
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 03 February 2022
Sec. Microbial Symbioses
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.825338
This article is part of the Research Topic Role of Microbiome in Disease Diagnostics and Therapeutics View all 25 articles
Trillions of microbes live within our bodies in a deep symbiotic relationship. Microbial populations vary across body sites, driven by differences in the environment, immunological factors, and interactions between microbial species. Major advances in genome sequencing enable a better understanding of microbiome composition. However, most of the microbial taxa and species of the human microbiome are still unknown. Without revealing the identity of these microbes as a first step, we cannot appreciate their role in human health and diseases. A shift in the microbial balance, termed dysbiosis, is linked to a broad range of diseases from simple colitis and indigestion to cancer and dementia. The last decade has witnessed an explosion in microbiome research that led to a better understanding of the microbiome structure and function. This understanding leads to potential opportunities to develop next-generation microbiome-based drugs and diagnostic biomarkers. However, our understanding is limited given the highly personalized nature of the microbiome and its complex and multidirectional interactions with the host. In this review, we discuss: (1) our current knowledge of microbiome structure and factors that shape the microbial composition, (2) recent associations between microbiome dysbiosis and diseases, and (3) opportunities of new microbiome-based therapeutics. We analyze common themes, promises, gaps, and challenges of the microbiome research.
The human microbiome is the collection of microbes, and their associated genes and secreted molecules that live on or inside the human body (Gilbert et al., 2018). The microbial communities within our bodies are highly personalized and are considered as unique to each individual as their fingerprints (Franzosa et al., 2015) and are even unique to each body site (Dekaboruah et al., 2020). Many factors shape microbial diversity and abundance such as diet, host genetics, diseases, drugs, and lifestyle (Yadav and Chauhan, 2021a). The collection of these factors results in a microbial signature, that is, either a balanced and diverse healthy microbiota or an imbalanced and dysbiotic composition. Microbial dysbiosis is associated with the onset and progression of many diseases such as inflammatory bowel diseases (IBDs), obesity, metabolic disorders, and mental disorders (Gorecki et al., 2019; Wei et al., 2020; Doelman et al., 2021). While there is a plethora of research linking the microbiome dysbiosis to a particular disease, little is known about the underlying mechanisms (Liang et al., 2019; Gomaa, 2020; Duran-Pinedo et al., 2021; Lau et al., 2021). It is logical to anticipate that missing or enriched microbial taxa might affect microbial interactions and secreted metabolites which in turn can change host metabolism and other body functions. Various factors contribute to this microbiome-host interactions including ecological, epigenetics, and genetics components (Yadav et al., 2018). Understanding how microbial metabolites influence the health or disease status would have a significant impact on treating diet related diseases (Yadav et al., 2018). Few examples of microbial metabolites are known to contribute to the health and well-being of the body such as short-chain fatty acids (SCFAs; Segain et al., 2000), which contribute to intestinal homeostasis, reduce inflammation, and decrease gut permeability. Other microbial metabolites mediate diseases such as lipopolysaccharides which increase inflammation (Lin et al., 2020) and colibactin, which is implicated in colon cancer (Wilson et al., 2019). Understanding the mechanisms and secreted molecules underlying microbiome-disease associations will lead to innovative therapeutic interventions (Yadav and Chauhan, 2021b). Much interest exists in the therapeutic potential of the microbiome (Knight, 2010; Raes, 2014). There is an intriguing interest in the use of probiotic microbes or fecal transplantation to restore the balance of gut microbes. Preliminary data suggest that probiotics can treat diseases beyond the stomach disorders such as cancer and Alzheimer’s. Although, the approach of developing microbiome-based therapeutics or adjuvants is promising, it is far more complicated than anticipated. The main challenge is the lack of causality direction. For example, we do not know if a change in microbiota drives the disease or the disease itself modulates the microbiome. Another obstacle is the highly dynamic and personalized nature of the microbiome, which makes developing drugs that are universally usable very challenging. In this review, we discuss the current knowledge of microbiome structure, function, and future applications. The microbiome research is gaining tremendous interest as documented by the explosion in publications with more than 20,000 articles published in 2020 alone. The review combines and critically discusses recent advances in the field and how these advances contributed to better understanding of structure and function of the microbiome. This understanding will lead to future promises of the microbiome therapeutics. We conclude with future directions and how to convert the basic science into translational medicine and developing of innovative microbiome-based therapy.
Multiple overlapping factors shape the microbiome composition. These factors include diet, stress, diseases, prescription and recreational drugs, smoking, drinking, aging, and others (Figure 1). Here, we discuss recent developments in revealing the implications of these factors and understanding the underlying mechanisms of how they modulate the microbiome.
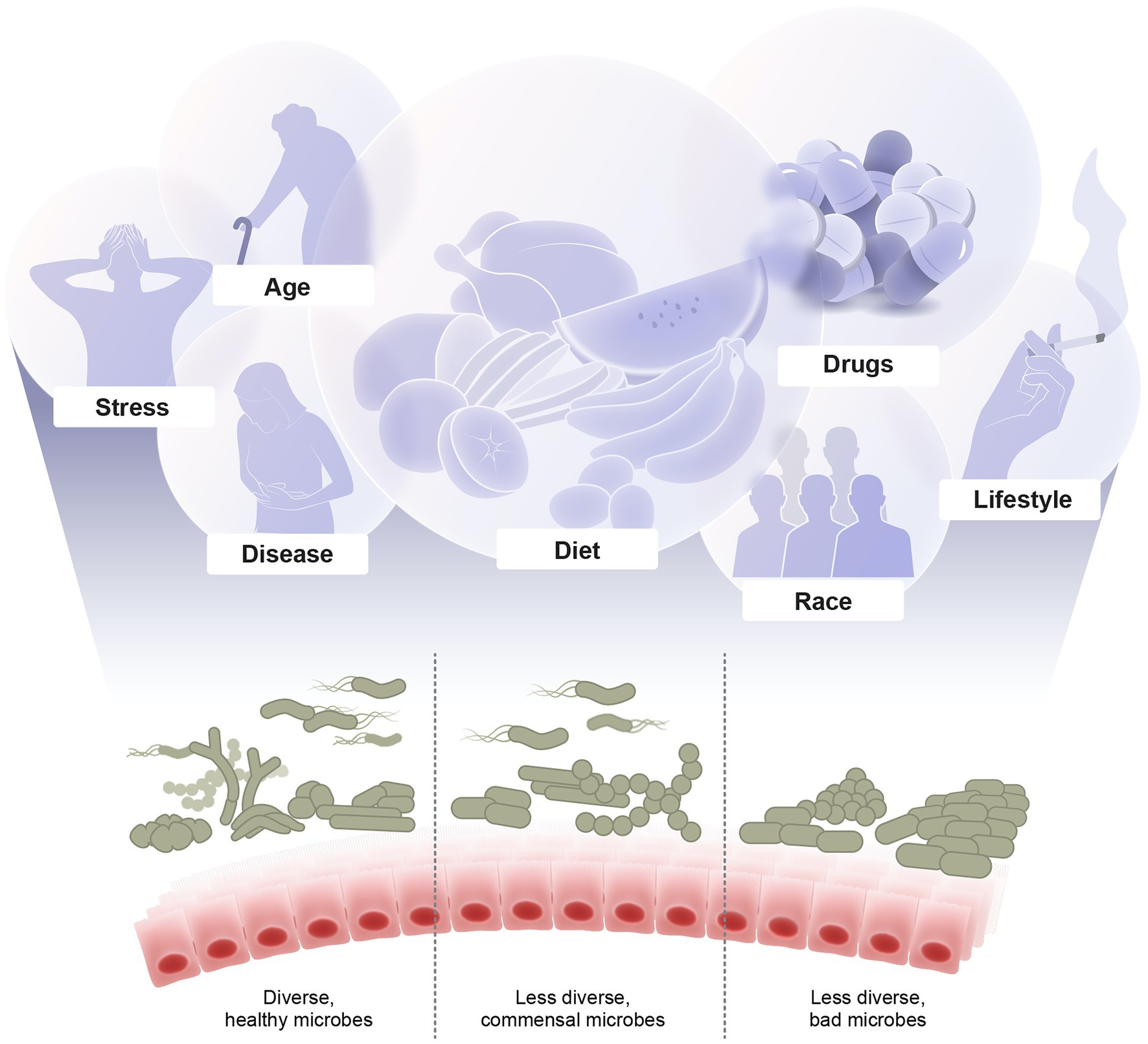
Figure 1. Major factors shaping the microbiome diversity. Illustrated are examples of multiple overlapping factors that shape the microbial composition including diet, stress, diseases, drugs, lifestyle, age, and host genetics. The outcome of these interacting factors are either healthy diverse, imbalanced, or dysbiotic microbiome which in turn affects the host health and diseases.
The interaction between diet, microbiome, and host is complex and multidirectional. Changing the diet regimen changes the microbiome composition. Different individuals’ microbiome responds differently to diet due to variations in food metabolism (Johnson et al., 2019). Similarly, different microbiome composition affects host metabolic capabilities. This bidirectional interaction further challenges our understanding of how the microbiome structure is shaped and the complexity of interacting factors from the external environment and host biology. Interestingly, changing the diet of immigrants from Asia to the United States is linked to an immediate and intense change in the microbiome structure with an impact on their health and development of obesity and its associated diseases (Vangay et al., 2018). The effect of diet on the onset and progression of certain diseases such as celiac disease and inflammatory bowel syndrome are now believed to be mediated by the microbiome and not a direct effect of the diet itself (Krishnareddy, 2019; Glassner et al., 2020). The beneficial effect of diet is also now thought to be mediated by the microbiome. For example, the health benefits of polyphenols are mediated through their promoting effect on Akkermansia muciniphila (Naito et al., 2018; Jayachandran et al., 2020). Similar links exist between dark chocolate and the microbiome structure. Dark chocolate is known for its anti-inflammatory properties due to its high content of antioxidant polyphenols (Magrone et al., 2017). A study showed that these antioxidants are poorly absorbed but could be fermented by colon bacteria such as Bifidobacterium into smaller polyphenolic polymers with anti-inflammatory properties that decrease cardiovascular inflammation (Wiese et al., 2019).
Evidence is mounting for the effect of diet on modulating the microbiome diversity, which in turn modulates the immune function and metabolic reactions. Fermented food such as kimchi, kombucha tea, and kefir shows positive enhancement of microbiome diversity associated with a significant decrease in inflammation biomarkers. Interestingly the affected biomarkers such as interleukin 6 are implicated in other diseases beyond the gastrointestinal tract (GIT) such as chronic stress, rheumatoid arthritis, and diabetes type 2 (Lau et al., 2021; Wastyk et al., 2021). This effect is observed in all participants of the cohort after only 10 weeks of incorporating fermented food into their diet. The uniqueness of this study is suggesting that only one change in the diet, and for a very short time, leads to a significant positive change in microbiome structure without the need to remodel the entire diet regime. An outstanding question regarding the mechanistic underpinning of the variation in response to different food remains.
In contrast to fermented food, the high fiber diet does not change the microbiome structure at the same time frame (Lau et al., 2021; Wastyk et al., 2021). One possible mechanism might be the lack of fiber degrading microbes. One of the well-known roles of gut microbiota is their ability to ferment indigestible dietary fibers to produce SCFAs. Another study shows that the incorporation of dietary fibers into the diet for 2 weeks only changes the microbiome composition in 20% of the studied subjects (Johnson et al., 2021; Lau et al., 2021). This observed effect is dependent on the original microbiome composition and the amount of fiber consumed (Johnson et al., 2021; Lau et al., 2021). The responder’s microbiome is enriched in Prevotella copri and shows less abundance of Bacteroides. These studies give us a new insight into the bidirectional interaction between dietary fiber and the microbiome. Curiously, we question whether adding probiotics will enable the existing microbiome to better benefit from dietary fibers or combining different food items can modulate microbiome response. In post-menopausal women, a very low-calorie diet, consisting of oats, greek yogurt, or fish, is associated with increases in Akkermansia sp., known for its ability to metabolize host glycan, and decreases in genera such as Roseburia, Ruminococcus, and Eubacterium. This microbiome shift is associated with high levels of SCFAs. Fecal transplant of this altered microbiome to germ-free (GF) mice resulted in the overabundance of the pathogen Clostridium difficile (von Schwartzenberg et al., 2021).
Similar to diet, nutrient supplements such as vitamins and minerals affect the microbiome composition (Merlen, 1986; Zimmer et al., 2012). In mice, low iron intake increases specific taxa such as Lachnospiraceae, Ruminococcaceae, and Rikenellaceae. These taxa are thought to regulate gut transport pathways to retain iron and decrease its excretion to maintain the iron level in the blood (Coe et al., 2021; Lau et al., 2021). Interestingly, Lactobacillus can sense iron deprivation and communicate the iron need to a murine host via the production of small-molecule mediators that interact with a hypoxia-inducible factor, HIF-2a (Das et al., 2020). Among these small molecules, reuterin and 1,3-diaminopropane (DAP) are shown to suppress intestinal HIF-2a activity both in vitro and in vivo. Specifically, reuterin and DAP prevent HIF-2a dimerization with aryl hydrocarbon receptor nuclear translocator (ARNT). It is interesting to speculate that iron deficiency selectively favors the growth of microbes and metabolites that exert high levels of HIF-2a inhibition. Microbial metabolites are known to affect the stability of HIF-2a, which in turns affects the intestinal hemostasias and gut barrier functions as comprehensively reviewed (Kumar et al., 2020). The microbiome can promote cellular iron storage via the induction of an iron storage protein, FTN, which also increases iron absorption (Vanoaica et al., 2010). It is still unclear though how microbes and their host compete during times of iron scarcity. Other studies show that high Na+ intake linked to hypertension alters gut microbial composition in mice increasing the relative abundance of several bacterial taxa such as Prevotella and Bacteroides. Prevotellaceae is linked to metabolic syndrome and chronic inflammation (Barbaro et al., 2017; Ferguson et al., 2019; Van Beusecum et al., 2019). High salt intake stimulates the production of pro-inflammatory markers such as IL-17, and, coupled with a defect in the regulation of natriuresis, which is associated with hypertension. Transplanting gut microbiomes from hypertensive subjects increases blood pressure in GF recipient mice, suggesting a causal role of the gut microbiome in the development of Na+ induced hypertension (Elijovich et al., 2020).
Stress remodels the gut microbiota and hence affects disease susceptibility (Kelly et al., 2015; Zijlmans et al., 2015). Studies show that social stress exposure decreases the abundance of microbes with anti-inflammatory activity such as para Bacteroides taxa (Maltz et al., 2018, 2019), which in turn decreases microbial anti-inflammatory metabolites such as SCFAs and contribute to a higher level of inflammation. Combined stress and infection or other inflammatory diseases worsen the outcome of the disease compared to non-stressed subjects. Consuming microbes known for anti-inflammatory activity might be beneficial for people with anxiety disorder and unmanageable stress levels.
Some studies found that depressed patients possess a higher level of Bacteroidales and a lower level of Lachnospiraceae (Naseribafrouei et al., 2014). Another study shows increased levels of Enterobacteriaceae and Alistipes in depressed patients (Jiang et al., 2015a). High stress during pregnancy is also associated with a change in vaginal Lactobacillus (Jašarević et al., 2015a). The consequences are also extended to the newborn babies from high-stress mothers, who show a higher abundance of Proteobacteria and lower abundances of Lactobacillus and Bifidobacteria (Zijlmans et al., 2015). This altered microbiome in infants results in a series of pathological conditions such as gut inflammation and the development of allergies (Zijlmans et al., 2015). Mouse offspring exposed to maternal stress had decreased levels of Bifidobacteria and Lactobacilli, associated with decreased cognitive functioning and increased anxiety (Golubeva et al., 2015; Jašarević et al., 2015b). Exposure to stressors can significantly change the microbial populations in the gastrointestinal tract of both humans and laboratory animals. These microbiota changes are more pronounced several days after exposure to a stressor than on the day of or the day after the exposure. In mice, exposure to stress results in microbiota shifts that are characterized by a lower abundance of Porphyromonadaceae, and increases in aerobic bacteria (Bailey et al., 2010). Stressed mice exhibited greater susceptibility to infections with the pathogen Citrobacter rodentium. Stressed mice show higher levels of tumor necrosis factor (TNF) and suppression of the anti-inflammatory cytokine IL-10 before the pathogen challenge, suggesting an immunomodulatory effect of dysbiosis microbiota, which renders the host more susceptible to infection (Bailey et al., 2010). Stress-induced inflammation affects the integrity of the tight junction resulting in leakage of molecules through gastrointestinal barriers and translocation of gut bacteria to the liver and spleen (Moussaoui et al., 2014). Probiotic strains such as Lactobacillus helveticus R0052, Bifidobacterium longum R0175 (Ait-Belgnaoui et al., 2012), and Lactobacillus farciminis (Ait-Belgnaoui et al., 2012) show promise in the treatment of stress-related disorders in mice.
It is commonly accepted that antibiotics, in a particularly broad-spectrum, alter the composition of gut microbiota which can last for months and even a year (Looft and Allen, 2012; Konstantinidis et al., 2020). Research shows that other non-antibiotics can also affect the composition and diversity of gut microbiota in population-based cohorts (Nichols et al., 2019; Papazafiropoulou, 2019). Drugs such as oral steroids, platelet aggregation inhibitors, antidepressants, and vitamin D supplements increase the abundance of Streptococcus salivarius (Vila et al., 2020; Weersma et al., 2020). Benzodiazepine was found to be associated with an increase in the abundance of Haemophilus parainfluenzae, a bacterium that has been reported to be more common in patients with Irritable Bowel Syndrome (Vila et al., 2020). Proton pump inhibitors selectively increase the abundances of Bifidobacterium dentium salivarius (Vila et al., 2020). SSRI antidepressants increase the abundance of Eubacterium ramulus, while tricyclic antidepressant increase the abundances of Clostridium leptum (Vila et al., 2020). Laxatives enrich Alistipes and Bacteroides species (Vila et al., 2020; Weersma et al., 2020). Steroids increase the abundance of Streptococcus mutans and B. dentium.
Addictive substances implicated in substance use disorder (SUD), such as opiates, cocaine, psychostimulants, and alcohol, also modulate the microbiome (Angoa-Pérez and Kuhn, 2021). Almost all research on SUD has primarily focused on the underlying CNS elements and ignored the microbiome link. Given that SUD affects millions every year, it is of intriguing importance to shed a light on the modulatory effect of abuse drugs on the gut microbiome. This research will help to understand subsequent consequences on the health and diseases of the host and to develop better therapeutics to tackle the SUD crisis. Some nutrients such as sugar and fats can exert an addictive effect like drugs of abuse. The similarity includes the development of dependence and withdrawal behavior and induction of dopamine release (Volkow et al., 2011). However, it is not known yet if the effect of diet on dopamine and brain reward mechanism is mediated by a change in the microbiome composition, which affects the gut-brain axis. Data collected from studying the effect of alcohol consumption on experimental animals show that alcohol can alter gut microbiota composition not only in high chronic doses but also even with mild and moderate consumption (Barr et al., 2018). Similar to stress, alcohol consumption can also increase intestinal permeability, alter the microbiome, and increase depression, these conditions are improved after withdrawal (Kelly et al., 2015).
Host genetics are the least understood factor in shaping the microbiota (Gaulke and Sharpton, 2018). Some studies show that genetic mutation results in atypical microbiota collection in mice (Buffington et al., 2021; Lau et al., 2021). A comparative analysis on 73 Argentinian participants living in United States and United Kingdom showed that the microbiome composition of obese patients showed a lower abundance of Porphyromonadaceae, Rikenellaceae, Bacteroidaceae, Tenericutes, and Verrucomicrobia. Other taxa such as Roseburia, Ruminococcus, Blautia producta, Eubacterium biforme, Clostridium lactatifermentans, and Ruminiclostridium showed higher abundance. While Lactobacillus remains unaltered. Interestingly, the gut microbiota of normal weight, overweight, and obese subjects clustered separately based on geography revealing a significant difference in their microbiota composition. For example, Ruminococcaceae taxa dominate samples collected in the United Kingdom, while Lachnospiraceae taxa are the most abundant taxa in samples collected from the United States. This microbial clustering results in a variation of the host metabolism and risk of developing diseases such as obesity (Lau et al., 2021; Pesoa et al., 2021). Residents of low-income countries show higher diversity in their microbiome compared to West countries. Interestingly, data show that immigrants from Southeast Asia to the United States lose 15% of their microbiome diversity immediately (Vangay et al., 2018). This loss of microbiome diversity is associated with the incidence of some diseases such as obesity and cardiovascular disorders. The loss of diversity might be due to a combination of factors such as the shift to a western diet with high calories, the drinking water, or the use of drugs and antibiotics (Vangay et al., 2018).
Understanding the development of microbiome profiles over time, as individuals age, remains an elusive challenge for researchers. In older populations especially, studies show associations between gut microbiome composition and physical fitness, diet, frailty, and mental capacity demonstrating the importance of a robust functioning microbiome (Lau et al., 2021; Wilmanski et al., 2021). Changes in the microbiome structure of older individuals have often been attributed to altered lifestyles, diets, reduced mobility, decreased immune function, reduced intestinal capability, changed gut morphology, increased use of medication and drugs, and recurrent infections (Kim and Benayoun, 2020). There remains some obscuring around the directions of causality in microbiome-aging interactions: are changes in gut microbial composition caused by reduced capability in older individuals, or are gut microbes the driving force behind symptoms of aging? The relationship between microbiome composition and aging is likely bi-directional, despite relationships of causality remaining ambiguous. However, patterns of association in the gut microbiomes of older individuals have been elucidated. Within three independent cohorts, comprising 9,000 individuals, researchers identified a pattern of healthy aging characterized by a depletion of core gut microbial taxa, namely Bacteroides, with healthier individuals having increasingly distinct microbiome compositions compared to other members within the study (Lau et al., 2021; Wilmanski et al., 2021). Another study, examining the microbiome compositions of “semi-supercentenarians,” individuals aged 105–109 years, found that aging is marked by increasing abundance of subdominant species, with decreases in dominant core microbiota within the Ruminococcaceae, Lachnospiraceae, and Bacteroidaceae families (Biagi et al., 2016). Semi-supercentenarians showed peculiarities in their microbiome compositions, indicating enrichment of health-associated bacterial groups such as Akkermansia and Bifidobacterium, which contributed to the maintenance of good health during aging. Another study examined the differences in the microbial composition of young adults, individuals in their seventies, and centenarians (Biagi et al., 2010). Results show that the microbiome composition of young adult and 70-year-old microbiomes are highly similar, while the centenarians experienced decreases in abundance of Faecalibacterium prausnitzii, with 10-fold increases in Eubacterium limosum. These shifts in microbial populations in centenarians are associated with increased inflammation. The social activity of elder peoples plays a role in shaping their microbiome composition. Elderly individuals living in long-term facilities showed a higher abundance of Bacteroidetes, Parabacteroides, Eubacterium, Anaerotruncus, Lactonifactor, and Coprobacillus. The elderly who has greater exposure to the outside communities showed a higher abundance of Firmicutes and Lachnospiraceae, which are associated with higher levels of SCFAs. High fiber diets are associated with more diverse microbiomes. One can postulate a link between the shift in microbial composition to age-related health deterioration. If proven true, this will open a very interesting avenue for microbiome-based therapeutics to slow down aging and related diseases (Claesson et al., 2012).
Lifestyle has a profound impact on microbiome composition and diversity. Lifestyle is a broad term that might include physical activity, smoking, drug abuse, and the surrounding environment. The level of physical activity, as well as the amount of exposure to pollutants, are all critical factors affecting microbiome diversity.
Exercise increases microbiome diversity (Dalton et al., 2019) and subsequently modulates the gut-brain access (Kim et al., 2018; Dalton et al., 2019). The microbiome of physically active individuals is more diverse with greater abundances of health-promoting bacteria including F. prausnitzii, Roseburia hominis, and A. muciniphila (Bressa et al., 2017). In young adults, a relationship between cardiorespiratory fitness and microbiota composition is also evident from the increased abundances of Firmicutes in fit young adults (Durk et al., 2019). A study wherein lean and obese individuals participated in 6 weeks of exercise training revealed that exercise induced a significant microbiome shift. This shift is characterized by higher abundances of SCFAs-producing bacteria such as Faecalibacterium and Lachnospiraceae and an increase in SCFAs in feces of non-obese participants (Allen et al., 2018).
Cigarette smoke is a complex mixture of more than 7,000 chemicals depositing directly to the lung and gut. These chemicals affect the microbiome composition increasing susceptibility to various diseases such as infections and inflammatory disorders (Rogers et al., 2012; Lee et al., 2018). Even E-cigarette significantly alter the oral microbiome with the increase in Veillonella and Haemophilus (Chopyk et al., 2021; Lau et al., 2021). Interestingly, the nose of smokers shows a higher colonization rate with Staphylococcus aureus in E-cigarette smokers (Chopyk et al., 2021; Lau et al., 2021). The gut microbiome of smokers is enriched in Bacteroides and depleted in Firmicutes and Proteobacteria when compared to non-smokers and past smokers (Lee et al., 2018). The microbiome of the small intestines shows a higher abundance of Firmicutes such as Veillonella and Streptococcus and a lower abundance of Prevotella and Neisseria. The abundance of Firmicutes is partially restored after stopping smoking (Shanahan et al., 2018). The oral microbiome is impacted by cigarette smoking with lower abundances of Proteobacteria, Capnocytophaga, Peptostreptococcus, and Leptotrichia and enrichment of Atopobium and Streptococcus (Wu et al., 2016). Depletions of bacterial taxa were associated with functional shifts in smokers’ oral microbiomes mainly the functions related to carbohydrate, energy expenditure, and xenobiotic metabolism. A study in mice shows that the change in microbiome extends to thirdhand smokers with a persistent effect on metabolism (He et al., 2021; Lau et al., 2021).
The environment alters the microbiome composition and subsequently mediates susceptibility to diseases (Littleford-Colquhoun et al., 2019). Westernization is linked to the decrease in microbiome diversity and increased incidence of diseases such as obesity and infectious diseases (Winglee et al., 2017). The difference in the environments includes multiple factors such as dietary culture, income, pollutions, and lifestyle. All of these variants change the microbiome. Young adults residing in Southern California, who are more exposed to atmospheric ozone, show lower gut microbial diversity with increased abundances of Bacteroides caecimuris (Fouladi et al., 2020). An interesting study profiled the microbiome from remains of ancient human feces and found a similarity to pre-industrial humans. Ancient microbiome shows higher abundances of Enterococcus, Treponema succinifaciens, and Ruminococcus champenellensis. These species might be linked to the consumption of complex carbohydrates (Lau et al., 2021; Wibowo et al., 2021). Humans in industrialized societies show microbiome enriched in A. muciniphila, Alisipes, and Bacteroides species. Industrialization is seemingly linked to higher abundances of antibiotic resistance genes compared to the microbiome of the pre-antibiotic era. A higher rate of horizontal gene transfer is also linked to the industrialization era, which might facilitate acquiring of new functions and adaptation to the change in the environment (Groussin et al., 2021; Lau et al., 2021). A pioneering study shows that the skin microbiome is altered in the urban population and this alteration changes the physiological response of the skin and is implicated in skin-related diseases (Lau et al., 2021; Wang et al., 2021c).
Recent advances in microbiome research deepen our understanding of microbiome function either in mediating or preventing diseases. Although lacking details of underpinning mechanisms, these studies still provide valuable insights on the depth and altitude of microbiome impact on our health and diseases. Methods used to analyze the microbiome are summarized (Table 1). Here, we discuss recent microbiome associations with diseases from inflammation to neurological and mental illness (Table 2; Figure 2).
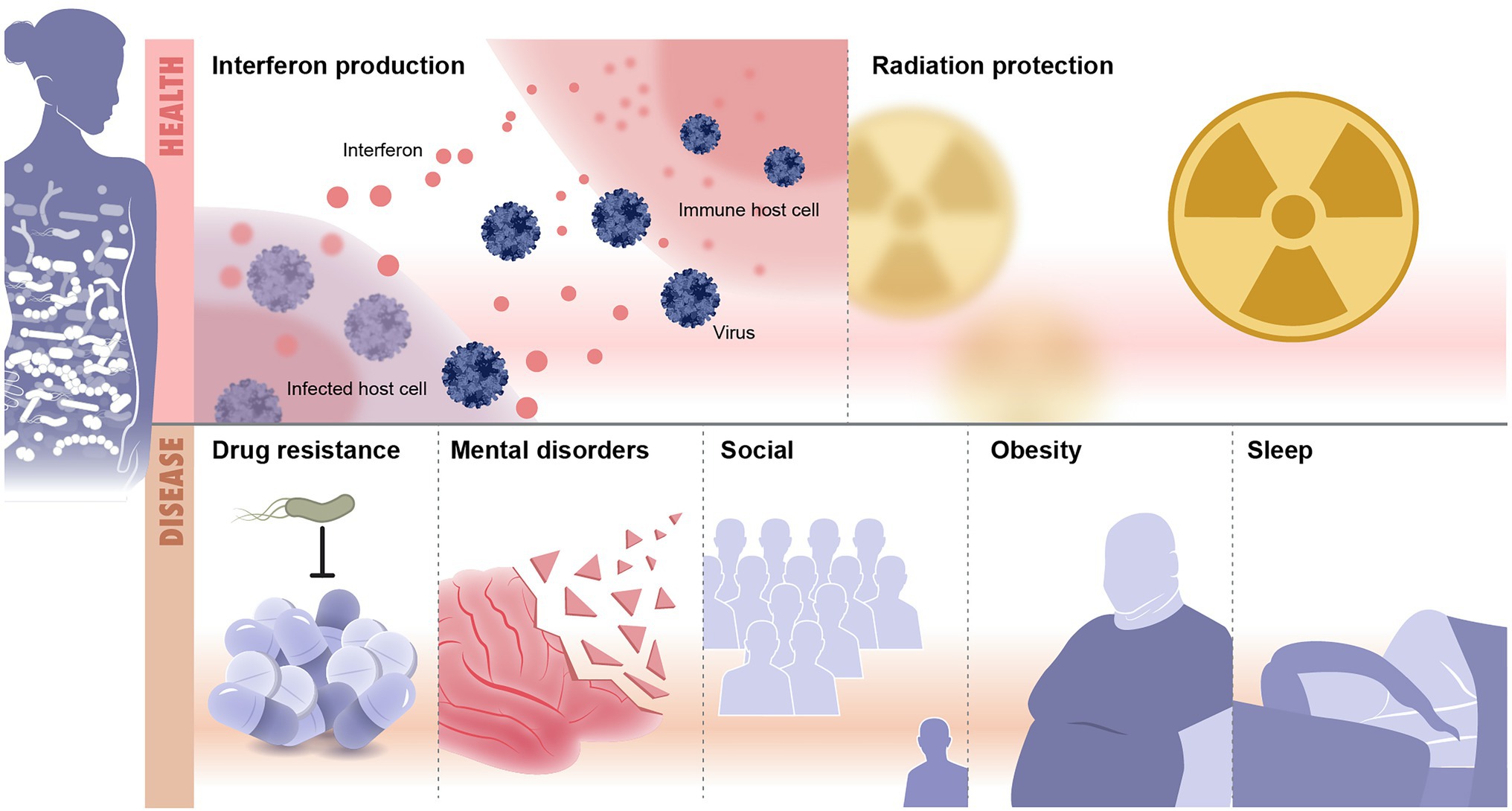
Figure 2. Microbiome and diseases association. Illustrated are examples of health and disease conditions that are linked to change in the microbiome. Health conditions associated with healthy microbiome include, as examples, protection from infections and radiation resistance. Diseases that result from imbalanced microbiomes are countless and include drug resistance, mental disorders, social diseases, obesity and metabolic syndrome, and sleep disorders.
The gut microbiome mediates mucosal immune response and directly implicates the inflammation process. Some microbiome species are known to produce SCFAs that decrease inflammation. Others are implicated in causing GIT inflammation that might progress into chronic conditions. Several review articles discussed the role of the microbiome in GIT inflammation and immune response (Abdulla et al., 2021; Sanders et al., 2021). Beyond GIT, inflammation is linked to intrauterine growth restriction (IUGR) in humans (Al-Azemi et al., 2017). However, there is no data available on the interplay between gut microbiota, proinflammatory cytokines, and IUGR (Fernandez-Gonzalez et al., 2020). Some data suggest that the administration of Lactobacillus strains improves glucose metabolism and decreases the risk of preeclampsia in pregnant women (Myhre et al., 2011). In livestock, high dietary fiber intake shows a significant decrease in inflammation and IUGR in pregnant animals (Liu et al., 2021). Identifying microbial strains that can improve pregnancy outcomes such as IUGR, pregnancy diabetes, preeclampsia, and other pregnancy complications is an important area of research that will inform advanced care regimens including the development of probiotics. Chronic inflammation is linked to the development of some cancers of the colon and pancreas.
Recent research demonstrates a link between dysbiosis in gut microbiota and cancer development (Ammer-Herrmenau et al., 2020; Lau et al., 2021; Matsukawa et al., 2021). Some microbes are more abundant in Pancreatic Ductal Adeno Carcinoma (PDAC) patients including Klebsiella pneumoniae, Clostridium bolteae, S. mutans, and Parabacteroides spp., Acinetobacter, and Pseudomonas (Thomas and Jobin, 2020; Lau et al., 2021; Matsukawa et al., 2021). The abundance of other microbes such as Firmicutes is significantly lower in PDAC patients (Lau et al., 2021; Zhou et al., 2021). Most PDAC patients suffer from a leaky gut which results in the translocation of harmful bacteria into the bloodstream (Wu et al., 2014). These bacteria can trigger an immune response through pathways that involve tumor-infiltrating lymphocytes (TILs) and their related cytokines, TLRs, innate immune cells, and others (Pagliari et al., 2018).TILs then produce pro inflammatory mediators that induce STAT3 and NF-kB pathways, which act as tumorigenic factors (Pagliari et al., 2018). This further increases cellular proliferation and suppresses apoptosis (Pagliari et al., 2018). Leaky gut is a potential possible mechanism for the development of PDAC. Fecal microbial transplant to overcome this immunosuppression might hold a future promise (Pitt et al., 2016).
Disturbance in the gut microbiota is associated with metabolic syndrome (Mazidi et al., 2016; Saklayen, 2018). Studies show that the gut microbial compositions in genetically obese mice are different compared to those lean and wild-type under the same polysaccharide-rich diet (Tseng and Chun-Ying, 2019). Obese mice show a reduced abundance of Bacteroidetes and enrichment of Firmicutes. A fecal transplant from obese mice to germ-free mice resulted in a significant increase in the total body fat (Turnbaugh et al., 2006). Obese individuals show higher concentrations of lipopolysaccharide (LPS) leaking from the intestinal mucosa (Lau et al., 2021; Zawada et al., 2021). LPS stimulates the secretion of the satiety hormone (the pancreatic peptide hormone, PYY), which slows down the motility of the digestive tract and thus affects food absorption (Lau et al., 2021; Zawada et al., 2021). Other studies show that obese individuals had a significantly high abundance of Firmicutes/Bacteroidetes ratio (Tseng and Chun-Ying, 2019; Lau et al., 2021; Palmas et al., 2021). Members of Firmicutes such as Ruminococcus gnavus increase the production of SCFAs increasing intestinal energy harvesting and hepatic lipogenesis after carbohydrate-rich diets (Lau et al., 2021; Palmas et al., 2021). Some probiotics such as Bifidobacterium animalis ssp. lactis and Lactobacillus gasseri show promise in decreasing inflammation and intestinal leakage (Tseng and Chun-Ying, 2019; Lau et al., 2021; Schütz et al., 2021).
Dysbiosis in the gut microbiome is associated with elevated blood pressure and consequently hypertension (Xu and Yang, 2020; Lau et al., 2021; Wang et al., 2021a). Hypertensive gut microbiomes had an increased abundance of opportunistic pathogens such as Klebsiella spp., Streptococcus spp., and Parabacteroides merdae (Yan et al., 2017). SCFAs producers such as Roseburia spp. and F. prausnitzii are lower in hypertensive patients (Yan et al., 2017). Previous studies showed a higher abundance of Klebsiella, Clostridium, and Streptococcus in patients with hypertension, which are known for their role as choline degraders (Hakenbeck et al., 2009; Craciun and Balskus, 2012; Kalnins et al., 2015). These patients also show overexpression of the choline utilization gene, cutC gene. The results suggest that choline intake and TMAO production might serve as diagnostic biomarkers for hypertension pathogenesis (Yan et al., 2017). Patients with coronary artery disease (CAD) showed an increase in Lactobacillales, and a decrease in Bacteroidetes (Yamashita et al., 2016), and a reduction in the abundance of Roseburia intestinalis and F. prausnitzii which are SCFAs producers (Jie et al., 2017; Xu and Yang, 2020). Reduction in secreted SCFAs leads to higher inflammation in CAD. In addition, the elevated gut permeability leads to an increased level of leaked LPS, which is associated with chronic inflammation in patients with post-myocardial infarction (Chambers et al., 2018; Zhou et al., 2018).
Understanding the gut-lung axes is crucial to tackling lung disorders such as inflammatory and infectious diseases. The gut microbiome modulates the disease susceptibility of the lung and the lung microbiome alters the proinflammatory function of the gut microbiome (van der Lelie and Taghavi, 2020; de Oliveira et al., 2021; Lau et al., 2021). Since the start of the COVID-19 pandemic, microbiome scientists race to understand the role of the microbiome in disease development and diagnosis. Gut dysbiosis is associated with the translocation of bacteria into the blood during COVID-19. This potentially causes a secondary infection that might be life-threatening. The researchers observed an abundance of opportunistic pathogenic bacteria in hospitalized COVID-19 including antimicrobial-resistant species (Lau et al., 2021; Venzon et al., 2021). Leakage of bacteria into the bloodstream following dysbiosis is critical to control and can drive detrimental effects. COVID infection is associated with diarrhea in 40% of the patients, which in turn changes the GIT microbiome and induces inflammatory cytokines. Ruminococcus gnavus is positively correlated with inflammatory biomarkers (Villapol, 2020). Some gut microbiota is more enriched in COVID patients such as R. gnavus, Ruminococcus torques, and Bacteroides dorei (Lau et al., 2021; Yeoh et al., 2021). In-depth shotgun metagenomic analysis on fecal samples of COVID-19 patients revealed that Coprobacillus, Clostridium ramosum, and Clostridium hathewayi correlate with the severity of the COVID infection (Villapol, 2020). Dysbiosis in the gut microbiota is linked to several other lung disorders including pulmonary fibrosis, asthma, and cystic fibrosis. Pulmonary fibrosis patients show a higher abundance of some gut microbes including Alloprevotella, Dubosiella, Helicobacter, OIsenella, Parasutterella, Rikenella, and Rikenllaceae. This microbial enrichment is associated with an elevated level of some diseases biomarkers including trigonelline, betaine, cytosine, thymidine, glycerophosphocholine, taurocholate, adenine, deoxyadenosine, and deoxycytidine (Gong et al., 2021; Lau et al., 2021). Asthmatic kids show a significant reduction in Bifidobacteria and an increase in Clostridia (Anand and Mande, 2018). Some studies suggest that the lung microbiome and gut microbiome are linked. For example, an infection with the influenza virus in the respiratory tract of mice models increases the proportion of Enterobacteriaceae and reduces Lactobacilli and Lactococci in the gut microbiome (Looft and Allen, 2012; Enaud et al., 2020). The colonization of the gut by species of C. difficile at the age of 1 month is associated with asthma and wheezing at the age of 6–7 (van Nimwegen et al., 2011). A pioneering study revealed a significant alveolar microbiome signature associated with several lung diseases including interstitial lung diseases, chronic obstructive pulmonary diseases, and sarcoidosis (Gupta et al., 2021; Lau et al., 2021). Interestingly, the authors show a correlation between disease-specific microbiomes and lung normal flora (Gupta et al., 2021; Lau et al., 2021). Disease-specific microbiome is a promising target toward developing microbiome-based diagnostic biomarkers (Gupta et al., 2021; Lau et al., 2021). Further research is needed to understand the mechanisms of action of this association, which will be significant in developing new microbiome therapeutics and diagnostic biomarkers.
The gut-brain axis is one of the hot topics in microbiome research (Salagre et al., 2017; Jang et al., 2020; Mayer et al., 2022). Microbiome associations are evident in many psychiatric and neurological disorders including anxiety, and depression.
The gut microbiome affects brain function early from the womb life. A study shows that maternal microbial alterations impact offspring brain maturation in mice (Buffington et al., 2016). Mice offspring that were fed high-fat diets were observed to have a 9-fold reduction in Lactobacillus reuteri and oxytocin-producing cells (Buffington et al., 2016). While mice offspring exposed to maternal stress show a low abundance of Bifidobacteria and Lactobacilli. This microbial shift is associated with a decrease in cognitive functioning and an increase in anxiety disorder (Golubeva et al., 2015). This study has further experimented in GF mice. GF mice showed anxious behavior and increased serotonin levels in the hippocampus compared to mice with normal flora. A genome-wide association study revealed 141 host genes and microbial taxa that are implicated in anxiety and depression in mice (Jin et al., 2021; Lau et al., 2021). The study shows that some of these host genes control the structure of the gut microbiome by modulating specific taxa and hence influence anxiety indirectly. The study suggests an interesting approach to consider both host and microbiome genes when assessing or treating anxiety (Jin et al., 2021; Lau et al., 2021). Microbiome taxa that are enriched in anxiety disorder include Ruminococcaceae, Clostridiaceae, and Clostridiales. While Bacteroidales and Bacteroidaceae showed lower abundance (Jin et al., 2021; Lau et al., 2021).
Research shows an association between depression and some microbial taxa such as Eggerthella, Subdoligranulum, Coprococcus, and Ruminococcaceae (Lau et al., 2021; Radjabzadeh et al., 2021). Some microbial species are involved in metabolic pathways linked to neurological functions such as glutamate, butyrate, serotonin, and gamma aminobutyric acid neurotransmitters (Lau et al., 2021; Radjabzadeh et al., 2021). These microbes include Lachnoclostridium, Sellimonas, Ruminococcaceae, Lachnospiraceae, Hungatella, Ruminococcus gavreauii, and Eubacterium ventriosum (Lau et al., 2021; Radjabzadeh et al., 2021). In another study, a microbiome analysis revealed that patients with major depressive disorders had an increased abundance of the genus Bacteroides and decreased abundance of Blautia and Eubacterium (Lau et al., 2021; Yang et al., 2021). Bacteroides species are also known for their regulation of metabolic pathways and the immune system (Maier et al., 2015). Bacteroides modulate cytokines production and increase inflammation (Schiepers et al., 2005). In contrast, Balutia species mediates beneficial anti-inflammatory effects (Bajaj et al., 2012). Another study showed that Clostridiales and Desulfovibrionaceae are enriched in depressed patients, while Bacteroidaceae show low abundance (Lau et al., 2021; Rhee et al., 2021). However, we still lack information about the directionality of this microbiome association and the underlying mechanism.
The gut microbiome composition is linked to sleep behavior (Maki et al., 2020). Disrupted sleep is linked to dysregulation in the immune system which leads to abnormal increases in inflammatory responses (Irwin, 2019). Sleep dysfunction can promote inflammation via two processes. First is through the upregulation of proinflammatory cytokines such as IL-6, tumor necrosis factor α (TNFα), and CRP (Matenchuk et al., 2020). The second is through the change in microbiome composition and function. In mice with sleep fragmentation (SF), the relative abundance of Firmicutes and the Firmicutes/Bacteroidetes ratio is reduced accompanying a decrease in butyrate-producing bacteria (Maki et al., 2020). Other studies found differences in Bacteroidetes, Firmicutes, Lachnospiraceae, or Blautia between short and normal sleepers (Anderson et al., 2017; Smith et al., 2019; Grosicki et al., 2020). Short sleepers showed a significantly lower abundance of Sutterella and a higher abundance of Pseudomonas (Agrawal et al., 2021; Lau et al., 2021). In a similar study, sleep-disturbed mice showed a shift in both the microbiome and metabolome. Firmicutes/Bacteroidetes ratio increased in the sleep-disrupted group along with a decrease in the Lactobacillus, Actinobacteria, and Bifidobacterium in comparison to control mice (Bowers et al., 2020). Studies investigated the role of probiotics in improving sleep with some promising preliminary results. For example, L. gasseri CP2305 shows some potential to improve the quality of sleep (Nishida et al., 2017; Matenchuk et al., 2020).
Fear is an emotional and behavioral response induced by a threat. Fear is considered an evolutionarily conserved mechanism that promotes survival (Carlson et al., 2021; Lau et al., 2021). A recent study found that babies with less balanced gut microbiomes show an increase in fear behavior in comparison to those with more balanced microbiomes (Carlson et al., 2021; Lau et al., 2021). Gut microbiome communities dominated by Bacteroides at the first year of age are associated with less non-social fear, while the lower abundance of Bacteroides and increased abundance of Veillonella, Dialister, Bifidobacterium, and Lactobacillus were linked to increased fear behavior (Carlson et al., 2021; Lau et al., 2021). In addition, the researchers found an association between the microbiome and the medial prefrontal cortex volume and amygdala volume at the first year of age. Infants who had an increased fear activity also had larger amygdala volumes (Carlson et al., 2021; Lau et al., 2021). Cutting the vagus nerve did not have any effect on the ability to extinguish fear in mice (Chu et al., 2019). The immune cells are the same in germ-free mice and antibiotic-treated mice. The fear response was associated with a reduction in some microbial metabolites in the cerebrospinal fluid, blood serum, and feces of GF mice. These metabolites include phenyl sulfate, pyrocatechol sulfate, and indoxyl sulfate (Chu et al., 2019). These findings suggest that microbiome metabolites might play a role in influencing brain activity, which then alters how mice extinguish fear memory. Administration of L. helveticus NS8 probiotic shows anxiolytic and antidepressant effects in rats with a restored level of corticosterone (Liang et al., 2015). This indicates that L. helveticus NS8 probiotics have a role in regulating the hypothalamic-pituitary adrenocortical (HPA) activation axis (Liang et al., 2015).
Children with autism spectrum disorder (ASD) show differential gut microbiome composition (Wang et al., 2011; Coury et al., 2012; Chaidez et al., 2014; Kheirouri et al., 2016). The microbiome in ASD shows a decrease in the abundance of Escherichia, Shigella, Veillonella, Akkermansia, Providencia, Dialister, Bifidobacterium, Streptococcus, Ruminococcaceae, Megasphaera, Eubacterium_coprostanol, Citrobacter, Ruminiclostridium, and Ruminiclostridium, while Eisenbergiella, Klebsiella, Faecalibacterium, and Blautia are significantly increased (Lau et al., 2021; Ye et al., 2021). Similarly, another research reveals that patients with ASD exhibit a lower Bacteroidetes/Firmicutes ratio (Kang et al., 2013). This alteration in the gut microbiome of ASD patients might be of diagnostic value for early detection of the disease. Children with ASD and gastrointestinal symptoms show higher levels of gut immune inflammation and gut dysbiosis (Hughes et al., 2018; Liu et al., 2021). Probiotics can reduce gut inflammation decreasing gut leakage, and downregulate inflammatory cytokines (Ng et al., 2019). The use of probiotics containing Lactobacillus and Bifidobacteria strains has been shown to ameliorate Gastrointestinal symptoms (Zhu et al., 2020). Another study showed that administration of L. reuteri improves social behaviors in autism model mice through signaling mechanisms via a nerve from the gut to the brain (Poutahidis et al., 2013; Sgritta et al., 2019).
Neurodegenerative diseases are characterized by the formation of amyloid plaque due to the aggregation of misfolded proteins in the neurons. To study the link between gut microbes and protein aggregation, Caenorhabditis elegans is used as a model organism. A study demonstrates that colonization of C. elegans gut with bacterial pathogens disrupted the intestines, muscles, and neurons and increased protein aggregation (Lau et al., 2021; Walker et al., 2021). Further investigation revealed that co-colonization with butyrogenic bacteria inhibited the aggregation of protein in C. elegans indicating that enteric bacteria play a role in the pathogenesis of neurodegenerative diseases such as Alzheimer’s disease (Lau et al., 2021; Walker et al., 2021). Other studies show that SCFAs production is altered in patients with neurodegenerative disease (Lau et al., 2021; Tran and Mohajeri, 2021). These metabolites might induce changes in the brain suppressing inflammation and cytokines production (Zhu et al., 2020). Emerging research investigates the use of probiotics as a therapeutic tool to suppress symptoms and delay the progression of Alzheimer’s (Peterson, 2020). Early results show that Lactobacillus, Bifidobacterium breve, and B. longum administration improved memory and learning dysfunction in animal models and patients with Alzheimer’s (O’Sullivan et al., 2011; Savignac et al., 2013; Peterson, 2020; Zhu et al., 2020). Microbiome composition is different in patients with dementia disorders compared to healthy populations. A study shows that Clostridiales, Lactobacillus, and Bacteroidales are enriched in patients (He et al., 2018a). This altered microbial composition is associated with an elevated high level of SCFAs and choline, the latter is a biomarker for membrane dysfunction (He et al., 2018b). Another study shows a significant reduction in Bacteroides and enrichment of Lactobacillales and Bifidobacterium in dementia patients (Saji et al., 2019). A possible mechanism for this observed link between gut microbiome composition and brain function might be the leaky gut. The leakage of harmful bacteria into the bloodstream affects mental health through immune regulation and oxidative stress (Belizário and Faintuch, 2018). Administration of probiotics might restore function or mediate symptoms of neurodegenerative diseases. Some preliminary data show that probiotics can improve cognitive function improving learning and memory in animal models (Lau et al., 2021; Ruiz-Gonzalez et al., 2021). However, future studies are necessary to understand the underlying mechanisms before proceeding to clinical trials in humans.
Commensal microbes are known to metabolize indigestible materials, defend against colonization of pathogens, and stimulate the immune system (Martín et al., 2013). Little is known about microbiome secreted molecules that drive the microbiome function and disease associations. Microbiome chemistry is what mediates microbe-host interaction and further disease susceptibility. Here, we discuss and highlight some microbiome known chemistry (Figure 3) and the future promise to develop microbiome-based therapeutics.
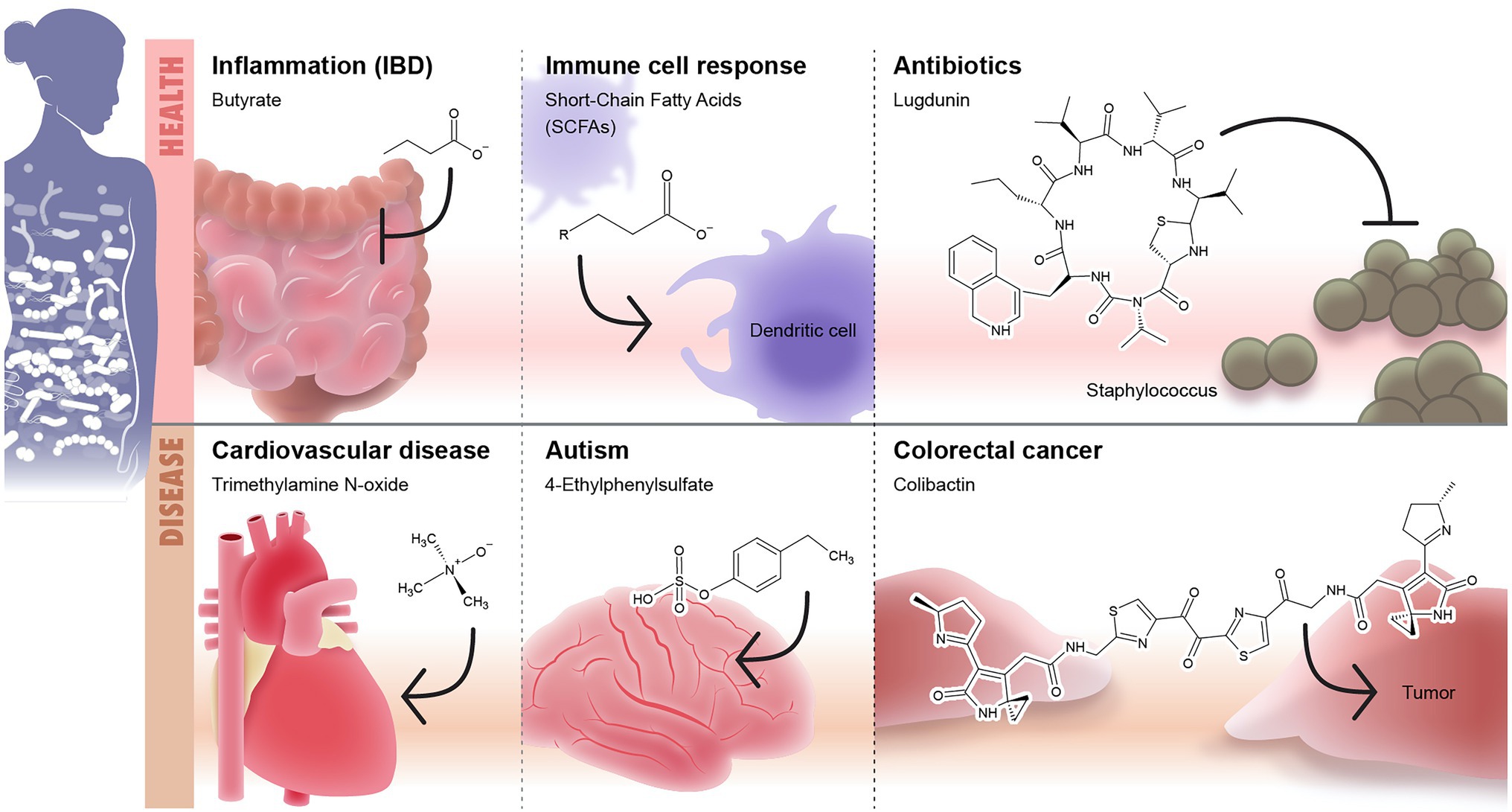
Figure 3. Microbiome-secreted molecules and their effect on human health and diseases. The first panel of the Illustration shows some examples of well-defined secreted molecules that affects human health including (1) short-chain fatty acids (SCFAs) such as butyrate which play anti-inflammatory role and modulate the intestinal immunity and (2) lugdunin as an example to microbiome-based antibiotic produced by nose microbiome and target Staphyloccous. The second panel shows examples of microbiome-based metabolites that are associated with onset or development of diseases including: (1) trimethylamine N-oxide (TMAO)/cardiovascular diseases, (2) 4-ethylphenylsulphate/autism, and (3) colibactin/colorectal cancer.
The gut microbiome utilizes dietary precursors such as choline, phosphatidylcholine, and L-carnitine to produce trimethylamine N-oxide (TMAO), which is a mediator between intestinal dysbiosis and vascular pathology (Lau et al., 2021; Sikora et al., 2021). TMAO mediates effective attraction between lipid membranes by partitioning unevenly between bulk and lipid domains (Sukenik et al., 2017). If there is damage in the intestinal barrier such in chronic psoriasis patients (Sikora et al., 2018, 2019a,b), TMA leaks into the systemic circulation and get transformed by hepatic enzymes into TMAO, which triggers a strong inflammatory response through activation of mitogen-activated protein kinase (MAPK) and subsequent cardiovascular diseases. TAMO producers include Clostridiaceae and Peptostreptococcaceae (Yamashita et al., 2016). Other research suggests that TMAO precursors drive toxic effects on the cardiovascular system (Jaworska et al., 2019). TMAO-induced inflammation could be suppressed by antibiotics (Janeiro et al., 2018). Zhu et al. (2020) showed that the formation of TMAO in microbes could be inhibited by 3,3-dimethyl-1-butanol (DMB). TMAO is further involved in the development of cancers and neurological disorders (Janeiro et al., 2018; Gatarek and Kaluzna-Czaplinska, 2021). TMAO is also linked to atherosclerosis and inhibition of hepatic bile acid synthesis; although, the underlying mechanism is lacking (Stubbs et al., 2016; Gatarek and Kaluzna-Czaplinska, 2021).
Colibactin is a microbial product associated with DNA damage leading to colorectal cancer (CRC; Putze et al., 2009; Arthur et al., 2012; Dziubańska-Kusibab et al., 2020). Colibactin is biosynthesized by deacetylation of the inactive precursor, precolibactin, by the peptidase enzyme, CIbP (Dziubańska-Kusibab et al., 2020). Colibactin alkylates DNA through its unique chemistry that includes a cyclopropane ring conjugated to an α,β-unsaturated imine. This creates adenine–colibactin adducts and then DNA crosslinks (Wilson et al., 2019; Xue et al., 2019; Dziubańska-Kusibab et al., 2020). Research predicts that these lesions in the DNA lead to mutations that promote CRC development (Dziubańska-Kusibab et al., 2020). In another study, colibactin showed an association with bowel cancer (Pleguezuelos-Manzano et al., 2020). The human cells treated with colibactin had twice the rate of DNA damage compared to the control group with some mutations that were only found in the colibactin-exposed cells (Pleguezuelos-Manzano et al., 2020). There were two mutation types: small indel signatures called ID-pks and a single base substitution signature called SBS-pks (Pleguezuelos-Manzano et al., 2020). This is evidence of the significant role of colibactin in GIT malignancy.
The microbial metabolite 4-Ethylphenyl sulfate (4EPS) plays a role in the development of autism-related behavioral disorders in mice (Hsiao et al., 2013). Several species of Clostridium can produce the precursors of 4EPS which is 4-ethylphenol (Nicholson et al., 2012). Mice that are treated with 4EPS exhibited anxiety-like behavior suggesting that elevated levels of 4EPS cause ASD-related behaviors; thus reinforcing the notion that the connection between the gut and the brain might be associated with autism (Hsiao et al., 2013). In addition, researchers found that the blood of mice with autism symptoms had levels of 4EPS that were about 46 times higher than that of the control group (Hsiao et al., 2013). In a similar study, 4-EPS induced ASD-like behaviors (Zhu et al., 2020). A lower abundance of Bacteroides is linked to Autism (Hsiao et al., 2013). Introducing Bacteroides fragilis to mice with autism-like symptoms improved the symptoms (Hsiao et al., 2013) demonstrating that the probiotic treatment with B. fragilis might be helpful as a therapeutic intervention for autism. Phenylketonuria (PKU) is associated with autism development (Khemir et al., 2016). PKU is a metabolic disease in which the body cannot break down phenylalanine (Xiong et al., 2015; Blasco et al., 2017; Glinton and Elsea, 2019). An elevated level of phenylalanine and phenylpyruvate causes brain damage with a significant reduction in serotonin and dopamine (Glinton and Elsea, 2019), which might contribute to the development of autism if left untreated (Thompson et al., 1993; de Groot et al., 2010).
Gut commensals produce SCFAs that inhibit pro-inflammatory responses mediated by intestinal macrophages (Chang et al., 2014). SCFAs production is altered in patients suffering from IBD (Zhuang et al., 2019). Butyrate is an example of SCFAs produced mainly by Firmicutes. Butyrate modulates the inflammatory immune response of intestinal macrophages. SCFAs modulate inflammation through multiple pathways include: (1) inhibiting adenylate cyclase which reduces the secondary messenger, cAMP (Houslay and Milligan, 1997; He et al., 2020), (2) activation of MAPK leading to an increase in Ca2+ concentration (Kimura et al., 2014; He et al., 2020), (3) stimulation of the release of anti-inflammatory interleukin 10 (IL-10) from the regulatory T cells, Tregs (Smith et al., 2013; He et al., 2020), (4) suppression of the expression of IL-6, IL-1β, and TNFα (Nakajima et al., 2017; Pirozzi et al., 2018; Mizuta et al., 2019; He et al., 2020), and (5) inhibiting histone deacetylase and downregulate lipopolysaccharide-induced pro-inflammatory mediators such as nitric oxide, IL-6, and IL-12 (Chang et al., 2014). Recent research shows that SCFAs bind to immune cell receptors in the respiratory tract and enhance lung antiviral response during infection with COVID-19 (de Oliveira et al., 2021; Lau et al., 2021). Beyond GIT, SCFAs are associated with changes in circulating immune cells and biomarkers, which are implicated in the development of multiple sclerosis (MS; Lau et al., 2021; Trend et al., 2021).
A common trait for any polymicrobial ecosystem is the production of antibiotics as an ecological fitness strategy to compete, survive, and thrive. The human microbiota is no different, dozens of antimicrobial compounds have been reported from the microbiome. Some examples of recently discovered antibiotics from the microbiome include Lugdunin, Lactocillin, cereulide, zwittermicin, tilivalline, and others (Table 3).
Lugdunin is a thiazolidine-containing cyclic peptide produced by Staphylococcus lugdunensis, a commensal of the human nose (Konai et al., 2020). Lugdunin is active against both methicillin-resistant S. aureus and vancomycin-resistant enterococci (Konai et al., 2020). Lugdunin also has an immunomodulatory activity (Bitschar et al., 2019). Pretreatment of primary human keratinocytes or mouse skin with the antimicrobial lugdunin resulted in a significant reduction of S. aureus colonization (Bitschar et al., 2019). Lugdunin increases the expression and release of LL-37 and CXCL8/MIP-2 in human keratinocytes and mouse skin resulting in the release of monocytes and neutrophils (Bitschar et al., 2019). Lactocillin is another thiopeptide antibiotic produced by L. gasseri, a commensal of the vaginal microbiome (Vásquez et al., 2002). Lactocillin is active against S. aureus, Enterococcus faecalis, Gardnerella vaginalis, and Corynebacterium aurimucosum (Donia et al., 2014).
Much interest exists in the potential of the evolved functions of the microbiome. A pioneering study aimed to computationally predict the function of microbes on earth estimates the presence of 35.5 million functions in bacteria of which only 0.02% are known (Starke et al., 2020). Despite the exploding body of research on the microbiome, our knowledge of its function and especially how it mediates health and diseases is still preliminary. The microbiome function is dependent on its structure and diversity, which is highly unique among individuals as shaped by multifactor. More dive into the individual’s unique microbiome might be a path to a personalized medicine approach. The drastic change in the microbiome of immigrants and the associated health consequences clearly demonstrates the structure and function dependency. Our knowledge of the change in microbiome composition with the onset or progression of diseases is based on association studies with a little dig into the mechanistic underpinning. The main concern in these studies is the lack of directionality of the microbiome disease relationship and the presence of other confounding factors. Recent research suggests that microbiome change in autism might be due to the picky eating habits restricting diet which in turn changes the microbiome composition (Lau et al., 2021; Yap et al., 2021). Another concern is that most association studies examine the change in the dominant microbial taxa, while recent studies show that rare microbes are the drivers of diseases. The use of microbiomes as probiotics or fecal transplant shows promise but with many challenges. Safety concerns are among the main challenges. One of the most studied and commercially available probiotic strains is Escherichia coli Nissle 1917 was found later to encode colibactin biosynthetic gene cluster implicated in CRC (Olier et al., 2012; Dziubańska-Kusibab et al., 2020). The second challenge is the efficacy and wide variation in response between individuals. In addition, introducing new microbes to an already established microbiome community comes with unpredicted outcomes include clearance from the body and failure to survive. Introducing the entire microbiome as in fecal transplant is another approach showing some efficacy especially in controlling recurrent C. difficile infection but with a complex and unpredictable outcome (Kazemian et al., 2020). The undesirable outcomes include the transfer of antibiotic resistance microbes (DeFilipp et al., 2019) and weight gain (Alang and Kelly, 2015). Recently the FDA issued a warning against the use of fecal transplant after two patients retracted antibiotic-resistant infections following administration of fecal transplant. A pioneering approach is designing of synthetic microbiome community with reproducible and controlled structure using an informatics approach (Clark et al., 2021; Lau et al., 2021). A synthetic microbiome community is drafted to supply a specific missing function such as butyrate synthesis for the treatment of inflammation (Clark et al., 2021; Lau et al., 2021). Personalized medicine based on microbiome signature of each individual is a direction that we should consider moving forward (Yadav et al., 2022). Microbiome signature could be used to assess diseases severity and prognosis, predict drug resistance or response rate, or even more importantly in diseases prevention (Behrouzi et al., 2019; Yadav et al., 2022). Certainly, the research on microbiome is exploding and the future of microbiome use in translation medicine is blooming.
FC and SH reviewed the literature, collected the data, contributed to writing, and developed the tables. WM reviewed the literature, collected the data, designed and developed the figures, wrote, and edited the manuscript. All authors contributed to the article and approved the submitted version.
This work is supported by a startup fund from Whitman College and funding from Murdock foundation.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdulla, M.-H., Agarwal, D., Singh, J. K., Traiki, T. B., Pandey, M. K., Ahmad, R., et al. (2021). Association of the microbiome with colorectal cancer development (review). Int. J. Oncol. 58:17. doi: 10.3892/ijo.2021.5197
Abee, T., Klaenhammer, T. R., and Letellier, L. (1994). Kinetic studies of the action of lactacin F, a bacteriocin produced by Lactobacillus johnsonii that forms poration complexes in the cytoplasmic membrane. Appl. Environ. Microbiol. 60, 1006–1013. doi: 10.1128/aem.60.3.1006-1013.1994
Agrawal, R., Ajami, N. J., Malhotra, S., Chen, L., White, D. L., Sharafkhaneh, A., et al. (2021). Habitual sleep duration and the colonic mucosa-associated gut microbiota in humans—a pilot study. Clocks Sleep 3, 387–397. doi: 10.3390/clockssleep3030025
Ait-Belgnaoui, A., Durand, H., Cartier, C., Chaumaz, G., Eutamene, H., Ferrier, L., et al. (2012). Prevention of gut leakiness by a probiotic treatment leads to attenuated HPA response to an acute psychological stress in rats. Psychoneuroendocrinology 37, 1885–1895. doi: 10.1016/j.psyneuen.2012.03.024
Al-Azemi, M., Raghupathy, R., and Azizieh, F. (2017). Pro-inflammatory and anti-inflammatory cytokine profiles in fetal growth restriction. Clin. Exp. Obstet. Gynecol. 44, 98–103. doi: 10.12891/ceog3295.2017
Alang, N., and Kelly, C. R. (2015). Weight gain after fecal microbiota transplantation. Open Forum Infect. Dis. 2:ofv004. doi: 10.1093/ofid/ofv004
Allen, J. M., Mailing, L. J., Niemiro, G. M., Moore, R., Cook, M. D., White, B. A., et al. (2018). Exercise alters gut microbiota composition and function in lean and obese humans. Med. Sci. Sports Exerc. 50, 747–757. doi: 10.1249/MSS.0000000000001495
Ammer-Herrmenau, C., Pfisterer, N., Weingarten, M. F. J., and Neesse, A. (2020). The microbiome in pancreatic diseases: recent advances and future perspectives. United European Gastroenterol. J. 8, 878–885. doi: 10.1177/2050640620944720
Anand, S., and Mande, S. S. (2018). Diet, microbiota and gut-lung connection. Front. Microbiol. 9:2147. doi: 10.3389/fmicb.2018.02147
Anderson, J. R., Ian Carroll, M., Azcarate-Peril, A., Rochette, A. D., Heinberg, L. J., Peat, C., et al. (2017). A preliminary examination of gut microbiota, sleep, and cognitive flexibility in healthy older adults. Sleep Med. 38, 104–107. doi: 10.1016/j.sleep.2017.07.018
Angoa-Pérez, M., and Kuhn, D. M. (2021). Evidence for modulation of substance use disorders by the gut microbiome: hidden in plain sight. Pharmacol. Rev. 73, 571–596. doi: 10.1124/pharmrev.120.000144
Arthur, J. C., Perez-Chanona, E., Mühlbauer, M., Tomkovich, S., Uronis, J. M., Fan, T.-J., et al. (2012). Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338, 120–123. doi: 10.1126/science.1224820
Bailey, M., Dowd, S., Parry, N., Galley, J., Schauer, D., and Lyte, M. (2010). Stressor exposure disrupts commensal microbial populations in the intestines and leads to increased colonization by Citrobacter rodentium. Infect. Immun. 78, 1509–1519. doi: 10.1128/IAI.00862-09
Bajaj, J. S., Hylemon, P. B., Ridlon, J. M., Heuman, D. M., Daita, K., White, M. B., et al. (2012). Colonic mucosal microbiome differs from stool microbiome in cirrhosis and hepatic encephalopathy and is linked to cognition and inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 303, G675–G685. doi: 10.1152/ajpgi.00152.2012
Barbaro, N. R., Foss, J. D., Kryshtal, D. O., Tsyba, N., Kumaresan, S., Xiao, L., et al. (2017). Dendritic cell amiloride-sensitive channels mediate sodium-induced inflammation and hypertension. Cell Rep. 21, 1009–1020. doi: 10.1016/j.celrep.2017.10.002
Barr, T., Sureshchandra, S., Ruegger, P., Zhang, J., Ma, W., Borneman, J., et al. (2018). Concurrent gut transcriptome and microbiota profiling following chronic ethanol consumption in nonhuman primates. Gut Microbes 9, 338–356. doi: 10.1080/19490976.2018.1441663
Behrouzi, A., Nafari, A. H., and Siadat, S. D. (2019). The significance of microbiome in personalized medicine. Clin. Transl. Med. 8:16. doi: 10.1186/s40169-019-0232-y
Beimfohr, C. (2016). A review of research conducted with probiotic E. coli marketed as symbioflor. Int. J. Bacteriol. 2016:3535621. doi: 10.1155/2016/3535621
Belizário, J. E., and Faintuch, J. (2018). Microbiome and gut dysbiosis. Exp. Supplement. 109, 459–476. doi: 10.1007/978-3-319-74932-7_13
Berg, G., Rybakova, D., Fischer, D., Cernava, T., Vergès, M.-C. C., Charles, T., et al. (2020). Microbiome definition re-visited: old concepts and new challenges. Microbiome 8:103. doi: 10.1186/s40168-020-00875-0
Beusecum, V., Justin, P., Barbaro, N. R., McDowell, Z., Aden, L. A., Xiao, L., et al. (2019). High salt activates CD11c+ antigen-presenting cells via SGK (serum glucocorticoid kinase) 1 to promote renal inflammation and salt-sensitive hypertension. Hypertension 74, 555–563. doi: 10.1161/HYPERTENSIONAHA.119.12761
Biagi, E., Franceschi, C., Rampelli, S., Severgnini, M., Ostan, R., Turroni, S., et al. (2016). Gut microbiota and extreme longevity. Curr. Biol. 26, 1480–1485. doi: 10.1016/j.cub.2016.04.016
Biagi, E., Nylund, L., Candela, M., Ostan, R., Bucci, L., Pini, E., et al. (2010). Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS One 5:e10667. doi: 10.1371/annotation/df45912f-d15c-44ab-8312-e7ec0607604d
Bitschar, K., Sauer, B., Focken, J., Dehmer, H., Moos, S., Konnerth, M., et al. (2019). Lugdunin amplifies innate immune responses in the skin in synergy with host- and microbiota-derived factors. Nat. Commun. 10:2730. doi: 10.1038/s41467-019-10646-7
Blasco, H., Veyrat-Durebex, C., Bertrand, M., Patin, F., Labarthe, F., Henique, H., et al. (2017). “A Multiplatform metabolomics approach to characterize plasma levels of phenylalanine and tyrosine in phenylketonuria,” in JIMD Reports. Vol. 32. eds. Morava E., Baumgartner M., Patterson M., Rahman S., Zschocke J., and Peters V. (Berlin, Heidelberg: Springer), 69–79.
Bowers, S. J., Vargas, F., González, A., He, S., Jiang, P., Dorrestein, P. C., et al. (2020). Repeated sleep disruption in mice leads to persistent shifts in the fecal microbiome and metabolome. PLoS One 15:e0229001. doi: 10.1371/journal.pone.0229001
Bressa, C., Bailén-Andrino, M., Pérez-Santiago, J., González-Soltero, R., Pérez, M., Montalvo-Lominchar, M. G., et al. (2017). Differences in gut microbiota profile between women with active lifestyle and sedentary women. PLoS One 12:e0171352. doi: 10.1371/journal.pone.0171352
Buffington, S. A., Di Prisco, G. V., Auchtung, T. A., Ajami, N. J., Petrosino, J. F., and Costa-Mattioli, M. (2016). Microbial reconstitution reverses maternal diet-induced social and synaptic deficits in offspring. Cell 165, 1762–1775. doi: 10.1016/j.cell.2016.06.001
Buffington, S. A., Dooling, S. W., Sgritta, M., Noecker, C., Murillo, O. D., Felice, D. F., et al. (2021). Dissecting the contribution of host genetics and the microbiome in complex behaviors. Cell 184, 1740.e16–1756.e16. doi: 10.1016/j.cell.2021.02.009
Bull, M., Plummer, S., Marchesi, J., and Mahenthiralingam, E. (2013). The life history of Lactobacillus acidophilus as a probiotic: a tale of revisionary taxonomy, misidentification and commercial success. FEMS Microbiol. Lett. 349, 77–87. doi: 10.1111/1574-6968.12293
Carlson, A. L., Kai Xia, M., Azcarate-Peril, A., Rosin, S. P., Fine, J. P., Wancen, M., et al. (2021). Infant gut microbiome composition is associated with non-social fear behavior in a pilot study. Nat. Commun. 12:3294. doi: 10.1038/s41467-021-23281-y
Cascales, E., Buchanan, S. K., Duché, D., Kleanthous, C., Lloubès, R., Postle, K., et al. (2007). Colicin biology. Microbiol. Mol. Biol. Rev. 71, 158–229. doi: 10.1128/MMBR.00036-06
Chaidez, V., Hansen, R. L., and Hertz-Picciotto, I. (2014). Gastrointestinal problems in children with autism, developmental delays or typical development. J. Autism Dev. Disord. 44, 1117–1127. doi: 10.1007/s10803-013-1973-x
Chambers, E. S., Preston, T., Frost, G., and Morrison, D. J. (2018). Role of gut microbiota-generated short-chain fatty acids in metabolic and cardiovascular health. Curr. Nutr. Rep. 7, 198–206. doi: 10.1007/s13668-018-0248-8
Chang, P. V., Hao, L., Offermanns, S., and Medzhitov, R. (2014). The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proce. Natl. Acad. Sci. U. S. A. 111, 2247–2252. doi: 10.1073/pnas.1322269111
Chopyk, J., Bojanowski, C. M., Shin, J., Moshensky, A., Fuentes, A. L., Bonde, S. S., et al. (2021). Compositional differences in the oral microbiome of E-cigarette users. Front. Microbiol. 12:599664. doi: 10.3389/fmicb.2021.599664
Chu, C., Murdock, M. H., Jing, D., Won, T. H., Chung, H., Kressel, A. M., et al. (2019). The microbiota regulate neuronal function and fear extinction learning. Nature 574, 543–548. doi: 10.1038/s41586-019-1644-y
Claesson, M. J., Jeffery, I. B., Conde, S., Power, S. E., O’Connor, E. M., Cusack, S., et al. (2012). Gut microbiota composition correlates with diet and health in the elderly. Nature 488, 178–184. doi: 10.1038/nature11319
Clark, R. L., Connors, B. M., Stevenson, D. M., Hromada, S. E., Hamilton, J. J., Amador-Noguez, D., et al. (2021). Design of synthetic human gut microbiome assembly and butyrate production. Nat. Commun. 12:3254. doi: 10.1038/s41467-021-22938-y
Coe, G. L., Pinkham, N. V., Celis, A. I., Johnson, C., DuBois, J. L., and Walk, S. T. (2021). Dynamic gut microbiome changes in response to low-iron challenge. Appl. Environ. Microbiol. 87, e02307–e02320. doi: 10.1128/AEM.02307-20
Coury, D. L., Ashwood, P., Fasano, A., Fuchs, G., Geraghty, M., Kaul, A., et al. (2012). Gastrointestinal conditions in children with autism spectrum disorder: developing a research agenda. Pediatrics 130, S160–S168. doi: 10.1542/peds.2012-0900N
Craciun, S., and Balskus, E. P. (2012). Microbial conversion of choline to trimethylamine requires a Glycyl radical enzyme. Proc. Natl. Acad. Sci. U. S. A. 109, 21307–21312. doi: 10.1073/pnas.1215689109
Dalton, A., Mermier, C., and Zuhl, M. (2019). Exercise influence on the microbiome-gut-brain axis. Gut Microbes 10, 555–568. doi: 10.1080/19490976.2018.1562268
Das, N. K., Schwartz, A. J., Barthel, G., Inohara, N., Liu, Q., Sankar, A., et al. (2020). Microbial metabolite signaling is required for systemic iron homeostasis. Cell Metab. 31, 115.e6–130.e6. doi: 10.1016/j.cmet.2019.10.005
DeFilipp, Z., Bloom, P. P., Soto, M. T., Mansour, M. K., Sater, M. R. A., Huntley, M. H., et al. (2019). Drug-resistant E. coli bacteremia transmitted by fecal microbiota transplant. N. Engl. J. Med. 381, 2043–2050. doi: 10.1056/NEJMoa1910437
de Groot, M. J., Hoeksma, M., Blau, N., Reijngoud, D. J., and van Spronsen, F. J. (2010). Pathogenesis of cognitive dysfunction in phenylketonuria: review of hypotheses. Mol. Genet. Metab. 99, S86–S89. doi: 10.1016/j.ymgme.2009.10.016
Dekaboruah, E., Suryavanshi, M. V., Chettri, D., and Verma, A. K. (2020). Human microbiome: an academic update on human body site specific surveillance and its possible role. Arch. Microbiol. 202, 2147–2167. doi: 10.1007/s00203-020-01931-x
de Oliveira, G. L. V., Oliveira, C. N. S., Pinzan, C. F., de Salis, L. V. V., and de Barros Cardoso, C. R. (2021). Microbiota modulation of the gut-lung axis in COVID-19. Front. Immunol. 12:635471. doi: 10.3389/fimmu.2021.635471
Dimitrijević, R., Stojanović, M., Zivković, I., Petersen, A., Jankov, R. M., Dimitrijević, L., et al. (2009). The identification of a low molecular mass bacteriocin, rhamnosin A, produced by Lactobacillus rhamnosus strain 68. J. Appl. Microbiol. 107, 2108–2115. doi: 10.1111/j.1365-2672.2009.04539.x
Doelman, A., Tigchelaar, S., McConeghy, B., Sinha, S., Keung, M. S., Manouchehri, N., et al. (2021). Characterization of the gut microbiome in a porcine model of thoracic spinal cord injury. BMC Genomics 22:775. doi: 10.1186/s12864-021-07979-3
Donia, M. S., Cimermancic, P., Schulze, C. J., Wieland, L. C., Brown, J. M., Mitreva, M., et al. (2014). A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell 158, 1402–1414. doi: 10.1016/j.cell.2014.08.032
Duran-Pinedo, A., Solbiati, J., Teles, F., Teles, R., Zang, Y., and Frias-Lopez, J. (2021). Long-term dynamics of the human oral microbiome during clinical disease progression. BMC Biol. 19:240. doi: 10.1186/s12915-021-01169-z
Durk, R. P., Castillo, E., Márquez-Magaña, L., Grosicki, G. J., Bolter, N. D., Matthew Lee, C., et al. (2019). Gut microbiota composition is related to cardiorespiratory fitness in healthy young adults. Int. J. Sport Nutr. Exerc. Metab. 29, 249–253. doi: 10.1123/ijsnem.2018-0024
Dziubańska-Kusibab, P. J., Berger, H., Battistini, F., Bouwman, B. A. M., Iftekhar, A., Katainen, R., et al. (2020). Colibactin DNA-damage signature indicates mutational impact in colorectal cancer. Nat. Med. 26, 1063–1069. doi: 10.1038/s41591-020-0908-2
Elijovich, F., Laffer, C. L., Sahinoz, M., Pitzer, A., Ferguson, J. F., and Kirabo, A. (2020). The gut microbiome, inflammation, and salt-sensitive hypertension. Curr. Hypertens. Rep. 22:79. doi: 10.1007/s11906-020-01091-9
Enaud, R., Prevel, R., Ciarlo, E., Beaufils, F., Wieërs, G., Guery, B., et al. (2020). The gut-lung axis in health and respiratory diseases: a place for inter-organ and inter-kingdom crosstalks. Front. Cell. Infect. Microbiol. 10:9. doi: 10.3389/fcimb.2020.00009
Faust, K., Fah Sathirapongsasuti, J., Izard, J., Segata, N., Gevers, D., Raes, J., et al. (2012). Microbial co-occurrence relationships in the human microbiome. PLoS Comput. Biol. 8:e1002606. doi: 10.1371/journal.pcbi.1002606
Ferguson, J. F., Aden, L. A., Barbaro, N. R., Van Beusecum, J. P., Xiao, L., Simmons, A. J., et al. (2019). High dietary salt–induced DC activation underlies microbial dysbiosis-associated hypertension. JCI Insight 4:e126241. doi: 10.1172/jci.insight.126241
Fernandez-Gonzalez, S., Ortiz-Arrabal, O., Torrecillas, A., Pérez-Cruz, M., Chueca, N., Gómez-Roig, M. D., et al. (2020). Study of the fetal and maternal microbiota in pregnant women with intrauterine growth restriction and its relationship with inflammatory biomarkers: a case-control study protocol (SPIRIT compliant). Medicine 99:e22722. doi: 10.1097/MD.0000000000022722
Flynn, S., van Sinderen, D., Thornton, G. M., Holo, H., Nes, I. F., and Kevin Collins, J. (2002). Characterization of the genetic locus responsible for the production of ABP-118, a novel bacteriocin produced by the probiotic bacterium Lactobacillus salivarius subsp. salivarius UCC118. Microbiology 148, 973–984. doi: 10.1099/00221287-148-4-973
Fouladi, F., Bailey, M. J., Patterson, W. B., Sioda, M., Blakley, I. C., Fodor, A. A., et al. (2020). Air pollution exposure is associated with the gut microbiome as revealed by shotgun metagenomic sequencing. Environ. Int. 138:105604. doi: 10.1016/j.envint.2020.105604
Franzosa, E. A., Huang, K., Meadow, J. F., Gevers, D., Lemon, K. P., Bohannan, B. J. M., et al. (2015). Identifying personal microbiomes using metagenomic codes. Proc. Natl. Acad. Sci. U. S. A. 112, E2930–E2938. doi: 10.1073/pnas.1423854112
Gaillard-Gendron, S., Vignon, D., Cottenceau, G., Graber, M., Zorn, N., van Dorsselaer, A., et al. (2000). Isolation, purification and partial amino acid sequence of a highly hydrophobic new microcin named microcin L produced by Escherichia coli. FEMS Microbiol. Lett. 193, 95–98. doi: 10.1111/j.1574-6968.2000.tb09408.x
Gatarek, P., and Kaluzna-Czaplinska, J. (2021). Trimethylamine N-oxide (TMAO) in human health. EXCLI J. 20, 301–319. doi: 10.17179/excli2020-3239
Gaulke, C. A., and Sharpton, T. J. (2018). The influence of ethnicity and geography on human gut microbiome composition. Nat. Med. 24, 1495–1496. doi: 10.1038/s41591-018-0210-8
Gilbert, J. A., Blaser, M. J., Gregory Caporaso, J., Jansson, J. K., Lynch, S. V., and Knight, R. (2018). Current understanding of the human microbiome. Nat. Med. 24, 392–400. doi: 10.1038/nm.4517
Glassner, K. L., Abraham, B. P., and Quigley, E. M. M. (2020). The microbiome and inflammatory bowel disease. J. Allergy Clin. Immunol. 145, 16–27. doi: 10.1016/j.jaci.2019.11.003
Glinton, K. E., and Elsea, S. H. (2019). Untargeted metabolomics for autism spectrum disorders: current status and future directions. Front. Psychol. 10:647. doi: 10.3389/fpsyt.2019.00647
Golubeva, A. V., Crampton, S., Desbonnet, L., Edge, D., O’Sullivan, O., Lomasney, K. W., et al. (2015). Prenatal stress-induced alterations in major physiological systems correlate with gut microbiota composition in adulthood. Psychoneuroendocrinology 60, 58–74. doi: 10.1016/j.psyneuen.2015.06.002
Gomaa, E. Z. (2020). Human gut microbiota/microbiome in health and diseases: a review. Antonie Van Leeuwenhoek 113, 2019–2040. doi: 10.1007/s10482-020-01474-7
Gong, G.-C., Song, S.-R., and Jin, S. (2021). Pulmonary fibrosis alters gut microbiota and associated metabolites in mice: an integrated 16S and metabolomics analysis. Life Sci. 264:118616. doi: 10.1016/j.lfs.2020.118616
Gorecki, A. M., Preskey, L., Bakeberg, M. C., Kenna, J. E., Gildenhuys, C., MacDougall, G., et al. (2019). Altered gut microbiome in Parkinson’s disease and the influence of lipopolysaccharide in a human α-synuclein over-expressing mouse model. Front. Neurosci. 13:839. doi: 10.3389/fnins.2019.00839
Goyal, D., Ali, S. A., and Singh, R. K. (2021). Emerging role of gut microbiota in modulation of neuroinflammation and neurodegeneration with emphasis on Alzheimer’s disease. Prog. Neuropsychopharmacol. Biol. Psychiatry 106:110112. doi: 10.1016/j.pnpbp.2020.110112
Grosicki, G. J., Riemann, B. L., Flatt, A. A., Valentino, T., and Lustgarten, M. S. (2020). Self-reported sleep quality is associated with gut microbiome composition in young, healthy individuals: a pilot study. Sleep Med. 73, 76–81. doi: 10.1016/j.sleep.2020.04.013
Groussin, M., Poyet, M., Sistiaga, A., Kearney, S. M., Moniz, K., Noel, M., et al. (2021). Elevated rates of horizontal gene transfer in the industrialized human microbiome. Cell 184, 2053.e18–2067.e18. doi: 10.1016/j.cell.2021.02.052
Guo, H., Chou, W.-C., Lai, Y., Liang, K., Tam, J. W., June Brickey, W., et al. (2020). Multi-omics analyses of radiation survivors identify radioprotective microbes and metabolites. Science 370:eaay9097. doi: 10.1126/science.aay9097
Gupta, S., Shariff, M., Chaturvedi, G., Sharma, A., Goel, N., Yadav, M., et al. (2021). Comparative analysis of the alveolar microbiome in COPD, ECOPD, sarcoidosis, and ILD patients to identify respiratory illnesses specific microbial signatures. Sci. Rep. 11:3963. doi: 10.1038/s41598-021-83524-2
Hakenbeck, R., Madhour, A., Denapaite, D., and Brückner, R. (2009). Versatility of choline metabolism and choline-binding proteins in Streptococcus pneumoniae and commensal streptococci. FEMS Microbiol. Rev. 33, 572–586. doi: 10.1111/j.1574-6976.2009.00172.x
He, Y., Kosciolek, T., Tang, J., Zhou, Y., Li, Z., Ma, X., et al. (2018a). Gut microbiome and magnetic resonance spectroscopy study of subjects at ultra-high risk for psychosis may support the membrane hypothesis. Eur. Psychiatry 53, 37–45. doi: 10.1016/j.eurpsy.2018.05.011
He, Y., Wei, W., Zheng, H.-M., Li, P., McDonald, D., Sheng, H.-F., et al. (2018b). Regional variation limits applications of healthy gut microbiome reference ranges and disease models. Nat. Med. 24, 1532–1535. doi: 10.1038/s41591-018-0164-x
He, J., Zhang, P., Shen, L., Niu, L., Tan, Y., Chen, L., et al. (2020). Short-chain fatty acids and their association with signalling pathways in inflammation, glucose and lipid metabolism. Int. J. Mol. Sci. 21:6356. doi: 10.3390/ijms21176356
He, L., Zhou, Y.-X., Zhang, Y., Hang, B., Chang, H., Schick, S. F., et al. (2021). Thirdhand cigarette smoke leads to age-dependent and persistent alterations in the cecal microbiome of mice. Microbiologyopen 10:e1198. doi: 10.1002/mbo3.1198
Houslay, M. D., and Milligan, G. (1997). Tailoring CAMP-signalling responses through isoform multiplicity. Trends Biochem. Sci. 22, 217–224. doi: 10.1016/S0968-0004(97)01050-5
Hsiao, E. Y., McBride, S. W., Hsien, S., Sharon, G., Hyde, E. R., McCue, T., et al. (2013). Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 155, 1451–1463. doi: 10.1016/j.cell.2013.11.024
Hughes, H. K., Rose, D., and Ashwood, P. (2018). The gut microbiota and dysbiosis in autism spectrum disorders. Curr. Neurol. Neurosci. Rep. 18:81. doi: 10.1007/s11910-018-0887-6
Lee, S. H., Yun, Y., Kim, S. J., Lee, E.-J., Chang, Y., Ryu, S., et al. (2018). Association between cigarette smoking status and composition of gut microbiota: population-based cross-sectional study. J. Clin. Med. 7:282. doi: 10.3390/jcm7090282
Irwin, M. R. (2019). Sleep and inflammation: partners in sickness and in health. Nat. Rev. Immunol. 19, 702–715. doi: 10.1038/s41577-019-0190-z
Janeiro, M. H., Ramírez, M. J., Milagro, F. I., Alfredo Martínez, J., and Solas, M. (2018). Implication of trimethylamine N-oxide (TMAO) in disease: potential biomarker or new therapeutic target. Nutrients 10:E1398. doi: 10.3390/nu10101398
Jang, S.-H., Woo, Y. S., Lee, S.-Y., and Bahk, W.-M. (2020). The brain-gut-microbiome axis in psychiatry. Int. J. Mol. Sci. 21:7122. doi: 10.3390/ijms21197122
Jašarević, E., Howerton, C. L., Howard, C. D., and Bale, T. L. (2015a). Alterations in the vaginal microbiome by maternal stress are associated with metabolic reprogramming of the offspring gut and brain. Endocrinology 156, 3265–3276. doi: 10.1210/en.2015-1177
Jašarević, E., Rodgers, A. B., and Bale, T. L. (2015b). A novel role for maternal stress and microbial transmission in early life programming and neurodevelopment. Neurobiol. Stress 1:81. doi: 10.1016/j.ynstr.2014.10.005
Jaworska, K., Bielinska, K., Gawrys-Kopczynska, M., and Ufnal, M. (2019). TMA (trimethylamine), but not its oxide TMAO (trimethylamine-oxide), exerts haemodynamic effects: implications for interpretation of cardiovascular actions of gut microbiome. Cardiovasc. Res. 115, 1948–1949. doi: 10.1093/cvr/cvz231
Jayachandran, M., Chung, S. S. M., and Baojun, X. (2020). A critical review of the relationship between dietary components, the gut microbe Akkermansia muciniphila, and human health. Crit. Rev. Food Sci. Nutr. 60, 2265–2276. doi: 10.1080/10408398.2019.1632789
Jiang, H., Ling, Z., Zhang, Y., Mao, H., Ma, Z., Yin, Y., et al. (2015). Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav. Immun. 48, 186–194. doi: 10.1016/j.bbi.2015.03.016
Jie, Z., Xia, H., Zhong, S.-L., Feng, Q., Li, S., Liang, S., et al. (2017). The gut microbiome in atherosclerotic cardiovascular disease. Nat. Commun. 8:845. doi: 10.1038/s41467-017-00900-1
Jin, X., Zhang, Y., Celniker, S. E., Xia, Y., Mao, J.-H., Snijders, A. M., et al. (2021). Gut microbiome partially mediates and coordinates the effects of genetics on anxiety-like behavior in collaborative cross mice. Sci. Rep. 11:270. doi: 10.1038/s41598-020-79538-x
Johnson, A., Houtti, M., Saboe, A., Koecher, K., Menon, R., and Knights, D. (2021). Whole wheat and bran cereal affects microbiome stability. Curr. Dev. Nutr. 5:1162. doi: 10.1093/cdn/nzab054_017
Johnson, A. J., Vangay, P., Al-Ghalith, G. A., Hillmann, B. M., Ward, T. L., Shields-Cutler, R. R., et al. (2019). Daily sampling reveals personalized diet-microbiome associations in humans. Cell Host Microbe 25, 789.e5–802.e5. doi: 10.1016/j.chom.2019.05.005
Kalnins, G., Kuka, J., Grinberga, S., Makrecka-Kuka, M., Liepinsh, E., Dambrova, M., et al. (2015). Structure and function of CutC choline lyase from human microbiota bacterium Klebsiella pneumoniae. J. Biol. Chem. 290, 21732–21740. doi: 10.1074/jbc.M115.670471
Kang, D.-W., Park, J. G., Ilhan, Z. E., Wallstrom, G., LaBaer, J., Adams, J. B., et al. (2013). Reduced incidence of Prevotella and other fermenters in intestinal microflora of autistic children. PLoS One 8:e68322. doi: 10.1371/journal.pone.0068322
Kawai, Y., Saito, T., Kitazawa, H., and Itoh, T. (1998). Gassericin A; an uncommon cyclic bacteriocin produced by Lactobacillus gasseri LA39 linked at N- and C-terminal ends. Biosci. Biotechnol. Biochem. 62, 2438–2440. doi: 10.1271/bbb.62.2438
Kazemian, N., Ramezankhani, M., Sehgal, A., Khalid, F. M., Kalkhoran, A. H. Z., Narayan, A., et al. (2020). The trans-kingdom battle between donor and recipient gut microbiome influences fecal microbiota transplantation outcome. Sci. Rep. 10:18349. doi: 10.1038/s41598-020-75162-x
Kelly, J. R., Kennedy, P. J., Cryan, J. F., Dinan, T. G., Clarke, G., and Hyland, N. P. (2015). Breaking down the barriers: the gut microbiome, intestinal permeability and stress-related psychiatric disorders. Front. Cell. Neurosci. 9:392. doi: 10.3389/fncel.2015.00392
Kheirouri, S., Kalejahi, P., and Noorazar, S. G. (2016). Plasma levels of serotonin, gastrointestinal symptoms, and sleep problems in children with autism. Turk. J. Med. Sci. 46, 1765–1772. doi: 10.3906/sag-1507-68
Khemir, S., Halayem, S., Azzouz, H., Siala, H., Ferchichi, M., Guedria, A., et al. (2016). Autism in phenylketonuria patients: from clinical presentation to molecular defects. J. Child Neurol. 31, 843–849. doi: 10.1177/0883073815623636
Kim, M., and Benayoun, B. A. (2020). The microbiome: an emerging key player in aging and longevity. Transl. Med. Aging 4, 103–116. doi: 10.1016/j.tma.2020.07.004
Kim, N., Yun, M., Young Joon, O., and Choi, H.-J. (2018). Mind-altering with the gut: modulation of the gut-brain axis with probiotics. J. Microbiol. 56, 172–182. doi: 10.1007/s12275-018-8032-4
Kimura, I., Inoue, D., Hirano, K., and Tsujimoto, G. (2014). The SCFA receptor GPR43 and energy metabolism. Front. Endocrinol. 5:85. doi: 10.3389/fendo.2014.00085
Knight, R. (2010). Translational medicine and the human microbiome. Genome Biol. 11:I15. doi: 10.1186/gb-2010-11-s1-i15
Konai, M. M., Barman, S., Acharya, Y., De, K., and Haldar, J. (2020). “Chapter 4—Recent development of antibacterial agents to combat drug-resistant gram-positive bacteria,” in Drug Discovery Targeting Drug-Resistant Bacteria. eds. Kesharwani P., Chopra S., and Dasgupta A. (United States: Academic Press), 71–104.
Konstantinidis, T., Tsigalou, C., Karvelas, A., Stavropoulou, E., Voidarou, C., and Bezirtzoglou, E. (2020). Effects of antibiotics upon the gut microbiome: a review of the literature. Biomedicines 8:502. doi: 10.3390/biomedicines8110502
Krishnareddy, S. (2019). The microbiome in celiac disease. Gastroenterol. Clin. North Am. 48, 115–126. doi: 10.1016/j.gtc.2018.09.008
Kubinyi, E., Rhali, S. B., Sándor, S., Szabó, A., and Felföldi, T. (2020). Gut microbiome composition is associated with age and memory performance in pet dogs. Animals 10:1488. doi: 10.3390/ani10091488
Kumar, T., Pandey, R., and Chauhan, N. S. (2020). Hypoxia inducible factor-1α: the curator of gut homeostasis. Front. Cell. Infect. Microbiol. 10:227. doi: 10.3389/fcimb.2020.00227
Lau, A. W. Y., Tan, L. T.-H., Mutalib, N.-S. A., Wong, S. H., Letchumanan, V., and Lee, L.-H. (2021). The chemistry of gut microbiome in health and diseases. Prog. Microbes Mol. Biol. 4:a000175. doi: 10.36877/pmmb.a0000175
Laviña, M., Gaggero, C., and Moreno, F. (1990). Microcin H47, a chromosome-encoded microcin antibiotic of Escherichia coli. J. Bacteriol. 172, 6585–6588. doi: 10.1128/jb.172.11.6585-6588.1990
Liang, D., Leung, R. K.-K., Guan, W., and William, W. A. (2019). Correction to: involvement of gut microbiome in human health and disease: brief overview, knowledge gaps and research opportunities. Gut Pathog. 11:57. doi: 10.1186/s13099-019-0339-0
Liang, S., Wang, T., Hu, X., Luo, J., Li, W., Wu, X., et al. (2015). Administration of Lactobacillus helveticus NS8 improves behavioral, cognitive, and biochemical aberrations caused by chronic restraint stress. Neuroscience 310, 561–577. doi: 10.1016/j.neuroscience.2015.09.033
Lin, T.-L., Shu, C.-C., Chen, Y.-M., Jang-Jih, L., Ting-Shu, W., Lai, W.-F., et al. (2020). Like cures like: pharmacological activity of anti-inflammatory lipopolysaccharides from gut microbiome. Front. Pharmacol. 11:554. doi: 10.3389/fphar.2020.00554
Littleford-Colquhoun, B. L., Weyrich, L. S., Kent, N., and Frere, C. H. (2019). City life alters the gut microbiome and stable isotope profiling of the eastern water dragon (Intellagama lesueurii). Mol. Ecol. 28, 4592–4607. doi: 10.1111/mec.15240
Liu, J., Liu, C., and Yue, J. (2021). Radiotherapy and the gut microbiome: facts and fiction. Radiat. Oncol. 16:9. doi: 10.1186/s13014-020-01735-9
Looft, T., and Allen, H. K. (2012). Collateral effects of antibiotics on mammalian gut microbiomes. Gut Microbes 3, 463–467. doi: 10.4161/gmic.21288
Magrone, T., Russo, M. A., and Jirillo, E. (2017). Cocoa and dark chocolate polyphenols: from biology to clinical applications. Front. Immunol. 8:677. doi: 10.3389/fimmu.2017.00677
Maier, E., Anderson, R. C., and Roy, N. C. (2015). Understanding how commensal obligate anaerobic bacteria regulate immune functions in the large intestine. Nutrients 7, 45–73. doi: 10.3390/nu7010045
Maifeld, A., Bartolomaeus, H., Löber, U., Avery, E. G., Steckhan, N., Markó, L., et al. (2021). Fasting alters the gut microbiome reducing blood pressure and body weight in metabolic syndrome patients. Nat. Commun. 12:1970. doi: 10.1038/s41467-021-22097-0
Maitre, Y., Micheneau, P., Delpierre, A., Mahalli, R., Guerin, M., Amador, G., et al. (2020). Did the brain and oral microbiota talk to each other? A review of the literature. J. Clin. Med. 9:E3876. doi: 10.3390/jcm9123876
Maki, K. A., Burke, L. A., Calik, M. W., Watanabe-Chailland, M., Sweeney, D., Romick-Rosendale, L. E., et al. (2020). Sleep fragmentation increases blood pressure and is associated with alterations in the gut microbiome and fecal metabolome in rats. Physiol. Genomics 52, 280–292. doi: 10.1152/physiolgenomics.00039.2020
Maltz, R. M., Keirsey, J., Kim, S. C., Mackos, A. R., Gharaibeh, R. Z., Moore, C. C., et al. (2018). Prolonged restraint stressor exposure in outbred CD-1 mice impacts microbiota, colonic inflammation, and short chain fatty acids. PLoS One 13:e0196961. doi: 10.1371/journal.pone.0196961
Maltz, R. M., Keirsey, J., Kim, S. C., Mackos, A. R., Gharaibeh, R. Z., Moore, C. C., et al. (2019). Social stress affects colonic inflammation, the gut microbiome, and short-chain fatty acid levels and receptors. J. Pediatr. Gastroenterol. Nutr. 68, 533–540. doi: 10.1097/MPG.0000000000002226
Martín, R., Miquel, S., Ulmer, J., Kechaou, N., Langella, P., and Bermúdez-Humarán, L. G. (2013). Role of commensal and probiotic bacteria in human health: a focus on inflammatory bowel disease. Microb. Cell Fact. 12:71. doi: 10.1186/1475-2859-12-71
Matenchuk, B. A., Mandhane, P. J., and Kozyrskyj, A. L. (2020). Sleep, circadian rhythm, and gut microbiota. Sleep Med. Rev. 53:101340. doi: 10.1016/j.smrv.2020.101340
Matsukawa, H., Iida, N., Kitamura, K., Terashima, T., Seishima, J., Makino, I., et al. (2021). Dysbiotic gut microbiota in pancreatic cancer patients form correlation networks with the oral microbiota and prognostic factors. Am. J. Cancer Res. 11, 3163–3175.
Mayer, E. A., Nance, K., and Chen, S. (2022). The gut–brain axis. Annu. Rev. Med. 73. doi: 10.1146/annurev-med-042320-014032 [Epub ahead of print]
Mazidi, M., Rezaie, P., Kengne, A. P., Mobarhan, M. G., and Ferns, G. A. (2016). Gut microbiome and metabolic syndrome. Diabetes Metab. Syndr. Clin. Res. Rev. 10, S150–S157. doi: 10.1016/j.dsx.2016.01.024
Merlen, J. F. (1986). Hermann Boerhaave, microcirculationist of the 17th century. J. Mal. Vasc. 11, 55–62.
Mizuta, K., Matoba, A., Shibata, S., Masaki, E., and Emala, C. W. (2019). Obesity-induced asthma: role of free fatty acid receptors. Jpn. Dent. Sci. Rev. 55, 103–107. doi: 10.1016/j.jdsr.2019.07.002
Moussaoui, N., Braniste, V., Ait-Belgnaoui, A., Gabanou, M., Sekkal, S., Olier, M., et al. (2014). Changes in intestinal glucocorticoid sensitivity in early life shape the risk of epithelial barrier defect in maternal-deprived rats. PLoS One 9:e88382. doi: 10.1371/journal.pone.0088382
Myhre, R., Brantsæter, A. L., Myking, S., Gjessing, H. K., Sengpiel, V., Meltzer, H. M., et al. (2011). Intake of probiotic food and risk of spontaneous preterm delivery. Am. J. Clin. Nutr. 93, 151–157. doi: 10.3945/ajcn.110.004085
Nagpal, R., Neth, B. J., Wang, S., Mishra, S. P., Craft, S., and Yadav, H. (2020). Gut mycobiome and its interaction with diet, gut bacteria and Alzheimer’s disease markers in subjects with mild cognitive impairment: a pilot study. EBioMedicine 59:102950. doi: 10.1016/j.ebiom.2020.102950
Naito, Y., Uchiyama, K., and Takagi, T. (2018). A next-generation beneficial microbe: Akkermansia muciniphila. J. Clin. Biochem. Nutr. 63, 33–35. doi: 10.3164/jcbn.18-57
Nakajima, A., Nakatani, A., Hasegawa, S., Irie, J., Ozawa, K., Tsujimoto, G., et al. (2017). The short chain fatty acid receptor GPR43 regulates inflammatory signals in adipose tissue M2-type macrophages. PLoS One 12:e0179696. doi: 10.1371/journal.pone.0179696
Naseribafrouei, A., Hestad, K., Avershina, E., Sekelja, M., Linløkken, A., Wilson, R., et al. (2014). Correlation between the human fecal microbiota and depression. Neurogastroenterol. Motil. 26, 1155–1162. doi: 10.1111/nmo.12378
Ng, Q. X., Loke, W., Venkatanarayanan, N., Lim, D. Y., Soh, A. Y. S., and Yeo, W. S. (2019). A systematic review of the role of prebiotics and probiotics in autism spectrum disorders. Medicina 55:129. doi: 10.3390/medicina55050129
Nichols, R. G., Peters, J. M., and Patterson, A. D. (2019). Interplay between the host, the human microbiome, and drug metabolism. Hum. Genomics 13:27. doi: 10.1186/s40246-019-0211-9
Nicholson, J. K., Holmes, E., Kinross, J., Burcelin, R., Gibson, G., Jia, W., et al. (2012). Host-gut microbiota metabolic interactions. Science 336, 1262–1267. doi: 10.1126/science.1223813
Nishida, K., Sawada, D., Kawai, T., Kuwano, Y., Fujiwara, S., and Rokutan, K. (2017). Para-psychobiotic Lactobacillus gasseri CP2305 ameliorates stress-related symptoms and sleep quality. J. Appl. Microbiol. 123, 1561–1570. doi: 10.1111/jam.13594
Olier, M., Marcq, I., Salvador-Cartier, C., Secher, T., Dobrindt, U., Boury, M., et al. (2012). Genotoxicity of Escherichia coli Nissle 1917 strain cannot be dissociated from its probiotic activity. Gut Microbes 3, 501–509. doi: 10.4161/gmic.21737
O’Sullivan, E., Barrett, E., Grenham, S., Fitzgerald, P., Stanton, C., Ross, R. P., et al. (2011). BDNF expression in the hippocampus of maternally separated rats: does bifidobacterium breve 6330 alter BDNF levels? Benefic. Microbes 2, 199–207. doi: 10.3920/BM2011.0015
Pagliari, D., Saviano, A., Newton, E. E., Serricchio, M. L., Dal Lago, A. A., Gasbarrini, A., et al. (2018). Gut microbiota-immune system crosstalk and pancreatic disorders. Mediat. Inflamm. 2018:e7946431. doi: 10.1155/2018/7946431
Palmas, V., Pisanu, S., Madau, V., Casula, E., Deledda, A., Cusano, R., et al. (2021). Gut microbiota markers associated with obesity and overweight in Italian adults. Sci. Rep. 11:5532. doi: 10.1038/s41598-021-84928-w
Papazafiropoulou, A. (2019). Anti-diabetic treatment leads to changes in gut microbiome. Front. Biosci. 24, 688–699. doi: 10.2741/4743
Patzer, S. I., Baquero, M. R., Bravo, D., Moreno, F., and Hantke, K. (2003). The colicin G, H and X determinants encode microcins M and H47, which might utilize the catecholate siderophore receptors FepA, Cir, Fiu and IroN. Microbiology 149, 2557–2570. doi: 10.1099/mic.0.26396-0
Pesoa, S. A., Portela, N., Fernández, E., Elbarcha, O., Gotteland, M., and Magne, F. (2021). Comparison of argentinean microbiota with other geographical populations reveals different taxonomic and functional signatures associated with obesity. Sci. Rep. 11:7762. doi: 10.1038/s41598-021-87365-x
Peterson, C. T. (2020). Dysfunction of the microbiota-gut-brain axis in neurodegenerative disease: the promise of therapeutic modulation with prebiotics, medicinal herbs, probiotics, and synbiotics. J. Evid. Based Integr. Med. 25:2515690X20957225. doi: 10.1177/2515690X20957225
Pirozzi, C., Francisco, V., Di Guida, F., Gómez, R., Lago, F., Pino, J., et al. (2018). Butyrate modulates inflammation in chondrocytes via GPR43 receptor. Cell. Physiol. Biochem. 51, 228–243. doi: 10.1159/000495203
Pitt, J. M., Vétizou, M., Boneca, I. G., Lepage, P., Chamaillard, M., and Zitvogel, L. (2016). Enhancing the clinical coverage and anticancer efficacy of immune checkpoint blockade through manipulation of the gut microbiota. Oncoimmunology 6:e1132137. doi: 10.1080/2162402X.2015.1132137
Pleguezuelos-Manzano, C., Puschhof, J., Huber, A. R., van Hoeck, A., Wood, H. M., Nomburg, J., et al. (2020). Mutational signature in colorectal cancer caused by genotoxic Pks+ E. coli. Nature 580, 269–273. doi: 10.1038/s41586-020-2080-8
Poutahidis, T., Kearney, S. M., Levkovich, T., Qi, P., Varian, B. J., Lakritz, J. R., et al. (2013). Microbial symbionts accelerate wound healing via the neuropeptide hormone oxytocin. PLoS One 8:e78898. doi: 10.1371/journal.pone.0078898
Putze, J., Hennequin, C., Nougayrède, J.-P., Zhang, W., Homburg, S., Karch, H., et al. (2009). Genetic structure and distribution of the colibactin genomic island among members of the family Enterobacteriaceae. Infect. Immun. 77, 4696–4703. doi: 10.1128/IAI.00522-09
Radjabzadeh, D., Bosch, J., Uitterlinden, A., Koos Zwinderman, M., Ikram, A., van Meurs, J., et al. (2021). “Gut microbiome-wide association study of depression.” [Preprint]. doi: 10.21203/rs.3.rs-570388/v1
Raes, J. (2014). The gut microbiome—a new target for understanding, diagnosing and treating disease. Arch. Public Health 72:K3. doi: 10.1186/2049-3258-72-S1-K3
Rea, M. C., Sit, C. S., Clayton, E., O’Connor, P. M., Whittal, R. M., Zheng, J., et al. (2010). Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. Proc. Natl. Acad. Sci. U. S. A. 107, 9352–9357. doi: 10.1073/pnas.0913554107
Rhee, S. J., Kim, H., Lee, Y., Lee, H. J., Park, C. H. K., Yang, J., et al. (2021). The association between serum microbial DNA composition and symptoms of depression and anxiety in mood disorders. Sci. Rep. 11:13987. doi: 10.1038/s41598-021-93112-z
Rogers, M. A. M., Todd Greene, M., Saint, S., Chenoweth, C. E., Malani, P. N., Trivedi, I., et al. (2012). Higher rates of Clostridium difficile infection among smokers. PLoS One 7:e42091. doi: 10.1371/journal.pone.0042091
Ruiz-Gonzalez, C., Roman, P., Rueda-Ruzafa, L., Rodriguez-Arrastia, M., and Cardona, D. (2021). Effects of probiotics supplementation on dementia and cognitive impairment: a systematic review and meta-analysis of preclinical and clinical studies. Prog. Neuropsychopharmacol. Biol. Psychiatry 108:110189. doi: 10.1016/j.pnpbp.2020.110189
Saji, N., Niida, S., Murotani, K., Hisada, T., Tsuduki, T., Sugimoto, T., et al. (2019). Analysis of the relationship between the gut microbiome and dementia: a cross-sectional study conducted in Japan. Sci. Rep. 9:1008. doi: 10.1038/s41598-018-38218-7
Saklayen, M. G. (2018). The global epidemic of the metabolic syndrome. Curr. Hypertens. Rep. 20:12. doi: 10.1007/s11906-018-0812-z
Salagre, E., Vieta, E., and Grande, I. (2017). The visceral brain: bipolar disorder and microbiota. Rev. Psiquiatr. Salud Ment. 10, 67–69. doi: 10.1016/j.rpsm.2017.02.001
Salomón, R. A., and Farías, R. N. (1992). Microcin 25, a novel antimicrobial peptide produced by Escherichia coli. J. Bacteriol. 174, 7428–7435. doi: 10.1128/jb.174.22.7428-7435.1992
Sanders, D. J., Inniss, S., Sebepos-Rogers, G., Rahman, F. Z., and Smith, A. M. (2021). The role of the microbiome in gastrointestinal inflammation. Biosci. Rep. 41:BSR20203850. doi: 10.1042/BSR20203850
Sani, G., Manchia, M., Simonetti, A., Janiri, D., Paribello, P., Pinna, F., et al. (2021). The role of gut microbiota in the high-risk construct of severe mental disorders: a mini review. Front. Psychol. 11:585769. doi: 10.3389/fpsyt.2020.585769
Savignac, H. M., Corona, G., Mills, H., Chen, L., Spencer, J. P. E., Tzortzis, G., et al. (2013). Prebiotic feeding elevates central brain derived neurotrophic factor, N-methyl-d-aspartate receptor subunits and d-serine. Neurochem. Int. 63, 756–764. doi: 10.1016/j.neuint.2013.10.006
Schiepers, O. J. G., Wichers, M. C., and Maes, M. (2005). Cytokines and major depression. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 29, 201–217. doi: 10.1016/j.pnpbp.2004.11.003
Schütz, F., Figueiredo-Braga, M., Barata, P., and Cruz-Martins, N. (2021). Obesity and gut microbiome: review of potential role of probiotics. Porto Biomed. J. 6:e111. doi: 10.1097/j.pbj.0000000000000111
Segain, J. P., Raingeard de la Blétière, D., Bourreille, A., Leray, V., Gervois, N., Rosales, C., et al. (2000). Butyrate inhibits inflammatory responses through NFkappaB inhibition: implications for Crohn’s disease. Gut 47, 397–403. doi: 10.1136/gut.47.3.397
Sgritta, M., Dooling, S. W., Buffington, S. A., Momin, E. N., Francis, M. B., Britton, R. A., et al. (2019). Mechanisms underlying microbial-mediated changes in social behavior in mouse models of autism spectrum disorder. Neuron 101, 246.e6–259.e6. doi: 10.1016/j.neuron.2018.11.018
Shanahan, E. R., Shah, A., Koloski, N., Walker, M. M., Talley, N. J., Morrison, M., et al. (2018). Influence of cigarette smoking on the human duodenal mucosa-associated microbiota. Microbiome 6:150. doi: 10.1186/s40168-018-0531-3
Sikora, M., Chrabąszcz, M., Maciejewski, C., Zaremba, M., Waśkiel, A., Olszewska, M., et al. (2018). Intestinal barrier integrity in patients with plaque psoriasis. J. Dermatol. 45, 1468–1470. doi: 10.1111/1346-8138.14647
Sikora, M., Chrabąszcz, M., Waśkiel-Burnat, A., Rakowska, A., Olszewska, M., and Rudnicka, L. (2019a). Claudin-3—a new intestinal integrity marker in patients with psoriasis: association with disease severity. J. Eur. Acad. Dermatol. Venereol. 33, 1907–1912. doi: 10.1111/jdv.15700
Sikora, M., Kiss, N., Stec, A., Giebultowicz, J., Samborowska, E., Jazwiec, R., et al. (2021). Trimethylamine N-oxide, a gut microbiota-derived metabolite, is associated with cardiovascular risk in psoriasis: a cross-sectional pilot study. Dermatol. Ther. 11, 1277–1289. doi: 10.1007/s13555-021-00547-3
Sikora, M., Stec, A., Chrabaszcz, M., Waskiel-Burnat, A., Zaremba, M., Olszewska, M., et al. (2019b). Intestinal fatty acid binding protein, a biomarker of intestinal barrier, is associated with severity of psoriasis. J. Clin. Med. 8:1021. doi: 10.3390/jcm8071021
Smith, R. P., Easson, C., Lyle, S. M., Kapoor, R., Donnelly, C. P., Davidson, E. J., et al. (2019). Gut microbiome diversity is associated with sleep physiology in humans. PLoS One 14:e0222394. doi: 10.1371/journal.pone.0222394
Smith, P. M., Howitt, M. R., Panikov, N., Michaud, M., Gallini, C. A., Bohlooly-Y, M., et al. (2013). The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573. doi: 10.1126/science.1241165
Solbach, M. D., Bonkowski, M., and Dumack, K. (2021). Novel endosymbionts in rhizarian amoebae imply universal infection of unrelated free-living amoebae by Legionellales. Front. Cell. Infect. Microbiol. 11:642216. doi: 10.3389/fcimb.2021.642216
Starke, R., Capek, P., Morais, D., Callister, S. J., and Jehmlich, N. (2020). The total microbiome functions in bacteria and fungi. J. Proteomics 213:103623. doi: 10.1016/j.jprot.2019.103623
Stubbs, J. R., House, J. A., Jacob Ocque, A., Zhang, S., Johnson, C., Kimber, C., et al. (2016). Serum trimethylamine-N-oxide is elevated in CKD and correlates with coronary atherosclerosis burden. J. Am. Soc. Nephrol. 27, 305–313. doi: 10.1681/ASN.2014111063
Sukenik, S., Dunsky, S., Barnoy, A., Shumilin, I., and Harries, D. (2017). TMAO mediates effective attraction between lipid membranes by partitioning unevenly between bulk and lipid domains. Phys. Chem. Chem. Phys. 19, 29862–29871. doi: 10.1039/C7CP04603K
Thomas, R. M., and Jobin, C. (2020). Microbiota in pancreatic health and disease: the next frontier in microbiome research. Nat. Rev. Gastroenterol. Hepatol. 17, 53–64. doi: 10.1038/s41575-019-0242-7
Thompson, A. J., Tillotson, S., Smith, I., Kendall, B., Moore, S. G., and Brenton, D. P. (1993). Brain MRI changes in phenylketonuria: associations with dietary status. Brain 116, 811–821. doi: 10.1093/brain/116.4.811
Toba, T., Samant, S. K., Yoshioka, E., and Itoh, T. (1991). Reutericin 6, a new bacteriocin produced by Lactobacillus reuteri LA 6. Lett. Appl. Microbiol. 13, 281–286. doi: 10.1111/j.1472-765X.1991.tb00629.x
Todokoro, D., Tomita, H., Inoue, T., and Ike, Y. (2006). Genetic analysis of bacteriocin 43 of vancomycin-resistant Enterococcus faecium. Appl. Environ. Microbiol. 72, 6955–6964. doi: 10.1128/AEM.00934-06
Tomita, H., Fujimoto, S., Tanimoto, K., and Ike, Y. (1996). Cloning and genetic organization of the bacteriocin 31 determinant encoded on the Enterococcus faecalis pheromone-responsive conjugative plasmid PYI17. J. Bacteriol. 178, 3585–3593. doi: 10.1128/jb.178.12.3585-3593.1996
Tran, S. M.-S., and Hasan Mohajeri, M. (2021). The role of gut bacterial metabolites in brain development, aging and disease. Nutrients 13:732. doi: 10.3390/nu13030732
Trend, S., Leffler, J., Jones, A. P., Cha, L., Gorman, S., Brown, D. A., et al. (2021). Associations of serum short-chain fatty acids with circulating immune cells and serum biomarkers in patients with multiple sclerosis. Sci. Rep. 11:5244. doi: 10.1038/s41598-021-84881-8
Tseng, C.-H., and Chun-Ying, W. (2019). The gut microbiome in obesity. J. Formos. Med. Assoc. 118, S3–S9. doi: 10.1016/j.jfma.2018.07.009
Turnbaugh, P. J., Ley, R. E., Mahowald, M. A., Magrini, V., Mardis, E. R., and Gordon, J. I. (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031. doi: 10.1038/nature05414
van der Lelie, D., and Taghavi, S. (2020). COVID-19 and the gut microbiome: more than a gut feeling. mSystems 5, e00453–e00520. doi: 10.1128/mSystems.00453-20
Vangay, P., Johnson, A. J., Ward, T. L., Al-Ghalith, G. A., Shields-Cutler, R. R., Hillmann, B. M., et al. (2018). US immigration westernizes the human gut microbiome. Cell 175, 962.e10–972.e10. doi: 10.1016/j.cell.2018.10.029
van Nimwegen, F. A., Penders, J., Stobberingh, E. E., Postma, D. S., Koppelman, G. H., Kerkhof, M., et al. (2011). Mode and place of delivery, gastrointestinal microbiota, and their influence on asthma and atopy. J. Allergy Clin. Immunol. 128, 948–955. doi: 10.1016/j.jaci.2011.07.027
Vanoaica, L., Darshan, D., Richman, L., Schümann, K., and Kühn, L. C. (2010). Intestinal ferritin H is required for an accurate control of iron absorption. Cell Metab. 12, 273–282. doi: 10.1016/j.cmet.2010.08.003
Vásquez, A., Jakobsson, T., Ahrné, S., Forsum, U., and Molin, G. (2002). Vaginal Lactobacillus flora of healthy Swedish women. J. Clin. Microbiol. 40, 2746–2749. doi: 10.1128/JCM.40.8.2746-2749.2002
Venzon, M., Bernard, L., Klein, J., Axelrad, J. E., Hussey, G. A., Sullivan, A. P., et al. (2021). “Gut microbiome dysbiosis during COVID-19 is associated with increased risk for bacteremia and microbial translocation.” BioRxiv [Preprint]. doi: 10.1101/2021.07.15.452246
Vila, V., Arnau, V. C., Sanna, S., Sinha, T., Imhann, F., Bourgonje, A. R., et al. (2020). Impact of commonly used drugs on the composition and metabolic function of the gut microbiota. Nat. Commun. 11:362. doi: 10.1038/s41467-019-14177-z
Villapol, S. (2020). Gastrointestinal symptoms associated with COVID-19: impact on the gut microbiome. Transl. Res. 226, 57–69. doi: 10.1016/j.trsl.2020.08.004
Volkow, N. D., Wang, G.-J., and Baler, R. D. (2011). Reward, dopamine and the control of food intake: implications for obesity. Trends Cogn. Sci. 15, 37–46. doi: 10.1016/j.tics.2010.11.001
von Schwartzenberg, R. J., Bisanz, J. E., Lyalina, S., Spanogiannopoulos, P., Ang, Q. Y., Cai, J., et al. (2021). Caloric restriction disrupts the microbiota and colonization resistance. Nature 595, 272–277. doi: 10.1038/s41586-021-03663-4
Walker, A. C., Bhargava, R., Vaziriyan-Sani, A. S., Pourciau, C., Donahue, E. T., Dove, A. S., et al. (2021). Colonization of the Caenorhabditis elegans gut with human enteric bacterial pathogens leads to proteostasis disruption that is rescued by butyrate. PLoS Pathog. 17:e1009510. doi: 10.1371/journal.ppat.1009510
Wang, P., Dong, Y., Jiao, J., Zuo, K., Han, C., Zhao, L., et al. (2021a). Cigarette smoking status alters dysbiotic gut microbes in hypertensive patients. J. Clin. Hypertens. 23, 1431–1446. doi: 10.1111/jch.14298
Wang, D. D., Nguyen, L. H., Li, Y., Yan, Y., Ma, W., Rinott, E., et al. (2021b). The gut microbiome modulates the protective association between a mediterranean diet and cardiometabolic disease risk. Nat. Med. 27, 333–343. doi: 10.1038/s41591-020-01223-3
Wang, L. W., Tancredi, D. J., and Thomas, D. W. (2011). The prevalence of gastrointestinal problems in children across the United States with autism spectrum disorders from families with multiple affected members. J. Dev. Behav. Pediatr. 32, 351–360. doi: 10.1097/DBP.0b013e31821bd06a
Wang, L., Yi-Ning, X., Chu, C.-C., Jing, Z., Chen, Y., Zhang, J., et al. (2021c). Facial skin microbiota-mediated host response to pollution stress revealed by microbiome networks of individual. mSystems 6:e0031921. doi: 10.1128/mSystems.00319-21
Wastyk, H. C., Fragiadakis, G. K., Perelman, D., Dahan, D., Merrill, B. D., Yu, F. B., et al. (2021). Gut-microbiota-targeted diets modulate human immune status. Cell 184, 4137.e14–4153.e14. doi: 10.1016/j.cell.2021.06.019
Weersma, R. K., Zhernakova, A., and Jingyuan, F. (2020). Interaction between drugs and the gut microbiome. Gut 69, 1510–1519. doi: 10.1136/gutjnl-2019-320204
Wei, Y., Li, Y., Yan, L., Sun, C., Miao, Q., Wang, Q., et al. (2020). Alterations of gut microbiome in autoimmune hepatitis. Gut 69, 569–577. doi: 10.1136/gutjnl-2018-317836
Wibowo, M. C., Yang, Z., Borry, M., Hübner, A., Huang, K. D., Tierney, B. T., et al. (2021). Reconstruction of ancient microbial genomes from the human gut. Nature 594, 234–239. doi: 10.1038/s41586-021-03532-0
Wiese, M., Bashmakov, Y., Chalyk, N., Nielsen, D. S., Krych, Ł., Kot, W., et al. (2019). Prebiotic effect of lycopene and dark chocolate on gut microbiome with systemic changes in liver metabolism, skeletal muscles and skin in moderately obese persons. Biomed. Res. Int. 2019:4625279. doi: 10.1155/2019/4625279
Wilmanski, T., Diener, C., Rappaport, N., Patwardhan, S., Wiedrick, J., Lapidus, J., et al. (2021). Gut microbiome pattern reflects healthy ageing and predicts survival in humans. Nat. Metab. 3, 274–286. doi: 10.1038/s42255-021-00348-0
Wilson, M. R., Jiang, Y., Villalta, P. W., Stornetta, A., Boudreau, P. D., Carrá, A., et al. (2019). The human gut bacterial genotoxin colibactin alkylates DNA. Science 363:eaar7785. doi: 10.1126/science.aar7785
Winglee, K., Howard, A. G., Sha, W., Gharaibeh, R. Z., Liu, J., Jin, D., et al. (2017). Recent urbanization in China is correlated with a westernized microbiome encoding increased virulence and antibiotic resistance genes. Microbiome 5:121. doi: 10.1186/s40168-017-0338-7
Wu, J., Peters, B. A., Dominianni, C., Zhang, Y., Pei, Z., Yang, L., et al. (2016). Cigarette smoking and the oral microbiome in a large study of American adults. ISME J. 10, 2435–2446. doi: 10.1038/ismej.2016.37
Wu, L. M., Sankaran, S. J., Plank, L. D., Windsor, J. A., and Petrov, M. S. (2014). Meta-analysis of gut barrier dysfunction in patients with acute pancreatitis. Br. J. Surg. 101, 1644–1656. doi: 10.1002/bjs.9665
Xiong, X., Sheng, X., Liu, D., Zeng, T., Peng, Y., and Wang, Y. (2015). A GC/MS-based metabolomic approach for reliable diagnosis of phenylketonuria. Anal. Bioanal. Chem. 407, 8825–8833. doi: 10.1007/s00216-015-9041-3
Xu, J., and Yang, Y. (2020). Implications of gut microbiome on coronary artery disease. Cardiovasc. Diagn. Ther. 10, 869–880. doi: 10.21037/cdt-20-428
Xue, M., Kim, C. S., Healy, A. R., Wernke, K. M., Wang, Z., Frischling, M. C., et al. (2019). Structure elucidation of colibactin and its DNA cross-links. Science 365:eaax2685. doi: 10.1126/science.aax2685
Yadav, M., and Chauhan, N. S. (2021a). Overview of the rules of the microbial engagement in the gut microbiome: a step towards microbiome therapeutics. J. Appl. Microbiol. 130, 1425–1441. doi: 10.1111/jam.14883
Yadav, M., and Chauhan, N. S. (2021b). Microbiome therapeutics: exploring the present scenario and challenges. Gastroenterol. Rep. goab046. doi: 10.1093/gastro/goab046
Yadav, M., Chauhan, N., Prasher, B., and Mukerji, M. (2022). “Dissecting human microbiome for personalized therapy,” in Reference Module in Food Science. ed. Smithers G. (USA: Elsevier).
Yadav, M., Verma, M. K., and Chauhan, N. S. (2018). A review of metabolic potential of human gut microbiome in human nutrition. Arch. Microbiol. 200, 203–217. doi: 10.1007/s00203-017-1459-x
Yamashita, T., Emoto, T., Sasaki, N., and Hirata, K.-i. (2016). Gut microbiota and coronary artery disease. Int. Heart J. 57, 663–671. doi: 10.1536/ihj.16-414
Yan, Q., Yifang, G., Li, X., Yang, W., Jia, L., Chen, C., et al. (2017). Alterations of the gut microbiome in hypertension. Front. Cell. Infect. Microbiol. 7:381. doi: 10.3389/fcimb.2017.00381
Yang, A. C., Kern, F., Losada, P. M., Agam, M. R., Maat, C. A., Schmartz, G. P., et al. (2021). Dysregulation of brain and choroid plexus cell types in severe COVID-19. Nature 595, 565–571. doi: 10.1038/s41586-021-03710-0
Yap, C. X., Henders, A. K., Alvares, G. A., Wood, D. L. A., Krause, L., Tyson, G. W., et al. (2021). Autism-related dietary preferences mediate autism-gut microbiome associations. Cell 184, 5916.e17–5931.e17. doi: 10.1016/j.cell.2021.10.015
Ye, F., Gao, X., Wang, Z., Cao, S., Liang, G., He, D., et al. (2021). Comparison of gut microbiota in autism spectrum disorders and neurotypical boys in China: a case-control study. Synth. Syst. Biotechnol. 6, 120–126. doi: 10.1016/j.synbio.2021.03.003
Yeoh, Y. K., Zuo, T., Lui, G. C.-Y., Zhang, F., Liu, Q., Li, A. Y., et al. (2021). Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut 70, 698–706. doi: 10.1136/gutjnl-2020-323020
Zawada, A., Rychter, A. M., Ratajczak, A. E., Lisiecka-Masian, A., Dobrowolska, A., and Krela-Kaźmierczak, I. (2021). Does gut-microbiome interaction protect against obesity and obesity-associated metabolic disorders? Microorganisms 9:18. doi: 10.3390/microorganisms9010018
Zhou, X., Li, J., Guo, J., Geng, B., Ji, W., Zhao, Q., et al. (2018). Gut-dependent microbial translocation induces inflammation and cardiovascular events after ST-elevation myocardial infarction. Microbiome 6:66. doi: 10.1186/s40168-018-0441-4
Zhou, W., Zhang, D., Li, Z., Jiang, H., Li, J., Ren, R., et al. (2021). The fecal microbiota of patients with pancreatic ductal adenocarcinoma and autoimmune pancreatitis characterized by metagenomic sequencing. J. Transl. Med. 19:215. doi: 10.1186/s12967-021-02882-7
Zhu, S., Jiang, Y., Kelin, X., Cui, M., Ye, W., Zhao, G., et al. (2020). The progress of gut microbiome research related to brain disorders. J. Neuroinflammation 17:25. doi: 10.1186/s12974-020-1705-z
Zhuang, X., Li, T., Li, M., Huang, S., Qiu, Y., Feng, R., et al. (2019). Systematic review and meta-analysis: short-chain fatty acid characterization in patients with inflammatory bowel disease. Inflamm. Bowel Dis. 25, 1751–1763. doi: 10.1093/ibd/izz188
Zijlmans, M. A. C., Katri Korpela, J., Riksen-Walraven, M., de Vos, W. M., and de Weerth, C. (2015). Maternal prenatal stress is associated with the infant intestinal microbiota. Psychoneuroendocrinology 53, 233–245. doi: 10.1016/j.psyneuen.2015.01.006
Keywords: human microbiome, disease associations, microbial diversity, probiotics, fecal transplant
Citation: Mousa WK, Chehadeh F and Husband S (2022) Recent Advances in Understanding the Structure and Function of the Human Microbiome. Front. Microbiol. 13:825338. doi: 10.3389/fmicb.2022.825338
Received: 30 November 2021; Accepted: 11 January 2022;
Published: 03 February 2022.
Edited by:
Nar Singh Chauhan, Maharshi Dayanand University, IndiaReviewed by:
Rajesh Pandey, CSIR-Institute of Genomics and Integrative Biology (CSIR-IGIB), IndiaCopyright © 2022 Mousa, Chehadeh and Husband. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Walaa K. Mousa, d2FsYWEubW91c2FAYWF1LmFjLmFl, orcid.org/0000-0003-3229-4499
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.