
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 14 March 2022
Sec. Microbiotechnology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.756209
This article is part of the Research Topic Trends in Applications and Improved Production of Biologically Active Metabolites using Microbial Fermentations View all 12 articles
 Zhumei Du1,2
Zhumei Du1,2 Seishi Yamasaki1
Seishi Yamasaki1 Tetsuji Oya1
Tetsuji Oya1 Damiao Nguluve3
Damiao Nguluve3 Denise Euridse3
Denise Euridse3 Benedito Tinga3
Benedito Tinga3 Felicidade Macome3
Felicidade Macome3 Yimin Cai1*
Yimin Cai1*To facilitate the use of woody plant (WP) as a natural biomass resource to address the shortage of feed for ruminants in the tropics, we use PacBio SMRT sequencing to explore the microbial co-occurrence network and silage fermentation of gliricidia and leucaena prepared with Napier grass (NG) and corn stover (CS) in Southern Africa. Based on dry matter, the crude protein contents of WP are as high as 25%. Compared with NG, the addition of CS speed up the dynamic succession of microorganisms in the silage fermentation process from Gram-negative bacteria to Gram-positive bacteria, and promoted Lactiplantibacillus plantarum to become the dominant community and enhanced the metabolic pathways of lactic acid and citric acid, thus improved the fermentation flavour and quality of WP silage. WP can be mixed with CS to make high-quality silage, which can alleviate the shortage of feed and promote local animal production.
To cope with insufficient feed caused by the rapid development of animal husbandry in the tropics, new and nutrient-rich locally available feed resources including natural woody plant (WP) have been investigated (Zhang et al., 2019). In addition to their contributions to a sustainable agricultural and animal ecosystem, tropical legume WP are rich sources of crude protein (CP) and minerals; they also promote biomass and animal production (Luscher et al., 2014).
Gliricidia [Gliricidia sepium (Jacq.) Kunth ex Walp.] is native to tropical, arid forests in Mexico and Central America. Beyond its region of origin, it is also grown in many tropical and subtropical regions, including the Caribbean, northern parts of South America, central Africa, portions of India, and Southeast Asia (Oliveira et al., 2018). Leucaena [Leucaena leucocephala (Lam.) de Wit] is a small, fast-growing mimosa tree native to southern Mexico and northern Central America (Belize and Guatemala), which has been naturalised throughout the tropics (Rengsirikul et al., 2011). The fresh branches and leaves of these WP are used as natural feed for grazing cattle because of their abundant nutrients, low lignin content, and good palatability (Speedy and Pugliese, 1992). Both types of WP have high biomass production capacity; the yield of their leaves can be as high as 20,000 kg-dry weight/ha/year. Under favourable climatic conditions, the yield can reach twice this level (Rajvanshi, 2019). Generally, the WP harvest time is concentrated in the rainy season, which is not conducive to hay preparation; however, silage fermentation is considered an ideal storage method. WP, like legume grass, have a high protein content, but they also have high moisture and low water-soluble carbohydrate (WSC) contents. Therefore, WP must be mixed with local gramineous forage or crop by-products to prepare silage.
Napier grass (NG, Pennisetum purpureum Schumach.) is the major feed resource in cattle production systems in Africa (Du et al., 2020). NG is widely planted in Africa because of its high biomass yield and adaptations for survival under a wide range of soil types, fertility levels, and weather conditions (Negawo et al., 2017). In contrast, corn (Zea mays L.) stover (CS) is the main crop by-product in Africa; dry stover is widely used for ruminant feed, but it is most often discarded in the field to be burned and used as fertiliser (Cai et al., 2020a). NG and CS are used to adjust the moisture content and increase the fermentation substrate level to optimise WP silage fermentation.
PacBio Single-Molecule Real-Time (SMRT) sequencing technology can cover the full read length of DNA fragments and obtain long sequence reads, providing microbial diversity information at the species level. This technology has been used to study the microbial community and fermentation mechanism in silage. However, there is limited information on how to improve the fermentation quality of WP silage in combination with local forage resources in Africa. To develop a high-quality WP preparation technology to alleviate the shortage of feed for ruminants and evaluate silage-related microbial communities, we used SMRT sequencing to assess the microbial community and co-occurrence network related to silage fermentation of WP prepared with NG and CS in Southern Africa.
The experiment was conducted at an experimental farm of the Agricultural Research Institute of Mozambique (IIAM, Matola, Mozambique) on February 26, 2019. NG was harvested from the first cutting at the early flowering stage. Corn was cultivated by a local farm in the same area, and fresh CS was collected after the harvest of corn cobs were harvested during the experiment. Gliricidia and leucaena grow naturally in hilly areas in the region (Matola, Mozambique). Young branches and leaves of both WP at the juvenile stage were obtained from first cuttings in different hilly areas with three replicates.
After harvest, gliricidia, leucaena, NG, and CS were immediately chopped into lengths of approximately 1–2 cm using a chopper (130DX; ARS Co., Ltd., Osaka, Japan). The chopped materials were homogenised according to the experimental design and the silages were prepared with gliricidia, leucaena, NG, CS, and their mixture. The mixing proportions of WP with NG or CS are 10, 25, and 50% based on a fresh matter (FM) basis. These homogenised materials were divided into two fractions. The first fraction was collected as fresh samples and placed into sterilised bags that were kept in an ice box. These were immediately transported to a laboratory to determine their lactic acid buffering capacity (LBC), chemical and protein compositions, energy, macro-minerals, and microbial community. The second fraction was used for making silage. As shown in Figure 1, the silages were prepared with three replicates by using polyethylene drum silos (20 L, Ka-Kosher Co., Ltd., Sinaloa, Mexico). Approximately 16 kg of mixture material was packed into the silo, all of the silos were then compacted to exclude air and storage was at ambient temperature (24–37°C). After 60 days of ensiling, silage samples (approximately 500 g) from each replicate from the top, middle, and bottom layers of the drum silos were taken and mixed thoroughly before taking subsamples. The fresh samples in each treatment were divided into three portions. The first is to store the samples (approximately 50 g) in a freezer at −80°C for future analysis of microbial community; the second is to dry the samples (approximately 200 g) for subsequent analysis of dry matter (DM), chemical and protein compositions, CP loss; and the third is to use 10 g samples to prepare the extract liquid to analyse the microbial population and fermentation quality of the silage.
The microbial population of materials and silages consisted of lactic acid bacteria (LAB), aerobic bacteria, coliform bacteria, yeasts, and moulds, which were measured by the plate counting method described by Cai et al. (1999). Samples (10 g) were blended with 90 mL of sterilised saline solution (8.50 g/L NaCl) and homogenised for 5 min in a Stomacher lab blender (400; Seward, United Kingdom). The resulting suspension was serially diluted from 10–1 to 10–8 with saline solution. A 0.05 mL aliquot from each diluted suspension was spread on agar plates. LAB were counted on Lactobacilli de Man, Rogosa, and Sharpe (MRS) agar medium (Difco Laboratories, Detroit, MI, United States) in an anaerobic box (TEHER Hard Anaerobox, ANX-1; Hirosawa Ltd., Tokyo, Japan). The LAB were identified by the Gram-positive and catalase-negative rods or cocci that produced lactic acid. Aerobic bacteria were grown on nutrient agar medium (Nissui-Seiyaku Co., Ltd., Tokyo, Japan) under aerobic conditions. Coliform bacteria were counted on blue light broth agar medium (Nissui-Seiyaku Co., Ltd., Tokyo, Japan), and their colonies were distinguished from other bacteria by the blue-colour colonies. All bacterial agar plates were incubated at 30°C for 2–3 days. Yeasts and moulds were counted on potato dextrose agar medium (Nissui-Seiyaku Co., Ltd., Tokyo, Japan) with sterilised tartaric acid solution (pH 3.5) at 30°C for 3–5 days of incubation. Yeasts and moulds were distinguished from other bacteria based on colony appearance and cell morphology. All the microbial colonies were reported as viable microbial numbers in colony-forming unit (cfu)/g of FM.
For SMRT sequencing analysis, the triplicate samples (10 g) were mixed with 90 mL of sterilised 0.85% NaCl solution and shaken at the speed of 250 rpm in a 4°C refrigerator for 45 min. The liquid mixture was filtered through a four-layer cheesecloth pre-autoclaved, and then the filtrate was centrifuged at 10,000 rmp for 10 min at 4°C to obtain microbial precipitate. Then the precipitated sample was used for DNA extraction by a DNA kit (D5625-01, Omega, Norcross, GA, United States) as described by Du et al. (2021a). The quality of the extracted DNA was monitored on 1% agarose gel electrophoresis and spectrophotometry (optical density at 260/280 nm ratio). All the DNA samples were stored at −20°C for future analysis.
The forward primer 27F and reverse primer 1492R were used to amplify the full-length 16S rRNA gene by PCR for SMRT sequencing (Du et al., 2021b). The PCR amplicons were purified using Agencourt AMPure XP Beads (Beckman Coulter, Indianapolis, IN, United States), and quantified by Qubit dsDNA HS Assay Kit and Qubit 3.0 fluorometer (Invitrogen, Thermo Fisher Scientific, Waltham, MA, United States). After the individual quantification step, amplicons were pooled in equal amounts. SMRTbell libraries were prepared from the amplified DNA by SMRTbell Express Template Prep Kit 2.0 according to the manufacturer’s instructions (PacBio, Menlo Park, CA, United States). Purified SMRTbell libraries from the pooled and barcoded samples were sequenced on a single PacBio Sequel II 8M cell using the Sequel II Sequencing kit 2.0.
Single-Molecule Real-Time sequencing was performed on a PacBio RS II instrument (Pacific Biosciences, Menlo Park, CA, United States) using P6-C4 chemistry (Mosher et al., 2013). Raw data were processed using the protocol RS_Readsofinsert.1 in SMRT Portal version 2.7 software (PacBio) (Du et al., 2021a). Low-quality sequences were removed using the Quantitative Insights Into Microbial Ecology (QIIME) package (version 1.7) (Caporaso et al., 2010a). Using 100% clustering of sequence identity, the extracted high-quality circular consensus sequence (CCS) were aligned to obtain representative sequences using Python nearest alignment space termination (PyNAST) and clustering and classification inference with U-statistics (UCLUST) analysis (Caporaso et al., 2010b; Edgar, 2010). Unique sequences were classified into operational taxonomic unit (OTU) based on a 99% threshold identity using the UCLUST algorithm (Lozupone and Knight, 2005). Potential chimeric sequences in the representative set of OTU were removed using the Chimera Slayer tool (Haas et al., 2011). The SILVA database version 132 was implemented to classify different OTU and annotate the taxonomic information for each OTU representative sequence based on Bergey’s taxonomy at the genus, family, order, class, and phylum levels, according to classification at an 80% minimum bootstrap threshold (Quast et al., 2013). OTU that occurred only once or twice were discarded. In order to describe the shared and unique microorganisms in all samples following the OTU clustering analyses, Venn diagrams were produced using open-source software package (version 1.2) of R statistical tools (Shade and Handselman, 2012). Since mixture silages showed similar bacterial community, we used gliricidia or leucaena + NG or CS (90 + 10) mixed silages as a representative for Venn diagram analysis. The relative abundances of different bacterial communities at the species level were also analysed for Windows statistical software package. Hierarchical cluster and heat map analyses were performed using R package pvclust (version 3.0.2) (Suzuki and Shimodaira, 2006; Mansfeldt et al., 2014). A microbial network analysis was drawn with Python language tool (Langfelder and Horvath, 2008).
The metabolic potential of the microbial community and the composition of functional genes were assigned to functional annotations of sequenced metagenomic sequences through 16S rRNA marker gene, and were postulated based on the Kyoto Encyclopedia of Genes and Genomes (KEGG). KEGG is utilised for bioinformatics research and education, including data analysis in genomics, metagenomics, metabolomics and other omics studies, modelling and simulation in systems biology, and translational research in drug development. The functional profiles and differences among different groups were analysed with phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt2) (Langille et al., 2013). Due to the same reason as the Venn diagram analysis, that is, two types of woody silage have similar fermentation characteristics, we only use gliricidia material and silage, and gliricidia silage and gliricidia + CS (90 + 10) mixed silage for metabolic pathway analysis.
The pre-ensiled materials and the silage were dried in an oven for 48 h at 65°C until a constant mass was attained. After drying, the samples were ground using a high speed vibrating sample mill (T1-200; for use with two containers with a working capacity of 50 mL; CMT Japan Co., Ltd., Yokohama, Japan). According to the methods of the AOAC International (2000), samples were analysed for DM (method 930.15), ash (method 923.03), CP (method 990.03), and ether extract (EE, method 920.39). The organic matter (OM) content was calculated as the weight lost after ashing. Neutral detergent fibre (NDF), acid detergent fibre (ADF), and acid detergent lignin (ADL) were determined as described by Van Soest et al. (1991). The NDF and ADF were expressed exclusive of residual ash. Heat stable amylase and sodium sulphite were used for the NDF procedure. ADL was analysed by solubilisation of cellulose with sulphuric acid. LBC was determined by titrating with NaOH from pH 4.0 to 6.0 (mmol/kg DM) after first reducing the pH to below 4.0 using HCl, as described by Muck et al. (1991). The WSC including glucose, sucrose, and fructose were determined by high performance liquid chromatography (HPLC, LC-2000 plus; JASCO Co., Tokyo, Japan) as described by Cai (2004). The analytical conditions were as follows: column, SC 1011 (8.0 mm × 30 cm, Shoko, Tokyo, Japan); oven temperature, 80°C; mobile phase, water; flow velocity, 1.0 mL/min; and detector, Jasco RI-1530.
Binding protein, effective protein, and neutral detergent insoluble protein (NDIP, indicates the CP contents of NDF residue) were analysed by the method of Cai et al. (2020b). CP loss (%) = (CP content of fresh material–CP content of silage)/CP content of fresh material × 100%. Gross energy (GE) was determined using an automatic oxygen bomb calorimeter (CA-4PJ; Shimadzu, Kyoto, Japan) (Gao et al., 2019). Digestible energy (DE) and metabolisable energy (ME) concentrations of the test ingredients were calculated using a difference procedure (Baker and Stein, 2009; Li et al., 2018). Herein, CP, EE, NDF, ADF, and ash are expressed as a percentage of DM, and the units for GE, DE, and ME are MJ/kg of DM. The macro-minerals contents of samples, including calcium, phosphorous, magnesium, and potassium, were measured using a wet-digestion method, and then analysed with an atomic absorption spectrophotometer (PerkinElmer, LAMBDA 1050, Yokohama, Japan) as described by Pequerul et al. (1993).
The terminal fermentation products of the silages were analysed using the method of cold-water extracts, as described by Cai (2004). The remaining wet silage sample (10 g) was homogenised in 90 mL of sterilised distilled water and kept in a refrigerator at 4°C for 24 h. Thereafter, the extract samples were filtered through quantitative ashless filter paper (circle size: 5A, 110 mm; Advantec Co., Ltd., Tokyo, Japan). The filtrate was used to determine pH, ammonia nitrogen (NH3-N), and organic acid (lactic acid, acetic acid, propionic acid, and butyric acid) contents. The pH was measured using a glass electrode pH meter (D-71; Horiba Co., Ltd., Kyoto, Japan). The NH3-N contents of silages were determined by steam distillation of the filtrates, as described by Cai (2004) using Kjeltec auto distillation (2200; Foss Tecator, Höganäs, Sweden). The silage filtrates were shaken with cation exchange resin (Amberlite, IR 120B H AG; Organo Corporation, Tokyo, Japan) and centrifuged at 6,500 × g and 4°C for 5 min. The supernatants were passed through a 0.45 mm filter under pressure, and the filtrates were then injected into an HPLC system (LC-2000 plus; JASCO Co., Tokyo, Japan) to determine organic acid contents in accordance with the methods described by Cai (2004). The HPLC system was equipped with a Shodex RSpak KC-811 column (8.0 mm × 30 cm; Showa Denko K. K., Tokyo, Japan) at an oven temperature of 60°C. The detector was a Jasco UV-2070 used at 450 nm with an eluent of 3 mM HClO4 and reagent of 0.2 mM bromothymol blue + 8 mM Na2HPO4 + 2 mM NaOH. The flow rate was 1.0 mL/min.
ANOVA was performed using the general linear model (GLM) procedure of Statistical Package for the Social Sciences (SPSS Version 19.0, SPSS Inc., Chicago, IL, United States) to examine the differences between samples, and significance was declared at P < 0.05. The LBC, microbial population, chemical and protein compositions, energy, and macro-minerals contents of samples were subjected to one-way ANOVA. Tukey’s honest significant difference (HSD) test was employed for different sample means (Steel and Torrie, 1980).
The hierarchical cluster and heat map analyses showed the correlation analyses of the bacterial community with lactic acid, LAB, pH, and NH3-N at species level, respectively. LAB, organic acid, pH, and NH3-N information are displayed horizontally, respectively, and the bacterial community information is displayed vertically. The corresponding value of the middle heat map is the Spearman correlation coefficient r, which ranges between −1 and 1, r < 0 indicates a negative correlation (blue), r > 0 indicates a positive correlation (red), and “*,” “**,” and “***” represent P < 0.05, P < 0.01, and P < 0.001, respectively. The network analysis showed the correlation networks among microorganisms at species levels. The circle represents the microorganism species, the circle size represents the average abundance of the species, the line represents the correlation between the two species, the thickness of the line represents the strength of the correlation, and the colour of the line: orange represents positive correlation, green means negative correlation.
The pH, LBC, microbial population, chemical and protein compositions, energy, and macro-mineral values of WP, NG, and CS before ensiling are shown in Table 1. The WSC content was lower (P < 0.001) and the LBC was higher (P < 0.001) in WP than in both forages. The WSC content was significantly lower (P < 0.001) in NG than in CS, but LBC showed the opposite pattern (P < 0.01). The LAB count was higher (P < 0.001) in CS than in WP and NG. Aerobic bacteria were predominant in all samples, and their counts were higher (all P < 0.05) than the counts of other microorganisms. The yeast count was lower (P < 0.05) in WP than in forages, whereas coliform bacteria showed the opposite pattern. The mould counts of all samples were similar at 3 lg cfu/g of FM. The DM contents in WP, NG, and CS ranged from 21.18 to 38.94%. The CP contents in gliricidia and leucaena were >25.91% of the contents in DM; the CP contents were higher (P < 0.001) in gliricidia and leucaena than in NG and CS. The EE and ADL contents were higher (P < 0.001) in WP than in NG and CS, but the NDF and ADF contents showed the opposite pattern. The effective protein, GE, DE, ME, calcium, magnesium, and potassium contents were higher (all P < 0.001) in gliricidia and leucaena than in NG and CS, whereas the binding protein and NDIP contents showed the opposite pattern. The phosphorous contents were similar in all samples.
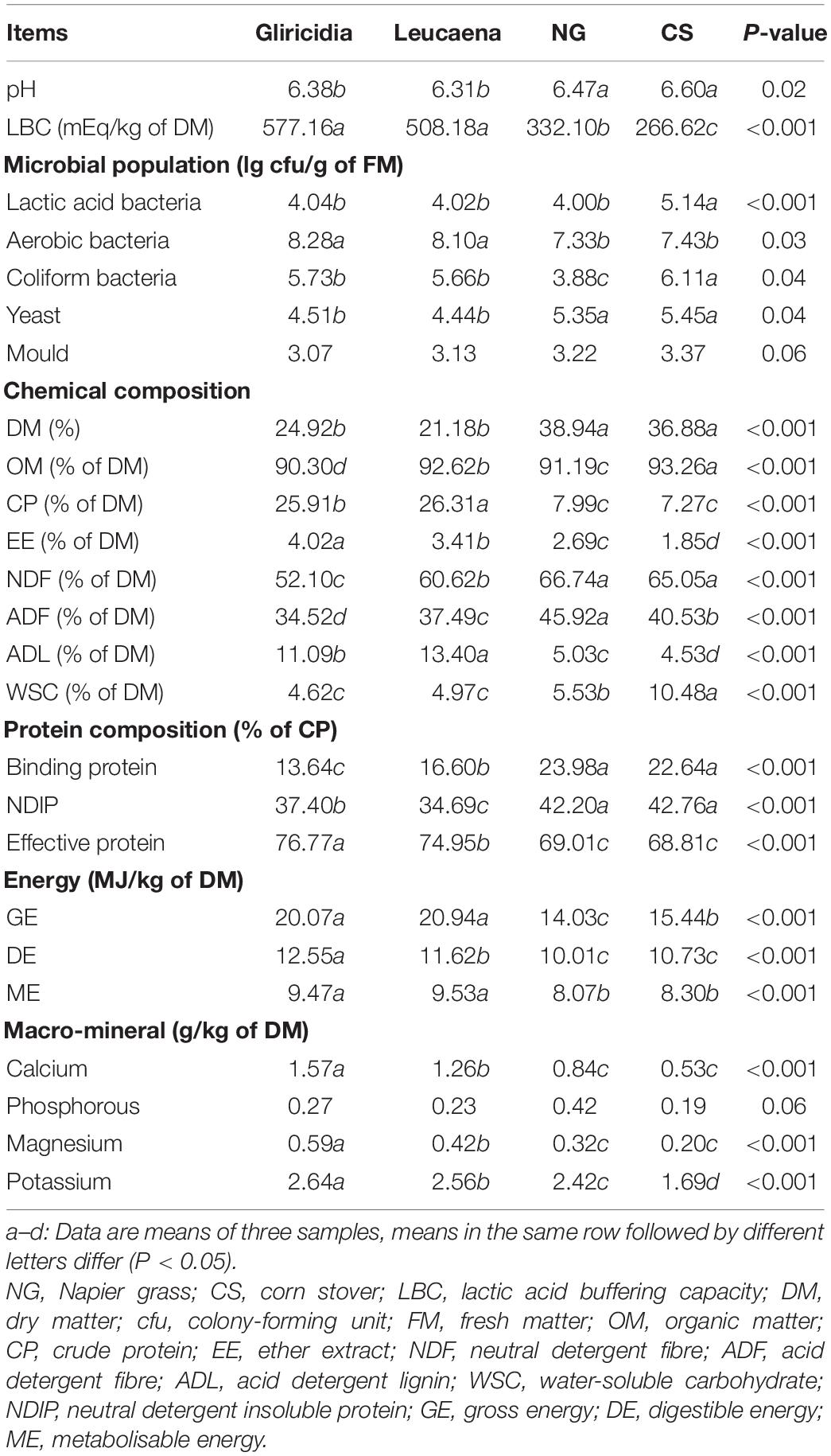
Table 1. pH, LBC, microbial population, chemical and protein compositions, energy, and macro-mineral of material.
The chemical and protein compositions and CP loss values of WP, NG, CS, and their mixture silages are shown in Table 2. For the WP mixture silages, as the proportion of NG or CS increased, the OM, CP, EE, and ADL contents decreased; conversely, the DM, NDF, and ADF contents and CP loss increased. The WP, NG, and their mixture silages had higher (all P < 0.05) CP losses than did the CS and its mixture silages.
The microbial population and fermentation quality of WP, NG, CS, and their mixture silages are shown in Table 3. Moulds were below the detectable level (<2 lg cfu/g of FM). LAB (7 lg cfu/g of FM) was predominant in CS and its mixture silages prepared with WP; LAB values were higher (all P < 0.05) in these samples than in other silages. Aerobic bacteria and coliform bacteria were present in WP, NG, and their mixture silages at 4–7 lg cfu/g of FM, but they were below detectable levels in CS and its mixture silages. Compared with NG, CS and its mixture silages had better fermentation quality with a lower (P < 0.001) pH and lower (both P < 0.001) acetic acid and NH3-N contents, as well as a higher (P < 0.05) lactic acid content. Propionic acid and butyric acid were produced in WP, NG, and their mixture silages, but they were below detectable levels in CS and its mixture silages.
A Venn diagram of the OTU at 97% sequence identity in WP prepared with NG and CS before and after ensiling is shown in Figure 2. The dominant microbiome of the gliricidia, NG, and CS materials (Figure 2A) contained 138 shared OTU, as well as 21, 48, and 100 unique OTU, respectively. The leucaena, NG, and CS materials (Figure 2B) contained 152 shared OTU and 25, 34, and 81 unique OTU, respectively. The WP silages shared 12 and 11 OTU with the gliricidia (Figure 2C) and Leucaena (Figure 2D) silages prepared with NG and CS, respectively. As seen in Figure 2C, the unique OTU ranged from 2 to 93 in CS, NG, gliricidia, and their mixture silages. As seen in Figure 2D, the unique OTU ranged from 2 to 50 in CS, NG, leucaena, and their mixture silages.
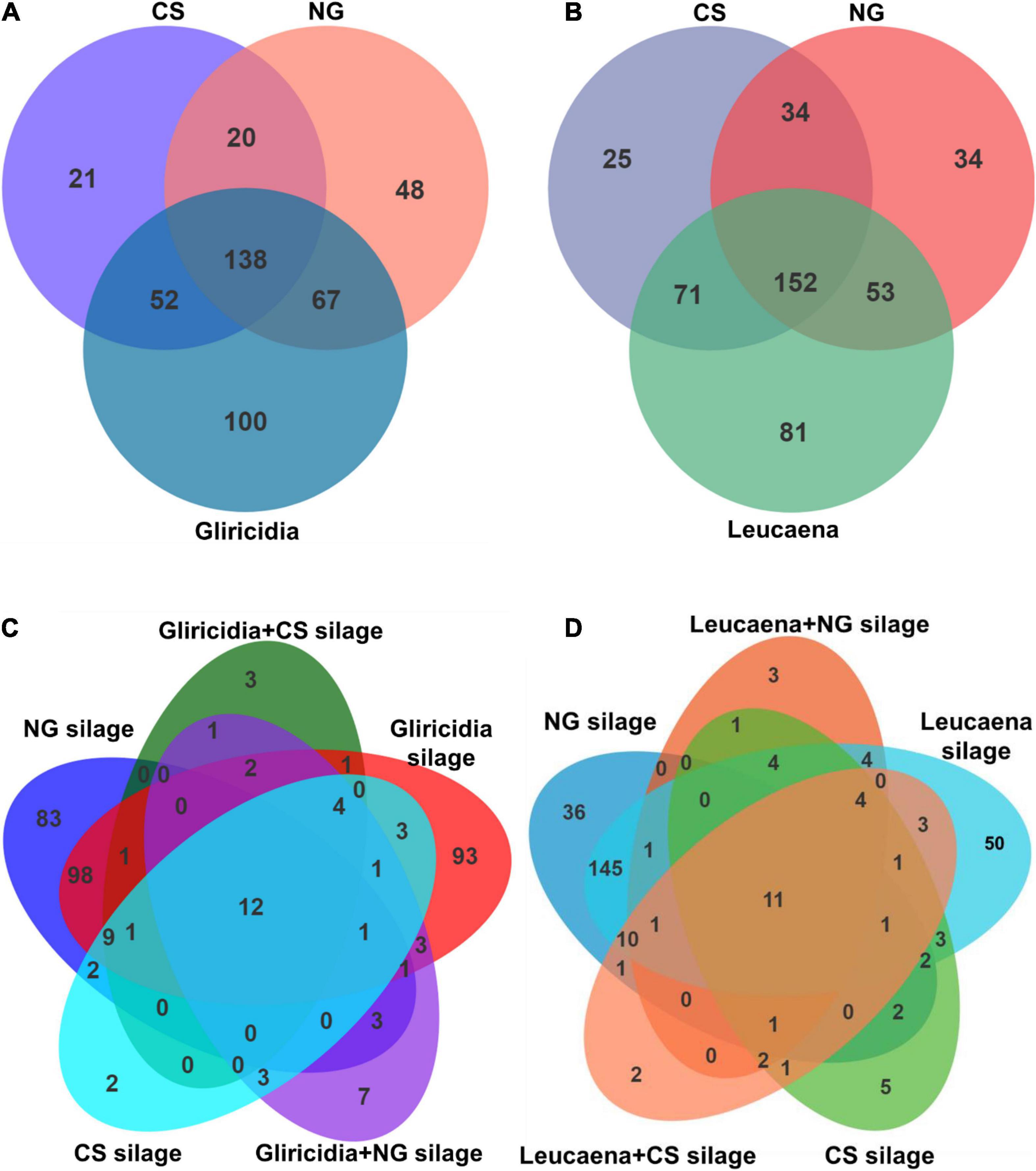
Figure 2. Venn diagram depicting unique or shared bacterial OTU (97% sequence identity) in gliricidia, leucaena, NG, CS materials and their mixture silages. (A) The number of OTU shared by gliricidia, NG, and CS before ensiling. (B) The number of OTU shared by leucaena, NG, and CS before ensiling. (C) The number of OTU shared by gliricidia, NG, and CS and their mixture silages. (D) The number of OTU shared by leucaena, NG, and CS and their mixture silages. OTU, operational taxonomic unit; NG, Napier grass; CS, corn stover. The mixing ratio of gliricidia or leucaena were 90% of the mixture silage based on a fresh matter.
Relative bacterial abundances at the species level in the WP, NG, and CS materials, and their mixture silages are shown in Figure 3. The dominant species in the WP material was Pantoea agglomerans. The dominant species in the NG material were Microbacterium trichothecenolyticum and P. agglomerans; the dominant species in CS material were M. trichothecenolyticum, Streptococcus sanguinis, and Methylobacterium adhaesivum. Lactiplantibacillus plantarum was present at a low level in all materials. After ensiling, L. plantarum was the predominant species in all silages, and its counts were higher (both P < 0.05) in NG and CS silages than in WP silage; CS silage had the highest number. Compared with the WP silages, the addition of NG and CS significantly increased (both P < 0.05) the proportion of L. plantarum in the mixture silages, with the highest relative abundances in the WP and CS mixture silages.
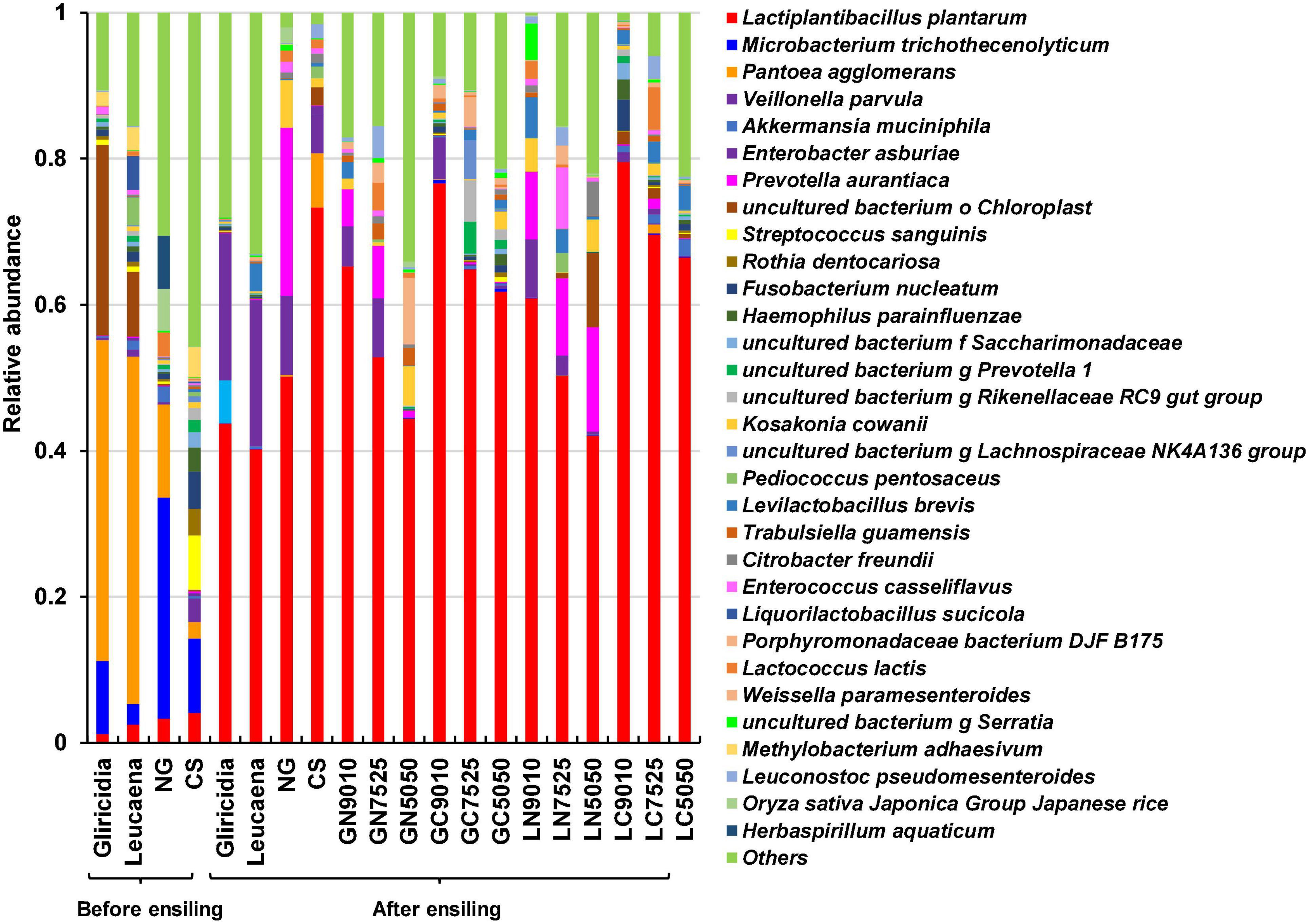
Figure 3. The relative abundances of the bacterial communities at the species levels in the gliricidia, Leucaena, NG, and CS materials, and their mixture silages. NG, Napier grass; CS, corn stover; GN, gliricidia + NG; GC, gliricidia + CS; LN, leucaena + NG; LC, leucaena + CS; 9010, 90 + 10%; 8020, 80 + 20%; 7030, 70 + 30%.
Correlation heatmap and hierarchical cluster analysis of the bacterial community at the species level and terminal fermentation products are shown in Figure 4. Lactic acid was positively correlated with Lactobacillus and Weissella species, such as L. plantarum, Levilactobacillus brevis, Limosilactobacillus fermentum, and Weissella paramesenteroides. LAB was positively correlated with L. plantarum, but negatively correlated with P. agglomerans. pH showed the opposite patterns. NH3-N content was negatively correlated with L. plantarum and positively correlated with Akkermansia muciniphila.
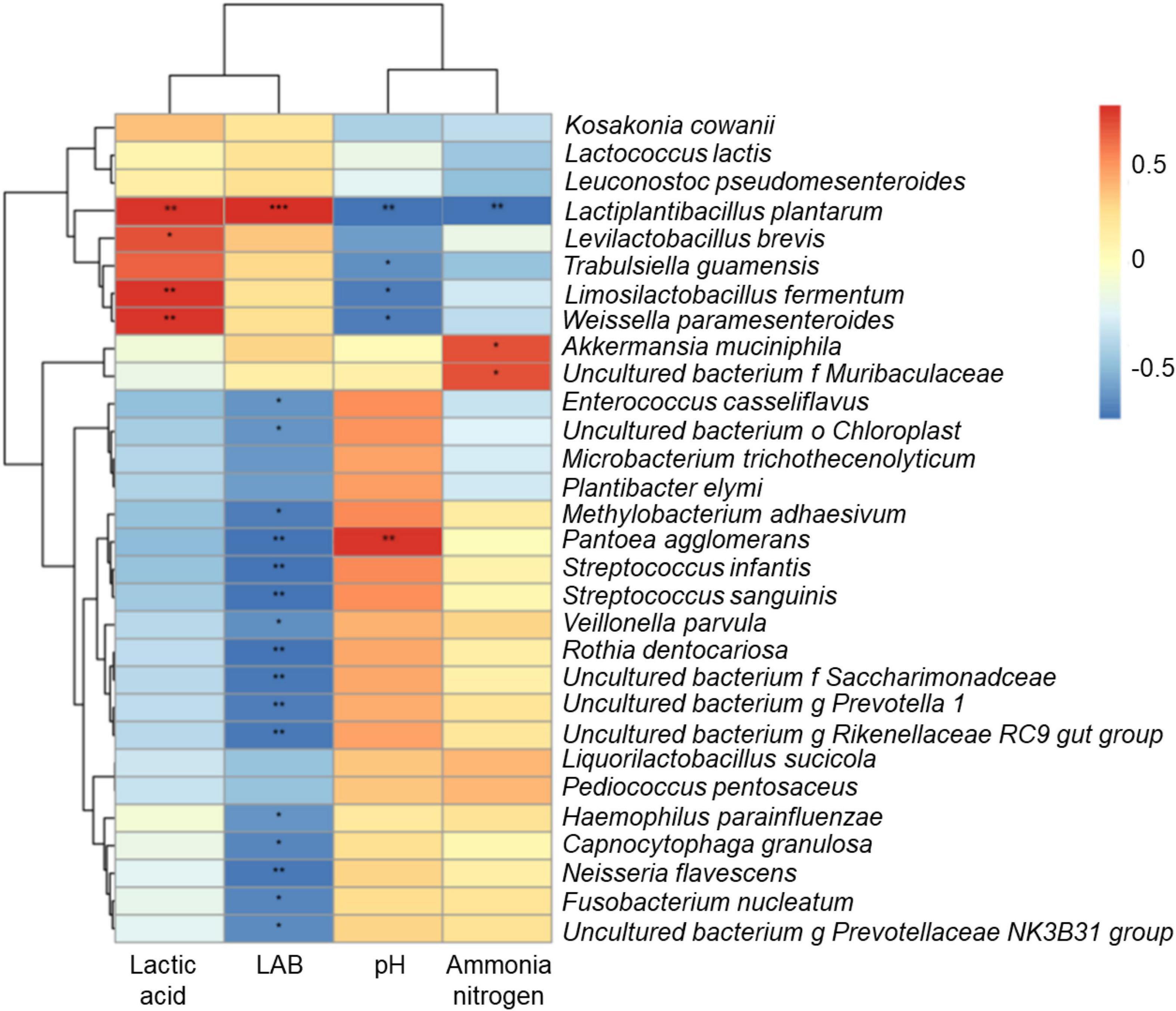
Figure 4. Correlation heatmap and hierarchical cluster analysis between bacterial community and terminal fermentation products at species level. LAB, lactic acid bacteria; NH3-N, ammonia nitrogen. *P < 0.05; **P < 0.01; ***P < 0.001.
The correlation networks among all microorganisms at the species level are shown in Figure 5. The most important species appeared to be L. plantarum, which was positively correlated with M. trichothecenolyticum and negatively correlated with both Kosakonia cowanii and Capnocytophaga granulosa. The second most important species was P. agglomerans, which was positively correlated with Porphyromonas pasteri and negatively correlated with M. adhaesivum.
The KEGG pathways enriched in gliricidia material and silage are shown in Figure 6A; the KEGG pathways enriched in gliricidia silage and gliricidia + CS mixed silage are shown in Figure 6B. Figure 6A shows that the carbohydrate metabolism, amino acid metabolism, and energy metabolism pathways were the predominant metabolic categories. Among them, the largest proportion of the metabolism pathway involved carbohydrate metabolism. The proportion of the carbohydrate metabolism pathway was higher (P < 0.01), whereas the proportions of amino acid and energy metabolism pathways were lower (both P < 0.01) in gliricidia silage than in gliricidia material. Figure 6B shows that the biosynthesis of secondary metabolites, biosynthesis of antibiotics, and microbial metabolism in diverse environments were the predominant metabolic categories in gliricidia silage and mixture silages. Compared with the gliricidia silage, the mixture silage had lower (P < 0.01) proportions of the above three metabolic pathways and a higher (P < 0.01) proportion of the tricarboxylic acid (TCA) cycle.
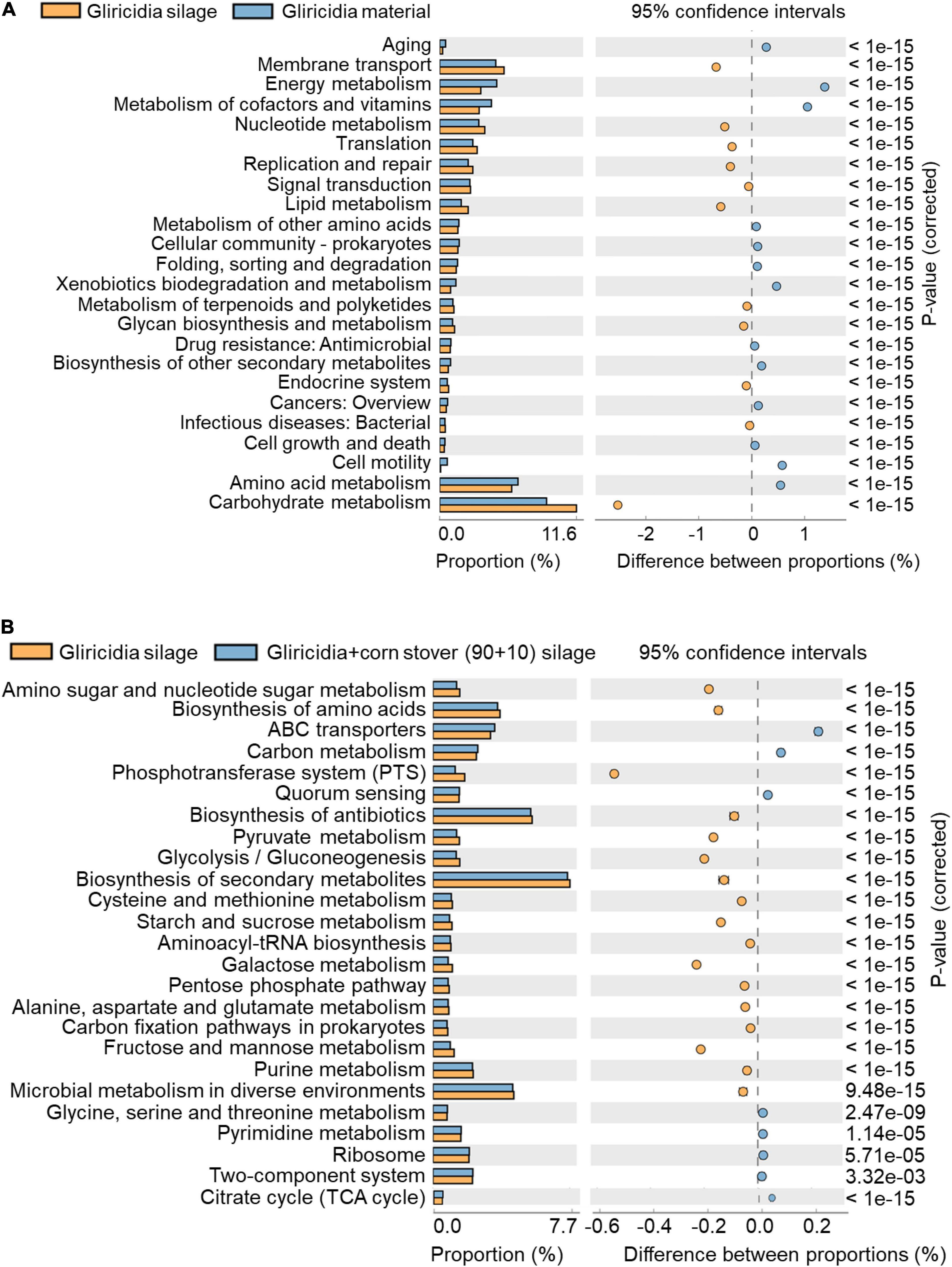
Figure 6. KEGG metabolic pathways in gliricidia material and silage (A), and gliricidia silage and gliricidia + CS (90 + 10) mixed silage (B). KEGG, Kyoto Encyclopedia of Genes and Genomes.
The desire to reduce feed cost and ensure ruminant production system profitability has prompted the development of WP resources as an alternative source of protein for sustainable animal production. WP are recommended as ruminant feed to improve forage quality, reduce the need to purchase concentrate, and decrease the cost of feeding animals. Generally, animals prefer forage with low fibre content and high NDF digestibility, as well as a high CP content (Egan, 1980), because this type of forage is associated with high animal intake and efficient assimilation of energy, minerals, and vitamins. In this study, as shown in Table 1, the WP material was rich in CP (>23.91% of DM) and protein (74.95% of CP); however, it had low fibre, binding protein, and NDIP contents. In addition, the WP material also had high contents of energy and macro-minerals. Therefore, the WP material contains high levels of digestible feed ingredients for use in livestock feed as a source of protein. It is difficult to produce good-quality silage using WP because of their high moisture content (Du et al., 2021b). In addition, the WSC, LAB, and LBC contents of materials affect silage fermentation (Cai et al., 2020a). The moisture content of WP material was >76%, possibly leading to poor fermentation. When NG and CS were added to WP, the moisture content was <70%, within the range (60–70%) suitable for fermentation (McDonald et al., 1991; Du et al., 2021b). A suitable moisture content inhibits the growth of harmful microorganisms such as Clostridium species; it also prevents butyric acid fermentation and protein decomposition (Cai et al., 2020b; Du et al., 2021a). For silage fermentation, other important factors are WSC > 5% of DM, LAB > 5 lg cfu/g of FM, and low LBC (Cai et al., 1999). In this study, the LAB count and WSC content in WP material were below these levels. This indicates that a small number of LAB is associated with poor silage fermentation, thus reducing fermentation quality. In addition, WP material has a high LBC value similar to the value of legume alfalfa, which hampers silage production (Xu et al., 2019). During ensiling, these cations neutralise the organic acids formed by silage fermentation, thus preventing pH reduction. Therefore, preparation of high-quality silage by direct ensiling of WP is problematic. NG and CS are locally available feed resources with good fermentation characteristics, such as low LBC count, high WSC, and optimal DM content. The addition of NG and CS can overcome some of the difficulties in WP silage fermentation.
Woody plant can usually be harvested multiple times during the rainy season. If it is prepared and stored in the manner used for hay, some nutrients will be lost during drying and storage (Zhang et al., 2019). After drying, the palatability and nutritional value of the leaves for livestock are greatly reduced (Du et al., 2021b). Silage can preserve the nutrients in fresh forage and ensure the supply of animal feed throughout the year (Yan et al., 2019). LAB have important roles in the preparation of silage, converting WSC into lactic acid, lowering the pH value, and inhibiting the growth of harmful bacteria; thus, they produce high-quality silage (Cai et al., 2020a). In this study (Tables 2, 3), particularly in the gliricidia or leucaena + CS (90 + 10) mixed silages, the LAB population significantly increased after 60 days of ensiling, inhibiting the growth of harmful bacteria. Adjustment of the moisture content and enhancement of fermentation substrate levels are methods that improve fermentation quality. Therefore, when using WP to prepare silage, the recommended grass or crop stover proportion is approximately 10%; this should be adjusted to 60–70% depending on moisture content. After harvest, fresh CS is generally rich in LAB and WSC, and the addition of CS can improve the fermentation quality of WP mixture silage. However, NG itself does not have good fermentation characteristics because of its low WSC content and low LAB count. The NG silage and WP + NG mixed silage did not show improved fermentation quality. Therefore, CS is recommended for the preparation of mixture silage with WP in Africa.
Silage fermentation is a dynamic process of microbial community succession and shifts in metabolite levels. Using traditional culture methods, only some microorganisms in silage can be isolated and identified; this hampers the analysis of fermentation mechanisms. Next-generation sequencing (NGS) technology has been used to evaluate silage microbial community, abundance, and diversity. It is a high-throughput approach that can detect a large number of microbial taxa. However, microorganisms can be identified only to the genus level, and the complete microbial community cannot be profiled based on partial 16S rRNA gene sequences (Bokulich and Mills, 2012; Mayo et al., 2014). The SMRT sequencing technology used in this study enables accurate evaluation of the microbial community and diversity within silage (Xu et al., 2019). As shown in Figure 1, compared with the NG and CS materials, the greater OTU numbers in gliricidia and leucaena may reflect epiphytic microorganisms on the surface, because of the high moisture content and rich nutrition of fresh branches and leaves. After ensiling, natural LAB create an acidic environment to inhibit core microorganisms, reducing their abundance and forming a unique microbial community in the silages.
Microbial community structure and function affect silage fermentation (McEniry et al., 2010; Guan et al., 2018; Cai et al., 2020b). As shown in Figure 2, the Gram-negative bacterium P. agglomerans was the major species in WP materials. This plant pathogen is often present in forage and is considered harmful in silage (Du et al., 2021a). The dominant species in the NG and CS materials was M. trichothecenolyticum, a Gram-positive and aerobic or weakly anaerobic non-spore-producing and acid-fast species. In contrast, the abundance of L. plantarum was low in all materials, indicating that WP were similar to forage and crop by-products; WP typically have a low abundance of LAB but a high abundance of undesirable bacteria before ensiling (Tian et al., 2020; Du et al., 2021b). Therefore, measures to control the microbial community are needed to improve silage fermentation.
Silage fermentation is conducted in an anaerobic, closed, solid-fermentation system (Cai et al., 1998). A higher relative abundance of LAB is frequently observed in this system, and the different microbial community structure have differential effects on the silage fermentation (Queiroz et al., 2013; Cai et al., 2020b). In this study (Figure 3), when WP and CS were mixed to prepare WP silage, the LAB can quickly become dominant bacteria and complete the succession process from Gram-negative bacteria to Gram-positive bacteria, indicating that mixture silage can effectively improve the fermentation quality by improving the microbial community. However, there was not great difference between the microbial communities in the NG silage and WP + NG mixed silages. A reasonable explanation for this phenomenon is that, compared with NG, CS contains more lactic acid bacteria, such as L. plantarum, which have stronger fermentation ability and acid tolerance. In the process of mixture silage fermentation, these epiphytic LAB existing on CS can quickly respond to the double pressure of anaerobic and acidic silage environment, carry out lactic acid fermentation and improve the fermentation quality of silage (Wang et al., 2019; Du et al., 2021a).
Microorganisms affect silage fermentation through a series of metabolites. For example, Lactobacillus species use WSC to produce lactic acid, while Enterobacter species ferment lactic acid into products such as acetic acid (McDonald et al., 1991), thereby influencing silage fermentation quality and aroma. In addition, these metabolites affect the microbial community structure. For example, lactic acid and acetic acid lower the pH and inhibit the growth of aerobic bacteria and moulds, thus affecting the aerobic stability of silage. Accordingly, silage fermentation is modulated by microbial community structure and metabolites, which interact during fermentation.
In this study, the production of lactic acid was attributed to Lactobacillus species. Lactobacilli are homofermentative type LAB that inhibit the activities of harmful microorganisms by rapid acidification in the later stage of silage fermentation (Cai, 2004; Mu et al., 2020). In the Figure 4, L. plantarum, L. brevis, L. fermentum, and W. paramesenteroides grew well and produced lactic acid in silage, inhibiting the growth of P. agglomerans by reducing the pH. The A. muciniphila competed for nutrients with L. plantarum during ensiling, while degrading proteins to produce NH3-N. However, silage fermentation by L. plantarum inhibited the growth of these bacteria and the production of NH3-N.
In the Figure 5, during silage fermentation, L. plantarum was positively correlated with M. trichothecenolyticum and negatively correlated with both K. cowanii and C. granulosa. The nature of this relationship is unclear; however, M. trichothecenolyticum is dependent on respiratory metabolism that produces acid from glucose and other sugars during the ensiling process. Therefore, L. plantarum inhibits the growth of other Gram-negative bacteria; it may promote the growth of M. trichothecenolyticum. The abundances of K. cowanii and C. granulosa were negatively correlated with the abundance of L. plantarum. These bacteria are often distributed on the surface of forage and compete with LAB for nutrients in the early stage of silage fermentation. As silage fermentation progresses, they are inhibited by the lactic acid from LAB. The P. agglomerans and P. pasteri are frequently present in silage fermentation; they produce n-butyric acid, acetic acid, propionic acid, isobutyric acid, isovaleric acid, and phenylacetic acid, thus reducing fermentation quality. Therefore, the two bacteria are positively correlated with each other, but both are negatively correlated with L. plantarum.
In the KEGG analysis, carbohydrate metabolism was the most important metabolic category in gliricidia materials and silage. Pyruvate metabolism, glycolysis, and butyrate metabolism were predominant components of the carbohydrate metabolic pathways, consistent with the main silage metabolism pathway reported by Du et al. (2021b). In this study (Figure 6), silage had a higher proportion of carbohydrate metabolism pathways than did the corresponding material, reflecting the community dynamics and metabolic activities of the major LAB. Amino acid metabolism can be oxidised into carbon dioxide and water through the tricarboxylic acid cycle, releasing energy (Halperin and Jungas, 1983). Therefore, fresh materials have higher rates of amino acid metabolism and energy metabolism pathways than do silages.
The roles of secondary metabolites and antibiotics in silage are unclear. Secondary metabolites and antibiotics may be produced by microorganisms—including bacteria, fungi, and actinomycetes—during silage fermentation, which interfere with the development of other cells. LAB grew more vigorously in mixture silage than in gliricidia silage, inhibiting the growth and metabolic activities of other microorganisms while suppressing the biosynthesis of secondary metabolites and antibiotics. However, the proportion of citric acid cycle was higher in the gliricidia + CS mixed silage than in gliricidia silage. In the citric acid cycle, the condensation reaction between oxaloacetate and the acetyl group of acetyl-CoA produces citric acid. Citric acid is used as a food acidifier, flavouring agent, and chelating agent. Citric acid-mediated adjustment of pH value can improve the performance of antioxidants, inhibit the activity of enzymes, and extend the shelf life of food (Ke et al., 2017). The WP and CS mixture silage has an increased citric acid content, which will improve its flavour and quality, facilitating long-term storage.
To improve fermentation quality and develop a preparation method for nutrient-rich WP, we used SMRT sequencing technology to study the microbial community and fermentation characteristics of WP silage following the addition of NG and CS in Southern Africa. WP have high CP, energy, and mineral contents; they offer ideal feed for ruminants. However, the preparation of good-quality WP silage is hampered by the low WSC content, low LAB count, and high LBC in WP. In PM silage prepared with NG and CS, the dominant microbial community shifted from Gram-negative bacteria to LAB, promoting lactic acid fermentation. L. plantarum was predominant, which improved silage flavour and quality. CS has good silage characteristics, helping to overcome the influence of unfavourable factors in silage fermentation. Addition of CS improves the microbial community structure and metabolic pathways of silage; thus, the combination of CS and WP is suitable for high-quality silage. WP offer a potential source of high-protein feed for ruminants, which can alleviate the feed shortage in Southern Africa.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
ZD and YC conceived and designed the study. ZD, SY, and TO carried out the experiments. ZD and DN carried out the data analysis. ZD, YC, DE, BT, and FM wrote and revised the manuscript. All authors read and approved the manuscript.
This work was supported by the project “Development of Sustainable Technologies to Increase Agricultural Productivity and Improve Food Security in Africa (Food Security in Africa),” Japan International Research Center for Agricultural Sciences (JIRCAS), Japan, and Instituto de Investigação Agrária de Moçambique (IIAM), Mozambique.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Japan International Research Center for Agricultural Sciences (JIRCAS, Japan) and Instituto de Investigação Agrária de Moçambique (IIAM, Mozambique). Plant samples imported from Mozambique to Japan were permitted by the Minister of Agriculture, Forestry and Fisheries of Japan under Act on Domestic Animal Infectious Diseases Control.
WP, woody plant; PM, paper mulberry; LBC, lactic acid buffer capacity; WSC, water-soluble carbohydrate; DM, dry matter; FM, fresh matter; OM, organic matter; CP, crude protein; EE, ether extract; NDF, neutral detergent fibre; ADF, acid detergent fibre; ADL, acid detergent lignin; NH3-N, ammonia nitrogen; LAB, lactic acid bacteria; OTU, operational taxonomic unit; cfu, colony-forming unit; NG, Napier grass; CS, corn stover.
AOAC International (2000). Official Methods of Analysis, 17th Edn. Arlington, VA: Association of Official Analytical Chemists.
Baker, K. M., and Stein, H. H. (2009). Amino acid digestibility and concentration of digestible and metabolizable energy in soybean meal produced from conventional, high-protein, or low-oligosaccharide varieties of soybeans and fed to growing pigs. J. Anim. Sci. 87, 2282–2290. doi: 10.2527/jas.2008-1414
Bokulich, N. A., and Mills, D. A. (2012). Next-generation approaches to the microbial ecology of food fermentations. BMB Rep. 45, 377–389. doi: 10.5483/bmbrep.2012.45.7.148
Cai, Y. (2004). “Analysis method for silage,” in Field and Laboratory Methods for Grassland Science, ed. Japanese Society of Grassland Science (Tokyo: Tosho Printing Co. Ltd), 279–282.
Cai, Y., Benno, Y., Ogawa, M., and Kumai, S. (1999). Effect of applying lactic acid bacteria isolated from forage crops on fermentation characteristics and aerobic deterioration of silage. J. Dairy Sci. 82, 520–526.
Cai, Y., Benno, Y., Ogawa, M., Ohmomo, S., Kumai, S., and Nakase, T. (1998). Influence of Lactobacillus spp. from an inoculant and of Weissella and Leuconostoc spp. from forage crops on silage fermentation. Appl. Environ. Microbiol. 64, 2982–2987. doi: 10.1128/AEM.64.8.2982-2987.1998
Cai, Y., Du, Z., Jethro, B. D., Nignan, M., and Yamasaki, S. (2020a). Analysis of main factors affecting silage fermentation of sorghum prepared with whole crop and stover in semiarid West Africa. Afr. J. Range For. Sci. 38, 169–178. doi: 10.2989/10220119.2020.1794959
Cai, Y., Du, Z., Yamasaki, S., Nguluve, D., Tinga, B., Macome, F., et al. (2020b). Community of natural lactic acid bacteria and silage fermentation of corn stover and sugarcane tops in Africa. Asian Australas. J. Anim. Sci. 33, 1252–1264. doi: 10.5713/ajas.19.0348
Caporaso, J. G., Bittinger, K., Bushman, F. D., DeSantis, T. Z., Andersen, G. L., and Knight, R. (2010a). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26, 266–267. doi: 10.1093/bioinformatics/btp636
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010b). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Du, Z., Sun, L., Chen, C., Lin, J., Yang, F., and Cai, Y. (2021a). Exploring microbial community structure and metabolic gene clusters during silage fermentation of paper mulberry, a high-protein woody plant. Anim. Feed Sci. Technol. 275:114766. doi: 10.1016/j.anifeedsci.2020.114766
Du, Z., Sun, L., Lin, Y., Yang, F., and Cai, Y. (2021b). The use of PacBio SMRT technology to explore the microbial network and fermentation characteristics of woody silage prepared with exogenous carbohydrate additives. J. Appl. Microbiol. 131, 2193–2211. doi: 10.1111/jam.15124
Du, Z., Yamasaki, S., Oya, T., Nguluve, D., Tinga, B., Macome, F., et al. (2020). Ensiling characteristics of total mixed ration prepared with local feed resources in Mozambique and their effects on nutrition value and milk production in Jersey dairy cattle. J. Anim. Sci. 91, e13370. doi: 10.1111/asj.13370
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Egan, A. (1980). Host animal-rumen relationships. Proc. Nutr. Soc. 39, 79–87. doi: 10.1079/pns19800011
Gao, J., Wang, P., Zhou, C., Li, P., Tang, H., Zhang, J., et al. (2019). Chemical composition and in vitro digestibility of corn stover during field exposure and the fermentation characteristics of silage prepared with microbial additives. Asian Australas. J. Anim. Sci. 32, 1854–1863. doi: 10.5713/ajas.18.0886
Guan, H., Yan, Y., Li, X., Li, X., Shuai, Y., Feng, G., et al. (2018). Microbial communities and natural fermentation of corn silages prepared with farm bunker-silo in Southwest China. Bioresour. Technol. 265, 282–290. doi: 10.1016/j.biortech.2018.06.018
Haas, B. J., Gevers, D., Earl, A. M., Feldgarden, M., Ward, D. V., Giannoukos, G., et al. (2011). Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21, 494–504. doi: 10.1101/gr.112730.110
Halperin, M. L., and Jungas, R. L. (1983). Metabolic production and renal disposal of hydrogen ions. Kidney Int. 24, 709–713. doi: 10.1038/ki.1983.217
Ke, W., Ding, W., Xu, D., Ding, L., Zhang, P., Li, F., et al. (2017). Effects of addition of malic or citric acids on fermentation quality and chemical characteristics of alfalfa silage. J. Dairy Sci. 100, 8958–8966. doi: 10.3168/jds.2017-12875
Langfelder, P., and Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559. doi: 10.1186/1471-2105-9-559
Langille, M. G. I., Zaneveld, J., Caporaso, J. G., McDonald, D., Knights, D., Reyes, J. A., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821. doi: 10.1038/nbt.2676
Li, E., Zhao, J., Liu, L., and Zhang, S. (2018). Digestible energy and metabolizable energy contents of konjac flour residues and ramie in growing pigs. Anim. Nutr. 4, 228–233. doi: 10.1016/j.aninu.2018.01.001
Lozupone, C., and Knight, R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005
Luscher, A., Mueller-Harvey, I., Soussana, J. F., Rees, R. M., and Peyraud, J. L. (2014). A review: potential of legume-based grassland–livestock systems in Europe. Grass Forage Sci. 69, 206–228. doi: 10.1111/gfs.12124
Mansfeldt, C. B., Rowe, A. R., Heavner, G. L. W., Zinder, S. H., and Richardson, R. E. (2014). Meta-analyses of Dehalococcoides mccartyi Strain 195 transcriptomic profiles identify a respiration rate-related gene expression transition point and interoperon recruitment of a key oxidoreductase subunit. Appl. Environ. Microb. 80, 6062–6072. doi: 10.1128/AEM.02130-14
Mayo, B., Rachid, C., Alegria, A., Leite, A., Peixoto, R., and Delgado, S. (2014). Impact of next generation sequencing techniques in food microbiology. Curr. Genomics 15, 293–309. doi: 10.2174/1389202915666140616233211
McDonald, P., Henderson, A. R., and Heron, S. J. E. (1991). The Biochemistry of Silage, 2nd Edn. Marlow: Chalcombe Publications.
McEniry, J., O’Kiely, P., Clipson, N. J. W., Forristal, P. D., and Doyle, E. M. (2010). Assessing the impact of various ensilage factors on the fermentation of grass silage using conventional culture and bacterial community analysis techniques. J. Appl. Microbiol. 108, 1584–1593. doi: 10.1111/j.1365-2672.2009.04557.x
Mosher, J. J., Bernberg, E. L., Shevchenko, O., Kan, J., and Kaplan, L. A. (2013). Efficacy of a 3rd generation high-throughput sequencing platform for analyses of 16S rRNA genes from environmental samples. J. Microbiol. Methods 95, 175–181. doi: 10.1016/j.mimet.2013.08.009
Mu, L., Xie, Z., Hu, L., Chen, G., and Zhang, Z. (2020). Cellulase interacts with Lactobacillus plantarum to affect chemical composition, bacterial communities, and aerobic stability in mixed silage of high-moisture amaranth and rice straw. Bioresour. Technol. 315:123772. doi: 10.1016/j.biortech.2020.123772
Muck, R. E., O’Kiely, P., and Wilson, R. K. (1991). Buffering capacities in permanent grasses. Irish J. Agr. Res. 30, 129–141.
Negawo, A. T., Teshome, A., Kumar, A., Hanson, J., and Jones, C. S. (2017). Opportunities for Napier grass (Pennisetum purpureum) improvement using molecular genetics. Agronomy 7:28. doi: 10.3390/agronomy7020028
Oliveira, A. P. D., Bagaldo, A. R., Loures, D. R. S., Bezerra, L. R., Moraes, S. A., Yamamoto, S. M., et al. (2018). Effect of ensiling gliricidia with cassava on silage quality, growth performance, digestibility, ingestive behavior and carcass traits in lambs. Anim. Feed Sci. Technol. 241, 198–209. doi: 10.1016/j.anifeedsci.2018.05.004
Pequerul, A., Pérez, C., Madero, P., Val, J., and Monge, E. (1993). “A rapid wet digestion method for plant analysis,” in Optimization of Plant Nutrition-Developments in Plant and Soil Sciences, eds M. A. C. Fragoso, M. L. Van Beusichem, and A. Houwers (Dordrecht: Springer), 3–6.
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Queiroz, O. C. M., Arriola, K. G., Daniel, J. L. P., and Adesogan, A. T. (2013). Effects of 8 chemical and bacterial additives on the quality of corn silage. J. Dairy Sci. 96, 5836–5843. doi: 10.3168/jds.2013-6691
Rajvanshi, A. K. (2019). Subabul Reloaded: How One Tree Could be the Answer to India’s Fodder, Fuel Needs. Bengaluru: The Better India.
Rengsirikul, K., Kanjanakuha, A., Ishii, Y., Kangvansaichol, K., Sripichitt, P., Punsuvon, V., et al. (2011). Potential forage and biomass production of newly introduced varieties of leucaena (Leucaena leucocephala (Lam.) de Wit.) in Thailand. Grass Sci. 57, 94–100. doi: 10.1111/j.1744-697X.2011.00213.x
Shade, A., and Handselman, J. (2012). Beyond the Venn diagram: the hunt for a core microbiome. Environ. Microbiol. 14, 4–12. doi: 10.1111/j.1462-2920.2011.02585.x
Speedy, A., and Pugliese, P. L. (1992). “Fodder trees and shrubs in range and farming systems in dry tropical africa,” in Legume Trees and Other Fodder Trees as Protein Sources for Livestock, eds A. W. Speedy and P.-L. Pugliese (Rome: Food and Agriculture Organization of the United Nations), 43–59.
Steel, R. G., and Torrie, J. H. (1980). Principles and Procedures of Statistics: A Biometrical Approach. New York, NY: McGraw Hill Company.
Suzuki, R., and Shimodaira, H. (2006). Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22, 1540–1542. doi: 10.1093/bioinformatics/btl117
Tian, H., Chen, Y., Zhu, N., Guo, Y., Deng, M., Liu, G., et al. (2020). Effect of Broussonetia papyrifera silage on the serum indicators, hindgut parameters and fecal bacterial community of Holstein heifers. AMB Express 10:197. doi: 10.1186/s13568-020-01135-y
Van Soest, P. J., Robertson, J. B., and Lewis, B. A. (1991). Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 74, 3583–3597. doi: 10.3168/jds.S0022-0302(91)78551-2
Wang, C., He, L., Xing, Y., Zhou, W., Yang, F., Chen, X., et al. (2019). Fermentation quality and microbial community of alfalfa and stylo silage mixed with Moringa oleifera leaves. Bioresour. Technol. 284, 240–247. doi: 10.1016/j.biortech.2019.03.129
Xu, D., Ding, W., Ke, W., Li, F., Zhang, P., and Guo, X. (2019). Modulation of metabolome and bacterial community in whole crop corn silage by inoculating homofermentative Lactobacillus plantarum and heterofermentative Lactobacillus buchneri. Front. Microbiol. 9:3299. doi: 10.3389/fmicb.2018.03299
Yan, Y., Li, X., Guan, H., Huang, L., Ma, X., Peng, Y., et al. (2019). Microbial community and fermentation characteristic of Italian ryegrass silage prepared with corn stover and lactic acid bacteria. Bioresour. Technol. 279, 166–173. doi: 10.1016/j.biortech.2019.01.107
Keywords: feed shortage, microbial co-occurrence network, natural biomass resource, silage fermentation, woody plant
Citation: Du Z, Yamasaki S, Oya T, Nguluve D, Euridse D, Tinga B, Macome F and Cai Y (2022) Microbial Co-occurrence Network and Fermentation Information of Natural Woody-Plant Silage Prepared With Grass and Crop By-Product in Southern Africa. Front. Microbiol. 13:756209. doi: 10.3389/fmicb.2022.756209
Received: 10 August 2021; Accepted: 27 January 2022;
Published: 14 March 2022.
Edited by:
Sailesh Malla, Chr. Hansen, DenmarkReviewed by:
Robin Anderson, Agricultural Research Service, United States Department of Agriculture, United StatesCopyright © 2022 Du, Yamasaki, Oya, Nguluve, Euridse, Tinga, Macome and Cai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yimin Cai, Y2FpQGFmZnJjLmdvLmpw
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.