- 1School of Environment, Harbin Institute of Technology, Harbin, China
- 2College of Resources and Environment, University of Chinese Academy of Sciences (UCAS), Beijing, China
An atrazine-utilizing bacterium, designated as ZY, was isolated from agricultural soil and identified as Paenarthrobacter ureafaciens. The P. ureafaciens ZY demonstrated a significant degradation capacity of atrazine, with the degradation efficiency of 12.5 mg L−1 h−1 in liquid media (at pH 7, 30°C, and the atrazine level of 100 mg L−1). The P. ureafaciens ZY contained three atrazine-degrading genes (i.e., trzN, atzB, and atzC) could metabolize atrazine to form cyanuric acid, which showed lower biotoxicity than the parent atrazine as predicted by Ecological Structure Activity Relationships model. A laboratory-scale pot experiment was performed to examine the degradation of atrazine by P. ureafaciens ZY inoculation and investigate its effects on the native microbial communities. The results exhibited that the P. ureafaciens ZY was conductive to the degradation of atrazine, increased the total soil phospholipid fatty acids at the atrazine level of 50, 70, and 100 mg kg−1. By using high-throughput sequencing analysis, Frateuria, Dyella, Burkholderia-Caballeronia-Paraburkholderia were considered as the most important indigenous atrazine-degrading microorganisms due to their relative abundances were positively correlated with the atrazine degradation rate. In addition, P. ureafaciens ZY also increased the abundance of atrazine-degrading genus Streptomyces and Bacillus, indicating that there may be a synergic relationship between them in the process of atrazine degradation. Our work provides a new insight between inoculums and native microorganisms on the degradation of atrazine.
1. Introduction
The extensive use of pesticides significantly contributes to agricultural production by protecting crops from pests and diseases (Sun et al., 2018). According to the database from the Food and Agriculture Organization (FAO) of the United Nations (FAO, 2021), the annual consumption of pesticides reached 1.77 million tons (equal to 3.35 kg ha−1) in China in 2019, which far exceeded the maximum pesticides usage allowed levels with 1.66 kg ha−1. In fact, as little as 1% of applied pesticides reaches the target weeds and pests, and a more significant proportion of pesticides residues in the soils will lead to toxic effects on the ecological system and human health (Liang et al., 2022; Sabzevari and Hofman, 2022). In addition, pesticides that can spread in specific environments threaten the living organisms and result in an ecosystem imbalance (Lee and Choi, 2020; Lykogianni et al., 2021).
Atrazine (2-chloro-4-ethylamino-6-isopropylamino-1,3,5-triazine), a synthetic herbicide, has been extensively applied for more than 50 years. Its application has been controversial due to its widespread, high fluidity, and persistence, resulting in its high residues in soils (Sun et al., 2017). However, China and the United States still widely use atrazine, even though it has been restricted in the European Union and Canada (de Albuquerque et al., 2020). Moreover, the toxic effects of atrazine, such as neurotoxic, endocrine disrupting, and cancerogenic have been reported (Hayes et al., 2002; Shenoy, 2012; Shan et al., 2021). Many studies have been illustrated the toxic effects of atrazine on various organisms, e.g., Caenorhabditis elegans (García-Espiñeira et al., 2018; Zhou et al., 2021), Chironomus tentans (Tyler Mehler et al., 2008), Daphnia carinata (He et al., 2012), Elodea canadensis (Brain et al., 2012), Lepomis macrochirus (Tyler Mehler et al., 2008), Litoditis marina (Francolino et al., 2021), Pimephales promelas (Dionne et al., 2021), Prochilodus lineatus (de Andrade et al., 2019), and so on. Considering the long-term use of atrazine and its toxic effects, it is of great concern to explore a high-efficiency and safe strategy to restore the atrazine-contaminated environments.
Attribute to the efforts of mitigating the toxic effects of atrazine, a number of strains that can degrade atrazine have been isolated and characterized, so far, which seems to be good from a cost-effective, quality, degradation efficiency, and safety point of view. These atrazine-degrading strains isolated from different sources were reported, mainly including Achromobacter (Tonelli Fernandes et al., 2018), Arthrobacter (Bazhanov et al., 2017; Zhao et al., 2018), Bacillus (Jakinala et al., 2019), Chelatobacter (Rousseaux et al., 2003), Citricoccus (Yang et al., 2018), Ensifer (Chen et al., 2017), Frankia (Rehan et al., 2014), Nocardia (Giardi et al., 1985), Nocardioides (Omotayo et al., 2013). Besides the high degradation efficiency in the media (Rostami et al., 2021), atrazine-degrading strains were also inoculated into the atrazine-contaminated soil to explore their biodegradation ability of atrazine (Rousseaux et al., 2003; Sebaï et al., 2011; Gao et al., 2018). According to Li et al. (2008), 98.6%, 96.2%, and 88.7% of atrazine were removed by Arthrobacter sp. AD26, A. aurescens TC1, and Pseudomonas sp. ADP in atrazine-contaminated soil (300 mg kg−1) after inoculation for 20 days, respectively. A 96.86% degradation percentage of atrazine was observed with Rhodobacter sphaeroides sp. W16 inoculation in the soil at the level of 100 mg kg−1 atrazine after 15 days (Du et al., 2011). But up to now, most of the studies still focus on the isolation of new atrazine degrader and evaluation of atrazine degradation. However, a few researches have reported the effects of atrazine degraders inoculation on indigenous bacteria community in soil. Therefore, besides isolating safe and efficient atrazine-degrading strains, evaluation of the effects of atrazine-degrading strains on native microbial communities in atrazine-contaminated soil for the further application of these atrazine degraders was also essential.
In this study, a strain with a high-efficiency degradation capacity of atrazine, P. ureafaciens ZY, was isolated from agricultural soil in northeast China. The different liquid media parameters (i.e., pH, temperature, atrazine concentration) that affect atrazine degradation were explored. The genes and metabolic pathways involved in atrazine degradation by strain P. ureafaciens ZY were elucidated. Besides, this work also investigated the degradation capacity of P. ureafaciens ZY in the soil and its effects on the dynamic of soil native microbiome through PLFAs and high-throughput sequencing analysis. Our work enriched the species of bacteria that could degrade atrazine and provided a theoretical basis for the remediation of atrazine-contaminated soils.
2. Materials and methods
2.1. Soil samples, chemicals, and media
Soil samples were collected from agricultural soil in the northeast of China (126°49′40″ E, 45°40′60” N) and were sieved to remove plant residues and stones for further study (The main reason we selected this area was that it had long grown corn, which had also led to the widespread use of atrazine). Atrazine was purchased from Shanghai Macklin Biochemical Co., Ltd. Cyanuric acid was purchased from Heowns Biochem Technologies, LLC, Tianjin. The liquid minimal salt media (MSM) amended with glucose as a carbon source was consisted of 2.4 g K2HPO4, 0.2 g MgSO4·7H2O, 1.2 g KH2PO4, 0.025 g CaCl2·2H2O, 0.008 g Fe2 (SO4)3, and 1.0 g glucose in 1,000 ml of deionized water at pH 7.0 (marked as MSMG in this study). The liquid LB media contained 10 g tryptone, 5 g yeast extract, and 10 g NaCl in 1,000 ml of deionized water at pH 7.0. Solid media was formed by adding 17 g L−1 agar into liquid media, and all media were sterilized at 121°C for 30 min for further used.
2.2. Isolation of atrazine-degrading strains
Soil samples (10 g) were added to 100 ml of MSMG (contained 100 mg L−1 atrazine) and incubated under anaerobic conditions at 30°C and shaken at 160 rpm for 7 days. Then, the enrichment culture (10 ml) was transferred into a fresh MSMG (contained 200 mg L−1 atrazine) and incubated for 7 days at the same conditions. The inoculation process was repeated five times until the final atrazine concentration reached 500 mg L−1. Subsequently, the culture was collected and plated onto MSMG agar plates contained 100 mg L−1 of atrazine and incubated at 30°C in the darkness. After 4 days of incubation, single colonies were selected aseptically and explored their degradation ability for atrazine. A strain designated as ZY with atrazine degradation activity was obtained for further analysis.
2.3. Identification and characterization of strain ZY
The identified experiments of strain ZY, including morphological observation, 16S rRNA gene sequence analysis, and Biology Gene III MicroPlate, were performed. Bacteria Genomic DNA Kit (TransGene Biotech Co., Ltd., Beijing, China) was used to extract the total genomic DNA of strain ZY. Strain ZY was identified via 16S rRNA gene sequencing, the oligonucleotide primers and the PCR cycle parameters used as previously described by Yang et al. (2018). Then the 16S rRNA was sequenced by Shihe Biotech Co., Ltd. (Harbin, China). The 16S rRNA of strain ZY was compared to those 16S rRNA available in the National Center for Biotechnology Information (NCBI) via Nucleotide BLAST. MEGA version 8.0 was used to construct a phylogenetic tree. BIOLOG Gen III MicroPlate (BIOLOG Inc., Hay ward, USA) was carried out to test carbon substrates utilization patterns of strain ZY by using automated tetrazolium-based microbial identification system produced by BIOLOG Inc. (Hay ward, USA; Mekonnen et al., 2019).
2.4. Inoculum preparation
The strain ZY was cultured overnight (18 h) in LB media at 30°C and shaken at 160 rpm. After centrifugation, the strain cells were collected and suspended three times in MSMG media. A final cells density of 1 × 108 CFU mL−1 (OD600nm = 1.0) determined by a UV-spectrophotometer (Shanghai Jinghua Technology Instrument Co., Ltd) was obtained for subsequent degradation experiments.
2.5. Atrazine degradation of strain ZY in MSMG media
Atrazine degradation in liquid MSMG media was tested at pH 3.0, 5.0, 7.0, 9.0, and 11.0. The effect of temperature on atrazine degradation was tested at 20, 25, 30, 35, and 40°C. The degradation effects of the initial atrazine concentration were tested at 10, 30, 50, 70, and 100 mg L−1. Only one parameter was changed during each culture. The MSMG suspension with OD600nm 1.0 were added into the fresh MSMG media at 1:50 (v:v) dosage of inoculation and cultured at 30°C. The OD600nm and atrazine in the culture were detected every 2 h. Atrazine and cyanuric acid were determined by ultra-performance liquid chromatography (UPLC) analysis (Yang et al., 2018). The media without inoculation was used as controls, and the atrazine degradation was less than 1.0% in the controls.
2.6. Analysis of atrazine-degrading genes
The strain ZY was analyzed by PCR for the presence of atrazine-degrading genes (i.e., atzA, atzB, atzC, atzD, atzE, atzF, trzN, and trzD). The primer pairs used in this study were described in Supplementary Table S2, and the reaction conditions referred to Sebaï et al. (2011). Heshi Biotech Co., Ltd. (Harbin, China) performed the sequencing of atrazine-degrading genes, and the sequence analysis was done via BLAST on NCBI databases.
2.7. Determination and identification of atrazine metabolites
To identify the atrazine metabolites produced by P. ureafaciens ZY, the MSMG culture collected at 6 h was analyzed using a rapid and reliable ultra-high performance liquid chromatography-quadrupole-time-of-flight tandem mass spectrometry (UPLC-QTOF-MS, 6545 Q-TOF, Agilent Technologies, U.S.) technique. Specific UPLC-Q-TOF-MS experimental parameters and the mass spectrometry methods were performed as described by previous studies (Tonelli Fernandes et al., 2018; Zhao et al., 2018).
2.8. Biodegradation of atrazine in soil
The 200 g soil sample (dry weight) was added with methanol-dissolved atrazine to reach final concentrations of 10, 30, 50, 70, and100 mg kg−1 (dry weight) and then was placed in a fume hood for 4 h to volatilize methanol. The soils were inoculated with strain ZY to reach the initial concentration of 4 × 106 CFU g−1, then passed through a mesh (2 mm) to achieve symmetrical. The treatments with ZY inoculums at 0, 10, 30, 50, 70, and 100 mg kg−1 atrazine were marked as P0, P10, P30, P50, P70 and P100, respectively. Soil samples containing atrazine at 0, 10, 30, 50, 70, and 100 mg kg−1 but without ZY inoculums were labeled as A0, A10, A30, A50, A70, A100, respectively. Then, soil samples were transferred to a plastic bowl (Φ 9 cm) and incubated in darkness at 25°C with 30% water-holding content adjusted by sterile distilled water. After inoculation for 1, 3, 5, and 7 d, soil samples were collected to analyze residual atrazine. In brief, 3 g of soil samples were added with 15 ml methanol for ultrasonic for 20 min, followed by centrifugation for supernatant collection, and the above operations were repeated three times (Luo et al., 2021). The received supernatant was concentrated by a rotary evaporator and dissolved by chromatographic methanol, followed by 0.22 μm filter membrane filtration for UPLC analysis.
2.9. The PLFAs analysis
The PLFAs analysis was used to assess soil microbial community structures. The PLFAs were extracted and analyzed using the methods reported by Frostegård et al. (1993) and Chaudhary et al. (2020). The Sherlock Microbial Identification System (SMIS), developed by MIDI Corporation (MIDI, Newark, Delaware, USA), was used to identify PLFAs in soil. All PLFAs classifications were shown in Supplementary Table S4 provided by MSIS.
2.10. Soil DNA extraction, sequencing and bioinformatics analysis
The Illumina MiSeq of PCR-amplified 16S rRNA was used to analysis the structure of soil microbial community. Total DNA of soil was extracted by using the Cetyltrimethyl Ammonium Bromide (CTAB) according to manufacturer’s instructions. The 16S rRNA gene fragment was amplified using a universal primer set 341F (5’-CCTACGGGNGGCWGCAG-3′) and 805R (5’-GACTACHVGGGTATCTAATCC-3′; Logue et al., 2016). Sequencing was performed by LC-Bio Technology Co., Ltd., Hang Zhou, Zhejiang Province, China. The raw reads have been submitted to NCBI with the SRA database accession of PRJNA909019.
All experiments were performed in triplicate, and the results were presented as arithmetic averages and standard deviations. The datas were analyzed with the SPSS 19.0 and Origin 18.0 software. The Ecological Structure Activity Relationships (ECOSAR) model was used to predict the toxicity of atrazine metabolites. Principal component analysis (PCA) was performed by CANOCO 5.0 software. Heat map analysis and network analysis were performed using the R package (v3.5.2).
3. Results and discussion
3.1. Isolation and characterization of strain ZY
A strain ZY with a strong atrazine-degrading ability was selected for further study. A light yellow, smooth, and round colony of strain ZY was observed in the LB plate (Supplementary Figure S1A). Its morphological characteristics indicated that strain ZY was a Gram positive, and rod-shaped bacteria (Supplementary Figures S1B, S2).
The 16S rRNA gene sequence of strain ZY was 1,376 bp (accession no. ON878081), very similar to those of accessible strains, e.g., P. ureafaciens NC (99.19%, accession no. NR_029281.1), P. nicotinovorans DSM 420 (98.37%, accession no. NR_026194.1), and P. histidinolovorans DSM 20115 (98.21%, accession no. NR_026234.1). In a phylogenetic tree constructed based on the similar 16S rRNA gene sequences of ZY (Figure 1), the strain ZY clustered with members of the genus Paenarthrobacter sp. and was preliminarily identified as a P. ureafaciens due to the closest relative with P. ureafaciens NC.
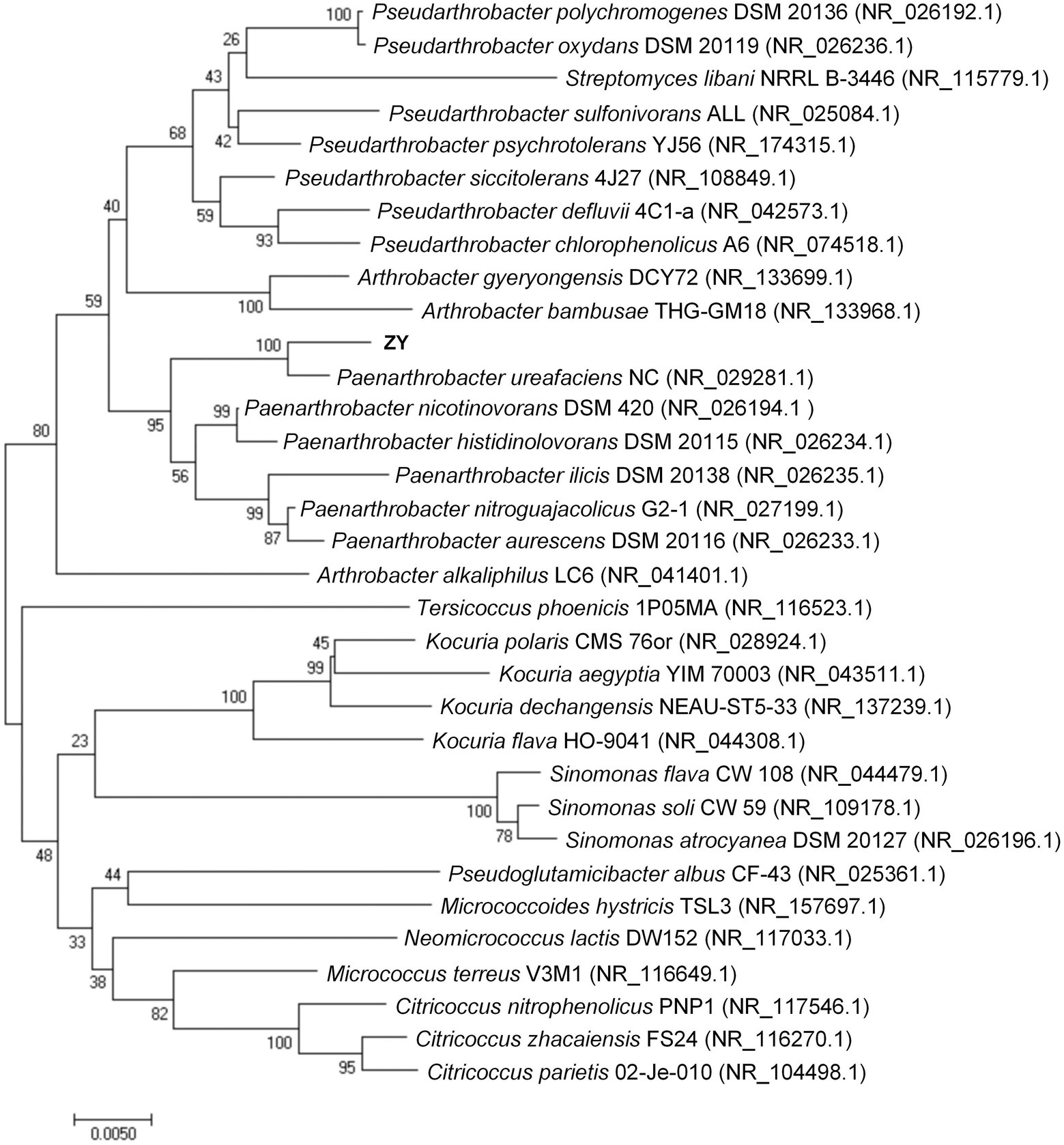
Figure 1. Phylogenetic tree based on the 16S rRNA gene sequences of strain ZY. The calculations were performed according to a neighbor-joining analysis (bootstrap number = 1,000), and the bar indicates 0.0050 substitution per nucleotide position.
Carbon utilization pattern of P. ureafaciens ZY was investigated via GEN III microplate after 48 h of incubation. The results have shown in Supplementary Table S3, Figure S3. Accordingly, P. ureafaciens ZY can metabolize D-fructose, D-mannose, D-galactose as carbon sources, but cannot assimilate dserine, troleandomycin, rifamycin, and minocycline (Supplementary Table S3).
3.2. The effects of culture conditions on the growth and atrazine degradation of Paenarthrobacter ureafaciens ZY
In the liquid MSMG, the effects of pH, temperature, and atrazine concentration on the growth and atrazine degradation of P. ureafaciens ZY were investigated.
3.2.1. pH
Environmental pH could influence the microbial remediation of contaminated sites (Siddique et al., 2002). The optimal pH range for the growth and atrazine degradation by Arthrobacter sp. strain HB-5 was 5.0–10.0 (Wang and Xie, 2012). Figure 2A displayed the growth and degradation of atrazine of P. ureafaciens ZY in MSMG (100 mg L−1 atrazine) at different pH levels for 6 h. After inoculation for 6 h, more than 64.83% of atrazine removal were observed at pH 5.0–11.0. At pH 3.0, P. ureafaciens ZY hardly grew and could not metabolize atrazine. The degradation rates of atrazine were 11.42, 12.11, 11.21, and 10.81 mg L−1 h−1 at pH ranging from 5 to 11, respectively. The pH range for the growth and atrazine degradation of P. ureafaciens ZY is wide and alkaliphilic, which also revealed that P. ureafaciens ZY had a great potential to rete atrazine-contaminated sites, particularly in alkaline sites. In addition, there was a significant correlation between the atrazine degradation percentage and OD600nm from pH 3 to 11 (r = 0.953, p < 0.01), suggesting that pH was an important factor affecting strain proliferation and atrazine degradation.
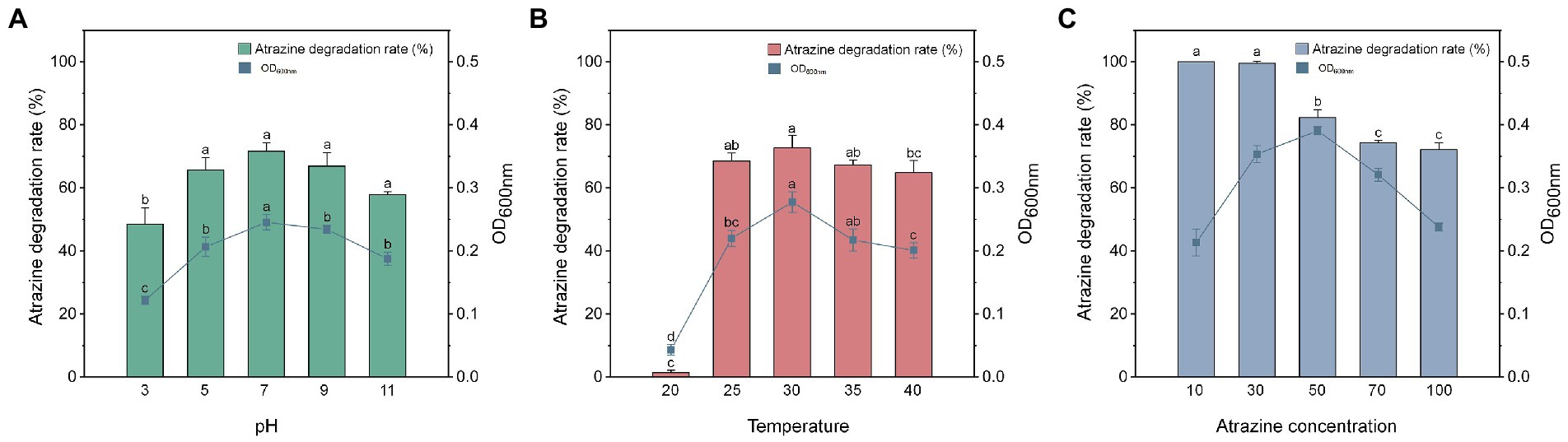
Figure 2. The effects of pH (A), temperature (B), and atrazine concentration (C) on the atrazine degradation and growth of Paenarthrobacter ureafaciens ZY.
3.2.2. Temperature
Temperature has played a key role in biodegradation efficiency of pollutants by microorganisms, and significantly influences the dissipation of s-triazine herbicides (Vela et al., 2004). Figure 2B showed the effects of temperature ranging from 20°C to 40°C on the degradation of 100 mg L−1 atrazine in MSMG (pH 7.0). With the increase of temperature, the degradation of atrazine enhanced and degradation rates were 8.08, 8.20, and 11.95 mg L−1 h−1 at 20, 25, and 30°C after 6 h, respectively. Notably, the degradation ability of atrazine were inhibited at 35°C and 40°C after 6 h. Yang et al. (2018) also found that the optimum temperature for the growth and degradation of Citricoccus sp. strain TT3 was 30°C, and the growth and atrazine degradation of Citricoccus sp. strain TT3 were inhibited at 37°C and 45°C. In addition, a significant correlation between the atrazine degradation percentage and OD600nm from 20 to 40°C (r = 0.869, p < 0.01) was observed, indicating that strain proliferation and atrazine degradation could be affected by the temperature.
3.2.3. Atrazine concentrations
The effects of atrazine concentrations at the levels of 10–100 mg L−1 on the growth and atrazine degradation of P. ureafaciens ZY were explored in MSMG at pH 7.0 and 30°C (Figure 2C). After 6 h inoculation of P. ureafaciens ZY, the degradation rates of atrazine were approximately 100, 99.57, 82.33, 74.28, and 72.17% at the atrazine levels of 10, 30, 50, 70, 100 mg L−1, respectively (Figure 2C). After the inoculation of 8 h, atrazine was completely degraded in all groups. According to OD600nm of P. ureafaciens ZY, with the increase of atrazine concentration (from 10 to 50 mg L−1), the growth of P. ureafaciens ZY was obviously promoted (Figure 2C). The concentrations of atrazine ranging from 70 to 100 mg L−1 inhibited the growth and atrazine degradation of P. ureafaciens ZY, which might due to the toxicity of high concentrations of atrazine or their metabolites to P. ureafaciens ZY (Zhao et al., 2018).
The degradation rate of P. ureafaciens ZY in 8 h at an atrazine concentration of 100 mg L−1 was 12.5 mg L−1 h−1, which was 27.78, 10.94, and 4.50 times greater than those of Rhodococcus sp. BCH2, Pseudomonas sp. EGD-AKN5, and Shewanella sp. YJY4, respectively (Supplementary Table S4). Even in close relatives Arthrobacter, P. ureafaciens ZY exhibited an increase in the degradation rate of 10.42 mg L−1 h−1 and 2.98 mg L−1 h−1 than those in Arthrobacter sp. DAT1 and Arthrobacter sp. ZXY-2 at the atrazine level of 100 mg L−1. Compared to the model atrazine-degrading bacterium Pseudomonas sp. ADP (Mandelbaum et al., 1995), P. ureafaciens ZY still shows an approximately 8.33 mg L−1 h−1 increase of degradation rate with 100 mg L−1 atrazine. To our best knowledge, P. ureafaciens ZY showed the highest degradation capacity of atrazine compared to the other reported atrazine-degrading strains at the atrazine level ranges from 50 to 100 mg L−1. These results illustrated that P. ureafaciens ZY had a great bioretion potential, especially for the heavily atrazine-contaminated sites.
3.3. Atrazine-degrading genes and the metabolites of atrazine
At present, atrazine degradation genes have been extensively investigated and proven to be highly conserved in many bacteria (de Souza et al., 1998; Singh and Jauhari, 2017). According to the different genetic compositions, atrazine-degrading bacteria are divided into two types. One type only contains atzA (trzN), atzB, and atzC, which could catalyze atrazine to produce cyanuric acid (Zhao et al., 2018). And the other type used extra genes atzD (trzD), atzE, and atzF to mineralize atrazine into CO2 and NH3 (Ma et al., 2017).
In this work, according to the primers in Supplementary Table S1, three genes including trzN, atzB, and atzC were obtained in P. ureafaciens ZY (Supplementary Figure S6). The amplified gene sequences were compared with those in NCBI database. The trzN (438 bp, ON954325) exhibited 100% identity with the trzN gene of Arthrobacter sp. AK-YN10 (HE716868.1) and Pseudomonas sp. ADP (FJ161692.1). The atzB (1,391 bp, ON911740) showed 100% homologous with the atzB gene of Arthrobacter sp. ZXY-2 (CP017421.1) and P. ureafaciens DnL1-1 (CP014574.1). The atzC (616 bp, ON954326) showed 99.67 and 99.67% homologous with the atzC gene of Pseudomonas sp. ADP (U66917.2) and Nocardioides sp. SP12 (AF537329.1). During the degradation process of atrazine by P. ureafaciens ZY, three metabolites of atrazine, were identified as hydroxyatrazine (m/z of 198.0), N-isopropylammelide (m/z of 171.0), and cyanuric acid (m/z of 128.0) by UPLC-Q-TOF-MS (Supplementary Figure S4). In Figure 3A, a putative atrazine degradation pathway was proposed: In the first step, hydrolytic dechlorination was induced by triazine hydrolase which endoded by trzN to yield hydroxyatrazine. Subsequently, hydroxyatrazine ethylaminohydrolase which encoded by atzB catalyzes hydroxyatrazine to form N-isopropylammelide. Finally, the N-isopropylammelide was eliminated from the s-triazine ring which induced by N-isopropylammelide isopropylamidohydrolase (encoded by atzC) to form cyanuric acid. Further determination of cyanuric acid content can also confirm the degradation path of atrazine (Figure 3B). The atrazine degradation pathway of P. ureafaciens ZY is similar to that in Arthrobacter sp. ZXY-2 (Zhao et al., 2018) and Citricoccus sp. strain TT3 (Yang et al., 2018), and different from other atrazine degraders which can mineralize atrazine, such as Ensifer sp. CX-T (Ma et al., 2017) and Pseudomonas sp. ADP (Martinez et al., 2001). By using the ECOSAR model, we also predicted the biotoxicity of the metabolites of atrazine, and the results were shown in Supplementary Table S5. Hydroxyatrazine, N-isopropylammelide, and cyanuric acid showed lower biotoxicity than its parent atrazine. Kolekar et al. (2019) also evaluated cytotoxicity tests by cell viability assay on HepG2 and indicated significant decrease in the toxicity of atrazine by the atrazine-degrading strains inoculation due to its effective degradation and formation of simpler and less/nontoxic metabolites (Hydroxyatrazine and N-isopropyllamilide). Intracellular reactive oxygen species levels and malondialdehyde content exhibited a significantly decreasing tendency after Escherichia coli exposure to intermediates (Hydroxyatrazine, N-isopropyllamilide, and cyanuric acid) of atrazine compared to parent atrazine (Yu et al., 2023). In conjunction, these results indicated that P. ureafaciens ZY had practical application value.
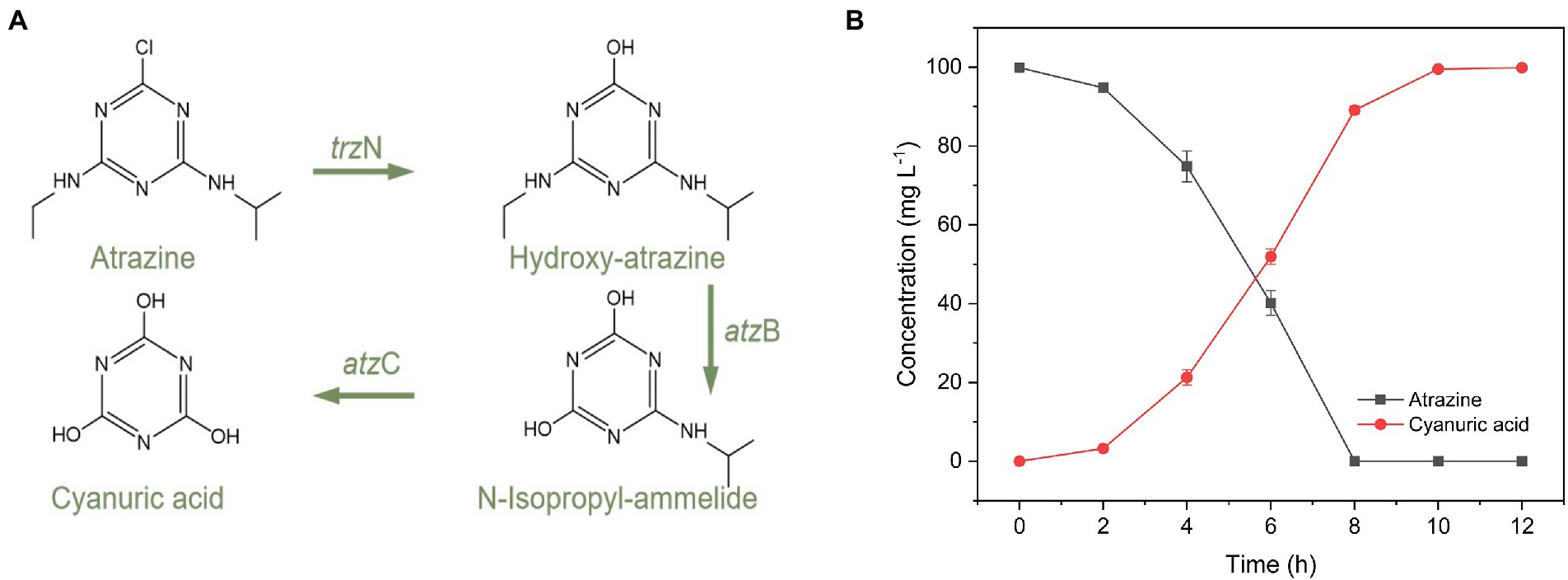
Figure 3. Concentration changes in atrazine and cyanuric acid (A). The proposed metabolic pathway of atrazine in P. ureafaciens ZY (B).
3.4. The removal of atrazine in soil with Paenarthrobacter ureafaciens ZY inoculation
The biodegradation of different concentrations of atrazine was further investigated in soil with or without P. ureafaciens ZY inoculation. Without P. ureafaciens ZY inoculation, the highest degradation of atrazine (17.66%) was found in A30 after 7 days. In the treatments of A10, A50, A70, and A100, the degradation of atrazine was 12.71%, 12.87%, 13.03%, and 7.44%, respectively (Figure 4A). Atrazine was not detected in A0 and P0. Complete degradation of atrazine at 10 and 30 mg kg−1 occurred within 5 days and 7 days in soil inoculated with P. ureafaciens ZY. In P50, P70, and P100, the degradation of atrazine at 7 days was 95.96%, 95.21%, and 85.31%, respectively (Figure 4B). In recent years, Paenarthrobacter has been screened and demonstrated strong atrazine-degrading ability. Jia et al. (2021) found that 95.9% of 5 mg kg−1 atrazine was removed from the soils when inoculated with Paenarthrobacter strain AT-5 with 7 days. Chen et al. (2021) also reported that 96.0% of atrazine had been degraded with Paenarthrobacter sp. W11 inoculation after 49 days of incubation in soils (50 mg kg−1 atrazine). These results indicated that P. ureafaciens had great potential for remediation of atrazine-contaminated soil.
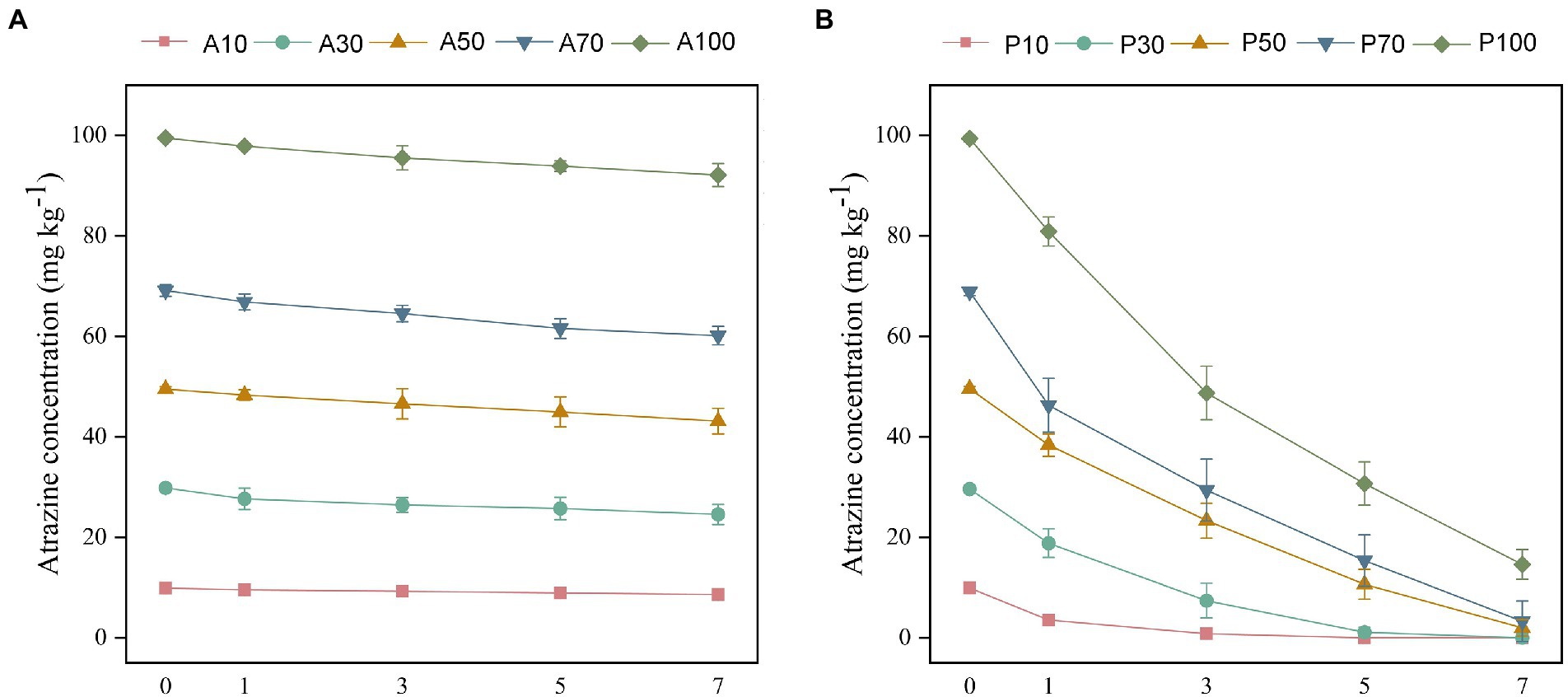
Figure 4. The concentrations of atrazine residues without P. ureafaciens ZY inoculation (A) and with P. ureafaciens ZY inoculation (B) treatments after 7 days. A10: 10 mg kg−1 atrazine-contaminated soil; A30: 30 mg kg−1 atrazine-contaminated soil; A50: 50 mg kg−1 atrazine-contaminated soil; A70: 70 mg kg−1 atrazine-contaminated soil; A100: 100 mg kg−1 atrazine-contaminated soil; P10: 10 mg kg−1 atrazine-contaminated soil with P. ureafaciens ZY inoculation; P30: 30 mg kg−1 atrazine-contaminated soil with P. ureafaciens ZY inoculation; P50: 50 mg kg−1 atrazine-contaminated soil with P. ureafaciens ZY inoculation; P70: 70 mg kg−1 atrazine-contaminated soil with P. ureafaciens ZY inoculation; P100: 100 mg kg−1 atrazine-contaminated soil with P. ureafaciens ZY inoculation.
In this work, the freshly added soils were used, and the addition of P. ureafaciens ZY showed a high degradation rate for atrazine in soil. Zhao et al. (2003) revealed that the degradation percentage of atrazine in the freshly added soils was significantly higher than that in the aged soils. The degradation efficiency decreased with the extension of inoculation time, and the half-life of atrazine inoculated in aged soil was longer than that in the freshly added soil. This can be explained by the fact that the bioavailability of atrazine in the freshly added soils is obviously increased compared to that in the aged soils (Yanze-Kontchou and Gschwind, 1995; Radosevich et al., 1997; Zhao et al., 2003).
3.5. The soil PLFAs dynamic following with Paenarthrobacter ureafaciens ZY inoculation
Figure 5 exhibited the abundance of PLFAs (i.e., the biomass of microorganisms) in different treatments. In non-inoculation treatments, the total PLFAs of A0 (123.026 nmol g−1) is higher than those in other treatments, especially in A70 (37.948 nmol g−1, p < 0.01) and A100 (14.579 nmol g−1, p < 0.01). Among them, the Gram-positive (Figure 5A), Gram-nagative (Figure 5B), Methanotroph (Figure 5C) and Actinomycetes (Figure 5E) also showed the same decrease trend. Previous studies have demonstrated that a specific inhibitory effect on soil microbes occurred by atrazine addition (Ros et al., 2006; Gao et al., 2018; Chen et al., 2021). Gao et al. (2018) also indicated that the utilization of carbon sources by microbial communities was decreased in atrazine-contaminated soil. The decline of soil microbial biomass might be caused by the accumulated metabolites, which might be more toxic to the microbes than the parent in the soil (Cheng et al., 2022). Seghers et al. (2003) revealed that atrazine significantly affected the structure and function of soil microbial community. Besides, the inhibition effects on the soil community and nitrification process were also reported (Chen et al., 2015).
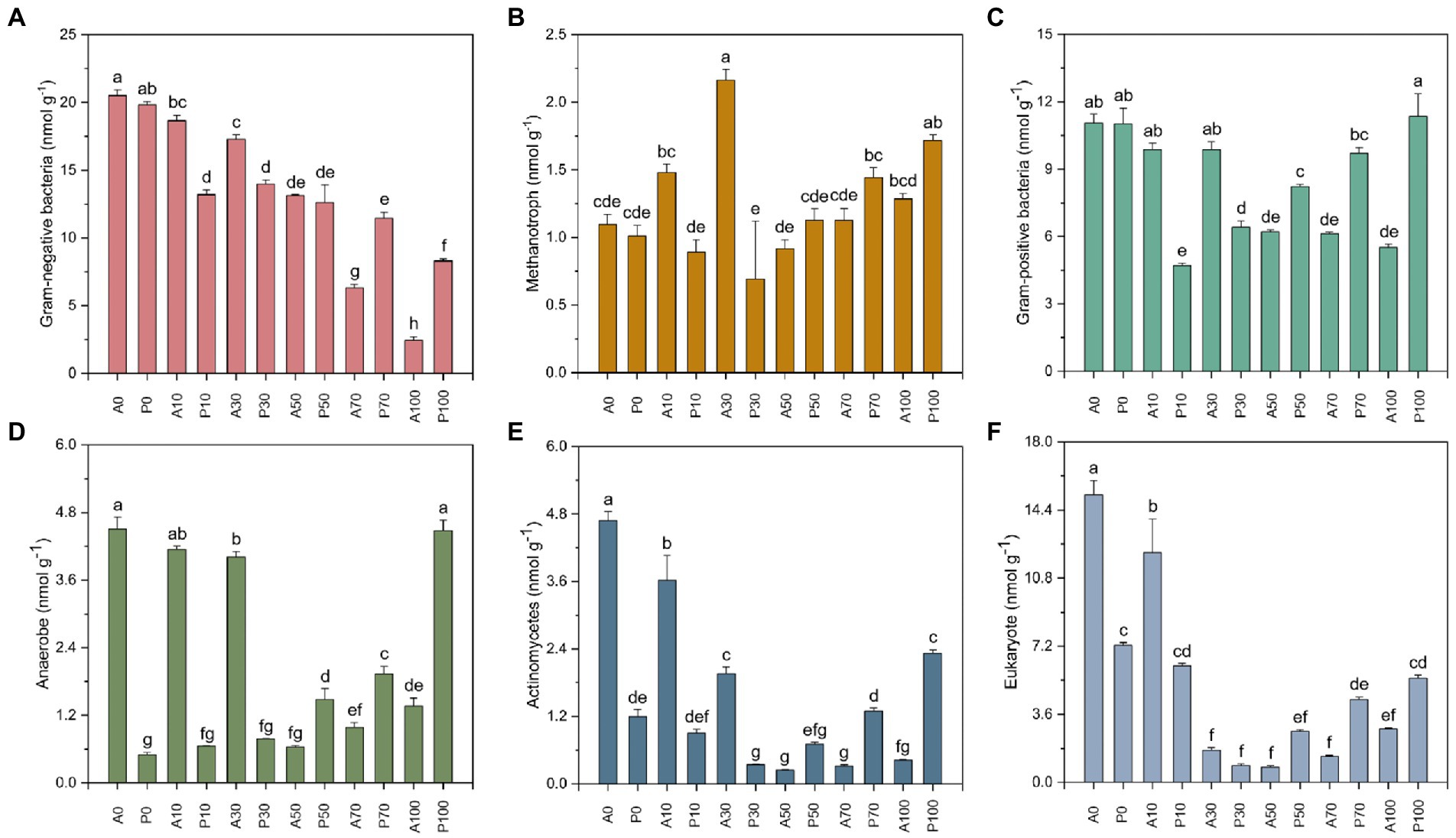
Figure 5. The PLFAs (nmol g−1) indicated by Gram-negative (A), Methanotroph (B), Gram-positive (C), Anaerobe (D), Actinobacteria (E), and Eukaryote (F) in different treatments on day 7. A0: Equivalent amount of methanol were added into native soil. A10: 10 mg kg−1 atrazine-contaminated soil; A30: 30 mg kg−1 atrazine-contaminated soil; A50: 50 mg kg−1 atrazine-contaminated soil; A70: 70 mg kg−1 atrazine-contaminated soil; A100: 100 mg kg−1 atrazine-contaminated soil; P0: Equivalent amount of methanol and P. ureafaciens ZY were added into native soil; P10: 10 mg kg−1 atrazine-contaminated soil with P. ureafaciens ZY inoculation; P30: 30 mg kg−1 atrazine-contaminated soil with P. ureafaciens ZY inoculation; P50: 50 mg kg−1 atrazine-contaminated soil with P. ureafaciens ZY inoculation; P70: 70 mg kg−1 atrazine-contaminated soil with P. ureafaciens ZY inoculation; P100: 100 mg kg−1 atrazine-contaminated soil with P. ureafaciens ZY inoculation.
At relatively low concentrations of 10–30 mg kg−1 of atrazine, the total PLFAs of the non-inoculated groups were higher than that in the inoculated groups, the decrease of PLFAs of 32.687 nmol g−1 and 19.858 nmol g−1 were found in A10 and A30 compared to P10 and P30. And the same decrease trend was observed in Gram-negative (Figure 5A), Methanotroph (Figure 5B), Gram-positive (Figure 5C), Anaerobe (Figure 5D), Actinomycetes (Figure 5E) and Eukaryote (Figure 5F), which might be caused by competition between P. ureafaciens ZY and indigenous microorganisms (Abtahi et al., 2020; Koolivand et al., 2022). On the contrary, the biostimulation of P. ureafaciens ZY led to a significant increase of PLFAs at atrazine concentration of 50–100 mg kg−1, especially for Gram-negative (Figure 5A), Gram-positive (Figure 5C), Anaerobe (Figure 5D), Actinomycetes (Figure 5E) in 100 mg kg−1 atrazine treatments. This result was consistent with Gao et al. (2018) and Chen et al. (2021), which was possibly related with two reasons First, with the continuous removal of atrazine after inoculation of P. ureafaciens ZY, the inhibition effect of atrazine on soil microorganisms gradually decreased (Ros et al., 2006). Secondly, most microorganisms in the soil could not have the abllity to metabolize atrazine and cannot directly absorb atrazine as a carbon source and nitrogen source (Singh and Jauhari, 2017). After the decomposition of atrazine by P. ureafaciens ZY, atrazine metabolites could be acted as nitrogen and carbon source by soil microorganisms to meet their growth and proliferation (Chen et al., 2021).
3.6. Effect of Paenarthrobacter ureafaciens ZY on soil bacterial community dynamics
The high-throughput sequencing was applied to investigate native microbial community dynamics at 7 days in different treatments. The OTU (Supplementary Figure S5A), Chao1 (Supplementary Figure S5B), Simpson (Supplementary Figure S5C) and Shannon (Supplementary Figure S5D) indexes of the bacterial community in A0 treatment were higher than that in other treatments at 7 days. These results indicated that atrazine and inoculation of P. ureafaciens ZY reduced the soil community richness and diversity. Jia et al. (2021) reported that the bioaugmentation of atrazine-contaminated soil with strain AT5 significantly decreased the bacterial richness and diversity, which might be due to the persistence and niche occupation of inoculation strain. In addition, P. ureafaciens ZY inoculation was also associated with interte production and nutrient consumption during the metabolism of atrazine, leading to the changes of soil micro-environment. Yang et al. (2021) have shown that the changes in the micro-environment could result in a significant decrease in bacterial richness and diversity. PCA analysis (Figure 6A) showed that both the bacterial community structure and the relative abundance of bacteria at the phylum level in A0 is similar to those in atrazine alone addition, and different from those in P. ureafaciens ZY inoculation treatments. It indicated that inoculums rather than atrazine was the main factor affecting the bacterial community structure (Jia et al., 2021).
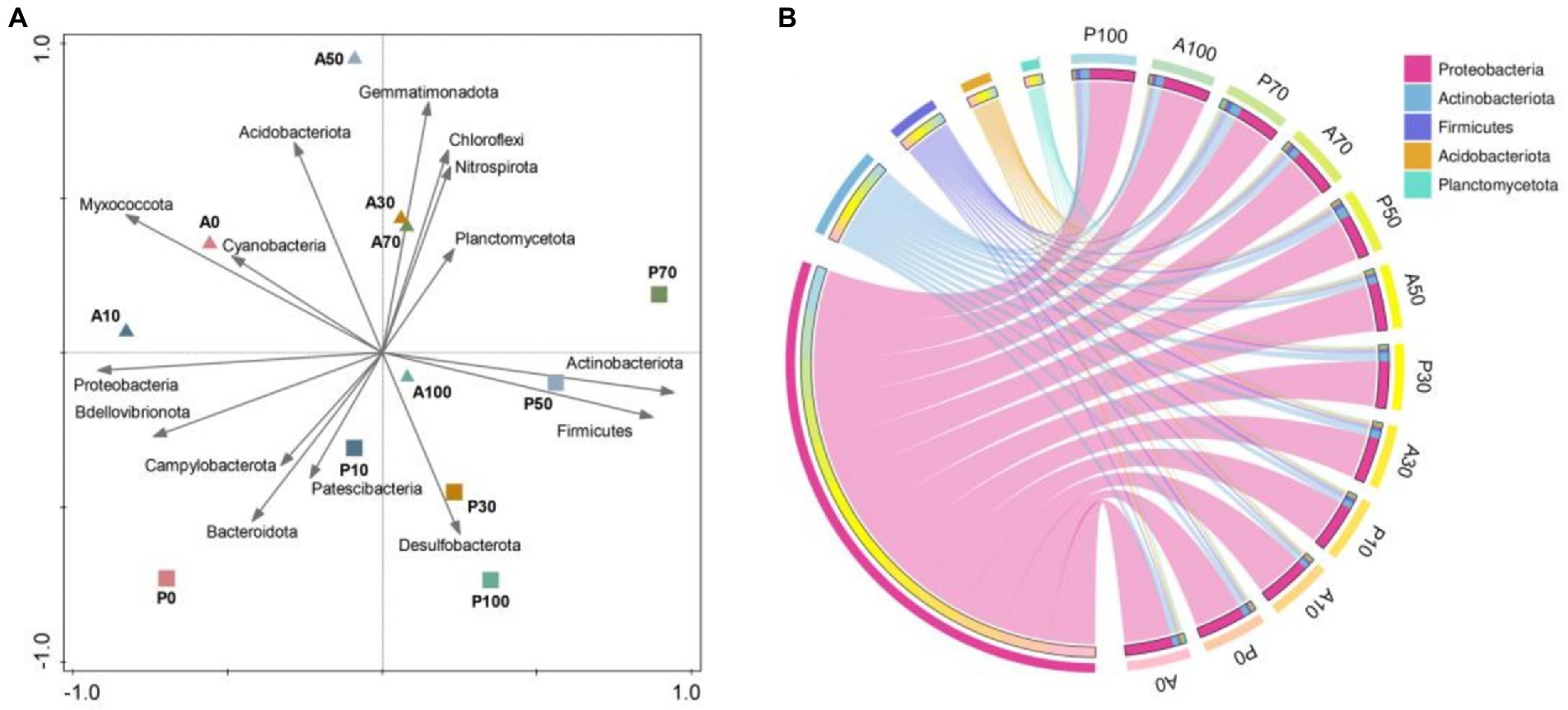
Figure 6. Effects of the different treatments on the soil bacterial community structure. Principal component analysis (PCA) based on the top 15 phylum of each sample (A); Chord diagram based on relative abundances of bacterial phylum in different treatments (B). A0: Equivalent amount of methanol were added into native soil. A10: 10 mg kg−1 atrazine-contaminated soil; A30: 30 mg kg−1 atrazine-contaminated soil; A50: 50 mg kg−1 atrazine-contaminated soil; A70: 70 mg kg−1 atrazine-contaminated soil; A100: 100 mg kg−1 atrazine-contaminated soil; P0: Equivalent amount of methanol and P. ureafaciens ZY were added into native soil; P10: 10 mg kg−1 atrazine-contaminated soil with P. ureafaciens ZY inoculation; P30: 30 mg kg−1 atrazine-contaminated soil with P. ureafaciens ZY inoculation; P50: 50 mg kg−1 atrazine-contaminated soil with P. ureafaciens ZY inoculation; P70: 70 mg kg−1 atrazine-contaminated soil with P. ureafaciens ZY inoculation; P100: 100 mg kg−1 atrazine-contaminated soil with P. ureafaciens ZY inoculation.
In our study, Proteobacteria, Actinobacteriota, Firmicutes, Acidobacteriota, Planctomycetota were the primary bacterial phyla in the soil samples (Figure 6B). Previous study found that Proteobacteria are the dominant microorganisms in atrazine-contaminated soils (Jia et al., 2021). The members of phylum, including Actinobacteria, Firmicutes, and Proteobacteria were reported to be the best sources for degradation of pesticides (Kumar et al., 2021). The reported atrazine degrading bacteria were mainly distributed in Actinobacteria, Firmicutes, and Proteobacteria, such as, Achromobacter (Proteobacteria), Arthrobacter/Citricoccus (Actinobacteria), Bacillus (Firmicutes), respectively (Tonelli Fernandes et al., 2018; Yang et al., 2018; Zhao et al., 2018; Jakinala et al., 2019).
Figure 7A shows the shift in the relative abundances of bacterial genera. The inoculated genus Paenarthrobacter was not been detected in the atrazine alone treatments due to its low abundance. The relative abundances of Paenarthrobacter were 1.08% (P0), 1.11% (P10), 2.56% (P30), 2.75% (P50), 4.39% (P70) and 3.30% (P100) at 7 days, respectively. These results showed that the inoculated P. ureafaciens ZY could survive and proliferate in atrazine-contaminated soils. In our study, the atrazine degradation rate was positively correlated with Frateuria (p < 0.01), Dyella (p < 0.05), Burkholderia-Caballeronia-Paraburkholderia (p < 0.05), and Paenarthrobacter (p < 0.01), respectively. Compared to A0, an increase of relative abundance of 16.71%–312.53% (Frateuria) and 7.09%–66.93% (Dyella) were found in other groups. Atrazine alone addition decreased the relative abundance by 1.49%–26.73% in Burkholderia-Caballeronia-Paraburkholderia, however, P. ureafaciens ZY increased the relative abundance of Burkholderia-Caballeronia-Paraburkholderia by 31.68%–221.78% compared to those in non-inoculation treatments. The previous studies had reported that Frateuria species could metabolize amine (Aoki et al., 1984) and Paraburkholderia lycopersici sp. nov., was involved in soil nitrogen metabolism (Tapia-García et al., 2020). Dyella also showed excellent ability to degrade pollutants (Li et al., 2009; Cai et al., 2022). In conclusion, Dyella, Frateuria and Burkholderia-Caballeronia-Paraburkholderia may be important genera involved in the metabolism of atrazine and its metabolites. In addition, as shown in Figure 7B. Paenarthrobacter ureafaciens ZY increased the abundance genus Streptomyces and Bacillus, and there is a positive correlation between P. ureafaciens ZY, Streptomyces and Bacillus. In the previous study, Bacillus showed atrazine degradation ability (Jakinala et al., 2019) and the inoculation of biomixtures with Streptomyces sp. M7 increased atrazine removal (Saez et al., 2022). These results suggested that there might be a synergistic relationship between P. ureafaciens ZY, Bacillus and Streptomyces during atrazine degradation.
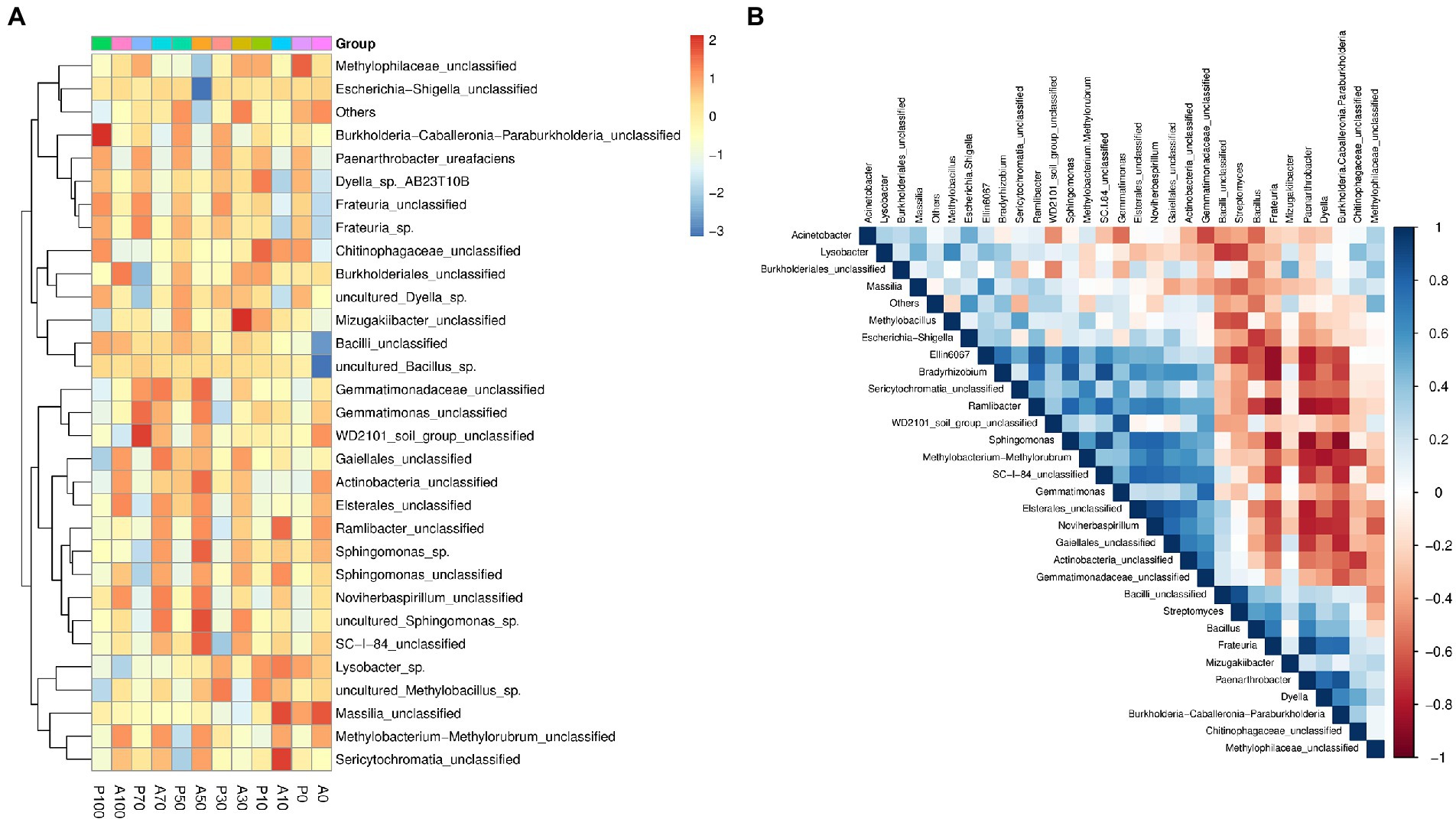
Figure 7. Heatmaps showing relative abundances of abundant genera (A); Spearman rank correlation obtained by the analysis of top 30 genera in different treatments at 7 days (B); Network A0: Equivalent amount of methanol were added into native soil. A10: 10 mg kg−1 atrazine-contaminated soil; A30: 30 mg kg−1 atrazine-contaminated soil; A50: 50 mg kg−1 atrazine-contaminated soil; A70: 70 mg kg−1 atrazine-contaminated soil; A100: 100 mg kg−1 atrazine-contaminated soil; P0: Equivalent amount of methanol and P. ureafaciens ZY were added into native soil; P10: 10 mg kg−1 atrazine-contaminated soil with P. ureafaciens ZY inoculation; P30: 30 mg kg−1 atrazine-contaminated soil with P. ureafaciens ZY inoculation; P50: 50 mg kg−1 atrazine-contaminated soil with P. ureafaciens ZY inoculation; P70: 70 mg kg−1 atrazine-contaminated soil with P. ureafaciens ZY inoculation; P100: 100 mg kg−1 atrazine-contaminated soil with P. ureafaciens ZY inoculation.
In addition, to further characterize the effects of P. ureafaciens ZY inoculation on soil native microbial community. The co-occurrence patterns of bacterial communities were assessed (Supplementary Figure S7). The more complex and well-connected co-occurrence networks were observed in atrazine alone treatments indicated by edge numbers and density with the value of 500 and 0.025 (Supplementary Table S8). This occurrence might be due to the stimulation of atrazine on the soil microbial diversity. The bacterial taxonomic composition of “hub nodes” in the network differed between atrazine alone treatments and P. ureafaciens ZY inoculation treatments. OTUs belonging to Sphingomonas in P. ureafaciens ZY inoculation treatments, and OTUs belonging to Gaiellales and Sericytochromatia in atrazine alone treatments were more connected with others (Supplementary Figure S7). Guo et al. (2022) found that hub species in microbial networks are recognized as mediators among associated microbiome. Trough network hubs, it follows that P. ureafaciens ZY inoculation may influence the assembly of soil native microbial community by regulating microbe-microbe interactions (Trivedi et al., 2020).
4. Conclusion
This study isolated and identified a novel atrazine degrader P. ureafaciens ZY. Paenarthrobacter ureafaciens ZY exhibited a significant degradation capacity of atrazine both in liquid media and soil. The P. ureafaciens ZY could metabolize atrazine to form cyanuric acid via related enzymes which encoded by trzN, atzB, and atzC. Besides, P. ureafaciens ZY increased the biomass of native bacterial communities and restored the microbial abundance and diversity in heavily atrazine-contaminated soil (50–100 mg kg−1 atrazine). In addition, Frateuria, Dyella, and Burkholderia-Caballeronia-Paraburkholderia showed significantly positive correlations with atrazine degradation rate, suggesting that they were also important participants in the atrazine degrading process. Moreover, there may be a synergic relationship between P. ureafaciens ZY, Streptomyces and Bacillus during atrazine degradation. Our work provides a theoretical basis for bioaugmentation in atrazine-contaminated soil.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number (s) can be found in the article/Supplementary material.
Author contributions
YZ: conceptualization, investigation, data curation, Writing—original draft. XL: supervision, funding acquisition, writing—review. YL: methodology, writing. HB: methodology, writing. JN: supervision, Writing—review and editing. GX: Writing—review and editing. All authors contributed to the article and approved the submitted version.
Funding
National Key Research and Development Program of China (2019YFC1906501 and 2018YFC1901100).
Acknowledgments
The authors greatly acknowledge the financial support from the National Key Research and Development Program of China (2019YFC1906501 and 2018YFC1901100).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1103168/full#supplementary-material
References
Abtahi, H., Parhamfar, M., Saeedi, R., Villaseñor, J., Sartaj, M., Kumar, V., et al. (2020). Effect of competition between petroleum-degrading bacteria and indigenous compost microorganisms on the efficiency of petroleum sludge bioretion: field application of mineral-based culture in the composting process. J. Environ. Manag. 258:110013. doi: 10.1016/j.jenvman.2019.110013
Aoki, K., Ohtsuka, K., Shinke, R., and Nishira, H. (1984). Rapid biodegradation of aniline by Frateuria species ANA-18 and its aniline metabolism. Agric. Biol. Chem. 48, 865–872. doi: 10.1080/00021369.1984.10866258
Bazhanov, D. P., Yang, K., Li, H. M., Li, C. Y., Li, J. S., Chen, X. F., et al. (2017). Colonization of plant roots and enhanced atrazine degradation by a strain of Arthrobacter ureafaciens. Appl. Microbiol. Biotechnol. 101, 6809–6820. doi: 10.1007/s00253-017-8405-3
Brain, R. A., Hoberg, J., Hosmer, A. J., and Wall, S. B. (2012). Influence of light intensity on the toxicity of atrazine to the submerged freshwater aquatic macrophyte Elodea canadensis. Ecotoxicol. Environ. Saf. 79, 55–61. doi: 10.1016/j.ecoenv.2011.12.001
Cai, Y. X., Yan, Z. Y., Ou, Y. J., Peng, B. S., Zhang, L. H., Shao, J. H., et al. (2022). Effects of different carbon sources on the removal of ciprofloxacin and pollutants by activated sludge: mechanism and biodegradation. J. Environ. Sci. (China) 111, 240–248. doi: 10.1016/j.jes.2021.03.037
Chaudhary, D. R., Rathore, A. P., and Sharma, S. (2020). Effect of halotolerant plant growth promoting rhizobacteria inoculation on soil microbial community structure and nutrients. Appl. Soil Ecol. 150:103461. doi: 10.1016/j.apsoil.2019.103461
Chen, S. M., Li, Y. Y., Fan, Z. W., Liu, F. M., Liu, H. T., Wang, L. C., et al. (2021). Soil bacterial community dynamics following bioaugmentation with Paenarthrobacter sp. W11 in atrazine-contaminated soil. Chemosphere 282:130976. doi: 10.1016/j.chemosphere.2021.130976
Chen, S. S., Yang, P. P., Kumar, J. R., Liu, Y., and Ma, L. M. (2017). Inconsistent carbon and nitrogen isotope fractionation in the biotransformation of atrazine by Ensifer sp. CX-T and Sinorihizobium sp. K. Int. Biodeterior. Biodegradation 125, 170–176. doi: 10.1016/j.ibiod.2017.09.014
Chen, Q. L., Yang, B. S., Wang, H., He, F., Gao, Y. C., and Scheel, R. (2015). Soil microbial community toxic response to atrazine and its residues under atrazine and lead contamination. Environ. Sci. Pollut. Res. 22, 996–1007. doi: 10.1007/s11356-014-3369-7
Cheng, X., Chen, K. X., Jiang, N. D., Wang, L., Jiang, H. Y., Zhao, Y. X., et al. (2022). Nitroreduction of imidacloprid by the Actinomycete Gordonia alkanivorans and the stability and scute toxicity of the nitroso metabolite. Chemosphere 291:132885. doi: 10.1016/j.chemosphere.2021.132885
de Albuquerque, F. P., de Oliveira, J. L., Moschini-Carlos, V., and Fraceto, L. F. (2020). An overview of the potential impacts of atrazine in aquatic environments: perspectives for tailored solutions based on nanotechnology. Sci. Total Environ. 700:134868. doi: 10.1016/j.scitotenv.2019.134868
de Andrade, L. L., do Espirito Santo Pereira, A., Fraceto, L. F., and dos Reis Martinez, C. B. (2019). Can atrazine loaded nanocapsules reduce the toxic effects of this herbicide on the fish Prochilodus lineatus? A multibiomarker approach. Sci. Total Environ. 663, 548–559. doi: 10.1016/j.scitotenv.2019.01.380
de Souza, M. L., Seffffernick, J., Martinez, B., Sadowsky, M. J., and Wackett, L. P. (1998). The atrazine catabolism genes atzABC are widespread and highly conserved. J. Bacteriol. 180, 1951–1954. doi: 10.1128/JB.180.7.1951-1954.1998
Dionne, E., Hanson, M. L., Anderson, J. C., and Brain, R. A. (2021). Chronic toxicity of technical atrazine to the fathead minnow (Pimephales promelas) during a full life-cycle exposure and an evaluation of the consistency of responses. Sci. Total Environ. 755:142589. doi: 10.1016/j.scitotenv.2020.142589
Du, J., Zhang, Y., Ma, Y. X., Li, J., and Zhang, Q. Y. (2011). Simulation study of atrazine-contaminated soil biodegradation by strain W16. Procedia Environ. Sci. 11, 1488–1492. doi: 10.1016/j.proenv.2011.12.224
Food and Agriculture Organization (FAO), (2021). FAOSTAT. Retrieved from: https://www.fao.org/faostat/en/#data/EF (Accessed March 1, 2022).
Francolino, B. Y., Valdes, Y., de Luna, C. A., de França, F. J. L., Moens, T., and dos Santos, G. A. P. (2021). Short-term lethal and sublethal atrazine effects on Litoditis marina: towards a nematode model for marine toxicity assessment? Ecol. Indic. 126:107642. doi: 10.1016/j.ecolind.2021.107642
Frostegård, Å., Bååth, E., and Tunlio, A. (1993). Shifts in the structure of soil microbial communities in limed forests as revealed by phospholipid fatty acid analysis. Soil Biol. Biochem. 25, 723–730. doi: 10.1016/0038-0717(93)90113-P
Gao, J. P., Song, P. P., Wang, G. Y., Wang, J. H., Zhu, L. S., and Wang, J. (2018). Responses of atrazine degradation and native bacterial community in soil to Arthrobacter sp. strain HB-5. Ecotoxicol. Environ. Saf. 159, 317–323. doi: 10.1016/j.ecoenv.2018.05.017
García-Espiñeira, M., Tejeda-Benitez, L., and Olivero-Verbel, J. (2018). Toxicity of atrazine-and alyphosate-based formulations on Caenorhabditis elegans. Ecotoxicol. Environ. Saf. 156, 216–222. doi: 10.1016/j.ecoenv.2018.02.075
Giardi, M. T., Giardina, M. C., and Filacchioni, G. (1985). Chemical and biological degradation of primary metabolism of atrazine by a Nocardia strain. Agricul. Biol. Chem. 49, 1551–1558. doi: 10.1080/00021369.1985.10866949
Guo, Q. X., Liu, L., Liu, J. T., Korpelainen, H., and Li, C. Y. (2022). Plant sex affects plant-microbiome assemblies of dioecious Populus cathayana trees under different soil nitrogen conditions. Microbiome 10:191. doi: 10.1186/s40168-022-01387-9
Hayes, T. B., Collins, A., Lee, M., Mendoza, M., Noriega, N., Stuart, A. A., et al. (2002). Hermaphroditic, demasculinized frogs after exposure to the herbicide atrazine at low ecologically relevant doses. Proc. Natl. Acad. Sci. U. S. A. 99, 5476–5480. doi: 10.1073/pnas.082121499
He, H. Z., Yu, J., Chen, G. K., Li, W. Y., He, J. B., and Li, H. S. (2012). Acute toxicity of butachlor and atrazine to freshwater green alga Scenedesmus obliquus and cladoceran Daphnia carinata. Ecotoxicol. Environ. Saf. 80, 91–96. doi: 10.1016/j.ecoenv.2012.02.009
Jakinala, P., Lingampally, N., Kyama, A., and Hameeda, B. (2019). Enhancement of atrazine biodegradation by marine isolate Bacillus velezensis MHNK1 in presence of surfactin lipopeptide. Ecotoxicol. Environ. Saf. 182:109372. doi: 10.1016/j.ecoenv.2019.109372
Jia, W. B., Li, N., Yang, T. N., Dai, W. X., Jiang, J. D., and Chen, K. (2021). Bioaugmentation of atrazine contaminated soil with Paenarthrobacter sp. strain AT-5 and its effect on the soil microbiome. Front. Microbiol. 12:771463. doi: 10.3389/fmicb.2021.771463
Kolekar, P. D., Patil, S. M., Suryavanshi, M. V., Suryawanshi, S. S., Khandare, R. V., Govindwar, S. P., et al. (2019). Microcosm study of atrazine bioremediation by indigenous microorganisms and cytotoxicity of biodegraded metabolites. J. Hazard. Mater. 374, 66–73. doi: 10.1016/j.jhazmat.2019.01.023
Koolivand, A., Abtahi, H., Parhamfar, M., Saeedi, R., Coulon, F., Kumar, V., et al. (2022). The effect of petroleum hydrocarbons concentration on competition between oil-degrading bacteria and indigenous compost microorganisms in petroleum sludge bioretion. Environ. Technol. Inno. 26:102319. doi: 10.1016/j.eti.2022.102319
Kumar, M., Yadav, A. N., Saxena, R., Paul, D., and Tomar, R. S. (2021). Biodiversity of pesticides degrading microbial communities and their environmental impact. Biocatal. Agric. Biotechnol. 31:101883. doi: 10.1016/j.bcab.2020.101883
Lee, G., and Choi, K. (2020). Adverse effects of pesticides on the functions of immune system. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 235:108789. doi: 10.1016/j.cbpc.2020.108789
Li, Q. Y., Li, Y., Zhu, X. K., and Cai, B. L. (2008). Isolation and characterization of atrazine-degrading Arthrobacter sp. AD26 and use of this strain in bioretion of contaminated soil. J. Environ. Sci. 20, 1226–1230. doi: 10.1016/S1001-0742(08)62213-5
Li, A., Qu, Y. Y., Zhou, J. T., and Gou, M. (2009). Isolation and characteristics of a novel biphenyl-degrading bacterial strain, Dyella ginsengisoli LA-4. J. Environ. Sci. (China) 21, 211–217. doi: 10.1016/S1001-0742(08)62253-6
Liang, Z., Abdelshafy, A. M., Luo, Z. S., Belwal, T., Lin, X. Y., Xu, Y. Q., et al. (2022). Occurrence, detection, and dissipation of pesticide residue in plant-derived foodstuff: a state-of-the-art review. Food Chem. 384:132494. doi: 10.1016/j.foodchem.2022.132494
Logue, J. B., Stedmon, C. A., Kellerman, A. M., Nielsen, N. J., Andersson, A. F., Laudon, H., et al. (2016). Experimental insights into the importance of aquatic bacterial community composition to the degradation of dissolved organic matter. ISME J. 10, 533–545. doi: 10.1038/ismej.2015.131
Lykogianni, M., Bempelou, E., Karamaouna, F., and Aliferis, K. A. (2021). Do pesticides promote or hinder sustainability in agriculture? The challenge of sustainable use of pesticides in modern agriculture. Sci. Total Environ. 795:148625. doi: 10.1016/j.scitotenv.2021.148625
Luo, S. W., Zhen, Z., Zhu, X. P., Ren, L., Wu, W. J., Zhang, W. J., et al. (2021). Accelerated atrazine degradation and altered metabolic pathways in goat manure assisted soil bioremediation. FEcotoxicol. Environ. Saf. 221:112432. doi: 10.1016/j.ecoenv.2021.112432
Ma, L. M., Chen, S. S., Yuan, J., Yang, P. P., Liu, Y., and Stewart, K. (2017). Rapid biodegradation of atrazine by Ensifer sp. strain and its degradation genes. Int. Biodeterior. Biodegrad. 116, 133–140. doi: 10.1016/j.ibiod.2016.10.022
Mandelbaum, R. T., Allan, D. L., and Wackett, L. P. (1995). Isolation and characterization of a Pseudomonas sp. that mineralizes the s-triazine herbicide atrazine. Appl. Environ. Microbiol. 61, 1451–1457. doi: 10.1128/aem.61.4.1451-1457.1995
Martinez, B., Tomkins, J., Wackett, L. P., Wing, R., and Sadowsky, M. J. (2001). Complete nucleotide sequence and organization of the atrazine catabolic plasmid pADP-1 from Pseudomonas sp. strain ADP. J. Bacteriol. 183, 5684–5697. doi: 10.1128/JB.183.19.5684-5697.2001
Mekonnen, E., Kebede, A., Tafesse, T., and Tafesse, M. (2019). Investigation of carbon substrate utilization patterns of three ureolytic bacteria. Biocatal. Agric. Biotechnol. 22:101429. doi: 10.1016/j.bcab.2019.101429
Omotayo, A. E., Ilori, M. O., Radosevich, M., and Amund, O. O. (2013). Metabolism of atrazine in liquid cultures and soil microcosms by Nocardioides strains isolated from a contaminated Nigerian agricultural soil. Soil Sediment Contam. 22, 365–375. doi: 10.1080/15320383.2013.733444
Radosevich, M., Traina, S. J., and Tuovinen, O. H. (1997). Atrazine mineralization in laboratory-aged soil microcosms inoculated with s-triazine-degrading bacteria. J. Environ. Qual. 26, 206–214. doi: 10.2134/jeq1997.00472425002600010030x
Rehan, M., Kluge, M., Fränzle, S., Kellner, H., Ullrich, R., and Hofrichter, M. (2014). Degradation of atrazine by Frankia alni ACN14a: gene regulation, dealkylation, and dechlorination. Appl. Microbiol. Biotechnol. 98, 6125–6135. doi: 10.1007/s00253-014-5665-z
Ros, M., Goberna, M., Moreno, J. L., Hernandez, T., García, C., Insam, H., et al. (2006). Molecular and physiological bacterial diversity of a semi-arid soil contaminated with different levels of formulated atrazine. Appl. Soil Ecol. 34, 93–102. doi: 10.1016/j.apsoil.2006.03.010
Rostami, S., Jafari, S., Moeini, Z., Jaskulak, M., Keshtgar, L., Badeenezhad, A., et al. (2021). Current methods and technologies for degradation of atrazine in contaminated soil and water: a review. Environ. Technol. Inno. 24:102019. doi: 10.1016/j.eti.2021.102019
Rousseaux, S., Hartmann, A., Lagacherie, B., Piutti, S., Andreux, F., and Soulas, G. (2003). Inoculation of an atrazine-degrading strain, Chelatobacter heintzii Cit1, in four different soils: effects of different inoculum densities. Chemosphere 51, 569–576. doi: 10.1016/S0045-6535(02)00810-X
Sabzevari, S., and Hofman, J. (2022). A worldwide review of currently used pesticides' monitoring in agricultural soils. Sci. Total Environ. 812:152344. doi: 10.1016/j.scitotenv.2021.152344
Saez, J. M., González, S. K., Ocante, T. A. L., Bigliardo, A. L., Briceño, G. E., and Benimeli, C. S. (2022). Actinobacteria bioaugmentation and substrate evaluation for biobeds useful for the treatment of atrazine residues in agricultural fields. J. Environ. Manag. 320:115870. doi: 10.1016/j.jenvman.2022.115870
Sebaï, T. E., Devers-Lamrani, M., Changey, F., Rouard, N., and Martin-Laurent, F. (2011). Evidence of atrazine mineralization in a soil from the Nile Delta: isolation of Arthrobacter sp. TES6, an atrazine-degrading strain. Int. Biodeterior. Biodegr. 65, 1249–1255. doi: 10.1016/j.ibiod.2011.05.011
Seghers, D., Verthé, K., Reheul, D., Bulcke, R., Siciliano, S. D., and Verstraete, W. (2003). Effect of long-term herbicide applications on the bacterial community structure and function in an agricultural soil. FEMS Microbiol. Ecol. 46, 139–146. doi: 10.1016/S0168-6496(03)00205-8
Shan, W. Q., Hu, W. Y., Wen, Y., Ding, X. W., Ma, X., Yan, W., et al. (2021). Evaluation of atrazine neurodevelopment toxicity in vitro-application of hESC-based neural differentiation model. Reprod. Toxicol. 103, 149–158. doi: 10.1016/j.reprotox.2021.06.009
Shenoy, K. (2012). Environmentally realistic exposure to the herbicide atrazine alters some sexually selected traits in male guppies. PLoS One 7:e30611. doi: 10.1371/journal.pone.0030611
Siddique, T., Okeke, B. C., Arshad, M., and Frankenberger, W. T. (2002). Temperature and pH effects on biodegradation of hexachlorocyclohexane isomers in water and a soil slurry. J. Agric. Food Chem. 50, 5070–5076. doi: 10.1021/jf0204304
Singh, S. N., and Jauhari, N. (2017). “Degradation of atrazine by plants and microbes” in Microbe-Induced Degradation of Pesticides. Environmental Science and Engineering. ed. S. Singh (Switzerland, FL: Springer), 213–225.
Sun, J. T., Pan, L. L., Zhan, Y., Tsang, D. C. W., Zhu, L. Z., and Li, X. D. (2017). Atrazine contamination in agricultural soils from the Yangtze River Delta of China and associated health risks. Environ. Geochem. Health 39, 369–378. doi: 10.1007/s10653-016-9853-x
Sun, S. X., Sidhu, V., Rong, Y. H., and Zheng, Y. (2018). Pesticide pollution in agricultural soils and sustainable remediation methods: a review. Curr. Pollut. Rep. 4, 240–250. doi: 10.1007/s40726-018-0092-x
Tapia-García, E. Y., Arroyo-Herrera, I., Rojas-Rojas, F. U., Ibarra, J. A., Vásquez-Murrieta, M. S., Martínez-Aguilar, L., et al. (2020). Paraburkholderia lycopersici sp. nov., a nitrogen-fixing species isolated from rhizoplane of Lycopersicon esculentum Mill. Var. Saladette in Mexico. Syst. Appl. Microbiol. 43:126133. doi: 10.1016/j.syapm.2020.126133
Tonelli Fernandes, A. F., Braz, V. S., Bauermeister, A., Rizzato Paschoal, J. A., Lopes, N. P., and Stehling, E. G. (2018). Degradation of atrazine by Pseudomonas sp. and Achromobacter sp. isolated from brazilian agricultural soil. Int. Biodeterior. Biodegr. 130, 17–22. doi: 10.1016/j.ibiod.2018.03.011
Trivedi, P., Leach, J. E., Tringe, S. G., Sa, T. M., and Singh, B. K. (2020). Plant-microbiome interactions: from community assembly to plant health. Nat. Rev. Microbiol. 18, 607–621. doi: 10.1038/s41579-020-00490-8
Tyler Mehler, W., Schuler, L. J., and Lydy, M. J. (2008). Examining the joint toxicity of chlorpyrifos and atrazine in the aquatic species: Lepomis macrochirus, Pimephales promelas and Chironomus tentans. Environ. Pollut. 152, 217–224. doi: 10.1016/j.envpol.2007.04.028
Vela, N., Navarro, G., Gimenez, M. J., and Navarro, S. (2004). Gradual fall of s-triazine herbicides in drinking and wastewater samples as influenced by light and temperature. Water. Air. Soil. Poll. 158, 3–19. doi: 10.1023/B:WATE.0000044827.22284.29
Wang, Q. F., and Xie, S. G. (2012). Isolation and characterization of a high-efficiency soil atrazine-degrading Arthrobacter sp. strain. Int. Biodeterior. Biodegra. 71, 61–66. doi: 10.1016/j.ibiod.2012.04.005
Yang, X. Y., Wei, H. Y., Zhu, C. X., and Geng, B. (2018). Biodegradation of atrazine by the novel Citricoccus sp. strain TT3. Ecotoxicol. Environ. Saf. 147, 144–150. doi: 10.1016/j.ecoenv.2017.08.046
Yang, H. X., Zhang, Y. L., Chuang, S. C., Cao, W. M., Ruan, Z. P., Xu, X. H., et al. (2021). Bioaugmentation of acetamiprid-contaminated soil with Pigmentiphaga sp. strain D-2 and its effect on the soil microbial community. Ecotoxicology 30, 1559–1571. doi: 10.1007/s10646-020-02336-8
Yanze-Kontchou, C., and Gschwind, N. (1995). Mineralization of the herbicide atrazine in soil inoculated with a pseudomonas strain. J. Agric. Food Chem. 43, 2291–2294. doi: 10.1021/JF00056A061
Yu, X. L., Jin, X., Liu, H., Yu, Y. Y., Tang, J., Zhou, R. J., et al. (2023). Enhanced degradation of atrazine through UV/bisulfite: mechanism, reaction pathways and toxicological analysis. Sci. Total Environ. 856:159157. doi: 10.1016/j.scitotenv.2022.159157
Zhao, S., Arthur, E. L., and Coats, J. R. (2003). Influence of microbial inoculation (pseudomonas sp. strain ADP), the enzyme atrazine chlorohydrolase, and vegetation on the degradation of atrazine and metolachlor in soil. J. Agric. Food Chem. 51, 3043–3048. doi: 10.1021/jf025954i
Zhao, X. Y., Wang, L., Ma, F., and Yang, J. X. (2018). Characterisation of an efficient atrazine-degrading bacterium, Arthrobacter sp. ZXY-2: an attempt to lay the foundation for potential bioaugmentation applications. Biotechnol. Biofuels 11:113. doi: 10.1186/s13068-018-1113-0
Keywords: atrazine, Paenarthrobacter ureafaciens, degradation, bioremediation, native microbial communities
Citation: Zhao Y, Li X, Li Y, Bao H, Nan J and Xu G (2023) Rapid biodegradation of atrazine by a novel Paenarthrobacter ureafaciens ZY and its effects on soil native microbial community dynamic. Front. Microbiol. 13:1103168. doi: 10.3389/fmicb.2022.1103168
Edited by:
Hao Zhou, Dalian University of Technology, ChinaReviewed by:
Xuwang Zhang, Dalian University of Technology, ChinaQiao Ma, Dalian Maritime University, China
Copyright © 2023 Zhao, Li, Li, Bao, Nan and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin Li,  bGl4aW53aW5kb3dzQDE2My5jb20=
bGl4aW53aW5kb3dzQDE2My5jb20=
 Yue Zhao
Yue Zhao Xin Li1*
Xin Li1*