- 1School of Biotechnology and Biomolecular Sciences, University of New South Wales, Sydney, NSW, Australia
- 2Faculty of Medicine, Monash University, Melbourne, VIC, Australia
- 3School of Medical Sciences, University of New South Wales, Sydney, NSW, Australia
- 4Gastrointestinal and Liver Unit, Prince of Wales Hospital, University of New South Wales, Sydney, NSW, Australia
Inflammatory bowel disease (IBD) is a chronic inflammatory condition of the gastrointestinal tract with unknown etiology. The pathogenesis of IBD results from immune responses to microbes in the gastrointestinal tract. Various bacterial species that are associated with human IBD have been identified. However, the microbes that trigger the development of human IBD are still not clear. Here we review bacterial species that are associated with human IBD and their pathogenic mechanisms to provide an updated broad understanding of this research field. IBD is an inflammatory syndrome rather than a single disease. We propose a three-stage pathogenesis model to illustrate the roles of different IBD-associated bacterial species and gut commensal bacteria in the development of human IBD. Finally, we recommend microbe-targeted therapeutic strategies based on the three-stage pathogenesis model.
Introduction
Inflammatory bowel disease (IBD) is a chronic inflammatory condition of the gastrointestinal tract (Ng et al., 2017). Crohn’s disease (CD) and ulcerative colitis (UC) are the two major clinical forms of IBD. IBD can occur in all age groups but most often in late adolescents and young adults (Johnston and Logan, 2008). The etiology of IBD is not completely understood. Studies suggest that environmental factors, gut microbes and genetics play a role in the development of IBD (Sairenji et al., 2017). The pathogenesis of IBD results from immune responses to microbes in the gastrointestinal tract, demonstrated by studies in patients with IBD and using animal models (De Souza and Fiocchi, 2016). The human gastrointestinal tract is colonized by millions of bacterial cells consisting of more than 500 bacterial species (Thursby and Juge, 2017). These microbes maintain a homeostatic relationship with the mucosal immune system in healthy individuals, thus referred to as commensal bacteria. The microbes that trigger the breakdown of the homeostasis between the mucosal immune system and the commensal microbes of the gut microbiota in patients with IBD are still not clear after decades of research. In this article, we review bacterial species that are associated with human IBD and their pathogenic mechanisms. We also propose a three-stage pathogenesis model to illustrate the roles of IBD-associated bacterial species and gut commensal bacteria in the development of human IBD. Finally, we recommend microbe-targeted therapeutic strategies based on the proposed three-stage pathogenesis model.
Interactions Between Gut Microbiota and the Mucosal Immune System in Homeostasis
There are dynamic interactions between gut microbes and the mucosal immune system in healthy individuals, however, such interactions do not induce inflammation. In order to better explain the roles of different bacterial species in the pathogenesis of IBD, we firstly summarize the main strategies that maintain the homeostasis between gut microbiota and the mucosal immune system in healthy conditions.
The first strategy is to limit the contact of luminal microbes with the host cells and tissues. This is achieved via the barrier formed by mucus, antimicrobial molecules secreted by intestinal epithelial cells, secretary IgA produced by plasma cells in the lamina propria and the intestinal epithelium (Bankaitis et al., 2018). The mucus that covers the small intestine is a loose and unattached layer with high concentrations of antibacterial molecules which limits the numbers of bacteria that can reach the epithelium and Peyer’s patches (Johansson et al., 2013; Sheng et al., 2013). The mucus that covers the large intestine has two layers. The inner mucus layer is firmly attached to intestinal epithelial cells and free of bacteria in healthy individuals. The outer mucus layer is loose and has bacterial colonization (Johansson et al., 2013). The intestinal epithelium contains four major types of cells that are derived from stem cells located in the crypts (Bankaitis et al., 2018). Most of the intestinal epithelial cells are enterocytes which are absorptive cells. The others are secretary cells including goblet cells, Paneth cells and enteroendocrine cells. Goblet cells secrete mucus. The epithelial cells are connected by tight junction proteins.
The second strategy is the strategically arranged innate pattern recognition receptors (PRRs) in the intestinal epithelium. Intestinal epithelial cells express many different types of PRRs, located on the surface or inside of cells. These receptors recognize and respond to intruding pathogens but maintain a tolerance to gut microbiota and food components when under a homeostatic state. Such a tolerance is maintained by the low expression of these receptors and their spatial location in healthy conditions (Santaolalla and Abreu, 2012). For example, Toll-like receptor-4 (TLR4) and MD-2 are expressed at a low level in normal intestinal epithelial cells and they do not respond to lipopolysaccharide (LPS), a major component of Gram-negative bacteria (Abreu et al., 2002). Furthermore, TLRs have a polarized distribution; they are located at the basolateral surfaces instead of apical surface, which further limits the response of the intestinal epithelium to products of commensal gut microbes that have crossed the mucus layer (Abreu et al., 2002; Burgueño and Abreu, 2020).
The third strategy is the unique regulatory network of the mucosal immune system. Macrophages in the lamina propria of the intestinal mucosa play an important role in both maintenance of intestinal homeostasis and defense of pathogens. Under homeostatic conditions, lamina propria macrophages uptake luminal bacteria or their products. These macrophages produce little or no pro-inflammatory cytokines after taking luminal antigens (Denning et al., 2007; Smith et al., 2011). Instead, they produce anti-inflammatory molecules such as interleukin (IL)-10 (Denning et al., 2007). Furthermore, CD11b+ F4/80+ CD11c–macrophages and CD103+ dendritic cells induce antigen-specific regulatory T cells (Treg) in the mesenteric lymph nodes (Denning et al., 2007; Bain and Schridde, 2018). The intestinal tissues contain high numbers of Forkhead box P3 (Foxp3) Treg cells that are important in preventing intestinal inflammation. Foxp3+ Treg cells control host immune responses through multiple mechanisms such as the production of immune suppression cytokines of tumor growth factor (TGF)-β and IL-10, changing antigen presenting cell functions such as down regulation of B7 molecules on dendritic cells (Harrison and Powrie, 2013).
Gut microbiota contributes to the development and function of the immune system. Animals raised in germ-free conditions where no live microorganisms were present had fewer and smaller Peyer’s patches and mesenteric lymph modes, reduced numbers of CD4+ T cells and intraepithelial CD8+ T cells in the lamina propria, and reduced numbers of IgA producing B cells and plasma cells (Round and Mazmanian, 2009). Isolated lymphoid follicles in the intestinal tract are formed and matured after birth, which are impaired as well in germ-free mice (Bouskra et al., 2008). Animals raised under germ-free conditions are less resistant to pathogen infections. Guinea pigs raised under conventional conditions resisted oral challenge with Shigella flexneri. However, germ-free guinea pigs receiving a similar challenge died (Sprinz et al., 1961). Germ-free mice also showed impaired abilities in eliminating Listeria monocytogenes after intraperitoneal infection and this may be due to lack of L-selectin +CD44+ T cells in sites of inflammation (Inagaki et al., 1996). These findings show that the products of gut microbiota continuously stimulate the mucosal immune system in homeostatic states, which serves as an important training strategy for host protection.
Bacterial Species Associated With Human Inflammatory Bowel Disease
Studies have searched for bacterial species that may play a role in the development of IBD. Here, we will review individual bacterial species that were found to be associated with human IBD according to the chronological order of the discovery year of their association with IBD. IBD-associated bacterial species with pathogenic mechanisms studied are listed in Table 1.
Clostridium difficile and Other Enteric Pathogen Infections in Inflammatory Bowel Disease
C. difficile is a Gram-positive spore-forming anaerobic bacterium, which is ubiquitous in nature and also colonizes the human intestinal tract (Ryan and Ray, 2004). C. difficile causes diarrhea and colitis, often in individuals who have been treated with antibiotics for other medical conditions.
The implications of C. difficile infection in the therapies for IBD relapses were suggested in 1980 (Lamont and Trnka, 1980). Later studies suggest that C. difficile may contribute to the relapse or trigger the development of IBD in some patients. Patients with IBD are at a higher risk of developing C. difficile infection. Studies of hospitalized patients from United States found that C. difficile was more common in patients with UC (3.9%) as compared to non-IBD patients (1.2%), and an adjusted odds ratio for C. difficile infection in patients with IBD, CD and UC were 2.9, 2.1, and 4.0, respectively (Meyer et al., 2004; Rodemann et al., 2007). Further studies from United States showed that enteric pathogens were detected in up to 20% of relapsed IBD patients with C. difficile being the most common pathogen (up to 90%) of the detected pathogens (Binion, 2012; Pant et al., 2013). IBD patients with C. difficile infection have worse outcomes and experience higher rates of recurrence (Issa et al., 2007; Jodorkovsky et al., 2010; Navaneethan et al., 2012). While these findings support a role of C. difficile infection in participating the pathogenesis in relapsed IBD, 10% of C. difficile infection occurred at the time of IBD diagnosis. This and the finding that IBD patients in remission had a significantly higher presence of toxigenic C. difficile in their intestinal tract as compared to healthy controls (8.2 vs. 1%) suggest that C. difficile infection may be a risk factor for the development of IBD (Clayton et al., 2009). A more recent study from the Netherlands employing both retrospective and prospective approaches found a low prevalence of C. difficile that was not associated with IBD. Their finding indicated that C. difficile is not a common trigger for exacerbations of IBD in clinical practice in the Netherlands (Masclee et al., 2013).
The major virulence factors of C. difficile are the two toxins encoded by tcdA and tcdB genes (McDonald et al., 2005; Belyi and Aktories, 2010; Aktories et al., 2017). These toxins inactivate host Rho and Ras family GTPase by glucosylation leading to the disruption of epithelial cytoskeleton and tight junctions, which may contribute to the increased epithelial permeability and luminal fluid accumulation associated with C. difficile infection (Kim et al., 2005). TcdA and TcdB toxins also cause cell death via caspase-dependent and caspase-independent apoptosis, further damaging the intestinal epithelial barrier (Hippenstiel et al., 2002; Qa’Dan et al., 2002; Matarrese et al., 2007; Nottrott et al., 2007). Both toxins also induce the production of proinflammatory cytokines in intestinal epithelial and immune cells, including IL-8, TNF-α, IL-1, and IL-6 (Savidge et al., 2003). By using multiomic analysis, it was found that the most discriminating metabolite between IBD and IBD infected with C. difficile was isocaproyltaurine, a covalent conjugate of a C. difficile fermentation product isocaproate and taurine from damaged tissue (Bushman et al., 2020).
In addition to C. difficile, other enteric pathogens were also detected in patients with flares of IBD. A recent study from United States found that compared to patients without IBD, patients with CD had a higher prevalence of norovirus (p = 0.05) and Campylobacter (p = 0.043). Compared to patients without IBD, patients with UC had a lower prevalence of norovirus (p = 0.049) and a higher prevalence of Campylobacter (p = 0.013), Plesiomonas (p = 0.049) and Escherichia coli (p < 0.001). However, these enteric infections did not impact IBD outcomes (Axelrad et al., 2018). A recent study from UK also reported that patients with IBD in primary care were more likely to present with common infections including gastrointestinal infections of C. difficile, Salmonella, Shigella and Campylobacter (Irving et al., 2021). Campylobacter species isolated in diagnostic laboratories are mainly Campylobacter jejuni.
Mycobacterium avium Subspecies paratuberculosis
M. avium is a mycobacterial species that is commonly present in the environment, which consists of four subspecies, including M. avium subspecies avium, M. avium subspecies paratuberculosis (MAP), M. avium subspecies hominissuis and M. avium subspecies silvaticum (Uchiya et al., 2017). MAP is present in about 70% of dairy herds without causing problems (Nielsen et al., 2000). However, it can cause Johne’s disease (chronic intestinal inflammation in distal small intestine) in cattle (Singh et al., 2014). Due to the similarities in histopathology between Johne’s disease and CD, it has been proposed that MAP may play a role in CD. In 1984, an unclassified mycobacterial species was isolated from gastrointestinal tissues of two patients with CD (Chiodini et al., 1984). A 7-day-old goat inoculated orally with 50 mg of the isolated organism developed a granulomatous lesion at the distal small intestine. The authors of this study concluded that their findings raise the possibility that a Mycobacterium plays an etiologic role in at least some cases of CD (Chiodini et al., 1984). Following this, multiple studies examined the association between MAP and CD, using bacterial cultivation or a PCR method targeting the insertion element (IS 900) in the genome of MAP. These studies generated controversial results and the details of these studies were previously reviewed by Naser et al. (2014).
Several clinical studies using anti-mycobacterial antibiotics in the treatment of CD reported beneficial effects. These studies used a combination of multiple antibiotics including rifabutin, clofazimine and clarithromycin, plus either ciprofloxacin, metronidazole, or ethambutol (Agrawal et al., 2020a,b). However, these studies did not identify whether patients included in the treatment were positive for MAP. Furthermore, the multiple antibiotics used in these studies would have had effects on other non-mycobacterial bacterial species. Although these studies did not prove MAP causes CD, they provided evidence supporting that bacterial species are involved in the pathogenesis of CD. Other studies also reported that different antibiotics such as nitroimidazoles and clofazimine also appear to be effective in the treatment of CD, further supporting that bacterial species are involved in the pathogenesis of CD (Feller et al., 2010).
The pathogenic mechanisms of MAP in Johne’s disease were studied. MAP is taken in by microfold (M) cells following recognition of the bacterium by the fibronectin receptor (Secott et al., 2004). MAP bacteria that travel across the intestinal epithelium are phagocytosed by macrophages via complement receptors. Infected macrophages form granulomas and become a latent infection or active disease (Birkness et al., 2007; Rice et al., 2019).
Enterotoxigenic Bacteroides fragilis
B. fragilis is a Gram-negative anaerobe, a commensal bacterium of the intestinal tract. However, a particular strain, namely enterotoxigenic B. fragilis strain, which produces an enterotoxin encoded by the bft gene, was reported to be associated with IBD disease activation or flare-up in year 2000 by a study from United States (Prindiville et al., 2000). The enterotoxin encoded by the bft gene is a secreted metalloprotease (Moncrief et al., 1995; Franco et al., 1997). A study in 2017 from Iran reported that bft gene was detected in 51.4% of intestinal biopsies from patients with UC and 1.6% non-IBD samples (Zamani et al., 2017). The B. fragilis enterotoxin cleaved the adherens junction protein E-cadherin and induced IL-8 production in intestinal epithelial cells (Wu et al., 1998, 2004; Kim et al., 2001). The enterotoxigenic B. fragilis strain induced persistent colitis with inflammatory cell infiltration, crypt abscesses and epithelial ulceration in wild type specific-pathogen free C57BL/6 mice and lethal colitis in germ-free C57BL/6 mice (Rhee et al., 2009). In multiple intestinal neoplasia mice, enterotoxigenic B. fragilis induced colitis via T17 response pathway and also induced colonic tumors (Wu et al., 2009). A recent study from the Netherlands reported a different result. They detected a higher prevalence of B. fragilis in patients with active CD as compared to patients in remission, however, bft-positive B. fragilis was not higher and they also found that bft-positive B. fragilis increased epithelial resistance rather than damaging the barrier (Becker et al., 2021).
Fusobacterium varium
F. varium is a Gram-negative bacterium. It was reported in 2002 from Japan that F. varium in colonic mucosa of patients with UC was significantly higher than that in CD and healthy controls detected using immunohistochemical method (Ohkusa et al., 2002). This study also detected a significantly higher antibodies to extracts of F. varium antigens (Ohkusa et al., 2002). In a following study, the same research group examined the cytotoxicity of supernatants of 20 bacterial species isolated from intestinal tissues of patients with UC. Only the supernatant of F. varium, which was found to contain high concentration of butyric acid, killed Vero cells (Ohkusa et al., 2003). Enemas containing F. varium culture supernatants given to mice induced colonic ulceration, crypt abscesses, inflammatory cell infiltration and apoptotic changes (Ohkusa et al., 2003). In a different study, Tahara et al. (2015) detected Fusobacterium in inflamed intestinal tissues of 152 Japanese UC patients using quantitative real-time PCR and found that Fusobacterium species were commonly present in patients with UC and was associated with mild clinical phenotypes of UC. However, the PCR primers that Tahara et al. (2015) used were not specific for F. varium.
A recent study in 2016 from Korea isolated Fusobacterium species from colonic biopsies of 51 (15.6%) patients with IBD (26 CD and 25 UC). Fusobacterium bacteria were isolated from eight of the 51 patients with IBD, however, only two were F. avium (Lee et al., 2016).
Adherent-Invasive Escherichia coli
E. coli is a Gram-negative facultative bacterium, which is a commensal bacterium of the human gut. However, some strains of E. coli are human enteric pathogens. The adherent-invasive Escherichia coli (AIEC) was reported to be associated with ileal CD in the adult population (Darfeuille-Michaud et al., 2004; Martin et al., 2004; Sasaki et al., 2007). AIEC strains were defined based on their ability to adhere to and invade intestinal epithelial cell lines assessed (Darfeuille-Michaud et al., 2004). Various cell lines such as Caco-2, intestine-407 and HT-29 cells were used in assessing AIEC strains. AIEC strains do not have specific virulence factors. Comparative genomic analysis of CD associated AIEC strains and non-AIEC strains did not identify any molecular marker that can differentiate AIEC strains from non-AIEC strains (O’Brien et al., 2017). In the past 20 years, extensive studies have been conducted to examine the pathogenic mechanisms of AIEC strains.
AIEC strains adhere to and invade intestinal epithelial cells. AIEC adhesion to intestinal epithelial cells was via interactions of the common type 1 pili adhesion FimH and carcinoembryonic antigen cell adhesion molecule 6 (CEACAM6) (Boudeau et al., 2001; Barnich et al., 2007; Carvalho et al., 2009; Dreux et al., 2013). CEACAM6 is a glycoprotein that is abnormally expressed in intestinal epithelial cells of patients with ileal CD (Barnich et al., 2007). AIEC strains also used long polar fimbriae to interact with M cells on the surface of Peyer’s patches, which facilitated the translocation of AIEC across monolayers of M cells (Chassaing et al., 2011).
AIEC strains resist the defense from macrophages. Macrophages play a critical role in clearance of bacterial infection. In macrophages, while AIEC bacteria targeted early by the autophagic machinery were degraded, those that escaped early uptake by autophagy were able to survive and replicate inside macrophages (Lapaquette et al., 2012). Impaired ATG16L1, IRGM, or NOD2, which are molecules affecting autophagy or bacterial sensing, favored AIEC persistence in macrophages and induced increased production of tumor necrosis factor (TNF)-α and IL-6 (Lapaquette et al., 2012). AIEC induced granulomas in vitro (Lapaquette et al., 2012). AIEC strains promote inflammation. AIEC strains induced production of cytokines such as IL-8, TNF-α, and IL-6 in both epithelial cells and macrophages (Subramanian et al., 2008; Lapaquette et al., 2012). Survival and replication of AIEC in macrophages did not cause macrophage death, but increased production of TNF-α and IL-6 (Lapaquette et al., 2012). AIEC strain LF82 induced intestinal inflammation in CEABAC10 mice, transgenic mice that express human CEACAMS (Carvalho et al., 2009). In conventional mice treated with streptomycin, AIEC strains also induced chronic intestinal inflammation and intestinal fibrosis (Small et al., 2013). CD8+ T cells showed a protective role in AIEC infection in streptomycin-treated mice, and depletion of CD8+ T cells increased cecal AIEC load, intestinal pathology, and fibrosis in C56BL/6 mice (Small et al., 2013).
Enteric Helicobacter Species
Helicobacters are Gram-negative bacterial species. H. pylori is the well-known human gastric pathogen. Some Helicobacter species were also isolated from intestinal tract of humans and animals, which are referred to as enteric Helicobacter species. By using a genus PCR, a study from Germany in 2004 detected a higher presence of Helicobacter species in colonic and ileal biopsies of patients with CD (12%), UC (17%) than controls (4%), however the difference did not reach statistical significance (Bohr et al., 2004). Sequencing of the genus PCR products showed that the detected Helicobacter DNAs were non-pylori Helicobacter species (Bohr et al., 2004). By using a Helicobacteraceae family specific PCR, a significantly higher presence of members of Helicobacteraceae were found in intestinal biopsies and fecal samples from children with CD (Zhang et al., 2006; Man et al., 2008; Kaakoush et al., 2010). Further identification of the PCR amplified DNA revealed that most of the detected members of Helicobacteraceae were enteric Helicobacter species (non-pylori Helicobacter species that colonize the intestinal tract). In intestinal biopsies and luminal washings collected from adult patients with IBD and controls, a study from UK detected non-pylori Helicobactor by PCR in 2.9% patients with IBD and 8.1% controls (Basset et al., 2004). Studies of detection of enteric Helicobacter species in IBD were mainly conducted by PCR. The lack of isolated enteric Helicobacter species from patients with IBD has prevented the study of pathogenic mechanisms of these bacterial species.
Campylobacter concisus
C. concisus is an oral bacterium and some strains can also colonize the intestinal tract (Zhang, 2015; Liu et al., 2018b). The association between C. concisus and pediatric CD was first reported using PCR detection of C. concisus 16S rRNA gene in intestinal biopsies and bacterial isolation (Zhang et al., 2009). The association between pediatric CD and C. concisus was then demonstrated using fecal samples (Man et al., 2010b). Later studies also reported the association between C. concisus and IBD, including both CD and UC, in the adult population (Mahendran et al., 2011; Mukhopadhya et al., 2011; Kirk et al., 2016). Genome analysis showed that C. concisus contains two genomospecies, and C. concisus strains isolated from intestinal tract of patients with IBD were predominately genomospecies 2 (Chung et al., 2016; Wang et al., 2017; Cornelius et al., 2021). Analysis of genomes of C. concisus strains isolated from patients with IBD and healthy controls also identified molecular markers for strains that were associated with severe CD and UC, these markers were from genomospecies 2 C. concisus strains (Liu et al., 2018a; Liu F. et al., 2020). Several studies also examined the pathogenic mechanisms that may contribute to the pathogenesis of IBD.
C. concisus damages the intestinal epithelial barrier. The bacterium induced apoptosis in human intestinal epithelial HT-29 cells following 48 h incubation and the expression of barrier-forming tight junction protein claudin-5 reduced to 66% (Nielsen et al., 2011). Tight junction protein redistribution caused by C. concisus was also observed (Man et al., 2010a; Nielsen et al., 2011). C. concisus strain adhered to and some strains were invasive to intestinal epithelial cells (Man et al., 2010a; Ismail et al., 2012). Some C. concisus strains have acquired prophages that contain the zot gene (Liu et al., 2016). The C. concisus Zot protein caused cell death in human intestinal epithelial Caco-2 cells (Mahendran et al., 2016).
C. concisus enhances the responses of intestinal epithelial cells and macrophages to commensal bacteria. Preincubation of THP-1 macrophage-like cells and HT-29 cells with C. concisus Zot protein significantly enhanced the production of TNF-α by these cells in response to E. coli (Mahendran et al., 2016). Incubation of HT-29 cells with C. concisus upregulated the expression of TLR4 and MD-2, which primarily recognize LPS (Ismail et al., 2013). LPS is a core component of the outer membrane of Gram-negative bacteria. A low level of expression of TLRs is a mechanism of maintain the tolerance to gut bacteria (Santaolalla and Abreu, 2012). Increased expression of TLR4 and MD-2 on intestinal epithelial cells may promote these cells to respond to commensal gut microbes that they usually do not respond under homeostasis, which causes intestinal inflammation. C. concisus has a spiral to curved shape, a morphology that better equips bacterial species with abilities to travel in mucus and get close to epithelium.
C. concisus induces intestinal inflammation. C. concisus induced the production of IL-8 in intestinal epithelial cells (Man et al., 2010a; Ismail et al., 2013), and the production of IL-8 and TNF-α in human macrophages (Man et al., 2010a; Mahendran et al., 2016).
C. concisus also has the potential to affect the adaptive immune system. C. concisus upregulated epithelial expression of the immune checkpoint protein PD-L1 in interferon (IFN)-γ sensitized HT-29 cells, suggesting that while C. concisus induces innate inflammatory responses, it has the potential to inhibit the function of T cells (Lee et al., 2021). However, the upregulated expression of PD-L1 was only examined at an mRNA level. Whether the upregulated PD-L1 has functional bioactivity at a protein level is not yet known.
An individual’s gastrointestinal environment has an impact on the enteric pathogenicity of C. concisus. The growth of C. concisus is highly dependent on the presence of H2 (Lee et al., 2014). In the human intestinal tract, the level of H2 gas generated by bacterial species is greatly affected by diet and gut bacterial composition, which may play a role in determining C. concisus intestinal colonization (Strocchi and Levitt, 1992).
Fusobacterium nucleatum
F. nucleatum is a Gram-negative anaerobe that colonizes the human oral cavity and intestinal tract. A study from Canada in 2011 reported a significantly higher isolation of F. nucleatum from intestinal biopsies of patients with IBD as compared with healthy controls (Strauss et al., 2011). Recently, F. nucleatum was reported to be abundant in intestinal tissues of patients with UC and IBD, and the abundance was associated with disease severity (Liu H. et al., 2020; Su et al., 2020).
The pathogenic mechanisms of F. nucleatum relating to IBD include damaging intestinal barrier and promoting inflammation. F. nucleatum strains isolated from inflamed biopsies of patients with IBD were more invasive to Caco-2 cells (Strauss et al., 2011). In mice with dextran sodium sulfate (DSS)-induced colitis, F. nucleatum promoted intestinal inflammation (Liu H. et al., 2020; Su et al., 2020). F. nucleatum damaged intestinal epithelial barrier by inducing autophagic epithelial cell death, affecting the expression and distribution of zonula occludens-1 (ZO-1) and occludin and skewing proinflammatory M1 macrophage (Liu L. et al., 2019; Liu H. et al., 2020; Su et al., 2020). In mice with DSS-induced colitis, F. nucleatum induced Th1 and Th17 subset T cell differentiations, promoted the secretion of proinflammatory cytokines such as TNF-α, IFN-γ, IL-1β, IL-6, and IL-17 (Liu L. et al., 2019).
F. nucleatum is also associated with colorectal cancer (Lee et al., 2019). Using colorectal cancer cell lines (HCT116, DLD1, SW480, and HT-29) and xenograft mice, it was shown that F. nucleatum promotes colorectal cancer cell growth by modulating E-cadherin and promotes cancer cell proliferation via FadA adhesin, activating TLR4 signaling to nuclear factor-kB (Rubinstein et al., 2013; Yang et al., 2017).
Changed Gut Microbiota in Inflammatory Bowel Disease
In recent years, numerous studies have compared the gut microbiota in patients with IBD and controls. These studies were recently reviewed by Pittayanon et al. (2020) in a systematic review published in Gastroenterology. Pittayanon et al. (2020) reviewed 48 such studies and found the following: (1) The diversity of microbiota was either decreased or not different in patients with IBD vs. controls. (2) For patients with CD, three studies reported a decrease of Christensenellaceae and Coriobacteriaceae as compared to controls. Faecalibacterium prausnitzii was reported to decrease in 6 of 11 studies. Two studies each of Actinomyces, Veillonella and E. coli revealed an increase of these organisms. (3) For patients with UC, Eubacterium rectale and Akkermansia were decreased in three studies, E. coli was increased in four of nine studies. (4) Pittayanon et al. (2020) stated that they cannot make conclusions due to inconsistent results and methods among studies.
Discussion
As stated above, several bacterial species were found to be positively associated with IBD, suggesting that they may play a role in the development of IBD.
IBD is not a single disease. The diagnosis of IBD is based on histopathology and inflammatory markers rather than microbes that trigger the development of the inflammatory conditions. Immune responses to different microbes may result in a similar histopathology. It is therefore not surprising that multiple bacterial species were found to be associated with IBD. Bacterial species that are associated with IBD were from different sources, including the oral cavity, intestinal tract, animal, or environment. C. concisus and F. nucleatum are oral bacterial species. The oral cavity is a constantly available source providing these organisms to the intestinal tract. In individuals with high numbers of C. concisus or F. nucleatum being translocated to the intestinal tract or with an intestinal microenvironment that is suitable for the growth of these organisms, C. concisus or F. nucleatum may reach sufficient numbers to cause pathogenic effects in the intestinal tract.
We propose a three-stage pathogenesis model to illustrate the roles of IBD-associated bacterial species and gut commensal bacterial species in the development of IBD (Figure 1). In the first stage of IBD development, the main players in inducing inflammation are the initiating microbes, which have the ability to overcome the mucosal defense system. Despite possessing different virulence factors, C. concisus, F. nucleatum, B. fragilis, F. varium, AIEC, C. difficile and many enteric pathogens share common pathogenic effects. They damage the intestinal epithelial cells and cause intestinal inflammation. Proinflammatory cytokines induced by these bacteria further damage the intestinal barrier (Liu F. et al., 2019). Most of the IBD-associated bacterial species are likely the initiating microbes contributing to stage 1 of IBD pathogenesis. Other enteric pathogens may also increase the risk of developing IBD if they cause prolonged infections.
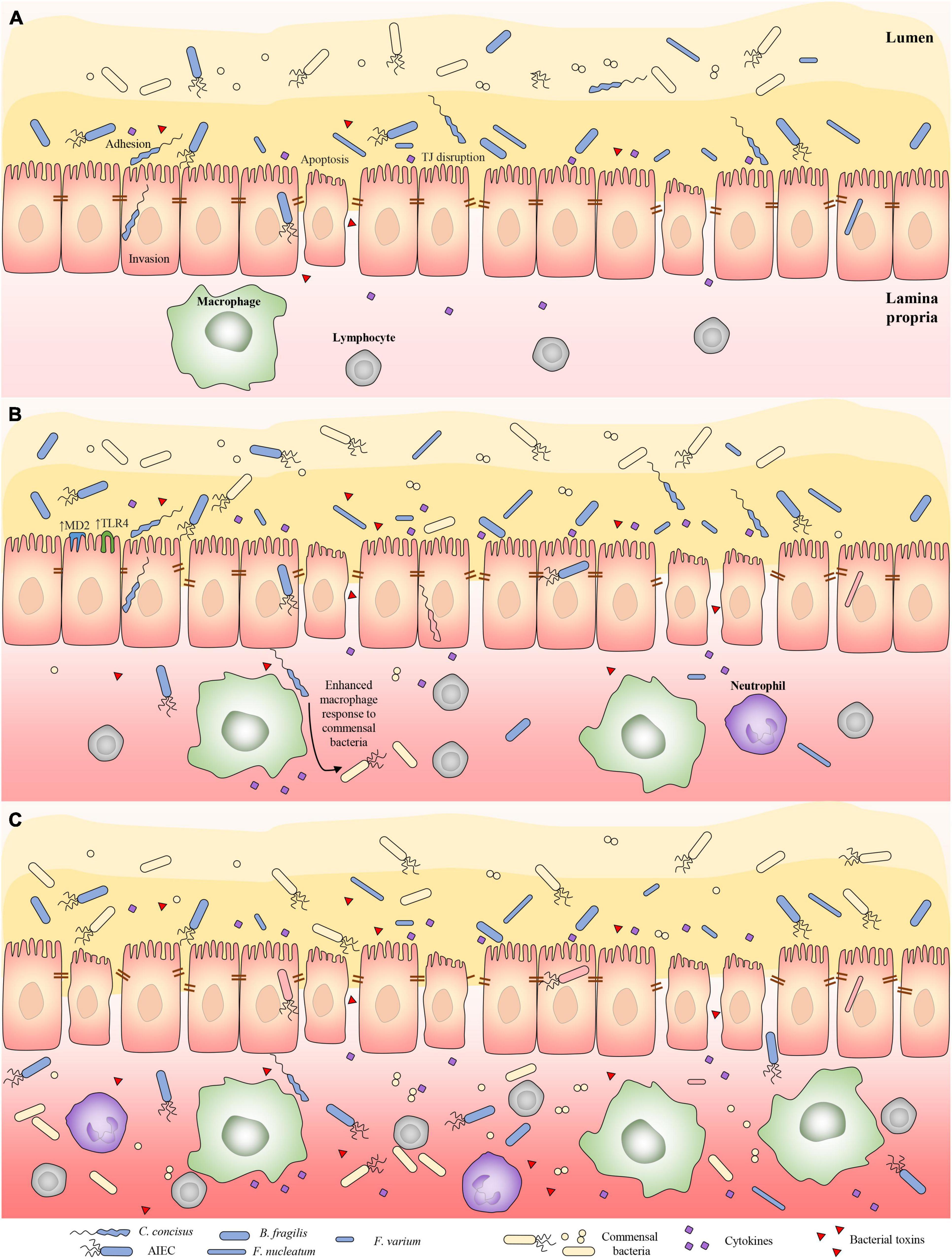
Figure 1. Three-stage pathogenesis model illustrating the role of initiating bacterial species and gut commensal bacterial species in contributing to the development of IBD. (A) In stage one, bacterial species that have abilities overcoming the mucosal defense cause damage to intestinal barrier and induce intestinal inflammation. Proinflammatory cytokines induced by these bacterial species further damage the intestinal barrier. Most of IBD-associated bacterial species in Table 1 belong to IBD-initiating bacterial species. Other enteric pathogens may also increase the risk of developing IBD if they cause prolonged infections. (B) In stage two, prolonged intestinal barrier damage caused by the IBD-initiating bacterial species and their abilities to enhance the response of mucosal immune system to gut commensal bacterial species lead to the breakdown of the homeostasis between the mucosal immune system and the gut microbiota, thus the gut commensal bacteria become part of the microbes that induce inflammation in the pathogenesis of IBD. (C) In the third stage of IBD pathogenesis, the gut commensal microbes are the main drivers of the intestinal inflammation due to their large amount, via both innate and adaptive immune responses. Inflammatory conditions are a changed ecosystem to gut microbes. Anaerobes are likely to disappear under an inflammatory environment due to their reduced ability to deal with oxidative stress as compared to facultative microbes. TJ, tight junction.
In the second stage of IBD development, both the initiating microbes and some gut commensal bacteria contribute to the pathogenesis. The continuous supply of C. concisus and F. nucleatum to the intestinal tract from the oral cavity and the fact that B. fragilis, AIEC and C. difficile are residing enteric microbes make it difficult to eliminate these bacteria by immune responses. These microbes are therefore likely to cause prolonged intestinal barrier damage and inflammation, which leads to a long-term increased passage of the gut commensal microbes and their products to intestinal tissues. Some of the initiating bacterial species also promote the responses of mucosal immune system to commensal bacteria. For example, C. concisus enhances the responses of macrophages to commensal bacterium E. coli and increases the expression of microbial recognition receptors on intestinal epithelial cells (Ismail et al., 2013; Mahendran et al., 2016). These effects eventually lead to the breakdown of the homeostasis between the mucosal immune system and the gut microbiota, thus some of the gut commensal bacteria become part of the microbes that induce inflammation in the pathogenesis of IBD (Figure 1).
In the third stage of IBD pathogenesis, the gut commensal microbes become the main drivers of the intestinal inflammation due to their large amount, via both innate and adaptive immune responses. Upon intestinal barrier damage, individual’s gut microbial communities determine the severity of intestinal inflammation, which has been clearly demonstrated using DSS colitis model (Roy et al., 2017). Bacterial species have different abilities in resistant inflammatory mediators released by the host. At this stage, some initiator microbes that have less abilities in dealing with oxygen stress such as C. concisus may not be able to survive long in an environment with severe inflammation (Yeow et al., 2020). The others such as E. coli thrive in the inflamed gut, due to its abilities to resist anti-bacterial molecules released by the host (Singh et al., 2015).
M. avium species is wildly present in the environment and in the gastrointestinal tract of animals. It usually only causes diseases in individuals with severely damaged immune system such as HIV patients with extremely low CD4+ T cells, underlying lung disease or sometimes in young children, and chronic obstructive pulmonary disease (Whiley et al., 2012). IBD most often occurs in young adults, which is a period with relatively strong immunity in the human life (Johnston and Logan, 2008). This disease pattern does not seem to support an initiating role of MAP in the pathogenesis of human IBD.
Numerous studies have examined the gut microbiota in patients IBD in the last decade. Most of these studies found reduced bacterial diversities and dysbiosis as compared to non-IBD controls. However, bacterial species contributing to the dysbiosis reported in different studies varied greatly. A recent systematic review analyzing 48 of such studies stated that they cannot draw conclusions due to inconsistent results and methods among studies (Pittayanon et al., 2020). Given the impacts of diet, inflammation, drugs, genetics, and many other factors on the growth of different bacterial species, the cross-feeding and inhibition between bacterial species in the gut microbiota, it is indeed a great challenge to find reliable relative gut bacterial abundance as markers to indicate IBD, which is not a single disease. The common feature of this group of disease is inflammation, which is caused by the same or different microbes. Inflammatory conditions are a changed ecosystem to gut microbes. The bacterial species in the human intestinal tract are largely anaerobes and facultative anaerobes. Anaerobes are likely to disappear under an inflammatory environment due to their limited ability to deal with oxidative stress as compared to facultative microbes. A recent study using a multiomics approach analyzing gut microbiota in patients with IBD and controls with 24 months follow up indeed has demonstrated this (Lloyd-Price et al., 2019). As mentioned above, with a damaged intestinal barrier caused by the IBD-associated bacterial species and the inflammatory cytokines, gut commensal bacterial species, can contribute to, and even become the main player in the pathogenesis of IBD due to their large amount at the third stage of IBD pathogenesis. Studies of gut microbiota predominantly used universal primers targeting 16S rRNA gene, which favors the amplification of dominant bacterial species and can only identify some bacteria to a species level. The minority bacterial species including some of the IBD-initiating bacterial species may not be detected using this approach. Indeed, the IBD-associated bacterial species listed in Table 1 were detected by specific PCR and bacterial isolation.
Current treatment for IBD mainly include anti-inflammatory drugs, immune suppressive drugs and monoclonal antibodies targeting TNF-α, IL-12, and integrin or immunosuppressive drugs. These drugs in general target the host immune response, except that immunosuppressive azathioprine and mercaptopurine have inhibitory effects to some bacterial species of the gastrointestinal tract including the IBD-associated C. concisus (Liu et al., 2017). For clinically diagnosed patients with IBD, most of them would be at the second or third stages of the proposed pathogenesis model. Given that microbes in the gastrointestinal tract, including both the gut commensal and the initiating microbes, are the drivers of the inflammation at these stages. It makes sense that strategies targeting these initiating microbes and commensal microbes should be included for the management of IBD. We recommend some anti-microbial strategies in additional to the currently used IBD therapies. The first strategy we suggest is to reduce the load of IBD-associated bacterial species in the oral cavity, which can in turn reduce the translocation of these bacteria to the intestinal tract. This can be achieved by physical or chemical procedures targeting bacteria in the oral cavity. The second strategy is to reduce interactions of gut commensal bacteria with the intestinal epithelium and cells of the mucosal immune system. Under chronic inflammatory conditions in the second and third stages of IBD pathogenesis, the relationship between commensal gut bacterial species and the host has changed. These microbes are not commensal to the host anymore but pathogens driving inflammation. Reduction of gut microbes and elimination of triggering microbes would be the most effective way to end the immune responses caused by them and restore the intestinal barrier. This may be best achieved by enema or other locally delivered antimicrobial agents targeting the lesion areas.
Author Contributions
LZ wrote the first version of the manuscript. FL generated the figure. FL, JX, SL, LL, and SR provided critical feedback and helped with editing the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Faculty Research Grant awarded to LZ from the University of New South Wales (Grant No. PS46772).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abreu, M. T., Arnold, E. T., Thomas, L. S., Gonsky, R., Zhou, Y., Hu, B., et al. (2002). TLR4 and MD-2 expression is regulated by immune-mediated signals in human intestinal epithelial cells. J. Biol. Chem. 277, 20431–20437. doi: 10.1074/jbc.M110333200
Agrawal, G., Clancy, A., Sharma, R., Huynh, R., Ramrakha, S., and Borody, T. (2020a). Targeted combination antibiotic therapy induces remission in treatment-naïve Crohn’s disease: a case series. Microorganisms 8:371. doi: 10.3390/microorganisms8030371
Agrawal, G., Hamblin, H., Clancy, A., and Borody, T. (2020b). Anti-mycobacterial antibiotic therapy induces remission in active paediatric Crohn’s disease. Microorganisms 8:1112. doi: 10.3390/microorganisms8081112
Aktories, K., Schwan, C., and Jank, T. (2017). Clostridium difficile toxin biology. Annu. Rev. Microbiol. 71, 281–307.
Axelrad, J. E., Joelson, A., Green, P. H., Lawlor, G., Lichtiger, S., Cadwell, K., et al. (2018). Enteric infections are common in patients with flares of inflammatory bowel disease. Am. J. Gastroenterol. 113, 1530–1539. doi: 10.1038/s41395-018-0211-8
Bain, C. C., and Schridde, A. (2018). Origin, differentiation, and function of intestinal macrophages. Front. Immunol. 9:2733. doi: 10.3389/fimmu.2018.02733
Bankaitis, E. D., Ha, A., Kuo, C. J., and Magness, S. T. (2018). Reserve stem cells in intestinal homeostasis and injury. Gastroenterology 155, 1348–1361. doi: 10.1053/j.gastro.2018.08.016
Barnich, N., Carvalho, F. A., Glasser, A.-L., Darcha, C., Jantscheff, P., Allez, M., et al. (2007). CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease. J. Clin. Investig. 117, 1566–1574. doi: 10.1172/JCI30504
Basset, C., Holton, J., Bazeos, A., Vaira, D., and Bloom, S. (2004). Are Helicobacter species and enterotoxigenic Bacteroides fragilis involved in inflammatory bowel disease? Dig. Dis. Sci. 49, 1425–1432. doi: 10.1023/b:ddas.0000042241.13489.88
Becker, H. E., Jamin, C., Bervoets, L., Boleij, A., Xu, P., Pierik, M. J., et al. (2021). Higher prevalence of Bacteroides fragilis in Crohn’s disease exacerbations and strain-dependent increase of epithelial resistance. Front. Microbiol. 12:598232. doi: 10.3389/fmicb.2021.598232
Belyi, Y., and Aktories, K. (2010). Bacterial toxin and effector glycosyltransferases. Biochim. Biophys. Acta Gen. Subj. 1800, 134–143. doi: 10.1016/j.bbagen.2009.07.022
Binion, D. G. (2012). Clostridium difficile infection in patients with inflammatory bowel disease. J. Gastroenterol. Hepatol. 8:615.
Birkness, K. A., Guarner, J., Sable, S. B., Tripp, R. A., Kellar, K. L., Bartlett, J., et al. (2007). An in vitro model of the leukocyte interactions associated with granuloma formation in Mycobacterium tuberculosis infection. Immunol. Cell Biol. 85, 160–168. doi: 10.1038/sj.icb.7100019
Bohr, U. R., Glasbrenner, B., Primus, A., Zagoura, A., Wex, T., and Malfertheiner, P. (2004). Identification of enterohepatic Helicobacter species in patients suffering from inflammatory bowel disease. J. Clin. Microbiol. 42, 2766–2768.
Boudeau, J., Barnich, N., and Darfeuille-Michaud, A. (2001). Type 1 pili-mediated adherence of Escherichia coli strain LF82 isolated from Crohn’s disease is involved in bacterial invasion of intestinal epithelial cells. Mol. Microbiol. 39, 1272–1284. doi: 10.1111/j.1365-2958.2001.02315.x
Bouskra, D., Brézillon, C., Bérard, M., Werts, C., Varona, R., Boneca, I. G., et al. (2008). Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature 456, 507–510. doi: 10.1038/nature07450
Burgueño, J. F., and Abreu, M. T. (2020). Epithelial toll-like receptors and their role in gut homeostasis and disease. Nat. Rev. Gastroenterol. Hepatol. 17, 263–278. doi: 10.1038/s41575-019-0261-4
Bushman, F. D., Conrad, M., Ren, Y., Zhao, C., Gu, C., Petucci, C., et al. (2020). Multi-omic analysis of the interaction between Clostridioides difficile infection and pediatric inflammatory bowel disease. Cell Host Microbe 28, 422.–433.
Carvalho, F. A., Barnich, N., Sivignon, A., Darcha, C., Chan, C. H., Stanners, C. P., et al. (2009). Crohn’s disease adherent-invasive Escherichia coli colonize and induce strong gut inflammation in transgenic mice expressing human CEACAM. J. Exp. Med. 206, 2179–2189. doi: 10.1084/jem.20090741
Chassaing, B., Rolhion, N., De Vallée, A., Sa’ad, Y. S., Prorok-Hamon, M., Neut, C., et al. (2011). Crohn disease–associated adherent-invasive E. coli bacteria target mouse and human Peyer’s patches via long polar fimbriae. J. Clin. Investig. 121, 966–975. doi: 10.1172/JCI44632
Chiodini, R. J., Van Kruiningen, H. J., Thayer, W. R., Merkal, R. S., and Coutu, J. A. (1984). Possible role of mycobacteria in inflammatory bowel disease. Dig. Dis. Sci. 29, 1073–1079.
Chung, H. K., Tay, A., Octavia, S., Chen, J., Liu, F., Ma, R., et al. (2016). Genome analysis of Campylobacter concisus strains from patients with inflammatory bowel disease and gastroenteritis provides new insights into pathogenicity. Sci, Rep. 6:38442.
Clayton, E. M., Rea, M. C., Shanahan, F., Quigley, E. M., Kiely, B., Hill, C., et al. (2009). The vexed relationship between Clostridium difficile and inflammatory bowel disease: an assessment of carriage in an outpatient setting among patients in remission. Am. J. Gastroenterol. 104, 1162–1169. doi: 10.1038/ajg.2009.4
Cornelius, A. J., Huq, M., On, S. L., French, N. P., Vandenberg, O., Miller, W. G., et al. (2021). Genetic characterisation of Campylobacter concisus: strategies for improved genomospecies discrimination. Syst. Appl. Microbiol. 44:126187. doi: 10.1016/j.syapm.2021.126187
Darfeuille-Michaud, A., Boudeau, J., Bulois, P., Neut, C., Glasser, A.-L., Barnich, N., et al. (2004). High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. J. Gastroenterol. 127, 412–421. doi: 10.1053/j.gastro.2004.04.061
De Souza, H. S., and Fiocchi, C. (2016). Immunopathogenesis of IBD: current state of the art. Nat. Rev. Gastroenterol. Hepatol. 13, 13–27. doi: 10.1038/nrgastro.2015.186
Denning, T. L., Wang, Y.-C., Patel, S. R., Williams, I. R., and Pulendran, B. (2007). Lamina propria macrophages and dendritic cells differentially induce regulatory and interleukin 17–producing T cell responses. Nat. Immunol. 8, 1086–1094. doi: 10.1038/ni1511
Dreux, N., Denizot, J., Martinez-Medina, M., Mellmann, A., Billig, M., Kisiela, D., et al. (2013). Point mutations in FimH adhesin of Crohn’s disease-associated adherent-invasive Escherichia coli enhance intestinal inflammatory response. PLoS Pathog. 9:e1003141. doi: 10.1371/journal.ppat.1003141
Feller, M., Huwiler, K., Schoepfer, A., Shang, A., Furrer, H., and Egger, M. (2010). Long-term antibiotic treatment for Crohn’s disease: systematic review and meta-analysis of placebo-controlled trials. Clin. Infect. Dis. 50, 473–480. doi: 10.1086/649923
Franco, A. A., Mundy, L. M., Trucksis, M., Wu, S., Kaper, J. B., and Sears, C. L. (1997). Cloning and characterization of the Bacteroides fragilis metalloprotease toxin gene. Infect. Immun. 65, 1007–1013. doi: 10.1128/iai.65.3.1007-1013.1997
Harrison, O. J., and Powrie, F. M. (2013). Regulatory T cells and immune tolerance in the intestine. Cold Spring Harb. Perspect. Biol. 5:a018341. doi: 10.1101/cshperspect.a018341
Hippenstiel, S., Schmeck, B., N’guessan, P. D., Seybold, J., Krüll, M., Preissner, K., et al. (2002). Rho protein inactivation induced apoptosis of cultured human endothelial cells. Am. J. Physiol. Lung. Cell Mol. Physiol. 283, L830–L838. doi: 10.1152/ajplung.00467.2001
Inagaki, H., Suzuki, T., Nomoto, K., and Yoshikai, Y. (1996). Increased susceptibility to primary infection with Listeria monocytogenes in germfree mice may be due to lack of accumulation of L-selectin+ CD44+ T cells in sites of inflammation. Infect. Immun. 64, 3280–3287. doi: 10.1128/iai.64.8.3280-3287.1996
Irving, P. M., De Lusignan, S., Tang, D., Nijher, M., and Barrett, K. (2021). Risk of common infections in people with inflammatory bowel disease in primary care: a population-based cohort study. BMJ Open Gastroenterol. 8:e000573. doi: 10.1136/bmjgast-2020-000573
Ismail, Y., Lee, H., Riordan, S. M., Grimm, M. C., and Zhang, L. (2013). The effects of oral and enteric Campylobacter concisus strains on expression of TLR4. MD-2, TLR2, TLR5 and COX-2 in HT-29 cells. PLoS One 8:e56888. doi: 10.1371/journal.pone.0056888
Ismail, Y., Mahendran, V., Octavia, S., Day, A. S., Riordan, S. M., Grimm, M. C., et al. (2012). Investigation of the enteric pathogenic potential of oral Campylobacter concisus strains isolated from patients with inflammatory bowel disease. PLoS One 7:e38217. doi: 10.1371/journal.pone.0038217
Issa, M., Vijayapal, A., Graham, M. B., Beaulieu, D. B., Otterson, M. F., Lundeen, S., et al. (2007). Impact of Clostridium difficile on inflammatory bowel disease. Clin. Gastroenterol. Hepatol. 5, 345–351.
Jodorkovsky, D., Young, Y., and Abreu, M. T. (2010). Clinical outcomes of patients with ulcerative colitis and co-existing Clostridium difficile infection. Dig. Dis. Sci. 55, 415–420. doi: 10.1007/s10620-009-0749-9
Johansson, M. E., Sjövall, H., and Hansson, G. C. (2013). The gastrointestinal mucus system in health and disease. Nat. Rev. Gastroenterol. Hepatol. 10, 352–361. doi: 10.1038/nrgastro.2013.35
Johnston, R. D., and Logan, R. F. (2008). What is the peak age for onset of IBD? Inflamm. Bowel. Dis. 14, S4–S5. doi: 10.1002/ibd.20545
Kaakoush, N. O., Holmes, J., Octavia, S., Man, S. M., Zhang, L., Castano-Rodriguez, N., et al. (2010). Detection of Helicobacteraceae in intestinal biopsies of children with Crohn’s disease. Helicobacter 15, 549–557. doi: 10.1111/j.1523-5378.2010.00792.x
Kim, H., Kokkotou, E., Na, X., Rhee, S. H., Moyer, M. P., Pothoulakis, C., et al. (2005). Clostridium difficile Toxin A–induced colonocyte apoptosis involves p53-dependent p21 (WAF1/CIP1) induction via p38 mitogen-activated protein kinase. Gastroenterology 129, 1875–1888. doi: 10.1053/j.gastro.2005.09.011
Kim, J., Oh, Y., Kim, Y., Oh, H., and Cho, Y. (2001). Polarized secretion of CXC chemokines by human intestinal epithelial cells in response to Bacteroides fragilis enterotoxin: NF-κB plays a major role in the regulation of IL-8 expression. Clin. Exp. Immunol. 123, 421–427. doi: 10.1046/j.1365-2249.2001.01462.x
Kirk, K. F., Nielsen, H. L., Thorlacius-Ussing, O., and Nielsen, H. (2016). Optimized cultivation of Campylobacter concisus from gut mucosal biopsies in inflammatory bowel disease. Gut Pathog. 8:27. doi: 10.1186/s13099-016-0111-7
Lamont, J. T., and Trnka, Y. (1980). Therapeutic implications of Clostridium difficile toxin during relapse of chronic inflammatory bowel disease. Lancet 315, 381–383. doi: 10.1016/s0140-6736(80)90939-3
Lapaquette, P., Bringer, M. A., and Darfeuille-Michaud, A. (2012). Defects in autophagy favour adherent-invasive Escherichia coli persistence within macrophages leading to increased pro-inflammatory response. Cell Microbiol. 14, 791–807. doi: 10.1111/j.1462-5822.2012.01768.x
Lee, H., Ma, R., Grimm, M. C., Riordan, S. M., Lan, R., Zhong, L., et al. (2014). Examination of the anaerobic growth of Campylobacter concisus strains. Int. J. Microbiol. 2014:476047.
Lee, S. A., Liu, F., Riordan, S., Lee, C. S., and Zhang, L. (2019). Global investigations of Fusobacterium nucleatum in human colorectal cancer. Front. Oncol. 9:566. doi: 10.3389/fonc.2019.00566
Lee, S. A., Liu, F., Yun, D. Y., Riordan, S. M., Tay, A. C. Y., Liu, L., et al. (2021). Campylobacter concisus upregulates PD-L1 mRNA expression in IFN-γ sensitized intestinal epithelial cells and induces cell death in esophageal epithelial cells. J. Oral. Microbiol. 13:1978732. doi: 10.1080/20002297.2021.1978732
Lee, Y., Eun, C. S., Lee, A. R., Park, C. H., and Han, D. S. (2016). Fusobacterium isolates recovered from colonic biopsies of inflammatory bowel disease patients in Korea. Ann. Lab. Med. 36, 387–389. doi: 10.3343/alm.2016.36.4.387
Liu, F., Chen, S., Luu, L. D. W., Lee, S. A., Tay, A. C. Y., Wu, R., et al. (2020). Analysis of complete Campylobacter concisus genomes identifies genomospecies features, secretion systems and novel plasmids and their association with severe ulcerative colitis. Microb. Genom. 6:mgen000457. doi: 10.1099/mgen.0.000457
Liu, F., Lee, H., Lan, R., and Zhang, L. (2016). Zonula occludens toxins and their prophages in Campylobacter species. Gut. Pathog. 8:43. doi: 10.1186/s13099-016-0125-1
Liu, F., Lee, S. A., Riordan, S. M., Zhang, L., and Zhu, L. (2019). Effects of anti-cytokine antibodies on gut barrier function. Mediators Inflamm 2019:7028253. doi: 10.1155/2019/7028253
Liu, F., Ma, R., Riordan, S. M., Grimm, M. C., Liu, L., Wang, Y., et al. (2017). Azathioprine, mercaptopurine, and 5-Aminosalicylic acid affect the growth of IBD-associated Campylobacter species and other enteric microbes. Front. Microbiol. 8:7028253.
Liu, F., Ma, R., Tay, C. Y. A., Octavia, S., Lan, R., Chung, H. K. L., et al. (2018a). Genomic analysis of oral Campylobacter concisus strains identified a potential bacterial molecular marker associated with active Crohn’s disease. Emerg. Microbes Infect. 7:64. doi: 10.1038/s41426-018-0065-6
Liu, F., Ma, R., Wang, Y., and Zhang, L. (2018b). The clinical importance of Campylobacter concisus and other human hosted Campylobacter species. Front. Cell Infect. Microbiol. 8:243. doi: 10.3389/fcimb.2018.00243
Liu, H., Hong, X. L., Sun, T. T., Huang, X. W., Wang, J. L., and Xiong, H. (2020). Fusobacterium nucleatum exacerbates colitis by damaging epithelial barriers and inducing aberrant inflammation. J. Dig. Dis. 21, 385–398. doi: 10.1111/1751-2980.12909
Liu, L., Liang, L., Liang, H., Wang, M., Lu, B., Xue, M., et al. (2019). Fusobacterium nucleatum aggravates the progression of colitis by regulating M1 macrophage polarization via AKT2 pathway. Front. Immunol. 10:1324. doi: 10.3389/fimmu.2019.01324
Lloyd-Price, J., Arze, C., Ananthakrishnan, A. N., Schirmer, M., Avila-Pacheco, J., Poon, T. W., et al. (2019). Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569, 655–662. doi: 10.1038/s41586-019-1237-9
Mahendran, V., Liu, F., Riordan, S., Grimm, M., Tanaka, M., and Zhang, L. (2016). Examination of the effects of Campylobacter concisus zonula occludens toxin on intestinal epithelial cells and macrophages. Gut. Pathog. 8:18. doi: 10.1186/s13099-016-0101-9
Mahendran, V., Riordan, S. M., Grimm, M. C., Tran, T. A., Major, J., Kaakoush, N. O., et al. (2011). Prevalence of Campylobacter species in adult Crohn’s disease and the preferential colonization sites of Campylobacter species in the human intestine. PLoS One 6:e25417. doi: 10.1371/journal.pone.0025417
Man, S. M., Kaakoush, N. O., Leach, S. T., Nahidi, L., Lu, H. K., Norman, J., et al. (2010a). Host attachment, invasion, and stimulation of proinflammatory cytokines by Campylobacter concisus and other non-Campylobacter jejuni Campylobacter species. J. Infect. Dis. 202, 1855–1865. doi: 10.1086/657316
Man, S. M., Zhang, L., Day, A. S., Leach, S. T., Lemberg, D. A., and Mitchell, H. (2010b). Campylobacter concisus and other Campylobacter species in children with newly diagnosed Crohn’s disease. Inflamm Bowel Dis. 16, 1008–1016.
Man, S. M., Zhang, L., Day, A. S., Leach, S., and Mitchell, H. (2008). Detection of enterohepatic and gastric Helicobacter species in fecal specimens of children with Crohn’s disease. Helicobacter 13, 234–238. doi: 10.1111/j.1523-5378.2008.00607.x
Martin, H. M., Campbell, B. J., Hart, C. A., Mpofu, C., Nayar, M., Singh, R., et al. (2004). Enhanced Escherichia coli adherence and invasion in Crohn’s disease and colon cancer. Gastroenterology 127, 80–93. doi: 10.1053/j.gastro.2004.03.054
Masclee, G. M., Penders, J., Jonkers, D. M., Wolffs, P. F., and Pierik, M. J. (2013). Is Clostridium difficile associated with relapse of inflammatory bowel disease? results from a retrospective and prospective cohort study in the Netherlands. Inflamm Bowel Dis. 19, 2125–2131. doi: 10.1097/MIB.0b013e318297d222
Matarrese, P., Falzano, L., Fabbri, A., Gambardella, L., Frank, C., Geny, B., et al. (2007). Clostridium difficile toxin B causes apoptosis in epithelial cells by thrilling mitochondria: involvement of ATP-sensitive mitochondrial potassium channels. J. Biol. Chem. 282, 9029–9041. doi: 10.1074/jbc.M607614200
McDonald, L. C., Killgore, G. E., Thompson, A., Owens, R. C. Jr., Kazakova, S. V., Sambol, S. P., et al. (2005). An epidemic, toxin gene–variant strain of Clostridium difficile. N. Engl. J. Med. 353, 2433–2441. doi: 10.1056/NEJMoa051590
Meyer, A. M., Ramzan, N. N., Loftus, E. V. Jr., Heigh, R. I., and Leighton, J. A. (2004). The diagnostic yield of stool pathogen studies during relapses of inflammatory bowel disease. J. Clin. Gastroenterol. 38, 772–775. doi: 10.1097/01.mcg.0000139057.05297.d6
Moncrief, J. S., Obiso, R. Jr., Barroso, L. A., Kling, J. J., Wright, R. L., Van Tassell, R. L., et al. (1995). The enterotoxin of Bacteroides fragilis is a metalloprotease. Infect. Immun. 63, 175–181. doi: 10.1128/iai.63.1.175-181.1995
Mukhopadhya, I., Thomson, J. M., Hansen, R., Berry, S. H., El-Omar, E. M., and Hold, G. L. (2011). Detection of Campylobacter concisus and other Campylobacter species in colonic biopsies from adults with ulcerative colitis. PLoS One 6:e21490. doi: 10.1371/journal.pone.0021490
Naser, S. A., Sagramsingh, S. R., Naser, A. S., and Thanigachalam, S. (2014). Mycobacterium avium subspecies paratuberculosis causes Crohn’s disease in some inflammatory bowel disease patients. World J. Gastroenterol. 20, 7403–7415.
Navaneethan, U., Mukewar, S., Gk Venkatesh, P., Lopez, R., and Shen, B. (2012). Clostridium difficile infection is associated with worse long term outcome in patients with ulcerative colitis. J. Crohn’s Colitis 6, 330–336. doi: 10.1016/j.crohns.2011.09.005
Ng, S. C., Shi, H. Y., Hamidi, N., Underwood, F. E., Tang, W., Benchimol, E. I., et al. (2017). Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet 390, 2769–2778. doi: 10.1016/S0140-6736(17)32448-0
Nielsen, H. L., Nielsen, H., Ejlertsen, T., Engberg, J., Günzel, D., Zeitz, M., et al. (2011). Oral and fecal Campylobacter concisus strains perturb barrier function by apoptosis induction in HT-29/B6 intestinal epithelial cells. PLoS One 6:e23858. doi: 10.1371/journal.pone.0023858
Nielsen, S., Thamsborg, S., Houe, H., and Bitsch, V. (2000). Bulk-tank milk ELISA antibodies for estimating the prevalence of paratuberculosis in Danish dairy herds. Prev. Vet. Med. 44, 1–7.
Nottrott, S., Schoentaube, J., Genth, H., Just, I., and Gerhard, R. (2007). Clostridium difficile toxin A-induced apoptosis is p53-independent but depends on glucosylation of Rho GTPases. Apoptosis 12, 1443–1453. doi: 10.1007/s10495-007-0074-8
O’Brien, C. L., Bringer, M.-A., Holt, K. E., Gordon, D. M., Dubois, A. L., Barnich, N., et al. (2017). Comparative genomics of Crohn’s disease-associated adherent-invasive Escherichia coli. Gut 66, 1382–1389. doi: 10.1136/gutjnl-2015-311059
Ohkusa, T., Okayasu, I., Ogihara, T., Morita, K., Ogawa, M., and Sato, N. (2003). Induction of experimental ulcerative colitis by Fusobacterium varium isolated from colonic mucosa of patients with ulcerative colitis. Gut 52, 79–83. doi: 10.1136/gut.52.1.79
Ohkusa, T., Sato, N., Ogihara, T., Morita, K., Ogawa, M., and Okayasu, I. (2002). Fusobacterium varium localized in the colonic mucosa of patients with ulcerative colitis stimulates species-specific antibody. J. Gastroenterol. Hepatol. 17, 849–853. doi: 10.1046/j.1440-1746.2002.02834.x
Pant, C., Anderson, M. P., Deshpande, A., Altaf, M. A., Grunow, J. E., Atreja, A., et al. (2013). Health care burden of Clostridium difficile infection in hospitalized children with inflammatory bowel disease. Inflamm Bowel Dis. 19, 1080–1085. doi: 10.1097/MIB.0b013e3182807563
Pittayanon, R., Lau, J. T., Leontiadis, G. I., Tse, F., Yuan, Y., Surette, M., et al. (2020). Differences in gut microbiota in patients with vs without inflammatory bowel diseases: a systematic review. Gastroenterology 158, 930.e1–946.e1. doi: 10.1053/j.gastro.2019.11.294
Prindiville, T. P., Sheikh, R. A., Cohen, S. H., Tang, Y. J., Cantrell, M. C., and Silva, J. Jr. (2000). Bacteroides fragilis enterotoxin gene sequences in patients with inflammatory bowel disease. Emerg. Infec. Dis. 6, 171–174. doi: 10.3201/eid0602.000210
Qa’Dan, M., Ramsey, M., Daniel, J., Spyres, L. M., Safiejko-Mroczka, B., Ortiz-Leduc, W., et al. (2002). Clostridium difficile toxin B activates dual caspase-dependent and caspase-independent apoptosis in intoxicated cells. Cell Microbiol. 4, 425–434. doi: 10.1046/j.1462-5822.2002.00201.x
Rhee, K.-J., Wu, S., Wu, X., Huso, D. L., Karim, B., Franco, A. A., et al. (2009). Induction of persistent colitis by a human commensal, enterotoxigenic Bacteroides fragilis, in wild-type C57BL/6 mice. Infect. Immun. 77, 1708–1718. doi: 10.1128/IAI.00814-08
Rice, J. H., Mcdaniel, M. M., Holland, A., and Eda, S. (2019). Modelling bovine granuloma formation in vitro upon infection with Mycobacterium avium subspecies paratuberculosis. Vet. Sci. 6:80. doi: 10.3390/vetsci6040080
Rodemann, J. F., Dubberke, E. R., Reske, K. A., Seo, D. H., and Stone, C. D. (2007). Incidence of Clostridium difficile infection in inflammatory bowel disease. Clin. Gastroenterol. Hepatol. 5, 339–344.
Round, J. L., and Mazmanian, S. K. (2009). The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9, 313–323. doi: 10.1038/nri2515
Roy, U., Gálvez, E. J. C., Iljazovic, A., Lesker, T. R., Błażejewski, A. J., Pils, M. C., et al. (2017). Distinct microbial communities trigger colitis development upon intestinal barrier damage via innate or adaptive immune cells. Cell Rep. 21, 994–1008. doi: 10.1016/j.celrep.2017.09.097
Rubinstein, M. R., Wang, X., Liu, W., Hao, Y., Cai, G., and Han, Y. W. (2013). Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe 14, 195–206. doi: 10.1016/j.chom.2013.07.012
Sairenji, T., Collins, K. L., and Evans, D. V. (2017). An update on inflammatory bowel disease. Prim Care 44, 673–692.
Santaolalla, R., and Abreu, M. T. (2012). Innate immunity in the small intestine. Curr. Opin. Gastroenterol. 28, 124–129. doi: 10.1097/mog.0b013e3283506559
Sasaki, M., Sitaraman, S. V., Babbin, B. A., Gerner-Smidt, P., Ribot, E. M., Garrett, N., et al. (2007). Invasive Escherichia coli are a feature of Crohn’s disease. Lab. Invest. 87, 1042–1054.
Savidge, T. C., Pan, W.-H., Newman, P., O’brien, M., Anton, P. M., and Pothoulakis, C. (2003). Clostridium difficile toxin B is an inflammatory enterotoxin in human intestine. Gastroenterology 125, 413–420. doi: 10.1016/s0016-5085(03)00902-8
Secott, T., Lin, T., and Wu, C. (2004). Mycobacterium avium subsp. paratuberculosis fibronectin attachment protein facilitates M-cell targeting and invasion through a fibronectin bridge with host integrins. Infect. Immun. 72, 3724–3732. doi: 10.1128/IAI.72.7.3724-3732.2004
Sheng, Y., Triyana, S., Wang, R., Das, I., Gerloff, K., Florin, T., et al. (2013). MUC1 and MUC13 differentially regulate epithelial inflammation in response to inflammatory and infectious stimuli. Mucosal Immunol. 6, 557–568. doi: 10.1038/mi.2012.98
Singh, S., Singh, P., Singh, A., Sohal, J., Kumar, N., Chaubey, K., et al. (2014). ‘Bio-Load’and bio-type profiles of Mycobacterium avium subspecies paratuberculosis infection in the domestic livestock population endemic for johne’s disease: a survey of 28 years (1985–2013) in India. Transbound Emerg. Dis. 61, 43–55. doi: 10.1111/tbed.12216
Singh, V., San Yeoh, B., Xiao, X., Kumar, M., Bachman, M., Borregaard, N., et al. (2015). Interplay between enterobactin, myeloperoxidase and lipocalin 2 regulates E. coli survival in the inflamed gut. Nat. Commun. 6, 1–11. doi: 10.1038/ncomms8113
Small, C.-L. N., Reid-Yu, S. A., Mcphee, J. B., and Coombes, B. K. (2013). Persistent infection with Crohn’s disease-associated adherent-invasive Escherichia coli leads to chronic inflammation and intestinal fibrosis. Nat. Commun. 4, 1–12. doi: 10.1038/ncomms2957
Smith, P., Smythies, L., Shen, R., Greenwell-Wild, T., Gliozzi, M., and Wahl, S. (2011). Intestinal macrophages and response to microbial encroachment. Mucosal Immunol. 4, 31–42. doi: 10.1038/mi.2010.66
Sprinz, H., Kundel, D. W., Dammin, G. J., Horowitz, R. E., Schneider, H., and Formal, S. B. (1961). The response of the germ-free guinea pig to oral bacterial challenge with Escherichia coli and Shigella flexneri: with special reference to lymphatic tissue and the intestinal tract. Am. J. Clin. Pathol. 39, 681–695.
Strauss, J., Kaplan, G. G., Beck, P. L., Rioux, K., Panaccione, R., Devinney, R., et al. (2011). Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm Bowel Dis. 17, 1971–1978. doi: 10.1002/ibd.21606
Strocchi, A., and Levitt, M. D. (1992). Factors affecting hydrogen production and consumption by human fecal flora. The critical roles of hydrogen tension and methanogenesis. J. Clin. Investig. 89, 1304–1311. doi: 10.1172/JCI115716
Su, W., Chen, Y., Cao, P., Chen, Y., Guo, Y., Wang, S., et al. (2020). Fusobacterium nucleatum promotes the development of ulcerative colitis by inducing the autophagic cell death of intestinal epithelial. Front. Cell Infect. Microbiol. 10:594806. doi: 10.3389/fcimb.2020.594806
Subramanian, S., Rhodes, J. M., Hart, A. C., Tam, B., Roberts, C. L., Smith, S. L., et al. (2008). Characterization of epithelial IL-8 response to inflammatory bowel disease mucosal E. coli and its inhibition by mesalamine. Inflamm Bowel Dis. 14, 162–175. doi: 10.1002/ibd.20296
Tahara, T., Shibata, T., Kawamura, T., Okubo, M., Ichikawa, Y., Sumi, K., et al. (2015). Fusobacterium detected in colonic biopsy and clinicopathological features of ulcerative colitis in Japan. Dig. Dis. Sci. 60, 205–210. doi: 10.1007/s10620-014-3316-y
Thursby, E., and Juge, N. (2017). Introduction to the human gut microbiota. Biochem. J. 474, 1823–1836. doi: 10.1042/bcj20160510
Uchiya, K.-I., Tomida, S., Nakagawa, T., Asahi, S., Nikai, T., and Ogawa, K. (2017). Comparative genome analyses of Mycobacterium avium reveal genomic features of its subspecies and strains that cause progression of pulmonary disease. Sci. Rep. 7, 1–14. doi: 10.1038/srep39750
Wang, Y., Liu, F., Zhang, X., Chung, H. K., Riordan, S. M., Grimm, M. C., et al. (2017). Campylobacter concisus genomospecies 2 is better adapted to the human gastrointestinal tract as compared with Campylobacter concisus genomospecies 1. Front. Physiol. 8:543. doi: 10.3389/fphys.2017.00543
Whiley, H., Keegan, A., Giglio, S., and Bentham, R. (2012). Mycobacterium avium complex–the role of potable water in disease transmission. J. Appl. Microbiol. 113, 223–232. doi: 10.1111/j.1365-2672.2012.05298.x
Wu, S., Lim, K.-C., Huang, J., Saidi, R. F., and Sears, C. L. (1998). Bacteroides fragilis enterotoxin cleaves the zonula adherens protein. E-cadherin. PNAS 95, 14979–14984. doi: 10.1073/pnas.95.25.14979
Wu, S., Powell, J., Mathioudakis, N., Kane, S., Fernandez, E., and Sears, C. L. (2004). Bacteroides fragilis enterotoxin induces intestinal epithelial cell secretion of interleukin-8 through mitogen-activated protein kinases and a tyrosine kinase-regulated nuclear factor-κB pathway. Infect. Immun. 72, 5832–5839. doi: 10.1128/IAI.72.10.5832-5839.2004
Wu, S., Rhee, K.-J., Albesiano, E., Rabizadeh, S., Wu, X., Yen, H.-R., et al. (2009). A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 15, 1016–1022. doi: 10.1038/nm.2015
Yang, Y., Weng, W., Peng, J., Hong, L., Yang, L., Toiyama, Y., et al. (2017). Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating Toll-like receptor 4 signaling to nuclear Factor–κB, and up-regulating expression of microRNA-21. Gastroenterology 152, 851.e24–866.e24. doi: 10.1053/j.gastro.2016.11.018
Yeow, M., Liu, F., Ma, R., Williams, T. J., Riordan, S. M., and Zhang, L. (2020). Analyses of energy metabolism and stress defence provide insights into Campylobacter concisus growth and pathogenicity. Gut. Pathog. 12:13. doi: 10.1186/s13099-020-00349-6
Zamani, S., Shariati, S. H., Zali, M. R., Aghdaei, H. A., Asiabar, A. S., Bokaie, S., et al. (2017). Detection of enterotoxigenic Bacteroides fragilis in patients with ulcerative colitis. Gut. Pathog. 9, 1–7.
Zhang, L. (2015). Oral Campylobacter species: initiators of a subgroup of inflammatory bowel disease? World J. Gastroenterol. 21, 9239–9244. doi: 10.3748/wjg.v21.i31.9239
Zhang, L., Day, A., Mckenzie, G., and Mitchell, H. (2006). Nongastric Helicobacter species detected in the intestinal tract of children. J. Clin. Microbiol. 44, 2276–2279. doi: 10.1128/JCM.02017-05
Keywords: inflammatory bowel disease, chronic inflammation, Campylobacter concisus, adherent-invasive Escherichia coli, Fusobacterium nucleatum, Mycobacterium avium subspecies paratuberculosis, Fusobacterium varium
Citation: Zhang L, Liu F, Xue J, Lee SA, Liu L and Riordan SM (2022) Bacterial Species Associated With Human Inflammatory Bowel Disease and Their Pathogenic Mechanisms. Front. Microbiol. 13:801892. doi: 10.3389/fmicb.2022.801892
Received: 26 October 2021; Accepted: 25 January 2022;
Published: 24 February 2022.
Edited by:
Honghua Hu, Macquarie University, AustraliaReviewed by:
Anna Aulicino, University of Oxford, United KingdomXiangdong Chen, China Pharmaceutical University, China
Copyright © 2022 Zhang, Liu, Xue, Lee, Liu and Riordan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Zhang, TC5aaGFuZ0B1bnN3LmVkdS5hdQ==
 Li Zhang
Li Zhang Fang Liu
Fang Liu Jessica Xue2
Jessica Xue2 Lu Liu
Lu Liu