- 1College of Resources, Sichuan Agricultural University, Chengdu, China
- 2Institute of Horticulture, Chengdu Academy of Agriculture and Forestry Sciences, Chengdu, China
- 3Safety and Environmental Protection Quality Supervision and Testing Research Institute, CNPC Chuanqing Drilling Engineering Co., Ltd., Guanghan, China
Aims: In garlic cultivation, long-time monoculture has resulted in continuous-cropping obstacles. However, the cause has not been studied to date.
Methods: We analyzed soils from garlic fields in Pengzhou, China, to determine continuous-cropping obstacle related changes in soil physicochemical properties and enzyme activities, and in the diversity and composition of bacterial and fungal communities. Furthermore, we examined the relationships between soil properties and the bacterial and fungal communities.
Results: The soil pH and the soil catalase, urease, invertase, and polyphenol oxidase activities were lower in the cropping obstacle soil than in the healthy control soil. The richness and diversity of the bacteria were lower in the cropping obstacle soil than in the control. The bacterial and fungal communities in the cropping obstacle soil were clearly different from those in the control soil. The differences in bacterial communities between the cropping obstacle soil and the control soil were associated with differences in pH and available potassium content. The taxa with higher relative abundances in the cropping obstacle soils included potential plant pathogens and the taxa with lower relative abundances included potential plant growth promoters.
Conclusion: The enrichment of plant pathogens and the depletion of plant growth promoting fungi may have contributed to the poor growth of garlic in the cropping obstacle soil. The enzyme activity and microbial community differences were associated with acidification that was likely an important factor in the deterioration of the soil ecological environment and the garlic cropping obstacle. The results provide information to guide agricultural practices in cultivating garlic.
Highlights
- Soil acidification was the primary factor correlating with garlic cropping obstacle.
- The activities of catalase, urease, invertase and polyphenol oxidase were lower in the cropping obstacle soil than in the healthy control soil.
- The enrichment of plant pathogens and the depletion of plant growth promoting fungi may have contributed to the garlic cropping obstacle.
- The assembly of bacterial communities was dominated by stochastic processes in garlic cropping obstacle soil.
Introduction
Garlic (Allium sativum L.) is rich in nutrients and has an appealing flavor and antibacterial properties; thus, it has been used as a seasoning, functional food, and traditional medicine for thousands of years worldwide (Corzo-Martinez et al., 2007; Liu et al., 2020). Currently, Asia, Europe, and Latin America are the main garlic production regions. In China, the garlic cropping area has reached 830 km2 and the annual yield is approximately 2.3 million tons (FAO, 2019). Growing demand for yields and the limited arable land have resulted in garlic production with high cropping intensity and monocultures over long periods. Generally, long-term monoculture continuous cropping decreased crop yield and quality a phenomenon known as cropping obstacle (Zhu et al., 2018). The cropping obstacle may include numerous biotic and abiotic factors, e.g., changes in microbial communities (Dong et al., 2017), enriched soil-borne plant pathogens (Liu et al., 2014), declines in soil enzyme activity (Fu et al., 2017), changes in soil physicochemical properties (Kaur and Singh, 2014; Perez-Brandan et al., 2014; Li et al., 2016), and autotoxicity of plants (van Wyk et al., 2017).
Soil microorganisms play an important role in agricultural production by maintaining soil quality and affecting nutrient cycling (Blagodatskaya and Kuzyakov, 2013; Sun et al., 2015; Beckers et al., 2017). In some instances, the interactions between soil microbial communities and soil properties modulate plant health through affecting pathogens (Wei et al., 2015). Cropping obstacles have been associated with soil bacterial and fungal communities (Gao et al., 2019, 2021; Xi et al., 2019). For example, decreases in diversity and abundance of beneficial soil microbes and an increase in pathogenic fungi such as Fusarium and Verticillium were associated with sugarcane cropping obstacle (Pang et al., 2021). However, the relationships between microbial communities and continuous cropping obstacles are not necessarily similar across diverse crops and environmental conditions, thus more studies are called for.
Soil enzymes, i.e., the extracellular enzymes produced by the microorganisms, are considered as indicators of soil fertility (Zeng et al., 2007; Paz-Ferreiro and Fu, 2016). The physicochemical properties of soil affect the composition of soil microbial communities and the enzyme activities (Liu et al., 2021). Among the soil physicochemical properties, soil pH is considered as a master variable in affecting the microbial communities (Fierer and Jackson, 2006). Evidently, the changes brought on by continuous cropping, e.g., the often detected acidification, have resulted in changes in the soil microbial communities and soil enzymatic activities (Gao et al., 2019, 2021; Zheng et al., 2019).
Pengzhou in the Chengdu Plain is one of the five major vegetable cultivation bases in China, with a garlic cultivation history of more than 30 years. In Pengzhou, garlic is commonly grown in rotation with spring rice. In recent years, the long-time monoculture has resulted in continuous-cropping obstacles. The symptoms of the obstacle include yellowing of the leaves starting from the tips and edges, stagnated growth of roots and plant, at its worst, dying of the plants. The continuous-cropping obstacles appear commonly on patches in a field and sometimes on an entire field.
To our knowledge, the continuous-cropping obstacles in garlic cultivation have not been studied to date. We analyzed soils from garlic fields in Pengzhou, with the aim to determine continuous-cropping obstacle related changes in (1) soil physicochemical properties and enzyme activities, and in (2) the diversity and composition of bacterial and fungal communities. Furthermore, we examined (3) the relationships between soil properties and the bacterial and fungal communities. We hypothesized that the continuous cropping obstacle would be associated with soil acidification that would further affect the soil microorganisms and decrease enzyme activities. The results provide information to guide agricultural practices in cultivating garlic.
Materials and Methods
Site Description and Field Sampling
The field experiment was performed in a garlic production area in Pengzhou (31°02′31″N-31°05′39″N, 103°50′51″E-103°59′42″E), Chengdu Plain, China, that is in the northern part of the subtropical humid climate zone, with a mean annual temperature of 15.7°C, precipitation of 960 mm and approximately 1,180 h of sunshine. The soil on the site is fertile paddy soil. Garlic was grown in rice-garlic rotation and covered with straw after sowing.
Samples were collected in March 2018 from seven fields with an area of 700–1,000 m2. In each field, three 60–100 m2 plots were randomly selected in areas with healthy garlic plants (CK) and in areas with cropping obstacles (D) (Figure 1). Five topsoil (0–20 cm) subsamples from a 10 cm circle around the plants were combined into a composite sample, an appropriate amount of homogenized soil was retained by quartering, and placed into sterile plastic bags that were sealed. The bags were kept on ice and transported to the laboratory immediately after sampling. Roots and other debris were removed from the soil samples and the samples were divided into two parts for immediate DNA extraction and physicochemical analyses.

Figure 1. (A) A garlic field with cropping obstacle (foreground) and healthy garlic plants (background). (B) Garlic plants grown in the healthy control soil (CK) and in the cropping obstacle soil (D).
Garlic Growth and Root Activity Analyses
The growth of the plants was assessed by measuring shoot and root lengths and the fresh weight of aboveground and root biomasses. Root activity was determined using 2,3,5-triphenyl tetrazolium chloride (TTC) staining (Comas et al., 2008).
Soil Physicochemical and Enzyme Activity Analyses
Prior the analyses, the soil samples were air-dried naturally in a cool and ventilated place. Soil pH was measured with a pH meter in a 1:2.5 soil to water slurry. Soil organic carbon content (SOC) was determined using the K2Cr2O7 oxidization method. Soil total (TN) and available nitrogen (AN), available phosphorus (AP) and available potassium (AK) contents were determined using Kjeldahl digestion, alkaline hydrolysis diffusion method and molybdenum blue method and flame photometry, respectively.
Soil enzyme activity analyses were performed as described earlier (Guan et al., 1986). Briefly, urease was assayed by colorimetric analysis of sodium phenate-sodium hypochlorite, acid phosphatase was colorimetrically estimated using disodium phenyl phosphate, invertase was colorimetrically determined by DNS based on the decreasing sugar content, phenol oxidase was determined by the pyrogallol colorimetric method, and catalase activity was assayed by potassium permanganate titration.
DNA Extraction, Amplification, and Sequencing
Total genomic DNA was extracted from 0.60 to 0.90 g fresh weight soils (corresponding to 0.50 g dry weight) using Fast DNA® SPIN for Soil Kit (MP BIO Laboratories, California, United States) according to the manufacturer’s instructions. The quantity and quality of extracted DNA were estimated using a NanoDrop NC2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States) and agarose gel electrophoresis, respectively. DNA samples were stored at −20°C.
The V3–V4 region of bacterial 16S rRNA gene was amplified using the primers 338F (5′-ACTCCTACGGGAGGCAGCA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′). The fungal ITS1 region was amplified using the primers ITS5F (5′-GGAAGTAAAAGTCGTAACAAGG-3′) and ITS1R (5′-GCTGCGTTCTTCATCGATGC-3′) using the Q5 High-Fidelity DNA Polymerase (New England Biolabs, Ipswich, MA). Sample-specific 7-bp barcodes were incorporated into the primers for multiplex sequencing. The reaction mixture of total volume 25 μl comprised the following: 5 μl of 5 × reaction buffer, 5 μl of 5 × GC buffer, 2 μl of 2.5 μM dNTPs, 1 μl of forward primer (10 μM), 1 μl of reverse primer (10 μM), 2 μl of DNA Template, 0.25 μl of Q5 DNA polymerase, and 8.75 μl of ddH2O. The polymerase chain reaction (PCR) was performed under the following cycling conditions: initial denaturation at 98°C (2 min), denaturation at 98°C (15 s), annealing at 55°C (30 s), extension at 72°C (30 s), and a final extension of 72°C for 5 min, for 25–30 cycles. PCR amplicons were purified with Vazyme VAHTSTM DNA Clean Beads (Vazyme, Nanjing, China) and quantified using the Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA, United States). Amplicons were pooled in equal amounts, and paired-end 2 × 250 bp sequencing was performed using the Illumina MiSeq platform with MiSeq Reagent Kit v3 at Shanghai Personal Biotechnology Co., Ltd. (Shanghai, China).
Sequence Analysis
Sequences were processed with QIIME 2 (Bolyen et al., 2019) according to the official tutorial1 with slight modifications. Briefly, raw sequence data were demultiplexed using the demux plugin followed by removing the primers with cut adapt plugin (Martin, 2011). Quality filtering, denoising, merging and chimera removal were performed using the DADA2 plugin (Callahan et al., 2016). Non-singleton amplicon sequence variants (ASVs) were aligned with mafft (Katoh et al., 2002) and used to construct a phylogeny with fasttree2 (Price et al., 2009). Taxonomy was assigned to ASVs using the classify-sklearn naïve Bayes taxonomy classifier in feature-classifier plugin (Bokulich et al., 2013) against the SILVA Release 132 and UNITE Release 8.0 databases (Koljalg et al., 2013). Taxonomic compositions were visualized using MEGAN and GraPhlAn (Asnicar et al., 2015). Chao1 and Shannon alpha diversity indices were calculated using the ASV table in QIIME2.
Statistical Analyses
Statistical analyses were performed using QIIME2, SPSS 21 (Version 21.0, SPSS Inc., Chicago, IL, United States) and R v.3.6.1.2 Differences in plant and soil properties were tested using Student’s t-test. The associations between garlic growth parameters and soil properties were analyzed using Pearson correlation. Differences in alpha diversity indices between treatments were tested using Kruskal-Wallis test and visualized as box plots. Beta diversity was analyzed based on Bray-Curtis dissimilarity and visualized using non-metric multidimensional scaling (NMDS) (Ramette, 2007). The differences in community compositions were tested using permutational multivariate analysis of variance (PERMANOVA) (McArdle and Anderson, 2001). Differential abundance of taxa was tested using linear discriminant analysis effect size (LEfSe) analysis (Segata et al., 2011). The relationships between environmental factors and microbial community structure were analyzed using distance-based redundancy analysis (dbRDA) in the “vegan” package in R v2.5.6 (Oksanen et al., 2020).
The weighted β nearest taxon index (βNTI) and Bray-Curtis-based Raup-Crick (RCbray) values were calculated via a null model methodology to differentiate the ecological processes that regulate microbial community assembly (Stegen et al., 2013, 2015). The βNTI was quantified by determination of the standard deviation between an observed level and the null distribution of the mean nearest taxon distance metric (βMNTD). The βMNTD and RCbray were calculated using the R packages “picante” and “vegan,” respectively (Kembel et al., 2010). Specifically, β-NTI > 2 indicated variable selection, and β-NTI < −2 indicated homogeneous selection; at |β-NTI| < 2, deterministic processes were associated with dispersal limitation when RCbray > 0.95, homogeneous dispersal when RCbray < −0.95, and undominated when |RCbray| < 0.95 (Stegen et al., 2015; Jiao et al., 2020; Luan et al., 2020).
Results
The Properties of Garlic Plants and Soils
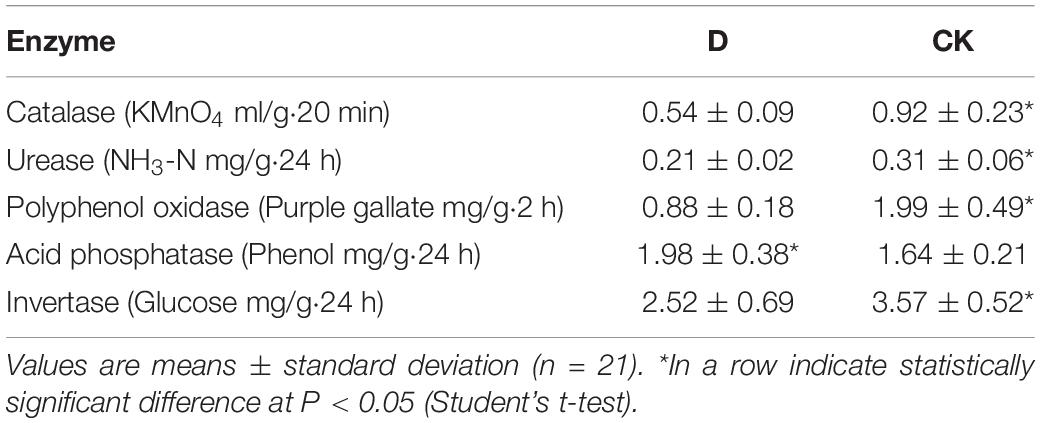
The biomasses, root and shoot lengths and root activities of cropping obstacle plants were lower than those of healthy plants, in which root activities decreased by 61.16% (P < 0.05) (Table 1). The pH was lower and AK content higher in the cropping obstacle soil than in the control (P < 0.05) (Table 2). Thus, the soil pH correlated positively and AK content negatively with the garlic growth parameters (P < 0.01) (Supplementary Table 1). The activities of soil catalase, urease, polyphenol oxidase and invertase were lower and the activity of acid phosphatase was higher in the cropping obstacle soil than in the control (P < 0.05) (Table 3).
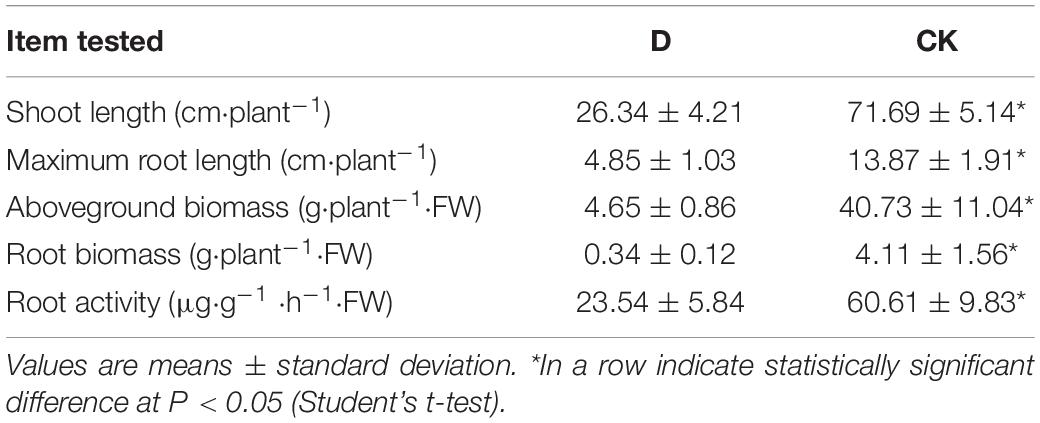
Table 1. The properties of garlic plants grown in cropping obstacle soil (D) and healthy control soil (CK).
Changes in Bacterial and Fungal Communities
The 974,584 16S rRNA gene and 1930,751 ITS sequences were grouped into 67,658 bacterial ASVs and 18,166 fungal ASVs. The richness and diversity indices of the bacteria were lower in the cropping obstacle soil than in the control (P < 0.05) (Figure 2).
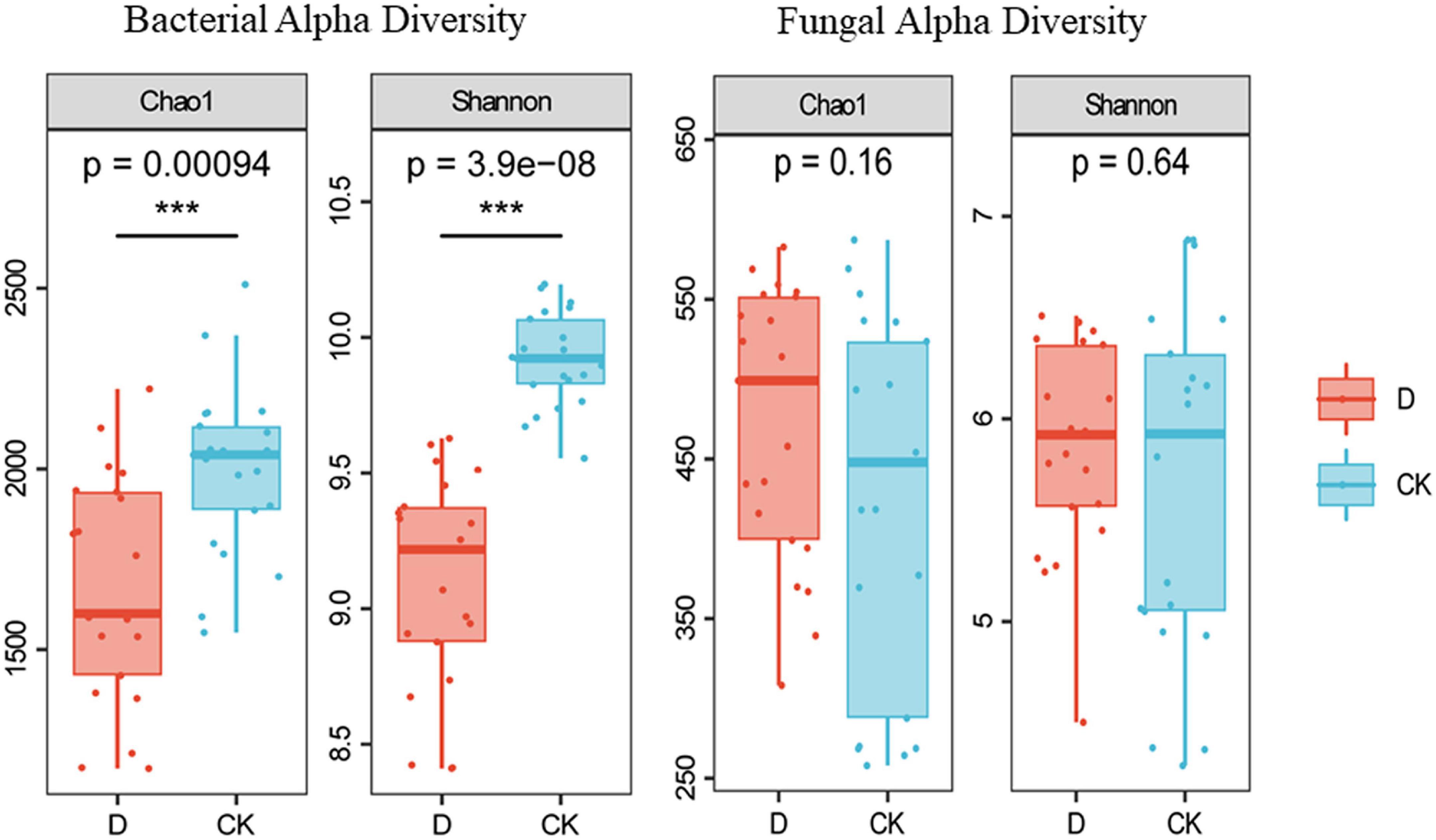
Figure 2. Alpha-diversity of bacterial and fungal communities in the garlic cropping obstacle soil (D) and healthy control soil (CK). *** Indicates statistically significant difference at P < 0.001.
At the phylum level, the bacterial sequences were classified into 36 phyla, out of which the relative abundances of Proteobacteria, Chloroflexi, Acidobacteria, and Actinobacteria were high (Figure 3A and Supplementary Table 2). Out of the 13 fungal phyla, the relative abundances of Ascomycota and Basidiomycota were high (Figure 3B and Supplementary Table 3). The bacterial sequences were classified into 893 genera and the fungal sequences into 453 genera (Figures 3C,D and Supplementary Tables 4, 5).
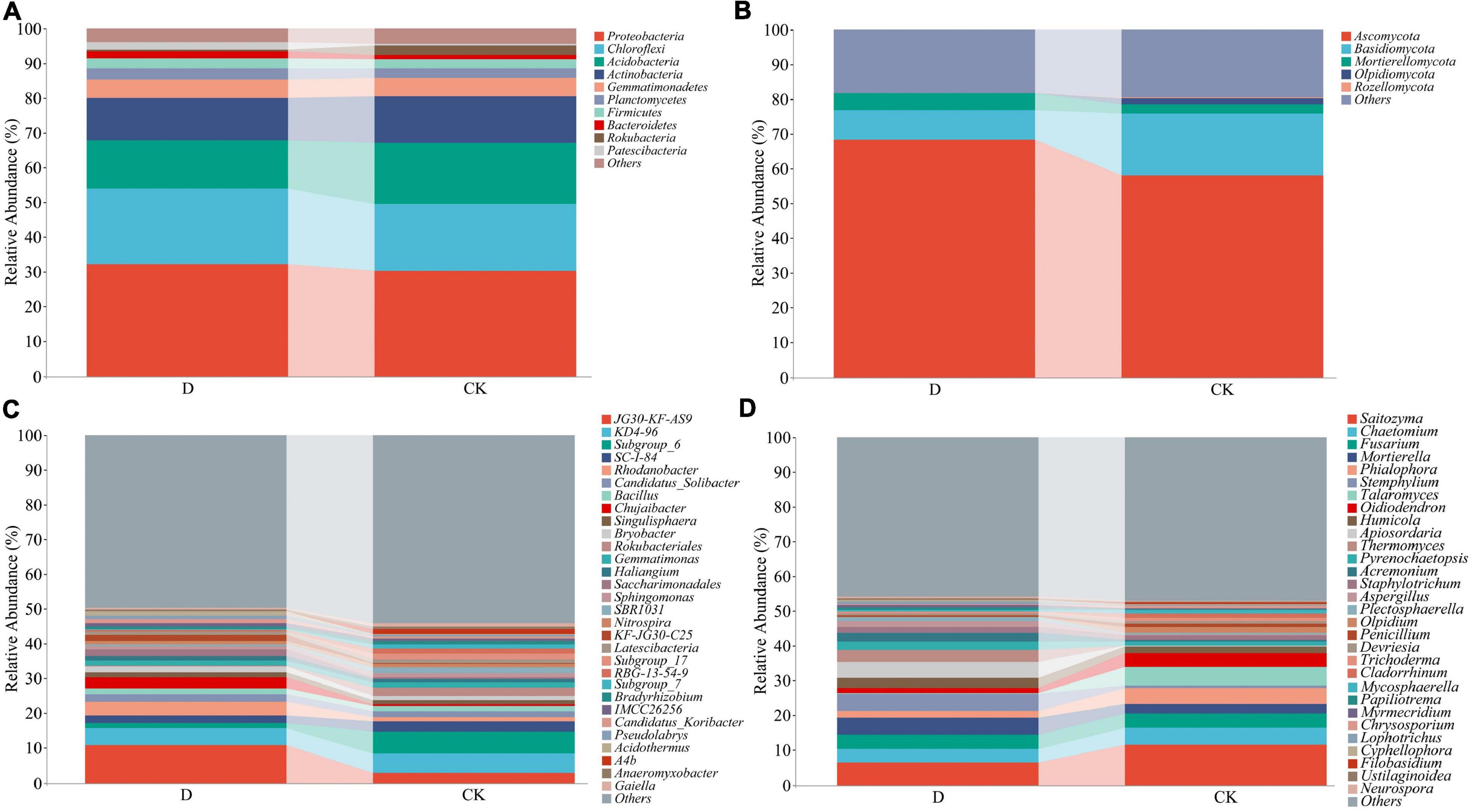
Figure 3. The relative abundances of bacterial and fungal phyla and genera in the garlic cropping obstacle soil (D) and healthy control soil (CK). (A) Bacterial phyla, (B) fungal phyla, (C) bacterial genera, (D) fungal genera.
Linear discriminant analysis (LDA) effect size (LEfSe) analysis was used to identify differentially abundant taxa. For the bacteria, the taxa with higher relative abundances in the cropping obstacle soil included phylum Chloroflexi, order Xanthomonadales and genera JG30-KF-AS9, Chujaibacter and Rhodanobacter; the taxa with lower relative abundances in the cropping obstacle soil included phyla Acidobacteria and Rokubacteria (Figure 4A). For the fungi, the taxa with higher relative abundances in the cropping obstacle soil included genera Stemphylium and Aspergillus; the taxa with lower relative abundances in the cropping obstacle soil included phylum Basidiomycota and genera Phialophora and Oidiodendron (Figure 4B).
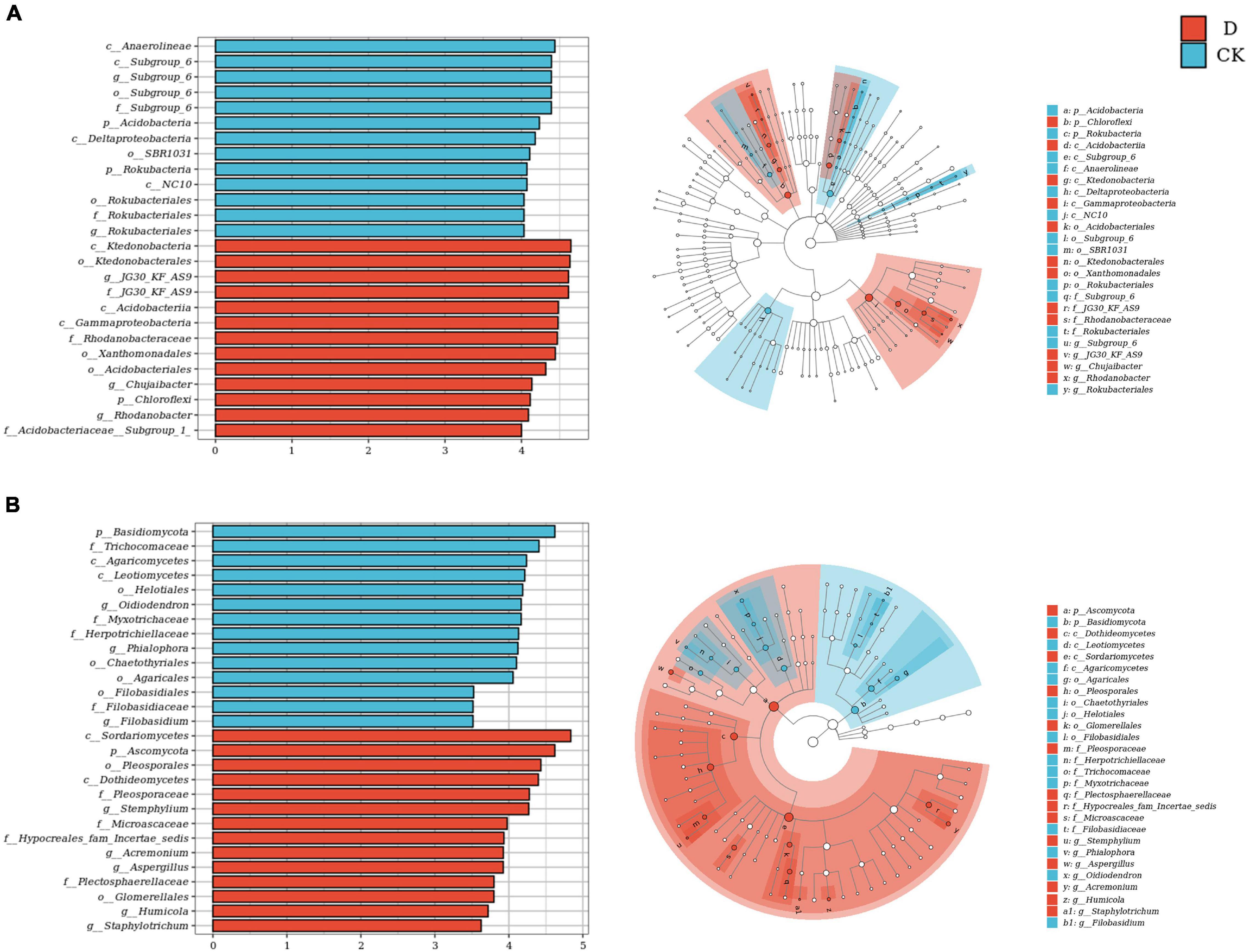
Figure 4. Differential abundances of (A) bacterial and (B) fungal taxa in the garlic cropping obstacle soil (D) and healthy control soil (CK). The histogram shows taxa with LDA scores ≥ 4.0 for bacterial, and LDA scores ≥ 3.5 for fungal in the linear discriminant (LDA) effect size analysis. The cladogram shows taxonomic differences between D and CK.
The Relationship Between the Microbial Community and Environmental Factors
In the non-metric multidimensional scaling (NMDS) based on the Bray-Curtis dissimilarity, the bacterial and fungal communities in the cropping obstacle soil were separated from those in the control soil (Figure 5). In addition, PERMANOVA indicated that the community compositions were different (P < 0.05) (Supplementary Table 6).
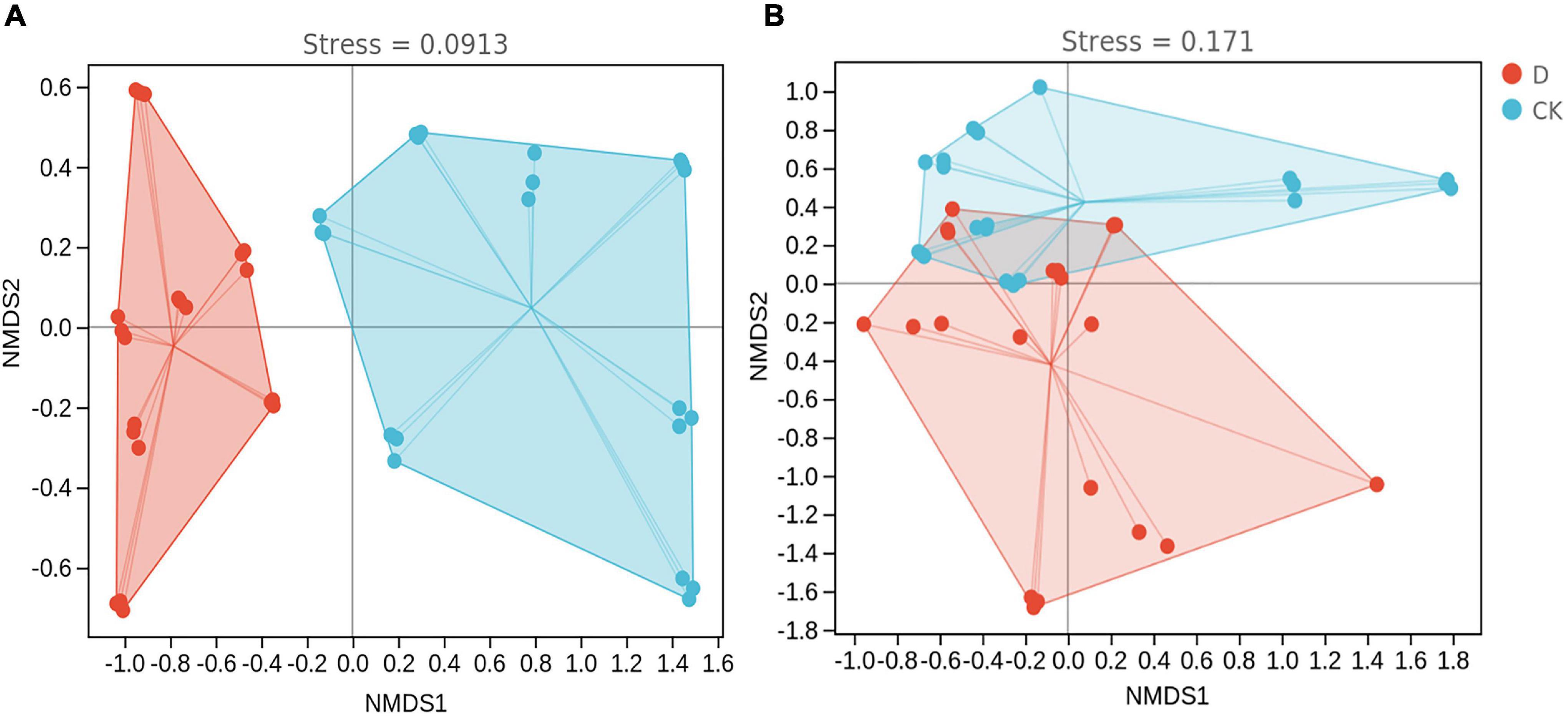
Figure 5. Non-metric multidimensional scaling of (A) bacterial and (B) fungal communities in the garlic cropping obstacle soil (D) and healthy control soil (CK).
Based on the distance-based redundancy analysis (dbRDA) analysis, the differences in bacterial community composition across samples were related to soil water content, pH, AP, AK, SOC, TN, polyphenol oxidase and acid phosphatase contents (P < 0.001) (Figure 6A and Supplementary Table 7). The differences in community composition between the garlic cropping obstacle soil and the control soil were associated with pH, AK, acid phosphatase, polyphenol oxidase and invertase (Figure 6A and Supplementary Table 7). The differences in fungal community composition across samples were associated with all the measured soil properties, but no clear treatment related associations were detected (Figure 6B and Supplementary Table 7).
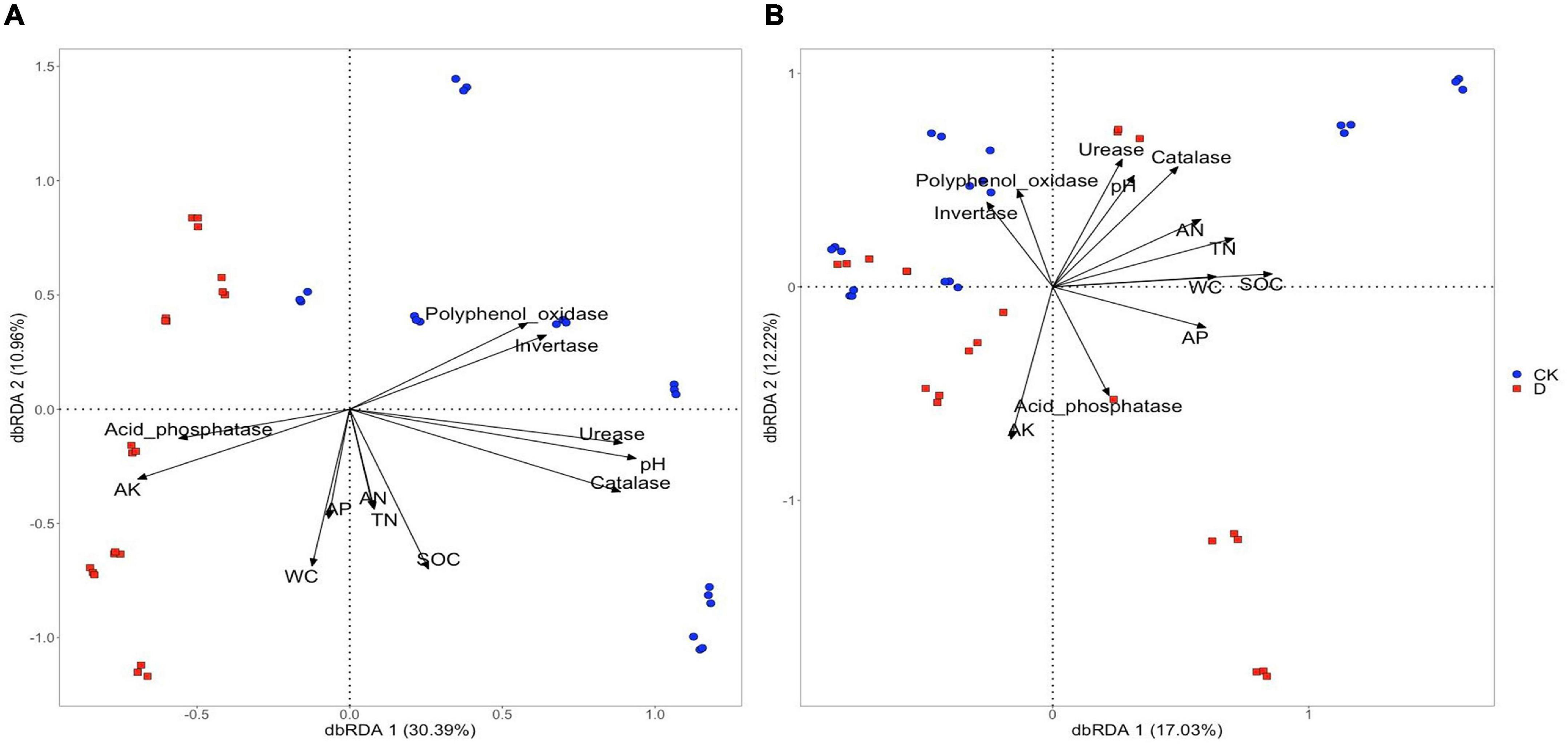
Figure 6. Distance-based redundancy analysis (dbRDA) of environmental factors with (A) bacterial and (B) fungal taxa in the garlic cropping obstacle soil (D) and healthy control soil (CK).
Assembly Processes of Soil Microbial Communities
In the bacterial communities, |β-NTI| > 2 accounted for 32.4% in the cropping obstacle soil and 61.9% in the control (Figure 7A), suggesting that the contribution of deterministic processes to community assembly were lower in the cropping obstacle soil. In the cropping obstacle soil, homogeneous dispersal accounted for 65.6% of the community assembly (Figure 7B). In the control soil, homogeneous selection accounted for 40.0% of the community assembly and variable selection accounted for 21.9%. In the fungal communities, |β-NTI| > 2 accounted for 53.8% in the cropping obstacle soil and 58.6% in the control, suggesting that the assembly processes were primarily deterministic with homogeneous selection as the dominant assembly process (Figure 7B).
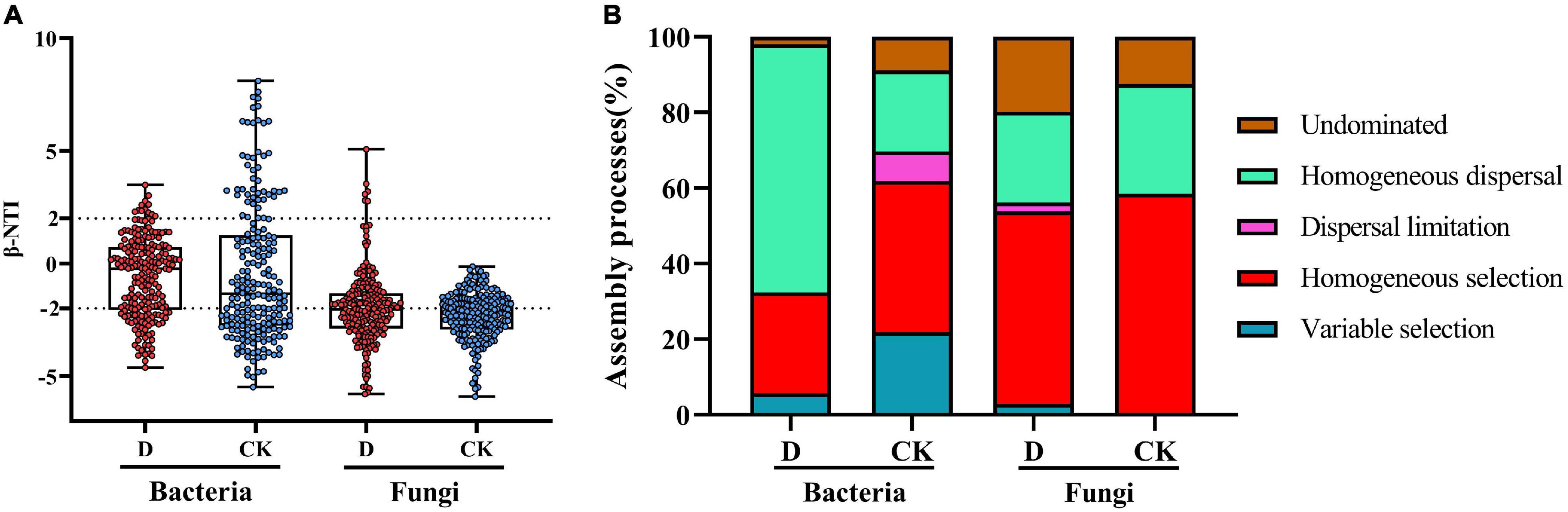
Figure 7. The microbial community assembly processes in the cropping obstacle soil (D) and in the healthy control soil (CK). (A) The values of the weighted beta nearest taxon index (βNTI). Horizontal dashed gray lines indicate upper and lower significance thresholds at βNTI = −2 and + 2, respectively. (B) The percentages of deterministic processes (homogeneous and variable selection), stochastic processes (dispersal limitation and homogeneous dispersal), and undominated processes.
Discussion
Continuous-cropping obstacle soils have received considerable attention in recent years. For example, the biotic and abiotic factors in cropping obstacle of peanut, strawberry and American ginseng have been determined (Li et al., 2014, 2018; Liu et al., 2021). To our knowledge, whether the conclusions based on other crops can be extrapolated to garlic cropping obstacle problem is still not known. We sampled soil and plants in fields with poorly growing garlic plants and noticed that in addition to the evidently lower aboveground biomass, the root biomass and root activity were also lower in the cropping obstacle soil than in the control soil with healthy garlic plants.
Soil properties can directly affect plant health (Wang et al., 2017). In agreement with previous studies (Li et al., 2018; Liu et al., 2021), compared to the control, the garlic cropping obstacle soil was characterized by lower pH and higher available potassium content. Acid stress in pH below 5.5 triggered sensitivity responses in roots, e.g., arrested root growth and death of root tip cells, and acidic soil limited plant growth and the uptake of nutrients from soil (Agegnehu et al., 2016; Gracas et al., 2021). The application of synthetic fertilizers with high level in rice-vegetable rotation has led to a soil acidification (Li et al., 2020; Shen et al., 2021) that may be an important cause for the garlic cropping obstacle. Although differences in the abiotic and biotic characteristic may exist even within close soil environments that share the same geographies (Fierer, 2017), what causes the patchiness of acidification requires further research.
Soil enzyme activities are employed as one of the important indicators of soil quality and fertility (Bastida et al., 2008). In long-period strawberry cropping, the soil enzyme activities decreased and the probability of diseases increased (Li et al., 2018). Similarly, compared to the control, the activities of soil urease, catalase, sucrase and polyphenol oxidase, i.e., enzymes that release soil nutrients for plants, were lower in the garlic cropping obstacle soil. Since soil enzyme activities and pH correlated strongly (Acosta-Martinez and Tabatabai, 2000), the acidification of the garlic cropping obstacle soil could have affected soil enzyme activity. As the soil enzymes mainly originate from soil microorganisms, changes in microbial metabolic activity, lower microbial abundance or changes in the microbial community composition (Zhao et al., 2009; Nannipieri et al., 2012) may have led to the lower enzyme activities.
Continuous cropping obstacles have been associated with lower microbial diversity, a decrease in beneficial microorganisms, and the enrichment of pathogenic microorganisms (Gao et al., 2019, 2021; Tan et al., 2021). According to the insurance hypothesis, biodiversity may act as a buffer against disturbances; in a diverse community, some species are likely to withstand disturbance and carry on functions (Yachi and Loreau, 1999). Alarmingly, in our study, both the richness and the diversity of the bacterial communities were lower in the cropping obstacle soil than in the control. Similar with (Shen et al., 2013), the lower bacterial alpha diversity in cropping obstacle soil may have been due to the lower pH.
Beta diversity analyses showed that the compositions of both the bacterial and fungal communities in the cropping obstacle soil were different from those in the control. Environmental factors affect the microbial communities in soil, with soil pH considered as the master variable in affecting bacterial communities (Fierer and Jackson, 2006). In our study, the differences in the bacterial communities between the cropping obstacle soil and the control were associated with differences in soil pH and AK content. Similarly, pH and AK content were among the main factors associated with bacterial community differences in continuously cropped potato fields (Zhao et al., 2020). Even though pH and nutrient contents affect fungal communities as well (Glassman et al., 2017; Li et al., 2022a), we found no clear associations between fungal community composition and soil properties.
Through studies on the community composition of bacteria and fungi, the differentially distributed taxa were identified by LEfSe analysis. The phylum Chloroflexi and the order Xanthomonadales were enriched in the cropping obstacle soil. Xanthomonadales include plant pathogens of significant economic and agricultural impact (Bayer-Santos et al., 2019). The higher relative abundance of Chloroflexi was mostly due to the uncultured genus JG30-KF-AS9 that can adapt to acidic soil and be detrimental to enzyme activities (Wang et al., 2019). Thus, the enrichment of JG30-KF-AS9 may be connected with the lower enzyme activities in the garlic cropping obstacle soil. Chujaibacter and Rhodanobacter which were actively developing in the presence of mineral fertilizers were acidophilic microorganisms participating in the nitrogen cycle (Semenov et al., 2020), it is speculated that the increase of these microorganisms may be related to soil acidification. Likewise, the fungal genera Stemphylium and Aspergillus that were enriched in the cropping obstacle soil include plant pathogens (Syed et al., 2020; Dumin et al., 2021). For example, Stemphylium spp. caused garlic leaf spot disease (Dumin et al., 2021). Genus Oidiodendron, one of the most widely investigated ericoid mycorrhizal fungi that had plant growth promoting characteristics (Wei et al., 2016; Baba et al., 2021), was depleted in the cropping obstacle soil. Thus, the enrichment of Xanthomonadales, Stemphylium and Aspergillus and depletion of Oidiodendron may have contributed to the poor growth of garlic in the cropping obstacle soil.
Uncovering the microbial community assembly processes is a challenging task (Stegen et al., 2013; Luan et al., 2020). Our results showed that the bacteria assembly processes in garlic cropping obstacle soil was governed by homogeneous dispersal, a stochastic process that homogenizes the bacterial community structure and causes low compositional turnover (Stegen et al., 2013). In the control, the community assembly was characterized by deterministic processes. Root exudates increase available resources that may facilitate the recruitment and selection of bacterial taxa (Li et al., 2022b), which may explain why deterministic processes governed assembly processes in the control soil with vigorously growing garlic plants.
Homogeneous selection was regarded as a factor leading to a stable state after disturbance in progressive succession of communities (Dini-Andreote et al., 2015). Together with the diversity and environmental factor association results, the dominance of homogeneous selection in both the cropping obstacle and control soils suggested that the fungal communities were more resilient than the bacterial communities.
Summary
Cropping obstacle soil was characterized by acidification and lower enzyme activities except for the activity of acid phosphatase. The lower bacterial richness and diversity, and the enrichment of plant pathogens may have contributed to the garlic cropping obstacle. The lower enzyme activity and microbial community differences were associated with lower pH in the cropping obstacle soil. The correlations among plant growth, soil properties and microbial communities cannot reveal cause and effect, thus discovering the exact mechanisms behind the cropping obstacle phenomenon require further controlled experiments.
Data Availability Statement
The data presented in the study are deposited in the National Center for Biotechnology Information (NCBI) with the accession number SRP348212.
Author Contributions
JY, QC, and ZS conceived and designed this experiment. JY, ZW, YL, and DC collected samples and carried out experiments. YG, KZ, XY, HL, and PP performed the bioinformatics and statistical analysis. JY, XZ, QC, and PP wrote and revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Key Research Project of Sichuan Province (No. 2019YFN0149) and the Professional Development Support Program of Sichuan Agricultural University.
Conflict of Interest
HL was employed by the CNPC Chuanqing Drilling Engineering Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We sincerely thank Chaoren Wang, Xiaoyu Zhou, and Die Liu for participating in the experiment. We appreciate technical support for data analysis provided by Shanghai Personalbio Technology Co., Ltd. and Xiangyang Shu.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.828196/full#supplementary-material
Footnotes
References
Acosta-Martinez, V., and Tabatabai, M. (2000). Enzyme activities in a limed agricultural soil. Biol. Fertil. Soils 31, 85–91. doi: 10.1007/s003740050628
Agegnehu, G., Nelson, P. N., and Bird, M. I. (2016). The effects of biochar, compost and their mixture and nitrogen fertilizer on yield and nitrogen use efficiency of barley grown on a Nitisol in the highlands of Ethiopia. Sci. Total Environ. 569, 869–879. doi: 10.1016/j.scitotenv.2016.05.033
Asnicar, F., Weingart, G., Tickle, T. L., Huttenhower, C., and Segata, N. (2015). Compact graphical representation of phylogenetic data and metadata with GraPhlAn. PeerJ 3:e1029. doi: 10.7717/peerj.1029
Baba, T., Hirose, D., Noma, S., and Ban, T. (2021). Inoculation with two Oidiodendron maius strains differentially alters the morphological characteristics of fibrous and pioneer roots of Vaccinium virgatum ‘Tifblue’ cuttings. Sci. Hortic. 281:109948. doi: 10.1016/j.scienta.2021.109948
Bastida, F., Zsolnay, A., Hernandez, T., and Garcia, C. (2008). Past, present and future of soil quality indices: a biological perspective. Geoderma 147, 159–171. doi: 10.1016/j.geoderma.2008.08.007
Bayer-Santos, E., Ceseti, L. D., Farah, C. S., and Alvarez-Martinez, C. E. (2019). Distribution, function and regulation of type 6 secretion systems of xanthomonadales. Front. Microbiol. 10:1635. doi: 10.3389/fmicb.2019.01635
Beckers, B., De Beeck, M. O., Weyens, N., Boerjan, W., and Vangronsveld, J. (2017). Structural variability and niche differentiation in the rhizosphere and endosphere bacterial microbiome of field-grown poplar trees. Microbiome 5:25. doi: 10.1186/s40168-017-0241-2
Blagodatskaya, E., and Kuzyakov, Y. (2013). Active microorganisms in soil: critical review of estimation criteria and approaches. Soil Biol. Biochem. 67, 192–211. doi: 10.1016/j.soilbio.2013.08.024
Bokulich, N. A., Subramanian, S., Faith, J. J., Gevers, D., Gordon, J. I., Knight, R., et al. (2013). Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10, 57–59. doi: 10.1038/nmeth.2276
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/Nmeth.3869
Comas, L., Eissenstat, D., and Lakso, A. (2008). Assesing root death and root system dynamics in a study of grape canopy pruning. New Phytol. 147, 171–178. doi: 10.1046/j.1469-8137.2000.00679.x
Corzo-Martinez, M., Corzo, N., and Villamiel, M. (2007). Biological properties of onions and garlic. Trends Food Sci. Technol. 18, 609–625. doi: 10.1016/j.tifs.2007.07.011
Dini-Andreote, F., Stegen, J. C., van Elsas, J. D., and Salles, J. F. (2015). Disentangling mechanisms that mediate the balance between stochastic and deterministic processes in microbial succession. Proc. Natl. Acad. Sci. U.S.A. 112, 1326–1332. doi: 10.1073/pnas.1414261112
Dong, L. L., Xu, J., Zhang, L. J., Yang, J., Liao, B. S., Li, X. W., et al. (2017). High-throughput sequencing technology reveals that continuous cropping of American ginseng results in changes in the microbial community in arable soil. Chin. Med. 12:18. doi: 10.1186/s13020-017-0139-8
Dumin, W., Park, M. J., Han, Y. K., Bae, Y. S., Park, J. H., and Back, C. G. (2021). First report of leaf spot disease caused by Stemphylium eturmiunum on garlic in Korea. Plant Dis. 106:318. doi: 10.1094/PDIS-03-21-0674-PDN
FAO (2019). Food and Agriculture Organization of the United Nations, FAOSTAT Statistics Database. Available online at: https://www.fao.org/faostat/en/#data
Fierer, N. (2017). Embracing the unknown: disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 15, 579–590.
Fierer, N., and Jackson, R. B. (2006). The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. U.S.A. 103, 626–631.
Fu, H. D., Zhang, G. X., Zhang, F., Sun, Z. P., Geng, G. M., and Li, T. L. (2017). Effects of continuous tomato monoculture on soil microbial properties and enzyme activities in a solar greenhouse. Sustainability 9:317. doi: 10.3390/su9020317
Gao, Z. Y., Han, M. K., Hu, Y. Y., Li, Z. Q., Liu, C. F., Wang, X., et al. (2019). Effects of continuous cropping of sweet potato on the fungal community structure in rhizospheric soil. Front. Microbiol. 10:2269. doi: 10.3389/fmicb.2019.02269
Gao, Z. Y., Hu, Y. Y., Han, M. K., Xu, J. J., Wang, X., Liu, L. F., et al. (2021). Effects of continuous cropping of sweet potatoes on the bacterial community structure in rhizospheric soil. BMC Microbiol. 21:2. doi: 10.1186/s12866-021-02194-2
Glassman, S. I., Wang, I. J., and Bruns, T. D. (2017). Environmental filtering by pH and soil nutrients drives community assembly in fungi at fine spatial scales. Mol. Ecol. 26, 6960–6973. doi: 10.1111/mec.14414
Gracas, J. P., Belloti, M., Lima, J. E., Peres, L. E. P., Burlat, V., Jamet, E., et al. (2021). Low pH-induced cell wall disturbances in Arabidopsis thaliana roots lead to a pattern-specific programmed cell death in the different root zones and arrested elongation in late elongation zone. Environ. Exp. Bot. 190:104596. doi: 10.1016/j.envexpbot.2021.104596
Guan, S. Y., Zhang, D., and Zhang, Z. (1986). Soil Enzyme and its Research Methods. Beijing: Agriculture Press, 274–297.
Jiao, S., Yang, Y. F., Xu, Y. Q., Zhang, J., and Lu, Y. H. (2020). Balance between community assembly processes mediates species coexistence in agricultural soil microbiomes across eastern China. ISME J. 14, 202–216. doi: 10.1038/s41396-019-0522-9
Katoh, K., Misawa, K., Kuma, K., and Miyata, T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Kaur, J., and Singh, J. P. (2014). Long-term effects of continuous cropping and different nutrient management practices on the distribution of organic nitrogen in soil under rice-wheat system. Plant Soil Environ. 60, 63–68. doi: 10.17221/440/2013-Pse
Kembel, S. W., Cowan, P. D., Helmus, M. R., Cornwell, W. K., Morlon, H., Ackerly, D. D., et al. (2010). Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26, 1463–1464. doi: 10.1093/bioinformatics/btq166
Koljalg, U., Nilsson, R. H., Abarenkov, K., Tedersoo, L., Taylor, A. F., Bahram, M., et al. (2013). Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 22, 5271–5277. doi: 10.1111/mec.12481
Li, Q., Zhang, D., Song, Z., Ren, L., Jin, X., Fang, W., et al. (2022a). Organic fertilizer activates soil beneficial microorganisms to promote strawberry growth and soil health after fumigation. Environ. Pollut. 295:118653. doi: 10.1016/j.envpol.2021.118653
Li, T., Li, Y., Wang, S., Wang, Z., Liu, Y., Wen, X., et al. (2022b). Assessing synergistic effects of crop rotation pattern, tillage practice, and rhizosphere effect on soil bacterial community structure and assembly in China’s Loess Plateau farmlands. Appl. Soil Ecol. 174:104411. doi: 10.1016/j.apsoil.2022.104411
Li, Q. Q., Li, A. W., Yu, X. L., Dai, T. F., Peng, Y. Y., Yuan, D., et al. (2020). Soil acidification of the soil profile across Chengdu Plain of China from the 1980s to 2010s. Sci. Total Environ. 698:134320. doi: 10.1016/j.scitotenv.2019.134320
Li, W. H., Liu, Q. Z., and Chen, P. (2018). Effect of long-term continuous cropping of strawberry on soil bacterial community structure and diversity. J. Integr. Agric. 17, 2570–2582. doi: 10.1016/S2095-3119(18)61944-6
Li, X. G., Ding, C. F., Zhang, T. L., and Wang, X. X. (2014). Fungal pathogen accumulation at the expense of plant-beneficial fungi as a consequence of consecutive peanut monoculturing. Soil Biol. Biochem. 72, 11–18. doi: 10.1016/j.soilbio.2014.01.019
Li, Z. G., Zu, C., Wang, C., Yang, J. F., Yu, H., and Wu, H. S. (2016). Different responses of rhizosphere and non-rhizosphere soil microbial communities to consecutive Piper nigrum L. monoculture. Sci. Rep. 6:35825. doi: 10.1038/srep35825
Liu, P. X., Weng, R., Sheng, X. J., Wang, X. L., Zhang, W. H., Qian, Y. Z., et al. (2020). Profiling of organosulfur compounds and amino acids in garlic from different regions of China. Food Chem. 305:125499. doi: 10.1016/j.foodchem.2019.125499
Liu, S., Wang, Z. Y., Niu, J. F., Dang, K. K., Zhang, S. K., Wang, S. Q., et al. (2021). Changes in physicochemical properties, enzymatic activities, and the microbial community of soil significantly influence the continuous cropping of Panax quinquefolius L. (American ginseng). Plant Soil 463, 427–446. doi: 10.1007/s11104-021-04911-2
Liu, X., Zhang, J. L., Gu, T. Y., Zhang, W. M., Shen, Q. R., Yin, S. X., et al. (2014). Microbial community diversities and taxa abundances in soils along a seven-year gradient of potato monoculture using high throughput pyrosequencing approach. PLoS One 9:86610. doi: 10.1371/journal.pone.0086610
Luan, L., Liang, C., Chen, L. J., Wang, H. T., Xu, Q. S., Jiang, Y. J., et al. (2020). Coupling bacterial community assembly to microbial metabolism across soil profiles. Msystems 5:e00298–20. doi: 10.1128/mSystems.00298-20
Martin, M. (2011). CUTADAPT removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
McArdle, B., and Anderson, M. (2001). Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology 82, 290–297. doi: 10.2307/2680104
Nannipieri, P., Giagnoni, L., Renella, G., Puglisi, E., Ceccanti, B., Masciandaro, G., et al. (2012). Soil enzymology: classical and molecular approaches. Biol. Fertil. Soils 48, 743–762. doi: 10.1007/s00374-012-0723-0
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2020). Vegan Community Ecology Package Version 2.5-7 November 2020.
Pang, Z., Dong, F., Liu, Q., Lin, W., Hu, C., and Yuan, Z. (2021). Soil metagenomics reveals effects of continuous sugarcane cropping on the structure and functional pathway of rhizospheric microbial community. Front. Microbiol. 12:627569. doi: 10.3389/fmicb.2021.627569
Paz-Ferreiro, J., and Fu, S. L. (2016). Biological indices for soil quality evaluation: perspectives and limitations. Land Degrad. Dev. 27, 14–25. doi: 10.1002/ldr.2262
Perez-Brandan, C., Huidobro, J., Grumberg, B., Scandiani, M. M., Luque, A. G., Meriles, J. M., et al. (2014). Soybean fungal soil-borne diseases: a parameter for measuring the effect of agricultural intensification on soil health. Can. J. Microbiol. 60, 73–84. doi: 10.1139/cjm-2013-0792
Price, M. N., Dehal, P. S., and Arkin, A. P. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. doi: 10.1093/molbev/msp077
Ramette, A. (2007). Multivariate analyses in microbial ecology. FEMS Microbiol. Ecol. 62, 142–160. doi: 10.1111/j.1574-6941.2007.00375.x
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60
Semenov, M. V., Krasnov, G. S., Semenov, V. M., and van Bruggen, A. H. C. (2020). Long-term fertilization rather than plant species shapes rhizosphere and bulk soil prokaryotic communities in agroecosystems. Appl. Soil Ecol. 154:103641. doi: 10.1016/j.apsoil.2020.103641
Shen, C. C., Xiong, J. B., Zhang, H. Y., Feng, Y. Z., Lin, X. G., Li, X. Y., et al. (2013). Soil pH drives the spatial distribution of bacterial communities along elevation on Changbai Mountain. Soil Biol. Biochem. 57, 204–211. doi: 10.1016/j.soilbio.2012.07.013
Shen, J., Tao, Q., Dong, Q., Luo, Y. L., Luo, J. P., He, Y. T., et al. (2021). Long-term conversion from rice-wheat to rice-vegetable rotations drives variation in soil microbial communities and shifts in nitrogen-cycling through soil profiles. Geoderma 404:115299. doi: 10.1016/j.geoderma.2021.115299
Stegen, J. C., Lin, X. J., Fredrickson, J. K., Chen, X. Y., Kennedy, D. W., Murray, C. J., et al. (2013). Quantifying community assembly processes and identifying features that impose them. ISME J. 7, 2069–2079. doi: 10.1038/ismej.2013.93
Stegen, J. C., Lin, X. J., Fredrickson, J. K., and Konopka, A. E. (2015). Estimating and mapping ecological processes influencing microbial community assembly. Front. Microbiol. 6:370. doi: 10.3389/fmicb.2015.00370
Sun, L., Gao, J. S., Huang, T., Kendall, J. R. A., Shen, Q. R., and Zhang, R. F. (2015). Parental material and cultivation determine soil bacterial community structure and fertility. FEMS Microbiol. Ecol. 91, 1–10. doi: 10.1093/femsec/fiu010
Syed, S., Tollamadugu, N. V. K. V. P., and Lian, B. (2020). Aspergillus and Fusarium control in the early stages of Arachis hypogaea (groundnut crop) by plant growth-promoting rhizobacteria (PGPR) consortium. Microbiol. Res. 240:126562. doi: 10.1016/j.micres.2020.126562
Tan, G., Liu, Y., Peng, S., Yin, H., Meng, D., Tao, J., et al. (2021). Soil potentials to resist continuous cropping obstacle: three field cases. Environ. Res. 200:111319. doi: 10.1016/j.envres.2021.111319
van Wyk, D. A. B., Adeleke, R., Rhode, O. H. J., Bezuidenhout, C. C., and Mienie, C. (2017). Ecological guild and enzyme activities of rhizosphere soil microbial communities associated with Bt-maize cultivation under field conditions in North West Province of South Africa. J. Basic Microbiol. 57, 781–792. doi: 10.1002/jobm.201700043
Wang, R., Zhang, H. C., Sun, L. G., Qi, G. F., Chen, S., and Zhao, X. Y. (2017). Microbial community composition is related to soil biological and chemical properties and bacterial wilt outbreak. Sci. Rep. 7:343. doi: 10.1038/s41598-017-00472-6
Wang, Z. B., Yang, Y., Xia, Y. Z., Wu, T., Zhu, J., Yang, J. M., et al. (2019). Time-course relationship between environmental factors and microbial diversity in tobacco soil. Sci. Rep. 9:19969. doi: 10.1038/s41598-019-55859-4
Wei, X., Chen, J., Zhang, C., and Pan, D. (2016). A new oidiodendron maius strain isolated from rhododendron fortunei and its effects on nitrogen uptake and plant growth. Front. Microbiol. 7:1327. doi: 10.3389/fmicb.2016.01327
Wei, Z., Yang, T., Friman, V. P., Xu, Y., Shen, Q., and Jousset, A. (2015). Trophic network architecture of root-associated bacterial communities determines pathogen invasion and plant health. Nat. Commun. 6:8413. doi: 10.1038/ncomms9413
Xi, H., Shen, J., Qu, Z., Yang, D., Liu, S., Nie, X., et al. (2019). Effects of long-term cotton continuous cropping on soil microbiome. Sci. Rep. 9:18297. doi: 10.1038/s41598-019-54771-1
Yachi, S., and Loreau, M. (1999). Biodiversity and ecosystem productivity in a fluctuating environment: the insurance hypothesis. Proc. Natl. Acad. Sci. U.S.A. 96, 1463–1468. doi: 10.1073/pnas.96.4.1463
Zeng, L. S., Liao, M., Chen, C. L., and Huang, C. Y. (2007). Effects of lead contamination on soil enzymatic activities, microbial biomass, and rice physiological indices in soil-lead-rice (Oryza sativa L.) system. Ecotoxicol. Environ. Safety 67, 67–74. doi: 10.1016/j.ecoenv.2006.05.001
Zhao, J., Zhang, D., Yang, Y. Q., Pan, Y., Zhao, D. M., Zhu, J. H., et al. (2020). Dissecting the effect of continuous cropping of potato on soil bacterial communities as revealed by high-throughput sequencing. PLoS One 15:e0233356. doi: 10.1371/journal.pone.0233356
Zhao, Y., Wang, P., Li, J., Chen, Y., Ying, X., and Liu, S. (2009). The effects of two organic manures on soil properties and crop yields on a temperate calcareous soil under a wheat–maize cropping system. Eur. J. Agron. 31, 36–42. doi: 10.1016/j.eja.2009.03.001
Zheng, Q., Hu, Y. T., Zhang, S. S., Noll, L., Bockle, T., Dietrich, M., et al. (2019). Soil multifunctionality is affected by the soil environment and by microbial Choo community composition and diversity. Soil Biol. Biochem. 136:107521. doi: 10.1016/j.soilbio.2019.107521
Keywords: garlic, cropping obstacles, soily acidification, soil enzyme activity, soil microbial community
Citation: Yu J, Liu Y, Wang Z, Huang X, Chai D, Gu Y, Zhao K, Yu X, Shuai Z, Liu H, Zhang X, Penttinen P and Chen Q (2022) The Cropping Obstacle of Garlic Was Associated With Changes in Soil Physicochemical Properties, Enzymatic Activities and Bacterial and Fungal Communities. Front. Microbiol. 13:828196. doi: 10.3389/fmicb.2022.828196
Received: 03 December 2021; Accepted: 28 February 2022;
Published: 30 March 2022.
Edited by:
Xun Wen Chen, Southern University of Science and Technology, ChinaReviewed by:
Xiaomin Li, South China Normal University, ChinaYu Shi, Institute of Soil Science (CAS), China
Copyright © 2022 Yu, Liu, Wang, Huang, Chai, Gu, Zhao, Yu, Shuai, Liu, Zhang, Penttinen and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Petri Penttinen, cGV0cmkucGVudHRpbmVuQGhlbHNpbmtpLmZp; Qiang Chen, Y3FpYW5nQHNpY2F1LmVkdS5jbg==
 Jinyang Yu
Jinyang Yu Yihao Liu1
Yihao Liu1 Yunfu Gu
Yunfu Gu Ke Zhao
Ke Zhao Xiumei Yu
Xiumei Yu Hanjun Liu
Hanjun Liu Xiaoping Zhang
Xiaoping Zhang Petri Penttinen
Petri Penttinen Qiang Chen
Qiang Chen