
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 12 April 2022
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.815600
This article is part of the Research Topic Role of Antimicrobials and Surfaces in Shaping the Tolerance, Evolution, Assembly and Biophysics of Bacterial Biofilms View all 6 articles
 Weiliang Zeng1
Weiliang Zeng1 Luozhu Feng2
Luozhu Feng2 Changrui Qian3
Changrui Qian3 Tao Chen1
Tao Chen1 Sipei Wang1
Sipei Wang1 Ying Zhang2
Ying Zhang2 Xiangkuo Zheng1
Xiangkuo Zheng1 Lingbo Wang1
Lingbo Wang1 Shixing Liu1
Shixing Liu1 Tieli Zhou1*
Tieli Zhou1* Yao Sun1*
Yao Sun1*Daptomycin is a last-line antibiotic used in the treatment of multidrug-resistant Enterococcus faecium infections. Alarmingly, daptomycin-resistant E. faecium isolates have emerged. In this study, we investigated the evolution and mechanisms of daptomycin resistance in clinical E. faecium isolates and the corresponding acquisition of collateral sensitivity (CS) as an evolutionary trade-off. We evolved daptomycin resistance in six daptomycin-susceptible E. faecium isolates to obtain daptomycin-resistant mutants. The six E. faecium strains successfully acquired high-level resistance to daptomycin in vitro, but this led to fitness costs in terms of growth, in vitro competition, and virulence. Mutations in liaFSR, yycFG, and cls; increased surface positive charge; thicker cell walls; and elevated expression of dltABCD and tagGH were observed in daptomycin-resistant mutants. Surprisingly, we observed the emergence of CS in SC1762 isolates after the induction of daptomycin resistance. Compared with parental strains, the SC1174-D strain (i.e., daptomycin-resistant mutant of SC1174; non-CS) showed significantly upregulated expression of the vanA gene cluster. However, in SC1762-D (i.e., daptomycin-resistant mutant of SC1762), all vanA cluster genes except the vanX gene were obviously downregulated. Further in silico analyses revealed that an IS1216E-based composite transposon was generated in SC1762-D, and it disrupted the vanH gene, likely affecting the structure and expression of the vanA gene cluster and resulting in resensitization to glycopeptides. Overall, this study reports a novel form of CS between daptomycin and glycopeptides in E. faecium. Further, it provides a valuable foundation for developing effective regimens and sequential combinations of daptomycin and glycopeptides against E. faecium.
Enterococcus faecium is a ubiquitous and opportunistic pathogen. It is a member of the ESKAPE (E. faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) family of pathogens, which are well-known for causing frequent and hard-to-treat healthcare-associated infections (Pendleton et al., 2013; Mulani et al., 2019). E. faecium can cause various infections in hospital and community settings, ranging from urinary tract infections (UTIs) to severe life-threatening infections such as bacteremia (Contreras et al., 2019). It has been reported that E. faecium infections are becoming increasingly common (Li et al., 2017). Vancomycin and teicoplanin were thought to be the most effective glycopeptides for treating Enterococcal infections. However, since vancomycin-resistant E. faecium (VREfm) was first identified in England and France in 1986 (O’Driscoll and Crank, 2015), such drug-resistant strains have been encountered worldwide and now represent a great public health threat due to their high mortality rates (Arias and Murray, 2012).
In 2003, daptomycin (DAP)—a cyclic lipopeptide antibacterial agent—was approved by the Food and Drug Administration for the treatment of complex skin and soft tissue infections, due to its rage of activity against methicillin-resistant S. aureus (MRSA) and vancomycin-resistant Enterococci (VRE) (Frankenfeld et al., 2018). However, with its extensive use in clinical settings, the number of treatment failures reported with DAP has been increasing (Jones et al., 2008; Lellek et al., 2015; Ji et al., 2020).
The mechanisms underlying DAP resistance in both S. aureus and Enterococci have been described. Studies have shown that gene mutations are responsible for DAP resistance. These include mutations in genes encoding cell-envelope homeostasis proteins, liaFSR (two-component regulatory system encoding genes), yycFG (i.e., three-component regulatory system encoding genes), and genes encoding enzymes associated with cell membrane phospholipid metabolism, cls (cardiolipin synthase), cfa (cyclopropane fatty acid synthetase), and mprF (multiple peptide resistance factor) (Munita et al., 2012; Werth et al., 2014; Lellek et al., 2015; Bayer et al., 2016; Prater et al., 2019; Ji et al., 2020). Changes in surface charge, membrane depolarization, and cell-wall thickness also contribute to DAP resistance (Jones et al., 2008; Arias et al., 2011; Miller et al., 2016; Kang et al., 2017). Although the mechanisms underlying DAP resistance are diverse, the dynamic evolution of acquired DAP resistance and the related mechanisms remain to be fully elucidated.
Under antimicrobial selection pressure, bacteria often make adaptive changes depending on the antibiotic environment. These changes also represent the evolutionary trade-off that bacteria make to acquire antibiotic resistance. It has been demonstrated that the acquisition of resistance to a specific antimicrobial can simultaneously increase or decrease a bacterium’s level of resistance to another antimicrobial. This process is called cross-resistance (CR) or collateral sensitivity (CS) (Roemhild et al., 2020). Notably, researchers have suggested that CS can be exploited to slow down or even reverse antibiotic resistance (Imamovic et al., 2018; Lázár et al., 2018). Hence, it is necessary to fully understand the mechanisms of CS, especially for a last-line-of-defense drug such as DAP. Nevertheless, there are few reports on CS for DAP. Canfield et al. (2021) reported that the sensitization of E. faecium to antimicrobials that act on the cell wall (e.g., ceftriaxone and ampicillin) and membrane (e.g., DAP) after the acquisition of phage resistance in E. faecium infected with lytic bacteriophages. Moreover, DAP can reverse rifampicin resistance in some strains of VRE through mechanisms that are currently unclear (Rand et al., 2007). Interestingly, we also observed the emergence of CS between DAP and glycopeptides in E. faecium after exposure to DAP. The aim of this study was to explore the evolution of DAP resistance and the corresponding CS in E. faecium with the aim of providing a basis for formulating more reasonable clinical treatment strategies and promoting the development of new antimicrobial combinations based on CS.
This study used strains obtained from the First Affiliated Hospital of Wenzhou Medical University. The Institutional Ethics Committee of the First Affiliated Hospital of Wenzhou Medical University exempted the study from ethics review and approval because the study was observational in nature and mainly focused on bacteria, with no patient-related interventions.
We collected and identified six DAP-susceptible (DAP-S) strains (i.e., SC1174, SC1379, SC1762, SC1543, SC1706, and SC1726), which were isolated between 2017 and 2018 from the First Affiliated Hospital of Wenzhou Medical University. These isolates were used as parental strains to generate DAP-resistant (DAP-R) mutants in vitro. Considering that calcium is necessary for the antimicrobial activity of DAP, both calcium (50 μg/ml) and magnesium ions (12.5 μg/ml) were added to Mueller-Hinton broth and Luria-Bertani (LB) broth in this study.
To examine the evolution of DAP resistance in E. faecium, an adaptive laboratory evolution experiment in vitro was performed in six parental strains, with slight modifications, and DAP-R mutants were obtained (Bhardwaj et al., 2018). The experimental design of the evolution experiment was shown in Supplementary Figure 1. Specifically, a single pure colony of the DAP-S parental strains was randomly picked, inoculated in 3 ml fresh LB broth, and allowed to grow to the logarithmic phase. Then, 30 μl of the overnight culture was transferred to 2.97 ml fresh LB broth containing graded concentrations of DAP: 1/2 × minimal inhibitory concentration (MIC), 1 × MIC, 2 × MIC, and 4 × MIC. Subsequently, the cultures were incubated at 37°C overnight without shaking. The tube with visible growth at the highest DAP concentration was used as the inoculum, which was cultured for 3 days in fresh LB broth with the DAP concentration, and then the inoculum was exposed to a higher concentration. The stability of the DAP-R mutants was confirmed using serial cultures in fresh LB broth without DAP (Li et al., 2017). The continuous passage of the parental strains in LB broth without DAP served as the control for this set of experiments.
According to the guidelines of the Clinical and Laboratory Standards Institute [CLSI] (2020), the MIC of DAP and commonly used antibiotics was determined for DAP-S and DAP-R strains using the broth microdilution and agar dilution methods. E. faecalis ATCC 29212 was used as the control strain. The results were also interpreted based on CLSI standards. The experiment was repeated in triplicate.
Genomic DNA was extracted from the experimental strains using a Biospin bacterial genome DNA extraction kit (Shanghai Boyun Biotech Co., Ltd. Shanghai, China.) and used as the template in polymerase chain reaction (PCR). DAP-resistance genes (i.e., liaFSR, yycFG, cls, and cfa) and glycopeptide-resistance gene (i.e., vanRSHAXYZ) were amplified using specific primers (Supplementary Table 1; Munita et al., 2012; Werth et al., 2014; Lellek et al., 2015). The PCR products were sequenced. In addition to the genome of the parental strains, nucleotide sequences were also compared with the genome of the standard strains E. faecium DO (accession number: CP003583) and E. faecium BM4147 (accession number: M97297) using BLAST1 (Qin et al., 2012; Cha et al., 2013).
The cytochrome C binding assay was performed using the parental strains and their mutants (Kang et al., 2017). Briefly, bacterial suspensions were allowed to regrow to the logarithmic phase after overnight culture. Then, cells were washed twice with 3-(N-Morpholino) propanesulfonic acid sodium salt (MOPS, pH = 7.0) after centrifugation, and the bacterial suspension was adjusted to an OD600 of 1.0. Cytochrome C was prepared in MOPS (5 mg/ml, pH = 7.0), and then, 100 μl of this solution was added to 900 μl of the bacterial suspension. After 30 min of incubation at room temperature, samples were centrifuged at 12,000 g for 5 min. The OD530 was measured spectrophotometrically. Lower levels of free cytochrome C in the supernatant indicated a greater net positive charge on the bacterial surface. The experiment was repeated at least three times.
Two parental strains (SC1174 and SC1762) and their evolved strains (SC1174-D and SC1762-D) were selected to measure the cell-wall thickness using transmission electron microscopy (TEM), as described previously (Arias et al., 2011). After overnight culture in LB broth, 1 ml of the bacterial suspension was pelleted and washed three times using 0.1 M Millonig’s phosphate buffer (pH = 7.0). The pellet was then resuspended in 1 ml glutaraldehyde and further processed in the Electron Microscope Room of Wenzhou Medical University. For each tested strain, the cell-wall thickness of at least 25 cells was measured at 40,000× and 60,000× magnification. Mean differences were compared using the Student’s t-test, with P-values of <0.05 indicating statistical significance.
The efflux pump inhibition test was performed to identify the relationship between efflux pumps and CS. The tested efflux pump inhibitors were as follows: carbonyl cyanide m-chlorophenylhydrazone (CCCP, 6 μg/ml), Phe-Arg-β-naphthylamide (PAβN) (20 μg/ml), omeprazole (OME, 100 μg/ml), reserpine (RES, 20 μg/ml), and chlorpromazine (CHL, 20 μg/ml). The MICs of E. faecium with or without the efflux pump inhibitors were compared to measure the efflux activities. Based on the literature, a positive phenotype for a particular strain was indicated by an MIC decrease of ≥4 after the supplementation of the efflux pump inhibitor (vs. without the efflux pump inhibitor) (Lin et al., 2020).
Total RNA was extracted with an RNeasy Mini Kit (QIAGEN, Valencia, CA, United States). Using a cDNA synthesis kit (Takara, Shiga, Japan), purified RNA was reverse transcribed to cDNA, which was used as a template for quantitative real-time PCR (qRT-PCR). The expression levels of dltABCD, tagGH, and vanRSHAXYZ genes were detected using their respective specific primers (Supplementary Table 1). The housekeeping gene 16S rRNA was used as the corresponding internal control, and transcript levels were calculated using the 2–Δ Δ Ct method. At least two independent runs were performed for each cDNA sample.
The genomic DNA of SC1762 and SC1762-D was extracted using an AxyPrep bacterial genomic DNA miniprep kit (Axygen Scientific, Union City, CA, United States). Whole-genome sequencing (WGS) was performed using a standard run on the Illumina HiSeq 2500 and Pacific Bioscience (PacBio) systems by the Shanghai Personal Biotechnology Co., Ltd. (Shanghai, China). Complete genomes were assembled by Canu (Koren et al., 2017) using long reads and then improved by Pilon (Walker et al., 2014) using short reads. Genomic sequences were annotated using Prokka (Seemann, 2014) and corrected using BLAST searches against the UniProtKB/Swiss-Prot, RefSeq, ISfinder (Siguier et al., 2006), and CARD (Jia et al., 2017) databases. Gene organization diagrams were generated using the Python script and modified with Inkscape2. The raw data from WGS were submitted and are accessible under NCBI accession numbers CP085894-CP085905 (SC1762) and CP085906-CP085917 (SC1762-D).
The growth rate of DAP-R mutants and the parental strains were investigated (Li et al., 2017). After growth in LB broth at 37°C for 24 h, cultures of these strains were diluted to an OD600 of 0.01 and incubated at 37°C for 24 h with agitation at 200 rpm. Growth curves were generated by plotting the changes in the OD600 value over time. The experiment was repeated in triplicate, and the average values were used for estimating growth parameters.
In vitro competition experiments were performed in triplicate to measure the fitness cost between parental strains and their mutants (Li et al., 2017). Cultures of exponentially growing cells (both parental strains and mutant strains) were adjusted to a dilution of 1 × 103 colony forming unit (CFU)/ml, and then equal volumes of the parental and mutant strains were combined. Subsequently, 1 ml of the mixture was added to 19 ml LB broth and cultured at 37°C with agitation at 200 rpm for 24 h, which corresponds to approximately 20 cell generations. Serial 10-fold dilutions were plated in duplicate on DAP-free LB agar and LB agar containing 4 μg/ml DAP (this concentration inhibited the growth of all parental strains). After overnight incubation at 37°C for 24 h, the CFUs of the DAP-R colonies and parental strains were counted. The competition index (CI) indicates the adaptive difference and was defined as the ratio between the number of CFUs from the parental strains and that from the mutants. The CI values were calculated for each independent competition assay, and the median values were obtained.
Quantifications of biofilm-bound crystal violet were performed for parental strains and evolved DAP-R mutants as previously reported, with slight modifications (Jung et al., 2019). For the biofilm formation assays, isolates were incubated at 37°C with shaking at 200 rpm overnight. Then, culture suspensions were diluted in fresh LB broth (1:100) and inoculated into a 96-well polystyrene microtiter plate (200 μl per well), which was incubated at 37°C for 48 h. After incubation, planktonic cells were removed, and 200 μl of 0.1% crystal violet was added to each well. The plate was incubated for 20 min at 37°C before it was washed twice with sterile water. Finally, 95% ethanol was used to dissolve the stained biofilm, and the absorbance at OD600 was measured for quantification. Untreated wells supplemented with sterile LB broth served as the control. All experiments were conducted in triplicate.
Galleria mellonella larvae were used as infection models to compare the virulence of parental strains and evolved DAP-R mutants; a slightly modified version of a previously described method was used (Taglialegna et al., 2019). Twelve caterpillars weighing 200–250 mg were randomly selected for testing each isolate. Then, 10 μl of bacterial suspension containing 108 CFU/ml or normal saline (NS) was injected into the last left proleg of the G. mellonella larvae using a 25 μl Hamilton precision syringe. Worms were incubated at 37°C in the dark, and their survival rates were observed and recorded every 24 h over a period of 6 days. The survival rates were assessed using the Kaplan–Meier analysis and the log-rank test. All experiments were performed in three independent replicates.
The paired Student’s t-test was performed using SPSS software version 26.0. The statistical analysis of growth rate was performed using the GraphPad Prism 8.0 software and one-way analysis of variance (ANOVA), and the log-rank test was used to analyze the survival rates of G. mellonella larvae infection models. P-values of < 0.05 were considered significant.
Daptomycin-resistant mutants were generated successfully through the adaptive laboratory evolution. The evolved DAP resistance remained stable after serial passages of E. faecium in a DAP-free growth medium. As shown in Figure 1, the DAP resistance of E. faecium increased with an increase in the induction concentration of DAP and the induction time. On the fourth day of DAP induction, DAP-R mutants were screened. After a 36-day induction period, the six mutants exhibited a 16–256× increase in the DAP MIC compared with their wild-type counterparts (Table 1). In summary, the DAP-R E. faecium mutants could be rapidly selected through serial passaging across a sub-lethal gradient of DAP.
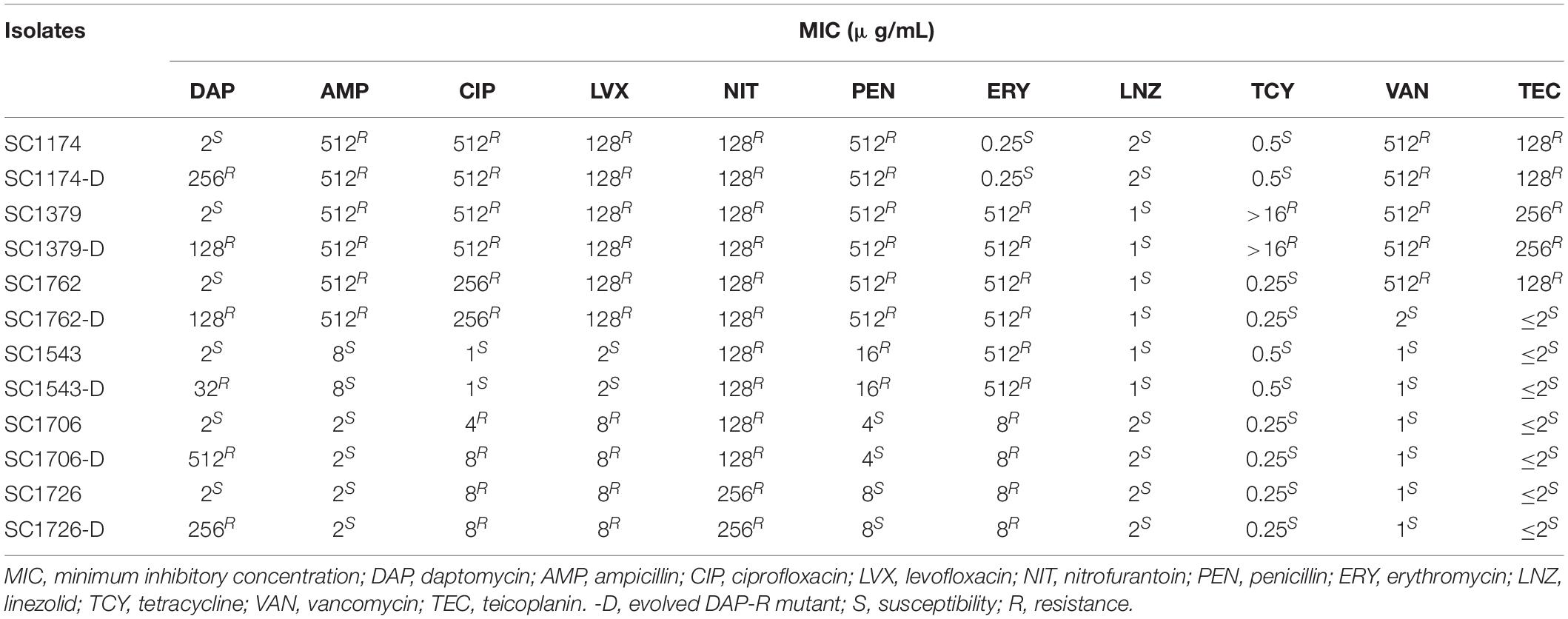
Table 1. Activity of daptomycin and other antibiotics against parental and mutant strains of Enterococcus faecium.
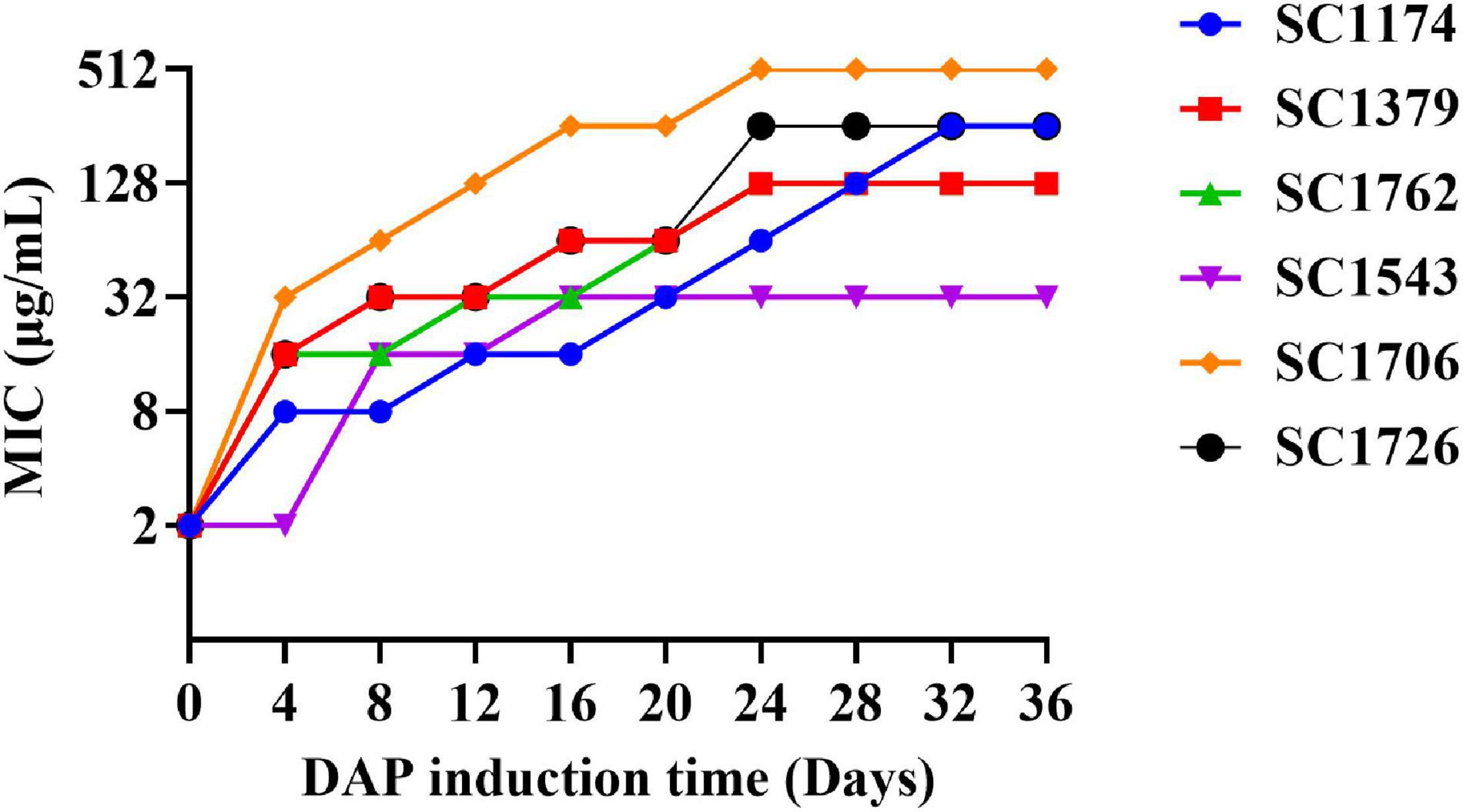
Figure 1. Enterococcus faecium adapts to daptomycin. The in vitro evolution of reduced daptomycin (DAP) susceptibility in evolved DAP-resistant (DAP-R) E. faecium through serial passages in increasing concentrations of DAP for a period of 36 days.
To determine whether the bacterial resistance phenotype can change after DAP induction, we first determined the susceptibility of the mutant strains to clinically relevant antibiotics (Table 1). Compared with the parental strain SC1762, the mutant strain SC1762-D exhibited a 64× increase in the DAP MIC, becoming clinically resistant to DAP. Interestingly, it also exhibited a 256× and 64× decrease in the MIC of vancomycin and teicoplanin, becoming clinically sensitive to these antibiotics, indicating CS between DAP and glycopeptides in E. faecium SC1762 and SC1762-D. Except for SC1762 and SC1762-D, as shown in Table 1, the antibiotic resistance phenotype of the evolved DAP-R strains was consistent with that of the corresponding parental strains.
We evaluated the mechanisms of DAP resistance in the evolved mutants. Results of PCR amplification showed that different strains had different gene mutation patterns (Figure 2 and Supplementary Figure 2). Mutations were identified in liaFSR, yycFG, and cls genes (except cfa), and the mutation sites were not exactly the same in different DAP-R mutants. Further, the mutation frequency for the same gene was also different in each mutant.
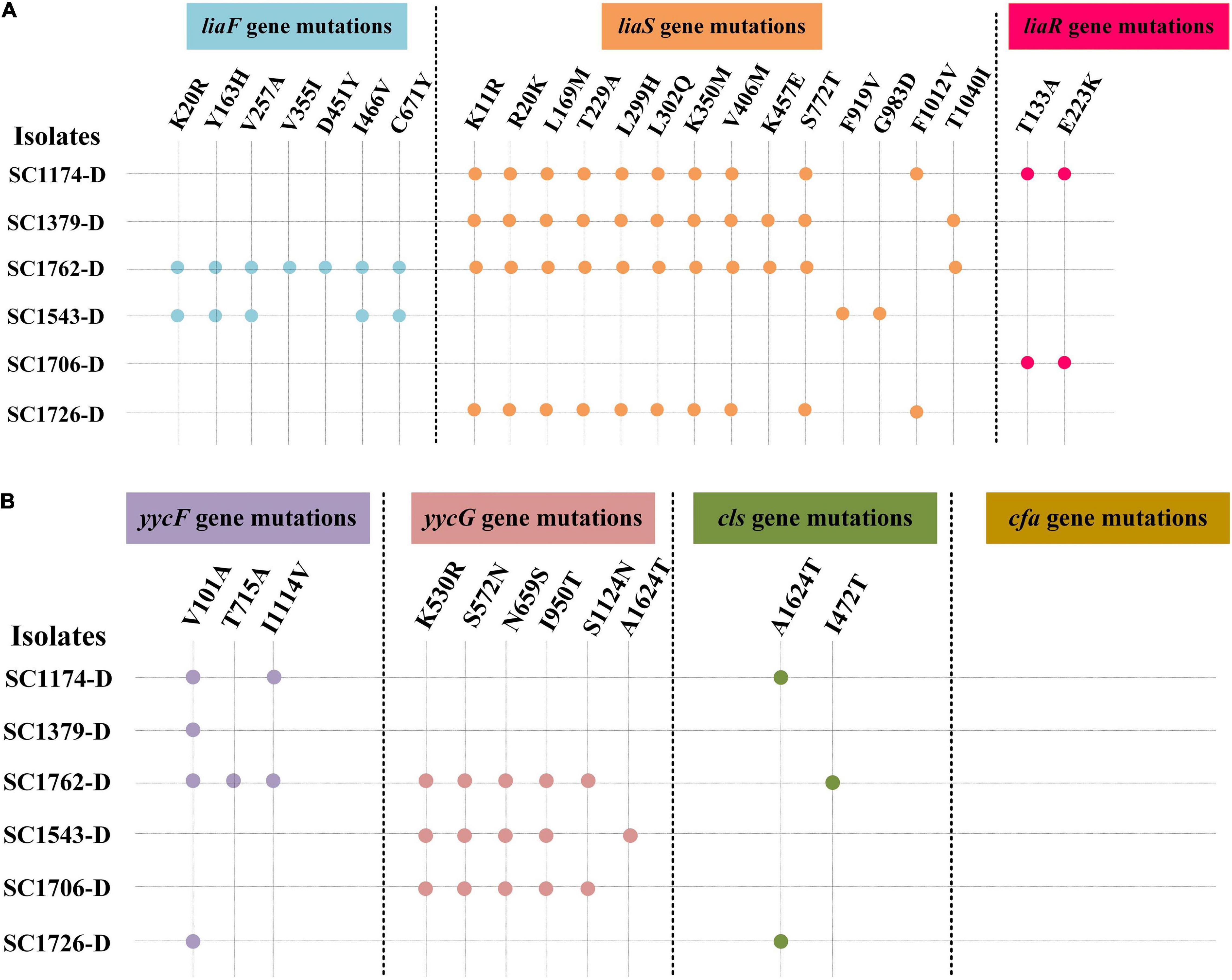
Figure 2. Genes mutations in daptomycin-resistant mutants. DAP resistance gene mutations in the evolved DAP-R mutants. -D, evolved DAP-R isolates. (A) liaFSR gene mutations; (B) yycFG, cls, and cfa gene mutations. Blue, orange, red, purple, pink, and green dots represent the mutation sites in the liaF, liaS, liaR, yycF, yycG, and cls genes, respectively.
Cytochrome C binding assays revealed that all DAP-R mutants, except for SC1379-D, had significantly higher positive charge on their surface than the corresponding parental strains (Figure 3). TEM data showed that the cell-wall thickness of SC1174-D and SC1762-D was greater than that of SC1174 (average thickness, 55.09 ± 6.55 nm vs. 19.97 ± 5.26 nm; P < 0.0001) and SC1762 (average thickness, 46.456 ± 9.08 nm vs. 18.21 ± 3.36 nm; P < 0.0001) (Figure 4), respectively. Collectively, these results suggested that the evolution of DAP resistance in vitro in E. faecium was associated with genetic mutations, changes in the positive charge on the cell surface, and the thickness of the cell wall.
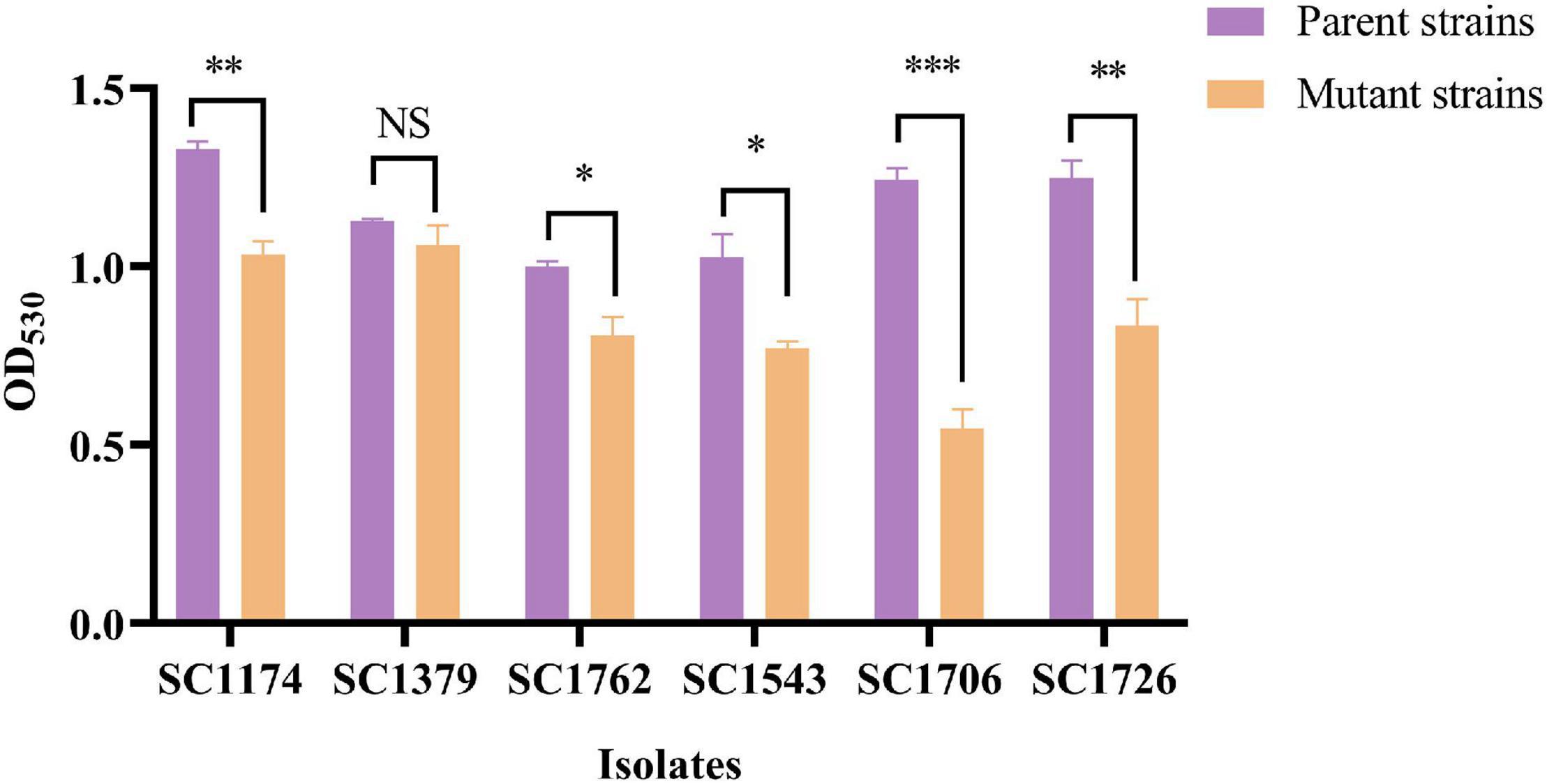
Figure 3. Relative positive surface charge examined based on cytochrome C binding. Evolved DAP-R mutants, except SC1379-D, had less free cytochrome C in the supernatant than their parental strains, indicating that they carried more net positive charge. Data are presented as mean values with error bars indicating standard deviations of the means of results from duplicate experiments, and P-values were calculated by the paired Student’s t-test (*P < 0.05; **P < 0.01; and ***P < 0.001; NS means no significant difference).
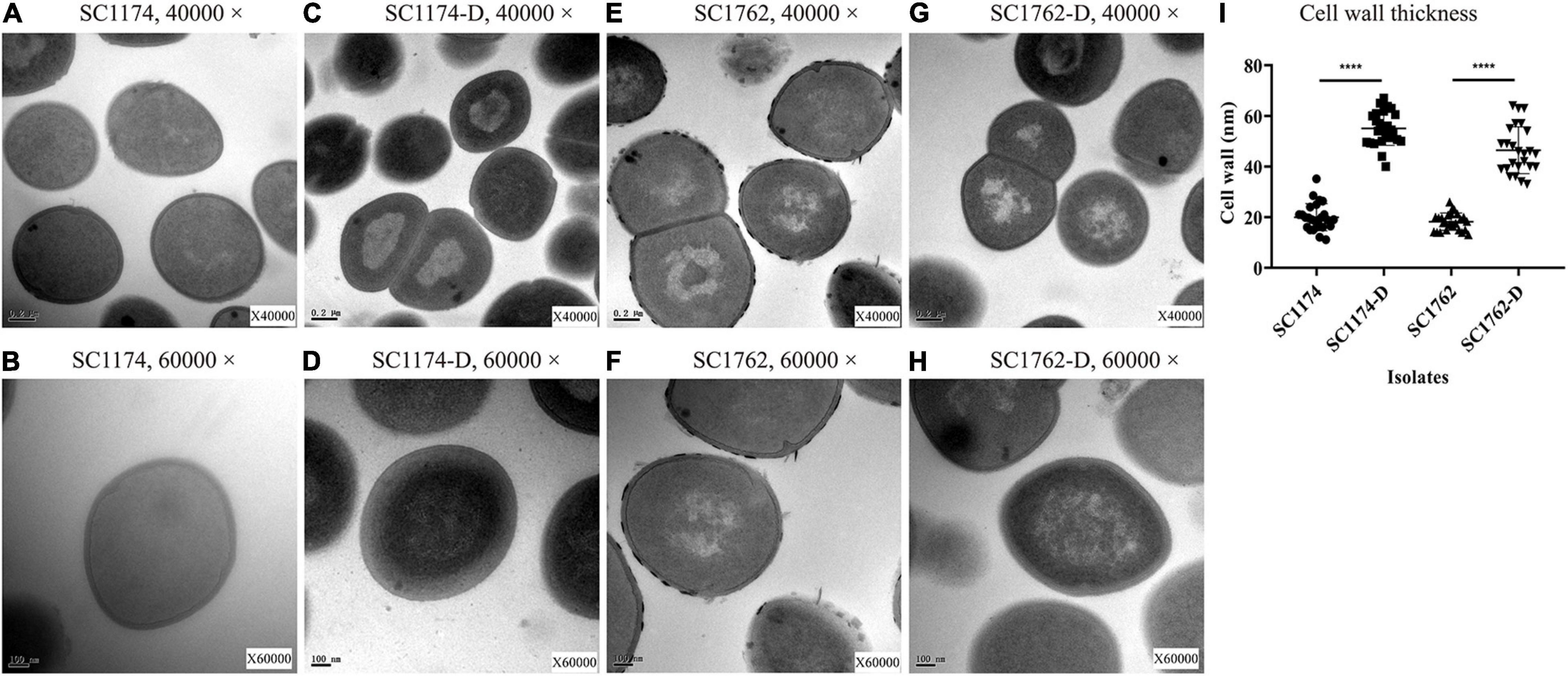
Figure 4. Cell-wall thickness examined using transmission electron microscopy. (A) SC1174, 40,000×; (B) SC1174, 60,000×; (C) SC1174-D, 40,000×; (D) SC1174-D, 60,000×; (E) SC1762, 40,000×; (F) SC1762, 60,000×; (G) SC1762-D, 40,000×; (H) SC1762-D, 60,000×; and (I) the cell-wall thickness of at least 25 cells was measured for SC1174, SC1174-D, SC1762, and SC1762-D. Data were presented as the mean and standard deviation. Asterisks indicate statistical significance using the paired Student’s t-test (****P < 0.0001).
We also determined the expression levels of surface positive charge-encoding genes dltABCD and cell-wall teichoic acid (WTA)-encoding genes tagGH in the parental strains and their mutants. Compared with the parental strains, all mutants except SC1379-D showed higher expression levels of dltABCD (Figure 5). Elevated tagGH expression was also observed among the mutants (Figure 5). These changes in gene expression could explain the increase in the positive surface charge and cell-wall thickness.
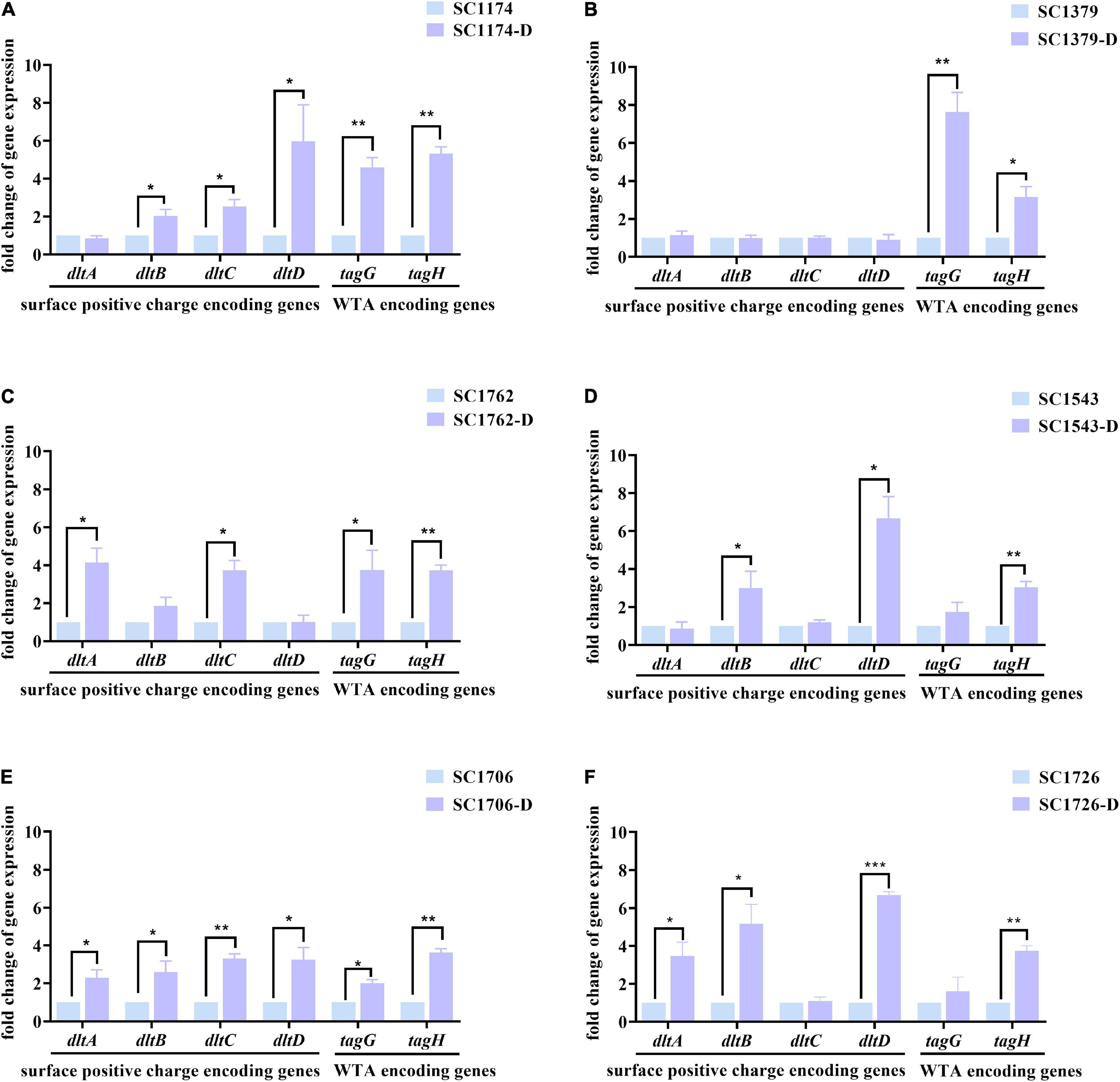
Figure 5. Relative expression levels of dltABCD and tagGH in the parental strains and DAP-R mutants. Values were normalized based on the internal control gene, 16S rRNA; data from the parental strain were normalized to 1 to allow the comparison of data across different samples. (A–F) show the expression levels of the surface positive charge-encoding genes in dltABCD and the cell-wall teichoic acid (WTA)-encoding genes in tagGH in SC1174, SC1379, SC1762, SC1543, SC1706, SC1726, and their corresponding evolved DAP-R mutants, respectively. Data represent the mean values from three independent experiments with error bars indicating standard deviations, and asterisks denote the significance of differences in expression by the paired Student’s t-test (*P < 0.05; **P < 0.01; and ***P < 0.001).
In order to explore the potential mechanisms of CS, the parental strain SC1762 and the DAP-R mutant SC1762-D (with CS) were used as the experimental group (CS group). Simultaneously, the parental strain SC1174 and DAP-R mutant SC1174-D (without CS) were randomly selected as the control group (non-CS group).
We first evaluated the mechanism of CS between DAP and glycopeptides using efflux pump inhibitors. We found that the DAP MIC of the four tested strains decreased by ≥fourfold after exposure to the efflux inhibitors CCCP and CHL; SC1174-D and SC1762-D had a positive efflux pump phenotype relative to their wild-type strains when an OME combination was used. Similarly, the MIC of vancomycin and teicoplanin also reduced by ≥fourfold among the four tested strains after exposure to CCCP. The efflux pump phenotype of both SC1174-D and SC1762-D was positive after the addition of PAβN, OME, RES, and CHL (Table 2). Taken together, these data suggested that there was no difference in the efflux pump phenotypes for DAP, teicoplanin, and vancomycin between the CS group and the non-CS group.
Genetic alterations and the relative expression levels of the vanA gene cluster were compared among the parental strains and DAP-R mutants using PCR and qRT-PCR. After comparisons with the standard strain E. faecium BM4147 and the corresponding parental strain, deletions or mutations in the vanA gene cluster were not detected in the CS and non-CS groups (data not shown). However, qRT-PCR data indicated that the expression of the vanA gene cluster was significantly higher in SC1174-D (Figure 6A) observably lower in SC1762-D (excluding vanX) than in the corresponding parental strains (Figure 6B).
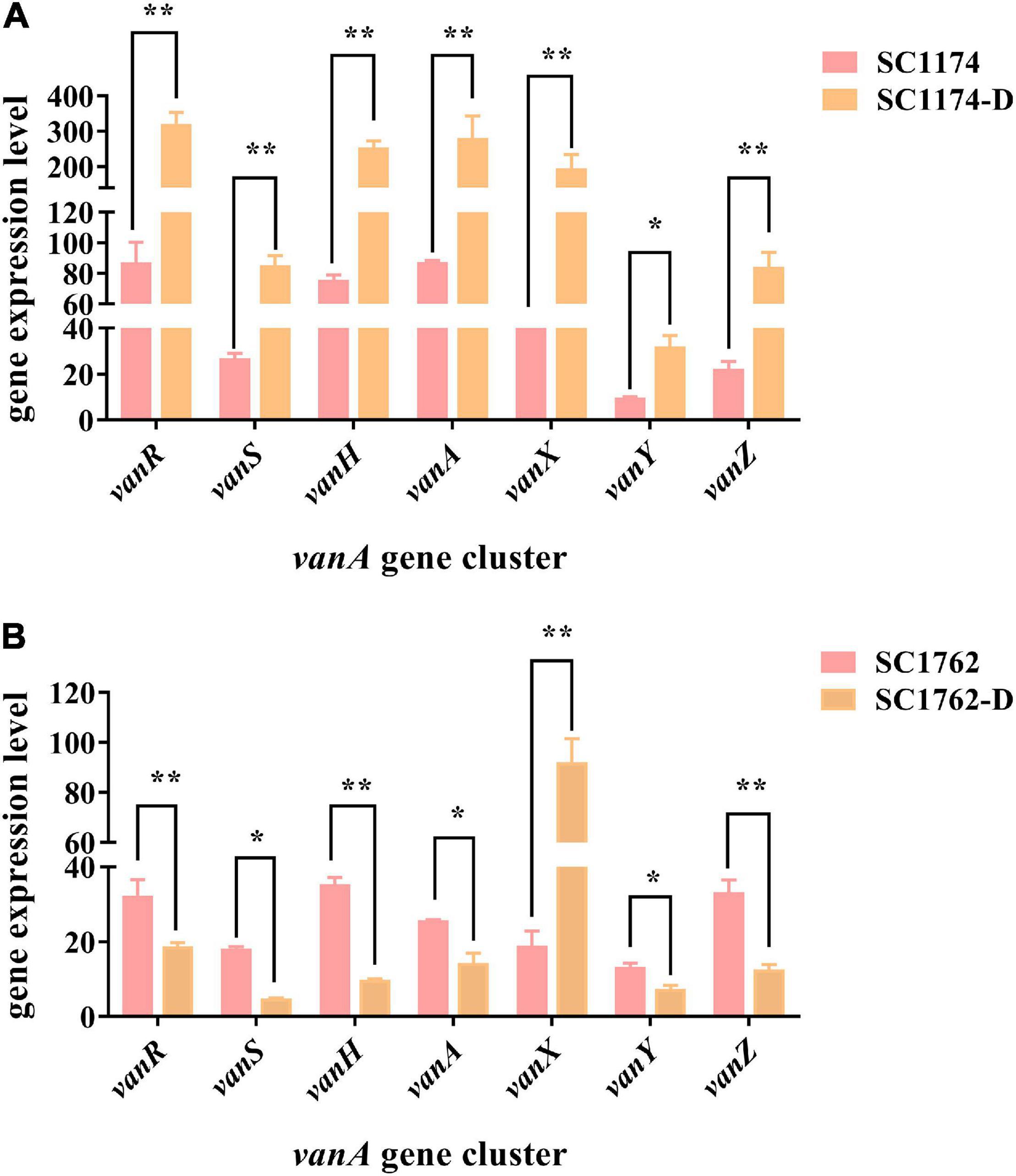
Figure 6. Relative expression levels of the vanA gene cluster in four E. faecium isolates. Expression levels of the vanA gene cluster in the non-CS group (A) and the CS group (B). Data represent the mean values from three independent experiments with error bars indicating standard deviations, and asterisks denote the significance of differences in expression by the paired Student’s t-test (*P < 0.05 and **P < 0.01).
Whole-genome sequencing was performed on SC1762 and SC1762-D to explain the reduced expression of the vanA gene cluster. Genomic features are shown in Supplementary Table 2. The plasmid (pSC1762-vanA) harboring the vanA gene cluster was 134,168 bp in length and shared the highest similarity (90% coverage, 99% identity) with pELF1 (accession number: LC495616) in GenBank (Supplementary Figure 3). The vanA gene cluster of SC1762 was located on a canonical Tn1546-like structure (Figure 7). However, the vanH gene in the mutant (pSC1762-D-vanA) was incomplete and truncated by an approximately 9 kb putative transposon (IS1216E-ISEfa4-repB-ISEfa7-IS1216E). The transposon was flanked by two copies of IS1216E in the same orientation. The sequences of IS1216E-ISEfa4 and ISEfa4-repB-ISEfa7-IS1216E were identical to those of Part II and Part I of pSC1762-vanA, respectively, suggesting a rearrangement of the IS-associated region (Figure 7). Furthermore, a 7-bp direct repeat (CTTTGGC), identical to the sequence (positions 75 to 81) of the vanH gene, was identified around the transposon. The results indicated a transposition process in the vanA gene cluster mediated by an IS1216E-based composite transposon.
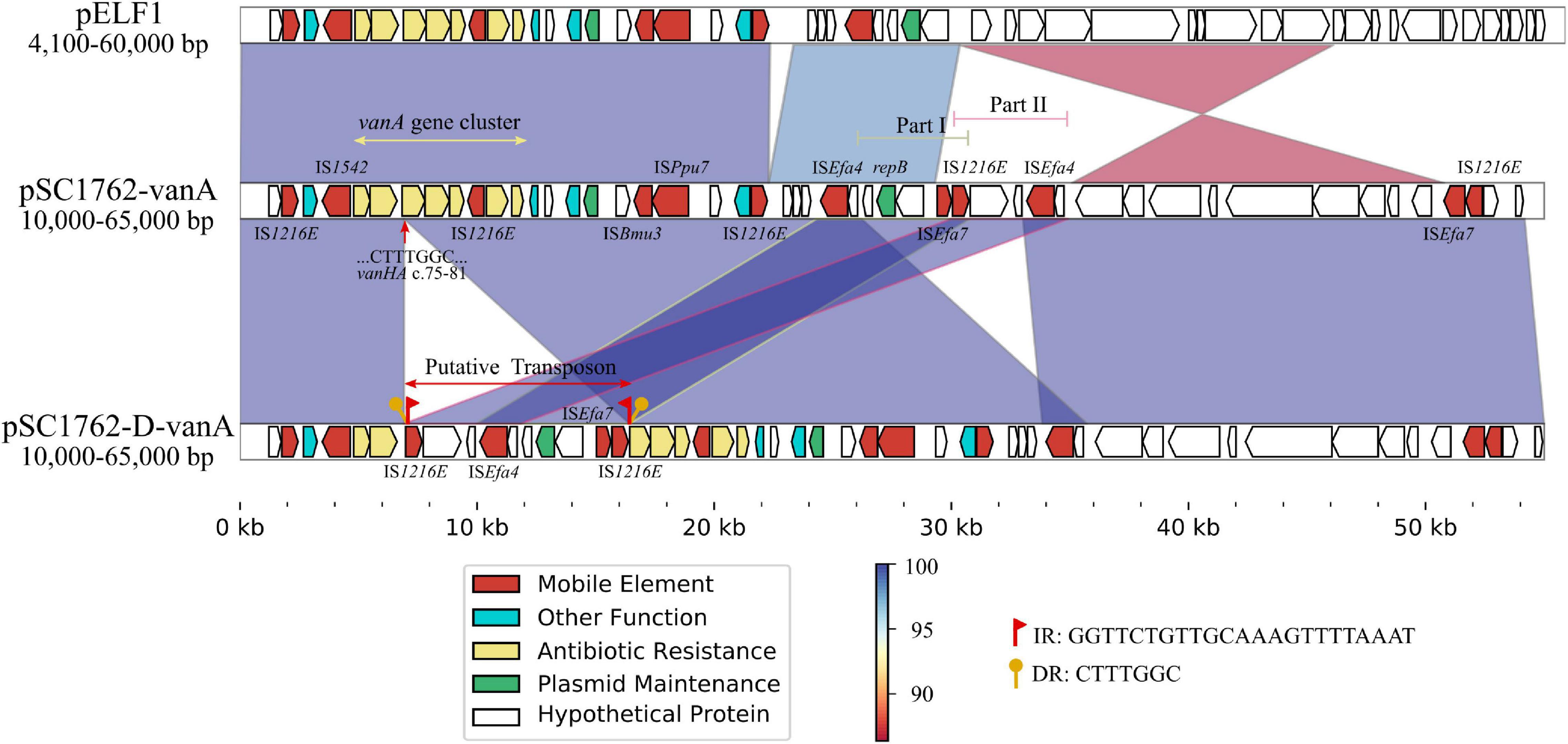
Figure 7. Comparative genetic structures and the genetic environment of the vanA gene cluster. The arrows represent sequence units or genes and are colored based on gene functional classification. Orthologous regions are connected and color coded.
To test the fitness difference between DAP-R mutants and their susceptible counterparts, we performed growth rate, in vitro competition, biofilm, and G. mellonella infection assays. The growth rate and bacterial density of the DAP-R mutants were lower than those of the original strains (Figure 8). Results of competition experiments indicated that DAP-R mutants showed a marked decrease in fitness (Figure 9A). Moreover, the biofilm-forming ability of evolved DAP-R strains was significantly higher than that of the corresponding parental strains; this difference was especially stark for SC1706-D (Figure 9B). To evaluate the virulence changes in the evolved strains, an infection model was generated using G. mellonella larvae. At 6 days post infection, the mortality of larvae infected with the DAP-R mutants was lower than that of larvae infected with the corresponding DAP-S strains, and no mortality was observed in the control injected with NS (Figure 10).
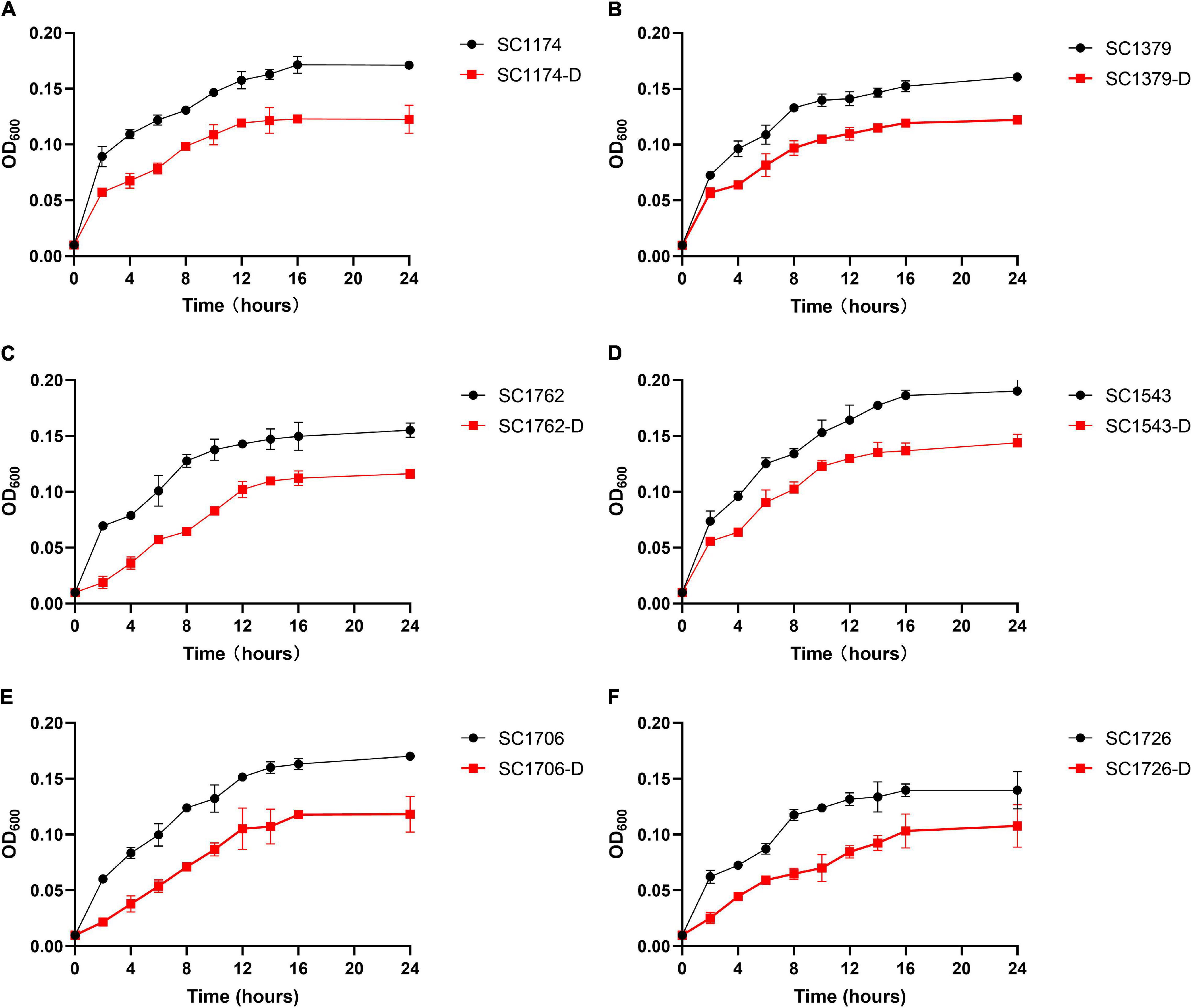
Figure 8. Bacterial growth curves for the parental strains and evolved strains. (A–F) show the growth curves for SC1174, SC1379, SC1762, SC1543, SC1706, SC1726, and their corresponding evolved DAP-R mutants, respectively. The standard deviation of three independent experiments is indicated in error bars. One-way analysis of variance (ANOVA) was used in statistical analysis (P < 0.05 vs. respective DAP-S parental strains).
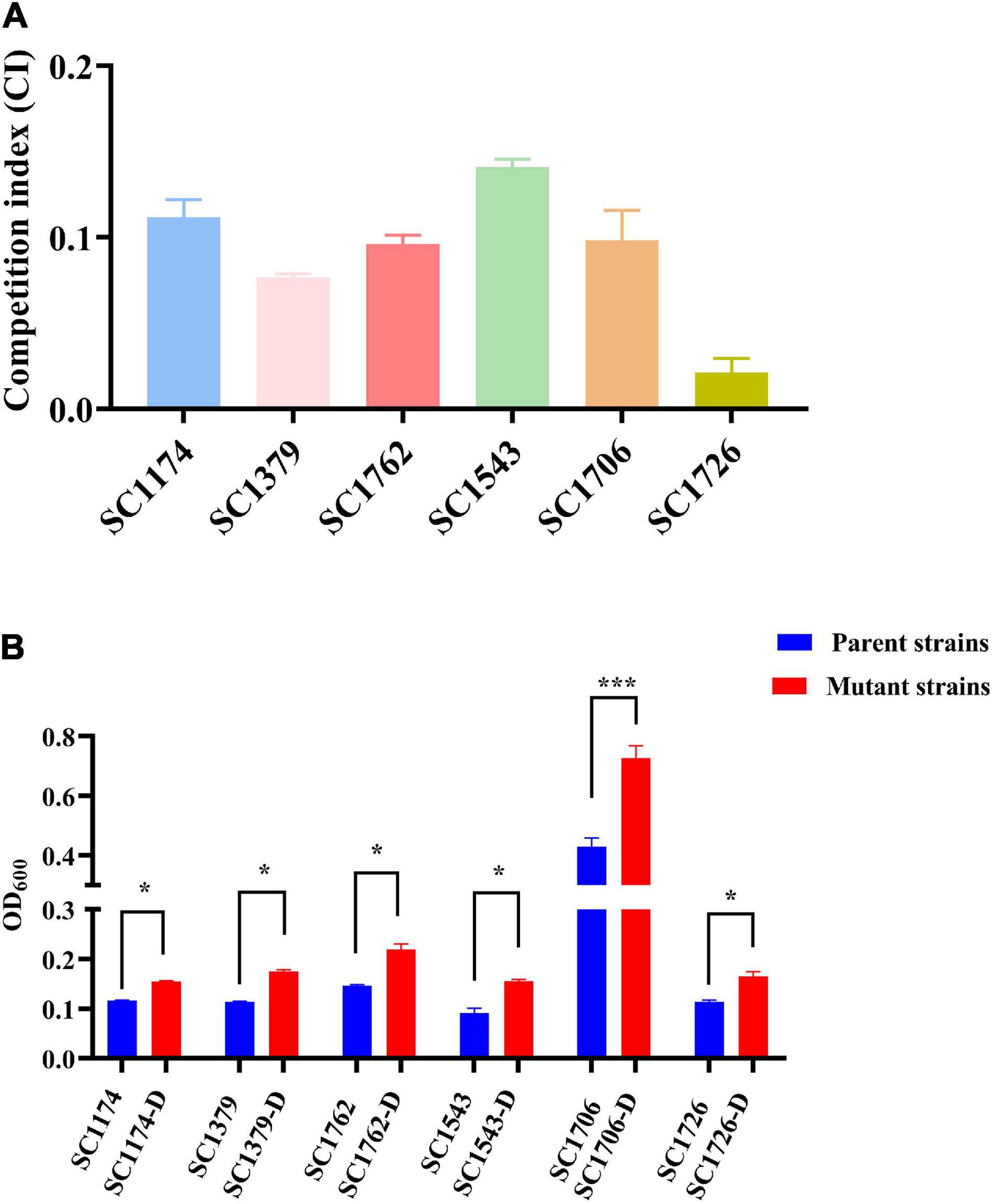
Figure 9. In vitro competition index and biofilm formation ability. (A) The in vitro CI in parental isolates and mutants. (B) Levels of biofilm formation in parental isolates and mutants. -D, evolved DAP-R mutants. The error bars show standard deviation from three independent experiments, and the absorbance value at OD600 was used to compare the biofilm formation ability of the DAP-S parental strains and respective evolved DAP-R strains with paired Student’s t-test (*P < 0.05 and ***P < 0.001).
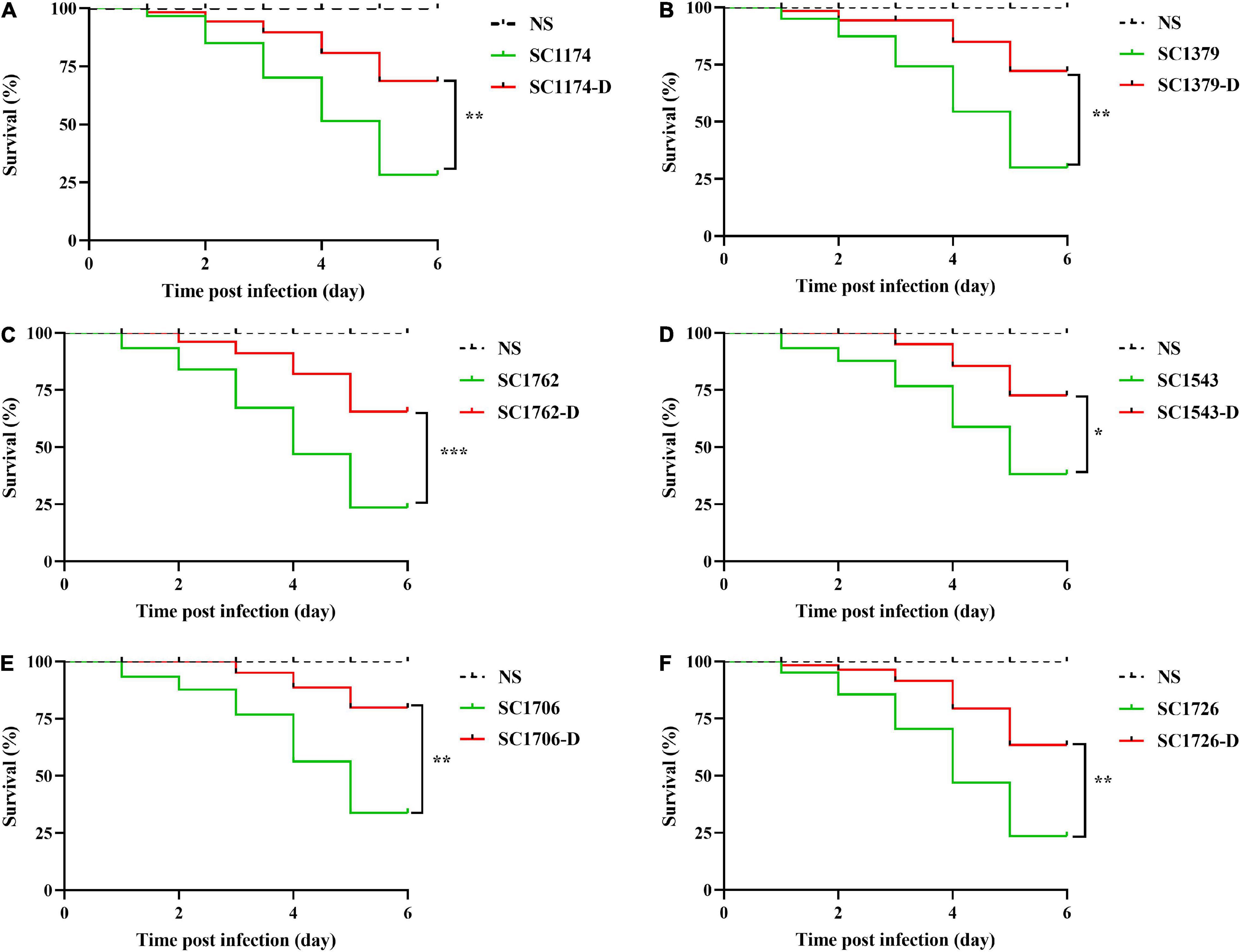
Figure 10. Infection of Galleria mellonella larvae. (A–F) show the survival curves of Galleria mellonella larvae infected with SC1174, SC1379, SC1762, SC1543, SC1706, SC1726, and their corresponding evolved DAP-R mutants, respectively. Survival data are plotted using the Kaplan–Meier method and expressed as percentage of survival vs. time. The corresponding P-values are given in the parenthesis for each group using the log-rank test (*P < 0.05; **P < 0.01; and ***P < 0.001).
With the extensive, continuous, and inappropriate use of antibiotics, bacterial resistance to antimicrobial agents has become increasingly common, posing a serious threat to public health around the world (Watkins and Bonomo, 2016). In this context, CS—a beneficial trade-off that allows the reversal of antibiotic resistance—has attracted the attention of researchers globally (Barbosa et al., 2017). A better understanding of the evolutionary processes related to antibiotic resistance and the molecular mechanisms of CS could potentially inform effective therapeutic strategies in the future. However, the evolution of DAP resistance and acquired CS are still poorly understood. In this study, we investigated the evolution and mechanisms of DAP resistance, and further explored the possible mechanisms underlying the reversal of vancomycin and teicoplanin resistance in the evolved DAP-R strain SC1762-D.
In this study, six wild-type strains rapidly and successfully developed stable DAP resistance under DAP exposure. This indicated that under clinical conditions, DAP doses and regimens should be controlled strictly while treating bacterial infections, even though DAP is a useful treatment option for patients with persistent infections. Laboratory evolution often leads to differences in physiological characteristics between adapted strains and parental strains, which are usually related to a fitness cost (Kang et al., 2017; Li et al., 2017). In our study, we identified reduced growth, in vitro competition, and virulence among the evolved DAP-R isolates, although biofilm formation increased and was associated with the acquisition of DAP resistance (Guo et al., 2012; Saffari et al., 2017).
Genetic alterations were analyzed to gain insights into the evolution and mechanisms of DAP resistance. Consistent with previous studies, mutations in liaFSR, yycFG, and cls were detected in evolved DAP-R E. faecium strains (Arias et al., 2011; Munita et al., 2012). As mentioned previously, cfa mutations are associated with DAP resistance; however, no mutation in cfa was detected in the evolved DAP-R isolates in this study. This may be due to differences in genetic heritability and backgrounds between strains. Moreover, the cfa mutation could be more likely involved in spontaneous resistance to DAP than in laboratory-evolved resistance. In our study, we observed that the mutation sites in DAP-R genes differed among evolved DAP-R mutants, illustrating the diversity and dynamicity of DAP-R evolution pathways in E. faecium. We also compared the relationship between genetic mutation sites and different DAP-R levels. As shown in Figure 2, the evolved DAP-R resistance strain SC1706-D with the highest resistance level (512 μg/ml) only had few mutation sites in two genes, which was less than other evolved DAP-R strains. Additionally, these mutation sites detected in SC1706-D also appeared in SC1174-D and SC1762-D. These data revealed that there was no significant correlation between genetic mutation sites and different DAP-R levels. In fact, except for genetic mutations, changes in the positive charge on the cell surface and the thickness of the cell wall also contributed to the evolution of DAP resistance, as mentioned earlier. In other words, the different sites of gene mutation may not be tightly related to the DAP resistant levels. Moreover, we detected a dramatic increase in the surface positive charge and cell-wall thickness in evolved DAP-R mutants compared with the parental strains, along with upregulation of dltABCD and/or tagGH. These findings are consistent with the results reported in previous studies (Mishra et al., 2012; Bayer et al., 2016; Miller et al., 2016).
Cross-resistance and collateral sensitivity are common evolutionary trade-offs during adaptive bacterial evolution (Lozano-Huntelman et al., 2020). The most clinically important trade-off for DAP resistance in the E. faecium strain SC1762-D was perhaps the resensitization to vancomycin and teicoplanin. In E. faecium, active efflux confers bacteria with the ability to counteract a wide range of antimicrobials, including glycopeptides (Nishioka et al., 2009). Azimi and Rastegar Lari (2017) reported that CS between aminoglycosides and beta-lactam antibiotics depends on active proton pumps. Thus, we hypothesized that DAP induction may inhibit efflux pump activity, preventing the pumping of intracellular glycopeptides out of the cell and restoring the sensitivity of SC1762-D to glycopeptides. Contrary to our expectations, there was no difference in the efflux pump phenotype between the CS group and non-CS group, indicating that efflux pump activity may not be involved in the CS between DAP and glycopeptides.
Further, we tested the hypothesis that mutations or expression-level changes in the vanA gene cluster drove the development of CS in SC1762-D. No deletion or mutation was observed in the vanA gene cluster in the mutant strains. Interestingly, in the CS group, the expression of the vanA gene cluster (except vanX) was significantly lower in SC1762-D than in the corresponding parental strain. However, in the non-CS group, i.e., SC1174-D, hyperexpression of this gene cluster was detected. These results suggested that the reduced expression of vanA gene cluster was mainly responsible for CS. In addition, we also tried to analyze the changes in the intergenic regions of the pSC1762-vanA and pSC1762-D-vanA plasmids, such as promoter activity, to explain the differences in vanA gene cluster expression. But no differences in promoter level or other differences (such as gene mutations and sequence deletions) were observed between the vanA gene clusters of SC1762 and SC1762-D, except for the insertion of IS elements.
A previous study reported that mobile genetic elements, such as IS elements, decrease or silence the expression of resistance-related genes (Sun et al., 2019). In this study, an IS1216E-based composite transposon was formed in the DAP-R mutant SC1762-D and disrupted the vanH gene, likely affecting the structure and expression of the vanA gene cluster. Vandecraen et al. (2017) confirmed that the insertion or transposition of IS elements is affected by pathogens and environmental signals, offering an adaptive strategy for bacteria and promoting genetic variability against environmental challenges. We found that IS insertion may be an adaptive strategy used by bacteria to survive under the selection pressure of DAP. However, unlike other vanA genes, vanX did not show decreased expression in SC1762-D. A possible explanation is that the function or activity of vanX was not influenced by the IS insertion (Monteiro da Silva et al., 2020).
The pSC1762-vanA plasmids carrying the vanA gene cluster have a high homology with the novel mobile linear pELF1 plasmid detected by Japanese scholars (Hashimoto et al., 2019). Hence, these two plasmids could also have the mobility and transferability observed in pELF1. Recent studies have confirmed that the vanA gene cluster can be disseminated clonally but also horizontally by plasmid dissemination or Tn1546 transposition between different genomic locations (Freitas et al., 2013; Lanza et al., 2015; Freitas et al., 2016; Arredondo-Alonso et al., 2021). Hence, it must be noted that pSC1762-vanA plasmids may confer the risk of antibiotic resistance transmission, and this should be monitored carefully.
Our research also has some limitations. Although mobile ISs were detected in both pSC1762-vanA and pSC1762-D-vanA plasmids, why IS insertion occurs in the vanA gene cluster and not in other resistance genes remains unclear. Moreover, it is not known whether the CS between DAP and glycopeptides is universal. In the future, we will further explore these questions in a focused manner.
In short, the findings of this investigation provide convincing evidence that E. faecium can easily acquire high-level DAP resistance in vitro. This resistance can be attributed to genetic mutations and changes in cell-wall thickness and cell-membrane charge. Further, this is the first study reporting CS between DAP and glycopeptides, which is related to the decreased expression of the vanA gene cluster owing to IS insertion. These results provide a proof-of-concept and support the sequential and combinational use of DAP with glycopeptides for the treatment of VRE infections.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
WZ conducted the experiments, analyzed the data, and wrote the manuscript. LF, CQ, and TC participated in experiments. SW, YZ, and XZ took part in analysis of results. LW and SL participated in the analysis of results. TZ and YS helped to design the study. All authors contributed to the article and approved the submitted version.
This work was financially supported by the National Natural Science Foundation of China (grant number 81802069).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We acknowledge the financial support of the National Natural Science Foundation of China (grant no. 81802069).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.815600/full#supplementary-material
Arias, C. A., and Murray, B. E. (2012). The rise of the Enterococcus: beyond vancomycin resistance. Nat. Rev. Microbiol. 10, 266–278. doi: 10.1038/nrmicro2761
Arias, C. A., Panesso, D., McGrath, D. M., Qin, X., Mojica, M. F., Miller, C., et al. (2011). Genetic basis for in vivo daptomycin resistance in enterococci. N. Engl. J. Med. 365, 892–900. doi: 10.1056/NEJMoa1011138
Arredondo-Alonso, S., Top, J., Corander, J., Willems, R. J. L., and Schürch, A. C. (2021). Mode and dynamics of vanA-type vancomycin resistance dissemination in Dutch hospitals. Genome Med. 13:9. doi: 10.1186/s13073-020-00825-3
Azimi, L., and Rastegar Lari, A. (2017). Collateral sensitivity between aminoglycosides and beta-lactam antibiotics depends on active proton pumps. Microb. Pathog. 112, 122–125. doi: 10.1016/j.micpath.2017.09.049
Barbosa, C., Trebosc, V., Kemmer, C., Rosenstiel, P., Beardmore, R., Schulenburg, H., et al. (2017). Alternative evolutionary paths to bacterial antibiotic resistance cause distinct collateral effects. Mol. Biol. Evol. 34, 2229–2244. doi: 10.1093/molbev/msx158
Bayer, A. S., Mishra, N. N., Cheung, A. L., Rubio, A., and Yang, S.-J. (2016). Dysregulation of mprF and dltABCD expression among daptomycin-non-susceptible MRSA clinical isolates. J. Antimicrob. Chemother. 71, 2100–2104. doi: 10.1093/jac/dkw142
Bhardwaj, P., Hans, A., Ruikar, K., Guan, Z., and Palmer, K. L. (2018). Reduced chlorhexidine and daptomycin susceptibility in vancomycin-resistant Enterococcus faecium after serial chlorhexidine exposure. Antimicrob. Agents Chemother. 62, e1235–e1217. doi: 10.1128/AAC.01235-17
Canfield, G. S., Chatterjee, A., Espinosa, J., Mangalea, M. R., Sheriff, E. K., Keidan, M., et al. (2021). Lytic bacteriophages facilitate antibiotic sensitization of Enterococcus faecium. Antimicrob. Agents Chemother. 65, e143–e121. doi: 10.1128/AAC.00143-21
Cha, J. O., Yoo, J. I., Kim, H. K., Kim, H. S., Yoo, J. S., Lee, Y. S., et al. (2013). Diversity of Tn1546 in vanA-positive Enterococcus faecium clinical isolates with VanA, VanB, and VanD phenotypes and susceptibility to vancomycin. J. Appl. Microbiol. 115, 969–976. doi: 10.1111/jam.12300
Clinical and Laboratory Standards Institute [CLSI] (2020). Performance Standards for Antimicrobial Susceptibility Testing; 29th Informational Supplement. CLSI Document M100-S30. Wayne, PA: Clinical and Laboratory Standards Institute.
Contreras, G. A., Munita, J. M., and Arias, C. A. (2019). Novel strategies for the management of vancomycin-resistant enterococcal infections. Curr. Infect. Dis. Rep. 21:22. doi: 10.1007/s11908-019-0680-y
Frankenfeld, C., Mittal, S., Melendez, Y., Mendez-Vigo, L., Lamp, K. C., Keller, K. N., et al. (2018). Daptomycin: a comparison of two intravenous formulations. Drug Des. Devel. Ther. 12, 1953–1958. doi: 10.2147/DDDT.S167010
Freitas, A. R., Novais, C., Tedim, A. P., Francia, M. V., Baquero, F., Peixe, L., et al. (2013). Microevolutionary events involving narrow host plasmids influences local fixation of vancomycin-resistance in Enterococcus populations. PLoS One 8:e60589. doi: 10.1371/journal.pone.0060589
Freitas, A. R., Tedim, A. P., Francia, M. V., Jensen, L. B., Novais, C., Peixe, L., et al. (2016). Multilevel population genetic analysis of vanA and vanB Enterococcus faecium causing nosocomial outbreaks in 27 countries (1986-2012). J. Antimicrob. Chemother. 71, 3351–3366. doi: 10.1093/jac/dkw312
Guo, B., Abdelraouf, K., Ledesma, K. R., Nikolaou, M., and Tam, V. H. (2012). Predicting bacterial fitness cost associated with drug resistance. J. Antimicrob. Chemother. 67, 928–932. doi: 10.1093/jac/dkr560
Hashimoto, Y., Taniguchi, M., Uesaka, K., Nomura, T., Hirakawa, H., Tanimoto, K., et al. (2019). Novel multidrug-resistant enterococcal mobile linear plasmid pELF1 encoding and gene clusters from a Japanese vancomycin-resistant enterococci isolate. Front. Microbiol. 10:2568. doi: 10.3389/fmicb.2019.02568
Imamovic, L., Ellabaan, M. M. H., Dantas Machado, A. M., Citterio, L., Wulff, T., Molin, S., et al. (2018). Drug-driven phenotypic convergence supports rational treatment strategies of chronic infections. Cell 172, 121.e–134.e. doi: 10.1016/j.cell.2017.12.012
Ji, S., Jiang, S., Wei, X., Sun, L., Wang, H., Zhao, F., et al. (2020). In-host evolution of daptomycin resistance and heteroresistance in methicillin-resistant Staphylococcus aureus strains from three endocarditis patients. J. Infect. Dis. 221(Suppl. 2), S243–S252. doi: 10.1093/infdis/jiz571
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., et al. (2017). CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573. doi: 10.1093/nar/gkw1004
Jones, T., Yeaman, M. R., Sakoulas, G., Yang, S.-J., Proctor, R. A., Sahl, H.-G., et al. (2008). Failures in clinical treatment of Staphylococcus aureus infection with daptomycin are associated with alterations in surface charge, membrane phospholipid asymmetry, and drug binding. Antimicrob. Agents Chemother. 52, 269–278. doi: 10.1128/AAC.00719-07
Jung, S., Park, O. J., Kim, A. R., Ahn, K. B., Lee, D., Kum, K. Y., et al. (2019). Lipoteichoic acids of lactobacilli inhibit Enterococcus faecalis biofilm formation and disrupt the preformed biofilm. J. Microbiol. 57, 310–315. doi: 10.1007/s12275-019-8538-4
Kang, K.-M., Mishra, N. N., Park, K. T., Lee, G.-Y., Park, Y. H., Bayer, A. S., et al. (2017). Phenotypic and genotypic correlates of daptomycin-resistant methicillin-susceptible Staphylococcus aureus clinical isolates. J. Microbiol. 55, 153–159. doi: 10.1007/s12275-017-6509-1
Koren, S., Walenz, B. P., Berlin, K., Miller, J. R., Bergman, N. H., and Phillippy, A. M. (2017). Canu: scalable and accurate long-read assembly via adaptive -mer weighting and repeat separation. Genome Res. 27, 722–736. doi: 10.1101/gr.215087.116
Lanza, V. F., Tedim, A. P., Martínez, J. L., Baquero, F., and Coque, T. M. (2015). The plasmidome of firmicutes: impact on the emergence and the spread of resistance to antimicrobials. Microbiol. Spectr. 3, las–0039–2014. doi: 10.1128/microbiolspec.PLAS-0039-2014
Lázár, V., Martins, A., Spohn, R., Daruka, L., Grézal, G., Fekete, G., et al. (2018). Antibiotic-resistant bacteria show widespread collateral sensitivity to antimicrobial peptides. Nat. Microbiol. 3, 718–731. doi: 10.1038/s41564-018-0164-0
Lellek, H., Franke, G. C., Ruckert, C., Wolters, M., Wolschke, C., Christner, M., et al. (2015). Emergence of daptomycin non-susceptibility in colonizing vancomycin-resistant Enterococcus faecium isolates during daptomycin therapy. Int. J. Med. Microbiol. 305, 902–909. doi: 10.1016/j.ijmm.2015.09.005
Li, S., Yin, Y., Chen, H., Wang, Q., Wang, X., and Wang, H. (2017). Fitness cost of daptomycin-resistant obtained from daptomycin selection pressure. Front. Microbiol. 8:2199. doi: 10.3389/fmicb.2017.02199
Lin, Z., Pu, Z., Xu, G., Bai, B., Chen, Z., Sun, X., et al. (2020). Omadacycline efficacy against Enterococcus faecalis isolated in China: activity, heteroresistance, and resistance mechanisms. Antimicrob. Agents Chemother. 64, e2097–e2019. doi: 10.1128/AAC.02097-19
Lozano-Huntelman, N. A., Singh, N., Valencia, A., Mira, P., Sakayan, M., Boucher, I., et al. (2020). Evolution of antibiotic cross-resistance and collateral sensitivity in using the mutant prevention concentration and the mutant selection window. Evol. Appl. 13, 808–823. doi: 10.1111/eva.12903
Miller, W. R., Bayer, A. S., and Arias, C. A. (2016). Mechanism of action and resistance to daptomycin in Staphylococcus aureus and enterococci. Cold Spring Harb. Perspect. Med. 6:a026997. doi: 10.1101/cshperspect.a026997
Mishra, N. N., Bayer, A. S., Tran, T. T., Shamoo, Y., Mileykovskaya, E., Dowhan, W., et al. (2012). Daptomycin resistance in enterococci is associated with distinct alterations of cell membrane phospholipid content. PLoS One 7:e43958. doi: 10.1371/journal.pone.0043958
Monteiro da Silva, B. N., Faria, A. R., Souza, S.d.S.R, Colodette, S. S., Morais, J. M., Teixeira, L. M., et al. (2020). Expression of VanA-type vancomycin resistance in a clinical isolate of Enterococcus faecium showing insertion of IS19 in the vanS gene. Int. J. Antimicrob. Agents 55:105897. doi: 10.1016/j.ijantimicag.2020.105897
Mulani, M. S., Kamble, E. E., Kumkar, S. N., Tawre, M. S., and Pardesi, K. R. (2019). Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: a review. Front. Microbiol. 10:539. doi: 10.3389/fmicb.2019.00539
Munita, J. M., Panesso, D., Diaz, L., Tran, T. T., Reyes, J., Wanger, A., et al. (2012). Correlation between mutations in liaFSR of Enterococcus faecium and MIC of daptomycin: revisiting daptomycin breakpoints. Antimicrob. Agents Chemother. 56, 4354–4359. doi: 10.1128/AAC.00509-12
Nishioka, T., Ogawa, W., Kuroda, T., Katsu, T., and Tsuchiya, T. (2009). Gene cloning and characterization of EfmA, a multidrug efflux pump, from Enterococcus faecium. Biol. Pharm. Bull. 32, 483–488. doi: 10.1248/bpb.32.483
O’Driscoll, T., and Crank, C. W. (2015). Vancomycin-resistant enterococcal infections: epidemiology, clinical manifestations, and optimal management. Infect. Drug Resist. 8, 217–230. doi: 10.2147/IDR.S54125
Pendleton, J. N., Gorman, S. P., and Gilmore, B. F. (2013). Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti Infect. Ther. 11, 297–308. doi: 10.1586/eri.13.12
Prater, A. G., Mehta, H. H., Kosgei, A. J., Miller, W. R., Tran, T. T., Arias, C. A., et al. (2019). Environment shapes the accessible daptomycin resistance mechanisms in Enterococcus faecium. Antimicrob. Agents Chemother. 63, e790–e719. doi: 10.1128/AAC.00790-19
Qin, X., Galloway-Peña, J. R., Sillanpaa, J., Roh, J. H., Nallapareddy, S. R., Chowdhury, S., et al. (2012). Complete genome sequence of Enterococcus faecium strain TX16 and comparative genomic analysis of Enterococcus faecium genomes. BMC Microbiol. 12:135. doi: 10.1186/1471-2180-12-135
Rand, K. H., Houck, H. J., and Silverman, J. A. (2007). Daptomycin-reversible rifampicin resistance in vancomycin-resistant Enterococcus faecium. J. Antimicrob. Chemother. 59, 1017–1020. doi: 10.1093/jac/dkm045
Roemhild, R., Linkevicius, M., and Andersson, D. I. (2020). Molecular mechanisms of collateral sensitivity to the antibiotic nitrofurantoin. PLoS Biol. 18:e3000612. doi: 10.1371/journal.pbio.3000612
Saffari, M., Karami, S., Firoozeh, F., and Sehat, M. (2017). Evaluation of biofilm-specific antimicrobial resistance genes in Pseudomonas aeruginosa isolates in Farabi Hospital. J. Med. Microbiol. 66, 905–909. doi: 10.1099/jmm.0.000521
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014
Sun, L., Qu, T., Wang, D., Chen, Y., Fu, Y., Yang, Q., et al. (2019). Characterization of vanM carrying clinical Enterococcus isolates and diversity of the suppressed vanM gene cluster. Infect. Genet. Evol. 68, 145–152. doi: 10.1016/j.meegid.2018.12.015
Taglialegna, A., Varela, M. C., Rosato, R. R., and Rosato, A. E. (2019). VraSR and virulence trait modulation during daptomycin resistance in methicillin-resistant infection. mSphere 4, e557–e518. doi: 10.1128/mSphere.00557-18
Vandecraen, J., Chandler, M., Aertsen, A., and Van Houdt, R. (2017). The impact of insertion sequences on bacterial genome plasticity and adaptability. Crit. Rev. Microbiol. 43, 709–730. doi: 10.1080/1040841X.2017.1303661
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963
Watkins, R. R., and Bonomo, R. A. (2016). Overview: global and local impact of antibiotic resistance. Infect. Dis. Clin. North Am. 30, 313–322. doi: 10.1016/j.idc.2016.02.001
Keywords: collateral sensitivity, resistance mechanisms, Enterococcus faecium, daptomycin, glycopeptides, fitness cost
Citation: Zeng W, Feng L, Qian C, Chen T, Wang S, Zhang Y, Zheng X, Wang L, Liu S, Zhou T and Sun Y (2022) Acquisition of Daptomycin Resistance by Enterococcus faecium Confers Collateral Sensitivity to Glycopeptides. Front. Microbiol. 13:815600. doi: 10.3389/fmicb.2022.815600
Received: 15 November 2021; Accepted: 10 March 2022;
Published: 12 April 2022.
Edited by:
Frank Schreiber, Federal Institute for Materials Research and Testing (BAM), GermanyReviewed by:
Ana P. Tedim, Institute of Health Sciences Studies of Castilla y León (IECSCYL), SpainCopyright © 2022 Zeng, Feng, Qian, Chen, Wang, Zhang, Zheng, Wang, Liu, Zhou and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yao Sun, c3VueWFvd3ptdUAxNjMuY29t; Tieli Zhou, d3l6dGxpQDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.