
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 20 June 2022
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.784628
This article is part of the Research TopicMALDI-TOF MS in Microbiological Diagnostics: Future Applications Beyond IdentificationView all 12 articles
The increasing spread of drug-resistant bacterial strains presents great challenges to clinical antibacterial treatment and public health, particularly with regard to β-lactamase-producing Enterobacteriaceae. A rapid and accurate detection method that can expedite precise clinical diagnostics and rational administration of antibiotics is urgently needed. Targeted proteomics, a technique involving selected reaction monitoring or multiple reaction monitoring, has been developed for detecting specific peptides. In the present study, a rapid single-colony-processing procedure combined with an improved parallel reaction monitoring (PRM) workflow based on HRAM Orbitrap MS was developed to detect carbapenemases (Klebsiella pneumoniae carbapenemase, KPC; imipenemase, IMP; Verona integron-encoded metallo-β-lactamase, VIM; New Delhi metallo-β-lactamase, NDM; and oxacillinase, OXA), extended spectrum β-lactamases (TEM and CTX-M), and AmpC (CMY-2) produced by Enterobacteriaceae. Specific peptides were selected and validated, and their coefficients of variation and stability were evaluated. In total, 188 Enterobacteriaceae strains were screened using the workflow. Fourteen out of total 19 peptides have 100% specificity; three peptides have specificity >95% and two peptides have specificity ranged from 74∼85%. On the sensitivity, only nine peptides have 95∼100% sensitivity. The other 10 peptides have sensitivity ranged from 27∼94%. Thus, a screening method based on peptide groups was developed for the first time. Taken together, this study described a rapid extraction and detection workflow for widespread β-lactamases, including KPC, IMP, VIM, NDM, OXA, CMY, CTX-M, and TEM, using single colonies of Enterobacteriaceae strains. PRM-targeted proteomics was proven to be a promising approach for the detection of drug-resistant enzymes.
With the extensive clinical use of carbapenems and β-lactam antibacterial drugs, the incidences of antibiotic resistance have increased. The increasing population of multi-drug resistant (MDR) and extensively drug-resistant strains accompanied with the rapid spread of antibiotic-resistance genes have posed great challenges to clinical anti-bacterial treatment and public health. In the list of bacteria for which new antibiotics are urgently needed released by the World Health Organization in 2017, carbapenem-resistant Enterobacteriaceae (CRE) and extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae, were included in the Critical group (Priority 1) (WHO, 2017). β-lactamases are a group of bacterial enzymes that can inactivate β-lactam antibiotics, resulting in the loss of antibacterial activity (Eliopoulos and Karen, 2001). β-lactamases are currently divided into four classes: A, B, C, and D according to the Ambler classification, based on their primary structure (Bush and Jacoby, 2010). Carbapenems are generally regarded as the last treatment choice for serious bacterial infections. Carbapenemases are β-lactamases with versatile hydrolytic capacities: the A and D class carbapenemases are serine-type hydrolases, such as Klebsiella pneumoniae carbapenemase (KPC), and oxacillinase (OXA). The B class carbapenemases are metallo-hydrolases, such as New Delhi metallo-β-lactamase (NDM), imipenemase (IMP), and Verona integron-encoded metallo-β-lactamase (VIM). ESBLs belong to the class A and D β-lactamases. ESBLs such as the TEM (ampicillin resistance) and CTX-M (cefotaxime resistance) groups belong to class A β-lactamases (Pitout et al., 2005). AmpC β-lactamases such as CMY-2 belong to class C of carbapenemases (Philippon et al., 2002). Enterobacteriaceae, including Escherichia coli, K. pneumoniae, and Enterobacter cloacae, which carry several types of β-lactamases, represent a great challenge to the clinical diagnostics and treatment of various infections (Duin, 2017).
Early diagnosis and effective drug treatment are key strategies to deal with the antibiotic-resistance problem (Rodríguez-Baño et al., 2018). Thus, rapid and accurate detection methods are urgently needed in clinical practice. A short testing time and accurate diagnosis will assist in providing an appropriate antibiotic treatment in time. For the last few decades, traditional approaches for the detection of β-lactamases, such as standard disk-diffusion procedure, broth microdilution, and agar dilution, have been used, which are time-consuming and can only determine the drug-resistance property but not the β-lactamase type. Synergy testing is accurate and allow to identify the carbapenemase classes, which is the info needed for therapeutic choices. Polymerase chain reaction (PCR) are very sensitive and fast and can be used also from primary samples (Dallenne et al., 2010). Whole genome sequencing, which can accurately identify the genotypes of β-lactamases, is still not practical for usual clinical application because of the high price. However, since drug-resistant enzymes are the products of regulated expression, the detection of a gene may not be correlated with the successful expression of the β-lactamases. Carba NP test based on the detection of enzyme activity (Vasoo et al., 2013) are rapid but not specific for a single type of carbapenemase. The mCIM and eCIM, phenotypic detection methods based on carbapenem inactivation methods, can detect carbapenemases in Enterobacteriaceae and Pseudomonas aeruginosa and differentiate metallo-beta-lactamases from serine carbapenemases in Enterobacteriaceae (Tsai et al., 2020).
The direct detection and quantification of β-lactamases have become easier with the development of protein detection and quantitation techniques in recent years. Lateral flow immunoassay methods based on antigen-antibody reaction (Boutal et al., 2017) are limited to the types of antibodies, but actually they cover the vast majority of carbapenemases found in clinical microbiology routine, beyond being extremely fast. For the past few decades, liquid chromatography with tandem mass spectrometry (LC-MS/MS) has been widely used in various fields of protein analysis, biochemical analysis, natural product analysis, and drug and food analysis among other areas (Suh et al., 2017). LC-MS/MS has gradually become one of the most popular analytical tools for protein detection. Shotgun proteomics has been used to identify wild-type and resistant strains of the pathogen Acinetobacter baumannii (Chang et al., 2013). Additionally, a capillary electrophoresis-electrospray ionization-tandem mass spectrometry bottom-up proteomics workflow has been established for the identification of OXA-48 and KPC (Fleurbaaij et al., 2014). MALDI-TOF MS has been used to identify for carbepanemase detection with different analytical approaches (Hleba et al., 2021). Additionally, the bottom-up proteomics approach has also been applied to identify CTX-M ESBLs (Fleurbaaij et al., 2017). Recently, targeted LC-MS/MS based on selected reaction monitoring (SRM) and multiple reaction monitoring (MRM) using triple quadrupole mass analyzer, and parallel reaction monitoring (PRM) techniques using on high resolution/accurate mass has been used in β-lactamase testing. Specific peptides of A. baumannii identified via LC-MS/MS profiling have been used to classify clinical isolates (Honghui et al., 2016). Targeted high-resolution MS assays have been developed for the detection of KPC, OXA-48, NDM, and VIM enzymes (Wang et al., 2017, 2019; Foudraine et al., 2019; Strich et al., 2019). However, there are still many β-lactamases that have not been detected by targeted proteomics. Traditional SRM-MS on a triple quadrupole mass spectrometer is limited in the complex sample analysis due to the mass filtering and low resolution quadrupole, and more method development time is needed to define transitions (precursor/product ion pairs). In our study, a system comprising a rapid sample-processing procedure combined with improved PRM using HRAM (High resolution accurate mass) Orbitrap MS was developed to detect carbapenemases (KPC, IMP, VIM, NDM, and OXA), ESBLs (TEM and CTX-M), and AmpC (CMY-2) using a single colony of Enterobacteriaceae strains.
A total of identified and subcultured 192 Enterobacteriaceae strains were used in this study (Supplementary Table 3), including K. pneumoniae American Type Culture Collection (ATCC) BAA-2146 and 191 clinically isolated strains. The selected strains comprised 73 E. coli, 83 K. pneumoniae, 25 E. cloacae strains, five Klebsiella oxytoca, and six Citrobacter freundii. Four of these isolates were used for the test development process, and 188 were used for the test validation phase. All the strains were cultured in Luria-Bertani agar plates at 37°C overnight. All the strains were stored in the Collection Center of Pathogen Microorganism of Chinese Academy of Medical Sciences in China.
Single colonies (diameter >2 mm) were picked using a micropipette tip or a 10 μL loop, and resuspended in 200 μL of 50 mM ammonium bicarbonate (MS grade; Merck, Germany), sonicated for 1 min (3 s of sonication, 6 s of rest), centrifuged at 12,000 × g for 2 min and heated at 95°C for 5 min, after which the buffer was removed using 10K Nanosep centrifugal device with Omega membrane (Pall Corporation, Port Washington, NY, United States). Ammonium bicarbonate buffer (50 mM) was added along with sequencing grade trypsin (Promega Corporation, Madison, WI, United States), and the solution was microwaved in a water bath followed by heat treatment at 55°C. The peptide concentration was measured using the Pierce™ Quantitative Colorimetric Peptide Assay kit (Thermo Fisher Scientific, Waltham, MA, United States).
Data-dependent analysis was performed on the Thermo Scientific Orbitrap Fusion Lumos platform coupled with an EASY-nLC 1200 system (Thermo Fisher Scientific, San Jose, CA, United States) to build a spectral library. The digests were separated by the trap column [ReproSil-Pur 120 C18-AQ (3 μm, Dr. Maisch GmbH, Ammerbuch, Germany); 20 × 0.05 mm] followed by a C18 column [ReproSil-Pur 120 C18 (1.9 μm, Dr. Maisch GmbH, Ammerbuch, Germany); 120 × 0.15 mm] at a flow rate of 600 μL/min. The solvent buffer A comprised water with 0.1% formic acid, and solvent B comprised 80% acetonitrile with 0.1% formic acid. After sample loading, the gradient was initiated with 11% of buffer B, and then increased from 11 to 13% of buffer B for 2 min. The gradient increased up to 32% of buffer B in 16 min, and then to 42% in 7 min. Finally, the gradient was increased to 95% of buffer B in 1 min and was maintained for 4 min. The MS parameters were as follows: MS1 (Orbitrap analysis; mass range, approximately 350–1,550 m/z; resolution, 120,000; AGC target, 5 × 105; RF lens, 50%; maximum injection time, 50 ms), MS2 [high-energy collisional dissociation (HCD) collision energy, 32%; maximum injection time, 22 ms; AGC, 5 × 104; isolation window, 1.6 Da; Orbitrap solution, 15,000]. For database analyses, these raw data were searched against a combined bacterial antimicrobial resistance database downloaded from the National Center for Biotechnology Information (NCBI; National Institutes of Health, Bethesda, MD, United States) using the Thermo Scientific™ Proteome Discoverer™ version 2.2 (PD2.2) software. Trypsin was specified for protein digestion with two missed cleavages allowed for each peptide. The search parameters were set as previously described (Espadas et al., 2017).
The protein sequences of KPC, VIM, IMP, NDM, OXA, TEM, CMY, and CTX-M were downloaded from the NCBI database1 to build an amino acid sequence library. Tryptic peptides were searched and aligned using the protein basic local alignment search tool (BLAST2) to ensure uniqueness. The peptides with missed-cleaved, two neighboring basic amino acids at either cleavage site (KK, RR, KR, and RK), and W and M residues were excluded.
Targeted proteomics was performed on the Thermo Scientific Orbitrap Fusion Lumos platform coupled with an EASY-nLC 1200 platform. QQDLVDYSPVSEK and VDAGDEQLER served as internal reference. The columns used and elution gradient were the same as mentioned above. PRM parameters: MS1 spectrum (Orbitrap analysis; resolution, 60,000; mass range, approximately 350–2000 m/z; RF lens, 30%; AGC target, 2.0 × 105; maximum injection time, 50 ms) and MS2 analysis (HCD; collision energy, 30%; AGC, 5.0 × 104; maximum injection time, 54 ms for specific peptides or 22 ms for synthetic isotope labeled (SIL) peptides; Orbitrap resolution, 30,000 for specific peptides or 7,500 for SIL; isolation window, 1.4 Da). Data on the peptides [retention time (RT), m/z, and charge] were imported into the MS method. For data analysis, the acquired data were analyzed using Skyline 20.1.0.155 (MacCoss Lab Software, University of Washington, Seattle, WA, United States) (MacLean et al., 2010). The amino acid sequences of the drug-resistant enzymes downloaded from the NCBI were imported as the background library. Data-dependent acquisition (DDA) raw data were imported to build the spectra database. After the amino acid sequences of specific and SIL were inserted, the PRM data were imported and for each targeted peptide, the ratio between the peak area of the endogenous peptide and that of the SIL was calculated, and the relative concentration of targeted peptides was calculated based on the SIL with fixed quantity. Whether the CRE/ESBL enzymes were defined as positive or negative depended on the peptide when the following criteria were met: an RT similar to that of the SIL, library dot product (dotp) > 0.8 and ratio dot products (rdotp) > 0.95. Coefficients of variation (CV) were calculated, and stability of the peptides was evaluated by performing three freeze-thaw cycles of the peptides and storage of the peptides for 0, 1, 3, and 4 days in the sample holder (10°C).
Multi-drug resistant genes were analyzed via PCR (Dallenne et al., 2010) using the GoTaq Green Master Mix (Promega). The primers and parameters used are listed in Supplementary Table 1.
Amino acid sequences of the enzymes were downloaded from the NCBI,3 and potential peptides were evaluated using PeptideCutter.4 In view of the varying responses of peptides analyzed via MS, four strains (Table 1) were used to evaluate the ionization capabilities of peptides via DDA, and the specificity of the peptides was assessed by performing BLASTp searches. Unique peptides with high signal stability, appropriate RT, and relatively stable amino acid residues were chosen as peptides markers for KPC, IMP, VIM, NDM, OXA, CMY, CTX-M, and TEM. Val (13C5, 15N), Gly (15N), and Ala (13C3, 15N) were used to label the peptides (Table 2). Other candidate peptides detected are listed in Supplementary Table 2. The data are deposited in the PRIDE repository, accession number PXD028791.
To develop a rapid method for peptide detection, a 30-min Nano LC-MS/MS method was developed, with parameters for peptide markers as shown in Table 2. Different Orbitrap resolutions for specific and SIL were used to improve identification speed, quantity and quality (Stopfer et al., 2021). Figure 1 shows the workflow of the rapid detection method for β-lactamases. The raw data were analyzed using Skyline, and the library dotp, rdotp, and R ratio values were exported. According to the library dotp, rdotp, and R ratio values obtained from the strains, the following rules were set: library dotp > 0.8, rdotp > 0.95, and RT similar to that of SIL (Figure 2). A wash procedure for 30 min was performed after each sample to avoid false-positive results caused by the carryover effect.
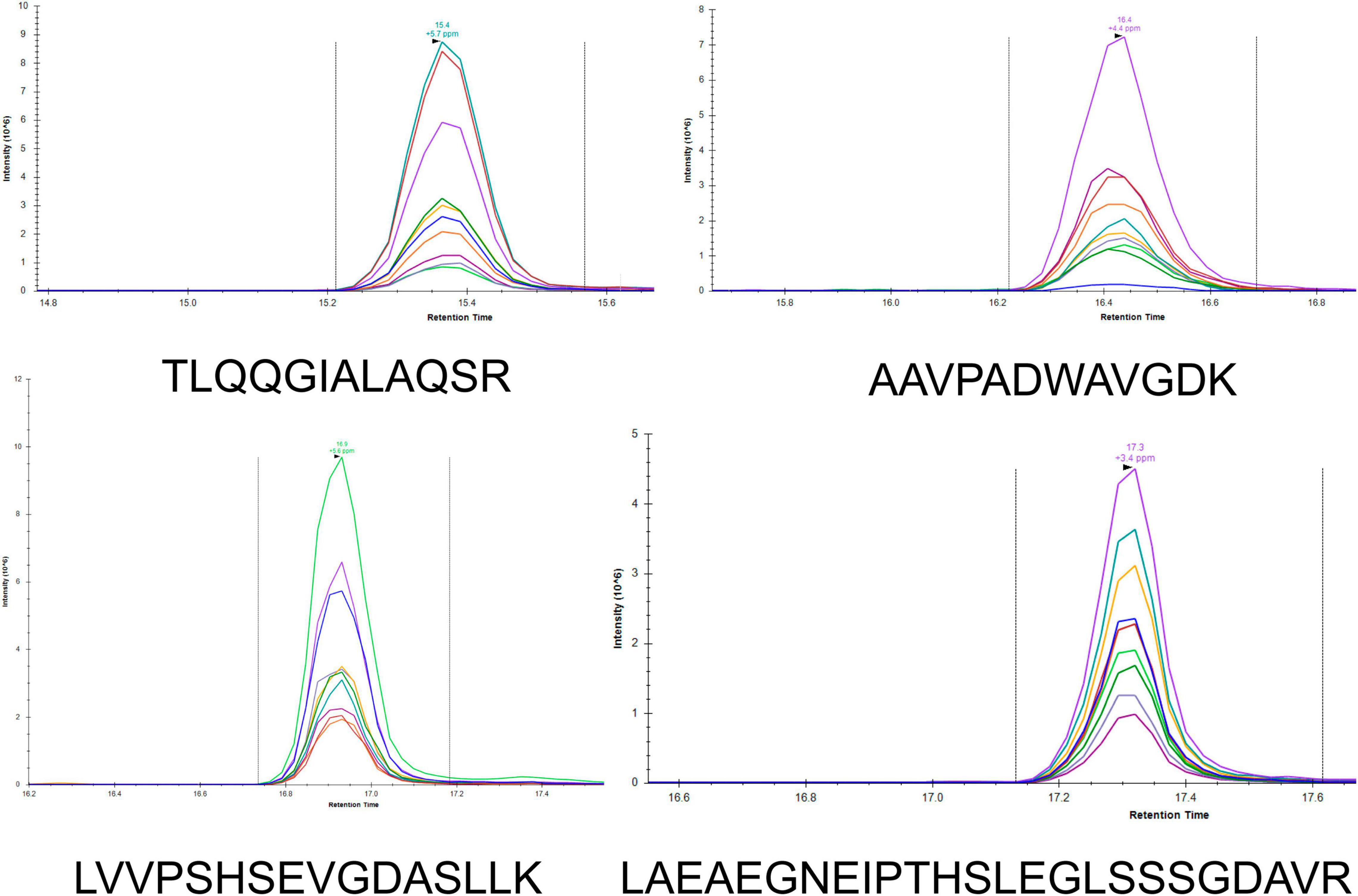
Figure 2. LC-MS/MS chromatograms of the peptides (TLQQGIALAQSR, AAVPADWAVGDK, LVVPSHSEVGDASLLK, and LAEAEGNEIPTHSLEGLSSSGDAVR) in Skyline. Different colors represent different fragment ions.
To obtain high-quality spectra while reducing the digestion time, protein solutions with trypsin were microwaved for 5, 10, and 15 min, as well as heated in a water bath at 55°C for 15, 30, and 45 min after being microwaved for 2 min separately. Peptides were collected and analyzed via Nano LC-MS/MS. The total processing time was <1 h. As shown in Supplementary Figure 1, the peptides could be detected for all digestion conditions even after being microwaved for 5 min. For most peptides, an increase in digestion time led to an increase in abundance; however, the digestion conditions had no effect on the peptides LAEAEGNEIPTHSLEGLSSSGDAVR, VQATNSFSGVNYWLVK, IINHNLPVK, and NSFGGVNYW LVK. To ensure that all peptides were identified under optimal conditions, microwave treatment for 2 min combined with heat treatment using a 55°C water bath for 30 min was performed in subsequent experiments.
The reproducibility of applications of the peptide markers was evaluated using four strains that were positive for KPC, IMP, VIM, NDM, OXA, CMY, CTX-M, and TEM based on the CV of six replicates in 1 day and in 3 different days (Table 3). The CVs of endogenous contents were determined using SIL (Table 2). The CVs of 16 peptides were <30%. While the CV of IINHNLPVK wasn’t acquired as signal miss in some of the samples. No carry-over effects were observed for any of the selected peptides.
Stability is an important property that should be evaluated in method development. The SIL were added to the peptide solutions of E. coli DH5α that did not contain β-lactamases. The peptide stability after three freeze-thaw cycles and stability in the sample holder were measured separately. Six replicates were performed for all experiments. As shown in Supplementary Figure 2, the contents of DGDELLLIDTAWGAK and VGYIELDLNSGK decreased significantly after three freeze-thaw cycles (<80%), whereas the contents of LIAQLGGPGGVTAFAR and APLILVTYFTQPQPK decreased slightly (approximately 80–90%). These results suggest that repeated freezing and thawing of these four peptides should be avoided during use. Regarding stability in the sample holder for 0, 1, 3, and 4 days, the content of DGDELLLIDTAWGAK decreased significantly (70%) in 1 day, and down to 6% on the fourth day (Supplementary Figure 3). The content of VGYIELDLNSGK decreased significantly (64%) on the third day, whereas the content of APLILVTYFTQPQPK reduced to approximately 82% on the fourth day. The results suggest that these peptides should be held for short durations in the sample holder.
After the preliminary evaluation of the rapid detection method, blinded testing of 188 clinical strains was conducted. Strains of E. coli, K. pneumoniae, E. cloacae strains, K. oxytoca, and C. freundii were included in the assays. As shown in Table 4, all the β-lactamases tested were detected both via LC-MS/MS and PCR. Most of the peptide markers for the β-lactamases showed 100% specificity except for LDEGVYVHTSFEEVNGWGVVPK(IMP, 85%), TEPTLNTAIPGDPR(CTX-M-1 group and 9 group, 97%), APLIL VTYFTQPQPK(CTX-M-1 group, 99%), SDLVNYNPIAEK (CTX-M-1 group, 95%), and VGYIELDLNSGK(TEM, 74%). Peptide markers for KPC(AAVPADWAVGDK), IMP-1, VIM-1, and NDM showed 100% sensitivity. However, the positive sensitivities for SQQQAGL LDTPIR(KPC), NSFGGVNYWLVK(IMP-4), LDEGVYVHTS FEEVNGWGVVPK(IMP), NNGLTEAWLESSLK(OXA-1), II NHNLPVK(OXA-1), ADIANNHPVTQQTLFELGSVSK(CMY-2), TLQQGIALAQSR(CMY-2), QLTLGHALGETQR(CTX-M-9 group), LIAQLGGPGGVTAFAR(CTX-M-9 group), TEPTLN TAIPGDPR(CTX-M-9 group, CTX-M-1 group partial), APLILVTYFTQPQPK(CTX-M-1 group), SDLVNYNPIAEK (CTX-M-1 group partial), and VGYIELDLNSGK(TEM) were 98% (49/50), 58% (14/24), 92% (24/26), 78% (14/18), 78% (14/18), 27% (6/22), 82% (18/22), 39% (27/70), 81% (57/70), 98% (116/118), 98% (57/58), 76% (44/58), and 74% (67/90), respectively. Peptide values were calculated using the labeled peptides that are listed in Table 4.
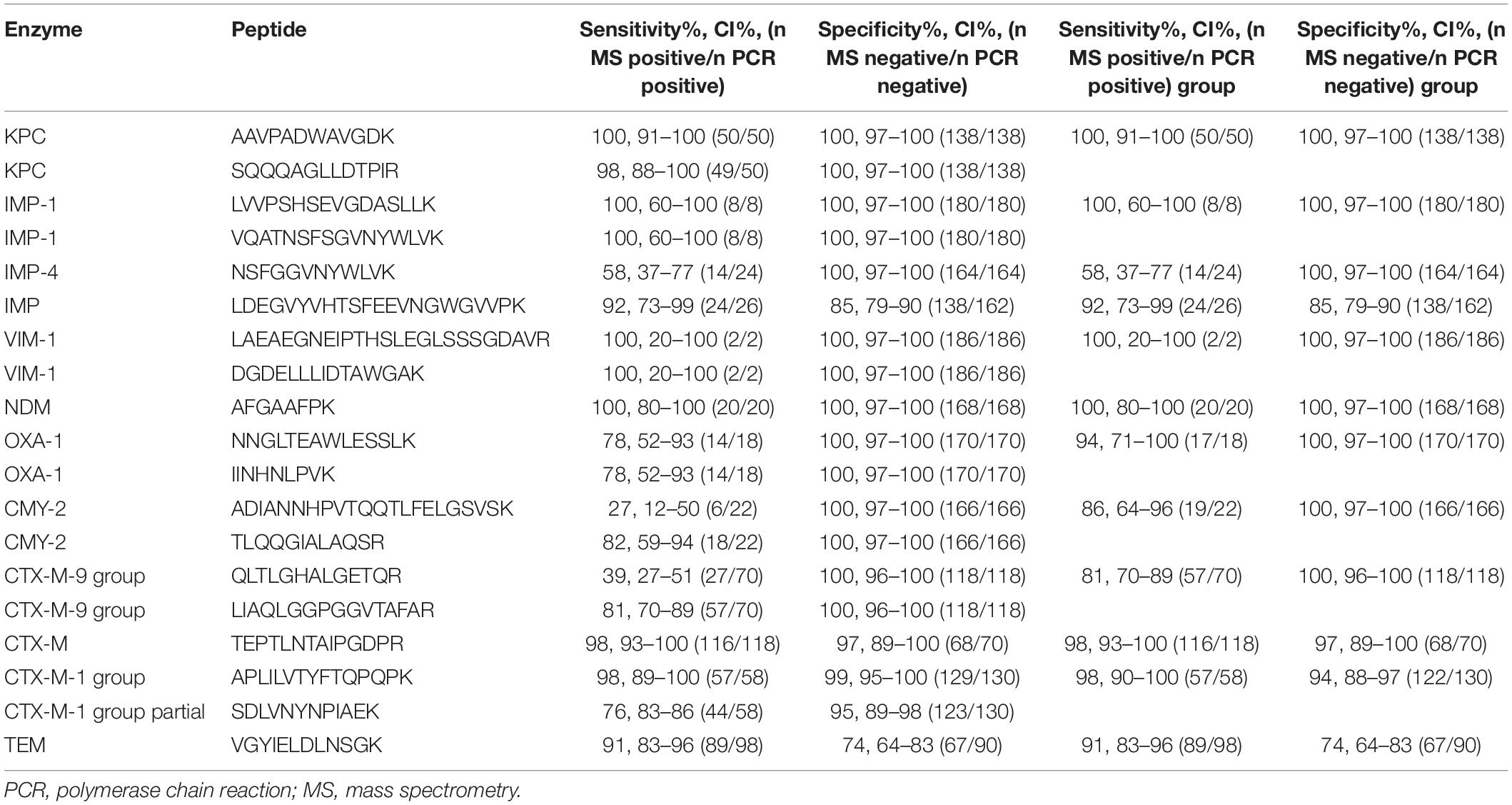
Table 4. Results of parallel reaction monitoring compared to those of polymerase chain reaction (PCR) in a validation set.
For the past few decades, β-lactam antibiotics have been one of the first-choice drugs for the treatment of several serious infectious diseases caused by Gram-negative bacteria (Bush and Bradford, 2016). However, the increasing rate of drug-resistance in bacteria has far exceeded the rate of development of new antibiotics at present. The spread of β-lactamases is mainly attributed to the presence of β-lactamase genes on plasmids, and the unreasonable use of antibiotics. Therefore, a timely detection of the type of β-lactamases will promote appropriate clinical antibiotic choice and inhibit the spread of drug-resistant genes (Iovleva and Doi, 2017). With the development of HRAM Orbitrap MS and supporting quantitative methods, the application of detection approaches based on specific peptides has gradually received attention. Initially, shotgun proteomics was used for β-lactamase detection and the proteins were detected in a single run; however, this depends on the database and results in a poor accuracy (Fleurbaaij et al., 2017). Subsequently, quantitative proteomics methods such as SRM and MRM have been applied to β-lactamases analysis, and detection based on specific peptides has proved to be feasible (Wang et al., 2017). Using LC-MS/MS to detect β-lactamases is a more direct approach than using PCR or disk-diffusion methods, and it is less hindered by multiple problems such as false-positive results. However, for peptide detection, the preparation procedure of peptides must be optimized to reduce the time taken.
In our study, we developed a rapid preparation procedure based on a single colony, thereby omitting the amplification process. Isolating bacteria using agar plates is the first step for isolating organisms from all types of clinical samples. As long as colonies are acquired, the identification process may be initiated. The procedures involving the reduction and alkylation of sulfhydryl groups were also removed as they do not significantly affect the digestion of targeted peptides. Moreover, a simplified Filter-aided sample preparation (FASP) method was used to ensure digestion efficiency. Previous studies have confirmed that microwave treatment for short periods can effectively digest the targeted peptides (Strich et al., 2019). In our study, we tested different digestion conditions for our selected peptides. Results showed that all the targeted peptides could be detected even after microwave treatment for only 5 min, which can greatly reduce the digestion time. For the LC-MS/MS procedure, a 30 min LC method combined with a PRM targeted method was used to identify the peptides. PRM can enable identification of multiple peptides in a high resolution and high mass accuracy mode (Navin, 2015). In contrast with a previous PRM detection method, we used a lower resolution for labeled peptides and higher resolution for targeted peptides to reduce the scanning time and improve the MS/MS quality as the concentration of SIL was high (Stopfer et al., 2021). By optimizing the detection methods, we have obtained a series of peptides with varying properties. Overall, a 30∼60 min preparation procedure, a 30 min LC-MS/MS procedure and a 10 min data processing were determined for the detection of β-lactamases.
For KPC, AAVPADWAVGDK and SQQQAGLLDTPIR were selected as peptides markers (Table 2 and Supplementary Table 2); LALEGLGVNGQ, LTLGSALAAPQR, and APIVLAVYTR were previously analyzed using Agilent 6540 Q-TOF (Wang et al., 2017). NALVPWSPISEK was detected for the first time but was not selected as a marker owing to its low dotp values. Both of the sensitivity and specificity of AAVPADWAVGDK was 100% as it exists in all the genotypes of blaKPC, indicating the possibility of becoming a peptide marker for KPC. But for SQQQAGLLDTPIR, the sensitivity was lower as the absence of blaKPC–13, blaKPC–45, andblaKPC–59. This study shows the first successful detection of IMP via LC-MS/MS. The results proved that LVVPSHSEVGDASLLK, VQATNSFSGVNYWLVK, and NSFGGVNYWLVK could effectively be used to identify IMP and distinguish the subtypes of blaIMP–1 and blaIMP–4. LDEGVYVHTSFEEVNGWGVVPK showed lower specificity for IMP detection. And for NSFGGVNYWLVK, it is inexplicable that the discrepant results were verified to be blaIMP–4 positive, while the Skyline map fragments irons of the discrepant results was different from the positive ones. This is the first time this phenomenon has been observed, and it may be related to the fragmentation of the peptide. Notably, LVVPSHSEAGDASLLK (IMP-4), whose A was replaced by V in IMP-1 (Δ14Da), was also detected and could be separated via high-resolution MS. LAEAEGNEIPTHSLEGLSSSGDAVR and DGDELLLIDTAWGAK were used to detect VIM-1, and both of them showed a better subtype coverage. However, as the lack of blaVIM–1 strains collected, only 2 blaVIM–1 positive strains were identified in 188 strains. With the continuous collection of blaVIM–1 positive strains, the effectiveness of detection by peptide markers can be better verified in the future. AFGAAFPK was selected as the marker as it exists in all NDM subtypes currently listed by the NCBI, and was also detected via the MRM targeted method (Wang et al., 2019). blaOXA is widely distributed in Enterobacteriaceae. Regarding the blaOXA–1 family, four peptides were detected, and NNGLTEAWLESSLK and IINHNLPVK, which were specific for the blaOXA–1 family were used for screening. However, according to the reproducibility results, the LC-MS/MS signal of IINHNLPVK was not stable in the detection process. blaCMY is an AmpC type ESBL gene. ADIANNHPVTQQTLFELGSVSK and TLQQGIALAQSR showed 27 and 82% sensitivity for the strains used in this study. However, for TEM (including TEM-1, TEM-2, and variants listed on the NCBI ANTIMICROBIAL RESISTANCE GENE database), LLTGELLTLASR, SALPAGWFIADK, IHYSQNDLVEYSPVTEK, QIAEIGASLIK, VDAGQEQLGR, VDAGQEQLGRR, and VGYIELDLNSGK were identified in DDA experiments (Supplementary Table 2). QIAEIGASLIK and VGYIELDLNSGK were choose as peptides markers, and QIAEIGASLIK was removed for its poor spectrum. For the 188 strains detected, VGYIELDLNSGK showed 91% sensitivity with lower specificity as 74% (67/90). Overall, this is the first study to use a PRM-based LC-MS/MS system to detect OXA, CMY, and TEM. blaCTX–M ESBL is a large group of ESBLs with an increasing number of subtypes. According to genetic structure, blaCTX–M enzymes are divided into four groups (CTX-M-1, CTX-M-2, CTX-M-9, and CTX-M-25) (Canton et al., 2012). Among them, the CTX-M-15 (CTX-M-1 group) and CTX-M-14 (CTX-M-9 group) are by far the most prevalent enzymes. In our study, only APLILVTYFTQPQPK showed 98% sensitivity for the CTX-M-1 group. QLTLGHALGETQR and LIAQLGGPGGVTAFAR only existed in certain strains of the CTX-M-9 group, whereas SDLVNYNPIAEK showed 76% sensitivity for the CTX-M-1 group. Additionally, TEPTLNTAIPGDPR could identify 98% of the strains containing CTX-M-1 or CTX-M-9 groups. The reason for the relatively lower sensitivity may be that the peptides are not the representative peptides for the entire genotype group, but only for the partial group due to the complexity of the variants. It may not be possible to use single peptide to achieve 100% sensitivity. As the overall detection sensitivity for each drug resistant enzyme is more important than the detection sensitivity of individual peptide, to overcome this complexity of huge number of variants, sensitivity and specificity based on peptides groups were calculated as shown in Table 4. The sensitivity of KPC, OXA, CMY-2, CTX-M-9 group, and CTX-M-1 peptides groups raised to 100, 94, 76, 81, and 98%. In addition, much more peptide markers need to be effectively detected and added for the group detection in the future study. In addition, several variants were observed for the peptides of blaCTX–M, such as LGVALIDTADNTQVLYR, LGVALINTADNTQTLYR, and LGVALINTADNSQILYR, which are similar in terms of m/z and RT, representing great challenges to detection via MS. Therefore, to improve the sensitivity of detection via LC-MS/MS, an overall analysis is suggested using all the peptides in one method. Further studies are required to identify specific peptides for one group or the entire blaCTX–M group.
The dotp is a measure of similarity between the fragment ratio of the endogenous and library peptide. The rdotp is a measure of similarity between the fragment ratio of the endogenous and SIL peptide. The criterium for judging positivity by a previous study was rdotp value > 0.95 (Foudraine et al., 2019). When we re-analyzed the results of the targeted proteomics experiment, we found that unified screening rules might not have been suitable for all peptides. For many peptides such as LDEGVYVHTSFEEVNGWGVVPK (IMP), DGDELLLIDTAWGAK (VIM-1), and TEPTLNTAIPGDPR (CTX-M-1 group partial), a rdotp value > 0.95 is not enough to screen for positive strains; therefore, library dotp values > 0.8 are required. For QLTLGHALGETQR (CTX-M-9), 100% (70/70) strains were positive for library dotp > 0.8 compared to PCR results; however, only 39% (27/70) strain was positive based on values of rdotp > 0.95 and library dotp > 0.8. For LIAQLGGPGGVTAFAR (CTX-M-9), 100% (70/70) strains were positive for library dotp > 0.8 compared to PCR results; however, only 81% (57/70) strains were positive based on values of rdotp > 0.95 and library dotp > 0.8. This phenomenon is partly due to the existence of variants.
Overall, directly identifying peptides via LC-MS/MS provides a new approach to detect β-lactamases. Our results showed the great potential of the rapid extraction and detection method in the detection of β-lactamases. Started by picking the colony, peptides were obtained by rapid ultrasonic lysis and rapid digestion (even in a microwave oven for 5 min), and then results were acquired by rapid LC-MS/MS analysis (30 min) and data processing. The application of targeted proteomics using single bacterial colonies and even blood samples in the future, will enable early clinical diagnostics and early treatment. However, there are several problems that need to be addressed. For example, at present, drug-resistant enzymes are named and classified based on gene sequence; however, the protein sequence is more decisive in determining function. Additionally, there is a lack of protein subtypes and classification based on amino acid sequences. Therefore, it is difficult to identify specific peptides for one group or one subtype. Variants are ubiquitous in β-lactamase enzymes such as blaCTX–M. The separation of variants with a similar m/z and RT through regular LC-MS/MS remains a large challenge.
In conclusion, the rapid and accurate identification of β-lactamases is of great significance to clinical diagnostics and treatment. This study describes a rapid extraction and detection workflow for widespread β-lactamases, including KPC, IMP, VIM, NDM, OXA, CMY, CTX-M, and TEM using single colonies of Enterobacteriaceae strains. PRM targeted proteomics was proven to be a promising approach for the detection of drug-resistant enzymes.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
XFY and YL: conceptualization and funding acquisition. YL, XH, XW, and JP: methodology. YL and GL: software. YL: validation, formal analysis, writing—original draft preparation, and supervision. XH: resources. XYY, XFY, and CL: writing—review and editing. GL: visualization. XFY: project administration. All authors have read and agreed to the published version of the manuscript.
This study was supported by the National Natural Science Foundation of China (32141003, 81803593, and 81621064), the CAMS Initiative for Innovative Medicine (2016-I2M-3-014), the National Mega-project for Innovative Drugs (2019ZX09721001), the MOST/MOF Funds for the National Science and Technology Resource Sharing Service Platform, China (2019-194-NPRC), and the Fundamental Research Funds for the Central Universities (3332018094).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We are very grateful to Dr. Hanxing Cheng and Dr. Yingzi Qi from Thermo Fisher Scientific for the help in PRM method development.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.784628/full#supplementary-material
Boutal, H., Naas, T., Devilliers, K., Oueslati, S., Dortet, L., Bernabeu, S., et al. (2017). Development and validation of a lateral flow immunoassay for rapid detection of NDM-Producing Enterobacteriaceae. J. Clin. Microbiol. 55, 2018–2029. doi: 10.1128/JCM.00248-17
Bush, K., and Bradford, P. A. (2016). β-Lactams and β-Lactamase inhibitors: an overview. Cold Spring Harb. Perspect. Med. 6:a025247. doi: 10.1101/cshperspect.a025247
Bush, K., and Jacoby, G. A. (2010). Updated functional classification of beta-lactamases. Antimicrob. Agents Chemother. 54, 969–976. doi: 10.1128/AAC.01009-09
Canton, R., Gonzalez-Alba, J. M., and Galan, J. C. (2012). CTX-M Enzymes: Origin and Diffusion. Front. Microbiol. 3:110. doi: 10.3389/fmicb.2012.00110
Chang, C. J., Lin, J. H., Chang, K. C., Lai, M. J., Rohini, R., and Hu, A. (2013). Diagnosis of β-Lactam resistance in Acinetobacter baumannii using shotgun proteomics and LC-Nano-Electrospray ionization ion trap mass spectrometry. Anal. Chem. 85, 2802–2808. doi: 10.1021/ac303326a
Dallenne, C., Costa, A. D., Decré, D., and Favier, C. (2010). Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65, 490–495. doi: 10.1093/jac/dkp498
Duin, V. D. (2017). Carbapenem-resistant Enterobacteriaceae: What we know and what we need to know. Virulence 8, 379–382. doi: 10.1080/21505594.2017.1306621
Eliopoulos, G. M., and Karen, B. (2001). New beta-lactamases in gram-negative bacteria: diversity and impact on the selection of antimicrobial therapy. Clin. Infect. Dis. 32, 1085–1089. doi: 10.1086/319610
Espadas, G., Borràs, E., Chiva, C., and Sabidó, E. (2017). Evaluation of different peptide fragmentation types and mass analyzers in data-dependent methods using an Orbitrap Fusion Lumos Tribrid mass spectrometer. Proteomics 17:1600416. doi: 10.1002/pmic.201600416
Fleurbaaij, F., Goessens, W., van Leeuwen, H. C., Kraakman, M. E. M., Bernards, S. T., Hensbergen, P. J., et al. (2017). Direct detection of extended-spectrum beta-lactamases (CTX-M) from blood cultures by LC-MS/MS bottom-up proteomics. Eur. J. Clin. Microbiol. Infect. Dis. 36, 1621–1628. doi: 10.1007/s10096-017-2975-y
Fleurbaaij, F., Heemskerk, A. A., Russcher, A., Klychnikov, O. I., Deelder, A. M., Mayboroda, O. A., et al. (2014). Capillary-electrophoresis mass spectrometry for the detection of carbapenemases in (multi-)drug-resistant Gram-negative bacteria. Anal. Chem. 86, 9154–9161. doi: 10.1021/ac502049p
Foudraine, D. E., Dekker, L. J. M., Strepis, N., Bexkens, M. L., and Goessens, W. H. F. (2019). Accurate detection of the four most prevalent carbapenemases in E. coli and K. pneumoniae by high-resolution mass spectrometry. Front. Microbiol. 10:2760. doi: 10.3389/fmicb.2019.02760
Hleba, L., Hlebova, M., Kovacik, A., Cubon, J., and Medo, J. (2021). Carbapenemase producing Klebsiella pneumoniae (KPC): What is the best MALDI-TOF MS Detection Method. Antibiotics 10:1549. doi: 10.3390/antibiotics10121549
Honghui, W., Drake, S. K., Chen, Y., Marjan, G., Margaret, T., Rosenberg, A. Z., et al. (2016). A novel peptidomic approach to strain typing of clinical Acinetobacter baumannii isolates using mass spectrometry. Clin. Chem. 62, 866–875.
Iovleva, A., and Doi, Y. (2017). Carbapenem-Resistant Enterobacteriaceae. Clin. Lab. Med. 37, 303–315. doi: 10.1016/j.cll.2017.01.005
Kwon, T., Jung, Y. H., Lee, S., Yun, M.-r., Kim, W., and Kim, D.-W. (2016). Comparative genomic analysis of Klebsiella pneumoniae subsp. pneumoniae KP617 and PittNDM01, NUHL24835, and ATCC BAA-2146 reveals unique evolutionary history of this strain. Gut Pathog. 8, 1–16. doi: 10.1186/s13099-016-0117-1
MacLean, B., Tomazela, D. M., Shulman, N., Chambers, M., Finney, G. L., Frewen, B., et al. (2010). Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968. doi: 10.1093/bioinformatics/btq054
Navin, R. (2015). Parallel reaction monitoring: a targeted experiment performed using high resolution and high mass accuracy mass spectrometry. Int. J. Mol. Sci. 16, 28566–28581. doi: 10.3390/ijms161226120
Philippon, A., Arlet, G., and Jacoby, G. A. (2002). Plasmid-determined AmpC-type beta-lactamases. Antimicrob. Agents Chemother. 46, 1–11. doi: 10.1128/aac.46.1.1-11.2002
Pitout, J. D., Nordmann, P., Laupland, K. B., and Poirel, L. (2005). Emergence of Enterobacteriaceae producing extended-spectrum beta-lactamases (ESBLs) in the community. J. Antimicrob. Chemother. 56, 52–59. doi: 10.1093/jac/dki166
Rodríguez-Baño, J., Gutiérrez-Gutiérrez, B., Machuca, I., and Pascual, A. (2018). Treatment of Infections Caused by Extended-Spectrum-Beta- Lactamase-, AmpC-, and Carbapenemase-Producing Enterobacteriaceae. Clin. Microbiol. Rev. 31:e00079-17. doi: 10.1128/CMR.00079-17
Stopfer, L. E., Flower, C. T., Gajadhar, A. S., Patel, B., Gallien, S., Lopez-Ferrer, D., et al. (2021). High-density, targeted monitoring of tyrosine phosphorylation reveals activated signaling networks in human tumors. Cancer Res. 81, 2495–2509. doi: 10.1158/0008-5472.CAN-20-3804
Strich, J. R., Wang, H., Cisse, O. H., Youn, J. H., Drake, S. K., Chen, Y., et al. (2019). Identification of the OXA-48 carbapenemase family by use of tryptic peptides and liquid Chromatography-Tandem mass spectrometry. J. Clin. Microbiol. 57:e01240-18. doi: 10.1128/JCM.01240-18
Suh, J. H., Makarova, A. M., Gomez, J. M., Paul, L. A., and Saba, J. D. (2017). An LC/MS/MS method for quantitation of chemopreventive sphingadienes in food products and biological samples. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 106, 292–299. doi: 10.1016/j.jchromb.2017.07.040
Tsai, Y. M., Wang, S., Chiu, H. C., Kao, C. Y., and Wen, L. L. (2020). Combination of modified carbapenem inactivation method (mCIM) and EDTA-CIM (eCIM) for phenotypic detection of carbapenemase-producing Enterobacteriaceae. BMC Microbiol. 20:315. doi: 10.1186/s12866-020-02010-3
Vasoo, S., Cunningham, S. A., Kohner, P. C., Simner, P. J., Mandrekar, J. N., Lolans, K., et al. (2013). Comparison of a novel, rapid chromogenic biochemical assay, the carba NP Test, with the modified hodge test for detection of Carbapenemase-Producing Gram-Negative Bacilli. J. Clin. Microbiol. 51, 3097–3101. doi: 10.1128/JCM.00965-13
Wang, H., Drake, S. K., Youn, J. H., Rosenberg, A. Z., Chen, Y., Gucek, M., et al. (2017). Peptide markers for rapid detection of KPC Carbapenemase by LC-MS/MS. Sci. Rep. 7:2531. doi: 10.1038/s41598-017-02749-2
Wang, H., Strich, J. R., Drake, S. K., Chen, Y., and Suffredini, A. F. (2019). Rapid Identification of New Delhi Metallo-β-Lactamase (NDM) Using Tryptic Peptides and LC-MS/MS. Antimicrob. Agents Chemother. 63:e00461-19. doi: 10.1128/AAC.00461-19
Keywords: Enterobacteriaceae, β-lactamases, specific peptides, detection, PRM
Citation: Lu Y, Hu X, Pang J, Wang X, Li G, Li C, Yang X and You X (2022) Parallel Reaction Monitoring Mass Spectrometry for Rapid and Accurate Identification of β-Lactamases Produced by Enterobacteriaceae. Front. Microbiol. 13:784628. doi: 10.3389/fmicb.2022.784628
Received: 12 January 2022; Accepted: 03 June 2022;
Published: 20 June 2022.
Edited by:
Karsten Becker, University Medicine Greifswald, GermanyReviewed by:
Robert A. Bonomo, United States Department of Veterans Affairs, United StatesCopyright © 2022 Lu, Hu, Pang, Wang, Li, Li, Yang and You. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuefu You, eHVlZnV5b3VAaG90bWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.