- 1College of Plant Protection, Southwest University, Chongqing, China
- 2Sichuan Company of China National Tobacco Corporation, Chengdu, China
Bacterial wilt, caused by the plant pathogen Ralstonia solanacearum, occurs more severely in acidified soil according to previous reports. However, R. solanacearum cannot grow well in acidic environments under barren nutrient culture conditions, especially when the pH is lower than 5. With the worsening acidification of farmland, further determination of how R. solanacearum adapts to the long-term acidic environment is worthwhile. In this study, experimental evolution was applied to evaluate the adaptability and mechanism of the R. solanacearum experimental population responding to long-term acid stress. We chose the CQPS-1 strain as the ancestor, and minimal medium (MM medium) with different pH values as the culture environment to simulate poor soil. After 1500 generations of serial passage experiments in pH 4.9 MM, acid-adapted experimental strains (denoted as C49 strains) were obtained, showing significantly higher growth rates than the growth rates of control experimental strains (serial passage experiment in pH 6.5 MM, denoted as C65 strains). Competition experiments showed that the competitive indices (CIs) of all selected clones from C49 strains were superior to the ancestor in acidic environment competitiveness. Based on the genome variation analysis and functional verification, we confirmed that loss of function in the phcA gene was associated with the acid fitness gain of R. solanacearum, which meant that the inactivation of the PhcA regulator caused by gene mutation mediated the population expansion of R. solanacearum when growing in an acidic stress environment. Moreover, the swimming motility of acid evolution strains and the phcA deletion mutant was significantly enhanced compared to CQPS-1. This work provided evidence for understanding the adaptive strategy of R. solanacearum to the long-term acidic environment.
Introduction
Ralstonia solanacearum, one of the top ten plant pathogenic bacteria in the world, is considered a variable compound species due to its large host range, wide distribution, and rich genetic diversity (Genin and Denny, 2012; Mansfield et al., 2012). Wide geographic distribution and host range make R. solanacearum face and adapt to various environments, such as poor nutrient environments and overly acidic environments. The soil environment is an important living place for R. solanacearum, especially in the fallow period. According to reports, R. solanacearum has a strong ability to survive in the soil (Graham and Lloyd, 1979).
Soil acidification is one of the most serious soil problems in the current ecological environment and agricultural production process (Guo et al., 2010; Kochian et al., 2015). Acidification not only affects plant growth and nutrient absorption but is also closely related to the occurrence of diseases. Previous studies have shown that soil acidification is related to the occurrence of bacterial wilt and the pathogenicity of R. solanacearum: lower pH in acidified soil results in more severe bacterial wilt, and the pathogenic genes of R. solanacearum are induced (Li et al., 2017). However, our previous research shows that the growth of R. solanacearum is inhibited when the pH is lower than 5.0 in culture with poor nutrient. With the continuous input of human activities such as nitrogen fertilizer, the problem of soil acidification has become increasingly serious (Zhalnina et al., 2015). The effect of a long-term acidic environment on R. solanacearum is worthy of attention but has not been reported thus far.
Evolutionary biologists have more specifically studied the limitations and potential of species to adapt to the environment, including abiotic stresses (such as salt stress, heavy metal stress, heat stress, pH stress, and drought or water stress), as well as when the environment changes how species react (Bijlsma and Loeschcke, 2005; Bridle and Vines, 2007; Flowers et al., 2010; Kooyers, 2015). Organisms can respond to these abiotic stresses in many ways. In the face of stress, organisms can adapt through phenotypic plasticity on the one hand, and change their phenotype to adapt to the abiotic environment; on the other hand, organisms can also respond to changes in genotypes that match abiotic conditions through evolutionary adaptation (Westeberhard, 1989; Kawecki and Ebert, 2004). When populations cannot quickly adapt or migrate (dispersion), these populations may be forced to extinction locally, and the remaining part of the surviving population will adapt to the environment and “evolve”. And thus, “experimental evolution and resequence” has a great potential to predict how natural populations will evolve and adapt to major changes in environmental conditions (Phillips and Burke, 2021). Genomes are a very useful resource to understand the evolutionary process and adaptability of the species. R. solanacearum strain GMI1000 was the first strain subject to whole genome analysis, after which a large number of strain genomes were published (Salanoubat et al., 2002; Peeters et al., 2013). A large amount of genomic information provides a basis for analyzing this population. More importantly, the extremely large possibility of variation within the genomes of the R. solanacearum species complex determines its strong adaptability and polymorphism (Salanoubat et al., 2002; Genin and Boucher, 2004). However, there is currently no information on the genomes of R. solanacearum strains adapted to acidic soil environments.
To explore how R. solanacearum survives and adapts to a low pH environment for a long time, the methods of experimental evolution and genome resequencing were selected for evaluation in this study, aiming to clarify the molecular mechanism of the R. solanacearum response to acid stress. The results showed that loss of function in the phcA gene was associated with the gain in acid fitness of R. solanacearum, which laid the foundation for theoretical research on the adaptive evolution of the R. solanacearum strain.
Materials and Methods
Minimal Medium With Different pH Values as the Acidic and Control Environment
This study simulated poor soil with minimal medium (MM medium). The MM medium is equivalent to 1/4 M63 medium (Boucher et al., 1985). The basic composition of MM medium contained FeSO4⋅7H2O, (NH4)2SO4, MgSO4, and KH2PO4. When cultivating R. solanacearum, glucose was used as the carbon source, supplemented at a final concentration of 20 mM. The different pH values of MM medium were adjusted by KOH. The pH of the MM medium was adjusted to 6.5 with KOH as the control environment and without KOH to adjust the pH to an acidic environment (pH 4.9) for the experiment.
Bacterial Strains and Culture Conditions
Ralstonia solanacearum strain CQPS-1 (phylotype I/biovar 3) with known genome information was used as the original strain in this study (Liu et al., 2017). R. solanacearum strain CQPS-1 was grown at 30°C in BG medium, B liquid medium, or MM medium with different pH values based on needs. Growth was detected by measuring the optical density (OD) at 600 nm. The bacterial growth curve was monitored according to Yang et al. (2016). Morphologies of colonies were observed on BG medium supplemented with triphenyltetrazolium chloride (TTC, 0.05 g⋅L–1).
Escherichia coli strains were grown at 37°C in Luria-Bertani (LB) medium. When necessary, antibiotics were added to the media at the following final concentrations: ampicillin (Amp, 100 mg⋅L–1), kanamycin (km, 50 mg⋅L–1), polymyxin B (PB, 50 mg⋅L–1), gentamycin (Gm, 20 mg⋅L–1).
Long-Term Experimental Evolution of Ralstonia solanacearum in an Acidic Environment
A serial passage experiment was performed on a clone of R. solanacearum CQPS-1 using MM medium with different pH values adjusted by KOH. At each SPE (serial passage experiment), bacteria were grown in 20 ml medium for 24 h at 30° with 180 r/min shaking, and then 100 μl of culture was transferred into 20 ml fresh medium (as shown in Figure 1). Each treatment had five independent cultures (equivalent to five parallel replicates). The number of bacterial generations at each SPE was estimated by comparing the number of cells at SPEn and the effective number of cells at SPEn+1, which was just an estimate, and the actual population generation should be larger than the estimated value population size. Finally, glycerol stock of strains stored at −80°C at point time, which can be recovered for relevant evaluation.
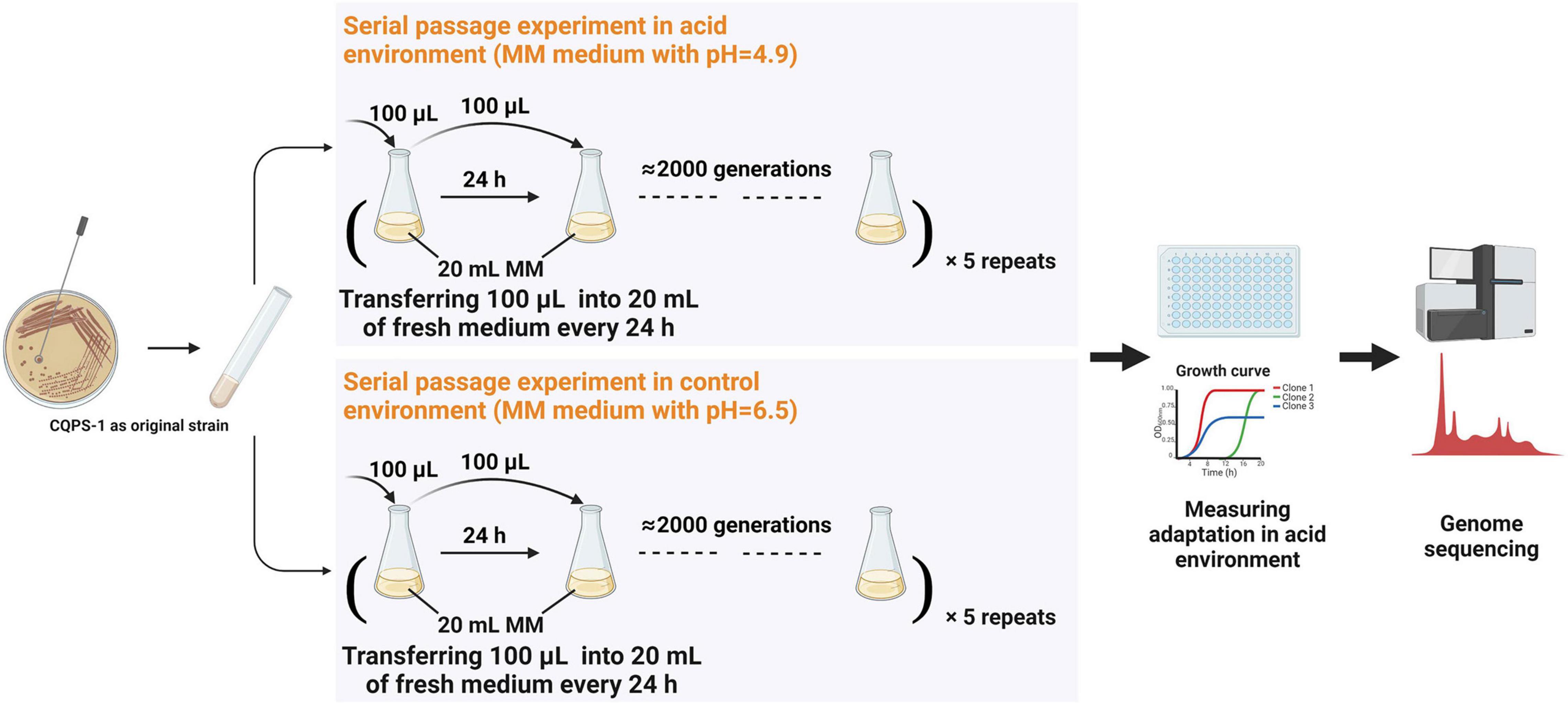
Figure 1. Long-term experimental evolution of CQPS-1 in an acidic environment. MM medium at pH 4.9 was used as the acidic environment, and MM medium (pH 6.5) was used as the control. Cultures were propagated daily by transferring 100 μL of each culture into 20 mL of fresh medium. The figure was created with BioRender.com.
Monitoring in vitro Growth Rates of Evolved Populations
The growth rates of evolved populations growing in an acidic environment were measured in MM cultures at pH 4.9. One clone was randomly isolated from every five independent populations generated by the serial passage experiment. Overnight-cultured bacterial suspension (1 μl) was inoculated in 199 μl MM medium (pH 4.9) in 96-well plates. Bacterial growth was monitored using a Multiskan GO microplate reader (Thermo Fisher Scientific). Measures of OD600 nm were performed every 5 min. The growth rates of each sample within 32 h were determined using the kinetic analysis module of Multiskan GO software. All samples were analyzed in triplicate and calculated to obtain an averaged value.
Bacterial Competition Assays in the Acidic Environment
The method used to estimate the fitness of R. solanacearum clones in acidic environments is based on competition assays with some modifications (Macho et al., 2010). Briefly, the acidic environment fitness of each evolved clone was measured by co-culturing with the ancestral clone in MM medium at pH 4.9. The distinction between experimental clones and original clones is based mainly on colony morphology: all the colonies after the serial passage experiment were small, dark red, with poor fluidity, while the colonies of the original strain CQPS-1 were cream-colored, irregularly shaped, and highly fluid (as shown in Supplementary Figure 1). A 108 colony forming unit (CFU)/ml bacterial suspension containing equal CFU of the experimental clone and the ancestral clone was inoculated in MM (pH 4.9) and grown at 30°C with 180 rpm shaking. After 24 h, serial dilutions of the inoculum were plated onto BG medium, to determine the bacterial ratio between the experimental and ancestral clones. The competitive index (CI) was defined as the experimental/ancestral-clone ratio divided by the ratio in the corresponding inoculum (Baumler et al., 1997; Guidot et al., 2014).
Genome Resequencing and Detection of Mutations
All genomes of strains were sequenced using Illumina technology. Libraries were constructed and sequenced by Biomarker Technologies Corporation using Illumina’s HiSeq-4000 with 2 × 150-bp paired-end reads, according to the Illumina company protocol. The quality of the raw reads was evaluated and filtered to obtain clean reads for subsequent bioinformatics analysis. Sequence data were mapped to the genome of the original strain CQPS-1 (accession numbers CP016914 and CP016915) (Liu et al., 2017). bwa software is used mainly for the comparison of the obtained short sequences with reference genomes (Li and Durbin, 2009). Picard1 was used to filter redundant reads. The HaplotypeCaller algorithm of GATK software and SnpEff have been used to detect and annotate SNPs and small InDels (Mckenna et al., 2010): gVCF was generated for each sample first, then population joint-genotype was performed; finally, the final set of variant sites was obtained after filtering. The filter parameters adopt the default values officially designated by GATK. All filtered polymorphisms were checked by PCR amplification and sequencing with Sanger technology using the primers reported in Supplementary Table 1.
Construction of Deletion Mutants and Complementation Analyses
Mutants with gene deletions were generated based on homologous recombination by pK18mobsacB as described by Zhang et al. (2015, 2018). Briefly, two flanking fragments of the target gene were amplified from the genomic DNA of CQPS-1. Then, a second round of polymerase chain reaction (PCR) was performed to generate the DNA fragment without the open-reading frame of the target gene. The generated DNA fragment was cloned into pK18mobsacB to obtain the target pK18 plasmid. After the sequence was validated, the target plasmid was delivered from E. coli strain S17-1 into R. solanacearum strain CQPS-1 by conjugative transfer. A deletion mutant was generated through consecutive homologous recombination events. The successful mutant was verified by PCR. The complementation analysis was performed with a pUC18-mini-Tn7T-Gm-based site-specific chromosomal integration system as previously described (Zhang et al., 2018; Liu et al., 2020). The primers used in this study are listed in Supplementary Table 1.
Swimming Motility
A swimming motility test was performed as previously reported (Liu et al., 2020). Briefly, overnight-cultured bacterial suspension (diluted to 1 × 108 CFU⋅ml–1) was stab-inoculated into MM medium semi agar (0.3%) plates (MSA medium) supplemented with 20 mM L-glutamate. The diameter of the swimming halo was measured after 72 h of incubation at 30°C. The experiment was repeated twice, with ten plates for each strain per replicate.
Statistical Analysis
The GraphPad Prism 8 software was used for statistical analysis. The data between two treatments were compared and statistically analyzed through the unpaired t-test. The data among multiple treatments were compared and statistically analyzed though oneway analysis of variance and Tukey’s HSD test.
Results
Growth of Original Strains in the Acidic Environment
Minimal medium (MM medium) was used to simulate poor soil in this study. MM medium with pH 4.9 was selected as the acidic environment, and the MM medium with pH 6.5 was selected as the control environment. The growth curves of the original strain CQPS-1 in these two media and the dynamics of pH during the growth process were evaluated (as shown in Figure 2). When growing in the control environment (pH 6.5 MM medium), the strain entered the logarithmic phase after 10 h of growth, reached the maximum OD value (OD600 nm = 1.04) at 30 h, then decreased slightly and remained stable; during growth in the control environment, the pH fluctuated at approximately 6.0, and at 28 h, the pH value was the lowest at 5.79. When growing in the acidic environment (pH 4.9 MM medium), the strain also began to grow rapidly after 10 h, reaching the maximum value at 30 h, but the maximum OD600 nm value was only 0.298, and then the value began to decline from 36 to 48 h, the OD600 nm at 48 h was only 0.109; approximately the pH during growth in the acidic environment, although the pH fluctuates slightly during the entire growth process, pH showed a downward trend as a whole (pH < 4.9), reaching the lowest point at 42 h (pH 3.35). The acidic environment is not conducive to the growth of R. solanacearum CQPS-1, and the pH value does not increase during the growth process, which can be used as a stress environment for the long-term experimental evolution of R. solanacearum.
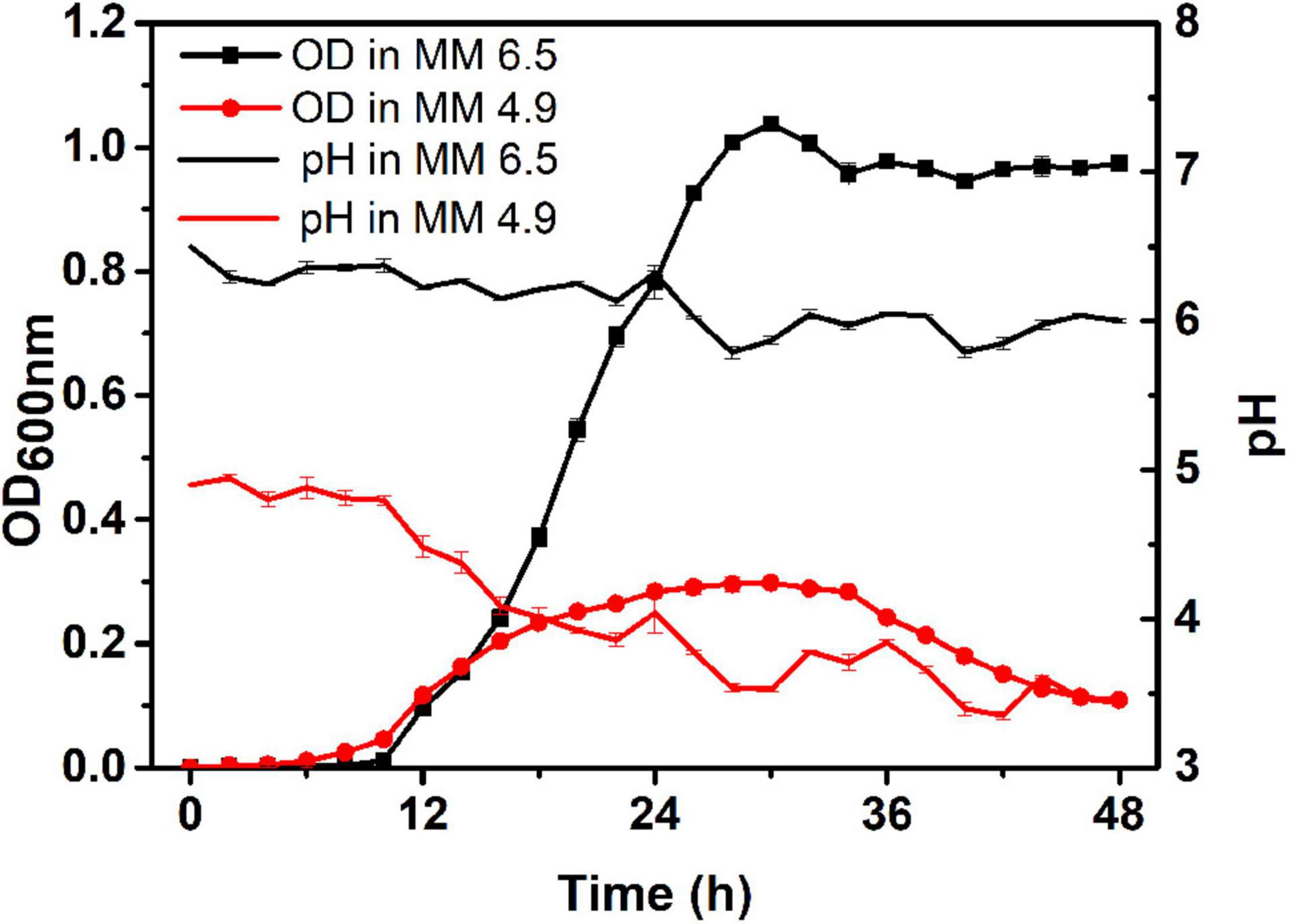
Figure 2. Growth curve and pH monitoring of CQPS-1 in MM medium with pH 6.5 or pH 4.9. MM 6.5 indicates the minimal medium with pH 6.5, MM 4.9 indicates the minimal medium with pH 4.9. Error bars indicate the standard error.
Serial Passage Experiment of Ralstonia solanacearum in an Acidic Environment
Long-term experimental evolution was performed in MM medium with pH 4.9 and pH 6.5 using the original strain CQPS-1, and the diagram of the experimental design is displayed in Figure 1. The strains grown in the acidic environment were denoted as C49, and the strains grown in the control environment were denoted as C65. Estimated by the plate count, R. solanacearum grew for approximately 6 generations when cultured in MM medium at pH 4.9 for 24 h and grew for approximately 7 generations when cultured in MM medium at pH 6.5 for 24 h. The time used for long-term serial culturing of 2000 generations was approximately 334 day in MM medium at pH 4.9, and 286 day in MM medium at pH 6.5.
After the long-term serial passage experiment, the morphologies of colonies changed on the BG plate containing TTC (shown in Supplementary Figure 1). All the colonies after the serial passage experiment were small, dark red, with poor fluidity, while the colonies of the original strain CQPS-1 were cream-colored, irregularly shaped, and highly fluid. The results indicated that after serial passage culturing in liquid MM medium, whether the pH was acidic or close to neutral, the bacterial colony morphology changed from the typical virulent type to the non-virulent type.
Long-Term Experimental Evolution of Ralstonia solanacearum in an Acidic Environment
The adaptation of the strains to the acidic environment was monitored by measuring the growth rate of the strain in MM medium at pH 4.9. As shown in Figure 3, when cultured for 1000 generations, the growth curves of some C49 strains (C49-3 and C49-5) showed a growth advantage compared to all C65 strains (Figure 3A). When growing to 1500 generations (Figure 3B), the overall growth curves of the C49 strains showed a better growth trend than the overall growth curves of the C65 strains. After 2000 generations of culture (Figure 3C), all C49 strains grew faster to the logarithmic phase and had higher growth than all C65 strains. Comparing the growth rates of C49 strains and C65 strains, 1000, 1500, and 2000 generations of C49 strains were 1.25-fold, 1.65-fold, and 1.22-fold larger than the growth rates of C65 strains, respectively, showing significant differences (Figures 3D–F).
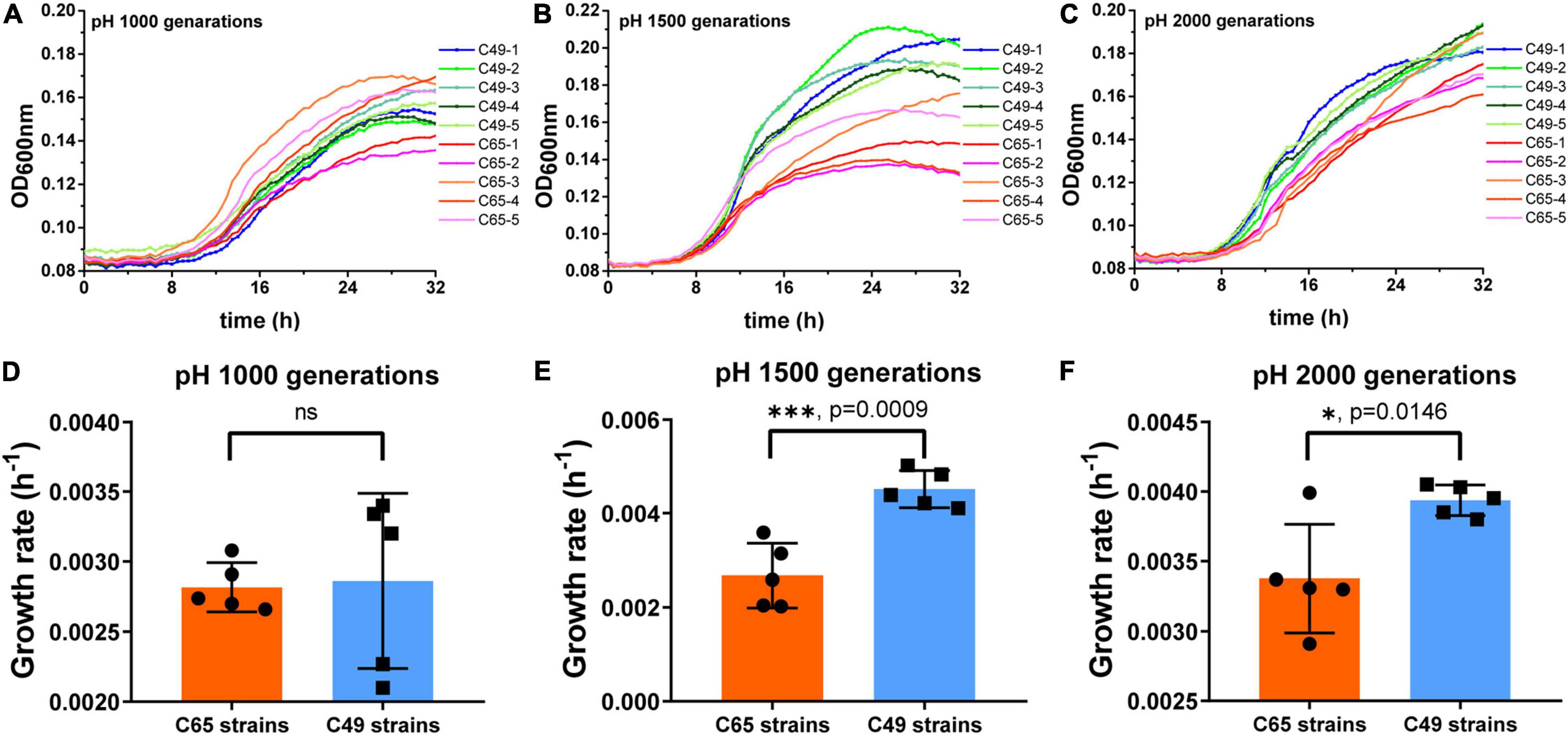
Figure 3. Growth curves and growth rates of 1000, 1500, and 2000 generations in MM medium (pH 4.9). Strains were cultured statically at 30°C in MM medium at pH 4.9. Bacterial growth was performed in 96-well plates and monitored using a Multiskan GO microplate reader (Thermo Fisher Scientific). The growth rates of each sample within 32 h were determined using the kinetic analysis module of Multiskan GO software. (A–C) Growth curves of 1000 generations, 1500 generations, and 2000 generations, respectively. (D–F) Growth rates of 1000 generations, 1500 generations, and 2000 generations. Asterisks indicate statistical significance (unpaired t-test).
Selection of Acid-Evolved Clones
Three clones were randomly isolated from each independent population generated by a long-term serial passage experiment from the control environment or the acidic environment to evaluate the fitness of the acidic environment. The acidic environment fitness of each selected clone was measured by co-culturing with the ancestral clone in MM medium at pH 4.9 using competition experiments by distinguishing colony morphology. A CI was used as a fitness estimator. The results are illustrated in Figure 4. The CIs of all selected clones from C49 strains were greater than 2 (Figure 4A). Therefore, the selected clones were superior to the original strain in acidic environment competitiveness. As shown in Figure 4B, the mean CI for the 2000 generations of C49 strains was 3.17, which was significantly higher than the mean CI for the 2000 generations of C65 strains (unpaired t-test analysis, p = 0.0006).
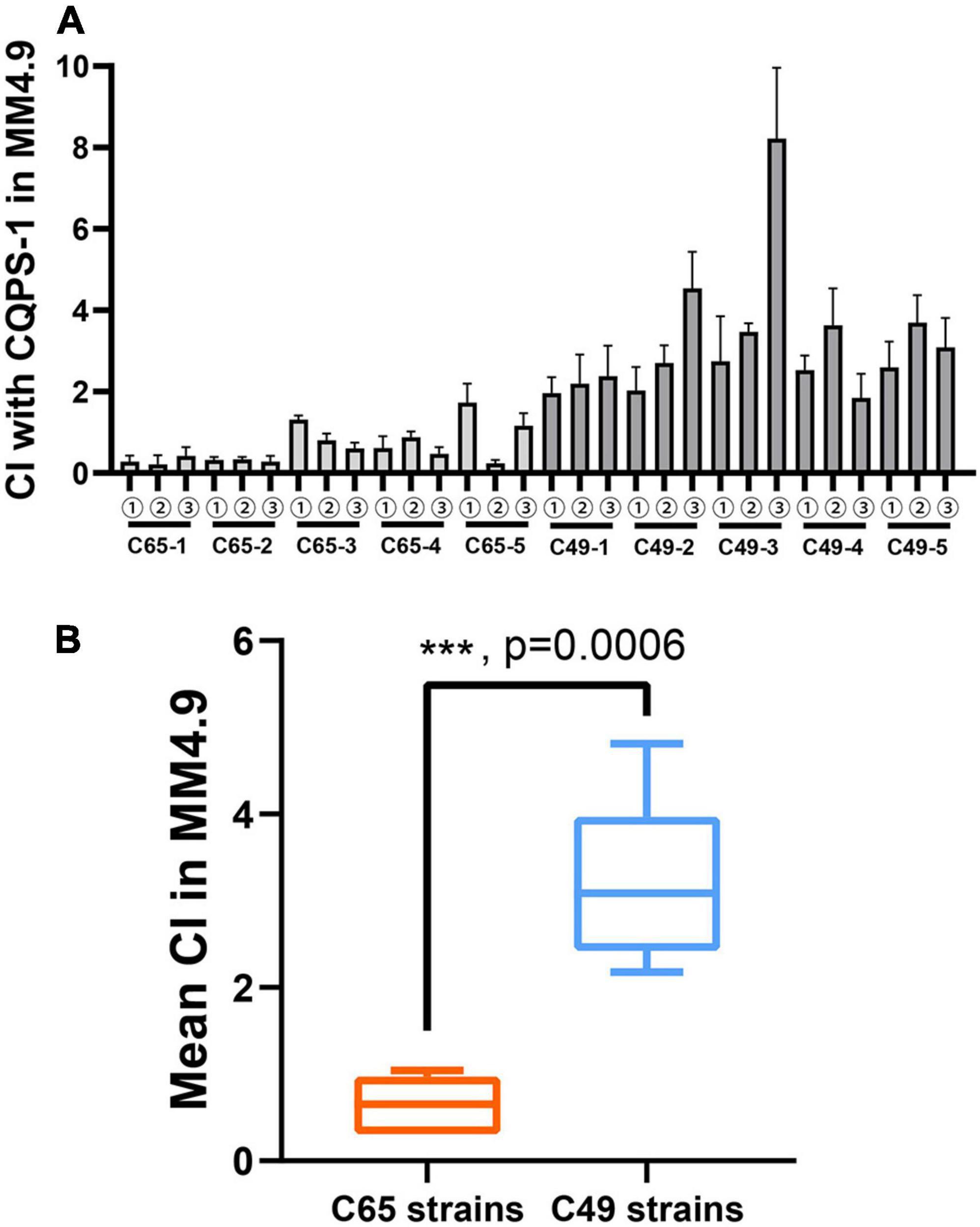
Figure 4. Competitive index (CI) values of experimental strains relative to the ancestral clone CQPS-1 growing in MM medium with pH 4.9 by distinguishing colony morphology. The CI was defined as the experimental/ancestral-clone ratio divided by the ratio in the corresponding inoculum. (A) Individual CI value of selected clones. ①-③ means three clones randomly isolated from each independent population generated by a long-term serial passage experiment from the control environment or the acidic environment. (B) Comparison of the mean CI of all selected clones from C49 strains and C65 strains. Asterisks indicate statistical significance (unpaired t-test).
Verification of Variant Gene Sequences of Experimental Strains
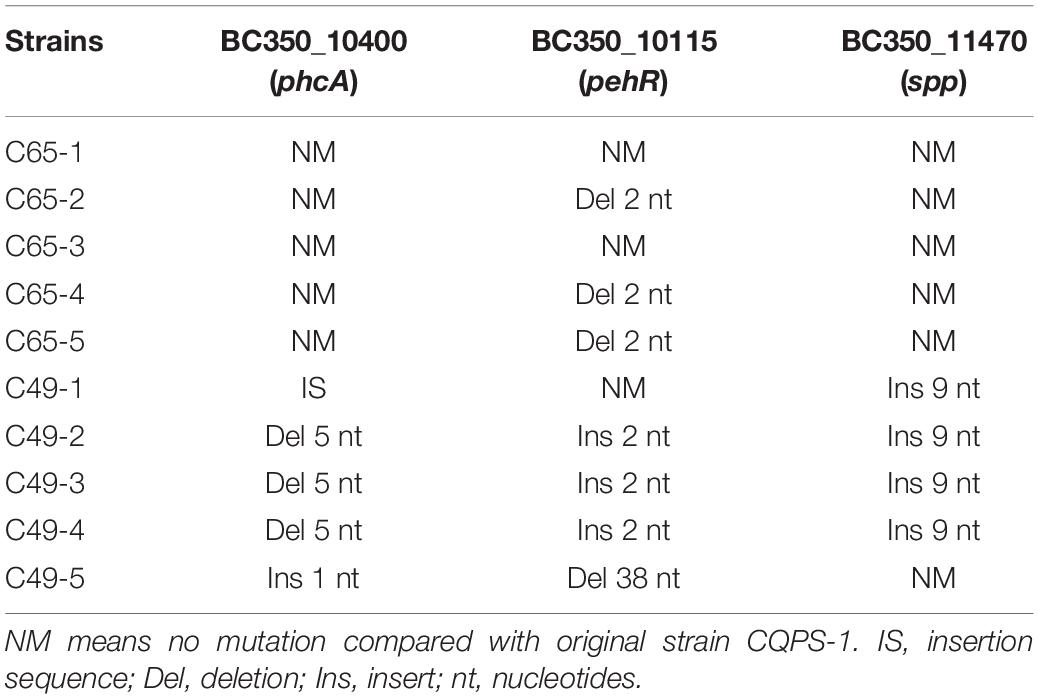
To determine the nature of genetic variations that occurred during the serial passage experiment of strain CQPS-1 in vitro, we sequenced the genomes of 10 clones based on the results of competition assays: 5 acid-evolved clones from C49 strains (C49-1③, C49-2③, C49-3③, C49-4②, C49-5②), and 5 control clones randomly selected from C65 strains (C65-1③, C65-2①, C65-3①, C65-4②, C65-5①). The details of the genome sequencing information for each sample are shown in Supplementary Table 2. The GC contents were 66.63–66.86%. The genome of the original strain CQPS-1 was used as the reference genome to analyze the variation of the experimental strains. More than 79 million paired-end reads (2 × 150 bp) were generated, leading to an approximately 300 × to 400 × total coverage of the reference genome (Supplementary Table 2). The comparison rates between the sample and the reference genome were 99.77–99.89%, and the genome coverage was 99.95%. Variations in the strains, including non-synonymous SNPs (single nucleotide polymorphisms) and small InDels (insertion-deletions), were counted (shown in Supplementary Table 3).
To screen for effective variations, the mutated gene that more than three genomes from five independent populations had mutations was selected. Then, the selected genes were verified by PCR amplification and Sanger sequencing in the C49 series and C65 series strains. Based on the sequencing and verification results, three genes were observed in parallel evolution: (1) gene ID BC350_10400 (transcriptional activator phcA); (2) BC350_10115 (response regulator transcription regulator protein pehR); and (3) BC350_11470, encoding a signal peptide protein of unknown function (shorthand “spp”). Table 1 lists all the genetic modifications identified in the sequenced clones compared with the CQPS-1 reference genome. The phcA gene of 5 clones from the long-term serial passage experiment of acidic environment mutated, although the mutation situation was different, and all the C65 series clones did not mutate. One IS (insertion sequence) movement was found in the phcA gene of C49-1, and 5 nucleotides were deleted in the phcA genes of C49-2, C49-3, and C49-4; moreover, 1 nucleotide was inserted in the phcA gene of C49-5. The pehR gene was mutated in both C49 series and C65 series clones, but the mutation sites were not the same (Table 1). In addition, 9 nucleotides were inserted in the spp gene of C49-1, C49-2, C49-3, and C49-4, and no mutation occurred in this gene of C49-5.
Loss of the phcA Gene Increased the Growth of Ralstonia solanacearum in MM Medium With pH 4.9
Based on the results of genome analysis, we can infer that the mutations of the phcA gene or spp gene may be related to the strain adapting to the acidic environment. To verify this possibility, we generated deletion mutants in the original strain CQPS-1 by gene knockout experiments. After deleting the spp gene, the growth of the mutant strain in the acidic environment (MM medium with pH 4.9) was no different from the growth of the mutant strain of the wild-type strain CQPS-1 (Supplementary Figure 2).
The growth rates of the ΔphcA mutant, C49 strains and the complemented phcA strains in MM medium at pH 4.9 were also evaluated (Figures 5A,B). At the end of incubation, the growth rates of the ΔphcA mutant and C49 strains were significantly higher than that of wild-type strain CQPS-1 (Figure 5A). The ΔphcA mutant complemented the phcA gene (under the control of its native promoter) growed as slow as the wild-type strain did (Figure 5A). Significantly, complementation of the C49 strains by insertion of a copy of the wild-type phcA locus and its promoter into the selectively neutral att site restored the mucoid colony morphology (Supplementary Figure 3), and showed a growth trend consistent with wild-type strain in MM medium at pH 4.9 (Figure 5B).
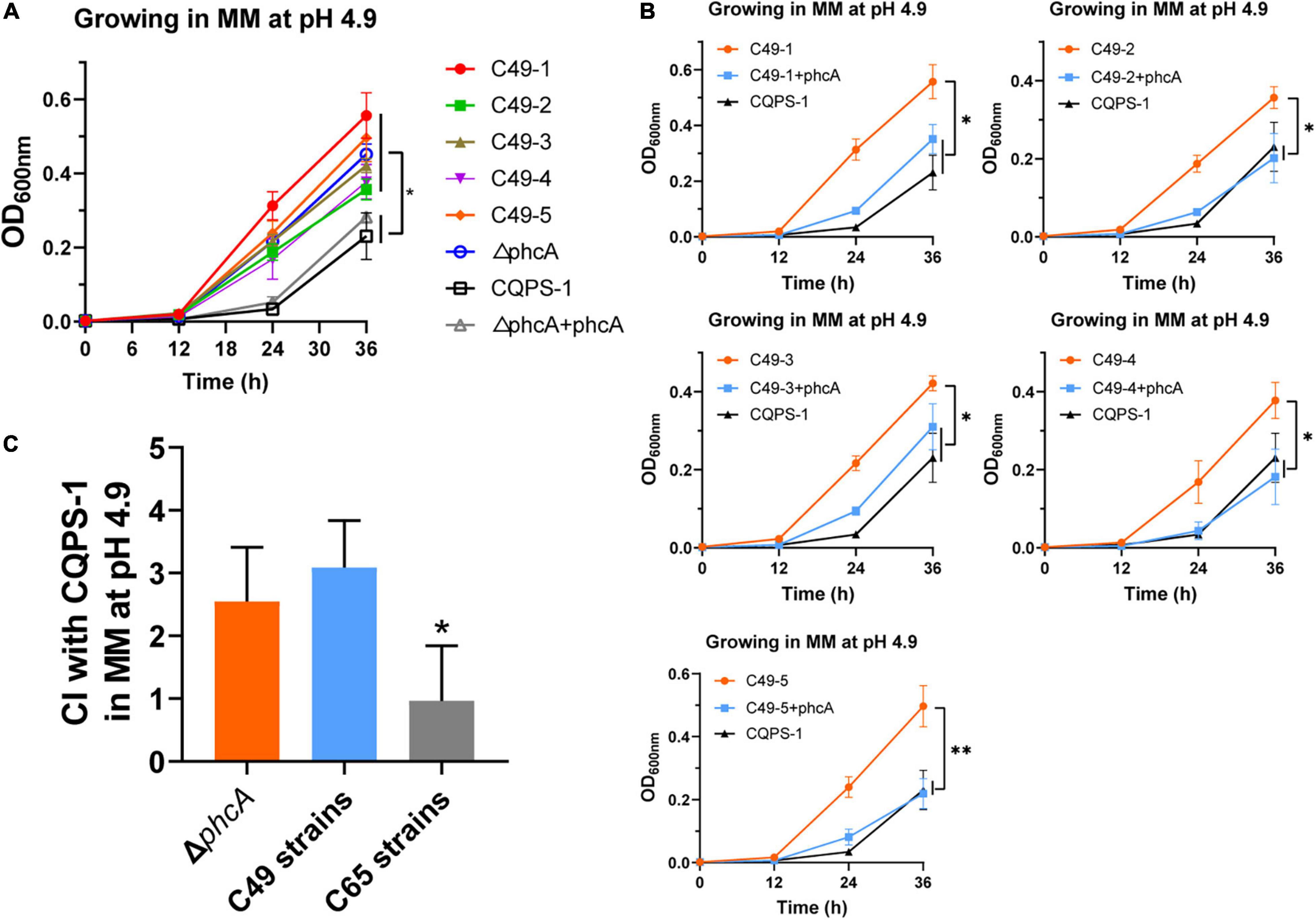
Figure 5. Growth curves and competition experiments of the phcA deletion mutant, C49 strains and the wild-type strain. (A) Growth curves of the phcA deletion mutant, C49 strains and the wild-type strain in MM medium at pH 4.9. (B) Growth curves of C49 strains after expressing an intact phcA gene by the complementary experiment. (C) The mean competitive index (CI) values of the phcA deletion mutant, acid-evolved clones, and control C65 clones relative to the wild-type CQPS-1 growing in MM medium at pH 4.9. The five selected clones of experimental C49 strains or C65 strains were used in the experiments. Asterisks indicate statistical significance (unpaired t-test, *p < 0.05 or **p < 0.01).
The competition experiments was performed to evaluate the acidic environment fitness of ΔphcA mutant in MM medium at pH 4.9 by co-culturing with the wide-type strain CQPS-1. As shown in Figure 5C, the mean CI of ΔphcA mutant was 2.55, which showed no significant difference when compared with the five acid-evolved clones (mean CI was 3.09), but showed significantly higher than the mean CI of five control C65 clones (mean CI was 0.97). The results showed that loss of function of the phcA gene was associated with the fitness gain of R. solanacearum in an acidic environment.
Swimming Motility of the Acid-Evolved Clones and the ΔphcA Mutant Enhanced
The swimming motility of the acid-evolved clones and the ΔphcA mutant was detected in MSA medium. The results 72 h after inoculation in MSA medium are shown in Figure 6. The mean swimming haloes produced by the acid-evolved clones (C49 strains), the control strains (the C65 strains), the ΔphcA mutant, and the original strain CQPS-1 were 32.74 ± 1.67 mm, 15.76 ± 0.94 mm, 32.88 ± 2.40 mm, and 19.74 ± 0.78 mm, respectively. Compared with the swimming haloes of CQPS-1, the haloes of the C49 strains were 1.66-fold increased, showing a significant difference (p < 0.01, ANOVA). There was no significant difference between swimming haloes of the C49 strains and the ΔphcA mutant. Complementation with the phcA gene (under the control of its native promoter) could completely restore the swimming motility of the ΔphcA mutant and C49 strains to that of wild-type strain CQPS-1 (Figure 6).
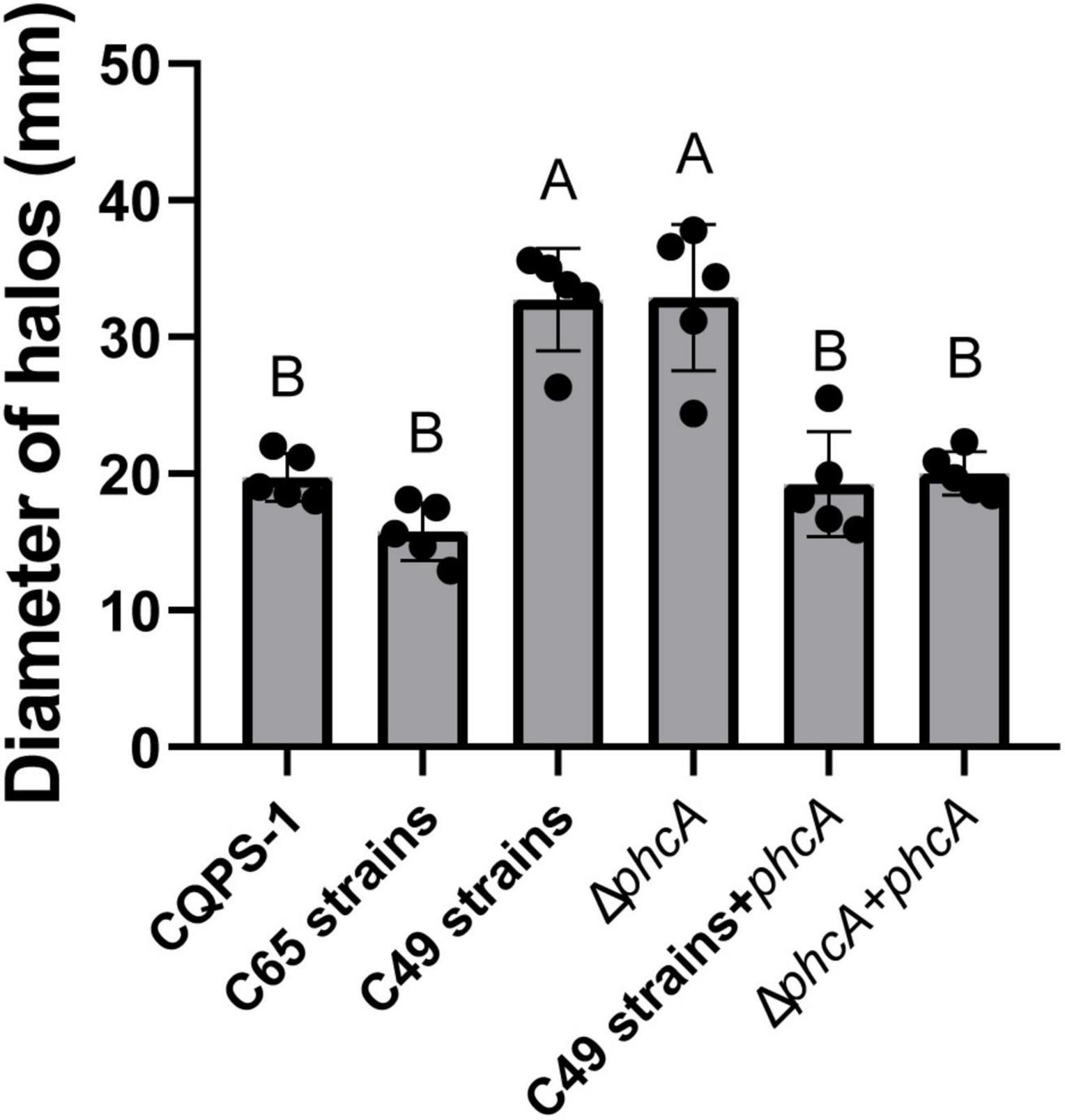
Figure 6. Swimming motility of the acid-evolved clones, the ΔphcA mutant, the control strains, and the original strain CQPS-1. Swimming motility assays were performed on MM medium semi agar (0.3%) plates (MSA medium) supplemented with 20 mM L-glutamate. Swimming halos observed after 72 h incubation at 30°C. The selected 5 “acid-evolved clones” and 5 “C65 control clones” were used as the C49 strains and C65 strains, respectively. Different letters indicated significant difference as determined by Tukey’s HSD (uppercase difference p-value < 0.01).
Discussion
Experimental evolution is an important tool for studying natural processes and molecular functions (Kassen, 2019; Remigi et al., 2019). This method can supplement traditional molecular genetics and enhance the understanding of the molecular processes of microbial cells. Under laboratory conditions, it is possible to quickly evaluate the influence of a single factor on the evolution of the experimental population and explore its adaptive molecular mechanism by controlling a single variable (Blount et al., 2012; Lenski, 2017). In addition, there are some experimental evolution studies implicating the same adaptive loci as those observed in natural populations (Hsu et al., 2021). This research performed long-term experimental evolution to study the process of R. solanacearum adapting to an acidic environment in the laboratory. More than 2000 generations of experimental strains were obtained by serial passage experiments. Combined with whole genome sequencing, we identified the molecular determinants that drive adaptation in R. solanacearum when grown in an acidic environment.
Experimental evolution has been applied in R. solanacearum in recent years, mainly to explore the genetic mechanism of the adaptability of the strain to the host (Guidot et al., 2014; Marchetti et al., 2014). Guidot et al. (2014) performed a serial passage experiment of R. solanacearum on its original hosts (hosts that can cause bacterial wilt) and distant hosts (plants whose strains can grow asymptomatically) to study the genetic bases of host adaptation. The results of phenotypic analysis showed that pathogenic bacteria can increase their adaptability for both the original and distant hosts. This research is a successful case of studying the adaptability process and mechanism of microorganisms to a controlled environment under laboratory conditions. Moreover, based on the results of genome resequencing, the function of efpR in plant adaptability was found (Guidot et al., 2014; Perrier et al., 2016), which means that the combination of experimental evolution and genome resequencing is a powerful method that can effectively provide a comprehensive understanding of the adaptation process of microbial genome changes in the experimental evolution of microorganisms. This study simulated poor soil with MM medium, and directly explored the relationship between the long-term effects of acid stress and R. solanacearum by experimental evolution. When the strains grew to more than 1000 generations, the acid experimental strains (C49 strains) showed significantly improved acid adaptability. The R. solanacearum strain was demonstrated to have highly variable plasticity and shows strong adaptability in adverse environments.
In soil, stationary broth culture, or long-term culture on agar plates, R. solanacearum spontaneously undergoes phenotypic conversion to the PC type, that is, changes from a fluid colony to a non-fluid colony (Kelman, 1954; Brumbley and Denny, 1990). The PC type is weakly pathogenic or non-pathogenic, but it can still colonize the host tissue without causing symptoms. Earlier reports pointed out that R. solanacearum has been cultured under laboratory conditions for a long period until it reaches a stable stage. After 20 days, the strain will undergo phenotypic conversion, and mucus-secreting and non-mucus-secreting strains coexist in the population (Zhu et al., 2010). Our results also show that under long-term experimental conditions, all experimental strains will transform into PC types. According to previous reports, spontaneous inactivation of phcA results in the phenotypic conversion of R. solanacearum (Brumbley et al., 1993; Jeong and Timmis, 2000). Combined with the results of this study, there are two situations in the spontaneous phenotypic conversion of R. solanacearum. One situation is that when the phenotype converts, the phcA gene is mutated. In this situation, mutations were IS movements (strain C49-1) or deletions (strains C49-2, C49-3, C49-4, C49-5), which resulted in modified coding sequences (CDS). The other situation is that there is no conversion in the phcA gene when the phenotype converts, such as strains in the C65 series (Table 1). However, whether the phenotypic conversion of the C65 series strains is due to gene regulation needs to be further verified.
PhcA, a LysR-type transcriptional regulator, plays an important role in the quorum sensing (QS) of R. solanacearum (Genin and Denny, 2012). QS regulates the growth patterns of R. solanacearum at different population densities. It has been reported that cells have little functional PhcA when the population is at low density, whereas cells at higher density have abundant functional PhcA (Genin and Denny, 2012). R. solanacearum can use many sources (carbon, nitrogen, sulfur, phosphorus and iron) more efficiently when it is at low cell density than in a high-cell-density mode (Khokhani et al., 2017). As previously reported, glucosidases, which convert complex sugars such as glycosides, glycans, and glycoconjugates to glucose, have more expression in ΔphcA cells than in wild-type cells (Khokhani et al., 2017). Glucose is an important carbon source for R. solanacearum. Rapid uptake of glucose may help R. solanacearum gain an advantage in competition. The “acid-evolved strains” with spontaneous mutation of PhcA have enhanced metabolic capacity and can rapidly utilize nutrients for its own growth in a barren acidic environment. Our study confirmes that the inactivation of the phcA gene is the reason why R. solanacearum can adapt to the long-term acidic environment (Figure 5).
The swimming motility of the acid-evolved strains was significantly enhanced compared with the swimming motility of the control strains and the original strain CQPS-1 (Figure 6). Swimming motility, one of the most important characteristics of R. solanacearum, allows R. solanacearum to effectively invade and colonize the host plant (Liu et al., 2001; Tans-Kersten et al., 2001, 2004; Dalsing and Allen, 2014). R. solanacearum has been reported to attach to the roots of the host faster after knocking out phcA (Khokhani et al., 2017). We could speculate that the “evolved-strains” of R. solanacearum would attach to the roots better due to the enhanced swimming motility when there is a host.
PhcA mediates a trade-off from maximizing growth to producing costly virulence factors (Peyraud et al., 2016; Khokhani et al., 2017; Lowe-Power et al., 2018). Although it is speculated that “acid-evolved strains” can achieve rapid colonization of hosts through better utilization of nutrients and superior motility, spontaneous mutation of PhcA would result in losses of pathogenicity, indicating that the strain may not cause host death. Interestingly, previous studies confirmed that the non-pathogenic form of R. solanacearum caused by mutations in the phcA gene could reverse the pathogenic form in planta or in the presence of root exudate of host plants (Poussier et al., 2003; Mori et al., 2012). Further speculation, after preferentially reaching the roots of the host, the “evolved-strains” may cause disease by phenotypic conversion after interacting with the host. This hypothesis needs further verification.
The spp gene deletion mutant of wild-type strain has been constructed, and the growth of the mutant in the acidic environment are not affected after the spp gene deletion (Supplementary Figure 2), indicating that the spp gene from wild-type strain has no direct relationship with the acidic adaptation. However, three codons have been inserted into this gene in the “acid-evolved strains” (Table 1), which means the protein could be still expressed. How the new spp protein functions after mutation still needs to be further explored.
Taken together, this work provided evidence for understanding the evolutionary dynamics of R. solanacearum in acidic environments. In short, R. solanacearum expands its population in an acidic environment by loss of function in the phcA gene. The abilities of acid-evolved clones to acquire after adapting to acidic environments remain to be further evaluated.
Data Availability Statement
The data of genome resequencing presented in the study are deposited in the Sequence Read Archive (SRA) of NCBI, accession number: PRJNA774485.
Author Contributions
YL and WD designed the research and drafted the manuscript. YL and XT conducted experiments and performed data analysis. YP, YD, and XL completed mutant construction, complementation analysis, and evaluation. JY and XL contributed to revising the manuscript. All authors made fundamental contributions to the manuscript, contributed to the interpretation of data, and approved the final manuscript.
Funding
This research was supported by Chongqing Postdoctoral special funding, Chongqing Postdoctoral Science Foundation (cstc2020jcyj-bshX0127 and cstc2021jcyj-bshX0013), the Key Project from China National Tobacco Corporation [110202001024 (JY-07)], the Key Project from Sichuan Company of China National Tobacco Corporation (SCYC201908 and SCYC202114), and the Key Project from Hunanxiangxi Branch of China National Tobacco Corporation (HN2020KJ03).
Conflict of Interest
JY is employed by Sichuan Branch of China Tobacco Corporation.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.829719/full#supplementary-material
Footnotes
References
Baumler, A. J., Tsolis, R. M., Valentine, P. J., Ficht, T. A., and Heffron, F. (1997). Synergistic effect of mutations in invA and lpfC on the ability of Salmonella typhimurium to cause murine typhoid. Infect. Immun. 65, 2254–2259. doi: 10.1128/iai.65.6.2254-2259.1997
Bijlsma, R., and Loeschcke, V. (2005). Environmental stress, adaptation and evolution: an overview. J. Evol. Biol. 18, 744–749. doi: 10.1111/j.1420-9101.2005.00962.x
Blount, Z. D., Barrick, J. E., Davidson, C. J., and Lenski, R. E. (2012). Genomic analysis of a key innovation in an experimental Escherichia coli population. Nature 489, 513–518. doi: 10.1038/nature11514
Boucher, C. A., Barberis, P. A., and Demery, D. A. (1985). Transposon mutagenesis of Pseudomonas solanacearum: isolation of Tn5-induced avirulent mutants. Microbiology 131, 2449–2457. doi: 10.1099/00221287-131-9-2449
Bridle, J. R., and Vines, T. H. (2007). Limits to evolution at range margins: when and why does adaptation fail? Trends Ecol. Evol. 22, 140–147. doi: 10.1016/j.tree.2006.11.002
Brumbley, S. M., Carney, B. F., and Denny, T. P. (1993). Phenotype conversion in Pseudomonas solanacearum due to spontaneous inactivation of PhcA, a putative lysr transcriptional regulator. J. Bacterio. 175, 5477–5487. doi: 10.1128/jb.175.17.5477-5487.1993
Brumbley, S. M., and Denny, T. P. (1990). Cloning of wild-type Pseudomonas solanacearum phcA, a gene that when mutated alters expression of multiple traits that contribute to virulence. J. Bacteriol. 172, 5677–5685. doi: 10.1128/jb.172.10.5677-5685.1990
Dalsing, B. L., and Allen, C. (2014). Nitrate assimilation contributes to ralstonia solanacearum root attachment. Stem colonization, and virulence. J. Bacteriol. 196, 949–960. doi: 10.1128/Jb.01378-13
Flowers, T. J., Galal, H. K., and Bromham, L. (2010). Evolution of halophytes: multiple origins of salt tolerance in land plants. Funct. Plant Biol. 37, 604–612. doi: 10.1071/FP09269
Genin, S., and Boucher, C. (2004). Lessons learned from the genome analysis of Ralstonia solanacearum. Annu. Rev. Phytopathol. 42, 107–134. doi: 10.1146/annurev.phyto.42.011204.104301
Genin, S., and Denny, T. P. (2012). Pathogenomics of the Ralstonia solanacearum species complex. Annu. Rev. Phytopathol. 50, 67–89. doi: 10.1146/annurev-phyto-081211-173000
Graham, J., and Lloyd, A. B. (1979). Survival of potato strain (Race-3) of Pseudomonas solanacearum in the deeper soil layers. Aust. J. of Agric. Res. 30, 489–496. doi: 10.1071/Ar9790489
Guidot, A., Jiang, W., Ferdy, J. B., Thébaud, C., Barberis, P., Gouzy, J., et al. (2014). Multihost experimental evolution of the pathogen Ralstonia solanacearum unveils genes involved in adaptation to plants. Mol. Biol. Evol. 31, 2913–2928. doi: 10.1093/molbev/msu229
Guo, J. H., Liu, X. J., Zhang, Y., Shen, J. L., Han, W. X., Zhang, W. F., et al. (2010). Significant Acidification in major Chinese croplands. Science 327, 1008–1010. doi: 10.1126/science.1182570
Hsu, S. K., Belmouaden, C., Nolte, V., and Schlotterer, C. (2021). Parallel gene expression evolution in natural and laboratory evolved populations. Mol. Ecol. 30, 884–894. doi: 10.1111/mec.15649
Jeong, E. L., and Timmis, J. N. (2000). Novel insertion sequence elements associated with genetic heterogeneity and phenotype conversion in Ralstonia solanacearum. J. Bacteriol. 182, 4673–4676. doi: 10.1128/Jb.182.16.4673-4676.2000
Kassen, R. (2019). Experimental evolution of innovation and novelty. Trends Ecol. Evol. 34, 712–722. doi: 10.1016/j.tree.2019.03.008
Kawecki, T. J., and Ebert, D. (2004). Conceptual issues in local adaptation. Ecol. Lett. 7, 1225–1241. doi: 10.1111/j.1461-0248.2004.00684.x
Kelman, A. (1954). The relationship of pathogenicity in Pseudomonas solanacearum to colony appearance on a tetrazolium medium. Phytopathology 44, 693–695.
Khokhani, D., Lowe-Power, T. M., Tran, T. M., and Allen, C. (2017). A single regulator mediates strategic switching between attachment/spread and growth/virulence in the plant pathogen Ralstonia solanacearum. mBio 8:e00895-17. doi: 10.1128/mBio.00895-17
Kochian, L. V., Pineros, M. A., Liu, J. P., and Magalhaes, J. V. (2015). Plant adaptation to acid soils: the molecular basis for crop aluminum resistance. Annu. Rev. Plant Biol. 66, 571–598. doi: 10.1146/annurev-arplant-043014-114822
Kooyers, N. J. (2015). The evolution of drought escape and avoidance in natural herbaceous populations. Plant Sci. 234, 155–162. doi: 10.1016/j.plantsci.2015.02.012
Lenski, R. E. (2017). Experimental evolution and the dynamics of adaptation and genome evolution in microbial populations. Isme J. 11, 2181–2194. doi: 10.1038/ismej.2017.69
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, S., Liu, Y., Wang, J., Yang, L., Zhang, S., Xu, C., et al. (2017). Soil acidification aggravates the occurrence of bacterial wilt in South China. Front. Microbiol. 8:703. doi: 10.3389/fmicb.2017.00703
Liu, H. L., Kang, Y. W., Genin, S., Schell, M. A., and Denny, T. P. (2001). Twitching motility of Ralstonia solanacearum requires a type IV pilus system. Microbiology 147, 3215–3229. doi: 10.1099/00221287-147-12-3215
Liu, Y., Tan, X., Cheng, H. J., Gong, J., Zhang, Y., Wang, D. B., et al. (2020). The cold shock family gene cspD3 is involved in the pathogenicity of Ralstonia solanacearum CQPS-1 to tobacco. Microb. Pathog. 142:104091. doi: 10.1016/j.micpath.2020.104091
Liu, Y., Tang, Y. M., Qin, X. Y., Yang, L., Jiang, G. F., Li, S. L., et al. (2017). Genome sequencing of Ralstonia solanacearum CQPS-1, a phylotype I strain collected from a highland area with continuous cropping of tobacco. Front. Microbiol. 8:974. doi: 10.3389/Fmicb.2017.00974
Lowe-Power, T. M., Khokhani, D., and Allen, C. (2018). How Ralstonia solanacearum exploits and thrives in the flowing plant xylem environment. Trends Microbiol. 26, 929–942. doi: 10.1016/j.tim.2018.06.002
Macho, A. P., Guidot, A., Barberis, P., Beuzon, C. R., and Genin, S. (2010). A competitive index assay identifies several ralstonia solanacearum type III effector mutant strains with reduced fitness in host plants. Mol. Plant Microbe Interact. 23, 1197–1205. doi: 10.1094/Mpmi-23-9-1197
Mansfield, J., Genin, S., Magori, S., Citovsky, V., Sriariyanum, M., Ronald, P., et al. (2012). Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 13, 614–629. doi: 10.1111/j.1364-3703.2012.00804.x
Marchetti, M., Jauneau, A., Capela, D., Remigi, P., Gris, C., Batut, J., et al. (2014). Shaping bacterial symbiosis with legumes by experimental evolution. Mol. Plant Microbe Interact. 27, 956–964. doi: 10.1094/Mpmi-03-14-0083-R
Mckenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. doi: 10.1101/gr.107524.110
Mori, T., Inada, T., Ogawa, K., Matsusaki, H., and Matsuzoe, N. (2012). Phenotypic conversion of Ralstonia Solanacearum in water extract of Solanum Toxicarium. J. Plant Pathol. 94, 535–542.
Peeters, N., Guidot, A., Vailleau, F., and Valls, M. (2013). Ralstonia solanacearum, a widespread bacterial plant pathogen in the post-genomic era. Mol. Plant Pathol. 14, 651–662. doi: 10.1111/mpp.12038
Perrier, A., Peyraud, R., Rengel, D., Barlet, X., Lucasson, E., Gouzy, J., et al. (2016). Enhanced in planta fitness through adaptive mutations in EfpR, a dual regulator of virulence and metabolic functions in the plant pathogen Ralstonia solanacearum. PLoS Pathog. 12:e1006044. doi: 10.1371/journal.ppat.1006044
Peyraud, R., Cottret, L., Marmiesse, L., Gouzy, J., and Genin, S. (2016). A resource allocation trade-off between Virulence and proliferation drives metabolic versatility in the plant pathogen Ralstonia solanacearum. PLoS Pathog. 12:e1005939. doi: 10.1371/journal.ppat.1005939
Phillips, M. A., and Burke, M. K. (2021). Can laboratory evolution experiments teach us about natural populations? Mol. Ecol. 30, 877–879. doi: 10.1111/mec.15790
Poussier, S., Thoquet, P., Trigalet-Demery, D., Barthet, S., Meyer, D., Arlat, M., et al. (2003). Host plant-dependent phenotypic reversion of Ralstonia solanacearum from non-pathogenic to pathogenic forms via alterations in the phcA gene. Mol. Microbiol. 49, 991–1003. doi: 10.1046/j.1365-2958.2003.03605.x
Remigi, P., Masson-Boivin, C., and Rocha, E. P. C. (2019). Experimental evolution as a tool to investigate natural processes and molecular functions. Trends Microbiol. 27, 623–634. doi: 10.1016/j.tim.2019.02.003
Salanoubat, M., Genin, S., Artiguenave, F., Gouzy, J., Mangenot, S., Arlat, M., et al. (2002). Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415, 497–502.
Tans-Kersten, J., Brown, D., and Allen, C. (2004). Swimming motility, a virulence trait of Ralstonia solanacearum, is regulated by FlhDC and the plant host environment. Mol. Plant Microbe Interact. 17, 686–695. doi: 10.1094/Mpmi.2004.17.6.686
Tans-Kersten, J., Huang, H. Y., and Allen, C. (2001). Ralstonia solanacearum needs motility for invasive virulence on tomato. J. Bacteriol. 183, 3597–3605. doi: 10.1128/Jb.183.12.3597-3605.2001
Westeberhard, M. J. (1989). Phenotypic plasticity and the origins of diversity. Annu. Rev. Ecol. Syst. 20, 249–278. doi: 10.1146/annurev.es.20.110189.001341
Yang, L., Ding, W., Xu, Y. Q., Wu, D. S., Li, S. L., Chen, J. N., et al. (2016). New insights into the antibacterial activity of hydroxycoumarins against Ralstonia solanacearum. Molecules 21:468. doi: 10.3390/Molecules21040468
Zhalnina, K., Dias, R., de Quadros, P. D., Davis-Richardson, A., Camargo, F. A. O., Clark, I. M., et al. (2015). Soil pH determines microbial diversity and composition in the park grass experiment. Microb. Ecol. 69, 395–406. doi: 10.1007/s00248-014-0530-2
Zhang, Y., Li, J., Zhang, W. Q., Shi, H. L., Luo, F., Hikichi, Y., et al. (2018). A putative LysR-type transcriptional regulator PrhO positively regulates the type III secretion system and contributes to the virulence of Ralstonia solanacearum. Mol. Pathol. 19, 1808–1819. doi: 10.1111/mpp.12660
Zhang, Y., Luo, F., Wu, D. S., Hikichi, Y., Kiba, A., Igarashi, Y., et al. (2015). PrhN, a putative marR family transcriptional regulator, is involved in positive regulation of type III secretion system and full virulence of Ralstonia solanacearum. Front. Microbiol. 6:357. doi: 10.3389/Fmicb.2015.00357
Keywords: Ralstonia solanacearum, experimental evolution, long-term acid stress, acid adaptation, phcA gene
Citation: Liu Y, Tan X, Pan Y, Yu J, Du Y, Liu X and Ding W (2022) Mutation in phcA Enhanced the Adaptation of Ralstonia solanacearum to Long-Term Acid Stress. Front. Microbiol. 13:829719. doi: 10.3389/fmicb.2022.829719
Received: 06 December 2021; Accepted: 03 May 2022;
Published: 26 May 2022.
Edited by:
William Martin, University of Dusseldorf Medical School, GermanyReviewed by:
Bo Zhu, Shanghai Jiao Tong University, ChinaTuan Minh Tran, University of South Alabama, United States
Lu Fan, Southern University of Science and Technology, China
Copyright © 2022 Liu, Tan, Pan, Yu, Du, Liu and Ding. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Ding, ZGluZ3dAc3d1LmVkdS5jbg==
 Ying Liu
Ying Liu Xi Tan1
Xi Tan1 Wei Ding
Wei Ding