- 1Department of Pharmaceutical Microbiology, Prof Dora Akunyili College of Pharmacy, Igbinedion University, Okada, Nigeria
- 2Institute of Medical Microbiology and Hygiene, Austrian Agency for Health and Food Safety, Vienna, Austria
There is a link between antibiotic resistance in humans, livestock and the environment. This study was carried out to characterize antibiotic resistant bovine and environmental Enterobacteriaceae isolates from Edo state, Nigeria. A total of 109 consecutive isolates of Enterobacteriaceae were isolated from March–May 2015 from 150 fecal samples of healthy bovine animals from three farms at slaughter in Edo state Nigeria. Similarly, 43 Enterobacteriaceae isolates were also obtained from a total of 100 environmental samples from different sources. Isolates were recovered and identified from samples using standard microbiological techniques. Recovered isolates were pre-identified by the Microbact Gram-Negative identification system and confirmed with Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry and ribosomal multilocus sequence typing (rMLST). Antibiotic susceptibility testing was carried out by Kirby-Bauer method for 14 antibiotics. Whole genome sequencing (WGS) was carried out for isolate characterization and identification of resistance determinants. Out of 109 animal and 43 environmental Enterobacteriaceae isolates, 18 (17%) and 8 (19%) isolates based on selection criteria showed antibiotic resistance and were further investigated by whole genome sequencing (WGS). Resistance genes were detected in all (100%) of the resistant bovine and environmental Enterobacteriaceae isolates. The resistance determinants included β-lactamase genes, aminoglycoside modifying enzymes, qnr genes, sulfonamide, tetracycline and trimethoprim resistance genes, respectively. Out of the 18 and 8 resistant animal and environmental isolates 3 (17%) and 2 (25%) were multidrug resistant (MDR) and had resistance determinants which included efflux genes, regulatory systems modulating antibiotic efflux and antibiotic target alteration genes. Our study shows the dissemination of antibiotic resistance especially MDR strains among Nigerian bovine and environmental Enterobacteriaceae isolates. The presence of these resistant strains in animals and the environment constitute a serious health concern indicated by the difficult treatment options of the infections caused by these organisms. To the best of our knowledge we report the first detailed genomic characterization of antibiotic resistance in bovine and environmental Enterobacteriaceae isolates for Nigeria.
Introduction
Members of the family Enterobacteriaceae are important pathogens of humans and animals causing various infections for example in humans which include septicemia, pneumonia, peritonitis etc. (Makharita et al., 2020). They have been reported to be the main causes of nosocomial infections (Centers for Disease Control and Prevention [CDC], 2013). In animals, members of the family Enterobacteriaceae can be divided into three groups depending on their pathogenicity. They include major animal pathogens like Escherichia coli, opportunistic pathogens that occasionally cause infections in animals like Proteus spp., Enterobacter spp., Citrobacter spp. and organisms of uncertain importance for animals, e.g., Erwina spp., Leclercia adecarboxylata (Veterian Key, 2016). Several virulence factors involved in disease occurrence in members of the Enterobacteriaceae family have been previously studied which include invasion, hemolysins, siderophores and toxins which are controlled by specific virulence genes (Pearce et al., 2006).
Antibiotic resistance is a serious health concern not only among human pathogens but also in isolates found in other habitats. Many resistant pathogenic bacteria and commensals are found in different hosts, or in the environment (Taylor et al., 2001). The connections between humans, animals, and their environment allow the transfer of bacteria and mobile genetic elements between different compartments (Woolhouse and Ward, 2013). Antibiotic resistance genes are emerging environmental contaminants, as they can be transferred to other bacteria present in their environment (Krishnasamy, 2018). Multidrug resistance has increased all over the world which is considered a public health threat. Several recent investigations have reported the emergence of multidrug resistant bacterial pathogen from different origins including humans, birds, cattle, and fish (Algammal et al., 2020, 2021a,b). This increases the need for routine application of antimicrobial susceptibility testing to detect the antibiotic of choice as well as screening of the emerging MDR strains. There are different mechanisms of resistance in members of the family Enterobacteriaceae. They include modification of the target, efflux phenomenon, impermeability, and enzymatic inactivation (Kotsyuba et al., 2014). Understanding resistance mechanisms in Enterobacteriaceae including how they develop and are transmitted is important for developing ways to monitor resistance spread effectively and also have effective treatment options. The various resistance determinants can be exploited to produce possible safe and potent novel antimicrobial drugs with new mechanisms of action.
Livestock animals are linked to humans through the food chain and the environment they share (Marshall and Levy, 2011). Over the years, antimicrobials have been applied for treatment of diseases in food producing animals, but also for growth promotion and prophylactic purposes. Multiple countries including developed and developing nations like Nigeria have introduced bans on the non-therapeutic use of antimicrobials. However, the coordinated surveillance and monitoring of antimicrobial use and resistance especially in developing countries like Nigeria is still limited, resulting in an increase of resistant pathogens. Most of the main classes of antibiotics used in human medicine are represented in a list of antibiotics tagged as “critically important” for livestock animals by the World Organization for Animal Health (OIE, 2007). Antibiotic residues are released into the environment as a result of inadequate absorption and metabolic activities carried out by food animals. Antimicrobials added to food for livestock animals can be disseminated as a result of leaching and release by urine and feces (Krishnasamy, 2018). The presence of antibiotic resistance in Enterobacteriaceae as well as other bacteria has been well documented in human, animal and environmental isolates in developed countries (Yates et al., 2004; Hoyle et al., 2005; Aarestrup et al., 2009; Fischer et al., 2012; Spoor et al., 2012). In low/middle income countries like Nigeria, research focus is majorly on antimicrobial resistance in human pathogens with only a few detailed reports on animal and environmental isolates (Adelowo and Fagade, 2009; Olowe et al., 2015; Ayeni et al., 2016; Ngbede et al., 2017; Adelowo et al., 2018; Shivakumaraswamy et al., 2019). The aim of this study was to investigate antibiotic resistance and characterize the resistance mechanism in Enterobacteriaceae isolated from bovine animals and the environment in Edo state Nigeria using whole genome sequencing.
Materials and Methods
Bacterial Isolation and Identification
A total of 109 consecutive isolates of Enterobacteriaceae were isolated from March–May 2015 from 150 fecal samples of healthy bovine animals at slaughter from three farms in Edo state Nigeria. The animals at slaughter were mostly Nigerian native cow breeds of both male and female sex. Further, 43 Enterobacteriaceae isolates were obtained from 100 environmental samples collected at six different sites in Edo state, Nigeria during the same period: 50 samples from refuse dump sites, 20 samples from flowing rivers, 20 samples from the soil and 10 samples from waste water. The environmental sampling sites were in close proximity with the animal farms. Isolates were identified using standard microbiological techniques (Cheesebrough, 2006). For the isolation and identification of the different members of Enterobacteriaceae, samples were inoculated on MacConkey agar plates (Oxoid, Hampshire, United Kingdom) and incubated for 24 h at 37°C. Distinct colonies were obtained from the agar plates and subcultured to obtain pure colonies. The Microbact Gram-Negative identification system (Oxoid, Basingstoke Hampshire, United Kingdom) was used in the preliminary identification of aerobic and facultative anaerobic Gram-negative bacteria (Enterobacteriaceae) (Mugg and Hill, 1981; Thomas, 1983). Organisms were identified based on pH change and substrate utilizations as established by previous reference methodologies (Cowan and Steel, 1977; Farmer 3rd et al., 1985; Balows et al., 1991). Preidentified isolates were further subcultured on Drigalski Lactose agar (Oxoid, Hampshire, United Kingdom) and subsequently confirmed by MALDI-TOF mass spectrometry (Bruker Daltonik GmbH, Bremen, Germany) analysis.
Antimicrobial Susceptibility Testing
The Kirby-Bauer susceptibility testing technique (Bauer et al., 1966) was performed and results were interpreted using European Committee on Antimicrobial Susceptibility Testing (EUCAST) criteria (European Committee on Antimicrobial Susceptibility Testing [EUCAST], 2015). The isolates were tested based on selection criteria with 14 antibiotics belonging to 11 classes of antimicrobial agents: carbapenems; meropenem (10 μg), ertapenem (10 μg), extended spectrum cephalosporins; ceftazidime (10 μg), cefotaxime (5 μg), cefepime (30 μg), penicillin + β-lactamase inhibitor; amoxicillin/clavulanic acid (30 μg), cephamycin; cefoxitin (30 μg), monobactam; aztreonam (30 μg), folate pathway inhibitors; trimethoprim (5 μg), fluoroquinolones; ciprofloxacin (5 μg), levofloxacin (5 μg), aminoglycosides; amikacin (30 μg), antipseudomonal penicillins + β-lactamase inhibitor; piperacillin/tazobactam (36 μg), phenicols; chloramphenicol (30 μg) (Oxoid, Basingstoke Hampshire, United Kingdom) on Mueller Hinton agar (Oxoid, Hampshire, United Kingdom) plates. These antibiotics are clinically important drugs both in the human and veterinary health sectors in Nigeria. Multidrug resistant isolates were identified among the tested isolates. Multidrug resistance was defined as non-susceptibility of an Enterobacteriaceae isolate to ≥1 agent of ≥3 antimicrobial classes (Magiorakos et al., 2012).
Whole Genome Sequencing
Whole genome sequencing (WGS) was carried out for 18 resistant/multidrug-resistant bovine animal isolates and 8 resistant/multidrug-resistant environmental isolates based on selection criteria. The MagAttract HMW DNA extraction kit (Qiagen, Hilden, Germany) was used for the extraction of genomic DNA (gDNA). Quantification of genomic DNA was carried out on a Qubit™ 2.0 Fluorometer using the dsDNA BR Assay kit (Invitrogen by Thermo Fisher Scientific, Waltham, MA, United States). Preparation of fragment libraries of the bacterial genomes was carried out using the Illumina Nextera XT DNA library preparation kit (Illumina Inc., San Diego, CA, United States). A paired end sequencing using a read length of 2 × 300 bp on an Illumina Miseq (Miseq v3.0, Illumina Inc., San Diego, CA, United States) was performed using Miseq reagent kit v3 containing the reagent cartridge and flow cell.
Raw reads (FASTQ files) were trimmed at their 5′ and 3′ ends until an average base quality of 30 was reached in a window of 20 bases, and Velvet version 1.1.04 (Zerbino, 2010) was used in carrying out the assembly using optimized k-mer size and coverage cutoff values based on the average contigs length with >1,000 bp. Species identification via MALDI-TOF MS was confirmed using ribosomal multilocus sequence typing (rMLST)1. Assembled genomes were uploaded to the ResFinder 2.1 web server2 (Zankari et al., 2012) and to the Comprehensive Antibiotic Resistance Database-Resistance Gene Identifier (CARD-RGI) (Jia et al., 2017) to identify antimicrobial resistance genes. ARGs were identified based on a minimum cutoff of 98% nucleotide identity for perfect or strict hits predicted by RGI. The Enterobase core genome multilocus sequence typing (cgMLST) scheme comprising 2,513 core genes3 was used for strain comparison using SeqSphere + version 7.7.5 (Ridom, Münster, Germany) as described recently (Bernreiter-Hofer et al., 2021; Cabal et al., 2021; Mišić et al., 2021). Multilocus sequence typing [MLST; MLST 1.8, Centre for Genomic Epidemiology (CGE), Lyngby DK] was used to type resistant isolates. Klebsiella variicola isolates were typed also by multilocus sequence typing [MLST Klebsiella variicola, Instituto Nacional de Salud Pública (INSP)4] (Barrios-Camacho et al., 2019). Plasmids on the draft genomes of the resistant Enterobacteriaceae isolates were analyzed and classified using PlasmidFinder 1.3 webtool5 based on a threshold of 95% ID (Carattoli et al., 2014). Plasmid replicons and Inc plasmid groups were identified.
Genomes From Sequence Read Archive
Three genomes of isolates in this study were downloaded from the Sequence Read Archive (SRA) Bioproject PRJEB20802 (Ayandiran et al., 2018).
Phylogenetic Analysis
Whole genome based cgMLST phylogenetic analysis including three genomes from Nigerian Escherichia coli poultry isolates retrieved from SRA with accession ERR1986373–ERR1986375 and all bovine Escherichia coli isolates (n = 12) in this study was carried out and a minimum spanning tree was calculated. The 15 isolates had eight sequence types (ST). In this study, Multilocus sequence typing (MLST) revealed seven STs (ST46, ST111, ST23, ST155, ST10, ST58, ST423) for Escherichia coli (n = 12) animal isolates (Supplementary Table 1).
Nucleotide Sequence Accession Numbers
This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession numbers JAIKTX000000000-JAIKUW000000000. The version described in this paper is version JAIKTX010000000-JAIKUW010000000.
Statistical Analyses
The correlation analyses between the tested antimicrobial agents as well as the phenotypic resistance pattern and the detected resistance genes were performed using SPSS statistical software package version 20.0 (IBM SPSS Inc., New York, NY, United States).
Results
Phenotypic Characteristics of Enterobacteriaceae in the Samples
The recovered Enterobacteriaceae isolates were pink (lactose fermenters) and colorless (non- lactose fermenters) colonies on MacConkey agar. Preliminary biochemical tests on the recovered isolates using the Microbact Gram-Negative identification system gave varying reactions to the different tests carried out. Biochemical tests included lysine decarboxylase, ornithine decarboxylase, H2S, Glucose, Mannitol, Xylose, ONPG, Indole, Urease, VP, Citrate and TDA. On Drigalski agar, Enterobacteriaceae isolates were yellow colonies (E. coli, Klebsiella, Enterobacter spp.) and blue-gray to blue-green colonies (Proteus, Serratia, Providencia spp.). Species identification using MALDI-TOF-MS assigned the 109 animal isolates to 9 species and the 43 environmental isolates to 10 species (Supplementary Tables 2, 3).
Antibiogram and Phenotypic Resistance Pattern of the Isolates
Out of the 109 animal and 43 environmental Enterobacteriaceae isolates, 18(17%) and 8(19%) isolates revealed antibiotic resistance in the Kirby-Bauer susceptibility testing technique (Supplementary Tables 4a,b). Species identification using MALDI-TOF-MS and ribosomal MLST assigned the 18 resistant animal isolates to 5 species and the 8 resistant environmental isolates to 7 species (Supplementary Table 5). Resistant animal isolates included Escherichia coli, Serratia marcescens, Klebsiella quasipneumoniae, Klebsiella variicola, Proteus terrae while the resistant environmental isolates included Enterobacter quasiroggenkampii, Enterobacter hormachei, Citrobacter koseri, Klebsiella quasipneumoniae, Klebsiella variicola, Proteus terrae, and Proteus faecis (Supplementary Table 5). The selected animal strains were resistant to trimethoprim (61%), cefoxitin (50%), amoxicillin-clavulanic acid (50%), while environmental strains were resistant to trimethoprim (75%) and cefoxitin (50%) (Supplementary Table 6).
Antimicrobial Resistance Genes of the Recovered Resistance Isolates
Whole genome sequencing revealed that all of the 18 resistant animal isolates harbored more than one resistance gene. Similarly, all the resistant environmental strains had multiple resistance genes detected by WGS.
The resistance determinants in the animal isolates included β-lactamase genes, blaOKP–B–1, blaOKP–B–7, blaTEM–1b, blaTEM–1c, blaLEN–16, blaLEN–17, blaAMPC1, blaAMPH; streptomycin resistance genes, strA and strB; aminoglycoside modifying enzymes, aadA1; fosfomycin resistance determinant, fosA, glpT; qnr genes, qnrD, qnrS1 and plasmid encoded efflux pump, oqxA; sulfonamide resistance genes, sul1, sul2, and sul3; tetracycline resistance gene, tet(A) and trimethoprim resistance genes, dfrA1, dfrA5, dfrA14. The resistance determinants in the environmental isolates included β-lactamase genes: blaOKP–B–2, blaACT–7, blaACT–25 blaCMG, blaCKO–1, blaTEM–1b, blaMAL–1, blaLEN–10, Sulfonamide resistance gene, sul2, phenicol resistance gene, catII, tetracycline resistance genes, tet(A), tet(D) and trimethoprim resistance gene, dfrA14. Other resistance determinants which included efflux genes emrA, emrB, emrK, emrY, msbA, regulatory systems modulating antibiotic efflux H-NS, marR, emrR, marA, CRP, antibiotic target alteration gene bacA, PmrF, eptA, EF-Tu mutants, ugd, were also detected in the antibiotic resistant animal and environmental isolates. Supplementary Table 7 shows the characteristics of the antibiotic resistant animal and environmental isolates. Figure 1 illustrates the distribution of the antimicrobial resistance genes among the recovered isolates. Ten plasmid incompatibility groups were identified among the animal Enterobacteriaceae isolates with IncF family types being predominant. Four plasmid incompatibility groups were identified among the antibiotic resistant environmental Enterobacteriaceae isolates with IncF family types also being predominant (Supplementary Table 1).
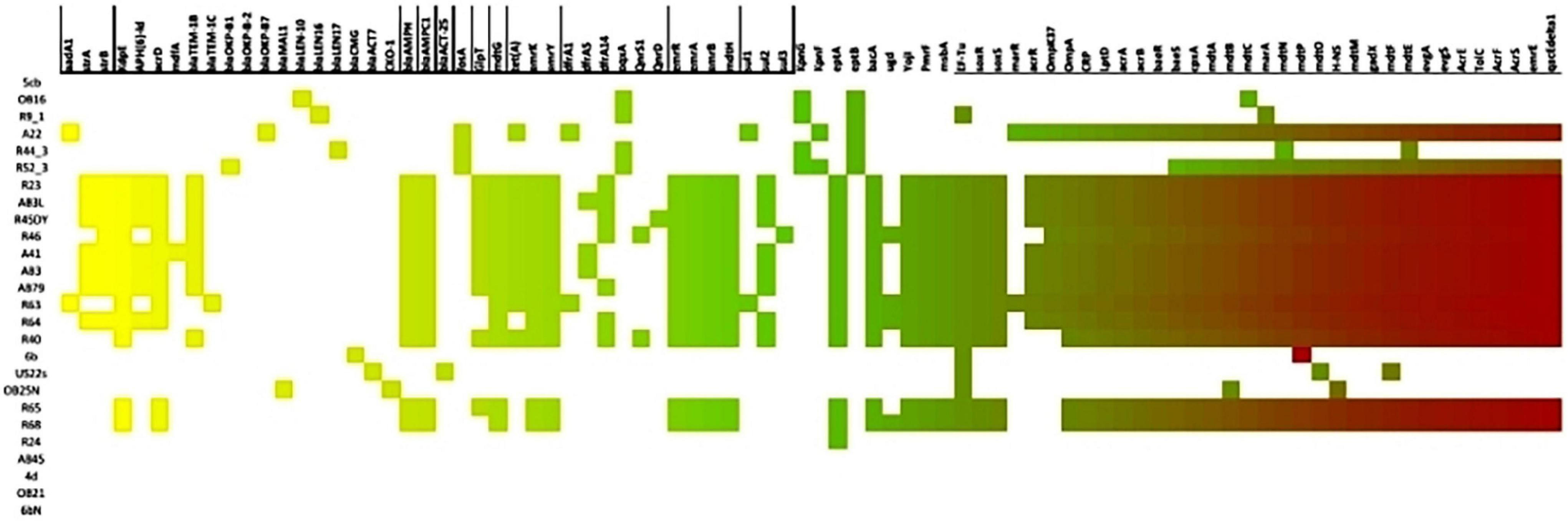
Figure 1. Heat map showing the distribution of antimicrobial resistance genes among the recovered isolates.
Correlation Between the Phenotypic and Genotypic Multidrug-Resistance Patterns
The antibiogram of the resistant recovered animal isolates showed that one isolate (6%) is multidrug-resistant (MDR) (MDR: non-susceptibility of the isolate to ≥1 agent of ≥3 antimicrobial classes) to cephalosporins: ceftazidime, cefotaxime, cephamycins: cefoxitin, monobactams: aztreonam, aminoglycosides: amikacin, antipseudomonal penicillins + β-lactamase inhibitor: piperacillin/tazobactam, folate pathway inhibitors: trimethoprim and had the regulatory system modulating antibiotic efflux CRP as resistance determinant. Out of the resistant animal isolates, two isolates (11%) were multidrug-resistant (MDR) to cephamycins: cefoxitin, folate pathway inhibitors: trimethoprim, penicillin + β-lactamase inhibitor: amoxicillin/clavulanic acid and had varying resistance determinants including regulatory systems modulating antibiotic efflux: CRP, H-NS, marR, emrR, marA, efflux genes: emrA, emrB, emrK, emrY, antibiotic target alteration genes: bacA, pmrF, eptA, EF-Tu mutants, beta-lactamase resistance genes: blaAMPC, blaAMPH, blaTEM1, sulfonamide resistant gene sul2, quinolone resistance gene qnrS1, trimethoprim resistant dihydrofolate reductase dfrA14 etc.
One of the resistant environmental isolates (13%) was multidrug-resistant to cephalosporins: ceftazidime, cefotaxime, cefepime, cephamycins: cefoxitin, monobactams: aztreonam, aminoglycosides: amikacin, antipseudomonal penicillins + β-lactamase inhibitor: piperacillin/tazobactam, folate pathway inhibitors: trimethoprim and had the regulatory system modulating antibiotic efflux CRP, beta-lactamase resistance genes: blaMAL1, blaCKO–1, elfamycin resistance: EF-Tu mutation as its resistance determinants while another isolate (13%) was multidrug resistant to cephalosporins: ceftazidime, cefotaxime, monobactams: aztreonam, aminoglycosides: amikacin, folate pathway inhibitors: trimethoprim and had the regulatory system modulating antibiotic efflux CRP as resistance determinant (Supplementary Table 8).
The correlation coefficient estimated between different tested antimicrobial agents as well as the phenotypic resistance pattern and the detected resistance genes showed a positive correlation, i.e., the correlation is statistically significant as calculated p-value is less than 0.05.
Genetic Comparison of Bovine Escherichia coli Isolates
CgMLST of the 15 isolates showed an allelic distance from one to 2028. Bovine Escherichia coli isolates in this study differed among each other in a minimum of one and a maximum of 1855 alleles. Based on the defined complex threshold (CT) of 20 allelic differences (Leopold et al., 2014) three clusters were obtained. The three Nigerian poultry isolates were closely related with a maximum allelic difference of 7 and were all located in cluster 1 and differed by a minimum of 1272 alleles from the bovine isolates from this study. Three bovine isolates (cluster 2, ST23) obtained from two different farms that were not in close proximity were closely related with a maximum allelic difference of 1. Two other bovine isolates (ST10) in this study from the same farm were also closely related with an allelic difference of 11 (Figure 2). The poultry isolates used in the strain comparison were obtained from a different state far from the farm locations in this study hence no close relatedness was observed.
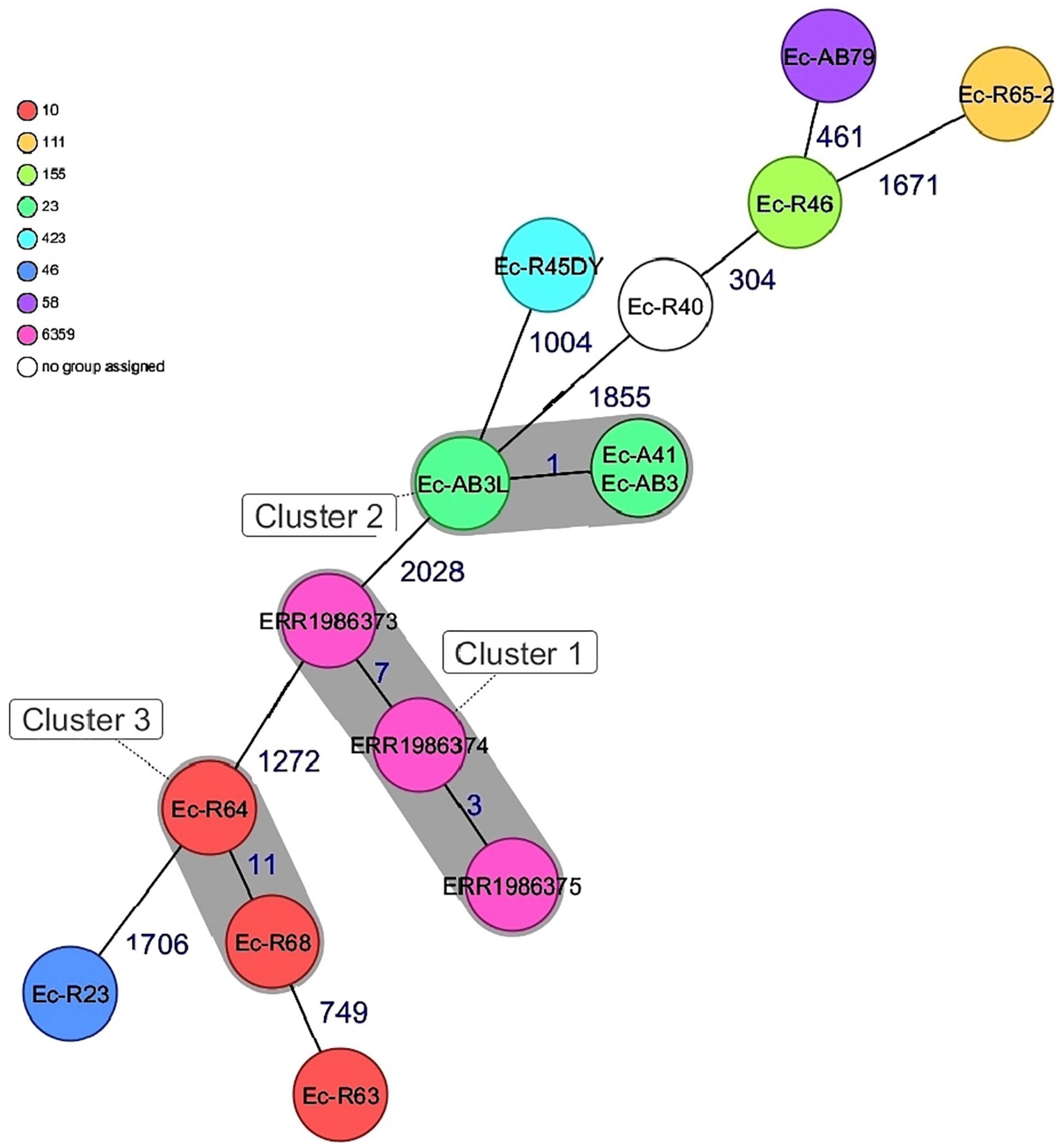
Figure 2. Minimum spanning tree for 15 E. coli isolates based on cgMLST of E.coli. Colors corresponds to the sequence types of the isolates. Each circle represents isolates with an allelic profile based on the sequences of 2520 core genome targets. Isolates with closely related genotypes were identified with a maximum of 11 allelic differences and are shaded in gray.
Multilocus Sequence Typing of Other Recovered Isolates
The animal Klebsiella variicola isolates had ST227 and 259 respectively. One animal Klebsiella quasipneumoniae had ST1136. The Proteus terrae, Serratia marcescens, and one Klebsiella quasipneumoniae animal isolate had previously unknown sequence types. One resistant E. coli animal isolate also had a new sequence type. The resistant Proteus terrae, Proteus faecis, Citrobacter koseri, Enterobacter hormachei, Enterobacter quasiroggenkampii, Klebsiella quasipneumoniae environmental isolates also had new sequence types (Supplementary Table 1). The Klebsiella. variicola environmental isolate was assigned to the new ST314.
Discussion
Enterobacteriaceae are typically found in animals and the environment (Otokunefor et al., 2018) and have been reported as reservoirs of resistance genes that could be passed to other bacterial cells in the system. The use of antibiotics in livestock production results in an increase in antibiotic resistant strains in people (Bevan et al., 2017). Sewage, humans, companion, and domestic animals and the industries have been previously reported as sources of resistant microorganisms to the environment in African settings (Moremi et al., 2017). Determination of antimicrobial resistance in bacteria using phenotypic characterization is very important for therapeutic purposes, but further genetic characterization may sometimes seems appropriate. Whole-genome sequencing plays a key role in enhancing our understanding of how bacteria evolve, are transmitted, and monitoring of antimicrobial resistance. Regardless of the source of isolation in this study, the phenotypic antimicrobial susceptibility testing revealed high frequency of resistance to trimethoprim and cefoxitin. Recently, plasmid mediated FOX-1 AmpC ß-lactamases were detected in Escherichia coli isolates from cows in Nigeria (Ejikeugwu et al., 2018). A recent review on antimicrobial use revealed a high level of antimicrobial usage of tetracyclines, aminoglycosides and penicillins in animal production systems in Africa because they belong to the cheapest antibiotics (Kimera et al., 2020). The previous report showed high prevalence of antimicrobial resistance including multidrug resistance in the environment. Reports from developing countries especially in Africa are of major public health concern as only limited diagnostic and therapeutic options are available (Kimera et al., 2020). The presence of antimicrobial-resistant (AMR) and multidrug- resistant (MDR) bacteria that colonize the gut of animals might play an important epidemiological role in the spread of antimicrobial resistance between livestock animals and humans, either directly or through consumption of contaminated food. Several mechanisms of bacterial resistance to antimicrobial agents have been previously reported (Kumar and Varela, 2013; Sun et al., 2014). They include antibiotic efflux, antibiotic target alteration, antibiotic inactivation, antibiotic target replacement and antibiotic target protection. In this study various resistance determinants that encode antimicrobial efflux pumps were detected both in the resistant and multidrug resistant isolates. Over-expression of such determinants has been connected to a rise of clinically important multidrug-resistant pathogens (Nikaido, 1994). The regulatory system modulating antibiotic efflux (Resistance-Nodulation-Division multidrug efflux pumps) for example has been previously reported as an important determinant of multidrug resistance in Gram-negative bacteria (Fernando and Kumar, 2013). Resistance determinants that encode multidrug efflux pumps of the major facilitator superfamily were also detected in this study and have also been previously reported to contribute to the emergence of multidrug-resistant organisms (Kumar et al., 2020). Results of this study underline the wide distribution and importance of this resistance mechanism.
Some African studies identified antimicrobial resistance genes [blaCTX–M, blaTEM, blaSHV, blaOXA, aac(6′)-lb-cr, tet(A), tet(B), sul1, sul2, qnr] in bacteria from humans, animals, and environmental sources (Hamza et al., 2016; Ribeiro et al., 2016; Eguale et al., 2017; Seni et al., 2018), indicating that drug resistant pathogens have high tendency to spread widely (De Boeck et al., 2012; Ekwanzala et al., 2018; Oloso et al., 2018). Results from this study correlate with the previous African reports as blaTEM, aph(6)-ld, tet(A), tet(D), sul1, sul2, qnr, dfrA1, dfrA5, dfrA14 were some identified resistance genes in the resistant Enterobacteriaceae isolates. Significantly, no extended spectrum beta-lactamase (ESBL) gene was detected in the bovine and environmental Enterobacteriaceae isolates. This is in contrast with a recent report on human clinical Enterobacteriaceae isolates collected from the same geographical location that had a high rate of ESBL genes detected among the isolates (Jesumirhewe et al., 2020).
The role of Enterobacteriaceae as a reservoir for ESBL genes and other resistance determinants is enhanced by the presence of IncF plasmid family type which evolves quickly by replicon diversification and acquisition of antibiotic resistance traits (Carattoli, 2013). Various plasmid replicon types detected among the resistant isolates in this study indicate their importance for dissemination of antibiotic resistance. For example, in this study, resistance genes mediating quinolone resistance and transmitted by plasmids, oqxA, qnrD, and qnrS1 were detected in the resistant animal and environmental isolates indicating that resistance to quinolones was promoted through the plasmid-mediated determinant. Plasmid-mediated quinolone resistance (PMQR) determinants play an important role in the transmission of resistance among bacterial isolates (Santhosh et al., 2017). The predominating presence of IncF plasmid replicon type in the animal and environmental isolates increases the possibility of acquiring more resistance determinants as well as the emergence of novel resistance determinants (Amos et al., 2014). Efflux pumps have been previously reported to be encoded by plasmid borne genetic elements which plays an important role in the transmission of multidrug-resistance (Nikaido, 1994; Nelson and Levy, 2011).
Most developing countries in Africa like Nigeria have been reported to lack effective antimicrobial surveillance systems or are at different stages of developing them (Alonso et al., 2017; Kimera et al., 2020). Monitoring the emergence and spread of antibiotic resistant isolates may assist in developing strategies for treatment and prevention of infections especially for animals which have limited choice of prophylactic and therapeutic antimicrobials. A systematic antimicrobial resistance surveillance using more advanced techniques like WGS is required especially in African settings to detect new antibiotic resistance mechanisms and to determine the virulence of multidrug resistance pathogens for risk assessment and prevention of infection (Hamza et al., 2016; Grundmann and Gelband, 2018). Data from WGS could assist in planning effective interventional measures.
This study provides detailed genomic characterization of antibiotic resistance in bovine animal and environmental Enterobacteriaceae isolates for Nigeria. A number of previous Nigerian studies described the detection, frequency/prevalence of resistance to antibiotics in livestock and the environment using phenotypical techniques (Ayandiran et al., 2014; Onuoha, 2017; Otokunefor et al., 2018; Ugwu et al., 2018). Only a few Nigerian studies have been able to explore the genetic mechanisms of antibiotic resistance in bovine animals and the environment (Olowe et al., 2015; Sharma et al., 2017; Adelowo et al., 2018; Ayandiran et al., 2018; Ejikeugwu et al., 2018) which is important to understand the dissemination of resistant isolates. Further studies to characterize prevalent clones and plasmids that harbor antibiotic resistance genes are required.
Whole genome based cgMLST phylogenetic analysis of the bovine Escherichia coli isolates suggest a probable clonal spread of the isolates of different sequence types and plasmid replicon types. ST10 is an important multilocus sequence type detected in three animal Escherichia coli isolates in this study. Previously, ST10 Escherichia coli have been detected in chicken and have been reported as antibiotic resistant ESBL-producers (Chen et al., 2016). Previous reports show Escherichia coli ST10 to be commonly associated with other animals and humans (Ewers et al., 2012; Chen et al., 2016). This is consistent with our results as the isolates were found in feces of bovine animals. One limitation of this study is that samples obtained were not sufficient to get a large number of isolates to analyze possible links between the bovine and environmental isolates. Although resistance determinants were detected in both the animal and environmental isolates it would be important to further analyze isolates to characterize the possible linkages between animals and the environment.
Conclusion
We report a wide dissemination of antibiotic resistant and multidrug resistant Nigerian bovine and environmental Enterobacteriaceae isolates. The emergence of these resistant strains is of public health concern indicated by the difficult treatment options in infections they cause. Very little data especially genotypic studies of isolates from animals and the environment exist for surveillance of antimicrobial resistance in most developing countries like Nigeria. It is necessary for developing countries like Nigeria to carry out surveillance systems involving a “One Health” approach which would be important to monitor transmission events. Support from global antimicrobial resistance networks should be sought to assist in developing and implementing antimicrobial resistance surveillance under one health approach.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
CJ conceived, designed, performed the experiments and wrote the manuscript. CJ, BS, FA, and WR analyzed the data. All the authors contributed to the article and approved the submitted version.
Funding
This work was supported by an ErnstMach grant 2015/2016 to CJ by the Austrian Federal Ministry of Science, Research and Economy (BMWFW). This work was supported financially by the Austrian Agency for Health and Food Safety (AGES), Vienna, Austria. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.793541/full#supplementary-material
Footnotes
- ^ https://pubmlst.org/species-id
- ^ http://www.genomicepidemiology.org
- ^ https://enterobase.readthedocs.io/en/latest/pipelines/escherichia-statistics.html
- ^ https://mlstkv.insp.mx/
- ^ http://cge.cbs.dtu.dk/services/PlasmidFinder
References
Aarestrup, F. M., Bager, F., Jensen, N. E., Madsen, M., Meyling, M., and Wegener, H. C. (2009). Resistance to antimicrobial agents used for animal therapy in pathogenic-, zoonotic- and indicator bacteria isolated from different food animals in Denmark: a baseline study for the Danish Integrated Antimicrobial Resistance Monitoring Programme (DANMAP). APMIS 160, 745–770. doi: 10.1111/j.1699-0463.1998.tb00222.x
Adelowo, O. O., and Fagade, O. E. (2009). The tetracycline resistance gene tet39 is present in both Gram-negative and Gram-positive bacteria from a polluted river, Southwestern Nigeria. Lett. Appl. Microbiol. 48, 167–172. doi: 10.1111/j.1472-765X.2008.02523.x
Adelowo, O. O., Vollmers, J., Mäusezahl, I., Kaster, A. K., and Müller, J. A. (2018). Detection of the carbapenemase gene blaVIM–5 in members of the Pseudomonas putida group isolated from polluted Nigerian wetlands. Sci. Rep. 8:15116. doi: 10.1038/s41598-018-33535-3
Algammal, A. M., Hashem, H. R., Alff, K. J., Hetta, H. F., Sheraba, N. S., Ramadan, H., et al. (2021a). atpD gene sequencing, multidrug resistance traits, virulence-determinants, and antimicrobial resistance genes of emerging XDR and MDR-Proteus mirabilis. Sci. Rep. 12:9476. doi: 10.1038/s41598-021-88861-w
Algammal, A. M., Hashem, H. R., Al-otaibi, A. S., Alfifi, K. J., El-dawody, E. M., Mahrous, E., et al. (2021b). Emerging MDR-Mycobacterium avium subsp. avium in house-reared domestic birds as the first report in Egypt. BMC Microbiol. 21:237. doi: 10.1186/s12866-021-02287-y
Algammal, A. M., Hetta, H. F., Batiha, G. E., Hozzein, W. N., El Kazzaz, W. M., Hashem, H. R., et al. (2020). Virulence-determinants and antibiotic-resistance genes of MDR-Escherichia coli isolated from secondary infections following FD-outbreak in cattle. Sci. Rep. 10:19779. doi: 10.1038/s41598-020-75914-9
Alonso, C. A., Zarazaga, M., Ben Sallem, R., Jouini, A., Ben Slama, K., and Torres, C. (2017). Antibiotic resistance in Escherichia coli in husbandry animals. the African perspective. Lett. Appl. Microbiol. 64, 318–334. doi: 10.1111/lam.12724
Amos, G. C. A., Hawkey, P. M., Gaze, W. H., and Wellington, E. M. (2014). Waste water effluent contributes to the dissemination of CTX-M-15 in the natural environment. J. Antimicrob. Chemother. 69, 1785–1791. doi: 10.1093/jac/dku079
Ayandiran, T. A., Ayandele, A. A., Dahunsi, S. O., and Ajala, O. O. (2014). Microbial assessment and prevalence of antibiotic resistance in polluted Oluwa River, Nigeria. Egyptian J. Aquatic Res. 40, 291–299. doi: 10.1016/j.ejar.2014.09.002
Ayandiran, T. O., Falgenhauer, L., Schmiede, J., Chakraborty, T., and Ayeni, F. A. (2018). High resistance to tetracycline and ciprofloxacin in bacteria isolated from poultry farms in Ibadan, Nigeria. J. Infect. Dev. Ctries 12, 462–470. doi: 10.3855/jidc.9862
Ayeni, F. A., Odumosu, B. T., Oluseyi, A. E., and Ruppitsch, W. (2016). Identification and prevalence of tetracycline resistance in Enterococci isolated from poultry in Ilishan, Ogun State, Nigeria. J. Pharm. Bioallied Sci. 8, 69–73. doi: 10.4103/0975-7406.171729
Balows, A., Hausler, W. J., Hermann, K. L., Isengerg, J. D., and Shadomy, H. J. (eds) (1991). Manual of Clinical Microbiology, 5th Edn. Washington, DC: American Society of Microbiology.
Barrios-Camacho, H., Aguilar-Vera, A., Beltran-Rojel, M., Aguilar-Vera, E., Duran-Bedolla, J., Rodriguez-Medina, N., et al. (2019). Molecular epidemiology of Klebsiella variicola obtained from different sources. Sci. Rep. 9:10610. doi: 10.1038/s41598-019-46998-9
Bauer, A. W., Kirby, M. M., Sherris, J. C., and Truck, M. (1966). Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 45, 493–496. doi: 10.1093/ajcp/45.4_ts.493
Bernreiter-Hofer, T., Schwarz, L., Müller, E., Cabal-Rosel, A., Korus, M., Misic, D., et al. (2021). The Pheno- and genotypic characterization of porcine Escherichia coli isolates. Microorganisms 9:1676. doi: 10.3390/microorganisms9081676
Bevan, E. R., Jones, A. M., and Hawkey, P. M. (2017). Global epidemiology of CTX-M β-lactamases: temporal and geographical shifts in genotype. J. Antimicrob. Chemother. 72, 2145–2155. doi: 10.1093/jac/dkx146
Cabal, A., Peischl, N., Rab, G., Stöger, A., Springer, B., Sucher, J., et al. (2021). Draft genome sequence of a multidrug-resistant Escherichia coli sequence Type 1193 pandemic clone isolated from wastewater in Austria. Microbiol. Resour. Announc. 10:e0076221. doi: 10.1128/MRA.00762-21
Carattoli, A. (2013). Plasmids and the spread of resistance. Int. J. Med. Microbiol. 303, 298–304. doi: 10.1016/j.ijmm.2013.02.001
Carattoli, A., Zankari, E., Garcia-Fernández, A., Larsen, M. V., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using Plasmid Finder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Centers for Disease Control and Prevention [CDC] (2013). Vital signs: carbapenem-resistant Enterobacteriaceae. MMWR. Morb. Mortal. Wkly Rep. 62, 165–170.
Cheesebrough, M. (2006). District Laboratory Practice in Tropical Countries, 2nd Edn. Cambridge: Cambridge University Press, 178–187.
Chen, P. A., Hung, C. H., Huang, P. C., Chen, J. R., Huang, I. F., Chen, W. L., et al. (2016). Characteristics of CTX-M extended-spectrum β-lactamase-producing Escherichia coli strains isolated from multiple rivers in Southern Taiwan. Appl. Environ. Microbiol. 82, 1889–1897. doi: 10.1128/AEM.0322-15
Cowan, S. T., and Steel, K. J. (1977). Manual for the Identification of Medical Bacteria, 2nd Edn. Cambridge: Cambridge University Press.
De Boeck, H., Lunguya, O., Muyembe, J. J., Glupczynski, Y., and Jacobs, J. (2012). Presence of extended-spectrum beta-lactamase-producing Enterobacteriaceae in waste waters, Kinshasa, the Democratic Republic of the Congo. Eur. J. Clin. Microbiol. Infect. Dis. 31, 3085–3088. doi: 10.1007/s10096-012-1669-8
Eguale, T., Birungi, J., Asrat, D., Njahira, M. N., Njuguna, J., Gebreyes, W. A., et al. (2017). Genetic markers associated with resistance to beta-lactam and quinolone antimicrobials in non-typhoidal Salmonella isolates from humans and animals in Central Ethiopia. Antimicrob. Resist. Infect. Control. 6:13. doi: 10.1186/s13756-017-0171-6
Ejikeugwu, C., Esinone, C., Iroha, I., and Adikwu, M. (2018). First detection of FOX-1 AmpC β-lactamase gene expression among Escherichia coli isolated from abattoir samples in Abakaliki, Nigeria. Oman Med. J. 33, 243–249. doi: 10.5001/omj.2018.44
Ekwanzala, M. D., Dewar, J. B., Kamika, I., and Momba, M. N. B. (2018). Systematic review in South Africa reveals antibiotic resistance genes shared between clinical and environmental settings. Infect. Drug Resist. 11, 1907–1920. doi: 10.2147/IDR.S170715
European Committee on Antimicrobial Susceptibility Testing [EUCAST] (2015). Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 5.0. Växjö: European Committee on Antimicrobial Susceptibility Testing.
Ewers, C., Bethe, A., Semmler, T., Guenther, S., and Wieler, L. H. (2012). Extended spectrum β-lactamase producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: a global perspective. Clin. Microbiol. Infect. 18, 646–655. doi: 10.1111/j.1469-0691.2012.03850.x
Farmer, J. J. 3rd, Davis, B. R., Hickman-Brenner, F. W., McWhorter, A., Huntley-Carter, G. P., Asbury, M. A., et al. (1985). Biochemical identification of new species and biogroups of Enterobacteriaceae isolated from clinical specimens. J. Clin. Micro. 21, 46–76. doi: 10.1128/jcm.21.1.46-76.1985
Fernando, D. M., and Kumar, A. (2013). Resistance-nodulation-division multidrug efflux pumps in gram-negative bacteria: role in virulence. Antibiotics 2, 163–181. doi: 10.3390/antibiotics2010163
Fischer, J., Rodrıguez, I., Schmoger, S., Friese, A., Roesler, U., Helmuth, R., et al. (2012). Salmonella enterica subsp. enterica producing VIM-1 carbapenemase isolated from livestock farms. J. Antimicrob. Chemother. 68, 478–480. doi: 10.1093/jac/dks393
Grundmann, H., and Gelband, H. (2018). Antimicrobial resistance surveillance with whole genome sequencing in Africa: it’s (about) time. Afr. J. Lab. Med. 7, 1–3. doi: 10.4102/ajlm.v7i2.761
Hamza, E., Dorgham, S. M., and Hamza, D. A. (2016). Carbapenemase-producing Klebsiella pneumoniae in broiler poultry farming in Egypt. Integr. Med. Res. 7, 8–10. doi: 10.1016/j.jgar.2016.06.004
Hoyle, D. V., Yates, C. M., Chase-Topping, M. E., Turner, E. J., Davies, S. E., and Low, J. C. (2005). Molecular epidemiology of antimicrobial resistant Escherichia coli strains in a cohort of newborn calves. Appl. Environ. Microbiol. 71, 6680–6688. doi: 10.1128/AEM.71.11.6680-6688.2005
Jesumirhewe, C., Springer, B., Allerberger, F., and Ruppitsch, W. (2020). Whole genome sequencing of extended-spectrum β-lactamase genes in Enterobacteriaceae isolates from Nigeria. PLoS One 15:e0231146. doi: 10.1371/journal.pone.0231146
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., et al. (2017). CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573. doi: 10.1093/nar/gkw1004
Kimera, Z. I., Mshana, S. E., and Rweyemamu, M. M. (2020). Antimicrobial use and resistance in food-producing animals and the environment: an African perspective. Antimicrob. Resist. Infect. Control. 9:37. doi: 10.1186/s13756-020-0697-x
Kotsyuba, K. R., Voronkova, O. S., Vinnikov, A. I., and Shevchenko, T. M. (2014). Mechanisms of antibiotic resistance of Enterobacteriaceae family representatives. Regulatory Mech. Biosyst. 5, 33–38. doi: 10.15421/021407
Krishnasamy, S. (2018). Antibiotic usage, residues and resistance genes from food animals to human and environment. J. Environ. Chem. Eng. 8:102221. doi: 10.1016/j.jece.2018.02.029
Kumar, S., Lekshmi, M., Parvathi, A., Ojha, M., Wenzel, N., and Varela, M. N. (2020). Functional and structural roles of the major facilitator superfamily bacterial multidrug efflux pumps. Microorganisms 8:266. doi: 10.3390/microorganisms8020266
Kumar, S., and Varela, M. F. (2013). “Molecular mechanisms of bacterial resistance to antimicrobial agents,” in Microbial Pathogens and Strategies for Combating Them: Science, Technology and Education, ed. A. Méndez-Vilas (Badajoz: Formatex Research Center), 522–534.
Leopold, S. R., Goering, R. V., Witten, A., Harmsen, D., and Mellmann, A. (2014). Bacterial whole-genome sequencing revisited: portable, scalable, and standardized analysis for typing and detection of virulence and antibiotic resistance genes. J. Clin. Microbiol. 52, 2365–2370. doi: 10.1128/JCM.00262-14
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Makharita, R. R., El-kholy, I., Hetta, H. F., Abdelaziz, M. H., Hagagy, F. I., Ahmed, A. A., et al. (2020). Antibiogram and genetic characterization of carbapenem-resistant gram-negative pathogens incriminated in healthcare-associated infections. Infect. Drug Resist. 13, 3991–4002. doi: 10.2147/IDR.S276975
Marshall, B. M., and Levy, S. B. (2011). Food animals and antimicrobials: impacts on human health. Clin. Microbiol. Rev. 4, 718–733. doi: 10.1128/CMR.00002-11
Mišić, D., Kiskaroly, F., Szostak, M. P., Cabal, A., Ruppitsch, W., Bernreiter-Hofer, T., et al. (2021). The first report of mcr-1-carrying Escherichia coli originating from animals in Serbia. Antibiotics 10:11063. doi: 10.3390/antibiotics10091063
Moremi, N., Claus, H., Vogel, U., and Mshana, S. E. (2017). Faecal carriage of CTX-M extended-spectrum beta-lactamase-producing Enterobacteriaceae among street children dwelling in Mwanza city, Tanzania. PLoS One 12:e184592. doi: 10.1371/journal.pone.0184592
Mugg, P. A., and Hill, A. (1981). Comparison of microbact 12E and 24E systems and the API 20E systems for the identification of Enterobacteriaceae. J. Hyg. Camb. 87, 287–297. doi: 10.1017/s0022172400069503
Nelson, M. L., and Levy, S. B. (2011). The history of the tetracyclines. Ann. N. Y. Acad. Sci. 1241, 17–32. doi: 10.1111/j.1749-6632.2011.06354.x
Ngbede, E. O., Raji, M. A., Kwanashie, C. N., and Kwaga, J. K. (2017). Antimicrobial resistance and virulence profile of Enterococci isolated from poultry and cattle sources in Nigeria. Trop. Anim. Health Prod. 49, 451–458. doi: 10.1007/s11250-016-1212-5
Nikaido, H. (1994). Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science 264, 382–388. doi: 10.1126/science.8153625
OIE (2007). List of Antimicrobials of Veterinary Importance. Available online at*: http://www.oie.int/fileadmin/Home/eng/Internationa_Standard_Setting/docs/pdf/OIE_list_antimicrobials.pdf (accessed January 26, 2021).
Oloso, N. O., Fagbo, S., Garbati, M., Olonitola, S. O., Awosanya, E. J., Aworh, M. K., et al. (2018). Antimicrobial resistance in food animals and the environment in Nigeria: a review. Int. J. Environ. Res. Public Health 15:1284. doi: 10.3390/ijerph15061284
Olowe, O. A., Adewumi, O., Odewale, G., Ojurongbe, O., and Adefioye, O. J. (2015). Phenotypic and molecular characterisation of extended-spectrum Beta-lactamase producing Escherichia coli obtained from animal fecal Samples in Ado Ekiti, Nigeria. J. Environ. Public Health 2015:497980. doi: 10.1155/2015/497980
Onuoha, S. C. (2017). The prevalence of antibiotic resistant diarrhogenic bacterial species in surface waters, South Eastern Nigeria. Ethiop J. Health Sci. 27, 319–330. doi: 10.4314/ejhs.v27i4.3
Otokunefor, K., Agbude, P., and Otokunefor, T. V. (2018). Non-clinical isolates as potential reservoirs of antibiotic resistance in port harcourt, Nigeria. Pan. Afr Med. J. 30:167. doi: 10.11604/pamj.2018.30.167.14261
Pearce, M. C., Evans, J., McKendrick, I. J., Smith, A. W., Knight, H. I., Mellor, D. J., et al. (2006). Prevalence and virulence factors of Escherichia coli serogroups O26, O103, O111, and O145 shed by cattle in Scotland. Appl. Environ. Microbiol. 72, 653–659. doi: 10.1128/AEM.72.1.653-659.2006
Ribeiro, T. G., Novais, A., Peixe, L., and Machado, E. (2016). Atypical epidemiology of CTX-M-15 among Enterobacteriaceae from a high diversity of non-clinical niches in Angola. J. Antimicrob. Chemother. 71, 1169–1173. doi: 10.1093/jac/dkv489
Santhosh, K. S., Deekshit, V. K., Venugopal, M. N., Karunasagar, I., and Karunasagar, I. (2017). Multiple antimicrobial resistance and novel point mutation in fluoroquinolone-resistant Escherichia coli isolates from Mangalore, India. Microb. Drug Resist. 23, 994–1001. doi: 10.1089/mdr.2016.0142
Seni, J., Moremi, N., Matee, M., van der Meer, F., DeVinney, R., Mshana, S., et al. (2018). Preliminary insights into the occurence of similar clones of extended-spectrum beta-lactamase-producing bacteria in humans, animals and the environment in Tanzania: a systematic review and meta-analysis between 2005 and 2006. Zoonoses Public Health 65, 1–10. doi: 10.1111/zph.12387
Sharma, P., Gupta, S. K., Adenipekun, E. O., Barrett, J. B., Hiott, L. M., Woodley, T. A., et al. (2017). Draft genome sequence analysis of multidrug-resistant Escherichia coli strains isolated in 2013 from humans and chickens in Nigeria. Genome Announc. 5, e1073–e1017. doi: 10.1128/genomeA.01073-17
Shivakumaraswamy, S. K., Deekshit, V. K., Vittal, R., Akhila, D. S., Mundanda, D. M., Mohan Raj, J. R., et al. (2019). Phenotypic & genotypic study of antimicrobial profile of bacteria isolates from environmental samples. Indian J. Med. Res. 149, 232–239. doi: 10.4103/ijmr.IJMR_2097_17
Spoor, L. E., McAdam, P. R., Weinert, L. A., Rambaut, A., Hasman, H., Aarestrup, F. M., et al. (2012). Livestock origin for a human pandemic clone of community associated methicillin-resistant Staphylococcus aureus. mBio 4, 1–6. doi: 10.1128/mBio.00356-13
Sun, J., Deng, Z., and Yan, A. (2014). Bacterial multidrug efflux pumps: mechanisms, physiology and pharmacological exploitations. Biochem. Biophys. Res. Commun. 453, 254–267. doi: 10.1016/j.bbrc.2014.05.090
Taylor, L. H., Latham, S. M., and Woolhouse, M. E. J. (2001). Risk factors for human disease emergence. Phil. Trans. R. Soc. Lond. B 356, 983–989. doi: 10.1098/rstb.2001.0888
Thomas, A. D. (1983). Evaluation of the API20E and the Microbact 24E systems for the evaluation of Pseudomonas pseudomallei. Vet. Microbiol. 8, 611–615.
Ugwu, M. C., Igbokwe, J. O., Okezie, U., Eze, P. M., Ejikeugwu, C. P., and Esimone, C. O. (2018). Prevalence of ESBLs and MBLs among Escherichia coli and Klebsiella pneumoniae isolates from a Nigerian Abattoir. J. Trop. Dis. 6:2. doi: 10.4172/2329-891X.1000261
Veterian Key (2016). Enterobacteriaceae Chapter 17. Available online at: https://veteriankey.com/enterobacteriaceae/ (accessed November 29, 2021).
Woolhouse, M. E., and Ward, M. J. (2013). Sources of antimicrobial resistance. Science 341, 1460–1461. doi: 10.1126/science.1243444
Yates, C. M., Pearce, M. C., Woolhouse, M. E. J., and Amyes, S. G. B. (2004). High frequency transfer and horizontal spread of apramycin resistance in calf faecal Escherichia coli. J. Antimicrob. Chemother. 54, 534–537. doi: 10.1093/jac/dkh353
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired resistance genes. J. Antimicrob. Chemother. 67, 22640–22644. doi: 10.1093/jac/dks261
Keywords: whole genome sequencing, Enterobacteriaceae, antibiotic resistance, bovine animals, environment
Citation: Jesumirhewe C, Springer B, Allerberger F and Ruppitsch W (2022) Genetic Characterization of Antibiotic Resistant Enterobacteriaceae Isolates From Bovine Animals and the Environment in Nigeria. Front. Microbiol. 13:793541. doi: 10.3389/fmicb.2022.793541
Received: 12 October 2021; Accepted: 10 January 2022;
Published: 25 February 2022.
Edited by:
Kristina Kadlec, Independent Researcher, Wunstorf, GermanyReviewed by:
Abdelazeem Algammal, Suez Canal University, EgyptJaroslav Hrabak, Charles University, Czechia
Copyright © 2022 Jesumirhewe, Springer, Allerberger and Ruppitsch. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christiana Jesumirhewe, ZWJhcnVub3NlbjIwMDJAeWFob28uY28udWs=
†These authors have contributed equally to this work
 Christiana Jesumirhewe
Christiana Jesumirhewe Burkhard Springer
Burkhard Springer Franz Allerberger
Franz Allerberger Werner Ruppitsch
Werner Ruppitsch