- 1State Key Laboratory of North China Crop Improvement and Regulation, Hebei Agricultural University, Baoding, China
- 2State Key Laboratory of Soil Erosion and Dryland Farming on the Loess Plateau, Institute of Soil and Water Conservation, Northwest A&F University, Xianyang, China
- 3College of Agronomy and Biotechnology, Hebei Normal University of Science and Technology, Qinhuangdao, China
Increased complexity of microbial networks can contribute to increased biodiversity and multifunctionality and thus crop productivity. However, it is not clear which combination ratio of regular and controlled-release urea will increase the soil microbial community complexity and improve maize yield in the North China Plain. To address this knowledge gap, a 2-year field experiment was conducted to explore the effects of the combination of regular (U) and controlled release (S) urea ratios [no fertilizer control (CT), regular urea alone (U), controlled-release urea alone (S), controlled-release urea mixed with regular urea 3:7 (SU3), controlled-release urea mixed with regular urea 5:5 (SU5), and controlled-release urea mixed with regular urea 7:3 (SU7)] on XianYu 688 yield and its rhizosphere and bulk soil microbial community composition and network complexity at different fertility stages. The combination of controlled-release and regular urea increased the N agronomic efficiency, N partial factors productivity, maize yield, and grain number per spike, with the maximum maize yield (9,186 kg ha–1) being achieved when the ratio of controlled-release urea to regular urea was 3:7 (SU3, p < 0.05). Maize yield increased by 13% in the SU3 treatment compared to the CT treatment. Rhizosphere soil microbial diversity remained stable at the silking stage of maize while increased at the physiological maturity stage of maize, with the increasing controlled-release to regular N fertilizer ratios (from 3:7 to 7:3, p < 0.05). This result suggests that a combination of regular and controlled-release N fertilizer can still substantially increase soil microbial diversity in the later stages of maize growth. The combination of controlled-release and regular urea is more effective in improving microbial network total links and average degree, and N agronomic efficiency (R2 = 0.79, p < 0.01), N partial factor productivity (R2 = 0.79, p < 0.01), spikes per unit area (R2 = 0.54, p < 0.05), and maize yield (R2 = 0.42, p < 0.05) increased with the microbial network complexity. This result indicates that the higher microbial network complexity is strongly associated with the higher N agronomic efficiency and N partial factors productivity and maize yield. In conclusion, the ratio of controlled-release to regular urea at SU3 not only increases the yield of maize and N agronomic efficiency but also enhances microbial diversity and network complexity in the North China Plain.
Introduction
As important decomposers of ecosystems, soil microorganisms are responsible for more than 90% of the decomposition of organic matter in ecosystems (Swift et al., 1979). They are involved in essential ecosystem functions (such as soil organic carbon storage, nutrient and hydrological cycling, decomposition, and terrestrial primary production) that are closely linked to global food supply and ecosystem services (Fan et al., 2020a; Jiao et al., 2021; Wang et al., 2021). For instance, soil microorganisms can mineralize mineral nutrients by releasing extracellular enzymes (Mooshammer et al., 2014), and after the microorganism dies, its cytosol enters the soil solution where it can be absorbed and used by plants to meet their vital activities (Yadav et al., 2021). Alternatively, plants can mineralize soil nutrients through the production of organic acids by their roots or induce the proliferation of microorganisms through root exudates (Jilling et al., 2021), thereby facilitating the mineralization of nutrients. In addition to their function of providing and mineralizing nutrients, soil microorganisms can also protect plants through a number of mechanisms (Yadav et al., 2021). For example, microorganisms produce glycosides that chelate iron in the soil and limit the uptake of this trace element by pathogens, hydrogen cyanide, antibiotics, hydrolytic enzymes, and the 1-aminocyclopropane-1-carboxylic acid deaminase exhibited by some specific microorganisms (Devi et al., 2020; Yadav et al., 2021). Therefore, soil microbial-plant as a system of interconnections, interactions and feedback, which in agricultural systems can improving sustainability and productivity (Mariotte et al., 2018; Sun Y. et al., 2021; Thakur et al., 2021).
In the agroecosystems, soil microbial communities coexist in multitrophic microbial food webs, which directly or indirectly affect crop health and yield (Nihorimbere et al., 2011; Fan et al., 2020b; Yu et al., 2021). Especially in the rhizosphere (root-soil interface), the zone of soil surrounding plant roots represents a hotspot of microbial and biochemical activity (York et al., 2016; Kuzyakov and Razavi, 2019; Li R. et al., 2021). Soil microorganisms are structured and form complex food webs, reflecting strong interrelationships within ecological networks, which is fundamental for characterizing species interactions and dynamics of ecosystems (Fan et al., 2021; Yuan et al., 2021). The high microbial diversity and network complexity can contribute to increased crop yields due to the important role of rhizosphere and bulk soil microorganisms in the mineralization of soil nutrients and the provision of nutrients for crop growth (Fan et al., 2020b; Li B. B. et al., 2021). However, soil microbial communities are very sensitive to soil disturbance and changes in the external environment (Wilkinson et al., 2002; Schneider et al., 2012), and changes in their composition and structure affect soil microbial enzyme secretion, respiratory metabolism, and other functions, which in turn affect soil nutrient cycling processes (Ren et al., 2021; Tekaya et al., 2021).
Numerous experiments have confirmed that fertilization management could alter the chemical properties of the soil solution, activate the microbial community, and significantly impact the microbial community structure and its complexity (Geisseler and Scow, 2014; Liu et al., 2019; Xiong et al., 2019; Fan et al., 2020a; Ji et al., 2021; Li B. B. et al., 2021; Zhang et al., 2021). For example, compared with individual applications, the combined inorganic fertilizers and cow manure led to the most resistant microbial community, which was associated with the highest levels of nutrient availability and plant production (Fan et al., 2020a). Ji et al. (2021) found that the high organic fertilizer treatments reduced the number of links in the microbial network at the genus level. Moreover, the network of organic fertilizer treatment soils contained more functionally interrelated microbial modules than soils with chemical fertilizer treatment, and the topological roles of characteristic microorganisms and key microbial organisms were significantly different between these two treatments (Gu et al., 2019). Li B. B. et al. (2021) found that a blend of regular and controlled-release urea increased the relative abundance of Penicillium and Aspergillus fungal genera and reduced the operational taxonomic unit (OTU) of nitrifying bacteria of the genus Nitrospira compared to regular urea treatment. The bacterial and fungi α diversity in a blend of regular and controlled-release urea treatment was lower than the regular urea treatment (Li R. et al., 2021). Lupwayi et al. (2010) found that the controlled-release urea increased microbial biomass carbon (MBC) or functional diversity more than urea in 3 site-years, but the opposite was observed in 1 site-year. This result shows that the effect of controlled-release urea and normal urea on microbial communities is influenced by the age of application of the fertilizer. Thereby, the effect of fertilizer application on microbial communities and their complexity is influenced by the type of fertilizer and its rate. Understanding the response of microbial diversity and community structural complexity to shifts in controlled-release and regular urea rates is urgent to our understanding of microbially mediated nutrient cycling processes in agroecosystems. However, there is a lack of understanding of how controlled-release urea combined with regular urea affects rhizosphere and bulk soil microbial diversity, community composition, and network complexity. The study of the mechanisms by which the diversity, stability, and complexity of soil microbial communities affect crop yield can provide a scientific basis for sustainable agricultural development.
Nitrogen (N) fertilizer (e.g., urea) has often been generously applied to increase crop biomass yield, which plays an important role in crop growth and enhances crop yield (Guo et al., 2021; Li R. et al., 2021). Although urea plays an important role in increasing crop yields, its rapid rate of decomposition means that full basal applications of N can easily cause N loss and increase the risk of environmental pollution, resulting in early crop failure and yield loss (Geisseler and Scow, 2014). However, there are difficulties in applying N fertilizer in the middle and late stages of crop life, particularly in some crops such as maize. Controlled-release urea is a new type of fertilizer that provides N to plants slowly, controls and regulates the release of N, and meets the nutritional requirements of crops (Grant et al., 2012). Compared with regular urea, controlled-release urea can reduce the damage to the crop root system in the pre-fertility stage, extend the nutrient absorption and utilization validity period, and has the advantages of N setback, improving the efficiency of N fertilizer utilization, reducing the number of manual fertilizer follow-ups, and improving economic efficiency (Blackshaw et al., 2011; Guo et al., 2017). Consequently, a blend of regular and controlled-release urea can address both the rapid loss of N fertilizer and the N requirements for later maize growth and development (Zheng et al., 2016; Zhang et al., 2021). For instance, a growing number of studies suggested that combining urea and controlled-release urea fertilizers improves maize yield and N use efficiency and reduces ammonia volatilization (Zheng et al., 2016; Zhang et al., 2019; Guo et al., 2021). However, the effect of controlled-release urea and conventional urea ratios on crop yield may vary from region to region as the effect of controlled-release urea and conventional urea ratios on crop yield is influenced by soil nutrient levels, the age of application, and microbial communities (Lupwayi et al., 2010; Li B. B. et al., 2021; Li R. et al., 2021). Currently, it is not clear what ratio of regular to controlled-release urea will promote increased maize yields and maintain healthy soil function by promoting the diversity of microbial community structure in the North China Plain. Additionally, the relationship between microbial diversity and community structural complexity and crop yield traits under changing ratios of controlled and regular urea is currently very poorly understood.
To address the above question, i.e., to characterize the effect of the ratio of regular and controlled-release urea [the treatments include no fertilizer control (CT), regular urea alone (U), controlled-release urea alone (S), controlled-release urea mixed with regular urea 3:7 (SU3), controlled-release urea mixed with regular urea 5:5 (SU5), and controlled-release urea mixed with regular urea 7:3 (SU7)] on maize yield and bacterial community structure and its network complexity, a 2-year field experiment was conducted in Xinji, Hebei, China. This study sought to reveal the mechanism of the effect of common N fertilizer and slow-acting urea application on the changes in microbial community composition and crop productivity. Therefore, the following three questions were addressed: (i) How do regular and controlled-release urea applications alone and their pairing ratios affect maize yield, spikes per unit area, grain number per spike, and hundred grains weight? (ii) How do bacterial diversity, community composition, and network complexity respond to changes in regular and controlled-release urea applications alone and their pairing ratios? (iii) What is the relationship between microbial network complexity and maize yield under N fertilizer treatments, and what ratio of regular to controlled-release urea would be more effective in improving maize yield and microbial network complexity?
Materials and Methods
Site Description and Experimental Design
The field experiment was performed in 2019 at the Experimental Station of Hebei Agricultural University, Xinji City, Hebei Province, China (43°31′N, 124°48′E). This area has a temperate semi-humid continental climate where the mean annual precipitation and mean annual temperature are 516.4 mm and 13.8°C. The soil texture at the research site is clay loam (ISSS Classification, International Soil Science Society). The basic physicochemical properties of soils are a bulk density of 1.47 g cm–3, soil organic matter of 18.47 g kg–1, total N of 1.25 g kg–1, alkali hydrolyzable N of 91.55 g kg–1, extractable Olsen phosphorus of 27.50 mg kg–1, and ammonium acetate extractable potassium of 145.79 mg kg–1, and pH of 7.8.
To study the effects of regular urea, controlled-release urea, and different ratios of regular urea to controlled-release urea on maize yield and soil microbial community structure at different maize growth stages, the maize variety XianYu 688 were selected as the test variety for a 2-year field experiment in 2019. The experimental design was a randomized group design with three field replications, in which the treatment plots were 8 m long and 3.6 m wide, and the maize seedlings were sown at a density of 675,000 plants hm–2, with 60 cm equal spacing. The fertilizer treatments included CT, U alone, S alone (the controlled-release urea was produced by Henan Xinlianxin Fertilizer Co., Ltd., with N, P2O5, and K2O contents of 43, 5, and 5%, respectively), SU3, SU5, and SU7, all at a rate of 180 kg hm–2. All were applied as a single base application before the sowing of summer maize. The same amount of phosphorus and potash was applied to each treatment, 90 kg ha–1 of P2O5 and 90 kg ha–1 of K2O. Other cultivation practices, such as removal of weeds, and chemical control of diseases and pests were performed using the recommended conventional approaches.
To investigate the effect of different N fertilizer types and their ratios on the microbial community structure of rhizosphere soil and bulk soil of maize at the silking and physiological maturity stage, the soil at different maize growth stages was collected. In each N fertilizer treatment plot, an “S”-shaped random sampling strategy with 5–6 small sampling locations was chosen. The soil samples from the 5–6 sampling points in each plot were mixed to obtain a representative sample of approximately 0.5 kg. A total of 72 representative samples [6 treatments × 2 stages × 3 replicates × 2 soil (rhizosphere soil and bulk soil) = 72 samples] were collected. After careful and thorough removal of plant tissues and residues from the soil, the collected samples were put into a foam box with an ice pack and moved to a refrigerator at −80°C within 24 h for DNA extraction.
Analysis
Soil microbial DNA was extracted from 0.5 g of well-mixed soil for each sample by grinding, freeze-thawing, and sodium dodecyl sulfate (SDS)-based cell lysis and was purified using the PowerMax Soil DNA Isolation Kit (MO BIO Laboratories, Carlsbad, CA, United States). The quality of extracted DNA was evaluated based on 260/280 nm (>1.8) and 260/230 nm (>1.7) ratios, obtained using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE, United States). The final DNA concentration was quantified using a Quant-IT Pico Green dsDNA Kit (Invitrogen Molecular Probes Inc., Oregon, United States). Although this extraction method is more labor-intensive and time-consuming, it is capable of recovering high-molecular-weight DNA with high yield and high quality from diverse representative soil samples (Yuan et al., 2021).
Primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′ and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) were used to amplify the V4 region of the bacterial 16S rRNA gene. PCR amplification and tag-encoded high-throughput sequencing of the 16S gene was performed by the Novogene Company (Beijing, China) using the Illumina HiSeq platform (PE 2500). QIIME 2.0 was used to generate raw sequence data from sequencing, and all raw reads were aligned to samples according to different barcodes (Caporaso et al., 2010). Both forward and reverse primers were trimmed. Paired-end reads of sufficient length were combined with at least 30 bp overlap into full-length sequences, and the average fragment length was 253 bp using the FLASH program (Reyon et al., 2012; Zhou et al., 2020). Unqualified sequences were filtered using the Btrim program, with a window size >20 as the threshold quality score. UPARSE was used to remove chimeric sequences and classify sequences into OTUs at a similarity of 97% (Edgar et al., 2011). All singleton OTUs were removed. Alpha diversity metrics including observed OTUs, Shannon, Simpson, Chao 1, and ACE indexes were then estimated in QIIME2.0. Bacterial representative sequences were assigned taxonomic information using the SILVA database as a reference (Pruesse et al., 2007).
Statistical Analysis
One-way ANOVA followed by the Duncan test (α = 0.05) was carried out using IBM SPSS Statistics 21 (SPSS, Chicago, IL, United States) software to test the significant differences in maize yield, spikes per unit area, grain number per spike and hundred grains weight, N agronomic efficiency, N partial factors productivity, and microbial α diversity (i.e., Shannon, Simpson, Chao1, and ACE index) at maize growth stage (silking and physiological maturity stages) under N fertilizer treatments [including CT, U, Controlled-release urea (S, and SU3, SU5, and SU7)]. Regression analysis was performed using IBM SPSS Statistics 20 software to assess the relationships among microbial network complexity maize yield, spikes per unit area, N agronomic efficiency, and N partial factors productivity under different N fertilizer treatments.
The N agronomic efficiency (Equation 1) and N partial factor productivity (Equation 2) were calculated according to methods described by Adiele et al. (2020) and Li et al. (2022):
N agronomic efficiency (kg–1) = (maize yield at Nx − maize yield at N0)/N rate (Equation 1)
N partial factors productivity (kg–1) = maize yield at Nx/N rate (Equation 2)
where the Nx is N treatment, N0 is an unfertilized plot, and N rate is the amount of fertilizer applied.
Network analysis is used to assess microbial community complexity (Zhou et al., 2020; Jiao et al., 2021). For the network analysis, relative abundance at the microbial phylum level was used to generate a correlation coefficient matrix. Then, a symmetric correlation matrix was constructed using interactions calculated between each of the two microbes using the Spearman correlation coefficient based on the R statistical environment. This correlation matrix was then transformed into a similar matrix by taking the absolute values. Networks were visualized using R statistical environment (Version 3.4.2, R Development Core Team, 2016) and Gephi 0.8.2.
Results
Changes in Maize Yield Traits Under Different N Treatment Ratios
Urea treatments significantly increased maize yield compared to the control treatment (Figure 1, p < 0.05), and the results suggested that N fertilizer application increased the crop yield in the North China Plain. However, the effect of urea application on maize yield is influenced by the ratio of controlled-release urea to regular urea. The SU3 treatment stimulates the increased maize yield compared with other N fertilizers (Figure 1, p < 0.05). In particular, compared to CT treatment, SU3 treatment increased maize yield by 13% (Figure 1, p < 0.05). This result indicates that a combination of controlled-release and regular urea is more effective in improving maize yields than the traditional regular and controlled-release urea applied separately. The results showed that the regular and controlled-release urea application increased the grain number per spike compared to the control treatment (Figure 1, p < 0.05). Additionally, the N agronomic efficiency and N partial factor productivity in SU3 treatment were nearly 2 times and 1.2 times higher than the CT treatment, respectively (Figure 1, p < 0.05). This result indicates that SU3 treatment is more effective in improving N agronomic efficiency and N partial factor productivity than CT and other treatments.
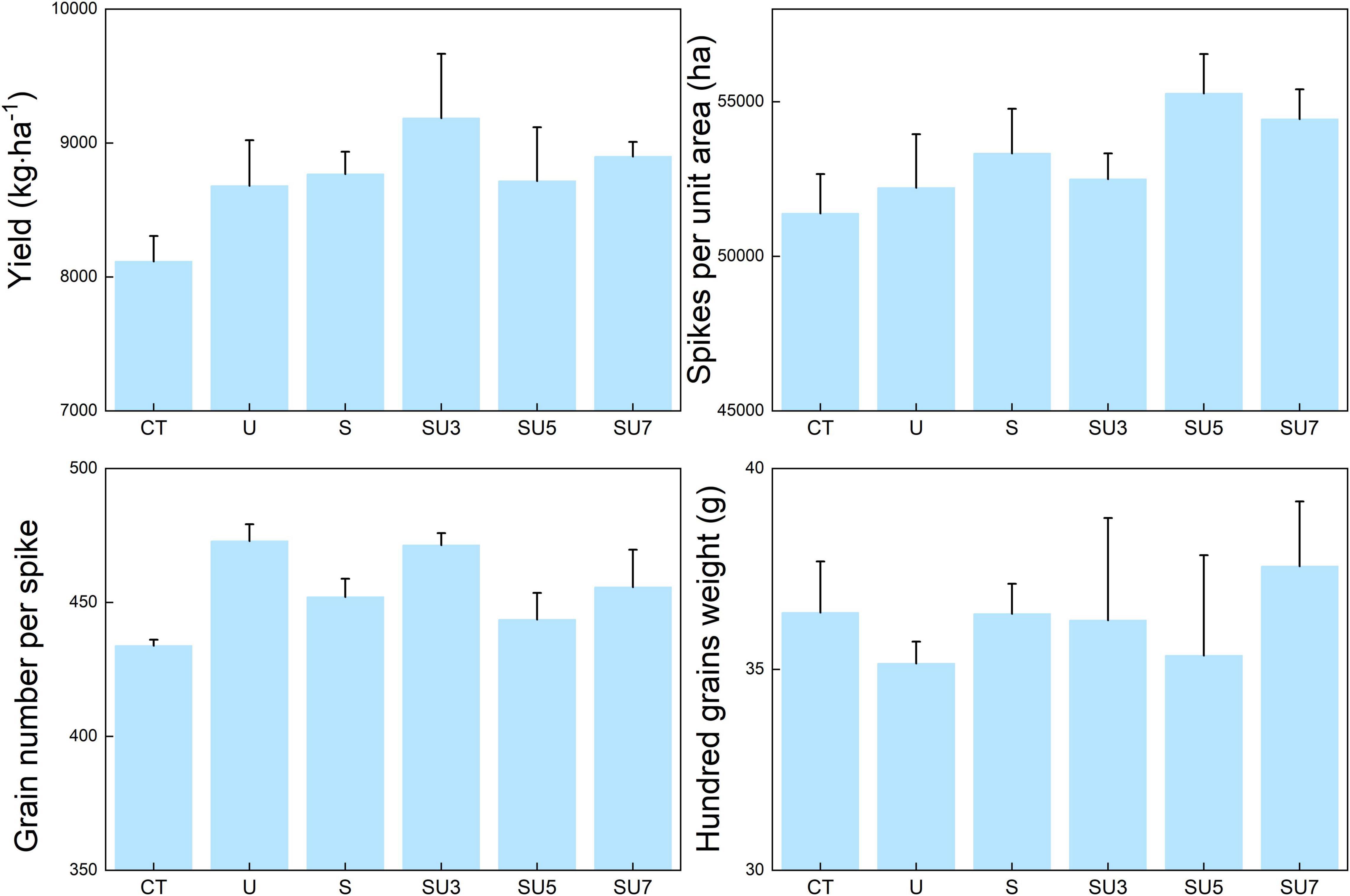
Figure 1. Effect of no fertilizer (CT), regular urea (U), controlled-release urea (S), and different ratios of controlled-release urea to regular urea (SU3, 3:7 ratio of controlled-release urea to regular urea; SU5, 5:5 ratio of controlled-release urea to regular urea; and SU7, 7:3 ratio of controlled-release urea to regular urea) on maize yield, spikes per unit area, grain number per spike and hundred grains weight, N agronomic efficiency, and N partial factors. Regular urea, controlled-release urea, and different ratios of regular urea and controlled-release urea are all applied at a rate of 180 kg ha– 1. Values are presented as the means ± standard error (SE). Lowercase and uppercase letters indicate significant differences between the nitrogen fertilizer treatment (p < 0.05).
Changes in Microbial Community Diversity and Composition Under Different N Treatments
Rhizosphere soil microbial diversity (i.e., Shannon, Simpson, Chao1, and ACE indexes) remains stable at the silking stage of maize with control and different controlled-release and regular urea ratios (Figure 2). However, results showed that the microbial diversity increased at the maize physiological maturity stage with the increasing controlled-release and regular urea ratios (controlled-release mixed with regular urea ratio increased from 3:7 to 7:3) (Figure 2, p < 0.05). This result demonstrated that a combination of controlled-release and regular urea can substantially increase soil microbial diversity in the later stages of maize growth (i.e., physiological maturity stage).
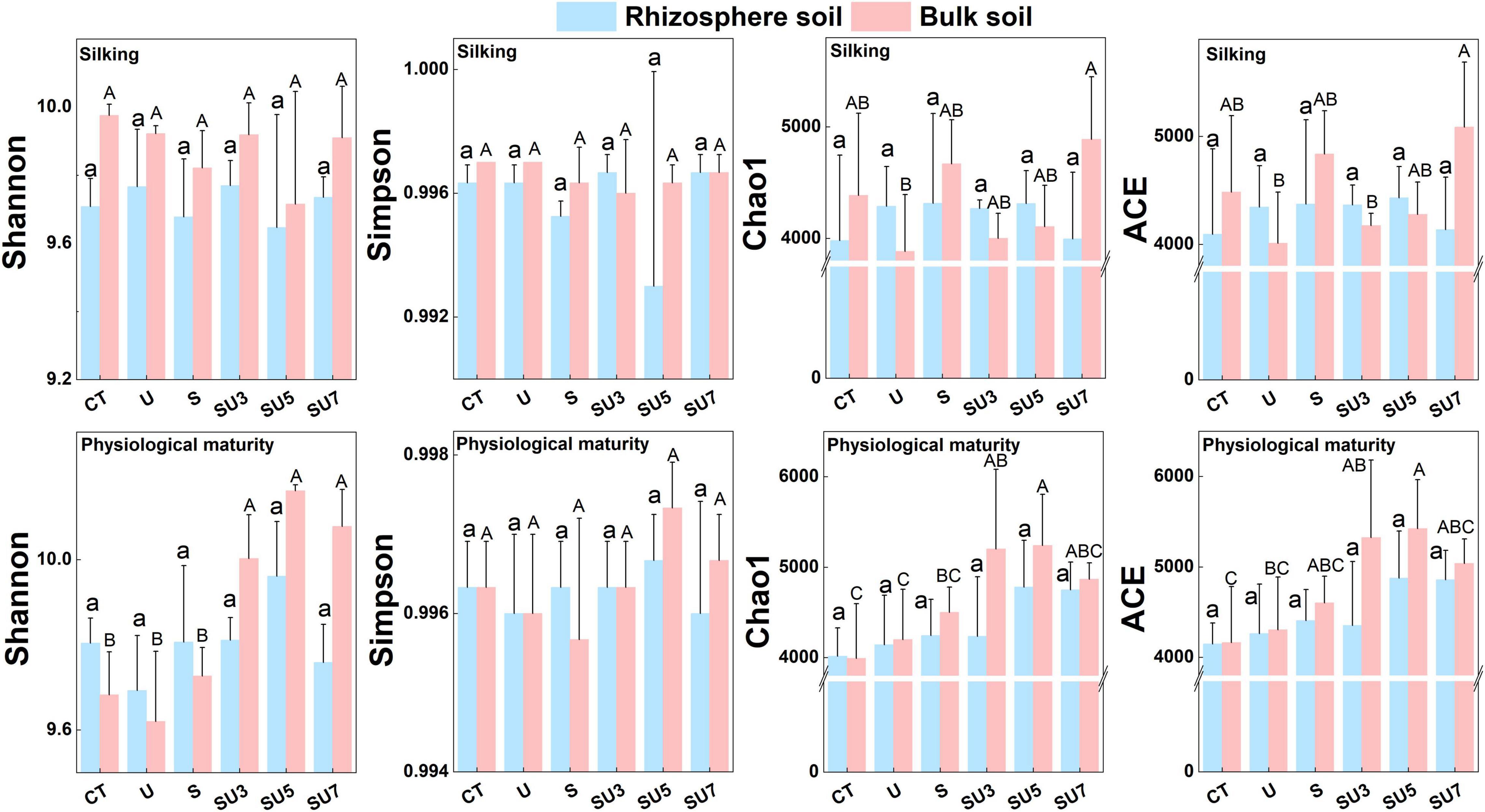
Figure 2. Effect of CT, U, S, SU3, SU5, and SU7 in rhizosphere and bulk soil bacterial α-diversity (Shannon, Simpson, Chao1, and ACE indexes) at different maize growing stages (i.e., silking and physiological maturity). Regular urea, controlled-release urea, and different ratios of regular urea and controlled-release urea are all applied at a rate of 180 kg ha– 1. Values are presented as the means ± SE. Lowercase and uppercase letters indicate significant differences between the nitrogen fertilizer treatment (p < 0.05).
In all treatments, proteobacteria were the most dominant microbial taxa (Figure 3), which in the maize silking stage was slightly higher than in the physiological maturity stage. In addition, the results showed that the relative abundance of proteobacteria in bulk soil decreased at the physiological maturity stage with the increasing controlled-release and regular urea (controlled-release mixed with regular urea ratio increased from 3:7 to 7:3). The relative abundance of Firmicutes and Bacteroidota increased in bulk soil at the physiological maturity stage compared to the silking stage (Figure 3). Simultaneous, soil microbial community structure was changed with the shift rhizosphere and bulk soil, maize silking, and physiological maturity stage (Figure 4).
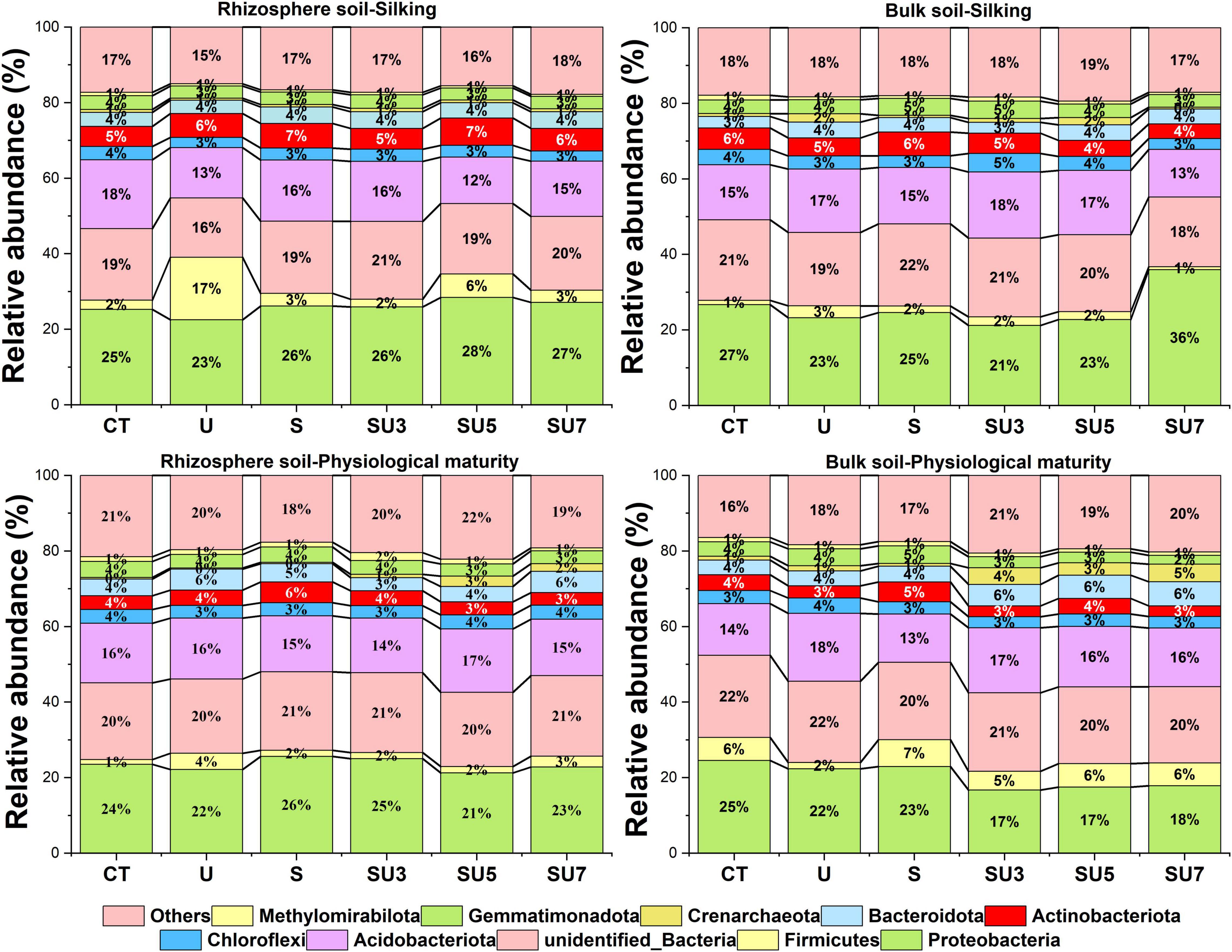
Figure 3. Effect of nitrogen fertilizer treatments (including CT, U, S, SU3, SU5, and SU7) on the microbial community composition of rhizosphere and bulk soil at different maize growth stages (i.e., silking and physiological maturity).
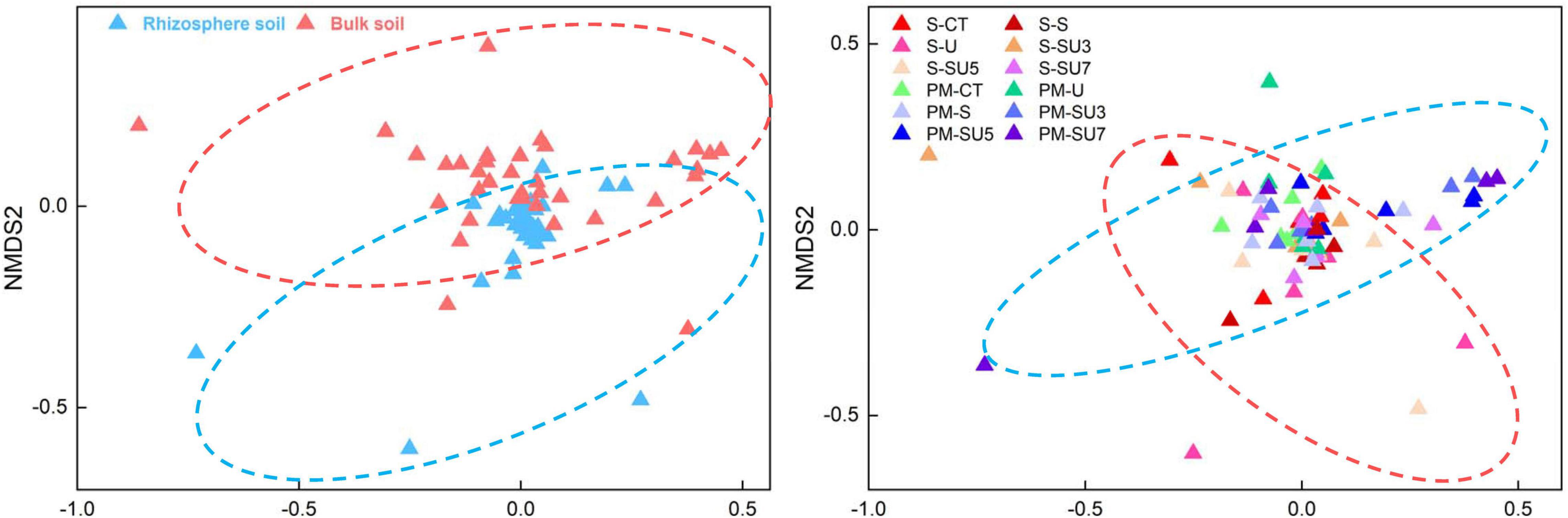
Figure 4. Effect of nitrogen fertilizer treatments (including CT, U, S, SU3, SU5, and SU7) on the NMDS of rhizosphere and bulk soil at different maize growth stages (i.e., silking and physiological maturity).
Shifts in Microbial Network Complexity in Different Maize Growth Stages Under N Treatment
The networks for rhizosphere and bulk soil at maize silking and physiological maturity stages were individually constructed and their topological properties were examined (Figures 5–8). Compared to the rhizosphere soil, the bulk soil in the maize silking stage has a higher microbial network total links and nodes (Figures 5, 6 and Table 1). In rhizosphere soil at the silking stage, the microbial network total links and nodes increased with the controlled-release to regular urea ratio, except the SU5 treatment (Figure 5). In bulk soil at the maize physiological maturity stage, the microbial network total links and average degree increased with the slow-release to regular N fertilizer ratios (Table 2). In particular, the combination of controlled-release and regular urea (i.e., SU3, 712, SU5, 453, and SU7, 504) had higher microbial network total links than the controlled-release and regular urea applied separately as well as the control treatment (Figure 8). This result suggests that the combination of controlled-release and regular urea is more effective in improving microbial network total links and an average degree in bulk soil at the maize physiological maturity stage than the traditional controlled-release and regular urea applied separately as well as the control treatment (Figures 7, 8 and Table 2).
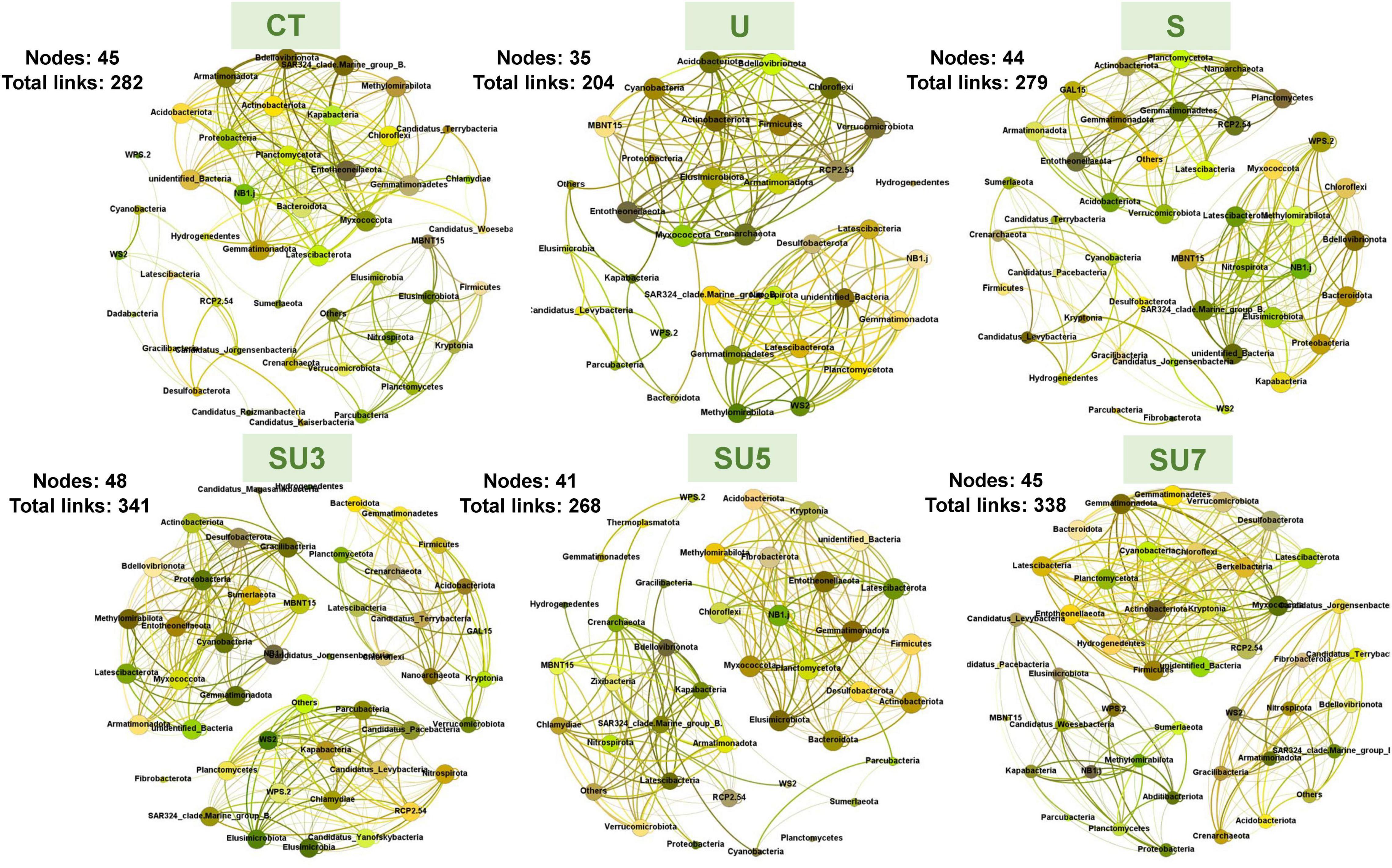
Figure 5. Overview of bacterial networks in the rhizosphere soil at the maize silking stage under different N fertilizer treatments (including CT, U, S, SU3, SU5, and SU7), with node size proportional to node connectivity. Node color represents various phylogenetic phyla.
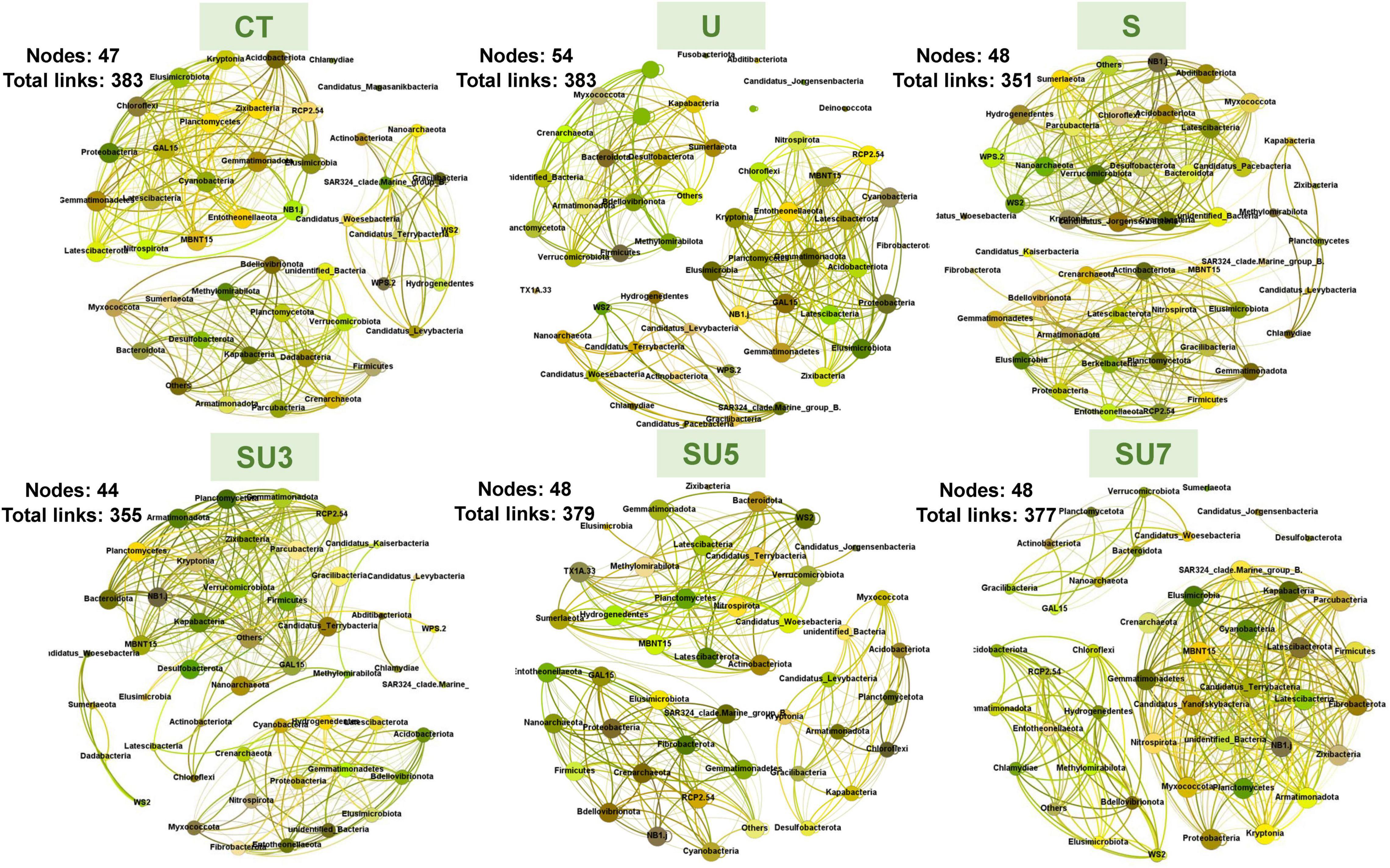
Figure 6. Overview of bacterial networks in the bulk soil at the maize silking stage under different N fertilizer treatments (including CT, U, S, SU3, SU5, and SU7), with node size proportional to node connectivity. Node color represents various phylogenetic phyla.
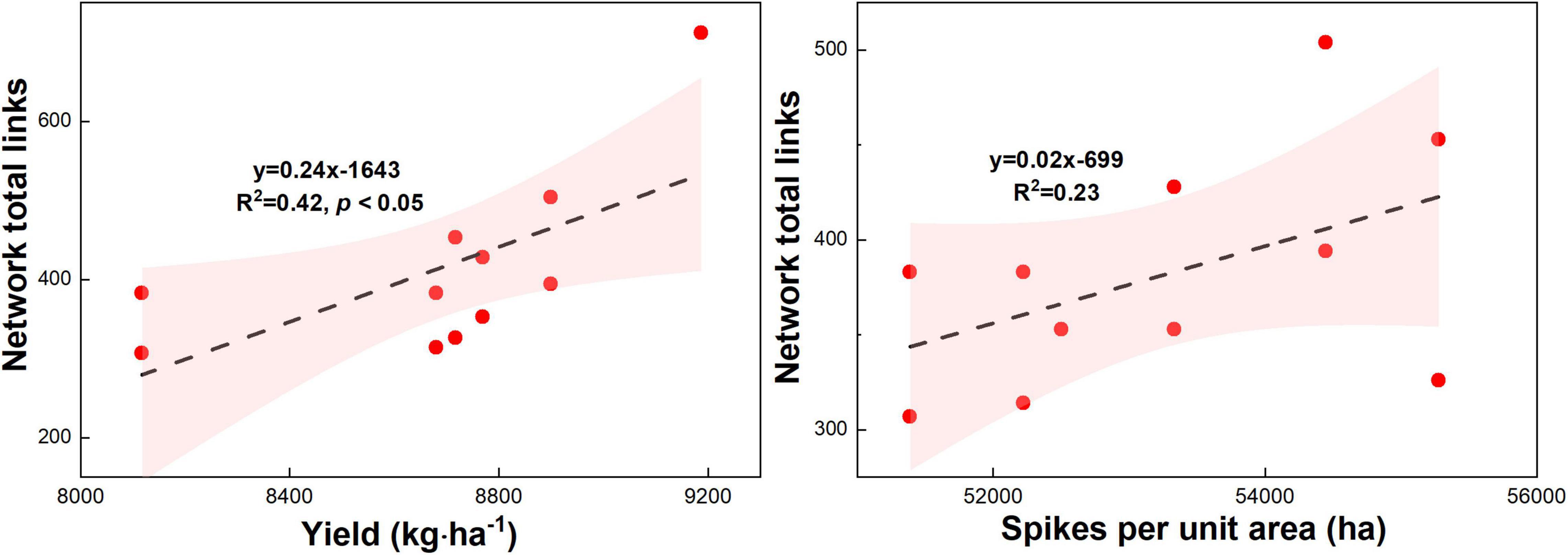
Figure 7. Overview of bacterial networks in the rhizosphere soil at the maize physiological maturity stage under different N fertilizer treatments (including CT, U, S, SU3, SU5, and SU7), with node size proportional to node connectivity. Node color represents various phylogenetic phyla.
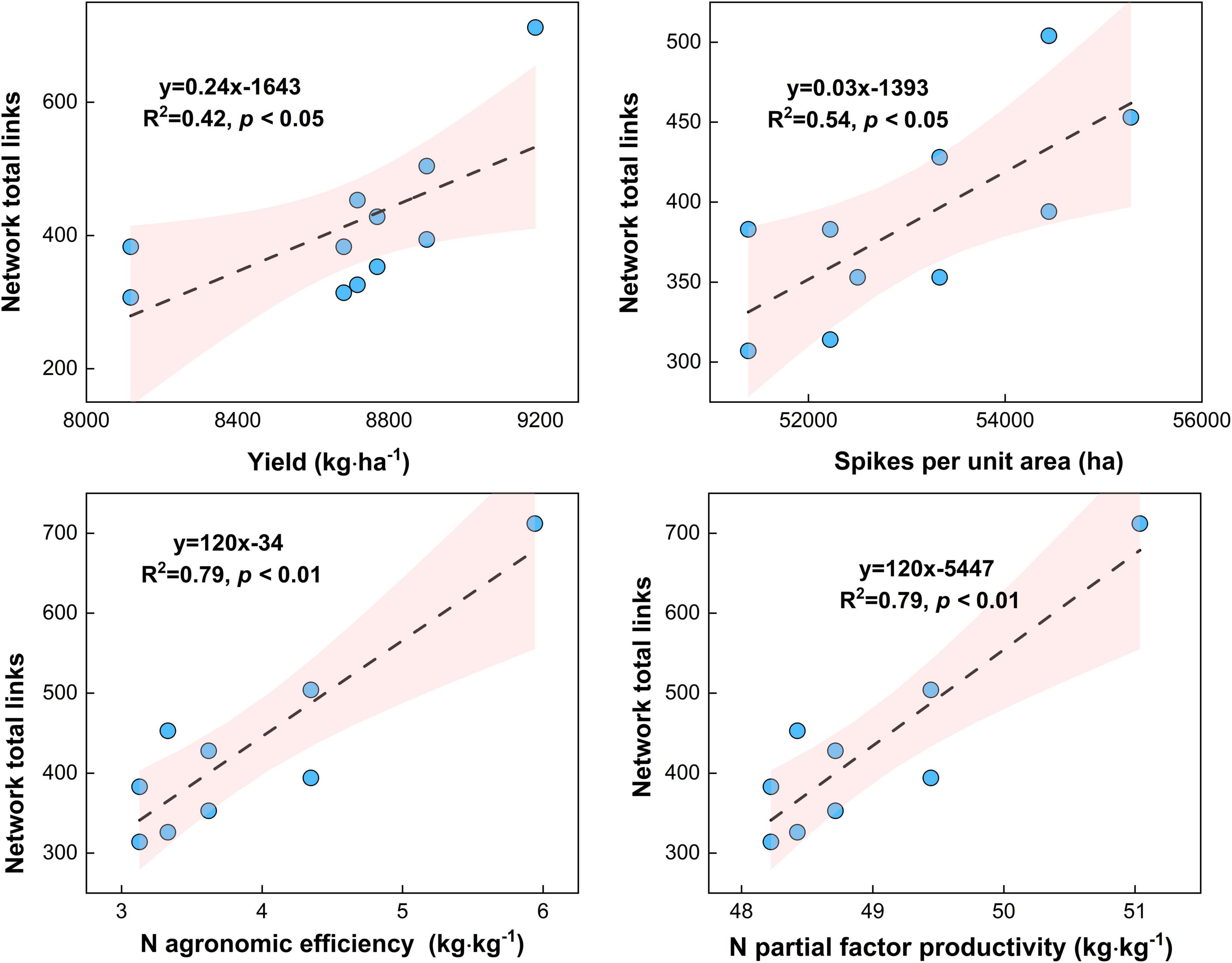
Figure 8. Overview of bacterial networks in the rhizosphere soil at the maize physiological maturity stage under different N fertilizer treatments (including CT, U, S, SU3, SU5, and SU7), with node size proportional to node connectivity. Node color represents various phylogenetic phyla.
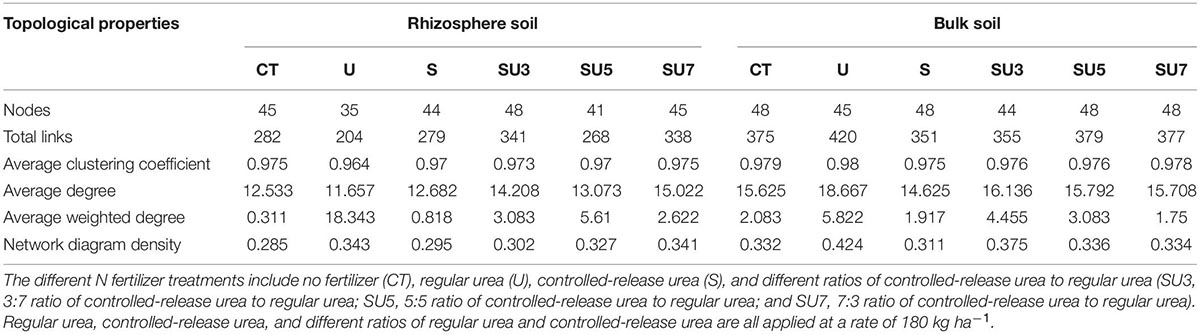
Table 1. Network topological characteristics in rhizosphere soils and bulk soils at the maize silking stage under different N fertilizer treatments.
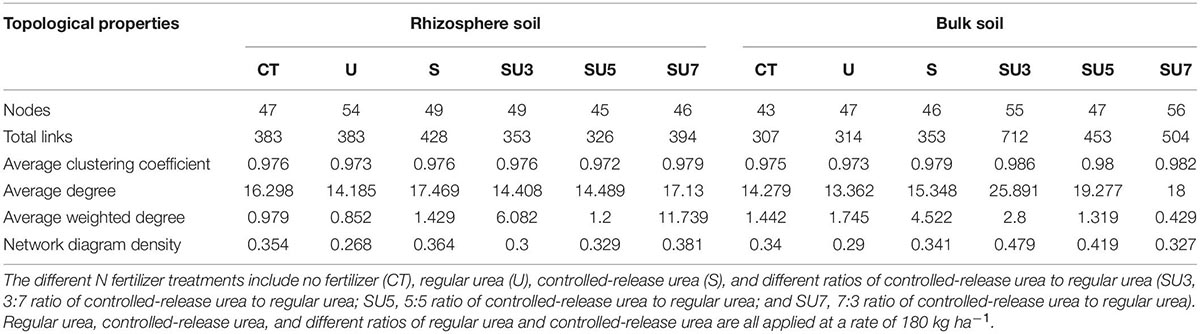
Table 2. Network topological characteristics in rhizosphere soils and bulk soils at the maize physiological maturity under different N fertilizer treatments.
Relationships Between the Microbial Network Complexity and the Maize Yield Traits and N Agronomic Efficiency and N Partial Factors Productivity
Maize yield (R2 = 0.45, p < 0.05) and spikes per unit area (R2 = 0.54, p < 0.05) increased with the microbial network complexity (i.e., total links) under different urea treatments (Figure 9, p < 0.05). This result highlights that the high microbial network complexity (i.e., microbial network total links) is closely associated with the higher maize yield. Meanwhile, the N agronomic efficiency (R2 = 0.79, p < 0.05) and N partial factor productivity (R2 = 0.79, p < 0.01) increased with the microbial network complexity (i.e., total links) under different urea treatments (Figure 9, p < 0.05). This result highlights that the high microbial network complexity (i.e., microbial network total links) can enhance the N agronomic efficiency and N partial factor productivity.
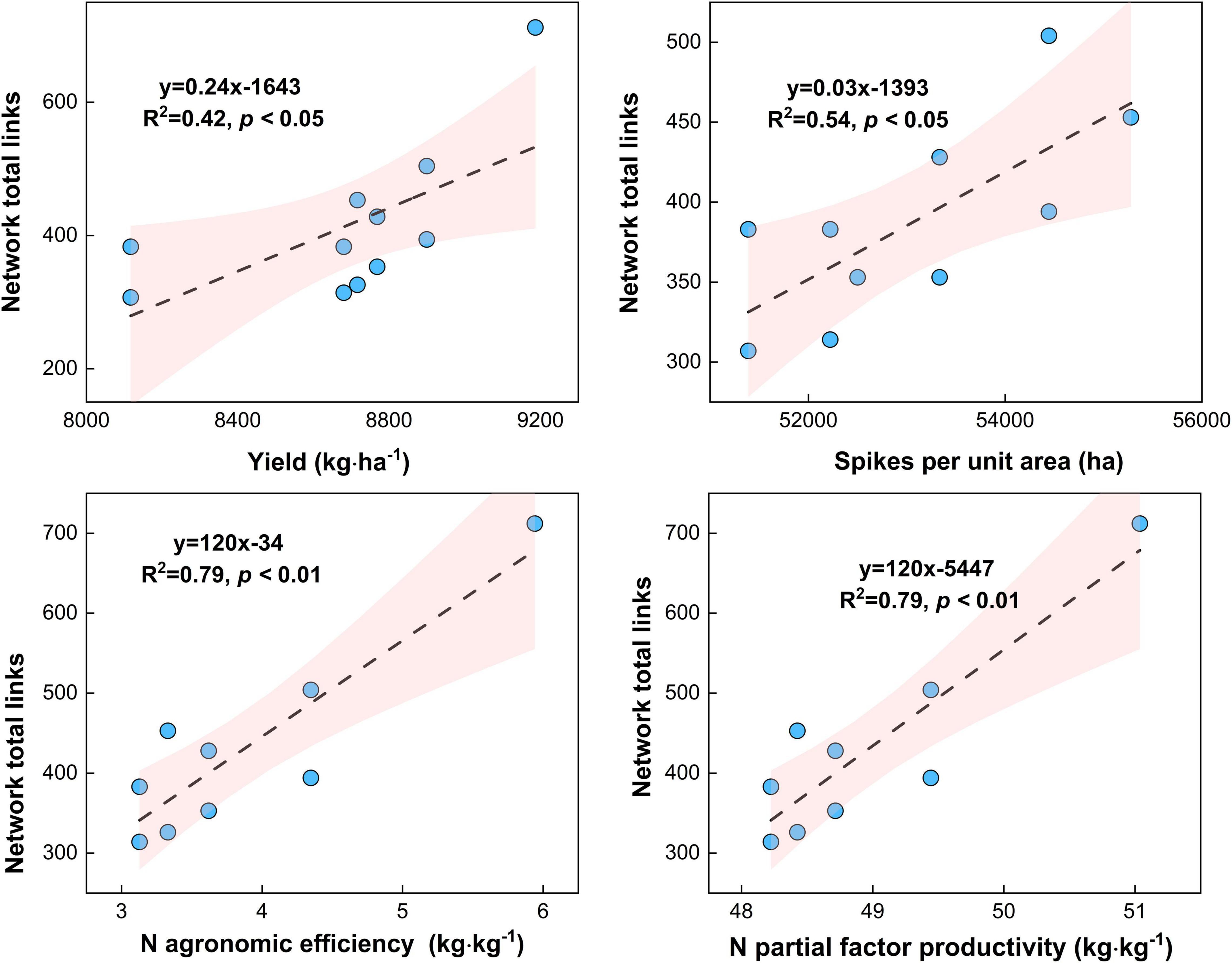
Figure 9. Relationship between microbial network complexity with maize yield, spikes per unit area, N agronomic efficiency, and N partial factor productivity under different N fertilizer treatments (including CT, U, S, SU3, SU5, and SU7).
Discussion
Effect of N Fertilizer Treatments on Maize Yield and Related Traits
Previous studies have shown that N fertilizer application can significantly contribute to increased maize yields (Zheng et al., 2016; Guo et al., 2017). In line with these studies, the results show that both regular and controlled-release urea alone and in combination can increase maize yield, spikes per area, and grain number per spike compared to the control treatment (Figure 1). However, the effects of N fertilizer on maize yield and spikes per area changed with the controlled-release urea and regular urea applied alone and their blending rates (Figure 1; Guo et al., 2021). Compared to the regular urea, the S, and SU3, both SU5 and SU7 can improve the maize yield (Figure 1). This might be due to the (i) excessive volatilization or loss of regular urea with precipitation leading to a potential loss in its contribution to maize growth and yield (Zhang et al., 2019); (ii) controlled-release urea is released into the soil solution at a rate that better matches the growth requirements of the crop and has a long fertilizer supply period (Shaviv, 2001; Li R. et al., 2021); and (iii) controlled-release urea could more efficiently improve the N use efficiency than regular urea (Zheng et al., 2016; Li et al., 2020). In this study, the SU3 treatment had the highest maize yield among the different blend ratios of regular and controlled-release urea (Figure 1). A similar result was confirmed by Guo et al. (2017, 2021), who found that the 3:7 blend ratio of controlled-release urea and regular urea was most effective in improving maize yield and NUE and mitigating NH3 volatilization and soil NO3-N leaching in Shaanxi province of China and Northwest China, respectively. This may be due to the fact that this ratio of controlled-release urea to regular urea both meets the nutrient requirements of maize in the early stages of fertility and provides a stable supply of nutrients for growth in the middle and late stages of fertility (Gao et al., 2021). Additionally, the N agronomic efficiency and N partial factor productivity were highest in the SU3 treatment, nearly 2 times and 1.2 times higher than the CT treatment, respectively (Figure 1, p < 0.05). This result suggests that SU3 treatment is more effective in improving N agronomic efficiency and N partial factor productivity than CT and other treatments. This is probably because this favorable ratio of controlled-release to regular urea both avoids rapid N losses and matches the fertilizer requirements of the maize growth stage, thus improving the N agronomic efficiency and N partial factor productivity (Zhang et al., 2022). Therefore, those results suggest that a 3:7 (SU3) blend of controlled-release urea and regular urea is the best ratio that maximizes summer maize yields and N agronomic efficiency in the North China Plain (Hebei Province).
Effect of N Fertilizer Treatments on Microbial Diversity, Composition, and Network Complexity
Soil microorganisms mediate many important biological processes for sustainable agriculture and it is widely recognized that fertilization activities have an impact on the community structure of microorganisms (Hamer et al., 2009; Geisseler and Scow, 2014; Sun R. et al., 2021). For example, Hamer et al. (2009) found that urea fertilization induced the alteration in microbial community composition from Gram-positive to Gram-negative bacteria and fungi. The microbial composition involved in the N-transforming process was altered by fertilization (Sun R. et al., 2021). However, in a wide range of soil and environmental conditions in western Canada, fertilizer application (e.g., controlled-release urea) had no significant effects on soil microorganisms in most cases, and the few significant effects were increases in microbial biomass or functional diversity (Lupwayi et al., 2010). In line with Lupwayi et al. (2010), those results show that different ratios of controlled-release urea and regular urea application had less effect on changes in microbial community composition, which did not change significantly at the silking and maturity stages of maize (Figure 3). This result demonstrated that the short-term applications of regular urea and controlled-release urea in the North China Plain did not alter the composition of soil microbial communities, and these fertilizers did not adversely affect most soil biological processes (Lupwayi et al., 2010). In addition, over 10 years (long-term) experiments also suggested that the N inputs on soil microbial community structure were minor and slightly different between prokaryotes and fungi (Tosi et al., 2021). This may be due to the fact that some microbial taxa can withstand quite challenging conditions (e.g., fertilization), which may play an important role in the ecological adaptation of microorganisms and plant growth (De Vries and Shade, 2013). Thus, microbial communities that maintain a stable pattern in response to fertilization may help to maintain food and fiber production in the presence of long-term nutrient fertilization (Fan et al., 2020a).
Although the application of controlled-release urea and regular urea and their different ratios did not alter the microbial community composition of the maize rhizosphere and bulk soil, which significantly affected microbial α diversity (Figure 2), the result showed that the microbial diversity (i.e., Shannon, Simpson, Chao1, and ACE indexes) remained stable at the silking stage of maize with control and different normal and slow-release N fertilizer ratios, whereas that increased at the maize physiological maturity stage with the increasing slow-release to regular N fertilizer ratios (from SU3 to SU7; Figure 2). This result suggests that both regular and controlled-release urea alone and in combination can contribute to an increase in microbial diversity. This may be due to the reduced leaching of soil N caused by the combined application of controlled-release urea and regular urea, which still increased soil NO3–-N and NH4+-N content in late crop growth (Zheng et al., 2016). The reduction in NO3–-N and NH4+-N uptake by the crop in late growth facilitated the use of these soluble nutrients by microorganisms, which in turn stimulated an increase in microbial biomass and diversity (Lupwayi et al., 2010; Huang et al., 2021). However, a previous study showed that the bacterial and fungi α diversity in a blend of regular to controlled-release urea treatment were lower than the regular urea treatment (Li B. B. et al., 2021). This study contradicts our findings, suggesting that the effect of the combination of controlled-release and conventional urea on microbial diversity may be influenced by regional conditions, such as nutrient availability or changes in the crop fertility stage (Figure 2; Zhang et al., 2022). For example, when nutrient availability in the soil decreases or when reduced nutrient levels may limit microbial proliferation and nutrient mineralization, competition between crops and microbes for N may reduce microbial diversity (Inselsbacher et al., 2010). Lupwayi et al. (2010) found that the controlled-release urea increased MBC or functional diversity more than urea in 3 site-years, but the opposite was observed in 1 site-year. This result shows that the effect of controlled-release urea and normal urea on microbial communities is influenced by the age of application of the fertilizer.
In the maize silking stage, the microbial network total links and an average degree in treatments of the combination of regular and controlled-release urea were higher than the regular and slow-release N fertilizer applied separately (Figure 5 and Table 1). Simultaneously, the microbial network total links increased with the controlled-release to regular urea ratios (Figure 8 and Table 2). This result demonstrated that the combination of regular and controlled-release urea is more effective in improving microbial network complexity than the traditional regular and slow-release N fertilizer applied separately. This is because the microbial network complexity is influenced by the type of fertilizer and the amount of fertilizer applied (Ji et al., 2021; Li B. B. et al., 2021). The blending of controlled-release urea and regular urea affects its network complexity by influencing the diversity of microorganisms. The SU3 treatment had the highest microbial network complexity in this study among the different blend ratios of regular and controlled-release urea (Figure 8). This result indicates that the SU3 treatment is the optimal treatment to improve the complexity of the soil microbial network in the North China Plain.
Relationships Between the Maize Yield and Microbial Network Complexity
Previous studies have shown that more complex systems with higher diversity are more productive because they are more resistant to environmental change (De Vries and Shade, 2013; Yuan et al., 2021). Additionally, the microbial network complexity enhances the link between biodiversity and multifunctionality in agricultural systems (Jiao et al., 2021). For example, the combination of inorganic fertilizer application and cow manure resulted in the most complex microbial community, and this network complexity was closely linked to high plant productivity and nutrient availability (Fan et al., 2020a). In this study, the application of controlled-release urea and regular urea and their different ratios resulted in more or fewer changes in microbial network complexity (Figure 5–8). Importantly, this result suggests that the maize yield increased with the microbial network complexity (i.e., total links) under different urea treatments (Figure 9, p < 0.05). The application SU3 led to the most resistant (i.e., higher total links) microbial community, which was associated with the highest levels of maize yield (Figures 1, 8). This result highlights that the high microbial network complexity (i.e., microbial network total links) resulted in a higher maize yield. This is mainly due to the (i) high microbial community network complexity (i.e., the most resistant microbial community) may lead to higher availability of nutrients for plants, which facilitate plants getting more nutrients and less competition from microbial species (Fan et al., 2020a), and (ii) the reduction in the relative abundance of potential plant fungal pathogens (Van Der Heijden et al., 2008). Moreover, the N agronomic efficiency and N partial factor productivity increased with the microbial network complexity (i.e., total links) under different urea treatments (Figure 9, p < 0.01). The SU3 treatment led to the most resistant (i.e., higher total links) microbial community, which was strongly associated with the highest levels of N agronomic efficiency and N partial factor productivity (Figures 1, 9). This result suggests that high microbial diversity and community complexity can significantly increase the N agronomic efficiency and N partial factor productivity of maize. This may be due to the fact that more complex microbial communities can accelerate soil nutrient cycling processes and enhance plant-microbial feedback (Fan et al., 2020a). Consequently, these findings suggest that the microbial network complexity is a critical factor in influencing maize production and fertilizer agronomic efficiency in an economically important crop system.
Conclusion
This result suggests that the short-term applications of regular urea and controlled-release urea and their combinations in the North China Plain did not alter the composition of soil microbial communities, but they did contribute to an increase in microbial diversity. A combination of controlled-release and regular urea is more effective in improving maize yields than the regular and controlled-release urea applied separately (p < 0.05). In particular, in this study, the SU3 treatment had the highest maize yield (9,186 kg ha–1) among the different blend ratios of regular and controlled release urea, which increased by 13% in the SU3 treatment compared to the CT treatment. Simultaneously, the microbial network total links increased with the controlled-release to regular urea ratios, and the SU3 treatment had the highest microbial network complexity among the different blend ratios of regular and controlled-release urea. The SU3 treatment had the highest N agronomic efficiency and N partial factor productivity of all treatments, nearly 2 and 1.2 times higher than the CT treatment, respectively (p < 0.05). The N agronomic efficiency, N partial factor productivity, spikes per unit area, and maize yield increased with the microbial network complexity (i.e., total links). This result indicates that the SU3 treatment is the optimal treatment to both improve the maize yield and microbial network complexity in the North China Plain. Therefore, this study provides solid evidence that the microbial network complexity is essential for supporting maize yield under different N fertilizer treatments.
Data Availability Statement
The data presented in the study are deposited in the Genome Sequence Archive (Genomics, Proteomics and Bioinformatics 2021) in National Genomics Data Center (Nucleic Acids Res 2022), China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences, accession numbers CRA005461 and CRA005462.
Author Contributions
Y-CZ: conceptualization and funding acquisition, methodology, project administration, supervision, validation, and writing-review and editing. YP: sample collection. X-LL and P-JT: formal analysis and investigation. P-TJ: data curation, visualization, and writing-original draft. XD and J-cZ: data collection and correlation analysis of hormone data. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by the National Key Research and Development Program of China (Grant No. 2018YFD0300503).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We sincerely acknowledge the reviewers for the critical comments on our original manuscript.
References
Adiele, J. G., Schut, A. G. T., van den Beuken, R. P. M., Ezui, K. S., Pypers, P., Ano, A. O., et al. (2020). Towards closing cassava yield gap in West Africa: agronomic efficiency and storage root yield responses to NPK fertilizers. Field Crops Res. 253:107820. doi: 10.1016/j.fcr.2020.107820
Blackshaw, R. E., Hao, X., Brandt, R. N., Clayton, G. W., Harker, K. N., O’Donovan, J. T., et al. (2011). Canola response to ESN and urea in a four-year no-till cropping system. Agron. J. 103, 92–99. doi: 10.2134/agronj2010.0299
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). Correspondence QIIME allows analysis of high- throughput community sequencing data intensity normalization improves color calling in SOLiD sequencing. Nat. Publ. Gr. 7, 335–336. doi: 10.1038/nmeth0510-335
De Vries, F. T., and Shade, A. (2013). Controls on soil microbial community stability under climate change. Front. Microbiol. 4:265. doi: 10.3389/fmicb.2013.00265
Devi, R., Kaur, T., Guleria, G., Rana, K. L., Kour, D., Yadav, N., et al. (2020). Fungal Secondary Metabolites and their Biotechnological Applications for Human Health. Amsterdam: Elsevier. doi: 10.1016/b978-0-12-820528-0.00010-7
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Fan, K., Delgado-Baquerizo, M., Guo, X., Wang, D., Zhu, Y. G., and Chu, H. (2020a). Microbial resistance promotes plant production in a four-decade nutrient fertilization experiment. Soil Biol. Biochem. 141:107679. doi: 10.1016/j.soilbio.2019.107679
Fan, K., Delgado-Baquerizo, M., Zhu, Y. G., and Chu, H. (2020b). Crop production correlates with soil multitrophic communities at the large spatial scale. Soil Biol. Biochem. 151:108047. doi: 10.1016/j.soilbio.2020.108047
Fan, K., Delgado-Baquerizo, M., Guo, X., Wang, D., Zhu, Y. G., and Chu, H. (2021). Biodiversity of key-stone phylotypes determines crop production in a 4-decade fertilization experiment. ISME J. 15, 550–561. doi: 10.1038/s41396-020-00796-8
Gao, Y., Song, X., Liu, K., Li, T., Zheng, W., Wang, Y., et al. (2021). Mixture of controlled-release and conventional urea fertilizer application changed soil aggregate stability, humic acid molecular composition, and maize nitrogen uptake. Sci. Total Environ. 789:147778. doi: 10.1016/j.scitotenv.2021.147778
Geisseler, D., and Scow, K. M. (2014). Long-term effects of mineral fertilizers on soil microorganisms - a review. Soil Biol. Biochem. 75, 54–63. doi: 10.1016/j.soilbio.2014.03.023
Grant, C. A., Wu, R., Selles, F., Harker, K. N., Clayton, G. W., Bittman, S., et al. (2012). Crop yield and nitrogen concentration with controlled release urea and split applications of nitrogen as compared to non-coated urea applied at seeding. Field Crops Res. 127, 170–180. doi: 10.1016/j.fcr.2011.11.002
Gu, S., Hu, Q., Cheng, Y., Bai, L., Liu, Z., Xiao, W., et al. (2019). Application of organic fertilizer improves microbial community diversity and alters microbial network structure in tea (Camellia sinensis) plantation soils. Soil Tillage Res. 195:104356. doi: 10.1016/j.still.2019.104356
Guo, J., Fan, J., Zhang, F., Yan, S., Zheng, J., Wu, Y., et al. (2021). Blending urea and slow-release nitrogen fertilizer increases dryland maize yield and nitrogen use efficiency while mitigating ammonia volatilization. Sci. Total Environ. 790:148058. doi: 10.1016/j.scitotenv.2021.148058
Guo, J., Wang, Y., Blaylock, A. D., and Chen, X. (2017). Mixture of controlled release and normal urea to optimize nitrogen management for high-yielding (>15 Mg ha-1) maize. Field Crops Res. 204, 23–30. doi: 10.1016/j.fcr.2016.12.021
Hamer, U., Potthast, K., and Makeschin, F. (2009). Urea fertilisation affected soil organic matter dynamics and microbial community structure in pasture soils of Southern Ecuador. Appl. Soil Ecol. 43, 226–233. doi: 10.1016/j.apsoil.2009.08.001
Huang, Y., Wang, Q., Zhang, W., Zhu, P., Xiao, Q., Wang, C., et al. (2021). Stoichiometric imbalance of soil carbon and nutrients drives microbial community structure under long-term fertilization. Appl. Soil Ecol. 168:104119. doi: 10.1016/j.apsoil.2021.104119
Inselsbacher, E., Hinko-Najera Umana, N., Stange, F. C., Gorfer, M., Schüller, E., Ripka, K., et al. (2010). Short-term competition between crop plants and soil microbes for inorganic N fertilizer. Soil Biol. Biochem. 42, 360–372. doi: 10.1016/j.soilbio.2009.11.019
Ji, L., Si, H., He, J., Fan, L., and Li, L. (2021). The shifts of maize soil microbial community and networks are related to soil properties under different organic fertilizers. Rhizosphere 19:100388. doi: 10.1016/j.rhisph.2021.100388
Jiao, S., Lu, Y., and Wei, G. (2021). Soil multitrophic network complexity enhances the link between biodiversity and multifunctionality in agricultural systems. Glob. Change Biol. 28, 140–153. doi: 10.1111/gcb.15917
Jilling, A., Keiluweit, M., Gutknecht, J., and Grandy, A. S. (2021). Priming mechanisms providing plants and microbes access to mineral-associated organic matter. Soil Biol. Biochem. 158:108265. doi: 10.1016/j.soilbio.2021.108265
Kuzyakov, Y., and Razavi, B. S. (2019). Rhizosphere size and shape: temporal dynamics and spatial stationarity. Soil Biol. Biochem. 135, 343–360. doi: 10.1016/j.soilbio.2019.05.011
Li, R., Gao, Y., Chen, Q., Li, Z., Gao, F., Meng, Q., et al. (2021). Blended controlled-release nitrogen fertilizer with straw returning improved soil nitrogen availability, soil microbial community, and root morphology of wheat. Soil Tillage Res. 212:105045. doi: 10.1016/j.still.2021.105045
Li, B. B., Roley, S. S., Duncan, D. S., Guo, J., Quensen, J. F., Yu, H. Q., et al. (2021). Long-term excess nitrogen fertilizer increases sensitivity of soil microbial community to seasonal change revealed by ecological network and metagenome analyses. Soil Biol. Biochem. 160:108349. doi: 10.1016/j.soilbio.2021.108349
Li, G., Zhao, B., Dong, S., Zhang, J., Liu, P., and Lu, W. (2020). Controlled-release urea combining with optimal irrigation improved grain yield, nitrogen uptake, and growth of maize. Agric. Water Manag. 227:105834. doi: 10.1016/j.agwat.2019.105834
Li, Y., Zheng, J., Wu, Q., Gong, X., Zhang, Z., Chen, Y., et al. (2022). Zeolite increases paddy soil potassium fixation, partial factor productivity, and potassium balance under alternate wetting and drying irrigation. Agric. Water Manag. 260:107294. doi: 10.1016/j.agwat.2021.107294
Liu, S., Zhou, L., Li, H, Zhao, X., Yang, Y., Zhu, Y., et al. (2019). Shrub encroachment decreases soil inorganic carbon stocks in Mongolian grasslands. J. Ecol. 1365–2745. doi: 10.1111/1365-2745.13298
Lupwayi, N. Z., Grant, C. A., Soon, Y. K., Clayton, G. W., Bittman, S., Malhi, S. S., et al. (2010). Soil microbial community response to controlled-release urea fertilizer under zero tillage and conventional tillage. Appl. Soil Ecol. 45, 254–261. doi: 10.1016/j.apsoil.2010.04.013
Mariotte, P., Mehrabi, Z., Bezemer, T. M., De Deyn, G. B., Kulmatiski, A., Drigo, B., et al. (2018). Plant–soil feedback: bridging natural and agricultural sciences. Trends Ecol. Evol. 33, 129–142. doi: 10.1016/j.tree.2017.11.005
Mooshammer, M., Wanek, W., Zechmeister-Boltenstern, S., and Richter, A. (2014). Stoichiometric imbalances between terrestrial decomposer communities and their resources: mechanisms and implications of microbial adaptations to their resources. Front. Microbiol. 5:22. doi: 10.3389/fmicb.2014.00022
Nihorimbere, V., Ongena, M., Smargiassi, M., and Thonart, P. (2011). Effet bénéfique de la communauté microbienne de la rhizosphère sur la croissance et la santé des plantes. Biotechnol. Agron. Soc. Environ. 15, 327–337.
Pruesse, E., Quast, C., Knittel, K., Fuchs, B. M., Ludwig, W., Peplies, J., et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188–7196. doi: 10.1093/nar/gkm864
R Development Core Team (2016). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Ren, J., Liu, X., Yang, W., Yang, X., Li, W., Xia, Q., et al. (2021). Rhizosphere soil properties, microbial community, and enzyme activities: short-term responses to partial substitution of chemical fertilizer with organic manure. J. Environ. Manage. 299:113650. doi: 10.1016/j.jenvman.2021.113650
Reyon, D., Tsai, S. Q., Khgayter, C., Foden, J. A., Sander, J. D., and Joung, J. K. (2012). FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol. 30, 460–465. doi: 10.1038/nbt.2170
Schneider, T., Keiblinger, K. M., Schmid, E., Sterflinger-Gleixner, K., Ellersdorfer, G., Roschitzki, B., et al. (2012). Who is who in litter decomposition Metaproteomics reveals major microbial players and their biogeochemical functions. ISME J. 6, 1749–1762. doi: 10.1038/ismej.2012.11
Shaviv, A. (2001). Advances in controlled release of fertilizers 71, before printing. Adv. Agron. 71, 1–49. doi: 10.1016/s0065-2113(01)71011-5
Sun, R., Wang, F., Hu, C., and Liu, B. (2021). Metagenomics reveals taxon-specific responses of the nitrogen-cycling microbial community to long-term nitrogen fertilization. Soil Biol. Biochem. 156:108214. doi: 10.1016/j.soilbio.2021.108214
Sun, Y. Y., Zhang, Q. X., Zhao, Y. P., Diao, Y. H., Gui, F. R., and Yang, G. Q. (2021). Beneficial rhizobacterium provides positive plant–soil feedback effects to Ageratina adenophora. J. Integr. Agric. 20, 1327–1335. doi: 10.1016/S2095-3119(20)63234-8
Swift, M. J., Heal, O. W., and Anderson, J. M. (1979). Decomposition in terrestrial ecosystems. Rev. Lit. Arts Am. 5, 12–24. doi: 10.1007/s00114-006-0159-1
Tekaya, M., Dahmen, S., Ben Mansour, M., Ferhout, H., Chehab, H., Hammami, M., et al. (2021). Foliar application of fertilizers and biostimulant has a strong impact on the olive (Olea europaea) rhizosphere microbial community profile and the abundance of arbuscular mycorrhizal fungi. Rhizosphere 19:100402. doi: 10.1016/j.rhisph.2021.100402
Thakur, M. P., van der Putten, W. H., Wilschut, R. A., Ciska Veen, G. F., Kardol, P., van Ruijven, J., et al. (2021). Plant–soil feedbacks and temporal dynamics of plant diversity–productivity relationships. Trends Ecol. Evol. 36, 651–661. doi: 10.1016/j.tree.2021.03.011
Tosi, M., Deen, W., Drijber, R., McPherson, M., Stengel, A., and Dunfield, K. (2021). Long-term N inputs shape microbial communities more strongly than current-year inputs in soils under 10-year continuous corn cropping. Soil Biol. Biochem. 160:108361. doi: 10.1016/j.soilbio.2021.108361
Van Der Heijden, M. G. A., Bardgett, R. D., and Van Straalen, N. M. (2008). The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 11, 296–310. doi: 10.1111/j.1461-0248.2007.01139.x
Wang, B., An, S., Liang, C., Liu, Y., and Kuzyakov, Y. (2021). Microbial necromass as the source of soil organic carbon in global ecosystems. Soil Biol. Biochem. 162:108422. doi: 10.1016/j.soilbio.2021.108422
Wilkinson, S. C., Anderson, J. M., Scardelis, S. P., Tisiafouli, M., Taylor, A., and Wolters, V. (2002). PLFA profiles of microbial communities in decomposing conifer litters subject to moisture stress. Soil Biol. Biochem. 34, 189–200. doi: 10.1016/S0038-0717(01)00168-7
Xiong, M., Sun, R., and Chen, L. (2019). A global comparison of soil erosion associated with land use and climate type. Geoderma 343, 31–39. doi: 10.1016/j.geoderma.2019.02.013
Yadav, A. N., Kour, D., Kaur, T., Devi, R., Yadav, A., Dikilitas, M., et al. (2021). Biodiversity, and biotechnological contribution of beneficial soil microbiomes for nutrient cycling, plant growth improvement and nutrient uptake. Biocatal. Agric. Biotechnol. 33:102009. doi: 10.1016/j.bcab.2021.102009
York, L. M., Carminati, A., Mooney, S. J., Ritz, K., and Bennett, M. J. (2016). The holistic rhizosphere: integrating zones, processes, and semantics in the soil influenced by roots. J. Exp. Bot. 67, 3629–3643. doi: 10.1093/jxb/erw108
Yu, L., Luo, S., Gou, Y., Xu, X., and Wang, J. (2021). Structure of rhizospheric microbial community and N cycling functional gene shifts with reduced N input in sugarcane-soybean intercropping in South China. Agric. Ecosyst. Environ. 314:107413. doi: 10.1016/j.agee.2021.107413
Yuan, M. M., Guo, X., Wu, L., Zhang, Y., Xiao, N., Ning, D., et al. (2021). Climate warming enhances microbial network complexity and stability. Nat. Clim. Change 11, 343–348. doi: 10.1038/s41558-021-00989-9
Zhang, G., Zhao, D., Liu, S., Liao, Y., and Han, J. (2022). Can controlled-release urea replace the split application of normal urea in China? A meta-analysis based on crop grain yield and nitrogen use efficiency. Field Crops Res. 275:108343. doi: 10.1016/j.fcr.2021.108343
Zhang, L., Liang, Z., Hu, Y., Schmidhalter, U., Zhang, W., Ruan, S., et al. (2021). Integrated assessment of agronomic, environmental and ecosystem economic benefits of blending use of controlled-release and common urea in wheat production. J. Clean. Prod. 287:125572. doi: 10.1016/j.jclepro.2020.125572
Zhang, W., Liang, Z., He, X., Wang, X., Shi, X., Zou, C., et al. (2019). The effects of controlled release urea on maize productivity and reactive nitrogen losses: a meta-analysis. Environ. Pollut. 246, 559–565. doi: 10.1016/j.envpol.2018.12.059
Zheng, W., Zhang, M., Liu, Z., Zhou, H., Lu, H., Zhang, W., et al. (2016). Combining controlled-release urea and normal urea to improve the nitrogen use efficiency and yield under wheat-maize double cropping system. Field Crops Res. 197, 52–62. doi: 10.1016/j.fcr.2016.08.004
Keywords: rhizosphere soil, controlled release urea, maize yield, microbial diversity, microbial network complexity, North China Plain
Citation: Ji P-t, Du X, Zhou J-c, Peng Y, Li X-l, Tao P-j and Zhang Y-c (2022) Network Analysis Reveals the Combination of Controlled-Release and Regular Urea Enhances Microbial Interactions and Improves Maize Yields. Front. Microbiol. 13:825787. doi: 10.3389/fmicb.2022.825787
Received: 30 November 2021; Accepted: 01 March 2022;
Published: 21 June 2022.
Edited by:
Fusheng Chen, Huazhong Agricultural University, ChinaReviewed by:
Huike Li, Northwest A&F University, ChinaKamlesh Kumar Meena, National Institute of Abiotic Stress Management (ICAR), India
Copyright © 2022 Ji, Du, Zhou, Peng, Li, Tao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yue-chen Zhang, amljdW5tb0AxMjYuY29t
 Peng-tao Ji
Peng-tao Ji Xiong Du1
Xiong Du1 Xiang-ling Li
Xiang-ling Li