
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 17 February 2022
Sec. Aquatic Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.826602
This article is part of the Research Topic Responses of Marine Microbes to Multiple Environmental Drivers of Global Change: the Interplay of Abiotic and Biotic Factors View all 23 articles
Phaeocystis globosa (P. globosa) is one of the dominant algae during harmful algal blooms (HABs) in coastal regions of Southern China. P. globosa exhibits complex heteromorphic life cycles that could switch between solitary cells and colonies. The ecological success of P. globosa has been attributed to its colony formation, although underlying mechanisms remain unknown. Here, we investigated different bacterial communities associated with P. globosa colonies and their influence on colony formation of two P. globosa strains isolated from coastal waters of Guangxi (GX) and Shantou (ST). Eight operational taxonomic units (OTUs) were observed in ST co-cultures and were identified as biomarkers based on Linear discriminant analysis Effect Size (LEfSe) analysis, while seven biomarkers were identified in P. globosa GX co-cultures. Bacterial communities associated with the P. globosa GX were more diverse than those of the ST strain. The most dominant phylum in the two co-cultures was Proteobacteria, within which Marinobacter was the most abundant genus in both GX and ST co-cultures. Bacteroidota were only observed in the GX co-cultures and Planctomycetota were only observed in the ST co-cultures. Co-culture experiments revealed that P. globosa colony formation was not influenced by low and medium cell densities of Marinobacter sp. GS7, but was inhibited by high cell densities of Marinobacter sp. GS7. Overall, these results indicated that the associated bacteria are selected by different P. globosa strains, which may affect the colony formation and development of P. globosa.
The Phaeocystis are globally distributed marine algae, which cause frequent coastal harmful algal blooms and play important roles in carbon and sulfur biogeochemical cycling (Schoemann et al., 2005; Verity et al., 2007). Owing to their negative effects on marine ecosystems, fisheries, and local economies, the bloom development of Phaeocystis has gained much attention in recent decades (Schoemann et al., 2005). Phaeocystis species, such as Phaeocystis globosa, Phaeocystis pouchetii, and Phaeocystis antarctica have all been reported to form extensive colony blooms in many regions including in tropic and polar waters (Schoemann et al., 2005).
Phaeocystis exhibit complex polymorphic life cycles involving solitary cell and colony stages, wherein solitary cells are generally 3–9 μm and colonies are usually several mm in diameter (Rousseau et al., 1994). Extraordinarily large Phaeocystis colonies (up to 3 cm in diameter) have also been found in coastal waters of South China (Qi et al., 2004; Smith et al., 2014).
Colony formation partially underlies the success of Phaeocystis in marine ecosystems (Rousseau et al., 1994; Hamm, 2000). Solitary cells are generally consumed by small grazers (Tang et al., 2001), while colonies are ingested by zooplankton to a lesser degree due to their tough exteriors and size mismatches between colonies and grazers (Hamm et al., 1999; Jakobsen and Tang, 2002). Thus, colony formation protects Phaeocystis cells from predation and thus significantly decreases mortality (Hamm et al., 1999). Several abiotic and biological factors have been proposed to affect the colony formation of Phaeocystis, such as light exposure (Wang et al., 2014), macronutrient levels (Wang et al., 2010), temperatures (Wang et al., 2010), and zooplankton grazing (Jakobsen and Tang, 2002), or combinations of these factors, while the molecular mechanisms underlying colony formation remain enigmatic (Verity et al., 2007).
Colonies largely comprise polysaccharides (Hamm et al., 1999), which provide a carbon source for surrounding bacterial populations (Dutz and Koski, 2006; Verity et al., 2007; Wemheuer et al., 2015). Previous studies have shown dynamic bacterial community compositions during P. globosa blooms (Li et al., 2020). For example, in the course of Phaeocystis blooms in the southern North Sea, bacterial diversity decreased significantly and Gammaproteobacteria became more abundant (Wemheuer et al., 2014, 2015). During the demise of a Phaeocystis spring bloom in the North Sea, Bacteroidota abundances increased sharply, which might be involved in mucopolysaccharide degradation (Alderkamp et al., 2006). Similar trends were also observed in the coastal blooms of South China. For instance, Li et al. (2020) found that bacterial diversity of free-living bacteria was lower during P. globosa marine blooms, while Marinobacterium, Erythrobacter, and Persicobacter became dominant in the terminal stage of P. globosa blooms. Zhu et al. (2021) suggested that seawater bacterial richness and diversity were significantly lower in comparison to P. globosa intracolonial fluids during P. globosa blooms (Zhu et al., 2021). Despite these observations, the potential effects of bacterioplankton on P. globosa colony formation remain enigmatic.
Here, bacterial communities associated with P. globosa isolated from coastal waters were investigated and their effects on P. globosa colony formation were explored via co-culture experiments. Specifically, this study aimed to address: (i) how different P. globosa strains influence bacterioplankton composition and diversity, (ii) the identity of bacterial species associated with P. globosa colony development, and (iii) the potential bacterial influences on P. globosa colony formation.
Phaeocystis globosa Guangxi (GX) and Shantou (ST) strains were isolated from coastal waters of Guangxi in 2017 and Shantou in 2003, respectively. Bacterial cells were isolated from the exponential stages of P. globosa GX and ST strain growth by serially diluting 1.0 ml aliquots of cultures into sterile seawater. After serial dilutions, 100 μl of the 10–2 to 10–7 dilutions were spread onto agar plates of marine agar 2216 (MA; BD Difco) (Yang et al., 2021). Plates were then incubated at 20°C for 7 days in the dark. Bacterial colonies exhibiting different morphological characteristics were isolated and stored in marine broth 2216 (MB, BD Difco) supplemented with 20% glycerol to form stocks that were stored at −80°C for future experiments (Xu et al., 2021a,b).
Bacteria grown on marine agar plates (MA; BD Difco) were incubated at 20°C in the dark with shaking at 200 revolutions per minute (rpm). To identify the isolated bacteria, single colonies were cultured in marine broth after incubating for 72 h in the dark, followed by centrifugation of cells at 10,000 rpm for 1 min. The centrifuged supernatants were removed and DNA was extracted from the pellets using a TIANGEN Bacterial DNA extraction kit according to the manufacturer’s instructions. The 16S rRNA genes from bacterial isolates were then amplified using universal 16S rRNA primers (27F, 1492R) (Frank et al., 2008) and a Green Tap amplification kit (Vazyme, China). Amplicons were sequenced at Sangon Biotech (Shanghai) Co., Ltd. (Shanghai, China). Marinobacter sp. GS7 cells were fixed using glutaraldehyde and then photographed using a scanning electron microscope (Zeiss ULTRA™ 55, Carl Zeiss Inc., Oberkochen, Germany).
Sequence alignments were generated for the 16S rRNA genes using the EzBioCloud server1 platform. Phylogenetic inference was then conducted using the Maximum-Likelihood method in Mega X and node support was evaluated with 1,000 bootstrap replicates (Kumar et al., 2018).
Cultures of P. globosa GX and ST strains were maintained in the exponential growth stage via regular dilution with f/2 medium, and were grown in f/2 medium at 20°C with 12 h light: 12 h dark diurnal cycles (100 μmol m–2 s–1) (Liang et al., 2020).
To generate axenic cultures, solitary P. globosa cells were separated by filtering culture stocks through 10 μm filters. The cells were centrifuged at 2,000 rpm for 5 min, then quickly rinsed with sterile f/2 medium twice and washed for 1 min in sterile media containing 20 μg ml–1 Triton X-100 (Amin et al., 2015). Solitary cells were subsequently washed off the filter by gentle shaking into sterile media containing a suite of antibiotics (per milliliter: 5 μg penicillin, 10 μg streptomycin, 0.1 mg kanamycin, and 1 mg ampicillin). Cells were then incubated in antibiotic-containing media for 48 h under equivalent growth conditions. Finally, 20 ml of antibiotic-treated cells were centrifuged at 2,000 rpm for 5 min, then washed twice with sterile f/2 medium by centrifuging at 2,000 rpm for 5 min and removal of supernatant fluid. The cells were then transferred to conventional f/2 media for 8 days, with four or five rounds of continual transfer. Bacterial contamination was checked via traditional agar plate culturing.
Bacteria were plated on fresh marine agar plates and grown from single colonies in marine broth by incubating for 72 h at 28°C in the dark with shaking at 200 rpm. Cells were then centrifuged at 8,000 rpm for 5 min and washed twice with sterile seawater, followed by diluting to a stock cell density of 101−1 × 107 cells/ml. Bacterial isolates and axenic P. globosa were then co-cultured using f/2 medium. The initial bacterial cell densities were 1 × 105, 1 × 106, and 1 × 107 cells/ml, while P. globosa cell densities were adjusted to 1 × 104 cells/ml to achieve starting bacterial: P. globosa ratios of 10:1, 100:1, and 1,000:1. Cultures with different starting ratios were designated as L (low cell density, 1 × 105 cells/ml), M (medium cell density, 1 × 106 cells/ml), and H (high cell density, 1 × 107 cells/ml). In addition, P. globosa GX and ST strain cultures without bacterial co-culture were used as controls. All treatments and controls were conducted in triplicate. Cultures were maintained under the same conditions as stock cultures for 10 days (Liang et al., 2020). The abundances of solitary and colonial P. globosa cells were then counted using an Olympus inverted microscope (CKX53, Japan).
A 400 ml sample of culture was used for DNA extractions, with three parallel replicates each. Samples were filtered with 0.22 μm Millipore filters to capture bacterial communities, followed by storage of filters at −20°C for subsequent DNA extraction. DNA extraction using the FastDNA Spin Kit for Soil (MP Biomedicals, Solon, OH, United States) according to the manufacturer’s instructions. DNA yield and purity were measured using a NanoDrop 2000 spectrophotometer (Thermo Scientific Inc., Waltham, MA, United States) and 1.0% agarose gel electrophoresis, respectively.
The V3–V4 hypervariable regions of bacterial 16S rRNA gene were amplified using the universal primer with 338F/806R (Xu et al., 2016). PCRs comprised TransStart® Fastpfu DNA Polymerase (TransGen Biotech, Beijing, China) and reactions were prepared according to the manufacturer’s instructions. PCR conditions included 95°C for 3 min followed by 27 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 45 s, with a final extension at 72°C for 10 min. High-throughput sequencing of amplicons was conducted at Shanghai Majorbio Bio-pharm Co., Ltd. (Shanghai, China) on the Illumina MiSeq platform (Illumina, San Diego, CA, United States) with 250 bp paired-end sequencing.
Sequences were merged using FLASH (version 1.2.7) (Magoc and Salzberg, 2011) and the raw FASTQ files were quality filtered using fastp (version 0.20.0) (Chen et al., 2018). Quality filtered sequences were aligned with the SLIVA alignment (Quast et al., 2013), and sequences annotated as chloroplasts or mitochondria were removed, followed by clustering of sequences into operational taxonomic units (OTUs) at the 97% nucleotide sequence similarity threshold with UPARSE (version 7.1) (Edgar, 2013). Linear discriminant analysis Effect Size (LEfSe) analysis (Segata et al., 2011) was used to explore potential bacterial biomarkers associated with different P. globosa strains. The potential functions of bacterial populations were evaluated with FAPROTAX (version 1.2.4) (Louca et al., 2016).
The OTU sequence numbers were normalized to an equal number by 31065 for the later statistical analysis. The α-diversity indexes for richness estimators (ACE and Chao1), diversity (Shannon and Simpson), and Good’s coverage were calculated by using “vegan” R package (Jari Oksanen et al., 2019). The NMDS (non-metric multidimensional scaling) were by ANOSIM with 999 permutations for Bray–Curtis dissimilarities by using vegan and ggplot2 R packages (Wickham, 2017; Jari Oksanen et al., 2019). Distance-based redundancy analysis (db-RDA) followed the ANOVA with 999 permutations by using vegan R package (Jari Oksanen et al., 2019). Using the variable inflation factor (VIF) index with a maximum cut-off score of 10 checked multicollinearity among solitary cells abundance, colonial abundance, and colony diameter of P. globosa.
Rarefaction curves and Good’s coverage index indicated that the level of sequencing conducted was adequate to recover most sample diversity for both the GX and ST strain co-cultures (Supplementary Figure 1). The ACE and Chao 1 richness index values for the GX co-culture bacterial community were much higher than those of the ST strain, indicating that bacterial richness was significantly different in the GX and ST strain co-cultures (ACE, p < 0.05, Chao 1, p < 0.01). Significant differences were not observed for the Shannon and Simpson indices when comparing the GX and ST communities (Figure 1A), which is consistent with their bacterial OTU compositions (ANOSIM, p > 0.05 for Bray–Curtis metrics) through the NMDS analysis (Figure 1B).
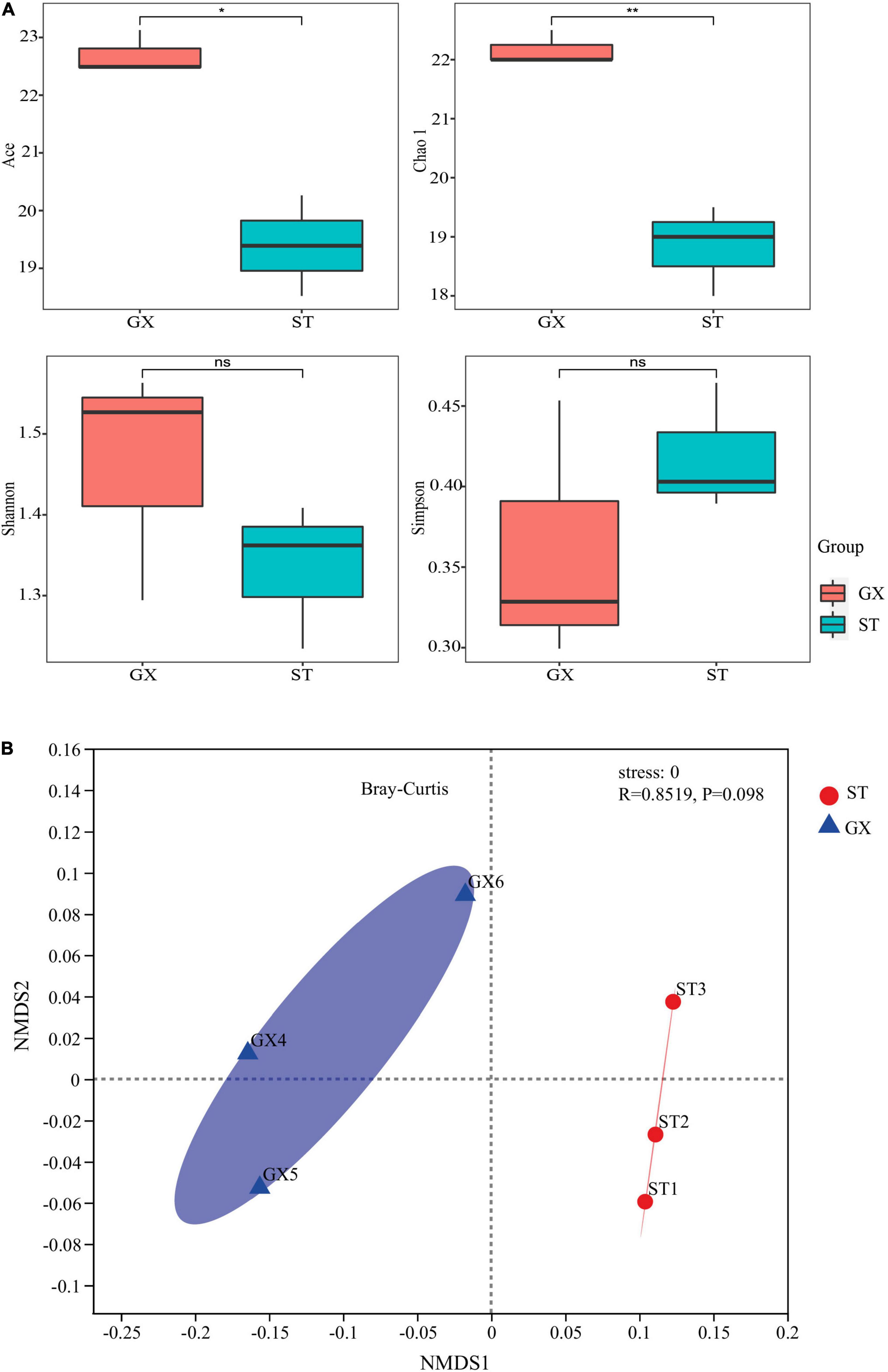
Figure 1. The diversity of bacterial communities associated with Phaeocystis globosa GX and ST strain cultures. (A) α-Diversity indices for the two co-culture communities. Statistical differences between pairs were assessed by t-tests. nsNot significant; *p < 0.05; **p < 0.01. (B) Non-parametric multidimensional scaling (NMDS) plots show variation in OTU compositions among the two co-cultures. GX, P. globosa GX strain co-culture; ST, P. globosa ST strain co-culture.
A total of 26 OTUs were identified from the 6 co-culture (ST1, ST2, ST and GX4, GX5, GX6) communities, comprising 5 phyla, 8 classes, 15 orders, 21 families, and 25 genera. OTU2 was the dominant OTU among all samples (Supplementary Table 1). Community composition was considerably similar in both GX and ST co-cultures (Figure 2A). At the phylum level, Proteobacteria (82.7% average relative abundance) dominated the bacterial communities in both co-cultures, followed by Bacteroidota (16.9%), which were more abundant in the GX co-cultures. Proteobacteria (98.8%) and Planctomycetota (1.2%) were more abundant in the ST co-cultures, while Bacteroidota was absent. Proteobacteria was the most abundant phylum for both GX and ST co-cultures (Figure 2B). Marinobacter, Marixanthomonas, and unclassified Alteromonadaceae were the dominant genera in the GX co-culture (abundances >5%), while Marinobacter, unclassified Alteromonadaceae, uncultured Hyphomonadaceae, and Labrenzia were the dominant genera in the ST co-culture. Although the bacterial compositions differed between the GX and ST co-cultures, Marinobacter dominated both systems, with 55.0% and 61.9% abundances in the GX and ST co-cultures, respectively (Figure 2C).
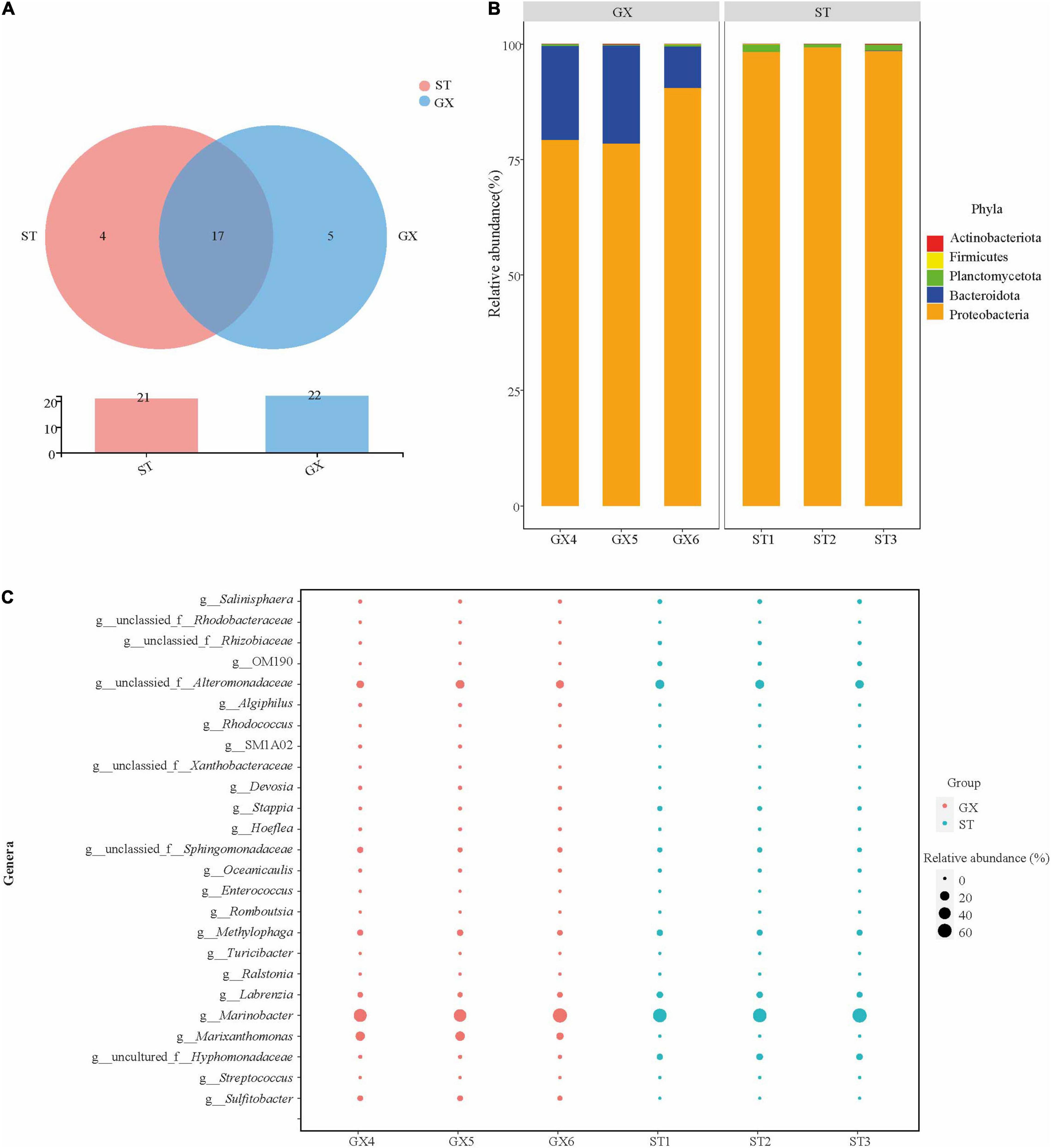
Figure 2. Composition of Phaeocystis globosa bacterial communities. (A) Shared and unique OTUs among co-cultures, visualized with Venn diagrams. (B) Co-culture bacterial community composition at the phylum level. (C) Co-culture bacterial community composition at the genus level. GX, P. globosa GX strain co-culture; ST, P. globosa ST strain co-culture.
Cladograms were used to depict the distributions of taxonomic groups (Figure 3), with LDA scores >2 being used to identify biomarkers with LEfSe analysis (Figure 4). More biomarkers (LDA > 2) were identified for the P. globosa ST co-cultures compared to the GX co-cultures. Specifically, LEfSe analysis indicated that the P. globosa ST co-cultures included eight biomarkers including OTU27 (p__Planctomycetota, c__OM190), OTU20 (g__Roseitalea), OTU23 (g__Cohaesibacter), OTU5 (g__Stappia), and OTU16 (g__Labrenzia) affiliated with Rhizobiales (p_Proteobacteria, c_Alphaproteobacteria); OTU18 (g__Rhodococcus) affiliated with the Corynebacteriales (p__Actinobacteriota, c__Actinobacteria); OTU25 affiliated with the Caulobacterales (p__Proteobacteria, c__Alphaproteobacteria); and OTU9 (g__unclassified_o__Salinisphaerales) affiliated with the Salinisphaerales (p__Proteobacteria, c__Gammaproteobacteria).
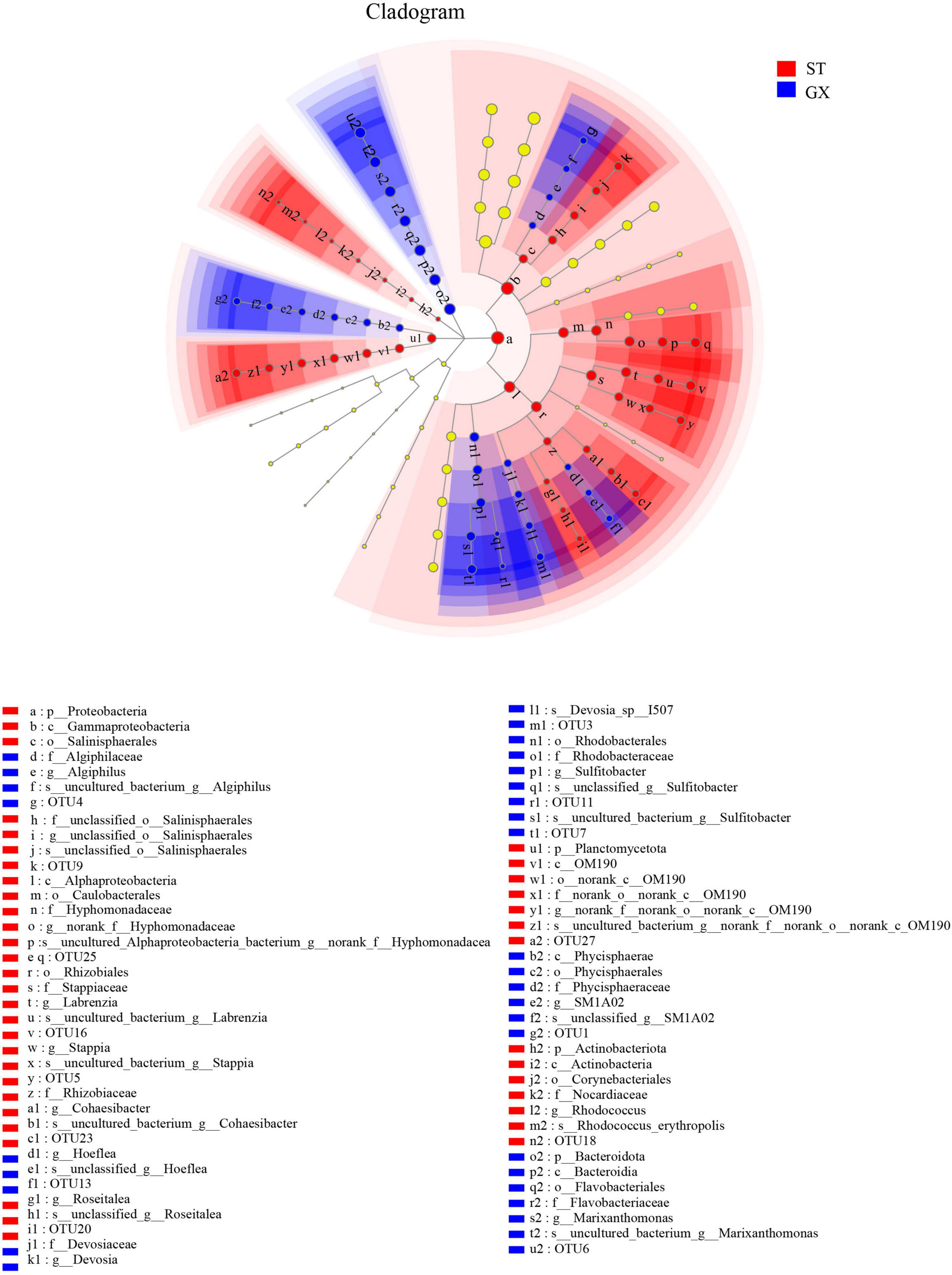
Figure 3. Cladogram showing the phylogenetic distributions of bacterial taxa associated with Phaeocystis globosa cultures. Circles indicate the taxonomic level ranging from the phylum to OTU levels. The diameter of each circle is proportional to the abundance of that group. GX, P. globosa GX strain co-culture; ST, P. globosa ST strain co-culture. p, phylum; o, order; f, family; g, genus; s, species.
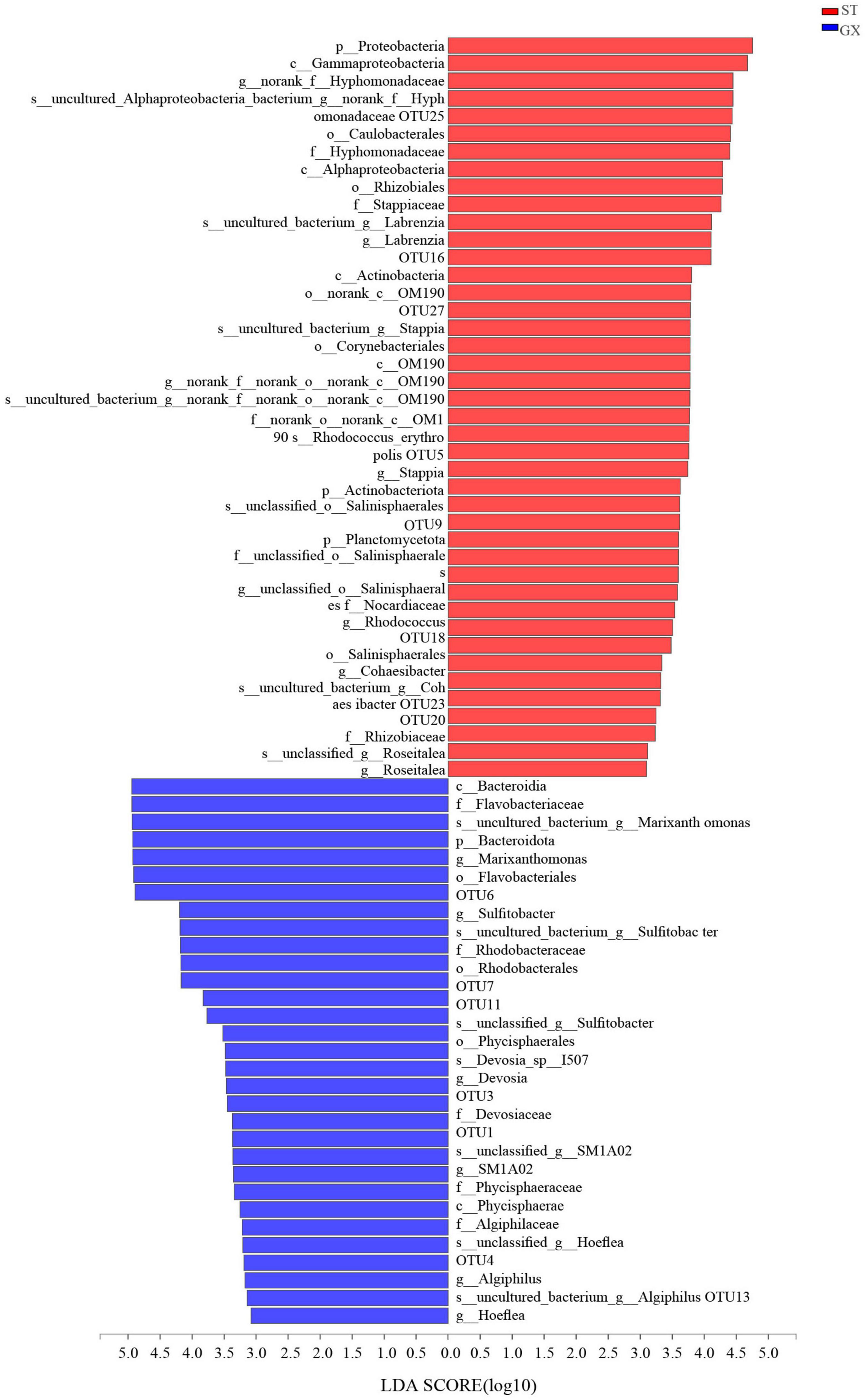
Figure 4. Indicator bacterial taxa with LDA scores >2. Different-colored regions of the bars in panel indicate taxa from different co-cultures. GX, P. globosa GX strain co-culture; ST, P. globosa ST strain co-culture. p, phylum; o, order; f, family; g, genus; s, species.
The P. globosa GX co-culture harbored seven biomarkers including OTU3 (g_Devosia) and OTU13 (g_Hoeflea) affiliated with the Rhizobiales (p_Proteobacteria, c_Alphaproteobacteria); OTU11 and OTU7 (g_Sulfitobacter) affiliated with the Rhodobacterales (p_Proteobacteria, c_Alphaproteobacteria); along with OTU6 (g__Marixanthomonas), OTU1 (g__SM1A02), and OTU4 (g__Algiphilus). Greater numbers of biomarkers affiliated with Rhizobiales were enriched in the P. globosa ST co-cultures compared to the GX co-culture, while one biomarker was affiliated with the Planctomycetota in P. globosa ST co-culture that was not observed in the GX co-cultures.
FAPROTAX was used to predict the potential metabolic functions of bacterioplankton populations based on 16S rRNA gene identities. Twelve functional groups were predicted from the 26 OTUs. Chemoheterotrophic (chemoheterotrophy and aerobic chemoheterotrophy) microbial populations were predicted as the most dominant groups, accounting for 67.7 and 70.3% of the P. globosa GX and ST co-cultures, respectively (Figure 5A). Hydrocarbon degradation-associated functional groups accounted for 24.3 and 26.0% of the P. globosa GX and ST co-cultures, respectively. OTU2 (Marinobacter) was present in all samples and was predicted to be involved in chemoheterotrophy, aerobic chemoheterotrophy, and hydrocarbon degradation (Figures 5B,C and Supplementary Table 2). Overall, functional predictions suggested that bacterial communities of P. globosa GX and ST co-cultures maintained similar metabolic functions, with the dominant functional profiles arising from Marinobacter.
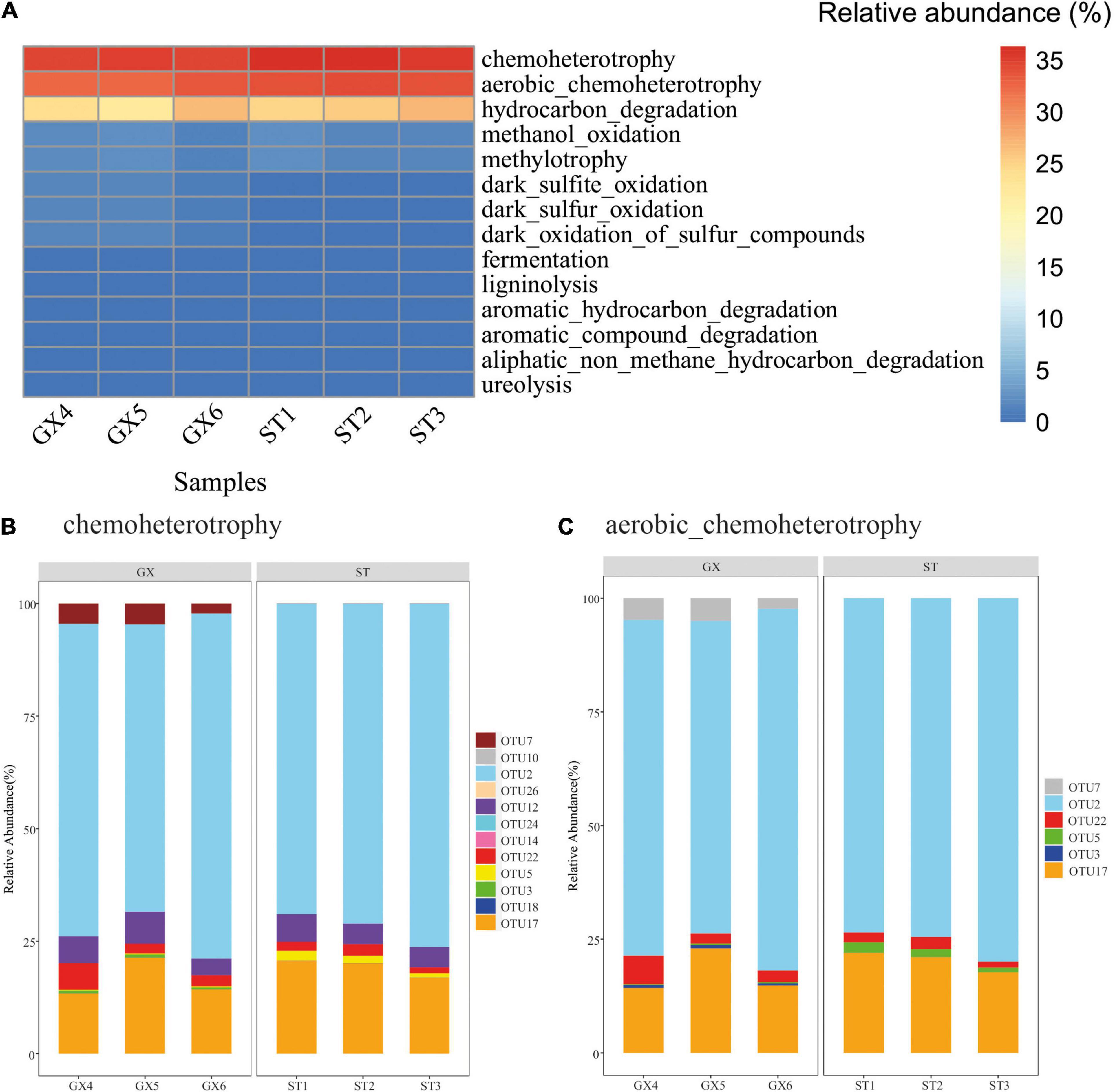
Figure 5. Predicted functional groups among the bacterial communities associated with Phaeocystis globosa cultures. (A) Relative abundances of functional classifications. (B) OTUs associated with predicted chemoheterotrophic functions. (C) OTUs associated with predicted aerobic chemoheterotrophic functions.
To identify strain-specific interactions between Phaeocystis and associated bacteria, ten and eight cultivable bacterial strains were isolated from the GX and ST co-cultures, respectively. The strains were identified as Alteromonas, Hoeflea, Labrenzia, Sulfitobacter, Oceanicaulis, and Marinobacter genera based on >97% similarity in 16S rRNA gene sequences.
Marinobacter GS7 cells are Gram-negative, rod-shaped, not flagellated, 1.5–2.5 μm in length, and 0.3–0.5 μm wide (Supplementary Figure 2). Phylogenetic analysis based on 16S rRNA gene sequences indicated that GS7 was closely related to Marinobacter shengliensis SL013A34A2 (98.6% 16S rRNA gene identity) (Supplementary Figure 3). GS7 formed a distinct phylogenetic cluster (16S rRNA genes exhibited 99.7% nucleotide similarity) with OTU2, which was the dominant taxa in GX and ST strain co-cultures (Supplementary Figure 4 and Supplementary Table 1).
We first determined that the growth rate of P. globosa was not influenced by the short-term removal of bacterial populations. Further, the growth rate was also unaffected when co-cultured with Marinobacter sp. GS7 at cell densities of 1 × 101 to 1 × 104 cells/ml. However, high cell densities of Marinobacter sp. GS7 significantly decreased (p < 0.01) the solitary cell abundances of P. globosa ST and GX strains (Figure 6A). In contrast, low and medium cell densities of Marinobacter sp. GS7 did not impact (p > 0.05) solitary cell abundances of the P. globosa ST and GX strains (Figure 6A). Addition of low and medium cell densities of Marinobacter sp. GS7 also did not impact (p > 0.05) P. globosa colony numbers for either strain. Rather, P. globosa GX and ST strains failed to form colonies when they were exposed to high cell densities of Marinobacter sp. GS7 (Figure 6B).
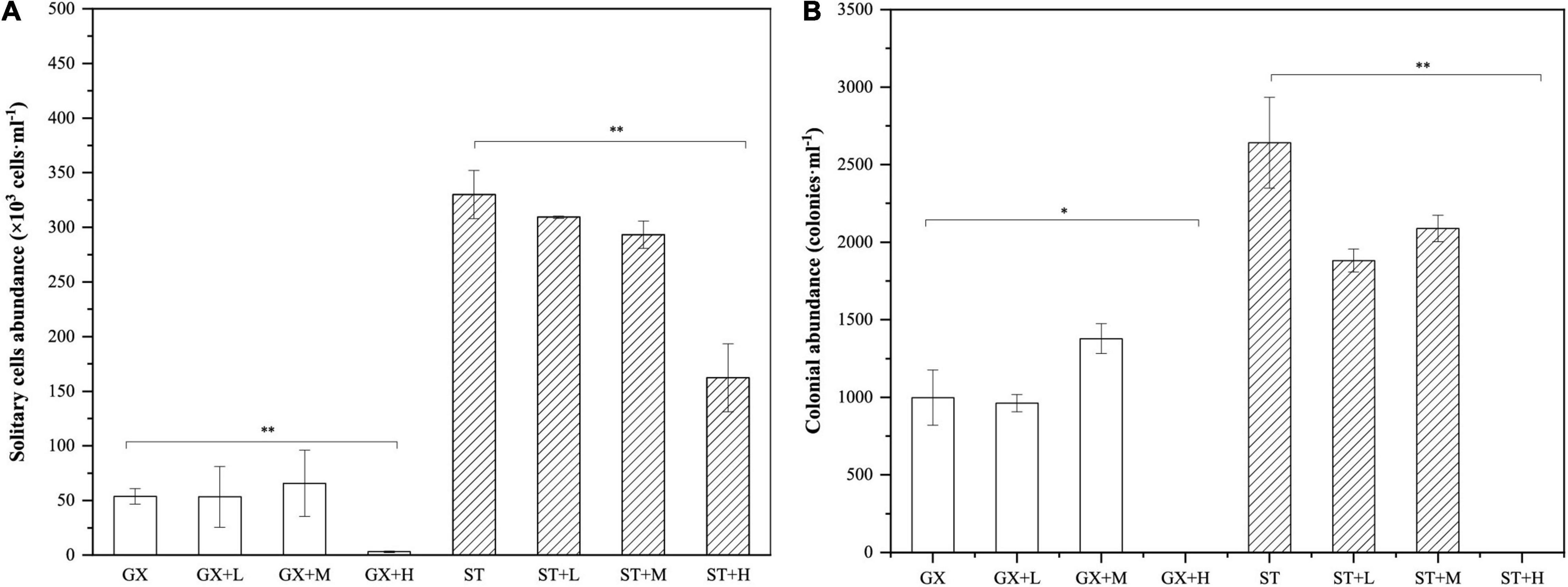
Figure 6. Growth characteristics of Phaeocystis globosa-Marinobacter sp. GS7 co-cultures. (A) The abundances of solitary P. globosa cells. (B) The abundances of P. globosa colonies. Each bar represents the mean of three biological replicates and error bars indicate standard deviations of the means. Statistical differences between treatments and controls were tested with t-tests. *p < 0.05; **p < 0.01. Non-significant comparisons are not marked. GX, P. globosa GX strain co-culture; ST, P. globosa ST strain co-culture. L, M, and H indicate the final bacterial cell densities of 1 × 105, 1 × 106, and 1 × 107 cells/ml, respectively.
Bacterial communities associated with field P. globosa blooms were studied for decades, while bacterial community assemblage and function associated with long-term P. globosa laboratory culture remain relatively understudied (Li et al., 2020; Rosenberg et al., 2021). Marine environmental conditions change dynamically across multiple temporal and spatial scales, thereby impacting bacterial communities (Rosenberg et al., 2021). These selective forces can drive divergent bacterial community succession in distinct natural marine environments (Jasti et al., 2005). In contrast to marine environments, laboratory culture conditions can be altered to modulate single variables including light, temperature, and the concentrations of dissolved inorganic and organic nutrients. In this study, only 26 bacterial OTUs were obtained from laboratory co-cultures, representing a simplified microbial community compared to communities in natural marine ecosystems. Many studies have shown that bacterial communities associated with P. globosa blooms are dominated by Proteobacteria and Bacteroidota (Brussaard et al., 2005; Buchan et al., 2014; Delmont et al., 2014; Wemheuer et al., 2015; Wohlbrand et al., 2017; Li et al., 2020). Moreover, Guangxi and Shantou are two distinct coastal systems, which are shaped by tides, weather, and local anthropogenic influences (Wang et al., 2013). For instance, the sea surface temperature and light intensity of Guangxi are usually higher than that of Shantou (Zhao et al., 2018), thus, primary production in Guangxi is relatively higher than Shantou over the whole year (Ma et al., 2021). These distinct local environments select for adapted P. globosa strains, which further select associated microbes. In laboratory cultures, this close phytoplankton-heterotroph association was maintained and enhanced after many generations. In this study, Proteobacteria were also the dominant bacteria phylum among all P. globosa long-term laboratory cultures, while Bacteroidota were not observed in the P. globosa ST strain co-culture. The abundances of Bacteroidota (OTU6) were positively correlated with colony abundances but were negatively correlated with solitary cell abundances by db-RDA analysis, suggesting Bacteroidota may be associated with P. globosa colonies. ST co-cultures are dominated by solitary cells in life history (Liang et al., 2020), which may not favor the attachment and growth of Bacteroidota, thus gradually lost during the P. globosa cultivation.
Since laboratory culture conditions of both strains were identical, the difference in bacterial community assemblage likely arose from selective forces imposed by differences in the two P. globosa strains. In addition, the most dominant bacterial genus, Marinobacter, as identified in this study, was not the most dominant one in previous studies which used P. globosa strains differ from the current study (Xu et al., 2021a,b; Zhu et al., 2021). These results suggest that different P. globosa strains might recruit different microbes, and stable association could be formed during the long-time laboratory co-culture (Kogure et al., 1982; Vaqué et al., 1990; Sapp et al., 2007).
In addition, db-RDA modeling was used to assess the associations of P. globosa solitary cell abundances and colony abundances as meaningful explanatory variables of bacterial community variation (Figure 7A). Bacterial communities of P. globosa GX cultures were associated with P. globosa colony abundances. In contrast, bacterial communities of P. globosa ST cultures differed from GX co-cultures and were associated with P. globosa solitary cell abundances. Thus, bacterial communities are impacted by P. globosa growth dynamics and strain-specific characteristics. These results are consistent with previous studies indicating that bacterial community composition is selected by characteristics of microalgal growth, but also by characteristics of different microalgal strains (Kogure et al., 1982; Vaqué et al., 1990; Sapp et al., 2007).
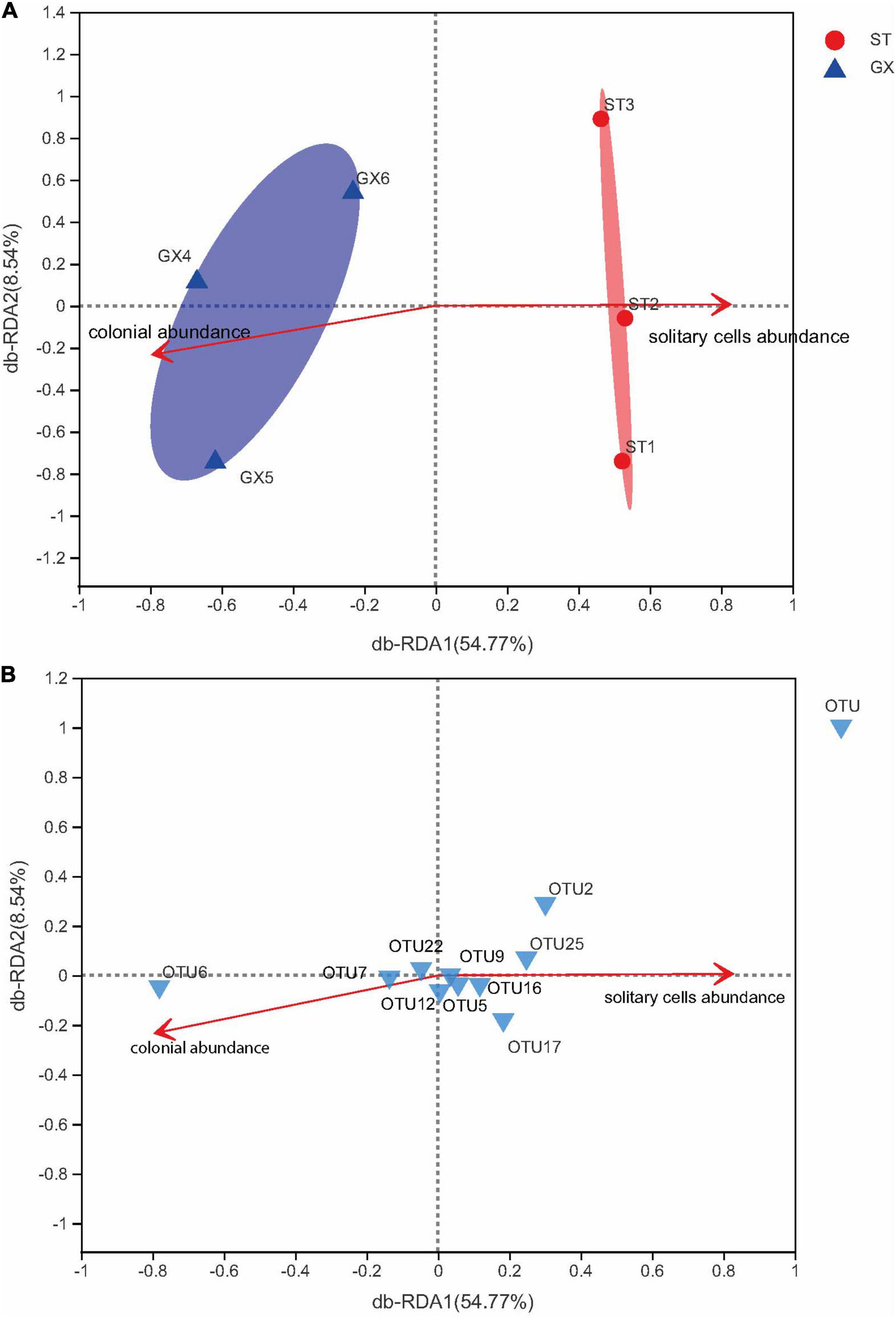
Figure 7. Distance-based redundancy analysis (db-RDA) ordination plots of bacterial community variation at the OTU level. (A) The relationships of Phaeocystis globosa growth indices and community composition. The relationships of P. globosa growth indices and the 10 most abundant OTUs (B). GX, P. globosa GX strain co-culture; ST, P. globosa ST strain co-culture.
In the present study, the potential functions of bacteria associated with P. globosa were evaluated using the FAPROTAX software. Chemoheterotrophy was the dominant predicted metabolic lifestyle among P. globosa co-culture bacteria, mostly contributed by Marinobacter. Likewise, db-RDA (Figure 7B) indicated that bacterial community composition (considering the 10 most abundant OTUs) varied with P. globosa growth. Specifically, the abundances of Marinobacter (OTU2) were positively correlated with solitary cell abundances but were negatively correlated with colony abundances, suggesting Marinobacter is associated with P. globosa growth.
Linear discriminant analysis Effect Size analysis indicated that the P. globosa GX and ST co-cultures were associated with different biomarkers. Interestingly, both P. globosa strains were associated with diverse N2-fixing bacteria, with five N2-fixing OTUs identified as biomarkers in the ST cultures, and two N2-fixing OTUs in the GX strain. These bacteria included Planctomycetota that encode nifD and nifH genes (Delmont et al., 2018) and Alphaproteobacteria (order Rhizobiales) that include N2-fixing rhizobia (Remigi et al., 2016; Zhang et al., 2021). Besides the N2-fixing bacterial biomarkers (Rhizobiales), P. globosa GX co-culture also harbored an OTU affiliated with Sulfitobacter which was not absent in the P. globosa ST co-culture. Previous studies Hypothesized that N2-fixing bacteria can fix nitrogen, which might release the nitrogen limitation of algae (Ferro et al., 2019; Tait et al., 2019). In addition, studies also showed that Sulfitobacter could enhance the growth of Pseudo-Nitzschia multiseries via secretion of the hormone indole-3-acetic acid (Amin et al., 2015; Seymour et al., 2017). Thus, long-term cultivation of P. globosa leads to the selection of bacteria that could be beneficial for the dominant microalgae.
A previous study also indicated that Marinobacter might be a potential bacterial bioindicator of P. globosa blooms (Li et al., 2020). Marinobacter exhibits high metabolic diversity, that can use a variety of carbon sources, and participate in important biogeochemical cycling processes (Bonis and Gralnick, 2015; Wang et al., 2015; Wemheuer et al., 2015). Besides, Marinobacter also plays a major role as a dominant Fe (II)-oxidizer in different environments (Bonis and Gralnick, 2015). Marinobacter can be found in single algal laboratory cultures, but also coexist with microalgae (Alavi et al., 2001; Hold et al., 2001b; Lupette et al., 2016).
An association of P. globosa solitary cell numbers and colony abundances was observed with Marinobacter sp. GS7 when grown in co-culture. Interestingly, co-culture with low and medium cell densities of Marinobacter did not lead to significant changes in P. globosa solitary cell and colony abundances, while high cell densities of Marinobacter sp. GS7 significantly impaired the growth and colony formation of P. globosa. The inhibition effect on P. globosa growth might be related to the physiological change of Marinobacter cells at high cell densities compared to low cell densities, such as chemotaxis, cell motility, and attachment (Sonnenschein Eva et al., 2012), or extracellular electron transfer (Eddie et al., 2021), or discharging of chemical mediators such as antibacterial and algicidal compounds (Imai et al., 1993; Mayali and Azam, 2004; Meyer et al., 2017; Cirri and Pohnert, 2019). Lifestyle change of Alteromonas, another opportunitroph frequently associated with marine phytoplankton, has been observed when co-culturing with Trichodesmium (Hou et al., 2018). Alteromonas cell motility and cellular activities were tightly regulated and coupled with the physiology of phytoplankton. Here the density-dependent inhibitory effect of Marinobacter on P. globosa growth might indicate a transition of their relationship from mutualism/commensalism to competition. When Marinobacter cell density was low, P. globosa exudates facilitated the heterotrophic growth of Marinobacter as important carbon sources (Fouilland et al., 2014). In return, Marinobacter may produce growth factors or secrete siderophores to promote the growth of associated phytoplankton (Töpel et al., 2019). At high cell densities, Marinobacter cells may compete with P. globosa for resources or compete with each other for attachment to phytoplankton, thus preventing P. globosa growth and colony formation. Previous studies reported that many heterotrophic bacteria could secrete chemicals inhibiting phytoplankton growth (Gallacher et al., 1997; Hold et al., 2001a; Kim et al., 2008; Yang et al., 2014). Some of the algicidal compounds are concentration dependent (Paul and Pohnert, 2011; Tan et al., 2016), which are likely to release at high bacterial cell densities (Mayali and Doucette, 2002; Roth et al., 2008) for competing nutrients (Meyer et al., 2017). Thus, it is possible that at high cell densities, algicidal compounds or other toxic substances produced by Marinobacter inhibited the growth and colony formation of P. globosa. Although we could not determine the exact chemicals in this study, it represents an interesting avenue for future research.
In this study, bacterial communities associated with two P. globosa strains and their influence on colony formation were evaluated. The P. globosa GX strain cultures showed higher bacterial community richness than the ST strain cultures. Overall community compositions and biomarker bacteria were also different between the two P. globosa strains, suggesting a strain-specific epibiont association. Co-culture experiments with P. globosa and Marinobacter sp. GS7 revealed that P. globosa formed colonies in the presence of low and medium cell densities of Marinobacter, but high cell densities of Marinobacter severely inhibited colony formation. In summary, these results support the hypothesis that bacteria communities associated with P. globosa strains are strain-specifically selected, and associated bacterioplankton could play key roles in P. globosa colony formation processes.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
SX, XW, Z-HW, and YW designed the experiment and prepared the manuscript. JL and FZ prepared the sampling and co-culture experiments. SX, KG, and SC conducted the sequencing and bioinformatics analyses. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (Nos. 41976082 and 42076141), Science & Technology Basic Resources Investigation Program of China (No. 2018FY100200), and the Strategic Priority Research Program of Chinese Academy of Sciences, grant no. XDB42000000.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank all people involved in this study. We also thank the funding program from the National Natural Science Foundation of China and Science & Technology Basic Resources Investigation Program of China.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.826602/full#supplementary-material
Alavi, M., Miller, T., Erlandson, K., Schneider, R., and Belas, R. (2001). Bacterial community associated with Pfiesteria-like dinoflagellate cultures. Environ. Microbiol. 3, 380–396. doi: 10.1046/j.1462-2920.2001.00207.x
Alderkamp, A.-C., Sintes, E., and Herndl, G. J. (2006). Abundance and activity of major groups of prokaryotic plankton in the coastal North Sea during spring and summer. Aquat. Microb. Ecol. 45, 237–246. doi: 10.3354/ame045237
Amin, S. A., Hmelo, L. R., Van Tol, H. M., Durham, B. P., Carlson, L. T., Heal, K. R., et al. (2015). Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 522, 98–101. doi: 10.1038/nature14488
Bonis, B. M., and Gralnick, J. A. (2015). Marinobacter subterrani, a genetically tractable neutrophilic Fe(II)-oxidizing strain isolated from the Soudan Iron Mine. Front. Microbiol. 6:719. doi: 10.3389/fmicb.2015.00719
Brussaard, C. P. D., Mari, X., Bleijswijk, J. D. L. V., and Veldhuis, M. J. W. (2005). A mesocosm study of Phaeocystis globosa (Prymnesiophyceae) population dynamics. Harmful Algae 4, 875–893. doi: 10.1016/j.hal.2004.12.012
Buchan, A., Lecleir, G. R., Gulvik, C. A., and Gonzalez, J. M. (2014). Master recyclers: features and functions of bacteria associated with phytoplankton blooms. Nat. Rev. Microbiol. 12, 686–698. doi: 10.1038/nrmicro3326
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Cirri, E., and Pohnert, G. (2019). Algae-bacteria interactions that balance the planktonic microbiome. New Phytol. 223, 100–106. doi: 10.1111/nph.15765
Delmont, T. O., Hammar, K. M., Ducklow, H. W., Yager, P. L., and Post, A. F. (2014). Phaeocystis antarctica blooms strongly influence bacterial community structures in the Amundsen Sea polynya. Front. Microbiol. 5:646. doi: 10.3389/fmicb.2014.00646
Delmont, T. O., Quince, C., Shaiber, A., Esen, O. C., Lee, S. T., Rappe, M. S., et al. (2018). Nitrogen-fixing populations of Planctomycetes and Proteobacteria are abundant in surface ocean metagenomes. Nat. Microbiol. 3, 804–813. doi: 10.1038/s41564-018-0176-9
Dutz, J., and Koski, M. (2006). Trophic significance of solitary cells of the prymnesiophyte Phaeocystis globosa depends on cell type. Limnol. Oceanogr. 51, 1230–1238. doi: 10.4319/lo.2006.51.3.1230
Eddie, B. J., Malanoski, A. P., Onderko, E. L., Phillips, D. A., and Glaven, S. M. (2021). Marinobacter atlanticus electrode biofilms differentially regulate gene expression depending on electrode potential and lifestyle. Biofilm 3:100051. doi: 10.1016/j.bioflm.2021.100051
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Fouilland, E., Tolosa, I., Bonnet, D., Bouvier, C., Bouvier, T., Bouvy, M., et al. (2014). Bacterial carbon dependence on freshly produced phytoplankton exudates under different nutrient availability and grazing pressure conditions in coastal marine waters. FEMS Microbiol. Ecol. 87, 757–769. doi: 10.1111/1574-6941.12262
Frank, J. A., Reich, C. I., Sharma, S., Weisbaum, J. S., Wilson, B. A., and Olsen, G. J. (2008). Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 74, 2461–2470. doi: 10.1128/AEM.02272-07
Gallacher, S., Flynn, K. J., Franco, J. M., Brueggemann, E., and Hines, H. (1997). Evidence for production of paralytic shellfish toxins by bacteria associated with Alexandrium spp.(Dinophyta) in culture. Appl. Environ. Microbiol. 63, 239–245. doi: 10.1128/aem.63.1.239-245.1997
Hamm, C. E. (2000). Architecture, ecology and biogeochemistry of Phaeocystis colonies. J. Sea Res. 43, 307–315. doi: 10.1016/S1385-1101(00)00014-9
Hamm, C. E., Simson, D. A., Merkel, R., and Smetacek, V. (1999). Colonies of Phaeocystis globosa are protected by a thin but tough skin. Mar. Ecol. Prog. Ser. 187, 101–111. doi: 10.3354/meps187101
Hold, G. L., Smith, E. A., Rappé, M. S., Maas, E. W., Moore, E. R. B., Stroempl, C., et al. (2001b). Characterisation of bacterial communities associated with toxic and non-toxic dinoflagellates: Alexandrium spp. and Scrippsiella trochoidea. FEMS Microbiol. Ecol. 37, 161–173. doi: 10.1111/j.1574-6941.2001.tb00864.x
Hold, G. L., Smith, E. A., Birkbeck, T. H., and Gallacher, S. (2001a). Comparison of paralytic shellfish toxin (PST) production by the dinoflagellates Alexandrium lusitanicum NEPCC 253 and Alexandrium tamarense NEPCC 407 in the presence and absence of bacteria. FEMS Microbiol. Ecol. 36, 223–234. doi: 10.1111/j.1574-6941.2001.tb00843.x
Hou, S., López-Pérez, M., Pfreundt, U., Belkin, N., Stüber, K., Huettel, B., et al. (2018). Benefit from decline: the primary transcriptome of Alteromonas macleodii str. Te101 during Trichodesmium demise. ISME J. 12, 981–996. doi: 10.1038/s41396-017-0034-4
Imai, I., Ishida, Y., and Hata, Y. (1993). Killing of marine phytoplankton by a gliding bacterium Cytophaga sp., isolated from the coastal sea of Japan. Mar. Biol. 116, 527–532. doi: 10.1007/bf00355470
Jakobsen, H. H., and Tang, K. W. (2002). Effects of protozoan grazing on colony formation in Phaeocystis globosa (Prymnesiophyceae) and the potential costs and benefits. Aquat. Microb. Ecol. 27, 261–273. doi: 10.3354/ame027261
Jari Oksanen, F. G. B., Michael Friendly, Roeland Kindt, Pierre Legendre, D. M., Minchin, P. R., O’hara, R. B., et al. (2019). Vegan: Community Ecology Package. R Package Version 2.5-5. Available online at: https://CRAN.R-project.org/package=vegan (accessed November 28, 2020).
Jasti, S., Sieracki, M. E., Poulton, N. J., Giewat, M. W., and Rooney-Varga, J. N. (2005). Phylogenetic diversity and specificity of bacteria closely associated with Alexandrium spp. and other phytoplankton. Appl. Environ. Microbiol. 71, 3483–3494. doi: 10.1128/AEM.71.7.3483-3494.2005
Kim, M.-J., Jeong, S.-Y., and Lee, S.-J. (2008). Isolation, identification, and algicidal activity of marine bacteria against Cochlodinium polykrikoides. J. Appl. Phycol. 20, 1069–1078. doi: 10.1007/s10811-008-9312-x
Kogure, K., Simidu, U., and Taga, N. (1982). Bacterial attachment to phytoplankton in sea water. J. Exp. Mar. Biol. Ecol. 56, 197–204.
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Li, N., Zhao, H., Jiang, G., Xu, Q., Tang, J., Li, X., et al. (2020). Phylogenetic responses of marine free-living bacterial community to Phaeocystis globosa bloom in beibu gulf, China. Front. Microbiol. 11:1624. doi: 10.3389/fmicb.2020.01624
Liang, D., Wang, X., Huo, Y., Wang, Y., and Li, S. (2020). Differences in the formation mechanism of giant colonies in two Phaeocystis globosa strains. Int. J. Mol. Sci. 21:5393. doi: 10.3390/ijms21155393
Louca, S., Parfrey, L. W., and Doebeli, M. (2016). Decoupling function and taxonomy in the global ocean microbiome. Science 353, 1272–1277. doi: 10.1126/science.aaf4507
Lupette, J., Lami, R., Krasovec, M., Grimsley, N., Moreau, H., Piganeau, G., et al. (2016). Marinobacter dominates the bacterial community of the Ostreococcus tauri phycosphere in culture. Front. Microbiol. 7:1414. doi: 10.3389/fmicb.2016.01414
Ma, C., Zhao, J., Ai, B., and Sun, S. (2021). Two-decade variability of sea surface temperature and chlorophyll-a in the Northern South China sea as revealed by reconstructed cloud-free satellite data. IEEE Trans. Geosci. Remote Sens. 59, 9033–9046. doi: 10.1109/TGRS.2021.3051025
Magoc, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Mayali, X., and Azam, F. (2004). Algicidal bacteria in the sea and their impact on algal blooms. J. Eukaryot. Microbiol. 51, 139–144. doi: 10.1111/j.1550-7408.2004.tb00538.x
Mayali, X., and Doucette, G. J. (2002). Microbial community interactions and population dynamics of an algicidal bacterium active against Karenia brevis (Dinophyceae). Harmful Algae 1, 277–293. doi: 10.1016/S1568-9883(02)00032-X
Meyer, N., Bigalke, A., Kaulfuß, A., and Pohnert, G. (2017). Strategies and ecological roles of algicidal bacteria. FEMS Microbiol. Rev. 41, 880–899. doi: 10.1093/femsre/fux029
Ferro, L., Colombo, M., Posadas, E., Funk, C., and Muñoz, R. (2019). Elucidating the symbiotic interactions between a locally isolated microalga Chlorella vulgaris and its co-occurring bacterium Rhizobium sp. in synthetic municipal wastewater. J. Appl. Phycol. 31, 2299–2310. doi: 10.1007/s10811-019-1741-1
Paul, C., and Pohnert, G. (2011). Interactions of the algicidal bacterium kordia algicida with diatoms: regulated protease excretion for specific algal lysis. PLoS One 6:e21032. doi: 10.1371/journal.pone.0021032
Qi, Y., Chen, J., Wang, Z., Xu, N., Wang, Y., Shen, P., et al. (2004). Some observations on harmful algal bloom (HAB) events along the coast of Guangdong, southern China in 1998. Hydrobiologia 512, 209–214. doi: 10.1023/B:HYDR.0000020329.06666.8c
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Remigi, P., Zhu, J., Young, J. P. W., and Masson-Boivin, C. (2016). Symbiosis within symbiosis: evolving nitrogen-fixing legume symbionts. Trends Microbiol. 24, 63–75. doi: 10.1016/j.tim.2015.10.007
Rosenberg, D. R., Haber, M., Goldford, J., Lalzar, M., Aharonovich, D., Al-Ashhab, A., et al. (2021). Particle-associated and free-living bacterial communities in an oligotrophic sea are affected by different environmental factors. Environ. Microbiol. 23, 4295–4308. doi: 10.1111/1462-2920.15611
Roth, P. B., Twiner, M. J., Mikulski, C. M., Barnhorst, A. B., and Doucette, G. J. (2008). Comparative analysis of two algicidal bacteria active against the red tide dinoflagellate Karenia brevis. Harmful Algae 7, 682–691. doi: 10.1016/j.hal.2008.02.002
Rousseau, V., Vaulot, D., Casotti, R., Cariou, V., Lenz, J., Gunkel, J., et al. (1994). The life cycle of Phaeocystis (Prymnesiophycaea): evidence and hypotheses. J. Mar. Syst. 5, 23–39. doi: 10.1016/0924-7963(94)90014-0
Sapp, M., Schwaderer, A. S., Wiltshire, K. H., Hoppe, H. G., Gerdts, G., and Wichels, A. (2007). Species-specific bacterial communities in the phycosphere of microalgae? Microb. Ecol. 53, 683–699. doi: 10.1007/s00248-006-9162-5
Schoemann, V., Becquevort, S., Stefels, J., Rousseau, V., and Lancelot, C. (2005). Phaeocystis blooms in the global ocean and their controlling mechanisms: a review. J. Sea Res. 53, 43–66. doi: 10.1016/j.seares.2004.01.008
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60
Seymour, J. R., Amin, S. A., Raina, J. B., and Stocker, R. (2017). Zooming in on the phycosphere: the ecological interface for phytoplankton-bacteria relationships. Nat. Microbiol. 2:17065. doi: 10.1038/nmicrobiol.2017.65
Smith, W. O., Liu, X., Tang, K. W., Delizo, L. M., Doan, N. H., Nguyen, N. L., et al. (2014). Giantism and its role in the harmful algal bloom species Phaeocystis globosa. Deep Sea Res. II Top. Stud. Oceanogr. 101, 95–106. doi: 10.1016/j.dsr2.2012.12.005
Sonnenschein Eva, C., Syit Desalegne, A., Grossart, H.-P., and Ullrich Matthias, S. (2012). Chemotaxis of Marinobacter adhaerens and its impact on attachment to the diatom Thalassiosira weissflogii. Appl. Environ. Microbiol. 78, 6900–6907. doi: 10.1128/AEM.01790-12
Tait, K., White, D. A., Kimmance, S. A., Tarran, G. A., Rooks, P. A., Jones, M., et al. (2019). Characterisation of bacteria from the cultures of a Chlorella strain isolated from textile wastewater and their growth enhancing effects on the axenic cultures of Chlorella vulgaris in low nutrient media. Algal Res. 44: 101666.
Tan, S., Hu, X., Yin, P., and Zhao, L. (2016). Photosynthetic inhibition and oxidative stress to the toxic Phaeocystis globosa caused by a diketopiperazine isolated from products of algicidal bacterium metabolism. J. Microbiol. 54, 364–375. doi: 10.1007/s12275-016-6012-0
Tang, K. W., Jakobsen, H. H., and Visser, A. W. (2001). Phaeocystis globosa (Prymnesiophyceae) and the planktonic food web: feeding, growth, and trophic interactions among grazers. Limnol. Oceanogr. 46, 1860–1870. doi: 10.4319/lo.2001.46.8.1860
Töpel, M., Pinder, M. I. M., Johansson, O. N., Kourtchenko, O., Godhe, A., and Clarke, A. K. (2019). Whole genome sequence of Marinobacter salarius strain SMR5, shown to promote growth in its diatom host. J. Genom. 7, 60–63. doi: 10.7150/jgen.39039
Vaqué, D., Duarte, C. M., and Marrasé, C. (1990). Influence of algal population dynamics on phytoplankton colonization by bacteria: evidence from two diatom species. Mar. Ecol. Prog. Ser. 65, 201–203.
Verity, P. G., Brussaard, C. P., Nejstgaard, J. C., Van Leeuwe, M. A., Lancelot, C., and Medlin, L. K. (2007). Current understanding of Phaeocystis ecology and biogeochemistry, and perspectives for future research. Biogeochemistry 83, 311–330. doi: 10.1007/s10533-007-9090-6
Wang, X., Tang, K. W., Wang, Y., and Smith, W. O. (2010). Temperature effects on growth, colony development and carbon partitioning in three Phaeocystis species. Aquat. Biol. 9, 239–249. doi: 10.3354/ab00256
Wang, X., Zheng, J., and Wang, Y. (2014). Effects of grazing on the colony formation in Phaeocystis globosa. J. Trop. Ecol. 5, 20–24. doi: 10.15886/j.cnki.rdswxb.2014.01.015
Wang, Y., Zhang, W., Lin, Y., Zheng, L., Cao, W., and Yang, J. (2013). Spatial pattern of the planktonic ciliate community and its relationship with the environment in spring in the northern Beibu Gulf, South China Sea. Oceanol. Hydrobiol. Stud. 42, 470–479. doi: 10.2478/s13545-013-0103-x
Wang, Z., Leary, D. H., Malanoski, A. P., Li, R. W., Hervey, W. J. T., Eddie, B. J., et al. (2015). A previously uncharacterized, nonphotosynthetic member of the Chromatiaceae is the primary CO2-fixing constituent in a self-regenerating biocathode. Appl. Environ. Microbiol. 81, 699–712. doi: 10.1128/AEM.02947-14
Wemheuer, B., Gullert, S., Billerbeck, S., Giebel, H. A., Voget, S., Simon, M., et al. (2014). Impact of a phytoplankton bloom on the diversity of the active bacterial community in the southern North Sea as revealed by metatranscriptomic approaches. FEMS Microbiol. Ecol. 87, 378–389. doi: 10.1111/1574-6941.12230
Wemheuer, B., Wemheuer, F., Hollensteiner, J., Meyer, F. D., Voget, S., and Daniel, R. (2015). The green impact: bacterioplankton response toward a phytoplankton spring bloom in the southern North Sea assessed by comparative metagenomic and metatranscriptomic approaches. Front. Microbiol. 6:805. doi: 10.3389/fmicb.2015.00805
Wickham, H. (2017). ggplot2 – Elegant Graphics for Data Analysis (2nd Edition). J. Stat. Softw. 77, 1–3. doi: 10.18637/jss.v077.b02
Wohlbrand, L., Wemheuer, B., Feenders, C., Ruppersberg, H. S., Hinrichs, C., Blasius, B., et al. (2017). Complementary metaproteomic approaches to assess the bacterioplankton response toward a phytoplankton spring bloom in the Southern North Sea. Front. Microbiol. 8:442. doi: 10.3389/fmicb.2017.00442
Xu, N., Tan, G., Wang, H., and Gai, X. (2016). Effect of biochar additions to soil on nitrogen leaching, microbial biomass and bacterial community structure. Eur. J. Soil Biol. 74, 1–8. doi: 10.1016/j.ejsobi.2016.02.004
Xu, S., He, C., Song, S., and Li, C. (2021a). Spatiotemporal dynamics of marine microbial communities following a Phaeocystis bloom: biogeography and co-occurrence patterns. Environ. Microbiol. Rep. 13, 294–308. doi: 10.1111/1758-2229.12929
Xu, S., Li, A., Zhang, M.-X., Yao, Q., and Zhu, H. (2021b). Lysobacter penaei sp. nov., isolated from intestinal content of a Pacific white shrimp (Penaeus vannamei). Int. J. Syst. Evol. Microbiol. 71:004593. doi: 10.1099/ijsem.0.004593
Yang, Q., Feng, Q., Zhang, B. P., Gao, J. J., Sheng, Z., Xue, Q. P., et al. (2021). Marinobacter alexandrii sp. nov., a novel yellow-pigmented and algae growth-promoting bacterium isolated from marine phycosphere microbiota. Antonie Van Leeuwenhoek 114, 709–718. doi: 10.1007/s10482-021-01551-5
Yang, X., Li, X., Zhou, Y., Zheng, W., Yu, C., and Zheng, T. (2014). Novel insights into the algicidal bacterium DH77-1 killing the toxic dinoflagellate Alexandrium tamarense. Sci. Total Environ. 482-483, 116–124. doi: 10.1016/j.scitotenv.2014.02.125
Zhang, X., Yu, J., Huang, Z., Li, H., Liu, X., Huang, J., et al. (2021). Enhanced Cd phytostabilization and rhizosphere bacterial diversity of Robinia pseudoacacia L. by endophyte Enterobacter sp. YG-14 combined with sludge biochar. Sci. Total Environ. 787:147660. doi: 10.1016/j.scitotenv.2021.147660
Zhao, Y., Peng, W., Guo, H., Chen, B., Zhou, Z., Xu, J., et al. (2018). Population genomics reveals genetic divergence and adaptive differentiation of Chinese Sea Bass (Lateolabrax maculatus). Mar. Biotechnol. 20, 45–59. doi: 10.1007/s10126-017-9786-0
Keywords: Phaeocystis globosa, associated bacteria, colony formation, Marinobacter, composition
Citation: Xu S, Wang X, Liu J, Zhou F, Guo K, Chen S, Wang Z-h and Wang Y (2022) Bacteria Associated With Phaeocystis globosa and Their Influence on Colony Formation. Front. Microbiol. 13:826602. doi: 10.3389/fmicb.2022.826602
Received: 01 December 2021; Accepted: 04 January 2022;
Published: 17 February 2022.
Edited by:
Yuanyuan Feng, Shanghai Jiao Tong University, ChinaReviewed by:
Qiang Zhang, Virginia Tech, United StatesCopyright © 2022 Xu, Wang, Liu, Zhou, Guo, Chen, Wang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhao-hui Wang, dHd6aEBqbnUuZWR1LmNu; Yan Wang, d2FuZ3lhbkBqbnUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.