- 1Department of Laboratory Medicine, Nanjing Drum Tower Hospital, The Affiliated Hospital of Nanjing University Medical School, Nanjing, Jiangsu, China
- 2Department of Acute Infectious Disease Control and Prevention, Jiangsu Provincial Center for Disease Control and Prevention, Nanjing, China
Objective: The worldwide dissemination of colistin-resistant Escherichia coli (E. coli) endangers public health. This study aimed to better understand the global genomic epidemiology of E. coli isolates carrying mobilized colistin resistance (mcr) genes, providing information to assist in infection and prevention.
Methods: Escherichia coli genomes were downloaded from NCBI, and mcr was detected using BLASTP. Per software was used to extract information on hosts, resources, collection data, and countries of origin from GenBank. Sequence types (STs), prevalence of plasmids, antimicrobial resistance genes (ARG), and virulence factors (VF) in these genomes were analyzed. Statistical analyses were performed to assess the relationships between mcr, ARGs, plasmids, and STs.
Results: In total, 778 mcr-positive isolates were identified. Four mcr variants were detected, with mcr-1 (86.1%) being the most widespread, followed by mcr-9 (5.7%), mcr-5 (4.4%), and mcr-3 (3.0%). Multiple ARGs were identified, with blaCTX–M (53.3%), fosA (28.8%), qnr (26.1%), blaNDM (19.8%), and aac (6’)-Ib-cr (14.5%) being the most common. Overall, 239 distinct STs were identified, of which ST10 (13.8%) was the most prevalent. A total of 113 different VFs were found, terC (99.9%) and gad (83.0%) were most frequently detected. Twenty types of plasmids were identified; IncFIB (64.1%), IncX (42.3%), and IncX (42.3%) were the most common replicons. IncI2 and IncX4 were frequently detected in mcr-1-positive isolates, whereas IncFII, IncI1-I, and IncHI2 were dominant plasmids in mcr-3, mcr-5, and mcr-9-positive isolates, respectively. A higher frequency of ARGs and VFs was observed among ST156 and ST131 isolates.
Conclusion: Our data indicated that more than half of the mcr-positive E. coli strains carried endemic ARGs and VFs. ST10 and ST156 isolates deserved further attention, given the rapid transmission of ST10 and the convergence of ARGs and VFs in ST156.
Introduction
Escherichia coli is a common pathogen responsible for multiple infections, including those in the urinary tract, bloodstream, and wounds, in humans and animals worldwide (Nang et al., 2019). It can easily acquire multi-drug resistance, owing to its ability to accumulate multiple resistance genes, primarily through horizontal gene transfer of mobile elements. Currently, its acquisition of genes encoding extended-spectrum β-lactamases (ESBLs), carbapenem-hydrolyzing ß-lactamase (CHßls), 16S rRNA methylases, plasmid-mediated quinolone resistance (PMQR), and plasmid-mediated glutathione S-transferase (PMGST) is of great clinical concern (Simoni et al., 2021; Yin et al., 2021; Guo et al., 2022), accelerating the evolution of drug resistance, making colistin the last-resort antibiotic for treating infections caused by multi-drug resistant bacteria. The use of colistin in livestock as a growth promoter has been banned in China (Wang et al., 2020) due to increasing polymyxin resistance. However, the use of colistin in clinical settings is increasing because of the emergence of increased multi-drug resistance (MDR), particularly carbapenem-resistant E. coli (CRE) (Liu et al., 2020); therefore, the emergence and spread of colistin-resistant E. coli are inevitable.
Colistins primarily bind to the outer membrane lipopolysaccharide through the interaction of their cationic residues with the phosphate groups of lipid A, changing the permeability of the cell envelope, leading to leakage of cell contents and bacterial death (Rodríguez-Santiago et al., 2021). However, mobile colistin resistance (mcr) genes have been reported to be the primary mechanism conferring resistance to colistin, although two-component systems (TCSs), such as PmrAB and PhoPQ; mutation(s) in the MgrB regulator; LPS modification (Bialvaei and Samadi Kafil, 2015); LPS overall charge changes, and the reduced affinity of colistin for the outer membrane, also contribute to colistin resistance based on chromosomal-encoded mechanisms (Zhang et al., 2021; Phuadraksa et al., 2022). The mcr genes encode transferable phosphoethanolamine transferases that modify the lipopolysaccharide of the bacterial outer membrane to weaken its binding to colistin (Yin et al., 2021). To date, more than 10 mcr variants (mcr-1 to mcr-10) have been identified, carried on different conjugative and non-conjugative plasmid backbones (Mmatli et al., 2022). These variants have been widely identified in many bacteria, including E. coli, Klebsiella, Enterobacter, Citrobacter, Proteus, Providencia, Salmonella, Pseudomonas, Acinetobacter, Aeromonas, Kluyvera, and Raoultella, as well as in diverse ecosystems, including soil, botanicals, wildlife, animal environments, and public places (Nang et al., 2019; Anyanwu et al., 2020). However, despite its status as the most common host for mcr genes, data on mcr distribution in E. coli are quite limited, although global surveillance showed an approximately 1.26% prevalence of mcr-positive E. coli among humans, animals, and environments (Dadashi et al., 2022; Mmatli et al., 2022), with the mcr-1, mcr-3, mcr-5, mcr-9, and mcr-10 variants most prevalent (Dadashi et al., 2022). These investigations all relied on PCR detection methods, with few data on the prevalence of mcr from global genomic databases.
Most importantly, mcr genes are carried by mobile genetic elements, especially conjugative plasmids, leading to their broad dissemination and establishment worldwide under the selective pressure of antimicrobial agents. Although isolates with mcr as well as other antimicrobial resistance genes (ARG), such as blaCHßLs, blaESBLs, PMQR, and fosA, have been frequently reported, posing clinical challenges (Haenni et al., 2016; Zhang W. et al., 2022), the prevalence of clinically endemic ARGs among mcr-positive strains remains unclear. Furthermore, a high diversity of plasmid reservoirs has been shown to be associated with mcr genes (Nang et al., 2019); however, the distribution of plasmid replicons among global mcr-positive E. coli and the associations between mcr and plasmid replicons remain unknown. Furthermore, multiple virulence factors (VFs) involved in bacterial adherence, invasion, immune modulation, effector delivery systems, and nutritional/metabolic factors have been identified in E. coli (Zhao et al., 2020; Kubelová et al., 2021). However, their distribution among mcr-positive E. coli and the association between VFs and ARGs require further investigation. Finally, the prevalence of VFs among internationally popular sequence types (STs) of mcr-positive E. coli is of vital importance. All of these will contribute to optimizing prevention and control measures to prevent further spread and outbreaks of mcr-positive infections.
In this study, we first investigated the distribution of mcr among global E. coli based on the whole genome sequencing (WGS) data from GenBank. We analyzed STs and the prevalence of ARGs, VFs, and plasmid replicons. Additionally, the distribution consistency of VFs and ARGs was tested, providing epidemiological data for the implementation of infection and prevention.
Materials and methods
Genomes
All E. coli genomes updated to 2021 were downloaded from NCBI using Aspera software in batches (Kim et al., 2018). As some of the genome-wide sequencing data from 2019 to 2021 have not yet been released when the study started, only strains identified from 1905 to 2019 were included in this study. Totally, 22,884 genomes were downloaded and tested for quality. The qualifying parameters were completeness >90% and contamination <5, with contig quantity ≤500 and N50 ≥40,000.
mcr identification
All mcr sequences were obtained from the NCBI Biological Resistance Reference Gene Database [(mcr)].1 BLASTP was performed, with thresholds set as expected value = 1e-5, coverage ≥50%, identity = 100%, and matching length = subject gene length.
Phylogenetic tree construction
Prokka was used to annotate the 778 E. coli isolates, and Roary software was adopted to obtain multiple sequence alignment files of 1,222 core genes (Zhou et al., 2021), which were further used to acquire the single nucleotide polymorphism information through SNP sites. Finally, RaxML was used to construct the maximum likelihood tree, and the evolution tree results were visualized by itol (Liu et al., 2022).
Sequence types
Sequence types of mcr-positive E. coli were identified using CLC workbench version 21.0.1. After consensus sequences were extracted, STs were analyzed using multilocus sequence typing (MLST) with E. coli (Oxford) as the reference database.
Antimicrobial resistance prevalence
The distribution of other ARGs was investigated using CLC workbench version 21.1. The fasta file was input into the files in CLC data using standard import, and after the consensus sequence was extracted, the prevalence of ARGs was analyzed using the ResFinder database for comparison. The results were exported as scv files, and further sorted for analysis.
Distributions of plasmid replicons and VFs
Genomes were submitted to the Center for Genomic Epidemiology. PlasmidFinder software2 was used to identify plasmid replicons, and Virulence Finder 2.03 was used to identify VFs.
Statistical analyses
Correlation analyses were performed using IBM SPSS 22.0. The distribution consistency of VFs and ARGs was tested by McNimar analysis, and a p-value of > 0.05 was taken as the consistency between them.
Results
General characteristics
Overall, 778 mcr-positive E. coli isolates were identified from 22,884 E. coli genomes (Supplementary Table 1). They were isolated in 25 countries covering 6 continents: including Asia (n = 555), South America (n = 119), Europe (n = 40), North America (n = 27), Africa (n = 17), and Oceania (n = 13). China (n = 401), Japan (n = 26), Thailand (n = 41), Laos (n = 22), Brazil (n = 37), Paraguay (n = 28), and the USA (n = 25) were the most common sources. From 1905 to 2011, only 13 mcr-positive isolates were submitted (Figure 1). The number of mcr-positive isolates gradually increased from 2012; and hundreds of genomes were submitted every year peaking in 2016 (n = 235).
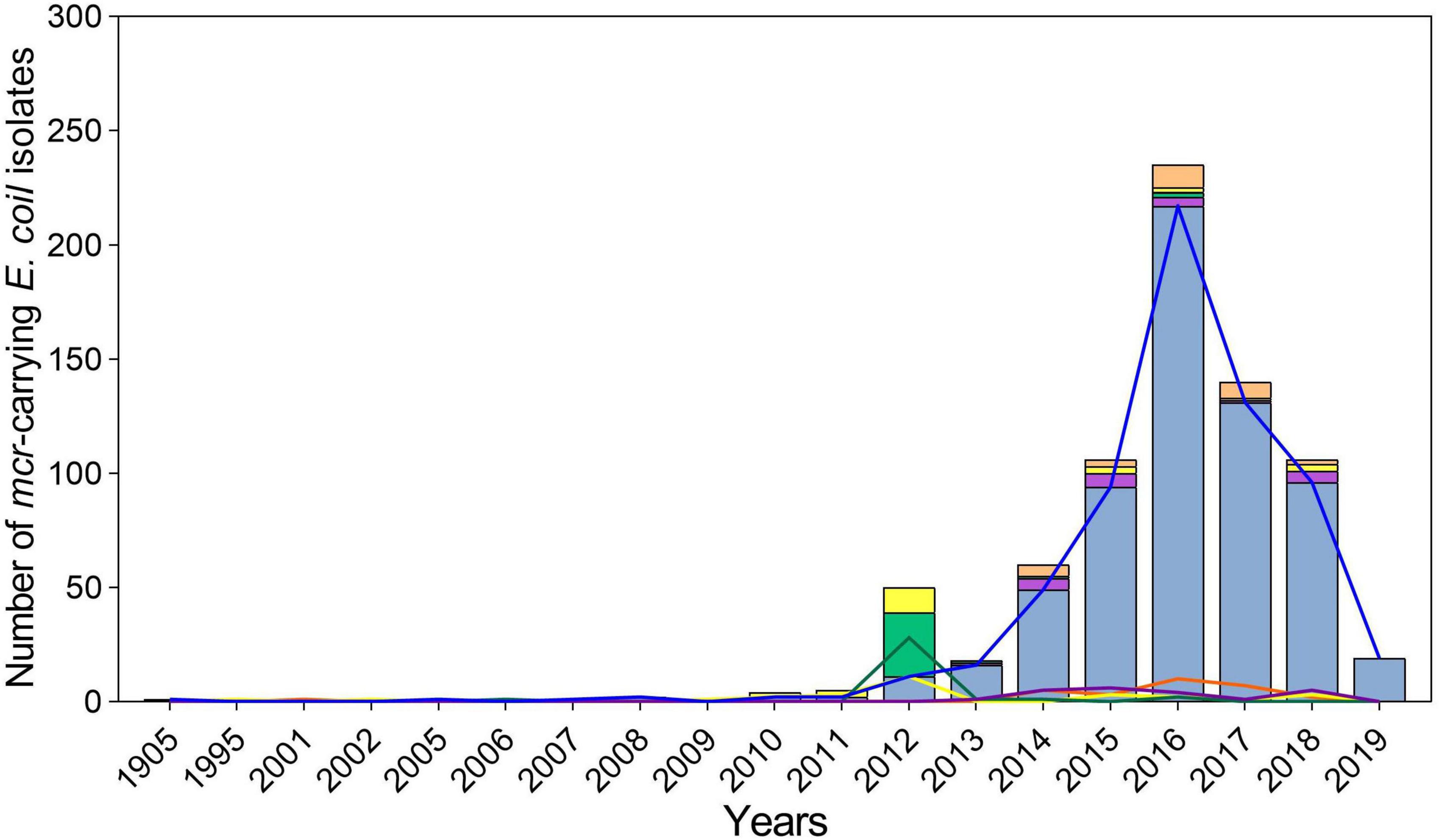
Figure 1. Characteristics of global spread of mcr-positive Escherichia coli per year. The light blue columns and blue lines represent mcr-1, the mauve columns and purple lines represent mcr-3, the light green columns and green lines represent mcr-5, the yellow columns and lines represent mcr-9, and the orange columns and lines represent the combination of varied mcr genes.
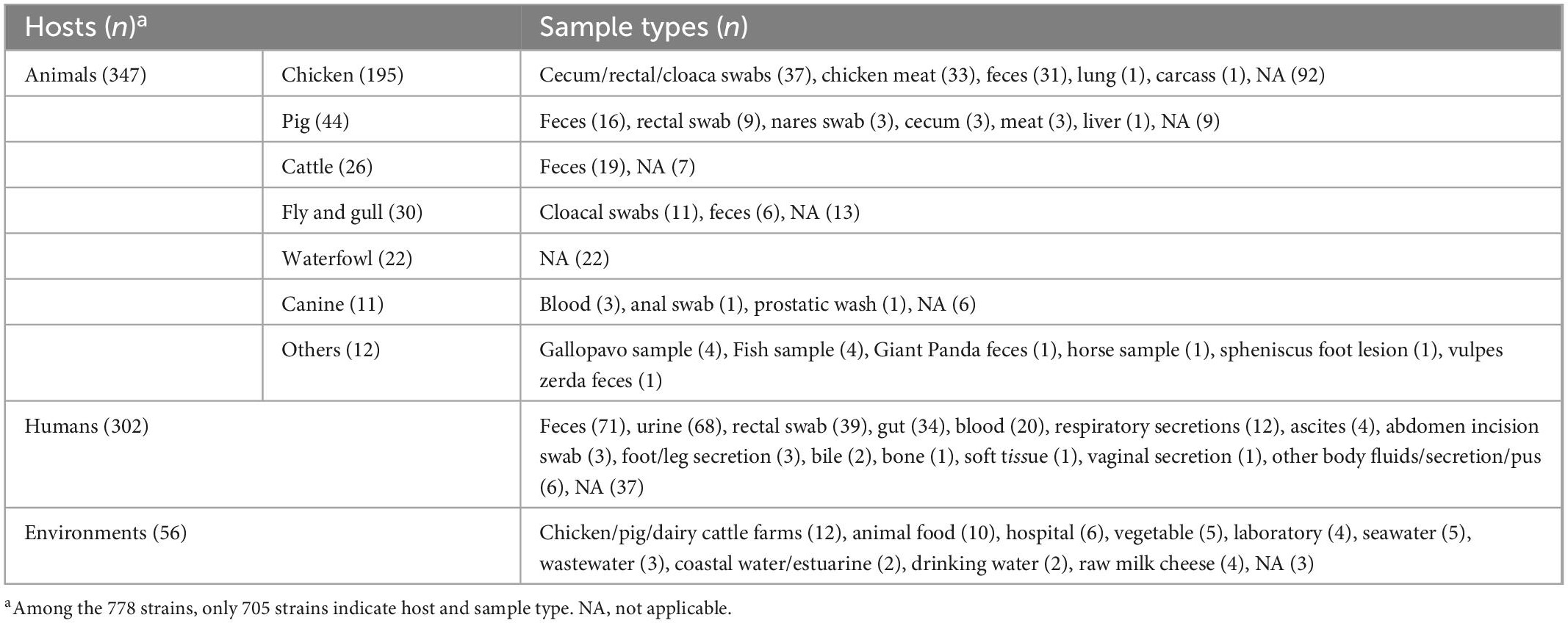
Regarding sources, we found that animals (n = 347, 44.6%) were the most common, with chickens (n = 202), pigs (n = 44), and cattle (n = 32) being the most common species. Feces (n = 72), cecum/rectal/cloaca swabs (n = 37), and meat (n = 33) were the dominant sources (Table 1). Humans accounted for 38.8% (n = 302) of all isolates taken from feces (n = 71), urine (n = 68), rectal swabs (n = 39), gut (n = 34), and blood (n = 20) (Table 1). Notably, mcr-positive strains were also found in the environment (n = 56, 7.2%), such as farms, hospitals, foods, and water.
mcr variant distribution
Four mcr variants were identified among the 778 isolates, with mcr-1 (n = 654, 86.1%) being the most prevalent one, followed by mcr-9 (n = 44, 5.7%), mcr-5 (n = 34, 4.4%), and mcr-3 (n = 23, 3.0%), besides, two or more mcr variants were found in 23 isolates. For subtypes, 1.1 (n = 633, 96.8%) was the most prevalent for mcr-1, with subtypes 1.5 and 1.7 also observed, accounting for 0.9% (n = 6) and 1.5% (n = 10) of mcr-1, respectively. Subtypes 3.1, 3.4, and 3.5 were common variants of mcr-3, accounting for 56.5% (n = 13), 13.0% (n = 3), and 21.7% (n = 5), respectively. No subtypes of mcr-5 and mcr-9 were detected. Notably, mcr-1, mcr-5, and mcr-9 were first identified in isolates collected in 1905, 1995, and 2001, respectively, before colistin was produced and used in veterinary and clinical settings. The mcr-3 variant emerged in 2013 and has begun to spread more recently (Figure 1). Geographically, Oceania variants were mostly mcr-9 (n = 12, 92.3%), whereas the most prevalent variants in South America were mcr-1 (n = 89, 74.8%) and mcr-5 (n = 30, 25.2%) (Figure 2). Both mcr-1 and mcr-9 were endemic in Europe, North America, and Africa, with mcr-1 accounting for 51.9%–88.2%. The mcr-1 (n = 523, 94.2%) and mcr-3 (n = 23, 4.1%) variants were most common in Asia (Figure 2).
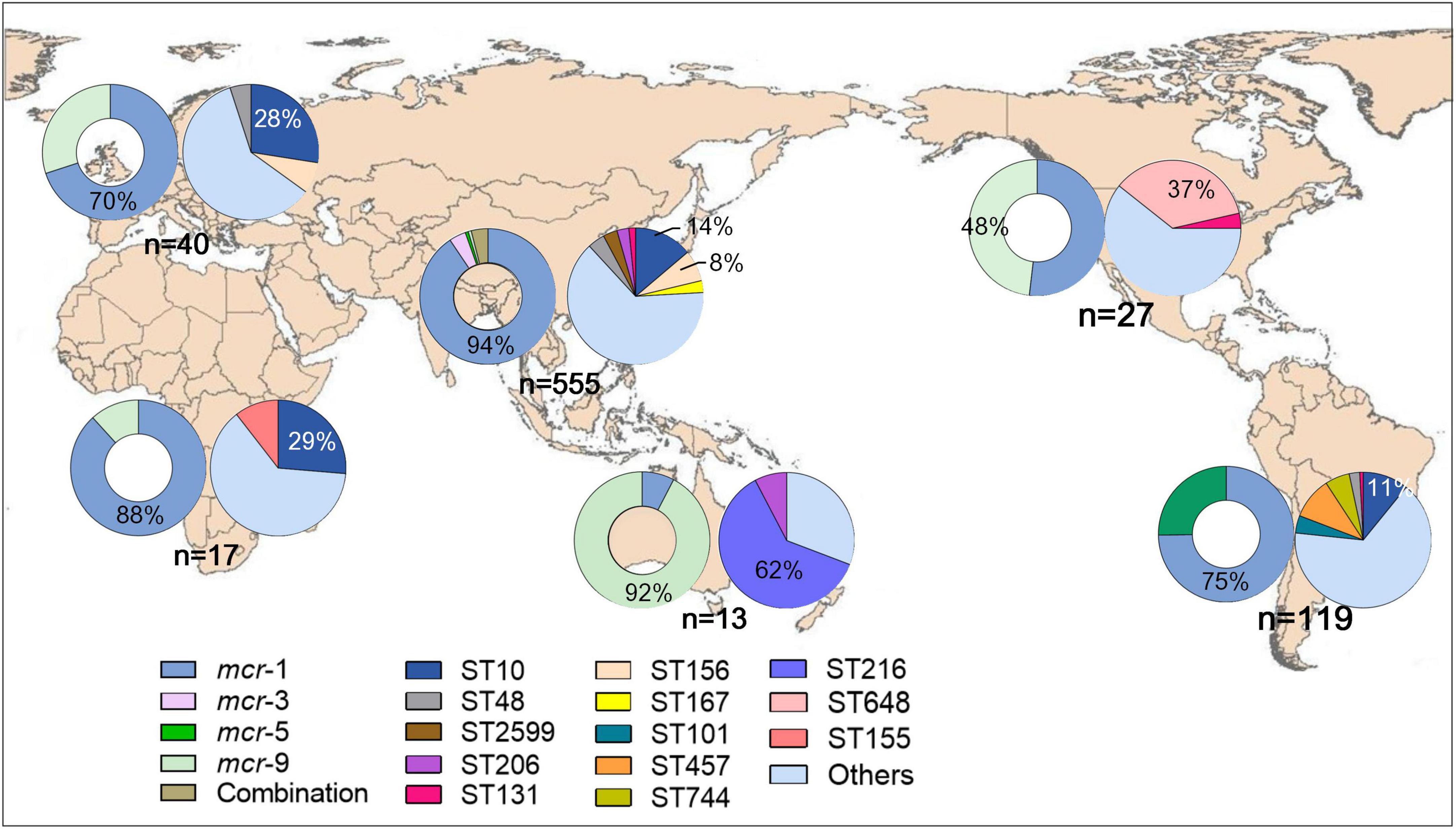
Figure 2. Geographical distribution of mcr and STs of mcr-positive Escherichia coli worldwide. The hollow and solid pie charts of each continent represent the distribution of mcr and STs, respectively.
Genetic relationship
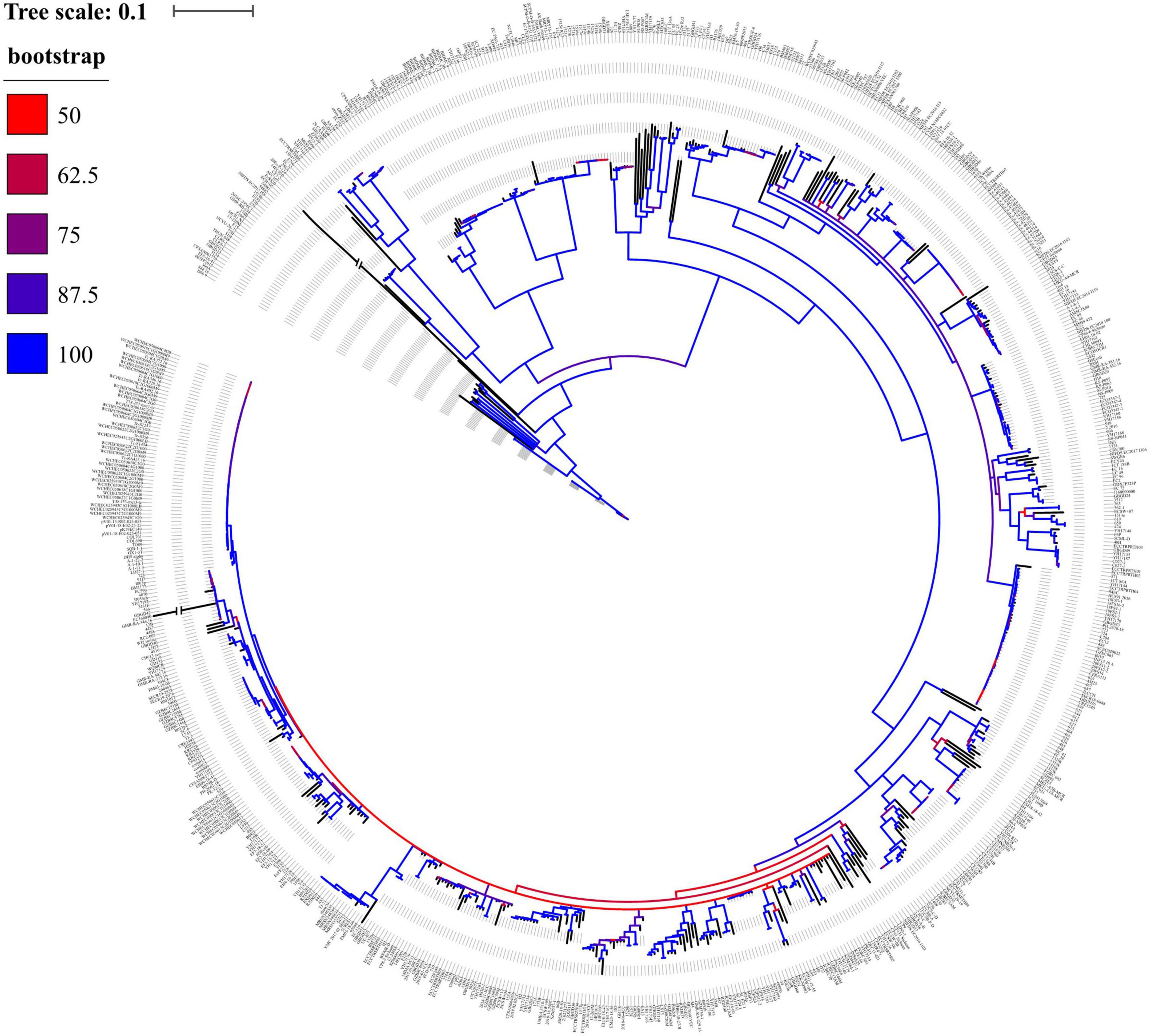
Overall, the phylogenetic tree displayed that 778 mcr-positive isolates were divided into 292 clades (Figure 3), indicating a genetic diversity of such strains. Notably, the biggest clade was composed of 55 isolates, of which 76.4% (n = 42) were ST10 from China and Colombia. Another large clade was composed of 48 isolates and 60.4% (n = 29) of them were ST156 from China, suggesting clonal dissemination of ST10 and ST156 mainly in China.
Antimicrobial resistance determinants
Multiple ARGs were identified in mcr-positive strains. The most common were ESBLs blaCTX–M (n = 415, 53.3%) and blaOXA (n = 114, 14.7%); CHßLs blaNDM (n = 154, 19.8%) and blaKPC (n = 15, 1.9%); pAmpCs blaCMY (46, 5.9%) and blaDHA (n = 7, 0.9%); 16s rRNA methylases rmtB (n = 88, 11.3%) and armA (n = 6, 0.8%); PMQRs aac (6’)-Ib-cr (n = 113, 14.5%), oqxAB (n = 56, 7.2%), and qnr (203, 26.1%); and the fosfomycin resistance gene fosA (n = 220, 28.3%) were detected. The common variant subtypes are shown in Figure 4A; blaCTX–M–55 (n = 117, 28.2%), blaCTX–M–14 (n = 94, 22.7%), and blaCTX–M–65 (n = 45, 10.8%) were dominant in blaCTX–M, whereas blaNDM–5 (n = 84, 54.6%), blaNDM–1 (n = 47, 30.5%), and blaNDM–9 (n = 19, 22.6%) were the most prevalent in blaNDM.
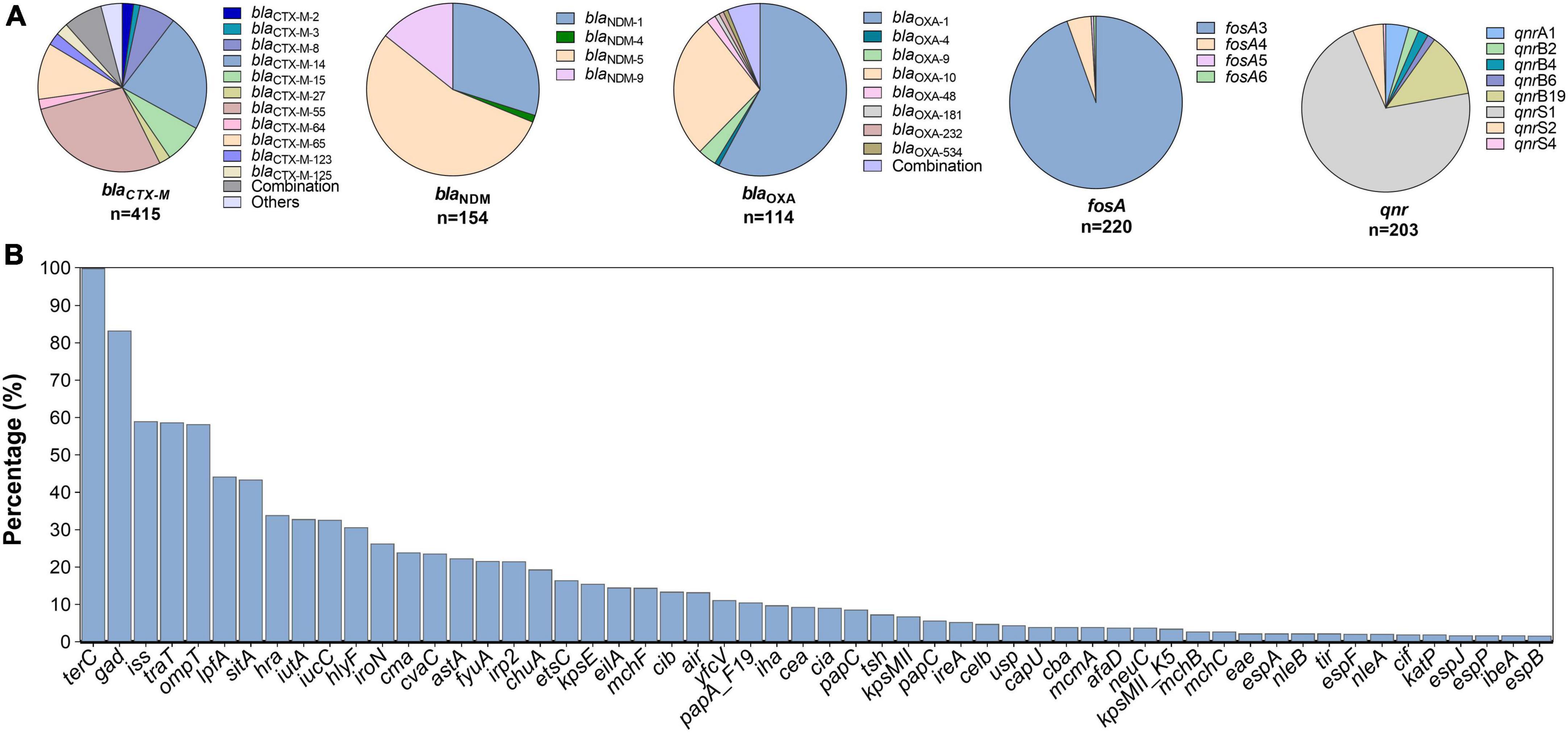
Figure 4. Subtypes of antimicrobial resistance genes (A) and prevalence of common virulent factors (B) among mcr-positive Escherichia coli.
Virulence factors
In total, 112 different VFs were identified, terC (n = 777, 99.9%) and gad (n = 646, 83.0%) being the most prevalent (Figure 4B). VFs involved in intestinal invasiveness and anti-phagocytosis, including sitA, iss, and iroN, were carried by 43.7% (340/778), 59.0% (459/778), and 26.4% of all isolates, respectively. Approximately 50% of the isolates contained traT, ompT, and lpfA with specific carrying rates of 58.61% (456/778), 57.84% (450/778), and 43.96% (342/778), respectively. Other VFs, such as hra (n = 264, 33.9%), iutA (n = 256, 32.9%), iucC (n = 256, 32.9%), and hlyF (n = 239, 30.7%), were also frequently detected. Other relatively less common VFs are shown in Figure 4B.
Sequence types
Overall, 239 distinct STs were identified, with ST10 (n = 107) being the most frequent. ST156 (n = 46), ST48 (n = 29), ST457 (n = 26), ST648 (n = 22), ST167 (n = 19), ST2599 (n = 19), ST101 (n = 18), ST206 (n = 16), ST617 (n = 15), ST744 (n = 13), ST155 (n = 12), ST2705 (n = 12), ST354 (n = 12), ST216 (n = 11), ST131 (n = 11), and ST69 (n = 10) were also detected. Other STs were detected (n < 10). Geographically, ST10 was the dominant type in Asia, Africa, Europe, and South America; ST156 was endemic in both Asia and Europe; and ST457 was the most common type in South America (Figure 2). The most common STs in Oceania and North America were ST216 and ST648, respectively (Figure 2).
Plasmid replicons
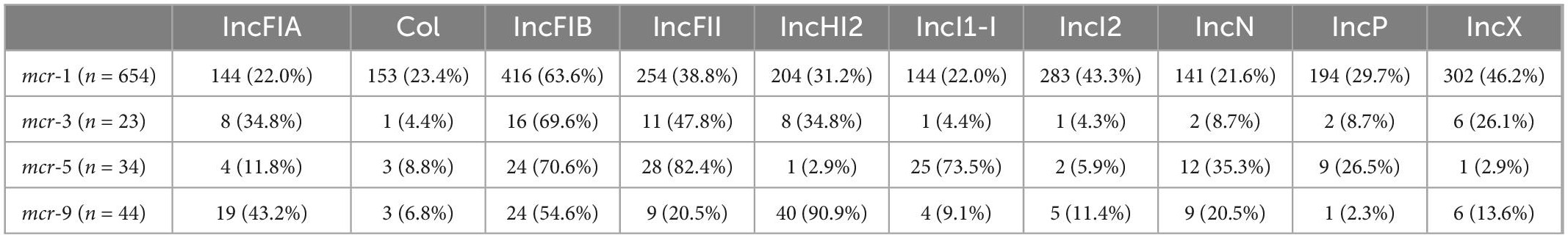
Of the 778 isolates, 20 types of plasmid replicons were identified with IncFIB (n = 499, 64.1%) being the most common replicon, followed by IncX (n = 329, 42.3%), IncFII (n = 316, 40.6%), IncI2 (n = 300, 38.6%), IncHI2 (n = 257, 33.0%), Col (n = 177, 22.8%), IncP (n = 210, 27.0%), IncFIA (n = 186, 23.9%), IncFIC (n = 150, 19.3%), IncI1-I (n = 179, 23.0%), IncY (n = 105, 13.5%), IncN (n = 170, 21.9%), and other more rare plasmid replicons (Table 2). Despite 50–70% of strains carrying IncFIB plasmids, several plasmids, such as IncI2, IncX, IncHI2, and IncFII, were relatively highly carried in mcr-1 strains, whereas plasmid IncFII was common in mcr-3 strains; 90.9% of mcr-9 strains carried IncHI2 and more than 70% of mcr-5 positive strains carried IncFII and IncI1-I plasmids (Table 2). The distribution of blaNDM was correlated with those of the plasmid replicons IncFIA, Col, IncI1-I, and IncN (p > 0.05). The prevalence of qnr was correlated with those of IncFIA, Col, IncI1-I, and IncP (p > 0.05). Additionally, the incidences of fosA, IncFIA, and IncP were correlated (p > 0.05) (Table 3). No significant differences were found in the distribution of some VFs and plasmid replicons, including the combination of hra, iutA, iucC, hlyF with IncHI2; sitA with IncX; lpfA with IncFII; and iroN with IncFIA, Col, IncI1-I, IncN, and IncP (Table 3).
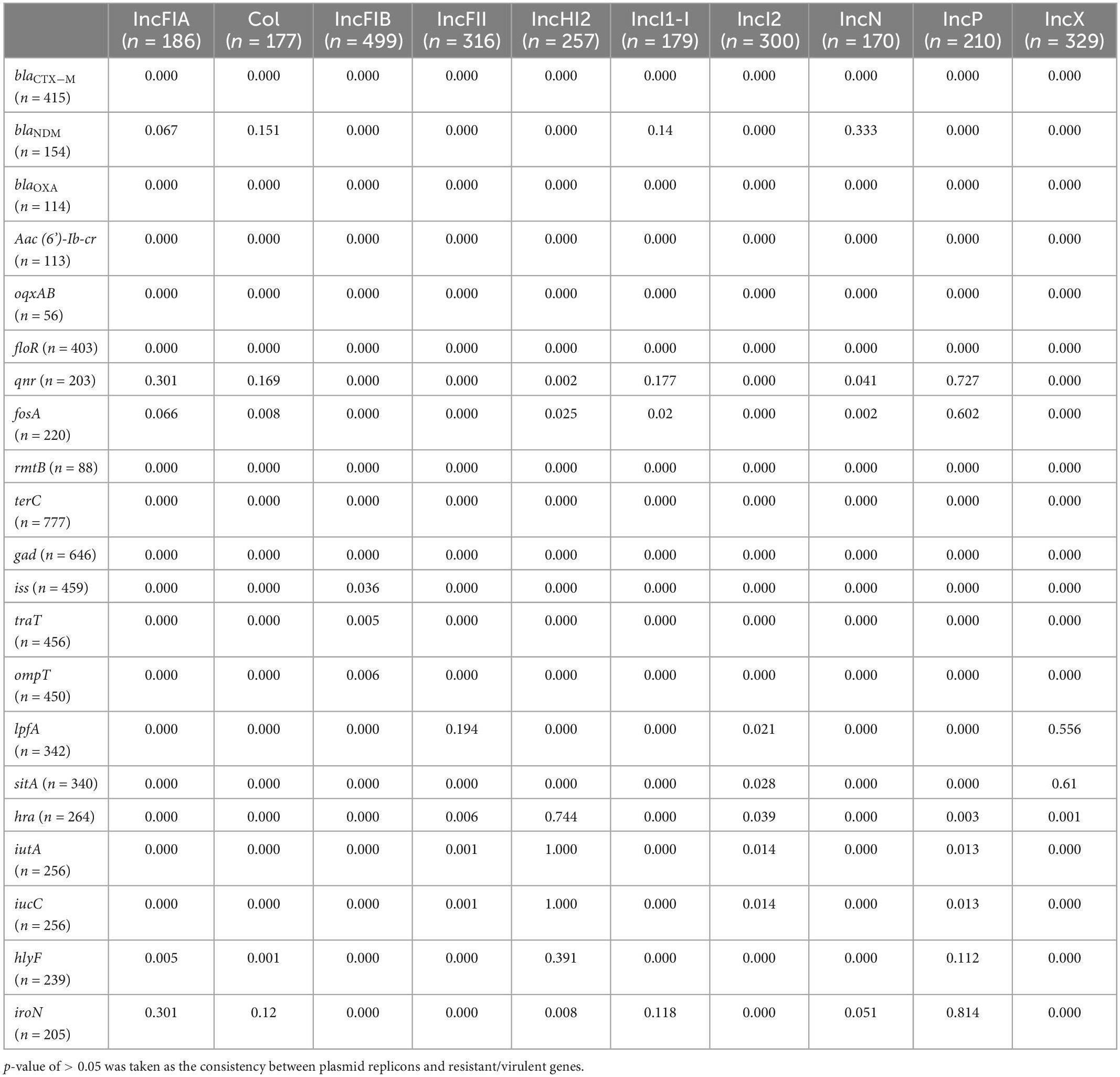
Table 3. P-values for differences in distribution of plasmid replicons and resistant/virulent genes.
The prevalence of ARGs and VFs among the ST10, ST156, ST457, ST648, ST216, and ST131 clones were compared. As the most prevalent ST worldwide, ST10 had a lower incidence of ARGs, whereas increased VFs and ARGs were observed among ST156 clones. For example, more than 50% of ST156 isolates carried blaCTX–M, blaNDM, aac (6’)-Ib-cr, oqxAB, floR, fosA, rmtB, lpfA, and Hra genes (Figure 4), indicating high virulence. Moreover, compared with ST648 and ST216, ST457 and ST131 were more commonly associated with VFs, including the traT, ompT, lpfA, sitA, iutA, iucC, hlyF genes, all of which have been implicated in bacterial survival, invasiveness, and adhesion (Figure 5).
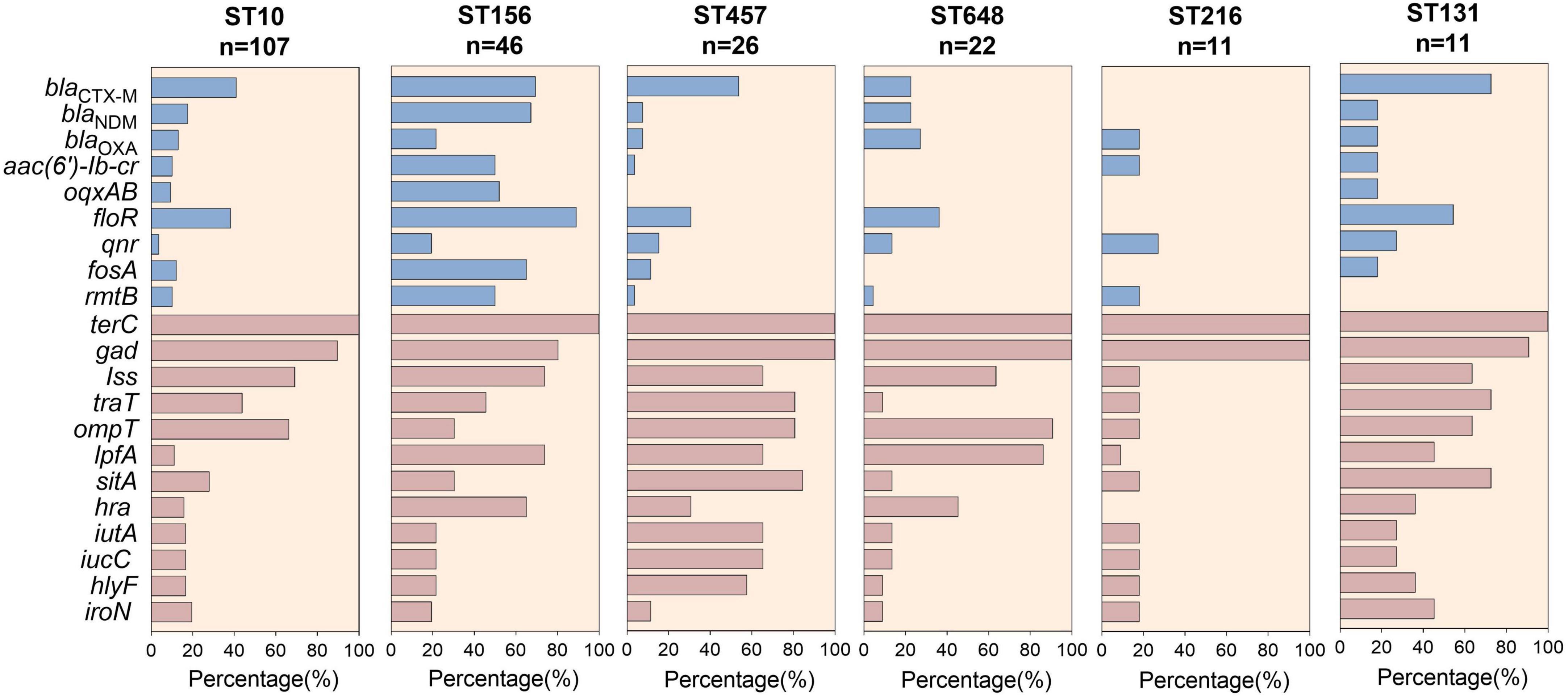
Figure 5. Carriage of antimicrobial resistance genes and virulent factors among globally prevalent ST strains.
Discussion
As the last-line defense against MDR E. coli infections, colistin has been widely used to treat infections caused by extensive-drug resistant (XDR) bacteria, such as CRE, albeit it has been banned for use in livestock as a growth promoter. The clinical utility of colistin is currently threatened by the global dissemination of mcr and its co-occurrence with other endemic ARGs, such as ESBL and CHßl, through different plasmids. Thus, understanding the prevalent characteristics of global mcr-positive E. coli isolates, including the prevalence of STs, endemic ARGs, VFs, and plasmid replicons, will be useful to guide clinical treatment for serious infections, as well as the implementation of preventative measures.
In this study, we observed a rapid increase in the genomes of global mcr-positive E. coli from 2014 to 2016. Since 2016, many countries have prohibited colistin as a growth promoter for livestock, including Brazil (November 2016), Thailand (February 2017), China (April 2017), Japan (July 2018), Malaysia (January 2019), Argentina (February 2019), and India (July 2019) (Center for Veterinary Drug Development, 2018; Shen et al., 2018; Wang et al., 2020). Although the number of mcr-positive E. coli genomes declined after 2016, given that some of the sequence data from 2019 to 2021 had not yet been released when we downloaded the data and conducted this study in 2021, the actual prevalence of mcr-positive strains worldwide should be more than the number we analyzed, so current global prevalence would likely be significantly higher. Additionally, as the poultry and swine industries accounted for 96% of total colistin sulfate livestock use (Wang et al., 2020), the presence of mcr in them provides evidence that colistin treatment has promoted the transmission of mcr, with livestock as the primary reservoir. Colistin-resistant E. coli can then spread by contaminating animal-derived food or contaminating crops by excrement to threaten public safety (Elbediwi et al., 2019).
The high incidence of mcr in our study is in accordance with a previous report (Dadashi et al., 2022), consistent with its identification as the globally predominant colistin-resistance gene. At present, mcr-1 could confer colistin resistance with broad minimal inhibitory concentrations (MICs) of 2–16 mg/L (Nang et al., 2019), for instance, 74% of mcr-1 positive E. coli isolates had MICs at 2 mg/L (Nguyen et al., 2022), while 52% of mcr-1 positive E. coli isolates in another study exhibited MICs of 4∼8 mg/L (Ćwiek et al., 2021). For other mcr genes, mcr-3 has been globally disseminated since it was first reported in Yin et al. (2017), and this gene inside the linker has been found to appear as a facilitator of colistin resistance (Xu et al., 2021). It also has been reported to facilitate evasion of host phagocytosis, but generally mediates low-level (≤8 mg/L) colistin resistance among Enterobacteriaceae (Yin et al., 2021). The mcr-9 variant was generally susceptible to colistin with MICs of ≤1 mg/L, because it did not confer colistin resistance itself, its colistin resistance replied to gene regulation (Nakamura et al., 2021), and mcr-5 was demonstrated to generate colistin resistance with MICs of 4–8 mg/L in S. enterica (Nang et al., 2019). Notably, more than 20% mcr-positive strains co-carried CHßLs, mainly blaNDM followed by blaKPC, indicating that almost one-fifth of the strains in our study may belong to CRE. In addition, more than 50% of the strains co-carried blaCTX–M, such a high prevalence of blaCTX–M showed that most of these strains belonged to MDR strains. Meanwhile, the co-carriage of 16s RNA methylase, PMQRs, and fosfomycin genes suggested that the mcr-positive strains were also reservoirs of numerous resistance genes (Guo et al., 2022).
The VF analysis showed that almost all mcr-positive strains also carried terC and gad genes. To the best of our knowledge, terC represents one of the key proteins in tellurite and colicine resistance, implicated in phage inhibition, resistance, and pathogenicity, and is widespread among bacterial species, particularly in pathogens (Peng et al., 2021; Rodríguez-Santiago et al., 2021). In contrast, gad has been reported to enzymatically decrease intracellular protons linked to E. coli flagellar motility (Yamanaka et al., 2022) and is involved in the colonization of the gastrointestinal tract. In addition, more than half of the strains contained iss, traT, and ompT, which participate in resistance to serum complement, outer membrane protease, and long polar fimbriae, respectively; these affect resistance to host innate immunity and facilitate colonization and adhesion. Moreover, iroN was detected in approximately 25% of all isolates and has been associated with increased 30-day mortality in patients with bacteremia (Hung et al., 2019).
We identified many STs, demonstrating the diversity of mcr-positive strains. The high prevalence of ST10 in our study was similar to that in a previous report (Dadashi et al., 2022), however, it was different from a report that ST131 was the most common ST for extraintestinal pathogenic E. coli (ExPEC) strains (Manges et al., 2019). To date, E. coli ST10 has been shown to be a high-risk mcr-1-positive isolate from cattle farm environments (Ali et al., 2021); its clonal spread with ∼50% fosfomycin resistance has been found among diarrheal calves in Xinjiang province, China (He et al., 2021). It also has been isolated from poultry in Poland (Ćwiek et al., 2021). The biological costs imposed by plasmid-mediated resistance and virulence affect its survival and spread (Yang et al., 2020), thus, the relatively rare ARGs and VFs within ST10 clones may explain its rapid spread.
Curiously, STs differed from continent to continent, and as frequent carriers for mcr-1 and blaNDM (Lin et al., 2020; Zhang X. et al., 2022), more ARGs and VFs were observed within the ST156 strain prevailing in Asia and Europe, indicating that it represents a major risk to public health, highlighting the need for increased surveillance. For example, E. coli ST156, carrying blaNDM–5, blaCTX–M–65, blaOXA–10, blaTEM–1, and mcr-1, has been found in clinical isolates in China (Lin et al., 2020; Zhang X. et al., 2022). Notably, ST648 has been reported as a high-risk, MDR, ESBL-producing strain in public aquatic environments (Furlan et al., 2020), but ST648 was not frequently detected in our study, with almost all of them being in North America. The differences in the distributions of ST clones may be related to antibiotic use, diet, and environmental factors. As the predominant ST among ExPEC isolates worldwide (Alangari et al., 2022), ST131 was shown to have higher virulence than other important ExPEC clones (Alqasim et al., 2020), whereas only 11 mcr-positive E. coli ST131 isolates were identified in this study, mainly from China, suggesting that ST10, rather than ST131 and ST648, is the main host for mcr, which may result from the fitness between the clones and mcr genes. Moreover, although ST216, with fewer VFs and ARGs, was dominant in Oceania, their association with IncHI2 plasmids may be of great concern (Tarabai et al., 2021).
The great diversity of plasmid replicons within mcr-positive strains indicates the great dissemination potential of ARGs and VFs. The high incidence of IncFIB is consistent with previous reports showing that IncFIB is common in Enterobacteriaceae (Khine et al., 2020). A previous study found that IncI2 and IncX conferred fitness advantages to host bacteria and outcompeted other plasmids (Bahador et al., 2018); the approximately 40% incidence of IncX and IncFII among mcr-positive strains suggests that most of the strains in this study have relatively high adaptability. Notably, IncI2 and IncX were common plasmids in mcr-1-positive strains, indicating that they may be dominant vectors driving mcr-1 transmission, but this finding of correlation analysis should be further confirmed. Considering the distribution deviation, we hypothesized that mcr-3 and mcr-9 were harbored by IncFII and IncHI2 plasmids, respectively, and mcr-5 may be borne on IncFII and IncI1-I plasmids. These findings were in accordance with previous reports that mcr-1 was carried by a range of plasmids such as IncHI2, IncI2, IncX4, IncFIA, and IncP (Haenni et al., 2016; Elbediwi et al., 2019; Liu et al., 2020; Sonnevend et al., 2022), and mcr-9 was always carried by IncHI2 (Simoni et al., 2021). Recently, blaNDM was reported to be disseminated via the IncP plasmid with a broad host range (Choudhury et al., 2019); moreover, most VFs that were associated with enteroinvasive E. coli, such as sit, iron, iss, iutA, ompT, and iroN, were reported to be located mainly on IncFIA/FIB and Col plasmids (Touzain et al., 2018; Hayashi et al., 2019). Both mcr-1 and blaCTX–M–1 were colocalized on IncHI2 and IncI2 plasmids in two studies (Haenni et al., 2016; Yin et al., 2021), which support the consistent distribution of plasmids and ARGs/VFs analyzed in our study, suggesting the dissemination potential of the related ARGs and VFs.
To the best of our knowledge, ours is the first study to evaluate the prevalence of global mcr-harboring E. coli isolates using WGS data. The 778 mcr-positive isolates, widely distributed among the six continents during 1905–2019, could be representative of the current global mcr-carriers. However, this study had some limitations. First, the actual resistance phenotypes of these strains were not available for assessment of differences between genotypes and phenotypes. Second, whether the resistance and mcr genes were carried on a single plasmid was unclear, so we cannot clearly illustrate the specific relationships between VFs and ARGs among plasmids. Third, our study depended on the submission of genomes to the database, so it could not represent areas where no data had been uploaded.
Collectively, our data found that mcr-1 is the most widespread gene that confers resistance to colistin. More than 50% of the global mcr-positive E. coli isolates had numerous ARGs, and the fifth strain belonged to CRE. Diverse STs have been identified, further attention should be focused on the ST10 and ST156 clones, considering the rapid transmission of ST10 and the convergence of ARGs and VFs within ST156.
Data availability statement
The original contributions presented in this study are included in this article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
XC and HS contributed to the experimental design of the study. JS, HZ, CL, HX, and CCL performed data acquisition and statistical analysis. JS, HZ, XC, and HS performed bioinformatics analysis and writing. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (81902124 and 82002205).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1105401/full#supplementary-material
Footnotes
- ^ https://www.ncbi.nlm.nih.gov/pathogens/isolates#/refgene/gene_family
- ^ https://cge.cbs.dtu.dk/services/PlasmidFinder/
- ^ https://cge.cbs.dtu.dk/services/VirulenceFinder/
References
Alangari, A., Jaffal, A. A., Alyousef, A. M., and Alyousef, A. A. (2022). Comparative metabolic characterization of extraintestinal pathogenic Escherichia coli blood isolates from Saudi Arabia. J. Trop. Med. 30:1745835. doi: 10.1155/2022/1745835
Ali, A., Fontana, H., Sano, E., Li, R., Humayon, M., Rahman, S., et al. (2021). Genomic features of a high-risk mcr-1.1-positive Escherichia coli ST10 isolated from cattle farm environment. Environ. Sci. Pollut. Res. Int. 28, 54147–54152. doi: 10.1007/s11356-021-15437-6
Alqasim, A., Abu Jaffal, A., and Alyousef, A. A. (2020). Prevalence and molecular characteristics of sequence type 131 clone among clinical uropathogenic Escherichia coli isolates in Riyadh, Saudi Arabia. Saudi J. Biol. Sci. 27, 296–302. doi: 10.1016/j.sjbs.2019.09.020
Anyanwu, M. U., Jaja, I. F., and Nwobi, O. C. (2020). Occurrence and characteristics of mobile colistin resistance (mcr) gene-containing isolates from the environment: A review. Int. J. Environ. Res. Public Health 17:1028. doi: 10.3390/ijerph17031028
Bahador, A., Farshadzadeh, Z., Raoofian, R., Mokhtaran, M., Pourakbari, B., Pourhajibagher, M., et al. (2018). Association of virulence gene expression with colistin-resistance in Acinetobacter baumannii: Analysis of genotype, antimicrobial susceptibility, and biofilm formation. Ann. Clin. Microbiol. Antimicrob. 17:24. doi: 10.1186/s12941-018-0277-6
Bialvaei, A. Z., and Samadi Kafil, H. (2015). Colistin, mechanisms and prevalence of resistance. Curr. Med. Res. Opin. 31, 707–721. doi: 10.1185/03007995.2015.1018989
Center for Veterinary Drug Development (2018). Development., Revocation of designation of virginiamycin, colistin sulfate, and decoquinate as feed additives. Ebetsu: Rakuno Gakuen University.
Choudhury, N. A., Paul, D., Das, B. J., Dhar Chanda, D., and Bhattacharjee, A. (2019). Adaptation of bla(NDM)through IncP plasmid within broad host range. Indian J. Med. Microbiol. 37, 527–530. doi: 10.4103/ijmm.IJMM_20_48
Ćwiek, K., Woźniak-Biel, A., Karwañska, M., Siedlecka, M., Lammens, C., Rebelo, A. R., et al. (2021). Phenotypic and genotypic characterization of mcr-1-positive multidrug-resistant Escherichia coli ST93, ST117, ST156, ST10, and ST744 isolated from poultry in Poland. Braz. J. Microbiol. 52, 1597–1609. doi: 10.1007/s42770-021-00538-8
Dadashi, M., Sameni, F., Bostanshirin, N., Yaslianifard, S., Khosravi-Dehaghi, N., Nasiri, M. J., et al. (2022). Global prevalence and molecular epidemiology of mcr-mediated colistin resistance in Escherichia coli clinical isolates: A systematic review. J. Glob. Antimicrob. Resist. 29, 444–461. doi: 10.1016/j.jgar.2021.10.022
Elbediwi, M., Li, Y., Paudyal, N., Pan, H., Li, X., Xie, S., et al. (2019). Global burden of colistin-resistant bacteria: Mobilized colistin resistance genes study (1980-2018). Microorganisms 7:461. doi: 10.3390/microorganisms7100461
Furlan, J. P. R., Savazzi, E. A., and Stehling, E. G. (2020). Widespread high-risk clones of multidrug-resistant extended-spectrum β-lactamase-producing Escherichia coli B2-ST131 and F-ST648 in public aquatic environments. Int. J. Antimicrob. Agents 56:106040. doi: 10.1016/j.ijantimicag.2020.106040
Guo, W. Y., Zhang, H., Cheng, M., Huang, M. R., Li, Q., Jiang, Y. W., et al. (2022). Molecular epidemiology of plasmid-mediated types 1 and 3 fimbriae associated with biofilm formation in multidrug resistant Escherichia coli from diseased food animals in Guangdong, China. Microbiol. Spectr. 10:e0250321. doi: 10.1128/spectrum.02503-21
Haenni, M., Poirel, L., Kieffer, N., Châtre, P., Saras, E., Métayer, V., et al. (2016). Co-occurrence of extended spectrum β lactamase and MCR-1 encoding genes on plasmids. Lancet Infect. Dis. 16, 281–282. doi: 10.1016/S1473-3099(16)00007-4
Hayashi, W., Tanaka, H., Taniguchi, Y., Iimura, M., Soga, E., Kubo, R., et al. (2019). Acquisition of mcr-1 and cocarriage of virulence genes in avian pathogenic Escherichia coli isolates from municipal wastewater influents in Japan. Appl. Environ. Microbiol. 85, e1661–19. doi: 10.1128/AEM.01661-19
He, W. Y., Zhang, X. X., Gao, G. L., Gao, M. Y., Zhong, F. G., Lv, L. C., et al. (2021). Clonal spread of Escherichia coli O101: H9-ST10 and O101: H9-ST167 strains carrying fosA3 and bla (CTX-M-14) among diarrheal calves in a Chinese farm, with Australian Chroicocephalus as the possible origin of E. coli O101: H9-ST10. Zool. Res. 42, 461–468. doi: 10.24272/j.issn.2095-8137.2021.153
Hung, W. T., Cheng, M. F., Tseng, F. C., Chen, Y. S., Shin-Jung Lee, S., Chang, T. H., et al. (2019). Bloodstream infection with extended-spectrum beta-lactamase-producing Escherichia coli: The role of virulence genes. J. Microbiol. Immunol. Infect. 52, 947–955. doi: 10.1016/j.jmii.2019.03.005
Khine, N. O., Lugsomya, K., Kaewgun, B., Honhanrob, L., Pairojrit, P., Jermprasert, S., et al. (2020). Multidrug resistance and virulence factors of Escherichia coli harboring plasmid-mediated colistin resistance: mcr-1 and mcr-3 genes in contracted pig farms in Thailand. Front. Vet. Sci. 7:582899. doi: 10.3389/fvets.2020.582899
Kim, T., Seo, H. D., Hennighausen, L., Lee, D., and Kang, K. (2018). Octopus-toolkit: A workflow to automate mining of public epigenomic and transcriptomic next-generation sequencing data. Nucleic Acids Res. 46:e53. doi: 10.3390/microorganisms9020308
Kubelová, M., Koláèková, I., Gelbíèová, T., Florianová, M., Kalová, A., and Karpíšková, R. (2021). Virulence properties of mcr-1-positive Escherichia coli isolated from retail poultry meat. Microorganisms 9:308.
Lin, Y., Yang, L., Lu, L., Wang, K., Li, J., Li, P., et al. (2020). Genomic features of an Escherichia coli ST156 strain harboring chromosome-located mcr-1 and plasmid-mediated bla(NDM-5). Infect. Genet. Evol. 85:104499. doi: 10.1016/j.meegid.2020.104499
Liu, J., Long, Z., Xue, L., and Li, C. (2022). First report of Didymella sinensis causing leaf blight on italian ryegrass in China. Plant Dis. doi: 10.1094/PDIS-08-22-1831-PDN (in press).
Liu, Y. Y., Zhou, Q., He, W., Lin, Q., Yang, J., and Liu, J. H. (2020). mcr-1 and plasmid prevalence in Escherichia coli from livestock. Lancet Infect. Dis. 20:1126. doi: 10.1016/S1473-3099(20)30697-6
Manges, A. R., Geum, H. M., Guo, A., Edens, T. J., Fibke, C. D., and Pitout, J. D. D. (2019). Global extraintestinal pathogenic Escherichia coli (ExPEC) lineages. Clin. Microbiol. Rev. 32, e00135–18. doi: 10.1128/CMR.00135-18
Mmatli, M., Mbelle, N. M., and Osei Sekyere, J. (2022). Global epidemiology, genetic environment, risk factors and therapeutic prospects of mcr genes: A current and emerging update. Front. Cell. Infect. Microbiol. 12:941358. doi: 10.3389/fcimb.2022.941358
Nakamura, A., Nakamura, T., Niki, M., Kuchibiro, T., Nishi, I., and Komatsu, M. (2021). Genomic characterization of ESBL- and carbapenemase-positive Enterobacteriaceae co-harboring mcr-9 in Japan. Front. Microbiol. 12:665432. doi: 10.3389/fmicb.2021.665432
Nang, S. C., Li, J., and Velkov, T. (2019). The rise and spread of mcr plasmid-mediated polymyxin resistance. Crit. Rev. Microbiol. 45, 131–161. doi: 10.1080/1040841X.2018.1492902
Nguyen, P. T. L., Ngo, T. H. H., Tran, T. M. H., Vu, T. N. B., Le, V. T., Tran, H. A., et al. (2022). Genomic epidemiological analysis of mcr-1-harboring Escherichia coli collected from livestock settings in Vietnam. Front. Vet. Sci. 9:1034610. doi: 10.3389/fvets.2022.1034610
Peng, W., Wang, Y., Fu, Y., Deng, Z., Lin, S., and Liang, R. (2021). Characterization of the tellurite-resistance properties and identification of the core function genes for tellurite resistance in Pseudomonas citronellolis SJTE-3. Microorganisms 10:95. doi: 10.3390/microorganisms10010095
Phuadraksa, T., Wichit, S., Arikit, S., Songtawee, N., and Yainoy, S. (2022). Co-occurrence of mcr-2 and mcr-3 genes on chromosome of multidrug-resistant Escherichia coli isolated from healthy individuals in Thailand. Int. J. Antimicrob. Agents 60:106662. doi: 10.1016/j.ijantimicag.2022.106662
Rodríguez-Santiago, J., Cornejo-Juárez, P., Silva-Sánchez, J., and Garza-Ramos, U. (2021). Polymyxin resistance in Enterobacterales: Overview and epidemiology in the Americas. Int. J. Antimicrob. Agents 58:106426. doi: 10.1016/j.ijantimicag.2021.106426
Shen, Y., Zhou, H., Xu, J., Wang, Y., Zhang, Q., Walsh, T. R., et al. (2018). Anthropogenic and environmental factors associated with high incidence of mcr-1 carriage in humans across China. Nat. Microbiol. 3, 1054–1062. doi: 10.1038/s41564-018-0205-8
Simoni, S., Mingoia, M., Brenciani, A., Carelli, M., Lleò, M. M., Malerba, G., et al. (2021). First IncHI2 plasmid carrying mcr-9.1, bla(VIM-1), and double copies of bla(KPC-3) in a multidrug-resistant Escherichia coli human isolate. mSphere 6:e0030221. doi: 10.1128/mSphere.00302-21
Sonnevend, Á, Alali, W. Q., Mahmoud, S. A., Ghazawi, A., Bharathan, G., Melegh, S., et al. (2022). Molecular characterization of MCR-1 producing enterobacterales isolated in poultry farms in the United Arab Emirates. Antibiotics (Basel) 11:305. doi: 10.3390/antibiotics11030305
Tarabai, H., Wyrsch, E. R., Bitar, I., Dolejska, M., and Djordjevic, S. P. (2021). Epidemic HI2 plasmids mobilising the carbapenemase gene bla(IMP-4) in Australian clinical samples identified in multiple sublineages of Escherichia coli ST216 colonising silver gulls. Microorganisms 9:567. doi: 10.3390/microorganisms9030567
Touzain, F., Le Devendec, L., de Boisséson, C., Baron, S., and Jouy, E. A. (2018). Perrin-Guyomard, Y. Blanchard, and I. Kempf, characterization of plasmids harboring blaCTX-M and blaCMY genes in E. coli from French broilers. PLoS One 13:e0188768. doi: 10.1371/journal.pone.0188768
Wang, Y., Xu, C., Zhang, R., Chen, Y., Shen, Y., Hu, F., et al. (2020). Changes in colistin resistance and mcr-1 abundance in Escherichia coli of animal and human origins following the ban of colistin-positive additives in China: An epidemiological comparative study. Lancet Infect. Dis. 20, 1161–1171. doi: 10.1016/S1473-3099(20)30149-3
Xu, Y., Chen, H., Zhang, H., Ullah, S., Hou, T., and Feng, Y. (2021). The MCR-3 inside linker appears as a facilitator of colistin resistance. Cell Rep. 35:109135. doi: 10.1016/j.celrep.2021.109135
Yamanaka, Y., Aizawa, S. I., and Yamamoto, K. (2022). The hdeD gene represses the expression of flagellum biosynthesis via LrhA in Escherichia coli K-12. J. Bacteriol. 204:e0042021. doi: 10.1128/JB.00420-21
Yang, Q. E., MacLean, C., Papkou, A., Pritchard, M., Powell, L., Thomas, D., et al. (2020). Compensatory mutations modulate the competitiveness and dynamics of plasmid-mediated colistin resistance in Escherichia coli clones. ISME J. 14, 861–865. doi: 10.1038/s41396-019-0578-6
Yin, W., Li, H., Shen, Y., Liu, Z., Wang, S., Shen, Z., et al. (2017). Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. mBio 8, e00543–17. doi: 10.1128/mBio.00543-17
Yin, W., Ling, Z., Dong, Y., Qiao, L., Shen, Y., Liu, Z., et al. (2021). Mobile colistin resistance enzyme MCR-3 facilitates bacterial evasion of host phagocytosis. Adv. Sci. (Weinh). 8:e2101336. doi: 10.1002/advs.202101336
Zhang, S., Abbas, M., Rehman, M. U., Wang, M., Jia, R., Chen, S., et al. (2021). Updates on the global dissemination of colistin-resistant Escherichia coli: An emerging threat to public health. Sci. Total Environ. 799:149280. doi: 10.1016/j.scitotenv.2021.149280
Zhang, W., Lu, X., Chen, S., Liu, Y., Peng, D., Wang, Z., et al. (2022). Molecular epidemiology and population genomics of tet(X4), bla(NDM) or mcr-1 positive Escherichia coli from migratory birds in southeast coast of China. Ecotoxicol. Environ. Saf. 244:114032. doi: 10.1016/j.ecoenv.2022.114032
Zhang, X., Fang, C., Zhang, J., Hua, W., He, R., and Zhou, M. (2022). Carbapenemase- and colistin resistant Escherichia coli strains from children in China: High genetic diversity and first report of bla (NDM-5), bla (CTX-M-65), bla (OXA-10), bla (TEM-1), and mcr-1.1 Genes Co-Occurrence in E. coli ST156. Infect. Drug Resist. 15, 5315–5320. doi: 10.2147/IDR.S378574
Zhao, F., Yang, H., Bi, D., Khaledi, A., and Qiao, M. (2020). A systematic review and meta-analysis of antibiotic resistance patterns, and the correlation between biofilm formation with virulence factors in uropathogenic E. coli isolated from urinary tract infections. Microb. Pathog. 144:104196. doi: 10.1016/j.micpath.2020.104196
Keywords: Escherichia coli, mcr, whole genome sequencing, antimicrobial resistance genes, sequence types
Citation: Shi J, Zhu H, Liu C, Xie H, Li C, Cao X and Shen H (2023) Epidemiological and genomic characteristics of global mcr-positive Escherichia coli isolates. Front. Microbiol. 13:1105401. doi: 10.3389/fmicb.2022.1105401
Received: 22 November 2022; Accepted: 28 December 2022;
Published: 18 January 2023.
Edited by:
Qiang Lin, University of Antwerp, BelgiumReviewed by:
Tao He, Jiangsu Academy of Agricultural Sciences (JAAS), ChinaUlises Garza-Ramos, National Institute of Public Health, Mexico
Copyright © 2023 Shi, Zhu, Liu, Xie, Li, Cao and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoli Cao,  Y2FvLXhpYW8tbGlAMTYzLmNvbQ==; Han Shen,
Y2FvLXhpYW8tbGlAMTYzLmNvbQ==; Han Shen,  c2hlbmhhbjEwMzY2QHNpbmEuY29t
c2hlbmhhbjEwMzY2QHNpbmEuY29t
†These authors have contributed equally to this work
 Jiping Shi
Jiping Shi Hong Zhu1†
Hong Zhu1† Xiaoli Cao
Xiaoli Cao Han Shen
Han Shen