- 1State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-Products, Institute of Food Sciences, Zhejiang Academy of Agricultural Sciences, Hangzhou, China
- 2Institute of Agro-Product Safety and Nutrition, Zhejiang Academy of Agricultural Sciences, Hangzhou, China
IR Biotyper (IRBT), which is a spectroscopic system for microorganism typing based on Fourier transform infrared (FTIR) technology, has been used to detect the spread of clones in clinical microbiology laboratories. However, the use of IRBT to detect probiotics has rarely been reported. Herein, we evaluated the discriminatory power of IRBT to type Lactiplantibacillus plantarum isolates at the strain level and explored its application potential in probiotic preliminary selection. Twenty Lactiplantibacillus isolates collected from pickled radishes during successive fermentation were used to test the robustness of IRBT at the strain level. IRBT was then compared with genotyping methods such as whole-genome sequencing (WGS), pulsed-field gel electrophoresis (PFGE), and multilocus sequence typing (MLST) to evaluate its discrimination power. IRBT distributed the 20 isolates into five clusters, with L. argentoratensis isolate C7-83 being the most distant from the other isolates, which belonged to L. plantarum. IRBT showed good reproducibility, although deviation in the discriminative power of IRBT was found at the strain level across laboratories, probably due to technical variance. All examined methods allowed bacterial identification at the strain level, but IRBT had higher discriminatory power than MLST and was comparable to the WGS and PFGE. In the phenotypic comparison study, we observed that the clustering results of probiotic physiological attributes (e.g., sensitivity to acid and bile salts, hydrophobicity of the cell surface, and resistance to antibiotics) were consistent with the typing results of IRBT. Our results indicated that IRBT is a robust tool for L. plantarum strain typing that could improve the efficiency of probiotic identification and preliminary screening, and can potentially be applied in probiotic traceability and quality control.
Introduction
Lactiplantibacillus plantarum (formerly Lactobacillus plantarum), which is a homofermentative lactic acid bacterium (LAB), has been widely applied as a model species for ecological, metabolic, and genetic studies in lactobacilli (Zheng et al., 2020). In addition, L. plantarum is used as a starter culture in multiple food and beverage fermentations and as a probiotic for both animals and humans, making it of considerable academic and economic importance (das Neves Selis et al., 2021). L. plantarum has been classified as a “nomadic” or “generalist” bacterium because of its diverse habitats (Martino et al., 2016; Choi et al., 2018; Yu et al., 2020, 2021). Consistent with its broad environmental and host ranges, L. plantarum strains have significant intraspecific genetic and phenotypic versatility due to strain-specific genes (Molenaar et al., 2005; Zheng et al., 2015; Duar et al., 2017; Salvetti et al., 2018; Cen et al., 2020; Pan et al., 2021). The latest taxonomic research has elevated L. plantarum subsp. plantarum and L. plantarum subsp. argentoratensis to the species level as L. plantarum and L. argentoratensis, respectively (Liu and Gu, 2020; Zheng et al., 2020).
Food and Agriculture Organization [FAO]/World Health Organization [WHO] (2002) published the “Guidelines for Evaluation of Probiotics in Food,” the selection criteria of which included host-related stress resistance, epithelial adhesion, antimicrobial activity, and safety assessment. These aspects serve to ensure that candidate probiotics can withstand unfavorable gastrointestinal conditions, colonize intestinal epithelial cells, and contribute to host health, leading to challenging probiotic screening processes based on stringent selection criteria. In most cases, the large number of probiotic candidates lead to the use of a “step-by-step approach” involving a series of in vitro required tests to progressively reduce the number of candidate probiotics (de Melo Pereira et al., 2018; Yoha et al., 2021). Because the properties and benefits of L. plantarum are strain-specific, an easy-to-use method for fast and effective L. plantarum strain typing is highly desirable in both academia and industry (Fuhren et al., 2020).
Whole genome sequencing (WGS), which provides more consistent genetic information is replacing pulsed-field gel electrophoresis (PFGE) as the new “gold standard” for identifying and classifying microorganism (Neoh et al., 2019). However, regardless of their discriminatory power, the high cost, laboriousness, and time consumption of the genotypic technologies prevent their routine implementation on a large-scale basis (Quintelas et al., 2018). In addition, phenotype prediction based on genetic data is not always straightforward, and the genetic single nucleotide polymorphism (SNP) thresholds for strain delineation in bacteriology have not been universally set (Van Rossum et al., 2020). In this context, spectroscopy-based techniques such as matrix-assisted laser desorption/ionization–time of flight mass spectrometry (MALDI-TOF MS), Raman spectroscopy, and Fourier transform infrared (FTIR) have emerged as more cost-effective, convenient, and faster alternatives in bacterial strain typing (Quintelas et al., 2018).
FTIR is based on using molecular vibration fingerprints to characterize microorganisms according to strain-specific absorbance patterns in the infrared spectrum (Dinkelacker et al., 2018). The main advantages of FTIR are that it is quick, inexpensive, laboratorial simplicity, and nondestructive, has high throughput, and provides relevant information about the biomolecular contents of microorganisms, including lipids, carbohydrates, proteins, and nucleic acids, derived from IR spectra (Quintelas et al., 2018). In particular, IR Biotyper (IRBT) (Bruker Daltonik GmbH, Bremen, Germany) based on FTIR technology was launched in 2017 and emerged as a very promising system in the field of microbial strain typing, with multiple reported successful applications (de las Rivas et al., 2006; Burckhardt et al., 2019; Martak et al., 2019; Hu et al., 2020; Deidda et al., 2021). The application of FTIR is not limited to clinical and epidemiological researches but also extends to the probiotic industry (Deidda et al., 2021). Discriminating probiotics at the strain level is challenging due to their high intraspecific variation, diverse modes of action, and required manufacturing and quality control processes. Therefore, strain-specific verification of a probiotic is fundamental to quarantine its stability, quality, safety, and efficacy. Whether FTIR technology can distinguish L. plantarum at the strain level needs to be further confirmed.
In this study, we aim to (1) test the strain-level typing capability of IRBT for members of the Lactiplantibacillus genus, (2) compare the discriminatory power and concordance of IRBT with those of other genotyping techniques (WGS, PFGE, and MLST), and (3) compare the fingerprint spectra generated by IRBT with the probiotic host-associated stress resistance and safety patterns to pave the way for the introduction of this new complementary phenotypic technique into routine probiotic screening processes.
Materials and Methods
Isolation and Identification of Lactiplantibacillus Strains
The sources of the isolates are listed in Table 1. Pickled radish brines from fermentation days 7 and 21 were collected in sterile bottles and transported to the laboratory for analyses. The suspensions were serially diluted with sterile 0.9% NaCl, pour-plated on de Man, Rogosa and Sharpe (MRS) agar (Oxoid, Basingstoke, United Kingdom) and incubated at 37°C under aerobic or anaerobic conditions for 48 ± 3 h. Single-colony isolates were selected randomly and repeatedly streaked for isolation on MRS prior to characterization. All isolate identities were confirmed at the species level by MALDI-TOF MS (MALDI Biotyper; Bruker Daltonik GmbH, Bremen, Germany). Since MALDI-TOF MS scores ≤2.0 may indicate false species identification, any isolates with scores below 2.0 were additionally confirmed by 16S rRNA gene sequencing analysis. Twenty isolates of L. plantarum were finally selected in this study. Until the WGS comparison, it was found that isolate C7-83 belonged to L. argentoratensis. Isolate R106 was deposited to the China General Microbiological Culture Collection Center (CGMCC) under the deposit number CGMCC 20370. Stock cultures of each isolate were frozen at −80°C in 20% (w/v) glycerol for further studies. For phenotypic and genotypic analysis, the Lactiplantibacillus isolates were routinely grown in MRS medium at 37°C without shaking.
IR Biotyper Analysis
All isolates were grown at 37°C in MRS medium sealed with Parafilm M for 48 ± 3 h. For IRBT analysis, loading samples was prepared according to Hu et al. (2020) by the modified H2O-EtOH method. First, a loopful of bacterial culture was collected and resuspended in 100 μL sterile H2O. After vortexing, 100 μL of 70% (vol/vol) ethanol was added, and the solution was mixed by pipetting to obtain a homogeneous suspension. Then, 15 μL of bacterial suspension was spotted onto the IRBT silicon sample plate and dried at 37°C until a dry film was formed. Three or four replicates were prepared for each sample. For each run, quality control was performed with the Infrared Test Standards (IRTS 1 and 2) in the IR Biotyper kit.
Spectra were recorded in transmission mode in the spectral range of 4,000–500 cm–1 (mid-IR) using an IR Biotyper spectrometer (Bruker Optics-Daltonics GmbH). The spectra were then acquired, visualized, and processed by OPUS v7.5 software (Bruker Optics GmbH). Then, second-derivative FTIR spectra in the polysaccharide absorption region (1,200–9,00 cm–1), which is the default setting of the manufacturer, were vector-normalized and used to amplify differences between isolates and correct variations related to spectral acquisition. Data that did not meet the default quality criteria were excluded from further analyses. Hierarchical clustering analysis (HCA) of the second-derivative IRBT to illustrate the relationships between individual strains was performed offline using IRBT Client Software v2.0 (Bruker Daltonik GmbH). Dendrograms were constructed using Euclidian distances and the average linkage clustering method. For each dataset explored, the IR Biotyper software automatically calculated a clustering cut-off value, which was the result of Simpson’s index of diversity and the mean coherence of the parameter defined by the user.
Reproducibility analysis of IRBT was performed since FTIR spectroscopy techniques are known to be sensitive to variations in culture media, incubation time, temperature, and hygrometry. We evaluated the reproducibility by analyzing the 20 isolates in two laboratories (the State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-Products, and the Clinical Microbiology Laboratory, School of Medicine, The Second Affiliated Hospital of Zhejiang University) independently three times on separate days.
Genotypic Analysis
Genotyping methods such as WGS, PFGE, and MLST were incorporated to compare their discrimination power and accuracy with IRBT (PFGE and MLST subsections refer to Supplementary Material).
Genomic DNA was extracted from the Lactiplantibacillus isolates using a bacterial genome extraction kit (GeneRay, Shanghai, China) according to the manufacturer’s instructions and subjected to whole-genome sequencing using a 150 bp paired library with the Illumina HiSeq 4000 platform at Novogene Bioinformatics Institute (Beijing, China). A total of 21.96 Gb (90.93% out of 24.15 Gb) of high-quality paired-end reads was retained for further analyses. Raw reads were trimmed and assembled into contigs using CLC Genomics Workbench CLCv.12.0.3 (CLC Bio, Aarhus, Denmark). The genomes were automatically annotated by the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) and deposited into GenBank BioProject PRJNA769251. Identification of acquired antimicrobial resistance genes was conducted using the ResFinder 4.0 databases1 (Bortolaia et al., 2020), and mobile genetic elements and their related antimicrobial resistance genes and virulence factors were identified by Mobile Element Finder2. Phylogenetic analysis of the genomes was performed by KSNP3.0 software using the maximum-likelihood method, with SNP detection based on k-mer analysis (Sommer, 2015). The pairwise SNP distance for 20 Lactiplantibacillus isolates was analyzed by the bioinformatics tool snp-dists 0.7.0 in the Bactopia Analysis Pipeline (Petit and Read, 2020). The whole-genome similarity between Lactiplantibacillus isolates was determined by the comparison of average nucleotide identity (ANI) values calculated using JSpecies software (Richter et al., 2016).
Phenotypic Analysis
To correlate IRBT types with phenotypic attributes, ten Lactiplantibacillus isolates (C7-85, C7-7, C7-39, C7-52, R62, R95, R47, R75, R98, and R106), which represented the five clusters, respectively, were further selected for probiotic physiological attributes characterization (Table 1).
Artificial Gastric Juice and Intestinal Juice Susceptibility
Gastric juice susceptibility (GJS) and bile salt susceptibility (BSS) were assessed according to previous studies with some modifications (Silva et al., 2013; Liu et al., 2021). Lactiplantibacillus cultures in the stationary phase were suspended in 0.9% saline at pH 7.0 (control), artificial gastric juice (NaCl 2 g/L, pepsin 3.2 g/L, pH 2.5) or artificial intestinal juice (NaHCO3 150 mM, trypsin 1.9 g/L, pH 8.0) and incubated at 37°C for 3 h. The samples were centrifuged, and the pellets were suspended in 1 mL MRS broth.
For GJS and BSS analyses, each culture was transferred to an Eppendorf microtube and diluted to 2% (v/v) in either MRS broth or MRS broth supplemented with 0.3% bile acids (Hopebio). Then, 200 μL bacterial suspensions were aliquoted into sterile 96-well microplates and incubated in a thermoregulated spectrophotometer (Microplate Spectrophotometer System SpectraMax i3x, Molecular Devices, Sunnyvale, CA, United States) for 18 h at 37°C. The absorbance was determined by the OD620 measured every 30 min. The growth inhibition percentage was calculated using GraphPad Prism 8.0 according to the formula (1–areaS/areaCT) × 100, where areaS and areaCT are the areas under the growth curve of the stressed strains (artificial gastric juice or bile salts) and controls, respectively. The strains were classified as resistant at GJS/BSS <40%, moderately resistant at 40% ≤ GJS/BSS ≤ 75% or susceptible at GJS/BSS >75%. The results were based on the average of three independent assays.
Surface Hydrophobicity Test
The hydrophobic/hydrophilic cell-surface properties of Lactiplantibacillus isolates were assessed by microbial adhesion to solvents (MATS) according to the methodology described by Silva et al. (2013) and Liu et al. (2021). Lactiplantibacillus cultures in the stationary phase were centrifuged, washed twice with PBS, and adjusted to an OD600 of 0.6 with 0.1 M KNO3 at pH 6.2 (A0). Next, 0.4 mL of xylene was mixed with 2.0 mL of each microbial suspension by vortexing for 2 min. The aqueous phase was removed, and the OD600 was measured (A1). MATS was calculated according to the formula (1 – A1/A0) × 100. The isolates were classified as hydrophobic at MATS >70%, amphiphilic at 30% ≤ MATS ≤ 70%, or hydrophilic at MATS <30%. The results were based on the average of three independent assays.
Antibiotic Susceptibility Test
The antibiotic susceptibility test was performed by the agar disk diffusion method (Sandes et al., 2017). Isolated Lactiplantibacillus strains were grown on MRS agar (Oxoid, Basingstoke, United Kingdom) under aerobiosis for 48 h at 37°C. Then, viable cell suspensions at a concentration of 108 (0.5 McFarland scale) were prepared using 0.85% buffered saline and spread onto MRS agar (Oxoid, Basingstoke, United Kingdom). Antibiotic disks were distributed on the surface of agar, and the plates were incubated for around 48 h at 37°C. Then, the diameters of the inhibition zones (mm) were recorded according to automatic inhibition zone measurement with a colony counter (Czone 8; Shineso Science & Technology Co., Ltd., Hangzhou, China). The 15 disks (Oxoid®, Basingstoke, United Kingdom) used contained amoxicillin-AMC (30 μg), erythromycin-E (15 μg), clindamycin-DA (2 μg), chloramphenicol-C (30 μg), tetracycline-TE (30 μg), gentamicin-CN (10 μg), ampicillin-AMP (10 μg), sulfamethoxazole-SXT (25 μg), ceftriaxone-CRO (30 μg), kanamycin-K (30 μg), streptomycin-S (10 μg), enrofloxacin-ENR (5 μg), penicillin-G-P (10 U), cefoxitin-FOX (30 μg), and quinupristin-QD (15 μg). Quality control of the antimicrobial disks was performed using Escherichia coli ATCC 25922. Lactiplantibacillus isolates were classified as resistant, moderately sensitive, or sensitive according to the cutoff levels proposed by Charteris et al. (1998). Aminoglycoside (gentamicin, kanamycin, streptomycin, and neomycin), ciprofloxacin, and trimethoprim antibiotic resistance is considered intrinsic to most Lactobacillus species. Besides, lactobacilli are susceptible to penicillin and β-lactams, chloramphenicol, tetracycline, erythromycin, linezolid, and quinupristin–dalfopristin (Campedelli et al., 2019).
Statistical Analysis
Non-hierarchical clustering (K-means) analysis and principal component analysis (PCA) based on data from gastric juice and bile salt susceptibility, surface hydrophobicity, and antibiotic susceptibility testing of the samples were conducted using R (version 4.0.2) to estimate and classify the probiotic phenotypes of Lactiplantibacillus isolates (R Core Team, 2020).
Results
IR Biotyper Analysis
The WGS typing results were used as a reference to assess the reproducibility of IRBT. For each dendrogram, the IRBT software automatically calculated a COV. The IRBT spectra in Figures 1A,B from the same laboratory clustered the 20 Lactiplantibacillus isolates (three technical replicates) into five different IR types (a∼e) (Supplementary Figure 1), corresponding to five different sequence types (1∼5) according to WGS, while inconsistent with those (four technical replicates) in Figure 1C from a different laboratory. The COV automatically calculated by the IRBT software for Figure 1C was 0.241, which distributed the 20 isolates into seven clusters that were not in agreement with the results of WGS. Therefore, the corrected COV 0.263, which was set on the “right-most” side of the distance between the two nodes (forming a new cluster and a higher distance value for the current nodes), was adopted in our study to avoid such interlaboratory deviation of discriminatory power of IRBT at the strain level. The corrected COVs (ranging from 0.241 to 0.263), all yielded results consistent with those of WGS, indicating good reproducibility and robustness of IRBT.
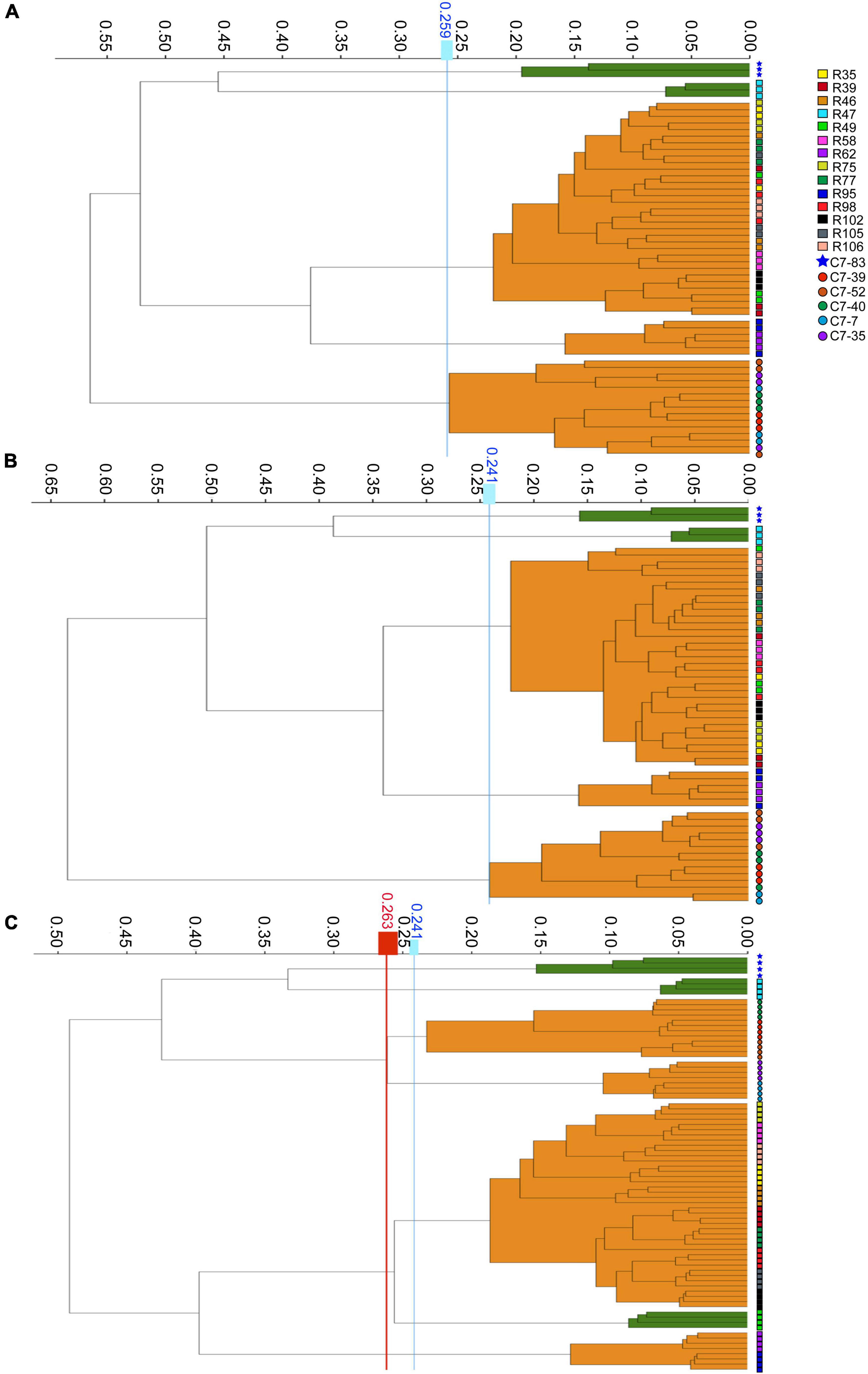
Figure 1. Dendrograms obtained by clustering the IRBT spectra of the twenty Lactiplantibacillus isolates. The vertical lines represent the cutoff values. (A–C) Represent clusters obtained on three independent days with corrected cutoff-off values of 0.259, 0.241, and 0.263, respectively. The IRBT spectra in (A,B) were obtained from the Zhejiang Academy of Agricultural Sciences, and the IRBT spectra in (C) were obtained from The Second Affiliated Hospital of Zhejiang University.
Comparison With Other Genotyping Methods
To generate a reference for the spectrum-based typing method, a whole-genome SNP-based phylogenetic comparison of 20 sequenced Lactiplantibacillus genomes was conducted. The number of SNPs identified by the kSNP that are unique to each node was annotated in the phylogenetic tree (Figure 2). WGS allowed the assignment of the 20 isolates to 5 lineages. Two WGS clusters contained only a single isolate, C7-83 and R47, whereas the other three clusters comprised 2–11 isolates. Within the same lineage, the maximum SNP differences between two isolates in lineage 4 was 76. Supplementary Table 1 shows the pairwise SNP distance matrix for the 20 isolates via a core-genome alignment. We found that intra-strain SNP threshold obtained by snp-dists was 12, inconsistent with the kSNP results. Therefore, using SNP threshold alone to assess whether bacteria originated from the same source can be misleading.
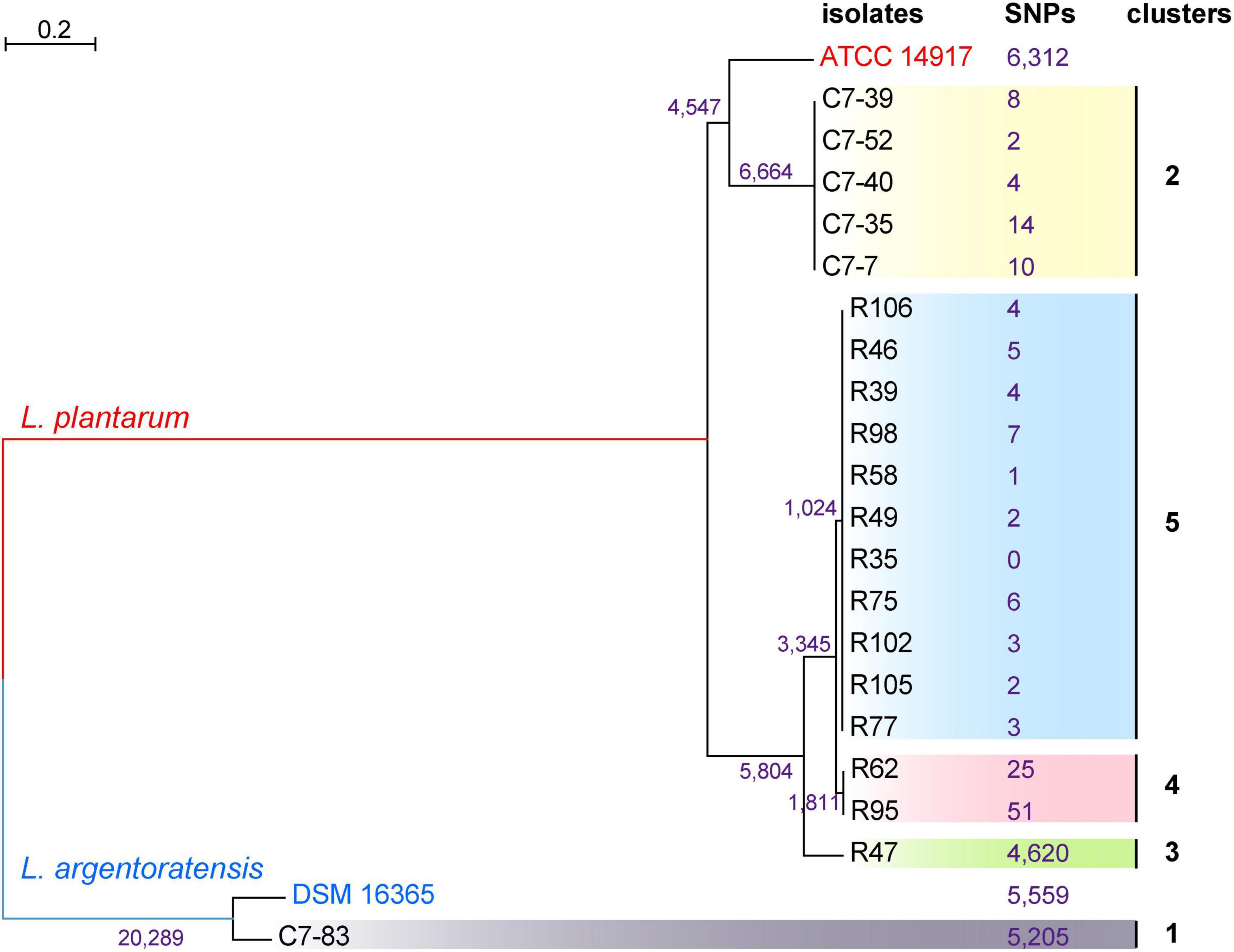
Figure 2. Phylogenetic tree of 20 Lactiplantibacillus isolates and two reference strains, L. argentoratensis DSM 16365 and L. plantarum ATCC 14817 based on whole-genome SNPs, showing five main clades. SNPs that unique to each node identified by the kSNP are marked.
The ANI values between the WGS data of each isolate and the reference genomes from two closely related species, L. plantarum and L. argentoratensis, were calculated (Figure 3). ANI value ≥95% indicates classification as the same species (Richter and Rosselló-Móra, 2009). All isolates showed ANI values >94.8% relative to the two references, with isolate C7-83 and the other 19 isolates displaying ANI values >98.6% relative to the reference strains L. argentoratensis DSM 16365 and L. plantarum ATCC 14817, respectively. ANI analysis confirmed that isolate C7-83 belongs to L. argentoratensis, whereas the other 19 isolates belong to L. plantarum. Hereinafter, the species attributions of the isolates used in this study are derived from the ANI calculations.

Figure 3. Heatmap of the interspecies genomic similarity based on the % ANI of 20 Lactiplantibacillus isolates and two reference strains, L. argentoratensis DSM 16365 and L. plantarum ATCC 14817. The bar on the right represents the color codes for ANI values from pairwise comparisons of isolates.
Comparison of IRBT spectroscopic typing with other genotypic typing results was shown in Table 2. The clustering results of IRBT, WGS, and PFGE were entirely consistent with each other, and all had higher discriminatory power than that of MLST. The MLST method failed to distinguish R62 and R95 from other L. plantarum strains.
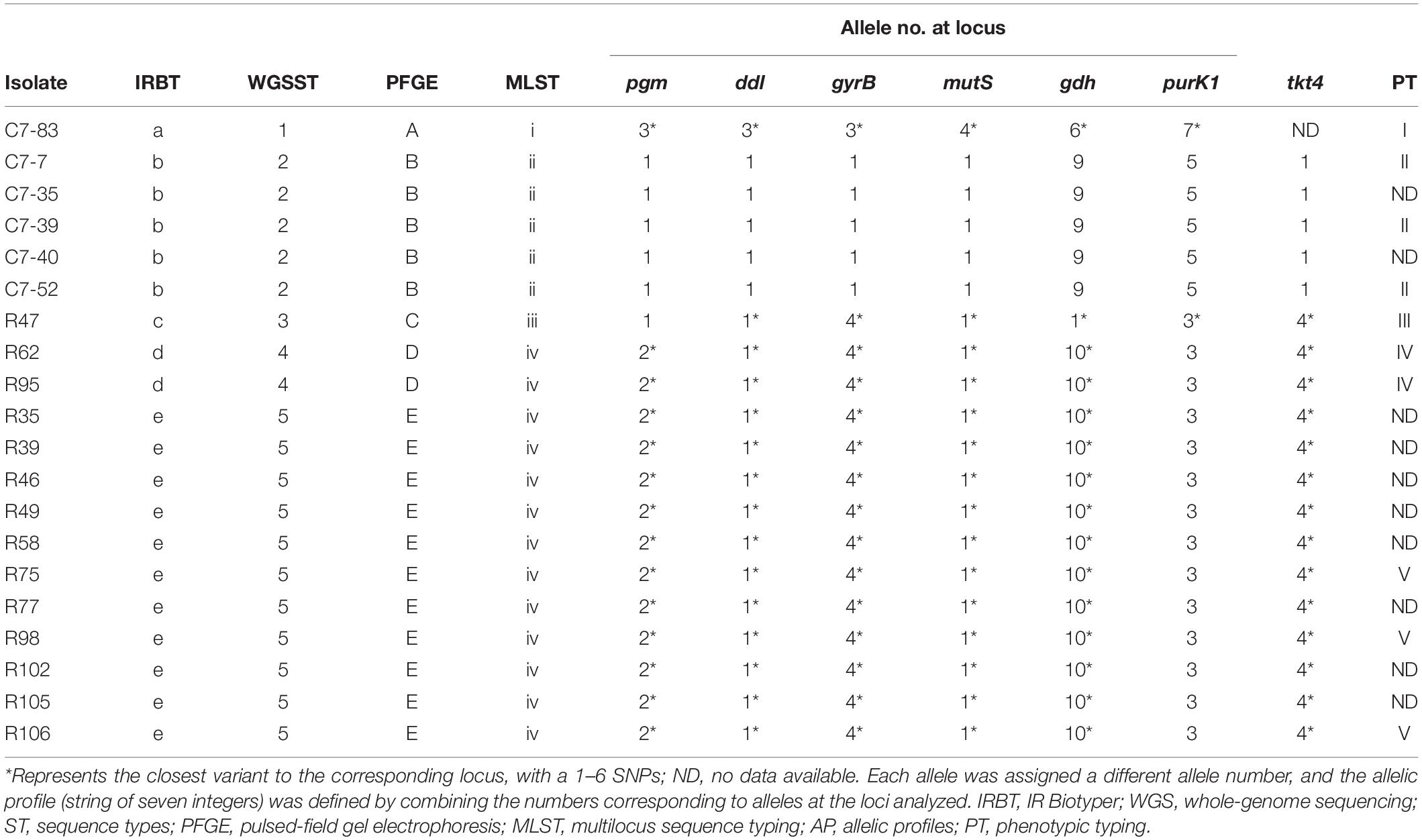
Table 2. Comparison of IRBT spectroscopic typing with other genotypic and phenotypic typing results.
Concordance With Physiological Attributes
Data from the 10 isolates regarding the phenotypes (acid and bile acid susceptibility, surface hydrophobicity, and antibiotic susceptibility) that associate with in vitro probiotic screening (Supplementary Tables 2, 3) were clustered and subjected to PCA analysis. As illustrated in Figure 4A, clustering analysis classified the 10 isolates into three major clusters and five subclusters (I–V). In accordance with the IRBT and genomic results, isolates C7-85 and R47 alone formed clusters I and III, respectively. Isolates R62 and R95 formed cluster IV. Cluster II included isolates C7-7, C7-39, and C7-52, whereas Cluster V contained isolates R75, R98, and R106.
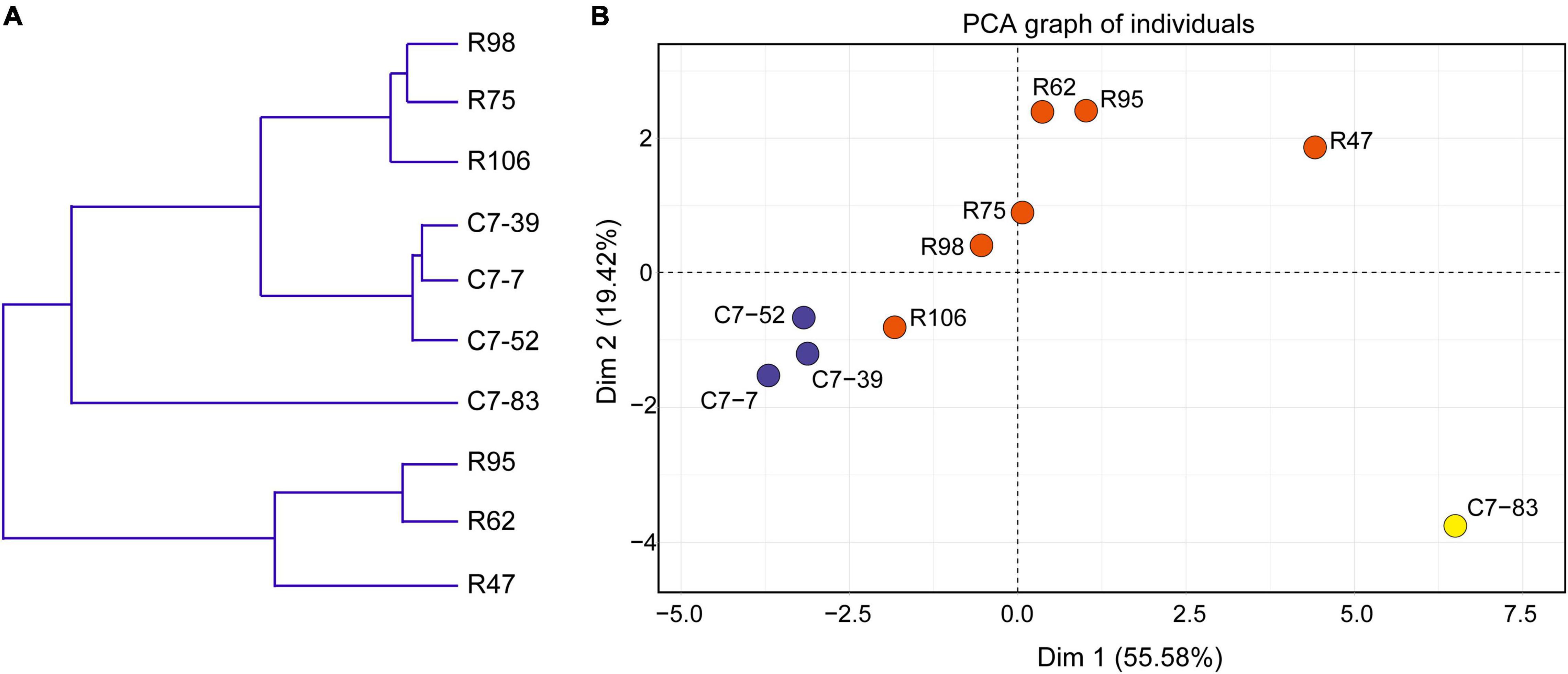
Figure 4. (A) Dendrogram of nonhierarchical clustering analysis and (B) score plot of principal component analysis for the ten Lactiplantibacillus isolates based on probiotic physiological data. Axis 1 and 2 accounted for 55.6% and 19.4% of the total variation present, respectively.
Furthermore, based on various physiological properties relevant to probiotic safety, gastrointestinal viability, and persistence, PCA was performed to assess the variability and similarities among the 10 isolates. As illustrated in Figure 4B, the first and second principal components represented 55.6 and 19.4% of the total 18 variables (GJS, BSS, MAST, and the diameters of the bacteriostatic circles of 15 antibiotics), respectively, and indicated the significant role of gastrointestinal tolerance proprieties in probiotic screening. Clear visual separation of the 10 isolates into five point sets was observed in the PCA plot, with isolate C7-83 being the most distant from the other isolates. These results indicated that differences in IRBT spectra can be reflected in probiotic related phenotypes and that the typing results from genotypic, phenotypic, and spectroscopic analyses were consistent (Table 2).
Discussion
To date, successful IRBT typing of probiotics at the strain level has only been reported for Bifidobacterium (Deidda et al., 2021). In lactic acid bacteria, only species-level typing of lactobacilli by FTIR has been reported (Oust et al., 2004; Bosch et al., 2006; Santos et al., 2015; Quintelas et al., 2018). The potential application of IRBT in probiotic industries has yet to be explored. The present study was the first to evaluate the ability of IRBT to type L. plantarum at the strain level. IRBT successfully distinguished the two closely related species within the Lactiplantibacillus genus, namely, L. plantarum and L. argentoratensis. Surprisingly, it also has potential to discriminate the different strains belonging to L. plantarum. The current definition of “strain” is confusing, and the genetic SNP thresholds for strain delineation have not been universally set in culture-centric microbiology. A “taxonomic strain” is defined as the descendants of a single isolation in pure culture and usually consisting of a succession of cultures ultimately derived from an initial single colony, whereas a “natural strain” is defined as a set of conspecific isolates with distinctive genotypic and/or phenotypic characteristics. Theoretically, it is possible to distinguish two “natural strains” based on a single nucleotide difference, even if no phenotypic differences are identified. By contrast, “taxonomic strains” can become phenotypically heterogeneous with as few as three mutations, but would still be called the same strain (Jackson et al., 2019; Van Rossum et al., 2020). Our study found that 20 L. plantarum isolates were consistently grouped into five IRBT spectral types, corresponding to five different WGS lineages, and five phenotypic patterns associated with probiotic screening.
Microorganisms, including probiotic, pathogenic or neutral strains, play a vital role in human health. On the one hand, certain pathogenic bacteria can cause a series of diseases after invading the host. On the other hand, symbiotic bacteria establish a mutually beneficial relationship with the host. Some probiotics may become adjuvant therapies in the treatment of certain diseases (Zhu and Liu, 2017; Atefi et al., 2021; Xu et al., 2021). Therefore, typing pathogenic and probiotic bacteria at the strain level is a tremendous challenge relevant to human health. However, in the field of microbial typing, IRBT is mainly used in routine clinical hygiene and infection control. The typing function of FTIR has been verified in a large number of gram-negative bacteria in the clinic, such as E. coli, Klebsiella oxytoca and Yersinia enterocolitica, as well as isolates that can cause outbreaks in hospitals such as Enterobacter cloacae, Klebsiella pneumoniae and Pseudomonas aeruginosa (Dieckmann et al., 2016; Dinkelacker et al., 2018; Quintelas et al., 2018; Martak et al., 2019; Novais et al., 2019; Hu et al., 2020). For gram-positive bacteria, the strain-level distinguishing ability of FTIR has been extensively explored in species that are mainly considered relevant to human diseases or food safety, such as Staphylococcus aureus and Listeria monocytogenes (Novais et al., 2019). However, it is necessary to consider not only the harmful factors of pathogenic bacteria but also the positive effects of probiotics. The quality and safety assessment of probiotics in foods and supplements is the responsibility of probiotic industries (Deidda et al., 2021). Due to the growing demand for probiotics, industries need to identify specific probiotics at the strain level quickly and accurately. Considering that the health benefits of probiotics are strain-specific, the European Food Safety Authority also requires the identification of probiotics at the strain level, which provides the basis for health claims (Scientific Opinion on the substantiation of health claims related to non characterised microorganisms pursuant to Article 13(1) of Regulation (EC) No 1924/2006, 2009). In clinical application scenarios, it is also important to consider both probiotic strain specificity and disease specificity to identify the appropriate probiotics that match specific diseases (McFarland et al., 2018). Therefore, the expanded application of IRBT in probiotic industries is urgently needed and necessary.
Lactiplantibacillus plantarum is a model species for research in lactobacilli, which comprise an important group of probiotics for both humans and animals (Martino et al., 2016; Choi et al., 2018; Teame et al., 2020; Yu et al., 2020, 2021). L. plantarum strains have diverse phenotypes and genotypes that facilitate a metabolic flexibility that allow them to colonize a variety of environments, including the human gastrointestinal tract (Pretzer et al., 2005; Marco et al., 2010; Fidanza et al., 2021). There have been reports indicating their potential beneficial effects on humans (Molenaar et al., 2005; Klarin et al., 2008; Siezen et al., 2010; Lewis-Mikhael et al., 2020; Zhao et al., 2021). Its excellent adaptability, extensive industrial applicability and powerful influence on human and animal physiology have made L. plantarum a microorganism of great interest to the academic community. Hence, we chose L. plantarum as the model bacterium to carry out the strain-level typing evaluation of IRBT, which will have great academic and industrial value.
The integration of hardware and software in IRBT provides a potential turn-key solution for routine use in probiotic industries. Determining a reference COV range is important in implementing IRBT as a new phenotypic typing method for routine use in probiotic typing and screening. The automatic COV is the compromise between finding as many clusters as possible that are pure and that a label is represented only by one cluster (Deidda et al., 2021). IRBT spectroscopy is known to be very sensitive to changes in culture medium, incubation time, temperature, and humidity. Different culture parameters will lead to distinct growth conditions among strains, which will have a significant impact on IRBT clustering results (Hu et al., 2020). In this research, fixed culture conditions for L. plantarum were used to obtain robust IRBT typing results within the same laboratory. However, upon introducing spectra from different laboratories, a higher COV and more branches of the clustering dendrogram for L. plantarum were observed when automatically calculated by the IRBT software. The deviation of IRBT clustering results across laboratories may have been caused by technical variances rather than biological variances for the following reasons: the frequency of spontaneous mutations in L. plantarum is lower than 10–7 mutants per plated cell (Nicoloff et al., 2005). Since strain transfer from one institution to another was controlled within three generations of bacterial growth, mutational events may have occurred in the spontaneous mutants even though this is unlikely. In addition, the IRBT spectroscopy of a particular strain is likely to remain consistent even with few numbers of mutations, for mutations in genes do not necessarily cause epigenetic changes; For reproducibility testing, more technical replicates of each isolate were conducted in the external than internal laboratories. Since the automatic COV tends to find as many clusters as possible (default algorithm of IRBT software), it may underestimate the original COV of the external laboratory, resulting in more clusters being generated than the internal laboratory; It is also worth noting that repeated experiments in different laboratories were not performed by the same technician. In fact, the incubation and preparation practices often have a much greater impact on the results than the drying of the same suspension on different spots. So, it is totally normal that different measurements of the same isolate are distributed farther than technical replica; Furthermore, subtle distinctions in IRBT device sensitivities could not be ruled out, although HCA analysis using Euclidean average linkage as exploration method was selected for all repeated experiments.
Previous studies have shown that various phenotypic and genotypic methods, such as MLST, Asc I-PFGE, and WGS, allow L. plantarum to be typed at the interspecies and intraspecies levels (Guidone et al., 2014; Martino et al., 2016; Evanovich et al., 2019; Manzoor and Tayyeb, 2019; Yu et al., 2021). Although these methods have completely updated our understanding of the bacterial population structure, they ignore the phenotypic variations caused by certain genetic changes, especially genotypic or phenotypic characteristics related to key macromolecules on the cell surface. In addition, genotyping techniques are usually time-consuming, labor-intensive, and expensive, limiting their wide application. In addition to DNA-based methods, IRBT has recently shown promising results for typing certain groups of gram-positive and gram-negative bacteria (Hu et al., 2020; Deidda et al., 2021). Comparison results in the present study demonstrated that IRBT had higher discriminatory power than MLST and had equivalent discriminatory power to that of the previous and new gold standards, PFGE and WGS, respectively. The MLST analysis failed to differentiate WGS sequence type 4 from 5 may have been due to the selection of insufficient numbers of alleles or inappropriate loci when performing MLST (Yu et al., 2020). In a typing study of Bifidobacteria, it is also proven that the resolution of IRBT is better than that of MLST, as IRBT could distinguish all strains of B. animalis subsp. lactis, while MLST fails to achieve this effect (Deidda et al., 2021).
Selecting probiotics from a large pool of isolates using a series of in vitro preliminary selections and followed by in vivo validation studies is laborious, expensive, time-consuming, and not easily achievable. In vitro selection is therefore the first approach used to select a few strains that can be further evaluated in vivo (Morelli, 2000). Due to the diverse ecological niches, phenotypes, and genotypes of L. plantarum, a large number of duplicates of the same strain may be isolated from the same niche, and obtaining new isolates from different habitats may also lead to multiple isolations of the same strain (Deidda et al., 2021). This makes the rigorous screening of probiotics from L. plantarum isolates very challenging. Therefore, it is particularly necessary to apply a fast, low-cost, and effective technology that can distinguish L. plantarum at the strain level to reduce the number of isolates to be considered before performing extensive screening and evaluation processes. PFGE is very laborious, whereas WGS is quite expensive, and both are time-consuming (2–3 days), especially when typing a large group of new isolates. Their applications in routine probiotic identification and typing may be restricted (Novais et al., 2019). In contrast, IRBT is a fast (within 3 h), inexpensive and high-throughput bacterial typing system. Moreover, the spectral typing from IRBT matched the probiotic phenotypic clustering results of the 10 representative Lactiplantibacillus isolates. Under the default IRBT settings, the observed spectral range focuses on the polysaccharide region (1,200–900 cm–1), which highlights changes in the carbohydrate composition of bacterial walls. These features may be closely related to acid and bile salt sensitivity, cell surface hydrophobicity, and antibiotic resistance, which are related to probiotic phenotypes (Kuda et al., 2013). Several studies have reported that the resistance of L. plantarum to acid, heat and other stresses is associated with cell membrane functions (Capozzi et al., 2011; Ricciardi et al., 2012). Therefore, due to the resulting reduced workload, probiotic screening efficiency would be greatly improved by the inclusion of IRBT before performing extensive preliminary selection processes. Based on our IRBT results in L. plantarum typing, a good database can be constructed for introduction into the routine procedures of probiotic screening and typing. One of the limitations of our study is the lack of inclusion of the type strain in IRBT, PFGE, and phenotypic studies. L. plantarum isolate R106 (CGMCC 20370) could be used for other researchers as a control in such studies.
Conclusion
In this study, we conducted the first investigation of L. plantarum strain typing based on IRBT. We demonstrated that IRBT could successfully strain-level type of L. plantarum, which has traditionally been used as a probiotic. A deviation in the discriminative power of IRBT was found interlaboratory at the strain level, probably caused by technical variance. Compared with other genotyping techniques, IRBT was equivalent to the previous and current gold standards, PFGE and WGS, in typing at the strain level and had higher discriminatory power than that of MLST. In the comparison with probiotic screening associated phenotyping, the clustering results of IRBT spectral types and phenotypes were consistent, revealing that these two attributes were closely related. Therefore, the typing results from spectroscopic, genotypic, and probiotic screening related phenotypic analyses were concordant. In addition, thanks to the great advantages of IRBT in DNA-based technology, such as ease of use, fast turnaround time, user-friendly software, and relatively low operating costs, IRBT is suitable for daily strain typing in probiotic laboratories and industries. IRBT not only could effectively improve the probiotic screening efficiency, but also has application potential in the traceability and quality control of probiotics. It is worth noting that the exchange of results between different laboratories based on IRBT requires a high level of standardization.
Data Availability Statement
The datasets presented in this study can be found in the BioProject PRJNA769251 at DDBJ/ENA/GenBank database. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
XL: conceptualization, writing – original draft, review, and editing, methodology, funding acquisition, and sources. BT: conceptualization, investigation, formal analysis, visualization, and writing – review and editing. JL: project administration, funding acquisition, and supervision. LZ and XW: methodology, investigation, and visualization. All authors contributed to the article and approved the submitted version.
Funding
This work was partially funded by the National Natural Science Foundation of China (Grant No. 32000079), Natural Science Foundation of Zhejiang Province (Grant No. LQ20C200009), Zhejiang “Qianjiang Talents Program” Category D Project and the Collaborative Extension Plan of Major Agricultural Technologies in Zhejiang Province (Grant No. 2021XTTGXM03). The funders had no influence on the study design; collection, analysis, and interpretation of data; writing of the manuscript; or decision to submit the manuscript for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We sincerely thank Yanyan Hu of Clinical Microbiology Laboratory, School of Medicine, The Second Affiliated Hospital of Zhejiang University for the independent verification of the IRBT cutoff value. We also thank the Vegetable Processing Laboratory of the Food Research Institute of Zhejiang Academy of Agricultural Sciences for sharing the L. plantarum isolates.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.823120/full#supplementary-material
Footnotes
References
Atefi, N., Fallahpour, M., Sharifi, S., Ghassemi, M., Roohaninasab, M., and Goodarzi, A. (2021). Probiotic as an adjuvant therapy in chronic urticaria: a blinded randomized controlled clinical trial. Eur. Ann. Allergy Clin. Immunol. [Online ahead of print] doi: 10.23822/eurannaci.1764-1489.200
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345
Bosch, A., Golowczyc, M. A., Abraham, A. G., Garrote, G. L., De Antoni, G. L., and Yantorno, O. (2006). Rapid discrimination of lactobacilli isolated from kefir grains by FT-IR spectroscopy. Int. J. Food Microbiol. 111, 280–287. doi: 10.1016/j.ijfoodmicro.2006.05.010
Burckhardt, I., Sebastian, K., Mauder, N., Kostrzewa, M., Burckhardt, F., and Zimmermann, S. (2019). Analysis of Streptococcus pneumoniae using Fourier-transformed infrared spectroscopy allows prediction of capsular serotype. Eur. J. Clin. Microbiol. Infect. Dis. 38, 1883–1890. doi: 10.1007/s10096-019-03622-y
Campedelli, I., Mathur, H., Salvetti, E., Clarke, S., Rea, M. C., Torriani, S., et al. (2019). Genus-wide assessment of antibiotic resistance in Lactobacillus spp. Appl. Environ. Microbiol. 85, e1738–e1718. doi: 10.1128/AEM.01738-18
Capozzi, V., Weidmann, S., Fiocco, D., Rieu, A., Hols, P., Guzzo, J., et al. (2011). Inactivation of a small heat shock protein affects cell morphology and membrane fluidity in Lactobacillus plantarum WCFS1. Res. Microbiol. 162, 419–425. doi: 10.1016/j.resmic.2011.02.010
Cen, S., Yin, R., Mao, B., Zhao, J., Zhang, H., Zhai, Q., et al. (2020). Comparative genomics shows niche-specific variations of Lactobacillus plantarum strains isolated from human, Drosophila melanogaster, vegetable and dairy sources. Food Biosci. 35:100581. doi: 10.1016/j.fbio.2020.100581
Charteris, W. P., Kelly, P. M., Morelli, L., and Collins, J. K. (1998). Antibiotic susceptibility of potentially probiotic Lactobacillus species. J. Food Prot. 61, 1636–1643. doi: 10.4315/0362-028X-61.12.1636
Choi, S., Jin, G. D., Park, J., You, I., and Kim, E. B. (2018). Pan-genomics of Lactobacillus plantarum revealed group-specific genomic profiles without habitat associations. J. Microbiol. Biotechnol. 28, 1352–1359. doi: 10.4014/jmb.1803.03029
das Neves Selis, N., de Oliveira, H. B. M., Leão, H. F., dos Anjos, Y. B., Sampaio, B. A., Correia, T. M. L., et al. (2021). Lactiplantibacillus plantarum strains isolated from spontaneously fermented cocoa exhibit potential probiotic properties against Gardnerella vaginalis and Neisseria gonorrhoeae. BMC Microbiol. 21:198. doi: 10.1186/s12866-021-02264-5
de las Rivas, B., Marcobal, A., and Muñoz, R. (2006). Development of a multilocus sequence typing method for analysis of Lactobacillus plantarum strains. Microbiology 152, 85–93. doi: 10.1099/mic.0.28482-0
de Melo Pereira, G. V., de Oliveira Coelho, B., Magalhães Júnior, A. I., Thomaz-Soccol, V., and Soccol, C. R. (2018). How to select a probiotic? A review and update of methods and criteria. Biotechnol. Adv. 36, 2060–2076. doi: 10.1016/j.biotechadv.2018.09.003
Deidda, F., Bozzi Cionci, N., Cordovana, M., Campedelli, I., Fracchetti, F., Di Gioia, D., et al. (2021). Bifidobacteria strain typing by fourier transform infrared spectroscopy. Front. Microbiol. 12:692975. doi: 10.3389/fmicb.2021.692975
Dieckmann, R., Hammerl, J. A., Hahmann, H., Wicke, A., Kleta, S., Dabrowski, P. W., et al. (2016). Rapid characterisation of: klebsiella oxytoca isolates from contaminated liquid hand soap using mass spectrometry, FTIR and Raman spectroscopy. Faraday Discuss. 187, 353–375. doi: 10.1039/c5fd00165j
Dinkelacker, A. G., Vogt, S., Oberhettinger, P., Mauder, N., Rau, J., Kostrzewa, M., et al. (2018). Typing and species identification of clinical klebsiella isolates by fourier transform infrared spectroscopy and matrixassisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 56, e843–e818. doi: 10.1128/JCM.00843-18
Duar, R. M., Lin, X. B., Zheng, J., Martino, M. E., Grenier, T., Pérez-Muñoz, M. E., et al. (2017). Lifestyles in transition: evolution and natural history of the genus Lactobacillus. FEMS Microbiol. Rev. 41, S27–S48. doi: 10.1093/femsre/fux030
Evanovich, E., De Souza Mendonça Mattos, P. J., and Guerreiro, J. F. (2019). Comparative genomic analysis of Lactobacillus plantarum: an overview. Int. J. Genomics. 2019:4973214. doi: 10.1155/2019/4973214
Fidanza, M., Panigrahi, P., and Kollmann, T. R. (2021). Lactiplantibacillus plantarum–nomad and ideal probiotic. Front. Microbiol. 12:712236. doi: 10.3389/fmicb.2021.712236
Food and Agriculture Organization [FAO]/World Health Organization [WHO] (2002). Guidelines for the Evaluation of Probiotics in Food (Working Group on Drafting Guidelines for the Evaluation of Probiotics in Food). Rome: Food and Agriculture Organization.
Fuhren, J., Schwalbe, M., Peralta-Marzal, L., Rösch, C., Schols, H. A., and Kleerebezem, M. (2020). Phenotypic and genetic characterization of differential galacto-oligosaccharide utilization in Lactobacillus plantarum. Sci. Rep. 10:21657. doi: 10.1038/s41598-020-78721-4
Guidone, A., Zotta, T., Ross, R. P., Stanton, C., Rea, M. C., Parente, E., et al. (2014). Functional properties of Lactobacillus plantarum strains: a multivariate screening study. LWT - Food Sci. Technol. 56, 69–76. doi: 10.1016/j.lwt.2013.10.036
Hu, Y., Zhou, H., Lu, J., Sun, Q., Liu, C., Zeng, Y., et al. (2020). Evaluation of the IR Biotyper for Klebsiella pneumoniae typing and its potentials in hospital hygiene management. Microb. Biotechnol. 14, 1343–1352. doi: 10.1111/1751-7915.13709
Jackson, S. A., Schoeni, J. L., Vegge, C., Pane, M., Stahl, B., Bradley, M., et al. (2019). Improving end-user trust in the quality of commercial probiotic products. Front. Microbiol. 10:739. doi: 10.3389/fmicb.2019.00739
Klarin, B., Molin, G., Jeppsson, B., and Larsson, A. (2008). Use of the probiotic Lactobacillus plantarum 299 to reduce pathogenic bacteria in the oropharynx of intubated patients: a randomised controlled open pilot study. Crit. Care. 12:R136. doi: 10.1186/cc7109
Kuda, T., Yazaki, T., Ono, M., Takahashi, H., and Kimura, B. (2013). In vitro cholesterol-lowering properties of Lactobacillus plantarum AN6 isolated from aji-narezushi. Lett. Appl. Microbiol. 57, 187–192. doi: 10.1111/lam.12094
Lewis-Mikhael, A. M., Davoodvandi, A., and Jafarnejad, S. (2020). Effect of Lactobacillus plantarum containing probiotics on blood pressure: a systematic review and meta-analysis. Pharmacol. Res. 153:104663. doi: 10.1016/j.phrs.2020.104663
Liu, D. D., and Gu, C. T. (2020). Proposal to reclassify Lactobacillus zhaodongensis, Lactobacillus zeae, Lactobacillus argentoratensis and Lactobacillus buchneri subsp. Silagei as Lacticaseibacillus zhaodongensis comb. nov., Lacticaseibacillus zeae comb. nov., Lactiplantibacillus argento. Int. J. Syst. Evol. Microbiol. 70, 6414–6417. doi: 10.1099/ijsem.0.004548
Liu, Z., Xu, C., Tian, R., Wang, W., Ma, J., Gu, L., et al. (2021). Screening beneficial bacteriostatic lactic acid bacteria in the intestine and studies of bacteriostatic substances. J. Zhejiang Univ. Sci. B. 22, 533–547. doi: 10.1631/jzus.B2000602
Manzoor, A., and Tayyeb, A. (2019). Functional probiotic attributes and gene encoding plantaracin among variant Lactobacillus Plantarum strains. Microb. Pathog. 131, 22–32. doi: 10.1016/j.micpath.2019.03.016
Marco, M. L., De Vries, M. C., Wels, M., Molenaar, D., Mangell, P., Ahrne, S., et al. (2010). Convergence in probiotic Lactobacillus gut-adaptive responses in humans and mice. ISME J. 4, 1481–1484. doi: 10.1038/ismej.2010.61
Martak, D., Valot, B., Sauget, M., Cholley, P., Thouverez, M., Bertrand, X., et al. (2019). Fourier-transform infra red spectroscopy can quickly type gram-negative bacilli responsible for hospital outbreaks. Front. Microbiol. 10:1440. doi: 10.3389/fmicb.2019.01440
Martino, M. E., Bayjanov, J. R., Caffrey, B. E., Wels, M., Joncour, P., Hughes, S., et al. (2016). Nomadic lifestyle of Lactobacillus plantarum revealed by comparative genomics of 54 strains isolated from different habitats. Environ. Microbiol. 18, 4974–4989. doi: 10.1111/1462-2920.13455
McFarland, L. V., Evans, C. T., and Goldstein, E. J. C. (2018). Strain-specificity and disease-specificity of probiotic efficacy: a systematic review and meta-analysis. Front. Med. 5:124. doi: 10.3389/fmed.2018.00124
Molenaar, D., Bringel, F., Schuren, F. H., De Vos, W. M., Siezen, R. J., and Kleerebezem, M. (2005). Exploring Lactobacillus plantarum genome diversity by using microarrays. J. Bacteriol. 187, 6119–6127. doi: 10.1128/JB.187.17.6119-6127.2005
Morelli, L. (2000). In vitro selection of probiotic lactobacilli: a critical appraisal. Curr. Issues Intest. Microbiol. 1, 59–67.
Neoh, H. M., Tan, X. E., Sapri, H. F., and Tan, T. L. (2019). Pulsed-field gel electrophoresis (PFGE): a review of the “gold standard” for bacteria typing and current alternatives. Infect. Genet. Evol. 74:103935. doi: 10.1016/j.meegid.2019.103935
Nicoloff, H., Elagöz, A., Arsène-Ploetze, F., Kammerer, B., Martinussen, J., and Bringel, F. (2005). Repression of the pyr operon in Lactobacillus plantarum prevents its ability to grow at low carbon dioxide levels. J. Bacteriol. 187, 2093–2104. doi: 10.1128/JB.187.6.2093-2104.2005
Novais, A., Freitas, A. R., Rodrigues, C., and Peixe, L. (2019). Fourier transform infrared spectroscopy: unlocking fundamentals and prospects for bacterial strain typing. Eur. J. Clin. Microbiol. Infect. Dis. 38, 427–448. doi: 10.1007/s10096-018-3431-3
Oust, A., Møretrø, T., Kirschner, C., Narvhus, J. A., and Kohler, A. (2004). Evaluation of the robustness of FT-IR spectra of lactobacilli towards changes in the bacterial growth conditions. FEMS Microbiol. Lett. 239, 111–116. doi: 10.1016/j.femsle.2004.08.024
Pan, Q., Cen, S., Yu, L., Tian, F., Zhao, J., Zhang, H., et al. (2021). Niche-Specific adaptive evolution of Lactobacillus plantarum strains isolated from human feces and paocai. Front. Cell. Infect. Microbiol. 10:615876. doi: 10.3389/fcimb.2020.615876
Petit, R. A., and Read, T. D. (2020). Bactopia: a flexible pipeline for complete analysis of bacterial genomes. mSystems 5, e190–e120. doi: 10.1128/msystems.00190-20
Pretzer, G., Snel, J., Molenaar, D., Wiersma, A., Bron, P. A., Lambert, J., et al. (2005). Biodiversity-based identification and functional characterization of the mannose-specific adhesin of Lactobacillus plantarum. J. Bacteriol. 187, 6128–6136. doi: 10.1128/JB.187.17.6128-6136.2005
Quintelas, C., Ferreira, E. C., Lopes, J. A., and Sousa, C. (2018). An overview of the evolution of infrared spectroscopy applied to bacterial typing. Biotechnol. J. 13:1700449. doi: 10.1002/biot.201700449
R Core Team (2020). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. Available online at: https://www.r-project.org/
Ricciardi, A., Parente, E., Guidone, A., Ianniello, R. G., Zotta, T., Sayem, S. M. A., et al. (2012). Genotypic diversity of stress response in Lactobacillus plantarum, Lactobacillus paraplantarum and Lactobacillus pentosus. Int. J. Food Microbiol. 157, 278–285. doi: 10.1016/j.ijfoodmicro.2012.05.018
Richter, M., and Rosselló-Móra, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 106, 19126–19131. doi: 10.1073/pnas.0906412106
Richter, M., Rosselló-Móra, R., Oliver Glöckner, F., and Peplies, J. (2016). JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics. 32, 929–931. doi: 10.1093/bioinformatics/btv681
Salvetti, E., Harris, H. M. B., Felis, G. E., and O’Toole, P. W. (2018). Comparative genomics of the genus Lactobacillus reveals robust phylogroups that provide the basis for reclassification. Appl. Environ. Microbiol. 84, e993–e918. doi: 10.1128/AEM.00993-18
Sandes, S., Alvim, L., Silva, B., Acurcio, L., Santos, C., Campos, M., et al. (2017). Selection of new lactic acid bacteria strains bearing probiotic features from mucosal microbiota of healthy calves: looking for immunobiotics through in vitro and in vivo approaches for immunoprophylaxis applications. Microbiol. Res. 200, 1–13. doi: 10.1016/j.micres.2017.03.008
Santos, M. I., Gerbino, E., Tymczyszyn, E., and Gomez-Zavaglia, A. (2015). Applications of infrared and raman spectroscopies to probiotic investigation. Foods. 4, 283–305. doi: 10.3390/foods4030283
Scientific Opinion on the substantiation of health claims related to non characterised microorganisms pursuant to Article 13(1) of Regulation (EC) No 1924/2006 (2009). Scientific Opinion on the substantiation of health claims related to non characterised microorganisms pursuant to Article 13(1) of Regulation (EC) No 1924/2006. EFSA J. 7:1247. doi: 10.2903/j.efsa.2009.1247
Siezen, R. J., Tzeneva, V. A., Castioni, A., Wels, M., Phan, H. T. K., Rademaker, J. L. W., et al. (2010). Phenotypic and genomic diversity of Lactobacillus plantarum strains isolated from various environmental niches. Environ. Microbiol. 12, 758–773. doi: 10.1111/j.1462-2920.2009.02119.x
Silva, B. C., Jung, L. R. C., Sandes, S. H. C., Alvim, L. B., Bomfim, M. R. Q., Nicoli, J. R., et al. (2013). In vitro assessment of functional properties of lactic acid bacteria isolated from faecal microbiota of healthy dogs for potential use as probiotics. Benef. Microbes. 4, 267–275. doi: 10.3920/BM2012.0048
Sommer, M. O. A. (2015). Advancing gut microbiome research using cultivation. Curr. Opin. Microbiol. 27, 127–132. doi: 10.1016/j.mib.2015.08.004
Teame, T., Wang, A., Xie, M., Zhang, Z., Yang, Y., Ding, Q., et al. (2020). Paraprobiotics and postbiotics of probiotic lactobacilli, their positive effects on the host and action mechanisms: a review. Front. Nutr. 7:570344. doi: 10.3389/fnut.2020.570344
Van Rossum, T., Ferretti, P., Maistrenko, O. M., and Bork, P. (2020). Diversity within species: interpreting strains in microbiomes. Nat. Rev. Microbiol. 18, 491–506. doi: 10.1038/s41579-020-0368-1
Xu, H. L., Zou, L. L., Chen, M. B., Wang, H., Shen, W. M., Zheng, Q. H., et al. (2021). Efficacy of probiotic adjuvant therapy for irritable bowel syndrome in children: a systematic review and meta-analysis. PLoS One 16:e0255160. doi: 10.1371/journal.pone.0255160
Yoha, K. S., Nida, S., Dutta, S., Moses, J. A., and Anandharamakrishnan, C. (2021). Targeted delivery of probiotics: perspectives on research and commercialization. Probiotics Antimicrob. Proteins 14, 15–48. doi: 10.1007/s12602-021-09791-7
Yu, A. O., Goldman, E. A., Brooks, J. T., Golomb, B. L., Yim, I. S., Gotcheva, V., et al. (2021). Strain diversity of plant-associated Lactiplantibacillus plantarum. Microb. Biotechnol. 14, 1990–2008. doi: 10.1111/1751-7915.13871
Yu, A. O., Leveau, J. H. J., and Marco, M. L. (2020). Abundance, diversity and plant-specific adaptations of plant-associated lactic acid bacteria. Environ. Microbiol. Rep. 12, 16–29. doi: 10.1111/1758-2229.12794
Zhao, W., Peng, C., Sakandar, H. A., Kwok, L. Y., and Zhang, W. (2021). Meta-Analysis: randomized trials of Lactobacillus plantarum on immune regulation over the last decades. Front. Immunol. 12:643420. doi: 10.3389/fimmu.2021.643420
Zheng, J., Ruan, L., Sun, M., and Gänzle, M. (2015). A genomic view of lactobacilli and pediococci demonstrates that phylogeny matches ecology and physiology. Appl. Environ. Microbiol. 81, 7233–7243. doi: 10.1128/AEM.02116-15
Zheng, J., Wittouck, S., Salvetti, E., Franz, C. M. A. P., Harris, H. M. B., Mattarelli, P., et al. (2020). A taxonomic note on the genus Lactobacillus: description of 23 novel genera, emended description of the genus Lactobacillus beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 70, 2782–2858. doi: 10.1099/ijsem.0.004107
Keywords: IR Biotyper, Lactiplantibacillus plantarum, probiotic screening, genotyping, phenotypic, strain typing
Citation: Li X, Zhu L, Wang X, Li J and Tang B (2022) Evaluation of IR Biotyper for Lactiplantibacillus plantarum Typing and Its Application Potential in Probiotic Preliminary Screening. Front. Microbiol. 13:823120. doi: 10.3389/fmicb.2022.823120
Received: 26 November 2021; Accepted: 24 February 2022;
Published: 24 March 2022.
Edited by:
Teresa Zotta, University of Basilicata, ItalyReviewed by:
Michael Gänzle, University of Alberta, CanadaPrabhu B. Patil, Institute of Microbial Technology (CSIR), India
Copyright © 2022 Li, Zhu, Wang, Li and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinjun Li, bGlqaW5qdW5AemFhcy5hYy5jbg==; Biao Tang, dGJfNDExQDE2My5jb20=
 Xiaoqiong Li
Xiaoqiong Li Liying Zhu1
Liying Zhu1 Jinjun Li
Jinjun Li Biao Tang
Biao Tang