- 1Fungal Genetics and Mycotoxicology Laboratory, Department of Microbiology, School of Life Sciences, Pondicherry University, Pondicherry, India
- 2Centre for Bioinformatics, School of Life Sciences, Pondicherry University, Pondicherry, India
- 3Institute of Bioinformatics, International Tech Park, Bengaluru, India
- 4Manipal Academy of Higher Education, Manipal, India
Ascomycetous fungi are found associated with a wide variety of substrates which range from fresh water to marine ecosystems, tropical to temperate forest soils and deserts, throughout the world over. These demystifying fungi exist as endophytes, pathogens and saprobes. They have been studied due to their ability to contaminate foods and feedstuffs, causing an elaboration of mycotoxins. The objectives of the study included extensive analyses of the morphological features of fungi, especially Aspergilli, which have been presented while studying them on specific mycological media. It is also an elaborate compilation of substantive macro- and micro-morphological characterization of different Aspergilli isolated from the spice Foeniculum vulgare used in India and other countries in the world. Further, a first of its kind attempt has been made to study their relative abundance and frequency of occurrence, molecular phylogeny and genetic relatedness to characterize the Aspergilli into specific sections, groups and clades. Single nucleotide polymorphism (SNP) analysis was carried out to evaluate the functional consequences of nucleotide variations, synonymous and non-synonymous mutations in the protein structure. The study resulted in a total of 3,506 Aspergillus isolates, which were obtained from seventy (70) fennel samples, representing 14 Aspergillus species. The two most frequently found species were A. niger and A. flavus with a relative abundance of 32.24 and 11.63%, respectively. The taxonomy and current placements have been reappraised with suggestions and prospects for future research from six sections namely Terrei, Flavi, Fumigati, Nidulantes, Nigri, and Versicolores. In addition, a total number of 27 isolates were studied and deposited at the National Centre for Biotechnology Information (NCBI) and five Aspergillus species have been identified and are being reported for the first time from the fennel seeds, based on partial sequence analysis of the official fungal barcode namely, Internal Transcribed Spacer (ITS) and a functional gene, beta tubulin gene locus, coupled with phenotypic characterization. SNPs for specific DNA regions have been used to identify variants in Aspergilli obtained from Indian fennel seeds for the first time. The need for a polyphasic approach of morphological identification and genetic characterization of Aspergilli from Foeniculum vulgare is addressed and presented here in adequate detail. Our current work makes extensive use of partial beta-tubulin gene sequences analyses to evaluate the association between SNPs in five Aspergillus species sections.
Introduction
Foeniculum vulgare Mill, commonly known as fennel in English and saunf in Hindi, belongs to the Umbelliferae (Apiaceae) family, is a flowering plant species, and is accredited by the International Code of Botanical Nomenclature (ICBN) (Badgujar et al., 2014). It is a perennial type of herb and is available in many regions like the United States, Northern Europe, southern Canada, Asia and Australia. This spice is flavorful, aromatic with culinary and medicinal uses. Fennel seeds are considered as a rich source of protein (2%), dietary fiber (10%), vitamin A (4.5%), thiamin (1%), vitamin C (14%) and dietary minerals like calcium (4%), iron (4%), magnesium (4%), and manganese (7%) with essential fatty acids. They are known to possess properties like antioxidant, antitumor, cryoprotective, hyper protective, hyperglycemic, and estrogenic activities (Javidnia et al., 2003; Samadi-Noshahr et al., 2021). It has a memory enhancing effect and can reduce stress (Koppula and Kumar, 2013). More importantly, F. vulgare has been used in the control of infectious diseases of bacterial, viral, fungal, mycobacterial and protozoan origin (Rather et al., 2016).
Statistics reveal that fennel production in India has doubled (58,265 tons) ever since the year 2000–2001 (27,332 tons), indicating the consumption trend in the sub-continent. Fennel is not just produced and processed in India, but it also exported. India exported around 23,562,460, and 20,295,380 million metric tons in the years 2018 and 2020–2021, respectively (Anonymous, 2020). The Ministry of Food Processing has estimated a loss of nearly 93 crores INR, due to harvest and post-harvest losses of India’s agricultural products (Moloney, 2019), fennel being one of them.
F. vulgare, like most cereals and grains can be infected by range of mycoflora, some of which may significantly damage the economic value of the crop. Aspergillus spp. infection and several other genera have been of specific concern in the recent years, because these fungal phytopathogens produce toxic metabolites (mycotoxins) which represent significant contaminants of food (Ahmad et al., 2014), feed (Streit et al., 2012), and agricultural commodities, spices being one of them (Makhlouf et al., 2019). The genus Aspergillus consists of a few over one hundred mold species, ever since its first characterization nearly 300 years ago. It is mitosporic and conidial group of fungi, where in some species exhibit teleomorphic stages and hence are classified under the division Ascomycota (Bennett, 2010). Apart from their wide use in the industry for benefits (Casas López et al., 2004), Aspergillus species are prolific producers of secondary metabolites known as mycotoxins (Goto et al., 1996). Some species of Aspergillus cause diseases in humans (Alshehri and Palanisamy, 2020; Kashyap, 2020), animals and birds (Arné et al., 2021). More than sixty Aspergillus sp. are pathogens which pose a major health concern (Pal et al., 2014). Aspergilli are the common contaminants of food and feedstuffs. Spices (for example, fennel seeds) occupy a prominent role in the day-to-day culinary preparations in India (Siruguri and Bhat, 2015) and are not free from being contaminated by a variety of mycotoxins. Post-harvest contamination and spoilage during storage deteriorates the nutritive value, make them unfit for consumption because of the production and elaboration of toxic secondary metabolites. The genus Aspergillus comprising of A. flavus, Aspergillus section Nigri, A. oryzae, A. parasiticus, A. terreus, and A. versicolor are frequent contaminants in agricultural commodities like paddy, milled rice, peanuts, maize, millets, and wheat. These Aspergilli produce several mycotoxins at different relative humidities and storage periods, namely aflatoxin B1, B2, G1, G2, ochratoxin A, and fumonisin B1 (Amadi and Adeniyi, 2009). Aflatoxins are toxigenic, carcinogenic, mutagenic, teratogenic, immunosuppressive, and are produced by Aspergilli as by-products which can also contaminate crops like peanut and cotton (Bhatnagar-Mathur et al., 2015; Soni et al., 2020) in addition to spices. Of the multitude of mycotoxins produced by Aspergillus species, aflatoxins B1 (Nurtjahja et al., 2019), fumonisin B2 (Han et al., 2017), and ochratoxin A (Magnoli et al., 2007), seem to be quite stable during storage of seeds and spices. Despite extensive toxicological studies with aflatoxins and other mycotoxins, the significance to human health, in case of several mycotoxins remains unclear. Synergistic interactions due to the co-occurrence of toxins is yet to be established.
Aspergillus species have medical and commercial importance. The genus Aspergillus has economic importance in different aspects like fermentation industry and enzyme production (Malathi and Chakraborty, 1991). Members of the genus produce a wide range of secondary metabolites, including lovastatin, a cholesterol-lowering drug, antitumor metabolites, etc. (Casas López et al., 2004). Few members of the genus are also the source of natural products which can be used to treat human diseases (Hiort et al., 2004).
Challenges in Identification of Aspergillus Species by Morphological Characterization
Mycologists have traditionally used phenotypic characteristics as a sole means for fungal identification. Classification of Aspergilli, which comprise highly speciose lineages, requires techniques such as fungal barcoding for accurate species identification. Aspergilli have been recognized since ancient times by the formation of a common morphological structure referred to as the “Aspergillum” which consists of an asexual reproductive and distinctive entity, with a characteristic stalk-like “conidiophore” bearing a bulbous vesicle, which in turn bears the phialides and metulae, on which chains of microconidia are arranged (Bennett, 2010) in basipetal succession. Morphological identification of Aspergillus spp. by using macro-morphological characters like colony diameter, color, obverse, and reverse colony characters, etc., and microscopic features like conidial heads, vesicles shape, length of vesicles, stipes, seriation, metula, conidia color, size shape and ornamentations, asci, ascospore size, shape, color, etc., would only help in the preliminary assignment of generic names, leading to an inadequacy in identification to assign species. The limitations posed by morphological identification are many. These tests are extremely time consuming, cumbersome, requiring skilled personnel and expertise in carrying out the whole process. Hence, fungal phylogeny which uses genetic and molecular tools, are being used alongside the conventional techniques to address the gap of misidentification (Raja et al., 2017).
To overcome this constraint, a set of adoptable standard procedures like the use of nuclear ribosomal genes (ITS) most commonly used in fungal identification, considered as the official barcoding marker and methods used for construction of phylogenetic tree, which facilitates species identification has been carried out (Raja et al., 2017). The ITS and beta-tubulin genes have not been systematically used in Aspergillus taxonomy from F. vulgare till date, and have thus been appropriately evaluated in this study. In addition, our purpose was to examine protein-coding functional sequences as a major genetic marker for assessing intraspecies diversity. Therefore, the beta-tubulin gene is being considered in the current study since researchers (Cleveland and Sullivan, 1985) have reported that multiple tubulin genes are required in all but the simplest eukaryotic organisms (Edgcomb et al., 2001), for the formation, maintenance and preservation of the essential microtubules. The beta-tubulin gene appears to occur as a single-copy (α1, α2, β1, γ tubulins) in Aspergillus niger, single-copy (α1, γ tubulins), two-copy (β1) or three-copy sequences (α2 tubulins) in Aspergillus nidulans (Zhao et al., 2014). Among the various genes with basic functionality of coding for proteins in all eukaryotes (Mages et al., 1995; Schütze et al., 1999), the beta-tubulin gene (Keeling et al., 2000; Ayliffe et al., 2001; Mukherjee et al., 2003; Juuti et al., 2005), is also essential for sexual development in A. nidulans (Kirk and Morris, 1991) and has received enormous attention due to its highly conserved nature. This is especially important while phylogenetic constructions are undertaken with a broad range of organisms or for a specific genus.
Single nucleotide polymorphisms (SNPs) are single-nucleotide substitutions of one base for another and are definitely the most prevalent set of genetic variety. There are two sorts of techniques to examine SNPs: genomic and functional. SNPs have a variety of consequences, for instance, drug resistance, effects on mRNA splicing, nucleocytoplasmic export, etc. There is no information available on SNPs in Aspergilli isolated from spices. The current analysis has made efforts to identify synonymous and non-synonymous mutations, to examine them evolutionarily, and to reclassify Aspergilli into their respective Sections for appropriate taxonomic identification.
With this background, the predominant objectives of this study constituted, the elaborate and detailed macro- and micro-morphological studies of Aspergillus species, molecular characterization of Aspergillus isolates obtained from Foeniculum vulgare samples using universal fungal bar-coding of the Internal Transcribed Spacer (ITS) and a functional genetic marker namely, beta–tubulin (β) gene sequences. This analysis was also performed with the primary objective of molecular fungal barcoding to substantiate our morphological analysis. Further, phylogenetic and single nucleotide polymorphism analyses have also been performed, for the first time from Aspergilli isolated from Indian fennel seed samples.
Materials and Methods
Collection of Samples
A total number of seventy (70) samples of the Indian spice Foeniculum vulgare were collected from different retail markets from several regions, namely New Delhi, Lucknow, Pinjore, West Midnapore, and Puducherry from India. These samples were from urban, semi-urban and rural zones from different geographical regions of India. The samples were purchased in portions of 100 g and stored in sterile polyethylene zip-lock pouches. All samples were adequately labeled with suitable codes. The pouches were then transported to the laboratory and analyzed sequentially at the earliest. Representative spice samples were stored at 4°C for further analysis.
Macro-Morphological Cultural Studies of Aspergilli
Standard methods of agar plating technique as described by the International Seed Testing Association (ISTA) were chosen for the mycofloral analyses of the Indian fennel samples. Briefly, 25 g of the spice samples was weighed, washed thrice with distilled water and then rinsed with 1% sodium hypochlorite (NaOCl) solution for one min to inactivate surface contaminants. Subsequently, the seeds were rinsed in distilled water to remove any adhering sodium hypochlorite. The seeds were briefly allowed to stand for about 5–10 min after they had been placed in sterile petriplates lined with sterile tissue paper and plated using standard procedures. The petri plates containing the seeds were incubated in upright position at 25–27°C for 7 days. The results were expressed as percentage of infection and the frequency of occurrence was calculated (Reddy et al., 2009; dos Santos-Ciscon et al., 2019).
Aspergillus isolates were obtained after plating Foeniculum vulgare spice samples on mycological/fungi-specific media namely fungal agar (FA) and potato dextrose agar (PDA) media (HiMedia, Mumbai, India) by following standard agar plating procedures (Xie et al., 2007; Hamzah et al., 2018). All media were prepared with the mandatory addition of a thermostable, antibacterial antibiotic namely chloramphenicol (Sigma-Aldrich, Steinhein, Germany) at the rate of 20 mg/1,000 mL of the fungi-specific media proposed. Briefly, representative samples were washed thrice with distilled water, dried for about 15 min and then plated @ 25 seeds per petri plate (100 × 15 mm, S-line, Borosil®, Mumbai, India), using aseptic techniques. The plates were then incubated at 25°C ± 2°C for 5–7 days with alternating periods of 12 h light and 12 h darkness. On the 7th day, Aspergillus colonies (green, greenish yellow, ochre/yellow, black, brownish-black, dark-brown, biscuit-brown, blue) were observed and isolated from the spice samples onto PDA medium. Spore suspensions of the Aspergilli were prepared and about 20 μL was transferred onto fresh PDA media. Colonies arising from single germinating spores were checked using Stereo Binocular Microscope (Magnus MSZ-Bi, Model: 13M1009), chosen for further studies and identified based on descriptions made in Fungal keys and Manuals (Thom and Raper, 1945; Varga and Samson, 2008) and Monographs (Refai et al., 2014).
Macro-Morphological Microscopic Studies of Aspergilli
Stereo binocular microscopic observations were carried out to detect presence of different species of Aspergilli and distinguish them from other fungal genera like Alternaria, Curvularia, Fusaria, Penicillia, Mucor, Mycelia sterilia, Trichothecium, etc., encountered during the analysis. Fungal flora were isolated from the fennel samples on potato dextrose agar (PDA) and czapek dox agar (CDA) media and identified using fungal manuals and keys (Thom and Raper, 1945; Refai et al., 2014). While mycoflora were being studied, emphasis was laid to the observation and isolation of Aspergilli. Macromorphological studies were carried out on aforesaid agar media. The isolates were inoculated either by single point or three-point inoculation method in glass petriplates (100 × 15 mm, S-line, Borosil®, Mumbai, India). After 7 days of incubation colony diameters, color, texture, sporulation pattern, obverse and reverse colony colors/characters and presence or absence ascomata (in older cultures incubated for 15–25 days) were determined.
Micro-Morphological Microscopic Studies of Aspergilli
The microscopic features of the isolated Aspergilli were sequentially studied using Light Microscopy (Olympus CH20i), Scanning Electron (SE; Hitachi, Model E-1010) and Differential Interference Contrast (DIC) microscopic analysis. Prominence was given to study characteristics like hyphal nature, septa, conidiophores, vesicle shape, sterigmata arrangement, and conidia formation. Standard protocols were followed while studying the microscopic features as outlined in identification Keys and Manuals (Thom and Raper, 1945; Varga and Samson, 2008). The light and scanning electron micrographs were taken at the Fungal Genetics and Mycotoxicology Laboratory, Department of Microbiology, Pondicherry University, while the DIC microscopic images (Nikon Upright Motorised Microscope, ECLIPSE Ni series, Nikon Corporation, Tokyo) were taken at the Fungal Biotechnology Laboratory, Department of Biotechnology, School of Life Sciences, Pondicherry University.
Preparation of Fungal Cultures for Genomic DNA Isolation
As and when microscopic analyses were being performed pure cultures of Aspergilli were inoculated onto PDA medium and incubated at 28°C for 7 days. Fungal mycelium from each petriplate was scraped off the medium using disposable sterile blades (Carbon steel Scalpel Blade No. 11, Olrada, India) and transferred to sterile pestle and mortar. The mycelial biomass (approximately close to 100 mg was collected) was used for DNA isolation.
Isolation of Fungal Genomic DNA
DNA was isolated from fungal isolates grown on synthetic PDA plates and DNA isolation was carried out using a Standard Kit (Gene JET plant Genomic DNA Purification Kit- K0791, Thermo Fisher Scientific, Vilnius, Lithuania). 100 mg of the fungal mycelial biomass was transferred to a sterile pestle and mortar. To this liquid nitrogen (200 mL) was carefully added along the walls of the mortar and crushed adequately in clockwise fashion for 5 min. The crushed mycelial biomass was immediately transferred to a new, sterile microcentrifuge tube (1.5 mL; Tarsons, Kolkata, India). Soon after, Lysis Buffer A (350 μL) and Lysis Buffer B (50 μL) were added in succession. A tiny quantity of RNase A (20 μL) was transferred to the microcentrifuge tubes containing the lysis buffers and crushed mycelial biomass. The specimen was then subjected to heat treatment for 10 min at 65°C using a sterile water bath with occasional stirring. After the heat treatment, the precipitation solution (130 μL) was added as per manufacturer’s instructions. The microcentrifuge tubes were adequately mixed and kept on frost for 5 min. The tubes were then spun at ≥ 20,000 g (≥ 14,000 rpm) for 5 min. The tubes were removed, the supernatant (precisely 450–550 μL) was collected and transferred to a clean microcentrifuge tube, and an equal volume of plant gDNA binding solution and 96% ethanol (400 μL) was added and vigorously mixed. Thereafter, 600–700 μL of the prepared solution was transferred, isolate-wise, to the new spin columns. The supernatant thus collected was centrifuged at 6,000 g (∼8,000 rpm) for about a min. Using the same column, the supernatant solution was removed and the remaining mixture was centrifuged for 1 min. The wash buffer I (500 μL) was added into the spin-columns and centrifuged at 8,000 g (∼10,000 rpm) for 1 min, followed by the addition of wash buffer II (500 μL) in the spin columns again. This time the spin columns were centrifuged at high speed of ≥ 20,000 g (≥ 14,000 rpm) for 3 min. The contents of the collection tube within the spin columns were discarded, and the column contents were transferred to fresh, new microcentrifuge tubes. For genomic DNA elution, the elution buffer (30 μL) was placed mid-point of the spin column layer, incubated at room temperature for 5 min. The microcentrifuge tubes were spun at a speed of 8,000 g (∼10,000 rpm) for 1 min. A second elution step was carried out using the elution buffer (20 μL). The purified DNA was stored at −20°C for further use.
Polymerase Chain Reaction of the Internal Transcribed Spacer and β-Tubulin Genes
A total number of 27 isolates were selected for a phylogenetic analysis of the ITS and β-tubulin gene using (Tam et al., 2014) as one of the references. Partial amplification of the Internal Transcribed Spacer (ITS) region and functional gene β-tubulin was performed using the primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) for ITS (Op De Beeck et al., 2014; Tam et al., 2014), bT2a (5′- GGTAACCAAATCGGTGCTGCTTTC-3′) and bT2b (5′- ACCCTCAGTGTAGTGACCCTTGGC-3′) for β-tubulin (Tam et al., 2014) respectively. Polymerase Chain Reaction (PCR) reactions were performed in a 25 μL volume, containing 12.5 μL (Taq DNA Polymerase 2x Master Mix RED, Ampliqon PCR Enzymes and Reagents, Stenhuggervej, Denmark), Forward primer (10 pmoL) 2.0 μL, Reverse primer (10 pmoL) 2.0 μL, Template DNA X μL (genomic DNA: 10–500 ng), Molecular biology grade water X μL (HiMedia®, Mumbai, India) for ITS gene, 12.5 μL (Taq DNA Polymerase 2x Master Mix RED, Ampliqon PCR Enzymes and Reagents), Forward primer (10 μM) 1.0 μL, Reverse primer (10 μM) 1.0 μL, Template DNA X μL (genomic DNA: 10–500 ng), Molecular biology grade water X μL (HiMedia®, Mumbai, India) for β-tubulin gene amplification. The cycling protocol consisted of an initial denaturation step of 94°C for 1 min 30 s, 32 cycles of denaturation at 95°C for 35 s, annealing for 55 s at 55°C for ITS and an initial denaturation step of 95°C for 3 min, 32 cycles of denaturation at 94°C for 30 s, annealing for 45 s at 65.5°C for β-tubulin and extension at 72°C for 1 min followed by a final extension of 10 min for ITS and 13 min for β-tubulin at 72°C. PCR products were sent to (Biokart India Pvt. Ltd., Bangalore, India) for purification and sequencing.
Phylogenetic Analyses
A consensus sequence for each locus of ITS and partial β-tubulin gene was generated using the forward and reverse sequence with CAP3 programme1 (Huang and Madan, 1999). Newly generated sequences have been deposited in GenBank2 (Table 1). A similarity search was performed for the newly generated sequences using BLASTn of the National Centre for Biotechnology Information.3 The homologous sequences for each isolate belonging to the corresponding species were retrieved from the GenBank database. The retrieved sequences were aligned using MAFTT v 74 (Katoh and Standley, 2013) with default parameters and alignments were edited with BioEdit v7.2.5.0 (Hall, 1999) and were manually adjusted as and when required. The low-quality bases at the start and end positions of the sequences were removed.
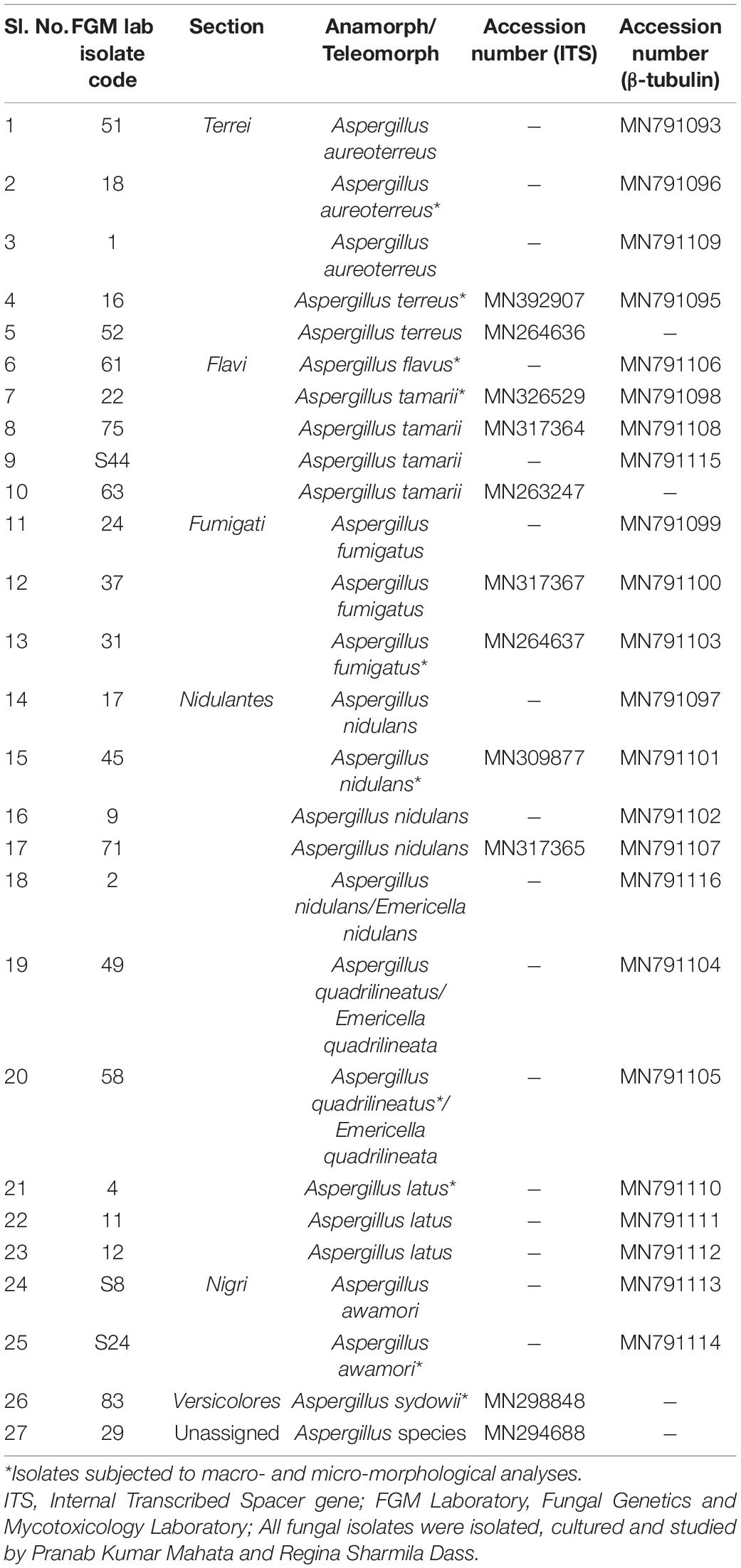
Table 1. Section-wise details of Aspergillus isolates used in the current study, GenBank accession numbers of ITS and β-tubulin gene sequences.
The retrieved homologous sequences for each genetic marker were aligned separately with ClustalX2 v2.1 (Thompson et al., 1997) using default parameters. Phylogenetic analyses of individual loci were performed by maximum parsimony (MP) method using PAUP v 4.0 beta win (Swofford and Sullivan, 2003) (heuristic search option with 1,000 random taxon additions and tree bisection and reconnection (TBR) as the branch swapping algorithm). The efficacy and the robustness of the parsimonious trees were evaluated with 1,000 bootstrap replications (Hillis and Bull, 1993). Further, the maximum likelihood (ML) method was used for the analysis of individual loci via the CIPRES Science Gateway.5 Analysis was carried out using RAxML v. 7.4.2 (Stamatakis et al., 2012). A general time-reversible model (GTR) was applied, including estimation of invariable sites and a discrete gamma distribution with four rate classes (GTRG + I). The reproducibility of the branches was evaluated by bootstrap analysis with 1,000 replicates.
The trees that were being generated were envisaged with FigTree v1.4.0 (Rambaut and Drummond, 2009). Bootstrap (bs) percentages of analysis were labeled at the nodes. Values less than 70% bs were removed. Branches with values 95% and above bs were thickened.
Single Nucleotide Polymorphisms
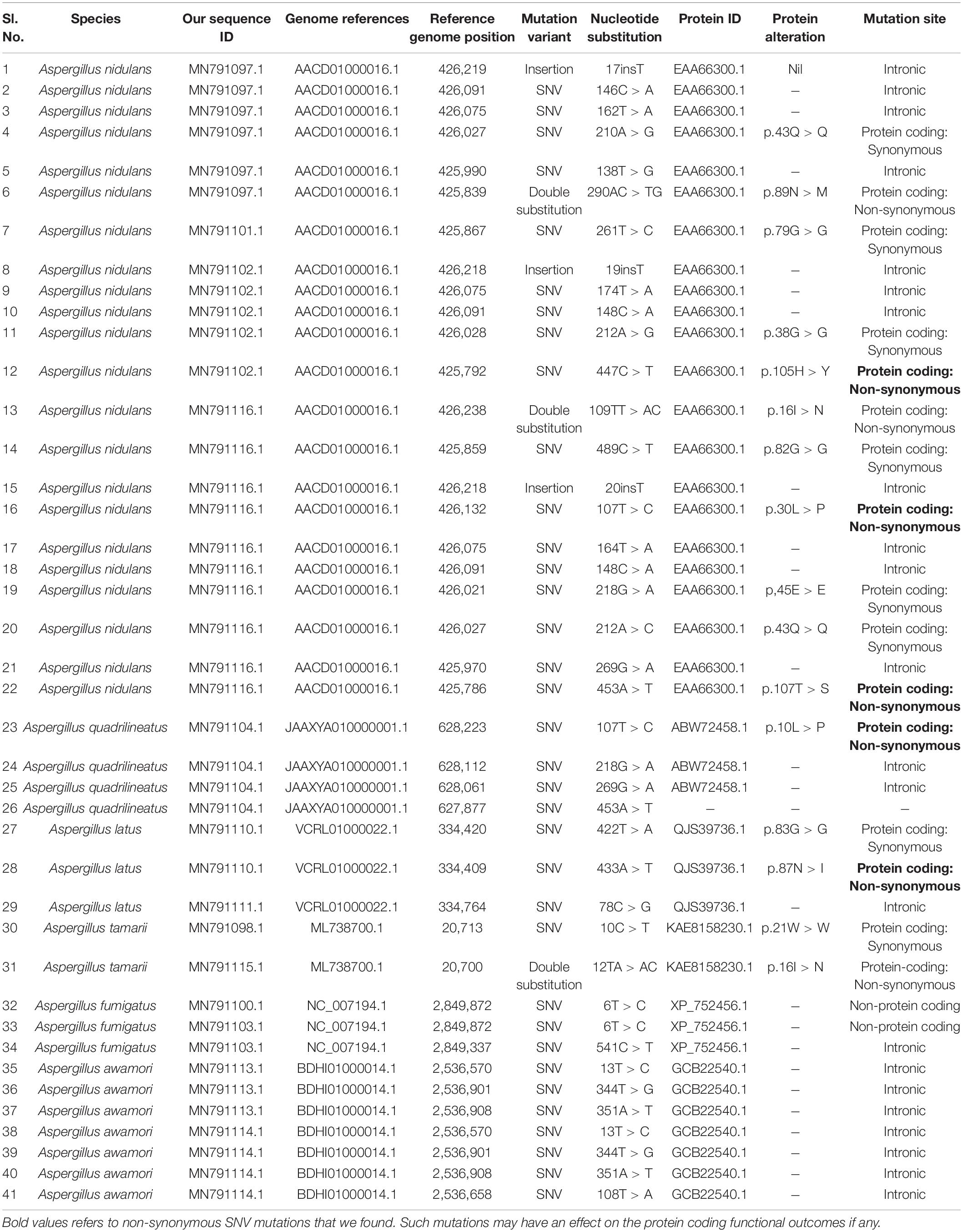
Each of the sequences of the fungal isolates listed in Table 1 were aligned with the reference genomes of respective species that are available on the NCBI using BLAST algorithm. Variants observed in these sequences were fetched manually and the corresponding variants on the protein sequences were annotated manually from the protein annotations of β-tubulin genes of the respective species.
Statistical Analysis
To learn more about the Aspergillus fungal isolates, we employed descriptive statistics on the data. The statistical studies were carried out using Origin 8.6 (Northampton, United States).
Results
Relative Abundance in Foeniculum vulgare
As shown in Figure 1 among all the isolates collected from F. vulgare (n = 70), we were able to identify fourteen (14) different species of the Aspergillus. Aspergillus niger (32.24%) ranked first, followed by A. flavus (11.63%), A. terreus (2.6%), A. nidulans (1.65%), A. tamarii (0.97%), A. species (0.96%), Emericella quadrilineata (0.45%), A. fumigatus (0.43%), A. latus (0.4%), A. aureoterreus (0.37%), A. awamori (0.37%), A. brasiliensis (0.29%), A. ochraceous (0.06%), and A. sydowii (0.03%).
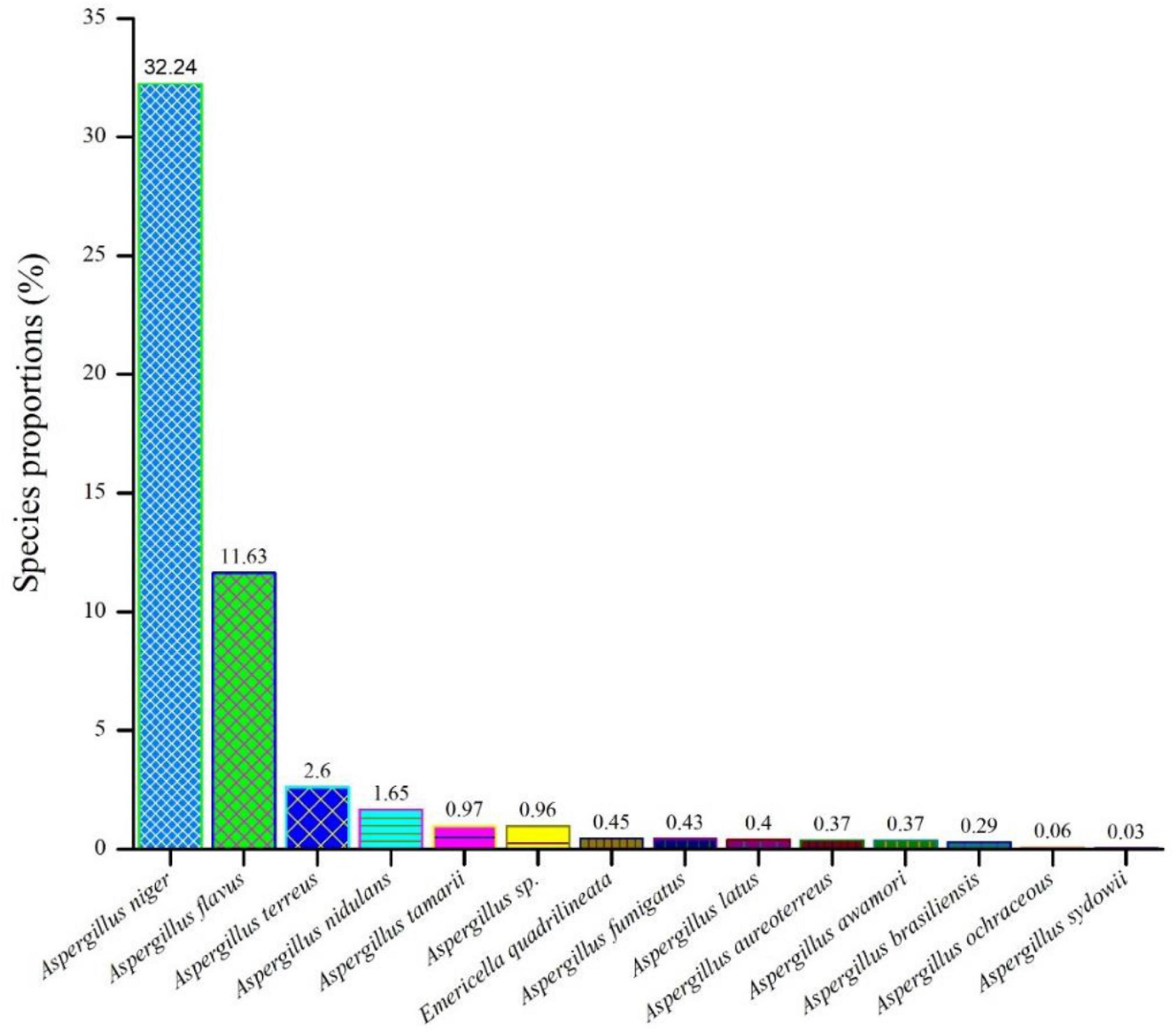
Figure 1. An assessment of the relative abundances of several Aspergillus species in the F. vulgare samples.
Aspergillus Diversity
The cultivation of fungi from seventy (70) fennel samples yielded a total of 3,506 Aspergillus isolates. Apart from Aspergilli, genera like Alternaria species, Curvularia species, Drechslera species, Fusarium species, Mucor species, Mycelia sterilia, Neurospora crassa, Penicillium species, Trichothecium species and Yeasts were also isolated based purely on morphological and microscopical analyses. The detailed cultural morphology and microscopic analyses of the Aspergilli has been carried out and has been presented with descriptions (Figures 2–5). A. niger (Relative abundance: 32.24%) followed by A. flavus (Relative abundance: 11.63%) were found to be the two most frequently occurring species. The fourteen (14) Aspergillus species were isolated from Indian fennel seeds, collected from different geographical zones of India, and studied in detail using morphological and genetic methods for molecular phylogeny. The Internal Transcribed Spacer gene of the ribosomal DNA (rDNA-ITS) sequencing identified 20 isolates under five Aspergillus Sections namely Terrei, Versicolores, Flavi, Fumigati and Nidulantes (Sub-genus II) and corresponding to the family Trichocomaceae. The isolates included A. nidulans, A. quadrilineatus, A. miyajii, A. rugulosus, A. terreus, A. tamarii, A. fumigatus, A. sydowii and Aspergillus species (Section Unassigned). Isolates sequenced for β-tubulin gene sequences revealed the identity of 23 Aspergilli with species namely A. nidulans, A. quadrilineatus, A. latus, A. terreus, A. aureoterreus, A. flavus, A. tamarii, A. fumigatus and A. awamori from five sections namely Nidulantes, Terrei, Flavi, Fumigati, and Nigri (Sub-genus I) with no known teleomorphs. The Aspergilli namely A. quadrilineatus, A. latus, A. aureoterreus, A. awamori and Aspergillus species are being reported for the very first time from fennel seeds. The details of the GenBank depositions are also shown (Table 1).
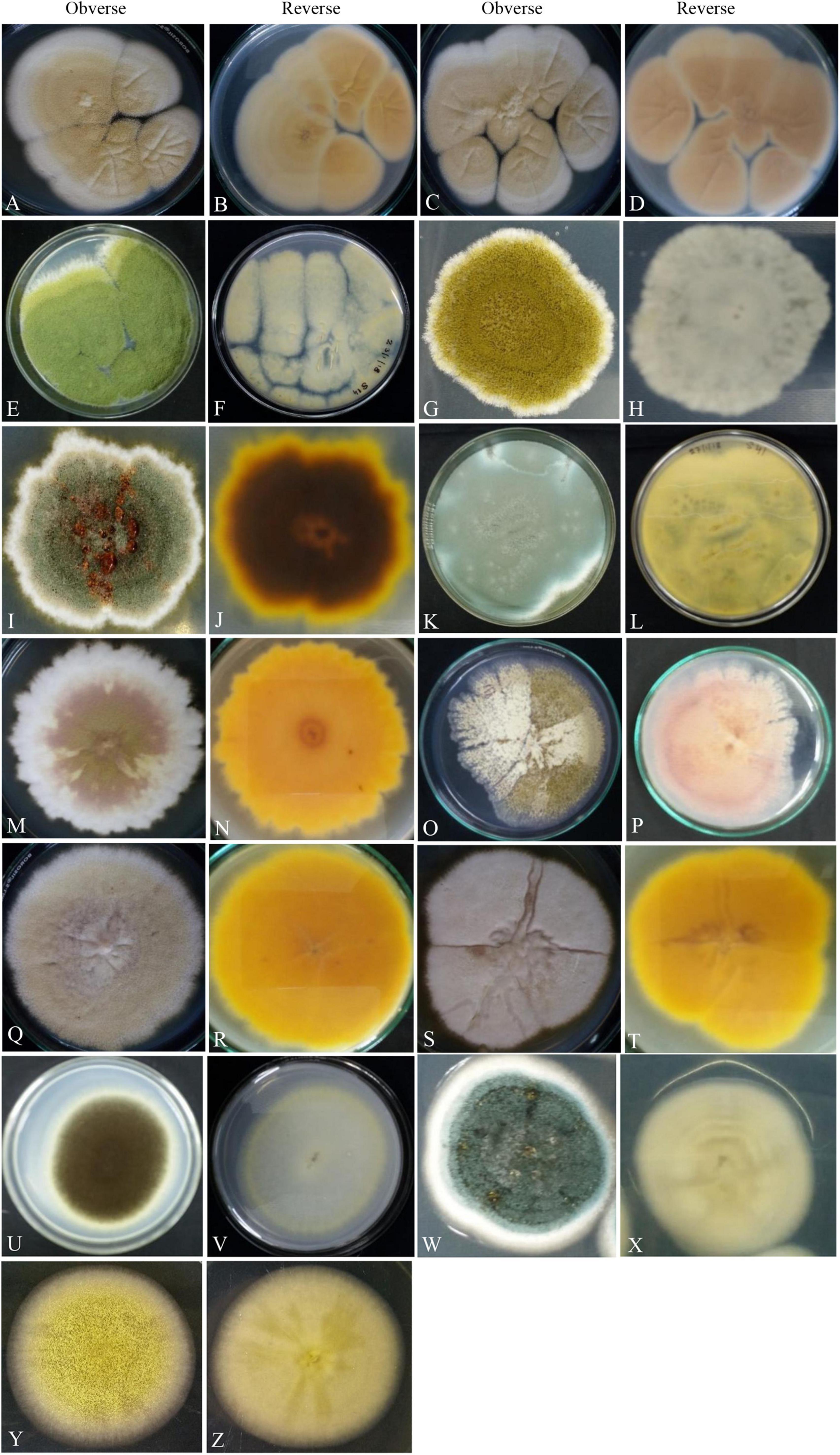
Figure 2. Photographs illustrating the macromorphology of strains of Aspergillus aureoterrus (A,B) (Acc. No. MN791096); Aspergillus terreus (C,D) (Acc. No. MN791095); Aspergillus flavus (E,F) (Acc. No. MN791106); Aspergillus tamarii (G,H) (Acc. No. MN791098); Aspergillus fumigatus (I,J) (Acc. No. MN791103); Aspergillus fumigatus (K,L) (Acc. No. MN791100); Aspergillus nidulans (M–P) (Acc. No. MN791101); Aspergillus quadrilineatus/Emericella quadrilineata (Q,R) (Acc. No. MN791105); Aspergillus latus (S,T) (Acc. No. MN791110); Aspergillus awamori (U,V) (Acc. No. MN791114); Aspergillus sydowii (W,X) (Acc. No. MN298848), and Aspergillus species (Y,Z) (Acc. No. MN294688) are shown. Images (O,P) of Aspergillus nidulans were incubated in CDA for 7 days, whereas all other isolates were kept in PDA.
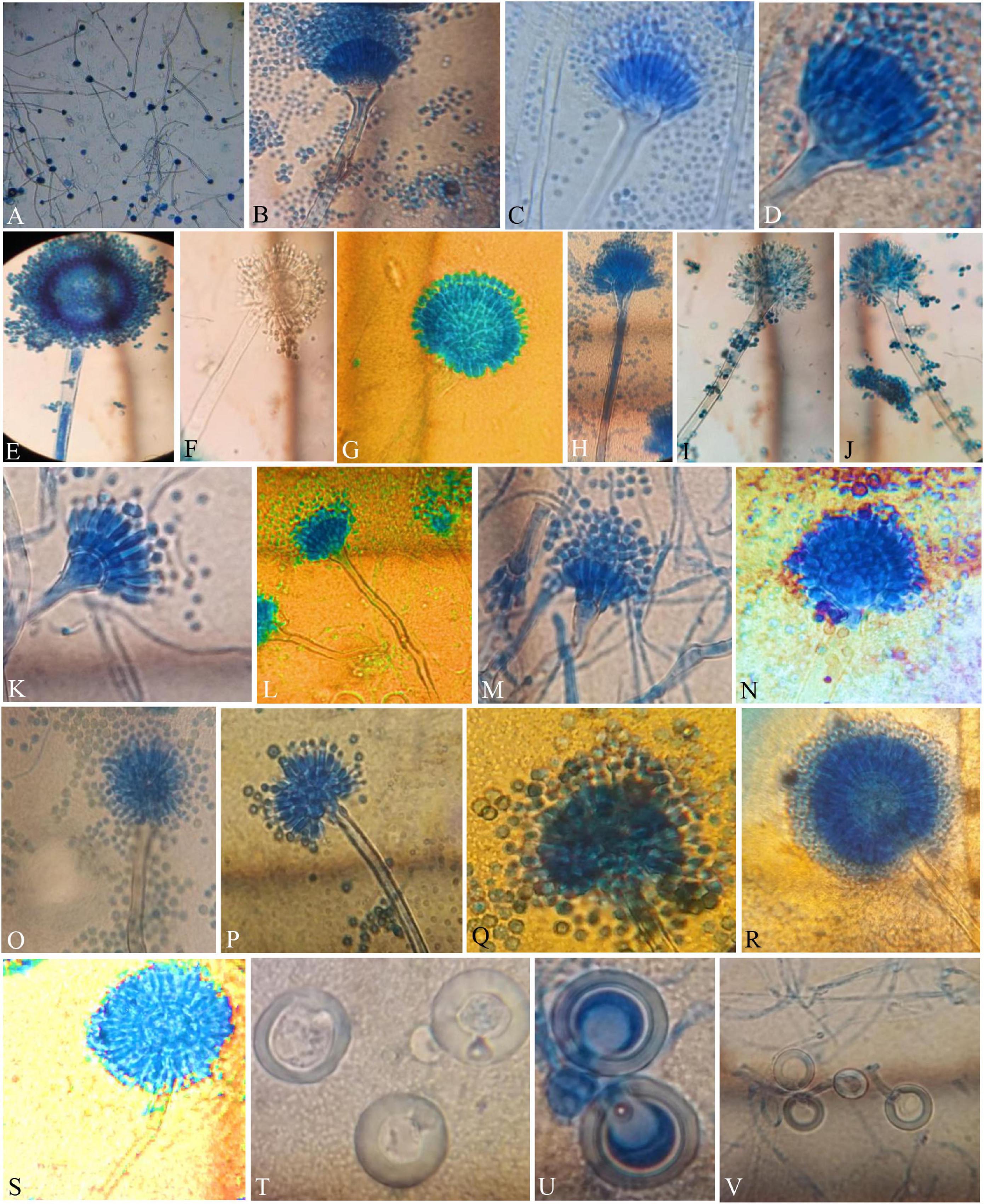
Figure 3. Compound Light Microscope (CLM) images of the hyphae, conidiophores, and Hülle cells, (A,B) hyphae and elongated conidiophores of A. aureoterreus; (C,D) enlarged conidiophores of A. terreus; (E,F) stained and unstained conidiophores of A. flavus; (G) enlarged conidial head of A. tamarii; (H–J) conidiophores with bottle-shaped vesicles, uniseriate sterigmata, and conidia of A. fumigatus; (K) enlarged conidiophores of A. nidulans; (L) conidiophores of E. quadrilineata; (M) conidiophores of A. latus; (N,O) A. awamori conidiophores; (P,Q) A. sydowii conidiophores; (R,S) Aspergillus species conidiophores, and (T–V) A. nidulans, E. quadrilineata, and A. latus Hülle cells, respectively.
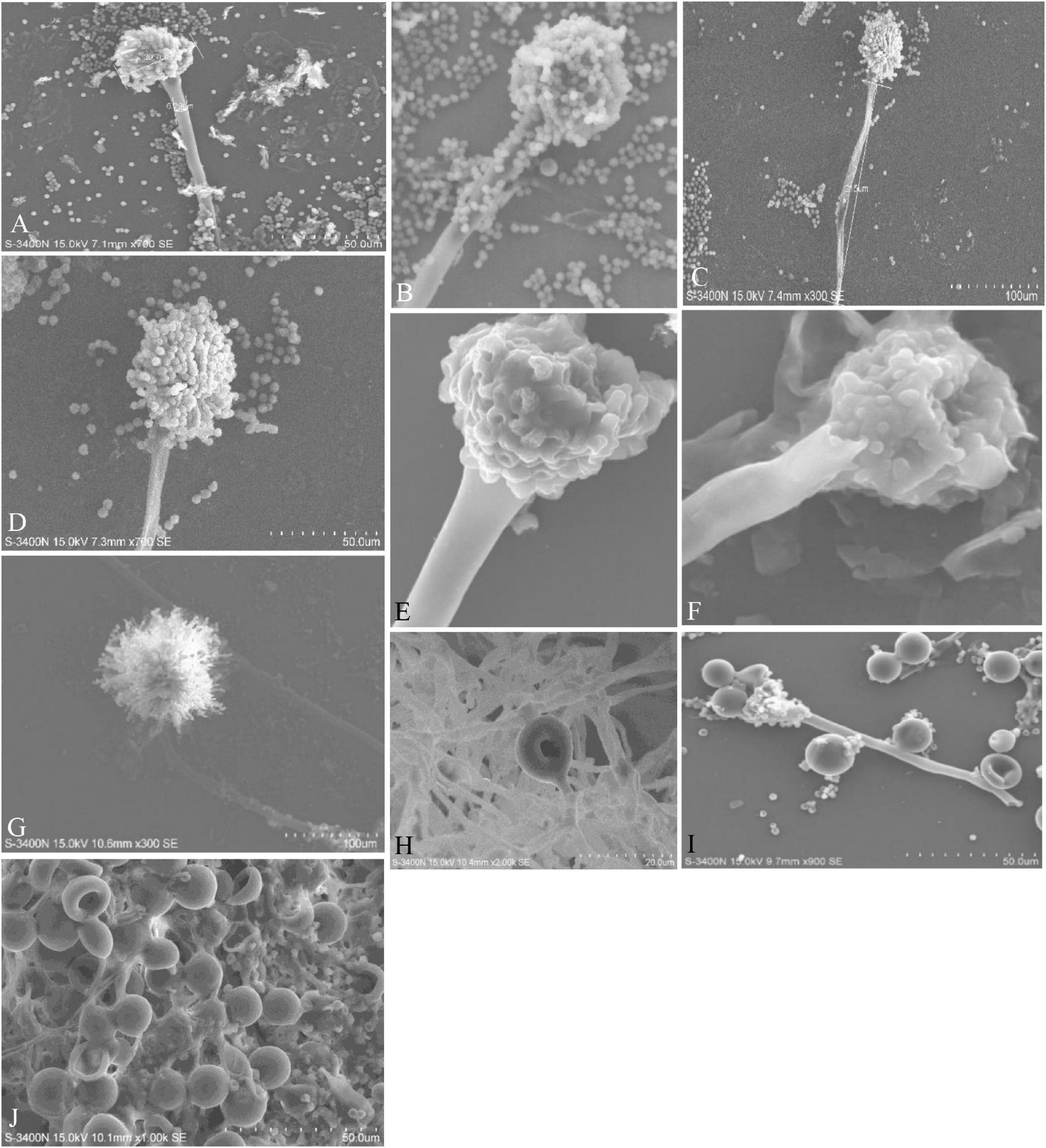
Figure 4. Conidiophores, vesicles, metulae, phialides, conidia, and Hülle cells of the strains are shown in Scanning Electron Microscope (SEM) images. (A) Conidiophores of A. aureoterreus; (B) conidiophores of A. terreus; (C,D) conidial head with chains of conidia of A. tamarii; (E) conidiophores of A. nidulans; (F) conidiophores of A. latus; (G) globose conidial heads, with metulae, phialides, chains of conidia and smooth conidiophores in A. awamori; (H,I) mycelium and Hülle cell development and Hülle cells of A. nidulans (J) Hülle cells formation in E. quadrilineata.
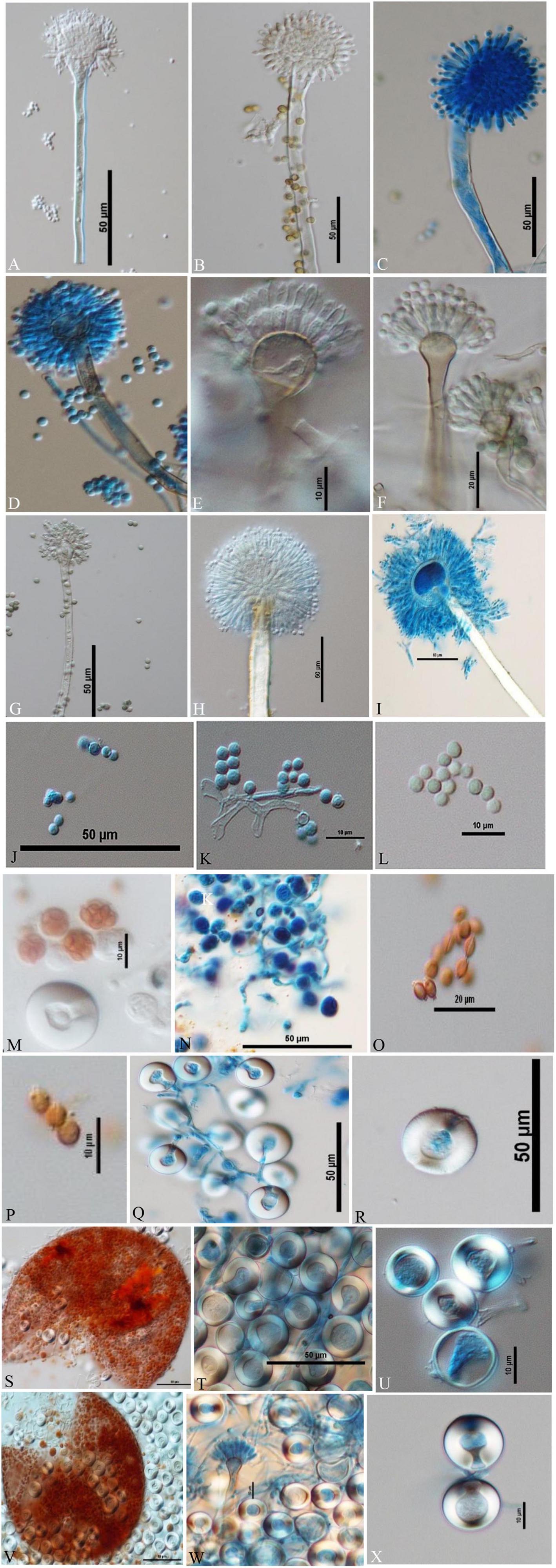
Figure 5. Conidiophores, conidia, asci, ascospores, cleistothecia, and H lle cells Differential Interference Contrast (DIC) images of Aspergilli (A) A. aureoterreus smooth, long, colourless conidiophores and columnar, biseriate conidial heads with hemispherical vesicles, metulae phialides, and conidia; (B,C) A. tamarii conidiophores containing globose, radiating and rough conidia, globose vesicles with uniseriate sterigmata; (D) A. nidulans smooth-walled, sinuate conidiophores and columnar conidial heads with small hemispherical vesicles, metulae, phialides, and conidia; (E) E. quadrilineata smooth, sinuate conidiophores and short, columnar conidial heads with hemispherical vesicles, metulae, phialides, and conidia; (F) A. latus smooth-walled, sinuate conidiophores and columnar conidial heads with small hemispherical vesicles, metulae, phialides, and conidia; (G) A. sydowii smooth, sinuous conidiophores and hemispherical conidial heads with globose to elliptical vesicles, metulae, phialides, and conidia; (H,I) Aspergillus species conidiophores with rough, pitted surfaces, globose conidial heads with globose, thinner vesicles, metulae, phialides, and conidia; (J–L) globose conidia of A. nidulans, E. quadrilineata, and A. latus, respectively; (M,N) asci of A. nidulans and A. latus; (O) lenticular ascospores of A. latus; (P) lenticular, smooth walled (unstained) ascospores of E. quadrilineata; (Q,R) Hülle cells of A. nidulans; (S–U) ruptured cleistothecium and Hülle cells of E. quadrilineata, and (V) in A. latus, ruptured cleistothecium showing asci, surrounded by Hülle cells, (W,X) Hülle cells separated from A. latus.
Phylogenetic Analyses
The PCR products of the ITS (Amplicon size: 600-bp) and β-tubulin (Amplicon size: 400-bp) genetic markers were analyzed (Figures 6A,B). Based on sequence analysis of ITS 1, ITS 4 and β-tubulin genes, eleven (11) monophyletic groups of Aspergilli have been well resolved into six Sections: Terrei, Flavi, Fumigati, Nidulantes, Nigri and Versicolores. The ITS marker, which is the most suitable, profoundly recommended and widely accepted fungal barcode (Schoch et al., 2012) served handy in the recognition, identification, and classification of species in their respective clades. Both ITS regions and β-tubulin gene phylogenetic trees are depicted in the phylograms. Data creation and post-sequencing of Aspergillus genes were examined, with results showing that the β-tubulin-based phylogenetic trees had a greater level of precision than those generated by the ITS regions. Analysis of β-tubulin-based phylogenetic trees for our isolates namely Aspergillus aureoterreus (Figure 7), Emericella quadrilineata (Figure 8), Aspergillus latus (Figure 9), and Aspergillus awamori (Figure 10) showed higher resolution. In addition, ITS-based phylogenetic trees for Aspergillus terreus, Aspergillus nidulans, Aspergillus sydowii, and Aspergillus sp., as well as β-tubulin-based phylogenetic trees for isolates of Aspergillus flavus, Aspergillus tamarii, and Aspergillus fumigatus, were created.
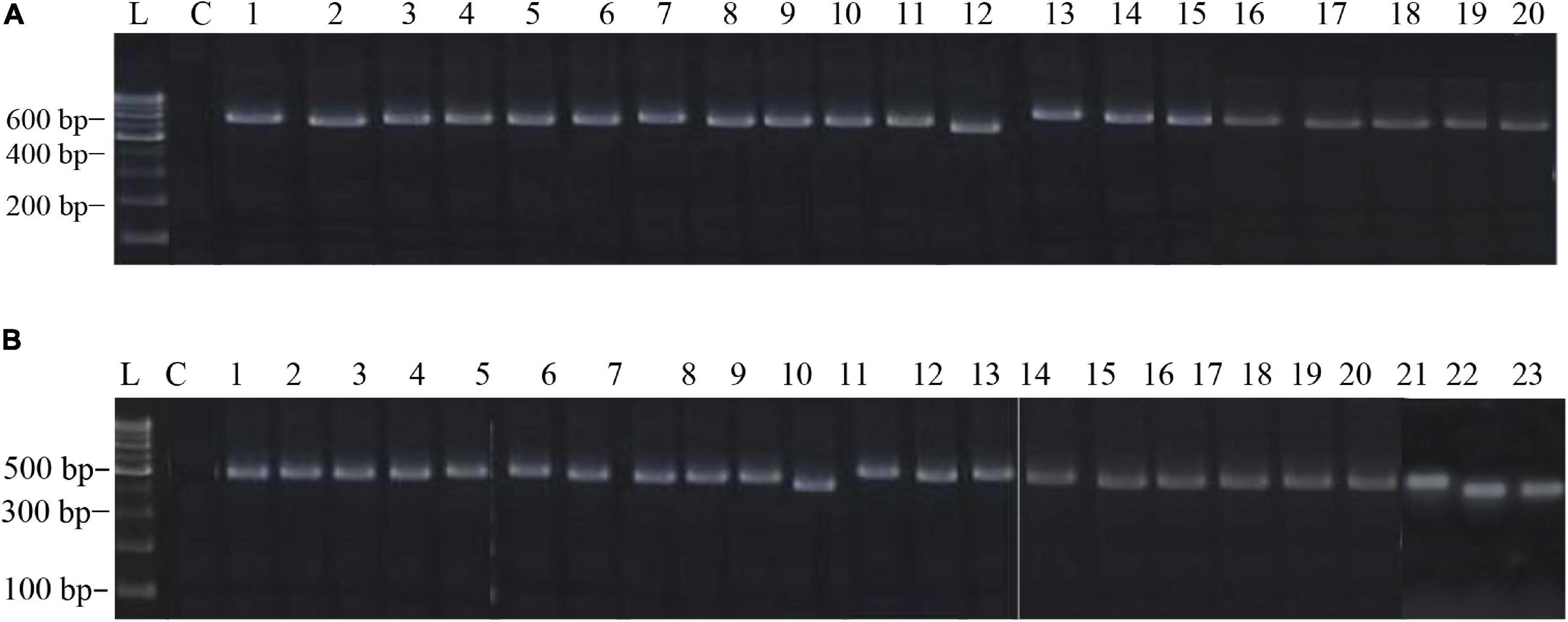
Figure 6. (A) The electrophoretogram shows the positive PCR amplification of Aspergillus ITS genes with species-specific amplicons. Here, L, 100-bp MBT049 HIMEDIA DNA ladder; C, PCR with a negative control; 1, 49-Aspergillus nidulans; 2, 58-Aspergillus nidulans; 3, 63-Aspergillus tamarii; 4, 31-Aspergillus fumigatus; 5, 37-Aspergillus fumigatus; 6, 45-Aspergillus nidulans; 7, 51-Aspergillus terreus; 8, 52-Aspergillus terreus; 9, 71-Aspergillus nidulans; 10, 75-Aspergillus tamarii; 11, 83-Aspergillus sydowii; 12, 17-Aspergillus miyajii; 13, 16-Aspergillus terreus; 14, 22-Aspergillus tamarii; 15, 29-Aspergillus sp.; 16, 4-Aspergillus quadrilineatus; 17, 2-Aspergillus rugulosus; 18, 1-Aspergillus terreus; 19, 11-Aspergillus nidulans; 20, 12-Aspergillus nidulans. (B) The electrophoretogram shows the positive PCR amplification of Aspergillus β-tubulin genes with species-specific amplicons. Here, L, 100-bp MBT049 HIMEDIA DNA ladder; C, PCR with a negative control; 1, 49-Emericella quadrilineata; 2, 58-Emericella quadrilineata; 3, 61-Aspergillus flavus; 4, 31-Aspergillus fumigatus; 5, 37-Aspergillus fumigatus; 6, 45-Aspergillus nidulans; 7, 51-Aspergillus aureoterreus; 8, 71-Aspergillus nidulans; 9, 75-Aspergillus tamarii; 10, 18-Aspergillus aureoterreus; 11, 17-Aspergillus nidulans; 12, 16-Aspergillus terreus; 13, 9-Aspergillus nidulans; 14, 24-Aspergillus fumigatus; 15, 22- Aspergillus tamarii; 16, 4-Aspergillus latus; 17, 2-Emericella nidulans; 18, 1-Aspergillus aureoterreus; 19, 11- Aspergillus latus; 20, 12-Aspergillus latus; 21, S8-Aspergillus awamori; 22, S24-Aspergillus awamori; 23, S44-Aspergillus tamarii.
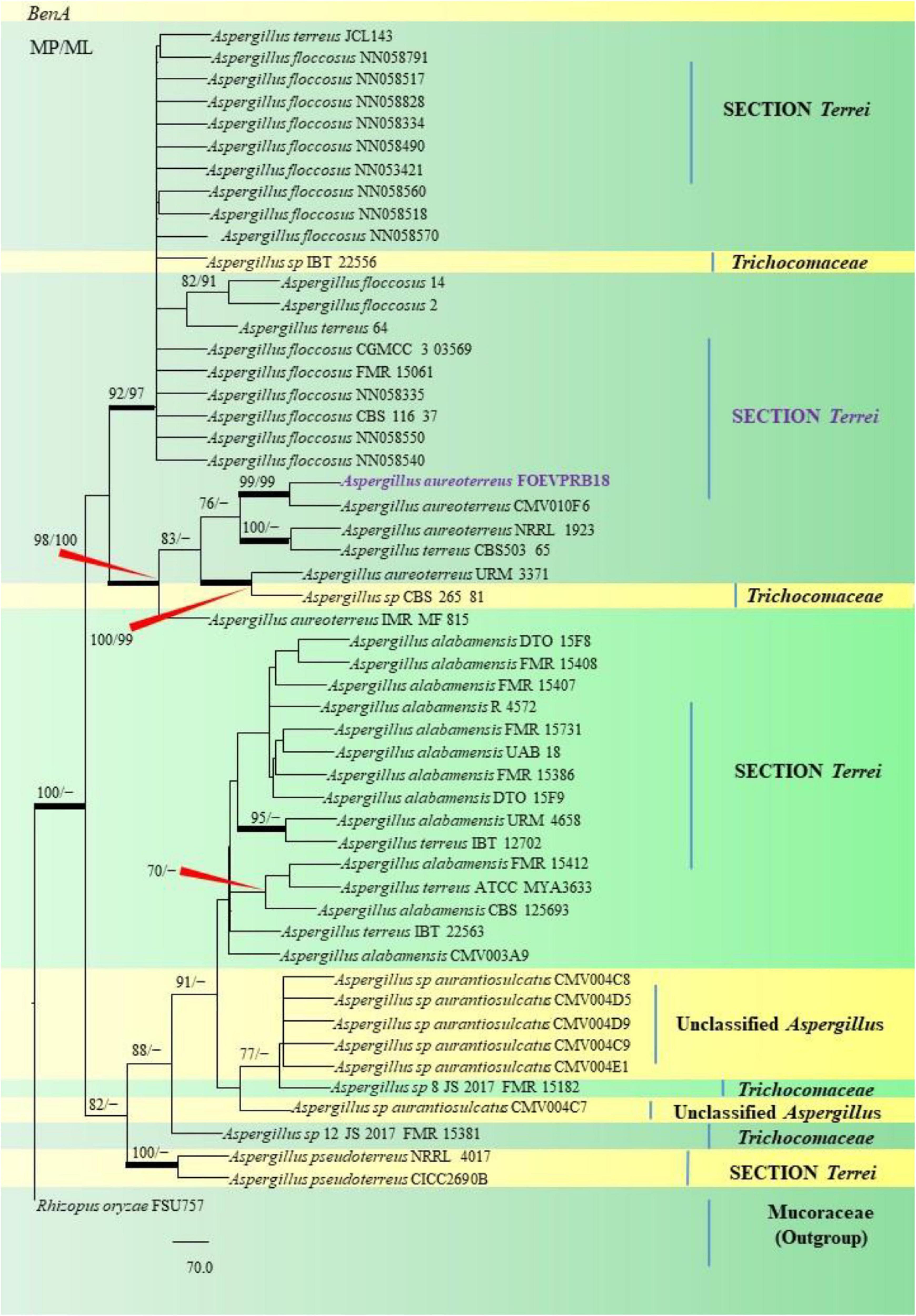
Figure 7. A phylogenetic tree (taxon branching diagram) presents the genetic analysis of the Aspergillus aureoterreus FOEVPRB18 clade belonging to the Section Terrei, exhibiting a maximum parsimony analysis of partial β-tubulin concatenated sequences, acquired. Bootstrap (bs) values ≥ 70% are designated at the nodes for maximum parsimony (MP) and maximum likelihood (ML) methods; branches with values ≥ 95% bs are bold black. A specific isolate code was mentioned as a suffix to the species name. Hyphen (-) represents the range of support values for parsimony and RAxML analyses of below 70%. Our isolate, which is marked in purple, was rooted using Rhizopus oryzae FSU757.
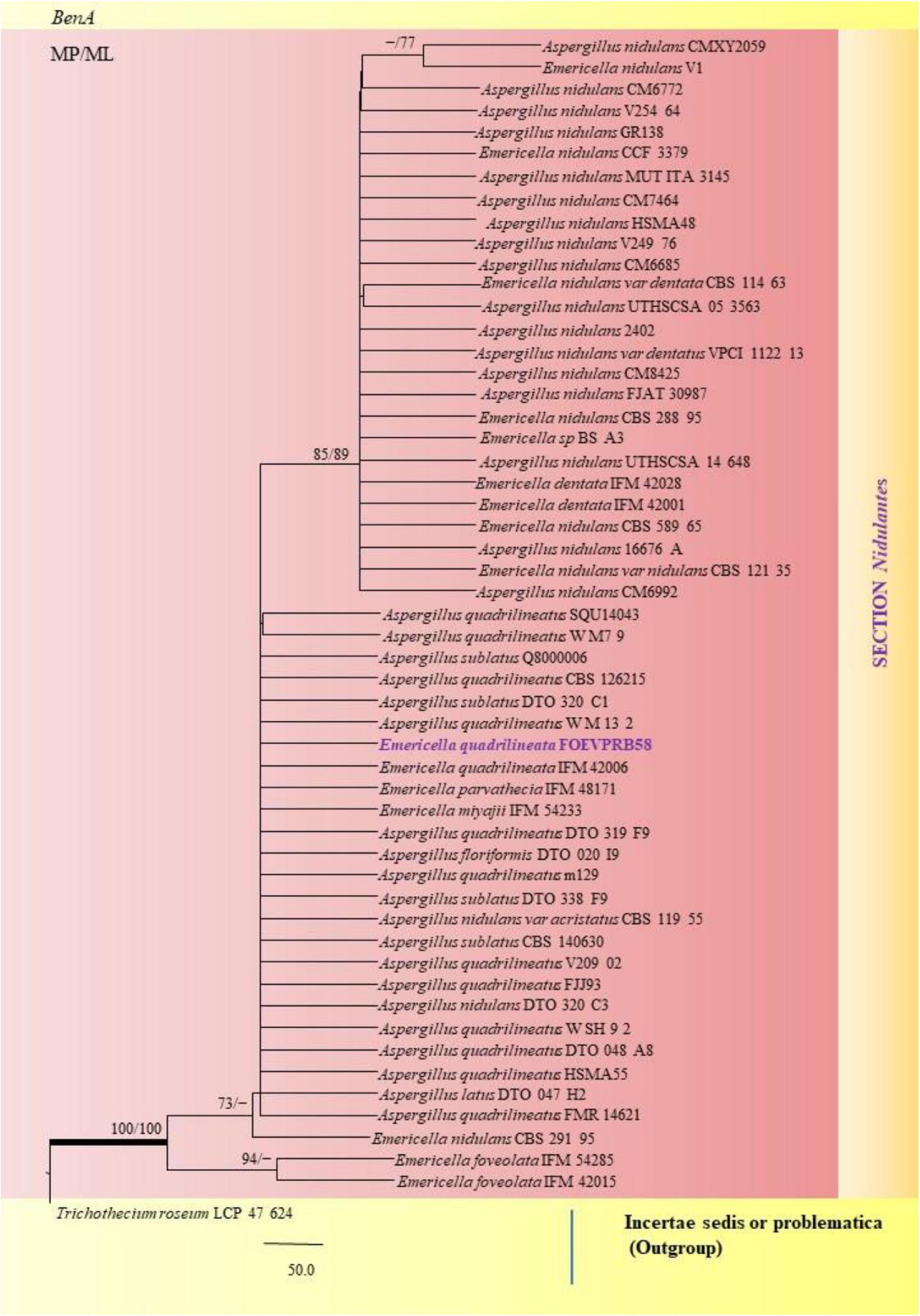
Figure 8. A phylogenetic tree (taxon branching diagram) which presents the genetic analysis of the Emericella quadrilineata FOEVPRB58 clade belonging to the Section Nidulantes, exhibiting a maximum parsimony analysis of partial β-tubulin concatenated sequences, acquired. Bootstrap (bs) values ≥ 70% are designated at the nodes for maximum parsimony (MP) and maximum likelihood (ML) methods; branches with values ≥ 95% bs are bold black. A specific isolate code was mentioned as a suffix to the species name. Hyphen (-) represents the range of support values for parsimony and RAxML analyses of below 70%. Our isolate, which is marked in purple, was rooted using Trichothecium roseum LCP 47 624.
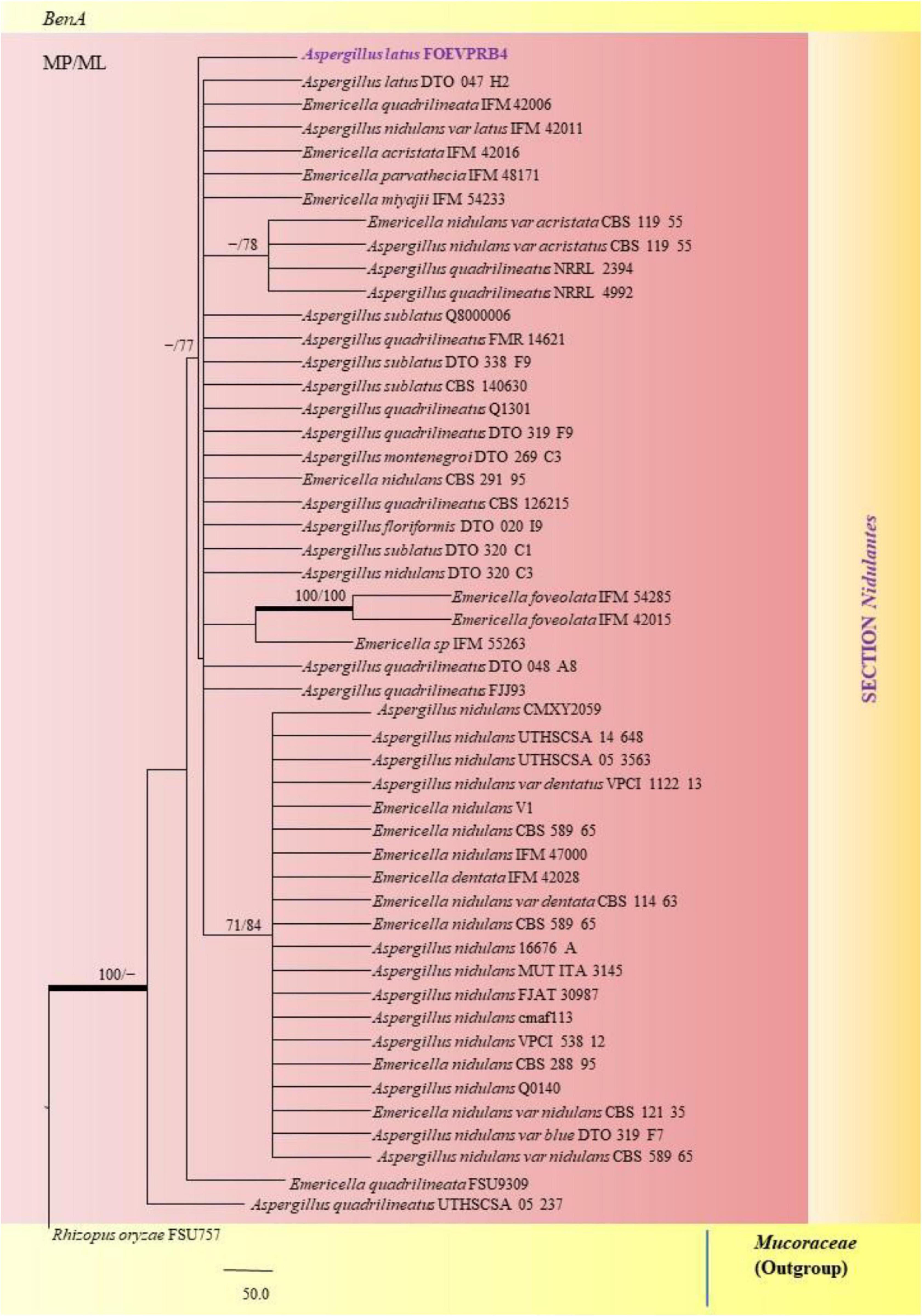
Figure 9. A phylogenetic tree (taxon branching diagram) which presents the genetic analysis of the Aspergillus latus FOEVPRB4 clade belonging to the Section Nidulantes, exhibiting a maximum parsimony analysis of partial β-tubulin concatenated sequences, acquired. Bootstrap (bs) values ≥ 70% are designated at the nodes for maximum parsimony (MP) and maximum likelihood (ML) methods; branches with values ≥ 95% bs are bold black. A specific isolate code was mentioned as a suffix to the species name. Hyphen (-) represents the range of support values for parsimony and RAxML analyses of below 70%. Our isolate, which is marked in purple, was rooted using Rhizopus oryzae FSU757.
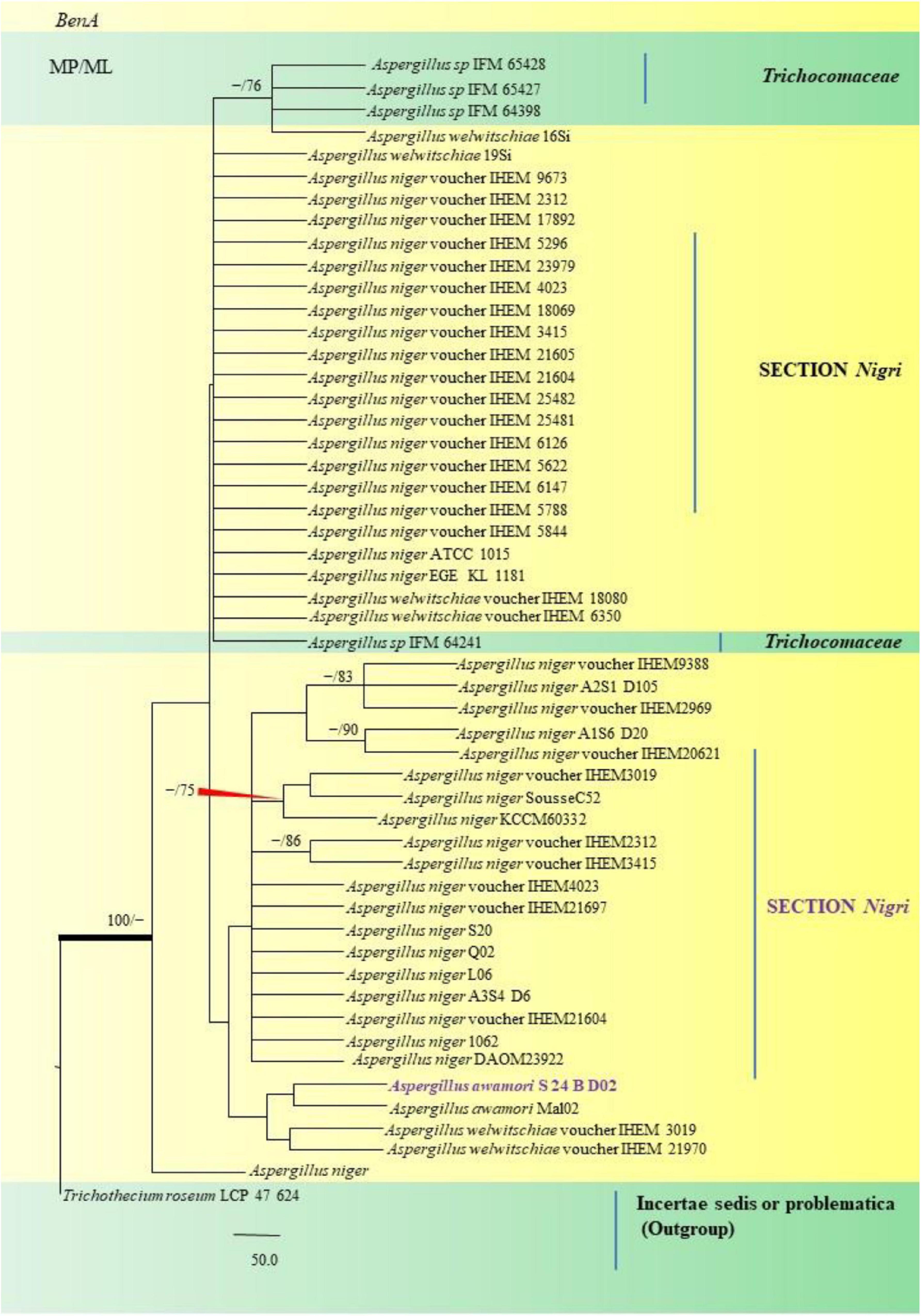
Figure 10. A phylogenetic tree (taxon branching diagram) which presents the genetic analysis of the Aspergillus awamori S 24 B D02 clade belonging to the Section Nigri, exhibiting a maximum parsimony analysis of partial β-tubulin concatenated sequences, acquired. Bootstrap (bs) values ≥ 70% are designated at the nodes for maximum parsimony (MP) and maximum likelihood (ML) methods; branches with values ≥ 95% bs are bold black. A specific isolate code was mentioned as a suffix to the species name. Hyphen (-) represents the range of support values for parsimony and RAxML analyses of below 70%. Our isolate, which is marked in purple, was rooted using Trichothecium roseum LCP 47 624.
Isolates studied in the present analysis, namely, A. aureoterreus FOEVPRB18 (Figure 7) was closely associated with the type strain A. aureoterreus CMV010F6 (MK451161), which has been deposited very recently at NCBI by researchers, while conducting studies to update the taxonomical status of Aspergillus species from South Africa (Visagie and Houbraken, 2020). The type strain A. terreus CVS503 65 (EU147717), was the most closely related species with our isolate (Figure 7), known to be a notorious fungus, playing a key role in human opportunistic pathogenesis, unpublished (Balajee et al., 2009). While A. aureoterreus CMV010F6 displayed 99.37% identity with our isolate A. aureoterreus FOEVPRB18, A. terreus CVS503 65 showed 96.32% identity.
Similarly, the clades A. flavus and A. tamarii were placed under the Flavi Section. A. flavus FOEVPRB61 occurred as a separate taxon although it was closely related to previously described clades of the same species (Figure 11). Partial ITS and β-tubulin gene sequences were used to arrive at species identity of the 27 Aspergillus isolates. Sequences from both the genetic markers gave almost the same identification. However, a doubtful analysis was experienced with Aspergillus isolate (ITS Barcode: MN294688; Section Unassigned), in determining a species name (Figure 12). Nevertheless, a detailed macro- and micro-morphological analysis of the unassigned fungal isolate is available. Although it emerged as a separate taxon during phylogenetic analysis, it showed 100% bs value (ML method) with Aspergillus sp. SS 30 1 studied by another research team from India, Direct Submission, Genetics, Agharkar Research Institute, Maharashtra, India (Chavan, 2020). This species was studied with Penicillium species clone 1 4 TSS as an outgroup. In most instances, our isolates were closely related to type strains which have been previously studied and described by providing appreciable support values.
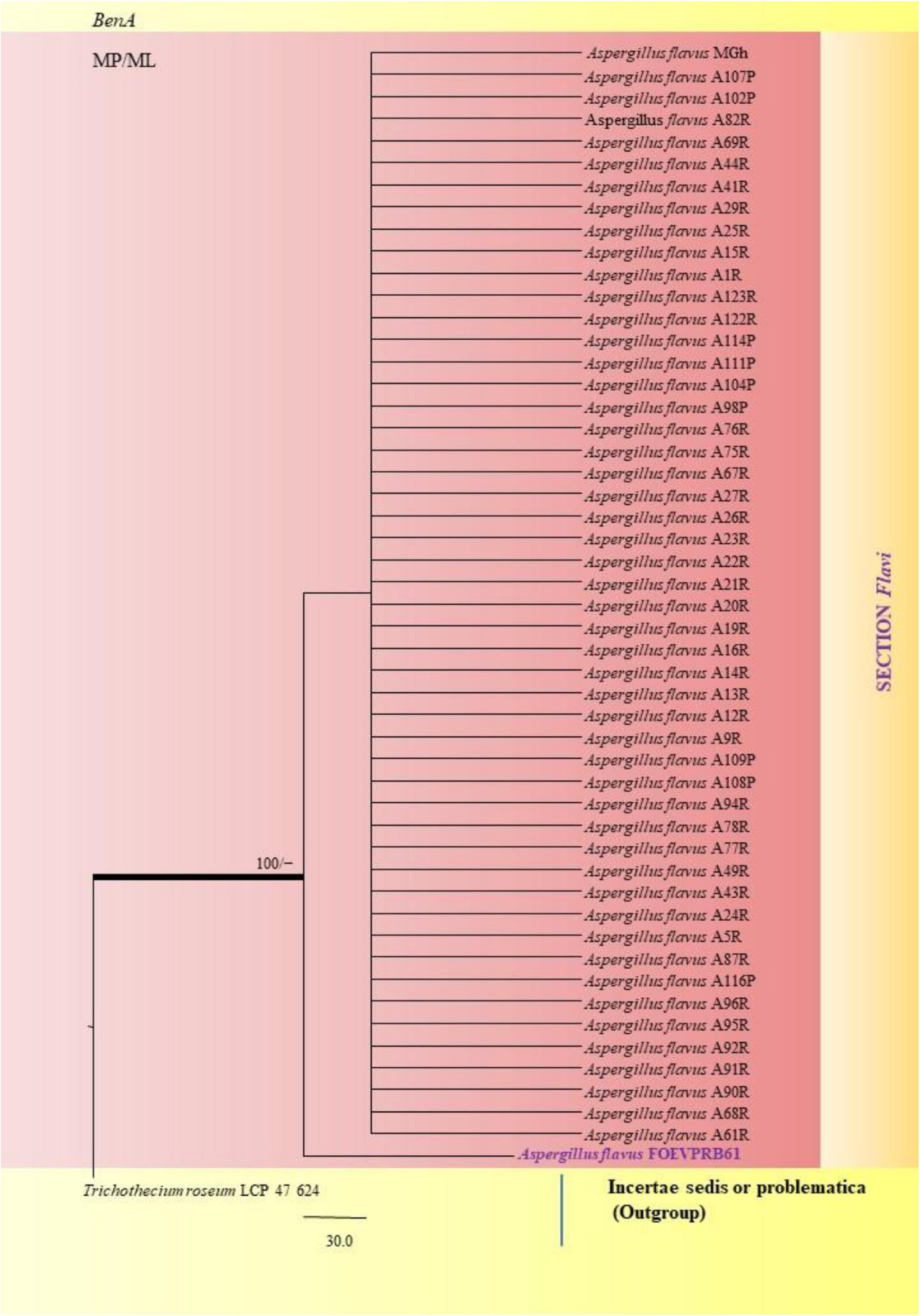
Figure 11. A phylogenetic tree (taxon branching diagram) which presents the genetic analysis of the Aspergillus flavus FOEVPRB61 clade belonging to the Section Flavi, exhibiting a maximum parsimony analysis of partial β-tubulin concatenated sequences, acquired. Bootstrap (bs) values ≥ 70% are designated at the nodes for maximum parsimony (MP) and maximum likelihood (ML) methods; branches with values ≥ 95% bs are bold black. A specific isolate code was mentioned as a suffix to the species name. Hyphen (-) represents the range of support values for parsimony and RAxML analyses of below 70%. Our isolate, which is marked in purple, was rooted using Trichothecium roseum LCP 47 624.
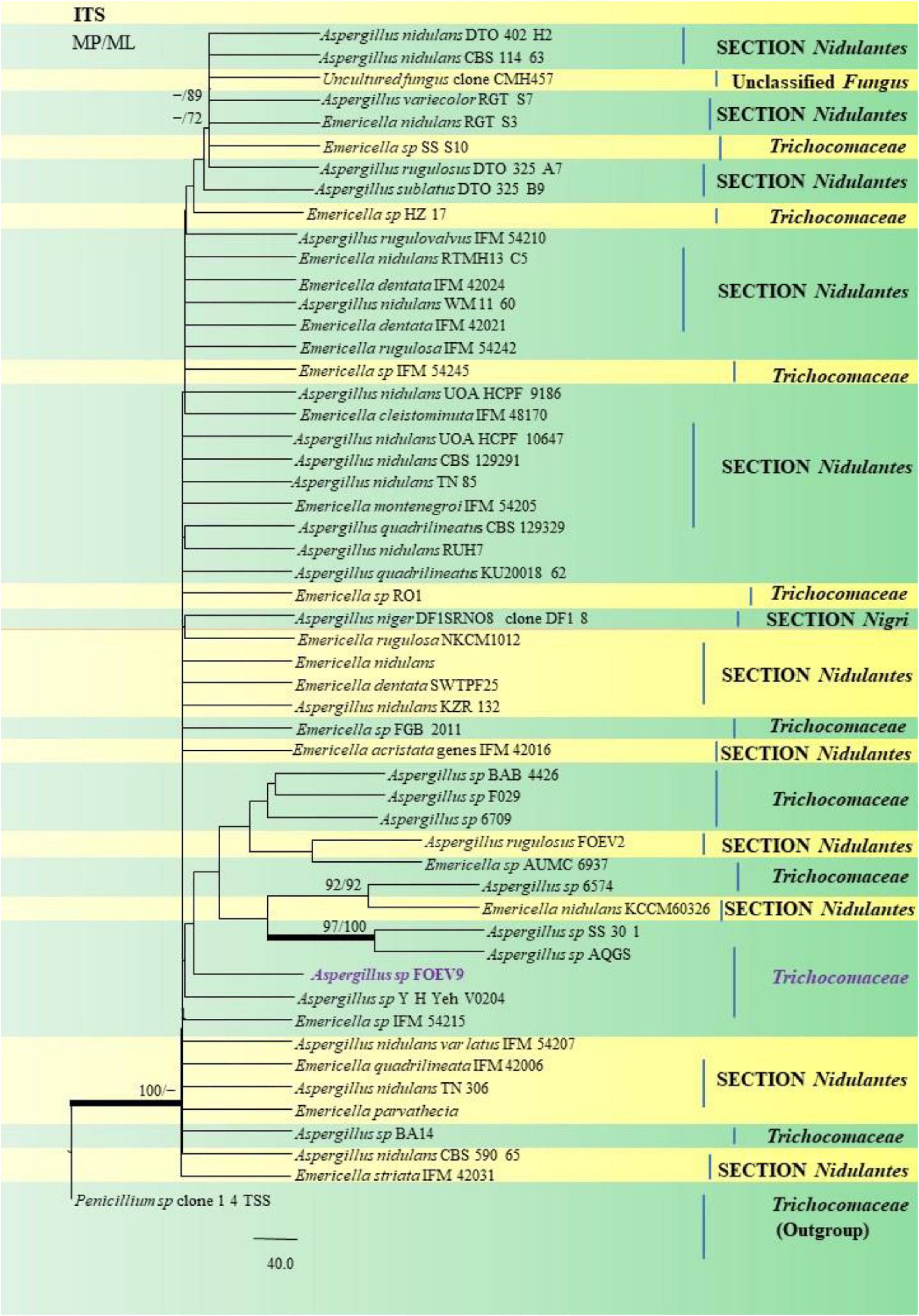
Figure 12. A phylogenetic tree (taxon branching diagram) which presents the genetic analysis of the Aspergillus sp. FOEV9 clade belonging to the family Trichocomaceae, exhibiting a maximum parsimony analysis of partial ITS 1 and ITS 4 concatenated sequences, acquired. Bootstrap (bs) values ≥ 70% are designated at the nodes for maximum parsimony (MP) and maximum likelihood (ML) methods; branches with values ≥ 95% bs are bold black. A specific isolate code was mentioned as a suffix to the species name. Hyphen (-) represents the range of support values for parsimony and RAxML analyses of below 70%. Our isolate, which is marked in purple, was rooted using Penicillium species clone 1 4 TSS.
In the Section Fumigati our isolate namely A. fumigatus FOEVPRB31 (Figure 13) was found to be a close relative of A. fumigatus 3, Direct Submission, Department of Dermatology, Nanjing University, China (Chen et al., 2018) and A. fumigatus CMXY2075, Direct Submission, Ecology of Clinical Fungi, Fungal Biodiversity Institute, Netherlands (Chen and Xu, 2018).
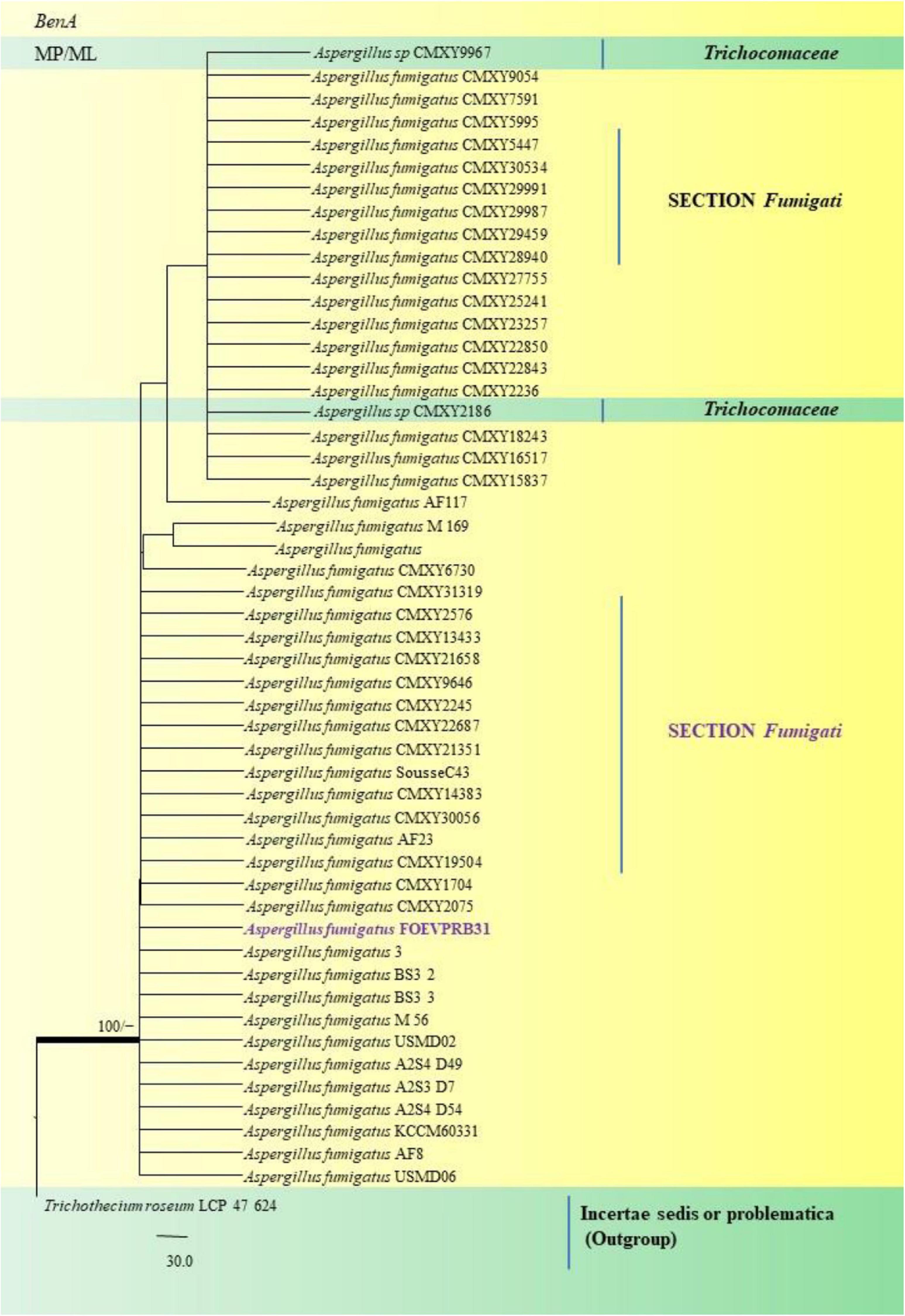
Figure 13. A phylogenetic tree (taxon branching diagram) which presents the genetic analysis of the Aspergillus fumigatusFOEVPRB31 clade belonging to the Section Fumigati exhibiting a maximum parsimony analysis of partial β-tubulin concatenated sequences, acquired. Bootstrap (bs) values ≥ 70% are designated at the nodes for maximum parsimony (MP) and maximum likelihood (ML) methods; branches with values ≥ 95% bs are bold black. A specific isolate code was mentioned as a suffix to the species name. Hyphen (-) represents the range of support values for parsimony and RAxML analyses of below 70%. Our isolate, which is marked in purple, was rooted using Trichothecium roseumLCP 47 624.
A. nidulans, A. quadrilineatus and A. latus were conveniently assigned in their clades under the Nidulantes Section. A. nidulans is phenotypically very similar to A. quadrilineatus except for differences in ascospore morphology (Chen et al., 2016). With respect to their phylogeny too they were situated in closely related clades, emerging from that of E. nidulans which is the sexual morph of A. nidulans (Figure 14). The sexual stages were appreciably documented in our isolate also, as realized by micro-morphological analyses. However, the concept of naming Aspergillus species with their sexual morphs is gradually becoming obsolete, due to major decisions taken at the meeting of the International Commission for Penicillium and Aspergillus (ICPA) in 2012.6 A. quadrilineatus and A. latus, which shared 99.76% identity with the type strain Emericella quadrilineata IFM 42006 (AB248335.1) and 99.55% identity with the type strain DTO 047 H2 (KU866810.1) respectively, were classified into well-defined clades with A. quadrilineatus, which formed discrete and conspicuous sexual stages and therefore is referred to as Emericella quadrilineata (Teleomorph). This was also in support of data generated through phylogenetic analysis of β-tubulin gene (Figure 8) being closely related to Emericella quadrilineata IFM 42006 (AB248335.1) studied previously (Matsuzawa et al., 2006). A. latus was found to emerge as a separate taxon (Figure 9) from A. quadrilineatus DTO 048 A8, Direct Submission, CBS-KNAW Fungal Biodiversity Centre (Chen et al., 2016) and displayed 100% bs value with Emericella foveolata IFM 54285 and E. foveolata IFM 42015 (Matsuzawa et al., 2006), which has also been studied under the Section Nidulantes while discerning evolutionary relatedness.
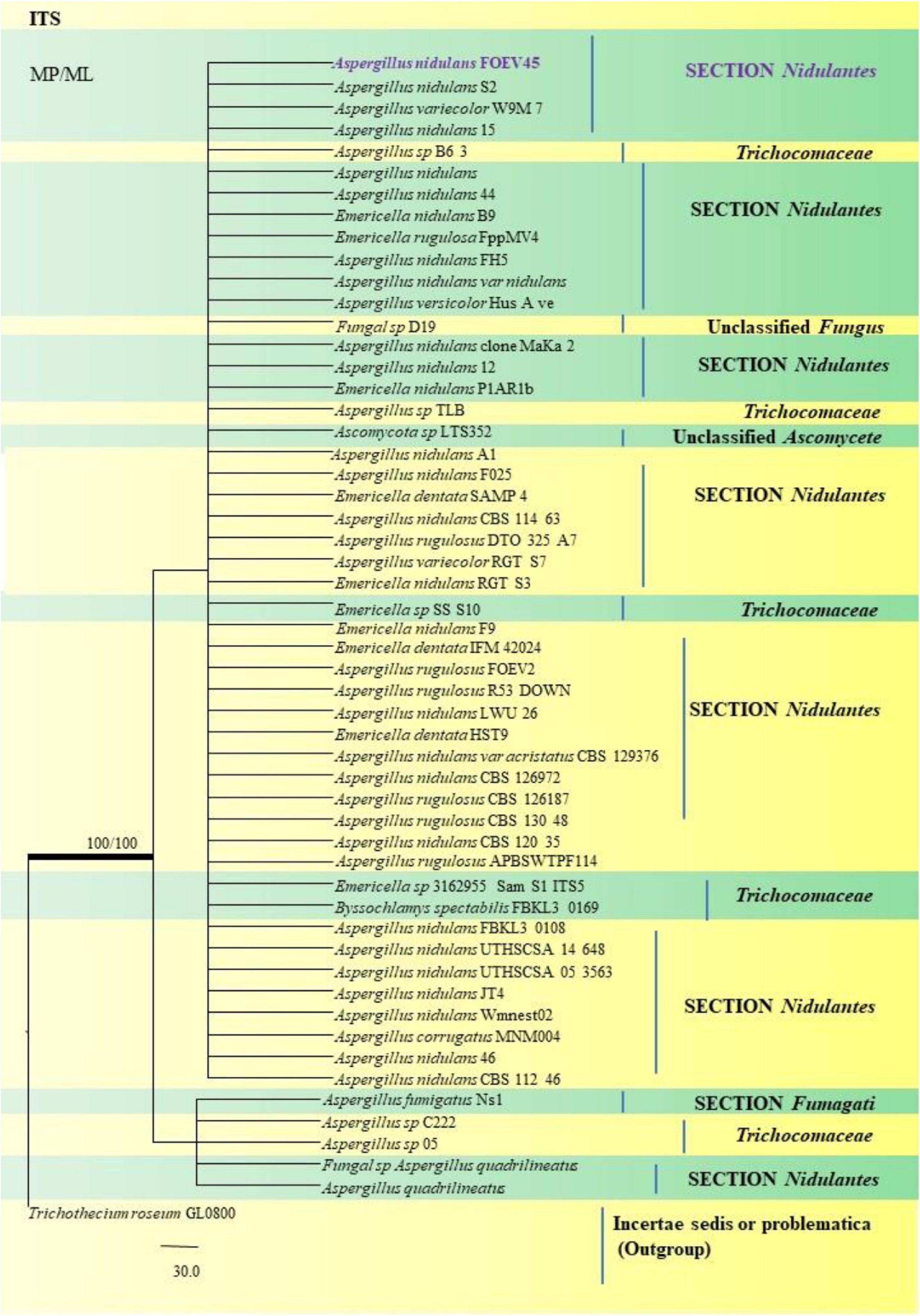
Figure 14. A phylogenetic tree (taxon branching diagram) which presents the genetic analysis of the Aspergillus nidulans FOEV45 clade belonging to the Section Nidulantes, exhibiting a maximum parsimony analysis of partial ITS 1 and ITS 4 concatenated sequences, acquired. Bootstrap (bs) values ≥ 70% are designated at the nodes for maximum parsimony (MP) and maximum likelihood (ML) methods; branches with values ≥ 95% bs are bold black. A specific isolate code was mentioned as a suffix to the species name. Hyphen (-) represents the range of support values for parsimony and RAxML analyses of below 70%. Our isolate, which is marked in purple, was rooted using Trichothecium roseum GL0800.
Aspergillus awamori S24 BD02 isolated from the fennel samples diverged as a clade closely related to A. awamori Mal02, Direct Submission, Department of Biotechnology, University of Verona, Italy (Andreolli et al., 2018), which in turn were related to A. niger studied collectively as black Aspergilli. Only one isolate namely A. sydowii FOEV83 was studied under the Section Versicolores. Studies inferred by phylogenetic analyses showed its close relatedness to A. sydowii DUCC5715, Direct Submission, Department of Microbiology, Dankook University, South Korea (Ahn et al., 2020), and a 100% bs value with A. versicolor M4 C4, Direct Submission, Postgraduate in Biological Sciences, University of Nayarit, Mexico (Bobadilla-Carrillo et al., 2020). This was especially true with cultural and morphological studies, where A. sydowii FOEV83 appeared very similar to A. versicolor by the characteristic pigmentation.
Single Nucleotide Polymorphisms
In order to study the variants in the β-tubulin genes, the sequences obtained from Sanger sequencing were aligned to the reference genome sequences of respective species. The alignment was carried out using BLASTn algorithm. The variants such as single nucleotide variants, substitutions, insertions, and deletions were identified manually. Based on the regions of coding sequence (CDS) of β-tubulin gene provided at the protein feature annotations, the effect of these mutations at protein level were manually evaluated. The site of variations at the genome and protein level are tabulated (Table 2). Once these sequences for each species had been aligned, we observed that they all share a high degree of identity and displayed few variants with the exceptions of MN791095, MN791096, MN791109, and MN791093 sequences. We found that five species MN791107: A. nidulans 4, MN791105: E. quadrilineata 2, MN791112: A. latus 3, MN791108: A. tamarii 2, and MN791099: A. fumigatus 1 did not exhibit variations with regard to the reference genome in their respective sections. Hence, these five species have been excluded (Table 2). Our SNP analysis showed a high number of intronic variances and protein coding synonymous variants (changes without coding sequences) that result in synonymous mutations. On the contrary, four non-synonymous (substituting amino acids with new ones) variants were identified- MN791102: A. nidulans 3, MN791116: E. nidulans 5, MN791104: E. quadrilineata 1, and MN791110: A. latus 1, all contain single nucleotide variations (SNVs) that alter protein sequences whose functional consequences has not been studied.
The SNP profiling analyses revealed five mis-sense mutations in the CDS (coding DNA sequence) regions of beta-tubulin gene in four Aspergillus isolates. Histidine was replaced by tyrosine at the 105th position in A. nidulans MN791102, leucine to proline and threonine to serine at positions 30 and 107, respectively, in E. nidulans MN791116. In E. quadrilineata MN791104 leucine was replaced to proline at the 10th position. Furthermore, analysis of A. latus MN791110 indicated that the amino acid asparagine was substituted with isoleucine at the 87th position.
Discussion
Spices occupy a very prominent role in the culinary preparations in India (Siruguri and Bhat, 2015). Based on the limited information and scientific evidence available through literature, fennel continues to be the most widely used herb in traditional medicine. F. vulgare is known to exhibit an array of pharmacological properties such as antimicrobial, antiviral, anti-inflammatory, anti-mutagenic, antipyretic, anti-spasmodic, anti-thrombotic, apoptotic, cardiovascular, chemomodulatory, anti-tumor, hepatoprotective, and hypoglycemic. Also, they’ve been linked to memory enhancement (Badgujar et al., 2014), which implies that they could be useful in the field of pharmaceutical biology for the development of new drugs to treat a wide range of ailments.
The use of spices in food has been practiced since time immemorial. Black pepper, cardamom, clove, coriander, cumin, ginger and other spices have been explored for their intrinsic antibacterial effects when used in foods, in addition to the flavors and aromas that they produce. In spite of the antibacterial compounds that they harbor, they are vulnerable to mycological deterioration, constantly being colonized by mycoflora like the Aspergilli, Penicillia, and Fusaria during pre- and post-harvest operations at the field.
Accurate identification of Aspergillus isolates up to the species level has become ecologically, epidemiologically, and pathologically significant because of the extensive damage that they cause in food crops (Battilani et al., 2016; Alshannaq and Yu, 2017), feed crops (Ráduly et al., 2020), and spices (Ali et al., 2015; Ssepuuya et al., 2018). Specific studies focused on mycological analyses have been conducted by research groups on spices like cumin, clove, black pepper, ginger, cardamom, coriander and cinnamon with dominancy of Aspergillus flavus and A. niger, with cumin being the most contaminated sample (Elshafie et al., 2002). Notable contamination by A. flavus, A. fumigatus, A. alutaceus, A. niger, and A. sulphureus has been observed in yet another study conducted using aniseed, rosemary and spice products (Ahene et al., 2011). Another research group (Hammami et al., 2014) examined fourteen distinct spice samples and reported the highest fungal contamination by Aspergillus flavus, A. nomius, and A. niger. The studies conducted using fennel seeds from India (Kulshrestha et al., 2014) have been sparse, with reports on Aspergillus contamination being investigated from fennel seeds, cumin and coriander seeds which are extensively used in culinary preparations in India. Two other research studies (Azzoune et al., 2015; Garcia et al., 2018) have been conducted in order to evaluate the mycotoxigenic potential of select Aspergilli, from nearly about two hundred spice samples of rosemary, fennel, cinnamon, clove, pepperoni, black and white pepper and oregano. Our study is the first of its kind to reveal a significant degree of Aspergillus contamination in the Indian subcontinent, with A. niger being the most frequently isolated species, followed by Mycelia sterilia, A. flavus, Mucor species, Penicillium species, and Yeasts, among others.
The Aspergilli are an all-pervasive and species substantial genus, containing more than three hundred thread-like filiform fungi (Anonymous, 2021). By far, the majority of Aspergilli from F. vulgare were found in the Nigri section in this study, which was collected from distinct geographical locations across India, including Delhi, Pondicherry, Rajasthan, Uttar Pradesh, and West Bengal. Isolates studied here, are predominantly predisposed with a toxigenic potential, known to cause a wide range of human and animal diseases, in addition to being plant pathogens.
Two Aspergilli, namely Aspergillus aureoterreus and A. terreus, were isolated from fennel samples classified under the Terrei Section. A. aureoterreus differs from Aspergillus terreus by the sequence analyses of the regions of the beta-tubulin genes (Samson et al., 2011). In the phylogenetic studies, the A. aureoterreus species formed a distinct lineage different from the A. terreus clade (Samson et al., 2011). The type-strain was isolated from Foeniculum vulgare (Sample No.18, place-Midnapore, West Bengal) and has been deposited in the National Center for Biotechnology Information (NCBI) GenBank with Accession No. MN791096.
A. sydowii has also been recognized as Emericella sydowii, a soil fungus, that could adulterate foodstuffs. It is also a pathogen known to cause human diseases like aspergillosis, onychomycosis, and keratomycosis (de Hoog et al., 2000). Apart from its disease-causing and toxigenic potential, A. sydowii has been the source of anticancer alkaloids. This species generates alkaloids, namely, 6-methoxyspirotryprostatin B, 18-oxotryprostatin A and 14-hydroxyterezine D (Zhang et al., 2008) and have potential cytotoxic activity against a549 cells (Nadumane et al., 2016).
Aspergillus flavus has received continual attention ever since its aflatoxigenic potential was discovered. The mold placed under the Flavi section, is known to be air-borne, thermotolerant and popularly as a saprophytic soil fungus. The fungus has been found all over the world (Ramírez-Camejo et al., 2012) and is abundant in temperate soil (Thom and Raper, 1945). A. flavus was found to be the most frequently occurring species in the fennel samples analyzed in the present study after A. niger and Mycelia sterilia, is known to be highly infectious with a broad range of infectivity and is able to contaminate agricultural crops during pre-harvest and post-harvest procedures (Bignell, 2010). In certain regions in Africa and Asia, AFs are appraised to be a major cause of severe acute sickness and loss of several lives per year (Wood, 1992; Alshannaq and Yu, 2017). The fungus is the most common source of aflatoxins in contaminated agricultural crops, can grow well in order to release aflatoxins frequently in almost any crop seed. The aflatoxins (AFs) are highly carcinogenic and immunosuppressive in nature, that has threatened global food safety, and has proven to be hazardous to animals, insects, and humans. It is an opportunistic fungal pathogen to humans as well as animals causing aspergillosis in immune-compromised hosts. In mammals, the pathogen can induce cancer of the liver (Amaike and Keller, 2011). Globally, over 4.5 billion people have been exposed to unmonitored levels of AFs (Alshannaq et al., 2018). Aflatoxin B1 (AFB1) is a highly toxic and the most potent natural compound to cause hepatocellular carcinoma in the universe. AFs are severely toxic, mutagenic, carcinogenic, teratogenic, immunosuppressants and are classified as Group1 carcinogens in human beings (Ostry et al., 2017) and hence are of special concern to human, animal and poultry health. A. tamarii is an epidemiological agent of human mycotic keratitis in India (Homa et al., 2019). Although it does not produce aflatoxins, it can induce elaboration of other toxic compounds like cyclopiazonic acid (Dorner et al., 1983; Ito, 1998), hence, posing a threat to human health.
Aspergillus fumigatus was studied under the Fumigati section and was isolated from fennel samples. With regard to the formation of sexual structures, sclerotia were not produced in the isolates studied by us. A. fumigatus has long been thought of reproduce only by asexual means until a detailed study was published by two researchers (Dyer and Paoletti, 2005). They elaborated on the possibility of the occurrence of sexual reproduction in A. fumigatus. Also, the detailed investigations and opinions of several mycologists have led to the findings, that point toward evidence that sexual reproduction may occur within populations of A. fumigatus. Teleomorphs of A. fumigatus are yet to be detected. However, it would be very significant to note that, the presence of sexual cycles would lead to an impending understanding of their biology and life cycles (Dyer and Paoletti, 2005). Other perspectives to understanding this species is its role as a human pathogen. A. fumigatus has been the highest prevailing aerial fungal parasite in immunodeficient hosts and is known to be associated with lethal systemic septicemia in multiple organs. It causes incursive infection in the lungs or long-term infection affecting the lungs, hypersensitive bronchopneumonia aspergillosis or allergic disorders in immune deficiencies (Hohl and Feldmesser, 2007; Segal, 2009). The disease can spread especially in individuals suffering from certain types of leukemia (Parahym et al., 2014) and those who are at the advanced stages of AIDS (Kaur et al., 2017), ailing with bronchial asthma (Kosmidis and Denning, 2015), fibrocystic disease of the pancreas (Düesberg et al., 2020), and in patients who have recovered from an influenza infection (Van De Veerdonk et al., 2017).
A. nidulans is a deuteromycetous member, which falls under the category of polyphyletic group of fungi whose sexual stages (teleomorph) are rare or unknown. In spite of this, mutants of A. nidulans are known to produce copious amounts of Hülle cells in the absence of sexual stages (Dyer and O’Gorman, 2012). A large number of species in the section Nidulantes have a sexual state whose name is Emericella, in the dual nomenclature system of fungi. If sexual stages are known and observed, the teleomorph has taxonomic precedence over the asexual stage, generally referred to as anamorph. Therefore, this species is most properly known as Emericella nidulans. Nevertheless, this fungal species will continue to be known in literature and the scientific or commercial industry for its inclusiveness as A. nidulans, which may be used here for both the anamorph and teleomorph interconvertibly. Asci contained eight-spored cells ascospores which were reddish/cherry-brown and lens-shaped and correlated with the original description by Thom and Raper (1945). A detailed microscopic description is available (Chen et al., 2016) for ascospore morphology of Aspergilli of the Nidulantes section. The ascospore color, ornamentation, shape and size are of particular value for differentiating species.
A. latus and A. quadrilineatus are being reported from F. vulgare for the very first time. Also, A. latus has been found to be phylogenetically similar to A. sublatus and A. montenegroi, thus considered synonymous. Ascospore morphology, which is a major and distinguishing feature while studying the sexual stages of different Aspergilli, were also documented in the current micromorphological analyses and were found to be orange/reddish brown with two pleated equatorial crests as studied recently and reported by Chen et al. (2016). The ascospores usually have two equatorial crests, whereas four crests can be seen in A. quadrilineatus by the aid of Scanning Electron Microscopy (SEM) and can sometimes be low or inconspicuous (Hubka et al., 2016). It has also been opined by Chen et al. (2016) that A. nidulans resembles A. quadrilineatus and is distinguishable in terms of its ascospore morphology. Phylogenetically A. miyajii has been found to be identical with A. quadrilineatus, A. parvathecius A. acristatus, and A. floriformis (Chen et al., 2016). A. awamori is being reported for the first and foremost time in fennel samples and is regarded as a domesticated form of A. niger (Samson et al., 2006) and its economic importance in Koji fermentation has found to be noteworthy (Kitamoto, 2002).
Nevertheless, the fungal phylogeny of Aspergillus species has undergone radical modifications over the course of over a hundred years. Phylogenetic analysis was especially useful when it was used to authenticate our morphological identification. In most instances, morphological identification and phylogenetic analyses were in agreement and helped in ascertaining species identity in A. awamori from the Nigri Section because of the striking phenotypic similarities to A. niger exhibited by our isolates. Phylogenetic studies were particularly useful while studying A. aureoterreus, A. latus and Emericella quadrilineata from Sections Terrei, and Nidulantes, respectively, along with A. awamori, which are being distinctly reported for the first time in F. vulgare. As a result, we were able to successfully distinguish them from closely related species and identify their Sections using the two genetic markers, which was precisely what we set out to do when this research was conceived.
Through phylogenetic analysis and tree construction, we were able to study twenty-two (22) Aspergillus species for the SNP analysis (Figure 15), classifying them into the Sections Nidulantes, Flavi, Nigri, Terrei, and Fumigati. Under the Nidulantes section, we could classify two A. latus (Isolate Nos. 1 and 2), five A. nidulans (Isolate Nos. 1, 2, 3, 4 and 5) and two E. quadrilineata (E. quadrilineata Isolate No. 1 and E. quadrilineata Isolate No. 2). Thus, the species of E. quadrilineata (Isolate No. 1) is rather closely connected to A. latus (Isolate No. 1) on one end, and A. nidulans (Isolate No. 3) is closely related to E. quadrilineata (Isolate No. 2) on the other end. F. vulgare samples from Pondicherry showed the presence of E. quadrilineata (Isolate No. 1) and A. latus (Isolate No. 1), and A. nidulans (Isolate No. 3) and E. quadrilineata (Isolate No. 2) were obtained from Rajasthan. While speaking about the diversity of fungi in the F. vulgare samples, three isolates of A. latus were studied: A. latus (Isolate Nos. 1 and 2) were obtained from Pondicherry, while A. latus (Isolate No. 3) was isolated from Jhargram, West Bengal, India. In the case of the occurrence of A. nidulans species, A. nidulans (Isolate Nos. 1, 2, 4, and 5) were from Pondicherry, whereas A. nidulans (Isolate No. 3) had been obtained from Rajasthan. This phylogenetic tree (Figure 15) of Aspergilli from F. vulgare samples from several regions demonstrate how closely related strains are distributed geographically. Additionally, in the section Flavi, three A. tamarii species appear, two of which, specifically (A. tamarii Isolate Nos. 1 and 2) were found to be more closely related and were isolated from Pondicherry spice samples. The A. tamarii (Isolate No. 3) from New Delhi emerged as a separate clade. Under the Nigri section, A. awamori species has been studied: A. awamori (FOEVPS8 1) was isolated from Pondicherry, while A. awamori (GLPL) was isolated from West Bengal. Three A. aureoterreus species and one A. terreus were grouped in the Terrei section. A. aureoterreus (Isolate No. 1) and A. terreus (Isolate No. 1) was recovered from a F. vulgare sample collected from New Delhi, A. aureoterreus (Isolate Nos. 2 and 3) were isolated from Pondicherry fennel seeds. A. fumigatus (Isolate Nos. 1 and 2) from Section Fumigati, were isolated from a Pondicherry fennel sample, whereas A. fumigatus (Isolate No. 3) which emerged as a separate branch was obtained from Lucknow. Strains are likely to represent a diverse set of features within the section under investigation because of the divergence between strains within clades.
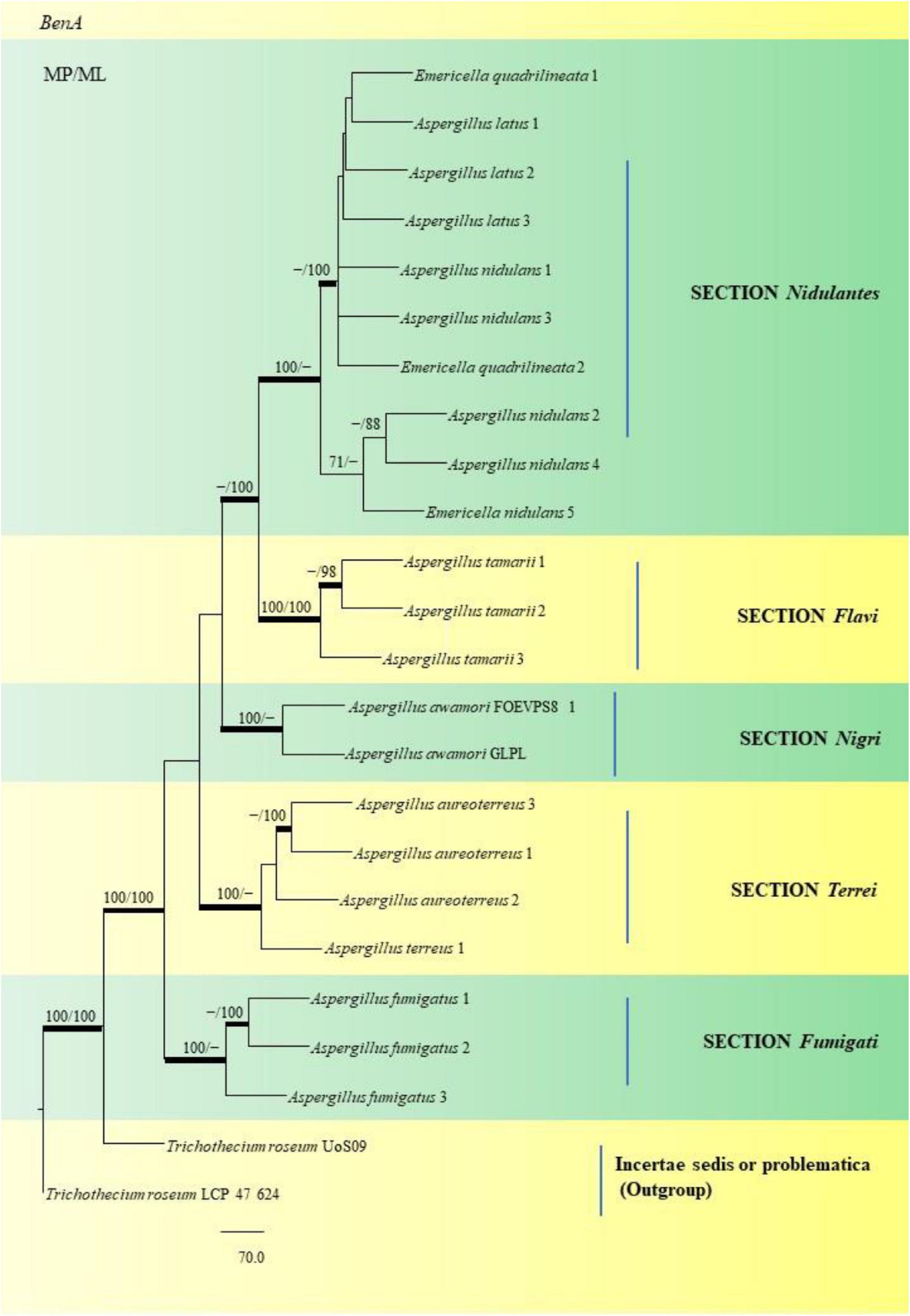
Figure 15. This phylogram depicts the beta-tubulin phylogenetic tree of 22 Aspergillus species in sections, and subsequent analysis of their single nucleotide polymorphisms is discussed below. Maximum parsimony analysis was used to create this tree, which incorporates partial β-tubulin concatenated sequences. Boot strap (bs) values ≥ 70% are designated at the nodes for maximum parsimony (MP) and maximum likelihood (ML) methods; branches with values ≥ 95% bs are bold black. Hyphen (-) indicates support values below 70% for both parsimony and RAxML analyses. Trichothecium roseum Uo S09 and Trichothecium roseum LCP 47 624 were used to root the isolates. The Fungal Genetics and Mycotoxicology (FGM) laboratory codes for the isolates are listed here. 49: E. quadrilineata 1, 4: A. latus 1, 11: A. latus 2, 12: A. latus 3: 17: A. nidulans 1, 9: A. nidulans 3, 58: E. quadrilineata 2, 45: A. nidulans 2, 71: A. nidulans 4, 2: E. quadrilineata, 22: A. tamarii 1, 75: A. tamarii 2, S44: A. tamarii 3, S8: A. awamori S8, S24: A. awamori GLPL, 51: A. aureoterreus 3, 18: A. aureoterreus 1, 1: A. aureoterreus 2, 16: A. terreus 1, 24: A. fumigatus 1, 37: A. fumigatus 2 and 31: A. fumigatus 3.
The SNV analysis reveals that several mutations were identified exclusively in the Nidulantes section. Strain E. nidulans 5 displayed most SNPs (Figure 15) suggesting that the Aspergilli identified under this section may contain an isolate that is distinct. This further divulges information that, there was considerable genetic heterogeneity between A. nidulans, E. quadrilineata and A. latus species. Additionally, the genetic divergence between distant isolates supports distribution of Aspergilli across geographic regions. Furthermore, as demonstrated in the Flavi, Fumigati, and Nigri sections, there is no variance since the genetic sequences employed as markers in this study are highly conserved in terms of SNV mutations, showing that most SNPs have a small effect on the protein structure. The non-synonymous SNPs that were identified in our study, may have significant favorable or negative outcomes and needs to be investigated. The variants may be unique and could be used as biological markers to study specific species. However, the presence of SNPs in Aspergillus species may help us understand the rationale behind genetic diversity with implications of functional consequences if any, through further explorations on their mycotoxigenic potential and related prospective studies. According to the present study, the highlights include Aspergillus diversity from fennel seeds, a polyphasic approach to reappraise Aspergilli into six sections using the official fungal barcode and functional genetic markers which have been performed in adequate detail, with β-tubulin coding sequences being the focus for SNP analysis. Substitutions, insertions and SNV-kind of mutations have been identified among the Aspergillus isolates considered in this study. The data represented is the first report to the best of our knowledge. The occurrence of Aspergilli may help us determine the health status of the spices or cereals as indicator organisms, to adopt suitable strategies for avoidance of contamination at the field and during storage.
Conclusion
The study was an elaborate investigation on the isolation and phenotypic characterization of Aspergilli from an Indian spice namely F. vulgare. While nearly 27 isolates have been adequately described with substantive morphological descriptions, attempts have been made to characterize these Aspergilli phylogenetically as well. A key strength of this endeavor is the number of fennel samples analyzed and Aspergilli being reported in a “first of it’ s kind” investigation. The research study constitutes a key milestone in analyses of evolutionary relationships of fungi, detection of single nucleotide polymorphisms in the target fungal isolates. This validates genetic information, which serves as an essentiality for existing and further research. Although, the study may have its limitations, we propose to expand on the number of isolates to be studied for SNPs and their possible impacts on protein structure and functional consequences. Studies on their mycotoxigenic potentials is currently underway. Clearly, with respect to the diversity of Aspergillus species F. vulgare, the study is the largest till date. The extensive use of partial beta-tubulin gene analyses to evaluate the association between SNPs in five Aspergillus species sections is one of the highlights. Our research outcomes suggest that the presence of Aspergilli in food stuffs may pose a considerable hazard and threat to human consumption. Hence, identification and characterization may serve as a key factor in designing strategies to control post-harvest contamination and elaboration of mycotoxins. Fungal genomics serves as a useful molecular tool for inquiries into fungal evolution, by detecting gene differences and gene structure, to deduce the genetic basis of fungal evolution. However, blending morphological characteristics with molecular phylogeny is considered equally important for taxonomic studies and is the norm today.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, MN791093; https://www.ncbi.nlm.nih.gov/genbank/, MN791096; https://www.ncbi.nlm.nih.gov/genbank/, MN791109; https://www.ncbi.nlm.nih.gov/genbank/, MN791095; https://www.ncbi.nlm.nih.gov/genbank/, MN791106; https://www.ncbi.nlm.nih.gov/genbank/, MN791098; https://www.ncbi.nlm.nih.gov/genbank/, MN791108; https://www.ncbi.nlm.nih.gov/genbank/, MN791115; https://www.ncbi.nlm.nih.gov/genbank/, MN791099; https://www.ncbi.nlm.nih.gov/genbank/, MN791100; https://www.ncbi.nlm.nih.gov/genbank/, MN791103; https://www.ncbi.nlm.nih.gov/genbank/, MN791097; https://www.ncbi.nlm.nih.gov/genbank/, MN791101; https://www.ncbi.nlm.nih.gov/genbank/, MN791102; https://www.ncbi.nlm.nih.gov/genbank/, MN791107; https://www.ncbi.nlm.nih.gov/genbank/, MN791116; https://www.ncbi.nlm.nih.gov/genbank/, MN791104; https://www.ncbi.nlm.nih.gov/genbank/, MN791105; https://www.ncbi.nlm.nih.gov/genbank/, MN791110; https://www.ncbi.nlm.nih.gov/genbank/, MN791111; https://www.ncbi.nlm.nih.gov/genbank/, MN791112; https://www.ncbi.nlm.nih.gov/genbank/, MN791113; https://www.ncbi.nlm.nih.gov/genbank/, MN791114; https://www.ncbi.nlm.nih.gov/genbank/, MN298848; https://www.ncbi.nlm.nih.gov/genbank/, MN309877; https://www.ncbi.nlm.nih.gov/genbank/, MN264637; https://www.ncbi.nlm.nih.gov/genbank/, MN326529; and https://www.ncbi.nlm.nih.gov/genbank/, MN39290.
Author Contributions
PM and RD engaged in the research project’s planning and design. PM carried out all the experiments, documented the findings, and wrote the manuscript. RD authored the results, drafted the manuscript, corrected the errors, and assisted in writing the manuscript. AP undertook the phylogenetic analysis, while BM performed the SNP analysis. All authors contributed to the article and approved the submitted version.
Conflict of Interest
BM was employed by the International Tech Park.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The Central Instrumentation Facility, Pondicherry University, is acclaimed for the services it provides. We sincerely acknowledge V. Venkateswara Sarma, Fungal Biotechnology Laboratory, Department of Biotechnology, School of Life Sciences, Pondicherry University, for providing the Differential Interference Contrast (DIC) microscope facility. In appreciation of Pondicherry University’s Non-NET University Fellowship, PKM extends his gratitude to the University.
Footnotes
- ^ http://doua.prabi.fr/software/cap3
- ^ https://www.ncbi.nlm.nih.gov/genbank/
- ^ https://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^ https://mafft.cbrc.jp/alignment/server/
- ^ https://www.phylo.org
- ^ http://www.aspergilluspenicillium.org/
References
Ahene, R. E., Odamtten, G. T., and Owusu, E. (2011). Fungal and bacterial contaminants of six spices and spice products in Ghana. Afr. J. Environ. Sci. Technol. 5, 633–640.
Ahmad, M. M., Ahmad, M., Ali, A., Hamid, R., Javed, S., and Abdin, M. Z. (2014). Detection of Aspergillus flavus and Aspergillus parasiticus from aflatoxin-contaminated peanuts and their differentiation using PCR-RFLP. Ann. Microbiol. 64, 1597–1605. doi: 10.1007/s13213-014-0803-5
Ahn, G., Kim, S., Lee, J.-E., Lee, S., Kim, S. H., and Kim, S.-R. (2020). Penicillium is Dominant Airborne Contaminant in Winter Seasons in Asthma Patients Houses: A Case Study in Korea. Available online at: https://www.ncbi.nlm.nih.gov/nuccore/MT582755 (accessed January 21, 2021).
Ali, N., Hashim, N. H., and Shuib, N. S. (2015). Natural occurrence of aflatoxins and ochratoxin A in processed spices marketed in Malaysia. Food Addit. Contam. Part A 32, 518–532. doi: 10.1080/19440049.2015.1011712
Alshannaq, A., and Yu, J. H. (2017). Occurrence, toxicity, and analysis of major mycotoxins in food. Int. J. Environ. Res. Public Health 14:632. doi: 10.3390/ijerph14060632
Alshannaq, A. F., Gibbons, J. G., Lee, M. K., Han, K. H., Hong, S. B., and Yu, J. H. (2018). Controlling aflatoxin contamination and propagation of Aspergillus flavus by a soy-fermenting Aspergillus oryzae strain. Sci. Rep. 8, 1–14. doi: 10.1038/s41598-018-35246-1
Alshehri, B., and Palanisamy, M. (2020). Evaluation of molecular identification of Aspergillus species causing fungal keratitis. Saudi J. Biol. Sci. 27, 751–756. doi: 10.1016/j.sjbs.2019.12.030
Amadi, J. E., and Adeniyi, D. O. (2009). Mycotoxin production by fungi isolated from stored grains. Afr. J. Biotechnol. 8, 1219–1221.
Amaike, S., and Keller, N. P. (2011). Aspergillus flavus. Annu. Rev. Phytopathol. 49, 107–133. doi: 10.1146/annurev-phyto-072910-095221
Andreolli, M., Milanesi, C., Faleri, C., Cresti, M., Lampis, S., Vallini, G., et al. (2018). Well Preserved Seeds of Apple Found in An Amphora Excavated From a Burned Roman Villa Despite the Deteriorating Microorganisms. Available online at: https://www.ncbi.nlm.nih.gov/nuccore/mh447369 (accessed March 22, 2021).
Anonymous (2020). Connect2india. Available online at: https://connect2india.com/global/Fennel-Seed-export-from-india/1 (accessed February 18, 2021).
Anonymous (2021). MycoCosm. Available online at: https://mycocosm.jgi.doe.gov/Aspquag1/Aspquag1.home.html (Accessed March 8, 2021).
Arné, P., Risco-Castillo, V., Jouvion, G., Le Barzic, C., and Guillot, J. (2021). Aspergillosis in wild birds. J. Fungi 7:241. doi: 10.3390/jof7030241
Ayliffe, M. A., Dodds, P. N., and Lawrence, G. J. (2001). Characterisation of a β-tubulin gene from Melampsora lini and comparison of fungal β-tubulin genes. Mycol. Res. 105, 818–826. doi: 10.1017/S0953756201004245
Azzoune, N., Mokrane, S., Riba, A., Bouras, N., Verheecke, C., Sabaou, N., et al. (2015). Contamination of common spices by aflatoxigenic fungi and aflatoxin B1 in Algeria. Qual. Assur. Saf. Crop. Foods 8, 137–144. doi: 10.3920/qas2014.0426
Badgujar, S. B., Patel, V. V., and Bandivdekar, A. H. (2014). Foeniculum vulgare Mill: a review of its botany, phytochemistry, pharmacology, contemporary application, and toxicology. Biomed. Res. Int. 2014:842674. doi: 10.1155/2014/842674
Balajee, S. A., Hurst, S. F., Baddley, J. W., Peterson, S. W., Nickle, D., Lass-Florl, C., et al. (2009). Molecular Phylogeny of the Opportunistic Fungal Pathogen Aspergillus terreus. Available online at: https://www.ncbi.nlm.nih.gov/nuccore/EU147717 (Accessed April 21, 2021).
Battilani, P., Toscano, P., Van der Fels-Klerx, H. J., Moretti, A., Leggieri, M. C., Brera, C., et al. (2016). Aflatoxin B 1 contamination in maize in Europe increases due to climate change. Sci. Rep. 6, 1–7. doi: 10.1038/srep24328
Bennett, J. W. (2010). “An overview of the genus Aspergillus,” in Aspergillus: Molecular Biology and Genomics, eds M. Machida and K. Gomi (Norfolk: Caister Academic Press), 1–17.
Bhatnagar-Mathur, P., Sunkara, S., Bhatnagar-Panwar, M., Waliyar, F., and Sharma, K. K. (2015). Biotechnological advances for combating Aspergillus flavus and aflatoxin contamination in crops. Plant Sci. 234, 119–132. doi: 10.1016/j.plantsci.2015.02.009
Bignell, E. (2010). Aspergillus: molecular biology and genomics. Biotechnol. J. 5, 336–337. doi: 10.1002/biot.201000025
Bobadilla-Carrillo, G. I., Magallon-Servin, P., Lopez-Vela, M., Ramirez-Ramirez, J. C., Gutierrez-Leyva, R., Palomino-Hermosillo, Y. A., et al. (2020). Characterization and Proliferation Capacity of Potentially Pathogenic Fungi in Marine and Freshwater Fish Commercial Feeds. Available online at: https://www.ncbi.nlm.nih.gov/nuccore/1799637099 (accessed April 21, 2021).
Casas López, J. L., Sánchez Pérez, J. A., Fernández Sevilla, J. M., Acién Fernández, F. G., Molina Grima, E., and Chisti, Y. (2004). Fermentation optimization for the production of lovastatin by Aspergillus terreus: use of response surface methodology. J. Chem. Technol. Biotechnol. 79, 1119–1126. doi: 10.1002/jctb.1100
Chavan, S. (2020). Molecular Identification of Fungi Causing Superficial Mycoses. Available online at: https://www.ncbi.nlm.nih.gov/nuccore/MT497447 (Accessed March 20, 2021).
Chen, A. J., Frisvad, J. C., Sun, B. D., Varga, J., Kocsubé, S., Dijksterhuis, J., et al. (2016). Aspergillus section Nidulantes (formerly Emericella): polyphasic taxonomy, chemistry and biology. Stud. Mycol. 84, 1–118. doi: 10.1016/j.simyco.2016.10.001
Chen, M., and Xu, Y. (2018). Phylogenetic Diversity and Antifungal Susceptibility of Clinical Aspergillus Isolates in Shanghai, China. Available online at: https://www.ncbi.nlm.nih.gov/nuccore/MG991348 (accessed January 20, 2021).
Chen, P., Liu, M., Zeng, Q., Zhang, Z., Kong, Q., Liu, W., et al. (2018). Uncovering New Mutations in the cyp51A Gene in Aspergillus Fumigatus Conferring Azole Resistance. Available online at: https://www.ncbi.nlm.nih.gov/nuccore/MH536090 (accessed February 26, 2021).
Cleveland, D. W., and Sullivan, K. F. (1985). Molecular biology and genetics of tubulin. Annu. Rev. Biochem. 54, 331–366. doi: 10.1146/annurev.bi.54.070185.001555
de Hoog, G. S., Guarro, J., Gené, J., and Figueras, M. J. (2000). Atlas of Clinical Fungi. Utrecht: Centraalbureau voor Schimmelcultures (CBS).
Dorner, J. W., Cole, R. J., Lomax, L. G., Gosser, H. S., and Diener, U. L. (1983). Cyclopiazonic acid production by Aspergillus flavus and its effects on broiler chickens. Appl. Environ. Microbiol. 46, 698–703. doi: 10.1128/aem.46.3.698-703.1983
dos Santos-Ciscon, B. A., van Diepeningen, A., da Cruz Machado, J., Dias, I. E., and Waalwijk, C. (2019). Aspergillus species from Brazilian dry beans and their toxigenic potential. Int. J. Food Microbiol. 292, 91–100. doi: 10.1016/j.ijfoodmicro.2018.12.006
Düesberg, U., Wosniok, J., Naehrlich, L., Eschenhagen, P., and Schwarz, C. (2020). Risk factors for respiratory Aspergillus fumigatus in German Cystic Fibrosis patients and impact on lung function. Sci. Rep. 10, 1–9. doi: 10.1038/s41598-020-75886-w
Dyer, P. S., and O’Gorman, C. M. (2012). Sexual development and cryptic sexuality in fungi: insights from Aspergillus species. FEMS Microbiol. Rev. 36, 165–192. doi: 10.1111/j.1574-6976.2011.00308.x
Dyer, P. S., and Paoletti, M. (2005). Reproduction in Aspergillus fumigatus: sexuality in a supposedly asexual species? Med. Mycol. 43, S7–S14. doi: 10.1080/13693780400029015
Edgcomb, V. P., Roger, A. J., Simpson, A. G., Kysela, D. T., and Sogin, M. L. (2001). Evolutionary relationships among “jakobid” flagellates as indicated by alpha-and beta-tubulin phylogenies. Mol. Biol. Evol. 18, 514–522. doi: 10.1093/oxfordjournals.molbev.a003830
Elshafie, A. E., Al-Rashdi, T. A., Al-Bahry, S. N., and Bakheit, C. S. (2002). Fungi and aflatoxins associated with spices in the Sultanate of Oman. Mycopathologia 155, 155–160. doi: 10.1023/A:1020427527963.pdf
Garcia, M. V., Parussolo, G., Moro, C. B., Bernardi, A. O., and Copetti, M. V. (2018). Fungi in spices and mycotoxigenic potential of some Aspergilli isolated. Food Microbiol. 73, 93–98. doi: 10.1016/j.fm.2018.01.013
Goto, T., Wicklow, D. T., and Ito, Y. (1996). Aflatoxin and cyclopiazonic acid production by a sclerotium-producing Aspergillus tamarii strain. Appl. Environ. Microbiol. 62, 4036–4038. doi: 10.1128/aem.62.11.4036-4038.1996
Hall, T. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Hammami, W., Fiori, S., Al Thani, R., Kali, N. A., Balmas, V., Migheli, Q., et al. (2014). Fungal and aflatoxin contamination of marketed spices. Food Control 37, 177–181. doi: 10.1016/j.foodcont.2013.09.027
Hamzah, T. N. T., Lee, S. Y., Hidayat, A., Terhem, R., Faridah-Hanum, I., and Mohamed, R. (2018). Diversity and characterization of endophytic fungi isolated from the tropical mangrove species, Rhizophora mucronata, and identification of potential antagonists against the soil-borne fungus, Fusarium solani. Front. Microbiol. 9:1707. doi: 10.3389/fmicb.2018.01707
Han, X., Jiang, H., Xu, J., Zhang, J., and Li, F. (2017). Dynamic Fumonisin B2 production by Aspergillus niger intented used in food industry in China. Toxins 9:217. doi: 10.3390/toxins9070217
Hillis, D. M., and Bull, J. J. (1993). An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 42, 182–192. doi: 10.1093/sysbio/42.2.182
Hiort, J., Maksimenka, K., Reichert, M., Perović-Ottstadt, S., Lin, W. H., Wray, V., et al. (2004). New natural products from the sponge-derived fungus Aspergillus niger. J. Nat. Prod. 67, 1532–1543. doi: 10.1021/np030551d
Hohl, T. M., and Feldmesser, M. (2007). Aspergillus fumigatus: principles of pathogenesis and host defense. Eukaryot. Cell 6, 1953–1963. doi: 10.1128/EC.00274-07
Homa, M., Manikandan, P., Szekeres, A., Kiss, N., Kocsubé, S., Kredics, L., et al. (2019). Characterization of Aspergillus tamarii strains from human keratomycoses: molecular identification, antifungal susceptibility patterns and cyclopiazonic acid producing abilities. Front. Microbiol. 10:2249. doi: 10.3389/fmicb.2019.02249
Huang, X., and Madan, A. (1999). CAP3: a DNA sequence assembly program. Genome Res. 9, 868–877. doi: 10.1101/gr.9.9.868
Hubka, V., Nováková, A., Peterson, S. W., Frisvad, J. C., Sklenář, F., and Matsuzawa, T. (2016). A reappraisal of Aspergillus section Nidulantes with descriptions of two new sterigmatocystin-producing species. Plant Syst. Evol. 302, 1267–1299. doi: 10.1007/s00606-016-1331-5
Ito, Y. (1998). Properties of Aspergillus tamarii, A. caelatus and related species from acidic tea field soils in Japan. Mycopathologia 144, 169–175. doi: 10.1023/A:1007021527106.pdf
Javidnia, K., Dastgheib, L., Samani, S. M., and Nasiri, A. (2003). Anti-hirsutism activity of fennel (fruits of Foeniculum vulgare) extract–a double-blind placebo-controlled study. Phytomedicine 10, 455–458. doi: 10.1078/094471103322331386
Juuti, J. T., Jokela, S., Tarkka, M. T., Paulin, L., and Lahdensalo, J. (2005). Two phylogenetically highly distinct β-tubulin genes of the basidiomycete Suillus bovinus. Curr. Genet. 47, 253–263. doi: 10.1007/s00294-005-0564-6
Kashyap, S. (2020). Approach to allergic bronchopulmonary aspergillosis (ABPA): quick review. EC Pul. Res. Med. 9, 90–101.
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kaur, R., Mehra, B., Dhakad, M. S., Goyal, R., and Dewan, R. (2017). Pulmonary aspergillosis as opportunistic mycoses in a cohort of human immunodeficiency virus-infected patients: report from a tertiary care hospital in North India. Int. J. Health Sci. 11:45.
Keeling, P. J., Luker, M. A., and Palmer, J. D. (2000). Evidence from beta-tubulin phylogeny that microsporidia evolved from within the fungi. Mol. Biol. Evol. 17, 23–31. doi: 10.1093/oxfordjournals.molbev.a026235
Kirk, K. E., and Morris, N. R. (1991). The tubB alpha-tubulin gene is essential for sexual development in Aspergillus nidulans. Genes Dev. 5, 2014–2023. doi: 10.1101/gad.5.11.2014
Kitamoto, K. (2002). Molecular biology of the Koji molds. Adv. Appl. Microbiol. 51, 129–154. doi: 10.1016/s0065-2164(02)51004-2
Koppula, S., and Kumar, H. (2013). Foeniculum vulgare Mill (Umbelliferae) attenuates stress and improves memory in wister rats. Trop. J. Pharm. Res. 12, 553–558. doi: 10.4314/tjpr.v12i4.17
Kosmidis, C., and Denning, D. W. (2015). The clinical spectrum of pulmonary aspergillosis. Thorax 70, 270–277. doi: 10.1136/thoraxjnl-2014-206291
Kulshrestha, P., Singh, C., Gupta, A., Mahajan, S., and Sharma, R. (2014). Mycoflora associated with spices. Int. J. Curr. Microbiol. Appl. Sci. 3, 741–746.
Mages, W., Cresnar, B., Harper, J. F., Brüderlein, M., and Schmitt, R. (1995). Volvox carteri α2-and β2-tubulin-encoding genes: regulatory signals and transcription. Gene 160, 47–54. doi: 10.1016/0378-1119(95)00178-9
Magnoli, C. E., Astoreca, A. L., Chiacchiera, S. M., and Dalcero, A. M. (2007). Occurrence of ochratoxin A and ochratoxigenic mycoflora in corn and corn-based foods and feeds in some South American countries. Mycopathologia 163, 249–260. doi: 10.1007/s11046-007-9005-z
Makhlouf, J., Carvajal-Campos, A., Querin, A., Tadrist, S., Puel, O., Lorber, S., et al. (2019). Morphologic, molecular and metabolic characterization of Aspergillus section Flavi in spices marketed in Lebanon. Sci. Rep. 9, 1–11. doi: 10.1038/s41598-019-41704-1
Malathi, S., and Chakraborty, R. (1991). Production of alkaline protease by a new Aspergillus flavus isolate under solid-substrate fermentation conditions for use as a depilation agent. Appl. Environ. Microbiol. 57, 712–716. doi: 10.1128/aem.57.3.712-716.1991
Matsuzawa, T., Yaguchi, T., Horie, Y., and Nshimura, K. (2006). Molecular Phylogenetics of the Genus Emericella and the Surface Structure of Ascospores. Available online at: https://www.ncbi.nlm.nih.gov/nuccore/AB248335 (accessed October 13, 2020).
Moloney, C. (2019). First Post. Available online at: https://www.firstpost.com/business/indias-major-agricultural-produce-losses-estimated-at-rs-92000-cr-2949002.html (accessed February 16, 2021).
Mukherjee, M., Hadar, R., Mukherjee, P. K., and Horwitz, B. A. (2003). Homologous expression of a mutated beta-tubulin gene does not confer benomyl resistance on Trichoderma virens. J. Appl. Microbiol. 95, 861–867. doi: 10.1046/j.1365-2672.2003.02061.x
Nadumane, V. K., Venkatachalam, P., and Gajaraj, B. (2016). “Aspergillus applications in cancer research,” in New and Future Developments in Microbial Biotechnology and Bioengineering, ed. V. K. Gupta (Amsterdam: Elsevier), 243–255.
Nurtjahja, K., Zuhra, C. F., Sembiring, H., Bungsu, A., Simanullang, J., Silalahi, J. E., et al. (2019). Fungal contamination spices from Indonesia with emphasis on Aspergillus flavus. Czech J. Food Sci. 37, 338–344. doi: 10.17221/18/2019-CJFS
Op De Beeck, M., Lievens, B., Busschaert, P., Declerck, S., Vangronsveld, J., and Colpaert, J. V. (2014). Comparison and validation of some ITS primer pairs useful for fungal metabarcoding studies. PLoS One 9:e97629. doi: 10.1371/journal.pone.0097629
Ostry, V., Malir, F., Toman, J., and Grosse, Y. (2017). Mycotoxins as human carcinogens—the IARC Monographs classification. Mycotoxin Res. 33, 65–73. doi: 10.1007/s12550-016-0265-7
Pal, M., Dave, P., and Manna, A. K. (2014). Emerging role of Aspergillus flavus in human and animal disorders. J. Mycopathol. Res. 52, 211–216.
Parahym, A. M. R. D. C., Neto, P. J. R., Silva, C. M. D., Gonçalves, S. S., and Motta, C. M. D. S. (2014). Fatal invasive Aspergillosis in acute lymphoblastic leukemia patient. J. Clin. Case Rep. 4:2. doi: 10.4172/2165-7920.1000368
Ráduly, Z., Szabó, L., Madar, A., Pócsi, I., and Csernoch, L. (2020). Toxicological and medical aspects of Aspergillus-derived mycotoxins entering the feed and food chain. Front. Microbiol. 10:2908. doi: 10.3389/fmicb.2019.02908
Raja, H. A., Miller, A. N., Pearce, C. J., and Oberlies, N. H. (2017). Fungal identification using molecular tools: a primer for the natural products research community. J. Nat. Products 80, 756–770. doi: 10.1021/acs.jnatprod.6b01085
Rambaut, A., and Drummond, A. J. (2009). FigTree v. 1.3.1. Computer Program and Documentation Distributed by the Author. Available online at: http://tree.bio.ed.ac.uk/software/ (accessed February 11, 2020).
Ramírez-Camejo, L. A., Zuluaga-Montero, A., Lázaro-Escudero, M., Hernández-Kendall, V., and Bayman, P. (2012). Phylogeography of the cosmopolitan fungus Aspergillus flavus: is everything everywhere? Fungal Biol. 116, 452–463. doi: 10.1016/j.funbio.2012.01.006
Rather, M. A., Dar, B. A., Sofi, S. N., Bhat, B. A., and Qurishi, M. A. (2016). Foeniculum vulgare: a comprehensive review of its traditional use, phytochemistry, pharmacology, and safety. Arab. J. Chem. 9, S1574–S1583. doi: 10.1016/j.arabjc.2012.04.011
Reddy, K. R. N., Reddy, C. S., and Muralidharan, K. (2009). Detection of Aspergillus spp. and aflatoxin B1 in rice in India. Food Microbiol. 26, 27–31. doi: 10.1016/j.fm.2008.07.013
Refai, M., El-Yazid, H. A., and Hassan, A. (eds) (2014). “Monograph on Aspergillus and aspergillosis in man, animals and birds,” in A Guide for Classification and Identification of Aspergilli, Diseases Caused by Them, Diagnosis and Treatment (Cambridge, MA: Academia Press).
Samadi-Noshahr, Z., Hadjzadeh, M. A. R., Moradi-Marjaneh, R., and Khajavi-Rad, A. (2021). The hepatoprotective effects of fennel seeds extract and trans-Anethole in streptozotocin-induced liver injury in rats. Food Sci. Nutr. 9, 1121–1131. doi: 10.1002/fsn3.2090
Samson, R. A., Hong, S. B., and Frisvad, J. C. (2006). Old and new concepts of species differentiation in Aspergillus. Med. Mycol. 44, S133–S148. doi: 10.1080/13693780600913224
Samson, R. A., Peterson, S. W., Frisvad, J. C., and Varga, J. (2011). New species in Aspergillus section terrei. Stud. Mycol. 69, 39–55. doi: 10.3114/sim.2011.69.04
Schoch, C. L., Seifert, K. A., Huhndorf, S., Robert, V., Spouge, J. L., Levesque, C. A., et al. (2012). Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. U.S.A. 109, 6241–6246. doi: 10.1073/pnas.1117018109
Schütze, J., Krasko, A., Custodio, M. R., Efremova, S. M., Müller, I. M., and Müller, W. E. (1999). Evolutionary relationships of Metazoa within the eukaryotes based on molecular data from Porifera. Proc. R. Soc. Lond. Ser. B Biol. Sci. 266, 63–73. doi: 10.1098/rspb.1999.0605
Segal, B. H. (2009). “Invasive aspergillosis in chronic granulomatous disease,” in Aspergillosis: From Diagnosis to Prevention, ed. A. C. Pasqualotto (London: Springer), 527–543.
Siruguri, V., and Bhat, R. V. (2015). Assessing intake of spices by pattern of spice use, frequency of consumption and portion size of spices consumed from routinely prepared dishes in southern India. Nutr. J. 14, 1–9. doi: 10.1186/1475-2891-14-7
Soni, P., Gangurde, S. S., Ortega-Beltran, A., Kumar, R., Parmar, S., Sudini, H. K., et al. (2020). Functional biology and molecular mechanisms of host-pathogen interactions for aflatoxin contamination in groundnut (Arachis hypogaea L.) and maize (Zea mays L.). Front. Microbiol. 11:227. doi: 10.3389/fmicb.2020.00227
Ssepuuya, G., Van Poucke, C., Ediage, E. N., Mulholland, C., Tritscher, A., Verger, P., et al. (2018). Mycotoxin contamination of sorghum and its contribution to human dietary exposure in four sub-Saharan countries. Food Addit. Contam. A 35, 1384–1393. doi: 10.1080/19440049.2018.1461253
Stamatakis, A., Aberer, A. J., Goll, C., Smith, S. A., Berger, S. A., and Izquierdo-Carrasco, F. (2012). RAxML-Light: a tool for computing terabyte phylogenies. Bioinformatics 28, 2064–2066. doi: 10.1093/bioinformatics/bts309
Streit, E., Schatzmayr, G., Tassis, P., Tzika, E., Marin, D., Taranu, I., et al. (2012). Current situation of mycotoxin contamination and co-occurrence in animal feed—Focus on Europe. Toxins 4, 788–809. doi: 10.3390/toxins4100788
Swofford, D. L., and Sullivan, J. (2003). “Phylogeny inference based on parsimony and other methods using PAUP*,” in The Phylogenetic Handbook: A Practical Approach to DNA and Protein Phylogeny, eds M. Salemi and A.-M. Vandamme (Cambridge, MA: Cambridge University Press).
Tam, E. W., Chen, J. H., Lau, E. C., Ngan, A. H., Fung, K. S., Lee, K. C., et al. (2014). Misidentification of Aspergillus nomius and Aspergillus tamarii as Aspergillus flavus: characterization by internal transcribed spacer, β-tubulin, and calmodulin gene sequencing, metabolic fingerprinting, and matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 52, 1153–1160. doi: 10.1128/JCM.03258-13
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Van De Veerdonk, F. L., Kolwijck, E., Lestrade, P. P., Hodiamont, C. J., Rijnders, B. J., Van Paassen, J., et al. (2017). Influenza-associated aspergillosis in critically ill patients. Am. J. Respir. Crit. Care Med. 196, 524–527. doi: 10.1164/rccm.201612-2540LE
Varga, J., and Samson, R. A. (eds) (2008). Aspergillus in the Genomic Era. Wageningen: Wageningen Academic Publishers.
Visagie, C. M., and Houbraken, J. (2020). Updating the taxonomy of Aspergillus in South Africa. Stud. Mycol. 95, 253–292. doi: 10.1016/j.simyco.2020.02.003
Wood, G. E. (1992). Mycotoxins in foods and feeds in the United States. J. Anim. Sci. 70, 3941–3949. doi: 10.2527/1992.70123941x
Xie, G. F., Li, W. J., Lu, J., Cao, Y., Fang, H., Zou, H. J., et al. (2007). Isolation and identification of representative fungi from Shaoxing rice wine wheat Qu using a polyphasic approach of culture-based and molecular-based methods. J. Inst. Brew. 113, 272–279. doi: 10.1002/j.2050-0416.2007.tb00287.x
Zhang, M., Wang, W. L., Fang, Y. C., Zhu, T. J., Gu, Q. Q., and Zhu, W. M. (2008). Cytotoxic alkaloids and antibiotic nordammarane triterpenoids from the marine-derived fungus Aspergillus sydowi. J. Nat. Prod. 71, 985–989. doi: 10.1021/np700737g
Keywords: fungal genomics, Aspergillus, polyphasic, phylogenetic analysis, single nucleotide polymorphisms (SNPs), single nucleotide variations (SNVs), non-synonymous and synonymous mutation
Citation: Mahata PK, Dass RS, Pan A and Muthusamy B (2022) Substantive Morphological Descriptions, Phylogenetic Analysis and Single Nucleotide Polymorphisms of Aspergillus Species From Foeniculum vulgare. Front. Microbiol. 13:832320. doi: 10.3389/fmicb.2022.832320
Received: 09 December 2021; Accepted: 24 January 2022;
Published: 18 February 2022.
Edited by:
Zheng Zhang, Shandong University, ChinaReviewed by:
Kamel A. Abd-Elsalam, Agricultural Research Center, EgyptFuguo Xing, Institute of Food Science and Technology, Chinese Academy of Agricultural Sciences (CAAS), China
Copyright © 2022 Mahata, Dass, Pan and Muthusamy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Regina Sharmila Dass, cmVnaW5hZGFzc0BnbWFpbC5jb20=
 Pranab Kumar Mahata
Pranab Kumar Mahata Regina Sharmila Dass
Regina Sharmila Dass Archana Pan
Archana Pan Babylakshmi Muthusamy
Babylakshmi Muthusamy