
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 25 February 2022
Sec. Virology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.825111
This article is part of the Research Topic Emerging and Re-emerging Viral Diseases View all 37 articles
 Baojing Lu1,2*†
Baojing Lu1,2*† Qi Tang1†
Qi Tang1† Qianyun Wang1
Qianyun Wang1 Xuejuan Liu1
Xuejuan Liu1 Hui Peng1
Hui Peng1 Binbin Zhu1
Binbin Zhu1 Li Xie3
Li Xie3 Zeng Li2
Zeng Li2 Hanzhong Wang4
Hanzhong Wang4 Zhenhua Zheng4
Zhenhua Zheng4 Linding Wang1*
Linding Wang1* Bao Li5*
Bao Li5*Enterovirus 71 (EV71) is one of the most important etiological agents for hand–foot–mouth disease. Compared with coxsackievirus A16 infection, EV71 infection is often associated with severe central nervous system complications, such as encephalitis, encephalomyelitis, and acute flaccid paralysis in infants and young children. In this study, we constructed a recombinant baculovirus with T7 ribonucleic acid polymerase under the control of a cytomegalovirus promoter and simultaneously engineered the T7 promoter upstream of a full-length EV71 complementary deoxyribonucleic acid. After transduction into mammalian cells, typical cytopathic effects (CPEs) and VP1 signals were detected in cells transfected with recombinant baculovirus. Additionally, viral particles located in the cytoplasm of human rhabdomyosarcoma cells (Rd) and Vero cells were observed by electron microscope, indicating that EV71 was recovered using a Bac-to-Bac expression system in vitro. After four passages, the rescued virus had a growth curve and plaque morphology similar to those of the parental virus. Furthermore, the Vp1 gene and the protein from the mouse brain were detected by reverse transcription polymerase chain reaction and immunohistochemistry after intracerebral injection of purified recombinant baculovirus. Typical CPEs were observed after inoculation of the supernatant from mouse brain to Rd cells, revealing a reconstruction of EV71 in vivo. Thus, we established a new approach to rescue EV71 based on a baculovirus expression system in vitro and in vivo, which may provide a safe and convenient platform for fundamental research and a strategy to rescue viruses that currently lack suitable cell culture and animal models.
Enterovirus 71 (EV71) belongs to the Enterovirus genus of the Picornaviridae family, which is a non-enveloped, positive, single-stranded ribonucleic acid virus, that is a common cause of hand–foot–mouth disease (HFMD) in young children (Mao et al., 2016), but occasionally it can also lead to severe diseases such as aseptic meningitis, poliomyelitis-like paralysis, and possibly fatal encephalitis (Solomon et al., 2010). Because of consecutive epidemics over the past years in the Asia-Pacific region including Singapore, Malaysia, Taiwan, and mainland China, this condition has attracted immense concern in global heath (Solomon et al., 2010). Although major efforts have been made to elucidate the molecular mechanisms, virus–host interactions, and immune evasion of EV71 infection, no approved antivirals and limited vaccine and animal models for EV71 have been developed (Mao et al., 2016; Kim et al., 2019; Wang et al., 2021).
Reverse genetics systems are one of the most important and powerful tools to study the molecular biology of viruses (Yu et al., 2019). With the outbreak of emerging viruses, such as SARS-CoV-2, MERS-CoV, or Ebola virus, this system will be important for functional genomics research and virus prevention (Cockrell et al., 2017; Cross and Geisbert, 2019; Thi Nhu Thao et al., 2020). A full-length cDNA clone was always used for the recovery of RNA viruses (Boyer and Haenni, 1994), and the first infectious RNA virus was successfully isolated from cDNA to generate poliovirus in 1981 (Racaniello and Baltimore, 1981). Since then, most positive-strand RNA viruses, such as hepatitis A virus, coxsackievirus B6, and norovirus, have been recovered based on the reverse genetics system (Tellier et al., 1996; Martino et al., 1999; Todd and Tripp, 2019). Arita et al. (2005) constructed an infectious clone of BrCr to explore the neurovirulence site of EV71, which was the first study of EV71 rescue to the best of our knowledge. Then various groups constructed infectious clones of EV71 using different strategies that showed potential advantages for gene mutation, animal model application, and vaccine production of rescue virus (Han et al., 2010; Meng et al., 2012; Zhang et al., 2020).
Various reverse genetics strategies are currently applied for virus recovery. Direct transfection of in vitro-synthesized RNA transcripts is a traditional and effective method for virus recovery especially for RNA viruses whose genome is smaller than 15 kb. Alternatively, a recombinant vaccinia virus with a T7 RNA polymerase (VacT7) system has been widely used for virus reconstruction, such as SARS-CoV (van den Worm et al., 2012), vesicular stomatitis virus (Lawson et al., 1995), and nodavirus (Lawson et al., 1995) because of its easy handling, higher expression level, and specificity. In these systems, the full-length cDNA molecule is placed downstream of the T7 or SP6 promoter, and the entire genomic RNA is either synthesized in vitro and the transcripts introduced into host cells, or the RNA is synthesized by the use of a T7 RNA polymerase-expressing helper virus. Therefore, the transformation efficiency, stability of RNA, and enzyme activity of T7 RNA polymerase are possible factors affecting the recovery of viruses in vitro.
The baculovirus expression vector is recognized as a useful viral vector not only for abundant expression of foreign proteins in insect cells but also for gene delivery into mammalian cells (Jarvis, 2009). Baculoviruses have many attractive features, such as easy manipulation, a large capacity for foreign DNAs, and a wide vertebrate host range (Jarvis, 2009). An additional advantage of the baculovirus expression system is that a single recombinant virus can express more than four foreign genes, and this system has also been widely used for drug screening, vaccine development, and gene therapy (Roldão et al., 2011). It is also an effective vector for constructing infectious clones. A T7 RNA polymerase recombinant baculovirus was explored in recovering poliovirus indicating possible recovery of infectious virus from cDNA clones (Yap et al., 1997). Delaney also applied recombinant HBV baculovirus to deliver the HBV genome to HepG2 cells (Delaney and Isom, 1998), and Lucifora produced infectious HBV by delivering a novel recombinant baculovirus to HepG2 cells in 2008 (Lucifora et al., 2008). To date, there has been no research on EV71 recovery by baculovirus expression systems.
In this study, we developed a new strategy for the recovery of EV71 by a baculovirus expression system. We constructed one recombinant baculovirus synthesizing T7 RNA polymerase under the control of the cytomegalovirus (CMV) promoter and simultaneously engineered the T7 promoter upstream of a full-length cDNA clone of BrCr for transcription. After transduction into mammalian cells and intracerebral injection of the baculovirus into suckling mice intracerebrally, infectious EV71 was recovered in vitro and in vivo. Our baculovirus-based reverse genetics approach may provide a new strategy for EV71 rescue and apply it in molecular biology and antiviral research.
Spodoptera frugiperda (Sf9) cells were cultured in Grace’s medium (Invitrogen, Carlsbad, CA, United States) supplemented with 10% (v/v) fetal bovine serum (FBS, Gibco) at 27°C; Vero and BHK cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies, Grand Island, NY, United States) supplemented with 10% heat-inactivated FBS at 37°C with 5% CO2. Human rhabdomyosarcoma cells (Rd) and human neuroblastoma cells (Sk) were grown in minimum essential medium (MEM; Life Technologies, Grand Island, NY, United States) containing 10% FBS at 37°C with 5% CO2. The EV71 BrCr strain was provided by the Institute of Medical Biology, Chinese Academy of Medical Sciences. The polyclonal antibody against EV71 VP1 for Western blotting was prepared by our laboratory, and the monoclonal antibody used for immunohistochemistry was purchased from NZK Biotech (Wuhan, China). β-actin antibody was purchased from Proteintech (Wuhan, China).
The donor plasmid pFB-CMV-Ef1-α was derived from pFastBacTM Dual (Invitrogen, Carlsbad, CA, United States) and replaced the p10 promoter (Pp10) and polyhedrin promoter (PpH) with cytomegalovirus and the Ef1-α promoter (Bai et al., 2008). The infectious cDNA clone pEV71-BrCr-TR (AB204852) used for PCR was kindly supplied by Dr. Arita (Arita et al., 2005). The pEV71-GFP-BrCr cDNA clone, in which a GFP gene was inserted between the 5′ UTR and N-terminus of the VP4 gene of pEV71-BrCr-TR was constructed as previously described (Shang et al., 2013). The T7 RNA polymerase gene was amplified by PCR from BL21 (DE3) using the specific primers T7-F: 5′-ATCGTCGACGCCACCATGAACACGATTAACATCG-3′ and T7-R: 5′-CAAAGCTTTTACGCGAACGCGAAGTCCGAC-3′ (underlined and italicized sequences are SalI and HindIII sites, respectively). After sequence confirmation, this gene was cloned into pFB-CMV-EF1-α using the SalI and HindIII sites and named as pFB-CMV-T7-EF1-α. The cDNA from the 5′ end to the nucleotide position 2,915 with a T7 promoter before the 5′ end of the BrCr genome was amplified using pEV71-BrCr-TR or pEV71-GFP-BrCr as the template, and the primers used were as follows: BrCr-2915-F: 5′-TCAATGCATGCTCGACTGGCTTATCGAAATTACG-3′ and BrCr-2915-R: 5′-TACCCGGGTGGAACAAACAT-3′ (underlined and italicized sequences were NsiI and SmaI sites, respectively). After digestion with NsiI and SmaI, the PCR product was cloned into pFB-CMV-T7-EF1-α. The rest of the cDNA of BrCr obtained from the pEV71-BrCr-TR was cloned into the above construct following the digestion by MIuI and SmaI, resulting in the donor bacmids pFB-T7-BrCr and pFB-T7-GFP-BrCr, which contained the full-length cDNA clone of BrCr. All sequences of the intermediate product were validated by sequencing before use in the subsequent cloning step.
The recombinant viruses (Ac-T7-BrCr, Ac-T7-GFP-BrCr) were generated by transfecting the indicated bacmids into Sf9 cells according to the manufacturer’s instructions (Bac-to-Bac baculovirus expression system, Invitrogen, Carlsbad, CA, United States). The recombinant baculovirus and wild-type virus were collected 60 h after infection, and cell debris was removed by centrifugation at 4,000 × g for 20 min. The supernatant was filtered through a 0.45-mm filter (Millipore, Burlington, VT, United States) and subsequently centrifuged at 25,000 rpm at 4°C for 90 min (BeckmanSW28 rotor) in 5 ml of a 30% (w/v) sucrose cushion in TE buffer (Wang et al., 2010). The virion pellet was resuspended in TE buffer, sucrose was removed by ultrafiltration (100 kDa, Millipore, Burlington, VT, United States), and the pellet was suspended in PBS and stored at –80°C. The viral titer was determined by the 50% tissue culture infectious dose (TCID50) assay using the Reed–Muench method (Svensson et al., 1999).
Mammalian cells were seeded in six-well culture dishes at a concentration of 2 × 105 cells per well. Eight hours after adherence, the medium was removed and replaced with virus (vAc-T7-BrCr, vAc-T7-GFP-BrCr, or wild-type AcMNPV) at a multiplicity of infection (MOI) of 50, and the cells were incubated for 6 h at 37°C. Then, the cells were washed and replaced with 2 ml of fresh medium.
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s protocol. First-strand cDNA was reverse transcribed from 2 μg of total RNA using the PrimeScript™ RT reagent kit with gDNA Eraser (TaKaRa, Dalian, China). Real-time PCR was conducted with SYBR Green Master Mix (Bio-Rad) on a CFX Connect Real-Time PCR Detection System from (Bio-Rad, Hercules, CA, United States). Specific primers used for qRT-PCR were previously described (Zheng et al., 2013), and GAPDH mRNA was measured as a control.
Cells were harvested and lysed with Western lysis buffer (Beyotime, Shanghai, China). After electrophoresis, proteins were transferred to PVDF membranes (Millipore, Burlington, VT, United States) for 30 min by semidry electrophoresis transfer (Bio-Rad). Then the membranes were blocked in 20 mM Tris–HCl buffer (pH 7.4) containing 37 mM NaCl with 5% BSA (Sigma-Aldrich, St. Louis, MO, United States) at 37°C for 1 h. After that, the film was incubated with polyclonal antibodies against VP1 (diluted 1:1,000 in TBS containing 0.1% Tween 20 and 2% BSA; our lab performed) and β-actin (1:1,000; Proteintech, Wuhan, China) overnight at 4°C. After three washes in TBST for 10 min each time, the membrane was incubated with AP-conjugated secondary antibody (Boster, Wuhan, China) for 2 h at room temperature. Finally, the signals were detected using a BCIP/NBT alkaline phosphatase color development kit (Beyotime, Shanghai, China).
Thirty-six hours post-transduction with vAc-T7-BrCr, Vero and Rd cells were fixed at 4°C with 2.5% (W/V) glutaraldehyde in 0.1 mol/L PBS (pH 7.2) overnight. Then the electron microscopy samples were prepared as previously described (Wang et al., 2010) and observed by transmission electron microscopy (FEI Tecnai G2, operated at 200 kV).
After transducting Vero cells with v-Ac-EV71, a rescued EV71 was obtained, and Rd cells were infected with the virus to amplify and obtain the F4 generation of virus. Rd cells were seeded in 12-well plates (2 × 105 cells per well, and the confluence reached approximately 60% the next day). The cells were separately infected with wild-type EV71 (WT EV71) or rescued EV71 (rEV71) at an MOI of 1. After 2 h of adsorption and washing with PBS twice, the supernatant was replaced with 2 ml of MEM + 10% FBS. Then the supernatant was collected at 6, 12, 16, 24, and 30 h postinfection, and viral titers were quantified by TCID50 assay.
Wild-type EV71 or rEV71 with the same TCID50 was diluted 1:10 by mixing 80 μl of virus sample with 720 μl of MEM. Two hundred microliters was added to individual wells of 12-well plates containing confluent Rd cells (2 × 105 cells per well, the plates were plated 1 day in advance, and a cell monolayer was formed 24 h later). After infection, the plates were adsorbed for 2 h and washed twice with PBS, and 1.5 ml of 2 × MEM (containing 4% fetal bovine serum) containing 1% agarose was added to individual wells of 12-well plates. Each well was fixed with 1 ml of crystal violet containing formaldehyde and dyed overnight after the plates were incubated at 37°C with 5% CO2 for 48 h. The numbers and morphology of plaques were recorded after the overlaying layer was flushed away with running water the next day.
One-day-old ICR mice were obtained from the Centers for Disease Prevention and Control of Hubei Province and maintained under specific pathogen-free (SPF) conditions. The mice were randomly divided into the experimental group and the control group (10 mice each). A total of 1 × 107 TCID50 purified recombinant baculoviruses Ac-T7-BrCr and AcMNPV in a volume of 20 μl or PBS was intracerebrally inoculated into mice using a 50-μl Hamilton injector (Hamilton Co., Reno, NV, United States). All experimental procedures were conducted according to the guidance of the Institutional Animal Care and Use Committee of Wuhan Institute of Virology, Chinese Academy of Sciences (No: WIVA07201502).
Brain samples were collected 7 days after inoculation. The samples were washed with sterilized PBS and dissected in half along the coronal line; one half was stored at –80°C, and the other was fixed in 4% paraformaldehyde for immunohistochemistry detection. For the RT-PCR assay, the samples were suspended in TRIzol for RNA extraction. Brain samples suspended in PBS were disrupted by a microelectric tissue homogenizer (Kimble, United States) and sterilized with a 0.22-μm filter for virus isolation in Rd cells.
Brain samples were fixed in 4% paraformaldehyde and embedded in paraffin. Sections (5 μm) were sliced by a section cutter (Leica, Wetzlar, Germany) and then immunostained overnight with anti-EV71 mAb (1:500) against VP1 at 4°C. Subsequently, the positive cells were visualized with a PV-6002 HRP-conjugated goat anti-mouse detection kit (Zhongshan, Beijing, China) and recorded by a light microscope. The immunofluorescence assay was carried out as previously reported (Pham et al., 2019; Tang et al., 2020). In short, the infected or non-infected Vero cells were washed by PBS three times, followed by fixing with 4% paraformaldehyde for 15 min and permeabilizing for 15 min with 0.5% Triton X-100. Subsequently, samples were incubated with VP1 antibody (1:200) at 4°C overnight. Finally, the positive cells were visualized with Alexa Fluor 647 Conjugate anti-mouse immunoglobulins (Cell Signaling Technology, Danvers, MA, United States) and DAPI (Beyotime, Shanghai, China) and recorded by a Leica confocal microscope.
The construction strategy for the transfer vector of pFB-T7-BrCr and pFB-T7-GFP-BrCr is as shown in Figure 1. Then recombinant bacmid DNA was transfected into Sf9 cells to produce recombinant baculovirus virus, and the virus Ac- T7-BrCr, which can express T7 RNA polymerase in mammalian cells and contains the T7 promoter before the genome of BrCr was constructed using a Bac-to-Bac expression system. Furthermore, to observe whether infectious EV71 can be synthesized after transduction, we constructed the baculovirus Ac-T7-GFP-BrCr carrying a GFP gene in the genome of EV71 as previously described (Shang et al., 2013).
Figure 1. Construction strategy of transfer vectors. The amplified gene of T7 RNA polymerase was inserted into pFB-CMV-EF1-α by SalI and HindIII. cDNA from the 5′ end to nucleotide position 2,915 with MIuI with a T7 promoter before the 5′ end of the BrCr genome was cloned into pFB-CMV-T7-EF1-α by NsiI and SmaI, and the rest of the cDNA was cloned by SmaI and MIuI.
To obtain the recovered infectious virus from the full-length cDNA clone, we transduced Vero cells with the baculoviruses Ac-T7-BrCr, Ac-T7-GFP-BrCr, or AcMNPV. Rescued EV71-induced cytopathic effects (CPEs) and fluorescence appeared 18 h posttransduction, and approximately 80% of the cells were rounded and lost normal cell morphology 48 h posttransduction compared with the controls. This result indicated that EV71 cDNA with a 5′-terminal T7 promoter was efficiently transcribed to generate infectious virus that replicated in permissive Vero cells and showed a specific CPE (Figure 2). At the same time, the culture medium from the transfected cells was collected, and viral titers were determined to be 3.6 × 106 and 1.8 × 106 TCID50/ml for Ac-T7-BrCr and Ac-T7-GFP-BrCr, respectively. To further confirm the recovered EV71, an immunofluorescence assay was performed to analyze the expression of viral structural protein VP1. The immunofluorescence assay results showed that, consistent with the VP1 expression of WT BrCr, the VP1 of recovered viruses was also detected (Supplementary Figure 1).
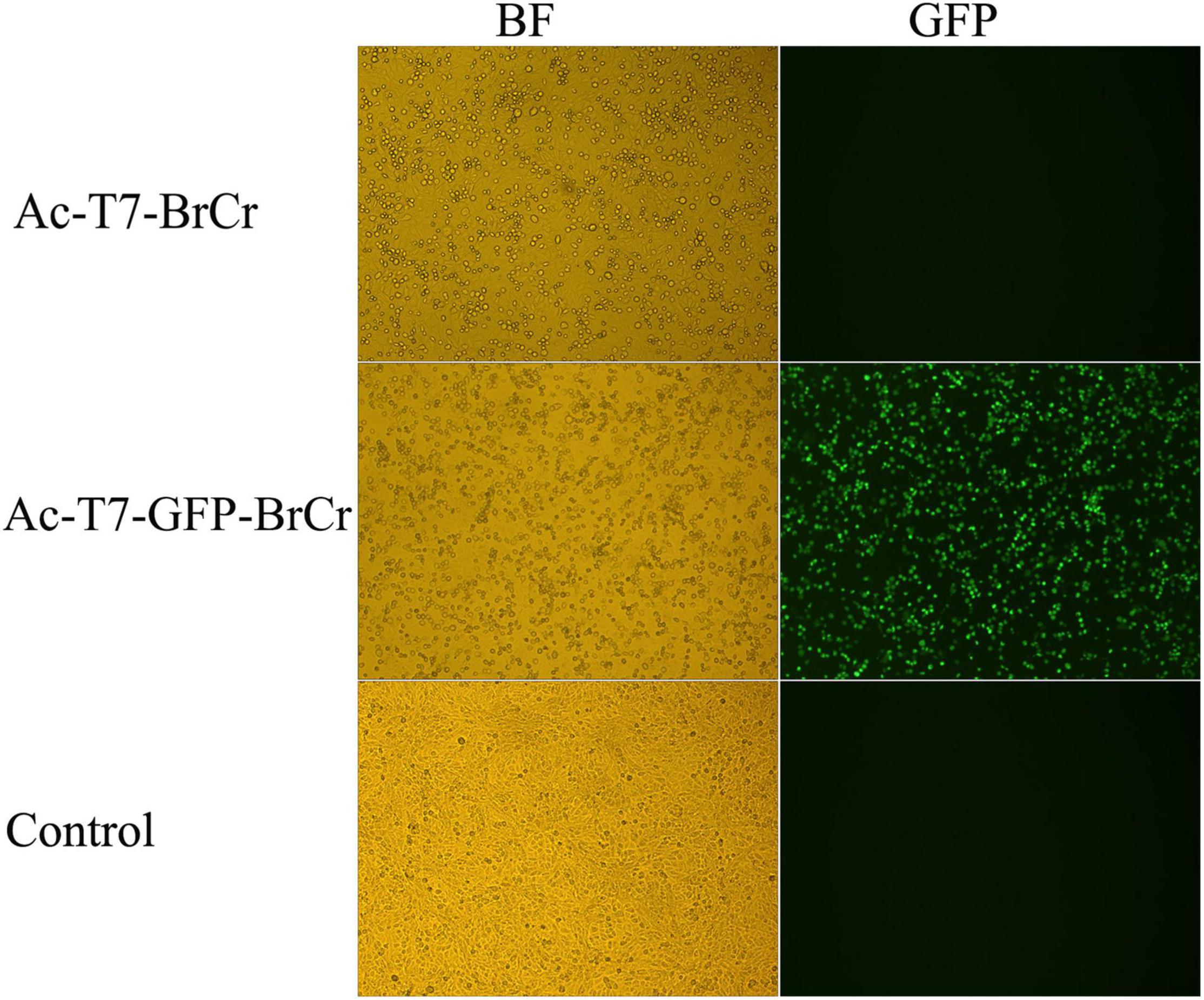
Figure 2. Enterovirus 71 (EV71) reconstruction in Vero cells. Vero cells were transduced with the baculoviruses Ac-pFB-T7-GFP-BrCr-BrCr, Ac-T7-GFP-BrCr, or wild-type AcMNPV. The left and right panels represent the same field of view. The left panels were visualized in bright field, and the right panels were visualized with a fluorescein isothiocyanate (FITC) filter set.
To confirm the EV71 recovery, we performed Western blotting to examine the expression of VP1, the major structural protein for EV71 in several mammalian cells (Figure 3A). Forty-eight hours posttransduction with Ac-T7-BrCr or AcMNPV at an MOI of 50, the debris of Vero, Rd, Sk, and BHK cells was collected for analysis. The results showed obvious signals in the Vero, Rd, and Sk cells, while an indistinguishable band was observed in the BHK cells. The cells transduced with wild-type AcMNPV were also analyzed as a negative control.
Figure 3. Confirmation of EV71 reconstruction in cell culture. (A) Expression of VP1 in mammalian cells transduced with vAc-T7-BrCr. Cells were harvested at 48 h post-transduction, subjected to 10% SDS–PAGE, and immunostained with anti-EV71-VP1 (upper panel) or anti-β-actin (lower panel) antibody. (B) mRNA analysis of Vp1 by real-time PCR after transduction with vAc-T7-BrCr in Vero, Rd, Sk, and BHK cells. All data are represented as the mean ± standard deviation of a set of triplicates. (C) Transmission electron microscopy images of the transduced cells. Thirty-six hours after transduction with vAc-T7-BrCr, Vero (left) and Rd cells (right) were fixed and examined for rescued virus. Viruses with a diameter of approximately 30 nm were clearly observed in the cytoplasm of the cells.
The generation of viral RNA was subsequently confirmed by real-time PCR. Consistent with the Western blotting results, as shown in Figure 3B, viral RNA in Vero cells transduced with Ac-T7-BrCr was clearly detected and was 10 times higher than that in Rd cells, but the other two cell lines showed detectable levels. This result revealed that recombinant baculovirus delivered the genome to the mammalian cells, but the efficiencies in these three EV71 susceptible cells were different.
Observation of viruses by electron microscopy was a direct evidence for rescued viruses. Therefore, Vero and Rd cells were examined using electron microscopy after transduction with baculovirus Ac-T7-BrCr. TEM images showed that large scales of viruses were in the cytoplasm and had a diameter of approximately 30 nm (Figure 3C), which was not observed in the control group that was transduced with wild-type baculovirus ACMNPV (Supplementary Figure 2).
To investigate whether rEV71 could produce infectious viruses, we used one step growth curve and plaque assay. As shown in Figure 4A, rEV71 showed proliferation rates similar to those of WT EV71 when an MOI of 1 was used to infect Rd cells. At 24 h postinoculation, both strains of EV71 reached peak viral titers, which subsequently declined at 30 h postinoculation. Additionally, similar plaque morphology and number were observed between WT EV71 and rEV71 (Figure 4B).
Figure 4. Characterization of rescued EV71. (A) Viral growth kinetics of rEV71 and WT EV71. (B) Plaque assays of rEV71 and WT EV71.
To identify the reconstruction of EV71 in vivo, we intracranially injected the baculoviruses Ac-T7-BrCr and AvMNPV into 1-day-old ICR mice. The transcription and translation of VP1 were detected by RT-PCR and immunohistochemistry methods 7 days after inoculation. A definite fragment was amplified from brain samples by RT-PCR, and the sequence was exactly identical to that of the parental virus (Figure 5A). Additionally, the immunohistochemical expression and localization of VP1 were noted in brown for tissues from the brains injected with Ac-T7-BrCr, while no signal was observed in the control group, which indicated the existence of the rescued virus (Figure 5B). Furthermore, at 36 h postincubation of purified supernatant from mouse brains with Rd cells, an obvious CPE was detected, which further demonstrated the infectivity of the rescued virus (Figure 5C).
Figure 5. Virus identification of inoculated mice. (A) Amplification and sequence confirmation of Vp1 from brain tissue by reverse transcription polymerase chain reaction (RT-PCR). (B) Immunohistochemical expression and localization of VP1. Numerous VP1 protein inclusions were observed in the cytoplasm. (C) Reinoculation of purified tissue supernatant into Rd cells and CPE appeared 36 h postinoculation.
The baculovirus expression system has been extensively applied not only for eukaryotic protein expression in insect cells but also for gene delivery to vertebrate cells, even primary and stem cells (Boyce and Bucher, 1996; Jarvis, 2009). Because of its high capacity for DNA, high transduction efficiency, and biosafety, this system has also been widely used for biotechnology, gene therapy, and vaccine research (Kost et al., 2005). Additionally, a lack of replication ability and low toxicity are the advantageous features for gene expression compared with the widely used vaccinia-T7 polymerase system (Fuerst et al., 1986).
Enterovirus 71 is one of the most common enteroviruses that cause hand–foot–mouth disease (HFMD) and even fatal encephalitis in young children. In addition to the development of antiviral treatment strategies, the major research focus is revealing the mechanisms of EV71 pathogenicity and fundamental questions in EV71 molecular biology and replication. Thus, the availability of full-length genomic cDNA clones for functional analysis of EV71 is a requirement. Furthermore, construction and characterization of an infectious cDNA clone of EV71 will contribute to research on the virus–host interaction, particularly when the virus does not have an efficient in vitro replication system (Delaney and Isom, 1998; Arita et al., 2005).
For the generation of infectious virus, the transcription plasmids for full-length cDNA clone of EV71 were linearized, followed by in vitro transcription by T7 RNA polymerase, and then the infectious RNA transcripts were purified and transfected into the indicated cells. However, the efficiency of transcription and RNA stability are possible obstacles for rescue of viruses. In this research, EV71 cDNA and the T7 RNA polymerase gene were cloned in one recombinant baculovirus (Figure 1), which strongly improved the efficiency of entering target cells. After transduction in mammalian cells, typical CPEs, such as cell rounding, fall off, floatation, etc., or fluorescence signals were visualized in the cells transduced with recombinant baculoviruses (Figure 2). Furthermore, we observed virus particles of approximately 30 nm located in the cytoplasm, which could not been seen in the control group transduced with wild-type baculovirus AcMNPV (Supplementary Figure 2), in Rd and Vero cells by electron microscopy. Then one-step growth curve and plaque assay were administrated to understand the infectivity of the progeny virus. Our results indicated that infectious EV71 was recovered and had similar growth kinetics and plaque morphology with wild-type EV71 (Figure 4). These results indicated that, compared with other strategies (Arita et al., 2005; Zhang et al., 2020; Yang et al., 2021), EV71 was rescued by a baculovirus expression system and displayed characteristics similar to those of the parental strain. Notably, this rescue system that delivered EV71 cDNA and the T7 RNA polymerase gene together by transduction into mammalian cells eliminated the requirement of additional T7 RNA polymerase and in vitro RNA synthesis, which increased the efficiency and cost savings of reverse genetics systems for basic research.
Baculoviruses with mammalian promoters have been harnessed for efficient gene delivery to mammalian cells and enter non-permissive cells for gene delivery (Mansouri et al., 2016; Ono et al., 2018; Hu et al., 2019). For some viruses that have a strict virus–host range, the major hurdles in research are no cell model systems in vitro and in vivo. For example, recombinant baculovirus was applied to HBV, and consistent HBV genes and infectious virions were observed upon transduction to liver cell lines (Delaney and Isom, 1998; Lucifora et al., 2008). Previous studies have reported that baculovirus systems can rescue viruses in mammalian cells, but the biological characteristics of the rescued viruses depend on themselves and are not related to baculoviruses (Yap et al., 1997). Here, we constructed EV71 infectious clones with a baculovirus system and detected the gene expression of rescued virus in different cells. The accumulation of viral RNA and protein expression of VP1 in Vero, Rd, Sk, and BHK cells after transduction with 50 MOI of baculovirus Ac-T7-BrCr were observed. The results showed a positive signal that varied in different target cells, whereas there was no signal in BHK (Figures 3A,B). Kost et al. (2005) reported that a defect in nuclear transport in certain cell types may block efficient baculovirus-mediated gene delivery. Therefore, the susceptibility to baculovirus transduction or the discrepancy in expression levels that differ in mammalian cells need to be further investigated. In line with previous studies, our results showed that Vero, Rd, and Sk cells were permissive for EV71 replication but not BHK cells (Yamayoshi et al., 2009), although these four cell lines were efficiently transduced by baculovirus (Liang et al., 2004). This result suggests that even though baculovirus has a high transduction efficiency of BHK, infectious EV71 cannot be obtained because of the lack of cell-to-cell spread; in a sense, this baculovirus system with broad host ranges may allow screening of permissive cells for viruses without cell culture or animal models.
In addition to in vitro gene expression, baculovirus has been widely used for gene delivery in vivo. Sarkis et al. (2000) reported that GFP was expressed in neural cells of mouse and rat brains after injection with baculovirus containing the GFP gene under the control of the cytomegalovirus promoter. Lehtolainen et al. (2002) showed unmodified baculovirus choroid plexus cells in rat ventricles. Hideki et al. also demonstrated that recombinant baculoviruses could efficiently transfer reporter genes not only into primary neural cells in vitro but also into the cerebrums of mice in vivo (Tani et al., 2003). It has been reported that complement restricts the baculovirus vector transcription when it is injected intravenously or directly into the liver (Sandig et al., 1996). Interestingly, infectious EV71 was successfully produced in an in vivo model by the baculovirus delivery system in our study (Figure 5). To the best of our knowledge, this is the first report that EV71 recombinant baculovirus can be delivered to, and replicate in, neurocyte after intracerebral injection of purified recombinant baculovirus (Figure 5B). The Vp1 gene and protein from the mouse brain were detected by RT-PCR and immunohistochemistry (Figure 5A). Moreover, a typical CPE was observed after inoculation of the supernatant from mouse brain with Rd cells (Figure 5C). All these results indicated that we successfully reconstructed EV71 by a baculovirus delivery system in vivo, which can provide a safe and convenient platform for the exploration of animal models and vaccines (Sarkis et al., 2000; Lehtolainen et al., 2002; Tani et al., 2003; Zhang et al., 2020).
In conclusion, we established a novel approach to rescue EV71 in vitro and in vivo based on a baculovirus expression system, which may provide with a safe and convenient platform for fundamental research and a strategy to rescue viruses that currently lack suitable cell culture and animal models.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The animal study was reviewed and approved by (No: WIVA07201502) the Institutional Animal Care and Use Committee of Wuhan Institute of Virology, Chinese Academy of Sciences.
HW, BLi, and LW were responsible for the design of the work. BLu and QT performed and drafted the manuscript. QT, QW, XL, ZL, and HP were responsible for the literature review, data analysis, and revision of the manuscript. All authors contributed to the article, read, and approved the final manuscript.
This work was supported by the Inflammation and Immune Mediated Diseases Laboratory of Anhui Province (IMMDL202007), National Natural Science Foundation of China (81401658 and 82102374), Scientific Research Project of Shenzhen Health and Family Planning System (SZBC2018009), Open Research Fund Program of National Biosafety Laboratory, Wuhan (NBL, 2018SPCAS002). Clinical Medicine Discipline Construction Project of Anhui Medical University (2021lcxk008).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors thank Xuefang An for the help of animal experiments and Arita for kindly providing the full-length BrCr cDNA clone.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.825111/full#supplementary-material
Supplementary Figure 1 | Immunofluorescence assay of rescued EV71 in Vero cells. Immunofluorescence assay was carried out to test the expression of viral structural protein VP1 (red) after transducting Vero cells with wide type AcMNPV (left), v-Ac-T7-BrCr (middle), and v-T7-GFP-BrCr (right).
Supplementary Figure 2 | Transmission electron microscopy images of Vero (left) and Rd (right) cells that transduced with the wide type baculovirus AcMNPV control.
Arita, M., Shimizu, H., Nagata, N., Ami, Y., Suzaki, Y., Sata, T., et al. (2005). Temperature-sensitive mutants of enterovirus 71 show attenuation in cynomolgus monkeys. J. Gen. Virol. 86, 1391–1401. doi: 10.1099/vir.0.80784-0
Bai, B., Lu, X., Meng, J., Hu, Q., Mao, P., Lu, B., et al. (2008). Vaccination of mice with recombinant baculovirus expressing spike or nucleocapsid protein of SARS-like coronavirus generates humoral and cellular immune responses. Mol. Immunol. 45, 868–875. doi: 10.1016/j.molimm.2007.08.010
Boyce, F. M., and Bucher, N. L. (1996). Baculovirus-mediated gene transfer into mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 93, 2348–2352. doi: 10.1073/pnas.93.6.2348
Boyer, J. C., and Haenni, A. L. (1994). Infectious transcripts and cDNA clones of RNA viruses. Virology 198, 415–426. doi: 10.1006/viro.1994.1053
Cockrell, A. S., Beall, A., Yount, B., and Baric, R. (2017). Efficient Reverse Genetic Systems for Rapid Genetic Manipulation of Emergent and Preemergent Infectious Coronaviruses. Methods Mol. Biol. 1602, 59–81. doi: 10.1007/978-1-4939-6964-7_5
Cross, R. W., and Geisbert, T. W. (2019). Use of reverse genetics to inform Ebola outbreak responses. Lancet Infect. Dis. 19, 925–927. doi: 10.1016/s1473-3099(19)30346-9
Delaney, W. E. T., and Isom, H. C. (1998). Hepatitis B virus replication in human HepG2 cells mediated by hepatitis B virus recombinant baculovirus. Hepatology 28, 1134–1146. doi: 10.1002/hep.510280432
Fuerst, T. R., Niles, E. G., Studier, F. W., and Moss, B. (1986). Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 83, 8122–8126. doi: 10.1073/pnas.83.21.8122
Han, J. F., Cao, R. Y., Tian, X., Yu, M., Qin, E. D., and Qin, C. F. (2010). Producing infectious enterovirus type 71 in a rapid strategy. Virol. J. 7:116. doi: 10.1186/1743-422x-7-116
Hu, L., Li, Y., Ning, Y. J., Deng, F., Vlak, J. M., Hu, Z., et al. (2019). The Major Hurdle for Effective Baculovirus Transduction into Mammalian Cells Is Passing Early Endosomes. J. Virol. 93, 1–18. doi: 10.1128/jvi.00709-19
Jarvis, D. L. (2009). “Chapter 14 Baculovirus–Insect Cell Expression Systems,” in Methods in Enzymology, eds R. R. Burgess and M. P. Deutscher (Cambridge: Academic Press), 191–222.
Kim, H. J., Son, H. S., Lee, S. W., Yoon, Y., Hyeon, J. Y., Chung, G. T., et al. (2019). Efficient expression of enterovirus 71 based on virus-like particles vaccine. PLoS One 14:e0210477. doi: 10.1371/journal.pone.0210477
Kost, T. A., Condreay, J. P., and Jarvis, D. L. (2005). Baculovirus as versatile vectors for protein expression in insect and mammalian cells. Nat. Biotechnol. 23, 567–575. doi: 10.1038/nbt1095
Lawson, N. D., Stillman, E. A., Whitt, M. A., and Rose, J. K. (1995). Recombinant vesicular stomatitis viruses from DNA. Proc. Natl. Acad. Sci. U. S. A. 92, 4477–4481. doi: 10.1073/pnas.92.10.4477
Lehtolainen, P., Tyynelä, K., Kannasto, J., Airenne, K. J., and Ylä-Herttuala, S. (2002). Baculoviruses exhibit restricted cell type specificity in rat brain: a comparison of baculovirus- and adenovirus-mediated intracerebral gene transfer in vivo. Gene Ther. 9, 1693–1699. doi: 10.1038/sj.gt.3301854
Liang, C. Y., Wang, H. Z., Li, T. X., Hu, Z. H., and Chen, X. W. (2004). High efficiency gene transfer into mammalian kidney cells using baculovirus vectors. Arch. Virol. 149, 51–60. doi: 10.1007/s00705-003-0197-3
Lucifora, J., Durantel, D., Belloni, L., Barraud, L., Villet, S., Vincent, I. E., et al. (2008). Initiation of hepatitis B virus genome replication and production of infectious virus following delivery in HepG2 cells by novel recombinant baculovirus vector. J. Gen. Virol. 89, 1819–1828. doi: 10.1099/vir.0.83659-0
Mansouri, M., Bellon-Echeverria, I., Rizk, A., Ehsaei, Z., Cianciolo Cosentino, C., Silva, C. S., et al. (2016). Highly efficient baculovirus-mediated multigene delivery in primary cells. Nat. Commun. 7:11529. doi: 10.1038/ncomms11529
Mao, Q. Y., Wang, Y., Bian, L., Xu, M., and Liang, Z. (2016). EV71 vaccine, a new tool to control outbreaks of hand, foot and mouth disease (HFMD). Expert Rev. Vaccines 15, 599–606. doi: 10.1586/14760584.2016.1138862
Martino, T. A., Tellier, R., Petric, M., Irwin, D. M., Afshar, A., and Liu, P. P. (1999). The complete consensus sequence of coxsackievirus B6 and generation of infectious clones by long RT-PCR. Virus Res. 64, 77–86. doi: 10.1016/s0168-1702(99)00081-7
Meng, T., Kiener, T. K., and Kwang, J. (2012). RNA polymerase I-driven reverse genetics system for enterovirus 71 and its implications for vaccine production. Virol. J. 9:238. doi: 10.1186/1743-422x-9-238
Ono, C., Okamoto, T., Abe, T., and Matsuura, Y. (2018). Baculovirus as a Tool for Gene Delivery and Gene Therapy. Viruses 10:510. doi: 10.3390/v10090510
Pham, L. V., Jensen, S. B., Fahnøe, U., Pedersen, M. S., Tang, Q., Ghanem, L., et al. (2019). HCV genotype 1-6 NS3 residue 80 substitutions impact protease inhibitor activity and promote viral escape. J. Hepatol. 70, 388–397. doi: 10.1016/j.jhep.2018.10.031
Racaniello, V. R., and Baltimore, D. (1981). Cloned poliovirus complementary DNA is infectious in mammalian cells. Science 214, 916–919. doi: 10.1126/science.6272391
Roldão, A., Vicente, T., Peixoto, C., Carrondo, M. J., and Alves, P. M. (2011). Quality control and analytical methods for baculovirus-based products. J. Invertebr. Pathol. 107, S94–S105. doi: 10.1016/j.jip.2011.05.009
Sandig, V., Hofmann, C., Steinert, S., Jennings, G., Schlag, P., and Strauss, M. (1996). Gene transfer into hepatocytes and human liver tissue by baculovirus vectors. Hum. Gene Ther. 7, 1937–1945. doi: 10.1089/hum.1996.7.16-1937
Sarkis, C., Serguera, C., Petres, S., Buchet, D., Ridet, J. L., Edelman, L., et al. (2000). Efficient transduction of neural cells in vitro and in vivo by a baculovirus-derived vector. Proc. Natl. Acad. Sci. U. S. A. 97, 14638–14643. doi: 10.1073/pnas.260472897
Shang, B., Deng, C., Ye, H., Xu, W., Yuan, Z., Shi, P. Y., et al. (2013). Development and characterization of a stable eGFP enterovirus 71 for antiviral screening. Antiviral Res. 97, 198–205. doi: 10.1016/j.antiviral.2012.12.010
Solomon, T., Lewthwaite, P., Perera, D., Cardosa, M. J., McMinn, P., and Ooi, M. H. (2010). Virology, epidemiology, pathogenesis, and control of enterovirus 71. Lancet Infect. Dis. 10, 778–790. doi: 10.1016/s1473-3099(10)70194-8
Svensson, L., Hjalmarsson, A., and Everitt, E. (1999). TCID50 determination by an immuno dot blot assay as exemplified in a study of storage conditions of infectious pancreatic necrosis virus. J. Virol. Methods 80, 17–24. doi: 10.1016/s0166-0934(99)00018-x
Tang, Q., Xu, Z., Jin, M., Shu, T., Chen, Y., Feng, L., et al. (2020). Identification of dibucaine derivatives as novel potent enterovirus 2C helicase inhibitors: in vitro, in vivo, and combination therapy study. Eur. J. Med. Chem. 202:112310. doi: 10.1016/j.ejmech.2020.112310
Tani, H., Limn, C. K., Yap, C. C., Onishi, M., Nozaki, M., Nishimune, Y., et al. (2003). In vitro and in vivo gene delivery by recombinant baculoviruses. J. Virol. 77, 9799–9808. doi: 10.1128/jvi.77.18.9799-9808.2003
Tellier, R., Bukh, J., Emerson, S. U., and Purcell, R. H. (1996). Amplification of the full-length hepatitis A virus genome by long reverse transcription-PCR and transcription of infectious RNA directly from the amplicon. Proc. Natl. Acad. Sci. U. S. A. 93, 4370–4373. doi: 10.1073/pnas.93.9.4370
Thi Nhu Thao, T., Labroussaa, F., Ebert, N., V’Kovski, P., Stalder, H., Portmann, J., et al. (2020). Rapid reconstruction of SARS-CoV-2 using a synthetic genomics platform. Nature 582, 561–565. doi: 10.1038/s41586-020-2294-9
Todd, K. V., and Tripp, R. A. (2019). Human Norovirus: experimental Models of Infection. Viruses 11:151. doi: 10.3390/v11020151
van den Worm, S. H., Eriksson, K. K., Zevenhoven, J. C., Weber, F., Züst, R., Kuri, T., et al. (2012). Reverse genetics of SARS-related coronavirus using vaccinia virus-based recombination. PLoS One 7:e32857. doi: 10.1371/journal.pone.0032857
Wang, J., Hu, Y., and Zheng, M. (2021). Enterovirus A71 antivirals: past, present, and future. Acta Pharm. Sin. B. doi: 10.1016/j.apsb.2021.08.017 [Epub ahead of print].
Wang, R., Deng, F., Hou, D., Zhao, Y., Guo, L., Wang, H., et al. (2010). Proteomics of the Autographa californica nucleopolyhedrovirus budded virions. J. Virol. 84, 7233–7242. doi: 10.1128/jvi.00040-10
Yamayoshi, S., Yamashita, Y., Li, J., Hanagata, N., Minowa, T., Takemura, T., et al. (2009). Scavenger receptor B2 is a cellular receptor for enterovirus 71. Nat. Med. 15, 798–801. doi: 10.1038/nm.1992
Yang, H., Zhao, X., Xun, M., Ma, C., and Wang, H. (2021). Reverse Genetic Approaches for the Generation of Full Length and Subgenomic Replicon of EV71 Virus. Front. Microbiol. 12:665879. doi: 10.3389/fmicb.2021.665879
Yap, C. C., Ishii, K., Aoki, Y., Aizaki, H., Tani, H., Shimizu, H., et al. (1997). A hybrid baculovirus-T7 RNA polymerase system for recovery of an infectious virus from cDNA. Virology 231, 192–200. doi: 10.1006/viro.1997.8537
Yu, J., Liu, R., Zhou, B., Chou, T. W., Ghedin, E., Sheng, Z., et al. (2019). Development and Characterization of a Reverse-Genetics System for Influenza D Virus. J. Virol. 93, e01186–19. doi: 10.1128/jvi.01186-19
Zhang, H., Song, Z., Zou, J., Feng, Y., Zhang, J., Ren, L., et al. (2020). An infectious clone of enterovirus 71(EV71) that is capable of infecting neonatal immune competent mice without adaptive mutations. Emerg. Microbes Infect. 9, 427–438. doi: 10.1080/22221751.2020.1729665
Keywords: enterovirus 71, recovery, baculovirus, infectious complementary deoxyribonucleic acid (cDNA) clone, in vitro and in vivo
Citation: Lu B, Tang Q, Wang Q, Liu X, Peng H, Zhu B, Xie L, Li Z, Wang H, Zheng Z, Wang L and Li B (2022) Recovery Infectious Enterovirus 71 by Bac-to-Bac Expression System in vitro and in vivo. Front. Microbiol. 13:825111. doi: 10.3389/fmicb.2022.825111
Received: 30 November 2021; Accepted: 19 January 2022;
Published: 25 February 2022.
Edited by:
Jun Wang, Rutgers, The State University of New Jersey, United StatesReviewed by:
Feifei Yin, Hainan Medical University, ChinaCopyright © 2022 Lu, Tang, Wang, Liu, Peng, Zhu, Xie, Li, Wang, Zheng, Wang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bao Li, bGliYW9oeUAxNjMuY29t; Linding Wang, d2FuZ2xpbmRpbmdAYWhtdS5lZHUuY24=; Baojing Lu, bHViYW9qaW5nQDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.