- 1Guangdong Provincial Key Laboratory for Plant Epigenetics, Longhua Bioindustry and Innovation Research Institute, College of Life Sciences and Oceanography, Shenzhen University, Shenzhen, China
- 2Key Laboratory of Optoelectronic Devices and Systems of Ministry of Education and Guangdong Province, College of Optoelectronic Engineering, Shenzhen University, Shenzhen, China
- 3Program in Plant Biology and Conservation, Northwestern University, Evanston, IL, United States
- 4Key Laboratory of Biodiversity and Biogeography, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China
- 5Plant Conservation Science, Chicago Botanic Garden, Glencoe, IL, United States
- 6Sam Mitchel Herbarium of Fungi, Denver Botanic Garden, Denver, CO, United States
Suillus is a genus of ectomycorrhizal fungi associated almost exclusively with Pinaceae. Lack of sample collections in East Asia and unresolved basal phylogenetic relationships of the genus are the major obstacles for better understanding the Suillus evolution. A resolved phylogeny of Suillus representing global diversity was achieved by sequencing multiple nuclear ribosomal and protein coding genes and extensive samples collected in East Asia. Fungal fossils are extremely rare, and the Eocene ectomycorrhizal symbiosis (ECM) fossil of Pinus root has been widely used for calibration. This study explored an alternative calibration scenario of the ECM fossil for controversy. Ancestral host associations of Suillus were estimated by maximum likelihood and Bayesian Markov chain Monte Carlo (MCMC) analyses, inferred from current host information from root tips and field observation. Host shift speciation explains the diversification of Suillus major clades. The three basal subgenera of Suillus were inferred to be associated with Larix, and diverged in early Eocene or Upper Cretaceous. In the early Oligocene or Paleocene, subgenus Suillus diverged and switched host to Pinus subgenus Strobus, and then switched to subgenus Pinus four times. Suillus subgenus Douglasii switched host from Larix to Pseudotsuga in Oligocene or Eocene. Increased species diversity occurred in subgenus Suillus after it switched host to Pinus but no associated speciation rate shifts were detected. Ancestral biogeographic distributions of Suillus and Pinaceae were estimated under the Dispersal Extinction Cladogenesis (DEC) model. Ancestral distribution patterns of Suillus and Pinaceae are related but generally discordant. Dispersals between Eurasia and North America explain the prevalence of disjunct Suillus taxa.
Introduction
The ectomycorrhizal symbiosis (ECM) is a common relationship between plants and fungi responsible for exchanging carbohydrates, mineral nutrients, and water (Landeweert et al., 2001; Nehls et al., 2010). Extant ectomycorrhizal fungi have evolved independently from over 80 saprotrophic ancestral lineages (Tedersoo and Smith, 2013, 2017; Martin et al., 2016).
The majority of ECM fungi are generalists, whereby one ECM fungal species can form an association with a wide variety of plant families (Bruns et al., 2002; Kennedy et al., 2003; Smith et al., 2009). However, a subset of ECM fungi is host-specific, i.e., they are associated with particular plant taxonomic groups. In contrast, their host plants are commonly generalists with the ability to recruit a broad spectrum of ECM fungal taxa (Ishida et al., 2007; Krpata et al., 2008; Deslippe et al., 2011; Leski and Rudawska, 2012). Species in the ECM genus Suillus have exceptionally high host fidelity to specific trees within the pine family (Pinaceae), with varying specificity from genera down to species (Thiers, 1975; Kretzer et al., 1996; Nguyen et al., 2016).
The evolutionary mechanisms explaining host specificity between symbiotic fungi and host plants include host shift speciation, coevolution, and cospeciation (Vienne et al., 2013). Host shift speciation has been confirmed in many ECM fungal lineages, including Strobilomyces, Leccinum, Hebeloma, Pisolithus, and Laccaria (Aanen et al., 2000; Martin et al., 2002; Bakker et al., 2004; Sato et al., 2017; Wilson et al., 2017). Diversification after host switching has also been documented in Strobilomyces, Leccinum, and Hebeloma, providing the ecological advantages of adapting to novel hosts and niches (Aanen et al., 2000; Bakker et al., 2004; Sato et al., 2017).
Being host-specific, most Suillus species are restricted to the Northern Hemisphere along with their Pinaceae hosts (Nguyen et al., 2016). Studies have shown that Suilloid taxa might have facilitated the invasion of Pinaceae into the Southern Hemisphere (Dickie et al., 2010; Hayward et al., 2015; Policelli et al., 2019). Diversity of Suillus species is severely underestimated in East Asia, with a significant lack of sampling hindering further biogeographic analyses (Wu et al., 2000; Mueller et al., 2001). Despite current efforts to discover novel species, the contemporary richness of the Suillus diversity in East Asia remains unevaluated (Verma and Reddy, 2015; Shi et al., 2016; Zhang et al., 2017).
This study compares the evolutionary relationships and distribution patterns of Suillus and their hosts Pinaceae throughout history. Multiple nuclear protein-coding genes of Suillus were collected, given that previous phylogenies based solely on ribosomal rRNA sequences could not resolve basal relationships of the genus (Kretzer et al., 1996; Wu et al., 2000; Mueller et al., 2001). To fill the sampling gap, many Suillus specimens were collected from East Asia, covering the geographic distributions of Pinaceae hosts, including approximately 26 putative undescribed species. We addressed three specific questions in this study, including (1) Can cospeciation or host shift speciation explain the diversification of Suillus major lineages? (2) Are geographic distribution patterns between Suillus and its host plants congruent? (3) Are the extensive speciation or diversification rate shifts driven by Suillus host switches or geographic range variation?
Materials and Methods
Specimens and Molecular Data Collection
Suillus specimens from North American and European herbaria were sampled for DNA extraction. To fill the geographic sampling gap, intensive field collections were conducted over 10 years in East Asia covering Pinaceae distribution ranges (Farjon, 1990, 2005). A list of 103 Suillus specimens representing approximately 86 species used in this study is given in Supplementary Table 1.
DNA extraction, PCR and cycle sequencing are as previously described (Zhang et al., 2017). For nuclear ribosomal rRNA internal transcribed spacers 1 and 2, the 5.8S rRNA gene and parts of the 28S rRNA genes, primers ITS-1F and ITS-4 were used (White et al., 1990; Gardes and Bruns, 1993). For some specimens with DNA degradation, amplification was carried out with internal primers ITS-2 and ITS-3 (White et al., 1990). For the nuclear partial 28S large subunit rRNA genes, primers LROR and LR5 were used (Vilgalys and Hester, 1990). For amplification of the nuclear protein coding translation elongation factor EF1-alpha (TEF1) partial gene, the following primers were used: primers TEF1-983f and TEF1-2212r, with additional internal primer TEF1-1567r (Rehner and Buckley, 2005), and one newly designed Suillus specific internal primer TEF1-Sintf (5′- TYR CAC AGC ATG MCA TGG TA -3′). For amplification of the nuclear protein coding RNA polymerase II largest subunit (RPB1) partial gene, the following primers were used: primers RPB1-Af and RPB1-Cr (Stiller and Hall, 1997; Matheny, 2005), with additional internal primer RPB1-Int2.2f (Binder et al., 2010) and RPB1-Int2.1r (Frøslev et al., 2005). For amplification of the nuclear protein coding RNA polymerase II second largest subunit (RPB2) partial gene, the primers were: RPB2-6F and RPB2-7.1r (Matheny et al., 2007), and newly designed Suillus specific internal primers RPB2-SintR (5′- CTC CRT CNT CNT CGC GRT AA -3′) and RPB2-SintF (5′- CAC GAC CRG CRT CYG TGT AY -3′).
Phylogenetic Analyses
An ITS phylogeny of Suillus was generated for identifying operation taxonomic units and for more information on host, distribution range and diversity. Rhizopogon abietis, Rhizopogon ochraceisporus, and Truncocolumella citrina were chosen as outgroups for the ITS dataset (Binder and Hibbett, 2006). Initial alignment for the ITS sequences was obtained in Mesquite 2.75 with manual adjustments (Edgar, 2004; Maddison and Maddison, 2011).
Phylogenetic tree of ITS sequences was inferred from Maximum likelihood (ML) and Bayesian methods performed on the CIPRES computing facility (Miller et al., 2015). Bootstrap analyses of ML were performed using RAxML 8.0.0 HPC2 on XSEDE with 1000 bootstrap replicates (Stamatakis, 2014). Bayesian phylogenetic analysis was performed using MrBayes 3.2.6 (Ronquist and Huelsenbeck, 2003; Ronquist et al., 2012). Number of substitution types Nst was set at 6, with 2 runs, 4 chains per run, each run searching for 1,000,000 generations sampling every 1000th generation. The first 10% of the sampled Bayesian trees of the analysis was discarded as the burnin. For convergence diagnosis, the estimated sample size (ESS) was above 200.
A supermatrix dataset was assembled with four loci including 28S, TEF1, RPB1, and RPB2. For the supermatrix dataset, each sample was selected from the ITS phylogeny to represent a unique haplotype. ITS sequences and introns of protein coding genes were not included because of their high level of sequence variability and poorer resolution at deeper nodes. Before concatenation, significant topological incongruence was evaluated among single gene phylogenies of 28S, TEF1, RPB1, and RPB2 using a cutoff of ≥ 70% maximum likelihood bootstrap (MLB) support and ≥ 0.98 Bayesian posterior probabilities (BPP). No major conflicts were detected for the inter-species evaluation. Rhizopogon nigrescens and Gomphidius roseus were chosen as outgroups for the multigene dataset (Binder and Hibbett, 2006).
Partitioning strategy and molecular models were searched using PartitionFinder V1.1.1 (Lanfear et al., 2012). The greedy algorithm was used to explore all nucleotide substitution models available under the Bayesian information criterion (BIC). Codon positions of protein coding genes were regarded for the partitioning analyses. Phylogenetic trees of the supermatrix were constructed with RAxML and Bayesian methods as described for the ITS phylogeny. Partitioning schemes and substitution models were set for the RAxML analysis under GTRGAMMA with 1000 bootstrap replicates. For the BI analysis, MrBayes 3.2.6 was implemented with the partitioned supermatrix and substitution models suggested by PartitionFinder (Ronquist et al., 2012).
Reconstructing Ancestral Host Associations
Current host associations of Suillus species were identified from global environmental samples, field observations and literature references. To be more conservative on host identifications, mycological references containing taxonomic uncertainties were not included. Host information for a certain Suillus species is documented if root tip samples were in the same OTUs defined by ≥ 70% MLB or ≥ 0.98 BPP in the ITS phylogeny. Field observations can provide reliable host information for Asian Suillus because almost each Pinus and Larix species grows in separate habitat (Farjon, 1990, 2005). Hosts of Suillus were classified to the generic level for Larix and Pseudotsuga, and to the subgeneric level for Pinus.
To reconstruct ancestral host associations for Suillus, ML and Bayesian Markov chain Monte Carlo (MCMC) analyses were conducted in BayesTraits v2.0 (Pagel and Meade, 2006). MCMC analyses were run for 1.01 × 106 iterations, sampling every 1,000, with the first 10,000 iterations discarded as a burn-in. The most probable host with a common ancestor was inferred if the acceptance rate is between 20–40% when the chain is at convergence.
Current Distributions of Suillus and Pinaceae
Collection site data were obtained from herbarium labels for Suillus taxa. Current ranges of Suillus species were also inferred from the global ITS phylogeny including environmental samples. Introduced taxa were excluded from the biogeographic analyses, e.g., Suillus luteus associated with introduced Pinus sylvestris in North America and S. lakei with introduced Pseudotsuga menziesii in Europe.
The current distributions of Pinaceae species were limited to their natural ranges (Farjon, 1990, 2005). Pinus, Larix, and Pseudotsuga distribution data across the world were also compiled from the global biodiversity information facility database (GBIF1.
Ancestral Range Estimation
To estimate ancestral ranges we used the Dispersal Extinction Cladogenesis (DEC) model of Lagrange implemented in R package BioGeoBEARS (Ree and Smith, 2008; Matzke, 2014). The founder-event dispersal parameter “j” was introduced for DEC assuming a cladogenetic event (Matzke, 2014).
The extant distributions of Suillus and Pinaceae were divided into four biogeographic units: (1) ENA: Eastern North America, east of the Rocky mountains, encompassing Canada to Florida; (2) WNA, Western North America, west of the Rocky mountains, encompassing Alaska through Central America; (3) EUA, encompassing Europe, Northern China and Central Asia; (4) In, Indo-Pacific, encompassing subtropical Southern China, Southeast Asia and the western Himalayas. We also refer the combined EUA and Indo-Pacific as Eurasia. Events including cladogenesis, anagenesis, and vicariance were interpreted based on the most probable range for each ancestral species.
Time-Scaled Phylogenies of Suillus and Pinaceae
A two-step calibration procedure was conducted following previous examples (Renner, 2005; Wilson et al., 2012, 2017). Step one of the BEAST analysis used two fungal fossils to calibrate the phylogeny of 37 Agaricomycete taxa, including 19 Suillus taxa. Taxonomic groups were defined in BEAUti including Agaricales, Boletales, Boletineae, Boletinus, Sclerodermatineae, “Marasmioid” fungi and Suillineae. Suillus taxa from five subgenera represent the genus. Two fossils in Agaricomycetes were utilized for calibration in step one. Archaeomarasmius leggetti from mid-Cretaceous amber of New Jersey resembles the extant genera Marasmius and Marasmiellus (Hibbett et al., 1997). Archaeomarasmius leggetti was regarded as the most recent common ancestor (MRCA) of Marasmius rotula and Mycena amabillisima and was calibrated at 90 Ma (million year ago) with a mean of 10 using a lognormal distribution (Renner, 2005; Wilson et al., 2012, 2017). The second fossil used for calibration is an ECM root of Pinus preserved in the Princeton chert of British Columbia in the early Eocene (c. 47.8 Ma) (Lepage et al., 1997; Pigg and Devore, 2016). The ECM fossil was used to calibrate the node for the Suillineae, including Suillus, Rhizopogon, and Gomphidius, and was calibrated using a lognormal distribution with an initial value 50 Ma and a log (mean) set 25. The same set of fossil calibrations was carried out in previous studies of Agaricomycetes (Floudas et al., 2012; Wilson et al., 2012, 2017). The Princeton chert fossil does not provide information on sporocarp morphology but the Pinus host is known. This study explores an alternative scenario: the fossil could be at clade(s) of Suillus associated with Pinus. Step two calibration inferred calibration points of major clades from step one to estimate divergence dates of the comprehensive Suillus phylogeny.
We used BEAUti v.1.8.2 to create XML files that incorporated the calibration priors, partitioning schemes and Bayesian parameters for analysis in BEAST v1.8.2 (Drummond et al., 2012). Bayesian parameters included a GTR + I + G model, Yule process speciation, and an uncorrelated lognormal relaxed clock model. The Bayesian Markov-chain Monte Carlo (MC3) analysis was run for 30 million generations, sampling every 1000th tree. Each analysis was run two times. The first 10% of the trees were removed as the burn-in and the remaining trees were combined using LogCombiner v1.8.2. A summary tree was produced using TreeAnnotator v1.8.2 (Drummond et al., 2012). Convergence, burn-in, means, medians and 95% highest posterior densities (HPDs) for nodes of interest were examined from BEAST logfiles using Tracer v1.6.0. Maximum clade credibility trees from the two steps were created using TreeAnnotator (BEAST package) and to summarize the posterior samples of trees produced by BEAST.
The calibrated phylogeny of Pinus was pruned from a published phylogeny of conifers (Leslie et al., 2012), which was generated from two nuclear genes (18S and a phytochrome gene, PHYP) and two chloroplast genes (matK and rbcL). It included 84% of Pinus global diversity. Fossils Larix altoborealis, Picea burtonii, and Tsuga swedaea within Pinaceae were used to calibrate the phylogeny (Leslie et al., 2012).
Diversification Rate Shifts
Lineage-through-time (LTT) plots were conducted in the R package APE for Suillus. For better comparison, Suillus/Pinus lineages and genus Pinus were plated on the same LTT plot (Paradis et al., 2004; Leslie et al., 2012).
To reveal speciation rate variations in the phylogeny of Suillus, Bayesian analysis of Macroevolutionary Mixtures (BAMM) v2.0 was utilized (Rabosky, 2014; Rabosky et al., 2014). BAMM reconstructs branch-specific evolutionary rates and allows rates to vary through time and among lineages. Without prior knowledge of the number and location of distinct regimes of diversification rates, BAMM simulates a posterior distribution of shift configurations on phylogenetic trees. The priors for the BAMM run was simulated by BAMMtools (Rabosky et al., 2014). Our sampled 66 OTUs are about 72% of the c. 92 known global Suillus OTUs. The employed incomplete sampling of 60% estimated unknown Suillus diversity from unsampled Pinaceae hosts. BAMM was run under the reversible-jump MCMC method with 10 million generations for the calibrated phylogenies of Pinaceae and Suillus. The first 10% was discarded as burn-in and the convergence was checked in coda (Plummer et al., 2008). BAMMtools visualized the output of BAMM to generate the mean phylorate plot and the 95% credible set of macroevolutionary rate configurations (credible shift sets). If zero rate shift was detected in the phylogeny, the expected number of shifts was not adjusted in BAMMtools to avoid type I error (Rabosky et al., 2014).
Results
Phylogenetic Analyses
A total of 393 sequences were generated (93 ITS, 89 28S, 90 TEF1, 61 RPB1, 60 RPB2), and 98 sequences were acquired from GenBank (94 ITS, 1 28S, 1 TEF1, 1 RPB1, 1 RPB2) (Supplementary Table 1). The ITS dataset was 834 bp in total length with 313 parsimony informative sites. We added 26 new OTUs representing new species from China and other studies (Verma and Reddy, 2015; Shi et al., 2016). RAxML and Bayesian analyses provided congruent results for the ITS phylogeny (Supplementary Figure 1). Deep relationships of the ITS phylogeny remain unresolved (Supplementary Figure 1). Host and geographic information was inferred from the ITS phylogeny and was applied to the Suillus species level (Supplementary Figure 1 and Supplementary Table 1). Matrices and phylogenetic trees are available in Treebase (number S210962).
For the multigene phylogeny, we sampled 66 OTUs (72% of known Suillus OTUs). Four OTUs contained multiple geographic representatives: Suillus brevipes, Suillus ampliporus, Suillus flavidus, and Suillus spectabilis. For the subsequent multigene analysis, taxa were selected from the ITS phylogeny to evenly represent the taxonomic and geographic diversity within Suillus (Supplementary Figure 1). The total length of the supermatrix was 3914 bp with 858 informative sites. The supermatrix excluding introns was 3138 bp with 626 informative sites and was partitioned as: (1) 28S and the first and second codon positions of TEF1, RPB1, and RPB2, with GTR + I + G as the best model; (2) the third codon positions of TEF1, RPB1, and RPB2 with GTR + G as the best model.
Phylogenies based on the supermatrices resolved the basal relationships of Suillus (Supplementary Figure 2), with five subgenera resolved and supported (Figure 1 and Supplementary Figure 2). The subgenus Boletinus (node D) was basal and sister to all other subgenera. In contrast, the subgenus Spectabilis (node F) was sister to a monophyletic clade (node G) containing all remaining subgenera. The subgenus Larigini was sister to the subgenus Douglasii, and the common ancestor of the two was sister to the subgenus Suillus. We recommend that two new monophyletic sections be recognized for the subgenus Suillus—I and II (Supplementary Figure 4; node M and L).
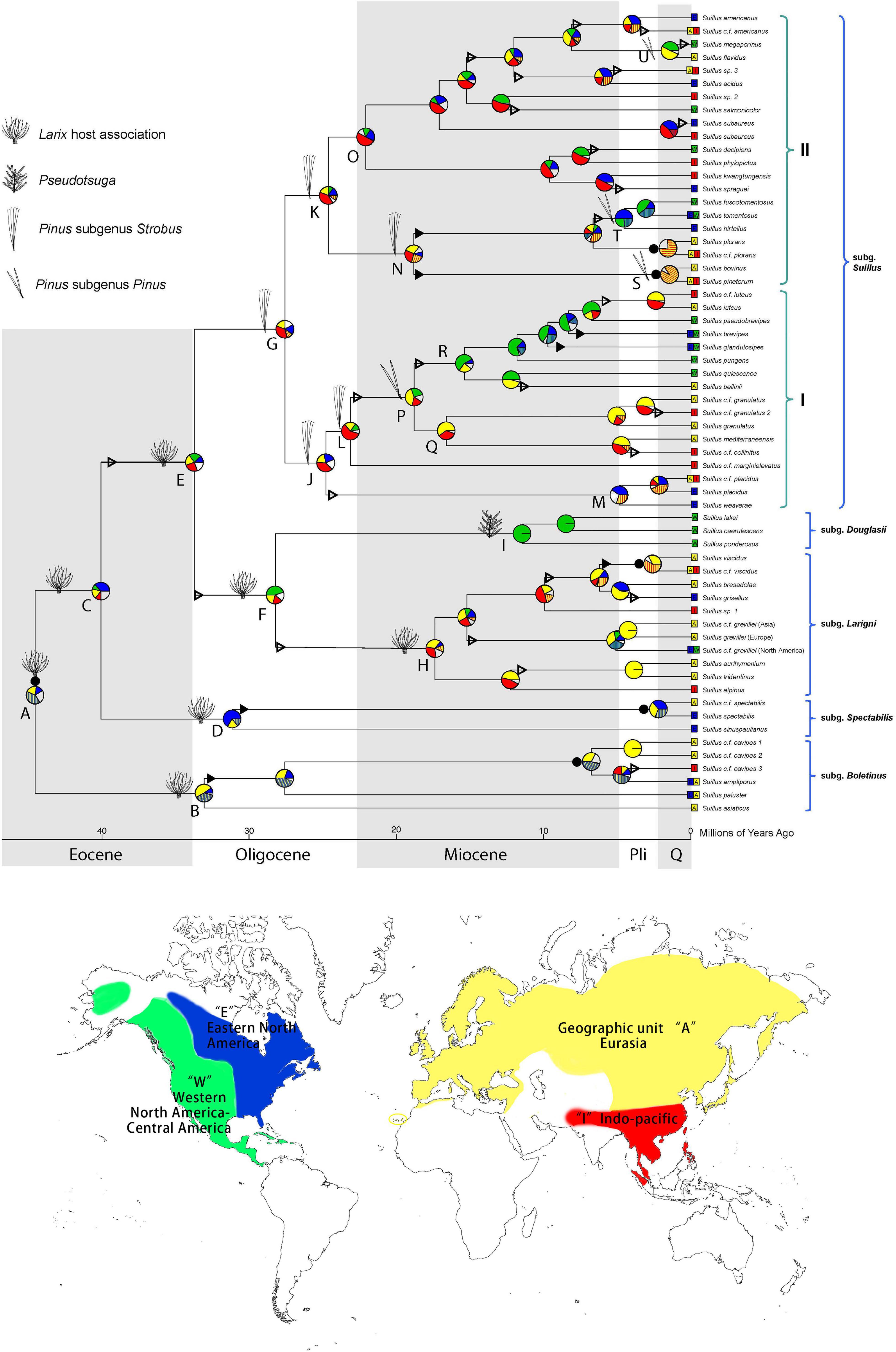
Figure 1. Historical biogeography of Suillus estimated by DEC + J model in BioGeoBEARS. Pie diagrams at each node denote geographical units or combination of units occupied by ancestral taxa. Geographic units are represented by different colors. Combined units are shown by hatching colors or by lettering. Width of pie wedges refers to the probability of that geographic unit or combination of units. White wedges indicate the sum of the units (or combined units) with individual probabilities < 15%. Triangles on branches denote “jump” dispersal events (founder effect cladogenesis), solid triangles indicate range expansion dispersal events (anagenesis), and solid circles by pie diagrams represent vicariant speciation events. Ancestral host associations were annotated at the nodes. Letters by key nodes are discussed in the text. Current geographic ranges of terminal taxa are indicated by colored boxes and lettering. Parentheses indicate the subgenera and section as annotated.
Evolutionary History and Host Associations of Suillus
Step one calibration under the Suillineae scenario at node A (Supplementary Figure 3A) indicated that the crown age of Suillus was c. 40.2 (31.9–50.8) Ma. Subgenera Boletinus, Spectabilis, and Larigini are all associated with Larix. Subgenus Douglasii switched host to Pseudotsuga at a stem age of c. 28.2 (median, 22.4–34.5) Ma in the mid-Oligocene (Supplementary Figure 4; node H). Another independent host switch, from Larix to Pinus, occurred in the late Eocene to early Oligocene, with a stem age of 33.7 (29.0–39.7) Ma (Supplementary Figure 4; node G). Ancestral host reconstruction revealed that the Pinus association initially occurred in Pinus subgenus Strobus, and later the subgenus Suillus independently switched to subgenus Pinus four separate times. The earliest switch to the subgenus Pinus was in section I at a stem age of 23.1 (18.6–27.8) Ma (Supplementary Figure 4; node N), leading to a relatively prosperous clade associated with subgenus Pinus. All other host switches to subgenus Pinus were in section II, among which the Suillus pinetorum and S. bovinus clade switched first in 18.8 (14.4–23.8) Ma (Supplementary Figure 4; node O). At node Q, a clade consisting of S. tomentosus, S. fuscotomentosus, and S. hirtellus switched hosts in the late Miocene at 6.6 (4.3–9.5) Ma. Lastly, at node R, the Suillus flavidus and S. megaporinus clade switched hosts in the late Miocene at 8.1 (5.0–11.6) Ma.
Suillus species switched to Pseudotsuga, while Pinus did not revert to the ancestral Larix association. Host associations with both subgenera of Pinus are reported for S. flavidus and S. acidus but should be further verified. For the North American S. subaureus, an initial association with Pinus subgenus Strobus is required for transferring into Quercus in later developmental stage, therefore Quercus was not included in the BayesTraits analysis (Lofgren et al., 2018). Further, Suillus sinuspaulianus was also excluded from the host reconstruction due to the uncertainty of its host association (Pomerleau and Smith, 1962; Kretzer et al., 1996).
Alternative Calibration Scenario of the Ectomycorrhizal Symbiosis Fungal Fossil
Ancestral host association supported the Suillus-Pinus association in the subgenus Suillus (Clade I, Supplementary Figure 4). The Pinus root ECM fossil can be regarded as the subgenus Suillus, thus provide an alternative calibration scenario (Lepage et al., 1997). The ECM fossil under the Suillus-Pinus scenario was calibrated at the stem of subgenus Suillus (Supplementary Figure 3B, node G). The new calibration revealed an earlier origin of Agaricomycetes in 299.2 (237.7–360.7) Ma versus 159.6 (112.8–262.9) Ma in the first scenario. It also indicated that Boletales originated 279.5 (220.2–337.5) Ma, much earlier than the estimations of scenario one and other studies (Supplementary Table 2). Further, it found that Suillus diverged from Rhizopogon in the Upper Cretaceous, and the crown age of Suillus was 71.1 (60.6–85.7) Ma (Supplementary Figure 3B and Supplementary Table 2). Suillus switched from Larix to Pinus in Paleocene 54.3 (50.4–62.2) Ma at node G; from Larix to Pseudotsuga in Eocene 46.5 (36.2–56.3) Ma at node H (Supplementary Figure 3B and Supplementary Table 2). Inclusive Suillus tree and ancestral host reconstruction under the Suillus-Pinus scenario are provided in Supplementary Figures 3, 5.
Biogeographic History of Suillus and Pinaceae
The ancestors of Larix and Suillus-Larix were circumboreally distributed (Figure 1 and Supplementary Figure 6). Extant Larix species have distinct ranges within each continent. Suillus has dispersed across the North American and Eurasian continents more frequently than Larix. Disjunct pairs of Suillus were discovered from Oligocene to as recent as the Suillus paluster and S. ampliporus species complexes (Figure 1). The ancestor of Pseudotsuga was circumboreally distributed and diverged into current species with disjunct distributions (Supplementary Figure 6). Three Suillus species were associated with Pseudotsuga menziesii in Western North America (WNA, Figure 1, clade K).
Distinct biogeographic patterns were found in Pinus but not in Suillus associated with Pinus. For the subgenus Strobus, Pinus section Parrya was limited to WNA (Supplementary Figure 6, clade H). In contrast, no Suillus specimens were found in association with Pinus section Parrya. The Pinus section Quinquefoliae originated in Eurasia and dispersed to the New World, with the most recent lineages returning to Eurasia (Supplementary Figure 6, clade I). Suillus that switched host from Larix to sect. Quinquefoliae were reconstructed in the circumboreal region (Figure 1, clade I), with their current distributions influenced by both vicariance and inter-continental dispersal events. Prevalent disjunct Eurasian and North American taxa include S. placidus, S. kwantungensis-S. spraguei, S. decipience-S. phylopictus, S. subaureus, and S. americanus. For the subgenus Pinus, section Pinus was distributed in Eurasia, with exception of Pinus resinosa in Eastern North America (ENA), and P. tropicalis in WNA. While the section Trifoliae was mainly distributed in WNA before dispersing to ENA three times for a quarter of its species. For Suillus associated with subgenus Pinus, section Suillus was originally located in the circumboreal region. Clade S retained a basal Eurasian species, dispersed to North America, and then dispersed back to Eurasia. Compared with Pinus, North American lineages in clade S shifted host from section Pinus to section Trifoliae and then to Pinus concomitant with inter-continental dispersal. Originating from a circumboreal ancestor associated with five-needle pines, clade O diverged into a few Eurasian species associated with Pinus section Pinus, and another clade T associated with Pinus section Trifoliae. Finally, clade U includes two disjunct species: Suillus megaporinus from WNA and S. flavidus from North Eurasia.
Speciation Rate Shifts in Suillus and Pinaceae
The LTT plot reveals the constant accumulation of Suillus lineages through time (Supplementary Figure 7). In the phylorate plot, diversification rate shifts were not detected for Suillus (Supplementary Figure 8). The second calibration scenario does not influence this result. Diversification rates (average range 0.12–0.14) were homogenous across Suillus lineages but varied through geological time along the phylogenetic tree branches.
Discussion
Lacking Cospeciation Patterns Between Suillus and Pinaceae
Cospeciation patterns were not identified between the Suillus subgenera and Pinaceae genera (Figure 2). Phylogenetic topologies were distinct between Suillus and Pinaceae. Basal subgenera of Suillus were all associated with Larix. Subgenera Larigni and Douglasii were sister clades, as were their hosts Larix and Pseudotsuga. Yet, given that only one species of Pseudotsuga was the host for subgenus Douglasii, this pattern was not congruent (Murata et al., 2013; Wen et al., 2014). Picea and Cathaya are not documented hosts for Suillus. Picea has been reported as one of the putative host genera for a species within the subgenus Spectabilis, but further studies are required to confirm this (Pomerleau and Smith, 1962). Pinus diverged into two subgenera, while subgenus Suillus diverged into two clades (I and II), which are not congruent with the host subgenera. If cospeciation had occurred between Suillus and Pinaceae, the phylogeny of Suillus should mirror that of Pinaceae in both topology and evolutionary time.
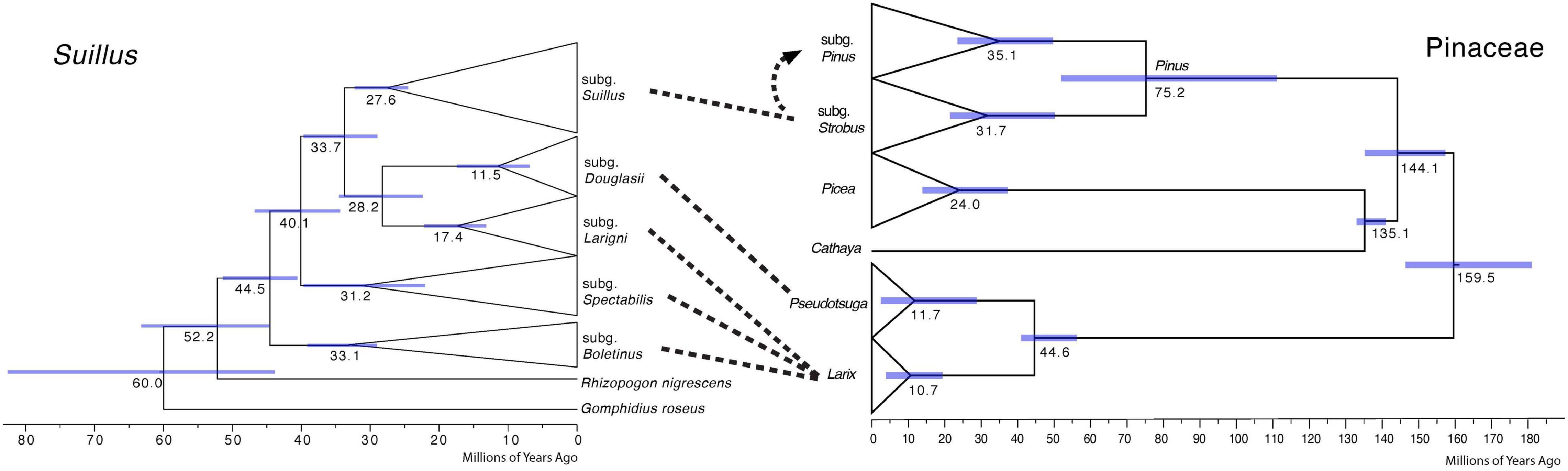
Figure 2. Comparison of calibrated phylogenies of Suillus and host Pinaceae genera. The phylogeny of Suillus is on the left, where terminal taxa are collapsed into triangles and only subgenera are shown. The phylogeny of Pinaceae is on the right, where host genera and two subgenera of Pinus are annotated. The phylogeny of Pinaceae is referred from Leslie et al. (2012). Dashed lines between the two phylogenies indicate host associations. Subgenus Suillus first established symbiosis with subgenus Strobus and then switched to subgenus Pinus as shown by a dashed arrow.
Host Shift Speciation of Suillus
Host shift speciation explained the evolutionary history of Suillus (Figure 1 and Supplementary Figure 4). To effectively adapt to a novel host, Suillus populations need to expand, accumulate variations, and experiment over sufficient time to successfully inoculate (Vienne et al., 2013). Host recognition of Suillus involves plant pathogenic pathways, and host switching might trigger plant defensive responses (Liao et al., 2016). Suillus basal lineages were associated with Larix for c. 20 million years before switching to the subgenus Strobus. Further, the subgenus Suillus was associated with the subgenus Strobus for c. 10 million years before switching to the subgenus Pinus. All these time periods were significantly longer under the second ECM fossil calibration scenario.
Suillus phylogeny did not mirror the phylogeny of Pinaceae, otherwise, host switching from Larix to a more closely related Pseudotsuga would have occurred before the switching to the Pinus subgenus Strobus, i.e., Picea and Cathaya would not have been skipped (Figure 2). The possibility cannot be excluded that extinction may help to explain the current host associations of Suillus, as Suillus could have switched to a now-extinct ancestor of Pseudotsuga, remaining only with Pseudotsuga menziesii (Murata et al., 2013; Wen et al., 2014).
A one-way direction of ECM fungal host subgenera level switching was first observed in Suillus, and this could be a unique trait in host-specific ECM fungi. After Suillus switched hosts from Larix to Pseudotsuga and the subgenus Strobus, they did not reverse to Larix. The same phenomenon is observed in the subgenus Suillus, whereby its host switched from the Pinus subgenus Strobus to the subgenus Pinus. It is plausible that the directional host switches reduce interspecific competition for ecological niches. Suillus species are pioneer ECM fungi for establishing plant seedlings and are generally not dominant in these underground communities (Gardes and Bruns, 1996; Zhou and Hogetsu, 2002; Policelli et al., 2019). Due to this ecological limitation, reducing interspecific competition is evolutionarily advantageous. Interspecific competition could also originate from another source: if different Suillus species inoculate the same host plant, they might compete for the same molecular pathways for symbiotic recognition and establishment (Liao et al., 2016).
Another question worth exploring is how Larix became the ancestral host for the genus Suillus. Truncocolumella, the sister genus of Suillus, is associated with Pseudotsuga and Tsuga (Zeller, 1940; Smith and Singer, 1959; Binder and Hibbett, 2006). The next closely related genus Rhizopogon, which is associated with Pinus, Picea, Pseudotsuga, Larix, Tsuga, Abies, and Picea of Pinaceae, as well as several Angiosperm genera (Molina and Trappe, 1994; Binder and Hibbett, 2006). Both host shift speciation and extinction may explain the basal association with Larix in the genus Suillus.
Uncertainty of the Ectomycorrhizal Symbiosis Fossil for Calibration
As a result of the ephemeral existence and soft tissue of fungal sporocarps, fungal fossils are extremely rare (Berbee and Taylor, 2010). The Eocene ECM fossil of Princeton chert has been widely used in mycological calibrations under the Suillineae scenario (Floudas et al., 2012; Wilson et al., 2012, 2017; Aime et al., 2018; Krah et al., 2018). But the application of this fossil for fungal calibration is controversial for the following reasons. The fossil was identified as a Pinus root tip with Suillineae ectomycorrhizae (Lepage et al., 1997). The fossil provided no sporocarp characters so its identity was uncertain. If host association of the ECM fossil is regarded as a prominent character, cautions must be made as other alternative calibration scenarios exist. We tried an alternative calibration scenario of the fossil in the Suillus-Pinus lineage. Yet different scenarios of the ECM fossil in the Rhizopogon-Pinus lineages have not yet been explored. As Suillus does not strictly cospeciate with Pinus, it is not applicable to compare the evolutionary history of Pinus and Suillus to find the preferred scenario.
Comparing Biogeographic Histories of Suillus and Pinaceae
The crown age of Pinaceae remains controversial, even using different fossils to calibrate (Wang et al., 2000; Lin et al., 2010; Leslie et al., 2012; Lu et al., 2014; Gernandt et al., 2016; Ran et al., 2018). As the original date of Suillus is still debated, rigid comparison between Suillus and Pinaceae referring to paleoclimate and geographic histories was refrained. Instead, general biogeographic patterns were compared with emphasis on the disjunct taxa of Eurasia-ENA and Eurasia-WNA. The biogeographic histories of Suillus and Pinaceae are generally discordant, as discussed below.
Consistent with other studies, the ancestor of Larix and Pseudotsuga was reconstructed to be within the circumboreal region (Supplementary Figure 6; Semerikov et al., 2003; Wei and Wang, 2003, 2004). However, other studies have recognized three biogeographic clades of Larix with phylogenetic supports: two Eurasian clades and one North American clade (Semerikov et al., 2003; Wei and Wang, 2003, 2004). No distinct geographic clades of Suillus associated with Larix were found in our study (Figure 1). Dispersions of Larix and Suillus were likely through the Bering land bridge (BLB) and the North Atlantic land bridge (NALB) (Wang and Ran, 2014; Jiang et al., 2019). North American and East Asian clades of Pseudotsuga are supported with phylogenetic data (Wei et al., 2010). Three Suillus species were in association with Pseudotsuga menziesii in WNA. With no ancient Suillus lineages found in the WNA, the host switching to Pseudotsuga could have assisted the Suillus dispersal to WNA. Suillus taxa were anticipated in Asian Pseudotsuga trees but were not found after extensive environmental sampling (Murata et al., 2013; Wen et al., 2014).
Vicariance plays a major role in shaping the Pinus biogeography, as all major clades of Pinus have distinct ranges (Eckert and Hall, 2006; Hao et al., 2015; Gallien et al., 2016). Long-distance dispersal across North America and Eurasia occurred approximately 3–5 times in Pinus. In contrast, inter-continental dispersal events influenced the evolution of the Suillus subgenus Suillus. North American and Eurasian disjunct taxa, arising in different geological epochs, were prevalent in subgenus Suillus. WNA is enriched with two Pinus sections and half of the Pinus diversity. The radiation of Pinus in WNA was shaped by the complex climatic history and orogeny of the Rocky Mountains and the Mexican highlands (Mastretta-Yanes et al., 2015; Antonelli et al., 2018; Hagen et al., 2019). In contrast, major clades of subgenus Suillus were not limited within biogeographic divisions, and the diversity of Suillus in WNA was relatively low. In addition, Suillus sporocarps were not yet identified from Pinus section Parrya, though Suillus was putatively reported from Pinus edulis root tip samples (Patterson et al., 2019). Suillus might have shifted to the WNA Pinus relatively recent in geological time; thus, climatic history and orogeny did not have the same effects on Suillus. Alternatively, the ability for Suillus to frequently disperse over long distances could have blurred their geographic boundaries.
Diversification Rate Shifts of Suillus
The BAMM analysis detected no speciation rate shifts within Suillus. Reported diversification rate shifts in fungi usually involve hyper diversified lineages initiated with a key innovation or migration into a significantly different environment (Kraichak et al., 2015; Sánchez-Ramírez et al., 2015; Wilson et al., 2017). Yet, Suillus host switches do not involve fundamental changes in living habit or adapting to very different environments. Host Pinus and the Suillus subgenus Suillus follow independent evolutionary trajectories. Overall, Suillus are almost absent from the Pinyon pines. Establishing associations with the subgenus Pinus lagged until the early Miocene or Eocene; therefore, Suillus likely missed the opportunity to diversify extensively in concert with subgenus Pinus.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
GM, P-GL, and RZ designed the study. RZ and XF-S performed the experiments. RZ and AW analyzed the data. RZ wrote the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Financial supports for this project were provided by a National Natural Science Foundation of China award to P-GL (NSFC-31270075) and by a National Science Foundation award to GM (DEB-0445216). Herbaria and personal collectors are appreciated for loaning specimens for DNA extraction and morphological examination: Field Museum of Natural History; University of California, Berkeley; Cryptogamic Herbarium, Kunming Institute of Botany; Royal Botanic Gardens, Kew; University of Tennessee Herbarium; Thomas D. Bruns, and Bradley R. Kropp. We thank Xiang-hua Wang, Wen-qing Zhao, Xiao-jin Wang, and Bo Li for their assistance in the field. Nikol G. Zdravkova is thanked for assistance in data analyses. This research was carried out in the Plant Genetics Laboratory at Chicago Botanic Garden, the Pritzker Laboratory for Molecular Systematics and Evolution at the Field Museum and the mycological laboratory at Kunming Institute of Botany.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.831450/full#supplementary-material
Footnotes
References
Aanen, D. K., Kuyper, T. W., Mes, T. H., and Hoekstra, R. F. (2000). The evolution of reproductive isolation in the ectomycorrhizal Hebeloma crustuliniforme aggregate (Basidiomycetes) in northwestern Europe: a phylogenetic approach. Evolution 54, 1192–1206. doi: 10.1111/j.0014-3820.2000.tb00554.x
Aime, M. C., Bell, C. D., and Wilson, A. W. (2018). Deconstructing the evolutionary complexity between rust fungi (Pucciniales) and their plant hosts. Stud. Mycol. 89, 143–152. doi: 10.1016/j.simyco.2018.02.002
Antonelli, A., Kissling, W. D., Flantua, S. G., Bermúdez, M. A., Mulch, A., Muellner-Riehl, A. N., et al. (2018). Geological and climatic influences on mountain biodiversity. Nat. Geosci. 11, 718–725. doi: 10.1038/s41561-018-0236-z
Bakker, H. C. D., Zuccarello, G. C., Kuyper, T. W., and Noordeloos, M. E. (2004). Evolution and host specificity in the ectomycorrhizal genus Leccinum. New Phytol. 163, 201–215. doi: 10.1111/j.1469-8137.2004.01090.x
Berbee, M. L., and Taylor, J. W. (2010). Dating the molecular clock in fungi–how close are we? Fungal Biol. Rev. 24, 1–16. doi: 10.1016/j.fbr.2010.03.001
Binder, M., and Hibbett, D. S. (2006). Molecular systematics and biological diversification of Boletales. Mycologia 98, 971–981. doi: 10.1080/15572536.2006.11832626
Binder, M., Larsson, K. H., Matheny, P. B., and Hibbett, D. S. (2010). Amylocorticiales ord. nov. and Jaapiales ord. nov.: early diverging clades of Agaricomycetidae dominated by corticioid forms. Mycologia 102, 865–880. doi: 10.3852/09-288
Bruns, T. D., Bidartondo, M. I., and Taylor, D. L. (2002). Host specificity in ectomycorrhizal communities: what do the exceptions tell us? Integr. Comp. Biol. 42, 352–359. doi: 10.1093/icb/42.2.352
Deslippe, J. R., Hartmann, M., Mohn, W. W., and Simard, S. (2011). Long-term experimental manipulation of climate alters the ectomycorrhizal community of Betula nana in Arctic tundra. Glob. Chang. Biol. 17, 1625–1636. doi: 10.1111/j.1365-2486.2010.02318.x
Dickie, I. A., Bolstridge, N., Cooper, J. A., and Peltzer, D. A. (2010). Co-invasion by Pinus and its mycorrhizal fungi. New Phytol. 187, 475–484. doi: 10.1111/j.1469-8137.2010.03277.x
Drummond, A. J., Suchard, M. A., Xie, D., and Rambaut, A. (2012). Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973. doi: 10.1093/molbev/mss075
Eckert, A. J., and Hall, B. D. (2006). Phylogeny, historical biogeography, and patterns of diversification for Pinus (Pinaceae): phylogenetic tests of fossil-based hypotheses. Mol. Phylogenet. Evol. 40, 166–182. doi: 10.1016/j.ympev.2006.03.009
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Farjon, A. (1990). Pinaceae: Drawings and Descriptions of the Genera Abies, Cedrus, Pseudolarix, Keteleeria, Nothotsuga, Tsuga, Cathaya, Pseudotsuga, Larix and Picea. Königstein: Koeltz Scientific Books.
Farjon, A. (2005). Pines: Drawings and Descriptions of the Genus Pinus. Leiden: Brill Academic Publishers.
Floudas, D., Binder, M., Riley, R., Barry, K., Blanchette, R. A., Henrissat, B., et al. (2012). The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336, 1715–1719. doi: 10.1126/science.1221748
Frøslev, T. G., Matheny, P. B., and Hibbett, D. S. (2005). Lower level relationships in the mushroom genus Cortinarius (Basidiomycota, Agaricales): a comparison of RPB1, RPB2, and ITS phylogenies. Mol. Phylogenet. Evol. 37, 602–618. doi: 10.1016/j.ympev.2005.06.016
Gallien, L., Saladin, B., Boucher, F. C., Richardson, D. M., and Zimmermann, N. E. (2016). Does the legacy of historical biogeography shape current invasiveness in pines. New Phytol. 209, 1096–1105. doi: 10.1111/nph.13700
Gardes, M., and Bruns, T. D. (1993). ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol. Ecol. 2, 113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x
Gardes, M., and Bruns, T. D. (1996). Community structure of ectomycorrhizal fungi in a pinus muricata forest: above- and below-ground views. Canadian Journal of Botany, 74, 1572–1583.
Gernandt, D. S., Holman, G., Campbell, C., Parks, M., Mathews, S., Raubeson, L. A., et al. (2016). Phylogenetics of extant and fossil Pinaceae: methods for increasing topological stability. Botany 94, 863–884. doi: 10.1139/cjb-2016-0064
Hagen, O., Vaterlaus, L., Albouy, C., Brown, A., Leugger, F., Onstein, R. E., et al. (2019). Mountain building, climate cooling and the richness of cold-adapted plants in the Northern Hemisphere. J. Biogeogr. 46, 1792–1807. doi: 10.1111/jbi.13653
Hao, Z., Liu, Y., Nazaire, M., Wei, X., and Wang, X. (2015). Molecular phylogenetics and evolutionary history of sect. Quinquefoliae (Pinus): implications for Northern Hemisphere biogeography. Mol. Phylogenet. Evol. 87, 65–79. doi: 10.1016/j.ympev.2015.03.013
Hayward, J., Horton, T. R., Pauchard, A., and Nuñez, M. A. (2015). A single ectomycorrhizal fungal species can enable a Pinus invasion. Ecology 96, 1438–1444. doi: 10.1890/14-1100.1
Hibbett, D. S., Grimaldi, D. A., and Donoghue, M. J. (1997). Fossil mushrooms from Miocene and Cretaceous ambers and the evolution of Homobasidiomycetes. Am. J. Bot. 84, 981–991. doi: 10.2307/2446289
Ishida, T. A., Nara, K., and Hogetsu, T. (2007). Host effects on ectomycorrhizal fungal communities: insight from eight host species in mixed conifer–broadleaf forests. New Phytol. 174, 430–440. doi: 10.1111/j.1469-8137.2007.02016.x
Jiang, D., Klaus, S., Zhang, Y. P., Hillis, D. M., and Li, J. T. (2019). Asymmetric biotic interchange across the Bering land bridge between Eurasia and North America. Natl. Sci. Rev. 6, 739–745. doi: 10.1093/nsr/nwz035
Kennedy, P. G., Izzo, A., and Bruns, T. D. (2003). There is high potential for the formation of common mycorrhizal networks between understorey and canopy trees in a mixed evergreen forest. J. Ecol. 91, 1071–1080. doi: 10.1046/j.1365-2745.2003.00829.x
Krah, F. S., Bässler, C., Heibl, C., Soghigian, J., Schaefer, H., and Hibbett, D. S. (2018). Evolutionary dynamics of host specialization in wood-decay fungi. BMC Evol. Biol. 18:119. doi: 10.1186/s12862-018-1229-7
Kraichak, E., Lucking, R., and Lumbsch, H. T. (2015). A unique trait associated with increased diversification in a hyperdiverse family of tropical lichen–forming fungi. Int. J. Plant Sci. 176, 597–606. doi: 10.1086/682061
Kretzer, A., Li, Y., Szaro, T., and Bruns, T. D. (1996). Internal transcribed spacer sequences from 38 recognized species of Suillus sensu lato: phylogenetic and taxonomic implications. Mycologia 88, 776–785. doi: 10.1080/00275514.1996.12026715
Krpata, D., Peintner, U., Langer, I., Fitz, W. J., and Schweiger, P. (2008). Ectomycorrhizal communities associated with Populus tremula growing on a heavy metal contaminated site. Fungal Biol. 112, 1069–1079. doi: 10.1016/j.mycres.2008.02.004
Landeweert, R., Hoffland, E., Finlay, R. D., Kuyper, T. W., and van Breemen, N. (2001). Linking plants to rocks: ectomycorrhizal fungi mobilize nutrients from minerals. Trends Ecol. Evol. 16, 248–254. doi: 10.1016/S0169-5347(01)02122-X
Lanfear, R., Calcott, B., Ho, S. Y. W., and Guindon, S. (2012). PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29, 1695–1701. doi: 10.1093/molbev/mss020
Lepage, B. A., Currah, R. S., Stockey, R. A., and Rothwell, G. W. (1997). Fossil ectomycorrhizae from the Middle Eocene. Am. J. Bot. 84, 410–412. doi: 10.2307/2446014
Leski, T., and Rudawska, M. (2012). Ectomycorrhizal fungal community of naturally regenerated European larch (Larix decidua) seedlings. Symbiosis 56, 45–53. doi: 10.1007/s13199-012-0164-4
Leslie, A. B., Beaulieu, J. M., Rai, H. S., Crane, P. R., Donoghue, M. J., and Mathews, S. (2012). Hemisphere-scale differences in conifer evolutionary dynamics. Proc. Natl. Acad. Sci. U. S. A. 109, 16217–16221. doi: 10.1073/pnas.1213621109
Liao, H., Chen, Y., and Vilgalys, R. (2016). Metatranscriptomic study of common and host-specific patterns of gene expression between pines and their symbiotic ectomycorrhizal fungi in the genus Suillus. PLoS Genet. 13:e1006575. doi: 10.1371/journal.pgen.1006348
Lin, C. P., Huang, J. P., Wu, C. S., Hsu, C. Y., and Chaw, S. M. (2010). Comparative chloroplast genomics reveals the evolution of Pinaceae genera and subfamilies. Genome Biol. Evol. 2, 504–517. doi: 10.1093/gbe/evq036
Lofgren, L., Nguyen, N. H., and Kennedy, P. G. (2018). Ectomycorrhizal host specificity in a changing world: can legacy effects explain anomalous current associations? New Phytol. 220, 1273–1284. doi: 10.1111/nph.15008
Lu, Y., Ran, J. H., Guo, D. M., Yang, Z. Y., and Wang, X. Q. (2014). Phylogeny and divergence times of Gymnosperms inferred from singlecopy nuclear genes. PLoS One 9:e107679. doi: 10.1371/journal.pone.0107679
Maddison, W. P., and Maddison, D. R. (2011). Mesquite: A Modular System for Evolutionary Analysis. Version 2.75. Available online at: http://mesquiteproject.org (assessed, 2015).
Martin, F., Diez, J., Dell, B., and Delaruelle, C. (2002). Phylogeography of the ectomycorrhizal Pisolithus species as inferred from nuclear ribosomal DNA ITS sequences. New Phytol. 153, 345–357. doi: 10.1046/j.0028-646X.2001.00313.x
Martin, F., Kohler, A., Murat, C., Veneaultfourrey, C., and Hibbett, D. S. (2016). Unearthing the roots of ectomycorrhizal symbioses. Nat. Rev. Microbiol. 14, 760–773. doi: 10.1038/nrmicro.2016.149
Mastretta-Yanes, A., Moreno-Letelier, A., Piñero, D., Jorgensen, T. H., and Emerson, B. C. (2015). Biodiversity in the Mexican highlands and the interaction of geology, geography and climate within the Trans-Mexican Volcanic Belt. J. Biogeogr. 42, 1586–1600. doi: 10.1111/jbi.12546
Matheny, P. B. (2005). Improving phylogenetic inference of mushrooms with RPB1 and RPB2 nucleotide sequences (Inocybe; Agaricales). Mol. Phylogenet. Evol. 35, 1–20. doi: 10.1016/j.ympev.2004.11.014
Matheny, P. B., Wang, Z., Binder, M., Curtis, J. M., Lim, Y. W., Nilsson, R. H., et al. (2007). Contributions of rpb2 and tef1 to the phylogeny of mushrooms and allies (Basidiomycota, Fungi). Mol. Phylogenet. Evol. 43, 430–451. doi: 10.1016/j.ympev.2006.08.024
Matzke, N. J. (2014). Model selection in historical biogeography reveals that founder-event speciation is a crucial process in island clades. Syst. Biol. 63, 951–970. doi: 10.1093/sysbio/syu056
Miller, M. A., Schwartz, T., Pickett, B. E., He, S., Klem, E. B., Scheuermann, R. H., et al. (2015). A RESTful API for access to phylogenetic tools via the CIPRES Science Gateway. Evol. Bioinform. Online 11, 43–48. doi: 10.4137/EBO.S21501
Molina, R., and Trappe, J. M. (1994). Biology of the ectomycorrhizal genus, Rhizopogon. New Phytol. 126, 653–675. doi: 10.1111/j.1469-8137.1994.tb02961.x
Mueller, G. M., Wu, Q., Huang, Y., Guo, S., Aldana-Gomez, R., and Vilgalys, R. (2001). Assessing biogeographic relationships between North American and Chinese macrofungi. J. Biogeogr. 28, 271–281. doi: 10.1046/j.1365-2699.2001.00540.x
Murata, M., Kinoshita, A., and Nara, K. (2013). Revisiting the host effect on ectomycorrhizal fungal communities: implications from host–fungal associations in relict Pseudotsuga japonica forests. Mycorrhiza 23, 641–653. doi: 10.1007/s00572-013-0504-0
Nehls, U., Göhringer, F., Wittulsky, S., and Dietz, S. (2010). Fungal carbohydrate support in the ectomycorrhizal symbiosis: a review. Plant Biol. 12, 292–301. doi: 10.1111/j.1438-8677.2009.00312.x
Nguyen, N. H., Vellinga, E. C., Bruns, T. D., and Kennedy, P. G. (2016). Phylogenetic assessment of global Suillus ITS sequences supports morphologically defined species and reveals synonymous and undescribed taxa. Mycologia 108, 1216–1228. doi: 10.3852/16-106
Pagel, M., and Meade, A. (2006). Bayesian analysis of correlated evolution of discrete characters by reversible-jump Markov chain Monte Carlo. Am. Nat. 167, 808–825. doi: 10.1086/503444
Paradis, E., Claude, J., and Strimmer, K. (2004). APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290. doi: 10.1093/bioinformatics/btg412
Patterson, A., Flores-Rentería, L., Whipple, A., Whitham, T., and Gehring, C. (2019). Common garden experiments disentangle plant genetic and environmental contributions to ectomycorrhizal fungal community structure. New Phytol. 221, 493–502. doi: 10.1111/nph.15352
Pigg, K. B., and Devore, M. L. (2016). A review of the plants of the Princeton chert (Eocene, British Columbia, Canada). Botany 94, 661–681. doi: 10.1139/cjb-2016-0079
Plummer, M., Best, N., Cowles, K., and Vines, K. (2008). Coda: Output Analysis and Diagnostics for MCMC. Available online at: https://cran.r-project.org/package=coda (assessed, 2015).
Policelli, N., Bruns, T. D., Vilgalys, R., and Nunez, M. A. (2019). Suilloid fungi as global drivers of pine invasions. New Phytol. 222, 714–725. doi: 10.1111/nph.15660
Pomerleau, R., and Smith, A. H. (1962). Fuscoboletinus, a new genus of the Boletales. Brittonia 14, 156–172. doi: 10.2307/2805220
Rabosky, D. L. (2014). Automatic detection of key innovations, rate shifts, and diversity-dependence on phylogenetic trees. PLoS One 9:e89543. doi: 10.1371/journal.pone.0089543
Rabosky, D. L., Grundler, M., Anderson, C., Shi, J. J., Brown, J. W., Huang, H., et al. (2014). BAMMtools: an R package for the analysis of evolutionary dynamics on phylogenetic trees. Methods Ecol. Evol. 5, 701–707. doi: 10.1111/2041-210X.12199
Ran, J. H., Shen, T. T., Wu, H., Gong, X., and Wang, X. Q. (2018). Phylogeny and evolutionary history of Pinaceae updated by transcriptomic analysis. Mol. Phylogenet. Evol. 129, 106–116. doi: 10.1016/j.ympev.2018.08.011
Ree, R. H., and Smith, S. A. (2008). Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst. Biol. 57, 4–14. doi: 10.1080/10635150701883881
Rehner, S. A., and Buckley, E. (2005). A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97, 84–98. doi: 10.1080/15572536.2006.11832842
Renner, S. S. (2005). Relaxed molecular clocks for dating historical plant dispersal events. Trends Plant Sci. 10, 550–558. doi: 10.1016/j.tplants.2005.09.010
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Ronquist, F., Teslenko, M., Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Sánchez-Ramírez, S., Etienne, R. S., and Moncalvo, J. (2015). High speciation rate at temperate latitudes explains unusual diversity gradients in a clade of ectomycorrhizal fungi. Evolution 69, 2196–2209. doi: 10.1111/evo.12722
Sato, H., Tanabe, A. S., and Toju, H. (2017). Host shifts enhance diversification of ectomycorrhizal fungi: diversification rate analysis of the ectomycorrhizal fungal genera Strobilomyces and Afroboletus with an 80-gene phylogeny. New Phytol. 214, 443–454. doi: 10.1111/nph.14368
Semerikov, V. L., Zhang, H., Sun, M., and Lascoux, M. (2003). Conflicting phylogenies of Larix (Pinaceae) based on cytoplasmic and nuclear DNA. Mol. Phylogenet. Evol. 27, 173–184. doi: 10.1016/S1055-7903(02)00447-5
Shi, X., Yu, F., Zhang, R., and Liu, P. (2016). Two new species of Suillus associated with larches in China. Mycotaxon 131, 305–315. doi: 10.5248/131.305
Smith, A. H., and Singer, R. (1959). Studies on secotiaceous fungi-IV: Gastroboletus, Truncocolumella and Chamonixia. Brittonia 11, 205–223. doi: 10.2307/2805006
Smith, M. E., Douhan, G. W., Fremier, A. K., and Rizzo, D. M. (2009). Are true multihost fungi the exception or the rule? Dominant ectomycorrhizal fungi on Pinus sabiniana differ from those on co-occurring Quercus species. New Phytol. 182, 295–299. doi: 10.1111/j.1469-8137.2009.02801.x
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stiller, J. W., and Hall, B. D. (1997). The origin of red algae: implications for plastid evolution. Proc. Natl. Acad. Sci. U. S. A. 94, 4520–4525. doi: 10.1073/pnas.94.9.4520
Tedersoo, L., and Smith, M. E. (2013). Lineages of ectomycorrhizal fungi revisited: foraging strategies and novel lineages revealed by sequences from belowground. Fungal Biol. Rev. 27, 83–99. doi: 10.1016/j.fbr.2013.09.001
Tedersoo, L., and Smith, M. E. (2017). “Ectomycorrhizal fungal lineages: detection of four new groups and notes on consistent recognition of ectomycorrhizal taxa in high-throughput sequencing studies,” in Biogeography Of Mycorrhizal Symbiosis, ed. L. Tedersoo (Berlin: Springer), 125–142. doi: 10.1007/978-3-319-56363-3_6
Thiers, H. D. (1975). The status of the genus Suillus in the United States. Nova Hedwigia 51, 247–278.
Verma, B., and Reddy, M. S. (2015). Suillus indicus sp. nov. (Boletales, Basidiomycota), a new boletoid fungus from northwestern Himalayas, India. Mycology 6, 35–41. doi: 10.1080/21501203.2014.988770
Vienne, D. M., Refregier, G., Lopezvillavicencio, M., Tellier, A., Hood, M. E., and Giraud, T. (2013). Cospeciation vs host-shift speciation: methods for testing, evidence from natural associations and relation to coevolution. New Phytol. 198, 347–385. doi: 10.1111/nph.12150
Vilgalys, R., and Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990
Wang, X. Q., and Ran, J. H. (2014). Evolution and biogeography of gymnosperms. Mol. Phylogenet. Evol. 75, 24–40. doi: 10.1016/j.ympev.2014.02.005
Wang, X., Tank, D. C., and Sang, T. (2000). Phylogeny and divergence times in Pinaceae: evidence from three genomes. Mol. Biol. Evol. 17, 773–781. doi: 10.1093/oxfordjournals.molbev.a026356
Wei, X. X., Yang, Z. Y., Li, Y., and Wang, X. Q. (2010). Molecular phylogeny and biogeography of Pseudotsuga (pinaceae): insights into the floristic relationship between taiwan and its adjacent areas. Mol. Phylogenet. Evol. 55, 776–785. doi: 10.1016/j.ympev.2010.03.007
Wei, X., and Wang, X. (2003). Phylogenetic split of Larix: evidence from paternally inherited cpDNA trnT-trnF region. Plant Syst. Evol. 239, 67–77. doi: 10.1007/s00606-002-0264-3
Wei, X., and Wang, X. (2004). Recolonization and radiation in Larix (Pinaceae): evidence from nuclear ribosomal DNA paralogues. Mol. Ecol. 13, 3115–3123. doi: 10.1111/j.1365-294X.2004.02299.x
Wen, Z., Murata, M., Xu, Z., Chen, Y., and Nara, K. (2014). Ectomycorrhizal fungal communities on the endangered Chinese Douglas-fir (Pseudotsuga sinensis) indicating regional fungal sharing overrides host conservatism across geographical regions. Plant Soil 387, 189–199. doi: 10.1007/s11104-014-2278-3
White, T. J., Bruns, T. D., Lee, S., and Taylor, J. W. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications, eds M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (Orlando: Academic Press), 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1
Wilson, A. W., Binder, M., and Hibbett, D. S. (2012). Diversity and evolution of ectomycorrhizal host associations in the Sclerodermatineae (Boletales, Basidiomycota). New Phytol. 194, 1079–1095. doi: 10.1111/j.1469-8137.2012.04109.x
Wilson, A. W., Hosaka, K., and Mueller, G. M. (2017). Evolution of ectomycorrhizas as a driver of diversification and biogeographic patterns in the model mycorrhizal mushroom genus Laccaria. New Phytol. 213, 1862–1873. doi: 10.1111/nph.14270
Wu, Q., Mueller, G. M., Lutzoni, F. M., Huang, Y., and Guo, S. (2000). Phylogenetic and biogeographic relationships of Eastern Asian and Eastern North American disjunct Suillus species (fungi) as inferred from nuclear ribosomal RNA ITS sequences. Mol. Phylogenet. Evol. 17, 37–47. doi: 10.1006/mpev.2000.0812
Zhang, R., Mueller, G. M., Shi, X., and Liu, P. (2017). Two new species in the Suillus spraguei complex from China. Mycologia 109, 296–307. doi: 10.1080/00275514.2017.1305942
Keywords: ancestral range, diversification rate, fossil calibration, host specificity, multigene phylogeny
Citation: Zhang R, Shi X-f, Liu P-g, Wilson AW and Mueller GM (2022) Host Shift Speciation of the Ectomycorrhizal Genus Suillus (Suillineae, Boletales) and Biogeographic Comparison With Its Host Pinaceae. Front. Microbiol. 13:831450. doi: 10.3389/fmicb.2022.831450
Received: 08 December 2021; Accepted: 24 February 2022;
Published: 30 March 2022.
Edited by:
Jesse L. Labbé, Invaio Sciences, United StatesReviewed by:
Tomás Allen Rush, Oak Ridge National Laboratory (DOE), United StatesMarisol Sanchez-Garcia, Swedish University of Agricultural Sciences, Sweden
Copyright © 2022 Zhang, Shi, Liu, Wilson and Mueller. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rui Zhang, cnVpekBzenUuZWR1LmNu; Gregory M. Mueller, Z211ZWxsZXJAY2hpY2Fnb2JvdGFuaWMub3Jn
 Rui Zhang1,2,3,4,5*
Rui Zhang1,2,3,4,5* Andrew W. Wilson
Andrew W. Wilson Gregory M. Mueller
Gregory M. Mueller