
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 28 February 2022
Sec. Microbial Symbioses
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.814864
This article is part of the Research Topic Insights in Microbial Symbioses: 2021 View all 7 articles
 Kuan Yan1,2
Kuan Yan1,2 Zihao Pei1,2
Zihao Pei1,2 Lina Meng1,2
Lina Meng1,2 Yu Zheng1,2
Yu Zheng1,2 Lian Wang1,2
Lian Wang1,2 Ruizhang Feng1,2
Ruizhang Feng1,2 Quanzi Li3
Quanzi Li3 Yang Liu4
Yang Liu4 Xianming Zhao1
Xianming Zhao1 Qin Wei1,2*†
Qin Wei1,2*† Ahmed H. El-Sappah1,2,5*†
Ahmed H. El-Sappah1,2,5*† Manzar Abbas1,2*†
Manzar Abbas1,2*†
Endophytic fungi act as seed endosymbiont, thereby playing a very crucial role in the growth and development of seeds. Seed-vectored endophytic fungi establish an everlasting association with seeds and travel from generation to generation. To explore the composition and diversity of endophytic fungi in Alpinia zerumbet seeds, high-throughput Illumina MiSeq sequencing was employed for the following stages: fruit formation period (YSJ1), young fruit period (YSJ2), early mature period (YSJ3), middle mature period (YSJ4), and late mature period (YSJ5). A total of 906,694 sequence reads and 745 operational taxonomic units (OTUs) were obtained and further classified into 8 phyla, 30 classes, 73 orders, 163 families, 302 genera, and 449 species. The highest endophytic fungal diversity was observed at YSJ5. The genera with the highest abundance were Cladosporium, Kodamaea, Hannaella, Mycothermus, Gibberella, Sarocladium, and Neopestalotiopsis. Functional Guild (FUNGuild) analysis revealed that endophytic fungi were undefined saprotroph, plant pathogens, animal pathogen–endophyte–lichen parasite–plant pathogen–wood saprotroph, and soil saprotrophs. Alternaria, Fusarium, Cladosporium, and Sarocladium, which are potential probiotics and can be used as biocontrol agents, were also abundant. This study is part of the Sustainable Development Goals of United Nations Organization (UNO) to “Establish Good Health and Well-Being.”
Plant endophytes are endosymbionts that are integral components of a plant microecosystem. The presence of these symbiotic endophytes has been reported among all plant tissues, including roots, root hairs, stems, branches, twigs, leaves, flowers, and seeds (Gond et al., 2015; White et al., 2015). During the long-term process of coevolution, endophytes have developed a symbiotic relationship with their hosts, such as plants, to enable better colonization of land (Johnston-Monje and Raizada, 2011). Endophytic fungi are members of a large class of endophytes that play a key role in plant resistance against both biotic and abiotic stresses; promote plant growth via the mobilization of nutrients, phosphate solubilization, and nitrogen fixation (Rosenblueth and Martínez-Romero, 2006); and produce biologically active compounds, such as phytohormones (Compant et al., 2010; Donoso et al., 2016). The absence of these symbiotic endophytes induces serious constraints on seed germination and seedling growth (Verma et al., 2017), while their presence improves seed germination and seedling growth (Donoso et al., 2016).
Seeds are important reproductive organs of plants that fall on land, the richest source of diverse microbes. On land, seeds gather multiple endophytic microbes on their coat, which may protect them from degradation or consumption (Dalling et al., 2011; Verma et al., 2017). Seed-derived endophytes have antibacterial, antianimal feeding, and growth-promoting effects on host seedling and can effectively increase the seed germination rate, seedling survival rate, seedling root length, seedling height, and biomass accumulation (Shade et al., 2017; Berg and Raaijmakers, 2018). Endophytic symbiotic fungi establish an irreversible long-lasting association, namely, seed-vectored endophytic fungi, which travels from generation to generation (Johnston-Monje and Raizada, 2011). In Eucalyptus, rice, tea, and maize, seed-derived endophytic microbes serve as a source of future microbiota of seedlings and develop a very pleasant aroma during pil fermentation of tea (Johnston-Monje and Raizada, 2011; Yan et al., 2021a,b). Therefore, research to disclose the complex diversity of the endophytes of plant seeds, especially for medicinal and spice plants species, is critical.
Alpinia zerumbet (Pers.) Burtt et Smith is an important aromatic plant in China and is widely cultivated in Southern Sichuan (Tao et al., 2009). The seeds of A. zerumbet are a rich source of volatile oil and possess beneficial biological activities, such as antibacterial, antioxidant, analgesic, and anti-inflammatory effects, thereby providing a soothing effect to the heart and brain (Shen et al., 2010). Although A. zerumbet is being extensively assessed for its volatile oil contents and biological activities, very little is known about endophytic microbes. Southern Sichuan has a unique geographical location and climatic conditions. As a result, the seeds of A. zerumbet grown in this area may harbor comparatively different endophytes. Evaluating microbial communities via traditional culturing technique does not reveal the complete microbial diversity of endophytes as only 0.1–10% of microbes are culturable. The composition and diversity of fungal communities can be determined using shotgun genomic sequencing or amplicon and sequencing of internal transcribed spacer (ITS) (De Filippis et al., 2017).
Seeds of A. zerumbet were collected from the southern Sichuan province of China and used as the material in this study. These seeds were employed to disclose the endophytic fungal community structure and diversity at five different growth stages. We deployed a robust culture-independent method, namely, high-throughput sequencing, to evaluate the community structure, diversity, and dominant genus of endophytic fungi. Our findings are helpful for revealing the correlation between the community structure and dynamic succession of endophytes at different developmental stages of A. zerumbet seeds. Accordingly, these results will open new research avenues, ultimately encouraging scientists to further determine the specific role of endophytes in volatile oil production in seeds of A. zerumbet.
Seeds of A. zerumbet were collected at five different growth stages, including fruit formation period (YSJ1), young fruit period (YSJ2), early mature period (YSJ3), middle mature period (YSJ4), and late mature period (YSJ5) in Yibin city, Sichuan Province, China (28°53′17″N, 104°43′7″E). Seeds were packed in small polythene bags, labeled, and immediately stored at 4°C to avoid decay or loss of fungal endophytes (Chang et al., 2018). Upon arrival at the laboratory, all appendages and soil patches on the surface of seeds were thoroughly washed with running tap water (Fareed et al., 2017). Seeds of each growth period were weighed (10 g, ∼100–120 seeds), packed in sterile bags, and stored at 4°C for future use as a research material. Surface sterilization of A. zerumbet seeds was performed via soaking in 75% ethanol for 30 s and 2% NaClO solution for 2 min with gentle shaking followed by three rounds of washing with sterile ddH2O. To confirm the surface sterilization of seeds, 20 μl of ddH2O from the final rinse was streaked on three plates containing a PDA medium and incubated at 28°C in a biochemical incubator in the dark (Abbas et al., 2016; El-Sappah et al., 2021).
To extract DNA from endophytes, 5 g of seeds was weighed on an electrical balance and immediately soaked in 25 ml of ddH2O in triplicate. Astragalus was divided into two halves using a sharp sterile blade, dipped in PBS solution, and placed on a magnetic stirrer for 30 min. After thorough stirring, the solution was filtered through a three-layered sterile gauze to remove coarse particles. The filtrate was subsequently centrifuged at 12,298 × g for 10 min at 4°C. The supernatant was discarded, and the precipitate of three replicates was mixed in one tube, which was further used for genomic DNA (gDNA) extraction using E.Z.N.A™ Fungal DNA Miniprep Kit (OMEGA, United States) according to the standard protocol (Peng et al., 2019). The quality of the extracted gDNA was confirmed by running 2 μl of each sample on 1% agarose gel and visualizing the gel under a UV light-installed gel documentation system, namely, iBright imaging system (iBright 1500, Thermo Fisher Scientific, United States) (Wang et al., 2018). The concentration of gDNA was measured using a NanoDrop 2,000 spectrophotometer at an absorption of A260 (2.1–42.5 ng/μl). The A. zerumbet seed sample of each stage was used for gDNA extraction in triplicate.
To amplify the ITS1 sequence of the fungal ITS region using PCR, the following primer pairs were employed: ITS1F (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS2R (5′-GCTGCGTTCTTCATCGATGC-3′) (Yan et al., 2021a). Each reaction comprised a 1 × polymerase chain reaction (PCR) buffer, 1 mM of dNTPs, 0.2 mM of each forward and reverse primer, 1.25 U of FastPfu DNA Polymerase (TransStart®, AP221-01, China), and 2 μl of DNA template, with a final volume of 25 μl achieved by adding ddH2O. The PCR conditions were as follows: initial denaturation at 95°C for 3 min, 32 cycles, denaturation at 95°C for 30 s, annealing at 55°C for 45 s, amplification at 68°C for 45 s, final amplification at 68°C for 20 min, and storage at 4°C for infinity (Abbas et al., 2020). To confirm amplification and amplicon size, 2 μl of each PCR product was separated on a 2% agarose gel and visualized under UV light. The PCR product was used to construct a gDNA library, which was further analyzed with robust and high-fidelity Illumina MiSeq™ PE300 sequencing.
Data for each sample were distinguished according to the sequence index and saved in fastq format. Each sample of MP or PE data had two files, fq1 and fq2, which contain the reads of both sequencing ends and the corresponding sequences. The data obtained by MiSeq sequencing were paired-end sequence data. First, according to the overlap relationship between PE reads, the paired reads were spliced (merges) into a sequence. Further, reads and their merging were quality controlled and filtered. The barcode and primer sequences were used to distinguish correct sequences and their direction to obtain optimized data. Raw reads were eliminated, duplicate reads were merged according to PE read overlap, reads quality was assessed, and splicing events were checked using fast length adjustment of short reads (FLASH-1.2.11) software. To avoid mixing samples, specific primer-based barcodes were assigned to each sample at the start and end of sequencing. Finally, we obtained enough high-quality reads; their directions were corrected and analyzed to disclose fungal diversity (Mukherjee et al., 2014).
To identify OTUs, phylogenetic clustering analysis was performed using the Uparse v7.0.10901 OTU clustering software tool at an identity threshold of 97% (Ye et al., 2017). The identification of chimeric sequences was performed using UCHIME v4.2 (Edgar et al., 2011) and subsequently erased from the raw data. A group of eight bases was called “word.” The total number of combinations was calculated according to the reference library and repeated 100 times. The combinations that appear 100 times were considered a bootstrap cutoff. Furthermore, the RDP classifier Bayesian algorithm was employed to measure the confidence score of the taxonomic assignment at each taxonomic level, which was 80%. Fungal community composition and scientific classifications of endophytes in A. zerumbet of each sample were analyzed at a standard classification system level, such as kingdom, phylum, class, order, family, genus, and species (Chen et al., 2016). The number of common and unique OTUs in each sample was counted, and a Venn diagram was constructed (Fouts et al., 2012).
Both alpha (α) and beta (β) diversities of endophytic fungi in A. zerumbet seeds were analyzed. The alpha diversity (α-diversity) of a microbial community involves the evaluation of sequencing depth, coverage, and comparison of abundance and diversity of seed-derived endophytes at different growth stages. We used Good’s coverage metric (C = 1-n1/N) to estimate sequence coverage, where n1 is the number of OTUs containing only one sequence and N the is total number of sequences obtained from one sample (He et al., 2020). In contrast, the beta diversity (β-diversity) involves the exploration of diversity among different species in a microbial community found at different habitats or communities. We calculated the α-diversity of three biological repeats of each sample by employing Chao 1, ACE, Shannon, and Simpson indices (Gihring et al., 2012; Rogers et al., 2016). To explore the Euclidean distances and dissimilarity index among all samples, variation in the composition of OTUs at an identity threshold of 97% was calculated in each sample by principal component analysis (PCA) and principal coordinate analysis (PCoA) (Calderón et al., 2016). To analyze the functional groups of endophytic fungi present in A. zerumbet seeds, all reads were analyzed in the FUNGuild database2 (Song et al., 2019). Comparative taxonomic analyses of the endophytic fungal community present in all samples were performed at each level of classification, and the R tool was used to construct both community structure diagrams and histograms (Lu et al., 2016; Zhou et al., 2017).
All data are expressed as mean values, and one-way analysis of variance (ANOVA) was used to conduct the analysis. Further, Tukey’s HSD test was performed to analyze variations among means of Shannon, Simpson, ACE, Chao, and diversity indices at a significance level of p < 0.05. All correlation and path coefficient analyses were performed with SPSS Statistics 20.0 software (SPSS Inc., Chicago, IL, United States) and Excel 2019.
Endophytes from A. zerumbet seeds were harvested, and high-fidelity MiSeq analysis was carried out after DNA extraction to discover the community structure of fungal endophytes. We obtained 171,166 reads from YSJ1, 176,695 from YSJ2, 171,928 from YSJ3, 201,842 from YSJ4, and 185,063 from YSJ5 (Table 1). From all seed samples collected at the different stages, 906,694 high-quality endophytic fungal genomic sequences were obtained with read lengths ranging from 140 to 527 bp (Table 1 and Supplementary Table 1). A group of eight bases was called “word,” and more than 60,000 combinations were obtained. The total combinations of sequences were calculated according to the “word” reference library and repeated 100 times. The number of calculations, which appeared in the same classification position for 100 times, was considered the bootstrap cutoff or confidence threshold, which was 80%. All classification results were obtained by following this bootstrap cutoff. A rarefaction curve was constructed to evaluate the sampling and sequencing depth of each sample (Figure 1). The rarefaction curves of all samples were flat and saturated, confirming that the sampling depth and sequence reads were at appropriate levels. The confidence in the endophytic fungal community structure was very high, which reflected the maximum inclusion of endophytic fungi in A. zerumbet seeds during sequencing.
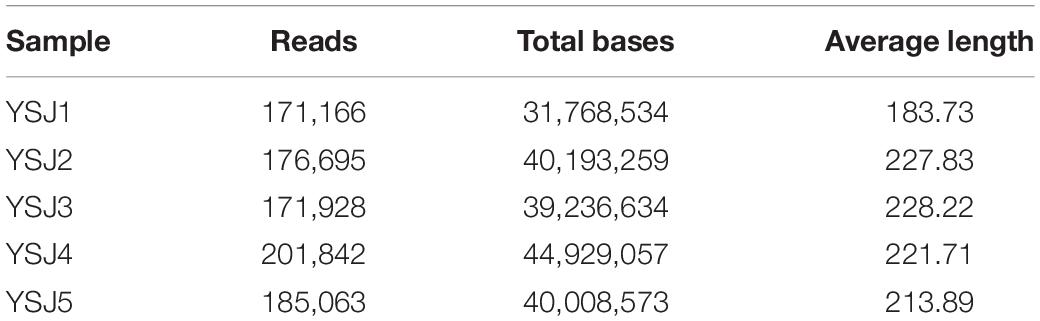
Table 1. Average internal transcribed spacer (ITS) reads of fungal endophytes in Alpinia zerumbet seeds at five stages.
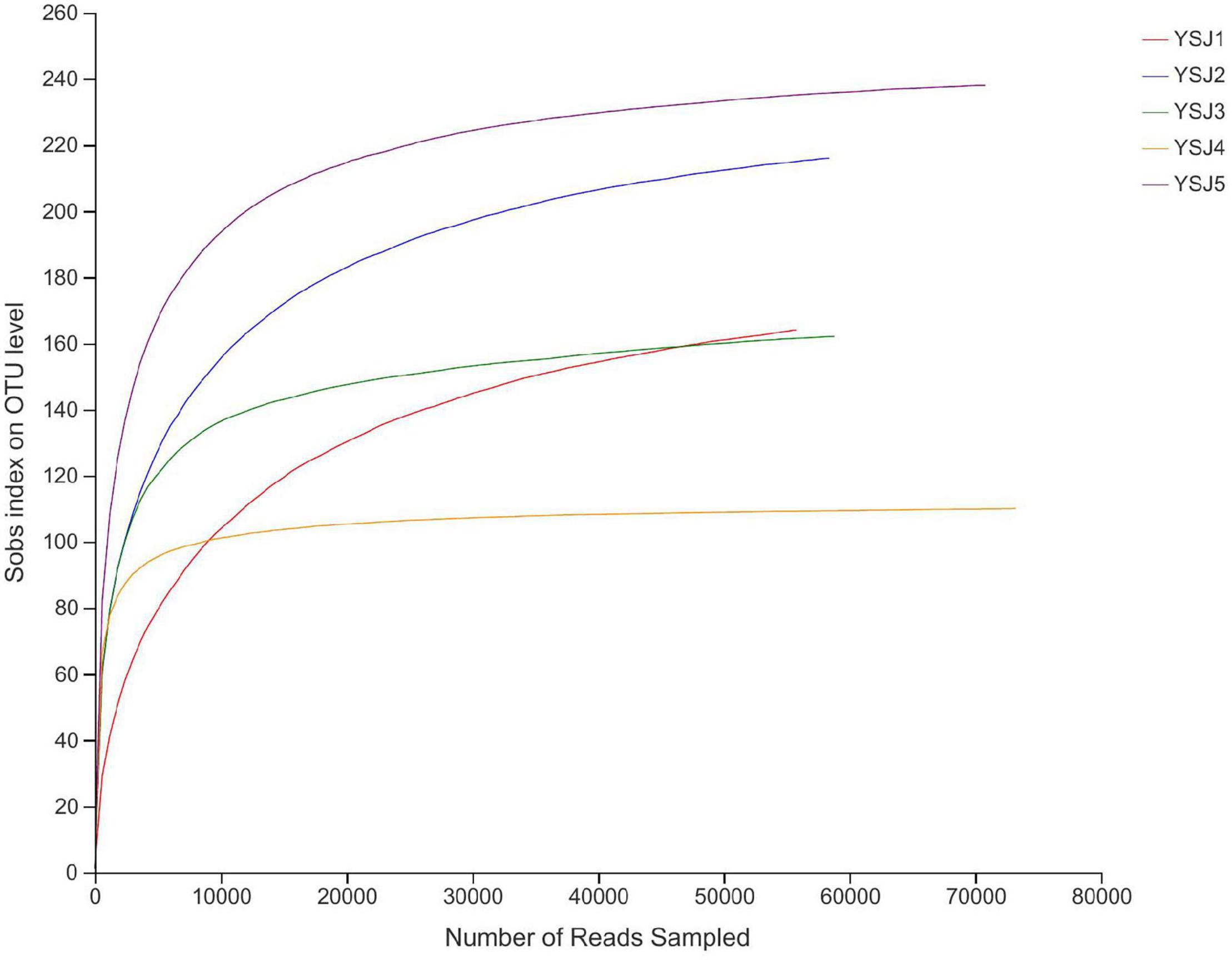
Figure 1. Rank abundance curves of all samples. The abscissa represents the rank number of operational taxonomic units (OTUs), and the ordinate represents the relative percentage of species at the classification level. The position of the abscissa of extension end point of the sample curve represents the number of species in each sample. Smooth curves indicate higher species diversity, while steep decline indicates a high proportion of bacterial strains and low fungal diversity.
A Venn diagram was constructed to depict similar and overlapping OTUs among all A. zerumbet seeds samples at different classification levels (Figure 2). A total of 741 OTUs were pooled through statistical analysis at a similarity threshold of 97% and further classified into 8 phyla, 30 classes, 73 orders, 163 families, 301 genera, and 446 species. Endophytic fungal OTUs obtained from each sample of A. zerumbet seeds collected at different stages were 397, 366, 293, 134, and 367, respectively (Supplementary Table 1). As displayed in the Venn diagram, 241 OTUs were common between YS1 and YS2, 177 OTUs were common between YS1 and YS3, 88 OTUs were common between YS1 and YS3, and 201 OTUs were common between YS1 and YS5. The unique OTUs among all samples collected at different stages were 95, 73, 60, 30, and 109, respectively, which were 23.9, 19.9, 20.5, 22.4, and 29.7% of the total OTUs. Evidently, only 62 endophytic fungal OTUs were common among all five groups, which were 8.3% of the total number of OTUs (Figure 2). A decreasing trend of endophytic fungal OTUs at the initial stage of growth of A. zerumbet seeds was observed; however, a gradual increase with an increase in growth was observed, indicating a significant change in the community structure of endophytic fungi.
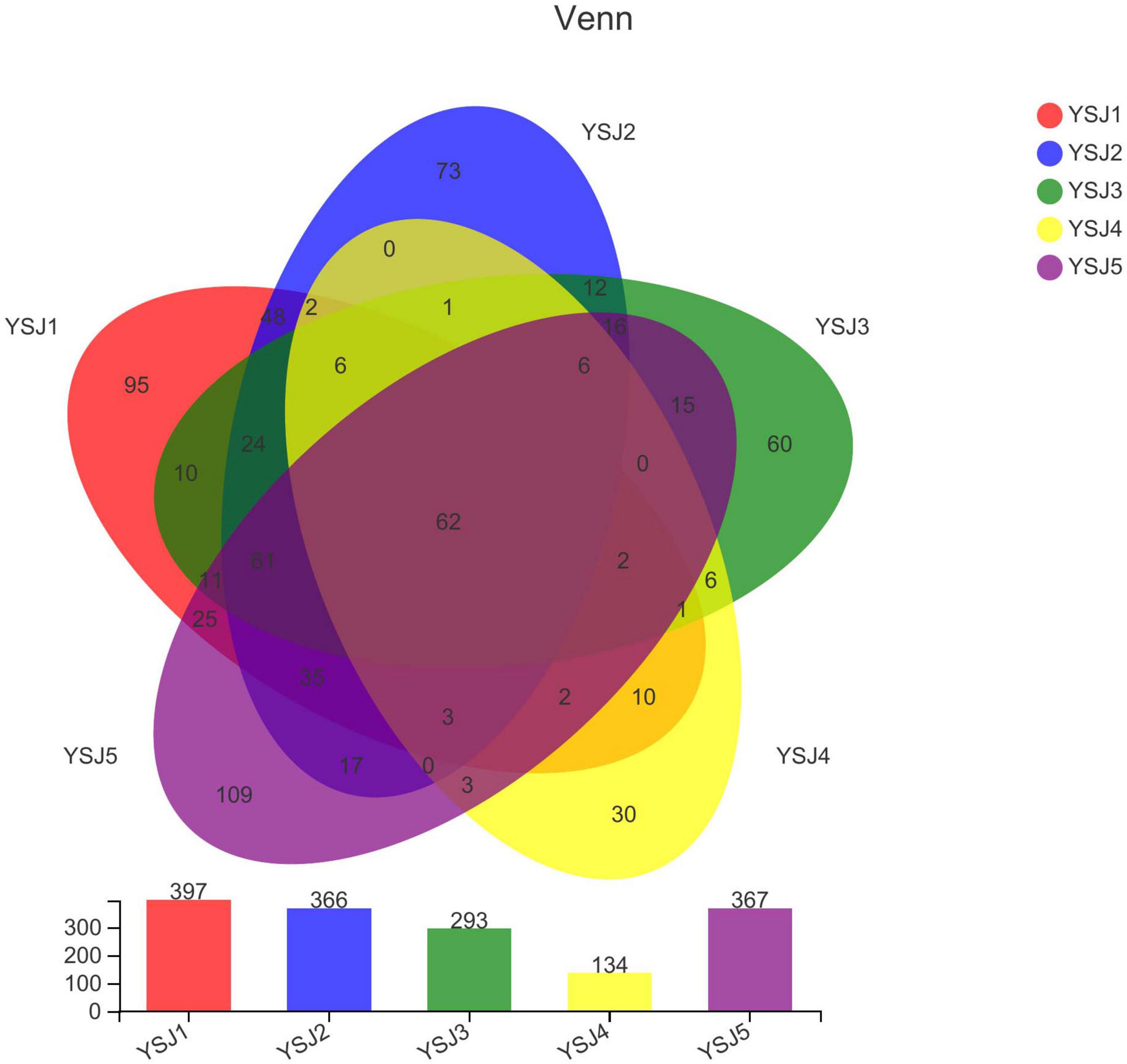
Figure 2. Venn diagram of OTUs. Different groups are represented by different colors, and numbers in overlapping portions represent the number of species common in all groups.
The coverage threshold of ITS sequences obtained from A. zerumbet seeds was > 0.9996, indicating that it represents the true fungal populations in the microbial community of each sample (Table 2). Higher Shannon index and lower Simpson index indicate the highest diversity index of fungal community in a sample. The highest endophytic fungal community diversity was observed in samples collected at the YSJ5 growth period followed by the YSJ1 period. In contrast, the lowest fungal community diversity was observed in samples collected at the YSJ4 growth stage. Similarly, Chao and ACE indices were employed to evaluate the richness of fungal communities in all samples (Table 2). We observed the highest endophytic fungal community abundance in YSJ5 samples followed by YSJ1 samples, and the lowest fungal community abundance in YSJ4 samples. These findings indicate that the richness and diversity of the endophytic fungi of A. zerumbet seeds were quite different at different growth stages, with an increase followed by a decrease (Table 2).
During taxonomic analysis, sequence reads were normalized at 43,847 minimum number of sequence reads per sample and a bar chart revealed 38 annotated genera of relatively high abundance and their taxonomic name, as well as relatively low abundant genera (Figure 3). Overall, the highly abundant endophytic fungal genera in all samples of A. zerumbet seeds were Cladosporium (5.64, 21.10, 34.36, 2.02, and 14.18%, respectively), Kodamaea (41.55, 4.22, < 0.01, < 0.01, and < 0.01%, respectively), Hannaella (20.36, 7.46, 4.94, 1.94, and 6.45%, respectively), Mycothermus (0.26, 0.43, 7.17, 23.57, and 0.45%, respectively), Gibberella (0.30, 9.60, 14.31, 3.51, and 1.59%, respectively), Sarocladium (0.31, 16.78, 5.90, 0.71, and 1.95%, respectively), and Neopestalotiopsis (0.43, 0.88, 2.70, 5.81, and 10.24%, respectively) (Figure 3). Evidently, the highest richness of fungal species was observed at YSJ5 followed by YSJ1. Cladosporium, Hannaella, and Gibberella had the highest abundance, while Plectosphaerella, Vishniacozyma, and Ramichloridium had the lowest abundance at each growth stage of A. zerumbet seeds. Kodamaea fungal genus had high abundance at YSJ1 and low abundance at YSJ3. Contrastingly, Neopetalotiopsis fungal genus had low abundance at YSJ2 and high abundance at YSJ5 (Figure 4). Thus, the growth pattern and abundance/richness of endophytic fungal genera were not the same at different growth stages of A. zerumbet seeds.
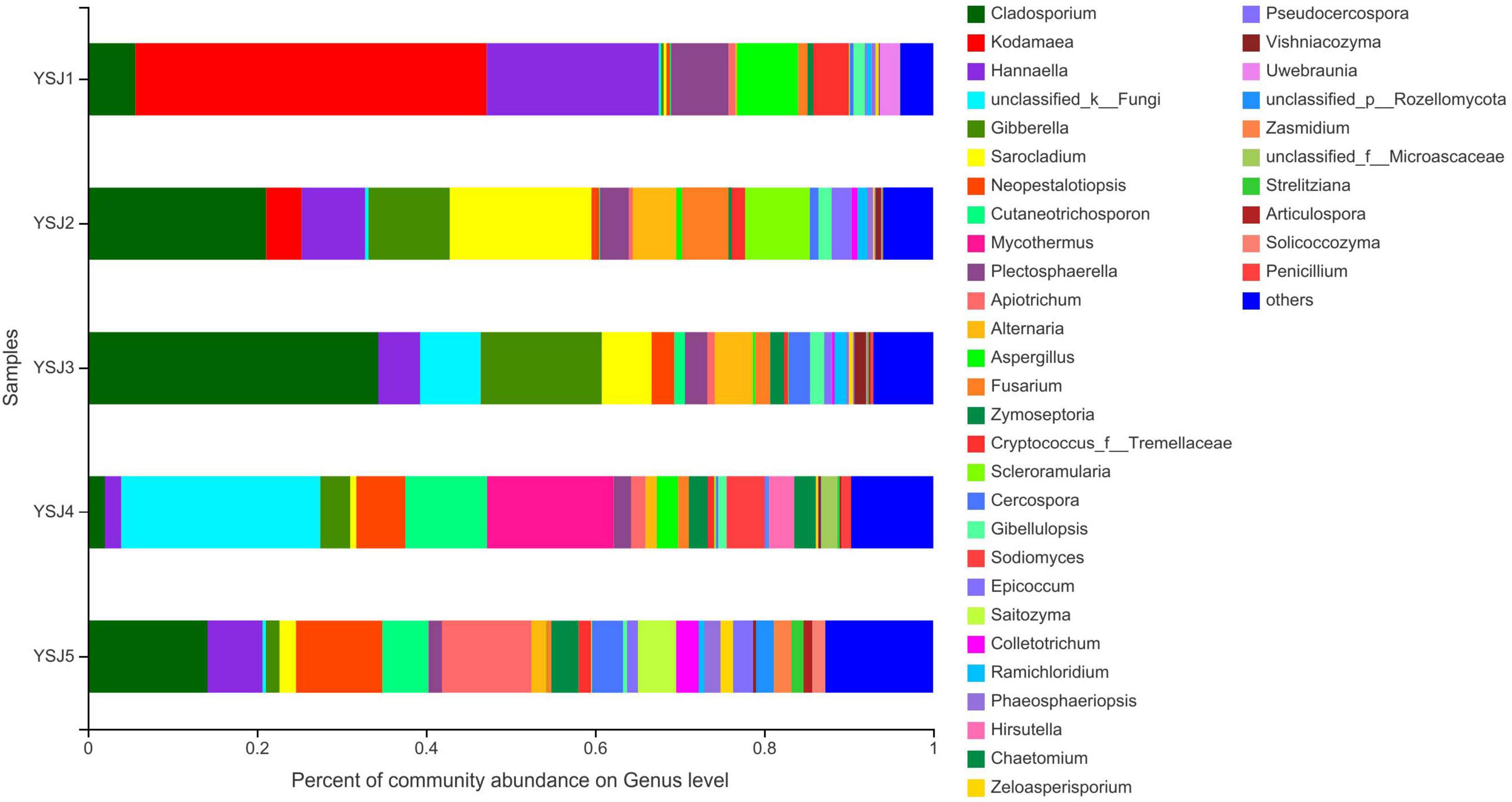
Figure 3. Fungal community structure bar plot analysis at the genus level. The ordinate represents sample name, and the abscissa represents the percentage of different species, which is represented by columns with different colors, sizes, and proportions of a species.
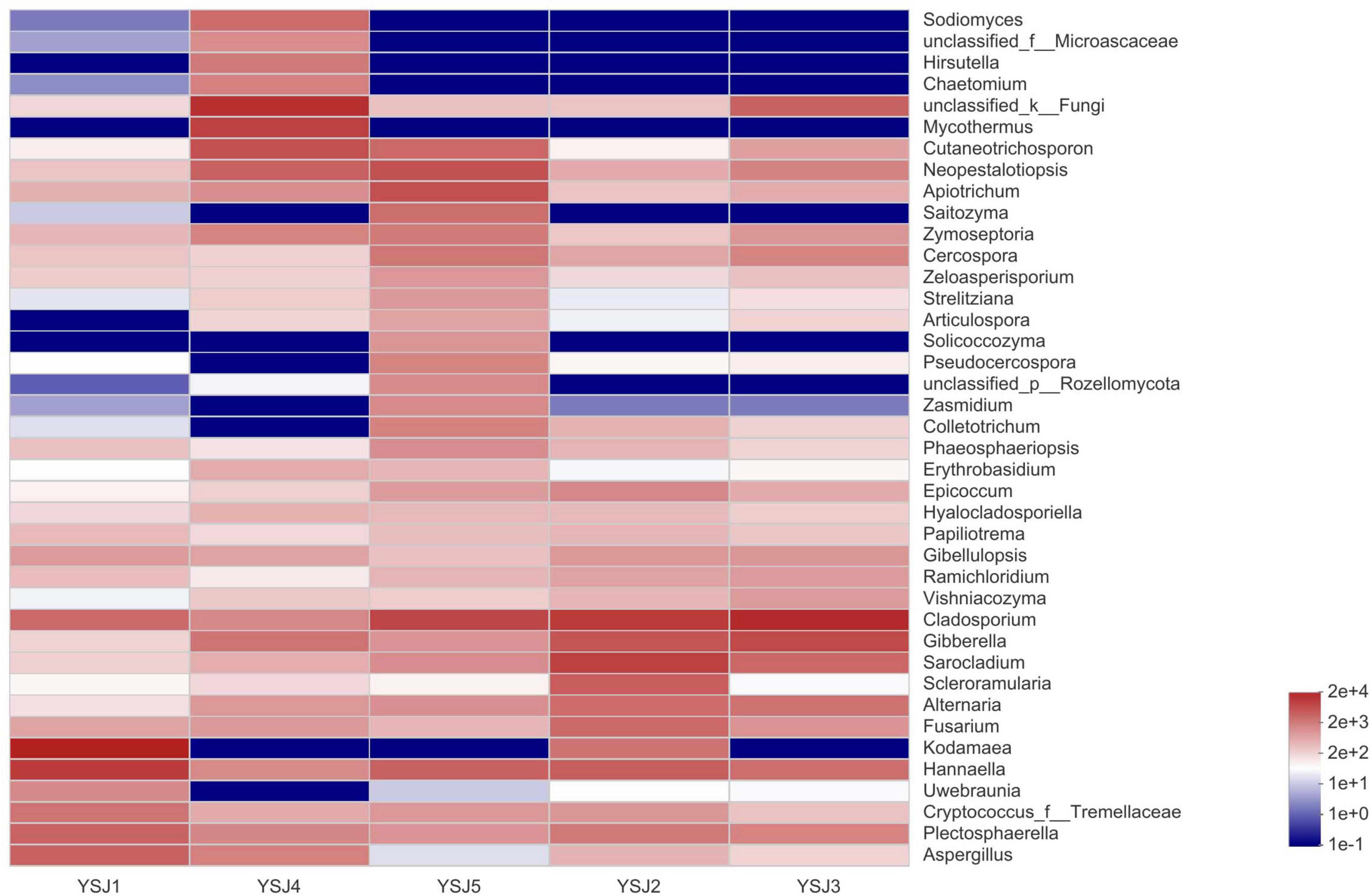
Figure 4. Community heatmap analysis at the genus level. The bar on the right side represents the abundance value by color gradient.
To explore the β-diversity, PCA (Figure 5) and PCoA (Figure 6) of samples of A. zerumbet seeds were performed. The highly abundant fungal genera present in all samples were mapped to construct PCA and PCoA maps, respectively. The sum of PC1 and PC2 was 92.83% in PCA and 94.59% in PCoA, indicating a high diversity in the fungal population structure in samples collected from A. zerumbet seeds. The samples collected at YSJ2, YSJ3, and YSJ4 were clustered closely but were far from samples collected at YSJ1 and YSJ5. Similarly, samples YSJ1 and YSJ5 were distantly clustered. These results proved the significant diversity among endophytic fungal communities in A. zerumbet seeds.
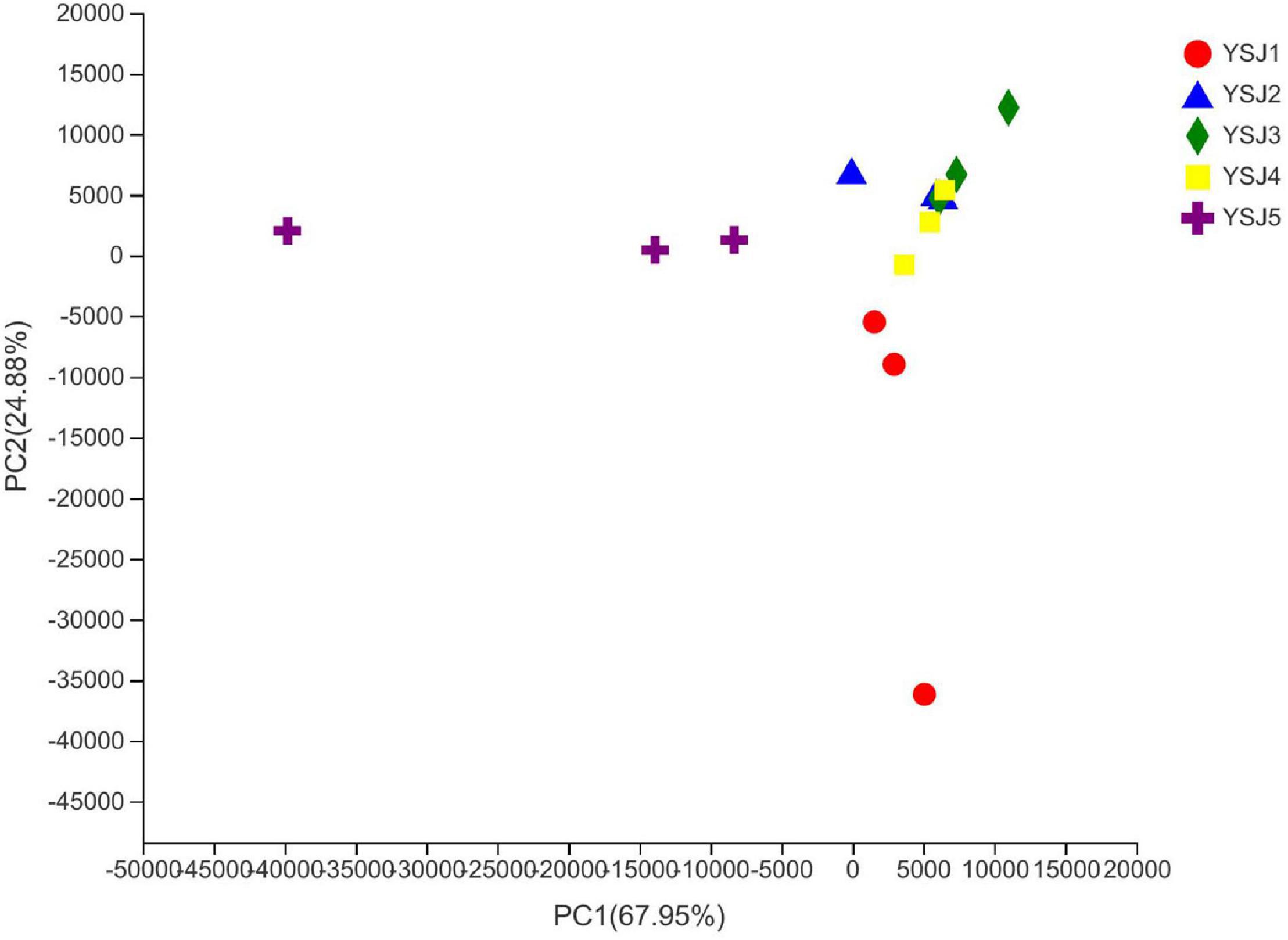
Figure 5. Multiple sample principal component analysis (PCA) of the OTU level. Both selected principal component axes are represented by the x-axis and y-axis, and percentage represents the difference in sample composition by principal component; scales of x-axis and y-axis represent relative distances. Samples are represented by different color points or shapes in different groups. Closeness of points or shapes represents the similarity level between fungal species composition.
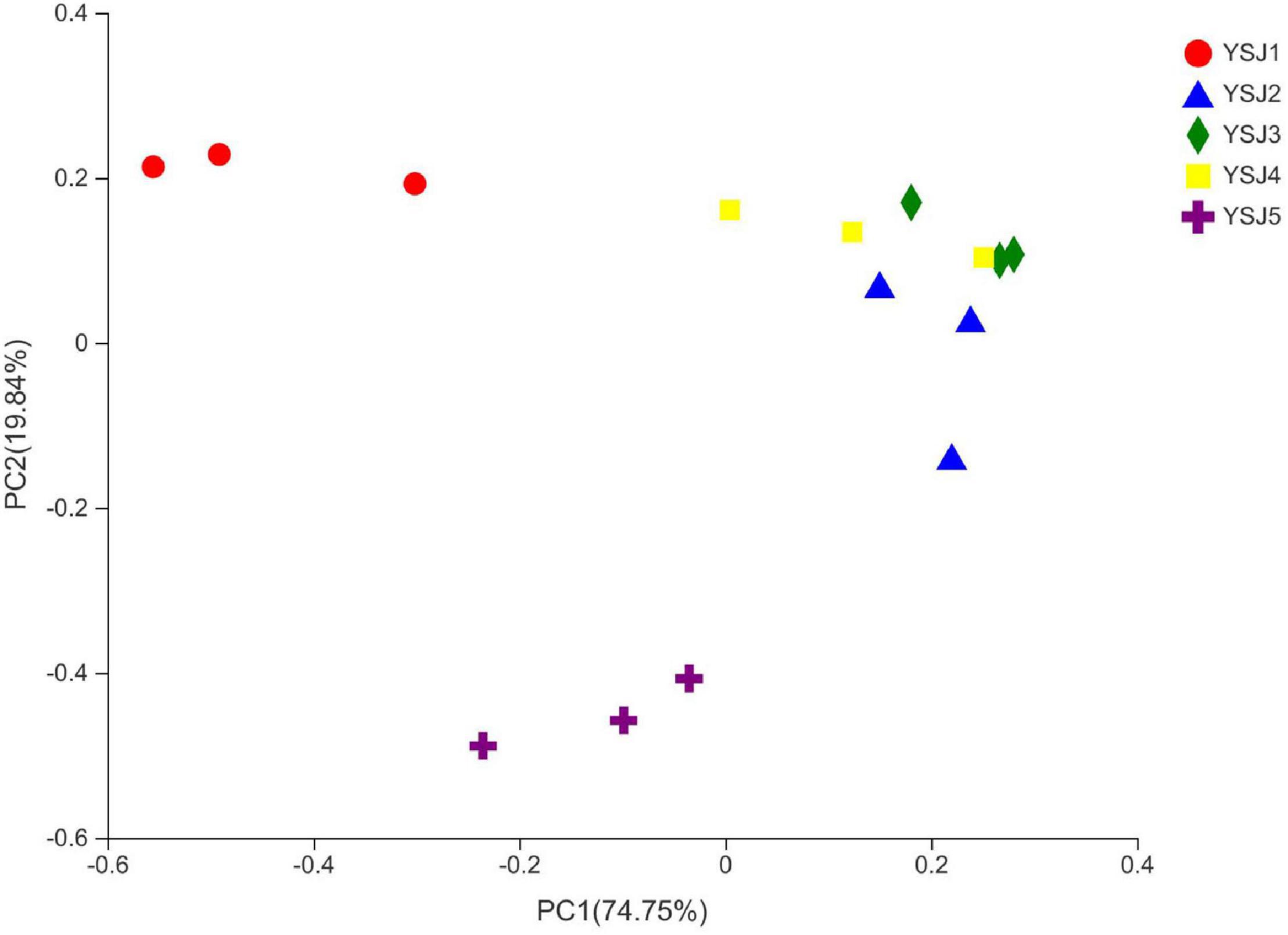
Figure 6. Multiple sample principal coordinate analysis (PCoA) of the OTU level. Both selected principal component axes are represented by the x-axis and y-axis, and percentage represents the difference in sample composition by principal component; scales of x-axis and y-axis represent relative distances. Samples are represented by different color points or shapes in different groups. Closeness of points or shapes represents similarity level between fungal species composition.
Functional Guild (FUNGuild) is a tool used for the classification and analysis of fungal communities in a microecological guild. Functional classification of fungi in a sample and abundance of fungal genera present in different samples are helpful to disclose their source and pathway (Schmidt et al., 2019). FUNGuild was used to classify fungi as undefined saprotroph, plant pathogens, animal pathogen–endophyte–lichen parasite–plant pathogen–wood saprotroph, soil saprotroph, and others (Figure 7). The source of A. zerumbet seeds-derived endophytic fungi differed at different growth stages; however, the environment was the richest source. The abundance of different types of fungal endophytes in samples collected from A. zerumbet seeds was altered at each growth stage from YSJ1 to YSJ5. Evidently, the samples collected at YSJ5 were highly abundant in endophytic fungi.
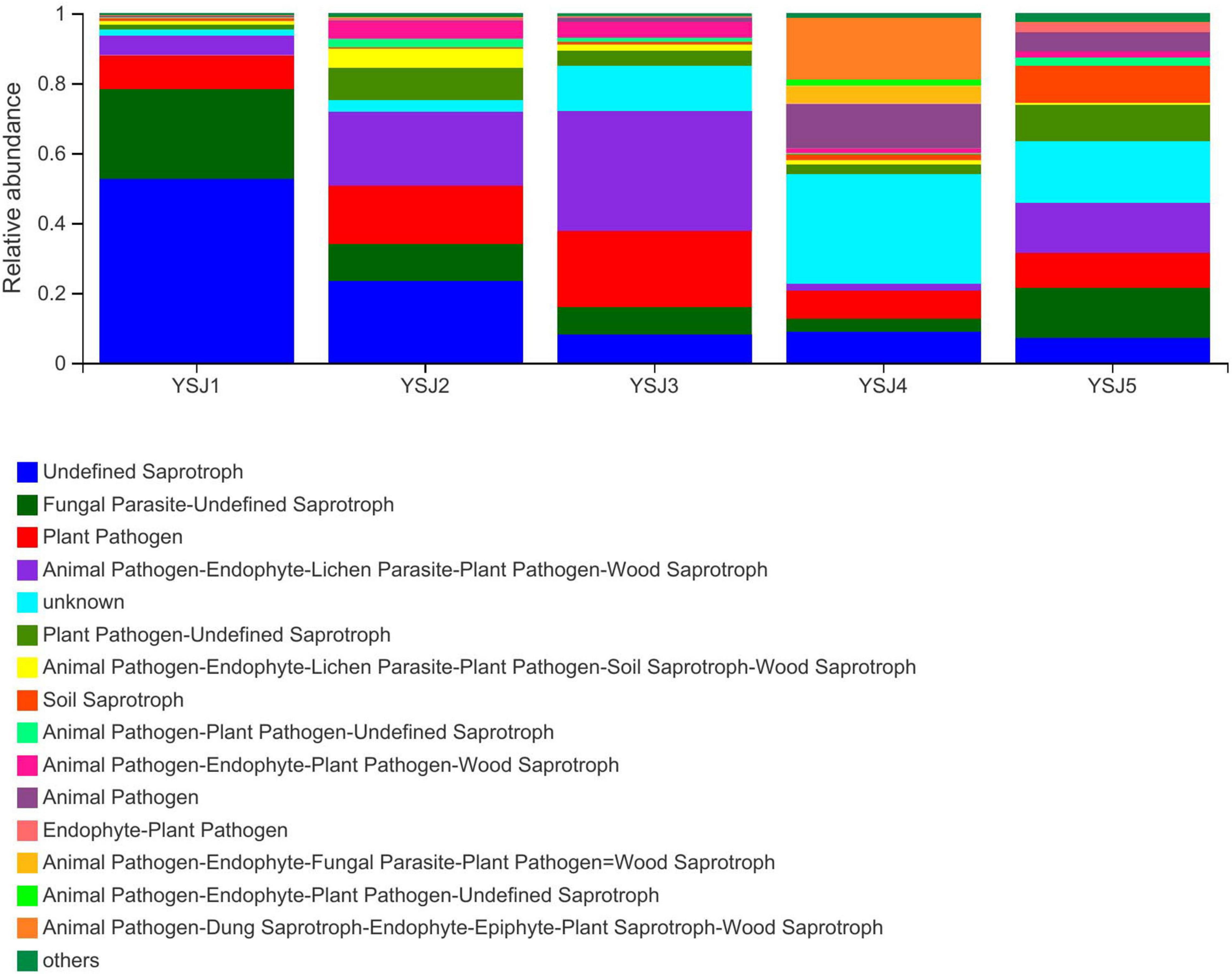
Figure 7. Functional Guild (FUNGuild) analysis of the fungal functional groups. Relative abundance of Guild in different groups or samples is depicted on the x-axis and groups or samples are presented on the y-axis. According to variation in the functional group, FUNGuild can calculate the abundance of each fungal species and its functional classification in each sample.
Normal germination of seeds of different plants is markedly dependent on the presence of diverse microbial communities (Mehra et al., 2017). Seed-derived microbial community is affected by soil, surrounding environment, host genotype, and seed quality (Klaedtke et al., 2016; Walitang et al., 2018). The diversity of microbial communities around seeds is affected by the physiological state of seed and significantly impacts seed health (Zhang et al., 2017). The presence of microbes in a plant rhizosphere has been widely assessed; however, little is known about seed-derived endophytes (Stroheker et al., 2018). Robust culture-independent Illumina high-throughput sequencing was employed to reveal the actual population, community structure, diversity, population succession dynamics, and correlation of endophytic fungi in A. zerumbet seeds at five different growth stages (YSJ1–YSJ5). A total of 906,694 high-fidelity fungal sequences were retrieved with an average length of 216.32 bp. A total of 745 OTUs were obtained, which were further divided into 8 phyla, 30 classes, 73 orders, 163 families, 302 genera, and 449 species. The following endophytic fungal genera, Gibberella, Aspergillus, Alternaria, Fusarium, Cladosporium, Kodamaea, and Hannaella, identified in A. zerumbet seeds were also isolated from the seeds of several species, such as Salvia miltiorrhiza (Barret et al., 2015; Chen et al., 2018).
Studies on different species of Podocarpus and Tinospora revealed that the geographical location and climatic conditions affect the endophytic fungal diversity in a given area (Joshee et al., 2009; Mishra et al., 2012). Additionally, metabolites stored in seeds are the richest source of nutrients and temperature for growth and reproduction of endophytic fungi (Abdallah et al., 2016). The diversity index of endophytic fungi in A. zerumbet seeds collected from the same area and different time periods decreased from YSJ2 to YSJ4 and then increased at YSJ5. This finding could be due to the availability of a favorable environment, such as temperature, water contents, and dry matter for fungal growth at YSJ1, which were depleted during YSJ2 to YSJ4 and stored at the final maturation stage, YSJ5 (Zhang et al., 2019). Different endophytes become dominant at different stages of seed germination due to their adaptive nature of variable internal seed environment, such as Kodamaea at YSJ1, Cladosporium at YSJ2 and YSJ3, Mycothermus at YSJ4, and Cladosporium at YSJ5; a similar trend was observed in rice seeds (Shahzad et al., 2017).
A mutual relationship exists between seeds and endophytes for their growth and the biosynthesis of secondary metabolites. In A. zerumbet seeds, we identified few common endophytes, such as Alternaria, Fusarium, Pythium, and Cladosporium, which were previously identified in tobacco seeds (Tang et al., 2002). The dominant genera Alternaria, Gibberella, and Aspergillus have certain antibacterial activities (Shan et al., 2019; Zhao et al., 2021). Cladosporium is a rich source of biologically active compounds (Salvatore et al., 2021). Sarocladium plays an efficient role in controlling soil-borne diseases (Błaszczyk et al., 2021). Fusarium plays a significant role in plant growth by depleting Cd2+, As3+, and Pb2+ contents and promoting seed germination (Jin et al., 2017). Colletotrichum harbors special metabolites that inhibit phosphatidylinositol 3-kinase (PI3Kα) (Yang et al., 2019). Aspergillus is famous for producing enzymes, such as cellulase, xylanase, manganese peroxidase (MnP), esterase (Batista-García et al., 2014), α-amylase of obligate halophilic, and lactase (Ali et al., 2015; Ma et al., 2017). These findings provide information that would be useful in further studies to determine the specific biological functions of A. zerumbet seed-derived endophytes.
Seeds detach from plants, disperse, and can gather pathogenic fungal microbes around them, which become active under favorable conditions, resulting in disease or decay (Song et al., 2017). Most of the endophytic fungi in A. zerumbet seeds were undefined saprotroph, plant pathogens, and animal pathogen–endophyte–lichen parasite–plant pathogen–wood saprotroph (Figure 7). The abundance of pathogens and saprotrophs indicates the presence of parasitized endophytes in A. zerumbet seeds. Noticeably, when FUNGuild was employed to explore ecological guilds to further parse OTUs of endophytic fungi in A. zerumbet seeds, some results were incomplete and uninterpretable. For example, Aspergillus candidus was only classified as a saprotroph; however, according to several reports, it is also an endophytic fungus in plants (Ma et al., 2017). Thus, the FUNGuild database needs to be updated.
We explored the endophytic fungi in A. zerumbet seeds by deploying culture-independent, robust high-throughput sequencing of the ITS region of fungi. Although the efficacy of ITS sequencing is not 100% due to the limitations of next-generation sequencing read length, this method covers a broad range of fungal genera relative to a traditional culturing technique. A. zerumbet seeds were also found to harbor endophytic fungal genera that are extensively distributed in all seeds, such as Altrnaria, Fusarium, Pythium, and Cladosporium. The abundance of endophytic fungi in A. zerumbet seeds may have some relation with the biosynthesis of an essential oil. Many potential probiotics were identified in A. zerumbet seeds, including Fusarium, Alternaria, Cryptococcus, Colletotrichum, and Aspergillus, which can be further used as probiotics and biocontrol agents.
The datasets presented in this study can be found in online repositories. The names of the repository and accession number(s) can be found below: European Nucleotide Archive (ENA) under accession number PRJEB49388 (ERP133892).
KY, MA, AE-S, and QW conceptualized the study and wrote the original draft. KY, ZP, LM, YZ, LW, RF, QL, YL, XZ, and MA draw the figures. KY, AE-S, QW, and MA did the editing and proofreading of the study. KY and MA supervised the study. KY was the project administrator. All authors contributed to the article and approved the submitted version.
KY acknowledges that this work was co-supported by the Sichuan Provincial Department of Science and Technology Project (Grant No. 2020JDRC0083) and the Scientific Research Project of Yibin University (Grant No. 2018RC09).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.814864/full#supplementary-material
Abbas, M., Nawaz, S., Ahmed, N., Abbas, G., Usman, M., Bashir, H. M. N., et al. (2016). Salicylic acid in amelioration of salt tolerance in wheat (Triticum aestivum L.) under in vitro conditions. Int. J. Res. Innov. Earth Sci. 3, 44–49.
Abbas, M., Peszlen, I., Shi, R., Kim, H., Katahira, R., Kafle, K., et al. (2020). Involvement of CesA4, CesA7-A/B and CesA8-A/B in secondary wall formation in Populus trichocarpa wood. Tree Physiol. 40, 73–89. doi: 10.1093/treephys/tpz020
Abdallah, R. A. B., Mokni-Tlili, S., Nefzi, A., Jabnoun-Khiareddine, H., and Daami-Remadi, M. (2016). Biocontrol of Fusarium wilt and growth promotion of tomato plants using endophytic bacteria isolated from Nicotiana glauca organs. Biol. Control 97, 80–88. doi: 10.1111/jph.12501
Ali, I., Akbar, A., Anwar, M., Prasongsuk, S., Lotrakul, P., and Punnapayak, H. (2015). Purification and characterization of a polyextremophilic α-amylase from an obligate halophilic Aspergillus penicillioides isolate and its potential for souse with detergents. Biomed Res. Int. 2015:245649. doi: 10.1155/2015/245649
Barret, M., Briand, M., Bonneau, S., Préveaux, A., Valière, S., Bouchez, O., et al. (2015). Emergence shapes the structure of the seed microbiota. Appl. Environ. Microbiol. 81, 1257–1266. doi: 10.1128/AEM.03722-14
Batista-García, R. A., Balcázar-López, E., Miranda-Miranda, E., Sánchez-Reyes, A., Cuervo-Soto, L., Aceves-Zamudio, D., et al. (2014). Characterization of lignocellulolytic activities from a moderate halophile strain of Aspergillus caesiellus isolated from a sugarcane bagasse fermentation. PLoS One 9:e105893. doi: 10.1371/journal.pone.0105893
Berg, G., and Raaijmakers, J. M. (2018). Saving seed microbiomes. ISME J. 12, 1167–1170. doi: 10.1038/s41396-017-0028-2
Błaszczyk, L., Waśkiewicz, A., Gromadzka, K., Mikołajczak, K., and Chełkowski, J. (2021). Sarocladium and Lecanicillium associated with maize seeds and their potential to form selected secondary metabolites. Biomolecules 11:98. doi: 10.3390/biom11010098
Calderón, K., Spor, A., Breuil, M. C., Bru, D., Bizouard, F., Violle, C., et al. (2016). Effectiveness of ecological rescue for altered soil microbial communities and functions. ISME J. 11, 272–283. doi: 10.1038/ismej.2016.86
Chang, E., Deng, N., Zhang, J., Liu, J., Chen, L., Zhao, X., et al. (2018). Proteome-level analysis of metabolism-and stress-related proteins during seed dormancy and germination in Gnetum parvifolium. J. Agric. Food Chem. 66, 3019–3029. doi: 10.1021/acs.jafc.7b05001
Chen, B., Teh, B. S., Sun, C., Hu, S., Lu, X., Boland, W., et al. (2016). Biodiversity and activity of the gut microbiota across the life history of the insect herbivore Spodoptera littoralis. Sci. Rep. 6:29505. doi: 10.1038/srep29505
Chen, H., Wu, H., Yan, B., Zhao, H., Liu, F., Zhang, H., et al. (2018). Core microbiome of medicinal plant Salvia miltiorrhiza seed: a rich reservoir of beneficial microbes for secondary metabolism? Int. J. Mol. Sci. 19:672. doi: 10.3390/ijms19030672
Compant, S., Clément, C., and Sessitsch, A. (2010). Plant growth-promoting bacteria in the rhizo-and endosphere of plants: their role, colonization, mechanisms involved and prospects for utilization. Soil Biol. Biochem. 42, 669–678. doi: 10.1016/j.soilbio.2009.11.024
Dalling, J. W., Davis, A. S., Schutte, B. J., and Elizabeth Arnold, A. (2011). Seed survival in soil: interacting effects of predation, dormancy and the soil microbial community. J. Ecol. 99, 89–95. doi: 10.1111/j.1365-2745.2010.01739.x
De Filippis, F., Laiola, M., Blaiotta, G., and Ercolini, D. (2017). Different amplicon targets for sequencing-based studies of fungal diversity. Appl. Environ. Microbiol. 83, e00905–e00917. doi: 10.1128/AEM.00905-17
Donoso, R., Leiva-Novoa, P., ZúIga, A., Timmermann, T., Recabarren-Gajardo, G., and González, B. (2016). Biochemical and genetic bases of indole-3-acetic acid (auxin phytohormone) degradation by the plant-growth-promoting rhizobacterium Paraburkholderia phytofirmans PsJN. Appl. Environ. Microbiol. 83:e1991-16. doi: 10.1128/AEM.01991-16
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
El-Sappah, A., Islam, M., Rather, S., Li, J., Yan, K., Xianming, Z., et al. (2021). Identification of novelroot-knot nematode (Meloidogyne incognita) resistant tomato genotypes. J. Anim. Plant Sci. 32, 1–17. doi: 10.36899/JAPS.2022.1.0407
Fareed, G., Atiq, M., Abbas, M., Usman, M., Abbas, G., and Qamar, S. H. (2017). Varietal reaction of cucumber (Cucumis sativus L.) germplasm for management of fusarium wilt of cucumber (FWC). Adv. Zool. Bot. 5, 1–3. doi: 10.13189/azb.2017.050101
Fouts, D. E., Lauren, B., Erin, B., Jason, I., and Granger, S. (2012). PanOCT: automated clustering of orthologs using conserved gene neighborhood for pan-genomic analysis of bacterial strains and closely related species. Nucleic Acids Res. 22:e172. doi: 10.1093/nar/gks757
Gihring, T. M., Green, S. J., and Schadt, C. W. (2012). Massively parallel rRNA gene sequencing exacerbates the potential for biased community diversity comparisons due to variable library sizes. Environ. Microbiol. 14, 285–290. doi: 10.1111/j.1462-2920.2011.02550.x
Gond, S. K., Bergen, M. S., Torres, M. S., and White, J. F. (2015). Endophytic Bacillus spp. produce antifungal lipopeptides and induce host defence gene expression in maize. Microbiol. Res. 172, 79–87. doi: 10.1016/j.micres.2014.11.004
He, Q., Wang, H., Chen, L., Gao, S., Zhang, W., Song, J., et al. (2020). Elevated salinity deteriorated enhanced biological phosphorus removal in an aerobic granular sludge sequencing batch reactor performing simultaneous nitrification, denitrification and phosphorus removal. J. Hazard. Mater. 390:121782. doi: 10.1016/j.jhazmat.2019.121782
Jin, H., Cheng, C., Xu, Q., Li, X., Liu, H., Mao, L., et al. (2017). Effects of endophytic fungi on tobacco growth and heavy metal content in leaves. Mycosystema 36, 186–192.
Johnston-Monje, D., and Raizada, M. N. (2011). Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology. PLoS One 6:e20396. doi: 10.1371/journal.pone.0020396
Joshee, S., Paulus, B. C., Park, D., and Johnston, P. R. (2009). Diversity and distribution of fungal foliar endophytes in New Zealand Podocarpaceae. Mycol. Res. 113, 1003–1015. doi: 10.1016/j.mycres.2009.06.004
Klaedtke, S., Jacques, M. A., Raggi, L., Préveaux, A., Bonneau, S., Negri, V., et al. (2016). Terroir is a key driver of seed-associated microbial assemblages. Environ. Microbiol. 18, 1792–1804. doi: 10.1111/1462-2920.12977
Lu, Y., Chen, J., Zheng, J., Hu, G., Wang, J., Huang, C., et al. (2016). Mucosal adherent bacterial dysbiosis in patients with colorectal adenomas. Sci. Rep. 6, 1–10. doi: 10.1038/srep26337
Ma, J., Zhang, X. L., Wang, Y., Zheng, J. Y., Wang, C. Y., and Shao, C. L. (2017). Aspergivones A and B, two new flavones isolated from a gorgonian-derived Aspergillus candidus fungus. Nat. Prod. Res. 31, 32–36. doi: 10.1080/14786419.2016.1207073
Mehra, S., Morrison, P., Coates, F., and Lawrie, A. (2017). Differences in carbon source utilisation by orchid mycorrhizal fungi from common and endangered species of Caladenia (Orchidaceae). Mycorrhiza 27, 95–108. doi: 10.1007/s00572-016-0732-1
Mishra, A., Gond, S. K., Kumar, A., Sharma, V. K., Verma, S. K., Kharwar, R. N., et al. (2012). Season and tissue type affect fungal endophyte communities of the Indian medicinal plant Tinospora cordifolia more strongly than geographic location. Microb. Ecol. 64, 388–398. doi: 10.1007/s00248-012-0029-7
Mukherjee, P. K., Chandra, J., Retuerto, M., Sikaroodi, M., Brown, R. E., Jurevic, R., et al. (2014). Oral mycobiome analysis of HIV-infected patients: identification of Pichia as an antagonist of opportunistic fungi. PLoS Pathog. 10:e1003996. doi: 10.1371/journal.ppat.1003996
Peng, X., Pang, H., Abbas, M., Yan, X., Dai, X., Li, Y., et al. (2019). Characterization of Cellulose synthase-like D (CSLD) family revealed the involvement of PtrCslD5 in root hair formation in Populus trichocarpa. Sci. Rep. 9, 1–9. doi: 10.1038/s41598-018-36529-3
Rogers, M. B., Firek, B., Shi, M., Yeh, A., Brower-Sinning, R., Aveson, V., et al. (2016). Disruption of the microbiota across multiple body sites in critically ill children. Microbiome 4:66. doi: 10.1186/s40168-016-0211-0
Rosenblueth, M., and Martínez-Romero, E. (2006). Bacterial endophytes and their interactions with hosts. Mol. Plant Microbe Interact. 19, 827–837. doi: 10.1094/MPMI-19-0827
Salvatore, M. M., Andolfi, A., and Nicoletti, R. (2021). The genus Cladosporium: a rich source of diverse and bioactive natural compounds. Molecules 26:3959. doi: 10.3390/molecules26133959
Schmidt, R., Mitchell, J., and Scow, K. (2019). Cover cropping and no-till increase diversity and symbiotroph: saprotroph ratios of soil fungal communities. Soil Biol. Biochem. 129, 99–109. doi: 10.1016/j.soilbio.2018.11.010
Shade, A., Jacques, M. A., and Barret, M. (2017). Ecological patterns of seed microbiome diversity, transmission, and assembly. Curr. Opin. Microbiol. 37, 15–22. doi: 10.1016/j.mib.2017.03.010
Shahzad, R., Waqas, M., Khan, A. L., Al-Hosni, K., Kang, S. M., Seo, C. W., et al. (2017). Indoleacetic acid production and plant growth promoting potential of bacterial endophytes isolated from rice (Oryza sativa L.) seeds. Acta Biol. Hung. 68, 175–186. doi: 10.1556/018.68.2017.2.5
Shan, T., Qin, K., Xie, Y., Zhang, W., Mao, Z., and Wang, J. (2019). Secondary metabolites of endophytic fungi isolated from Casuarina equisetifolia and their bioactivities. J. South China Agric. Univ. 40, 67–74.
Shen, X. C., Hu, H. S., and Xiao, H. T. (2010). GC-MS analysis of chemical constituents of the essential oil from different parts of Alpinia zerumbet (Pers.) Burtt et Smith. Chinese J. Pharmaceut. Anal. 30, 1399–1403.
Song, H., Singh, D., Tomlinson, K. W., Yang, X., Ogwu, M. C., Slik, J. F., et al. (2019). Tropical forest conversion to rubber plantation in southwest China results in lower fungal beta diversity and reduced network complexity. FEMS Microbiol. Ecol. 95:fiz092. doi: 10.1093/femsec/fiz092
Song, Z., Kennedy, P. G., Liew, F. J., and Schilling, J. S. (2017). Fungal endophytes as priority colonizers initiating wood decomposition. Funct. Ecol. 31, 407–418. doi: 10.1111/1365-2435.12735
Stroheker, S., Dubach, V., and Sieber, T. N. (2018). Competitiveness of endophytic Phialocephala fortinii sl–Acephala applanata strains in Norway spruce roots. Fungal Biol. 122, 345–352. doi: 10.1016/j.funbio.2018.02.005
Tang, Z. X., Fang, D. H., and Bai, Y. F. (2002). Detection and analysis of tobacco seed internal fungi in yunnan. J. Yunnan Agric. Univ. 17, 445–445. doi: 10.1006/jfls.2001.0409
Tao, L., Shen, X. C., Peng, J., and Zhi, N. (2009). Analysis on volatile oils from different parts of Alpinia zerumbet by GC-MS. Chinese Traditional Patent Medicine 31, 909–911.
Verma, S. K., Kingsley, K., Irizarry, I., Bergen, M., Kharwar, R., and White, J. (2017). Seed-vectored endophytic bacteria modulate development of rice seedlings. J. Appl. Microbiol. 122, 1680–1691. doi: 10.1111/jam.13463
Walitang, D. I., Kim, C. G., Kim, K., Kang, Y., Kim, Y. K., and Sa, T. (2018). The influence of host genotype and salt stress on the seed endophytic community of salt-sensitive and salt-tolerant rice cultivars. BMC Plant Biol. 18:51. doi: 10.1186/s12870-018-1261-1
Wang, J., Abbas, M., Wen, Y., Niu, D., Wang, L., Sun, Y., et al. (2018). Selection and validation of reference genes for quantitative gene expression analyses in black locust (Robinia pseudoacacia L.) using real-time quantitative PCR. PLoS One 13:e0193076. doi: 10.1371/journal.pone.0193076
White, J. F., Chen, Q., Torres, M. S., Mattera, R., Irizarry, I., Tadych, M., et al. (2015). Collaboration between grass seedlings and rhizobacteria to scavenge organic nitrogen in soils. AoB Plants 7:lu093. doi: 10.1093/aobpla/plu093
Yan, K., Abbas, M., Meng, L., Cai, H., Peng, Z., Li, Q., et al. (2021a). Analysis of the fungal diversity and community structure in sichuan dark tea during pile-fermentation. Front. Microbiol. 12:706714. doi: 10.3389/fmicb.2021.706714
Yan, K., Yan, L., Meng, L., Cai, H., Duan, A., Wang, L., et al. (2021b). Comprehensive analysis of bacterial community structure and diversity in Sichuan dark tea (Camellia sinensis). Front. Microbiol. 12:735618. doi: 10.3389/fmicb.2021.735618
Yang, Z. D., Li, Z. J., Zhao, J. W., Sun, J. H., Yang, L. J., and Shu, Z. M. (2019). Secondary metabolites and PI3K inhibitory activity of Colletotrichum gloeosporioides, a fungal endophyte of Uncaria rhynchophylla. Curr. Microbiol. 76, 904–908. doi: 10.1007/s00284-019-01707-7
Ye, J., Joseph, S. D., Ji, M., Nielsen, S., Mitchell, D. R., Donne, S., et al. (2017). Chemolithotrophic processes in the bacterial communities on the surface of mineral-enriched biochars. ISME J. 11, 1087–1101. doi: 10.1038/ismej.2016.187
Zhang, J., Zhang, C., Yang, J., Zhang, R., Gao, J., Zhao, X., et al. (2019). Insights into endophytic bacterial community structures of seeds among various Oryza sativa L. rice genotypes. J. Plant Growth Regul. 38, 93–102. doi: 10.1007/s00344-018-9812-0
Zhang, Z., Ci, D., Zhang, G., Ding, H., Yang, J., Dai, L., et al. (2017). Diversity of microbial community structure in the spermosphere of saline-alkali soil in shandong area. Acta Microbiol. Sin. 57, 582–596.
Zhao, S., Wang, B., Tian, K., Ji, W., Zhang, T., Ping, C., et al. (2021). Novel metabolites from the Cercis chinensis derived endophytic fungus Alternaria alternata ZHJG5 and their antibacterial activities. Pest Manag. Sci. 77, 2264–2271. doi: 10.1002/ps.6251
Keywords: Alpinia zerumbet seed, endophytic fungi, high-throughput sequencing, probiotics, community structure
Citation: Yan K, Pei Z, Meng L, Zheng Y, Wang L, Feng R, Li Q, Liu Y, Zhao X, Wei Q, El-Sappah AH and Abbas M (2022) Determination of Community Structure and Diversity of Seed-Vectored Endophytic Fungi in Alpinia zerumbet. Front. Microbiol. 13:814864. doi: 10.3389/fmicb.2022.814864
Received: 14 November 2021; Accepted: 06 January 2022;
Published: 28 February 2022.
Edited by:
Zhiyong Li, Shanghai Jiao Tong University, ChinaReviewed by:
Ivica Z. Dimkić, University of Belgrade, SerbiaCopyright © 2022 Yan, Pei, Meng, Zheng, Wang, Feng, Li, Liu, Zhao, Wei, El-Sappah and Abbas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Manzar Abbas, YWJiYXMyNDcyQGhvdG1haWwuY29t; Ahmed H. El-Sappah, YWhtZWRfZWxzYXBwYWgyMDA2QHlhaG9vLmNvbQ==; Qin Wei, d2VpcWluMjAwMS02N0AxNjMuY29t
†These authors share co-corresponding authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.