- 1State Key Laboratory of Biocontrol, Guangdong Key Laboratory of Pharmaceutical Functional Genes, School of Life Sciences, Southern Marine Science and Engineering Guangdong Laboratory (Zhuhai), Higher Education Mega Center, Sun Yat-sen University, Guangzhou, China
- 2Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 3Department of Pediatrics, The Third Affiliated Hospital of Sun Yat-sen University, Guangzhou, China
- 4Faculty of Health Sciences, University of Macau, Taipa, Macau SAR, China
Antibiotic-resistant Vibrio alginolyticus becomes a worldwide challenge threatening both human health and food safety. The approach in managing such infection is largely absent, despite the fact that the mechanisms of antibiotic resistance have been extensively investigated. Metabolic modulation has been documented to be a novel approach in improving antibiotic efficacy. In this study, we characterize the metabolic signature of V. alginolyticus exposed to 0.3 or 0.5 μg/ml of ofloxacin (OFX). By profiling the metabolome, we find that bacteria treated by the two different concentrations of OFX generate different metabolic signatures. While a part of these metabolites was shared by both groups, the other metabolites represent their own signatures. The pathway enrichment analysis demonstrates that the pyruvate cycle is disrupted in the bacteria treated by the 0.3 μg/ml OFX as compared to those by the 0.5 μg/ml. Importantly, the disruption of pyruvate cycle confers the capability of bacteria to survive under 0.5 μg/ml of antibiotic stress. Further analysis identifies that the fatty acid biosynthesis is elevated in bacteria treated by 0.3 μg/ml OFX, and inhibition on fatty acid completely prevents the bacteria from survival even under such dose of antibiotic stress. Our study suggests that bacteria adapt to antibiotic stress by modulating the metabolic flux for survival, which could be targeted to increase antibiotic efficacy.
Introduction
Vibrio spp., a group of Gram-negative bacteria, are prevalent in freshwater, estuarine, and marine environments (Baker-Austin et al., 2017). These bacteria are highly adaptable to different environments and tolerant to various adverse conditions (Baker-Austin et al., 2018). They represent one of the dominant species in seawater. Some of the Vibrios, such as Vibrio alginolyticus, V. parahemolyticus, and V. vulnificus, are pathogenic to both aquatic animals and human beings (Ina-Salwany et al., 2019). V. alginolyticus, for example, infects a wide range of economical aquatic animals including shrimp, fish, and shellfish. Infected fish are commonly associated with red spots on the ventral and lateral areas, swollen and dark skin lesions, and the release of blood exudate, known as vibriosis (Novoa et al., 1992). In addition, V. alginolyticus also causes bacteremia in fish that are exemplified as exophthalmos, ascites, and inflated spleen (Gong et al., 2020). Moreover, V. alginolyticus can rapidly kill the fish host before any symptom can be identified. V. alginolyticus can cause ear and wound infection in humans after being exposed to contaminated water (Jones et al., 2013). Therefore, V. alginolyticus is a harmful species that causes huge economic loss in aquaculture and presents a potential threat to human health.
Antibiotics is the primary choice to treat V. alginolyticus infection. Quinolone is a representative group of antibiotics that have been widely used in aquaculture in the past decades, owning to its high killing efficacy to different species of the genus Vibrio (Backhaus et al., 2000). The mechanism underlying the quinolone-mediated killing of bacteria is that it works as an inhibitor to the DNA gyrase, which belongs to the type II topoisomerases that are mainly involved in the topological transition of DNA (Reece and Maxwell, 1991). A quinolone antibiotic can result in the inhibition on DNA replication, repairment, recombination, and transposition, and thus disrupt RNA and protein biosynthesis (Paton and Reeves, 1988; Just, 1993), leading to a bactericidal effect (Baquero and Levin, 2021). However, the overuse of antibiotics including quinolones in aquaculture generates antibiotic-resistant bacteria. Clinically, multidrug-resistant Vibrios were frequently isolated from a variety of seafood in many countries, including China, Korea, and the United States (Zhao et al., 2011; Gong et al., 2020). Quinolone-associated resistance mainly results from the genetic mutations on the quinolone resistance-determining region (QRDR) of the gyrase gene, the decrease in membrane permeability, the increase in efflux pump, or plasmid-mediated resistance (Okuda et al., 1999; Otsuka et al., 2005; Xia et al., 2013; Hamed et al., 2018; Zhou et al., 2019).
Antibiotic resistance is strongly linked to bacterial metabolism (Stokes et al., 2019). Lab-evolved and clinic isolated kanamycin-resistant bacterial strains have different metabolomes to kanamycin-sensitive strain (Peng et al., 2015), even though the resistant mechanism was undefined. The identification of glucose, alanine, fructose, and glutamate as decreased metabolites in abundance in antibiotic-resistant bacteria indicates that metabolites could be potential regulators of antibiotic resistance. The exogenous supplementation of these substances could reverse antibiotic resistance through promoting antibiotic uptake; thus, a high concentration of antibiotics could be fluxed into the bacteria and exceed the threshold that bacteria can tolerate. By this way, the kanamycin-resistant bacteria were resensitized to kanamycin (Peng et al., 2015; Su et al., 2015, 2018; Cheng et al., 2019; Zhang et al., 2020). Similarly, a recent report identified that glutamine and inosine were decreased in abundance in multidrug-resistant bacteria. The exogenous supplementation of glutamine and inosine potentially augmented the antibiotic efficacy, especially for β-lactams. These two metabolites increased antibiotic influx into the bacteria that overcome the effect by an efflux pump and hydrolysis by β-lactams (Zhao et al., 2021). In addition, metabolites, such as glucose and alanine, not only promote antibiotic killing but also promote the generation of reactive oxygen species, which is downregulated in antibiotic-resistant V. alginolyticus; the generation of reactive oxygen species (ROS) promotes killing (Ye et al., 2018; Zhang et al., 2019, 2020). Moreover, the deletion of sodium-pumping complex (NAQ) complex confers antibiotic resistance in V. alginolyticus, but it can also be reversed through metabolite supplementation (Jiang et al., 2020). Metabolite-enhanced antibiotic killing is applicable not only to antibiotic resistance but also to antibiotic tolerance (Vega et al., 2012). Therefore, the identification of metabolites able to significantly reverse antibiotic resistance is urgently needed. In this study, we monitored the global metabolic responses of V. alginolyticus to ofloxacin (OFX) and found that OFX-mediated killing required the activation of pyruvate cycle and the inhibition of fatty acid synthesis.
Materials and Methods
Bacterial Strains and Culture Conditions
V. alginolyticus ATCC 33787 was grown at 30°C in Luria–Bertani (LB) broth (pH 7.0) supplemented with 3% sodium chloride. The overnight culture was diluted 1:100 (v/v) in fresh LB broth (supplemented with 0, 0.3, or 0.5 μg/ml OFX) at 30°C for 2 h. The bacteria were harvested by centrifugation at 8,000 rpm for 3 min, and the cells were washed and resuspended in sterile saline and adjusted to an optical density (OD600) of 1.0. To build a growth curve, the bacterial growth was examined by measuring the OD600 of the culture at the indicated time. To study the effect of antibiotics, inhibitors of pyruvate cycle, or fatty acid biosynthesis on bacterial growth, the bacterial growth was examined by measuring the OD600 of the bacterial cultures at 10 h in medium with OFX, malonate, or triclosan (Sangon Biotech, Shanghai, China). At least three biologic replicates were performed.
Measurement of the Minimum Inhibitory Concentration
The Minimum Inhibitory Concentration (MIC) was determined by antimicrobial susceptibility testing following the previous description. In brief, an overnight bacterial culture grown in 3% NaCl was diluted 1:100 (v/v) in fresh LB medium and cultured at 30°C, to an OD600 of 0.5. Bacterial cells equivalent to 105 colony-forming unit (CFU) were dispensed into each well of a 96-well microtiter polystyrene tray that contained a series of 2-fold dilutions of antibiotic. Following 16 h incubation at 30°C, the bacterial growth was examined by measuring the OD600 of the culture. The MIC was defined as the lowest antibiotic concentration that inhibited visible growth. Three biological repeats were carried out.
Metabolomics Analysis
Bacterial sample preparation was carried out as previously described (Du et al., 2017). The bacteria were harvested by centrifugation at 8,000 rpm for 3 min, and the cells were washed and resuspended in sterile saline and adjusted to 1.0 at OD600. About 10 ml bacteria with OD600 = 1.0 were quenched with cold methanol and sonicated for 10 min at 200 W. Samples were centrifuged with12,000 rpm at 4°C for 10 min. A supernatant, containing 1 mg/ml ribitol (Sigma-Aldrich, St. Louis, MO, United States) as internal analytical standard, was transferred into a new tube and dried by a vacuum centrifugation device (LABCONCO). The dried extracts were then incubated with 80 μl methoxyamine hydrochloride (20 mg/ml, Sigma-Aldrich, St. Louis, MO, United States) in pyridine (Sigma-Aldrich, St. Louis, MO, United States) for 180 min at 37°C, and the derivatization was done with an identical volume of N-methyl-N-(trimethylsilyl) trifluoroacetamide (Sigma-Aldrich, St. Louis, MO, United States) for another 45 min. Samples were centrifuged at 12,000 rpm for 15 min, and the supernatant was transferred into new tubes. Gas chromatography–mass spectrometry (GC-MS) analysis was performed with an Agilent GC-MS instrument. By using spectral matching and retention time (RT) indexes from the National Institute of Standards and Technology (NIST) library of the NIST MS search 2.0, metabolites were identified. Metabolites were determined after removing known artificial peaks and merging the identical compounds. The resulting data matrix was normalized by the concentrations of added internal standards and the total intensity. Normalized peak intensities formed a single matrix with Rt-m/z pairs (retention time–mass charge ratio pairs) for each file in the data set. The matrix can be used for further analysis. According to a reference distribution, Z score analysis was used to scale each metabolite. Statistical difference was obtained by the Kruskal–Wallis test and Mann–Whitney test using solutions statistical package for the social sciences (SPSS) 13.0. Values with p <0.01 were considered as significant. Hierarchical clustering was completed in the R platform with the package gplots1 using the distance matrix. Multivariate statistical analysis included principal component analysis (PCA) and orthogonal partial least square discriminant analysis (OPLS-DA) implemented with SIMCA 12.0.1 (Umetrics, Umeå, Sweden). Control scaling was selected prior to fitting. All variables were mean centered and scaled to the Pareto variance of each variable. PCA was used to reduce the high dimension of the data set. We analyzed the differential metabolites to their respective biochemical pathways as outlined in the MetaboAnalyst 3.01 (Bala et al., 2021). Pathways were enriched by p-value < 0.05.
Real-Time Quantitative PCR
Quantitative real-time PCR (qRT-PCR) was carried out as previously described (Li et al., 2016). About 1 ml of bacterial culture with OD600 = 1.0 was harvested. The total RNA of each sample was isolated with Trizol (Invitrogen, Carlsbad, CA, United States). Reverse transcription- PCR was conducted by using an EvoM-MLV RT kit with gDNA clean for quantitative PCR (qPCR) (AG11705; Accurate Biotechnology, Guangdong, China) with 1 mg of total RNA according to the manufacturer’s instructions. qRT-PCR was performed in 384-well plates with a total volume of 10 μl containing 5 μl 2 × SYBR green premix pro Taq HS qPCR kit (AG11701; Accurate Biotechnology, Guangdong, China), 2.6 μl H2O, 2 μl cDNA template, and 0.2 μl each of forward and reverse primers (10 mM). The primers are listed in Supplementary Table 1. All samples were tested in biological triplicate and run on the LightCycler 480 system (Roche, Mannheim, Germany) according to the manufacturer’s instructions, and four independent samples were assayed for both the control group and the test group. The cycling parameters were 95°C for 30 s to activate the polymerase; 40 cycles of 95°C for 10 s; and 56°C for 30 s. Fluorescence measurements were performed at 72°C for 1 s during each cycle. Cycling was terminated at 95°C with a calefactive velocity of 5°C/s to obtain a melting curve. Data are shown as the relative mRNA expression compared with control with the endogenous reference 16S rRNA gene.
Measurement of the Activity of Enzymes in the Pyruvate Cycle
The determination of pyruvate dehydrogenase (PDH), alpha-ketoglutarate dehydrogenase (KGDH), succinate dehydrogenase (SDH), and malate dehydrogenase (MDH) activity was carried out as previously described (Cheng et al., 2017). The activity of phosphoenolpyruvate carboxykinase (PEPCK) enzymatic activity was performed through the PEPCK Assay Kit according to the manufacturer’s manual (Suzhou Comin Biotechnology, Suzhou, China). V. alginolyticus cultured in medium with OFX was collected and washed three times with sterile saline. The bacterial cells were suspended in × 1 PBS (pH 7.0) to OD600 = 1.0. Samples of 30 ml were collected by centrifugation at 8,000 rpm for 5 min. Pellets were resuspended in phosphate-buffered saline (PBS), and the cells were lysed by sonication for 10 min (200 W total power with 35% output, 2 s pulse, 3 s pause over ice). The solution was then centrifuged with 12,000 rpm at 4°C for 10 min to remove insoluble materials. The protein concentration of the supernatant was quantified by the BCA protein concentration determination kit (Beyotime, P0009). Then, 200 μg proteins were used for the determination of enzyme activity.
Nicotinamide Adenine Dinucleotide – Hydrogen Measurement
Nicotinamide adenine dinucleotide – hydrogen measurement (NADH) measurement was carried out as described previously (Su et al., 2018). V. alginolyticus cultured in the presence of different concentrations of OFX were collected, washed, and resuspended in NADH extraction buffer. Then, extracts were heated at 60°C for 5 min. After the NAD extraction buffer was added, the extracts were vortexed and centrifuged at 12,000 rpm for 5 min. Supernatant was then collected for the measurement of NADH with the EnzyChrom NAD/NADH Assay Kit, following the user manual (BioAssay Systems).
Measurement of Adenosine Triphosphate
The adenosine triphosphate (ATP) level in the cells was determined by a BacTiter-Glo™ Microbial Cell Viability Assay (Cat. G8231; Promega, Madison, WI, United States) as previously described (Cheng et al., 2019). In brief, bacterial cells were harvested after being treated by centrifugation at 8,000 rpm for 3 min and were then washed twice with sterile saline. Cells were resuspended with saline solution and adjusted to OD600 ≈0.2. Then, 50 μl samples were added to the well in a 96-well plate and mixed with an equal volume of the kit solution. The absorbance of the solution was measured using VICTOR X5 (PerkinElmer, Turku, Finland) according to the manufacturer’s instructions. The concentration of ATP was calculated according to the standard curve of ATP.
Results
Responses of Vibrio alginolyticus to Different Concentrations of Ofloxacin
To determine the optimal time point to analyze the metabolic response of bacteria to ofloxacin stress, V. alginolyticus was exposed to either 0.3 or 0.5 μg/ml OFX, corresponding to 1 or 1.6 × MIC of the parental strain, respectively (Figure 1A). In a time-dependent killing experiment, the number of viable bacteria was consistent at different time points during the whole course of antibiotic treatment by 0.3 μg/ml. However, the OD600 was increased at the first 7 h and began to decline slightly afterward (Figures 1B,C). In contrast, the number of viable bacteria dropped significantly when the bacteria were exposed to 0.5 μg/ml OFX, even though the OD600 was increased at the first 3 h (Figures 1B,C). The discrepancy of optical density (OD) value and viable counts might be due to the effect of cell filamentation at low antibiotic concentration where a filament with many cells produces a single colony. These data suggest that bacteria mounted a differential response to antibiotic stress as early as 1 h post-treatment of OFX. These differences became more significant at 2 h post-treatment of OFX that either continued to grow (control) or kept at a constant level of growth (0.3 μg/ml), or decreased the growth (0.5 μg/ml). So, 2 h antibiotic stress gives a good time point for the following analysis when the bacteria begin to die but OD is similar among the three groups. More importantly, the bacteria being treated with 0.3 μg/ml had a constant viability throughout the time-course experiment, suggesting that a survival mechanism might be present as compared to the bacteria being treated with 0.5 μg/ml.
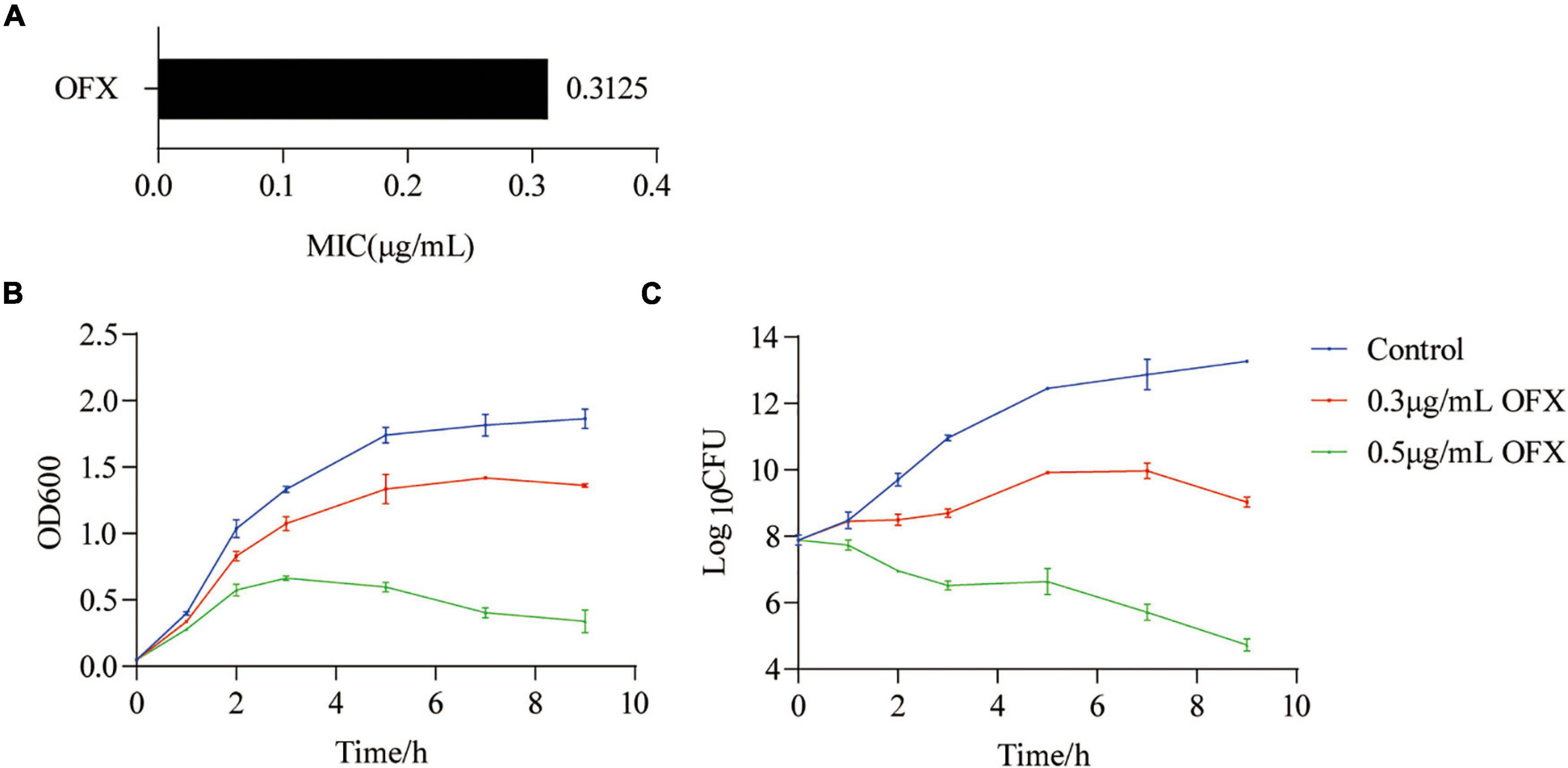
Figure 1. Killing curves of ofloxacin on V. alginolyticus. (A) MIC of V. alginolyticus. (B) Growth curve of V. alginolyticus in medium with the indicated concentrations of OFX. (C) CFU of (A). Each data point represents means ± standard errors of the means (SEM) from three replicates.
Metabolomic Profiles of Vibrio alginolyticus to Ofloxacin Exposure
Bacteria culture was collected 2 h post-treatment of OFX for gas chromatography–mass spectrometry (GC-MS)-based metabolomic profiling for all of the three groups. Four biological replicates were included in each group, and each biological replicate had two technique replicates, altogether generating a total of 24 data points. The reproducibility of the data was demonstrated by Pearson correlation coefficients, showing low variation between the replicates (the relative standard deviation: 1.47% ± 0.03%) (Figure 2A). These results suggested that the differential abundance of the metabolites observed was attributed to different treatment but not to technique issues. Therefore, a total of 229 peaks were aligned, and the metabolites were identified by searching against NIST. The internal standard, ribitol, and any other known solvents were excluded. Finally, 60 metabolites were identified from all the three samples (Figure 2B). The identified metabolites belong to the functional categories of carbohydrate (14.75%), amino acids (27.87%), lipids (34.43%), nucleotides (9.84%), and unknown metabolites (13.11%), respectively (Figure 2C). These data together suggest that V. alginolyticus mounts metabolic shifts under ofloxacin stress.
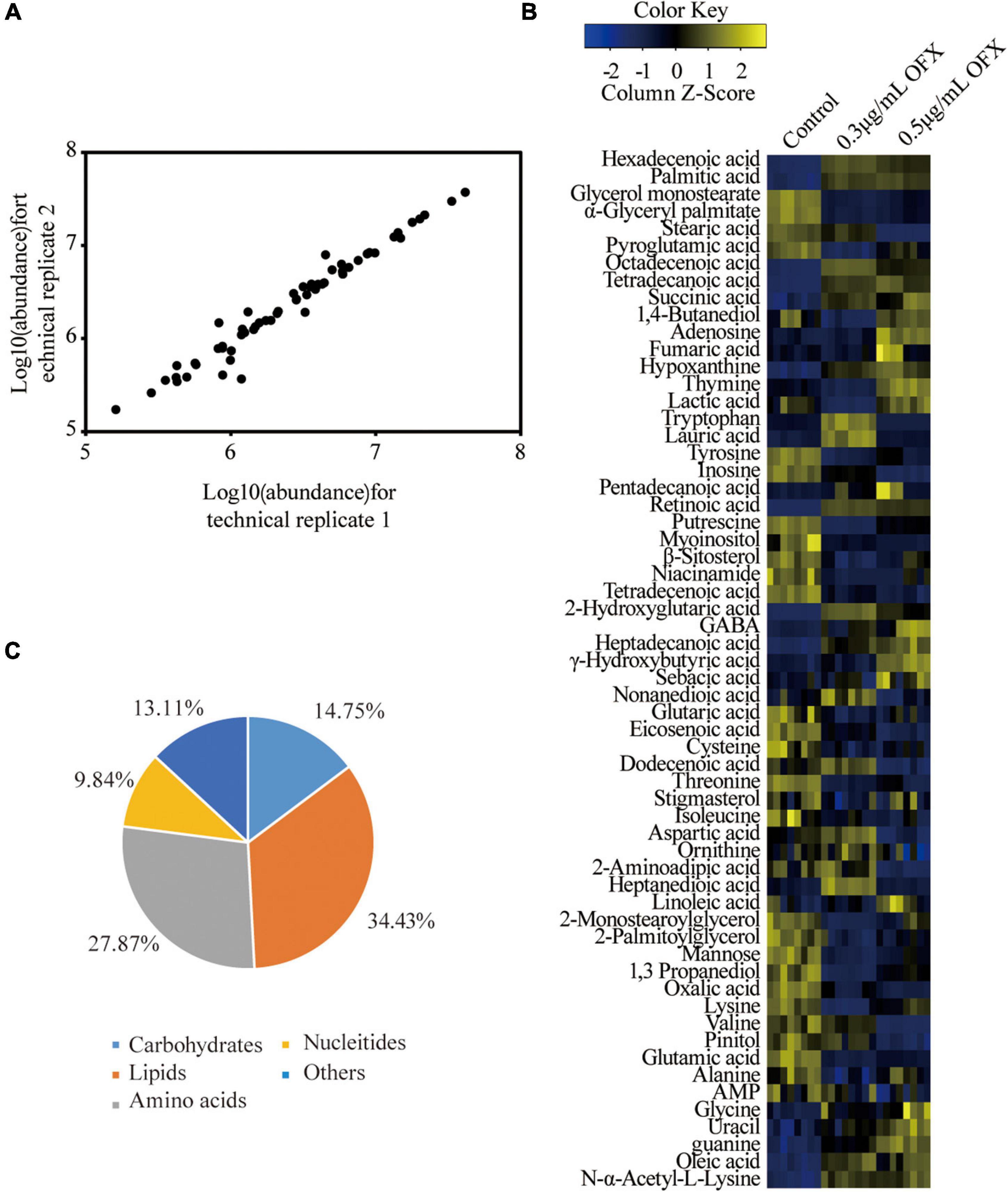
Figure 2. Metabolomic profiles of V. alginolyticus in response to OFX. (A) Heat map of identified metabolites. Scale is shown at bottom, where blue to yellow represents low-to-high abundance. (B) Reproducibility of metabolomic profiling. (C) Functional categories of the identified metabolites.
Metabolomic Signature of Vibrio alginilyticus to Different Concentrations of Ofloxacin
The different patterns in the growth of V. alginotlyticus exposed at different concentrations of OFX suggest that the bacteria treated by 0.3 μg/ml OFX may adapt differential metabolic responses to allow the bacteria to keep growing. Thus, a protective metabolic mechanism may exist to cope with OFX stress. To identify the metabolites with differential abundance, the Mann–Whitney test (Wilcoxon rank sum test) was adopted. A total of 51 and 55 differential metabolites were identified in the bacteria treated by 0.3 and 0.5 μg/ml OFX, corresponding to 0.5 and 0.69% false discovery rate, respectively, as compared to the control group (Figure 3A). The z-score, which demonstrates the distribution of the abundance of each metabolite around the mean, spanned from –6.63 to 89.50 in the 0.3 μg/ml group and from –6.94 to 80.00 in the 0.5 μg/ml. Accordingly, the abundance of 23 metabolites were increased and 28 metabolites were decreased in the 0.3 μg/ml group; 25 metabolites were increased and 32 metabolites were decreased in the 0.5 μg/ml group (Figure 3B). We analyzed the altered metabolites according to their functional categories and found that the differential metabolites under the category of lipids account for the most number of altered metabolites, followed by amino acids, then carbohydrates and other unknown metabolites, and nucleotides (Figure 3C). These data demonstrated that lipid metabolism and amino acid metabolism were significantly altered during the exposure. In addition, the number of altered metabolites between 0.3 and 0.5 μg/ml were different (Figure 3D). It is notable that the number of increased metabolites under the category of lipids in bacteria treated by 0.3 μg/ml OFX was more than the decreased metabolites. In contrast, a higher number of decreased metabolites under the category of lipids was observed in the bacteria treated by the 0.5 μg/ml OFX (Figure 3D).
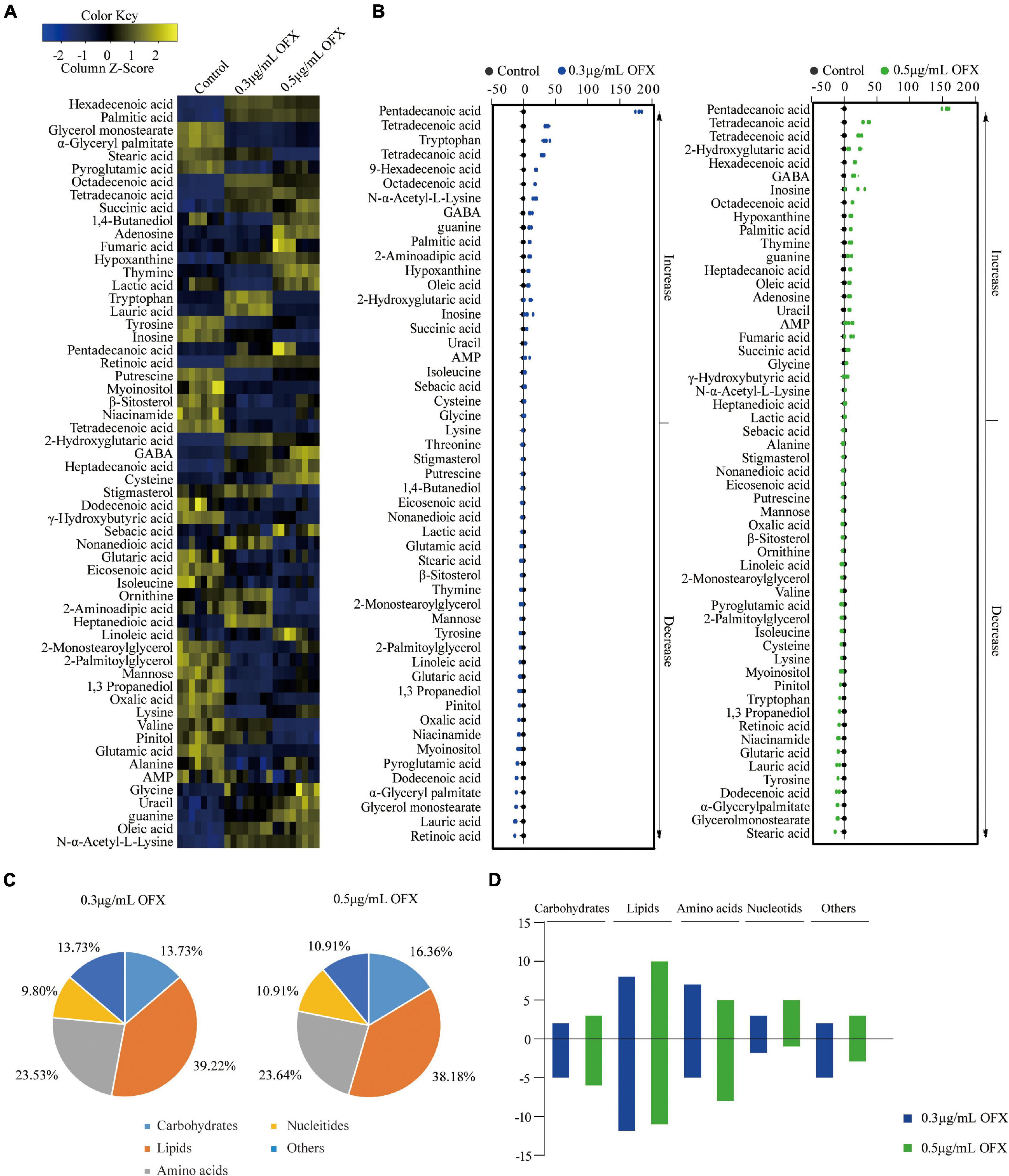
Figure 3. Differential abundance of metabolites in response to OFX. (A) Heat map showing relative abundance of metabolites (Wilcoxon P < 0.05) in control, 0.3 μg/ml OFX, and 0.5 μg/ml OFX groups. Scale is shown at bottom, where blue to yellow represents low-to-high abundance. (B) Z-score plot of differential metabolites based on control. Z-score varied between –6.63 and 89.50 for 0.3 μg/ml OFX to control, between –6.94 and 80.00 for 0.5 μg/ml OFX to control. Each point represents one metabolite in one technical repeat and is colored by sample types. (C) Percentage of differential abundance of metabolites in different functional categories. (D) The number of metabolites increased and decreased in different categories.
Thus, bacteria mount a differential metabolic response to 0.3 and 0.5 μg/ml OFX treatment.
Pathway Analysis of Differential Metabolites Upon Ofloxacin Treatment
Pathway enrichment analysis is of great importance to dissect the metabolic states influenced by antibiotics (Peng et al., 2015; Cheng et al., 2019; Zhao et al., 2021), despite the fact that the metabolites identified by GC-MS are biased toward organic acids. Thus, to get an idea about the difference between the 0.3 and 0.5 μg/ml groups, we further analyzed the differential metabolites. Among the 59 differential metabolites found in the bacteria treated by 0.3 and the 0.5 μg/ml OFX, 47 metabolites were shared by the two groups, where the abundance of 17 metabolites were increased; 24 metabolites were decreased in both of the groups. The abundance of four metabolites, tryptophan, isoleucine, sebacic acid, and L-cysteine, were increased in the 0.3 μg/ml group but decreased in the 0.5 μg/ml group. Another two metabolites, lactic acid and thymine, were decreased in the 0.3 μg/ml group but increased in the 0.5 μg/ml group (Figure 4A). Four metabolites, with one increased and three decreased metabolites, were specific to the 0.3 μg/ml group, and eight metabolites, with five increased and three decreased metabolites, were specific to the 0.5 μg/ml group (Figure 4A). The altered metabolites shared by both groups were analyzed by pathway analysis. A total of eight pathways were identified including alanine, aspartate, and glutamate metabolism; arginine and proline metabolism; arginine biosynthesis; lysine degradation; butanoate metabolism; glutathione metabolism; aminoacyl–tRNA biosynthesis; and the biosynthesis of unsaturated fatty acids (Figure 4B). It is interesting to note that the amino acids were significantly downregulated in the 0.5 μg/ml group but only a part of them was downregulated in the 0.3 μg/ml group, since the biosynthesis of amino acids was highly dependent on central carbon metabolism. This result suggests that central carbon metabolism was likely disrupted so that it fails to provide a sufficient substrate for macromolecule synthesis. In correlation with the decrease in the biosynthesis of amino acids, the intermediates metabolite (fumaric acid and succinic acid) of the pyruvate cycle, a novel metabolic cycle that provides energy to bacteria (Su et al., 2018; Zhang et al., 2020), was accumulated. This accumulation was much more significant in the 0.5 μg/ml group. The metabolite, γ-aminobutyric acid (GABA), that refills the succinate, was also accumulated in a dose-dependent manner (Figure 4C). However, the biosynthesis of amino acids was decreased in a dose-dependent manner. Since the altered metabolites of the pyruvate cycle in both of the 0.3 and 0.5 μg/ml groups showed the same trend of change, we postulate that pyruvate cycle might be critical for the survival under OFX stress.
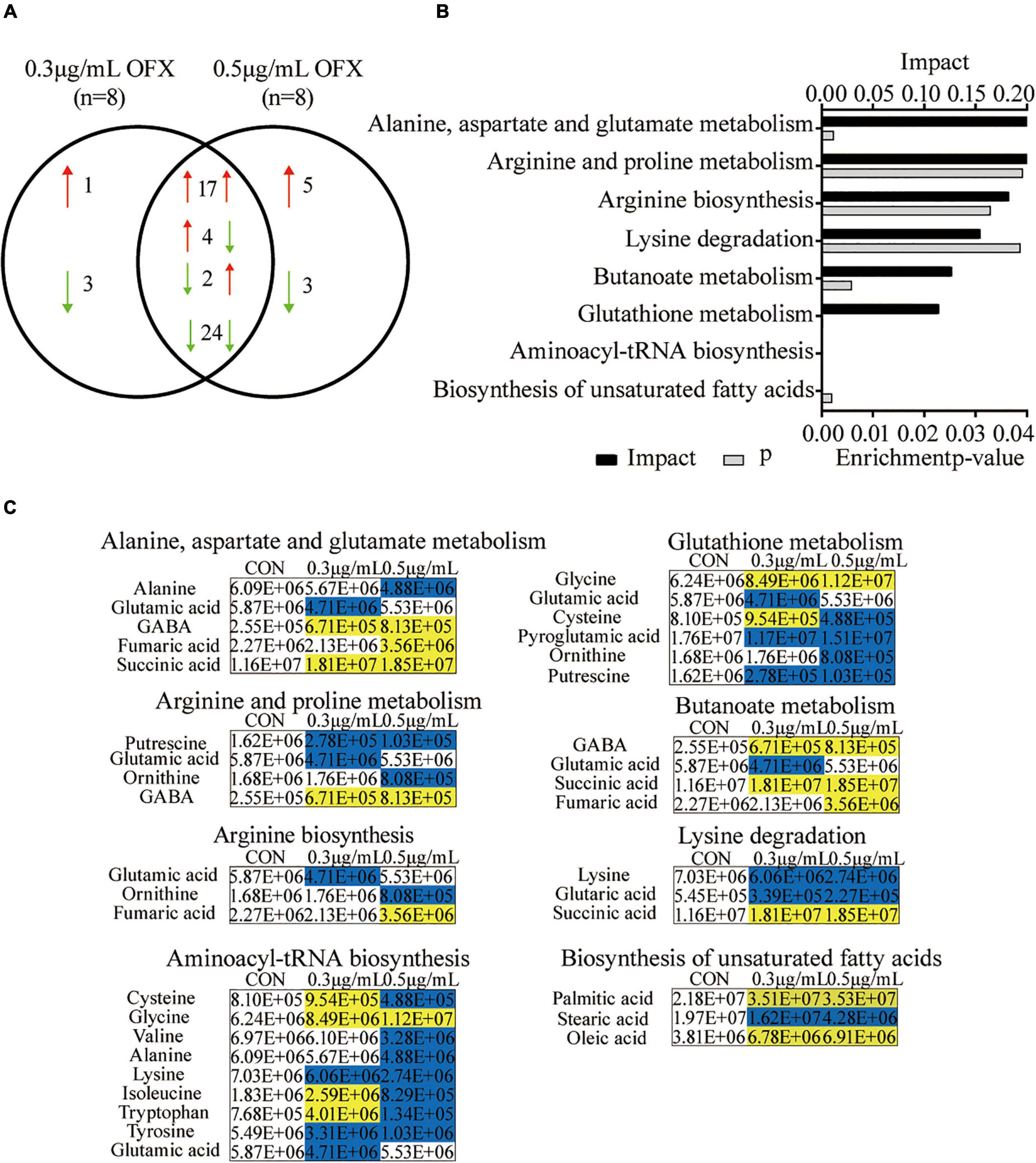
Figure 4. Pathway analysis of differential metabolites upon OFX treatment (A) Metabolites that were shared between the control, 0.3 μg/ml OFX, and 0.5 μg/ml OFX groups and their unique metabolites. (B) Enriched pathways by the shared metabolites. The metabolites of differential abundance were selected and analyzed in MetaboAnalyst to enrich pathways. Seven pathways that had significant difference (P < 0.05) were enriched and sorted by their weights (impact). (C) Differential metabolites in the enriched pathways.
Disruption of Pyruvate Cycle Inhibits Ofloxacin-Induced Cell Death
To confirm our hypothesis that pyruvate cycle was critical for OFX-mediated cell death, we quantify the enzymatic activity of pyruvate cycle in the bacteria treated by 0.3 and 0.5 μg/ml treatment, respectively, at 2 h post-treatment of OFX, the time point in collecting samples for GC-MS analysis (Figure 1B). Five enzymes including two pyruvate cycle-specific enzymes, PDH, and PEPCK and three enzymes in the canonical TCA cycle, succinate dehydrogenase (SDH), α-ketoglutarate dehydrogenase (α-KGDH), and MDH were included. Interestingly, the activity of PDH was increased in both of the groups and no significant difference was observed between the 0.3 and 0.5 μg/ml groups (Figure 5A). The SDH activity remained unchanged at the 0.3 μg/ml group but dramatically increased in the bacteria treated by 0.5 μg/ml OFX (Figure 5B), which was corroborated by the observation that fumarate was accumulated (Figure 4C). Interestingly, the α-KGDH was increased in a dose-dependent manner (Figure 5C), consistent with the gradual increase of the succinate level in the metabolomic data. The MDH activity increased 1.95-fold in the 0.3 μg/ml group, and 3.02-fold in the 0.5 μg/ml group (Figure 5D). The activity of PEPCK was unchanged at the 0.3 μg/ml group but increased in the bacteria treated by 0.5 μg/ml OFX (Figure 5E). As pyruvate cycle provides NADH, the aberrant pyruvate cycle suggests that it might be associated with the dysregulated redox potential that contributes to the ultimate cell death. Indeed, the ratio of NAD+/NADH was decreased in a dose-dependent manner (Figure 5F), indicating the preferred conversion of NAD+ to NADH. Consistently, the level of ATP content was also increased (Figure 5G). These data together suggest that the functional pyruvate cycle is associated with the promotion of OFX-mediated killing. Supportively, the block of the pyruvate cycle with malonate (20 mM), the inhibitor of succinate dehydrogenase, completely rescued the bacteria from the killing by either 0.3 or 0.5 μg/ml OFX, implying that pyruvate cycle was critical for OFX-mediated killing (Figures 5H,I).
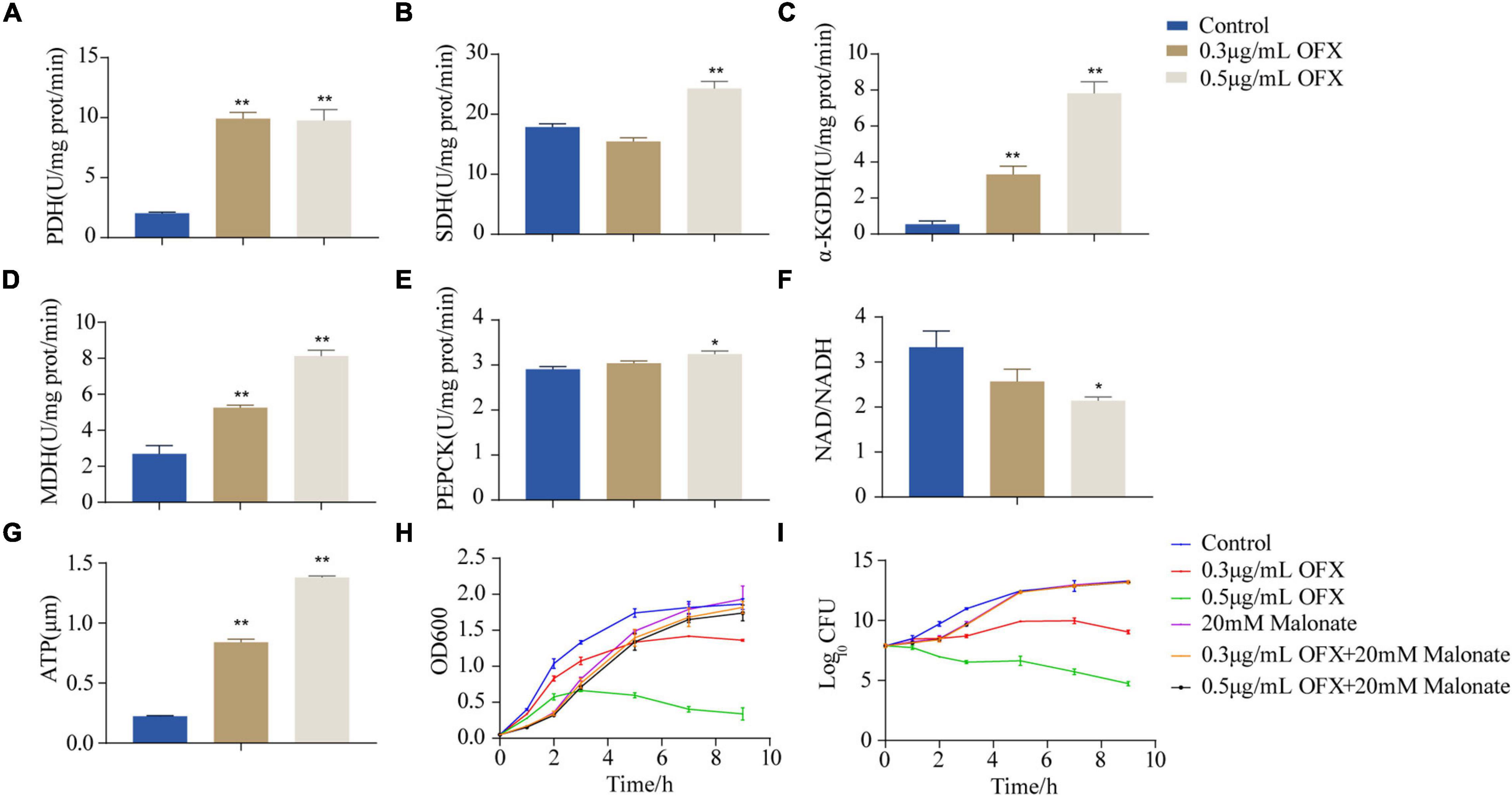
Figure 5. Disruption of pyruvate cycle inhibit OFX-induced cell death. (A) Activity of pyruvate dehydrogenase of pyruvate cycle upon OFX treatments. (B) Activity of succinate dehydrogenase of pyruvate cycle upon OFX treatments. (C) Activity of α-ketoglutarate dehydrogenase of pyruvate cycle upon OFX treatments. (D) Activity of malate dehydrogenase of pyruvate cycle upon OFX treatments. (E) Activity of phosphoenolpyruvate carboxykinase of pyruvate cycle upon OFX treatments. (F) NAD/NADH of V. alginolyticus in medium with the indicated concentrations of OFX. (G) ATP of V. alginolyticus in medium with the indicated concentrations of OFX. (H) Growth curve of V. alginolyticus in medium with the indicated concentrations of OFX or 20 mM malonate. (I) CFU of (H). The statistical analyses were performed by one-way ANOVA, followed by LSD multiple comparison test, *P<0.05; **P<0.01. Error bars represent means ± SEM. Each experiment was performed in triplicate.
Identification of Crucial Metabolites Using Multivariate Data Analysis
To identify the metabolites that distinguish the 0.3 μg/ml group and 0.5 μg/ml group, OPLS-DA was adopted to recognize the sample patterns. The three groups, namely, the control, 0.3 μg/ml group, and 0.5 μg/ml group, were clearly separated from each other (Figure 6A). In detail, t[1] component separated the control group from the other two groups, whereas t[2] separated the 0.3 μg/ml group from the 0.5 μg/ml group (Figure 6A). S-plot, which allows the visualization both of covariance and correlation between the metabolites and the modeled class designation (Kalesinskas et al., 2018), was then used to identify the crucial biomarkers. As compared to the control group, the level of hexadecenoic acid, octadecenoic acid, palmitic acid, tetradecanoic acid, succinic acid and N-α-acetyl-L-lysine, stearic acid, oxalic acid, pyroglutamic acid, and 1, 3-propanediol were the most significant metabolites [variable important in projection (VIP) > 1, p < 0.05] in the 0.3 μg/ml group (Figure 6B). On the other hand, stearic acid, glycine, lysine, valine, tyrosine, 2-aminoadipic acid, tryptophan, isoleucine, ornithine, thymine, adenosine, lactic acid, 1, 4-butanediol, fumaric acid, and uracil were most significant metabolites in the 0.5 μg/ml group (VIP > 1, p < 0.05) (Figure 6C). Those crucial biomarkers were displayed as scatter plot in Supplementary Figure 1. We were particularly interested in the metabolite whose contents were increased in the 0.3 μg/ml group but decreased in the 0.5 μg/ml group since the bacteria in the 0.3 μg/ml group were surviving, whereas the bacteria in 0.5 μg/ml group died massively. By analyzing these metabolites, we may identify the metabolites that guarantee the cell survival. Such metabolites included isoleucine, 2- aminoadipic acid, N-α-acetyl-L-lysine, hexadecenoic acid, octadecenoic acid, and tryptophan (Figure 6D). The presence of two fatty acids implied that fatty acid synthesis might be important for cell survival.
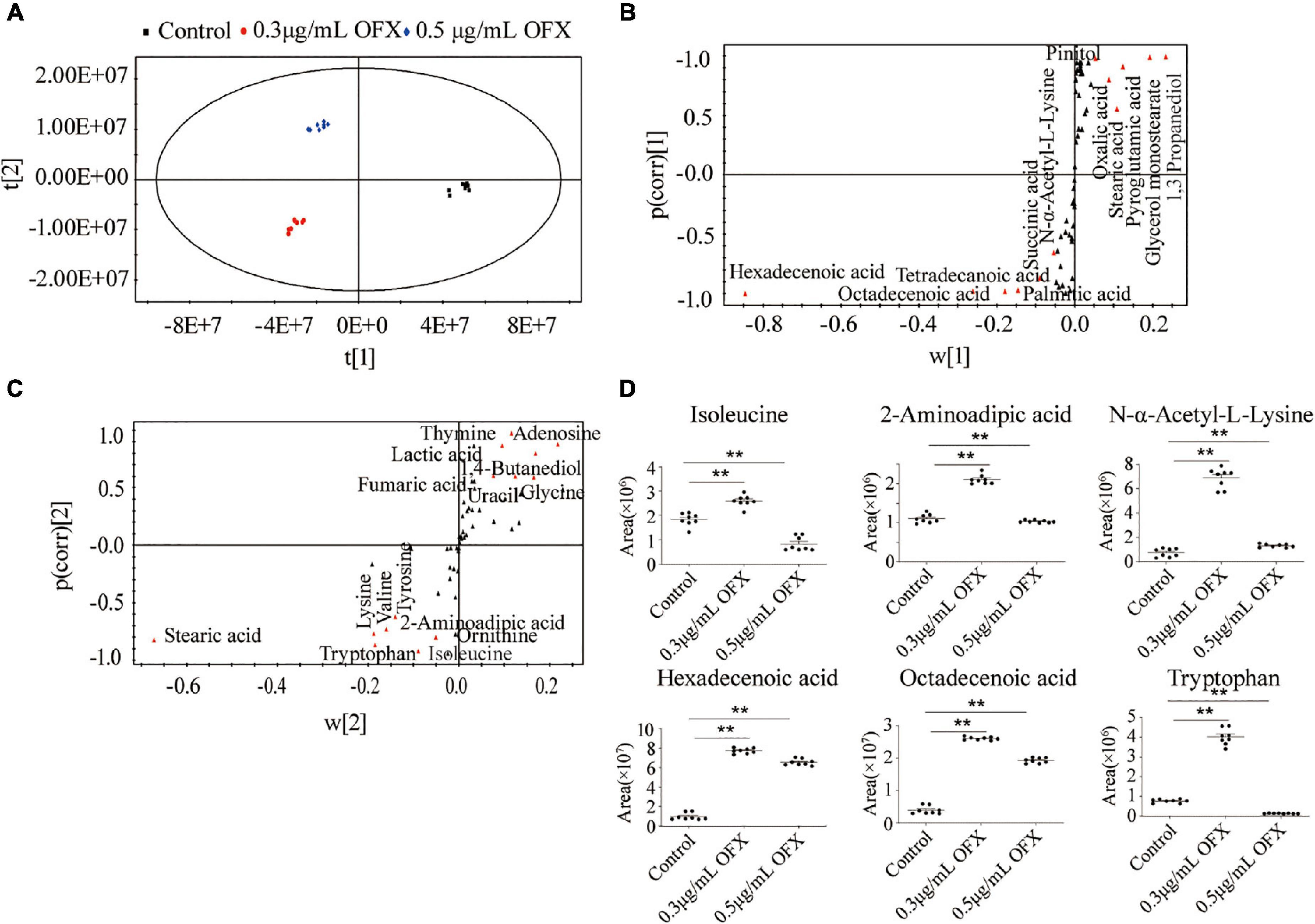
Figure 6. Identification of crucial metabolites using multivariate data analysis. (A) Principal-component analysis of control, 0.3 μg/ml OFX, and 0.5 μg/ml OFX groups according to the treatment set. Each dot represents the technological replicate analysis of samples in the plot. (B) S-plot generates from OPLS-DA. Predictive component p[1] and correlation p(corr)[1] differentiate control from 0.3 and 0.5 μg/ml OFX. (C) S-plot generates from OPLS-DA. Predictive component p[2] and correlation p(corr)[2] separate 0.3 μg/ml OFX from 0.5 μg/ml OFX. Triangles represent metabolites. Potential biomarkers are highlighted in red. (D) Abundance of crucial metabolites in control and 0.3 and 0.5 μg/ml OFX. Statistical analysis was performed with Mann–Whitney U-test. **P < 0.01.
Fatty Acids Synthesis Is Important for Bacteria Survival Under Ofloxacin Stress
To identify whether fatty acid biosynthesis contribute to bacterial survival under OFX stress, the genes of the fatty acid synthesis was quantified (Figure 7A). The expression of all of the genes, except fabF, were increased upon OFX stress. The increase of fabZ expression was shown an OFX-dose-dependent manner, whereas fabV, fabA, and tesA increased most significantly in the 0.3 μg/ml group than those in the 0.5 μg/ml group (Figure 7A). To further confirm our observation, the expression of unsaturated fatty acid pathway, AT730_18405, AT730_17775, AT730_00125, and AT730_05425, was quantified by qRT-PCR. It was shown that the expression of AT730_00125 were increased more significant in 0.3 μg/ml group than that in the 0.5 μg/ml group, and that AT730_17775 was decreased in both groups (Figure 7B). To confirm the role of fatty acid synthesis in OFX stress, bacteria were treated by 0.3 or 0.5 μg/ml OFX in the presence triclosan and 2-aminooxazole, two inhibitors of fatty acid biosynthesis. Triclosan is a specific inhibitor to 3-oxoacyl-[acyl-carrier-protein] synthase II (Tkachenko et al., 2007). While 2-aminooxazole selectively targets acetyl-CoA carboxylase (Polyak et al., 2012). Triclosan alone could delay the growth of bacteria. The growth curve for bacteria treated by 0.3 or 0.5 μg/ml OFX were similar to that in Figure 1A. However, the presence of triclosan completely abolished the growth of both of the OFX-treated groups (Figures 7C,D). In addition, 2-aminooxazole alone has no impact on the growth of bacteria, but it decreases the survival upon OFX treatment (Figures 7E,F). The two inhibitors have different levels of synergistic effects, which may be due to their different targets. These data together suggest that fatty acid biosynthesis is important to cope with the OFX stress.
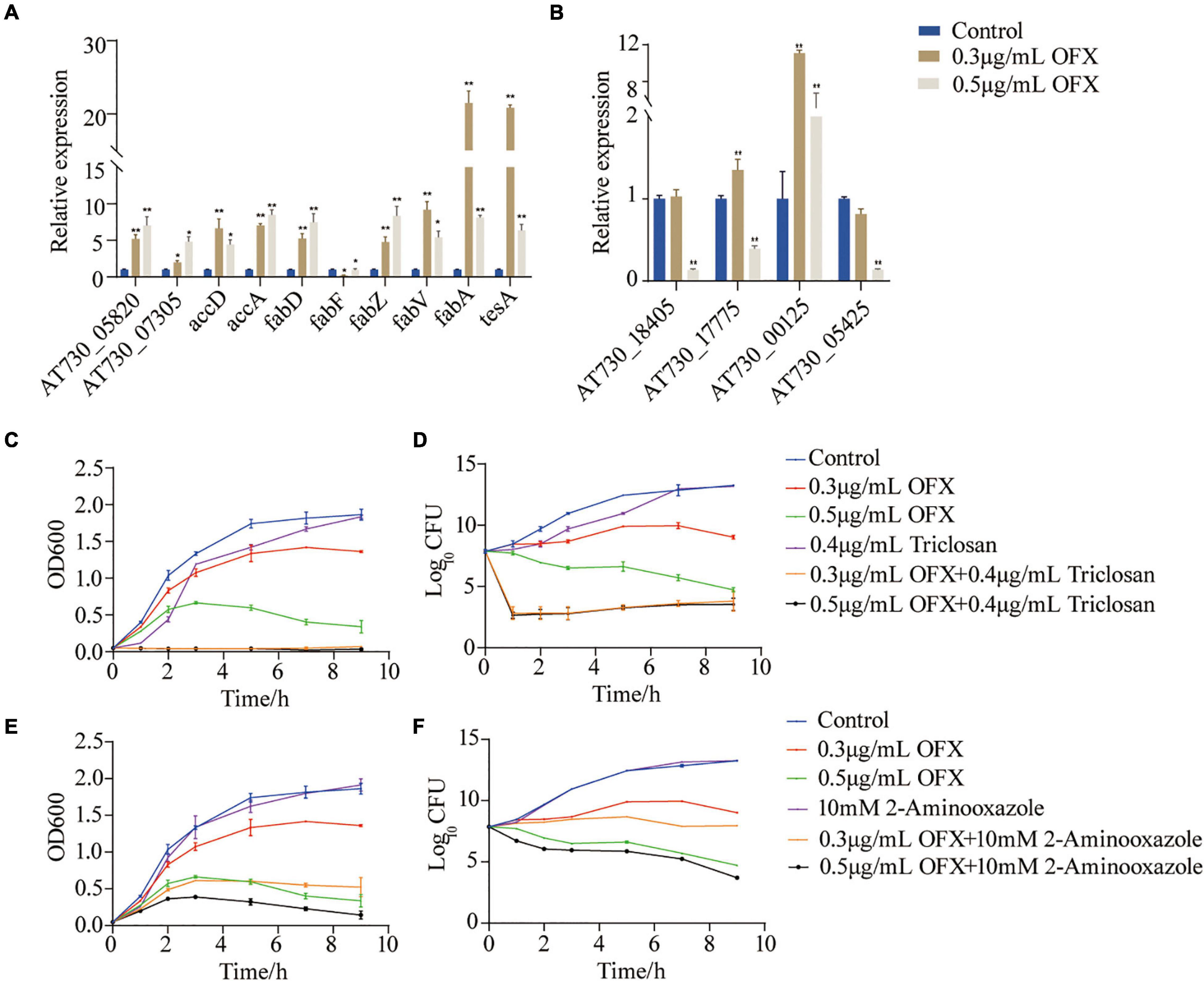
Figure 7. Fatty acid synthesis is a survival factor under OFX stress (A) Quantification of gene expression of fatty acid treated by indicated concentrations of OFX for 2 h. (B) Quantification of gene expression of unsaturated fatty acids treated by indicated concentrations of OFX for 2 h. (C) Growth curve of V. alginolyticus in medium with the indicated concentrations of OFX or 0.4 μg/ml triclosan. (D) CFU of (C). (E) Growth curve of V. alginolyticus in medium with the indicated concentrations of OFX or 10 mM 2-aminooxazole. (F) CFU of (E). The statistical analyses were performed with Mann–Whitney U-test unless otherwise indicated. *P<0.05; **P<0.01. Error bars represent means ± standard errors of the means (SEM). All of the experiments were repeated at least three times.
Discussion
Tackling antibiotic resistance is an emerging topic throughout the world and requires urgent attention in both the policies and management of existing antibiotic-resistant bacteria (Tetteh et al., 2020). The development of a novel strategy in eliminating antibiotic-resistant bacteria has been researched for decades. The screening of novel compounds by high-throughput screening (Penchovsky and Stoilova, 2013) or artificial intelligence-based screening (Lau et al., 2021) or cultures from the unculturable bacteria (Lodhi et al., 2018) aids the finding of new chemicals that kill antibiotic-resistant bacteria without detectable resistance. These studies shed light on the innovation of new drugs that probably compensate for the currently exhausted antibiotic reservoir. However, the pipeline is still too long, and it takes around 10 years for an antibiotic to be successfully approved for clinical usage (Bush et al., 2011). Meanwhile, attempts on other methods, such as probiotic, bacteriophage-based approach, anti-virulence, and immunomodulatory molecules, have been proposed and studied for decades, but none is prompting in the clinic as well as not applicable in aquaculture. Therefore, resensitizing antibiotic-resistant bacteria to current available antibiotics becomes an urgent task that can overcome the shortage in the effective management approaches on combating antibiotic resistance.
Metabolism-based strategy offers a novel perspective to deal with antibiotic-resistant bacteria. Based on the hypothesis that metabolic state determines antibiotic efficacy, we and others have shown that metabolites, including alanine, glucose, fructose, glutamate, pyruvate, thymine, and serine, can reprogram the metabolomes of the antibiotic-resistant bacteria and resensitize the antibiotic-resistant bacteria to antibiotics via increasing antibiotic uptake, counteracting reactive oxygen species, generating nitric oxide, or interfering with the antibiotic binding to the target (Peng et al., 2015; Su et al., 2015; Perez-Morales and Bustamante, 2016; Ye et al., 2018; Zhang et al., 2020; Kuang et al., 2021). These data highlight that the metabolome of the bacteria can be remodeled through exogenous supplementation of metabolites, which is promising to be used to treat infectious diseases in clinic. Moreover, these metabolism-based methods were also applicable to fight serum-resistant bacteria. It can be used to boost host immunity to eliminate serum-resistant bacteria or directly enhance immunity including phagocytosis (Cheng et al., 2019; Yang et al., 2021a,b, c).
Unlike the above-mentioned studies, this study explores the metabolic feature of V. alginolyticus upon the treatment by different concentrations of OFX. As compared to the antibiotic-resistant bacteria, the metabolic profile was much more dynamic during antibiotic stress. The expression of the pyruvate cycle genes or for fatty acid biosynthesis genes was quite different at various time-point post-treatments of OFX. One interesting observation here was that the bacteria treated by both concentrations of OFX had a higher level of pyruvate cycle and lower level of fatty acid synthesis. Despite small differences in concentrations, it is interesting to note that the bacteria mount such huge differences in the growth curve and enzymatic activity. This might be due to whether the intracellular concentration of antibiotics can exceed the threshold for the bacteria. Previous studies demonstrate that a minor difference of the intracellular concentration of antibiotics can tell apart the antibiotic-sensitive and antibiotic-resistant bacteria (Peng et al., 2015). The bacteria exposed to 0.5 μg/ml OFX might accumulate a higher level of antibiotics in the same period of time as to those exposed to 0.3 μg/ml OFX. Thus, it is highly possible that bacteria may require a decision-making to flux into the pyruvate cycle or to block the fatty acid synthesis, which needs to be further addressed (Figure 8). Moreover, another noticeable effect is that the bacteria at the MIC (0.3 μg/ml) can continue to grow, as shown in Figure 1. It is possible that the stage and doses of bacteria mattered. For the MIC experiments, 5 × 104 CFU grown at logarithmic-phase bacteria was used, and kept in the incubator without shaking. But for the antibiotic stress experiment, overnight bacterial were diluted at 1:100 (v/v) in fresh medium to a final concentration as high as 107 CFU, and more importantly, the diluted culture was kept with shaking. These differences may explain why the bacteria can still grow at MIC value.
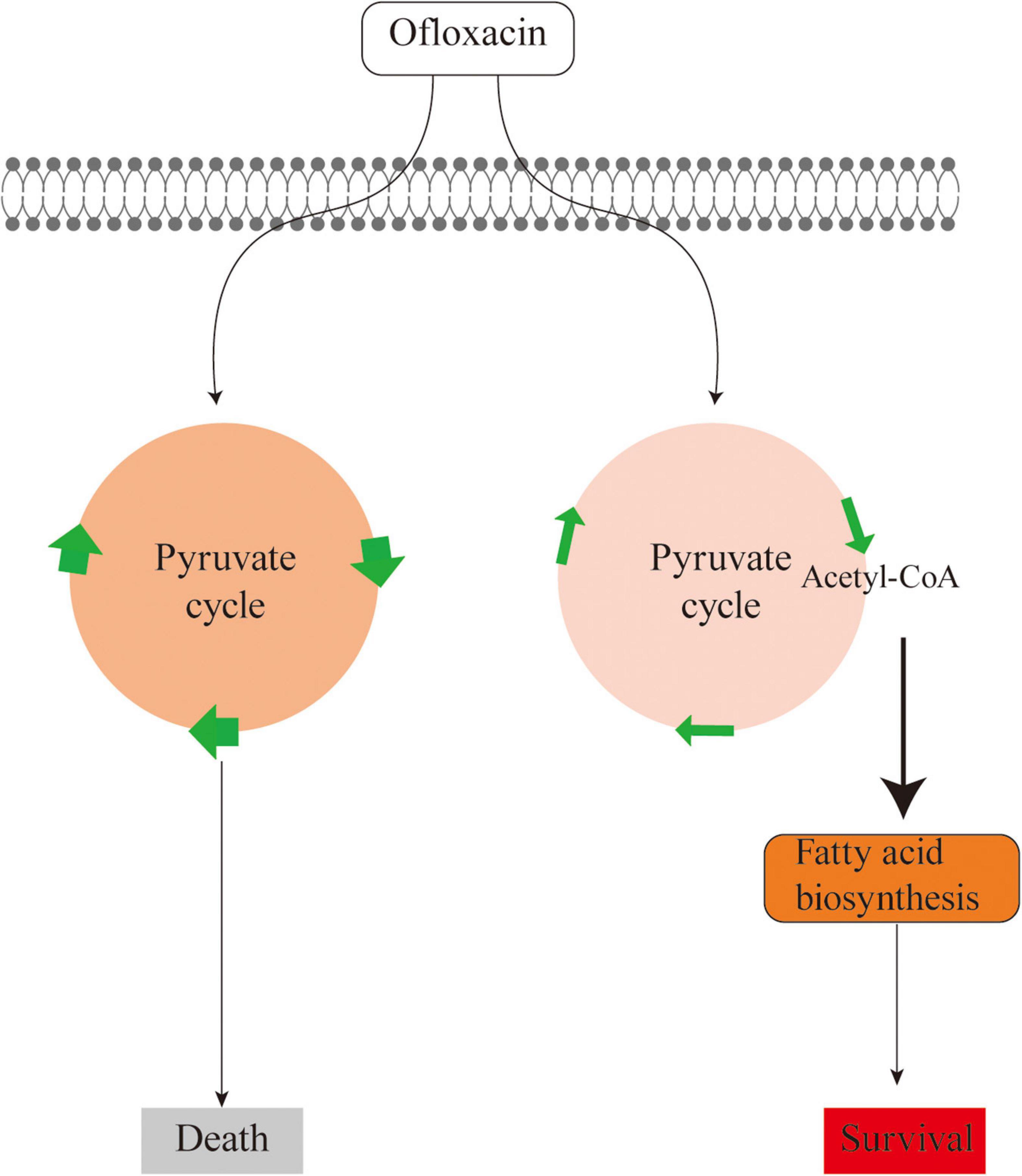
Figure 8. Sketch diagram describing a mechanism by which V. alginolyticus adapts to antibiotic stress.
Increased fatty acid biosynthesis has been studied in antibiotic-resistant bacteria. A recent report demonstrated that increased fatty acid biosynthesis can decrease the membrane fluidity and restrict the influx of antibiotics in norfloxacin-resistant bacteria (Su et al., 2021). Nevertheless, enhanced fatty acid biosynthesis was also reported in bacteria that are resistant to ceftazidime, levofloxacin, methicillin, and ciprofloxacin (Konai and Haldar, 2017; Cheng et al., 2018; Liu et al., 2019; Su et al., 2021). In addition, the inhibition of fatty acid synthesis can revoke the antibiotic sensitivity by two–fivefold (Su et al., 2021). It is interesting to observe that increased fatty acid biosynthesis promotes bacterial growth during antibiotic stress. The increase in fatty acid biosynthesis might be a strategy deployed by bacteria to maintain the membrane integrity as the membrane could be disrupted by antibiotic-induced ROS (Konai and Haldar, 2017; Ye et al., 2021). The altered transcription of fatty acid biosynthesis might be attributed to the antibiotic effect that reduces the transcription and translation of lipoproteins supporting cell membrane structure (Braun and Hantke, 2019). Therefore, the co-occurrence of increased fatty acid biosynthesis in both antibiotic stress and antibiotic-resistant bacteria imply that bacteria may coordinate fatty acid biosynthesis during the regime of antibiotic treatment. Thus, searching for effective inhibitors in inhibiting fatty acid biosynthesis may be a potential strategy in preventing the evolution of antibiotic-resistant bacteria during antibiotic treatment.
However, the limitation of this study is that we only chose two concentrations of antibiotics that may not be able to cover up other mechanisms employed by the bacteria to cope with antibiotics. It is certain to use a wider range of antibiotic concentrations, ranging from sublethal doses to lethal doses, to obtain a comprehensive view of the metabolic response upon antibiotic stress in the future study. In addition, we also cannot exclude the possibility that OFX triggered such metabolic changes. The viability of bacteria treated with 0.3 μg/ml OFX being constant may be due to the apoptotic-like bacterial death, which requires energy, and the following blockage of pyruvate cycle may deplenish the energy for cell death (Lewis, 2000). It is therefore necessary to identify the enzymes, for example, being targeted by OFX in addition to DNA gyrase.
In summary, our study demonstrates that V. alginolyticus mounts a differential metabolic response to different concentrations of OFX, which is associated with the consequences of treatment. By metabolomic profiling, pathway, and multivariate analysis, we found the metabolic flux to pyruvate cycle and fatty acid synthesis plays important roles. The inhibition of fatty acid biosynthesis was found to increase antibiotic efficacy. Thus, our study expands the understanding of metabolism-based approach to fight against antibiotic resistance.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
YY, YPY and HY conducted the experiments. YY, YPY and ZGC performed data analysis. YY, JZ, and BP interpreted the data. BP wrote the manuscript, conceptualized, and designed the project. All authors reviewed the manuscript and acknowledged the contributions.
Funding
This work was sponsored by grants from National Natural Science Foundation of China (Nos. 31872602, 32061133007, and 31822058), project supported by Innovation Group Project of Southern Marine Science and Engineering Guangdong Laboratory (Zhuhai) (No. 311020006), and The Youth Talent Support Program of Guangdong Province (No. 2017GC010617) to BP.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.818923/full#supplementary-material
Footnotes
References
Backhaus, T., Scholze, M., and Grimme, L. H. (2000). The single substance and mixture toxicity of quinolones to the bioluminescent bacterium Vibrio fischeri. Aquat Toxicol. 49, 49–61. doi: 10.1016/s0166-445x(99)00069-7
Baker-Austin, C., Oliver, J. D., Alam, M., Ali, A., Waldor, M. K., Qadri, F., et al. (2018). Vibrio spp. infections. Nat. Rev. Dis. Primers 4:8. doi: 10.1038/s41572-018-0005-8
Baker-Austin, C., Trinanes, J., Gonzalez-Escalona, N., and Martinez-Urtaza, J. (2017). Non-cholera vibrios: the microbial barometer of climate change. Trends Microbiol. 25, 76–84. doi: 10.1016/j.tim.2016.09.008
Bala, C. G., Rusu, A., Ciobanu, D., Bucsa, C., and Roman, G. (2021). Amino acid signature of oxidative stress in patients with type 2 diabetes: targeted exploratory metabolomic research. Antioxidants (Basel) 10:610. doi: 10.3390/antiox10040610
Baquero, F., and Levin, B. R. (2021). Proximate and ultimate causes of the bactericidal action of antibiotics. Nat. Rev. Microbiol. 19, 123–132. doi: 10.1038/s41579-020-00443-1
Braun, V., and Hantke, K. (2019). Lipoproteins: structure, function, biosynthesis. Subcell Biochem. 92, 39–77. doi: 10.1007/978-3-030-18768-2_3
Bush, K., Courvalin, P., Dantas, G., Davies, J., Eisenstein, B., Huovinen, P., et al. (2011). Tackling antibiotic resistance. Nat. Rev. Microbiol. 9, 894–896. doi: 10.1038/nrmicro2693
Cheng, Z. X., Gong, Q. Y., Wang, Z., Chen, Z. G., Ye, J. Z., Li, J., et al. (2017). Edwardsiella tarda tunes tricarboxylic acid cycle to evade complement-mediated killing. Front. Immunol. 8:1706. doi: 10.3389/fimmu.2017.01706
Cheng, Z. X., Guo, C., Chen, Z. G., Yang, T. C., Zhang, J. Y., Wang, J., et al. (2019). Glycine, serine and threonine metabolism confounds efficacy of complement-mediated killing. Nat. Commun. 10:3325. doi: 10.1038/s41467-019-11129-5
Cheng, Z. X., Yang, M. J., Peng, B., Peng, X. X., Lin, X. M., and Li, H. (2018). The depressed central carbon and energy metabolisms is associated to the acquisition of levofloxacin resistance in Vibrio alginolyticus. J. Proteomics 181, 83–91. doi: 10.1016/j.jprot.2018.04.002
Du, C. C., Yang, M. J., Li, M. Y., Yang, J., Peng, B., Li, H., et al. (2017). Metabolic mechanism for l-leucine-induced metabolome to eliminate streptococcus iniae. J. Proteome Res. 16, 1880–1889. doi: 10.1021/acs.jproteome.6b00944
Gong, Q. Y., Yang, M. J., Yang, L. F., Chen, Z. G., Jiang, M., and Peng, B. (2020). Metabolic modulation of redox state confounds fish survival against Vibrio alginolyticus infection. Microb Biotechnol. 13, 796–812. doi: 10.1111/1751-7915.13553
Hamed, S. M., Elkhatib, W. F., El-Mahallawy, H. A., Helmy, M. M., Ashour, M. S., and Aboshanab, K. M. A. (2018). Multiple mechanisms contributing to ciprofloxacin resistance among gram negative bacteria causing infections to cancer patients. Sci. Rep. 8:12268. doi: 10.1038/s41598-018-30756-4
Ina-Salwany, M. Y., Al-Saari, N., Mohamad, A., Mursidi, F. A., Mohd-Aris, A., Amal, M. N. A., et al. (2019). Vibriosis in fish: a review on disease development and prevention. J. Aquat. Anim Health 31, 3–22. doi: 10.1002/aah.10045
Jiang, M., Kuang, S. F., Lai, S. S., Zhang, S., Yang, J., Peng, B., et al. (2020). Na(+)-NQR confers aminoglycoside resistance via the regulation of l-alanine metabolism. mBio 11:e02186-20. doi: 10.1128/mBio.02086-20
Jones, E. H., Feldman, K. A., Palmer, A., Butler, E., Blythe, D., and Mitchell, C. S. (2013). Vibrio infections and surveillance in maryland, 2002-2008. Public Health Rep. 128, 537–545. doi: 10.1177/003335491312800613
Just, P. M. (1993). Overview of the fluoroquinolone antibiotics. Pharmacotherapy 13(2 Pt 2), 4S–17S.
Kalesinskas, L., Cudone, E., Fofanov, Y., and Putonti, C. (2018). S-plot2: rapid visual and statistical analysis of genomic sequences. Evol. Bioinform. Online 14:1176934318797354. doi: 10.1177/1176934318797354
Konai, M. M., and Haldar, J. (2017). Fatty acid comprising lysine conjugates: anti-MRSA agents that display in vivo efficacy by disrupting biofilms with no resistance development. Bioconjug Chem. 28, 1194–1204. doi: 10.1021/acs.bioconjchem.7b00055
Kuang, S. F., Chen, Y. T., Chen, J. J., Peng, X. X., Chen, Z. G., and Li, H. (2021). Synergy of alanine and gentamicin to reduce nitric oxide for elevating killing efficacy to antibiotic-resistant Vibrio alginolyticus. Virulence 12, 1737–1753. doi: 10.1080/21505594.2021.1947447
Lau, H. J., Lim, C. H., Foo, S. C., and Tan, H. S. (2021). The role of artificial intelligence in the battle against antimicrobial-resistant bacteria. Curr. Genet. 67, 421–429. doi: 10.1007/s00294-021-01156-5
Lewis, K. (2000). Programmed death in bacteria. Microbiol. Mol. Biol. Rev. 64, 503–514. doi: 10.1128/MMBR.64.3.503-514.2000
Li, W., Yao, Z., Sun, L., Hu, W., Cao, J., Lin, W., et al. (2016). Proteomics analysis reveals a potential antibiotic cocktail therapy strategy for aeromonas hydrophila infection in biofilm. J. Proteome Res. 15, 1810–1820. doi: 10.1021/acs.jproteome.5b01127
Liu, S. R., Peng, X. X., and Li, H. (2019). Metabolic mechanism of ceftazidime resistance in Vibrio alginolyticus. Infect Drug Resist 12, 417–429. doi: 10.2147/IDR.S179639
Lodhi, A. F., Zhang, Y., Adil, M., and Deng, Y. (2018). Antibiotic discovery: combining isolation chip (iChip) technology and co-culture technique. Appl. Microbiol. Biotechnol. 102, 7333–7341. doi: 10.1007/s00253-018-9193-0
Novoa, B., Nunñez, S., Fernández-Puentes, C., Figueras, A. J., and Toranzo, A. E. (1992). Epizootic study in a turbot farm: bacteriology, virology, parasitology and histology. Aquaculture 107, 253–258. doi: 10.1016/0044-8486(92)90074-U
Okuda, J., Hayakawa, E., Nishibuchi, M., and Nishino, T. (1999). Sequence analysis of the gyrA and parC homologues of a wild-type strain of Vibrio parahaemolyticus and its fluoroquinolone-resistant mutants. Antimicrob Agents Chemother 43, 1156–1162. doi: 10.1128/AAC.43.5.1156
Otsuka, M., Yasuda, M., Morita, Y., Otsuka, C., Tsuchiya, T., Omote, H., et al. (2005). Identification of essential amino acid residues of the NorM Na+/multidrug antiporter in Vibrio parahaemolyticus. J. Bacteriol. 187, 1552–1558. doi: 10.1128/JB.187.5.1552-1558.2005
Paton, J. H., and Reeves, D. S. (1988). Fluoroquinolone antibiotics. microbiology, pharmacokinetics and clinical use. Drugs 36, 193–228. doi: 10.2165/00003495-198836020-00004
Penchovsky, R., and Stoilova, C. C. (2013). Riboswitch-based antibacterial drug discovery using high-throughput screening methods. Expert. Opin. Drug Dis. 8, 65–82. doi: 10.1517/17460441.2013.740455
Peng, B., Su, Y. B., Li, H., Han, Y., Guo, C., Tian, Y. M., et al. (2015). Exogenous alanine and/or glucose plus kanamycin kills antibiotic-resistant bacteria. Cell Metab. 21, 249–262. doi: 10.1016/j.cmet.2015.01.008
Perez-Morales, D., and Bustamante, V. H. (2016). The global regulatory system Csr senses glucose through the phosphoenolpyruvate: carbohydrate phosphotransferase system. Mol. Microbiol. 99, 623–626. doi: 10.1111/mmi.13285
Polyak, S. W., Abell, A. D., Wilce, M. C., Zhang, L., and Booker, G. W. (2012). Structure, function and selective inhibition of bacterial acetyl-coa carboxylase. Appl. Microbiol. Biotechnol. 93, 983–992. doi: 10.1007/s00253-011-3796-z
Reece, R. J., and Maxwell, A. (1991). DNA gyrase: structure and function. Crit. Rev. Biochem. Mol. Biol. 26, 335–375. doi: 10.3109/10409239109114072
Stokes, J. M., Lopatkin, A. J., Lobritz, M. A., and Collins, J. J. (2019). Bacterial metabolism and antibiotic efficacy. Cell Metab. 30, 251–259. doi: 10.1016/j.cmet.2019.06.009
Su, Y. B., Kuang, S. F., Ye, J. Z., Tao, J. J., Li, H., Peng, X. X., et al. (2021). Enhanced biosynthesis of fatty acids is associated with the acquisition of ciprofloxacin resistance in edwardsiella tarda. mSystems 6:e0069421. doi: 10.1128/mSystems.00694-21
Su, Y. B., Peng, B., Han, Y., Li, H., and Peng, X. X. (2015). Fructose restores susceptibility of multidrug-resistant Edwardsiella tarda to kanamycin. J. Proteome Res. 14, 1612–1620. doi: 10.1021/pr501285f
Su, Y. B., Peng, B., Li, H., Cheng, Z. X., Zhang, T. T., Zhu, J. X., et al. (2018). Pyruvate cycle increases aminoglycoside efficacy and provides respiratory energy in bacteria. Proc. Natl. Acad. Sci. U.S.A. 115, E1578–E1587. doi: 10.1073/pnas.1714645115
Tetteh, J. N. A., Matthaus, F., and Hernandez-Vargas, E. A. (2020). A survey of within-host and between-hosts modelling for antibiotic resistance. Biosystems 196:104182. doi: 10.1016/j.biosystems.2020.104182
Tkachenko, O., Shepard, J., Aris, V. M., Joy, A., Bello, A., Londono, I., et al. (2007). A triclosan-ciprofloxacin cross-resistant mutant strain of Staphylococcus aureus displays an alteration in the expression of several cell membrane structural and functional genes. Res. Microbiol. 158, 651–658. doi: 10.1016/j.resmic.2007.09.003
Vega, N. M., Allison, K. R., Khalil, A. S., and Collins, J. J. (2012). Signaling-mediated bacterial persister formation. Nat. Chem. Biol. 8, 431–433. doi: 10.1038/nchembio.915
Xia, R., Ren, Y., and Xu, H. (2013). Identification of plasmid-mediated quinolone resistance qnr genes in multidrug-resistant gram-negative bacteria from hospital wastewaters and receiving waters in the Jinan area. China. Microb. Drug Resist. 19, 446–456. doi: 10.1089/mdr.2012.0210
Yang, D. X., Yang, H., Cao, Y. C., Jiang, M., Zheng, J., and Peng, B. (2021a). Succinate promotes phagocytosis of monocytes/macrophages in teleost fish. Front. Mol. Biosci. 8:644957. doi: 10.3389/fmolb.2021.644957
Yang, D. X., Yang, M. J., Yin, Y., Kou, T. S., Peng, L. T., Chen, Z. G., et al. (2021b). Serine metabolism tunes immune responses to promote oreochromis niloticus survival upon edwardsiella tarda infection. mSystems 6:e0042621. doi: 10.1128/mSystems.00426-21
Yang, M. J., Jiang, M., Peng, X. X., and Li, H. (2021c). Myo-inositol restores tilapia’s ability against infection by aeromonas sobria in higher water temperature. Front. Immunol. 12:682724. doi: 10.3389/fimmu.2021.682724
Ye, J., Su, Y., Peng, X., and Li, H. (2021). Reactive oxygen species-related ceftazidime resistance is caused by the pyruvate cycle perturbation and reverted by Fe(3 +) in edwardsiella tarda. Front. Microbiol. 12:654783. doi: 10.3389/fmicb.2021.654783
Ye, J. Z., Su, Y. B., Lin, X. M., Lai, S. S., Li, W. X., Ali, F., et al. (2018). Alanine enhances aminoglycosides-induced ROS production as revealed by proteomic analysis. Front. Microbiol. 9:29. doi: 10.3389/fmicb.2018.00029
Zhang, S., Wang, J., Jiang, M., Xu, D., Peng, B., Peng, X. X., et al. (2019). Reduced redox-dependent mechanism and glucose-mediated reversal in gentamicin-resistant Vibrio alginolyticus. Environ. Microbiol. 21, 4724–4739. doi: 10.1111/1462-2920.14811
Zhang, S., Yang, M. J., Peng, B., Peng, X. X., and Li, H. (2020). Reduced ROS-mediated antibiotic resistance and its reverting by glucose in Vibrio alginolyticus. Environ. Microbiol. 22, 4367–4380. doi: 10.1111/1462-2920.15085
Zhao, F., and Zhou, D.-q, Cao, H.-h, Ma, L.-p, and Jiang, Y.-h (2011). Distribution, serological and molecular characterization of Vibrio parahaemolyticus from shellfish in the eastern coast of China. Food Control 22, 1095–1100. doi: 10.1016/j.foodcont.2010.12.017
Zhao, X. L., Chen, Z. G., Yang, T. C., Jiang, M., Wang, J., Cheng, Z. X., et al. (2021). Glutamine promotes antibiotic uptake to kill multidrug-resistant uropathogenic bacteria. Sci. Transl. Med. 13:eabj0716. doi: 10.1126/scitranslmed.abj0716
Keywords: Vibrio alginolyticus, ofloxacin, metabolomics, pyruvate cycle, fatty acid synthesis
Citation: Yin Y, Yin Y, Yang H, Chen Z, Zheng J and Peng B (2022) Vibrio alginolyticus Survives From Ofloxacin Stress by Metabolic Adjustment. Front. Microbiol. 13:818923. doi: 10.3389/fmicb.2022.818923
Received: 20 November 2021; Accepted: 19 January 2022;
Published: 16 March 2022.
Edited by:
Jose L. Martinez, Spanish National Research Council (CSIC), SpainReviewed by:
Volker Behrends, University of Roehampton London, United KingdomFernando Baquero, Ramón y Cajal Institute for Health Research, Spain
Copyright © 2022 Yin, Yin, Yang, Chen, Zheng and Peng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bo Peng, cGVuZ2IyNkBtYWlsLnN5c3UuZWR1LmNu
 Yue Yin
Yue Yin Yuanpan Yin1,2
Yuanpan Yin1,2 Hao Yang
Hao Yang Zhuanggui Chen
Zhuanggui Chen Jun Zheng
Jun Zheng Bo Peng
Bo Peng