- 1State Key Laboratory of Trauma, Burns and Combined Injury, Department of Wound Infection and Drug, Daping Hospital, Army Medical University, Chongqing, China
- 2Department of Spine Surgery, Center of Orthopedics, Daping Hospital, Army Medical University, Chongqing, China
- 3Trauma Center, The Second Affiliated Hospital of Hainan Medical University, Haikou, China
- 4State Key Laboratory of Trauma, Burn and Combined Injuries, Medical Center of Trauma and War Injuries, Daping Hospital, Army Medical University, Chongqing, China
Background: Host-microbiota crosstalk has been implicated in multiple host metabolic pathway axes that regulate intestinal barrier function. Although constitutive cytochrome P4501A1 (CYP1A1) expression perturbs the microbiome-derived autoregulatory loop following enteric infection, little is known about the role of host CYP1A1 in modulating gut microbiome-mediated signaling during methicillin-resistant Staphylococcus aureus (MRSA)-induced abdominal sepsis and its effects on intestinal barrier integrity.
Methods: Abdominal sepsis was induced by the intraperitoneal injection of MRSA in mice. The effect of CYP1A1 deficiency on gut barrier integrity was investigated using RNA sequencing, microbiome analyses, and targeted metabolomics. The microbiota-produced metabolites were validated in patients with sepsis and persistent MRSA infection.
Results: Mice lacking CYP1A1 exhibited an altered gut microbiome, a reduced metabolic shift from lysine to cadaverine in the caecal contents and antimicrobial molecule production (Retnlb, Gbp7, and Gbp3), and they were protected against gut barrier disruption when subjected to MRSA challenge. These beneficial effects were validated in aryl hydrocarbon receptor (AHR) knockout (KO) mice by cohousing with CYP1A1 KO mice and abrogated after supplementation with cadaverine or Enterococcus faecalis, the primary microbiota genus for cadaverine synthesis. Antibiotic-driven gut dysbacteriosis impaired the survival benefit and disrupted the intestinal barrier integrity in CYP1A1 KO mice after MRSA infection. Furthermore, increased cadaverine levels in feces and serum were detected in critically ill patients with gut leakiness during persistent MRSA infection, whereas cadaverine was not detected in healthy controls. Additionally, microbiota-derived cadaverine induced enterocyte junction disruption by activating the histamine H4 receptor/nuclear factor-κB/myosin light-chain kinase signaling pathway.
Conclusion: This study revealed the unexpected function of host CYP1A1 in microbiota-mediated cadaverine metabolism, with crucial consequences for dysbacteriosis following MRSA-induced abdominal sepsis, indicating that inhibiting CYP1A1 or blocking cadaverine-histamine H4 receptor signaling could be a potential therapeutic target against abdominal sepsis.
Clinical Trial Registration: [http://www.chictr.org.cn/index.aspx], identifier [ChiCTR1800018646].
Introduction
Sepsis is a multifactorial syndrome caused by systemic or localized infections, leading to life-threatening organ dysfunction (Cheung et al., 2021). Abdominal sepsis is attributed to primary or secondary peritonitis triggered by Gram-positive or Gram-negative bacteria (Martin-Loeches et al., 2019), including methicillin-resistant Staphylococcus aureus (MRSA). Abnormal MRSA colonization in the gut markedly increases the risk of MRSA-associated enteritis, and the resistance gene can be transferred to commensal microbes (Huttner et al., 2012; Nataraj and Mallappa, 2021). Paradoxically, combination antibiotic therapy for MRSA salvage treatment will aggravate intestinal disorders. Furthermore, antibiotics do not exert their effect in a vacuum but within a complex host (Kullar et al., 2016). Thus, using novel host-based therapeutic strategies to remodel the health-promoting gut microbiota for MRSA-induced dysbacteriosis is a promising direction.
The gut microbiota composition is subject to the host genome from birth and participates in multiple host metabolic pathway axes that regulate intestinal barrier function (Nicholson et al., 2012; Sharkey et al., 2018). Gut barrier integrity is seriously disrupted during severe peritonitis-induced sepsis (Zhou and Verne, 2018). Thus, the abdominal sepsis-induced dysbiotic intestinal microenvironment leading to increased epithelial permeability and gut barrier disruption may facilitate pathogenic bacterial colonization. Specifically, as a gatekeeper against bacterial translocation, the intestinal epithelial barrier relies on enterocyte junctions to maintain integrity (Sharkey et al., 2018). However, a gap exists in understanding the host–microbiota interaction involved in regulating the intestinal epithelial cell (IEC) junctions during abdominal sepsis. Notably, constitutive expression of cytochrome P4501A1 (CYP1A1) throughout intestinal epithelial cells increases the susceptibility to enteric infection (Schiering et al., 2017), and whether the host CYP1A1–gut microbiota axis affects IEC barrier disruption against abdominal sepsis remains unclear.
Our previous study demonstrated that CYP1A1 mRNA expression levels are elevated in PBMCs of patients with sepsis, and strongly correlate with sequential organ failure assessment (SOFA) scores (Tian et al., 2020). Similarly, polymorphisms in the CYP1A1 gene partially contribute to the increased vulnerability to community-acquired pneumonia (CAP) and nosocomial pneumonia (NP) (Salnikova et al., 2013; Zhao et al., 2017). Compounds that inhibit the activity or expression of CYP1A1 exert anti-inflammatory, antitumor, anti-mycoplasma, anti-Citrobacter rodentium, and anti-parasitic effects (Westerink et al., 2008; Wincent et al., 2012; Fang et al., 2016). Furthermore, germ-free mice (microbiota deficient from birth) or those treated with antibiotics demonstrated a lower level of CYP1A1 expression (Korecka et al., 2016; Fu et al., 2017; Wang et al., 2018). Diurnal 6-formylindolo[3,2-b]carbazole/aryl hydrocarbon receptor (AHR)/CYP1A1 feedback controls the intestinal immune system to coordinate the diurnal rhythmicity of gut microbiota (Rannug, 2020). In this context, unsurprisingly, a crosstalk between CYP1A1 and the resident gut microbiome may exist. Several studies have suggested that constitutive CYP1A1 expression rapidly metabolizes AHR ligands in the gut lumen, indirectly impairing AHR activation in the intestinal immune system following enteric infection (Schiering et al., 2017; Stockinger et al., 2021). Intriguingly, we previously reported the possible AHR-independent regulation of CYP1A1 expression and CYP1A1-induced proinflammatory responses during the progression of inflammation and sepsis (Tian et al., 2020). However, we did not explore whether the interaction between CYP1A1 and microbe-mediated signaling is independent of AHR. Thus, the possibility of non-AHR-dependent action on the intestinal barrier integrity in MRSA-induced abdominal sepsis remains to be determined.
In the present study, we sought to investigate the role of host CYP1A1 as a gut flora modulator and its impact on microbiota-mediated metabolic signaling during abdominal sepsis-induced intestinal epithelial disruption. Our data showed that compared to Cyp1a1+/+ mice, Cyp1a1–/– mice showed attenuated acute MRSA-induced gut barrier disruption, partially explained by the absence of severe microbial dysbiosis and harmful gut microbial metabolites. We further demonstrated that the increased abundance of Enterococcus faecalis impaired MRSA-induced intestinal barrier dysfunction, primarily via cadaverine production and the degradation of IEC junctions. Furthermore, host CYP1A1–microbiota crosstalk involving the gut microbiome-cadaverine-enterocyte junction axis could be transferred to AHR KO mice through microbiota horizontal transmission.
Materials and Methods
Animal Experiments
Male C57BL/6 mice [wild-type (WT), 6–8 weeks old, 20–23 g] were obtained from the Experimental Animal Center of Army Medical Center. Whole-body Cyp1a1-knockout (KO) (Cyp1a1–/–) mice with a C57BL/6 background were born from heterozygotes at the Army Medical Center of PLA (Chongqing, China) transgenic animal breeding facility. The mice were separated into different cages based on their genotype (Cyp1a1+/+, Cyp1a1±, and Cyp1a1–/–) and sex after weaning. Two or three mice were housed in the same cage following same-sex and same-genotype feeding rules. Considering the existence of parental effects, littermates of the same sex and genotype were randomly assigned to different cages. Male C57BL/6 Ahr± mice were purchased from the Jackson Laboratory. The Ahr–/– mice were born from heterozygotes at our facility. To assess gut microbiota-mediated signaling in a non-AHR-dependent manner in Cyp1a1–/– mice, we performed a cohousing experiment. The Ahr–/– mice were cohoused with Cyp1a1+/+ mice and Cyp1a1–/– mice at a 1:1 ratio after weaning for 10 weeks. All the mice were housed in autoclaved cages under a 12 h light/dark cycle and 25°C conditions. The reagents used and their concentrations for the in vivo experiments were as follows: cadaverine (250 mg/kg; Sigma–Aldrich, St. Louis, MO, United States) and JNJ7777120 (150 mg/kg; Selleck, Houston, TX, United States). The mice were treated daily with 4 × 107 colony-forming units (CFUs) of E. faecalis (ATCC47077, Manassas, VA, United States) by oral gavage. The mouse experiments were approved by the Laboratory Animal Welfare and Ethics Committee of Third Military Medical University (AMUWEC20201455).
Methicillin-Resistant Staphylococcus aureus-Induced Peritonitis in a Mouse Abdominal Sepsis Model
The MRSA strain was obtained from a clinical laboratory of the Army Medical Center of PLA. The MRSA was routinely incubated in 15 ml of Luria-Bertani liquid medium (10 g/L of NaCl, 5 g/L of Yeast Extract, and 10 g/L of tryptone) on a shaker at 37°C overnight at 200 rpm. The bacterial solution was collected the next day, and each well was detected at 570 nm after doubling dilution using an enzyme-labeled instrument (BioTek, Winooski, VT, United States) to determine the concentration. The MRSA [3 × 108 CFUs/mouse] suspended in sterile saline was injected intraperitoneally into mice to establish the abdominal sepsis model.
Human Subjects
Human blood and fecal sample collection and the study procedures were approved by the Ethics Committee of Army Medical Center of PLA, China (ChiCTR1800018646), and informed consent forms were completed by all the patients and healthy volunteers. For this prospective clinical study, 18 patients were diagnosed with sepsis according to 2016 Surviving Sepsis Campaign guidelines (Rhodes et al., 2017), and 20 healthy controls were recruited after screening at the Army Medical Center of PLA from December 2019 to January 2021. All patients with sepsis met the following criteria: (1) age ≥ 18 years; (2) length of ICU stay ≥ 48 h; (3) patients with continuous indwelling catheters, and MRSA detected in specimens submitted within 48 h. Patients who abandoned the treatment were transferred to another hospital or had a clinical infectious disease (e.g., HBV and HIV infection) were excluded from this study. Healthy control subjects were enrolled in the Physical Examination OPC of Army Medical Center for blood and fecal sample collection. For all patients with sepsis and MRSA infection, a dynamic collection of samples was conducted during the infection period. Whole blood samples were quickly sent to the laboratory for serum extraction within 2 h after collection, and fecal samples were placed in liquid nitrogen immediately.
Pathogen Load Detection
The method to determine the bacterial load in the peripheral blood has been described previously (Tian et al., 2020). Briefly, blood was drawn from the caudal vein of the mice 12 h after MRSA challenge (n = 3/group) and was diluted 100-fold with 1 ml of sterile Luria-Bertani agar plates. The number of bacterial colonies, CFUs, was counted after 18 h of sterile incubation at 37°C.
Intestinal Epithelial Cell Preparation
Primary intestinal epithelial cells (IECs) were carefully isolated from mice according to the established protocol provided by Dávalos-Salas et al. (2019). Briefly, the ileum opened longitudinally were washed in cold 1 × phosphate-buffered saline (supplemented with 1 mM of phenylmethylsulfonyl fluoride and 1% of penicillin-streptomycin solution) in a 10 cm Petri dish and moved to a 50 ml centrifuge tube with tissue digestion buffer (5% of fetal bovine serum, Hanks’ Balanced Salt Solution, 15 mM of EDTA, and RNA later) by incubation in a 37°C shaker, at 200 rpm, and for 30 min. The tissues were then transferred to a preheated collagenase XI, neutral protease I, and DNase I solution in a 37°C shaker for 20 min. IEC pellets were collected after centrifugation for 5 min at 4°C at 1,200 rpm.
Ileum Transcriptome Sequencing and Analysis
Total RNA was extracted from the ileal tissues of the Cyp1a1+/+_con, Cyp1a1+/+_MRSA, Cyp1a1–/–_con, and Cyp1a1–/–_MRSA groups using TRIzol (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s instructions, and each group was prepared using four parallel replicates. RNA sequencing was performed by the Beijing Genomics Institute Sequencing and Microarray Facility using the BGIseq500 platform (BGI-Shenzhen, China). Differential expression analysis was performed using DESeq2 (v1.4.5) (Love et al., 2014) with a Q-value ≤ 0.05. To perform gene set analysis, Gene Ontology (GO)1 and Kyoto Encyclopedia of Genes and Genomes (KEGG)2 enrichment analyses of annotated differentially expressed genes (DEGs) were performed using Phyper3 based on the hypergeometric test. The significance levels of terms and pathways were corrected using the Q-value with a rigorous threshold (Q-value ≤ 0.05) and the q-value calibration package4. All the data were analyzed using the online Dr. Tom network platform of BGI5.
Ultra-High-Performance Liquid Chromatography Analysis of the Cadaverine Concentration in the Serum and Caecal Contents
All the samples from humans and mice were extracted and derivatized as reported previously (Liu et al., 2013; Levy et al., 2015) with a few modifications. Briefly, a 100 μL aliquot of the clear supernatant (or standard solution) was transferred to an Eppendorf tube and then mixed with 50 μL of 20 mg/mL of dansyl chloride in acetonitrile and 50 μL of 0.1 mol/L sodium carbonate after a 1 h incubation at 40°C in the dark. Dansyl derivatives were added to 50 μL of 0.1% of formic acid in water, vortexed for 15 s, and centrifuged at 12,000 rpm and 4°C for 10 min. An 80 μL aliquot of the clear supernatant was transferred to an autosampler vial for UHPLC–MS/MS analysis. The UHPLC separation was performed using an Agilent 1290 Infinity II series UHPLC system (Agilent Technologies, Santa Clara, CA, United States) equipped with a Waters ACQUITY UPLC HSS T3 column (100 × 2.1 mm, 1.8 μm). Mobile phase A was 10 mmol/L ammonium formate and 0.1% of formic acid in water, and mobile phase B was acetonitrile. The column temperature was set at 35°C. The autosampler temperature was set at 4°C, and the injection volume was 2 μL. An Agilent 6460 triple quadrupole mass spectrometer (Agilent Technologies, Santa Clara, CA, United States) equipped with an AJS electrospray ionization interface was applied for assay development. Agilent MassHunter Workstation Software (B.08.00; Agilent Technologies, Santa Clara, CA, United States) was used for MRM data acquisition and processing. The signal-to-noise ratios (S/Ns) were used to determine the lower limits of detection and quantitation. The precision of the quantitation was measured as the relative standard deviation, determined by injecting analytical replicates of a quality control (QC) sample. The accuracy of quantitation was evaluated by the analytical recovery of the QC samples. The percent recovery was calculated as [(mean observed concentration)/(spiked concentration)] × 100%.
Microbial Biosynthesis of Cadaverine (LdcC and CadA)
Caecal content samples were collected immediately after sacrifice in RNase-free cryotubes, snap-frozen in liquid nitrogen, and stored at −80°C. Total caecal microbial DNA was extracted using the QIAamp® Fast DNA Stool Mini Kit (51604, Inc., Duesseldorf, Germany) according to the manufacturer’s instructions. The abundance of LdcC and CadA from mouse caecal DNA samples were determined by quantitative polymerase chain reaction (qPCR). The LdcC gene was amplified using the forward primer CGGCCCTTATAACCTGCTGTTTC and reverse primer CC TTGTGCCAGATCCTGAATACG. The CadA gene was amplified using the forward primer GTCTGTGCGGCGTTATTTTTGAC and reverse primer CACCCAGCGCATATTCAAAGAAG.
16S rDNA Sequencing and Analysis
Caecal content samples were collected immediately after sacrifice in RNase-free cryotubes, snap-frozen in liquid nitrogen, and stored at −80°C. Total caecal microbial DNA was extracted using the E.Z.N.A. ®Stool DNA Kit (D4015; Omega, Inc., Alpharetta, GA, United States) according to the manufacturer’s instructions. The DNA extraction quality and concentration were detected by 1% agarose gel electrophoresis and UV spectrophotometry. The 16S rDNA genes were amplified using specific primers with barcodes on the 5′ ends targeting the V3–V4 region (Logue et al., 2016): 341 forward primer CCTACGGGNGGCWGCAG – 805 reverse primer GACTACHVGGGTATCTAATCC. The PCR products were purified using AMPure XT beads (Beckman Coulter Genomics, Danvers, MA, United States) and quantified using Qubit (Invitrogen, Carlsbad, CA, United States). The size and quantity of the amplicon library were assessed using an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, United States) and the Library Quantification Kit for Illumina (Kapa Biosystems, Woburn, MA, United States), respectively. The libraries were sequenced using the NovaSeq PE250 platform. The sequencing depth was approximately 50,000 tags. Quality filtering of the raw reads was performed under specific filtering conditions to obtain high-quality clean tags according to fqtrim (v0.94). Chimeric sequences were filtered using Vsearch software (v2.3.4). Paired-end reads were assembled using fast length adjustment of short reads (FLASH, v1.2.8). Alpha diversity and beta diversity were calculated by normalizing to the same sequences randomly using QIIME2. Next, according to the SILVA (release 132) classifier, feature abundance was normalized using the relative abundance of each sample. BLAST (NCBI, United States) was used for sequence alignment, and the feature sequences were annotated using the SILVA database for each representative sequence. The total raw read counts and processed read counts for all samples were 27,72,681 and 22,86,029, respectively.
Diamine Oxidase and Lysine Decarboxylase Activity Quantification
Serum from humans was used to detect diamine oxidase (DAO) enzyme activity using enzyme-linked immunosorbent assay (ELISA), as described by Honzawa et al. (2011). The human blood samples were kept at room temperature for 1 h before centrifugation for 20 min at 1,000 × g and were immediately stored at −80°C for subsequent quantification.
After sonication of the mouse caecal contents, the intestinal bacterial cell supernatant was used to determine lysine decarboxylase (LDC) enzyme activity using ELISA, as previously described (Lee and Cho, 2001). Feces samples (100 mg) suspended in 2.5 ml of cold phosphate-buffered saline were centrifuged at 1,000 × g for 20 min to obtain the supernatant and were centrifuged again (4,000 rpm, 20 min, 4°C) to obtain the bacterial cells. These cells resuspended in lysis solution (50 mM of Tris-HCl at pH 7.4, 2 mM of EDTA, and 100 mM of NaCl) were crushed using an ultrasonic cell disruptor (BRANSON, Branson, MO, United States) under the following conditions: 20 kHz, 150 W, and crushing for 60 s three times, 60 s apart at 4°C.
Fluorescein Isothiocyanate-Dextran Permeability Assay of the Ileum
In vivo leakage of mouse intestinal barrier function was measured using fluorescein isothiocyanate (FITC)-dextran (FD4; Sigma–Aldrich, St. Louis, MO, United States) at a 500 mg/kg dose as described previously (Natividad et al., 2018). The mice were orally administered 0.3 ml of FD4, 8 h before sacrifice, and serum samples were dynamically collected 6, 12, and 24 h after MRSA-induced sepsis. The FITC-dextran levels in serum were determined using a fluorescence microplate reader (BioTek, Winooski, VT, United States), setting an excitation wavelength of 470 nm and emission wavelength of 530 nm.
Immunofluorescence Analysis
The method for cell immunofluorescence was described previously (Zhu et al., 2018). The primary antibodies used were mouse anti-CYP1A1 (Santa Cruz; sc-25304; 1:100) and mouse anti-AHR (Gene Tex; GTX22770; 1:100). The method for ileum tissue immunofluorescence was described previously (Kumar et al., 2021). The primary antibodies used were rabbit anti-ZO-1 (Invitrogen; PA5-85256; 1:100) and rabbit anti-occludin (Invitrogen; 40-4700; 1:100). The secondary antibodies used were goat anti-Mouse IgG (H + L) highly cross-adsorbed secondary antibodies (Invitrogen; Alexa Fluor Plus 488 and 647; 1:400). All the images were captured using an FV3000 confocal microscope (Olympus, Tokyo, Japan).
Histopathological Examination
Ileum tissues were sectioned at 5 μm of thickness and stained with hematoxylin and eosin (H&E) using standard protocols. The histological scores were evaluated using a scoring system. The sections primarily contained inflammatory cell infiltrate, epithelial damage, and mucosal architecture (Martin et al., 2021).
Quantitative Reverse Transcription PCR
Total RNA was extracted using the PureLink RNA Kit (Invitrogen, Carlsbad, CA, United States; 12183018A). The cDNA was synthesized by reverse transcription using the PrimeScript RT Reagent Kit (Takara Biotechnology Co. Ltd., Tokyo, Japan). The expressions of CYP1A1, zonula occludens (ZO)-1, occludin, Retnlb, Gbp-3, Gbp-7, HRH1, HRH2, HRH3, HRH4, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNAs were amplified and quantified in triplicate using 1 μg of cDNA and SYBR Premix (TaKaRa Bio, Tokyo, Japan) and the BioRad CFX96 biosystem (Table 1).
Western Blotting
Nuclear protein and cytoplasmic protein were extracted from lysed primary IECs using an extraction kit (P0028; Beyotime, Shanghai, China) and phenylmethylsulfonyl fluoride (100 mM; ST506, Beyotime, Shanghai, China). Total proteins were isolated from the ileal tissues using the Minute™ Total Protein Extraction Kit for Intestines (Invent, Eden Prairie, MN, United States). The protein lysates prepared above were electrophoresed on 10% or 6% stain-free polyacrylamide gels (Bio-Rad, Hercules, CA, United States), electrotransferred to 0.2 μm or 0.45 μm of polyvinylidene difluoride (PVDF) membranes (Millipore, Chicago, IL, United States), incubated with the primary and secondary antibodies (cell signaling technology; 1:2000) in sequence, and subjected to signal visualization using an enhanced chemiluminescent detection system (Bio-Rad, Hercules, CA, United States). Band intensities were quantified using ImageJ software (NIH, Bethesda, MD, United States). The primary antibodies used were mouse anti-CYP1A1 (Santa Cruz; sc-25304; 1:100), rabbit anti-ZO-1 (Invitrogen; PA5-85256; 1:1000), rabbit anti-E-cadherin (Cell Signaling Technology; 3195; 1:1000), rabbit anti-occludin (Invitrogen; 40-4700; 1:1000), rabbit anti-HRH4 (Bioss; bs-10165R; 1:300), rabbit anti-MLCK (Invitrogen; PA5-79716; 1:1000), rabbit anti-phospho-MLC2 (Cell Signaling Technology; 3671; 1:1000), rabbit anti-p65 (Cell Signaling Technology; 8242; 1:1000), rabbit anti-PCNA (Cell Signaling Technology; 13110; 1:1000), rabbit anti-phospho-IκBα (Cell Signaling Technology; 2859; 1:1000), and rabbit anti-β-actin (Cell Signaling Technology; 4970; 1:1000). Secondary antibodies were purchased from Cell Signaling Technology (Danvers, MA, United States).
Statistical Analysis
The data are presented as means ± standard error (SEM) of three independent experiments. The survival rates were analyzed using the log-rank test. Multiple group comparisons were performed using a one-way analysis of variance. All statistical analyses were performed using Prism 9.0 (GraphPad Software, United States), and P-values < 0.05 were considered statistically significant.
Results
CYP1A1 Deletion Promotes Survival and Attenuates Intestinal Epithelial Dysfunction in Methicillin-Resistant Staphylococcus aureus-Induced Sepsis
We have previously demonstrated that CYP1A1 is overexpressed in peritoneal macrophages isolated from mice following lipopolysaccharide or Escherichia coli challenge (Tian et al., 2020). Similarly, the current study showed that CYP1A1 mRNA expression was altered in the ileal epithelium of MRSA-induced bacterial septic mice, increased markedly within 6 h, and then decreased (Figure 1A). Additionally, a significant increase was found in the expression of the CYP1A1 protein at 12 h post-MRSA infection (p-Mi) (Figure 1B). Consistent with these findings, the level of CYP1A1 was upregulated by confocal imaging (Figure 1C) in IECs isolated from WT mouse ileum at 12 h p-Mi relative to controls.
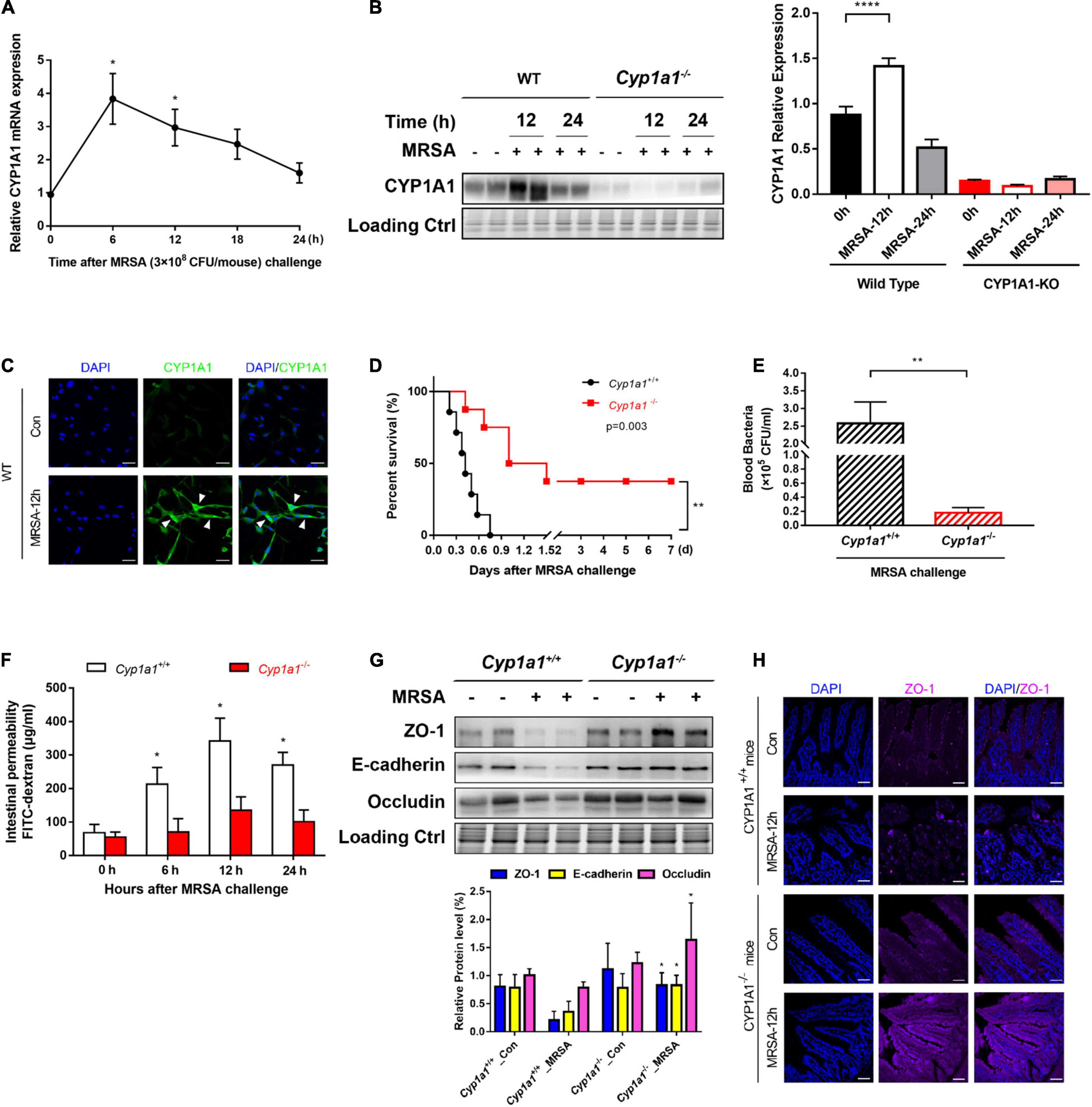
Figure 1. Cyp1a1–/– mice are protected from methicillin-resistant Staphylococcus aureus (MRSA)-induced sepsis and intestinal barrier disruption. (A) mRNA levels of CYP1A1 at 12 h post-MRSA infection (p-Mi) in the ileal epithelium of wild type (WT) mice (n = 3–4). (B) Immunoblot and densitometry plots of MRSA-induced CYP1A1 levels in the ileal epithelium from CYP1A1-knockout (Cyp1a1–/–) mice (n = 5–6) or WT mice (n = 5–6). (C) Representative immunostaining of IECs CYP1A1 (green) from WT mouse ileum, DAPI (blue) before (Control; Con) or at 12 h p-Mi (MRSA-12 h). (D) Survival of Cyp1a1+/+ (n = 10) and Cyp1a1–/– (n = 9) mice subjected to MRSA from three independent experiments. (E) Cyp1a1+/+ (n = 3) and Cyp1a1–/– (n = 3) bacterial colony forming units (CFUs) in the peripheral blood were calculated at 12 h p-Mi from two independent experiments. (F) Intestinal permeability was measured at 12 h p-Mi from three independent experiments (n = 8 mice/group). (G) Immunoblot and densitometry plots of cell junction proteins in the ileal epithelium from two mice before or 12 h p-Mi (Cyp1a1+/+ mice, n = 5–6; Cyp1a1–/– mice, n = 5–6). Differences were analyzed using one-way ANOVA. (H) Immunofluorescence staining of ileum sections of Cyp1a1+/+ mice and Cyp1a1–/– mice for ZO-1 (purple) and DAPI (blue). Scale bar: 40 μm. Error bars, ± SEM. *P < 0.05, **P < 0.01, ****P < 0.0001.
To further ascertain the role of Cyp1a1 in the development of MRSA-induced sepsis, we used Cyp1a1–/– and wild-type littermates (Cyp1a1+/+ mice). Cyp1a1+/+ littermates succumbed to sepsis, whereas Cyp1a1–/– mice showed markedly reduced mortality after MRSA challenge (hazard ratio = 4.739; 95% CI: 1.23–18.25; P = 0.003) (Figure 1D). Furthermore, compared to Cyp1a1–/– mice at 12 h p-Mi, Cyp1a1+/+ mice had a significantly higher bacterial load in the peripheral blood (P = 0.002) (Figure 1E). Considering that the intestinal mucosal barrier is the first line of defense against invading bacteria (Turner, 2009), intestinal permeability and key cell-to-cell junctions were investigated. The expression of ZO-1, occludin, and E-cadherin was significantly increased in the ileal epithelium from Cyp1a1–/– infected mice compared with that from Cyp1a1+/+ littermates (Figure 1G). Additionally, the effect observed at the cell junction level was similar to that at the damaged intestinal barrier, a finding consistent with the decrease in FD-4 in Cyp1a1+/+-infected mice (Figure 1F). Immunostaining also showed increased expression of ZO-1 in the ileum of Cyp1a1–/– infected mice (Figure 1H). These data suggest that the loss of CYP1A1 renders mice less susceptible to MRSA-induced abdominal sepsis through intestinal hypo-permeability.
Transcriptomic Changes in the Ileum Are Regulated by CYP1A1
To investigate the potential mechanisms by which Cyp1a1–/– mice facilitate enterocyte barrier improvements following MRSA-induced sepsis, RNA sequencing of ileal tissues from Cyp1a1+/+ and Cyp1a1–/– mice in the absence and presence of MRSA was performed. A total of 238 DEGs were identified between the Cyp1a1–/–_MRSA and Cyp1a1+/+_MRSA groups, comprising 65 downregulated genes and 173 upregulated genes (Figures 2A,B). The GO analysis of upregulated genes in the Cyp1a1–/–_MRSA group revealed several DEGs encoding antibacterial proteins (Figures 2C,D). This finding further indicated that a series of cell adhesion molecule (CAM)-related genes, such as Tjp1, Ocln, Cldn4, Cdh1, and Spn, were upregulated in the Cyp1a1+/+_MRSA group (Figure 2D). Similarly, our data confirmed that both the mRNA and protein levels of cell junction markers, such as ZO-1, E-cadherin, and occludin, were increased in Cyp1a1+/+-infected mice (Figure 1G), accompanied by high mRNA levels of host defense pathway molecules, such as Retnlb, Gbp7, and Gbp3 (data not shown). Furthermore, KEGG pathway analysis illustrated that these downregulated genes in the Cyp1a1–/–_MRSA group were primarily related to metabolic pathways, including lysine degradation, retinol metabolism, steroid hormone biosynthesis, and histidine metabolism (Figures 2E,F).
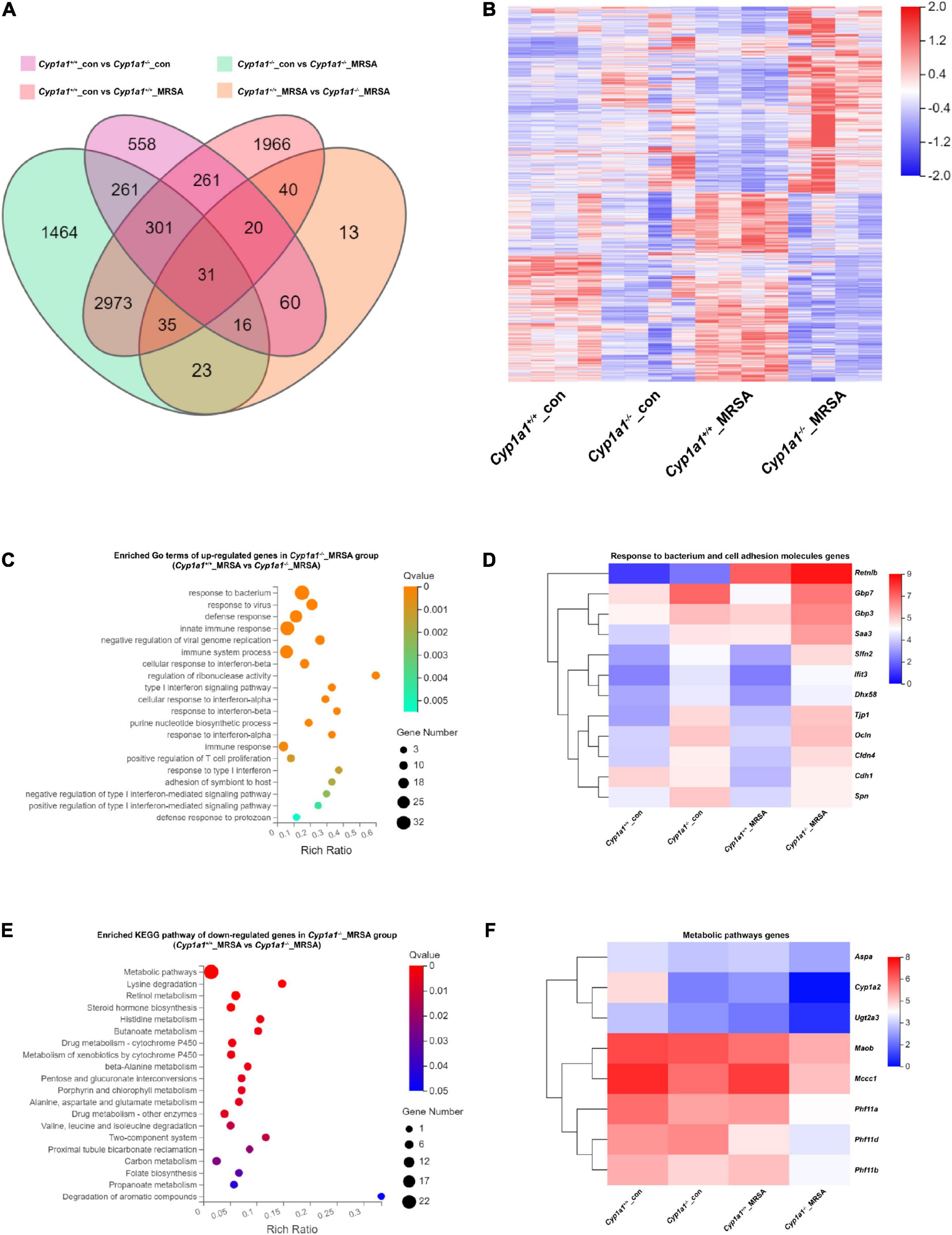
Figure 2. Ileum RNA profile comparison between Cyp1a1+/+ mice and Cyp1a1–/– mice at 12 h p-Mi. (A) Venn diagram summarizing the expression of genes in ileum tissues from Cyp1a1+/+ mice and Cyp1a1–/– mice with or without MRSA infection, as determined by RNA-seq data. (B) Heatmap of the differentially expressed genes (DEGs) in ileum tissues from mice as in (A). (C) Functional annotation of genes by the top 20 gene ontology (GO) terms upregulated in Cyp1a1–/– mice at 12 p-Mi. (D) Heatmap of the expression of the DEGs encoding bacterial response and cell adhesion molecules. (E) Functional annotation of genes by the top 20 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways downregulated in Cyp1a1–/– mice at 12 h p-Mi. (F) Heatmap of the expression of the DEGs encoding metabolic pathways.
Loss of CYP1A1 Reduces the Metabolic Shift From Lysine to Cadaverine in the Caecal Contents During Methicillin-Resistant Staphylococcus aureus-Induced Sepsis
Cadaverine is produced by the decarboxylation of lysine by the LDC enzyme via the gut microbiome (Figure 3A). To further characterize the metabolic role of CYP1A1 during MRSA-induced sepsis, the cadaverine concentration and LDC activity were measured in the caecal contents and the serum of infected Cyp1a1+/+ and Cyp1a1–/– mice by HPLC analysis and ELISA, respectively. Cadaverine levels and LDC activity in the caecal contents at 6 h p-Mi were mildly increased in Cyp1a1+/+ and Cyp1a1–/– mice. However, within 24 h, the cadaverine level gradually returned to normal in Cyp1a1–/– mice but not in Cyp1a1+/+ mice with MRSA infection (Figures 3B,D). Correspondingly, Cyp1a1–/– mice had a lower level of cadaverine in the serum following MRSA challenge than Cyp1a1+/+ mice (Figure 3C). To further address whether intestinal cadaverine biosynthesis was modified in Cyp1a1–/– mice, the abundance of the LdcC and CadA genes encoding LDC in fecal DNA was assessed. As expected, Cyp1a1–/– mice had a lower abundance of the CadA and LdcC genes than Cyp1a1+/+ mice in fecal DNA at 12 h p-Mi (Figures 3E,F). Subsequently, cadaverine pretreatment (250 mg/kg) of Cyp1a1–/– mice for 5 days impaired survival following MRSA challenge compared with that of the vehicle-treated control group; Cyp1a1+/+ mice also showed the same trend (Figure 3G). At the same time, a consistent decrease in the mRNA expression of cell junctions, such as Zo1 and higher intestinal permeability, was observed in Cyp1a1–/– mice treated with cadaverine (Figures 3H,I). These data demonstrated that cadaverine pretreatment could abolish the protective effects on the gut barrier function in Cyp1a1–/– mice.
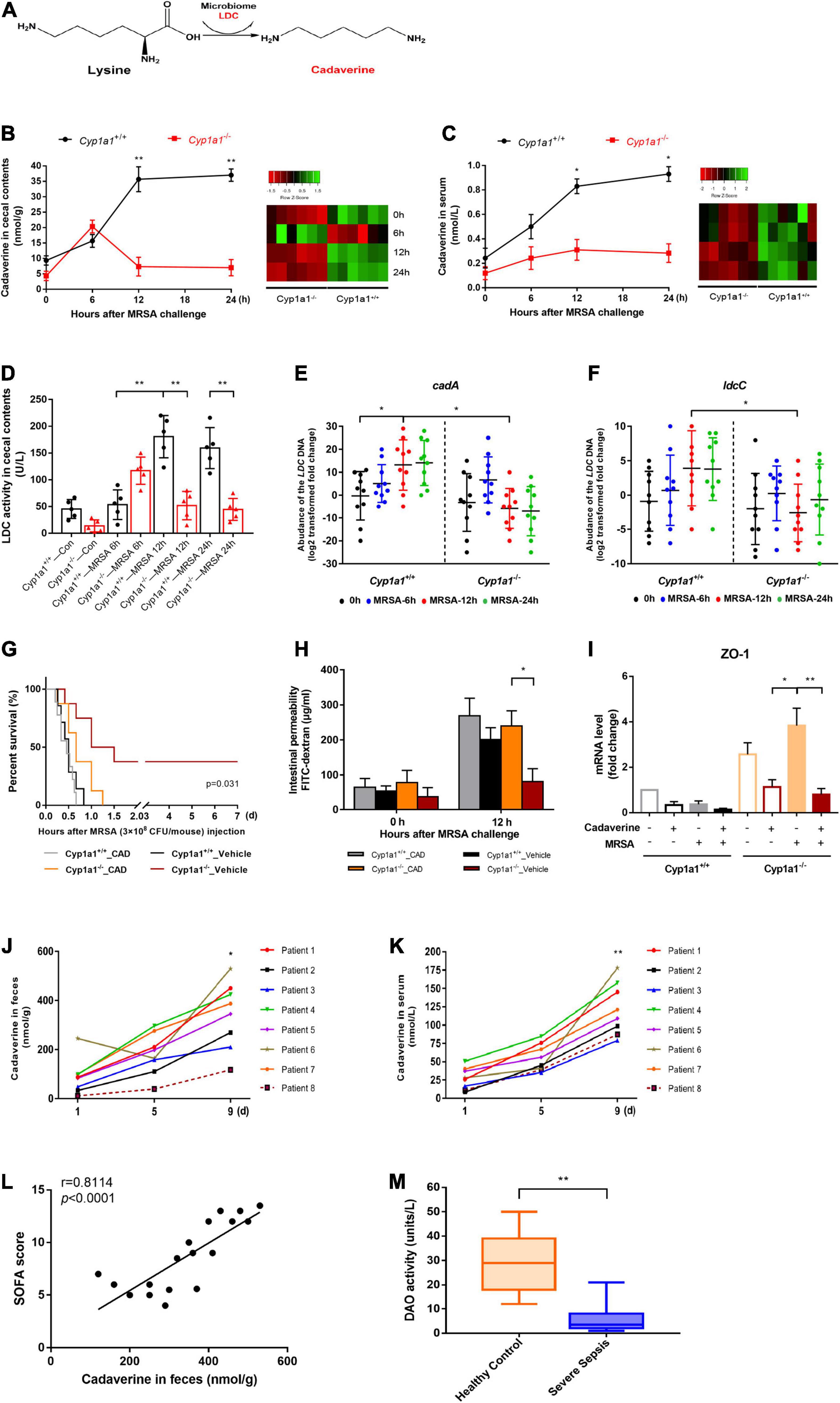
Figure 3. Caecal cadaverine is elevated during MRSA-induced sepsis. (A) Schematic diagram of cadaverine (CAD) biosynthesis from L-lysine. CAD levels in (B) the caecal contents and (C) serum (n = 9) were measured by UHPLC–MS following MRSA challenge. (D) Lysine decarboxylase (LDC) enzyme activity and RT–qPCR analysis of (E) cadA and (F) ldcC within caecal DNA samples. CAD administration to Cyp1a1–/– mice led to (G) reduced survival; (H) a higher FITC-dextran concentration in serum, and (I) reduced mRNA expression of ZO-1 in the ileal epithelium, as shown by RT–qPCR. CAD levels in (J) feces and (K) serum from patients with sepsis and MRSA infection (n = 8). (L) Correlation between the fecal CAD level and sequential organ failure assessment (SOFA) score. The Spearman correlation coefficient was used for evaluation. (M) Serum diamine oxidase (DAO) enzyme activity from patients with sepsis and MRSA infection (n = 18) and healthy controls (n = 20). Error bars, ± SEM. *P < 0.05, **P < 0.01.
The functional relevance of cadaverine levels in fecal and serum samples from patients with sepsis and MRSA infection and healthy control individuals was assessed. Similar to the mouse results, increased cadaverine levels were consistently detected in the fecal and serum samples from critically ill patients with persistent MRSA infection (Figures 3J,K), whereas cadaverine was not detected in the healthy controls. The fecal cadaverine level was further positively correlated with the SOFA score (Figure 3L). Since biopsy samples of the ileum from critically ill patients could not be easily obtained, DAO activity in serum was used to indirectly reflect the intestinal permeability of the small intestine (Honzawa et al., 2011). Serum DAO activity in patients with sepsis and MRSA infection was significantly lower than that in the healthy control subjects (Figure 3M). Collectively, the results indicate that critically ill patients with a “leaky gut” have higher cadaverine levels.
Decreased Cadaverine Levels Are Associated With an Altered Microbiome in Cyp1a1-Knockout Mice With Methicillin-Resistant Staphylococcus aureus Infection
Alterations from lysine to cadaverine profiles are closely related to the involvement of gut bacteria (Kovács et al., 2019). To correlate the specific bacterial taxa with cadaverine levels following MRSA-induced sepsis, we performed 16S ribosomal DNA sequencing and clarified the caecal microbial diversity and composition of Cyp1a1+/+ and Cyp1a1–/– mice in the presence or absence of MRSA challenge at 24 h. The unweighted UniFrac-based 3D-PCoA of all caecal microbiota revealed distinct segregation among these groups (Figure 4C, Supplementary Table 3), although no significant difference was found in the Shannon diversity of the gut flora between MRSA groups (Figure 4B, Supplementary Table 1). However, the bacterial richness was enormously decreased in Cyp1a1+/+ mice compared with that in Cyp1a1–/– mice under basal conditions (Figure 4A, Supplementary Table 1). In particular, extreme dysbiosis was found at the phylum level namely, an increase in Proteobacteria and a loss of Bacteroidetes in Cyp1a1+/+ mice compared to those in Cyp1a1–/– mice at 24 h p-Mi (Figure 4D). Notably, Cyp1a1–/– mice demonstrated significantly fewer cadaverine-producing taxa than Cyp1a1+/+ mice, as shown by the LEfSe analysis presented by a cladogram. The main differences between the Cyp1a1+/+_MRSA and Cyp1a1–/–_MRSA groups were a reduction in the abundance of E. faecalis (phylum Firmicutes), Escherichia shigella (phylum Proteobacteria), and Rikenellaceae (phylum Bacteroidetes) along with an increase in Odoribacter (phylum Bacteroidetes), Marinifilaceae (phylum Bacteroidetes), Rikenella (phylum Bacteroidetes), Intestinimonas (phylum Firmicutes), Oscillibacter (phylum Firmicutes), Verrucomicrobiae (phylum Verrucomicrobia), Ruminococcaceae (phylum Verrucomicrobia), and Lachnospiraceae (phylum Firmicutes) (Figure 4E, Supplementary Table 2). These results demonstrate that CYP1A1 deficiency plays an essential role in remodeling gut microbiota composition and reducing the abundance of opportunistic pathogens and flora imbalance following MRSA challenge.
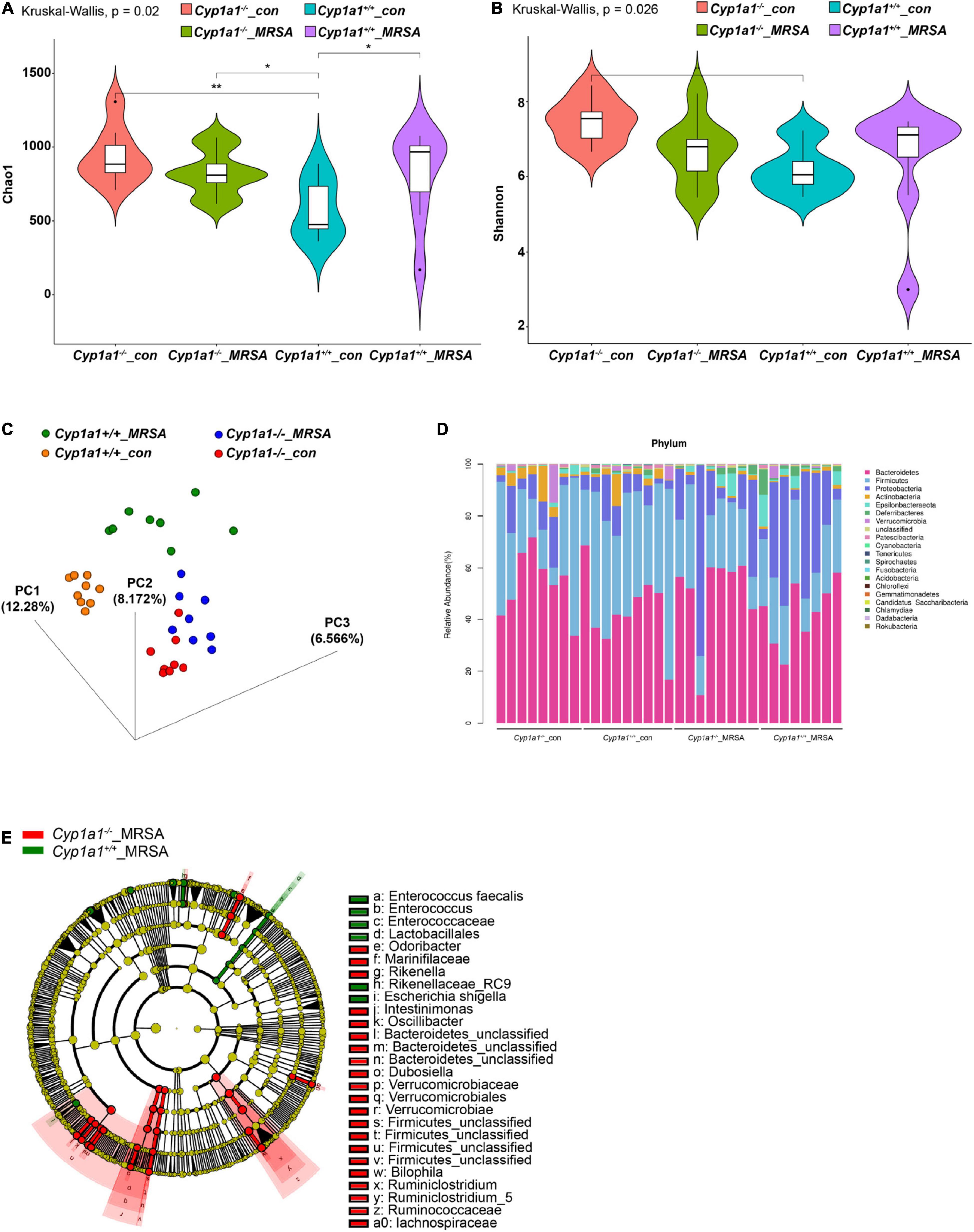
Figure 4. Cyp1a1 deficiency alters the gut microbiota composition. (A) Chao 1 index and (B) Shannon index measuring bacterial richness and diversity by 16S rDNA sequencing in the caecal content of Cyp1a1+/+ mice and Cyp1a1–/– mice with or without MRSA infection (n = 8–9). (C) The unweighted UniFrac-based 3D-PCoA showed obvious differences in the gut microbiota composition between mice in (A) (n = 8–9). (D) Bar chart of the relative microbiome abundance at the phylum level from mice as in (A). (E) The cladogram based on LEfSe analysis shows bacterial taxa (class, order, family, and genus) differentially abundant in Cyp1a1+/+ mice and Cyp1a1–/– mice with MRSA infection. Green indicates increased abundance in the Cyp1a1+/+_MRSA group; red indicates increased abundance in the Cyp1a1–/–_MRSA group. *P < 0.05, **P < 0.01.
Antibiotic Administration-Induced Intestinal Dysbacteriosis Disrupts Intestinal Barrier Integrity in Cyp1a1-Knockout Mice
To further validate whether the protective effect of Cyp1a1–/– mice is partly mediated by the gut microbiome, we administered a broad-spectrum of antibiotic cocktail (ABX) containing vancomycin (0.5 g/L; Sigma), ampicillin (1 g/L; Sigma), neomycin (1 g/L; Sigma), and metronidazole (1 g/L; Sigma) to Cyp1a1+/+ mice and Cyp1a1–/– mice twice a day for 2 weeks before abdominal sepsis modeling. Importantly, the survival benefit of Cyp1a1–/– mice was significantly lost after ABX-induced intestinal dysbacteriosis, indicating that the intact gut microbiota in Cyp1a1–/– mice are required to combat MRSA-induced abdominal sepsis (Supplementary Figure 1A). Additionally, the level of cadaverine in the caecal contents was markedly increased in ABX-treated Cyp1a1+/+ mice and Cyp1a1–/– mice compared to that in control mice that were not administered ABX (Supplementary Figure 1B). This finding agrees with the expectation that ABX perturbs gut microbiota and leads to metabolic disorders (de Jong et al., 2020). Furthermore, immunostaining analysis of ileal tissue confirmed the decrease in ZO-1 expression in Cyp1a1–/– mice that received ABX when subjected to MRSA infection (Supplementary Figure 1C). Taken together, these results further support that the ABX-driven gut microbiome perturbation to Cyp1a1–/– mice negates the survival benefit and disrupts intestinal barrier integrity after MRSA-induced sepsis.
Enterococcus faecalis Counteracts the Original Intestinal Barrier Protection in Cyp1a1-Knockout Mice With Methicillin-Resistant Staphylococcus aureus Infection
Enterococcus is one of the most cadaverine-producing commensal bacteria in the gut. Thus, its abnormal colonization can disrupt the intestinal microecology due to the accumulation of harmful microbial products (Zitvogel and Kroemer, 2019). As expected, the abundance of E. faecalis was positively correlated with the cadaverine concentration in Cyp1a1+/+ and Cyp1a1–/– mouse intestines (r = 0.9123, p < 0.0001), suggesting the role of E. faecalis in the shift from lysine production toward cadaverine production (Figure 5A and Supplementary Figures 2A,B). Since the data pointed to the abundance of E. faecalis in Cyp1a1+/+ mice, which succumbed to sepsis, we investigated the direct influence of E. faecalis treatment on gut barrier integrity. Therefore, we treated Cyp1a1+/+ mice and Cyp1a1–/– mice daily with a commercial strain of E. faecalis (4 × 107 CFU/mouse) or saline for 10 weeks by oral gavage before MRSA challenge (Figure 5B). All the animals receiving E. faecalis died within 24 h p-Mi, whereas the survival of Cyp1a1–/– mice not receiving E. faecalis was extended (Figure 5C). Furthermore, E. faecalis treatment significantly increased intestinal permeability, accompanied by a reduction in Zo1 and Ocln mRNA expression in the ileal epithelium and increased histopathology scores for the ileum in both Cyp1a1+/+ mice and Cyp1a1–/– mice following MRSA challenge (Figures 5D–H). E. faecalis treatment also effectively increased the cadaverine levels in the caecal contents from Cyp1a1+/+ and Cyp1a1–/– mice (Figure 5I). Thus, E. faecalis displays a disruptive genotype that causes gut leakiness in mice.
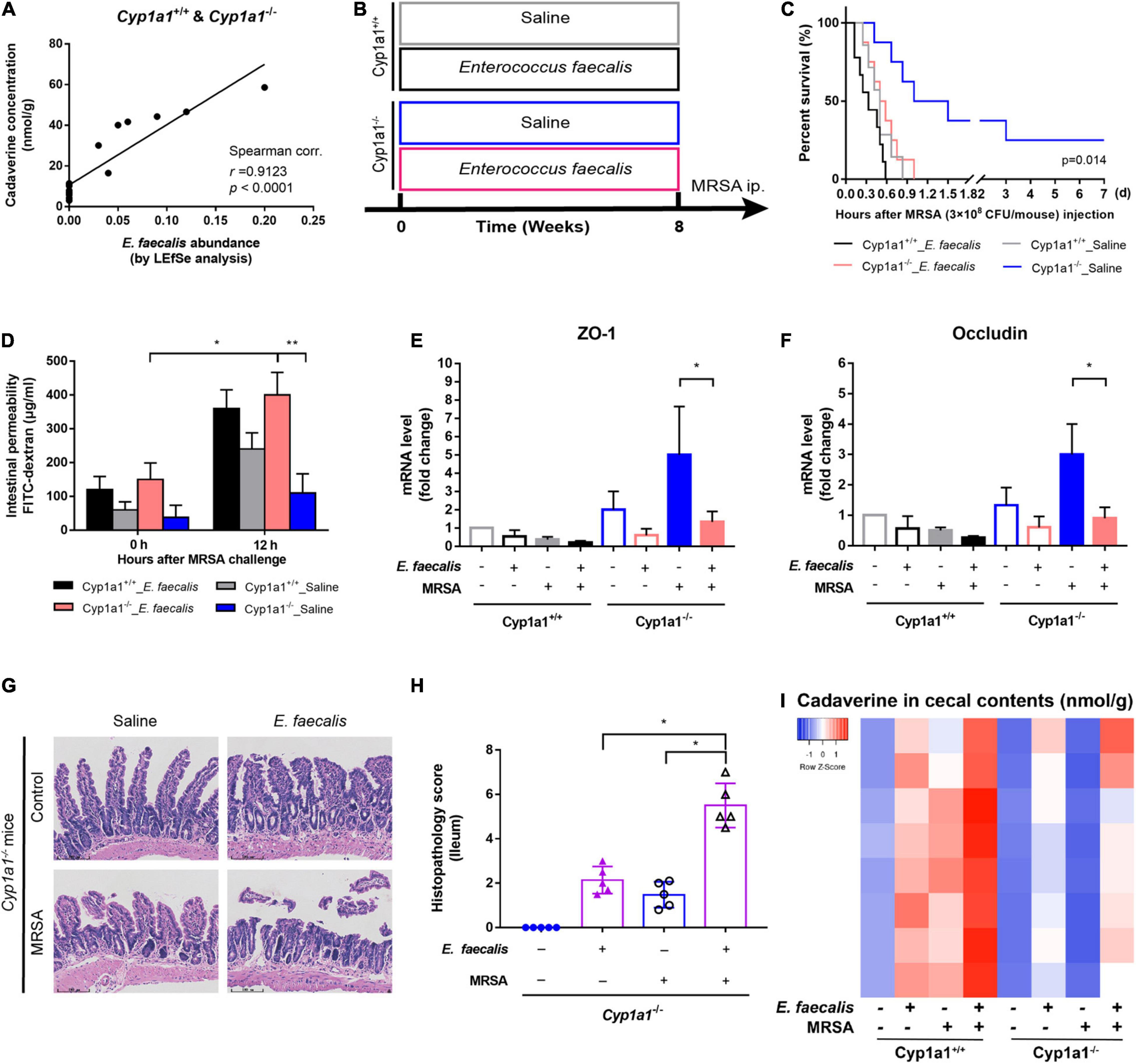
Figure 5. Cadaverine-producing Enterococcus faecalis causes gut barrier disruption and increased mortality. (A) Correlation analysis between the cadaverine (CAD) level and paired E. faecalis abundance in the caecal contents from Cyp1a1+/+ mice and Cyp1a1–/– mice with MRSA infection. (B) Cyp1a1+/+ mice and Cyp1a1–/– mice were treated daily with saline or E. faecalis (4 × 107 CFU) by oral gavage for 8 weeks. E. faecalis treatment of Cyp1a1–/– mice led to (C) increased mortality, (D) higher fluorescein isothiocyanate (FITC)-dextran concentration in serum, and reduced mRNA levels of (E) zonula occludens (ZO)-1 and (F) occludin in the ileum epithelium. (G) Representative H&E staining in the ileum from Cyp1a1–/– mice with or without E. faecalis treatment. Scale bar, 100 μm. (H) Histological scores of the ileum in each group (n = 4–5). (I) Heatmap of the caecal contents from each group. Error bars, ±SEM. *P < 0.05, **P < 0.01.
Microbiota-Derived Cadaverine Induces Enterocyte Junction Disruption by Activating the HRH4/NF-κB/Myosin Light Chain Kinase Signaling Pathway
Based on the functional screening of small molecules, cadaverine was recognized as a bacterial metabolite likely responsible for histamine receptor (HRH) family member HRH4 agonism (Colosimo et al., 2019). Consistently, only HRH4 was markedly increased in the ileal epithelium of Cyp1a1+/+ mice compared with that in Cyp1a1–/– mice following MRSA infection, with no significant changes in other receptors, such as HRH1, HRH2, and HRH3 (Figures 6A,B). Intestinal barrier loss reflects the increased permeability of the cell-to-cell tight junction due to myosin light chain kinase (MLCK) activation and myosin II regulatory light chain (MLC2) phosphorylation (Otani and Furuse, 2020). We observed increased MLCK and p-MLC2 protein expression in IECs from Cyp1a1+/+ mice at 12 h p-Mi, accompanied by a substantial decrease in the expression of ZO-1 and E-cadherin. In contrast, decreased MLCK and phospho-MLC2 expression and a concomitant increased ZO-1 and E-cadherin expression were observed in IECs from Cyp1a1–/– mice at 12 h p-Mi (Figures 6C–E). NF-κB p65 can bind to the MLCK promoter region to cause MLCK-mediated MLC phosphorylation (Lawrence, 2009; Han et al., 2017), and participates in histamine-mediated signaling (Huang et al., 2011). Therefore, we evaluated the activation of NF-κB and found that nuclear expression of p65 and cytoplasmic expression of phospho-IκBα were markedly increased in IECs from Cyp1a1+/+-infected mice, whereas Cyp1a1–/–-infected mice showed a low level of p65 translocation to the nucleus (Figures 6F,G). Furthermore, the effect of the HRH4 antagonist JNJ7777120 (150 mg/kg) on cadaverine-induced junctional protein disruption was examined. This cadaverine-induced decrease in ZO-1 and E-cadherin in the ileal epithelium was significantly reversed by JNJ7777120 in Cyp1a1+/+-infected mice (Figure 6J). HRH4 signaling blockade by JNJ7777120 also reduced the nuclear translocation of NF-κB p65 and cytosolic level of phospho-IκBα in the IECs of Cyp1a1+/+-infected mice (Figures 6H,I). Collectively, these results indicate that the binding of cadaverine to HRH4 induces gut barrier dysfunction by activating NF-κB/MLCK signaling.
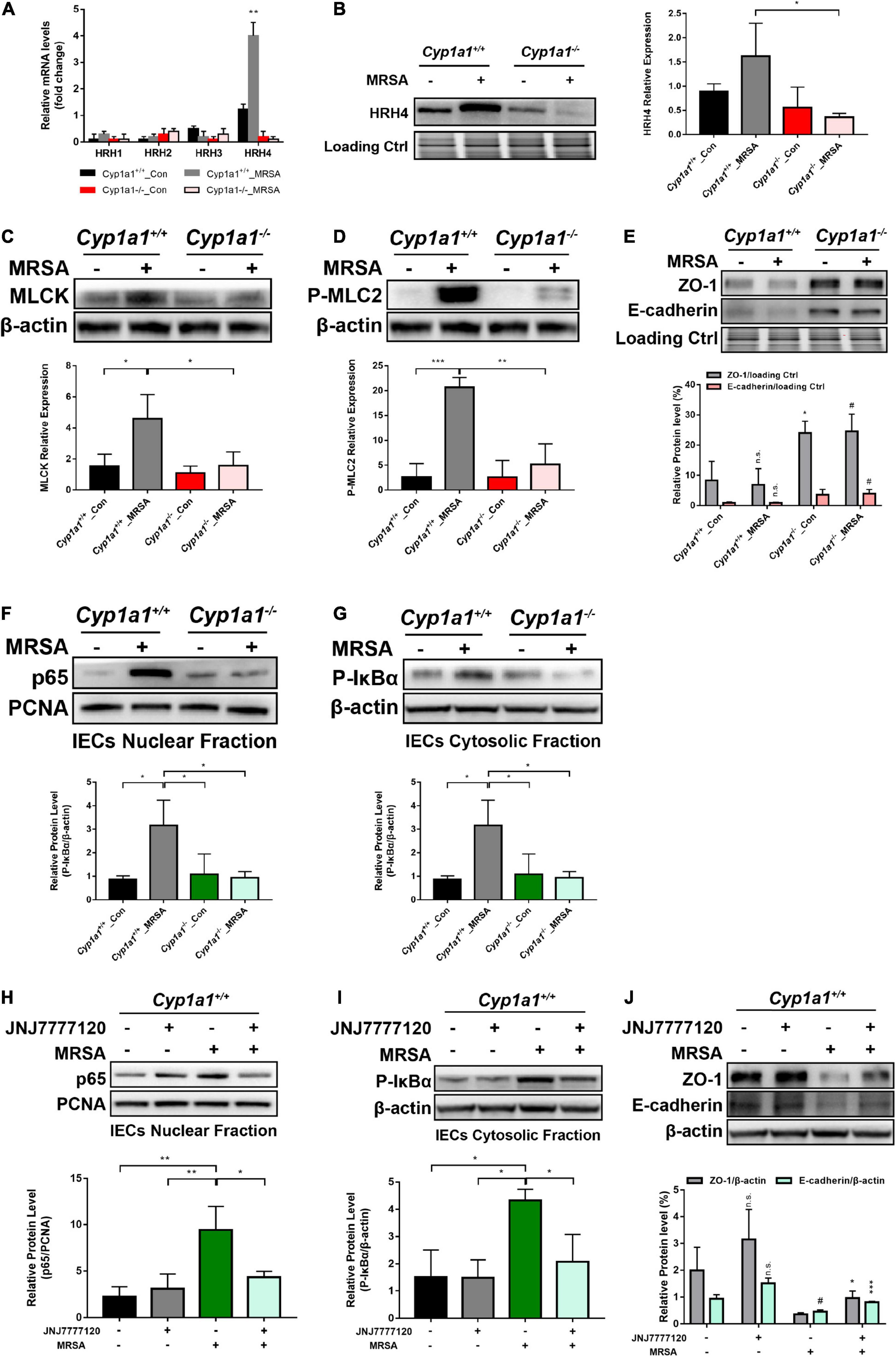
Figure 6. Microbiota-derived CAD induces gut barrier disruption through HRH4/NF-κB/MLCK activation. (A) Relative mRNA expression of HRH1, HRH2, HRH3, and HRH4 in the ileal epithelium from each group (n = 3–4). (B) Immunoblot and densitometry plots of the MRSA-induced HRH4 levels in the ileal epithelium from each group (n = 5–6). (C,D) Immunoblot and densitometry plots of MLCK (C) and P-MLC2 (D) in whole-cell lysates of IECs from each group (n = 5–6). (E) Immunoblot and densitometry plots of tight junction proteins in the ileal epithelium from each group (n = 6–7). (F,G) Immunoblot and densitometry plots of the p65 levels in the nuclear extracts (F) and those of P-IκBα levels in the cytosolic extracts (G) of IECs from two mice before or at 12 h p-Mi (Cyp1a1+/+ mice, n = 6–7; Cyp1a1–/– mice, n = 6–7). PCNA is a nuclear loading control. (H–J) Immunoblot and densitometry plots of the p65 levels in the nuclear extracts (H), P-IκBα levels in the cytosolic extracts (I), and tight junction protein levels in whole-cell lysates (J) of IECs from each group (n = 6–7). Error bars, ± SEM. Differences were analyzed using one-way ANOVA. *P < 0.05, **P < 0.01, ***P < 0.001, n.s. not significant. #Comparison between Cyp1a1+/+_MRSA and Cyp1a1–/–_MRSA groups or between JNJ7777120–MRSA+ and JNJ7777120+MRSA+ groups. #P < 0.05.
CYP1A1 Deficiency Ameliorates Gut Barrier Dysfunction Induced by Commensal Bacteria in an Aryl Hydrocarbon Receptor-Independent Manner
Since feedback control of AHR-CYP1A1 signaling is required to regulate intestinal immune functions (Schiering et al., 2017), we analyzed the expression of CYP1A1 and AHR in IECs from AHR KO and CYP1A1 KO mice. The increased level of CYP1A1 was confirmed by western blot analysis in Ahr–/– mouse IECs following MRSA challenge as early as 6 h and was restored to normal levels at 24 h p-Mi (Figure 7A). Additionally, CYP1A1 deletion did not show a prominent effect on the nuclear translocation of AHR in IECs but was slightly accumulated in the cytoplasm after MRSA infection as demonstrated by immunofluorescence assay (Figure 7B). Furthermore, pathway analysis of our RNA-seq data did not detect AHR-related pathways in Cyp1a1–/– mice with MRSA infection (Figure 2E). Previously, we found that using a CYP1A1 inhibitor improved the survival and bacterial clearance in septic mice (Tian et al., 2020). Notably, we also found that the key enterocyte junction proteins, such as ZO-1, E-cadherin, and occludin, were markedly increased in the ileal epithelium of Ahr–/– mice after treatment with bergamottin, a competitive inhibitor of CYP1A1 (Figures 7C,D).
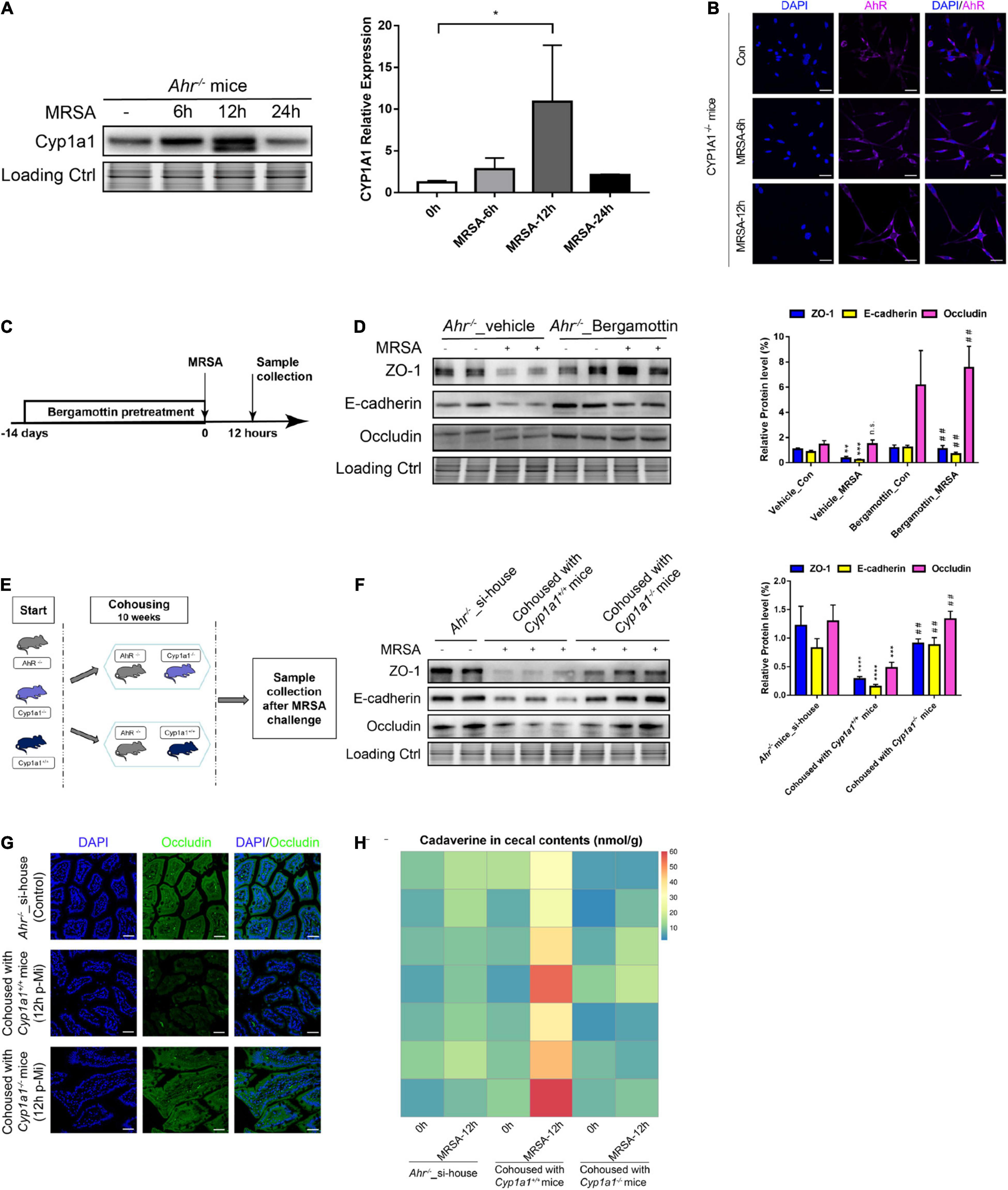
Figure 7. Gut barrier function induced by commensal bacteria in an AHR-independent manner. (A) Immunoblot and densitometry plots of MRSA-induced CYP1A1 levels in the ileal epithelium from Ahr–/– mice (n = 5–6). (B) Representative immunostaining of IEC AHR (purple) and DAPI (blue) from Cyp1a1–/– mouse ileum before (Control; Con) or 6 h (MRSA-6h) and 12 h p-Mi (MRSA-12h). (C) Experimental design. Ahr–/– mice were fed bergamottin for 14 days before the MRSA challenge. Ileum mucosa collection at 12 h p-Mi for immunoblotting. (D) Immunoblot and densitometry plots of cell junction proteins in the ileal epithelium from Ahr–/–_Vehicle mice (n = 7–8) or Ahr–/–_Bergamottin mice (n = 7–8). (E) Experimental design. Ahr–/– mice were cohoused with Cyp1a1+/+ mice and Cyp1a1–/– mice before MRSA challenge for 10 weeks. Sample collection at 12 h p-Mi. (F) Immunoblot and densitometry plots of cell junction proteins in the ileal epithelium from Ahr–/–_si-house mice (n = 7–8), Ahr–/– mice cohoused with Cyp1a1+/+ mice (n = 8–10) or Ahr–/– mice cohoused with Cyp1a1–/– mice (n = 8–10). (G) Immunofluorescence staining of ileum sections for occludin (green) and DAPI (blue) from each group. Scale bar: 40 μm. (H) Heatmap of the caecal contents from each group. Error bars, ±SEM. *P < 0.05. si-house, only Ahr–/– mice were housed in a cage; cohouse, Ahr–/– mice were housed in a cage with Cyp1a1+/+ mice or Cyp1a1–/– mice separately. Differences were analyzed using one-way ANOVA. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, n.s., not significant. ##Comparison between Ahr–/–_Vehicle-MRSA and Ahr–/–_Bergamottin-MRSA groups, or Ahr–/– mice cohoused with Cyp1a1+/+ mice and Ahr–/– mice cohoused with Cyp1a1–/– mice. ##P < 0.01.
Since the gut microbiota confers gut barrier dysfunction by metabolizing lysine to cadaverine in the gut of Cyp1a1+/+ mice under MRSA infection, we cohoused Ahr–/– mice with Cyp1a1+/+ mice and Cyp1a1–/– mice to investigate whether direct gut microbiota-mediated signaling occurs in a non-AHR dependent manner following MRSA challenge (Figure 7E). As shown in Figure 7F, increased expression of ZO-1, E-cadherin, and occludin in the ileal epithelium was detected in Ahr–/– mice after cohousing with Cyp1a1–/– mice for 10 weeks but not after cohousing with Cyp1a1+/+ mice. Additionally, immunostaining revealed similar changes in occludin expression in the ileum of Ahr–/– mice cohoused with Cyp1a1–/– mice, suggesting an improved epithelial barrier after floral transfer from Cyp1a1–/– mice (Figure 7G). Therefore, we asked whether AHR-KO mice at 10 weeks post-cohousing contributed to the deregulation of lysine degradation. The increased level of cadaverine at 12 h p-Mi in Ahr–/– mice after cohousing with Cyp1a1+/+ mice but not with Cyp1a1–/– mice suggests that unique bacterial taxa in Cyp1a1–/– mice induce events that promote cell-to-cell junctions of enterocytes (Figure 7H). These data highlight that although AHR plays a key role in safeguarding the barrier function, the microbiome-metabolite axis may govern the effect on the gut barrier in Cyp1a1–/– mice independent of AHR.
Discussion
Gut barrier dysfunction is considered both a result and cause of sepsis development (Haussner et al., 2019). For example, sepsis changes from the symbiotic intestinal microenvironment to a dysbiotic environment with a predominance of pathogenic bacteria and harmful metabolites (Fay et al., 2017; Adelman et al., 2020). Decreased production of beneficial microbial products directly interacts with IECs and impacts host health (Ghosh et al., 2021). CYP1A1 plays a critical role in regulating the inflammatory response and phagocytosis in individuals with sepsis and may modulate the gut microbiota (Obata et al., 2020; Tian et al., 2020). A recent study showed that dietary phytochemicals strengthen the epithelial barrier by inhibiting CYP1A1 and causing alterations in the microbiome composition (Rannug, 2020). The research covered in this report showed that genetic ablation of CYP1A1 contributes to intestinal barrier protection against MRSA-induced sepsis by shifting the microbiota composition to decrease cadaverine metabolite levels in a non-AHR-dependent manner. Additionally, the above gut microbiota-mediated protective effects are negated in the presence of antibiotics. Furthermore, oral supplementation with cadaverine or a commercial strain of E. faecalis counteracts the original protection conferred to Cyp1a1 KO mice with MRSA infection.
Emerging evidence suggests that polymorphisms in the CYP1A1 gene partly contribute to the increased vulnerability to CAP and NP (Salnikova et al., 2013; Zhao et al., 2017). However, the detailed effect of CYP1A1 on intestinal barrier function in sepsis caused by drug-resistant bacteria remains to be explored. Constitutive CYP1A1 overexpression results in reduced peristaltic activity in the colon and increased susceptibility to enteric infection (Schiering et al., 2017; Obata et al., 2020). Motivated by these discoveries, we analyzed the transcriptome of ileal sections by RNA-seq and identified a series of downregulated genes in the Cyp1a1–/–_MRSA group that were primarily enriched in metabolic pathways. Among these pathways, we focused on lysine degradation, whose expression was strongly downregulated by CYP1A1 deficiency. Cadaverine production from lysine degradation by commensal bacteria requires the specific bacterial enzyme, LDC. Therefore, the limiting factor may not be the substrate lysine but the presence of bacteria that produce cadaverine, particularly Enterococcus, Streptococcus, and Escherichia/Shigella (Mu et al., 2016). Notably, we described for the first time the gut microbiota in Cyp1a1 KO mice under both basal conditions and MRSA-induced sepsis conditions, confirming the previously held notion that inhibiting CYP1A1 contributes to maintaining homeostasis by changing microbiota-associated cascade signaling (Rannug, 2020).
Although numerous studies have performed phenotypic characterization of bacterial aetiologies, less is known about gut microbiome changes after MRSA-induced sepsis. The main differences we observed after MRSA-induced sepsis in this study were an increased abundance of E. faecalis and E. shigella in Cyp1a1+/+ mice, but these changes were not observed in Cyp1a1–/– mice. These findings in Cyp1a1+/+ mice are consistent with the evidence supporting that gut microbiota alteration in patients with sepsis is characterized by significant increases in the genus, Enterococcus (Wilmore et al., 2018; Liu et al., 2021). In particular, enteric dysbiosis dominated by Enterococcus has been reported to be a high-risk factor for mortality among patients with sepsis (Adelman et al., 2020; Agudelo-Ochoa et al., 2020). Furthermore, the reduction in helpful metabolites, such as butyrate, a health-promoting effector mediating anti-inflammatory signals in the intestine, has been specifically associated with the presence of Enterococcus species (Wolff et al., 2018; Agudelo-Ochoa et al., 2020). These data suggest that opportunistic pathogens are increased while health-promoting bacteria and metabolites are reduced following MRSA infection. The loss of CYP1A1, which protects against bacterial sepsis, limits the abundance of E. faecalis and increases taxa associated with metabolic health, such as Intestinimonas butyriciproducens, under MRSA challenge. Although the precise mechanisms by which host genetics influence the composition of gut microbes must be fully elucidated, our data suggest a dynamic host–microbiome relationship within CYP1A1 and specific taxa. Additional work on genome-wide association is required to understand how CYP1A1 deficiency modulates the gut microbiota against MRSA-induced sepsis.
Additionally, mice fed with the same diet have different microbial metabolites, likely explaining interindividual differences in the gut microbiome (Smits et al., 2016). Using UHPLC–MS analysis, we found that the gut microbial metabolite cadaverine was accumulated in Cyp1a1+/+ mice with MRSA infection due to the abundance of E. faecalis. Genetic deletion of CYP1A1 not only suppressed E. faecalis but also reduced the level of cadaverine after MRSA challenge. Cadaverine was originally identified as a death-associated odor (Hussain et al., 2013) and a harmful metabolite in the gut. The supplementation of cadaverine in Cyp1a1–/– mice with MRSA increased mortality and disrupted cell junctions, suggesting that the high level of cadaverine was harmful to the maintenance of host health. In parallel, this phenomenon did not appear to be limited to mice, as higher cadaverine levels were also detected in fecal and serum samples from patients with sepsis and persistent MRSA infection but absent in healthy volunteers. Additionally, serum DAO activity in patients with sepsis and MRSA infection was significantly lower than that in healthy controls, indicating that critically ill patients with gut leakiness had higher cadaverine levels. Furthermore, the elevated cadaverine level in feces was positively correlated with the SOFA score in these patients. Despite the limited number of participants, our findings support the view that patients with traumatic brain injuries, severe liver insufficiency, and Crohn’s disease have higher cadaverine levels (Pelletti et al., 2019; Diederen et al., 2020).
Excess antibiotic use in critically ill patients can also lead to severe microbial dysbiosis due to the combined effect of detrimental metabolite production and intestinal microenvironment disturbance. Recently, cadaverine had a dose-dependent cytotoxic effect toward HT29 intestinal cells in in vitro cultures (Del Rio et al., 2019); however, the specific mechanism underlying this effect remains unknown. Further study revealed that cadaverine might be responsible for HRH4 agonism (Colosimo et al., 2019). Consistently, a higher level of cadaverine in Cyp1a1+/+ infected mice had a high affinity for HRH4 compared with that in Cyp1a1–/– mice. Although few studies have described the effects of cadaverine and its receptors on the epithelial function, the efficacy of antihistamines in the airway epithelial barrier dysfunction might reflect HRH4-mediated actions (De Benedetto et al., 2015). In addition to the activation of the MAPK and JAK/STAT signaling pathways by HRH4 (Morse et al., 2001; Horr et al., 2006; Michel et al., 2008), we observed that the cadaverine-HRH4 interaction activated classic NF-κB signaling, which facilitated MLCK-mediated major TJ protein (ZO-1 and occludin) and AJ protein (E-cadherin) disruption in Cyp1a1+/+ mice with MRSA infection. Thus, the cadaverine-induced signaling cascade is complex, and the heterogeneity in histamine receptors leads to the activation of multiple downstream signaling pathways (De Benedetto et al., 2015). The potential ability to suppress CYP1A1 or inhibit intestinal cadaverine-HRH4 signaling to enhance IEC junctions, or a combination of these two strategies, warrants further study to develop more effective sepsis treatment.
Interestingly, another novel finding from this study was that CYP1A1 KO-induced alterations in the intestinal flora and metabolites reduced the disruption of IEC junctions in mice, an effect that was transmissible by cohousing with Ahr–/– mice. The AHR plays a protective role in regulating homeostatic processes at barrier sites, particularly in the intestinal microenvironment (Stockinger et al., 2021). Accordingly, AHR-deficient mice exhibit enhanced susceptibility to severe colitis and Citrobacter rodentium infection (Li et al., 2011; Rannug, 2020). Although genetic deletion of CYP1A1 delays natural ligand metabolism, resulting in increased AHR signaling (Schiering et al., 2017), the nuclear translocation of AHR was not induced in the IECs of ileum from Cyp1a1–/– mice under MRSA challenge in the present study. Two potential reasons could explain this finding: (1) this action may be enabled by the AHR non-genomic pathway rather than the classic genomic pathway, which is primarily characterized by AHR nuclear translocation, or (2) IECs act as sensors for AHR ligands and then transmit signals across the epithelial layer to mucosal immune cells (Schiering et al., 2017; Stockinger et al., 2021). In the absence of gut bacteria that produce AHR ligands or a lack of endogenous ligands, a quasi-AHR-deficient state could exist persistently unless exogenous dietary supplements are used (Pernomian et al., 2020). We cohoused AHR-KO and CYP1A1-KO mice to directly explore the impact of unique metabolites generated by intestinal bacteria on gut barrier function, regardless of host genetic defects in AHR. Specific bacterial taxa exchanged horizontally from Cyp1a1+/+ mice produced a higher level of cadaverine with increased intestinal permeability and gut barrier disruption after MRSA challenge, whereas these effects were significantly reversed in Ahr–/– mice cohoused with Cyp1a1–/– mice. Consistent with human studies, the degree of horizontal transmission predicted bacterial genera with pathogenic representatives responsible for enteric infection (Moeller et al., 2018). Furthermore, these results provide indications for the future: (1) Donors need equal and careful screening before clinical fecal microbiota transplantation (FMT) in sepsis. Although FMT has been applied in patients with sepsis to correct dysbiosis, recent studies have reported that E. coli infections acquired via FMT can cause death (DeFilipp et al., 2019). Given the gut leakiness observed in Ahr–/– mice after cohousing with Cyp1a1+/+ mice, genetic polymorphisms, such as CYP1A1, may need to be examined in donors before FMT. (2) Although the present study demonstrates novel AHR-independent effects on the microbial-metabolite axis, it does not rule out a significant role of AHR in coordinating the intestinal epithelium and mucosal immune system via an unidentified mechanism. Therefore, further study will be required to dissect the effect of host–microbe interactions on the mucosal barrier using specific deletion of AHR and CYP1A1 in the intestinal epithelial cells from transgenic mice.
However, this study has limitations that should be mentioned. We only investigated the impact of the whole-body deletion of CYP1A1 on microbiota-mediated metabolic signaling during abdominal sepsis-induced intestinal epithelial disruption. Since enteric stromal and immune cells in the gut also express CYP1A1, we could not rule out the possibility that these cells might also contribute to the host–microbiome crosstalk. Therefore, a mouse model of selective CYP1A1 deletion in IECs will be needed to fully demonstrate the role of the CYP1A1-microbiota metabolic axis in sepsis-induced epithelial barrier dysfunction. Additionally, primary IECs from patients with sepsis are difficult to obtain in clinical practice, and the usage of human intestinal organoid culture model seems to be a better experimental solution in follow-up studies. Although the present study suggests that host CYP1A1 plays a direct or indirect role in microbiota-mediated cadaverine metabolism during MRSA-induced sepsis, other mechanisms may exist, with or without the involvement of the microbiome. Finally, due to the fragile gut microbial equilibrium in patients with severe sepsis, further studies are needed to clarify whether a similar mechanism participates in patients with sepsis and MRSA infection.
Conclusion
This study reveals an unexpected mechanism of host CYP1A1 in microbiota-mediated cadaverine metabolism during MRSA-induced abdominal sepsis, through which host–microbiota metabolic axes directly affect enterocyte junction disruption in an AHR-independent manner. Therefore, suppressing CYP1A1 or blocking intestinal cadaverine-HRH4 signaling by modulating host–microbiome crosstalk represents a promising strategy to maintain gut barrier homeostasis to treat abdominal sepsis.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found here: Accession to cite for transcriptomics SRA data: PRJNA805025. Accession to cite for 16S rDNA SRA data: PRJNA804256.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethics Committee of Army Medical Center of PLA. The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by Laboratory Animal Welfare and Ethics Committee of Third Military Medical University.
Author Contributions
HL and XM conceived and designed the study. XM, HJ, and XC conducted most of the experiments and wrote the manuscript. WD and WT helped with protein extraction and western blotting analysis. GL and JZ assisted with cellular immunofluorescence. XuY provided three models of transgenic mice. FW and WL helped with animal experiments. XiY assisted with clinical data collection. HL interpreted the data and supervised the work. All authors read and approved the final manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (81871612), Chinese Basic Science Program (19SWAQ18), Chinese Science and Technology Basic Enhancement Program (2021GFKJ01), and 2020 Chongqing Talent Program (2020CQYC01). The funders did not have any role in the study design, data collection and analyzes, or writing this manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Mercedes Dávalos-Salas (Olivia Newton-John Cancer Research Institute, Australia) for technical support on IEC preparation. We thank Marinda Ma (Wageningen University & Research, Netherlands) for manuscript drafting instruction and language review. We also thank Shanghai BIOTREE Biological Technology Co., Ltd. (Shanghai, China) for metabolomics analysis and LC-Bio Technology Co., Ltd. (Hangzhou, China) for 16S rDNA analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.802409/full#supplementary-material
Supplementary Figure 1 | Antibiotic administration reverses the protection of intestinal barrier integrity in Cyp1a1-knockout mice. (A) Survival of Cyp1a1+/+ (n = 8) and Cyp1a1–/– (n = 8) mice in the presence or absence of antibiotic cocktail (ABX) and subjected to MRSA from two independent experiments (*P < 0.05; log-rank analysis). (B) Heatmap of the caecal contents from each group. (C) Immunofluorescence staining of ileum sections for ZO-1 (green) and DAPI (blue) from each group (scale bar: 50 μm); white boxes indicate the magnified areas (scale bar: 20 μm).
Supplementary Figure 2 | Spearman’s correlation analysis. Correlation analysis between the CAD level and paired E. faecalis abundance in the caecal contents from Cyp1a1+/+ mice (A) and Cyp1a1–/– mice (B) with MRSA infection.
Supplementary Table 1 | Alpha diversity between Cyp1a1+/+ and Cyp1a1–/– mice in the presence or absence of MRSA challenge at 24 h.
Supplementary Table 2 | Features profiling between Cyp1a1+/+ and Cyp1a1–/– mice in the presence or absence of MRSA challenge at 24 h.
Supplementary Table 3 | The abundance-unweighted Unifrac distance matrix between Cyp1a1+/+ and Cyp1a1–/– mice in the presence or absence of MRSA challenge at 24 h.
Abbreviations
ABX, antibiotic cocktail; AHR, aryl hydrocarbon receptor; CAMs, cell adhesion molecules; CAP, community-acquired pneumonia; CFU, colony-forming unit; CYP1A1 cytochrome P450family 1, subfamily A, polypeptide 1; DAO, diamine oxidase; DEGs, differentially expressed genes; ELISA, enzyme-linked immunosorbent assay; FITC, fluorescein isothiocyanate; FMT, fecal microbiota transplantation; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GO, gene ontology; H&E, hematoxylin and eosin; HRH, histamine receptor; IECs, intestinal epithelial cells; KEGG, Kyoto encyclopedia of genes and genomes; KO, knockout; LDC, lysine decarboxylase; MLCK, myosin light chain kinase; MLC2, myosin II regulatory light chain; MODS, multiple organ dysfunction syndrome; MRSA, methicillin-resistant Staphylococcus aureus; NP, nosocomial pneumonia; p-Mi, post-MRSA infection; QC, quality control; qPCR, quantitative polymerase chain reaction; SEM, standard error; SOFA, sequential organ failure assessment; UHPLC, ultra-high-performance liquid chromatography; WT, wild type; ZOs, zonula occludens.
Footnotes
- ^ http://www.geneontology.org/
- ^ https://www.kegg.jp/
- ^ https://en.wikipedia.org/wiki/Hypergeometric_distribution
- ^ https://bioconductor.org/packages/release/bioc/html/qvalue.html
- ^ http://report.bgi.com
References
Adelman, M. W., Woodworth, M. H., Langelier, C., Busch, L. M., Kempker, J. A., Kraft, C. S., et al. (2020). The gut microbiome’s role in the development, maintenance, and outcomes of sepsis. Crit. Care 24:278. doi: 10.1186/s13054-020-02989-1
Agudelo-Ochoa, G. M., Valdés-Duque, B. E., Giraldo-Giraldo, N. A., Jaillier-Ramírez, A. M., Giraldo-Villa, A., Acevedo-Castaño, I., et al. (2020). Gut microbiota profiles in critically ill patients, potential biomarkers and risk variables for sepsis. Gut Microbes 12:1707610. doi: 10.1080/19490976.2019.1707610
Cheung, G., Bae, J. S., Liu, R., Hunt, R. L., Zheng, Y., and Otto, M. (2021). Bacterial virulence plays a crucial role in MRSA sepsis. PLoS Pathog. 17:e1009369. doi: 10.1371/journal.ppat.1009369
Colosimo, D. A., Kohn, J. A., Luo, P. M., Piscotta, F. J., Han, S. M., Pickard, A. J., et al. (2019). Mapping interactions of microbial metabolites with human G-protein-coupled receptors. Cell Host Microbe 26, 273–282.e7. doi: 10.1016/j.chom.2019.07.002
Dávalos-Salas, M., Montgomery, M. K., Reehorst, C. M., Nightingale, R., Ng, I., Anderton, H., et al. (2019). Deletion of intestinal Hdac3 remodels the lipidome of enterocytes and protects mice from diet-induced obesity. Nat. Commun. 10:5291. doi: 10.1038/s41467-019-13180-8
De Benedetto, A., Yoshida, T., Fridy, S., Park, J. E., Kuo, I. H., and Beck, L. A. (2015). Histamine and skin barrier: are histamine antagonists useful for the prevention or treatment of atopic dermatitis. J. Clin. Med. 4, 741–755. doi: 10.3390/jcm4040741
de Jong, S. E., Olin, A., and Pulendran, B. (2020). The impact of the microbiome on immunity to vaccination in humans. Cell Host Microbe 28, 169–179. doi: 10.1016/j.chom.2020.06.014
DeFilipp, Z., Bloom, P. P., Torres Soto, M., Mansour, M. K., Sater, M., Huntley, M. H., et al. (2019). Drug-resistant E. coli bacteremia transmitted by fecal microbiota transplant. N. Engl. J. Med. 381, 2043–2050.
Del Rio, B., Redruello, B., Linares, D. M., Ladero, V., Ruas-Madiedo, P., Fernandez, M., et al. (2019). The biogenic amines putrescine and cadaverine show in vitro cytotoxicity at concentrations that can be found in foods. Sci. Rep. 9:120.
Diederen, K., Li, J. V., Donachie, G. E., de Meij, T. G., de Waart, D. R., Hakvoort, T., et al. (2020). Exclusive enteral nutrition mediates gut microbial and metabolic changes that are associated with remission in children with Crohn’s disease. Sci. Rep. 10:18879. doi: 10.1038/s41598-020-75306-z
Fang, X., Zhao, W., Xu, J., Tu, F., Wang, X., Li, B., et al. (2016). CYP1A1 mediates the suppression of major inflammatory cytokines in pulmonary alveolar macrophage (PAM) cell lines caused by Mycoplasma hyponeumoniae. Dev. Comp. Immunol. 65, 132–138. doi: 10.1016/j.dci.2016.06.023
Fay, K. T., Ford, M. L., and Coopersmith, C. M. (2017). The intestinal microenvironment in sepsis. Biochim. Biophys. Acta Mol. Basis Dis. 1863, 2574–2583. doi: 10.1016/j.bbadis.2017.03.005
Fu, Z. D., Selwyn, F. P., Cui, J. Y., and Klaassen, C. D. (2017). RNA-Seq profiling of intestinal expression of xenobiotic processing genes in germ-free mice. Drug Metab. Dispos. 45, 1225–1238. doi: 10.1124/dmd.117.077313
Ghosh, S., Whitley, C. S., Haribabu, B., and Jala, V. R. (2021). Regulation of intestinal barrier function by microbial metabolites. Cell. Mol. Gastroenterol. Hepatol. 11, 1463–1482.
Han, J., Li, J. H., Bai, G., Shen, G. S., Chen, J., Liu, J. N., et al. (2017). Acanthopanax senticosus polysaccharides-induced intestinal tight junction injury alleviation via inhibition of NF-κB/MLCK pathway in a mouse endotoxemia model. World J. Gastroenterol. 23, 2175–2184. doi: 10.3748/wjg.v23.i12.2175
Haussner, F., Chakraborty, S., Halbgebauer, R., and Huber-Lang, M. (2019). Challenge to the intestinal mucosa during sepsis. Front. Immunol. 10:891. doi: 10.3389/fimmu.2019.00891
Honzawa, Y., Nakase, H., Matsuura, M., and Chiba, T. (2011). Clinical significance of serum diamine oxidase activity in inflammatory bowel disease: importance of evaluation of small intestinal permeability. Inflamm. Bowel Dis. 17, E23–E25. doi: 10.1002/ibd.21588
Horr, B., Borck, H., Thurmond, R., Grösch, S., and Diel, F. (2006). STAT1 phosphorylation and cleavage is regulated by the histamine (H4) receptor in human atopic and non-atopic lymphocytes. Int. Immunopharmacol. 6, 1577–1585. doi: 10.1016/j.intimp.2006.06.005
Huang, W. C., Chai, C. Y., Chen, W. C., Hou, M. F., Wang, Y. S., Chiu, Y. C., et al. (2011). Histamine regulates cyclooxygenase 2 gene activation through Orai1-mediated NFκB activation in lung cancer cells. Cell Calcium 50, 27–35. doi: 10.1016/j.ceca.2011.04.004
Hussain, A., Saraiva, L. R., Ferrero, D. M., Ahuja, G., Krishna, V. S., Liberles, S. D., et al. (2013). High-affinity olfactory receptor for the death-associated odor cadaverine. Proc. Natl. Acad. Sci. U.S.A. 110, 19579–19584. doi: 10.1073/pnas.1318596110
Huttner, B., Robicsek, A. A., Gervaz, P., Perencevich, E. N., Schiffer, E., Schrenzel, J., et al. (2012). Epidemiology of methicillin-resistant Staphylococcus aureus carriage and MRSA surgical site infections in patients undergoing colorectal surgery: a cohort study in two centers. Surg. Infect. 13, 401–405. doi: 10.1089/sur.2011.107
Korecka, A., Dona, A., Lahiri, S., Tett, A. J., Al-Asmakh, M., Braniste, V., et al. (2016). Bidirectional communication between the aryl hydrocarbon receptor (AhR) and the microbiome tunes host metabolism. NPJ Biofilms Microbiomes 2:16014. doi: 10.1038/npjbiofilms.2016.14
Kovács, T., Mikó, E., Vida, A., Sebő, É., Toth, J., Csonka, T., et al. (2019). Cadaverine, a metabolite of the microbiome, reduces breast cancer aggressiveness through trace amino acid receptors. Sci. Rep. 9:1300. doi: 10.1038/s41598-018-37664-7
Kullar, R., Sakoulas, G., Deresinski, S., and van Hal, S. J. (2016). When sepsis persists: a review of MRSA bacteraemia salvage therapy. J. Antimicrob. Chemother. 71, 576–586. doi: 10.1093/jac/dkv368
Kumar, A., Priyamvada, S., Ge, Y., Jayawardena, D., Singhal, M., Anbazhagan, A. N., et al. (2021). A novel role of SLC26A3 in the maintenance of intestinal epithelial barrier integrity. Gastroenterology 160, 1240–1255.e3. doi: 10.1053/j.gastro.2020.11.008
Lawrence, T. (2009). The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb. Perspect. Biol. 1:a001651.
Lee, Y. S., and Cho, Y. D. (2001). Identification of essential active-site residues in ornithine decarboxylase of Nicotiana glutinosa decarboxylating both L-ornithine and L-lysine. Biochem. J. 360, 657–665. doi: 10.1042/0264-6021:3600657
Levy, M., Thaiss, C. A., Zeevi, D., Dohnalová, L., Zilberman-Schapira, G., Mahdi, J. A., et al. (2015). Microbiota-modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell 163, 1428–1443. doi: 10.1016/j.cell.2015.10.048
Li, Y., Innocentin, S., Withers, D. R., Roberts, N. A., Gallagher, A. R., Grigorieva, E. F., et al. (2011). Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell 147, 629–640. doi: 10.1016/j.cell.2011.09.025
Liu, R., Li, Q., Ma, R., Lin, X., Xu, H., and Bi, K. (2013). Determination of polyamine metabolome in plasma and urine by ultrahigh performance liquid chromatography-tandem mass spectrometry method: application to identify potential markers for human hepatic cancer. Anal. Chim. Acta 791, 36–45. doi: 10.1016/j.aca.2013.06.044
Liu, W., Cheng, M., Li, J., Zhang, P., Fan, H., Hu, Q., et al. (2021). Classification of the gut microbiota of patients in intensive care units during development of sepsis and septic shock. Genomics Proteomics Bioinformatics. 18, 696–707. doi: 10.1016/j.gpb.2020.06.011
Logue, J. B., Stedmon, C. A., Kellerman, A. M., Nielsen, N. J., Andersson, A. F., Laudon, H., et al. (2016). Experimental insights into the importance of aquatic bacterial community composition to the degradation of dissolved organic matter. ISME J. 10, 533–545. doi: 10.1038/ismej.2015.131
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Martin, O., Bergonzini, A., Lopez Chiloeches, M., Paparouna, E., Butter, D., Theodorou, S., et al. (2021). Influence of the microenvironment on modulation of the host response by typhoid toxin. Cell Rep. 35:108931. doi: 10.1016/j.celrep.2021.108931
Martin-Loeches, I., Timsit, J. F., Leone, M., de Waele, J., Sartelli, M., Kerrigan, S., et al. (2019). Clinical controversies in abdominal sepsis. Insights for critical care settings. J. Crit. Care 53, 53–58. doi: 10.1016/j.jcrc.2019.05.023
Michel, I., Borck, H., McElligott, S., Krieg, C., and Diel, F. (2008). Histamine receptor H4R-selective ligands influence the STAT6 transcription activation domain (TAD) and the DNA-binding. Inflamm. Res. 57(Suppl. 1) S47–S48. doi: 10.1007/s00011-007-0623-1
Moeller, A. H., Suzuki, T. A., Phifer-Rixey, M., and Nachman, M. W. (2018). Transmission modes of the mammalian gut microbiota. Science 362, 453–457. doi: 10.1126/science.aat7164
Morse, K. L., Behan, J., Laz, T. M., West, R. E. Jr., Greenfeder, S. A., Anthes, J. C., et al. (2001). Cloning and characterization of a novel human histamine receptor. J. Pharmacol. Exp. Ther. 296, 1058–1066.
Mu, C., Yang, Y., Luo, Z., Guan, L., and Zhu, W. (2016). The colonic microbiome and epithelial transcriptome are altered in rats fed a high-protein diet compared with a normal-protein diet. J. Nutr. 146, 474–483. doi: 10.3945/jn.115.223990
Nataraj, B. H., and Mallappa, R. H. (2021). Antibiotic resistance crisis: an update on antagonistic interactions between probiotics and methicillin-resistant Staphylococcus aureus (MRSA). Curr. Microbiol. 78, 2194–2211. doi: 10.1007/s00284-021-02442-8
Natividad, J. M., Agus, A., Planchais, J., Lamas, B., Jarry, A. C., Martin, R., et al. (2018). Impaired aryl hydrocarbon receptor ligand production by the gut microbiota is a key factor in metabolic syndrome. Cell Metab. 28, 737–49e4. doi: 10.1016/j.cmet.2018.07.001
Nicholson, J. K., Holmes, E., Kinross, J., Burcelin, R., Gibson, G., Jia, W., et al. (2012). Host-gut microbiota metabolic interactions. Science 336, 1262–1267.
Obata, Y., Castaño, Á., Boeing, S., Bon-Frauches, A. C., Fung, C., Fallesen, T., et al. (2020). Neuronal programming by microbiota regulates intestinal physiology. Nature 578, 284–289. doi: 10.1038/s41586-020-1975-8
Otani, T., and Furuse, M. (2020). Tight junction structure and function revisited. Trends Cell Biol. 30, 805–817.
Pelletti, G., Garagnani, M., Barone, R., Boscolo-Berto, R., Rossi, F., Morotti, A., et al. (2019). Validation and preliminary application of a GC-MS method for the determination of putrescine and cadaverine in the human brain: a promising technique for PMI estimation. Forensic Sci. Int. 297, 221–227. doi: 10.1016/j.forsciint.2019.01.025
Pernomian, L., Duarte-Silva, M., and de Barros Cardoso, C. R. (2020). The aryl hydrocarbon receptor (AHR) as a potential target for the control of intestinal inflammation: insights from an immune and bacteria sensor receptor. Clin. Rev. Allergy Immunol. 59, 382–390. doi: 10.1007/s12016-020-08789-3
Rannug, A. (2020). How the AHR became important in intestinal homeostasis-A diurnal FICZ/AHR/CYP1A1 feedback controls both immunity and immunopathology. Int. J. Mol. Sci. 21:5681. doi: 10.3390/ijms21165681
Rhodes, A., Evans, L. E., Alhazzani, W., Levy, M. M., Antonelli, M., Ferrer, R., et al. (2017). Surviving sepsis campaign: international guidelines for management of sepsis and septic shock: 2016. Crit. Care Med. 45, 486–552.
Salnikova, L. E., Smelaya, T. V., Moroz, V. V., Golubev, A. M., and Rubanovich, A. V. (2013). Functional polymorphisms in the CYP1A1, ACE, and IL-6 genes contribute to susceptibility to community-acquired and nosocomial pneumonia. Int. J. Infect. Dis. 17, e433–e442. doi: 10.1016/j.ijid.2013.01.005
Schiering, C., Wincent, E., Metidji, A., Iseppon, A., Li, Y., Potocnik, A. J., et al. (2017). Feedback control of AHR signaling regulates intestinal immunity. Nature 542, 242–245.
Sharkey, K. A., Beck, P. L., and McKay, D. M. (2018). Neuroimmunophysiology of the gut: advances and emerging concepts focusing on the epithelium. Nat. Rev. Gastroenterol. Hepatol. 15, 765–784. doi: 10.1038/s41575-018-0051-4
Smits, S. A., Marcobal, A., Higginbottom, S., Sonnenburg, J. L., and Kashyap, P. C. (2016). Individualized responses of gut microbiota to dietary intervention modeled in humanized mice. mSystems 1:e00098-16. doi: 10.1128/mSystems.00098-16
Stockinger, B., Shah, K., and Wincent, E. (2021). AHR in the intestinal microenvironment: safeguarding barrier function. Nat. Rev. Gastroenterol. Hepatol. 18, 559–570. doi: 10.1038/s41575-021-00430-8
Tian, L. X., Tang, X., Zhu, J. Y., Luo, L., Ma, X. Y., Cheng, S. W., et al. (2020). Cytochrome P450 1A1 enhances inflammatory responses and impedes phagocytosis of bacteria in macrophages during sepsis. Cell Commun. Signal. 18:70.
Turner, J. R. (2009). Intestinal mucosal barrier function in health and disease. Nat. Rev. Immunol. 9, 799–809. doi: 10.1038/nri2653
Wang, J., Wang, P., Tian, H., Tian, F., Zhang, Y., Zhang, L., et al. (2018). Aryl hydrocarbon receptor/IL-22/Stat3 signaling pathway is involved in the modulation of intestinal mucosa antimicrobial molecules by commensal microbiota in mice. Innate Immun. 24, 297–306. doi: 10.1177/1753425918785016
Westerink, W. M., Stevenson, J. C., and Schoonen, W. G. (2008). Pharmacologic profiling of human and rat cytochrome P450 1A1 and 1A2 induction and competition. Arch. Toxicol. 82, 909–921.
Wilmore, J. R., Gaudette, B. T., Gomez Atria, D., Hashemi, T., Jones, D. D., Gardner, C. A., et al. (2018). Commensal microbes induce serum IgA responses that protect against polymicrobial sepsis. Cell Host Microbe 23, 302–311.e3. doi: 10.1016/j.chom.2018.01.005
Wincent, E., Bengtsson, J., Mohammadi Bardbori, A., Alsberg, T., Luecke, S., Rannug, U., et al. (2012). Inhibition of cytochrome P4501-dependent clearance of the endogenous agonist FICZ as a mechanism for activation of the aryl hydrocarbon receptor. Proc. Natl. Acad. Sci. U.S.A. 109, 4479–4484. doi: 10.1073/pnas.1118467109
Wolff, N. S., Hugenholtz, F., and Wiersinga, W. J. (2018). The emerging role of the microbiota in the ICU. Crit. Care 22:78. doi: 10.1186/s13054-018-1999-8
Zhao, J., Zhang, W., Shen, L., Yang, X., Liu, Y., and Gai, Z. (2017). Association of the ACE, GSTM1, IL-6, NOS3, and CYP1A1 polymorphisms with susceptibility of mycoplasma pneumoniae pneumonia in Chinese children. Medicine 96:e6642. doi: 10.1097/MD.0000000000006642
Zhou, Q., and Verne, G. N. (2018). Intestinal hyperpermeability: a gateway to multi-organ failure. J. Clin. Invest. 128, 4764–4766. doi: 10.1172/JCI124366
Zhu, J., Luo, L., Tian, L., Yin, S., Ma, X., Cheng, S., et al. (2018). Aryl hydrocarbon receptor promotes IL-10 expression in inflammatory macrophages through Src-STAT3 signaling pathway. Front. Immunol. 9:2033. doi: 10.3389/fimmu.2018.02033
Keywords: CYP1A1, gut microbiota, intestinal barrier, cadaverine, enterocytes junction, sepsis, methicillin-resistant Staphylococcus aureus
Citation: Ma X, Jin H, Chu X, Dai W, Tang W, Zhu J, Wang F, Yang X, Li W, Liu G, Yang X and Liang H (2022) The Host CYP1A1-Microbiota Metabolic Axis Promotes Gut Barrier Disruption in Methicillin-Resistant Staphylococcus aureus-Induced Abdominal Sepsis. Front. Microbiol. 13:802409. doi: 10.3389/fmicb.2022.802409
Received: 15 November 2021; Accepted: 08 March 2022;
Published: 27 April 2022.
Edited by:
Gee W. Lau, University of Illinois at Urbana-Champaign, United StatesReviewed by:
You-Hee Cho, CHA University, South KoreaChristopher Gaulke, University of Illinois at Urbana-Champaign, United States
Lokesh Sharma, Yale University, United States
Copyright © 2022 Ma, Jin, Chu, Dai, Tang, Zhu, Wang, Yang, Li, Liu, Yang and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huaping Liang, MTM2MzgzNTY3MjhAMTYzLmNvbQ==
 Xiaoyuan Ma
Xiaoyuan Ma Huaijian Jin1,2
Huaijian Jin1,2 Huaping Liang
Huaping Liang