
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 06 April 2022
Sec. Evolutionary and Genomic Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.817494
This article is part of the Research Topic Host-Microbe Interaction and Coevolution View all 11 articles
 Jingnan Lv1†
Jingnan Lv1† Jie Zhu1†
Jie Zhu1† Ting Wang2†
Ting Wang2† Xiaofang Xie1
Xiaofang Xie1 Tao Wang1
Tao Wang1 Zhichen Zhu1
Zhichen Zhu1 Liang Chen3,4
Liang Chen3,4 Fengyun Zhong5*
Fengyun Zhong5* Hong Du1*
Hong Du1*Hypervirulent Klebsiella pneumoniae (hvKP) is an evolving infectious pathogen associated with high mortality. The convergence of hypervirulence and multidrug resistance further challenges the clinical treatment options for K. pneumoniae infections. The QseBC two-component system (TCS) is a component of quorum-sensing regulatory cascade and functions as a global regulator of biofilm growth, bacterial motility, and virulence in Escherichia coli. However, the functional mechanisms of QseBC in hvKP have not been reported, and we aim to examine the role of QseBC in regulating virulence in hvKP strain ATCC43816. The CRISPR-Cas9 system was used to construct qseB, qseC, and qseBC knockout in ATCC43816. No significant alterations in the growth and antibiotic susceptibility were detected between wild-type and mutants. The deletion of qseC led to an increase of biofilm formation, resistance to serum killing, and high mortality in the G. mellonella model. RNAseq differential gene expression analysis exhibited that gene-associated biofilm formation (glgC, glgP, glgA, gcvA, bcsA, ydaM, paaF, ptsG), bacterial type VI secretion system (virB4, virB6, virB10, vgrG, hcp), and biosynthesis of siderophore (entC, entD, entE) were significantly upregulated in comparison with the wild-type control. In addition, qseB, ygiW (encode OB-family protein), and AraC family transcriptional regulator IT767_23090 genes showed highest expressions in the absence of QseC, which might be related to increased virulence. The study provided new insights into the functional importance of QseBC in regulating the virulence of hvKP.
Hypervirulent Klebsiella pneumoniae (hvKP) is a gram-negative opportunistic pathogen that has become a worldwide concern due to increasing cases of life-threatening infections in healthy individuals (Harada et al., 2019; Russo and Marr, 2019; Zafar et al., 2019). HvKP is such an invasive strain in part because of its virulence factors that protect it from immunological responses to survive. These virulence factors include capsule, siderophore, fimbriae, biofilm, but the mechanisms of hypervirulence are not fully defined (Choby et al., 2020; Zhu et al., 2021).
Two-component systems (TCS) are dominant bacterial signal-transduction systems (Groisman, 2016). Quorum sensing is a ubiquitous chemical communication process that bacteria use to response to environmental cues (Abisado et al., 2018). The QseBC two-component system is associated with quorum sensing and functions as a regulator of virulence in a wide range of species (Weigel and Demuth, 2016). QseC, a membrane-bound sensor kinase, senses signals (epinephrine/norepinephrine/autoinducer-3) and phosphorylates the response regulator QseB to control the virulence of enterohemorrhagic Escherichia coli (EHEC) (Sperandio et al., 2002; Clarke et al., 2006). Activation of QseBC results in enhanced motility and upregulation of flagellar genes in Salmonella Typhimurium (Bearson and Bearson, 2008). The qseC mutant is attenuated for virulence in rabbits (EHEC) and piglets (S. Typhimurium) (Merighi et al., 2009). Likewise, this function of QseBC had been reported in rare species, such as Edwardsiella tarda, Aggregatibacter actinomycetemcomitans, Aeromonas hydrophila. Disruption of qseB and qseC exhibited impaired flagellar motilities as well as in vivo competitive abilities of Edwardsiella tarda (Wang et al., 2011). In the oral pathogen Aggregatibacter actinomycetemcomitans, activation of QseBC by autoinducer-2 (AI-2) resulted in increased biofilm growth (Novak et al., 2010). Moreover, Khajanchi et al. (2012) found that the decrease in virulence of Aeromonas hydrophilaΔqseB were associated with reduced production of protease and cytotoxic enterotoxin. However, the regulon and function of QseBC in hvKP is currently unknown. In order to explore the role of QseBC in, qseB and qseC single and double knock-out mutants of the hvKP strain ATCC43816 were constructed and studied.
Strains, plasmids and primers used in this study are listed in Supplementary Table 1. The ST439 K2 hypervirulent K. pneumoniae ATCC43816 was obtained from the American Type Culture Collection (ATCC). E. coli DH5α and K. pneumoniae strains were grown aerobically in lysogeny broth (LB) medium (1% tryptone, 0.5% yeast, 1%NaCl). Strains with temperature-sensitive plasmid pCasKP and L-arabinose-inducible expression plasmid pBAD24 were grown at 30°C and in the presence of 0.2% arabinose, respectively. Selective antibiotics were added at the following concentrations: rifampicin 100 μg/ml, apramycin 30 μg/ml, and ampicillin 100 μg/ml.
The hvKP ATCC43816 qseB, qseC, and qseBC mutants were constructed using the CRISPR/Cas9 mediated genome-editing system (Wang et al., 2018). The plasmid pCasKP was electroporated into ATCC43816, followed by selection on agar containing 30 μg/ml apramycin and PCR confirmation. The 20-nt base-pairing region (N20) of an sgRNA was designed through the online web server.1 Primers HindIII-QseB/QseC-N20-For and XbaI-gRNA-Rev were used to amplify the sgRNA fragment from plasmid pSGKP-Rif. The PCR product was subsequently ligated into HindIII and XbaI digested pSGKP-Rif to construct the final pSGKP-Rif with targeted sgRNA (pSGKP-QseB/QseC-N20). We utilized the linear homologous DNA fragment as the repair templates. The linear double-stranded DNA (dsDNA) and the pSGKP-QseB/QseC-N20 plasmid were co-transformed into the L-arabinose-induced pCasKP-positive ATCC43816 to generate the qseB, qseC, and qseBC deletions. The cultures were plated on LB agar plates containing 30 μg/ml apramycin and 100 μg/ml rifampicin, and the mutants were confirmed in selected colonies by both PCR and sequencing.
The qseC gene was amplified from the chromosomal DNA of ATCC43816 using primers QseC-C-For and QseC-C-Rev. The PCR products were then assembled into the multiple cloning site (MCS) of vector pBAD24 using the ClonExpress ® Ultra One Step Cloning Kit (Vazyme Biotech Co., Ltd., China). The recombinant plasmid pBAD24-qseC was transformed into E. coli DH5α, and then primers pBAD24-For and pBAD24-Rev were used to confirm positive colonies on LB agar with 100 μg/ml ampicillin. The qseC mutant was transformed with the pBAD24-qseC plasmid and named as ΔqseC-pCqseC. ATCC43816 and ΔqseC carrying the empty vector pBAD24 were used as control.
Growth curves of ATCC43816, ΔqseB, ΔqseC, ΔqseBC, ATCC43816-pBAD24, ΔqseC-pBAD24, and ΔqseC-pCqseC were determined by subculturing in LB medium and growth. Briefly, overnight cultures of ATCC43816 and mutants were diluted 1:100 in 20 ml of fresh LB broth and grown at 37°C with shaking at 250 rpm. The cell density was detected per hour by OD600 measurements. The experiment was conducted with three independent cultures.
Antimicrobial susceptibility testing was performed using a BD Phoenix™ 100 Automated Microbiology System. Results were interpreted according to the Clinical and Laboratory Standards Institute (CLSI M100, 30th edition) breakpoints.
Biofilm formation was estimated by crystal violet (CV) assay as previously described (Khajanchi et al., 2012; Fu et al., 2018). ATCC43816, ΔqseB, ΔqseC, ΔqseBC, ATCC43816-pBAD24, ΔqseC-pBAD24, and ΔqseC-pCqseC were grown overnight in LB broth and then sub-cultured 1:100 into fresh LB, and grown for 6 h; 20 μl of culture was transferred to a 96-cell polystyrene microtiter plate containing 180 μl brain heart infusion (BHI). Afterward, the plate was incubated at 37°C for 36 h. The medium was then decanted and planktonic cells were washed off gently with sterile water. Adhered biofilms were stained for 30 min at room temperature with 250 μl 1% CV (Beyotime, China), and the purple area was dissolved with 33% glacial acetic acid. Finally, biofilm quantification was measured by using microtiter plate reader (TECAN Infinite M200 Pro NanoQuant) at a wavelength of 595 nm. Absorbance data were obtained from three replicate experiments. In addition, we performed the above procedure by using plastic centrifuge tubes to visualize results of biofilm formation.
Overnight cultures of ATCC43816, ΔqseB, ΔqseC, ΔqseBC, ATCC43816-pBAD24, ΔqseC-pBAD24, and ΔqseC-pCqseC grown in LB were sub-cultured 1:100 and grown to late exponential phase; 250 μl bacteria (105CFU/ml) were incubated with 750 μl serum for 2 h. Samples were serially diluted and plated onto LB agar, and counted colonies were incubated overnight.
G. mellonella larvae were purchased from Tianjin Huiyude Biotech Company, Tianjin, China. Each group included 40 larvae, and bacterial inoculum (105CFU/larva) was administered as 10 μl injection into the left proleg. Saline inoculated larvae were included as negative control. Larvae were incubated at 37°C, in the dark, and scored every 24 h for survival or death (no response to stimulation recorded as dead). The killing assays were performed in triplicate using three different batches of G. mellonella larvae.
The RNA-sequencing was performed by Shanghai Majorbio Bio-pharm Technology Co., Ltd. Total RNA was extracted from the late-exponentially growing ATCC43816, ΔqseB, ΔqseC, and ΔqseBC using TRIzol ® Reagent (Invitrogen) followed by genomic DNA removal using DNase (Takara). RNAseq libraries were prepared according TruSeq™ RNA sample preparation Kit from Illumina (San Diego, CA, United States). The ribosomal RNA (rRNA) was removed using Ribo-Zero Magnetic Kit (epicenter). Paired-end RNA sequencing was performed with the Illumina HiSeq × TEN (2 × 150 bp read length).
The data were analyzed with the free online platform of Majorbio Cloud Platform.2 Raw sequencing reads were trimmed and filtered for high quality reads before being mapped to ATCC43816 genomes (Accession: PRJNA675363) using Bowtie2.3 RSEM4 was used to quantify expression levels of genes and transcripts. Differentially expressed genes (DEGs) were identified using DESeq2 packages5. The differential expression was considered as statistically significant if p < 0.05 and |log2(fold change)| > 2. KEGG and GO enrichment was conducted using KOBAS 2.06 and Goatools.7
Overnight culture of ATCC43816, ΔqseB, ΔqseC, ΔqseBC, ATCC43816-pBAD24, ΔqseC-pBAD24, and ΔqseC-pCqseC were diluted 1:100 in LB media and grown for 6 h to late exponential phase. Cells were collected and treated with Trizol (Sigma) to extract total RNA, followed by DNase treatment as above. The transcript levels of genes were tested by quantitative reverse-transcription PCR (qRT-PCR). HiScript III RT SuperMix kit and Taq Pro Universal SYBR qPCR Master Mix kit (Vazyme Biotech Co., Ltd., China) was used according to the manufacturer’s instructions. Differences in gene expression were normalized with the expression of 16S rRNA and calculated by the 2–ΔΔCt method. All of the qRT-PCR assays were repeated at least three times.
The Student t-test was used to detect the differences between groups in the experiments. Survival curves were assessed by log-rank rest. p < 0.05 was considered statistically significant.
The raw sequencing files in this study are available at the NCBI Sequence Read Archive (SRA) under BioProject PRJNA784457.
To investigate the roles of QseBC in hvKP, ΔqseB, ΔqseC, and ΔqseBC were constructed by CRISPR/Cas9-mediated genome editing. The correct mutants were confirmed by PCR and Sanger sequencing. The colony morphologies of the three mutants on LB agar plates were similar to that of the wild-type. The growth curves of three mutants in LB broth over a period of 24 h were also similar to that of the wild-type strain (Figure 1). Antimicrobial susceptibility testing showed that ATCC43816, ΔqseB, ΔqseC, and ΔqseBC were susceptible to all tested antibacterial drugs. There was no significant difference of the minimum inhibitory concentration (MIC) values in ATCC43816, ΔqseB, ΔqseC, and ΔqseBC against 18 antibiotics examined (Supplementary Table 2). These results indicated that deletion of qseB, qseC, and qseBC did not affect growth and antimicrobial susceptibility of hvKP ATCC43816.
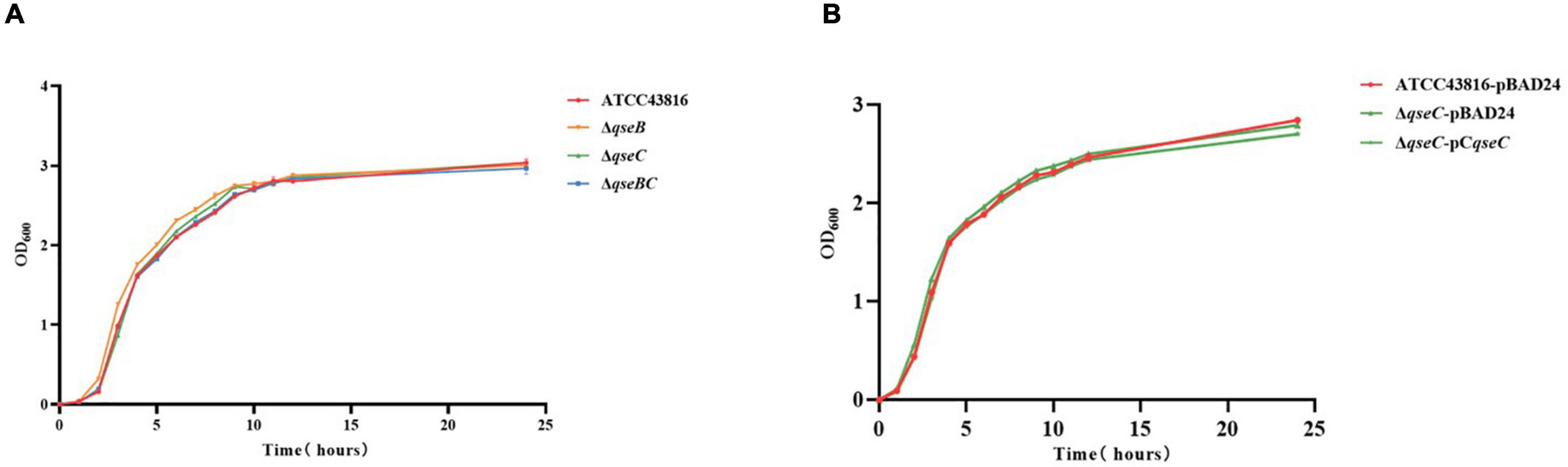
Figure 1. (A) The growth rates of ATCC43816, ΔqseB, ΔqseC and ΔqseBC were similar by measuring the optical density (OD600) at per hour over a period of 24 h. (B) Growth curves of ATCC43816-pBAD24, ΔqseC-pBAD24 and the complement strain ΔqseC-pCqseC.
That QseBC regulated biofilm formation had been reported in E. coli (Sharma and Casey, 2014; Gou et al., 2019). Here, the CV staining assay was applied to examine whether QseBC affected biofilm formation in hvKP ATCC43816. As shown in Figure 2, ΔqseC formed a significantly increased, solid-surface-associated biofilm in the polystyrene plate, with about a twofold increase in the CV staining when compared with the biofilm formed by ATCC43816. Compensation of qseC partially decreased the biofilm formation (Figures 3B,C). Since growth was not altered after qseC-deletion (Figure 1), the biofilm changes were not a result of growth differences. However, ΔqseB and ΔqseBC produced a similar level of biofilms to that from the wild-type strain. These data suggested to us that QseC negatively controlled biofilm formation in hvKP ATCC43816.
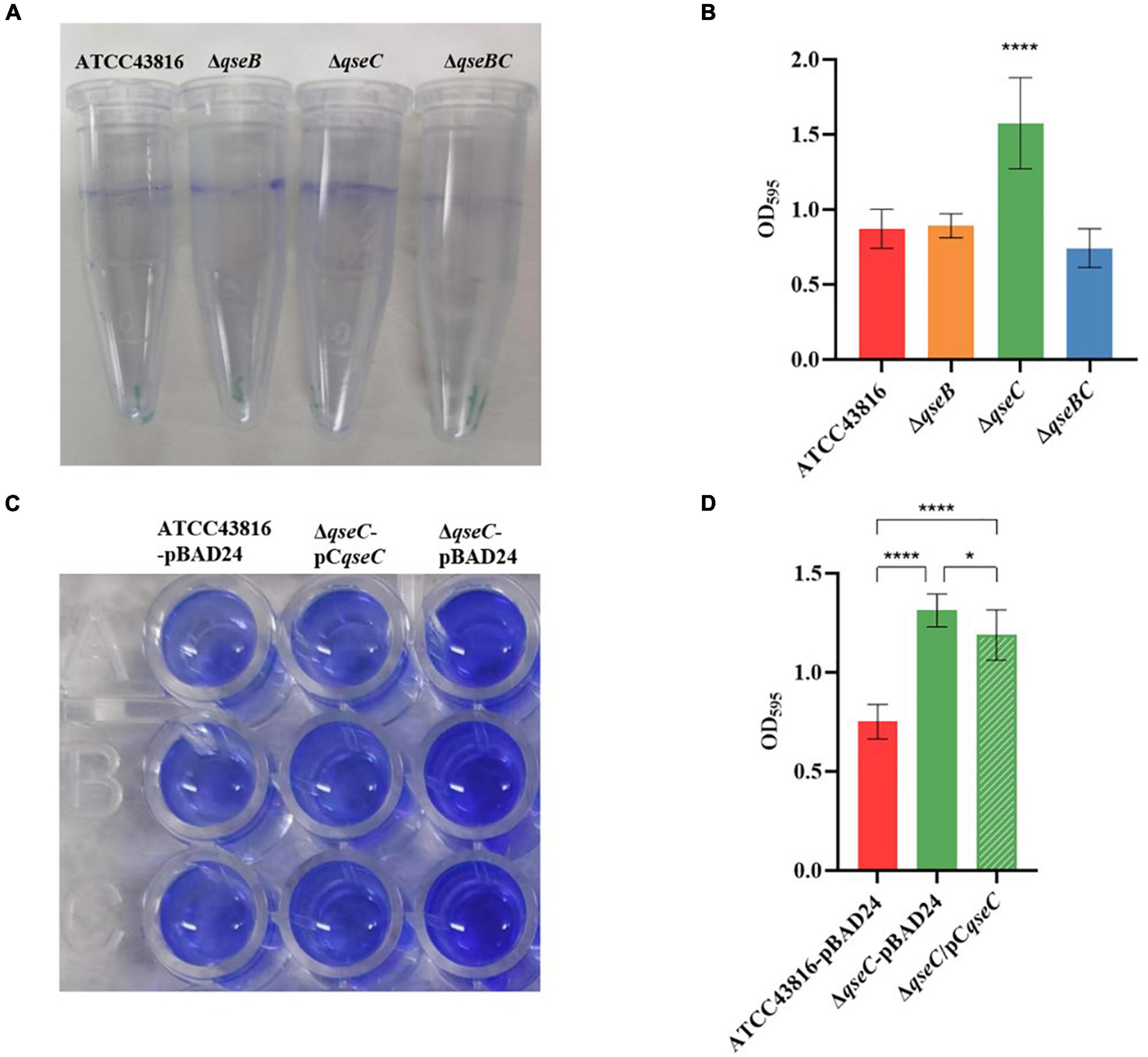
Figure 2. (A) Detection of biofilm formed by ATCC43816, ΔqseB, ΔqseC and ΔqseBC. Image of biofilm stained with 1% CV and adhered on plastic centrifuge tubes. (B) Biofilm were stained with 1% CV and washed with sterile water. The extracted color was dissolved with 33% glacial acetic acid and measured at OD595. Asterisks (****) represent statistically significant differences in biofilm formation between ATCC43816 and its qseCmutant (P < 0.0001, t test). (C) Image of biofilm formed by ATCC43816-pBAD24, ΔqseC-pBAD24 and the complement strain ΔqseC-pCqseC. Biofilm were stained with 1% CV, washed with sterile water and then dissolved with 33% glacial acetic acid. (D) The extracted color was measured at OD595 to show biofilm production. Asterisks (****) represent statistically significant differences in biofilm formation between ATCC43816-pBAD24 and ΔqseC-pBAD24/ΔqseC-pCqseC (P < 0.0001, t test). The biofilm formation of ΔqseC-pCqseC was significantly lower compared with that of ΔqseC-pBAD24 (P = 0.0366, t test).
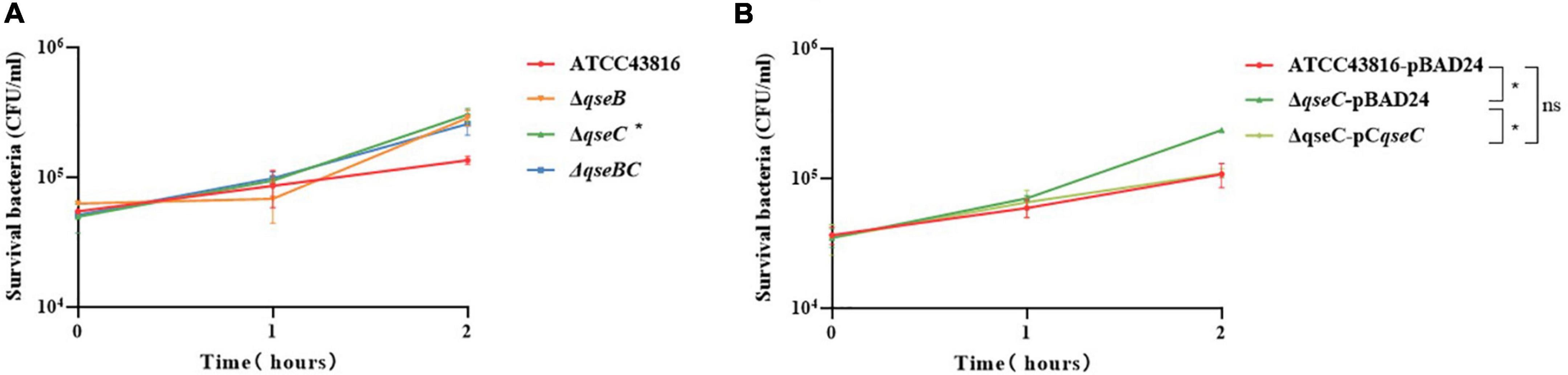
Figure 3. (A) Effect of human serum on ATCC43816, ΔqseB, ΔqseC and ΔqseBC. The bacteria were mixed with serum from healthy human volunteers for 2 hours. ΔqseC had significantly higher resistance when compared to control ATCC43816 (P = 0.0438, two tailed unpaired t test). (B) Effect of human serum on ATCC43816-pBAD24, ΔqseC-pBAD24 and the complement strain ΔqseC-pCqseC. ΔqseC-pBAD24 had significantly higher resistance when compared to control ATCC43816-pBAD24 (P = 0.0327, two tailed unpaired t test).
Serum killing assay was used to study serum resistance in the wild-type and mutants in vitro. Viable counts of both wild-type strain and mutants were > 100% throughout the time period, suggesting that they all had a high level of defense against serum bactericidal activity (Figure 3A). A significant difference was identified between ATCC43816 and ΔqseC. After 2 h of incubation, the mean survival count of ΔqseC showed 3.02 × 105 CFU/ml and exhibited ∼3 times higher than that of ATCC43816 and complemented strain ΔqseC-pCqseC (Figure 3B), while serum-resistant levels of ΔqseB and ΔqseBC had no significant difference with that of ATCC43816 in serum killing assay.
The Galleria mellonella infection model has been suggested to be a valid method for evaluating K. pneumoniae pathogenicity (Insua et al., 2013). To examine the influence of QseBC on in vivo virulence of hvKP ATCC43816, the G. mellonella killing assay was then performed. The qseB and qseBC mutants remained as viable as ATCC43816 in experimentally infected G. mellonella. After 72 h of infection, the survival rate of ΔqseC was about less than 20%. By contrast, the survival rate of ATCC43816, qseB, and qseBC mutants was about 40%, which was significantly higher than that of the ΔqseC (Figure 4A). As shown in Figure 4B, the survival rate was significantly increased in ΔqseC-pCqseC relative to ΔqseC. These data demonstrated that ΔqseC had relatively higher in vivo virulence in comparison to the wild-type strain.
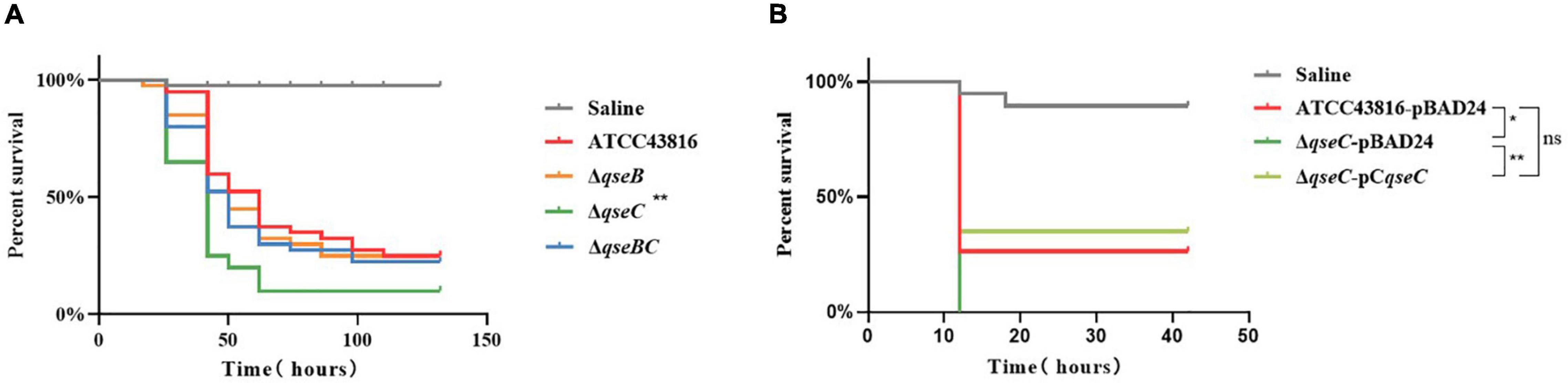
Figure 4. (A) Larvae survival of ATCC43816, ΔqseB, ΔqseC and ΔqseBC. Galleria mellonella were injected with 105 CFU wild-type and mutants (40 larvae per group). The survival of G. mellonella infected with ΔqseC was significantly lower compared with that of the wild-type group (P = 0.0012, Log-rank test). (B) Larvae survival of ATCC43816-pBAD24, ΔqseC-pBAD24 and the complement strain ΔqseC-pCqseC. The survival of G. mellonella infected with ΔqseC-pCqseC was significantly higher than that of ΔqseC-pBAD24 (P = 0.0040, Log-rank test).
To explore possible transcriptomic contributions of QseBC to virulence in hvKP ATCC43816, we examined the gene expression profiles of the late-exponentially growing ATCC43816, ΔqseB, ΔqseC, and ΔqseBC. We identified differentially expressed genes (DEGs) by comparing the RNA-sequencing data with the control strain ATCC43816. DEGs were identified by adjusted p < 0.05 and |log2(fold change)| > 2. Compared to the wild-type strain, the number of DEGs in ΔqseB, ΔqseC, and ΔqseBC groups was 58 (49 upregulated and 9 downregulated genes), 216 (186 upregulated and 30 downregulated genes), and 193 (171 upregulated and 22 downregulated genes), respectively (Supplementary Tables 3–5). In our analysis, we detected 12 DEGs that were shared by ΔqseB and ΔqseC, 13 DEGs shared by ΔqseB and ΔqseBC, 54 DEGs shared by ΔqseC and ΔqseBC, and 21 DEGs shared by all three mutants (Figure 5A). Notably, among the 129 DEGs unique to ΔqseC, IT767_23090 (AraC family transcriptional regulator family) and ygiW showed the highest expression levels (up to 300-fold increase). Compared with ATCC43816, ΔqseC apparently had a significantly high-expression of the qseB gene. The gene expressions of selected genes were confirmed by qRT-PCR and the qRT-PCR showed similar results as the RNA-seq analysis (Figures 6, 7A). The transcript levels of qseB, ygiW, and araC were restored in ΔqseC-pCqseC (Figure 7B).
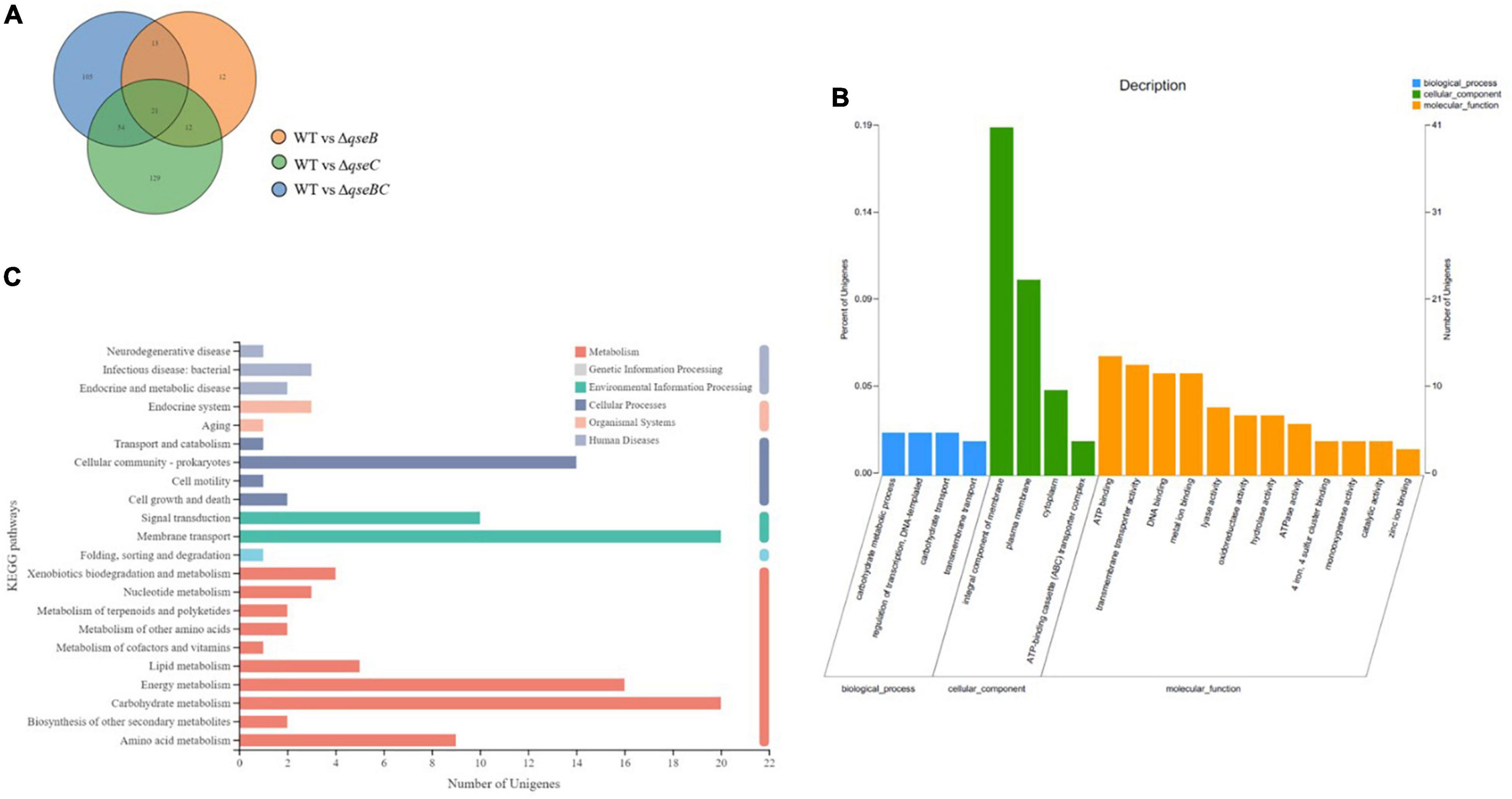
Figure 5. Analysis of total RNA sequencing. (A) The Venn diagram shows the overlapped DEGs numbers across ΔqseB, ΔqseC and ΔqseBC. (B) Significantly enriched GO terms of DGEs in the qseCmutant. (C) Significantly enriched KEGG pathways of DEGs in the qseCmutant.
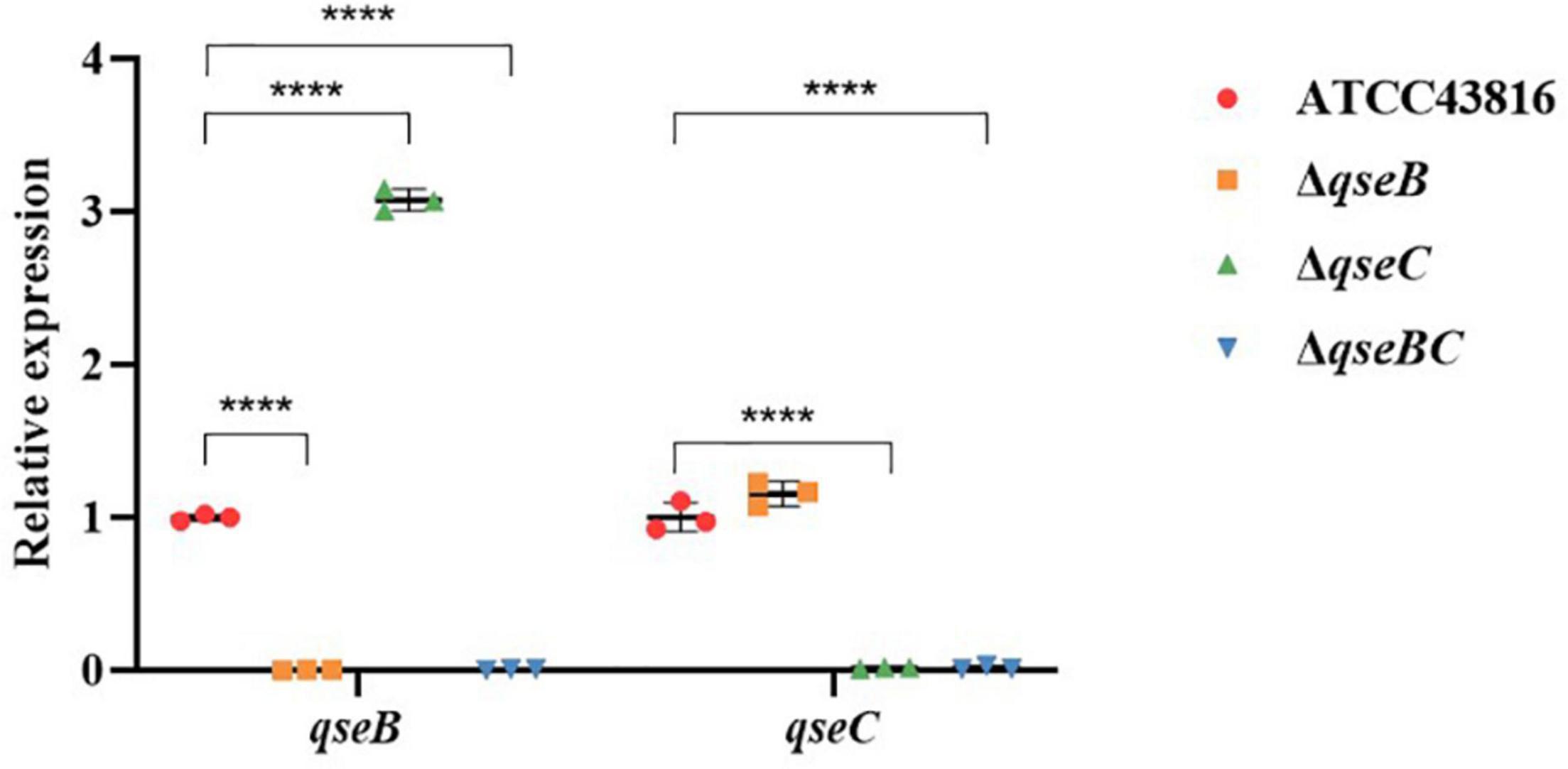
Figure 6. Expression of genes qseB, qseC in ATCC43816, ΔqseB, ΔqseC and ΔqseBC. Cultures of the wild-type and mutants were grown in LB medium to late-exponential phase, and expressions of genes qseC, qseB were measured by qRT-PCR and normalized to the expression in the wild-type strain. Data were mean ± standard deviation of three replicates. Significant expression differences between the wild-type andmutants were determined using two-way ANOVA, ****P < 0.0001.
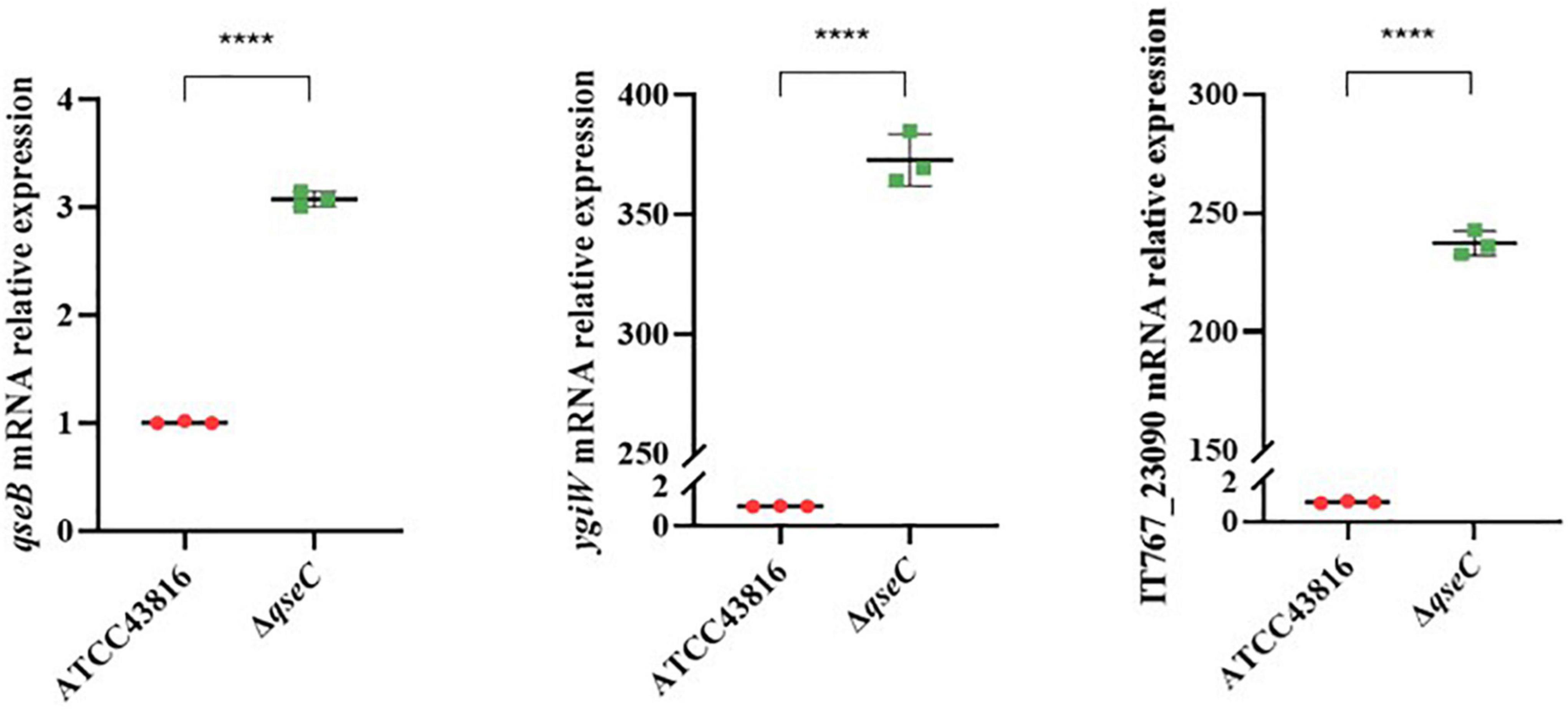
Figure 7. (A) Expression of genes qseB, ygiW, IT767_23090 in ATCC43816 and ΔqseC. Cultures of ATCC43816 and ΔqseC were grown in LB medium to late-exponential phase, and expressions of genes qseB, ygiW, IT767_23090 were measured by qRT-PCR and normalized to the expression in the wild-type strain. Data were mean ± standard deviation of three replicates. Significant expression differences between the wild-type and qseCmutant were determined using t test. ****P < 0.0001. (B) Expression of genes qseB, ygiW, IT767_23090 in ATCC43816-pBAD24, ΔqseC-pBAD24 and the complement strain ΔqseC-pCqseC. Data were mean ± standard deviation of three replicates. Significant expression differences between the wild-type and qseCmutant were determined using t test. *P < 0.05, ***P < 0.001, ****P < 0.0001.
The 216 DEGs of ΔqseC group were performed by Gene Ontology (GO) enrichment analysis. Twenty significantly enriched GO terms from three categories (biological process, cellular component, and molecular function) were shown in Figure 5B. Most DEGs were enriched in two GO terms of cellular component, integral component of membrane and plasma membrane. In addition, DGEs were also subject to KEGG pathway enrichment analysis. Cellular community, signal transduction, membrane transport, energy metabolism, carbohydrate metabolism, and amino acid metabolism were significantly enriched in KEGG pathways (Figure 5C). Among the cellular community pathway, genes associated biofilm formation (glgC, glgP, glgA, gcvA, bcsA, ydaM, paaF, ptsG) in ΔqseC were significantly up-regulated as comparison with the control, which is consistent with the increased biofilm production described above. Moreover, several up-regulated DEGs in pathway involved in virulence, including the bacterial secretion system (virB4, virB6, virB10, vgrG, hcp) and biosynthesis of the siderophore group (entC, entD, entE), were identified. The upregulation of these genes may be associated with the increased virulence observed in the in vivo and in vitro models, which deserve further studies.
In this study, we investigated the effect of QseBC on the pathogenic bacteria virulence and biofilm formation in hvKp strain ATCC43816. The results showed that qseC-deletion increased serum resistance in the serum killing assay, but ΔqseB and ΔqseBC showed no significant difference compared with the wild-type strain. The results suggested that ΔqseC could increase serum resistance and possibly enhances the virulence of hvKP. In order to further test this hypothesis, we conducted the in vivo G. mellonella killing assay. The results showed that there was a significantly higher virulence of the qseC mutant strain when compared to the control ATCC43816. The results of biofilm formation testing also showed that there was a significantly increased biofilm production in ΔqseC, while the levels of biofilm production in ΔqseB and ΔqseBC mutants were similar to those from the wild-type strain. These data suggested that the QseC negatively controlled biofilm formation in hvKP ATCC43816, and ΔqseC, but not ΔqseB and ΔqseBC, had higher serum resistance and in vivo virulence in the G. mellonella model. Several previous studies have demonstrated that QseC has a regulatory function on the motility, biofilm formation, and virulence in some other pathogens. For example, Yang et al. (2014) found that motility was diminished and biofilm formation was decreased in an E. coli qseC mutant. However, our study found that QseC is negatively correlated with the virulence in ATCC43816. Our result appears to be contradicted with the results for E. coli, and an explanation is the TCS QseBC system may behave differently among different species.
In order to further explore the mechanism how QseC negatively regulate the virulence, transcriptional profiling was implemented. The RNA-seq results suggested that in ΔqseC, the gene associated with biofilm formation (glgC, glgP, glgA, gcvA, bcsA, ydaM, paaF, ptsG), bacterial type VI secretion system (virB4, virB6, virB10, vgrG, hcp), and biosynthesis of siderophore (entC, entD, entE) were significantly up-regulated in comparison to the control. It is worth noting that deletion of qseC resulted in the highest overexpression of ygiW and qseB. In the oral pathogen Aggregatibacter actinomycetemcomitans, the qseBC genes are co-expressed with ygiW, and the transcription of ygiW-qseBC operon is directed by the promoter in upstream region of ygiW that contains QseB binding sites (Juárez-Rodríguez et al., 2013, 2014). However, analysis of the ATCC43816 genome (accession no. CP009208) showed that ygiW was transcribed from the opposite strand as qseBC (Weigel and Demuth, 2016), which also suggested that the gene expression regulation for qseBC may be different in A. actinomycetemcomitans and K. pneumoniae. In the absence of the sensor protein QseC, QseB regulator cannot receive the signal of automatic phosphorylation. Therefore, the expression of qseB is supposed to be downregulated due to the lack of autophosphorylation signals from QseC. However, in our study, the expression of qseB was significantly upregulated by approximately four times, indicating that QseB could accept autophosphorylation signals from other sources. For example, Guckes et al. (2013) reported that in uropathogenic E. coli, polymyxin resistance (Pmr) sensor kinase PmrB could active QseB in the absence of QseC. Interestingly, ΔqseB and ΔqseBC have similar levels of biofilm formation, serum resistance, and G. mellonella killing effects to those from the wild-type strain. We therefore hypothesized that in the presence of both QseB and QseC, QseC works as sensor and receives signal, and transfers the phosphate groups to the intracellular response regulator QseB. The phosphorylated QseB directly regulates the expression of iron carrier-related genes, and other virulence-related genes, leading to the changes of bacterial virulence and biofilm formation. In the absence of QseC, QseB could compensatorily receive phosphate groups from other sensor kinase, while the compensatory pathway may be inhibited in the normal condition. When qseB was knocked out (both in ΔqseB and ΔqseBC), the qseB regulatory pathway was interrupted, and ΔqseB and ΔqseBC behaved similarly in the in vivo and in vitro models.
We also analyzed the differential gene expressions between ΔqseB, ΔqseC, ΔqseBC. IT767_23090, belonging to AraC family transcriptional regulator family, was upregulated by 305-fold. In Bacillus subtilis, the AraC family transcriptional regulatory protein PrkC is used to phosphorylate proteins in the metabolic pathway (Libby et al., 2015). In Vibrio cholerae, AraC family transcribed regulated protein ToxT promotes virulence by activating cholera toxin and coordinately regulating the expression of fimbriae (Thomson et al., 2015). The AraC family transcribed regulatory protein RarA leads to multiple drug resistance by regulating the expression of active efflux aggregation in K. pneumoniae (Jiménez-Castellanos et al., 2016). The study by Chen et al. (2015) showed that the AraC family transcription regulator protein RSP increases the expression of Staphylococcus aureus virulence by upregulating the accessory gene regulator (Agr) controlled toxins phenol-soluble modules (PSMs) and alpha-toxin. As such, we suspected that ΔqseC may in part work through upregulating the AraC family transcriptional regulator IT767_23090 to confer to higher virulence in hvKP ATCC43816. However, further studies are needed to determine the role of IT767_23090 in the increased virulence in ΔqseC.
In summary, our data showed that ΔqseC leads to the high expression of qseB, ygiW, IT767_23090, and genes associated biofilm formation, bacterial type VI secretion system, and biosynthesis of siderophore, which might contribute to the increased of virulence and biofilm formation. This research provides new insights into the role of QseBC in biofilm formation and virulence of hvKP ATCC43816.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
JL, JZ, and TW contributed equally to this work. The order of co-first authors was determined by discussion and mutual agreement between three co-first authors. HD and FZ are corresponding authors. All authors contributed to the article and approved the submitted version.
This study was supported by the grants from the Natural Science Fund of China (82002204), the Science Foundation of Jiangsu Province Health Department (ZDB2020014), the Science Foundation of Suzhou Health Department (LCZX202106), and the Medical and Health Science and Technology Plan Project of Suzhou National New and Hi-Tech Industrial Development Zone (2019Q013).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.817494/full#supplementary-material
Abisado, R. G., Benomar, S., Klaus, J. R., Dandekar, A. A., and Chandler, J. R. (2018). Bacterial quorum sensing and microbial community interactions. mBio 9:e02331-17.
Bearson, B. L., and Bearson, S. M. (2008). The role of the QseC quorum-sensing sensor kinase in colonization and norepinephrine-enhanced motility of Salmonella enterica serovar Typhimurium. Microb. Pathog. 44, 271–278. doi: 10.1016/j.micpath.2007.10.001
Chen, Z., Wang, Y., Tian, L., Zhu, X., Li, L., Zhang, B., et al. (2015). First report in China of Enterobacteriaceae clinical isolates coharboring blaNDM-1 and blaIMP-4 drug resistance genes. Microb. Drug Resist. 21, 167–170. doi: 10.1089/mdr.2014.0087
Choby, J. E., Howard-Anderson, J., and Weiss, D. S. (2020). Hypervirulent Klebsiella pneumoniae - clinical and molecular perspectives. J. Intern. Med. 287, 283–300.
Clarke, M. B., Hughes, D. T., Zhu, C., Boedeker, E. C., and Sperandio, V. (2006). The QseC sensor kinase: a bacterial adrenergic receptor. Proc. Natl. Acad. Sci. U. S. A. 103, 10420– 10425.
Fu, L., Huang, M., Zhang, X., Yang, X., Liu, Y., Zhang, L., et al. (2018). Frequency of virulence factors in high biofilm formation bla(KPC-2) producing Klebsiella pneumoniae strains from hospitals. Microb. Pathog. 116, 168–172. doi: 10.1016/j.micpath.2018.01.030
Gou, Y., Liu, W., Wang, J. J., Tan, L., Hong, B., Guo, L., et al. (2019). CRISPR-Cas9 knockout of qseB induced asynchrony between motility and biofilm formation in Escherichia coli. Can. J. Microbiol. 65, 691–702. doi: 10.1139/cjm-2019-0100
Groisman, E. A. (2016). Feedback control of two-component regulatory systems. Annu. Rev. Microbiol. 70, 103–124. doi: 10.1146/annurev-micro-102215-095331
Guckes, K. R., Kostakioti, M., Breland, E. J., Gu, A. P., Shaffer, C. L., Martinez, C. R. III, et al. (2013). Strong cross-system interactions drive the activation of the QseB response regulator in the absence of its cognate sensor. Proc. Natl. Acad. Sci. U. S. A. 110, 16592–16597. doi: 10.1073/pnas.1315320110
Hao, M., He, Y., Zhang, H., Liao, X. P., Liu, Y. H., Sun, J., et al. (2020). crispr-cas9-mediated carbapenemase gene and plasmid curing in carbapenem-resistant Enterobacteriaceae. Antimicrob. Agents Chemother. 64:e00843-20. doi: 10.1128/AAC.00843-20
Harada, S., Aoki, K., Yamamoto, S., Ishii, Y., Sekiya, N., Kurai, H., et al. (2019). Clinical and molecular characteristics of Klebsiella pneumoniae isolates causing bloodstream infections in Japan: occurrence of hypervirulent infections in health care. J. Clin. Microbiol. 57:e01206-19. doi: 10.1128/JCM.01206-19
Insua, J. L., Llobet, E., Moranta, D., Pérez-Gutiérrez, C., Tomás, A., Garmendia, J., et al. (2013). Modeling Klebsiella pneumoniae pathogenesis by infection of the wax moth Galleria mellonella. Infect. Immun. 81, 3552–3565. doi: 10.1128/IAI.00391-13
Jiménez-Castellanos, J. C., Wan Ahmad Kamil, W. N., Cheung, C. H., Tobin, M. S., Brown, J., Isaac, S. G., et al. (2016). Comparative effects of overproducing the AraC-type transcriptional regulators MarA, SoxS, RarA and RamA on antimicrobial drug susceptibility in Klebsiella pneumoniae. J. Antimicrob. Chemother. 71, 1820–1825. doi: 10.1093/jac/dkw088
Juárez-Rodríguez, M. D., Torres-Escobar, A., and Demuth, D. R. (2013). ygiW and qseBC are co-expressed in Aggregatibacter actinomycetemcomitans and regulate biofilm growth. Microbiology 159, 989–1001. doi: 10.1099/mic.0.066183-0
Juárez-Rodríguez, M. D., Torres-Escobar, A., and Demuth, D. R. (2014). Transcriptional regulation of the Aggregatibacter actinomycetemcomitans ygiW-qseBC operon by QseB and integration host factor proteins. Microbiology 160, 2583–2594. doi: 10.1099/mic.0.083501-0
Khajanchi, B. K., Kozlova, E. V., Sha, J., Popov, V. L., and Chopra, A. K. (2012). The two-component QseBC signalling system regulates in vitro and in vivo virulence of Aeromonas hydrophila. Microbiology 158, 259–271. doi: 10.1099/mic.0.051805-0
Libby, E. A., Goss, L. A., and Dworkin, J. (2015). The Eukaryotic-like Ser/Thr Kinase PrkC regulates the essential WalRK two-component system in Bacillus subtilis. PLoS Genet. 11:e1005275. doi: 10.1371/journal.pgen.1005275
Merighi, M., Septer, A. N., Carroll-Portillo, A., Bhatiya, A., Porwollik, S., Mcclelland, M., et al. (2009). Genome-wide analysis of the PreA/PreB (QseB/QseC) regulon of Salmonella enterica serovar Typhimurium. BMC Microbiol. 9:42. doi: 10.1186/1471-2180-9-42
Novak, E. A., Shao, H., Daep, C. A., and Demuth, D. R. (2010). Autoinducer-2 and QseC control biofilm formation and in vivo virulence of Aggregatibacter actinomycetemcomitans. Infect Immun 78, 2919–2926. doi: 10.1128/IAI.01376-09
Russo, T. A., and Marr, C. M. (2019). Hypervirulent Klebsiella pneumoniae. Clin. Microbiol. Rev. 32:e00001-19.
Sharma, V. K., and Casey, T. A. (2014). Escherichia coli O157:H7 lacking the qseBC-encoded quorum-sensing system outcompetes the parental strain in colonization of cattle intestines. Appl. Environ. Microbiol. 80, 1882–1892. doi: 10.1128/AEM.03198-13
Sperandio, V., Torres, A. G., and Kaper, J. B. (2002). Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol. Microbiol. 43, 809–821. doi: 10.1046/j.1365-2958.2002.02803.x
Thomson, J. J., Plecha, S. C., and Withey, J. H. (2015). A small unstructured region in Vibrio cholerae ToxT mediates the response to positive and negative effectors and ToxT proteolysis. J. Bacteriol. 197, 654–668. doi: 10.1128/JB.02068-14
Wang, X., Wang, Q., Yang, M., Xiao, J., Liu, Q., Wu, H., et al. (2011). QseBC controls flagellar motility, fimbrial hemagglutination and intracellular virulence in fish pathogen Edwardsiella tarda. Fish Shellfish Immunol. 30, 944–953. doi: 10.1016/j.fsi.2011.01.019
Wang, Y., Wang, S., Chen, W., Song, L., Zhang, Y., Shen, Z., et al. (2018). CRISPR-Cas9 and CRISPR-assisted cytidine deaminase enable precise and efficient genome editing in Klebsiella pneumoniae. Appl. Environ. Microbiol. 84:e01834-18. doi: 10.1128/AEM.01834-18
Weigel, W. A., and Demuth, D. R. (2016). QseBC, a two-component bacterial adrenergic receptor and global regulator of virulence in Enterobacteriaceae and Pasteurellaceae. Mol. Oral Microbiol. 31, 379–397. doi: 10.1111/omi.12138
Yang, K., Meng, J., Huang, Y. C., Ye, L. H., Li, G. J., Huang, J., et al. (2014). The role of the QseC quorum-sensing sensor kinase in epinephrine-enhanced motility and biofilm formation by Escherichia coli. Cell Biochem. Biophys. 70, 391–398. doi: 10.1007/s12013-014-9924-5
Zafar, S., Hanif, S., Akhtar, H., and Faryal, R. (2019). Emergence of hypervirulent K. pneumoniae causing complicated UTI in kidney stone patients. Microb. Pathog. 135:103647. doi: 10.1016/j.micpath.2019.103647
Keywords: hypervirulent, Klebsiella pneumoniae, QseBC, two-component system, biofilm formation
Citation: Lv J, Zhu J, Wang T, Xie X, Wang T, Zhu Z, Chen L, Zhong F and Du H (2022) The Role of the Two-Component QseBC Signaling System in Biofilm Formation and Virulence of Hypervirulent Klebsiella pneumoniae ATCC43816. Front. Microbiol. 13:817494. doi: 10.3389/fmicb.2022.817494
Received: 18 November 2021; Accepted: 26 January 2022;
Published: 06 April 2022.
Edited by:
Wei Huang, Johns Hopkins University, United StatesReviewed by:
Yiquan Zhang, Jiangnan University, ChinaCopyright © 2022 Lv, Zhu, Wang, Xie, Wang, Zhu, Chen, Zhong and Du. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Du, aG9uZ19kdUAxMjYuY29t; Fengyun Zhong, emZ5c3pAMTI2LmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.