- 1UCD School of Veterinary Medicine, University College Dublin, Dublin, Ireland
- 2Hokkaido University International Institute for Zoonosis Control, Sapporo, Japan
- 3UCD School of Agriculture and Food Science, University College Dublin, Dublin, Ireland
- 4Division of Bioresources, Research Center for Zoonosis Control, Hokkaido University, Sapporo, Japan
- 5UCD Conway Institute, University College Dublin, Dublin, Ireland
The Mycobacterium tuberculosis complex (MTBC) contains the causative agents of tuberculosis (TB) in mammals. The archetypal members of the MTBC, Mycobacterium tuberculosis and Mycobacterium bovis, cause human tuberculosis and bovine tuberculosis, respectively. Although M. tuberculosis and M. bovis share over 99.9% genome identity, they show distinct host adaptation for humans and animals; hence, while the molecular basis of host adaptation is encoded in their genomes, the mechanistic basis of host tropism is still unclear. Exploration of the in vitro phenotypic consequences of known genetic difference between M. bovis and M. tuberculosis offers one route to explore genotype–phenotype links that may play a role in host adaptation. The TbD1 (“Mycobacterium tuberculosis deletion 1 region”) locus encompasses the mmpS6 and mmpL6 genes. TbD1 is absent in M. tuberculosis “modern” lineages (Lineages 2, 3, and 4) but present in “ancestral” M. tuberculosis (Lineages 1 and 7), Mycobacterium africanum lineages (Lineages 5 and 6), newly identified M. tuberculosis lineages (Lineages 8 and 9), and animal adapted strains, such as M. bovis. The function of TbD1 has previously been investigated in M. tuberculosis, where conflicting data has emerged on the role of TbD1 in sensitivity to oxidative stress, while the underlying mechanistic basis of such a phenotype is unclear. In this study, we aimed to shed further light on the role of the TbD1 locus by exploring its function in M. bovis. Toward this, we constructed an M. bovis TbD1 knockout (ΔTbD1) strain and conducted comparative transcriptomics to define global gene expression profiles of M. bovis wild-type (WT) and the ΔTbD1 strains under in vitro culture conditions (rolling and standing cultures). This analysis revealed differential induction of a hypoxia-driven copper response in WT and ΔTbD1 strains. In vitro phenotypic assays demonstrated that the deletion of TbD1 sensitized M. bovis to H2O2 and hypoxia-specific copper toxicity. Our study provides new information on the function of the TbD1 locus in M. bovis and its role in stress responses in the MTBC.
Introduction
Tuberculosis (TB) caused by Mycobacterium tuberculosis is one of the leading causes of death worldwide due to a single infectious agent. TB killed an estimated 1.5 million people in 2020 alone, with an increase in TB deaths for the first time in over a decade due to the disruption to TB control programs caused by the COVID-19 pandemic [World Health Organization [WHO], 2021]. Bovine tuberculosis (bTB) is mainly caused by Mycobacterium bovis, however, M. bovis can also infect and sustain in many other domesticated and wild mammalian hosts, such as goats, badgers, deer, and feral water buffalo (Gormley and Corner, 2018). M. bovis is also a zoonotic risk in terms of transmission of infection from animals to humans, albeit M. bovis has a limited transmission capacity between immune competent humans compared to M. tuberculosis (Francis, 1950; Magnus, 1966). On the other hand, modern strains of M. tuberculosis appear attenuated in cattle compared to M. bovis and seem unable to sustain in cattle populations, although there are reported cases of potential reverse zoonotic transmission of M. tuberculosis to animals (Whelan et al., 2010; Ameni et al., 2011). The archetypal MTBC strains M. tuberculosis H37Rv and M. bovis AF2122/97 show more than 99.9% genome sequence identity, in spite of the distinct host preference displayed by the bacilli (Garnier et al., 2003). Elucidating the molecular basis of MTBC host preference will help to reveal not only the evolutionary steps that led to the emergence of these pathogens but may also reveal new ways to control TB in animals and humans.
The M. tuberculosis specific deletion one region (TbD1) is a 2153 bp DNA region that was originally identified through comparative genomics between the M. tuberculosis H37Rv genome with other MTBC members (Brosch et al., 2002). TbD1 was used as a marker of so-called “ancient” and “modern” lineages, whereby MTBC strains having TbD1 intact were deemed as having the ancestral configuration of this locus. The TbD1 locus encodes two proteins: the putative membrane protein MmpS6 and a transmembrane transport protein MmpL6. MmpL are “mycobacterial membrane protein large” proteins that belong to the resistance-nodulation-cell division (RND) efflux pump families and perform functions as diverse as transport of lipids, uptake of nutrients, iron acquisition, or extrusion of toxic compounds (Wells et al., 2013; Delmar et al., 2015; Melly and Purdy, 2019). The genome of M. tuberculosis H37Rv encodes 13 MmpL proteins (Cole et al., 1998) which have been classified into hydrophobe/amphiphile efflux (HAE) subfamilies of RND based on phylogenetic comparisons; hence, MmpL1, 2, 4, 5, 6, 8, 9, 10, and 12 belong to the HAE2 family while MmpL3, 7, and 11 belong to HAE3 family (Sandhu and Akhter, 2015). In Escherichia coli the HAE-1 RND transporter AcrB is only active in conjunction with membrane fusion proteins (MFPs) and outer membrane factors (OMFs) that form an elongated tripartite complex, AcrAB(ZZ)-TolC (Kobylka et al., 2020). In mycobacteria, the MmpS proteins are speculated to function in a similar way to MFPs, but their precise role remains to be defined (Deshayes et al., 2010; Wells et al., 2013).
Five RND transporters with known structures are AcrB, CusA, MexB, ZneA, and MtrD. These proteins have 12-helix transmembrane domains with a periplasmic N-terminal porter (PN) subdomain located between TM1 and TM2, and a periplasmic C-terminal porter (PC) subdomain inserted between TM7 and TM8 (Murakami et al., 2002; Sennhauser et al., 2009; Long et al., 2010; Su et al., 2012; Nakashima et al., 2013; Pak et al., 2013). In M. tuberculosis H37Rv, MmpL1, 2, 3, 4, 5, 7, 8, 9, 10, 11, and 12 have 11–12 TMs and one or two large periplasmic loops that make up the porter domain. In all human-adapted modern M. tuberculosis lineages, including major epidemic strains such as those of Lineage 2 (Beijing) and Lineage 4 (Euro-American Haarlem) (Hershberg et al., 2008; Wirth et al., 2008), TbD1 is deleted with the mmpL6 remnant encoding a protein with only five TMs that lacks the periplasmic porter domain. This suggests that MmpL6 is not functional in TbD1-deficient mycobacterial strains. However, M. tuberculosis strains from lineages with an intact TbD1 locus, as well as animal-adapted lineages including M. bovis, encode MmpL6 harboring 12 TMs, and two porter subdomains: PN and PC (Sandhu and Akhter, 2015).
It has been suggested that the loss of TbD1 gave a selective advantage to modern M. tuberculosis strains (Reiling et al., 2013; Bottai et al., 2020). This is supported by the fact that modern strains trigger an attenuated inflammatory host response and have an enhanced ability to grow in human macrophages, as well as an association with increased bacillary loads in the lungs of infected mice compared to ancestral strains that contain an intact TbD1 locus (Portevin et al., 2011; Reiling et al., 2013; Bottai et al., 2020). Also, distinct geographic distributions for ancestral and modern strains have been found by several studies, e.g., ancestral strains in Lineage 1 are found mainly in countries bordering the Indian Ocean, while modern strains have a wider global distribution (Merker et al., 2015).
There have been two main studies that have explored the function of the TbD1 locus. Arumugam et al. (2019) showed that oxidative stress can trigger the expression of mmpS6 and mmpL6 genes, and the presence of the intact TbD1 locus afforded a higher tolerance to oxidative stress for M. tuberculosis. On the other hand, Bottai et al. (2020) presented evidence that loss of TbD1 conferred protection against oxidative stress.
We sought to explore the function of TbD1 in M. bovis as a route to determine its function in the archetypal animal-adapted tubercule bacillus. We constructed M. bovis TbD1 knockout and complemented mutants and explored transcriptomic alterations between the TbD1 mutant and wild-type (WT) under different in vitro culture conditions. These analyses revealed differential expression of genes involved in hypoxia-driven copper stress response between the M. bovis TbD1 mutant and WT. Subsequent in vitro experiments revealed a potential link between the TbD1 locus in M. bovis with a hypoxia-specific response to copper and oxidative stress. This work therefore reveals new insight into the functional significance of genetic variations between members of the MTBC, linking variation in the TbD1 locus to pathogen responses to in vivo relevant stresses.
Materials and Methods
Bacterial Strains and Culture Conditions
Escherichia coli strains that were used for plasmid propagation in MultiSite Gateway cloning (Life Technologies/Invitrogen/Thermo Fisher Scientific, Loughborough, United Kingdom) procedures were grown in LB medium or LB agar plates supplemented with selected antibiotics. Ampicillin (50 μg/ml), zeocin (25 μg/ml), and hygromycin (50 μg/ml) were added as required. BCG Denmark was grown in liquid Middlebrook 7H9 medium (Becton Dickinson, New Jersey, NJ, United States) supplemented with 0.05% Tween 80, 0.2% glycerol, 0.5% bovine serum albumin, 0.2% glucose, and 0.085% NaCl or 7H11 agar plates (Becton Dickinson, New Jersey, United States) supplemented with 0.2% glycerol, 0.5% BSA, 0.2% glucose, and 0.085% NaCl. M. bovis AF2122/97 was grown in the 7H9 and 7H11 media as described above, with 40 mM sodium pyruvate (Sigma-Aldrich, Ireland). When required, kanamycin, hygromycin, or zeocin were added to growth media to a final concentration of 50, 50, or 25 μg/ml, respectively. Standing cultures for RNA extraction were grown in 30 ml of 7H9 in 50 ml tubes (Sarstedt) with the caps tightly screwed, without shaking at 37°C. Rolling cultures for RNA extraction were grown in 30 ml of 7H9 in 850 cm2 roller bottles (Cellmaster), rolling at 2–3 rpm at 37°C. Sauton’s medium was prepared using 4 g L-asparagine, 2 g citric acid, 0.5 g KH2PO4, 0.5 g MgSO4, 0.05 g ferric ammonium citrate, 0.1 ml of 0.01% ZnSO4, 60 ml glycerol, 2.5 ml of 20% Tween 80 in 900 ml deionized water and adjust pH to 7.0 with 1 M NaOH, adding 40 mM sodium pyruvate for M. bovis AF2122/97. The strains used in this study are listed in Supplementary Table 1.
Mutant Generation and Complementation
Mycobacterium bovis AF2122/97 and BCG Demark ΔTbD1 mutants were constructed using the Che9c recombineering system (van Kessel and Hatfull, 2007). Briefly, pNitET-SacB-kan plasmids were transformed into M. bovis strains by electroporation to create recombinogenic M. bovis strains. During the allelic exchange process, the hygromycin cassette from the allelic exchange substrate (AES) that was constructed via MultiSite Gateway (Life Technologies/Invitrogen/Thermo Fisher Scientific, Loughborough, United Kingdom) replaced TbD1. Knockouts were selected on 7H11 plates containing hygromycin. PCR was used for initial mutant screening for verification of the TbD1 deletion and presence of the hygromycin cassette. The left and right junction arms of the hygromycin cassette in ΔTbD1 were amplified by PCR and Sanger sequenced to rule out rearrangements in neighbor genes. The spontaneous loss of pNitET-SacB-kan plasmid was also verified by PCR. The complemented M. bovis strains were constructed using integrative plasmid pDE43-MCZ-TbD1 that were also produced using the Gateway system; these plasmids integrate into conserved attB site on the mycobacterial genome (Saviola and Bishai, 2004), allowing stable expression of the TbD1 locus under the native promoter. The colonies were checked by PCR to verify the rescue of the TbD1. The plasmids and primers used in this study are listed in Supplementary Table 1.
RNA Extraction, RNA-seq, and RT-qPCR
For RNA-seq, M. bovis AF2122/97 WT and ΔTbD1 strains were grown in triplicate in 7H9 medium in rolling or standing conditions to OD600 = 0.4–0.6. RNA samples were extracted as previously described (Malone et al., 2018). RNA concentration was determined using the NanoDrop One spectrometer (Thermo Scientific, Massachusetts, MA, United States) and RNA integrity number (RIN) was determined by RNA 6000 Nano kit (Agilent Technologies, Cork, Ireland) on the Bioanalyser 2100 (Agilent Technologies, Cork, Ireland). All RNA samples were shown to have RIN values over 8.5 and were sent for RNA-seq. Following rRNA removal using the Ribo-Zero rRNA kit (Illumina, California, CA, United States), strand-specific RNA libraries were prepared with an insert size of 250–300 bp using the NEBNext Ultra II Directional RNA Library Prep Kit (New England Biolabs, United Kingdom) and sequencing was performed by the commercial supplier (Novogene, Cambridge, United Kingdom) on an Illumina NovaSeq 6000 machine. Single-end, strand-specific 150 bp reads data were generated for M. bovis AF2122/97 WT and ΔTbD1 strains grown in standing condition and paired-end, strand-specific 150 bp reads data were generated for M. bovis AF2122/97 WT and ΔTbD1 strains grown in rolling condition. For RT-qPCR, RNA samples were extracted, and 1 μg of RNA was reverse transcribed to single-stranded cDNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems/Thermo Fisher Scientific, Warrington, United Kingdom) according to the manufacturer’s guidelines, minus RT controls and no-template controls were included. The cDNA was diluted to 2 ng/μl RNA equivalents. For qPCR (20.0 μl final volume), the master mix was prepared using 1.2 μl of forward/reverse primer (300 nM final), 10 μl SYBR Green Mix (Applied Biosystems/Thermo Fisher Scientific, Warrington, United Kingdom), 2.6 μl nuclease-free H2O, and 5 μl cDNA template. Duplicate qPCR was performed for each sample on the 7500 Fast Real-Time PCR System (Applied Biosystems/Thermo Fisher Scientific, Warrington, United Kingdom) with an initial denaturation at 95°C for 10 min, with 40 cycles of denaturation (95°C for 15 s), and a combined annealing/extension (60°C for 1 min), followed by a final melt curve, to confirm primer specificity. The qPCR data was analyzed using the qBASE + qbasePLUS software package.
Differential Gene Expression Analysis of Mycobacterium bovis AF2122/97
Raw reads were processed firstly by FastQC tool (Babraham Bioinformatics, 2019a – FastQC A Quality Control tool for High Throughput Sequence Data) to check the quality and then Trim Galore (Babraham Bioinformatics, 2019b – Trim Galore!) was used to trim off reads with low-quality base calls and adapters. Bwa-mem (Li, 2013) were used to do the alignment between the raw reads with M. bovis AF2122/97 reference genome. FeatureCounts (Liao et al., 2014) was used for reads counting for each gene. The R package DESeq2 (Love et al., 2014) was used for differentially expressed gene (DEG) analysis with fold-change and adjusted p-value threshold. The analyses of protein–protein interaction and functional enrichment were performed by the STRING and were created by Cytoscape (Shannon et al., 2003; Szklarczyk et al., 2019).
Drop Assays and Copper Challenge
For drop assays, BCG Denmark WT and TbD1 mutants were scaled up to OD600 = 0.8–1.0 as described above. Cultures were then pelleted by centrifugation and washed twice with Sauton’s media and resuspended in Sauton’s media to OD600 = 0.1. Ten-fold serial dilutions were conducted with each culture on a 96-well plate. Six-microliters drops were spotted onto 7H11 agar plates with increasing concentrations (25, 100, and 150 μM) of CuSO4 in replicates and incubated at 37°C for 14–16 days. For copper challenge in liquid media, cultures were collected, washed, and resuspended as described above and 150 μM CuSO4 was added. The OD600 were then read constantly over 14 days. To check the cell viability after copper stress was imposed on M. bovis AF2122/97, the strains were grown in 7H9 with 40 mM sodium pyruvate to OD600 = ∼0.8 and cultures were then pelleted by centrifugation, washed twice with Sauton’s media and resuspended in Sauton’s media to OD600 = 0.1. Cultures then were maintained in the absence (control) or presence of 200 μM CuSO4 in standing conditions for 10 days. Cultures were plated out at day 10 on 7H11 plates. CFU were determined after 2–3 weeks incubation. The viability was expressed as a percentage of survival, calculated as the ratio between the CFU recovered from cultures exposed to 200 μM CuSO4 over those obtained from unexposed cultures.
H2O2 Challenge and CFU Determination
Mycobacterium bovis AF2122/97 WT and TbD1 mutants were cultured in 7H9 medium to mid-log phase and diluted to OD600 = 0.1. Ten millimolars and 40 mM of H2O2 were added and incubated at 37°C for 1 h and cultures were plated on 7H11 plates. After 2–4 weeks incubation at 37°C, CFU were enumerated. Susceptibility was expressed as a percentage of survival, calculated as the ratio between the CFU recovered from cultures exposed to stress over those obtained in unexposed cultures, multiplied by 100.
Results
Generation and Verification of Mycobacterium bovis TbD1 Mutants
TbD1 mutants were constructed in two M. bovis genetic backgrounds: M. bovis AF2122/97 and M. bovis BCG Denmark. ΔTbD1 mutants were constructed using the Che9c recombineering system, with TbD1 replaced by a hygromycin resistance cassette via allelic exchange (Figure 1A; van Kessel and Hatfull, 2007). PCR amplification of the TbD1 internal region verified the deletion of TbD1 and the replacement with the hygromycin cassette (Figure 1B). PCR amplification of the flanking regions and Sanger sequencing of the PCR fragments verified that there were no off-target mutations in the flanking regions. To perform trans-complementation of the TbD1 locus back into ΔTbD1 strains we constructed an integrative plasmid (pDE43-MCZ-TbD1) with expression of the TbD1 locus driven by native mmpS6 promoter. The complemented mutant strains, termed ΔTbD1:TbD1, were verified by PCR to confirm the genomic integration of the TbD1 locus (Figure 1B).
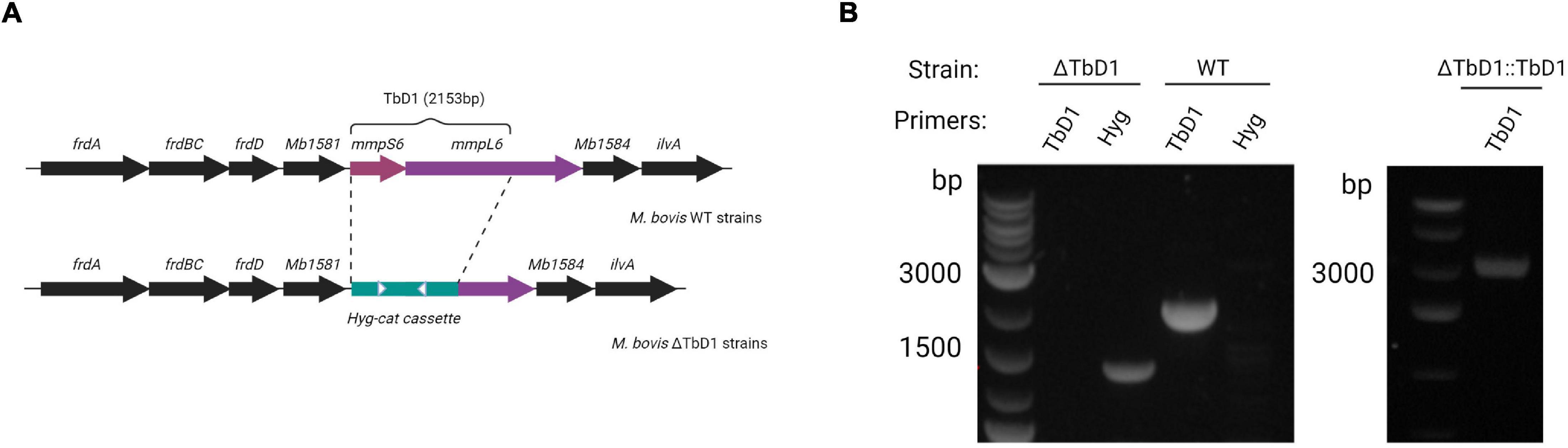
Figure 1. Construction of M. bovis ΔTbD1 mutant and complemented ΔTbD1:TbD1 strain. (A) Diagram showing the TbD1 and flanking genes. The TbD1 region was replaced by a 3.5 kB hyg-cat cassette (green bar) to generate M. bovis ΔTbD1 strains. White arrowheads indicate primer targets (Hyg F&R) to verify the Hyg cassette. (B) Agarose gel electrophoresis image after PCR with primers targeting TbD1 specific internal region and primers targeting hyg-cat cassette to verify the knockout of TbD1 and the presence of hyg-cat cassette on BCG Denmark ΔTbD1 strains and PCR targeting TbD1 locus to verify the complementation of ΔTbD1 on ΔTbD1:TbD1 strains.
Comparative Transcriptome Analysis of Mycobacterium bovis AF2122/97 Under Standing and Rolling Culture Conditions
Standing culture conditions are known to induce a low-oxygen environment and hypoxic response in M. bovis BCG (Florczyk et al., 2001). Thus, we analyzed RNA-seq data from M. bovis AF2122/97 WT strains under rolling and standing conditions as a simple system in which to study what effects these different culture conditions would have on M. bovis global gene transcription, and how this might impact transcription at the TbD1 locus.
By analysis of global transcription patterns from M. bovis AF2122/97 under standing and rolling culture conditions we obtained 179 DEGs after filtering with fold-change and p-value thresholds (|log2FC|> 2; Padj.<0.05). DEGs were then converted to M. tuberculosis H37Rv orthologs for comparison to M. tuberculosis annotations and the previous literature. Many of the DEGs that related to hypoxia/low oxygen responses (acr/hspX/Rv2031c and acg/Rv2032) were highly expressed in M. bovis AF2122/97 WT under standing as compared to rolling growth conditions. Among the DEGs, genes for some well-known transcription factors that mediate responses of M. tuberculosis to hypoxia, e.g., dosR, Rv0081, Rv0324, and kmtR were also found to be upregulated in M. bovis standing cultures (Galagan et al., 2013). Gene ontology (GO) functional analysis of the DEGs enriched GO terms included Steroid biosynthetic process (GO:0006694), Cholesterol catabolic process (GO:0006707), Cholesterol metabolic process (GO:0008203), Alcohol catabolic process (GO:0046164), Sterol metabolic process (GO:0016125), Steroid metabolic process (GO:0008202), Lipid catabolic process (GO:0016042), Organic hydroxy compound (GO:1901615), and Alcohol metabolic process (GO:0006066) (Figure 2B and Supplementary Tables 3, 6). The upregulation of genes under these GO terms showed an induction of lipid metabolism in M. bovis cultured under standing conditions.
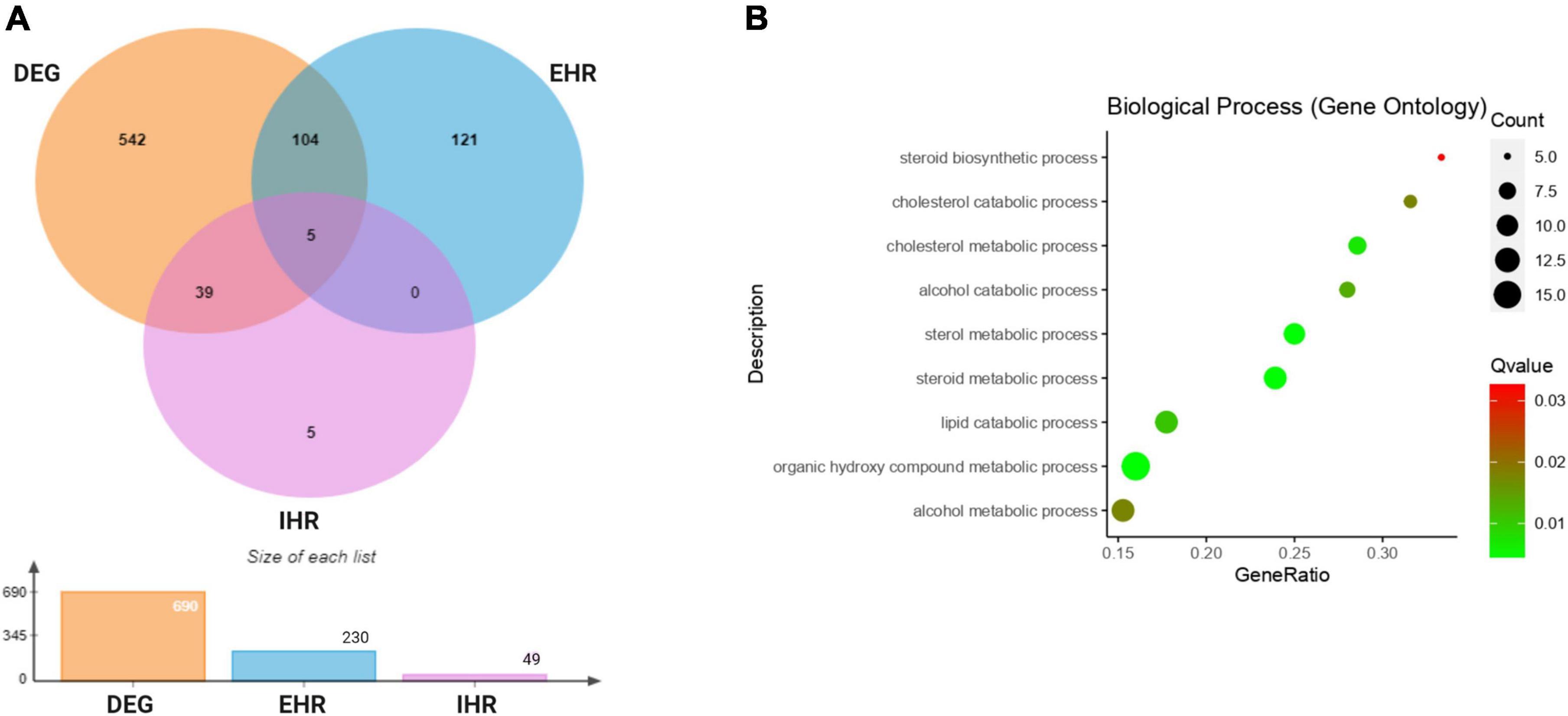
Figure 2. Comparative transcriptome analysis of M. bovis AF2122/97 WT under standing and rolling culture conditions. (A) Venn diagram showing the overlap of the genes shared among differentially expressed genes (DEG: orange)in M. bovis AF2122/97 standing cultures as compared to rolling cultures, enduring hypoxic response (EHR: blue) and initial hypoxic response (IHR: purple) gene lists. (B) Gene ontology (GO) enrichment analysis for differentially expressed genes in standing M. bovis AF2122/97. The gene ratio refers to the ratio of the number of genesin the GO entry following enrichment to the total number of genes in the GO entry. An increased gene ratio indicates greater enrichment. Lower Q value indicates higher significance.
Rustad et al. (2008) identified 230 enduring hypoxic response (EHR) genes that were stably induced by hypoxic stress and largely independent of the DosR regulon. To allow direct comparison with their data, we thus used the same log2FC threshold of |log2FC|> 1 and Padj.< 0.05 as they employed. These filters identified 109 genes from our DEG that overlapped with EHR genes (230 genes in total) and 44 overlapping genes with DosR regulated initial hypoxic response (IHR) genes (49 genes in total), showing a significant overlap between our standing conditions and hypoxic conditions (Figure 2A and Supplementary Table 2). This again indicated that the M. bovis AF2122/97 standing cultures had undergone hypoxic responses. Notably, the expression of mmpL6 was approximately twofold higher in standing as compared to rolling conditions, indicating that expression of the TbD1 locus is induced in standing culture conditions.
The Effects of TbD1 on Global Gene Expression of Mycobacterium bovis AF2122/97 Under Standing and Rolling Growth Conditions
The standing and rolling culture conditions provided us with an in vitro system to explore transcriptome differences between M. bovis AF2122/97 WT and ΔTbD1 strains. A principal-component analysis (PCA) plot (Figure 3A) of gene expression between M. bovis AF2122/97 WT and ΔTbD1 revealed separation of WT and ΔTbD1 standing culture groups, showing that the gene expression profiles of WT vs. ΔTbD1 M. bovis AF2122/97 were only significantly different when the bacilli were grown under standing conditions.
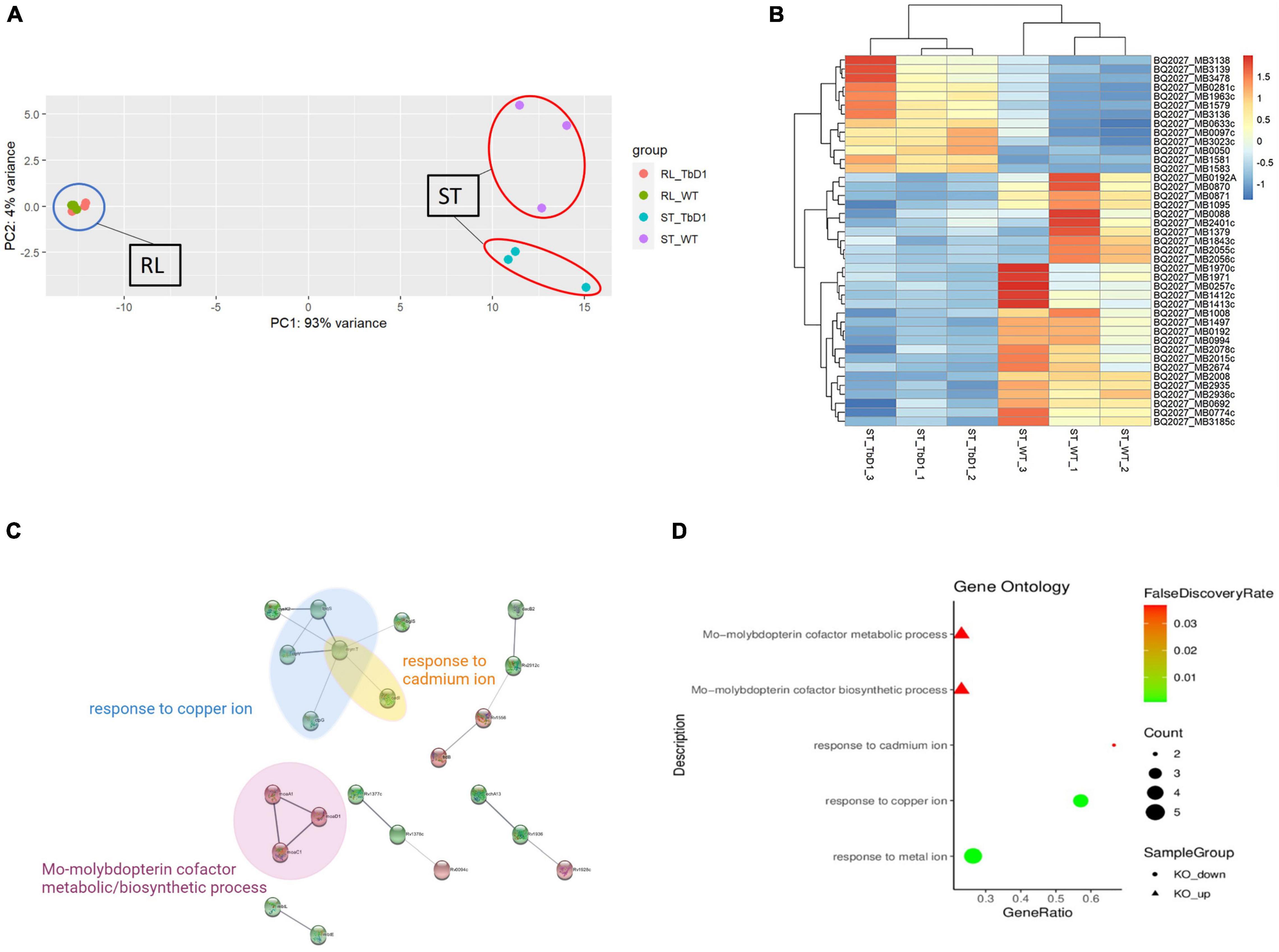
Figure 3. Comparative transcriptome analysis of M. bovis AF2122/97 WT and ΔTbD1 under standing growth condition and rolling growth condition. (A) Principal-component analysis (PCA) of the WT and ΔTbD1 M. bovis AF2122/97 transcriptomes from two different culture conditions: blue circle indicates the rolling condition and red circles indicate the standing condition. Each point represents a strain sample as indicated in the legend on the right. (B) Hierarchical clustering-heatmap of the 41 differentially expressed genes in M. bovis AF2122/97 ΔTbD1. Upregulated expression is presented in red and downregulated expression is presented in blue. Each row represents a gene, and each column represents a strain sample. (C) Protein–protein interaction network of the differentially expressed genes. Red nodes represent upregulated genes, while green nodes represent downregulated genes. The edges indicate the association between genes and edge width indicates the score of confidence of an interaction on the available evidence from STRING (the wider the edge, the higher scores of the interaction). (D) Gene ontology (GO) enrichment analysis for differentially expressed genes in M. bovis AF2122/97 ΔTbD1 grown in standing conditions. The GO enrichment terms from enrichment analysis of the upregulated (triangle) and downregulated (circle) genes are presented. The gene ratio refers to the ratio of the number of genes in the GO entry following enrichment to the total number of genes in the GO entry. An increased gene ratio indicates greater enrichment. Lower false discovery rate indicates higher significance.
Given that mmpL6 gene was upregulated in standing condition as stated above, indicated a “switch-on” of TbD1 locus and associated pathways by these conditions, we thus focused on standing cultures specifically to study the effects of the TbD1 locus on global gene expression in M. bovis AF2122/97. There were 41 DEGs meeting the thresholds applied (|log2FC|> 1 and Padj.< 0.05) in M. bovis AF2122/97 ΔTbD1 cultured with standing conditions (Figure 3B). The DEGs were converted to M. tuberculosis H37Rv orthologs to facilitate input into the STRING database (Szklarczyk et al., 2019) and hence allow analysis of the protein–protein interactions and GO enrichment in the DEG (Figure 3C and Supplementary Table 4). The enriched GO terms indicated that knockout of TbD1 led to differential expression of genes involved in Response to copper ion (GO:0046688), Response to cadmium ion (GO:0046686), Response to metal ion (GO:0010038), Mo-molybdopterin cofactor biosynthetic process (GO:0006777), and Mo-molybdopterin cofactor metabolic process (GO:0019720) (Figures 3C,D). Four (mymT, lpqS, ctpV, and ctpG) of the seven genes from the copper ion response GO term (GO:0046688) were significantly downregulated in the M. bovis AF2122/97 ΔTbD1 under standing growth conditions. Apart from the enriched genes mentioned above, other copper-responsive genes regulated by CsoR and RicR were significantly downregulated in the ΔTbD1 mutant under standing conditions (Supplementary Table 5).
TbD1 Locus Is Involved in Mycobacterium bovis AF2122/97 Hypoxia-Specific Copper Response
Based on the transcriptomic data that revealed differential regulation of genes implicated in copper stress responses in the ΔTbD1 mutant, we then explored whether the TbD1 locus was implicated in the maintenance of copper homeostasis in M. bovis AF2122/97 when grown under our standing culture conditions. We thus grew M. bovis AF2122/97 WT, ΔTbD1, and ΔTbD1:TbD1 strains to OD600 = ∼0.8; cultures were washed and resuspended in Sauton’s media to OD600 = 0.1, then 200 μM of CuSO4 added to the “treated” group and incubated for another 10 days maintaining standing culture conditions. CFU was determined and percentage survival was calculated based on CFU relative to the non-CuSO4 treated control group. The results showed that TbD1 intact WT and complemented M. bovis AF2122/97 ΔTbD1 (WT and ΔTbD1:TbD1) strains had significantly higher viability at 200 μM CuSO4 after 10-day incubation (Figure 4) than M. bovis AF2122/97 ΔTbD1. These results identified that the TbD1 locus of M. bovis AF2122/97 is important in protection against excess copper in a hypoxic environment.
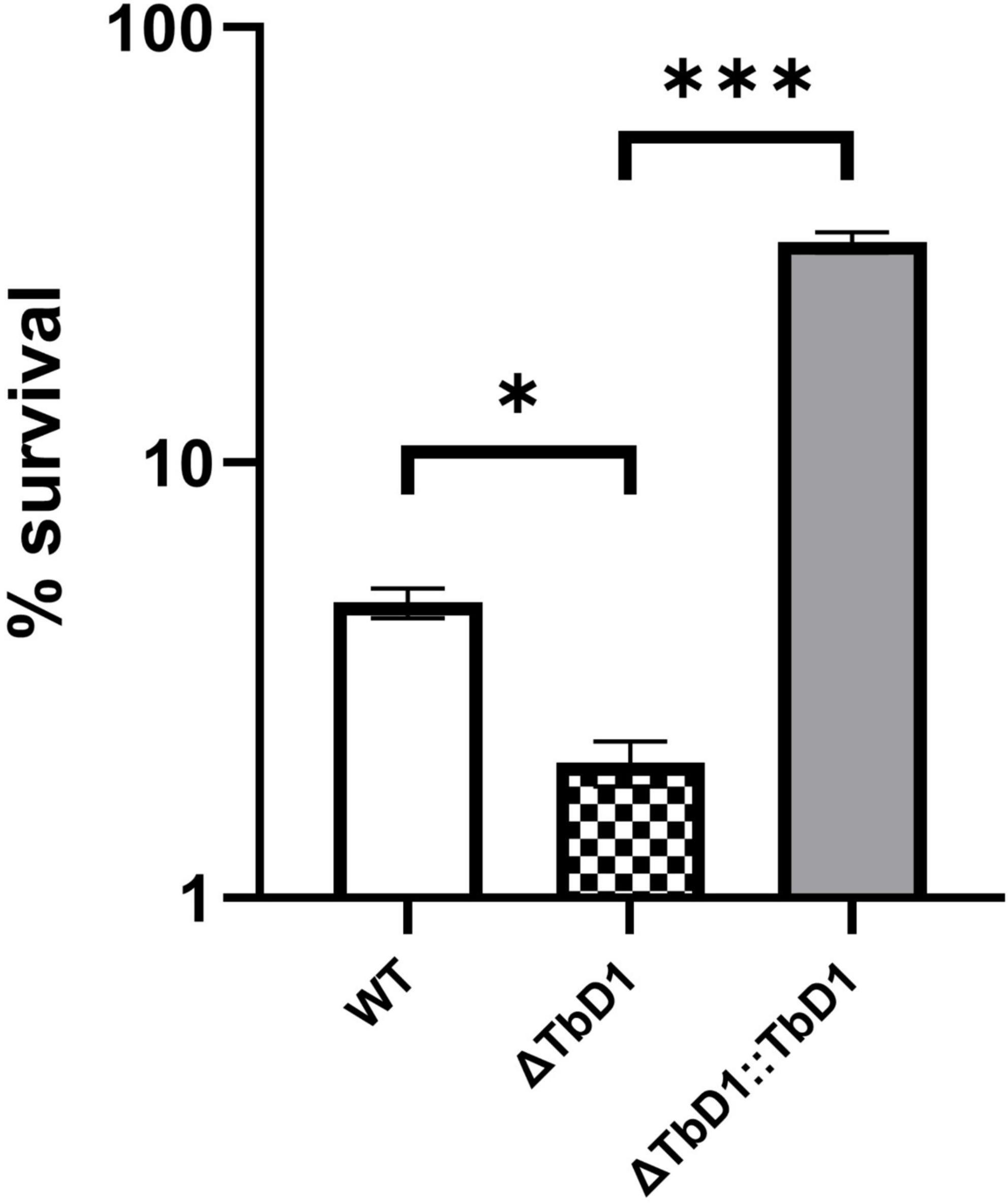
Figure 4. Copper stress experiments with M. bovis AF2122/97. Copper stress experiments were performed on M. bovis AF2122/97 WT, ΔTbD1, and ΔTbD1:TbD1 strains to assess the ability of TbD1 mutants to survive at high concentration of Copper ions. Two hundred micromolars of CuSO4 was added into standing cultures for 10 days without agitation and then cultures were plated on 7H11 plates with trace amounts of Copper. CFU were then counted after 2–3 weeks of incubation and survival percentages were determined relative to the non-CuSO4 control. The data are shown as means ± SD and are representative of three independent experiments. Statistical significance of differences in survival percentages calculated using one-way ANOVA followed by the Dunnett’s post hoc test for multiple-comparisons (*p < 0.05; ***p < 0.001).
TbD1 Locus Is Involved in H2O2 Resistance in Mycobacterium bovis AF2122/97
Previous publications have presented conflicting conclusions in terms of oxidative stress tolerance of M. tuberculosis with an intact or deleted TbD1 locus. We therefore conducted oxidative stress challenges on our M. bovis AF2122/97 WT and TbD1 mutants, using rolling cultures to match methods used in these previous publications. Forty millimolars H2O2 was added to mid-log phase cultures of M. bovis AF2122/97 WT, ΔTbD1, and ΔTbD1:TbD1 strains and incubated for 1 h at 37°C, with percentage survival determined based on CFU relative to the control (no treatment). The results showed that M. bovis AF2122/97 with an intact TbD1 locus had significantly improved survival under H2O2 stress (Figure 5). Interestingly, we identified that M. bovis AF2122/97 had higher resistance to H2O2 stress than M. tuberculosis H37Rv. A previous study reported that M. tuberculosis H37Rv showed approximately 10% survival after 1 h exposure to a 10 mM H2O2 challenge (Bottai et al., 2020); our work confirmed this level of M. tuberculosis H37Rv survival to 10 mM H2O2, while we found that M. bovis AF2122/97 challenged with 10 mM H2O2 showed survival that was similar to the non-H2O2 treated control group (Supplementary Figure 1). Previously, Forrellad et al. (2019) found a higher resistance of M. bovis NCTC10772 to H2O2 relative to M. tuberculosis CDC1551, while Firmani and Riley (2002) reported that M. bovis Ravanel was more resistant than M. tuberculosis H37Rv to 5 mM H2O2. The gene encoding catalase-peroxidase, katG, has a SNP at position 463 that distinguishes M. bovis (463L) from M. tuberculosis H37Rv (463R) and other Lineage 4 strains (Heym et al., 1995; Stucki et al., 2016). Previous functional studies have, however, shown that this SNP does not lead to altered KatG activity (Saint-Joanis et al., 1999) excluding it as being important in H2O2 sensitivity. Hence, our data extends previous findings on H2O2 sensitivity, showing that M. bovis AF2122/97 is more resistant to H2O2 stress than M. tuberculosis H37Rv.
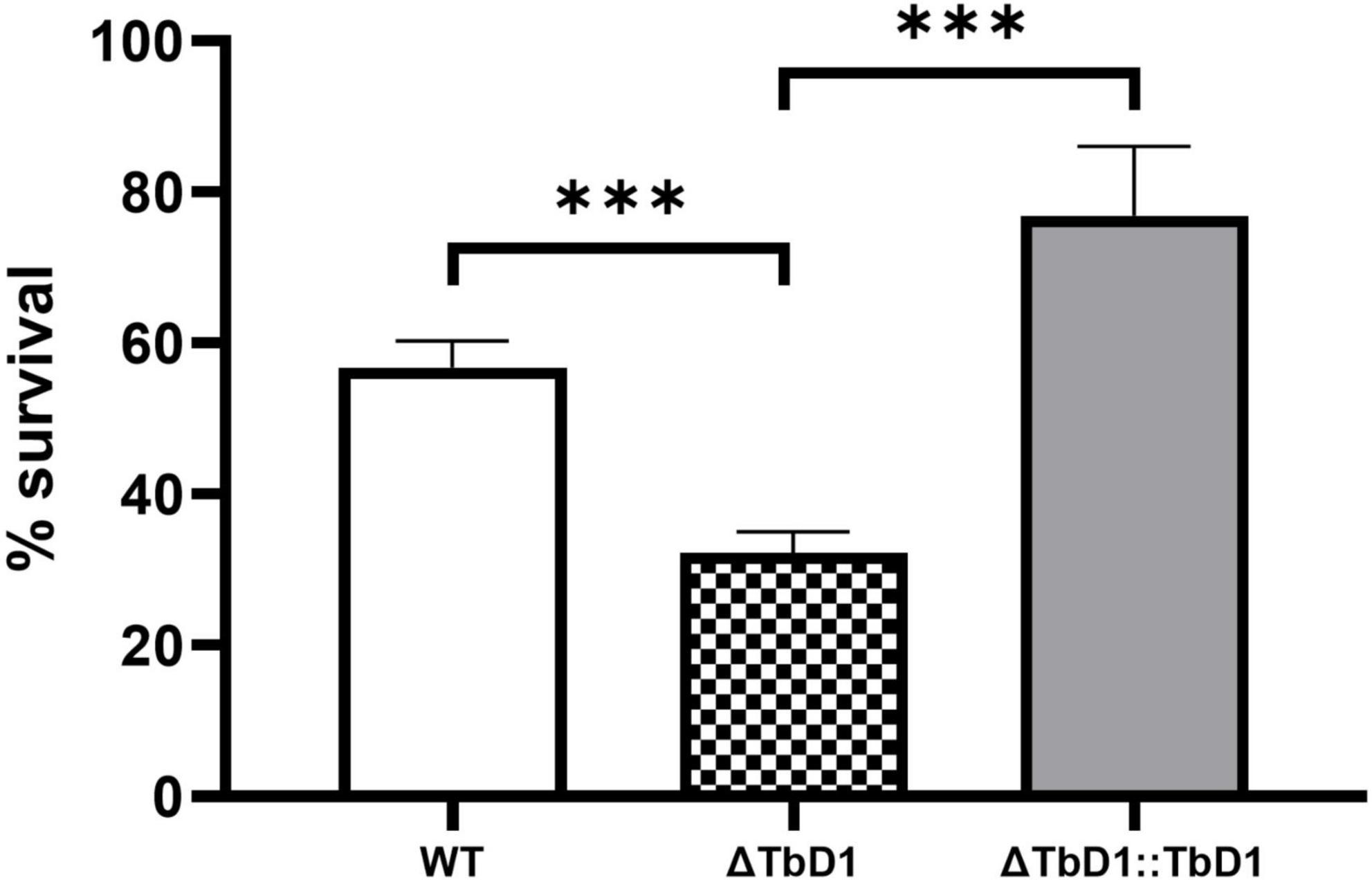
Figure 5. Oxidative stress experiments of M. bovis AF2122/97. H2O2 stress experiments with M. bovis AF2122/97 WT, ΔTbD1, and ΔTbD1:TbD1 strains to assess the survival ability of strains to 40 mM H2O2. Forty millimolars H2O2 was added into standing cultures for 1 h and then cultures were plated on 7H11 plates. CFU were then counted after 2–3 weeks of incubation and survival percentages were determined relative to the non-treated control. The data are shown as means ± SD and are representative of three independent experiments. Statistical significance of differences in survival percentages calculated using one-way ANOVA followed by the Dunnett’s post hoc test for multiple-comparisons (***p < 0.001).
RT-qPCR on BCG Denmark Further Validated RNA-seq Results and Highlighted the TbD1 Mediated Hypoxia-Specific Copper Pathway in Mycobacterium bovis
Our RNA-seq data was obtained from M. bovis AF2122/97 WT and ΔTbD1 mutant strains. As a parallel way to validate the impact of the loss of TbD1 and the DEG findings, we constructed an independent mutant in the attenuated M. bovis strain BCG Denmark. RT-qPCR targeting several DEGs from the M. bovis AF2122/97 RNA-seq analysis (Supplementary Table 1) was performed on M. bovis BCG Denmark WT and ΔTbD1 strains. In agreement with our results from M. bovis AF2122/97, the expression of mmpS6 and mmpL6 in the BCG Denmark WT were approximately two to threefold higher in response to standing growth conditions than when grown rolling (Figures 6A,B). Our RT-qPCR results also showed induction of TbD1 expression by Triclosan induced oxidative stress in M. bovis AF2122/97 (Supplementary Figure 2), in agreement with a previous study in M. tuberculosis (Betts et al., 2003); hence our findings demonstrate that the TbD1 locus responds to changes in the redox status of M. bovis. The expression of two copper-responsive genes, cysK2 (Figure 6C) and ctpV (Figure 6D) were also determined in BCG Denmark WT and ΔTbD1 in response to standing vs. rolling culture conditions. While there was no significant differential expression of cysK2 and ctpV between BCG Denmark WT and ΔTbD1 strains when grown under rolling conditions, the two genes were significantly downregulated in BCG Denmark ΔTbD1 as compared to WT when grown using standing conditions. Thus, the RT-qPCR results on BCG Denmark WT and ΔTbD1 concurred with the M. bovis AF2122/97 RNA-seq data, revealing a link between the TbD1 locus in redox-state sensing and copper homeostasis.
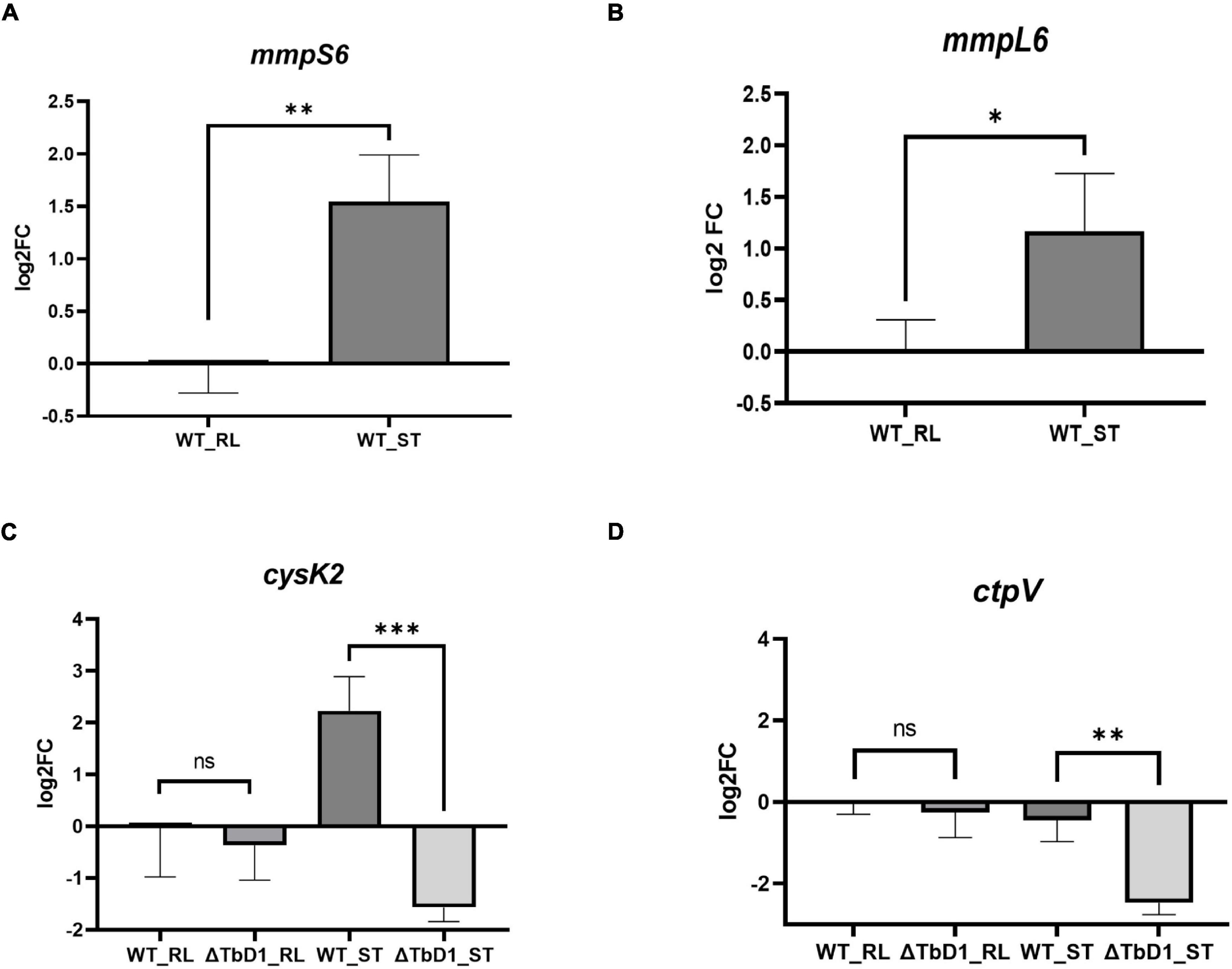
Figure 6. RT-qPCR validation on selected genes on BCG Denmark WT and ΔTbD1 under rolling and standing culture conditions. Log2 fold-change (log2FC) values were generated by comparing the expression of genes of BCG Denmark WT and ΔTbD1 strains under different culture conditions vs. WT under rolling condition control using 2 –ΔΔCt method, and expression levels of (A) mmpS6, (B) mmpL6, (C) cysK2, and (D) ctpV genes have shown. The data shown are means ± SD of gene expression from three independent biological replicates with duplicates, normalized with respect to 16S rRNA. Statistical significances of differences were calculated by Student’s t-test (*p < 0.05; **p < 0.01; ***p < 0.001; ns, not significant) are shown.
TbD1 Locus and Hypoxia-Specific Copper Sensitivity in BCG Denmark
We next sought to use our BCG Denmark WT and ΔTbD1 strains to assess copper sensitivity and compare with our previous findings with M. bovis AF2122/97 WT and ΔTbD1. Analysis of BCG Denmark WT and ΔTbD1 strains in a basal Sauton’s liquid media supplemented with a range of copper concentrations revealed that the growth of BCG Denmark ΔTbD1 was inhibited by 150 μM CuSO4 (Figure 7A). Analysis of growth on solid media was assessed by drop assays on 7H11 agar plates containing 25, 100, and 150 μM CuSO4. These assays showed no apparent difference in growth of BCG Denmark WT, ΔTbD1, and ΔTbD1:TbD1 on plates with a low concentration of CuSO4 (25 μM) while BCG Denmark ΔTbD1 had limited growth at 150 μM CuSO4 with a smaller colony size. The BCG Denmark ΔTbD1 complemented with the TbD1 locus driven from its native promoter showed increased growth at 150 μM CuSO4 (Figure 7B).
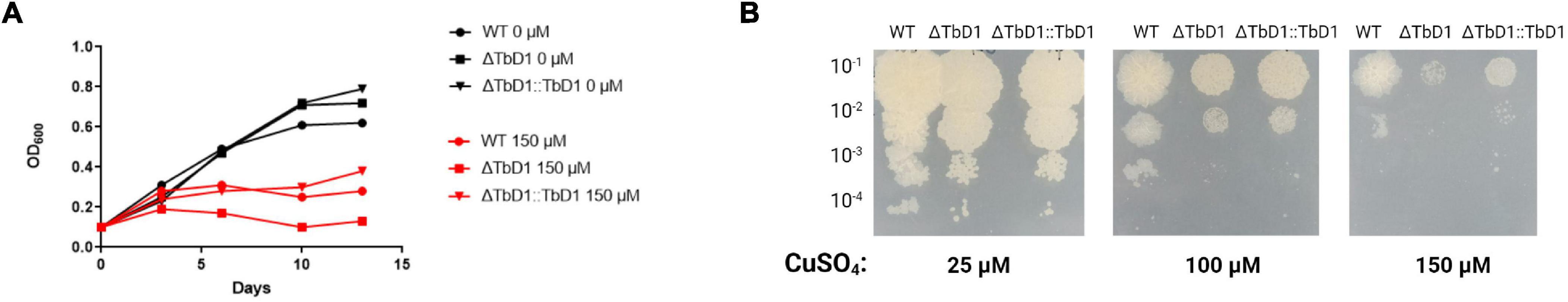
Figure 7. Contribution of TbD1 on BCG Denmark to Copper sensitivity. (A) BCG Denmark WT (circles), ΔTbD1 (squares), and ΔTbD1:TbD1 (triangles) strains were grown at 37°C in standing Sauton’s liquid media, and CuSO4 was added. Cultures were challenged with 0 μM CuSO4 (black symbols) or 150 μM CuSO4 (red symbols). Absorbance at 600 nm (OD600) was measured for 2 weeks. The data are representative of three independent experiments. (B) Drop assay determining the Copper resistance of TbD1 mutants. Serial dilutions (10-fold) of BCG Denmark WT, ΔTbD1, and ΔTbD1:TbD1 cultures were spotted onto 7H11 plates containing 25, 100, or 150 μM CuSO4. Data are representative of two independent experiments with replicates.
BCG Denmark ΔTbD1 Strains Expressed a Significantly Lower Level of Copper-Responsive Genes Under Copper Stress in Standing Condition and Contributed to Its Copper Sensitivity Phenotypes
To explore the sensitivity phenotypes that we observed in M. bovis AF2122/97 ΔTbD1 and BCG Denmark ΔTbD1 when grown under conditions of excess copper, we checked the expression level of several copper-responsive genes in standing BCG Denmark cultures after cultures had been incubated with 100 μM of CuSO4 for 3 h. To determine the candidate reference genes for this analysis the geNorm algorithm (Vandesompele et al., 2002) was used to examine the stability of eight potential reference genes identified from the literature (Supplementary Table 1). The geNorm analysis identified that optimal normalization would be obtained using the geometric mean of ftsZ and fbpB, and these were subsequently used in our gene expression analysis. We used cultures grown in basal Sauton’s media (without added CuSO4) as controls. Results showed that when cultured using standing conditions without copper, expression of cysK2 and ctpV were not significantly different among BCG Denmark WT, ΔTbD1, and ΔTbD1:TbD1 strains. However, when cultured with a high concentration of copper (100 μM), the expression of cysK2 and ctpV were significantly lower in BCG Denmark ΔTbD1 as compared to WT. Complementation of BCG Denmark ΔTbD1 with the WT locus (ΔTbD1:TbD1 strain) restored expression of cysK2 and ctpV to WT levels (Figure 8). Hence, sensitivity phenotypes to copper stress that were observed in M. bovis ΔTbD1 strains were due to disturbed copper homeostasis caused by TbD1 knockout.
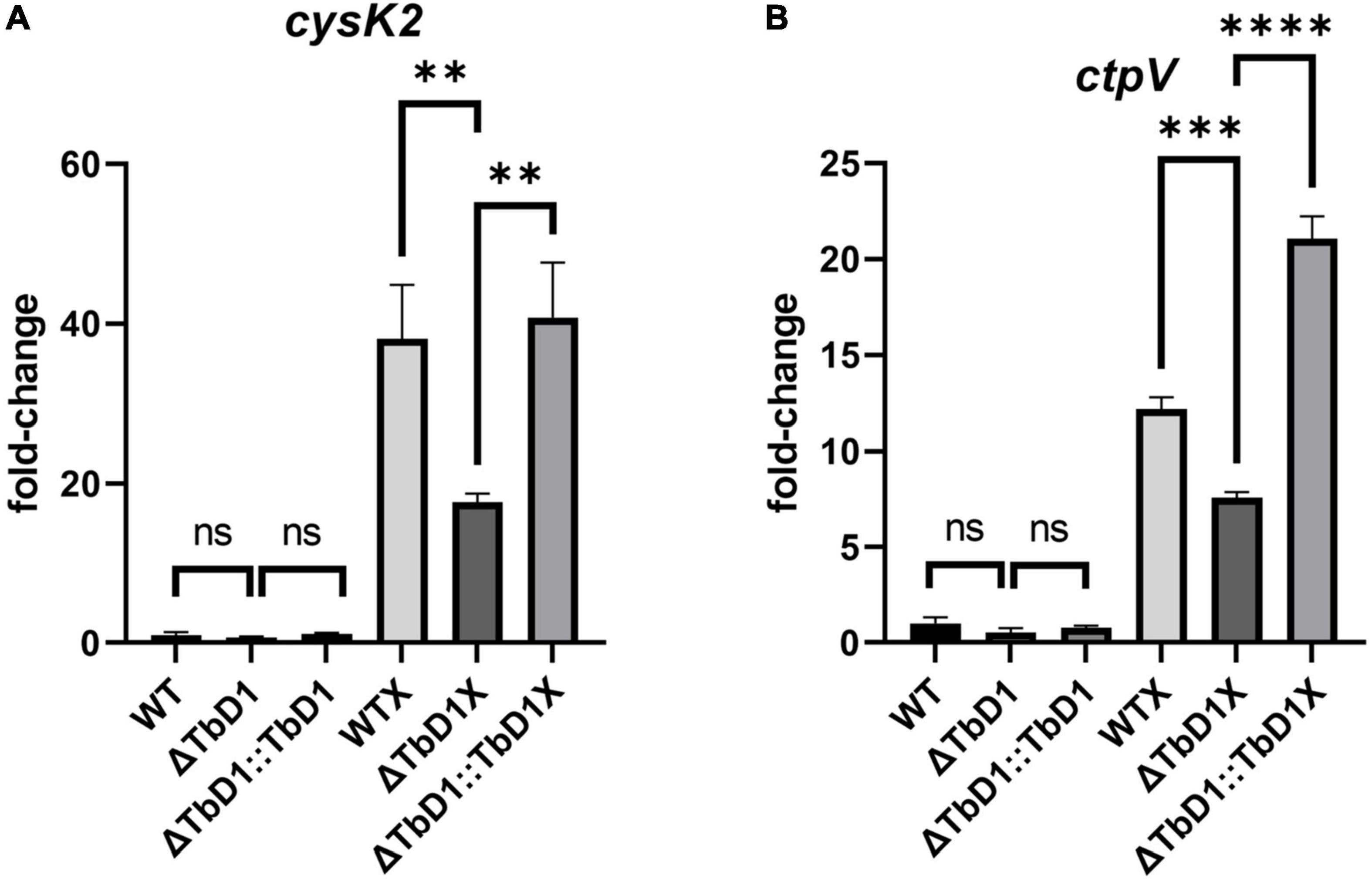
Figure 8. BCG Denmark ΔTbD1 copper response. BCG Denmark ΔTbD1 expressed copper-responsive genes at a significantly lower level under copper stress in standing culture conditions. Fold-change values were generated by comparing the expression of genes of WT, ΔTbD1, and ΔTbD1:TbD1 BCG Denmark strains treated with 100 μM CuSO4 for 3 h (marked as X) or untreated culture conditions vs. WT untreated control using 2 –ΔΔCt method, and expression levels of (A) cysK2 gene and (B) ctpV gene have shown. The data shown are means ± SD of gene expression from three independent biological replicates with duplicates, normalized with respect to two reference genes ftsZ and fbpB. Statistical significances of differences were calculated by one-way ANOVA followed by the Dunnett’s post hoc test for multiple-comparisons (**p < 0.01; ***p < 0.001; ****p < 0.0001; ns, not significant) are shown.
Discussion
The function of TbD1 in M. tuberculosis strains has revealed conflicting results on the role of the locus in the oxidative stress response (Arumugam et al., 2019; Bottai et al., 2020). Hence while Arumugam et al. (2019) found that M. tuberculosis strains with an intact TbD1 locus had increased resistance to oxidative stress, Bottai et al. (2020) found that loss of the TbD1 locus conferred M. tuberculosis strains with increased resistance to oxidative stress. The reasons for these divergent findings are not immediately apparent, but may reflect differences in experimental protocols or strain genetic backgrounds. Our work did not seek to directly address these discrepancies, but rather to explore the function of TbD1 in M. bovis as a route to assess the locus’s functional role in animal-adapted tubercle bacilli. Hence, we used M. bovis WT and mutant strains as a basis for comparative transcriptomic studies. These studies indicated that in M. bovis, TbD1 mediated hypoxia-dependent copper response pathways.
Copper is an essential micronutrient, but it can be toxic at high concentrations as it catalyzes the formation of potent oxidants, hydroxyl radicals, from hydrogen peroxide via the Fenton reaction; these oxidants inhibit metabolic processes and damage DNA, lipids, and proteins (Kumar et al., 2011). A previous study described the transcriptional profile of M. tuberculosis after 3 h exposure to 500 μM CuCl2; 11 out of 30 copper-induced genes overlapped with genes identified in another study of cultures exposed to H2O2, showing a correlation between the oxidative and copper stress responses (Schnappinger et al., 2003; Ward et al., 2008). Copper can also interact with sulfur atoms in Fe–S cluster proteins and replace enzyme metal cofactors, thereby inactivating them at a high kinetic rate (Pope et al., 2013). M. tuberculosis is known to elaborate three independent pathways in response to excess copper. One is McdB which exports copper to the extracellular environment. The other two pathways are regulated by Cu+-responsive transcriptional repressors CsoR and RicR; RicR regulates genes controlled by six different promoters, while CsoR regulates a single four-gene operon (Darwin, 2015).
Our comparative transcriptomic study of M. bovis WT and ΔTbD1 mutant identified that multiple CsoR and RicR regulated genes were significantly downregulated in the ΔTbD1 mutant as compared to WT when strains were grown under standing culture conditions (that contain trace amounts of copper). These downregulated genes included cysK2 which encodes an L-cysteine synthase that is involved in the biosynthesis of S-sulfocysteine, a precursor of mycothiol which acts as a redox buffer (Steiner et al., 2014). Other downregulated genes were ctpV which encodes a P1B-type ATPase that pumps copper cations to the cellular periplasm (Ward et al., 2010), and mymT that encodes a metallothionein that can bind up to seven copper ions with high avidity and hence increases copper tolerance (Rowland and Niederweis, 2012). Notably, we found that other enriched GO terms in our comparative analysis between mutant and WT included the Cadmium ion response GO term, a finding that might be explained by cross talk between copper ion response and cadmium ion response. Hence two genes in the Cadmium ion response GO term encoded CadI, which encodes a cadmium-induced protein, was found to be the highest induced gene after copper exposure (Hotter et al., 2001; Ward et al., 2008), and MymT which is a metallothionein that is induced by cadmium and copper (Gold et al., 2008). Interestingly, cadI, cysK2, and ctpG were also found to be induced by 0.5 mM of zinc in M. tuberculosis (Botella et al., 2011), showing the overlap between divalent metal ion responses in mycobacteria. Other GO terms that were revealed through DEG analysis related to Mo-molybdopterin cofactor metabolic/biosynthetic process (moaA1, moaC1, and moaD1) which are involved in the early steps of Mo-molybdopterin cofactor biosynthesis. Early steps in MoCo biosynthesis are linked to copper, iron, and metal homeostasis, as well as being linked with sulfur and cysteine metabolism (Williams et al., 2014). It is also possible that due to the downregulation of cysK2, accumulated thiosulfate stimulated the feedback expression of Moa1 locus via sulfur assimilation.
The effects of different culture conditions (standing vs. rolling) seen in our study highlights the importance of culture conditions when defining in vitro mycobacterial mutant phenotypes. Rolling is a common in vitro culture method for mycobacteria as it minimizes aerosol generation but maximizes aeration and yield. However, mycobacterial pathogens would not normally encounter such high aeration rates in vivo, but instead low oxygen tension is the more usual state during infection (Rustad et al., 2009). Hypoxia-based in vitro models are used as dormancy models and are thought to mirror some aspects of in vivo latent infection, with hypoxia being a key host-induced stress that limits the growth of tubercle bacilli and induces non-replicating persistence (Wayne and Sohaskey, 2001). Furthermore, in the context of reduced oxygen environments in vivo, it is known that hypoxia acts in concert with copper stress in the host macrophage (Via et al., 2008). The level of intraphagosomal copper concentration increases from 25 to 500 μM after phagocytosis of M. tuberculosis, but no such increase occurs after phagocytosis of less virulent mycobacterial species such as M. avium and M. smegmatis (Wagner et al., 2005). What’s more, previous studies have shown that copper exhibits greater toxicity under hypoxic environments because of the reduction of Cu2+ to more labile Cu+ under such conditions, and that the host macrophage increases copper uptake under hypoxia through upregulation of the macrophage copper importer Cu+ transport protein 1 (CTR1) (Beswick et al., 1976; White et al., 2009). Indeed, the effect is not specific to mycobacteria; Macomber and colleagues found that E. coli strains were more sensitive to copper under hypoxia (Macomber and Imlay, 2009). Also, previous studies proved that Saccharomyces cerevisiae and E. coli accumulate higher concentrations of copper in anaerobic cultures as compared with aerobic cultures (Strain and Culotta, 1996; Outten et al., 2001). Our findings corroborate the linkage of reduced oxygen to increased copper toxicity, with increased in vitro copper sensitivity of M. bovis ΔTbD1 when grown with standing culture conditions in liquid media and in drop assays on 7H11 plates. On this latter point of growth on solid media, it has been suggested (Beswick et al., 1976) that colonies on plates could experience reduced oxygen availability even with aerobically incubation, which may again potentiate the action of copper.
It is known that M. tuberculosis shifts to use lipids, including cholesterol, as a primary source of nutrition in the host (Bloch and Segal, 1956; McKinney et al., 2000; Pandey and Sassetti, 2008; Marrero et al., 2010). Furthermore, acidic pH and hypoxia are known to be a common stresses imposed on tubercle bacilli during infection (Wayne and Sohaskey, 2001). Notably, in vitro assays that seek to mimic in vivo stresses commonly encountered by M. tuberculosis have been shown to alter carbon metabolic flux, i.e., a recent study showed that lipids are the preferred primary carbon source in an acidic environment (Gouzy et al., 2021). GO analysis revealed an induction of genes involved in lipid metabolism in M. bovis AF2122/97 WT cultured under standing conditions. It’s thus perhaps not surprising that in our RNA-seq results, in vitro hypoxia was seen to trigger induction of M. bovis AF2122/97 genes involved in lipid-based metabolism; this likely shows that M. bovis senses hypoxia as a signal to adapt to the in vivo host environment. However, there were no significant DEGs for lipid metabolism between the WT and M. bovis ΔTbD1 mutant involved in either standing or rolling conditions. This observation suggests that the MmpS/L6 transport system is not involved in lipid transport per se. Indeed, the MmpL family has been shown to have diverse functions including uptake of nutrients, iron acquisition, or extrusion of toxins as stated. Furthermore, Bottai et al. (2020) used lipidomics assays on a set of WT and TbD1 mutants in an attempt to identify TbD1-associated lipids, but no clear difference between strains were observed.
It is also worth highlighting the AAC-AAG SNP in mmpL6 which is only present in animal adapted strains. This SNP causes a non-synonymous N551K change located in the long α-helical hairpin on the PC subdomain of the protein (Supplementary Figure 1). Based on previous studies of RND transporters from Gram-negative bacteria, the PC subdomain is thought to contain binding sites for the exported ligands and have a role in substrate binding, hence determining substrate specificity (Elkins and Nikaido, 2002; Tikhonova et al., 2002). The presence of this SNP may have no functional significance, or may indicate subtle differences in substrate specificity of MmpL6 in animal-adapted lineages.
The mechanisms of copper acquisition and transport in mycobacteria are poorly identified, and copper chaperones have not been identified. As a way to conceptualize our findings, we hypothesize that the TbD1 locus may mediate a hypoxia-specific copper response by translocation of an (as yet unknown) copper chaperone across the cytoplasmic membrane. This potential system would have the chaperone transport copper ions into the cytoplasm with subsequent protein–protein interaction between the chaperone-copper complex and RicR/CsoR, allowing the expression of downstream proteins and promote adaptive changes in intracellular copper homeostasis (Figure 9). Notably mmpl3 was found to be significantly downregulated under 0.5 mM zinc, while copper-responsive genes (i.e., lpqS, cysK2, and mymT) were induced in an mmpl3 depleted M. tuberculosis mutant (Botella et al., 2011; Degiacomi et al., 2017). This indicates that other MmpL proteins are also be involved in metal responses, and suggests a possible fruitful area for future studies.
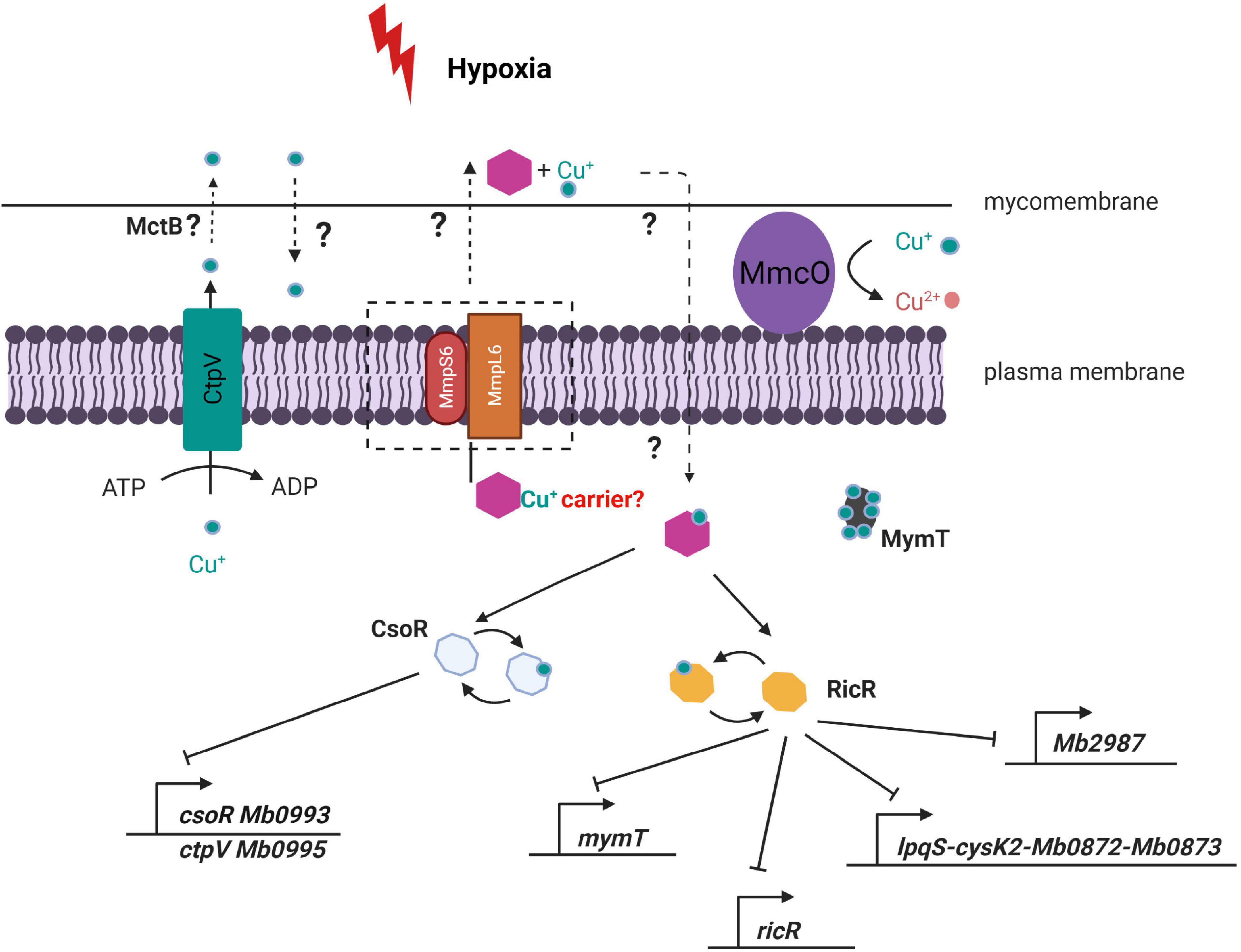
Figure 9. Model of potential consequences of ΔTbD1 disrupted copper homeostasis under hypoxia. The module suggests that the TbD1 locus mediates a hypoxia-specific copper response via translocation of an unidentified copper chaperone (purple hexagon) across the cytoplasmic membrane. The chaperone then transports copper ions into the cytoplasm with subsequent protein–protein interaction between the chaperone-copper complex and RicR/CsoR, allowing the expression of downstream proteins and promoting adaptive changes in intracellular copper homeostasis.
In conclusion, our findings provide insight into the function of the TbD1 locus in M. bovis and reveal a role in hypoxia-specific copper detoxification. Further experimentation will be required to elucidate whether the presence of an intact TbD1 locus favors M. bovis in adaptation to its preferred animal host.
Data Availability Statement
The raw sequencing data are available in the Sequence Read Archive (SRA) under the BioProject ID PRJNA774648.
Author Contributions
RM and SG designed the experiments and wrote the manuscript. RM performed the experimental work and data analyses. DF and GG contributed to bioinformatics analyses. JB contributed to RT-qPCR work. YS and CN contributed to experimental advice and interpretation. All authors approved the final submission.
Funding
RM was funded by a China Scholarship Council (CSC) grant from the Ministry of Education the People’s Republic of China. Work was also supported by funding from Science Foundation Ireland (SFI/15/IA/3154) to SG, and through the Global Station for Zoonosis Control, Hokkaido University, Japan.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Daria Bottai for advice and discussion.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.817952/full#supplementary-material
References
Ameni, G., Vordermeier, M., Firdessa, R., Aseffa, A., Hewinson, G., Gordon, S. V., et al. (2011). Mycobacterium tuberculosis infection in grazing cattle in central Ethiopia. Vet. J. 188, 359–361. doi: 10.1016/j.tvjl.2010.05.005
Arumugam, P., Shankaran, D., Bothra, A., Gandotra, S., and Rao, V. (2019). The MmpS6-MmpL6 operon is an oxidative stress response system providing selective advantage to Mycobacterium tuberculosis in stress. J. Infect. Dis. 219, 459–469. doi: 10.1093/infdis/jiy526
Babraham Bioinformatics (2019a). FastQC A Quality Control tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed June 12, 2019).
Babraham Bioinformatics (2019b). Trim Galore!. Available online at: https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed June 12, 2019).
Beswick, P. H., Hall, G. H., Hook, A. J., Little, K., McBrien, D. C. H., and Lott, K. A. K. (1976). Copper toxicity: evidence for the conversion of cupric to cuprous copper in vivo under anaerobic conditions. Chem. Biol. Interact. 14, 347–356. doi: 10.1016/0009-2797(76)90113-7
Betts, J. C., McLaren, A., Lennon, M. G., Kelly, F. M., Lukey, P. T., Blakemore, S. J., et al. (2003). Signature gene expression profiles discriminate between isoniazid-, thiolactomycin-, and triclosan-treated Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 47, 2903–2913. doi: 10.1128/AAC.47.9.2903-2913.2003
Bloch, H., and Segal, W. (1956). Biochemical differentiation of Mycobacterium tuberculosis grown in vivo and in vitro. J. Bacteriol. 72, 132–141. doi: 10.1128/jb.72.2.132-141.1956
Botella, H., Peyron, P., Levillain, F., Poincloux, R., Poquet, Y., Brandli, I., et al. (2011). Mycobacterial p(1)-type ATPases mediate resistance to zinc poisoning in human macrophages. Cell Host Microbe 10, 248–259. doi: 10.1016/j.chom.2011.08.006
Bottai, D., Frigui, W., Sayes, F., Di Luca, M., Spadoni, D., Pawlik, A., et al. (2020). TbD1 deletion as a driver of the evolutionary success of modern epidemic Mycobacterium tuberculosis lineages. Nat. Commun. 11:684. doi: 10.1038/s41467-020-14508-5
Brosch, R., Gordon, S. V., Marmiesse, M., Brodin, P., Buchrieser, C., Eiglmeier, K., et al. (2002). A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. U.S.A. 99, 3684–3689. doi: 10.1073/pnas.052548299
Cole, S. T., Brosch, R., Parkhill, J., Garnier, T., Churcher, C., Harris, D., et al. (1998). Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544. doi: 10.1038/31159
Darwin, K. H. (2015). Mycobacterium tuberculosis and copper: a newly appreciated defense against an old foe? J. Biol. Chem. 290, 18962–18966. doi: 10.1074/jbc.R115.640193
Degiacomi, G., Benjak, A., Madacki, J., Boldrin, F., Provvedi, R., Palù, G., et al. (2017). Essentiality of mmpL3 and impact of its silencing on Mycobacterium tuberculosis gene expression. Sci. Rep. 7:43495. doi: 10.1038/srep43495
Delmar, J. A., Chou, T.-H., Wright, C. C., Licon, M. H., Doh, J. K., Radhakrishnan, A., et al. (2015). Structural basis for the regulation of the MmpL transporters of Mycobacterium tuberculosis. J. Biol. Chem. 290, 28559–28574. doi: 10.1074/jbc.M115.683797
Deshayes, C., Bach, H., Euphrasie, D., Attarian, R., Coureuil, M., Sougakoff, W., et al. (2010). MmpS4 promotes glycopeptidolipids biosynthesis and export in Mycobacterium smegmatis. Mol. Microbiol. 78, 989–1003. doi: 10.1111/j.1365-2958.2010.07385.x
Elkins, C. A., and Nikaido, H. (2002). Substrate specificity of the RND-type multidrug efflux pumps AcrB and AcrD of Escherichia coli is determined predominantly by two large periplasmic loops. J. Bacteriol. 184, 6490–6498. doi: 10.1128/JB.184.23.6490-6499.2002
Firmani, M. A., and Riley, L. W. (2002). Reactive nitrogen intermediates have a bacteriostatic effect on Mycobacterium tuberculosis in vitro. J. Clin. Microbiol. 40, 3162–3166. doi: 10.1128/JCM.40.9.3162-3166.2002
Florczyk, M. A., McCue, L. A., Stack, R. F., Hauer, C. R., and McDonough, K. A. (2001). Identification and characterization of mycobacterial proteins differentially expressed under standing and shaking culture conditions, including Rv2623 from a novel class of putative ATP-binding proteins. Infect. Immun. 69, 5777–5785. doi: 10.1128/IAI.69.9.5777-5785.2001
Forrellad, M. A., Vázquez, C. L., Blanco, F. C., Klepp, L. I., García, E. A., Rocha, R. V., et al. (2019). Rv2617c and P36 are virulence factors of pathogenic mycobacteria involved in resistance to oxidative stress. Virulence 10, 1026–1033. doi: 10.1080/21505594.2019.1693714
Francis, J. (1950). Control of infection with the bovine tubercle bacillus. Lancet 1, 34–39. doi: 10.1016/s0140-6736(50)90236-4
Galagan, J. E., Minch, K., Peterson, M., Lyubetskaya, A., Azizi, E., Sweet, L., et al. (2013). The Mycobacterium tuberculosis regulatory network and hypoxia. Nature 499, 178–183. doi: 10.1038/nature12337
Garnier, T., Eiglmeier, K., Camus, J., Medina, N., Mansoor, H., Pryor, M., et al. (2003). The complete genome sequence of Mycobacterium bovis. Proc. Natl. Acad. Sci. U.S.A. 100, 7877–7882. doi: 10.1073/pnas.1130426100
Gold, B., Deng, H., Bryk, R., Vargas, D., Eliezer, D., Roberts, J., et al. (2008). Identification of a copper-binding metallothionein in pathogenic mycobacteria. Nat. Chem. Biol. 4, 609–616. doi: 10.1038/nchembio.109
Gormley, E., and Corner, L. A. L. (2018). Wild animal tuberculosis: stakeholder value systems and management of disease. Front. Vet. Sci. 5:327. doi: 10.3389/fvets.2018.00327
Gouzy, A., Healy, C., Black, K. A., Rhee, K. Y., and Ehrt, S. (2021). Growth of Mycobacterium tuberculosis at acidic pH depends on lipid assimilation and is accompanied by reduced GAPDH activity. Proc. Natl. Acad. Sci. U.S.A. 118:e2024571118. doi: 10.1073/pnas.2024571118
Hershberg, R., Lipatov, M., Small, P. M., Sheffer, H., Niemann, S., Homolka, S., et al. (2008). High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol. 6:e311. doi: 10.1371/journal.pbio.0060311
Heym, B., Alzari, P. M., Honore, N., and Cole, S. T. (1995). Missense mutations in the catalase-peroxidase gene, katG, are associated with isoniazid resistance in Mycobacterium tuberculosis. Mol. Microbiol. 15, 235–245. doi: 10.1111/j.1365-2958.1995.tb02238.x
Hotter, G. S., Wilson, T., and Collins, D. M. (2001). Identification of a cadmium-induced gene in Mycobacterium bovis and Mycobacterium tuberculosis. FEMS Microbiol. Lett. 200, 151–155. doi: 10.1111/j.1574-6968.2001.tb10707.x
Kobylka, J., Kuth, M. S., Müller, R. T., Geertsma, E. R., and Pos, K. M. (2020). AcrB: a mean, keen, drug efflux machine. Ann. N.Y. Acad. Sci. 1459, 38–68. doi: 10.1111/nyas.14239
Kumar, A., Farhana, A., Guidry, L., Saini, V., Hondalus, M., and Steyn, A. J. C. (2011). Redox homeostasis in mycobacteria: the key to tuberculosis control? Expert Rev. Mol. Med. 13:e39. doi: 10.1017/S1462399411002079
Li, H. (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv [Preprint]. arXiv:1303.3997.
Liao, Y., Smyth, G. K., and Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Long, F., Su, C., Zimmermann, M. T., Boyken, S. E., Rajashankar, K. R., Jernigan, R. L., et al. (2010). Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport. Nature 467, 484–488. doi: 10.1038/nature09395
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Macomber, L., and Imlay, J. A. (2009). The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc. Natl. Acad. Sci. U.S.A. 106, 8344–8349. doi: 10.1073/pnas.0812808106
Magnus, K. (1966). Epidemiological basis of tuberculosis eradication. 3. Risk of pulmonary tuberculosis after human and bovine infection. Bull. World Health Organ. 35, 483–508.
Malone, K. M., Rue-Albrecht, K., Magee, D. A., Conlon, K., Schubert, O. T., Nalpas, N. C., et al. (2018). Comparative ’omics analyses differentiate Mycobacterium tuberculosis and Mycobacterium bovis and reveal distinct macrophage responses to infection with the human and bovine Tubercle bacilli. Microb. Genomics 4:e000163. doi: 10.1099/mgen.0.000163
Marrero, J., Rhee, K. Y., Schnappinger, D., Pethe, K., and Ehrt, S. (2010). Gluconeogenic carbon flow of tricarboxylic acid cycle intermediates is critical for Mycobacterium tuberculosis to establish and maintain infection. Proc. Natl. Acad. Sci. U.S.A. 107, 9819–9824. doi: 10.1073/pnas.1000715107
McKinney, J. D., Höner zu Bentrup, K., Muñoz-Elías, E. J., Miczak, A., Chen, B., Chan, W. T., et al. (2000). Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406, 735–738. doi: 10.1038/35021074
Melly, G., and Purdy, G. E. (2019). MmpL proteins in physiology and pathogenesis of M. tuberculosis. Microorganisms 7, 1–16. doi: 10.3390/microorganisms7030070
Merker, M., Blin, C., Mona, S., Duforet-Frebourg, N., Lecher, S., Willery, E., et al. (2015). Evolutionary history and global spread of the Mycobacterium tuberculosis Beijing lineage. Nat. Genet. 47, 242–249. doi: 10.1038/ng.3195
Murakami, S., Nakashima, R., Yamashita, E., and Yamaguchi, A. (2002). Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419, 587–593. doi: 10.1038/nature01050
Nakashima, R., Sakurai, K., Yamasaki, S., Hayashi, K., Nagata, C., Hoshino, K., et al. (2013). Structural basis for the inhibition of bacterial multidrug exporters. Nature 500, 102–106. doi: 10.1038/nature12300
Outten, F. W., Huffman, D. L., Hale, J. A., and O’Halloran, T. V. (2001). The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J. Biol. Chem. 276, 30670–30677. doi: 10.1074/jbc.M104122200
Pak, J. E., Ekendé, E. N., Kifle, E. G., O’Connell, J. D., De Angelis, F., Tessema, M. B., et al. (2013). Structures of intermediate transport states of ZneA, a Zn(II)/proton antiporter. Proc. Natl. Acad. Sci. U.S.A. 110, 18484–18489. doi: 10.1073/pnas.1318705110
Pandey, A. K., and Sassetti, C. M. (2008). Mycobacterial persistence requires the utilization of host cholesterol. Proc. Natl. Acad. Sci. U.S.A. 105, 4376–4380. doi: 10.1073/pnas.0711159105
Pope, C. R., De Feo, C. J., and Unger, V. M. (2013). Cellular distribution of copper to superoxide dismutase involves scaffolding by membranes. Proc. Natl. Acad. Sci. U.S.A. 110, 20491–20496. doi: 10.1073/pnas.1309820110
Portevin, D., Gagneux, S., Comas, I., and Young, D. (2011). Human macrophage responses to clinical isolates from the Mycobacterium tuberculosis complex discriminate between ancient and modern lineages. PLoS Pathog. 7:e1001307. doi: 10.1371/journal.ppat.1001307
Reiling, N., Homolka, S., Walter, K., Brandenburg, J., Niwinski, L., Ernst, M., et al. (2013). Clade-specific virulence patterns of Mycobacterium tuberculosis complex strains in human primary macrophages and aerogenically infected mice. mBio 4:e00250-13. doi: 10.1128/mBio.00250-13
Rowland, J. L., and Niederweis, M. (2012). Resistance mechanisms of Mycobacterium tuberculosis against phagosomal copper overload. Tuberculosis 92, 202–210. doi: 10.1016/j.tube.2011.12.006
Rustad, T. R., Harrell, M. I., Liao, R., and Sherman, D. R. (2008). The enduring hypoxic response of Mycobacterium tuberculosis. PLoS One 3:e1502. doi: 10.1371/journal.pone.0001502
Rustad, T. R., Sherrid, A. M., Minch, K. J., and Sherman, D. R. (2009). Hypoxia: a window into Mycobacterium tuberculosis latency. Cell. Microbiol. 11, 1151–1159. doi: 10.1111/j.1462-5822.2009.01325.x
Saint-Joanis, B., Souchon, H., Wilming, M., Johnsson, K., Alzari, P. M., and Cole, S. T. (1999). Use of site-directed mutagenesis to probe the structure, function and isoniazid activation of the catalase/peroxidase, KatG, from Mycobacterium tuberculosis. Biochem. J. 338(Pt 3), 753–760.
Sandhu, P., and Akhter, Y. (2015). The internal gene duplication and interrupted coding sequences in the MmpL genes of Mycobacterium tuberculosis: towards understanding the multidrug transport in an evolutionary perspective. Int. J. Med. Microbiol. 305, 413–423. doi: 10.1016/j.ijmm.2015.03.005
Saviola, B., and Bishai, W. R. (2004). Method to integrate multiple plasmids into the mycobacterial chromosome. Nucleic Acids Res. 32:e11. doi: 10.1093/nar/gnh005
Schnappinger, D., Ehrt, S., Voskuil, M. I., Liu, Y., Mangan, J. A., Monahan, I. M., et al. (2003). Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198, 693–704. doi: 10.1084/jem.20030846
Sennhauser, G., Bukowska, M. A., Briand, C., and Grütter, M. G. (2009). Crystal structure of the multidrug exporter MexB from Pseudomonas aeruginosa. J. Mol. Biol. 389, 134–145. doi: 10.1016/j.jmb.2009.04.001
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Steiner, E. M., Böth, D., Lössl, P., Vilaplana, F., Schnell, R., and Schneider, G. (2014). CysK2 from Mycobacterium tuberculosis is an O-phospho-L-serine-dependent S-sulfocysteine synthase. J. Bacteriol. 196, 3410–3420. doi: 10.1128/JB.01851-14
Strain, J., and Culotta, V. C. (1996). Copper ions and the regulation of Saccharomyces cerevisiae metallothionein genes under aerobic and anaerobic conditions. Mol. Gen. Genet. 251, 139–145. doi: 10.1007/BF02172911
Stucki, D., Brites, D., Jeljeli, L., Coscolla, M., Liu, Q., Trauner, A., et al. (2016). Mycobacterium tuberculosis lineage 4 comprises globally distributed and geographically restricted sublineages. Nat. Genet. 48, 1535–1543. doi: 10.1038/ng.3704
Su, C., Long, F., Lei, H., Bolla, J. R., Do, S. V., Rajashankar, K. R., et al. (2012). Charged amino acids (R83, E567, D617, E625, R669, and K678) of CusA are required for metal ion transport in the Cus efflux system. J. Mol. Biol. 422, 429–441. doi: 10.1016/j.jmb.2012.05.038
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613. doi: 10.1093/nar/gky1131
Tikhonova, E. B., Wang, Q., and Zgurskaya, H. I. (2002). Chimeric analysis of the multicomponent multidrug efflux transporters from gram-negative bacteria. J. Bacteriol. 184, 6499–6507. doi: 10.1128/JB.184.23.6499-6507.2002
van Kessel, J. C., and Hatfull, G. F. (2007). Recombineering in Mycobacterium tuberculosis. Nat. Methods 4, 147–152. doi: 10.1038/nmeth996
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034
Via, L. E., Lin, P. L., Ray, S. M., Carrillo, J., Allen, S. S., Eum, S. Y., et al. (2008). Tuberculous granulomas are hypoxic in guinea pigs, rabbits, and nonhuman primates. Infect. Immun. 76, 2333–2340. doi: 10.1128/IAI.01515-07
Wagner, D., Maser, J., Moric, I., Boechat, N., Vogt, S., Gicquel, B., et al. (2005). Changes of the phagosomal elemental concentrations by Mycobacterium tuberculosis Mramp. Microbiology 151, 323–332. doi: 10.1099/mic.0.27213-0
Ward, S. K., Abomoelak, B., Hoye, E. A., Steinberg, H., and Talaat, A. M. (2010). CtpV: a putative copper exporter required for full virulence of Mycobacterium tuberculosis. Mol. Microbiol. 77, 1096–1110. doi: 10.1111/j.1365-2958.2010.07273.x
Ward, S. K., Hoye, E. A., and Talaat, A. M. (2008). The global responses of Mycobacterium tuberculosis to physiological levels of copper. J. Bacteriol. 190, 2939–2946. doi: 10.1128/JB.01847-07
Wayne, L. G., and Sohaskey, C. D. (2001). Nonreplicating persistence of Mycobacterium tuberculosis. Annu. Rev. Microbiol. 55, 139–163. doi: 10.1146/annurev.micro.55.1.139
Wells, R. M., Jones, C. M., Xi, Z., Speer, A., Danilchanka, O., Doornbos, K. S., et al. (2013). Discovery of a siderophore export system essential for virulence of Mycobacterium tuberculosis. PLoS Pathog. 9:e1003120. doi: 10.1371/journal.ppat.1003120
Whelan, A. O., Coad, M., Cockle, P. J., Hewinson, G., Vordermeier, M., and Gordon, S. V. (2010). Revisiting host preference in the Mycobacterium tuberculosis complex: experimental infection shows M. tuberculosis H37Rv to be avirulent in cattle. PLoS One 5:e8527. doi: 10.1371/journal.pone.0008527
White, C., Kambe, T., Fulcher, Y. G., Sachdev, S. W., Bush, A. I., Fritsche, K., et al. (2009). Copper transport into the secretory pathway is regulated by oxygen in macrophages. J. Cell Sci. 122, 1315–1321. doi: 10.1242/jcs.043216
Williams, M., Mizrahi, V., and Kana, B. D. (2014). Molybdenum cofactor: a key component of Mycobacterium tuberculosis pathogenesis? Crit. Rev. Microbiol. 40, 18–29. doi: 10.3109/1040841X.2012.749211
Wirth, T., Hildebrand, F., Allix-Béguec, C., Wölbeling, F., Kubica, T., Kremer, K., et al. (2008). Origin, spread and demography of the Mycobacterium tuberculosis complex. PLoS Pathog. 4:e1000160. doi: 10.1371/journal.ppat.1000160
Keywords: tuberculosis, transcriptomic (RNA-seq), Mycobacterium bovis, phenotype, hypoxia, TbD1
Citation: Ma R, Farrell D, Gonzalez G, Browne JA, Nakajima C, Suzuki Y and Gordon SV (2022) The TbD1 Locus Mediates a Hypoxia-Induced Copper Response in Mycobacterium bovis. Front. Microbiol. 13:817952. doi: 10.3389/fmicb.2022.817952
Received: 18 November 2021; Accepted: 10 February 2022;
Published: 14 April 2022.
Edited by:
Mireia Coscolla, University of Valencia, SpainReviewed by:
Tiago Beites, Cornell University, United StatesElizabeth Sawyer, University of Westminster, United Kingdom
Susanne Homolka, Research Center Borstel (LG), Germany
Copyright © 2022 Ma, Farrell, Gonzalez, Browne, Nakajima, Suzuki and Gordon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stephen V. Gordon, c3RlcGhlbi5nb3Jkb25AdWNkLmll
†Present address: Gabriel Gonzalez, National Virus Reference Laboratory, University College Dublin, Dublin, Ireland
 Ruoyao Ma
Ruoyao Ma Damien Farrell1
Damien Farrell1 Gabriel Gonzalez
Gabriel Gonzalez John A. Browne
John A. Browne Yasuhiko Suzuki
Yasuhiko Suzuki Stephen V. Gordon
Stephen V. Gordon