- 1School of Life Sciences, University of Hawai‘i at Mānoa, Honolulu, HI, United States
- 2School of Ocean and Earth Science and Technology, University of Hawai‘i at Mānoa, Honolulu, HI, United States
Zinc ion (Zn2+) is an essential micronutrient and a potent antioxidant. However, Zn2+ is often limited in the environment. Upon Zn2+ limitation, Mycolicibacterium (basonym: Mycobacterium) smegmatis (Msm) undergoes a morphogenesis, which relies on alternative ribosomal proteins (AltRPs); i.e., Zn2+-independent paralogues of Zn2+-dependent ribosomal proteins. However, the underlying physiological changes triggered by Zn2+ limitation and how AltRPs contribute to these changes were not known. In this study, we expand the knowledge of mechanisms utilized by Msm to endure Zn2+ limitation, by comparing the transcriptomes and proteomes of Zn2+-limited and Zn2+-replete Msm. We further compare, corroborate and contrast our results to those reported for the pathogenic mycobacterium, M. tuberculosis, which highlighted conservation of the upregulated oxidative stress response when Zn2+ is limited in both mycobacteria. By comparing the multi-omics analysis of a knockout mutant lacking AltRPs (ΔaltRP) to the Msm wild type strain, we specify the involvement of AltRPs in the response to Zn2+ limitation. Our results show that AltRP expression in Msm does not affect the conserved oxidative stress response during Zn2+ limitation observed in mycobacteria, but AltRPs do significantly impact expression patterns of numerous genes that may be involved in morphogenesis or other adaptive responses. We conclude that AltRPs are not only important as functional replacements for their Zn2+-dependent paralogues; they are also involved in the transcriptomic response to the Zn2+-limited environment.
Introduction
Zinc ion (Zn2+) is an important micronutrient, but many bacteria encounter Zn2+-limited environments (Corbin et al., 2008; Alloway, 2009). Acting as a cofactor to many proteins (Andreini et al., 2006) and as a potent antioxidant (Powell, 2000), the presence or absence of Zn2+ can affect bacterial physiology (Lim et al., 2013; Dow and Prisic, 2018; Velasco et al., 2018). For example, in the human pathogen Mycobacterium tuberculosis (Mtb), Zn2+-limitation induces vast changes in expression patterns of the transcriptome and proteome, correlating with increased resistance to oxidative stress and increased virulence of Zn2+-limited Mtb (Dow et al., 2021). A marked feature of Zn2+-limited Mtb was upregulation of the oxidative stress response, evidenced by increased expression of key antioxidant enzymes and genes involved in DNA repair (Dow et al., 2021).
The adaptations to Zn2+ limitation could be driven by the fact that Mtb transits through a Zn2+-depleted niche (i.e., caseum and sputum) during active tuberculosis (Dow et al., 2021). However, environmental bacteria such as Mycolicibacterium (basonym: Mycobacterium) smegmatis (Msm) may also encounter Zn2+ limitation, and so these adaptations could span beyond the context of the infection. Determining if these adaptations to Zn2+ limitation are unique to tuberculosis for Mtb or conserved across mycobacteria will indicate the importance of this adaptive response in (myco)bacteria.
We anticipate vast and unique changes in gene and protein expression patterns in response to Zn2+ limitation in Msm compared to Mtb, especially considering that the previously observed morphological changes (elongated cells with condensed DNA) in Zn2+-limited Msm do not occur in Mtb (Dow and Prisic, 2018; Dow et al., 2021). Moreover, the Zn2+-dependent morphogenesis in Msm relies on expression of Zn2+-independent alternative ribosomal proteins (AltRPs) (Dow and Prisic, 2018). The conserved mycobacterial altRP operon contains four genes encoding AltRPs that replace their primary homologues in the ribosomes of Zn2+-limited mycobacteria (Dow and Prisic, 2018; Li et al., 2018; Tobiasson et al., 2019). Although implicated in shaping mycobacterial physiology, the exact role of AltRPs is unclear (Dow and Prisic, 2018). Because AltRPs are incorporated into functional ribosomes that engage in active translation (Tobiasson et al., 2019), it has been suggested that AltRP-containing ribosomes may selectively translate certain mRNAs (Chen et al., 2020). Therefore, the “failure” of a mutant lacking the altRP operon to undergo the Zn2+-dependent morphogenesis could be due to a checkpoint “failure” in translation, since this strain can only build ribosomes with Zn2+-dependent (or “primary,” Prim) paralogs. Tracking gene expression patterns (i.e., transcript/protein pairs) in the wild type and a mutant lacking AltRPs (ΔaltRP) during Zn2+-limitation will further enlighten the role of AltRPs in mycobacterial physiology.
In this study, we employ a multi-omics analysis of Zn2+-limited Msm and compare the results to those described for Mtb to determine whether the link between Zn2+-limitation and specific transcriptional responses, particularly regulation of the oxidative stress response, is conserved in mycobacteria. We further describe the effects of Zn2+-limitation in Msm beyond those that are shared with Mtb. Given the role of AltRPs in the Zn2+-dependent morphogenesis in Msm, we compare the transcriptome and proteome of the ΔaltRP mutant and the wild type, to inform how AltRP expression contributes to the adaptation to Zn2+-limitation in mycobacteria. Together, this multi-omics analysis of Msm grown in Zn2+-replete media (ZRM) and Zn2+-limited media (ZLM), along with the ΔaltRP mutant and the in trans complement strain (ΔaltRP/c), also grown in ZLM, provides molecular details on how Zn2+ and AltRP expression contributes to mycobacterial physiology.
Materials and Methods
Media, Strains and Bacterial Growth
All chemicals were purchased from Thermo Fisher Scientific™ or VWR™, unless otherwise noted. Middlebrook 7H9 (Difco) broth supplemented with ADC-T (0.5% albumin, 0.2% glucose, 0.085% NaCl, 0.05% Tween 80) was used to maintain Msm strains. Preparation of chemically defined Sauton’s medium (0.05% KH2PO4, 0.05% MgSO47H2O, 0.2% citric acid, 0.005% ferric ammonium citrate, 6% glycerol, 0.4% asparagine, 0.05% Tween 80, pH 7.4) was done with care to avoid Zn2+ contamination from glassware as previously described (Dow et al., 2021). Omitting ZnSO4 in Sauton’s Zn2+-limited medium (ZLM) yields a final Zn2+ concentration around 100 nM, detected by ICP-MS (Dow et al., 2021). For Zn2+-replete medium (ZRM), ZnSO4 was added to ZLM at 6 μM concentration.
Wild type Msm (mc2 155), the deletion mutant of the altRP operon (ΔaltRP) and its complement strain (ΔaltRP/c) were constructed for a previously published study (Dow and Prisic, 2018). Zn2+-limitation was achieved through growth in ZLM to stationary phase as previously described (Dow and Prisic, 2018), at which time cells were harvested for analysis (i.e., 72 h after inoculation). Zn2+-replete cells (i.e., wild type grown in ZRM) were harvested at the same time as the Zn2+-limited cells.
Isolation of RNA and Proteins
From 50 mL cultures, 25 mL of each culture was transferred to 50 mL conical tubes and cells were pelleted with centrifugation at 3,000 × g for 10 mins at 4°C. The supernatant was discarded, and cell pellets were resuspended in 1 mL TRIzol™ reagent (Invitrogen™, Cat. #: 15596026) and transferred to a 2 mL screw-cap tube containing 200 μL of 0.1 mm zirconia beads (BioSpec, Cat. #: 11079101z). Bacterial cell pellets in TRIzol™ were lysed via beating 3 times for 45 seconds at 7000 rpm in a MagNA Lyser (Roche) with cooling on ice for 3 mins between cycles. Screw cap tubes were spun at 12,000 × g for 5 mins at 4°C and the supernatant was transferred to new 1.5 mL tubes. RNA and proteins were extracted from TRIzol™ supernatants concurrently, following the manufacturer’s protocol. Proteins were resuspended in 9.5 M urea and 2% CHAPS buffer, pH 9.1 and stored at −80°C.
Precipitated RNA from the aqueous TRIzol™ fraction was resuspended in nuclease free water and purified using High Pure RNA Isolation kit (Roche, Cat. #: 11828665001) with omission of the on-column DNase digest step. Purified RNA was quantified using the ratio of absorbance at 260 and 280 nm using a Nanodrop™ (Thermo Fisher Scientific™) spectrophotometer and digested in solution twice with TURBO™ DNase (Invitrogen™, Cat. #: AM2239) following the manufacturer’s directions. DNase-digested RNA was re-purified using the same kit.
High-Throughput RNA Sequencing and Differential Expression Analysis
Ribosomal RNA was depleted from the samples using Ribo-Zero plus rRNA depletion kit (Illumina®, Cat. #:20037135) before being sequenced on an Illumina® MiSeq platform for generation of 150 base pair paired-end reads. RNA-seq was performed by GENEWIZ® (South Plainfield, NJ 07080). Analysis of raw read data was achieved following the pipeline: fastQC (Andrews, 2010) of raw reads in .fastq format, trimmomatic (Bolger et al., 2014), fastQC of trimmed reads, Bowtie 2 (Langmead and Salzberg, 2012) alignment of trimmed files and featureCounts (Liao et al., 2014) to define tagwise abundances for coding and non-coding RNAs from BAM files. Reference genome files were obtained from NCBI using the M. smegmatis mc2 155 reference genome assembly (ASM1500v1.39). We manually curated certain gene annotations in the tables when the gene was mentioned in the text but did not have an associated gene annotation in the reference file. The gene names were inferred from orthologues with gene annotations in Mtb, or from gene names reported in the literature for Msm. Homologues between Msm and Mtb were obtained from Mycobrowser, Release 3 (Kapopoulou et al., 2011). Data analysis was performed through the command line using the high-performance computing cyberinfrastructure from the University of Hawaii Information Technology Services.
Genes with tagwise abundances of less than 10 counts in all strains and conditions were considered lowly expressed and excluded from the differential expression analysis. The tagwise abundances were normalized using trimmed means of M-values (TMM) method in edgeR to obtain logCPM values for each feature (Robinson et al., 2010) and the quantile-adjusted conditional maximum likelihood (qCML) linear modeling approach of normalized gene abundances was used to determine the common dispersion using limma (Ritchie et al., 2015). Differential gene expression of the linearized model was determined using an “exact-like test” that utilizes a negative binomial distribution model and Benjamini-Hochberg false discovery rate (FDR) approximation to control for the family wise error rate (Ritchie et al., 2015). Significantly differentially expressed genes (DE genes) were defined as having an absolute fold change (absFC) greater than 2 and an FDR (adjusted P-value) less than 5% (FDR < 0.05, absFC > 2). A log2-fold change (lfc) threshold of 1 was applied using treat function in limma to ensure the false discovery rate (FDR) is controlled using the Benjamini-Hochberg procedure for multiple testing correction while only considering genes with changes in expression levels above the threshold (absFC > 2) (Ritchie et al., 2015). RNA sequence analysis using the edgeR-limma workflow was based on the previously published script (Law et al., 2018) and was performed using R version 4.1.0 (R Core Team, 2019).
Enrichment analysis for gene ontology (GO) terms from DE genes in was achieved using DAVID with a p-Value < 0.05 used to define significantly enriched GO terms unless otherwise stated (Ashburner et al., 2000; Huang et al., 2009a,b). The circle plots were created using the R package GOplot version 1.0.2 (Walter et al., 2015). The color of the bars in the inner circle of the circle plots and the bar graphs is determined by the z-score which indicates whether the given biological process is more likely to be decreased or increased in the dataset and is given as follows:
Whereas up and down are the number of DE genes that are upregulated or downregulated, respectively in the comparison and count is the total number of genes in the GO term. Superimposition of DE genes onto the global metabolic network and carbon metabolism pathways was achieved using KEGG (Kanehisa, 2000; Kanehisa et al., 2019).
Protein Profiling Using Label-Free Quantitative Mass Spectrometry and Differential Expression Analysis
Total cellular protein was isolated from the organic fraction from TRIzol™ extractions above, and proteins were purified as previously reported (Prisic et al., 2015). Protein concentration of solubilized pellets was determined with the DC assay (Bio-Rad). Protein (100 μg) was digested using filter-aided sample preparation (FASP) method (Wiśniewski and Rakus, 2014) and digested peptides were resuspended in 5% acetonitrile/5% formic acid and analyzed by UC Davis Genome Center – Proteomics Core by LC-MS/MS on a Q Exactive™ Plus Orbitrap Mass spectrometer in conjunction with Proxeon Easy-nLC II HPLC (Thermo Fisher Scientific) and Proxeon nanospray source following the Core’s standard protocol. Buffer A was 0.2% formic acid in water and buffer B was 0.2% formic acid in acetonitrile. The following method was used: flow rate was 2 μL/min, 10 min buffer A, 90 min 5-35% gradient buffer B, 5 min 95% buffer B, 5 min 5% buffer B. Mass spectra in .raw format was converted to .mzXML format using MSConvert from ProteoWizard (ProteoWizard 3.0.19317.0ef6e44d0). Tandem mass spectra were extracted, and MS/MS analyzed with X! Tandem for peptide identification using the following search parameters: cysteine alkylation – iodoacetamide; digestion – trypsin; fixed modifications – carbamidomethylation (C); variable modifications – oxidation (M), deamidation (N,Q), phosphorylation (S,T,Y), and acetylation (K); precursor mass tolerance – 20ppm and fragment mass tolerance – 10ppm (The GPM, thegpm.org; version X! Tandem Alanine (2017.2.1.4)) (Craig and Beavis, 2004). Peptides were searched in X! Tandem for protein identification using a custom .fasta file of protein sequences from M. smegmatis mc2 155 reference genome assembly (ASM1500v1.39), obtained from NCBI. The reverse sequences of all proteins were included in the .fasta file as decoy sequences. It has been demonstrated that multidimensional proteomic datasets from label-free quantitation experiments have a mean-dispersion relationship that can be modeled in edgeR, and as such we used the same edgeR-limma workflow and cutoff values to define significance as applied to the RNA-seq dataset for differential expression analysis of the proteome using spectral counts from X! Tandem as tagwise abundances (Branson and Freitas, 2016; Law et al., 2018). The PLS analysis of the transcriptome and proteome including the arrow plot was done in R version 4.1.0 (R Core Team, 2019) using the mixOmics package (Rohart et al., 2017).
Results
Global Changes in the Msm Transcriptome and Proteome Upon Zn2+ Limitation
Our first goal was to define the transcriptional response to Zn2+ limitation by comparing expression patterns of Msm grown in ZLM (i.e., Sauton’s medium without Zn2+ supplementation) and ZRM (i.e., Sauton’s medium with standard 6 μM Zn2+ supplementation) using high-throughput RNA sequencing (RNA-seq). After removing genes with no (n = 139) and low (n = 172) expression values across both conditions, we detected 95% of coding and non-coding transcripts in the Msm genome. Transcript abundances were analyzed to determine differential expression (DE) patterns using an absolute fold-change cut-off of two (log2 fold-change, lfc = 1), yielding 1,084 genes that are upregulated and 742 genes that are downregulated at the transcript level in the Zn2+-limited condition (i.e., wild type grown in ZLM vs. wild type grown in ZRM) (Figure 1A). As expected, we observed upregulation of genes in the Msm Zur regulon (Novichkov et al., 2013; Supplementary Table S1) and, as observed in Mtb (Dow et al., 2021), hundreds of genes not under control of Zur were also upregulated (or downregulated) in the Zn2+-limited condition (Supplementary Table S2). Transcriptional changes during gradual Zn2+ limiting conditions when bacteria are grown in ZLM (i.e., without addition of a Zn2+ chelator) had a strong overlap with previous reports for acute Zn2+ depletion through addition of the Zn2+-specific chelator TPEN; 74% of DE genes in TPEN-treated Msm followed the same expression levels in the wild type ZLM vs. wild type ZRM comparison (Goethe et al., 2020).
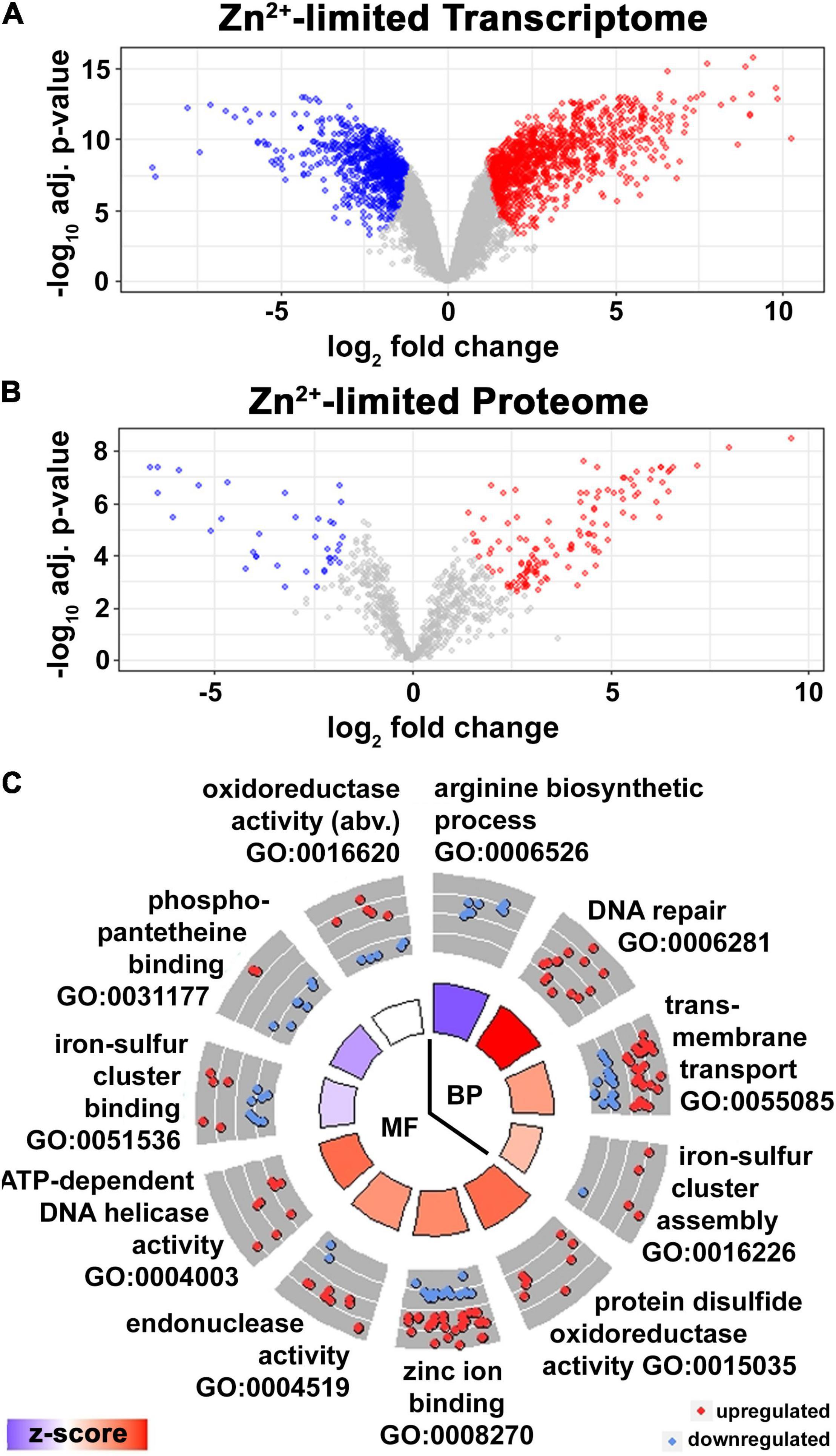
Figure 1. Adaptive response to Zn2+ limitation in Msm. (A) Volcano plot of DE transcripts (absolute lfc > 1, adj. p-Value < 0.05) in wild type Msm in ZLM vs. ZRM. Transcripts (data points) colored in red, blue, and gray are significantly upregulated, downregulated or not significantly regulated in ZLM vs. ZRM, respectively. Three independent biological replicates for each condition were used in the analysis. (B) Volcano plot of DE proteins (absolute lfc > 1, adj. p-Value < 0.05) in wild type Msm in ZLM vs. ZRM. Proteins (data points) colored in red, blue, and gray are significantly upregulated, downregulated or not significantly regulated in ZLM vs. ZRM, respectively. Three independent biological replicates for each condition were used in the analysis. (C) Circle plot showing GO terms of biological processes (BP) and molecular functions (MF) significantly enriched in the DE transcripts in wild type ZLM vs. ZRM (p-Value < 0.05). Each slice of the circle is labeled with the enriched GO term, the outer circle shows a scatter plot of lfc values for DE genes with the given GO annotation, where red circles display upregulated and blue circles downregulated genes in wild type ZLM vs. ZRM. The color of bars in the inner circle correlates with the z-score which indicates whether the given biological process is more likely to be increased (red) or decreased (blue) in the dataset and the height of the bars represents the significance for the enriched term (larger bars have smaller p-Value). When necessary, the full-length description of some GO terms are abbreviated (abv.) for clarity.
In Mtb, analysis of the proteome supported the transcriptomic changes observed, and a conserved response to Zn2+-limiting conditions was described from the overlap of DE transcripts and proteins (Dow et al., 2021). Taking the same approach here, we probed the proteomes of wild type Msm grown in ZRM and ZLM using semi-quantitative spectral counting (SpC) from shotgun proteomics. After removing lowly expressed proteins having fewer than 10 SpC, a total of 804 proteins were identified (Supplementary Table S3). After imparting an absolute cut-off value of two (lfc = 1) to define DE, 162 proteins were found to be DE in the Zn2+-limited condition (i.e., wild type grown in ZLM vs. ZRM) (Figure 1B and Supplementary Table S3). Just under 50% of these genes (n = 77) had overlap between the expression levels of the protein and the transcript (Supplementary Table S4). To describe the functionality of the DE genes with a conserved response at the level of the transcriptome and proteome in the Zn2+-limited condition, we conducted an enrichment analysis for gene ontology (GO) terms. Enriched GO terms yielded processes involved in translation, metal ion transport, oxidoreductase activity and structural constituent of the ribosome. The list of genes with conserved expression levels of the transcript and protein was too short to provide a high-power analysis, so we expanded the enrichment analysis to include all DE proteins. This slightly expanded the analysis with significantly enriched GO terms including processes involved in rRNA binding and N-acetyltransferase activity.
While the overlap of DE transcripts and proteins demonstrates the robust cellular response to Zn2+-limiting conditions, the low resolution at the level of the proteome using the shotgun proteomics approach allowed us to evaluate only 9% of the changes detected at the level of the transcriptome. Therefore, to expand the enrichment analysis for GO terms and to provide a greater understanding of the functionality of DE genes in the Zn2+-limited condition, we conducted the enrichment analysis on the list of DE transcripts. The enrichment analysis covered 1824 DE genes, resulting in 728 genes in 12 functional annotations (GO terms of “biological processes” and “molecular function”) that were enriched in the Zn2+-limited condition (Figure 1C). The circle plot shows downregulation of processes involved in arginine biosynthesis and phosphopantetheine binding during Zn2+ limitation. On the other hand, processes involved in DNA maintenance (i.e., DNA repair, endonuclease activity, and DNA helicase activity) as well as protein disulfide oxidoreductase activity were the most upregulated processes during Zn2+ limitation. Similar results were apparent in enriched KEGG terms; “homologous recombination” was the most upregulated term, while “alanine, aspartate and glutamate metabolism” and “oxidative phosphorylation” were downregulated terms in wild type ZLM vs. wild type ZRM comparison (Supplementary Figure S1). In conclusion, Zn2+ limitation in Msm triggers a robust transcriptional response, including genes that are required for redox homeostasis and repairing damaged proteins and DNA. The transcriptomic response is reflected in the proteome, however, less than half of the DE proteins positively correlated with the transcriptome suggesting post-transcriptional or translational processes may significantly contribute to the cellular response to Zn2+-limitation.
Upregulated Oxidative Stress Response Is Conserved in Zn2+-Limited Mycobacteria
The concentration of Zn2+ available during growth has a drastic impact on the transcriptomes of mycobacteria; Msm differentially regulated 26% and Mtb regulated 17% of their respective transcriptomes during Zn2+-limiting conditions. The transcriptomics analysis of Zn2+-limited Msm yielded similar results to those described for Zn2+-limited Mtb, specifically regarding increased DNA repair processes (Dow et al., 2021). Over half of the genes in Mtb have orthologues in Msm; 403 of the orthologous genes were DE in Zn2+-limited Mtb (Dow et al., 2021). Of those genes, 41% (n = 167) were also DE in Msm, but only 30% (n = 122) were DE in the same direction (i.e., up- or down- regulated) in both species in the Zn2+-limited condition (Supplementary Table S5). An enrichment of GO terms in the orthologous DE genes with the same regulation level in Msm and Mtb highlighted that processes involved in DNA replication, recombination and repair were upregulated during Zn2+ limitation in both mycobacteria (Figure 2A and Supplementary Table S5). The response to damaged DNA is further implicated by upregulation of the gene encoding the DNA-damage sensing alternative sigma factor SigG (Smollett et al., 2011) in both Msm (MSMEG_0219) and Mtb (Dow et al., 2021; Supplementary Table S5). Furthermore, like Mtb (Dow et al., 2021), Msm also upregulates key antioxidant enzymes including catalase-peroxidase (katG, MSMEG_6384), alkylhydroperoxidase (ahpC, MSMEG_4891) and thioredoxin-disulfide reductase (trxB, MSMEG_6933) in Zn2+-limited conditions, the latter two being upregulated at the protein level as well (Supplementary Tables S3, S5). The shift from fatty acid biosynthesis to degradation and decreased oxidative phosphorylation described in Mtb (Dow et al., 2021) was also evident in Msm when projecting DE genes belonging to the KEGG term “metabolic pathways” onto a global metabolic network (Supplementary Figure S2). In sum, the overlap in the transcriptomics analysis of Zn2+-limited Msm with a comparable analysis in Mtb (Dow et al., 2021) highlights conservation of the upregulated oxidative stress response during Zn2+-limiting conditions in mycobacteria.
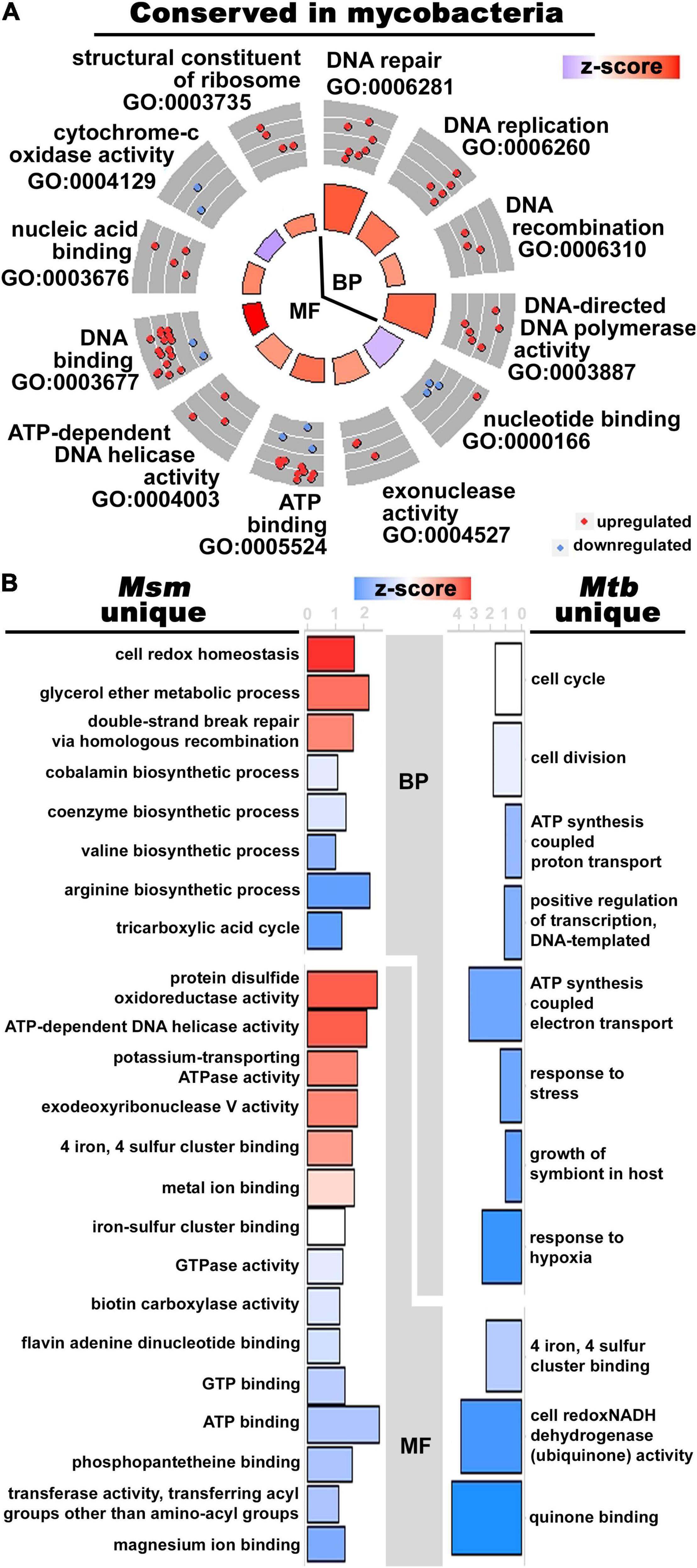
Figure 2. The alignment and discordance of DE orthologous genes in Zn2+-limited Msm and Mtb. (A) Circle plot showing enriched GO terms in the orthologous genes with shared differential expression patterns in response to Zn2+ limitation in mycobacteria. The enrichment was conducted using the locus tags from Msm. See Figure 1C for circle plot explanation. (B) Inflected vertical bar graphs of enriched GO terms in the orthologous mycobacterial genes that were uniquely regulated in Zn2+-limited Msm (left) and Mtb (right). The enrichments were conducted using the locus tags of the respective mycobacterial species (i.e., MSMEG tags for unique DE genes in Msm and Rv tags for unique DE genes in Mtb). The bars are colored by z-score as explained for the circle plot in Figure 1C. The axis of each bar graph is the -log10 (p-Value) of the enriched term. Due to the restriction of only considering orthologous genes, significantly enriched GO terms were defined with p-Value < 0.1 for all analyses in this figure.
Unique Regulation of Certain Processes in Zn2+-Limited Mycobacteria
While there was an overlap of specific processes in Zn2+-limited Msm and Mtb (Dow et al., 2021), the expression patterns of genes uniquely DE in either mycobacteria experiencing Zn2+-limitation was much greater than those that were shared. To begin, 442 orthologous genes were DE in Msm, but were not DE in Mtb (Dow et al., 2021; Supplementary Table S6). Enriched GO terms show approximately equal distribution of processes that are up- and down- regulated in this list of genes. Upregulated processes unique to Zn2+-limited Msm include redox homeostasis, protein disulfide oxidoreductase activity and processes involved with DNA repair (Figure 2B), possibly indicating a more robust oxidative stress response in Zn2+-limited Msm compared to Mtb. Downregulated processes include arginine and valine biosynthesis, the tricarboxylic acid cycle, and genes involved in binding to magnesium, ATP and GTP (Figure 2B).
There were 236 DE orthologous genes in Mtb (Dow et al., 2021) that were not DE in Msm, and 5 of these genes were not detected (i.e., did not pass low expression filter) in Msm (Supplementary Table S7). Among these genes, 64% are downregulated and 36% are upregulated in the Zn2+ limited condition in Mtb (Dow et al., 2021), however, the enriched processes associated with these genes are exclusively downregulated (Figure 2B and Supplementary Table S7). The functionally enriched genes are involved in processes of quinone binding, NADH dehydrogenase activity, growth of symbiont in host, and the response to hypoxia and stress (Figure 2B). Notably, the two-component response regulator mtrA and four genes in the mammalian cell entry operon 1 (mce1C-F) were uniquely downregulated in Zn2+-limited Mtb (Supplementary Table S7). A large portion of the uniquely regulated genes in Mtb are directly involved in processes relevant to the host-pathogen interaction.
In addition to the uniquely DE genes described above, 45 orthologous genes were discordantly regulated in Zn2+-limited Msm and Mtb (Dow et al., 2021; Supplementary Table S8). Enriched GO terms in the genes that were downregulated in Msm but upregulated in Mtb (Dow et al., 2021) were involved in “de novo” UMP and pyrimidine biosynthesis, and arginine biosynthesis (Supplementary Figure S3). On the other hand, enriched GO terms that were upregulated in Msm but downregulated in Mtb (Dow et al., 2021) include processes involved in ergothioneine biosynthesis from histidine, oxidoreductase activity and ribonucleoside-diphosphate reductase activity using thioredoxin disulfide as acceptor (Supplementary Figure S3). Genes with the most remarkable levels of discordant regulation include ferredoxin, i.e., fdxA (MSMEG_1124) and lipase, i.e., lipU (MSMEG_5271), which were both significantly downregulated in Mtb (Dow et al., 2021), but upregulated in Msm (Supplementary Table S8). Furthermore, the gene encoding the WhiB6 transcription factor is highly upregulated in Mtb (Dow et al., 2021), but not DE in Msm (MSMEG_0051), whereas the opposite is true for WhiB7 (MSMEG_1953) (Supplementary Tables S6, S7). Altogether, these results suggest that, in response to Zn2+-limitation, mycobacteria may have evolved unique regulation strategies where Mtb regulates processes relevant to the context of infection and Msm regulates processes to support cell growth such as maintaining redox homeostasis, repairing damaged biomolecules and modulating central carbon metabolism.
Msm Regulates Central Carbon Metabolism During Zn2+ Limitation
It was curious to see differential regulation of many genes involved in central metabolism in Zn2+-limited Msm. A general downregulation of metabolic genes might be anticipated in Zn2+-limited Msm since cell density is much higher in ZRM than ZLM (Dow and Prisic, 2018), a phenotype not observed in Mtb (Dow et al., 2021), but this was not the trend observed. In fact, of the DE genes belonging to metabolic pathways as defined by KEGG, 40% are upregulated and 60% are downregulated in the Zn2+-limited condition. Analysis of the DE genes involved in carbon metabolism showed a specific downregulation of genes encoding enzymes involved in the non-oxidative arm of the pentose phosphate pathway (PPP) along with two downstream enzymes involved in glycolysis, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), i.e., gap (MSMEG_3084) and phosphoglycerate kinase, i.e., pgk (MSMEG_3085), during Zn2+ limitation (Supplementary Figure S4). The oxidative arm of the PPP (ox-PPP) is the predominant producer of cellular NADPH; redirection of glycolytic flux through the ox-PPP can increase the antioxidant defenses that combat oxidative stress (Mullarky and Cantley, 2015). Redirection of glycolytic flux through the ox-PPP can also be achieved by inhibition of downstream glycolytic enzymes such as GAPDH (Mullarky and Cantley, 2015). The changes in expression patterns of genes that divert glycolytic flux through the ox-PPP for increased NADPH production in Zn2+-limited Msm further implicate the relevance of combating oxidative stress in the Zn2+-limited environment.
Deletion of the altRP Operon Significantly Impacts the Transcriptome and Proteome
Msm grown in ZLM build active Alt ribosomes (i.e., ribosomes containing AltRPs), and AltRPs are required for the morphogenesis of Zn2+-limited Msm (Dow and Prisic, 2018; Tobiasson et al., 2019). Since the ΔaltRP mutant grows in ZLM, but is unable to grow in the presence of TPEN (Dow and Prisic, 2018), it is possible to investigate the involvement of AltRPs during the response to gradual Zn2+ limitation in ZLM. To specify the role of AltRPs in the Zn2+-dependent phenotype, we investigated the transcriptomes of the ΔaltRP mutant and the in trans complement strain (ΔaltRP/c) grown in ZLM compared to the wild type grown in ZRM (Supplementary Table S9). Principal component analysis (PCA) shows that Zn2+ is the leading dimension explaining 71% of variance in the transcriptomics dataset, while genotype (i.e., deletion of AltRPs) explains only 18% (Supplementary Figure S5). As in the wild type ZLM vs. wild type ZRM comparison, we defined DE using a lfc = 1 cut-off, resulting in 1,835 and 1,490 DE genes in ΔaltRP and ΔaltRP/c when compared to the wild type grown in ZRM, respectively (Supplementary Table S9). To highlight gene expression patterns that are affected by deletion of AltRPs (i.e., the altRP operon), we looked for overlap in DE genes of the wild type, ΔaltRP and ΔaltRP/c strains grown in ZLM, compared to the wild type grown in ZRM. The Venn diagram in Figure 3A shows that 56% of the DE genes in the wild type ZLM vs. wild type ZRM comparison were also DE in the ΔaltRP and ΔaltRP/c strains (n = 1028). This data shows, in agreement with the PCA, that majority of DE genes are driven by Zn2+ concentration and not the presence or absence of AltRPs.
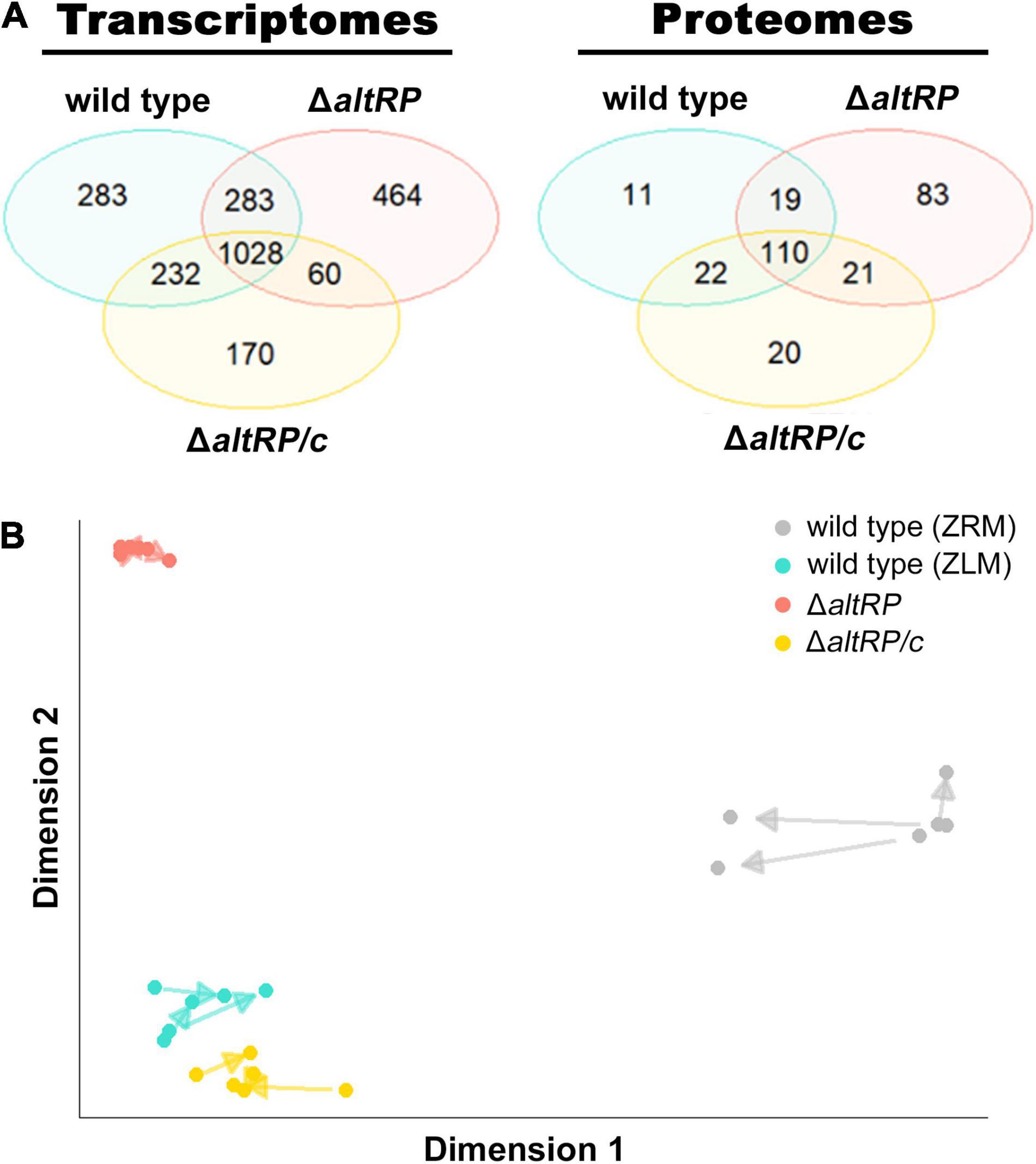
Figure 3. Transcriptomes and proteomes of Zn2+-limited wild type, ΔaltRP and ΔaltRP/c compared to the Zn2+-replete wild type. (A) Venn diagrams showing the overlap of DE genes (left) and proteins (right) in the wild type, ΔaltRP, and ΔaltRP/c (grown in ZLM) when compared to the wild type grown in ZRM. (B) Arrow plot from PLS analysis of the transcriptomes and proteomes of the wild type, ΔaltRP, ΔaltRP/c (grown in ZLM) and the wild type grown in ZRM. The representation space maximizes the covariance of the components, and as such the scale is arbitrary and absolute values do not affect the alignment.
Because of the potential role for AltRPs in translation (Chen et al., 2020), the next step was to define the correlation between the transcriptome and proteome of Zn2+-limited Msm, and to determine if the ΔaltRP mutant follows patterns observed in the wild type. Accordingly, in addition to probing the proteomes of the wild type grown in ZRM and ZLM, we also included the ΔaltRP mutant and its complement strain (ΔaltRP/c) grown in ZLM. After removing proteins with fewer than 10 SpC across all strains and imparting an absolute cut-off value of two (lfc = 1) to define DE, we identified 233 and 173 DE proteins in ΔaltRP and ΔaltRP/c respectively, when compared to the wild type grown in ZRM (Supplementary Table S3). Again, Zn2+ is the leading dimension describing 63% and genotype accounting for 19% of variance in the dataset (Supplementary Figure S5). The Venn diagram in Figure 3A shows the overlap of DE proteins in the Zn2+-limited strains compared to the wild type grown in ZRM. Similar to the transcriptomes, most DE proteins (68%, n = 110) in the wild type ZLM vs. wild type ZRM comparison were conserved across all Zn2+-limited strains (i.e., wild type, ΔaltRP and ΔaltRP/c), but again, the ΔaltRP mutant had a considerable number of DE proteins (n = 83) unique to this comparison.
A multivariate analysis using partial least squares (PLS) regression, which maximizes the covariance between components from two datasets (e.g., transcriptomics and proteomics) (Wold et al., 2001), was used to create an arrow plot overlapping the covariance together in one dimension. In the arrow plot, each arrow connects the transcriptome (tail) and the proteome (head) from the same sample. Shorter arrows indicate better agreement found by PLS between both datasets. From the arrow plot in Figure 3B, we see that for all strains, the proteome is closely related to the transcriptome and there is preservation of the dimensions driving variance in both datasets. Notably, while the Zn2+-replete condition had the most variation between the transcriptome and proteome, the ΔaltRP mutant had the least amount of variation among the Zn2+-limited samples, which may indicate altered processes of post-transcriptional or post-translational regulation in the ΔaltRP mutant.
Discordance Between Transcript and Protein Abundance in the Wild Type and the ΔaltRP Mutant
Based on the multi-omics analysis and given the potential role for AltRPs in the process of translation (Chen et al., 2020), we wanted to further investigate the variance between the transcriptome and proteome with respect to AltRP expression. To this end, we investigated examples of discordant regulation between a transcript and the protein it encodes in the wild type ZLM vs. wild type ZRM comparison (i.e., in wild type cells with Alt ribosomes). In our dataset, discordant regulation could be observed in one of two ways, either the protein is underrepresented compared to its transcript, or the protein is overrepresented compared to its transcript. Genes with underrepresented proteins could have discordant regulation levels in one of the following three ways: [1] the transcript is upregulated and the protein is not DE (n = 68), [2] the transcript is upregulated and the protein is downregulated (no examples) or [3] the transcript is not DE and the protein is downregulated (n = 24) (Supplementary Table S10). On the other hand, genes where the protein is overrepresented compared to its transcript could have discordant regulation levels in one of the following three ways: [1] the transcript is downregulated and the protein is not DE (n = 82), [2] the transcript is downregulated and the protein is upregulated (n = 5) or [3] the transcript is not DE and the protein is upregulated (n = 55) (Supplementary Table S11). In all these comparisons, the gene was only considered if both the transcript and the protein were detected.
Next, to divulge to role of AltRPs in discordant gene expression, we turned to look at expression patterns of the genes described above in the ΔaltRP vs. wild type grown in ZRM comparison. If an Alt ribosome-dependent regulation (e.g., selective translation by Alt ribosomes) is responsible for a discordant regulation of gene products, we would expect that the ΔaltRP strain would not have this discordance. Starting with the proteins that are underrepresented compared to their transcripts, we found that more than a half of them follow the same discordance in the wild type and the ΔaltRP, and therefore are not candidates for Alt-dependent translational regulation (Supplementary Table S10). Moreover, one-fifth of the genes were not transcriptionally regulated in the same way in the wild type and the ΔaltRP mutant, and most of these genes were not DE in the ΔaltRP mutant, so comparison of transcript-protein abundances for these genes was inconclusive. However, a quarter of the genes had discordant regulation in the wild type that could be attributed to the presence of AltRPs. One set of genes had significantly upregulated transcripts in both the wild type and ΔaltRP mutant, and although at the protein level they were not DE in the wild type, they were significantly upregulated in the ΔaltRP mutant (Table 1). Notable proteins on this list are involved in Fe-S assembly (SufC/D), Clp protease (ClpP) and catalase peroxidase (KatG). Another set of genes included nine examples that were not DE at the transcript level in wild type or ΔaltRP, and were significantly downregulated at the protein level in wild type, but were not DE in ΔaltRP (Table 1). Interestingly, this set of genes encodes five ribosomal proteins and two proteins involved in translation (IF-2 and DnaJ chaperone).
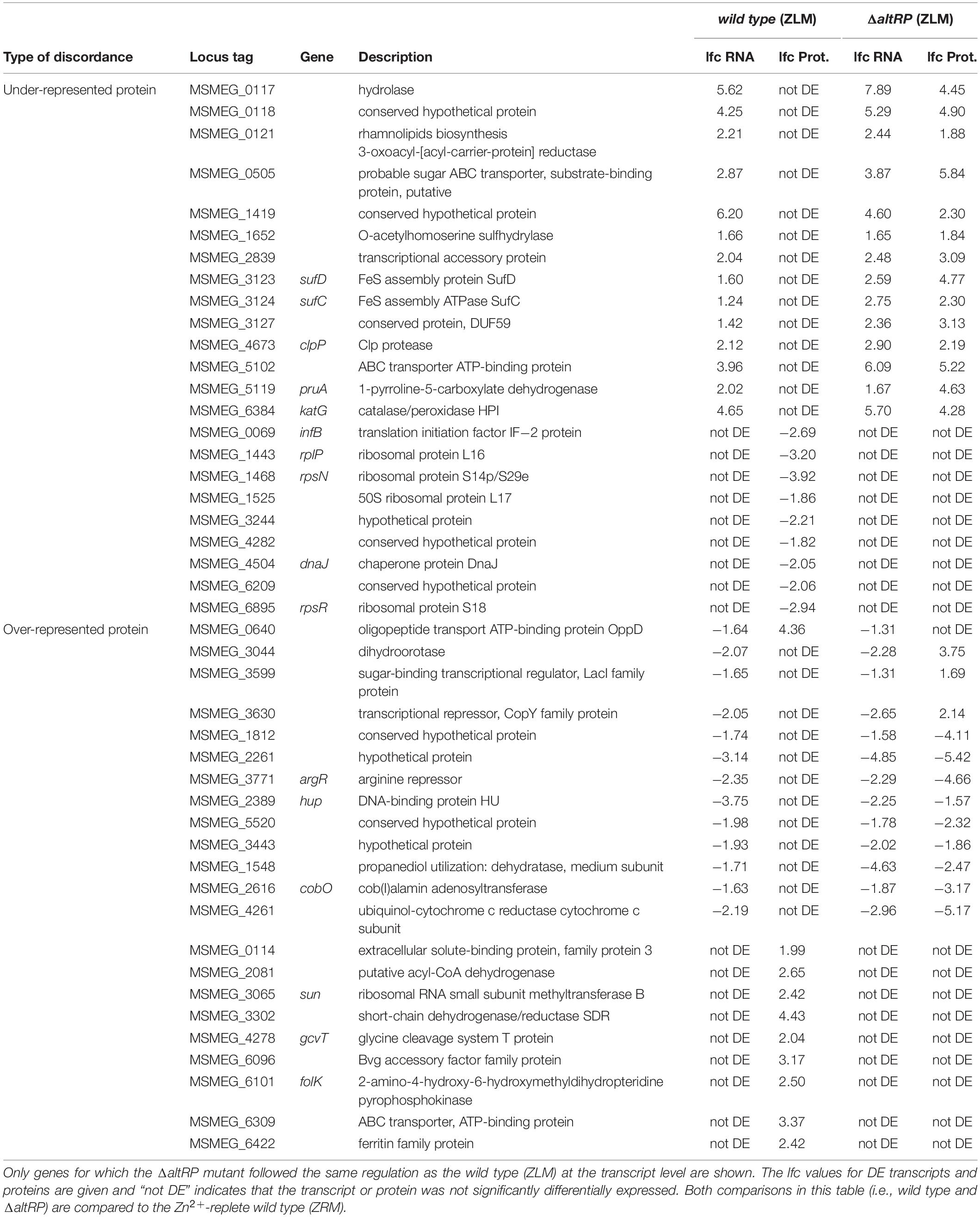
Table 1. Genes with discordant regulation between the transcript (RNA) and protein (Prot.) in Zn2+-limited wild type (ZLM) and the overlap with ΔaltRP.
Moving on to the genes where the protein was overrepresented compared to its transcript in the wild type ZLM vs. wild type ZRM comparison, we also found that half of these genes follow the same discordant regulation levels in ΔaltRP vs. wild type ZRM (Supplementary Table S11). Another one-third of the genes did not follow the same regulation at the transcript level in the ΔaltRP mutant as observed in the wild type grown in ZLM, again making the comparison of AltRP-dependent discordant expression for these genes inconclusive (Supplementary Table S11). The final one-sixth of the genes had discordant regulation that could be attributed to AltRPs (Table 1). With a few exceptions, the regulation levels in the ΔaltRP mutant mirrored the wild type but did not meet the threshold for significance (Supplementary Table S11). Notable examples include the ABC transporter ATP-binding protein (MSMEG_6309), which was significantly overrepresented at the protein level in the wild type but not in the ΔaltRP mutant, and the gene encoding the oligopeptide transport ATP-binding protein OppD (MSMEG_0640), showing strong upregulation at the protein level only in the wild type. Interestingly, these proteins that are significantly overrepresented only in the wild type background are involved in transporting molecules into the cell.
Of note in these comparisons, is that about half of the discordant examples of regulation levels were preserved in the ΔaltRP background, and there were relatively few examples where the discordance found in wild type was drastically different in the ΔaltRP background. Additionally, many of these genes did not agree at the transcript level between wild type and ΔaltRP. In nearly all these cases, the protein expression level followed that of the transcript in the ΔaltRP mutant. These results, along with those presented in the arrow plot in Figure 3B, indicate that changes in protein expression patterns between the wild type and the ΔaltRP mutant are strongly driven by alterations in the transcriptomes of these two strains. While we did identify potential examples of regulation by Alt ribosomes through investigation of discordant expression levels (Table 1), they were relatively rare and are probably not directly and/or solely responsible for the different phenotypes observed in the wild type and the ΔaltRP mutant experiencing Zn2+ limitation.
Increased Oxidative Stress Response During Zn2+ Limitation Is Independent of AltRP Expression
From the multi-omics analysis of the wild type and the ΔaltRP mutant, we observed that deletion of AltRPs causes a significant perturbation to the transcriptome, which partially, correlates with changes to the proteome. However, the Venn diagrams in Figure 3A also show a conserved response to Zn2+ limitation in the wild type, ΔaltRP mutant and the complement strain (ΔaltRP/c). Like the observed overlap in transcript and protein expression levels in the wild type, just over 50% of the proteins with concordant regulation in all strains (i.e., at the intersection of all strains in both Venn diagrams in Figure 3A) overlapped with concordantly regulated transcripts in all strains (Supplementary Table S12). Again, pathway analysis using proteomics was not possible due to limited resolution, so we used transcriptomics data to understand which processes employed during Zn2+ limitation are independent of AltRP expression. We selected the DE genes common to all three strains grown in ZLM when compared to the wild type grown in ZRM (union of all comparisons) from the Venn diagram in Figure 3A (n = 1028) to conduct an enrichment analysis for GO terms. The enrichment analysis covered 1027 shared DE genes, yielding 421 genes with 11 functional annotations of “biological processes” and “molecular functions” that were significantly enriched in all strains grown in ZLM (regardless of deletion of the altRP operon) when compared to the wild-type Zn2+-replete condition (Figure 4A).
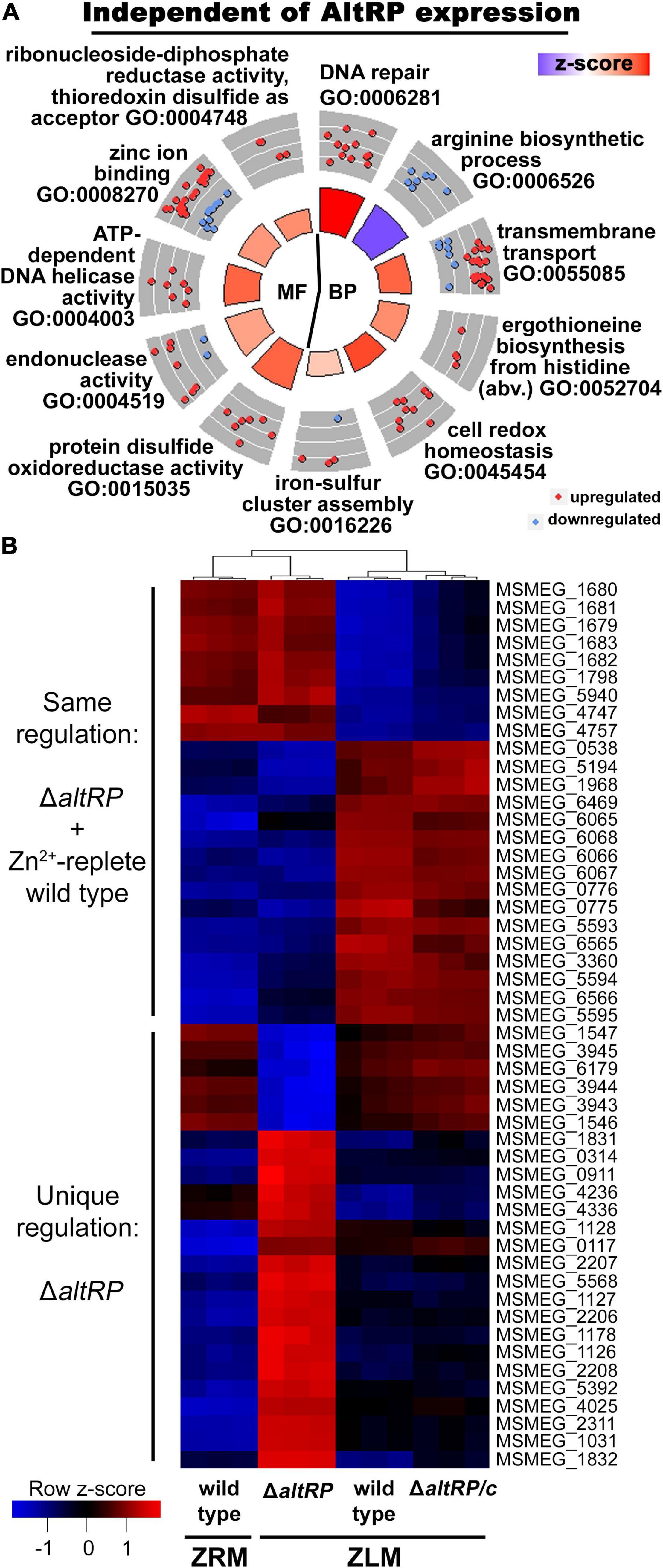
Figure 4. Differentially expressed genes independent (A) and dependent (B) of AltRP expression during Zn2+-limitation. (A) Circle plot showing enriched GO terms in the DE genes shared between the wild type and the ΔaltRP mutant during Zn2+ limitation (i.e., both grown in ZLM and compared to the wild type grown in ZRM). DE analysis was conducted on three independent biological replicates for each strain. See Figure 1C for circle plot explanation. (B) Heatmap showing relative transcript expression levels in all strains and conditions for the top 50 DE genes (by adj.P.val) between the ΔaltRP vs. wild type in ZLM comparison. For each strain or condition, the relative expression for each independent biological replicate is shown in the column.
As predicted by the fact that over half of the DE genes in the wild type ZLM vs. wild type ZRM comparison were preserved across all Zn2+-limited strains, the GO enrichment analysis has significant overlap with DE genes in Zn2+-limited wild type (Figures 1C, 4A). In all Zn2+-limited strains, independent of AltRP expression, processes involved in DNA repair and iron-sulfur cluster assembly are upregulated, and the arginine biosynthetic process is downregulated (Figure 4A). Additionally, key antioxidant enzymes upregulated in Zn2+-limited wild type were also upregulated in ΔaltRP and ΔaltRP/c, including catalase-peroxidase (katG), alkylhydroperoxidase (ahpC) and thioredoxin (trx and trxB) (Supplementary Table S9). Among these genes, ahpC, trx, and the DNA repair protein recA were DE at the level of the transcriptome and proteome in all Zn2+-limited strains (Supplementary Table S3). The GO enrichment of DE genes shared between the wild type, ΔaltRP and ΔaltRP/c strains in Zn2+-limiting conditions when compared to the Zn2+-replete wild type (i.e., wild type grown in ZRM), and the overlap of specific genes at the protein level, shows that the increased oxidative stress response upon Zn2+-limiting conditions occurs independently of AltRP expression in Msm.
AltRP-Dependent Response During Zn2+-Limitation Includes Downregulated Cell Division and Nucleotide Biosynthesis and Altered Respiration
Beyond the conserved response to Zn2+ limitation in all strains, the multi-omics analysis shows that the altRP operon deletion did impart significant changes at the level of the transcriptome and proteome. To further define the impact of lacking AltRPs during Zn2+ limitation, we investigated the DE genes that were not conserved between the ΔaltRP mutant and the wild type in ZLM. First, from the Venn diagram of transcripts in Figure 3A, we see the ΔaltRP mutant has many unique DE genes, but also, nearly as many genes are only DE in the wild type and ΔaltRP/c strains. The 232 genes at the intersection of the wild type and ΔaltRP/c comparisons in Figure 3A were interesting because they represented processes that were only DE in the Zn2+-limited condition when AltRPs are present (Supplementary Table S13). Genes that are exclusively downregulated in Alt ribosome-containing cells are involved in processes of [1] cell division including the chromosomal replication initiator protein dnaA (MSMEG_6947), sepF (MSMEG_4219), pbpB (MSMEG_4233) and amiB (MSMEG_1679), [2] respiration through succinate dehydrogenase, sdhCD (MSMEG_1671 and MSMEG_1672), and [3] purine nucleotide biosynthesis, purA (MSMEG_0759), nadBC (MSMEG_3200 and MSMEG_3201) and aspA (MSMEG_1677). Genes exclusively upregulated in Alt ribosome-containing cells are involved in [1] respiration, through NADH dehydrogenase complex, nuoB and nuoE subunits (MSMEG_2062 and MSMEG_2059) and the non-proton pumping (type II) NADH dehydrogenase protein ndh (MSMEG_3621), and [2] regulation of gene expression including the SigE anti-sigma factor rseA (MSMEG_5071) and the SigH anti-sigma factor rshA (MSMEG_1915). All in all, processes associated with cell division and nucleotide biosynthesis are downregulated and respiration is altered only in the presence of AltRPs during Zn2+ limitation.
AltRP-Expressing Cells Alter Nucleotide Acquisition Strategy Through Induction of GOGAT and Decreased Expression of Purine Import Mechanisms
The Venn diagrams in Figure 3A demonstrate unique regulation of a set of genes and proteins in the wild type and the complement, but not the ΔaltRP mutant in Zn2+-limiting conditions. Additionally, among the Zn2+-limited strains, the ΔaltRP mutant also has many unique DE genes, compared to the other two strains that express AltRPs. This led us to investigate the direct effects of lacking AltRP expression in a Zn2+-limited environment by comparing transcriptomes of the ΔaltRP mutant vs. wild type in ZLM which yielded 472 DE genes, with 61% being complemented in ΔaltRP/c (Supplementary Table S14). The heatmap in Figure 4B shows the relative transcript expression levels in all strains and conditions for the top 50 DE genes between the ΔaltRP vs. wild type in ZLM, all of these genes were complemented in ΔaltRP/c. Two patterns emerge in the heatmap (Figure 4B); a set of genes that are either up- or down- regulated uniquely in the ΔaltRP compared to all other strains (including wild type grown in ZRM) (n = 25), and a set of genes that are coordinately up- or down- regulated in the ΔaltRP mutant and the Zn2+-replete wild type (grown in ZRM) when compared to the wild type and ΔaltRP/c grown in ZLM (n = 25) (Supplementary Table S15).
Since the ΔaltRP phenotype more closely resembled that of the Zn2+-replete wild type (Dow and Prisic, 2018), we first looked at the DE genes from the heatmap that are shared between the ΔaltRP mutant and wild type grown in ZRM (Figure 4B and Supplementary Table S15). In this list, five of the DE genes correlated with DE proteins (all have concordant regulation levels), including the altRP operon and a MarR regulatory protein (MSMEG_0538). Many of the topmost differentially regulated genes are ordered, and thus appear to be in operons (64%), and three genes are transcriptional regulators (n = 3). A set of ordered genes that were downregulated in ΔaltRP and the wild type grown in ZRM encode pyruvate dehydrogenase (MSMEG_5593), ferredoxin-dependent glutamate synthase, aka GOGAT (MSMEG_5594) and the MarR transcriptional regulatory protein (MSMEG_5595). A set of ordered genes that were upregulated in ΔaltRP and the wild type grown in ZRM encodes an amidase (MSMEG_1679), a cytosine/purine/uracil/thiamine/allantoin permease family protein (MSMEG_1683) and an endoribonuclease L-PSP superfamily protein (MSMEG_1681). The L-PSP superfamily belongs to the widely distributed YER057c/YjgF/UK114 family of proteins conserved among all domains of life and serving myriad functions; with a proposed role in purine regulation in bacteria (Sinha et al., 1999). Interestingly, these processes converge at the level of nucleotide metabolism; GOGAT shifts the fate of glutamine nitrogen away from the anaplerotic reactions of the TCA cycle to nucleotide biosynthesis, while import mechanisms for nucleotide precursors are differentially regulated. Together, the expression patterns unique to strains that build primary ribosomes (i.e., ΔaltRP and the wild type grown in ZRM), suggests that AltRP expression alters the route to nucleotide acquisition.
Glyoxylate Shunt Is Induced via Expression of Isocitrate Lyase in the ΔaltRP Mutant
Finally, the ΔaltRP mutant displayed a set of genes in the heatmap that were DE only in this genetic background (Figure 4B and Supplementary Table S15). Again, many of the genes appear to be in operons and four of the genes are transcriptional regulators. At the protein level, six DE proteins were detected and five were DE and correlated concordantly with DE genes; one of the DE proteins was not complemented in ΔaltRP/c (MSMEG_0117) (Supplementary Table S15). The ΔaltRP mutant increased expression of isocitrate lyase (MSMEG_0911), also DE at the protein level, and glucose-6-phosphate 1-dehydrogenase (MSMEG_0314) (Supplementary Table S15). Downregulated expression of coenzyme B12-dependent glycerol dehydrogenase was observed at the transcript (MSMEG_1546 and MSMEG_1547) and protein level (MSMEG_1546) (Supplementary Table S15). Projecting the DE genes involved in carbon metabolism onto a network shows that, in addition to the selective downregulation of the non-oxidative arm of the PPP observed in the wild type, the ΔaltRP mutant also upregulates key enzymes of the oxidative arm (Supplementary Figure S6). Additionally, the two-component response regulator DevR (MSMEG_3944) was strongly downregulated in the ΔaltRP mutant at both the transcript and protein level (Supplementary Table S15). All in all, deletion of AltRPs influenced expression patterns of genes involved in central carbon metabolism, including induction of the glyoxylate shunt through isocitrate lyase, and numerous transcriptional regulators during Zn2+ limitation.
Discussion
This multi-omics analysis of Zn2+-limited Msm shows that Zn2+ limitation dramatically impacts the transcriptome and proteome while highlighting the contribution of alternative ribosomal proteins (AltRPs) in the response to Zn2+ limitation. Furthermore, this study compares, corroborates and contrasts the findings from a similar study in Zn2+-limited Mtb (Dow et al., 2021). Zn2+-limited mycobacteria (i.e., Mtb and Msm) upregulate key antioxidant enzymes (e.g., catalase-peroxidase, alkylhydroperoxidase and thioredoxins) and many genes involved in homologous recombination and DNA repair. Both species also downregulate fatty acid biosynthesis and oxidative phosphorylation during Zn2+ limitation; mechanisms that are presumed to conserve reducing power (NADPH) and decrease endogenous oxidant production (Dow et al., 2021). These congruencies signify the importance of the oxidative stress response during Zn2+-limiting conditions in both pathogenic and non-pathogenic mycobacteria, possibly implicating its importance in other bacteria. Moreover, we demonstrate that the upregulated oxidative stress response in Zn2+-limited conditions does not depend on the expression of AltRPs in Msm. Finally, we describe the processes that are affected by expression of AltRPs during Zn2+-limitation, further signifying the role of these Zn2+-independent ribosomal protein paralogs in mycobacterial physiology (Figure 5).
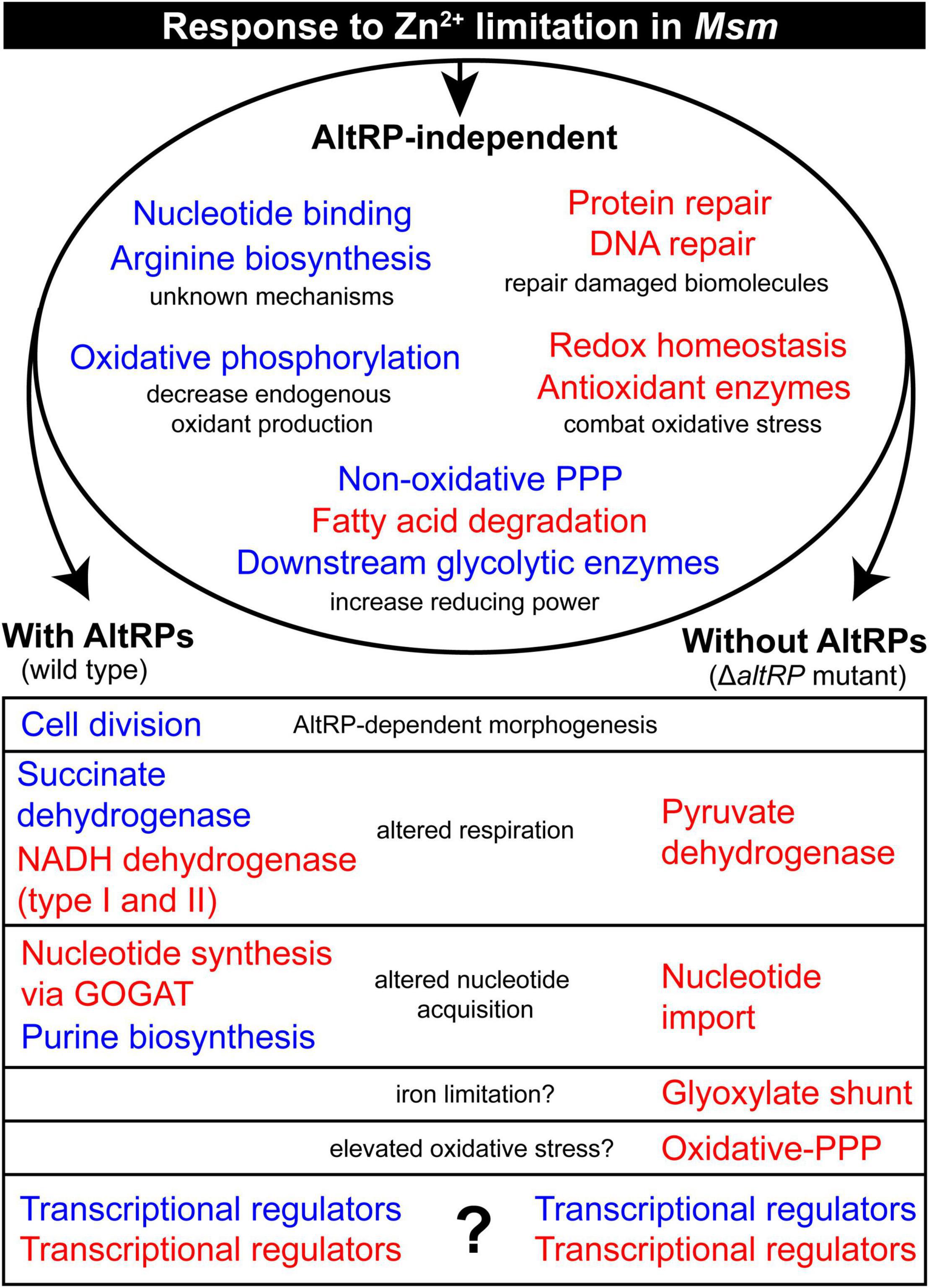
Figure 5. Summary of the molecular response to Zn2+-limitation in Msm, including AltRP-specific effects. The model shows the shared and unique physiological response of the wild type and the ΔaltRP mutant grown under Zn2+-limiting conditions when compared to the Zn2+-replete wild type. The multi-omics data was simplified into general biological processes which are colored in red and blue to indicate up-regulation and down-regulation of process during Zn2+ limitation, respectively. The descriptors in black give the presumed reason for the differential regulation of the process in the Zn2+-limited environment, or the relevance of the differences between the wild type and ΔaltRP mutant.
The oxidative stress response is a conserved feature in Zn2+-limited mycobacteria and closely resembles the transcriptomic response to mycobacteria treated with oxidizing agents (Voskuil et al., 2011; Li et al., 2015). Mycobacteria exhibit two DNA damage response pathways; the LexA/RecA-dependent “SOS response” and a LexA/RecA-independent pathway (Müller et al., 2018). From this study, we conclude Zn2+ limitation likely induces the LexA/RecA-independent pathway of DNA repair, considering 83% of genes in the PafBC regulon (Müller et al., 2018) are upregulated in Zn2+-limited Msm. Because the SOS response is associated with bacterial persistence (Podlesek and Žgur Bertok, 2020), this finding has important implications for the role of Zn2+-limitation in mycobacterial physiology. Beyond the conserved response, Mtb and Msm exhibited many unique regulatory strategies for orthologous genes when facing Zn2+ limitation. Notably, Mtb downregulates key processes relevant to infection (e.g., mce1C-F and mtrA). Interestingly, decreased expression levels of these genes correlate with a hypervirulent phenotype (Shimono et al., 2003; Fol et al., 2006), and are in agreement with the increased virulence observed in our previous mouse infection study of Zn2+-limited Mtb (Dow et al., 2021). On the other hand, Zn2+-limited Msm specifically downregulates genes binding ATP and GTP and amino acid biosynthesis, while upregulating processes involved in redox homeostasis and repairing oxidized proteins and damaged DNA (beyond the DNA repair genes also upregulated in Mtb). Accordingly, the fast-growing Msm appears to have a more robust oxidative stress response in ZLM compared to its slow-growing relative. This observation could be explained by the different growth rates of these mycobacteria, especially considering that in the same media (i.e., ZLM), Zn2+-limited Msm has much lower cell density compared to the Zn2+-replete condition (Dow and Prisic, 2018), but Zn2+ supplementation has no effect on growth of Mtb (Dow et al., 2021). Moreover, Zn2+-limited Msm attains a considerably higher cell density than Mtb (in stationary phase of the respective growth curves in ZLM) (Dow and Prisic, 2018; Dow et al., 2021). Therefore, due to the increased biomass and biological demand for Zn2+, Msm likely experiences more severe Zn2+ limitation than Mtb in the same growth condition (i.e., ZLM). Logically, this extends to Msm enduring more severe oxidative stress and is a likely explanation for the more robust oxidative stress response observed in Msm.
This analysis of Zn2+-limited Msm expands the mechanisms relevant to combating oxidative stress that were previously described in Mtb (Dow et al., 2021), while highlighting divergent features of these mycobacteria when facing Zn2+-limitation. One such example is the differential regulation of ergothioneine biosynthesis, which was upregulated in Msm, but downregulated in Mtb during Zn2+ limitation. Ergothioneine is believed to function as an antioxidant, although its deletion from Msm did not affect survival upon exposure to oxidizing agents (Sao Emani et al., 2013). In addition, its biosynthesis in Mtb appears to be dictated by nutrient starvation, not oxidative stress (Richard-Greenblatt et al., 2015). Although we hypothesize that Msm experiences more severe oxidative stress in ZLM, thus providing credence to the specific induction of ergothioneine in Msm, this observation could, again, point to the presumed difference in nutrient demands for Msm and Mtb grown in ZLM. Another divergent example is the unique upregulation of pyrimidine and arginine biosynthesis in Zn2+-limited Mtb. Arginine limitation triggers oxidative damage and sterilizes Mtb (Tiwari et al., 2018), and our results suggest this might be a weakness unique to Mtb, since arginine biosynthesis was, in contrast, downregulated in Zn2+-limited Msm and therefore likely does not serve the same protective role during oxidative stress in Msm as it does in Mtb. Finally, Zn2+ limitation differentially altered the expression of certain transcription factors in mycobacteria, with the gene encoding WhiB6 induced in Mtb and the gene encoding WhiB7 induced in Msm. Both transcription factors have been implicated in maintaining redox balance (Burian et al., 2012; Chawla et al., 2012); however, WhiB6 also links to virulence in Mtb (Chen et al., 2016) and WhiB7 links to iron limitation in Msm (Geiman et al., 2006). The selective induction of different whiB-like genes shown here suggests mycobacteria have adapted altered usage of specific regulatory networks in response to limited Zn2+. In sum, while both mycobacteria upregulated strategies to combat oxidative stress upon Zn2+ limitation, there is evidence that Mtb has also evolved to use Zn2+ specifically as a cue in the context of infection.
Both mycobacteria altered carbon metabolism in response to Zn2+ limitation. However, they did so in unique ways, albeit presumably, to the same end. While Zn2+-limited Msm downregulated genes in the non-oxidative PPP and the downstream glycolytic enzymes gap and pgk, Zn2+-limited Mtb upregulated key enzymes in the oxidative PPP directly, i.e., glucose-6-phosphate dehydrogenase (g6pd) and 6-phosphogluconate dehydrogenase (6pgd), while downregulating phosphofructokinase-1 (pfkA), the first committed step in glycolysis (Dow et al., 2021). Both of these strategies allow glycolytic flux to be diverted through the oxidative PPP arm, which increases NADPH production to fuel cellular antioxidant systems (Mullarky and Cantley, 2015). Presumably, these divergent mechanisms work to achieve the same goal of increased NADPH production in Zn2+-limited mycobacteria, which is required to reduce thioredoxin and maintain antioxidant reducing power (Wong et al., 2017). The different routes taken to redirect glycolytic flux to combat oxidative stress (Mullarky and Cantley, 2015) could again be reflective of different growth characteristics. Msm, having a greater need to reduce cellular carbon for biomass, cannot afford an overall decrease in glycolytic flux and so depends on downregulating the non-oxidative PPP arm and subsequent glycolytic enzymes. Mtb on the other hand, could take a more direct approach to increasing flux through the oxidative PPP arm directly, by upregulating these enzymes and creating a bottleneck at commitment to glycolysis. These strategies reflect unique mechanisms to divert glycolysis for increased reducing power during oxidative stress via metabolic reprogramming in mycobacteria.
To further elucidate the role of AltRPs in the Zn2+-dependent response in Msm, we first investigated the concordance between gene expression patterns at the level of the transcriptome and proteome since AltRPs are implicated in conferring selectivity in the process of translation (Chen et al., 2020). We found that, regardless of AltRP expression, most proteins follow the expression pattern of the transcript. Notable exceptions involve protease systems. Although significantly upregulated at the transcript level in both wild type and ΔaltRP, the ClpP1 peptidase component of the Clp protease (MSMEG_4673) (Akopian et al., 2012) was only upregulated at the protein level in ΔaltRP. Furthermore, components the ClpB-DnaK bi-chaperone system, which reactivates aggregated proteins (Yin et al., 2021), showed opposing discordant regulation. ClpB subunit (MSMEG_0732) was upregulated at the transcript level only in wild type and upregulated at the protein level only in ΔaltRP. Beyond that, although not differentially expressed at the transcript level in ether wild type or ΔaltRP, DnaJ (a cochaperone that chauffeurs clients to the DnaK system) (Fay and Glickman, 2014) was downregulated at the protein level in wild type only. While our results do not necessarily implicate the direct involvement of AltRPs in the discordant regulation of these protease systems, they do indicate that AltRP expression may help preserve the fidelity of translation during Zn2+-limiting conditions, since there was an overabundance of the ClpP protease and protein delivery to the bi-chaperone system in the ΔaltRP background. Underrepresented protein expression of translation initiation factor (IF-2) in AltRP-expressing cells suggests that one possible mechanism by which Alt-ribosomes may contribute to protein fidelity is by decreasing the rate of translation, perhaps to enable more efficient nascent protein folding. Indeed Alt-ribosomes are shown to exhibit a relative initiation defect and are suggested to be “slow and accurate” compared to primary ribosomes (Chen et al., 2020). This assumption is further supported by the increased abundance of trigger factor protein (MSMEG_4674), a ribosome associated chaperone, in the ΔaltRP mutant.
By directly comparing the transcriptomes and proteomes of the ΔaltRP mutant and the wild type, we were able to describe several processes that were differentially regulated upon the expression of AltRPs in the context of Zn2+-limitation (Figure 5). Remarkably, the ΔaltRP mutant utilized the glyoxylate shunt via induction of isocitrate lyase (MSMEG_0911) while concurrently upregulating succinate dehydrogenase, an enzyme required in both the TCA cycle and the electron transport chain. During iron limitation, due to decreased activity of NADH dehydrogenase (containing seven Fe-S clusters), induction of the glyoxylate shunt may help rearrange bacterial metabolism to increase the efficiency of electron flow (Koedooder et al., 2018). Interestingly, turnover of Fe-S proteins appears to be elevated in the absence of AltRPs, since only the ΔaltRP mutant upregulated the SUF operon at the protein level. This observation suggests that AltRPs may play a role in iron homeostasis during Zn2+-limiting conditions, which is consistent with the iron-limiting phenotype observed in the ΔaltRP mutant (Chen et al., 2020). Accordingly, it’s tempting to speculate that the glyoxylate shunt is linked to iron-limiting conditions in the ΔaltRP mutant, and by extension, that AltRP expression helps maintain iron homeostasis during Zn2+ limitation in mycobacteria. It is important to note, however, that the glyoxylate shunt is one of a few specific features of the ΔaltRP mutant in Msm that mirrors the phenotype of Zn2+-limited (but AltRP-expressing) Mtb. Other overlapping phenotypes include downregulation of the arginine repressor (argR), the gene encoding the two-component response regulator DevR, and upregulation of key enzymes in the ox-PPP. These observations suggest the possibly that AltRPs have evolved to have unique roles in different mycobacterial species and highlights the need to decipher the exact role of AltRPs in pathogenic and non-pathogenic (or slow- and fast-growing) mycobacteria.
To that end, this multi-omics analysis shed light onto possible mechanisms that could be involved in the AltRP-dependent phenotype observed in Zn2+-limited Msm, specifically with regards to the elongated cell length phenotype (Figure 5). Certain processes involved in cell division were only differentially regulated during Zn2+ limitation when AltRPs were expressed. This includes the chromosomal replication initiation factor (dnaA) and the septum formation components (sepF and pbpB), which were downregulated during Zn2+ limitation, but not in the ΔaltRP mutant. Further, the ΔaltRP mutant upregulated expression of the gene encoding the transcriptional regulator WhmD (MSMEG_1831) compared to the wild type and complement strains in ZLM. WhmD has a role in septum formation and cell division, and decreased expression leads to elongated cells with diminished septum formation (Gomez and Bishai, 2000). Furthermore, two cell wall hydrolases, amidase AmiB (MSMEG_1679) and hydrolase (MSMEG_0117), were among the topmost upregulated genes in ΔaltRP compared to the wild type and complement strains. Peptidoglycan hydrolases open the mesh-like peptidoglycan cell wall for insertion of new material (Kieser and Rubin, 2014), and AmiB is responsible for cell separation in E. coli (Uehara et al., 2010). Consistently, AmiB is strongly downregulated only in elongated cells (i.e., Zn2+-limited, AltRP-expressing cells). There were numerous processes involved in cell division that are uniquely regulated in the ΔaltRP mutant compared to the other Zn2+-limited strains that could explain the failure of the ΔaltRP mutant to elongate upon Zn2+ limitation.
This study demonstrates the benefit of the model mycobacterium, Msm, in evaluating the contribution of AltRPs in bacterial physiology, due the role of AltRP expression in the Zn2+-limited phenotype in Msm. Using this system, we were able to monitor changes in expression patterns of numerous genes and proteins that correlate with the phenotypic observations associated with AltRP expression in the context of Zn2+ limitation. It was intriguing that most of the differential regulation between the wild type and the ΔaltRP mutant was at the level of the transcriptome, and it is not clear how AltRPs would regulate the expression of these genes. Interestingly, the expression levels of many transcriptional regulators were altered by the presence or absence of AltRPs, offering a potential explanation for the different transcriptional responses observed (Figure 5). These changes could stem from differences in post-transcriptional or post-translational processes in Zn2+-limited cells containing Alt- or Prim- ribosomes (i.e., wild type and ΔaltRP, respectively), as a direct effect of selective translation, AltRP-specific interactions with other cellular components, or other unknown mechanisms. Indeed, deciphering mechanisms of AltRP-dependent gene regulation is a notable pursuit. All in all, we conclude that AltRPs are not only important as functional replacements for their Zn2+-dependent paralogues; they are also involved in the transcriptomic response to the Zn2+-limited environment.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: Transcriptomics data (RNA-seq) are available from the GEO database (GSE188233) and proteomics data are available from the PRIDE database (PXD029588).
Author Contributions
AD and SP designed and conceptualized the study and edited the final manuscript. AD performed all experiments, did the bioinformatics analysis, and wrote the original manuscript. AB obtained tagwise counts from raw RNA-seq data. EM obtained spectral counts from raw proteomics data. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Science Foundation (NSF) CAREER Award 1844854 (www.nsf.gov) to SP.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
This is publication number 159 from the School of Life Sciences, University of Hawai‘i at Mānoa.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.811774/full#supplementary-material
References
Akopian, T., Kandror, O., Raju, R. M., UnniKrishnan, M., Rubin, E. J., and Goldberg, A. L. (2012). The active ClpP protease from M. tuberculosis is a complex composed of a heptameric ClpP1 and a ClpP2 ring. EMBO J. 31:1529. doi: 10.1038/EMBOJ.2012.5
Alloway, B. (2009). Soil factors associated with zinc deficiency in crops and humans. Environ. Geochem. Health 31, 537–548. doi: 10.1007/s10653-009-9255-4
Andreini, C., Banci, L., Bertini, I., and Rosato, A. (2006). Zinc through the three domains of life. J. Proteome Res. 5, 3173–3178. doi: 10.1021/pr0603699
Andrews, S. (2010). Fastqc: A Quality Control Tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed October 7, 2018).
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene Ontology: tool for the unification of biology. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Branson, O. E., and Freitas, M. A. (2016). Tag-count analysis of large-scale proteomic data. J. Proteome Res. 15, 4742–4746. doi: 10.1021/acs.jproteome.6b00554
Burian, J., Ramón-García, S., Sweet, G., Gómez-Velasco, A., Av-Gay, Y., and Thompson, C. J. (2012). The mycobacterial transcriptional regulator whib7 gene links redox homeostasis and intrinsic antibiotic resistance *. J. Biol. Chem. 287, 299–310. doi: 10.1074/JBC.M111.302588
Chawla, M., Parikh, P., Saxena, A., Munshi, M., Mehta, M., Mai, D., et al. (2012). Mycobacterium tuberculosis WhiB4 regulates oxidative stress response to modulate survival and dissemination in vivo. Mol. Microbiol. 85, 1148–1165. doi: 10.1111/j.1365-2958.2012.08165.x
Chen, Y.-X., Xu, Z., Ge, X., Sanyal, S., Lu, Z. J., and Javid, B. (2020). Selective translation by alternative bacterial ribosomes. Proc. Natl. Acad. Sci. U.S.A. 117, 19487–19496. doi: 10.1073/pnas.2009607117
Chen, Z., Hu, Y., Cumming, B. M., Lu, P., Feng, L., Deng, J., et al. (2016). Mycobacterial WhiB6 differentially regulates ESX-1 and the dos regulon to modulate granuloma formation and virulence in zebrafish. Cell Rep. 16, 2512–2524. doi: 10.1016/j.celrep.2016.07.080
Corbin, B. D., Seeley, E. H., Raab, A., Feldmann, J., Miller, M. R., Torres, V. J., et al. (2008). Metal chelation and inhibition of bacterial growth in tissue abscesses. Science 319, 962–965. doi: 10.1126/science.1152449
Craig, R., and Beavis, R. C. (2004). TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467. doi: 10.1093/bioinformatics/bth092
Dow, A., and Prisic, S. (2018). Alternative ribosomal proteins are required for growth and morphogenesis of Mycobacterium smegmatis under zinc limiting conditions. PLoS One 13:e0196300. doi: 10.1371/journal.pone.0196300
Dow, A., Sule, P., O’Donnell, T. J., Burger, A., Mattila, J. T., Antonio, B., et al. (2021). Zinc limitation triggers anticipatory adaptations in Mycobacterium tuberculosis. PLoS Pathog. 17:e1009570. doi: 10.1371/journal.ppat.1009570
Fay, A., and Glickman, M. S. (2014). An essential nonredundant role for mycobacterial dnak in native protein folding. PLoS Genet. 10:e1004516. doi: 10.1371/journal.pgen.1004516
Fol, M., Chauhan, A., Nair, N. K., Maloney, E., Moomey, M., Jagannath, C., et al. (2006). Modulation of Mycobacterium tuberculosis proliferation by MtrA, an essential two-component response regulator. Mol. Microbiol. 60, 643–657. doi: 10.1111/j.1365-2958.2006.05137.x
Geiman, D. E., Raghunand, T. R., Agarwal, N., and Bishai, W. R. (2006). Differential gene expression in response to exposure to antimycobacterial agents and other stress conditions among seven Mycobacterium tuberculosis whiB-like genes. Antimicrob. Agents Chemother. 50, 2836–2841. doi: 10.1128/AAC.00295-06
Goethe, E., Laarmann, K., Lührs, J., Jarek, M., Meens, J., Lewin, A., et al. (2020). Critical role of zur and SmtB in zinc homeostasis of Mycobacterium smegmatis. mSystems 5:e00880-19. doi: 10.1128/mSystems.00880-19
Gomez, J. E., and Bishai, W. R. (2000). whmD is an essential mycobacterial gene required for proper septation and cell division. Proc. Natl. Acad. Sci. U.S.A. 97:8554. doi: 10.1073/PNAS.140225297
Huang, D. W., Sherman, B. T., and Lempicki, R. A. (2009a). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13. doi: 10.1093/nar/gkn923
Huang, D. W., Sherman, B. T., and Lempicki, R. A. (2009b). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. doi: 10.1038/nprot.2008.211
Kanehisa, M. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
Kanehisa, M., Sato, Y., Furumichi, M., Morishima, K., and Tanabe, M. (2019). New approach for understanding genome variations in KEGG. Nucleic Acids Res. 47, D590–D595. doi: 10.1093/nar/gky962
Kapopoulou, A., Lew, J. M., and Cole, S. T. (2011). The mycobrowser portal: a comprehensive and manually annotated resource for mycobacterial genomes. Tuberculosis 91, 8–13. doi: 10.1016/j.tube.2010.09.006
Kieser, K. J., and Rubin, E. J. (2014). How sisters grow apart: mycobacterial growth and division. Nat. Rev. Microbiol. 12, 550–562. doi: 10.1038/nrmicro3299
Koedooder, C., Guéneuguès, A., Van Geersdaële, R., Vergé, V., Bouget, F.-Y., Labreuche, Y., et al. (2018). The role of the glyoxylate shunt in the acclimation to iron limitation in marine heterotrophic bacteria. Front. Mar. Sci. 5:435. doi: 10.3389/FMARS.2018.00435
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Law, C. W., Alhamdoosh, M., Su, S., Dong, X., Tian, L., Smyth, G. K., et al. (2018). RNA-seq analysis is easy as 1-2-3 with limma, Glimma and edgeR. F1000Res. 5:1408. doi: 10.12688/f1000research.9005.3
Li, X., Wu, J., Han, J., Hu, Y., and Mi, K. (2015). Distinct responses of Mycobacterium smegmatis to exposure to low and high levels of hydrogen peroxide. PLoS One 10:e0134595. doi: 10.1371/journal.pone.0134595
Li, Y., Sharma, M. R., Koripella, R. K., Yang, Y., Kaushal, P. S., Lin, Q., et al. (2018). Zinc depletion induces ribosome hibernation in mycobacteria. Proc. Natl. Acad. Sci. U.S.A. 115, 8191–8196. doi: 10.1073/pnas.1804555115
Liao, Y., Smyth, G. K., and Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Lim, C. K., Hassan, K. A., Penesyan, A., Loper, J. E., and Paulsen, I. T. (2013). The effect of zinc limitation on the transcriptome of Pseudomonas protegens Pf-5. Environ. Microbiol. 15, 702–715. doi: 10.1111/j.1462-2920.2012.02849.x
Mullarky, E., and Cantley, L. C. (2015). “Diverting glycolysis to combat oxidative stress,” in Innovative Medicine, eds K. Nakao, N. Minato, and S. Uemoto (Tokyo: Springer), 3–23. doi: 10.1007/978-4-431-55651-0_1
Müller, A. U., Imkamp, F., and Weber-Ban, E. (2018). The mycobacterial LexA/RecA-independent DNA damage response is controlled by PafBC and the pup-proteasome system. Cell Rep. 23, 3551–3564. doi: 10.1016/j.celrep.2018.05.073
Novichkov, P. S., Kazakov, A. E., Ravcheev, D. A., Leyn, S. A., Kovaleva, G. Y., Sutormin, R. A., et al. (2013). RegPrecise 3.0 – A resource for genome-scale exploration of transcriptional regulation in bacteria. BMC Genomics 14:745. doi: 10.1186/1471-2164-14-745
Podlesek, Z., and Žgur Bertok, D. (2020). The DNA damage inducible SOS response is a key player in the generation of bacterial persister cells and population wide tolerance. Front. Microbiol. 11:785. doi: 10.3389/fmicb.2020.01785
Powell, S. R. (2000). The antioxidant properties of zinc. J. Nutr. 130, 1447S–1454S. doi: 10.1093/jn/130.5.1447S
Prisic, S., Hwang, H., Dow, A., Barnaby, O., Pan, T. S., Lonzanida, J. A., et al. (2015). Zinc regulates a switch between primary and alternative S18 ribosomal proteins in Mycobacterium tuberculosis. Mol. Microbiol. 97, 263–280. doi: 10.1111/mmi.13022
R Core Team (2019). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Richard-Greenblatt, M., Bach, H., Adamson, J., Peña-Diaz, S., Li, W., Steyn, A. J. C., et al. (2015). Regulation of ergothioneine biosynthesis and its effect on Mycobacterium tuberculosis Growth and Infectivity *. J. Biol. Chem. 290, 23064–23076. doi: 10.1074/JBC.M115.648642
Ritchie, M. E., Phipson, B., Wu, D., Hu, Y., Law, C. W., Shi, W., et al. (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e47. doi: 10.1093/nar/gkv007
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Rohart, F., Gautier, B., Singh, A., and Cao, K.-A. L. (2017). mixOmics: an R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 13:e1005752. doi: 10.1371/JOURNAL.PCBI.1005752
Sao Emani, C., Williams, M. J., Wiid, I. J., Hiten, N. F., Viljoen, A. J., Pietersen, R.-D. D., et al. (2013). Ergothioneine is a secreted antioxidant in Mycobacterium smegmatis. Antimicrob. Agents Chemother. 57, 3202–3207. doi: 10.1128/AAC.02572-12
Shimono, N., Morici, L., Casali, N., Cantrell, S., Sidders, B., Ehrt, S., et al. (2003). Hypervirulent mutant of Mycobacterium tuberculosis resulting from disruption of the mce1 operon. Proc. Natl. Acad. Sci. U.S.A. 100, 15918–15923. doi: 10.1073/pnas.2433882100
Sinha, S., Rappu, P., Lange, S. C., Mantsala, P., Zalkin, H., and Smith, J. L. (1999). Crystal structure of Bacillus subtilis YabJ, a purine regulatory protein and member of the highly conserved YjgF family. Proc. Natl. Acad. Sci. U.S.A. 96, 13074–13079. doi: 10.1073/pnas.96.23.13074
Smollett, K. L., Dawson, L. F., and Davis, E. O. (2011). SigG does not control gene expression in response to DNA Damage in Mycobacterium tuberculosis H37Rv. J. Bacteriol. 193, 1007–1011. doi: 10.1128/JB.01241-10
Tiwari, S., van Tonder, A. J., Vilchèze, C., Mendes, V., Thomas, S. E., Malek, A., et al. (2018). Arginine-deprivation–induced oxidative damage sterilizes Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 115, 9779–9784. doi: 10.1073/pnas.1808874115
Tobiasson, V., Dow, A., Prisic, S., and Amunts, A. (2019). Zinc depletion does not necessarily induce ribosome hibernation in mycobacteria. Proc. Natl. Acad. Sci. U.S.A. 116, 2395–2397. doi: 10.1073/pnas.1817490116
Uehara, T., Parzych, K. R., Dinh, T., and Bernhardt, T. G. (2010). Daughter cell separation is controlled by cytokinetic ring-activated cell wall hydrolysis. EMBO J. 29, 1412–1422. doi: 10.1038/emboj.2010.36
Velasco, E., Wang, S., Sanet, M., Fernández-Vázquez, J., Jové, D., Glaría, E., et al. (2018). A new role for Zinc limitation in bacterial pathogenicity: modulation of α-hemolysin from uropathogenic Escherichia coli. Sci. Rep. 8:6535. doi: 10.1038/s41598-018-24964-1
Voskuil, M. I., Bartek, I. L., Visconti, K., and Schoolnik, G. K. (2011). The response of Mycobacterium Tuberculosis to reactive oxygen and nitrogen species. Front. Microbiol. 2:105. doi: 10.3389/fmicb.2011.00105
Walter, W., Sánchez-Cabo, F., and Ricote, M. (2015). GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics 31, 2912–2914. doi: 10.1093/bioinformatics/btv300
Wiśniewski, J. R., and Rakus, D. (2014). Multi-enzyme digestion FASP and the ‘Total Protein Approach’-based absolute quantification of the Escherichia coli proteome. J. Proteomics 109, 322–331. doi: 10.1016/j.jprot.2014.07.012
Wold, S., Sjöström, M., and Eriksson, L. (2001). PLS-regression: a basic tool of chemometrics. Chemom. Intell. Lab. Syst. 58, 109–130. doi: 10.1016/S0169-7439(01)00155-1
Wong, C. F., Shin, J., Subramanian Manimekalai, M. S., Saw, W. G., Yin, Z., Bhushan, S., et al. (2017). AhpC of the mycobacterial antioxidant defense system and its interaction with its reducing partner Thioredoxin-C. Sci. Rep. 7:5159. doi: 10.1038/s41598-017-05354-5
Keywords: mycobacteria, ribosomal proteins, zinc, Mycobacterium smegmatis, multi-omics, transcriptome, proteome, oxidative stress response
Citation: Dow A, Burger A, Marcantonio E and Prisic S (2022) Multi-Omics Profiling Specifies Involvement of Alternative Ribosomal Proteins in Response to Zinc Limitation in Mycobacterium smegmatis. Front. Microbiol. 13:811774. doi: 10.3389/fmicb.2022.811774
Received: 09 November 2021; Accepted: 04 January 2022;
Published: 10 February 2022.
Edited by:
Nelson da Cruz Soares, University of Sharjah, United Arab EmiratesReviewed by:
Rosario Duran, Institut Pasteur de Montevideo, UruguayJianping Xie, Southwest University, China
Scarlet Sara Shell, Worcester Polytechnic Institute, United States
Copyright © 2022 Dow, Burger, Marcantonio and Prisic. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sladjana Prisic, cHJpc2ljQGhhd2FpaS5lZHU=
 Allexa Dow
Allexa Dow Andrew Burger2
Andrew Burger2 Endrei Marcantonio
Endrei Marcantonio Sladjana Prisic
Sladjana Prisic