
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 09 March 2022
Sec. Microbial Symbioses
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.806882
 Guo-Dong Dai1
Guo-Dong Dai1 Hong-Bin Yan1
Hong-Bin Yan1 Li Li1
Li Li1 Lin-Sheng Zhang1
Lin-Sheng Zhang1 Zhan-Long Liu2
Zhan-Long Liu2 Sheng-Zhi Gao3
Sheng-Zhi Gao3 John Asekhaen Ohiolei1
John Asekhaen Ohiolei1 Yao-Dong Wu1
Yao-Dong Wu1 Ai-Min Guo1
Ai-Min Guo1 Bao-Quan Fu1,4
Bao-Quan Fu1,4 Wan-Zhong Jia1,4*
Wan-Zhong Jia1,4*
In the present study, a new species of the genus Moniliformis species is described taxonomically in the mitochondrial genomic context. The parasite was found in a plateau zokor captured in a high-altitude area of Xiahe County of Gansu Province, China. The mitochondrial (mt) genome length of this new species was 14,066 bp comprising 36 genes and 2 additional non-coding regions (SNR and LNR), without atp8. The molecular phylogeny inferred by the cytochrome c oxidase subunit I gene (cox1) and the18S ribosomal RNA gene (18S rDNA) sequences showed that the parasite as a sister species to other Moniliformis spp. and was named Moniliformis sp. XH-2020. The phylogeny of the concatenated amino acid sequences of the 12 protein-coding genes (PCGs) showed Moniliformis sp. XH-2020 in the same cluster as Macracanthorhynchus hirudinaceus and Oncicola luehei confirming the cox1 and 18S rDNA phylogenetic inference. In addition, the entire mt genome sequenced in this study represents the first in the order Moniliformida, providing molecular material for further study of the phylogeny of the class Archiacanthocephala. Moreover, the species of this class, use arthropods as intermediate hosts and mammals as definitive hosts and are agents of acanthocephaliasis, a zoonosis in humans. Therefore, this study not only expands the host range among potential wild animal hosts for Archiacanthocephalans which is of great ecological and evolutionary significance but also has important significance for the research of zoonotic parasitic diseases.
Acanthocephalans are obligate endoparasites of vertebrates that complete their life cycle with the participation of arthropods (Kennedy, 2006). At present, there are 1,330 verified acanthocephalans across four classes. Moniliformis sp. XH-2020 identified in this study is within the order Moniliformida Schmidt (1972). So far, about 20 species exist in the order Moniliformida (Amin, 2013), its main definitive and intermediate hosts are rodents and insects respectively. Carnivores such as the families Canidae, Felidae and Mustelidae (Kozlov, 1977), as well as birds and hedgehogs (Khaldi et al., 2012), can serve as facultative definitive hosts. Human cases of infection have been reported (Berenji et al., 2007). The parasite develops into an adult and completes its life cycle only after the intermediate host of the infected larva is preyed on by the definitive host (Khalaf et al., 2021). Moniliformis sp. XH-2020 was found in the small intestine of the plateau zokor, Eospalax fontanierii baileyi Thomas, 1911 (Rodentia: Spalacidae), which is a typical subterranean rodent (Su et al., 2015, 2018). Eospalax fontanierii mainly inhabits high altitude areas like the Qinghai-Tibet Plateau (average elevation 4,500 m) and southwestern part of Gansu province (average elevation 2,500 m). The plateau region has a special ecological environment with less anthropogenic activities, therefore, many parasites maintain their life cycle by taking advantage of the predator-prey relationship between wild animals.
Taxonomically, Moniliformis sp. XH-2020 belongs to Acanthocephala; Archiacanthocephala; Moniliformida; Moniliformidae. However, the taxonomic classification of acanthocephalans has not been completely resolved due to the limited species and genetic information available. Moreover, accurate classification of acanthocephalans based on morphology and ecology alone remains a major challenge (Amin, 2013). In particular, the monophyletic or paraphyletic issues of Palaeacanthocephala and the phylogenetic position of Polyacanthocephala based on nuclear ribosomal DNA remains unresolved (Gazi et al., 2016). Interestingly, studies on mt genome have demonstrated its potentials in resolving taxonomic challenges due to the high mutation rate, maternal inheritance, and highly conserved characteristics (Jia et al., 2012).
There are very few studies on Moniliformidae parasitism in rodents in China. Elsewhere, more studies continue to report new species of acanthocephalans (Amin et al., 2016, 2019; Guerreiro Martins et al., 2017; Gomes et al., 2020; Lynggaard et al., 2021) indicating a high species richness. However, no complete mitochondrial (mt) genome sequence is available for this order, hence, besides analysis of the taxonomic status of Moniliformis spp. and description of the mitochondrial genomic characteristic we also provide the complete mt genome sequence while investigating the presence of Moniliformis spp. in plateau zokors captured in the high altitude areas of Gansu province. This study further expands the definitive host range of Moniliformidae in China as well as the growing number of acanthocephalans, which has reference value for the control of acanthocephaliasis. Meanwhile, the complete mitochondrial genome obtained in this study provides molecular material for further molecular evolutionary analysis of Moniliformis spp.
A total of 1,426 plateau zokors (Eospalax fontanierii baileyi) were caught by mousetraps in Xiahe County (102°52′ E; 35°2′ N; altitude at 3,105 m), Luqu County (102°58′ E; 34°48′ N; altitude at 3,114 m) and Hezuo City (103°22′ E; 35°18′ N; altitude at 2,968 m) in Gansu Province, China. This was achieved during the population control program that is conducted annually to protect the grassland ecology from the destructive impact of rodents on grassland vegetation. After trapping, carcasses were dissected and examined based on common predilection site of helminth parasites (including the liver, lungs, intestines, thoracic cavity, and abdomen). The organs and bodies were then disposed according to local guidelines and regulations. Encountered toruliform parasites were cleaned using phosphate buffer saline and preserved in 70% alcohol for identification.
The specimens were collected as they fell off the intestinal wall during the examination of the intestinal content and fixed in 70% alcohol. This was followed by morphological identification of the worms. In particular, the size, color, shape, nodal and somatic segments of the worm were observed. For scanning electron microscopy (SEM), specimens previously fixed in 70% ethanol were dehydrated in an ascending ethanol series (80, 90, 100%). Then the scolex and tail of the parasite were cut off and were processed for SEM following standard methods, including critical point-dried (CPD) with CO2 in sample baskets, mounted with silver adhesive tape on aluminum stubs and sputter-coated with a 20 nm layer of gold for 3 min using a Polaron #3500 sputter coater. Samples were then examined using an Apreo S microscope (Apreo S SEM, Thermo Scientific, United States) under low vacuum conditions using 20 Kv at the Electron Microscopy Centre of Lanzhou University. Finally, digital images were stored on a USB for further use.
Host liver and worm segments were selected for genomic DNA (gDNA) extraction, and the specific operation steps were carried out according to the manufacturer’s instructions (DNeasy Blood & Tissue Kit, Germany). The 18S rDNA gene, cox1 gene, the complete mt genome of the acanthocephalans and plateau zokor’s cox1 gene were downloaded from NCBI GenBank database1 to serve as reference sequences (Supplementary Table 1). Using Oligo 6.0, primers targeting the cox1 gene of the host were designed to identify the zokor species while the 18S rDNA was used to identify the parasite specimens. PCR was performed in a final reaction volume of 50 μl (HIQ Pfu Master Mix polymerase 25 μl, ddH2O 19 μl, forward primers 2 μl, reverse primers 2 μl, gDNA template 2 μl) using the following amplification program: predenaturation at 98°C for 3 min, denaturation at 98°C for 10 sec, annealing at 56°C for 20 sec, extension at 72°C for 70 sec, final extension at 72°C for 5 min. All PCR products were visualized in 1% agarose gel electrophoresis and sequenced for BLASTn. The BLASTn result of the parasite specimen demonstrated similarity to the order Archiacanthocephala that comprised two species (M. hirudinaceus and O. luehei) with complete mt genome sequences, which were used as reference sequences to design 9 overlapping primers (Table 1) to amplify the complete mt genome sequences of the current specimen. All primers in this study were synthesized by Tsingke Biological Technology Company (Xi’an, China).

Table 1. List of primer pairs used in the amplification of the complete mitochondrial genome and gene of the parasite and the host’s cox1 gene.
The DNAstar software package was used to assemble the 9 overlapping sequence reads. Using the two reference species, we performed a preliminary mt genome annotation using the online program GeSeq2 (Tillich et al., 2017). The annotated results were further checked by SnapGene software, and manual modifications were carried out according to the reference sequences. The online tool ARWEN3 was used to predict tRNA genes (Laslett and Canback, 2008). VARNA software4 was used to draw the secondary structures of tRNA (Darty et al., 2009). Nucleic acid sequence, codon usage, amino acid composition and base ratio were all completed in MEGA7 (version 7.0.26) (Kumar et al., 2016).
For the evolutionary analysis of the identified parasite specimen, this study used three gene datasets: the cox1 sequences of 61 acanthocephalans, the concatenated amino acid sequences of the 12 mt PCGs of 16 acanthocephalans and the 18S rDNA gene of 23 acanthocephalans, all of which represented the four classes (Archiacanthocephala, Palaeacanthocephala, Eoacanthocephala and Polyacanthocephala) (Muhammad et al., 2019). Philodina citrina Ehrenberg, 1832 (Rotifera: Philodina) was used as an outgroup. Sequence alignment was completed using MAFFT (Katoh and Standley, 2013). The software trimAl was used to remove poorly aligned regions from the alignment to improve the quality of subsequent analyses (Capella-Gutiérrez et al., 2009). DAMBE v. 7.2.1365 was used to verify the replacement saturation of the sequence after alignment (Xia, 2018). The phylogenetic analysis for three datasets was carried out using Bayesian Inference (BI), and Maximum-Likelihood (ML) method was further used to verify the phylogenetic results. For Bayesian Inference (BI) (Ronquist and Huelsenbeck, 2003), settings for the cox1 dataset were lser rates = gamma, prset aamodelpr = Mixed, mcmc ngen = 4,000,000, samplefreg = 1,000, nchains = 4; and for 18S rDNA gene and concatenated amino acid sequences of the12 PCGs, lser nst = 6, rates = invgamma, mcmc ngen = 1,000,000, samplefreg = 1,000, nchains = 4. The analysis continued until the average standard deviation of split frequencies was lower than 0.01. On completion, 25% were discarded as burn-in. The phylograms were viewed and embellished using the online tool: iTOL6 (Letunic and Bork, 2019). For Maximum-Likelihood (ML) inference, the online program IQ-TREE7 was used (Trifinopoulos et al., 2016).
The plateau zokor has a thick and round body with a short snout, small eyes, short tail, and strong limbs. The adult coat is grayish-brown from head to tail, darker gray on the ventral surface than on the back, bluish-gray or dark gray on the juveniles and the semi-adults. Adult body length is about 160–235 mm and weighs about 173–490 g (Figure 1A). Three cases of infection were detected in 1,426 individuals (the infection rate was 0.21%) of the plateau zokor in the high elevation pastoral areas of Gansu, during May 2020. The thorny-headed worm identified in this study is closely related to species of the family Moniliformidae. The parasite is cylindrical and toruliform in shape, with light yellow coloration and measures 5–10 cm in length (Figure 1B). Light microscopy revealed that the parasite possessed a small proboscis with many hooks and toruliform segments (Figures 1C,D).
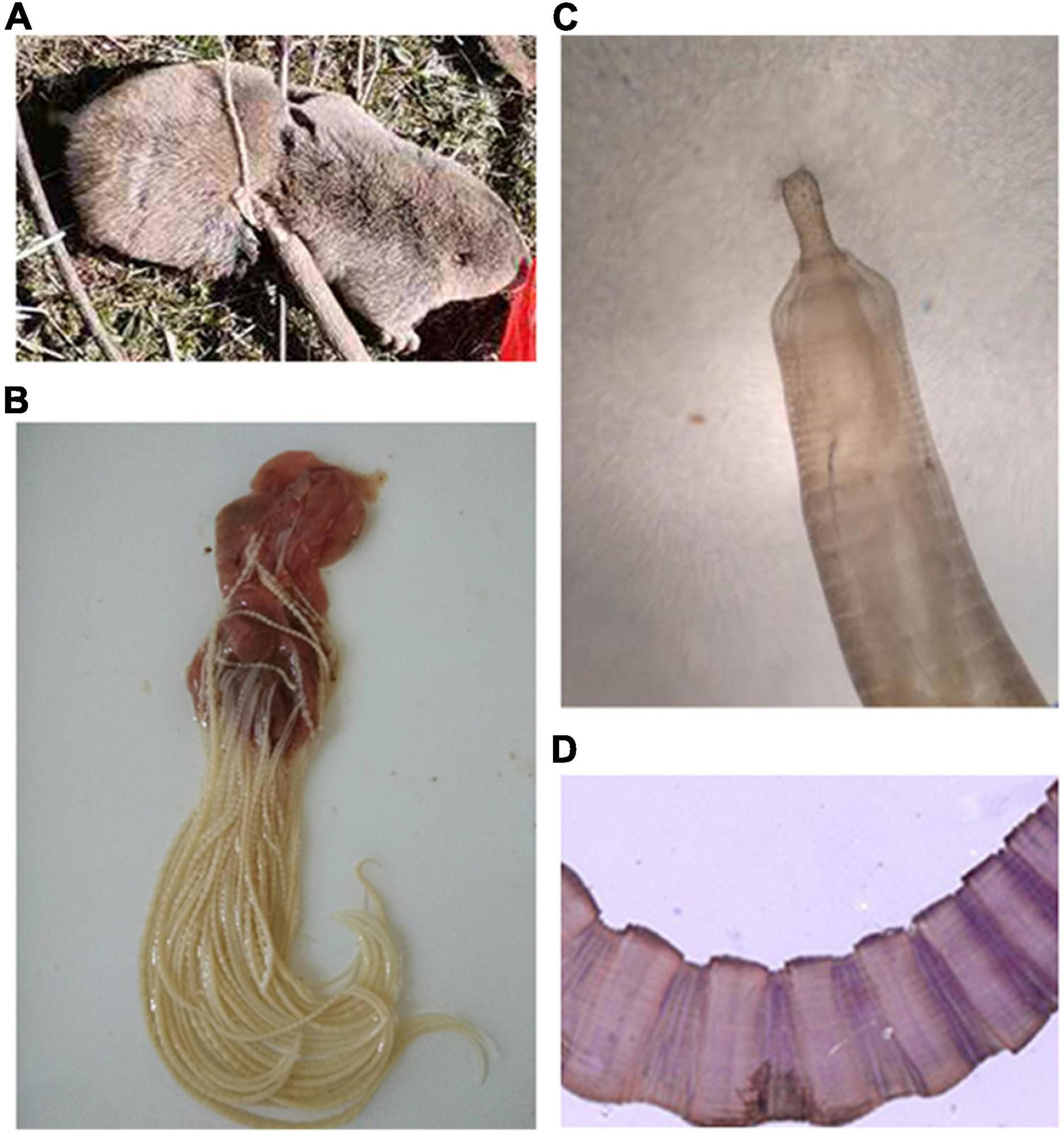
Figure 1. Morphological characteristics of the hosts and Moniliformis sp. XH-2020. (A) The captured plateau zokor. (B) Parasite adsorbed in the small intestinal wall after repeated washes with PBS buffer. (C,D) The structure of proboscis and segments of the parasite as seen under a light microscope (100×).
The SEM image of the anterior and posterior regions are shown in Figures 2A–D. Considering morphological limitations and overlapping characteristics among similar organisms, the host and parasite species were further investigated using genetic molecular markers.
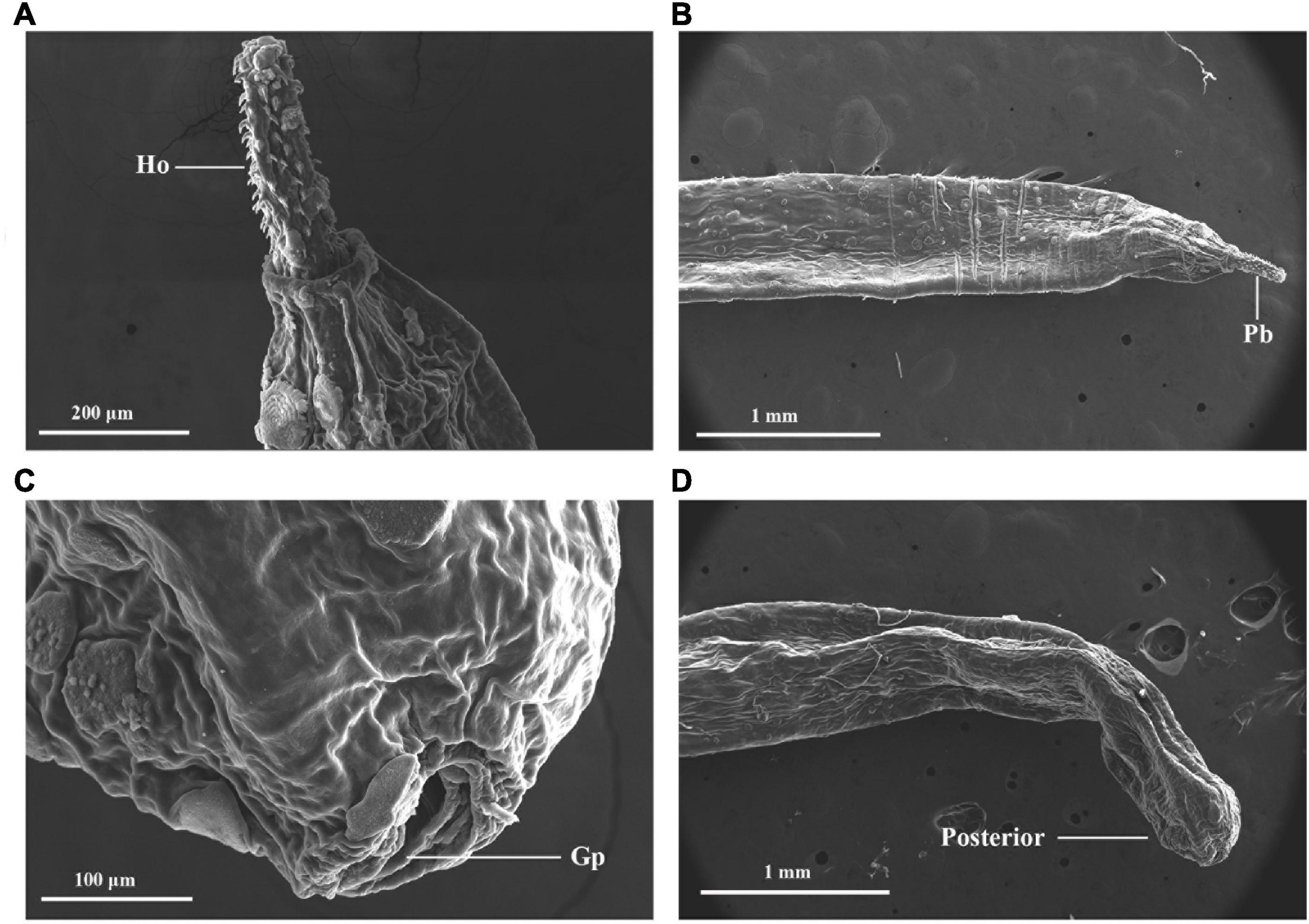
Figure 2. External morphology of Moniliformis sp. XH-2020 via SEM: (A) anterior end of the adult with proboscis; (B) lateral view of the proboscis with hooks, (C) posterior end of the adult, (D) posterior end showing a terminal gonopore. Pb, proboscis; Ho, hook; Gp, gonopore.
In this study, the partial nucleotide sequences of the host cox1 gene (1,299 bp) and 18S rDNA gene (1,414 bp) of the parasite were used for identification. The BLASTn results confirmed the plateau zokor as Eospalax fontanierii baileyi with a 100% identity while the parasite demonstrated 98.87–100% identity with the genus Moniliformis, but the mitochondrial cox1 gene was only 76.12–87% similar to other acanthocephalans in GenBank. Therefore, in order to accurately describe the taxonomic status of the specimen, the reference sequences of cox1, 18S rDNA and the entire mt genome of other acanthocephalans were downloaded from GenBank followed by phylogenetic analysis.
The complete mt genome sequence was manually assembled using the nine overlapping mtDNA fragments by joining the 5’ and 3’ ends of two adjacent fragments. A total length of 14,066 bp mtDNA was realized after manual assemblage. The 36 genes of the parasite mt genome includes 12 PCGs (cox1-3, nad1-6, atp6, nad4L and cytb), 22 transfer RNA genes (tRNAs), 2 ribosomal RNA genes (rrnL and rrnS), and additional 2 non-coding regions (long non-coding region LNR and short non-coding region, SNR), without atp8 (Figure 3). The length of rrnS was 631 bp, located between trnM and trnF genes while rrnL was 949 bp, and was located between trnY and trnL1 genes. SNR (306 bp) was located between trnI and trnM while LNR (401 bp) was located between trnW and trnV. The 22 distinct nucleotide sequences ranging from 50 to 66 bp in size for the acanthocephalan were predicted to fold into cloverleaf-like secondary structure of tRNAs with the exception of trnS2, which lack a dihydrouridine (DHU) arm (Supplementary Figure 1). Meanwhile, many tRNA secondary structures predicted in this study lack TΨC arm, which is similar to other mtDNA studies of acanthocephalans (Steinauer et al., 2005; Gazi et al., 2012, 2015, 2016; Pan and Nie, 2013, 2014; Weber et al., 2013; Muhammad et al., 2019). Because the mt genome is a circular type of DNA molecule, the cox1 gene was artificially set as the starting point of the entire genome following existing mt genome data of acanthocephalans (Gazi et al., 2012; Weber et al., 2013; Wey-Fabrizius et al., 2013).
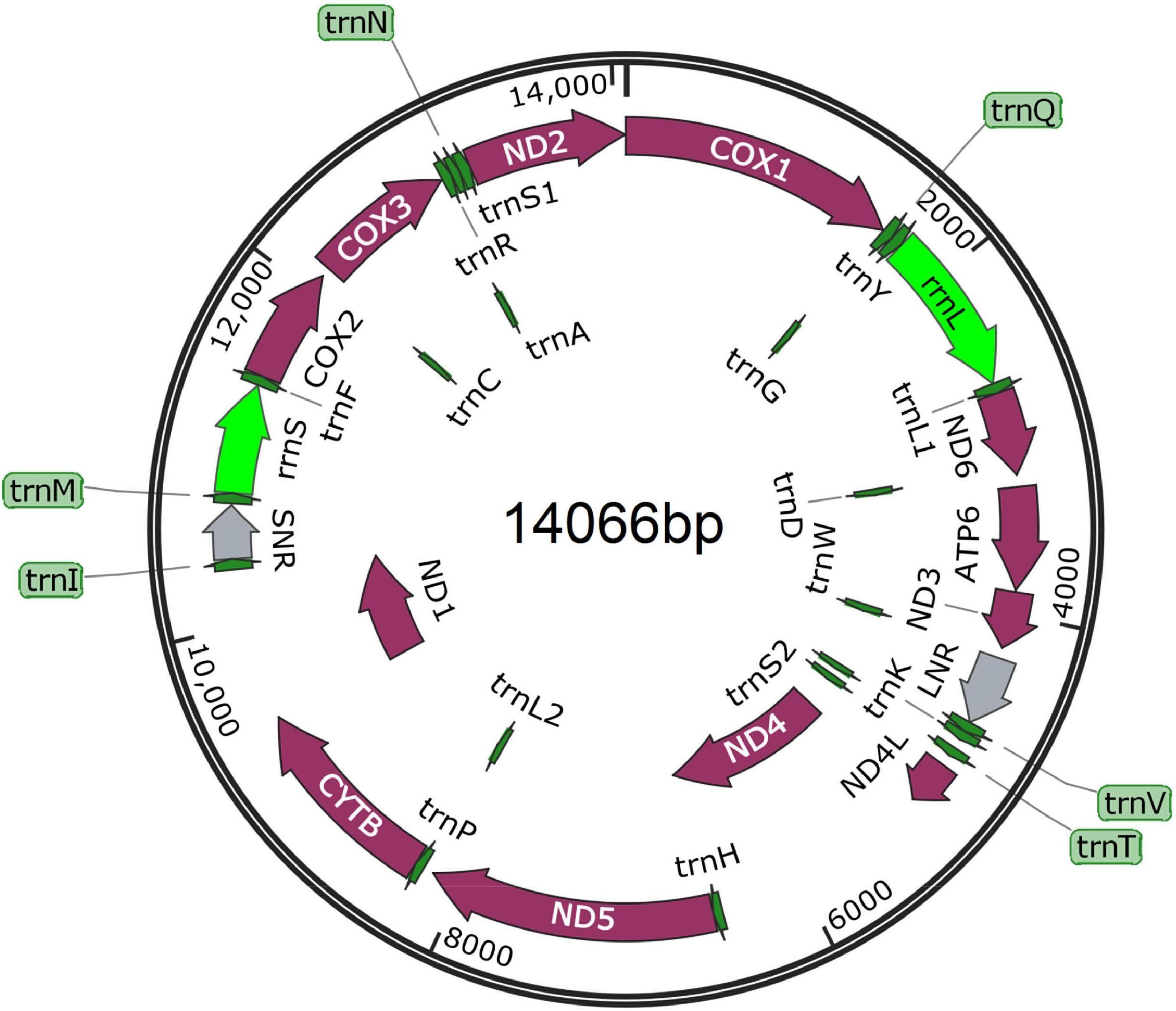
Figure 3. Composition of the complete mitochondrial genome of Moniliformis sp. XH-2020. All genes are arranged clockwise with the names of the genes annotated. LNR and SNR represent long non-coding and short non-coding regions.
Named temporarily as Moniliformis sp. XH-2020, the nucleotide composition analysis of the complete mt genome showed the following: T = 41.2%, C = 8.5%, A = 25.0% and G = 25.3%, with an overall A + T content of 66.2%. The A + T content of the protein-coding mt genes was 65.8%, which is a more than that of O. luehei (58.8%), slightly more than P. celatus (61.2%) and M. hirudinaceus (63.9%), but less than that of Leptorhynchoides thecatus (71.6%), Paratenuisentis ambiguus (67.1%). The results of the nucleotide composition demonstrate the complete mt genome of Moniliformis sp. XH-2020 is a T-rich mtDNA and excessively favors A + T content. The total number of triplet codons of the 12 PCGs in the first position was 3,486 bp, where T account for 33.6%, C 7.7%, A 26.7% and G 32.0% (Supplementary Table 2).
The total length of the 12 PCGs was 10,728 bp, encoding 3,564 amino acids. Nad5 gene had the longest sequence (1,641 bp), followed by cox1 (1,590 bp), with nad4L (261 bp) the shortest. Using the invertebrate mitochondrial code, the relative synonymous codon usage (RSCU) parameter was used to compute codon usage bias, the results demonstrate TTT as the most frequently used codon, with an RSCU value of 1.85, while the codon with the lowest frequency was CTC, with an RSCU of 0.01. The CGC codon was absent (Supplementary Table 3). Among the amino acid sequences, Val amino acid appeared frequently (16.44%) with preference for GTT, GTA and GTG codons, followed by Leu (14.14%) (TTA and TTG) and the least frequent was Gln amino acid (0.61%) (CAG). Initiation/termination codons are shown in Table 2 with more genes utilizing the GTG start codons and TAA stop codons.
Phylogenetic analysis of cox1 gene and 18S rDNA gene inferred by the BI method showed that Moniliformis sp. XH-2020 formed a monophyletic group with representative species of the class Archiacanthocephala with strong nodal support values (cox1 BPP = 1, 18S rDNA BPP = 1) (Figures 4, 5), and was consistent with the results of ML trees (cox1 ML-BP = 100, 18S rDNA ML-BP = 100) (Supplementary Figures 2, 3). Meanwhile, BI tree depicts a monophyletic Archiacanthocephala as the most basal and sister to the other three classes, which sit well with the results of previous investigations (Verweyen et al., 2011; Weber et al., 2013; Gazi et al., 2016; Song et al., 2016; Muhammad et al., 2019). The BI tree of cox1 gene showed that Moniliformis sp. XH-2020 formed a sister relationship with representative species of the order Moniliformida and forms a monophyletic group (cox1 BPP = 0.99) (Figure 4). Similarly, the phylogenetic tree of 18S rDNA gene (including BI and ML trees) also depicted monophyly (Figure 5 and Supplementary Figure 3). Species in the class Archiacanthocephala also formed a monophyletic group both in BI and ML inference of the cox1 and 18S rDNA genes (cox1 BPP = 1.00, 18S rDNA BPP = 1). In the BI tree of 18S rDNA gene, Mediorhynchus grandis in the order Gigantorhynchida formed a clade with Moniliformis moniliformis (Figure 5), this relationship is consistent with previous phylogenetic results of small subunits of ribosome DNA (SSU) (Lynggaard et al., 2021).
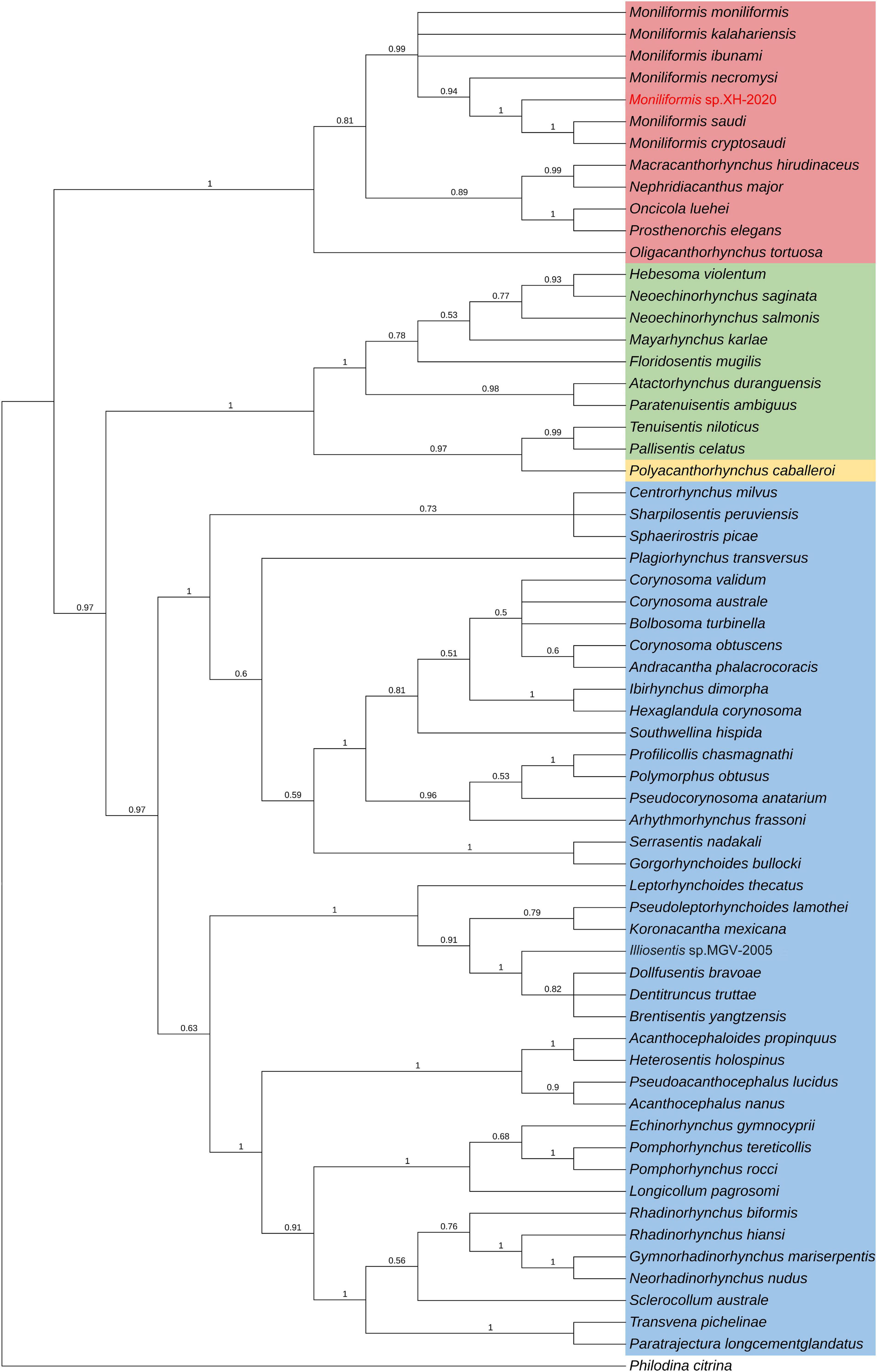
Figure 4. Phylogenetic relationship of Moniliformis sp. XH-2020 with other acanthocephalans based on cox1 gene by Bayesian inference. Node values are Bayesian posterior probabilities (BPP), the red color indicates the class Archiacanthocephala, the green color indicates the class Eoacanthocephala, the blue color indicates the class Palaeacanthocephala and the yellow color indicates the class Polyacanthocephala.
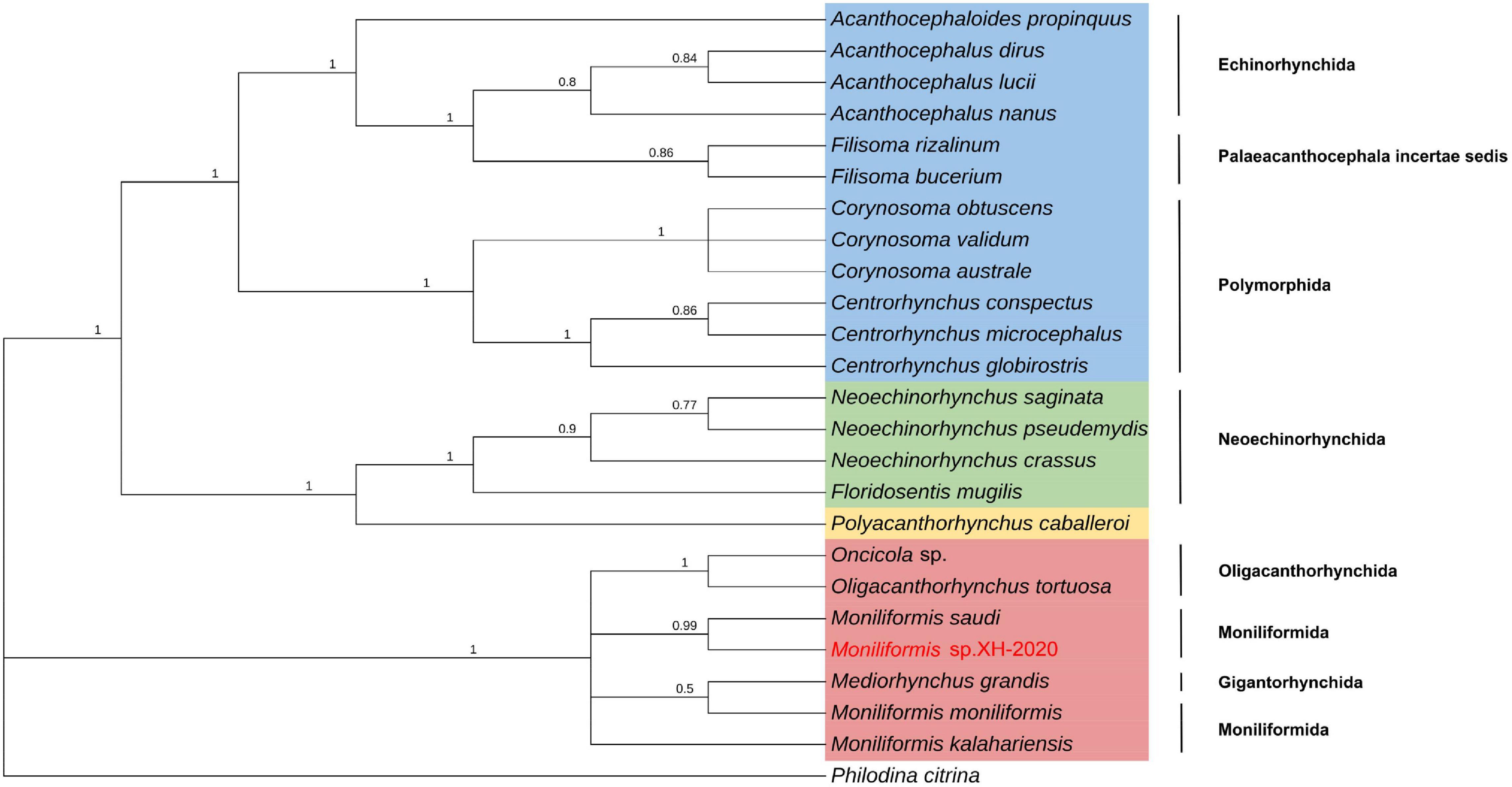
Figure 5. Molecular evolutionary analysis of acanthocephalans based on 18S rDNA gene by Bayesian inference. Node values are Bayesian posterior probabilities (BPP), the red color indicates the class Archiacanthocephala, the green color indicates the class Eoacanthocephala, the blue color indicates the class Palaeacanthocephala and the yellow color indicates the class Polyacanthocephala.
The class Palaeacanthocephala represented by 40 taxa were divided into two clusters in BI trees (Figures 4, 5). For the phylogenetic results of cox1, a cluster was formed by members of the order Polymorphida, Palaeacanthocephala incertae sedis and Echinorhynchida. In this cluster, Sharpilosentis peruviensis of the order Palaeacanthocephala incertae sedis and two species of the order Echinorhynchida (Serrasentis nadakali and Gorgorhynchoides bullock) formed a monophyletic group with members of the order Polymorphida (Figure 4), similar to ML tree output (Supplementary Figure 2). Another paraphyletic cluster was constituted by members of the order Echinorhynchida and Palaeacanthocephala incertae sedis (Figure 4). Moroever, similar observations has also been made (Garcia-Varela and Nadler, 2005; Verweyen et al., 2011; Radwan, 2012). For the class Polyacanthocephala, the representative species Polyacanthorhynchus caballeroi formed a monophyletic group with members of class Eoacanthocephala both in BI and ML trees (Figures 4, 5 and Supplementary Figures 2, 3), in contrast to previous morphological classification that placed Polyacanthocephala species within the Palaeacanthocephala (Schmidt and Canaris, 1967; Bullock, 1969; Amin, 1985). The taxonomic status of the class Polyacanthocephala remains controversial due to the classification limitations and unverified species. Particularly, the question of whether it represents a separate class has become a long-held taxonomic issue.
The widespread application of mt markers and evidence from previous studies (Verweyen et al., 2011; Martins et al., 2017) suggest that a revisit to the current taxonomic classification is warranted as there seems to be a lack of phylogenetic resolution and consistency between species of certain orders. Case in point, the phylogenetic relationship between species of the class Palaeacanthocephala, and the taxonomic status of the class Polyacanthocephala. Therefore, effective molecular markers and sufficient effective species are urgently needed to reclassify acanthocephalans in the future. In the phylogenetic analysis of species, researchers are increasingly interested in mitochondrial gene markers because of its maternal inheritance, accumulation of spontaneous mutation characteristics, and rate of evolution (Brown et al., 1979; Jia et al., 2012). Compared with mt genes, 18S rDNA gene is relatively conserved and is one of the molecular markers commonly used in phylogenetic analysis (Amin et al., 2019). Therefore, the combination of mitochondrial genes and nuclear genes as molecular markers for species classification may provide unique insights into species classification.
To further verify the phylogeny reconstructed by cox1 and 18S rDNA gene, the phylogenetic relationship was inferred by the concatenated amino acid sequences of the 12 PCGs (3,641 characters of the amino acid sequence) of 18 taxa in this study (16 acanthocephalans, 1 rotifera and the current isolate) (Figure 6). In BI and ML trees, the four classes were identified as three major monophyletic clades with strong nodal support values; Palaeacanthocephala BPP = 1.00, ML-BP = 100, Archiacanthocephala BPP = 1.00, ML-BP = 100,
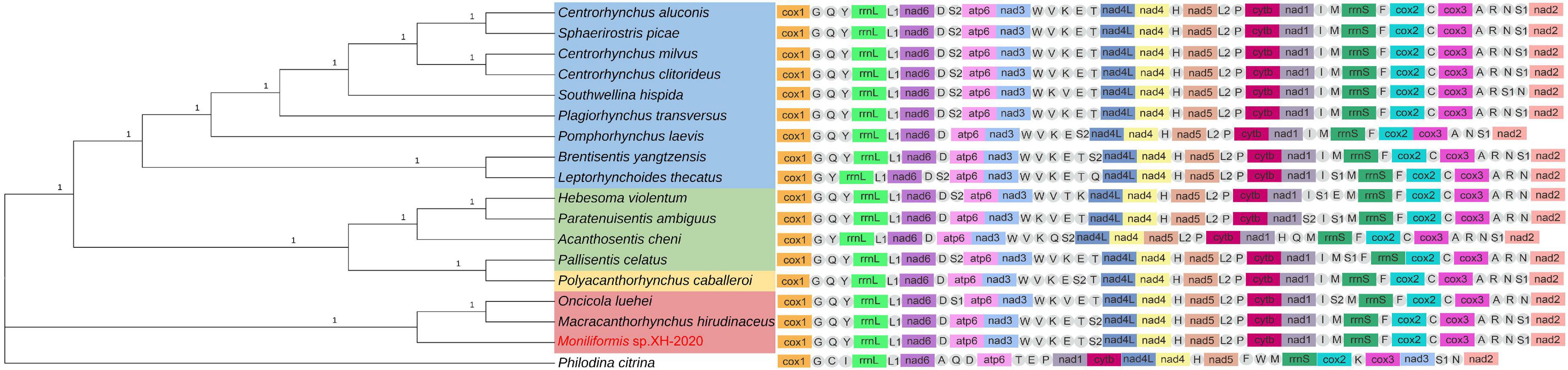
Figure 6. Phylogenetic relationship of Moniliformis sp. XH-2020 with other acanthocephalans based on the concatenated amino acid sequences of 12 PCGs by Bayesian inference and mt gene order. Node values are Bayesian posterior probabilities (BPP), the red color indicates the class Archiacanthocephala, the green color indicates the class Eoacanthocephala, the blue color indicates the class Palaeacanthocephala, the yellow color indicates the class Polyacanthocephala, the tRNAs are labeled by single-letter code for the corresponding amino acid.
Eoacanthocephala and Polyacanthocephala, BPP = 1.00, ML-BP = 100 (Figure 6 and Supplementary Figure 4). The monophyletic Archiacanthocephala group contained the current isolate Moniliformis sp. XH-2020 and formed a sister taxa (BPP = 1.00, ML-BP = 100) with M. hirudinaceus and O. luehei of the family Oligacanthorhynchidae, and the tree topologies of these two families is consistent with previous study (Gomes et al., 2020).
The class Palaeacanthocephala is the largest acanthocephalan class in species abundance and represetend by three orders. The representative species formed a monophyletic group with high nodal support values (BPP = 1.00, ML-BP = 100), although inconsistent with the cox1 and 18S rDNA phyolgentic inference, this unresolved discrepancy is probably a result of the limited number of complete mt geneome sequences used for reconstruction due to scarcity of complete mt genome of acanthocephalans in GenBank database. Although Centrorhynchus aluconis, C. clitorideus and C. milvus belonged to the same genus Centrorhynchus in order Polymorphida, C. aluconis alongside Sphaerirostris picae of a different genus, formed a clade (BPP = 1.00, ML-BP = 100) and the other two Centrorhynchus spp. formed a clade (BPP = 1.00, ML-BP = 100). This result is inconsistent with previous phylogenetic analysis (Gazi et al., 2016; Song et al., 2016), that placed Southwellina hispida, C. aluconis and Plagiorhynchus transversus in a monophyletic group based on the 12 protein-coding genes. Also, three Echinorhynchid species (Pomphorhynchus laevis, Brentisentis yangtzensis and L. thecatus) were not monophyletic, while B. yangtzensis and L. thecatus formed a well-supported clade (BPP = 1.00 ML-BP = 100). P. caballeroi was nested within Eoacanthocephala, forming a clade with Pallisentis celatus (BPP = 1.00, ML-BP = 100) and in agreement with the phylogenetic result of cox1 and 18S rDNA genes. Nonetheless, due to insufficient species sequence information, the problem of whether both groups are monophyletic or paraphyletic remains a subject of future investigation.
The order of mt gene arrangement was relatively consistent with other closely related species which is an important evidence of relatedness. All acanthocephalans with complete mt genome information in GenBank, a total of 17 species from all four classes and a Bdelloidea species as an outgroup, were selected for comparison of mt gene order (Figure 6). The result showed that the sequence of the 12 PCGs and 2 ribosomal RNA genes were identical, while the position of tRNAs varied with species. Three gene blocks (nad2-cox1-trnG, trnY-rrnL-trnL1-nad6 and nad4L-nad4-trnH-nad5) were found to be present in the mt genomes of all acanthocephalans, and is consistent with the results of previous studies (except for trnN-nad2-cox1-trnG) (Gazi et al., 2012, 2015; Figure 6). The nad1 and nad3 genes in the outgroup P. chitrina were located differently from the acanthocephalan, which provides additional evidence supporting the relatedness among acanthocephalan species. The gene order of all Palaeacanthocephala group species were relatively conserved, with a few species demonstrating different tRNA gene positions. In particular, the mt gene arrangement of C. luconis, S. picae, C. litorideus, and C. melvus were identical, which further supported the phylogenetic analysis demonstrating evolution from the same ancestors. But the P. laevis lacks trnR, trnC and trnT genes, and there were translocations of three tRNAs (trnQ, trnS2, and trnS1) between B. yangtzensis and L. thecatus. Among the three species in the class Archiacanthocephala, M. hirudinaceus and Moniliformis sp. XH-2020 showed identical genetic order, similar to B. yangtzensis of the class Palaeacanthocephala. The tRNA genes of the O. luehe, another species of the class Archiacanthocephala, vary greatly in the position of the mt genome. There were many differences observed in the mt genome order between species of the class Eoacanthocephala in contrast with the phylogenetic results. P. caballeroi, the only representative species of the class Polyacanthocephala, has an almost identical gene arrangement as P. celatus (except for translocations of two tRNAs [trnS1 and trnS2]), and is consistent with the sister-relationship shared in the phylogeny which further strengthens the relationship between Eoacanthocephala and Polyacanthocephala. Although gene arrangement in the mtDNA can reflect the relationship between species to an extent, the lack of mitochondrial genome data remains a major contributing factor to the taxonomic challenges of acanthocephalans and thus, warrant more studies.
Besides the genetic evidence and description of a putatively new species of Monoliformis in a wild rodent (Eospalax fontanierii baileyi) in China, this study provides the complete mitochondrial genome representing the order Moniliformida for the first time which will serve as reference molecular material for the accurate classification of acanthocephalans in the future and in understanding the transmission and host range of members of Moniliformida.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, OK415026, 18SrDNA gene accession number OM388438.
The animal study was reviewed and approved by all animals were handled in strict accordance with good animal practice according to the Animal Ethics Procedures and Guidelines of the People’s Republic of China, and the study was approved by the Animal Ethics Committee of Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences (No. LVRIAEC2012-007).
G-DD performed the experiments. LL, H-BY, W-ZJ, and B-QF conceived and designed the experiments. L-SZ, Z-LL, S-ZG, and A-MG completed the collection of samples. JO and Y-DW performed the data analyses. All authors contributed to the article and approved the submitted version.
This work was funded by Cultivation of Achievements of State Key Laboratory of Veterinary Etiological Biology (SKLVEB2020CGPY01) and Central Public-Interest Scientific Institution Basal Research Fund (1610312020016).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.806882/full#supplementary-material
Amin, O. M. (1985). “Classification,” in Biology of the Acanthocephala, eds B. B. Nickol and D. W. T. Crompton (Cambridge, U.K: Cambridge University Press), 27–72.
Amin, O. M. (2013). Classification of the Acanthocephala. Folia Parasitol. 60, 273–305. doi: 10.14411/fp.2013.031
Amin, O. M., Heckmann, R. A., Osama, M., and Evans, R. P. (2016). Morphological and molecular descriptions of Moniliformis saudi sp. n. (Acanthocephala: Moniliformidae) from the desert hedgehog, Paraechinus aethiopicus (Ehrenberg) in Saudi Arabia, with a key to species and notes on histopathology. Folia Parasitol. 26:2016.014. doi: 10.14411/fp.2016.014
Amin, O. M., Heckmann, R. A., Sharifdini, M., and Albayati, N. Y. (2019). Moniliformis cryptosaudi n. sp. (Acanthocephala: Moniliformidae) from the Long-eared Hedgehog Hemiechinus auritus (Gmelin) (Erinaceidae) in Iraq; A Case of Incipient Cryptic Speciation Related to M. saudi in Saudi Arabia. Acta Parasitol. 64, 195–204. doi: 10.2478/s11686-018-00021-9
Berenji, F., Fata, A., and Hosseininejad, Z. (2007). A case of Moniliformis moniliformis (Acanthocephala) infection in Iran. Korean J. Parasitol. 45, 145–148. doi: 10.3347/kjp.2007.45.2.145
Brown, W. M., George, M. J., and Wilson, A. C. (1979). Rapid evolution of animal mitochondrial DNA. Proc. Natl. Acad. Sci. U. S. A. 76, 1967–1971. doi: 10.1073/pnas.76.4.1967
Bullock, W. L. (1969). “Morphological features as tools and as pitfalls in acanthocephalan systematics,” in Problems in Systematics of Parasites, ed. G. D. Schmidt (Baltimore, FL: University Park Press), 9–43.
Capella-Gutiérrez, S., Silla-Martínez, J. M., and Gabaldón, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Darty, K., Denise, A., and Ponty, Y. (2009). VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics 25, 1974–1975. doi: 10.1093/bioinformatics/btp250
Garcia-Varela, M., and Nadler, S. A. (2005). Phylogenetic relationships of Palaeacanthocephala (Acanthocephala) inferred from SSU and LSU rDNA gene sequences. J. Parasitol. 91, 1401–1409. doi: 10.1645/GE-523R.1
Gazi, M., Kim, J., García-Varela, M., Park, C., Littlewood, D. T. J., and Park, J. K. (2016). Mitogenomic phylogeny of Acanthocephala reveals novel class relationships. Zool. Scr. 45, 437–454. doi: 10.1111/zsc.12160
Gazi, M., Kim, J., and Park, J. K. (2015). The complete mitochondrial genome sequence of Southwellina hispida supports monophyly of Palaeacanthocephala (Acanthocephala: Polymorphida). Parasitol. Int. 64, 64–68. doi: 10.1016/j.parint.2015.01.009
Gazi, M., Sultana, T., Min, G. S., Park, Y. C., Garcia-Varela, M., Nadler, S. A., et al. (2012). The complete mitochondrial genome sequence of Oncicola luehei (Acanthocephala: Archiacanthocephala) and its phylogenetic position within Syndermata. Parasitol. Int. 61, 307–316. doi: 10.1016/j.parint.2011.12.001
Gomes, A. P. N., Costa, N. A., Gentile, R., Vilela, R. V., and Maldonado, A. (2020). Morphological and genetic description of Moniliformis necromysi sp. n. (Archiacanthocephala) from the wild rodent Necromys lasiurus (Cricetidae: Sigmondontinae) in Brazil. J. Helminthol. 94:e138. doi: 10.1017/S0022149X20000188
Guerreiro Martins, N. B., Del Rosario Robles, M., and Navone, G. T. (2017). A new species of Moniliformis from a Sigmodontinae rodent in Patagonia (Argentina). Parasitol. Res. 116, 2091–2099. doi: 10.1007/s00436-017-5508-9
Jia, W. Z., Yan, H. B., Lou, Z. Z., Ni, X. W., Dyachenko, V., Li, H. M., et al. (2012). Mitochondrial genes and genomes support a cryptic species of tapeworm within Taenia taeniaeformis. Acta Trop. 123, 154–163. doi: 10.1016/j.actatropica.2012.04.006
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kennedy, C. (2006). “Biogeography and distribution,” in Ecology of the Acanthocephala, ed. C. R. Kennedy (Cambridge: Cambridge University Press), 28–51. doi: 10.1017/CBO9780511541902
Khalaf, A. K., Swadi, B. F., and Mahmoudvand, H. (2021). Morphological characterization of Moniliformis moniliformis isolated from an Iraqi patient. J. Parasit. Dis. 45, 128–130. doi: 10.1007/s12639-020-01287-5
Khaldi, M., Torres, J., Samso, B., Miquel, J., Biche, M., Benyettou, M., et al. (2012). Endoparasites (helminths and coccidians) in the hedgehogs Atelerix algirus and Paraechinus aethiopicus from Algeria. Afr. Zool. 47, 48–54. doi: 10.3377/004.047.0114
Kozlov, D. P. (1977). Key to the helminths of carnivores in the USSR. Opreditel’ gel’mintov khishchnykh mlekopitayushchikh SSSR. Moscow, Russia: Nauka, 276.
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Laslett, D., and Canback, B. (2008). ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 24, 172–175. doi: 10.1093/bioinformatics/btm573
Letunic, I., and Bork, P. (2019). Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, 256–259. doi: 10.1093/nar/gkz239
Lynggaard, C., García-Prieto, L., Guzmán-Cornejo, C., and García-Varela, M. (2021). Description of a new species of Moniliformis (Acanthocephala: Moniliformidae) from Peromyscus hylocetes (Rodentia: Cricetidae) in Mexico. Parasitol. Int. 83:102315. doi: 10.1016/j.parint.2021.102315
Martins, N. B. G., Robles, M. D. R., and Navone, G. T. (2017). A new species of Moniliformis from a Sigmodontinae rodent in Patagonia (Argentina). Parasitol. Res. 116, 2091–2099.
Muhammad, N., Suleman, Ma, J., Khan, M. S., Wu, S. S., Zhu, X. Q., et al. (2019). Characterization of the complete mitochondrial genome of Centrorhynchus milvus (Acanthocephala: Polymorphida) and its phylogenetic implications. Infect. Genet. Evol. 75:e103946. doi: 10.1016/j.meegid.2019.103946
Pan, T. S., and Nie, P. (2013). The complete mitochondrial genome of Pallisentis celatus (Acanthocephala) with phylogenetic analysis of acanthocephalans and rotifers. Folia Parasitol. 60, 181–191. doi: 10.14411/fp.2013.021
Pan, T. S., and Nie, P. (2014). The cloning of the mitochondrial genome of Hebesoma violentum (acanthocephala) and the phylogenetic analysis of acanthocephalans. Acta Hydrobiol. Sin. 38, 351–361. doi: 10.7541/2014.50
Radwan, N. A. (2012). Phylogenetic analysis of Sphaerirostris picae (Acanthocephala: Centrorhynchidae) based on large and small subunit ribosomal DNA gene. Int. J. Parasitol. Res. 4, 106–110.
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Schmidt, G. D. (1972). Revision of the class Archiacanthocephala Meyer, 1931 (Phylum Acanthocephala), with emphasis on Oligacanthorhynchidae. J. Parasitol. 58, 290–297.
Schmidt, G. D., and Canaris, A. G. (1967). Acanthocephala from Kenya with descriptions of two new species. J. Parasitol. 53, 634–637. doi: 10.2307/3276730
Song, R., Zhang, D., Deng, S. M., Ding, D. M., Liao, F. C., and Liu, L. S. (2016). The complete mitochondrial genome of Acanthosentis cheni (Acanthocephala: Quadrigyridae). Mitochondrial DNA Part B Resour. 1, 797–798. doi: 10.1080/23802359.2016.1197076
Steinauer, M. L., Nickol, B. B., Broughton, R., and Ortí, G. (2005). First sequenced mitochondrial genome from the phylum Acanthocephala (Leptorhynchoides thecatus) and its phylogenetic position within Metazoa. J. Mol. Evol. 60, 706–715. doi: 10.1007/s00239-004-0159-8
Su, J. H., Aryal, A., Nan, Z. B., and Ji, W. H. (2015). Climate change-induced range expansion of a subterranean rodent: implications for rangeland management in Qinghai-Tibetan Plateau. PLoS One 10:e138969. doi: 10.1371/journal.pone.0138969
Su, J. H., Hegab, I. M., Ji, W. H., and Nan, Z. B. (2018). Function-related drivers of skull morphometric variation and sexual size dimorphism in a subterranean rodent, Plateau Zokor (Eospalax baileyi). Ecol. Evol. 8, 4631–4643. doi: 10.1002/ece3.3986
Tillich, M., Lehwark, P., Pellizzer, T., Ulbricht-Jones, E. S., Fischer, A., Bock, R., et al. (2017). GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45, 6–11. doi: 10.1093/nar/gkx391
Trifinopoulos, J., Nguyen, L. T., von Haeseler, A., and Minh, B. Q. (2016). W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44, 232–235. doi: 10.1093/nar/gkw256
Verweyen, L., Klimpel, S., and Palm, H. W. (2011). Molecular phylogeny of the Acanthocephala (class Palaeacanthocephala) with a paraphyletic assemblage of the orders Polymorphida and Echinorhynchida. PLoS One 6:e28285. doi: 10.1371/journal.pone.0028285
Weber, M., Wey-Fabrizius, A. R., Podsiadlowski, L., Witek, A., Schill, R. O., Sugar, L., et al. (2013). Phylogenetic analyses of endoparasitic Acanthocephala based on mitochondrial genomes suggest secondary loss of sensory organs. Mol. Phylogenet. Evol. 66, 182–189. doi: 10.1016/j.ympev.2012.09.017
Wey-Fabrizius, A. R., Podsiadlowski, L., Herlyn, H., and Hankeln, T. (2013). Platyzoan mitochondrial genomes. Mol. Phylogenet. Evol. 69, 365–375. doi: 10.1016/j.ympev.2012.12.015
Keywords: Moniliformis, mitochondrial genome, 18S rDNA gene, gene annotation, phylogeny, cox1 gene
Citation: Dai G-D, Yan H-B, Li L, Zhang L-S, Liu Z-L, Gao S-Z, Ohiolei JA, Wu Y-D, Guo A-M, Fu B-Q and Jia W-Z (2022) Molecular Characterization of a New Moniliformis sp. From a Plateau Zokor (Eospalax fontanierii baileyi) in China. Front. Microbiol. 13:806882. doi: 10.3389/fmicb.2022.806882
Received: 01 December 2021; Accepted: 17 January 2022;
Published: 09 March 2022.
Edited by:
David William Waite, Ministry for Primary Industries, New ZealandReviewed by:
Yi-Tian Fu, Hunan Agricultural University, ChinaCopyright © 2022 Dai, Yan, Li, Zhang, Liu, Gao, Ohiolei, Wu, Guo, Fu and Jia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wan-Zhong Jia, amlhd2FuemhvbmdAY2Fhcy5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.