- 1Department of Food Biosciences, Teagasc Food Research Centre, Co. Cork, Ireland
- 2Unit for Food Microbiology, Institute for Food Safety, Food Technology and Veterinary Public Health, University of Veterinary Medicine, Vienna, Austria
- 3Austrian Competence Center for Feed and Food Quality, Safety and Innovation, Tulln, Austria
Listeria monocytogenes is an environmentally adapted saprophyte that can change into a human and animal bacterial pathogen with zoonotic potential through several regulatory systems. In this review, the focus is on the occurrence of Listeria sensu stricto and sensu lato in different ecological niches, the detection methods, and their analytical limitations. It also highlights the occurrence of L. monocytogenes genotypes in the environment (soil, water, and wildlife), reflects on the molecular determinants of L. monocytogenes for the saprophytic lifestyle and the potential for antibiotic resistance. In particular, the strain-specific properties with which some genotypes circulate in wastewater, surface water, soil, wildlife, and agricultural environments are of particular interest for the continuously updating risk analysis.
The Genus Listeria
Listeria spp. are Gram-positive, facultative anaerobic, non-spore forming, and catalase positive rods (Rocourt and Buchrieser, 2007). The genus currently includes 28 species and six subspecies with taxonomic names effectively published under the rules of the international code of nomenclature of bacteria. Orsi and co-workers separated the species into two phenotypically and genotypically distinct groups: Listeria sensu lato and Listeria sensu stricto (Orsi and Wiedmann, 2016). The first group is comprised of a pathogenic saprophytes present in the environment and includes L. grayi (subsp. grayi, and murrayi; Rocourt et al., 1992), L. rocourtiae (Leclercq et al., 2010), L. fleischmannii (subsp. fleischmannii and coloradonensis; Bertsch et al., 2013; den Bakker et al., 2013), L. weihenstephanensis (Halter et al., 2013), L. floridensis, L. aquatica, L. cornellensis, L. riparia, L. grandensis (den Bakker et al., 2014), L. booriae, L. newyorkensis (Weller et al., 2015a), L. costaricensis (Núñez-Montero et al., 2018), L. goaensis (Doijad et al., 2018), L. thailandensis (Leclercq et al., 2019), L. valentina (Quereda et al., 2020), L. portnoyi, and L. rustica (Carlin et al., 2021a).
The Listeria sensu stricto group includes the species L. innocua (Seeliger, 1981), L. welshimeri (Rocourt and Grimont, 1983), L. seeligeri (Rocourt and Grimont, 1983), L. marthii (Graves et al., 2010), L. farberi, L. immobilis, L. cossartiae (subsp. cossartiae and cayugensis; Carlin et al., 2021a,b), and the two pathogenic Listeria species L. monocytogenes (Murray et al., 1926; Pirie, 1940) and L. ivanovii (subsp. ivanovii and londoniensis; Seeliger et al., 1984; Boerlin et al., 1992). These two species are known to be pathogenic for both humans and animals (biohazard level 2), nevertheless L. ivanovii has only been associated with human disease very rarely (Guillet et al., 2010).
Generally, Listeria sensu stricto are able to grow at low temperatures (<4°C) and are motile due to the presence of flagella (Walker et al., 1990; Orsi and Wiedmann, 2016). Fully formed flagella are usually observed at temperatures between 20°C and 28°C since flagellin expression, required for assembly of flagella, is temperature dependent and reduced at 37°C (Peel et al., 1988). The regulation of this expression has been shown to be different between L. innocua and L. monocytogenes. Listeria innocua is motile at 37°C; however, only extremely rarely isolates of L. monocytogenes show reduced repression of flagellin production with consequent maintenance of motility at this temperature (Kathariou et al., 1995).
L. monocytogenes is known to cause both sporadic or epidemic listeriosis in high-risk groups (elderly, neonates, pregnant women, and immunocompromised people) and its incidence has been steadily increasing in the EU/EEA. There are two courses of listeriosis: a non-invasive and an invasive form. Non-invasive listeriosis (febrile gastroenteritis) is a mild form of the disease that mainly affects healthy people. The hospitalization rate for the invasive form of listeriosis is 95% and includes symptoms as sepsis, meningitis, rhomboencephalitis, and abortion (Allerberger and Wagner, 2010; EFSA and ECDC, 2019). Listeriosis is one of the most serious foodborne diseases under surveillance as it may lead to high morbidity and mortality rates in high-risk groups (20–30%; Watson, 2009; Allerberger and Wagner, 2010; EFSA and ECDC, 2015; EFSA, 2018). The transmission to consumers often occurs via contaminated food (Wesley, 2007). This review provides an overview of the current knowledge on the lifestyle of the genus Listeria in the environment addressing particular factors relevant to food safety.
Methodologies for Recovery of Listeria monocytogenes and Other Listeria species From Environments
Isolation of L. monocytogenes from a wide range of environmental samples from food processing plants, farms, or natural environments is challenging due to the numerous background microbiota (e.g., aerobic spore-formers, Enterococci, lactic acid bacteria, coryneform bacteria, Pseudomonas, and Enterobacteriaceae) that can act as competitors and the environmental factors that cause stress during enrichment (Stessl et al., 2009; Zilelidou et al., 2016). There are several protocols for the detection of L. monocytogenes in environmental samples; what they all have in common is the critical step of the enrichment process where the number of L. monocytogenes is increased to allow detection. Generally, an efficient enrichment process for the recovery of L. monocytogenes from the environment uses a two-phase process with a smaller number of selective additives in the first step. Incubation of Listeria enrichment broths at low temperatures has long been common practice (Gray et al., 1948; Gray, 1960), relying on the psychrotrophic nature of Listeria to perform selection. Van Renterghem et al. (1991) did not achieve a satisfying recovery of L. monocytogenes by cold enrichment techniques. The same was observed by Hayes et al. (1991), who compared the cold enrichment technique with the standardized U.S. Department of Agriculture (USDA) method and realized that the USDA method showed significantly better isolation rates (96 versus 59%). Moreover, some members of the Listeria sensu lato group (e.g., Listeria floridensis, Listeria aquatica, and L. fleischmanii) will not grow below 7°C, which can be particularly relevant if a complex consortium of Listeria sensu stricto and Listeria sensu lato is to be successfully detected (den Bakker et al., 2014; Barre et al., 2016).
A detailed characterization of the media used for Listeria enrichment and its evolution can be found in the review by Curtis and Lee (1995).
Currently, the U.S.A. Food and Drug Administration (FDA) Bacteriological Analytical Manual method for the detection of L. monocytogenes in food and environmental samples recommends a four-hour pre-enrichment at 30°C in Buffered Listeria enrichment broth (BLEB), followed by the addition of the selective supplements and a further incubation period for 24 to 48 h (Hitchins et al., 2017).
The United States Department of Agriculture (USDA) recommends the use of the media developed at the University of Vermont (UVM) to perform the enrichment. A first enrichment step in UVM-I broth with incubation at 30°C for 20–26 h depending on the sample matrix is recommended, followed by a second enrichment in morpholine propane sulfonic acid buffered Listeria enrichment broth (MOPS-BLEB) at 35°C for 18–24 h (USDA, 2019). In the European Union, ISO standards are more frequently used and are the base for legal compliance. According to ISO standard 11,290-1 (ISO 11290-1, 2017), Listeria detection is performed using a two-step enrichment procedure: a 24 ± 2 h incubation of the sample in Half-strength Fraser at 30°C is followed by a secondary enrichment in Fraser for 24 ± 2 h at 37°C. A description of the media composition and selective agents as well as the various differential media used for plating and identification can be found in a review by Law et al. (2015).
The impact of the methodology used for enrichment is still a matter of current research as it has been repeatedly seen that the performance of enrichment media is affected by the nature of the sample and greatly influences the time required for L. monocytogenes detection (Salazar et al., 2020). Etty et al. (2019) have shown that Listeria enrichment broth (LEB) allowed a better growth of L. monocytogenes, when compared to Fraser broth and UVM, due to the presence of dextrose that promotes the expression of the p60 protein observed to be proportional to the cellular growth.
The selective enrichment step is further complicated in challenging environments such as soils, where bacterial competition may play a role and lead to the loss of L. monocytogenes and other Listeria species (Keys et al., 2016; Zilelidou et al., 2016). It has even been observed that certain L. monocytogenes genotypes outcompete each other: In selective enrichment UVM, lineage I strains are displaced by lineage II strains (Etty et al., 2019). Zilelidou et al. (2016) using a limited number of strains, described a strain-specific dominance in the ISO protocol; however, no correlation to lineages was established. No serotype nor lineage bias was observed in buffered Listeria enrichment broth (BLEB) in a study by Gorski et al. (2006).
Keys et al. (2016) obtained the highest Listeria populations in selective enrichments by adding sodium chloride and lithium chloride at higher temperatures (35°C versus 30°C). The authors pointed out the limited number of selective substances for use in Listeria-specific enrichment media and thus resulting in only limited selective formulations. There is a need for optimization of Listeria selective enrichments to improve recovery from the increasing diversity of Listeria species, especially in samples with a high baseline microbial load.
A recent publication by Carlin et al. (2021c) has looked into the performance of enrichment broths used in reference methods (FDA, USDA, and ISO) given the increased number of Listeria species described in the last years. This study expanded on the work by Barre et al. (2016) increasing the number of different species and evaluating the USDA and FDA broth on top of the ISO standard as well as three selective and differential agars. The authors observed a weak ability to recover new sensu lato Listeria species for the ISO, and both for ISO and USDA enrichment procedures, the growth of other sensu lato Listeria species could lead to the outgrowth of two sensu stricto Listeria species. Only, the BLEB broth used in the FDA method was capable of recovering all 19 species tested by Carlin et al. (2021c). These authors also observed atypical colony morphologies and growth inhibition on selective and differential agars used in the three studied methods for both sensu stricto and sensu lato species highlighting the potential for false negative results, hence recommending the inclusion of a comprehensive Listeria species panel for method evaluations (Carlin et al., 2021c). These results can have a large impact in the adequate detection of Listeria and therefore prompt the need for an exhaustive and inter-laboratorial study for following the growth of standardized strains, representative of all the different species, throughout each of the steps of the different methods.
The direct application of molecular methods has also been widely explored. However, Locatelli et al. (2013) showed that the sole application of molecular detection methods after direct DNA extraction from soil samples underestimated the presence of L. monocytogenes in soil compared to the cultivation-based method. Further characterization of Listeria isolates, including species differentiation, was often performed by sigB sequencing for allele typing in combination with multiplex PCR (Doumith et al., 2004; Nightingale et al., 2007).
New approaches combining quasimetagenomic approaches with microbiome analyses based on 16 S RNA sequencing provide good insights into the co-selection of L. monocytogenes in the presence of a diverse background flora (e.g., Serratia, Pseudomonas, and Enterococcus, Bacillus) during selective enrichment. Wagner et al. (2021) tended to use a shortened enrichment time of 4 h for molecular detection to fully capture L. monocytogenes genotype diversity based on molecular methods.
In source tracking approaches in contaminated ice cream applying the latter novel tools, Ottesen et al. (2020) showed that L. monocytogenes was inhibited by the competitive flora (Anoxybacillus and Geobacillus) until 20 h of incubation. The number of species decreased with increasing the enrichment time, and L. monocytogenes and Rothia mucilaginosa were the only species detected at all-time points (Commichaux et al., 2021).
The Prevalence of Listeria in the Environment
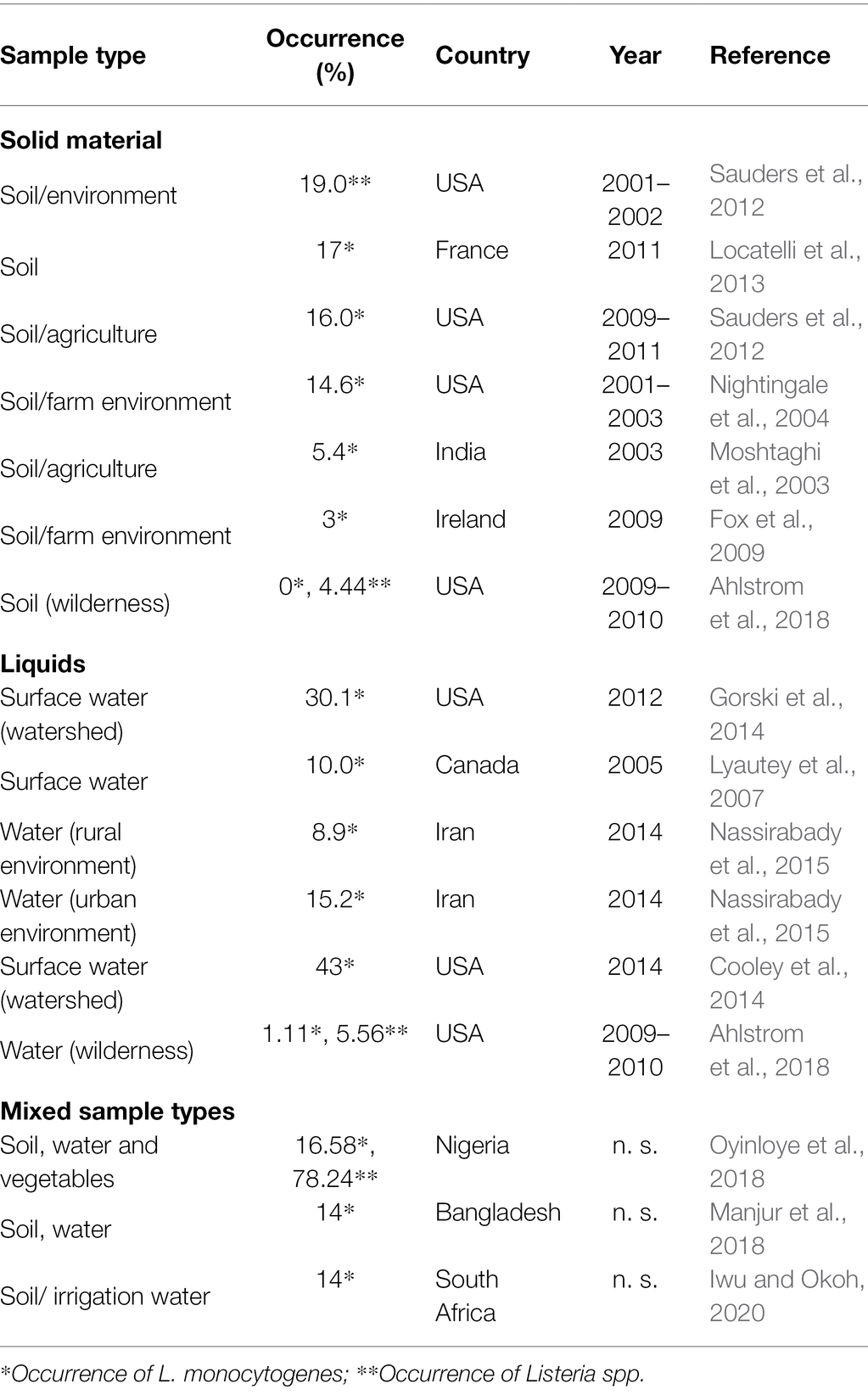
As with some other Gram-positive microorganisms, such as spore-formers, the occurrence of Listeria species is related to habitat, such as water, soil, and decaying vegetation (Sauders and Wiedmann, 2007), although prevalence remains variable. As stated before, one of the factors for this variability is attributed to the use of different enrichment protocols, but the wide range of environmental parameters, such as climatic conditions and soil composition, also determine the survival of a strain. The microbiota, mesobiota, and macrobiota present in the environment were observed to be critical. Vivant et al. (2013) compiled relevant cases of the occurrence of L. monocytogenes between 1971 and 2013. The authors reviewed the literature on the influence of the soil characteristics in the population dynamics and conclude that the short-term survival variability can be attributed to the soil chemical properties, such as the basic cation saturation ratio, whereas the long-term survival variability is attributed to the soil texture, namely, clay content. The pH of the soil is a major factor influencing the survival of L. monocytogenes in soil. Table 1 updates the review by Vivant et al. (2013) covering the period of years 2002 to 2020. Prevalence data in these studies indicate L. monocytogenes presence at the limit of detection and up 19% in soil and between 8.9 and 43% in water. In a nationwide survey in the United States of America, a 31% Listeria prevalence was detected out of 1,004 investigated soil samples, with 594 Listeria spp. isolates with different sigB allele types. Variables related to soil properties, climate, and land use were evaluated, and salinity, molybdenum concentration, and moisture were identified as the most important variables in explaining the presence or absence of Listeria (Liao et al., 2021).
Chapin et al. (2014) studied the ecology of Listeria and found that in the case of L. monocytogenes, prevalence was not only related to seasons and geographic factors, but that meteorological factors and proximity to water and pastures also played an important role. By analyzing five sampling sites in New York State, the author described a prevalence of L. monocytogenes in natural and agricultural environment that ranged from 8.0% (59/734) to 15.0% (88/588). The prevalence of Listeria spp. was significantly higher in summer (37%) than in fall and spring in production areas.
It has been hypothesized that the ability of the genus to adapt to cold environments would contribute to a higher detection at high altitudes. However, Linke et al. (2014) observed that the overall prevalence of Listeria spp. at altitudes above 1,500 m was very low (2.6%) and the few isolation events were +shown to originate from members of the L. ivanovii and L. seeligeri species. Factors other than higher elevation, such as UV radiation, intense freeze–thaw cycles, a shorter growing season, and lower human and animal densities with consequently less runoff bringing Listeria spp. into the soil, created harsher environmental conditions (Lyautey et al., 2007; Strawn et al., 2013; Cooley et al., 2014). The latter research on natural reservoirs of Listeria species in environmental ecosystems also confirmed the importance of proximity to water sources by finding higher isolation rates of L. monocytogenes and L. innocua in samples collected from floodplains of the Danube and Schwarza rivers in Austria (Linke et al., 2014).
The presence of water was also identified as a relevant risk factor in soil samples by Weller et al. (2015b). The authors observed that, in field crops environments, among other factors, the period between irrigation and sampling was statistically significant for detection probability, with 3 days having an odds ratio of about 39 for L. monocytogenes and about 5 for Listeria spp., respectively, compared to a period of 10 days or more. However, Castro-Ibáñez et al. (2015) found that, unlike other foodborne pathogens, the time following a flooding event did not affect the risk of L. monocytogenes contamination in lettuce fields and hypothesized that this was due to the low level of contamination with this pathogen in floodwater.
Isolation of the various Listeria species differed significantly between natural and urban environments, with L. seeligeri and L. welshimeri being more abundant in the former and L. monocytogenes and L. innocua in the latter (Sauders et al., 2012).
These differences have been explained by factors such as fecal excretion of Listeria spp. by wildlife or soil properties that favor the survival of certain species (Sauders and Wiedmann, 2007; Hellström et al., 2008). The ability of different Listeria species to utilize different carbon sources also plays a role. The fact that L. ivanovii and L. seeligeri are able to utilize xylose from the decomposition of cellulose may allow these species to survive longer in soil where this nutrient is abundant (Sauders and Wiedmann, 2007). Nevertheless, the association between Listeria and soil was noted in a study conducted in urban areas, where the highest prevalence was found in samples taken from treaded shoe soles rather than smooth treaded soles (12.5–20.7% vs. 0–6.6%), and near urban parks rather than shopping malls (Schoder et al., 2015).
Listeria monocytogenes Genotypes Prevalence in the Environment
Various typing systems are available for genetic characterization of L. monocytogenes. Serotyping, based on somatic and flagellar antigen differences, distinguishes 14 serotypes (Farber and Peterkin, 1991; Yin et al., 2019). Four genetic lineages can be discriminated by sequence-based methods. Pulsed-field gel electrophoresis (PFGE) is a gel-based subtyping method with superior discriminatory power. Multilocus sequence typing (MLST) is a sequence-based method that focuses on the typing of seven housekeeping genes and allows assignment to sequence types and clonal complexes. The most internationally recognized scheme was proposed by Ragon et al. (2008), an adaptation of the initially proposed scheme by Salcedo et al. (2003). This typing method is still currently used (Kaszoni-Rückerl et al., 2020; Stessl et al., 2021).
Most listeriosis outbreaks are caused by serotypes 1/2a, 1/2b, 1/2c, and 4b (Cossart, 2011). Isolates of genetic lineage I are associated with sporadic and epidemic listerioses in humans, whereas isolates of lineage III are more commonly associated with listerioses in animals (Orsi et al., 2011; Buchanan et al., 2017).
L. monocytogenes genetic lineage I and II are the most prevalent Listeria MLST profiles in the BIGSdb-Listeria (Moura et al., 2016; Figure 1). Genetic lineage I is more present among human isolates, and lineage II is often assigned to the food and environmental niche (Lomonaco et al., 2015). However, when analyzing this type of data, it is important to bear in mind that the database may be biased as its representativeness is unknown.
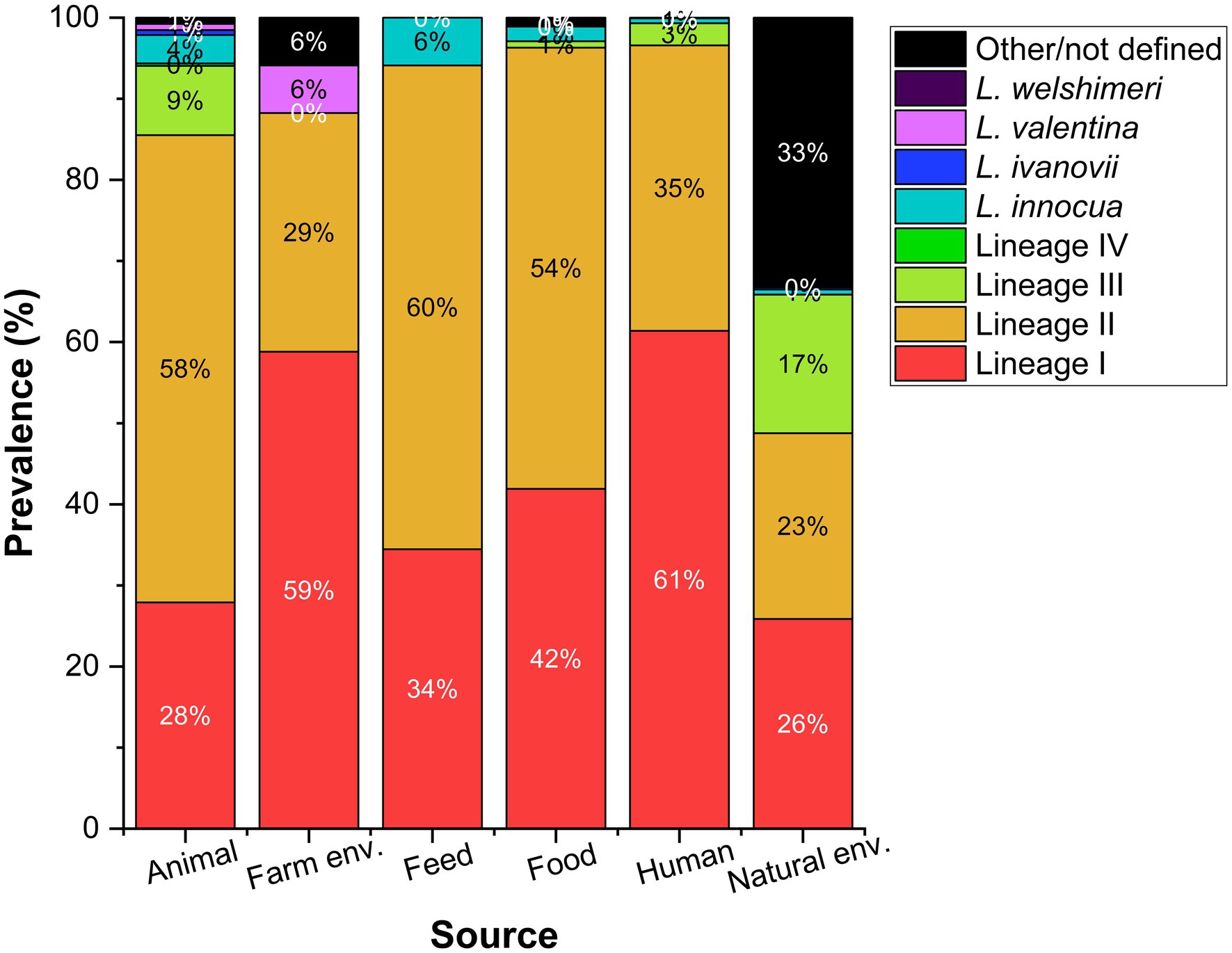
Figure 1. Prevalence reported as percentage of Listeria species including L. monocytogenes genetic lineages (I–IV) according to different sources. Data collected from the BIGSdb-Listeria (Moura et al., 2016) on August 18th 2021 for a total of 656, 17, 119, 1,105, 1,435, and 773 isolates from each source.
Recombination does not occur uniformly throughout the Listeria genome, nor across genetic lineages. den Bakker et al. (2008) identified a higher recombination rate in genetic lineage II, more successful in the adaption to different environments and the survival of stress factors, than lineage I. These authors tested for point mutations and recombination using the 7-locus MLST scheme and found that the occurrence of recombination was about six times more frequent in lineage II than in lineage I, when compared to the occurrence of point mutations. These differences between lineages contribute to the differential detection of lineages I and II in environmental isolates. Linke et al. (2014) and Stea et al. (2015) found a nearly identical distribution of these genetic lineages in samples collected in Austria and Canada, respectively.
Zamudio et al. (2020) compared food and human isolates from Switzerland, Germany, and Netherlands with a total of 708 genomes using a cgMLST scheme including 1701 genes (Ruppitsch et al., 2015; Halbedel et al., 2018). These authors observed that sublineage allelic diversity within lineages II and I was greater using a cgMLST approach within lineage I, than within lineage II. However, when this analysis was performed considering SNPs differences within strains, genetic lineage I had a lower genetic diversity (with about 5,000 SNPs difference), while genetic lineage II had about 15,000 SNPs difference.
The most abundant L. monocytogenes clonal complexes (CCs) assigned to each genetic lineage according to the BIGSdb-Listeria (Moura et al., 2016) on August 18th 2021 are shown in Figure 2. We draw the reader’s attention to the unknown representativeness of data included in publicly available databases as analyzed in Figure 1.
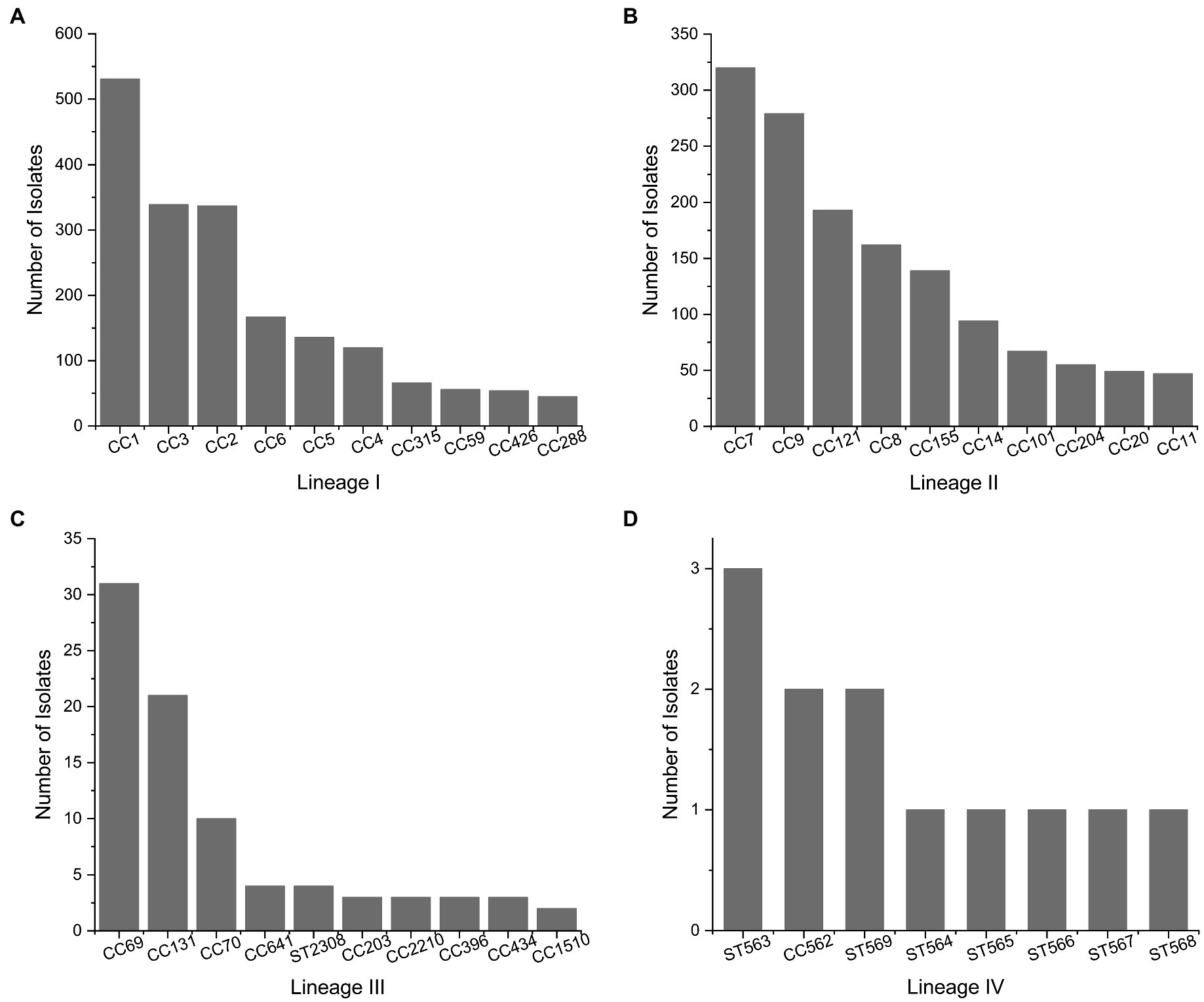
Figure 2. Most Abundant clonal complexes of L. monocytogenes for genetic lineages. (A) Lineage I, (B) lineage II, (C) lineage III, and (D) lineage IV. Data collected from the BIGSdb-Listeria (Moura et al., 2016) on August 18th 2021 for a total of 2087, 2025, 283, and 12 isolates in lineages I, II, III, and IV, respectively.
The distribution of L. monocytogenes in certain areas is influenced by the progressive expansion of genotypes harboring plasmids, transposons, prophages, and Listeria genomic islands (e.g., LGI2 for arsenic resistance; Lee et al., 2017; Muhterem-Uyar et al., 2018). Heavy metal resistance is closely linked to the ability of L. monocytogenes to cause disease, especially in 4b isolates. CC2 and CC1 harbor the highest prevalence of arsenic resistant isolates, followed by CC4, CC315, and CC9.
Most CC8 isolates contain Listeria genomic island 1, which contains the emrE gene and increases tolerance to quaternary ammonium compounds (QACs; Kovacevic et al., 2016). Certain ST121 harbor the transposon Tn6188, which is also involved in sublethal adaptation to QACs (Müller et al., 2013). Some key genetic features of L. monocytogenes are conserved in certain genotypes, such as stress survival islet 1 (SSI-1) and SSI-2, which enhance adaptation to acidic/bile and alkaline/oxidative stress, respectively (Ryan et al., 2010; Harter et al., 2017). The presence of SSI-1 is clonal among L. monocytogenes CCs 3, 5, 88, 224, and 315 (genetic lineage I) and CC7, 8, 9, 155, 193, 199, 204, and 321 (genetic lineage II; Hingston et al., 2017; Malekmohammadi et al., 2017). Clonality has also been observed for SSI-2, which is present in L. monocytogenes CC121 and as a genetic insert common in L. innocua strains (Hein et al., 2011).
Current genomic studies of Listeria sensu stricto and senu lato are enriching knowledge of the diversity and adaptability of phylotypes and genotypes, and some inferences can be made about L. monocytogenes. In fact, Liao et al. (2021) compiled a US-wide atlas of 1854 Listeria isolated from soil and determined through whole-genome analysis that the “cosmopolitan” phylotypes had a more open pangenome and were affected by positive selection, which was reflected in their core genome.
Molecular Determinants of Listeria monocytogenes Saprophytic Lifestyle
L. monocytogenes can survive and multiply in the complex and diverse environment of soil. Soil is usually composed of organic matter, plant roots, decaying vegetation, minerals, and a variety of microorganisms and invertebrates. The availability of nutrients, water, pH, and interactions among organisms living in the soil determine gene expression. The importance of soil and water sources to the genus Listeria is reinforced by the fact that newly described Listeria species and subspecies were often associated with these niches (Carlin et al., 2021a).
In general, sigma factors play an important role in metabolism and the transition to a pathogenic lifestyle. In particular, the SigB regulon (σB) has been identified as an important regulon controlling several functions, including stress response and virulence (Dorey et al., 2019). A recent systematic literature review on this topic identified 304 genes with σB-dependent consensus promoters encoding various functions, including stress response (e.g., osmotic, oxidative, and acid stress; n = 73) and virulence (n = 51; Liu et al., 2019).
The SigB controlled stress response mechanisms facilitates the adaptation of L. monocytogenes to the soil environment. Using a sigB deletion mutant, Gorski et al. (2011) observed the role of SigB, in the initial adaptation phase as well as long-term survival in soil associated with transition to and survival in stationary phase. It has been clearly demonstrated that SigB regulation is essential for the survival of L. monocytogenes in nature, although the factors activated by SigB under these severe conditions remain to be deciphered (Dorey et al., 2019).
The accessory gene regulator (Agr) system is an auto-inducible transcriptional regulator associated with virulence as well as biofilm formation. The Agr system is organized as a four-gene operon, agrBDCA. By using deletion mutants on agrA (response-negative mutant) or the signal pro-peptide AgrD (signal-negative mutant), Vivant et al. (2014b) demonstrated that the Agr communication system is required for optimal survival of L. monocytogenes in soil, especially when complex microbial communities are active. The transcriptome of an agrA deletion mutant also differs significantly from the wild type under soil conditions, in contrast to abiotic conditions. Vivant et al. (2014a) observed that 578 protein-coding genes related to the cell envelope and cellular processes, as well as several non-coding RNAs, were differentially transcribed. In addition, the Agr system positively regulates the chitinase expression (Paspaliari et al., 2014). Transcriptional microarray analysis during growth of Listeria in soil extracts revealed that other enzymes involved in catabolism and nutrient acquisition mechanisms also exhibited higher levels of transcription to respond to soil nutrient scarcity and to allow utilization of abundant carbon sources such as cellulose or chitin (Piveteau et al., 2011).
The SigB and Agr systems act synergistically, as the ability to colonize grass roots under sterile soil conditions was significantly impaired in the double mutant compared to the wild type and each of the single mutants. Under biotic conditions, strains viability decreased over time for both wild type and mutants, although this effect was stronger in the double mutant (Marinho et al., 2020).
Brunhede et al. (2020) demonstrated that the LisRK two-component system is important for growth in sterile soils and survival in non-sterile soils. The authors observed reduced fitness in two different soil types (silty clay and loamy sandy soil) of isogenic mutants of lisR and lisK compared to the wild-type strain. Loamy sandy soils showed a greater decrease in bacterial population when the experiment was conducted in non-sterile soil. A mutant lacking all seven sRNA-coding genes of the lhrC family was used to show that it is involved in the regulation of L. monocytogenes gene expression in soil. While residing as a saprophyte in the environment, L. monocytogenes may be uptaken by susceptible hosts and become an intracellular pathogen.
The key transcriptional regulators PrfA and SigB play a central role in mediating the switch between these two lifestyles. Detailed information on the interaction between these regulators can be found in a recent report by Gaballa et al. (2019).
Vivant et al. (2017) studied the changes in the transcriptome of L. monocytogenes CIP 110868 when adapted to lagoon effluents and soil. Genes involved in mobility, chemotaxis, carbohydrate transport, and the cell envelope were upregulated, especially in lagoon effluents.
Tan et al. (2016) studied the importance of L. monocytogenes attachment to plant cell walls and identified the role of pectin and xyloglucan important for the attachment process. Overall, the attachment was dependent on strain-specific properties but not directly influenced by the strain concentration.
Natural environments can be a reservoir for particularly virulent strains. Internalin A (InlA), which plays an important role in L. monocytogenes virulence, is intact in 90% of isolates from natural environments (Gorski et al., 2016), in contrast to isolates from food processing, where approximately 35% of isolates have a truncated InlA gene (Kovacevic et al., 2013). In food, processing environments and food L. monocytogenes sublineages (SLs) dominantly present are SL9, SL121, among others (SL101 and SL5), which harbor genes relevant for Benzalkonium chloride tolerance. Interestingly, 31% L. monocytogenes isolates indicated truncated inlA genes (Hurley et al., 2019).
Linke et al. (2014) observed in a subset of 16 isolates from soil that about two-thirds of the isolates had a truncated inlA gene, while one-third had an intact inlA gene. This is particularly relevant to adaptation to natural environmental stresses, as Hingston et al. (2017) observed that when L. monocytogenes with a full-length inlA gene were compared to those with a premature stop codon, the former was found to have significantly improved cold tolerance.
Raschle et al. (2021) reported hypervirulent L. monocytogenes CC1, CC4, CC6, and CC412 in river ecosystems. Therefore, the latter L. monocytogenes genotypes with intact inlA/B and prfA genes and LIP-4 (CC4) also have a reservoir in surface waters, which should be considered as a risk for transmission to humans and animals.
Antimicrobial Tolerance and Resistance in environmental Listeria monocytogenes
Currently, there are informative publications that critically illuminate the subject of L. monocytogenes and the tolerance or potential resistance to antimicrobials (Yin et al., 2019; Baquero et al., 2020). There are some papers that have focused on antimicrobial resistance in L. monocytogenes in the past and have used the term resistance without EUCAST or CLSI clinical breakpoints being available for each of these test substances. In the case of L. monocytogenes, the use of the term “increased tolerance” when observing increased MIC levels to a test substance without existing clinical breakpoints is certainly more appropriate. According to EUCAST, clinical breakpoints are available for penicillins, meropenem, erythromycin, and trimethoprim-sulfamethoxazole.1 Some authors expand the range of clinical breakpoints with the available ones of enterococci or staphylococci (Conter et al., 2009; Bertsch et al., 2014).
The number of antimicrobials to which L. monocytogenes was more tolerant or even resistant in literature were tetracycline, clindamycin, ampicillin, and erythromycin (Supplementary Table S1). The percentage of tolerant L. monocytogenes strains to antimicrobials varies among studies, but in some studies, all test strains were more tolerant to at least one antimicrobial agent.
L. monocytogenes is usually susceptible to penicillins, trimethoprim, aminoglycosides, macrolides, and vancomycin (Troxler et al., 2000; Bertsch et al., 2014), but most strains show native resistance to cephalosporins, first generation quinolones (nalidixic acid and cinoxacin), sulfamethoxazole, fosfomycin, oxacillin, and lincosamides (Olaimat et al., 2018).
The majority (88.5%) of L. monocytogenes isolated in a study by Linke et al. (2014) indicated a higher minimum inhibitory concentration (MIC) to cefotaxime and 35% of isolates to ceftriaxone. These results are also consistent with those of Troxler et al. (2000) who described a natural resistance of most Listeria strains to modern cephalosporins. Soni et al. (2014) also observed a higher tolerance to cefoxitin, a second-generation cephalosporin. In addition, 60% of the soil isolates were more tolerant to ciprofloxacin.
L. monocytogenes isolated from soil, water, and vegetables in Nigeria were reported to be resistant to the β-lactam group (ampicillin and amoxicillin) and to more than one antibiotic class (Oyinloye et al., 2018). Manjur et al. (2018) found a high percentage of strains resistant to ampicillin and penicillin (71%) and 100% resistance to erythromycin in urban soil samples. Recent articles recommend retesting the strain panel, because ampicillin resistance occurs in only a small percentage of isolates (<1%), due to possible horizontal gene transfer (HGT) of E. faecium (Baquero et al., 2020).
In some studies, certain L. monocytogens sequence types (STs) indicated increased tolerance or resistance to antibiotics: Yan et al. (2019) reported antimicrobial resistance (tetracycline, erythromycin, chloramphenicol, and trimethoprim-sulfamethoxazole) in ST3, ST8, ST9, ST155, and ST515 food isolates. The predominant ST9 indicated resistance acquired by horizontal gene transfer. Certain genotypes present in soil samples (ST1, ST2, ST6, ST7, ST20, ST37, and ST91) showed an increased tolerance to cephalosporine. ST7, ST20, ST2, and ST6 were more tolerant to erythromycin and ST20 to linezolid (Linke et al., 2014).
Antimicrobial increased tolerance or even resistance in L. monocytogenes can be caused by efflux pumps or the acquisition of genetic elements, like plasmids and transposons (Matereke and Okoh, 2020). Some of the resistances, such as those against fosfomycin, depend on PrfA-dependent cell uptake mechanisms and may only occur under lab conditions, because PrfA is downregulated when nutrients such as phosphorylated sugars are available (Scortti et al., 2006).
Kremer et al. (2017) described a plasmid and efflux transporter in L. monocytogenes that lead to benzalkonium chloride tolerance in combination with higher MICs for gentamicin and amoxicillin. Since benzalkonium chloride is used for disinfection in food processing plants, co-selection of resistance mechanisms could lead to difficulties in listeriosis treatment. Oliveira et al. (2017) described that L. monocytogenes strains adapted to subinhibitory concentrations of resveratrol, a natural product used as a food preservative, were more tolerant to heat and acid stress.
Strains exposed to environmental stress may be resistant to higher levels of antibiotics due to stress adaptation (Lungu et al., 2011). Accumulation of heavy metals, such as mercury, cadmium zinc, arsenic, copper, and lead in the environment, leads to co-selection of antibiotic resistance in Gram-positive bacteria. Metal-related antibiotic-resistant determinants that reside on the same mobile genetic elements can be horizontally transferred from the bacteria in which they evolved (e.g., staphylococci, enterococci, and lactic acid bacteria) to related pathogens such as L. monocytogenes (Imran et al., 2019; Parsons et al., 2020).
Recently, microplastics polluting the marine environment were found to harbor metals, antibiotics, and pathogenic bacteria and provide a favorable environment for metal-induced co-selection of multidrug-resistant pathogens (Imran et al., 2019). Zhao et al. (2019) investigated antibiotic resistance genes (ARGs) in metal-contaminated urban and suburban soils collected in the western part of Northern Ireland and detected 164 ARGs. Sample types included stream water and sediment, heavy mineral concentrate, surface, and deep soil (Smyth, 2007). The authors detected an average of 3.4 × 107 ARG gene copies per gram of soil and observed a correlation between the presence of heavy metals in soil and the spread of ARGs through horizontal gene transfer.
This indicates the need to further elucidate the mechanisms behind antimicrobial resistance and cross-resistance and to raise awareness of the impact of the use of preservatives, disinfectants, and environmental contaminants.
Listeria in the Environment and its Entrance Into Food Processing better facilities
Hof (2003) provided an overview of the history and epidemiology of listeriosis and referred to the work of Schlech et al. (1983) as a milestone in listeriosis research, because it established the link between the disease and a contaminated food—coleslaw. Indeed, Listeria that occur in the natural environment often find their way into food processing facilities and the food chain through raw materials. Proximity to wildlife, birds and livestock and fecal contamination are closely related to the prevalence of L. monocytogenes during crop production. As mentioned earlier, pre-harvest conditions, such as manure application and irrigation water quality, not only determine the prevalence of Listeria in produce, but also favor the presence of Listeria at levels that are not manageable and controllable with established practices in the industry.
Several crops that were contaminated before harvest have been implicated in foodborne outbreaks, namely, mushrooms, celery, melons, apples, lettuce, and corn (European Food Safety Authority and European Centre for Disease Prevention and Control, 2018; Centers for Disease Control and Prevention, 2021). Increased rainfall or flooding is associated with an increased prevalence of zoonotic pathogens in production environments and also in natural environments (Linke et al., 2014; Weller et al., 2015b). The contamination of fresh vegetables with L. monocytogenes could be reduced by minimizing irrigating prior to harvesting. The presence of L. monocytogenes on seeds facilitates its entry into the plants during its germination. Therefore, seed decontamination is a potential promising method to reduce microbial internalization and diffusion in crops (Miceli and Settanni, 2019).
Practices such as sanitizing food and disinfecting food-contact surfaces are necessary to ensure food safety and, if done inadequately, can lead to contamination of produce during subsequent operations such as cutting and slicing (Zhu et al., 2017). Marik et al. (2020) highlight the importance of produce surface topography and its background microbiota of L. monocytogenes growth and survival characteristics on intact fruit and vegetable surfaces during postharvest handling.
Listeria is ingested not only by wild animals but also by farm animals and excreted asymptomatically without causing clinical signs. In most cases, L. monocytogenes can be isolated from the intestines of healthy deer and wild boar (Weindl et al., 2016) but also cattle, sheep, and pigs (Iida et al., 1991; Hurtado et al., 2017). Palacios-Gorba et al. (2021) monitored the presence of Listeria in 19 dairy farms during seven consecutive seasons from autumn 2018 to spring 2020 in cattle, goat, and sheep farms. The authors detected the presence of Listeria in 18 dairy farms with L. monocytogenes being the second most prevalent species after L. innocua. Moreover, the authors observed that within L. monocytogenes 70% of the isolates belonged to lineages I. The authors observed as the most prevalent SLs and CCs for animal samples SL1/CC1, SL219/CC4, SL26/CC26, and SL87/CC87 and for the environmental samples SL666/CC666. Therefore, Listeria can be shed from farm animals in the agricultural environment and introduced into the food-processing environment if adequate hygiene barriers are not in place. Food processing plant employees can be a source of contamination of the food processing environment due to inadequate hygiene practices or lack of or inadequate protective equipment (Stessl et al., 2020). However, practices in the industry, in animal harvesting and processing plants, are particularly delicate. More than the apparent cleanliness of the animal’s hide, the processes of skinning and evisceration are crucial to the contamination of a carcass with soil and fecal matter, which can then lead to permanent contamination of the processing environment (Demaître et al., 2021).
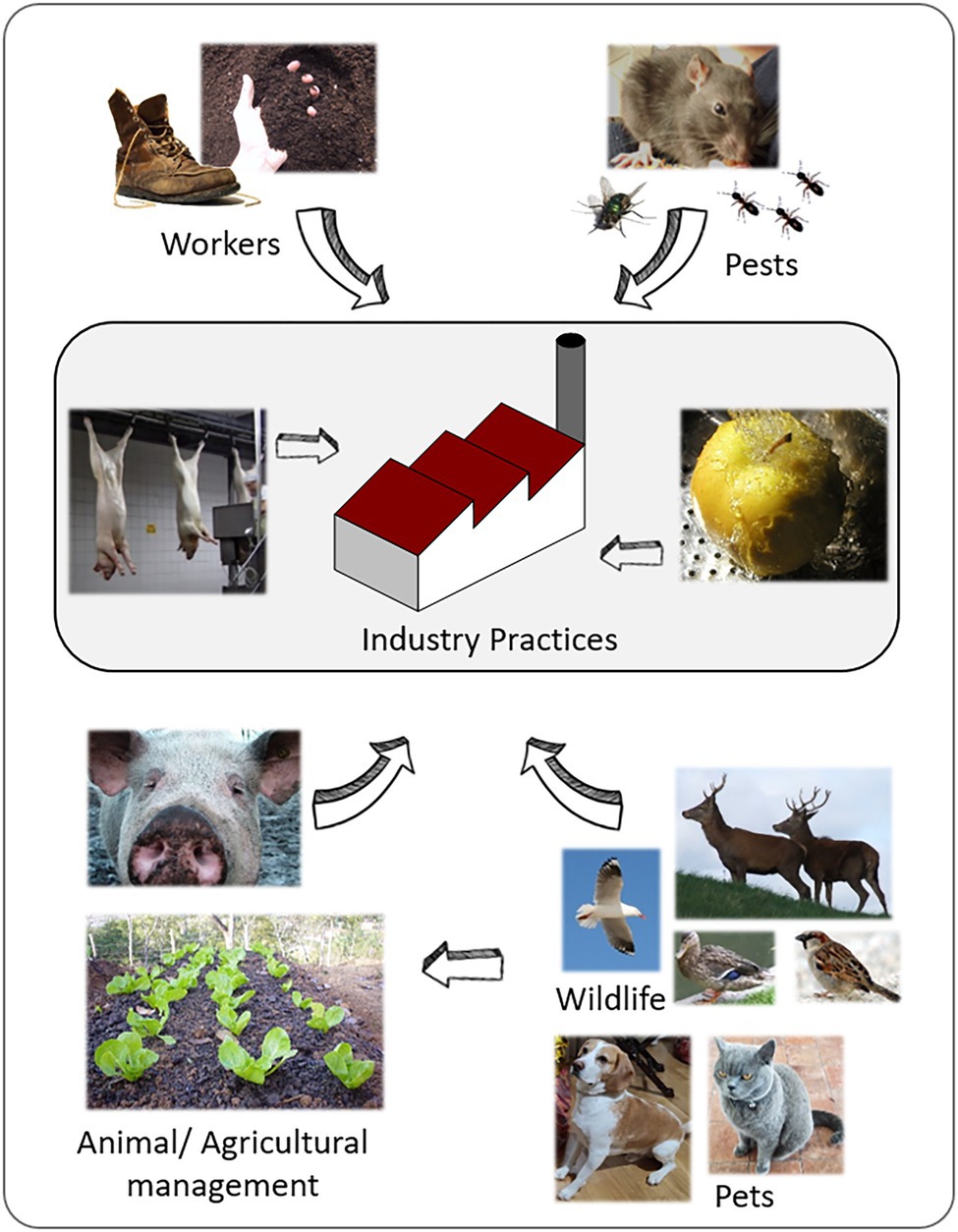
Many biotic vectors can play an important role in the transmission of Listeria between the environment and food processing facilities, either through direct interaction with the food or animals or as a direct source in a food processing environment. Rodents and insects such as flies and cockroaches have been shown to be a source of Listeria (Wang et al., 2017; Li et al., 2018). Wang et al. (2017) observed that Listeria, especially L. monocytogenes ST87, is present at low levels in the intestinal tracts of wild rodents in the natural environment of China. This is highly relevant as ST87 is one of the most common STs present in the food chain and human listeriosis in China (Li et al., 2018). Figure 3 summarizes the most common routs of entry of environmental Listeria into food processing facilities. In detail, the importance of food handling practices within the food facility, the compliance to good manufacturing practices in terms of worker hygiene and pest control, and the role L. monocytogenes transmission by wild and domestic animals. The latter might contaminate raw material or food processing environment. Furthermore, environmental factors (e.g., the quality of irrigation water) influence the produce safety.
Conclusion
The genus Listeria currently comprised by 28 species just and as many other Gram-positive microorganisms its occurrence is associated to the natural environmental habitats such as soil and decaying vegetation. However, Listeria’s prevalence in the saprosphere is greatly dependent on many biotic and abiotic factor. Among those factors, the soil type and its characteristics such as pH and mineral composition are determinant. The proximity to water sources is also extremely relevant for the population dynamics. Other factors such as meteorological factors, geographic, and seasonal variations are referred to as relevant. Many biotic factors related to human and other animal density and vegetation have been suggested as explanations for the presence or absence of Listeria. Differences in isolation among the different species are often attributed to differential fecal shedding and nutrient usage ability. Moreover, many are the molecular determinants that condition the fitness to the saprophytic lifestyle. The SigB regulon, the Agr system, and the LisRK system determine the fate of Listeria in the environment along with the PrfA regulon, which is determinant for the transition to intracellular pathogen lifestyle.
Exposure to environmental stresses, including heavy metals, is of particular importance because it determines the fate of mobile genetic elements that may be related not only to environmental fitness but also to antibiotic resistance, which is an obvious problem for human and animal health. Within the genus, L. monocytogenes is a relevant human pathogen that is frequently transmitted through food and causes sporadic and epidemic listeriosis in high-risk groups. This foodborne pathogen finds its way from the saprosphere into the food chain by a wide range of routs that range from the raw materials that may be naturally contaminated or become contaminated by inadequate procedures, the presence of wildlife, pets or pest naturally carriers, or by bad practices of food processing facilities workers.
Only the full awareness of the factors that determine the presence of Listeria in the natural environment can lead to the improvement of the agronomical and zootechnical practices capable of preventing the entrance of Listeria in the primary production. Improving industrial practices, including creating appropriate training for personnel that will effectively prevent the occurrence and selection of this microorganism in a food environment, must also be based on a thorough knowledge of the saprophytic lifestyle of Listeria.
Author Contributions
AL and KL prepared the manuscript, figures, and tables. BS and MW designed the review, raised funds, supervised the work, and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was partly funded by the Austrian Competence Centre for Feed and Food Quality, Safety and Innovation (FFoQSI). The COMET-K1 competence centre FFoQSI is funded by the Austrian ministries BMVIT and BMDW, and the Austrian provinces Niederoesterreich, Upper Austria and Vienna, within the scope of COMET—Competence 301 Centers for Excellent Technologies. The COMET program is handled by the Austrian Research Promotion Agency FFG. AL is funded by the Research Leaders 2025 postdoctoral funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement number 754380.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The data on Listeria species presented on Figure 1 and clonal complexes of L. monocytogenes presented on Figure 2 are publicly available at BIGSdb-Listeria. We thank the Institute Pasteur teams for the curation and maintenance of BIGSdb-Pasteur databases at http://bigsdb.pasteur.fr.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.789801/full#supplementary-material
Supplementary Table S1 | Percentage of antibiotic tolerance or resistance of Listeria monocytogenes mentioned in literature in regards to sample type and geographic origin.
Footnotes
References
Ahlstrom, C. A., Manuel, C. S., Den Bakker, H. C., Wiedmann, M., and Nightingale, K. K. (2018). Molecular ecology of Listeria spp., Salmonella, Escherichia coli O157:H7 and non-O157 Shiga toxin-producing E. coli in pristine natural environments in Northern Colorado. J. Appl. Microbiol. 124, 511–521. doi: 10.1111/jam.13657
Allerberger, F., and Wagner, M. (2010). Listeriosis: a resurgent foodborne infection. Clin. Microbiol. Infect. 16, 16–23. doi: 10.1111/j.1469-0691.2009.03109.x
Baquero, F., Lanza, V. F., Duval, M., and Coque, T. M. (2020). Ecogenetics of antibiotic resistance in Listeria monocytogenes. Molecular Microbiology 113, 570–579. doi: 10.1111/mmi.14454
Barre, L., Angelidis, A. S., Boussaid, D., Brasseur, E. D., Manso, E., and Gnanou Besse, N. (2016). Applicability of the EN ISO 11290-1 standard method for Listeria monocytogenes detection in presence of new listeria species. Int. J. Food Microbiol. 238, 281–287. doi: 10.1016/j.ijfoodmicro.2016.09.028
Bertsch, D., Muelli, M., Weller, M., Uruty, A., Lacroix, C., and Meile, L. (2014). Antimicrobial susceptibility and antibiotic resistance gene transfer analysis of foodborne, clinical, and environmental Listeria spp. isolates including Listeria monocytogenes. Microbiology 3, 118–127. doi: 10.1002/mbo3.155
Bertsch, D., Rau, J., Eugster, M. R., Haug, M. C., Lawson, P. A., Lacroix, C., et al. (2013). Listeria fleischmannii sp. nov., isolated from cheese. Int. J. Syst. Evol. Microbiol. 63, 526–532. doi: 10.1099/ijs.0.036947-0
Boerlin, P., Rocourt, J., Grimont, F., Grimont, P. A. D., Jacquet, C., and Piffaretti, J. C. (1992). Listeria ivanovii subsp. Londoniensis subsp. nov. Int. J. Syst. Bacteriol. 42, 69–73. doi: 10.1099/00207713-42-1-69
Brunhede, M. Z., Santos, T. D., Gal, L., Garmyn, D., Kallipolitis, B. H., and Piveteau, P. (2020). LisRK is required for optimal fitness of Listeria monocytogenes in soil. FEMS Microbiol. Lett. 367:fnaa188. doi: 10.1093/femsle/fnaa188
Buchanan, R. L., Gorris, L. G., Hayman, M. M., Jackson, T. C., and Whiting, R. C. (2017). A review of Listeria monocytogenes: an update on outbreaks, virulence, dose-response, ecology, and risk assessments. Food Control 75, 1–13. doi: 10.1016/j.foodcont.2016.12.016
Carlin, C. R., Liao, J., Weller, D., Guo, X., Orsi, R., and Wiedmann, M. (2021a). Listeria cossartiae sp. Nov., Listeria immobilis sp. nov., Listeria portnoyi sp. nov. and Listeria rustica sp. nov., isolated from agricultural water and natural environments. Int. J. Syst. Evol. Microbiol. 71:004795. doi: 10.1099/ijsem.0.004795
Carlin, C. R., Liao, J., Weller, D. L., Guo, X., Orsi, R., and Wiedmann, M. (2021b). Corrigendum to: Listeria cossartiae sp. nov., Listeria farberi sp. nov., Listeria immobilis sp. nov., Listeria portnoyi sp. nov. and Listeria rustica sp. nov., isolated from agricultural water and natural environments. Int. J. Syst. Evol. Microbiol. 71:13210. doi: 10.1099/ijsem.0.004885
Carlin, C. R., Roof, S., and Wiedmann, M. (2021c). An assessment of reference method selective broths and plating media using 19 Listeria spp. highlights the importance of including diverse species in Listeria spp. method evaluations. J. Food Prot. doi: 10.4315/JFP-21-293 [Epub ahead of print]
Castro-Ibáñez, I., Gil, M. I., Tudela, J. A., and Allende, A. (2015). Microbial safety considerations of flooding in primary production of leafy greens: a case study. Food Res. Int. 68, 62–69. doi: 10.1016/j.foodres.2014.05.065
Centers for Disease Control and Prevention (2021). Listeria Outbreaks. Available at: https://www.cdc.gov/listeria/outbreaks/index.html (Accessed February 15, 2022).
Chapin, T. K., Nightingale, K. K., Worobo, R. W., Wiedmann, M., and Strawn, L. K. (2014). Geographical and meteorological factors associated with isolation of Listeria species in New York State produce production and natural environments. J. Food Prot. 77, 1919–1928. doi: 10.4315/0362-028X.JFP-14-132
Commichaux, S., Javkar, K., Ramachandran, P., Nagarajan, N., Bertrand, D., Chen, Y., et al. (2021). Evaluating the accuracy of Listeria monocytogenes assemblies from quasimetagenomic samples using long and short reads. BMC Genomics 22:389. doi: 10.1186/s12864-021-07702-2
Conter, M., Paludi, D., Zanardi, E., Ghidini, S., Vergara, A., and Ianieri, A. (2009). Characterization of antimicrobial resistance of foodborne Listeria monocytogenes. Int. J. Food Microbiol. 128, 497–500. doi: 10.1016/j.ijfoodmicro.2008.10.018
Cooley, M. B., Quiñones, B., Oryang, D., Mandrell, R. E., Gorski, L., Mclean, R. J. C., et al. (2014). Prevalence of Shiga toxin producing Escherichia coli, Salmonella enterica, and Listeria monocytogenes at public access watershed sites in a California Central Coast agricultural region. Front. Cell. Infect. Microbiol. 4:30. doi: 10.3389/fcimb.2014.00030
Cossart, P. (2011). Illuminating the landscape of host-pathogen interactions with the bacterium Listeria monocytogenes. Proc. Natl. Acad. Sci. U. S. A. 108, 19484–19491. doi: 10.1073/pnas.1112371108
Curtis, G. D. W., and Lee, W. H. (1995). Culture media and methods for the isolation of Listeria monocytogenes. Int. J. Food Microbiol. 26, 1–13. doi: 10.1016/0168-1605(93)E0027-O
Demaître, N., De Reu, K., Haegeman, A., Schaumont, D., De Zutter, L., Geeraerd, A., et al. (2021). Study of the transfer of Listeria monocytogenes during the slaughter of cattle using molecular typing. Meat Sci. 175:108450. doi: 10.1016/j.meatsci.2021.108450
den Bakker, H. C., Didelot, X., Fortes, E. D., Nightingale, K. K., and Wiedmann, M. (2008). Lineage specific recombination rates and microevolution in Listeria monocytogenes. BMC Evol. Biol. 8:277. doi: 10.1186/1471-2148-8-277
den Bakker, H. C., Manuel, C. S., Fortes, E. D., Wiedmann, M., and Nightingale, K. K. (2013). Genome sequencing identifies Listeria fleischmannii subsp. Coloradonensis subsp. nov., isolated from a ranch. Int. J. Syst. Evol. Microbiol. 63, 3257–3268. doi: 10.1099/ijs.0.048587-0
den Bakker, H. C., Warchocki, S., Wright, E. M., Allred, A. F., Ahlstrom, C., Manuel, C. S., et al. (2014). Listeria floridensis sp. nov., Listeria aquatica sp. nov., Listeria cornellensis sp. nov., Listeria riparia sp. nov. and Listeria grandensis sp. nov., from agricultural and natural environments. Int. J. Syst. Evol. Microbiol. 64, 1882–1889. doi: 10.1099/ijs.0.052720-0
Doijad, S. P., Poharkar, K. V., Kale, S. B., Kerkar, S., Kalorey, D. R., Kurkure, N. V., et al. (2018). Listeria goaensis sp. Nov. Int. J. Syst. Evol. Microbiol. 68, 3285–3291. doi: 10.1099/ijsem.0.002980
Dorey, A., Marinho, C., Piveteau, P., and O’Byrne, C. (2019). Role and regulation of the stress activated sigma factor sigma B (σB) in the saprophytic and host-associated life stages of Listeria monocytogenes. Adv. Appl. Microbiol. 106, 1–48. doi: 10.1016/bs.aambs.2018.11.001
Doumith, M., Buchrieser, C., Glaser, P., Jacquet, C., and Martin, P. (2004). Differentiation of the major Listeria monocytogenes Serovars by multiplex PCR. J. Clin. Microbiol. 42, 3819–3822. doi: 10.1128/JCM.42.8.3819-3822.2004
EFSA (2018). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J. 16:5500. doi: 10.2903/j.efsa.2018.5500
EFSA and ECDC (2015). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2014. EFSA J. 13:4329. doi: 10.2903/j.efsa.2015.4139
EFSA and ECDC (2019). The European Union one health 2018 Zoonoses report. EFSA J. 17:e05926. doi: 10.2903/j.efsa.2019.5926
Etty, M. C., D’Auria, S., Fraschini, C., Salmieri, S., and Lacroix, M. (2019). Effect of the optimized selective enrichment medium on the expression of the p60 protein used as Listeria monocytogenes antigen in specific sandwich ELISA. Res. Microbiol. 170, 182–191. doi: 10.1016/j.resmic.2019.03.004
European Food Safety Authority and European Centre for Disease Prevention and Control (2018). Multi-country outbreak of Listeria monocytogenes serogroup IVb, multi-locus sequence type 6, infections probably linked to frozen corn. EFSA Support. Publ. 15.
Farber, J. M., and Peterkin, P. I. (1991). Listeria monocytogenes, a food-borne pathogen. Microbiol. Mol. Biol. Rev. 55, 476–511. doi: 10.1128/mr.55.3.476-511.1991
Fox, E., O’mahony, T., Clancy, M., Dempsey, R., O’brien, M., and Jordan, K. (2009). Listeria monocytogenes in the Irish Dairy Farm Environment. Available at: http://meridian.allenpress.com/jfp/article-pdf/72/7/1450/1680255/0362-028x-72_7_1450.pdf (Accessed February 15, 2022).
Gaballa, A., Guariglia-oropeza, V., Wiedmann, M., and Boor, J. (2019). Cross talk between SigB and PrfA in Listeria monocytogenes facilitates transitions between extra- and intracellular environments. Microbiol. Mol. Biol. Rev. 83, 1–25. doi: 10.1128/MMBR.00034-19
Gorski, L., Duhé, J. M., and Flaherty, D. (2011). The sigma B operon is a determinant of fitness for a Listeria monocytogenes serotype 4b strain in soil. Foodborne Pathog. Dis. 8, 699–704. doi: 10.1089/fpd.2010.0752
Gorski, L., Flaherty, D., and Mandrell, R. E. (2006). Competitive fitness of Listeria monocytogenes serotype 1/2a and 4b strains in mixed cultures with and without food in the U.S. Food and Drug Administration Enrichment Protocol. Appl. Environ. Microbiol. 72, 776–783. doi: 10.1128/AEM.72.1.776-783.2006
Gorski, L., Parker, C. T., Liang, A. S., Walker, S., and Romanolo, K. F. (2016). The majority of genotypes of the virulence gene inlA are intact among natural watershed isolates of Listeria monocytogenes from the Central California Coast. PLoS One 11:e0167566. doi: 10.1371/journal.pone.0167566
Gorski, L., Walker, S., Liang, A. S., Nguyen, K. M., Govoni, J., Carychao, D., et al. (2014). Comparison of subtypes of Listeria monocytogenes isolates from naturally contaminated watershed samples with and without a selective secondary enrichment. PLoS One 9:e92467. doi: 10.1371/journal.pone.0092467
Graves, L. M., Helsel, L. O., Steigerwalt, A. G., Morey, R. E., Daneshvar, M. I., Roof, S. E., et al. (2010). Listeria marthii sp. nov., isolated from the natural environment, Finger Lakes National Forest. Int. J. Syst. Evol. Microbiol. 60, 1280–1288. doi: 10.1099/ijs.0.014118-0
Gray, M. L. (1960). Isolation of Listeria monocytogenes from oat silage. Science 132, 1767–1768. doi: 10.1126/science.132.3441.1767
Gray, M. L., Stafseth, H. J., Thorp, F., Sholl, L. B., and Riley, W. F. (1948). A new technique for isolating Listerellae from the bovine brain. J. Bacteriol. 55, 471–476. doi: 10.1128/jb.55.4.471-476.1948
Guillet, C., Join-Lambert, O., Le Monnier, A., Leclercq, A., Mechaï, F., Mamzer-Bruneel, M. F., et al. (2010). Human listeriosis caused by Listeria ivanovii. Emerg. Infect. Dis. 16, 136–138. doi: 10.3201/eid1601.091155
Halbedel, S., Prager, R., Fuchs, S., Trost, E., Werner, G., and Flieger, A. (2018). Whole-genome sequencing of recent Listeria monocytogenes isolates from Germany reveals population structure and disease clusters. J. Clin. Microbiol. 56, e00119–e00118. doi: 10.1128/JCM.00119-18
Halter, E. L., Neuhaus, K., and Scherer, S. (2013). Listeria weihenstephanensis sp. nov., isolated from the water plant Lemna trisulca taken from a freshwater pond. Int. J. Syst. Evol. Microbiol. 63, 641–647. doi: 10.1099/ijs.0.036830-0
Harter, E., Wagner, E. M., Zaiser, A., Halecker, S., Wagner, M., and Rychli, K. (2017). Stress survival islet 2, predominantly present in Listeria monocytogenes strains of sequence type 121, is involved in the alkaline and oxidative stress responses. Appl. Environ. Microbiol. 83, e00827–e00817. doi: 10.1128/AEM.00827-17
Hayes, P. S., Graves, L. M., Ajello, G. W., Swaminathan, B., Weaver, R. E., Wenger, J. D., et al. (1991). Comparison of cold enrichment and U.S. Department of Agriculture methods for isolating Listeria monocytogenes from naturally contaminated foods. Appl. Environ. Microbiol. 57, 2109–2113. doi: 10.1128/aem.57.8.2109-2113.1991
Hein, I., Klinger, S., Dooms, M., Flekna, G., Stessl, B., Leclercq, A., et al. (2011). Stress survival islet 1 (SSI-1) survey in Listeria monocytogenes reveals an insert common to Listeria innocua in sequence type 121 L. monocytogenes strains. Appl. Environ. Microbiol. 77, 2169–2173. doi: 10.1128/AEM.02159-10
Hellström, S., Kiviniemi, K., Autio, T., and Korkeala, H. (2008). Listeria monocytogenes is common in wild birds in Helsinki region and genotypes are frequently similar with those found along the food chain. J. Appl. Microbiol. 104, 883–888. doi: 10.1111/j.1365-2672.2007.03604.x
Hingston, P., Chen, J., Dhillon, B. K., Laing, C., Bertelli, C., Gannon, V., et al. (2017). Genotypes associated with Listeria monocytogenes isolates displaying impaired or enhanced tolerances to cold, salt, acid, or desiccation stress. Front. Microbiol. 8:369. doi: 10.3389/fmicb.2017.00369
Hitchins, A. D., Jinneman, K., and Chen, Y. (2017). BAM Chapter 10: Detection of Listeria monocytogenes in Foods and Environmental Samples, and Enumeration of Listeria monocytogenes in Foods | FDA. Available at: https://www.fda.gov/food/laboratory-methods-food/bam-chapter-10-detection-listeria-monocytogenes-foods-and-environmental-samples-and-enumeration (Accessed February 19, 2021).
Hof, H. (2003). History and epidemiology of listeriosis. FEMS Immunol. Med. Microbiol. 35, 199–202. doi: 10.1016/S0928-8244(02)00471-6
Hurley, D., Luque-Sastre, L., Parker, C. T., Huynh, S., Eshwar, A. K., Nguyen, S. V., et al. (2019). Whole-genome sequencing-based characterization of 100 Listeria monocytogenes isolates collected from food processing environments over a four-year period. mSphere 4, e00252–e00219. doi: 10.1128/mSphere.00252-19
Hurtado, A., Ocejo, M., and Oporto, B. (2017). Salmonella spp. and Listeria monocytogenes shedding in domestic ruminants and characterization of potentially pathogenic strains. Vet. Microbiol. 210, 71–76. doi: 10.1016/j.vetmic.2017.09.003
Iida, T., Kanzaki, M., Maruyama, T., Inoue, S., and Kaneuchi, C. (1991). Prevalence of Listeria monocytogenes in intestinal contents of healthy animals in Japan. J. Vet. Med. Sci. 53, 873–875. doi: 10.1292/jvms.53.873
Imran, M., Das, K. R., and Naik, M. M. (2019). Co-selection of multi-antibiotic resistance in bacterial pathogens in metal and microplastic contaminated environments: an emerging health threat. Chemosphere 215, 846–857. doi: 10.1016/j.chemosphere.2018.10.114
ISO 11290-1 (2017). Microbiology of the food chain — Horizontal method for the detection and enumeration of Listeria monocytogenes and of Listeria spp. — Part 1: Detection method. Geneva, Switzerland: International Organization for Standardization.
Iwu, C. D., and Okoh, A. I. (2020). Characterization of antibiogram fingerprints in Listeria monocytogenes recovered from irrigation water and agricultural soil samples. PLoS One 15:e0228956. doi: 10.1371/journal.pone.0228956
Kaszoni-Rückerl, I., Mustedanagic, A., Muri-Klinger, S., Brugger, K., Wagner, K. H., Wagner, M., et al. (2020). Predominance of distinct Listeria innocua and Listeria monocytogenes in recurrent contamination events at dairy processing facilities. Microorganisms 8:234. doi: 10.3390/microorganisms8020234
Kathariou, S., Kanenaka, R., Allen, R. D., Fok, A. K., and Mizumoto, C. (1995). Repression of motility and flagellin production at 37°C is stronger in Listeria monocytogenes than in the nonpathogenic species Listeria innocua. Can. J. Microbiol. 41, 572–577. doi: 10.1139/m95-076
Keys, A. L., Hitchins, A. D., and Derike Smiley, R. (2016). Contribution of selective conditions to microbial competition in four Listeria selective enrichment formulations HHS public access. J. Food Prot. 79, 1904–1910. doi: 10.4315/0362-028X.JFP-16-216
Kovacevic, J., Arguedas-Villa, C., Wozniak, A., Tasara, T., and Allen, K. J. (2013). Examination of food chain-derived Listeria monocytogenes strains of different serotypes reveals considerable diversity in inlA genotypes, mutability, and adaptation to cold temperatures. Appl. Environ. Microbiol. 79, 1915–1922. doi: 10.1128/AEM.03341-12
Kovacevic, J., Wałecka-Zacharska, E., Kitts, D. D., Ziegler, J., Gilmour, M. W., Wałecka-Zacharska, E., et al. (2016). Tolerance of Listeria monocytogenes to quaternary ammonium sanitizers is mediated by a novel efflux pump encoded by emrE. Appl. Environ. Microbiol. 82, 939–953. doi: 10.1128/AEM.03741-15
Kremer, P. H. C., Lees, J. A., Koopmans, M. M., Ferwerda, B., Arends, A. W. M., Feller, M. M., et al. (2017). Benzalkonium tolerance genes and outcome in Listeria monocytogenes meningitis. Clin. Microbiol. Infect. 23, 265.e1–265.e7. doi: 10.1016/j.cmi.2016.12.008
Law, J. W. F., Ab Mutalib, N. S., Chan, K. G., and Lee, L. H. (2015). An insight into the isolation, enumeration, and molecular detection of Listeria monocytogenes in food. Front. Microbiol. 6:1227. doi: 10.3389/fmicb.2015.01227
Leclercq, A., Clermont, D., Bizet, C., Grimont, P. A. D., Le Flèche-Matéos, A., Roche, S. M., et al. (2010). Listeria rocourtiae sp. nov. Int. J. Syst. Evol. Microbiol. 60, 2210–2214. doi: 10.1099/ijs.0.017376-0
Leclercq, A., Moura, A., Vales, G., Tessaud-Rita, N., Aguilhon, C., and Lecuit, M. (2019). Listeria thailandensis sp. nov. Int. J. Syst. Evol. Microbiol. 69, 74–81. doi: 10.1099/ijsem.0.003097
Lee, S., Ward, T. J., Jima, D. D., Parsonsa, C., and Kathariou, S. (2017). The arsenic resistance-associated listeria genomic Island LGI2 exhibits sequence and integration site diversity and a propensity for three Listeria monocytogenes clones with enhanced virulence. Appl. Environ. Microbiol. 83, e01189–e01117. doi: 10.1128/AEM.01189-17
Li, H., Wang, P., Lan, R., Luo, L., Cao, X., Wang, Y., et al. (2018). Risk factors and level of Listeria monocytogenes contamination of raw pork in retail markets in China. Front. Microbiol. 9:1090. doi: 10.3389/fmicb.2018.01090
Liao, J., Guo, X., Weller, D. L., Pollak, S., Buckley, D. H., Wiedmann, M., et al. (2021). Nationwide genomic atlas of soil-dwelling Listeria reveals effects of selection and population ecology on pangenome evolution. Nat. Microbiol. 6, 1021–1030. doi: 10.1038/s41564-021-00935-7
Linke, K., Rückerl, I., Brugger, K., Karpiskova, R., Walland, J., Muri-Klinger, S., et al. (2014). Reservoirs of Listeria species in three environmental ecosystems. Appl. Environ. Microbiol. 80, 5583–5592. doi: 10.1128/AEM.01018-14
Liu, Y., Orsi, R. H., Gaballa, A., Wiedmann, M., Boor, K. J., and Guariglia-Oropeza, V. (2019). Systematic review of the Listeria monocytogenes σ regulon supports a role in stress response, virulence and metabolism. Future Microbiol. 14, 801–828. doi: 10.2217/fmb-2019-0072
Locatelli, A., Depret, G., Jolivet, C., Henry, S., Dequiedt, S., Piveteau, P., et al. (2013). Nation-wide study of the occurrence of Listeria monocytogenes in French soils using culture-based and molecular detection methods. J. Microbiol. Methods 93, 242–250. doi: 10.1016/j.mimet.2013.03.017
Lomonaco, S., Nucera, D., and Filipello, V. (2015). The evolution and epidemiology of Listeria monocytogenes in Europe and the United States. Infect. Genet. Evol. 35, 172–183. doi: 10.1016/j.meegid.2015.08.008
Lungu, B., O’Bryan, C. A., Muthaiyan, A., Milillo, S. R., Johnson, M. G., Crandall, P. G., et al. (2011). Listeria monocytogenes: antibiotic resistance in food production. Foodborne Pathog. Dis. 8, 569–578. doi: 10.1089/fpd.2010.0718
Lyautey, E., Lapen, D. R., Wilkes, G., McCleary, K., Pagotto, F., Tyler, K., et al. (2007). Distribution and characteristics of Listeria monocytogenes isolates from surface waters of the south Nation River watershed, Ontario, Canada. Appl. Environ. Microbiol. 73, 5401–5410. doi: 10.1128/AEM.00354-07
Malekmohammadi, S., Kodjovi, K. K., Sherwood, J., and Bergholz, T. M. (2017). Genetic and environmental factors influence Listeria monocytogenes nisin resistance. J. Appl. Microbiol. 123, 262–270. doi: 10.1111/jam.13479
Manjur, M. S. E., Siddique, S., and Ahmed, S. (2018). Multi-drug resistant pathogenic Listeria monocytogenes in surface water and soil samples of Dhaka city. Bangladesh J. Microbiol. 33, 39–42. doi: 10.3329/bjm.v33i1.39602
Marik, C. M., Zuchel, J., Schaffner, D. W., and Strawn, L. K. (2020). Growth and survival of Listeria monocytogenes on intact fruit and vegetable surfaces during postharvest handling: a systematic literature review. J. Food Prot. 83, 108–128. doi: 10.4315/0362-028X.JFP-19-283
Marinho, C. M., Garmyn, D., Gal, L., Brunhede, M. Z., O’Byrne, C., and Piveteau, P. (2020). Investigation of the roles of Agra and σb regulators in Listeria monocytogenes adaptation to roots and soil. FEMS Microbiol. Lett. 367:fnaa036. doi: 10.1093/femsle/fnaa036
Matereke, L. T., and Okoh, A. I. (2020). Listeria monocytogenes virulence, antimicrobial resistance and environmental persistence: a review. Pathogens, 9, 528. doi: 10.3390/pathogens9070528
Miceli, A., and Settanni, L. (2019). Influence of agronomic practices and pre-harvest conditions on the attachment and development of Listeria monocytogenes in vegetables. Ann. Microbiol. 69, 185–199. doi: 10.1007/s13213-019-1435-6
Moshtaghi, H., Garg, S. R., and Mandokhot, U. V. (2003). Prevalence of Listeria in soil. Indian J. Exp. Biol. 41, 1466–1468.
Moura, A., Criscuolo, A., Pouseele, H., Maury, M. M., Leclercq, A., Tarr, C., et al. (2016). Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat. Microbiol. 2:16185. doi: 10.1038/nmicrobiol.2016.185
Muhterem-Uyar, M., Ciolacu, L., Wagner, K. H., Wagner, M., Schmitz-Esser, S., and Stessl, B. (2018). New aspects on Listeria monocytogenes ST5-ECVI predominance in a heavily contaminated cheese processing environment. Front. Microbiol. 9:64. doi: 10.3389/fmicb.2018.00064
Müller, A., Rychli, K., Muhterem-Uyar, M., Zaiser, A., Stessl, B., Guinane, C. M., et al. (2013). Tn6188 - A novel transposon in Listeria monocytogenes responsible for tolerance to benzalkonium chloride. PLoS One 8:e76835. doi: 10.1371/journal.pone.0076835
Murray, E. G. D., Webb, R. A., and Swann, M. B. R. (1926). A disease of rabbits characterised by a large mononuclear leucocytosis, caused by a hitherto undescribed bacillus Bacterium monocytogenes (n.sp.). J. Pathol. Bacteriol. 29, 407–439. doi: 10.1002/path.1700290409
Nassirabady, N., Meghdadi, H., and Alami, A. (2015). Isolation of Listeria monocytogenes of Karun River (enviromental sources rural and urban) by culture and PCR assay. Int. J. Enteric Pathog. 3:21829. doi: 10.17795/ijep21829
Nightingale, K., Bovell, L., Grajczyk, A., and Wiedmann, M. (2007). Combined sigB allelic typing and multiplex PCR provide improved discriminatory power and reliability for Listeria monocytogenes molecular serotyping. J. Microbiol. Methods 68, 52–59. doi: 10.1016/j.mimet.2006.06.005
Nightingale, K. K., Schukken, Y. H., Nightingale, C. R., Fortes, E. D., Ho, A. J., Her, Z., et al. (2004). Ecology and transmission of Listena monocytogenes infecting ruminants and in the farm environment. Appl. Environ. Microbiol. 70, 4458–4467. doi: 10.1128/AEM.70.8.4458-4467.2004
Núñez-Montero, K., Leclercq, A., Moura, A., Vales, G., Peraza, J., Pizarro-Cerdá, J., et al. (2018). Listeria costaricensis sp. Nov. Int. J. Syst. Evol. Microbiol. 68, 844–850. doi: 10.1099/ijsem.0.002596
Olaimat, A. N., Al-Holy, M. A., Shahbaz, H. M., Al-Nabulsi, A. A., Abu Ghoush, M. H., Osaili, T. M., et al. (2018). Emergence of antibiotic resistance in Listeria monocytogenes isolated from food products: a comprehensive review. Compr. Rev. Food Sci. Food Saf. 17, 1277–1292. doi: 10.1111/1541-4337.12387
Oliveira, A. R., Domingues, F. C., and Ferreira, S. (2017). The influence of resveratrol adaptation on resistance to antibiotics, benzalkonium chloride, heat and acid stresses of Staphylococcus aureus and Listeria monocytogenes. Food Control 73, 1420–1425. doi: 10.1016/j.foodcont.2016.11.011
Orsi, R. H., de Bakker, H. C., and Wiedmann, M. (2011). Listeria monocytogenes lineages: genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 301, 79–96. doi: 10.1016/j.ijmm.2010.05.002
Orsi, R. H., and Wiedmann, M. (2016). Characteristics and distribution of Listeria spp., including Listeria species newly described since 2009. Appl. Microbiol. Biotechnol. 100, 5273–5287. doi: 10.1007/s00253-016-7552-2
Ottesen, A., Ramachandran, P., Chen, Y., Brown, E., Reed, E., and Strain, E. (2020). Quasimetagenomic source tracking of Listeria monocytogenes from naturally contaminated ice cream. BMC Infect. Dis 20:83. doi: 10.1186/s12879-019-4747-z
Oyinloye, M. A., David, O. M., Muhammad, A., and Famurewa, O. (2018). Occurrence and molecular characterisation of Listeria species in Some fresh-cut vegetables and environmental samples. J. Adv. Microbiol. 12, 1–12. doi: 10.9734/JAMB/2018/43046
Palacios-Gorba, C., Moura, A., Gomis, J., Leclercq, A., Gómez-Martín, Á., Bracq-Dieye, H., et al. (2021). Ruminant-associated Listeria monocytogenes isolates belong preferentially to dairy-associated hypervirulent clones: a longitudinal study in 19 farms. Environ. Microbiol. 23, 7617–7631. doi: 10.1111/1462-2920.15860
Parsons, C., Lee, S., and Kathariou, S. (2020). Dissemination and conservation of cadmium and arsenic resistance determinants in Listeria and other gram-positive bacteria. Mol. Microbiol. 113, 560–569. doi: 10.1111/mmi.14470
Paspaliari, D. K., Mollerup, M. S., Kallipolitis, B. H., Ingmer, H., and Larsen, M. H. (2014). Chitinase expression in Listeria monocytogenes is positively regulated by the Agr system. PLoS One 9:e95385. doi: 10.1371/journal.pone.0095385
Peel, M., Donachie, W., and Shaw, A. (1988). Temperature-dependent expression of flagella of Listeria monocytogenes studied by electron microscopy, SDS-PAGE and Western blotting. J. Gen. Microbiol. 134, 2171–2178. doi: 10.1099/00221287-134-8-2171
Piveteau, P., Depret, G., Pivato, B., Garmyn, D., and Hartmann, A. (2011). Changes in gene expression during adaptation of Listeria monocytogenes to the soil environment. PLoS One 6:e24881. doi: 10.1371/journal.pone.0024881
Quereda, J. J., Leclercq, A., Moura, A., Vales, G., Gómez-Martín, Á., García-Muñoz, Á., et al. (2020). Listeria valentina sp. nov., isolated from a water trough and the faeces of healthy sheep. Int. J. Syst. Evol. Microbiol. 70, 5868–5879. doi: 10.1099/ijsem.0.004494
Ragon, M., Wirth, T., Hollandt, F., Lavenir, R., Lecuit, M., Monnier, A. Le, et al. (2008). A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 4, e1000146. doi: 10.1371/journal.ppat.1000146
Raschle, S., Stephan, R., Stevens, M. J., Cernela, N., Zurfluh, K., Muchaamba, F., et al. (2021). Environmental dissemination of pathogenic Listeria monocytogenes in flowing surface waters in Switzerland. Sci. Rep. 11:9066. doi: 10.1038/s41598-021-88514-y
Rocourt, J., Boerlin, P., Grimont, F., Jacquet, C., and Piffaretti, J. C. (1992). Assignment of Listeria grayi and Listeria murrayi to a single species, Listeria grayi, with a revised description of Listeria grayi. Int. J. Syst. Bacteriol. 42, 171–174. doi: 10.1099/00207713-42-1-171
Rocourt, J., and Buchrieser, C. (2007). “The genus Listeria and Listeria monocytogenes: phylogenetic position, taxonomy, and identification” in Listeria, Listeriosis, and Food Safety. eds. E. T. Ryser and E. H. Marth (Boca Raton: CRC Press), 19–38.
Rocourt, J., and Grimont, P. A. D. (1983). Listeria welshimeri sp. nov. and Listeria seeligeri sp. nov. Int. J. Syst. Bacteriol. 33, 866–869. doi: 10.1099/00207713-33-4-866
Ruppitsch, W., Pietzka, A., Prior, K., Bletz, S., Fernandez, H. L., Allerberger, F., et al. (2015). Defining and evaluating a core genome multilocus sequence typing scheme for whole-genome sequence-based typing of Listeria monocytogenes. J. Clin. Microbiol. 53, 2869–2876. doi: 10.1128/JCM.01193-15
Ryan, S., Begley, M., Hill, C., and Gahan, C. G. M. (2010). A five-gene stress survival islet (SSI-1) that contributes to the growth of Listeria monocytogenes in suboptimal conditions. J. Appl. Microbiol. 109, 984–995. doi: 10.1111/j.1365-2672.2010.04726.x
Salazar, J. K., Fay, M., Eckert, C., Stewart, D., Cranford, V., and Tortorello, M. L. (2020). Evaluation of methods of enrichment and compositing of environmental samples for detection of Listeria monocytogenes. J. Food Prot. 84, 639–646. doi: 10.4315/JFP-20-276
Salcedo, C., Arreaza, L., Alcalá, B., and de la Fuente, L., A., V. J. (2003). Development of a multilocus sequence typing method for analysis of Listeria monocytogenes clones. J. Clin. Microbiol. 41, 757–762. doi: 10.1128/JCM.41.2.757-762.2003
Sauders, B. D., Overdevest, J., Fortes, E., Windham, K., Schukken, Y., Lembo, A., et al. (2012). Diversity of Listeria species in urban and natural environments. Appl. Environ. Microbiol. 78, 4420–4433. doi: 10.1128/AEM.00282-12
Sauders, B. D., and Wiedmann, M. (2007). “Ecology of Listeria species and L. monocytogenes in the natural environment” in Listeria, Listeriosis, and Food Safety. eds. E. T. Ryser and E. H. Marth (Boca Raton, FL: CRC Press), 21–53.
Schlech, W. F., Lavigne, P. M., Bortolussi, R. A., Allen, A. C., Haldane, E. V., Wort, A. J., et al. (1983). Epidemic Listeriosis — evidence for transmission by food. N. Engl. J. Med. 308, 203–206. doi: 10.1056/NEJM198301273080407
Schoder, D., Schmalwieser, A., Szakmary-Brändle, K., Stessl, B., and Wagner, M. (2015). Urban prevalence of Listeria spp. and Listeria monocytogenes in public lavatories and on shoe soles of facility patrons in the European Capital City Vienna. Zoonoses Public Health 62, 179–186. doi: 10.1111/zph.12121
Scortti, M., Lacharme-Lora, L., Wagner, M., Chico-Calero, I., Losito, P., and Vázquez-Boland, J. A. (2006). Coexpression of virulence and fosfomycin susceptibility in Listeria: molecular basis of an antimicrobial in vitro–in vivo paradox. Nat. Med. 12, 515–517. doi: 10.1038/nm1396
Seeliger, H. P. R. (1981). Apathogene listerien: L. innocua sp.n. (Seeliger et Schoofs, 1977). Zentralbl. Bakteriol. B 249, 487–493. doi: 10.1016/S0174-3031(81)80108-4
Seeliger, H. P. R., Rocourt, J., Schrettenbrunner, A., Grimont, P. A. D., and Jones, D. (1984). Listeria ivanovii sp. nov. Int. J. Syst. Bacteriol. 34, 336–337. doi: 10.1099/00207713-34-3-336
Smyth, D. (2007). Methods used in the Tellus Geochemical Mapping of Northern Ireland. British Geological Survey. OR/07/022, 1–89.
Soni, D. K., Singh, M., Singh, D. V., and Dubey, S. K. (2014). Virulence and genotypic characterization of Listeria monocytogenes isolated from vegetable and soil samples. BMC Microbiol. 14:241. doi: 10.1186/s12866-014-0241-3
Stea, E. C., Purdue, L. M., Jamieson, R. C., Yost, C. K., and Hansen, L. T. (2015). Comparison of the prevalences and diversities of Listeria species and Listeria monocytogenes in an urban and a rural agricultural watershed. Appl. Environ. Microbiol. 81, 3812–3822. doi: 10.1128/AEM.00416-15
Stessl, B., Luf, W., Wagner, M., and Schoder, D. (2009). Performance testing of six chromogenic ALOA-type media for the detection of Listeria monocytogenes. J. Appl. Microbiol. 106, 651–659. doi: 10.1111/j.1365-2672.2008.04039.x
Stessl, B., Szakmary-Brändle, K., Vorberg, U., Schoder, D., and Wagner, M. (2020). Temporal analysis of the Listeria monocytogenes population structure in floor drains during reconstruction and expansion of a meat processing plant. Int. J. Food Microbiol. 314:108360. doi: 10.1016/j.ijfoodmicro.2019.108360
Stessl, B., Wagner, M., and Ruppitsch, W. (2021). “Multilocus sequence typing (MLST) and whole genome sequencing (WGS) of Listeria monocytogenes and Listeria innocua BT - Listeria monocytogenes: methods and protocols,” in Methods in Molecular Biology. eds. E. M. Fox, H. Bierne, and B. Stessl (New York, NY: Springer US), 89–103.
Strawn, L. K., Gröhn, Y. T., Warchocki, S., Worobo, R. W., Bihn, E. A., and Wiedmann, M. (2013). Risk factors associated with salmonella and Listeria monocytogenes contamination of produce fields. Appl. Environ. Microbiol. 79, 7618–7627. doi: 10.1128/AEM.02831-13
Tan, M. S., Rahman, S., and Dykes, G. A. (2016). Pectin and xyloglucan influence the attachment of Salmonella enterica and Listeria monocytogenes to bacterial cellulose-derived plant cell wall models. Appl. Environ. Microbiol. 82, 680–688. doi: 10.1128/AEM.02609-15
Troxler, R., von Graevenitz, A., Funke, G., Wiedemann, B., and Stock, I. (2000). Natural antibiotic susceptibility of Listeria species: L. grayi, L. innocua, L. ivanovii, L. monocytogenes, L. seeligeri and L. welshimeri strains. Clin. Microbiol. Infect. 6, 525–535. doi: 10.1046/j.1469-0691.2000.00168.x
USDA (2019). Laboratory Guidebook Notice of Change Title: Isolation and Identification of Listeria monocytogenes from Red Meat, Poultry, Ready-To-Eat Siluriformes (Fish) and Egg Products, and Environmental Samples. Available at: https://www.fsis.usda.gov/wps/wcm/connect/1710bee8-76b9-4e6c-92fc-fdc290dbfa92/mlg-8.pdf?MOD=AJPERES (Accessed February 19, 2021).
Van Renterghem, B., Huysman, F., Rygole, R., and Verstraete, W. (1991). Detection and prevalence of Listeria monocytogenes in the agricultural ecosystem. J. Appl. Bacteriol. 71, 211–217. doi: 10.1111/j.1365-2672.1991.tb04450.x
Vivant, A. L., Desneux, J., Pourcher, A. M., and Piveteau, P. (2017). Transcriptomic analysis of the adaptation of Listeria monocytogenes to lagoon and soil matrices associated with a piggery environment: comparison of expression profiles. Front. Microbiol. 8:1811. doi: 10.3389/fmicb.2017.01811
Vivant, A.-L., Garmyn, D., Gal, L., Hartmann, A., and Piveteau, P. (2014a). Survival of Listeria monocytogenes in soil requires AgrA-mediated. Appl. Environ. Microbiol. 81, 5073–5084. doi: 10.1128/AEM.04134-14
Vivant, A.-L., Garmyn, D., Gal, L., and Piveteau, P. (2014b). The Agr communication system provides a benefit to the populations of Listeria monocytogenes in soil. Front. Cell. Infect. Microbiol. 4:160. doi: 10.3389/fcimb.2014.00160
Vivant, A.-L., Garmyn, D., and Piveteau, P. (2013). Listeria monocytogenes, a down-to-earth pathogen. Front. Cell. Infect. Microbiol. 3:87. doi: 10.3389/fcimb.2013.00087
Wagner, E., Fagerlund, A., Langsrud, S., Møretrø, T., Jensen, M. R., and Moen, B. (2021). Surveillance of Listeria monocytogenes: early detection, population dynamics, and quasimetagenomic sequencing during selective enrichment. Appl. Environ. Microbiol. 87, e01774–e01721. doi: 10.1128/AEM.01774-21
Walker, S. J., Archer, P., and Banks, J. G. (1990). Growth of Listeria monocytogenes at refrigeration temperatures. J. Appl. Bacteriol. 68, 157–162. doi: 10.1016/B978-0-12-385007-2.00012-7
Wang, Y., Lu, L., Lan, R., Salazar, J. K., Liu, J., Xu, J., et al. (2017). Isolation and characterization of Listeria species from rodents in natural environments in China. Emerg. Microbes Infect. 6:e44. doi: 10.1038/emi.2017.28
Watson, R. (2009). Listeriosis remains a cause for concern in Europe. BMJ 338:b319. doi: 10.1136/bmj.b2488
Weindl, L., Frank, E., Ullrich, U., Heurich, M., Kleta, S., Ellerbroek, L., et al. (2016). Listeria monocytogenes in different specimens from healthy red deer and wild boars. Foodborne Pathog. Dis. 13, 391–397. doi: 10.1089/fpd.2015.2061
Weller, D., Andrus, A., Wiedmann, M., and den Bakker, H. C. (2015a). Listeria booriae sp. nov. and Listeria newyorkensis sp. nov., from food processing environments in the USA. Int. J. Syst. Evol. Microbiol. 65, 286–292. doi: 10.1099/ijs.0.070839-0
Weller, D., Wiedmann, M., and Strawn, L. K. (2015b). Irrigation is significantly associated with an increased prevalence of Listeria monocytogenes in produce production environments in New York state. J. Food Prot. 78, 1132–1141. doi: 10.4315/0362-028X.JFP-14-584
Wesley, I. V. (2007). “Listeriosis in Animals,” in Listeria, Listeriosis, and Food Safety. eds. E. T. Ryser and E. H. Marth (Boca Raton, FL: CRC Press), 55–84.
Yin, Y., Yao, H., Doijad, S., Kong, S., Shen, Y., Cai, X., et al. (2019). A hybrid sub-lineage of Listeria monocytogenes comprising hypervirulent isolates. Nature Commun. 10:4283. doi: 10.1038/s41467-019-12072-1
Zamudio, R., Haigh, R. D., Ralph, J. D., De Ste Croix, M., Tasara, T., Zurfluh, K., et al. (2020). Lineage-specific evolution and gene flow in Listeria monocytogenes are independent of bacteriophages. Environ. Microbiol. 22, 5058–5072. doi: 10.1111/1462-2920.15111
Zhao, Y., Cocerva, T., Cox, S., Tardif, S., Su, J. Q., Zhu, Y. G., et al. (2019). Evidence for co-selection of antibiotic resistance genes and mobile genetic elements in metal polluted urban soils. Sci. Total Environ. 656, 512–520. doi: 10.1016/j.scitotenv.2018.11.372
Zhu, Q., Gooneratne, R., and Hussain, M. A. (2017). Listeria monocytogenes in fresh produce: outbreaks, prevalence and contamination levels. Foods 6:21. doi: 10.3390/foods6030021
Zilelidou, E., Manthou, E., and Skandamis, P. (2016). Growth differences and competition between Listeria monocytogenes strains determine their predominance on ham slices and lead to bias during selective enrichment with the ISO protocol. Int. J. Food Microbiol. 235, 60–70. doi: 10.1016/j.ijfoodmicro.2016.07.016
Keywords: Listeria monocytogenes, saprophyte, environment, genotypes, soil, water
Citation: Lourenco A, Linke K, Wagner M and Stessl B (2022) The Saprophytic Lifestyle of Listeria monocytogenes and Entry Into the Food-Processing Environment. Front. Microbiol. 13:789801. doi: 10.3389/fmicb.2022.789801
Edited by:
Arun K. Bhunia, Purdue University, United StatesReviewed by:
Atin Datta, United States Food and Drug Administration, United StatesAlexandre Leclercq, Institut Pasteur, France
Moutong Chen, Guangdong Academy of Science, China
Copyright © 2022 Lourenco, Linke, Wagner and Stessl. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Beatrix Stessl, QmVhdHJpeC5TdGVzc2xAdmV0bWVkdW5pLmFjLmF0
 Antonio Lourenco
Antonio Lourenco Kristina Linke2
Kristina Linke2 Martin Wagner
Martin Wagner Beatrix Stessl
Beatrix Stessl