
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 24 May 2022
Sec. Terrestrial Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.830905
Beneficial bacteria in the rhizosphere are known to trigger faster and stronger plant immune responses to biotic and abiotic stressors. In the present study, we aimed to test the hypothesis that a rhizosphere microbiome transplant (RMT) may improve the immune response and reduce the disease rates of barley (Hordeum vulgare). This hypothesis was tested in a greenhouse system with the powdery mildew-causing fungus Blumeria graminis f. sp. hordei (Bgh). Detached rhizosphere microbiome from barley grown in a field soil was transplanted to barley seedlings grown in potting soil with reduced microbial diversity. Saline-treated plants served as control. At the three-leaf stage, barley was infected with Bgh. Decreased susceptibility to Bgh was observed for barley treated with the RMT as displayed by lower Bgh pustule counts in a detached leaf assay. A trend toward enhanced relative transcript abundances of the defense-related genes PR1b and PR17b was observed in leaves, 24 h after the Bgh challenge, when compared to the control. Moreover, 10 days after the Bgh challenge, the barley rhizosphere microbiome was harvested and analyzed by sequencing of 16S rRNA gene amplicons. The microbial community composition was significantly influenced by the RMT and displayed higher microbial diversity compared to the control. Furthermore, microbial beta-diversity and predicted functional profiles revealed a treatment-dependent clustering. Bacterial isolates from the RMT showed in vitro plant beneficial traits related to induced resistance. Our results showed that transplantation of a rhizosphere microbiome could be a sustainable strategy to improve the health of plants grown in potting soil with low microbial diversity under greenhouse conditions.
Plants together with their associated microorganisms build the plant holobiont (Vandenkoornhuyse et al., 2015). The genomes of these associated microorganisms are often called the second genome of the plant (Berendsen et al., 2012), as they encode important plant-related functions involved in the nutrient supply or protection against biotic and abiotic stressors (Vandenkoornhuyse et al., 2015; Berg et al., 2021). In the rhizosphere, the small zone of soil influenced by the plant root (Hiltner, 1904; Hartmann et al., 2008), the microbiome is shaped by many factors such as soil type, plant species and developmental stage, agricultural management, and geographic location (Gomes et al., 2001; Berg and Smalla, 2009; Alegria Terrazas et al., 2020; Bziuk et al., 2021).
Beneficial microorganisms in the rhizosphere can have plant growth-promoting (PGP) activity or direct antagonistic activity against soil-borne pathogens, or they can induce resistance in foliar tissues to prepare the plant for a potential future pathogen attack (reviewed by Mendes et al., 2013; Pieterse et al., 2014; Berg et al., 2017). The plant’s immune system can be enhanced by microbial secondary metabolites, such as siderophores, N-acyl homoserine lactones (AHLs), 2,4-diacetylphloroglucinol (DAPG), or the phytohormone indole-3-acetic acid (IAA) (Pieterse et al., 2014; Mauch-Mani et al., 2017; Gouda et al., 2018; Vannier et al., 2019). Beneficial microbes are considered important alternatives to chemical plant protection strategies for the improvement of plant performance and health in agricultural systems (Syed Ab Rahman et al., 2018). In terms of the enhanced plant defense response, a plethora of studies already reported positive effects of beneficial strains on several plant species (reviewed in Mauch-Mani et al., 2017; Syed Ab Rahman et al., 2018; Berg et al., 2021). Indeed, there are already commercially available products (e.g., Mülner et al., 2020). For barley (Hordeum vulgare), the model crop used in the present study, several studies reported an enhanced plant defense response triggered by AHL-producing bacteria (Han et al., 2016; Shrestha et al., 2019; Wehner et al., 2019).
Nowadays, not only single microbial biocontrol strains are used to improve plant performance and health, but also microbial consortia. The use of microbial consortia or synthetic communities (SynComs) gained attention during the last decade as promising alternatives to chemical pesticides and fertilizers (Sarma et al., 2015; Syed Ab Rahman et al., 2018; Vannier et al., 2019), and it is assumed that the enhancement of plant resistance is improved compared to single microbial inoculants (Woo and Pepe, 2018; Niu et al., 2020). Microbial consortia can enhance a plant’s resistance against biotic stressors, for instance bacterial (Gu et al., 2020) or fungal pathogens (Niu et al., 2017; Carrión et al., 2019), but they also confer tolerance toward abiotic factors like cold stress (Moradtalab et al., 2020). Furthermore, the plant itself can select a beneficial microbial consortium by recruitment of specific microbes as reported in a study on Arabidopsis leaf challenge with downy mildew-causing Oomycete Hyaloperonospora arabidopsidis (Berendsen et al., 2018). The challenge led to a proliferation of a consortium in the rhizosphere consisting of Microbacterium, Stenotrophomonas, and Xanthomonas, and the following generation of plants grown in this soil showed enhanced resistance to the disease compared to previously unplanted bulk soils (Berendsen et al., 2018). Additionally, the selection for a beneficial rhizosphere microbial community was shown to depend on the level of plant resistance (Mendes et al., 2018).
One of the best described natural microbiome-based plant defenses is suppressive soils (reviewed by Schlatter et al., 2017). Suppressive soils can inhibit the growth and activity of soil-borne pathogens due to a specific structure of the microbial community. A conducive soil can become suppressive by soil inoculation of 1–10% of a suppressive soil to the conducive soil (Schlatter et al., 2017). Moreover, soil inoculation was also reported to confer an aboveground resistance in chrysanthemum plants against thrips (Pineda et al., 2020). In the case of aboveground pests and phytopathogens, it was shown that the plants’ immune response additionally varies depending on the soil used as growth substrate (Chialva et al., 2018; Bziuk et al., 2021). In a previous study, we investigated the immune response in barley grown in a field vs. potting soil to Blumeria graminis f. sp. hordei (Bgh). We observed that barley grown in autoclaved or non-autoclaved potting soils showed considerably higher Bgh challenge rates than those grown in field soils, which made us assume that the rhizosphere microbiome might play an important role as a driver for better plant performance (Bziuk et al., 2021). However, soils used for barley growth differed also in soil texture and physicochemical properties (Bziuk et al., 2021), therefore, the exact role of the microbiome remained unclear.
To exclude the influence of different soil physicochemical properties mentioned above (Bziuk et al., 2021), we inoculated barley seedlings grown in autoclaved standard potting soil with the rhizosphere microbiome transplant (RMT) from barley grown in field soil (Figure 1). Plants treated with saline served as a negative control. In addition, barley was inoculated with Ensifer meliloti Rm2011 expR+ (M. McIntosh; further called E. meliloti expR+), producing the AHL molecule N-3-oxo-tetradecanoyl-L-homoserine lactone oxo-C14-HSL (Zarkani et al., 2013). This strain was already shown to induce resistance against Bgh in barley, in an AHL-dependent manner (Hernández-Reyes et al., 2014; Shrestha et al., 2019). As a control for inoculation with E. meliloti expR+, we used the E. meliloti Rm2011 pBBR2attM strain (Zarkani et al., 2013; further called E. meliloti attM), which carries a plasmid with the lactonase gene attM originating from Agrobacterium tumefaciens that hydrolyzes the lactone ring of AHL molecules (Zarkani et al., 2013). To evaluate the plant’s immune response, we infected barley with the powdery mildew-causing fungus Bgh. We investigated the relative transcript abundance of the defense-related genes PR1b and PR17b in leaves by qRT-PCR and determined the Bgh pustule number in a detached leaf assay. Further, we examined the rhizosphere microbial community sampled 10 days after Bgh challenge based on 16S rRNA gene amplicon sequencing in terms of composition and predicted functions. Furthermore, bacterial isolates obtained from the RMT were characterized in vitro for potential plant beneficial functions. Our results strengthen the notion that microbial diversity is an important factor shaping the plant’s defense response.
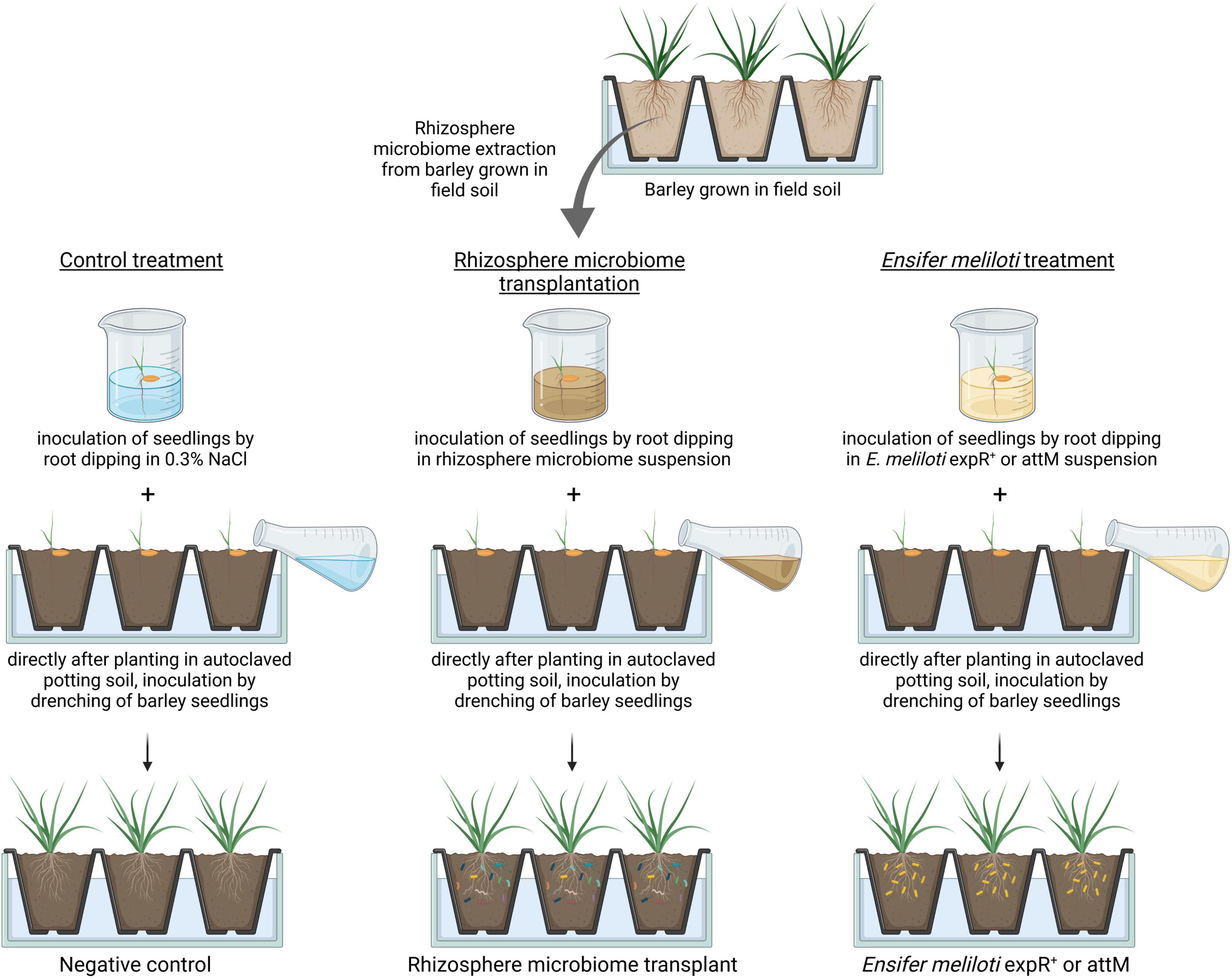
Figure 1. Experimental design for rhizosphere microbiome transplantation. The barley cultivar “Golden Promise” was grown in field soil under greenhouse conditions. At the three-leaf stage (BBCH13), the rhizosphere microbiome was harvested and a rhizosphere microbiome suspension was prepared. Barley seedlings were dipped into the rhizosphere microbiome suspension, planted into autoclaved potting soil, and then additionally drenched with the rhizosphere microbiome suspension. Dipping of seedlings prior to drenching was as well performed for control plants with saline (0.3% NaCl) and for plants treated with either of the two Ensifer meliloti strains expR+ and attM. The figure was created with biorender.com.
Barley (Hordeum vulgare) cultivar Golden Promise seeds (Simpsons Malt Limited, Berwick-upon-Tweed, United Kingdom) were germinated on wet filter paper for 3 days. Seedlings (one per pot) were planted in approximately 100 g of sieved (2 mm) field soil. Plants were further grown under greenhouse conditions at 18°C and 16/8 h (day/night) photoperiod and watered with tap water according to demand. The field soil used for this experiment was taken from a long-term field experiment in Bernburg, Germany. Agricultural management previously applied was conventional moldboard plow (20–30 cm depth), additionally to a standard N-fertilization of 200 kg/ha N and fungicide application (Deubel et al., 2011; Bziuk et al., 2021). In the three-leaf stage (BBCH13; according to Meier, 2018), rhizospheres were harvested with 0.3% NaCl as described in a later section. Rhizosphere suspensions were pooled and diluted in a ratio of 1:2 with 0.3% NaCl (saline) to establish the RMT. Four replicates of rhizospheres were additionally sampled to extract total community DNA for 16S rRNA gene amplicon sequencing as described in a later section.
Barley seeds were pre-germinated as described above, and the seedlings were soaked for a few seconds in the RMT. Afterward, they were planted into approximately 100 g of autoclaved (3 times at 134°C for 10 min with always 1 day in between to reduce microbial diversity) potting soil (Fruhstorfer Erde, Hawita Gruppe GmbH, Vechta, Germany) 1:2 (w/w) mixed with sand (Sahara Spielsand, WECO GmbH, Leer, Germany). After planting, barley seedlings were additionally drenched with 10 ml RMT. Control plants were treated as described above with 0.3% NaCl (saline; Figure 1). Four unplanted pots drenched with 10 ml RMT or 0.3% NaCl (saline) served as bulk soil control. A dilution series of the RMT was plated on Reasoner’s 2A agar (R2A) medium supplemented with 100 μg/ml cycloheximide to reduce fungal growth, and the colony forming units (CFUs) were determined after 24 and 48 h and 6 days.
Additionally to the treatments of RMT and 0.3% NaCl control, barley plants were inoculated with the resistance-inducing strain E. meliloti expR+ (Figure 1; Hernández-Reyes et al., 2014; Shrestha et al., 2019). As a control for this additional treatment, the E. meliloti strain attM was used, which expresses a lactonase (AttM from A. tumefaciens) that hydrolyzes the AHL molecule oxo-C14-HSL, the base for E. meliloti expR+-induced resistance in barley.
Overnight cultures of E. meliloti attM and expR+ in tryptone yeast (TY) medium were diluted to an OD600 of 0.6 and grown until an OD600 of 0.8 for the best AHL production. The cell cultures were centrifuged at 10,000 × g for 10 min and washed with 0.3% NaCl. The bacterial cells were resuspended with 0.3% NaCl to an OD600 of 0.1 corresponding to 108 CFU/ml. Barley seedlings were treated with either of the two bacterial inocula described above. A dilution series of the inocula were plated on TY agar medium supplemented with 250 μg/ml streptomycin for E. meliloti expR+ and with 250 μg/ml streptomycin and 100 μg/ml kanamycin for E. meliloti attM to control the inoculum CFU.
The challenge rate of Bgh was determined by counting macroscopically visible Bgh colonies, called pustules, in three independent detached leaf assays. Barley second leaves (BBCH13) were placed with the adaxial side up, on 0.8% water agar plates and positioned inside an challenge tower. The leaves were infected with approximately 250 fresh conidia of Bgh per cm2. The procedure of Bgh challenge was described in detail by Bziuk et al. (2021). Briefly, Bgh conidia were blown from already mildewed barley plants inside the challenge tower via pressurized air and were allowed to settle down on the plates containing the leaves for about 10 min. Counting chambers were used to determine the conidia density. The plates containing the leaves were incubated for 5 days in a growth chamber at 20°C, 16/8 h day/night photoperiod, and 60% humidity. On day 5 of incubation, Bgh pustules were counted on 5 cm leaf length under a binocular. The number of pustules was divided by the leaf area and normalized to the Bgh inoculation density. Eight replicates per treatment were used in the first repetition and 10 replicates each for the second and third repetition. Significance was tested with SAS 9.4 (SAS Institute, Cary, NC, United States), and p ≤ 0.05 was assumed as different. The obligate biotrophic leaf fungus B. graminis (DC) Speer f. sp. hordei EM Marchal race A6 was maintained on barley cultivar Igri.
Quantification of the plant immune response of barley to Bgh was performed measuring the relative transcript abundance of the defense-related genes PR1b and PR17b, commonly used as indicator genes for defense responses in barley (Tufan et al., 2017; Shrestha et al., 2019). First, whole plants were infected with Bgh as described above. Samples of the second and third leaves of the whole infected and uninfected barley were taken before and 24 h after Bgh exposure. Three replicates per treatment were taken for each of the four individual experiments. RNA was extracted from leaf samples as described by Bziuk et al. (2021). A quantitative reverse transcription (qRT)-PCR was performed for HvUBQ60 (5′-CAGTAGTGGCGGTCGAAGTG-3′ and 5′ACC CTCGCCGACTACAACAT-3′; Gen-Bank M60175.1) and the Pathogenesis-Related (PR) genes PR1b (5′-GGACTACGACTACG GCTCCA-3′ and 5′-GGCTCGTAGTTGCAGGTGAT-3′) and PR17b (5′-CGAGGTTCCTCGACTACTGC-3′ and 5′-ATCAC ATTCAGCCTCCGAAC-3′; Shrestha et al., 2019). The relative transcript abundance of PR1b and PR17b was normalized to HvUBQ60 expression and the 0 h time point resulting in the 2–ΔΔ CT value (Livak and Schmittgen, 2001). Significance was tested with Student’s t-test, and p ≤ 0.05 was assumed as different.
Four replicate plants of barley grown in field soil were used for RMT preparation (described above), as well as four replicate plants grown in potting soil (10 days after exposure to Bgh) for each treatment (RMT or control, Bgh-infected, or uninfected), were used for rhizosphere sampling. First, loosely attached soil was removed by shaking the plant, additionally, a tweezer was used to remove bigger potting soil particles. Afterward, the complete root system with more tightly bound soil was transferred to a 50 ml tube and nine volumes of 0.85% NaCl were added. After 1-min vortexing to detach the rhizosphere the roots were removed. Rhizosphere suspensions were centrifuged for 20 min at 8,500 g and the supernatant was discarded. Bulk soil samples were taken from the respective unplanted pots by mixing the soil carefully. Rhizosphere and bulk soil pellets were stored at –20°C until total community DNA extraction.
For total community DNA extraction, 0.5 g (wet weight) of barley rhizosphere or bulk soil samples were treated with the FastDNA Spin Kit for Soil (MP Biomedicals, Eschwege, Germany) according to the manufacturer’s instructions.
Amplicon sequencing libraries of rhizosphere and bulk soil samples were prepared in a two-step PCR targeting the V3-V4 region of the 16S rRNA gene. The first PCR was performed using the Uni341F (5′-CCTAYGGGRBGCASCAG-3′) and Uni806R (5′-GGACTACHVGGGTWTCTAAT-3′) primers (Yu et al., 2005; Caporaso et al., 2011; Sundberg et al., 2013) as described by Bziuk et al. (2021). First PCR products were purified using the HighPrep PCR clean-up (MagBio Genomics, Gaithersburg, MD, United States) in a 0.65:1 (beads:PCR reaction) volumetric ratio. The second PCR was used to add Illumina sequencing adapters and sample-specific dual indices (IDT Integrated DNA Technologies, Coralville, IA, United States) using PCRBIO HiFi (PCR Biosystems Ltd., London, United Kingdom) for 15 amplification cycles. Second PCR products were purified as described above, and further processing of the purified PCR products was performed as described in the study by Bziuk et al. (2021). The resulting library was sequenced on an Illumina MiSeq platform at the Section of Microbiology, University of Copenhagen (Denmark) using Reagent Kit v2 (2 × 250 cycles) (Illumina, San Diego, CA, United States) and including 1st and 2nd PCR negative controls, as well as a mock control.
Amplicon sequencing data analysis was performed as previously described in the study by Bziuk et al. (2021). Briefly, primer sequences were removed using Cutadapt v.2.3 (Martin, 2011), and amplicon sequence variants (ASV) were generated using the DADA2 version 1.10.0 (Callahan et al., 2016) plugin for QIIME2 (Bolyen et al., 2019) and using q2-feature-classifier classify-sklearn module trained with SILVA SSU rel. 132 database (Quast et al., 2013).
The ASV dataset was further cleaned using the package phyloseq (McMurdie and Holmes, 2013) of RStudio (RStudio, Boston, MA, United States) version R3.6.3. ASV affiliated to chloroplasts (28 ASV representing 0.986% of total reads), mitochondria (four ASV representing 0.289% of total reads), and unassigned Kingdom or Phylum (30 ASV representing 0.048% of total reads) were removed from the dataset. The dataset was also filtered based on the 16S rRNA V3-V4 region size keeping only ASV of a length between 350 and 440 bp (21 ASV were removed). DECONTAM (Davis et al., 2018) was used to identify potential contaminations as determined by the prevalence of ASV in the negative controls (from 1st and 2nd PCR) revealing no true contaminants. After cleaning, 97.67% of the dataset was kept. Rhizosphere samples obtained a median of 12 204 reads and 323.5 ASV per sample. Bulk soils contained a median of 7470.5 reads and 271 ASV per sample.
Alpha-diversity indices were calculated based on read count data 100 times randomly subsampled to the lowest number of sequences (6946) using RStudio R3.6.3 packages multcomp (Hothorn et al., 2008) and vegan (Oksanen et al., 2019). Three samples were excluded from the analysis due to a low amount of sequence reads. Analysis of variance (ANOVA) and permutational analysis of variance (PERMANOVA) was performed with vegan package and based on read count data, the principle component analysis (PCA) based on relative abundances. PERMANOVA was conducted based on Bray–Curtis dissimilarity indices (10,000 permutations). Treatment-dependent changes in the relative abundances of microbial genera were examined using the packages edgeR (Robinson et al., 2010) and phyloseq (McMurdie and Holmes, 2013) with a likelihood ratio test under negative binomial distribution and generalized linear models (FDR-corrected; p ≤ 0.05). The heatmap visualizing relative abundances of selected genera was created using the pheatmap package (Kolde, 2015). Functional predictions of the microbial community based on the 16S rRNA amplicon data were performed using Tax4Fun2 (Wemheuer et al., 2020) with the Ref99NR reference dataset.
Bacterial isolates were randomly picked from plates with 20–100 colonies obtained after plating a serial dilution of the RMT on the R2A medium and incubation at 28°C for 6 days. Genomic DNA was extracted from 10 representative RMT isolates using the Genomic DNA Extraction Kit (Qiagen, Hilden, Germany) and the Silica Bead DNA Gel Extraction Kit (Thermo Fisher Scientific, Waltham, MA, United States). A BOX-PCR fingerprint was conducted for each isolate (Martin et al., 1992) and taxonomic affiliation was determined based on sequencing (Macrogen Europe B.V., Amsterdam, Netherlands) of a partial 16S rRNA gene fragment (Heuer et al., 2009). The sequences were trimmed and assembled using CLC MainWorkbench 20.0.3 (Qiagen, Aarhus, Denmark) and taxonomic affiliation was determined based on the NCBI database (NCBI, Bethesda, MD, United States).
Bacterial isolates of the RMT were tested in vitro for potential plant beneficial activities: protease, β-1,3-glucanase, and cellulase (Weinert et al., 2010), as well as chitinase activity (Berg et al., 2001). Further assays tested for ACC deaminase and IAA production (Koo et al., 2010), production of siderophores (Schwyn and Neilands, 1987), and AHLs using the sensor strains Chromobacterium violaceum VIR07 and cv026 (Durán et al., 2016).
The challenge rate determined in the detached leaf assay by Bgh pustule counts revealed significantly decreased counts for RMT-treated barley compared to the control (Figure 2). As an internal control, we used E. meliloti expR+ and attM strains, as described in the section “Materials and Methods.” Plants treated with E. meliloti expR+ showed a decreased challenge rate compared to E. meliloti attM. Although the decreased challenge rate of E. meliloti expR+ was not significant compared to E. meliloti attM when all treatments were statistically analyzed together (Tukey’s test; Figure 2), it was highly significant in a single comparison with E. meliloti expR+ vs. attM (Student’s t-test, p = 0.003). Bgh challenge rates revealed similar levels for RMT-treated barley and the internal positive control E. meliloti expR+.

Figure 2. Bgh challenge rate decreased after rhizosphere microbiome transplantation. Effects of inoculation on the relative amount of Bgh pustules per cm2 were determined in a detached leaf assay. Barley was either treated with the rhizosphere microbiome transplant (RMT) from field soil or saline (0.3% NaCl) as control (Ctrl). Inoculation of the Ensifer meliloti (E.m.) strains expR+ (resistance inducing) and attM (control) was used as internal positive control. Bars represent mean ± SE of four independent experiments (n = 10, except for the first repetition n = 8). Different letters indicate significant differences (Tukey’s test; p ≤ 0.05).
Relative transcript abundances of the two defense-related genes PR1b and PR17b showed similar levels for the uninfected plants in all treatments (Figures 3A,B). After the Bgh challenge, relative transcript abundances of PR1b and PR17b were enhanced in all treatments investigated, but only E. meliloti expR+ treated samples displayed significantly higher relative transcript abundance for both genes. Barley plants that had received the RMT showed higher relative transcript abundances compared to the control, although not significant due to the high variability between the individual experiments.
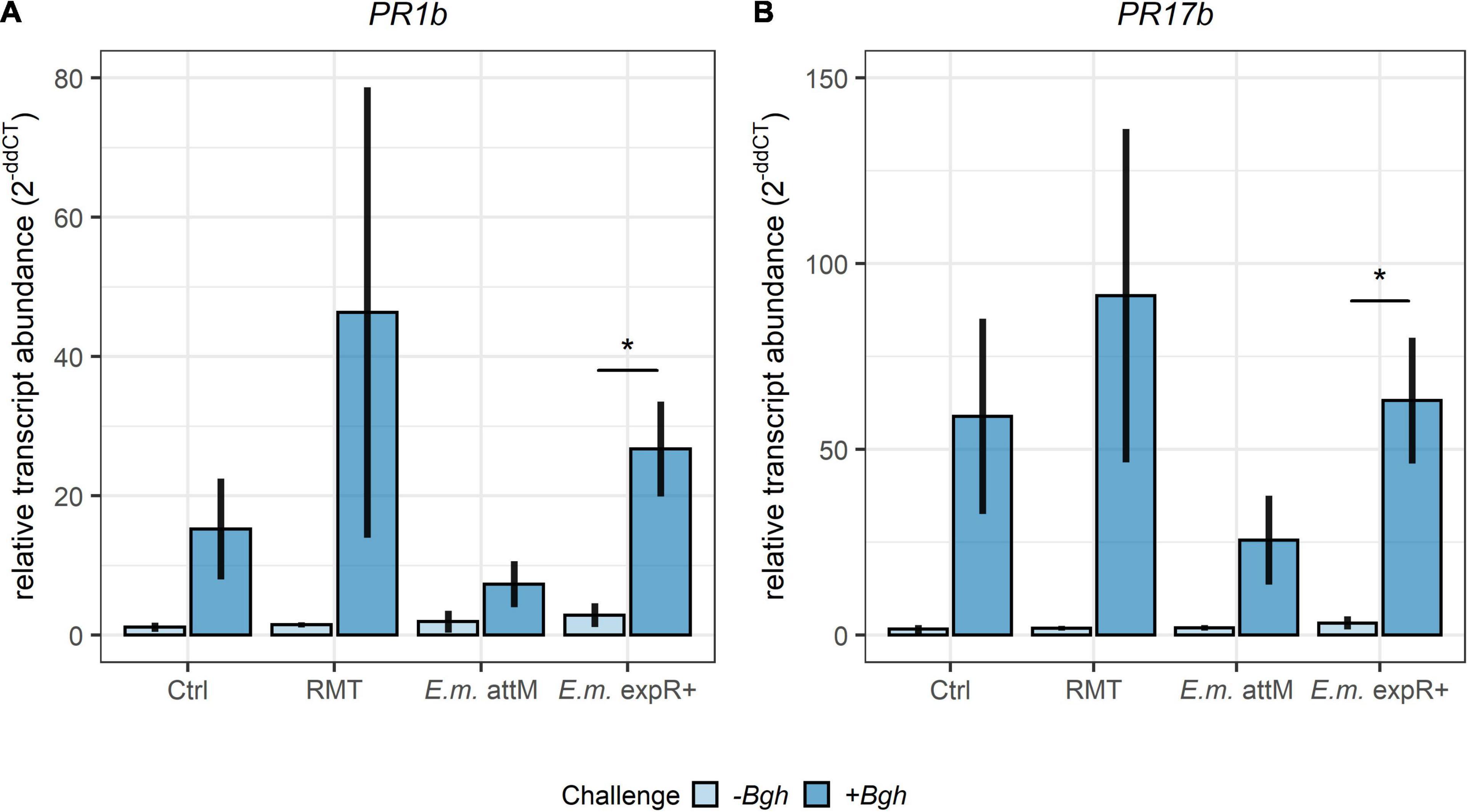
Figure 3. Higher relative transcript abundances of defense-related genes after Bgh challenge of barley treated with the rhizosphere microbiome transplant. Relative transcript abundances measured by qRT-PCR of the defense-related genes (A) PR1b and (B) PR17b in barley leaves 24 h after Bgh challenge. Barley was treated with the rhizosphere microbiome transplant (RMT), saline (0.3% NaCl) as control (Ctrl) or either of the two E. meliloti (E.m.) strains expR+ and attM. Bars represent the mean ± SE of three individual experiments (with each n = 3 per treatment). Transcript abundance was normalized to UBQ transcript abundance and the 0 h time point. Significant differences were tested by Student’s t-test (p ≤ 0.05).
The microbial diversity and community composition of barley rhizosphere were analyzed across treatments [i.e., rhizosphere (RS), RMT-treated and control (Ctrl), either Bgh-infected (+Bgh) or uninfected (–Bgh), bulk soil (BS) and field rhizosphere], and a total of 330,701 reads were obtained that assigned to 3779 ASV. However, three samples from different treatments had to be excluded from the analysis due to low read numbers (RS Ctrl, BS Ctrl, BS RMT). The rarefaction curve of the remaining samples (Supplementary Figure 1) showed a sufficient library size to cover the microbial diversity.
The RMT treatment increased microbial diversity in barley rhizosphere and in bulk soil, compared to the control (Figure 4 and Supplementary Table 1). For instance, Species richness was 1.5 × higher in RMT-treated bulk soil than in the control soil (Supplementary Table 1). Aboveground Bgh exposure did not affect the Species richness or Shannon diversity in the rhizosphere of barley treated with RMT or the control. ANOVA of alpha-diversity indices revealed a significant influence of the RMT treatment on both Species richness and Shannon diversity (p ≤ 0.001). Pielou’s evenness was also affected by RMT (p ≤ 0.001) with higher evenness for RMT-treated samples (Supplementary Table 1).

Figure 4. Microbial alpha-diversity is higher after rhizosphere microbiome transplantation. Shannon index of rhizosphere microbiome transplant (RMT) or control (Ctrl) of barley rhizosphere and bulk soil samples. Barley was additionally infected with Bgh. Different letters indicate significant differences (Tukey’s test p ≤ 0.05) that were determined for rhizosphere and bulk soil samples independently. Means ± SD value are written in Supplementary Table 1.
Visualization of the microbial beta-diversity by PCA (Figure 5A) revealed a clear separation along the first axis into RMT and control, for both rhizosphere and bulk soil samples. Along the second axis, the microhabitats rhizosphere and bulk soil were separated. There was no clear distinction between barley infected with Bgh or not for both RMT and control rhizospheres, as the respective samples clustered together.
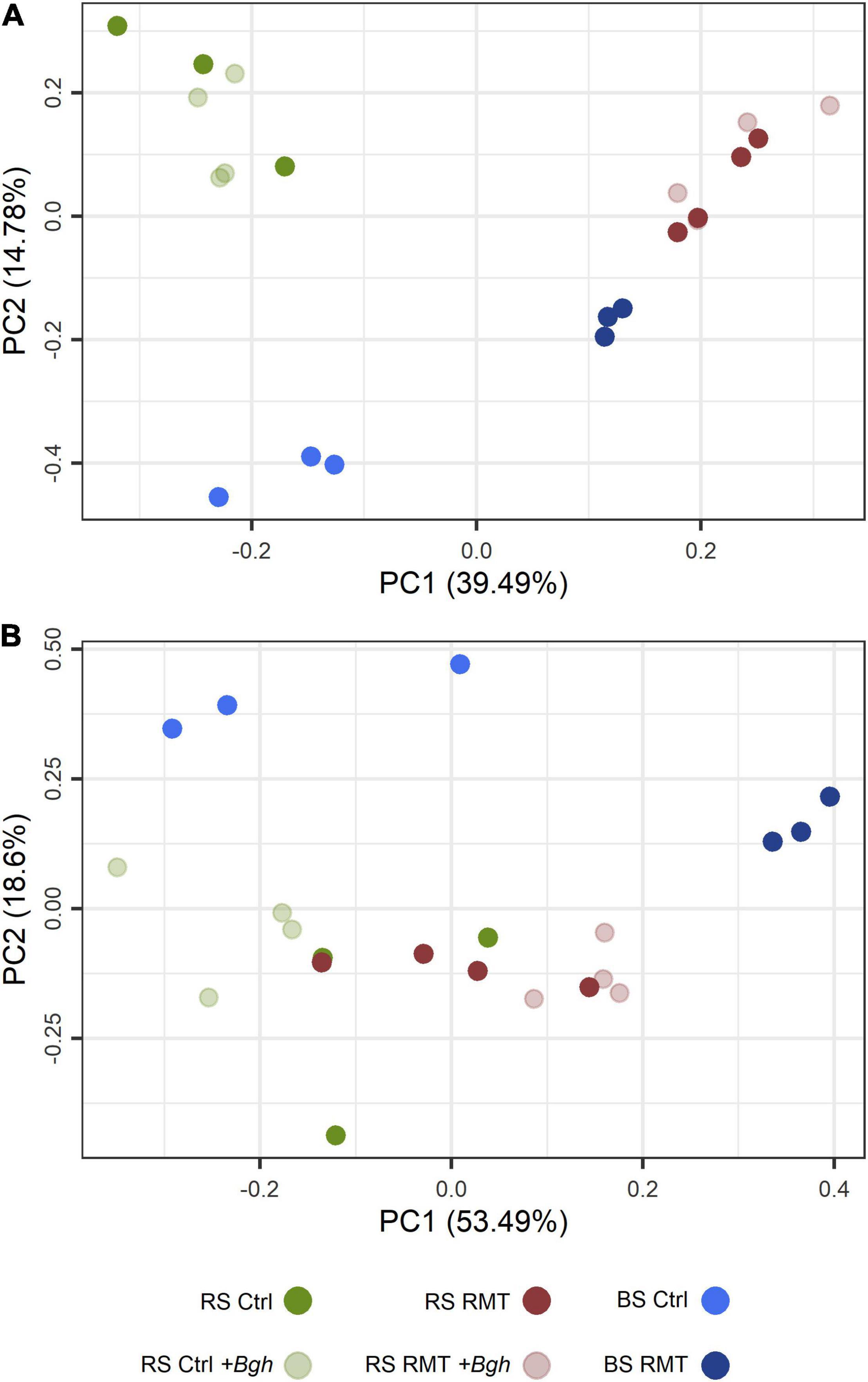
Figure 5. Community composition and predicted functional profile of barley rhizosphere and bulk soil microbial communities. (A) Microbial community composition of barley rhizosphere (RS) and bulk soil (BS) was visualized by constrained principal component analysis (PCA) based on the relative abundances of rhizosphere microbiome transplant (RMT) and control (Ctrl) amplicon sequence variants (ASV), either infected with Bgh or not. (B) Predicted KEGG Orthologs (KO) for each microbial community of RS or BS, treated with either RMT or as Ctrl with and without Bgh infected, visualized by PCA based on the relative abundance of each predicted KO.
The effect of the RMT treatment on the microbial community composition was verified by PERMANOVA (R2 = 0.46; p ≤ 0.001; Table 1), as well as the effect of the microhabitat (R2 = 0.13; p ≤ 0.001). Bgh challenge did not affect the microbial community composition in the rhizosphere of RMT-treated barley (R2 = 0.17; p = 0.22), but interestingly, Bgh infected significantly influenced the control (R2 = 0.27; p ≤ 0.05).
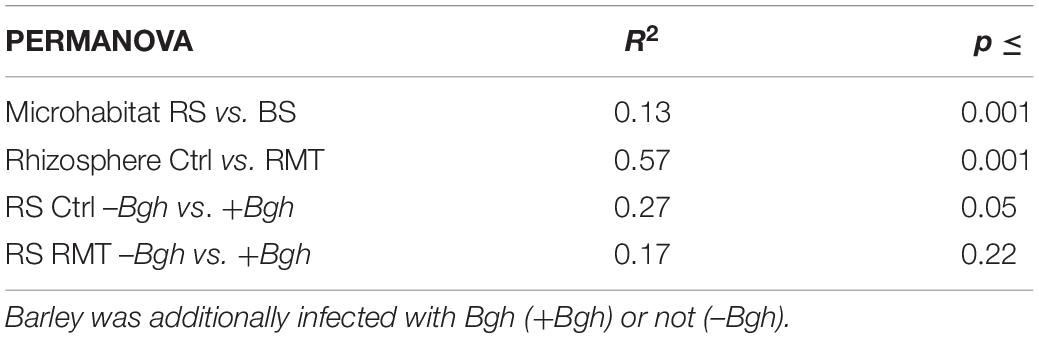
Table 1. PERMANOVA of barley rhizosphere (RS) and bulk soil (BS) samples, either treated with rhizosphere microbiome transplant (RMT) or as control (Ctrl).
Dominant phyla in the rhizosphere and bulk soil samples of both RMT and control samples were Proteobacteria, Bacteroidetes, Firmicutes, and Actinobacteria (Table 2). Notable differences in relative abundances between RMT and control samples were detected in the rhizosphere of uninfected and Bgh-infected barley for Firmicutes that decreased after transplantation (RMT 9.0/6.5% vs. Ctrl 17.2/15.5%) and Actinobacteria that increased after transplantation (RMT 6.4/6.5% vs. Ctrl 1.3/2.8%). Furthermore, increased relative abundance after RMT treatment was also observed for the lower abundant Verrucomicrobia (RMT 2.4/1.3% vs. Ctrl 1.1/1.4%), as well as for Patescibacteria (RMT 2.2/3.7% vs. Ctrl 0.1/0.1%). Verrucomicrobia was the only phylum that also displayed significant differences for RMT-treated and Bgh-infected barley rhizosphere (–Bgh 2.4% vs. +Bgh 1.3%).
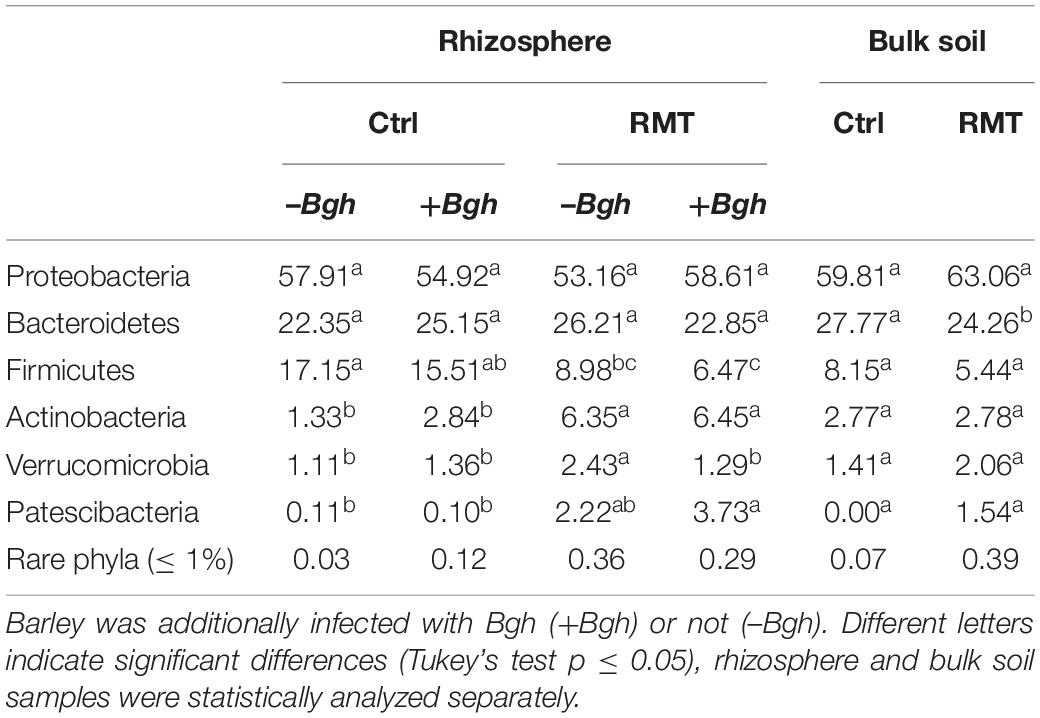
Table 2. Relative abundances of microbial phyla present in barley rhizosphere and bulk soil samples, either treated with rhizosphere microbiome transplant (RMT) or as control (Ctrl).
Some highly abundant taxa present in the rhizosphere of barley grown in field soil representing the RMT were not able to establish in the rhizosphere of barley grown in potting soil: Nitrososphaeraceae, Acidobacteria subgroup 6 and Nitrospira (Figure 6A). Other taxa were well transferred by RMT, like Chitinophagaceae or Sphingomonas, as these taxa had similar relative abundances in both barley rhizosphere from field and RMT-treated, whereas they were less abundant in the rhizosphere of control plants (Figure 6A). Some taxa that were highly abundant in the rhizosphere from barley grown in field soil that served as RMT had similar relative abundances in the RMT-treated rhizosphere, such as Bacillus or Paenibacillus, but were even more abundant in the control.
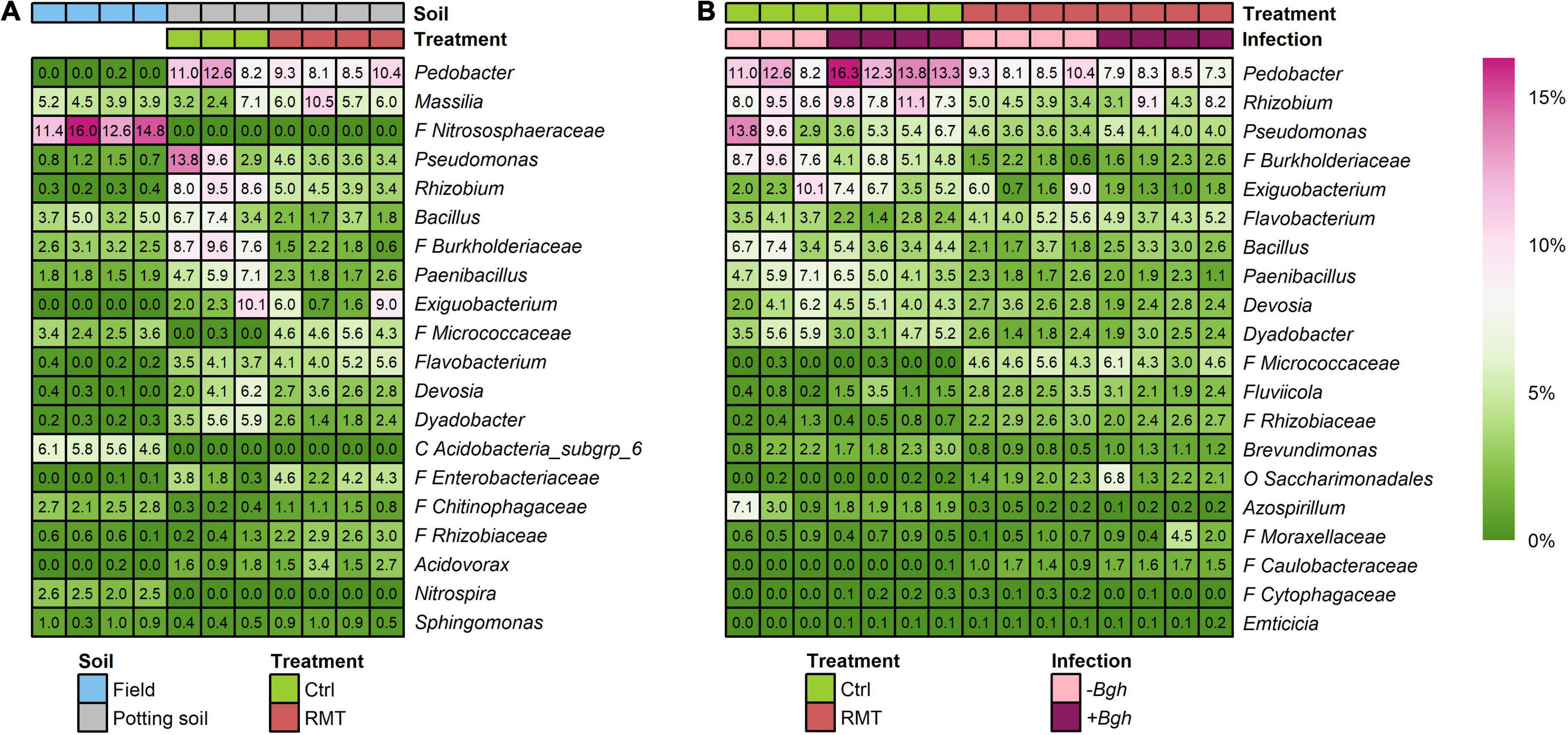
Figure 6. Relative abundances of taxa. (A) Abundant taxa present in the rhizosphere of barley grown in field soil representing the rhizosphere microbiome transplant (RMT), rhizosphere of RMT-treated and control (Ctrl) plants grown in potting soil with reduced microbial diversity. (B) Abundant taxa differing between Ctrl and RMT rhizospheres as determined with a likelihood ratio test under negative binomial distribution and generalized linear models (FDR-corrected; p ≤ 0.05). Additionally, relative abundances of Fluviicola, Emticicia, and Cytophagaceae were differing due to Bgh challenge in Ctrl samples. F, family; O, order; C, class.
Several taxa were highly abundant in both rhizosphere and bulk soil samples of all potting soil samples (RMT/Ctrl), such as Pedobacter, Rhizobium, Massilia, or Pseudomonas. The RMT treatment influenced the relative abundances of several highly abundant taxa (Figure 6B and Supplementary Table 3): unclassified members of Micrococcaceae and Rhizobiaceae were significantly higher in relative abundance for both uninfected and Bgh-infected samples in the rhizosphere of RMT-treated barley, whereas in the control Burkholderiaceae, Paenibacillus, Dyadobacter, and Azospirillium had higher relative abundances. Fluviicola showed higher relative abundance in the rhizosphere of uninfected RMT-treated barley compared to the control, whereas for uninfected control rhizosphere samples Rhizobium, Pseudomonas, Bacillus, and Brevundimonas displayed higher relative abundances. The influence of RMT in Bgh-infected rhizosphere samples revealed a higher relative abundance of Flavobacterium, Saccharimonadales, and Moraxellaceae compared to the control. For the Bgh-infected control plants, Pedobacter, Exiguobacterium, and Devosia had higher relative abundances compared to the respective RMT samples.
Interestingly, the unclassified members of Micrococcaceae that were highly abundant in RMT-treated rhizospheres (4.8%), were almost not detected in the control (0.1%). The relative abundance of Micrococcaceae in barley rhizosphere from field soil that served as RMT was about 3.0%. The respective sequences of the unclassified Micrococcaceae ASV were blasted to the NCBI database for further taxonomic affiliation. All eight ASV belonged to either Arthrobacter or Pseudarthrobacter (Supplementary Table 2).
Differences in relative abundance due to the Bgh challenge were not detected for RMT-treated barley. Only in the rhizosphere of the control, the three taxa were significantly increased in relative abundance after exposure to Bgh: Fluviicola, Emticicia, and Cytophagaceae (Figure 6B).
In order to investigate the putative functions of members of the microbial communities, a functional prediction profile was established using Tax4Fun2 (Wemheuer et al., 2020). In total, 22,831 Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthologs (KOs) were predicted among the different communities in rhizosphere and bulk soil samples.
Predicted functional profiles of the RMT-treated or control bulk soil communities clustered separately as revealed by PCA based on the relative abundances of predicted KOs (Figure 5B). In contrast, the predicted functional profiles for the rhizosphere of RMT and control plants were found to be similar. Bgh challenge seemed to influence the functional prediction profile of RMT and control rhizospheres, as observed in the PCA (Figure 5B). PERMANOVA was used to verify the differences visualized in the PCA. The predicted functional profiles were significantly different between the microhabitat rhizosphere or bulk soil (R2 = 0.20, p ≤ 0.001), as well as between the rhizospheres of RMT-treated and control plants (R2 = 0.40, p ≤ 0.001). Although the rhizospheres of uninfected RMT and control plants grouped closely together in the PCA, they were significantly different (R2 = 0.43, p ≤ 0.05). The highest influence on the predicted functional profile was exerted by challenge with Bgh (R2 = 0.71, p ≤ 0.05).
The prediction of functions based on 16S rRNA gene amplicon data might give an insight into the potential functional capabilities of a microbial community, but it remains unclear whether these functions are actually exhibited. Therefore, 10 randomly chosen bacterial isolates obtained from the RMT were characterized to verify their potential plant beneficial characteristics. BOX-PCR fingerprints of bacterial RMT isolates showed diverse patterns suggesting high diversity within the isolates. Among the randomly selected isolates, nine out of 10 belonged to different genera (Supplementary Table 4), based on partial 16S rRNA gene sequence annotation using the NCBI database: Microbacterium, Brevibacterium, Pantoea, Arthrobacter, Mucilaginibacter, Flavobacterium, Pseudarthrobacter, Bacillus, and Curtobacterium. Bacillus, Flavobacterium, and members of Micrococcaceae (Arthrobacter and Pseudarthrobacter) were also of higher abundance in the 16S rRNA amplicon data. Several potential plant beneficial characteristics were detected by means of in vitro assays (Figure 7). Protease activity and IAA production were characteristics presented by most of the isolates (6/10). Lytic enzyme activities demonstrated by several isolates were β-1,3-glucanase (Arthrobacter, Curtobacterium, and Flavobacterium), cellulase (Mucilaginibacter, Flavobacterium, and Pseudarthrobacter), and ACC deaminase (Microbacterium, Arthrobacter, and Pseudarthrobacter). Siderophore production was presented by Pantoea, Flavobacterium, Arthrobacter, and Curtobacterium. Pantoea was the only isolate showing AHL production and the ability to solubilize phosphate. Microbacterium was the only isolate able to degrade chitin. Brevibacterium and Bacillus showed only protease activity. In summary, representative bacterial isolates of the RMT were taxonomically diverse and showed in vitro potential plant beneficial properties.
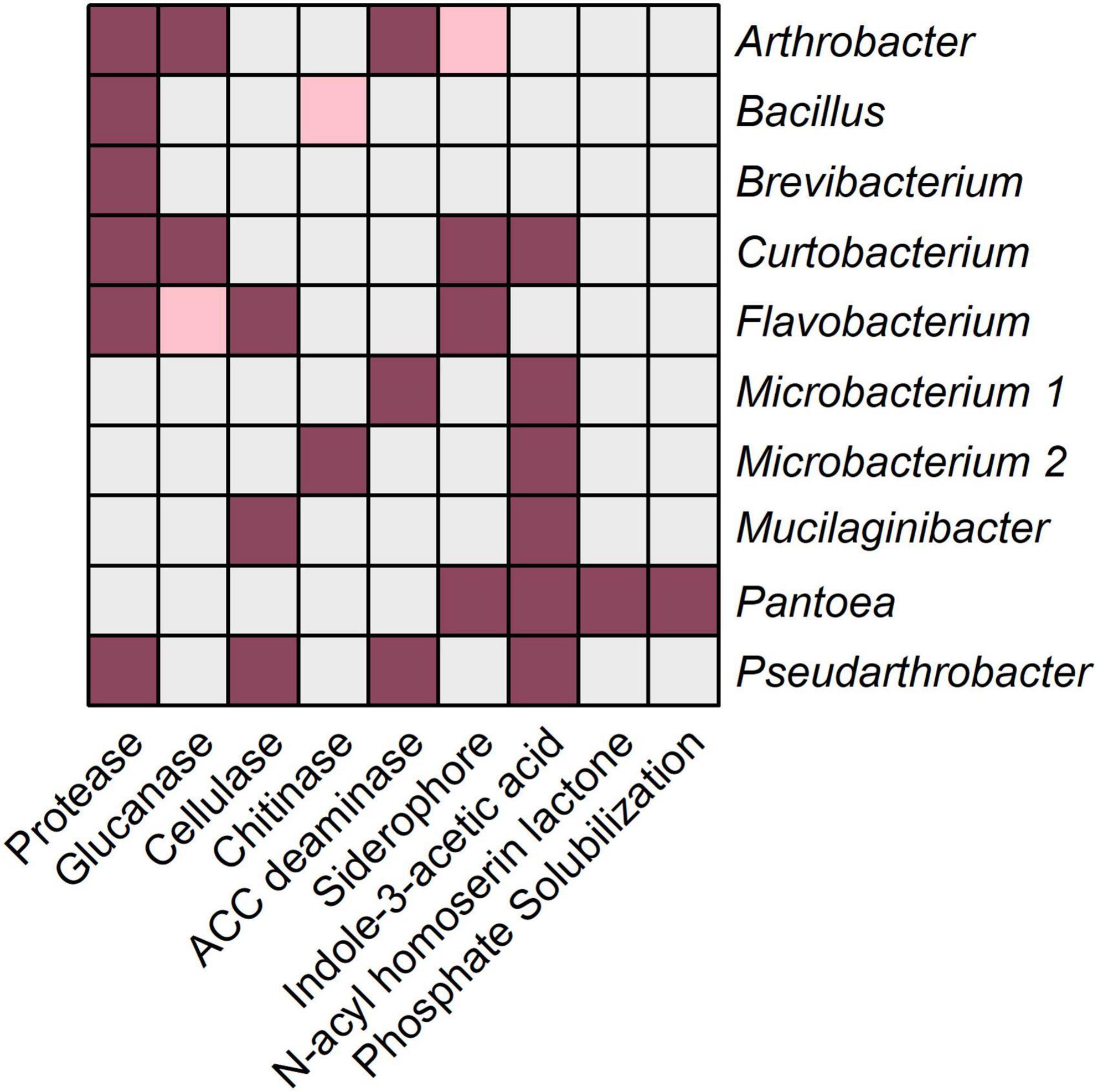
Figure 7. In vitro functional characteristics of bacterial isolates obtained from the rhizosphere microbiome transplant. Ten representative bacterial isolates obtained from the rhizosphere microbiome transplant were tested in vitro for their potential plant beneficial characteristics in different bioassays described in the section “Materials and Methods.” Gray color represents a negative result, pink color represents a slight positive result and violet color represents a positive result.
Microbiome modulation is thought to be an efficient method to improve plant performance and health (Berg et al., 2021). In the present study, we demonstrated that transplantation of a rhizosphere microbiome from barley grown in field soil to barley grown in potting soil with reduced microbial diversity significantly decreased the susceptibility to the aboveground leaf pathogen Bgh. A higher belowground microbial diversity does not only suppress soil-borne pathogens or invaders as reported before (Kusstatscher et al., 2019; Schierstaedt et al., 2020) but can also decrease the susceptibility toward foliar pathogens. Our results showed that the transplantation of a rhizosphere microbiome with high microbial diversity may be an important factor shaping the plant defense response.
Numerous studies already reported on the importance of microbial diversity for belowground plant protection against pathogens, for instance in suppressive soils (reviewed by Berg et al., 2017; Schlatter et al., 2017). Although single microorganisms or synthetic communities were described to confer resistance to aboveground pathogens via induced systemic resistance or priming (e.g., reviewed by Mauch-Mani et al., 2017; Vannier et al., 2019), only a few studies suggested the positive impact of the whole soil or rhizosphere microbiome on resistance to aboveground phytopathogens or pests (Chialva et al., 2018; Pineda et al., 2020; Bziuk et al., 2021), however, in most cases, different soils were used. Here, we showed that microbiome modulation decreased barleys’ susceptibility to powdery mildew, excluding the influence of different physicochemical properties from diverse soils.
Although the relative transcript abundances of the defense-related genes PR1b and PR17b were increased after Bgh challenge in RMT-treated barley compared to the control, due to high variation between the three independent experimental repetitions, we cannot draw conclusions on their significance. However, the relative transcript abundance of those genes was significantly increased in plants inoculated with E. meliloti expR+, after infected with Bgh. Previous studies indicated that E. meliloti expR+-induced resistance is based on the production of the AHL oxo-C14-HSL and could be mimicked using the synthetic molecule, in a phenomenon called AHL-priming (Hernández-Reyes et al., 2014). In the latter study, the authors observed a significant increase in relative transcript abundance of PR1b in plants exposed to oxo-C14-HSL and infected with Bgh after 24 h. Here, we assessed a greenhouse-based system. In contrast to already published Bgh pustule counts, transcript abundance data for AHL-primed barley after the Bgh challenge were available only from in vitro systems. Compared to the in vitro system of Hernández-Reyes et al. (2014), our results suggest that relative transcript abundance data generated from in vivo, greenhouse-grown barley inoculated with E. meliloti might not be consistent enough, probably due to the high variability of the gene expression level in several independent experiments. Nevertheless, the trend toward higher relative transcript abundances of defense-related genes in RMT-treated barley, together with the significantly decreased Bgh challenge rate, provided evidence of a decreased susceptibility given by the RMT.
The observation that the microbial alpha-diversity increased in RMT-treated rhizosphere and bulk soil was not surprising, as a highly diverse microbiome from field soil was transferred to potting soil with reduced microbial diversity (Figure 8). The reduced number of Bgh pustules, combined with the trend toward increased gene expression in RMT-treated barley suggests that the plants were prepared for a potential pathogen attack. As a consequence of the modulation of the rhizosphere microbiome by RMT, no influence on the community composition was visible after the Bgh challenge and no recruitment of specific microbes is needed, as these might already be present. The significant influence of the Bgh challenge on the microbial community composition of control samples, together with the lower alpha-diversity indices in the control, suggests that the low diverse microbiome in potting soil was disturbed and on contrary, the higher diversity in the RMT-treated rhizosphere buffered the impact of the aboveground challenge. Similar observations were reported before, such as the success of an invading species in soil was linked to the community diversity and evenness (De Roy et al., 2013; Mallon et al., 2015; Schierstaedt et al., 2020). Berg et al. (2017) suggested as well that diseases are often correlated with microbial dysbiosis or reduced soil diversity. Berg et al. (2021) suggested the “Anna Karenina Principle” for the plant microbiome that was previously considered by Zaneveld et al. (2017) for the animal microbiome. The principle describes that dysbiotic plants vary more in their microbiome composition compared to healthy plants (Berg et al., 2021). Healthy plants grown in soil with comparable high microbial diversity might have better options to select plant beneficial microbes or functional traits compared to unhealthy, dysbiotic plants.
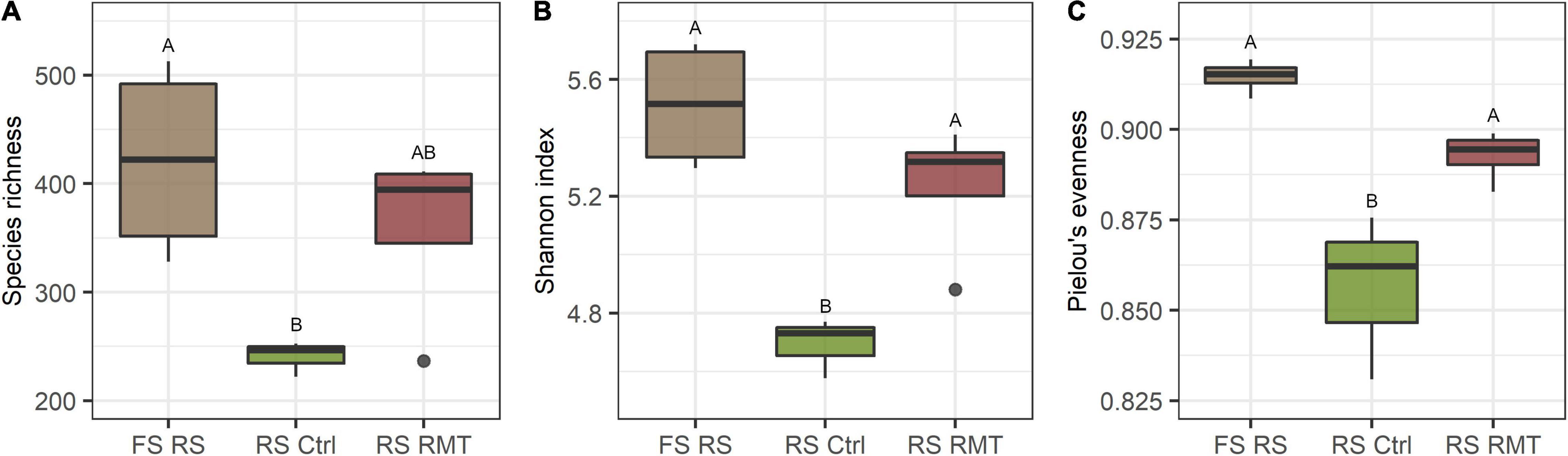
Figure 8. Comparison of microbial diversity between barley rhizosphere from field soil, rhizosphere microbiome transplant, and control treatments. (A) Species richness, (B) Shannon index, and (C) Pielou’s evenness of barley rhizosphere (RS) from field soil (FS) that served as rhizosphere microbiome transplant (RMT), additionally to rhizosphere from barley grown in potting soil treated with RMT or the control (Ctrl). Different letters indicate significant differences (Tukey’s test; p ≤ 0.05).
Furthermore, we detected taxa with treatment-dependent relative abundances, also in response to the Bgh challenge (Figure 6B). In Bgh-infected control samples, the genus Fluviicola was about four times higher compared to uninfected barley (relative abundance 1.9 vs. 0.5%). Fluviicola (Flavobacteriales) was first described by O’Sullivan et al. (2005). By now, the type strain is sequenced (Woyke et al., 2011), but only four species of the genus Fluviicola are described. There is not much knowledge of the environmental and functional role of Fluviicola in a microbial community, but there are reports on different contaminated soils and thus it might be involved in the degradation of the respective contaminants (Song et al., 2016; Lu and Lu, 2018; Jia et al., 2019; Revelo Romo et al., 2019). This is the first time that ASV affiliated to this genus were detected in the rhizosphere of barley infected aboveground with Bgh and, although not measured in the present study, it is assumed that released phenolic compounds, typically involved in plant defense, (Mandal et al., 2010) might have triggered the proliferation of Fluviicola sp. Other taxa with significantly higher relative abundances after the Bgh challenge in the rhizosphere of the control samples belonged to the family Cytophagaceae, whose members are able to degrade cellulose (Stanier, 1940). Degradation of cellulose seems plausible as the challenge results in increased amounts of dead plant material that can serve as a nutrient source. One member of Cytophagaceae, the genus Emiticicia, revealed higher relative abundances after Bgh challenge, however, not much is known about this genus (Saha and Chakrabarti, 2006). The results do not suggest taxa responsible for plant protection or microbial dysbiosis, but rather point toward increased microbial degradation.
The predicted functional profiles revealed that the different bulk soil microbial communities were also distinct in their predicted functional capabilities, likely due to the RMT treatment. In the uninfected rhizosphere (RMT/Ctrl; –Bgh), microbial communities were significantly different in their structure, but probably displayed similar major functions (Figure 5B). After the Bgh challenge (RMT/Ctrl; +Bgh), the predicted functional profile of rhizosphere samples showed higher separation between RMT and control (Figure 5B; PERMANOVA), suggesting a change in predicted functional capabilities after the pathogen challenge. Taken together, RMT mainly shifted the microbial community structure but the Bgh challenge mainly shifted the predicted functional profile.
The 16S rRNA gene amplicon sequencing method used in the present study has limitations, therefore, bacterial isolates obtained from the RMT were investigated for potential plant beneficial functions. Interestingly, some isolates belonged to highly abundant members of the RMT-treated rhizosphere, such as Arthrobacter and Pseudarthrobacter (Micrococcaceae). Unclassified Micrococcaceae displayed increased relative abundances in RMT-treated rhizosphere compared to the control based on the 16S rRNA amplicon data. The respective ASV sequences revealed high sequence similarity to the genera Arthrobacter or Pseudarthrobacter (Supplementary Table 2). Micrococcaceae were also highly abundant in the respective rhizosphere from field soil used for RMT (relative abundance field soil: 3.0%; Ctrl: 0.1%; RMT: 4.8%). All obtained bacterial isolates showed at least one potential plant beneficial property, nevertheless, isolates affiliated with Pantoea, Curtobacterium, as well as Arthrobacter, Pseudarthrobacter, and Flavobacterium displayed the highest number of potential plant beneficial properties. Strains of the genus Arthrobacter are already known for their plant-beneficial capabilities, for instance, the production of ACC deaminase (Barnawal et al., 2017) and siderophores (Sharma et al., 2016), as also observed in the present study. Furthermore, Arthrobacter species were found to enhance the salt tolerance of wheat plants (Barnawal et al., 2017; Safdarian et al., 2019) and showed antagonistic activity against several fungal plant pathogens (Pylak et al., 2020), suggesting a high potential for plant protection.
The present study showed that microbiome modulation by RMTs might be a sustainable strategy to improve the health of plants grown in low diversity soil, such as potting soils that are used for greenhouse production. Plants grown in potting soil showed higher disease incidence than previously reported for barley infected with Bgh. Treatment of barley grown in potting soil with RMT improved plant health by decreasing the challenge rate of Bgh. The rhizosphere microbiome of RMT-treated barley showed a higher microbial diversity and a higher relative abundance of potentially plant beneficial taxa such as Arthrobacter and Pseudarthrobacter. The present study provides unequivocal evidence for the important role of the rhizosphere microbiome and its natural diversity for plant health.
The data presented in the study are deposited in the Sequence Read Archive of NCBI, BioProject accession number: PRJNA777052 (as a reviewer, you can use this link: https://dataview.ncbi.nlm.nih.gov/object/PRJNA777052?reviewer=dm8chdu2vvu4n0aqfva7994oul).
NB and KS designed the study. NB performed the experiments and conducted the statistical analyses. SS coordinated the sequencing facility at KU. LM carried out the amplicon sequencing. LM and NB performed the bioinformatic analyses of amplicon sequencing data. NB wrote the manuscript with the contributions of KS and AS. All authors read and approved the final manuscript.
This study was supported by the German Federal Ministry of Education and Research (BMBF) in the framework of the project PrimedPlant (grant no. FKZ031B0196B).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Desirée Lauterbach for her excellent technical assistance during all experimental steps. Further, we thank Benjamin Straube for functional characterization of the isolates and Ichie Ojiro for help with the detached leaf assay. We also thank Simpsons Malt Limited for providing barley seeds, Anette Deubel for supplying the field soil and Ilse-Marie Jungkurth for proofreading. Further, we would like to thank the Biocomputing Core Facility from the Department of Biology at the University of Copenhagen for ASV generation in amplicon sequencing analyses computation.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.830905/full#supplementary-material
Alegria Terrazas, R., Balbirnie-Cumming, K., Morris, J., Hedley, P. E., Russell, J., Paterson, E., et al. (2020). A footprint of plant eco-geographic adaptation on the composition of the barley rhizosphere bacterial microbiota. Sci. Rep. 10:12916. doi: 10.1038/s41598-020-69672-x
Barnawal, D., Bharti, N., Pandey, S. S., Pandey, A., Chanotiya, C. S., and Kalra, A. (2017). Plant growth-promoting rhizobacteria enhance wheat salt and drought stress tolerance by altering endogenous phytohormone levels and TaCTR1/TaDREB2 expression. Physiol. Plant 161, 502–514. doi: 10.1111/ppl.12614
Berendsen, R. L., Pieterse, C. M. J., and Bakker, P. A. H. M. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Berendsen, R. L., Vismans, G., Yu, K., Song, Y., Jonge, R., de, et al. (2018). Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J. 2018:1. doi: 10.1038/s41396-018-0093-1
Berg, G., Fritze, A., Roskot, N., and Smalla, K. (2001). Evaluation of potential biocontrol rhizobacteria from different host plants of Verticillium dahliae Kleb. J. Appl. Microbiol. 91, 963–971. doi: 10.1046/j.1365-2672.2001.01462.x
Berg, G., Köberl, M., Rybakova, D., Müller, H., Grosch, R., and Smalla, K. (2017). Plant microbial diversity is suggested as the key to future biocontrol and health trends. FEMS Microbiol. Ecol. 93:50. doi: 10.1093/femsec/fix050
Berg, G., Kusstatscher, P., Abdelfattah, A., Cernava, T., and Smalla, K. (2021). Microbiome modulation - toward a better understanding of plant microbiome response to microbial inoculants. Front. Microbiol. 12:650610. doi: 10.3389/fmicb.2021.650610
Berg, G., and Smalla, K. (2009). Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 68, 1–13. doi: 10.1111/j.1574-6941.2009.00654.x
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Bziuk, N., Maccario, L., Douchkov, D., Lueck, S., Babin, D., Sørensen, S. J., et al. (2021). Tillage shapes the soil and rhizosphere microbiome of barley - but not its susceptibility towards Blumeria graminis f. sp. hordei. FEMS Microbiol. Ecol 2021:18. doi: 10.1093/femsec/fiab018
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U S A 108, (Suppl. 1), 4516–4522. doi: 10.1073/pnas.1000080107
Carrión, V. J., Perez-Jaramillo, J., Cordovez, V., Tracanna, V., Hollander, M., de, et al. (2019). Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science 366, 606–612. doi: 10.1126/science.aaw9285
Chialva, M., Di Salvioli Fossalunga, A., Daghino, S., Ghignone, S., Bagnaresi, P., Chiapello, M., et al. (2018). Native soils with their microbiotas elicit a state of alert in tomato plants. New Phytol. 220, 1296–1308. doi: 10.1111/nph.15014
Davis, N. M., Proctor, D. M., Holmes, S. P., Relman, D. A., and Callahan, B. J. (2018). Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6:226. doi: 10.1186/s40168-018-0605-2
De Roy, K., Marzorati, M., Negroni, A., Thas, O., Balloi, A., Fava, F., et al. (2013). Environmental conditions and community evenness determine the outcome of biological invasion. Nat. Commun. 4:1383. doi: 10.1038/ncomms2392
Deubel, A., Hofmann, B., and Orzessek, D. (2011). Long-term effects of tillage on stratification and plant availability of phosphate and potassium in a loess chernozem. Soil Tillage Res. 117, 85–92. doi: 10.1016/j.still.2011.09.001
Durán, N., Justo, G. Z., Durán, M., Brocchi, M., Cordi, L., Tasic, L., et al. (2016). Advances in Chromobacterium violaceum and properties of violacein-Its main secondary metabolite: a review. Biotechnol. Adv. 34, 1030–1045. doi: 10.1016/j.biotechadv.2016.06.003
Gomes, N. C. M., Heuer, H., Schönfeld, J., Costa, R., Mendonça-Hagler, L., and Smalla, K. (2001). Bacterial diversity of the rhizosphere of maize (Zea mays) grown in tropical soil studied by temperature gradient gel electrophoresis. Plant Soil. 232, 167–180. doi: 10.1023/A:1010350406708
Gouda, S., Kerry, R. G., Das, G., Paramithiotis, S., Shin, H.-S., and Patra, J. K. (2018). Revitalization of plant growth promoting rhizobacteria for sustainable development in agriculture. Microbiol. Res. 206, 131–140. doi: 10.1016/j.micres.2017.08.016
Gu, S., Yang, T., Shao, Z., Wang, T., Cao, K., Jousset, A., et al. (2020). Siderophore-mediated interactions determine the disease suppressiveness of microbial consortia. mSystems 5:19. doi: 10.1128/mSystems.00811-19
Han, S., Li, D., Trost, E., Mayer, K. F., Vlot, A. C., Heller, W., et al. (2016). Systemic responses of barley to the 3-hydroxy-decanoyl-homoserine lactone producing plant beneficial endophyte Acidovorax radicis N35. Front. Plant Sci. 7:1868. doi: 10.3389/fpls.2016.01868
Hartmann, A., Rothballer, M., and Schmid, M. (2008). Lorenz Hiltner, a pioneer in rhizosphere microbial ecology and soil bacteriology research. Plant Soil 312, 7–14. doi: 10.1007/s11104-007-9514-z
Hernández-Reyes, C., Schenk, S. T., Neumann, C., Kogel, K.-H., and Schikora, A. (2014). N-acyl-homoserine lactones-producing bacteria protect plants against plant and human pathogens. Microb. Biotechnol. 7, 580–588. doi: 10.1111/1751-7915.12177
Heuer, H., Kopmann, C., Binh, C. T. T., Top, E. M., and Smalla, K. (2009). Spreading antibiotic resistance through spread manure: characteristics of a novel plasmid type with low %G+C content. Environ. Microbiol. 11, 937–949. doi: 10.1111/j.1462-2920.2008.01819.x
Hiltner, L. (1904). Über neuere Erfahrungen und Probleme auf dem Gebiet der Bodenbakteriologie und unter besonderer Berücksichtigung der Gründüngung und Brache. Arb. DLG 1904, 59–78.
Hothorn, T., Bretz, F., and Westfall, P. (2008). Simultaneous inference in general parametric models. Biom J. 50, 346–363. doi: 10.1002/bimj.200810425
Jia, S., Wu, J., Ye, L., Zhao, F., Li, T., and Zhang, X.-X. (2019). Metagenomic assembly provides a deep insight into the antibiotic resistome alteration induced by drinking water chlorination and its correlations with bacterial host changes. J. Hazard Mater. 379:120841. doi: 10.1016/j.jhazmat.2019.120841
Kolde, R. (2015). pheatmap: Pretty Heatmaps. Available online at: http://CRAN.R-project.org/package=pheatmap (Accessed September 02, 2021).
Koo, S.-Y., Hong, S. H., Ryu, H. W., and Cho, K. (2010). Plant growth-promoting trait of rhizobacteria isolated from soil contaminated with petroleum and heavy metals. J. Microbiol. Biotechnol. 20, 587–593. doi: 10.4014/jmb.0907.07017
Kusstatscher, P., Cernava, T., Harms, K., Maier, J., Eigner, H., Berg, G., et al. (2019). Disease incidence in sugar beet fields is correlated with microbial diversity and distinct biological markers. Phytobiomes J. 3, 22–30. doi: 10.1094/PBIOMES-01-19-0008-R
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, X.-M., and Lu, P.-Z. (2018). Response of microbial communities to pesticide residues in soil restored with Azolla imbricata. Appl. Microbiol. Biotechnol. 102, 475–484. doi: 10.1007/s00253-017-8596-7
Mallon, C. A., van Elsas, J. D., and Salles, J. F. (2015). Microbial invasions: the process, patterns, and mechanisms. Trends Microbiol. 23, 719–729. doi: 10.1016/j.tim.2015.07.013
Mandal, S. M., Chakraborty, D., and Dey, S. (2010). Phenolic acids act as signaling molecules in plant-microbe symbioses. Plant Signal Behav. 5, 359–368. doi: 10.4161/psb.5.4.10871
Martin, B., Humbert, O., Camara, M., Guenzi, E., Walker, J., Mitchell, T., et al. (1992). A highly conserved repeated DNA element located in the chromosome of Streptococcus pneumoniae. Nucleic Acids Res. 20, 3479–3483. doi: 10.1093/nar/20.13.3479
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17:10. doi: 10.14806/ej.17.1.200
Mauch-Mani, B., Baccelli, I., Luna, E., and Flors, V. (2017). Defense priming: an adaptive part of induced resistance. Annu. Rev. Plant Biol. 68, 485–512. doi: 10.1146/annurev-arplant-042916-041132
McMurdie, P. J., and Holmes, S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217
Meier, U. (2018). Growth stages of mono- and dicotyledonous plants: BBCH Monograph. Open Agrar Repositorium. Quedlinburg, Germany.
Mendes, L. W., Raaijmakers, J. M., Hollander, M., de, Mendes, R., and Tsai, S. M. (2018). Influence of resistance breeding in common bean on rhizosphere microbiome composition and function. ISME J. 12, 212–224. doi: 10.1038/ismej.2017.158
Mendes, R., Garbeva, P., and Raaijmakers, J. M. (2013). The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 37, 634–663. doi: 10.1111/1574-6976.12028
Moradtalab, N., Ahmed, A., Geistlinger, J., Walker, F., Höglinger, B., Ludewig, U., et al. (2020). Synergisms of microbial consortia, N forms, and micronutrients alleviate oxidative damage and stimulate hormonal cold stress adaptations in maize. Front. Plant Sci. 11:396. doi: 10.3389/fpls.2020.00396
Mülner, P., Schwarz, E., Dietel, K., Junge, H., Herfort, S., Weydmann, M., et al. (2020). Profiling for bioactive peptides and volatiles of plant growth promoting strains of the Bacillus subtilis complex of industrial relevance. Front. Microbiol. 11:1432. doi: 10.3389/fmicb.2020.01432
Niu, B., Paulson, J. N., Zheng, X., and Kolter, R. (2017). Simplified and representative bacterial community of maize roots. Proc. Natl. Acad. Sci. U S A 114, E2450–E2459. doi: 10.1073/pnas.1616148114
Niu, B., Wang, W., Yuan, Z., Sederoff, R. R., Sederoff, H., Chiang, V. L., et al. (2020). Microbial interactions within multiple-strain biological control agents impact soil-borne plant disease. Front. Microbiol. 11:585404. doi: 10.3389/fmicb.2020.585404
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2019). vegan: Community Ecology Package. Available online at: https://CRAN.R-project.org/package=vegan (accessed September 15, 2020).
O’Sullivan, L. A., Rinna, J., Humphreys, G., Weightman, A. J., and Fry, J. C. (2005). Fluviicola taffensis gen. nov., sp. nov., a novel freshwater bacterium of the family Cryomorphaceae in the phylum ‘Bacteroidetes’. Int. J. Syst. Evol. Microbiol. 55, 2189–2194. doi: 10.1099/ijs.0.63736-0
Pieterse, C. M. J., Zamioudis, C., Berendsen, R. L., Weller, D. M., van Wees, S. C. M., and Bakker, P. A. H. M. (2014). Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 52, 347–375. doi: 10.1146/annurev-phyto-082712-102340
Pineda, A., Kaplan, I., Hannula, S. E., Ghanem, W., and Bezemer, T. M. (2020). Conditioning the soil microbiome through plant-soil feedbacks suppresses an aboveground insect pest. New Phytol. 226, 595–608. doi: 10.1111/nph.16385
Pylak, M., Oszust, K., and Frąc, M. (2020). Searching for new beneficial bacterial isolates of wild raspberries for biocontrol of phytopathogens-antagonistic properties and functional characterization. Int. J. Mol. Sci. 21:21249361. doi: 10.3390/ijms21249361
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Revelo Romo, D. M., Hurtado Gutiérrez, N. H., Ruiz Pazos, J. O., Pabón Figueroa, L. V., and Ordóñez Ordóñez, L. A. (2019). Bacterial diversity in the Cr(VI) reducing biocathode of a Microbial Fuel Cell with salt bridge. Rev. Argent Microbiol. 51, 110–118. doi: 10.1016/j.ram.2018.04.005
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Safdarian, M., Askari, H., Shariati, J. V., and Nematzadeh, G. (2019). Transcriptional responses of wheat roots inoculated with Arthrobacter nitroguajacolicus to salt stress. Sci. Rep. 9:1792. doi: 10.1038/s41598-018-38398-2
Saha, P., and Chakrabarti, T. (2006). Emticicia oligotrophica gen. nov., sp. nov., a new member of the family ‘Flexibacteraceae’, phylum Bacteroidetes. Int. J. Syst. Evol. Microbiol. 56, 991–995. doi: 10.1099/ijs.0.64086-0
Sarma, B. K., Yadav, S. K., Singh, S., and Singh, H. B. (2015). Microbial consortium-mediated plant defense against phytopathogens: Readdressing for enhancing efficacy. Soil Biol. Biochem. 87, 25–33. doi: 10.1016/j.soilbio.2015.04.001
Schierstaedt, J., Jechalke, S., Nesme, J., Neuhaus, K., Sørensen, S. J., Grosch, R., et al. (2020). Salmonella persistence in soil depends on reciprocal interactions with indigenous microorganisms. Environ. Microbiol. 2020, 14972. doi: 10.1111/1462-2920.14972
Schlatter, D., Kinkel, L., Thomashow, L., Weller, D., and Paulitz, T. (2017). Disease suppressive soils: new insights from the soil microbiome. J. Phytopathol. 107, 1284–1297. doi: 10.1094/PHYTO-03-17-0111-RVW
Schwyn, B., and Neilands, J. B. (1987). Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160, 47–56. doi: 10.1016/0003-2697(87)90612-9
Sharma, M., Mishra, V., Rau, N., and Sharma, R. S. (2016). Increased iron-stress resilience of maize through inoculation of siderophore-producing Arthrobacter globiformis from mine. J. Basic Microbiol. 56, 719–735. doi: 10.1002/jobm.201500450
Shrestha, A., Elhady, A., Adss, S., Wehner, G., Böttcher, C., Heuer, H., et al. (2019). Genetic differences in barley govern the responsiveness to N -acyl homoserine lactone. Phytobiomes J. 3, 191–202. doi: 10.1094/PBIOMES-03-19-0015-R
Song, Y., Li, Y., Zhang, W., Wang, F., Bian, Y., Boughner, L. A., et al. (2016). Novel biochar-plant tandem approach for remediating hexachlorobenzene contaminated soils: proof-of-concept and new insight into the rhizosphere. J. Agric. Food Chem. 64, 5464–5471. doi: 10.1021/acs.jafc.6b01035
Stanier, R. Y. (1940). Studies on the Cytophagas. J. Bacteriol. 40, 619–635. doi: 10.1128/jb.40.5.619-635.1940
Sundberg, C., Al-Soud, W. A., Larsson, M., Alm, E., Yekta, S. S., Svensson, B. H., et al. (2013). 454 pyrosequencing analyses of bacterial and archaeal richness in 21 full-scale biogas digesters. FEMS Microbiol Ecol 85, 612–626. doi: 10.1111/1574-6941.12148
Syed Ab Rahman, S. F., Singh, E., Pieterse, C. M. J., and Schenk, P. M. (2018). Emerging microbial biocontrol strategies for plant pathogens. Plant Sci. 267, 102–111. doi: 10.1016/j.plantsci.2017.11.012
Tufan, F., Uçarlı, C., Tunalı, B., and Gürel, F. (2017). Analysis of early events in barley (Hordeum vulgare L.) roots in response to Fusarium culmorum infection. Eur. J. Plant Pathol. 148, 343–355. doi: 10.1007/s10658-016-1087-3
Vandenkoornhuyse, P., Quaiser, A., Duhamel, M., Le Van, A., and Dufresne, A. (2015). The importance of the microbiome of the plant holobiont. New Phytol. 206, 1196–1206. doi: 10.1111/nph.13312
Vannier, N., Agler, M., and Hacquard, S. (2019). Microbiota-mediated disease resistance in plants. PLoS Pathog. 15:e1007740. doi: 10.1371/journal.ppat.1007740
Wehner, G., Kopahnke, D., Richter, K., Kecke, S., Schikora, A., and Ordon, F. (2019). Priming is a suitable strategy to enhance resistance towards leaf rust in barley. Phytobiomes J. 3, 46–51. doi: 10.1094/PBIOMES-09-18-0041-R
Weinert, N., Meincke, R., Gottwald, C., Heuer, H., Schloter, M., Berg, G., et al. (2010). Bacterial diversity on the surface of potato tubers in soil and the influence of the plant genotype. FEMS Microbiol. Ecol. 74, 114–123. doi: 10.1111/j.1574-6941.2010.00936.x
Wemheuer, F., Taylor, J. A., Daniel, R., Johnston, E., Meinicke, P., Thomas, T., et al. (2020). Tax4Fun2: prediction of habitat-specific functional profiles and functional redundancy based on 16S rRNA gene sequences. Environ. Microbiome 15:11. doi: 10.1186/s40793-020-00358-7
Woo, S. L., and Pepe, O. (2018). Microbial consortia: promising probiotics as plant biostimulants for sustainable agriculture. Front. Plant Sci. 9:1801. doi: 10.3389/fpls.2018.01801
Woyke, T., Chertkov, O., Lapidus, A., Nolan, M., Lucas, S., Del Rio, T. G., et al. (2011). Complete genome sequence of the gliding freshwater bacterium Fluviicola taffensis type strain (RW262). Stand Genomic Sci 5, 21–29.
Yu, Y., Lee, C., Kim, J., and Hwang, S. (2005). Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol. Bioeng. 89, 670–679. doi: 10.1002/bit.20347
Zaneveld, J. R., McMinds, R., and Vega Thurber, R. (2017). Stress and stability: applying the Anna Karenina principle to animal microbiomes. Nat. Microbiol. 2:17121. doi: 10.1038/nmicrobiol.2017.121
Keywords: Hordeum vulgare, Blumeria graminis f. sp. hordei, plant health, microbial community, rhizosphere, microbial transplant, decreased susceptibility
Citation: Bziuk N, Maccario L, Sørensen SJ, Schikora A and Smalla K (2022) Barley Rhizosphere Microbiome Transplantation – A Strategy to Decrease Susceptibility of Barley Grown in Soils With Low Microbial Diversity to Powdery Mildew. Front. Microbiol. 13:830905. doi: 10.3389/fmicb.2022.830905
Received: 07 December 2021; Accepted: 26 April 2022;
Published: 24 May 2022.
Edited by:
Jeanette M. Norton, Utah State University, United StatesReviewed by:
Antonin Dreiseitl, Agricultural Research Institute Kromeriz, CzechiaCopyright © 2022 Bziuk, Maccario, Sørensen, Schikora and Smalla. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kornelia Smalla, a29ybmVsaWEuc21hbGxhQGp1bGl1cy1rdWVobi5kZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.