
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 06 May 2022
Sec. Microbiotechnology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.763014
 Cheng-Gang Ren1,2
Cheng-Gang Ren1,2 Cun-Cui Kong1
Cun-Cui Kong1 Zheng-Yi Liu1,2
Zheng-Yi Liu1,2 Zhi-Hai Zhong1,2
Zhi-Hai Zhong1,2 Jian-Chao Yang3
Jian-Chao Yang3 Xiao-Li Wang4
Xiao-Li Wang4 Song Qin1,2*
Song Qin1,2*
Soil salinity adversely affects plant growth and has become a major limiting factor for agricultural development worldwide. There is a continuing demand for sustainable technology innovation in saline agriculture. Among various bio-techniques being used to reduce the salinity hazard, symbiotic microorganisms such as rhizobia and arbuscular mycorrhizal (AM) fungi have proved to be efficient. These symbiotic associations each deploy an array of well-tuned mechanisms to provide salinity tolerance for the plant. In this review, we first comprehensively cover major research advances in symbiont-induced salinity tolerance in plants. Second, we describe the common signaling process used by legumes to control symbiosis establishment with rhizobia and AM fungi. Multi-omics technologies have enabled us to identify and characterize more genes involved in symbiosis, and eventually, map out the key signaling pathways. These developments have laid the foundation for technological innovations that use symbiotic microorganisms to improve crop salt tolerance on a larger scale. Thus, with the aim of better utilizing symbiotic microorganisms in saline agriculture, we propose the possibility of developing non-legume ‘holobionts’ by taking advantage of newly developed genome editing technology. This will open a new avenue for capitalizing on symbiotic microorganisms to enhance plant saline tolerance for increased sustainability and yields in saline agriculture.
The current world population of 7.8 billion is expected to reach 9.8 billion in 2050, increase by 25% in the next 30 years (Figure 1A). Global food production will need to increase as well. Historically, the highest global population growth rates, with increases of over 1.8% per year, occurred between 1955 and 1975, peaking to 2.1% between 1965 and 1970 (UN, 2019). In roughly coincident time frames, scientific and technical advances induced a series of innovations in farming that increased crop yields dramatically and were later known as the “Green Revolution” (GR, the 1950s to 1970s) (Wu et al., 2020). Part of the core operation of the GR was carrying out large-scale monoculture and using chemical pesticides, herbicides, and fertilizers (Liu et al., 2014; Nicolopoulou-Stamati et al., 2016). This farming method successfully increased the grain yield of the main crops (Figure 1B). However, its adverse consequences are becoming increasingly apparent. The inevitable consequence of large-scale monoculture is deep-rooted perennial species are replaced by shallow-rooted, annual species. It will increase leakage and groundwater recharge, leading to dissolved salts move toward the soil surface. Eventually, the soil becomes salinized (FAO, 2015). Going hand-in-hand, the increasing use of synthetic fertilizers poses hidden dangers to sustainable agricultural development and food security worldwide (Figure 1C). Therefore, achieving food security for the growing population amidst the gradual salinization of farmland is one of the most important missions for modern agriculture.
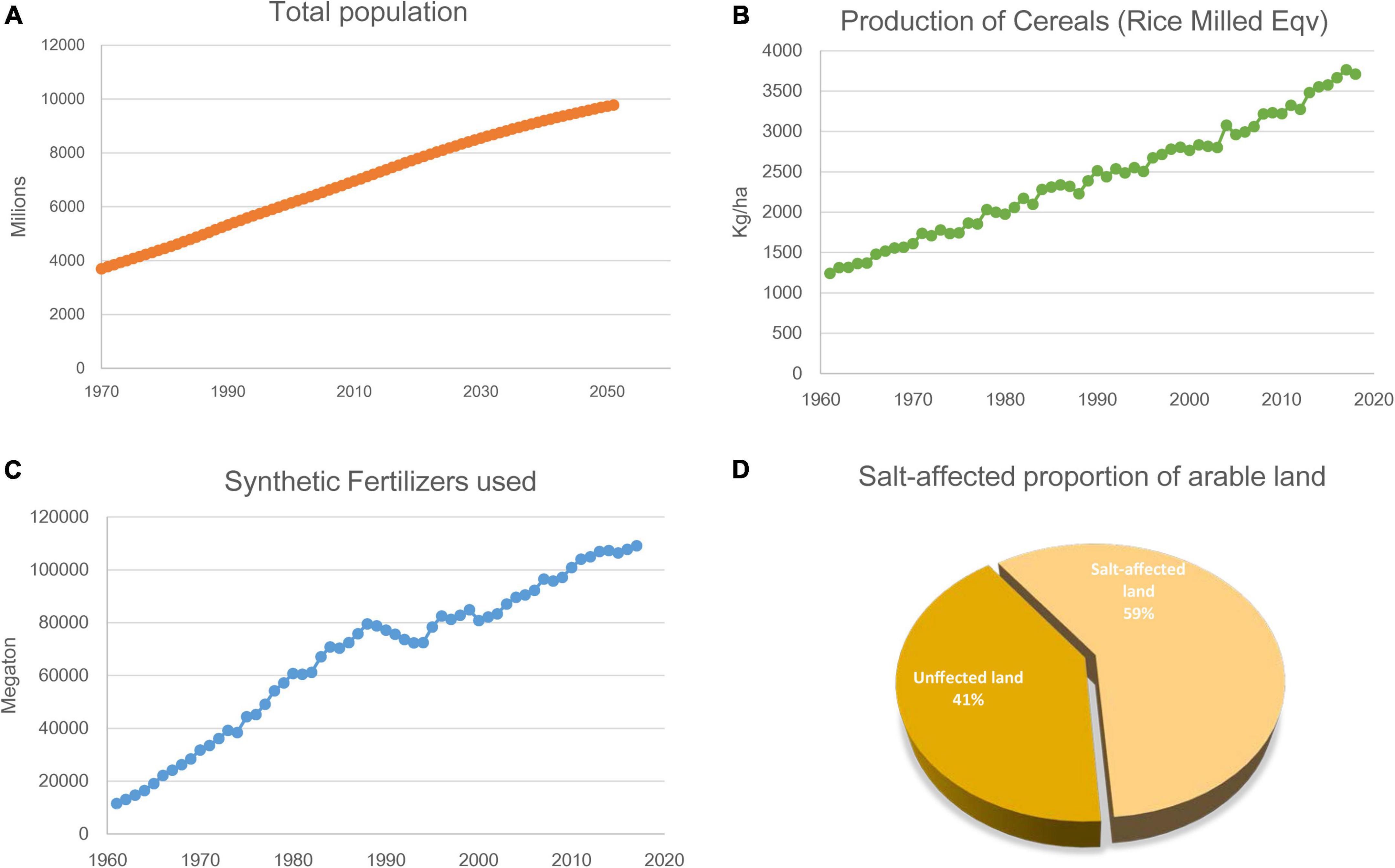
Figure 1. Global population, crop yield, synthetic fertilizers used and salt-affected proportion of arable land. (A) World population from 1970 to 2060. (B) Production of Cereals (Rice Milled Eqv) of the world since 1961 to 2018. (C) Consumption of synthetic nitrogen fertilizers of the world since 1961 to 2018. (D) Salt-affected proportion of arable land. All data taken from FAO-STAT (http://www.fao.org/statistics/en/). Figures ellaborated by the authors from FAOSTAT data.
It is not economically or environmentally feasible to expand traditional agricultural practices to meet future demand. Therefore, there is an urgent need for alternative technologies to sustainably meet global food security requirements. One way to increase sustainable crop yields is to amplify the role of plant–microbe symbiosis. Symbiotic microorganisms such as arbuscular mycorrhizal (AM) fungi and rhizobia can significantly improve crop growth and vigor, nutrient utilization efficiency, and biological/abiotic stress resistance. If these effects could be used in saline agriculture, they could increase agricultural productivity and food quality sustainably, thereby bringing positive environmental, social, and economic results.
Since the beginning of industrialization, human activities have severely damaged the natural hydrological balance in many regions of the world. These activities affect the natural distribution of salt in various surface landscapes and ultimately lead to the deterioration of the natural and agricultural environment. On a global scale, soil salinization has become a growing threat to food production amidst increasing climate change. Soil salinization is commonly caused by climate changes (primary) or anthropogenic activity (secondary). Primary processes include weathering of mother rock, seawater deposition, and atmospheric deposition. Secondary processes include inadequate drainage, brackish water irrigation, and long-term continuous agricultural irrigation (Rengasamy, 2010). The area of primary salinization is estimated to be slightly under 1 billion ha. Secondary salinization has occurred on around 77 million ha, of which 58% is in irrigated areas; as much as 20% of all irrigated areas are estimated to be salt-affected within India, Pakistan, China, Iraq, and Iran (FAO, 2015; Abbaspour and Ashraf Vaghefi, 2019). About 5.2 billion ha of the world’s agricultural land is already salt-affected and not suitable for conventional crop farming (Figure 1D; Ahmed et al., 2016).
Soil salinization is caused by the excessive accumulation of ions in the soil, including calcium, magnesium, sodium, sulfate, and chloride ions, resulting in plant growth inhibition. Excessive salt interferes with the absorption of nutrients and water by plants, thereby disrupting the physiological processes necessary for plant growth and development (Munns, 2002). Therefore, salinization is an important factor causing land degradation and a major threat to non-renewable soil resources. Unfortunately, the salinization of farmland is continuing and is estimated to be expanding by 0.3–1.5 million hectares every year, resulting in crop yield losses in these areas of more than 20% (Porcel et al., 2011). Worldwide soil salinization will have a double impact on social and economic progress. With the continuous salinization of arable land, agricultural income and the world food supply will eventually suffer. It is estimated that 12–27.3 billion US dollars are lost annually due to reductions in crop productivity (Qadir et al., 2014).
Soil salinity affects the germination and growth of plants, and excessive salinity can cause severe yield reductions (Evelin et al., 2009). Excessive salinity has three negative effects on plants. First, the toxic effects of specific ions such as sodium and chlorine inhibit protein synthesis and damaged organelles, enzyme structures, and the system on which photosynthesis and respiration depend. Second, excessive salt can hinder nutrient absorption and/or transportation to shoots, resulting in nutrient deficiency in plants (Marschner, 2002; Evelin et al., 2009). Finally, too much salt in the soil reduces its osmotic potential and hinders water absorption by the root system, leading to physiological drought in the plant. In this state, the plant must reduce its internal osmotic potential to prevent water from entering the soil from the roots. Due to their immobility, when facing constant environmental stress plants not only develop their adaptive mechanisms, but also co-evolve with soil microorganisms to develop complex mechanisms to resist stress. For example, the interaction with symbiotic soil microorganisms such as rhizobium and AM fungi have a great impact on the salt stress tolerance of plants (Table 1; Marschner, 2002; Porcel et al., 2011; Ngom et al., 2016).
AM fungi can establish a symbiotic relationship with approximately 80% of terrestrial plant species, including crops (Berruti et al., 2016). These microsymbionts exist by obtaining nutrients from plants, and can effectively help plants to obtain water and nutrients needed for growth from the soil. In addition, they can improve the ability of plants to resist abiotic stress (Shabala and Pottosin, 2014). In brief, AM fungi increase the osmotic potential of root cells by enhancing the host plant’s water absorption, nutrient intake, and accumulation of osmotic adjustment substances, thereby reducing salt stress on the host plant. Studies have shown that the formation of arbuscular mycorrhizae can reduce the absorption of Cl– ions by root tissues and at the same time prevent the transfer of Na+ to upper stem and leaf tissues under high salinity (Evelin et al., 2009). Under natural conditions, AM fungi can survive in high-concentration saltwater environments (Evelin et al., 2009). For instance, AM fungi were found in the heavily saline-alkali soil of the Tabriz Plain, Iran, with a soil salinity up to 92.0 dS/m (Aliasgharzad et al., 2001). The effect of AM fungi on plant salt tolerance has been studied in many plants, including giant reed (Romero-Munar et al., 2019), Sesbania (Kong et al., 2017; Ren et al., 2018), Zea mays (Sheng et al., 2011; Krishnamoorthy et al., 2016), cucumber (Hashem et al., 2018), olive (Porras-Soriano et al., 2009), Chrysanthemum morifolium (Wang et al., 2018), durum wheat (Fileccia et al., 2017), rice (Porcel et al., 2016), desert grass (Hashem et al., 2015) and tomato (Khalloufi et al., 2017). In mycorrhizal plants, AM fungi improved salt tolerance, helped maintain normal growth and yield under salt stress (Elhindi et al., 2017; Fileccia et al., 2017; Wang et al., 2018), nutrient absorption capacity (Krishnamoorthy et al., 2016; Elhindi et al., 2017), photosynthesis capability (Hashem et al., 2015; Shamshiri and Fattahi, 2016; Chen et al., 2017), and proline content, and promoted the accumulation of soluble sugars in roots. Under salt stress, the colonization of arbuscular mycorrhizae significantly increased the biomass of Sesbania (Kong et al., 2017); at 100 mM salinity, the biomass increased by 431%. It has also been reported that AM fungus inoculation has a similar growth-promoting effect on sweet sorghum, and can promote better biomass production than in plants without AM fungus inoculation in a salt environment (Wang et al., 2019). These benefits of AM fungi under saline conditions depend on the symbiotic associations formed by specific strains and plants (Table 1); therefore, it is necessary to select efficient fungal strains for certain plants.
Rhizobium is a genus of Gram-negative multi-source soil bacteria that can form nodules on the roots of legumes. These bacteria exist in special root nodule cells and provide nitrogen for plant growth by fixing N2 from the atmosphere, while at the same time the plant provides a carbon source for their growth (Pawlowski and Demchenko, 2012). Many studies have shown that inoculating suitable rhizobia strains can increase the dry weight of legumes under salt stress conditions, including Sesbania cannabina (Ren et al., 2016), Stylosanthes guianensis (Dong et al., 2017), chickpea (Deepika and Satyavir, 2015), pigeon pea (Bano et al., 2015), and soybean (Egamberdieva et al., 2017). This growth-promoting effect comes from an effective symbiotic relationship. Ethane reduction activity can be detected even under high salt conditions, but depends on the specific rhizobia–legume symbiosis combination (Bala et al., 1990; Elsheikh and Wood, 1995). Studies have shown that under salt stress conditions, salt-tolerant rhizobia strains can form a functional symbiosis with S. cannabina, and soybean (Ren et al., 2016; Egamberdieva et al., 2017), while salt-sensitive strains cannot. These results indicate that the inoculation of salt-tolerant rhizobia can improve biological nitrogen fixation under salt stress conditions. Numerous studies have shown that fast-growing rhizobia are more salt-tolerant than slow-growing rhizobia. Strains of the genus Rhizobium are generally more salt-tolerant than those of the genus Bradyrhizobium. Therefore, inoculating symbiotic strains with stronger salt tolerance under salt stress conditions may better promote the growth and yield of host plants (Zahran, 2001).
Several salt-tolerant rhizobia have been isolated that can tolerate high salt environments (Oshone et al., 2013; Srivastava et al., 2013). Some of these strains can grow at NaCl concentrations exceeding 350 mM (Dong et al., 2017). The salt tolerance of rhizobia is related to the accumulation of various osmotic adjustment substances in their cells (Del Cerro et al., 2019). These osmotic regulators include K+, glutamic acid, proline, glycine betaine, proline betaine, trehalose, dipeptide N-acetyl glutamine, and poly β-hydroxybutyrate. Their protective effects on rhizobia cells under high-salt conditions have been reported one after another. Studies have found that in R. meliloti salt-tolerant strains, the glycine betaine content is higher than insensitive strains (Dong et al., 2017). It is also believed that IAA synthesis by rhizobia can prevent the harmful effects of salinity. Bianco and Defez (2009) reported an IAA over-yielding mutant of Sinorhizobium meliloti, which significantly increased the tolerance of Medicago truncatula to salt stress. Compared with the wild-type strain, the proline content and accumulation of antioxidant enzymes were higher in plants inoculated with the mutants. Inoculation with these symbiotic bacteria can help the host plants effectively resist salt stress. For example, it has been reported that inoculation of B. japonicum S2492 significantly increased the dry weight, plant height, and yield (> 35%) of soybeans in arid saline soil (Egamberdiyeva et al., 2004).
In summary, previous studies have shown that selected AM fungi and rhizobia strains that are compatible with plants can be used to improve the salt tolerance of crops and plants used for saline soil remediation. However, for crops used in saline agriculture, a lack of compatibility with these microsymbionts is likely to become the bottleneck of this new agricultural technology. To fully exploit the beneficial effects of plant–microsymbiont associations, we need to understand the molecular mechanism of symbiosis between plants and these symbionts (Figure 2).
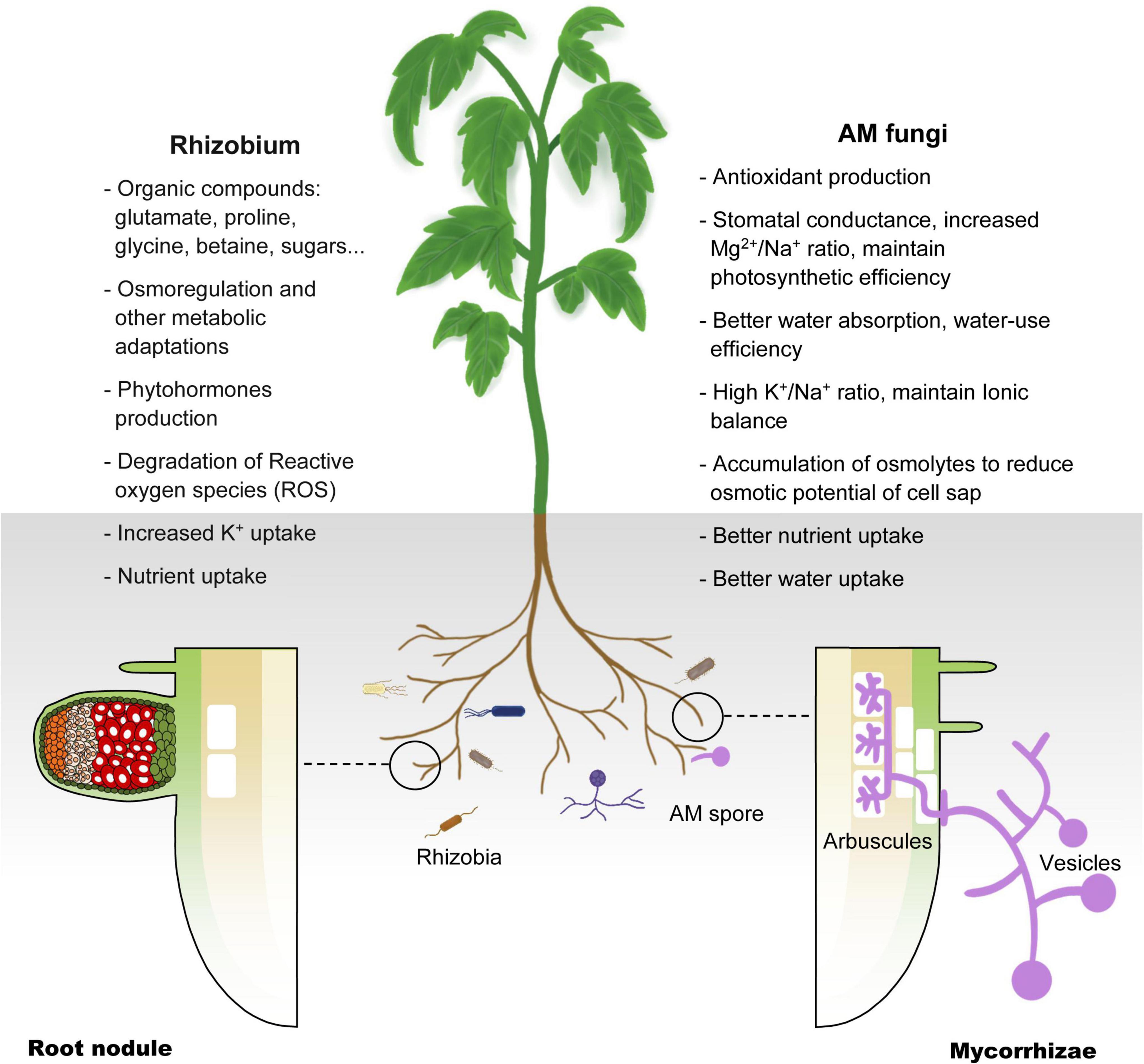
Figure 2. Schematic illustration of the mechanisms deployed by AM fungi and Rhizobium in host plant coping with salinity stress. Salinity impedes plant absorption of water and nutrients, resulting in physiological drought. AM and Rhizobium help plants in salt stress by improving osmoregulation, antioxidant production, K+ uptake and other nutrient uptake (see text).
Symbiotic microorganisms that promote plant abiotic (salt) stress resistance occur naturally and exist widely, such as AM fungi and Rhizobium. However, traditional crop breeding techniques, including genetic engineering, domestication and crossbreeding, do not consider the role of symbiotic microorganisms in promoting stress resistance. Moreover, the application of new genetic engineering techniques in breeding overlooks the perspective of plant–microbial symbiosis. Fortunately, research on plant–microbial symbiosis has been continuous and fruitful. There has been considerable research progress on key genes that regulate the process of symbiosis establishment between plants and microorganisms. Among them, the most exciting one is a common symbiotic pathway may be exist in plants (Oldroyd, 2013). Kistner et al. (2005) found that a single gene mutation can inhibit both bacterial and fungal infection of plant root tissue. At the same time, numerous studies of Ca2+ signaling in nuclei have shown that it can mediate calcium oscillations of varying intensity (Yuan et al., 2017; Poovaiah and Du, 2018; Plasencia et al., 2021), to activate different downstream pathways. This may be the reason why CCaMK protein, core component in the common symbiotic pathway, can be activated differentially, e.g., mycorrhizal or nodular formation (Russo et al., 2013). These findings could become a key starting point for the use of new gene-editing technologies to engineer non-legumes to establish better symbiotic relationships with versatile symbiotic microorganisms.
Because legumes can establish symbiotic relationships with AM fungi and rhizobia at the same time, people have carried out extensive research on legumes from the perspective of symbiotic molecular mechanisms. In the past few decades, genetic studies on legumes and AM fungi have successively identified the genetic components necessary for the establishment of a symbiotic relationship. These genes constitute the molecular basis of the relationship between most terrestrial plants, including gramineous plants, and microsymbionts, and are now collectively referred to as the common symbiotic pathway (MacLean et al., 2017; de Bruijn, 2020; Figure 3). Studies have found that AM fungi use the so-called ‘Myc factor’ to stimulate plant roots to begin a dialogue and eventually form a symbiotic relationship (Rasmussen et al., 2016; Pimprikar and Gutjahr, 2018). The chemical composition of the Myc factor is lipochitooligosaccharides (LCOs), secreted by AM fungi and released into the rhizosphere of plants. The Myc factor is sensed by the LysM receptor kinase present on the plant root cell membrane (Gough et al., 2018; He et al., 2019). The transduction of Myc factor signals into plant cells triggers the AM symbiosis signaling pathway. The currently identified components of this pathway are an LRR receptor kinase (MtDMI2/LjSYMRK), a nuclear cation channel protein (MtDMI1/LjPOLLUX, LjCASTOR), a nucleoporin protein (LjNUP85, LjNUP133, NENA), a calcium pump protein (MtMCA8), a calcium-dependent and calmodulin-dependent protein kinase (MtDMI3/LjCCaMK) and its interacting protein components (MtIPD3/LjCYCLOPS) and two GRAS-family transcription factors, NSP2 and RAM1 (Venkateshwaran et al., 2012; Binder and Parniske, 2013; Xue et al., 2015; Gough et al., 2018; Grosche et al., 2018; Hakoshima, 2018; Li et al., 2018; Pimprikar and Gutjahr, 2018). Studies have proved that the above components are all necessary for the establishment of symbiosis. Mutant plants containing non-functional genes cannot form a sound symbiotic structure. Many related studies have found that most terrestrial plants (including non-legumes such as corn and rice), and even some lower plants (such as mosses), contain AM symbiotic signaling pathway-related genes. This phenomenon shows that plants have evolved for symbiosis and that the emergence of the ‘molecular machinery’ related to symbiosis in plants has a very ancient origin (Rimington et al., 2018; Delaux and Schornack, 2021).
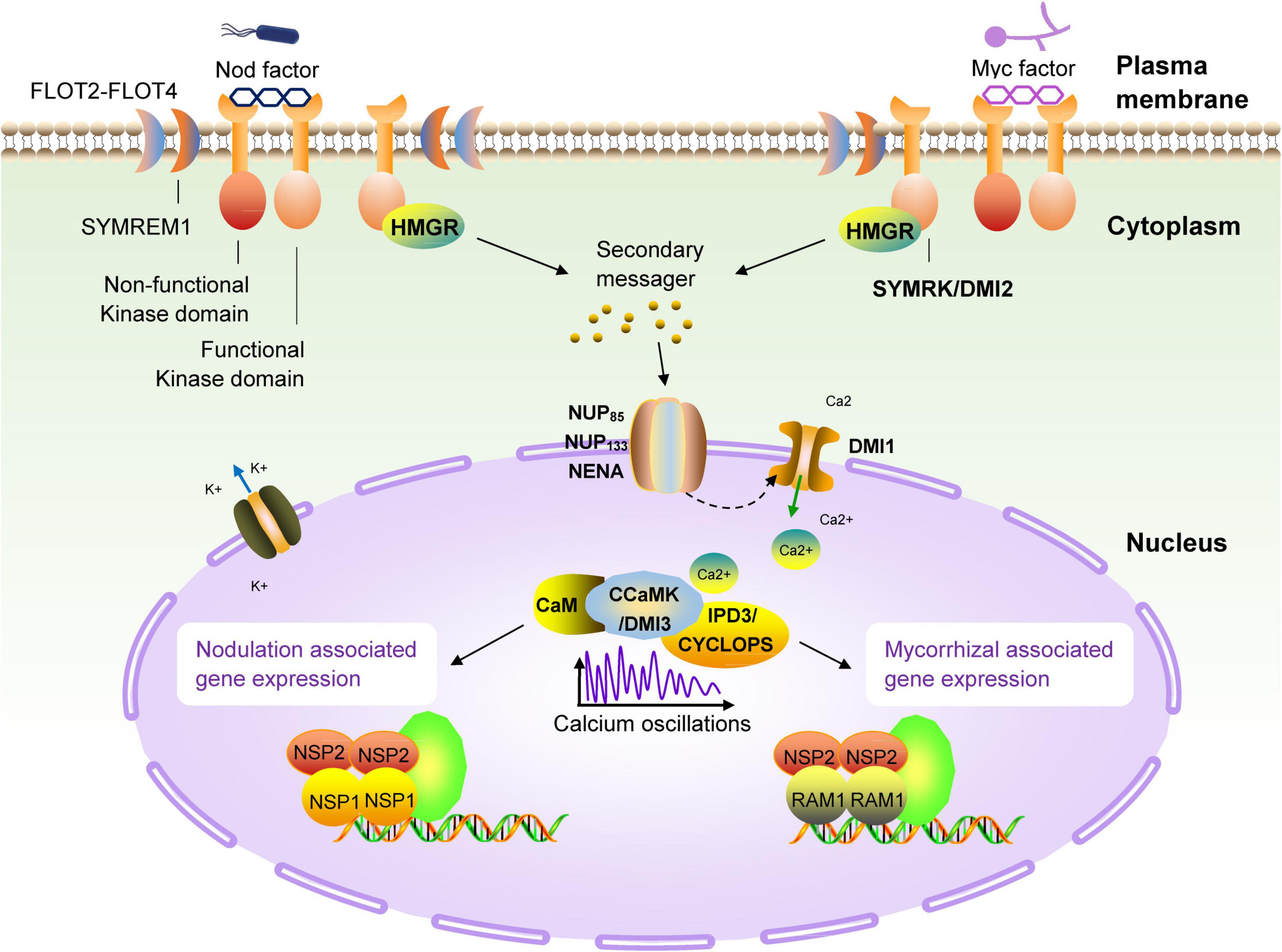
Figure 3. Schematic diagram of the common symbiotic pathway (SYM) of nodulation and arbuscular mycorrhizal signals in legumes. The first known molecular component in the symbiotic pathway (SYM) signaling pathway is the receptor-like kinase DMI2 of M. truncatula (homologous to SYMRK of L. japonicus) (Kevei et al., 2007; Antolín-Llovera et al., 2014). The direct target of DMI2 is unclear, but it can be determined that the signal is transmitted to the nuclear membrane by a secondary messenger. There are three nucleoporins (NUP85, NUP133 and NENA) in L. japonicus, which continue to transmit symbiotic signals to the nucleus (Genre and Russo, 2016). There is a recognized cation channel DMI1 on the nuclear membrane, the results have confirmed the role of DMI1 in the formation of Ca2+ oscillations, which has also been confirmed to participate in the conduction of this signal into the nucleus (Charpentier et al., 2008). In the nucleus, the calmodulin-dependent kinase DMI3 (CCaMK in L. japonicus) is responsible for decoding the induced calcium oscillation signal (Gleason et al., 2006; Tirichine et al., 2006), and then, together with IPD3 (homolog of CYCLOPS in L. japonicus) (Yano et al., 2008; Singh et al., 2014), it takes over the upstream signal and activates a set of downstream transcriptional regulators (NSP1, NSP2, and RAM1) (Kaló et al., 2005; Smit et al., 2005; Hirsch et al., 2009; Pimprikar et al., 2016), these regulatory factors regulates the expression of genes related to nodulation and arbuscular mycorrhiza in the terminal, respectively.
Leguminous plants are the second most diverse group among terrestrial plants. In addition to symbiosis with AM fungi, they can also establish effective symbiotic relationships with Rhizobium species. This relationship appears in the form of root nodules. In rare cases, the symbiosis can form stem nodules, such as in Azorhizobium caulinodans and Sesbania (Lee et al., 2008; Liu et al., 2017). Nodules are a group of highly specialized plant cells wrapped under the epidermis of plant roots. Their purpose is to provide shelter for the bacteria. Inside, the rhizobia efficiently fix nitrogen from the atmosphere and exchange nutrients with the plant, forming a close relationship. Related genetic studies on legumes have shown that the ancient SYM pathway has further specialized in legumes. The nodulation ability of some legume mutants with defective AM symbiosis as mentioned above is also affected, which indicates the high degree of homology of the SYM pathway in the establishment of different forms of symbiosis between plants and microorganisms (Kistner et al., 2005). In addition to the known members of the SYM pathway, other molecular components related to the establishment of symbiosis have high homology. For example, NSP1 is an important component of rhizobia–legume symbiosis, its original copy (RAM1) is also critical in AM symbiosis, and it seems to belong to a broader SYM pathway (Gobbato et al., 2012; Fonouni-Farde et al., 2016; Floss et al., 2017; Lace and Ott, 2018). Detailed studies have discovered more and more key molecular components involved in the symbiosis of rhizobia and plants, and gradually revealed an ancient truth: these newly discovered components are part of or modified from the SYM pathway. The symbiotic interaction between legumes and rhizobia begins with the host’s perception of microbial signal molecules called ‘nodulation factors’ (NFs) released by rhizobia (Buhian and Bensmihen, 2018). Notably, Nod factors are also LCOs, which are recognized and bound by LysM-type receptor kinases in legumes (Buendia et al., 2018; Gough et al., 2018; Bozsoki et al., 2020). Studies have shown that NFs directly binds to the NF receptors NFR5 and NFR1, which are located on the cell membrane of Lotus japonicus. The dissociation constant (Kd) value of NF-ligand binding is in the nanomolar range (about 10 nM for NFR5 and about 5 nM for NFR1), which is close to the concentration known to trigger the onset of symbiosis (Broghammer et al., 2012). The signal cascade required for nodulation is eventually triggered by the perception of LCOs by these plasma membrane receptors and triggers a series of rapid reactions in the host cell, including the formation of ion currents, alkalization of the cytoplasm, production of reactive oxygen species, and nuclear and perinuclear calcium oscillations (Cardenas et al., 2008; Chabaud et al., 2011; Naffah de Souza et al., 2017).
The abovementioned research progress shows that symbiotic genes related to the SYM pathway existed in the common ancestor of land plants, and their functions have remained fundamentally unchanged during the evolution of land plants (Wang et al., 2010; Radhakrishnan et al., 2019). Moreover, increasing numbers of related studies have revealed that this common symbiotic pathway (SYM) is highly conserved in different symbiotic relationships, and even the interactions of plants with microbes other than mycorrhiza and nodules are associated with components of the SYM pathway (Chiu and Paszkowski, 2020). The discovery of the SYM pathway and continuing research progress provide a promising strategy for genetically engineering symbiotic molecular components into plants, to utilize symbiotic microorganisms to improve the vigor and salt resistance of host plants.
In the past two decades, research on clarifying the molecular mechanism of plant–microbe symbiosis has not only successfully identified individual key genes involved in the establishment of symbiosis through reverse genetics but also made much progress through high-throughput genomics and proteomics quantitative strategies (Genre and Russo, 2016; Lardi and Pessi, 2018; Rehman et al., 2019; Chiu and Paszkowski, 2020; diCenzo et al., 2020; Tong et al., 2020). With the completion of genome sequencing for a large number of plants (including legumes and non-leguminous crops), it has become more feasible to apply the “-omics” strategy to the study of symbiosis (Lardi and Pessi, 2018). Regarding the use of proteomics and phosphorylation proteomics to study the symbiosis signal of legumes, Rose et al. (2012) used a proteomics method based on deep non-targeted mass spectrometry and found that the phosphorylation status of 13,506 phosphorylation sites of 7,739 M. truncatula proteins changed rapidly under NF treatment. The study also found that in the early stage of symbiotic signal transmission (within 1 h after NF induction), the types and contents of proteins did not change. The rapid cellular and molecular response induced by NF mainly relied on post-translational modifications, such as phosphorylation, rather than protein synthesis or degradation (Rose et al., 2012). This study also employed a genetics method to discover some new symbiosis-related candidate genes using a limited number of key gene loss-of-function mutants in the SYM pathway, which may be involved in symbiosis signaling, cell cycle regulation, and root hair growth and infection (Rose et al., 2012). Similarly, Nguyen et al. (2012) analyzed the phosphoproteome during the rhizobial colonization of soybean root hair cells to understand the molecular mechanism of nodule formation. Their study identified 1,126 phosphorylated soybean proteins and detected 1,659 phosphorylation sites, and in soybean plants inoculated with Bradyrhizobium japonicum, 273 phosphorylation sites of 240 phosphorylated proteins were significantly altered, many of which were the same phosphorylation sites identified by Rose et al. (2012). In addition, a genome-wide transcriptome study aimed at elucidating the Myc factor signal transduction process in M. truncatula found that all kinds of symbiotic LCO analogs upregulated a common set of genes (Czaja et al., 2012). On an even larger scale, the three plant systems Casuarina glauca, M. truncatula and Oryza sativa were used to compare the transcriptomes of genes involved in rhizobial nodulation, actinomycete nodulation, and arbuscular mycorrhiza formation. The results showed that a group of common genes was regulated in these three endosymbiosis processes (Tromas et al., 2012). These systemic approaches together provide a rapid means of filling the gaps in our knowledge on symbiotic signal transduction as well as potentially pointing us in new directions with genes and proteins not previously thought to be involved. “Multi-omics approaches” together enable the rapid description of the mechanism of symbiotic signal transduction, potentially providing a blueprint for genetically engineering crops to utilize the potential of symbiotic microorganisms to promote growth and resist stress.
Global human activities and climate change are intensifying, leading to further intensification of cultivated land salinization. To expand the planting area on saline soils, increase the net yield of crops, and avoid the impact of negative environments on crop yields while reducing the dependence of large-scale shallow-root monocropping crops on chemical fertilizers, a promising strategy is to make full use of growth- and salt resistance-promoting symbiotic microorganisms such as AM fungi and rhizobia. To achieve this goal, we need to fully understand the molecular mechanism of symbiosis between plants (especially legumes) and microorganisms. One specific method is to use the symbiosis mechanism of SYM (the relevant molecular components of this mechanism already exist in most land plants) together with genetic engineering methods to modify the main food crops (legumes and non-legumes) so that they can establish better symbiotic relationships with microsymbionts, and achieve high-quality microsymbiosis through screening to construct a plant ‘holobiont’ (Figure 4). This will allow us to make full use of the growth-promoting and anti-stress functions of microsymbionts to enable the sustainable development of saline agriculture under future climate change conditions. Because most terrestrial plants (including cereals) already contain the relevant molecular components of the SYM symbiosis mechanism, realizing the perception of rhizobial signals is the key first step to initiating the SYM pathway and achieving infection in non-legume plants. Studies on Parasponia have shown that successful cell colonization can enable a certain degree of nitrogen fixation without the formation of complex nodule organs (Geurts et al., 2012), which implies that the microsymbiont and the host are also carrying out the exchange of nutrients and physiological signals in an incomplete symbiotic relationship with only infection achieved. Unlike NF signaling, the receptors specifically used for Myc factor sensing have not yet been determined. Although the LysM type receptor kinase LYR1 is strongly upregulated during AM symbiosis in M. truncatula (Arrighi et al., 2006; Benedito et al., 2008; Young et al., 2011), there is still a lack of direct experimental evidence to support its role in Myc factor perception. Myc factors can trigger calcium oscillation in both legumes and non-legume dicots, but there is no such response in monocots. However, studies have shown that there is crosstalk between the NF and Myc factor signaling pathways in the same plant (Groth et al., 2010; Gully et al., 2018; Suzaki et al., 2019). This phenomenon indicates that it is possible to modify the signal receptors in plants to make them sense the NF and Myc factors at the same time. In the natural environment, only a few monocotyledonous and dicotyledonous plants can form close and effective associations with rhizobium. However, most of the research aimed at using symbiotic microorganisms to promote stress resistance in plants has focused on screening out more effective microbial strains. The specific molecular mechanism encoded in the plant genome tightly controls the interaction with these beneficial microsymbionts. We have shown in this review that plants continue to use this ancient and extensive symbiotic molecular machinery (the SYM pathway) in their evolution to recruit AM fungi and a variety of tolerance-promoting bacteria to adapt to environmental stress. This fact demonstrates that utilizing microsymbiosis by artificially modifying the key components of the SYM pathway in target crops is an achievable goal. For example, the transformation of economically valuable non-legume crops to expand beneficial microsymbiosis in the soil will have a huge impact on the sustainability of food amidst the gradual salinization of arable land. Therefore, the combination of ongoing high-throughput research and in-depth genetic analysis will be of vital importance for engineering salt tolerance-promoting associations in non-leguminous crops and particularly in cereals.
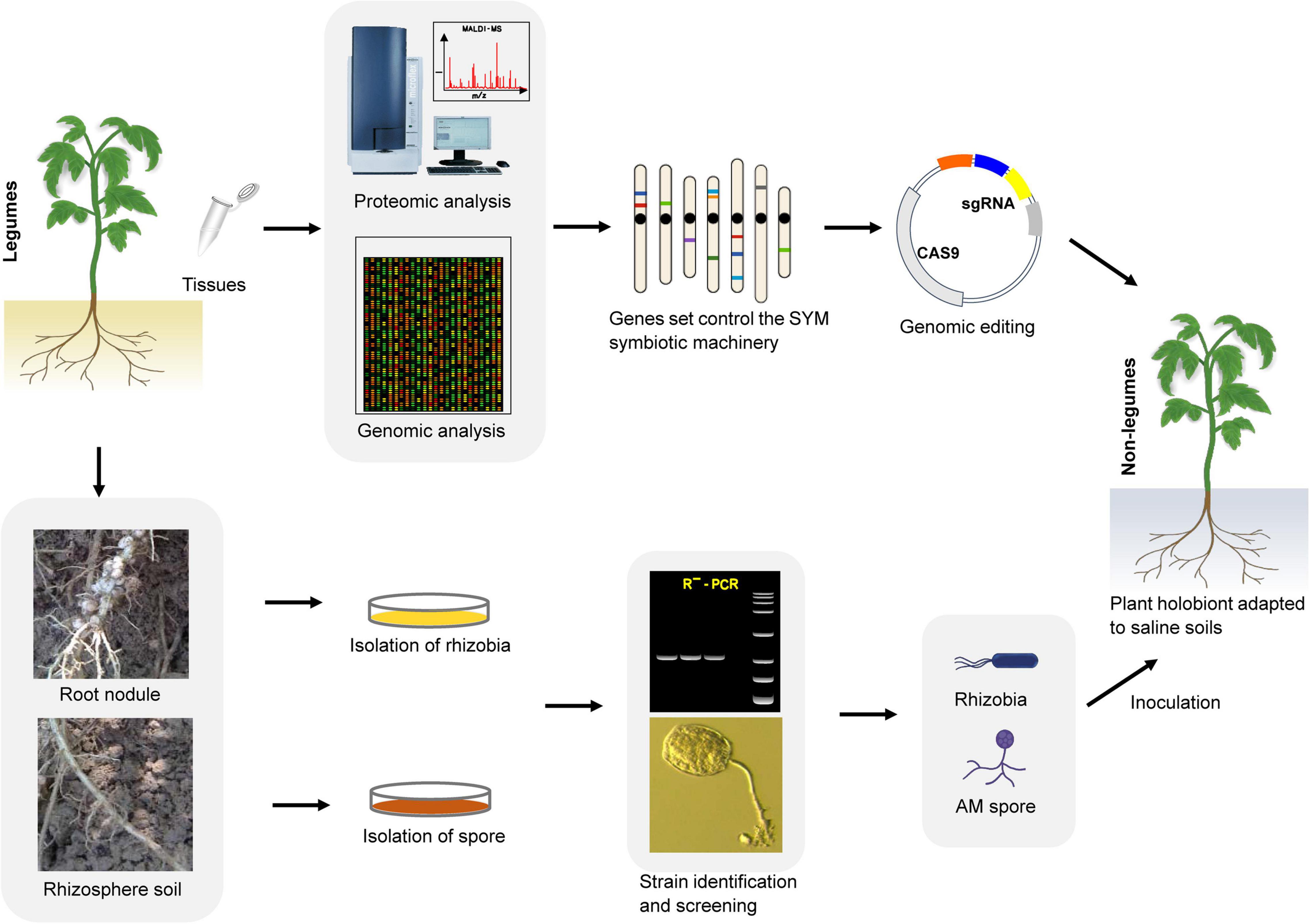
Figure 4. An integrated tool for developing plant ‘holobiont’ adapted to saline. First, through multi-omics approaches to identify ‘anchor genes’ in the SYM pathway. Then, using genome editing tool such as CRISPR/Cas9 to engineer non-leguminous crops to associate better with rhizobia and AM fungi utilize the SYM symbiotic machinery. In parallel, screening of highly efficient salt-tolerant improving strains from the rhizosphere of saline-alkali soil plants. At last, in combing genetically modified crops that are easy to establish symbiotic relationships with specific AM fungi and rhizobia strains to develop a plant ‘holobiont’, which can better adapt to the salt soil environment.
It is clear that expansion of conventional agricultural practices to meet future demands amidst the gradual salinization of farmland is neither economically nor environmentally feasible. Therefore, saline agriculture urgently needs innovations to follow on from the Green Revolution to sustainably meet the demand for global food security. One way to improve the production efficiency of crops under saline conditions is to expand the growth-promoting and stress-resistance effects of microsymbionts. These microsymbionts include AM fungi and a variety of rhizobia that can improve crop growth and vitality, nutrient utilization efficiency, and biotic/abiotic stress tolerance. However, traditional crop breeding technologies (including genetic engineering technology, domestication, and hybrid breeding) rarely consider the role of symbiotic microorganisms in promoting plant stress resistance, and breeding using new genetic engineering technologies overlooks the perspective of plant–microbe symbiosis. Fortunately, however, research on plant–microbe symbiotic relationships have been continuous and fruitful. In the past few decades, researchers have successfully identified the key molecular components of arbuscular mycorrhiza and nodule formation with plants through genetic studies. These genes are collectively referred to as the SYM pathway. Through multi-omics methods, analysis of these key symbiotic components has continuously deepened. Based on these findings, we may use new gene-editing technologies such as the clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein 9 (Cas9) system to customize plant utilization of the SYM symbiotic machinery (which is present already in most land plants and, in particular, in cereals), in combination with AM fungi and rhizobia strains to develop salt-tolerant plant ‘holobiont’. These developments will open a new avenue for capitalizing on symbiotic microorganisms to strengthen plant salt resistance. Agricultural practices and production efficiency under saline conditions will be greatly improved to meet the increasing global food demand.
C-GR analyzed the data and wrote the manuscript. Z-YL, C-CK, Z-HZ, X-LW, J-CY and SQ participated in revisions of the manuscript. All authors have read the manuscript and approved the final version of the manuscript. Special thanks to Wang Yu for his hand drawing in Figure 1.
This work was financed by the Key Research and Development Plan of Shandong Province (2019GSF109104), Key Research and Development Program of Yantai (2019MSGY123 and 2020MSGY068), Shandong Natural Science Foundation (ZR2021MC106), and Innovation project of Shandong Academy of Agricultural Sciences (CXGC2022C06).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbaspour, K., and Ashraf Vaghefi, S. (2019). Harmonized World Soil Database in SWAT Format. Los Angeles, CA: PANGAEA.
Abdel-Fattah, G. M., and Asrar, A.-W. A. (2011). Arbuscular mycorrhizal fungal application to improve growth and tolerance of wheat (Triticum aestivum L.) plants grown in saline soil. Acta Physiol. Plant. 34, 267–277. doi: 10.1007/s11738-011-0825-6
Ahmed, M. Z., Gul, B., Khan, M. A., and Watanabe, K. N. (2016). “1 - characterization and function of sodium exchanger genes in Aeluropus lagopoides under NaCl stress,” in Halophytes for Food Security in Dry Lands, eds M. A. Khan, M. Ozturk, B. Gul, and M. Z. Ahmed (San Diego, CA: Academic Press), 1–16. doi: 10.1016/b978-0-12-801854-5.00001-7
Aliasgharzad, N., Rastin, S. N., Towfighi, H., and Alizadeh, A. (2001). Occurrence of arbuscular mycorrhizal fungi in saline soils of the Tabriz Plain of Iran in relation to some physical and chemical properties of soil. Mycorrhiza 11, 119–122. doi: 10.1007/s005720100113
Antolín-Llovera, M., Ried, M. K., and Parniske, M. (2014). Cleavage of the SYMBIOSIS RECEPTOR-LIKE KINASE ectodomain promotes complex formation with Nod factor receptor 5. Curr. Biol. 24, 422–427. doi: 10.1016/j.cub.2013.12.053
Arrighi, J. F., Barre, A., Ben Amor, B., Bersoult, A., Soriano, L. C., Mirabella, R., et al. (2006). The Medicago truncatula lysin [corrected] motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 142, 265–279. doi: 10.1104/pp.106.084657
Bala, N., Sharma, P. K., and Lakshminarayana, K. (1990). Nodulation and nitrogen fixation by salinity-tolerant rhizobia in symbiosis with tree legumes. Agric. Ecosyst. Environ. 33, 33–46. doi: 10.1016/0167-8809(90)90142-z
Bano, A., and Fatima, M. (2009). Salt tolerance in Zea mays (L). following inoculation with Rhizobium and Pseudomonas. Biol. Fertil. Soils 45, 405–413. doi: 10.1007/s00374-008-0344-9
Bano, D. A., Singh, R. K., Waza, S. A., and Singh, N. P. (2015). Effect of cowpea Bradyrhizobium (RA-5) and Burkholderia cepacia (RRE-5) on growth parameters of pigeonpea under salt stress conditions. J. Pure Appl. Microbiol. 9, 2539–2545.
Benedito, V. A., Torres-Jerez, I., Murray, J. D., Andriankaja, A., Allen, S., Kakar, K., et al. (2008). A gene expression atlas of the model legume Medicago truncatula. Plant J. 55, 504–513. doi: 10.1111/j.1365-313X.2008.03519.x
Berruti, A., Lumini, E., Balestrini, R., and Bianciotto, V. (2016). Arbuscular mycorrhizal fungi as natural biofertilizers: let’s benefit from past successes. Front. Microbiol. 6:1559. doi: 10.3389/fmicb.2015.01559
Bianco, C., and Defez, R. (2009). Medicago truncatula improves salt tolerance when nodulated by an indole-3-acetic acid-overproducing Sinorhizobium meliloti strain. J. Exp. Bot. 60, 3097–3107. doi: 10.1093/jxb/erp140
Binder, A., and Parniske, M. (2013). Analysis of the Lotus japonicus nuclear pore NUP107-160 subcomplex reveals pronounced structural plasticity and functional redundancy. Front. Plant Sci. 4:552. doi: 10.3389/fpls.2013.00552
Bozsoki, Z., Gysel, K., Hansen, S. B., Lironi, D., Krönauer, C., Feng, F., et al. (2020). Ligand-recognizing motifs in plant LysM receptors are major determinants of specificity. Science 369, 663–670. doi: 10.1126/science.abb3377
Broghammer, A., Krusell, L., Blaise, M., Sauer, J., Sullivan, J. T., Maolanon, N., et al. (2012). Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc. Natl. Acad. Sci. U.S.A. 109, 13859–13864. doi: 10.1073/pnas.1205171109
Buendia, L., Girardin, A., Wang, T., Cottret, L., and Lefebvre, B. (2018). LysM receptor-like kinase and LysM receptor-like protein families: an update on phylogeny and functional characterization. Front. Plant Sci. 9:1531. doi: 10.3389/fpls.2018.01531
Buhian, W. P., and Bensmihen, S. (2018). Mini-review: nod factor regulation of phytohormone signaling and homeostasis during rhizobia-legume symbiosis. Front. Plant Sci. 9:1247. doi: 10.3389/fpls.2018.01247
Cardenas, L., Martinez, A., Sanchez, F., and Quinto, C. (2008). Fast, transient and specific intracellular ROS changes in living root hair cells responding to Nod factors (NFs). Plant J. 56, 802–813. doi: 10.1111/j.1365-313X.2008.03644.x
Chabaud, M., Genre, A., Sieberer, B. J., Faccio, A., Fournier, J., Novero, M., et al. (2011). Arbuscular mycorrhizal hyphopodia and germinated spore exudates trigger Ca2+ spiking in the legume and nonlegume root epidermis. New Phytol. 189, 347–355. doi: 10.1111/j.1469-8137.2010.03464.x
Charpentier, M., Bredemeier, R., Wanner, G., Takeda, N., Schleiff, E., and Parniske, M. (2008). Lotus japonicus CASTOR and POLLUX are ion channels essential for perinuclear calcium spiking in legume root endosymbiosis. Plant Cell 20, 3467–3479. doi: 10.1105/tpc.108.063255
Chen, J., Zhang, H., Zhang, X., and Tang, M. (2017). Arbuscular mycorrhizal symbiosis alleviates salt stress in black locust through improved photosynthesis, water status, and K(+)/Na(+) homeostasis. Front. Plant Sci. 8:1739. doi: 10.3389/fpls.2017.01739
Chiu, C. H., and Paszkowski, U. (2020). Receptor-like kinases sustain symbiotic scrutiny. Plant Physiol. 182, 1597–1612. doi: 10.1104/pp.19.01341
Czaja, L. F., Hogekamp, C., Lamm, P., Maillet, F., Martinez, E. A., Samain, E., et al. (2012). Transcriptional responses toward diffusible signals from symbiotic microbes reveal MtNFP- and MtDMI3-dependent reprogramming of host gene expression by arbuscular mycorrhizal fungal lipochitooligosaccharides. Plant Physiol. 159, 1671–1685. doi: 10.1104/pp.112.195990
Dardanelli, M. S., Fernández De Córdoba, F. J., Espuny, M. R., Rodríguez Carvajal, M. A., Soria Díaz, M. E., Gil Serrano, A. M., et al. (2008). Effect of Azospirillum brasilense coinoculated with Rhizobium on Phaseolus vulgaris flavonoids and Nod factor production under salt stress. Soil Biol. Biochem. 40, 2713–2721. doi: 10.1016/j.soilbio.2008.06.016
de Bruijn, F. J. (ed.). (2020). “The common symbiotic signaling pathway (CSSP or SYM),” in The Model Legume Medicago truncatula (New York, NY: Wiley), 521–521. doi: 10.1002/9781119409144.part8
Deepika, C., and Satyavir, S. S. (2015). Inducing salinity tolerance in chickpea (Cicer arietinum L.) by inoculation of 1-aminocyclopropane-1-carboxylic acid deaminase-containing Mesorhizobium strains. Afr. J. Microbiol. Res. 9, 117–124. doi: 10.5897/ajmr2014.7087
Del Cerro, P., Megias, M., Lopez-Baena, F. J., Gil-Serrano, A., Perez-Montano, F., and Ollero, F. J. (2019). Osmotic stress activates nif and fix genes and induces the Rhizobium tropici CIAT 899 Nod factor production via NodD2 by up-regulation of the nodA2 operon and the nodA3 gene. PLoS One 14:e0213298. doi: 10.1371/journal.pone.0213298
Delaux, P.-M., and Schornack, S. (2021). Plant evolution driven by interactions with symbiotic and pathogenic microbes. Science 371:eaba6605. doi: 10.1126/science.aba6605
diCenzo, G. C., Tesi, M., Pfau, T., Mengoni, A., and Fondi, M. (2020). Genome-scale metabolic reconstruction of the symbiosis between a leguminous plant and a nitrogen-fixing bacterium. Nat. Commun. 11:2574. doi: 10.1038/s41467-020-16484-2
Diouf, D., Duponnois, R., Tidiane Ba, A., Neyra, M., and Lesueur, D. (2006). Symbiosis of Acacia auriculiformis and Acacia mangium with mycorrhizal fungi and Bradyrhizobium spp. improves salt tolerance in greenhouse conditions. Funct. Plant Biol. 32, 1143–1152. doi: 10.1071/FP04069
Dong, R., Zhang, J., Huan, H., Bai, C., Chen, Z., and Liu, G. (2017). High salt tolerance of a Bradyrhizobium strain and its promotion of the growth of Stylosanthes guianensis. Int. J. Mol. Sci. 18:1625. doi: 10.3390/ijms18081625
Egamberdieva, D., Wirth, S., Jabborova, D., Räsänen, L. A., and Liao, H. (2017). Coordination between Bradyrhizobium and Pseudomonas alleviates salt stress in soybean through altering root system architecture. J. Plant Interact. 12, 100–107. doi: 10.1080/17429145.2017.1294212
Egamberdiyeva, D., Qarshieva, D., and Davranov, K. (2004). The use of Bradyrhizobium to enhance growth and yield of soybean in calcareous soil in Uzbekistan. J. Plant Growth Regul. 23, 54–57.
Elhindi, K. M., El-Din, A. S., and Elgorban, A. M. (2017). The impact of arbuscular mycorrhizal fungi in mitigating salt-induced adverse effects in sweet basil (Ocimum basilicum L.). Saudi J. Biol. Sci. 24, 170–179. doi: 10.1016/j.sjbs.2016.02.010
Elsheikh, E. A. E., and Wood, M. (1995). Nodulation and N2 fixation by soybean inoculated with salt-tolerant rhizobia or salt-sensitive bradyrhizobia in saline soil. Soil Biol. Biochem. 27, 657–661. doi: 10.1016/0038-0717(95)98645-5
Estrada, B., Aroca, R., Maathuis, F. J., Barea, J. M., and Ruiz-Lozano, J. M. (2013). Arbuscular mycorrhizal fungi native from a Mediterranean saline area enhance maize tolerance to salinity through improved ion homeostasis. Plant Cell Environ. 36, 1771–1782. doi: 10.1111/pce.12082
Evelin, H., Kapoor, R., and Giri, B. (2009). Arbuscular mycorrhizal fungi in alleviation of salt stress: a review. Ann. Bot. 104, 1263–1280. doi: 10.1093/aob/mcp251
FAO (2015). Status of the World’s Soil Resources (SWSR) – Main Report. Rome: Food and Agriculture Organization of the United Nations.
Fileccia, V., Ruisi, P., Ingraffia, R., Giambalvo, D., Frenda, A. S., and Martinelli, F. (2017). Arbuscular mycorrhizal symbiosis mitigates the negative effects of salinity on durum wheat. PLoS One 12:e0184158. doi: 10.1371/journal.pone.0184158
Floss, D. S., Gomez, S. K., Park, H. J., Maclean, A. M., Muller, L. M., Bhattarai, K. K., et al. (2017). A transcriptional program for arbuscule degeneration during AM symbiosis is regulated by MYB1. Curr. Biol. 27, 1206–1212. doi: 10.1016/j.cub.2017.03.003
Fonouni-Farde, C., Tan, S., Baudin, M., Brault, M., Wen, J., Mysore, K. S., et al. (2016). DELLA-mediated gibberellin signalling regulates Nod factor signalling and rhizobial infection. Nat. Commun. 7:12636. doi: 10.1038/ncomms12636
Fukami, J., De La Osa, C., Ollero, F. J., Megías, M., and Hungria, M. (2018). Co-inoculation of maize with Azospirillum brasilense and Rhizobium tropici as a strategy to mitigate salinity stress. Funct. Plant Biol. 45, 328–339. doi: 10.1071/FP17167
Genre, A., and Russo, G. (2016). Does a common pathway transduce symbiotic signals in plant-microbe interactions? Front. Plant Sci. 7:96. doi: 10.3389/fpls.2016.00096
Geurts, R., Lillo, A., and Bisseling, T. (2012). Exploiting an ancient signalling machinery to enjoy a nitrogen fixing symbiosis. Curr. Opin. Plant Biol. 15, 438–443. doi: 10.1016/j.pbi.2012.04.004
Giri, B., and Mukerji, K. G. (2004). Mycorrhizal inoculant alleviates salt stress in Sesbania aegyptiaca and Sesbania grandiflora under field conditions: evidence for reduced sodium and improved magnesium uptake. Mycorrhiza 14, 307–312. doi: 10.1007/s00572-003-0274-1
Gleason, C., Chaudhuri, S., Yang, T., Muñoz, A., Poovaiah, B. W., and Oldroyd, G. E. (2006). Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature 441, 1149–1152. doi: 10.1038/nature04812
Gobbato, E., Marsh, J. F., Vernie, T., Wang, E., Maillet, F., Kim, J., et al. (2012). A GRAS-type transcription factor with a specific function in mycorrhizal signaling. Curr. Biol. 22, 2236–2241. doi: 10.1016/j.cub.2012.09.044
Gough, C., Cottret, L., Lefebvre, B., and Bono, J. J. (2018). Evolutionary history of plant LysM receptor proteins related to root endosymbiosis. Front. Plant Sci. 9:923. doi: 10.3389/fpls.2018.00923
Grosche, C., Genau, A. C., and Rensing, S. A. (2018). Evolution of the symbiosis-specific GRAS regulatory network in bryophytes. Front. Plant Sci. 9:1621. doi: 10.3389/fpls.2018.01621
Groth, M., Takeda, N., Perry, J., Uchida, H., Draxl, S., Brachmann, A., et al. (2010). NENA, a Lotus japonicus homolog of Sec13, is required for rhizodermal infection by arbuscular mycorrhiza fungi and rhizobia but dispensable for cortical endosymbiotic development. Plant Cell 22, 2509–2526. doi: 10.1105/tpc.109.069807
Gully, D., Czernic, P., Cruveiller, S., Mahe, F., Longin, C., Vallenet, D., et al. (2018). Transcriptome profiles of nod factor-independent symbiosis in the tropical legume Aeschynomene evenia. Sci. Rep. 8:10934. doi: 10.1038/s41598-018-29301-0
Hakoshima, T. (2018). Structural basis of the specific interactions of GRAS family proteins. FEBS Lett. 592, 489–501. doi: 10.1002/1873-3468.12987
Hammer, E. C., and Rillig, M. C. (2011). The influence of different stresses on glomalin levels in an arbuscular mycorrhizal fungus–salinity increases glomalin content. PLoS One 6:e28426. doi: 10.1371/journal.pone.0028426
Hashem, A., Abd_Allah, E. F., Alqarawi, A. A., Aldubise, A., and Egamberdieva, D. (2015). Arbuscular mycorrhizal fungi enhances salinity tolerance of Panicum turgidum Forssk by altering photosynthetic and antioxidant pathways. J. Plant Interact. 10, 230–242. doi: 10.1080/17429145.2015.1052025
Hashem, A., Alqarawi, A. A., Radhakrishnan, R., Al-Arjani, A. F., Aldehaish, H. A., Egamberdieva, D., et al. (2018). Arbuscular mycorrhizal fungi regulate the oxidative system, hormones and ionic equilibrium to trigger salt stress tolerance in Cucumis sativus L. Saudi J. Biol. Sci. 25, 1102–1114. doi: 10.1016/j.sjbs.2018.03.009
He, J., Zhang, C., Dai, H., Liu, H., Zhang, X., Yang, J., et al. (2019). A LysM receptor heteromer mediates perception of arbuscular mycorrhizal symbiotic signal in rice. Mol. Plant 12, 1561–1576. doi: 10.1016/j.molp.2019.10.015
Hirsch, S., Kim, J., Muñoz, A., Heckmann, A. B., Downie, J. A., and Oldroyd, G. E. (2009). GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 21, 545–557. doi: 10.1105/tpc.108.064501
Kaló, P., Gleason, C., Edwards, A., Marsh, J., Mitra, R. M., Hirsch, S., et al. (2005). Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308, 1786–1789. doi: 10.1126/science.1110951
Kevei, Z., Lougnon, G., Mergaert, P., Horváth, G. V., Kereszt, A., Jayaraman, D., et al. (2007). 3-hydroxy-3-methylglutaryl coenzyme a reductase 1 interacts with NORK and is crucial for nodulation in Medicago truncatula. Plant Cell 19, 3974–3989. doi: 10.1105/tpc.107.053975
Khalloufi, M., Martínez-Andújar, C., Lachaâl, M., Karray-Bouraoui, N., Pérez-Alfocea, F., and Albacete, A. (2017). The interaction between foliar GA3 application and arbuscular mycorrhizal fungi inoculation improves growth in salinized tomato (Solanum lycopersicum L.) plants by modifying the hormonal balance. J. Plant Physiol. 214, 134–144. doi: 10.1016/j.jplph.2017.04.012
Kistner, C., Winzer, T., Pitzschke, A., Mulder, L., Sato, S., Kaneko, T., et al. (2005). Seven Lotus japonicus genes required for transcriptional reprogramming of the root during fungal and bacterial symbiosis. Plant Cell 17, 2217–2229. doi: 10.1105/tpc.105.032714
Kong, C.-C., Ren, C.-G., Li, R.-Z., Xie, Z.-H., and Wang, J.-P. (2017). Hydrogen peroxide and strigolactones signaling are involved in alleviation of salt stress induced by arbuscular mycorrhizal fungus in Sesbania cannabina seedlings. J. Plant Growth Regul. 36, 734–742. doi: 10.1007/s00344-017-9675-9
Krishnamoorthy, R., Kim, K., Subramanian, P., Senthilkumar, M., Anandham, R., and Sa, T. (2016). Arbuscular mycorrhizal fungi and associated bacteria isolated from salt-affected soil enhances the tolerance of maize to salinity in coastal reclamation soil. Agric. Ecosyst. Environ. 231, 233–239. doi: 10.1016/j.agee.2016.05.037
Lace, B., and Ott, T. (2018). Commonalities and differences in controlling multipartite intracellular infections of legume roots by symbiotic microbes. Plant Cell Physiol. 59, 661–672. doi: 10.1093/pcp/pcy043
Lardi, M., and Pessi, G. (2018). Functional genomics approaches to studying symbioses between legumes and nitrogen-fixing rhizobia. High Throughput 7:15. doi: 10.3390/ht7020015
Lee, K. B., De Backer, P., Aono, T., Liu, C. T., Suzuki, S., Suzuki, T., et al. (2008). The genome of the versatile nitrogen fixer Azorhizobium caulinodans ORS571. BMC Genomics 9:271. doi: 10.1186/1471-2164-9-271
Li, H., Chen, M., Duan, L., Zhang, T., Cao, Y., and Zhang, Z. (2018). Domain swap approach reveals the critical roles of different domains of SYMRK in root nodule symbiosis in Lotus japonicus. Front. Plant Sci. 9:697. doi: 10.3389/fpls.2018.00697
Li, T., Liu, R.-J., He, X.-H., and Wang, B.-S. (2012). Enhancement of superoxide dismutase and catalase activities and salt tolerance of euhalophyte Suaeda salsa L. by mycorrhizal fungus Glomus mosseae. Pedosphere 22, 217–224. doi: 10.1016/s1002-0160(12)60008-3
Liu, H., Wang, X., Qi, H., Wang, Q., Chen, Y., Li, Q., et al. (2017). The infection and impact of Azorhizobium caulinodans ORS571 on wheat (Triticum aestivum L.). PLoS One 12:e0187947. doi: 10.1371/journal.pone.0187947
Liu, Y., Pan, X., and Li, J. (2014). A 1961–2010 record of fertilizer use, pesticide application and cereal yields: a review. Agron. Sustain. Dev. 35, 83–93. doi: 10.1007/s13593-014-0259-9
MacLean, A. M., Bravo, A., and Harrison, M. J. (2017). Plant signaling and metabolic pathways enabling arbuscular mycorrhizal symbiosis. Plant Cell 29, 2319–2335. doi: 10.1105/tpc.17.00555
Marschner, H. (2002). “14 - effect of internal and external factors on root growth and development,” in Marschner’s Mineral Nutrition of Higher Plants, 2nd Edn, ed. H. Marschner (San Diego, CA: Academic Press), 508–536. doi: 10.1016/b978-012473542-2/50016-x
Munns, R. (2002). Comparative physiology of salt and water stress. Plant Cell Environ. 25, 239–250. doi: 10.1046/j.0016-8025.2001.00808.x
Naffah de Souza, C., Breda, L. C. D., Khan, M. A., De Almeida, S. R., Camara, N. O. S., Sweezey, N., et al. (2017). Alkaline pH promotes NADPH oxidase-independent neutrophil extracellular trap formation: a matter of mitochondrial reactive oxygen species generation and citrullination and cleavage of histone. Front. Immunol. 8:1849. doi: 10.3389/fimmu.2017.01849
Ngom, M., Oshone, R., Diagne, N., Cissoko, M., Svistoonoff, S., Tisa, L. S., et al. (2016). Tolerance to environmental stress by the nitrogen-fixing actinobacterium Frankia and its role in actinorhizal plants adaptation. Symbiosis 70, 17–29. doi: 10.1007/s13199-016-0396-9
Nguyen, T. H., Brechenmacher, L., Aldrich, J. T., Clauss, T. R., Gritsenko, M. A., Hixson, K. K., et al. (2012). Quantitative phosphoproteomic analysis of soybean root hairs inoculated with Bradyrhizobium japonicum. Mol. Cell. Proteomics 11, 1140–1155. doi: 10.1074/mcp.M112.018028
Nicolopoulou-Stamati, P., Maipas, S., Kotampasi, C., Stamatis, P., and Hens, L. (2016). Chemical pesticides and human health: the urgent need for a new concept in agriculture. Front. Public Health 4:148. doi: 10.3389/fpubh.2016.00148
Oldroyd, G. E. (2013). Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 11, 252–263. doi: 10.1038/nrmicro2990
Oshone, R., Mansour, S. R., and Tisa, L. S. (2013). Effect of salt stress on the physiology of Frankia sp strain CcI6. J. Biosci. 38, 699–702. doi: 10.1007/s12038-013-9371-2
Pandey, R., and Garg, N. (2017). High effectiveness of Rhizophagus irregularis is linked to superior modulation of antioxidant defence mechanisms in Cajanus cajan (L.) Millsp. genotypes grown under salinity stress. Mycorrhiza 27, 669–682. doi: 10.1007/s00572-017-0778-8
Pandey, R. P., Srivastava, A. K., Gupta, V. K., O’donovan, A., and Ramteke, P. W. (2018). Enhanced yield of diverse varieties of chickpea (Cicer arietinum L.) by different isolates of Mesorhizobium ciceri. Environ. Sustain. 1, 425–435. doi: 10.1007/s42398-018-00039-9
Pawlowski, K., and Demchenko, K. N. (2012). The diversity of actinorhizal symbiosis. Protoplasma 249, 967–979. doi: 10.1007/s00709-012-0388-4
Pimprikar, P., Carbonnel, S., Paries, M., Katzer, K., Klingl, V., Bohmer, M. J., et al. (2016). A CCaMK-CYCLOPS-DELLA complex activates transcription of RAM1 to regulate arbuscule branching. Curr. Biol. 26, 987–998. doi: 10.1016/j.cub.2016.01.069
Pimprikar, P., and Gutjahr, C. (2018). Transcriptional regulation of arbuscular mycorrhiza development. Plant Cell Physiol. 59, 673–690. doi: 10.1093/pcp/pcy024
Plasencia, F. A., Estrada, Y., Flores, F. B., Ortíz-Atienza, A., Lozano, R., and Egea, I. (2021). The Ca2+ sensor Calcineurin B–like protein 10 in plants: emerging new crucial roles for plant abiotic stress tolerance. Front. Plant Sci. 11:599944. doi: 10.3389/fpls.2020.599944
Poovaiah, B. W., and Du, L. (2018). Calcium signaling: decoding mechanism of calcium signatures. New Phytol. 217, 1394–1396. doi: 10.1111/nph.15003
Porcel, R., Aroca, R., Azcon, R., and Ruiz-Lozano, J. M. (2016). Regulation of cation transporter genes by the arbuscular mycorrhizal symbiosis in rice plants subjected to salinity suggests improved salt tolerance due to reduced Na(+) root-to-shoot distribution. Mycorrhiza 26, 673–684. doi: 10.1007/s00572-016-0704-5
Porcel, R., Aroca, R., and Ruiz-Lozano, J. M. (2011). Salinity stress alleviation using arbuscular mycorrhizal fungi. A review. Agron. Sustain. Dev. 32, 181–200. doi: 10.1007/s13593-011-0029-x
Porras-Soriano, A., Soriano-Martín, M. L., Porras-Piedra, A., and Azcón, R. (2009). Arbuscular mycorrhizal fungi increased growth, nutrient uptake and tolerance to salinity in olive trees under nursery conditions. J. Plant Physiol. 166, 1350–1359. doi: 10.1016/j.jplph.2009.02.010
Qadir, M., Quillérou, E., Nangia, V., Murtaza, G., Singh, M., Thomas, R. J., et al. (2014). Economics of salt-induced land degradation and restoration. Nat. Resour. Forum 38, 282–295. doi: 10.1111/1477-8947.12054
Radhakrishnan, G. V., Keller, J., Rich, M. K., Vernié, T., Mbadinga Mbaginda, D. L., Vigneron, N., et al. (2019). An ancestral signalling pathway is conserved in intracellular symbioses-forming plant lineages. Nat. Plants 6, 280–289. doi: 10.1038/s41477-020-0613-7
Rasmussen, S. R., Fuchtbauer, W., Novero, M., Volpe, V., Malkov, N., Genre, A., et al. (2016). Intraradical colonization by arbuscular mycorrhizal fungi triggers induction of a lipochitooligosaccharide receptor. Sci. Rep. 6:29733. doi: 10.1038/srep29733
Rehman, H. M., Cheung, W. L., Wong, K. S., Xie, M., Luk, C. Y., Wong, F. L., et al. (2019). High-throughput mass spectrometric analysis of the whole proteome and secretome from Sinorhizobium fredii strains CCBAU25509 and CCBAU45436. Front. Microbiol. 10:2569. doi: 10.3389/fmicb.2019.02569
Ren, C.-G., Bai, Y.-J., Kong, C.-C., Bian, B., and Xie, Z.-H. (2016). Synergistic interactions between salt-tolerant rhizobia and arbuscular mycorrhizal fungi on salinity tolerance of Sesbania cannabina plants. J. Plant Growth Regul. 35, 1098–1107. doi: 10.1007/s00344-016-9607-0
Ren, C. G., Kong, C. C., and Xie, Z. H. (2018). Role of abscisic acid in strigolactone-induced salt stress tolerance in arbuscular mycorrhizal Sesbania cannabina seedlings. BMC Plant Biol. 18:74. doi: 10.1186/s12870-018-1292-7
Rengasamy, P. (2010). Soil processes affecting crop production in salt-affected soils. Funct. Plant Biol. 37, 613–620. doi: 10.1071/fp09249
Rimington, W. R., Pressel, S., Duckett, J. G., Field, K. J., Read, D. J., and Bidartondo, M. I. (2018). Ancient plants with ancient fungi: liverworts associate with early-diverging arbuscular mycorrhizal fungi. Proc. R. Soc. B Biol. Sci. 285:20181600. doi: 10.1098/rspb.2018.1600
Romero-Munar, A., Baraza, E., Gulias, J., and Cabot, C. (2019). Arbuscular mycorrhizal fungi confer salt tolerance in giant reed (Arundo donax L.) plants grown under low phosphorus by reducing leaf Na(+) concentration and improving phosphorus use efficiency. Front. Plant Sci. 10:843. doi: 10.3389/fpls.2019.00843
Rose, C. M., Venkateshwaran, M., Volkening, J. D., Grimsrud, P. A., Maeda, J., Bailey, D. J., et al. (2012). Rapid phosphoproteomic and transcriptomic changes in the rhizobia-legume symbiosis. Mol. Cell. Proteomics 11, 724–744. doi: 10.1074/mcp.M112.019208
Russo, G., Spinella, S., Sciacca, E., Bonfante, P., and Genre, A. (2013). Automated analysis of calcium spiking profiles with CaSA software: two case studies from root-microbe symbioses. BMC Plant Biol. 13:224. doi: 10.1186/1471-2229-13-224
Saghafi, D., Ghorbanpour, M., Shirafkan Ajirloo, H., and Asgari Lajayer, B. (2019). Enhancement of growth and salt tolerance in Brassica napus L. seedlings by halotolerant Rhizobium strains containing ACC-deaminase activity. Plant Physiol. Rep. 24, 225–235. doi: 10.1007/s40502-019-00444-0
Sepúlveda-Caamaño, M., Gerding, M., Vargas, M., Moya-Elizondo, E., Oyarzúa, P., and Campos, J. (2018). Lentil (Lens culinaris L.) growth promoting rhizobacteria and their effect on nodulation in coinoculation with rhizobia. Arch. Agron. Soil Sci. 64, 244–256. doi: 10.1080/03650340.2017.1342034
Shabala, S., and Pottosin, I. (2014). Regulation of potassium transport in plants under hostile conditions: implications for abiotic and biotic stress tolerance. Physiol. Plant. 151, 257–279. doi: 10.1111/ppl.12165
Shamshiri, M. H., and Fattahi, M. (2016). Effects of arbuscular mycorrhizal fungi on photosystem II activity of three pistachio rootstocks under salt stress as probed by the OJIP-test. Russ. J. Plant Physiol. 63, 101–110. doi: 10.1134/s1021443716010155
Sheng, M., Tang, M., Zhang, F., and Huang, Y. (2011). Influence of arbuscular mycorrhiza on organic solutes in maize leaves under salt stress. Mycorrhiza 21, 423–430. doi: 10.1007/s00572-010-0353-z
Singh, S., Katzer, K., Lambert, J., Cerri, M., and Parniske, M. (2014). CYCLOPS, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe 15, 139–152. doi: 10.1016/j.chom.2014.01.011
Smit, P., Raedts, J., Portyanko, V., Debellé, F., Gough, C., Bisseling, T., et al. (2005). NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308, 1789–1791. doi: 10.1126/science.1111025
Srivastava, A., Singh, S. S., and Mishra, A. K. (2013). Sodium transport and mechanism(s) of sodium tolerance in Frankia strains. J. Basic Microbiol. 53, 163–174. doi: 10.1002/jobm.201100586
Suzaki, T., Takeda, N., Nishida, H., Hoshino, M., Ito, M., Misawa, F., et al. (2019). LACK OF SYMBIONT ACCOMMODATION controls intracellular symbiont accommodation in root nodule and arbuscular mycorrhizal symbiosis in Lotus japonicus. PLoS Genet. 15:e1007865. doi: 10.1371/journal.pgen.1007865
Tirichine, L., Imaizumi-Anraku, H., Yoshida, S., Murakami, Y., Madsen, L. H., Miwa, H., et al. (2006). Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature 441, 1153–1156. doi: 10.1038/nature04862
Tong, W., Li, X., Wang, E., Cao, Y., Chen, W., Tao, S., et al. (2020). Genomic insight into the origins and evolution of symbiosis genes in Phaseolus vulgaris microsymbionts. BMC Genomics 21:186. doi: 10.1186/s12864-020-6578-0
Tromas, A., Parizot, B., Diagne, N., Champion, A., Hocher, V., Cissoko, M., et al. (2012). Heart of endosymbioses: transcriptomics reveals a conserved genetic program among arbuscular mycorrhizal, actinorhizal and legume-rhizobial symbioses. PLoS One 7:e44742. doi: 10.1371/journal.pone.0044742
Venkateshwaran, M., Cosme, A., Han, L., Banba, M., Satyshur, K. A., Schleiff, E., et al. (2012). The recent evolution of a symbiotic ion channel in the legume family altered ion conductance and improved functionality in calcium signaling. Plant Cell 24, 2528–2545. doi: 10.1105/tpc.112.098475
Wang, B., Yeun, L. H., Xue, J. Y., Liu, Y., Ane, J. M., and Qiu, Y. L. (2010). Presence of three mycorrhizal genes in the common ancestor of land plants suggests a key role of mycorrhizas in the colonization of land by plants. New Phytol. 186, 514–525. doi: 10.1111/j.1469-8137.2009.03137.x
Wang, F., Sun, Y., and Shi, Z. (2019). Arbuscular mycorrhiza enhances biomass production and salt tolerance of sweet sorghum. Microorganisms 7:289. doi: 10.3390/microorganisms7090289
Wang, Y., Wang, M., Li, Y., Wu, A., and Huang, J. (2018). Effects of arbuscular mycorrhizal fungi on growth and nitrogen uptake of Chrysanthemum morifolium under salt stress. PLoS One 13:e0196408. doi: 10.1371/journal.pone.0196408
Wu, K., Wang, S., Song, W., Zhang, J., Wang, Y., Liu, Q., et al. (2020). Enhanced sustainable green revolution yield via nitrogen-responsive chromatin modulation in rice. Science 367:eaaz2046. doi: 10.1126/science.aaz2046
Wu, N., Li, Z., Liu, H., and Tang, M. (2015). Influence of arbuscular mycorrhiza on photosynthesis and water status of Populus cathayana Rehder males and females under salt stress. Acta Physiol. Plant. 37:183.
Xue, L., Cui, H., Buer, B., Vijayakumar, V., Delaux, P. M., Junkermann, S., et al. (2015). Network of GRAS transcription factors involved in the control of arbuscule development in Lotus japonicus. Plant Physiol. 167, 854–871. doi: 10.1104/pp.114.255430
Yano, K., Yoshida, S., Müller, J., Singh, S., Banba, M., Vickers, K., et al. (2008). CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc. Natl. Acad. Sci. U.S.A. 105, 20540–20545. doi: 10.1073/pnas.0806858105
Young, N. D., Debelle, F., Oldroyd, G. E., Geurts, R., Cannon, S. B., Udvardi, M. K., et al. (2011). The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480, 520–524. doi: 10.1038/nature10625
Yuan, P., Jauregui, E., Du, L., Tanaka, K., and Poovaiah, B. W. (2017). Calcium signatures and signaling events orchestrate plant-microbe interactions. Curr. Opin. Plant Biol. 38, 173–183. doi: 10.1016/j.pbi.2017.06.003
Zahir, Z. A., Shah, M. K., Naveed, M., and Akhter, M. J. (2010). Substrate-dependent auxin production by Rhizobium phaseoli improves the growth and yield of Vigna radiata L. under salt stress conditions. J. Microbiol. Biotechnol. 20, 1288–1294. doi: 10.4014/jmb.1002.02010
Keywords: symbiosis, sustainable agriculture, saline soil, plant ‘holobiont’, common symbiotic pathway
Citation: Ren C-G, Kong C-C, Liu Z-Y, Zhong Z-H, Yang J-C, Wang X-L and Qin S (2022) A Perspective on Developing a Plant ‘Holobiont’ for Future Saline Agriculture. Front. Microbiol. 13:763014. doi: 10.3389/fmicb.2022.763014
Received: 23 August 2021; Accepted: 28 March 2022;
Published: 06 May 2022.
Edited by:
Marina G. Kalyuzhanaya, San Diego State University, United StatesReviewed by:
Joanna Dames, Rhodes University, South AfricaCopyright © 2022 Ren, Kong, Liu, Zhong, Yang, Wang and Qin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Song Qin, U3FpbkB5aWMuYWMuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.