
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol. , 03 May 2022
Sec. Microbial Physiology and Metabolism
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.757711
This article is part of the Research Topic Regulation of Prokaryotic Cell Division View all 17 articles
FtsZ is the cytoskeletal protein that organizes the formation of the septal ring and orchestrates bacterial cell division. Its association to the membrane is essential for its function. In this mini-review I will address the question of how this association can interfere with the structure and dynamic properties of the filaments and argue that its dynamics could also remodel the underlying lipid membrane through its activity. Thus, lipid rearrangement might need to be considered when trying to understand FtsZ’s function. This new element could help understand how FtsZ assembly coordinates positioning and recruitment of the proteins forming the septal ring inside the cell with the activity of the machinery involved in peptidoglycan synthesis located in the periplasmic space.
Bacterial division requires a profound morphological change in the cell. It involves the concerted action in space and time of different cellular elements: chromosomes divide and distribute between the new cells as proteins forming the divisome localize in the center and promote active membrane deformation and peptidoglycan synthesis required to separate the cell into two new ones (Mahone and Goley, 2020).
The study of this fascinating field is considered to have started with the first description of the FtsZ ring in 1991 made by Bi and Lutkenhaus (1991). Since then, we have gained huge insight into how this complex process takes place. A combination of genetic and biochemical studies in vitro and in vivo have provided knowledge of the essential proteins participating and how they interact and assemble. There are several reviews summarizing our current knowledge of the formation of the septal ring (Adams and Errington, 2009; Mingorance et al., 2010; Den Blaauwen and Luirink, 2019; Du and Lutkenhaus, 2019; Mahone and Goley, 2020; McQuillen and Xiao, 2020; Barrows and Goley, 2021). Advances in single molecule fluorescence, applied both to reconstituted systems (Caldas et al., 2019; García-Soriano et al., 2020) and to whole cells (McCausland et al., 2021; Squyres et al., 2021; Yang et al., 2021), have allowed a more detailed analysis of the structural rearrangements occurring during this active energy-consuming process. These studies have revealed a highly dynamic and concerted interaction between septal ring and peptidoglycan synthesis proteins (Bisson-Filho et al., 2017; Yang et al., 2017; McCausland et al., 2021; Squyres et al., 2021).
FtsZ is the cytoskeletal protein that organizes the formation of the septal ring and orchestrates the division process (Margolin, 2005; Huang et al., 2013; Ortiz et al., 2015; Barrows and Goley, 2021). Since its first description (Dai and Lutkenhaus, 1991; Pla et al., 1991; de Boer et al., 1992; RayChaudhuri and Park, 1992), much work has focused in understanding how it behaves in vitro and how it interacts with other proteins. We know that membrane association is essential for its function and that, even when attached through an inserted transmembrane region, in the absence of any of the proteins present in vivo, it displays a dynamic rich behavior and deforms membranes (Osawa et al., 2009; Caldas et al., 2019; Ramirez-Diaz et al., 2021).
We have identified a large catalog of proteins participating in the formation of the septal ring, but we still have very few cues about how they communicate to coordinate cell division. Information is transferred between proteins located inside the membrane, that actively condense to form the septal ring, and proteins whose function involves communication and transfer of material across the membrane. Peptidoglycan building blocks are exported to the periplasmic space for the peptidoglycan synthetic machinery to work (Ruiz, 2016). Direct protein-protein interactions are frequently evoked as the main element guiding the concerted process, but it is difficult to imagine that the fluid membrane that hosts many of the proteins involved plays no role, as is frequently depicted in illustrations.
Membranes are not passive elements (Kalappurakkal et al., 2020), particularly when associated to active cytoskeletal proteins that form polymeric filamentous structures that reorganize by dissipating energy consumed from ATP or GTP hydrolysis (Litschel et al., 2021). The long-range order of cytoskeletal filaments is ideal for building sufficiently large structures responsive to multiple inputs that can generate mechanical forces. It is therefore not surprising that several bacterial cytoskeletal proteins play essential roles in cell division (Shih and Rothfield, 2006; Cabeen and Jacobs-Wagner, 2010; Ingerson-Mahar and Gitai, 2012; Pilhofer and Jensen, 2013; Busiek and Margolin, 2015; Siliakus et al., 2017). Some are involved in DNA segregation, ParM for example, and others, such as eukaryotic actin and tubulin homologues MreB, FtsA, and FtsZ, are associated to the membrane and play a major role in maintaining cell shape and remodeling the membrane.
Studies in eukaryotes have shown that active cytoskeletal proteins regulate lipid and protein distribution and membrane mechanical properties, originating non-equilibrium distributions and emerging properties (Gowrishankar et al., 2012; Longo, 2018; Steinkühler et al., 2019; Shaw et al., 2020). The actin cortex may influence plasma membrane organization either by direct interaction with lipids and proteins or through the flow of actin filaments generated by myosin-induced stresses. This activity can also drive actin-associated membrane components out of equilibrium inducing active processes such as persistent advection, clustering, and anomalous density fluctuations, even though measurable hydrodynamic flows in the plasma membrane of unconnected components are not present. The remodeling of the actomyosin layer also affects phase-segregation in the membrane bilayer. It has been experimentally shown that actin activity changes the size and dynamics of the formed domains and that, in addition, membrane domains influence the actin organization (Köster et al., 2016). Lipid domains, referred to as rafts, play an important role in regulating functions such as membrane trafficking (Redpath et al., 2020) and response of membrane receptors (Gowrishankar et al., 2012). Other membrane associated GTP self-aggregating proteins such as dynamins (Kalia and Frost, 2019) and septins (Lam and Calvo, 2019), also reshape the lipid membrane in eukaryotic cells by undergoing GTP-induced conformational changes, although the details of how this happens are still to be elucidated (Pannuzzo et al., 2018). Thus, the current picture of a cell membrane, based on experiments mostly carried out in eukaryotes, is that of a composite material in which lipids and proteins interact to transfer information across membrane components (Sezgin et al., 2017).
In this minireview I will focus on FtsZ polymerization on membranes, but I will first briefly update what we know about bacterial membranes to highlight the evidence that indicates that we should extrapolate what we have learned about cytoskeletal membrane associated proteins in eukaryotes to better understand what is happening in the bacterial membrane during cell division.
To what extent is the presence of the membrane surface and the confinement it imposes to the FtsZ filaments relevant to reveal the rich dynamic behavior observed? Can we also expect bacterial membranes to be responsive to the activity of membrane associated cytoskeletal proteins, as has been shown for eukaryotic cytoskeleton? Could protein modulated membrane properties participate in coordinating the complex interactions acting on the cell division machinery? These are the questions I will address. First, I will briefly summarize advances in our understanding of bacterial membrane organization. I will then briefly review what we know about the active and dynamic FtsZ filaments and their interaction with the membrane. In the discussion I will argue that it might be interesting to attend the role the lipid membrane could play in regulating the process in order to fully understand how bacterial cell division occurs.
Bacterial membranes have been less subject to biophysical studies than eukaryotic membranes. Their small size and the complexity rendered by the presence of a peptidoglycan layer make their characterization difficult. However, spatial and temporal reorganization of lipids and proteins on bacterial surfaces take place during complex functions, just as happens in eukaryotes. High resolution optical fluorescence microscopy has confirmed that their inner membrane shares the lateral heterogeneity and complexity found in eukaryotic cells (Dempwolff et al., 2016; Raghunathan and Kenworthy, 2018). Domains and lipid rafts in bacterial membranes are highly organized with specific lipids and associated proteins forming specialized subcellular compartments. They have been referred to as functional membrane microdomains (FMM) to differentiate them from the eukaryotic lipids rafts containing cholesterol, not present in bacteria. The lateral segregation has been observed to be associated to the presence of anionic lipids, squalene, phosphatidyl-ethanolamine, and proteins such as flotillins (Nishibori et al., 2005; Mileykovskaya and Dowhan, 2009; Bramkamp and Lopez, 2015; Lin and Weibel, 2016; Strahl and Errington, 2017; Yokoyama and Matsui, 2020).
Membrane structure and dynamics are interdependent. Structural heterogeneity has consequences in the heterogeneous dynamics of proteins on the membrane (Nenninger et al., 2014; Adhyapak et al., 2020; Pluhackova and Horner, 2021). Therefore, membrane composition also plays a regulatory role in cell physiology. Stress is known to trigger metabolic pathways that sense threats and mount a protective response involving modification of cell wall composition through the regulation of biosynthetic pathways of their components (Rowlett et al., 2017; Willdigg and Helmann, 2021). There is evidence that certain lipids are enriched in the division site (Mileykovskaya and Dowhan, 2005), indicating that the division process is no exception.
In summary, experimental results indicate that non-random distribution of proteins and lipids also play an important role in protein-protein interactions in bacteria (Raghunathan and Kenworthy, 2018; Zeno et al., 2020), as is well established for eukaryotic membranes.
FtsZ is a soluble 40.3 kDa protein that binds and hydrolyzes GTP and shows a striking structural similarity to tubulins (Erickson, 1997). Its crystal structure was determined in 1998 (Löwe and Amos, 1998) and since then, much biochemical work on the isolated protein in solution has described its polymerization properties. Lateral interactions between individual filaments participate in bundling (Hörger et al., 2008a; Milam et al., 2012), affect their arrangement on surfaces (Márquez et al., 2017) and are strongly modulated by the presence of different ions or crowding agents (Erickson et al., 1996; Popp et al., 2009; Figure 1). The presence of curvature in the filaments has been controversial. In analogy to tubulin, it was first considered that the presence of GDP induced the curved conformation whereas the GTP loaded monomers formed straight filaments (Lu et al., 2000; Hsin et al., 2012). However, crystal structures of monomers containing GDP and GTP were not able to confirm this association of the nucleotide phosphorylation state with the degree of curvature (Oliva et al., 2004), and conditions in which the monomers contained essentially phosphorylated nucleotide were observed to be highly curved (Mateos-Gil et al., 2012b; Loose and Mitchison, 2013; Ramirez-Diaz García-Soriano et al., 2018). More recent structural characterizations, coming both from structural and molecular dynamics simulations, have depicted highly flexible monomer in which the relative orientation of the C-terminal and N-terminal domains, linked through a helix 7, can change up to nearly 30 degrees (Martín-Galiano et al., 2010; Matsui et al., 2012; Fujita et al., 2017; Wagstaff et al., 2017). Two monomer conformations have been described, open and close, also referred to as relaxed and tense, that appear to be associated to whether the monomer is isolated or polymerized (Schumacher et al., 2020; Figure 1). Molecular dynamics simulations have also revealed the presence of a significant angle between monomers that confers a twist to the filaments (Hsin et al., 2012; Gonzalez de Prado Salas et al., 2014; Lv et al., 2021). Experimental observations have confirmed that filaments on surfaces indeed manifest this twist (Arumugam et al., 2012; Gonzalez de Prado Salas et al., 2014; Ramirez-Diaz et al., 2021).
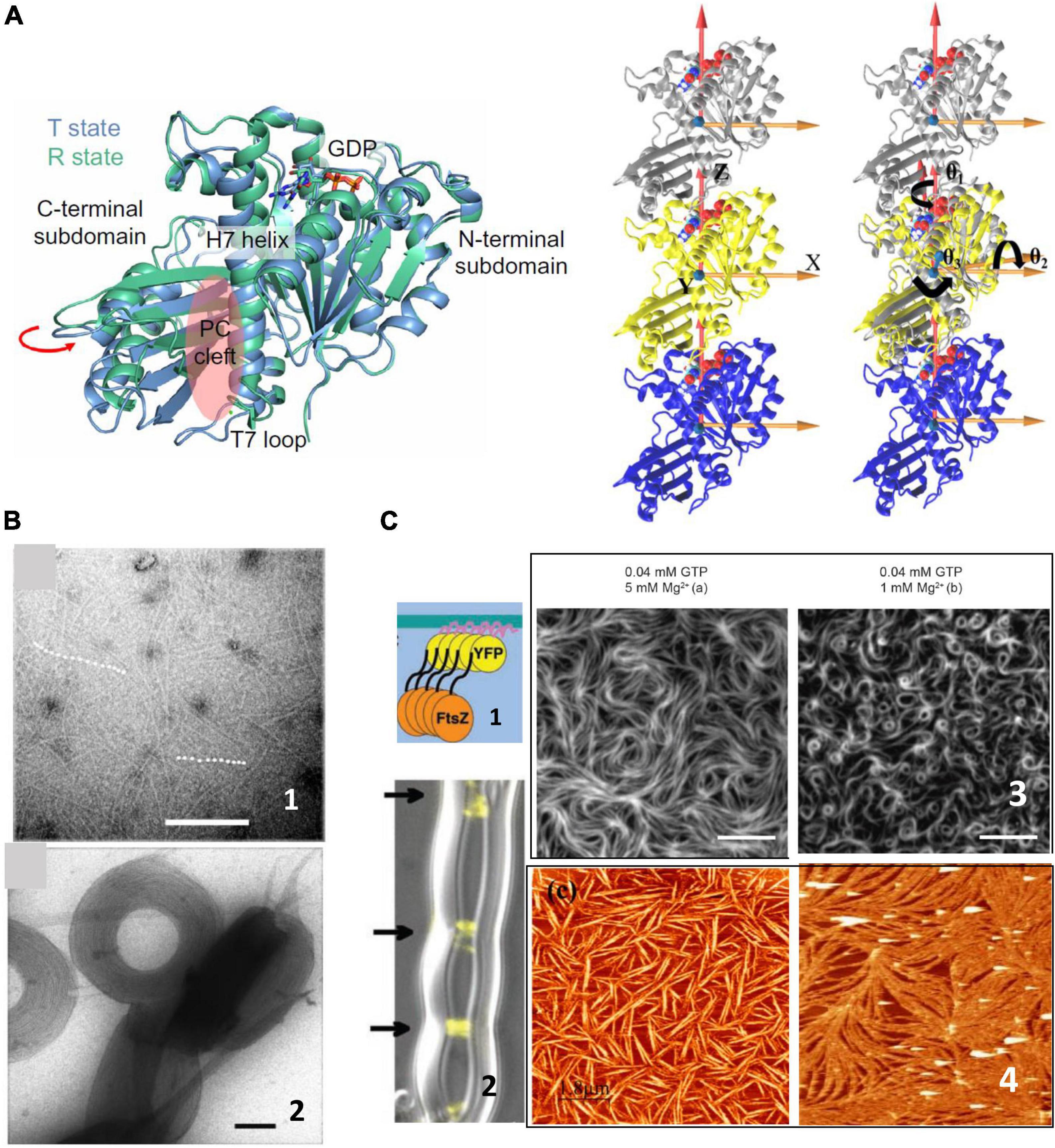
Figure 1. FtsZ structure and polymerization in solution. (A) Left shows the two crystal structures of FtsZ from Staphylococcus aureus corresponding to the T and R state conformations in the same crystal, indicating the structural equilibrium of the two states (adapted from Fujita et al., 2017). The trimers on the right show the definitions of bending and twisting angles obtained from molecular dynamics simulations. The left trimer shows the coordinate system and the one on the right shows the bending angles, θ1 (rotations around the Z axis), θ2 (rotations around the X axis), and θ3 (rotations around the Y axis), tracked by calculating the rotations of the top (silver) subunit to align the initial reference frame of the middle subunit (adapted from Lv et al., 2021). (B) Shows representative negative stained TEM images of FtsZ filaments in solution (1) and aggregates formed in the presence of crowding agents (2). Scale bar is 100 nm (adapted from Popp et al., 2009). (C) Shows surface confined FtsZ polymers. (1) Show a chimeric FtsZ containing a fluorescent protein and a membrane targeting sequence (MTS) on liposomes (2) (adapted from Osawa et al., 2008) and on planar supported membranes (3) (adapted from Ramirez-Diaz García-Soriano et al., 2018). Scale bars are 5 μm. (4) Shows Atomic Force Microscopy images of FtsZ filaments attached covalently to a lipid membrane (left) (adapted from Encinar et al., 2013) or bound through the protein ZipA (right) (adapted from Mateos-Gil et al., 2012a). Scale bars are 2 μm.
Early studies showed that FtsZ polymers are dynamic, both in vivo and in vitro (Stricker et al., 2002; Chen and Erickson, 2005, 2009; Chen et al., 2005), where filaments studied in solution revealed instability due to continuous monomer exchange associated to GTP hydrolysis (Romberg and Levin, 2003). Concentration dependent cooperative polymerization was detected quite early and opened questions regarding how individual filaments, the structures mainly observed at the time, could account for cooperativity, more compatible with the existence of double filaments (Chen et al., 2005; Erickson, 2017). Later cooperativity was attributed to a conformational switch (Huecas and Andreu, 2003) in the monomer that facilitates polymerization, in line with the increasing evidence that monomers are flexible (Martín-Galiano et al., 2010).
All together, the picture that emerged from biochemical and structural studies of filaments forming in solution was of soft, flexible, and polymorphic assemblies that can adopt a large range of shapes easily malleable through environmental conditions (Figure 1). A yet open challenge is to understand how such dynamic structures are responsible for coordinating and maintaining the formation of the septal ring when associated to the membrane.
The dynamic behavior of FtsZ showed unexpected complexity when filaments were observed associated to a surface. Although some biochemical studies in solution gave hints that polymer growth could be directional (Osawa and Erickson, 2005; Redick et al., 2005), it was not until single molecule fluorescence allowed visualizing polymers associated to membranes, both in vivo and in vitro, that dynamics of FtsZ filaments was clearly observed. Several groups confirmed that inside the cell FtsZ moves along the cell circumference in a treadmilling fashion along the cell-wall remodeling machinery (Bisson-Filho et al., 2017; Yang et al., 2017; Perez et al., 2019). Studies in vitro, also showed that FtsZ filaments on supported membranes manifested unexpected cooperative behavior based on treadmilling (Loose and Mitchison, 2013), forming vortices moving in opposite directions depending on the orientation of the monomers on the surface (Ramirez-Diaz García-Soriano et al., 2018). This behavior does not require any additional proteins, only a chimeric FtsZ containing a fluorescent protein and a membrane targeting sequence (MTS) incorporated at one end to allow for membrane attachment (Ramirez-Diaz García-Soriano et al., 2018; Figure 1). Addition of filament binding proteins such as ZapA modulate precision and robustness of the collective behavior (Caldas et al., 2019), but not the essential traits, suggesting that FtsZ itself contains the required information to treadmill and rearrange on surfaces.
Experiments have shown that surface attachment regulates filament dynamics, as had been previously predicted from theoretical models (Mateos-Gil et al., 2019). Removing the C-terminal flexible linker abolishes treadmilling (García-Soriano et al., 2020). Filaments bind to several membrane associated regulatory proteins, such as FtsA, ZipA, or MinC, through the C-terminal constant region located at the end of a non-structured C-terminal linker (Buske and Levin, 2013; Gardner et al., 2013). This disordered region can vary in length between species but is required for proper cell division (Ohashi et al., 2002; Buske et al., 2015; Cohan et al., 2020). One interpretation is that regulating the distance and attachment strength through a flexible spacer is important for modulating the dynamic assembly. In some organisms, FtsZ binds to the membrane through ZipA, which provides an additional non-structured region that sits between the FtsZ monomer and the surface (Ohashi et al., 2002; Mateos-Gil et al., 2016). We can only speculate about why these flexible spacers and distance from the membrane are important for FtsZ function. We do not know if they affect the monomer conformational switch that has been proposed to regulate polymer dynamics. Another possibility is that attachment strength modulates filament twist and thus filament mechanical properties, as has been suggested by theory (Gonzalez de Prado Salas et al., 2014; Mateos-Gil et al., 2019). Modulating the compactness of this variable region through the binding of regulatory proteins could be a way to affect filament dynamics and structure. Filaments grow from both ends, without treadmilling (Márquez et al., 2019), when they are firmly attached to a membrane with a defined orientation, confirming that surface attachment and orientation have a strong impact on their structure and dynamics on membranes.
Surface confinement and lipid type modulate both filament dynamics and shape (Mateos-Gil et al., 2012a; Encinar et al., 2013). Filament’s preferential curvature, lateral interactions and twist (Hörger et al., 2008a,b; Paez A. et al., 2009; Paez P. et al., 2009; Mateos-Gil et al., 2019; Márquez et al., 2019) are revealed differently in solution and under confinement on a two dimensional surface. Theoretical models that include flexible monomer attachment and torsion provide testable hypothesis indicating that only these two elements are enough to induce an asymmetry in the filament ends accessibility to monomer addition that would generate different growth speed on each end, which manifests as treadmilling (Mateos-Gil et al., 2019). Asymmetric confinement to a surface is also likely to regulate the conformation of the flexible monomers and thus polymer dynamics.
Filaments on the surface play a GTP-dependent active role on membrane deformation, in spite of their continuous monomer exchange. The debate of whether the force needed to constrict the membrane during cell division comes exclusively from FtsZ filaments or from the peptidoglycan synthetic machinery is still open, although there is no question that at least some membrane deformation is provided by the protein activity (Xiao and Goley, 2016). Filaments create concave or convex deformations or inwards cone structures emerging from the membrane surface (Xiao and Goley, 2016), depending on whether they interact through the N-terminal or the C-terminal end of the protein, indicating drilling-like inward forces (Osawa et al., 2008, 2009). One available hypothesis to explain how flexible, dynamic filaments deform the membrane (Xiao and Goley, 2016) is that torsion provides partly the flexibility found in solution or on surfaces where the attachment is loose. Tight anchoring could stiffen the filaments (Gonzalez de Prado Salas et al., 2014), allowing them to readily deform the membrane and exert force (Mateos-Gil et al., 2019).
One question that is seldom addressed is how the activity of the filaments and the monomer exchange due to GTP hydrolysis affects the structure and integrity of the membrane. FtsZ has a GTP hydrolysis rate comparable to monomer turnover time, and releases the hydrolyzed nucleotide very fast (Romberg and Mitchison, 2004). This rapid release of products implies that the energy from GTP hydrolysis is quickly dissipated, in contrast to microtubules, where it is stored as strain in the polymer (Caplow et al., 1994) and released during microtubule disassembly to perform mechanical work.
How does this dissipated energy affect the membrane? There is experimental evidence that FtsZ filaments attached to a supported lipid bilayer affect the shape of lipid domains and that the segregated lipids also influence the distribution and shape of the filaments (González de Prado Salas et al., 2015). Modeling this behavior requires considering filament torsion, monomer exchange, lateral interactions, and preferential curvature (Gonzalez de Prado Salas et al., 2014), and the tendency of the lipids to segregate into domains (Figure 2). Given the complex composition of the lipids of the bacterial inner membrane, it is not difficult to imagine that this lipid-protein interplay will be relevant also in the living cell and that the membrane components will respond to the remodeling of the filaments on its surface. Experiments in model membranes provide evidence that this is indeed the case. Langmuir monolayers of E. coli lipids indicate that this lipid mixture is easily stretchable (López-Montero et al., 2008) and that the presence of active FtsZ filaments attached through ZipA further softened the membrane (López-Montero et al., 2012, 2013). The energy dissipated by GTP hydrolysis could be transmitted to the membrane affecting the distribution and size of lipid domains in the inner side of the E. coli membrane (Zerrouk et al., 2008). This domain reorganization could affect membrane plasticity and modulate its local mechanical properties (López-Montero et al., 2010). Figure 2C shows a cartoon of how the tension created by filament formation could affect lipid domain redistribution on the membrane.
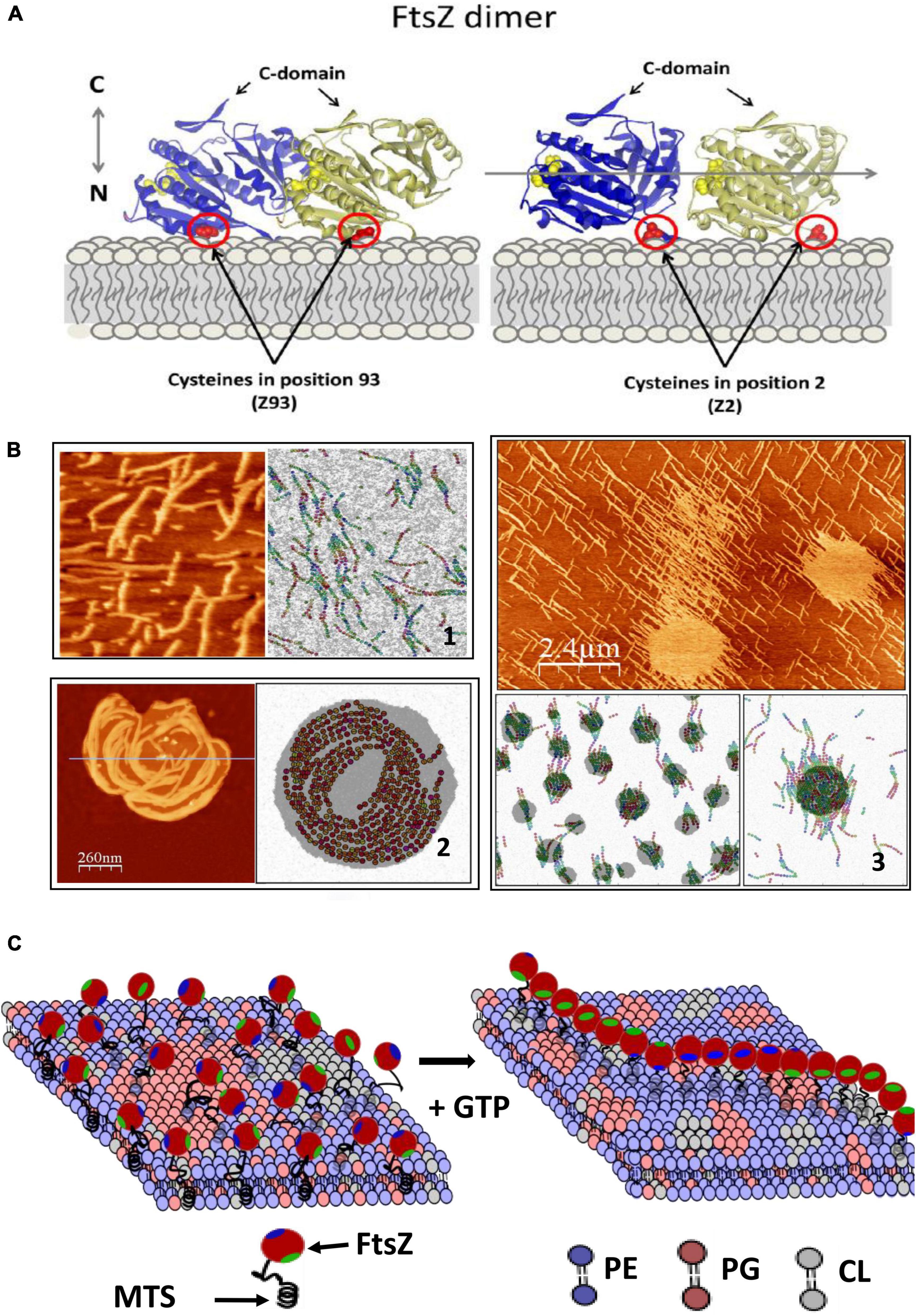
Figure 2. FtsZ activity on the lipid membrane. (A) Shows how E.coli FtsZ monomers are oriented when attached covalently to a maleimide lipid (DSPE-MAL) in the bilayer through a cysteine placed in different positions. (B) Shows 3 examples of structures observed with AFM and snapshots of MC simulations of the model that describes the system including three terms: the dynamic interactions between protein monomers, the interactions between lipid components, and a mixed term considering protein–lipid interactions. Including torsion of the monomers within the filament in the model is necessary to account for the observed filament shapes. (1) Shows mutant E93C above lipid segregation temperature; (2) shows mutant F2C below lipid segregation temperature and (3) mutant E93C at higher DSPE-MAL concentration (adapted from González de Prado Salas et al., 2015). (C) Shows a schematic illustration of how polymerization of FtsZ could affect the membrane. The association is through a membrane targeting sequence (MTS) localized in the C-terminal region (green). The components are the main lipids present in the E.coli inner membrane (phosphatidylethanolamine, PE, phsophatidylglycerol, PG and cardiolipin, CL). Tension created by filament torsion and preferential curvature restrictions upon surface confinement could affect lipid reorganization.
Condensation of FtsZ filaments in the central position of the inner cell membrane orchestrates the complex process of cell division. This event transmits information along and across the membrane surface through its interaction with many other proteins located both in the membrane as well as protruding from either side, toward the periplasmic space or toward the intracellular space. The aim of this mini-review has been to try to stitch together information that has been gathered from different approaches to the study of FtsZ polymers. One of the important challenges that remains in the field is closing the gap between the information provided by various experimental approaches that access different spatial and temporal scales. Whereas molecular dynamics simulations describe a highly flexible monomer, it is difficult to associate these conformational switches to the cooperative polymerization and treadmilling assembly observed at surfaces. We do not know either how the functionally important C-terminal non-structured region is modulating monomer-monomer interactions or their association to membrane bound anchoring proteins. Individual monomer restructuring undergone during the nucleotide hydrolysis cycle could be affected by surface confinement on a charged lipid surface. However, it is experimentally difficult to associate monomeric conformations to the collective behavior of the filaments observed on the membrane. In order to reconcile all the information available we will most likely need to incorporate additional techniques such as solid state NMR or other spectroscopic techniques to look in more detail at the structural rearrangement of the proteins and the lipids during the ring formation process.
Another issue that I have stressed is the potential importance of the lipid rearrangement due to the activity of FtsZ. There is another example of a membrane associated cytoskeletal protein participating in bacterial division for which there is evidence that its function is associated to membrane properties. MreB is a membrane associated actin homolog required in rod-shaped bacteria for cell shape maintenance and for peptidoglycan synthesis (Dempwolff et al., 2011; Nurse and Marians, 2013; Lee et al., 2020). Its direct interaction with FtsZ is required for septum synthesis and cell division in Escherichia coli (Varma and Young, 2009; Fenton and Gerdes, 2013). It has recently been shown that phospholipid composition and membrane fluidity affect the localization of MreB, which in turn affects peptidoglycan synthesis (Kurita et al., 2020). It has also been shown that MreB promotes membrane fluidity and affects membrane protein localization (Strahl et al., 2014; Oswald et al., 2016). This further illustrates that local modulation of membrane properties might be a strategy to convey information between different proteins involved in the various functions active during cell division.
There is enough accumulated evidence indicating that proteins and lipids in membranes participate in a concerted fashion to generate complex functions. Active energy consuming proteins near a membrane invest part of this energy to transform membrane structure and properties. Although most of the experimental evidence of these interactions comes from studies done in eukaryotic cells, observations of FtsZ on lipid membranes indicate that in prokaryotic cells this two-way communication between protein activity and membrane properties also exists. Considering the mutual influence between the collective properties of active proteins and membrane lipids offers a wider conceptual framework to understand the intricate regulation of different protein activities during cell division. It is likely to provide additional elements to understand this complex yet fascinating process.
MV wrote the article.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The author acknowledges Open Access funding provided thanks to the CRUE-CSIC agreement with Frontiers and financial support from Comunidad de Madrid/FEDER, EU through project SINOXPHOS-CM (S2018/BAA-4403) and MCIU/AEI/FEDER, EU, for funding project RTI2018-095090-B-I00. The author also thank Prof. Pedro Tarazona for fruitful discussions.
Adams, D. W., and Errington, J. (2009). Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat. Rev. Microbiol. 7, 642–653. doi: 10.1038/nrmicro2198
Adhyapak, P., Srivatsav, A. T., Mishra, M., Singh, A., Narayan, R., and Kapoor, S. (2020). Dynamical organization of compositionally distinct inner and outer membrane lipids of Mycobacteria. Biophys. J. 118, 1279–1291. doi: 10.1016/j.bpj.2020.01.027
Arumugam, S., Chwastek, G., Fischer-Friedrich, E., Ehrig, C. I, and Schwille, P. (2012). Surface topology engineering of membranes for the mechanical investigation of the tubulin homologue FtsZ. Angew. Chem. Int. Ed. 51, 11858–11862. doi: 10.1002/anie.201204332
Barrows, J. M., and Goley, E. D. (2021). FtsZ dynamics in bacterial division: what, how, and why? Curr. Opin. Cell Biol. 68, 163–172. doi: 10.1016/j.ceb.2020.10.013
Bi, E., and Lutkenhaus, J. (1991). FtsZ ring structure associated with division in Escherichia coli. Nature 354:161. doi: 10.1038/354161a0
Bisson-Filho, A. W., Hsu, Y. P., Squyres, G. R., Kuru, E., Wu, F., Jukes, C., et al. (2017). Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science 355:739. doi: 10.1126/science.aak9973
Bramkamp, M., and Lopez, D. (2015). Exploring the existence of lipid rafts in bacteria. Microbiol. Mol. Biol. Rev. 79, 81–100. doi: 10.1128/mmbr.00036-14
Busiek, K. K., and Margolin, W. (2015). Bacterial actin and tubulin homologs in cell growth and division. Curr. Biol. 25, R243–R254. doi: 10.1016/j.cub.2015.01.030
Buske, P. J., and Levin, P. A. (2013). A flexible C-terminal linker is required for proper FtsZ assembly in vitro and cytokinetic ring formation in vivo. Mol. Microbiol. 89, 249–263. doi: 10.1111/mmi.12272
Buske, P. J., Mittal, A., Pappu, R. V., and Levin, P. A. (2015). An intrinsically disordered linker plays a critical role in bacterial cell division. Semin. Cell Dev. Biol. 37, 3–10. doi: 10.1016/j.semcdb.2014.09.017
Cabeen, M. T., and Jacobs-Wagner, C. (2010). The bacterial cytoskeleton. Annu. Rev. Genet. 44, 365–392. doi: 10.1146/annurev-genet-102108-134845
Caldas, P., López-Pelegrín, M., Pearce, D. J. G., Budanur, N. B., Brugués, J., and Loose, M. (2019). Cooperative ordering of treadmilling filaments in cytoskeletal networks of FtsZ and its crosslinker ZapA. Nat. Commun. 10:5744. doi: 10.1038/s41467-019-13702-4
Caplow, M., Ruhlen, R. L., and Shanks, J. (1994). The free energy for hydrolysis of a microtubule-bound nucleotide triphosphate is near zero: all of the free energy for hydrolysis is stored in the microtubule lattice. J. Cell Biol. 127, 779–788. doi: 10.1083/JCB.127.3.779
Chen, Y., and Erickson, H. P. (2005). Rapid in vitro assembly dynamics and subunit turnover of FtsZ demonstrated by fluorescence resonance energy transfer. J. Biol. Chem. 280, 22549–22554. doi: 10.1074/JBC.M500895200
Chen, Y., and Erickson, H. P. (2009). FtsZ filament dynamics at steady state: subunit exchange with and without nucleotide hydrolysis. Biochemistry 48, 6664–6673. doi: 10.1021/bi8022653
Chen, Y., Bjornson, K., Redick, S. D., and Erickson, H. P. (2005). A rapid fluorescence assay for FtsZ assembly indicates cooperative assembly with a dimer nucleus. Biophys. J. 88, 505–514. doi: 10.1529/BIOPHYSJ.104.044149
Cohan, M. C., Eddelbuettel, A. M. P., Levin, P. A., and Pappu, R. V. (2020). Dissecting the functional contributions of the intrinsically disordered C-terminal tail of Bacillus subtilis FtsZ. J. Mol. Biol. 432, 3205–3221. doi: 10.1016/j.jmb.2020.03.008
Dai, K., and Lutkenhaus, J. (1991). FtsZ is an essential cell division gene in Escherichia coli. J. Bacteriol. 173, 3500–3506. doi: 10.1128/jb.173.11.3500-3506.1991
de Boer, P., Crossley, R., and Rothfield, L. (1992). The essential bacterial cell division protein FtsZ is a GTPase. Nature 359, 254–256. doi: 10.1038/359254a0
Dempwolff, F., Reimold, C., Reth, M., and Graumann, P. L. (2011). Bacillus subtilis MreB orthologs self-organize into filamentous structures underneath the cell membrane in a heterologous cell system. PLoS One 6:e27035. doi: 10.1371/journal.pone.0027035
Dempwolff, F., Schmidt, F. K., Hervás, A. B., Stroh, A., Rösch, T. C., Riese, C. N., et al. (2016). Super resolution fluorescence microscopy and tracking of bacterial flotillin (Reggie) paralogs provide evidence for defined-sized protein microdomains within the bacterial membrane but absence of clusters containing detergent-resistant proteins. PLoS Genet. 12:e1006116. doi: 10.1371/journal.pgen.1006116
Den Blaauwen, T., and Luirink, J. (2019). Checks and balances in bacterial cell division. mBio 10:e00149-19. doi: 10.1128/mBio.00149-19
Du, S., and Lutkenhaus, J. (2019). At the heart of bacterial cytokinesis: the Z ring. Trends Microbiol. 27, 781–791. doi: 10.1016/j.tim.2019.04.011
Encinar, M., Kralicek, A. V., Martos, A., Krupka, M., Cid, S., Alonso, A., et al. (2013). Polymorphism of FtsZ filaments on lipid surfaces: role of monomer orientation. Langmuir 29, 9436–9446. doi: 10.1021/la401673z
Erickson, H. P. (1997). FtsZ, a tubulin homolog, in prokaryote cell division. Trends Cell Biol. 7, 362–367. doi: 10.1016/s0962-8924(97)01108-2
Erickson, H. P. (2017). The discovery of the prokaryotic cytoskeleton: 25th anniversary. Mol. Biol. Cell 28, 357–358. doi: 10.1091/mbc.E16-03-0183
Erickson, H. P., Taylor, D. W., Taylor, K. A., and Bramhill, D. (1996). Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc. Natl. Acad. Sci. U.S.A. 93, 519–523. doi: 10.1073/pnas.93.1.519
Fenton, A. K., and Gerdes, K. (2013). Direct interaction of FtsZ and MreB is required for septum synthesis and cell division in Escherichia coli. EMBO J. 32, 1953–1965. doi: 10.1038/emboj.2013.129
Fujita, J., Harada, R., Maeda, Y., Saito, Y., Mizohata, E., Inoue, T., et al. (2017). Identification of the key interactions in structural transition pathway of FtsZ from Staphylococcus aureus. J. Struct. Biol. 198, 65–73. doi: 10.1016/j.jsb.2017.04.008
García-Soriano, D. A., Heermann, T., Raso, A., Rivas, G., and Schwille, P. (2020). The speed of FtsZ treadmilling is tightly regulated by membrane binding. Sci. Rep. 10:10447. doi: 10.1038/s41598-020-67224-x
Gardner, K. A. J. A., Moore, D. A., and Erickson, H. P. (2013). The C-terminal linker of Escherichia coli FtsZ functions as an intrinsically disordered peptide. Mol. Microbiol. 89, 264–275. doi: 10.1111/mmi.12279
González de Prado Salas, P., Encinar, M., Alonso, A., Vélez, M., and Tarazona, P. (2015). Modeling the interplay between protein and lipid aggregation in supported membranes. Chem. Phys. Lipids 185, 141–152. doi: 10.1016/j.chemphyslip.2014.06.006
Gonzalez de Prado Salas, P., Hörger, I., Martín-García, F., Mendieta, J., Alonso, Á, Encinar, M., et al. (2014). Torsion and curvature of FtsZ filaments. Soft Matter 10, 1977–1986. doi: 10.1039/C3SM52516C
Gowrishankar, K., Ghosh, S., Saha, S., Rumamol, C., Mayor, S., and Rao, M. (2012). Active remodeling of cortical actin regulates spatiotemporal organization of cell surface molecules. Cell 149, 1353–1367. doi: 10.1016/j.cell.2012.05.008
Hörger, I., Velasco, E., Mingorance, J., Rivas, G., Tarazona, P., and Vélez, M. (2008a). Langevin computer simulations of bacterial protein filaments and the force-generating mechanism during cell division. Phys. Rev. E Stat. Nonlinear Soft Matter Phys. 77:011902. doi: 10.1103/PhysRevE.77.011902
Hörger, I., Velasco, E., Rivas, G., Vélez, M., and Tarazona, P. (2008b). FtsZ bacterial cytoskeletal polymers on curved surfaces: the importance of lateral interactions. Biophys. J. 94, L81–L83. doi: 10.1529/biophysj.107.128363
Hsin, J., Gopinathan, A., and Huang, K. C. (2012). Nucleotide-dependent conformations of FtsZ dimers and force generation observed through molecular dynamics simulations. Proc. Natl. Acad. Sci. U.S.A. 109, 9432–9437. doi: 10.1073/pnas.1120761109
Huang, K.-H., Durand-Heredia, J., and Janakiraman, A. (2013). FtsZ ring stability: of bundles, tubules, crosslinks, and curves. J. Bacteriol. 195, 1859–1868. doi: 10.1128/JB.02157-12
Huecas, S., and Andreu, J. M. (2003). Energetics of the cooperative assembly of cell division protein FtsZ and the nucleotide hydrolysis switch. J. Biol. Chem. 278, 46146–46154. doi: 10.1074/jbc.M307128200
Ingerson-Mahar, M., and Gitai, Z. (2012). A growing family: the expanding universe of the bacterial cytoskeleton. FEMS Microbiol. Rev. 36, 256–267. doi: 10.1111/j.1574-6976.2011.00316.x
Kalappurakkal, J. M., Sil, P., and Mayor, S. (2020). Toward a new picture of the living plasma membrane. Protein Sci. 29, 1355–1365. doi: 10.1002/pro.3874
Kalia, R., and Frost, A. (2019). Open and cut: allosteric motion and membrane fission by dynamin superfamily proteins. Mol. Biol. Cell 30:2097. doi: 10.1091/MBC.E16-10-0709
Köster, D. V., Husain, K., Iljazi, E., and Mayor, S. (2016). Actomyosin dynamics drive local membrane component organization in an in vitro active composite layer. Proc. Natl. Acad. Sci. U.S.A. 113, E1645–E1654. doi: 10.1073/pnas.1514030113
Kurita, K., Kato, F., and Shiomi, D. (2020). Alteration of membrane fluidity or phospholipid composition perturbs rotation of MreB complexes in Escherichia coli. Front. Mol. Biosci. 7:582660. doi: 10.3389/fmolb.2020.582660
Lam, M., and Calvo, F. (2019). Regulation of mechanotransduction: emerging roles for septins. Cytoskeleton (Hoboken) 76, 115–122. doi: 10.1002/CM.21485
Lee, J., Cox, J. V., and Ouellette, S. P. (2020). Critical role for the extended N terminus of chlamydial MreB in directing its membrane association and potential interaction with divisome proteins. J. Bacteriol. 202:e00034-20. doi: 10.1128/JB.00034-20
Lin, T. Y., and Weibel, D. B. (2016). Organization and function of anionic phospholipids in bacteria. Appl. Microbiol. Biotechnol. 100, 4255–4267. doi: 10.1007/s00253-016-7468-x
Litschel, T., Kelley, C. F., Holz, D., Adeli Koudehi, M., Vogel, S. K., Burbaum, L., et al. (2021). Reconstitution of contractile actomyosin rings in vesicles. Nat. Commun. 12:2254. doi: 10.1038/s41467-021-22422-7
Longo, M. L. (2018). Preface to emergence of complex behavior in biomembranes. Biochim. Biophys. Acta Biomembr. 1860, 1955–1956. doi: 10.1016/j.bbamem.2018.08.015
Loose, M., and Mitchison, T. J. (2013). The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns. Nat. Cell Biol. 16, 38–46. doi: 10.1038/ncb2885
López-Montero, I., Arriaga, L. R., Monroy, F., Rivas, G., Tarazona, P., and Vélez, M. (2008). High fluidity and soft elasticity of the inner membrane of Escherichia coli revealed by the surface rheology of model langmuir monolayers. Langmuir 24, 4065–4076. doi: 10.1021/la703350s
López-Montero, I., Arriaga, L. R., Rivas, G., Vélez, M., and Monroy, F. (2010). Lipid domains and mechanical plasticity of Escherichia coli lipid monolayers. Chem. Phys. Lipids 163, 56–63. doi: 10.1016/J.CHEMPHYSLIP.2009.10.002
López-Montero, I., López-Navajas, P., Mingorance, J., Vélez, M., Vicente, M., and Monroy, F. (2013). Membrane reconstitution of FtsZ-ZipA complex inside giant spherical vesicles made of E. coli lipids: large membrane dilation and analysis of membrane plasticity. Biochim. Biophys. Acta Biomembr. 1828, 687–698. doi: 10.1016/j.bbamem.2012.11.003
López-Montero, I., Mateos-Gil, P., Sferrazza, M., Navajas, P. L., Rivas, G., Vélez, M., et al. (2012). Active membrane viscoelasticity by the bacterial FtsZ-division protein. Langmuir 28, 4744–4753. doi: 10.1021/la204742b
Löwe, J., and Amos, L. C. (1998). Crystal structure of the bacterial cell-division protein FtsZ. Nature 391, 203–206. doi: 10.1038/34472
Lu, C. L., Reedy, M., and Erickson, H. P. (2000). Straight and curved conformations of FtsZ are regulated by GTP hydrolysis. J. Bacteriol. 182, 164–170. doi: 10.1128/jb.182.1.164-170.2000
Lv, D., Li, J., and Ye, S. (2021). The assembly switch mechanism of FtsZ filament revealed by all-atom molecular dynamics simulations and coarse-grained models. Front. Microbiol. 12:639883. doi: 10.3389/fmicb.2021.639883
Mahone, C. R., and Goley, E. D. (2020). Bacterial cell division at a glance. J. Cell Sci. 133:jcs237057. doi: 10.1242/jcs.237057
Margolin, W. (2005). FtsZ and the division of prokaryotic cells and organelles. Nat. Rev. Mol. Cell Biol. 6, 862–871. doi: 10.1038/nrm1745
Márquez, I. F., Mateos-Gil, P., Shin, J. Y., Lagos, R., Monasterio, O., and Vélez, M. (2017). Mutations on FtsZ lateral helix H3 that disrupt cell viability hamper reorganization of polymers on lipid surfaces. Biochim. Biophys. Acta Biomembr. 1859, 1815–1827. doi: 10.1016/j.bbamem.2017.06.009
Márquez, I., Díaz-Haro, G., and Vélez, M. (2019). Surface orientation and binding strength modulate shape of FtsZ on lipid surfaces. Int. J. Mol. Sci. 20:2545. doi: 10.3390/ijms20102545
Martín-Galiano, A. J., Buey, R. M., Cabezas, M., and Andreu, J. M. (2010). Mapping flexibility and the assembly switch of cell division protein FtsZ by computational and mutational approaches. J. Biol. Chem. 285, 22554–22565. doi: 10.1074/jbc.M110.117127
Mateos-Gil, P., Márquez, I., López-Navajas, P., Jiménez, M., Vicente, M., Mingorance, J., et al. (2012a). FtsZ polymers bound to lipid bilayers through ZipA form dynamic two dimensional networks. Biochim. Biophys. Acta Biomembr. 1818, 806–813. doi: 10.1016/j.bbamem.2011.12.012
Mateos-Gil, P., Paez, A., Hörger, I., Rivas, G., Vicente, M., Tarazona, P., et al. (2012b). Depolymerization dynamics of individual filaments of bacterial cytoskeletal protein FtsZ. Proc. Natl. Acad. Sci. U.S.A. 109, 8133–8138. doi: 10.1073/pnas.1204844109
Mateos-Gil, P., Tarazona, P., and Vélez, M. (2019). Bacterial cell division: modeling FtsZ assembly and force generation from single filament experimental data. FEMS Microbiol. Rev. 039, 73–87. doi: 10.1093/femsre/fuy039
Mateos-Gil, P., Tsortos, A., Velez, M., and Gizeli, E. (2016). Monitoring structural changes in intrinsically disordered proteins using QCM-D: application to the bacterial cell division protein ZipA. Chem. Commun. 52, 6541–6544. doi: 10.1039/C6CC02127A
Matsui, T., Yamane, J., Mogi, N., Yamaguchi, H., Takemoto, H., Yao, M., et al. (2012). Structural reorganization of the bacterial cell-division protein FtsZ from Staphylococcus aureus. Acta Crystallogr. Sect. D 68, 1175–1188. doi: 10.1107/S0907444912022640
McCausland, J. W., Yang, X., Squyres, G. R., Lyu, Z., Bruce, K. E., Lamanna, M. M., et al. (2021). Treadmilling FtsZ polymers drive the directional movement of sPG-synthesis enzymes via a Brownian ratchet mechanism. Nat. Commun. 12:609. doi: 10.1038/s41467-020-20873-y
McQuillen, R., and Xiao, J. (2020). Insights into the structure, function, and dynamics of the bacterial cytokinetic FtsZ-ring. Annu. Rev. Biophys. 49, 309–341. doi: 10.1146/annurev-biophys-121219-081703
Milam, E., Osawa, M., and Erickson, H. P. (2012). Negative-stain electron microscopy of inside-out FtsZ rings reconstituted on artificial membrane tubules show ribbons of protofilaments. Biophys. J. 103, 59–68. doi: 10.1016/j.bpj.2012.05.035
Mileykovskaya, E., and Dowhan, W. (2005). Role of membrane lipids in bacterial division-site selection. Curr. Opin. Microbiol. 8, 135–142. doi: 10.1016/j.mib.2005.02.012
Mileykovskaya, E., and Dowhan, W. (2009). Cardiolipin membrane domains in prokaryotes and eukaryotes. Biochim. Biophys. Acta Biomembr. 1788, 2084–2091. doi: 10.1016/j.bbamem.2009.04.003
Mingorance, J., Rivas, G., Vélez, M., Gómez-Puertas, P., and Vicente, M. (2010). Strong FtsZ is with the force: mechanisms to constrict bacteria. Trends Microbiol. 18, 348–356. doi: 10.1016/j.tim.2010.06.001
Nenninger, A., Mastroianni, G., Robson, A., Lenn, T., Xue, Q., Leake, M. C., et al. (2014). Independent mobility of proteins and lipids in the plasma membrane of Escherichia coli. Mol. Microbiol. 92, 1142–1153. doi: 10.1111/mmi.12619
Nishibori, A., Kusaka, J., Hara, H., Umeda, M., and Matsumoto, K. (2005). Phosphatidylethanolamine domains and localization of phospholipid synthases in Bacillus subtilis membranes. J. Bacteriol. 187, 2163–2174. doi: 10.1128/JB.187.6.2163-2174.2005
Nurse, P., and Marians, K. J. (2013). Purification and characterization of Escherichia coli MreB protein. J. Biol. Chem 288, 3469–3475. doi: 10.1074/jbc.M112.413708
Ohashi, T., Hale, C. A., de Boer, P. A. J., and Erickson, H. P. (2002). Structural evidence that the P/Q Domain of ZipA is an unstructured, flexible tether between the membrane and the C-Terminal FtsZ-binding domain. J. Bacteriol. 184, 4313–4315. doi: 10.1128/jb.184.15.4313-4315.2002
Oliva, M. A., Cordell, S. C., and Lowe, J. (2004). Structural insights into FtsZ protofilament formation. Nat. Struct. Mol. Biol. 11, 1243–1250. doi: 10.1038/nsmb855
Ortiz, C., Natale, P., Cueto, L., and Vicente, M. (2015). The keepers of the ring: regulators of FtsZ assembly. FEMS Microbiol. Rev. 40, 57–67. doi: 10.1093/femsre/fuv040
Osawa, M., and Erickson, H. P. (2005). Probing the domain structure of FtsZ by random truncation and insertion of GFP. Microbiology 151, 4033–4043. doi: 10.1099/mic.0.28219-0
Osawa, M., Anderson, D. E., and Erickson, H. P. (2008). Reconstitution of contractile FtsZ Rings in liposomes. Science 320, 792–794. doi: 10.1126/science.1154520
Osawa, M., Anderson, D. E., and Erickson, H. P. (2009). Curved FtsZ protofilaments generate bending forces on liposome membranes. EMBO J. 28, 3476–3484. doi: 10.1038/emboj.2009.277
Oswald, F., Varadarajan, A., Lill, H., Peterman, E. J. G., and Bollen, Y. J. M. (2016). MreB-dependent organization of the E. coli cytoplasmic membrane controls membrane protein diffusion. Biophys. J. 110, 1139–1149. doi: 10.1016/j.bpj.2016.01.010
Paez, A., Tarazona, P., Mateos-Gil, P., and Vélez, M. (2009). Self-organization of curved living polymers: FtsZ protein filaments. Soft Matter 5, 2625–2637. doi: 10.1039/b902935d
Paez, P., Mateos-Gil Hörger, I., Mingorance, J., Rivas, G., Vicente, M., Vélez, M., et al. (2009). Simple modeling of FtsZ polymers on flat and curved surfaces: correlation with experimental in vitro observations. PMC Biophys. 2:8. doi: 10.1186/1757-5036-2-8
Pannuzzo, M., McDargh, Z. A., and Deserno, M. (2018). The role of scaffold reshaping and disassembly in dynamin driven membrane fission. Elife 7:e39441. doi: 10.7554/ELIFE.39441
Perez, A. J., Cesbron, Y., Shaw, S. L., Bazan Villicana, J., Tsui, H. T., Boersma, M. J., et al. (2019). Movement dynamics of divisome proteins and PBP2x: FtsW in cells of Streptococcus pneumoniae. Proc. Natl. Acad. Sci. U.S.A. 116, 3211–3220. doi: 10.1073/pnas.1816018116
Pilhofer, M., and Jensen, G. J. (2013). The bacterial cytoskeleton: more than twisted filaments. Curr. Opin. Cell Biol. 25, 125–133. doi: 10.1016/j.ceb.2012.10.019
Pla, J., Sanchez, M., Palacios, P., Vicente, M., and Aldea, M. (1991). Preferential cytoplasmic location of FtsZ, a protein essential for Escherichia coli septation. Mol. Microbiol. 5, 1681–1686. doi: 10.1111/j.1365-2958.1991.tb01915.x
Pluhackova, K., and Horner, A. (2021). Native-like membrane models of E. coli polar lipid extract shed light on the importance of lipid composition complexity. BMC Biol. 19:4. doi: 10.1186/s12915-020-00936-8
Popp, D., Iwasa, M., Narita, A., Erickson, H. P., and Maéda, Y. (2009). FtsZ condensates: an in vitro electron microscopy study. Biopolymers 91, 340–350. doi: 10.1002/BIP.21136
Raghunathan, K., and Kenworthy, A. K. (2018). Dynamic pattern generation in cell membranes: Current insights into membrane organization. Biochim. Biophys. Acta Biomembr. 1860, 2018–2031. doi: 10.1016/j.bbamem.2018.05.002
Ramirez-Diaz García-Soriano, D. A., Raso, A., Mücksch, J., Feingold, M., Rivas, G., and Schwille, P. (2018). Treadmilling analysis reveals new insights into dynamic FtsZ ring architecture. PLoS Biol. 16:e2004845. doi: 10.1371/journal.pbio.2004845
Ramirez-Diaz, D. A., Merino-Salomón, A., Meyer, F., Heymann, M., Rivas, G., Bramkamp, M., et al. (2021). FtsZ induces membrane deformations via torsional stress upon GTP hydrolysis. Nat. Commun. 12:3310. doi: 10.1038/s41467-021-23387-3
RayChaudhuri, D., and Park, J. T. (1992). Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature 359, 251–254. doi: 10.1038/359251a0
Redick, S. D., Stricker, J., Briscoe, G., and Erickson, H. P. (2005). Mutants of FtsZ targeting the protofilament interface: effects on cell division and GTPase activity. J. Bacteriol. 187, 2727–2736. doi: 10.1128/JB.187.8.2727-2736.2005
Redpath, G. M. I., Betzler, V. M., Rossatti, P., and Rossy, J. (2020). Membrane heterogeneity controls cellular endocytic trafficking. Front. Cell Dev. Biol. 8:757. doi: 10.3389/fcell.2020.00757
Romberg, L., and Levin, P. A. (2003). Assembly dynamics of the bacterial cell division protein FtsZ: poised at the edge of stability. Annu. Rev. Microbiol. 57, 125–154. doi: 10.1146/annurev.micro.57.012903.074300
Romberg, L., and Mitchison, T. J. (2004). Rate-limiting guanosine 5′-triphosphate hydrolysis during nucleotide turnover by FtsZ, a prokaryotic tubulin homologue involved in bacterial cell division. Biochemistry 43, 282–288. doi: 10.1021/bi035465r
Rowlett, V. W., Mallampalli, V. K. P. S., Karlstaedt, A., Dowhan, W., Taegtmeyer, H., Margolin, W., et al. (2017). Impact of membrane phospholipid alterations in Escherichia coli on cellular function and bacterial stress adaptation. J. Bacteriol. 199:e00849-16. doi: 10.1128/JB.00849-16
Ruiz, N. (2016). Filling holes in peptidoglycan biogenesis of Escherichia coli. Curr. Opin. Microbiol. 34, 1–6. doi: 10.1016/j.mib.2016.07.010
Schumacher, M. A., Ohashi, T., Corbin, L., and Erickson, H. P. (2020). High-resolution crystal structures of Escherichia coli FtsZ bound to GDP and GTP. Acta Crystallogr. Sect. F Struct. Biol. Commun. 76, 94–102. doi: 10.1107/S2053230X20001132
Sezgin, E. I, Mayor, S., and Eggeling, C. (2017). The mystery of membrane organization: composition, regulation and roles of lipid rafts. Nat. Rev. Mol. Cell Biol. 18, 361–374. doi: 10.1038/nrm.2017.16
Shaw, T. R., Ghosh, S., and Veatch, S. L. (2020). Critical phenomena in plasma membrane organization and function. Annu. Rev. Phys. Chem. 72, 541–564. doi: 10.1146/annurev-physchem-090419
Shih, Y.-L., and Rothfield, L. (2006). The bacterial cytoskeleton. Microbiol. Mol. Biol. Rev. 70, 729–754. doi: 10.1128/mmbr.00017-06
Siliakus, M. F., van der Oost, J., and Kengen, S. W. M. (2017). Adaptations of archaeal and bacterial membranes to variations in temperature, pH and pressure. Extremophiles 21, 651–670. doi: 10.1007/s00792-017-0939-x
Squyres, G. R., Holmes, M. J., Barger, S. R., Pennycook, B. R., Ryan, J., Yan, V. T., et al. (2021). Single-molecule imaging reveals that Z-ring condensation is essential for cell division in Bacillus subtilis. Nat. Microbiol. 6, 553–562. doi: 10.1038/s41564-021-00878-z
Steinkühler, J., Sezgin, E., Urbančič, I., Eggeling, C., and Dimova, R. (2019). Mechanical properties of plasma membrane vesicles correlate with lipid order, viscosity and cell density. Commun. Biol. 2:337. doi: 10.1038/s42003-019-0583-3
Strahl, H., and Errington, J. (2017). bacterial membranes: structure, domains, and function. Annu. Rev. Microbiol. 71, 519–538. doi: 10.1146/annurev-micro-102215
Strahl, H., Bürmann, F., and Hamoen, L. W. (2014). The actin homologue MreB organizes the bacterial cell membrane. Nat. Commun. 5:3442. doi: 10.1038/ncomms4442
Stricker, J., Maddox, P., Salmon, E. D., and Erickson, H. P. (2002). Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proc. Natl. Acad. Sci. U.S.A. 99, 3171–3175. doi: 10.1073/pnas.052595099
Varma, A., and Young, K. D. (2009). In Escherichia coli, MreB and FtsZ direct the synthesis of lateral cell wall via independent pathways that require PBP 2. J. Bacteriol. 191, 3526–3533. doi: 10.1128/JB.01812-08
Wagstaff, M., Tsim, M., Oliva, M. A., García-Sanchez, A., Kureisaite-Ciziene, D., Andreu, J. M., et al. (2017). A polymerization-associated structural switch in ftsz that enables treadmilling of model filaments. mBio 8:e00254-17. doi: 10.1128/mBio.00254-17
Willdigg, J. R., and Helmann, J. D. (2021). Mini review: bacterial membrane composition and its modulation in response to stress. Front. Mol. Biosci. 8:634438. doi: 10.3389/fmolb.2021.634438
Xiao, J., and Goley, E. D. (2016). Redefining the roles of the FtsZ-ring in bacterial cytokinesis. Curr. Opin. Microbiol. 34, 90–96. doi: 10.1016/j.mib.2016.08.008
Yang, X., Lyu, Z., Miguel, A., McQuillen, R., Huang, K. C., and Xiao, J. (2017). GTPase activity–coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science 355:744. doi: 10.1126/science.aak9995
Yang, X., McQuillen, R., Lyu, Z., Phillips-Mason, P., De La Cruz, A., McCausland, J. W., et al. (2021). A two-track model for the spatiotemporal coordination of bacterial septal cell wall synthesis revealed by single-molecule imaging of FtsW. Nat. Microbiol. 6, 584–593. doi: 10.1038/s41564-020-00853-0
Yokoyama, H., and Matsui, I. (2020). The lipid raft markers stomatin, prohibitin, flotillin, and HflK/C (SPFH)-domain proteins form an operon with NfeD proteins and function with apolar polyisoprenoid lipids. Crit. Rev. Microbiol. 46, 38–48. doi: 10.1080/1040841X.2020.1716682
Zeno, W. F., Day, K. J., Gordon, V. D., and Stachowiak, J. C. (2020). Principles and applications of biological membrane organization. Annu. Rev. Biophys. 49, 19–39. doi: 10.1146/annurev-biophys-121219-081637
Keywords: bacterial division, bacterial cytoskeletal proteins, FtsZ, lipid membrane, polymerization
Citation: Vélez M (2022) How Does the Spatial Confinement of FtsZ to a Membrane Surface Affect Its Polymerization Properties and Function? Front. Microbiol. 13:757711. doi: 10.3389/fmicb.2022.757711
Received: 12 August 2021; Accepted: 27 January 2022;
Published: 03 May 2022.
Edited by:
Cara C. Boutte, University of Texas at Arlington, United StatesReviewed by:
Ramanujam Srinivasan, National Institute of Science Education and Research (NISER), IndiaCopyright © 2022 Vélez. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marisela Vélez, bWFyaXNlbGEudmVsZXpAaWNwLmNzaWMuZXM=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.