
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 18 April 2022
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.831037
This article is part of the Research Topic Improving Antimicrobial Peptides Translational Potential Through Peptidomimetics View all 6 articles
The uncontrollable spread of superbugs calls for new approaches in dealing with microbial-antibiotic resistance. Accordingly, the anti-virulence approach has arisen as an attractive unconventional strategy to face multidrug-resistant pathogens. As an emergent strategy, there is an imperative demand for discovery, design, and development of anti-virulence drugs. In this regard, peptidomimetic compounds could be a valuable source of anti-virulence drugs, since these molecules circumvent several shortcomings of natural peptide-based drugs like proteolytic instability, immunogenicity, toxicity, and low bioavailability. Some emerging evidence points to the feasibility of peptidomimetics to impair pathogen virulence. Consequently, in this review, we shed some light on the potential of peptidomimetics as anti-virulence drugs to overcome antibiotic resistance. Specifically, we address the anti-virulence activity of peptidomimetics against pathogens’ secretion systems, biofilms, and quorum-sensing systems.
The WHO has recognized antimicrobial resistance as a serious global public health threat (World Health Organization, 2021). It is estimated that about 700,000 deaths by antibiotic-resistant infections take place annually, but this quantity could rise to millions in the next few years (Dadgostar, 2019; World Health Organization, 2019). Antibiotic resistance negatively affects medical procedures like surgery, organ transplantation, and chemotherapy (World Health Organization, 2021). The emergence and spread of superbugs have imposed the necessity to look for non-conventional therapy to face this challenge. In this regard, anti-virulence therapy has emerged as an alternative approach to overcome the emergence and spread of antimicrobial-resistant pathogens (Fleitas Martínez et al., 2019a; Krell and Matilla, 2021). Anti-virulence therapy aims to reduce the virulence capacity of pathogens through interference with their virulence factors or virulence-associated processes, trying not to affect the pathogen’s fitness (Fleitas Martínez et al., 2019a; Krell and Matilla, 2021). Such an approach could exert less selective pressure on the selection of resistant pathogens and could make pathogens more susceptible to the host immune system (Liu et al., 2008; Dickey et al., 2017). Since anti-virulence therapy targets virulence factors associated with the pathogen, it should work in a pathogen-specific manner, which should minimize the host-microbiota damage (Dickey et al., 2017).
As a non-traditional therapeutic approach anti-virulence therapy faces translational challenges, so it has been envisioned as an adjunctive therapy more than an alternative therapy to antibiotics (Theuretzbacher and Piddock, 2019; Moir et al., 2021). In this regard, anti-virulence drugs when used in combination with an antibiotic could increase the antibiotics efficacy and diminish their cytotoxic effects (De La Fuente-Núñez et al., 2015; Dickey et al., 2017; Topa et al., 2020). In immunocompromised patients, the pathogens could persist after anti-virulence treatment, due to anti-virulence drugs not kill the pathogens. In this context, the use of combined therapy with antibiotics could be desirable (Hotinger et al., 2021a). Despite being conceived to exert less selective pressure, anti-virulence drugs are not free from the development of resistance by pathogens (Maeda et al., 2012; Maura et al., 2016).
On the other hand, as an emergent strategy, there is an urgent demand for discovery, design, and development of anti-virulence drugs. Natural peptide-based drugs have been identified as promising anti-virulence agents; however, these types of drugs tend to be associated with proteolytic instability, immunogenicity, toxicity, and low bioavailability, which could limit their applicability as anti-virulence agents (Overhage et al., 2008; Kudryashova et al., 2017; Larzabal et al., 2019; Lenci and Trabocchi, 2020; Rezende et al., 2021). In this context, peptidomimetic compounds could be a valuable source of anti-virulence drugs, since they can overcome some of the shortcomings associated with natural peptide-based drugs (De La Fuente-Núñez et al., 2015; Duprez et al., 2015). Although peptidomimetics have been exploited mainly as antimicrobial agents, experimental evidence also supports their potential as anti-virulence drugs (De La Fuente-Núñez et al., 2015; Duprez et al., 2015; Lam et al., 2017; Hershkovits et al., 2019; Cherrak et al., 2021; Lam et al., 2021). Consequently, in this review, we will address some bacterial virulence-associated processes in which peptidomimetics have shown their anti-virulence potential (Figure 1). The activity of peptidomimetics will be described in relation to bacterial secretion systems, bacterial biofilms, and quorum sensing systems, which are intimately linked with bacterial virulence.
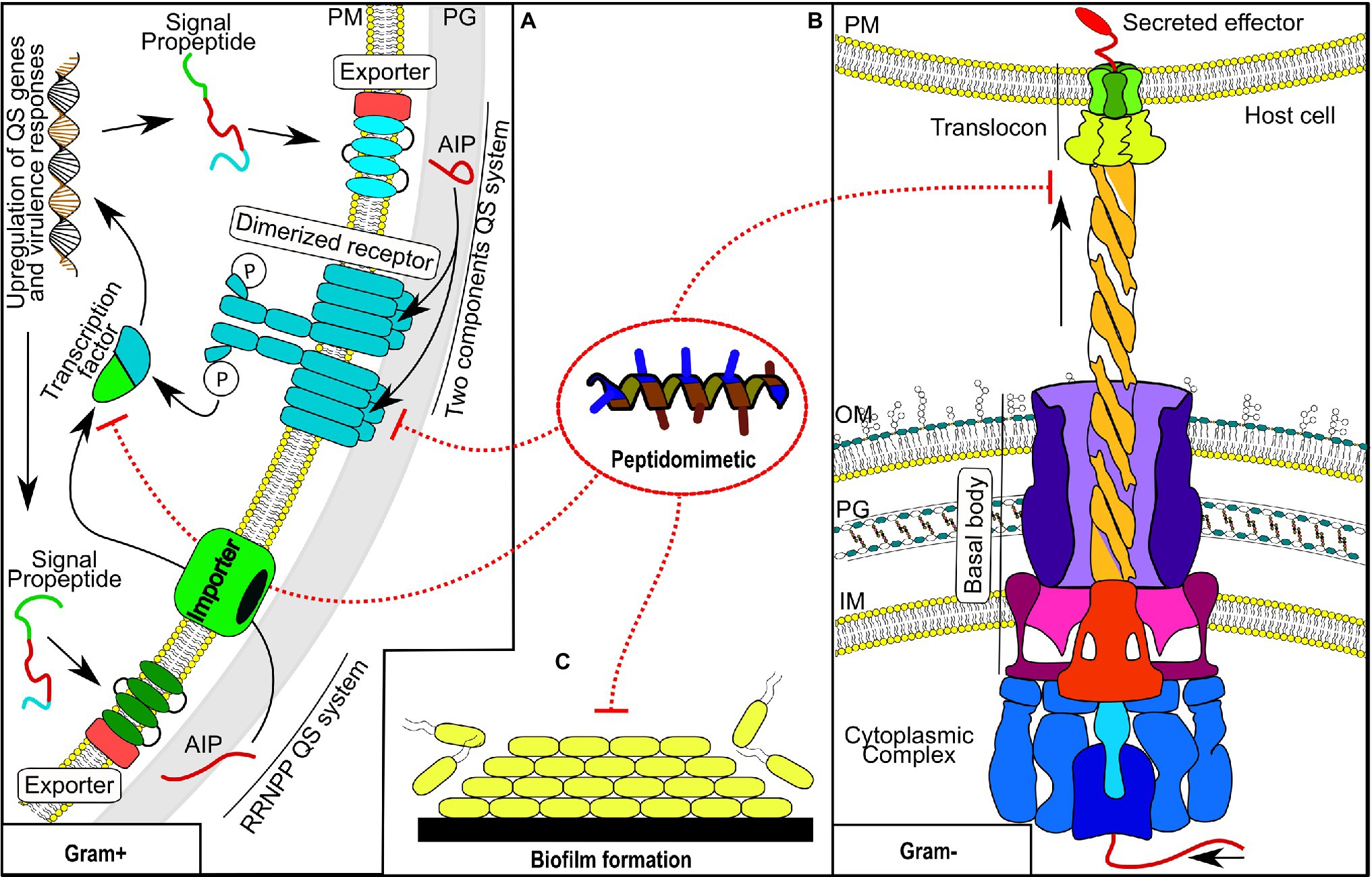
Figure 1. Bacterial virulence-associated processes targeted by peptidomimetics. (A) Two quorum-sensing systems from Gram-positive bacteria. (B) A representative Gram-negative secretion system. (C) Any phase of biofilm formation. QS, quorum sensing; PM, phospholipid membrane; PG, peptidoglycan cell wall; AIP, autoinducing peptides; OM, outer membrane; and IM, inner membrane.
Bacterial pathogens use secretion systems to secrete virulence factors that allow them to manipulate and colonize the host environment (Grishin et al., 2021). Experimental evidence supports that interference with bacterial secretion systems could be a feasible strategy to affect the virulence of relevant bacterial pathogens. It has been observed that mutant strains in secretion system components are less virulent than the wild-type strains in animal infection models (Wang et al., 2016; Sanchez-Villamil et al., 2020; Spencer et al., 2021; Taylor et al., 2021). Moreover, the inhibition of secretion systems by drugs also attenuated the pathogen’s virulence in vivo (Sheremet et al., 2018; Aburto-Rodríguez et al., 2021; Jia et al., 2021; Zhang et al., 2021; Zigangirova et al., 2021).
Biofilms are a form of bacterial growth that ensures pathogens’ persistence and dissemination (Lebeaux et al., 2014; Batoni et al., 2016; Sahoo et al., 2021). Biofilms are produced by a wide diversity of microbes growing in monomicrobial or polymicrobial communities enclosed in a protective matrix (Preda and Săndulescu, 2019; Luo et al., 2021). Biofilm development is a regulated process, in which second-messengers and quorum-sensing regulatory systems are involved (Mahto et al., 2021). The growth in biofilms confers high resistance against antibiotics and immune system clearance due to the convergence of multiple resistance mechanisms (Taylor et al., 2014; Batoni et al., 2016; Uruén et al., 2021). It is estimated that biofilms are involved in ~80% of all chronic infections and ~65% of all bacterial infections (Preda and Săndulescu, 2019). Biofilm-associated infections are challenged to treat, so several therapeutic strategies are being explored to treat these types of infections (Lebeaux et al., 2014; Fu et al., 2021; Sahoo et al., 2021).
Quorum sensing systems are signaling networks that regulate bacterial behavior and virulence genes expression (LaSarre and Federle, 2013). These communicational systems are intimately linked to bacterial biofilm development since they influence several stages of the biofilm life cycle (Lazar et al., 2021; Mahto et al., 2021). Therefore, interference with quorum-sensing systems is one of the explored strategies to overcome biofilm development by pathogenic bacteria (Brackman and Coenye, 2015; Paluch et al., 2020; Fu et al., 2021). The functionality of quorum-sensing systems resides on basic components that are druggable and tend to be absent in mammals (LaSarre and Federle, 2013; Fleitas Martínez et al., 2019b; Ellermann and Sperandio, 2020). Similarly to the observed in bacterial secretion systems, mutant strains in quorum sensing system components could become avirulent strains in infection models (Askoura et al., 2021; Kumar et al., 2021). Studies focused on quenching quorum sensing systems with inhibitors have demonstrated that targeting these communication systems is a promising strategy to face resistant pathogens (Sully et al., 2014; Daly et al., 2015; Parlet et al., 2019; Salam et al., 2021; Wang et al., 2021).
In order to invade, colonize, replicate, and settle successfully in the host, bacterial pathogens use an array of secreted effectors that allow them to manipulate the host environment (Grishin et al., 2021). These secreted effectors are regulated and efficiently delivered by specialized secretion systems (Green and Mecsas, 2016; Grishin et al., 2021). The secretion systems are closely linked with several bacterial pathogenic processes including host invasion and colonization, host immune response evasion, persistence, cytotoxicity and cellular damage, spread, and biofilm formation (Cianciotto and White, 2017; Gorasia et al., 2020; Whelan et al., 2020; Hajra et al., 2021; Rivera-Calzada et al., 2021; Viana et al., 2021; Yu et al., 2021). Consequently, these secretion systems are important for pathogen virulence, making them a target to be explored in anti-virulence therapy (Kimura et al., 2011; Sun et al., 2014; Shaffer et al., 2016; Morgan et al., 2017; Lettl et al., 2020; Cherrak et al., 2021). Currently, nine bacterial secretion systems (I-IX) have been described, from which those majorly targeted by peptidomimetics have been systems III (T3SS) and VI (T6SS) (Lam et al., 2017; Cherrak et al., 2021; Lam et al., 2021).
T3SS is a secretion system associated with flagellum components (Flagellar-T3SS) transport in motile bacteria, but also with virulence effectors transport in pathogenic Gram-negative bacteria through the Non-Flagellar-T3SS system (NF-T3SS) (Hajra et al., 2021). NF-T3SS is a multiprotein complex that transports bacterial virulence effectors directly from the pathogen cytosol into the host cell through a needle-like structure that traverses pathogen and host cell membranes (Hajra et al., 2021; Hotinger et al., 2021b). Structurally, NF-T3SS is composed of five major elements that include the cytoplasmic complex, export apparatus, basal body, needle, and translocon (Hajra et al., 2021; Hotinger et al., 2021b).
Since NF-T3SS is a multiprotein complex, protein–protein interactions could be a key process for the secretion system assembly and functionality (Hajra et al., 2021; Hotinger et al., 2021b). Consequently, interference with the protein–protein interaction network could be a strategy for the development of anti-virulence drugs that target NF-T3SS. In this regard, Stone et al. (2011) showed that a peptidomimetic based on the interaction between proteins CdsN (Type 3 system ATPase) and CdsL in Chlamydia pneumoniae inhibited the invasion of HeLa cells by this pathogen in a dose-dependent manner. This peptidomimetic was inspired by one of the binding CdsL domains present in CdsN. Specifically, peptide TRFARA (CdsL binding sequence in CdsN 265–270) was extended with a CdsN sequence that flanked it. Moreover, it was fused at the N-terminal with a membrane transport sequence (YGRKKRRQRRR), and cysteine residues were also added. These modifications yielded the peptidomimetic YGRKKRRQRRRCVVLMMDSVTRFARALC. The membrane transport sequence inclusion in the peptidomimetic facilitated its penetration of C. pneumoniae infectious elementary bodies (Stone et al., 2011).
Synthetic coiled-coil peptides also interfered with the assembly and functionality of NF-T3SS in enteropathogenic Escherichia coli (EPEC), enterohemorrhagic E. coli O157:H7 (EHEC), and the murine pathogen Citrobacter rodentium (Larzabal et al., 2010, 2013). These peptides appear to act by disrupting the oligomerization of the NF-T3SS system EspA protein by interacting with the C-terminal coiled-coil domain of EspA (Larzabal et al., 2019). Coiled-coil peptides protected mice from C. rodentium infection, avoiding intestinal damage (Larzabal et al., 2013). Although they are not peptidomimetics, the promising selective inhibitory activity against NF-T3SS exerted by coiled-coil peptides, as well as their tolerability and safety in animal models, make them a valuable scaffold for the development and design of novel peptidomimetic with enhanced anti-virulence activity.
Cyclic peptomers (cyclic peptide-peptoid hybrid molecules) are a new NF-T3SS peptidomimetic inhibitors class derived from epiphepropeptin D (EpD), a stereoisomer of the cyclic natural hexapeptide phepropeptin D. They selectively inhibit NF-T3SS activity without affecting bacterial growth or flagellar motility and without exerting considerable toxic effects on mammalian cells (Figure 2; Lam et al., 2017). Cyclic peptomers with single peptoid substitutions on the cyclic peptide backbone significantly inhibited the Yersinia pseudotuberculosis NF-T3SS system (Lam et al., 2017). However, the introduction of multiple peptoid substitutions at positions 1, 2, 3, and 4 on the peptide backbone yielded new peptomers with enhanced Y. pseudotuberculosis NF-T3SS system dose-dependent inhibition (Figure 2; Lam et al., 2017). Cyclic peptomers also inhibited the Pseudomonas aeruginosa NF-T3SS system in a dose-dependent fashion. Specifically, the peptomers EpD-3’N, EpD-1,2 N, EpD-1,2,3’N, EpD-1,2,4’N, and EpD-1,2,3´,4’N were the most active against Y. pseudotuberculosis and P. aeruginosa NF-T3SS systems (Figure 2; Lam et al., 2017). Moreover, cyclic peptomers inhibited the activity of Y. pseudotuberculosis effector proteins in HeLa cells as well as their translocation into CHO-K1 cells via NF-T3SS. Particularly, EpD-3’N and EpD-1,2,4’N exerted inhibitory activity of Y. pseudotuberculosis effector proteins in a range of concentrations of 3.75–60 and 1.875–60 μM, respectively, whereas EpD-1,2,3’N (1.875–15 μM), EpD-1,2,3´,4’N (1.875–15 μM), EpD-4’N (1.875–30 μM), and EpD-1,3’N (1.875–7.5 μM) exerted some inhibitory activity at a narrower concentration range. Nevertheless, some of these peptomers at concentrations greater than or equal to 15 μM appear to produce undesirable effects on the host actin cytoskeleton (Lam et al., 2017).
A promising cyclic peptomer (4EpDN) selectively inhibited NF-T3SS systems belonging to different T3SS families and different pathogens, such as Y. pseudotuberculosis, Yersinia enterocolitica, P. aeruginosa, Salmonella enterica serovar Thyphimurium, and Chlamydia trachomatis, without affecting bacterial growth (Lam et al., 2021). 4EpDN was derived from the previously reported peptomer EpD-1,2 N (Figure 2) via stereochemistry scan. Specifically, 4EpDN and EpD-1,2 N differ at positions 3 and 5 by D, L-leucine stereoisomers (Figure 2). Immunofluorescence microscopy analysis of 4EpDN-treated Y. pseudotuberculosis showed a reduction in the number of YscF punctates on bacteria (YscF is a T3SS needle subunit in Yersinia), suggesting that 4EpDN probably operates by interfering with the assembly or stability of T3SS needles, which impair the secretion of virulence factors. A relevant fact was that 4EpDN inhibited chlamydial infection of HeLa cells (Lam et al., 2021). In general, the experimental evidence provided by these studies suggests that cyclic peptomers are promising anti-virulence agents that act as broad-spectrum NF-T3SS inhibitors.
The type VI secretion system (T6SS) could also be targeted by peptidomimetics (Cherrak et al., 2021). T6SS has been associated with virulence in several bacterial pathogens, including some ESKAPE group members (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, P. aeruginosa, and Enterobacter cloacae), which are a serious threat for the human health due to the development of high resistance levels against a broad spectrum of antibiotics (Boyer et al., 2009; Wang et al., 2015; Kim et al., 2017; Zhou et al., 2020). Like to NF-T3SS, T6SS is a multiprotein complex for whose assembly the protein–protein interactions could be essential (Cherrak et al., 2021). Therefore, the T6SS protein–protein interaction network could be exploited for the development of anti-virulence drugs. A rationally designed cycled peptide has recently been reported that interferes with the interaction between TssK (trimer) and TssG, which impaired the T6SS baseplate biogenesis in the enteroaggregative E. coli (EAEC) (Cherrak et al., 2021). TssK and TssG interact via two-foot regions (foot 1 and 2) in TssG and by an N-terminal β sandwich shoulder domain (NTD) in TssK. The foot regions on TssG have a triangular fold and contain a conserved sequence pattern, LGXXXX1LGXXXX2LG (LG repeats), which harbor conserved hydrophobic amino acid residues (L or M) in certain positions, followed by small or basic residues (G, S, R, and K). The XXXX1 sequence is variable in composition and length, whereas the XXXX2 sequence is less variable. The TssK NTD domain has a C3-symmetric shape and harbors hydrophobic cavities involved in the interactions with the LG repeats motif of TssG. Hydrophobic interactions are essential for TssK-TssG interaction stability. Based on this structural information, a biomimetic cyclic peptide (BCP) (SRPVMG-SRPVMG-SRPVMG) was designed, mimicking the interaction interface between TssG foot domains and the TssK NTD domain (Cherrak et al., 2021). BCP was designed from TssG foot 1 and cycled via head-to-tail cyclization. The BCP cyclic structure allowed it to fit better with the C3-symmetric shape of the TssK NTD domain. In vitro assays showed that BCP binds to TssK and interferes with TssKFGE complex formation (baseplate wedge complex). Moreover, in permeabilized EAEC bacteria treated with BCP, toxic effects were not observed, and there was a reduction in the number of TssK-sfGFP containing foci. Interestingly, it was observed that pathogenic bacteria containing T6SS systems show high conservation of the TssK motif targeted by BCP. The TssK-TssG interacting interface is also conserved. Such facts suggest that BCP could exert a broad-spectrum inhibitory activity or could be a scaffold for anti-T6SS broad-spectrum drug development (Cherrak et al., 2021). Despite its promising activity, BCP needs improvement in issues associated with cell membrane diffusion, affinity, and stability.
Bacterial secretion systems have been recognized as an attractive target to develop anti-virulence drugs. A diversity of compounds that inhibit bacterial secretion systems attenuated pathogen virulence in animal models (Hudson et al., 2007; Larzabal et al., 2013; Sawa et al., 2014; Berube et al., 2017; Sheremet et al., 2018; Feng et al., 2019; Ngo et al., 2019). It suggests that these systems are a feasible target and that compounds that target them could act as anti-virulence drugs. Most of the experimental evidence about peptidomimetic inhibitory activities against bacterial secretion systems reside on in vitro assays. Therefore, the design and development of studies in animal models are imperative to have convincing evidence of their anti-virulence activity.
Autoinducer peptides (AIPs) are components of a signaling system used by some pathogenic microorganisms, mainly Gram-positive bacteria, to invade their hosts (LaSarre and Federle, 2013). AIPs are short peptides whose amino acid content varies between seven to twelve residues. In some cases, their cyclized C-terminal is a structural requirement for activity (LaSarre and Federle, 2013; Khan et al., 2015). Peptidomimetics have been studied with regard to peptide-based quorum sensing (QS) with two principal objectives. Peptidomimetics were used to identify structure–activity relationships (SARs) of natural AIPs and to develop new protease-resistant compounds. Peptide-based QS comprises two system mechanisms typical of Gram-positive bacteria (Figure 1), whose final function is to modulate virulence, communication, and/or antibiotic resistance through the expression or repression of proteins (McBrayer et al., 2020). The first one, which has been studied extensively (Li and Rebuffat, 2020), is called the two-component pathway system (Wu et al., 2020). Typically, such a system comprise two transmembrane proteins responsible for AIP secretion and later interaction for activity, respectively (Khan et al., 2015; Wu et al., 2020). It has a transcription factor responsible for the QS system expression, which is activated after phosphorylation by a dimerized receptor (Khan et al., 2015; McBrayer et al., 2020). Some categories of this system are described as the accessory gene regulator (Agr system) from S. aureus (Jenul and Horswill, 2019), the competence operon (Com system) from Streptococcus pneumoniae (Chandler and Morrison, 1987), and the system regulator (Fsr system) from Enterococcus faecalis (Qin et al., 2000). The second system involves the pathway of response regulators (RRNPP) (Perez-Pascual et al., 2016). The identified RRNPP are (PlcR) and (NprR) from Bacillus cereus (Uehara et al., 1974; Agaisse et al., 1999), (Rap) from Bacillus subtilis (Jiang et al., 2000), (PrgX) from E. faecalis (Kozlowicz et al., 2006), and (Rgg) from Streptococcus (Ibrahim et al., 2007).
The pathway of two components typically includes some membranal proteins (proteases and signal transduction) system, which allows maturation of macrocyclic peptides and extracellular signalization, respectively (Wu et al., 2020). For Staphylococci spp., the AgrA response regulator allows P2 and P3 promotors to be acted on, which upregulate the RNAII (agr operon) and RNAIII effector molecules, respectively (Jenul and Horswill, 2019). RNAIII is responsible for virulence compound expression (Morfeldt et al., 1995). The agr operon transcription translates AgrA-D proteins responsible for autoinducing and intra- or interspecific communication (Novick et al., 1995). The AgrD protein is the immature AIP (Ji et al., 1995), which should be cleaved and secreted by Sps and AgrB membranal proteins, respectively (Thoendel and Horswill, 2009, 2013). When the mature AIP is exported, it acts on the AgrC receptor, initializing a phosphorylation signal from the AgrC receptor to the AgrA regulator (Novick et al., 1995; Lina et al., 1998). The phosphorylated AgrA can upregulate P2 and P3 promotors to close the cycle (Koenig et al., 2004).
There are four (I-IV) types of AIP in Staphylococcus species (Jenul and Horswill, 2019). For S. aureus, each AIP is an activator of its cognate receptor (AgrC I-IV) but an inactivator for other receptors (Wang and Muir, 2016). Only AIP IV is an agonist on both AgrC I and IV receptors. However, for S. epidermidis, AIP I is an AgrC I activator, but an AgrC II-III antagonist, while AIPs II-III are AgrC II-III activators, but AgrC I antagonists (West et al., 2021). Studies have shown that AIPs’ agonism or antagonism activity could be correlated to twist the linker interdomains (between sensor and HK) of the ArgC receptor (Wang et al., 2017b). Activation or inhibition performed by ArgC receptors could be governed by the opposite rotation of such a linker promoted by AIP binding (Wang and Muir, 2016). Some AIP features are responsible for peptide/argC interaction. In principle, natural AIPs should have their C-terminal extremity cyclized (Khan et al., 2015). It is presumed that macrocyclic structure favors circular and planar disposition of side-chains in AIP amino acids for ArgC interaction (Horswill and Gordon, 2020). Unfortunately, for synthetic AIPs, macrocycle synthesis seems to be an obstacle to clinical development (Gordon, 2020). However, some linear peptidomimetics (peptide and peptoid mixture) were shown to be effective competitive antagonists of the AgrC receptor (Karathanasi et al., 2018). In that case, linear structures decrease synthesis steps, costs of production, and they may also become more stable. Interestingly, the peptide counterparts (A3, D3, and G3) of prominent peptidomimetics showed no anti-virulence activity (Karathanasi et al., 2018). This could be explained by the structural conformation of the peptides in solution (Tal-Gan et al., 2016). While natural peptides could adopt β-sheet or α-helical structures, these peptoids would interrupt secondary structures, facilitating specific disposition of the side-chain length necessary for receptor interaction. Nevertheless, this is not a general rule. For the S. pneumoniae Com system, AIPs (named CSP1 and CSP2) showed some degree of helicity necessary for AIP/cognate receptor interaction (Yang et al., 2017). The fundamental activity may occur due to a hydrophobic patch induced by secondary peptide structure (McBrayer et al., 2020). An example of this phenomenon is CSP2-d10, whose D-amino acid did not interrupt such a hydrophobic patch and therefore maintained the agonistic activity (Yang et al., 2017).
For S. epidermidis, some amino acids in the endocyclic segment (9, 10, 11, and 12) are important for AIP II-III/ArgC interactions, while exocyclic amino acids seem to be efficient for AIP switch activity (agonism/antagonism) (West et al., 2021). For AIP I/ArgC interaction, amino acids positioned at 3, 5, 6, 7, and 8 were seen to be important for AIP I/ArgC interaction, and the third residue was responsible for switch activity (Yang et al., 2016). Peptidomimetics involving ArgC interactions are shown in Table 1. For S. aureus, for example, a D-enantiomer scan in AIP III revealed that (Ile1 and Asp2) substitutions were potent inhibitors of AgrC I-IV receptors, while other substitutions showed a preference for some receptors or low activity on them (Tal-Gan et al., 2013). Interestingly, three acetylated derivatives of the same AIP showed more potent antagonism on the four-receptor type than D-enantiomer counterparts (Tal-Gan et al., 2013). AIP acetylation on the N-terminal had previously been identified. This modification, substituting extracyclic amino acids in AIP II, showed to be a shift that would turn agonist AIPs into antagonists in the AgrC system (Lyon et al., 2002). It is suggested that the exocyclic tail from AIPs should be extended far from the macrocycle for agonist activity, while in antagonist activity, the tail should be folded over the macrocycle (Horswill and Gordon, 2020).
It is not surprising that some modified AIPs without extracyclic amino acids show antagonist activity with similar potency to those natural agonists (Tal-Gan et al., 2016). A study of 63 peptidomimetics was developed. Among them, three acetylated homologues of AIP II, with two hydrophilic intracyclic amino acids substituted by methylene (n = 5–8), showed antagonist activity on AgrC I-IV (Vasquez et al., 2017). A battery of synthetic analogues was developed from a peptidomimetic (n7OFF) designed by the authors (Table 1). Among them, five peptidomimetics showed prominent anti-virulence activity, of which Bnc3 was the most potent (Vasquez and Blackwell, 2019). These last investigations involved chemical modifications into the AIPs. So far, antagonistic activity of peptidomimetics has been developed from D-enantiomers, acetylated peptoids, linearized AIPs, or those with chemical modifications. Also, peptidomimetics with additional modification showed an effect on S. aureus (Table 1). The lipopeptide-like AIP, decanoyl-ornithyl-ornithyl-dodecanoyl-ornithyl-amide (C10OOc12O), had anti-virulence activity on the Agr-MRSA system (Hershkovits et al., 2019). A current study made on other Staphylococci spp. shows that a modified AIP makes the difference in AIP/receptor activity. D-enantiomers of AIP III, from S. epidermidis, were tested on AgrC system. Here, the Ser10 epimer held its agonist activity, but when the substitution was on the Lys4 epimer, the peptide showed a strong antagonist activity (West et al., 2021). Other substitutions by D-enantiomers in AIPs II and III reported in this work did not show positive results.
SAR studies with peptidomimetics in Com and Fsr systems appear less in the literature. These systems are composed of homologous proteins of the agr system (Gray et al., 2013; Li and Rebuffat, 2020). A histidine kinase (ComD and FsrC, respectively) is responsible for AIP interaction and the signaling for transcription regulation (Gray et al., 2013). In a competence-inducing QS system, cyclic structure in AIPs seems not to be a requirement (LaSarre and Federle, 2013). Like AIP in the agr system, the competence stimulating peptide (CSP) is responsible for targeting ComD receptors (Novick and Geisinger, 2008). In such a system, there are two CSP types (CSP1 and CSP2) for their respective cognate targets (Pozzi et al., 1996). A systematic D-enantiomer scan was done using CSP1 and CSP2 on receptors from S. pneumoniae (Table 1). Among the variants, some CSP1 or CSP2 analogs enhanced the agonistic activity, but only a CSP2 variant (CSP2-E1ADD10) had potent antagonistic activity on ComD2 (Yang et al., 2017). Curiously, unlike AIPs in Staphylococci spp., each CSP type is not an antagonist of its noncognate receptor. Subsequent modifications led to the most potent antagonist targeting both ComD1 and ComD2 receptors (Koirala and Tal-Gan, 2020). This and another 13 analogs, with norvaline and D-enantiomer substitutions in the fourth and tenth positions, respectively, showed antagonistic activity on the ComD2 receptor (Koirala and Tal-Gan, 2020). The last of them to be developed, CSP1-E1A-cyc (Dap6E10), increased its resistance to degradation, and was nontoxic in vivo (Table 1). CSP1-E1A-cyc and another cyclized analog that has the Lys4 substituted with 2,3-diaminopropionic acid showed ComD1 and ComD2 antagonism (Yang et al., 2020). Furthermore, D-enantiomers or N-methyl analogs hold agonistic activity on S. mutans ComD receptors (Table 1). Importantly, the 18-CSP(DL4) epimer showed enhanced activity in comparison with its natural counterpart (Bikash et al., 2018). Moreover, none of the N-methyl peptidomimetics surpassed the original analog’s activity (Bikash and Tal-Gan, 2019). ComD inhibition activity by these peptidomimetics was not detected (Bikash and Tal-Gan, 2019).
Some SAR studies were made with the Fsr system from E. faecalis (Table 1). In this system, the AIP named gelatinase biosynthesis-activating pheromone (GBAP) is responsible for FsrC receptors’ interaction (Qin et al., 2000). Like AIPs in Staphylococci spp., GBAPs are cyclized on their carboxyl extremity (LaSarre and Federle, 2013). From three libraries made with modified GBAP (D-enantiomers and alanine scanned and acetylated), some analogs were discovered with equivalent or more potent agonistic activity, but others with low antagonistic activity (McBrayer et al., 2017). To investigate the best analog (Ac-GBAP-desQ1N2), an N-methyl scan was performed. Only the Ac-GBAP-Des(Q1N2)[N-MeF7] analog showed similar agonistic activity (McBrayer et al., 2019). Fortunately, for this system, some antagonistic candidates were developed (Table 1). ZBzl-YAA5911, an analog with N-terminal benzyloxycarbonylated (Z) and the Tyr5 added to the benzyl (Bzl) group, was the most potent both in vitro and in vivo (Nakayama et al., 2013).
The RRNPP pathway system has a mechanism like the two-component pathway system. In this system, AIP is translated, exported, and internalized again for target interaction (Perez-Pascual et al., 2016). It is also involved in intra- and interspecific communications (Verdugo-Fuentes et al., 2020). Unlike the pathway of the two-component system, previously mentioned, this system imports the respective AIP to target intracellular factors (Perez-Pascual et al., 2016; McBrayer et al., 2020). Few SAR studies have been done using this system. Due to the intracellular target localization, research into AIPs from the RRNPP pathway may be more difficult. An additional challenge could be the design of peptides to move across the membrane (Wu et al., 2020). Several compounds, which target diverse points in the AIP system, have been investigated to achieve QS modulation in Gram-positive bacteria (Fleitas Martínez et al., 2019b). For example, to inhibit the AIP/cognate receptor interaction of the Rgg system, some drug-like molecules have been showed to be successful (Aggarwal et al., 2015). However, these molecules do not have a peptide nature. Among the PlcR, NprR, Rap, PrgX, and Rgg systems identified until now (Perez-Pascual et al., 2016; McBrayer et al., 2020), only PlcR was approached from this perspective (Table 1). The AIP (named PapR from this system) is responsible for the tetratricopeptide repeat (TPR)-type regulatory domain of PlcR interaction (Bouillaut et al., 2008). For Bacillus cereus, a D-enantiomer scan of PapR7 showed two analogs with potent antagonistic activity (Yehuda et al., 2018). Another analog displayed more potent agonistic activity than its original counterpart. Overall, for the RRNPP pathway, it is necessary to increase studies to reveal molecular mechanisms for AIPs/cognate receptor interaction. Importantly, attention is called to develop peptidomimetics for anti-virulence activity on pathogenic bacteria.
Several peptidomimetics have been used as pharmacological tools to test AIPs-QS system interactions (Table 1). Most of them showed promising in vitro activities. Unfortunately, few works identified the efficacy of these peptidomimetics in vivo. Three peptidomimetics that exerted anti-virulence activities were reported, curiously, belonging to Fsr and Com systems. ZBzl-YAA5911 protected against the retinal damage in a rabbit endophthalmitis model (Nakayama et al., 2013), while GBAP-N5[YBzl]M11A, another GBAP analog, inhibited the formation of biofilm by wild-type E. faecalis (McBrayer et al., 2018). CSP1-E1Acyc(Dap6E10), an AIP from the Com system, attenuated the mortality caused by S. pneumoniae in an acute pneumonia mouse model (Yang et al., 2020). The studies suggest that peptidomimetics that exert a quorum-sensing inhibitory activity can confer protection against pathogens. However, more research must be conducted on this topic.
Bacterial biofilms are a predominant type of bacterial growth, where a defensive extracellular polymeric matrix composed mainly of proteins, DNA, RNA, polysaccharides, water, and ions protect the bacteria (Sahoo et al., 2021). The persistence, dissemination, and virulence of bacterial pathogens are facilitated by growing in biofilms (Sahoo et al., 2021). The high antibiotic resistance in biofilms is associated with the convergence of several intrinsic, acquired and adaptive resistance mechanisms (Taylor et al., 2014; Uruén et al., 2021). These mechanisms comprise reduced penetration and sequestering of antimicrobials by the extracellular matrix, high levels of degradative enzymes, horizontal resistance gene transfer, and metabolically heterogeneous bacterial population including persisters (Taylor et al., 2014; Bellich et al., 2018). To develop anti-virulence therapies, biofilms are attractive targets, due to their involvement in virulence and antibiotic resistance.
The treatment of biofilm-associated infections with conventional antimicrobials is very challenging since biofilms are highly heterogeneous entities (structurally, temporarily, and spatially), where multiple resistance mechanisms converge (Taylor et al., 2014; Batoni et al., 2016). Consequently, alternative therapies are being explored against biofilms, and anti-virulence therapy is one of them (Fleitas Martínez et al., 2019a; Xu et al., 2021). Anti-virulence therapy aims to impair biofilm formation and development, trying to exert a marginal effect on pathogen viability (Fleitas Martínez et al., 2019a). In this regard, some peptidomimetics have shown anti-biofilm activity at sub-inhibitory concentrations [in reference to minimum inhibitory concentration (MIC)] (De La Fuente-Núñez et al., 2015).
The first step to biofilm formation is the reversible deposition and attachment of planktonic bacterial cells on surfaces. The blockage of this initial step could impair biofilm establishment. Four synthetic polyurethanes (mLys/mPhe, mLys/mAla, mArg/mAla, and mArg/mPhe) that mimic anti-biofilm peptides were reported (Table 2; Vishwakarma et al., 2021). Each synthetic polyurethane consisted of charged and hydrophobic pendant groups with a charge-to-hydrophobicity ratio of 60/40. The charged pendant groups mLys and mArg structurally mimic lysine and arginine amino acids, respectively, whereas hydrophobic pendant groups mPhe and mAla structurally mimic phenylalanine and alanine amino acids, respectively (Figure 3; Vishwakarma et al., 2021). Polyurethane antimicrobial activity appears to be affected in enriched mediums like Lysogeny Broth (LB) and Muller Hinton Broth (MH) due to complexation with protein present at media and poor solubility at concentration higher than 250 μg.ml−1. At 0.25 x Minimum Biofilm Inhibitory Concentration (MBIC), synthetic polyurethanes notoriously affected the attachment of P. aeruginosa PAO1 to a glass surface when compared with the cationic cyclic AMP polymyxin B (~0.5× MBIC) and ciprofloxacin (0.5× MBIC). The most active synthetic polyurethane was mLys/mPhe. Zeta-potential measurements of synthetic polyurethane-treated cells suggest that these peptidomimetics modify the superficial charge of P. aeruginosa PAO1. The changes in bacterial surface charge induced by the synthetic polyurethanes may constitute one of the mechanisms by which they interfere with the bacterial adherence to the abiotic surfaces since electrostatic interactions between bacteria and surfaces are important to the initial attachment (Tuson and Weibel, 2013). Experimental evidence based on membrane permeability assay using N-phenyl-1-naphthylamine (NPN) points to non-membranolytic interactions between synthetic polyurethanes and bacteria, because polyurethanes do not provoke bacterial membrane permeabilization (Vishwakarma et al., 2021). Consequently, the authors suggested that, at low concentrations, synthetic polyurethanes inhibit biofilm formation by interacting with the bacterial surface in such a way that bacterial attachment is impaired without killing the bacteria (Vishwakarma et al., 2021). An interesting observation was that the attached P. aeruginosa PAO1 cells, when treated with ciprofloxacin, showed a filamentous appearance, which has been associated with antibiotic resistance development (Bos et al., 2015). However, the synthetic polyurethane-treated cells do not show this filamentous phenotype. Further experimental evidence showed that mLys/mAla and mLys/mPhe stimulated twitching motility, whereas mArg/mAla and mArg/mPhe noticeably reduced swarming motility. Twitching motility stimulation and swarming motility inhibition have been identified as mechanisms through which AMPs exert their anti-biofilm activity (Overhage et al., 2008; De La Fuente-Núñez et al., 2012). Synthetic polyurethanes appear to interfere with pathogen biofilm development through modulation of bacteria-surface interactions, and their anti-biofilm activity is exerted without toxicity toward mammalian cells, which made them attractive anti-virulence agents (Table 2).
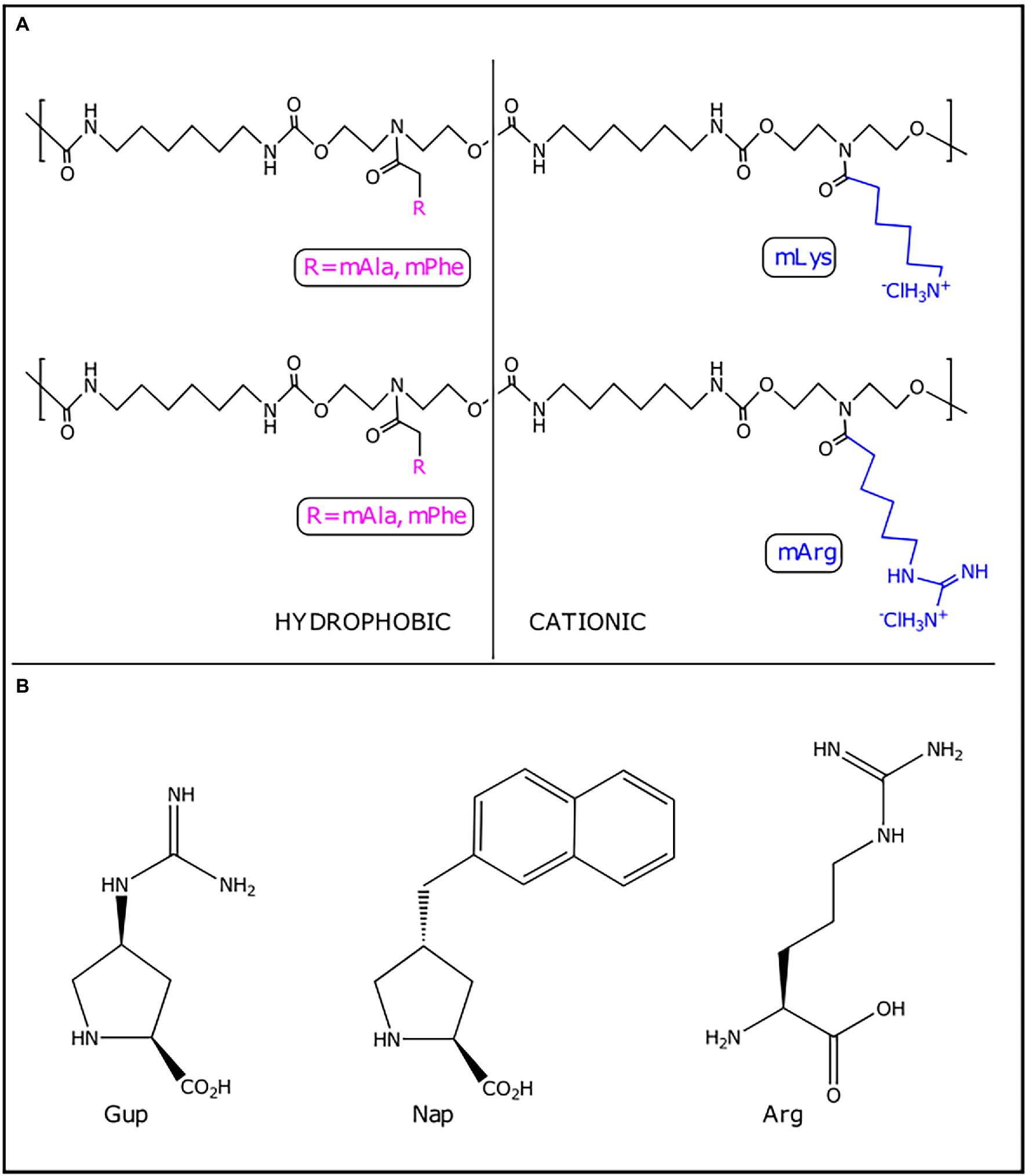
Figure 3. Peptidomimetics with anti-biofilm activity. (A) Synthetic polyurethanes. (B) Key residues in functionalized proline-rich peptides. Gup, guanidinoproline; Arg, arginine; and Nap, naphthylproline.
Nal-P-113 (AKR-Nal-NalGYKRKF-Nal-NH2) is a salt-resistant peptidomimetic derived from the peptide P-113 via substitution of their histidine residues by β-naphthylalanine residues (Yu et al., 2011). Nal-P-113 appears to exert anti-biofilm activity against Porphyromonas gingivalis W83 biofilms through different mechanisms, dependent on peptide concentrations (Wang et al., 2017a). At high concentrations, Nal-P-113 exerts its anti-biofilm activity by killing the bacteria, whereas at sub-inhibitory concentrations (respective to planktonic growth), the exerted anti-biofilm activity appears to involve several molecular pathways, as suggested by gene expression data from microarray gene analysis between Nal-P-113-treated P. gingivalis W83 and non-treated bacteria (Wang et al., 2017a). In the Nal-P-113-treated bacteria, a significant downregulation was observed in genes that encode for cationic outer membrane proteins OmpH-1 and OmpH-2, which are recognized as virulence factors and have been detected in another study with upregulated expression on biofilms (Romero-Lastra et al., 2017). The fimB (PG2133) gene that encodes for FimB (Major Fimbrium Anchoring Subunit) was downregulated. FimB is involved in biofilm formation and adhesion onto host and bacterial cells (Uniprot, 2021). Then, sub-inhibitory concentrations of Nal-P-113 appear to induce surface changes in P. gingivalis that affect the establishment of biofilms. In line with this, scanning electron microscopy analysis of Nal-P-113-treated bacteria showed their inability to fuse together (Wang et al., 2017a). Furthermore, other sets of downregulated genes encode for ABC transporters. Previous experimental evidence suggests that ABC transporters could be involved in cell-to-surface and cell-to-cell interactions, the authors suggested that downregulation of these ABC transporter-encoding genes could also influence the inability of the Nal-P-113-treated bacteria to form biofilms (Kolenbrander et al., 1994; Matthysse et al., 1996; Hinsa et al., 2003). Genes PG1551 and PG1552, which encode for HmuY and HmuR proteins, were downregulated. HmuY and HmuR constitute a system that allows P. gingivalis uptake and use of heme (Olczak et al., 2008). Consequently, Nal-P-113-treated bacteria showed reduced usage of hemin in comparison with the control; since hemin availability appears to influence P. gingivalis capability to form biofilms (Dashper et al., 2012); the downregulated expression of PG1551 and PG1552 could be a pathway through which Nal-P-113 disturbs biofilm formation. It was observed that in Nal-P-113-treated bacteria, the transposase-encoding genes were downregulated; however, when bacteria are formerly treated with H2O2, the subsequent treatment with Nal-P-113 did not inhibit biofilm formation. Since earlier studies showed that, in P. gingivalis, H2O2 induces the expression of transposase genes, the results suggest that transposase genes’ downregulation by Nal-P-113 is important for its anti-biofilm activity (Diaz et al., 2006; McKenzie et al., 2015).
Another explored approach to hamper biofilms via peptidomimetics is by interfering with signaling molecules, especially alarmones, which are ribonucleotides or ribonucleotide derivatives involved in the bacterial adaptive response to environmental stresses. Pioneering work by De la Fuente-Núñez et al. (2015) showed the anti-biofilm potential of the peptidomimetics DJK-5 and DJK-6 (Table 2; De La Fuente-Núñez et al., 2015). Both DJK-5 and DJK-6 are D-enantiomers related to IDR-1018, a peptide that exerted an anti-biofilm activity via sequestering and further triggering the degradation of alarmones [(p)ppGpp] (De la Fuente-Núñez et al., 2014). Similarly to IDR-1018, DJK-5 and DJK-6 appear to exert their anti-biofilm activity by promoting the degradation of [(p)ppGpp]. DJK-5 and DJK-6 exerted potent in vitro broad-spectrum biofilm inhibitory activity against the wild-type and antibiotic-resistant strains of P. aeruginosa, E. coli, A. baumannii, K. pneumoniae, S. enterica, and Burkholderia cenocepacia (De La Fuente-Núñez et al., 2015). Moreover, the in vivo potential of DJK-5 and DJK-6 was demonstrated in biofilm-infection models, since both peptidomimetics protected Caenorhabditis elegans and Galleria mellonella against P. aeruginosa PAO1 lethal infections without exerting toxic effects. An important finding was that the combination of DJK-5 and DJK-6 with ceftazidime, imipenem, ciprofloxacin, or tobramycin inhibited biofilm formation. This allowed a reduction in the quantities of antibiotic needed, since the peptidomimetic-antibiotic interactions were often synergistic, near synergistic, or additive (De La Fuente-Núñez et al., 2015). Similarly, the peptidomimetic-antibiotic combination tended to be effective in eradicating mature biofilms (De La Fuente-Núñez et al., 2015). Further experimental evidence showed the potential of the combination of DJK-6 with β-lactam antibiotics to treat biofilms produced by carbapenemase-producing K. pneumoniae (KPC) clinical isolates (Ribeiro et al., 2015).
Studies focused on cutaneous abscess in mice shed additional evidence of the potential of DJK-5 as an anti-virulence drug either as monotherapy or in combination with antibiotics (Mansour et al., 2016; Pletzer et al., 2017, 2018). A stringent response of bacterial pathogens like MRSA and P. aeruginosa appears to be linked to abscess pathology. In line with this, in CA-MRSA USA300 and P. aeruginosa LESB58-infected mice treated with DJK-5, there was a reduction in the pathological manifestations of abscess (Mansour et al., 2016). It was also observed that DJK-5 treatment repressed the production of phenol-soluble modulin (PSM) toxins by S. aureus, which could contribute to virulence attenuation since PMS toxins mediate tissue damage and immune response evasion (Mansour et al., 2016). A later study demonstrated the effectiveness of DJK-5 in treating cutaneous abscesses caused by P. aeruginosa strains. DJK-5-treated animals showed reduced dermonecrosis and bacterial counts at the abscess site (Pletzer et al., 2017). A remarkable observation was that DJK-5 suppressed rpoZ-spoT operon in vivo expression. Since rpoZ (ω subunit of the RNA polymerase) could influence relA (encode for RelA, the enzyme that synthesizes pppGpp) expression, it was hypothesized that in addition to binding and triggering ppGpp degradation, DJK-5 could also disrupt ppGpp levels through the downregulation of relA expression (Pletzer et al., 2017). Furthermore, based on a cutaneous mouse infection model, more evidence of the anti-virulence potential of DJK-5 was added, since animals treated with DJK-5 tended to have reduced bacterial load in the infection site. Combinatory DJK-5 therapy with ciprofloxacin, meropenem, erythromycin, gentamicin, and vancomycin diminished abscess sizes and/or reduced bacterial load in murine cutaneous abscesses caused by E. coli and ESKAPE pathogens (Pletzer et al., 2018). It was proposed that DJK-5 could synergize with antibiotics via disrupting the stringent-stress response and breaking the permeability barrier, which could facilitate antibiotic penetration (Pletzer et al., 2018).
Peptidomimetics that target the (p)ppGpp-mediated stringent response could be promising anti-virulence drugs to cope with biofilm-associated infections. The stringent response appears to be involved in biofilm development, a rise in persistence, abscess formation, and host-environment adaptation (Alford et al., 2019; Kundra et al., 2020). Although biofilms and abscesses are physiologically different, both are high-density bacterial infections that tend to be recalcitrant to antibiotic therapy (Mansour et al., 2016; Pletzer et al., 2018). Moreover, the (p)ppGpp-mediated stringent response is broadly distributed in bacteria and absent in mammals, which made it an attractive target for the development of drugs with broad-spectrum activity and low toxicity (De la Fuente-Núñez et al., 2014; Pletzer and Hancock, 2016).
In addition to (p)ppGpp, cyclic di-guanosine (c-di-GMP) is another second-messenger nucleotide that have been targeted by peptidomimetics (Foletti et al., 2018; Hee et al., 2020). Recently, functionalized proline-rich peptides with the capacity to bind selectively to c-di-GMP showed anti-biofilm activity against P. aeruginosa biofilms (Foletti et al., 2018). The peptides that bonded to c-di-GMP with higher affinity and selectivity contained three cationic residues of guanidinoproline (Gup) and/or arginine (Arg) and the aromatic group naphthyl (Nap) (Figure 3). This arrangement of aromatic and cationic residues is essential for the binding to c-di-GMP through π-π, electrostatic and H-bonding interactions. The peptides were assayed to evaluate their biofilm inhibitory capacity, and Ac-Gup-Gup-Nap-Arg-NH2 and Ac-Nap-Gup-Gup-Nap-Arg-NH2 were the most active peptides (Table 2; Foletti et al., 2018). Another study reported a shortened and modified c-di-GMP-sequestering peptide (CSP4) derived from CleD (a CheY-like c-di-GMP effector protein) (Hee et al., 2020). CSP4 is 19 amino acids residues in length, with its N-terminal acetylated and its C-terminal amidated, which bind to c-di-GMP specifically and with high affinity. Although CSP4 binds to c-di-GMP, when it was expressed in P. aeruginosa either alone or fused with Venus fluorescent protein, no effect was observed on biofilm formation. However, the expression of another c-di-GMP-sequestering peptide variant (CSP2) as a fused protein with maltose-binding protein (MBP) and hexahistidine (H6) (H6-MBP-CSP2) inhibited the biofilm formation (Hee et al., 2020). Further studies should be addressed to verify if c-di-GMP-sequestering peptides could penetrate the biofilm structure and sequester c-di-GMP.
Similarly to (p)ppGpp, peptidomimetics that target c-di-GMP, lowering its levels, could be a promising anti-virulence strategy, since c-di-GMP plays a critical role in the virulence of several medically important pathogens (Hall and Lee, 2018; Valentini and Filloux, 2019). However, it is important to consider that the effect of c-di-GMP levels on virulence could be specific to pathogens and type of infection (Hall and Lee, 2018). An additional element that made c-di-GMP an interesting target is that it is not biosynthesized by humans (Andersen et al., 2021).
Currently, antimicrobial resistance is a major concern for human health. The emergence and spread of multidrug resistant pathogens are forcing the search for non-conventional therapeutic strategies. In this regard, anti-virulence therapy is one of the non-conventional approaches that is being explored, and it aims to disrupt pathogen virulence but tries to exert marginal selective pressure. As a nascent strategy, it is necessary to discover, design, and develop anti-virulence compounds. Natural peptide-based drugs act as anti-virulence compounds but present some pharmacological shortcomings. Instead, peptidomimetics have improved pharmacologic properties, so they could be a valuable source of anti-virulence drugs. Although peptidomimetics have been widely explored as antimicrobials, experimental evidence supports their potential as anti-virulence drugs. Among the virulence factors and virulence-associated processes targeted by peptidomimetics are bacterial secretion systems, quorum-sensing systems, and biofilms. Bacterial secretion systems are an attractive target for the development of anti-virulence drugs, due to their link with virulence-associated processes, such as host invasion and immune system evasion. Moreover, their wide distribution in bacterial pathogens makes it possible to develop pathogen-specific anti-virulence drugs. Due to their multiprotein composition, protein–protein interactions are a particularly important process in secretion system assembly and function; consequently, protein–protein interactions could be a target to be exploited for the development of novel peptidomimetics with anti-virulence activity. Another key target for the development of peptidomimetic-based anti-virulence drugs is quorum-sensing systems, since they regulate the expression of several virulence factors. Several peptidomimetics have been designed to target quorum-sensing systems dependent on autoinducing peptides. Although peptidomimetics have been mainly involved in SAR studies, these promising molecules could be interesting for pharmacological development due to their advantages. Interestingly, in some cases, peptidomimetics keep their specificity on bacterial receptors and enhance agonistic activity, but most importantly, they can antagonize several receptors simultaneously. In SAR studies, the quorum-sensing system most targeted by peptidomimetics has been the Agr system from Staphylococci spp., whereas the RRNPP pathway has been poorly explored. Consequently, few peptidomimetics have been designed to target the RRNPP system, which reveals the lack of knowledge about this system’s action mechanism. However, for the peptide-based QS pathway, several peptidomimetic candidates appear to be promising for subsequent studies and biotechnological development.
Peptidomimetics are also an emerging approach to tackle biofilms, showing promising results. However, in the context of anti-virulence therapy, it is necessary to understand how the anti-biofilm activity of peptidomimetics could influence the production of virulence factors, since virulence factor production may be affected by the type of bacterial growth (Chua et al., 2014; Guilhen et al., 2016; Quinn et al., 2021). In this context, for example, the disintegration of biofilms could yield biofilm-dispersed cells with virulence factor profiles different to biofilm and planktonic growth states (Chua et al., 2014; Guilhen et al., 2016, 2017). Biofilm-dispersed bacteria could have a superior capacity for colonization and evasion of host immune response, thus increasing their virulence (Chua et al., 2014; Guilhen et al., 2017, 2019).
Overall, peptidomimetics appear to be promising anti-virulence drugs. However, it is necessary to get inside their anti-virulence mechanisms and it is important to develop more studies focused on assessing their efficacy in vivo.
HD performed figure design. All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
This work was supported by the CAPES, CNPq, FUNDECT, and FAPDF.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aburto-Rodríguez, N. A., Muñoz-Cázares, N., Castro-Torres, V. A., González-Pedrajo, B., Díaz-Guerrero, M., García-Contreras, R., et al. (2021). Anti-pathogenic properties of the combination of a T3SS inhibitory halogenated pyrrolidone with C-30 furanone. Molecules 26:7635. doi: 10.3390/molecules26247635
Agaisse, H., Gominet, M., Økstad, O. A., Kolstø, A. B., and Lereclus, D. (1999). PlcR is a pleiotropic regulator of extracellular virulence factor gene expression in Bacillus thuringiensis. Mol. Microbiol. 32, 1043–1053. doi: 10.1046/j.1365-2958.1999.01419.x
Aggarwal, C., Jimenez, J. C., Lee, H., Chlipala, G. E., Ratia, K., and Federle, M. J. (2015). Identification of quorum-sensing inhibitors disrupting signaling between Rgg and short hydrophobic peptides in streptococci. MBio 6, e00393–e00415. doi: 10.1128/mBio.00393-15
Alford, M. A., Pletzer, D., and Hancock, R. E. (2019). Dismantling the bacterial virulence program. Microb. Biotechnol. 12, 409–413. doi: 10.1111/1751-7915.13388
Andersen, J. B., Kragh, K. N., Hultqvist, L. D., Rybtke, M., Nilsson, M., Jakobsen, T. H., et al. (2021). Induction of native c-di-GMP phosphodiesterases leads to dispersal of Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 65, e02431–e02420. doi: 10.1128/AAC.02431-20
Askoura, M., Almalki, A. J., Lila, A. S. A., Almansour, K., Alshammari, F., Khafagy, E.-S., et al. (2021). Alteration of Salmonella enterica virulence and host pathogenesis through targeting sdiA by using the CRISPR-Cas9 system. Microorganisms 9:2564. doi: 10.3390/microorganisms9122564
Batoni, G., Maisetta, G., and Esin, S. (2016). Antimicrobial peptides and their interaction with biofilms of medically relevant bacteria. Biochim. Biophys. Acta Biomembr. 1858, 1044–1060. doi: 10.1016/j.bbamem.2015.10.013
Bellich, B., Lagatolla, C., Tossi, A., Benincasa, M., Cescutti, P., and Rizzo, R. (2018). Influence of bacterial biofilm polysaccharide structure on interactions with antimicrobial peptides: a study on Klebsiella pneumoniae. Int. J. Mol. Sci. 19:1685. doi: 10.3390/ijms19061685
Berube, B. J., Murphy, K. R., Torhan, M. C., Bowlin, N. O., Williams, J. D., Bowlin, T. L., et al. (2017). Impact of type III secretion effectors and of phenoxyacetamide inhibitors of type III secretion on abscess formation in a mouse model of Pseudomonas aeruginosa infection. Antimicrob. Agents Chemother. 61, e01202–e01217. doi: 10.1128/AAC.01202-17
Bikash, C. R., Hamry, S. R., and Tal-Gan, Y. (2018). Structure–activity relationships of the competence stimulating peptide in Streptococcus mutans reveal motifs critical for membrane protease SepM recognition and ComD receptor activation. ACS Infect. Dis. 4, 1385–1394. doi: 10.1021/acsinfecdis.8b00115
Bikash, C. R., and Tal-Gan, Y. (2019). Identification of highly potent competence stimulating peptide-based quorum sensing activators in Streptococcus mutans through the utilization of N-methyl and reverse alanine scanning. Bioorg. Med. Chem. Lett. 29, 811–814. doi: 10.1016/j.bmcl.2019.01.029
Bos, J., Zhang, Q., Vyawahare, S., Rogers, E., Rosenberg, S. M., and Austin, R. H. (2015). Emergence of antibiotic resistance from multinucleated bacterial filaments. Proc. Natl. Acad. Sci. 112, 178–183. doi: 10.1073/pnas.1420702111
Bouillaut, L., Perchat, S., Arold, S., Zorrilla, S., Slamti, L., Henry, C., et al. (2008). Molecular basis for group-specific activation of the virulence regulator PlcR by PapR heptapeptides. Nucleic Acids Res. 36, 3791–3801. doi: 10.1093/nar/gkn149
Boyer, F., Fichant, G., Berthod, J., Vandenbrouck, Y., and Attree, I. (2009). Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics 10:104. doi: 10.1186/1471-2164-10-104
Brackman, G., and Coenye, T. (2015). Quorum sensing inhibitors as anti-biofilm agents. Curr. Pharm. Des. 21, 5–11. doi: 10.2174/1381612820666140905114627
Chandler, M. S., and Morrison, D. A. (1987). Competence for genetic transformation in Streptococcus pneumoniae: molecular cloning of com, a competence control locus. J. Bacteriol. 169, 2005–2011. doi: 10.1128/jb.169.5.2005-2011.1987
Cherrak, Y., Filella-Merce, I., Schmidt, V., Byrne, D., Sgoluppi, V., Chaiaheloudjou, R., et al. (2021). Inhibiting type VI secretion system activity with a biomimetic peptide designed to target the baseplate wedge complex. MBio 12, e01348–e01321. doi: 10.1128/mBio.01348-21
Chua, S. L., Liu, Y., Yam, J. K. H., Chen, Y., Vejborg, R. M., Tan, B. G. C., et al. (2014). Dispersed cells represent a distinct stage in the transition from bacterial biofilm to planktonic lifestyles. Nat. Commun. 5:4462. doi: 10.1038/ncomms5462
Cianciotto, N. P., and White, R. C. (2017). Expanding role of type II secretion in bacterial pathogenesis and beyond. Infect. Immun. 85, e00014–e00017. doi: 10.1128/IAI.00014-17
Dadgostar, P. (2019). Antimicrobial resistance: implications and costs. Infect. Drug Resist. 12, 3903–3910. doi: 10.2147/IDR.S234610
Daly, S. M., Elmore, B. O., Kavanaugh, J. S., Triplett, K. D., Figueroa, M., Raja, H. A., et al. (2015). ω-Hydroxyemodin limits Staphylococcus aureus quorum sensing-mediated pathogenesis and inflammation. Antimicrob. Agents Chemother. 59, 2223–2235. doi: 10.1128/AAC.04564-14
Dashper, S. G., Pan, Y., Veith, P. D., Chen, Y.-Y., Toh, E. C., Liu, S. W., et al. (2012). Lactoferrin inhibits Porphyromonas gingivalis proteinases and has sustained biofilm inhibitory activity. Antimicrob. Agents Chemother. 56, 1548–1556. doi: 10.1128/AAC.05100-11
De La Fuente-Núñez, C., Korolik, V., Bains, M., Nguyen, U., Breidenstein, E. B., Horsman, S., et al. (2012). Inhibition of bacterial biofilm formation and swarming motility by a small synthetic cationic peptide. Antimicrob. Agents Chemother. 56, 2696–2704. doi: 10.1128/AAC.00064-12
De la Fuente-Núñez, C., Reffuveille, F., Haney, E. F., Straus, S. K., and Hancock, R. E. (2014). Broad-spectrum anti-biofilm peptide that targets a cellular stress response. PLoS Pathog. 10:e1004152. doi: 10.1371/journal.ppat.1004152
De La Fuente-Núñez, C., Reffuveille, F., Mansour, S. C., Reckseidler-Zenteno, S. L., Hernández, D., Brackman, G., et al. (2015). D-enantiomeric peptides that eradicate wild-type and multidrug-resistant biofilms and protect against lethal Pseudomonas aeruginosa infections. Chem. Biol. 22, 196–205. doi: 10.1016/j.chembiol.2015.01.002
Diaz, P. I., Slakeski, N., Reynolds, E. C., Morona, R., Rogers, A. H., and Kolenbrander, P. E. (2006). Role of oxyR in the oral anaerobe Porphyromonas gingivalis. J. Bacteriol. 188, 2454–2462. doi: 10.1128/JB.188.7.2454-2462.2006
Dickey, S. W., Cheung, G. Y., and Otto, M. (2017). Different drugs for bad bugs: antivirulence strategies in the age of antibiotic resistance. Nat. Rev. Drug Discov. 16, 457–471. doi: 10.1038/nrd.2017.23
Duprez, W., Bachu, P., Stoermer, M. J., Tay, S., McMahon, R. M., Fairlie, D. P., et al. (2015). Virtual screening of peptide and peptidomimetic fragments targeted to inhibit bacterial dithiol oxidase DsbA. PLoS One 10:e0133805. doi: 10.1371/journal.pone.0133805
Ellermann, M., and Sperandio, V. (2020). Bacterial signaling as an antimicrobial target. Curr. Opin. Microbiol. 57, 78–86. doi: 10.1016/j.mib.2020.08.001
Feng, C., Huang, Y., He, W., Cheng, X., Liu, H., Huang, Y., et al. (2019). Tanshinones: first-in-class inhibitors of the biogenesis of the type 3 secretion system needle of Pseudomonas aeruginosa for antibiotic therapy. ACS Cent. Sci. 5, 1278–1288. doi: 10.1021/acscentsci.9b00452
Fleitas Martínez, O., Cardoso, M. H., Ribeiro, S. M., and Franco, O. L. (2019a). Recent advances in anti-virulence therapeutic strategies with a focus on dismantling bacterial membrane microdomains, toxin neutralization, quorum-sensing interference and biofilm inhibition. Front. Cell. Infect. Microbiol. 9:74. doi: 10.3389/fcimb.2019.00074
Fleitas Martínez, O., Rigueiras, P. O., Pires, A. D. S., Porto, W. F., Silva, O. N., De la Fuente-Nunez, C., et al. (2019b). Interference with quorum-sensing signal biosynthesis as a promising therapeutic strategy against multidrug-resistant pathogens. Front. Cell. Infect. Microbiol. 8:444. doi: 10.3389/fcimb.2018.00444
Foletti, C., Kramer, R. A., Mauser, H., Jenal, U., Bleicher, K. H., and Wennemers, H. (2018). Functionalized proline-rich peptides bind the bacterial second messenger c-di-GMP. Angew. Chem. Int. Ed. 57, 7729–7733. doi: 10.1002/anie.201801845
Fu, J., Zhang, Y., Lin, S., Zhang, W., Shu, G., Lin, J., et al. (2021). Strategies for interfering with bacterial early stage biofilms. Front. Microbiol. 12:675843. doi: 10.3389/fmicb.2021.675843
Gorasia, D. G., Veith, P. D., and Reynolds, E. C. (2020). The type IX secretion system: advances in structure, function and organisation. Microorganisms 8:1173. doi: 10.3390/microorganisms8081173
Gordon, C. P. (2020). Synthetic strategies to access staphylococcus auto-inducing peptides as quorum sensing modulators. Org. Biomol. Chem. 18, 379–390. doi: 10.1039/c9ob02038a
Gray, B., Hall, P., and Gresham, H. (2013). Targeting agr-and agr-like quorum sensing systems for development of common therapeutics to treat multiple gram-positive bacterial infections. Sensors 13, 5130–5166. doi: 10.3390/s130405130
Green, E. R., and Mecsas, J. (2016). Bacterial secretion systems: an overview. Microbiol. Spectr. 4:13. doi: 10.1128/microbiolspec.VMBF-0012-2015
Grishin, A., Voth, K., Gagarinova, A., and Cygler, M. (2021). Structural biology of the invasion arsenal of gram-negative bacterial pathogens. FEBS J. doi: 10.1111/febs.15794 [Epub ahead of print].
Guilhen, C., Charbonnel, N., Parisot, N., Gueguen, N., Iltis, A., Forestier, C., et al. (2016). Transcriptional profiling of Klebsiella pneumoniae defines signatures for planktonic, sessile and biofilm-dispersed cells. BMC Genomics 17:237. doi: 10.1186/s12864-016-2557-x
Guilhen, C., Forestier, C., and Balestrino, D. (2017). Biofilm dispersal: multiple elaborate strategies for dissemination of bacteria with unique properties. Mol. Microbiol. 105, 188–210. doi: 10.1111/mmi.13698
Guilhen, C., Miquel, S., Charbonnel, N., Joseph, L., Carrier, G., Forestier, C., et al. (2019). Colonization and immune modulation properties of Klebsiella pneumoniae biofilm-dispersed cells. NPJ Biofilms Microbiomes 5, 1–11. doi: 10.1038/s41522-019-0098-1
Hajra, D., Nair, A. V., and Chakravortty, D. (2021). An elegant nano-injection machinery for sabotaging the host: role of type III secretion system in virulence of different human and animal pathogenic bacteria. Phys. Life Rev. 38, 25–54. doi: 10.1016/j.plrev.2021.05.007
Hall, C. L., and Lee, V. T. (2018). Cyclic-di-GMP regulation of virulence in bacterial pathogens. Wiley Interdiscip. Rev. RNA 9:e1454. doi: 10.1002/wrna.1454
Hee, C.-S., Habazettl, J., Schmutz, C., Schirmer, T., Jenal, U., and Grzesiek, S. (2020). Intercepting second-messenger signaling by rationally designed peptides sequestering c-di-GMP. Proc. Natl. Acad. Sci. 117, 17211–17220. doi: 10.1073/pnas.2001232117
Hershkovits, A. S., Pozdnyakov, I., Meir, O., and Mor, A. (2019). Sub-inhibitory membrane damage undermines Staphylococcus aureus virulence. Biochim. Biophys. Acta Biomembr. 1861, 1172–1179. doi: 10.1016/j.bbamem.2019.04.002
Hinsa, S. M., Espinosa-Urgel, M., Ramos, J. L., and O'Toole, G. A. (2003). Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol. Microbiol. 49, 905–918. doi: 10.1046/j.1365-2958.2003.03615.x
Horswill, A. R., and Gordon, C. P. (2020). Structure–activity relationship studies of small molecule modulators of the staphylococcal accessory gene regulator. J. Med. Chem. 63, 2705–2730. doi: 10.1021/acs.jmedchem.9b00798
Hotinger, J. A., Morris, S. T., and May, A. E. (2021a). The case against antibiotics and for anti-virulence therapeutics. Microorganisms 9:2049. doi: 10.3390/microorganisms9102049
Hotinger, J. A., Pendergrass, H. A., and May, A. E. (2021b). Molecular targets and strategies for inhibition of the bacterial type III secretion system (T3SS); inhibitors directly binding to T3SS components. Biomol. Ther. 11:316. doi: 10.3390/biom11020316
Hudson, D. L., Layton, A. N., Field, T. R., Bowen, A. J., Wolf-Watz, H., Elofsson, M., et al. (2007). Inhibition of type III secretion in Salmonella enterica serovar Typhimurium by small-molecule inhibitors. Antimicrob. Agents Chemother. 51, 2631–2635. doi: 10.1128/AAC.01492-06
Ibrahim, M., Guillot, A., Wessner, F., Algaron, F., Besset, C., Courtin, P., et al. (2007). Control of the transcription of a short gene encoding a cyclic peptide in Streptococcus thermophilus: a new quorum-sensing system? J. Bacteriol. 189, 8844–8854. doi: 10.1128/JB.01057-07
Jenul, C., and Horswill, A. R. (2019). Regulation of Staphylococcus aureus virulence. Microbiol. Spectr. 7:29. doi: 10.1128/microbiolspec.GPP3-0031-2018
Ji, G., Beavis, R. C., and Novick, R. P. (1995). Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc. Natl. Acad. Sci. 92, 12055–12059. doi: 10.1073/pnas.92.26.12055
Jia, P., Zhang, Y., Xu, J., Zhu, M., Peng, S., Zhang, Y., et al. (2021). IMB-BZ as an inhibitor targeting ESX-1 secretion system to control mycobacterial infection. J. Infect. Dis. 225, 608–616. doi: 10.1093/infdis/jiab486
Jiang, M., Shao, W., Perego, M., and Hoch, J. A. (2000). Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol. Microbiol. 38, 535–542. doi: 10.1046/j.1365-2958.2000.02148.x
Karathanasi, G., Bojer, M. S., Baldry, M., Johannessen, B. A., Wolff, S., Greco, I., et al. (2018). Linear peptidomimetics as potent antagonists of Staphylococcus aureus agr quorum sensing. Sci. Rep. 8, 1–11. doi: 10.1038/s41598-018-21951-4
Khan, B. A., Yeh, A. J., Cheung, G. Y., and Otto, M. (2015). Investigational therapies targeting quorum-sensing for the treatment of Staphylococcus aureus infections. Expert Opin. Investig. Drugs 24, 689–704. doi: 10.1517/13543784.2015.1019062
Kim, J., Lee, J.-Y., Lee, H., Choi, J. Y., Kim, D. H., Wi, Y. M., et al. (2017). Microbiological features and clinical impact of the type VI secretion system (T6SS) in Acinetobacter baumannii isolates causing bacteremia. Virulence 8, 1378–1389. doi: 10.1080/21505594.2017.1323164
Kimura, K., Iwatsuki, M., Nagai, T., Matsumoto, A., Takahashi, Y., Shiomi, K., et al. (2011). A small-molecule inhibitor of the bacterial type III secretion system protects against in vivo infection with Citrobacter rodentium. J. Antibiot. 64, 197–203. doi: 10.1038/ja.2010.155
Koenig, R. L., Ray, J. L., Maleki, S. J., Smeltzer, M. S., and Hurlburt, B. K. (2004). Staphylococcus aureus AgrA binding to the RNAIII-agr regulatory region. J. Bacteriol. 186, 7549–7555. doi: 10.1128/JB.186.22.7549-7555.2004
Koirala, B., and Tal-Gan, Y. (2020). Development of Streptococcus pneumoniae pan-group quorum-sensing modulators. Chembiochem. 21, 340–345. doi: 10.1002/cbic.201900365
Kolenbrander, P. E., Andersen, R. N., and Ganeshkumar, N. (1994). Nucleotide sequence of the Streptococcus gordonii PK488 coaggregation adhesin gene, scaA, and ATP-binding cassette. Infect. Immun. 62, 4469–4480. doi: 10.1128/iai.62.10.4469-4480.1994
Kozlowicz, B. K., Dworkin, M., and Dunny, G. M. (2006). Pheromone-inducible conjugation in Enterococcus faecalis: a model for the evolution of biological complexity? Int. J. Med. Microbiol. 296, 141–147. doi: 10.1016/j.ijmm.2006.01.040
Krell, T., and Matilla, M. A. (2021). Antimicrobial resistance: progress and challenges in antibiotic discovery and anti-infective therapy. Microb. Biotechnol. 15, 70–78. doi: 10.1111/1751-7915.13945
Kudryashova, E., Seveau, S. M., and Kudryashov, D. S. (2017). Targeting and inactivation of bacterial toxins by human defensins. Biol. Chem. 398, 1069–1085. doi: 10.1515/hsz-2017-0106
Kumar, L., Brenner, N., Brice, J., Klein-Seetharaman, J., and Sarkar, S. K. (2021). Cephalosporins interfere with quorum sensing and improve the ability of Caenorhabditis elegans to survive Pseudomonas aeruginosa infection. Front. Microbiol. 12:598498. doi: 10.3389/fmicb.2021.598498
Kundra, S., Colomer-Winter, C., and Lemos, J. A. (2020). Survival of the fittest: the relationship of (p) ppGpp with bacterial virulence. Front. Microbiol. 11:3124. doi: 10.3389/fmicb.2020.601417
Lam, H. N., Lau, T., Lentz, A., Sherry, J., Cabrera-Cortez, A., Hug, K., et al. (2021). Developing cyclic peptomers as broad-spectrum type III secretion system inhibitors in gram-negative bacteria. Antimicrob. Agents Chemother. 65, e01690–e01620.
Lam, H., Schwochert, J., Lao, Y., Lau, T., Lloyd, C., Luu, J., et al. (2017). Synthetic cyclic peptomers as type III secretion system inhibitors. Antimicrob. Agents Chemother. 61, e00060–e00117. doi: 10.1128/AAC.00060-17
Larzabal, M., Baldoni, H. A., Suvire, F. D., Curto, L. M., Gomez, G. E., Da Silva, W. M., et al. (2019). An inhibitory mechanism of action of coiled-coil peptides against type three secretion system from enteropathogenic Escherichia coli. J. Pept. Sci. 25:e3149. doi: 10.1002/psc.3149
Larzabal, M., Mercado, E. C., Vilte, D. A., Salazar-Gonzalez, H., Cataldi, A., and Navarro-Garcia, F. (2010). Designed coiled-coil peptides inhibit the type three secretion system of enteropathogenic Escherichia coli. PLoS One 5:e9046. doi: 10.1371/journal.pone.0009046
Larzabal, M., Zotta, E., Ibarra, C., Rabinovitz, B. C., Vilte, D. A., Mercado, E. C., et al. (2013). Effect of coiled-coil peptides on the function of the type III secretion system-dependent activity of enterohemorragic Escherichia coli O157: H7 and Citrobacter rodentium. Int. J. Med. Microbiol. 303, 9–15. doi: 10.1016/j.ijmm.2012.12.001
LaSarre, B., and Federle, M. J. (2013). Exploiting quorum sensing to confuse bacterial pathogens. Microbiol. Mol. Biol. Rev. 77, 73–111. doi: 10.1128/MMBR.00046-12
Lazar, V., Holban, A. M., Curutiu, C., and Chifiriuc, M. C. (2021). Modulation of quorum sensing and biofilms in less investigated gram-negative ESKAPE pathogens. Front. Microbiol. 12:676510. doi: 10.3389/fmicb.2021.676510
Lebeaux, D., Ghigo, J.-M., and Beloin, C. (2014). Biofilm-related infections: bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol. Mol. Biol. Rev. 78, 510–543. doi: 10.1128/MMBR.00013-14
Lenci, E., and Trabocchi, A. (2020). Peptidomimetic toolbox for drug discovery. Chem. Soc. Rev. 49, 3262–3277. doi: 10.1039/D0CS00102C
Lettl, C., Schindele, F., Testolin, G., Bär, A., Rehm, T., Brönstrup, M., et al. (2020). Inhibition of type IV secretion activity and growth of helicobacter pylori by cisplatin and other platinum complexes. Front. Cell. Infect. Microbiol. 10:602958. doi: 10.3389/fcimb.2020.602958
Li, Y., and Rebuffat, S. (2020). The manifold roles of microbial ribosomal peptide–based natural products in physiology and ecology. J. Biol. Chem. 295, 34–54. doi: 10.1074/jbc.REV119.006545
Lina, G., Jarraud, S., Ji, G., Greenland, T., Pedraza, A., Etienne, J., et al. (1998). Transmembrane topology and histidine protein kinase activity of AgrC, the agr signal receptor in Staphylococcus aureus. Mol. Microbiol. 28, 655–662. doi: 10.1046/j.1365-2958.1998.00830.x
Liu, C.-I., Liu, G. Y., Song, Y., Yin, F., Hensler, M. E., Jeng, W.-Y., et al. (2008). A cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulence. Science 319, 1391–1394. doi: 10.1126/science.1153018
Luo, A., Wang, F., Sun, D., Liu, X., and Xin, B. (2021). Formation, development, and cross-species interactions in biofilms. Front. Microbiol. 12:757327. doi: 10.3389/fmicb.2021.757327
Lyon, G. J., Wright, J. S., Muir, T. W., and Novick, R. P. (2002). Key determinants of receptor activation in the agr autoinducing peptides of Staphylococcus aureus. Biochemistry 41, 10095–10104. doi: 10.1021/bi026049u
Maeda, T., García-Contreras, R., Pu, M., Sheng, L., Garcia, L. R., Tomás, M., et al. (2012). Quorum quenching quandary: resistance to antivirulence compounds. ISME J. 6, 493–501. doi: 10.1038/ismej.2011.122
Mahto, K. U., Kumari, S., and Das, S. (2021). Unraveling the complex regulatory networks in biofilm formation in bacteria and relevance of biofilms in environmental remediation. Crit. Rev. Biochem. Mol. Biol. 56, 1–28. doi: 10.1080/10409238.2021.2015747
Mansour, S. C., Pletzer, D., de la Fuente-Núñez, C., Kim, P., Cheung, G. Y., Joo, H.-S., et al. (2016). Bacterial abscess formation is controlled by the stringent stress response and can be targeted therapeutically. EBioMedicine 12, 219–226. doi: 10.1016/j.ebiom.2016.09.015
Matthysse, A. G., Yarnall, H. A., and Young, N. (1996). Requirement for genes with homology to ABC transport systems for attachment and virulence of Agrobacterium tumefaciens. J. Bacteriol. 178, 5302–5308. doi: 10.1128/jb.178.17.5302-5308.1996
Maura, D., Ballok, A. E., and Rahme, L. G. (2016). Considerations and caveats in anti-virulence drug development. Curr. Opin. Microbiol. 33, 41–46. doi: 10.1016/j.mib.2016.06.001
McBrayer, D. N., Cameron, C. D., Gantman, B. K., and Tal-Gan, Y. (2018). Rational design of potent activators and inhibitors of the Enterococcus faecalis fsr quorum sensing circuit. ACS Chem. Biol. 13, 2673–2681. doi: 10.1021/acschembio.8b00610
McBrayer, D. N., Cameron, C. D., and Tal-Gan, Y. (2020). Development and utilization of peptide-based quorum sensing modulators in gram-positive bacteria. Org. Biomol. Chem. 18, 7273–7290. doi: 10.1039/D0OB01421D
McBrayer, D. N., Gantman, B. K., Cameron, C. D., and Tal-Gan, Y. (2017). An entirely solid phase peptide synthesis-based strategy for synthesis of gelatinase biosynthesis-activating pheromone (GBAP) analogue libraries: investigating the structure–activity relationships of the Enterococcus faecalis quorum sensing signal. Org. Lett. 19, 3295–3298. doi: 10.1021/acs.orglett.7b01444
McBrayer, D. N., Gantman, B. K., and Tal-Gan, Y. (2019). N-methylation of amino acids in gelatinase biosynthesis-activating pheromone identifies key site for stability enhancement with retention of the Enterococcus faecalis fsr quorum sensing circuit response. ACS Infect. Dis. 5, 1035–1041. doi: 10.1021/acsinfecdis.9b00097
McKenzie, R., Aruni, W., Johnson, N., Robles, A., Dou, Y., Henry, L., et al. (2015). Metabolome variations in the Porphyromonas gingivalis vim A mutant during hydrogen peroxide-induced oxidative stress. Mol. Oral Microbiol. 30, 111–127. doi: 10.1111/omi.12075
Moir, D. T., Opperman, T. J., Aron, Z. D., and Bowlin, T. L. (2021). Adjunctive therapy for multidrug-resistant bacterial infections: type III secretion system and efflux inhibitors. Drug Discov. Today 26, 2173–2181. doi: 10.1016/j.drudis.2021.03.031
Morfeldt, E., Taylor, D. V., Von Gabain, A., and Arvidson, S. (1995). Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 14, 4569–4577. doi: 10.1002/j.1460-2075.1995.tb00136.x
Morgan, J. M., Duncan, M. C., Johnson, K. S., Diepold, A., Lam, H., Dupzyk, A. J., et al. (2017). Piericidin A1 blocks Yersinia Ysc type III secretion system needle assembly. mSphere 2, e00030–e00117. doi: 10.1128/mSphere.00030-17
Nakayama, J., Yokohata, R., Sato, M., Suzuki, T., Matsufuji, T., Nishiguchi, K., et al. (2013). Development of a peptide antagonist against fsr quorum sensing of Enterococcus faecalis. ACS Chem. Biol. 8, 804–811. doi: 10.1021/cb300717f
Ngo, T.-D., Plé, S., Thomas, A., Barette, C., Fortuné, A., Bouzidi, Y., et al. (2019). Chimeric protein–protein interface inhibitors allow efficient inhibition of type III secretion machinery and Pseudomonas aeruginosa virulence. ACS Infect. Dis. 5, 1843–1854. doi: 10.1021/acsinfecdis.9b00154
Novick, R. P., and Geisinger, E. (2008). Quorum sensing in Staphylococci. Annu. Rev. Genet. 42, 541–564. doi: 10.1146/annurev.genet.42.110807.091640
Novick, R., Projan, S., Kornblum, J., Ross, H., Ji, G., Kreiswirth, B., et al. (1995). The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol. Gen. Genet. MGG 248, 446–458. doi: 10.1007/BF02191645
Olczak, T., Sroka, A., Potempa, J., and Olczak, M. (2008). Porphyromonas gingivalis HmuY and HmuR: further characterization of a novel mechanism of heme utilization. Arch. Microbiol. 189, 197–210. doi: 10.1007/s00203-007-0309-7
Overhage, J., Campisano, A., Bains, M., Torfs, E. C., Rehm, B. H., and Hancock, R. E. (2008). Human host defense peptide LL-37 prevents bacterial biofilm formation. Infect. Immun. 76, 4176–4182. doi: 10.1128/IAI.00318-08
Paluch, E., Rewak-Soroczyńska, J., Jędrusik, I., Mazurkiewicz, E., and Jermakow, K. (2020). Prevention of biofilm formation by quorum quenching. Appl. Microbiol. Biotechnol. 104, 1871–1881. doi: 10.1007/s00253-020-10349-w
Parlet, C. P., Kavanaugh, J. S., Crosby, H. A., Raja, H. A., El-Elimat, T., Todd, D. A., et al. (2019). Apicidin attenuates MRSA virulence through quorum-sensing inhibition and enhanced host defense. Cell Rep. 27, 187.e6–198.e6. doi: 10.1016/j.celrep.2019.03.018
Perez-Pascual, D., Monnet, V., and Gardan, R. (2016). Bacterial cell–cell communication in the host via RRNPP peptide-binding regulators. Front. Microbiol. 7:706. doi: 10.3389/fmicb.2016.00706
Pletzer, D., and Hancock, R. E. (2016). Antibiofilm peptides: potential as broad-spectrum agents. J. Bacteriol. 198, 2572–2578. doi: 10.1128/JB.00017-16
Pletzer, D., Mansour, S. C., and Hancock, R. E. (2018). Synergy between conventional antibiotics and anti-biofilm peptides in a murine, sub-cutaneous abscess model caused by recalcitrant ESKAPE pathogens. PLoS Pathog. 14:e1007084. doi: 10.1371/journal.ppat.1007084
Pletzer, D., Wolfmeier, H., Bains, M., and Hancock, R. E. (2017). Synthetic peptides to target stringent response-controlled virulence in a Pseudomonas aeruginosa murine cutaneous infection model. Front. Microbiol. 8:1867. doi: 10.3389/fmicb.2017.01867
Pozzi, G., Masala, L., Iannelli, F., Manganelli, R., Havarstein, L., Piccoli, L., et al. (1996). Competence for genetic transformation in encapsulated strains of Streptococcus pneumoniae: two allelic variants of the peptide pheromone. J. Bacteriol. 178, 6087–6090. doi: 10.1128/jb.178.20.6087-6090.1996
Preda, V. G., and Săndulescu, O. (2019). Communication is the key: biofilms, quorum sensing, formation and prevention. Discover 7:e100. doi: 10.15190/d.2019.13
Qin, X., Singh, K. V., Weinstock, G. M., and Murray, B. E. (2000). Effects of Enterococcus faecalis fsr genes on production of gelatinase and a serine protease and virulence. Infect. Immun. 68, 2579–2586. doi: 10.1128/IAI.68.5.2579-2586.2000
Quinn, R. J., Barraza, I., García-Diéguez, L., Pajon, C., Krausfeldt, L. E., Ibrahim, K., et al. (2021). Periodically disturbing the spatial structure of biofilms can affect the production of an essential virulence factor in Pseudomonas aeruginosa. mSystems 6:e0096121. doi: 10.1128/mSystems.00961-21
Rezende, S. B., Oshiro, K. G., Júnior, N. G., Franco, O. L., and Cardoso, M. H. (2021). Advances on chemically modified antimicrobial peptides for generating peptide antibiotics. Chem. Commun. 57, 11578–11590. doi: 10.1039/D1CC03793E
Ribeiro, S. M., De La Fuente-Núñez, C., Baquir, B., Faria-Junior, C., Franco, O. L., and Hancock, R. E. (2015). Antibiofilm peptides increase the susceptibility of carbapenemase-producing Klebsiella pneumoniae clinical isolates to β-lactam antibiotics. Antimicrob. Agents Chemother. 59, 3906–3912. doi: 10.1128/AAC.00092-15
Rivera-Calzada, A., Famelis, N., Llorca, O., and Geibel, S. (2021). Type VII secretion systems: structure, functions and transport models. Nat. Rev. Microbiol. 19, 567–584. doi: 10.1038/s41579-021-00560-5
Romero-Lastra, P., Sanchez, M., Ribeiro-Vidal, H., Llama-Palacios, A., Figuero, E., Herrera, D., et al. (2017). Comparative gene expression analysis of Porphyromonas gingivalis ATCC 33277 in planktonic and biofilms states. PLoS One 12:e0174669. doi: 10.1371/journal.pone.0174669
Sahoo, A., Swain, S. S., Behera, A., Sahoo, G., Mahapatra, P. K., and Panda, S. K. (2021). Antimicrobial peptides derived from Insects offer a novel therapeutic option to combat biofilm: a review. Front. Microbiol. 12:661195. doi: 10.3389/fmicb.2021.661195
Salam, A. M., Porras, G., Cho, Y.-S. K., Brown, M. M., Risener, C. J., Marquez, L., et al. (2021). Castaneroxy A from the leaves of Castanea sativa inhibits virulence in Staphylococcus aureus. Front. Pharmacol. 12:1487. doi: 10.3389/fphar.2021.640179
Sanchez-Villamil, J. I., Tapia, D., Borlee, G. I., Borlee, B. R., Walker, D. H., and Torres, A. G. (2020). Burkholderia pseudomallei as an enteric pathogen: identification of virulence factors mediating gastrointestinal infection. Infect. Immun. 89, e00654–e00720. doi: 10.1128/IAI.00654-20
Sawa, T., Ito, E., Nguyen, V. H., and Haight, M. (2014). Anti-PcrV antibody strategies against virulent Pseudomonas aeruginosa. Hum. Vaccines Immunother. 10, 2843–2852. doi: 10.4161/21645515.2014.971641
Shaffer, C. L., Good, J. A., Kumar, S., Krishnan, K. S., Gaddy, J. A., Loh, J. T., et al. (2016). Peptidomimetic small molecules disrupt type IV secretion system activity in diverse bacterial pathogens. MBio 7, e00221–e00316. doi: 10.1128/mBio.00221-16
Sheremet, A. B., Zigangirova, N. A., Zayakin, E. S., Luyksaar, S. I., Kapotina, L. N., Nesterenko, L. N., et al. (2018). Small molecule inhibitor of type three secretion system belonging to a class 2, 4-disubstituted-4H-[1, 3, 4]-thiadiazine-5-ones improves survival and decreases bacterial loads in an airway Pseudomonas aeruginosa infection in mice. Biomed. Res. Int. 2018:5810767. doi: 10.1155/2018/5810767
Spencer, B. L., Tak, U., Mendonça, J. C., Nagao, P. E., Niederweis, M., and Doran, K. S. (2021). A type VII secretion system in group B Streptococcus mediates cytotoxicity and virulence. PLoS Pathog. 17:e1010121. doi: 10.1371/journal.ppat.1010121
Stone, C. B., Bulir, D. C., Emdin, C. A., Pirie, R. M., Porfilio, E. A., Slootstra, J. W., et al. (2011). Chlamydia pneumoniae CdsL regulates CdsN ATPase activity, and disruption with a peptide mimetic prevents bacterial invasion. Front. Microbiol. 2:21. doi: 10.3389/fmicb.2011.00021
Sully, E. K., Malachowa, N., Elmore, B. O., Alexander, S. M., Femling, J. K., Gray, B. M., et al. (2014). Selective chemical inhibition of agr quorum sensing in Staphylococcus aureus promotes host defense with minimal impact on resistance. PLoS Pathog. 10:e1004174. doi: 10.1371/journal.ppat.1004174
Sun, K., Bröms, J., Lavander, M., Gurram, B. K., Enquist, P.-A., Andersson, C. D., et al. (2014). Screening for inhibition of Vibrio cholerae VipA-VipB interaction identifies small-molecule compounds active against type VI secretion. Antimicrob. Agents Chemother. 58, 4123–4130. doi: 10.1128/AAC.02819-13
Tal-Gan, Y., Ivancic, M., Cornilescu, G., and Blackwell, H. E. (2016). Characterization of structural elements in native autoinducing peptides and non-native analogues that permit the differential modulation of AgrC-type quorum sensing receptors in Staphylococcus aureus. Org. Biomol. Chem. 14, 113–121. doi: 10.1039/C5OB01735A
Tal-Gan, Y., Stacy, D. M., Foegen, M. K., Koenig, D. W., and Blackwell, H. E. (2013). Highly potent inhibitors of quorum sensing in Staphylococcus aureus revealed through a systematic synthetic study of the group-III autoinducing peptide. J. Am. Chem. Soc. 135, 7869–7882. doi: 10.1021/ja3112115
Taylor, J. C., Gao, X., Xu, J., Holder, M., Petrosino, J., Kumar, R., et al. (2021). A type VII secretion system of Streptococcus gallolyticus subsp. gallolyticus contributes to gut colonization and the development of colon tumors. PLoS Pathog. 17:e1009182. doi: 10.1371/journal.ppat.1009182
Taylor, P. K., Yeung, A. T., and Hancock, R. E. (2014). Antibiotic resistance in Pseudomonas aeruginosa biofilms: towards the development of novel anti-biofilm therapies. J. Biotechnol. 191, 121–130. doi: 10.1016/j.jbiotec.2014.09.003
Theuretzbacher, U., and Piddock, L. J. (2019). Non-traditional antibacterial therapeutic options and challenges. Cell Host Microbe 26, 61–72. doi: 10.1016/j.chom.2019.06.004
Thoendel, M., and Horswill, A. R. (2009). Identification of Staphylococcus aureus AgrD residues required for autoinducing peptide biosynthesis. J. Biol. Chem. 284, 21828–21838. doi: 10.1074/jbc.M109.031757
Thoendel, M., and Horswill, A. R. (2013). Random mutagenesis and topology analysis of the autoinducing peptide biosynthesis proteins in S taphylococcus aureus. Mol. Microbiol. 87, 318–337. doi: 10.1111/mmi.12100
Topa, S. H., Palombo, E. A., Kingshott, P., and Blackall, L. L. (2020). Activity of cinnamaldehyde on quorum sensing and biofilm susceptibility to antibiotics in Pseudomonas aeruginosa. Microorganisms 8:455. doi: 10.3390/microorganisms8030455
Tuson, H. H., and Weibel, D. B. (2013). Bacteria–surface interactions. Soft Matter 9, 4368–4380. doi: 10.1039/c3sm27705d
Uehara, H., Yoneda, Y., Yamane, K., and Maruo, B. (1974). Regulation of neutral protease productivity in Bacillus subtilis: transformation of high protease productivity. J. Bacteriol. 119, 82–91. doi: 10.1128/jb.119.1.82-91.1974
Uniprot (2021). Available at: https://www.uniprot.org/uniprot/Q7MT55 (Accessed November 25, 2021).
Uruén, C., Chopo-Escuin, G., Tommassen, J., Mainar-Jaime, R. C., and Arenas, J. (2021). Biofilms as promoters of bacterial antibiotic resistance and tolerance. Antibiotics 10:3. doi: 10.3390/antibiotics10010003
Valentini, M., and Filloux, A. (2019). Multiple roles of c-di-GMP signaling in bacterial pathogenesis. Annu. Rev. Microbiol. 73, 387–406. doi: 10.1146/annurev-micro-020518-115555
Vasquez, J. K., and Blackwell, H. E. (2019). Simplified autoinducing peptide mimetics with single-nanomolar activity against the Staphylococcus aureus AgrC quorum sensing receptor. ACS Infect. Dis. 5, 484–492. doi: 10.1021/acsinfecdis.9b00002
Vasquez, J. K., Tal-Gan, Y., Cornilescu, G., Tyler, K. A., and Blackwell, H. E. (2017). Simplified AIP-II peptidomimetics are potent inhibitors of Staphylococcus aureus AgrC quorum sensing receptors. ChemBioChem 18, 413–423. doi: 10.1002/cbic.201600516
Verdugo-Fuentes, A., Gastélum, G., Rocha, J., and de la Torre, M. (2020). Multiple and overlapping functions of quorum sensing proteins for cell specialization in Bacillus species. J. Bacteriol. 202, e00721–e00819. doi: 10.1128/JB.00721-19
Viana, F., Peringathara, S. S., Rizvi, A., and Schroeder, G. N. (2021). Host manipulation by bacterial type III and type IV secretion system effector proteases. Cell. Microbiol. 23:e13384. doi: 10.1111/cmi.13384
Vishwakarma, A., Dang, F., Ferrell, A., Barton, H. A., and Joy, A. (2021). Peptidomimetic polyurethanes inhibit bacterial biofilm formation and disrupt surface established biofilms. J. Am. Chem. Soc. 143, 9440–9449. doi: 10.1021/jacs.1c02324
Wang, S., Feng, Y., Han, X., Cai, X., Yang, L., Liu, C., et al. (2021). Inhibition of virulence factors and biofilm formation by Wogonin attenuates pathogenicity of Pseudomonas aeruginosa PAO1 via targeting pqs quorum-sensing system. Int. J. Mol. Sci. 22:12699. doi: 10.3390/ijms222312699
Wang, Y., Hu, M., Liu, Q., Qin, J., Dai, Y., He, L., et al. (2016). Role of the ESAT-6 secretion system in virulence of the emerging community-associated Staphylococcus aureus lineage ST398. Sci. Rep. 6:25163. doi: 10.1038/srep25163
Wang, H.-Y., Lin, L., Tan, L.-S., Yu, H.-Y., Cheng, J.-W., and Pan, Y.-P. (2017a). Molecular pathways underlying inhibitory effect of antimicrobial peptide Nal-P-113 on bacteria biofilms formation of Porphyromonas gingivalis W83 by DNA microarray. BMC Microbiol. 17:37. doi: 10.1186/s12866-017-0948-z
Wang, B., and Muir, T. W. (2016). Regulation of virulence in Staphylococcus aureus: molecular mechanisms and remaining puzzles. Cell Chem. Biol. 23, 214–224. doi: 10.1016/j.chembiol.2016.01.004
Wang, T., Si, M., Song, Y., Zhu, W., Gao, F., Wang, Y., et al. (2015). Type VI secretion system transports Zn2+ to combat multiple stresses and host immunity. PLoS Pathog. 11:e1005020. doi: 10.1371/journal.ppat.1005020
Wang, B., Zhao, A., Xie, Q., Olinares, P. D., Chait, B. T., Novick, R. P., et al. (2017b). Functional plasticity of the AgrC receptor histidine kinase required for staphylococcal virulence. Cell Chem. Biol. 24, 76–86. doi: 10.1016/j.chembiol.2016.12.008
West, K. H., Shen, W., Eisenbraun, E. L., Yang, T., Vasquez, J. K., Horswill, A. R., et al. (2021). Non-native peptides capable of pan-activating the agr quorum sensing system across multiple specificity groups of Staphylococcus epidermidis. ACS Chem. Biol. 16, 1070–1078. doi: 10.1021/acschembio.1c00240
Whelan, R., McVicker, G., and Leo, J. C. (2020). Staying out or going in? The interplay between type 3 and type 5 secretion systems in adhesion and invasion of enterobacterial pathogens. Int. J. Mol. Sci. 21:4102. doi: 10.3390/ijms21114102
World Health Organization (2019). New Report Calls for Urgent Action to Avert Antimicrobial Resistance Crisis. Available at: https://www.who.int/news/item/29-04-2019-new-report-calls-for-urgent-action-to-avert-antimicrobial-resistance-crisis (Accessed November 25, 2021).
World Health Organization (2021). Antimicrobial Resistance. Available at: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (Accessed November 25, 2021).
Wu, S., Liu, J., Liu, C., Yang, A., and Qiao, J. (2020). Quorum sensing for population-level control of bacteria and potential therapeutic applications. Cell. Mol. Life Sci. 77, 1319–1343. doi: 10.1007/s00018-019-03326-8
Xu, Q., Hu, X., and Wang, Y. (2021). Alternatives to conventional antibiotic therapy: potential therapeutic strategies of combating antimicrobial-resistance and biofilm-related infections. Mol. Biotechnol. 63, 1103–1124. doi: 10.1007/s12033-021-00371-2
Yang, Y., Koirala, B., Sanchez, L. A., Phillips, N. R., Hamry, S. R., and Tal-Gan, Y. (2017). Structure–activity relationships of the competence stimulating peptides (CSPs) in Streptococcus pneumoniae reveal motifs critical for intra-group and cross-group ComD receptor activation. ACS Chem. Biol. 12, 1141–1151. doi: 10.1021/acschembio.7b00007
Yang, Y., Lin, J., Harrington, A., Cornilescu, G., Lau, G. W., and Tal-Gan, Y. (2020). Designing cyclic competence-stimulating peptide (CSP) analogs with pan-group quorum-sensing inhibition activity in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. 117, 1689–1699. doi: 10.1073/pnas.1915812117
Yang, T., Tal-Gan, Y., Paharik, A. E., Horswill, A. R., and Blackwell, H. E. (2016). Structure–function analyses of a Staphylococcus epidermidis autoinducing peptide reveals motifs critical for AgrC-type receptor modulation. ACS Chem. Biol. 11, 1982–1991. doi: 10.1021/acschembio.6b00120
Yehuda, A., Slamti, L., Bochnik-Tamir, R., Malach, E., Lereclus, D., and Hayouka, Z. (2018). Turning off Bacillus cereus quorum sensing system with peptidic analogs. Chem. Commun. 54, 9777–9780. doi: 10.1039/c8cc05496g
Yu, H.-Y., Tu, C.-H., Yip, B.-S., Chen, H.-L., Cheng, H.-T., Huang, K.-C., et al. (2011). Easy strategy to increase salt resistance of antimicrobial peptides. Antimicrob. Agents Chemother. 55, 4918–4921. doi: 10.1128/AAC.00202-11
Yu, K.-W., Xue, P., Fu, Y., and Yang, L. (2021). T6SS mediated stress responses for bacterial environmental survival and host adaptation. Int. J. Mol. Sci. 22:478. doi: 10.3390/ijms22020478
Zhang, P., Guo, Q., Wei, Z., Yang, Q., Guo, Z., Shen, L., et al. (2021). Baicalin represses type three secretion system of Pseudomonas aeruginosa through PQS system. Molecules 26:1497. doi: 10.3390/molecules26061497
Zhou, M., Lan, Y., Wang, S., Liu, Q., Jian, Z., Li, Y., et al. (2020). Epidemiology and molecular characteristics of the type VI secretion system in Klebsiella pneumoniae isolated from bloodstream infections. J. Clin. Lab. Anal. 34:e23459. doi: 10.1002/jcla.23459
Keywords: peptidomimetics, anti-virulence, biofilms, quorum sensing, secretion systems, resistance
Citation: Martínez OF, Duque HM and Franco OL (2022) Peptidomimetics as Potential Anti-Virulence Drugs Against Resistant Bacterial Pathogens. Front. Microbiol. 13:831037. doi: 10.3389/fmicb.2022.831037
Received: 07 December 2021; Accepted: 28 February 2022;
Published: 18 April 2022.
Edited by:
Alfredo M. Angeles-Boza, University of Connecticut, United StatesReviewed by:
John N. Alumasa, The Pennsylvania State University (PSU), United StatesCopyright © 2022 Martínez, Duque and Franco. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Octávio Luiz Franco, b2NmcmFuY29AZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.