
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 11 April 2022
Sec. Microbial Symbioses
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.785415
This article is part of the Research Topic Exploring the Insect Microbiome: The Potential Future Role in Biotechnology Industry View all 8 articles
 Xiayu Li1†
Xiayu Li1† Yong Zhang1†
Yong Zhang1† Linlin Zhou1
Linlin Zhou1 Tian Gao2
Tian Gao2 Yu Zhao1
Yu Zhao1 Song Liu2
Song Liu2 Qingqing Su1
Qingqing Su1 Chaoling Wei2
Chaoling Wei2 Yunqiu Yang2*
Yunqiu Yang2* Yanhua Long1*
Yanhua Long1*The gut bacteria of insects play an important role in their nutrition, maintenance, and ecological adaption. Ectropis grisescens is the most important leaf-feeding pest in tea gardens in China. In order to explore whether E. grisescens adaptation under starvation stress is related to its gut bacteria, we used a culture-independent method to compare the composition and diversity of their gut bacteria under starvation treatment. The results revealed no significant changes in core gut bacteria composition and diversity within 24 h of starvation. However, non-core gut bacterial Bacillus increased significantly under starvation conditions. B. cereus strain EG-Q3 isolated from the gut of E. grisescens in carbon source-selected medium showed the ability to degrade fat bodies from E. grisescens in vitro and in vivo. Moreover, the fat-lowering ratio of E. grisescens fed with B. cereus strain EG-Q3 (6.76 ± 1.281%) was significantly higher than that of the control group (3.96 ± 0.801%, t = 4.15, df = 8, p < 0.01) after starvation for 4 h. These findings suggest that non-core gut bacterial B. cereus strain EG-Q3 contributes to host adaptation to starvation. Together, this research provides evidence that E. grisescens may benefit from non-core gut bacteria under starvation conditions.
Insects are consumers, and cannot produce the organic matter necessary to satisfy their metabolic needs. Due to seasonal and climate change, food distribution is inconsistent throughout a year, and most insects will be subject to hunger stress during some stage of their lives (Rotkopf et al., 2013). Hunger stress has adverse effects including decreased metabolism (Hietakangas and Cohen, 2009) and reproductive ability (Billings et al., 2018), and may even result in death (Yang et al., 2016). Therefore, insects can enter an anti-stress state to adapt to those adverse conditions. For example, when larvae of Drosophila melanogaster (Diptera: Drosophilidae) and Harmonia axyridis (Coleoptera: Coccinellidae) are nutritionally challenged, they exhibit cannibalistic behavior (Ahmad et al., 2015). Larvae may enter the pupal stage earlier and may remain in this stage longer with increasing days of starvation (Ballard et al., 2008). Insufficient food also limits insect reproduction (Zhang et al., 2015; Ojima et al., 2018) because reducing reproductive investment under starvation conditions can increase somatic cell maintenance (Elkin and Reid, 2005; García-Roger et al., 2006; Billings et al., 2018). Notably, the studies cited above focused on insect adaptation to hunger stress from only the perspective of their behavior. Other studies have shown that insects secrete Adipokinetic hormone to degrade fat and increase energy metabolism in poor nutritional states (Parkash et al., 2012; Rovenko et al., 2015; Kim and Rulifson, 2004; Lee and Park, 2004).
The gut microbiota is associated with many essential host physiological functions (Sommer and Bäckhed, 2013; Xu et al., 2019; Grenier and Leulier, 2020). With the advent of sequencing technology, many bacteria impacting insect life activities have been found in the guts of a variety of insects (Dillon et al., 2008; Hu et al., 2013; Fang et al., 2013; Ma et al., 2021). Some studies have focused on how insects adapt to different types of stress through gut microbes. For example, D. melanogaster reared at high temperatures became more cold-tolerant after receiving transplants of gut bacteria from flies which were reared at low temperatures (Moghadam et al., 2017). Bemisia tabaci that contained scattered Rickettsia phenotype showed significantly higher thermotolerance (Brumin et al., 2011). In addition, Bactrocera dorsalis inoculated with isolated Citrobacter sp. showed greater resistance to trichlorphon, confirming the key role of Citrobacter sp. in insecticide resistance (Brumin et al., 2011; Cheng et al., 2017). These illustrate the roles of gut microbes in host adaptation to different types of stress, however, whether gut bacteria contribute to host responses to starvation stress remains unknown.
The tea Geometrid moth Ectropis grisescens (Warren) (Lepidoptera: Geometridae) is one of the most destructive tea plant pests throughout the world (Zhang, 2001; Antony et al., 2011). These chewing defoliators have both high fecundity and gluttony, and typically cause significant losses to tea crops in terms of both yield and quality (Antony et al., 2011; Nakai and Lacey, 2017). E. grisescens often cope with food shortages, therefore, exploring how E. grisescens adapt to starvation stress using gut bacteria may provide novel insight. Here, we compared the diversity and abundance of gut bacteria in E. grisescens during different periods of starvation and identified a bacterial strain (EG-Q3) with the ability to degrade fat. In addition, we tested the fat body degradation ability of this strain in vitro and in vivo. Our results will expand our understanding of the physiological roles of gut microbiota.
Ectropis grisescens eggs were obtained from stock cultures from the State Key Laboratory of Tea Plant Biology and Utilization, Anhui Agricultural University, Hefei, China (31.86°°N, 117.27°°E). The collected larvae were reared on tea leaves in transparent boxes in a controlled climate room (22°C ± 1°C; relative humidity 75 ± 10%; 16 h light:8 h dark photoperiod). The tea leaves were cut from branches of tea plants using scissors and inserted into floral foam for storage (Zhang et al., 2021).
Larvae were reared on tea leaves until they reached the fifth instar. Then, 250 healthy 5th-instar larvae were selected for starvation treatment for 0, 4, 8, 12, or 24 h.
Ten 5th-instar larvae were pooled to into biological replicates and five biological replicates were established per group. Larvae were surface-sterilized by dipping in 75% ethanol for 15 s and then rinsing twice with sterile water for 15 s each time. Dissecting scissors were used to cut laterally behind the head capsule, and the gut was removed from the cuticle with larval forceps. The whole gut, including contents, was collected and placed in a 2.0 ml microcentrifuge tube for DNA extraction.
Total genomic DNA was extracted from samples using a QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany) (Mirsepasi et al., 2014). The quality of the DNA was assessed using electrophoresis in 1.2% (w/v) agarose gel prior to amplification and sequencing.
Gut bacteria were analyzed by sequencing the V3-V4 region of the 16S ribosomal RNA gene (16S rRNA) using the Illumina NovaSeq platform (Illumina, San Diego, CA, United States). Genomic DNA samples were subjected to PCR for amplification of the V3-V4 regions of the 16S rRNA using the universal primers 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′). All PCR reactions consisted of 15 μl of Phusion R High-Fidelity PCR Master Mix (New England Biolabs, Beverly, MA, United States), 2 μM forward and reverse primers, and 10 ng of template DNA. The thermal cycling conditions were as follows: 98°C for 1 min followed by 30 cycles at 98°C for 10 s, 50°C for 30 s, 72°C for 30 s, and 72°C for 5 min. The mixture of PCR products was then purified with a Gel Extraction Kit (Qiagen). After PCR amplification, the samples were sequenced on the Illumina NovaSeq platform and 250-bp paired-end reads were generated. All sequences are available as SRA files at the National Center for Biotechnology Information Sequence Read Archive database (NCBI-SRA) under bioProject PRJNA720281 (SRA accession numbers: SAMN18644841).
The sequences were analyzed using the QIIME software package and were used to compare the relative abundances of bacterial taxa (Caporaso et al., 2010b). Operational taxonomic units (OTUs) were assigned with a 97% similarity cutoff using UCLUST version 1.2.22 (Edgar, 2010). The representative sequence (the sequence with the highest relative abundance) for each OTU was used to build the overall OTU table. The taxonomic classifications of each microbial OTU were assigned using Ribosomal Database Project classifier PyNast and SILVA and UNITE as bacterial 16S rRNA databases (Caporaso et al., 2010). Abundances of OTUs were normalized using a standard value for the sample with the fewest sequences. Subsequent analyses of alpha diversity were performed using this normalized output.
The alpha- and beta-diversity indices were calculated using QIIME V1.7.0. The alpha diversity of the gut bacteria complexes was calculated as two diversity indices (Shannon-Wiener and Simpson’s) and two richness estimators (ACE and Chao 1 index). Rarefaction curves were used to verify the quality and depth of sampling. Principal coordinate analysis (PCoA) with weighted and unweighted UniFrac distance metrics was used to detect differences among microbial community structures (Lozupone and Knight, 2005).
Fifty 4th-instar larvae were collected and divided into five groups. Each group was starved for 0, 4, 8, 12, or 24 h, and then immediately soaked in 70% ethanol for 3 min to remove surface bacteria. The dissected guts of the larvae were divided among five grinders with 1 ml of sterile water each. The homogenate was added to 100 ml enrichment medium (Yeast extract 0.20 g/L, NaCl 0.50 g/L, Na2HPO4 3.50 g/L, KH2PO4 1.50 g/L, MgSO4⋅7H2O 0.50 g/L, Olive oil 10 ml/L) and then incubated at 200 rpm overnight at 37°C. Twenty-four hours later, 1 ml of the enriched liquid was incubated in new enrichment medium for a second round of enrichment. This step was repeated twice. Finally, 100 μl of enriched liquid was coated on a Rhodamine B tablet (Jette and Ziomek, 1994) and cultured at 30°C for 3 days. The grown colonies were purified three times. The transparent circles from cultured strains were observed by irradiating plates with UV light at 350 nm (Kouker and Jaeger, 1987), and the diameter of each fluorescence circle and colony was measured using Vernier calipers. Strains with certain fat degradation ability were selected based on their H/C value, which represent the ratio of hydrolysis circle diameter (H) to colony diameter (C).
Lipase producing bacteria isolated from E. grisescens were identified using 16S rDNA gene sequencing and a series of physiological and biochemical tests. A phylogenetic tree of those 16S rDNA sequences was constructed using MAGA 7.0.
The isolated strain, EG-Q3, was incubated in LB medium overnight at 200 rpm and 37°C. The bacterial suspension was then centrifuged to remove the LB medium. The pellet of bacteria cells was then washed with ddH2O to further remove residual medium. The bacterial cells were diluted with ddH2O to a concentration of OD600 = 1.0. Ten test tubes containing 5 ml MSM medium (NaCl 1.00 g/L, (NH4)2SO4 1.00 g/L, K2HPO4 1.50 g/L, KH2PO4 0.50 g/L, MgSO4⋅7H2O 0.50 g/L, pH 7.0–7.5) were divided into three experimental groups (i, ii, and iii) and a blank control. Each experiment was replicated three times. Fat bodies dissected from five E. grisescens larvae were mashed in a beaker containing 20 ml ddH2O. Then, 1 ml fat body homogenate was aspirated into MSM mediums of groups i and iii using pipettes. The above EG-Q3 suspension (OD600 = 1.0) was inoculated into sterilized MSM mediums of groups ii and iii at 4% inoculum volume. All of the test tubes were incubated for 3 days at 200 rpm and 37°C.
Bacterial suspension of EG-Q3 (OD600 = 1.0) was applied to the surface of tea leaves evenly with a brush and allowed to air dry. Some tea leaves were treated at this step using sterile water instead of bacteria suspension. This treatment was repeated three times. One hundred and thirty-eight 2nd-instar larvae with similar body weight (mean ± SD; 11.021 ± 0.512 mg) were divided into control (I) and experimental (II) groups, and were placed in disposable Petri dishes with tea leaves and fed for 3 days. The tea leaves were renewed daily. In order to observe the colonization of bacteria, nine larvae were randomly selected from each group and their guts were diluted and applied to MYP medium (the appropriate medium for B. cereus) (Peng et al., 2001). Then, the remaining 120 larvae were subjected to starvation stress. Each treatment contained five biological replicates with ten larvae per replicate. The body mass of larvae from the two groups was measured after 4 and 12 h starvation using an electric microbalance.
The Illumina NovaSeq sequencing of the bacterial 16S rRNA amplicons from E. grisescens with after 0, 4, 8, 12, and 24 h starvation yielded 2,244,492 raw reads in total. After quality filtering and read merging, a total of 1,853,092 high-quality sequences remined. Rarefaction curves clearly demonstrated that the sampling efforts were adequate to fully represent the richness of the gut microbial communities (Figure 1).
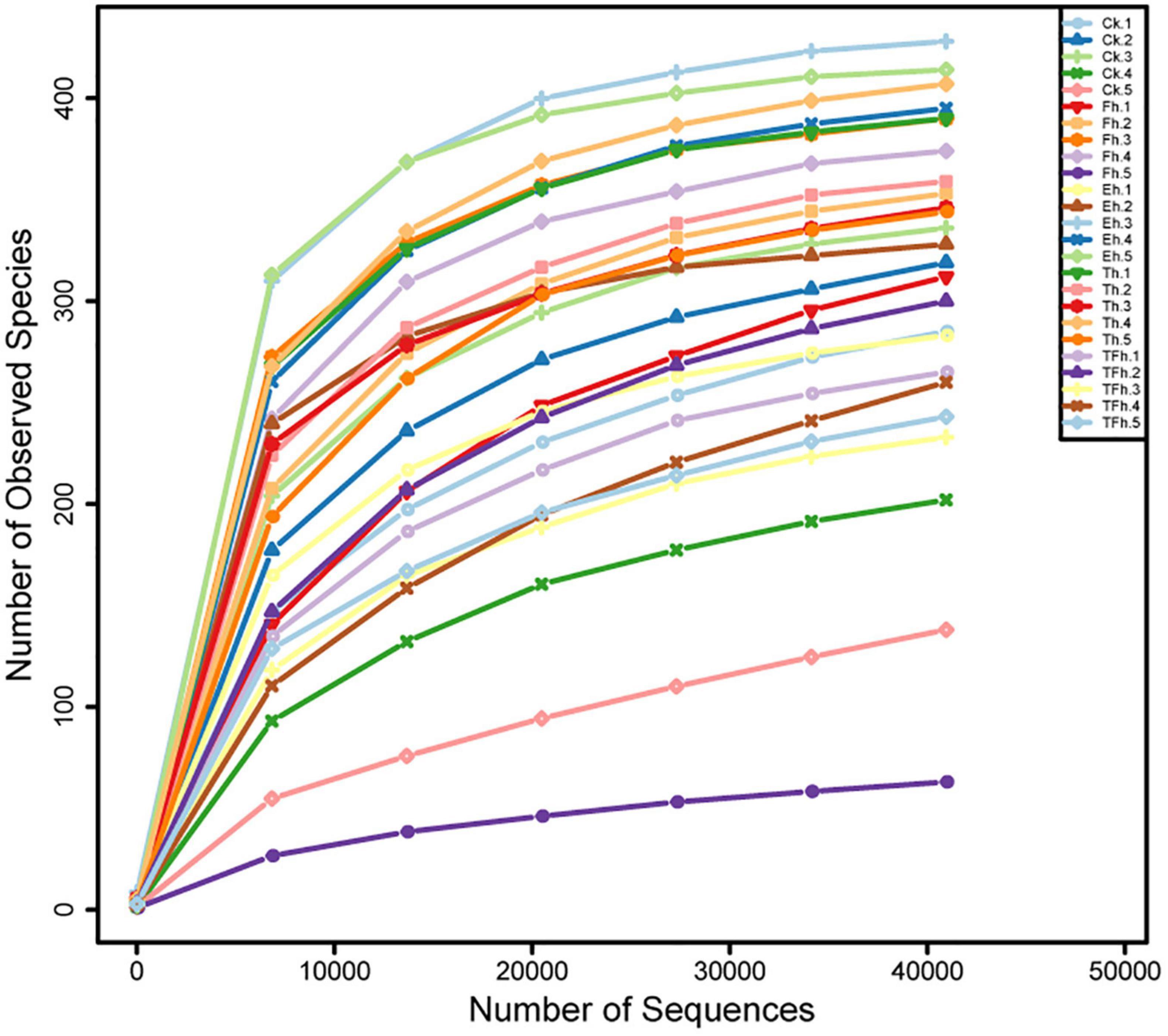
Figure 1. Rarefaction curves based on the numbers of operational taxonomic units (OTUs) observed in the gut microbial communities of E. grisescens larvae after 0 h (CK), 4 h (Fh), 8 h (Eh), 12 h (Th), and 24 h (TFh) starvation treatment.
The OTU richness (ACE and Chao 1 index) of the gut bacteria did not differ significantly among groups (Figures 2A,B). However, the OTU diversity (Shannon index) of the gut bacteria of larvae exposed to starvation for 24 h (TFh; 1.52 ± 0.27) differed from that of the gut bacteria of larvae exposed to starvation for 8 h (Eh; 2.98 ± 0.68; t = 3.99; df = 8; p < 0.001) and 12 h (Th; 2.71 ± 1.14; t = 2.02; df = 8; p < 0.5). In addition, the OTU diversity (Simpson index) of the gut bacteria of TFh (0.38 ± 0.10) differed from that of Eh (0.65 ± 0.17; t = 2.66; df = 8; p < 0.5) (Figures 2C,D).
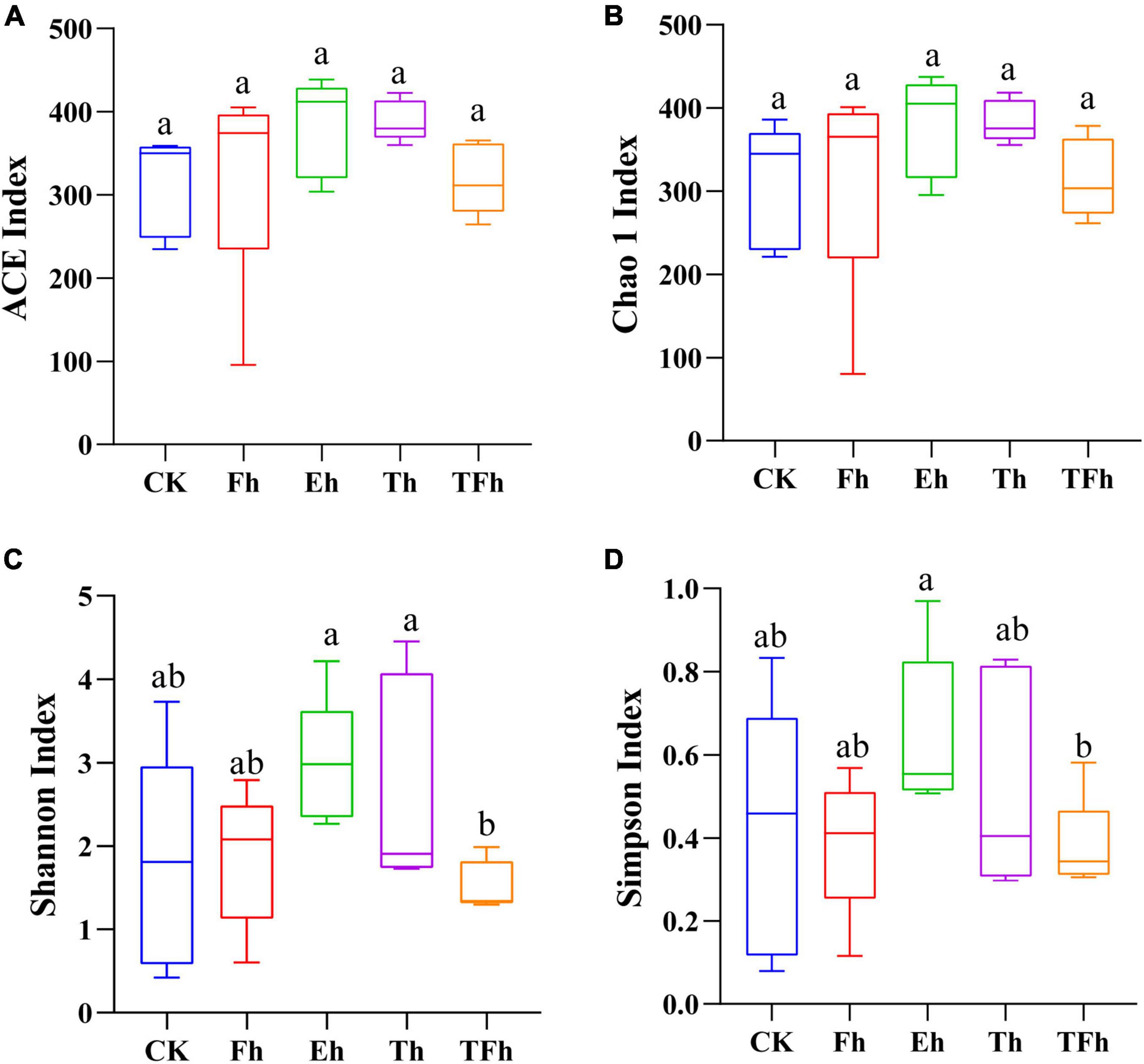
Figure 2. The alpha-diversity based on ACE (A), Chao 1 (B), Shannon (C), and Simpson (D) indices of the gut microbial community of E. grisescens. Significant differences were detected using unpaired two-tailed t-tests. CK: no starvation treatment; Fh: 4 h starvation treatment; Eh: 8 h starvation treatment; Th: 12 h starvation treatment; TFh: 24 h starvation treatment.
The PCoA analysis using distances based on weighted and unweighted UniFrac showed that the gut bacterial communities of E. grisescens larvae were not distinctive (Figures 3A,B).
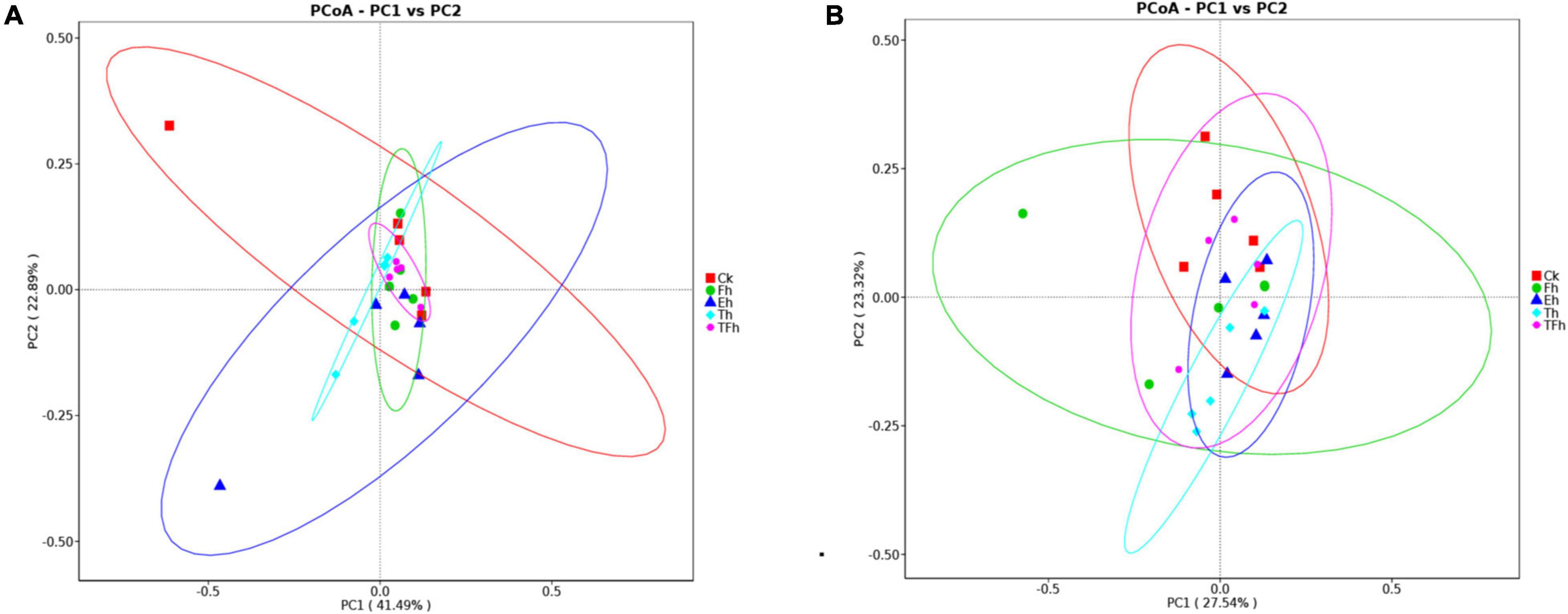
Figure 3. Principal coordinate analysis (PCoA) of bacteria using distances based on weighted (A) and unweighted (B) UniFrac values.
Three hundred sixty-four OTUs were shared by all groups, and accounted for 86.87, 81.79, 79.30, 80.35, and 81.80% of the total OTUs in CK, Fh, Eh, Th, and TFh, respectively (Figure 4A). The dominant bacterial phyla in each group were Firmicutes (50.89–78.12%), Proteobacteria (12.27–25.01%), and Bacteroidetes (0.38–14.95%) (Figure 4B). The dominant bacterial genera in each group were Enterococcus (64.19–79.90%) and Wolbachia (2.13–13.11%) (Figure 4C). The relative abundance of Bacillus in CK (0.01 ± 0.17%) differed significantly from that in Fh (0.64 ± 0.14%; t = 8.51; df = 8; p < 0.0001), Eh (0.32 ± 0.12%; t = 5.06; df = 8; p < 0.001), and Th (0.16 ± 0.14%; t = 2.01; df = 8; p < 0.05) (Figure 4D).
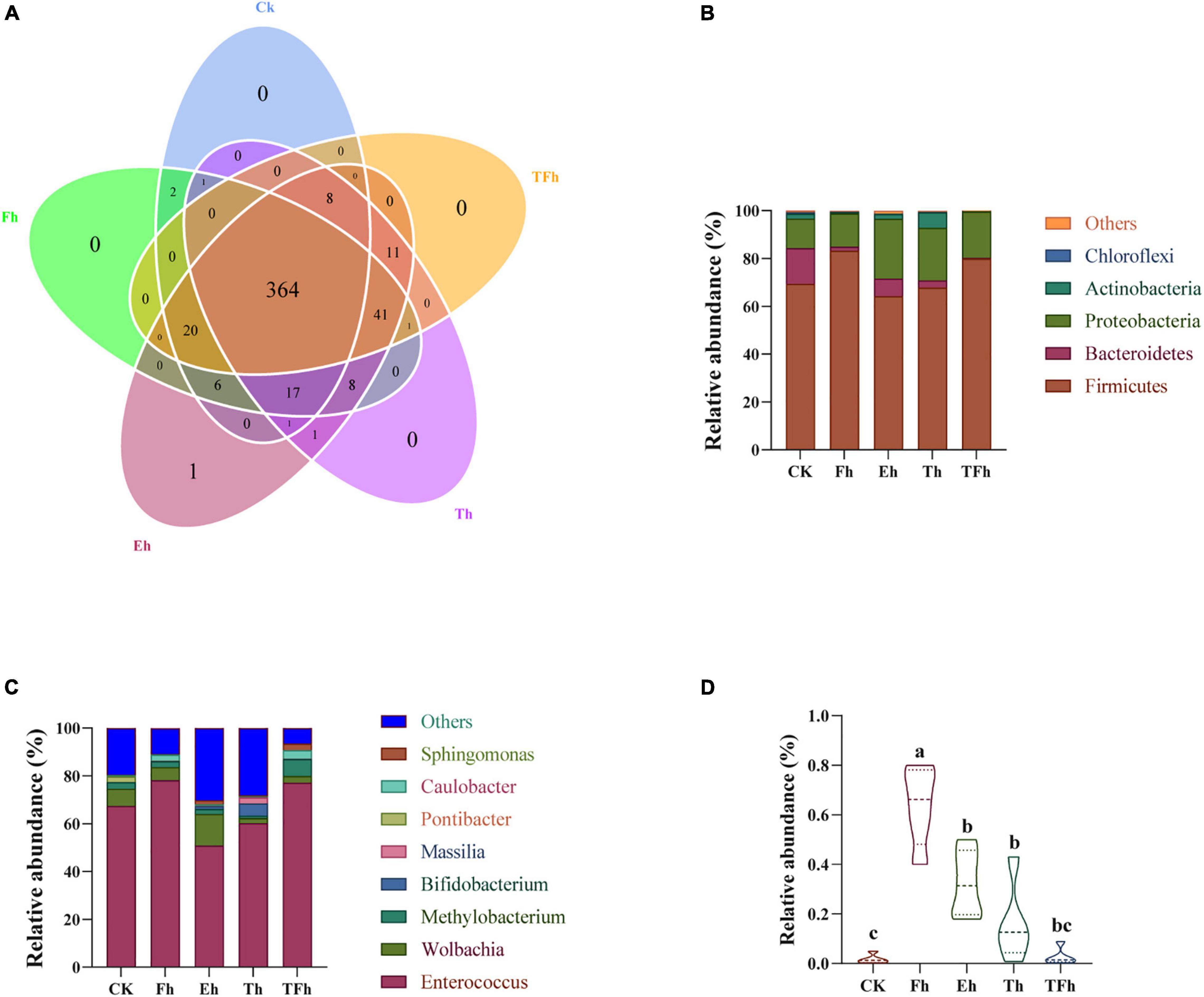
Figure 4. Venn diagram (A) of the operational taxonomic units (OTUs) in CK, Fh, Eh, Th, and TFh. Relative abundances are shown for bacteria at the phylum (B) and genus (C) levels. CK: no starvation treatment; Fh: 4 h starvation treatment; Eh: 8 h starvation treatment; Th: 12 h starvation treatment; TFh: Twenty-four hours starvation treatment. (D) The relative abundance of Bacillus.
The gut contents of larvae subjected to starvation for different amounts of time were cultivated on Rhodamine B selective medium, and five potential lipase-producing strains were identified based on colony characteristics (Figure 5A). These strains were named EG-Q1, EG-Q2, EG-Q3, EG-Q4, and EG-Q5.
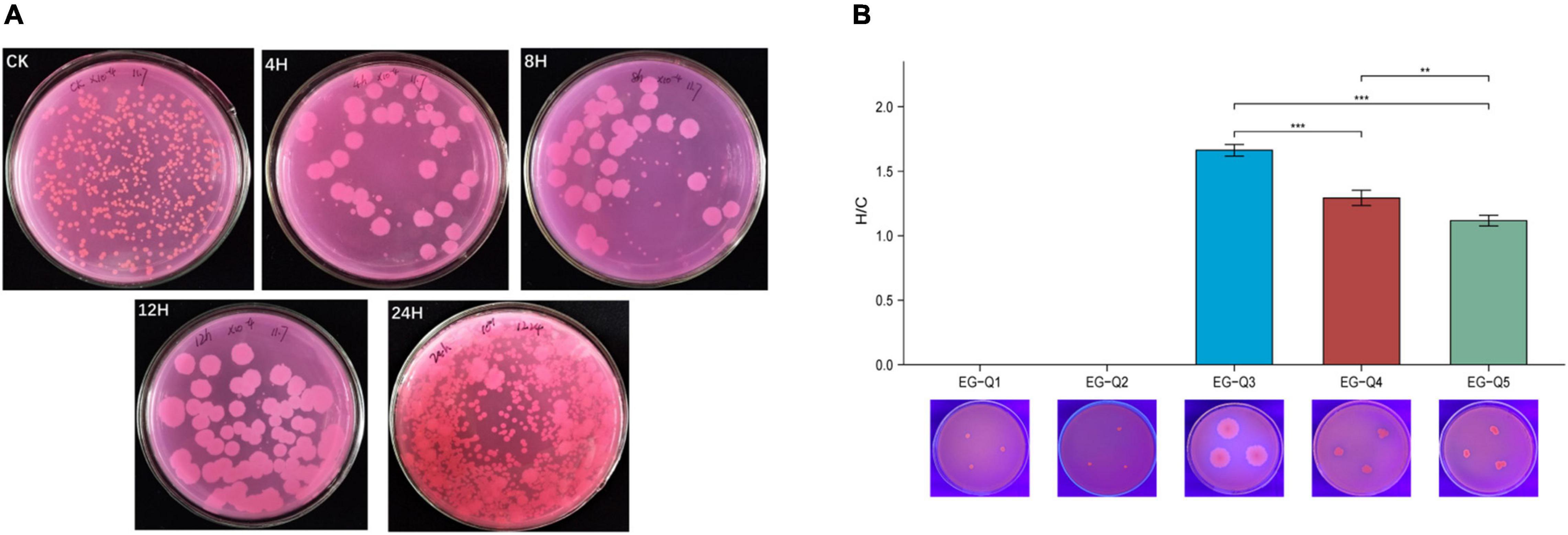
Figure 5. (A) The growth of intestinal commensal bacteria on Rhodamine B medium, (B) The H/C values and hydrolysis circles of five screened strains.
Isolates of the five strains were inoculated on selective medium again in order to compare their abilities to degrade lipids. Although EG-Q1 and EG-Q2 could be cultured on the selective medium, obvious hydrolysis circles were not observed. The H/C value of EG-Q3 (1.66 ± 0.05) was significantly higher than that of the other strains (Figure 5B). Therefore, EG-Q3 was selected for further in vivo and in vitro tests.
The three isolates producing transparent circles (EG-Q3, EG-Q4, and EG-Q5) were rod shaped Gram-positive bacteria (Supplementary Figure). Through combination with the phylogenetic tree constructed from 16S rDNA sequences of the five strains (Figure 6), EG-Q3 was identified as B. cereus. The genome sequence of the isolated strain was registered in GenBank (the Accession Number: MZ497322.1).
In in vitro tests of fat body degradation (Figure 7), we found that fat bodies floating in suspension were quite obvious and unbroken in group i. In group ii, although EG-Q3 was inoculated into the MSM medium, the turbidity of the medium did not change, and resembled group CK. However, the fat bodies in group iii were broken and had nearly disappeared, and the MSM medium became significantly turbid. This indicated that EG-Q3 can use fat bodies for proliferation.
To further determine whether bacteria EG-Q3 could proliferate in the larval gut, tea leaves coated with EG-Q3 were fed to E. grisescens. After 3 days of supplementation, EG-Q3 could be detected in the guts of larvae (Figure 8). There was no difference in the body weights of the two groups of insects before starvation (Figure 9A). After confirming that EG-Q3 can colonize larval guts, the larvae were starved and weighed regularly. We found that in group I, the body mass of larvae fed with EG-Q3 decreased significantly after 4 h of starvation (from 67.014 ± 5.318 mg to 62.514 ± 5.519 mg), and the fat-lowering ratio after 4 h of starvation was 6.76%, nearly twice that of group II (3.96%) (Figure 9B). With the extension of starvation time, the weight of the larvae further decreased. The fat-lowering ratio increased to 13.85% after 12 h starvation (control group 11.05%).
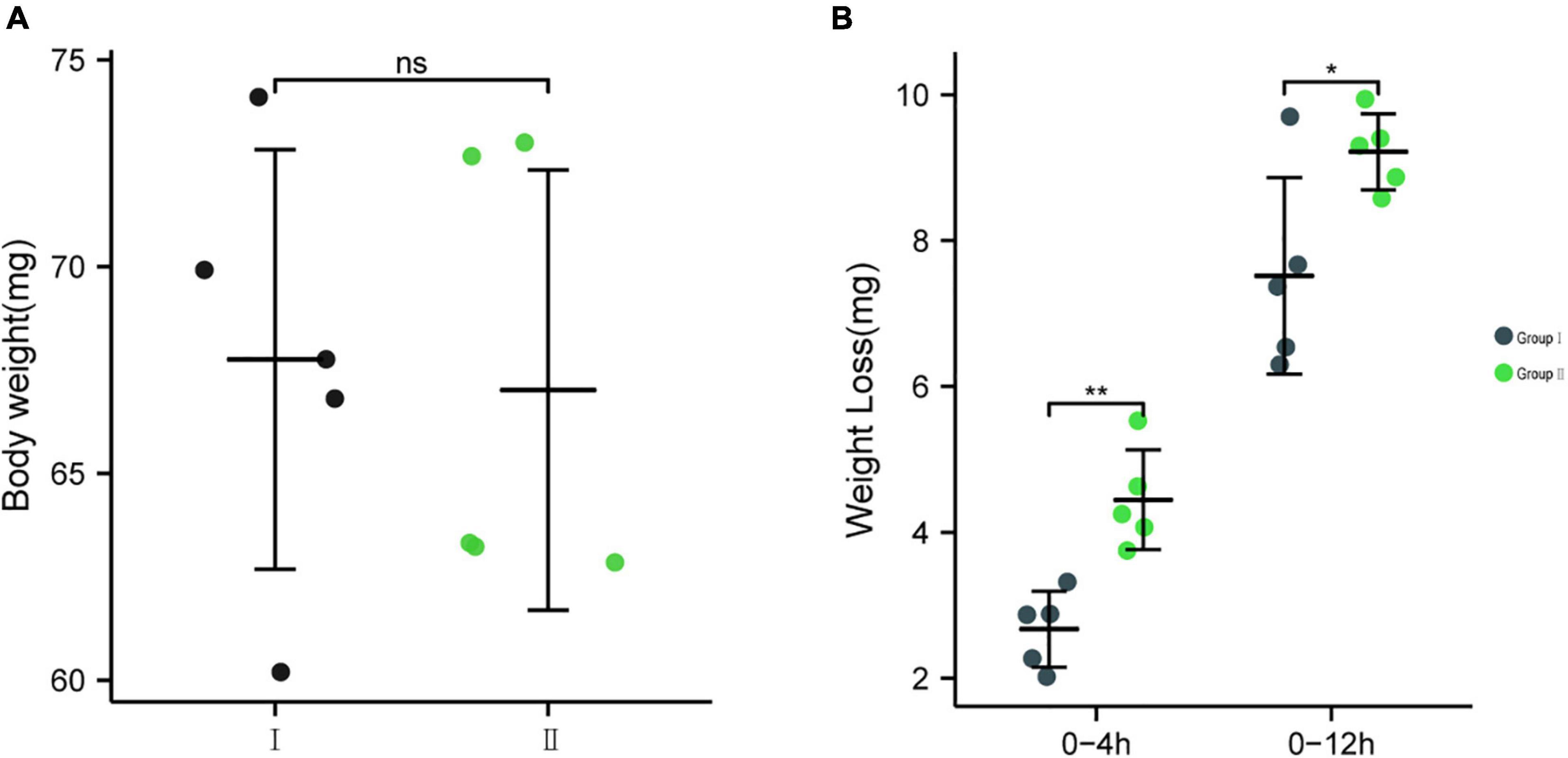
Figure 9. (A) The body weights of the two groups of insects before starvation, (B) The effect of feeding EG-Q3 on body mass. Independent sample t-tests were used to evaluate significance. Data are presented as mean ± SD (n = 5). * indicates significance difference between two groups: *, p < 0.05; **, p < 0.01.
Gut bacteria are of increasing interest to entomologists due to their role in mediating insects’ fitness. In this study, a culture-independent approach was used to compare the composition and diversity of gut bacteria under starvation treatment. The results revealed no significant change in gut bacteria composition and diversity within 24 h of starvation. Simultaneously, the composition of core gut bacteria of E. grisescens observed in this study was consistent with previous reports (Zhang et al., 2019, 2021). However, the abundance of a non-core bacterial Bacillus changed significantly. Therefore, the role of Bacillus in the starvation of E. grisescens attracted our attention. In this study, we reported the first isolation of B. cereus (EG-Q3) associated with host lipid metabolism from the gut of the tea pest E. grisescens.
As a multifunctional biocatalyst, lipase has excellent application value and prospects in industries such as biodiesel, food and beverage, leather, textile, detergent, pharmaceutical, and medical (Prem et al., 2020). In industry, bacteria and fungi are the best biological sources of lipase (Boutaiba et al., 2006; Moura et al., 2015; Elif Demirkan et al., 2021). Compared to Achromobacter, Alcaligenes, Arthrobacter, Burkholderia, Chromobacterium, Geobacillus, and Pseudomonas, Bacillus plays a pivotal role in secreting lipase (Sarmah et al., 2018; Elif Demirkan et al., 2021). Since the leaves that Lepidopteran insects feed on contain fat or fatty acid, it is possible to screen for lipase-producing strains in their intestinal tracts. Nine lipase-producing bacterial strains, including Bacillus, Brevibacterium, Corynebacterium, Staphylococcus, Klebsiella, and Stenotrophomonas were obtained from the intestines of Bombyx mori (Wei et al., 2011). This is similar to the bacterial strains observed in the guts of E. grisescens. In addition, many lipase-producing strains were detected in the guts of Antheraea assamensis, Helicoverpa armigera, and Plutella xylostella, demonstrating their importance to host nutrition (Gandotra et al., 2016). However, the increased abundance of Bacillus and its functions under starvation stress have not previously been reported.
Previous studies have investigated insect behavior and their physiological responses to starvation stress (Ojima et al., 2018; Billings et al., 2018; Rovenko et al., 2015). With the development of sequencing technology and the rise of research in the field of gut microbes, researchers have found that intestinal microbes play an irreplaceable role in lipid metabolism. Previous studies conducted on germ free mice (GF-mice) and conventionally raised mice (CONV-R) found that intestinal bacteria help hosts to accumulate fat from food (Backhed et al., 2004). In-depth molecular research has shown that intestinal microbes can affect lipid metabolism by regulating adipose-related genes expression in mice (Zhao et al., 2018). Furthermore, a non-dominant intestinal bacterium of Niletilapia, Citrobacter spp., has been shown to aid the host in harvesting energy from a high-fat diet (Zhang et al., 2020). Here, we used deep sequencing technology and found that the significant change in EG-Q3 abundance in the gut of E. grisescens under starvation stress is related to lipid metabolism. This suggests that expanding the study of lipid metabolism in entomology may have benefits. This is the first study to explore the relationship between gut bacteria or non-core bacteria and insect lipid metabolism. In this study, 16S rRNA amplicon sequencing analysis showed that the abundance of Bacillus increased over time in the gut of starvation-stressed hosts and reached its maximum after 4 h of starvation. This implies that Bacillus participates in host lipid metabolism under starvation stress. In the in vivo test, group II significantly reduced body weight after 4 h of starvation relative to group I, likely due to the colonization of EG-Q3 in the gut. Therefore, EG-Q3 from Bacillus played an important role by helping E. grisescens consume its own fat bodies to obtain energy during the period of starvation. However, the difference in the rate of weight loss between the two groups decreased after 12 h. This may have been a result of proliferation of the small amount of B. cereus in normal intestines after starvation. This explanation is consistent with the changes in gut symbiotic bacterial communities found in our bioinformatics analysis.
In conclusion, the role of symbiotic bacteria should not be neglected in future studies of insect adaptation to starvation. Strengthening the research on the role of symbiotic bacteria and exploring intestinal bacteria related to stress resistance of other insects is conducive to a comprehensive and in-depth understanding of insect adaptation.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA720281.
YL, YY, and YoZ designed the project. XL, QS, TG, and LZ performed the experiments. YoZ and SL helped the analysis of sequencing data. XL and YoZ drafted the manuscript. YL, YuZ, and CW revised the manuscript. All authors contributed to the study conception and design and approved the final version for submission.
This study was supported by the National Key Research and Development Program of China (2019YFD1001601) and the National Natural Science Foundation of China (Grant Nos. 32072421 and 31870635).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.785415/full#supplementary-material
Ahmad, M., Chaudhary, S. U., Afzal, A. J., and Tariq, M. (2015). Starvation-induced dietary behaviour in Drosophila melanogaster larvae and adults. Sci. Rep. 5:14285. doi: 10.1038/srep14285
Antony, B., Sinu, P. A., and Das, S. (2011). New record of nucleopolyhe droviruses in tea looper caterpillars in India. J. Invertebr. Pathol. 108, 63–67. doi: 10.1016/j.jip.2011.06.007
Backhed, F., Ding, H., Wang, T., Hooper, L. V., Koh, G. Y., Nagy, A., et al. (2004). The gut microbiota as an environmental factor that regulates fat storage. P. Nati. Acad. Sci. U.S.A 101, 15718–15723. doi: 10.1073/pnas.0407076101
Ballard, J. W., Melvin, R. G., and Simpson, S. J. (2008). Starvation resistance is positively correlated with body lipid proportion in five wild caught Drosophila simulans populations. J. Insect. Physiol. 54, 1371–1376. doi: 10.1016/j.jinsphys.2008.07.009
Billings, A. C., Schultz, K. E., Hernandez, E. A., Jones, W. E., and Price, D. K. (2018). Male courtship behaviors and female choice reduced during experimental starvation stress. Behav. Ecol. 30, 231–239. doi: 10.1093/beheco/ary144
Boutaiba, S., Bhatnagar, T., Hacene, H., Mitchell, D. A., and BaRatti, J. C. (2006). Preliminary characterisation of a lipolytic activity from an extremely halophilic archaeon. Natronococcus sp. J. Mol. Catal. B: Enzym. 41, 21–26. doi: 10.1016/j.molcatb.2006.03.010
Brumin, M., Kontse, Da Lov, S., and Ghanim, M. (2011). Rickettsia influences thermotolerance in the whitefly Bemisia tabaci B biotype. Insect. Sci. 18, 57–66. doi: 10.1111/j.1744-7917.2010.01396.x
Caporaso, J. G., Bittinger, K., Bushman, F. D., DeSantis, T. Z., Andersen, G. L., and Knight, R. (2010). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26, 266–267. doi: 10.1093/bioinformatics/btp636
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010b). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Cheng, D., Guo, Z., Riegler, M., Xi, Z., Liang, G., and Xu, Y. (2017). Gut symbiont enhances insecticide resistance in a significant pest, the oriental fruit fly Bactrocera dorsalis (hendel). Microbiome 5:13. doi: 10.1186/s40168-017-0236-z
Dillon, R. J., Webster, G., Weightman, A. J., Dillon, V. M., Blanford, S., and Charnley, A. K. (2008). Composition of Acridid gut bacterial communities as revealed by 16S rRNA gene analysis. J. Invertebr. Pathol. 97, 265–272. doi: 10.1016/j.jip.2007.09.010
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Elif Demirkan, E., Aybey, Çetinkaya, A., and Abdou, M. (2021). Lipase from new isolate Bacillus cereus ATA179: optimization of production conditions, partial purification, characterization and its potential in the detergent industry. Turk. J. Biol. 45, 287–300. doi: 10.3906/biy-2101-22
Elkin, C. M., and Reid, M. L. (2005). Low energy reserves and energy allocation decisions affect reproduction by Mountain Pine Beetles. Dendroctonus ponderosae. Funct. Ecol. 19, 102–109. doi: 10.1111/j.0269-8463.2005.00935.x
Fang, W., Fang, Z., Liu, Z., Yuan, J., Zhang, X., Peng, H., et al. (2013). Phylogenetic analysis of bacterial community in the gut of American cockroach (Periplaneta americana). Acta. Microbiol. Sin. 53, 984–994.
Gandotra, S., Bhuyan, P. M., Gogoi, D. K., Kumar, A., and Subramanian, S. (2016). Screening of nutritionally important gut bacteria from the lepidopteran insects through qualitative enzyme assays. P. Natl. A. Sci. India. B. 88, 329–337. doi: 10.1007/s40011-016-0762-7
García-Roger, E. M., Martínez, A., and Serra, M. (2006). Starvation tolerance of rotifers produced from parthenogenetic eggs and from diapausing eggs: a life table approach. J. Plankton. Res. 28, 257–265. doi: 10.1093/plankt/fbi062
Grenier, T., and Leulier, F. (2020). How commensal microbes shape the physiology of Drosophila melanogaster. Curr. Opin. Insect. Sci. 41, 92–99. doi: 10.1016/j.cois.2020.08.002
Hietakangas, V., and Cohen, S. M. (2009). Regulation of tissue growth through nutrient sensing. Ann. Rev. Genet. 43, 389–410. doi: 10.1146/ANNUREV-GENET-102108-134815
Hu, X., Wang, C., Chen, H., and Ma, J. (2013). Differences in the structure of the gut bacteria communities in development stages of the Chinese white pine beetle (Dendroctonus armandi). Int. J. Mol. Sci. 14, 21006–21020. doi: 10.3390/ijms141021006
Jette, J. F., and Ziomek, E. (1994). Determination of lipase activity by a rhodamine-triglyceride-agarose assay. Anal. Biochem. 219, 256–260. doi: 10.1006/abio.1994.1265
Kim, S. K., and Rulifson, E. J. (2004). Conserved mechanisms of glucose sensing and regulation by Drosophila corpora cardiaca cells. Nature 431, 316–320. doi: 10.1038/nature02897
Kouker, G., and Jaeger, K. E. (1987). Specific and sensitive plate assay for bacterial lipases. Appl. Environ. Microb. 53, 211–213. doi: 10.1016/S0065-2164(08)70081-9
Lee, G., and Park, J. H. (2004). Hemolymph sugar homeostasis and starvation induced hyperactivity affected by genetic manipulations of the adipokinetic hormone-encoding gene in Drosophila melanogaster. Genetics 167, 311–323. doi: 10.1534/genetics.167.1.311
Lozupone, C., and Knight, R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235. doi: 10.1128/aem.71.12.8228-8235.2005
Ma, M., Tu, C., Luo, J., Lu, M., Zhang, S., and Xu, L. (2021). Metabolic and immunological effects of gut microbiota in leaf beetles at the local and systemic levels. Integr. Zool. 16, 313–323. doi: 10.1111/1749-4877.12528
Mirsepasi, H., Persson, S., Struve, C., Andersen, L., Petersen, A. M., and Krogfelt, K. A. (2014). Microbial diversity in fecal samples depends on DNA extraction method: easymag DNA extraction compared to qiaamp DNA stool mini kit extraction. BMC. Res. Notes 7:1–50. doi: 10.1186/1756-0500-7-50
Moghadam, N. N., Thorshauge, P. M., Kristensen, T. N., Jonge, N. D., and Nielsen, J. L. (2017). Strong responses of Drosophila melanogaster microbiota to developmental temperature. Fly 12, 1–12. doi: 10.1080/19336934.2017.1394558
Moura, M. V., da Silva, G. P., de Oliveira, Machado, A. C., Torres, F. A., Freire, D. M., and Almeida, R. V. (2015). Displaying lipase B from Candida antarctica in Pichia pastoris using the yeast surface display approach: prospection of a new anchor and characterization of the whole cell biocatalyst. PloS. One 10:e0141454. doi: 10.1371/journal.pone.0141454
Nakai, M., and Lacey, L. A. (2017). ““microbial control of insect pests of tea and coffee,”,” in Microbial Control of Insect and Mite Pests, ed. L. A. Lacey (Cambridge: Academic Press), 223–235. doi: 10.1016/B978-0-12-803527-6.00015-9
Ojima, N., Hara, Y., Ito, H., and Yamamoto, D. (2018). Genetic dissection of stress-induced reproductive arrest in Drosophila melanogaster females. PloS Genet. 14:e1007434. doi: 10.1371/journal.pgen.1007434
Parkash, R., Aggarwal, D. D., Ranga, P., and Singh, D. (2012). Divergence of larval resource acquisition for water conservation and starvation resistance in Drosophila melanogaster. J. Comp. Physiol. B 182, 625–640. doi: 10.1007/s00360-011-0641-8
Peng, H., Ford, V., and Franpton, E. W. (2001). Isolation and enumeration of Bacillus cereus from foods on a novel chromogenic plating medium. Food Microbiol. 18, 231–238. doi: 10.1006/fmic.2000.0369
Prem, C., Ranjan, S., and Pankaj, K. A. (2020). Microbial lipases and their industrial applications: a comprehensive review. Microb. Cell. Fact 19:169. doi: 10.1186/s12934-020-01428-8
Rotkopf, R., Alcalay, Y., Bar-Hanin, E., Barkae, E. D., and Ovadia, O. (2013). Slow growth improves compensation ability: examining growth rate and starvation endurance in pit-building antlions from semi-arid and hyper-arid regions. Evol. Ecol. 27, 1129–1144. doi: 10.1007/s10682-013-9644-0
Rovenko, B. M., Perkhulyn, N. V., Gospodaryov, D. V., Sanz, A., Lushchak, O. V., and Lushchak, V. I. (2015). High consumption of fructose rather than glucose promotes a diet-induced obese phenotype in Drosophila melanogaster. Comp. Biochem. Physiol. Part A. 180, 75–85. doi: 10.1016/j.cbpa.2014.11.008
Sarmah, N., Revathi, D., Sheelu, G., Yamuna, K. R., Sridhar, S., Mehtab, V., et al. (2018). Recent advances on sources and industrial applications of lipase. Biotechnol. Prog 34, 5–28. doi: 10.1002/btpr.2581
Sommer, F., and Bäckhed, F. (2013). The gut microbiota—masters of host development and physiology. Nat. Rev. Microbiol. 11, 227–238. doi: 10.1038/nrmicro2974
Wei, F., Wang, X. Q., Wei, Z., Liu, G. Y., and Wan, Y. J. (2011). Isolation and characterization of lipase-producing bacteria in the intestine of the silkworm. Bombyx mori, reared on different forage. J. Insect. Sci. 11, 135–131. doi: 10.1673/031.011.13501
Xu, L., Liu, Y., Xu, S., and Lu, M. (2019). Gut commensal bacteria in biological invasions. Integr. Zool. 14, 613–618. doi: 10.1111/1749-4877.12385
Yang, W. K., Tang, F. F., Liu, Z. H., Zhong, J., and Dong, Z. P. (2016). Effects of starvation on DefensinA and DefensinB expression in silkworm, Bombyx mori. Southwest. Chin. J. Agricul. Sci. 29, 3019–3022. doi: 10.16213/j.cnki.scjas.2016.12.044
Zhang, H. H. (2001). Fauna of tea geometrid in China. Chin. J. Tea. Sci 2, 157–160. doi: 10.13305/j.cnki.jts.2001.02.019
Zhang, H. H., Chen, J. H., Ji, Q. E., and Luo, M. J. (2015). Overview in the study and application of the influencing factors on oviposition behavior of insects. J. Environ. Entomol. 37, 432–440. doi: 10.3969/j.issn.1674-0858.2015.02.31
Zhang, M. L., Li, M., Sheng, Y., Tan, F., and Du, Z. Y. (2020). Citrobacter species increase energy harvest by modulating intestinal microbiota in fish: nondominant species play important functions. mSystems 5, e303–e320. doi: 10.1128/mSystems.00303-20
Zhang, Y., Liu, S., Jiang, R., Zhang, C., Gao, T., Wang, Y., et al. (2021). Wolbachia strain wGri from the tea geometrid moth Ectropis grisescens contributes to its host’s fecundity. Front. Microbiol. 12:694466. doi: 10.3389/fmicb.2021.694466
Zhang, Y., Zhao, T. Y., Deng, J. D., Long, Y. H., Zhang, L. W., and Yang, Y. Q. (2019). Positive effects of the tea catechin (-)-epigallocatechin-3-gallate on gut bacteria and fitness of Ectropis obliqua Prout (Lepidoptera: Geometridae). Sci. Rep. 9:5021. doi: 10.1038/s41598-019-41637-9
Keywords: gut bacteria, starvation, lipid metabolism, physiological adaptation, mechanism
Citation: Li X, Zhang Y, Zhou L, Gao T, Zhao Y, Liu S, Su Q, Wei C, Yang Y and Long Y (2022) Bacillus cereus (EG-Q3) in the Gut of Ectropis grisescens Contributes to Host Response to Starvation Conditions. Front. Microbiol. 13:785415. doi: 10.3389/fmicb.2022.785415
Received: 29 September 2021; Accepted: 11 March 2022;
Published: 11 April 2022.
Edited by:
Min Lu, Institute of Zoology (CAS), ChinaReviewed by:
Waqas Wakil, University of Agriculture, Faisalabad, PakistanCopyright © 2022 Li, Zhang, Zhou, Gao, Zhao, Liu, Su, Wei, Yang and Long. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanhua Long, bG9uZ3lhbmh1YUBhaGF1LmVkdS5jbg==; Yunqiu Yang, eXlxX2x5aEAxNjMuY29t
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.