
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 05 April 2022
Sec. Infectious Agents and Disease
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.833655
This article is part of the Research Topic Invasive candidiasis – Pathogenic Mechanisms, Host Immune Responses and Therapeutic strategies View all 6 articles
Candida albicans, an important fungal pathogen of humans, displays different morphologies, such as yeast, pseudo-hyphae and hyphae, which are recognized unequally by phagocytic cells of the innate immune response. Once C. albicans cells invade host tissues, immune cells such as macrophages are attracted to the site of infection and activated to recognize, engulf and kill the pathogen. We have investigated this fungal cell-macrophage interface by using high-throughput screening of the C. albicans GRACE library to identify genes that can influence this interaction and modify the kinetics of engulfment. Compared with the wild-type (WT) strain, we identified generally faster rates of engulfment for those fungal strains with constitutive pseudo-hyphal and hyphal phenotypes, whereas yeast-form-locked strains showed a reduced and delayed recognition and internalization by macrophages. We identified a number of GRACE strains that showed normal morphological development but exhibited different recognition and engulfment kinetics by cultured macrophages and characterized two mutants that modified interactions with the murine and human-derived macrophages. One mutant inactivated an uncharacterized C. albicans open reading frame that is the ortholog of S. cerevisiae OPY1, the other inactivated CaKRE1. The modified interaction was monitored during a 4 h co-culture. Early in the interaction, both opy1 and kre1 mutant strains showed reduced recognition and engulfment rates by macrophages when compared with WT cells. At fungal germ tube initiation, the engulfment kinetics increased for both mutants and WT cells, however the WT cells still showed a higher internalization by macrophages up to 2 h of interaction. Subsequently, between 2 and 4 h of the interaction, when most macrophages contain engulfed fungal cells, the engulfment kinetics increased for the opy1 mutant and further decreased for the kre1 mutant compared with Ca-WT. It appears that fungal morphology influences macrophage association with C. albicans cells and that both OPY1 and KRE1 play roles in the interaction of the fungal cells with phagocytes.
Fungal infections are a significant threat to human health. The diploid commensal fungus Candida albicans represents an important pathogen worldwide, and Candidiasis is able to cause as high as 40% mortality in immunocompromised patients (Horn et al., 2009). As a common member of the human microbiota, C. albicans will interact with other microbes as well as with host tissues (Höfs et al., 2016). Candida can live as a commensal on mucosal tissues such as the oral cavity, where it remains generally harmless and can persist long term (Naglik et al., 2011). A key aspect that influences C. albicans success as either a commensal or pathogen is its relationship with the host immune response (Netea et al., 2008). Candidiasis, the clinically defined systemic fungal disease, occurs when the normal host–fungal equilibrium is disrupted by overgrowth of typically invasive hyphal cells. Different conditions can trigger this behaviour, such as immune suppression by chemotherapy or HIV infection, or in organ transplant patients with suppressed immune responses (Fidel, 2002). When any of these scenarios take place, changes in gene expression in coordination with morphological remodelling can occur in the fungal cells and facilitate the transition from commensal to pathogen (Gow et al., 2002; Hube, 2004; Sudbery et al., 2004).
In response to an increased fungal burden, the immune response typically involves leukocytes, present in different tissues, as the first line of defense (Cheng et al., 2012; Qin et al., 2016). The interface of C. albicans and phagocytes has been well documented as one of the most studied host-pathogen interactions (Urban et al., 2009; Lewis et al., 2012; Erwig and Gow, 2016). Once the early immune response is triggered by the fungal cells, the first responders to the insult that arrive to control and clear the fungus correspond to leukocytes such as neutrophils and macrophages (Romani, 2011; Jimenez-Lopez and Lorenz, 2013). These phagocytic immune cells detect the presence of C. albicans by recognizing the pathogen’s surface structures, such as the cell wall, which contains several molecules that act as pathogen-associated molecular patterns (PAMPs). This initiates the induction of pattern recognition receptors (PRRs) on immune cells that facilitate the internalization of Candida cells by phagocytes (Rubin-Bejerano et al., 2007; Hall et al., 2013). This process may end up with the successful killing of the internalized fungal cells, or with a win by the fungus through proliferation and escape from the phagocytes, and subsequent further invasion of mucosal tissues.
This host–fungal interplay has been well documented, and several genes and pathways that are crucial for both phagocyte and fungal cell function have been characterized using genetic, microscopic and chemical approaches (Lorenz et al., 2004; Netea et al., 2006; McKenzie et al., 2010). An important characteristic of C. albicans influencing engulfment by phagocytic cells is its morphological state; morphology has been described as one the most crucial virulence features of this fungus (Saville et al., 2003; Hall, 2015). The commensal, typically yeast form cells of C. albicans seem generally to be compatible with host/fungal homeostasis and avoid triggering major induction of the host immune response. However, increased proliferation, and the switch to the hyphal state can alert the host immune response to initiate control over this microbial pathogen by phagocytic cells including neutrophils and macrophages (Lewis et al., 2012).
Although there is considerable information on this interface between the fungal cell and the phagocyte cell, many aspects of the C. albicans genetic circuits and molecular pathways required to manage the host response and control its own viability in the mammalian host are still incompletely understood. We conducted a screening of the interactions between a collection of C. albicans conditional mutants and cultured macrophages to investigate the kinetics of this fungal-macrophage cell interface and identify new C. albicans genes that may be involved in the interactions with macrophages. From our initial screening, we saw that the engulfment process of macrophage for C. albicans depends on fungal morphology, with yeast-locked mutants less engulfed than mutants that morphologically behaved as the wild-type, and with filamentation-locked mutants more highly engulfed. Furthermore, we identified, among those mutants that show a morphological pattern equivalent to wild-type, a number of candidates that showed modified kinetics of recognition and engulfment by macrophages. This will expand our knowledge of this opportunistic pathogenic fungus and provide new insights into the host immune response against C. albicans.
The GRACE™ (gene replacement and conditional expression) library (Roemer et al., 2003), was used as a source of C. albicans genes to screen for modifications in the interaction of the fungal cells with macrophages. This collection has about 2500 diploid strains, which were engineered by replacing the native promoter of one allele of each manipulated gene with a TET (tetracycline)-regulated-promoter and deleting the other copy of the gene, replacing it with an auxotrophic marker. Each modified gene can thus be conditionally repressed when cells are placed are under TET-promoter repression (Nakayama et al., 2000). Briefly, the library was streaked from frozen stocks on YPD agar plates (2% agar, 2% peptone, 1% yeast extract, 2% dextrose) supplemented with 50 mg/l uridine. Then, individual colonies for each mutant were grown in a pre-culture in liquid YPD media supplemented with uridine O/N at 30°C with 220 rpm shaking. Subsequently, an aliquot was measured at OD600 and a dilution to an OD600 of 0.3 was re-inoculated in 5ml of YPD uridine media containing 20 μg/ml of doxycycline and grown O/N at 30°C at 220 rpm, then diluted again to OD600 of 0.3 and grown for a further 3 h in YPD uridine with 20 μg/ml of doxycycline. Finally, cells were washed, counted and diluted for assessment for macrophage interaction.
The RAW 264.7 murine macrophage cell line was kindly provided by Dr. Albert Descoteaux (INRS-Armand-Frappier, Laval, QC, Canada). Macrophages were cultured in DMEM medium supplemented with 10% FBS, penicillin/streptomycin and HEPES. Macrophages were seeded at a 2 × 105 density and grown for 48 h at 37°C and 5% CO2. Once cells reached 80% confluency, macrophages were collected with Trypsin-EDTA and then centrifuged for 10 min at 10,000 × rpm, at room temperature. Cells were then stained with trypan blue and counted, and an aliquot of 1.2 × 106 viable cells/ml was prepared for further macrophage engulfment assays with the fungal cells.
The protocol was approved by the Concordia University Human Research Ethics committee (certificate 30009292) and signed consent was obtained prior to the study. Venous blood was drawn from a human participant into ten heparinized vacutainer tubes. The participant was healthy as assessed by self-reporting of their health condition. In brief, the peripheral blood mononuclear cells (PBMC) were obtained using Ficoll-Hypaque density centrifugation techniques as previously described (Tabatabaei Shafiei et al., 2014). Monocytes were obtained using EasySep™ human CD14 positive selection kit according to manufacturer’s instructions. Subsequently, the PBMCs were cultured and differentiated to macrophages as described (Leascoat et al., 2018). Briefly, monocytes were cultured and differentiated into macrophages for 6 days using M-CSF (50 ng/ml) in RPMI medium, supplemented with 10% heat-inactivated calf bovine serum (CBS) and pen/strep. At day 6, differentiated macrophages were collected and counted for further engulfment assays testing C. albicans cells.
An aliquot of each GRACE mutant and the parental strain (CAI4), was spun down, the media removed, and the cells washed three times with PBS (phosphate buffer saline). Then the fungal cell concentration was adjusted to 1 × 108 cells/ml (Tóth et al., 2004). Next, fungal cells were stained with 50 mg/ml of Calcofluor White (CFW) and incubated for 10 min at RT (Williams and Lewis, 2011). The fungal cells were then washed three times in PBS, and finally a 1/100 dilution in PBS for each strain was prepared for the assay. For the initial screening, both macrophage and fungal cells of each strain were mixed together at a ratio 1:4 (Candida cells: macrophage), in wells of a 96-well plate and then visualized with the high-content screening microscope ImageXpress XSL wide-field (Molecular Devices, Sunnyvale, CA, United States). The 96 well plate was placed in a chamber equilibrated at 37°C and 5% CO2. Images were captured at 40X objective magnification on two channels (transmitted light, and DAPI). Every live-imaging interaction between macrophage and fungal cells was set to take images every 5 min during a 4 h total running time. Subsequently a time-lapse video was generated using the MetaXpress high content imaging acquisition and analysis software (Version 6.1.1, Molecular Devices). At least 300 macrophage per wide field (out of 6 wide fields per assay) were counted, including those with and without fungal cells internalized over the entire 4 h of the assay. Fungal cells (either budding, germ-tube forming or filament forms) engulfed by macrophages were counted and averaged for every six time-points corresponding to 30 min or 0.5 h as expressed in the figures, and normalized to the ratio: [number of fungal cells at time point X/number of fungal cells for every time point]/10 macrophages to generate an engulfment index (EI) (Tóth et al., 2004). The average macrophage uptake was calculated as the ratio of macrophages with engulfed fungal cells (stained or fluorescence signal) divided by the number of macrophages without fungal engulfment for 12 time points corresponding to 1 h of interaction, counted over six wide-field images, from three independent assays. Further time-lapse video data were edited using image-J software. Macrophage engulfment kinetic curves, histograms and statistical analysis were analyzed with GraphPad Prism (Version 6.0).
From the initial screening of the Candida-macrophage interaction, we identified strains that showed a lower rate of engulfment compared with wild-type strain although they underwent an apparently normal yeast-hyphal transition. Selected genes were deleted by the transient CRISPR-Cas9 method applied in C. albicans (Min et al., 2016; see Table 1 about the strains used or generated in this study). These deletions were made in a CAI4 strain tagged with GFP (Marcil et al., 2002). Each component for CRISPR was independently amplified from the pV1093 plasmid (Vyas et al., 2015). For the repair template, the SAT-1 cassette, containing the nourseothricin (NAT) gene was engineered by fusion PCR, adding approximately 300 bp of the flanking sequence of the gene of interest upstream and downstream of NAT to direct gene replacement of the gene of interest Table 2 (primers description). The single-guide RNA (sgRNA) cassette was also generated from the same pV1093 plasmid. Both sequence specific and chimeric primers for fusion PCR were designed as follows (Vyas et al., 2015) (see Table 2 for details on each specific sgRNA for each deleted gene). A first round of PCR was run in order to generate two amplicons: The first one included a forward oligo, starting at a 800 bp up in the SNR52 promoter was used with a reverse primer containing a specific 20 bp RNA guide, either targeting the OPY1 or KRE1 gene, followed by the end of the SNR52 promoter. The second amplicon of 800 bp was amplified with a forward primer beginning with a sgRNA scaffold for each targeted gene, followed by the end of the ENO1 terminator, and a reverse primer including the downstream ENO1 terminator. A second step for extension of the chimeric primers was run in order to fuse both PCR products (Vyas et al., 2015). Finally, a nested PCR amplification with an outer upstream SRN52 forward primer and a downstream reverse primer from the ENO1 terminator, generated a nearly 1425 bp sgRNA cassette (Huang et al., 2021).
Transformation was carried out as described (Walther and Wendland, 2003) and modified cells were selected on YPD-NAT plates (200 μg/ml). Subsequently, resistant colonies were screened by colony PCR for NAT insertion at the desired locus to confirm gene deletion and replacement by the resistance gene marker. We made a separate construct in the CAI4 background strain by adding an RFP-tag as follows: primers including Mlu1 forward and Nhe1 reverse restriction sites (see list of oligos on Table 2) were designed to amplify a 711 bp mCherry (RFP) gene from plasmid pHES262 (pADH1-Msn2-mCherry) (Stewart-Ornstein et al., 2017). Further, a CIP10Act-cyc plasmid (Murad et al., 2000) was digested with Mlu1/Nhe1 restriction enzymes and the mCherry (RFP) fragment was ligated into the plasmid. Subsequently the RFP-expressing plasmid was transformed into CAI4 cells at the RP10 locus and the red fluorescent signal was verified by microscopy.
Detailed kinetics of macrophage and specific C. albicans knock-out strains were measured using a Nikon Eclipse TiE inverted epifluorescence microscope, including bright field illumination and fluorescent filters GFP/Alexa 488 and Cy3/Alexa 546/555/568, with cell growth chambers supplied with 5% CO2 and held at 37°C. Macrophages and GFP-tagged C. albicans mutants, in the presence or not of WT C. albicans tagged with RFP, were grown in the same conditions and prepared for visualization of the interactions as previously described. A 4 h long imaging profile was recorded for the interaction between macrophage and fungal cells. Then the kinetics of this interaction were analyzed under the same parameters used for the high-throughput imaging analysis.
For the initial screening of the kinetics of interaction between C. albicans cells and macrophages, 1,227 strains from the GRACE library (Figure 1A) were investigated for their interaction with cultured mouse macrophage cells (see Supplementary Table 1). We identified three defined groups of C. albicans mutants based on morphology; the yeast-locked mutants that typically remained non-proliferating for the entire assay; a large group of mutants termed like-wild-type (like-WT) mutants that develop as normal Candida cells, initiating germ-tube formation with normal kinetics; and a third group corresponding to mutants with a constitutive filamentous phenotype that continue growing and extending their hyphal structures. Most of the mutants cultured from the GRACE library correspond to like-WT (93%), then a smaller group of 72 constitutive filamentous mutants (5.4%) as well as two dozen yeast-locked mutants that accounted for 1.6% of the total number of mutants screened. The overall average engulfment index over the 4 h interaction period shows clear differences related to the morphology phenotype of the GRACE mutants. The yeast-locked mutants were poorly recognised by the macrophages, reaching just an average engulfment index (EI), established over the entire 4 h of interaction, of 0.60 ± 1.1; this was followed by the like-WT mutants with an EI of 2.65 ± 0.5, and with the filamentous mutants showing the highest rate (EI of 4.0 ± 0.7) over this period (Figure 1A).
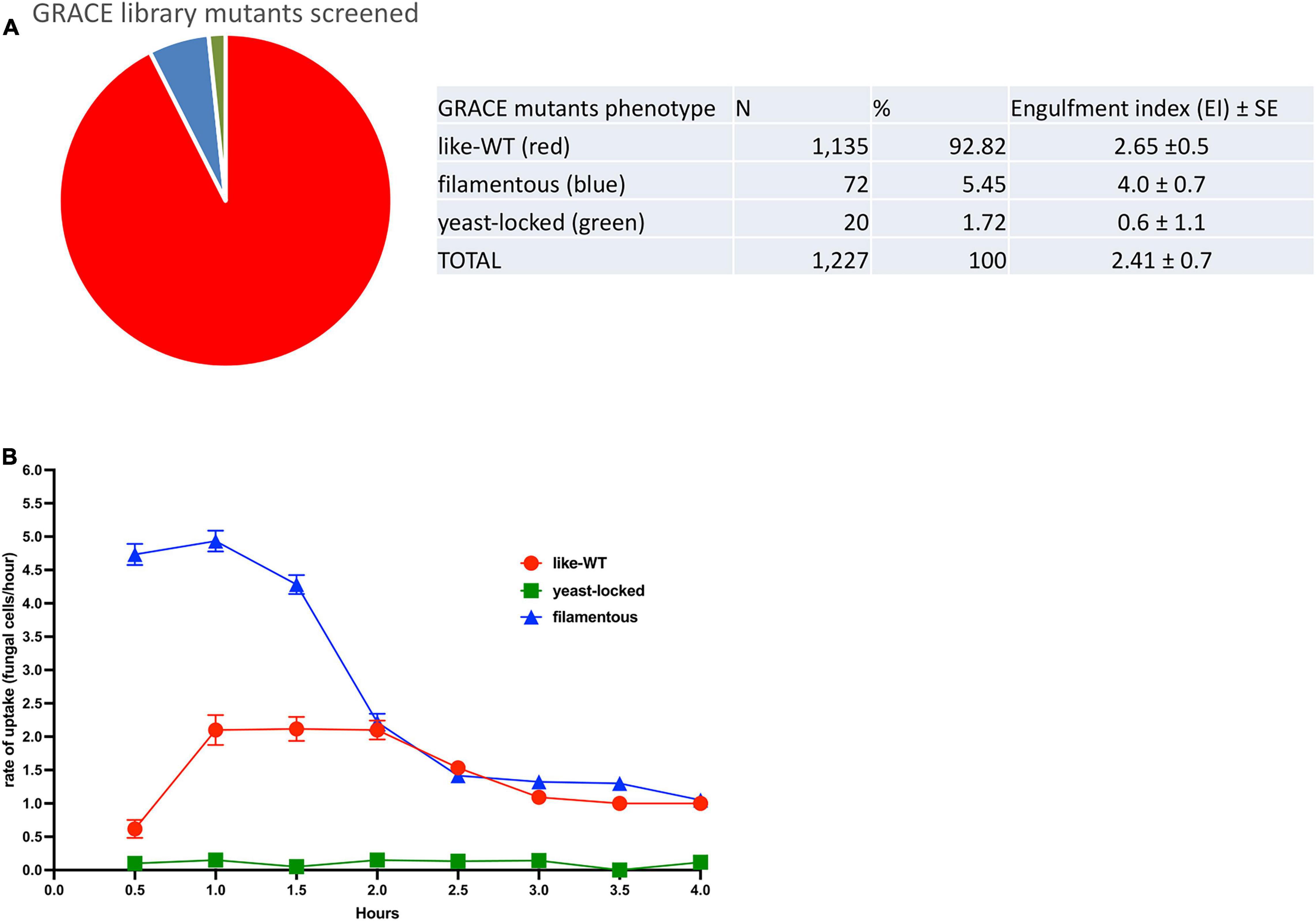
Figure 1. GRACE mutants-macrophage engulfment screening. (A) A total of 1227 mutants were screened for their interaction with macrophages. Three distinct groups were identified in this analysis that correlate with the fungal cell morphology. The main group found corresponds to strains like-wild-type where the mutation did not perturb the normal serum-induced morphological transition (92.82%), followed by filamentous mutants (5.45%) and yeast-locked mutants (1.72%). The overall engulfment rate observed by 10 macrophages/h was estimated for each mutant group: yeast-locked cells showed the lower engulfment rate (0.60 ± 1.1), followed by like-WT (wild-type) mutants (2.65 ± 0.5) and the filamentous mutants with the higher rate (4.0 ± 0.7). Average ± SE. (B) Kinetics of engulfment by murine macrophages, show a distinctive path for each phenotypic mutant group. Filamentous mutants (blue line) showed a higher engulfment kinetics over the first 2 h compared with the trace driven by the like-WT mutants (red line), which does an increasing trace from 0.5 to 1 h and then continued steady up to 2 h. After this time, the kinetic of both mutant groups, declined progressively up to the 4 h of the assay. In contrast the kinetic of mutants corresponding to the yeast-locked shape (green line), remained equally flat for over the entire 4 h of interaction between macrophage and fungal cells, indicating an almost absent recognition and internalization by the macrophages.
These differences are confirmed in the kinetics of interaction between specific C. albicans mutants and macrophages, monitored at specific timepoints (Figure 1B). At the early timepoints, the rates of engulfment by macrophages for the GRACE mutants are markedly different for each phenotypic group. Over the first 1.5 h of interaction, there was a steady and consistently higher rate of engulfment for the filamentous mutants. By contrast the like-WT mutants displayed an increasing rate of engulfment up to 1 h and then reached a continuous plateau. After 2 h of interaction, for both filamentous and like-WT mutants, the rate of engulfment by the macrophages tend to drop and to do so progressively until the end of the assay. We also noticed from our observations throughout the time-lapse videos that after 3 h of interaction a noticeable fraction of the fungal cells of both filamentous and like-WT mutants were growing out from the macrophages, either breaking out or dramatically stretching the macrophage membrane. In the case of filamentous strains, our observations tracking the recognition and engulfment of macrophages suggested that macrophages were ineffective at engulfing the tips of the filamentous mutants and arresting their growth (see representative Supplementary Video 3).
By contrast, the kinetics of engulfment for the yeast-locked mutants did not vary for the entire 4 h of incubation, and the macrophages barely engulfed these Candida yeast-cells. Many of these yeast-locked mutants correspond to essential genes encoding a variety of functions in C. albicans (see Supplementary Table 1). As many of these null mutants are involved in transcription, translation and chromosome segregation, it is likely that many cellular functions are absent in these mutants, interfering with development as a normal fungal cell, and affecting the production of surface antigens and fungal-derived molecules important in triggering macrophage engulfment.
Overall, most of the mutants screened showed similar kinetics to the CAI4-wild-type strain (tagged in red) for interaction with and internalization by the macrophages. However, in the group of like-WT mutants we found a few strains that had a morphological development (from budding yeast, germ tube formation and hyphal development) similar to the wild-type, but which showed a clearly reduced rate of engulfment by the macrophages (see Table 3). Two of the mutated genes in the strains identified in this class were the ortholog of the Saccharomyces cerevisiae gene KRE1 (Killer toxin REsistant 1), and an uncharacterized gene ORF19.4245. We examined this uncharacterized gene in more detail. A BLAST alignment search and further structural assessment identified this candidate as the likely ortholog of the S. cerevisiae gene OPY1. This identification was based on the structural alignment analysis, which showed similarly positioned and conserved Pleckstrin-homology (PH) domains in both the C. albicans and S. cerevisiae proteins (Figure 2). Functional analysis of the OPY1 gene in S. cerevisiae suggests it plays a role in regulating the phosphatydilinositol-4,5-biphosphate [(PtdIns 4,5)P2] lipid content through interaction with the PtdIns(4)P 5-kinase, Mss4 (Ling et al., 2012).
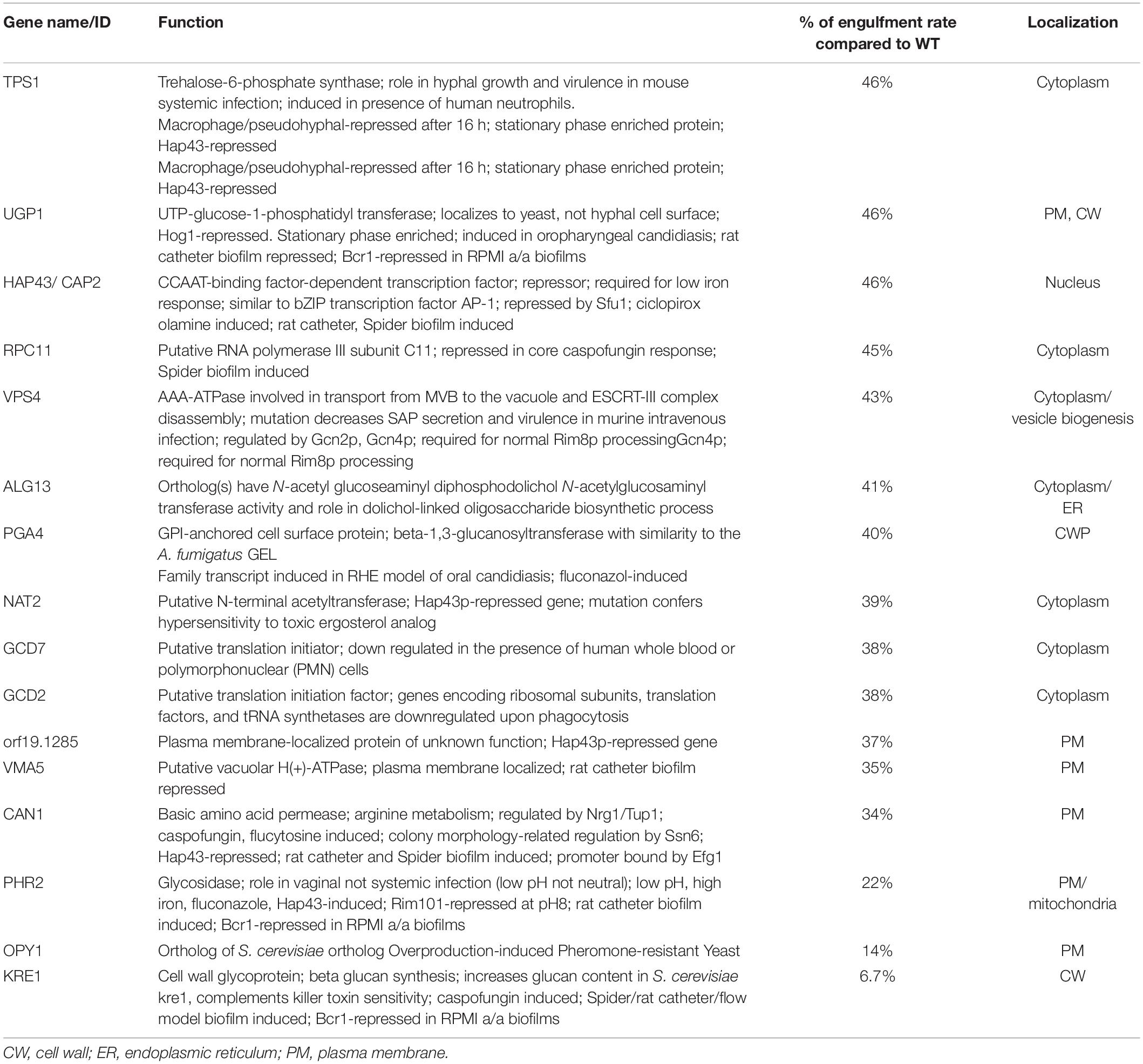
Table 3. GRACE mutant hits with phenotype as like-WT, with reduced rate of macrophage engulfment compared with CAI4-WT, and their putative localisation on C. albicans, as described on the Candida Genome data base.

Figure 2. Pairwise protein alignment between S. cerevisiae Opy1 (Sc-Opy1) and C. albicans Orf19.4245. Amino acid identity was found to be 20%. Underlined sequences correspond to predicted Pleckstrin homology (PH) domains in Ca-Orf19.4245. Identities are highlighted with bold letters; identities within the PH domains are bold in red. Alignment made with Global alignment software, available on BLAST ® from the NCBI website (https://blast.ncbi.nlm.nih.gov/Blast.cgi#). Protein sequence scanned and predicted with SMART bioinformatics software (http://smart.embl.de).
Following the establishment at the differences in the kinetics of engulfment between these two Candida mutants and WT-RFP, we tested if the presence of phagocytes could produce any differences in the growth and development of either the knock-out or the parental strains. We separately incubated each of the Δ/Δ kre1-GFP, Δ/Δopy1-GFP and WT-RFP strains in the fungal-macrophage medium; all strains developed normally from yeast, through germ tube formation and finally into pseudo-hyphae/filamentous fungal cells (Figure 3). When analyzed in detail, we did not find differences in the growth and normal development of the fungal cells in the presence or absence of the murine phagocytes.
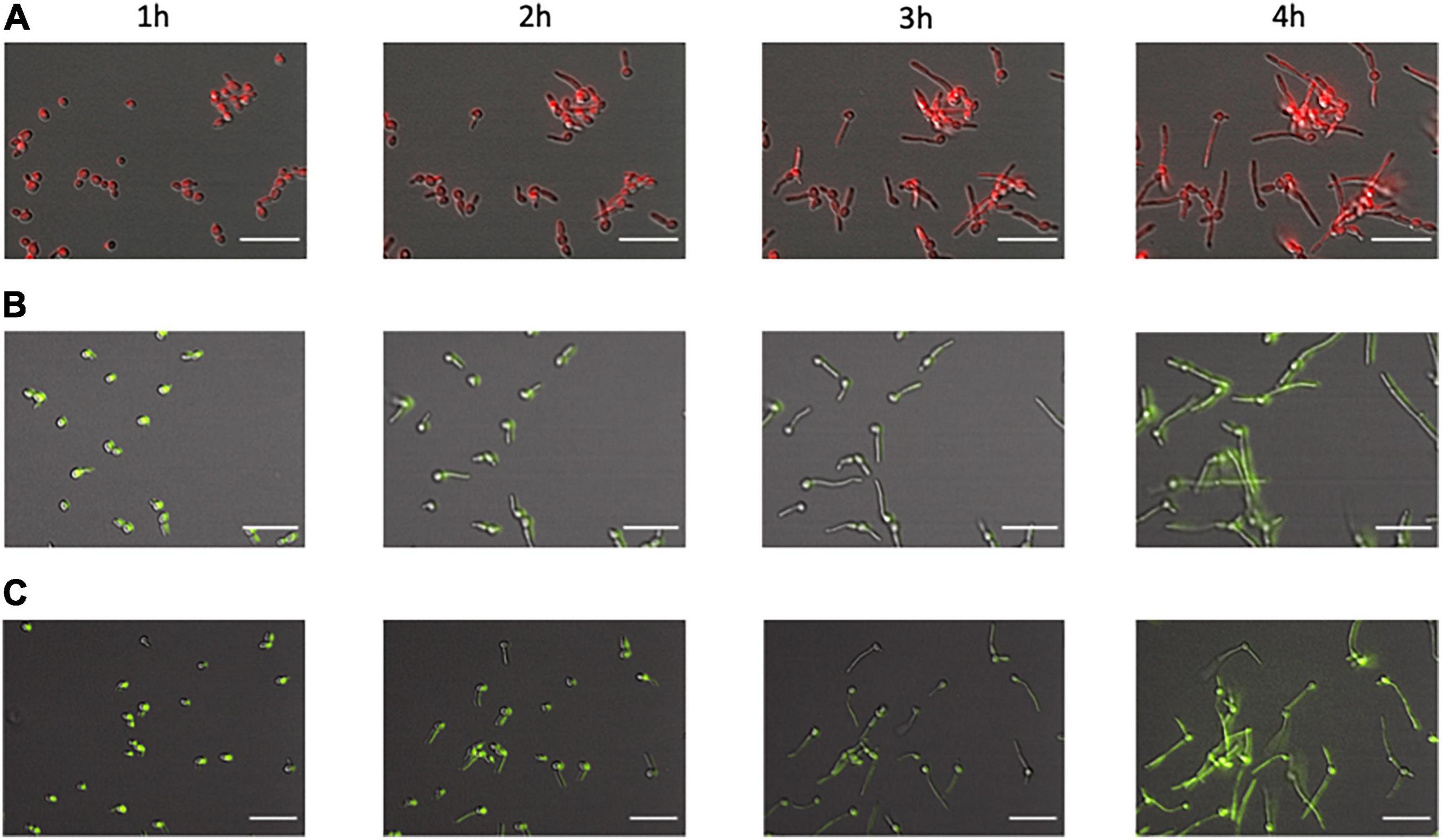
Figure 3. Development of the C albicans morphological phenotype over time. We compared fungal cells from WT-RFP (A), Δ/Δopy1-GFP (B), and Δ/Δkre1-GFP (C). All cells, regardless of phenotype or tag, showed the same morphological pattern. At 1 h of incubation most of them are still in the budding form with a few showing germ tube initiations. At 2 h there are frequent germ tubes growing from the original yeast cells. At 3 and 4 h there are extended filamentous cells and frequent initiations of a second filament from an original yeast cell. Scale bar = 20 μm.
We constructed non-conditional null mutant strains Δ/Δopy1 and Δ/Δkre1 and assessed their kinetics of engulfment by macrophages; these strains confirmed the different pattern of engulfment relative to the wild-type. Figure 4 shows a direct comparison between the wild-type-CAI4-RFP C. albicans cells (WT-RFP) and the GFP tagged Δ/Δopy1 mutant. We observed that macrophages recognized and internalized fungal cells at early time points (0.5 h–0.9 h), with a marked preference for WT-RFP compared with Δ/Δopy1 mutant (Figure 4A, top panel). This trend on the preference of WT-RFP cells over the Δ/Δopy1 mutant by the macrophages changed throughout the imaging of the fungal cell-macrophage interaction (Figure 4B). The kinetics clearly showed a lower engulfment rate on the Δ/Δopy1 mutant at the initial time points (0.5, 0.9, 1 h) and then, coinciding with germ tube formation, the engulfment for Δ/Δopy1 mutant increased slightly over the WT-RFP cells, and finally both strains exhibit similar kinetics in the recognition and internalization by the macrophages (Figures 4B,C). We also imaged the same assay including WT-RFP and the background WT-GFP (Figure 4A lower panel). For these controls, the kinetics of engulfment by the macrophages are similar, and display a few initial internalizations by the macrophages (0.5, 0.9 h) and then continuous engulfment kinetics as seen for the WT-RFP and Δ/Δopy1.
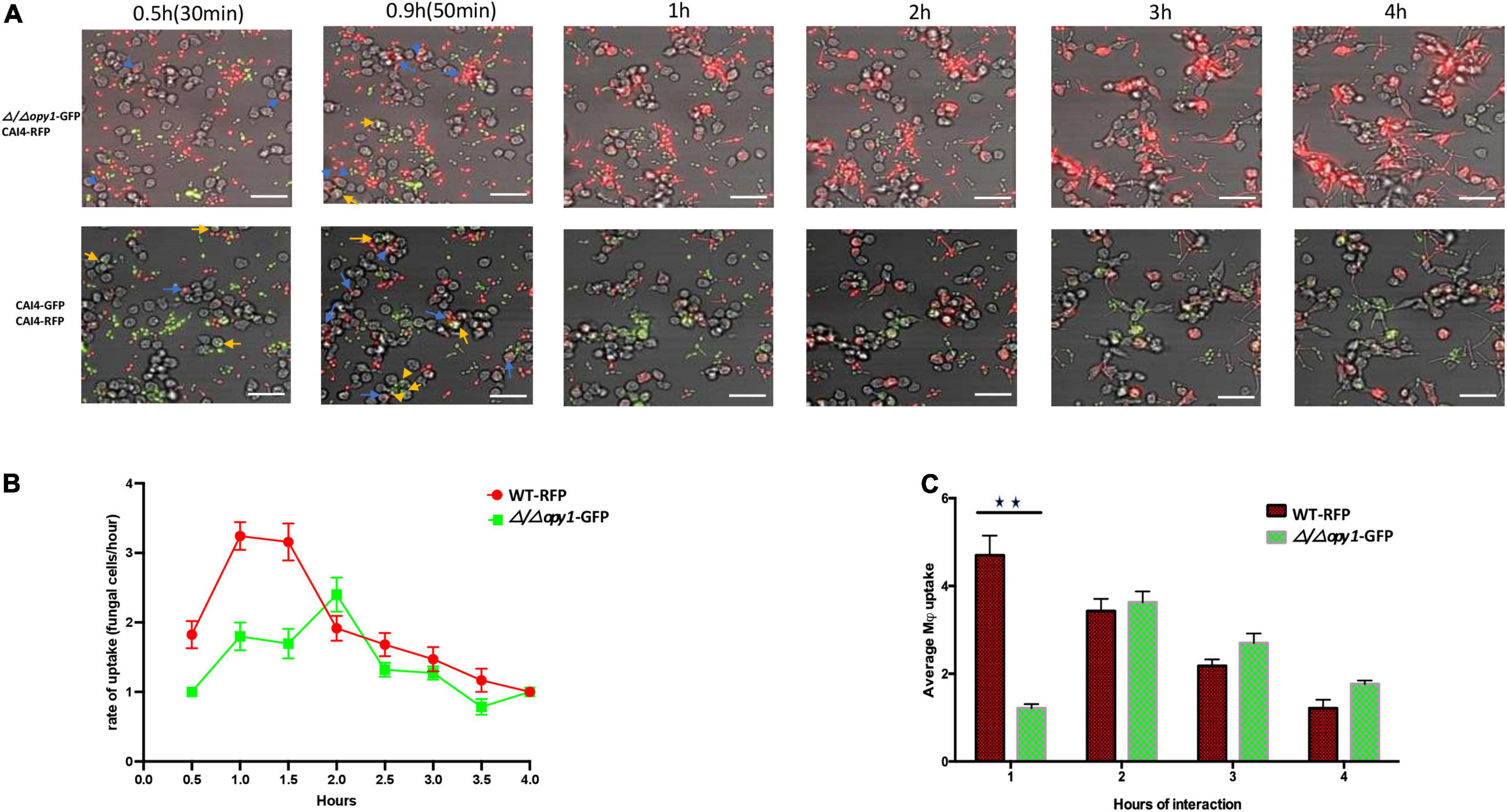
Figure 4. Kinetics of engulfment by macrophages of fluorescently-tagged C. albicans cells, with red (WT-RFP) and green tagged cells (Δ/Δ/opy1-GFP mutant) incubated together. RAW 264.7 macrophages were incubated for 4 h with C. albicans wild-type and mutants. (A) Microscopy images depicting the kinetics of macrophage engulfment of red-fluorescing CAI-4-RFP WT cells and green-fluorescing Δ/Δopy1-GFP mutant cells. Blue arrows indicate WT-RFP cells engulfed by macrophages and yellow arrows indicate (Δ/Δopy1-GFP mutant cells engulfed by macrophages. (B) The mutant (Δ/Δ/opy1-GFP) showed a delayed engulfment compared with the WT strain. (C) This is reflected in the average engulfment rate every hour of interaction when compared with the WT-RFP values; the mutant shows significantly reduced engulfment at the 1 h time point, Blue arrows indicate fungal cell engulfment by macrophages at different time points. Statistical test by two-way ANOVA. **p-value > 0.001. Scale bar = 20 μm.
We performed the same direct comparison assay for the GFP tagged Δ/Δkre1 mutant and WT-RFP cells in their interaction with the macrophages (Figures 5A–C). We also found an early and clear preference of the macrophages to engulf WT-RFP cells over Δ/Δkre1-GFP mutants (Figure 5A top panel). In this case the kinetics of engulfment by the macrophages remained biased to the WT-RFP Candida cells compared with the Δ/Δopy11-GFP mutants (Figure 5B) for the entire 4 h of imaging assay. This was supported with the average uptake by macrophages of fungal cells, with a non-significant but slightly higher rate for the WT-RFP over the Δ/Δ kre1-GFP mutants (Figure 5C). In comparison with the assay including both CAI4-RFP and CAI4-GFP (Figure 5A lower panel), the macrophages recognized and internalized both WT-tagged cells with no significant preference as seen previously (Figure 5A lower panel), and the engulfment kinetics remained equivalent through the 4 h of interaction.
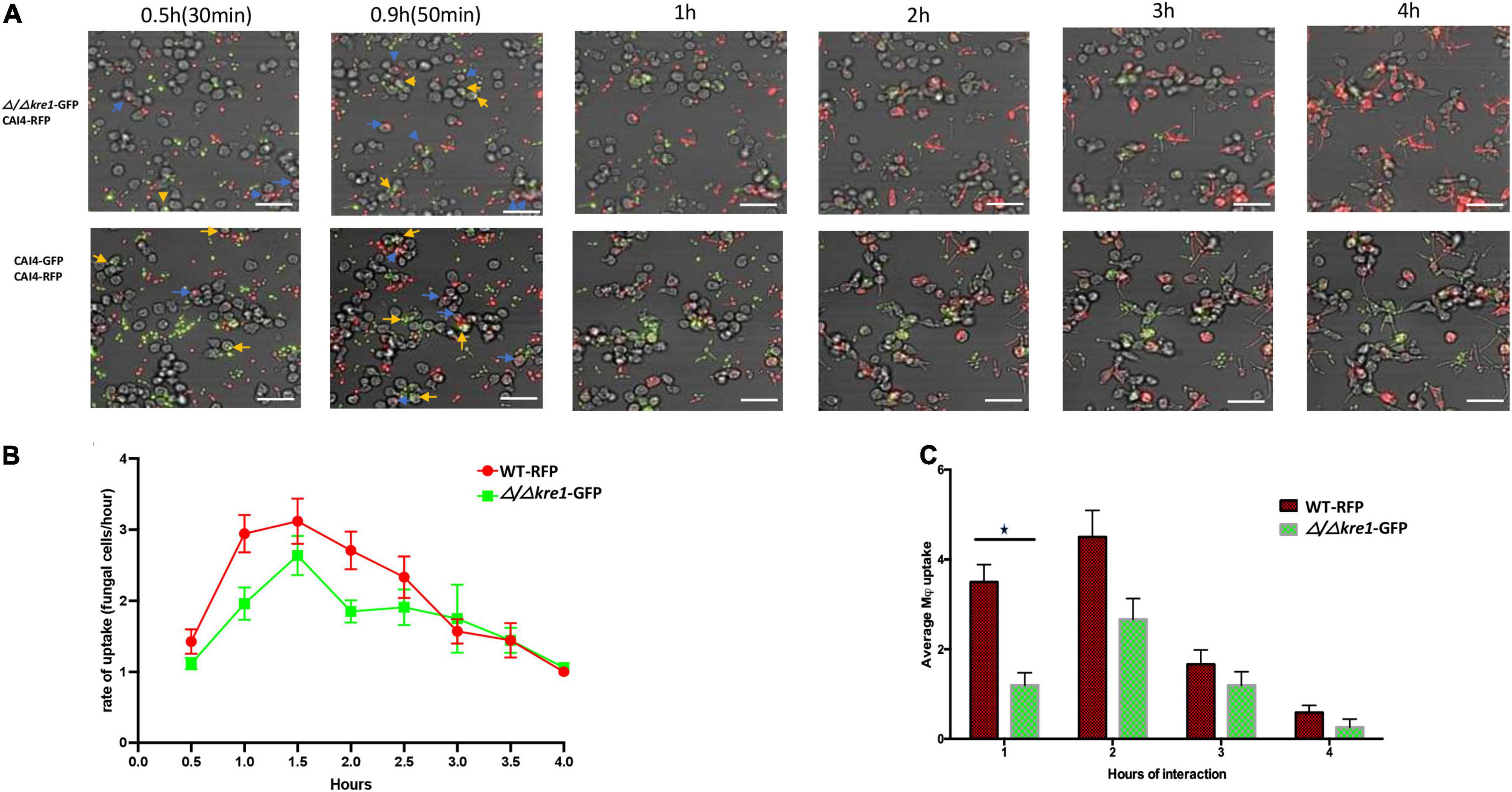
Figure 5. Kinetics of engulfment by macrophages of fluorescently-tagged C. albicans cells, with red (WT-RFP) and green tagged cells (Δ/Δkre1-GFP mutant) incubated together. RAW 264.7 macrophages were incubated for 4 h with C. albicans wild-type and mutants. (A) Microscopy images depicting the kinetics of macrophage engulfment of red-fluorescing CAI-4-RFP WT cells and green-fluorescing Δ/Δkre1-GFP mutant cells. Blue arrows indicate WT-RFP cells engulfed by macrophages and yellow arrows indicate Δ/Δkre1-GFP mutant cells engulfed by macrophages. (B) The mutant (Δ/Δkre-GFP) showed reduced kinetics of engulfment compared with the WT strain. (C) This is reflected by reduced rates at all time points when compared with the WT-RFP values. Blue arrows indicate fungal cell engulfment by macrophages at different time points. Statistical test by two-way ANOVA. *p-value > 0.005. Scale bar = 20 μm.
We also tested the kinetics of macrophage engulfment when incubating either mutants or WT-RFP with the macrophages separately (see Supplementary Figures 1, 2). In both cases, the engulfment patterns of the mutants were very similar to that seen in the mixed cultures. The Δ/Δopy1-GFP mutants showed a delayed recognition and engulfment by the macrophages, most dramatically at the initial time point of 0.5 h; engulfment then increased up to the 2 h point of interaction (Supplementary Figure 1B) and then decreased gradually until 4 h. By contrast, the WT-RFP engulfment pattern showed a higher rate of engulfment over the first 1 h of interaction and then dropped steadily down until the end of the 4 h of interaction. The average fungal cell uptake by macrophages per hour also shows greater internalization of WT-RFP cells compared with the Δ/Δopy1-GFP mutant, which later increased after the 2 h up to the end of the 4 h of interaction (Supplementary Figure 1C), behaving similarly to the kinetics of internalization (Supplementary Figure 1B). The Δ/Δ kre1-GFP mutants incubated separately from the WT-RFP cells also showed a pattern of engulfment by the mouse macrophages similar to that in the mixed cultures. In this case both Δ/Δ kre1-GFP and WT-RFP cells had similar patterns of engulfment through the monitored period, however the WT-RFP always displayed a higher rate compared with the Δ/Δ kre1-GFP mutants at each time point (Supplementary Figures 2A,B). At 2.5 h the kinetics of engulfment change due to a drop in macrophage internalization, and then continue decreasing until 4 h of interaction. We also observed that after 3 h of the interaction, many fungal cells once engulfed by the macrophages were able to extend out through the phagocytes once they initiated filamentation. Overall, the average uptake by the macrophages correlates with the kinetics, showing a higher rate-for the WT-RFPs compared with the Δ/Δ kre1-GFP mutants over the 4 h interaction assay (Supplementary Figure 2C).
We initially used murine RAW264.7 macrophages to study the C. albicans GRACE library of conditional mutants and to investigate in detail two hits, KRE1 and the uncharacterized ORF19.4245, identified as the ortholog of S. cerevisiae OPY1 from the original screening. To further investigate these fungal-macrophage interactions we repeated the assay with both Δ/Δ kre1-GFP and Δ/Δ opy1-GFP using human monocyte-derived macrophages. We tested each GFP-tagged mutant in the presence of WT-RFP and the human monocyte-derived macrophages (Figure 6). In order to test the ability of the new phagocytic cell line and avoid any bias from the background CAI-4 we first incubated the monocytes with both WT-RFP and WT-GFP (Figure 6A). After the 4 h incubation assay, we found that both WTs produced similar kinetics of engulfment with increasing engulfment from 0.5 h reaching a peak between 1 and 1.5 h to then slowly decreasing to the 4 h time point (Figure 6D). When we tested the Δ/Δ kre1-GFP or the Δ/Δ opy1-GFP mutants along with WT-RFP cells and these human monocyte-derived macrophages, we saw different kinetics of engulfment for both mutants compared with the WT-RFP. Whereas the WT-RPF kinetics remain similar to the control pattern, increasing in the period 0.5–1.5 h and then dropping slowly until 4 h (Figures 6E,F), the kinetics for the mutants Δ/Δ opy1-GFP or Δ/Δ kre1-GFP exhibited reduced macrophage recognition compared with the WT-RFP. The most significant difference occurred at 2 h of the assay, when both mutants displayed a peak of macrophage engulfment, and coinciding with the drop of internalization of the WT-RFP, suggesting a delayed response. When we analysed the average macrophage uptake, again we found substantial differences between WT-RPF and Δ/Δ opy1-GFP (Figure 6E) and Δ/Δ kre1-GFP (Figure 6F) at 1h of incubation, but little difference after that point. Thus, it appears that both mutants reduce macrophage internalization relative to the WT-RFP cells.
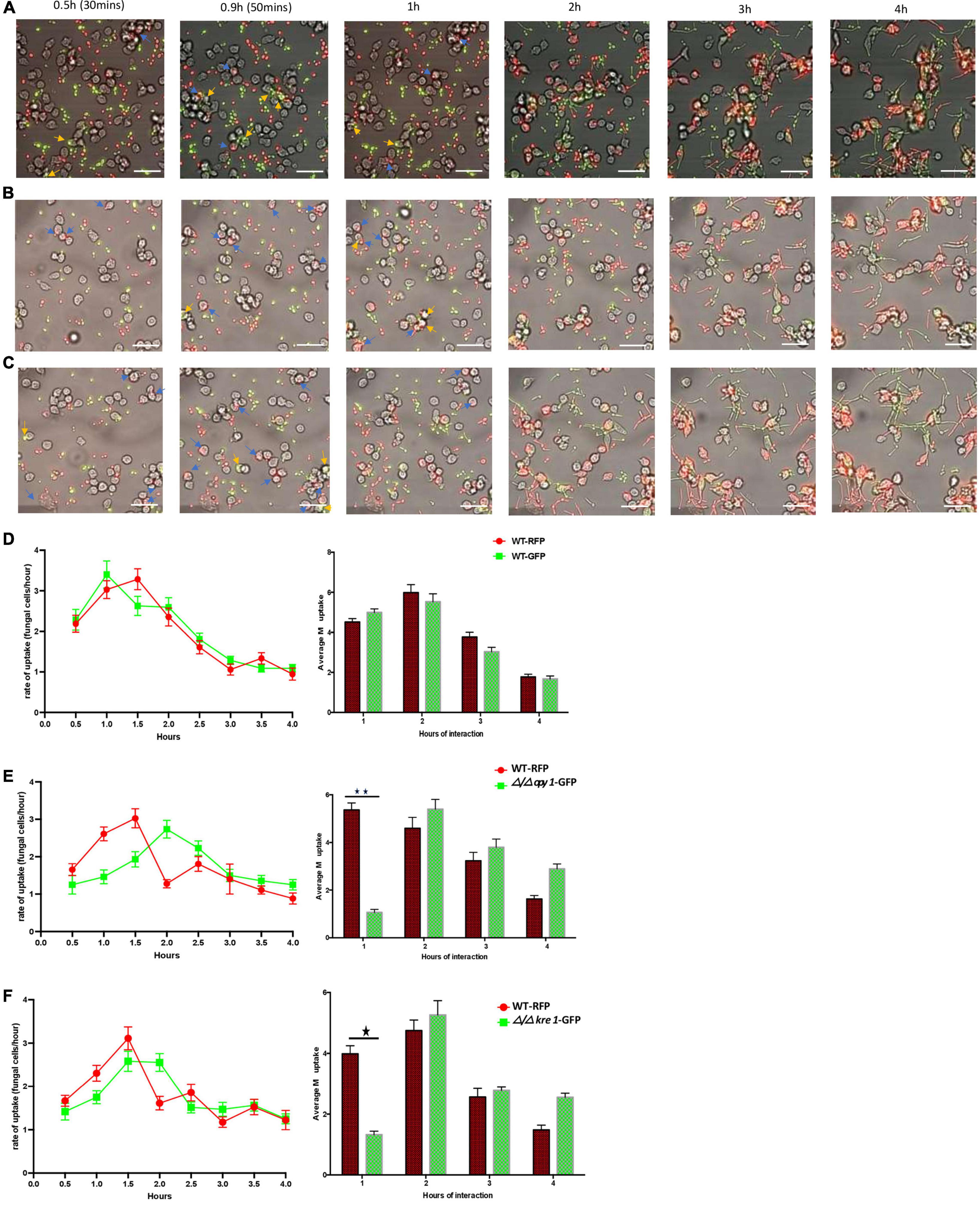
Figure 6. Kinetics of engulfment by macrophages of C. albicans (WT and mutants) incubated together with human monocyte-derived macrophages. (A) Kinetics of engulfment of CAI-4 WT cells tagged with GFP (green) or RFP (red). Compared with the WT strain [either RFP or RFP tagged on (D)], both mutants [Δ/Δopy1-GFP on (E)] and Δ/Δkre1-GFP (F) produced distinct and reduced engulfment kinetics, different macrophage uptake, specially marked at 1h of interaction. Blue arrows indicate WT-RFP cells and yellow engulfed of WT-GFP by macrophages. (B) Kinetics of engulfment displayed by macrophages for Δ/Δopy1-GFP (green) and WT-RFP (red). Blue arrows indicate WT-RFP cells engulfed by macrophages and yellow arrows indicate Δ/Δopy1-GFP mutant cells engulfed by macrophages (C). The engulfment kinetics by macrophages of Δ/Δkre1-GFP (green) and WT-RFP (red). Both mutants produced distinct and reduced engulfment kinetics compared with the WT strain. Blue arrows indicate WT-RFP cells engulfed by macrophages and yellow arrows indicate Δ/Δkre1-GFP mutant cells engulfed by macrophages. This is reflected as well in the average engulfment rate every hour of interaction when they are compared with the WT-RFP values. Statistical test by two-way ANOVA. *p-value > 0.005. **p-value > 0.001. With this add, both p-values are incorporated on the legend and makes it clear and consistent with the histograms on (E) and (F). Scale bar = 20 μm.
How the pathogenic fungal microbe, Candida albicans, regulates its behaviour to face the immune response represented by phagocytes, is a matter that remains under intensive investigation. In the present study, we performed a genetic screen of C. albicans mutants and assessed their interaction with macrophages by monitoring the kinetics of this interaction from initial recognition to internalization of the fungal cells. Our screening of the GRACE library suggests that the interaction between C. albicans cells and macrophages relies significantly on the morphology of the fungal cells. There was a clear preference in the recognition and engulfment by macrophages of mutants with a filamentous phenotype. Less effectively engulfed were mutants that develop similar to WT cells, starting as budding cells, but initiating germ tube formation followed by hyphal extension in the serum-containing media growing the macrophage cells. Finally, cells that remained locked in the yeast form were poorly recognized and engulfed.
Our present observations are consistent with other studies (Kumamoto and Vinces, 2005; Lewis et al., 2012), and have extended the analysis to over 1,200 mutants from the GRACE library. Several lines of evidence suggest that aspects of the cell surface of hyphal cells are an important signal for macrophage recognition. First, hyphal-locked mutants are efficiently engulfed from the initiation of the interaction, while yeast-locked cells remain recalcitrant to engulfment. Second, the majority of the mutant cells that undergo the yeast-hyphal transition are more efficiently engulfed after the initiation of germ tube development. Finally, two mutants that reduce the efficiency of engulfment in cells that undergo a classic germ tube initiation and hyphal transition are implicated in aspects of cell wall and cell membrane formation.
In the group of budding-arrested mutants, the poor rate of macrophage engulfment is interesting, because many of these variants lack functions critical for fungal cells, such as translation initiation factors and elements required for chromosome segregation. It is possible that these budding-defective mutants are poorly recognized and internalized by macrophages, due to reduced cell growth. Alternatively, many of these yeast-locked mutants may be lacking the production of fungal-derived biomolecules such as farnesol (Ghosh et al., 2010), which have been linked to immune cell activation, pathogen recognition and internalization (Blasi et al., 1992; Ghosh et al., 2010; Mukaremera et al., 2017). These characteristics, perhaps, would make these mutants less visible to the macrophages.
In contrast, a rapid rate of engulfment was identified for those mutants with a constitutive filamentation phenotype when they were examined for their interaction with macrophages. In this group there are genes encoding proteins involved in the biogenesis of ribosomal subunits and translation factors, as well as various enzymes that regulate oxidative stress, ATP-hydrolysis and amino acid biosynthesis. The marked preference of macrophages for this group of mutants may have several explanations; in particular these filamentous cells may offer a wide set of cell wall antigens with the potential to be recognized by immune cells (Cassone et al., 1987; Urban et al., 2003; Shen et al., 2015). Typically, once macrophages have internalized pathogens, they activate the phagosome to trigger destruction of the engulfed cells (Bain et al., 2014), In our observations, macrophages were able to internalize hyper-filamentous strains from the GRACE library screened in our study as seen before (Marcil et al., 2002). We also noticed that some hyphal mutants produced elongated filaments in different planes and directions, making them inaccessible to complete engulfment by the macrophages (Supplementary Video 3).
At later timepoints (3 h) we observed that some of the WT Candida and the KO mutants were able to break through from macrophages at close to 10% of the engulfment events, compared with a less than 5% for the hyper-filamentous mutants screened in our study. This partial lack of killing capacity of macrophages when they face longer hyphal cells or elongated fungal cells inside them, relies on the inhibition in forming and maturing the phagosome (Bain et al., 2014; Maxson et al., 2018), the organelle responsible for inactivating microorganisms internalized by macrophages. Usually, Candida cells can escape from phagocytes by affecting inflammasome-dependent pyroptosis at early time points, based on the multiplicity of infection (MOI) (Wellington et al., 2014) and ultimately lysing the macrophage as the fungal cells continue to grow. Under our conditions, a MOI of 1:4 (Candida: macrophages), there was a delayed effect on the macrophage pyroptosis compared with previous publications that used higher number of Candida cells (MOI 10) (Wellington et al., 2014; Ding et al., 2021). Once fungal cells escape the phagocytes there will be an impact on the macrophage uptake capacity at later time points that may influence the macrophage uptake index, which decreases substantially in the last 2h of interaction in our assay, as visualized in Supplementary Videos 4–12.
In our tested portion of the GRACE library collection, we found that after gene repression almost 80% of the strains displayed the typical growth pattern of C. albicans WT cells. Even in the repressed state, these cells develop as morphologically normal, starting as yeast cells, then initiating germ tube formation and ultimately forming hyphal or pseudo-hyphal forms in the presence of macrophages. This group of strains were differentially recognized over the course of the morphological transitions; the budding forms were poorly recognized, while after germ tube initiation (after 50 min) and later filamentous extension, cells were efficiently internalized, reaching their maximum engulfment by macrophages between the first and second hour (Figure 1). However, within this class of cells we identified a set of repressed genes that generated, in their interface with macrophages, reduced engulfment kinetics compared with the WT (Table 3). Among these mutants, a few genes are involved in metabolic pathways (TPS1, UGP1, ALG13), transcription (HAP43/CAP2, RCP11), translation initiation (GCD7, GCD2), vesicle trafficking (VPS4), while many of the genes found with lower kinetics are linked to the plasma membrane and cell wall (Zaragoza et al., 1998; Lorenz et al., 2004; Bruno et al., 2006; Hall and Gow, 2013).
We have focused on two genes whose inactivation triggered reduced engulfment rates in the interaction with the macrophages with no evident impact on the morphological transition: KRE1 and OPY1. We created null mutants by CRISPR gene editing and challenged these mutants in the fungal-phagocyte engulfment assay. We confirmed that, as previously observed from the GRACE library conditionally inactivated strains, both Δ/Δ opy1 and Δ/Δ kre1 mutants have a distinctive kinetics in the interaction with macrophages compared to the WT. When we investigated the fungal-macrophage kinetics during the first two hours of interaction when most of the engulfment occur, the Δ/Δ kre1 mutant showed a reduced kinetics compared to the WT, but somewhat higher compared with our observation of the KRE1-GRACE mutant (data not shown). Kre1 (killer resistant 1) is part of a family of glycoproteins anchored in the cell wall, originally described in Saccharomyces cerevisiae (Breing et al., 2004), to be involved in β-1,6-glucan synthesis (Boone et al., 1990). Although the mutant Δ/Δ kre1 may produce an altered β-1,6-glucan distribution, the sugar cell wall constitution in C. albicans has been described with a more abundant composition of β-1,3-glucan rather than β-1,6-glucan (Boone et al., 1991). It is possible that alteration of β-1,6-glucan synthesis at the cell wall in C. albicans may provide variable recognition by macrophages, because of the constant remodeling of this fungal cell structure enriched by other sugars and proteins that serve as pathogen-associated molecular patterns (PAMPS) (Klis et al., 2001; Hopke et al., 2016). Overall, when we tested the Δ/Δ kre1-GFP mutant with either murine or human-derived macrophages, there was a consistent decrease in the engulfment rate for this mutant, mainly in the first 2h of interaction, compared with the Candida WT-RFP cells (Figures 5, 6C,F and Supplementary Figures 1B, 2B).
The Δ/Δopy1 null mutant exhibited lower and delayed internalization by macrophages, also consistent with the GRACE library strain. This identifies a new gene, predicted to be involved in phospho-inositol metabolism, in macrophage engulfment of C. albicans cells. How this gene influences macrophage recognition is unclear. Its ortholog in S. cerevisiae (ScOpy1) has been described as regulating the phosphatydilinositol-4,5-biphosphate [(PtdIns 4,5)P2] lipid content (35) through direct binding with the kinase Mss4 to regulate PtdIns (4,5)P2, at the inner leaf of the plasma membrane in yeast. PtdIns (4,5)P2 represent signaling lipids that are implicated in eukaryotic cells in endocytosis, actin binding in the cytoskeleton and cell volume maintenance (Ene et al., 2015; Hammond, 2016). In addition, PtdIns (4,5)P2 are essential lipids that maintain plasma membrane components such as proteins that insert into the lipid bilayer and constitute the scaffold for the cell wall outer structure (Hammond, 2016). We observed at early time points in the fungal cell-macrophage interaction, when fungal cells were in the budding form, macrophages have a marked preference for Candida WT rather than the Δ/Δopy1 strain. The absence of Opy1 may alter the plasma membrane or may affect and modify the proper building and arrangement of cell wall components such as chitin and β-glucan-anchored proteins linked to glycophosphatidylinositol (GPI) (McLaughlin and Murray, 2005; Badrane et al., 2016). Over the middle part of the interaction (1–2 h) during germ tube extension and pseudohyphe growth, the kinetics of the recognition and engulfment of macrophages for Δ/Δopy1 mutants was delayed and reduced compared with Candida WT cells. This was seen by our wide-field microscopy visualization as well as in the automated counting generated in the cell sorter experiment. During the end of the assay (2–4 h), there was much less difference in the phagocyte’s internalization of filamentous Δ/Δ opy1 and WT cells, suggesting the impact of the modulation in PtdIns (4,5)P2 is primarily limited to the earlier events.
This phenotype observed in Δ/Δ opy1 mutants, was confirmed when we challenged these cells in presence of macrophages and a few Candida WT cells, and it was observed the predilection of the macrophage to take in the WT cells over the Δ/Δ opy1 mutants. All this suggests that Opy1 function regulating PtdIns (4,5)P2 levels at the plasma membrane has a significant consequence for the initial recognition by macrophages and that phosphotidyl-inosositides function in the fungal-macrophage interplay. This conclusion it may have implications for our understanding of the C albicans structural arrangements that drive the immune response from phagocytes. This interaction may depend not just on the cell wall components and fungal released molecules (Richard and Plaine, 2007; Chaffin, 2008), but also on the addition of PtdIns (4,5)P2 lipids to ensure normal functioning on the outer layers of C. albicans allowing the pathogen cells to “be seen” and ingested by macrophages at early stages of C. albicans infection.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
The studies involving human participants were reviewed and approved by Concordia University Human Research Ethics Committee. 7141 Sherbrooke Street West, H4B1R6, QC, Canada. The patients/participants provided their written informed consent to participate in this study.
MW and PG: rational, experimental design, and manuscript writing. PG and PD: experimental work. PG: data curation and analysis. All authors contributed to the article and approved the submitted version.
This work was supported with a FRSQ post-doctoral fellowship to PG and NSERC CRC Tier 1 grant # 950-228957, and CIHR MOP-45216 to MW.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors would like to acknowledge the help from Dr. Albert Descoteaux from Institute Armand-Frappier, in Laval, QC for providing the murine macrophage cell line used in this study. Also, we acknowledge Susan Sillaots for the C. albicans CAI4-RFP cells and Chris Law at CMCI from Concordia University, for technical support on imaging experiments.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.833655/full#supplementary-material
Supplementary Figure 1 | The kinetics of engulfment displayed by macrophages of ΔΔ/opy1-GFP (green) and WT-RFP (red) separately.
Supplementary Figure 2 | The kinetics of engulfment displayed by macrophages of ΔΔ/kre1-GFP (green) and WT-RFP (red) separately.
Supplementary Table 1 | Of C. albicans GRACE conditional mutants screened in this study.
Supplementary Video 1 | Video for GRACE library screening with the 274.RAW-macrophages 40X magnification. Representative of a yeast-locked mutant stained with Calcofluor (CW).
Supplementary Video 2 | Video for GRACE library screening with the 274.RAW-macrophages 40X magnification. Representative of like-WT mutant stained with Calcofluor (CW).
Supplementary Video 3 | Video for GRACE library screening with the 274.RAW-macrophages 40X magnification. Representative of a filamentous mutants stained with Calcofluor (CW).
Supplementary Video 4 | Video for OPY1 together with WT-RFP in presence of RAW267.4 macrophages. Time-lapse video (4 h) on macrophage/fungal cell interactions. Mutants Δ/Δopy1-GFP or Δ/Δkre1-GFP and WT-RFP in presence of 274.RAW-macrophages. 40X magnification.
Supplementary Video 5 | Video for KRE1 together with WT-RFP in presence of RAW267.4 macrophages. Time-lapse video (4 h) on macrophage/fungal cell interactions. Mutants Δ/Δopy1-GFP or Δ/Δkre1-GFP and WT-RFP in presence of 274.RAW-macrophages. 40X magnification.
Supplementary Video 6 | Video for OPY1 engulfment kinetics separated. Time-lapse video (4 h) on Macrophage/fungal cell interactions. Mutants Δ/Δopy1-GFP or Δ/Δkre1-GFP and WT-RFP in presence of 274.RAW-macrophages. 40X magnification.
Supplementary Video 7 | Video for WT-RFP engulfment kinetics separated for comparison with (SV-6) Δ/Δopy1-GFP. Time-lapse video (4 h) on macrophage/fungal cell interactions. CAI4 WT-RFP in presence of 274.RAW-macrophages. 40X magnification.
Supplementary Video 8 | Video for KRE1 engulfment kinetics separated. Time-lapse video (4 h) on macrophage/fungal cell interactions. Mutants Δ/Δopy1-GFP or Δ/Δkre1-GFP and WT-RFP in presence of 274.RAW-macrophages. 40X magnification.
Supplementary Video 9 | Video for WT-RFP engulfment kinetics separated for comparison with (Supplementary Video 6) Δ/Δkre1-GFP. Time-lapse video (4 h) on macrophage/fungal cell interactions. CAI4 WT-RFP in presence of 274.RAW-macrophages. 40X magnification.
Supplementary Video 10 | Video for OPY1 together with human monocyte-derived macrophages. Time-lapse videos (4 h) on Macrophage/fungal cell interactions: WT-RFP cells and mutant Δ/Δopy1-GFP in presence of human-derived macrophages. 40X magnification.
Supplementary Video 11 | Video for KRE1 together with human monocyte-derived macrophages. Time-lapse videos (4 h) on Macrophage/fungal cell interactions: WT-RFP cells and mutant Δ/Δkre1-GFP in presence of human-derived macrophages. 40X magnification.
Supplementary Video 12 | Video for CAI4 background strains, WT-GFP and WT-RFP together with human monocyte-derived macrophages. Time-lapse videos (4 h) on Macrophage/fungal cell interactions. 40X magnification.
Supplementary Video 13 | Video for non-stained WT-Candida and macrophages as control for the screening of Candida GRACE strains stained with Calcofluor White (CFW) and interaction with macrophages.
Badrane, H., Nguyen, M. H., and Clancy, C. J. (2016). Highly dynamic and specific phosphatidylinositol 4,5-bisphosphate, septin, and cell wall integrity pathway responses correlate with Caspofungin activity against Candida albicans. Antimicrob. Agents Chemother. 60, 3591–3600. doi: 10.1128/AAC.02711-15
Bain, J. M., Louw, J., Lewis, L. E., Okai, B., Walls, C. A., Ballou, E. R., et al. (2014). Candida albicans hypha formation and mannan masking of β-glucan inhibit macrophage phagosome maturation. mBio 5:e01874. doi: 10.1128/mBio.01874-14
Blasi, E., Pitzurra, L., Puliti, M., Lanfrancone, L., and Bistoni, F. (1992). Early differential molecular response of a macrophage cell line to yeast and hyphal forms of Candida albicans. Infect. Immun. 60, 832–837. doi: 10.1128/iai.60.3.832-837.1992
Boone, C., Sdicu, A., Laroche, M., and Bussey, H. (1991). Isolation from Candida albicans of a functional homolog of the Saccharomyces cerevisiae KRE1 gene, which is involved in cell wall beta-glucan synthesis. J. Bacteriol. 173, 6859–6864. doi: 10.1128/jb.173.21.6859-6864.1991
Boone, C., Sommer, S. S., Hensel, A., and Bussey, H. (1990). Yeast KRE genes provide evidence for a pathway of cell wall beta-glucan assembly. J. Cell Biol. 110, 1833–1843. doi: 10.1083/jcb.110.5.1833
Breing, F., Schleinkover, K., and Schmitt, M. J. (2004). Yeast Kre1p is GPI-anchored and involved in both cell wall assembly and architecture. Microbiology 150, 3209–3218. doi: 10.1099/mic.0.27175-0
Bruno, V. M., Kalachikov, S., Subaran, R., Nobile, C. J., Kyratsous, C., and Mitchell, A. P. (2006). Control of the C. albicans cell wall damage response by transcriptional regulator Cas5. PLoS Pathog. 2:e21. doi: 10.1371/journal.ppat.0020021
Cassone, A., Marconi, P., and Bistoni, F. (1987). Cell wall of Candida albicans and host response. Crit. Rev. Microbiol. 15, 87–95. doi: 10.3109/10408418709104451
Chaffin, W. L. (2008). Candida albicans cell wall proteins. Microbiol. Mol. Biol. Rev. 72, 495–544. doi: 10.1128/MMBR.00032-07
Cheng, S.-C., Joosten, L. A. B., Kullberg, B.-J., and Netea, M. G. (2012). Interplay between Candida albicans and the mammalian innate host defense. Infect. Immun. 80, 1304–1313. doi: 10.1128/IAI.06146-11
Ding, X., Kambara, H., Guo, R., Kanneganti, A., Acosta-Zaldívar, M., Li, J., et al. (2021). Inflammasome-mediated GSDMD activation facilitates escape of Candida albicans from macrophages. Nat. Commun. 12:6699. doi: 10.1038/s41467-021-27034-9
Ene, I. V., Walker, L. A., Schiavone, M., Lee, K. K., Martin-Yken, H., Dague, E., et al. (2015). Cell wall remodeling enzymes modulate fungal cell wall elasticity and osmotic stress resistance. mBio 6:e00986. doi: 10.1128/mBio.00986-15
Erwig, L. P., and Gow, N. A. R. (2016). Interactions of fungal pathogens with phagocytes. Nat. Rev. Microbiol. 14, 163–176. doi: 10.1038/nrmicro.2015.21
Fonzi, W. A., and Irwin, M. Y. (1993). Isogenic strain construction and gene mapping in Candida albicans. Genetics 134, 717–738. doi: 10.1093/genetics/134.3.717
Ghosh, S., Howe, N., Volk, K., Tati, S., Nickerson, K. W., and Petro, T. M. (2010). Candida albicans cell wall components and farnesol stimulate the expression of both inflammatory and regulatory cytokines in the murine RAW264.7 macrophage cell line: Candida albicans and macrophage cytokines. FEMS Immunol. Med. Microbiol. 60, 63–73. doi: 10.1111/j.1574-695X.2010.00717.x
Gow, N. A. R., Brown, A. J. P., and Odds, F. C. (2002). Fungal morphogenesis and host invasion. Curr. Opin. Microbiol. 5, 366–371. doi: 10.1016/S1369-5274(02)00338-7
Hall, R. A. (2015). Dressed to impress: impact of environmental adaptation on the Candida albicans cell wall. Mol. Microbiol. 97, 7–17. doi: 10.1111/mmi.13020
Hall, R. A., Bates, S., Lenardon, M. D., Maccallum, D. M., Wagener, J., Lowman, D. W., et al. (2013). The Mnn2 mannosyltransferase family modulates mannoprotein fibril length, immune recognition and virulence of Candida albicans. PLoS Pathog. 9:e1003276. doi: 10.1371/journal.ppat.1003276
Hall, R. A., and Gow, N. A. R. (2013). Mannosylation in Candida albicans: role in cell wall function and immune recognition. Mol. Microbiol. 90, 1147–1161. doi: 10.1111/mmi.12426
Hammond, G. R. V. (2016). Does PtdIns(4,5)P2 concentrate so it can multi-task? Biochem. Soc. Trans. 44, 228–233. doi: 10.1042/BST20150211
Höfs, S., Mogavero, S., and Hube, B. (2016). Interaction of Candida albicans with host cells: virulence factors, host defense, escape strategies, and the microbiota. J. Microbiol. 54, 149–169. doi: 10.1007/s12275-016-5514-0
Hopke, A., Nicke, N., Hidu, E. E., Degani, G., Popolo, L., and Wheeler, R. T. (2016). Neutrophil attack triggers extracellular trap-dependent Candida cell wall remodeling and altered immune recognition. PLoS Pathog. 12:e1005644. doi: 10.1371/journal.ppat.1005644
Horn, D. L., Neofytos, D., Anaissie, E. J., Fishman, J. A., Steinbach, W. J., Olyaei, A. J., et al. (2009). Epidemiology and outcomes of Candidemia in 2019 patients: data from the prospective antifungal therapy alliance registry. Clin. Infect. Dis. 12, 1695–1703. doi: 10.1086/599039
Huang, M. Y., Cravener, M. C., and Mitchell, A. P. (2021). Targeted genetic changes in Candida albicans using transient CRISPR-Cas9 expression. Curr. Protoc. 1:e19. doi: 10.1002/cpz1.19
Hube, B. (2004). From commensal to pathogen: stage- and tissue-specific gene expression of Candida albicans. Curr. Opin. Microbiol. 7, 336–341. doi: 10.1016/j.mib.2004.06.003
Jimenez-Lopez, C., and Lorenz, M. C. (2013). Fungal immune evasion in a model host pathogen interaction: Candida albicans versus macrophages. PLoS Pathog. 9:e1003741. doi: 10.1371/journal.ppat.1003741
Klis, F. M., de Groot, P., and Hellingwerf, K. (2001). Molecular organization of the cell wall of Candida albicans. Med. Mycol. 39(Suppl. 1), 1–8. doi: 10.1080/mmy.39.1.1.8-0
Kumamoto, C. A., and Vinces, M. D. (2005). Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence. Cell. Microbiol. 7, 1546–1554. doi: 10.1111/j.1462-5822.2005.00616.x
Leascoat, A., Ballerie, A., Augagneur, Y., Morzadec, C., Vernhet, L., Fardel, O., et al. (2018). Distinct properties of human M-CSF and GM-CSF monocyte-derived macrophages to stimulate pathological lung conditions in vitro: application to systemic and inflammatory disorders with pulmonary involvement. Int. J. Mol. Sci. 19:894. doi: 10.3390/ijms19030894
Lewis, E. L., Bain, J. M., Lowes, C., Gillespie, C., Rudkin, F. M., Gow, N. A. R., et al. (2012). Stage specific assessment of Candida albicans phagocytosis by macrophages identifies cell wall composition and morphogenesis as key determinants. PLoS Pathog. 8:e1002578. doi: 10.1371/journal.ppat.1002578
Ling, Y., Stefan, C. J., MacGurn, J. A., Audhya, A., and Emr, S. D. (2012). The dual PH domain protein Opy1 functions as a sensor and modulator of PtdIns(4,5)P2 synthesis: opy1 regulates Mss4 lipid kinase activity. EMBO J. 31, 2882–2894. doi: 10.1038/emboj.2012.127
Lorenz, M. C., Bender, J. A., and Fink, G. R. (2004). Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot. Cell 3, 1076–1087. doi: 10.1128/EC.3.5.1076-1087.2004
Marcil, A., Harcus, D., Thomas, D. Y., and Whiteway, M. (2002). Candida albicans killing by RAW 264.7 mouse macrophage cells: effects of candida genotype, infection ratios, and gamma interferon treatment. Infect. Immun. 70:63192911. doi: 10.1128/IAI.70.11.6319-6329.2002
Maxson, M. E., Naj, X., O’Meara, T. R., Plumb, J. D., Cowen, L. E., and Grinstein, S. (2018). Integrin-based diffusion barrier separates membranes domains enabling the formation of microbiostatic frustrated phagosome. eLife 7:e34798. doi: 10.7554/eLife.34798
McKenzie, C. G. J., Koser, U., Lewis, L. E., Bain, J. M., Mora-Montes, H. M., Barker, R. N., et al. (2010). Contribution of Candida albicans cell wall components to recognition by and escape from murine macrophages. Infect. Immun. 78, 1650–1658. doi: 10.1128/IAI.00001-10
McLaughlin, S., and Murray, D. (2005). Plasma membrane phosphoinositide organization by protein electrostatics. Nature 438, 605–611. doi: 10.1038/nature04398
Min, K., Ichikawa, Y., Woolford, C. A., and Mitchell, A. P. (2016). Candida albicans gene deletion with a transient CRISPR-Cas9 system. mSphere 1:e00130-16. doi: 10.1128/mSphere.00130-16
Mukaremera, L., Lee, K. K., Mora-Montes, H. M., and Gow, N. A. R. (2017). Candida albicans yeast, pseudohyphal, and hyphal morphogenesis differentially affects immune recognition. Front. Immunol. 8:629. doi: 10.3389/fimmu.2017.00629
Murad, A. M. A., Lee, P. R., Broadbent, I. D., Barelle, C. J., and Brown, A. J. P. (2000). CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast 16, 325–327. doi: 10.1002/1097-0061(20000315)16:4<325::AID-YEA538>3.0.CO;2-#
Naglik, J. R., Moyes, D., Wächtler, B., and Hube, B. (2011). Candida albicans interactions with epithelial cells and mucosal immunity. Microbes Infect. 13, 963–976. doi: 10.1016/j.micinf.2011.06.009
Nakayama, H., Mio, T., Nagahashi, S., Kokado, M., Arisawa, M., and Aoki, Y. (2000). Tetracycline-regulatable system to tightly control gene expression in the pathogenic fungus Candida albicans. Infect. Immun. 68, 6712–6719. doi: 10.1128/IAI.68.12.6712-6719.2000
Netea, M. G., Gordon, D. B., Kullberg, B. J., and Gow, N. A. R. (2008). An integrated model of the recognition of Candida albicans by the innate immune system. Nat. Rev. Microbiol. 6, 67–78. doi: 10.1038/nrmicro1815
Netea, M. G., Gow, N. A. R., Munro, C. A., Bates, S., Collins, C., Ferwerda, G., et al. (2006). Immune sensing of Candida albicans requires cooperative recognition of mannans and glucans by lectin and toll-like receptors. J. Clin. Invest. 116, 1642–1650. doi: 10.1172/JCI27114
Qin, Y., Zhang, L., Xu, Z., Zhang, J., Jiang, Y.-Y., Cao, Y., et al. (2016). Innate immune cell response upon Candida albicans infection. Virulence 5, 512–526. doi: 10.1080/21505594.2016.1138201
Richard, M. L., and Plaine, A. (2007). Comprehensive analysis of glycosylphosphatidylinositol-anchored proteins in Candida albicans. Eukaryot. Cell 6, 119–133. doi: 10.1128/EC.00297-06
Roemer, T., Jiang, B., Davison, J., Ketela, T., Veillette, K., Breton, A., et al. (2003). Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery: C. albicans essential gene identification and antifungal drug discovery. Mol. Microbiol. 50, 167–181. doi: 10.1046/j.1365-2958.2003.03697.x
Romani, L. (2011). Immunity to fungal infections. Nat. Rev. Immunol. 11, 275–288. doi: 10.1038/nri2939
Rubin-Bejerano, I., Abeijon, C., Magnelli, P., Grisafi, P., and Fink, G. R. (2007). Phagocytosis by human neutrophils is stimulated by a unique fungal cell wall component. Cell Host Microbe 2, 55–67. doi: 10.1016/j.chom.2007.06.002
Saville, S. P., Lazzell, A. N., Monteagudo, C., and Lopez-Ribot, J. L. (2003). Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot. Cell 2, 1053–1060. doi: 10.1128/EC.2.5.1053-1060.2003
Shen, H., Chen, S. M., Liu, W., Zhu, F., He, L. J., Zhang, J. D., et al. (2015). Abolishing cell wall glycosylphosphatidylinositol-anchored proteins in Candida albicans enhances recognition by host dectin-1. Infect. Immun. 83, 2694–2704. doi: 10.1128/IAI.00097-15
Stewart-Ornstein, J., Chen, S., Bhatnagar, R., Weissman, J. S., and El-Samad, H. (2017). Model-guided optogenetic study of PKA signaling in budding yeast. Mol. Biol. Cell 28, 221–227. doi: 10.1091/mbc.e16-06-0354
Sudbery, P., Gow, N., and Berman, J. (2004). The distinct morphogenic states of Candida albicans. Trends Microbiol. 12, 317–324. doi: 10.1016/j.tim.2004.05.008
Tabatabaei Shafiei, M., Carvajal Gonczi, C. M., Rahman, M. S., East, A., François, J., and Darlington, P. J. (2014). Detecting glycogen in peripheral blood mononuclear cells with periodic acid schiff staining. J. Vis. Exp. 94:52199. doi: 10.3791/52199
Tóth, R., Tóth, A., Papp, C., Jankovics, F., Vágvölgyi, C., Alonso, M. F., et al. (2004). Kinetic studies of Candida parapsilosis phagocytosis by macrophages and detection of intracellular survival mechanisms. Front. Microbiol. 5:633. doi: 10.3389/fmicb.2014.00633
Urban, C., Sohn, K., Lottspeich, F., Brunner, H., and Rupp, S. (2003). Identification of cell surface determinants in Candida albicans reveals Tsa1p, a protein differentially localized in the cell. FEBS Lett. 544, 228–235. doi: 10.1016/S0014-5793(03)00455-1
Urban, C. F., Ermert, D., Schmid, M., Abu-Abed, U., Goosmann, C., Nacken, W., et al. (2009). Neutrophil extracellular traps contain calprotectin, a cytosolic protein complex involved in host defense against Candida albicans. PLoS Pathog. 5:e1000639. doi: 10.1371/journal.ppat.1000639
Vyas, V. K., Inmaculada Barrasa, M., and Fink, G. R. (2015). A Candida albicans CRISPR system permits genetic engineering of essential genes and gene families. Sci. Adv. 1:e1500248. doi: 10.1126/sciadv.1500248
Walther, A., and Wendland, J. (2003). An improved transformation protocol for the human fungal pathogen Candida albicans. Curr. Genet. 42, 339–343. doi: 10.1007/s00294-002-0349-0
Wellington, M., Koselny, K., Sutterwala, F. S., and Krysan, D. (2014). Candida albicans triggers NLRP3-mediated pyroptosis in macrophages. Eukaryot. Cell 13, 329–340. doi: 10.1128/EC.00336-13
Williams, D., and Lewis, M. (2011). Pathogenesis and treatment of oral candidosis. J. Oral. Microbiol. 3:5771. doi: 10.3402/jom.v3i0.5771
Keywords: Candida albicans, macrophages, immune response, engulfment kinetics, fungal morphology, plasma membrane, cell wall
Citation: Godoy P, Darlington PJ and Whiteway M (2022) Genetic Screening of Candida albicans Inactivation Mutants Identifies New Genes Involved in Macrophage-Fungal Cell Interactions. Front. Microbiol. 13:833655. doi: 10.3389/fmicb.2022.833655
Received: 11 December 2021; Accepted: 08 March 2022;
Published: 05 April 2022.
Edited by:
Melissa DePaoli Johnson, Duke University, United StatesReviewed by:
Mira Edgerton, University at Buffalo, United StatesCopyright © 2022 Godoy, Darlington and Whiteway. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Malcolm Whiteway, bWFsY29sbS53aGl0ZXdheUBjb25jb3JkaWEuY2E=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.