesylated L-HDAg to enter into nucleus through the nuclear pore complex and the positively charged RBD drive them bind to HDV RNA. Nonetheless, the farnesyl group on L-HDAg is necessary for the combination between HDV RNP and HBsAg-containing envelope (Hwang and Lai, 1993), and is also required in HDV RNA amplification inhibition (Sato et al., 2004).
After the start of RNA editing, the amount of L-HDAg in the nucleus increases in a time-dependent manner until the balance between the two HDAg is achieved. A small amount of L-HDAg is sufficient to inhibit the replication of gRNA. When L-HDAg exceeds S-HDAg, it inhibits the amplification of agRNA and mRNA (Modahl and Lai, 2000).
As the amount of L-HDAg increases in the nucleus, the proportion of L-HDAg monomers on the newly synthesized HDV gRNP is also increased. Considering the number of S-HDAg and L-HDAg are equivalent in circulating HDV (Bergmann and Gerin, 1986), we speculate that when the number of two HDAg is almost equal in a single HDV RNP, L-HDAg NES becomes the dominant signal, driving the egress of HDV gRNP from the nucleus through the nuclear pore complex (Lee et al., 2001). S-HDAg and L-HDAg cannot leave the nucleus in the absence of HDV RNA, presumably because binding to the RNA is required to expose L-HDAg NES (Tavanez et al., 2002).
Experiments have shown that despite the binding to HDAg, agRNA cannot leave the nucleus, most of the agRNA remains in the nucleus, but the amount of HDV gRNA in the cytoplasm and nucleus is almost equal (Macnaughton and Lai, 2002). However, edited-gRNA is exported from the nucleus and is eventually detected in the patient’s plasma (Homs et al., 2016; Sopena et al., 2018; Dziri et al., 2021). After leaving the nucleus, driven by farnesyl group on L-HDAg, HDV gRNP and edited-gRNP (but not agRNP) anchor to the endoplasmic reticulum membrane, where HBsAg is produced (Sureau and Negro, 2016).
After assembling with HBsAg-containing envelope, HDV is secreted via HBV budding mechanism. As helper virus, HBV only mediates HDV de novo entry and secretion.
In HBV mono-infection, HBsAg is overexpressed in the endoplasmic reticulum and partially buds to the multivesicular body to bind to the HBV core particles (Niklasch et al., 2021). Mature HBV particles contain an equimolar proportion of the three forms of HBsAg (Sureau et al., 1993) and are secreted through the endosomal sorting complex required for transport-dependently multivesicular body (ESCRT/MVB) pathway (Lambert et al., 2007; Watanabe et al., 2007). The remaining excess of HBsAg is dimerized by protein disulfide isomerase in the endoplasmic reticulum compartment, from where it migrates to the pre-Golgi membrane as transmembrane dimer and self-assembles to form subviral particle (SVP) (Patient et al., 2007). These SVPs are 22 nm diameter spheres, composed of S-HBsAg and small amount of M-HBsAg, and do not contain viral genome (Sureau et al., 1993). After assembly, they are secreted through the Golgi apparatus classic constitutive secretory pathway (Patient et al., 2007). Since there is no L-HBsAg in sphere-SVPs, they cannot compete with HBV for hNTCP receptors. Via ESCRT/MVB pathway, Some HBsAg self-assembles to form filament-SVPs, which contain a much larger amount of the L-HBsAg compared to sphere-SVPs, and are secreted through ESCRT/MVB pathway like infectious viral particles (Jiang et al., 2015). The titer of SVP (1012–1013/mL) in serum of chronic hepatitis B patients is much higher than infectious HBV particle (108–109/mL) (Sureau and Negro, 2016), which is considered a strategy for HBV to evade the immune response.
The envelope composition of HDV is similar to sphere-SVPs, S-HBsAg accounts for about 94%, and M-HBsAg accounts for about 5%. However, unlike sphere-SVPs that do not contain L-HBsAg, HDV envelope contains about 1% L-HBsAg (Bonino et al., 1986). A small number of L-HBsAg (about 3-4/virus) is sufficient for HDV infectivity (Le Duff et al., 2009). At the onset of acute infection, the titer of HDV is 1010–1011/mL serum (Sureau and Negro, 2016). Considering that the titer of circulating HDV is higher than HBV, approaching of SVPs, and the composition of HDV envelope is similar to sphere-SVPs, it could be speculated that HDV uses the sphere-SVPs secretion pathway, Golgi apparatus classic constitutive secretory pathway for its secretion.
When HDV RNP combines with HBsAg, the assembly signal of HDV RNP is located at the L-HDAg C-terminal farnesyl group (O’Malley and Lazinski, 2005), and the assembly signal of HBsAg is located at the TRM on the S region C-terminal matrix domain (Komla-Soukha and Sureau, 2006). The aromatic residues W196, W199, and W201, essential for TRM assembly capacity, are conserved in the envelope protein of the orthohepadnavirus (Komla-Soukha and Sureau, 2006). Owing to evolutionary conservation, other orthohepadnaviruses [such as woodchuck hepatitis virus (Lukash et al., 2019), the woolly monkey HBV (Barrera et al., 2004), and the tent-making bat HBV (Drexler et al., 2013)] also provide helper functions to HDV experimentally, except the envelope protein of duck hepatitis B virus as it lacks an HDV matrix domain (Sureau and Negro, 2016).
Without M-HBsAg and L-HBsAg expression, S-HBsAg drives HDV assembly and secretion, but the HDV only enveloped by S-HBsAg is not infectious (Sureau et al., 1993). L-HBsAg drives infection, and S-HBsAg drives assembly and secretion, while M-HBsAg may be dispensable (Sureau et al., 1994). Therefore, the infectivity of CHD patient’s serum may not be proportional solely to its HDV titer, but also affected by the number of L-HBsAg on HDV (Sureau et al., 1993). Except for the number of HDV and L-HBsAg, the genotypes of HBV and HDV also affect the infectivity. In vitro tests have shown that the genotype of HBV determines the number of HDV infected cells and the number of HDV genomes that successfully start replication after entry (Freitas et al., 2014). In addition, the combination of different HBV and HDV genotypes determine the efficacy of HDV egress and entry (Wang et al., 2021).
After co-transfecting plasmids encoding HDV and E1E2 (hepatitis C virus (HCV) envelope protein) into HCV host cells, HDV replicates and binds to E1E2, then is secreted and enters into other cells via E1E2 (Perez-Vargas et al., 2019). But clinically, HDV proliferation can’t rely on HCV instead of HBV, there are very few HBV DNA-negative, HCV RNA-positive, and HDV RNA-positive patients (Chemin et al., 2021; Roggenbach et al., 2021). These few cases could be ascribed to occult HBV infection (Raimondo et al., 2019).
Without HBsAg expression, HDV persists in humanized mice for at least 6 weeks, leading to host cell detachment and death due to rapid virus replication, or be converted by HBV infection to develop coinfection (Giersch et al., 2014). Cell death may lead to cell lysis with exposure of cell debris and newly synthesized HDV RNP, but the unenveloped HDV RNP cannot invade the host cell. Except for disappearance and conversion, HDV spreads through hepatocyte proliferation (Giersch et al., 2019). Recently, we have discovered HDV-like agents in several taxa (termite, fish, toad, newt, snake, bird, woodchuck, rat, bat, and deer), and these agents lack the association with a respective animal hepadnavirus (Wille et al., 2018; Chang et al., 2019; Hetzel et al., 2019; Paraskevopoulou et al., 2020; Bergner et al., 2021; Iwamoto et al., 2021). This implies that HDV may not be a satellite virus initially, and the adaptation of HDV to HBsAg was an occasional and recent evolutionary event that provides HDV hepatotropism and accelerates the spread of HDV in humans (Netter et al., 2021; Zhang and Urban, 2021).
The entire life cycle of HDV (de novo entry, RNA amplification, HDAg production, assembly, and secretion) and the regions that drive each step are illustrated in Figure 5.
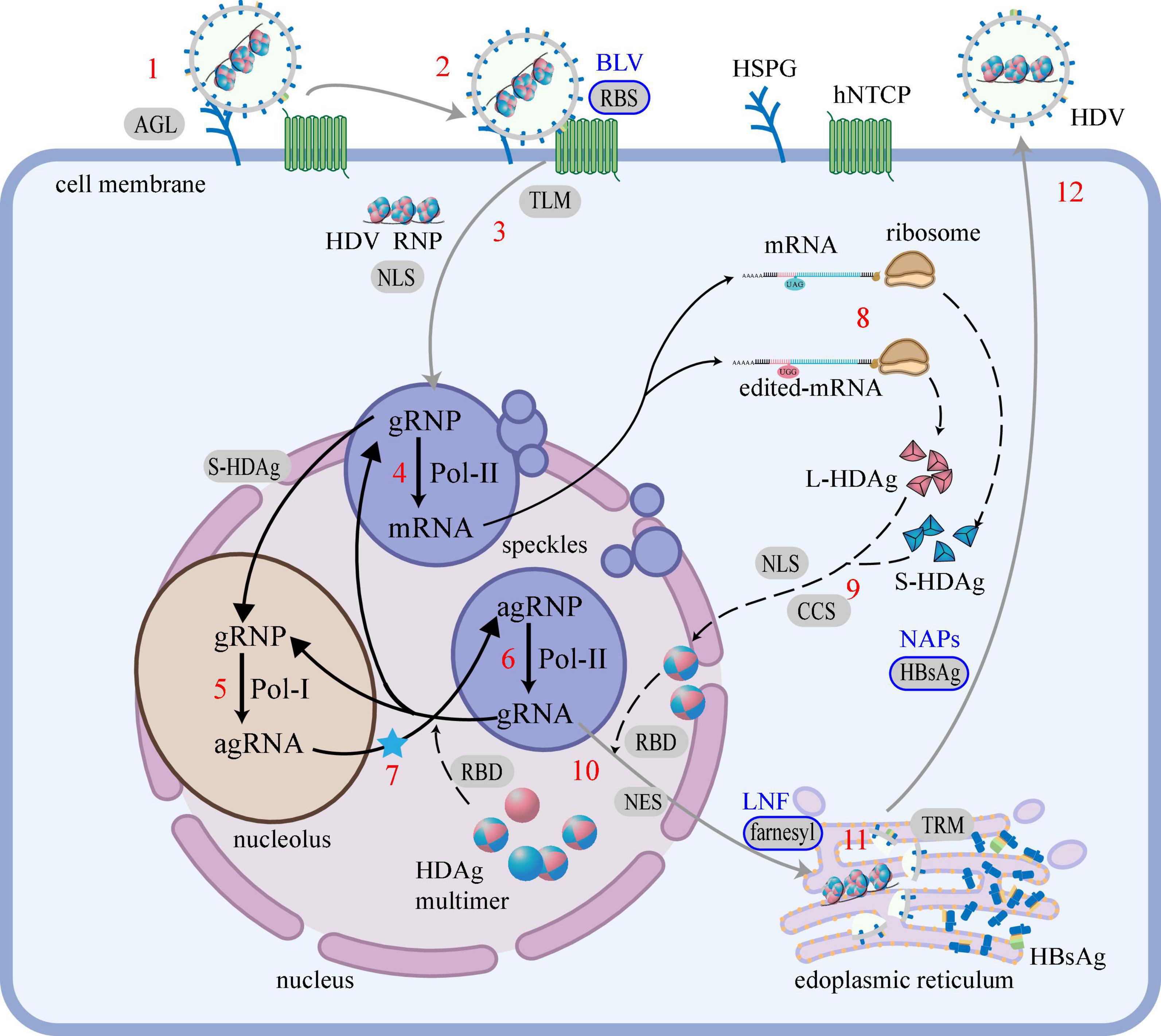
Figure 5. HDV life cycle and the regions that drive each step. The life cycle of HDV is mainly divided into 5 steps: (i) De novo entry, (1) attachment; (2) recognition; (3) entry into cells; (ii) RNA amplification, (4) mRNA transcription; (5) agRNA replication; (6) gRNA replication; (7) RNA editing (blue star); (iii) HDAg production, (8) translation; (9) modification; (iv) Assembly, (10) HDV RNP assembly; (11) HDV particles assembly; (v) Secretion, (12) secretion. Black words mark cell or virus components, red numbers mark life cycle steps, words on the gray background mark the regions that drive the step and the blue border marks the regions that have been applicated to HDV therapy, blue words mark the corresponding therapies. The gray line represents the migration of the virus, the solid black line represents the migration of the RNA, and the black dashed line represents the migration of the protein. Regions of HDAg: RBD, RNA-binding domain; CCS, Coiled-coil sequence; NLS, Nuclear localization signal; NES, Nuclear export signal; farnesyl, farnesyl lipid group. Regions of HBsAg: RBS, Receptor-binding site; TLM, Translocation motif; AGL, Antigenic loop; TRM, Try-rich motif. Cell or virus components: HSPG, heparan sulfate proteoglycans; hNTCP, human sodium taurocholate coreceptor peptide; gRNA, genomic RNA; agRNA, antigenomic RNA.
Hepatitis delta virus replication triggers host cell to produce IFN-β and IFN-λ (Zhang et al., 2018), which are secreted out of the cell. Both secreted IFNs and administered IFNs are recognized by cognate receptors [such as interferon alpha receptor (IFNAR), interferon lambda receptor (IFNLR), and interleukin 10 receptor (IL10R)] on the surface of cells. The recognition induces IFN-stimulated genes, which inhibit viral replication, lower the cellular permissiveness, and activate the adaptive immune response (Zhang and Urban, 2020, 2021).
Interferon-α and Interferon-β belong to type I IFN, and they activate IFN-stimulated genes after being recognized by IFNAR1/IFNAR2, which are ubiquitously expressed on eukaryotic cells membrane (Sommereyns et al., 2008). Compared to standard IFN-α, pegylated IFN-α (Peg-IFN-α) has a longer plasma half-life, allowing a once-a-week administration, better compliance and efficiency. A meta-analysis showed that after administration of standard IFN-α for 12–48 weeks and 24–72 weeks of treatment-free follow-up, 17.4% of CHD patients had undetectable HDV RNA (Abbas et al., 2011); while after administration of Peg-IFN-α for 48 and 24 weeks of treatment-free follow-up, 31.0% of CHD patients had undetectable HDV RNA (Wedemeyer et al., 2011). Both standard IFN-α and Peg-IFN-α lead to frequent and sometimes severe adverse reactions, such as flu-like symptoms, asthenia, weight loss, alopecia, thrombocytopenia, and leukopenia (Abbas et al., 2011; Wedemeyer et al., 2011).
Interferon-λ is a type III IFN that activates IFN-stimulated genes after being recognized by IFNLR1/IL10R2, which are preferentially expressed in the epithelial tissues of the lung, liver, and gut (Sommereyns et al., 2008). The specificity of receptor expression imparts IFN-λ improved liver targeting and higher tolerability than IFN-α, resulting in fewer systemic adverse reactions and comparable antiviral efficacy. The common on-treatment adverse reactions included flu-like symptoms and elevated transaminase levels (Etzion et al., 2019). In the humanized mouse model, standard IFN-λ effectively reduced all intrahepatic markers of HDV infection (Giersch et al., 2017). After administration of Peg-IFN-λ for 48 and 24 weeks of treatment-free follow-up, 35.7% of CHD patients had undetectable HDV RNA (Etzion et al., 2019).
Bulevirtide (formerly known as Myrcludex B) is a myristoylated synthetic lipopeptide, with the same amino acids sequence as the N-terminal 47 aa in L-HBsAg RBS. Via binding to hNTCP, Bulevirtide competes with L-HBsAg RBS and inhibits the entry of HBV and HDV into hepatocytes. The effect of Bulevirtide monotherapy is not satisfactory, after administration of Bulevirtide for 48 weeks, only 13.3% of CHD patients had undetectable HDV RNA. However, the effect of administrating Bulevirtide with other drugs is excellent. When Bulevirtide and Peg-IFN-α are administered in combination for 48 weeks, 80.0% of CHD patients had undetectable HDV RNA, and after an additional 24 weeks of treatment-free follow-up, still, 53.3% of CHD patients had undetectable HDV RNA. Hitherto, Bulevirtide is administered daily by subcutaneous injections; oral preparations are still under development. The drug is generally well-tolerated, and the most frequent adverse reactions are increased total bile salts (asymptomatic and reversible after the treatment is discontinued) and injection site reactions (Kang and Syed, 2020).
Lonafarnib is a farnesyl transferase inhibitor that prevents the farnesylation of L-HDAg, thus blocking the combination between HDV RNP and HBsAg. Lonafarnib should be administered with other antiviral drugs to achieve the best effect. When Lonafarnib and Ritonavir (RTV) are administered in combination for 24 weeks, 38.5% of CHD patients had undetectable HDV RNA (Lok et al., 2021). When Lonafarnib, Ritonavir, and Peg-IFN-λ are administered in combination for 24 weeks, 50.0% of CHD patients had undetectable HDV RNA, and after another 24 weeks of treatment-free follow-up, 22.7% of CHD patients still had undetectable HDV RNA. Lonafarnib is associated with significant adverse reactions, such as vomiting, diarrhea, infection, nausea, decreased appetite, fatigue, and upper respiratory tract infections (Dhillon, 2021).
Nucleic acid polymers (NAPs) are phosphorothioated oligonucleotides, which means in every phosphodiester linkage of the oligonucleotides, one of non-bridging oxygen atom is replaced by a sulfur atom. In local hydrophobic microenvironment, the free electron in the non-bridging sulfur atom can transit from the unshared (charged) state to a shared (uncharged) state with the central phosphorous atom. The transition ability of the sulfur atom allows NAPs to adopt a localized hydrophobic property along their long axis and form a hydrophobic surface, the rest part of the NAPs forms a hydrophilic surface, altogether confer NAPs amphipathic property (Vaillant, 2019).
The host protein interaction with NAPs that drives their antiviral effects is not yet clear. Current study suggests via folding together of the hydrophobic surfaces, amphipathic NAPs bind amphipathic α-helices in class I viral glycoproteins, which are present on the surface of many enveloped viruses (Blanchet et al., 2019). This may explain why NAPs possess broad spectrum antiviral ability (Vaillant et al., 2006; Cardin et al., 2009). NAPs inhibit secretion of HBsAg from hepatocytes and block replenishment of HBsAg in the circulation, allowing host mediated clearance. Unlike nucleoside/nucleotide analogs that possess antiviral activity in HBV infection but are not effective against HDV infection (Yurdaydin et al., 2008; Abbas et al., 2016), NAPs are effective therapy for HDV infection.
REP 2139-Ca as a kind of NAPs has been administrated to CHD patients in the REP301 and REP 301-LTF study. After administration of REP 2139-Ca for 15 weeks, followed by a combination of REP 2139-Ca and Peg-IFN-α for 15 weeks, 83.3% of CHD patients had undetectable HDV RNA. Continuing to administrate Peg-IFN-α for 33 weeks, 75.0% of CHD patients had undetectable HDV RNA (Bazinet et al., 2017). After another 18 months of treatment-free follow-up, 58.3% of CHD patients still had undetectable HDV RNA (Bazinet et al., 2018). The most frequent adverse reactions observed during REP 2139-Ca monotherapy were pyrexia and chills, and less frequently conjunctival hyperemia, headache, and asthenia near the end of infusion (Bazinet et al., 2017).
The detail clinical data of the above-mentioned four types of drugs are summarized in Table 2.
Hepatitis delta virus is a robust pathogen that spreads widely. Since an ideal therapy is yet unavailable, in-depth exploration is essential. The present review summarizes the effects of the specific regions of HDAg and HBsAg on HDV life cycle: (i) De novo entry. After attaching to HSPG via AGL, HDV is recognized by hNTCP via RBS to enter into hepatocyte. In cytoplasm, HDV RNP is released via TLM, and enters into nucleus via NLS; (ii) RNA amplification. S-HDAg activates the amplification of mRNA and gRNA, stabilizes agRNA and promotes its cleavage and self-ligation; (iii) HDAg production. After modification, mediated by CCS and NLS, HDAg oligomerizes and re-enters the nucleus; (iv) Assembly. HDV RNA wraps around HDAg multimers to form HDV gRNP via RBD, NES drives the egress of HDV gRNP from the nucleus. After leaving the nucleus, HDV gRNP anchors to the endoplasmic reticulum membrane via farnesyl group, and binds to HBsAg-containing envelope via the interaction between TRM and farnesyl group to form HDV; (v) Secretion. Driven by HBsAg-containing envelope, HDV secretes via the Golgi apparatus classic constitutive secretory pathway.
Some regions have been applicated for HDV therapy: (i) RBS: Bulevirtide is an L-HBsAg RBS analog, competing with RBS for specific receptor hNTCP to block HDV entry; (ii) Farnesyl group: Lonafarnib is a farnesyl transferase inhibitor, preventing the farnesylation of L-HDAg, thus blocking the combination between HDV RNP and HBsAg; (iii) HBsAg: NAPs are phosphorothioated oligonucleotides that bind to amphipathic α-helices of HBsAg, preventing the secretion of HBsAg. Except these three regions, several other regions are potential therapeutic targets. For example, blocking NLS or NES of HDAg will prevent HDAg nucleocytoplasmic shuttling, blocking CCS or RBD of HDAg will prevent the assembly among HDAg or between HDAg and RNA, and blocking TRM of HBsAg will prevent the assembly between HDV RNP and HBsAg-containing envelope.
There are still several mechanisms underlying the HDV life cycle are yet ill-understood, including but not limited to the following: the assembly structure of HDV RNA and HDAg; similarities and differences between HBV and HDV during the de novo entry; key factors driving HDV to remove the HBsAg-containing envelope and expose HDV RNP; method to change the HDV RNP dominant signal from farnesyl group (anchor to endoplasmic reticulum) in assembly stage to NLS (enter into the nucleus) in new round of the de novo entry; whether and how Pol-III participates in the amplification of HDV RNA; the domains or signals of S-HDAg involved in the migration of gRNP from the nucleoplasm to the nucleolus during RNA amplification; enzymes catalyzing the connection of HDV RNA linear monomers to form circular monomers. Investigating HDV proliferation mechanisms and the functions of these specific regions on HDAg and HBsAg will help us to discover new treatment strategies and develop new drugs associated with better therapeutic effects, fewer adverse reactions, and lower recurrence rates.
JZ: data curation, formal analysis, writing-original draft, and writing-review and editing. XG: data curation, writing-original draft, and writing-review and editing. JD: writing-review and editing. HX: formal analysis, validation, and writing-review and editing. JN: funding acquisition, supervision, and writing-review and editing. XC: data curation, supervision, validation, and writing-review and editing. All authors contributed to the article and approved the submitted version.
The present study was supported by the National Natural Science Foundation of China (grant no. 81970519).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Xiaomei Wang and Chang Wang for their valuable comments on this manuscript.
Abbas, Z., Khan, M. A., Salih, M., and Jafri, W. (2011). Interferon alpha for chronic hepatitis D. Cochrane Database Syst. Rev. 2011:Cd006002. doi: 10.1002/14651858.CD006002.pub2
Abbas, Z., Memon, M. S., Umer, M. A., Abbas, M., and Shazi, L. (2016). Co-treatment with pegylated interferon alfa-2a and entecavir for hepatitis D: a randomized trial. World J. Hepatol. 8, 625–631. doi: 10.4254/wjh.v8.i14.625
Abeywickrama-Samarakoon, N., Cortay, J. C., Sureau, C., Müller, S., Alfaiate, D., Guerrieri, F., et al. (2020). Hepatitis Delta virus histone mimicry drives the recruitment of chromatin remodelers for viral RNA replication. Nat. Commun. 11:419. doi: 10.1038/s41467-020-14299-9
Barrera, A., Guerra, B., Lee, H., and Lanford, R. E. (2004). Analysis of host range phenotypes of primate hepadnaviruses by in vitro infections of hepatitis D virus pseudotypes. J. Virol. 78, 5233–5243. doi: 10.1128/jvi.78.10.5233-5243.2004
Bass, B. L. (2002). RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 71, 817–846. doi: 10.1146/annurev.biochem.71.110601.135501
Bazinet, M., Pântea, V., Cebotarescu, V., Cojuhari, L., Jimbei, P., Albrecht, J., et al. (2017). Safety and efficacy of REP 2139 and pegylated interferon alfa-2a for treatment-naive patients with chronic hepatitis B virus and hepatitis D virus co-infection (REP 301 and REP 301-LTF): a non-randomised, open-label, phase 2 trial. Lancet Gastroenterol. Hepatol. 2, 877–889. doi: 10.1016/s2468-1253(17)30288-1
Bazinet, M., Pantea, V., Cebotarescu, V., Cojuhari, L., Jimbei, P., and Vaillant, A. (2018). Establishment of persistent functional remission of HBV and HDV infection following REP 2139 and pegylated interferon alpha 2a therapy in patients with chronic HBV/HDV co-infection: 18 month follow-up results from the REP 301-LTF study. J. Hepatol. 68:S509. doi: 10.1016/s0168-8278(18)31266-2
Bergmann, K. F., and Gerin, J. L. (1986). Antigens of hepatitis delta virus in the liver and serum of humans and animals. J. Infect. Dis. 154, 702–706. doi: 10.1093/infdis/154.4.702
Bergner, L. M., Orton, R. J., Broos, A., Tello, C., Becker, D. J., Carrera, J. E., et al. (2021). Diversification of mammalian deltaviruses by host shifting. Proc. Natl. Acad. Sci. U S A. 118:e2019907118. doi: 10.1073/pnas.2019907118
Blanchet, M., Sinnathamby, V., Vaillant, A., and Labonté, P. (2019). Inhibition of HBsAg secretion by nucleic acid polymers in HepG2.2.15?cells. Antiviral Res. 164, 97–105. doi: 10.1016/j.antiviral.2019.02.009
Blanchet, M., and Sureau, C. (2007). Infectivity determinants of the hepatitis B virus pre-S domain are confined to the N-terminal 75 amino acid residues. J. Virol. 81, 5841–5849. doi: 10.1128/jvi.00096-07
Bonino, F., Heermann, K. H., Rizzetto, M., and Gerlich, W. H. (1986). Hepatitis delta virus: protein composition of delta antigen and its hepatitis B virus-derived envelope. J. Virol. 58, 945–950. doi: 10.1128/jvi.58.3.945-950.1986
Bonino, F., Hoyer, B., Ford, E., Shih, J. W., Purcell, R. H., and Gerin, J. L. (1981). The delta agent: HBsAg particles with delta antigen and RNA in the serum of an HBV carrier. Hepatology 1, 127–131. doi: 10.1002/hep.1840010207
Botelho-Souza, L. F., Vasconcelos, M. P. A., Dos Santos, A. O., Salcedo, J. M. V., and Vieira, D. S. (2017). Hepatitis delta: virological and clinical aspects. Virol. J. 14:177. doi: 10.1186/s12985-017-0845-y
Bruss, V., Hagelstein, J., Gerhardt, E., and Galle, P. R. (1996). Myristylation of the large surface protein is required for hepatitis B virus in vitro infectivity. Virology 218, 396–399. doi: 10.1006/viro.1996.0209
Bulla, G. A., and Siddiqui, A. (1989). Negative regulation of the hepatitis B virus pre-S1 promoter by internal DNA sequences. Virology 170, 251–260. doi: 10.1016/0042-6822(89)90373-5
Cao, D., Haussecker, D., Huang, Y., and Kay, M. A. (2009). Combined proteomic-RNAi screen for host factors involved in human hepatitis delta virus replication. RNA 15, 1971–1979. doi: 10.1261/rna.1782209
Cardin, R. D., Bravo, F. J., Sewell, A. P., Cummins, J., Flamand, L., Juteau, J. M., et al. (2009). Amphipathic DNA polymers exhibit antiviral activity against systemic murine cytomegalovirus infection. Virol. J. 6:214. doi: 10.1186/1743-422x-6-214
Casey, J. L. (2012). Control of ADAR1 editing of hepatitis delta virus RNAs. Curr. Top. Microbiol. Immunol. 353, 123–143. doi: 10.1007/82_2011_146
Chang, J., Nie, X., Chang, H. E., Han, Z., and Taylor, J. (2008). Transcription of hepatitis delta virus RNA by RNA polymerase II. J. Virol. 82, 1118–1127. doi: 10.1128/jvi.01758-07
Chang, W. S., Pettersson, J. H., Le Lay, C., Shi, M., Lo, N., Wille, M., et al. (2019). Novel hepatitis D-like agents in vertebrates and invertebrates. Virus Evol. 5:vez021. doi: 10.1093/ve/vez021
Chemin, I., Pujol, F. H., Scholtès, C., Loureiro, C. L., Amirache, F., Levrero, M., et al. (2021). Preliminary evidence for hepatitis delta virus exposure in patients who are apparently not infected with hepatitis B virus. Hepatology 73, 861–864. doi: 10.1002/hep.31453
Chen, Y. S., Huang, W. H., Hong, S. Y., Tsay, Y. G., and Chen, P. J. (2008). ERK1/2-mediated phosphorylation of small hepatitis delta antigen at serine 177 enhances hepatitis delta virus antigenomic RNA replication. J. Virol. 82, 9345–9358. doi: 10.1128/jvi.00656-08
Drexler, J. F., Geipel, A., König, A., Corman, V. M., van Riel, D., Leijten, L. M., et al. (2013). Bats carry pathogenic hepadnaviruses antigenically related to hepatitis B virus and capable of infecting human hepatocytes. Proc. Natl. Acad. Sci. U S A. 110, 16151–16156. doi: 10.1073/pnas.1308049110
Dziri, S., Rodriguez, C., Gerber, A., Brichler, S., Alloui, C., Roulot, D., et al. (2021). Variable in vivo hepatitis D Virus (HDV) RNA editing rates according to the HDV genotype. Viruses 13:1572. doi: 10.3390/v13081572
Etzion, O., Hamid, S. S., Lurie, Y., Gane, E., Bader, N., Yardeni, D., et al. (2019). End of study results from LIMT HDV study: 36% durable virologic response at 24 weeks post-treatment with pegylated interferon lambda monotherapy in patients with chronic hepatitis delta virus infection. J. Hepatol. 70, E32–E32. doi: 10.1016/s0618-8278(19)30058-1
Fattovich, G., Giustina, G., Christensen, E., Pantalena, M., Zagni, I., Realdi, G., et al. (2000). Influence of hepatitis delta virus infection on morbidity and mortality in compensated cirrhosis type B the European concerted action on viral hepatitis (Eurohep). Gut 46, 420–426. doi: 10.1136/gut.46.3.420
Freitas, N., Abe, K., Cunha, C., Menne, S., and Gudima, S. O. (2014). Support of the infectivity of hepatitis delta virus particles by the envelope proteins of different genotypes of hepatitis B virus. J. Virol. 88, 6255–6267. doi: 10.1128/jvi.00346-14
Fukano, K., Tsukuda, S., Oshima, M., Suzuki, R., Aizaki, H., Ohki, M., et al. (2018). Troglitazone impedes the oligomerization of sodium taurocholate cotransporting polypeptide and entry of hepatitis B virus into hepatocytes. Front. Microbiol. 9:3257. doi: 10.3389/fmicb.2018.03257
Giersch, K., Bhadra, O. D., Volz, T., Allweiss, L., Riecken, K., Fehse, B., et al. (2019). Hepatitis delta virus persists during liver regeneration and is amplified through cell division both in vitro and in vivo. Gut 68, 150–157. doi: 10.1136/gutjnl-2017-314713
Giersch, K., Helbig, M., Volz, T., Allweiss, L., Mancke, L. V., Lohse, A. W., et al. (2014). Persistent hepatitis D virus mono-infection in humanized mice is efficiently converted by hepatitis B virus to a productive co-infection. J. Hepatol. 60, 538–544. doi: 10.1016/j.jhep.2013.11.010
Giersch, K., Homs, M., Volz, T., Helbig, M., Allweiss, L., Lohse, A. W., et al. (2017). Both interferon alpha and lambda can reduce all intrahepatic HDV infection markers in HBV/HDV infected humanized mice. Sci. Rep. 7:3757. doi: 10.1038/s41598-017-03946-9
Glenn, J. S., and White, J. M. (1991). trans-dominant inhibition of human hepatitis delta virus genome replication. J. Virol. 65, 2357–2361. doi: 10.1128/jvi.65.5.2357-2361.1991
Greco-Stewart, V. S., Schissel, E., and Pelchat, M. (2009). The hepatitis delta virus RNA genome interacts with the human RNA polymerases I and III. Virology 386, 12–15. doi: 10.1016/j.virol.2009.02.007
Griffin, B., Chasovskikh, S., Dritschilo, A., and Casey, J. (2014). Hepatitis delta antigen requires a flexible quasi-double-stranded RNA structure to bind and condense hepatitis delta virus RNA in a ribonucleoprotein complex. J. Virol. 88, 7402–7411. doi: 10.1128/jvi.00443-14
Gudima, S., Chang, J., Moraleda, G., Azvolinsky, A., and Taylor, J. (2002). Parameters of human hepatitis delta virus genome replication: the quantity, quality, and intracellular distribution of viral proteins and RNA. J. Virol. 76, 3709–3719. doi: 10.1128/jvi.76.8.3709-3719.2002
Gudima, S., Meier, A., Dunbrack, R., Taylor, J., and Bruss, V. (2007). Two potentially important elements of the hepatitis B virus large envelope protein are dispensable for the infectivity of hepatitis delta virus. J. Virol. 81, 4343–4347. doi: 10.1128/jvi.02478-06
Harichandran, K., Shen, Y., Stephenson Tsoris, S., Lee, S. C., and Casey, J. L. (2019). Hepatitis delta antigen regulates mRNA and antigenome RNA levels during hepatitis delta virus replication. J. Virol. 93, e1989–e1918. doi: 10.1128/jvi.01989-18
Herrscher, C., Pastor, F., Burlaud-Gaillard, J., Dumans, A., Seigneuret, F., Moreau, A., et al. (2020). Hepatitis B virus entry into HepG2-NTCP cells requires clathrin-mediated endocytosis. Cell Microbiol. 22:e13205. doi: 10.1111/cmi.13205
Hetzel, U., Szirovicza, L., Smura, T., Prähauser, B., Vapalahti, O., Kipar, A., et al. (2019). Identification of a novel deltavirus in boa constrictors. mBio 10:e00014–e19. doi: 10.1128/mBio.00014-19
Homs, M., Rodriguez-Frias, F., Gregori, J., Ruiz, A., Reimundo, P., Casillas, R., et al. (2016). Evidence of an exponential decay pattern of the hepatitis delta virus evolution rate and fluctuations in quasispecies complexity in long-term studies of chronic delta infection. PLoS One 11:e0158557. doi: 10.1371/journal.pone.0158557
Hsu, C. W., Juang, H. H., Kuo, C. Y., Li, H. P., Iang, S. B., Lin, S. H., et al. (2019). Structural pattern differences in unbranched rod-like RNA of hepatitis delta virus affect RNA editing. Viruses 11;934. doi: 10.3390/v11100934
Hu, Q., Zhang, F., Duan, L., Wang, B., Ye, Y., Li, P., et al. (2020). E-cadherin plays a role in hepatitis B virus entry through affecting glycosylated sodium-taurocholate cotransporting polypeptide distribution. Front. Cell. Infect. Microbiol. 10:74. doi: 10.3389/fcimb.2020.00074
Huang, W. H., Chen, C. W., Wu, H. L., and Chen, P. J. (2006). Post-translational modification of delta antigen of hepatitis D virus. Curr. Top. Microbiol. Immunol. 307, 91–112. doi: 10.1007/3-540-29802-9_5
Huang, W. H., Mai, R. T., and Lee, Y. H. (2008). Transcription factor YY1 and its associated acetyltransferases CBP and p300 interact with hepatitis delta antigens and modulate hepatitis delta virus RNA replication. J. Virol. 82, 7313–7324. doi: 10.1128/jvi.02581-07
Huang, W. H., Yung, B. Y., Syu, W. J., and Lee, Y. H. (2001). The nucleolar phosphoprotein B23 interacts with hepatitis delta antigens and modulates the hepatitis delta virus RNA replication. J. Biol. Chem. 276, 25166–25175. doi: 10.1074/jbc.M010087200
Hwang, S. B., and Lai, M. M. (1993). Isoprenylation mediates direct protein-protein interactions between hepatitis large delta antigen and hepatitis B virus surface antigen. J. Virol. 67, 7659–7662. doi: 10.1128/jvi.67.12.7659-7662.1993
Iwamoto, M., Saso, W., Nishioka, K., Ohashi, H., Sugiyama, R., Ryo, A., et al. (2020). The machinery for endocytosis of epidermal growth factor receptor coordinates the transport of incoming hepatitis B virus to the endosomal network. J. Biol. Chem. 295, 800–807. doi: 10.1074/jbc.AC119.010366
Iwamoto, M., Saso, W., Sugiyama, R., Ishii, K., Ohki, M., Nagamori, S., et al. (2019). Epidermal growth factor receptor is a host-entry cofactor triggering hepatitis B virus internalization. Proc. Natl. Acad. Sci. U S A. 116, 8487–8492. doi: 10.1073/pnas.1811064116
Iwamoto, M., Shibata, Y., Kawasaki, J., Kojima, S., Li, Y. T., Iwami, S., et al. (2021). Identification of novel avian and mammalian deltaviruses provides new insights into deltavirus evolution. Virus Evol. 7:veab003. doi: 10.1093/ve/veab003
Jiang, B., and Hildt, E. (2020). Intracellular trafficking of HBV particles. Cells 9:2023. doi: 10.3390/cells9092023
Jiang, B., Himmelsbach, K., Ren, H., Boller, K., and Hildt, E. (2015). Subviral hepatitis B virus filaments, like infectious viral particles, are released via multivesicular bodies. J. Virol. 90, 3330–3341. doi: 10.1128/jvi.03109-15
Kang, C., and Syed, Y. Y. (2020). Bulevirtide: first approval. Drugs 80, 1601–1605. doi: 10.1007/s40265-020-01400-1
Komla-Soukha, I., and Sureau, C. (2006). A tryptophan-rich motif in the carboxyl terminus of the small envelope protein of hepatitis B virus is central to the assembly of hepatitis delta virus particles. J. Virol. 80, 4648–4655. doi: 10.1128/jvi.80.10.4648-4655.2006
Lambert, C., Döring, T., and Prange, R. (2007). Hepatitis B virus maturation is sensitive to functional inhibition of ESCRT-III, Vps4, and gamma 2-adaptin. J. Virol. 81, 9050–9060. doi: 10.1128/jvi.00479-07
Le Duff, Y., Blanchet, M., and Sureau, C. (2009). The pre-S1 and antigenic loop infectivity determinants of the hepatitis B virus envelope proteins are functionally independent. J. Virol. 83, 12443–12451. doi: 10.1128/jvi.01594-09
Lee, C. H., Chang, S. C., Chen, C. J., and Chang, M. F. (1998). The nucleolin binding activity of hepatitis delta antigen is associated with nucleolus targeting. J. Biol. Chem. 273, 7650–7656. doi: 10.1074/jbc.273.13.7650
Lee, C. H., Chang, S. C., Wu, C. H., and Chang, M. F. (2001). A novel chromosome region maintenance 1-independent nuclear export signal of the large form of hepatitis delta antigen that is required for the viral assembly. J. Biol. Chem. 276, 8142–8148. doi: 10.1074/jbc.M004477200
Lee, C. Z., Lin, J. H., Chao, M., McKnight, K., and Lai, M. M. (1993). RNA-binding activity of hepatitis delta antigen involves two arginine-rich motifs and is required for hepatitis delta virus RNA replication. J. Virol. 67, 2221–2227. doi: 10.1128/jvi.67.4.2221-2227.1993
Lee, C. Z., and Sheu, J. C. (2008). Histone H1e interacts with small hepatitis delta antigen and affects hepatitis delta virus replication. Virology 375, 197–204. doi: 10.1016/j.virol.2008.02.003
Lempp, F., Ni, Y., and Urban, S. (2016). Hepatitis delta virus: insights into a peculiar pathogen and novel treatment options. Nat. Rev. Gastroenterol. Hepatol. 13, 580–589. doi: 10.1038/nrgastro.2016.126
Lempp, F. A., Wiedtke, E., Qu, B., Roques, P., Chemin, I., Vondran, F. W. R., et al. (2017). Sodium taurocholate cotransporting polypeptide is the limiting host factor of hepatitis B virus infection in macaque and pig hepatocytes. Hepatology 66, 703–716. doi: 10.1002/hep.29112
Li, Y. J., Stallcup, M. R., and Lai, M. M. (2004). Hepatitis delta virus antigen is methylated at arginine residues, and methylation regulates subcellular localization and RNA replication. J. Virol. 78, 13325–13334. doi: 10.1128/jvi.78.23.13325-13334.2004
Löffler-Mary, H., Dumortier, J., Klentsch-Zimmer, C., and Prange, R. (2000). Hepatitis B virus assembly is sensitive to changes in the cytosolic S loop of the envelope proteins. Virology 270, 358–367. doi: 10.1006/viro.2000.0268
Lok, A. S., Negro, F., Asselah, T., Farci, P., and Rizzetto, M. (2021). Endpoints and new options for treatment of chronic hepatitis D. Hepatology 74, 3479–3485. doi: 10.1002/hep.32082
Lukash, T., Freitas, N., Menne, S., and Gudima, S. O. (2019). Down-regulation of hepatitis delta virus super-infection in the woodchuck model. Virology 531, 100–113. doi: 10.1016/j.virol.2019.03.002
Macnaughton, T. B., and Lai, M. M. (2002). Genomic but not antigenomic hepatitis delta virus RNA is preferentially exported from the nucleus immediately after synthesis and processing. J. Virol. 76, 3928–3935. doi: 10.1128/jvi.76.8.3928-3935.2002
Macnaughton, T. B., and Lai, M. M. (2006). HDV RNA replication: ancient relic or primer? Curr. Top. Microbiol. Immunol. 307, 25–45. doi: 10.1007/3-540-29802-9_2
Macovei, A., Petrareanu, C., Lazar, C., Florian, P., and Branza-Nichita, N. (2013). Regulation of hepatitis B virus infection by Rab5, Rab7, and the endolysosomal compartment. J. Virol. 87, 6415–6427. doi: 10.1128/jvi.00393-13
Meier, A., Mehrle, S., Weiss, T. S., Mier, W., and Urban, S. (2013). Myristoylated PreS1-domain of the hepatitis B virus L-protein mediates specific binding to differentiated hepatocytes. Hepatology 58, 31–42. doi: 10.1002/hep.26181
Modahl, L. E., and Lai, M. M. (2000). The large delta antigen of hepatitis delta virus potently inhibits genomic but not antigenomic RNA synthesis: a mechanism enabling initiation of viral replication. J. Virol. 74, 7375–7380. doi: 10.1128/jvi.74.16.7375-7380.2000
Mu, J. J., Chen, D. S., and Chen, P. J. (2001). The conserved serine 177 in the delta antigen of hepatitis delta virus is one putative phosphorylation site and is required for efficient viral RNA replication. J. Virol. 75, 9087–9095. doi: 10.1128/jvi.75.19.9087-9095.2001
Mu, J. J., Tsay, Y. G., Juan, L. J., Fu, T. F., Huang, W. H., Chen, D. S., et al. (2004). The small delta antigen of hepatitis delta virus is an acetylated protein and acetylation of lysine 72 may influence its cellular localization and viral RNA synthesis. Virology 319, 60–70. doi: 10.1016/j.virol.2003.10.024
Negro, F., Korba, B. E., Forzani, B., Baroudy, B. M., Brown, T. L., Gerin, J. L., et al. (1989). Hepatitis delta virus (HDV) and woodchuck hepatitis virus (WHV) nucleic acids in tissues of HDV-infected chronic WHV carrier woodchucks. J. Virol. 63, 1612–1618. doi: 10.1128/jvi.63.4.1612-1618.1989
Netter, H. J., Barrios, M. H., Littlejohn, M., and Yuen, L. K. W. (2021). Hepatitis delta virus (HDV) and delta-like agents: insights into their origin. Front. Microbiol. 12:652962. doi: 10.3389/fmicb.2021.652962
Ni, Y., Lempp, F. A., Mehrle, S., Nkongolo, S., Kaufman, C., Fälth, M., et al. (2014). Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology 146, 1070–1083. doi: 10.1053/j.gastro.2013.12.024
Niklasch, M., Zimmermann, P., and Nassal, M. (2021). The hepatitis B virus nucleocapsid-dynamic compartment for infectious virus production and new antiviral target. Biomedicines 9:1577. doi: 10.3390/biomedicines9111577
O’Malley, B., and Lazinski, D. W. (2005). Roles of carboxyl-terminal and farnesylated residues in the functions of the large hepatitis delta antigen. J. Virol. 79, 1142–1153. doi: 10.1128/jvi.79.2.1142-1153.2005
Paraskevopoulou, S., Pirzer, F., Goldmann, N., Schmid, J., Corman, V. M., Gottula, L. T., et al. (2020). Mammalian deltavirus without hepadnavirus coinfection in the neotropical rodent Proechimys semispinosus. Proc. Natl. Acad. Sci. U S A. 117, 17977–17983. doi: 10.1073/pnas.2006750117
Patient, R., Hourioux, C., Sizaret, P. Y., Trassard, S., Sureau, C., and Roingeard, P. (2007). Hepatitis B virus subviral envelope particle morphogenesis and intracellular trafficking. J. Virol. 81, 3842–3851. doi: 10.1128/jvi.02741-06
Perez-Vargas, J., Amirache, F., Boson, B., Mialon, C., Freitas, N., Sureau, C., et al. (2019). Enveloped viruses distinct from HBV induce dissemination of hepatitis D virus in vivo. Nat. Commun. 10:2098. doi: 10.1038/s41467-019-10117-z
Poisson, F., Roingeard, P., Baillou, A., Dubois, F., Bonelli, F., Calogero, R. A., et al. (1993). Characterization of RNA-binding domains of hepatitis delta antigen. J. Gen. Virol. 74(Pt 11), 2473–2478. doi: 10.1099/0022-1317-74-11-2473
Polaris Observatory Collaborators (2018). Global prevalence, treatment, and prevention of hepatitis B virus infection in 2016: a modelling study. Lancet. Gastroenterol. Hepatol. 3, 383–403. doi: 10.1016/s2468-1253(18)30056-6
Polo, J. M., Jeng, K. S., Lim, B., Govindarajan, S., Hofman, F., Sangiorgi, F., et al. (1995). Transgenic mice support replication of hepatitis delta virus RNA in multiple tissues, particularly in skeletal muscle. J. Virol. 69, 4880–4887. doi: 10.1128/jvi.69.8.4880-4887.1995
Rabe, B., Glebe, D., and Kann, M. (2006). Lipid-mediated introduction of hepatitis B virus capsids into nonsusceptible cells allows highly efficient replication and facilitates the study of early infection events. J. Virol. 80, 5465–5473. doi: 10.1128/jvi.02303-05
Raimondo, G., Locarnini, S., Pollicino, T., Levrero, M., Zoulim, F., and Lok, A. S. (2019). Update of the statements on biology and clinical impact of occult hepatitis B virus infection. J. Hepatol. 71, 397–408. doi: 10.1016/j.jhep.2019.03.034
Reid, C. E., and Lazinski, D. W. (2000). A host-specific function is required for ligation of a wide variety of ribozyme-processed RNAs. Proc. Natl. Acad. Sci. U S A. 97, 424–429. doi: 10.1073/pnas.97.1.424
Riccitelli, N., and Lupták, A. (2013). HDV family of self-cleaving ribozymes. Prog. Mol. Biol. Transl. Sci. 120, 123–171. doi: 10.1016/b978-0-12-381286-5.00004-4
Roggenbach, I., Chi, X., Lempp, F. A., Qu, B., Walter, L., Wu, R., et al. (2021). HDV seroprevalence in HBsAg-positive patients in china occurs in hotspots and is not associated with HCV mono-infection. Viruses 13:1799. doi: 10.3390/v13091799
Ryu, W., Netter, H., Bayer, M., and Taylor, J. (1993). Ribonucleoprotein complexes of hepatitis delta virus. J. Virol. 67, 3281–3287. doi: 10.1128/jvi.67.6.3281-3287.1993
Sato, S., Cornillez-Ty, C., and Lazinski, D. W. (2004). By inhibiting replication, the large hepatitis delta antigen can indirectly regulate amber/W editing and its own expression. J. Virol. 78, 8120–8134. doi: 10.1128/jvi.78.15.8120-8134.2004
Sheu, G. T. (2002). Initiation of hepatitis delta virus (HDV) replication: HDV RNA encoding the large delta antigen cannot replicate. J. Gen. Virol. 83(Pt 10), 2507–2513. doi: 10.1099/0022-1317-83-10-2507
Siddiqui, A., Jameel, S., and Mapoles, J. (1986). Transcriptional control elements of hepatitis B surface antigen gene. Proc. Natl. Acad. Sci. U S A. 83, 566–570. doi: 10.1073/pnas.83.3.566
Sommereyns, C., Paul, S., Staeheli, P., and Michiels, T. (2008). IFN-lambda (IFN-lambda) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 4:e1000017. doi: 10.1371/journal.ppat.1000017
Sopena, S., Godoy, C., Tabernero, D., Homs, M., Gregori, J., Riveiro-Barciela, M., et al. (2018). Quantitative characterization of hepatitis delta virus genome edition by next-generation sequencing. Virus Res. 243, 52–59. doi: 10.1016/j.virusres.2017.10.003
Steiner, S., Huebner, M. T., and Dreesman, G. R. (1974). Major polar lipids of hepatitis B antigen preparations: evidence for the presence of a glycosphingolipid. J. Virol. 14, 572–577. doi: 10.1128/jvi.14.3.572-577.1974
Stockdale, A., Kreuels, B., Henrion, M., Giorgi, E., Kyomuhangi, I., de Martel, C., et al. (2020). The global prevalence of hepatitis D virus infection: systematic review and meta-analysis. J. Hepatol. 73, 523–532. doi: 10.1016/j.jhep.2020.04.008
Stoeckl, L., Funk, A., Kopitzki, A., Brandenburg, B., Oess, S., Will, H., et al. (2006). Identification of a structural motif crucial for infectivity of hepatitis B viruses. Proc. Natl. Acad. Sci. U S A. 103, 6730–6734. doi: 10.1073/pnas.0509765103
Sureau, C., Guerra, B., and Lanford, R. E. (1993). Role of the large hepatitis B virus envelope protein in infectivity of the hepatitis delta virion. J. Virol. 67, 366–372. doi: 10.1128/jvi.67.1.366-372.1993
Sureau, C., Guerra, B., and Lee, H. (1994). The middle hepatitis B virus envelope protein is not necessary for infectivity of hepatitis delta virus. J. Virol. 68, 4063–4066. doi: 10.1128/jvi.68.6.4063-4066.1994
Sureau, C., and Negro, F. (2016). The hepatitis delta virus: Replication and pathogenesis. J. Hepatol. 64, S102–S116. doi: 10.1016/j.jhep.2016.02.013
Sureau, C., and Salisse, J. (2013). A conformational heparan sulfate binding site essential to infectivity overlaps with the conserved hepatitis B virus a-determinant. Hepatology 57, 985–994. doi: 10.1002/hep.26125
Tavanez, J. P., Cunha, C., Silva, M. C., David, E., Monjardino, J., and Carmo-Fonseca, M. (2002). Hepatitis delta virus ribonucleoproteins shuttle between the nucleus and the cytoplasm. RNA 8, 637–646. doi: 10.1017/s1355838202026432
Taylor, J. (2015). Hepatitis D virus replication. Cold Spring Harb. Perspect. Med. 5:a021568. doi: 10.1101/cshperspect.a021568
Tseng, C. H., Cheng, T. S., Shu, C. Y., Jeng, K. S., and Lai, M. M. (2010). Modification of small hepatitis delta virus antigen by SUMO protein. J. Virol. 84, 918–927. doi: 10.1128/jvi.01034-09
Vaillant, A. (2019). REP 2139: antiviral mechanisms and applications in achieving functional control of HBV and HDV infection. ACS Infect. Dis. 5, 675–687. doi: 10.1021/acsinfecdis.8b00156
Vaillant, A., Juteau, J. M., Lu, H., Liu, S., Lackman-Smith, C., Ptak, R., et al. (2006). Phosphorothioate oligonucleotides inhibit human immunodeficiency virus type 1 fusion by blocking gp41 core formation. Antimicrob. Agents Chemother. 50, 1393–1401. doi: 10.1128/aac.50.4.1393-1401.2006
Verrier, E. R., Colpitts, C. C., Bach, C., Heydmann, L., Weiss, A., Renaud, M., et al. (2016). A targeted functional RNA interference screen uncovers glypican 5 as an entry factor for hepatitis B and D viruses. Hepatology 63, 35–48. doi: 10.1002/hep.28013
Wadkins, T. S., Perrotta, A. T., Ferré-D’Amaré, A. R., Doudna, J. A., and Been, M. D. (1999). A nested double pseudoknot is required for self-cleavage activity of both the genomic and antigenomic hepatitis delta virus ribozymes. RNA 5, 720–727. doi: 10.1017/s1355838299990209
Wang, W., Lempp, F. A., Schlund, F., Walter, L., Decker, C. C., Zhang, Z., et al. (2021). Assembly and infection efficacy of hepatitis B virus surface protein exchanges in 8 hepatitis D virus genotype isolates. J. Hepatol. 75, 311–323. doi: 10.1016/j.jhep.2021.03.025
Watanabe, T., Sorensen, E. M., Naito, A., Schott, M., Kim, S., and Ahlquist, P. (2007). Involvement of host cellular multivesicular body functions in hepatitis B virus budding. Proc. Natl. Acad. Sci. U S A. 104, 10205–10210. doi: 10.1073/pnas.0704000104
Wedemeyer, H., Yurdaydìn, C., Dalekos, G. N., Erhardt, A., Çakaloğlu, Y., Değertekin, H., et al. (2011). Peginterferon plus adefovir versus either drug alone for hepatitis delta. N. Engl. J. Med. 364, 322–331. doi: 10.1056/NEJMoa0912696
Wille, M., Netter, H. J., Littlejohn, M., Yuen, L., Shi, M., Eden, J. S., et al. (2018). A divergent hepatitis D-like agent in birds. Viruses 10:720. doi: 10.3390/v10120720
Xia, Y., Chang, M., Wei, D., Govindarajan, S., and Lai, M. (1990). Heterogeneity of hepatitis delta antigen. Virology 178, 331–336. doi: 10.1016/0042-6822(90)90415-n
Xia, Y. P., and Lai, M. M. (1992). Oligomerization of hepatitis delta antigen is required for both the trans-activating and trans-dominant inhibitory activities of the delta antigen. J. Virol. 66, 6641–6648. doi: 10.1128/jvi.66.11.6641-6648.1992
Xia, Y. P., Yeh, C. T., Ou, J. H., and Lai, M. M. (1992). Characterization of nuclear targeting signal of hepatitis delta antigen: nuclear transport as a protein complex. J. Virol. 66, 914–921. doi: 10.1128/jvi.66.2.914-921.1992
Yamaguchi, Y., Filipovska, J., Yano, K., Furuya, A., Inukai, N., Narita, T., et al. (2001). Stimulation of RNA polymerase II elongation by hepatitis delta antigen. Science 293, 124–127. doi: 10.1126/science.1057925
Yamaguchi, Y., Mura, T., Chanarat, S., Okamoto, S., and Handa, H. (2007). Hepatitis delta antigen binds to the clamp of RNA polymerase II and affects transcriptional fidelity. Genes Cells 12, 863–875. doi: 10.1111/j.1365-2443.2007.01094.x
Yan, H., Zhong, G., Xu, G., He, W., Jing, Z., Gao, Z., et al. (2012). Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife 1:e00049. doi: 10.7554/eLife.00049
Yang, S., Shen, Z., Kang, Y., Sun, L., Viswanathan, U., Guo, H., et al. (2021). A putative amphipathic alpha helix in hepatitis B virus small envelope protein plays a critical role in the morphogenesis of subviral particles. J. Virol. 95, e2399–e2320. doi: 10.1128/jvi.02399-20
Yurdaydin, C., Bozkaya, H., Onder, F. O., Sentürk, H., Karaaslan, H., Akdoğan, M., et al. (2008). Treatment of chronic delta hepatitis with lamivudine vs lamivudine + interferon vs interferon. J. Viral. Hepat. 15, 314–321. doi: 10.1111/j.1365-2893.2007.00936.x
Zhang, Z., Filzmayer, C., Ni, Y., Sültmann, H., Mutz, P., Hiet, M. S., et al. (2018). Hepatitis D virus replication is sensed by MDA5 and induces IFN-β/λ responses in hepatocytes. J. Hepatol. 69, 25–35. doi: 10.1016/j.jhep.2018.02.021
Zhang, Z., and Urban, S. (2020). Interplay between Hepatitis D Virus and the Interferon Response. Vijruses 12:1334. doi: 10.3390/v12111334
Zhang, Z., and Urban, S. (2021). New insights into HDV persistence: The role of interferon response and implications for upcoming novel therapies. J. Hepatol. 74, 686–699. doi: 10.1016/j.jhep.2020.11.032
Keywords: hepatitis delta virus, single-stranded RNA, antigen, viral structures, virus life cycle, therapy
Citation: Zi J, Gao X, Du J, Xu H, Niu J and Chi X (2022) Multiple Regions Drive Hepatitis Delta Virus Proliferation and Are Therapeutic Targets. Front. Microbiol. 13:838382. doi: 10.3389/fmicb.2022.838382
Received: 17 December 2021; Accepted: 11 March 2022;
Published: 06 April 2022.
Edited by:
Julie Lucifora, Institut National de la Santé et de la Recherche Médicale (INSERM), FranceReviewed by:
Eloi R. Verrier, INSERM UMR_S1110 Institute de Recherche sur les Maladies Virales et Hepatiques, FranceCopyright © 2022 Zi, Gao, Du, Xu, Niu and Chi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junqi Niu, anVucWluaXVAamx1LmVkdS5jbg==; Xiumei Chi, Y2hpeG1Aamx1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.