
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 07 March 2022
Sec. Terrestrial Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.797444
This article is part of the Research Topic Rhizosphere Microbiology: Toward a Clean and Healthy Soil Environment View all 25 articles
Soil contamination by heavy metals, particularly mercury (Hg), is a problem that can seriously affect the environment, animals, and human health. Hg has the capacity to biomagnify in the food chain. That fact can lead to pathologies, of those which affect the central nervous system being the most severe. It is convenient to know the biological environmental indicators that alert of the effects of Hg contamination as well as the biological mechanisms that can help in its remediation. To contribute to this knowledge, this study conducted comparative analysis by the use of Shotgun metagenomics of the microbial communities in rhizospheric soils and bulk soil of the mining region of Almadén (Ciudad Real, Spain), one of the most affected areas by Hg in the world The sequences obtained was analyzed with MetaPhlAn2 tool and SUPER-FOCUS. The most abundant taxa in the taxonomic analysis in bulk soil were those of Actinobateria and Alphaproteobacteria. On the contrary, in the rhizospheric soil microorganisms belonging to the phylum Proteobacteria were abundant, evidencing that roots have a selective effect on the rhizospheric communities. In order to analyze possible indicators of biological contamination, a functional potential analysis was performed. The results point to a co-selection of the mechanisms of resistance to Hg and the mechanisms of resistance to antibiotics or other toxic compounds in environments contaminated by Hg. Likewise, the finding of antibiotic resistance mechanisms typical of the human clinic, such as resistance to beta-lactams and glycopeptics (vancomycin), suggests that these environments can behave as reservoirs. The sequences involved in Hg resistance (operon mer and efflux pumps) have a similar abundance in both soil types. However, the response to abiotic stress (salinity, desiccation, and contaminants) is more prevalent in rhizospheric soil. Finally, sequences involved in nitrogen fixation and metabolism and plant growth promotion (PGP genes) were identified, with higher relative abundances in rhizospheric soils. These findings can be the starting point for the targeted search for microorganisms suitable for further use in bioremediation processes in Hg-contaminated environments.
Mercury (Hg) is a highly toxic element that severely affects ecosystems (Hsu-Kim et al., 2018; Liu et al., 2018). It has the capacity to enter and biomagnify in the food chain and therefore affects human health even at low concentrations (Bjørklund et al., 2019). The accumulation of Hg can lead to pathologies, with those affecting the central nervous system being the most serious, such as Minamata syndrome (Gil-Hernández et al., 2020; Marumoto et al., 2020). The presence of Hg in various ecosystems is widely described. Exceptionally, environments with extremely high concentrations of this heavy metal have been described, such as those detected in the Almadén mercury mining region (> 8889 μg/g) (US Environmental Protection Agency, 2011).
The presence of Hg in soils conditions the development of organisms that inhabit it, with bacterial communities being one of the most vulnerable groups. Some of the bacterial species capable of resisting the presence of this pollutant could be suitable in processes of remediating affected soils, this is why there is a growing scientific interest in knowing the composition of these Hg-tolerant edaphic communities (Zhao et al., 2021). There are several references to the usefulness of these techniques in soil samples (Li et al., 2018; Westmann et al., 2018; Castillo Villamizar et al., 2019; Nelkner et al., 2019) and, in particular, in soils contaminated with different toxins (Garrido-Sanz et al., 2018; Kumar et al., 2018; Thomas et al., 2019).
Metagenomics consist of the complete study of genetic material extracted from a sample. Various metagenomic methods based on either DNA amplification and sequencing or DNA fragmentation and alignment are currently available (Giagnoni et al., 2018). One of the main metagenomic techniques, based on sequencing, is the creation of genetic libraries. The bioinformatic analysis of the data obtained allows us to reconstruct the metabolism of the organisms that make up the community and to predict their potential functional roles in the ecosystem through the so-called “environmental gene labels” (Youngblut et al., 2020). This field has also been called environmental genomics, ecogenomics or community genomics (Riesenfeld et al., 2004; Cordier et al., 2021). Methods based on functional analysis of DNA libraries from the entire microbial community of a particular medium can be a great source for the discovery of new genes (Handelsman, 2004; Douglas et al., 2020; Moon et al., 2020), study of unculturable microorganisms (Handelsman, 2004; Tyson et al., 2004; Cycil et al., 2020) and creation of genomic libraries (Enagbonma et al., 2020). This approach has also been used successfully in the study of antibiotic resistance in complex communities (de Abreu et al., 2020). One of the most widely used methods of study in metagenomics, and used in this work, is the “Shotgun metagenomics” technique. It consists of purifying the sample’s DNA and randomly fragmenting it into many small sequences that align into consensus sequences. These sequences are processed through analysis programs that allow taxonomic and functional identification of the sample’s DNA. SUPER-FOCUS is a bioinformatic tool which use the non-annotated sequences to predict potential functional activity, this allows study the whole metagenome as one unit and reveling the functional potential profile of the whole community (Chacón-Vargas et al., 2020; Collins et al., 2021).
To this end, this study aims to: (1) Find the taxonomic proportion and composition of the microbiological community of the soils of Almadén. (2) It seeks to provide an interpretation of the ecological behavior of the community, analyzing its functional potential information with SUPER-FOCUS.
The samples analyzed came from the mining district of Almadén, Ciudad Real (Spain), and were collected during the spring season Specifically, the slope “S” of Cerro Buitrones was sampled from the so-called “Plot 6” (38°46′25.1″N 4°51′03.9″W), described by other authors in previous studies (Millán et al., 2007). The concentration of Hg in this plot was 1710 mg/kg total Hg, 0.609 mg/kg soluble Hg and 7.3 mg/kg exchangeable Hg. Soil samples for the metagenomic study were obtained from the rhizosphere and bulk soil, together with plants described by Robas et al. (2021a): Rumex induratus Boiss. and Reut., Rumex bucephalophorus L., Avena sativa L., Medicago sativa L. and Vicia benghalensis L. (Robas et al., 2021a).
To obtain samples of rhizospheric soil (RS), the root of each plant specimen was gently shaken in order to remove soil fractions that were not tightly adhering to the root. The part of the soil attached to the root was then carefully separated to make up 2 g per plant. The five rhizospheric fractions were then combined into a single sample, in order to obtain10 g of soil that was homogenized for further metagenomic study. The 10 g of bulk soil (BS) was obtained in the same way, by sampling 2 g of soil near each plant, avoiding the rhizospheric fraction. Each sample was divided into 3 technical replicates for the metagenomic analysis.
The DNA was purified by the “DNeasy Power Soil Pro Kit” (Qiagen, United States) following the manufacturer’s instructions. An enzyme lysis step with lysozyme was included in order to obtain the highest and best amount of total bacterial DNA. Purified DNA was quantified using PicoGreenTM (Thermo Fisher Scientific, United States) from 40 pg. The genetic libraries were constructed using mechanical fragmentation and adaptors ligation by TruSeq (Illumina ®, United States) methodology. The metagenome sequences obtained were assembled using metaSPAdes tool (Nurk et al., 2017).
DNA isolated from BS and RS samples was used for metagenomic analysis. These samples were processed and sequenced with Shotgun using Illumina ® MiSeq desktop using the 2 × 250 bp paired-end reagent V2 Kits (Illumina ®, United States) technology with a standard quantification pattern. Bioinformatic analysis and quality control were performed using the Fast QC tool (Andrews, 2017). Q-score was used to predict the probability of an error in base-calling. Over 75% of bases > Q30 averaged across the entire run was considered acceptable. Raw sequence reads underwent quality trimming using Trimmomatic to remove adaptor contaminants and low-quality reads (Li et al., 2010).
The MetaPhlAn2 (Metagenomic Phylogenetic Analysis v2) tool was used for taxonomic analysis (Segata et al., 2012; Truong et al., 2015). This is a computational tool for profiling the composition of microbial communities from metagenomic shotgun sequencing data. MetaPhlAn relies on unique clade-specific marker genes identified from ∼17,000 reference genomes (∼13,500 bacterial and archaeal, ∼3,500 viral, and ∼110 eukaryotic) to make taxonomic predictions. It was used bowtie2 –bt2_ps “very-sensitive” preset parameters, –tax_lev “a” for prediction of all taxonomic levels, “–min_cu_len 2000” for minimum total nucleotide length for the markers in the clade, and “–stat_q 0,1” for quantile value.
The SUPER–FOCUS tool (SUbsystems Profile by databasE Reduction) was used for the functional potential analysis of the data obtained by Shotgun metagenomics. FOCUS uses a reference database to identify subsystems (predicted protein groups with similar potential function) (Silva et al., 2016). This tool reports functional annotation using CD-HIT and with the SEED database, we reduced the references of the data set (Overbeek et al., 2004; Aziz et al., 2012). SUPER-FOCUS identifies the taxonomic profile of the data and creates a database with the subsystems for predicted organism. Metagenomic data was aligned against the database using RAPSearch2 (Zhao et al., 2012). Sequences with e-values ≤ 1e-5, a minimum identity of 60%, and an alignment length ≥ 15 amino acids were retained. This database categorizes information into three levels of detail: “level I” (large functional potential groups), “level II” (families of potential functional activities) and “level III” and SEED (specific potential functional role and the protein to which the sequences belong) (Silva et al., 2016).
For the statistical analysis, SPSS v.27.0 program (Version 27.0 IBM Corp, Armonk, NY, United States) was used. In order to evaluate the quality of the technical replicates in each soil a Pearson correlation (r) of the percent genus abundances was done. Simpson and Shannon diversity index were also calculated with the relative abundances obtained from the taxonomical analysis to assess the ecological richness between BS and RS.
In the metagenomic DNA extraction and sequencing of RS and BS samples, were generated a total of 15,939,287 and 15,826,564 raw reads across the three technical replicates respectively, maintaining the 98.1% (RS) and 95.3% (BS) of the sequences after QC. The sequences were aligned and analyzed in two steps, taxonomical identification and functional annotation of the sequences. Species abundance between technical replicates was highly correlated (all comparisons r > 0.99 with Pearson correlation test). Processing a larger number of samples would allow obtaining statistically more complete information and reducing the limitation of the results in subsequent studies. The following results are presented divide by taxonomic identification and functional potential analysis.
Taxonomic profile and relative abundances of the microbial community in RS and RF was analyzed using MetaPhlAn2. The identification of organisms is based on the assignment of the gene pool to a taxon. Comparing the BS and RS samples, a difference in abundance between viruses and bacteria was observed with an apparent 21 and 79% relative abundance respectively in RS and a 2 and 98% relative abundance respectively in BS.
Figure 1 shows the relative abundances of different bacterial groups. Examining the results obtained in BS, shows that the most abundant group is Actinobacteria. However, the best represented taxon in RS is Alpharoteobacteria. Acidobacteria and Cyanobacteria only appear in BS, and Betaproteobacteria and Gammaproteobacteria are only represented in RS.
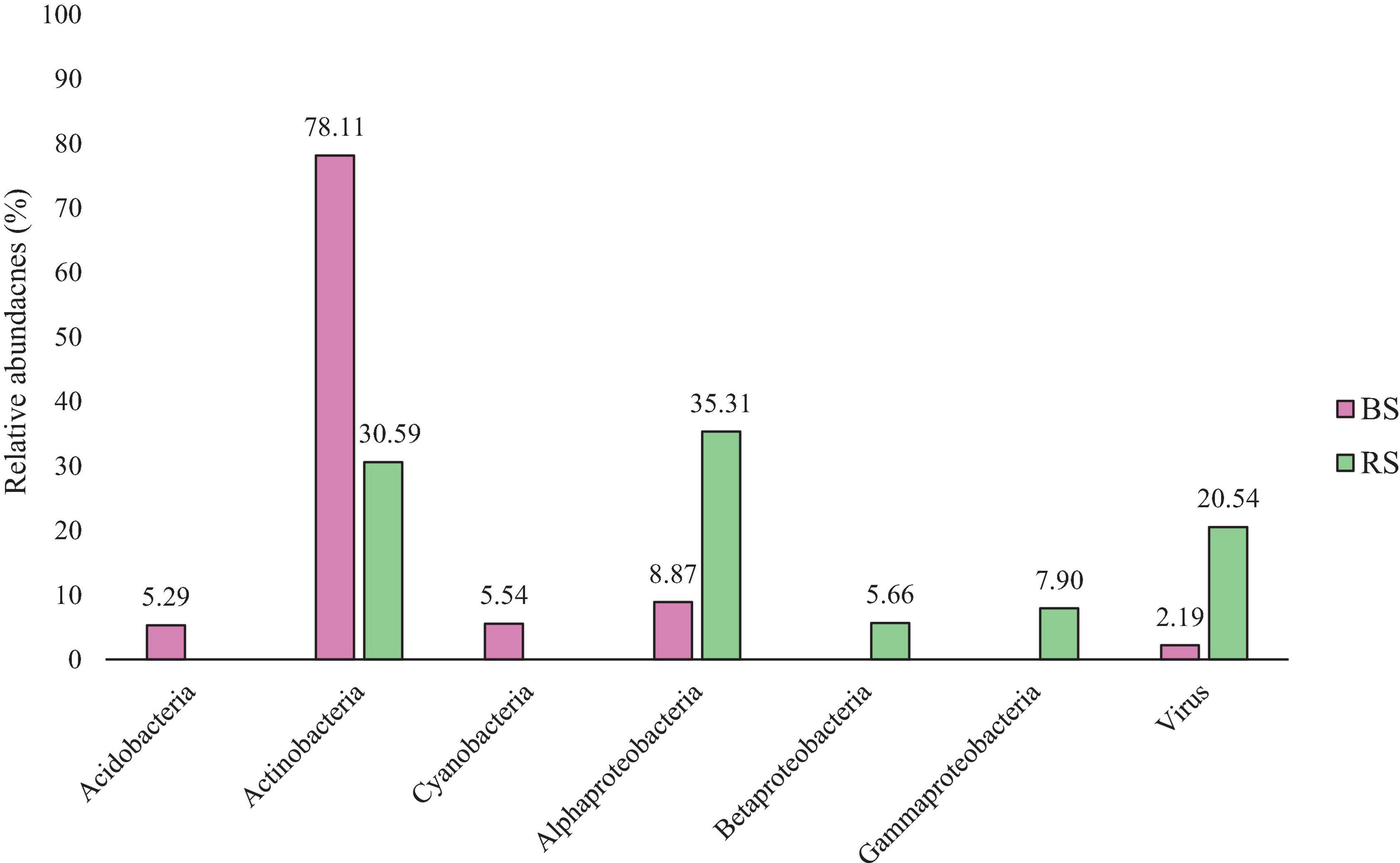
Figure 1. Comparative of the Relative abundances by taxonomic level of Class in BS (Pink - Bulk Soil) and RS (Green - Rhizospheric Soil). Data in the figure show how the relative abundances of the diferent classes are further grouped according to their presence in RS or BS.
Figure 2 shows the results of the relative abundances for the species taxon. Both samples seem to have a high diversity levels [Simpson’s diversity (D) and Shannon’s diversity (H)], being RS diversity levels (DRS = 0.14, HRS = 9.18) higher than BS (DBS = 0.4, HBS = 4.3). In RS, 73.42% of the genetic material was identified, leaving 26.58% unidentified (Figure 2 and Supplementary Table 1). Similarly, in BS 38.87% were identified. 61.13% of this DNA belongs to the various taxa (Figure 2 and Supplementary Table 1).
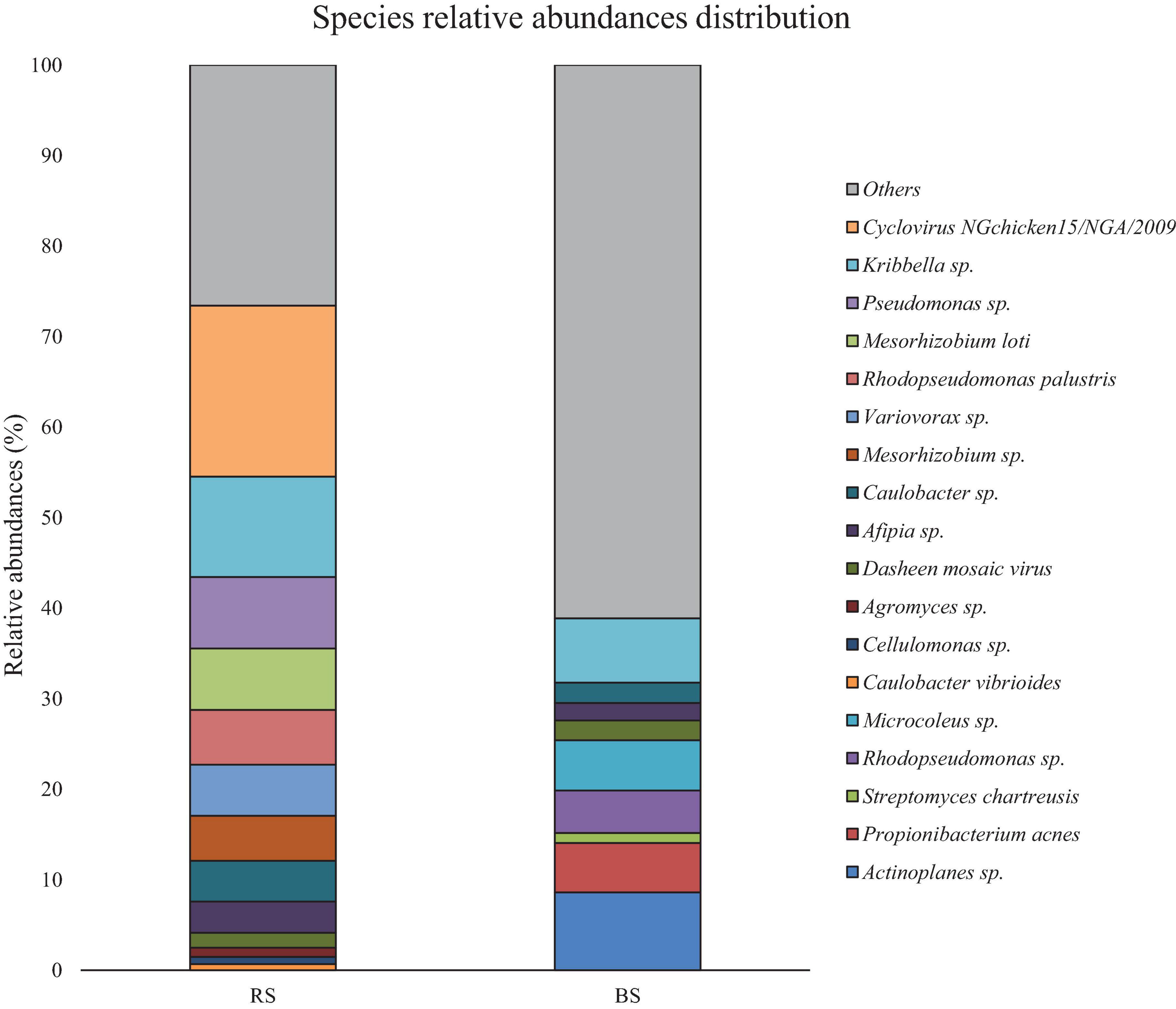
Figure 2. Comparative of the population relative abundances by taxonomic level of species in RS and BS. A pool of species could not be identifided with the sequences present in the metagenome in both soil.
The relative abundance of Kribbella sp. stands out in both samples, having a greater representation in RS. Similarly, the high representation of Pseudomonas sp. and Mesorhizobium sp. in RS (Figure 2) stands out. Likewise, the presence of strains of the genera Actinoplanes sp., Microcoleus sp. and Propionibacterium sp. seems to be especially abundant in BS (Figure 2).
Viral DNAs have been identified as Cyclovirus NGChicken15/NGA/2009, with 18.88% representation in RS. Less abundant in both samples was the mosaic Dasheen virus.
The functional analysis with SUPER-FOCUS present three levels (I-III) and predicted proteins (SEED) with the non-annotated metagenome sequences that allows pool the sequences by potential functional activity clusters. The two metagenomes obtained were assembled using MetaSpades. A total of 813,375 and 676,195 contigs were obtained, respectively.
In the analysis of level I (more general group of potential functional activity), the sequences pooled on the same potential functional activity were ordered according to their relative abundance. In this way, the functional potential content of the samples was ranked, reflecting the abundance of the different subsystems.
Were found 35 functional potential groups (Supplementary Table 2). The most abundant subsystems are those which seem to be related to basal metabolism and basic functions for the survival of bacteria, such as the carbon cycle, amino acid synthesis, and functional activities involved in breathing, among others, while subsystems encompassing more specific characteristics (such as virulence or photosynthesis) are less represented.
It is interesting that the functional potential cluster endowment of “stress response” presents 4.45% RS and 4.36% BS abundances. This functional potential group is among the 10 most abundant, which indicates the high environmental pressure suffered by bacteria in soils contaminated with Hg.
At this level (Figure 3), the potential functional activities were collected in a more concrete way, ordering them according to their relative abundances by families of same potential functional activities. Were found at this level 192 functional potential clusters (Supplementary Table 3).
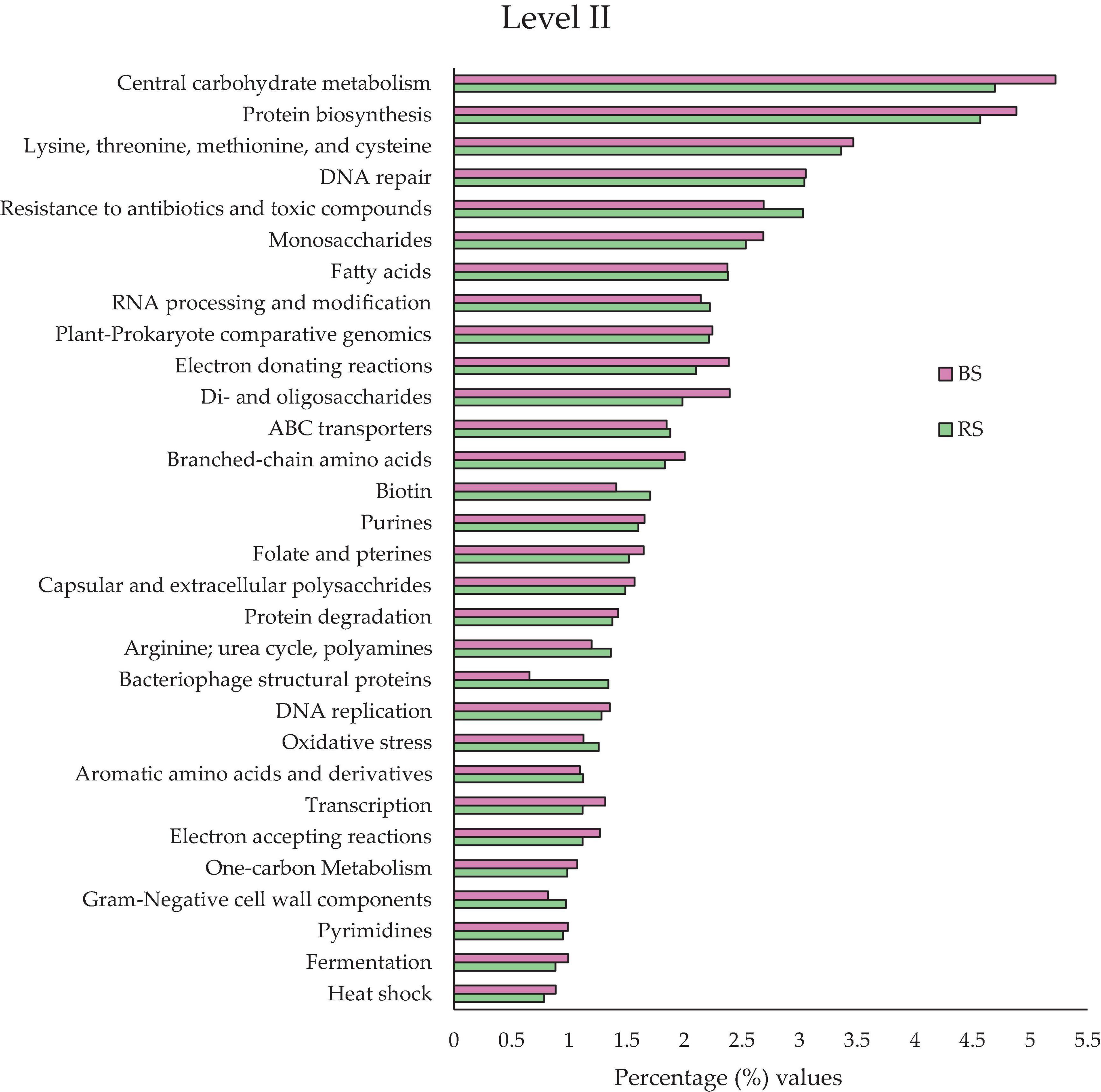
Figure 3. Comparison of the relative abundances of the 30 most abundant functional types in BS (pink bars) and RS (green bars) in level II, all the information at that level could be found in Supplementary Table 3. All the values are expressed in percentages.
Some potential functional activities are especially relevant and could allow for explaining the biological behavior of the Hg-tolerant edaphic communities. The following activities stand out (Figure 3): “Resistance to antibiotics and toxic compounds” (3.03% RS and 2.96% BS), “ABC type conveyors” (1.88% RS and 1.85% BS) and “oxidative stress” (1.26% RS and 1.13% BS).
The third level identifies the potential functional role to which the sequences under study belong, revealing the potential function within the metabolism of the bacteria. On level III, 1,186 functional potential groups were found and 24,441 functional potential annotations on SEED were done (Supplementary Tables 4, 5).
The 35 most abundant subsystems belong to a division of the subfamilies found in levels I and II. Furthermore, most of the subfamilies of biological interest in this study seems to be represented with lower relative abundance percentages.
At this level, there are several subsystems that group the resistance to heavy metals and to Hg, in particular. Among them we find various predicted proteins of the mer operon, Hg-reductases and ABC-type transporters associated with resistance to heavy metals.
Noteworthy is the large number of subsystems could be linked to resistance to various antibiotics, including beta-lactamases, predicted proteins regulating the BlaR1 family, various proteins of multiple resistance systems, multiple resistance systems linked to the MexAB-OprM and MexEF-OprN complex, fluoroquinolone resistance, vancomycin resistance, mdtABCD flow pump cluster and bacitracin stress response.
There are also subsystems involved in the biological cycle of nitrogen (N). Particularly important are those linked to the nitrogenase complex for atmospheric N fixation. Likewise, the potential functional activities responsible for the promotion of plant growth are represented as “production of auxins,” “metabolism of ethylene,” “siderophore production,” and “phosphate solubilization.” With a major relative abundance in RS.
In the soils analyzed, representatives of six taxonomic phylum appear: Acidobacteria, Actinobacteria, Cyanobacteria, Alphaproteobacteria, Betaproteobacteria, and Gammaproteobacteria, whose analysis and discussion of the findings are described below. The results obtained show how the taxonomical diversity varies between bulk soil and rhizospheric soil, being positive selected by the plant root those that have a potential benefit to the plant, like proteobacteria group (Figure 1 and Table 1) (Moulin et al., 2001; Zai et al., 2021).
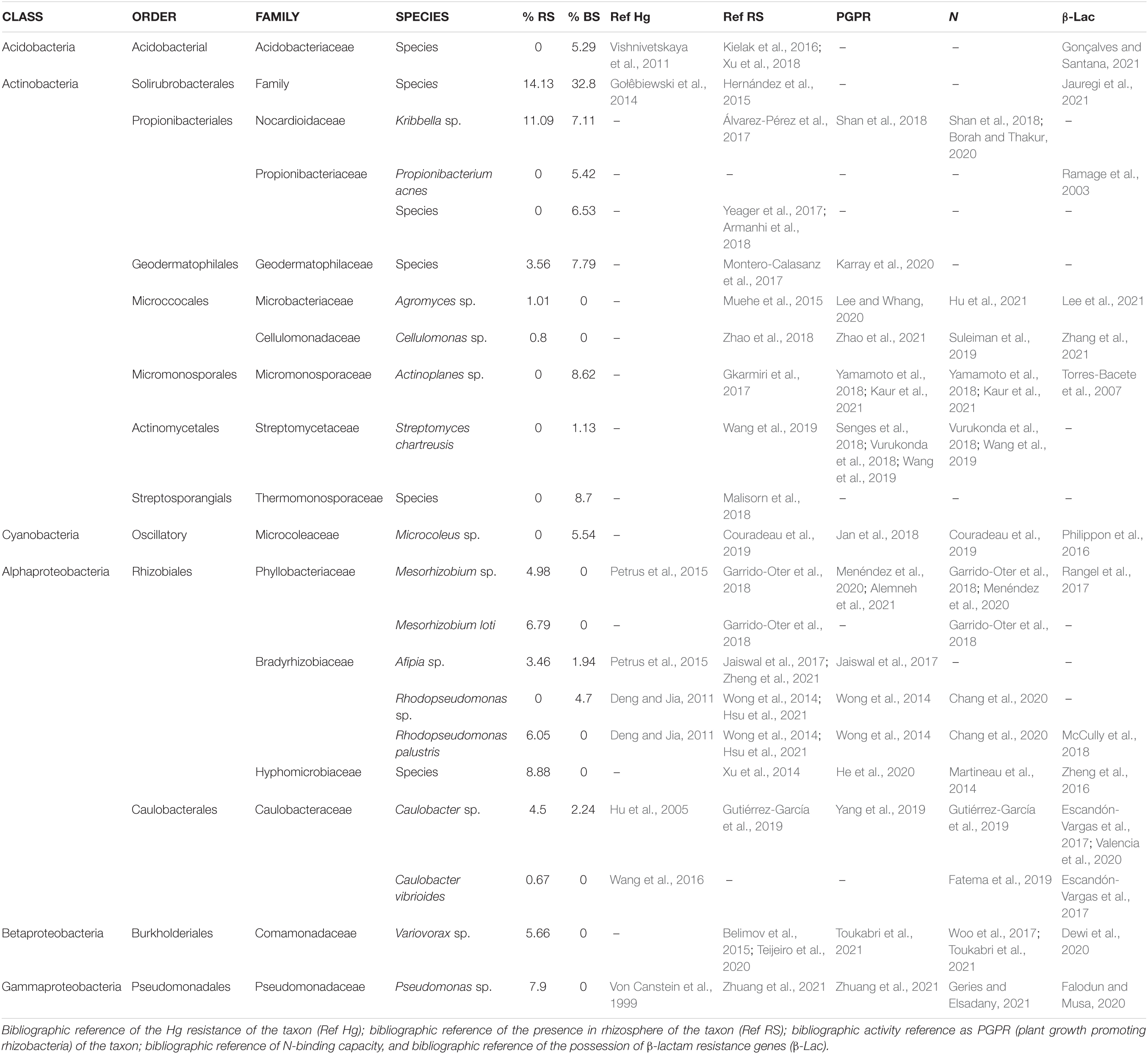
Table 1. General table of bacterial taxonomic identification, together with their relative abundances in BS and RS.
Acidobacteria are a phylum within the bacteria domain of species ubiquitous in the soil (Kalam et al., 2020). In the present study, the sample was identified up to the family taxon Acidobacteriaceae (Table 1) (Foesel et al., 2016). In the same way as in our results, the predilection of this bacterial group for the bulk soil has also been described in comparison with the rhizosphere (Kielak et al., 2016; Conradie and Jacobs, 2020). Several authors have also discussed the potential of this bacterial class for the degradation of various pollutants (Feng et al., 2021; Gonçalves and Santana, 2021) and its potential biotechnological application in Hg degradation (Vishnivetskaya et al., 2011; McDaniel et al., 2020).
Actinobacteria are a phylum and class of Gram-positive bacteria and are the group with the highest representation in BS and the second most represented in RS. This taxon is particularly interesting because of its wide biotechnological potential. Most antibiotics and many of the compounds used in the production of medicines come from the species in this class (Hopwood, 2007). There are recent studies of the relationship of these bacteria with resistance to various heavy substances (Yun et al., 2020), among which are the mer genes of resistance to Hg (Christakis et al., 2021). In this manuscript, representatives have been found in seven of the orders pertaining to this class (Table 1).
Two families and two species of the Propionibacteriales order were identified. Propionibacterium acnes is first described as Hg resistant as well as its presence in rhizospheric soils.
Kribbella sp. is linked to both types of soil. Its resistance to Hg is not included in the scientific literature, although some authors have described strains of this genus resistant to other heavy metal divalent cations like Cu, Ni, and Cd (Chanda et al., 2017; Rosenfeld et al., 2018). Other authors describe the capacity of produce siderophores by some species (Acquah et al., 2020). The presence of this bacteria in a Hg-contaminated soil and the data found on the bibliography suggests postulating that species as a good target to look for strains with possible biotechnological potential for use in soil bioremediation.
Within the Geodermatophilales order, the Geodermatophilaceae family present in both types of soils was identified (Table 1). Although bacteria associated with the rhizosphere of some plant species have been described in the literature (Montero-Calasanz et al., 2017), it was found with a greater abundance in BS in the present study. There are references to some species in this family as resistant to heavy metals (Kou et al., 2018), although no evidence of their description as resistant to Hg has been found.
In the order of Microccocales, two families and two species were detected (Table 1), Agromyces sp. and Cellulomonas sp. Neither of these two species is cited as resistant to Hg, a fact that is first described in this study. Its resistance to other heavy metals is described (Corretto et al., 2017; Brookshier et al., 2018). The presence of Agromyces sp. associated with Arabidopsis halleri roots in the extraction of heavy metals by phytoextraction is not known (Muehe et al., 2015); the role of Cellulomonas sp. in plant rhizome as a producer of IAA is known (Zhao et al., 2018). The findings on these two bacteria are postulated by organisms with a possible biotechnological potential to look for strains for use in soil bioremediation in this study, despite their low representation in the data obtained (Table 1).
In the Micromonosporal order, a single species Actinoplanes sp. was detected, only in BS, being described as good PGPR (Table 1) (Gkarmiri et al., 2017; Yamamoto et al., 2018). Works such as that of Wang et al. (2019) have demonstrated the tolerance to various heavy metals by Actinoplanes sp.; no resistance references to Hg were found. The resistance to Hg of these species it’s first described in this study.
The genus Streptomyces (Actinomicetales) is one of the most studied due to its high biotechnological and industrial potential, since a large quantity of antibiotics currently used clinically are derived from these bacteria (Hopwood, 2007). Its resistance to Hg has been referenced for more than 20 years (Ravel et al., 1998). Streptomyces chartreusis has been described as good PGPR (Table 1) (Senges et al., 2018; Vurukonda et al., 2018; Wang et al., 2019). This is the first time S. chartreusis has been referred to in a soil contaminated with Hg. The potential PGPR and its remediation capability noted in the literature, along with its presence in the study soil, suggests S. chartreusis as a good target for search strains that could be used in the remediation of this environment.
Cyanobacteria (bacteria domain) include bacteria capable of performing oxygenic photosynthesis. It’s a phylum that have an extensive ecological distribution, as well it’s needed to take in account that the plots sampled are located in rainwater leaching areas and close to water sources, so it understandable to find this phylum on the sample (Mehetre et al., 2022). The species Microcoleus sp. was detected (Table 1). Microcoleus sp. has not been described as resistant to Hg. Godoy-Lozano et al. (2018) shows the high potential of this species to degrade various heavy metals. The data collected in the literature and their presence in these soils are indicative of their potential as research targets for its possible use in the bioremediation of Hg.
Mesorhizobium sp. and Mesorhizobium loti (Phyllobacteriaceae) have been studied for their ability to form nodules in plant roots and fix N (Garrido-Oter et al., 2018). They are of great interest for their close relationship with the plant root and have been described as resistant to various heavy metals, such as Cd or Pb (Fan et al., 2018), highlighting their strong potential for use in soil remediation.
Afipia sp., Rhodopseudomonas sp. and Rhodopseudomonas palustris (Bradyrhizobiaceae): Afipia sp. arouses interest for its potential use in remediation of soils contaminated with Hg (Petrus et al., 2015). Rhodopseudomonas sp. and Rhodopseudomonas palustris are described as PGPR (Wong et al., 2014; Hsu et al., 2021). Some authors (Batool and Rehman, 2017; Xiao et al., 2021) relate the PGPR capacity of Rhodopseudomonas sp. and R. palustris to their ability to remediate Hg-contaminated soils.
Calubacter sp. and Caulobacter vibrioides (Caulobacterales) have been described as resistant to various heavy metals because they host divalent cation ATPase transporters on their membranes (Hu et al., 2005). As for C. vibrioides, it has solubilizing activity of selenium used in processes of detoxification of Hg (Wang et al., 2016). As well, this bacterias appear in the bibliography described as nitrogen fixers (Gutiérrez-García et al., 2019). For these reasons, these bacteria have a biotechnological interest for use in future bioremediation pathways.
For this Class, only one specie could be identified. Variovorax sp. (Burkholderiales) has been characterized by Belimov et al. (2015), noted for its activity as PGPR and its resistance to various heavy metals, although not appearing in the literature as resistant to Hg. Having found this species in RS with high concentration of Hg postulates it as a good candidate to further studies to find strains from this species with potential use in bioremediation.
Gammaproteobacteria are a diverse class of Gram-negative bacteria with a wide biological distribution. Pseudomonas sp. have a high biotechnological interest and have been studied for their characteristics as PGPR (Yasmeen et al., 2021). The bioremediation capacity of Hg is well known (González et al., 2021; Imron et al., 2021; Robas et al., 2021a). Therefore, a study of its potential use in the remediation of contaminated soils and look for non-phytopathogenic specific strains is of interest.
Two viral families were identified in the metagenome, Circoviridae and Potyviridae. The first is a family that is distributed in the environment infecting mainly vertebrates (Delwart and Li, 2012). Only the species Cyclovirus NGChicken15/NGA/2009was identified, it is a virus typically pathogenic from farm birds. It is interesting and remarkable that of the 21% of the relative taxonomic abundance of viruses in the RS sample, 18.88% belong to a single virus that is very far from its natural host. Due to the ubiquity of this virus and its presence in our sample only in RS we suspect that it could be a virus very similar to NGchicken15 that is infecting the plant or any of the bacteria in the rhizosphere such as prophage.
From the family Potyviridae was identified the species Dasheen mosaic virus, very common species pathogenic of vegetables (Francki et al., 2012) with a similar relative abundance in both soils (1.66% RS and 2.19% BS) It is not strange to find this species both in the fraction of SL and RF given that the samples were collected at a time of high growth and therefore of a greater transmissibility of this virus to plants and to the soils.
Functional potential bioinformatic analysis allows the identification of potential metabolic activities and processes. In this way, it is possible to sort the potential functions of the microbial community according to their abundance. However, minority activities should be taken into account as long as they allow the biological behavior of these communities to be interpreted. In this sense, most of the functional potential clusters identified, and sequences associated to a protein belong to basal metabolism. However, the mechanisms of Hg resistance, those involved in oxidative stress, N metabolism and PGPR activity are more important from an ecological and functional potential point of view. In addition, sequences associated to a protein have been found whose presence in soils can only be interpreted as indicating biological contamination (Li et al., 2020; Stange and Tiehm, 2020). Since there is no antibiotic pressure on the analyzed soils, the mechanisms of resistance commonly described in clinical studies should not be detected in environmental samples. For this reason, in the area of “One Health” they deserve to be analyzed and interpreted as bioindicators of biological pollution. Microbial soil communities can act as reservoirs from which information can be transferred horizontally to potential pathogens, becoming a threat to human, animal and environmental health (Wang et al., 2021).
Soils, especially those under high environmental pressure, act as a natural reservoir of resistance to existing antibiotics or provide the potential to host bacteria of clinical importance (Yan et al., 2020; Liao et al., 2021). Several studies show the existence of a co-choice between the presence of various toxic compounds in the environment and the selection of antibiotic resistance naturally, together with co-resistance to heavy metals and antibiotics (Yan et al., 2020; Mazhar et al., 2021; Robas et al., 2021b).
At level II this functional cluster were more represented in RS than in BS. At level III of the metagenome study, several specific role clusters for resistance to rare antibiotics in the natural environment have been identified (Supplementary Table 12), such as β-lactamases and mechanisms associated with resistance to beta-lactams and predicted proteins associated with gene responses to these antibiotics BlaR1 and MecR1 (Silveira et al., 2018).
Likewise, the presence of various transporters dependent on ATP of Pb, Cb, Cu, and Hg in the group of functional activities regulating the beta-lactams BlaR1 was found, and a direct relationship between the presence of these heavy metals and β-lactam resistance. Among the predicted proteins related with β-lactam resistance genes isolated in this study, some belonging to classes A, C, and D were identified.
Class A includes several subsystems, all fundamentally linked to ampicillin resistance, that are rarely used clinically owing to numerous described resistances that exist for this antibiotic (Kaye et al., 2000; Rice et al., 2001). CTX-M-16 was found, which gives greater catalytic power than other cefotaximes (Bonnet et al., 2001). The finding in the present work of resistance mechanisms to these antibiotics was evidence of the selection of resistance mechanisms of clinical origin, especially when occurring in semi-synthetic compounds that do not occur in nature.
The AmpC-type β-lactamases are of great clinical importance because they are hydrolyzed penicillins, cefamycins, oxyminocephalosporins and monobactams, although they are not active against fourth-generation carbapenemic cephalosporins (Jacoby, 2009; Aguirre et al., 2020). The relevance of these genes is provided by their high transmissibility, since many are found in plasmids (Ku et al., 2019; Rensing et al., 2019).
The MexAB-OprM and MexEF-OprN complexes are a protein assembly of membrane transporters that provide multi-resistance to antibiotics, identified primarily in Pseudomonas sp. and are highly linked to multi-resistance in P. aeruginosa (Ma et al., 2021). As shown in Table 1, there is high probability that these genes that could confer multi-resistance can be associated with the Pseudomonas identified in this manuscript. In the analysis of the sequences, were predicted proteins from the MexT regulator was found, which is inactivated by some Hg resistance genes, making the strains carrying the MexEF-OprN complex sensitive to carbapenems (Köhler et al., 1999).
At level III, sequences related to RND transporters have been found in multi-resistance efflux pumps functional potential cluster. Among these, there is a large representation of MexAB-OprM and MexEF-OprN, along with MexCD-OprJ antibiotic transporters, related not only to antibiotic resistance but also mediate processes of quorum sensing (Alcalde-Rico et al., 2018). Other antibiotic resistance predicted proteins found were those from AcrAB-TolC, a system responsible for the expulsion of antibiotics, such as penicillin G, cloxacillin, naphthyllin, macrolides, novobiocin, linezolid, and fusidic acid derivatives; this system is commonly associated with E. coli (Anes et al., 2015; Byrd et al., 2021). Proteins from MdtABC genes have also been predicted in SEED, related TolC, which give resistance to novobiocin, quinolones and phosphomycin, among others (Anes et al., 2015). CmeABC membrane transporters were also found, related to Campylobacter jejuni’s resistance to a wide variety of antibiotics (Lin et al., 2002; Yan et al., 2006), although authors such as Stopnisek et al. (2016) have reported the presence of these genes in other species within the Rhizobiales order.
Some proteins directly related to antibiotic multi-resistance mechanisms have been predicted with the metagenome sequences in Gram-positive bacteria cluster. The MdtRP operon of Bacillus sp. confers resistance to several antibiotics such as novobiocin, streptomycin and actinomycin, and is regulated by the MarR repressor (Gupta et al., 2019; Warmbold et al., 2020).
Another of the functional subsystems identified in level III and SEED is related to tripartite protein expulsion systems in Gram-negative bacteria. Similar to the already noted MexAB-OprM or AcrAB-TolC, they belong to membrane transporters RND (Daury et al., 2016). No specific identification of any of these sequences was achieved beyond identifying them as antibiotic ejection systems within the RND group and related to tripartite proteins such as MdtABC, and ejection proteins from genes such as TolC or OprN. This highlights a variety of resistance systems that bacteria can possess in a hostile environment, and which are still unknown.
An important functional cluster related to fluoroquinolone resistance has been found. These are synthesis compounds, not found naturally in environmental samples. Hooper (2000) describes how fluoroquinolone resistances are acquired by mutations in the genes of Topoisomerases II and IV, predicted proteins present in the metagenome sequences of the samples analyzed at level III and SEED. Similarly, pumps from Lde genes were found, which are specific to fluoroquinolones (Godreuil et al., 2003).
A functional cluster and predicted proteins related with vancomycin resistance were also found. The proteins predicted VanA, VanB, VanH, VanR, VanS, VanW, VanX and VanZ, all of which were directly related to vancomycin resistance (Qureshi et al., 2014; Stogios and Savchenko, 2020) and VanZ, which in turn gave teicoplanin resistance (Qureshi et al., 2014) and VanW whose role in resistance mechanisms is still unknown. These genes are usually grouped by their function and level of resistance, thus taking the VanAB, VanHAX, and VanRS groups (Bugg et al., 1991; Arthur et al., 1992). The presence of some sequences that predict proteins, such as those related with VanAB genes, appearing in plasmids has been studied (Ishihara et al., 2013; Qureshi et al., 2014; Sivertsen et al., 2016), giving this resistance greater potential to be transmitted to other species. Several proteins related with that regulation of the activity of this resistance were also found, such as VanRS and a histidin-kinase system that activates the resistance system (Arthur and Quintiliani, 2001; Qureshi et al., 2014; Stogios and Savchenko, 2020).
Another group of resistance functional potential cluster was that corresponding to bacitracin resistance. The predicted proteins in SEED correspond to ABC flow pumps associated with the Bacillus genus such as BceAB, BceR, YvcPR, YxdM, YclH, YknY, BseL and LiaRS (Mascher et al., 2004; Kingston et al., 2014). Some of these genes have also been reported in other Gram-positives, such as some species of Enterococcus and Clostridium (Charlebois et al., 2012; Zhou et al., 2019).
The mechanisms of resistance to Hg are widely described in the microbial world. For this reason, it is not uncommon to detect various predicted proteins associated with genes from operon mer (MerC, MerE, and MerT) (Gionfriddo et al., 2020) and mercuroreductases in the samples analyzed (Supplementary Table 6). In the same way, some resistance and transport mechanisms for divalent cations (Co, Zn, and Cd primarily) (Supplementary Table 6) were found, revealing the probably participation of that mechanisms in the resistance to Hg of the microorganisms. Likewise, ABC-type transporters (ATP-binding cassette), capable of providing resistance to bacteria against various toxins (Acar et al., 2020; Thomas and Tampé, 2020), account for almost 50% of membrane transporters in level II. At SEED level it can be found some ABC and RND efflux systems related with resistance with divalent cations and heavy metals (Supplementary Table 6). These transporters are widely distributed throughout the metagenome and primarily associated with resistance systems, acting as efflux pumps for different toxic compounds (Supplementary Table 3).
Environmental factors are known to cause oxidative stress to the colonizing microorganisms of contaminated soils. In order to colonize these environments, bacteria need effective biological mechanisms. Stress response genes give microorganism methods of adaptability to host situations and environments that may affect their normal development (Chakraborty and Kenney, 2018; Dweba et al., 2018; Kokou et al., 2018). These potential functional activities are found with a high relative abundance (Supplementary Table 7), as the soil is a “nutritional desert” and has the abiotic stress of high Hg concentration. These factors are in a similar proportion in both samples; it seems that BS and RS exert a different environmental pressure on organisms in terms of stress. Various ABC transport mechanisms have been developed, which function as part of the stress response machine in a hostile environment (Teichmann et al., 2018; Van Goethem et al., 2018). As well some sequences related with the carbon starvation, like proteins of Slp (Alexander and St John, 1994) and Sspa (Yin et al., 2021) were found. And functional potential clusters related with weather conditions (Cold and heat shock, desiccation stress and osmotic stress) (Supplementary Table 7).
N is a limiting macronutrient for the proliferation of microorganisms and the growth of plants. Therefore, the enzymes involved in the synthesis of nitrogen compounds usable by plants are relevant. In the present study, in level I was found a functional potential cluster that involves the N metabolism (Supplementary Table 8). Several sequences have been associated with proteins related to the assimilation of nitrate and nitrite as the Nar (NarA, NarD, NarE, NarK, NarL, and NarP) (Fukuda et al., 2015). Among the N metabolism, proteins from Nir denitrification genes were identified. Some of these sequences, related with genes such as NirT and Nos, are involved in N monoxide denitrification and formation (Bergaust et al., 2012; Belbahri et al., 2017). At level III, was alsa found a cluster related with the nitrogen fixation, fundamentally from the operon Nif were identified, which codes for the nitrogenase complex, fundamental in the fixation of atmospheric N in the soil (Di Cesare et al., 2018; Dasgupta et al., 2021; Supplementary Table 8).
Plant growth-promoting bacteria are characterized by their ability to produce and/or adapt to a hostile environment, such as auxin production, phosphate solubilization, ACC degradation, and siderophore production. Several sequences clustered in potential functional activities involved in the synthesis of auxins, an important factor promoting the growth of plants, have been identified (Mishra et al., 2021). Specifically, 3-indolacetic acid (IAA) is related to a large number of processes that improve plant quality. Some proteins from the principal biosynthetic routes of IAA, indole-3-pyruvate and indole-3-acetamide were predicted (Nascimento et al., 2021), such as proteins IorA and IorB (Supplementary Table 9). Likewise, IAA has the capacity to improve the tolerance of plants to adverse conditions and stress by heavy metals (Ma et al., 2011; Nazli et al., 2021). Some authors (Ma et al., 2011; Zainab et al., 2020) have established a relationship between rhizospheric bacteria producing IAA and significant improvement together with greater speed in phytoextraction of heavy substances and recovery of contaminated soils.
Plants and microorganisms compete for phosphorus present in the environment; therefore, the solubilization of phosphorus by microorganisms contributes to the promotion of plant growth (Pereira et al., 2020). A large number of proteins genes have been predicted for acid phosphatases, phytases, gluconate dehydrogenases, ketogluconate dehydrogenases, and glucose-1-dehydrogenase (Supplementary Table 10), which are encompassed in the context of phosphate solubilization (Suleman et al., 2018).
The predicted proteins of the AcdS genes of the ACC deaminase (1-aminocyclopropane-1-carboxylate desaminase) found in our metagenome interfere with the synthesis of ethylene in the plant by degradation of a metabolic precursor, thereby reducing stress in the tissues (Glick, 2014; del Carmen Orozco-Mosqueda et al., 2020). Ethylene is a marker of plant stress and senescence, so this enzyme helps plants withstand stressful environments, such as soils contaminated with heavy metals.
A wide variety of siderophore functional potential clusters were also identified (Supplementary Table 11), it can be found at level II a functional potential cluster related with siderophores (Supplementary Table 3). Siderophores act as metal chelators favoring, among other potential functions, the absorption of iron and its entry into the food chain. Plants that are able to use bacterial siderophores as a source of iron (Wang et al., 1993) increase their chances of survival and adaptation to contaminated environments.
Several conclusions can be drawn from this study.
In the taxonomic analyze, the most abundant microbial genome analyzed belongs to the bacteria domain. The prevalent taxa are those of Actinobateria and Alphaproteobacteria. Betaproteobacteria and Gammaproteobacteria seems to be intimately linked to rhizospheric soil (RS). Likewise, Cyanobacteria and Acidobacteria only have representation in bulk soil (BS). Similarly, the genome belonging to the Domain Eukarya involved in the potential functional activity of microbial activity was detected. The viral genomes present in the sample are interpreted as prophages.
On the functional potential profiling, the presence of antibiotic resistance mechanisms and other toxic compounds could confirm previous studies pointing to their co-selection in Hg-contaminated environments. The finding of resistance mechanisms proper to human clinical, evidence of biological contamination, suggests that these environments may behave as reservoirs. Although, the presence of Hg resistance functional clusters and involved in the response to oxidative stress are present as a minority; however, their biological significance justifies the behavior of the microbial community. The abundance of PGP and N-fixation functional activities detected in the metagenome sequences may be an opportunity for further selection of both effective bioremediation strains and genes for promoting biotechnological use in the production of GMOs.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
AP and PJ supervised the project and acquired funding for this research. All authors designed the experiments, made intellectual contributions, conducted the experiments, analyzed the data, wrote and edited the manuscript.
This research has been funded by Fundación Universitaria San Pablo CEU (Microbiology SAI) and Banco Santander, grant number FUSP-BS-PPC01/2014.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.797444/full#supplementary-material
Supplementary Table 1 | Relative abundances at all taxonomical levels (Metaphlan data table).
Supplementary Table 2 | Relative abundances of SUPERFOCUS functional level I.
Supplementary Table 3 | Relative abundances of SUPERFOCUS functional level II.
Supplementary Table 4 | Relative abundances of SUPERFOCUS functional level III.
Supplementary Table 5 | Relative abundances of SUPERFOCUS at SEED function level.
Supplementary Table 6 | Relative abundances of SUPERFOCUS functional levels I–III and SEED of resistance to heavy metals potential functional activities.
Supplementary Table 7 | Relative abundances of SUPERFOCUS functional levels I–III and SEED of stress response and oxidative stress potential functional activities.
Supplementary Table 8 | Relative abundances of SUPERFOCUS functional levels I–III and SEED of nitrogen metabolism potential functional activities.
Supplementary Table 9 | Relative abundances of SUPERFOCUS functional levels I–III and SEED of IAA potential functional activities.
Supplementary Table 10 | Relative abundances of SUPERFOCUS functional levels I–III and SEED of ACCd potential functional activities.
Supplementary Table 11 | Relative abundances of SUPERFOCUS functional levels I–III and SEED of siderophores potential functional activities.
Supplementary Table 12 | Relative abundances of SUPERFOCUS functional levels I–III and SEED of resistance to antibiotics potential functional activities.
Acar, B., Rose, J., Fas, B. A., Ben-Tal, N., Lewinson, O., and Haliloglu, T. (2020). Distinct allosteric networks underlie mechanistic speciation of ABC transporters. Structure 28, 651–663. doi: 10.1016/j.str.2020.03.014
Acquah, K. S., Beukes, D. R., Warner, D. F., Meyers, P. R., Sunassee, S. N., Maglangit, F., et al. (2020). Novel South African rare actinomycete Kribbella speibonae strain SK5: a prolific producer of hydroxamate siderophores including new dehydroxylated congeners. Molecules 25:2979. doi: 10.3390/molecules25132979
Aguirre, L., Vidal, A., Seminati, C., Tello, M., Redondo, N., Darwich, L., et al. (2020). Antimicrobial resistance profile and prevalence of extended-spectrum beta-lactamases (ESBL), AmpC beta-lactamases and colistin resistance (mcr) genes in Escherichia coli from swine between 1999 and 2018. Porcine Health Manag. 6:8. doi: 10.1186/s40813-020-00146-2
Alcalde-Rico, M., Olivares-Pacheco, J., Alvarez-Ortega, C., Cámara, M., and Martínez, J. L. (2018). Role of the multidrug resistance efflux pump MexCD-OprJ in the Pseudomonas aeruginosa quorum sensing response. Front. Microbiol. 9:2752. doi: 10.3389/fmicb.2018.02752
Alemneh, A. A., Zhou, Y., Ryder, M. H., and Denton, M. D. (2021). Large-scale screening of rhizobacteria to enhance the chickpea-Mesorhizobium symbiosis using a plant-based strategy. Rhizosphere 18:100361. doi: 10.1016/j.rhisph.2021.100361
Alexander, D. M., and St John, A. C. (1994). Characterization of the carbon starvation-inducible and stationary phase-inducible gene slp encoding an outer membrane lipoprotein in Escherichia coli. Mol. Microbiol. 11, 1059–1071. doi: 10.1111/j.1365-2958.1994.tb00383.x
Álvarez-Pérez, J. M., González-García, S., Cobos, R., Olego, M. Á., Ibañez, A., Díez-Galán, A., et al. (2017). Use of endophytic and rhizosphere actinobacteria from grapevine plants to reduce nursery fungal graft infections that lead to young grapevine decline. Appl. Environ. Microbiol. 83:e01564-17. doi: 10.1128/AEM.01564-17
Andrews, S. (2017). FastQC: a Quality Control Tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed August 22, 2021).
Anes, J., McCusker, M. P., Fanning, S., and Martins, M. (2015). The ins and outs of RND efflux pumps in Escherichia coli. Front. Microbiol. 6:587. doi: 10.3389/fmicb.2015.00587
Armanhi, J. S. L., de Souza, R. S. C., Damasceno, N., Damasceno, N. B., de Araújo, L. M., Imperial, J., et al. (2018). A community-based culture collection for targeting novel plant growth-promoting bacteria from the sugarcane microbiome. Front. Plant Sci. 8:2191. doi: 10.3389/fpls.2017.02191
Arthur, M., Molinas, C., and Courvalin, P. (1992). The VanS-VanR two-component regulatory system controls synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J. Bacteriol. 174, 2582–2591. doi: 10.1128/jb.174.8.2582-2591.1992
Arthur, M., and Quintiliani, R. Jr. (2001). Regulation of VanA-and VanB-type glycopeptide resistance in enterococci. Antimicrob. Agents Chemother. 45, 375–381. doi: 10.1128/aac.45.2.375-381.2001
Aziz, R. K., Devoid, S., Disz, T., Edwards, R. A., Henry, C. S., Olsen, G. J., et al. (2012). SEED servers: high-performance access to the SEED genomes, annotations, and metabolic models. PLoS One 7:e48053. doi: 10.1371/journal.pone.0048053
Batool, K., and Rehman, Y. (2017). Arsenic-redox transformation and plant growth promotion by purple nonsulfur bacteria Rhodopseudomonas palustris CS2 and Rhodopseudomonas faecalis SS5. Biomed Res. Int. 2017:6250327. doi: 10.1155/2017/6250327
Belbahri, L., Chenari Bouket, A., Rekik, I., Alenezi, F. N., Vallat, A., Luptakova, L., et al. (2017). Comparative genomics of Bacillus amyloliquefaciens strains reveals a core genome with traits for habitat adaptation and a secondary metabolites rich accessory genome. Front. Microbiol. 8:1438. doi: 10.3389/fmicb.2017.01438
Belimov, A. A., Puhalsky, I. V., Safronova, V. I., Shaposhnikov, A. I., Vishnyakova, M. A., Semenova, E. V., et al. (2015). Role of plant genotype and soil conditions in symbiotic plant-microbe interactions for adaptation of plants to cadmium-polluted soils. Water Air Soil Pollut. 226:264.
Bergaust, L., van Spanning, R. J., Frostegård, Å., and Bakken, L. R. (2012). Expression of nitrous oxide reductase in Paracoccus denitrificans is regulated by oxygen and nitric oxide through FnrP and NNR. Microbiology 158, 826–834. doi: 10.1099/mic.0.054148-0
Bjørklund, G., Tinkov, A. A., Dadar, M., Rahman, M. M., Chirumbolo, S., Skalny, A. V., et al. (2019). Insights into the potential role of mercury in Alzheimer’s disease. J. Mol. Neurosci. 67, 511–533. doi: 10.1007/s12031-019-01274-3
Bonnet, R., Dutour, C., Sampaio, J., Chanal, C., Sirot, D., Labia, R., et al. (2001). Novel cefotaximase (CTX-M-16) with increased catalytic efficiency due to substitution Asp-240→ Gly. Antimicrob. Agents Chemother. 45, 2269–2275. doi: 10.1128/AAC.45.8.2269-2275.2001
Borah, A., and Thakur, D. (2020). Phylogenetic and functional characterization of culturable endophytic actinobacteria associated with Camellia spp. for growth promotion in commercial tea cultivars. Front. Microbiol. 11:318. doi: 10.3389/fmicb.2020.00318
Brookshier, A., Santo Domingo, J., Kourtev, P., and Learman, D. (2018). Draft genome sequences of two Bacillus sp. strains and four Cellulomonas sp. strains isolated from heavy-metal-contaminated soil. Microbiol. Resour. Announc. 7:e01063-18. doi: 10.1128/MRA.01063-18
Bugg, T. D., Wright, G. D., Dutka-Malen, S., Arthur, M., Courvalin, P., and Walsh, C. T. (1991). Molecular basis for vancomycin resistance in Enterococcus faecium BM4147: biosynthesis of a depsipeptide peptidoglycan precursor by vancomycin resistance proteins VanH and VanA. Biochemistry 30, 10408–10415. doi: 10.1021/bi00107a007
Byrd, B. A., Zenick, B., Rocha-Granados, M. C., Englander, H. E., Hare, P. J., LaGree, T. J., et al. (2021). The AcrAB-TolC efflux pump impacts persistence and resistance development in stationary-phase Escherichia coli following delafloxacin treatment. Antimicrob. Agents Chemother. 65:e0028121. doi: 10.1128/AAC.00281-21
Castillo Villamizar, G. A., Nacke, H., Boehning, M., Herz, K., and Daniel, R. (2019). Functional metagenomics reveals an overlooked diversity and novel features of soil-derived bacterial phosphatases and phytases. mBio 10:e01966-18. doi: 10.1128/mBio.01966-18
Chacón-Vargas, K., Torres, J., Giles-Gómez, M., Escalante, A., and Gibbons, J. G. (2020). Genomic profiling of bacterial and fungal communities and their predictive functionality during pulque fermentation by whole-genome shotgun sequencing. Sci. Rep. 10:15115. doi: 10.1038/s41598-020-71864-4
Chakraborty, S., and Kenney, L. J. (2018). A new role of OmpR in acid and osmotic stress in Salmonella and E. coli. Front. Microbiol. 9:2656. doi: 10.3389/fmicb.2018.02656
Chanda, D., Sharma, G., Jha, D., and Hijri, M. (2017). “Tolerance of microorganisms in soil contaminated with trace metals: an overview,” in Recent Advances in Applied Microbiology, ed. P. Shukla (Berlin: Springer), 165–193. doi: 10.1007/978-981-10-5275-0_8
Chang, T.-H., Wang, R., Peng, Y.-H., Chou, T.-H., Li, Y.-J., and Shih, Y. (2020). Biodegradation of hexabromocyclododecane by Rhodopseudomonas palustris YSC3 strain: a free-living nitrogen-fixing bacterium isolated in Taiwan. Chemosphere 246:125621. doi: 10.1016/j.chemosphere.2019.125621
Charlebois, A., Jalbert, L.-A., Harel, J., Masson, L., and Archambault, M. (2012). Characterization of genes encoding for acquired bacitracin resistance in Clostridium perfringens. PLoS One 7:e44449. doi: 10.1371/journal.pone.0044449
Christakis, C. A., Barkay, T., and Boyd, E. S. (2021). Expanded diversity and phylogeny of mer genes broadens mercury resistance paradigms and reveals an origin for MerA among thermophilic Archaea. Front. Microbiol. 12:682605. doi: 10.3389/fmicb.2021.682605
Collins, F. W., Walsh, C. J., Gomez-Sala, B., Guijarro-García, E., Stokes, D., Jakobsdóttir, K. B., et al. (2021). The microbiome of deep-sea fish reveals new microbial species and a sparsity of antibiotic resistance genes. Gut Microbes 13, 1–13. doi: 10.1080/19490976.2021.1921924
Conradie, T., and Jacobs, K. (2020). Seasonal and agricultural response of Acidobacteria present in two fynbos rhizosphere soils. Diversity 12:277. doi: 10.3390/d12070277
Cordier, T., Alonso-Sáez, L., Apothéloz-Perret-Gentil, L., Aylagas, E., Bohan, D. A., Bouchez, A., et al. (2021). Ecosystems monitoring powered by environmental genomics: a review of current strategies with an implementation roadmap. Mol. Ecol. 30, 2937–2958. doi: 10.1111/mec.15472
Corretto, E., Antonielli, L., Sessitsch, A., Compant, S., Höfer, C., Puschenreiter, M., et al. (2017). Complete genome sequence of the heavy metal resistant bacterium Agromyces aureus AR33 T and comparison with related Actinobacteria. Stand. Genomic Sci. 12:2. doi: 10.1186/s40793-016-0217-z
Couradeau, E., Giraldo-Silva, A., De Martini, F., and Garcia-Pichel, F. (2019). Spatial segregation of the biological soil crust microbiome around its foundational cyanobacterium, Microcoleus vaginatus, and the formation of a nitrogen-fixing cyanosphere. Microbiome 7:55. doi: 10.1186/s40168-019-0661-2
Cycil, L. M., DasSarma, S., Pecher, W., McDonald, R., AbdulSalam, M., and Hasan, F. (2020). Metagenomic insights into the diversity of halophilic microorganisms indigenous to the Karak Salt Mine, Pakistan. Front. Microbiol. 11:1567. doi: 10.3389/fmicb.2020.01567
Dasgupta, D., Panda, A. K., Mishra, R., Mahanty, A., De Mandal, S., and Bisht, S. S. (2021). “Nif genes: tools for sustainable agriculture,” in Recent Advancement in Microbial Biotechnology, eds S. De Mandal and A. K. Passari (Amsterdam: Elsevier), 413–434. doi: 10.1016/b978-0-12-822098-6.00012-4
Daury, L., Orange, F., Taveau, J.-C., Verchere, A., Monlezun, L., Gounou, C., et al. (2016). Tripartite assembly of RND multidrug efflux pumps. Nat. Commun. 7:10731. doi: 10.1038/ncomms10731
de Abreu, V. A., Perdigão, J., and Almeida, S. (2020). Metagenomic approaches to analyze antimicrobial resistance: an overview. Front. Genet. 11:575592. doi: 10.3389/fgene.2020.575592
del Carmen Orozco-Mosqueda, M., Glick, B. R., and Santoyo, G. (2020). ACC deaminase in plant growth-promoting bacteria (PGPB): an efficient mechanism to counter salt stress in crops. Microbiol. Res. 235:126439. doi: 10.1016/j.micres.2020.126439
Delwart, E., and Li, L. (2012). Rapidly expanding genetic diversity and host range of the Circoviridae viral family and other Rep encoding small circular ssDNA genomes. Virus Res. 164, 114–121. doi: 10.1016/j.virusres.2011.11.021
Deng, X., and Jia, P. (2011). Construction and characterization of a photosynthetic bacterium genetically engineered for Hg2+ uptake. Bioresour. Technol. 102, 3083–3088. doi: 10.1016/j.biortech.2010.10.051
Dewi, D. A. R., Götz, B., and Thomas, T. (2020). Diversity and genetic basis for carbapenem resistance in a coastal marine environment. Appl. Environ. Microbiol. 86:e02939-19. doi: 10.1128/AEM.02939-19
Di Cesare, A., Cabello-Yeves, P. J., Chrismas, N. A., Sánchez-Baracaldo, P., Salcher, M. M., and Callieri, C. (2018). Genome analysis of the freshwater planktonic Vulcanococcus limneticus sp. nov. reveals horizontal transfer of nitrogenase operon and alternative pathways of nitrogen utilization. BMC Genomics 19:259. doi: 10.1186/s12864-018-4648-3
Douglas, G. M., Maffei, V. J., Zaneveld, J. R., Yurgel, S. N., Brown, J. R., Taylor, C. M., et al. (2020). PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 38, 685–688. doi: 10.1038/s41587-020-0548-6
Dweba, C. C., Zishiri, O. T., and El Zowalaty, M. E. (2018). Methicillin-resistant Staphylococcus aureus: livestock-associated, antimicrobial, and heavy metal resistance. Infect. Drug Resist. 11, 2497–2509. doi: 10.2147/IDR.S175967
Enagbonma, B. J., Ajilogba, C. F., and Babalola, O. O. (2020). Metagenomic profiling of bacterial diversity and community structure in termite mounds and surrounding soils. Arch. Microbiol. 202, 2697–2709. doi: 10.1007/s00203-020-01994-w
Escandón-Vargas, K., Reyes, S., Gutiérrez, S., and Villegas, M. V. (2017). The epidemiology of carbapenemases in Latin America and the Caribbean. Expert Rev. Anti Infect. Ther. 15, 277–297.
Falodun, O. I., and Musa, I. B. (2020). Pseudomonas species from cattle dung producing extended spectrum and metallo beta-lactamases. Eur. J. Biol. Res. 10, 1–10.
Fan, M., Xiao, X., Guo, Y., Zhang, J., Wang, E., Chen, W., et al. (2018). Enhanced phytoremdiation of Robinia pseudoacacia in heavy metal-contaminated soils with rhizobia and the associated bacterial community structure and function. Chemosphere 197, 729–740. doi: 10.1016/j.chemosphere.2018.01.102
Fatema, K., Mahmud, N. U., and Islam, M. T. (eds). (2019). “Beneficial effects of weed endophytic bacteria: diversity and potentials of their usage in sustainable agriculture,” in Agron Crops (New York, NY: Springer International Publisher), 349–364.
Feng, L., Song, J., Gu, H., and Zhen, X. (2021). Mechanism of contaminant removal by algae-bacteria symbiosis in a PBR system during the treatment of anaerobic digestion effluents. Agric. Water Manag. 247:106556. doi: 10.1016/j.agwat.2020.106556
Foesel, B. U., Mayer, S., Luckner, M., Wanner, G., Rohde, M., and Overmann, J. (2016). Occallatibacter riparius gen. nov., sp. nov. and Occallatibacter savannae sp. nov., acidobacteria isolated from Namibian soils, and emended description of the family Acidobacteriaceae. Int. J. Syst. Evol. Microbiol. 66, 219–229. doi: 10.1099/ijsem.0.000700
Francki, R. I. B., Fauquet, C., Knudson, D., and Brown, F. (2012). Classification and Nomenclature of Viruses: Fifth Report of the International Committee on Taxonomy of Viruses. Virology Division of the International Union of Microbiological Societies. Berlin: Springer Science & Business Media.
Fukuda, M., Takeda, H., Kato, H. E., Doki, S., Ito, K., Maturana, A. D., et al. (2015). Structural basis for dynamic mechanism of nitrate/nitrite antiport by NarK. Nat. Commun. 6:7097. doi: 10.1038/ncomms8097
Garrido-Oter, R., Nakano, R. T., Dombrowski, N., Ma, K.-W., Team, T. A., McHardy, A. C., et al. (2018). Modular traits of the Rhizobiales root microbiota and their evolutionary relationship with symbiotic rhizobia. Cell Host Microbe 24, 155–167. doi: 10.1016/j.chom.2018.06.006
Garrido-Sanz, D., Manzano, J., Martín, M., Redondo-Nieto, M., and Rivilla, R. (2018). Metagenomic analysis of a biphenyl-degrading soil bacterial consortium reveals the metabolic roles of specific populations. Front. Microbiol. 9:232. doi: 10.3389/fmicb.2018.00232
Geries, L., and Elsadany, A. Y. (2021). Maximizing growth and productivity of onion (Allium cepa L.) by Spirulina platensis extract and nitrogen-fixing endophyte Pseudomonas stutzeri. Arch. Microbiol. 203, 169–181. doi: 10.1007/s00203-020-01991-z
Giagnoni, L., Arenella, M., Galardi, E., Nannipieri, P., and Renella, G. (2018). Bacterial culturability and the viable but non-culturable (VBNC) state studied by a proteomic approach using an artificial soil. Soil Biol. Biochem. 118, 51–58. doi: 10.1016/j.soilbio.2017.12.004
Gil-Hernández, F., Gómez-Fernández, A. R., la Torre-Aguilar, M. J., Pérez-Navero, J. L., Flores-Rojas, K., Martín-Borreguero, P., et al. (2020). Neurotoxicity by mercury is not associated with autism spectrum disorders in Spanish children. Ital. J. Pediatr. 46:19. doi: 10.1186/s13052-020-0780-1
Gionfriddo, C. M., Stott, M. B., Power, J. F., Ogorek, J. M., Krabbenhoft, D. P., Wick, R., et al. (2020). Genome-resolved metagenomics and detailed geochemical speciation analyses yield new insights into microbial mercury cycling in geothermal springs. Appl. Environ. Microbiol. 86:e00176-20. doi: 10.1128/AEM.00176-20
Gkarmiri, K., Mahmood, S., Ekblad, A., Alström, S., Högberg, N., and Finlay, R. (2017). Identifying the active microbiome associated with roots and rhizosphere soil of oilseed rape. Appl. Environ. Microbiol. 83:e01938-17. doi: 10.1128/AEM.01938-17
Glick, B. R. (2014). Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 169, 30–39. doi: 10.1016/j.micres.2013.09.009
Godoy-Lozano, E. E., Escobar-Zepeda, A., Raggi, L., Merino, E., Gutierrez-Rios, R. M., Juarez, K., et al. (2018). Bacterial diversity and the geochemical landscape in the Southwestern Gulf of Mexico. Front. Microbiol. 9:2528. doi: 10.3389/fmicb.2018.02528
Godreuil, S., Galimand, M., Gerbaud, G., Jacquet, C., and Courvalin, P. (2003). Efflux pump Lde is associated with fluoroquinolone resistance in Listeria monocytogenes. Antimicrob. Agents Chemother. 47, 704–708. doi: 10.1128/AAC.47.2.704-708.2003
Gołêbiewski, M., Deja-Sikora, E., Cichosz, M., Tretyn, A., and Wróbel, B. (2014). 16S rDNA pyrosequencing analysis of bacterial community in heavy metals polluted soils. Microb. Ecol. 67, 635–647. doi: 10.1007/s00248-013-0344-7
Gonçalves, O. S., and Santana, M. F. (2021). The coexistence of monopartite integrative and conjugative elements in the genomes of Acidobacteria. Gene 777:145476. doi: 10.1016/j.gene.2021.145476
González, D., Blanco, C., Probanza, A., Jiménez, P. A., and Robas, M. (2021). Evaluation of the PGPR capacity of four bacterial strains and their mixtures, tested on Lupinus albus var. Dorado seedlings, for the bioremediation of mercury-polluted soils. Processes 9:1293. doi: 10.3390/pr9081293
Gupta, A., Pande, A., Sabrin, A., Thapa, S. S., Gioe, B. W., and Grove, A. (2019). MarR family transcription factors from Burkholderia species: hidden clues to control of virulence-associated genes. Microbiol. Mol. Biol. Rev. 83:e00039-18. doi: 10.1128/MMBR.00039-18
Gutiérrez-García, K., Bustos-Díaz, E. D., Corona-Gómez, J. A., Ramos-Aboites, H. E., Sélem-Mojica, N., Cruz-Morales, P., et al. (2019). Cycad coralloid roots contain bacterial communities including cyanobacteria and Caulobacter spp. that encode niche-specific biosynthetic gene clusters. Genome Biol. Evol. 11, 319–334. doi: 10.1093/gbe/evy266
Handelsman, J. (2004). Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 68, 669–685. doi: 10.1128/MMBR.68.4.669-685.2004
He, S., Li, Y., Guo, H., Lu, L., and Yang, C. (2020). Combined effect of ryegrass and Hyphomicrobium sp. GHH on the remediation of EE2-Cd co-contaminated soil. J. Soils Sediments 20, 425–434.
Hernández, M., Dumont, M. G., Yuan, Q., and Conrad, R. (2015). Different bacterial populations associated with the roots and rhizosphere of rice incorporate plant-derived carbon. Appl. Environ. Microbiol. 81, 2244–2253. doi: 10.1128/AEM.03209-14
Hooper, D. C. (2000). Mechanisms of action and resistance of older and newer fluoroquinolones. Clin. Infect. Dis. 31, S24–S28. doi: 10.1086/314056
Hopwood, D. A. (2007). Streptomyces in Nature and Medicine: the Antibiotic Makers. Oxford: Oxford University Press.
Hsu, S.-H., Shen, M.-W., Chen, J.-C., Lur, H.-S., and Liu, C.-T. (2021). The photosynthetic bacterium Rhodopseudomonas palustris strain ps3 exerts plant growth-promoting effects by stimulating nitrogen uptake and elevating auxin levels in expanding leaves. Front. Plant Sci. 12:573634. doi: 10.3389/fpls.2021.573634
Hsu-Kim, H., Eckley, C. S., Achá, D., Feng, X., Gilmour, C. C., Jonsson, S., et al. (2018). Challenges and opportunities for managing aquatic mercury pollution in altered landscapes. Ambio 47, 141–169. doi: 10.1007/s13280-017-1006-7
Hu, H., Li, H., Hao, M., Ren, Y., Zhang, M., and Liu, R. (2021). Nitrogen fixation and crop productivity enhancements co-driven by intercrop root exudates and key rhizosphere bacteria. J. Appl. Ecol. 58, 2243–2255. doi: 10.1111/1365-2664.13964
Hu, P., Brodie, E. L., Suzuki, Y., McAdams, H. H., and Andersen, G. L. (2005). Whole-genome transcriptional analysis of heavy metal stresses in Caulobacter crescentus. J. Bacteriol. 187, 8437–8449. doi: 10.1128/JB.187.24.8437-8449.2005
Imron, M. F., Kurniawan, S. B., and Abdullah, S. R. S. (2021). Resistance of bacteria isolated from leachate to heavy metals and the removal of Hg by Pseudomonas aeruginosa strain FZ-2 at different salinity levels in a batch biosorption system. Sustain. Environ. Res. 31:14.
Ishihara, S., Bitner, J. J., Farley, G. H., and Gillock, E. T. (2013). Vancomycin-resistant gram-positive cocci isolated from the saliva of wild songbirds. Curr. Microbiol. 66, 337–343. doi: 10.1007/s00284-012-0278-1
Jaiswal, A. K., Elad, Y., Paudel, I., Graber, E. R., Cytryn, E., and Frenkel, O. (2017). Linking the belowground microbial composition, diversity and activity to soilborne disease suppression and growth promotion of tomato amended with biochar. Sci. Rep. 7:44382. doi: 10.1038/srep44382
Jan, Z., Ali, S., Sultan, T., Khan, M. J., Shah, Z., and Khan, F. (2018). Impact of different strains of cyanobacteria on rice crop growth and nutrients uptake under saline soil condition. Sarhad J. Agric. 34, 450–458.
Jauregi, L., Epelde, L., Alkorta, I., and Garbisu, C. (2021). Antibiotic resistance in agricultural soil and crops associated to the application of cow manure-derived amendments from conventional and organic livestock farms. Front. Vet. Sci. 8:633858. doi: 10.3389/fvets.2021.633858
Kalam, S., Basu, A., Ahmad, I., Sayyed, R., El Enshasy, H. A., Dailin, D. J., et al. (2020). Recent understanding of soil Acidobacteria and their ecological significance: a critical review. Front. Microbiol. 11:580024. doi: 10.3389/fmicb.2020.580024
Karray, F., Gargouri, M., Chebaane, A., Mhiri, N., Mliki, A., and Sayadi, S. (2020). Climatic aridity gradient modulates the diversity of the rhizosphere and endosphere bacterial microbiomes of Opuntia ficus-indica. Front. Microbiol. 11:1622. doi: 10.3389/fmicb.2020.01622
Kaur, I., Gaur, V. K., Regar, R. K., Roy, A., Srivastava, P. K., Gaur, R., et al. (2021). Plants exert beneficial influence on soil microbiome in a HCH contaminated soil revealing advantage of microbe-assisted plant-based HCH remediation of a dumpsite. Chemosphere 280:130690. doi: 10.1016/j.chemosphere.2021.130690
Kaye, K. S., Harris, A. D., Gold, H., and Carmeli, Y. (2000). Risk factors for recovery of ampicillin-sulbactam-resistant Escherichia coli in hospitalized patients. Antimicrob. Agents Chemother. 44, 1004–1009. doi: 10.1128/AAC.44.4.1004-1009.2000
Kielak, A. M., Barreto, C. C., Kowalchuk, G. A., van Veen, J. A., and Kuramae, E. E. (2016). The ecology of Acidobacteria: moving beyond genes and genomes. Front. Microbiol. 7:744. doi: 10.3389/fmicb.2016.00744
Kingston, A. W., Zhao, H., Cook, G. M., and Helmann, J. D. (2014). Accumulation of heptaprenyl diphosphate sensitizes Bacillus subtilis to bacitracin: implications for the mechanism of resistance mediated by the BceAB transporter. Mol. Microbiol. 93, 37–49. doi: 10.1111/mmi.12637
Köhler, T., Epp, S. F., Curty, L. K., and Pechère, J.-C. (1999). Characterization of MexT, the regulator of the MexE-MexF-OprN multidrug efflux system of Pseudomonas aeruginosa. J. Bacteriol. 181, 6300–6305. doi: 10.1128/JB.181.20.6300-6305.1999
Kokou, F., Sasson, G., Nitzan, T., Doron-Faigenboim, A., Harpaz, S., Cnaani, A., et al. (2018). Host genetic selection for cold tolerance shapes microbiome composition and modulates its response to temperature. eLife 7:e36398. doi: 10.7554/eLife.36398
Kou, S., Vincent, G., Gonzalez, E., Pitre, F. E., Labrecque, M., and Brereton, N. J. (2018). The response of a 16S ribosomal RNA gene fragment amplified community to lead, zinc, and copper pollution in a Shanghai field trial. Front. Microbiol. 9:366. doi: 10.3389/fmicb.2018.00366
Ku, Y.-H., Lee, M.-F., Chuang, Y.-C., and Yu, W.-L. (2019). Detection of plasmid-mediated β-lactamase genes and emergence of a novel AmpC (CMH-1) in Enterobacter cloacae at a medical Center in Southern Taiwan. J. Clin. Med. 8:8. doi: 10.3390/jcm8010008
Kumar, V., AlMomin, S., Al-Aqeel, H., Al-Salameen, F., Nair, S., and Shajan, A. (2018). Metagenomic analysis of rhizosphere microflora of oil-contaminated soil planted with barley and alfalfa. PLoS One 13:e0202127. doi: 10.1371/journal.pone.0202127
Lee, J.-C., and Whang, K.-S. (2020). Agromyces humi sp. nov., actinobacterium isolated from farm soil. Int. J. Syst. Evol. Microbiol. 70, 5032–5039. doi: 10.1099/ijsem.0.004376
Lee, S. A., Heo, J., Kim, M. A., Tamura, T., Saitou, S., Kwon, S.-W., et al. (2021). Protaetiibacter larvae sp. nov. and Agromyces intestinalis sp. nov., isolated from the gut of larvae of Protaetia brevitarsis seulensis, reclassification of Lysinimonas yzui as Pseudolysinimonas yzui comb. nov. and emended description of the genus Pseudolysinimonas. Int. J. Syst. Evol. Microbiol. 71:004669. doi: 10.1099/ijsem.0.004669
Li, H.-Y., Wang, H., Wang, H.-T., Xin, P.-Y., Xu, X.-H., Ma, Y., et al. (2018). The chemodiversity of paddy soil dissolved organic matter correlates with microbial community at continental scales. Microbiome 6:187.
Li, L.-G., Huang, Q., Yin, X., and Zhang, T. (2020). Source tracking of antibiotic resistance genes in the environment—challenges, progress, and prospects. Water Res. 185:116127. doi: 10.1016/j.watres.2020.116127
Li, R., Zhu, H., Ruan, J., Qian, W., Fang, X., Shi, Z., et al. (2010). De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 20, 265–272. doi: 10.1101/gr.097261.109
Liao, H., Li, X., Yang, Q., Bai, Y., Cui, P., Wen, C., et al. (2021). Herbicide selection promotes antibiotic resistance in soil microbiomes. Mol. Biol. Evol. 38, 2337–2350. doi: 10.1093/molbev/msab029
Lin, J., Michel, L. O., and Zhang, Q. (2002). CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob. Agents Chemother. 46, 2124–2131. doi: 10.1128/AAC.46.7.2124-2131.2002
Liu, Y.-R., Delgado-Baquerizo, M., Bi, L., Zhu, J., and He, J.-Z. (2018). Consistent responses of soil microbial taxonomic and functional attributes to mercury pollution across China. Microbiome 6:183. doi: 10.1186/s40168-018-0572-7
Ma, Y., Prasad, M., Rajkumar, M., and Freitas, H. (2011). Plant growth promoting rhizobacteria and endophytes accelerate phytoremediation of metalliferous soils. Biotechnol. Adv. 29, 248–258. doi: 10.1016/j.biotechadv.2010.12.001
Ma, Z., Xu, C., Zhang, X., Wang, D., Pan, X., Liu, H., et al. (2021). A MexR mutation which confers aztreonam resistance to Pseudomonas aeruginosa. Front. Microbiol. 12:659808. doi: 10.3389/fmicb.2021.659808
Malisorn, K., Kanchanasin, P., Phongsopitanun, W., and Tanasupawat, S. (2018). Actinomadura rhizosphaerae sp. nov., isolated from rhizosphere soil of the plant Azadirachta indica. Int. J. Syst. Evol. Microbiol. 68, 3012–3016. doi: 10.1099/ijsem.0.002940
Martineau, C., Villeneuve, C., Mauffrey, F., and Villemur, R. (2014). Complete genome sequence of Hyphomicrobium nitrativorans strain NL23, a denitrifying bacterium isolated from biofilm of a methanol-fed denitrification system treating seawater at the Montreal Biodome. Genome Announc. 2:e01165-13. doi: 10.1128/genomeA.01165-13
Marumoto, M., Sakamoto, M., Marumoto, K., Tsuruta, S., and Komohara, Y. (2020). Mercury and selenium localization in the cerebrum, cerebellum, liver, and kidney of a minamata disease case. Acta Histochem. Cytochem. 53, 147–155. doi: 10.1267/ahc.20-00009
Mascher, T., Zimmer, S. L., Smith, T.-A., and Helmann, J. D. (2004). Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob. Agents Chemother. 48, 2888–2896. doi: 10.1128/AAC.48.8.2888-2896.2004
Mazhar, S. H., Li, X., Rashid, A., Su, J., Xu, J., Brejnrod, A. D., et al. (2021). Co-selection of antibiotic resistance genes, and mobile genetic elements in the presence of heavy metals in poultry farm environments. Sci. Total Environ. 755:142702. doi: 10.1016/j.scitotenv.2020.142702
McCully, A. L., Behringer, M. G., Gliessman, J. R., Pilipenko, E. V., Mazny, J. L., Lynch, M., et al. (2018). An Escherichia coli nitrogen starvation response is important for mutualistic coexistence with Rhodopseudomonas palustris. Appl. Environ. Microbiol. 84:e00404-18. doi: 10.1128/AEM.00404-18
McDaniel, E. A., Peterson, B. D., Stevens, S. L., Tran, P. Q., Anantharaman, K., and McMahon, K. D. (2020). Expanded phylogenetic diversity and metabolic flexibility of mercury-methylating microorganisms. mSystems 5:e00299-20. doi: 10.1128/mSystems.00299-20
Mehetre, G. T., Zothanpuia, Deka, P., Carrie, W., and Lalrokimi, and Singh, B. P. (2022). “Chapter6 - Thermophilic and thermotolerant cyanobacteria: environmental and biotechnological perspectives,” in Cyanobacterial Lifestyle and its Applications in Biotechnology, eds P. Singh, M. Fillat, and A. Kumar (Cambridge, MA: Academic Press), 159–178. doi: 10.1016/B978-0-323-90634-0.00014-7
Menéndez, E., Pérez-Yépez, J., Hernández, M., Rodríguez-Pérez, A., Velázquez, E., and León-Barrios, M. (2020). Plant growth promotion abilities of phylogenetically diverse Mesorhizobium strains: effect in the root colonization and development of tomato seedlings. Microorganisms 8:412. doi: 10.3390/microorganisms8030412
Millán, R., Carpena, R., Schmid, T., Sierra, M., Moreno, E., Peñalosa, J., et al. (2007). Rehabilitación de suelos contaminados con mercurio: estrategias aplicables en el área de Almadén. Rev. Ecosistemas 16, 56–66.
Mishra, B. S., Sharma, M., and Laxmi, A. (2021). Role of sugar and auxin crosstalk in plant growth and development. Physiol. Plant [Online ahead of print]. doi: 10.1111/ppl.13546
Montero-Calasanz, M., del, C., Meier-Kolthoff, J. P., Zhang, D.-F., Yaramis, A., Rohde, M., et al. (2017). Genome-scale data call for a taxonomic rearrangement of Geodermatophilaceae. Front. Microbiol. 8:2501. doi: 10.3389/fmicb.2017.02501
Moon, K., Jeon, J. H., Kang, I., Park, K. S., Lee, K., Cha, C.-J., et al. (2020). Freshwater viral metagenome reveals novel and functional phage-borne antibiotic resistance genes. Microbiome 8:75. doi: 10.1186/s40168-020-00863-4
Moulin, L., Munive, A., Dreyfus, B., and Boivin-Masson, C. (2001). Nodulation of legumes by members of the β-subclass of Proteobacteria. Nature 411, 948–950. doi: 10.1038/35082070
Muehe, E. M., Weigold, P., Adaktylou, I. J., Planer-Friedrich, B., Kraemer, U., Kappler, A., et al. (2015). Rhizosphere microbial community composition affects cadmium and zinc uptake by the metal-hyperaccumulating plant Arabidopsis halleri. Appl. Environ. Microbiol. 81, 2173–2181. doi: 10.1128/AEM.03359-14
Nascimento, F. X., Glick, B. R., and Rossi, M. J. (2021). Multiple plant hormone catabolism activities: an adaptation to a plant-associated lifestyle by Achromobacter spp. Environ. Microbiol. Rep. 13, 533–539. doi: 10.1111/1758-2229.12987
Nazli, F., Wang, X., Ahmad, M., Hussain, A., Dar, A., Nasim, M., et al. (2021). Efficacy of indole acetic acid and exopolysaccharides-producing Bacillus safensis strain FN13 for inducing Cd-stress tolerance and plant growth promotion in Brassica juncea (L.). Appl. Sci. 11:4160.
Nelkner, J., Henke, C., Lin, T. W., Pätzold, W., Hassa, J., Jaenicke, S., et al. (2019). Effect of long-term farming practices on agricultural soil microbiome members represented by metagenomically assembled genomes (MAGs) and their predicted plant-beneficial genes. Genes 10:424. doi: 10.3390/genes10060424
Nurk, S., Meleshko, D., Korobeynikov, A., and Pevzner, P. A. (2017). metaSPAdes: a new versatile metagenomic assembler. Genome Res. 27, 824–834. doi: 10.1101/gr.213959.116
Overbeek, R., Disz, T., and Stevens, R. (2004). The SEED: a peer-to-peer environment for genome annotation. Commun. ACM 47, 46–51. doi: 10.1145/1029496.1029525
Pereira, N. C. M., Galindo, F. S., Gazola, R. P. D., Dupas, E., Rosa, P. A. L., and Mortinho, E. S. (2020). Corn yield and phosphorus use efficiency response to phosphorus rates associated with plant growth promoting bacteria. Front. Environ. Sci. 8:40. doi: 10.3389/fenvs.2020.00040
Petrus, A. K., Rutner, C., Liu, S., Wang, Y., and Wiatrowski, H. A. (2015). Mercury reduction and methyl mercury degradation by the soil bacterium Xanthobacter autotrophicus Py2. Appl. Environ. Microbiol. 81, 7833–7838. doi: 10.1128/AEM.01982-15
Philippon, A., Slama, P., Dény, P., and Labia, R. (2016). A structure-based classification of class A β-lactamases, a broadly diverse family of enzymes. Clin. Microbiol. Rev. 29, 29–57. doi: 10.1128/CMR.00019-15
Qureshi, N. K., Yin, S., and Boyle-Vavra, S. (2014). The role of the Staphylococcal VraTSR regulatory system on vancomycin resistance and vanA operon expression in vancomycin-resistant Staphylococcus aureus. PLoS One 9:e85873. doi: 10.1371/journal.pone.0085873
Ramage, G., Tunney, M. M., Patrick, S., Gorman, S. P., and Nixon, J. R. (2003). Formation of Propionibacterium acnes biofilms on orthopaedic biomaterials and their susceptibility to antimicrobials. Biomaterials 24, 3221–3227. doi: 10.1016/s0142-9612(03)00173-x
Rangel, W., de, M., de Oliveira Longatti, S. M., Ferreira, P. A., Bonaldi, D. S., Guimaraes, A. A., et al. (2017). Leguminosae native nodulating bacteria from a gold mine as-contaminated soil: multi-resistance to trace elements, and possible role in plant growth and mineral nutrition. Int. J. Phytoremediation 19, 925–936. doi: 10.1080/15226514.2017.1303812
Ravel, J., Schrempf, H., and Hill, R. T. (1998). Mercury resistance is encoded by transferable giant linear plasmids in two Chesapeake Bay Streptomyces strains. Appl. Environ. Microbiol. 64, 3383–3388. doi: 10.1128/AEM.64.9.3383-3388.1998
Rensing, K. L., Abdallah, H., Koek, A., Elmowalid, G. A., Vandenbroucke-Grauls, C. M., Al Naiemi, N., et al. (2019). Prevalence of plasmid-mediated AmpC in Enterobacteriaceae isolated from humans and from retail meat in Zagazig, Egypt. Antimicrob. Resist. Infect. Control 8:45. doi: 10.1186/s13756-019-0494-6
Rice, L. B., Carias, L. L., Hutton-Thomas, R., Sifaoui, F., Gutmann, L., and Rudin, S. D. (2001). Penicillin-binding protein 5 and expression of ampicillin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 45, 1480–1486. doi: 10.1128/AAC.45.5.1480-1486.2001
Riesenfeld, C. S., Schloss, P. D., and Handelsman, J. (2004). Metagenomics: genomic analysis of microbial communities. Annu. Rev. Genet. 38, 525–552. doi: 10.1146/annurev.genet.38.072902.091216
Robas, M., Jiménez, P. A., González, D., and Probanza, A. (2021a). Bio-mercury remediation suitability index: a novel proposal that compiles the PGPR features of bacterial strains and its potential use in phytoremediation. Int. J. Environ. Res. Public Health 18:4213. doi: 10.3390/ijerph18084213
Robas, M., Probanza, A., González, D., and Jiménez, P. A. (2021b). Mercury and antibiotic resistance co-selection in Bacillus sp. isolates from the Almadén mining district. Int. J. Environ. Res. Public Health 18:8304. doi: 10.3390/ijerph18168304
Rosenfeld, C. E., James Bruce, R., Santelli Cara, M., and Löffler Frank, E. (2018). Persistent bacterial and fungal community shifts exhibited in selenium-contaminated reclaimed mine soils. Appl. Environ. Microbiol. 84:e01394-18. doi: 10.1128/AEM.01394-18
Segata, N., Waldron, L., Ballarini, A., Narasimhan, V., Jousson, O., and Huttenhower, C. (2012). Metagenomic microbial community profiling using unique clade-specific marker genes. Nat. Methods 9, 811–814. doi: 10.1038/nmeth.2066
Senges, C. H., Al-Dilaimi, A., Marchbank, D. H., Wibberg, D., Winkler, A., Haltli, B., et al. (2018). The secreted metabolome of Streptomyces chartreusis and implications for bacterial chemistry. Proc. Natl. Acad. Sci. U.S.A. 115, 2490–2495. doi: 10.1073/pnas.1715713115
Shan, W., Zhou, Y., Liu, H., and Yu, X. (2018). Endophytic actinomycetes from tea plants (Camellia sinensis): isolation, abundance, antimicrobial, and plant-growth-promoting activities. Biomed Res. Int. 2018:1470305. doi: 10.1155/2018/1470305
Silva, G. G. Z., Green, K. T., Dutilh, B. E., and Edwards, R. A. (2016). SUPER-FOCUS: a tool for agile functional analysis of shotgun metagenomic data. Bioinformatics 32, 354–361. doi: 10.1093/bioinformatics/btv584
Silveira, M. C., Azevedo da Silva, R., Faria da Mota, F., Catanho, M., Jardim, R., Guimarães, A. C. R., et al. (2018). Systematic identification and classification of β-lactamases based on sequence similarity criteria: β-lactamase annotation. Evol. Bioinform. 14:1176934318797351. doi: 10.1177/1176934318797351
Sivertsen, A., Pedersen, T., Larssen, K. W., Bergh, K., Rønning, T. G., Radtke, A., et al. (2016). A silenced vanA gene cluster on a transferable plasmid caused an outbreak of vancomycin-variable enterococci. Antimicrob. Agents Chemother. 60, 4119–4127. doi: 10.1128/AAC.00286-16
Stange, C., and Tiehm, A. (2020). Occurrence of antibiotic resistance genes and microbial source tracking markers in the water of a karst spring in Germany. Sci. Total Environ. 742:140529. doi: 10.1016/j.scitotenv.2020.140529
Stogios, P. J., and Savchenko, A. (2020). Molecular mechanisms of vancomycin resistance. Protein Sci. 29, 654–669.
Stopnisek, N., Zühlke, D., Carlier, A., Barberán, A., Fierer, N., Becher, D., et al. (2016). Molecular mechanisms underlying the close association between soil Burkholderia and fungi. ISME J. 10, 253–264. doi: 10.1038/ismej.2015.73
Suleiman, M., Quoreshi, A., Bhat, N., Manuvel, A., and Sivadasan, M. (2019). Divulging diazotrophic bacterial community structure in Kuwait desert ecosystems and their N2-fixation potential. PLoS One 14:e0220679. doi: 10.1371/journal.pone.0220679
Suleman, M., Yasmin, S., Rasul, M., Yahya, M., Atta, B. M., and Mirza, M. S. (2018). Phosphate solubilizing bacteria with glucose dehydrogenase gene for phosphorus uptake and beneficial effects on wheat. PLoS One 13:e0204408. doi: 10.1371/journal.pone.0204408
Teichmann, L., Kümmel, H., Warmbold, B., and Bremer, E. (2018). OpuF, a new Bacillus compatible solute ABC transporter with a substrate-binding protein fused to the transmembrane domain. Appl. Environ. Microbiol. 84:e01728-18. doi: 10.1128/AEM.01728-18
Teijeiro, R. G., Belimov, A. A., and Dodd, I. C. (2020). Microbial inoculum development for ameliorating crop drought stress: a case study of Variovorax paradoxus 5C-2. New Biotechnol. 56, 103–113. doi: 10.1016/j.nbt.2019.12.006
Thomas, C., and Tampé, R. (2020). Structural and mechanistic principles of ABC transporters. Annu. Rev. Biochem. 89, 605–636. doi: 10.1146/annurev-biochem-011520-105201
Thomas, F., Corre, E., and Cébron, A. (2019). Stable isotope probing and metagenomics highlight the effect of plants on uncultured phenanthrene-degrading bacterial consortium in polluted soil. ISME J. 13, 1814–1830. doi: 10.1038/s41396-019-0394-z
Torres-Bacete, J., Hormigo, D., Stuart, M., Arroyo, M., Torres, P., Castillón, M. P., et al. (2007). Newly discovered penicillin acylase activity of aculeacin A acylase from Actinoplanes utahensis. Appl. Environ. Microbiol. 73, 5378–5381. doi: 10.1128/AEM.00452-07
Toukabri, W., Ferchichi, N., Hlel, D., Jadlaoui, M., Kheriji, O., Mhamdi, R., et al. (2021). Response of intercropped barley and fenugreek to mono-and co-inoculation with Sinorhizobium meliloti F42 and Variovorax paradoxus F310 under contrasting agroclimatic regions. Arch. Microbiol. 203, 1657–1670. doi: 10.1007/s00203-020-02180-8
Truong, D. T., Franzosa, E. A., Tickle, T. L., Scholz, M., Weingart, G., Pasolli, E., et al. (2015). MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat. Methods 12, 902–903. doi: 10.1038/nmeth.3589
Tyson, G. W., Chapman, J., Hugenholtz, P., Allen, E. E., Ram, R. J., Richardson, P. M., et al. (2004). Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428, 37–43. doi: 10.1038/nature02340
US Environmental Protection Agency (2011). 2010 Biennial National Listing of Fish Advisories. Washington, DC: United States Environmental Protection Agency.
Valencia, A. O., Braz, V. S., Magalhães, M., and Galhardo, R. S. (2020). Role of error-prone DNA polymerases in spontaneous mutagenesis in Caulobacter crescentus. Genet. Mol. Biol. 43:e20180283. doi: 10.1590/1678-4685-GMB-2018-0283
Van Goethem, M. W., Pierneef, R., Bezuidt, O. K., Van De Peer, Y., Cowan, D. A., and Makhalanyane, T. P. (2018). A reservoir of ‘historical’ antibiotic resistance genes in remote pristine Antarctic soils. Microbiome 6:40. doi: 10.1186/s40168-018-0424-5
Vishnivetskaya, T. A., Mosher, J. J., Palumbo, A. V., Yang, Z. K., Podar, M., Brown, S. D., et al. (2011). Mercury and other heavy metals influence bacterial community structure in contaminated Tennessee streams. Appl. Environ. Microbiol. 77, 302–311. doi: 10.1128/AEM.01715-10
Von Canstein, H., Li, Y., Timmis, K., Deckwer, W.-D., and Wagner-Dobler, I. (1999). Removal of mercury from chloralkali electrolysis wastewater by a mercury-resistant Pseudomonas putida strain. Appl. Environ. Microbiol. 65, 5279–5284. doi: 10.1128/AEM.65.12.5279-5284.1999
Vurukonda, S. S. K. P., Giovanardi, D., and Stefani, E. (2018). Plant growth promoting and biocontrol activity of Streptomyces spp. as endophytes. Int. J. Mol. Sci. 19:952. doi: 10.3390/ijms19040952
Wang, F., Fu, Y.-H., Sheng, H.-J., Topp, E., Jiang, X., Zhu, Y.-G., et al. (2021). Antibiotic resistance in the soil ecosystem: a one health perspective. Curr. Opin. Environ. Sci. Health 20:100230. doi: 10.1016/j.coesh.2021.100230
Wang, Y., Brown, H., Crowley, D., and Szaniszlo, P. (1993). Evidence for direct utilization of a siderophore, ferrioxamine B, in axenically grown cucumber. Plant Cell Environ. 16, 579–585. doi: 10.1111/j.1365-3040.1993.tb00906.x
Wang, Y., Qin, Y., Kot, W., Zhang, F., Zheng, S., Wang, G., et al. (2016). Genome sequence of selenium-solubilizing bacterium Caulobacter vibrioides T5M6. Genome Announc. 4:e01721-15. doi: 10.1128/genomeA.01721-15
Wang, Z., Solanki, M. K., Yu, Z.-X., Yang, L.-T., An, Q.-L., Dong, D.-F., et al. (2019). Draft genome analysis offers insights into the mechanism by which Streptomyces chartreusis WZS021 increases drought tolerance in sugarcane. Front. Microbiol. 9:3262. doi: 10.3389/fmicb.2018.03262
Warmbold, B., Ronzheimer, S., Freibert, S.-A., Seubert, A., Hoffmann, T., and Bremer, E. (2020). Two MarR-type repressors balance precursor uptake and glycine betaine synthesis in Bacillus subtilis to provide cytoprotection against sustained osmotic stress. Front. Microbiol. 11:1700. doi: 10.3389/fmicb.2020.01700
Westmann, C. A., Alves, L., de, F., Silva-Rocha, R., and Guazzaroni, M.-E. (2018). Mining novel constitutive promoter elements in soil metagenomic libraries in Escherichia coli. Front. Microbiol. 9:1344. doi: 10.3389/fmicb.2018.01344
Wong, W.-T., Tseng, C.-H., Hsu, S.-H., Lur, H.-S., Mo, C.-W., Huang, C.-N., et al. (2014). Promoting effects of a single Rhodopseudomonas palustris inoculant on plant growth by Brassica rapa chinensis under low fertilizer input. Microbes Environ. 29, 303–313. doi: 10.1264/jsme2.me14056
Woo, H. L., DeAngelis, K. M., Teshima, H., Davenport, K., Daligault, H., Erkkila, T., et al. (2017). High-quality draft genome sequences of four lignocellulose-degrading bacteria isolated from Puerto Rican forest soil: Gordonia sp., Paenibacillus sp., Variovorax sp., and Vogesella sp. Genome Announc. 5:e00300-17. doi: 10.1128/genomeA.00300-17
Xiao, X., Zhu, Y., Gao, Y., Fu, J., Zhao, Y., and Zhao, L. (2021). Inoculation of paddy soils with Rhodopseudomonas palustris enhanced heavy metal immobilisation. Plant Soil Environ. 67, 55–60.
Xu, J., Zhang, Y., Zhang, P., Trivedi, P., Riera, N., Wang, Y., et al. (2018). The structure and function of the global citrus rhizosphere microbiome. Nat. Commun. 9:4894. doi: 10.1038/s41467-018-07343-2
Xu, L., Zeng, X.-C., Nie, Y., Luo, X., Zhou, E., Zhou, L., et al. (2014). Pontibacter diazotrophicus sp. nov., a novel nitrogen-fixing bacterium of the family Cytophagaceae. PLoS One 9:e92294. doi: 10.1371/journal.pone.0092294
Yamamoto, K., Shiwa, Y., Ishige, T., Sakamoto, H., Tanaka, K., Uchino, M., et al. (2018). Bacterial diversity associated with the rhizosphere and endosphere of two halophytes: Glaux maritima and Salicornia europaea. Front. Microbiol. 9:2878. doi: 10.3389/fmicb.2018.02878
Yan, C., Wang, F., Liu, H., Liu, H., Pu, S., Lin, F., et al. (2020). Deciphering the toxic effects of metals in gold mining area: microbial community tolerance mechanism and change of antibiotic resistance genes. Environ. Res. 189:109869. doi: 10.1016/j.envres.2020.109869
Yan, M., Sahin, O., Lin, J., and Zhang, Q. (2006). Role of the CmeABC efflux pump in the emergence of fluoroquinolone-resistant Campylobacter under selection pressure. J. Antimicrob. Chemother. 58, 1154–1159. doi: 10.1093/jac/dkl412
Yang, E., Sun, L., Ding, X., Sun, D., Liu, J., and Wang, W. (2019). Complete genome sequence of Caulobacter flavus RHGG3 T, a type species of the genus Caulobacter with plant growth-promoting traits and heavy metal resistance. 3 Biotech 9:42. doi: 10.1007/s13205-019-1569-z
Yasmeen, T., Aziz, A., Tariq, M., Arif, M. S., Shahzad, S. M., Riaz, M., et al. (2021). “Pseudomonas as plant growth-promoting bacteria and its role in alleviation of abiotic stress,” in Plant Growth-Promoting Microbes for Sustainable Biotic and Abiotic Stress Management, eds H. I. Mohamed, H. D. Saad El-Beltagi, and K. A. Abd-Elsalam (Berlin: Springer), 157–185. doi: 10.1007/978-3-030-66587-6_7
Yeager, C. M., Gallegos-Graves, L. V., Dunbar, J., Hesse, C. N., Daligault, H., and Kuske, C. R. (2017). Polysaccharide degradation capability of Actinomycetales soil isolates from a semiarid grassland of the Colorado Plateau. Appl. Environ. Microbiol. 83:e03020-16. doi: 10.1128/AEM.03020-16
Yin, J., Ding, M., Zha, F., Zhang, J., Meng, Q., and Yu, Z. (2021). Stringent starvation protein regulates prodiginine biosynthesis via affecting siderophore production in Pseudoalteromonas sp. strain R3. Appl. Environ. Microbiol. 87:e02949-20. doi: 10.1128/AEM.02949-20
Youngblut, N. D., De la Cuesta-Zuluaga, J., Reischer, G. H., Dauser, S., Schuster, N., Walzer, C., et al. (2020). Large-scale metagenome assembly reveals novel animal-associated microbial genomes, biosynthetic gene clusters, and other genetic diversity. mSystems 5:e01045-20. doi: 10.1128/mSystems.01045-20
Yun, B.-R., Malik, A., and Kim, S. B. (2020). Genome based characterization of Kitasatospora sp. MMS16-BH015, a multiple heavy metal resistant soil actinobacterium with high antimicrobial potential. Gene 733:144379. doi: 10.1016/j.gene.2020.144379
Zai, X., Luo, W., Bai, W., Li, Y., Xiao, X., Gao, X., et al. (2021). Effect of root diameter on the selection and network interactions of root-associated bacterial microbiomes in Robinia pseudoacacia L. Microb. Ecol. 82, 391–402. doi: 10.1007/s00248-020-01678-4
Zainab, N., Din, B. U., Javed, M. T., Afridi, M. S., Mukhtar, T., Kamran, M. A., et al. (2020). Deciphering metal toxicity responses of flax (Linum usitatissimum L.) with exopolysaccharide and ACC-deaminase producing bacteria in industrially contaminated soils. Plant Physiol. Biochem. 152, 90–99. doi: 10.1016/j.plaphy.2020.04.039
Zhang, S.-S., Xu, J.-F., Sun, X.-L., Guo, W., and Liu, Z.-S. (2021). Cellulomonas taurus sp. nov., a novel bacteria with multiple hydrolase activity isolated from livestock, and potential application in wastewater treatment. Antonie Van Leeuwenhoek 114, 527–538. doi: 10.1007/s10482-021-01538-2
Zhao, K., Li, J., Zhang, X., Chen, Q., Liu, M., Ao, X., et al. (2018). Actinobacteria associated with Glycyrrhiza inflata Bat. are diverse and have plant growth promoting and antimicrobial activity. Sci. Rep. 8:13661. doi: 10.1038/s41598-018-32097-8
Zhao, M. M., Kou, J., Chen, Y., Xue, L., Fan, T. T., and Wang, S. (2021). Bioremediation of wastewater containing mercury using three newly isolated bacterial strains. J. Clean. Prod. 299:126869.
Zhao, Y., Tang, H., and Ye, Y. (2012). RAPSearch2: a fast and memory-efficient protein similarity search tool for next-generation sequencing data. Bioinformatics 28, 125–126. doi: 10.1093/bioinformatics/btr595
Zheng, B., Jiang, X., Xu, Z., Fang, Y., and Li, L. (2016). Characterization of a novel metallo-β-lactamases fold hydrolase from Pelagibacterium halotolerans, a marine halotolerant bacterium isolated from East China Sea. Extremophiles 20, 37–44. doi: 10.1007/s00792-015-0795-5
Zheng, Y.-X., Yang, M., Wu, J.-G., Wang, J.-M., Xu, Y.-L., Yan, D., et al. (2021). Study on interaction between bacterial community and microecological environment in rhizosphere soil of tobacco root rot caused by Fusarium solani. Mater. Express 11, 166–173.
Zhou, L., Wang, L., Tian, P., Bao, T., Li, L., and Zhao, X. (2019). The LiaFSR and BsrXRS systems contribute to bile salt resistance in Enterococcus faecium isolates. Front. Microbiol. 10:1048. doi: 10.3389/fmicb.2019.01048
Keywords: PGPR, antibiotics, bioremediation, shotgun metagenomics, co-selection
Citation: González D, Robas M, Fernández V, Bárcena M, Probanza A and Jiménez PA (2022) Comparative Metagenomic Study of Rhizospheric and Bulk Mercury-Contaminated Soils in the Mining District of Almadén. Front. Microbiol. 13:797444. doi: 10.3389/fmicb.2022.797444
Received: 18 October 2021; Accepted: 17 January 2022;
Published: 07 March 2022.
Edited by:
Wei Zhang, Michigan State University, United StatesReviewed by:
Rakshak Kumar, Institute of Himalayan Bioresource Technology (CSIR), IndiaCopyright © 2022 González, Robas, Fernández, Bárcena, Probanza and Jiménez. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniel González, ZGFuaWVsLmdvbnphbGV6cmVndWVyb0BjZXUuZXM=
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.