- 1Bioproduction Research Institute, National Institute of Advanced Industrial Science and Technology, Tsukuba, Japan
- 2Bioinformation and DDBJ Center, National Institute of Genetics, Mishima, Japan
- 3Research Center of Genetic Resources, Core Technology Research Headquarters, National Agriculture and Food Research Organization, Tsukuba, Japan
- 4Institute of Livestock and Grassland Science, National Agriculture and Food Research Organization, Nasushiobara, Japan
- 5Faculty of Life and Environmental Sciences, University of Tsukuba, Tsukuba, Japan
Bile salt hydrolase (BSH) enzymes produced by intestinal Lactobacillus species have been recognized as major targets for probiotic studies owing to their weight-loss and cholesterol-lowering effects. In this study, we isolated a highly thermostable BSH with broad substrate specificity, designed as LapBSH (BSH from a probiotic bacterium, Lactobacillus paragasseri JCM 5343T). The recombinant LapBSH protein clearly hydrolyzed 12 different substrates, including primary/secondary, major/minor, and taurine/glycine-conjugated bile salts in mammalian digestive tracts. Intriguingly, LapBSH further displayed a highly thermostable ability among all characterized BSH enzymes. Indeed, this enzyme retained above 80% of its optimum BSH activity even after 6 h of incubation at 50–90°C. LapBSH also exerted a functionally stable activity and maintained above 85% of its original activity after pre-heating at 85°C for 2 h. Therefore, LapBSH is a very unique probiotic enzyme with broad substrate specificity and high thermostability. The strain itself, JCM 5343T, was also found to exhibit high heat-resistance ability and could form colonies even after exposure to 85°C for 2 h. As thermostable enzyme/bacterium offers industrial and biotechnological advantages in terms of its productivity and stability improvements, both thermostable LapBSH and thermotolerant L. paragasseri JCM 5343T could be promising candidates for future probiotic research.
Introduction
Bile salts synthesized in the mammalian liver from cholesterol are surface-active and amphipathic steroid detergents that play an essential role in lipid digestion and absorption in mammalian gastrointestinal tracts. Bile salts are widely known as antimicrobial agents that inhibit bacterial growth by damaging the cell membranes of intestinal bacteria (Urdaneta and Casadesús, 2017). Some intestinal bacteria can overcome the toxicity of bile salts by producing bile salt hydrolase (BSH, EC 3.5.1.24), a key enzyme that can hydrolyze and deconjugate glycine or taurine from the cholesterol core of the bile acids (Begley et al., 2006). BSH enzymes also confer significant beneficial impacts on human health (e.g., serum cholesterol reduction) (Begley et al., 2006), in addition to their bile detoxification ability in probiotic lactic acid bacteria. A variety of intestinal bacteria have been found to exert BSH activity (Begley et al., 2006; Jones et al., 2008; O’Flaherty et al., 2018). Furthermore, Lactobacillus species are known to contribute to most of the total BSH activity in mammalian small intestines (Chand et al., 2017).
To date, several genes encoding BSH have been isolated from Lacticaseibacillus casei (Zhang et al., 2009), Limosilactobacillus fermentum (Kumar et al., 2013), Lactiplantibacillus plantarum (Christiaens et al., 1992; Lambert et al., 2008; Ren et al., 2011; Dong et al., 2012, 2013; Gu et al., 2014), Limosilactobacillus reuteri (Taranto et al., 1999), Lacticaseibacillus rhamnosus (Kaya et al., 2017), Ligilactobacillus salivarius (Wang et al., 2012; Bi et al., 2013), and some Lactobacillus spp. including Lactobacillus acidophilus (McAuliffe et al., 2005), Lactobacillus gasseri (Rani et al., 2017; Kusada et al., 2021), Lactobacillus johnsonii (Lundeen and Savage, 1990, 1992; Elkins et al., 2001; Oh et al., 2008; Chae et al., 2013), and Lactobacillus paragasseri (Kusada et al., submitted). The substrate specificity of BSH enzymes may be strain specific; however, most of the characterized BSH enzymes prefer to hydrolyze glycine-conjugated bile salts rather than taurine-conjugated ones (Dong and Lee, 2018). Furthermore, few BSH enzymes from L. johnsonii and L. salivarius have more specificity to taurine-conjugated bile salts (Oh et al., 2008; Chae et al., 2013), whereas some BSH enzymes from L. acidophilus and L. plantarum are known to hydrolyze both glycine- and taurine-conjugated bile salts (McAuliffe et al., 2005; Gu et al., 2014).
In general, most previously identified BSH enzymes function optimally under mesophilic conditions (approximately around 30–55°C) (Dong and Lee, 2018), except only one BSH enzyme from L. johnsonii PF01 (LjBSHC), which displays optimum activity at 70°C (Chae et al., 2013). Apart from the intestinal bacteria, a thermophilic Brevibacillus spp. isolated from hot spring water possesses a BSH enzyme with the optimum temperature of 60°C (Sridevi et al., 2009); however, the gene encoding the BSH enzyme has never been isolated and characterized. Thus, thermostable BSH enzyme remains largely unknown. Of note, thermostable enzyme and its thermotolerant host bacterium extend their potential application in biotechnological industries, which often require high heat treatment process, because (i) enzyme reaction at high temperature leads to improvement of productivity by reducing the viscosity of the reaction solution, increasing the solubility of substrates, and eliminating the potential risk of contamination, including pathogens (Lasa and Berenguer, 1993; Haki and Rakshit, 2003), and (ii) thermostable enzymes generally tend to be stable under harsh industrial process conditions (e.g., acids, alkalis, and organic solvents) (Lasa and Berenguer, 1993; Haki and Rakshit, 2003). Based on these biotechnological advantages, we assume that a thermostable BSH enzyme and a thermotolerant BSH-producing lactic acid bacterium could be potential new targets for future probiotic studies and could be employed in the food/pharmaceutical industries.
In the present study, we explored a new candidate of bsh gene from L. paragasseri JCM 5343T, a BSH-producing probiotic candidate isolated from human feces (Tanizawa et al., 2018). Intriguingly, we succeeded in discovering a novel BSH enzyme (LapBSH) with unique features, such as broad substrate specificity and highly thermostable activity among all BSHs identified, by cloning and expressing the gene from the strain JCM 5343T and conducting a biochemical characterization of its enzymatic functions.
Materials and Methods
Bacterial Strains Used in This Study
A probiotic lactic acid bacterium L. paragasseri strain JCM 5343T (= KCTC 3172T = ATCC 4963T) (Tanizawa et al., 2018) was purchased from the Japan Collection of Microorganisms (JCM, RIKEN BRC, Ibaraki, Japan). Strain JCM 5343T was cultivated using de Man-Rogosa-Sharpe medium (MRS, Difco Laboratories, Detroit, MI, United States) with headspace gas of N2/CO2 (80:20, v/v) at 37°C under anaerobic conditions. The Escherichia coli strain, DH5α (GMbiolab, Taichung, Taiwan), was used as the host strain for gene cloning, while the E. coli strain Origami™ 2 (DE3) (Novagen, Madison, WI, United States) was used as the host strain for the gene expression experiment. Both E. coli strains were cultured on Luria-Bertani (LB) agar or in LB broth (Nacalai Tesque Inc., Kyoto, Japan) supplemented with 100 μg/ml ampicillin (Sigma-Aldrich, Saint Louis, MO, United States) at 37°C.
Cloning and Heterologous Expression of a Putative Bile Salt Hydrolase Gene
Based on the sequence analyses and homology searches using NCBI BLAST program1, UniProt BLAST tool2, InterProScan3, Pfam4, and SignalP-5.0 server5, a gene encoding a putative BSH was screened from the complete genome sequence of strain JCM 5343T (accession number AP018549) (Tanizawa et al., 2018). A putative bsh gene candidate (called lapBSH, locus_tag = “LpgJCM5343_07940”) was amplified with PrimeSTAR HS DNA Polymerase (TaKaRa, Tokyo, Japan) using following primer set: CG GGATCCTGTACCTCAATTATTTATGATTCAAAC (BamHI site underlined) and CGGAATTCATTTTGATAGTTAATA TGTTGCTTTTC (EcoRI site underlined). The PCR cycling program was follows: initial denaturation at 98°C for 5 min, followed by 40 cycles at 98°C for 10 s and 68°C for 60 s. Gene cloning, overexpression, and protein purification of lapBSH were performed according to our previous study (Kusada et al., 2017), with slight modifications. In brief, the PCR product was purified and sub-cloned into an expression vector pCold II (TaKaRa). Thereafter, the resulting plasmid was transformed into E. coli Origami™2 (DE3) competent cells. Isopropyl-β-D-thiogalactopyranoside (IPTG, Nacalai Tesque) was added to the culture medium at a final concentration of 100 μM when the OD600 of the culture was approximately 0.5. After the addition of IPTG, the E. coli cells were incubated overnight at 15°C with shaking. The cells were pelleted by centrifugation at 5,800 × g for 10 min, suspended in lysis buffer (20 mM Tris-HCl, 200 mM NaCl, 10% glycerin, pH 6.0), and disrupted by sonication using an ultrasonic disintegrator (Sonicator BRANSON Sonifer 250, Branson, Danbury, CT, United States) in an ice-water bath. The cell debris was pelleted by centrifugation, and the resulting supernatant was further purified using a standard nickel affinity chromatography method based on a previous study (Kusada et al., 2017). The purified LapBSH solution was dialyzed with semipermeable membrane (Spectra/Por 3 membrane MWCO: 3,500, Repligen, Waltham, MA, United States) at 4°C and further concentrated using VIVASPIN Turbo 4 centrifugal filter units (MWCO: 10,000 PES membrane, Sartorius, Göttingen, Germany). To verify the purity and molecular weight of a recombinant LapBSH protein, sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and native-PAGE were performed using Mini-PROTEAN TGX precast polyacrylamide gel (Bio-Rad, Hercules, CA, United States). The gel was stained with QC Colloidal Coomassie Stain (Bio-Rad) via gentle agitation. To determine the effect of pH on oligomeric states of LapBSH protein, we used Good’s buffer solutions and adjusted the pH values (pH 3.0–10.0) of LapBSH before native-PAGE analysis.
Bile Salt Hydrolase Activity
The BSH assay was performed using the purified LapBSH protein as described previously (Allain et al., 2018; Kusada et al., 2021). In brief, the purified LapBSH protein (100 μg/100 μl) was mixed with 0.24 mg/100 μl of the selected conjugated bile salts and incubated at 37°C. The bile salt hydrolysis reaction was terminated by adding 15% trichloroacetic acid solution (FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan), and the proteins were removed by centrifugation at 10,000 × g for 15 min at 20°C. The supernatant was then mixed with 0.3 M borate buffer with 1% SDS (pH 9.5) and 0.3% 2,4,6-trinitrobenzenesulfonic acid solution (Tokyo Kasei Kogyo Co., Ltd., Tokyo, Japan). This mixture was statically incubated for 30 min at room temperature in the dark. Thereafter, 0.6 mM HCl was added to stop the reaction. The released glycine or taurine was measured at 416 nm using a SPARK 10M multimode microplate reader (TECAN, Männedorf, Switzerland). As a negative control, each bile salt solution was mixed with buffer instead of LapBSH protein (no enzyme control). Three independent experiments were performed (total n = 24). Student’s t-test was performed using GraphPad Prism version 8.0 software program (GraphPad Software, San Diego, CA, United States). For all analyses, a p-value less than 0.05 (P < 0.05) was defined as statistically significant. The tested substrates were glycocholic acid (GCA, Sigma-Aldrich), glycochenodeoxycholic acid (GCDCA, Sigma-Aldrich), glycodeoxycholic acid (GDCA, Sigma-Aldrich), glycoursodeoxycholic acid (GUDCA, Tokyo Kasei Kogyo), glycolithocholic acid (GLCA, Cayman Chemical, Ann Arbor, MI, United States), glycohyodeoxycholic acid (GHDCA, FUJIFILM Wako Pure Chemical Corporation), taurocholic acid (TCA, Nacalai Tesque), taurochenodeoxycholic acid (TCDCA, Sigma-Aldrich), taurodeoxycholic acid (TDCA, Nacalai Tesque), tauroursodeoxycholic acid (TUDCA, Sigma-Aldrich), taurolithocholic acid (TLCA, Cayman Chemical), and taurohyodeoxycholic acid (THDCA, FUJIFILM Wako Pure Chemical Corporation). Furthermore, we determined the effects of pre-treatment with EDTA on LapBSH activity. Briefly, the purified LapBSH protein was mixed with EDTA at a final concentration of 5 mM. After, the pre-treated LapBSH enzymes were mixed with 12 substrates, and BSH activity was detected as described above.
Biochemical Characterization of LapBSH
Studies on the optimum enzymatic condition of LapBSH were performed as described previously (Rani et al., 2017; Kusada et al., 2021). We selected TDCA as a representative substrate and mixed it with purified LapBSH (100 μg/100 μl) at various temperatures (10–90°C, in intervals of 10°C) and pH values (pH 3.0–10.0, in intervals of pH 1.0). After 6 h of incubation, the released taurine was detected as described above. We used Good’s buffer solution to determine the effects of pH on the enzyme activity of LapBSH, acetate buffer (CH3COONa 3H2O) pH 3.0–4.0, MES buffer (C6H13NO4S H2O) pH 5.0–6.0, HEPES buffer (C8H18N2O4S) pH 7.0–8.0, and CAPS (C9H19NO3S) pH 9.0–10.0. In addition, the thermostability of LapBSH protein was evaluated after incubation at different temperatures (55–100°C, in intervals of 5°C) for 2 h and cooling to a room temperature. The resulting pre-heated LapBSH was mixed with TDCA for 6 h, and the released taurine was detected as described above. All experiments were performed with eight technical replicates (n = 8).
Inhibitory Effects of Metal Ions on LapBSH Activity
We determined the effects of metal ions on LapBSH activity according to a previous study (Rani et al., 2017). Briefly, the purified LapBSH protein was pre-incubated with nine metal ions (CuCl2, CuSO4, MnCl2, MnSO4, MgCl2, MgSO4, ZnCl2, ZnSO4, and CaCl2) at a final concentration of 5 mM for 30 min at 37°C. After the pre-incubated LapBSH enzymes were mixed with TDCA and incubated at 37°C for 6 h, the released taurine was detected as described above. LapBSH protein without pre-incubation with metal ion was used as a control (no inhibition control). All experiments were performed with eight technical replicates (n = 8).
Gas Chromatograph Mass Spectrometry Analysis
The penicillin acylase activity of purified LapBSH protein was demonstrated by gas chromatograph mass spectrometry (GC-MS) analysis as described previously (Kusada et al., 2017; Yasutake et al., 2017). Briefly, 10 mM of penicillin G (Nacalai Tesque) solution was mixed with the purified LapBSH and buffer as a no-enzyme control and incubated at 37°C. After incubation, the digestion mixtures were extracted three times with equal volumes of ethyl acetate (FUJIFILM Wako Pure Chemical Corporation). Thereafter, the organic phase was evaporated to dryness in vacuum for 10 min (EYELA, Tokyo, Japan). The resulting sample was re-dissolved in methanol (FUJIFILM Wako Pure Chemical Corporation) and introduced onto a SHIMADZU GCMS-QP5050 system (Shimadzu Co., Ltd., Kyoto, Japan) equipped with a DB-5 capillary column (30 m × 0.25 mm, 0.25-μm film thickness; Agilent Technologies, Palo Alto, CA, United States).
Conserved Amino Acid Sequence Analysis
A multiple-amino-acid sequence alignment analysis was performed using the CLUSTAL W2 program and GENETYX-MAC software version 20.1.1 (GENETYX, Tokyo, Japan). The amino acid sequence of LapBSH was aligned with that of characterized BSHs from lactic acid bacteria.
Thermal Resistance Test of Lactobacillus paragasseri
Strain JCM 5343T was cultured in MRS broth at 37°C under anaerobic conditions. Full-grown cultures were heat-treated at temperatures ranging from 50°C to 75°C with different incubation times according to the Food Sanitation Law in Japan. After heat treatment, the cultures were streaked on MRS agar with or without 0.25% TDCA and incubated anaerobically at 37°C. The JCM 5343T culture was also heat-treated at 85°C for 30–150 min (in intervals of 30 min), and the resulting cultures were spotted on MRS agar plates and cultivated at 37°C under anaerobic conditions. L. gasseri JCM 1131T and L. helveticus JCM 1004 obtained from the JCM were cultured and heat-treated as described above to serve as heat-sensitive controls. Moreover, the number of viable colony-forming-units (CFU) was compared before and after heat-treatment at 50–80°C (in intervals of 10°C) for 30 min. All experiments were performed with three technical replicates (n = 3).
Results and Discussion
Sequence Analyses of a Putative Bile Salt Hydrolase Gene
Based on sequence analyses and homology searches, we found a putative BSH gene (called lapBSH) in the L. paragasseri JCM 5343T genome (Supplementary Figure 1). To obtain a recombinant LapBSH protein, we constructed a heterologous lapBSH gene expression system using E. coli Origami™ 2 (DE3) and purified the His-tagged LapBSH protein by nickel affinity chromatography. Based on SDS-PAGE analysis, the molecular weight of purified LapBSH protein was approximately 40.0 kDa in size (Supplementary Figure 2A), which is almost consistent with its calculated molecular mass based on the amino acid sequence of LapBSH (326 amino acids).
The amino acid sequence of LapBSH showed a significantly high sequence similarity with previously identified BSH (LgBSH) from L. gasseri FR4 (99.39%) (Rani et al., 2017), followed by BSHs from L. johnsonii strains (84.38 and 84.06%) and L. acidophilus strains (62.50 and 60.82%). LapBSH further exhibited moderate similarity to BSHs from L. plantarum strains (52.22 and 51.90%) and L. salivarius strains (45.17–47.66%). Interestingly, LapBSH showed a relatively low sequence similarity (39.32%) with LpBSH, the other BSH isolated from the same host strain, JCM 5343T (Kusada et al., submitted). These sequence analyses suggest that LapBSH may be a potential BSH enzyme; however, its substrate specificity and biochemical characteristics remain unclear since biochemical features of BSHs are often in discord with their high sequence similarities (Lundeen and Savage, 1990, 1992; Elkins et al., 2001; Oh et al., 2008). Accordingly, we attempted to identify and compare the enzymatic characteristics of LapBSH with experimentally identified BSHs, especially in LgBSH and thermostable BSHs.
Bile Salt Hydrolase Activity of a Recombinant LapBSH
To determine the BSH activity and substrate specificity of LapBSH, we used 12 different mammalian bile salts, including glycine-conjugated (GCA, GCDCA, GDCA, GUDCA, GLCA, and GHDCA) and taurine-conjugated (TCA, TCDCA, TDCA, TUDCA, TLCA, and THDCA) bile salts, by detecting the release of glycine or taurine from the hydrolysis of conjugated bile salts. As shown in Figure 1, LapBSH exhibited a notable hydrolysis activity toward all conjugated bile salts tested. In particular, this enzyme displayed high hydrolyzing activity toward six human major conjugated bile salts (TCA, TCDCA, TDCA, GCA, GCDCA, and GDCA), which constitute almost all of the conjugated bile salts in human intestine, and three minor conjugated bile salts (GHDCA, TUDCA, and THDCA) (Figure 1). LapBSH also showed lower but significant BSH activity toward other three minor substrates (TLCA, GUDCA, and GLCA) (Figure 1). A very small amount of GHDCA, TUDCA, GUDCA, TLCA, and GLCA have been found in humans (Sacquet et al., 1983; Mims and Hercules, 2003; Moreira et al., 2017; Semba et al., 2017), whereas THDCA has been absent in humans but present in rodents (Schmidt et al., 2019). Such findings suggest that LapBSH has broad substrate specificity toward minor as well as major bile salts. Therefore, LapBSH could act as a functional BSH enzyme and provide ecological advantages related to the bile detoxification for the host, L. paragasseri JCM 5343T, to enhance its survivability in human intestine. Of note, we confirmed the almost equal or slightly increased LapBSH activity when pre-treated with 5 mM EDTA (Figure 1), owing to the removal of metal ions (BSH inhibitors) in the buffers.
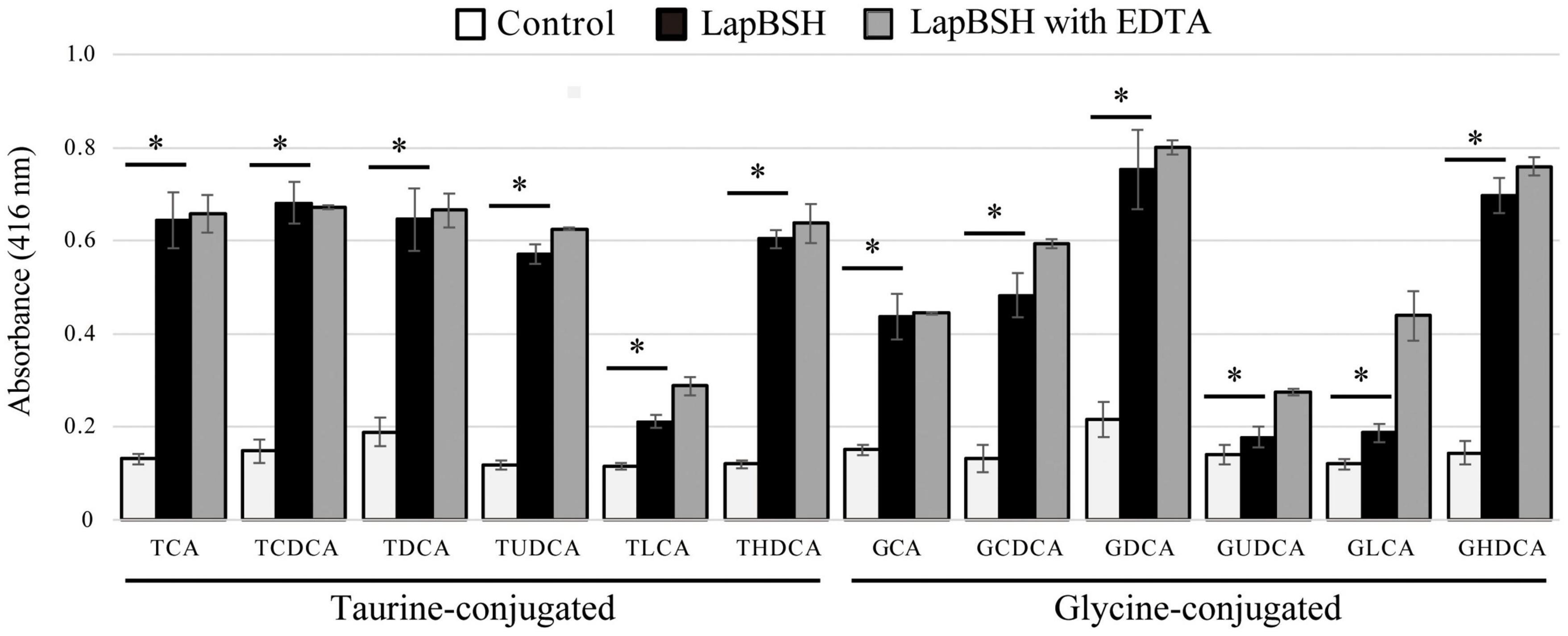
Figure 1. Bile salt hydrolase (BSH) activity and substrate specificity of LapBSH. The tested substrates were glycocholic acid (GCA), glycochenodeoxycholic acid (GCDCA), glycodeoxycholic acid (GDCA), glycoursodeoxycholic acid (GUDCA), glycolithocholic acid (GLCA), glycohyodeoxycholic acid (GHDCA), taurocholic acid (TCA), taurochenodeoxycholic acid (TCDCA), taurodeoxycholic acid (TDCA), tauroursodeoxycholic acid (TUDCA), taurolithocholic acid (TLCA), and taurohyodeoxycholic acid (THDCA). Values represent the mean of three independent experiments (each n = 8). Error bars represent standard deviation (SD). *A p-value less than 0.05 (P < 0.05) was defined as statistically significant.
To date, many previously characterized BSHs from lactic acid bacteria (e.g., Bifidobacterium spp. and Lactobacillus spp.) have been reported to hydrolyze six major human-conjugated bile salts (TCA, TCDCA, TDCA, GCA, GCDCA, and GDCA) as well as LapBSH. However, the hydrolysis activity of these identified BSHs toward the residual six minor bile salts (TUDCA, TLCA, THDCA, GUDCA, GLCA, and GHDCA) has not been fully revealed. However, three known BSHs (BlBSH from B. longum and LjBSHA and LjBSHC from L. johnsonii) can hydrolyze TUDCA and THDCA, while LapBSH was capable of hydrolyzing all of them. Besides, LgBSH sharing high sequence similarity with LapBSH could hydrolyze four major bile salts (GCA, GDCA TCA, and TDCA) (Rani et al., 2017); however, its hydrolytic activity toward other residual substrates (TCDCA, TUDCA, TLCA, THDCA, GCDCA GUDCA, GLCA, and GHDCA) is unknown. Furthermore, LgBSH prefers to hydrolyze glycine-conjugated bile salts rather than taurine-conjugated bile salts (Rani et al., 2017), while LapBSH can hydrolyze both glycine- and taurine-conjugated bile salts, indicating that LapBSH and LgBSH have different substrate specificities, regardless of their high sequence similarities. Therefore, LapBSH is a new BSH enzyme with broad substrate specificity that can deconjugate 12 different bile salts, including primary/secondary, major/minor, and taurine/glycine-conjugated bile salts in mammalian digestive tracts.
Penicillin Acylase Activity of LapBSH
In addition to BSH activity, we further determined whether recombinant LapBSH could hydrolyze β-lactam antibiotics, as some previously identified BSHs also showed penicillin acylase activity and the capability of hydrolyzing penicillin (Rossocha et al., 2005; Chand et al., 2017; Rani et al., 2017). For verification, GC-MS analyses were performed to detect phenylacetic acid generated via the hydrolysis of penicillin G according to our previous study (Kusada et al., 2017). We observed a significant peak with GC retention time of 5.299 min after the recombinant LapBSH was mixed with penicillin G solution (Supplementary Figure 3A). Furthermore, mass spectrometry of the 5.299-min GC fraction revealed the M-H ion at m/z 136 (Supplementary Figure 3B), aligning with the molecular weight of phenylacetic acid. Of note, this significant peak was not detected when penicillin G solution was incubated with buffer (data not shown). These findings clearly indicate that recombinant LapBSH also exhibits penicillin acylase activity as well as BSH activity and further emphasize its broad substrate specificity. It is noteworthy that LgBSH also exhibited penicillin acylase activity as well as LapBSH (Rani et al., 2017), suggesting that the amino acid residue(s) involved in their bi-functionality (BSH and penicillin acylase) would be conserved in these enzymes.
Biochemical Characterization of LapBSH
We determined the effects of pH and temperature on the enzyme activity of LapBSH as described in the experimental procedures. The maximum BSH activity of LapBSH was observed at pH 6.0 (Figure 2A). LapBSH exhibited a stable activity and retained above 80% of its original activity at pH 3.0–7.0; however, a significant decrease in enzyme activities was observed at pH values higher than 8.0. As for optimum temperature, the maximum BSH activity of LapBSH was observed at 37°C (Figure 2B). More importantly, LapBSH retained above 80% of its residual activity in high temperature ranges of 50–90°C (Figure 2B), strongly suggesting that LapBSH could have high thermostability. Indeed, LapBSH was functionally stable with above 85 and 65% retention of its original activity when the proteins were pre-heated at 85°C and 90°C for 2 h, respectively (Figure 2C).
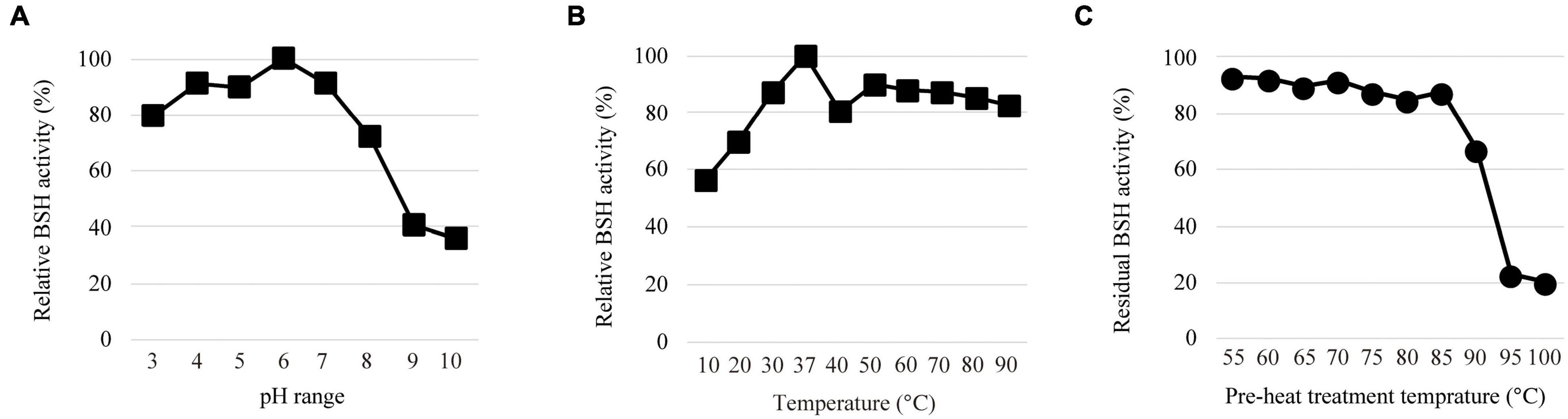
Figure 2. Biochemical features of LapBSH. (A) The effect of pH (pH 3.0–pH 10.0), (B) temperature (10°–90°C), and (C) pre-heat treatment (55°–100°C) on BSH activity toward TDCA. Each value represents the mean of eight technical replicates (n = 8). Maximum activity was recorded as 100%. Error bars indicate SD.
Of note, LgBSH, the most closely related enzyme to LapBSH, displayed optimal activity at pH 5.5 and 52°C, and enzyme activity significantly decreased with increasing temperatures (e.g., approximately 50 and 10% of its original activity at 60°C and 70°C, respectively) (Rani et al., 2017). Such finding indicates that LgBSH has little or no thermostability, and the biochemical characteristics of LapBSH and LgBSH markedly differed despite a sequence similarity above 99%. We further compared the thermostable activity of LapBSH with two experimentally identified thermostable BSHs. Sridevi et al. (2009) reported that the BSH from Brevibacillus sp. was active in the temperature range from 30°C to 80°C (optimum at 60°C); however, its residual BSH activity decreased by about 35% of its original activity at 70°C after 1 h of pre-incubation (Sridevi et al., 2009). Chae et al. (2013) also demonstrated that the optimum activity of BSH from L. johnsonii PF01 (LjBSHC) occurred at 70°C, and its residual BSH activity at 90°C decreased below 20% of its original activity (Chae et al., 2013).
To clarify whether LapBSH was denatured by heat treatment, native-PAGE analysis of LapBSH was performed. As shown in Supplementary Figure 2B, we confirmed obvious bands of LapBSH proteins after heat treatment at 90°C for up to 120 min, suggesting that a part of LapBSH proteins remain at 90°C. Furthermore, based on the native-PAGE analysis, the molecular weight of native LapBSH was approximately 160 kDa in size (Supplementary Figure 2B), which corresponded to its tetramer. We further confirmed that oligomerization states of LapBSH were affected under acidic (pH 3.0 and 4.0) and alkaline pH conditions (pH 9.0 and 10.0) (Supplementary Figure 2C), suggesting that pH condition contributes to electrostatic interaction at the interface between subunits of LapBSH. Xu et al. (2016) reported that two BSH molecules (derived from L. salivarius) packed as a dimer in the asymmetric unit (Xu et al., 2016). This suggests that the presence of a higher assembly form (e.g., tetramer assembled as a pair of dimers) may cause a large shift in the stability of LapBSH, because oligomerization of proteins may enhance the cooperative interaction between subunits and lead to increased stability against heat denaturation (Goodsell and Olson, 2000). Maes et al. (1999) reported that some other thermophilic enzymes exist as higher-order association states compared with their mesophilic analogues. In fact, triosephosphate isomerases (TIM) from three hyperthermophilic microorganisms (Thermotoga maritima, Pyrococcus woesei, and Methanothermus fervidus) are known to form tetrameric assemblies (Kohlhoff et al., 1996; Bell et al., 1998; Maes et al., 1999), whereas all the other known bacterial and eukaryotic TIMs form dimeric complexes (Kohlhoff et al., 1996). Furthermore, Villeret et al. (1998) reported that heat stabilization of the ornithine carbamoyltransferase (OTCase) dodecamer in Pyrococcus furiosus mainly results from hydrophobic interfaces between four trimers, while other mesophilic OTCases are homotrimeric (Villeret et al., 1998). Based on these previous studies, we assume that a higher assembly form accounts for the thermostability of LapBSH. Furthermore, a future study regarding a thermal denaturation curve of LapBSH could reveal a better understanding of its reversibility and cooperativity of stabilizing factors contributing to the high stability. Thus, to the best of our knowledge, LapBSH is a highly thermostable BSH among all of the previously identified BSH enzymes.
Inhibitory Assay of LapBSH Activity
We determined the effects of nine metal ions (CuCl2, CuSO4, MnCl2, MnSO4, MgCl2, MgSO4, ZnCl2, ZnSO4, and CaCl2) on LapBSH activity. These compounds were reported to inhibit BSH activity (Rani et al., 2017), though their inhibition mechanisms are largely unknown. Among these chemicals tested, copper compounds (CuCl2 and CuSO4) and zinc compounds (ZnCl2 and ZnSO4) were found to inhibit more than 80 and 65% of LapBSH activity, respectively (Figure 3). Furthermore, other manganese, magnesium, and calcium compounds were found to display a lower inhibitory activity (approximately 11–20%) of LapBSH (Figure 3). Interestingly, although LapBSH has a high sequence similarity with LgBSH from L. gasseri FR4 (Rani et al., 2017), the inhibitory effects of metal ions on the BSH activity of these two BSHs were also significantly different. In fact, manganese compounds (MnCl2 and MnSO4) significantly inhibited LgBSH (>70% inhibition) but did not notably inhibit LapBSH (<20% inhibition) activities. In contrast, ZnSO4 showed relatively low (<40% inhibition) and high (>70% inhibition) inhibitory activities toward LgBSH and LapBSH, respectively (Rani et al., 2017). It has also been found that MnCl2 and MnSO4 clearly inhibited the enzyme activity (68.1 and 83.1%, respectively) of BSH from L. salivarius NRRL B-30514 (Wang et al., 2012). Furthermore, Tanaka et al. (2000) demonstrated that MgSO4 and CaCl2 significantly inhibited the enzyme activity of BSH from Bifidobacterium longum SBT2928 (84 and 94% inhibition, respectively) (Tanaka et al., 2000), while MgSO4 and CaCl2 rarely inhibited the BSH activity of LapBSH (17.41 and 11.58%, respectively). Finally, a strong enzyme inhibition (95.2%) of BSH from L. plantarum CK102 in the presence of CaCl2 has been reported (Ha et al., 2006). Based on the findings of previous studies and this study, the effects of metal ion inhibitors on BSH activity may vary among BSHs.
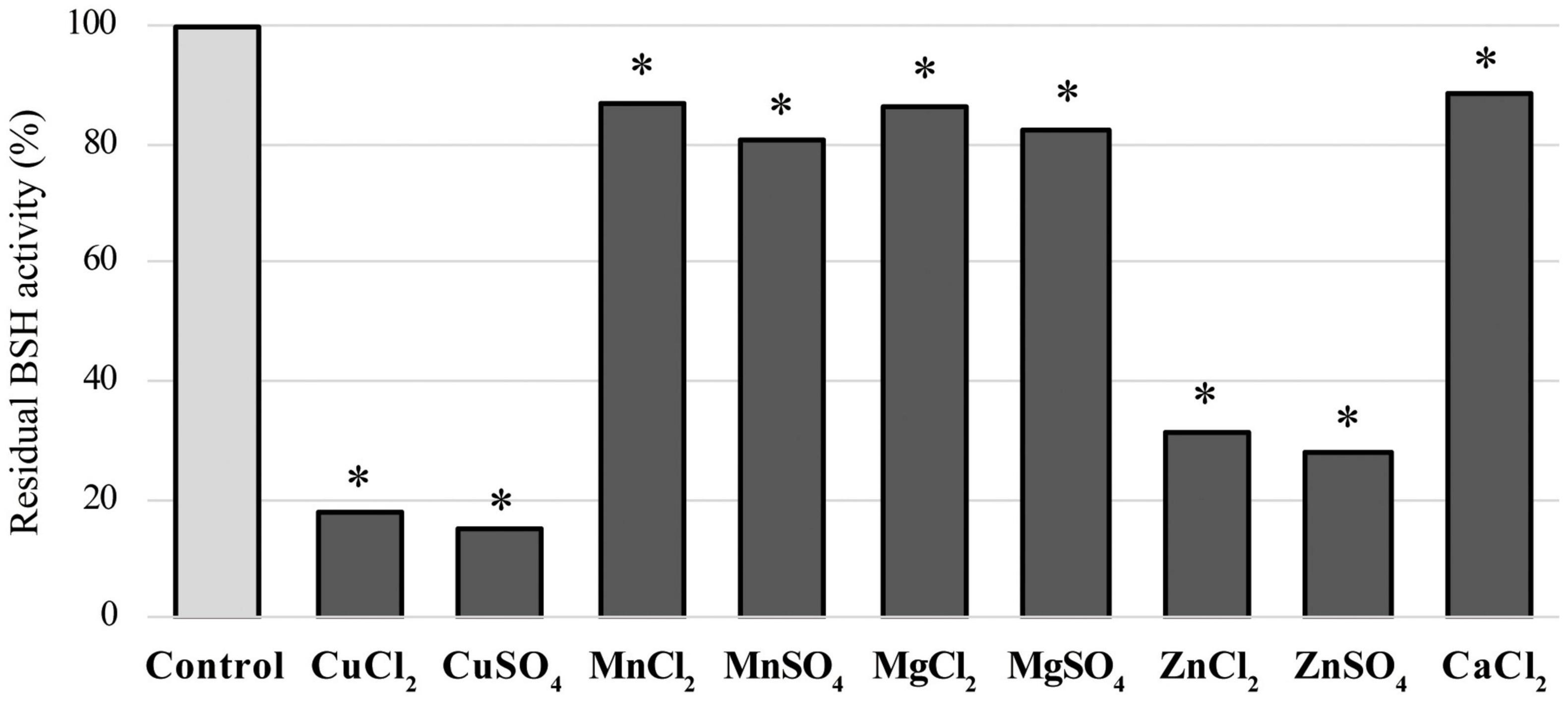
Figure 3. Effects of ion compounds on LapBSH activity. LapBSH proteins were pre-incubated with each of the nine metal ions at a final concentration of 5 mM for 30 min at 37°C. BSH activities were measured based on the activity of the pre-incubated LapBSH enzymes toward TDCA. Values represent the mean of eight technical replicates (n = 8). Maximum activity was recorded as 100%. Error bars indicate SD. *A p-value less than 0.05 (P < 0.05) was defined as statistically significant.
Multiple Sequence Alignment Analysis
The amino acid sequence of LapBSH was aligned with five BSHs known to hydrolyze both glycine- and taurine-conjugated bile salts (GC/TC) and one BSH that can hydrolyze only taurine-conjugated bile salts (TC) (Figure 4). The five residues (Cys-2, Arg-17, Asp-20, Asn-172, and Arg-225 in LapBSH) associated with active site and catalytic reaction were completely conserved in all proteins (Figure 4) (Dong and Lee, 2018). Among these conserved residues, N-terminal cysteine residue (Cys-2) is commonly known to play a critical role as a catalytic nucleophile in this protein family (Rossocha et al., 2005). However, seven amino acid residues (Leu-19, Lsy-59, Phe-66, Phe-130, Lys-133, Leu-138, and Trp-140 in LapBSH; indicated by green asterisk) that are involved in the potential substrate specificity (Rossocha et al., 2005; Dong and Lee, 2018) were not fully conserved among the proteins (Figure 4). Furthermore, another seven amino acid residues (Leu-21, Ile-23, Phe-25, Ile-57, Leu-64, Leu-133, and Ser-136 in LapBSH; indicated by purple asterisk) (Kumar et al., 2006; Dong and Lee, 2018) and one residue (Gln-134 in LapBSH; indicated by pink asterisk) (Rani et al., 2017; Dong and Lee, 2018) responsible for the putative substrate specificity were not fully conserved (Figure 4). These results suggest the presence of different amino acid residue(s) involved in substrate specificity.
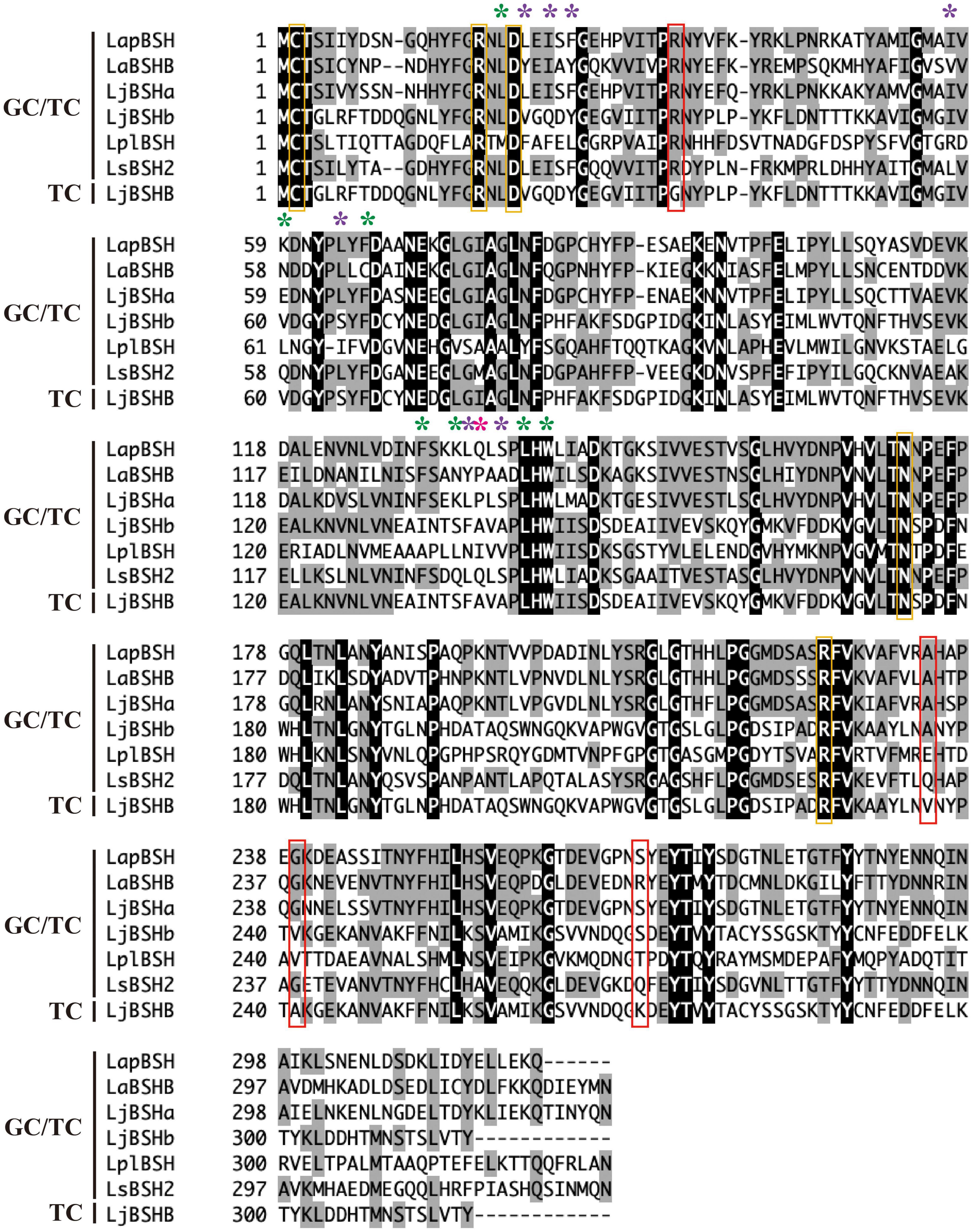
Figure 4. Multiple alignment of the amino acid sequences of LapBSH and other BSHs from Lactobacillus species. The black shading and gray shading represent identical and similar amino acid residues, respectively. The conserved residues (Cys, Arg, Asp, and Asn) associated with the predicted active site are indicated in orange boxes. Amino acid residues involved in the potential substrate specificity of known BSHs (CpBSH, BlBSH, and LgBSH) are indicated by green, purple, and pink asterisks, respectively (Rossocha et al., 2005; Kumar et al., 2006; Rani et al., 2017; Dong and Lee, 2018). Four amino acid residues that are not conserved in LjBSHB (TC) are indicated in red boxes. Abbreviated as follows: LapBSH (BBD48441), LaBSHB (AAV42923), LjBSHa (AAG22541), LjBSHb (AAC34381), LplBSH (AGG13404), LsBSH2 (ACL98205), and LjBSHB (EF536029).
The substrate preference of BSH enzymes vary among Lactobacillus strains of the same species. Indeed, LjBSHb from L. johnsonii 100-100 hydrolyzes both GC and TC (Lundeen and Savage, 1990, 1992; Elkins et al., 2001), while LjBSHB from L. johnsonii PF01 hydrolyzes only TC (Oh et al., 2008). However, very interestingly, these two BSHs (LjBSHb and LjBSHB) show very high amino acid sequence similarity (98.73%), and only four amino acid residues are substituted (Supplementary Figure 4A). Therefore, we assume that these four amino acid residues may be related to the substrate specificity. Indeed, we discovered that four residues of LjBSHB (TC) were not conserved in other BSHs (GC/TC), including LapBSH. In particular, Gly-35 of LjBSHB was completely substituted by Arg in other BSHs (GC/TC) (Figure 4), inferring that these residues may possibly be associated with the potential substrate preference of BSH enzymes.
LapBSH showed a high sequence similarity (99.39%) to LgBSH from L. gasseri FR4 (Rani et al., 2017), and only two amino acid substitutions (M97V and S178G) were confirmed (Supplementary Figure 4B). Based on the phylogenetic analysis, LapBSH and LgBSH were confirmed to be closely related with each other and were clearly categorized into the same BSH subgroup (Supplementary Figure 5). However, LapBSH and LgBSH differ mostly in terms of substrate specificity, inhibitory effects of metal ions on BSH activity, optimum temperature, and thermostability as described above (see Supplementary Table 1). Wright (1991) reported that asparagine and glutamine residues are susceptible to deamidation related to protein breakdown and aging in high-temperature environments, and this deamidation reaction would be facilitated by serine residues within the same protein (Wright, 1991). Anbar et al. (2010) demonstrated that a single mutation (S329G) dramatically increased the thermostability of endoglucanase in a thermophilic anaerobic bacterium, Clostridium thermocellum (Anbar et al., 2010). However, two mutations (M97V and S178G) may not be the only reason for the overall heat stabilization in LapBSH: changes of the Gibbs free energy and melting temperature by point substitutions are usually not large. In addition to two mutations, we speculate that the oligomerization of LapBSH is involved in its thermostability due to an increase in the cooperative interaction between two subunits, leading to structural stabilization as mentioned above. To clarify this point, future crystallographic study and site-directed mutagenesis analysis of LapBSH would enable a further understanding of its thermostability.
Thermotolerance Ability of Lactobacillus paragasseri JCM 5343T
We determined whether strain JCM 5343T has thermotolerance ability. According to the Food Sanitation Law in Japan, full-grown cultures of strain JCM 5343T were heat treated at different temperatures (50–75°C) for different incubation times (57 s to 5 h). We confirmed that strain JCM 5343T could grow on MRS agar under all culture conditions tested (Supplementary Figure 6), although members of the Lactobacillus genera are generally known to be heat sensitive. Of note, BSH activity and bile salt resistance ability of strain JCM 5343T were maintained after exposure to all heat-treatment conditions (Supplementary Figure 6). We further determined the effects of heat treatment on L. paragasseri JCM 5343T by CFU counting. The mean CFU/milliliter in the starting sample was (8.8 ± 0.5) × 109. After heat treatment at 50°C and 60°C, the CFU decreased slightly to (3.4 ± 0.4) × 109 and (5.8 ± 1.1) × 107, respectively. After heat treatment at 70°C and 80°C, the CFU was largely decreased (Supplementary Table 2). Surprisingly, strain JCM 5343T could form colonies reproducibly after heat treatment at 85°C for 120 min (Figure 5), though only a few colonies survived each time. As the other two well-known probiotic Lactobacillus species [L. gasseri JCM 1131T (Kusada et al., 2021) and L. helveticus JCM 1004 (Pan et al., 2005)] were strictly heat sensitive and could not form colonies after heat treatment at 85°C (Supplementary Figure 7), L. paragasseri JCM 5343T may have higher thermostability than other probiotic Lactobacillus species.
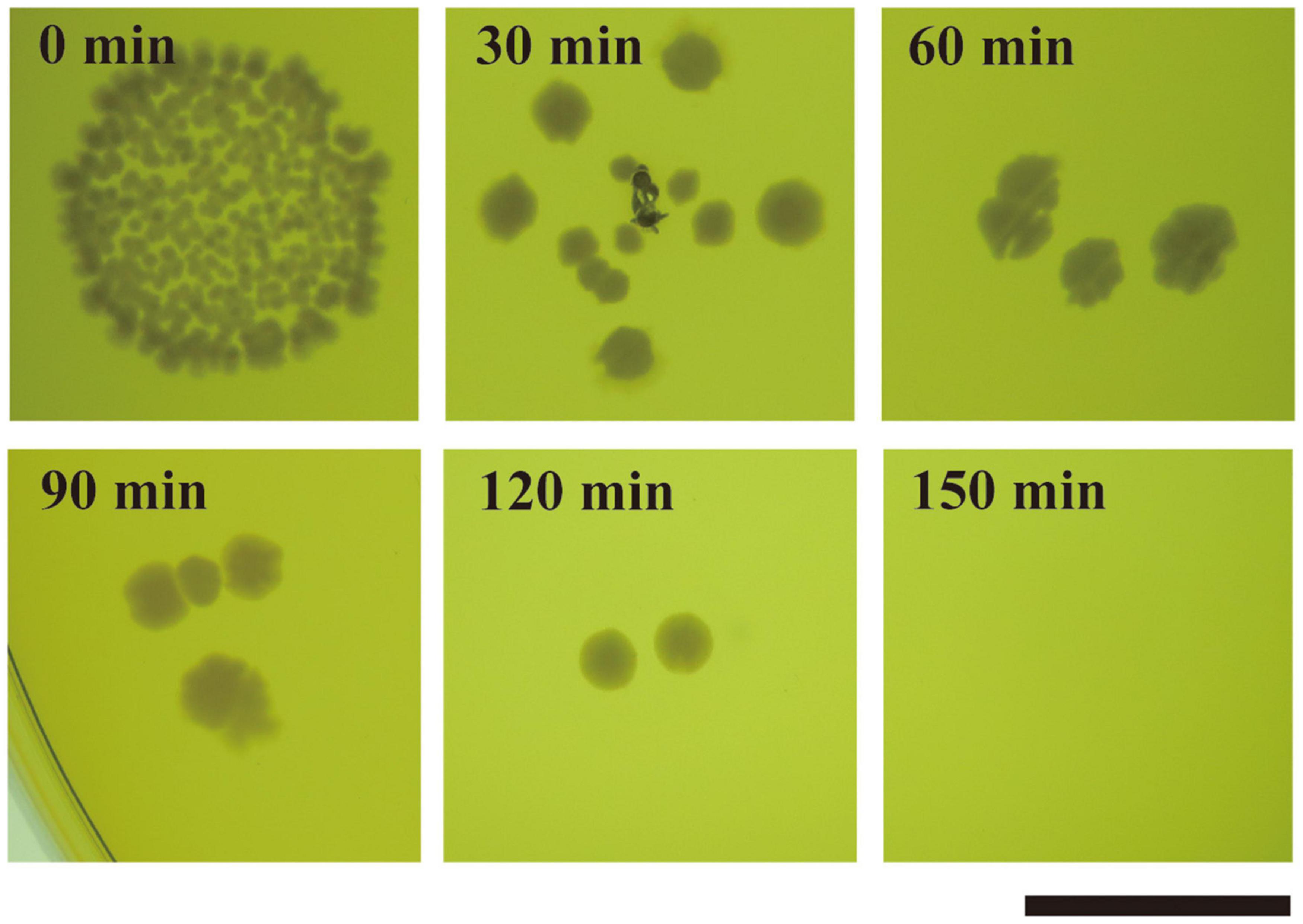
Figure 5. Thermal resistance assay of L. paragasseri JCM 5343T. Strain JCM 5343T culture was heat treated at 85°C for 30–150 min (intervals of 30 min). After heat treatment, the resulting cultures were spotted on de Man-Rogosa-Sharpe medium (MRS) agar plates and incubated at 37°C under anaerobic conditions. Scale bar indicates 1 cm.
As mentioned above, we demonstrated that not only LapBSH but also its host microorganism (L. paragasseri JCM 5343T) retained high thermostability. The thermostable LapBSH enzyme found in the present study may largely contribute to the BSH activity and bile salt resistance ability of strain JCM 5343T under heat-treatment conditions, as strain JCM 5343T has the other BSH enzyme (LpBSH) found in our previous study; this BSH enzyme displayed BSH activity only under mesophilic conditions (up to 40°C) and had a significant reduction in enzyme activities at temperatures higher than 50°C (Kusada et al., submitted). The extension of the industrial application of both thermostable LapBSH and its thermotolerant host L. paragasseri JCM 5343T would be of great advantage in oppressive biotechnological processes, including high temperature, acidic/alkaline pH, and organic solvents. A further investigation is needed to verify whether LapBSH/JCM 5343T can function and maintain their probiotic effects under high-temperature conditions in vivo and in vitro.
Conclusion
In the present study, we discovered a novel BSH enzyme (LapBSH) from L. paragasseri JCM 5343T and verified its two unique enzymatic traits. The first unique feature of the LapBSH enzyme is its broad substrate specificity. Indeed, our extended analysis demonstrated that the LapBSH enzyme could hydrolyze up to 12 different conjugated bile salts, including primary/secondary bile salts (e.g., GCA and GDCA), taurine/glycine-conjugated bile salts (e.g., TCA and GCA), and major/minor bile salts in human intestine (e.g., GCA and GHDCA). This enzymatic feature could confer bile-detoxification ability on the host bacterium, L. paragasseri JCM 5343T, to enhance its survivability in mammalian intestine. The other outstanding feature of LapBSH is thermostability. Although few thermostable BSH enzymes have been reported to date, LapBSH exhibited markedly higher thermostability than the other BSHs identified. Furthermore, we demonstrated that strain JCM 5343T displayed high heat-resistance ability and could grow after exposure to 85°C for 2 h, although Lactobacillus species are known to display heat susceptibility. Taken together, our findings suggest that both LapBSH and L. paragasseri JCM 5343T could be unprecedented candidates for future probiotic research, as thermostable enzyme and thermotolerant bacterium have significant industrial and biotechnological advantages for improving enzymatic productivity and stability relative to their mesophilic counterparts.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
HK, MA, MT, and HT made significant contributions in the conceptualization and investigation. HK and HT performed the data curation, formal analysis, funding acquisition, supervision, and writing—original draft. HT was the project administrator. All authors contributed to the article and approved the submitted version.
Funding
HK and HT were funded by the JSPS KAKENHI grant number 19K16633 and AMED PRIME grant number JP18gm6010019, respectively. This work was partially supported by the JST ERATO grant number JPMJER1502 and MEXT KAKENHI (Post-Koch Ecology, grant numbers 19H05679 and 19H05683).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.810872/full#supplementary-material
Footnotes
- ^ http://www.ncbi.nlm.nih.gov/
- ^ https://www.uniprot.org/blast/
- ^ http://www.ebi.ac.uk/interpro/search/sequence-search
- ^ http://pfam.xfam.org/
- ^ http://www.cbs.dtu.dk/services/SignalP/
References
Allain, T., Chaouch, S., Thomas, M., Vallee, I., Buret, A. G., Langella, P., et al. (2018). Bile-salt-hydrolases from the probiotic strain Lactobacillus johnsonii La1 mediate anti-giardial activity in vitro and in vivo. Front. Microbiol. 8:2707. doi: 10.3389/fmicb.2017.02707
Anbar, M., Lamed, R., and Bayer, E. A. (2010). Thermostability enhancement of Clostridium thermocellum cellulosomal endoglucanase Cel8A by a single glycine substitution. ChemCatChem 2, 997–1003.
Begley, M., Hill, C., and Gahan, C. G. (2006). Bile salt hydrolase activity in probiotics. Appl. Environ. Microbiol. 72, 1729–1738. doi: 10.1128/aem.72.3.1729-1738.2006
Bell, G. S., Russell, R. J., Kohlhoff, M., Hensel, R., Danson, M. J., Hough, D. W., et al. (1998). Preliminary crystallographic studies of triosephosphate isomerase (TIM) from the hyperthermophilic Archaeon Pyrococcus woesei. Acta Crystallogr. D. Biol. Crystallogr. 54, 1419–1421. doi: 10.1107/s0907444998004910
Bi, J., Fang, F., Lu, S., Du, G., and Chen, J. (2013). New insight into the catalytic properties of bile salt hydrolase. J. Mol. Catal. B Enzym 96, 46–51. doi: 10.1371/journal.pone.0114379
Chae, J. P., Valeriano, V. D., Kim, G. B., and Kang, D. K. (2013). Molecular cloning, characterization and comparison of bile salt hydrolases from Lactobacillus johnsonii PF01. J. Appl. Microbiol. 114, 121–133. doi: 10.1111/jam.12027
Chand, D., Avinash, V. S., Yadav, Y., Pundle, A. V., Suresh, C. G., and Ramasamy, S. (2017). Molecular features of bile salt hydrolases and relevance in human health. Biochim. Biophys. Acta. Gen. Subj. 1861, 2981–2991. doi: 10.1016/j.bbagen.2016.09.024
Christiaens, H., Leer, R. J., Pouwels, P. H., and Verstraete, W. (1992). Cloning and expression of a conjugated bile acid hydrolase gene from Lactobacillus plantarum by using a direct plate assay. Appl. Environ. Microbiol. 58, 3792–3798. doi: 10.1128/aem.58.12.3792-3798.1992
Dong, Z., and Lee, B. H. (2018). Bile salt hydrolases: structure and function, substrate preference, and inhibitor development. Protein Sci. 27, 1742–1754. doi: 10.1002/pro.3484
Dong, Z., Zhang, J., Lee, B., Li, H., Du, G., and Chen, J. (2012). A bile salt hydrolase gene of Lactobacillus plantarum BBE7 with high cholesterol-removing activity. Eur. Food Res. Technol. 235, 419–427.
Dong, Z., Zhang, J., Lee, B. H., Li, H., Du, G., and Chen, J. (2013). Secretory expression and characterization of a bile salt hydrolase from Lactobacillus plantarum in Escherichia coli. J. Mol. Catal. B Enzym. 93, 57–64.
Elkins, C. A., Moser, S. A., and Savage, D. C. (2001). Genes encoding bile salt hydrolases and conjugated bile salt transporters in Lactobacillus johnsonii 100-100 and other Lactobacillus species. Microbiology 147, 3403–3412. doi: 10.1099/00221287-147-12-3403
Goodsell, D. S., and Olson, A. J. (2000). Structural symmetry and protein function. Annu. Rev. Biophys. Biomol. Struct. 29, 105–153. doi: 10.1146/annurev.biophys.29.1.105
Gu, X.-C., Luo, X.-G., Wang, C.-X., Ma, D.-Y., Wang, Y., He, Y.-Y., et al. (2014). Cloning and analysis of bile salt hydrolase genes from Lactobacillus plantarum CGMCC No. 8198. Biotechnol. Lett. 36, 975–983. doi: 10.1007/s10529-013-1434-9
Ha, C.-G., Cho, J.-K., Chai, Y.-G., Ha, Y., and Shin, S.-H. (2006). Purification and characterization of bile salt hydrolase from Lactobacillus plantarum CK 102. J. Microbiol. Biotechnol. 16, 1047–1052.
Haki, G. D., and Rakshit, S. K. (2003). Developments in industrially important thermostable enzymes: a review. Bioresour. Technol. 89, 17–34. doi: 10.1016/s0960-8524(03)00033-6
Jones, B. V., Begley, M., Hill, C., Gahan, C. G., and Marchesi, J. R. (2008). Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc. Natl. Acad. Sci. U.S.A. 105, 13580–13585. doi: 10.1073/pnas.0804437105
Kaya, Y., Kök, M. Ş, and Öztürk, M. (2017). Molecular cloning, expression and characterization of bile salt hydrolase from Lactobacillus rhamnosus E9 strain. Food Biotechnol. 31, 128–140. doi: 10.1080/08905436.2017.1303778
Kohlhoff, M., Dahm, A., and Hensel, R. (1996). Tetrameric triosephosphate isomerase from hyperthermophilic Archaea. FEBS Lett. 383, 245–250. doi: 10.1016/0014-5793(96)00249-9
Kumar, R., Rajkumar, H., Kumar, M., Varikuti, S. R., Athimamula, R., Shujauddin, M., et al. (2013). Molecular cloning, characterization and heterologous expression of bile salt hydrolase (Bsh) from Lactobacillus fermentum NCDO394. Mol. Biol. Rep. 40, 5057–5066. doi: 10.1007/s11033-013-2607-2
Kumar, R. S., Brannigan, J. A., Prabhune, A. A., Pundle, A. V., Dodson, G. G., Dodson, E. J., et al. (2006). Structural and functional analysis of a conjugated bile salt hydrolase from Bifidobacterium longum reveals an evolutionary relationship with penicillin V acylase. J. Biol. Chem. 281, 32516–32525. doi: 10.1074/jbc.M604172200
Kusada, H., Morinaga, K., and Tamaki, H. (2021). Identification of bile salt hydrolase and bile salt resistance in a probiotic bacterium Lactobacillus gasseri JCM1131T. Microorganisms 9:1011. doi: 10.3390/microorganisms9051011
Kusada, H., Tamaki, H., Kamagata, Y., Hanada, S., and Kimura, N. (2017). A novel quorum-quenching N-acylhomoserine lactone acylase from Acidovorax sp. strain MR-S7 mediates antibiotic resistance. Appl. Environ. Microbiol 83, e00080-17. doi: 10.1128/AEM.00080-17
Lambert, J. M., Bongers, R. S., De Vos, W. M., and Kleerebezem, M. (2008). Functional analysis of four bile salt hydrolase and penicillin acylase family members in Lactobacillus plantarum WCFS1. Appl. Environ. Microbiol. 74, 4719–4726. doi: 10.1128/AEM.00137-08
Lasa, I., and Berenguer, J. (1993). Thermophilic enzymes and their biotechnological potential. Microbiologia 9, 77–89.
Lundeen, S. G., and Savage, D. C. (1990). Characterization and purification of bile salt hydrolase from Lactobacillus sp. strain 100-100. J. Bacteriol. 172, 4171–4177.
Lundeen, S. G., and Savage, D. C. (1992). Multiple forms of bile salt hydrolase from Lactobacillus sp. strain 100-100. J. Bacteriol. 174, 7217–7220.
Maes, D., Zeelen, J. P., Thanki, N., Beaucamp, N., Alvarez, M., Thi, M. H., et al. (1999). The crystal structure of triosephosphate isomerase (TIM) from Thermotoga maritima: a comparative thermostability structural analysis of ten different TIM structures. Proteins 37, 441–453.
McAuliffe, O., Cano, R. J., and Klaenhammer, T. R. (2005). Genetic analysis of two bile salt hydrolase activities in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 71, 4925–4929. doi: 10.1128/AEM.71.8.4925-4929.2005
Mims, D., and Hercules, D. (2003). Quantification of bile acids directly from urine by MALDI-TOF-MS. Anal. Bioanal. Chem. 375, 609–616. doi: 10.1007/s00216-003-1771-y
Moreira, S., Fonseca, I., Nunes, M. J., Rosa, A., Lemos, L., Rodrigues, E., et al. (2017). Nrf2 activation by tauroursodeoxycholic acid in experimental models of Parkinson’s disease. Exp. Neurol. 295, 77–87. doi: 10.1016/j.expneurol.2017.05.009
O’Flaherty, S., Briner Crawley, A., Theriot, C. M., and Barrangou, R. (2018). The Lactobacillus bile salt hydrolase repertoire reveals niche-specific adaptation. mSphere 3, e140–e118. doi: 10.1128/mSphere.00140-18
Oh, H. K., Lee, J. Y., Lim, S. J., Kim, M. J., Kim, G. B., Kim, J. H., et al. (2008). Molecular cloning and characterization of a bile salt hydrolase from Lactobacillus acidophilus PF01. J. Microbiol. Biotechnol. 18, 449–456.
Pan, D., Luo, Y., and Tanokura, M. (2005). Antihypertensive peptides from skimmed milk hydrolysate digested by cell-free extract of Lactobacillus helveticus JCM1004. Food Chem. 91, 123–129. doi: 10.1016/j.foodchem.2004.05.055
Rani, R. P., Anandharaj, M., and Ravindran, A. D. (2017). Characterization of bile salt hydrolase from Lactobacillus gasseri FR4 and demonstration of its substrate specificity and inhibitory mechanism using molecular docking analysis. Front. Microbiol. 8:1004. doi: 10.3389/fmicb.2017.01004
Ren, J., Sun, K., Wu, Z., Yao, J., and Guo, B. (2011). All 4 Bile salt hydrolase proteins are responsible for the hydrolysis activity in Lactobacillus plantarum ST-III. J. Food Sci. 76, M622–M628. doi: 10.1111/j.1750-3841.2011.02431.x
Rossocha, M., Schultz-Heienbrok, R., Von Moeller, H., Coleman, J. P., and Saenger, W. (2005). Conjugated bile acid hydrolase is a tetrameric N-terminal thiol hydrolase with specific recognition of its cholyl but not of its tauryl product. Biochemistry 44, 5739–5748. doi: 10.1021/bi0473206
Sacquet, E., Parquet, M., Riottot, M., Raizman, A., Jarrige, P., Huguet, C., et al. (1983). Intestinal absorption, excretion, and biotransformation of hyodeoxycholic acid in man. J. Lipid. Res. 24, 604–613.
Schmidt, A., Joussen, S., Hausmann, R., Grunder, S., and Wiemuth, D. (2019). Bile acids are potent inhibitors of rat P2X2 receptors. Purinergic. Signal. 15, 213–221. doi: 10.1007/s11302-019-09657-2
Semba, R. D., Gonzalez-Freire, M., Moaddel, R., Trehan, I., Maleta, K. M., Khadeer, M., et al. (2017). Environmental enteric dysfunction is associated with altered bile acid metabolism. J. Pediatr. Gastroenterol. Nutr. 64, 536–540. doi: 10.1097/MPG.0000000000001313
Sridevi, N., Srivastava, S., Khan, B. M., and Prabhune, A. A. (2009). Characterization of the smallest dimeric bile salt hydrolase from a thermophile Brevibacillus sp. Extremophiles 13, 363–370. doi: 10.1007/s00792-008-0224-0
Tanaka, H., Hashiba, H., Kok, J., and Mierau, I. (2000). Bile salt hydrolase of Bifidobacterium longum-biochemical and genetic characterization. Appl. Environ. Microbiol. 66, 2502–2512. doi: 10.1128/AEM.66.6.2502-2512.2000
Tanizawa, Y., Tada, I., Kobayashi, H., Endo, A., Maeno, S., Toyoda, A., et al. (2018). Lactobacillus paragasseri sp. nov., a sister taxon of Lactobacillus gasseri, based on whole-genome sequence analyses. Int. J. Syst. Evol. Microbiol. 68, 3512–3517. doi: 10.1099/ijsem.0.003020
Taranto, M., Sesma, F., and De Valdez, G. F. (1999). Localization and primary characterization of bile salt hydrolase from Lactobacillus reuteri. Biotechnol. Lett. 21, 935–938.
Urdaneta, V., and Casadesús, J. (2017). Interactions between bacteria and bile salts in the gastrointestinal and hepatobiliary tracts. Front. Med. 4:163. doi: 10.3389/fmed.2017.00163
Villeret, V., Clantin, B., Tricot, C., Legrain, C., Roovers, M., Stalon, V., et al. (1998). The crystal structure of Pyrococcus furiosus ornithine carbamoyltransferase reveals a key role for oligomerization in enzyme stability at extremely high temperatures. Proc. Natl. Acad. Sci. U.S.A. 95, 2801–2806. doi: 10.1073/pnas.95.6.2801
Wang, Z., Zeng, X., Mo, Y., Smith, K., Guo, Y., and Lin, J. (2012). Identification and characterization of a bile salt hydrolase from Lactobacillus salivarius for development of novel alternatives to antibiotic growth promoters. Appl. Environ. Microbiol. 78, 8795–8802. doi: 10.1128/AEM.02519-12
Wright, H. T. (1991). Nonenzymatic deamidation of asparaginyl and glutaminyl residues in proteins. Crit. Rev. Biochem. Mol. Biol. 26, 1–52. doi: 10.3109/10409239109081719
Xu, F., Guo, F., Hu, X. J., and Lin, J. (2016). Crystal structure of bile salt hydrolase from Lactobacillus salivarius. Acta Crystallogr. F. Struct. Biol. Commun. 72, 376–381. doi: 10.1107/S2053230X16005707
Yasutake, Y., Kusada, H., Ebuchi, T., Hanada, S., Kamagata, Y., Tamura, T., et al. (2017). Bifunctional quorum-quenching and antibiotic-acylase MacQ forms a 170-kDa capsule-shaped molecule containing spacer polypeptides. Sci. Rep. 7:8946. doi: 10.1038/s41598-017-09399-4
Keywords: bile salt hydrolase, Lactobacillus paragasseri, probiotics, substrate specificity, thermostability
Citation: Kusada H, Arita M, Tohno M and Tamaki H (2022) Isolation of a Highly Thermostable Bile Salt Hydrolase With Broad Substrate Specificity From Lactobacillus paragasseri. Front. Microbiol. 13:810872. doi: 10.3389/fmicb.2022.810872
Received: 08 November 2021; Accepted: 10 January 2022;
Published: 17 February 2022.
Edited by:
Arun K. Bhunia, Purdue University, United StatesReviewed by:
Neelja Singhal, University of Delhi, IndiaJai K. Kaushik, National Dairy Research Institute (ICAR), India
Copyright © 2022 Kusada, Arita, Tohno and Tamaki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hiroyuki Kusada, a3VzYWRhLWhpcm95dWtpQGFpc3QuZ28uanA=; Hideyuki Tamaki, dGFtYWtpLWhpZGV5dWtpQGFpc3QuZ28uanA=
 Hiroyuki Kusada
Hiroyuki Kusada Masanori Arita
Masanori Arita Masanori Tohno
Masanori Tohno Hideyuki Tamaki
Hideyuki Tamaki