
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 02 June 2022
Sec. Microbe and Virus Interactions with Plants
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.799396
Biotrophic plant pathogenic fungi are widely distributed and are among the most damaging pathogenic organisms of agriculturally important crops responsible for significant losses in quality and yield. However, the pathogenesis of obligate parasitic pathogenic microorganisms is still under investigation because they cannot reproduce and complete their life cycle on an artificial medium. The successful lifestyle of biotrophic fungal pathogens depends on their ability to secrete effector proteins to manipulate or evade plant defense response. By integrating genomics, transcriptomics, and effectoromics, insights into how the adaptation of biotrophic plant fungal pathogens adapt to their host populations can be gained. Efficient tools to decipher the precise molecular mechanisms of rust–plant interactions, and standardized routines in genomics and functional pipelines have been established and will pave the way for comparative studies. Deciphering fungal pathogenesis not only allows us to better understand how fungal pathogens infect host plants but also provides valuable information for plant diseases control, including new strategies to prevent, delay, or inhibit fungal development. Our review provides a comprehensive overview of the efforts that have been made to decipher the effector proteins of biotrophic fungal pathogens and demonstrates how rapidly research in the field of obligate biotrophy has progressed.
Plants are threatened by a variety of microorganisms during their growth, including biotrophs, necrotrophs, or hemi-biotrophs (Glazebrook, 2005). Different microorganisms use different strategies to infect and damage plants. Gratifying progress has been made so far in the pathogenesis of necrotrophic or hemi-biotrophic microbes, and the pre-infection pathways and pathogenesis of necrotrophic microbes have been identified. However, the pathogenesis of obligate parasitic pathogenic microorganisms is difficult because they cannot reproduce and complete their life cycle on artificial media. Biotrophic plant fungal pathogens are among the ten pathogens considered most important internationally to plant pathology (Dean et al., 2012). Obligate parasitic plant fungi are a large group among which rust and powdery mildew are the two largest groups. Rust fungi represent one of the most important fungal orders, consisting of more than 8,000 known species (Aime et al., 2014; Lorrain et al., 2019). These fascinating parasites attack a wide range of plants and cause severe damage (Lorrain et al., 2019). It is typical of many powdery mildews where the fungi form a powdery coating of white spores on the leaf surface and some common examples include powdery mildew of cereals and grasses (Erysiphe graminis) and gooseberry (Sphaerotheca mors-uvae) (Moore, 2020). The disease causes high yield losses of up to 30% (Hückelhoven, 2005).
All pathogenic fungi, despite their different infection and nutrient acquisition mechanisms, can be recognized by the defense system of the plant resulting in activation of the host defense system. A strong host response to fungal infection is facilitated by the activation of local and systemic responses due to innate immunity, over a prolonged period of time (Schwessinger and Ronald, 2012). The ability of plants to effectively respond to pathogen infection preliminarily depends on pre-formed mechanisms of defense, which include pre-formed barriers such as cuticle and phytoanticipins. Cuticle is an important barrier to pathogen penetration in which cell wall and cuticle thickness influence resistance against pathogens. In some plants, adult plant resistance is linked to the decreased ability of fungal pathogens to penetrate through the thicker and tougher cell walls (Guest and Brown, 1997). Cuticular wax is composed of cutin and wax which is deposited on the leaf surface hence water cannot be retained on it and this will complicate the germination of spores in the absence of water. Phytoanticipins act as pre-formed constitutive chemical barriers against microbial attack. Some of the phytoanticipins like glucosinolates and cyanogenic glycosides exist as inactive precursor stores in healthy tissues and are only activated as a result of tissue damage (Tiku, 2020). Pattern-recognition receptors (PRRs) localized on the host membrane recognize PAMPs (pathogen-associated molecular patterns) within the host apoplast during pathogen infection, which in turn activates PAMP-triggered immunity (PTI). Biotrophic plant fungal pathogens suppress PTI components by secreting virulence factors known as effectors into the host cells thereby causing diseases (Martel et al., 2021). In response, host plants evolved another layer of immunity in which the plant intracellular nucleotide-binding/leucine rich-repeat (NB-NLR) receptors and resistance (R) proteins specifically detect cognate avirulence factors and trigger a robust defense response called effector-triggered immunity (ETI). Direct or indirect effector recognition activates NRLs and this results in an array of induced mechanisms including reactive oxygen species (ROS) production, hypersensitive response, generation of phytoalexins, and accumulation of pathogenesis-related proteins (Amil-Ruiz et al., 2011). An integrated signaling network mediates the two-tier defense system and this system largely shares the downstream signaling machinery (Tsuda and Katagiri, 2010). How pathogens overcome the host’s plant two-tiered defense system is a hot topic in plant pathology (Harris et al., 2020). How the biotrophic plant pathogenic fungi can successfully infect the host and establish a parasitic relationship in which they can manipulate the host physiology and defense system is still the main area of work of many researchers worldwide.
The inability to culture or genetically modify obligate biotrophic fungal pathogens in vitro complicates the study of the molecular bases of rust fungal pathogenicity. With technological advances and the use of new experimental methods, transcriptome sequencing and metabolomics are increasing our understanding of infection strategies, the functions of biotrophic fungal effectors, and the mechanisms underlying pathogen-host interactions. Researchers have endeavored to discover the pathogenic mechanisms and in particular to study the functional characteristics of effectors secreted by biotrophic fungal pathogens through gene silencing, RNAi and overexpression, either in their host or in a heterologous system (Jaswal et al., 2020; Prasad et al., 2020). In this review, we describe the definitions, examples, and characteristic features of biotrophic fungal pathogens, and infection strategies of biotrophic plant fungi, the effectors of biotrophic fungal pathogens, and how they manipulate the plant immune system, and summarize the major research advances on biotrophic fungal pathogens.
Apart from the use of the term biotroph in several fields of research, there is still no clear definition of this terminology. A biotroph is characterized by an exceptional lifestyle that supports nutrient acquisition from living host cells and is completely dependent on the host for successful completion of the life cycle (Stotz et al., 2014; Gebrie, 2016). Biotrophs are pathogenic organisms that are completely dependent on the host cells for their nutrient acquisition and can secrete effectors to suppress or regulate plant basal defense. They form some sophisticated structures such as appressoria, infection peg, and haustoria to facilitate infection of epidermal cells and hyphae for nutrient uptake without damaging the host cells and with different forms in different Eumycota (Mendgen and Hahn, 2002; Delaye et al., 2013). Their lifestyle has complicated the detailed analysis of the molecular mechanisms underlying their pathogenicity, i.e., host evasion or suppression of plant immunity (Duplessis et al., 2011). Biotrophs generally include fungus rusts (Basidiomycetes), powdery mildew pathogens (Ascomycetes), and Oomycetes (downy mildew and white rusts) as obligate biotrophs. The biotrophic plant fungal pathogens consist of the two major groups of rust and powdery mildew. Rust fungi are among the most serious pathogens of major crops worldwide such as wheat, barley, soybean, maize, millet, flax, as well as coffee, etc. (Fisher et al., 2012). These include Puccinia striiformis f. sp. tritici (Pst), Puccinia graminis f. sp. tritici (Pgt), and Puccini triticina (Pt) which cause stripe rust, stem rust and leaf rust on wheat respectively, Melamspora lini (M. lini) (which cause flax rust), Phakospora pachyrhizi (which cause Asian soybean rust), Hemileia vastatrix (which cause coffee leaf rust), and Melampsora larici-populina, which cause defoliating poplar leaf rust disease. All these pathogens cause high yield losses of more than 15–∼80% if not controlled (Lawrence et al., 2007; Kelly et al., 2015). Powdery mildew fungi are widespread and vital fungal pathogens that infect over 10,000 plant species worldwide and cause yield losses of up to 40% in various frugal crops such as wheat and barley, as well as vegetable crops and fruit trees (Takamatsu, 2014).
One of the key features of obligate biotrophic fungal plant pathogens is their full dependence on the living host plant tissues, that is, they can only feed, grow, and reproduce on their living host (Latijnhouwers et al., 2003; Voegele and Mendgen, 2011; Kemen et al., 2015; Dracatos et al., 2018; Tang et al., 2018). They are highly and irreversibly adapted to their host plant. This implies that, generally, obligate biotrophic fungal pathogens has been defined by their inability to grow on artificial media. They have narrow, specialized host ranges and they enter their host through natural openings or by direct penetration (Moore, 2020). Biotrophic fungal pathogens are characterized by specialized structures called the haustoria that serve for nutrient absorption, and secretion of effector proteins to the host cytoplasm via the extrahaustorial matrix (Figure 1). The extrahaustorial matrix, a gel-matrix enriched in proteins and carbohydrates from both the pathogen and the host, is essential in maintaining the biotrophic lifestyle including plant pathogen recognition evasion (Perfect and Green, 2001; Green et al., 2002; Caillaud et al., 2014; Oliveira-Garcia and Valent, 2015). Although hemi-biotrophic fungal pathogens form haustoria during their biotrophic phase, the haustoria of the biotrophic fungal pathogens have a neck ring that seals the interface between the host and pathogen plasm membrane, disconnecting the extrahaustorial matrix from the plant and fungal apoplast and establishing a biotroph-specific compartment (Kemen et al., 2015; Figure 1). It is a characteristic of biotrophic plant pathogens that they are able to alter and reprogram the host metabolism (Kleemann et al., 2012). The ability of these pathogens to secrete effector proteins in planta is crucial in this context. They possess interfacial layers comprised of carbohydrates and proteins that separate fungal and plasma membranes. The characterization of the biotrophic plant fungal pathogens is based the genetic analysis of disease resistance with plants, how defense against fungal pathogens is regulated, and is also based on genome derived analysis of carbohydrate-active enzyme gene content. They secrete a limited amount of lytic enzymes to promote successful penetration of the host cell without any toxin production (Kemen et al., 2015). Host viability is essential for biotrophic plant fungal pathogens. Although biotrophic plant fungal pathogens are ubiquitous and have different lifestyles, they share some important common characteristics (Table 1).
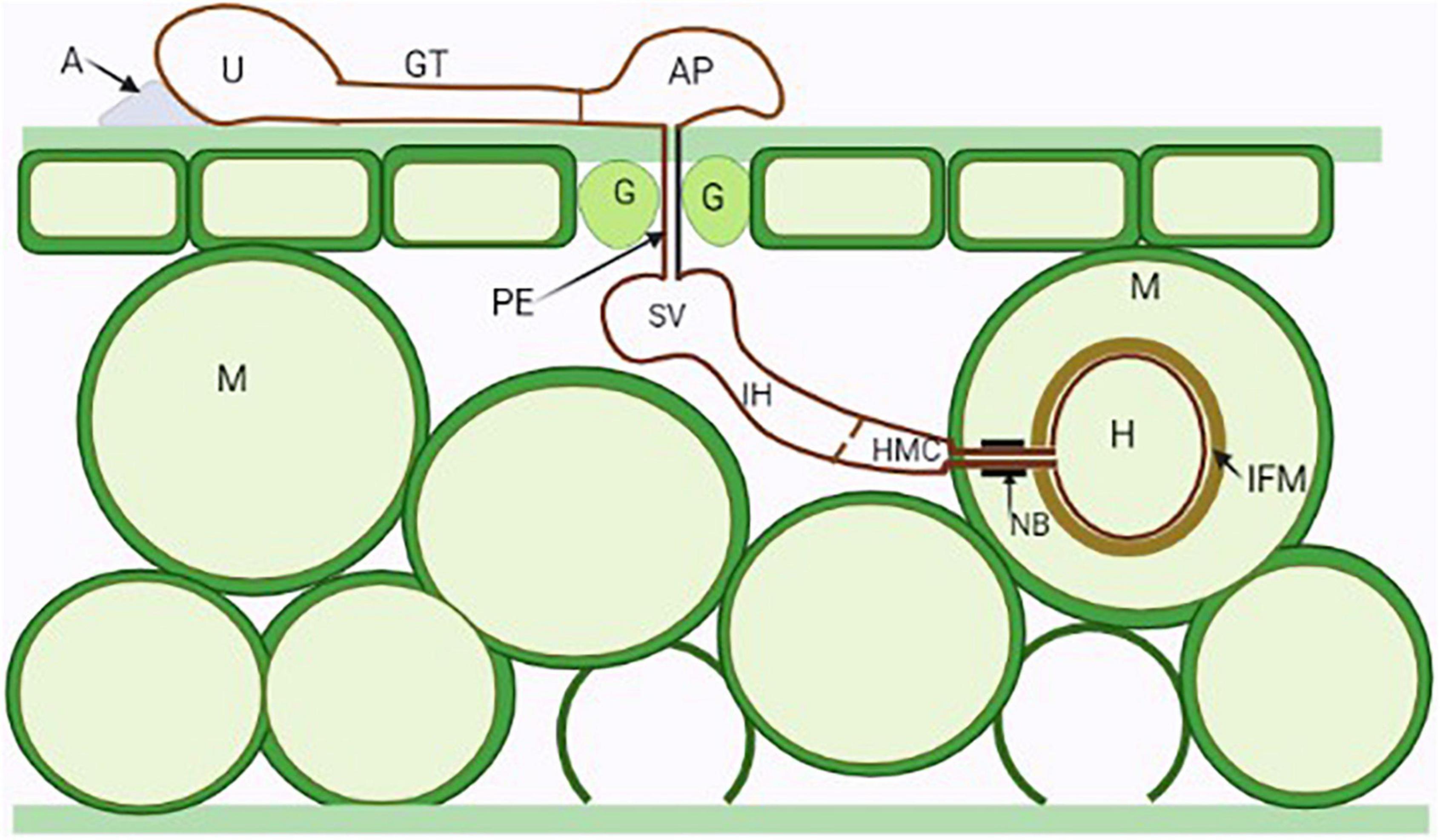
Figure 1. Illustration of the general infection process of a dikaryotic stage rust fungus. A urediospore (U) attached to an adhesion pad (A) germinate forming germ tubes (GT) which sense the topography of host cuticle and develop appressoria (AP) above stomata (Guard cells—G). Penetration occurs through stomata by the penetration hyphae (PE) into the substomatal spaces where the fungus differentiates into substomatal vesicles (SV) and elongates into an intercellular hypha (IH) which comes into contact with the host mesophyll cells (M) and develops haustorial mother cells (HMC). Following this, haustorial formation is initiated, neckbands (NB) are formed around the site of penetration of the mesophyll cell and an interfacial matrix (IFM) develop between the haustoria cell wall and the cell plasma membrane.
Biotrophic fungi employ various strategies to avoid triggering host defense responses. These include forming specialized infection structures to avoid host recognition, facilitating attachment, penetration, and propagation to successfully establish pathogenesis (Oliveira-Garcia and Valent, 2015).
Organisms that cause rusts and powdery mildews employ either direct penetration or penetration through the stomatal openings from the appressorium. In plant-pathogen interactions, the ability of the fungal spore to adhere to the host plant is considered a fundamental requirement for spore germination, germ tube elongation, and appressorium formation (Tucker and Talbot, 2001). Most powdery mildew fungal species use their infection structures called appressoria to penetrate directly into cuticle and cell wall of the host plant to colonize exclusively the epidermal cells of the plant, whereas most rust fungi form their appressorium over the stomata to penetrate the intercellular spaces of the mesophyll, potentially bypassing epidermal defense responses. Chemical or physical signals from the host plant including ethylene signals (Sharma and Gautam, 2019), topographic signals (Jones et al., 2001), cutin monomers (Sharma and Gautam, 2019), and substratum hydrophobicity (Kamakura et al., 2002) trigger the formation of appressoria.
In wheat rust, for example, the dikaryotic urediospores germinate within a short time once they land on the wheat epidermis when the humidity meets the germination requirements and the germ tube extends perpendicular to the vein and is directed toward stomatal cells of the leaf surface (Figure 1). An appressorium then forms above the stomata, followed by the development of a penetration peg to invade the host (Voegele et al., 2009). The formation of the penetration peg is initiated by haustorial mother cells, from which some lytic enzymes are secreted to promote successful penetration of the host cell wall, followed by the differentiation of the haustorium in the host cells (Kim, 2015). The continued growth of the peg within the substomatal chamber penetrates the host tissues (Chethana et al., 2021), and the cavity forms a vesicle (substomatal vesicle), which in turn forms one or more infection hyphae. Uromyces fabae secretes cell wall-degrading enzymes (pectate lyases, pectin esterases, and cellulases) under strict progressive regulation, only after the formation of the haustorial mother cells or infection hyphae and appressoria formation (Deising et al., 1995). The sequential production of these enzymes is directly related to the extreme local requirements for host cell entry during haustorium formation inside the leaf (Mendgen and Hahn, 2002).
Rust fungi possess monokaryotic and dikaryotic forms, all of which produce haustoria during their infection process. Dikaryotic haustoria arise from haustorial mother cells and have a haustorial neck with a haustorial body associated with the intercellular hyphae (Harder and Chong, 1984). The dikaryotic nature of fungal rust implies that they harbor significant genetic variation that is shared between the two haplotypes (Figueroa et al., 2020). Monokaryotic haustoria are terminal intercellular hyphae that have a septum near the penetration site (Gold and Mendgen, 1984; Harder and Chong, 1984; Heath, 1995). A distinct dense-neckband separates the extrahaustorial matrix of the rust fungus from the apoplasm (Figure 1). The formation of haustoria induces significant re-organization of host cytoskeleton, nuclear DNA and endomembrane system (Kobayashi et al., 1994; Heath, 1997; Voegele and Mendgen, 2003). The haustoria are not only a sophisticated structure for nutrient acquisition, but also an intense site where all secreted proteins, including effectors, are expressed (Garnica et al., 2014), and it has also been speculated that they are essential for signal transduction, communication, and evasion of host recognition (Hahn and Mendgen, 2001; Voegele and Mendgen, 2003, 2011). This implies that any interaction in obligate biotrophs involves exclusive and specific differentiation of infection structures, and that the exact specialization that occurs is highly dependent on the host and pathogen in question (Van Sumere et al., 1957). The urediospores of the soybean rust fungus Phakopsora pachyrhizi unwaveringly penetrate the host epidermis and form a highly turgid appressorium (Goellner et al., 2010; Loehrer et al., 2014). The physical force exerted by P. pachyrhizi appressoria based on turgor pressure is 3.9 or 5.5 μN, the osmotic potential is 5.13 MPa, and sugar alcohols are the predominant osmolytes within these cells (Loehrer et al., 2014). The turgor pressure of glycerol creates the mechanical strength of the appressorium through the breakdown of lipid and glycogen stores within as well as melanin in the cell walls of the appressoria (Latijnhouwers et al., 2003; Loehrer et al., 2014).
Powdery mildew fungal pathogens infect the aerial parts of higher plants and rarely kill their hosts, but their pathogenesis causes major yield losses mainly due to withdrawal of nutrients from the host plant, reduction of photosynthesis, stunted growth, and increased respiration and transpiration (Wu et al., 2021). Landing of conidial spores on the host surface is followed by attachment and penetration into the host cuticle and cell wall. They also form some sophisticated structures such as the appressorium, which is a key structure for invasion (Hückelhoven, 2005), and the hook-shaped appressorium grows over the cell surface of the epidermis. Meanwhile, powdery mildew germ tubes penetrate directly the host plant cell wall using both enzymatic and mechanical power forces (appressoria) (Green et al., 2002). Cell wall-degrading enzymes of plant pathogenic fungi play a fundamental role during infection by disrupting the host cell defense system and thereby acting as virulence or pathogenicity factors in some plant-pathogen interactions (Wood, 1960; Köller et al., 1982). During Bgh-barely interactions, Bgh accumulates 3-hydroxykynurenine, a redox-active substance that facilitates cross-linking of the pathogen to its host surface (Wilson et al., 2003). Bgh also expresses a secreted catalase that is essential in the removal of hydrogen peroxide produced by the host to cross-link its cell wall for penetration resistance, promoting invasive growth and spread during host infection (Zhang et al., 2004). Successful penetration results in the development of a penetration peg that penetrates directly into the epidermal cuticle of the host plant leading to the formation of haustoria which invaginates the host plasma membrane and are important for nutrient acquisition without damaging the host. The haustoria of powdery mildew formed in the epidermal cells of the host are ultimate unicellular structures consisting of a globular central body with projecting filamentous lobes (Gil and Gay, 1977).
Biotrophic plant pathogenic fungi can also attack their host by modulating pH which creates a favorable environment for infection, thus promoting pathogenicity (Strange, 2007). In addition, biotrophic pathogenic fungi produce some secondary metabolites such as siderophores, and pigments that promote their virulence and can also produce or modify hormones of the host plant such as; auxins (IAA), gibberellic acids (GA), jasmonic acid (JA), cytokinins, and abscisic acid (ABA), thus disrupting the corresponding plant hormone signaling pathways (Patkar et al., 2015; Shen et al., 2018). Wheat stem rust fungus specifically produces tryptophan 2-monooxygenase in the sucker, which causes the wheat cells to produce a large amount of indole acetic acid, which in turn regulates host disease resistance and promotes pathogen infection (Yin et al., 2014; Prasad et al., 2019). In research on the pathogenicity of Pt, adenylate cyclase (AC), a signaling molecule, was found to influence the pathogenicity of the leaf rust pathogen (Panwar et al., 2013).
Invading pathogens also require some sophisticated strategies to overcome the host defense system. For successful infection and establishment of compatible interactions leading to proliferation, biotrophic fungi must counteract PTI (Dangl and Jones, 2001) to evade the host defense system, and to facilitate this, the pathogen secretes several effector proteins. The pathogen’s effector proteins are distinctive recognition targets for the host defense system, whereas components of the host defense signals or their receptors are the main target of effectors during infection. Currently, effectors are considered as a group of proteins without mutual conservation, that are uniquely responsible for some crucial functions such as tethering the host defense machinery by producing cytotoxicity to facilitate pathogen progression (Jaswal et al., 2020). After invasion of the host, effectors suppress the host’s basal defense and manipulate its physiology to promote their invasive growth and multiplication. Several biotrophic fungal effectors exploit this mode of action by sequestrating chitin and also make use of other various mechanisms to subvert PTI (Table 2). Effector genes are subject to high selection pressures, and therefore evolve rapidly, giving them a particular mode of action that allows them to manipulate the host’s defense system and physiology. For a strong defense system against invading pathogens, the host harbors R-genes that recognize pathogen effectors and transforms a virulent pathogen into an avirulent one (Jones and Dangl, 2006). Avirulence (Avr) genes encoded effector proteins are recognized by host R genes (Flor, 1971). However, in various pathosystems there is an indirect interaction between R and Avr gene products and this follows a guard or decoy model (Jones and Dangl, 2006).
Effectors are mostly expressed in the haustoria and some expressed in infection hyphae of obligate biotrophs and can be categorized according to their specific site of action in the host, i.e., apoplastic and cytoplasmic effectors (Kamoun, 2006; Tanaka et al., 2020). Effectors of biotrophic fungal pathogens are either delivered in the cytoplasm of the host plant where they unswervingly manipulate processes in cells of the host (cytoplasmic effectors), or they may remain in the apoplast space to protect fungal cells from host plant defense components (apoplastic effectors) (Tanaka et al., 2020). Apoplastic effectors, which are important for full virulence, can inhibit host plant enzymes such as the peroxidases, chitinases, and proteases that can be harmful to the pathogen. Cytoplasmic effectors are delivered directly into the host plant cytoplasm, either by infection vesicles or haustoria, which are specialized structures (Kamoun, 2006), and accumulate in a biotrophic interfacial complex, some of them subsequently targeting specific compartments of the plant cell. They are recognized by intracellular R proteins of the cytoplasmic nucleotide-binding site leucine-rich repeat (NBS-LRR) and activate HR, which curbs invasive growth of the pathogen (Jones and Dangl, 2006; Dodds and Rathjen, 2010). Biotrophic plant fungal pathogens have also evolved some sophisticated mechanisms to direct their effectors to the host chloroplast by mimicking host sorting signals (Kretschmer et al., 2019). Effectors target the chloroplast where they suppress host plant basal immunity by suppressing cell death, decreasing the expression of defense-related genes and callose deposition, and causing accumulation of ROS (Petre et al., 2015; Xu et al., 2019).
Some biotrophic fungal effector proteins function as RNAi suppressors and it has been speculated that conserved proteins are the main target of these suppressors so that they can deactivate the host RNAi machinery. For example, the Pgt effector PgtSR1 suppresses host RNA silencing and impedes basal plant defense by swaying the spread of small RNAs that function as regulators of host defense (Yin et al., 2019). Host RNAi suppression machinery leads to inadequate regulation of various host defense pathways facilitating successful infection. Degradation of host RNA by biotrophic fungal effectors also helps in manipulating the host defense machinery (Jaswal et al., 2020). Pst02549 accumulates in processing bodies (P-bodies) and is involved in the uncapping, degradation, and storage of messenger RNA through the formation of protein complexes, thus facilitating infection (Petre et al., 2016). Effectors of biotrophic fungi also bind directly to the host DNA promoter, thereby regulating or altering its transcriptional processes, resulting in defective defense genes (Ahmed et al., 2018). Moreover, the effectors of biotrophic fungi camouflage the host as modulators, leading to the diversion of the metabolic flux of many compounds, resulting in a deficiency of precursors or compounds responsible for host defense (Tanaka et al., 2014; van der Linde and Göhre, 2021). Some well-known other examples are: CSEP0055 (Zhang et al., 2012), BEC3, BEC4 (Schmidt et al., 2014), CSEP0105, CSEP0162 (Ahmed et al., 2015), BEC1054 (Pennington et al., 2016), BEC1019 (Zhang et al., 2019), CSEP0027 (Yuan et al., 2021), CSEP0139 and CSEP0182 (Li et al., 2021) from Bgh and AvrPm3a1/f1 from Bgt (Bourras et al., 2015; Parlange et al., 2015). The effector protein PNPi of Pt acts directly on wheat TaNPR1 to inhibit resistance of wheat to leaf rust (Bi et al., 2020). AvrPm3a2/f2 and AvrPm2 of Bgt (Bourras et al., 2015, 2018), Avra1, Avra13 of Bgh (Lu et al., 2016), and SvrPma1/f1 suppress recognition of Avr genes (Parlange et al., 2015; Bourras et al., 2016). A substantial number of candidate effectors from biotrophic fungal pathogens that are well-known to infect crop plants have been predicted, cloned, and characterized (Table 2). From the current studies on biotrophic plant fungal pathogen effectors, it can be concluded that, effectors play important roles in pathogenicity by subverting PTI, suppressing cell death signaling activities, promoting virulence during infection, and some of these effectors are also required for penetration and formation of infection structures such as haustoria. This clearly asserts that biotrophic plant fungal pathogen effectors employ various mechanisms to either interfere with host PTI or ETI to enhance their pathogenicity.
Genome analysis has the potential to open up new paths for the discovery of new virulence factors in biotrophic plant fungal pathogens. More information about the genome and genes related with pathogenicity can be obtained by genome sequencing. Plant-pathogen interactions studies have been transformed by genomic technologies. With the advent of next generation sequencing technology (NGS), which has reduced the cost of sequencing, a reasonable number of genomes of biotrophic fungal pathogens have recently become available. In the case of wheat rust fungi, the first pathogen genome to be sequenced was Pgt, which had a size of 88.6 Mb (Duplessis et al., 2011). This was followed by whole genome sequencing of Pst. The genome of the US Pst race PST-130 was the first to be sequenced and the assembled genome was 64.8 Mb in size (Table 3). A Chinese Pst race CYR32 provided the subsequent genome with a size of 100 Mb and a large sequencing depth (Zheng et al., 2013). Another US pathotype PST-78 provided the third genome sequence of high quality, which was 117.31 Mb in size (Cuomo et al., 2017). Three draft genome sequences have been reported for Pt to date including one from an isolate of extensively virulent race 77 with a size of 95.22 Mb (Kiran et al., 2016), another from an isolate of narrowly avirulent race 106 with a size of 105.07 Mb (Kiran et al., 2016) and the last from race 1 (BBBD), the most primitive race recognized in North America, with a size of 135.34 Mb (Cuomo et al., 2017; Table 3). Subsequent sequencing of five Australian Pgt isolates (Upadhyaya et al., 2014) and 10 Pst isolates from different countries was performed (Cantu et al., 2013; Zheng et al., 2013) and sequence analysis revealed large genome heterozygosity of dikaryotic rust fungi. Sequencing of the genome of the wheat powdery mildew fungus Bgt revealed that it is 180 Mb in size and that 7,588 protein-coding genes are reported to be encoded in this genome (Table 3). Sequencing of three other isolates, 94,202, JIW2, and 70 from Switzerland, England, and Israel respectively, was also performed and found that the Bgt genome exhibits a high degree of adaptability and flexibility, indicating that this biotrophic pathogen has evolved uniquely (Wicker et al., 2013). When whole genome sequencing of wheat and barley powdery mildews was analyzed, a radical decrease in gene content was observed compared to other ascomycetes, and an expansion of the putative gene complement was observed (Spanu et al., 2010; Wicker et al., 2013).
During plant-pathogen interactions, the expression profiles of secreted proteins are characteristically complicated. Fungal structural differentiation, competing microbes, and cell-to-cell communication secrete various non-effectors that are essential for the colonization and fortification of biotrophic plant fungal pathogens. Consequently, the search for suitable effectors for subsequent experimental validation is economical for ensuing experimental corroboration. Discovery of biotrophic plant fungal effectors is classically performed using an integration of experimental techniques like genome-wide association study (Sánchez-Vallet et al., 2018; Richards et al., 2019), comparative genomics and phenotype association (Williams et al., 2016; Plissonneau et al., 2017; Wu et al., 2017, 2020; Beckerson et al., 2019; Chen et al., 2019; Mousavi-Derazmahalleh et al., 2019), proteomics (Gawehns et al., 2015; Mesarich et al., 2018), and transcriptomics (Gervais et al., 2017; Jones et al., 2019; Elmore et al., 2020; Human et al., 2020). Recently developed machine learning algorithms for detecting proteins with effector-like features have opened up new avenues for refining effector prediction pipelines. FunEffector-Pred (Wang et al., 2020) and EffectorP (Sperschneider et al., 2016, 2018b) utilize amino acid molecular weight, charge, frequencies, and other protein features to directly predict effector-like proteins. Tools like EffectorP and FunEffector-Pred, when combined with secretion prediction, may provide a more robust alternative to basic hard filters. Signal peptides of effectors can be predicted using some online searches. These tools involve integration of various taught algorithms and are usually very sensitive and accurate (Sonah et al., 2016). Since both transmembrane proteins and effector proteins contain hydrophobic segments, it is of great importance to distinguish the two and effectors have been found to contain a shorter hydrophobic portion compared to transmembrane proteins. Various online tools and WEB servers have been used for transmembrane domain prediction. These include TMHMM server v.2.0, and others (Sonah et al., 2016). YLoc, WoLF PSORT, Phobius, and SignalP are very helpful in predicting signal peptides as well as extracellular localization of candidate effectors. Prediction of the localization of candidate effectors from the plant cytoplasm, nucleus, mitochondria, and chloroplasts has been greatly improved by the development of the tools ApoplastP (Sperschneider et al., 2017; Sperschneider et al., 2018a) and LOCALIZER (Sperschneider et al., 2018b), and these tools are also beneficial for evaluating candidates but do not always predict effector candidature (Jones et al., 2021). However, computer-based prediction tools do not provide concrete results because they focus on a few features such as sequence similarities or small size. Therefore, it is necessary to include additional materials such as in-planta expression data and any other relevant information to improve the predictability of effectors of a pathogen of concern. The increased integration of next-generation sequencing and high-throughput genotype technologies has highly accelerated the identification of candidate secreted effector proteins (CSEPs) encoding Avr proteins in powdery mildews (Bourras et al., 2018). Bulk segregant analysis (BSA) (Bourras et al., 2015), map-based cloning, genome-wide association studies (GWAS) (Praz et al., 2017), and RNAseq based GWAS (Lu et al., 2016) were used in the identification of powdery mildew Avrs (Bourras et al., 2018). These methods were used in the cloning and characterization of four avirulence effectors: AvrPm3a2/f2, and AvrPm2 from Bgt which are recognized by Pm3a/f, and Pm2 wheat R genes, respectively (Bourras et al., 2015; Parlange et al., 2015; Lu et al., 2016; Praz et al., 2017), and Bgh Avra1 and Avra13 which are recognized by Mla1 and Mla13 barley R genes respectively (Lu et al., 2016), and also SvrPm3a1/f1 which suppresses Avr recognition (Bourras et al., 2015; Parlange et al., 2015). Due to the complexity of rust structure and large genome, RNA-seq is considered an effective method for identifying CSEPs. 16,399 and 17,773 secreted proteins were found from the sequencing of Mlp 98AG31 and Pgt SCCL transcriptome, respectively (Duplessis et al., 2011). Sequencing of differential virulence stripe rust races PST-87/7 and PST-08/21 from United Kingdom, identified only five candidate effector proteins with polymorphism among 2,999 secreted proteins (Cantu et al., 2013). Six different Pt races that interacted with wheat, 6 days post-inoculation were sequenced using Illumina platform and 532 secreted protein candidates were predicted, of which 456 were present in the assays for all races, and 12 Avr candidate genes were predicted (Bruce et al., 2014). The efficiency in predicting CSEPs is improved by integrated software that includes EffectorP v2.0, SignalP v4.1, TMHMM v2.0, and TargetP v1.1 for the analysis of RNAseq data. Recently, Zhang and colleagues used these softwares to screen CSEPs in wheat Pt KHTT, THSN, and JHKT isolates and obtained a total of 635 candidate effector proteins with small size (50-422 amino acids) and different sequences. Pt CSEPs were found to possess various family domains including thaumatin family, glycosyl hydrolase family, Barwin family, and others, and different motifs were also detected, such as RxLR, YxSL[R/K], [L/I]xAR, [Y/F/W]xC, G[I/F/Y] [A/L/S/T]R, and three de novo motifs (Zhang et al., 2020). For the identification of genes involved in Pt pathogenicity and virulence, Wei et al. (2020) performed RNA-seq at 144hpi to analyze the differentially expressed genes between THTT and THTS Pt pathotypes, to gain more insight into the molecular basis of Pt-wheat interaction. mRNA was sequenced and a total of 2,784 DGEs were detected, of which 45 genes were specifically expressed in THTS and 44 differentially expressed effector candidates were observed in both isolates. Analysis of the obtained results indicated that, although the two pathotypes of THTT and THTS contribute similar virulence to wheat, many genes are involved in the interaction with susceptible wheat cultivar Thatcher. This also demonstrates that the pathogenicity of rust is very complicated (Wei et al., 2020). In another recent study, genome-wide association was used in profiling of effector candidate associated with the infection process of the Pt pathotype PHTTP (P). Zhao et al. (2020) established that, 363 secreted proteins were encoded by upregulated genes, with only 79 of them also predicted as possible effectors by EffectorP. Using Regex and hmmsearch, 719 RXLR-like, 138Y/F/WxC, 19PNPi-like, 19CRN-like, and 9 CFEM effector candidate were identified from the deduced database, including transcriptome data from this study and data based on the Pt 1-1 BBBD Race 1 genome. Of the 19 PNPi-like candidate effectors, four of them possessing DPBB_ 1 conserved domain demonstrated physical interactions with wheat NPR1 protein in yeast two-hybrid assay. Only seven CFEM and nine Y/F/WxC candidate effectors were transiently expressed in Nicotiana benthamiana. None of the predicted effector candidates showed suppression or induction of cell death triggered by the protein BAX, but acceleration of cell death and enhanced ROS accumulation was only shown by the expression of PTTG_08198, a CFEM effector candidate (Zhao et al., 2020). AvrSr35, AvrSr50, and AvrSr27 were cloned, and mutation, RNA-seq and genome sequencing were performed on Pgt isolates (Chen et al., 2017; Salcedo et al., 2017; Upadhyaya et al., 2021). AvrSr35, AvrSr50, and AvrSr27 discovery did not only provide new tools for identifying Pgt avirulence gene and characterizing the molecular determinants of immunity in wheat, but also revealed the complexity of Pgt pathogenicity mechanism.
A comparative genomics approach was integrated with association analysis to identify candidate effector genes corresponding to Lr20 in phenotypically paired Australian Pt isolates. This study employed whole-genome sequencing to analyze twenty Pt isolates consisting of 10 phenotype-matched pairs with different Lr20 pathogenicity. This was the first study to integrate phenotype-genotype associations with effector prediction in Pt genomes and the approach used could circumvent the technical difficulties of working with obligate Pt and accelerate the identification of avirulence genes (Wu et al., 2017). Wu et al. (2020) carried long-read-based de novo genomy assembly and comparative genomics of wheat leaf rust pathotype Pt104 and identified candidates for avirulence genes. They established that AvrLr2a canditae GN104ID162_007386, AvrLrka candidate GN1041D162_024924, and the AvrLr26 candidate GN104ID162_020918 had corresponding orthologs of PTTG_07365, PTTG_28070, and PTTG_1194 in the Pt race 1 genome, respectively (Wu et al., 2020). In support of their findings, a proteomics study of Pt race 1 haustoria predicted that these Pt race 1 orthologs were also candidate effectors (Rampitsch et al., 2015).
The use of natural pathosystems to confirm biotrophic fungal effectors has been challenging, leading to the use of alternative hosts for their expression. This is now the most economical, rapid and applicable approach to validate biotrophic fungal effectors. In addition, N. benthamiana has been found to be an appropriate expression system for the obligate biotrophic fungal pathogens, particularly for validating rust effector proteins. This has also been successfully used in the identification of rust effectors acting at many sites in the plant cell compartments. In a recent study, the transient expression system of mesophyll protoplasts was found to be an important and useful tool for studying signal transduction in pathogen-model plant interactions (Su et al., 2021). Chitin and flg22 can be used as triggers for the PTI response in wheat leaves and TaPr-1-14 can also be used as a marker gene for detecting the PTI response. Screening and identification of Pst effector proteins can be performed rapidly and efficiently using a transient wheat protoplast expression system (Su et al., 2021). Out of 39 haustorial effector genes that were successfully screened and cloned using the protoplast transient expression system, three haustorial effector proteins PSEC2, PSEC17, and PSEC45 were identified and had the ability to inhibit wheat response to PTI. The three effector proteins were highly expressed during wheat infection and parasitism and were found to be localized in wheat protoplast somatic cytoplasm and nucleus (Su et al., 2021). The number of effectors identified recently has been greatly accelerated by access to fungal transcriptomic and genomic sequences. The triumph in identifying effectors could envisage their virulence-promoting or host-manipulating function, but, their exact function can only be guessed after identifying their interacting partners. Various approaches including co-immunoprecipitation (Co-IP) assays, yeast-two-hybrid (Y2H) approaches and pull-down assays of the target can be used to validate effector proteins and their interacting targets (Prasad et al., 2019; Jaswal et al., 2020). Advances in genomics studies paved a way for the identification of a repertoire of candidate effectors while some computational approaches coupled with high-throughput tools continue to help in the functional characterization of the effectors. A comprehensive understanding of pathosystems effector biology and the breeding of resistant crops has been facilitated by the discovery of a repertoire of effectors (Prasad et al., 2019).
The study of biotrophic pathogens has made considerable progress following the advent of the genomics era. However, most disease-causing organisms that grow on plants or have a reciprocal relationship with the host cannot be cultured. The lack of an effective genetic transformation system has hindered the progress in understanding the pathogenicity mechanisms of the biotrophic fungi. Little is known about how the biotrophic fungal pathogens absorb their nutrients from living host cells, and the genetic factors that determine the host exclusivity. How are the effector proteins delivered into the host cytoplasm and what is the functional compartment of the effectors? What is the target in the host? What affects the signal system pathway? A detailed answer to these questions is required to understand the pathogenicity mechanisms of biotrophic plant fungal pathogens. The mining and identification of candidate effectors and related genes are important first step toward functional assays to decipher their contribution to pathogenicity. The main research methods used in the study of biotrophic plant fungal pathogens include genome sequencing, transcriptome analysis, software analysis, and heterologous expression. Although they are more efficient than ever, they need to be improved, especially the need to develop a highly effective genetic transformation system. The establishment of a genetic transformation system for biotrophic fungi will greatly promote the study of pathogenic mechanisms of obligate parasites and the application of the corresponding genes. In order to broaden the toolbox to study biotrophic plant fungal pathogens, new methodologies and techniques such as for the visualization of plant-pathogens interactions must be developed. Rust–plant interactions still require effective techniques for unraveling their specific molecular mechanisms. Standardized routines in genomics needs to be developed as well as constructing functional pipelines to improve comparative studies. New techniques must be developed for the study of the dynamics of biotrophic interactions, such as metabolic pathways and fluxes, in situ.
Biotrophic plant fungal pathogens differ from the necrotrophs and hemi-biotrophs in their infection strategies and pathogenicity mechanisms, and are currently the focus of scientific research in molecular plant pathology because their sophisticated infection strategies make them a constant threat to many crops around the world. Normally, they require specialized infection structures to avoid host recognition and to facilitate their attachment and penetration. They also require a limited amount of enzymes to promote pathogen penetration and infection. The haustorium is the most important structure. Haustoria are considered not only as means of nutrition acquisition for the invasive growth of biotrophic plant fungal pathogens but also as an arsenal of effectors to bypass the host surveillance system and undermine the host’s multilayered defenses. The molecular bases of obligate biotrophy have been revealed by comparative genomic analyzes and transcriptome analyzes and these may pave the way for establishing biotrophic in vitro culture protocols for biotrophic plant fungal pathogens by imitating the absorption state of nutrients. Biotrophic plant fungal pathogens effectors are fundamental for establishing compatible interactions with the host plant. Moreover, they are undoubtedly virulence factors that are secreted to suppress or regulate host defenses by targeting host cognate proteins associated with resistance and modulating host cellular responses, thereby promoting virulence and pathogenicity. Due to the parasitic nature of biotrophic plant fungal pathogens, the lack of an effective transformation system affects the analysis of the functions of pathogenic fungi effector proteins and pathogenic genes. However, the current heterologous expression system and host-induced gene silencing technology still have some targeting and genetic background effects, which may affect the accurate analysis of the biological function of genes. Therefore, it is necessary to develop an efficient transformation system in the future to provide a good approach for gene function analysis of the biotrophic pathogenic fungi. Understanding effectors provide a fascinating illustration of the remarkably sophisticated molecular dialog that exists between biotrophic plant fungal pathogens and their hosts. By integrating genomics, transcriptomics and effectoromics, insights into the adaptation of biotrophic plant fungal pathogens to their host populations can be gained. Functional characterization of effectors during host-pathogen interactions will provide the basis for future studies on the role of effectors and haustoria, for understanding pathogenesis mechanisms, and for developing effective management strategies to control diseases caused by pathogenic rust fungi and powdery mildew.
JM and WY conceptualized the manuscript, went through all the research manuscripts, and finalized the manuscript. JM researched on the topic and wrote the review. All authors revised the manuscript and approved it for publication.
This work was funded by the Natural Science Foundation of China (Nos. 301571956, 301871915, and 32172367), Hebei Province Natural Science Foundation (No. C2020204071), National Key R&D Research Program of China (Nos. 2013CB127702 and 2017YFD020170700), Modern Agricultural Industry System of Wheat Industry in Hebei Province (No. HBCT2018010204).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Avr, avirulence; AC, adenylate cyclase; CSEPs, candidate secreted effector proteins; DAMPs, damaged-associated molecular patterns; ETI, effector-triggered immunity; PAMPs, pathogen-associated molecular patterns; PCD, programmed cell death; PRRs, pathogen recognition receptors; PTI, pathogen-associated molecular pattern triggered immunity; ROS, reactive oxygen species.
Ahmed, A. A., Pedersen, C., Schultz-Larsen, T., Kwaaitaal, M., Jørgensen, H. J., and Thordal-Christensen, H. (2015). The barley powdery mildew candidate secreted effector protein CSEP0105 inhibits the chaperone activity of a small heat shock protein. Plant Physiol. 168, 321–333. doi: 10.1104/pp.15.00278
Ahmed, A. A., Pedersen, C., and Thordal-Christensen, H. (2016). The barley powdery mildew effector candidates CSEP0081 and CSEP0254 promote fungal infection success. PLoS One. 11:e0157586. doi: 10.1371/journal.pone.0157586
Ahmed, H., Howton, T. C., Sun, Y., Weinberger, N., Belkhadir, Y., and Mukhtar, M. S. (2018). Network biology discovers pathogen contact points in host protein-protein interactomes. Nat. Commun. 9:2312. doi: 10.1038/s41467-018-04632-8
Aime, M., Toome, M., and McLaughlin, D. (2014). “The pucciniomycotina,” in The Mycota VII Part A, eds D. MacLaughlin and J. W. Spatafora (Berlin: Springer), 271–294.
Amil-Ruiz, F., Blanco-Portales, R., Muñoz-Blanco, J., and Caballero, J. L. (2011). The strawberry plant defense mechanism: a molecular review. Plant Cell Physiol. 52, 1873–1903. doi: 10.1093/pcp/pcr136
Beckerson, W. C., Rodríguez de la Vega, R. C., Hartmann, F. E., Duhamel, M., Giraud, T., Perlin, M. H., et al. (2019). Cause and effectors: whole-genome comparisons reveal shared but rapidly evolving effector sets among host-specific plant-castrating fungi. Am. Soc. Microbiol. 10:e02391-19. doi: 10.1128/mBio.02391-19
Belkhadir, Y., Subramaniam, R., and Dangl, J. L. (2004). Plant disease resistance protein signaling: NBS–LRR proteins and their partners. Curr. Opin. Plant Biol. 7, 391–399. doi: 10.1016/j.pbi.2004.05.009
Bi, W., Zhao, S., Zhao, J., Su, J., Yu, X., Liu, D., et al. (2020). Rust effector PNPi interacting with wheat TaPR1a attenuates plant defense response. Phytopathol. Res. 2:34. doi: 10.1186/s42483-020-00075-6
Bourras, S., McNally, K. E., Ben-David, R., Parlange, F., Roffler, S., Praz, C. R., et al. (2015). Multiple avirulence loci and allele-specific effector recognition control the Pm3 race-specific resistance of wheat to powdery mildew. Plant Cell 27, 2991–3012. doi: 10.1105/tpc.15.00171
Bourras, S., McNally, K. E., Müller, M. C., Wicker, T., and Keller, B. (2016). Avirulence genes in cereal powdery mildews: the gene-for-gene hypothesis 2.0. Front. Plant Sci. 7:241. doi: 10.3389/fpls.2016.00241
Bourras, S., Praz, C. R., Spanu, P. D., and Keller, B. (2018). Cereal powdery mildew effectors: a complex toolbox for an obligate pathogen. Curr. Opin. Microbiol. 46, 26–33. doi: 10.1016/j.mib.2018.01.018
Bruce, M., Neugebauer, K. A., Joly, D. L., Migeon, P., Cuomo, C. A., Wang, S., et al. (2014). Using transcription of six Puccinia triticina races to identify the effective secretome during infection of wheat. Front. Plant Sci. 4:520. doi: 10.3389/fpls.2013.00520
Caillaud, M.-C., Wirthmueller, L., Sklenar, J., Findlay, K., Piquerez, S. J. M., Jones, A. M. E., et al. (2014). The plasmodesmal protein PDLP1 localises to haustoria-associated membranes during downy mildew infection and regulates callose deposition. PLoS Pathog. 10:e1004496. doi: 10.1371/journal.ppat.1004496
Cantu, D., Govindarajulu, M., Kozik, A., Wang, M., Chen, X., Kojima, K. K., et al. (2011). Next generation sequencing provides rapid access to the genome of Puccinia striiformis f. sp. tritici, the causal agent of wheat stripe rust. PLoS One 6:e24230. doi: 10.1371/journal.pone.0024230
Cantu, D., Segovia, V., MacLean, D., Bayles, R., Chen, X., Kamoun, S., et al. (2013). Genome analyses of the wheat yellow (stripe) rust pathogen Puccinia striiformis f. sp. tritici reveal polymorphic and haustorial expressed secreted proteins as candidate effectors. BMC Genomics 14:270. doi: 10.1186/1471-2164-14-270
Chen, J., Upadhyaya, N. M., Ortiz, D., Sperschneider, J., Li, F., Bouton, C., et al. (2017). Loss of AvrSr50 by somatic exchange in stem rust leads to virulence for Sr50 resistance in wheat. Science 358, 1607–1610. doi: 10.1126/science.aao4810
Chen, J., Wu, J., Zhang, P., Dong, C., Upadhyaya, N. M., Zhou, Q., et al. (2019). De Novo genome assembly and comparative genomics of the barley leaf rust pathogen Puccinia hordei identifies candidates for three avirulence genes. G3 (Bethesda) 9, 3263–3271. doi: 10.1534/g3.119.400450
Cheng, Y., Wu, K., Yao, J., Li, S., Wang, X., Huang, L., et al. (2017). PSTha5a23, a candidate effector from the obligate biotrophic pathogen Puccinia striiformis f. sp. tritici, is involved in plant defense suppression and rust pathogenicity. Environ. Microbiol. 19, 1717–1729. doi: 10.1111/1462-2920.13610
Chethana, K. W. T., Jayawardena, R. S., Chen, Y.-J., Konta, S., Tibpromma, S., Abeywickrama, P. D., et al. (2021). Diversity and function of appressoria. Pathogens 10:746. doi: 10.3390/pathogens10060746
Cuomo, C. A., Bakkeren, G., Khalil, H. B., Panwar, V., Joly, D., Linning, R., et al. (2017). Comparative analysis highlights variable genome content of wheat rusts and divergence of the mating loci. G3 (Bethesda) 7, 361–376. doi: 10.1534/g3.116.032797
Dangl, J. L., and Jones, J. D. G. (2001). Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. doi: 10.1038/35081161
de Carvalho, M. C. d. C. G., Costa Nascimento, L., Darben, L. M. Polizel-Podanosqui, A. M. Lopes-Caitar, V. S., Qi, M., et al. (2017). Prediction of the in planta Phakopsora pachyrhizi secretome and potential effector families. Mol. Plant Pathol. 18, 363–377. doi: https://doi.org/10.1111/mpp.12405
De Guillen, K., Lorrain, C., Tsan, P., Barthe, P., Petre, B., Saveleva, N., et al. (2019). Structural genomics applied to the rust fungus Melampsora larici-populina reveals two candidate effector proteins adopting cystine knot and NTF2-like protein folds. Sci. Rep. 9:18084. doi: 10.1038/s41598-019-53816-9
Dean, R., Van Kan, J. A. L., Pretorius, Z. A., Hammond-Kosack, K. E., Di Pietro, A., Spanu, P. D., et al. (2012). The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. doi: 10.1111/j.1364-3703.2011.00783.x
Deising, H., Rauscher, M., Haug, M., and Heiler, S. (1995). Differentiation and cell wall degrading enzymes in the obligately biotrophic rust fungus Uromyces viciae-fabae. Can. J. Bot. 73, 624–631. doi: 10.1139/b95-304
Delaye, L., García-Guzmán, G., and Heil, M. (2013). Endophytes versus biotrophic and necrotrophic pathogens—are fungal lifestyles evolutionarily stable traits? Fungal Divers. 60, 125–135. doi: 10.1007/s13225-013-0240-y
Dodds, P. N., and Rathjen, J. P. (2010). Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11, 539–548. doi: 10.1038/nrg2812
Dracatos, P. M., Haghdoust, R., Singh, D., and Park, R. F. (2018). Exploring and exploiting the boundaries of host specificity using the cereal rust and mildew models. New Phytol. 218, 453–462. doi: 10.1111/nph.15044
Duplessis, S., Cuomo, C. A., Lin, Y.-C., Aerts, A., Tisserant, E., Veneault-Fourrey, C., et al. (2011). Obligate biotrophy features unravelled by the genomic analysis of rust fungi. Proc. Natl. Acad. Sci. U.S.A. 108:9166. doi: 10.1073/pnas.1019315108
Elmore, M. G., Banerjee, S., Pedley, K. F., Ruck, A., and Whitham, S. A. (2020). De novo transcriptome of Phakopsora pachyrhizi uncovers putative effector repertoire during infection. Physiol. Mol. Plant Pathol. 110:101464. doi: 10.1016/j.pmpp.2020.101464
Figueroa, M., Dodds, P. N., and Henningsen, E. C. (2020). Evolution of virulence in rust fungi – multiple solutions to one problem. Curr. Opin. Plant Biol. 56, 20–27. doi: 10.1016/j.pbi.2020.02.007
Fisher, M. C., Henk, D. A., Briggs, C. J., Brownstein, J. S., Madoff, L. C., McCraw, S. L., et al. (2012). Emerging fungal threats to animal, plant and ecosystem health. Nature 484, 186–194. doi: 10.1038/nature10947
Flor, H. H. (1971). Current status of the gene-for-gene concept. Ann. Rev. Phytopathol. 9, 275–296. doi: 10.1146/annurev.py.09.090171.001423
Gaouar, O., Morency, M.-J., Letanneur, C., Seguin, A., and Germain, H. (2016). The 124202 candidate effector of Melampsora larici-populina interacts with membranes in Nicotiana and Arabidopsis. Can. J. Plant Pathol. 38, 197–208. doi: 10.1080/07060661.2016.1153523
Garnica, D. P., Nemri, A., Upadhyaya, N. M., Rathjen, J. P., and Dodds, P. N. (2014). The ins and outs of rust haustoria. PLoS Pathog. 10:e1004329. doi: 10.1371/journal.ppat.1004329
Gawehns, F., Ma, L., Bruning, O., Houterman, P. M., Boeren, S., Cornelissen, B. J., et al. (2015). The effector repertoire of Fusarium oxysporum determines the tomato xylem proteome composition following infection. Front. Plant Sci. 6:967. doi: 10.3389/fpls.2015.00967
Gebrie, S. (2016). Biotrophic fungi infection and plant defense mechanism. J. Plant Pathol. Microbiol. 7:378. doi: 10.4172/2157-7471.1000378
Gervais, J., Plissonneau, C., Linglin, J., Meyer, M., Labadie, K., Cruaud, C., et al. (2017). Different waves of effector genes with contrasted genomic location are expressed by Leptosphaeria maculans during cotyledon and stem colonization of oilseed rape. Mol. Plant Pathol. 18, 1113–1126. doi: 10.1111/mpp.12464
Gil, F., and Gay, J. (1977). Ultrastructural and physiological properties of the host interfacial components of haustoria of Erysiphe pisi in vivo and in vitro. Physiol. Plant Pathol. 10, 1–12.
Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. doi: 10.1146/annurev.phyto.43.040204.135923
Goellner, K., Loehrer, M., Langenbach, C., Conrath, U., Koch, E., and Schaffrath, U. (2010). Phakopsora pachyrhizi, the causal agent of Asian soybean rust. Mol. Plant Pathol. 11, 169–177. doi: 10.1111/j.1364-3703.2009.00589.x
Gold, R. E., and Mendgen, K. (1984). Cytology of basidiospore germination, penetration, and early colonization of Phaseolus vulgaris by Uromyces appendiculatus var. appendiculatus. Can. J. Bot. 62, 1989–2002. doi: 10.1139/b84-271
Green, J., Carver, T., and Gurr, S. (2002). “The formation and function of infection feeding structures,” in Powdery Mildews: A Comprehensive Treatise, eds R. R. Belanger, W. R. Bushnell, A. J. Dik, and T. L. W. Carver (St. Paul, MN: American Phytopathological Society), 66–82.
Guest, D., and Brown, J. (1997). “Plant defences against pathogens,” in Plant Pathogens and Plant Diseases, eds J. F. Brown and J. H. Ogle (Armidale, NSW: Rockvale Publications).
Hahn, M., and Mendgen, K. (1997). Characterization of in planta-induced rust genes isolated from a haustorium-specific cDNA library. Mol. Plant Microbe Interact. 10, 427–437. doi: 10.1094/mpmi.1997.10.4.427
Hahn, M., and Mendgen, K. (2001). Signal and nutrient exchange at biotrophic plant-fungus interfaces. Curr. Opin. Plant Biol. 4, 322–327. doi: 10.1016/s1369-5266(00)00180-1
Hammond-Kosack, K. E., and Parker, J. E. (2003). Deciphering plant–pathogen communication: fresh perspectives for molecular resistance breeding. Curr. Opin. Biotechnol. 14, 177–193. doi: 10.1016/S0958-1669(03)00035-1
Harder, D. E., and Chong, J. (1984). “Structure and physiology of haustoria,” in The Cereal Rusts, vol 1, Origins, Specificity, Structure and Physiology, eds W. R. Bushell and A. R. Roelfs (Orlando, FL: Academic Press), 431–476.
Harris, J. M., Balint-Kurti, P., Bede, J. C., Day, B., Gold, S., Goss, E. M., et al. (2020). What are the top 10 unanswered questions in molecular plant-microbe interactions? Mol. Plant Microbe Interact. 33, 1354–1365. doi: 10.1094/MPMI-08-20-0229-CR
Heath, M. C. (1995). Signal exchange between higher plants and rust fungi. Can. J. Bot. 73, 616–623. doi: 10.1139/b95-303
Heath, M. C. (1997). Signalling between pathogenic rust fungi and resistant or susceptible host plants. Ann. Bot. 80, 713–720. doi: 10.1006/anbo.1997.0507
Hückelhoven, R. (2005). Powdery mildew susceptibility and biotrophic infection strategies. FEMS Microbiol. Lett. 245, 9–17. doi: 10.1016/j.femsle.2005.03.001
Human, M. P., Berger, D. K., and Crampton, B. G. (2020). Time-course RNAseq reveals exserohilum turcicum effectors and pathogenicity determinants. Front. Microbiol. 11:360. doi: 10.3389/fmicb.2020.00360
Jaswal, R., Kiran, K., Rajarammohan, S., Dubey, H., Singh, P. K., Sharma, Y., et al. (2020). Effector biology of biotrophic plant fungal pathogens: current advances and future prospects. Microbiol. Res. 241:126567. doi: 10.1016/j.micres.2020.126567
Jones, D. A. B., John, E., Rybak, K., Phan, H. T. T., Singh, K. B., Lin, S. Y., et al. (2019). A specific fungal transcription factor controls effector gene expression and orchestrates the establishment of the necrotrophic pathogen lifestyle on wheat. Sci. Rep. 9:15884. doi: 10.1038/s41598-019-52444-7
Jones, D. A. B., Rozano, L., Debler, J. W., Mancera, R. L., Moolhuijzen, P. M., and Hane, J. K. (2021). An automated and combinative method for the predictive ranking of candidate effector proteins of fungal plant pathogens. Sci. Rep. 11:19731. doi: 10.1038/s41598-021-99363-0
Jones, H., Whipps, J., and Gurr, S. (2001). The tomato powdery mildew fungus Oidium neolycopersici. Mol. Plant Pathol. 2, 303–309. doi: 10.1046/j.1464-6722.2001.00084.x
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kamakura, T., Yamaguchi, S., Saitoh, K., Teraoka, T., and Yamaguchi, I. (2002). A novel gene, CBP1, encoding a putative extracellular chitin-binding protein, may play an important role in the hydrophobic surface sensing of Magnaporthe grisea during appressorium differentiation. Mol. Plant Microbe Interact. 15, 437–444. doi: 10.1094/mpmi.2002.15.5.437
Kamoun, S. (2006). A catalogue of the effector secretome of plant pathogenic oomycetes. Annu. Rev. Phytopathol. 44, 41–60. doi: 10.1146/annurev.phyto.44.070505.143436
Kelly, H. Y., Dufault, N. S., Walker, D. R., Isard, S. A., Schneider, R. W., Giesler, L. J., et al. (2015). From select agent to an established pathogen: the response to Phakopsora pachyrhizi (Soybean rust) in North America. Phytopathology 105, 905–916. doi: 10.1094/phyto-02-15-0054-fi
Kemen, A. C., Agler, M. T., and Kemen, E. (2015). Host–microbe and microbe–microbe interactions in the evolution of obligate plant parasitism. New Phytol. 206, 1207–1228. doi: 10.1111/nph.13284
Kim, K. W. (2015). Ultrastructure of the rust fungus Puccinia miscanthi in the teliospore stage interacting with the biofuel plant Miscanthus sinensis. Plant Pathol. J. 31, 299–304. doi: 10.5423/PPJ.NT.04.2015.0051
Kiran, K., Rawal, H. C., Dubey, H., Jaswal, R., Bhardwaj, S. C., Prasad, P., et al. (2017). Dissection of genomic features and variations of three pathotypes of Puccinia striiformis through whole genome sequencing. Sci. Rep. 7:42419. doi: 10.1038/srep42419
Kiran, K., Rawal, H. C., Dubey, H., Jaswal, R., Devanna, B. N., Gupta, D. K., et al. (2016). Draft genome of the wheat rust pathogen (Puccinia triticina) unravels genome-wide structural variations during evolution. Genome Biol. Evol. 8, 2702–2721. doi: 10.1093/gbe/evw197
Kleemann, J., Rincon-Rivera, L. J., Takahara, H., Neumann, U., Ver Loren van Themaat, E., van der Does, H. C., et al. (2012). Sequential delivery of host-induced virulence effectors by appressoria and intracellular hyphae of the phytopathogen Colletotrichum higginsianum. PLoS Pathog. 8:e1002643. doi: 10.1371/journal.ppat.1002643
Kobayashi, I., Kobayashi, Y., and Hardham, A. R. (1994). Dynamic reorganization of microtubules and microfilaments in flax cells during the resistance response to flax rust infection. Planta 195, 237–247. doi: 10.1007/BF00199684
Köller, W., Allan, C. R., and Kolattukudy, P. E. (1982). Role of cutinase and cell wall degrading enzymes in infection of Pisum sativum by Fusarium solani f. sp. pisi. Physiol. Plant Pathol. 20, 47–60. doi: 10.1016/0048-4059(82)90023-6
Kretschmer, M., Damoo, D., Djamei, A., and Kronstad, J. (2019). Chloroplasts and plant immunity: where are the fungal effectors? Pathogens 9:19. doi: 10.3390/pathogens9010019
Latijnhouwers, M., de Wit, P. J., and Govers, F. (2003). Oomycetes and fungi: similar weaponry to attack plants. Trends Microbiol. 11, 462–469. doi: 10.1016/j.tim.2003.08.002
Lawrence, G. J., Dodds, P. N., and Ellis, J. G. (2007). Rust of flax and linseed caused by Melampsora lini. Mol. Plant Pathol. 8, 349–364. doi: 10.1111/j.1364-3703.2007.00405.x
Li, X., Jin, C., Yuan, H., Huang, W., Liu, F., Fan, R., et al. (2021). The barley powdery mildew effectors CSEP0139 and CSEP0182 suppress cell death and promote B. graminis fungal virulence in plants. Phytopathol. Res. 3:7. doi: 10.1186/s42483-021-00084-z
Liu, C., Pedersen, C., Schultz-Larsen, T., Aguilar, G. B., Madriz-Ordeñana, K., Hovmøller, M. S., et al. (2016). The stripe rust fungal effector PEC6 suppresses pattern-triggered immunity in a host species-independent manner and interacts with adenosine kinases. New Phytol. doi: 10.1111/nph.14034
Loehrer, M., Botterweck, J., Jahnke, J., Mahlmann, D. M., Gaetgens, J., Oldiges, M., et al. (2014). In vivo assessment by mach–zehnder double-beam interferometry of the invasive force exerted by the Asian soybean rust fungus (Phakopsora pachyrhizi). New Phytol. 203, 620–631. doi: 10.1111/nph.12784
Lorrain, C., Gonçalves dos Santos, K. C., Germain, H., Hecker, A., and Duplessis, S. (2019). Advances in understanding obligate biotrophy in rust fungi. New Phytol. 222, 1190–1206. doi: 10.1111/nph.15641
Lu, X., Kracher, B., Saur, I. M., Bauer, S., Ellwood, S. R., Wise, R., et al. (2016). Allelic barley MLA immune receptors recognize sequence-unrelated avirulence effectors of the powdery mildew pathogen. Proc. Natl. Acad. Sci. U.S.A. 113, E6486–E6495. doi: 10.1073/pnas.1612947113
Marone, D., Russo, M. A., Laidò, G., De Leonardis, A. M., and Mastrangelo, A. M. (2013). Plant nucleotide binding site–leucine-rich repeat (NBS-LRR) genes: active guardians in host defense responses. Int. J. Mol. Sci. 14, 7302–7326. doi: 10.3390/ijms14047302
Martel, A., Ruiz-Bedoya, T., Breit-McNally, C., Laflamme, B., Desveaux, D., and Guttman, D. S. (2021). The ETS-ETI cycle: evolutionary processes and metapopulation dynamics driving the diversification of pathogen effectors and host immune factors. Curr. Opin. Plant Biol. 62:102011. doi: 10.1016/j.pbi.2021.102011
Mendgen, K., and Hahn, M. (2002). Plant infection and the establishment of fungal biotrophy. Trends Plant Sci. 7, 352–356. doi: 10.1016/s1360-1385(02)02297-5
Mesarich, C. H., Ökmen, B., Rovenich, H., Griffiths, S. A., Wang, C., Karimi Jashni, M., et al. (2018). Specific hypersensitive response-associated recognition of new apoplastic effectors from Cladosporium fulvum in wild tomato. Mol. Plant Microbe Interact. 31, 145–162. doi: 10.1094/mpmi-05-17-0114-fi
Miller, M. E., Zhang, Y., Omidvar, V., Sperschneider, J., Schwessinger, B., Raley, C., et al. (2018). De novo assembly and phasing of dikaryotic genomes from two isolates of Puccinia coronata f. sp. avenae, the causal agent of oat crown rust. Am. Soc. Microbiol. 9:e01650-17. doi: 10.1128/mBio.01650-17
Moore, D. (2020). 21st Century Guidebook to Fungi. Reviews and Contents, 2nd Edn. Cambridge: Cambridge University Press.
Mousavi-Derazmahalleh, M., Chang, S., Thomas, G., Derbyshire, M., Bayer, P. E., Edwards, D., et al. (2019). Prediction of pathogenicity genes involved in adaptation to a lupin host in the fungal pathogens Botrytis cinerea and Sclerotinia sclerotiorum via comparative genomics. BMC Genomics 20:385. doi: 10.1186/s12864-019-5774-2
Nemri, A., Saunders, D., Anderson, C., Upadhyaya, N., Win, J., Lawrence, G., et al. (2014). The genome sequence and effector complement of the flax rust pathogen Melampsora lini. Front. Plant Sci. 5:98. doi: 10.3389/fpls.2014.00098
Ni, D., Lumyong, S., Hyde, K., Bulgakov, T., Phillips, A., and Yan, J. (2016). Mycosphere essays 9: defining biotrophs and hemibiotrophs. Mycosphere 7, 545–559. doi: 10.5943/mycosphere/7/5/2
Oliveira-Garcia, E., and Valent, B. (2015). How eukaryotic filamentous pathogens evade plant recognition. Curr. Opin. Microbiol. 26, 92–101. doi: 10.1016/j.mib.2015.06.012
Ozketen, A. C., Andac-Ozketen, A., Dagvadorj, B., Demiralay, B., and Akkaya, M. S. (2020). In-depth secretome analysis of Puccinia striiformis f. sp. tritici in infected wheat uncovers effector function. bioRxiv [Preprint]. doi: 10.1101/2020.01.11.897884
Panwar, V., McCallum, B., and Bakkeren, G. (2013). Endogenous silencing of Puccinia triticina pathogenicity genes through in planta-expressed sequences leads to the suppression of rust diseases on wheat. Plant J. 73, 521–532. doi: 10.1111/tpj.12047
Parlange, F., Roffler, S., Menardo, F., Ben-David, R., Bourras, S., McNally, K. E., et al. (2015). Genetic and molecular characterization of a locus involved in avirulence of Blumeria graminis f. sp. tritici on wheat Pm3 resistance alleles. Fungal Genet. Biol. 82, 181–192. doi: 10.1016/j.fgb.2015.06.009
Patkar, R. N., Benke, P. I., Qu, Z., Chen, Y. Y., Yang, F., Swarup, S., et al. (2015). A fungal monooxygenase-derived jasmonate attenuates host innate immunity. Nat. Chem. Biol. 11, 733–740. doi: 10.1038/nchembio.1885
Pawlowski, M. L., and Hartman, G. L. (2016). Infection mechanisms and colonization patterns of fungi associated with soybean. Fungal Pathogen. 25:25. doi: 10.5772/62305
Pennington, H. G., Gheorghe, D. M., Damerum, A., Pliego, C., Spanu, P. D., Cramer, R., et al. (2016). Interactions between the powdery mildew effector BEC1054 and barley proteins identify candidate host targets. J. Proteome Res. 15, 826–839. doi: 10.1021/acs.jproteome.5b00732
Perfect, S., and Green, J. (2001). Infection structures of biotrophic and hemibiotrophic fungal plant pathogens. Mol. Plant Pathol. 2, 101–108. doi: 10.1046/j.1364-3703.2001.00055.x
Petre, B., Lorrain, C., Saunders, D., Win, J., Sklenar, J., Duplessis, S., et al. (2015). Rust fungal effectors mimic host transit peptides to translocate into chloroplasts. Cell. Microbiol. 18, 453–465. doi: 10.1111/cmi.12530
Petre, B., Saunders, D. G., Sklenar, J., Lorrain, C., Krasileva, K. V., Win, J., et al. (2016). Heterologous expression screens in Nicotiana benthamiana identify a candidate effector of the wheat yellow rust pathogen that associates with processing bodies. PLoS One 11:e0149035. doi: 10.1371/journal.pone.0149035
Pliego, C., Nowara, D., Bonciani, G., Gheorghe, D., Xu, R., Surana, P., et al. (2013). Host-induced gene silencing in barley powdery mildew reveals a class of ribonuclease-like effectors. Mol. Plant Microbe Interact. 26, 633–642. doi: 10.1094/MPMI-01-13-0005-R
Plissonneau, C., Benevenuto, J., Mohd-Assaad, N., Fouché, S., Hartmann, F. E., and Croll, D. (2017). Using population and comparative genomics to understand the genetic basis of effector-driven fungal pathogen evolution. Front. Plant Sci. 8:119. doi: 10.3389/fpls.2017.00119
Porto, B. N., Caixeta, E. T., Mathioni, S. M., Vidigal, P. M. P., Zambolim, L., Zambolim, E. M., et al. (2019). Genome sequencing and transcript analysis of Hemileia vastatrix reveal expression dynamics of candidate effectors dependent on host compatibility. PLoS One 14:e0215598. doi: 10.1371/journal.pone.0215598
Prasad, P., Savadi, S., Bhardwaj, S. C., Gangwar, O. P., and Kumar, S. (2019). Rust pathogen effectors: perspectives in resistance breeding. Planta 250, 1–22. doi: 10.1007/s00425-019-03167-6
Prasad, P., Savadi, S., Bhardwaj, S. C., and Gupta, P. K. (2020). The progress of leaf rust research in wheat. Fungal Biol. 124, 537–550. doi: 10.1016/j.funbio.2020.02.013
Praz, C. R., Bourras, S., Zeng, F., Sánchez-Martín, J., Menardo, F., Xue, M., et al. (2017). AvrPm2 encodes an RNase-like avirulence effector which is conserved in the two different specialized forms of wheat and rye powdery mildew fungus. New Phytol. 213, 1301–1314. doi: 10.1111/nph.14372
Qi, M., Link, T. I., Müller, M., Hirschburger, D., Pudake, R. N., Pedley, K. F., et al. (2016). A small cysteine-rich protein from the Asian soybean rust fungus, Phakopsora pachyrhizi, suppresses plant immunity. PLoS Pathog. 12:e1005827. doi: 10.1371/journal.ppat.1005827
Qi, T., Guo, J., Liu, P., He, F., Wan, C., Islam, M. A., et al. (2019). Stripe rust effector PstGSRE1 disrupts nuclear localization of ROS-promoting transcription factor TaLOL2 to defeat ROS-induced defense in wheat. Mol. Plant 12, 1624–1638. doi: 10.1016/j.molp.2019.09.010
Qi, Y., Wei, J., Zhang, N., Yang, W., and Liu, D. (2020). Puccinia triticina effector protein Pt18906 triggered two-layer defense reaction in TcLr27+31. Sci. Agric. Sin. 53, 2371–2384. doi: 10.3864/j.issn.0578-1752.2020.12.006
Rahman, M. S., Madina, M. H., Plourde, M. B., dos Santos, K. C., Huang, X., Zhang, Y., et al. (2021). The fungal effector Mlp37347 alters plasmodesmata fluxes and enhances susceptibility to pathogen. Microorganisms 9:1232. doi: 10.3390/microorganisms9061232
Rampitsch, C., Günel, A., Beimcik, E., and Mauthe, W. (2015). Proteome of monoclonal antibody-purified haustoria from Puccinia triticina race-1. Proteomics 15, 1307–1315. doi: 10.1002/pmic.201400241
Richards, J. K., Stukenbrock, E. H., Carpenter, J., Liu, Z., Cowger, C., Faris, J. D., et al. (2019). Local adaptation drives the diversification of effectors in the fungal wheat pathogen Parastagonospora nodorum in the United States. PLoS Genet. 15:e1008223. doi: 10.1371/journal.pgen.1008223
Rochi, L., Diéguez, M. J., Burguener, G., Darino, M. A., Pergolesi, M. F., Ingala, L. R., et al. (2018). Characterization and comparative analysis of the genome of Puccinia sorghi Schwein, the causal agent of maize common rust. Fungal Genet. Biol. 112, 31–39. doi: 10.1016/j.fgb.2016.10.001
Salcedo, A., Rutter, W., Wang, S., Akhunova, A., Bolus, S., Chao, S., et al. (2017). Variation in the AvrSr35 gene determines Sr35 resistance against wheat stem rust race Ug99. Science 358, 1604–1606. doi: 10.1126/science.aao7294
Sánchez-Vallet, A., Hartmann, F. E., Marcel, T. C., and Croll, D. (2018). Nature’s genetic screens: using genome-wide association studies for effector discovery. Mol. Plant Pathol. 19, 3–6. doi: 10.1111/mpp.12592
Schmidt, S. M., Kuhn, H., Micali, C., Liller, C., Kwaaitaal, M., and Panstruga, R. (2014). Interaction of a Blumeria graminis f. sp. hordei effector candidate with a barley ARF-GAP suggests that host vesicle trafficking is a fungal pathogenicity target. Mol. Plant Pathol. 15, 535–549. doi: 10.1111/mpp.12110
Schwessinger, B., and Ronald, P. C. (2012). Plant innate immunity: perception of conserved microbial signatures. Annu. Rev. Plant Biol. 63, 451–482. doi: 10.1146/annurev-arplant-042811-105518
Schwessinger, B., Sperschneider, J., Cuddy, W. S., Garnica, D. P., Miller, M. E., Taylor, J. M., et al. (2018). A near-complete haplotype-phased genome of the dikaryotic wheat stripe rust fungus Puccinia striiformis f. sp. tritici reveals high interhaplotype diversity. Am. Soc. Microbiol. 9:1232. doi: 10.1128/mBio.02275-17
Scott, K. J. (1972). Obligate parasitism by phytopathogenic fungi. Biol. Rev. 47, 537–572. doi: 10.1111/j.1469-185X.1972.tb01081.x
Segovia, V., Bruce, M., Shoup Rupp, J. L., Huang, L., Bakkeren, G., Trick, H. N., et al. (2016). Two small secreted proteins from Puccinia triticina induce reduction of ß-glucoronidase transient expression in wheat isolines containing Lr9, Lr24 and Lr26. Can. J. Plant Pathol. 38, 91–102. doi: 10.1080/07060661.2016.1150884
Sharma, N., and Gautam, A. (2019). Early pathogenicity events in plant pathogenic fungi: a comprehensive review. Biol. Forum Int. J. 11, 24–34.
Shen, Q., Liu, Y., and Naqvi, N. I. (2018). Fungal effectors at the crossroads of phytohormone signaling. Curr. Opin. Microbiol. 46, 1–6. doi: 10.1016/j.mib.2018.01.006
Sonah, H., Deshmukh, R. K., and Bélanger, R. R. (2016). Computational prediction of effector proteins in fungi: opportunities and challenges. Front. Plant Sci. 7:126. doi: 10.3389/fpls.2016.00126
Spanu, P., Abbott, J., Amselem, J., Burgis, T., Soanes, D., Stüber, K., et al. (2010). Genome expansion and gene loss in powdery mildew fungi reveal trade-offs in extreme parasitism. Science 330, 1543–1546. doi: 10.1126/science.1194573
Sperschneider, J., Catanzariti, A.-M., DeBoer, K., Petre, B., Gardiner, D. M., Singh, K. B., et al. (2017). LOCALIZER: subcellular localization prediction of both plant and effector proteins in the plant cell. Sci. Rep. 7:44598. doi: 10.1038/srep44598
Sperschneider, J., Dodds, P. N., Gardiner, D. M., Singh, K. B., and Taylor, J. M. (2018b). Improved prediction of fungal effector proteins from secretomes with EffectorP 2.0. Mol. Plant Pathol. 19, 2094–2110. doi: 10.1111/mpp.12682
Sperschneider, J., Dodds, P. N., Singh, K. B., and Taylor, J. M. (2018a). ApoplastP: prediction of effectors and plant proteins in the apoplast using machine learning. New Phytol. 217, 1764–1778. doi: 10.1111/nph.14946
Sperschneider, J., Gardiner, D. M., Dodds, P. N., Tini, F., Covarelli, L., Singh, K. B., et al. (2016). EffectorP: predicting fungal effector proteins from secretomes using machine learning. New Phytol. 210, 743–761. doi: 10.1111/nph.13794
Stotz, H. U., Mitrousia, G. K., de Wit, P. J. G. M., and Fitt, B. D. L. (2014). Effector-triggered defence against apoplastic fungal pathogens. Trends Plant Sci. 19, 491–500. doi: 10.1016/j.tplants.2014.04.009
Strange, R. N. (2007). Phytotoxins produced by microbial plant pathogens. Nat. Product Rep. 24, 127–144. doi: 10.1039/b513232k
Su, Y., Chen, Y., Chen, J., Zhang, Z., Guo, J., Cai, Y., et al. (2021). Effectors of Puccinia striiformis f. sp. tritici suppressing the pathogenic-associated molecular pattern-triggered immune response were screened by transient expression of wheat protoplasts. Int. J. Mol. Sci. 22:4985. doi: 10.3390/ijms22094985
Takamatsu, S. (2014). Molecular phylogeny reveals phenotypic evolution of powdery mildews (Erysiphales, Ascomycota). J. Gen. Plant Pathol. 79, 218–226. doi: 10.1007/s10327-013-0447-5
Tan, M.-K., Collins, D., Chen, Z., Englezou, A., and Wilkins, M. R. (2014). A brief overview of the size and composition of the myrtle rust genome and its taxonomic status. Mycology 5, 52–63. doi: 10.1080/21501203.2014.919967
Tanaka, S., Brefort, T., Neidig, N., Djamei, A., Kahnt, J., Vermerris, W., et al. (2014). A secreted Ustilago maydis effector promotes virulence by targeting anthocyanin biosynthesis in maize. Elife 3:e01355. doi: 10.7554/eLife.01355
Tanaka, S., Gollin, I., Rössel, N., and Kahmann, R. (2020). The functionally conserved effector Sta1 is a fungal cell wall protein required for virulence in Ustilago maydis. New Phytol. 227, 185–199. doi: 10.1111/nph.16508
Tang, C., Xu, Q., Zhao, M., Wang, X., and Kang, Z. (2018). Understanding the lifestyles and pathogenicity mechanisms of obligate biotrophic fungi in wheat: the emerging genomics era. Crop J. 6, 60–67. doi: 10.1016/j.cj.2017.11.003
Tao, F., Hu, Y., Su, C., Li, J., Guo, L., Xu, X., et al. (2020). Revealing differentially expressed genes and identifying effector proteins of Puccinia striiformis f. sp. tritici in response to high-temperature seedling plant resistance of wheat based on transcriptome sequencing. mSphere 5:e00096-20. doi: 10.1128/mSphere.00096-20
Tiku, A. R. (2020). “Antimicrobial compounds (phytoanticipins and phytoalexins) and their role in plant defense,” in Co-Evolution of Secondary Metabolites, eds J.-M. Mérillon and K. G. Ramawat (Cham: Springer International Publishing), 845–868.
Tsuda, K., and Katagiri, F. (2010). Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 13, 459–465. doi: 10.1016/j.pbi.2010.04.006
Tucker, S. L., and Talbot, N. J. (2001). Surface attachment and pre-penetration stage development by plant pathogenic fungi. Annu. Rev. Phytopathol. 39, 385–417. doi: 10.1146/annurev.phyto.39.1.385
Upadhyaya, N. M., Garnica, D. P., Karaoglu, H., Sperschneider, J., Nemri, A., Xu, B., et al. (2014). Comparative genomics of Australian isolates of the wheat stem rust pathogen Puccinia graminis f. sp. tritici reveals extensive polymorphism in candidate effector genes. Front. Plant Sci. 5:759. doi: 10.3389/fpls.2014.00759
Upadhyaya, N. M., Mago, R., Panwar, V., Hewitt, T., Luo, M., Chen, J., et al. (2021). Genomics accelerated isolation of a new stem rust avirulence gene–wheat resistance gene pair. Nat. Plants 7, 1220–1228. doi: 10.1038/s41477-021-00971-5
van der Linde, K., and Göhre, V. (2021). How do smut fungi use plant signals to spatiotemporally orientate on and in planta. J. Fungi (Basel) 7:107. doi: 10.3390/jof7020107
Van Sumere, C. F., Van, S.-D., Preter, C., and Ledingham, G. A. (1957). Cell-wall-splitting enzymes of Puccinia graminis var. tritici. Can. J. Microbiol. 3, 761–770. doi: 10.1139/m57-086
Voegele, R., Hahn, M., and Mendgen, K. (2009). “The uredinales: cytology, biochemistry, and molecular biology,” in The Mycota, V. Plant Relationships, ed. H. B. Deising (Berlin: Springer Verlag), 69–98.
Voegele, R. T., and Mendgen, K. (2003). Rust haustoria: nutrient uptake and beyond. New Phytol. 159, 93–100. doi: 10.1046/j.1469-8137.2003.00761.x
Voegele, R. T., and Mendgen, K. W. (2011). Nutrient uptake in rust fungi: how sweet is parasitic life? Euphytica 179, 41–55. doi: 10.1007/s10681-011-0358-5
Wang, C., Wang, P., Han, S., Wang, L., Zhao, Y., and Juan, L. (2020). FunEffector-Pred: identification of fungi effector by activate learning and genetic algorithm sampling of imbalanced data. IEEE Access 8, 57674–57683. doi: 10.1109/ACCESS.2020.2982410
Wang, X., Zhai, T., Zhang, X., Tang, C., Zhuang, R., Zhao, H., et al. (2021). Two stripe rust effectors impair wheat resistance by suppressing import of host Fe–S protein into chloroplasts. Plant Physiol. 187, 2530–2543. doi: 10.1093/plphys/kiab434
Wei, J., Cui, L., Zhang, N., Du, D., Meng, Q., Yan, H., et al. (2020). Puccinia triticina pathotypes THTT and THTS display complex transcript profiles on wheat cultivar Thatcher. BMC Genetics 21:48. doi: 10.1186/s12863-020-00851-5
Wicker, T., Oberhaensli, S., Parlange, F., Buchmann, J. P., Shatalina, M., Roffler, S., et al. (2013). The wheat powdery mildew genome shows the unique evolution of an obligate biotroph. Nat. Genet. 45, 1092–1096. doi: 10.1038/ng.2704
Williams, A. H., Sharma, M., Thatcher, L. F., Azam, S., Hane, J. K., Sperschneider, J., et al. (2016). Comparative genomics and prediction of conditionally dispensable sequences in legume–infecting Fusarium oxysporum formae speciales facilitates identification of candidate effectors. BMC Genomics 17:191. doi: 10.1186/s12864-016-2486-8
Wilson, T. J., Thomsen, K. K., Petersen, B. O., Duus, J., and Oliver, R. P. (2003). Detection of 3-hydroxykynurenine in a plant pathogenic fungus. Biochem. J. 371(Pt 3), 783–788. doi: 10.1042/bj20021797
Wood, R. K. S. (1960). Pectic and cellulolytic enzymes in plant disease. Annu. Rev. Plant Physiol. 11, 299–322. doi: 10.1146/annurev.pp.11.060160.001503
Wu, J. Q., Dong, C., Song, L., and Park, R. F. (2020). Long-read–based de novo genome assembly and comparative genomics of the wheat leaf rust pathogen Puccinia triticina identifies candidates for three avirulence genes. Front. Genet. 11:521. doi: 10.3389/fgene.2020.00521
Wu, J. Q., Sakthikumar, S., Dong, C., Zhang, P., Cuomo, C. A., and Park, R. F. (2017). Comparative genomics integrated with association analysis identifies candidate effector genes corresponding to Lr20 in phenotype-paired Puccinia triticina isolates from Australia. Front. Plant Sci. 8:148. doi: 10.3389/fpls.2017.00148
Wu, X., Bian, Q., Gao, Y., Ni, X., Sun, Y., Xuan, Y., et al. (2021). Evaluation of resistance to powdery mildew and identification of resistance genes in wheat cultivars. PeerJ 9:e10425. doi: 10.7717/peerj.10425
Xia, C., Wang, M., Yin, C., Cornejo, O. E., Hulbert, S. H., and Chen, X. (2018). Genomic insights into host adaptation between the wheat stripe rust pathogen (Puccinia striiformis f. sp. tritici) and the barley stripe rust pathogen (Puccinia striiformis f. sp. hordei). BMC Genomics 19:664. doi: 10.1186/s12864-018-5041-y
Xu, Q., Tang, C., Wang, X., Sun, S., Zhao, J., Kang, Z., et al. (2019). An effector protein of the wheat stripe rust fungus targets chloroplasts and suppresses chloroplast function. Nat. Commun. 10:5571. doi: 10.1038/s41467-019-13487-6
Yang, Q., Huai, B., Lu, Y., Cai, K., Guo, J., Zhu, X., et al. (2020). A stripe rust effector Pst18363 targets and stabilises TaNUDX23 that promotes stripe rust disease. New Phytol. 225, 880–895. doi: 10.1111/nph.16199
Yin, C., Park, J. J., Gang, D. R., and Hulbert, S. H. (2014). Characterization of a tryptophan 2-monooxygenase gene from Puccinia graminis f. sp. tritici involved in auxin biosynthesis and rust pathogenicity. Mol. Plant Microbe Interact. 27, 227–235. doi: 10.1094/mpmi-09-13-0289-fi
Yin, C., Ramachandran, S. R., Zhai, Y., Bu, C., Pappu, H. R., and Hulbert, S. H. (2019). A novel fungal effector from Puccinia graminis suppressing RNA silencing and plant defense responses. New Phytol. 222, 1561–1572. doi: 10.1111/nph.15676
Yuan, H., Jin, C., Pei, H., Zhao, L., Li, X., Li, J., et al. (2021). The powdery mildew effector CSEP0027 interacts with barley catalase to regulate host immunity. Front. Plant Sci. 12:733237. doi: 10.3389/fpls.2021.733237
Zhang, W.-J., Pedersen, C., Kwaaitaal, M., Gregersen, P. L., Mørch, S. M., Hanisch, S., et al. (2012). Interaction of barley powdery mildew effector candidate CSEP0055 with the defence protein PR17c. Mol. Plant Pathol. 13, 1110–1119. doi: 10.1111/j.1364-3703.2012.00820.x
Zhang, Y., Wei, J., Qi, Y., Li, J., Amin, R., Yang, W., et al. (2020). Predicating the effector proteins secreted by Puccinia triticina through transcriptomic analysis and multiple prediction approaches. Front. Microbiol. 11:538032. doi: 10.3389/fmicb.2020.538032
Zhang, Y., Xu, K., Yu, D., Liu, Z., Peng, C., Li, X., et al. (2019). The highly conserved barley powdery mildew effector BEC1019 confers susceptibility to biotrophic and necrotrophic pathogens in wheat. Int. J. Mol. Sci. 20:4376. doi: 10.3390/ijms20184376
Zhang, Z., Henderson, C., and Gurr, S. J. (2004). Blumeria graminis secretes an extracellular catalase during infection of barley: potential role in suppression of host defence. Mol. Plant Pathol. 5, 537–547. doi: 10.1111/j.1364-3703.2004.00251.x
Zhao, M., Wang, J., Ji, S., Chen, Z., Xu, J., Tang, C., et al. (2018). Candidate effector Pst_8713 impairs the plant immunity and contributes to virulence of Puccinia striiformis f. sp. tritici. Front. Plant Sci. 9:1294. doi: 10.3389/fpls.2018.01294
Zhao, S., Shang, X., Bi, W., Yu, X., Liu, D., Kang, Z., et al. (2020). Genome-wide identification of effector candidates with conserved motifs from the wheat leaf rust fungus Puccinia triticina. Front. Microbiol. 11:1188. doi: 10.3389/fmicb.2020.01188
Keywords: biotrophs, plant fungal pathogens, effectors, pathogen-host interaction, pathogenicity
Citation: Mapuranga J, Zhang N, Zhang L, Chang J and Yang W (2022) Infection Strategies and Pathogenicity of Biotrophic Plant Fungal Pathogens. Front. Microbiol. 13:799396. doi: 10.3389/fmicb.2022.799396
Received: 22 October 2021; Accepted: 19 April 2022;
Published: 02 June 2022.
Edited by:
Jesús Navas-Castillo, La Mayora Experimental Station (CSIC), SpainReviewed by:
Xiaojie Wang, Northwest A&F University, ChinaCopyright © 2022 Mapuranga, Zhang, Zhang, Chang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenxiang Yang, d2VueGlhbmd5YW5nMjAwM0AxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.