
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 30 May 2022
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.810312
Antibiotic resistance represents one of the most pressing concerns facing public health today. While the current antibiotic resistance crisis has been driven primarily by the anthropogenic overuse of antibiotics in human and animal health, recent efforts have revealed several important environmental dimensions underlying this public health issue. Antibiotic resistant (AR) microbes, AR genes, and antibiotics have all been found widespread in natural environments, reflecting the ancient origins of this phenomenon. In addition, modern societal advancements in sanitation engineering (i.e., sewage treatment) have also contributed to the dissemination of resistance, and concerningly, may also be promoting the evolution of resistance to water treatment. This is reflected in the recent characterization of naturalized wastewater strains of Escherichia coli—strains that appear to be adapted to live in wastewater (and meat packing plants). These strains carry a plethora of stress-resistance genes against common treatment processes, such as chlorination, heat, UV light, and advanced oxidation, mechanisms which potentially facilitate their survival during sewage treatment. These strains also carry an abundance of common antibiotic resistance genes, and evidence suggests that resistance to some antibiotics is linked to resistance to treatment (e.g., tetracycline resistance and chlorine resistance). As such, these naturalized E. coli populations may be co-evolving resistance against both antibiotics and water treatment. Recently, extraintestinal pathogenic strains of E. coli (ExPEC) have also been shown to exhibit phenotypic resistance to water treatment, seemingly associated with the presence of various shared genetic elements with naturalized wastewater E. coli. Consequently, some pathogenic microbes may also be evolving resistance to the two most important public health interventions for controlling infectious disease in modern society—antibiotic therapy and water treatment.
The world is currently in an antibiotic resistance crisis. The severity of this crisis is best exemplified by bacterial pathogens of pandemic concern, including the multidrug-resistant and extremely drug-resistant Mycobacterium tuberculosis, the extended-spectrum beta-lactamase (ESBL) and carbapenem-resistant Enterobacteriaceae (CRE), as well as several microbes collectively known as the “ESKAPE” pathogens, including vancomycin-resistant Enterococcus faecium (VRE), methicillin-resistant Staphylococcus aureus (MRSA), Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp. (Davies and Davies, 2010; Ventola, 2015; Tacconelli et al., 2018; Jernigan et al., 2020). The rapid growth of antibiotic resistance has placed an incredible burden on the health system (US Department of Health and Human Services and CDC, 2019; Serra-Burriel et al., 2020; Nelson et al., 2021) and without urgent action, estimates suggest that drug resistant infections will cost $100 trillion dollars (USD) and cause the deaths of 10 million people annually by 2050 (O’Neill, 2016). Indeed, the world appears to be headed toward a “post-antibiotic era” (Kwon and Powderly, 2021), where many infectious diseases may become effectively untreatable and routine medical procedures too dangerous to perform without robust anti-infection preventative therapies.
With the looming threat of a post-antibiotic era, increased efforts have been redirected toward identifying and mitigating the major drivers of antibiotic resistance. While the evolution and transmission of antibiotic resistance have been particularly well-characterized within human (Shallcross, 2014; Center for Disease Dynamics, Economics and Policy, 2015; Ventola, 2015; Holmes et al., 2016; Pal et al., 2016) and animal contexts (Cabello et al., 2013; Center for Disease Dynamics, Economics and Policy, 2015; Martin et al., 2015; Holmes et al., 2016; Pal et al., 2016), the role of the environment in the current antibiotic resistance crisis has received considerably less attention (McCubbin et al., 2021). The focus on human/animal health is understandable and justifiable given that acquired resistance is largely driven by anthropogenic use of antimicrobials (Skurnik et al., 2006; Grall et al., 2015), but recent efforts have been made to address the environmental dimensions of the evolution and dissemination of antibiotic resistance (Martinez, 2009; Wright, 2010; Finley et al., 2013; Perry and Wright, 2013; Berendonk et al., 2015; Bengtsson-Palme et al., 2018; Stanton et al., 2020). Nevertheless, most One Health approaches designed to combat antibiotic resistance continue to focus primarily on mitigating antibiotic resistance-promoting activities and promoting stewardship in human, agricultural, and aquacultural sectors (Hoelzer et al., 2017; Tang et al., 2017; Scott et al., 2018; Nathwani et al., 2019).
Despite the under-representation of environmental health perspectives in One Health approaches to antibiotic resistance (Iossa and White, 2018; McCubbin et al., 2021), both natural and man-made environments closely interface with the cycling of antibiotic resistance across human and animal sectors. Although this is most evident in “antibiotic pollution” of the environment due to anthropogenic use of antibiotics (Kraemer et al., 2019), the natural environment also serves as an important reservoir of antibiotic resistant microbes and their resistance genes (Huijbers et al., 2015). In particular, “naturalized” microbial populations—microbes that have evolved to primarily inhabit non-host environments (Ishii et al., 2006; Devane et al., 2020)—may play an important role in the origin, evolution, and dissemination of antibiotic resistance. Reflecting this, sewage/wastewater treatment plants, have been identified as an important “hotspot” for antibiotic resistance, and in this review, we examine the natural wastewater microbiome as a potential reservoir for the emergence of antibiotic resistance in the microbial world. As a model of our understanding, we describe the evolutionary emergence of naturalized wastewater Escherichia coli. These strains of E. coli appear to have evolved to live and survive in modern-day, engineered wastewater treatment plants. They display a certain level of resistance to wastewater treatment (i.e., conventional treatment as well as disinfection; chlorination, advanced oxidants, and heat) and carry an abundance of antibiotic resistance genes (Zhi et al., 2017, 2019), implicating their role as a potential reservoir for both antibiotic and wastewater treatment resistance. Similarly, some pathotypes of E. coli, such as the extra-intestinal pathogenic E. coli (ExPEC) also appear to resist wastewater treatment and exhibit similar genetic properties as wastewater naturalized strains (Zhi et al., 2020). Concerningly, the evidence suggests that microbes may have evolved adaptations against two critical public health interventions that are fundamental for the control of infectious disease in modern society—water treatment/sanitation and antibiotic therapy.
It is undeniable that, since the “golden era” of antibiotic discovery (Aminov, 2010), the widespread use of antibiotics in clinical and agricultural contexts have primarily driven the current antibiotic resistance crisis (Shallcross, 2014; Martin et al., 2015). However, it is important to note that the current crisis merely represents a relatively recent development in the long history of antibiotic resistance in the microbial world. Indeed, the anthropogenic adoption of antibiotics has simply exerted an extreme global selective pressure promoting the emergence and dissemination of antibiotic resistance in microbes at an unprecedented scale.
Considering this, antibiotic resistance can be thought of as either intrinsic, which refers to the basal level of resistance provided by the inherent structural or biochemical characteristics of the bacterial cell prior to the widespread use of antibiotics, or acquired, which mainly refers to the recent transfer of genetic determinants conferring resistance to bacterial populations, particularly pathogens, and in response to the selective pressure exerted by the rise in antibiotic use (Martinez, 2009). The concept of intrinsic resistance directly implicates the environment in the evolution of antibiotic resistance, especially considering that microbial communities have inhabited natural environments for billions of years before the first appearance of humans or animals. Reflecting this, it is now recognized that various antibiotic resistance determinants have ancient, often environmental, origins (Mindlin et al., 2008; Martinez, 2009; D’Costa et al., 2011; Perron et al., 2015).
Antibiotic resistance is fundamentally a natural phenomenon, and this is particularly apparent in naturalized microbial communities. Microbial populations that are “naturalized” refer to those subgroups that have evolved to preferentially survive, inhabit, and grow in external, non-host environments as their primary niche (Ishii et al., 2006; Devane et al., 2020). Some naturalized microbes belong to host-associated taxa, but appear to have diverged from their host-derived counterparts very recently, or in some cases millions of years ago (Walk et al., 2009; Staley et al., 2014; Walk, 2015), and include organisms such as E. coli (Ishii et al., 2006; Jang et al., 2017). Reflecting their evolutionary divergence, these naturalized populations have been found to be genotypically distinct from host-associated lineages (Ishii et al., 2006; Tymensen et al., 2015; Zhi et al., 2019), and appear to have evolved various adaptive mechanisms to thrive outside of the host niche. Importantly, this may include adaptive mechanisms associated with antibiotic resistance, as antibiotics present in the environment play important roles in the dynamics and composition of these microbial populations (Gao et al., 2020).
The connection between natural microbes and antibiotic resistance is immediately evident in their role as producers of antibiotic compounds. Many of the major classes of antibiotics—including those under considerable threat in the current resistance crisis—originated in microbes naturally occurring in the environment, particularly those inhabiting soil environments (Chandra and Kumar, 2017). The antibiotic potential of naturally-produced bioactive molecules was recognized early during antibiotic discovery, such as seen in the discovery of penicillin by Fleming from the soil-derived Penicillium mold (Fleming, 1929). Recognizing the wealth of potential antibiotic compounds present in soil microbial communities, Waksman pioneered comprehensive screening protocols on the soil dwelling filamentous actinomycetes, which led to the discovery of antibiotics such as streptomycin and neomycin, the first antibiotic active agents against tuberculosis (Schatz et al., 1944; Waksman and Lechevalier, 1949). Waksman’s work not only established actinomycetes as prolific antibiotic producers, but also specifically identified the genus Streptomyces as an important producer of antibacterial products (Waksman et al., 2010). Reflecting this, actinomycetes and streptomycetes have been found to possess remarkably large genomes, and typically harbor genomic regions specifically dedicated to the biosynthesis of various natural products (Traxler and Kolter, 2015), demonstrating the vast antibiotic producing potential of natural, and specifically soil-dwelling, microbes. Indeed, the majority of antibiotics that have been discovered since the “golden era” have either been characterized as, or were derived from, natural products produced primarily by actinomycetes, as well as other environmental bacteria or environmental fungi (Hutchings et al., 2019).
As antibiotics serve inhibitory, antibacterial purposes in clinical and laboratory settings, it was immediately assumed that these bioactive products would also mediate inter-species competition and “microbial warfare” in natural environments (Allen et al., 2010), which would in turn select for the evolution of resistance against these bioactive compounds. Although the antibacterial effects of some natural microbial products have been demonstrated (Currie et al., 1999; Neeno-Eckwall et al., 2001), naturally-produced antibiotic compounds are typically produced and released at concentrations significantly lower than the minimum inhibitory concentrations typically used in clinical medicine. Furthermore, as the in situ physiologic concentrations of naturally-produced antibiotic compounds have yet to be consistently measured, the antibacterial functions attributed to these compounds in natural contexts remain unclear. Despite this, it is well understood that sub-inhibitory concentrations of antibiotics still generate genotypic and phenotypic variability upon which selection can act (López and Blázquez, 2009; Laureti et al., 2013), which can in turn promote the development of antibiotic resistance in natural microbial communities (Andersson and Hughes, 2014; Murray et al., 2018).
Alternative functions for the bioactive small molecules produced by natural microbial communities have been proposed. In particular, a growing body of evidence indicates that many natural microbial products exhibit hormesis (Davies et al., 2006), in which they exhibit a dose-dependent antibiotic effect at high concentrations but appear to play distinct functions at in situ concentrations present in natural environments. For example, at lower concentrations, naturally-produced bioactive molecules appear to activate intercellular signaling pathways important in microbial community dynamics, particularly in modulating gene expression related to quorum sensing, biofilm formation, adherence, and virulence (Traxler and Kolter, 2015; Murray et al., 2018). Reflecting this, some of the earliest antibiotic substances isolated from P. aeruginosa were later found to consist of quorum-sensing molecules that simultaneously displayed potent antibacterial activities (Kaufmann et al., 2005; Dubern and Diggle, 2008). As such, natural bioactive compounds may be involved in metabolic signaling networks in microbial communities, whereby the receptors for these molecules involve the same cellular structures that are targeted for inhibition during antibiotic use (Allen et al., 2010). Recently, an alternative function for naturally-produced antibiotic compounds in predation dynamics have been proposed, as it has been suggested that antibiotic production may be the result of predation-driven selection during nutritional and evolutionary bottlenecks (Leisner et al., 2016).
The finding that antibiotics appear to mediate other functions beyond exerting antibacterial effects in natural environments suggests that they were distributed throughout the environment long before their introduction for therapeutic and prophylactic purposes. Indeed, resistance-conferring genes and mutations have been shown to impact metabolism, allowing particular strains to utilize various alternative peptide, phosphorous, and carbon sources for nutrition (Allen et al., 2010). As such, the same evolutionary events that confer resistance in clinical or laboratory contexts may alternatively promote metabolic flexibility in the environment. Similarly, extensive gene knockout analyses have revealed a myriad of genes that, while primarily involved in other functions, contribute to an antibiotic resistance phenotype (Gomez and Neyfakh, 2006; Breidenstein et al., 2008; Duo et al., 2008; Tamae et al., 2008). Conversely, studies have shown that microbes can carry a plethora of resistance genes, but concomitantly, are susceptible to antibiotic treatment (Zhi et al., 2019), suggesting that these genes may provide other important functions within the life history of a microbe. Considering the prevalence of antibiotic resistance genes in the microbial world, particularly in environmental microbial communities, resistance determinants have likely evolved in response to a variety of selection pressures prior to the discovery and widespread use of antibiotics as therapeutics.
Consequently, microbial populations found in environmental matrices appear to demonstrate a basal level of resistance against a single antibiotic or a group of antibiotics. For instance, a comprehensive resistance screen of morphologically distinct spore-forming actinomycetes against 21 antibiotics conducted by D’Costa et al. (2006) identified several strains that were resistant to an average of 7 or 8 antibiotics across the major classes, indicating that these strains were naturally multidrug resistant. Naturally-resistant microorganisms within these microbial communities may then serve as reservoirs of antibiotic resistance, and resistance-adjacent, genes, especially if they become incorporated into mobile genetic elements that can be horizontally acquired by microbes of clinical concern. Reflecting this, Forsberg et al. (2012) characterized soil bacteria that shared a remarkably similar antibiotic resistome compared to various human pathogens. Screening of the multidrug-resistant soil bacteria revealed several resistance cassettes against antibiotics including beta-lactams, aminoglycosides, amphenicols, sulfonamides, and tetracyclines, containing genes with 100% identity to those found in various human pathogens. As the sequence similarity encompassed mobilization sequences, the resistance elements in question may have been laterally transferred to the pathogenic microbes. Importantly, the antibiotic resistome of environmental microbial communities extends beyond known resistance genes and may include novel resistance determinants that have yet to be characterized. Indeed, comprehensive screens of resistance genes and mobile genetic elements in environmental, human, and animal microbiomes have revealed relatively low abundances of known antibiotic resistance elements in natural environments, but the high taxonomic and genetic diversity identified in environmental microbiomes indicate that they may harbor a vast collection of currently unknown resistance genes that could be transferred to pathogens (Pal et al., 2016; Steen et al., 2019).
Overall, there remains relatively little evidence for the direct transfer of antibiotic resistance genes between the environmental resistome and clinical pathogens. Although there is great potential for the horizontal transfer of resistance genes, transfer events are only made evident when the genes become clinically relevant. As such, while several transfer events may occur, only a select few of the transferred genes will become fixed in a pathogenic population as natural selection will simultaneously act at the level of the gene itself and on whether the gene is maintained following the transfer event. Nevertheless, certain antibiotic resistance genes, especially those of particular concern in this antibiotic resistance crisis, appear to have origins in environmental microbes. One such class of antibiotic resistance genes include the beta-lactamases, which are estimated to have originated over 2 billion years ago (Hall and Barlow, 2004). Interestingly, several beta-lactamase families have been estimated to have mobilized from bacterial chromosomes onto plasmids millions of years ago (Barlow and Hall, 2002). For instance, despite their recent emergence and proliferation within clinical pathogens, certain beta-lactamases within the OXA family appear to have originated from the free-living environmental Shewanella species. Indeed, the beta-lactamase OXA-48, which mediates carbapenem resistance in Enterobacteriaceae, appears to have originated from the chromosomal blaOXA-54 gene found in Shewanella oneidensis, which is commonly found in lake sediments (Poirel et al., 2004). Similarly, the beta-lactamase OXA-181 appears to have originated from chromosomal genes carried by Shewanella xiamenensis, an environmental microbe commonly found in marine and freshwater environments (Potron et al., 2011). This also appears to be the case for class A beta-lactamases, which can hydrolyze a wide range of extended spectrum cephalosporins and monobactams, rendering many antibiotics ineffective (Cantón et al., 2012). While there are several families of class A extended spectrum beta-lactamases, the CTX-M family has received particular attention due to its explosive dissemination worldwide in a “CTX-M pandemic” (Cantón and Coque, 2006). Interestingly, several CTX-M beta-lactamase clusters appear to have been mobilized from chromosomal beta-lactamase genes harbored by various free-living Kluyvera species that are ubiquitous in soil, water, and sewage environments (Watson et al., 2017). Indeed, the chromosomal beta-lactamase gene kluC found in Kluyvera cryocrescens is thought to be the ancestor to the CTX-M-1 cluster (Decousser et al., 2001) and kluA from K. ascorbata may have given rise to the CTX-M-2 cluster (Humeniuk et al., 2002), while the genes kluG, kluY, and blaCTX-M-78 from K. georgiana appear to be the likely origin of the CTX-M-8 (Poirel et al., 2002), CTX-M-9 (Olson et al., 2005), and CTX-M-25 (Rodríguez et al., 2010) clusters, respectively.
Kluyvera species have also been implicated in the origin of other resistance genes, such as the fosfomycin-resistance gene fosA4 (Rodriguez et al., 2018). A similar environmental origin has been proposed for the qnr family of genes that confer resistance to quinolones. Indeed, qnr-like genes have been characterized in environmental microbes such as waterborne Vibrionaceae (Poirel et al., 2005a) and the marine and freshwater-dwelling Shewenella algae (Poirel et al., 2005b). Interestingly, when these environmental qnr genes were transformed into E. coli, they conferred resistance to quinolone antibiotics (Poirel et al., 2005a). As this evidence indicates, although they may not directly pose a risk to the health of humans and animals, microorganisms of environmental origin, including natural and recently naturalized microbes, may act as important reservoirs for the mobilization of key antibiotic resistance genes that, when acquired by pathogenic populations, can render antibiotic therapies ineffective.
A better understanding of the environmental influences on the rapid growth of antibiotic resistance has led to the identification of environmental hotspots in the current antibiotic resistance crisis. In particular, wastewater treatment plants have received considerable attention as important drivers of antibiotic resistance, especially as a conduit for the dissemination of antibiotic resistance from anthropogenic activities to natural environments (Figure 1). Indeed, antibiotics, antibiotic resistant microbes, and their associated resistance genes are widespread in wastewater matrices (Schwartz et al., 2003; Baquero et al., 2008), such that their subsequent release via effluent discharge represents an important source of antibiotic pollution in the surrounding environment. As such, despite the recent technological emergence of wastewater treatment plants to manage the sanitary wastes of densely populated societies (i.e., within last century), wastewater treatment plants have been identified as important “hotspots” in the dissemination of antibiotic resistance (Michael et al., 2013; Rizzo et al., 2013).
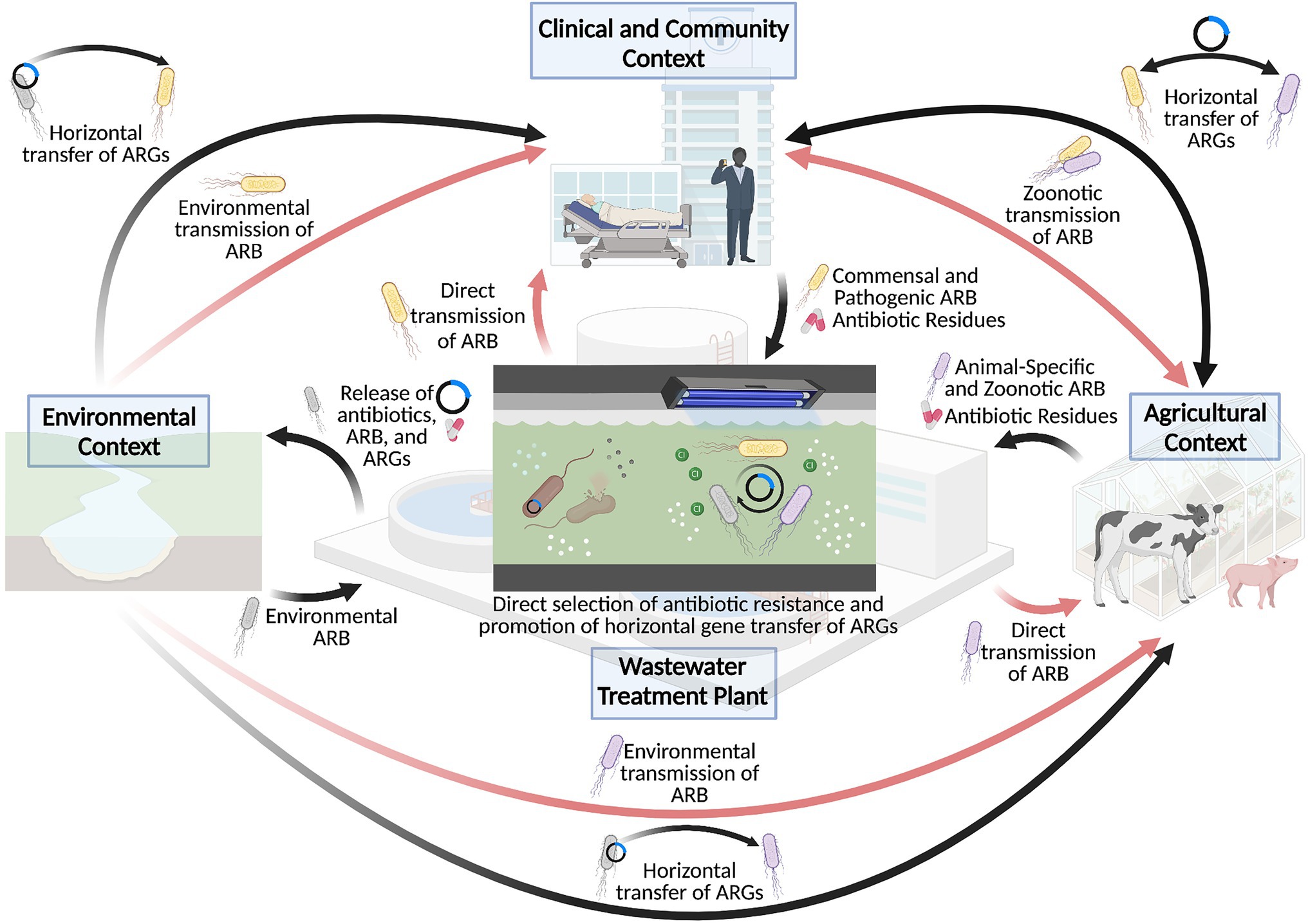
Figure 1. Wastewater treatment plants as a hotspot for the dissemination and selection of antibiotic resistance. Wastewater treatment plants have played an important role in driving the antibiotic resistance crisis, particularly through cycling antibiotics, antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARG) between clinical, agricultural, and environmental sectors, as depicted by the black arrows. This in turn contributes to the transmission of antibiotic resistant bacteria to human and animal hosts, as shown in the red arrows. Importantly, the conditions of the wastewater treatment plant may also directly contribute to the selection of antibiotic resistance or promote the mobilization of resistance genes within the wastewater matrix, which further exacerbates the antibiotic resistance crisis. Created with BioRender.com.
Although water treatment leverages a variety of processes to eliminate the various microbiological, chemical, and physical contaminants prevalent in wastewater, conventional treatment regimens do not reliably eliminate all antibiotics and other pharmaceutical compounds from the wastewater matrix (Michael et al., 2013). Biological processes, for instance, which include sorption to activated sludge and microbiological degradation, appear to be quite variable in their capacity to eliminate different classes of antibiotics. While several studies have demonstrated the efficient removal of various antibiotics through biological means, including beta-lactams (Watkinson et al., 2009), tetracyclines (Yang et al., 2005), sulfonamides (Yang et al., 2005; Li et al., 2009), and quinolones (Zorita et al., 2009), removal efficiencies overall vary widely across studies and water treatment processes. Indeed, other studies have demonstrated an incomplete removal of the same antibiotics in response to biological processes (Brown et al., 2006; Cha et al., 2006), whereas other antibiotic classes such as macrolides appear to be recalcitrant to removal through biological treatment (Li and Zhang, 2011). Studies evaluating the removal efficiencies of oxidative and disinfection treatments also yielded varied results. Ozonation, for instance, appears to remove a significant degree of several antibiotic classes, though the exact efficiency depended on the specific antibiotic (Michael et al., 2013). Similarly, chlorination has been demonstrated to remove certain antibiotics such as cephalexin with high efficiency, while exhibiting incomplete removal of various other compounds including the human-derived erythromycin metabolite erythromycin-H2O, as well as antibiotics such as sulfamethoxazole and trimethoprim (Li and Zhang, 2011). Moreover, while ultraviolet (UV) irradiation has been shown to remove antibiotics including sulfonamides and quinolones at high efficiencies, macrolides and tetracycline were largely resilient and were removed at significantly lower rates (Kim et al., 2009).
Ultimately, although different treatment processes may collectively contribute to the efficient removal of each antibiotic class, the wide range observed in the removal efficiencies of each antibiotic in response to different treatment process, and the variability in treatment programs used across wastewater treatment plants, indicates that water treatment overall does not completely remove antibiotics from the wastewater matrix. Reflecting this, antibiotics have been increasingly identified in treated wastewater effluents (Zhang and Li, 2011; Östman et al., 2017; Rodriguez-Mozaz et al., 2020) and appear to be particularly high in effluents from the pharmaceutical industry and hospitals, and which have been shown to contribute to hotspots of antibiotic resistance in natural environments (Karkman et al., 2019). Furthermore, while antibiotics themselves appear to be widespread in wastewater matrices, research on the fate of antibiotics during water treatment do not typically address the antibiotic metabolites that are produced following human and animal use (Michael et al., 2013). As such, the selection potential of discharged effluents may be greater than current estimates suggest. Given that current policies surrounding antibiotic resistance and antibiotic pollution do not address the impact of antibiotics present in treated effluents, wastewater treatment plants will continue to represent an important source of antibiotic pollution promoting the selection of antibiotic resistance in natural environments (Barancheshme and Munir, 2018; Kraemer et al., 2019).
Beyond antibiotics, wastewater treatment plants are also implicated in the large-scale release of antibiotic resistance genes into downstream environments. The removal efficiency for resistance genes during water treatment is similarly variable across treatment processes and water treatment programs (Wang and Chen, 2020). For instance, while some studies demonstrated that biological membrane processes could efficiently remove tetracycline (tetW and tetO) and sulfonamide (sul1) resistance genes (Li and Zhang, 2011), others found that biological processes such as biological filter and anaerobic techniques did not significantly reduce any resistance genes during water treatment (Yuan et al., 2016). Furthermore, while ozonation was found to reliably reduce antibiotic resistance genes, disinfection processes, such as chlorination and UV irradiation, were found to be comparatively inefficient, exhibiting negligible removal rates for various resistance genes (McKinney and Pruden, 2012; Stange et al., 2019). Indeed, the finding that certain resistance genes have even been enriched following water treatment (Laht et al., 2014; Bengtsson-Palme et al., 2016) suggests that, like antibiotics, the associated resistance genes may also persist during water treatment. In a survey of antibiotic resistance genes across wastewater samples, Szczepanowski et al. (2009) identified 123 clinically-relevant resistance genes in the final effluents. Similarly, Mao et al. (2015) identified several genes conferring resistance against tetracycline, sulfonamides, quinolones, and erythromycin that were discharged at higher rates in the final effluents than their initial prevalence in influents prior to treatment.
The wide prevalence of antibiotic resistance genes in wastewater raises the possibility of discharged effluents introducing resistance genes to downstream environmental microbial communities. This prospect is particularly concerning considering that these resistance genes may be mobile (Davies and Davies, 2010). For instance, using a high-throughput array to quantify antibiotic resistance genes and their mobilization potential during wastewater treatment, Karkman et al. (2016) found that water treatment enriched various genes involved in gene mobilization, including a Tn25-type transposase gene and various clinical class 1 integrons—elements that are implicated in the evolution and dissemination of antibiotic resistance (Babakhani and Oloomi, 2018; Souque et al., 2021). Similarly, using a metagenomic approach, Che et al. (2019) identified a wide prevalence of antibiotic resistance genes conferring resistance to aminoglycosides, beta-lactams, tetracycline, sulfonamides, chloramphenicol, trimethoprim, and quinolones in wastewater effluents, of which over half appeared to be harbored on mobile genetic elements such as plasmids and integrative and conjugative elements. As multi-drug resistant plasmids related to environmental bacteria have also been identified in wastewater effluents (Szczepanowski et al., 2004), wastewater discharge may introduce and promote the horizontal exchange of resistance genes with naturalized microbial communities in environments downstream of wastewater treatment plants.
The wide range of antibiotic resistance genes and mobile genetic elements that are released following water treatment points to the presence of an extensive and highly mobile resistome in wastewater. With such an accessible reservoir of mobile antibiotic resistance genes, the importance of wastewater treatment plants in the antibiotic resistance crisis may extend well beyond the dissemination of antibiotic resistant-determining contaminants in downstream environments. In fact, the wastewater matrix itself may be particularly hospitable for the acquisition and dissemination of antibiotic resistance.
Effectively representing an “endpoint” in the urban water cycle, wastewater treatment plants collect antibiotic-impacted waste from various human, animal, and environmental sources prior to treatment. As a result, wastewater matrices constitute a complex mix of environmental, clinical, and agricultural microorganisms, infused in a concoction of toxic contaminants including biocides, heavy metals, toxic organics, antibiotics, and other pharmaceuticals (Östman et al., 2017). Concerningly, antibiotic resistant bacteria themselves are major constituents of wastewater microbiomes (Fouz et al., 2020). A wide range of antibiotic resistant microbes, including clinically relevant populations such as ESBL-producing Enterobacteriaceae (Amador et al., 2015; Galler et al., 2018; Scaccia et al., 2020; Zhi et al., 2020), vancomycin-resistant Enterococcus (Iweriebor et al., 2015; Nishiyama et al., 2015; Galler et al., 2018; Gouliouris et al., 2019), and MRSA (Galler et al., 2018), appear to be widespread in wastewater matrices, representing a potential source of antibiotic resistant bacteria that can be released into downstream environments through effluent discharge.
Beyond this resistant subpopulation, however, the wastewater matrix may also be particularly conducive for the evolution and transmission of antibiotic resistance within the wastewater treatment plant itself. Wastewater matrices comprise a particularly nutrient-rich environment that can support a complex microbiome of human, animal, and environmental microbes. Considering that this microbiome is accompanied by an extensive, highly mobile resistome (Schlüter et al., 2007; Popowska and Krawczyk-Balska, 2013; Ju et al., 2019; Rodríguez et al., 2021) of clinically relevant and novel environment resistance genes, various selection pressures can act in parallel to promote the development of resistance in microbes present in the wastewater matrix. Intuitively, the high concentrations of antibiotics in wastewaters, particularly from pharmaceutical industries (Larsson et al., 2007; Larsson, 2014; Bengtsson-Palme and Larsson, 2016), can strongly select for antibiotic resistance within the wastewater matrix. Outside of this context, however, the concentrations of antibiotics in wastewater treatment plants do not typically exceed the minimum inhibitory concentrations that define resistance in clinical and laboratory settings (Szczepanowski et al., 2009; Ju et al., 2019). Despite this, sub-inhibitory concentrations of antibiotics can still select for resistance, and they appear to be a particularly important driver in doing so (Gullberg et al., 2011; Marshall and Levy, 2011; Andersson and Hughes, 2012; Bengtsson-Palme and Larsson, 2016). As an example, various antibiotics, even at sub-lethal doses, have been shown to activate the bacterial SOS response (Miller et al., 2004; Mesak et al., 2008; Baharoglu and Mazel, 2011), which can generate genetic diversity upon which selection can act and promote the development of adaptive mechanisms (Podlesek and Žgur Bertok, 2020). Although the SOS response does not necessarily accelerate the evolution of antibiotic resistance, it does appear to mediate adaptation by increasing bacterial fitness to the presence of sub-inhibitory concentrations of antibiotics (Torres-Barceló et al., 2015). Furthermore, the SOS response has been linked to increased rates of gene mobilization (Beaber et al., 2004; Guerin et al., 2009), which may further promote the dissemination of antibiotic resistance across microbial populations. As studies have demonstrated that wastewater microbial communities can undergo an extensive degree of mobile genetic exchange, including across phyla (Li et al., 2018), the presence of antibiotics in wastewater appears to exert a strong selective pressure for the dissemination of antibiotic resistance within the wastewater microbiome (Figure 1).
Beyond direct selection, the wastewater matrix and the treatment processes themselves provide other selective pressures that can simultaneously promote the development of antibiotic resistance (Figure 1). The high concentrations of biocides and heavy metals in wastewater matrices (Östman et al., 2017), for instance, have been implicated in the co-evolution of antibiotic resistance alongside metal resistance (Pal et al., 2015; Wales and Davies, 2015). This can occur through the co-localization of biocide- and metal-resistance genes with antibiotic resistance genes on the same genetic elements, such that the selection of one resistance gene will lead to the selection of the whole genetic element. While the previous studies have characterized such genetic elements in E. coli strains derived from host populations carrying both antibiotic resistance and metal resistance genes (Wyrsch et al., 2019; Zingali et al., 2020), these elements may also be present within the wastewater environment. Reflecting this, in an analysis of the plasmid metagenome from wastewater treatment plants, Li et al. (2015) identified a broad spectrum of both antibiotic and metal resistance genes. While several plasmids harboring antibiotic resistance genes or metal resistance genes were characterized, one plasmid was found to harbor both kinds of resistance genes concurrently. Alternatively, cross-resistance can occur through the broad-spectrum activity of resistance mechanisms conferring protection against multiple hazardous substrates at a time. Multi-drug efflux pumps, for instance, appear to mediate the efflux of toxic compounds in general, which includes antibiotics but can also apply to biocides and heavy metals (Blanco et al., 2016). As such, the harsh conditions of the wastewater matrix may be particularly conducive for the selection of antibiotic resistance in the wastewater microbiome.
The treatment processes employed during wastewater treatment may also select for antibiotic resistance, particularly in those microbial populations that are adept at surviving or even growing in this matrix. For instance, UV irradiation has been shown to activate the SOS response (Aksenov, 1999), which can promote the mobilization of resistance genes throughout the surviving population. Furthermore, chlorine disinfection has been shown to promote the transfer of resistance genes across microbial genera by enhancing the natural competency (i.e., uptake of naked DNA) of cells for mobile genetic elements that can transform the surviving population (Jin et al., 2020). As such, the wastewater treatment plant appears to be a particularly conducive environment for the selection of antibiotic resistance. Importantly, although studies have yet to identify distinct selection forces operating in wastewater treatment plants (Bengtsson-Palme et al., 2016), the selection of antibiotic resistance genes during treatment has been observed (Liu et al., 2018; Pazda et al., 2020), reinforcing the idea that wastewater treatment plants represent important hotspots for the evolution and selection of antibiotic resistance.
Characterization of the resistomes found in wastewater treatment plants suggest that naturalized microbes adapted to wastewater matrices may represent an important source of resistance genes contributing to the dissemination of antibiotic resistance in the microbial world. Unfortunately, the majority of the research has focused on clinical microbes in wastewater, even though clinical subpopulations are only transiently present in wastewater matrices due to their removal during the course of water treatment (Lu et al., 2015). In contrast, naturalized wastewater microbes may represent a persistent source of resistance genes driving the evolution of antibiotic resistance within the wastewater treatment plant.
A series of studies conducted by Zhi et al. (2016a, 2017, 2019) have characterized novel clonal strains of E. coli that appear to have evolved to survive and replicate in wastewater and sewage environments as their primary niche. These findings point to an interesting prospect concerning the life history of naturalized microbes and their role in the evolution of antibiotic resistance. Typically, natural microbial populations are conventionally understood as “ancient” microbial lineages that predate the existence of human or animal hosts; however, the characterization of naturalized wastewater E. coli suggests that host-derived bacterial populations may also evolve toward the adaptation and colonization of newly engineered environments. Notably, naturalized wastewater strains of E. coli have been primarily isolated from wastewater treatment plants and appear to be globally distributed (Zhi et al., 2016a, 2019). Considering that large-scale engineered wastewater treatment systems are a relatively new technology for sanitary management (becoming widespread only in the last century), it can be argued these naturalized wastewater strains represent a recently emerged subpopulation of E. coli that evolved to exploit a new engineered environment for its growth and survival.
Genetically and phenotypically, these naturalized E. coli strains were found to be distinct from both commensal and pathogenic intestinal (enteric) E. coli and the cryptic Escherichia lineages (Zhi et al., 2017, 2019; Wang et al., 2020). These strains were originally isolated from sewage samples treated with chlorine at doses sufficient to inactivate 99.99% of the E. coli population in the sample (4 log10 reduction), and therefore represented the chlorine resistant population found in sewage. Using a novel bioinformatic, logic regression-based approach to search for unique DNA sequence biomarkers in intergenic regions that define host/niche specificity (Zhi et al., 2015, 2016b), Zhi et al. (2016a) identified naturalized wastewater strains harboring a unique single-nucleotide polymorphic (SNP) biomarker in the csgBAC-csgDEFG and asnS-ompF intergenic loci, which was not found in 780 strains of enteric E. coli from humans and animals or the cryptic Escherichia clades (Zhi et al., 2016a). Of the chlorine tolerant population collected from treated sewage, 82% of all isolates recovered possessed this intergenic SNP biomarker specific for wastewater. Furthermore, 75% of these intergenic SNP biomarker positive strains were also found to harbor a unique insertion element, IS30, located specifically within the uspC–flhDC intergenic locus (GenBank accession number: ON075843). Subsequent whole genome analysis revealed that many of the isolates appeared to be clonal, even though they were isolated from different treatment plants in Canada (Zhi et al., 2019), and clonal-relatedness even extended to wastewater strains found in the United States and Switzerland (Zhi et al., 2019). PCR assays targeting the uspC–IS30–flhDC marker could easily distinguish these naturalized strains from their enteric counterparts (Zhi et al., 2016a). Despite this, the naturalized strains were found to: (a) be biochemically similar to enteric E. coli according to phenotypic biochemical analyses (i.e., Vitek 2® Automated Bacterial Analysis); (b) display a typical growth morphology on selective media commonly used to identify E. coli in clinical and environmental laboratories; and (c) phylogenomically group with enteric E. coli strains, away from the cryptic Escherichia clades when Escherichia albertii was used as the outgroup (Zhi et al., 2016a, 2019). Interestingly, while many E. coli strains found in water environments are classically associated with phylogroup B1 (Berthe et al., 2013; Touchon et al., 2020; Nowicki et al., 2021), the naturalized wastewater strains formed a separate cluster within phylogroup A within two sequence types, ST399 and ST635 (Zhi et al., 2019), suggesting that these strains may have undergone an evolutionary divergence from enteric strains of E. coli.
Up until very recently, and across the vast publicly-available genome database on E. coli, the uspC-IS-30-flhDC marker was only ever observed in E. coli strains isolated from wastewater treatment plants. However, Yang et al. (2021) recently identified E. coli strains carrying this biomarker in naturalized E. coli strains found in meat processing plants in Canada. This finding raises the prospect that these strains may have evolved to become naturalized populations in engineered environments, particularly those environments that employ disinfection processes.
Importantly, naturalized wastewater strains are characterized by several remarkable phenotypic adaptations for surviving the wastewater treatment/disinfection processes. In comparison to enteric strains, naturalized wastewater E. coli were found to be phenotypically resistant to various physiochemical water treatment processes, displaying 100,000-fold greater resistance to chlorination, an enhanced capacity for biofilm formation, greater resistance to advanced oxidants, and an extreme heat-resistant phenotype (Zhi et al., 2016a, 2017; Wang et al., 2020). The degree of heat resistance observed in these naturalized wastewater strains was far greater than what had been described for other heat-resistant strains of E. coli, designating them “extremely” heat resistant (Wang et al., 2020). This extreme heat resistance phenotype was linked to a genomic island known as the locus of heat resistance (LHR; Wang et al., 2020). Since heat is often used as a disinfection strategy in meat packing plants and for sewage treatment (i.e., composting of biosolids), maintenance of the LHR could be an adaptive solution for these naturalized strains to survive these environments. In addition, the LHR was also found to concurrently confer resistance to chlorine and advanced oxidants (Wang et al., 2020), processes that are common in water treatment. In turn, resistance to chlorine and oxidants recapitulates the fact that chlorine-based disinfectants are often used for sanitizing surfaces in meat packing plants. Interestingly, the LHR is flanked by transposons and has been found in other strains of E. coli (Mercer et al., 2015, 2017), suggesting that microbes have evolved diverse strategies to deal with multiple stressors present in a given environment.
The genomes of naturalized wastewater E. coli strains, as well as those found in meat packing plants, were also shown to be enriched for various stress-resistance genes compared to their enteric counterparts, likely enhancing their survival against treatment processes (Zhi et al., 2019; Yang et al., 2021). Interestingly, a significant number of stress/repair genes appear to have been acquired through horizontal gene transfer and are not commonly found in enteric E. coli strains (Zhi et al., 2019). Moreover, the genomes of these naturalized strains contain an over-abundance of stress-resistance genes (and environmental adaptation genes) compared to enteric strains (Zhi et al., 2019), including those associated with:
a. DNA repair systems linked to damage caused by UV irradiation (i.e., a common wastewater treatment practice);
b. type I and III restriction-modification systems, which cleave foreign DNA and are likely critical for survival in a sewage/wastewater environment that is abundant in diverse bacteriophages and conjugative plasmids (Dupuis et al., 2013);
c. various oxidoreductases/glutathione transferase systems associated with resistance to oxidative stress, and implicated in resistance against UV, chlorine, and hydrogen peroxide (Patridge and Ferry, 2006; Allocati et al., 2009; Khemiri et al., 2014; Lemire et al., 2017);
d. toxin-antitoxin systems that modulate nutrient starvation and intra-species competition in population-dense environments with limited resources (Christensen et al., 2001, 2003);
e. heteromeric transposase elements known to be important for environmental adaptation by promoting horizontal gene transfer and integration of genomic islands among bacteria (Peters et al., 2014); and
f. heavy metal resistance genes that may promote survival in response to the high concentrations of the many heavy metals present in wastewater matrices (Östman et al., 2017; Feng et al., 2018).
Interestingly, functional genomics revealed that, compared to enteric E. coli, these naturalized wastewater strains simultaneously lacked various genes implicated in host colonization processes (i.e., adhesin synthesis genes) and pathogenesis (i.e., type IV secretion system genes; Zhi et al., 2019; Yang et al., 2021). Collectively, the genetic, phylogenomic, phenotypic, and ecotypic evidence suggest that naturalized wastewater-specific E. coli strains appear to have evolved to exploit wastewater/sewage treatment plants (and potentially meat packing plants) as their primary niche for growth and survival (Figure 2A), and concomitantly abandoned life in the gut of humans and animals. Indeed, their evolution from a host-colonizing to naturalized population is supported by phylogenomic analyses that demonstrate their evolutionary relationship to, and divergence from, enteric E. coli strains within phylogroup A (Zhi et al., 2019).
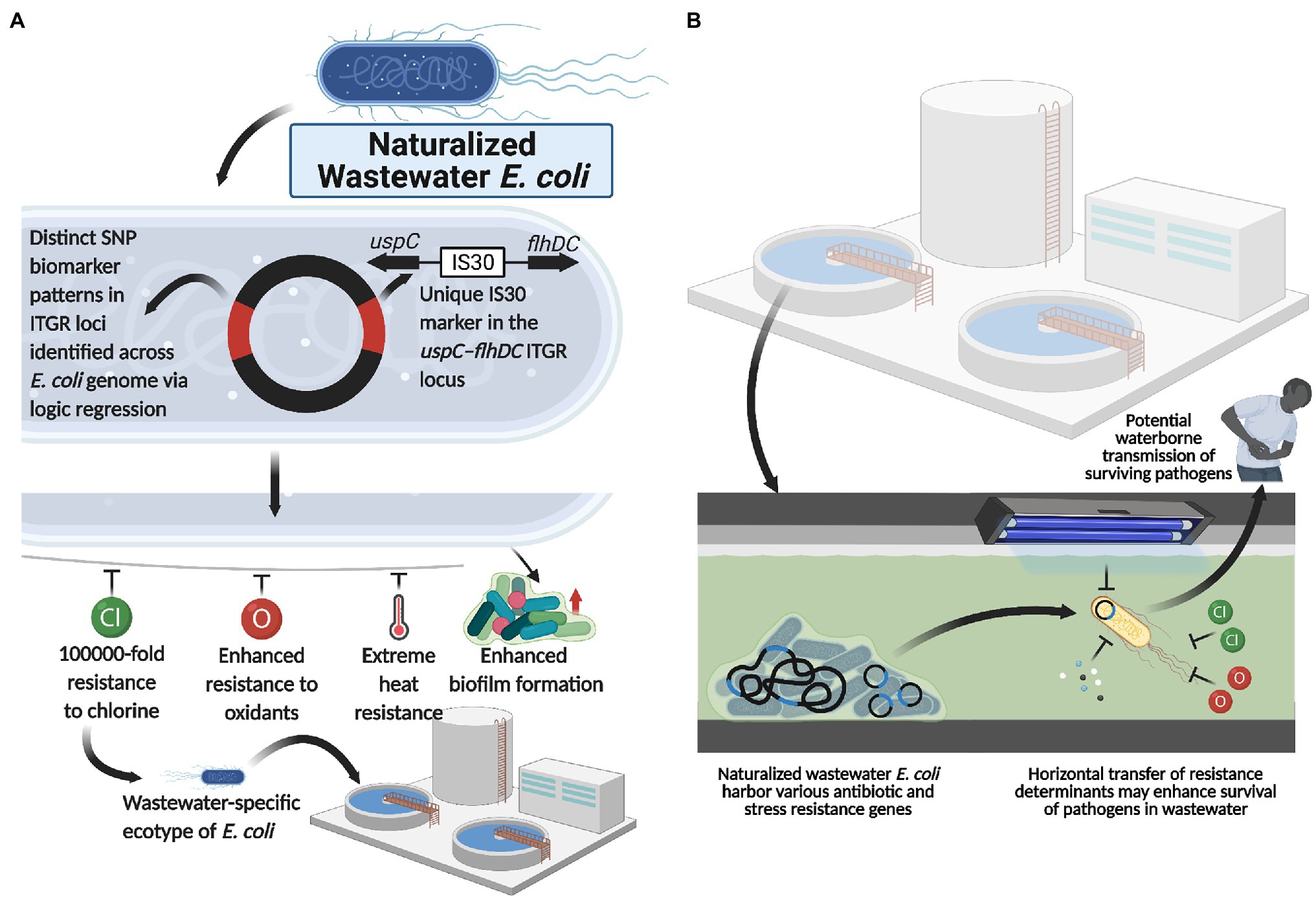
Figure 2. Naturalized wastewater E. coli as a reservoir for antibiotic and water treatment resistance. (A) Naturalized wastewater E. coli strains harboring unique SNP ITGR biomarkers and the uspC–IS30–flhDC genetic marker are characterized by various phenotypic adaptations, including extreme heat, chlorine and oxidant resistance as well as enhanced biofilm formation capacity, which appear to underlie their recent naturalization into wastewater matrices. In line with this, these naturalized strains harbor an abundance of antibiotic and stress resistance genes that may be mobilized and acquired by other microbes in the wastewater matrix, including pathogenic strains. Importantly, this suggests that (B) naturalized wastewater microbes may serve as reservoirs for both antibiotic resistance and water treatment resistance, enhancing the survival and transmission of pathogens present in wastewater. Created with BioRender.com.
Interestingly, water treatment resistant naturalized E. coli also possessed a plethora of antibiotic resistance (AR) genes and for which a common backbone of 47 AR genes were shared across the genomes of all naturalized strains isolated from wastewater treatment plants in Canada, Switzerland, and the United States (Table 1). The global distribution of a common backbone of AR genes in naturalized strains suggests that these AR-genes may play an important and adaptive role in their survival in the wastewater matrix, possibly during secondary treatment (i.e., a stage of intense microbial competition, predation, etc.) or in response to contaminants such as heavy metals or other noxious compounds. Notably, the vast majority of this shared repertoire of AR genes include multi-drug efflux pumps (Table 1), which presumably mediate broad-spectrum activity against a plethora of hazardous substrates that may be present in wastewater (Blanco et al., 2016). Interestingly, despite possessing a plethora of antibiotic resistance genes, the strains themselves were shown to be susceptible to most antibiotics when tested at clinical breakpoints (Zhi et al., 2019). As such, the specific roles that these resistance genes play in the life history and survival of the naturalized wastewater strains remains to be determined, but could involve endogenous resistance to bioactive molecules from other microbes, hormesis, quorum sensing, and/or intercellular signaling (as discussed in the previous section of this review). Importantly, several studies have revealed a possible link between antibiotic resistance and water treatment resistance. For instance, Huang et al. (2013) demonstrated a phenotypic relationship between chlorine resistance and tetracycline resistance, suggesting that chlorine treatment may promote the selection of E. coli isolates highly resistant to tetracycline. Additionally, Liu et al. (2018), demonstrated that the use of chlorine dioxide during water treatment resulted in the selection of macrolide (ermB), tetracycline (tetA, tetB, and tetC), sulfonamide (sul1, sul2, and sul3), β-lactam (ampC), aminoglycoside [aph(2′)-Id], rifampicin (katG), and vancomycin (vanA) resistance genes in treated wastewater effluent samples. Interestingly, three of these genes, including ermB, ampC, and sul1, were harbored by the naturalized wastewater E. coli strains described by Zhi et al. (2019). Collectively, these data suggest that AR genes in naturalized strains may enhance survival against specific wastewater treatment processes, which can lead to their enrichment across the treatment process (Laht et al., 2014; Mao et al., 2015; Bengtsson-Palme et al., 2016). Alternatively, as these strains appear to be adapted to wastewater matrices, their endogenous growth can lead to the amplification of harbored AR genes within the wastewater matrix—such that these strains act as a persistent reservoir of genetic elements that may simultaneously confer antibiotic resistance and promote survival within wastewater (Figure 2B).
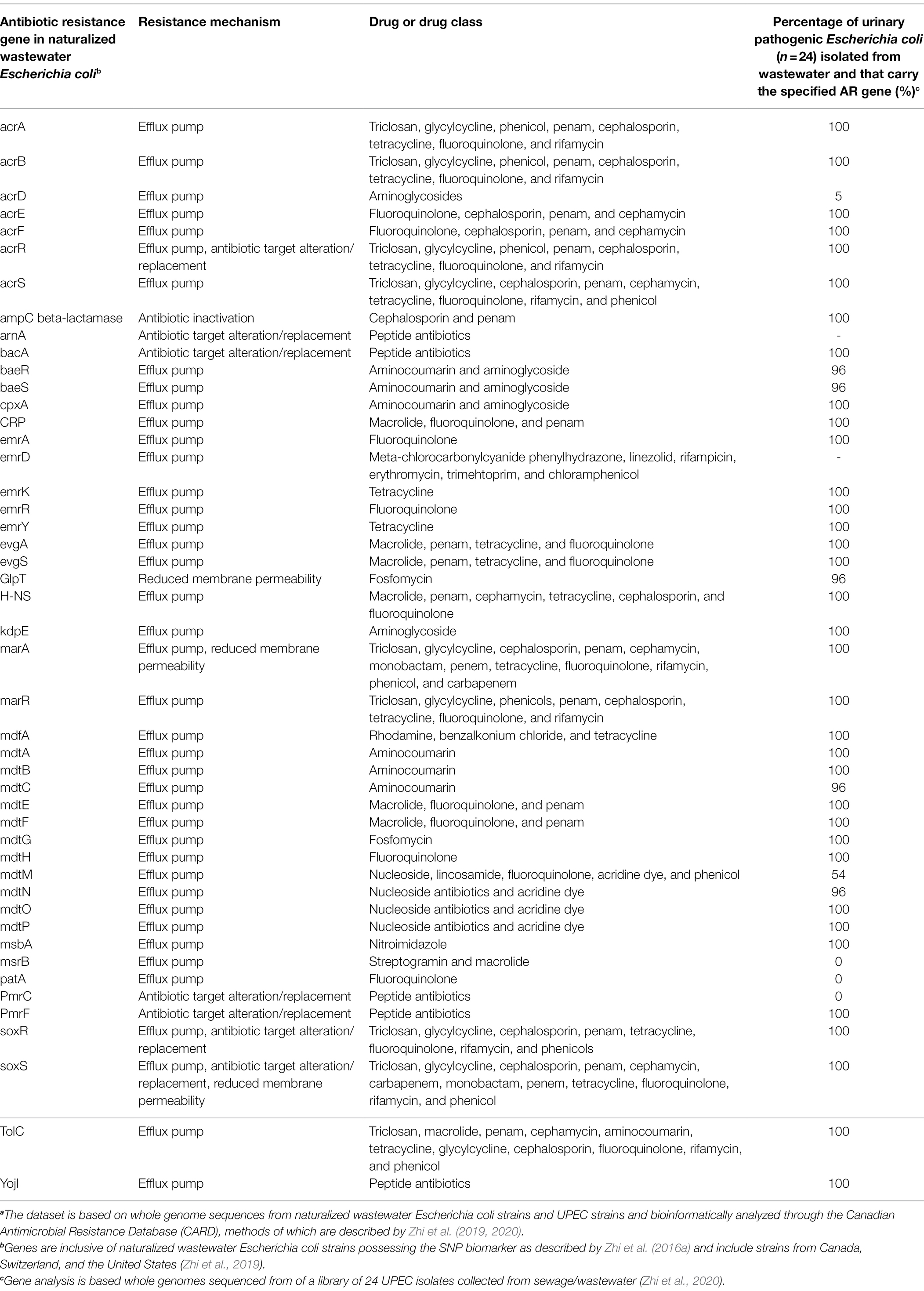
Table 1. A list of the common suite of antibiotic resistance genes found in naturalized wastewater Escherichia coli strains (based on Zhi et al., 2019) implicating their importance in promoting survival and resistance to wastewater treatment and a comparison of the carriage of these genes in water treatment resistant extra-intestinal pathogenic Escherichia coli isolated from wastewater (based on Zhi et al., 2020).a
Beyond their intrinsic resistance genes, recent data also suggests that naturalized wastewater E. coli may be permissive and competent at acquiring antibiotic resistance genes from clinical strains. For example, an E. coli isolate collected from a hospital wastewater system in Switzerland by Zurfluh et al. (2017a, b), which was subsequently shown to be a naturalized wastewater strain by Zhi et al. (2019), possessed various carbapenem resistance genes, including a plasmid-encoded 16S rRNA methyltransferase gene (rmtB) and the β-lactamase blaTEM-1b—common constituents in extended spectrum β-lactamase (ESBL) clinical isolates of E. coli. Interestingly, naturalized wastewater strains collected in other studies lacked these particular genes (Zhi et al., 2019); however, strains from Zhi et al. (2019) were isolated from municipal sewage, whereas Zurfluh et al. (2017b) isolated their naturalized E. coli strain directly from a hospital sewage/wastewater system. Considering that antibiotic residues in untreated hospital wastewater are much greater than those typically seen in municipal wastewater (Karthikeyan and Meyer, 2006), heightened exposure to antibiotics in hospital wastewater may provide the necessary selective pressure for the retention of some AR genes (i.e., carbapenem resistance genes and beta-lactams) in naturalized wastewater E. coli strains (Zurfluh et al., 2017b). As such, there is evidence to suggest that naturalized wastewater E. coli strains may be amenable to the acquisition and transfer of resistance genes, including the possibility of acquiring conjugative plasmids from enteric strains and/or the uptake of free AR genes from the wastewater matrix (i.e., transformation).
The abundance of AR genes and the resistance of naturalized wastewater E. coli strains to water treatment is cause for concern for several reasons. Firstly, although naturalized wastewater E. coli isolates may not be a direct threat to humans (i.e., not capable of infecting humans given their specificity for wastewater), they represent an important and persistent reservoir of AR genes and water treatment resistance genes. Secondly, sewage/wastewater may act as an amplification matrix for AR and water treatment resistance genes through the endogenous growth and replication of these naturalized strains within these matrices. Thirdly, there exists the possibility of horizontal gene transfer of AR genes and possibly water treatment resistance genes from naturalized wastewater microbes to environmental microbes, a phenomenon that has been previously demonstrated (Marcinek et al., 1998; Karkman et al., 2018). Fourthly, and most importantly, since naturalized wastewater E. coli appear to display some resistance to water treatment, and for which resistance might be related to AR-genes or other transmissible elements (e.g., the LHR), naturalized strains represent an important genetic reservoir for the potential emergence of pathogenic E. coli resistant to both water treatment and antibiotics—a concerning prospect considering that water treatment and sanitation represent the greatest human achievements for the control of infectious disease in modern society.
A growing body of evidence also points to the possibility that naturalized wastewater E. coli strains may be playing an important role in the emergence of certain pathotypes of E. coli in wastewater. Reflecting this, several studies have demonstrated that extraintestinal pathogenic subpopulations of E. coli appear to be differentially capable of surviving wastewater treatment (Table 2). For instance, Anastasi et al. (2013), observed that various strains of pathogenic E. coli differentially survive wastewater treatment processes, and notably, the surviving population was predominated by ExPEC pathotypes, in particular, the urinary pathogenic E. coli (UPEC). In another study, Anastasi et al. (2010) observed that almost 60% of isolates surviving wastewater treatment possessed one or more virulence genes associated with the UPEC pathotype (papA/H, papE/F, papC, hlyA, cnf1, and iroN). Similarly, using a virulence gene profiling approach, Adefisoye and Okoh (2016) observed that >41.7% of E. coli isolates from treated wastewater effluents represented potential UPEC strains. Likewise, Calhau et al. (2015), identified UPEC in wastewater effluents based on the presence of UPEC-associated virulence genes (i.e., iutA, papA/H, and sfa) and pathogenicity islands (i.e., PAI IV536, PAI ICFT073, PAI IICFT073), leading to the observation that UPEC strains dominated the E. coli population in hospital treated wastewaters, and included extended spectrum beta-lactamase (ESBL) producing strains belonging to the pandemic-associated sequence type (ST) 131 lineage.
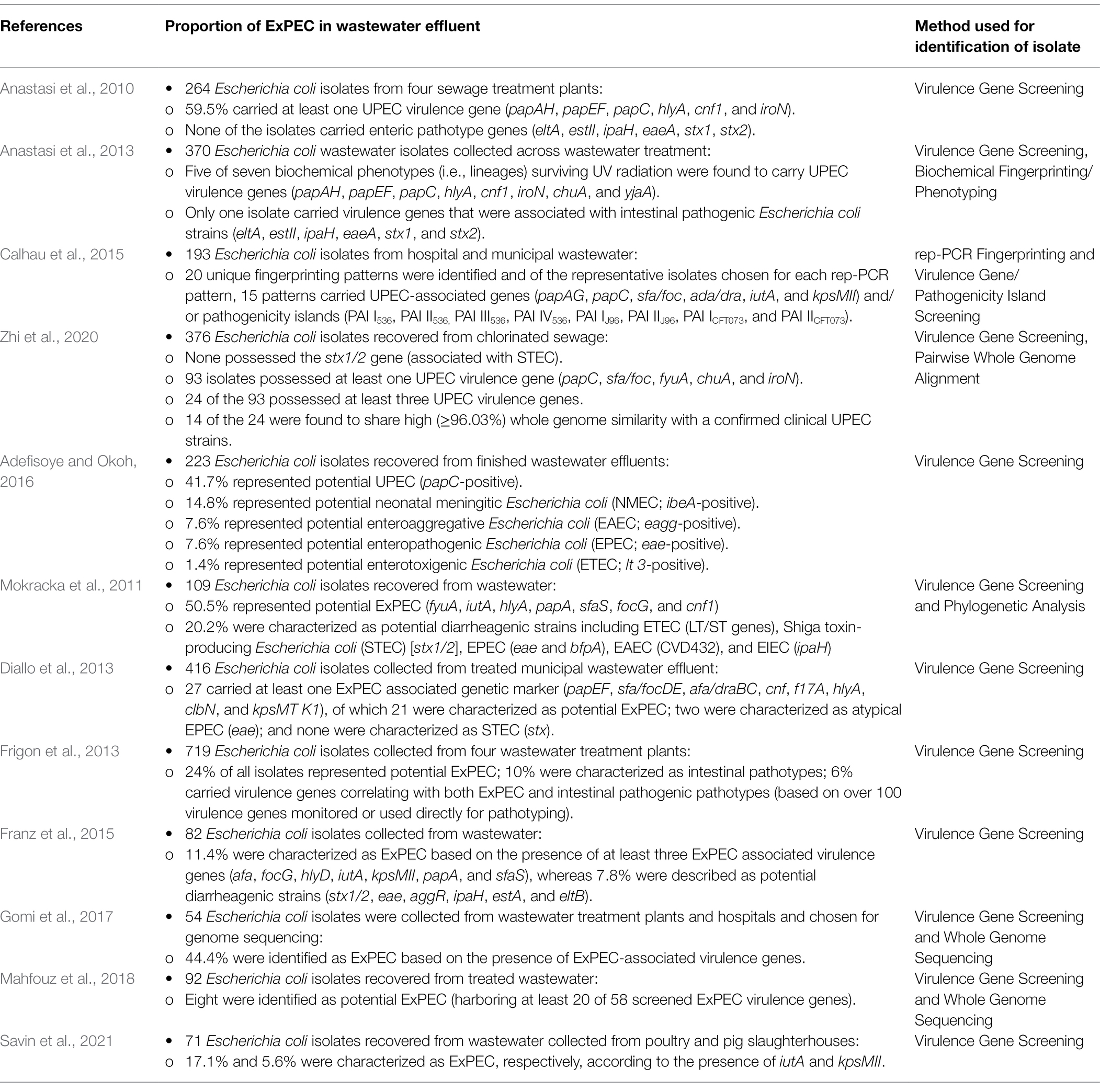
Table 2. Summary of studies evaluating the prevalence of extraintestinal pathogenic Escherichia coli (ExPEC) in treated wastewater.
In line with these findings, a comprehensive genomic analysis performed by Zhi et al. (2020) identified UPEC strains as common constituents within treated sewage and wastewater effluents, and E. coli strains recovered from wastewater matrices were found to cluster within several clinically relevant sequence types including ST131, ST95, ST127, and ST640. Several wastewater strains were found to be virtually identical to clinical UPEC strains across the whole genomes (i.e., as high as 99.49% genome similarity), and many carried the exact same complement of virulence genes as their clinical counterparts. Concerningly, the wastewater-derived UPEC isolates also carried a plethora of antibiotic resistance genes, and several isolates were identified as belonging to the pandemic O25b-ST131 clonal group representing potential ESBL-producing strains. The remarkable similarity between the wastewater isolates and clinical UPEC strains, coupled with the findings of others, led the authors to conclude that UPEC strains possess a differential capacity to survive full scale wastewater treatment (Zhi et al., 2020). While these findings do not discount the survival of intestinal commensal and pathogenic E. coli during water treatment (Omar and Barnard, 2014), the evidence seems to indicate that ExPEC strains are disproportionately represented in the surviving population of E. coli in wastewater (Table 2), and which has led some authors to suggest that “higher-than-expected” levels of ExPEC infection may exist in the community or that ExPEC strains naturally occur in wastewater (Paulshus et al., 2019).
Interestingly, several lines of evidence point toward a close relationship between naturalized wastewater E. coli strains and emerging water treatment-resistant ExPEC populations. As previously outlined, all naturalized wastewater E. coli strains share a common complement of antibiotic resistant genes (Table 1), potentially implying their importance in mediating survival in a wastewater matrix (Zhi et al., 2019). Remarkably, UPEC strains also possess many of these antibiotic resistance genes (Table 1), suggesting some common mechanisms that may underlie the differential survival of UPEC and naturalized strains during wastewater treatment. Although naturalized wastewater E. coli strains appear to lack certain genes associated with adhesion/colonization and pathogenesis in the gut, they possessed upward of 36 different UPEC-related virulence genes, including: (a) the ferrienterobactin multi-enzyme sythetase complex and transport system; (b) various UPEC fimbrial genes; (c) the curli virulence gene (csgB); (d) the ompA outer membrane protein gene; and (e) the flagellar biosynthesis protein encoded by flhA (Zhi et al., 2019). Some naturalized strains also possessed the yersiniabactin siderophore biosynthetic pathway and receptor/transport system found in many clinical UPEC, and some strains also possessed the UPEC virulence genes fyuA and irp1/2 (Zhi et al., 2019). It is interesting to note that the virulence genes highlighted above have been identified as being important in the survival of UPEC in the extraintestinal environment, particularly in the nutrient-deprived and osmotically-stressed environment of the urinary tract characterized by low solute concentrations and repetitive flushing (Mann et al., 2017). Many of the shared virulence genes between UPEC and naturalized strains are iron-sequestering genes (e.g., ferribactin, yersiniabactin, and enterobactin-related genes and transport pathways), and known to be important for UPEC’s survival in iron-deficient urine (Terlizzi et al., 2017). Considering that wastewater is similarly characterized by a limited availability of iron at dilute concentrations, these genes may play a similar role for the naturalized strains in the wastewater environment. Notably, many of these so-called UPEC-related virulence genes were also found in the genomes of environmental cryptic clades of Escherichia (Zhi et al., 2019), providing further support for the idea that the primary function of these genes may be for survival in environmental niches (i.e., urine, water, sewage, and wastewater). It is also interesting to note that naturalized wastewater strains possess the UPEC-associated virulence gene irp, and this gene has been linked to protection against protozoan grazing and predation (Adiba et al., 2010). Consequently, these so-called “UPEC virulence genes” may actually represent environmental adaptation genes necessary for the survival of E. coli outside of the intestinal environment.
There is also growing speculation that water may be an important, and underestimated, vehicle of transmission for ExPEC (Graham et al., 2021). Indeed, urinary tract infections, the vast majority of which are caused by UPEC, have been epidemiologically-linked to exposures associated with contaminated recreational water (Søraas et al., 2013). Similarly, in a study by Tanner et al. (2019), ESBL-producing Enterobacteriaceae, including E. coli, were found in 6.4% of treated drinking water samples that failed bacteriological water quality parameters in the United States (i.e., total coliforms), leading the authors to conclude that treated drinking water may be an underestimated vehicle for transmission of ExPEC in the community.
Other than the indirect evidence provided above that sheds some light on the phenotypic (i.e., ability to survive wastewater treatment) and genotypic relationship (i.e., shared AR and virulence genes) between naturalized wastewater strains of E. coli and ExPEC, there is no definitive evidence (as of yet) that clearly demonstrates genetic compatibility between naturalized E. coli and ExPEC strains and the transfer of AR or water treatment-resistant genes between these populations. Nevertheless, and as demonstrated throughout this paper, wastewater is known to be an environmental “hotspot” for AR bacteria, AR genes and the selection forces that drive evolutionary emergence of antibiotic resistance. The story of naturalized wastewater E. coli and the potential emergence of water treatment and antibiotic resistance in pathogenic populations is concerning, especially considering that E. coli is only one species present in a vast wastewater microbiome. Indeed, the very same natural selection forces could be driving the naturalization of other microbial populations in wastewater and subsequent evolution of water treatment resistance in other pathogenic populations, such as the ESKAPE pathogens. Clearly, the impact to public health could be substantial and more research is needed.
The widespread prevalence of antibiotic resistance in the microbial world represents one of the most critical problems facing public health today; however, the genetic determinants underlying the current resistance crisis appear to have their origins in ancient, naturalized microbes, indicating that naturalized microbial populations have played key roles in the origin, dissemination, and evolution of antibiotic resistance. The recent discovery of naturalized wastewater-specific strains of E. coli bring to light an even more ominous specter that antibiotic resistance could be linked to water treatment resistance. Importantly, water treatment represents the single greatest public health intervention for control of infectious diseases in modern society, and the idea that microbes may be breaching this barrier, is very concerning, and more research investment and policy action is needed in this area.
DY and NN wrote the bulk of the manuscript with scientific and editorial insights from SO, SZ, and KR. DY, SZ, and KR generated research data regarding naturalized wastewater E. coli. SO and NN provided the funding support. All authors contributed to the article and approved the submitted version.
Funding was provided by a grant to SO and NN from the Albert Ministry of Jobs, Economy, and Innovation (Major Innovation Fund) for the Antimicrobial Resistance – One Health Consortium, the Canadian Institutes for Health Research (CIHR; to NN), the Natural Sciences and Engineering Research Council (NSERC; to NN), Alberta Innovates (to NN and SO), National Natural Sciences Foundation of China (grant number: 82073514), and Ningbo Natural Sciences Foundation (grant number: 202003N4114).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adefisoye, M. A., and Okoh, A. I. (2016). Identification and antimicrobial resistance prevalence of pathogenic Escherichia coli strains from treated wastewater effluents in eastern cape, South Africa. Microbiology 5, 143–151. doi: 10.1002/mbo3.319
Adiba, S., Nizak, C., van Baalen, M., Denamur, E., and Depaulis, F. (2010). From grazing resistance to pathogenesis: The coincidental evolution of virulence factors. PLoS One 5, 1–10. doi: 10.1371/journal.pone.0011882
Aksenov, S. V. (1999). Induction of the SOS response in ultraviolet-irradiated Escherichia coli analyzed by dynamics of LexA, RecA and SulA proteins. J. Biol. Phys. 25, 263–277. doi: 10.1023/a:1005163310168
Allen, H. K., Donato, J., Wang, H. H., Cloud-Hansen, K. A., Davies, J., and Handelsman, J. (2010). Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 8, 251–259. doi: 10.1038/nrmicro2312
Allocati, N., Federici, L., Masulli, M., and Di Ilio, C. (2009). Glutathione transferases in bacteria. FEBS J. 276, 58–75. doi: 10.1111/j.1742-4658.2008.06743.x
Amador, P. P., Fernandes, R. M., Prudêncio, M. C., Barreto, M. P., and Duarte, I. M. (2015). Antibiotic resistance in wastewater: occurrence and fate of Enterobacteriaceae producers of class A and class C β-lactamases. J. Environ. Sci. Heal. - Part A Toxic/Hazardous Subst. Environ. Eng. 50, 26–39. doi: 10.1080/10934529.2015.964602
Aminov, R. I. (2010). A brief history of the antibiotic era: lessons learned and challenges for the future. Front. Microbiol. 1, 1–7. doi: 10.3389/fmicb.2010.00134
Anastasi, E. M., Matthews, B., Gundogdu, A., Vollmerhausen, T. L., Ramos, N. L., Stratton, H., et al. (2010). Prevalence and persistence of Escherichia coli strains with uropathogenic virulence characteristics in sewage treatment plants. Appl. Environ. Microbiol. 76, 5882–5886. doi: 10.1128/AEM.00141-10
Anastasi, E. M., Wohlsen, T. D., Stratton, H. M., and Katouli, M. (2013). Survival of Escherichia coli in two sewage treatment plants using UV irradiation and chlorination for disinfection. Water Res. 47, 6670–6679. doi: 10.1016/j.watres.2013.09.008
Andersson, D. I., and Hughes, D. (2012). Evolution of antibiotic resistance at non-lethal drug concentrations. Drug Resist. Updat. 15, 162–172. doi: 10.1016/j.drup.2012.03.005
Andersson, D. I., and Hughes, D. (2014). Microbiological effects of sublethal levels of antibiotics. Nat. Rev. Microbiol. 12, 465–478. doi: 10.1038/nrmicro3270
Babakhani, S., and Oloomi, M. (2018). Transposons: the agents of antibiotic resistance in bacteria. J. Basic Microbiol. 58, 905–917. doi: 10.1002/jobm.201800204
Baharoglu, Z., and Mazel, D. (2011). Vibrio cholerae triggers SOS and mutagenesis in response to a wide range of antibiotics: a route towards multiresistance. Antimicrob. Agents Chemother. 55, 2438–2441. doi: 10.1128/AAC.01549-10
Baquero, F., Martínez, J. L., and Cantón, R. (2008). Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol. 19, 260–265. doi: 10.1016/j.copbio.2008.05.006
Barancheshme, F., and Munir, M. (2018). Strategies to combat antibiotic resistance in the wastewater treatment plants. Front. Microbiol. 8:2603. doi: 10.3389/fmicb.2017.02603
Barlow, M., and Hall, B. G. (2002). Phylogenetic analysis shows that the OXA β-lactamase genes have been on plasmids for millions of years. J. Mol. Evol. 55, 314–321. doi: 10.1007/s00239-002-2328-y
Beaber, J. W., Hochhut, B., and Waldor, M. K. (2004). SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427, 72–74. doi: 10.1038/nature02241
Bengtsson-Palme, J., Hammarén, R., Pal, C., Östman, M., Björlenius, B., Flach, C. F., et al. (2016). Elucidating selection processes for antibiotic resistance in sewage treatment plants using metagenomics. Sci. Total Environ. 572, 697–712. doi: 10.1016/j.scitotenv.2016.06.228
Bengtsson-Palme, J., Kristiansson, E., and Larsson, D. G. J. (2018). Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiol. Rev. 42, 68–80. doi: 10.1093/femsre/fux053
Bengtsson-Palme, J., and Larsson, D. G. J. (2016). Concentrations of antibiotics predicted to select for resistant bacteria: proposed limits for environmental regulation. Environ. Int. 86, 140–149. doi: 10.1016/j.envint.2015.10.015
Berendonk, T. U., Manaia, C. M., Merlin, C., Fatta-Kassinos, D., Cytryn, E., Walsh, F., et al. (2015). Tackling antibiotic resistance: The environmental framework. Nat. Rev. Microbiol. 13, 310–317. doi: 10.1038/nrmicro3439
Berthe, T., Ratajczak, M., Clermont, O., Denamur, E., and Petit, F. (2013). Evidence for coexistence of distinct Escherichia coli populations in various aquatic environments and their survival in estuary water. Appl. Environ. Microbiol. 79, 4684–4693. doi: 10.1128/AEM.00698-13
Blanco, P., Hernando-Amado, S., Reales-Calderon, J. A., Corona, F., Lira, F., Alcalde-Rico, M., et al. (2016). Bacterial multidrug efflux pumps: much more than antibiotic resistance determinants. Microorganisms 4, 1–19. doi: 10.3390/microorganisms4010014
Breidenstein, E. B. M., Khaira, B. K., Wiegand, I., Overhage, J., and Hancock, R. E. W. (2008). Complex ciprofloxacin resistome revealed by screening a Pseudomonas aeruginosa mutant library for altered susceptibility. Antimicrob. Agents Chemother. 52, 4486–4491. doi: 10.1128/AAC.00222-08
Brown, K. D., Kulis, J., Thomson, B., Chapman, T. H., and Mawhinney, D. B. (2006). Occurrence of antibiotics in hospital, residential, and dairy effluent, municipal wastewater, and the Rio Grande in New Mexico. Sci. Total Environ. 366, 772–783. doi: 10.1016/j.scitotenv.2005.10.007
Cabello, F. C., Godfrey, H. P., Tomova, A., Ivanova, L., Dölz, H., Millanao, A., et al. (2013). Antimicrobial use in aquaculture re-examined: its relevance to antimicrobial resistance and to animal and human health. Environ. Microbiol. 15, 1917–1942. doi: 10.1111/1462-2920.12134
Calhau, V., Mendes, C., Pena, A., Mendonça, N., and Da Silva, G. J. (2015). Virulence and plasmidic resistance determinant of Escherichia coli isolated from municipal and hospital wastewater treatment plants. J. Water Health 13, 311–318. doi: 10.2166/wh.2014.327
Cantón, R., and Coque, T. M. (2006). The CTX-M β-lactamase pandemic. Curr. Opin. Microbiol. 9, 466–475. doi: 10.1016/j.mib.2006.08.011
Cantón, R., González-Alba, J. M., and Galán, J. C. (2012). CTX-M enzymes: origin and diffusion. Front. Microbiol. 3:110. doi: 10.3389/fmicb.2012.00110
Center for Disease Dynamics, Economics and Policy. (2015). State of the World’s Antibiotics, 2015. CDDEP. Available at: https://cddep.org/publications/state_worlds_antibiotics_2015/ (Accessed September 16, 2021).
Cha, J. M., Yang, S., and Carlson, K. H. (2006). Trace determination of β-lactam antibiotics in surface water and urban wastewater using liquid chromatography combined with electrospray tandem mass spectrometry. J. Chromatogr. A 1115, 46–57. doi: 10.1016/j.chroma.2006.02.086
Chandra, N., and Kumar, S. (2017). “Antibiotics producing soil microorganisms,” in Antibiotics and Antibiotics Resistance Genes in Soils, (eds.) M. Z. Hashmi, et al. Springer International Publishing AG, 1–18. doi: 10.1007/978-3-319-66260-2_1
Che, Y., Xia, Y., Liu, L., Li, A. D., Yang, Y., and Zhang, T. (2019). Mobile antibiotic resistome in wastewater treatment plants revealed by Nanopore metagenomic sequencing. Microbiome 7:44. doi: 10.1186/s40168-019-0663-0
Christensen, S. K., Mikkelsen, M., Pedersen, K., and Gerdes, K. (2001). RelE, a global inhibitor of translation, is activated during nutritional stress. Proc. Natl. Acad. Sci. U. S. A. 98, 14328–14333. doi: 10.1073/pnas.251327898
Christensen, S. K., Pedersen, K., Hansen, F. G., and Gerdes, K. (2003). Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol. Biol. 332, 809–819. doi: 10.1016/S0022-2836(03)00922-7
Currie, C., Scott, J., Summerbell, R., and Malloch, D. (1999). Fungus-growing ants use antibiotic-producing bacteria to control garden pests. Nature 398, 701–704. doi: 10.1038/19519
D’Costa, V. M., King, C. E., Kalan, L., Morar, M., Sung, W. W. L., Schwarz, C., et al. (2011). Antibiotic resistance is ancient. Nature 477, 457–461. doi: 10.1038/nature10388
D’Costa, V. M., McGrann, K. M., Hughes, D. W., and Wright, G. D. (2006). Sampling the antibiotic resistome. Science 311, 374–377. doi: 10.1126/science.1120800
Davies, J., and Davies, D. (2010). Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 74, 417–433. doi: 10.1128/mmbr.00016-10
Davies, J., Spiegelman, G. B., and Yim, G. (2006). The world of subinhibitory antibiotic concentrations. Curr. Opin. Microbiol. 9, 445–453. doi: 10.1016/j.mib.2006.08.006
Decousser, J. W., Poirel, L., and Nordmann, P. (2001). Characterization of a chromosomally encoded extended-spectrum class A β-lactamase from Kluyvera cryocrescens. Antimicrob. Agents Chemother. 45, 3595–3598. doi: 10.1128/AAC.45.12.3595-3598.2001
Devane, M. L., Moriarty, E., Weaver, L., Cookson, A., and Gilpin, B. (2020). Fecal indicator bacteria from environmental sources; strategies for identification to improve water quality monitoring. Water Res. 185:116204. doi: 10.1016/j.watres.2020.116204
Diallo, A. A., Brugère, H., Kérourédan, M., Dupouy, V., Toutain, P. L., Bousquet-Mélou, A., et al. (2013). Persistence and prevalence of pathogenic and extended-spectrum beta-lactamase-producing Escherichia coli in municipal wastewater treatment plant receiving slaughterhouse wastewater. Water Res. 47, 4719–4729. doi: 10.1016/j.watres.2013.04.047
Dubern, J. F., and Diggle, S. P. (2008). Quorum sensing by 2-alkyl-4-quinolones in Pseudomonas aeruginosa and other bacterial species. Mol. Biosyst. 4, 882–888. doi: 10.1039/b803796p
Duo, M., Hou, S., and Ren, D. (2008). Identifying Escherichia coli genes involved in intrinsic multidrug resistance. Appl. Microbiol. Biotechnol. 81, 731–741. doi: 10.1007/s00253-008-1709-6
Dupuis, M. È., Villion, M., Magadán, A. H., and Moineau, S. (2013). CRISPR-Cas and restriction-modification systems are compatible and increase phage resistance. Nat. Commun. 4, 1–7. doi: 10.1038/ncomms3087
Feng, J., Chen, X., Jia, L., Liu, Q., Chen, X., Han, D., et al. (2018). Effluent concentration and removal efficiency of nine heavy metals in secondary treatment plants in Shanghai, China. Environ. Sci. Pollut. Res. Int. 25, 17058–17065. doi: 10.1007/s11356-018-1915-4
Finley, R. L., Collignon, P., Larsson, D. G. J., Mcewen, S. A., Li, X. Z., Gaze, W. H., et al. (2013). The scourge of antibiotic resistance: the important role of the environment. Clin. Infect. Dis. 57, 704–710. doi: 10.1093/cid/cit355
Fleming, A. (1929). On the antibacterial action of cultures of a Penicillium with special reference to their use in the isolation of B. influenzae. Brit. J. Exp. Pathol. 10, 226–236.
Forsberg, K. J., Reyes, A., Wang, B., Selleck, E. M., Sommer, M. O. A., and Dantas, G. (2012). The shared antibiotic resistome of soil bacteria and human pathogens. Science 337, 1107–1111. doi: 10.1126/science.1220761
Fouz, N., Pangesti, K. N. A., Yasir, M., Al-Malki, A. L., Azhar, E. I., Hill-Cawthorne, G. A., et al. (2020). The contribution of wastewater to the transmission of antimicrobial resistance in the environment: implications of mass gathering settings. Trop. Med. Infect. Dis. 5:33. doi: 10.3390/tropicalmed5010033
Franz, E., Veenman, C., Van Hoek, A. H. A. M., Husman, A. D. R., and Blaak, H. (2015). Pathogenic Escherichia coli producing extended-spectrum β-lactamases isolated from surface water and wastewater. Sci. Rep. 5, 1–9. doi: 10.1038/srep14372
Frigon, D., Biswal, B. K., Mazza, M., Masson, L., and Gehr, R. (2013). Biological and physiochemical wastewater treatment processes reduce the prevalence of virulent Escherichia coli. Appl. Environ. Microbiol. 79, 835–844. doi: 10.1128/AEM.02789-12
Galler, H., Feierl, G., Petternel, C., Reinthaler, F. F., Haas, D., Habib, J., et al. (2018). Multiresistant bacteria isolated from activated sludge in Austria. Int. J. Environ. Res. Public Health 15:479. doi: 10.3390/ijerph15030479
Gao, Q., Dong, Q., Wu, L., Yang, Y., Hale, L., Qin, Z., et al. (2020). Environmental antibiotics drives the genetic functions of resistome dynamics. Environ. Int. 135:105398. doi: 10.1016/j.envint.2019.105398
Gomez, M. J., and Neyfakh, A. A. (2006). Genes involved in intrinsic antibiotic resistance of Acinetobacter baylyi. Antimicrob. Agents Chemother. 50, 3562–3567. doi: 10.1128/AAC.00579-06
Gomi, R., Matsuda, T., Matsumura, Y., Yamamoto, M., Tanaka, M., Ichiyama, S., et al. (2017). Occurrence of clinically important lineages, including the sequence type 131 C1-M27 subclone, among extended-spectrum-ß-lactamase-producing Escherichia coli in wastewater. Antimicrob. Agents Chemother. 61, 1–9. doi: 10.1128/AAC.00564-17
Gouliouris, T., Raven, K. E., Moradigaravand, D., Ludden, C., Coll, F., Blane, B., et al. (2019). Detection of vancomycin-resistant Enterococcus faecium hospital-adapted lineages in municipal wastewater treatment plants indicates widespread distribution and release into the environment. Genome Res. 29, 626–634. doi: 10.1101/gr.232629.117
Graham, J. P., Amato, H. K., Mendizabal-Cabrera, R., Alvarez, D., and Ramay, B. M. (2021). Waterborne urinary tract infections: have we overlooked an important source of exposure? Am. J. Trop. Med. Hyg. 105, 12–17. doi: 10.4269/ajtmh.20-1271
Grall, N., Barraud, O., Wieder, I., Hua, A., Perrier, M., Babosan, A., et al. (2015). Lack of dissemination of acquired resistance to b-lactams in small wild mammals around an isolated village in the Amazonian forest. Environ. Microbiol. Rep. 7, 698–708. doi: 10.1111/1758-2229.12289
Guerin, É., Cambray, G., Sanchez-Alberola, N., Campoy, S., Erill, I., Re, S. Da, et al. (2009). The SOS response controls integron recombination. Science 324, 1034. doi: 10.1126/science.1172914
Gullberg, E., Cao, S., Berg, O. G., Ilbäck, C., Sandegren, L., Hughes, D., et al. (2011). Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog. 7, e1002158–e1002159. doi: 10.1371/journal.ppat.1002158
Hall, B. G., and Barlow, M. (2004). Evolution of the serine β-lactamases: past, present and future. Drug Resist. Updat. 7, 111–123. doi: 10.1016/j.drup.2004.02.003
Hoelzer, K., Wong, N., Thomas, J., Talkington, K., Jungman, E., and Coukell, A. (2017). Antimicrobial drug use in food-producing animals and associated human health risks: what, and how strong, is the evidence? BMC Vet. Res. 13, 1–38. doi: 10.1186/s12917-017-1131-3
Holmes, A. H., Moore, L. S. P., Sundsfjord, A., Steinbakk, M., Regmi, S., Karkey, A., et al. (2016). Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 387, 176–187. doi: 10.1016/S0140-6736(15)00473-0
Huang, J. J., Hu, H. Y., Wu, Y. H., Wei, B., and Lu, Y. (2013). Effect of chlorination and ultraviolet disinfection on tetA-mediated tetracycline resistance of Escherichia coli. Chemosphere 90, 2247–2253. doi: 10.1016/j.chemosphere.2012.10.008
Huijbers, P. M. C., Blaak, H., De Jong, M. C. M., Graat, E. A. M., Vandenbroucke-Grauls, C. M. J. E., and De Roda Husman, A. M. (2015). Role of the environment in the transmission of antimicrobial resistance to humans: a review. Environ. Sci. Technol. 49, 11993–12004. doi: 10.1021/acs.est.5b02566
Humeniuk, C., Arlet, G., Gautier, V., Grimont, P., Labia, R., and Philippon, A. (2002). β-Lactamases of Kluyvera ascorbata, probable progenitors of some plasmid-encoded CTX-M types. Antimicrob. Agents Chemother. 46, 3045–3049. doi: 10.1128/AAC.46.9.3045-3049.2002
Hutchings, M., Truman, A., and Wilkinson, B. (2019). Antibiotics: past, present and future. Curr. Opin. Microbiol. 51, 72–80. doi: 10.1016/j.mib.2019.10.008
Iossa, G., and White, P. C. L. (2018). The natural environment: a critical missing link in national action plans on antimicrobial resistance. Bull. World Health Organ. 96, 858–860. doi: 10.2471/BLT.18.210898
Ishii, S., Ksoll, W. B., Hicks, R. E., and Sadowsky, M. J. (2006). Presence and growth of naturalized Escherichia coli in temperate soils from Lake Superior watersheds. Appl. Environ. Microbiol. 72, 612–621. doi: 10.1128/AEM.72.1.612-621.2006
Iweriebor, B. C., Gaqavu, S., Obi, L. C., Nwodo, U. U., and Okoh, A. I. (2015). Antibiotic susceptibilities of enterococcus species isolated from hospital and domestic wastewater effluents in alice, eastern cape province of South Africa. Int. J. Environ. Res. Public Health 12, 4231–4246. doi: 10.3390/ijerph120404231
Jang, J., Hur, H. G., Sadowsky, M. J., Byappanahalli, M. N., Yan, T., and Ishii, S. (2017). Environmental Escherichia coli: ecology and public health implications—a review. J. Appl. Microbiol. 123, 570–581. doi: 10.1111/jam.13468
Jernigan, J. A., Hatfield, K. M., Wolford, H., Nelson, R. E., Olubajo, B., Reddy, S. C., et al. (2020). Multidrug-resistant bacterial infections in U.S. hospitalized patients, 2012–2017. N. Engl. J. Med. 382, 1309–1319. doi: 10.1056/nejmoa1914433
Jin, M., Liu, L., Wang, D., Yang, D., Liu, W., Yin, J., et al. (2020). Chlorine disinfection promotes the exchange of antibiotic resistance genes across bacterial genera by natural transformation. ISME J. 14, 1847–1856. doi: 10.1038/s41396-020-0656-9
Ju, F., Beck, K., Yin, X., Maccagnan, A., McArdell, C. S., Singer, H. P., et al. (2019). Wastewater treatment plant resistomes are shaped by bacterial composition, genetic exchange, and upregulated expression in the effluent microbiomes. ISME J. 13, 346–360. doi: 10.1038/s41396-018-0277-8
Karkman, A., Do, T. T., Walsh, F., and Virta, M. P. J. (2018). Antibiotic-resistance genes in waste water. Trends Microbiol. 26, 220–228. doi: 10.1016/j.tim.2017.09.005
Karkman, A., Johnson, T. A., Lyra, C., Stedtfeld, R. D., Tamminen, M., Tiedje, J. M., et al. (2016). High-throughput quantification of antibiotic resistance genes from an urban wastewater treatment plant. FEMS Microbiol. Ecol. 92, 1–7. doi: 10.1093/femsec/fiw014
Karkman, A., Pärnänen, K., and Larsson, D. G. J. (2019). Fecal pollution can explain antibiotic resistance gene abundances in anthropogenically impacted environments. Nat. Commun. 10, 80–88. doi: 10.1038/s41467-018-07992-3
Karthikeyan, K. G., and Meyer, M. T. (2006). Occurrence of antibiotics in wastewater treatment facilities in Wisconsin, USA. Sci. Total Environ. 361, 196–207. doi: 10.1016/j.scitotenv.2005.06.030
Kaufmann, G. F., Sartorio, R., Lee, S. H., Rogers, C. J., Meijler, M. M., Moss, J. A., et al. (2005). Revisiting quorum sensing: discovery of additional chemical and biological functions for 3-oxo-N-acylhomoserine lactones. Proc. Natl. Acad. Sci. U. S. A. 102, 309–314. doi: 10.1073/pnas.0408639102
Khemiri, A., Carrière, M., Bremond, N., Ben Mlouka, M. A., Coquet, L., Llorens, I., et al. (2014). Escherichia coli response to uranyl exposure at low pH and associated protein regulations. PLoS One 9:e89863. doi: 10.1371/journal.pone.0089863
Kim, I., Yamashita, N., and Tanaka, H. (2009). Performance of UV and UV/H2O2 processes for the removal of pharmaceuticals detected in secondary effluent of a sewage treatment plant in Japan. J. Hazard. Mater. 166, 1134–1140. doi: 10.1016/j.jhazmat.2008.12.020
Kraemer, S. A., Ramachandran, A., and Perron, G. G. (2019). Antibiotic pollution in the environment: From microbial ecology to public policy. Microorganisms 7, 1–24. doi: 10.3390/microorganisms7060180
Kwon, J. H., and Powderly, W. G. (2021). The post-antibiotic era is here. Science 373:471. doi: 10.1126/science.abl5997
Laht, M., Karkman, A., Voolaid, V., Ritz, C., Tenson, T., Virta, M., et al. (2014). Abundances of tetracycline, sulphonamide and beta-lactam antibiotic resistance genes in conventional wastewater treatment plants (WWTPs) with different waste load. PLoS One 9, 1–8. doi: 10.1371/journal.pone.0103705
Larsson, D. G. J. (2014). Pollution from drug manufacturing: review and perspectives. Philos. Trans. R. Soc. B Biol. Sci. 369:20130571. doi: 10.1098/rstb.2013.0571
Larsson, D. G. J., de Pedro, C., and Paxeus, N. (2007). Effluent from drug manufactures contains extremely high levels of pharmaceuticals. J. Hazard. Mater. 148, 751–755. doi: 10.1016/j.jhazmat.2007.07.008
Laureti, L., Matic, I., and Gutierrez, A. (2013). Bacterial responses and genome instability induced by subinhibitory concentrations of antibiotics. Antibiotics 2, 100–114. doi: 10.3390/antibiotics2010100
Leisner, J. J., Jørgensen, N. O. G., and Middelboe, M. (2016). Predation and selection for antibiotic resistance in natural environments. Evol. Appl. 9, 427–434. doi: 10.1111/eva.12353
Lemire, J., Alhasawi, A., Appanna, V. P., Tharmalingam, S., and Appanna, V. D. (2017). Metabolic defence against oxidative stress: the road less travelled so far. J. Appl. Microbiol. 123, 798–809. doi: 10.1111/jam.13509
Li, L., Dechesne, A., He, Z., Madsen, J. S., Nesme, J., Sørensen, S. J., et al. (2018). Estimating the transfer range of plasmids encoding antimicrobial resistance in a wastewater treatment plant microbial community. Environ. Sci. Technol. Lett. 5, 260–265. doi: 10.1021/acs.estlett.8b00105
Li, A. D., Li, L. G., and Zhang, T. (2015). Exploring antibiotic resistance genes and metal resistance genes in plasmid metagenomes from wastewater treatment plants. Front. Microbiol. 6, 1–11. doi: 10.3389/fmicb.2015.01025
Li, B., and Zhang, T. (2011). Mass flows and removal of antibiotics in two municipal wastewater treatment plants. Chemosphere 83, 1284–1289. doi: 10.1016/j.chemosphere.2011.03.002
Li, B., Zhang, T., Xu, Z., and Fang, H. H. P. (2009). Rapid analysis of 21 antibiotics of multiple classes in municipal wastewater using ultra performance liquid chromatography-tandem mass spectrometry. Anal. Chim. Acta 645, 64–72. doi: 10.1016/j.aca.2009.04.042
Liu, S. S., Qu, H. M., Yang, D., Hu, H., Liu, W. L., Qiu, Z. G., et al. (2018). Chlorine disinfection increases both intracellular and extracellular antibiotic resistance genes in a full-scale wastewater treatment plant. Water Res. 136, 131–136. doi: 10.1016/j.watres.2018.02.036
López, E., and Blázquez, J. (2009). Effect of subinhibitory concentrations of antibiotics on intrachromosomal homologous recombination in Escherichia coli. Antimicrob. Agents Chemother. 53, 3411–3415. doi: 10.1128/AAC.00358-09
Lu, X., Zhang, X. X., Wang, Z., Huang, K., Wang, Y., Liang, W., et al. (2015). Bacterial pathogens and community composition in advanced sewage treatment systems revealed by metagenomics analysis based on high-throughput sequencing. PLoS One 10, 1–15. doi: 10.1371/journal.pone.0125549
Mahfouz, N., Caucci, S., Achatz, E., Semmler, T., Guenther, S., Berendonk, T. U., et al. (2018). High genomic diversity of multi-drug resistant wastewater Escherichia coli. Sci. Rep. 8, 8928–8912. doi: 10.1038/s41598-018-27292-6
Mann, R., Mediati, D. G., Duggin, I. G., Harry, E. J., and Bottomley, A. L. (2017). Metabolic adaptations of Uropathogenic E. coli in the urinary tract. Front. Cell. Infect. Microbiol. 7:241. doi: 10.3389/fcimb.2017.00241
Mao, D., Yu, S., Rysz, M., Luo, Y., Yang, F., Li, F., et al. (2015). Prevalence and proliferation of antibiotic resistance genes in two municipal wastewater treatment plants. Water Res. 85, 458–466. doi: 10.1016/j.watres.2015.09.010
Marcinek, H., Wirth, R., Muscholl-Silberhorn, A., and Gauer, M. (1998). Enterococcus faecalis gene transfer under natural conditions in municipal sewage water treatment plants. Appl. Environ. Microbiol. 64, 626–632. doi: 10.1128/aem.64.2.626-632.1998
Marshall, B. M., and Levy, S. B. (2011). Food animals and antimicrobials: impacts on human health. Clin. Microbiol. Rev. 24, 718–733. doi: 10.1128/CMR.00002-11
Martin, M. J., Thottathil, S. E., and Newman, T. B. (2015). Antibiotics overuse in animal agriculture: a call to action for health care providers. Am. J. Public Health 105, 2409–2410. doi: 10.2105/AJPH.2015.302870
Martinez, J. L. (2009). The role of natural environments in the evolution of resistance traits in pathogenic bacteria. Proc. R. Soc. B Biol. Sci. 276, 2521–2530. doi: 10.1098/rspb.2009.0320
McCubbin, K. D., Anholt, R. M., de Jong, E., Ida, J. A., Nóbrega, D. B., Kastelic, J. P., et al. (2021). Knowledge gaps in the understanding of antimicrobial resistance in Canada. Front. Public Health 9, 1–14. doi: 10.3389/fpubh.2021.726484
McKinney, C. W., and Pruden, A. (2012). Ultraviolet disinfection of antibiotic resistant bacteria and their antibiotic resistance genes in water and wastewater. Environ. Sci. Technol. 46, 13393–13400. doi: 10.1021/es303652q
Mercer, R., Nguyen, O., Ou, Q., McMullen, L., and Gänzle, M. G. (2017). Functional analysis of genes comprising the locus of heat resistance in Escherichia coli. Appl. Environ. Microbiol. 83, 1–13. doi: 10.1128/AEM.01400-17
Mercer, R. G., Zheng, J., Garcia-Hernandez, R., Ruan, L., Gänzle, M. G., and McMullen, L. M. (2015). Genetic determinants of heat resistance in Escherichia coli. Front. Microbiol. 6, 1–13. doi: 10.3389/fmicb.2015.00932
Mesak, L. R., Miao, V., and Davies, J. (2008). Effects of subinhibitory concentrations of antibiotics on SOS and DNA repair gene expression in Staphylococcus aureus. Antimicrob. Agents Chemother. 52, 3394–3397. doi: 10.1128/AAC.01599-07
Michael, I., Rizzo, L., McArdell, C. S., Manaia, C. M., Merlin, C., Schwartz, T., et al. (2013). Urban wastewater treatment plants as hotspots for the release of antibiotics in the environment: a review. Water Res. 47, 957–995. doi: 10.1016/j.watres.2012.11.027
Miller, C., Thomsen, L. E., Gaggero, C., Mosseri, R., Ingmer, H., and Cohen, S. N. (2004). SOS response induction by β-lactams and bacterial defense against antibiotic lethality. Science 305, 1629–1631. doi: 10.1126/science.1101630
Mindlin, S. Z., Soina, V. S., Ptrova, M. A., and Gorlenko, Z. M. (2008). Isolation of antibiotic resistance bacterial strains from East Siberia permafrost sediments. Genetika 44, 36–44.
Mokracka, J., Koczura, R., Jabłońska, L., and Kaznowski, A. (2011). Phylogenetic groups, virulence genes and quinolone resistance of integron-bearing Escherichia coli strains isolated from a wastewater treatment plant. Antonie Van Leeuwenhoek 99, 817–824. doi: 10.1007/s10482-011-9555-4
Murray, A. K., Zhang, L., Yin, X., Zhang, T., Buckling, A., Snape, J., et al. (2018). Novel insights into selection for antibiotic resistance in complex microbial communities. MBio 9, e00969–e00918. doi: 10.1128/mBio.00969-18
Nathwani, D., Varghese, D., Stephens, J., Ansari, W., Martin, S., and Charbonneau, C. (2019). Value of hospital antimicrobial stewardship programs [ASPs]: a systematic review. Antimicrob. Resist. Infect. Control 8, 1–13. doi: 10.1186/s13756-019-0471-0
Neeno-Eckwall, E. C., Kinkel, L. L., and Schottel, J. L. (2001). Competition and antibiosis in the biological control of potato scab. Can. J. Microbiol. 47, 332–340. doi: 10.1139/cjm-47-4-332
Nelson, R. E., Hatfield, K. M., Wolford, H., Samore, M. H., Scott, R. D. II, Reddy, S. C., et al. (2021). National estimates of healthcare costs associated with multidrug-resistant bacterial infections among hospitalized patients in the United States. Clin. Infect. Dis. 72, S17–S26. doi: 10.1093/cid/ciaa1581
Nishiyama, M., Iguchi, A., and Suzuki, Y. (2015). Identification of Enterococcus faecium and Enterococcus faecalis as vanC-type vancomycin-resistant enterococci (VRE) from sewage and river water in the provincial city of Miyazaki, Japan. J. Environ. Sci. Heal. - Part A Toxic/Hazardous Subst. Environ. Eng. 50, 16–25. doi: 10.1080/10934529.2015.964599
Nowicki, S., DeLaurent, Z. R., De Villiers, E. P., Githinji, G., and Charles, K. J. (2021). The utility of Escherichia coli as a contamination indicator for rural drinking water: evidence from whole genome sequencing. PLoS One 16, e0245910–e0245923. doi: 10.1371/journal.pone.0245910
O’Neill, J. (2016). Review on Antimicrobial Resistance: Tackling drug-resistant infections globally. Available at: https://amr-review.org/ (Accessed September 16, 2021).
Olson, A. B., Silverman, M., Boyd, D. A., McGeer, A., Willey, B. M., Pong-Porter, V., et al. (2005). Identification of a progenitor of the CTX-M-9 group of extended-spectrum β-lactamases from Kluyvera georgiana isolated in Guyana. Antimicrob. Agents Chemother. 49, 2112–2115. doi: 10.1128/AAC.49.5.2112-2115.2005
Omar, K. B., and Barnard, T. G. (2014). Detection of diarrhoeagenic Escherichia coli in clinical and environmental water sources in South Africa using single-step 11-gene m-PCR. World J. Microbiol. Biotechnol. 30, 2663–2671. doi: 10.1007/s11274-014-1690-4
Östman, M., Lindberg, R. H., Fick, J., Björn, E., and Tysklind, M. (2017). Screening of biocides, metals and antibiotics in Swedish sewage sludge and wastewater. Water Res. 115, 318–328. doi: 10.1016/j.watres.2017.03.011
Pal, C., Bengtsson-Palme, J., Kristiansson, E., and Larsson, D. G. J. (2015). Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genomics 16, 964–914. doi: 10.1186/s12864-015-2153-5
Pal, C., Bengtsson-Palme, J., Kristiansson, E., and Larsson, D. G. J. (2016). The structure and diversity of human, animal and environmental resistomes. Microbiome 4, 54–15. doi: 10.1186/s40168-016-0199-5
Patridge, E. V., and Ferry, J. G. (2006). WrbA from Escherichia coli and Archaeoglobus fulgidus is an NAD(P)H:quinone oxidoreductase. J. Bacteriol. 188, 3498–3506. doi: 10.1128/JB.188.10.3498-3506.2006
Paulshus, E., Thorell, K., Guzman-Otazo, J., Joffre, E., Colque, P., Kühn, I., et al. (2019). Repeated isolation of extended-spectrum-lactamase-positive Escherichia coli sequence types 648 and 131 from community wastewater indicates that sewage systems are important sources of emerging clones of antibiotic-resistant bacteria. Antimicrob. Agents Chemother. 63, e00823–e008219. doi: 10.1128/AAC.00823-19
Pazda, M., Rybicka, M., Stolte, S., Bielawski, K. P., Stepnowski, P., Kumirska, J., et al. (2020). Identification of selected antibiotic resistance genes in two different wastewater treatment plant systems in Poland: A preliminary study. Molecules 25, 1–18. doi: 10.3390/molecules25122851
Perron, G. G., Whyte, L., Turnbaugh, P. J., Goordial, J., Hanage, W. P., Dantas, G., et al. (2015). Functional characterization of bacteria isolated from ancient arctic soil exposes diverse resistance mechanisms to modern antibiotics. PLoS One 10, 1–19. doi: 10.1371/journal.pone.0069533
Perry, J. A., and Wright, G. D. (2013). The antibiotic resistance “mobilome”: searching for the link between environment and clinic. Front. Microbiol. 4, 1–7. doi: 10.3389/fmicb.2013.00138
Peters, J. E., Fricker, A. D., Kapili, B. J., and Petassi, M. T. (2014). Heteromeric transposase elements: generators of genomic islands across diverse bacteria. Mol. Microbiol. 93, 1084–1092. doi: 10.1111/mmi.12740
Podlesek, Z., and Žgur Bertok, D. (2020). The DNA damage inducible SOS response is a key player in the generation of bacterial persister cells and population wide tolerance. Front. Microbiol. 11, 1–8. doi: 10.3389/fmicb.2020.01785
Poirel, L., Héritier, C., and Nordmann, P. (2004). Chromosome-encoded ambler class D β-lactamase of Shewanella oneidensis as a progenitor of carbapenem-hydrolyzing oxacillinase. Antimicrob. Agents Chemother. 48, 348–351. doi: 10.1128/AAC.48.1.348-351.2004
Poirel, L., Kämpfer, P., and Nordmann, P. (2002). Chromosome-encoded ambler class a β-lactamase of Kluyvera georgiana, a probable progenitor of a subgroup of CTX-M extended-spectrum β-lactamases. Antimicrob. Agents Chemother. 46, 4038–4040. doi: 10.1128/AAC.46.12.4038-4040.2002
Poirel, L., Liard, A., Rodriguez-Martinez, J. M., and Nordmann, P. (2005a). Vibrionaceae as a possible source of Qnr-like quinolone resistance determinants. J. Antimicrob. Chemother. 56, 1118–1121. doi: 10.1093/jac/dki371
Poirel, L., Rodriguez-Martinez, J. M., Mammeri, H., Liard, A., and Nordmann, P. (2005b). Origin of plasmid-mediated quinolone resistance determinant QnrA. Antimicrob. Agents Chemother. 49, 3523–3525. doi: 10.1128/AAC.49.8.3523-3525.2005
Popowska, M., and Krawczyk-Balska, A. (2013). Broad-host-range IncP-1 plasmids and their resistance potential. Front. Microbiol. 4, 1–8. doi: 10.3389/fmicb.2013.00044
Potron, A., Poirel, L., and Nordmann, P. (2011). Origin of OXA-181, an emerging carbapenem-hydrolyzing oxacillinase, as a chromosomal gene in Shewanella xiamenensis. Antimicrob. Agents Chemother. 55, 4405–4407. doi: 10.1128/AAC.00681-11
Rizzo, L., Manaia, C., Merlin, C., Schwartz, T., Dagot, C., Ploy, M. C., et al. (2013). Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Sci. Total Environ. 447, 345–360. doi: 10.1016/j.scitotenv.2013.01.032
Rodriguez, M. M., Ghiglione, B., Power, P., Naas, T., and Gutkind, G. (2018). Proposing Kluyvera georgiana as the origin of the plasmid-mediated resistance gene fosA4. Antimicrob. Agents Chemother. 62, 1–5. doi: 10.1128/AAC.00710-18
Rodríguez, M. M., Power, P., Sader, H., Galleni, M., and Gutkind, G. (2010). Novel chromosome-encoded CTX-M-78 β-lactamase from a Kluyvera georgiana clinical isolate as a putative origin of CTX-M-25 subgroup. Antimicrob. Agents Chemother. 54, 3070–3071. doi: 10.1128/AAC.01615-09
Rodríguez, E. A., Ramirez, D., Balcázar, J. L., and Jiménez, J. N. (2021). Metagenomic analysis of urban wastewater resistome and mobilome: a support for antimicrobial resistance surveillance in an endemic country. Environ. Pollut. 276:116736. doi: 10.1016/j.envpol.2021.116736
Rodriguez-Mozaz, S., Vaz-Moreira, I., Varela Della Giustina, S., Llorca, M., Barceló, D., Schubert, S., et al. (2020). Antibiotic residues in final effluents of European wastewater treatment plants and their impact on the aquatic environment. Environ. Int. 140:105733. doi: 10.1016/j.envint.2020.105733
Savin, M., Bierbaum, G., Kreyenschmidt, J., Schmithausen, R. M., Sib, E., Schmoger, S., et al. (2021). Clinically relevant Escherichia coli isolates from process waters and wastewater of poultry and pig slaughterhouses in Germany. Microorganisms 9, 1–17. doi: 10.3390/microorganisms9040698
Scaccia, N., Vaz-Moreira, I., and Manaia, C. M. (2020). Persistence of wastewater antibiotic resistant bacteria and their genes in human fecal material. FEMS Microbiol. Ecol. 96:fiaa058. doi: 10.1093/femsec/fiaa058
Schatz, A., Bugle, E., and Waksman, S. A. (1944). Streptomycin, a substance exhibiting antibiotic activity against Gram-positive and Gram-negative bacteria. Proc. Soc. Exp. Biol. Med. 55, 66–69. doi: 10.3181/00379727-55-14461
Schlüter, A., Szczepanowski, R., Pühler, A., and Top, E. M. (2007). Genomics of IncP-1 antibiotic resistance plasmids isolated from wastewater treatment plants provides evidence for a widely accessible drug resistance gene pool. FEMS Microbiol. Rev. 31, 449–477. doi: 10.1111/j.1574-6976.2007.00074.x
Schwartz, T., Kohnen, W., Jansen, B., and Obst, U. (2003). Detection of antibiotic-resistant bacteria and their resistance genes in wastewater, surface water, and drinking water biofilms. FEMS Microbiol. Ecol. 43, 325–335. doi: 10.1016/S0168-6496(02)00444-0
Scott, A. M., Beller, E., Glasziou, P., Clark, J., Ranakusuma, R. W., Byambasuren, O., et al. (2018). Is antimicrobial administration to food animals a direct threat to human health? A rapid systematic review. Int. J. Antimicrob. Agents 52, 316–323. doi: 10.1016/j.ijantimicag.2018.04.005
Serra-Burriel, M., Keys, M., Campillo-Artero, C., Agodi, A., Barchitta, M., Gikas, A., et al. (2020). Impact of multi-drug resistant bacteria on economic and clinical outcomes of healthcare-associated infections in adults: systematic review and meta-analysis. PLoS One 15, e0227139–e0227114. doi: 10.1371/journal.pone.0227139
Shallcross, L. J. (2014). Antibiotic overuse: A key driver of antimicrobial resistance. Br. J. Gen. Pract. 64, 604–605. doi: 10.3399/bjgp14X682561
Skurnik, D., Ruimy, R., Andremont, A., Amorin, C., Rouquet, P., Picard, B., et al. (2006). Effect of human vicinity on antimicrobial resistance and integrons in animal faecal Escherichia coli. J. Antimicrob. Chemother. 57, 1215–1219. doi: 10.1093/jac/dkl122
Søraas, A., Sundsfjord, A., Sandven, I., Brunborg, C., and Jenum, P. A. (2013). Risk factors for community-acquired urinary tract infections caused by ESBL-producing enterobacteriaceae -A case-control study in a low prevalence country. PLoS One 8, 1–7. doi: 10.1371/journal.pone.0069581
Souque, C., Escudero, J. A., and Maclean, R. C. (2021). Integron activity accelerates the evolution of antibiotic resistance. Elife 10, 1–47. doi: 10.7554/eLife.62474
Staley, C., Dunny, G. M., and Sadowsky, M. J. (2014). Environmental and Animal-Associated Enterococci. 1st Edn. Elsevier Inc. doi: 10.1016/B978-0-12-800261-2.00004-9
Stange, C., Sidhu, J. P. S., Toze, S., and Tiehm, A. (2019). Comparative removal of antibiotic resistance genes during chlorination, ozonation, and UV treatment. Int. J. Hyg. Environ. Health 222, 541–548. doi: 10.1016/j.ijheh.2019.02.002
Stanton, I. C., Bethel, A., Leonard, A. F. C., Gaze, W. H., and Garside, R. (2020). What is the research evidence for antibiotic resistance exposure and transmission to humans from the environment? A systematic map protocol. Environ. Evid. 9, 12–18. doi: 10.1186/s13750-020-00197-6
Steen, A. D., Crits-Christoph, A., Carini, P., DeAngelis, K. M., Fierer, N., Lloyd, K. G., et al. (2019). High proportions of bacteria and archaea across most biomes remain uncultured. ISME J. 13, 3126–3130. doi: 10.1038/s41396-019-0484-y
Szczepanowski, R., Krahn, I., Linke, B., Goesmann, A., Pühler, A., and Schlüter, A. (2004). Antibiotic multiresistance plasmid pRSB101 isolated from a wastewater treatment plant is related to plasmids residing in phytopathogenic bacteria and carries eight different resistance determinants including a multidrug transport system. Microbiology 150, 3613–3630. doi: 10.1099/mic.0.27317-0
Szczepanowski, R., Linke, B., Krahn, I., Gartemann, K. H., Gützkow, T., Eichler, W., et al. (2009). Detection of 140 clinically relevant antibiotic-resistance genes in the plasmid metagenome of wastewater treatment plant bacteria showing reduced susceptibility to selected antibiotics. Microbiology 155, 2306–2319. doi: 10.1099/mic.0.028233-0
Tacconelli, E., Carrara, E., Savoldi, A., Harbarth, S., Mendelson, M., Monnet, D. L., et al. (2018). Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327. doi: 10.1016/S1473-3099(17)30753-3
Tamae, C., Liu, A., Kim, K., Sitz, D., Hong, J., Becket, E., et al. (2008). Determination of antibiotic hypersensitivity among 4,000 single-gene-knockout mutants of Escherichia coli. J. Bacteriol. 190, 5981–5988. doi: 10.1128/JB.01982-07
Tang, K. L., Caffrey, N. P., Nóbrega, D. B., Cork, S. C., Ronksley, P. E., Barkema, H. W., et al. (2017). Restricting the use of antibiotics in food-producing animals and its associations with antibiotic resistance in food-producing animals and human beings: a systematic review and meta-analysis. Lancet Planet. Heal. 1, e316–e327. doi: 10.1016/S2542-5196(17)30141-9
Tanner, W. D., VanDerslice, J. A., Goel, R. K., Leecaster, M. K., Fisher, M. A., Olstadt, J., et al. (2019). Multi-state study of Enterobacteriaceae harboring extended-spectrum beta-lactamase and carbapenemase genes in U.S. drinking water. Sci. Rep. 9:3938. doi: 10.1038/s41598-019-40420-0
Terlizzi, M. E., Gribaudo, G., and Maffei, M. E. (2017). UroPathogenic Escherichia coli (UPEC) infections: virulence factors, bladder responses, antibiotic, and non-antibiotic antimicrobial strategies. Front. Microbiol. 8:1566. doi: 10.3389/fmicb.2017.01566
Torres-Barceló, C., Kojadinovic, M., Moxon, R., and Maclean, R. C. (2015). The SOS response increases bacterial fitness, but not evolvability, under a sublethal dose of antibiotic. Proc. R. Soc. B Biol. Sci. 282:20150885. doi: 10.1098/rspb.2015.0885
Touchon, M., Perrin, A., De Sousa, J. A. M., Vangchhia, B., Burn, S., O’Brien, C. L., et al. (2020). Phylogenetic background and habitat drive the genetic diversification of Escherichia coli. PLoS Genet. 16, e1008866–e1008843. doi: 10.1371/journal.pgen.1008866
Traxler, M. F., and Kolter, R. (2015). Natural products in soil microbe interactions and evolution. Nat. Prod. Rep. 32, 956–970. doi: 10.1039/c5np00013k
Tymensen, L. D., Pyrdok, F., Coles, D., Koning, W., Mcallister, T. A., Jokinen, C. C., et al. (2015). Comparative accessory gene fingerprinting of surface water Escherichia coli reveals genetically diverse naturalized population. J. Appl. Microbiol. 119, 263–277. doi: 10.1111/jam.12814
US Department of Health and Human Services, and CDC. (2019). Antibiotic Resistance Threats in the United States. Centers Dis. Control Prev., 1–150. Available at: https://www.cdc.gov/drugresistance/biggest_threats.html (Accessed September 16, 2021).
Ventola, C. L. (2015). The antibiotic resistance crisis—Part 1: Causes and threats. Pharm. Ther. 40, 277–283. doi: 10.5796/electrochemistry.82.749
Waksman, S. A., and Lechevalier, H. A. (1949). Neomycin, a new antibiotic active against streptomycin-resistant bacteria, including tuberculosis organisms. Science 109, 305–307. doi: 10.1126/science.109.2830.305
Waksman, S. A., Schatz, A., and Reynolds, D. M. (2010). Production of antibiotic substances by actinomycetes. Ann. N. Y. Acad. Sci. 1213, 112–124. doi: 10.1111/j.1749-6632.2010.05861.x
Wales, A. D., and Davies, R. H. (2015). Co-selection of resistance to antibiotics, biocides and heavy metals, and its relevance to foodborne pathogens. Antibiotics 4, 567–604. doi: 10.3390/antibiotics4040567
Walk, S. T. (2015). The “cryptic” Escherichia. EcoSal Plus 6:26435255. doi: 10.1128/ecosalplus.esp-0002-2015
Walk, S. T., Alm, E. W., Gordon, D. M., Ram, J. L., Toranzos, G. A., Tiedje, J. M., et al. (2009). Cryptic lineages of the genus Escherichia. Appl. Environ. Microbiol. 75, 6534–6544. doi: 10.1128/AEM.01262-09
Wang, J., and Chen, X. (2020). Removal of antibiotic resistance genes (ARGs) in various wastewater treatment processes: an overview. Crit. Rev. Environ. Sci. Technol. 52, 571–630. doi: 10.1080/10643389.2020.1835124
Wang, Z., Fang, Y., Zhi, S., Simpson, D. J., Gill, A., Mcmullen, L. M., et al. (2020). The locus of heat resistance confers resistance to chlorine and other oxidizing chemicals in Escherichia coli. Appl. Environ. Microbiol. 86, 1–16. doi: 10.1128/AEM.02123-19
Watkinson, A. J., Murby, E. J., Kolpin, D. W., and Costanzo, S. D. (2009). The occurrence of antibiotics in an urban watershed: From wastewater to drinking water. Sci. Total Environ. 407, 2711–2723. doi: 10.1016/j.scitotenv.2008.11.059
Watson, J. R., Wallihan, R., and Antonara, S. (2017). Less Commonly Encountered Enterobacteriaceae. 5th Edn. Elsevier Inc. doi: 10.1016/B978-0-323-40181-4.00142-0
Wright, G. D. (2010). Antibiotic resistance in the environment: a link to the clinic? Curr. Opin. Microbiol. 13, 589–594. doi: 10.1016/j.mib.2010.08.005
Wyrsch, E. R., Reid, C. J., DeMaere, M. Z., Liu, M. Y., Chapman, T. A., Roy Chowdhury, P., et al. (2019). Complete sequences of multiple-drug resistant IncHI2 ST3 plasmids in Escherichia coli of porcine origin in Australia. Front. Sustain. Food Syst. 3, 1–10. doi: 10.3389/fsufs.2019.00018
Yang, S., Cha, J., and Carlson, K. (2005). Simultaneous extraction and analysis of 11 tetracycline and sulfonamide antibiotics in influent and effluent domestic wastewater by solid-phase extraction and liquid chromatography-electrospray ionization tandem mass spectrometry. J. Chromatogr. A 1097, 40–53. doi: 10.1016/j.chroma.2005.08.027
Yang, X., Tran, F., Zhang, P., and Wang, H. (2021). Genomic and phenotypic analysis of heat and sanitizer resistance in Escherichia coli from beef in relation to the locus of heat resistance. Appl. Environ. Microbiol. 87:e0157421. doi: 10.1128/AEM.01574-21
Yuan, Q. B., Guo, M. T., Wei, W. J., and Yang, J. (2016). Reductions of bacterial antibiotic resistance through five biological treatment processes treated municipal wastewater. Environ. Sci. Pollut. Res. 23, 19495–19503. doi: 10.1007/s11356-016-7048-8
Zhang, T., and Li, B. (2011). Occurrence, transformation, and fate of antibiotics in municipal wastewater treatment plants. Crit. Rev. Environ. Sci. Technol. 41, 951–998. doi: 10.1080/10643380903392692
Zhi, S., Banting, G., Li, Q., Edge, T. A., Topp, E., Sokurenko, M., et al. (2016a). Evidence of naturalized stress-tolerant strains of Escherichia coli in municipal wastewater treatment plants. Appl. Environ. Microbiol. 82, 5505–5518. doi: 10.1128/AEM.00143-16
Zhi, S., Banting, G. S., Ruecker, N. J., and Neumann, N. F. (2017). Stress resistance in naturalised waste water E. coli strains. J. Environ. Eng. Sci. 12, 42–50. doi: 10.1680/jenes.16.00021
Zhi, S., Banting, G., Stothard, P., Ashbolt, N. J., Checkley, S., Meyer, K., et al. (2019). Evidence for the evolution, clonal expansion and global dissemination of water treatment-resistant naturalized strains of Escherichia coli in wastewater. Water Res. 156, 208–222. doi: 10.1016/j.watres.2019.03.024
Zhi, S., Li, Q., Yasui, Y., Banting, G., Edge, T. A., Topp, E., et al. (2016b). An evaluation of logic regression-based biomarker discovery across multiple intergenic regions for predicting host specificity in Escherichia coli. Mol. Phylogenet. Evol. 103, 133–142. doi: 10.1016/j.ympev.2016.07.016
Zhi, S., Li, Q., Yasui, Y., and Edge, T., E. Topp, and Neumann, N. F. (2015). Assessing host-specificity of Escherichia coli using a supervised learning logic-regression-based analysis of single nucleotide polymorphisms in intergenic regions. Mol. Phylogenet. Evol. 92. 72–81. doi: 10.1016/j.ympev.2015.06.007
Zhi, S., Stothard, P., Banting, G., Scott, C., Huntley, K., Ryu, K., et al. (2020). Characterization of water treatment-resistant and multidrug-resistant urinary pathogenic Escherichia coli in treated wastewater. Water Res. 182:115827. doi: 10.1016/j.watres.2020.115827
Zingali, T., Chapman, T. A., Webster, J., Chowdhury, P. R., and Djordjevic, S. P. (2020). Genomic characterisation of a multiple drug resistant IncHI2 ST4 plasmid in Escherichia coli ST744 in Australia. Microorganisms 8, 1–13. doi: 10.3390/microorganisms8060896
Zorita, S., Mårtensson, L., and Mathiasson, L. (2009). Occurrence and removal of pharmaceuticals in a municipal sewage treatment system in the south of Sweden. Sci. Total Environ. 407, 2760–2770. doi: 10.1016/j.scitotenv.2008.12.030
Zurfluh, K., Bagutti, C., Brodmann, P., Alt, M., Schulze, J., Fanning, S., et al. (2017a). Wastewater is a reservoir for clinically relevant carbapenemase- and 16s rRNA methylase-producing Enterobacteriaceae. Int. J. Antimicrob. Agents 50, 436–440. doi: 10.1016/j.ijantimicag.2017.04.017
Keywords: antibiotic resistance, water treatment resistance, genome, wastewater, naturalized Escherichia coli
Citation: Yu D, Ryu K, Zhi S, Otto SJG and Neumann NF (2022) Naturalized Escherichia coli in Wastewater and the Co-evolution of Bacterial Resistance to Water Treatment and Antibiotics. Front. Microbiol. 13:810312. doi: 10.3389/fmicb.2022.810312
Received: 08 November 2021; Accepted: 09 May 2022;
Published: 30 May 2022.
Edited by:
Teresa M. Coque, Ramón y Cajal Institute for Health Research, SpainReviewed by:
Olga Cristina Nunes, University of Porto, PortugalCopyright © 2022 Yu, Ryu, Zhi, Otto and Neumann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Norman F. Neumann, bmZuZXVtYW5AdWFsYmVydGEuY2E=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.