
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 11 January 2023
Sec. Food Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1108890
This article is part of the Research Topic Exploring the Role of Microorganisms in Silages: Species, Communities, Interactions, and Functional Characteristics View all 35 articles
Introduction: Ensiling whole-crop oat (Avena sativa L.) has attracted a growing interest in the Qinghai-Tibet Plateau. The study aimed to investigate the microbial community and chemical composition of fresh and ensiling oat harvested from six different elevations of the Qinghai-Tibet Plateau.
Method: The oat (A. sativa L. cv. Qingyin No. 1) was planted in six different sites across Qinghai-Tibet Plateau (BM, Bomi County; BY, Bayi County; DZ, Dazi County; BR, Biru County; SC, Suo County; SN, Seni County), where the elevations were in the range of 2,800–4,500 m above sea level (a. s. l.). Oat was harvested at the milk stage and ensiled for 90 days.
Results: The highest crude protein (CP) and lowest water-soluble carbohydrate (WSC) were observed in fresh oat of SN and BM, respectively, however, no distinct gradient trend in WSC and CP concentrations along the elevation gradient. The lowest LAB counts in fresh oat from the highest elevational regions of SN. After 90 days of ensiling, the pH in all oat silages was lower than 4.2, and silages from SC and SN showed a lower pH and butyric acid concentration, and higher lactic acid (LA) concentration than silages of other regions. The oat silage from BR showed the lowest LA concentration and the highest pH. The bimodal distributions of fungal and bacterial richness in fresh oat along the elevation gradient were observed, while the elevation gradients did not affect the fungal Shannon index in fresh oat. Dioszegia, Cladosporium, and Vishniacozyma were the prevalent fungal genus in fresh oat, while Wickerhamomyces, Candida, and Saccharomyces dominated the fungal communities of silages. Wickerhamomyces and Candida were the dominant genera in oat silages from BM and SC, respectively. Erwinia, Paenibacillus, Pseudomonas, Leuconostoc, and Exiguobacterium dominated the bacterial community of fresh oat, while Lactobacillus and Kosakonia were the dominant bacterial genus in oat silages. Pantoea was the most dominant bacterial genus in fresh oat from low-elevational regions (BM, BY, and DZ). Oat from SN exhibited the best fermentation quality although fresh oat of SN hosted the lowest LAB counts, indicating that high-efficient LAB might be present in fresh oat sampled from high altitudes.
The Qinghai-Tibet Plateau is the largest and highest plateau with a unique and fragile ecosystem because of high ultraviolet radiation, low precipitation, and low temperatures, which restrained crop growth (Wu et al., 2022). Tibetan living on the Qinghai-Tibetan Plateau rely for survival on the yaks (Bos grunniens) since ancient times (Guo et al., 2014), which is a unique bovine well adapted to the harsh alpine environmental conditions and extensive grazing management all year round. However, the dramatic climate change and intensive grazing activities in past decades have caused gradual rangeland degradation, reducing the availability of herbage (Ding et al., 2014). The reallocation and transporting of forage became an effective management practice to prevent grassland degradation and ensure the sustainability of the livestock industry. Cultivation and ensiling forage crops in arable regions of the Qinghai-Tibet Plateau would provide the available forage resources for the spatiotemporal allocation of forages.
Oat (Avena sativa L.) is a common forage crop cultivated on the Qinghai-Tibet Plateau because of its strong drought resistance, short growth cycle, high forage yield, good palatability, and high nutritional value. Ensiling whole-crop oat has attracted a growing interest in the Qinghai-Tibet Plateau, and oat silage became an important relief and emergency fodder to alleviate the forage shortage of cattle and sheep during the cold season. Ensiling fermentation is a traditional technique to preserve the original composition of nutrients based on LA fermentation under anaerobic conditions. The harsh natural environment and unique climates might contribute to the difference in the epiphytic microbial community and chemical composition of forages between the Qinghai-Tibet Plateau and other regions worldwide (Ding et al., 2020). Moreover, the large elevation gradient across the Qinghai-Tibet Plateau enlarged the discrepancy in microbial and chemical composition in crops cultivated at different elevations. Previous studies revealed the altitudinal distribution patterns of phyllosphere microbial communities and silage fermentation of the wild grass of Kobresia pygmaea and Elymus nutans along the elevation gradient on the Qinghai-Tibet Plateau (Ding et al., 2020; Yang X. et al., 2022). However, there are limited studies on the altitudinal distribution patterns of phyllosphere microbial communities and chemical composition in cultivated grass, which might be different from that of wild grass because more human activities were involved in the cultivation during the crop growth.
The study aimed to investigate the microbial community and chemical composition of fresh and ensiling oat harvested from six different elevations of the Qinghai-Tibet Plateau. It was hypothesized that the microbial community and chemical composition were different among elevations, which contributed to the discrepancy in the fermentation quality of oat silage harvested from six regions of the Qinghai-Tibetan Plateau.
The oat was grown at six different sites across arable regions of Qinghai-Tibet Plateau (BM, Bomi County; BY, Bayi County; DZ, Dazi County; BR, Biru County; SC, Suo County; SN, Seni County), where the elevation was in the range of 2,800–4,500 m above sea level (a. s. l.). The geographic location and climate characteristics of these sites are shown in Supplementary Table S1. The oat variety of Avena sativa L. cv. Qingyin No. 1 was planted in six sites and underwent similar field management measures. Oat was harvested at the milk stage from 20 August to 5 September 2021. Three plots (100 m × 50 m block) were set for each site, and the oat from each plot was chopped in 1–2 cm lengths and packed into polyethylene plastic bags (dimensions 270 mm × 300 mm) followed by vacuum-sealing. The three plots served as replications for each elevation. The bags were taken to Lhasa and stored for 90 days at an ambient temperature (10–24°C). The fresh forages from each plot were sampled for chemical analyses, microbial counting, and DNA extraction.
The silage bags were opened on d 90 of ensiling, and all silages of each bag were put into an ethanol-sterilized plastic container and mixed thoroughly. Then all silages were divided into 4 sub-samples. The first sub-sample was dried at 65°C in a forced-air oven for 48 h to determine DM contents, the dried samples were ground through a 1-mm sieve by a laboratory knife grinder. The ground samples were preserved for chemical analyses. Total nitrogen (TN) content was analyzed with a Kjeltec 8,400-Analyzer (FOSS Analytical AB, Höganäs, Sweden), and the crude protein (CP) was calculated by TN × 6.25 (Krishnamoorthy et al., 1982). The water-soluble carbohydrate (WSC) content was quantified according to the method of Thomas (1977). The neutral detergent fiber (NDF) and acid detergent fiber (ADF) were quantified according to the Van Soest procedures (Van Soest et al., 1991).
The second sub-sample (20 g wet basis) was extracted with 60 ml distilled water at 4°C for 24 h. The solution was filtered through four layers of medical gauze and Whatman filter paper (Hangzhou Xinhua Co., LTD., China), followed by measuring pH immediately using a pH electrode (Hanna Instruments Italia Srl, Padua, Italy). Then silage extract (5 ml) was centrifuged at 12000× g for 10 min at 4°C, and the supernatant was filtered through a 0.45 μm membrane for organic acid and ethanol analyses according to Yang X. et al. (2022). Ammonia N of silage extract was measured by the phenol-hypochlorite method according to Broderick and Kang (1980).
The third sub-sample (10 g) was homogenized with 90 ml of sterile sodium chloride solution (0.85%) for 1 min, followed by filtering through four layers of cheesecloth. Then the filtrate was 10-fold serially diluted for microbial counting, and another solution was used for microbial DNA extraction. The LAB was counted on deMan, Rogosa, and Sharp agar after 48 h of anaerobic incubation at 37°C. The number of yeasts and molds was determined on Potato Dextrose Agar (PDA) medium after 48–72 h of aerobic incubation at 28°C. Enterobacteriaceae were counted on purple-red bile glucose agar after 24 h of incubation at 37°C under aerobic conditions. Aerobic bacteria were counted on the nutrient agar medium (Qingdao Haibo Biotechnology Co., Ltd) after aerobic incubation for 48 h at 37°C. All of the microbial data were transformed to log10 for presentation and statistical analysis.
Microbial DNA was extracted using the Fast DNA SPIN Kit for Soil (MP Biomedicals, Solon, OH, USA). The bacterial 16S rRNA V3–V4 and fungal ITS regions were amplified with primers 338F-806R and ITS1F-ITS2aR, respectively. After purification, the purified PCR amplicons were paired-end sequenced using the Illumina MiSeq PE300 platform (Illumina Inc., San Diego, CA, USA). All raw reads were checked to discard low-quality sequences (quality scores <20) using FLASH (Version 1.2.11) and QIIME (Version 1.7.0). Operation taxonomic units (OTUs) were clustered based on a 97% sequence similarity cutoff using UPARSE (Version 7.1).1 Then the chimeric sequences were identified and removed using UCHIME. Bacterial and fungal compositions were analyzed at genus levels using the Silva and Unite database with a confidence threshold of 70%, respectively. Alpha-diversity estimates (Chao1, and Shannon) and beta-diversity evaluation, based on principal coordinate analysis (PCoA), were performed using the Phyloseq and Vegan packages on R. All DNA sequences have been deposited in the NCBI Short Read Archive database under BioProject PRJNA910079.
The experiment was a completely randomized design, all data from chemical composition, microbial populations, microbial diversity, and fermentation were analyzed by one-way analysis of variance (ANOVA) using the GLM procedure of SAS (version 9.3; SAS Institute Inc., Cary, NC). A polynomial contrast was used to test the linear or quadratic effects of elevation gradient on the parameters measured. The differences between means were assessed by Tukey’s multiple comparisons (p < 0.05).
The chemical composition and microbial populations of fresh oat harvested from different sites of the Qinghai-Tibet Plateau are listed in Table 1. The DM contents of fresh oat ranged from 232 to 310 g kg−1 of fresh weight (FW), and the elevation gradient exhibited a linear effect on DM content (p < 0.001). Among all fresh forages, both NDF (p = 0.008) and ADF (p = 0.003) in fresh oat harvested from BR were the highest. The CP content was affected by elevation gradient with a linear and quadratic effect (p < 0.001). The elevation gradient exhibited a quadratic effect on WSC content (p < 0.001). The elevation gradient significantly affected the numbers of LAB with linear and quadratic effects (p < 0.001). The numbers of yeast and aerobic bacteria were significantly affected by the elevation gradient (p < 0.01).
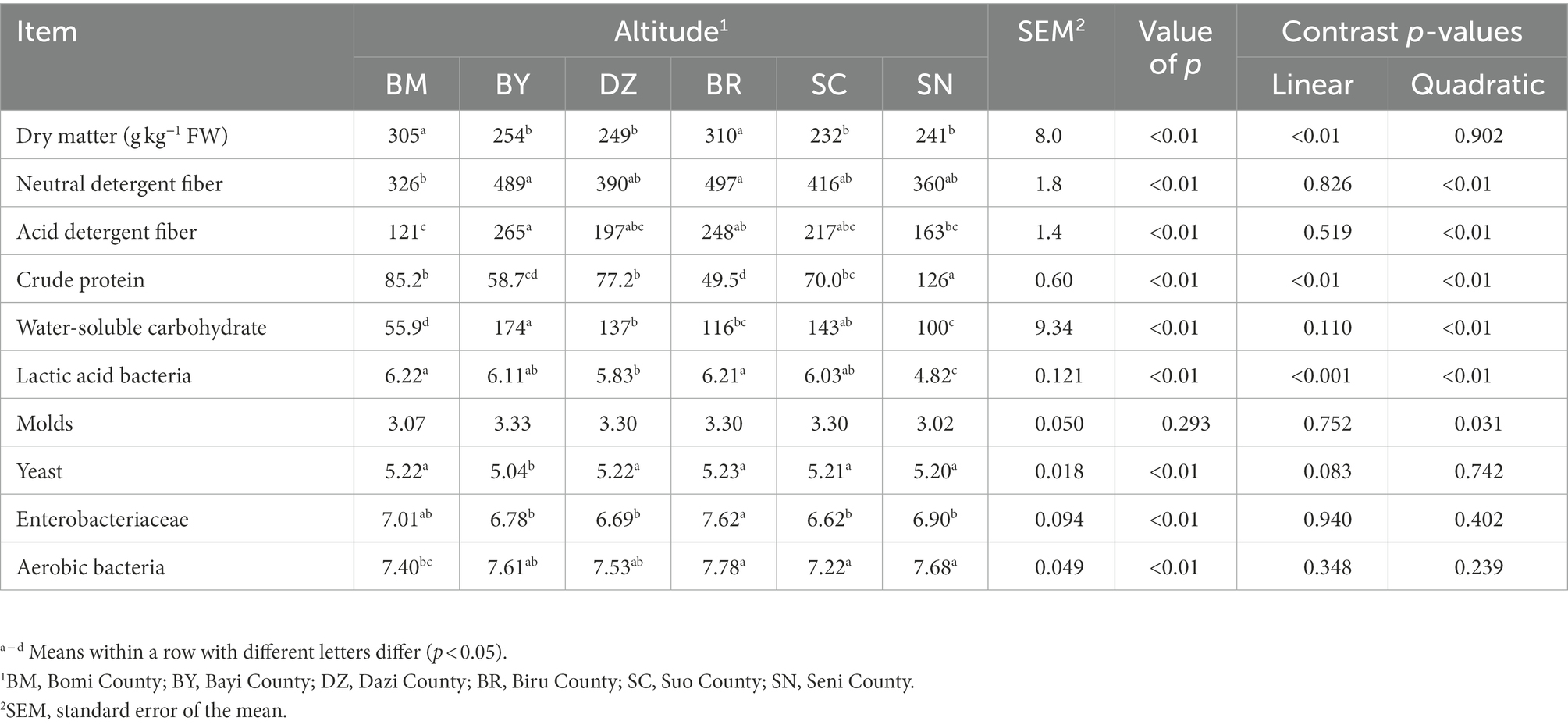
Table 1. The chemical composition (g/kg DM basis unless stated otherwise) and microbial populations (log10 cfu/g FW) of fresh oat (Avena sativa L.).
The fermentation quality, microbial population, and chemical composition of oat silage are listed in Table 2. There was a quadratic effect (p < 0.01) of elevation gradient on the DM and WSC contents. The DZ and SC silages had the lowest DM content, while the BR silage had the highest WSC content among all silages. The ammonia N concentration exhibited a quadratic (p < 0.01) response to the elevation gradient with the lowest value in BR silage. The amounts of LAB (p = 0.01) and aerobic bacteria exhibited a quadratic response to the elevation gradient (p < 0.01).
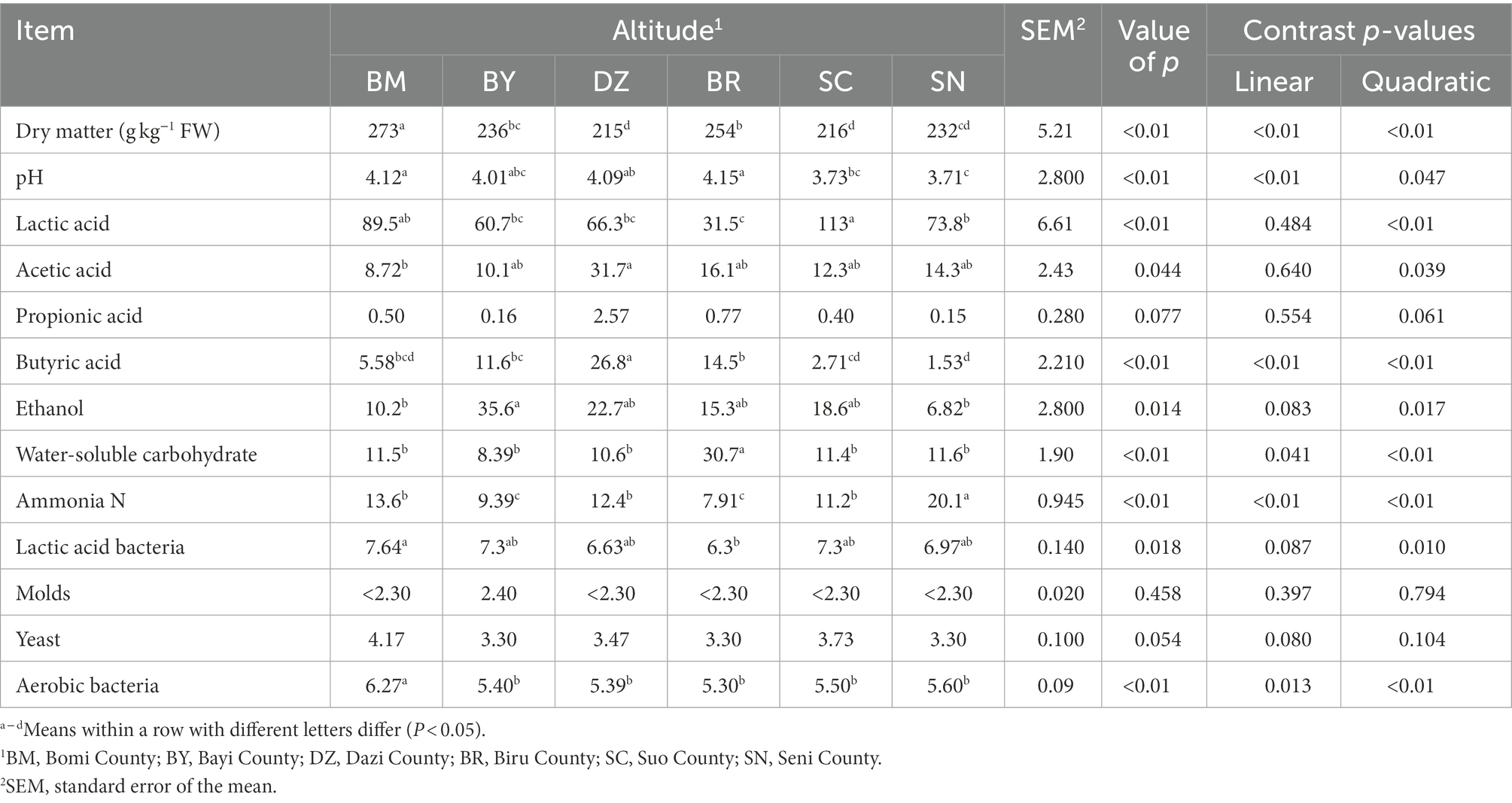
Table 2. The chemical composition, fermentation quality (g/kg DM basis unless stated otherwise), and microbial populations (log 10 cfu/g FW) of ensiled oat (Avena sativa L.).
The silage pH decreased in linear (p < 0.01) and quadratic (p = 0.047) manners along the elevation gradient. The LA concentration was affected by elevation gradient (p < 0.01) with the highest LA values in SC. The AA concentration exhibited a quadratic (p < 0.01) response to the elevation gradient with the highest AA concentration in DZ. The elevation gradient quadratically affected (p < 0.01) the butyric acid concentration. The ethanol concentration exhibited a quadratic (p = 0.017) response to the elevation gradient with the highest value in BY.
We obtained 2,662,561 and 2,876,965 reads by amplicon sequencing of bacterial 16S rRNA V3-V4 and fungal ITS regions, respectively. The greater coverage (>99%) and plateaued rarefaction curves for all samples indicated that the sequencing depth was adequate for reliable analysis of the bacterial and fungal community. The α-diversity indexes of the bacterial and fungal communities are shown in Figure 1; Supplementary Table S2. The bacterial Shannon index in fresh oat of BR was the lowest among all fresh materials while the bacterial Shannon index in oat silage of SN was the lowest among all silages. The bacterial Chao1 index decreased after 90 days of ensiling. Both fresh and ensiled oat from BR showed the lowest bacterial Chao1 index. The fungal Shannon was similar (p > 0.05) among all fresh materials while that in BM and SC silages was lower than in other silages. Fresh oat from BY and BR showed the highest and lowest Chao 1 index among all fresh materials while the elevation gradient did not affect (p > 0.05) the fungal Chao 1 index of silages.
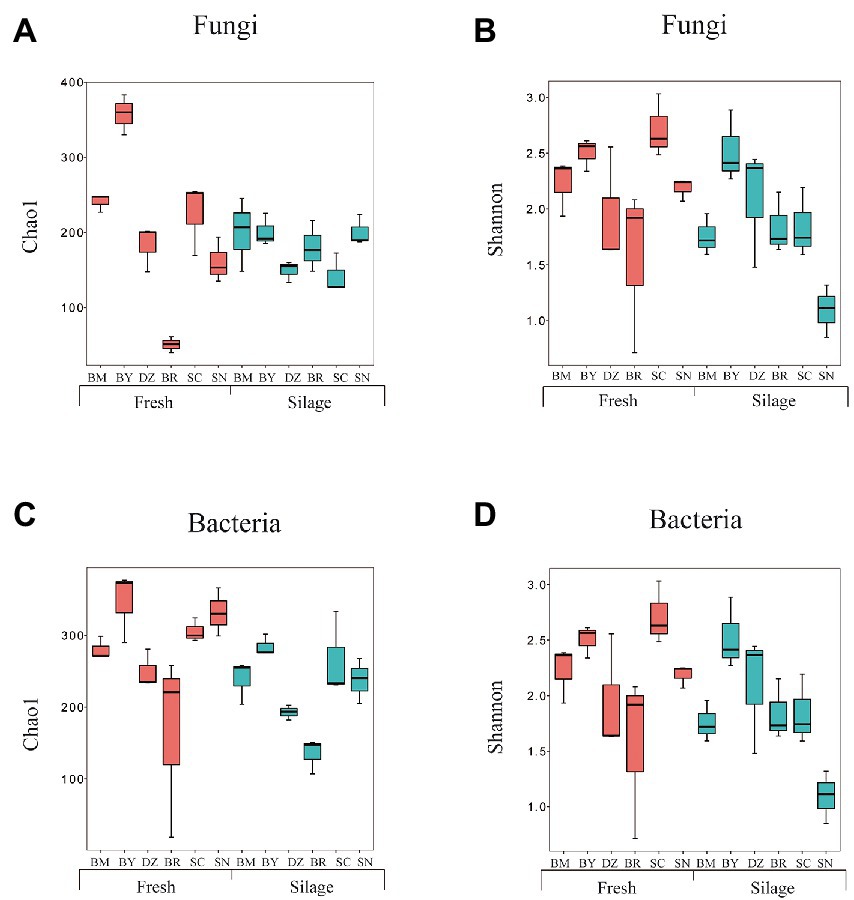
Figure 1. Alpha diversity of fungal (A,B) and bacterial (C,D) community diversities of fresh and ensiled oat along the elevation gradient on the Tibetan Plateau. BM, Bomi County; BY, Bayi County; DZ, Dazi County; BR, Biru County; SC, Suo County; SN, Seni County.
The PCoA plot on the unweighted UniFrac showed the separation of bacterial and fungal communities of fresh and ensiling oat (Figure 2). Two principal components accounted for 54.07% of the variation in taxonomic composition among samples, the PC1 and PC2 axis explained 35.18 and 18.89% of the total variation of the bacterial community, respectively (Figure 2B). The fresh oat and 90-days silages were separated in the plot. Among fresh oat, oat harvested from SN was separated from oat of other regions. The silages of SN and SC were clustered together and separated from oat silages from other regions. Two principal components (PC1 and PC2) accounted for 37.42% of the variation in taxonomic composition among samples (22.21 and 15.21%, respectively) for the fungal community (Figure 2A). The fresh forages were clustered in the second and third quadrants, while all silages were dispersed in the first and fourth quadrants.
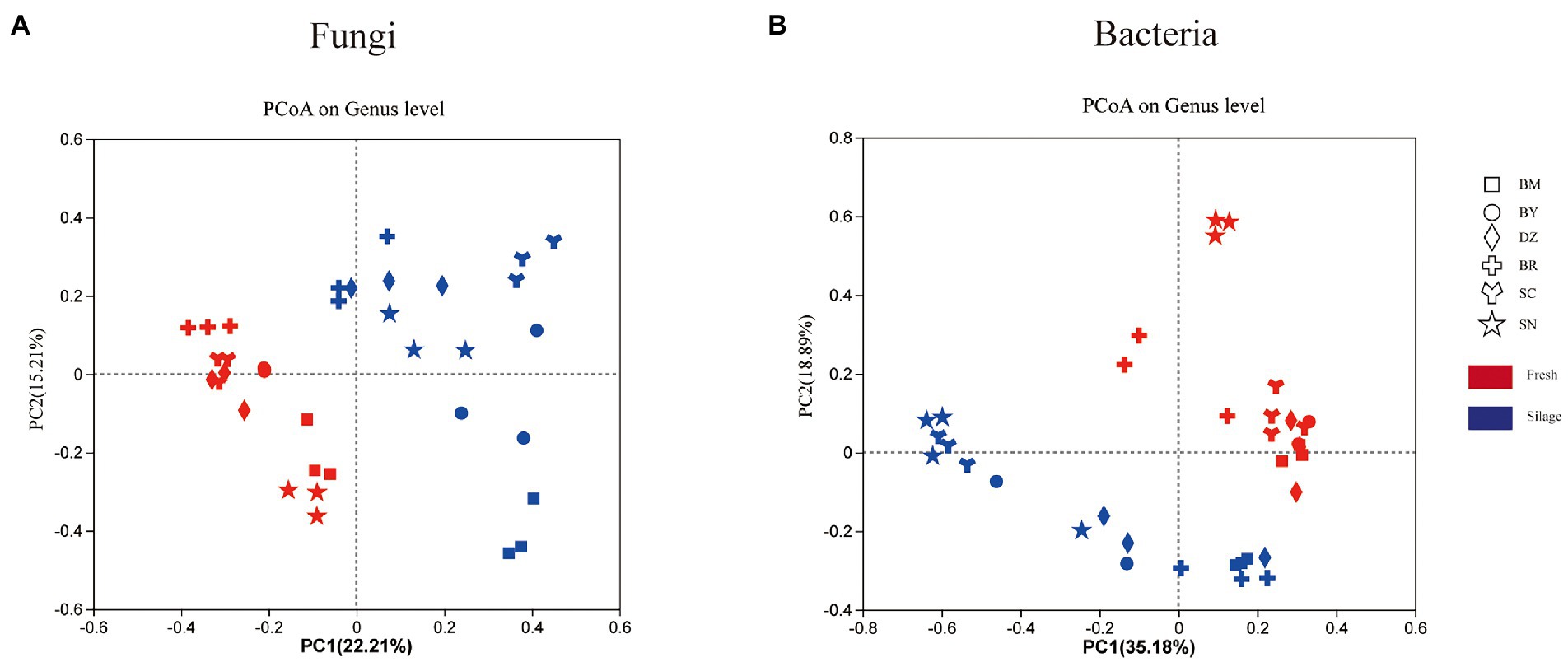
Figure 2. The unweighted Principal coordinate analysis (PCoA) of fungal (A) and bacterial (B) communities of fresh and ensiled oat along the elevation gradient on the Tibetan Plateau. BM, Bomi County; BY, Bayi County; DZ, Dazi County; BR, Biru County; SC, Suo County; SN, Seni County.
The bacterial community composition in fresh and ensiled oat at the genus level are shown in Figure 3. Erwinia, Paenibacillus, Pseudomonas, Leuconostoc, and Exiguobacterium dominated the bacterial community of fresh oat, while Lactobacillus and Kosakonia were the dominant bacterial genus in oat silages (Figure 3B). Pantoea was the most dominant bacterial genus in fresh oat harvested from low elevational regions (BM, BY, and DZ), and its relative abundances (RA) decreased to 4.9, 17.7, and 3.1% in BR, SC, and SN, respectively. After 90 days of ensiling, the RA of Pantoea marked declined except for the oat silage of BR. The RA of Lactobacillus in fresh oat was lower than 5%, it markedly increased and became the most dominant genus in oat silages of SC (71.2%) and SN (83.6%) after 90 days of ensiling. Serratia was the prevalent bacterial genus in fresh oat, and its RA significantly increased in oat silages of low-elevational regions (BM, BY, DZ, and BR) after 90 days of ensiling. Erwinia was present in all fresh oat, and its RA decreased after 90 days of ensiling. Paenibacillus, Pseudomonas, and Exiguobacterium were detectable in all fresh oat and disappeared in all silages. Leuconostoc was the most dominant genus in fresh oat from BR, while it was undetectable in fresh oat from other elevations.
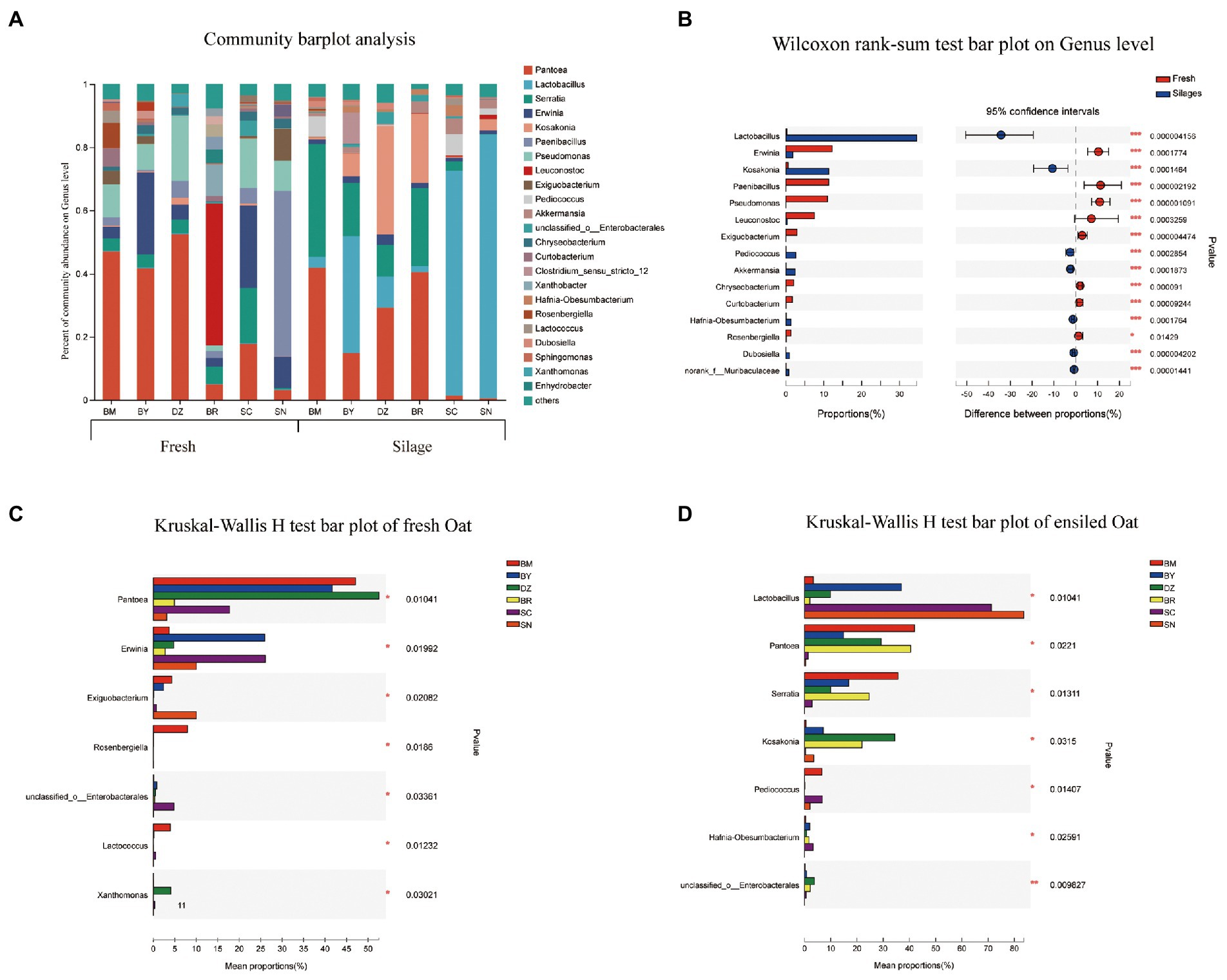
Figure 3. Bacterial community composition of fresh and ensiled oat along the elevation gradient on the Tibetan Plateau. (A) The bar plots of bacterial community composition of fresh and ensiled oat at the genus level. (B) Extended error bar plot showing the most abundant Bacterial genus that had significant differences between fresh and ensiled oat. Positive differences in mean relative abundance indicate the Bacterial genus overrepresented in fresh oat, while negative differences indicate greater abundance in oat silages. (C) Relative abundances of bacterial genera showed significant differences among fresh oat. A one-way ANOVA was used to evaluate the significance of differences between the indicated groups. *p < 0.05,**p < 0.01,***p < 0.001. (D) Relative abundances of bacterial genera showed significant differences among oat silages. A one-way ANOVA was used to evaluate the significance of differences between the indicated groups. *p < 0.05,**p < 0.01,***p < 0.001.
The fungal community composition at the genus level in fresh and ensiled oat are shown in Figure 4. Dioszegia, Cladosporium, and Vishniacozyma were the prevalent fungal genus in fresh oat with different RA among elevation gradients. The RA of Dioszegia was increased from 2.6% in fresh oat of BM to 44.9% in fresh oat of SC. Cladosporium was the most dominant fungal genus in BY and decreased along the elevation gradient. Vishniacozyma was the most dominant fungal genus in BM, accounting for RA of 23.1%. After 90 days of ensiling, all 3 of Dioszegia, Cladosporium, and Vishniacozyma were still detectable in silages, however, the dominant role was replaced by other fungal genera. Wickerhamomyces, Candida, and Saccharomyces dominated the fungal communities of silages although they present in very low abundance in fresh oat (Figure 4B). Wickerhamomyces and Candida were the dominant genera in ensiling oat from BM and SC, respectively.
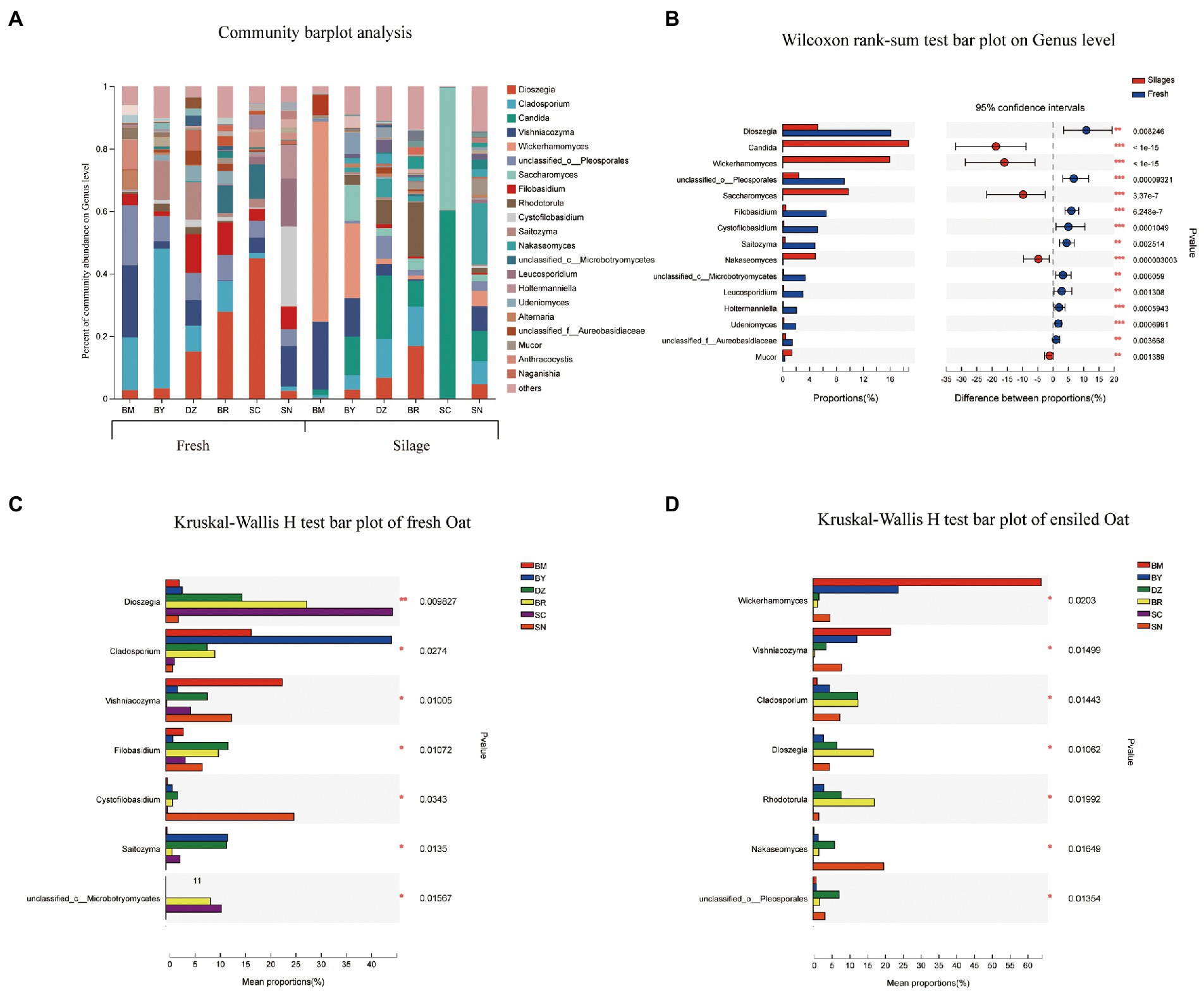
Figure 4. Fungal community composition of fresh and ensiled oat along the elevation gradient on the Tibetan Plateau. (A) The bar plots of fungal community composition of fresh and ensiled oat at the genus level. (B) Extended error bar plot showing the most abundant Fungal genus that had significant differences between fresh and ensiled oat. Positive differences in mean relative abundance indicate the Fungal genus overrepresented in fresh oat, while negative differences indicate greater abundance in oat silages. (C) Relative abundances of fungal genera showed significant differences among fresh oat. A one-way ANOVA was used to evaluate the significance of differences between the indicated groups. *p < 0.05,**p < 0.01,***p < 0.001. (D) Relative abundances of fungal genera showed significant differences among oat silages. A one-way ANOVA was used to evaluate the significance of differences between the indicated groups. *p < 0.05,**p < 0.01,***p < 0.001.
The elevation is a complicated, indirect gradient along which several environmental variables shift, resulting in a basic diversity gradient trend in plant biogeography (Afzal et al., 2021). The low temperature at high elevational regions restrains plant respiration and results in the accumulation of carbohydrates, proteins, and ether extract in the cell protoplasm, which will be beneficial to decrease its freezing point and enhance its adaptive resistance to cold (Li et al., 2014). Plants growing at higher elevations require relatively more energy to respond to serious environmental stress (Marini et al., 2009). Ding et al. (2020) observed an increase in WSC and CP in fresh E. nutans along the increasing elevation. The highest CP and lowest WSC were observed in fresh oat of SN and BM, respectively, however, we did not find a distinct gradient trend in WSC and CP concentrations along the elevation gradient. In the present study, oat is a cultivated forage crop for livestock in the Qinghai-Tibetan plateau, its chemical compositions are not only affected by climate induced by elevation gradients but also influenced by the human activities. Fu et al. (2022) studied the effects of climate change and human activities on forage nutritional quality, they found that the effects of climate change and human activities on nutritional quality of forage were indistinguishable across the whole Tibet, however, human activities altered the sensitivities of forage nutritional quality to climate change.
Sufficient WSC and LAB are the crucial factors for silage fermentation, the WSC in all fresh oat except for BM was above 100 g/kg, which is higher than the recommended level of WSC for well silage fermentation (Woolford, 1984). The LAB counts in fresh oat were higher than 4.8 log10 cfu/g FW, which is close to the recommended counts of LAB (105 cfu/g FM) for well-fermented silages (You et al., 2021).
The lowest LAB counts in fresh oat from the highest elevational regions of SN might be related to the harsh environment because microbes must overcome the stress induced by the extreme climate of high-altitude regions. Some microorganisms still could adapt to these extreme conditions including low temperatures, high levels of solar radiation, periodic freeze–thaw cycles, and nutrient limitations. Liu et al. (2022) reported that nearly 1,000 microbe species were discovered in ‘extreme’ Tibetan glaciers, and 82% of the genomes were novel species.
After 90 days of ensiling, the pH in all oat silages was lower than 4.2, indicating all oat silage exhibited well fermentation quality. Of six oat silages, silages from SC and SN showed the lowest pH and butyric acid concentration, and higher LA concentration. Oat from SN exhibited the best fermentation quality although the fresh oat of SN hosted the lowest LAB counts, indicating that high-efficient LAB might be present in fresh oat sampled from high altitudes. Ding et al. (2020) also observed a higher LA concentration in E. nutans silages sampled from Naqu (altitude of 4,752 m) than in other low altitudes (<4,228 m), and they attributed the high LA concentration to the greater LA-fermentation efficiency of LAB on this region. The oat silage from BR showed the lowest LA concentration and the highest pH, which were in line with its highest residual WSC concentration after 90 days of ensiling, indicating the complex epiphytic microbial composition might retard the LA fermentation.
In the present study, butyric acid was observed in all silages, indicating all oat silages had undergone a clostridial fermentation. The production of butyric acid usually results in high losses of dry matter and digestible energy (Vissers et al., 2007). High butyric acid sometimes could induce ketosis in lactating cows. Silage with a high butyric acid is also less palatable and can also promote the onset of ketosis in lactating cows. The oat silage from DZ showed the highest butyric acid concentrations, which was related to its lowest DM content. Buxton and Muck (2003) reported that moist silage (>70% moisture) usually is associated with poor fermentation dominated by undesirable butyric acid-forming bacteria. The highest ethanol content in oat silage of BY might be related to the high WSC in fresh oat. de Oliveira et al. (2016) reported that in some cases, the higher WSC content of forage could cause acid silage and increase ethanol contents due to yeast activity, however, excess ethanol negatively affected the silage quality because it resulted in a decrease in DM intake.
The elevational patterns for plant and animal diversity generally follow a certain pattern, however, microbes do not follow similar and clear elevational diversity patterns with plants and animals (Fierer et al., 2011). The inconsistency of phyllosphere microbial diversity patterns across elevations could be attributed to changing environmental factors induced by elevational variables (Yang N. et al., 2022). Yang X. et al. (2022) found that the phyllosphere bacterial and fungal richness showed a unimodal distribution, phyllosphere fungal diversity decreased while the phyllosphere bacterial diversity increased along the elevational gradient. We observed the bimodal distribution of fungal and bacterial richness in fresh oat along the elevation gradient (2852–4447 m a.s.l). Vacher et al. (2016) demonstrated that the higher dispersal ability of bacteria and higher sensitivity of fungi to temperature variations might account for the bigger response of phyllosphere fungal communities to elevation gradient than bacterial communities. In the present study, the elevation gradients did not affect the fungal Shannon index in fresh oat (p > 0.05), which was consistent with the reports by Yang X. et al. (2022), who also detected similar soil fungal Shannon index across elevation gradients.
In the study, Pantoea dominated (>41%) the fresh oat of lower elevations (BM, BY, and DZ). This is in line with the reports of Ding et al. (2020), who reported that Pseudomonas and Pantoea were the dominant bacterial genera in fresh E. nutans of Tianzhu (2,965 a.s.l.). Cheng et al. (2022) also found that Pantoea was the main microorganisms of fresh oat harvested from Hongyuan on the Qinghai-Tibetan Plateau, where the altitude is 3,500 m a.s.l. Keshri et al. (2019) reported that Pantoea (34.7%) was the dominant genera in freshly chopped wheat plants, the RA of Pantoea increased to 46.3% after 6 h of ensiling, but then decreased continuously during silage maturation and they represented only 0.02% of the overall population at the terminal stage of 90 days. After 90 days of ensiling, Pantoea in oat silages from BM, BY, and DZ decreased to the second dominant bacterial genus. Pantoea species are gram-negative bacteria from the Enterobacteriaceae family, generally associated with plants, and are recognized as undesirable bacteria of silages because they can compete for nutrients with LAB (Li et al., 2018). In the study, Pantoea were not suppressed and remained at the high RA in BM, BY, DZ, and BR, however, it decreased to 1.4 and 0.4% in SC and SN, respectively. This was related to the flourishing of Lactobacillus, which confirmed the previous assumption that high-efficient LAB might be present in fresh oat sampled from high altitudes.
Lactobacillus was the minor genus in all fresh oat before ensiling, but its RA increased to 71.2 and 83.6% in oat silages of SC and SN, respectively. This was in line with the high acid concentration and low pH in oat silages of SC and SN. Previous studies also reported that Lactobacillus dominated the terminal silages (Guan et al., 2018; Hisham et al., 2022).
Serratia was the prevalent bacterial genus while Kosakonia was the minor bacterial genus in fresh oat, however, their RA increased during 90 days of ensiling. Little is known about the role of Serratia during ensiling, Li H. Z. et al. (2022) reported that Serratia could grow and survive under anaerobic conditions, and observed the increment of Serratia after ensiling. Duan et al. (2020) reported that Serratia spoiled chicken breasts were effectively inhibited by an antimicrobial substance produced by Lactobacillus. In the present study, the lower RA of Serratis in silage of SC and SN might be related to the dominant bacteria of Lactobacillus. Li H. Z. et al. (2022) also reported that silages with a higher RA of Lactobacillus showed a lower RA of Serratia and they proclaimed that Serratia was acid intolerant. Kosakonia is a genus of the Enterobacteriaceae family and has been observed by many researchers (Xiong et al., 2022). Kosakonia has been proven to have the ability to reduce ammonia nitrogen and volatile chemicals in silage (Zhang et al., 2021). In the present study, the lower ammonia N in oat silages of BY, DZ, and BR might be attributed to the higher RA of Kosakonia than other silages.
Erwinia was present in all fresh oat, however, certain species of Erwinia do not grow below pH 5.0 (Ogunade et al., 2018), supporting the decline of Erwinia after 90 days of ensiling in the present study. Enterobacteria, including Erwinia herbicola and Rahnella aquitilis, often were observed as dominant bacteria in fresh crops, however, they would be superseded by other genera during ensiling such as Escherichia coli, Hafnia alvei, and Serratia fonticola (Driehuis and Elferink, 2000).
Paenibacillus, Pseudomonas, and Exiguobacterium were detectable in all fresh oat, however, they disappeared in all silages. The genera of Paenibacillus, Pseudomonas, and Exiguobacterium were found in cold habitats (Rawat et al., 2020), they usually were inhibited during ensiling because of their intolerance to low pH (Borreani et al., 2013). Keshri et al. (2019) reported that Pantoea, Weissella, Pseudomonas, Exiguobacterium, and Paenibacillus dominated the bacterial community of fresh whole-crop wheat, however, they were replaced by Lactobacillus in the terminal silage.
Leuconostoc was the most dominant genus in fresh oat of BR, however, it was undetectable in fresh oat of other elevations. Cai et al. (1998) reported Leuconostocs were the most numerous and widely distributed on forage crops and silage. Leuconostoc could grow vigorously during the early stage of ensiling to initiate the silage fermentation, creating an aerobic and acidic environment for the proliferation of Lactobacilli. In the present study, it is not clear why Leuconostoc was the most dominant genus in fresh oat from BR rather than other elevations.
The study revealed the change in the fungal community between fresh and ensiled oat, the RA of Dioszegia in fresh oat increased with the elevation gradient and peaked at fresh oat from SC. This might be attributed to their cold-adapted properties. Trochine et al. (2017) isolated several species of the Dioszegia genus from glacier surface snow and found these yeasts showed strong cold-adapted capacity. After 90 days of ensiling, the RA of Dioszegia decreased, indicating they are unadaptable to the ensiling environment. Duniere et al. (2017) reported that the potential mycotoxigenic fungi of Cladosporium were the core microbiome in fresh small grains, but their RA markedly declined during ensiling. In the present study, the RA of Cladosporium spp. decreased during 90 days of ensiling. Vishniacozyma (also known as Cryptococcus) has been isolated from soil and wheat, but there are limited reports regarding this genus in silage. It is reported that Vishniacozyma could assimilate lactic acid and D-lactose (Tian et al., 2022; Li X. et al., 2022). Vishniacozyma was observed in both fresh and ensiled oat in the study, indicating the fresh oat might be subjected to soil contamination before ensiling. Filobasidium was detected in all fresh oat with the highest RA in fresh oat of DZ and BR, however, it markedly decreased after 90 days of ensiling. This is in agreement with the reports of Xu et al. (2019), who found that Filobasidium was an abundant genus in fresh corn and declined with the ensiling progress. Because of its psychrophilic abilities, Cystofilobasidium was detected in fresh oat harvested from the highest elevations with the lowest average annual temperature (Nakagawa et al., 2005). Wickerhamomyces, Candida, Saccharomyces, and Rhodotorula were the minor genus in fresh oat before ensiling, however, they became the major fungal community after 90 days of ensiling. Yang X. et al., (2022) studied the phyllosphere microbial communities and silage fermentation of K. pygmaea on the Tibetan Plateau and found that the fungal composition markedly changed: Schizophyllum, Phodotorula, Rhodotorula, Candida, and Issatchenkia became the most dominant fungal genera after 60 days of ensiling. Duniere et al. (2017) reported that members of the Saccharomycetales including Candida, Wickerhamomyces, or Saccharomyces were the predominant fungi in terminal silage. Wickerhamomyces are present in diverse habitats and frequently associated with the processing of food and grain products. Agarussi et al. (2022) reported that the predominance of fungi of Wickerhamomyces during the fermentation of sorghum grains was due to their tolerance to extreme environmental conditions. Saccharomycetales spp. dominated the fungal community after 90 days of ensiling of small grain (Duniere et al., 2017). Yang X. et al. (2022) also reported that Rhodotorula became the dominant fungal genus in K. pygmaea 60-days silages harvested from 5,000 m a. s. l. This might be related to their cold-adaptability, Hu et al. (2015) isolated cold-tolerant yeast of Rhodotorula mucilaginosa from soil samples collected on the Qinghai-Tibet Plateau.
The highest crude protein (CP) and lowest water-soluble carbohydrate (WSC) were observed in fresh oat from SN and BM, respectively, however, no distinct gradient trend in WSC and CP concentrations along the elevation gradient. The bimodal distributions of fungal and bacterial richness in fresh oat along the elevation gradient were observed, while the elevation gradients did not affect the fungal Shannon index in fresh oat. Oat from SN exhibited the best fermentation quality although the fresh oat of SN hosted the lowest LAB counts, indicating that high-efficient LAB might be present in fresh oat sampled from high altitudes. The high-efficient LAB can be further isolated from the Qinghai-Tibet Plateau, especially the higher elevational regions where the microorganisms hold a strong tolerance to the extreme environment.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
TS: conceiving the idea, designing the experiment, and writing, reviewing, and editing. YB and ZYa: sampling and analysis, data analysis, writing, reviewing, and editing. ZYu: formal analysis, methodology, review, and editing. RS and PX: writing, reviewing, and editing. ZYu, RS, and KF: sampling, analysis, and editing. All co-authors participated in discussions and revised the manuscript, contributed to the article, and approved the submitted version.
This research was supported by the National Natural Science Foundation of China (31960355 and 32160780), The Tibetan Key R&D Program (XZ202001ZY0047N), and the State Key Laboratory of Barley and Yak Germplasm Resources and Genetics Improvement (Tibet Academy of Agricultural and Animal Husbandry Sciences) (XZNKY-2019-C-007K09).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1108890/full#supplementary-material
Afzal, S., Nesar, H., Imran, Z., and Ahmad, W. (2021). Altitudinal gradient affect abundance, diversity and metabolic footprint of soil nematodes in Banihal-pass of Pir-Panjal mountain range. Sci. Rep. 11:16214. doi: 10.1038/s41598-021-95651-x
Agarussi, M. C. N., Pereira, O. G., Pimentel, F. E., Azevedo, C. F., Da Silva, V. P., and Silva, F. F. E. (2022). Microbiome of rehydrated corn and sorghum grain silages treated with microbial inoculants in different fermentation periods. Sci. Rep. :12:16864. doi: 10.1038/s41598-022-21461-4
Borreani, G., Dolci, P., Tabacco, E., and Cocolin, L. (2013). Aerobic deterioration stimulates outgrowth of spore-forming Paenibacillus in corn silage stored under oxygen-barrier or polyethylene films. J. Dairy Sci. 96, 5206–5216. doi: 10.3168/jds.2013-6649
Broderick, G. A., and Kang, J. H. (1980). Automated simultaneous determination of ammonia and total amino acids in ruminal fluid and in vitro media. J. Dairy Sci. 63, 64–75. doi: 10.3168/jds.S0022-0302(80)82888-8
Buxton, D. R., and Muck, R. E. (2003). Silage Science and Technology. Madison, WI: American Society of Agronomy
Cai, Y., Benno, Y., Ogawa, M., Ohmomo, S., Kumai, S., and Nakase, T. (1998). Influence of Lactobacillus spp. from an inoculant and of Weissella and Leuconostoc spp. from forage crops on silage fermentation. Appl. Environ. Microbiol. 64, 2982–2987. doi: 10.1128/AEM.64.8.2982-2987.1998
Cheng, Q., Chen, L., Chen, Y., Li, P., and Chen, C. (2022). Effects of LAB inoculants on the fermentation quality, chemical composition, and bacterial Community of oat Silage on the Qinghai-Tibetan plateau. Microorganisms 10:787. doi: 10.3390/microorganisms10040787
De Oliveira, J. S., Santos, E. M., and Dos Santos, A. P. M. (2016). “Intake and digestibility of silages” in Advances in Silage Production and Utilization. eds. T. Silva and E. Santos (London: IntechOpen).
Ding, Z., Bai, J., Xu, D., Li, F., Zhang, Y., and Guo, X. S. (2020). Microbial community dynamics and natural fermentation profiles of ensiled alpine grass Elymus nutans prepared from different regions of the Qinghai-Tibetan plateau. Front. Microbiol. 11:11. doi: 10.3389/fmicb.2020.00855
Ding, L. M., Wang, Y. P., Brosh, A., Chen, J. Q., Gibb, M. J., Shang, Z. H., et al. (2014). Seasonal heat production and energy balance of grazing yaks on the Qinghai-Tibetan plateau. Anim. Feed Sci. Tech. 198, 83–93. doi: 10.1016/j.anifeedsci.2014.09.022
Driehuis, F., and Elferink, S. J. W. H. O. (2000). The impact of the quality of silage on animal health and food safety: a review. Vet. Quart. 22, 212–216. doi: 10.1080/01652176.2000.9695061
Duan, X., Duan, S., Wang, Q., Ji, R., Cao, Y., and Miao, J. (2020). Effects of the natural antimicrobial substance from lactobacillus paracasei FX-6 on shelf life and microbial composition in chicken breast during refrigerated storage. Food Control 109:106906. doi: 10.1016/j.foodcont.2019.106906
Duniere, L., Xu, S., Long, J., Elekwachi, C., Wang, Y., Turkington, K., et al. (2017). Bacterial and fungal core microbiomes associated with small grain silages during ensiling and aerobic spoilage. BMC Microbiol. 17:50. doi: 10.1186/s12866-017-0947-0
Fierer, N., Mccain, C. M., Meir, P., Zimmermann, M., Rapp, J. M., Silman, M. R., et al. (2011). Microbes do not follow the elevational diversity patterns of plants and animals. Ecology 92, 797–804. doi: 10.1890/10-1170.1
Fu, G., Wang, J., and Li, S. (2022). Response of forage nutritional quality to climate change and human activities in alpine grasslands. Sci. Total Environ. 845:157552. doi: 10.1016/j.scitotenv.2022.157552
Guan, H., Yan, Y., Li, X., Li, X., Shuai, Y., Feng, G., et al. (2018). Microbial communities and natural fermentation of corn silages prepared with farm bunker-silo in Southwest China. Bioresour. Technol. 265, 282–290. doi: 10.1016/j.biortech.2018.06.018
Guo, X., Long, R., Kreuzer, M., Ding, L., Shang, Z., Zhang, Y., et al. (2014). Importance of functional ingredients in yak Milk-derived food on health of Tibetan nomads living under high-altitude stress: a review. Crit. Rev. Food Sci. Nutr. 54, 292–302. doi: 10.1080/10408398.2011.584134
Hisham, M. B., Hashim, A. M., Hanafi, N. M., Rahman, N. A., Mutalib, N. E. A., Tan, C. K., et al. (2022). Bacterial communities associated with silage of different forage crops in Malaysian climate analysed using 16S amplicon metagenomics. Sci. Rep 12:7107
Hu, H., Yan, F., Wilson, C., Shen, Q., and Zheng, X. (2015). The ability of a cold-adapted Rhodotorula mucilaginosa strain from Tibet to control blue mold in pear fruit. Antonie Van Leeuwenhoek 108, 1391–1404. doi: 10.1007/s10482-015-0593-1
Keshri, J., Chen, Y. R., Pinto, R., Kroupitski, Y., Weinberg, Z. G., and Saldinger, S. S. (2019). Bacterial dynamics of wheat silage. Front. Microbiol. 10:16. doi: 10.3389/fmicb.2019.01532
Krishnamoorthy, U., Muscato, T. V., Sniffen, C. J., and Van Soest, P. J. (1982). Nitrogen fractions in selected feedstuffs. J. Dairy Sci. 65, 217–225. doi: 10.3168/jds.S0022-0302(82)82180-2
Li, Y., Da Silva, E. B., Li, J., and Kung, L. (2022a). Effect of homo-fermentative lactic acid bacteria inoculants on fermentation characteristics and bacterial and fungal communities in alfalfa silage. Fermentation 8:621. doi: 10.3390/fermentation8110621
Li, X., Yang, Y., Ma, L., Sun, X., Yang, S., Kong, X., et al. (2014). Comparative proteomics analyses of Kobresia pygmaea adaptation to environment along an elevational gradient on the central Tibetan plateau. PLoS One 9:e98410. doi: 10.1371/journal.pone.0098410
Li, H. L., Zeng, T. R., Du, Z. C., Dong, X. T., Xin, Y. F., Wu, Y. S., et al. (2022b). Assessment on the fermentation quality and bacterial Community of Mixed Silage of Faba bean with forage wheat or oat. Front. Microbiol. 13:875819. doi: 10.3389/fmicb.2022.875819
Li, P., Zhang, Y., Gou, W., Cheng, Q., Bai, S., and Cai, Y. (2018). Silage fermentation and bacterial community of bur clover, annual ryegrass and their mixtures prepared with microbial inoculant and chemical additive. Anim. Feed Sci. Tech 247, 285–293.
Liu, Y., Ji, M., Yu, T., Zaugg, J., Anesio, A. M., Zhang, Z., et al. (2022). A genome and gene catalog of glacier microbiomes. Nat. Biotechnol. 40, 1341–1348. doi: 10.1038/s41587-022-01367-2
Marini, L., Gaston, K. J., Prosser, F., and Hulme, P. E. (2009). Contrasting response of native and alien plant species richness to environmental energy and human impact along alpine elevation gradients. Glob. Ecol. Biogeogr. 18, 652–661. doi: 10.1111/j.1466-8238.2009.00484.x
Nakagawa, T., Nagaoka, T., Miyaji, T., and Tomizuka, N. (2005). Cold-active polygalacturonase from psychrophilic-basidiomycetous yeast Cystofilobasidium capitatum strain PPY-1. Biosci. Biotechnol. Biochem. 69, 419–421. doi: 10.1271/bbb.69.419
Ogunade, I. M., Jiang, Y., Pech Cervantes, A. A., Kim, D. H., Oliveira, A. S., Vyas, D., et al. (2018). Bacterial diversity and composition of alfalfa silage as analyzed by Illumina MiSeq sequencing: effects of Escherichia coli O157:H7 and silage additives. J. Dairy Sci. 101, 2048–2059. doi: 10.3168/jds.2017-12876
Rawat, J., Yadav, N., and Pande, V. (2020). “Chapter 7 – Role of rhizospheric microbial diversity in plant growth promotion in maintaining the sustainable agrosystem at high altitude regions” in Recent Advancements in Microbial Diversity. eds. S. De Mandal and P. Bhatt (Cambridge, MA: Academic Press), 147–196.
Thomas, T. A. (1977). An automated procedure for the determination of soluble carbohydrates in herbage. J. Sci. Food Agr. 28, 639–642. doi: 10.1002/jsfa.2740280711
Tian, H., Shi, R. F., Wei, H. Y., Lu, N. N., Sun, W. L., Ren, H. W., et al. (2022). Effects of rumen fluid as bioaugmentation additive on fungal diversity dynamics during sweet sorghum ensiling. Bioresources 17, 2977–2996. doi: 10.15376/biores.17.2.2977-2996
Trochine, A., Turchetti, B., Vaz, A. B. M., Brandao, L., Rosa, L. H., Buzzini, P., et al. (2017). Description of Dioszegia patagonica sp. nov., a novel carotenogenic yeast isolated from cold environments. Int. J. Syst. Evol. Microbiol. 67, 4332–4339. doi: 10.1099/ijsem.0.002211
Vacher, C., Cordier, T., and Vallance, J. (2016). Phyllosphere fungal communities differentiate more thoroughly than bacterial communities along an elevation gradient. Microbial. Ecol. 72, 1–3. doi: 10.1007/s00248-016-0742-8
Van Soest, P. J., Robertson, J. B., and Lewis, B. A. (1991). Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 74, 3583–3597. doi: 10.3168/jds.S0022-0302(91)78551-2
Vissers, M. M., Driehuis, F., Te Giffel, M. C., De Jong, P., and Lankveld, J. M. (2007). Concentrations of butyric acid bacteria spores in silage and relationships with aerobic deterioration. J. Dairy Sci. 90, 928–936. doi: 10.3168/jds.S0022-0302(07)71576-X
Wu, J., Wang, G., Chen, W., Pan, S., and Zeng, J. (2022). Terrain gradient variations in the ecosystem services value of the Qinghai-Tibet Plateau, China. Glob. Ecol. Conservat. 34:e02008. doi: 10.1016/j.gecco.2022.e02008
Xiong, Y., Xu, J., Guo, L., Chen, F., Jiang, D., Lin, Y., et al. (2022). Exploring the effects of different bacteria additives on fermentation quality, microbial community and in vitro gas production of forage oat silage. Animals 12:1122. doi: 10.3390/ani12091122
Xu, S., Yang, J., Qi, M., Smiley, B., Rutherford, W., Wang, Y., et al. (2019). Impact of Saccharomyces cerevisiae and Lactobacillus buchneri on microbial communities during ensiling and aerobic spoilage of corn silage. J. Anim. Sci. 97, 1273–1285. doi: 10.1093/jas/skz021
Yang, N., Li, X., Liu, D., Zhang, Y., Chen, Y., Wang, B., et al. (2022). Diversity patterns and drivers of soil bacterial and fungal communities along elevational gradients in the Southern Himalayas, China. Appl. Soil Ecol. 178:104563. doi: 10.1016/j.apsoil.2022.104563
Yang, X., Bao, Y., Shao, T., Wang, W., Ma, P., Wang, W., et al. (2022). Altitudinal distribution patterns of phyllosphere microbial communities and their contribution to silage fermentation of Kobresia pygmaea along the elevation gradient on the Tibetan Plateau. Front. Microbiol. 13:13. doi: 10.3389/fmicb.2022.874582
You, S. H., Du, S., Ge, G. T., Wan, T., and Jia, Y. S. (2021). Microbial community and fermentation characteristics of native grass prepared without or with isolated lactic acid bacteria on the Mongolian plateau. Front. Microbiol. 12:12. doi: 10.3389/fmicb.2021.731770
Keywords: elevational gradients, fermentation quality, microbial community, oat, Tibetan Plateau
Citation: Bao Y, Yangzong Z, Yuan Z, Shi R, Feng K, Xin P and Song T (2023) The microbial communities and natural fermentation quality of ensiling oat (Avena sativa L.) harvest from different elevations on the Qinghai-Tibet Plateau. Front. Microbiol. 13:1108890. doi: 10.3389/fmicb.2022.1108890
Received: 01 December 2022; Accepted: 19 December 2022;
Published: 11 January 2023.
Edited by:
Siran Wang, Nanjing Agricultural University, ChinaReviewed by:
Yanlin Xue, Inner Mongolia Academy of Agriculture and Animal Husbandry Science, ChinaCopyright © 2023 Bao, Yangzong, Yuan, Shi, Feng, Xin and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tianzeng Song, ✉ c29uZ3RpYW56ZW5nMTIzQHNpbmEuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.