
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 18 February 2022
Sec. Infectious Agents and Disease
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.822243
This article is part of the Research Topic Streptococci in Infectious Diseases – Pathogenic Mechanisms and Host Immune Responses View all 9 articles
 Dag Harald Skutlaberg1,2*
Dag Harald Skutlaberg1,2* Harald G. Wiker1,2
Harald G. Wiker1,2 Haima Mylvaganam2
Haima Mylvaganam2  The INFECT Study Group
The INFECT Study Group Anna Norrby-Teglund3
Anna Norrby-Teglund3 Steinar Skrede1,4
Steinar Skrede1,4Objectives: Biofilm formation has been demonstrated in muscle and soft tissue samples from patients with necrotizing soft tissue infection (NSTI) caused by Streptococcus pyogenes, but the clinical importance of this observation is not clear. Although M-protein has been shown to be important for in vitro biofilm formation in S. pyogenes, the evidence for an association between emm type and biofilm forming capacity is conflicting. Here we characterize the biofilm forming capacity in a collection of S. pyogenes isolates causing NSTI, and relate this to emm type of the isolates and clinical characteristics of the patients.
Methods: Bacterial isolates and clinical data were obtained from NSTI patients enrolled in a multicenter prospective observational study. Biofilm forming capacity was determined using a microtiter plate assay.
Results: Among 57 cases, the three most frequently encountered emm types were emm1 (n = 22), emm3 (n = 13), and emm28 (n = 7). The distribution of biofilm forming capacity in emm1 was qualitatively (narrow-ranged normal distribution) and quantitatively (21/22 isolates in the intermediate range) different from other emm types (wide ranged, multimodal distribution with 5/35 isolates in the same range as emm1). There were no significant associations between biofilm forming capacity and clinical characteristics of the patients.
Conclusions: The biofilm forming capacity of emm1 isolates was uniform and differed significantly from other emm types. The impact of biofilm formation in NSTI caused by S. pyogenes on clinical outcomes remains uncertain.
Streptococcus pyogenes causes a broad spectrum of disease manifestations ranging from mild, superficial infections to life-threatening invasive diseases, as well as post-streptococcal sequelae. This pathogen accounts for more than half a billion new cases and more than half a million deaths annually on a global scale (World Health Organization, 2005). Invasive S. pyogenes infections have a reported annual incidence of around 3 per 100,000 persons in the Scandinavian countries (Darenberg et al., 2007; Luca-Harari et al., 2008; Naseer et al., 2016). Necrotizing soft tissue infections (NSTIs) are acute, life-threatening conditions characterized by rapid and extensive destruction of the deep soft tissue (Sartelli et al., 2018). S. pyogenes is the most common pathogen in monomicrobial NSTIs (Skrede et al., 2020).
Biofilms are aggregates of microorganisms adhering to each other and to a surface and embedded within an extracellular matrix consisting of polysaccharides, proteins and nucleic acids. Microorganisms growing within a biofilm are largely protected against the host’s immune systems and the effect of antibiotics (Kumar et al., 2017).
S. pyogenes biofilm like structures have been demonstrated in vitro (Baldassarri et al., 2006; Lembke et al., 2006; Koller et al., 2010) as well as in vivo (Siemens et al., 2016). M protein, encoded by the emm gene, has an important role in S. pyogenes biofilm formation (Cho and Caparon, 2005; Courtney et al., 2009). However, there is conflicting evidence for a potential association between emm type and biofilm formation (Baldassarri et al., 2006; Koller et al., 2010; Ogawa et al., 2011; Wozniak et al., 2017). Other surface associated molecules known to be involved in S. pyogenes biofilm formation include pili (Manetti et al., 2007), streptococcal collagen like protein 1 (Scl1) (Oliver-Kozup et al., 2011) and hyaluronic acid capsule (Cho and Caparon, 2005). The clinical relevance of the S. pyogenes biofilm phenotype in mild infections of the upper respiratory tract and skin is recognized (Fiedler et al., 2015). In a previous study, we documented biofilm formation in vivo in NSTIs caused by S. pyogenes with biofilm demonstrated in tissue biopsies in over 30% of the cases (Siemens et al., 2016).
The aim of this study was to characterize in vitro biofilm forming capacity of S. pyogenes causing monomicrobial NSTI. We also wanted to explore possible associations between biofilm forming capacity and emm type of the isolates and clinical characteristics, including outcome, of the patients.
Patients with confirmed monomicrobial NSTI caused by S. pyogenes prospectively enrolled in the INFECT project (ClinicalTrials.gov, NCT01790698), were included in this study. Demographic and clinical data were obtained from the INFECT trial database, described in detail elsewhere (Madsen et al., 2018).
Bacteria were isolated from sterile sites, including deep tissue or blood, and stored as frozen stock cultures at −80°C. Identification was performed using matrix-assisted laser desorption/ionization time-of-flight (MALDI-ToF) mass spectrometry. emm-typing and multi-locus sequence-typing (MLST) was done as described earlier (Bruun et al., 2020). Phylogenetic analysis was performed using CSI Phylogeny 1.4 available at Center for Genomic Epidemiology website (Kaas et al., 2014). The phylogenetic tree was edited in Geneious 9.1.7.1 Genomic sequences was retrieved from the European Nucleotide Archive (ENA database) under the BioProject PRJNA524111. One of the isolates (from case 2006) was used as a reference and included in every microtiter plate of the biofilm assay.
Necrotizing soft tissue infection is defined as necrotic or deliquescent soft tissue with widespread undermining of surrounding tissue as observed perioperative by the surgeon (Madsen et al., 2018).
Sepsis-related Organ Failure Assessment (SOFA) score is a tool to evaluate morbidity in critical ill patients based on respiratory-, circulatory-, renal-, coagulation-, hepatic-, and central nervous system function (Vincent et al., 1996).
Biofilms were formed according to a customization of a static biofilm culture model (Kwasny and Opperman, 2010). In short, portions of frozen stock cultures were first plated on sheep blood agar (SBA) and incubated overnight. A single colony from this first subculture was streaked on a fresh SBA plate and incubated for 18 ± 1.5 h. Colonies from second cultures were suspended in Tryptic Soy Broth containing 1% glucose (TSBG). The suspension was adjusted to a turbidity equivalent to a bacterial cell density of approximately 108 colony forming units per ml (CFU/ml).
Wells of flat bottomed, 96 well, microtiter plates (Nunc A/S, Roskilde, Denmark, catalog no. 167008) were inoculated with 200 μl of a 10–1 dilution of this suspension. Inoculated microtiter plates were covered by a lid and placed at 4°C for 4 h (±20 min) in order to let the bacteria sediment without significant multiplication. The plates were then incubated at 37°C in ambient air for 18 h (±25 min).
Details of the microtiter plate lay out are shown in Supplementary Figure 1. In order to reduce evaporation from inoculated wells, all wells at the edge of the microtiter plate were filled with sterile TSBG. Six of these wells were used for blank correction of optical density (OD) measurements.
At the end of incubation, the growth medium was removed by gentle aspiration. Bacteria loosely attached to the biofilms were removed by careful addition and subsequent aspiration of 300 μl Phosphate-buffered saline (pH 7.2). This rinse procedure was performed twice. Biofilms were heat fixed at 60°C for at least 90 min and thereafter stained using 0.06% crystal violet solution (50 μl) for 5 min at room temperature. Unbound stain was removed by gentle aspiration and the wells were rinsed five times as described above. Biofilms were then dried by leaving the plates without lid at room temperature for at least 60 min. Stain bound to biofilm was eluted by incubating the biofilms with 200 μl 30% acetic acid for 5 min. One hundred μl of thoroughly mixed eluate were transferred to a fresh microtiter plate and optical density was measured at a wavelength of 600 nm (OD600) using Synergy H1 microplate reader (BioTek, Winoosky, VT, United States). The mean blank-corrected OD600-value of eluate from six wells from each isolate served as a measure of biofilm forming capacity.
Viable bacterial density of the inocula was retrospectively verified by quantitative culture. If inoculum density was outside an acceptance range of 5.0 × 106–1.5 × 107 CFU/ml, the OD600-result was discarded and the actual isolate was re-tested.
Normally distributed continuous variables are presented as mean [95% confidence interval (CI)] and compared using one-way ANOVA, while categorical variables are presented as numbers (percentages) and compared using Fisher’s exact test (two-tailed). Statistical significance was assessed after controlling the false discovery rate using the Benjamini-Hochberg procedure with a false discovery rate of 0.1. Quartiles were determined using the weighted average method. Between-run precision of the biofilm assay was determined using blank-corrected mean OD600-values of the reference isolate and expressed as coefficient of variation (CV). Statistical analyses were performed using IBM SPSS Statistics Version 24 (IBM Corp., Armonk, NY, United States). Power analyses were performed using G∗Power 3.1.9.2.
One-hundred-and-fourteen (28%) of 409 patients included in the INFECT study had a monomicrobial infection with S. pyogenes. When this study was conducted, bacterial isolates were available from 59 cases. Of these, two cases were excluded because the density of bacterial inocula was repeatedly below the lower acceptance limit in the biofilm assay. Demographic and clinical data of the 57 included cases, are summarized in Table 1. The three most prevalent emm types, emm1, emm3, and emm28, comprised nearly three quarters of all isolates (Table 2). All emm1 isolates belonged to sequence type (ST) 28 and all emm28 isolates belonged to ST 52. The emm3 isolates were either ST 15 (n = 8) or ST 315 (n = 5), which differ from each other with a single base-pair in a single locus. With a few exceptions, the isolates within each emm type clustered close together (Figure 1).
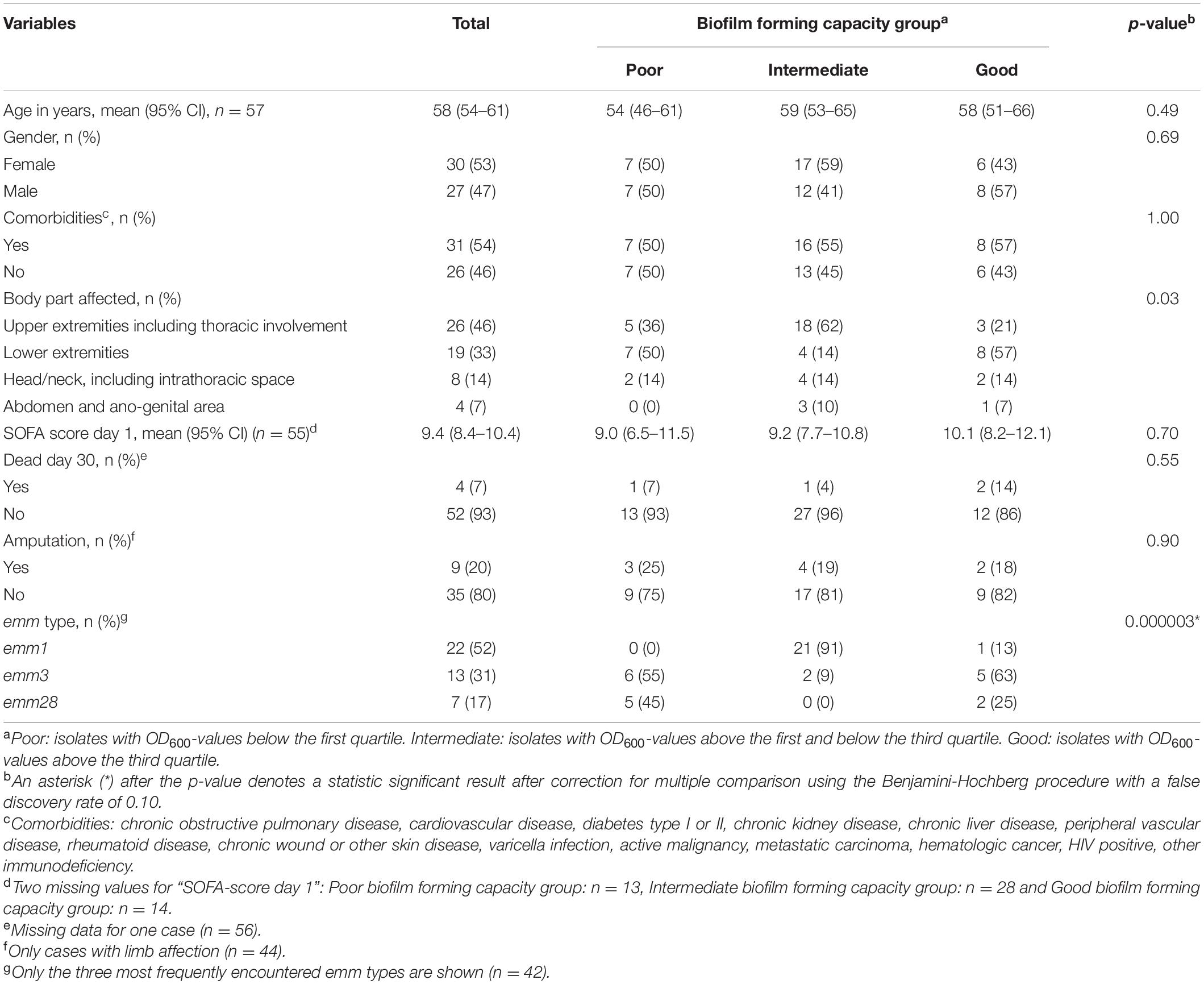
Table 1. Biofilm forming capacity in relation to patient demographics, clinical variables and emm type.
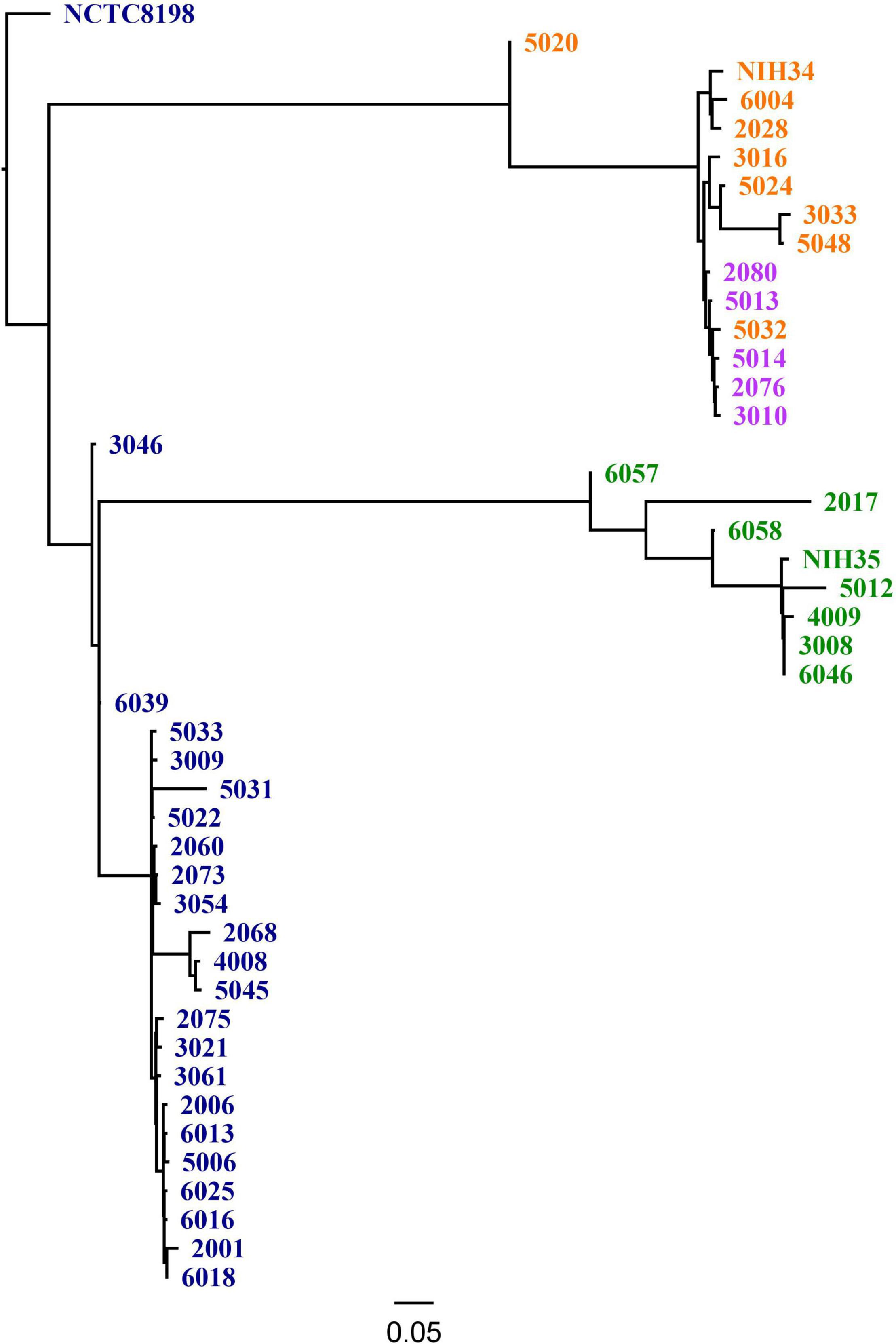
Figure 1. Single nucleotide polymorphism (SNP) based Phylogeny using CSI Phylogeny 1.4, including the three most common emm types in the study and reference strains [NCTC8198 (GenBank accession LN831034.1), NIH34 (GenBank accession AP023387.1) and NIH35 (GenBank accession AP023388.1)]. The color indicates emm- and ST type, including dark blue: emm1 (ST 28); purple: emm3 (ST 15); orange: emm3 (ST 315) and green: emm28 (ST 52).
Between-run CV for the biofilm forming capacity assay was 12.2% (data not shown). Biofilm forming capacity had a wide range (OD600-values from 0.01 to 1.31) and a multimodal distribution (Figure 2). However, all emm1 isolates (n = 22) clustered closely within a narrow range (OD600-values from 0.32 to 0.65) and had a normally distributed biofilm forming capacity. Isolates other than emm1 showed great variations in biofilm forming capacity, and only 14% (5/35) of these had biofilm forming capacity within the same range as the emm1 isolates (Figure 3).
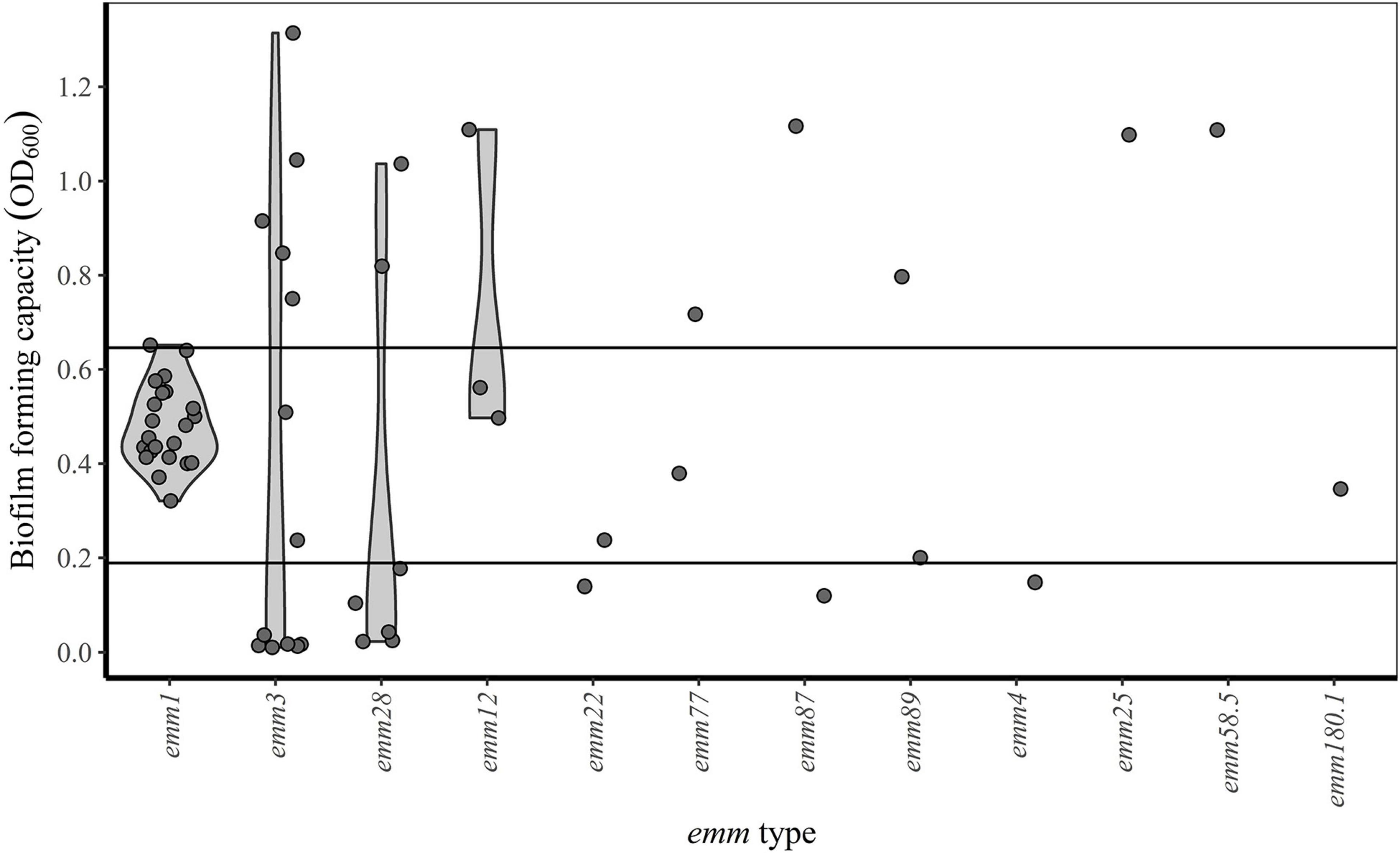
Figure 3. emm type and biofilm forming capacity. Horizontal lines indicate the first (OD600 = 0.19) and third (OD600 = 0.65) quartiles. Isolates with OD600-values below the first quartile, between the first and third quartile and above the third quartile are categorized as poor, intermediate and good biofilm formers, respectively.
For comparative statistical analyses, isolates were classified according to their biofilm forming capacity: isolates with OD600-values below the first quartile (OD600 = 0.19), between the first and third quartile (OD600 = 0.65) and above the third quartile were categorized as poor, intermediate and good biofilm formers respectively (Table 1). The only emm1 isolate categorized as a good biofilm former, clustered together with the other emm1 isolates, but had an OD600 value slightly above the third quartile (Figure 3). While the emm3 isolates were distributed across the whole range of biofilm forming capacity, the emm28 isolates were either poor or good biofilm formers (Table 1 and Figure 3). For the remaining emm types there were too few observations to deduce a pattern (Figure 3).
Eight of the bacterial strains included in the present study are isolated from clinical biopsies thoroughly characterized with regard to in vivo biofilm formation by Siemens et al. (2016). The results from our in vitro model corresponded well to the presence of biofilm in vivo, as assessed by confocal laser scan microscopy and scanning electron microscopy (Figure 4).
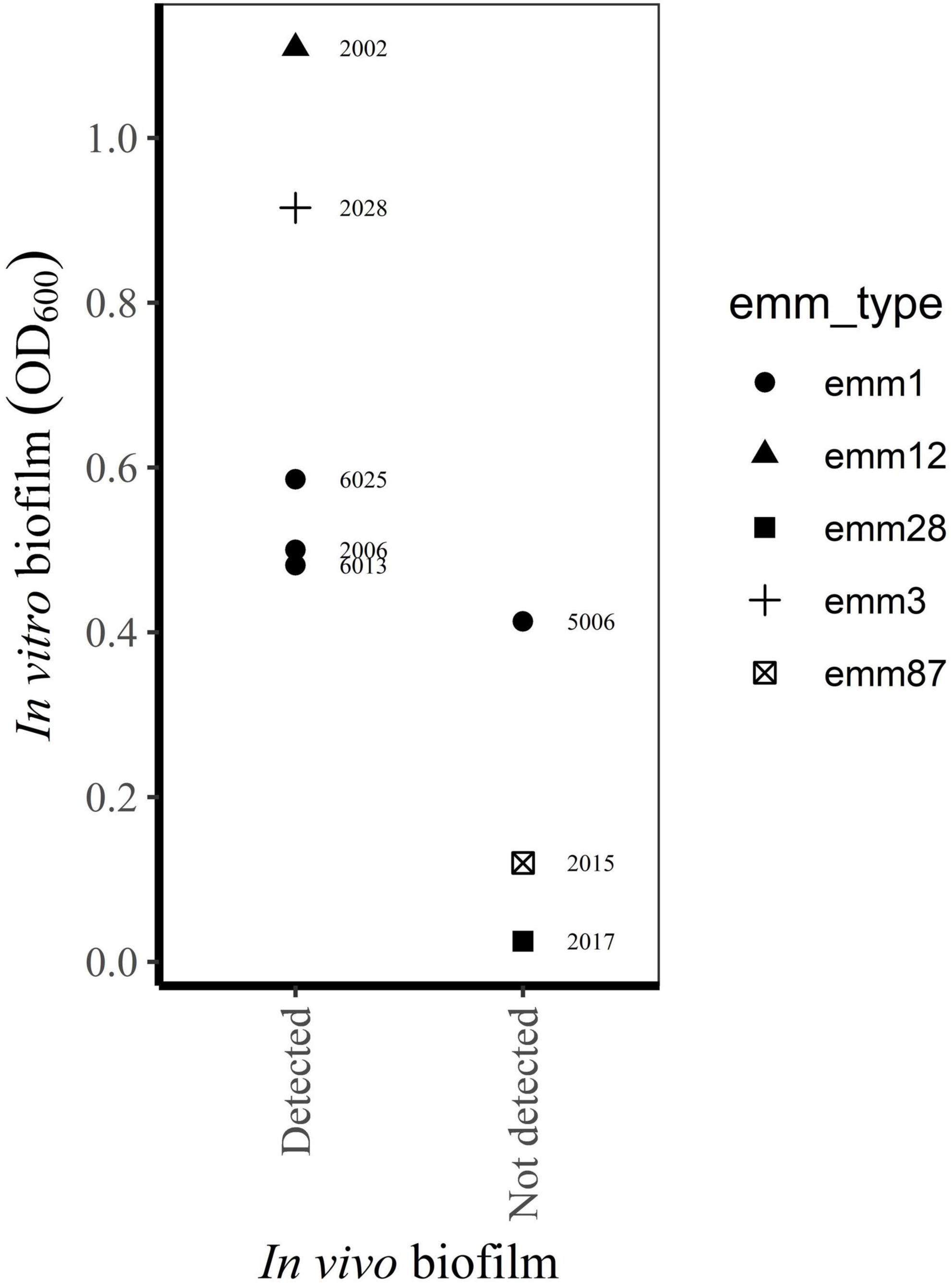
Figure 4. Relationship between biofilm in vivo (Siemens et al., 2016) and in vitro biofilm forming capacity of bacterial isolates from the same biopsies (this study).
The biofilm forming capacity of emm1 isolates was uniform and differed significantly from other emm types. However, the associations between biofilm forming capacity and the different clinical variables were all non-significant.
The distribution of emm types in the present study fits well with results from previous prevalence reports of invasive streptococcal infections in the Scandinavian countries (Gherardi et al., 2018). Both the close relationship between emm type and sequence type (MLST) as well as the genomic stability among emm1 isolates are in accordance with other reports (Enright et al., 2001; Barnett et al., 2018; Coppens et al., 2019; Li et al., 2020). Our finding that most emm1 isolates are better biofilm formers than the majority of emm28 isolates, is consistent with previous studies (Baldassarri et al., 2006; Koller et al., 2010; Ogawa et al., 2011). The only study among these with a sufficient number of emm3 isolates, found a more homogenous distribution of biofilm forming capacity within this emm type compared to our data (Baldassarri et al., 2006). Other studies addressing the relationship between emm type and biofilm formation includes no or just a few emm1, emm3, and/or emm28 isolates (Lembke et al., 2006; Thenmozhi et al., 2011; Wozniak et al., 2017), making a comparison to our results unreliable.
Lack of association between biofilm formation and clinical characteristics of patients with S. pyogenes NSTI is in accordance with earlier findings from our group (Siemens et al., 2016). Both studies use material from the INFECT project (Madsen et al., 2018), but whereas the previous study was based on assessment of biofilms in tissue biopsies (Siemens et al., 2016), the present study focuses on biofilm formation in an in vitro model. In addition, our study includes more cases (n = 57) than the previous study (n = 31). We are not aware of any other studies addressing the clinical significance of biofilm formation by S. pyogenes in NSTIs.
The main strengths of the present study are the prospective inclusion of cases, strict inclusion criteria and standardized collection of clinical data and bacterial isolates. As far as we know, this is the largest collection of S. pyogenes isolated from prospectively enrolled NSTI patients. Still the sample size is too small to conclude on a relationship between biofilm forming capacity and clinical characteristics of the patients, and this is a limitation in our study. Post-hoc power analyses of the different clinical variables, estimates powers of < 0.1, given a small effect size (Cohen’s w = 0.1), a significance level of p = 0.05 and the actual sample size (n = 57). Further, the categorization of bacterial isolates into biofilm forming groups may lead to misclassifications due to analytical inaccuracy. An examination of OD600-values close to the cut-off values, taking the between-run CV into account, reveals seven isolates (including four emm1 and one emm28 isolate) where analytical imprecision could have influenced categorization. However, these possible misclassifications will not affect the conclusion on an association between emm type and biofilm formation. Other limitations of our study is the in vitro design, disregarding the influence of host factors on biofilm formation, as well as the narrow spectrum of conditions used in the biofilm model. However, concordance between the in vitro biofilm forming capacity of selected isolates in this study and the presence of biofilm in vivo in corresponding biopsies (Siemens et al., 2016), indicates the usefulness of the in vitro model to predict biofilm formation in infected tissue (Figure 4).
Clonal relationship is a possible explanation of the uniform biofilm formation capacity demonstrated among emm1 isolates in this study. In contrast, emm3 isolates, display great variations in biofilm formation capacity despite close genetic relatedness within this emm type.
Some genetic variants are under positive selection in subsets of isolates (Lefebure and Stanhope, 2007; Carroll et al., 2011; Oliver-Kozup et al., 2011; Shea et al., 2011; Olsen et al., 2012), implying a beneficial effect in specific ecological niches. Importantly, genetic variants affecting the expression of virulence factors known to be involved in biofilm formation, may be unevenly distributed between isolates with the same emm type. Examples of this are surface associated structures like pili (Manetti et al., 2007; Koller et al., 2010), Streptococcal collagen like protein 1 (Scl1) (Oliver-Kozup et al., 2011) and hyaluronic acid capsule (Cho and Caparon, 2005; Shea et al., 2011), as well as the secreted streptococcal cysteine proteinase B (SpeB) (Carroll et al., 2011; Olsen et al., 2012).
Future research based on genomics may reveal the virulence factors’ contribution to the variations in biofilm forming capacity observed in the present study. However, genes may be variably expressed under different conditions or due to mutations in regulatory parts of the genome. Great differences between transcriptomes and proteomes at different time points of S. pyogenes biofilm formation, suggesting a substantial regulation by non-transcriptional mechanisms, underscores the importance of assessment at both the RNA and protein levels (Freiberg et al., 2016). Comparison of qualitative and quantitative expression of different genes, as well as detection of post-translational variants of proteins expressed during biofilm formation, could reveal components hitherto unknown to be important in streptococcal biofilm formation.
In summary, we have described the biofilm forming capacity in a collection of S. pyogenes isolated from patients with NSTI. We have demonstrated that invasive emm1 isolates have a distinctive biofilm forming capacity compared to other invasive emm types, but were unable to detect associations of biofilm formation in NSTI to clinical outcomes.
Trond Bruun, Oddvar Oppegaard, Eivind Rath, Torbjørn Nedrebø, Ole Hyldegaard, Michael Nekludov, Mattias Svensson, Ylva Karlsson, Per Arnell, and Anshu Babbar. Affiliations for members of the Infect Study Group are detailed in the Supplementary Material.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
The studies involving human participants were reviewed and approved by the Regional Committee for Ethics in Medical Research (2012/2227) in Western Norway, the Danish Ethical Committee (1211709), the Swedish Ethical Committee (Dnr 930-12), and the Danish Data Protection Agency (30-0900). The patients/participants provided their written informed consent to participate in this study.
AN-T and SS: conceptualization. DS, SS, HW, and HM: methodology. DS: validation, formal analysis, investigation, writing original draft, and visualization. DS, SS, HW, HM, and AN-T: writing—review and editing. SS: project administration. AN-T, SS, and DS: funding acquisition. All authors contributed to the article and approved the submitted version.
This study was supported by Swedish Research Council (Grant 2014-8609-117204-47), FP7-HEALTH [Grant agreement 305340 (INFECT)], Horizon 2020 Framework program [Grant agreement ERAPerMed Project 2018-151 (PerMIT)], NORDFORSK [Project no. 90456 (PerAID)], and Norwegian Surveillance System for Antimicrobial Drug Resistance (NORM) (Grant 16_13).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We acknowledge the contributions of clinical-, laboratory-, and research personnel at each of the INFECT study sites. We also thank all patients who, with their consent to participate in the INFECT research project, has made this study possible.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.822243/full#supplementary-material
Baldassarri, L., Creti, R., Recchia, S., Imperi, M., Facinelli, B., Giovanetti, E., et al. (2006). Therapeutic failures of antibiotics used to treat macrolide-susceptible Streptococcus pyogenes infections may be due to biofilm formation. J. Clin. Microbiol. 44, 2721–2727. doi: 10.1128/JCM.00512-06
Barnett, T. C., Bowen, A. C., and Carapetis, J. R. (2018). The fall and rise of Group A Streptococcus diseases. Epidemiol. Infect. 147:e4. doi: 10.1017/S0950268818002285
Bruun, T., Rath, E., Bruun Madsen, M., Oppegaard, O., Nekludov, M., Arnell, P., et al. (2020). Risk factors and Predictors of Mortality in Streptococcal Necrotizing Soft-Tissue Infections: a Multicenter Prospective Study. Clin. Infect. Dis. 72, 293–300. doi: 10.1093/cid/ciaa027
Carroll, R. K., Shelburne, S. A. III, Olsen, R. J., Suber, B., Sahasrabhojane, P., Kumaraswami, M., et al. (2011). Naturally occurring single amino acid replacements in a regulatory protein alter streptococcal gene expression and virulence in mice. J. Clin. Invest. 121, 1956–1968. doi: 10.1172/JCI45169
Cho, K. H., and Caparon, M. G. (2005). Patterns of virulence gene expression differ between biofilm and tissue communities of Streptococcus pyogenes. Mol. Microbiol. 57, 1545–1556. doi: 10.1111/j.1365-2958.2005.04786.x
Coppens, J., Xavier, B. B., Loens, K., Lammens, C., Ieven, M., Matheeussen, V., et al. (2019). Remarkable Genome Stability among emm1 Group A Streptococcus in Belgium over 19 Years. Genome. Biol. Evol. 11, 1432–1439. doi: 10.1093/gbe/evz093
Courtney, H. S., Ofek, I., Penfound, T., Nizet, V., Pence, M. A., Kreikemeyer, B., et al. (2009). Relationship between expression of the family of M proteins and lipoteichoic acid to hydrophobicity and biofilm formation in Streptococcus pyogenes. PLoS One 4:e4166. doi: 10.1371/journal.pone.0004166
Darenberg, J., Luca-Harari, B., Jasir, A., Sandgren, A., Pettersson, H., Schalen, C., et al. (2007). Molecular and clinical characteristics of invasive group A streptococcal infection in Sweden. Clin. Infect. Dis. 45, 450–458. doi: 10.1086/519936
Enright, M. C., Spratt, B. G., Kalia, A., Cross, J. H., and Bessen, D. E. (2001). Multilocus sequence typing of Streptococcus pyogenes and the relationships between emm type and clone. Infect. Immun. 69, 2416–2427. doi: 10.1128/IAI.69.4.2416-2427.2001
Fiedler, T., Koller, T., and Kreikemeyer, B. (2015). Streptococcus pyogenes biofilms-formation, biology, and clinical relevance. Front. Cell Infect. Microbiol. 5:15. doi: 10.3389/fcimb.2015.00015
Freiberg, J. A., Le Breton, Y., Tran, B. Q., Scott, A. J., Harro, J. M., Ernst, R. K., et al. (2016). Global Analysis and Comparison of the Transcriptomes and Proteomes of Group A Streptococcus Biofilms. mSystems 1, e149–e116. doi: 10.1128/mSystems.00149-16
Gherardi, G., Vitali, L. A., and Creti, R. (2018). Prevalent emm Types among Invasive GAS in Europe and North America since Year 2000. Front. Public Health 6:59. doi: 10.3389/fpubh.2018.00059
Koller, T., Manetti, A. G., Kreikemeyer, B., Lembke, C., Margarit, I., Grandi, G., et al. (2010). Typing of the pilus-protein-encoding FCT region and biofilm formation as novel parameters in epidemiological investigations of Streptococcus pyogenes isolates from various infection sites. J. Med. Microbiol. 59, 442–452. doi: 10.1099/jmm.0.013581-0
Kumar, A., Alam, A., Rani, M., Ehtesham, N. Z., and Hasnain, S. E. (2017). Biofilms: survival and defense strategy for pathogens. Int. J. Med. Microbiol. 307, 481–489. doi: 10.1016/j.ijmm.2017.09.016
Kwasny, S. M., and Opperman, T. J. (2010). Static biofilm cultures of Gram-positive pathogens grown in a microtiter format used for anti-biofilm drug discovery. Curr. Protoc. Pharmacol. 13A:18. doi: 10.1002/0471141755.ph13a08s50
Kaas, R. S., Leekitcharoenphon, P., Aarestrup, F. M., and Lund, O. (2014). Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS One 9:e104984. doi: 10.1371/journal.pone.0104984
Lefebure, T., and Stanhope, M. J. (2007). Evolution of the core and pan-genome of Streptococcus: positive selection, recombination, and genome composition. Genome. Biol. 8:R71. doi: 10.1186/gb-2007-8-5-r71
Lembke, C., Podbielski, A., Hidalgo-Grass, C., Jonas, L., Hanski, E., and Kreikemeyer, B. (2006). Characterization of biofilm formation by clinically relevant serotypes of group A streptococci. Appl. Environ. Microbiol. 72, 2864–2875. doi: 10.1128/AEM.72.4.2864-2875.2006
Li, Y., Rivers, J., Mathis, S., Li, Z., Velusamy, S., Nanduri, S. A., et al. (2020). Genomic Surveillance of Streptococcus pyogenes Strains Causing Invasive Disease, United States, 2016–2017. Front. Microbiol. 11:1547. doi: 10.3389/fmicb.2020.01547
Luca-Harari, B., Ekelund, K., Van Der Linden, M., Staum-Kaltoft, M., Hammerum, A. M., and Jasir, A. (2008). Clinical and epidemiological aspects of invasive Streptococcus pyogenes infections in Denmark during 2003 and 2004. J. Clin. Microbiol. 46, 79–86. doi: 10.1128/JCM.01626-07
Madsen, M. B., Skrede, S., Bruun, T., Arnell, P., Rosen, A., Nekludov, M., et al. (2018). Necrotizing soft tissue infections - a multicentre, prospective observational study (INFECT): protocol and statistical analysis plan. Acta Anaesthesiol. Scand 62, 272–279. doi: 10.1111/aas.13024
Manetti, A. G., Zingaretti, C., Falugi, F., Capo, S., Bombaci, M., Bagnoli, F., et al. (2007). Streptococcus pyogenes pili promote pharyngeal cell adhesion and biofilm formation. Mol. Microbiol. 64, 968–983. doi: 10.1111/j.1365-2958.2007.05704.x
Naseer, U., Steinbakk, M., Blystad, H., and Caugant, D. A. (2016). Epidemiology of invasive group A streptococcal infections in Norway 2010-2014: A retrospective cohort study. Eur. J. Clin. Microbiol. Infect. Dis. 35, 1639–1648. doi: 10.1007/s10096-016-2704-y
Ogawa, T., Terao, Y., Okuni, H., Ninomiya, K., Sakata, H., Ikebe, K., et al. (2011). Biofilm formation or internalization into epithelial cells enable Streptococcus pyogenes to evade antibiotic eradication in patients with pharyngitis. Microb. Pathog. 51, 58–68. doi: 10.1016/j.micpath.2011.03.009
Oliver-Kozup, H. A., Elliott, M., Bachert, B. A., Martin, K. H., Reid, S. D., Schwegler-Berry, D. E., et al. (2011). The streptococcal collagen-like protein-1 (Scl1) is a significant determinant for biofilm formation by group A Streptococcus. BMC Microbiol. 11:262. doi: 10.1186/1471-2180-11-262
Olsen, R. J., Laucirica, D. R., Watkins, M. E., Feske, M. L., Garcia-Bustillos, J. R., Vu, C., et al. (2012). Polymorphisms in regulator of protease B (RopB) alter disease phenotype and strain virulence of serotype M3 group A Streptococcus. J. Infect. Dis. 205, 1719–1729. doi: 10.1093/infdis/jir825
Sartelli, M., Guirao, X., Hardcastle, T. C., Kluger, Y., Boermeester, M. A., Rasa, K., et al. (2018). 2018 WSES/SIS-E consensus conference: recommendations for the management of skin and soft-tissue infections. World J. Emerg. Surg. 13:58. doi: 10.1186/s13017-018-0219-9
Shea, P. R., Beres, S. B., Flores, A. R., Ewbank, A. L., Gonzalez-Lugo, J. H., Martagon-Rosado, A. J., et al. (2011). Distinct signatures of diversifying selection revealed by genome analysis of respiratory tract and invasive bacterial populations. Proc. Natl. Acad. Sci. U.S.A. 108, 5039–5044. doi: 10.1073/pnas.1016282108
Siemens, N., Chakrakodi, B., Shambat, S. M., Morgan, M., Bergsten, H., Hyldegaard, O., et al. (2016). Biofilm in group A streptococcal necrotizing soft tissue infections. JCI Insight 1:e87882. doi: 10.1172/jci.insight.87882
Skrede, S., Bruun, T., Rath, E., and Oppegaard, O. (2020). Microbiological Etiology of Necrotizing Soft Tissue Infections. Adv. Exp. Med. Biol. 1294, 53–71. doi: 10.1007/978-3-030-57616-5_5
Thenmozhi, R., Balaji, K., Kumar, R., Rao, T. S., and Pandian, S. K. (2011). Characterization of biofilms in different clinical M serotypes of Streptococcus pyogenes. J. Basic Microbiol. 51, 196–204. doi: 10.1002/jobm.201000006
Vincent, J.-L., Moreno, R., Takala, J., Willatts, S., De Mendonça, A., Bruining, H., et al. (1996). The SOFA (Sepsis-related Organ Failure Assessment) score to describe organ dysfunction/failure. Intensiv. Care Med. 22, 707–710. doi: 10.1007/bf01709751
World Health Organization. (2005). The Current Evidence for the Burden of Group A Streptococcal Diseases. Switzerland: World Health Organization.
Keywords: Streptococcus pyogenes, necrotizing soft tissue infection (NSTI), biofilms, M-protein, emm1
Citation: Skutlaberg DH, Wiker HG, Mylvaganam H, The INFECT Study Group, Norrby-Teglund A and Skrede S (2022) Consistent Biofilm Formation by Streptococcus pyogenes emm 1 Isolated From Patients With Necrotizing Soft Tissue Infections. Front. Microbiol. 13:822243. doi: 10.3389/fmicb.2022.822243
Received: 25 November 2021; Accepted: 27 January 2022;
Published: 18 February 2022.
Edited by:
Simone Bergmann, Technische Universitat Braunschweig, GermanyReviewed by:
Mattias Collin, Lund University, SwedenCopyright © 2022 Skutlaberg, Wiker, Mylvaganam, The INFECT Study Group, Norrby-Teglund and Skrede. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dag Harald Skutlaberg, ZGFnLmhhcmFsZC5za3V0bGFiZXJnQGhlbHNlLWJlcmdlbi5ubw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.