- 1Department of Entomology, RH Smith Faculty of Agriculture, Food and Environment, Hebrew University of Jerusalem, Rehovot, Israel
- 2Department of Entomology, Newe-Ya’ar Research Center, ARO, Ramat-Yishay, Israel
- 3Department of Evolution and Environmental Biology, University of Haifa, Haifa, Israel
- 4The Institute of Plant Sciences, The Volcani Center, ARO, Rishon LeZion, Israel
Many arthropods host bacterial symbionts, some of which are known to influence host nutrition and diet breadth. Omnivorous bugs of the genus Macrolophus (Heteroptera: Miridae) are mainly predatory, but may also feed on plants. The species M. pygmaeus and M. melanotoma (=M. caliginosus) are key natural enemies of various economically important agricultural pests, and are known to harbor two Rickettsia species, R. bellii and R. limoniae. To test for possible involvement of symbiotic bacteria in the nutritional ecology of these biocontrol agents, the abundance, phylogeny, and distribution patterns of the two Rickettsia species in M. pygmaeus and M. melanotoma were studied. Both of the Rickettsia species were found in 100 and 84% of all tested individuals of M. pygmaeus and M. melanotoma, respectively. Phylogenetic analysis showed that a co-evolutionary process between Macrolophus species and their Rickettsia is infrequent. Localization of R. bellii and R. limoniae has been detected in both female and male of M. pygmaeus and M. melanotoma. FISH analysis of female gonads revealed the presence of both Rickettsia species in the germarium of both bug species. Each of the two Rickettsia species displayed a unique distribution pattern along the digestive system of the bugs, mostly occupying separate epithelial cells, unknown caeca-like organs, the Malpighian tubules and the salivary glands. This pattern differed between the two Macrolophus species: in M. pygmaeus, R. limoniae was distributed more broadly along the host digestive system and R. bellii was located primarily in the foregut and midgut. In contrast, in M. melanotoma, R. bellii was more broadly distributed along the digestive system than the clustered R. limoniae. Taken together, these results suggest that Rickettsia may have a role in the nutritional ecology of their plant-and prey-consuming hosts.
1. Introduction
Insects are known to form successful long-term symbioses with endosymbiotic bacteria (Harris et al., 2010; Gupta and Nair, 2020). Such bacteria may be referred to as primary symbionts, if they are obligatory and thus essential for the survival of the host (Sudakaran et al., 2017), or as secondary symbionts, if they are not involved in functions essential for host survival or reproduction. Facultative association with secondary symbionts may affect host biology and ecology by influencing host fitness through altered traits and capabilities (Sudakaran et al., 2017).
Rickettsia (Alphaproteobacteria: Rickettsiales) are gram-negative obligate intracellular bacteria found within eukaryotic cells; they have a variety of interactions with their arthropod hosts (Weinert et al., 2015; El Karkouri et al., 2022). Rickettsia can be found in many insect tissues, including Malpighian tubules, gut compartments, oocytes, and, rarely, sperm cells (Machtelinckx et al., 2012; Watanabe et al., 2014; Dally et al., 2020). While Rickettsia were initially recognized as human and animal pathogens transmitted by blood-feeding arthropods such as ticks, mites, lice and fleas, in recent decades most species and strains have been shown to be non-pathogenic endosymbionts of arthropods (Weinert et al., 2015; Pilgrim et al., 2021).
Most insect hosts of Rickettsia belong to the orders Hemiptera, Coleoptera, Diptera and Hymenoptera (Weinert et al., 2015), in all of which the bacteria have a positive effect on host survival and reproductive success. In the pea aphid Acyrthosiphon pisum (Hemiptera: Aphididae), for instance, Rickettsia improve host resistance to a pathogenic Pandora fungus (Lukasik et al., 2013). In the sweet potato whitefly Bemisia tabaci (Hemiptera: Aleyrodidae), Rickettsia provide protection against Pseudomonas syringae infection (Hendry et al., 2014), and enhance fertility, longevity and development (Himler et al., 2011). Additionally, these bacteria stimulate oogenesis in the booklouse Liposcelis bostrychophila (Psocoptera: Liposcelididae) (Perotti et al., 2006). Rickettsia may also have negative effects on their hosts, for example by slowing development (Semiatizki et al., 2020) or inducing male-killing in the host (Werren et al., 1994; Giorgini et al., 2010).
Within the suborder Heteroptera (Order Hemiptera), the family Miridae includes some 10,000 species with a wide range of feeding habits including herbivory, carnivory, and omnivory (Wheeler, 2002). Several members of this family, such as Nesidiocoris tenuis, Macrolophus pygmaeus and M. melanotoma, serve as biological control agents against key crop pests (Schaefer and Panizzi, 2000; Castañé et al., 2011), yet these mainly predaceous species may also feed on plant materials (Perdikis and Lykouressis, 2000). The potential of these predators to reduce agricultural yields when prey is scarce has limited their use in biological control programs (Castañé et al., 2011; Moerkens et al., 2016). It is therefore important to explore the presence and role of symbionts in the nutritional ecology of these omnivorous biological control agents.
Omnivores, like most insects, serve as hosts to symbiotic bacteria. In previous studies, M. pygmaeus was found to harbor Wolbachia and two species of Rickettsia, R. bellii and R. limoniae. In contrast, M. melanotoma was observed to house only Wolbachia and R. limoniae. Regarding symbiont distribution, Wolbachia are located in the ovaries of M. pygmaeus, where they induce cytoplasmic incompatibility (Machtelinckx et al., 2009, 2012). The two Rickettsia species, in contrast, were found in our previous study to be distributed in both the digestive and reproductive systems of M. pygmaeus, each displaying a unique cellular occupancy and a specific distribution pattern along the digestive system compartment (Dally et al., 2020). The significance of these differences in distribution is not yet clear, but a link to the omnivorous host diet has been proposed. The current study revealed the presence of both Rickettsia species, R. bellii and R. limoniae, in M. melanotoma, in contrast with the earlier findings of Machtelinckx et al. (2012). Accordingly, our objective is to further explore the role of Rickettsia in omnivore diet and nutrition. To this end, the occurrence of Rickettsia species in M. melanotoma was described, their localization patterns were compared to those found in M. pygmaeus, and Rickettsia abundance and phylogeny in the two hosts were determined.
2. Materials and methods
2.1. Insect origin, DNA extraction, and verification of insect identity
For this study, 82 females of M. melanotoma were collected from Dittrichia viscosa (Asteraceae) in various locations in northern and south-central Israel (see Supplementary additional file 1, table of collection sites of M. melanotoma females in Israel). The collected insects were placed immediately in 100% ethanol, and then stored at-20°Cuntil analysis. A culture of M. pygmaeus was established in February 2018 with 30 adult females and 20 adult males obtained from a commercial biological control company (BioBee Sde Eliyahu Ltd., Israel), with occasional infusion of additional insects from the same source (for details see Dally et al., 2020). DNA was extracted from individual insects using the Nucleospin Tissue XS Kit (“Macherey-Nagel,” Switzerland), following the manufacturer’s instructions.
The identity of field-collected M. melanotoma males and females was determined under a stereoscopic microscope, based on morphological characters presented by Martinez-Cascales et al. (2006). In addition, the mitochondrial cytochrome oxidase I (COI) gene fragment was used to verify species identity. COI was amplified from all individuals and sequenced individually using LCO1490 and HCO2198 primers (Table 1). Sterilized water and DNA of Bemisia tabaci served as negative and positive controls, respectively. PCR procedures were carried out following the protocol described by Dally et al. (2020), and sequencing was performed using an automatic sequencer (ABI 3700 DNA analyser, Macrogen Inc.). The resulting sequences were compared with known sequences in the databases, using BLAST searches, and deposited in NCBI GenBank, under accession numbers OQ374915-OQ374918, OQ374921-OQ374924, OQ398710, OQ398711,OQ271380-OQ271381 and OQ410975.
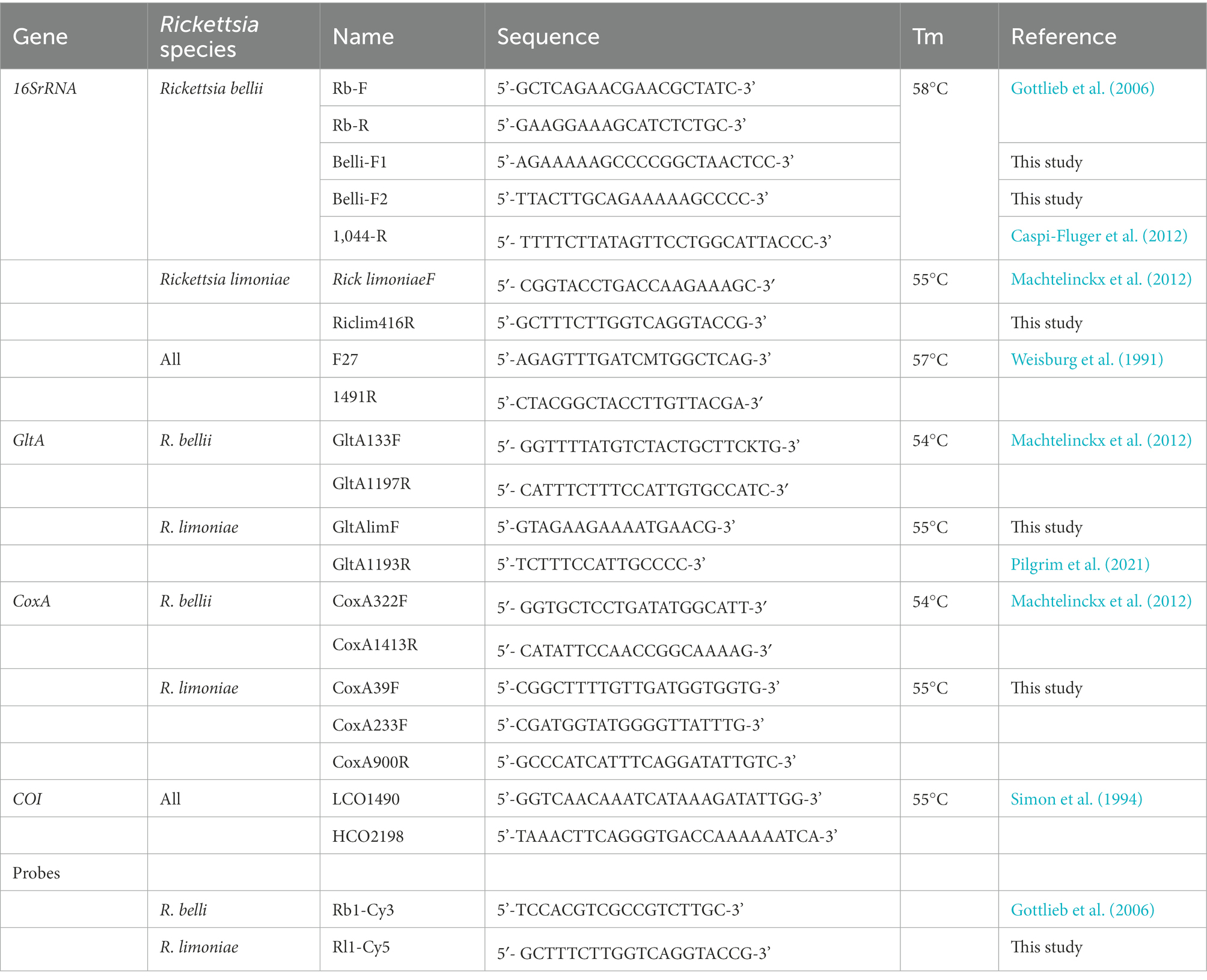
Table 1. Primer and probe sequences used in this study for PCR analyses and fluorescence in situ hybridization of Rickettsia endosymbionts.
2.2. Rickettsia prevalence in Macrolophus melanotoma
The abundance of R. bellii and R. limoniae in M. pygmaeus was reported by Dally et al. (2020). PCR was used to assess the prevalence of the two Rickettsia species in M. melanotoma. The DNA extracted from each of the collected individuals was screened with species-specific primers for the 16S rRNA gene of R. bellii and R. limoniae (as describe above; Table 1). DNA of M. pygmaeus harboring the bacteria served as a positive control for both bacterial species, following Dally et al. (2020).
2.3. Phylogenetic analysis
2.3.1. Phylogenetic analysis of Rickettsia
The phylogenetic relationships between the two Rickettsia species were inferred from concatenated sequences of a minimum of two of the following three genes: 16S rRNA, GltA (citrate synthase) and CoxA (cytochrome oxidase c subunit 1) (primer pairs are detailed in Table 1). Consensus sequences were obtained using DNAman software and were deposited in NCBI GenBank, under accession numbers OQ374915-OQ374918, OQ374921-OQ374924, OQ398710, OQ398711,OQ271380-OQ271381 and OQ410975. Representative sequences were chosen from Pilgrim et al. (2021) (see Supplementary additional files 2, 3; accession numbers used for Rickettsia phylogenetic analyses). The total length of concatenated sequences was in the range of ~2,100–3,000 bp. Multiple sequence alignment was conducted by MAFFT using default parameters, and a maximum likelihood tree was constructed with the GTR substitution model using PhyML, with branch support measured by approximate likelihood ratio tests (SH-aLRT; (Guindon et al., 2010). Rickettsia japonica (accession number AP017600.1) was used as an outgroup for both R. limoniae and R. bellii trees.
2.3.2. Phylogenetic analysis of Macrolophus spp.
The phylogenetic relationships between the two studied Macrolophus species and other members of the Miridae were investigated using the mitochondrial cytochrome oxidase subunit I (COI) gene. A ~ 650 bp-long fragment was sequenced using the primers LCO1490, HCO2198 (Table 1). Representative sequences were chosen from a BLASTN search targeted to Miridae (taxid no. 30083) (See Supplementary additional file 4; accession numbers used for Mitochondrial COI phylogenetic analyses). Multiple sequence alignment was conducted by MAFFT using default parameters, and a maximum likelihood tree was constructed with the GTR substitution model using PhyML, with 100 replicates for bootstrap support. Orius laevigatus (Heteroptera: Anthocoridae) was used as an outgroup.
2.4. Morphology of the gut, ovaries, and salivary glands
To characterize the morphology of various relevant M. melanotoma organs, more than 40 adult females and 40 adult males were dissected under a stereomicroscope. Dissections were photographed with a 3-D digital microscope (see description in Dally et al., 2020).
2.5. Localization of Rickettsia bellii and Rickettsia limoniae in Macrolophus melanotoma
Fluorescent in situ hybridization (FISH) was performed to determine the location of R. limoniae and R. bellii in the reproductive organs, digestive tract and salivary glands. The protocol described by Gottlieb et al. (2006) was followed with slight modifications for 50 mounted samples (see Dally et al., 2020). Images were acquired using an OLYMPUS IX 81 (Japan) inverted laser scanning confocal microscope (FLUOVIEW 500) equipped with 405, 561, 640 nm laser lines, a UplanApo 10 x/0.4 NA dry objective, and PlanApo 40 x/0.9 NA and 60 ×/1.0 NA water immersion objectives (see description in Dally et al., 2020). Confocal optical sections were obtained at increments of 5 μm, 2.5 μm, 1.3 μm and 0.8 μm for 10x, 20x, 40x and 60x objectives, respectively.
3. Results
3.1. Species verification
BLAST searches to all consensus sequences of the COI gene of the field-collected M. melanotoma and laboratory M. pygmaeus exhibited over 99.5% sequence similarity to available sequences of the matching Macrolophus species, thus verifying species identification.
3.2. Rickettsia prevalence in Macrolophus melanotoma
Out of the 82 M. melanotoma adults screened by diagnostic PCR, 84% (n = 69) were found to be positive for both Rickettsia species. R. bellii was detected alone in 10% (n = 8) of the samples, R. limoniae alone was detected in 1% (n = 1), and no Rickettsia were found in 5% (n = 4) of the adults. As previously reported, in M. pygmaeus both Rickettsia species were present in 100% of the tested adults (Dally et al., 2020).
3.3. Phylogenetic analysis
The aim of the analysis was to examine the overall phylogenetic assignment of the studied Macrolophus strains and Rickettsia symbionts, and to assess whether any co-evolutionary pattern could be identified between the symbionts and their corresponding hosts.
3.3.1. Macrolophus species
Analysis of the phylogenetic relationships between the two studied Macrolophus species and other members of the Miridae confirmed the BLAST search results and morphological identification; each of them clustered together with other haplotypes of the relevant species, and the overall tree was fitted to the taxonomic assignments of other Miridae members (Figure 1).
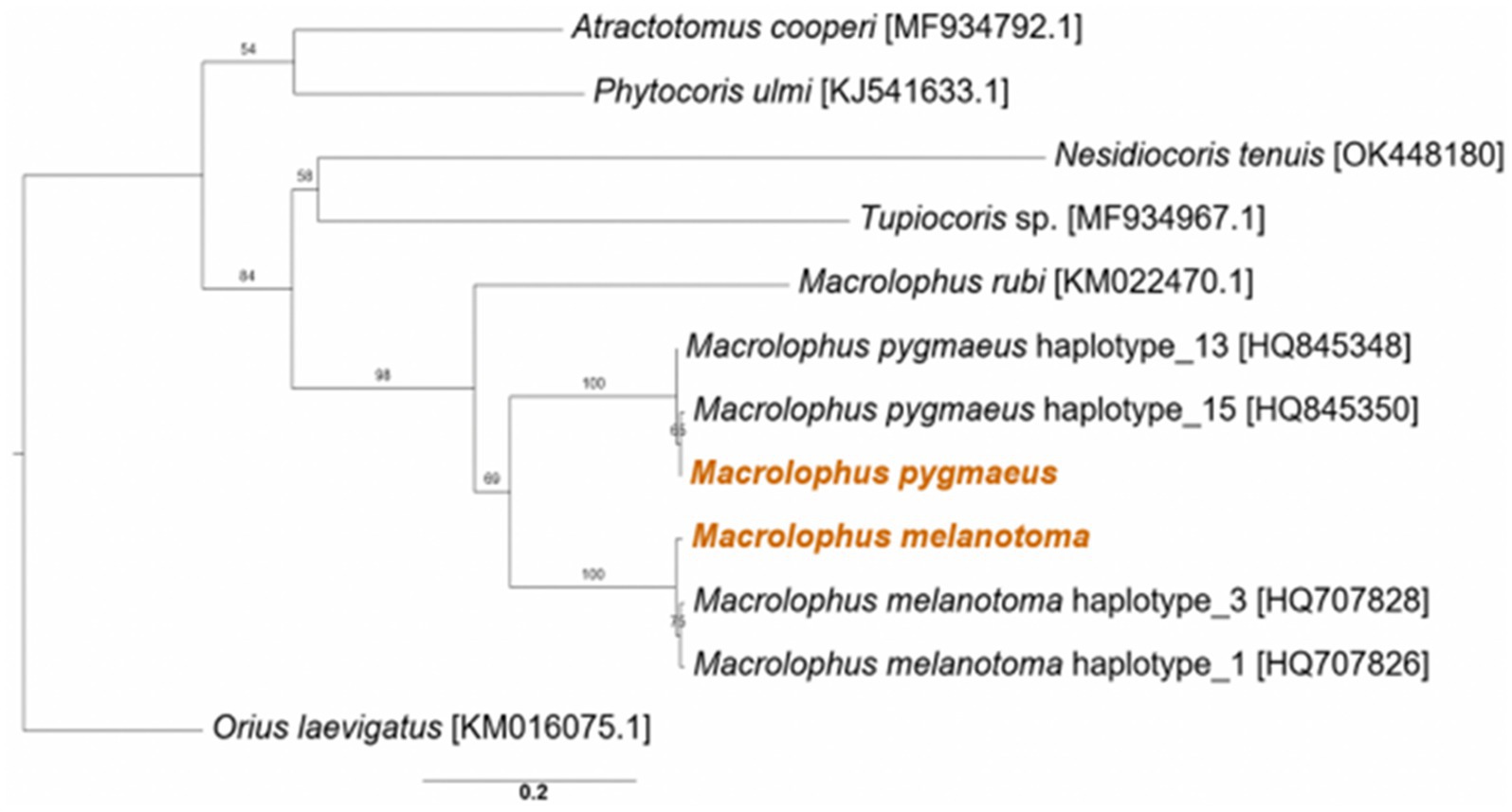
Figure 1. Maximum likelihood phylogeny of Macrolophus spp. from the present study (colored) and other species in the Miridae family, based on the COI gene. Accession numbers of the reference sequences are in brackets. The outgroup is Orius laevigatus (Hemiptera: Anthocoridae). Bootstrap support values of 100 repeats are indicated. Scale bar indicates 0.2 substitutions per site.
3.3.2. Rickettsia species
The three genes from each of the Rickettsia species sequenced from M. melanotoma specimens collected in all five localities in Israel, had over 99% similarity between them, indicating that M. melanotoma from all locations is infected with the same strains of R. bellii and the same strains of R. limoniae. Accordingly, we constructed consensus sequences for each of the Rickettsia strains from all field collected specimens of Macrolophus. Concatenate sequences of three R. limoniae and R. bellii genes from each Macrolophus species were constructed to assess whether any co-evolutionary pattern could be identified between the symbionts and their corresponding hosts. R. limoniae inhabiting M. pygmaeus and M. melanotoma showed greater similarity to each other than to any of the other chosen reference sequences (Figure 2A). Pairwise alignment between them was 96%, while alignment with all other sequences ranged from 70 to 92% (alignment with the outgroup was 54%). The studied strains clustered together with an R. limoniae strain from Deronectes platynotus, a dytiscid beetle. Strains from other heteropterans, including a strain from M. pygmaeus obtained by Pilgrim et al. (2021), were distributed along the branches of the tree, without a pattern resembling hosts’ phylogeny.
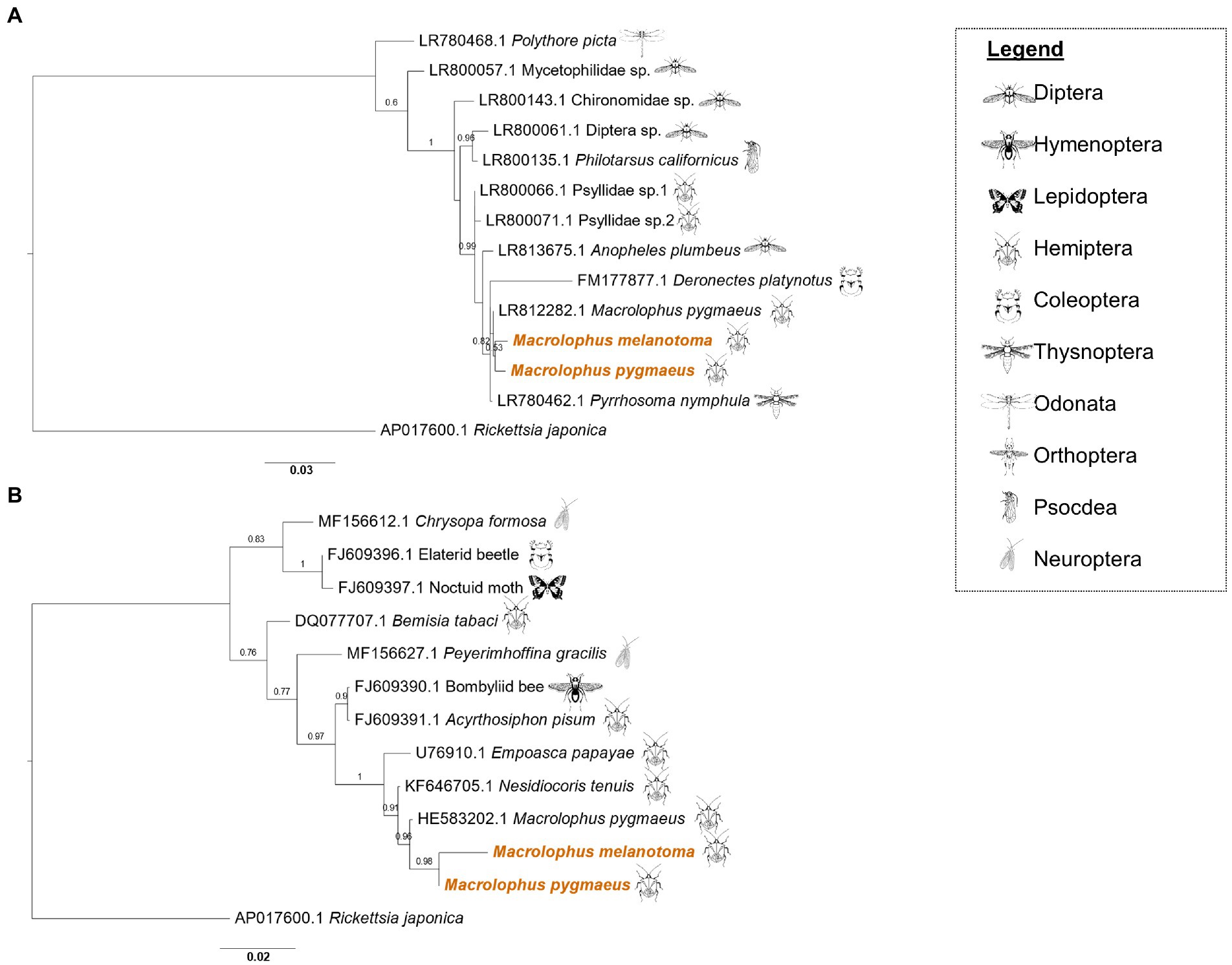
Figure 2. Maximum likelihood phylogeny of Rickettsia limoniae and Rickettsia bellii from Macrolophus melanotoma and Macrolophus pygmaeus. (A) Phylogeny of R. limoniae in the present study (colored) and other insect species, inferred from concatenated sequences of 16S rRNA, GltA, and CoxA genes. Scale bar = 0.03 substitutions per site. (B) Phylogeny of R. bellii. The analysis of all R. bellii strains was inferred from concatenated sequences of 16S rRNA, GltA and CoxA genes, except that the concatenate of M. melanotoma lacks the 16S rRNA sequence. Names at the tips indicate the insect host of the specific strain, and numbers indicate the accession number of the 16S rRNA gene. Accession numbers of the two other genes included in the concatenate can be found in Supplementary files 2 and 3. Scale bar = 0.02 substitutions per site. Insect icons indicate the insect order of the host.
As in R. limoniae, R. bellii clustered together on the phylogenetic tree (Figure 2B). Pairwise alignment of the M. pygmaeus strain to others on the tree ranged from 62% (Bemisia tabaci [DQ077707.1]) to 98% (M. pygmaeus from another study [HE583203.1]). Interestingly, while the CoxA and GltA sequences of the R. bellii symbiont in M. melanotoma were similar to other sequences from the bellii group, the 16S rRNA gene was more similar to sequences obtained from the limoniae group (Supplementary Figure 1).
3.4. Morphology of the gut, ovary, and salivary gland
Microscopic observation of the ovaries and digestive system of M. melanotoma revealed similarity to those previously described in M. pygmaeus (Figures 3A–C; Dally et al., 2020). The digestive tract consists of a tubular foregut with a direct opening to the mouth; a large sac-like anterior part of the midgut; a second tubular region of the midgut; a soft, somewhat swollen third midgut region; and the posterior fourth midgut region, also moderately swollen, which connects to the hindgut at the point of attachment of the Malpighian tubules (Figures 3B,C). Two caeca-like organs, which appear to be larger in males than in females, are connected to the posterior end of the fourth midgut region in the area of the Malpighian tubule openings (M4 in Figures 3B,C).
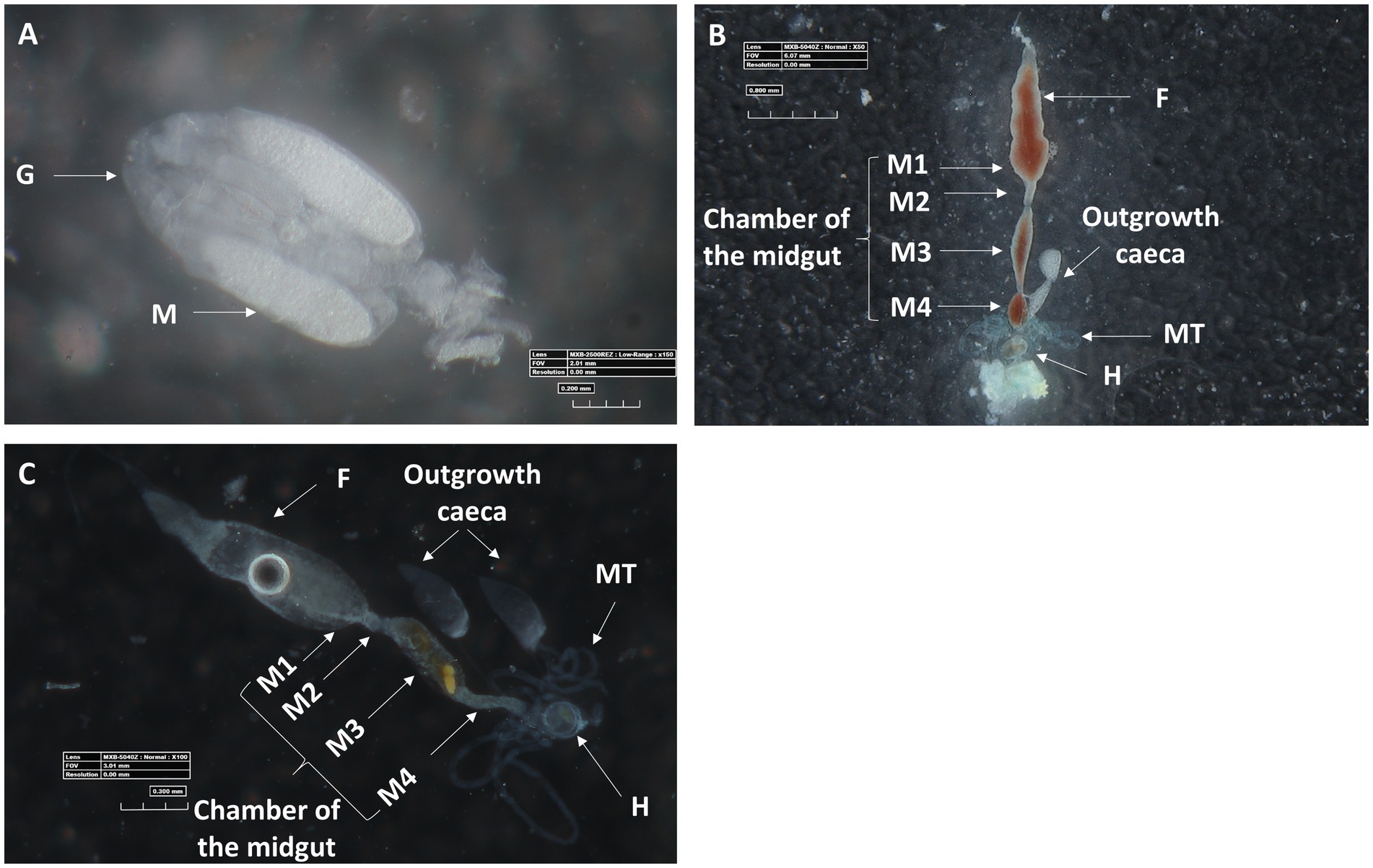
Figure 3. Three-D digital microscopic images of M. melanotoma telotrophic ovary, and female and male digestive systems. (A) An isolated ovary with several ovarioles; M, mature oocyte; G, Germarium, (B) Isolated female and (C) male digestive systems; F, foregut; M1, midgut first region; M2, midgut second region; M3, midgut third region; M4, midgut fourth region (with outgrowth caeca); MT, Malpighian tubules; H, hindgut.
Microscopic observations further revealed the structural configuration of the salivary glands of female M. melanotoma and M. pygmaeus. Mirids, like all terrestrial heteropterans, have a pair of salivary glands located in the thorax, next to the alimentary canal (Wheeler, 2002). The system appeared to have similar morphology in both bug species, with two symmetrical salivary glands, each composed of an anterior lobe, a posterior lobe, and a salivary duct (Figure 4A).
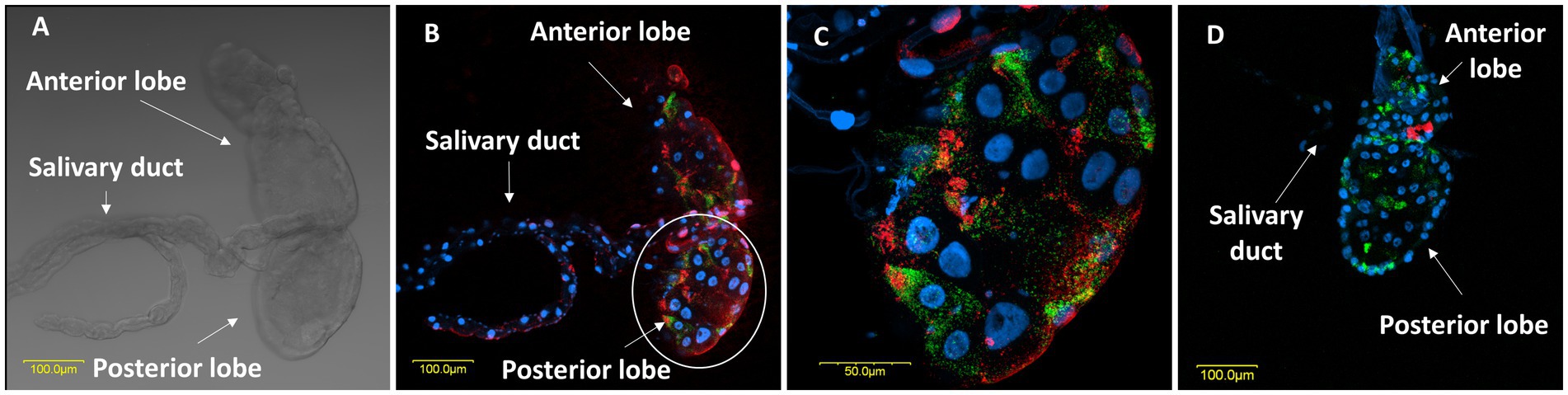
Figure 4. (A) A light transmitted microscopic image of a salivary gland in a M. melanotoma female, showing the anterior and posterior lobes of the salivary gland, and the salivary duct. FISH of the salivary glands of M. melanotoma (B) and M. pygmaeus (D) females. DNA in blue, R. bellii in red, and R. limoniae in green. (C) Enlarged region of the posterior lobe of the salivary gland of M. melanotoma female (white circle in B). Images (B–D) represent serial Z section of 15 μm, 27 μm, and 20 μm, respectively.
3.5. Localization of Rickettsia bellii and Rickettsia limoniae in Macrolophus pygmaeus and Macrolophus melanotoma
In situ hybridization targeting bacterial 16S rRNA allowed visualization of the two Rickettsia species within the ovaries, digestive tract and salivary gland of M. pygmaeus and M. melanotoma. In the ovaries, R. bellii and R. limoniae were found to be concentrated mainly in the germarium (Figure 5A) and scattered therein (Figures 5A,B).
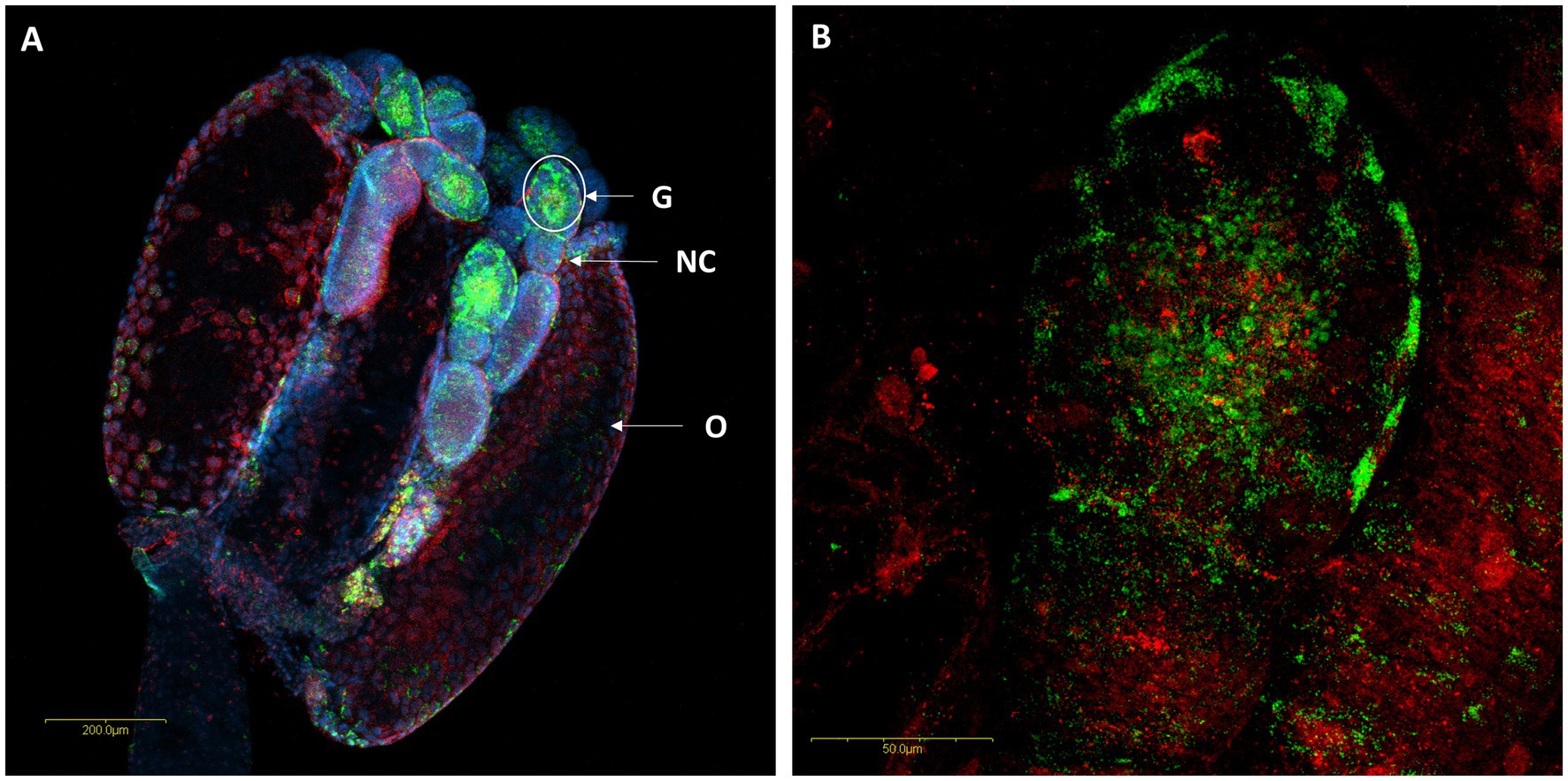
Figure 5. FISH of M. melanotoma telotrophic ovarioles with R. bellii specific probes (red), R. limoniae specific probes (green), and DNA dye (blue). (A) Ovary with several ovarioles, R. bellii and R. limoniae are concentrated in the germarium; G, Germarium; NC, Nurse cells; O, Oocyte. (B) Enlarged region of the germarium (white circle in A). Images (A,B) represent serial Z section of 40 μm and 26 μm, respectively.
A comparison of M. pygmaeus and M. melanotoma revealed the presence of large numbers of the two Rickettsia species throughout the digestive system in both females and males, with each Macrolophus species displaying a unique distribution pattern. In M. pygmaeus, R. limoniae was more broadly distributed along the host digestive system, while R. bellii was located primarily in the foregut and the midgut (Figure 6). In M. melanotoma, in contrast, R. bellii was more broadly distributed along the digestive system, while R. limoniae was clustered (Figures 7, 8). In both bug species, the two Rickettsia were usually isolated in separate host cells; they were, however, infrequently found sharing a common epithelial cell (Figure 8A2). FISH targeting bacterial 16S rRNA visualized R. limoniae and R. bellii in the caeca-like organs, in both females and males (Figures 7A2, 8A3). Likewise, FISH analysis detected R. bellii and R. limoniae within the Malpighian tubules of females and males of both bug species (Figures 7A3, 8A5).
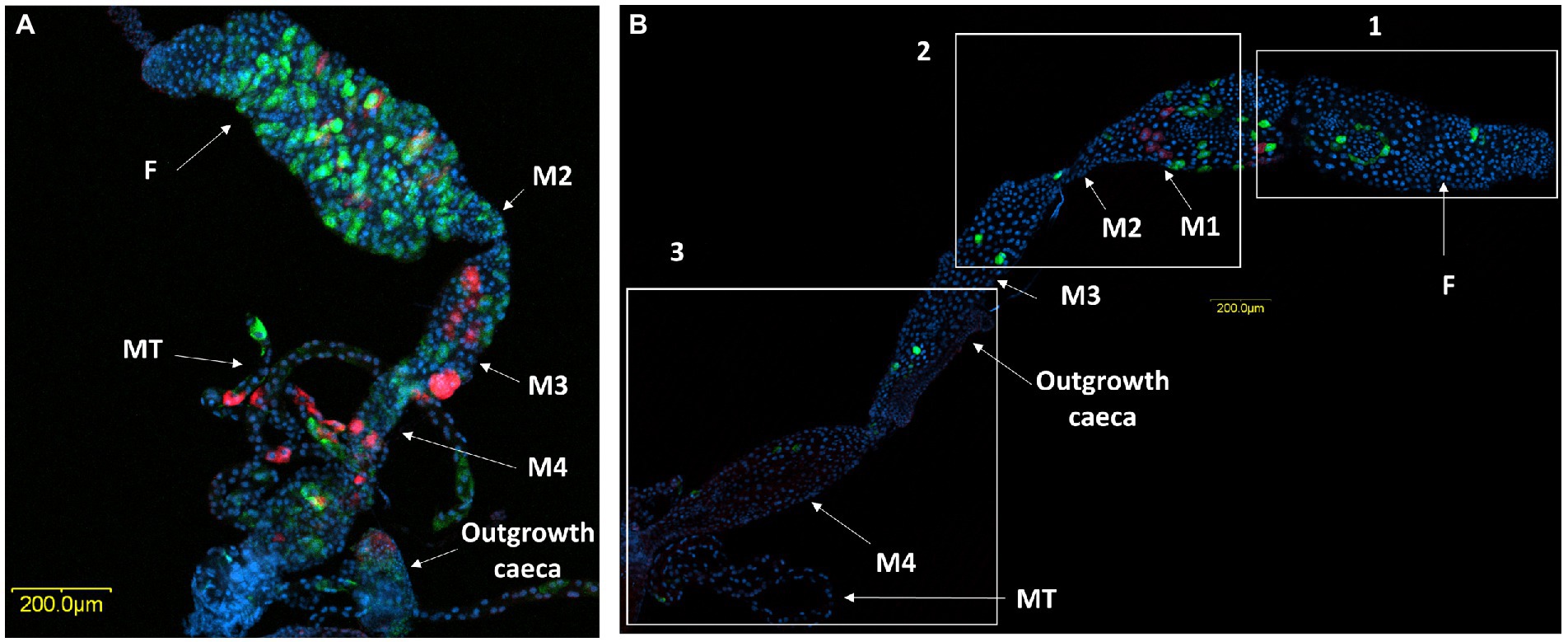
Figure 6. FISH of M. pygmaeus digestive system (DNA in blue). Male (A) and female (B) digestive systems, R. bellii (red), R. limoniae (green). Reconstruction: three frames of the same gut (B); rectangle 1 picture number 1, rectangle 2 picture number 2, rectangle 3 picture number 3. Images A and B number 1, B number 2, B number 3 represent serial Z section of 45 μm, 25 μm, 55 μm and 60 μm respectively; F, foregut; M1, midgut first region; M2, midgut second region; M3, midgut third region (with outgrowth caeca); M4, midgut fourth region; MT, Malpighian tubules.
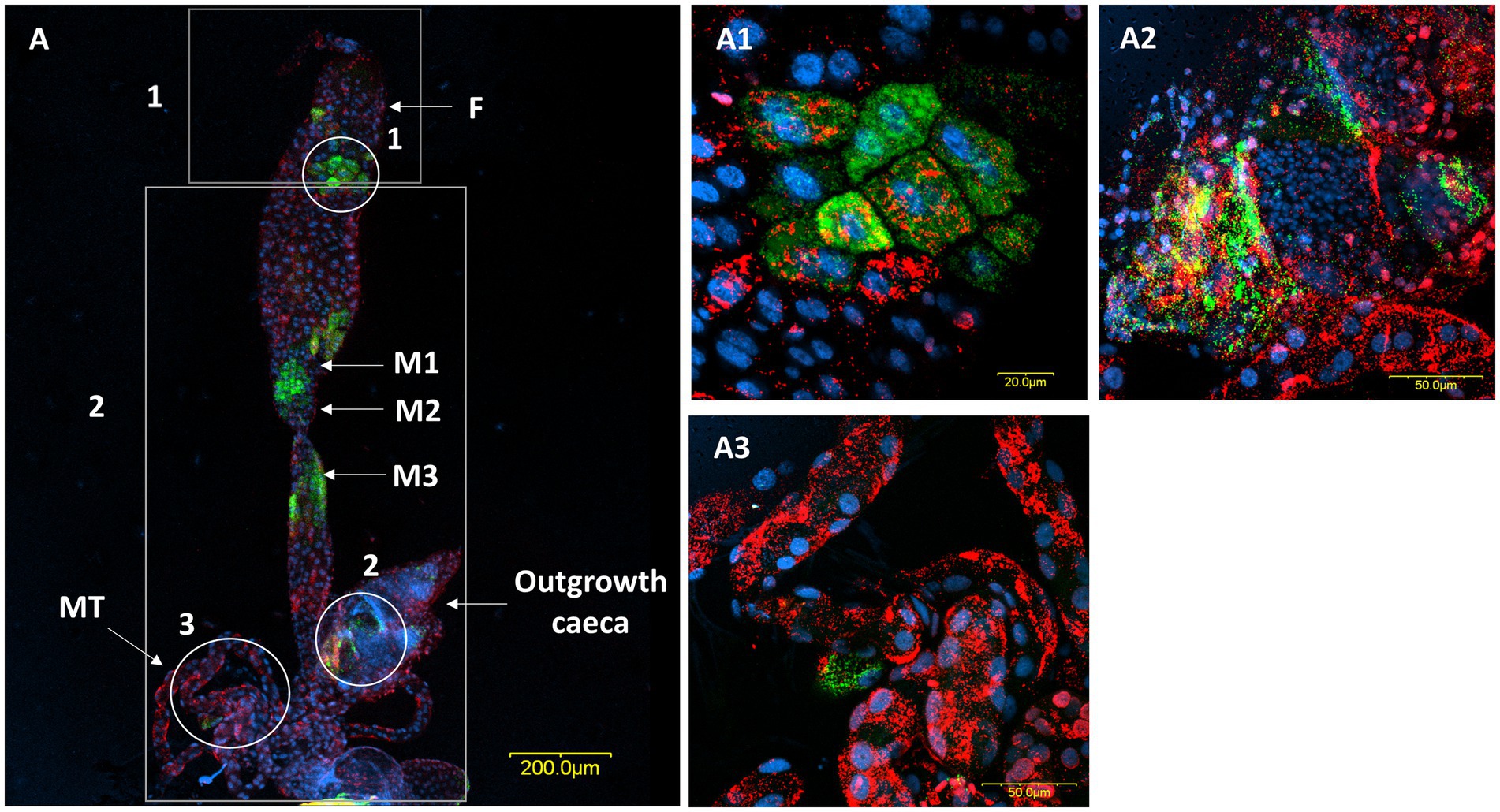
Figure 7. FISH of a male digestive system (DNA in blue). (A) The digestive system, R. bellii (red), R. limoniae (green). Reconstruction: two frames of the same gut (A); rectangle 1 picture number 1, rectangle 2 picture number 2. (A1) Enlarged region from the foregut (see circle 1 in A). (A2) Enlargement of the outgrowth caeca region, in the gut tissue of the fourth midgut region (see circle 2 in A). (A3) Enlarged region of Malpighian tubules (see circle 3 in A). Images A number 1, A number 2, and (A1–A3) represent serial Z section of 40 μm, 40 μm, 6.4 μm, 11.2 μm, 28 μm respectively; F, foregut; M1, midgut first region; M2, midgut second region; M3, midgut third region; MT, Malpighian tubules.
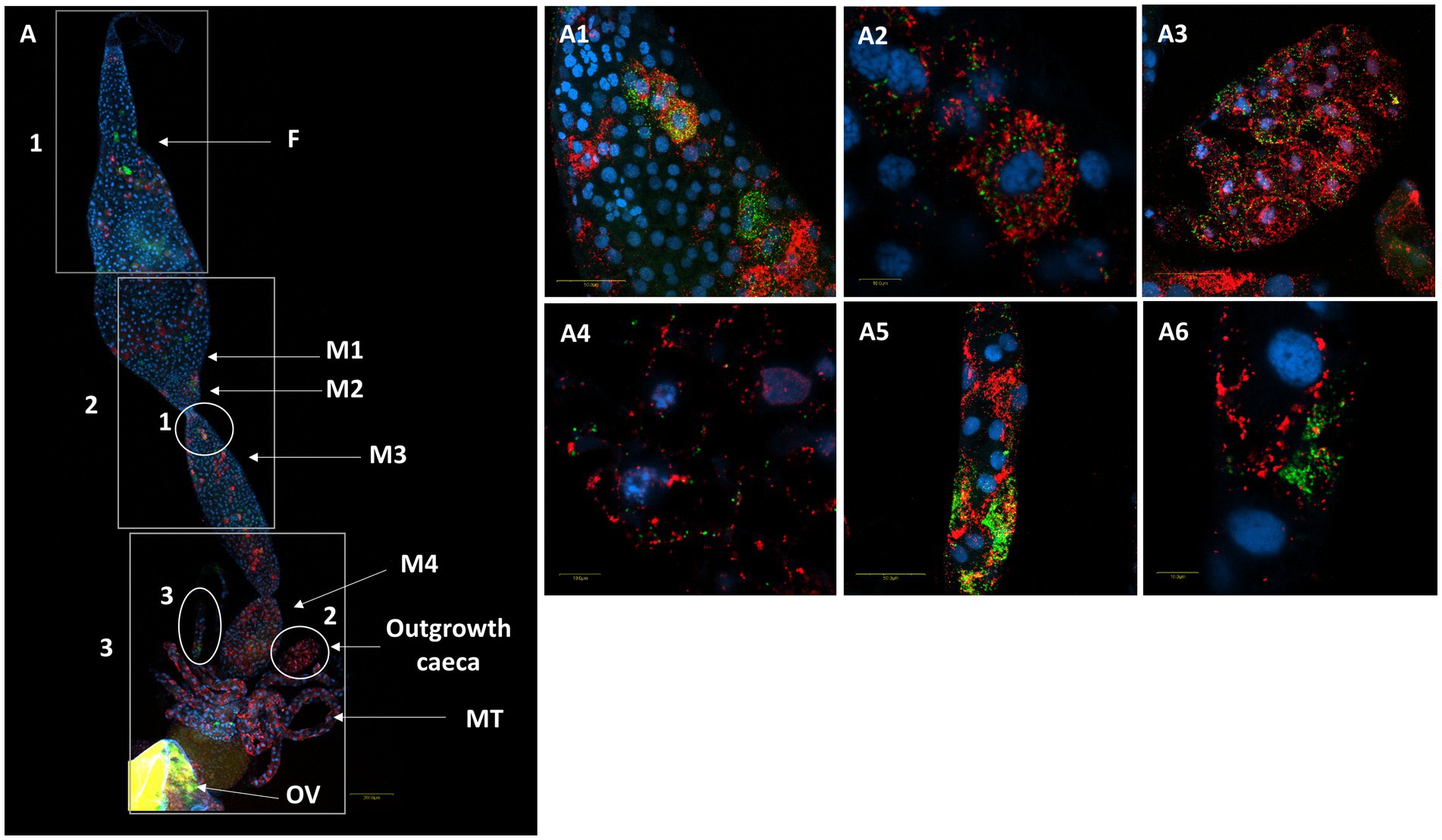
Figure 8. FISH of a female M. melanotoma digestive system (DNA in blue). (A) The whole digestive tract, R. bellii (red), R. limoniae (green), reconstructed from 3 images along the gut. Reconstruction: three frames of the same gut (A); rectangle 1 picture number 1, rectangle 2 picture number 2, rectangle 3 picture number 3 with two outgrowth caeca from the gut tissue of the fourth midgut region. (A1,A2) Enlarged region of the midgut (circle 1 in A). (A3,A4) Enlarged region of the two outgrowth caeca (circle 2 in A). (A5,A6) Enlarged region of the Malpighian tubules (circle 3 in A). Images A number 1, A number 2, A number 3 and (A1–A6) represent serial Z section of 40 μm, 40 μm, 35 μm, 21 μm, 1 μm, 27 μm, 1 μm, 22 μm, and 1 μm respectively; F, foregut; M1, midgut first region; M2, midgut second region; M3, midgut third region; M4, midgut fourth region (with outgrowth caeca); MT, Malpighian tubules; OV, ovipositor.
FISH analysis revealed the presence of R. bellii and R. limoniae in the anterior and posterior lobes of the salivary glands as well as the salivary duct in both species, with unique distribution patterns in each Macrolophus species. In M. melanotoma, R. bellii was more broadly distributed throughout the salivary gland, whereas R. limoniae appeared to be more clustered. In M. pygmaeus, this distribution pattern is reversed (Figure 4).
4. Discussion
In this study we detected, characterized and compared two symbiotic Rickettsia species, R. limoniae and R. bellii, inhabiting two Macrolophus species, M. pygmaeus and M. melanotoma. Phylogenetic analyses showed that R. limoniae from the two bugs studied most closely resemble each other, and share higher sequence similarity than to the symbiont sequenced from M. pygmaeus reported from the United Kingdom. Further, these R. limoniae sequences cluster closer to a bacterium sequenced from an Odonata species, than to those found in various other Hemiptera. A similar phylogenetic pattern was obtained for R. bellii, but based on the genes sequenced, it seems there is a clade that includes the symbiont found in Hemiptera, which all cluster together, with the exception of a strain from Bemisia tabaci. Altogether, the emerging pattern agrees with previous studies that suggest occasional horizontal transfer of Rickettsia (Nováková and Šmajs, 2019). Such a transfer may occur via feeding, either directly by the consumption of infected prey, or indirectly via feeding on host plants shared with other phytophagous insects. It is also possible that additional insect species feeding on the same host plants might take up microorganisms transferred by the bug to the plant. Rickettsia have been shown to be acquired from environmental sources by two species of Spalangia (Hymenoptera: Pteromalidae), S. endius and S. cameroni (Tzuri et al., 2021). Furthermore, Chrostek et al. (2017) reviewed the transmission of Rickettsia, Wolbachia, and Cardinium through plants by the leafhopper Euscelidius variegatus. These findings indicate that a co-evolutionary process between Macrolophus species and their symbiotic Rickettsia is unlikely.
In M. melanotoma, R. bellii is clustered with the bellii group based on the CoxA and GltA genes, but is more similar to the R. limoniae group according to the 16S rRNA gene phylogeny. These results suggest a possible recombination between the two Rickettsia species (Jiggins, 2006). Recombination events are not rare in Rickettsia genomes (Wu et al., 2009; Merhej and Raoult, 2011), and play an important role in the evolution of these bacteria, notably by enabling them to adapt to new hosts (Thomas, 2016).
The two Rickettsia species were documented in all individuals of M. pygmaeus (Dally et al., 2020) and had a high rate of occurrence (84%) in screened M. melanotoma, adults. This is contrary to the findings of a previous study, in which only R. limoniae, was detected in M. melanotoma (Machtelinckx et al., 2012). This difference may stem from the different collection sites (Israel in the current study while Machtelinckx et al., 2012 collected in Greece and Italy), which may have various environmental conditions, selection pressures or infection histories. Moreover, this may be an indication of horizontal transfer of Rickettsia between the mirid species. Finding the two Rickettsia species in the salivary glands of their mirid hosts lends support to the possibility that the bacteria may be transferred to, and acquired from, their host’s food source. It is increasingly noted that bacteria can colonize the salivary glands of insects; this may have significant implications for plant–insect interactions, particularly for disease transmission by herbivorous insects (Kaiser et al., 2010; Body et al., 2013). Two well-documented examples are Citrus greening, the most destructive citrus disease in the world, caused by Candidatus Liberibacter asiaticus which resides in the salivary glands of the vector, the Asian citrus psyllid Diaphorina citri (Ammar et al., 2011), and Flavescence dorèe, a severe grapevine disease caused by Candidatus Phytoplasma vitis found in salivary glands of the leafhopper Scaphoideus titanus (Cicadellidae) (Marzorati et al., 2006). The endosymbiotic bacterium Cardinium was found to be injected into the plant by this leafhopper vector without any notable influence (Gonella et al., 2015). It can thus be seen that bacteria transmitted by herbivores to their host plants are not necessarily pathogenic. In some cases, bacterial symbionts have been shown to alter plant metabolic reconfiguration in a way that better meets insect nutritional needs. The endosymbiont Wolbachia has been shown to be transmitted by larvae of the leaf-mining moth Phyllonorycter blancardella to the leaves of apple seedlings (Malus domestica), where the bacterium alters the phytohormonal profile of the leaves, creating an optimal microenvironment for its host (Kaiser et al., 2010; Body et al., 2013). Similar transmission was reported from another hemipteran, the sweet potato whitefly Bemisia tabaci, which transmits and acquires Rickettsia through the phloem of cotton plants (Caspi-Fluger et al., 2012).
The distribution of the two Rickettsia species in M. melanotoma ovarioles resembles their localization in M. pygmaeus (Machtelinckx et al., 2012; Dally et al., 2020). In both cases, the symbionts were found primarily in the germarium, strongly suggesting that the bacteria are transmitted vertically from the mother to her offspring via the egg (transovarial transmission), a common transmission pathway in many symbiont-host systems (Monti, 2017).
The two Rickettsia species displayed a unique distribution pattern in the two studied Macrolophus species. In M. melanotoma, R. bellii was distributed throughout the entire digestive tract, while R. limoniae appeared mainly in the foregut and the midgut. In M. pygmaeus, on the other hand, the distribution of R. bellii was more restricted than that of R. limoniae. Many phenotypic, genetic and physiological factors may lead to such variation in Rickettsia distribution in the gut. In addition, environmental factors such as seasonal conditions and host plants may also be involved. In the present study, for example, M. pygmaeus was reared in the laboratory on frozen Ceratitis capitata eggs and tomato seedlings under optimal temperature conditions, whereas M. melanotoma was collected in the spring from Dittrichia viscosa plants. Because species in the Miridae, especially those of the omnivorous Dicyphini tribe, are the best-known group of arthropods specialized for foraging, feeding, and oviposition on sticky plants (Wheeler and Krimmel, 2015), the difference in the food plant of the two studied hosts may influence Rickettsia involvement in the bugs’ nutritional ecology, and their distribution pattern might be influenced by the food source.
Mutualistic microbes often inhabit the caeca connected to the most anterior portion of the midgut (Hosokawa et al., 2016; Nardi et al., 2019), yet to the best of our knowledge such caeca structures have not been reported from any mirid species studied so far. Instead, our earlier work on M. pygmaeus (Dally et al., 2020), as well as the present study, revealed the presence of a paired organ at the posterior end of the midgut. Although this caecum-like organ appears to differ morphologically between the two studied Macrolophus species, as well as between males and females, it harbors both Rickettsia species in separate cells or, more rarely, together in the same cell. The function of these caeca-like organs is as yet unknown.
Obligate blood-feeders such as ticks, bed bugs, and tsetse flies have evolved to rely on microbial endosymbionts to supplement several B vitamins – such as biotin (B7), folate (B9), and riboflavin (B2) – that are deficient in blood. For example, the endosymbiont Rickettsia buchneri provides its tick hosts with biotin and folate, essential components for cell growth in all eukaryotic and prokaryotic organisms (Rio et al., 2016; Narasimhan et al., 2021).
In many hemipterans, symbiotic bacteria found on the midgut epithelia supply their hosts with essential nutrients such as amino acids and vitamins (Kikuchi et al., 2009), and recycle metabolic wastes of the host (Ohbayashi et al., 2019). The presence of Rickettsia in the digestive tract of both M. pygmaeus and M. melanotoma, as well as other omnivorous mirids, such as the midgut epithelial cells of Stenotus binotatus (Chang and Musgrave, 1970) and the lumen of the digestive tract of Nesidiocoris tenuis (Caspi-Fluger et al., 2014), may be indicative of the nutritional role filled by Rickettsia symbionts in this group. However, the omnivorous habit of these bugs ensures a balanced diet, so other phenotypes could not be ruled out. Examples could be found in Liu and Guo (2019), which reviewed the roles Rickettsia and Wolbachia play in protecting hosts from stresses caused by natural enemies, heat, and toxins. Moreover, M. pygmaeus became more sensitive to freezing conditions when all three symbionts, Wolbachia, R. bellii and R. limoniae were removed, but it is unclear the absence of which symbiont causes the negative effect (Maes et al., 2012).
Coinfections are well known for Wolbachia (Ant and Sinkins, 2018) but have been less commonly recorded for other symbionts. The presence of both R. bellii and R. limoniae in the two studied bug species is, however, in agreement with the widespread occurrence of co-infections in the Torix group of Rickettsia (Pilgrim et al., 2021). Co-infection with Rickettsia from the Torix group was also observed in the two damselflies Coenagrion puella and Coenagrion pulchellum (Thongprem et al., 2021).
In conclusion, the presence of two Rickettsia species with specific distribution patterns that differed between two Macrolophus host species was described. These symbiont species are phylogenetically similar and were found in the vast majority of the field-collected host bugs. As sequencing the genome of the two Rickettsia species could elucidate the role of such symbionts in the feeding habits of their hosts, it would be warranted to expand this research by combining lab experiments on feeding behavior with investigations into molecular genetics and bioinformatics.
Data availability statement
The data presented in the study are deposited in the GenBank repository, accession numbers OQ374915-OQ374918, OQ374921-OQ374924, OQ398710, OQ398711, OQ271380-OQ271381 and OQ410975.
Author contributions
MD: wrote the manuscript, performed, and designed the work. YZ: performed the phylogenetic analysis and wrote sections of the manuscript. EB: captured FISH images, and wrote the FISH method section. NM-D: contributed to the phylogenetic analysis section. MC and EZ-F: contributed to conception and design of the study. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This research was supported by the Israel Science Foundation (grant no. 397/21 to EZ-F).
Acknowledgments
We thank Murad Ghanim and Shira Gal for technical help; Yuval Gottlieb, Elad Chiel, and Ruth Ann Yonah for valuable comments on drafts of the manuscript; and BioBee Sde Eliyahu Ltd. for providing a laboratory strain of M. pygmaeus.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1107153/full#supplementary-material
References
Ammar, E. D., Shatters, R. G., and Hall, D. G. (2011). Localization of candidatus Liberibacter asiaticus, associated with citrus huanglongbing disease, in its psyllid vector using fluorescence in situ hybridization. J. Phytopathol. 159, 726–734. doi: 10.1111/j.1439-0434.2011.01836.x
Ant, T. H., and Sinkins, S. P. (2018). A Wolbachia triple-strain infection generates self-incompatibility in Aedes albopictus and transmission instability in Aedes aegypti. Parasit. Vectors 11, 295–297. doi: 10.1186/s13071-018-2870-0
Body, M., Kaiser, W., Dubreuil, G., Casas, J., and Giron, D. (2013). Leaf-miners co-opt microorganisms to enhance their nutritional environment. J. Chem. Ecol. 39, 969–977. doi: 10.1007/s10886-013-0307-y
Caspi-Fluger, A., Inbar, M., Mozes-Daube, N., Katzir, N., Portnoy, V., Belausov, E., et al. (2012). Horizontal transmission of the insect symbiont Rickettsia is plant-mediated. Proc. R. Soc. B Biol. Sci. 279, 1791–1796. doi: 10.1098/rspb.2011.2095
Caspi-Fluger, A., Inbar, M., Steinberg, S., Friedmann, Y., Freund, M., Mozes-Daube, N., et al. (2014). Characterization of the symbiont Rickettsia in the mirid bug Nesidiocoris tenuis (Reuter) (Heteroptera: Miridae). Bull. Entomol. Res. 104, 681–688. doi: 10.1017/S0007485314000492
Castañé, C., Arnó, J., Gabarra, R., and Alomar, O. (2011). Plant damage to vegetable crops by zoophytophagous mirid predators. Biol. Control 59, 22–29. doi: 10.1016/j.biocontrol.2011.03.007
Chang, K. P., and Musgrave, A. J. (1970). Ultrastructure of Rickettsia-like microorganisms in the midgut of a plant bug, Stenotus binotatus jak. (Heteroptera: Miridae). Can. J. Microbiol. 16, 621–622. doi: 10.1139/m70-104
Chrostek, E., Pelz-Stelinski, K., Hurst, G. D., and Hughes, G. L. (2017). Horizontal transmission of intracellular insect symbionts via plants. Front. Microbiol. 8:2237. doi: 10.3389/fmicb.2017.02237
Dally, M., Lalzar, M., Belausov, E., Gottlieb, Y., Coll, M., and Zchori-Fein, E. (2020). Cellular localization of two Rickettsia symbionts in the digestive system and within the ovaries of the mirid bug, Macrolophous pygmaeus. Insects 11, 1–15. doi: 10.3390/insects11080530
El Karkouri, K., Ghigo, E., Raoult, D., and Fournier, P. E. (2022). Genomic evolution and adaptation of arthropod-associated Rickettsia. Sci. Rep. 12, 3807–3815. doi: 10.1038/s41598-022-07725-z
Giorgini, M., Bernardo, U., Monti, M. M., Nappo, A. G., and Gebiola, M. (2010). Rickettsia symbionts cause parthenogenetic reproduction in the parasitoid wasp Pnigalio soemius (hymenoptera: Eulophidae). Appl. Environ. Microbiol. 76, 2589–2599. doi: 10.1128/AEM.03154-09
Gonella, E., Pajoro, M., Marzorati, M., Crotti, E., Mandrioli, M., Pontini, M., et al. (2015). Plant-mediated interspecific horizontal transmission of an intracellular symbiont in insects. Sci. Rep. 5, 1–10. doi: 10.1038/srep15811
Gottlieb, Y., Ghanim, M., Chiel, E., Gerling, D., Portnoy, V., Steinberg, S., et al. (2006). Identification and localization of a Rickettsia sp. in Bemisia tabaci (Homoptera: Aleyrodidae). Environ. Microbiol. 72, 3646–3652. doi: 10.1128/AEM.72.5.3646-3652.2006
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Gupta, A., and Nair, S. (2020). Dynamics of insect–microbiome interaction influence host and microbial symbiont. Front. Microbiol. 11:1357. doi: 10.3389/fmicb.2020.01357
Harris, H. L., Brennan, L. J., Keddie, B. A., and Braig, H. R. (2010). Bacterial symbionts in insects: balancing life and death. Symbiosis 51, 37–53. doi: 10.1007/s13199-010-0065-3
Hendry, T. A., Hunter, M. S., and Baltrus, D. A. (2014). The facultative symbiont Rickettsia protects an invasive whitefly against entomopathogenic Pseudomonas syringae strains. Appl. Environ. Microbiol. 80, 7161–7168. doi: 10.1128/AEM.02447-14
Himler, A. G., Adachi-Hagimori, T., Bergen, J. E., Kozuch, A., Kelly, S. E., Tabashnik, B. E., et al. (2011). Rapid spread of a bacterial symbiont in an invasive whitefly is driven by fitness benefits and female bias. Science 332, 254–256. doi: 10.1126/science.1199410
Hosokawa, T., Matsuura, Y., Kikuchi, Y., and Fukatsu, T. (2016). Recurrent evolution of gut symbiotic bacteria in pentatomid stinkbugs. Zool. Lett. 2, 24–29. doi: 10.1186/s40851-016-0061-4
Jiggins, F. M. (2006). Adaptive evolution and recombination of Rickettsia antigens. J. Mol. Evol. 62, 99–110. doi: 10.1007/s00239-005-0080-9
Kaiser, W., Huguet, E., Casas, J., Commin, C., and Giron, D. (2010). Plant green-island phenotype induced by leaf-miners is mediated by bacterial symbionts. Proc. R. Soc. B Biol. Sci. 277, 2311–2319. doi: 10.1098/rspb.2010.0214
Kikuchi, Y., Hosokawa, T., Nikoh, N., Meng, X. Y., Kamagata, Y., and Fukatsu, T. (2009). Host-symbiont co-speciation and reductive genome evolution in gut symbiotic bacteria of acanthosomatid stinkbugs. BMC Biol. 7, 1–22. doi: 10.1186/1741-7007-7-2
Liu, X. D., and Guo, H. F. (2019). Importance of endosymbionts Wolbachia and Rickettsia in insect resistance development. Curr. Opin. Insect Sci. 33, 84–90. doi: 10.1016/j.cois.2019.05.003
Lukasik, P., Guo, H., Van Asch, M., Ferrari, J., and Godfray, H. C. J. (2013). Protection against a fungal pathogen conferred by the aphid facultative endosymbionts Rickettsia and Spiroplasma is expressed in multiple host genotypes and species and is not influenced by co-infection with another symbiont. J. Evol. Biol. 26, 2654–2661. doi: 10.1111/jeb.12260
Machtelinckx, T., Van Leeuwen, T., Van De Wiele, T., Boon, N., De Vos, W. H., Sanchez, J. A., et al. (2012). Microbial community of predatory bugs of the genus Macrolophus (Hemiptera: Miridae). BMC Microbiol. 12, 1–14. doi: 10.1186/1471-2180-12-S1-S9
Machtelinckx, T., Van Leeuwen, T., Vanholme, B., Gehesquière, B., Dermauw, W., Vandekerkhove, B., et al. (2009). Wolbachia induces strong cytoplasmic incompatibility in the predatory bug Macrolophus pygmaeus. Insect Mol. Biol. 18, 373–381. doi: 10.1111/j.1365-2583.2009.00877.x
Maes, S., Machtelinckx, T., Moens, M., Grégoire, J. C., and De Clercq, P. (2012). The influence of acclimation, endosymbionts and diet on the supercooling capacity of the predatory bug Macrolophus pygmaeus. BioControl 57, 643–651. doi: 10.1007/s10526-012-9446-2
Martinez-Cascales, J. I., Cenis, J. L., Cassis, G., and Sanchez, J. A. (2006). Species identity of Macrolophus melanotoma (costa 1853) and Macrolophus pygmaeus (Rambur 1839) (Insecta: Heteroptera: Miridae) based on morphological and molecular data and bionomic implications. Insect Syst. Evol. 37, 385–404. doi: 10.1163/187631206788831470
Marzorati, M., Alma, A., Sacchi, L., Pajoro, M., Palermo, S., Brusetti, L., et al. (2006). A novel Bacteroidetes symbiont is localized in Scaphoideus titanus, the insect vector of Flavescence Dorée in Vitis vinifera. Appl. Environ. Microbiol. 72, 1467–1475. doi: 10.1128/AEM.72.2.1467-1475.2006
Merhej, V., and Raoult, D. (2011). Rickettsial evolution in the light of comparative genomics. Biol. Rev. 86, 379–405. doi: 10.1111/j.1469-185X.2010.00151.x
Moerkens, R., Berckmoes, E., Van Damme, V., Ortega-Parra, N., Hanssen, I., Wuytack, M., et al. (2016). High population densities of Macrolophus Pygmaeus on tomato plants can cause economic fruit damage: interaction with Pepino mosaic virus? Pest Manag. Sci. 72, 1350–1358. doi: 10.1002/ps.4159
Monti, M. (2017). Oocytes - maternal information and functions. Eur. J. Basic Appl. Histochem. 61:2849. doi: 10.4081/ejh.2017.2849
Narasimhan, S., Swei, A., Abouneameh, S., Pal, U., Pedra, J. H. F., and Fikrig, E. (2021). Grappling with the tick microbiome. Trends Parasitol. 37, 722–733. doi: 10.1016/j.pt.2021.04.004
Nardi, J. B., Miller, L. A., and Bee, C. M. (2019). Interfaces between microbes and membranes of host epithelial cells in hemipteran midguts. J. Morphol. 280, 1046–1060. doi: 10.1002/jmor.21000
Nováková, M., and Šmajs, D. (2019). Rickettsial endosymbionts of ticks. Ticks Tick-Borne Pathogens, 81–88. doi: 10.5772/intechopen.80767
Ohbayashi, T., Futahashi, R., Terashima, M., Barrière, Q., Lamouche, F., Takeshita, K., et al. (2019). Comparative cytology, physiology and transcriptomics of Burkholderia insecticola in symbiosis with the bean bug Riptortus pedestris and in culture. ISME J. 13, 1469–1483. doi: 10.1038/s41396-019-0361-8
Perdikis, D., and Lykouressis, D. (2000). Effects of various items, host plants, and temperatures on the development and survival of Macrolophus pygmaeus Rambur (Hemiptera: Miridae). Biol. Control 17, 55–60. doi: 10.1006/bcon.1999.0774
Perotti, M. A., Clarke, H. K., Turner, B. D., and Braig, H. R. (2006). Rickettsia as obligate and mycetomic bacteria. FASEB J. 20, 2372–2374. doi: 10.1096/fj.06-5870fje
Pilgrim, J., Thongprem, P., Davison, H. R., Siozios, S., Baylis, M., Zakharov, E. V., et al. (2021). Torix Rickettsia are widespread in arthropods and reflect a neglected symbiosis. Gigascience 10, 1–19. doi: 10.1093/gigascience/giab021
Rio, R. V. M., Attardo, G. M., and Weiss, B. L. (2016). Grandeur alliances: symbiont metabolic integration and obligate arthropod hematophagy. Trends Parasitol. 32, 739–749. doi: 10.1016/j.pt.2016.05.002
Schaefer, C. W., and Panizzi, A. R. (2000). Economic Importance of Heteroptera. Washington, D.C: CRC Press. 37–83.
Semiatizki, A., Weiss, B., Bagim, S., Rohkin-Shalom, S., Kaltenpoth, M., and Chiel, E. (2020). Effects, interactions, and localization of Rickettsia and Wolbachia in the house fly parasitoid, Spalangia endius. Microb. Ecol. 80, 718–728. doi: 10.1007/s00248-020-01520-x
Simon, C., Frati, F., Beckenbach, A., Crespi, B., Liu, H., and Flook, P. (1994). Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann. Entomol. Soc. Am. 87, 651–701. doi: 10.1093/aesa/87.6.651
Sudakaran, S., Kost, C., and Kaltenpoth, M. (2017). Symbiont acquisition and replacement as a source of ecological innovation. Trends Microbiol. 25, 375–390. doi: 10.1016/j.tim.2017.02.014
Thomas, S. (2016). Rickettsiales: Biology, Molecular Biology, Epidemiology, and Vaccine Development. New York: Springer. 2–23.
Thongprem, P., Davison, H. R., Thompson, D. J., Lorenzo-Carballa, M. O., and Hurst, G. D. D. (2021). Incidence and diversity of Torix Rickettsia–Odonata symbioses. Microb. Ecol. 81, 203–212. doi: 10.1007/s00248-020-01568-9
Tzuri, N., Caspi-Fluger, A., Betelman, K., Rohkin Shalom, S., and Chiel, E. (2021). Horizontal transmission of microbial symbionts within a guild of fly parasitoids. Microb. Ecol. 81, 818–827. doi: 10.1007/s00248-020-01618-2
Watanabe, K., Yukuhiro, F., Matsuura, Y., Fukatsu, T., and Noda, H. (2014). Intrasperm vertical symbiont transmission. Proc. Natl. Acad. Sci. U. S. A. 111, 7433–7437. doi: 10.1073/pnas.1402476111
Weinert, L. A., Morand, S., and Krasnov, B. R. and, and Littlewood, D. T. J. (2015). The Diversity and Phylogeny of Rickettsia. Parasite Diversity and Diversification: Evolutionary Ecology Meets Phylogenetics. Cambridge: Cambridge University Press.
Weisburg, W. G., Barns, S. M., Pelletier, D. A., and Lane, D. J. (1991). 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703. doi: 10.1128/jb.173.2.697-703.1991
Werren, J. H., Hurst, G. D. D., Zhang, W., Breeuwer, J. A. J., Stouthamer, R., and Majerus, M. E. N. (1994). Rickettsial relative associated with male killing in the ladybird beetle (Adalia bipunctata). J. Bacteriol. 176, 388–394. doi: 10.1128/jb.176.2.388-394.1994
Wheeler, A. G. (2002). Biology of the Plant Bugs (Hemiptera: Miridae): Pests, Predators, Opportunists. New York: Cornell University Press. 13–15, 125–131.
Wheeler, A. G., and Krimmel, B. A. (2015). Mirid (Hemiptera: Heteroptera) specialists of sticky plants: adaptations, interactions, and ecological implications. Annu. Rev. Entomol. 60, 393–414. doi: 10.1146/annurev-ento-010814-020932
Keywords: FISH, Macrolophus melanotoma, Macrolophus pygmaeus, omnivory, Rickettsia bellii, Rickettsia limoniae
Citation: Dally M, Izraeli Y, Belausov E, Mozes-Daube N, Coll M and Zchori-Fein E (2023) Rickettsia association with two Macrolophus (Heteroptera: Miridae) species: A comparative study of phylogenies and within-host localization patterns. Front. Microbiol. 13:1107153. doi: 10.3389/fmicb.2022.1107153
Edited by:
Takema Fukatsu, National Institute of Advanced Industrial Science and Technology, JapanReviewed by:
Olivier Duron, Centre National de la Recherche Scientifique, FranceJun-Bo Luan, Shenyang Agricultural University, China
Copyright © 2023 Dally, Izraeli, Belausov, Mozes-Daube, Coll and Zchori-Fein. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Einat Zchori-Fein, ✉ ZWluYXRAYWdyaS5nb3YuaWw=
 Maria Dally
Maria Dally Yehuda Izraeli
Yehuda Izraeli Eduard Belausov
Eduard Belausov Netta Mozes-Daube2
Netta Mozes-Daube2 Moshe Coll
Moshe Coll Einat Zchori-Fein
Einat Zchori-Fein