
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol. , 25 April 2022
Sec. Evolutionary and Genomic Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.835313
This article is part of the Research Topic Host Microbiome Interactions in Chronic Lifestyle Disorders and Complex Diseases in Humans View all 6 articles
 Angitha N. Nath1†
Angitha N. Nath1† R. J. Retnakumar1,2†
R. J. Retnakumar1,2† Ashik Francis1
Ashik Francis1 Prakash Chhetri3
Prakash Chhetri3 Namrata Thapa3
Namrata Thapa3 Santanu Chattopadhyay1*
Santanu Chattopadhyay1*
It is increasingly being recognized that severe gastroduodenal diseases such as peptic ulcer and gastric cancer are not just the outcomes of Helicobacter pylori infection in the stomach. Rather, both diseases develop and progress due to the perfect storms created by a combination of multiple factors such as the expression of different H. pylori virulence proteins, consequent human immune responses, and dysbiosis in gastrointestinal microbiomes. In this mini review, we have discussed how the genomes of H. pylori and other gastrointestinal microbes as well as the genomes of different human populations encode complex and variable virulome–immunome interplay, which influences gastroduodenal health. The heterogeneities that are encrypted in the genomes of different human populations and in the genomes of their respective resident microbes partly explain the inconsistencies in clinical outcomes among the H. pylori-infected people.
No pathogenic bacteria created a bigger spark among microbiologists, gastroenterologists, and science enthusiasts than Helicobacter pylori upon its discovery from the human stomach. Once the colonization of this bacterium in the harshly acidic human gastric milieu was confirmed by histology and culture by Robin Warren and Barry Marshall in 1983, within the next 20 years, the total number of publications on it surpassed the total number of publications on Salmonella, which was discovered in 1855 (Warren and Marshall, 1983; Harry et al., 2001). Once the role of H. pylori as the causative agent of gastritis, peptic ulcer disease (PUD), and gastric cancer (GC) was firmly established, the World Health Organization (WHO) classified it as Class I carcinogen (first among all bacteria) in 1994 (IARC, 1994); gastritis and PUD became curable with triple therapy (a proton pump inhibitor and two antibiotics); gastric mucosa-associated lymphoid tissue (MALT) lymphoma became the first malignancy to be reversed with antimicrobial agents (Malfertheiner et al., 2014); and both Warren and Marshall were awarded the Nobel Prize in Physiology and Medicine in 2005. However, at the same time, it was also realized that H. pylori colonization in the human stomach is remarkably common and is not just restricted to patients suffering from gastric and duodenal diseases. A study showed that in 2017, 4.4 billion people (57.9% of the global population at that time) were infected with H. pylori (Hooi et al., 2017). Although H. pylori remains colonized in the stomach of a major fraction of the population, only a small subset of people, typically 10–20% of the infected population, suffer from severe gastroduodenal diseases such as PUD and GC, but the reasons for the inconsistencies in clinical outcomes were not precisely understood (Bauer and Meyer, 2011). Data emerged in the past four decades suggest us to appreciate that PUD and GC may have multiple and complex etiologies such as H. pylori infection, polymorphisms in human cytokine genes, dysbiosis in the gastric and intestinal microbiome, the influence of geography, climate, and altitude, lifestyles such as diet, smoking, and alcohol consumption, and perturbations that are imposed by different medicines (Alexander et al., 2021). In this mini review, we discussed how complex and functional host–microbe interplays that determine the gastroduodenal health are encoded in the human genome and in the genomes of trillions of microbes (including H. pylori) that populate the human gastrointestinal tract.
H. pylori is one of the oldest human pathogens known. A 5,300-year-old iceman mummy (named Ötzi), excavated from the Italian Alps, had H. pylori DNA in the stomach (Maixner et al., 2016). Interestingly, unlike most human pathogens, H. pylori exclusively colonizes humans (Mendall and Northfield, 1995). H. pylori colonization is acquired during the initial years of life by an intrafamilial manner through oral/fecal–oral route and remains colonized for decades before causing any severe diseases (Brown, 2000). Most (80–90%) of the infections, however, either remain asymptomatic and unnoticed or cause noticeable gastritis. Typically, the antral predominant gastritis is associated with H. pylori-induced gastrin secretion followed by increased H+ secretion by parietal cells and predisposes to PUD. However, in direct contrast, long-term H. pylori infection leads to hypochlorhydria due to decreased H+ secretion by the parietal cells and this allows the growth of several other bacteria (discussed later) (McColl et al., 1998). This cascade of events leads to atrophic gastritis, which eventually progresses to intestinal metaplasia, intraepithelial neoplasia, and GC (Barra et al., 2021).
Continuous changes, as part of evolution within the human stomach, have occurred and accumulated in different H. pylori genomes for thousands of years along with human migrations, which has started 60,000 years ago in different clades, followed by settlements in different geographical regions (Correa and Piazuelo, 2012). The modern H. pylori strains, which have successfully “coevolved” with humans, are classified into distinct populations—hpAfrica1 (subpopulations—hspWAfrica and hspSAfrica), hpAfrica2, hpEastAsia (subpopulations—hspAmerind, hspEAsia, and hspMaori), hpEurope, hpNEAfrica, hpAsia2, and hpSahul (Falush et al., 2003; Linz et al., 2007; Moodley et al., 2009). The above H. pylori populations exhibit distinct geographical predisposal. hpEurope is distributed in the Middle East, India, Iran, and Europe. hspWAfrica is present in Western Africa, while hspSAfrica and hpAfrica2 are present in South Africa. hpNEAfrica circulates in Nilo-Saharan speakers of northern Nigeria, Sudan, Ethiopia, and Somalia. hspEAsia is distributed among East Asians, while hspMaori is seen in Taiwan’s Aboriginal, Melanesian, and Polynesian populations. hspAmerind is present among Native Americans. hpAsia2 is seen in populations of Malaysia, Thailand, Bangladesh, and northern India. Strains from Papua New Guinea, Aboriginals, and Australia belong to the hpSahul population (Correa and Piazuelo, 2012).
Overall, H. pylori is one of the most genetically diverse species among bacterial pathogens (Suzuki et al., 2012). Its diversity is due to a higher level of spontaneous mutations occurring within the restricted gastric niche, a higher frequency of horizontal gene transfer, and natural competence (Israel et al., 2001). Impaired DNA repair, integration of acquired DNA into the “plasticity zones,” and higher intraspecific recombination also contributed to diverse genetic forms (Fernandez-Gonzalez and Backert, 2014). H. pylori remains a very successful human pathogen for centuries with a considerably lower proportion of terminal clinical outcomes and higher self-propagation across generations with a plethora of virulence factors that facilitate chronic colonization in the human stomach, where the pH is nearly 2. H. pylori virulence factors include urease, flagella, adhesins, and several effector proteins that lead to pathogenesis. One of the major effector proteins of H. pylori is the oncoprotein CagA, encoded by the cagA gene present in the 40 kb cag pathogenicity island (cagPAI). Inside the gastric cell, phosphorylated CagA interacts with Src homology-2 (SH2) domains of the host proteins such as CSK, Grb2, and SHP2, leading to altered cell proliferation and differentiation, cytoskeletal changes, and increased proinflammatory cytokines (IL-8) secretion via the NF-κB pathway (Papadakos et al., 2013; Hatakeyama, 2014). Another effector protein, the VacA, gets internalized by binding to the receptor protein tyrosine phosphatases (RPTPα and RPTPβ) and low-density lipoprotein receptor-related protein-1 (LRP1), resulting in cell vacuolation and cell deaths by apoptosis, necrosis, and autophagy (Foegeding et al., 2016; Chauhan et al., 2019).
The capabilities of the H. pylori strains to establish colonization and to induce pathogenic alterations in the stomach are greatly determined by the allelic types of virulence genes, which vary with geography (Table 1). For vacA, the alleles observed are s1 (with subtypes s1a and s1b) and s2 for the vacAs region; i1 (with subtypes i1a and i1b) and i2 for the vacAi region; m1 (with subtypes m1a, m1b, and m1c) and m2 for the vacAm region; c1 and c2 for the vacAc region; and d1 and d2 for the vacAd region (Trang et al., 2016; Alexander et al., 2021). CagA protein also shows distinct variations between strains circulating in different populations. The Western H. pylori strains carry CagA EPIYA-C, while the EPIYA-D is typically expressed by East Asian strains (Figure 1; Higashi et al., 2002). The vacAs1i1m1cagA + (particularly of East Asian-type CagA) strains are associated with aggressive clinical outcomes as compared to vacAs2m2cagA − strains. The diversities in virulence encoded in the H. pylori genomes in different populations are the major determinants of different clinical manifestations in different populations (Table 1; Chang et al., 2018). For example, the prevalence of GC is highest in East Asian countries, but is remarkably low in African countries (Rawla and Barsouk, 2019).

Table 1. Variations in H. pylori genotype, microbiome, and host genes and association with gastric diseases.
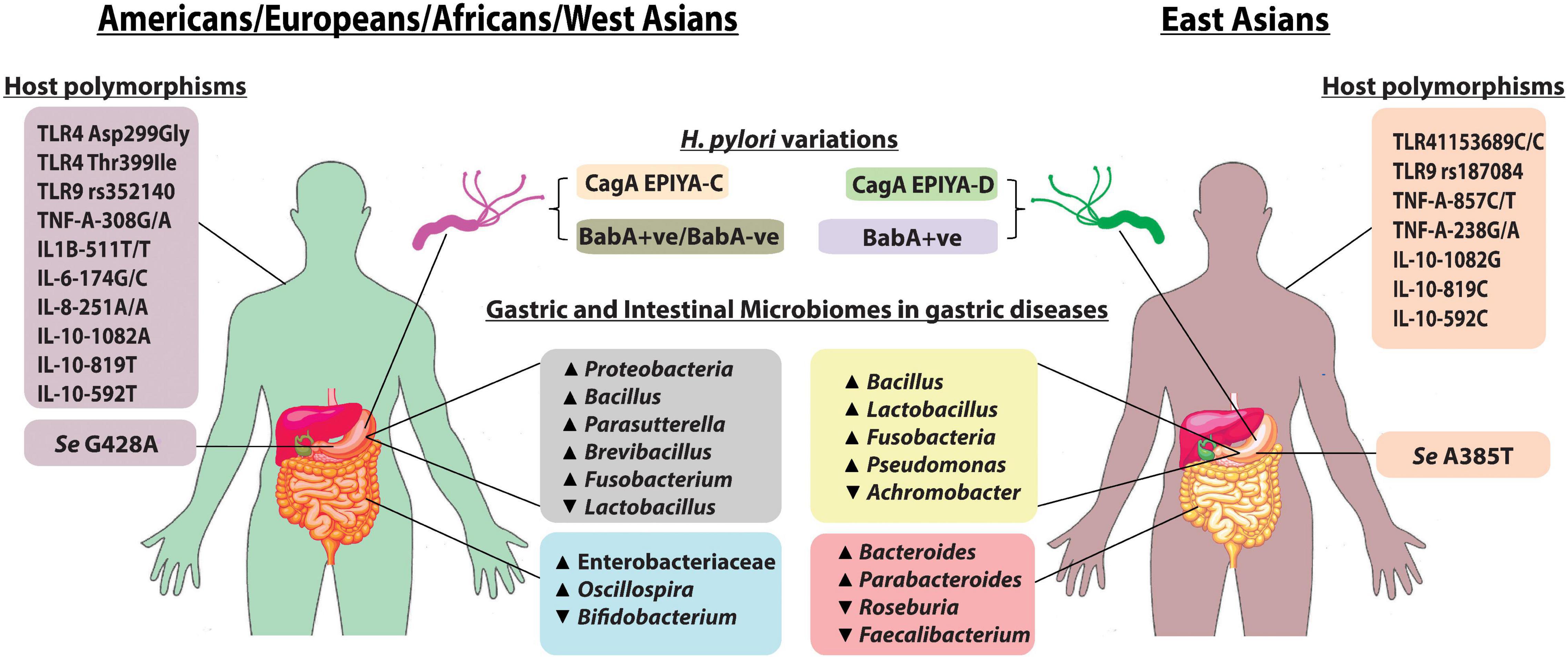
Figure 1. Pattern of gastric and intestinal microbiomes, host polymorphisms, and H. pylori type between East Asia and the rest of the world: the more virulent H. pylori CagA EPIYA-D is mostly found in East Asia when compared with H. pylori CagA EPIYA-C, which is predominant in the Western world (Yamaoka, 2008). Almost all strains from East Asia express BabA, while in West, BabA-positive strains as well as a minority of BabA-negative strains are present. FUT2, encoded by the Se gene, is required for the synthesis of LeB antigen (BabA-binding host molecule). Mutations in the Se genes, Se G428A and Se A385T, are found to inactivate and to reduce the activity of FUT2, respectively (Anstee, 2010). The alteration in gastric and intestinal microbiome composition is observed in H. pylori infection, and gastric diseases also exhibit regional variations as shown in the figure. Additionally, specific host immune gene polymorphisms in different populations predispose an individual susceptibility to H. pylori colonization and gastric diseases.
Every living human body carries its unique microbiome composed of a few trillion microbes along with their respective genomes that express different proteins for carrying out metabolic functions (Cho and Blaser, 2012; Blaser, 2014; Gilbert et al., 2018). Every anatomical niche of a healthy individual has a distinct microbiome that helps in various physiological processes (Kennedy and Chang, 2020). Conversely, enrichment of a few bacterial taxa, which leads to dysbiosis in the microbiome, is deleterious to human health (Turnbaugh et al., 2007; Hullar et al., 2014). The microbiome composition of each niche for each individual varies due to several factors such as antibiotic usage, geography, diet, lifestyle, and H. pylori infection (Alexander et al., 2021).
Chronic colonization of the H. pylori in the stomach alters the local immune response, leading to dysbiosis in the gastric microbiome (Brawner et al., 2014). Proteobacteria and Firmicutes are predominant in the gastric microbiome of H. pylori-positive individuals, while Actinobacteria, Firmicutes, and Bacteroidetes were dominant in H. pylori-negative individuals (Andersson et al., 2008). Significant variations in the relative abundances of genera such as Veillonella, Granulicatella, Neisseria, Fusobacterium, Prevotella, Actinomyces, and Streptococcus were also observed between H. pylori-positive and H. pylori-negative individuals (Klymiuk et al., 2017). The gastric microbiome composition of patients with H. pylori-associated gastritis was almost exclusively dominated by H. pylori, whereas a high microbial diversity was observed in H. pylori-negative gastritis individuals and in normal individuals (Parsons et al., 2017; Gantuya et al., 2019). Notably, in a study from Malaysia, Streptococcus was isolated at a significantly higher frequency in PUD cases (Khosravi et al., 2014). On the other hand, in the gastric microbiome of patients with GC, the abundance of Helicobacteraceae was lower than that in patients with chronic gastritis (Eun et al., 2014). For patients with advanced stages of GC, a distinct lower abundance of H. pylori, but higher abundances of Prevotella, Streptococcus, Veillonella, and Lactobacillus, in the gastric microbiome was observed (Dicksved et al., 2009). An increased abundance of the family Lachnospiraceae and Lactobacillus coleohominis along with a decreased count of Neisseria, Porphyromonas, and Streptococcus sinensis showed an association with GC (Aviles-Jimenez et al., 2014). On the other hand, GC patients from China had enrichment of species such as Slackia exigua, Streptococcus anginosus, Peptostreptococcus stomatis, Dialister pneumosintes, and Parvimonas micra in the stomach (Coker et al., 2018). Furthermore, an overgrowth of nitrate-reducing bacteria in the atrophic stomach is attributed to the development of gastric cancer via the accumulation of N-nitroso compounds (Barra et al., 2021).
Even though H. pylori colonization is restricted to the stomach, an alteration in the normal intestinal microbiome is observed during H. pylori infection in the murine model and in human (Kienesberger et al., 2016; Dash et al., 2019). Although the mechanism remains poorly studied, it is possibly due to the H. pylori-induced hypochlorhydria and altered gastrointestinal immunity. H. pylori infection is associated with increased diversity in the intestinal microbiome (Yang et al., 2019; Devi et al., 2021). Children with H. pylori-associated gastritis showed a higher abundance of Parabacteroides and Bacteroides, and a lower abundance of Faecalibacterium and Roseburia, than the healthy control group (Benavides-Ward et al., 2018). In India, individuals with H. pylori-associated diseases showed a higher Oscillospira abundance in the intestinal microbiome, while their Bifidobacterium abundance was remarkably low (Devi et al., 2021). A lower Bifidobacterium abundance in the intestine was also observed among Finnish patients with GC along with a higher abundance of Enterobacteriaceae (Table 1; Sarhadi et al., 2021). The use of probiotics such as Bifidobacterium and Lactobacillus has been shown to moderately improve H. pylori eradication and reduce the side effects of antibiotics (Zhang et al., 2015). It is known that probiotic strains of Lactobacillus and Bifidobacterium also impart a protective effect against H. pylori infection (Yang et al., 2021). L. acidophilus and L. bulgaricus decrease H. pylori adhesion to the gastric epithelial cells. Also, L. bulgaricus suppresses the secretion of proinflammatory cytokine IL-8 by gastric epithelial cells (Song et al., 2019).
Apart from H. pylori infection, gastrointestinal microbiome composition is affected by geographical variations and ethnicity, which indirectly influence the progression of gastric diseases (Figure 1; Gupta et al., 2017). Gastric microbiome analysis showed a higher abundance of Proteobacteria in Europeans, while a higher abundance of Firmicutes was observed in Asians. GC cohort from South Korea had a higher abundance of Lactobacillus, followed by Fusobacterium, and a lower abundance of Achromobacter, while Vietnamese cohorts had an opposite trend (Cavadas et al., 2020). Bacillus and Pseudomonas were found to be dominant in GC cohorts from both regions. Patients with GC from the United States and Europe had a relatively higher abundance of Bacillus, Parasutterella, Brevibacillus, and Fusobacterium. Patients with GC from the United States also had a lower abundance of Lactobacillus (Cavadas et al., 2020). Although the link between dysbiosis and gastroduodenal diseases is noticeable, the functional mechanisms involved in the process remained poorly described to date.
H. pylori is present in the human stomach since ancient times, but only a subset of the H. pylori-infected population is genetically predisposed to gastroduodenal diseases. Colonization of H. pylori in the human stomach is recognized by the body with pathogen recognition receptors (PRR) such as nucleotide-binding oligomerization domain (NOD) and Toll-like receptor (TLR) and eventually leads to the expression of cytokines, such as tumor necrosis factor (TNF) and interleukin 8 (IL-8) (Deforge and Remick, 1991; Amarante-Mendes et al., 2018). Genome-wide association study showed that polymorphisms in the PRR and cytokine genes among individuals from different geographical and ethnic backgrounds critically affect the immune response to H. pylori and clinical outcomes (Table 1 and Figure 1; Mommersteeg et al., 2018). A case-control study from Saudi Arabia showed that patients with TLR4-rs4986790 (A > G), TLR4-rs4986791 (C > T), and TLR10-rs10004195 (A > T) have a significant association with H. pylori infection, and TLR9-rs352140 (C > T) is connected with H. pylori-associated chronic gastritis (Eed et al., 2020). However, for people with Chinese ethnicity, the CC genotype of TLR4-rs11536889 and TLR9-rs187084 (T > C) is associated with an increased risk of GC, while TLR4-rs1927911, rs10759931, and rs10116253 were found to confer protection against GC (Castaño-Rodríguez et al., 2013). Also, a Chinese population with TLR10-rs10004195 polymorphism exhibited protection against H. pylori infection (Tang et al., 2015). A significant association has been identified between H. pylori-related GC and TLR4 SNPs, Asp299Gly, and Thr399Ile in a Caucasian population (Cheng et al., 2007). NOD1 796G > A polymorphism is linked to gastric mucosal inflammation in H. pylori-infected Korean population, while NOD1 796A/A genotype increases risk of gastric atrophy and antral intestinal metaplasia in a Turkish population (Kara et al., 2010; Kim et al., 2013). TNF-A-308G/A polymorphism increases the risk of GC in Caucasians, while TNF-A-857C/T and TNF-A-238G/A polymorphism increases the risk of gastric tumorigenesis in Asians (Yang et al., 2014; de Brito et al., 2018). Europeans with IL-1B-511T/-31T/IL-1RN*2 have a high risk of GC, while in a Kazakh population, IL-1B-511T/T and IL-1B-31C/C increase the risk of gastritis (El-Omar et al., 2000; Kulmambetova et al., 2014). However, in India, IL-1B-511TT genotype was higher in H. pylori-infected patients with PUD (Chakravorty et al., 2006). IL-6-174 G/G polymorphism in a Brazilian population is associated with a higher GC risk, while IL-8-251 A/A shows a higher risk of PUD (Gatti et al., 2007; Ramis et al., 2017). It was observed that IL-10-592T, IL-10-819T, and IL-10-1082A alleles increased the risk of GC in Caucasians, while IL-10-592C, IL-10-819C, and IL-10-1082G alleles were associated with GC risk in Asians (Kim et al., 2014). The TT genotype of IL-10-819C/T was shown to confer protection against GC in Mexican and Asian populations (Xue et al., 2012; Martínez-Campos et al., 2019).
Since the origin of anatomically modern humans in Africa and their subsequent migration, parallel evolutions and diversifications have also occurred to trillions of microbes (including H. pylori), which remained inhabited on and in the human body over the entire periods of human migrations and settlements. The pattern of genetic distance between different H. pylori strains from different populations reflects the migration pattern and its coevolution with its host (Falush et al., 2003; Domínguez-Bello et al., 2008). H. pylori remains attached to the human stomach with its adhesins. The blood group antigen-binding adhesin (BabA) on the bacterial surface binds to a difucosylated ABO/Lewis b (LeB) antigen present on the surface of human gastric epithelial cells (Huang et al., 2016). Both H. pylori BabA and human LeB are diverse proteins, which show remarkable variations with geography and ethnicity that subsequently affects colonization and clinical outcomes (Figure 1). Similarly, colonizing in the human gastrointestinal tract by members of the microbiome depends on the respective adhesin–receptor interactions, which are yet to be described. Also, while all virulence genes within the genomes of different H. pylori strains are well studied, the total virulence-associated proteins encoded in the genomes of all members of the gastric and intestinal microbiome, the virulome, which must have an effect on the gastric epithelium, are completely overlooked till date.
Like microbial virulome, human immunome, the total immune response genes present to protect against the invading pathogens, is also not sufficiently understood. It is known that the gastric niche contains several PRRs such as TLR and NLR along with antimicrobial peptides and mucins (Peek et al., 2010). The presence of antimicrobial peptides such as cathelicidins, hepcidins, and defensins and O-glycosylated protein mucin plays an important role in protecting the gastric epithelium from bacterial colonization (McGuckin et al., 2011; Li and Yu, 2020). A recent study also demonstrated the importance of galectin-3 in gastric epithelium against H. pylori infection (Park et al., 2016).
The virulome–immunome interplay is inevitable and possibly contributes to determining the clinical outcomes in the context of H. pylori infection and microbiome alteration. Bacterial pathogens are capable of modulating the host immune responses and cause damage. For example, Propionibacterium acnes (associated with lymphocytic gastritis) is known to produce short-chain fatty acids (SCFAs) such as propionate and butyrate that induce NKG2D–NKG2DL (natural killer group 2 member D) and the proinflammatory cytokine IL-15 that promote the progression to GC (Montalban-Arques et al., 2016). Further studies are necessary to understand how the gastrointestinal virulome manipulates the human immunome in the context of PUD and GC.
PUD and GC are complex diseases that develop with the influence of multiple factors. All major contributory factors–the H. pylori virulence, the gastrointestinal microbiome along with their virulome, and the microbe-responsive human immunome–show tremendous unevenness among different individuals and among different geographic regions, which is also linked to human migrations and settlements. The present inconsistencies that we observe in the clinical outcomes within the H. pylori-infected population settled in different locations have their roots in the combined evolutions of human immunome along with H. pylori virulence and gastrointestinal virulome, which is being continued for at least 60,000 years.
Human gastrointestinal health is undeniably the consequence of dynamic interplay between the gastrointestinal virulome and the host immunome. Recent studies suggest that a shift in this equilibrium has far-reaching effects on the progression of gastroduodenal diseases. Engineering the gastrointestinal microbiome by interventions like probiotics to modulate the host immune response may turn out to be an efficient strategy for the management of a spectrum of gastroduodenal diseases in the future, particularly in this era of growing antimicrobial resistance. However, further multidisciplinary approaches are required for uncovering the complex mechanisms so that more specific and effective microbiome-based therapies can be designed in the future.
SC conceived the idea. AN, RR, AF, and PC contributed to writing the manuscript. NT and SC edited the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by a grant (MED/2017/46) from the Department of Biotechnology (DBT) to SC as well as by the institutional support from Rajiv Gandhi Centre for Biotechnology, an autonomous institute under the Department of Biotechnology, Government of India.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank T. R. Santhosh Kumar, Scientist-G and Dean, Rajiv Gandhi Centre for Biotechnology, for his valuable suggestions and inputs. We also thank Chandrabhas Narayana, Director, Rajiv Gandhi Centre for Biotechnology, for institutional support.
Alexander, S. M., Retnakumar, R. J., Chouhan, D., Devi, T. N. B., Dharmaseelan, S., Devadas, K., et al. (2021). Helicobacter pylori in human stomach: the inconsistencies in clinical outcomes and the probable causes. Front. Microbiol. 12:713955. doi: 10.3389/fmicb.2021.713955
Amarante-Mendes, G. P., Adjemian, S., Branco, L. M., Zanetti, L. C., Weinlich, R., and Bortoluci, K. R. (2018). Pattern recognition receptors and the host cell death molecular machinery. Front. Immunol. 9:2379. doi: 10.3389/fimmu.2018.02379
Andersson, A. F., Lindberg, M., Jakobsson, H., Bäckhed, F., Nyrén, P., and Engstrand, L. (2008). Comparative analysis of human gut microbiota by barcoded pyrosequencing. PloS One 3:e2836. doi: 10.1371/journal.pone.0002836
Anstee, D. J. (2010). The relationship between blood groups and disease. Blood 115, 4635–4643. doi: 10.1182/blood-2010-01-261859
Aviles-Jimenez, F., Vazquez-Jimenez, F., Medrano-Guzman, R., Mantilla, A., and Torres, J. (2014). Stomach microbiota composition varies between patients with non-atrophic gastritis and patients with intestinal type of gastric cancer. Sci. Rep. 4:4202. doi: 10.1038/srep04202
Barra, W. F., Sarquis, D. P., Khayat, A. S., Khayat, B. C. M., Demachki, S., Anaissi, A. K. M., et al. (2021). Gastric cancer microbiome. Pathobiology 88, 156–169.
Bauer, B., and Meyer, T. F. (2011). The human gastric pathogen Helicobacter pylori and its association with gastric cancer and ulcer disease. Ulcers 2011. doi: 10.1155/2011/340157
Benavides-Ward, A., Vasquez-Achaya, F., Silva-Caso, W., Aguilar-Luis, M. A., Mazulis, F., Urteaga, N., et al. (2018). Helicobacter pylori and its relationship with variations of gut microbiota in asymptomatic children between 6 and 12 years. BMC Res. Notes 11:468. doi: 10.1186/s13104-018-3565-5
Blaser, M. J. (2014). The microbiome revolution. J. Clin. Invest. 124, 4162–4165. doi: 10.1172/jci78366
Brawner, K. M., Morrow, C. D., and Smith, P. D. (2014). Gastric microbiome and gastric cancer. Cancer J. 20, 211. doi: 10.1097/ppo.0000000000000043
Brown, L. M. (2000). Helicobacter pylori: epidemiology and routes of transmission. Epidemiol. Rev. 22, 283–297. doi: 10.1093/oxfordjournals.epirev.a018040
Castaño-Rodríguez, N., Kaakoush, N. O., Goh, K.-L., Fock, K. M., and Mitchell, H. M. (2013). The role of TLR2, TLR4 and CD14 genetic polymorphisms in gastric carcinogenesis: a case-control study and meta-analysis. PLoS One 8:e60327. doi: 10.1371/journal.pone.0060327
Cavadas, B., Camacho, R., Ferreira, J. C., Ferreira, R. M., Figueiredo, C., Brazma, A., et al. (2020). Gastric microbiome diversities in gastric cancer patients from Europe and Asia mimic the human population structure and are partly driven by microbiome quantitative trait loci. Microorganisms 8:1196. doi: 10.3390/microorganisms8081196
Chakravorty, M., Ghosh, A., Choudhury, A., Santra, A., Hembrum, J., and Roychoudhury, S. (2006). Interaction between IL1B gene promoter polymorphisms in determining susceptibility to Helicobacter pylori associated duodenal ulcer. Hum. Mutat. 27, 411–419. doi: 10.1002/humu.20299
Chang, W.-L., Yeh, Y.-C., and Sheu, B.-S. (2018). The impacts of H. pylori virulence factors on the development of gastroduodenal diseases. J. Biomed. Sci. 25, 1–9. doi: 10.1186/s12929-018-0466-9
Chauhan, N., Tay, A. C. Y., Marshall, B. J., and Jain, U. (2019). Helicobacter pylori VacA, a distinct toxin exerts diverse functionalities in numerous cells: an overview. Helicobacter 24, e12544. doi: 10.1111/hel.12544
Cheng, P.-L., Eng, H.-L., Chou, M.-H., You, H.-L., and Lin, T.-M. (2007). Genetic polymorphisms of viral infection-associated toll-like receptors in Chinese population. Transl. Res. 150, 311–318. doi: 10.1016/j.trsl.2007.03.010
Cho, I., and Blaser, M. J. (2012). The human microbiome: at the interface of health and disease. Nat. Rev. Genet. 13, 260–270. doi: 10.1038/nrg3182
Coker, O. O., Dai, Z., Nie, Y., Zhao, G., Cao, L., Nakatsu, G., et al. (2018). Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut 67, 1024–1032. doi: 10.1136/gutjnl-2017-314281
Correa, P., and Piazuelo, M. B. (2012). Evolutionary history of the Helicobacter pylori genome: implications for gastric carcinogenesis. Gut Liver 6, 21. doi: 10.5009/gnl.2012.6.1.21
Dash, N. R., Khoder, G., Nada, A. M., and Al Bataineh, M. T. (2019). Exploring the impact of Helicobacter pylori on gut microbiome composition. PLoS One 14:e0218274. doi: 10.1371/journal.pone.0218274
de Brito, B. B., Da Silva, F. A. F., and De Melo, F. F. (2018). Role of polymorphisms in genes that encode cytokines and Helicobacter pylori virulence factors in gastric carcinogenesis. World J. Clin. Oncol. 9:83. doi: 10.5306/wjco.v9.i5.83
Deforge, L., and Remick, D. G. (1991). Kinetics of TNF, IL-6, and IL-8 gene expression in LPS-stimulated human whole blood. Biochem. Biophys. Res. Commun. 174, 18–24. doi: 10.1016/0006-291x(91)90478-p
Devi, T. B., Devadas, K., George, M., Gandhimathi, A., Chouhan, D., Retnakumar, R., et al. (2021). Low bifidobacterium abundance in the lower gut microbiota is associated with Helicobacter pylori-related gastric ulcer and gastric cancer. Front. Microbiol. 12:171. doi: 10.3389/fmicb.2021.631140
Dicksved, J., Lindberg, M., Rosenquist, M., Enroth, H., Jansson, J. K., and Engstrand, L. (2009). Molecular characterization of the stomach microbiota in patients with gastric cancer and in controls. J. Med. Microbiol. 58, 509–516. doi: 10.1099/jmm.0.007302-0
Domínguez-Bello, M. G., Pérez, M. E., Bortolini, M. C., Salzano, F. M., Pericchi, L. R., Zambrano-Guzman, O., et al. (2008). Amerindian Helicobacter pylori strains go extinct, as European strains expand their host range. PLoS One 3:e3307. doi: 10.1371/journal.pone.0003307
Eed, E. M., Hawash, Y. A., Khalifa, A. S., Alsharif, K. F., Alghamdi, S. A., Almalki, A. A., et al. (2020). Association of toll-like receptors 2, 4, 9 and 10 genes polymorphisms and Helicobacter pylori-related gastric diseases in Saudi patients. Indian J. Med. Microbiol. 38, 94–100. doi: 10.4103/ijmm.IJMM_20_164
El-Omar, E. M., Carrington, M., Chow, W.-H., Mccoll, K. E., Bream, J. H., Young, H. A., et al. (2000). Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature 404, 398–402. doi: 10.1038/35006081
Eun, C. S., Kim, B. K., Han, D. S., Kim, S. Y., Kim, K. M., Choi, B. Y., et al. (2014). Differences in gastric mucosal microbiota profiling in patients with chronic gastritis, intestinal metaplasia, and gastric cancer using pyrosequencing methods. Helicobacter 19, 407–416. doi: 10.1111/hel.12145
Falush, D., Wirth, T., Linz, B., Pritchard, J. K., Stephens, M., Kidd, M., et al. (2003). Traces of human migrations in Helicobacter pylori populations. Science 299, 1582–1585. doi: 10.1126/science.1080857
Fernandez-Gonzalez, E., and Backert, S. (2014). DNA transfer in the gastric pathogen Helicobacter pylori. J. Gastroenterol. 49, 594–604. doi: 10.1007/s00535-014-0938-y
Foegeding, N. J., Caston, R. R., Mcclain, M. S., Ohi, M. D., and Cover, T. L. (2016). An overview of Helicobacter pylori VacA toxin biology. Toxins 8:173. doi: 10.3390/toxins8060173
Gantuya, B., El-Serag, H. B., Matsumoto, T., Ajami, N. J., Oyuntsetseg, K., Azzaya, D., et al. (2019). Gastric microbiota in Helicobacter pylori-negative and-positive gastritis among high incidence of gastric cancer area. Cancers 11, 504. doi: 10.3390/cancers11040504
Gatti, L. L., Burbano, R. R., Zambaldi-Tunes, M., De-Lábio, R. W., De Assumpção, P. P., De Arruda Cardoso-Smith, M., et al. (2007). Interleukin-6 polymorphisms, Helicobacter pylori infection in adult Brazilian patients with chronic gastritis and gastric adenocarcinoma. Arch. Med. Res. 38, 551–555. doi: 10.1016/j.arcmed.2006.12.011
Gilbert, J. A., Blaser, M. J., Caporaso, J. G., Jansson, J. K., Lynch, S. V., and Knight, R. (2018). Current understanding of the human microbiome. Nat. Med. 24, 392–400. doi: 10.1038/nm.4517
Gupta, V. K., Paul, S., and Dutta, C. (2017). Geography, ethnicity or subsistence-specific variations in human microbiome composition and diversity. Front. Microbiol. 8:1162. doi: 10.3389/fmicb.2017.01162
Harry, L. T., Mobley, G. L. M., and Hazell, S. L. (2001). “Overview,” in Helicobacter pylori: Physiology and Genetics, eds G. L. M. Harry, L. T. Mobley, and S. L. Hazell (Washington, DC: ASM Press).
Hatakeyama, M. (2014). Helicobacter pylori CagA and gastric cancer: a paradigm for hit-and-run carcinogenesis. Cell Host Microbe 15, 306–316. doi: 10.1016/j.chom.2014.02.008
Higashi, H., Tsutsumi, R., Fujita, A., Yamazaki, S., Asaka, M., Azuma, T., et al. (2002). Biological activity of the Helicobacter pylori virulence factor CagA is determined by variation in the tyrosine phosphorylation sites. Proc. Natl. Acad. Sci. U.S.A. 99, 14428–14433. doi: 10.1073/pnas.222375399
Hishida, A., Matsuo, K., Goto, Y., Naito, M., Wakai, K., Tajima, K., et al. (2011). Combined effect of miR-146a rs2910164 G/C polymorphism and Toll-like receptor 4+ 3725 G/C polymorphism on the risk of severe gastric atrophy in Japanese. Dig. Dis. Sci. 56, 1131–1137. doi: 10.1007/s10620-010-1376-1
Hooi, J. K., Lai, W. Y., Ng, W. K., Suen, M. M., Underwood, F. E., Tanyingoh, D., et al. (2017). Global prevalence of Helicobacter pylori infection: systematic review and meta-analysis. Gastroenterology 153, 420–429.
Huang, Y., Wang, Q.-L., Cheng, D.-D., Xu, W.-T., and Lu, N.-H. (2016). Adhesion and invasion of gastric mucosa epithelial cells by Helicobacter pylori. Front. Cell. Infect. Microbiol. 6:159. doi: 10.3389/fcimb.2016.00159
Hullar, M. A., Burnett-Hartman, A. N., and Lampe, J. W. (2014). Gut microbes, diet, and cancer. Adv. Nutr. Cancer 159, 377–399. doi: 10.1007/978-3-642-38007-5_22
Israel, D. A., Salama, N., Krishna, U., Rieger, U. M., Atherton, J. C., Falkow, S., et al. (2001). Helicobacter pylori genetic diversity within the gastric niche of a single human host. Proc. Natl. Acad. Sci. U.S.A. 98, 14625–14630. doi: 10.1073/pnas.251551698
Kara, B., Akkiz, H., Doran, F., Bayram, S., Erken, E., Gumurdullu, Y., et al. (2010). The significance of E266K polymorphism in the NOD1 gene on Helicobacter pylori infection: an effective force on pathogenesis? Clin. Exp. Med. 10, 107–112. doi: 10.1007/s10238-009-0077-6
Keikha, M., Ali-Hassanzadeh, M., and Karbalaei, M. (2020). Association of Helicobacter pylori vacA genotypes and peptic ulcer in Iranian population: a systematic review and meta-analysis. BMC Gastroenterol. 20:266. doi: 10.1186/s12876-020-01406-9
Kennedy, M. S., and Chang, E. B. (2020). The microbiome: composition and locations. Prog. Mol. Biol. Transl. Sci. 176, 1–42. doi: 10.1016/bs.pmbts.2020.08.013
Khosravi, Y., Dieye, Y., Poh, B. H., Ng, C. G., Loke, M. F., Goh, K. L., et al. (2014). Culturable bacterial microbiota of the stomach of Helicobacter pylori positive and negative gastric disease patients. Sci. World J. 2014:610421. doi: 10.1155/2014/610421
Kienesberger, S., Cox, L., Livanos, A., Zhang, X.-S., Chung, J., Perez-Perez, G., et al. (2016). Gastric Helicobacter pylori infection affects local and distant microbial populations and host responses. Cell Rep. 14, 1395–1407. doi: 10.1016/j.celrep.2016.01.017
Kim, E. J., Lee, J. R., Chung, W. C., Jung, S. H., Sung, H. J., Lee, Y. W., et al. (2013). Association between genetic polymorphisms of NOD 1 and Helicobacter pylori-induced gastric mucosal inflammation in healthy Korean population. Helicobacter 18, 143–150. doi: 10.1111/hel.12020
Kim, J., Kim, Y., and Lee, K.-A. (2014). Ethnic differences in gastric cancer genetic susceptibility: allele flips of interleukin gene. World J. Gastroenterol. 20:4558. doi: 10.3748/wjg.v20.i16.4558
Klymiuk, I., Bilgilier, C., Stadlmann, A., Thannesberger, J., Kastner, M.-T., Högenauer, C., et al. (2017). The human gastric microbiome is predicated upon infection with Helicobacter pylori. Front. Microbiol. 8:2508. doi: 10.3389/fmicb.2017.02508
Kulmambetova, G. N., Imanbekova, M. K., Logvinenko, A. A., Sukashev, A. T., Filipenko, M. L., and Ramanculov, E. M. (2014). Association of cytokine gene polymorphisms with gastritis in a Kazakh population. Asian Pac. J. Cancer Prev. 15, 7763–7768. doi: 10.7314/apjcp.2014.15.18.7763
Li, Q., and Yu, H. (2020). The role of non-H. pylori bacteria in the development of gastric cancer. Am. J. Cancer Res. 10:2271.
Linz, B., Balloux, F., Moodley, Y., Manica, A., Liu, H., Roumagnac, P., et al. (2007). An African origin for the intimate association between humans and Helicobacter pylori. Nature 445, 915–918. doi: 10.1038/nature05562
Maixner, F., Krause-Kyora, B., Turaev, D., Herbig, A., Hoopmann, M. R., Hallows, J. L., et al. (2016). The 5300-year-old Helicobacter pylori genome of the Iceman. Science 351, 162–165. doi: 10.1126/science.aad2545
Malfertheiner, P., Link, A., and Selgrad, M. (2014). Helicobacter pylori: perspectives and time trends. Nat. Rev. Gastroenterol. Hepatol. 11, 628–638. doi: 10.1038/nrgastro.2014.99
Martínez-Campos, C., Torres-Poveda, K., Camorlinga-Ponce, M., Flores-Luna, L., Maldonado-Bernal, C., Madrid-Marina, V., et al. (2019). Polymorphisms in IL-10 and TGF-β gene promoter are associated with lower risk to gastric cancer in a Mexican population. BMC Cancer 19:453. doi: 10.1186/s12885-019-5627-z
Mayerle, J., Den Hoed, C. M., Schurmann, C., Stolk, L., Homuth, G., Peters, M. J., et al. (2013). Identification of genetic loci associated with Helicobacter pylori serologic status. JAMA 309, 1912–1920. doi: 10.1001/jama.2013.4350
McColl, K., El-Omar, E., and Gillen, D. (1998). Interactions between H. pylori infection, gastric acid secretion and anti-secretory therapy. Br. Med. Bull. 54, 121–138. doi: 10.1093/oxfordjournals.bmb.a011663
McGuckin, M. A., Lindén, S. K., Sutton, P., and Florin, T. H. (2011). Mucin dynamics and enteric pathogens. Nat. Rev. Microbiol. 9, 265–278. doi: 10.1038/nrmicro2538
Mommersteeg, M. C., Yu, J., Peppelenbosch, M. P., and Fuhler, G. M. (2018). Genetic host factors in Helicobacter pylori-induced carcinogenesis: Emerging new paradigms. Biochim. Biophys. Acta 1869, 42–52. doi: 10.1016/j.bbcan.2017.11.003
Montalban-Arques, A., Wurm, P., Trajanoski, S., Schauer, S., Kienesberger, S., Halwachs, B., et al. (2016). Propionibacterium acnes overabundance and natural killer group 2 member D system activation in corpus-dominant lymphocytic gastritis. J. Pathol. 240, 425–436. doi: 10.1002/path.4782
Moodley, Y., Linz, B., Yamaoka, Y., Windsor, H. M., Breurec, S., Wu, J.-Y., et al. (2009). The peopling of the Pacific from a bacterial perspective. Science 323, 527–530. doi: 10.1126/science.1166083
Papadakos, K. S., Sougleri, I. S., Mentis, A. F., Hatziloukas, E., and Sgouras, D. N. (2013). Presence of terminal EPIYA phosphorylation motifs in Helicobacter pylori CagA contributes to IL-8 secretion, irrespective of the number of repeats. PLoS One 8:e56291. doi: 10.1371/journal.pone.0056291
Park, A.-M., Hagiwara, S., Hsu, D. K., Liu, F.-T., and Yoshie, O. (2016). Galectin-3 plays an important role in innate immunity to gastric infection by Helicobacter pylori. Infect. Immun. 84, 1184–1193. doi: 10.1128/IAI.01299-15
Parsons, B. N., Ijaz, U. Z., D’amore, R., Burkitt, M. D., Eccles, R., Lenzi, L., et al. (2017). Comparison of the human gastric microbiota in hypochlorhydric states arising as a result of Helicobacter pylori-induced atrophic gastritis, autoimmune atrophic gastritis and proton pump inhibitor use. PLoS Pathog. 13:e1006653. doi: 10.1371/journal.ppat.1006653
Peek, R. M. Jr., Fiske, C., and Wilson, K. T. (2010). Role of innate immunity in Helicobacter pylori-induced gastric malignancy. Physiol. Rev. 90, 831–858. doi: 10.1152/physrev.00039.2009
Ramis, I. B., Vianna, J. S., Gonçalves, C. V., Von Groll, A., Dellagostin, O. A., and Da Silva, P. E. A. (2017). Polymorphisms of the IL-6, IL-8 and IL-10 genes and the risk of gastric pathology in patients infected with Helicobacter pylori. J. Microbiol. Immunol. Infect. 50, 153–159. doi: 10.1016/j.jmii.2015.03.002
Rawla, P., and Barsouk, A. (2019). Epidemiology of gastric cancer: global trends, risk factors and prevention. Prz. Gastroenterol. 14:26. doi: 10.5114/pg.2018.80001
Sahara, S., Sugimoto, M., Vilaichone, R.-K., Mahachai, V., Miyajima, H., Furuta, T., et al. (2012). Role of Helicobacter pylori cagA EPIYA motif and vacA genotypes for the development of gastrointestinal diseases in Southeast Asian countries: a meta-analysis. BMC Infect. Dis. 12:223. doi: 10.1186/1471-2334-12-223
Sarhadi, V., Mathew, B., Kokkola, A., Karla, T., Tikkanen, M., Rautelin, H., et al. (2021). Gut microbiota of patients with different subtypes of gastric cancer and gastrointestinal stromal tumors. Gut Pathog. 13:11. doi: 10.1186/s13099-021-00403-x
Saxena, A., Shukla, S., Prasad, K., and Ghoshal, U. (2011). Virulence attributes of Helicobacter pylori isolates & their association with gastroduodenal disease. Indian J. Med. Res. 133:514.
Song, H., Zhou, L., Liu, D., Ge, L., and Li, Y. (2019). Probiotic effect on Helicobacter pylori attachment and inhibition of inflammation in human gastric epithelial cells. Exp. Ther. Med. 18, 1551–1562. doi: 10.3892/etm.2019.7742
Sugimoto, M., and Yamaoka, Y. (2009). The association of vacA genotype and Helicobacter pylori-related disease in Latin American and African populations. Clin. Microbiol. Infect. 15, 835–842. doi: 10.1111/j.1469-0691.2009.02769.x
Suzuki, R., Shiota, S., and Yamaoka, Y. (2012). Molecular epidemiology, population genetics, and pathogenic role of Helicobacter pylori. Infect. Genet. Evol. 12, 203–213. doi: 10.1016/j.meegid.2011.12.002
Tang, F.-B., Li, Z.-X., Wang, Y.-M., Zhang, L., Ma, J.-L., Zhou, T., et al. (2015). Toll-like receptor 1 and 10 polymorphisms, Helicobacter pylori susceptibility and risk of gastric lesions in a high-risk Chinese population. Infect. Genet. Evol. 31, 263–269. doi: 10.1016/j.meegid.2015.02.005
Trang, T. T. H., Thanh Binh, T., and Yamaoka, Y. (2016). Relationship between vacA types and development of gastroduodenal diseases. Toxins 8:182. doi: 10.3390/toxins8060182
Turnbaugh, P. J., Ley, R. E., Hamady, M., Fraser-Liggett, C. M., Knight, R., and Gordon, J. I. (2007). The human microbiome project. Nature 449, 804–810.
Van Doorn, L. J., Figueiredo, C., Mégraud, F., Pena, S., Midolo, P., Queiroz, D. M. D. M., et al. (1999). Geographic distribution of vacA allelic types of Helicobacter pylori. Gastroenterology 116, 823–830. doi: 10.1016/s0016-5085(99)70065-x
Warren, J. R., and Marshall, B. (1983). Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet 321, 1273–1275. doi: 10.1016/s0140-6736(83)92719-8
Xue, H., Lin, B., An, J., Zhu, Y., and Huang, G. (2012). Interleukin-10-819 promoter polymorphism in association with gastric cancer risk. BMC Cancer 12:102. doi: 10.1186/1471-2407-12-102
Yamaoka, Y. (2008). Roles of Helicobacter pylori BabA in gastroduodenal pathogenesis. World J. Gastroenterol. 14, 4265. doi: 10.3748/wjg.14.4265
Yang, J., Zhou, X., Liu, X., Ling, Z., and Ji, F. (2021). Role of the gastric microbiome in gastric cancer: from carcinogenesis to treatment. Front. Microbiol. 12:641322. doi: 10.3389/fmicb.2021.641322
Yang, J.-P., Hyun, M.-H., Yoon, J.-M., Park, M.-J., Kim, D., and Park, S. (2014). Association between TNF-α-308 G/A gene polymorphism and gastric cancer risk: a systematic review and meta-analysis. Cytokine 70, 104–114. doi: 10.1016/j.cyto.2014.07.005
Yang, L., Zhang, J., Xu, J., Wei, X., Yang, J., Liu, Y., et al. (2019). Helicobacter pylori infection aggravates dysbiosis of gut microbiome in children with gastritis. Front. Cell. Infect. Microbiol. 9:375. doi: 10.3389/fcimb.2019.00375
Keywords: H. pylori, genome, microbiome, peptic ulcer disease, gastric cancer, virulome, immunome
Citation: Nath AN, Retnakumar RJ, Francis A, Chhetri P, Thapa N and Chattopadhyay S (2022) Peptic Ulcer and Gastric Cancer: Is It All in the Complex Host–Microbiome Interplay That Is Encoded in the Genomes of “Us” and “Them”? Front. Microbiol. 13:835313. doi: 10.3389/fmicb.2022.835313
Received: 14 December 2021; Accepted: 18 March 2022;
Published: 25 April 2022.
Edited by:
Souvik Mukherjee, National Institute of Biomedical Genomics (NIBMG), IndiaReviewed by:
Sunil D. Saroj, Symbiosis International University, IndiaCopyright © 2022 Nath, Retnakumar, Francis, Chhetri, Thapa and Chattopadhyay. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Santanu Chattopadhyay, c2FudGFudUByZ2NiLnJlcy5pbg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.