- 1Key Laboratory of Exploration and Utilization of Aquatic Genetic Resources, Ministry of Education, Shanghai Ocean University, Shanghai, China
- 2International Research Center for Marine Biosciences, Ministry of Science and Technology, Shanghai Ocean University, Shanghai, China
Hepcidin is a small peptide composed of signal peptide, propeptide, and the bioactive mature peptide from N terminal to C terminal. Mature hepcidin is an antibacterial peptide and iron regulator with eight highly conserved cysteines forming four intramolecular disulfide bonds, giving it a β sheet hairpin-like structure. Hepcidin homologs are found in a variety of vertebrates, especially fish, and their diversity may be associated with different habitats and different levels of pathogens. Dissostichus mawsoni, an Antarctic notothenioid fish that lives in the coldest water unlike most places of the world, with at least two hepcidin variants with eight cysteines. We confirmed the formation process of activated mature hepcidins from D. mawsoni in Chinese hamster ovary (CHO) cell line, obtained recombinant hepcidin protein from prokaryotes, and characterized its binding ability and antibacterial activity against varying bacteria. The expression of hepcidin in CHO cell line showed that the prepropeptide of Dmhep_8cysV1 and Dmhep_8cysV2 cleavage into smaller mature peptide. The antibacterial assay and flow cytometry showed that Dmhep_8cysV1, Dmhep_8cysV2, and Drhep bound to different bacteria and killed them with different minimum inhibitory concentration. These data suggest that hepcidin plays an important role in the innate immunity of D. mawsoni and is of great value in improving resistance to pathogens.
Introduction
Since the 1950s, antibiotics have been widely used around the world to fend off pathogens. Unfortunately, multidrug-resistant microbes have evolved at an unprecedented rate and are insensitive to many current treatment (Pei et al., 2019). In the 21st century, people have increasingly turned to study plants and animals in the hope of identifying and characterizing antimicrobial peptides with broad-spectrum antimicrobial activity and unique membrane-directed mechanism to replace traditional antibiotics (Iman et al., 2019). So far, thousands of antimicrobial peptides have been discovered from different species; however, their use has not been widely promoted because of their high production costs.
Hepcidins, a group of cysteine-rich peptides, are widespread in all kinds of animals. It was initially identified as a natural antibacterial peptide from human urine and highly expressed in liver (Krause et al., 2000; Park et al., 2001); subsequently, it was discovered to be an important iron homeostasis regulator in mammals (Fleming and Sly, 2001; Nicolas et al., 2001; Weinstein et al., 2002). Hepcidin polypeptide has been demonstrated to regulate iron release from cells by binding to its receptor, an iron transport membrane protein, ferroportin, which results in its degradation (Nemeth et al., 2004; De Domenico et al., 2007; Fleming, 2008). However, the detailed mechanism of the antibacterial function of hepcidin still remains to be studied further.
Hepcidins have been identified and predicted in many vertebrates, including fishes, amphibians, reptiles, mammals, and birds. All known hepcidin peptides, containing 84–91 amino acids (aa), comprise signal peptide, propeptide, and the bioactive mature peptide from N terminal to C terminal. There are eight conserved cysteine residues in almost all hepcidin mature peptides, which form four intramolecular disulfide bonds that stabilize a βsheet hairpin-like structure (Hunter et al., 2002; Lauth et al., 2005; Ganz and Nemeth, 2006). Researchers have demonstrated that the disulfide bonds are necessary for hepcidin to kill bacteria (Hocquellet et al., 2012; Lin et al., 2014). There are many reported researches focused on fish hepcidin. Cysteine-rich hepcidins have been isolated and identified in fish including large yellow croaker (Pseudosciaena crocea) and bass (Shike et al., 2002; Zhang et al., 2009). In addition, the antibacterial activities of recombinant or synthesized hepcidin from at least 21 fish species have been tested, including large yellow croaker (P. crocea) (Wang et al., 2009), Japanese flounder (Paralichthys olivaceus) (Srinivasulu et al., 2008), tilapia (Oreochromis mossambicus) (Huang et al., 2007), Nile tilapia (Oreochromis niloticus) (Abdelkhalek et al., 2020; Yin et al., 2022), gilthead seabream (Sparus aurata) (Cuesta et al., 2008), orange-spotted grouper (Epinephelus coioides) (Zhou et al., 2011), black porgy (Acanthopagrus schlegelii) (Yang et al., 2011), medaka (Oryzias melastigmus) (Cai et al., 2012), Chinese rare minnow (Gobiocypris rarus) (Ke et al., 2015), convict cichlid (Amatitlania nigrofasciata) (Chi et al., 2015), caspian trout (Salmo caspius) (Iman et al., 2019), trout (álvarez et al., 2014), zebrafish (Danio rerio) (Lin et al., 2014), pallas (Brachymystax lenok) (Xu et al., 2018), medium carp (Puntius sarana) (Das et al., 2015), brown trout (Salmo trutta) (Huang et al., 2019), roughskin sculpin (Trachidermus fasciatus) (Liu et al., 2017), simensis crocodile (Crocodylus siamensis) (Hao et al., 2012), and hybrid fish (Carassius auratus) (Luo et al., 2020), channel catfish (Ictalurus punctatus) (Tao et al., 2014), Mudskipper (Boleophthalmus pectinirostris) (Chen et al., 2018), and there is evidence that the antibacterial function of hepcidin is conserved in fish. Moreover, hepcidin sequences have been cloned from various fishes without testing their antibacterial activity, for example, olive flounder (P. olivaceus) (Kim et al., 2005), Atlantic cod (Gadus morhua) (Solstad et al., 2008), and Antarctic tooth fish (Dissostichus mawsoni) (Xu et al., 2008).
Although hepcidin gene widely is distributed in vertebrates, and its number varies across species, from one to seven. It has been identified that there is only one hepcidin gene in human, two in mice, five in an Antarctic fish D. mawsoni (Xu et al., 2008), and seven in black sea bream (Yang et al., 2007). Among them the D. mawsoni (NCBI: txid6530, Fishbase ID: 7039) widely is distributed in freezing waters of high-latitude Antarctic coasts, as far south as 77.5°S (McMurdo Sound), the southern limit of Antarctic marine life. Despite that D. mawsoni grows slow in the cold habitat, it can still grow to enormous size (2 m in length and 140 kg in weight), which means they have a strong immune system (Chen et al., 2019). Five variants of hepcidin were identified in D. mawsoni genome by sequencing (NCBI: EU221590.1, EU221595.1, EU221596.1, EU221602.1, and EU221603.1), and their amino acid sequences are shown in Supplementary Figure 1. One of the conserved variants is named Dmhep_8cysV1 (EU221590), whereas the rest are newly discovered in genome, such as Dmhep_8cysV2 (EU221595). However, as a common freshwater model organism, there is only one conserved hepcidin in the genome of D. rerio (NCBI: txid7955, Fishbase ID: 1822), named Drhep (NCBI: NC_007127.7). Studies have shown that zebrafish recombinant hepcidin peptide can inhibit the growth of Escherichia coli, Vibrio anguillarum, Staphylococcus aureus, and Bacillus subtilis (Lin et al., 2014). However, whether non-conserved hepcidin from D. mawsoni has antibacterial activity remains unclear. Therefore, our study determined and compared the antibacterial activity of hepcidins in D. mawsoni and D. rerio.
Materials and Methods
Fish and Bacterial Strains
All zebrafish and D. mawsoni procedures were approved by Shanghai Ocean University Animal Care and Use Committee. The AB zebrafish strains were purchased from Shanghai Institute of Biology, Chinese Academy of Sciences. The light cycle was maintained at 10-h light off and 14-h light on. The D. mawsoni was collected from McMurdo Sound. Aeromonas hydrophila, E. coli, Edwardsiella tarda, S. aureus, and Streptococcus agalactiae are cultured in Luria broth (LB) medium at their optimum growth temperature.
RNA Isolation and Tissue Expression Analysis
Various tissues of adult fish were dissected according to the method as previously described (Tripti and Mary, 2010). Tissues were rapidly frozen in liquid nitrogen and stored at −80°C until use. Total RNA for cloning and quantitative reverse transcription polymerase chain reaction (qRT-PCR) was extracted from fish brain, liver, gill, ovary, muscle, skin, heart, gut, kidney, and spleen using TRIzol reagent (Invitrogen) following the manufacturer’s instructions. RNA quality was assessed by electrophoresis on a 1% agarose gel, and RNA concentration was estimated using a Nanodrop 2000 spectrophotometer (Thermo Fisher).
One microgram of total RNA was reverse transcribed using PrimerScript™ RT reagent Kit with gDNA Erase (Takara). qRT-PCR was carried out with FastStart SYBR Green Master (Roche), and each reaction was triplicated to avoid possible random variations. Primers were designed according to various hepcidin sequences obtained from the National Center for Biotechnology Information (NCBI). Primers are shown in Table 1.
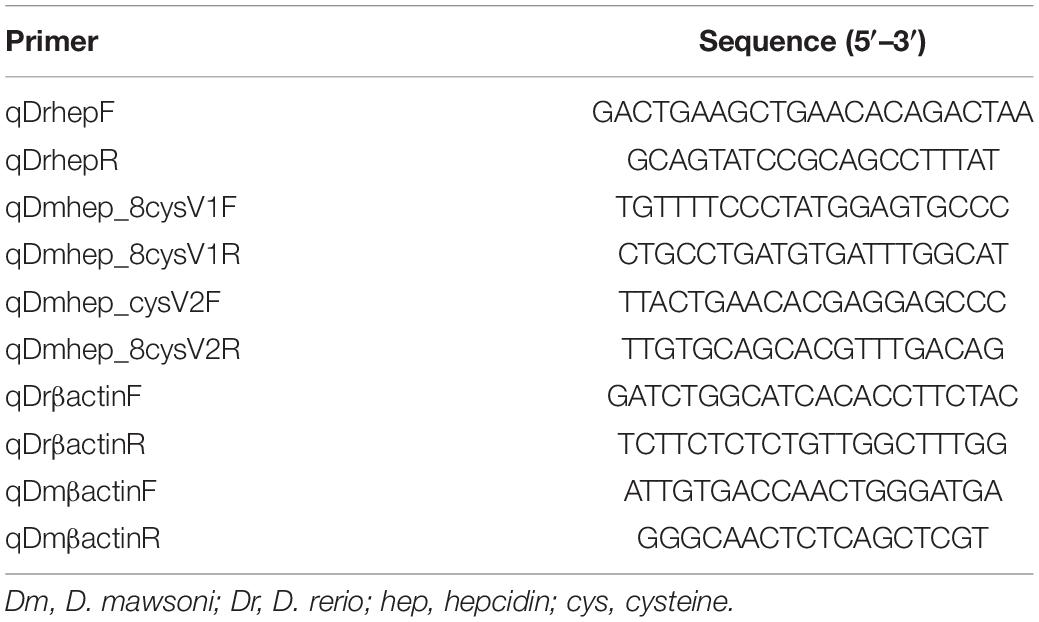
Table 1. Primers for quantification of D. mawsoni and D. rerio hepcidin mRNA by real time quantitative PCR.
Sequence and Phylogenetic Analyses
The hepcidin coding sequences (CDS) sequences were analyzed using DNAMAN8 software, and the deduced amino acid sequences were translated using online website, https://web.expasy.org/translate/. The theoretical isoelectric point and molecular weight of the protein were calculated using online website, https://web.expasy.org/compute_pi/. The domain prediction of hepcidin amino acid sequence was executed in SMART.1 The deduced amino acid sequences of hepcidin in other species were identified in NCBI GenBank.2 Multiple sequence alignment was created with DNAMAN8 software.
Recombinant Plasmid Construction
Hepcidin CDS sequences were inserted into a plasmid with zebrafish β-actin promoter at EcoRI and BamHI sites. EGFP and mCherry DNA sequence was inserted into CDS regions following 5′ terminal of propeptide and 3′ terminal of mature peptide by PCR and In-Fusion Cloning, respectively. These plasmids were transfected into Chinese hamster ovary (CHO) cells using transfection reagent (Thermo Fisher). The hepcidin mature sequences were inserted into pHis-TEV using NcoI and XhoI enzyme digesting sites. The recombinant plasmid with His tag sequence at the 5′ terminal of mature hepcidin sequence can be transformed into E. coli to produce hepcidin protein controlled by T7 promoter. All the recombinant plasmids were verified by sequencing.
Expression of Hepcidin in Chinese Hamster Ovary Cell Line
Seed CHO cells had 70–90% confluence for transfection. Diluting 6 μL turbofect reagent and 4 μg plasmid in 100 μL Opti-MEM medium (Gibco), respectively. Then, the two diluting solutions were mixed and incubate for 15 min at room temperature. The mixture was added to cells and shaken gently. The cells were imaged under the fluorescence microscopy 72 h after transfection.
Expression and Identification of Hepcidin in Escherichia coli
The recombinant pHis-TEV plasmids were transformed into BL21(DE3) and cultured in ampicillin resistance LB plate at 37°C. A single colony containing various recombinant plasmid was transferred into 4 mL ampicillin (20 μg/mL)–resistant LB medium and cultured with 200 revolutions/min (rpm) shaking at 37°C for 12 h. Three milliliters of the cultures was transferred into 300 mL fresh LB medium containing ampicillin and incubated at 37°C until the OD600 reached 0.5, and then isopropyl thiogalactopyranoside was added with a final concentration 0.05 and 0 mM, respectively. The cultures were grown at 18°C for 16 h to induce the expression of the recombinant protein better. The cells were collected, centrifuged at 6,000 g at 4°C for 10 min, and resuspended in phosphate-buffered saline (PBS) buffer (Hyclone) for 15 min with 100-W ultrasound in an ice water bath for completely crack of the bacteria. The lysate was centrifuged at 12,000 g at 4°C for 20 min, and the precipitation was resuspended in 8 M urea (Sigma), and the supernatant was collected by centrifugation at 4°C after 10 min in ice bath with 100-W ultrasound. Then 20 μL of PBS lysate and urea lysate were respectively taken for Western blot detection. The total protein sample was separated by 15% sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis gel and transferred to 0.22-μm polyvinylidene difluoride membranes (Millipore Corporation) and then blocked with TBST buffer (50 mM Tris–HCl, 150 mM NaCl, and 0.1% Tween20, pH 7.4) containing 5% non-fat milk (wt/vol) at 4°C overnight. After washing with TBST three times for 30 min, the membrane was incubated with mouse anti-His Ab (1:1,000) for 2 h at room temperature. The membrane was washed thrice with TBST for 30 min and then incubated with the horseradish peroxidase–conjugated goat anti-mouse immunoglobulin G Ab (1:3,000) for 1 h at room temperature. After washing with TBST, the membrane was visualized with SuperSignal™ West Pico PLUS Chemluminescent Substrate (Thermo) and imaged using Amersham imager 600 (GE).
Purification of Recombinant Hepcidin Protein
The 8 M urea lysate was purified using a protein purification system (AKTA Pure, GE) with Ni-NTA Resin column. Equal volume of 8 M urea containing 0.4% PEG2000, 0.2 mM GSSG, 2 mM GSH, 2% L-arginine, and 10% glycerol was added into the eluted protein. Then, the eluted protein was treated with gradient dialysis in buffer (0.2% PEG2000, 0.1 mM GSSG, 2 mM GSH, 5% glycerol, 1% L-arginine, 0.1 M EDTA, and 6 or 4 or 3 or 2 or 1 or 0 M urea) of different urea concentrations. Finally, various hepcidin proteins were concentrated by using the concentration tube under 3,000 rpm at 4°C. The concentrated proteins were detected using Coomassie blue staining.
Antibacterial Assay
Antibacterial activity experiment was performed using microtiter broth dilution method; 100 μL test bacteria strains were diluted to approximately 3 × 106 colony-forming units/mL, and then 100 μL diluted peptide was added to 96-well microplates, and PBS (pH 7.4) was used as a control. The mix was incubated in a 37°C incubator. The bacterial growth curve was plotted by measuring the absorbance at 600 nm (OD600). The OD600 values were measured and recorded, which began when bacteria and peptide were mixed, starting with 0 h and then every hour for 12 h. Survival rate was calculated as the cell density in the presence of hepcidin peptides compared with the cell density of control. Assay was performed three times, and all data are presented as mean ± SD.
Flow Cytometry Analysis of Bacterial Immunofluorescence
The 3 × 105 bacteria cells were treated with 22 μM Dmhep_8cysV1 protein, 19 μM Dmhep_8cysV2 protein, and 18 μM Drhep protein for 1 h, respectively. Then, bacteria cells were collected at 3,000 rpm for 5 min and washed with PBS buffer twice in PBS (pH 7.4). Then, 100 μL 75% ethanol was added to the bacteria, incubated for 30 min, centrifuged, discarded the supernatant, and washed cells with PBS twice. One hundred microliters of 0.1% Triton X-100 was added and incubated for 45 min; the cells were washed three times in PBS. The cells were incubated in PBS containing 100 μg/mL lysozyme and 5 mM EDTA for 30 min; the cells were washed three times in PBS. One hundred microliters 0.5% blocking reagent solution (Sigma) was added and incubated for 30 min; 1 μL mouse anti-His Ab (1:100) was directly added and shaken for 2 h at 37°C. The suspension was centrifuged and discarded; the cells were washed three times in PBST (PBS containing 0.05% Tween-20); 100 μL 0.5% blocking buffer was added containing goat anti-mouse Alexa Fluor 488 ab (1:500) for 30 min at 37°C. The suspension was centrifuged and discarded; the cells were washed three times in PBST and once in PBS; the cells were resuspended in PBS. Finally, the fluorescence signal of cells was analyzed is using Accuri C6 (Becton, Dickinson and Company, BD). The data were analyzed with FlowJo software.
Results
Phylogenetic Analysis of Different Hepcidin Variants of Dissostichus mawsoni and Danio rerio
To determine the phylogenetic relation of hepcidins from D. mawsoni and D. rerio, a phylogenetic tree was constructed with CDS sequence of hepcidins of teleost and other species (Figure 1). The results show D. rerio and Cyprinidae hepcidin in a clade position, whereas two hepcidins from D. mawsoni (Dmhep_8cysV1 and Dmhep_8cysV2) were in the same branch position with those of Notothenia angustata (Nototheniidae), respectively. The phylogenetic relationship of hepcidins between D. mawsoni and D. rerio was relatively distant.
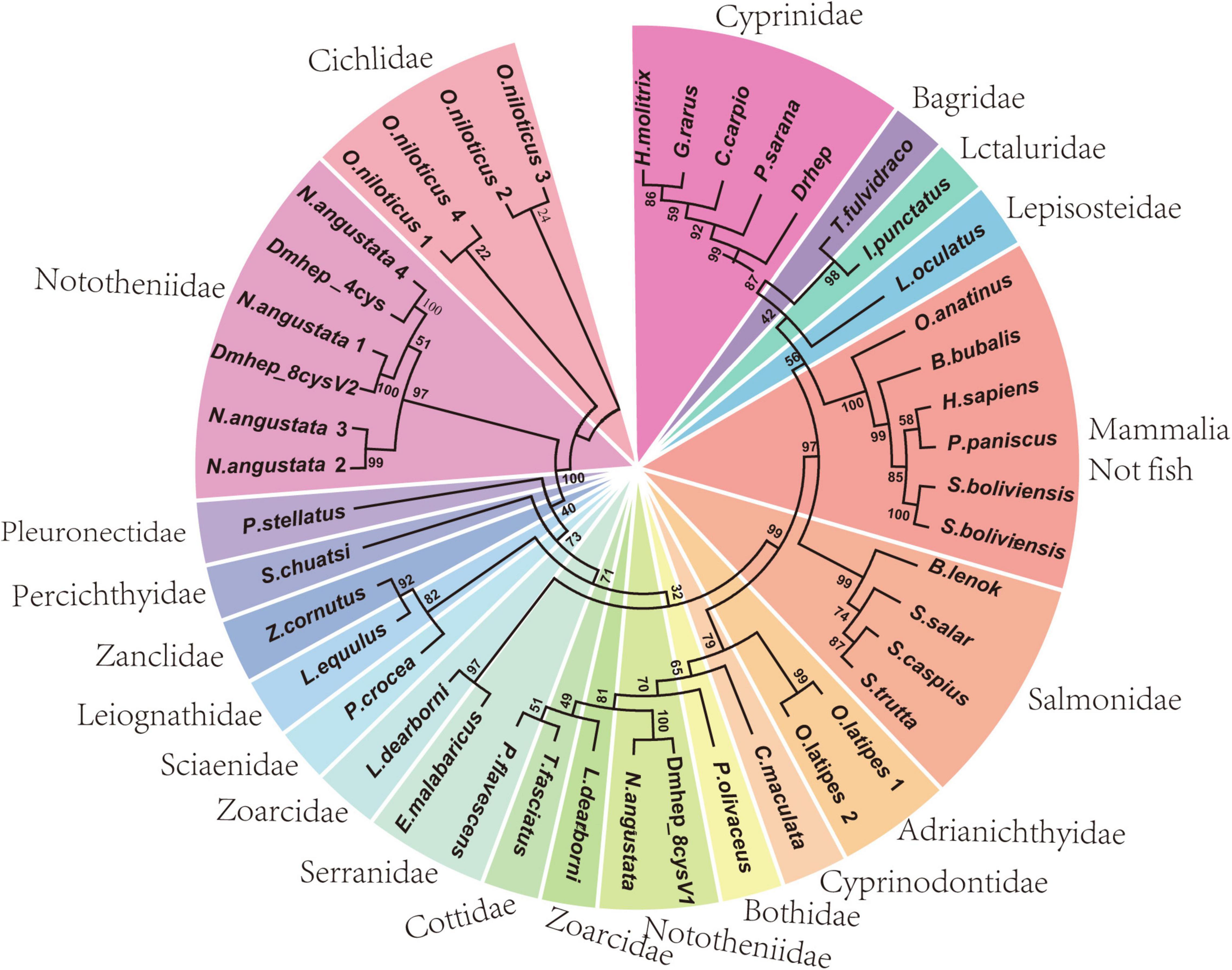
Figure 1. Phylogenetic analysis of hepcidins from teleost and other vertebrates. The phylogenetic trees constructed by neighborhood join method in MEGA 7.0. The numbers given are frequency (%) of a given branch in 1,000 bootstrap replications.
Sequence Alignment of Different Hepcidin Variants of Dissostichus mawsoni and Danio rerio
Whole genome sequencing analysis revealed that the hepcidin gene of D. mawsoni is distributed at both genomic loci A and B, and the most different between them is the number of cysteines, ranging from three to eight (Figure 2A). The hepcidin genes of D. mawsoni named Dmhep_8cysV1 are located at locus A and Dmhep_8cysV2 at locus B, respectively, and the only one from D. rerio named Drhep is also at locus B. There are eight cysteines in all of the three. Their Genbank accession numbers are EU221590.1, EU221595.1, and NC_007127.7. Their CDSs were cloned, sequenced, and aligned. The CDS lengths of Dmhep_8cysV1, Dmhep_8cysV2, and Drhep were 273, 267, and 276 bp, respectively. The full lengths of the three hepcidin peptides are 90, 88, and 91 aa, although their signal peptides are 24 aa, the mature region contain 25, 23, and 24 aa, respectively. And the predicted molecular weights of their mature peptide were 2.96, 2.93, and 2.89 KD, and the theoretical isoelectric points were 8.76, 9.37, and 8.94, respectively. Amino acid sequence alignment shows that Dmhep_8cysV2 is significantly different from the other two, especially from the mature region. However, the mature region of Dmhep_8cysV1 is highly similar to that of Drhep, suggesting that their functions and biological activities may be very similar.
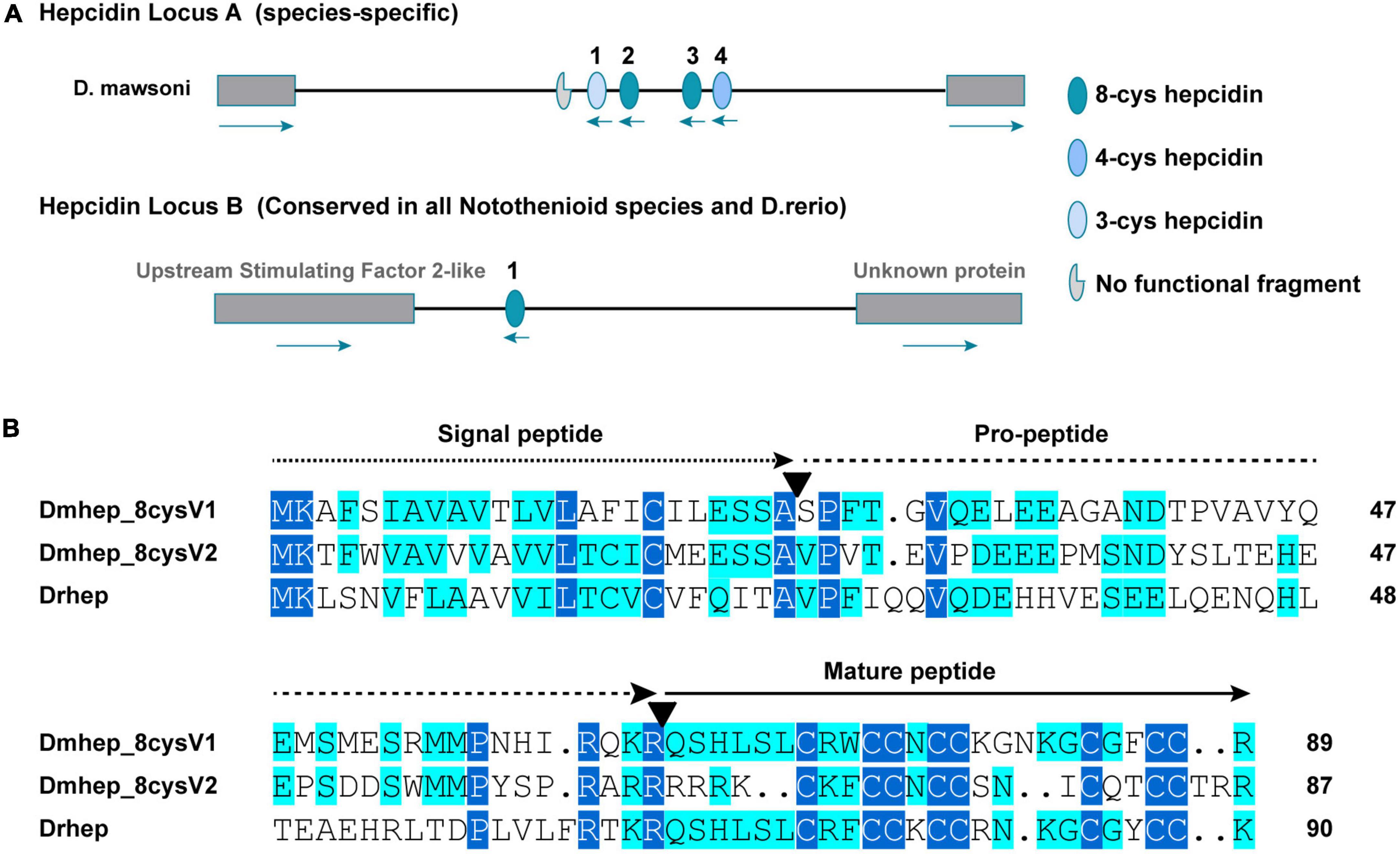
Figure 2. Sequence characteristics of hepcidin variants. (A) The distribution of conserved and non-conserved hepcidin variants in the genome. (B) Multiple amino acid alignment and comparison of the different hepcidin variants from D. mawsoni (Dmhep_8cysV1 and Dmhep_8cysV2) and D. rerio (Drhep). The three functional regions of the prepropeptide are represented by different lines at the top of each region. The number of amino acids for each variant is shown at the end of each sequence. The putative cleavage sites of the signal peptide and the mature hepcidin are marked with inverted triangles. Levels of amino acid conservation are highlighted by dark blue (identical) and light blue (two variations).
Hepcidin mRNA Expression in Dissostichus mawsoni and Danio rerio
To detect the tissue expression level of hepcidin, mRNA expression was detected in 10 tissues collected from D. rerio and D. mawsoni, respectively, including brain, liver, gill, ovary, muscle, skin, heart, gut, kidney, and spleen (Figure 3). Obviously, hepcidin mRNA expression was the highest in liver of the two fishes. The mRNA expression level was very low in other tissues, except spleen, which was the secondary tissue with high expression of Drhep, which is responsible for immune and hematopoietic response.
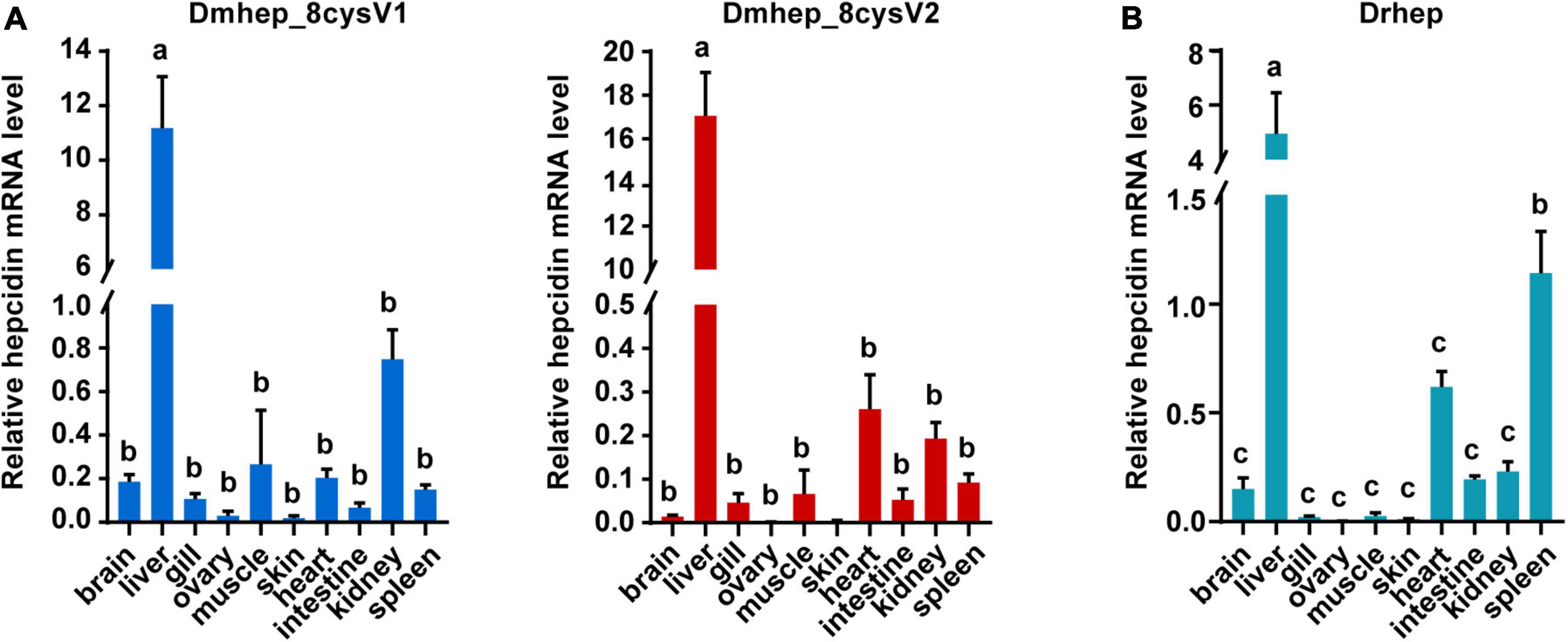
Figure 3. The relative mRNA expression of hepcidin variants in different tissues of D. mawsoni (A) and D. rerio (B). Error bars, mean ± SD, n = 3 (biological replicates). The same letters (a, b, and c) indicate no significant difference between different tissues, and different letters indicate statistical difference (p < 0.05) between different tissues.
Hepcidin Prepropeptides Were Cleaved Into Mature Peptide and Propeptide in vitro
The premature peptide motif “RXK/RR” has been hypothesized to be the cleavage site of furin-like endoproteases, but it has not been demonstrated that the prepeptide and mature peptide can be separated in living cell automatically. To identify that propeptide and mature peptide could be cleaved in live cell, green fluorescent protein tag and red fluorescent protein tag were fused with N-terminal of propeptide and C-terminal of mature peptide, respectively (Figure 4A). The recombinant hepcidin plasmid (Dmhep_8cysV1 and Dmhep_8cysV2) was transfected and expressed in CHO cells; microscopic imaging revealed that green fluorescent protein and red fluorescent protein were separated (Figure 4B), whereas the two fluorescent proteins of control plasmid without hepcidin sequence were completely fused in CHO cells (Figure 4B). Taken together, the results reveal that hepcidin prepropeptides were cleaved into mature peptide and propeptide in vitro.
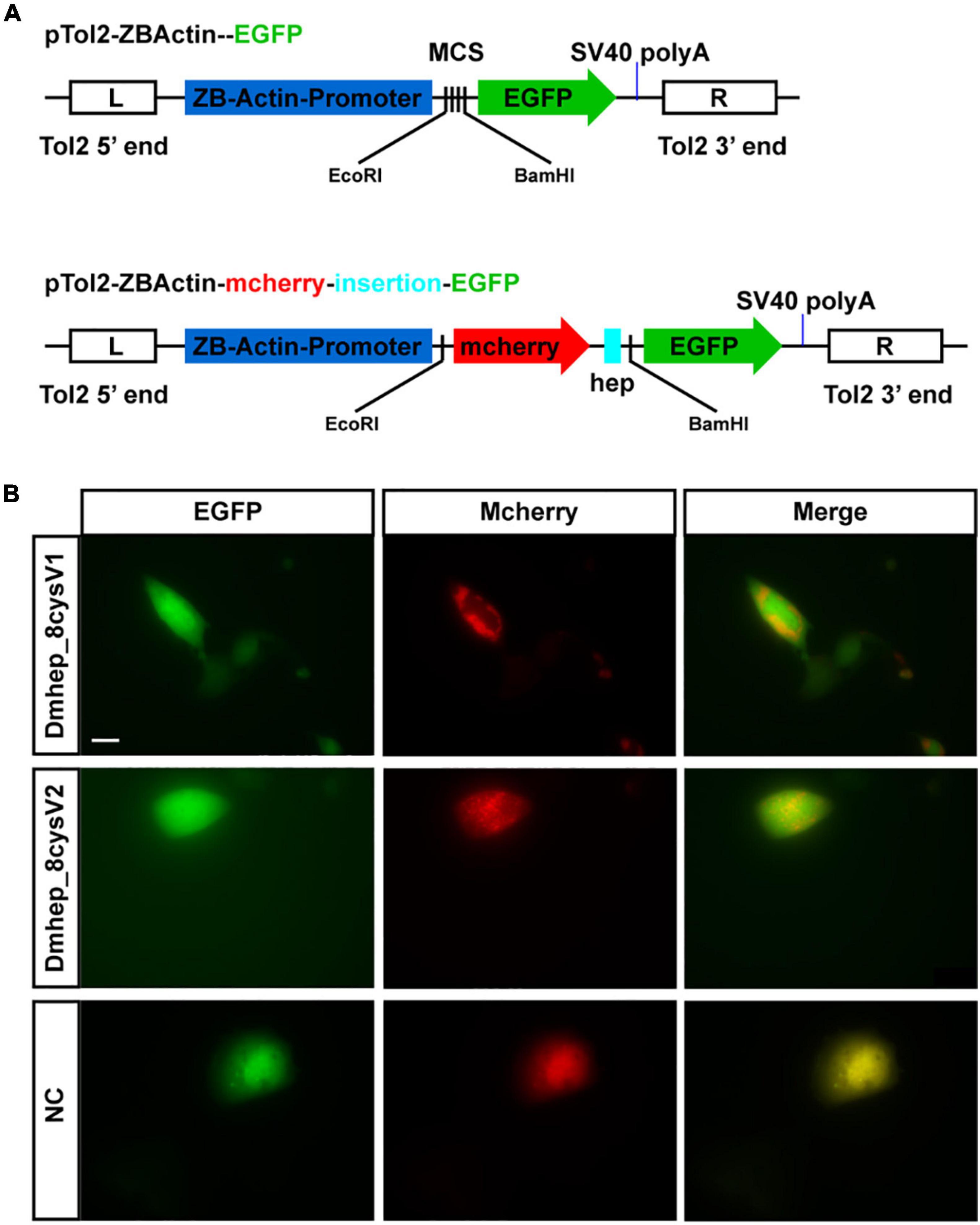
Figure 4. Hepcidin prepropeptide were cleaved into mature peptide and propeptide in CHO cell line. (A) The structure of recombinant plasmid with or without CDS of hepcidin and fluorescent protein. (B) The distribution of red and green fluorescent protein in CHO cell line. NC, negative control.
The Antibacterial Activity of Recombinant Protein of Different Hepcidin Variants
To figure out whether functional divergence occurred in these sequentially diverse hepcidin variants, we examined their antibacterial activity. Drhep, Dmhep_8cysV1, and Dmhep_8cysV2 recombinant proteins purified by His tag show a single band about 10 kDa on an SDS polyacrylamide gel by Coomassie blue staining and Western blot (Figure 5).
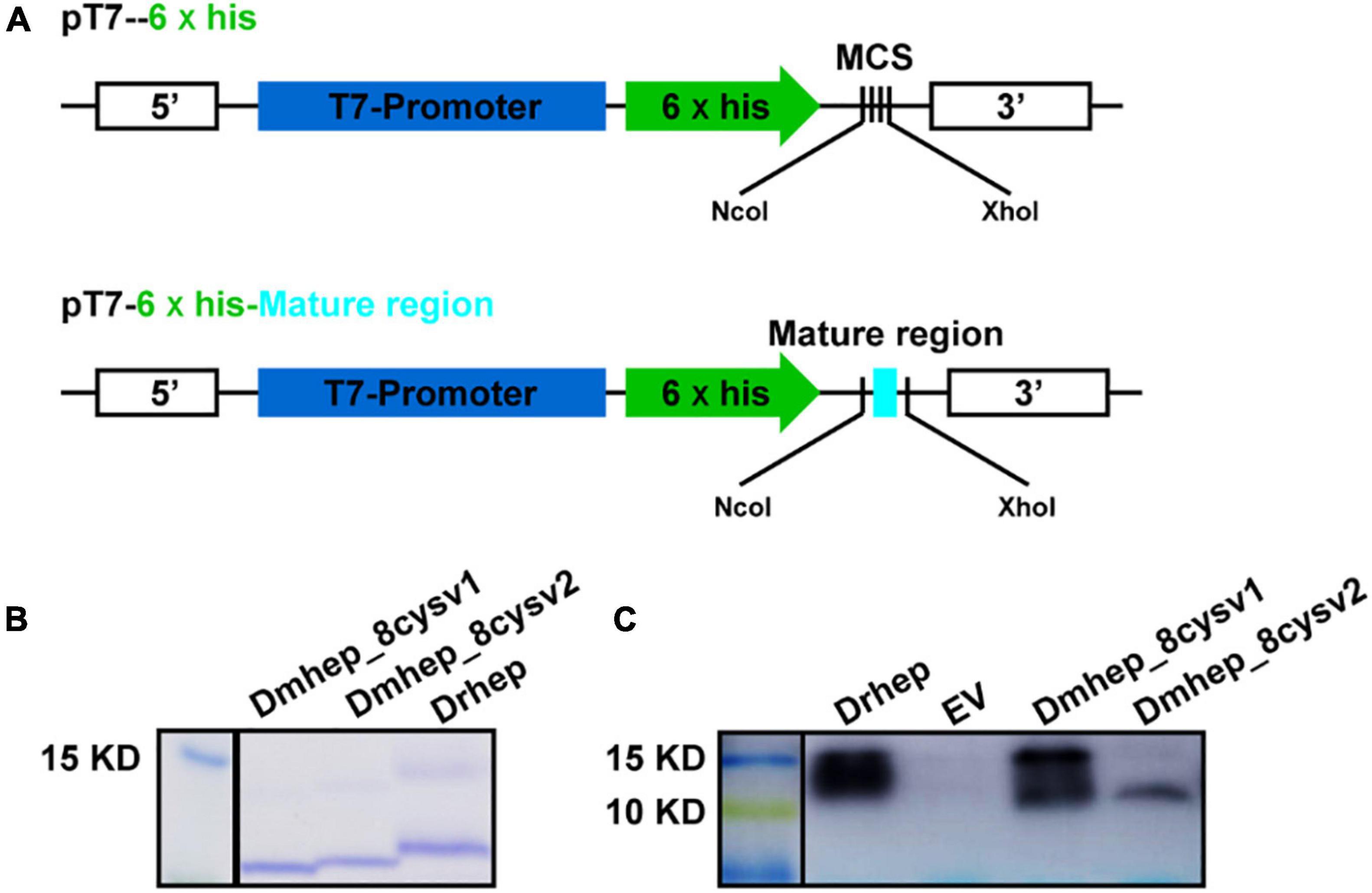
Figure 5. Expression and purification of hepcidin in E. coli. (A) The structure of recombinant plasmid with or without mature region of hepcidin. (B) Purified hepcidin peptide detected by Coomassie blue staining. (C) Purified hepcidin peptide detected by Western blot.
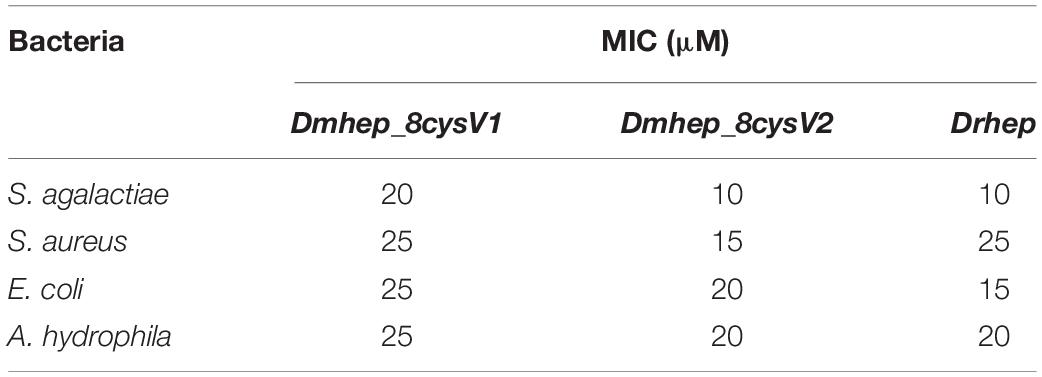
The purified recombinant protein was constructed in prokaryotic E. coli and tested for its antibacterial activities against Gram-positive and Gram-negative bacteria. All three recombinant hepcidin peptides showed antibacterial activities. The minimum inhibitory concentration (MIC) of Drhep against E. coli, S. agalactiae, A. hydrophila, and S. aureus were 15, 10, 20, and 25 μM, respectively. The MICs of Dmhep_8cysV1 against E. coli, S. agalactiae, A. hydrophila, and S. aureus were 25, 20, 25, and 25 μM, respectively. The MICs of Dmhep_8cysV2 against E. coli, S. agalactiae, A. hydrophila, and S. aureus were 20, 10, 20, and 15 μM, respectively (Table 2). The three hepcidins from D. mawsoni and D. rerio had different degrees of antibacterial activity overall.
Hepcidin Protein Can Bind to Various Bacteria
Flow cytometry analysis of bacterial immunofluorescence confirmed hepcidin peptide kills bacteria by binding to them. The result showed that all three hepcidin proteins can bind to the bacteria at a concentration of 20 μM (Figure 6). The binding capacity of Dmhep_8cysV2 to E. coli was stronger than that of Dmhep_8cysV1 (Figure 6A), and the binding capacity of Dmhep_8cysV2 was the same as that of Drhep. Dmhep_8cysV2 also had better binding ability to S. aureus than the other two (Figure 6B).
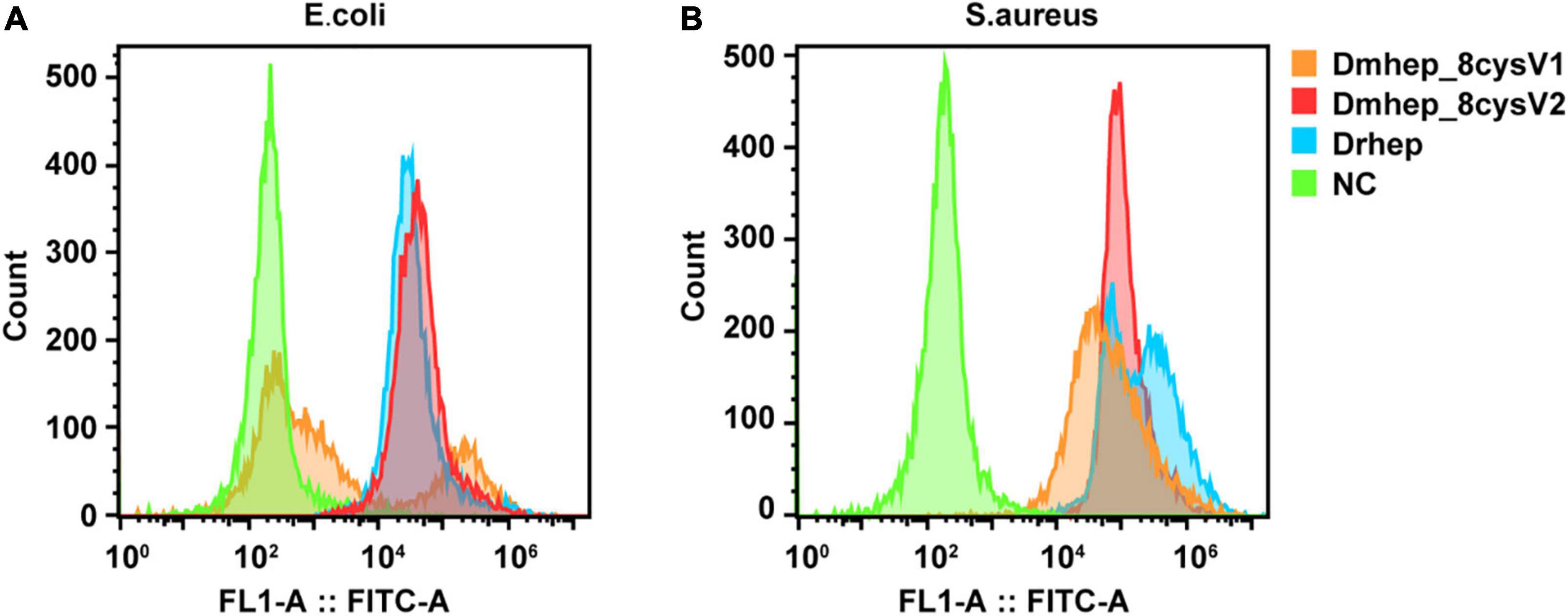
Figure 6. Various hepcidin peptides were detected by flow cytometry to bind to E. coli (A) and S. aureus (B). Orange represents Dmhep_8cysV1 (20 μM), red represents Dmhep_8cysV2 (20 μM), blue represents Drhep (20 μM), and green represents NC (negative control, PBS).
Discussion
Antimicrobial peptides are one of the important components of fish living in microbe-rich environment (Samuthirapandian et al., 2010). The AMP (antimicrobe peptide) hepcidin widely exists in fish and plays an important role in innate immune system of fish (Shi and Camus, 2006). In recent years, a large number of studies have been carried out on the antibacterial activity of AMP, especially on hepcidin of various vertebrates, looking forward to find a substitute for antibiotic. The sequences and structure domains of hepcidin variants from D. mawsoni were analyzed; their recombinant peptides were obtained, and their antimicrobial activities were tested in this study.
The encoded prepropeptide contains 84 aa in human, but only 20- to 25-aa peptides were initially found in urine (Park et al., 2001), meaning that the prepropeptide was cleaved to form a mature peptide. The conserved arginine/lysine propeptide cleavage recognition site (RXRR/RXKR) of furin-like endoproteases was found in all three hepcidins (Figure 2B). Dmhep_8cysV1 and Dmhep_8cysV2 propeptides separate into two parts in CHO cells (Figure 4B), probably because arginine residues at their carboxyl terminal are cleaved by widespread furin, similar to viral glycoproteins (Garten et al., 1994). This procedure is common with posttranslational modification of membrane and secretory proteins to form a functional peptide. In addition, it is assumed that the full hepcidin sequence supplies a codon long enough to ensure that the translation process is performed smoothly and correctly.
Sequence alignment showed that the mature region of Dmhep_8cysV1 is highly parallel to Drhep, but Dmhep_8cysV2 had low similarities with the other two mature regions except for the eight cysteines in this region. It is believed that the mature peptide of teleost fish is highly conserved (Ke et al., 2015), and the newly evolved Dmhep_8cysV2 in the genome enables D. mawsoni to acquire hepcidin sequence diversity. Therefore, whether the diverse sequences of hepcidin are helpful to increase the immunity ability for D. mawsoni is still unclear. The result showed that mature hepcidin could inhibit all kinds of bacteria. In this study, the recombinant peptide of D. mawsoni and hepcidin of D. rerio both inhibited the growth of Gram-positive and Gram-negative bacteria. However, Dmhep_8cysV2 had a lower MIC against S. aureus than both the other two. This may relate to the origin of Dmhep_8cysV2, as it is a newly evolved site located at genomic locus A, rather than the ancient locus B such as Dmhep_8cysV1 and Drhep. In addition, the amino acid sequence showed that Dmhep_8cysV2 was significantly different from Dmhep_8cysV1 and Drhep, which is attributed to the differences in antibacterial ability, for which the latter two show more similarities than the former. Different hepcidin variants have different MIC values for some bacteria, but these prove that hepcidin has a broad-spectrum antibacterial ability.
Given the special habitat of D. mawsoni and its various hepcidin variants, the additional function of hepcidins deserves further study and exploration. Many studies have shown that hepcidin mature peptides form a characteristic antiparallel β-sheet conformation with eight, six, or four conserved cysteines (De Domenico et al., 2008). At the same time, there was evidence that proved hepcidin lost antibacterial function if all the eight cysteines were replaced by alanine (Lin et al., 2014). In fact, there are three hepcidins in D. mawsoni, except for the two mentioned in this article, with the third one containing only four cysteines. Therefore, the relationship between the structure and function of different hepcidins variants remains to be explored.
In addition, although many hepcidins have a certain degree of antibacterial activity, how they kill bacteria is still not clear. Previous studies have demonstrated that peptides with β sheet structure bind to bacterial membrane to form complexes that kill microorganisms (Huang, 2000; Chen et al., 2003). Flow cytometry showed that hepcidin from D. mawsoni and D. rerio could bind to bacteria and may be the complexes formed in this process. However, structure of the complexes is still unclear, and whether there are other molecules in bacterial cells that can target hepcidin needs further study.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics Statement
The animal study was reviewed and approved by the Shanghai Ocean University Animal Care and Use Committee.
Author Contributions
LC managed the project. ML designed and performed experiments and wrote the manuscript. RH contributed to interpretation of data and edits to the manuscript. WY contributed to RNA and strain preparations. WL and QX performed fish and tissue collections and sample preparations. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Key Research and Development Program of China (2018YFD0900601), The Belt and Road international joint laboratory construction projects of science and technology innovation action plan from the Science and Technology Commission of Shanghai Municipality (19590750500), and Polar Innovation Platform Grant (JDXT-2019-01).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Jun Zou for helping us collect the A. hydrophila. We thank Lang Gui for providing the S. agalactiae.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.834477/full#supplementary-material
Footnotes
References
Abdelkhalek, N. K., Risha, E., El-Adl, M. A., Salama, M. F., and Dawood, M. A. (2020). Antibacterial and antioxidant activity of clove oil against Streptococcus iniae infection in Nile tilapia (Oreochromis niloticus) and its effect on hepatic hepcidin expression. Fish Shellfish Immunol. 104, 478–488. doi: 10.1016/j.fsi.2020.05.064
álvarez, C. A., Guzmán, F., Cárdenas, C., Marshall, S. H., and Mercado, L. (2014). Antimicrobial activity of trout hepcidin. Fish Shellfish Immunol. 41, 93–101. doi: 10.1016/j.fsi.2014.04.013
Cai, L., Cai, J. J., Liu, H. P., Fan, D. Q., Peng, H., and Wang, K. J. (2012). Recombinant medaka (Oryzias melastigmus) pro-hepcidin: Multifunctional characterization. Comp. Biochem. Physiol. Biochen. Mol. Biol. 161, 140–147. doi: 10.1016/j.cbpb.2011.10.006
Chen, F. Y., Lee, M. T., and Huang, H. (2003). Evidence for Membrane Thinning Effect as the Mechanism for Peptide-Induced Pore Formation. Biophys. J. 84, 3751–3758. doi: 10.1016/S0006-3495(03)75103-0
Chen, J., Nie, L., and Chen, J. (2018). Mudskipper (Boleophthalmus pectinirostris) Hepcidin-1 and Hepcidin-2 Present Different Gene Expression Profile and Antibacterial Activity and Possess Distinct Protective Effect against Edwardsiella tarda Infection. Probiot. Antimicrob. Prot. 10, 176–185. doi: 10.1007/s12602-017-9352-0
Chen, L., Lu, Y., Li, W., Ren, Y., Yu, M., Jiang, S., et al. (2019). The genomic basis for colonizing the freezing Southern Ocean revealed by Antarctic toothfish and Patagonia robalo genomes. GigaScience. 8:giz016. doi: 10.1093/gigascience/giz016
Chi, J. R., Liao, L. S., Wang, R. G., Jhu, C. S., Wu, J. L., and Hu, S. Y. (2015). Molecular cloning and functional characterization of the hepcidin gene from the convict cichlid (Amatitlania nigrofasciata) and its expression pattern in response to lipopolysaccharide challenge. Fish Physiol. Biochem. 41, 449–461. doi: 10.1007/s10695-014-9996-6
Cuesta, A., Meseguer, J., and Esteban, M. A. (2008). The antimicrobial peptide hepcidin exerts an important role in the innate immunity against bacteria in the bony fish gilthead seabream. Mol. Immunol. 45, 2333–2342. doi: 10.1016/j.molimm.2007.11.007
Das, A., Mohapatra, A., and Sahoo, P. K. (2015). Cloning and Characterization of Antimicrobial Peptide, Hepcidin in Medium Carp, Puntius sarana. Int. J. Peptide Res. Therap. 21, 139–147. doi: 10.1007/s10989-014-9438-4
De Domenico, I., Nemeth, E., Nelson, J. M., Phillips, J. D., Ajioka, R. S., and Kay, M. S. (2008). RETRACTED: The Hepcidin-Binding Site on Ferroportin Is Evolutionarily Conserved. Cell Metab. 8, 146–156. doi: 10.1016/j.cmet.2008.07.002
De Domenico, I., Ward, D. M., Patti, M. C., Jeong, S. Y., David, S., and Musci, G. (2007). Ferroxidase activity is required for the stability of cell surface ferroportin in cells expressing GPI-ceruloplasmin. EMBO J. 26, 2823–2231. doi: 10.1038/sj.emboj.7601735
Fleming, M. D. (2008). The regulation of hepcidin and its effects on systemic and cellular iron metabolism. Hematology 1, 151–158. doi: 10.1182/asheducation-2008.1.151
Fleming, R. E., and Sly, W. S. (2001). Hepcidin: A putative iron-regulatory hormone relevant to hereditary hemochromatosis and the anemia of chronic disease. Proc. Natl. Acad. Sci. 98, 8160–8162. doi: 10.1073/pnas.161296298
Ganz, T., and Nemeth, E. (2006). Regulation of iron acquisition and iron distribution in mammals. Biochim. Biophys. Acta. 1763, 690–699. doi: 10.1016/j.bbamcr.2006.03.014
Garten, W., Hallenberger, S., Ortmann, D., Schäfer, W., Vey, M., and Angliker, H. (1994). Processing of viral glycoproteins by the subtilisin-like endoprotease furin and its inhibition by specific peptidylchloroalkylketones. Biochimie. 76, 217–225. doi: 10.1016/0300-9084(94)90149-x
Hao, J., Li, Y. W., Xie, M. Q., and Li, A. X. (2012). Molecular cloning, recombinant expression and antibacterial activity analysis of hepcidin from Simensis crocodile (Crocodylus siamensis). Comp. Biochem. Physiol. Biochen. Mol. Biol. 163, 309–315. doi: 10.1016/j.cbpb.2012.08.002
Hocquellet, A., Senechal, C., and Garbay, B. (2012). Importance of the disulfide bridges in the antibacterial activity of human hepcidin. Peptides. 36, 303–307. doi: 10.1016/j.peptides.2012.06.001
Huang, H. W. (2000). Action of Antimicrobial Peptides: Two-State Model. Biochenistry 39, 8347–8352. doi: 10.1021/bi000946l
Huang, P. H., Chen, J. Y., and Kuo, C. M. (2007). Three different hepcidins from tilapia, Oreochromis mossambicus: Analysis of their expressions and biological functions. Mol. Immunol. 44, 1922–1934. doi: 10.1016/j.molimm.2006.09.031
Huang, T., Gu, W., Wang, B., Zhang, Y., Cui, L., and Yao, Z. (2019). Identification and expression of the hepcidin gene from brown trout (Salmo trutta) and functional analysis of its synthetic peptide. Fish Shellfish Immunol. 87, 243–253. doi: 10.1016/j.fsi.2019.01.020
Hunter, H. N., Fulton, D. B., Ganz, T., and Vogel, H. J. (2002). The solution structure of human hepcidin, a peptide hormone with antimicrobial activity that is involved in iron uptake and hereditary hemochromatosis. J. Biol. Chem. 277, 37597–37603. doi: 10.1074/jbc.M205305200
Iman, S., Reza, K. M., Saman, H., Hamed, P., and Michael, W. (2019). Cloning, characterization and tissue-specific expression of the antimicrobial peptide hepcidin from caspian trout (Salmo caspius) and the antibacterial activity of the synthetic peptide. Fish Shellfish Immunol. 90, 288–296. doi: 10.1016/j.fsi.2019.05.010
Ke, F., Wang, Y., Yang, C. S., and Xu, C. (2015). Molecular cloning and antibacterial activity of hepcidin from Chinese rare minnow (Gobiocypris rarus). Electr. J. Biotechnol. 18, 169–174. doi: 10.1016/j.dci.2020.103845
Kim, Y. O., Hong, S., Nam, B. H., Lee, J. H., Kim, K. K., and Lee, S. J. (2005). Molecular Cloning and Expression Analysis of Two Hepcidin Genes from Olive Flounder Paralichthys olivaceus. Biosci. Biotechnol. Biochem. 69, 1411–1414. doi: 10.1271/bbb.69.1411
Krause, A., Neitz, S., Magert, H. J., Schulz, A., Forssmann, W. G., and Schulz-Knappe, P. (2000). LEAP-1, a novel highly disulfide-bonded human peptide, exhibits antimicrobial activity. FEBS lett. 480, 147–150. doi: 10.1016/s0014-5793(00)01920-7
Lauth, X., Babon, J. J., Stannard, J. A., Singh, S., Nizet, V., and Carlberg, J. M. (2005). Bass hepcidin synthesis, solution structure, antimicrobial activities and synergism, and in vivo hepatic response to bacterial infections. J. Biol. Chem. 280, 9272–9282. doi: 10.1074/jbc.M411154200
Lin, W., Liu, S., Hu, L., and Zhang, S. (2014). Characterization and bioactivity of hepcidin-2 in zebrafish: Dependence of antibacterial activity upon disulfide bridges. Peptides. 57, 36–42. doi: 10.1016/j.peptides.2014.04.014
Liu, Y., Han, X., Chen, X., Yu, S., Chai, Y., and Zhai, T. (2017). Molecular characterization and functional analysis of the hepcidin gene from roughskin sculpin (Trachidermus fasciatus). Fish Shellfish Immunol. 68, 349–358. doi: 10.1016/j.fsi.2017.07.044
Luo, S. W., Luo, Z. Y., Yan, T., Luo, K. K., Feng, P. H., and Liu, S. J. (2020). Antibacterial and immunoregulatory activity of a novel hepcidin homologue in diploid hybrid fish (Carassius auratus cuvieri ♀× Carassius auratus red var ♂). Fish Shellfish Immunol. 98, 551–563. doi: 10.1016/j.fsi.2020.01.038
Nemeth, E., Tuttle, M. S., Powelson, J., Vaughn, M. B., Donovan, A., and Ward, D. M. (2004). Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science 306, 2090–2093. doi: 10.1126/science.1104742
Nicolas, G., Bennoun, M., Devaux, I., Beaumont, C., Grandchamp, B., and Kahn, A. (2001). Lack of hepcidin gene expression and severe tissue iron overload in upstream stimulatory factor 2 (USF2) knockout mice. Porc. Natl. Acad. Sci. U.S.A. 98, 8780–8785. doi: 10.1073/pnas.151179498
Park, C. H., Valore, E. V., Waring, A. J., and Ganze, T. (2001). Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J Biol Chem. 276, 7806–7810. doi: 10.1074/jbc.M008922200
Pei, J., Chen, D., Jin, W., Geng, J., Wang, W., and Zhang, S. (2019). Structure and mode of action of a novel antibacterial peptide from the blood of Andrias davidianus. Lett. Appl. Microbiol. 69, 312–317. doi: 10.1111/lam.13219
Samuthirapandian, R., Kaliyaperumal, K., Rameshkumar, G., and Ajith Kumar, T. T. (2010). Antimicrobial Peptides from the Marine Fishes. Res. J. Immunol. 3, 146–156. doi: 10.3923/rji.2010.146.156
Shi, J., and Camus, A. C. (2006). Hepcidins in amphibians and fishes: Antimicrobial peptides or iron-regulatory hormones? Dev. Comp. Immunol. 30, 746–755. doi: 10.1016/j.dci.2005.10.009
Shike, H., Lauth, X., Westerman, M., Ostland, V., Carlberg, J., Van Olst, J., et al. (2002). Bass hepcidin is a novel antimicrobial peptide induced by bacterial challenge. Euro. J. Biochem. 269, 2232–2237. doi: 10.1046/j.1432-1033.2002.02881.x
Solstad, T., Larsen, A. N., Seppola, M., and Jørgensen, T. (2008). Identification, cloning and expression analysis of a hepcidin cDNA of the Atlantic cod (Gadus morhua L.). Fish Shellfish Immunol. 2008. 25, 298–310. doi: 10.1016/j.fsi.2008.05.013
Srinivasulu, B., Syvitski, R., Seo, J. K., Mattatall, N. R., Knickle, L. C., and Douglas, S. E. (2008). Expression, purification and structural characterization of recombinant hepcidin, an antimicrobial peptide identified in Japanese flounder, Paralichthys olivaceus. Prot. Express. Purification 61, 36–44. doi: 10.1016/j.pep.2008.05.012
Tao, Y., Zhao, D. M., and Wen, Y. (2014). Expression, purification and antibacterial activity of the channel catfish hepcidin mature peptide. Prot. Express. Purification. 94, 73–78. doi: 10.1016/j.pep.2013.11.001
Tripti, G., and Mary, C. M. (2010). Dissection of Organs from the Adult Zebrafish. J. Visualiz. Exp. 37, e1717–e1722. doi: 10.3791/1717
Wang, K. J., Cai, J. J., Cai, L., Qu, H. D., Yang, M., and Zhang, M. (2009). Cloning and expression of a hepcidin gene from a marine fish (Pseudosciaena crocea) and the antimicrobial activity of its synthetic peptide. Peptides 30, 638–646. doi: 10.1016/j.peptides.2008.12.014
Weinstein, D. A., Roy, C. N., Fleming, M. D., Loda, M. F., Wolfsdorf, J. I., and Andrews, N. C. (2002). Inappropriate expression of hepcidin is associated with iron refractory anemia: implications for the anemia of chronic disease. Blood 100, 3776–3781. doi: 10.1182/blood-2002-04-1260
Xu, G., Huang, T., Gu, W., Zhang, Y., Yao, Z., and Zhao, C. (2018). Characterization, expression, and functional analysis of the hepcidin gene from Brachymystax lenok. Dev. Comp. Immunol. 89, 131–140. doi: 10.1016/j.dci.2018.08.013
Xu, Q., Christina Cheng, C.-H., Hu, P., Ye, H., Chen, Z., and Cao, L. (2008). Adaptive Evolution of Hepcidin Genes in Antarctic Notothenioid Fishes. Mol. Biol. Evol. 25, 1099–1112. doi: 10.1093/molbev/msn056
Yang, M., Chen, B., Cai, J. J., Peng, H., Cai, L., and Yuan, J. J. (2011). Molecular characterization of hepcidin AS-hepc2 and AS-hepc6 in black porgy (Acanthopagrus schlegelii): Expression pattern responded to bacterial challenge and in vitro antimicrobial activity. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 158, 155–163. doi: 10.1016/j.cbpb.2010.11.003
Yang, M., Wang, K. J., Chen, J. H., Qu, H. D., and Li, S. J. (2007). Genomic organization and tissue-specific expression analysis of hepcidin-like genes from black porgy (Acanthopagrus schlegelii B). Fish Shellfish Immunol. 23, 1060–1071. doi: 10.1016/j.fsi.2007.04.011
Yin, X. X., Chen, N., Mu, L. L., Bai, H., Wu, H., and Ye, J. M. (2022). Identification and characterization of hepcidin from Nile Tilapia (Oreochromis niloticus) in response to bacterial infection and iron overload. Aquaculture 546:737317. doi: 10.1016/j.aquaculture.2021.737317
Zhang, J., Yan, Q., Ji, R., Zou, W., and Guo, G. J. (2009). Isolation and characterization of a hepcidin peptide from the head kidney of large yellow croaker. Pseudosciaena crocea. Fish & Shellfish Immunol. 26, 864–870. doi: 10.1016/j.fsi.2009.03.014
Keywords: hepcidin, Antarctic notothenioid fish, antibacterial peptide, recombinant protein, antibacterial activity
Citation: Liu M, Hu R, Li W, Yang W, Xu Q and Chen L (2022) Identification of Antibacterial Activity of Hepcidin From Antarctic Notothenioid Fish. Front. Microbiol. 13:834477. doi: 10.3389/fmicb.2022.834477
Received: 05 January 2022; Accepted: 25 February 2022;
Published: 12 April 2022.
Edited by:
Octavio Luiz Franco, Catholic University of Brasilia (UCB), BrazilReviewed by:
Kostas Pantopoulos, McGill University, CanadaFanny Guzmán, Pontificia Universidad Católica de Valparaíso, Chile
Copyright © 2022 Liu, Hu, Li, Yang, Xu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liangbiao Chen, bGJjaGVuQHNob3UuZWR1LmNu
 Mingli Liu
Mingli Liu Ruiqin Hu
Ruiqin Hu Wenhao Li
Wenhao Li Wenyi Yang
Wenyi Yang Qianghua Xu
Qianghua Xu Liangbiao Chen
Liangbiao Chen