
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 11 March 2022
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.703113
This article is part of the Research TopicCarbapenemases in Gram-negative Bacteria: A Global Health Threat and Therapeutic ChallengeView all 19 articles
 Tran Thi Thuy Duong1†
Tran Thi Thuy Duong1† Ya-Min Tsai2†
Ya-Min Tsai2† Li-Li Wen2
Li-Li Wen2 Hui-Chuan Chiu2
Hui-Chuan Chiu2 Pek Kee Chen1
Pek Kee Chen1 Tran Thi Dieu Thuy1
Tran Thi Dieu Thuy1 Pei-Yun Kuo1
Pei-Yun Kuo1 Jazon Harl Hidrosollo1
Jazon Harl Hidrosollo1 Shining Wang1
Shining Wang1 Yen-Zhen Zhang1
Yen-Zhen Zhang1 Wei-Hung Lin3,4
Wei-Hung Lin3,4 Ming-Cheng Wang3,5
Ming-Cheng Wang3,5 Cheng-Yen Kao1*
Cheng-Yen Kao1*Enterobacterales clinical isolates are now being resistant to clinically achievable concentrations of most commonly used antibiotics that makes treatment of hospitalized patients very challenging. We hereby determine the molecular characteristics of carbapenemase genes in carbapenem-resistant Enterobacterales (CRE) isolates in Taiwan. A total of 455 CRE isolates were identified between August 2011 to July 2020. Minimum inhibitory concentrations for selected carbapenems were tested using Vitek 2, and carbapenemase genes were determined using polymerase chain reaction in combination with sequencing. Phenotypic detection of carbapenemase was determined by modified carbapenem inactivation method (mCIM) and EDTA-modified carbapenem inactivation method (eCIM) to validate our PCR screening results. Pulsed-field gel electrophoresis (PFGE) was used to determine the clonality of carbapenemase-producing Enterobacterales (CPE) isolates, and the transferability of carbapenemase-carrying plasmids was determined by conjugation assays. A slight increase in carbapenem-resistant E. coli (CREC) was observed, however, the prevalence of carbapenem-resistant K. pneumoniae (CRKP) was steady, during 2011–2020. The dominant species among our CRE was K. pneumoniae (270/455, 59.3%), followed by E. coli (81/455, 17.8%), Morganella morganii (32/455, 7.0%), and Enterobacter cloacae (25/455, 5.5%). From 2011 to 2020, the total percentage of CPE increased steadily, accounting for 61.0% of CRE in 2020. Moreover, 122 of 455 CRE isolates (26.8%) were CPE. Among the CPE isolates, the dominant carbapenemase gene was blaOXA–48–like (54/122, 44.3%), and the second most common carbapenemase gene was blaKPC–2 (47/122, 38.5%). The sensitivity and specificity for mCIM to detect carbapenemase in the 455 isolates were both 100% in this study. The PFGE results showed that 39 carbapenemase-producing E. coli and 69 carbapenemase-producing K. pneumoniae isolates carrying blaKPC–2 and/or blaNDM–5 could be classified into 5 and 12 clusters, respectively. In conclusion, our results showed an increase in CPE isolates in Taiwan. Moreover, the distribution of carbapenemase and antimicrobial susceptibility in CPE were associated with PFGE typing.
Enterobacterales are Gram-negative, facultatively anaerobic, non-spore-forming rods, and one of the most common causes of nosocomial infections. Successive studies have demonstrated increasing antibiotic resistance among clinical Enterobacterales isolates, and high proportions of Enterobacterales isolates are now non-susceptible to clinically achievable concentrations of most commonly used antibiotics, such as broad-spectrum cephalosporins (Iredell et al., 2016; De Oliveira et al., 2020). Carbapenems are considered effective antimicrobial options for the treatment of critically ill patients with a variety of bacterial infections due to their broadest spectrum among β-lactam antibiotics and their relative resistance to hydrolysis by most β-lactamases (Barry et al., 1985). However, carbapenem resistance rates in Enterobacterales isolates have increased worldwide over the past decade (Logan and Weinstein, 2017; Lutgring, 2019).
The presence of innate resistance mechanisms and the acquisition of clusters of foreign resistance genes (e.g., plasmid, transposon, or integron) that promote survival of Enterobacterales under antibiotic treatment and host selection pressures are associated with the rapid emergence of multidrug-resistant (MDR) or extensively drug-resistant (XDR) Enterobacterales worldwide (Iredell et al., 2016; Kopotsa et al., 2019). Reduced expression or mutations in porins, overexpression of efflux pumps, and the presence of β-lactamases, play a critical role in resistance to carbapenems (Poirel et al., 2004; Iredell et al., 2016; Sugawara et al., 2016; De Oliveira et al., 2020). The first reported plasmid-mediated carbapenemase gene, K. pneumoniae carbapenemase (blaKPC), was identified in K. pneumoniae in 2001 (Yigit et al., 2001) and became the predominant carbapenemase in K. pneumoniae (Chen L. et al., 2014; Chen et al., 2018). blaNDM is the second most common carbapenemase found among carbapenem-resistant Enterobacterales (CRE) in China and is more prevalent in E. coli (Zhang et al., 2018). Therefore, the study aimed to investigate the molecular epidemiology of CRE in a regional hospital in Taiwan during 2011–2020.
Enterobacterales were isolated at En Chu Kong Hospital (ECKH), from 2011 August to 2020 July. En Chu Kong hospital, located at Sanxia district, New Taipei city, is an approximate 500-bed capacity regional teaching hospital (include three buildings: Fuxing building, Zhongshan building, and outpatient department building). The ECKH provides comprehensive medical services from fetus to the elderly, from acute trauma to hospice care, and from precision medicine to community health. These isolates were identified in the clinical laboratory by colony morphology, Gram stain, biochemical tests, and the Vitek 2 system (bioMérieux, Marcy-l’Étoile, France) according to the manufacturer’s recommendations. Non-duplicate 27,585 E. coli and 11,582 K. pneumoniae were collected in this study. The susceptibility of Enterobacterales isolates to third-generation cephalosporins (ceftazidime or ceftriaxone, 30 μg/disc, BD BBL™ Sensi-Disc™, Sparks, MD, United States) was determined by the disk diffusion method on Mueller-Hinton (MH) agar plates according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (M100-S30) (CLSI, 2020). Third-generation cephalosporin-resistant isolates were also tested for susceptibility to carbapenems, including imipenem, ertapenem, meropenem, and doripenem (10 μg/disc, BD BBL, United States). A total of 455 CRE isolates were identified and stored at −80°C in tryptic soy broth (TSB) containing 20% glycerol (v/v) until use.
Bacterial genomic DNA was isolated from bacteria grown overnight at 37°C in 3°ml LB broth. The bacterial culture was centrifuged at 12,000 rpm for 1 min, and the supernatant was removed. Crude DNA extracts were obtained by suspending the pellet in 300 μl distilled water and boiling at 95°C for 10min, followed by centrifugation at 12,000 rpm for 5 min. The DNA-containing supernatant was transferred to a new eppendorf tube, and DNA samples were stored at 4°C until testing. PCR amplification for detection of β-lactamase genes (blaKPC, blaNDM, blaIMP, blaVIM, blaOXA–48, blaGES, blaIMI, blaSME, blaSPM, blaSIM, blaDIM, and blaGIM) was performed on a GeneExplorer Thermal Cycler (BIOER, China) with the Fast-Run™ 2× Taq Master Mix (Protech, Taipei, Taiwan). Primers and PCR procedures used for the detection of β-lactamase genes have been described in previous studies (Yigit et al., 2001; Ellington et al., 2007; Doyle et al., 2012; Mlynarcik et al., 2016). PCR products were analyzed by electrophoresis using 1.2% agarose gels in 0.5× Tris-borate-EDTA (TBE) buffer. Gels were stained with ethidium bromide (EtBr), and PCR products were visualized using UV transilluminator. Clinical K. pneumoniae isolates carrying blaKPC, blaNDM, blaIMP, blaVIM, and blaOXA–48 were used as PCR positive controls. The PCR products of blaGES, blaIMI, blaSME, blaSPM, blaSIM, blaDIM, or blaGIM, with relevant expected sizes were verified by sequencing due to the lack of relative control strains.
The modified carbapenem inactivation method (mCIM) and EDTA-modified carbapenem inactivation method (eCIM) were performed on CRE isolates according to the previous study to detect the presence of carbapenemase (Sfeir et al., 2019; Tsai et al., 2020). Briefly, a 1-μl loopful of bacteria was resuspended in a 2-ml tube containing TSB. Another 1-μl loopful of bacteria was resuspended in a 2-ml tube containing TSB supplemented with EDTA at a final concentration of 5 mM. A meropenem disk was placed in each tube, and the tubes were incubated at 35°C for 4 h ± 15 min. The disks were then removed and placed onto MH agar plates freshly plated with a 0.5 McFarland suspension of a carbapenem-susceptible E. coli strain ATCC 25922. Plates were incubated at 35°C for 16 to 20 h, and mCIM and eCIM results were interpreted as previously described (Sfeir et al., 2019). In accordance with CLSI guidelines (CLSI, 2020), K. pneumoniae ATCC BAA-1706 (carbapenemase negative), K. pneumoniae ATCC BAA-1705 (blaKPC positive), and K. pneumoniae ATCC BAA-2146 (blaNDM positive) were used as internal controls for mCIM and eCIM testing. The mCIM and eCIM tests were replicated by two independent investigators to ensure reproducibility.
Minimum inhibitory concentrations (MICs) for cefmetazole, cefotaxime, ceftazidime, cefepime, imipenem, ertapenem, meropenem, amikacin, gentamicin, ciprofloxacin, levofloxacin, tigecycline, colistin, and trimethoprim for CPE isolates were determined with Vitek 2 using the card AST-N322 according to the manufacturer’s instructions. E. coli ATCC 25922 was used as a quality control strain. Antibiotic susceptibility (except tigecycline) was interpreted according to CLSI guidelines (M100-S30) (Zhang et al., 2018). The results of tigecycline susceptibility were interpreted according to the breakpoints of the U.S. Food and Drug Administration (FDA) (≥ 8.0 μg/ml, resistant; 4.0 μg/ml, intermediate; ≤ 2.0 μg/ml, susceptible).
Pulsed-field gel electrophoresis was performed to determine the clonality of CPE isolates according to a previous study (Li et al., 2018). Briefly, PFGE of XbaI-digested genomic DNA was performed using a CHEF Mapper XA instrument (Bio-Rad Laboratories, Inc., Hercules, CA, United States) with the following parameters: separation on a 1% agarose gel (Seakem Gold agarose; FMC Bio Products) in 0.5× TBE buffer for 19 h at 14°C with pulse times ranging from 5 to 35 s at 6 V/cm. Gels were stained with EtBr and photographed with UV transillumination. PFGE profiles were analyzed and compared using the GelCompar II software, version 2.0 (Unimed Healthcare, Inc., Houston, TX, United States). The PFGE patterns were interpreted according to a previous study (Bando et al., 2009) and the isolates having > 80% pattern similarity were assigned to the same cluster.
The liquid mating-out assay was performed to transfer carbapenemase genes from CPE isolates to the rifampicin- and streptomycin-resistant E. coli C600 strain as previously described (Kao et al., 2016). All isolates tested were sensitive to rifampicin or streptomycin at the concentration of 256 μg/ml. Therefore, transconjugants were selected on LB plates with 256 μg/ml rifampicin (Sigma-Aldrich, United States) or 256 μg/ml streptomycin (Sigma-Aldrich, United States) in combination with 1 μg/ml meropenem. The conjugation assay was performed in triplicate to determine the transferability of the plasmid.
A Cochran–Armitage test was used to evaluate trends in CREC, CRKP, CPE, CPEC, and CPKP over time. Statistical analyses were performed using the JMP software (SAS Institute Inc., Cary, NC, United States). A p-value < 0.05 is statistically significant.
The dominant species among our CRE was K. pneumoniae (270/455, 59.4%), followed by E. coli (80/455, 17.6%), Morganella morganii (32/455, 7.0%), and Enterobacter cloacae (25/455, 5.5%) (Table 1). Thus, 0.003% (80/27,585) E. coli and 2.323% (270/11,582) K. pneumoniae showed resistance to carbapenem. A slight increase in carbapenem-resistant E. coli (CREC) was observed during 2011–2020 (p > 0.05) (Figure 1A). In contrast, the prevalence of carbapenem-resistant K. pneumoniae (CRKP) was steady (p > 0.05) (Figure 1B). In addition, 205 (44.9%) and 169 (37.0%) CRE strains were isolated from urine and sputum, respectively (Table 1). CRKP was most frequently isolated from sputum (127/270, 47.0%), followed by urine (107/270, 39.6%). In contrast, CREC was most frequently isolated from urine (54/80, 67.5%), followed by sputum (12/80, 15.0%) (Table 1).
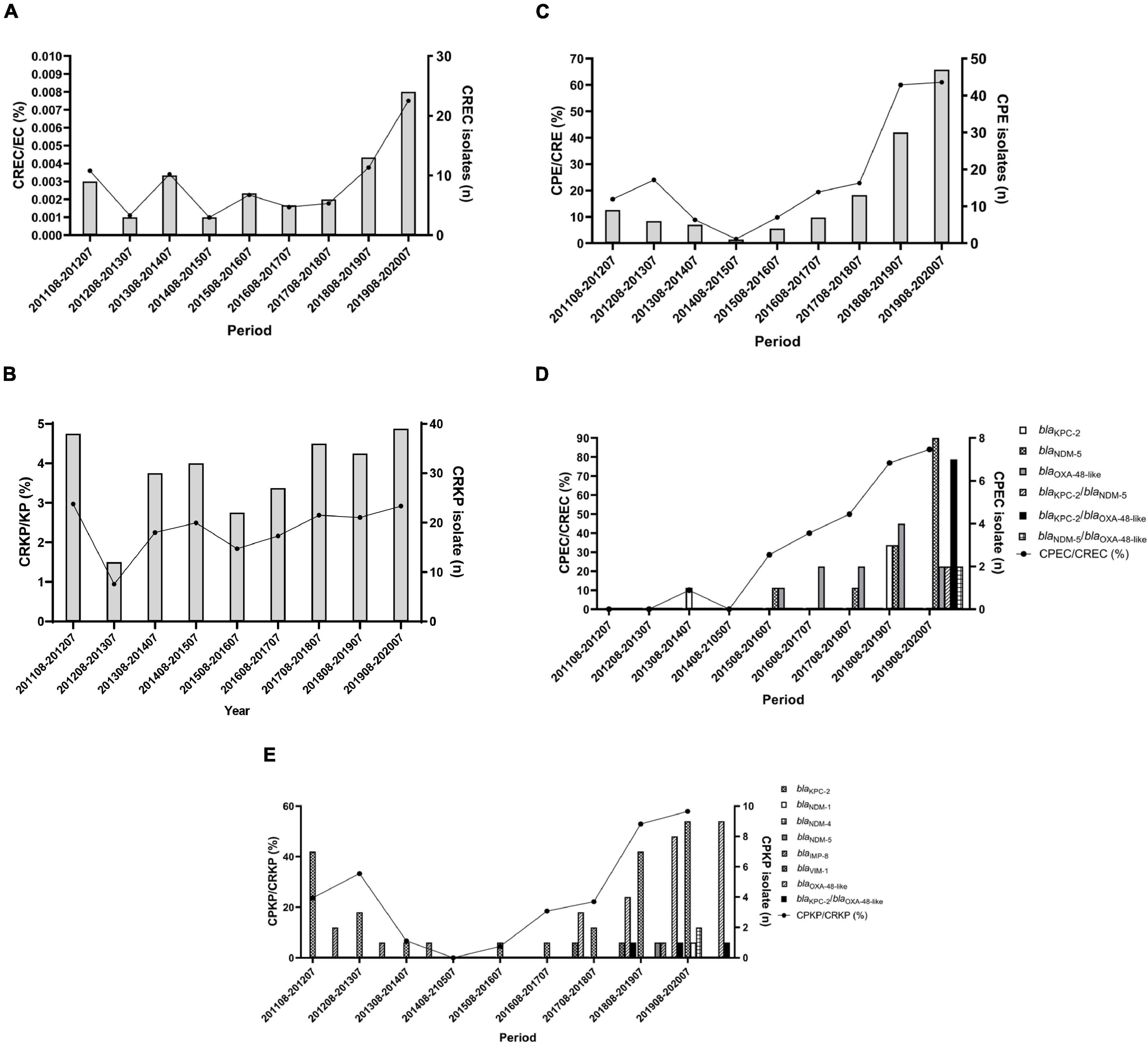
Figure 1. Distribution of carbapenemase-producing Enterobacterales during 2011–2020. (A) Annual proportions and numbers of carbapenem-resistant E. coli among all E. coli. (B) Annual proportions and numbers of carbapenem-resistant K. pneumoniae among all K. pneumoniae. (C) Annual proportions and numbers of carbapenemase-producers among CRE. (D,E) Annual proportions and numbers of carbapenemase-producers among carbapenem-resistant E. coli (D) and K. pneumoniae (E). The percentage of isolates is plotted as a line graph on the primary axis while the number of isolates is plotted as bars on the secondary axis.
PCR was used to detect the presence of carbapenemase genes, and the results showed that 122 of 455 CRE isolates (26.8%) were CPE (Table 2). No carbapenemase genes were detected in carbapenem-resistant Providencia rettgeri, Providencia stuartii, and Serratia marcescens. In addition, no GES-, IMI-, SME-, SPM-, SIM-, DIM-, and GIM-producers were identified in our CRE isolates. Overall, the dominant carbapenemase gene was blaOXA–48–like (54/122, 44.3%) among CPE isolates, and the second most common carbapenemase gene was blaKPC–2 (47/122, 38.5%) (Table 2). In addition, we found metallo-carbapenemases NDM, IMP, and VIM in 23 (18 blaNDM–5, 3 blaNDM–1, and 2 blaNDM–4), 9 (9 blaIMP–8), and 4 (blaVIM–1) CPE isolates, respectively (Table 2). Importantly, 15 CPE isolates carrying more than one carbapenemase gene were identified during 2017–2020. Seven E. coli isolates and 3 K. pneumoniae isolates had both blaOXA–48–like and blaKPC–2 (Table 2). Coexistence of blaKPC–2/blaNDM–5 and blaOXA–48–like/blaNDM–5 was found in 2 and 2 carbapenem-resistant E. coli, respectively. One Klebsiella oxytoca isolate had both blaVIM–1 and blaNDM–1 (Table 2).
Among the CRE isolates, 39 (39/80, 48.8%) E. coli and 69 (69/270, 25.6%) K. pneumoniae isolates were CPE. The dominant carbapenemase gene was blaOXA–48–like (20/39, 51.3%) among carbapenemase-producing E. coli (CPEC) isolates, and the second most common carbapenemase gene was blaNDM–5 (17/39, 43.6%). In contrast, blaKPC–2 was found dominant among carbapenemase-producing K. pneumoniae (CPKP) isolates (34/69, 49.3%), followed by blaOXA–48–like (27/69, 39.1%) (Table 2).
It was previously reported that eCIM in combination with the mCIM is efficient for identifying CPE (Sfeir et al., 2019). Therefore, phenotypic detection mCIM/eCIM was performed on our 455 CRE isolates to validate PCR results for carbapenemase gene detection. In this study, the sensitivity and specificity for the mCIM to detect carbapenemase in 455 CRE isolates were both 100% which is consistent with our previous report (Tsai et al., 2020).
Interestingly, we found that four isolates containing metallo-carbapenemase NDM-5 and non-metallo-carbapenemases (OXA-48 or KPC-2) showed inconsistent mCIM/eCIM results (Table 3). E. coli isolate 488, an NDM-5 and OXA-48 producer, showed a false-negative result in mCIM/eCIM (Table 3). In contrast, E. coli isolate 514 with blaNDM–5 and blaOXA–48 showed a positive mCIM/eCIM result (Table 3). In addition, E. coli 571 and 572 with blaKPC–2/blaNDM–5 also showed positive results by mCIM/eCIM (Table 3).
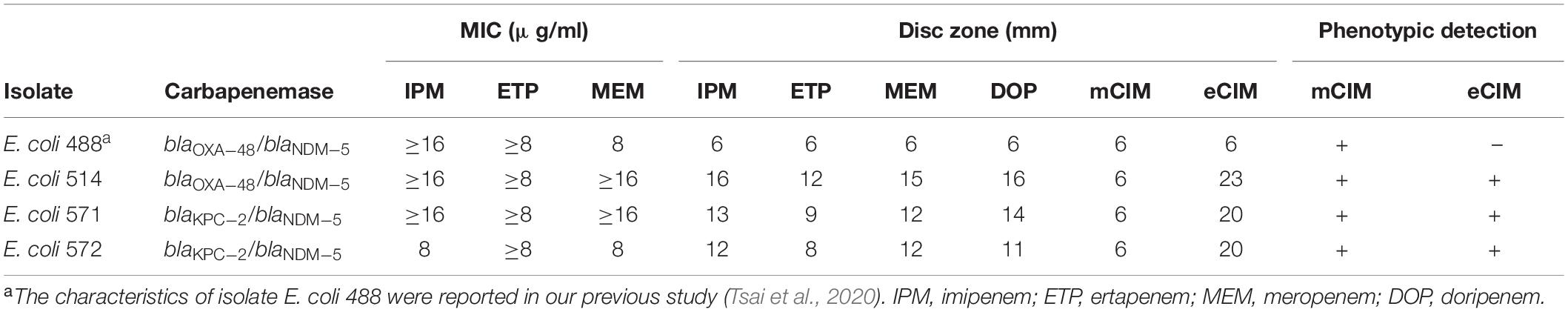
Table 3. Characteristics of three isolates that contained both metallo-carbapenemases and non-metallo-carbapenemases.
From 2011 to 2020, the total percentage of CPE increased steadily, accounting for 61.0% of CRE in 2020 (16.7% in August 2011–July 2012) (p < 0.0001) (Figure 1C). E. coli and K. pneumoniae isolates were dominant in our CPE collection (Table 2), so we aimed to further characterize the molecular epidemiology of CPEC and CPKP isolates. Among CPEC isolates, we found a dramatic increase in blaNDM–5 and blaKPC–2/blaOXA–48–like in 2020 (p < 0.0001) (Figure 1D). In contrast to CPEC, blaKPC–2 and blaOXA–48–like were predominant in CPKP isolates in 2020 (p < 0.0001) (Figure 1E).
The clonality of 39 CPEC and 69 CPKP isolates carrying carbapenemase KPC-2, NDM, and OXA-48 was further determined by PFGE (Figure 2). The PFGE patterns of 30 E. coli CPE isolates were assigned to five clusters based on > 80% pattern similarity (Figure 2A). All isolates in cluster 1 (n = 3) contained blaKPC–2, isolates in cluster 2 (n = 5) contained blaoxa–48, and isolates in cluster 5 (n = 7) contained both blaKPC–2 and blaoxa–48 (Figure 2A). In contrast, isolates from clusters 3 (n = 12) and 4 (n = 3) contained blaNDM–5 (two isolates contained blaNDM–5 and blaKPC–2; one isolate contained blaNDM–5 and blaoxa–48) (Figure 2A). Interestingly, isolates belonging to clusters 2 or 5 were resistant to gentamycin (Figure 2A). Although all 39 CPEC were resistant to ciprofloxacin (MIC ≥ 4 μg/ml) and levofloxacin (MIC ≥ 8 μg/ml), these isolates were sensitive to tigecycline (MIC ≤ 0.5 μg/ml) and colistin (MIC ≤ 0.5 μg/ml) (Figure 2A).
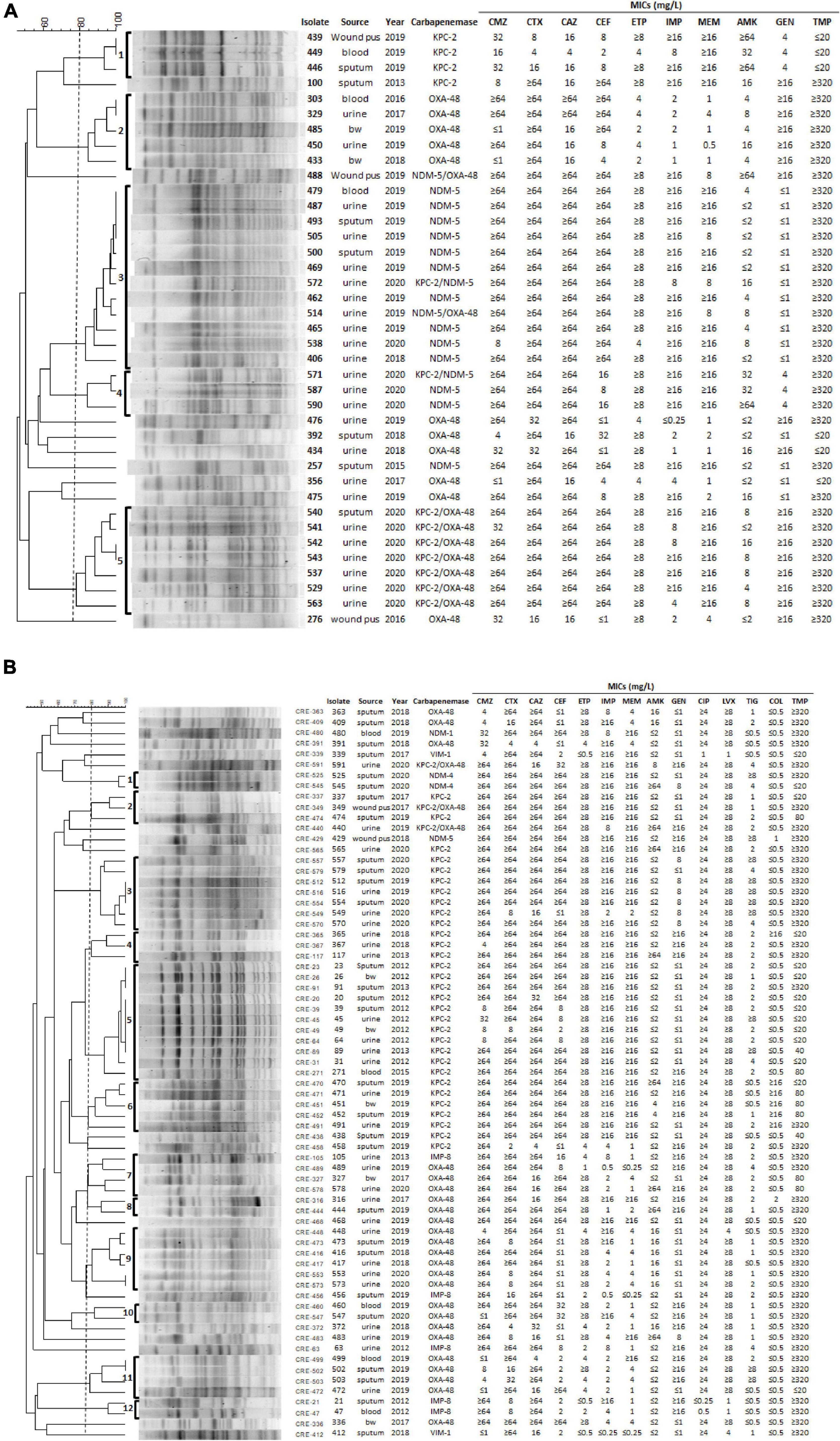
Figure 2. PFGE, the origin of isolate, year, MICs of antibiotics, and carbapenemase genes in 39 E. coli (A) and 69 K. pneumoniae (B) isolates. (A) All CPEC had the following MICs of antibiotics: ampicillin ≥ 32 μg/ml, piperacillin > 128 μg/ml, cefazolin ≥ 64 μg/ml, ciprofloxacin ≥ 4 μg/ml, levofloxacin ≥ 8 μg/ml, tigecycline ≤ 0.5 μg/ml, and colistin ≤ 0.5 μg/ml. (B) All CPKP had MICs of the antibiotics as follows, ampicillin ≥ 32 μg/ml and cefazolin ≥ 64 μg/ml. bw, bronchial washing; CMZ, cefmetazole; CTX, cefotaxime; CAZ, ceftazidime; CEF, cefepime; IPM, imipenem; ETP, ertapenem; MEM, meropenem; AMK, amikacin; GEN, gentamicin; CIP, ciprofloxacin; LVX, levofloxacin; TIG, tigecycline; COL, colistin; TMP, trimethoprim.
The PFGE patterns of 69 CPKP isolates were assigned to 12 clusters based on > 80% pattern similarity (Figure 2B). All isolates in clusters 2 (n = 3), 3 (n = 7), 4 (n = 3), 5 (n = 11), and 6 (n = 5) contained blaKPC–2, whereas isolates in clusters 8 (n = 2), 9 (n = 6), 10 (n = 2), and 11 (n = 4) contained blaoxa–48 (Figure 2B). Clusters 1 (n = 2) and 12 (n = 2) isolates contained blaNDM–4 and blaIMP–8, respectively (Figure 2B). Only isolates in cluster 12 were susceptible to both ciprofloxacin and levofloxacin (Figure 2B). In addition, 6 and 11 CPKP were resistant to colistin (MIC ≥ 4 μl/ml) and tigecycline (MIC ≥ 8 μl/ml), respectively (Figure 2B). The PFGE results indicate that the distribution of carbapenemase and antimicrobial susceptibility in CPE were associated with PFGE typing (Figure 2).
A total of 37 CPEC and 53 CPKP isolates carrying carbapenemase were further analyzed with conjugation assays to determine whether there were horizontally spread carbapenemase-carrying plasmids in Taiwan (18 CPE isolates showed resistance to rifampicin were excluded in this assay). Transfer of carbapenemase gene by conjugation to E. coli C600 was successful in 4 NDM-5-producing CPEC (isolates 257, 462, 500, and 505) and 1 IMP-8-producing CPKP (isolate 21). However, all blaKPC–2- and blaOXA–48-carrying plasmids did not show transferability.
In this study, we isolated 455 CRE isolates from a regional teaching hospital in Taiwan (2011 August to 2020 July) and present their characteristics. Our results showed that 122 of 455 CRE isolates were CPE (Table 2). No carbapenemase genes were detected in our carbapenem-resistant P. rettgeri, P. stuartii, and S. marcescens. However, we could not rule out the presence of other carbapenemases in these isolates.
The dominant carbapenemase gene among our CPE isolates was blaOXA–48, and the second most common carbapenemase gene was blaKPC–2 (Table 2). Chiu et al. (2018) showed a sharp increase in the annual prevalence rate of OXA-48-like producers among Taiwanese CPE isolates between 2012 and 2015. In 2017, blaOXA–48 was detected in 18.2% of CPE in Taiwan (Jean et al., 2018). Moreover, Wu et al. (2021) reported that the blaKPC–2 was the most common carbapenemase gene in CRKP isolated from patients with bacteremia at a hospital in northern Taiwan from 2013 to 2018. A 22-year (1998–2019) observation to determine the evolution of carbapenemase genes in K. pneumoniae in Taiwan discovered that the endemicity has changed from blaIMP–8, blaNDM–1, and blaVIM–1 to the most common blaKPC–2 and rapidly emerging blaOXA–48 (Lai and Yu, 2021). These results are consistent with our finding that the distribution of blaOXA–48–like was dramatically increased after 2018. Therefore, whether there is a circulation of OXA-48-producing plasmids/isolates in Taiwan is worth continually monitoring. Surprisingly, a very low conjugation rate of carbapenemase genes-carrying plasmids was observed in this study. Therefore, taxonomic relatedness and recipient strain used for conjugation tests may limit the conjugation in liquid matings (Chen Y. T. et al., 2014; Alderliesten et al., 2020).
Interestingly, the data from Surveillance of Multicentre Antimicrobial Resistance in Taiwan (SMART) with a multicenter collection of bacteremic isolates of E. coli (n = 423) and K. pneumoniae (n = 372) showed the carbapenem resistance rates were 1.2% (5/423) in E. coli and 7.5% (28/372) in K. pneumoniae (Liu et al., 2020). Moreover, carbapenemase genes were detected in 67.8% K. pneumoniae isolates (19/28). Among the CRKP isolates, 57.1% (16/28) harbored blaKPC (Liu et al., 2020). However, in 2019, we found OXA-48-like was the dominant carbapenemase in CRKP. These results suggested the difference in the distribution of carbapenemase genes in CRKP isolated from different specimens and regions. Moreover, previous studies showed that most KPC-2 producers in CRKP were ST11 (Liu et al., 2020; Wu et al., 2021). Therefore, the ST type of OXA-48-like producing CRKP is worth further investigating.
We also found a dramatic increase in blaNDM–5 and blaKPC–2/blaOXA–48–like from 2018 to 2020 in CREC when compared to the years before 2018 (Figure 1). Huang et al. (2021) reported an increase of NDM-producing E. coli in northern Taiwan during 2016 to 2018. Importantly, in all five blaNDM–5-positive isolates, the blaNDM–5 gene was located in a ∼46 kb IncX3 plasmid that were nearly identical to each other (Huang et al., 2021). These five blaNDM–5-containing plasmids are similar to pP785-NDM5 from China (Ho et al., 2018). These results suggest the dissemination of a specific IncX3 blaNDM–5-containing plasmid in Taiwan. Seven CPEC isolates having blaKPC–2/blaOXA–48–like were collected from August 2019 to July 2020 in this study. The clonality of these seven strains remain unclear and worth investigating to determine whether is a specific CPEC clone outbreak in the hospital.
In addition, we found 15 CPE isolates carrying more than one carbapenemase gene during 2017–2020 (Table 2). Whether these carbapenemase genes were located on a single plasmid is worth investigating. We also found that four isolates containing metallo-carbapenemase NDM-5 and non-metallo-carbapenemases (OXA-48 or KPC-2) showed inconsistent mCIM/eCIM results (Table 3). These results raised the possibility that different expression levels of carbapenemase genes were present in these isolates, thus affecting the phenotypic detection of mCIM/eCIM. In addition, it remains to be investigated whether the genotypes of the carbapenemase genes in these isolates affect their enzymatic activity.
The emergence of CRE strains that also exhibit resistance to colistin and tigecycline has become a major clinical concern, as these two antibiotics are used as first-line for the treatment of CRE infections (Doi, 2019). Our results showed that all CPEC were sensitive to colistin and tigecycline (Figure 2A), while 6 and 11 CPKP were resistant to colistin (MIC ≥ 4 μl/ml) and tigecycline (MIC ≥ 8 μl/ml), respectively (Figure 2B). The mechanisms responsible for colistin and tigecycline resistance of these CPKP isolates remain to be studied. In addition, all CPKP isolates (n = 5) in cluster 6 were resistant to colistin and 5 of 7 isolates in cluster 3 were resistant to tigecycline (Figure 2B). The characteristics of the isolates in clusters 3 and 6 are worthy of future study.
Hentschke et al. (2010) demonstrated the induction of AcrAB-mediated multidrug resistance by prior treatment with multiple antibiotics. Kanwar et al. (2018) also revealed the occurrence of treatment-emergent colistin-resistant KPC-producing K. pneumoniae after 8 days of colistin-based combination therapy due to disruption of mgrB. Studies with clinical isolates provided evidence of an association between the emergence of colistin and tigecycline resistance in CRKP and frequent antibiotic use (van Duin et al., 2014; Du et al., 2018; Kanwar et al., 2018). Therefore, it is worth investigating whether the patient received long-term antibiotics, including tigecycline or colistin, after the diagnosis of CRKP infection, inducing resistance in this study.
Salipante et al. (2015) reported that whole-genome sequencing (WGS) has significantly improved resolving power for strain typing compared to PFGE. However, the present barriers to the universal adoption of WGS by clinical laboratories include relatively high costs of instrumentation and a lack of bioinformatic expertise (Salipante et al., 2015). Therefore, in this study, we performed PFGE to determine the clonality of 39 CPEC and 69 CPKP isolates carrying carbapenemase KPC-2, NDM, and OXA-48.
In conclusion, our longitudinal collection of isolates showed the increase of CPE in CRE isolates in Taiwan during 2011–2020, and the dominant carbapenemase gene was blaOXA–48–like, followed by blaKPC–2, among our CPE isolates. Moreover, we found the carbapenemase distribution and antimicrobial susceptibility in CPE were associated with PFGE typing. Although the analysis of our study was restricted to a single hospital as opposed to population-based, the continued epidemiological surveillance and control of antimicrobial prescribing and consumption would reduce the prevalence of drug-resistant organisms and the spread of antibiotic resistance.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Y-MT, H-CC, and L-LW contributed to the collection of isolates. TD, PC, Y-MT, H-CC, P-YK, TT, JH, SW, Y-ZZ, W-HL, and M-CW performed the experiments and interpreted the results of bacterial identification, antibiotic susceptibility tests, mCIM/eCIM tests, carbapenemases detection, and conjugation assays. C-YK was responsible for data analysis, manuscript writing, designed the study, and responsible for overall management and planning. All authors read and approved the final manuscript.
This work was supported by the Ministry of Science and Technology, Taiwan (Grant Number 109-2320-B- -036-MY3); the En Chu Kong Hospital, Taiwan (Grant Number ECKH_W11001); the Yen Tjing Ling Medical Foundation, Taiwan (Grant Number CI-110-13); and the National Yang-Ming University-Far Eastern Memorial Hospital Joint Research Program, Taiwan (Grant Number 110DN14).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alderliesten, J. B., Duxbury, S. J. N., Zwart, M. P., de Visser, J., Stegeman, A., and Fischer, E. A. J. (2020). Effect of donor-recipient relatedness on the plasmid conjugation frequency: a meta-analysis. BMC Microbiol. 20:135. doi: 10.1186/s12866-020-01825-4
Bando, S. Y., Andrade, F. B., Guth, B. E., Elias, W. P., Moreira-Filho, C. A., and Pestana de Castro, A. F. (2009). Atypical enteropathogenic Escherichia coli genomic background allows the acquisition of non-EPEC virulence factors. FEMS Microbiol. Lett. 299, 22–30. doi: 10.1111/j.1574-6968.2009.01735.x
Barry, A. L., Jones, R. N., Thornsberry, C., Ayers, L. W., and Kundargi, R. (1985). Imipenem (N-formimidoyl thienamycin): in vitro antimicrobial activity and beta-lactamase stability. Diagn. Microbiol. Infect. Dis. 3, 93–104. doi: 10.1016/0732-8893(85)90017-3
Chen, C. M., Guo, M. K., Ke, S. C., Lin, Y. P., Li, C. R., and VyNguyen, H. T. (2018). Emergence and nosocomial spread of ST11 carbapenem-resistant Klebsiella pneumoniae co-producing OXA-48 and KPC-2 in a regional hospital in Taiwan. J. Med. Microbiol. 67, 957–964. doi: 10.1099/jmm.0.000771
Chen, L., Mathema, B., Chavda, K. D., DeLeo, F. R., Bonomo, R. A., and Kreiswirth, B. N. (2014). Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol. 22, 686–696. doi: 10.1016/j.tim.2014.09.003
Chen, Y. T., Lin, J. C., Fung, C. P., Lu, P. L., Chuang, Y. C., Wu, T. L., et al. (2014). KPC-2-encoding plasmids from Escherichia coli and Klebsiella pneumoniae in Taiwan. J. Antimicrob. Chemother. 69, 628–631.
Chiu, S. K., Ma, L., Chan, M. C., Lin, Y. T., Fung, C. P., Wu, T. L., et al. (2018). Carbapenem nonsusceptible klebsiella pneumoniae in taiwan: dissemination and increasing resistance of carbapenemase producers during 2012-2015. Sci. Rep. 8:8468. doi: 10.1038/s41598-018-26691-z
De Oliveira, D. M. P., Forde, B. M., Kidd, T. J., Harris, P. N. A., Schembri, M. A., and Beatson, S. A. (2020). Antimicrobial resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 33, 181–119.
Doi, Y. (2019). Treatment Options for Carbapenem-resistant Gram-negative Bacterial Infections. Clin. Infect. Dis. 69, S565–S575. doi: 10.1093/cid/ciz830
Doyle, D., Peirano, G., Lascols, C., Lloyd, T., Church, D. L., and Pitout, J. D. (2012). Laboratory detection of Enterobacteriaceae that produce carbapenemases. J. Clin. Microbiol. 50, 3877–3880.
Du, X., He, F., Shi, Q., Zhao, F., Xu, J., Fu, Y., et al. (2018). The rapid emergence of tigecycline resistance in blaKPC-2 harboring klebsiella pneumoniae, as mediated in vivo by mutation in teta during tigecycline treatment. Front. Microbiol. 9:648. doi: 10.3389/fmicb.2018.00648
Ellington, M. J., Kistler, J., Livermore, D. M., and Woodford, N. (2007). Multiplex PCR for rapid detection of genes encoding acquired metallo-beta-lactamases. J. Antimicrob. Chemother. 59, 321–322. doi: 10.1093/jac/dkl481
Hentschke, M., Wolters, M., Sobottka, I., Rohde, H., and Aepfelbacher, M. (2010). ramR mutations in clinical isolates of Klebsiella pneumoniae with reduced susceptibility to tigecycline. Antimicrobial. Agents Chemother. 54, 2720–2723. doi: 10.1128/AAC.00085-10
Ho, P. L., Wang, Y., Liu, M. C., Lai, E. L., Law, P. Y., Cao, H., et al. (2018). IncX3 Epidemic plasmid carrying blaNDM-5 in Escherichia coli from swine in multiple geographic areas in china. Antimicrobial. Agents Chemother. 62, e2295–e2217. doi: 10.1128/AAC.02295-17
Huang, Y. S., Sai, W. C., Li, J. J., Chen, P. Y., Wang, J. T., Chen, Y. T., et al. (2021). Increasing New Delhi metallo-beta-lactamase-positive Escherichia coli among carbapenem non-susceptible Enterobacteriaceae in Taiwan during 2016 to 2018. Sci. Rep. 11:2609. doi: 10.1038/s41598-021-82166-8
Iredell, J., Brown, J., and Tagg, K. (2016). Antibiotic resistance in Enterobacteriaceae: mechanisms and clinical implications. BMJ 352:6420.
Jean, S. S., Lee, N. Y., Tang, H. J., Lu, M. C., Ko, W. C., and Hsueh, P. R. (2018). Carbapenem-resistant enterobacteriaceae infections: taiwan aspects. Front. Microbiol. 9:2888. doi: 10.3389/fmicb.2018.02888
Kanwar, A., Marshall, S. H., Perez, F., Tomas, M., Jacobs, M. R., Hujer, A. M., et al. (2018). Emergence of resistance to colistin during the treatment of bloodstream infection caused by klebsiella pneumoniae carbapenemase-producing klebsiella pneumoniae. Open Forum. Infect. Dis. 5:54. doi: 10.1093/ofid/ofy054
Kao, C. Y., Udval, U., Huang, Y. T., Wu, H. M., Huang, A. H., Bolormaa, E., et al. (2016). Molecular characterization of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella spp. isolates in Mongolia. J. Microbiol. Immunol. Infect. 49, 692–700. doi: 10.1016/j.jmii.2015.05.009
Kopotsa, K., Osei Sekyere, J., and Mbelle, N. M. (2019). Plasmid evolution in carbapenemase-producing Enterobacteriaceae: a review. Ann. NY Acad. Sci. 1457, 61–91. doi: 10.1111/nyas.14223
Lai, C. C., and Yu, W. L. (2021). Klebsiella pneumoniae harboring carbapenemase genes in taiwan: its evolution over 20 Years, 1998-2019. Int. J. Antimicrob. Agents 58:106354. doi: 10.1016/j.ijantimicag.2021.106354
Li, H. Y., Kao, C. Y., Lin, W. H., Zheng, P. X., Yan, J. J., Wang, M. C., et al. (2018). Characterization of CRISPR-Cas systems in clinical klebsiella pneumoniae isolates uncovers its potential association with antibiotic susceptibility. Front. Microbiol. 9:1595. doi: 10.3389/fmicb.2018.01595
Liu, P. Y., Lee, Y. L., Lu, M. C., Shao, P. L., Lu, P. L., Chen, Y. H., et al. (2020). National surveillance of antimicrobial susceptibility of bacteremic gram-negative bacteria with emphasis on community-acquired resistant isolates: report from the 2019 surveillance of multicenter antimicrobial resistance in taiwan (SMART). Antimicrobial. Agents Chemother. 64, e1089–e1020. doi: 10.1128/AAC.01089-20
Logan, L. K., and Weinstein, R. A. (2017). The epidemiology of carbapenem-resistant enterobacteriaceae: the impact and evolution of a global menace. J. Infect. Dis. 215, S28–S36. doi: 10.1093/infdis/jiw282
Lutgring, J. D. (2019). Carbapenem-resistant Enterobacteriaceae: An emerging bacterial threat. Semin. Diagn. Pathol. 36, 182–186. doi: 10.1053/j.semdp.2019.04.011
Mlynarcik, P., Roderova, M., and Kolar, M. (2016). Primer evaluation for pcr and its application for detection of carbapenemases in enterobacteriaceae. Jundishapur J. Microbiol. 9:e29314. doi: 10.5812/jjm.29314
Poirel, L., Heritier, C., Spicq, C., and Nordmann, P. (2004). In vivo acquisition of high-level resistance to imipenem in Escherichia coli. J. Clin. Microbiol. 42, 3831–3833.
Salipante, S. J., SenGupta, D. J., Cummings, L. A., Land, T. A., Hoogestraat, D. R., and Cookson, B. T. (2015). Application of whole-genome sequencing for bacterial strain typing in molecular epidemiology. J. Clin. Microbiol. 53, 1072–1079. doi: 10.1128/JCM.03385-14
Sfeir, M. M., Hayden, J. A., Fauntleroy, K. A., Mazur, C., Johnson, J. K., Simner, P. J., et al. (2019). EDTA-Modified carbapenem inactivation method: a phenotypic method for detecting metallo-beta-lactamase-producing enterobacteriaceae. J. Clin. Microbiol. 57, e1757–e1718. doi: 10.1128/JCM.01757-18
Sugawara, E., Kojima, S., and Nikaido, H. (2016). Klebsiella pneumoniae major porins ompk35 and ompk36 allow more efficient diffusion of beta-lactams than their escherichia coli homologs ompf and OMpC. J. Bacteriol. 198, 3200–3208. doi: 10.1128/JB.00590-16
Tsai, Y. M., Wang, S., Chiu, H. C., Kao, C. Y., and Wen, L. L. (2020). Combination of modified carbapenem inactivation method (mCIM) and EDTA-CIM (eCIM) for phenotypic detection of carbapenemase-producing Enterobacteriaceae. BMC Microbiol. 20:315. doi: 10.1186/s12866-020-02010-3
van Duin, D., Cober, E. D., Richter, S. S., Perez, F., Cline, M., Kaye, K. S., et al. (2014). Tigecycline therapy for carbapenem-resistant Klebsiella pneumoniae (CRKP) bacteriuria leads to tigecycline resistance. Clin. Microbiol. Infect. 20, 1117–1120. doi: 10.1111/1469-0691.12714
Wu, A. Y., Chang, H., Wang, N. Y., Sun, F. J., and Liu, C. P. (2021). Clinical and molecular characteristics and risk factors for patients acquiring carbapenemase-producing and non-carbapenemase-producing carbapenem-nonsusceptible-Enterobacterales bacteremia. J. Microbiol. Immunol. Infect. 9, 235–238. doi: 10.1016/j.jmii.2021.10.008
Yigit, H., Queenan, A. M., Anderson, G. J., Domenech-Sanchez, A., Biddle, J. W., and Steward, C. D. (2001). Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45, 1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001
Keywords: carbapenem-resistant Enterobacterales (CRE), KPC-2, OXA-48, NDM, pulsed-field gel electrophoresis (PFGE), carbapenemase
Citation: Duong TTT, Tsai Y-M, Wen L-L, Chiu H-C, Chen PK, Thuy TTD, Kuo P-Y, Hidrosollo JH, Wang S, Zhang Y-Z, Lin W-H, Wang M-C and Kao C-Y (2022) A Longitudinal Nine-Year Study of the Molecular Epidemiology of Carbapenemase-Producing Enterobacterales Isolated From a Regional Hospital in Taiwan: Predominance of Carbapenemase KPC-2 and OXA-48. Front. Microbiol. 13:703113. doi: 10.3389/fmicb.2022.703113
Received: 30 April 2021; Accepted: 23 February 2022;
Published: 11 March 2022.
Edited by:
Che-Hsin Lee, National Sun Yat-sen University, TaiwanReviewed by:
Miriam Cordovana, Bruker Daltonik GmbH, GermanyCopyright © 2022 Duong, Tsai, Wen, Chiu, Chen, Thuy, Kuo, Hidrosollo, Wang, Zhang, Lin, Wang and Kao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cheng-Yen Kao, a2FvY3lAeW0uZWR1LnR3
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.