- 1Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China
- 2Department of Otolaryngology, Huazhong University of Science and Technology Union Shenzhen Hospital, Shenzhen, China
- 3School of Medicine, Southern University of Science and Technology, Shenzhen, China
- 4Center for Translational Medicine Research and Development, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China
- 5Department of Pathogen Biology, Shenzhen Center for Disease Control and Prevention, Shenzhen, China
- 6Department of Critical Care Medicine, Shenzhen Second People’s Hospital, The First Affiliated Hospital of Shenzhen University, Shenzhen, China
Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV), Middle East Respiratory Syndrome (MERS), and the recent SARS-CoV-2 are lethal coronaviruses (CoVs) that have caused dreadful epidemic or pandemic in a large region or globally. Infections of human respiratory systems and other important organs by these pathogenic viruses often results in high rates of morbidity and mortality. Efficient anti-viral drugs are needed. Herein, we firstly take SARS-CoV-2 as an example to present the molecular mechanism of CoV infection cycle, including the receptor binding, viral entry, intracellular replication, virion assembly, and release. Then according to their mode of action, we provide a summary of anti-viral peptides that have been reported in peer-reviewed publications. Even though CoVs can rapidly evolve to gain resistance to the conventional small molecule drugs, peptide-based inhibitors targeting various steps of CoV lifecycle remain a promising approach. Peptides can be continuously modified to improve their antiviral efficacy and spectrum along with the emergence of new viral variants.
Introduction
Coronaviruses are membrane enveloped virus particles, which contain a single-stranded positive-sense ribonucleic acid (RNA) genome and a matrix of RNA-associated capsid proteins (Zhou et al., 2020; Li et al., 2022). Taxonomically, four genera are classified within the coronaviridae family, including alpha-, beta-, gamma-, and delta-coronaviruses. Among them, seven alpha-and beta-CoV species have been identified as zoonotic coronaviruses (HCoVs). The highly pathogenic members are Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV), Middle East Respiratory Syndrome Coronavirus (MERS-CoV), and the recently emerged SARS-CoV-2, all of which are capable of causing severe respiratory tract infections and acute respiratory distress syndrome (ARDS). Infections by the intensively pathogenic HCoVs, especially SARS-CoV-2, have been the top concern of public health in recent years. The other HCoVs, HCoV-229E, HCoV-NL63, HCoV-OC43, and HCoV-HKU1 that normally cause mild respiratory illness have circulated within human populations for centuries. Although numerous drugs and vaccines have been developed and applied to combating SARS-CoV-2 or subsequent variants, drug resistance raises great concern (Rawson et al., 2020; Tannock et al., 2020; Kasuga et al., 2021; Şimşek-Yavuz and Komsuoğlu elikyurt, 2021). For example, the SARS-CoV-2 B.1.617.2 (delta) variant can rapidly gain resistance to monoclonal antibody after treatment (Rockett et al., 2022). The more recent B.1.1.529 (Omicron) variant is highly resistant to the majority of existing SARS-CoV-2 neutralizing antibodies (Cao et al., 2022; Hoffmann et al., 2022) as well as mRNA vaccines (Cele et al., 2022; Edara et al., 2022). Therefore, effective broad-spectrum antiviral therapeutics are still needed.
Recent observations indicated that peptides of diverse sources (either natural or synthetic) represent a class of promising antivirals. Peptides are small fragments of proteins typically comprising of 2–50 amino acid residues. These peptides achieve viral inhibition through various modes of actions, including direct binding to virions or host cell-surface receptors, blocking viral entry, interfering enzymatic activity to inhibit intracellular replication, and indirectly modulating immune responses(Schütz et al., 2020; Ghosh and Weinberg, 2021; Heydari et al., 2021). Compared to the conventional small molecule drugs, peptide synthesis can be quickly launched and modified (Vagner et al., 2008; Gao et al., 2018). More importantly, the chemical composition makes peptides highly specific and effective to their targets, even at nanomolar or picomolar concentrations (Cao et al., 2020; Schütz et al., 2020; Heydari et al., 2021; Shah et al., 2022; Yang et al., 2022).
Herein, we take SARS-CoV-2 as an instance to introduce the structural and functional properties of coronaviruses, and the viral infection process. Then, a state-of-the-art overview is provided to summarize recent researches that report the anti-CoV efficacy of peptides and their potentials in clinical use.
CoV genome structure and viral infection mechanism
Structural and functional dissection of SARS-CoV-2 genome encoded proteins
The full-length genome of SARS-CoV-2 consists of 29,870 bases with a 5′-cap and a 3′- poly(A) tail of variable length (Wu et al., 2020; Zhu N. et al., 2020; Figure 1A). Three functional types of proteins are encoded by the viral genome (Bai et al., 2022), including (1) structural proteins spike (S), membrane (M), envelop (E), and nucleocapsid (N) that constitute virions; (2) non-structural proteins that are mainly responsible for proteolysis and RNA synthesis; and (3) accessory proteins that are mainly involved in immune evasion (Figure 1B).
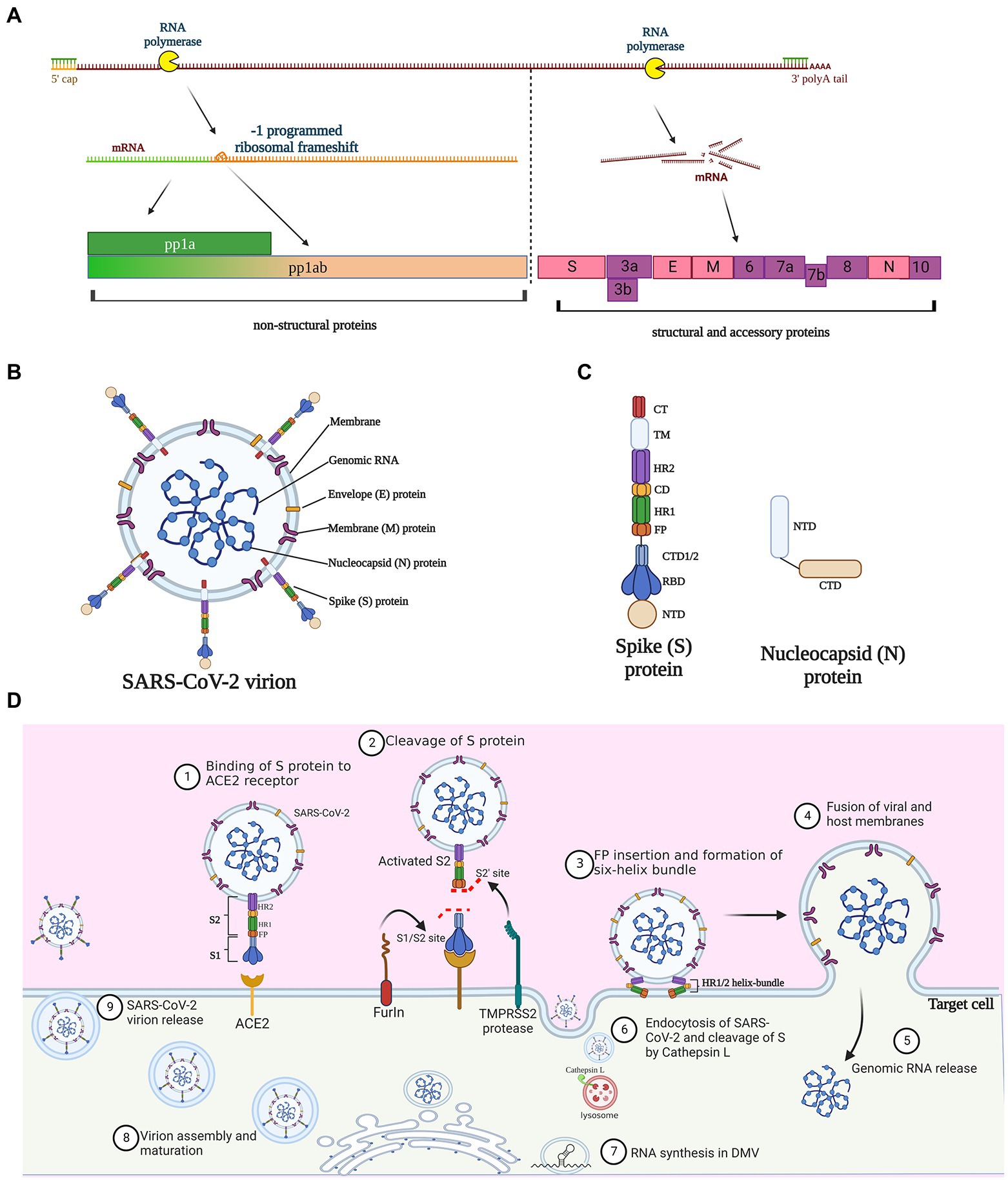
Figure 1. Molecular and structural bases of SARS-CoV-2 infection. (A) Proteins encoded by SARS-CoV-2 genome. Three quarters of the genome at the 5′-terminus encode the replicase polyproteins pp1a and pp1ab, which can be cleaved to generate 16 nonstructural proteins; pp1ab is derived from minus 1 site programmed ribosomal frameshift at the stop codon during the synthesis of pp1a. The 3′-terminus one quarter of the viral genome encode four structural proteins and accessory proteins. S, spike; E, envelope; M, membrane; and N, nucleocapsid. (B) Schematic diagram of SARS-CoV-2 virion structure. (C) Protein structure of spike protein and nucleocapsid protein. NTD, N-terminal domain; RBD, receptor-binding domain; CTD, C-terminal domain; FP, fusion peptide; HR, heptad repeat; CD, connector domain; TM, transmembrane domain; and CT, cytoplasmic tail. (D) The infection lifecycle of SARS-CoV-2. TMPRSS2, Type II transmembrane serine protease; ACE2, angiotensin-converting enzyme 2.
The 5′-proximal three quarters of the genome encode the replicases pp1a and pp1ab (Figure 1A), which can be further cleaved by the virus-encoded proteases papain-like protease (PLpro) and chymotrypsin-like or main protease (Mpro), to generate 16 nonstructural proteins (NSP 1–16). The yield balance between pp1a and pp1ab is controlled through a fine-tuning regulatory mechanism named programmed ribosomal frameshifting, which has been nicely summarized elsewhere (Malone et al., 2022). Nonstructural proteins have multiple roles in genome replication, transcription, viral morphogenesis, and dysregulation of host immune responses (Malone et al., 2022; Yan et al., 2022). For example, when mature NSP1 is released from the replicase polyproteins following proteolytic cleavage, it rapidly induces host mRNA degradation and shuts down translation of host proteins (Huang et al., 2011; Thoms et al., 2020), while the other NSPs come to form the replication-transcription complex (RTC). NSP12, in synergy with its auxiliary co-factors NSP7 and NSP8, constitutes the RNA-dependent RNA polymerase complex (RdRp) and serves as the replication/transcription machinery to replicate viral genome, rather than host polymerase (Kirchdoerfer and Ward, 2019; Yan et al., 2021).
The 3′-proximal one quarter of the viral genome is transcribed into a nested set of sub-genomic RNAs that are in turn translated to structural proteins and accessory proteins. As with all CoVs, SARS-CoV-2 structure proteins include S, M, E, and N proteins. The S protein, protruding from the viral surface, binds to the angiotensin-converting enzyme 2 (ACE2) to initiate viral entry into host cells, a vital process for CoV infection (Huang et al., 2020; Walls et al., 2020). Thus, as the most easily accessible but also an indispensable viral component, the S protein has become an attractive target of anti-coronavirus peptides in a vast number of researches (Huang et al., 2020; Schütz et al., 2020). Structurally, S protein possesses two subunits, S1 and S2. The S1 subunit consists of N-and C-terminal domains and an important receptor-binding domain (RBD), while the S2, involved in membrane fusion and viral entry, contains a fusion peptide (FP), two heptapeptide repeat (HR1 and HR2), a transmembrane (TM), and cytoplasmic (CT) domains (Huang et al., 2020; Figure 1C). The E protein is a transmembrane protein responsible for viral assembly, budding, morphogenesis, and trafficking (Schoeman and Fielding, 2019). E protein directly contributes to the viral pathogenesis since it not only activates the host NACHT, LRR, and PYD domain-containing protein 3 (NLRP3) inflammasome (Nieto-Torres et al., 2015), but also undermines the tight junction protein complex of the lung epithelium (Chai et al., 2021; Javorsky et al., 2021). The M protein is the major component of the viral envelop, conferring the virion size and spherical structure. M protein is involved in interaction and trafficking of multiple viral proteins, as well as assembly and release of virion particles (Yan et al., 2022). The SARS-CoV M protein can stimulate the host to produce a specific CD8+ T cell immune response (Li et al., 2021). Owning to the high sequence identity (90.5%) of the M protein gene between SARS-CoV-2 and SARS-CoV (Mahtarin et al., 2022), the SARS-CoV-2 M protein is likely to have similar immunogenic effects (Su et al., 2021). The main role of the N protein is binding to genomic RNA to form a ribonucleoprotein complex, which is related to viral replication and assembly (Mcbride et al., 2014; Guo et al., 2016). Compared to the other structure proteins, the gene encoding N protein is highly conserved and stable with few mutations over time (Hodge et al., 2021; Figure 1C). The C-terminal region of N protein favors viral immune evasion by antagonizing the host interferon-beta (IFN-β) pathway (Lu et al., 2011). Given these basic findings, the N protein is a great potential target for diagnosis and therapy against CoV infection.
Eleven genes encoding accessory proteins also locate within the 3′- proximal part of SARS-CoV-2 genome and they are interlaced with structural protein genes. Although the characterization of these accessory proteins is relatively limited, they appear to have important roles in pathogenesis and immune evasion, rather than virus replication (Redondo et al., 2021). Mutations are frequently detected in accessory proteins among variants of concern, indicative of increasing transmissibility and immune evasion (Shang et al., 2020). In light of their frequent mutations, accessory proteins might not be favorable targets of the broad-spectrum anti-coronavirus peptides. Functional analysis of those proteins substantiates the bioinformatic indication. Through diverse strategies, the accessory proteins, ORF3b (Konno et al., 2020), ORF6 (Miorin et al., 2020), ORF7a (Cao et al., 2021), and ORF8 (Lei et al., 2020), can antagonize the type I IFN response, an important host defense reaction against viral infection.
Infection mechanism of SARS-CoV-2—binding, entry, intracellular replication, virion assembly, and release
The SARS-CoV-2 infection involves multiple steps (Figure 1D). Initially, the S protein is cleaved and activated by the host proprotein convertase furin, leaving the protruding extracellular S1 subunit and the transmembrane S2 subunit non-covalently bounded (Peacock et al., 2021). The cleavage exposes the RBD in S1, which directly interacts with the peptidase domain of ACE2 and induces drastic transformational alteration of S2 (Cai et al., 2020; Liu et al., 2020). The cleavage of S2 by Type II transmembrane serine protease (TMPRSS2) further exposes the fusion peptide, thus facilitating its insertion into cellular membrane (Fraser et al., 2022; Iwata-Yoshikawa et al., 2022). Simultaneously, the HR1 and HR2 in S2 form a six-helix bundle fusion core, which acts as a hinge to bring the viral and host cell membrane in close proximity (Yao H. et al., 2020; Xia et al., 2020b). Alternatively, the pH-dependent enzyme cathepsin L can also implement the cleavage of S2 when viral entry is dependent on endocytosis (Matsuyama et al., 2020; Hoffmann et al., 2020b). After the membrane fusion or endocytosis, the SARS-CoV-2 gRNAs are released into cytosol, and soon translated into two replicase polyproteins pp1a and pp1ab, by hijacking the host cell ribosomes. pp1a and pp1ab are digested by the viral proteases, Mpro and PLpro, into 16 non-structural proteins, which further form the RTCs for RNA synthesis (Malone et al., 2022). NSP3 and NSP4 drive the rearrangement of the endoplasmic reticulum (ER) into double membrane vesicles (DMVs; Snijder and Limpens, 2020), where the RTCs produce new gRNA and a set of sub-genomic mRNAs that are finally translated into four structural proteins and a few accessory proteins. SARS-CoV-2 assembly commences as the gRNAs are coated with nucleocapsid proteins, resulting in RNA-nucleocapsid complexes that bud into the endoplasmic reticulum-Golgi intermediate compartment (ERGIC) to form mature virions (Boson et al., 2021). Finally, the virus particles are released via the budding of the Golgi apparatus and exocytosis of the cell membrane for a new round of infections.
Peptides working at different infection stages are potent anti-CoV agents
Peptides targeting initial binding of S protein to the ACE2 receptor
Targeting the RBD domain in S protein to inhibit its binding to ACE2 has been so far an intensively popular strategy against CoVs (Figure 2A). The charged amino acids between residues 22 and 57 of ACE2 are predicted to be the critical interaction site (Han et al., 2006). In an early work, two peptides P4 and P5 that mimicked this region can bind to SARS-CoV S1 RBD and inhibit pseudo-virion infection with a high half-maximal-inhibitory concentration (IC50) of 50 and 6 μM, respectively. Interestingly, another peptide comprised of two discontinuous segments of ACE2 (a.a. 22–44 and 351–357) showed higher antiviral efficacy (IC50: 0.1 μM) in a HeLa cell model (Han et al., 2006). The S proteins of SARS-CoV and SARS-CoV-2 share 76% sequence homology while their RBDs share 75% similarity (Jaimes et al., 2020). Although SARS-CoV-2 has greater ACE2 binding affinity (Wrapp et al., 2020) and higher transmissibility (Zhou et al., 2020), the high sequence similarity indicates that peptides effectively blocking the S1 RBD of SARS-CoV might also inhibit SARS-CoV-2 infection. To address the SARS-CoV-2 infection, a series of RBD-targeting peptides have been synthesized or discovered (Cao et al., 2020; Jaiswal and Kumar, 2020; Tavassoly et al., 2020; Wang et al., 2021). Using ACE2 as the scaffold, researchers synthesized two peptides AHB1 and AHB2, which neutralized SARS-CoV-2 with IC50 values of 35 and 16 nM, respectively (Cao et al., 2020). Surprisingly, another two peptides (LCB1 and LCB3) based on de-novo sequencing of the RBD-binding motifs showed a much higher potency in preventing SARS-CoV-2 infection of mammalian Vero-E6 cells, with IC50 values of 23.54 and 48.1 pM, respectively (Cao et al., 2020). Except the abovementioned synthetic peptides, a natural peptide produced by airway epithelium, human cathelicidin LL37, can bind to the S1 RBD and inhibit SARS-CoV-2 S pseudo-virion infection with a IC50 value of 4.74 μg/ml (Wang et al., 2021). Notably, the RBD is not the exclusive ACE2-interaction site, since peptides targeting other regions in S1 were also able to neutralize SARS-CoV (Zheng et al., 2005) and, thus potentially SARS-CoV-2.
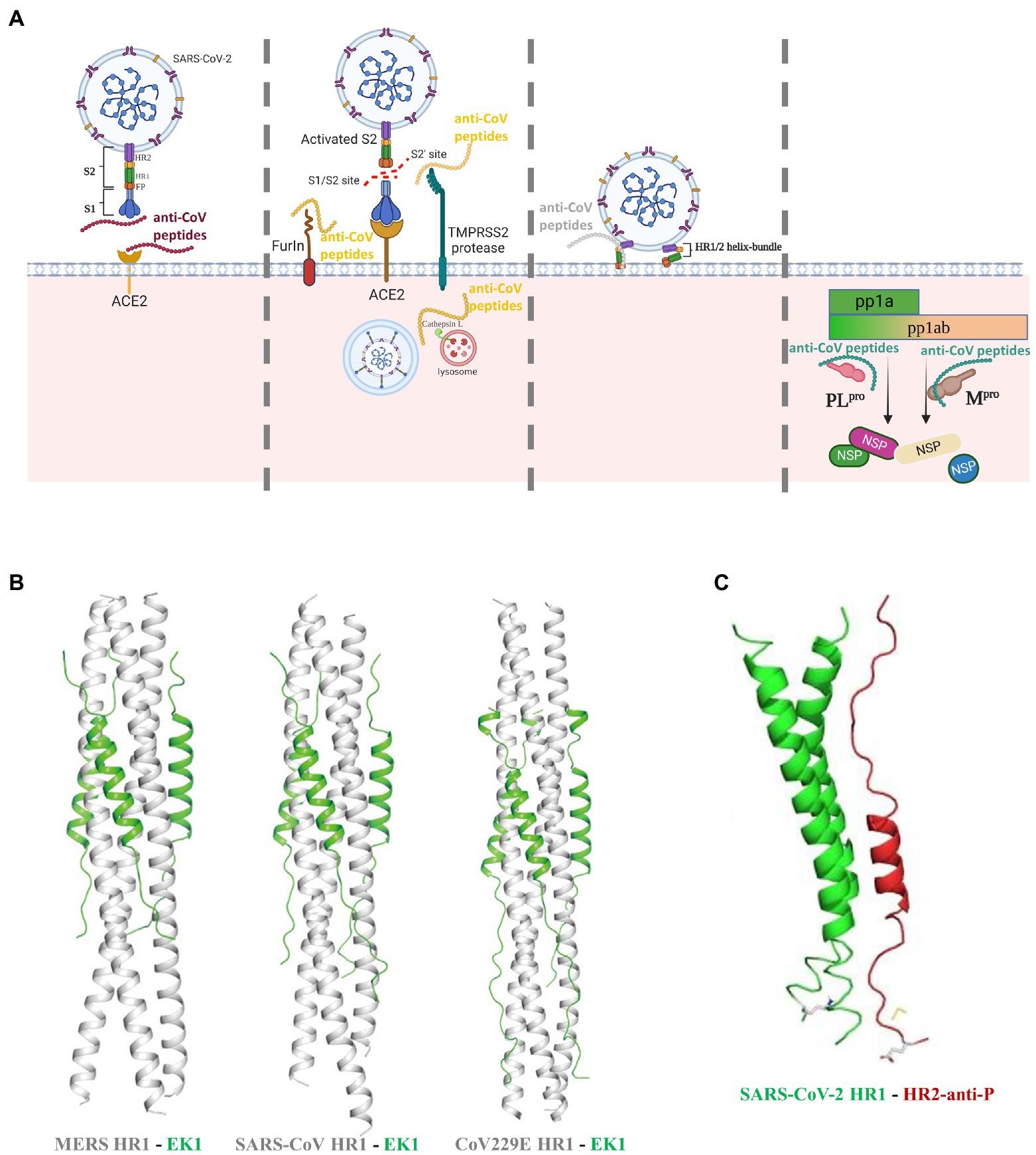
Figure 2. Peptides as potent inhibitors against coronavirus. (A) Anti-CoV peptides can work with diverse modes of actions by targeting the binding of S protein to host receptor, or the proteolytic processing of S proteins by host proteases, or the host-viral membrane fusion, or the viral proteases that are responsible for cleavage of replicase polyproteins into non-structural proteins. (B) The X-ray crystal structures of membrane-fusion inhibitory peptide EK1 shown as green in complex with the S protein HR1 regions of various CoVs shown as white (Xia et al., 2019b). (C) The X-ray crystal structures of membrane-fusion inhibitory peptide HR2-anti-P shown as red binding to the SARS-CoV-2 HR1 region shown as green (Ling et al., 2020).
HCoV-NL63, SARS-CoV, and SARS-CoV-2 employ ACE2 as the entry receptor, thus peptides targeting ACE2 can shield the binding of S proteins (Figure 2A). Based on this, different inhibitors cloaking the ACE2 have been therefore synthesized, but their antiviral efficacy varies considerably, with IC50 values ranging from nanomolar to millimolar concentrations (Huang et al., 2003; Hu et al., 2005; Ho et al., 2006; Struck et al., 2012). Of note, recent studies discovered that some natural peptides can also target ACE2 to inhibit SARS-CoV-2 infection (Wang et al., 2020; Beddingfield et al., 2021). Human defensin 5 (HD5), a natural lectin-like α-defensin produced by the Paneth cells, can stably bind to ACE2 and prevent infection of S protein-expressing pseudo-virions at concentrations as low as 10 μg/ml (Wang et al., 2020). ATN-161, a fibronectin derivative, might also bind to ACE2 to significantly reduce SARS-CoV-2 infection (IC50: 3 μM; Beddingfield et al., 2021). However, clinical use of such ACE2-blocking peptides or drugs warrants serious attention, as ACE2 belongs to the renin-angiotensiongen system where it promotes vasodilation, while interference might have life-threatening side effects.
Peptides targeting proteolytic activation of S protein
As mentioned above, cleavage and activation of S proteins by host proteases are crucial to establish CoV infection, while peptides targeting them can efficiently inhibit viral infection (Figure 2A). Although proteins of multiple viruses can be activated by the proprotein convertase furin (Volchkov et al., 1998; Sugrue et al., 2001; Braun and Sauter, 2019), this feature distinguishes SARS-CoV-2 from SARS-CoV (Matsuyama et al., 2018; Schütz et al., 2020). Two recent studies showed that treatment by the furin inhibitors decanoyl-RVKR-chloromethylketone and MI-1851 can abolish furin cleavage and inhibit SARS-CoV-2 infection of mammalian cells (Bestle et al., 2020; Cheng et al., 2020). Different from furin, TMPRSS2 is involved in proteolytic activation of more CoV species (Shen et al., 2017; Böttcher-Friebertshäuser, 2018), while inhibitors blocking its enzymatic activity could be promising broad-spectrum antivirals. A previous work demonstrated that three different TMPRSS2 inhibitors strongly prevented SARS-CoV-2 and SARS-CoV multiplication in Calu-3 cells in a dose-dependent manner (Bestle et al., 2020). More excitingly, Shapira and coworkers recently synthesized a more potent TMPRSS2 inhibitor N-0385, which can act as a pan-SARS-CoV-2 prophylactic and therapeutic agent and inhibit the cellular entry of multiple SARS-CoV-2 variants of concern at nanomolar concentrations (Shapira et al., 2022). Cellular entry of CoVs might occur through either membrane fusion or endocytosis. In the latter case, the S protein should be activated by proteolysis of cathepsin L, a lysosome-associated protease (Gomes et al., 2020; Zhao M. M. et al., 2021). Blocking the cathepsin L to inhibit SARS-CoV-2 has been tested. P9, a derivate peptide of mouse β-defensin-4, exhibited broad antiviral activities against SARS-CoV, SARS-CoV-2, MERS-CoV and influenza virus, via interfering cathepsin L and preventing endosomal acidification (Zhao et al., 2016, 2020). In the follow-up works, this peptide was further optimized into P9R and 8P9R, which showed higher level of anti-SARS-CoV-2 potency, with IC50 values of 0.9 and 0.3 μg/ml, respectively (Zhao et al., 2020; Zhao H. et al., 2021). In sum, inhibition of host proteases seems a promising antiviral strategy. However, as with the case of ACE2, interference of the physiologically relevant proteases may induce unwanted adverse effect. This calls for sufficient trials in the future to test the potential cytotoxicity and global impact if the protease inhibitors are to be applied to clinical use.
Peptides targeting membrane fusion process
In the past 2 decades, a vast number of studies have synthesized diverse anti-viral peptides that target the membrane fusion step (Schütz et al., 2020; Heydari et al., 2021). These fusion inhibitors have been so far the most extensively studied and the most promising ones to be translated into therapeutic peptides. Mechanistically, they were designed to mimic one of the HR regions in the S2 subunit, interact with the complementary HR, block formation of the HR1-HR2 helix bundle, and thus interfere with virus-host membrane fusion (Figure 2A). Comparatively, peptides derived from HR1 (that target HR2) appear often poorly active (Bosch et al., 2004; Liu et al., 2004; Xia et al., 2020b), likely owing to their propensity to self-aggregation. A large number of peptides have been synthesized to address the previous SARS-CoV and MERS-CoV pandemics, mostly showing strong anti-CoV activity with IC50 values ranging in micromolar concentrations (Barnard et al., 2004; Liu et al., 2004; Yuan et al., 2004; Zheng et al., 2005; Chu et al., 2008; Ujike et al., 2008; O'keefe et al., 2010; Lu et al., 2014; Channappanavar et al., 2015; Zhao et al., 2016; Sun et al., 2017; Wang et al., 2018; Huang et al., 2019; Xia et al., 2019a). Since the HR1 amino acid sequences of SARS-CoV and SARS-CoV-2 share 92.6% similarity and their HR2 is almost identical (Xia et al., 2020b), peptides derived from SARS-CoV HR2 are very likely to inhibit SARS-CoV-2 infection. In agreement with this suggestion, a pan-coronavirus fusion inhibitor EK1, which was previously identified as a potent antiviral agent against SARS-CoV and MERS-CoV (Figure 2B), also reduced SARS-CoV-2 infection of TMPRSS2-negative Vero-E6 cells with an IC50 value of 2.5 μM (Xia et al., 2019b, 2020a). Of interest, the IC50 value was 10-fold lower in TMPRSS2-positive Caco-2 cells (Conzelmann et al., 2020; Xia et al., 2020a), reinforcing again the importance of S protein processing in CoV infection. Computational analysis is a powerful tool in designing the potent SARS-CoV-2 inhibitors. For instance, at the onset of the COVID-19 pandemic, researchers used the in silico approaches to design a potent HR1-targeting peptide to prevent membrane fusion (Figure 2C; Ling et al., 2020). In another work, two potent peptides Fp13-HR1 and Fp14-HR1, that were screened from 17 SARS-CoV HR2-derived fusion inhibitors, were predicted to have a high binding affinity to SARS-CoV-2 HR1, thus they might be effective fusion inhibitors of SARS-CoV-2 (Efaz et al., 2021). To improve antiviral efficacy and stability, rational modifications of the existing inhibitory peptides are also of great importance. This has been nicely exemplified by the modification of the abovementioned EK1 into EK1C4 by linking a cholesterol group to the C-terminus. The optimized peptide displayed more than 19–190-fold potency in preventing the infection of several CoVs, including SARS-CoV-2 (Xia et al., 2020a). Another representative is IPB-02, which was modified from the HR1-targeting peptide IPB-01 by conjugation of a cholesterol group. The refined peptide showed stronger antiviral effect against SARS-CoV-2 with the IC50 value decreasing from 22 to 0.08 μM (Zhu Y. et al., 2020). Optimization of fusion inhibitors is not limited to linkage of current peptides to functional groups. Although previously reported HR1-derived peptide inhibitors exhibit poor inhibitory activities, foldon-mediated trimerization of the C-terminus conferred a HR1-derived peptide with higher inhibitory activity against SARS-CoV-2, SARS-CoV-2 variants of concern (VOCs), SARS-CoV, and MERS-CoV (Bi et al., 2022). Moreover, an inspirational work identified that the extended N-terminus of HR2 also involves in interacting with HR1, and a synthetic peptide including this region achieved single-digit nanomolar inhibition of several SARS-CoV-2 variants (Yang et al., 2022).
Peptides targeting intracellular replication and assembly of coronavirus
Coronavirus infection can also be impeded intracellularly. For example, several chemical compounds Boceprevir, GC-376, calpain inhibitors II and XII had a wide range antiviral activity, via a dual mechanism of action by targeting both viral Mpro and host cell cathepsin L (Fu et al., 2020; Hu et al., 2020; Ma et al., 2020). These findings indicated that the viral components necessary for RNA replication and assembly are also favorable targets for the anti-CoV peptide design (Figure 2A). Indeed, an early study showed that a Mpro-targeting octapeptide impeded replication of the SARS-CoV at the concentration of 1 mg·L−1 (Gan et al., 2006). Another research reported that Cbz-AVLQ-CN, a broad-spectrum peptide, effectively inhibited six different CoV species with IC50 values of 1.3–4.6 μM (Chuck et al., 2014). Interestingly, several active peptides that can bind to both Mpro and monoamine oxidase A of SARS-CoV-2 can be generated from hydrolysis of fish proteins, representing potential inhibitors from food source against CoV (Yao Y. et al., 2020). Blocking other enzymes of CoVs is an alternative strategy. Two synthetic peptides K29 and K12 could markedly inhibit the activity of SARS-CoV nsp16 (methytransferase) in a dose-dependent manner, thus disrupting its role in viral RNA synthesis, but viral inhibition assays are still needed (Ke et al., 2012).
In addition to S protein, the N protein might also be a rational target for the anti-CoV peptide design. The C-terminal domains (CTDs) of the N proteins mediate the self-association of the protein to form high-order oligomers, and deletion of 13 amino acids in the HCoV-229E N protein CTD appeared incapable of forming a high degree of oligomerization (Chang et al., 2005). In line with this notion, a C-terminal tail peptide N377–389 interfered with the oligomerization of the CTD of HCoV-229E N protein and inhibited viral replication at 300 μM (Lo et al., 2013). This finding provides insights that blocking the formation of the N protein-RNA higher-order oligomers and in turn the virion assembly can contribute to viral inhibition.
Perspectives and conclusion
To conclude, peptides that can target various steps in CoV lifecycle have shown great potential in combating CoV infection. In some cases, the peptide-based inhibitors seem to have lower possibility to cause drug resistance (Zhao et al., 2020), and they rarely induce detectable cytotoxicity (Gan et al., 2006). Some peptides exhibit significantly strong and broad-spectrum effect against multiple CoV species. More importantly, combination of peptides with different antiviral mechanisms could generate synergistic impact (Bestle et al., 2020; Hoffmann et al., 2020a). However, to translate the peptides into clinical therapeutics, they should be safe and stable in vivo, while many works need more effort on this aspect. Therefore, a future perspective is to refine current peptides to be more effective and long-lasting, as with the case of EK14C, which was subject to two rounds of optimization from OC43-HR2P. To this end, a database containing comprehensive and precise information of 214 unique anti-CoV peptides would contribute to more rational design or modification (Zhang et al., 2022).
Author contributions
MT, XZ, YH, and LL devised the framework, wrote and revised the manuscript. WC, JQ, SG, SL, LL, and MT contributed to literature search and gave insightful suggestions in revising this work. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (81900071), the Fellowship of China Postdoctoral Science Foundation (2022M713287), the Medical Research Foundation of Guangdong Province (A2022046), and the Shenzhen Science and Technology Innovation Commission for Research and Development Projects (JSGG20200807171603039, JSGG20191118161401741, CYJ20210324112607020 and zJCYJ20220530141616037).
Acknowledgments
BioRender was used to create schematic representations.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Bai, C., Zhong, Q., and Gao, G. F. (2022). Overview of SARS-CoV-2 genome-encoded proteins. Sci. China Life Sci. 65, 280–294. doi: 10.1007/s11427-021-1964-4
Barnard, D. L., Hubbard, V. D., Burton, J., Smee, D. F., Morrey, J. D., Otto, M. J., et al. (2004). Inhibition of severe acute respiratory syndrome-associated coronavirus (SARSCoV) by calpain inhibitors and β-D-N4-Hydroxycytidine. Antivir. Chem. Chemother. 15, 15–22. doi: 10.1177/095632020401500102
Beddingfield, B. J., Iwanaga, N., Chapagain, P. P., Zheng, W., Roy, C. J., Hu, T. Y., et al. (2021). The integrin binding peptide, ATN-161, as a novel therapy for SARS-CoV-2 infection. JACC. Basic Transl. Sci. 6, 1–8. doi: 10.1016/j.jacbts.2020.10.003
Bestle, D., Heindl, M. R., Limburg, H., Van Lam, T., Pilgram, O., Moulton, H., et al. (2020). TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci. Alliance 3:e202000786. doi: 10.26508/lsa.202000786
Bi, W., Chen, G., and Dang, B. (2022). Novel engineered SARS-CoV-2 HR1 Trimer exhibits improved potency and broad-Spectrum activity against SARS-CoV-2 and its variants. J. Virol. 96, e00681–e00622. doi: 10.1128/jvi.00681-22
Bosch, B. J., Martina, B. E. E., Van Der Zee, R., Lepault, J., Haijema, B. J., Versluis, C., et al. (2004). Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc. Natl. Acad. Sci. U. S. A. 101, 8455–8460. doi: 10.1073/pnas.0400576101
Boson, B., Legros, V., Zhou, B., Siret, E., Mathieu, C., Cosset, F.-L., et al. (2021). The SARS-CoV-2 envelope and membrane proteins modulate maturation and retention of the spike protein, allowing assembly of virus-like particles. J. Biol. Chem. 296:100111. doi: 10.1073/pnas.0400576101
Böttcher-Friebertshäuser, E. (2018). “Membrane-anchored serine proteases: host cell factors in proteolytic activation of viral glycoproteins” in Activation of Viruses by Host Proteases, ed. Böttcher-Friebertshäuser, E., Garten, W. and Klenk, H. (Cham, FL: Springer). 153–203.
Braun, E., and Sauter, D. (2019). Furin-mediated protein processing in infectious diseases and cancer. Clin. Transl. Immunol. 8:e1073. doi: 10.1002/cti2.1073
Cai, Y., Zhang, J., Xiao, T., Peng, H., Sterling, S. M., Walsh, R. M., et al. (2020). Distinct conformational states of SARS-CoV-2 spike protein. Science 369, 1586–1592. doi: 10.1126/science.abd4251
Cao, L., Goreshnik, I., Coventry, B., Case, J. B., Miller, L., Kozodoy, L., et al. (2020). De novo design of picomolar SARS-CoV-2 miniprotein inhibitors. Science 370, 426–431. doi: 10.1126/science.abd9909
Cao, Z., Xia, H., Rajsbaum, R., Xia, X., Wang, H., and Shi, P.-Y. (2021). Ubiquitination of SARS-CoV-2 ORF7a promotes antagonism of interferon response. Cell. Mol. Immunol. 18, 746–748. doi: 10.1038/s41423-020-00603-6
Cao, Y., Wang, J., Jian, F., Xiao, T., Song, W., Yisimayi, A., et al. (2022). Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 602, 657–663. doi: 10.1038/s41586-021-04385-3
Cele, S., Jackson, L., Khoury, D. S., Khan, K., Moyo-Gwete, T., Tegally, H., et al. (2022). Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature 602, 654–656. doi: 10.1038/s41586-021-04387-1
Chai, J., Cai, Y., Pang, C., Wang, L., Mcsweeney, S., Shanklin, J., et al. (2021). Structural basis for SARS-CoV-2 envelope protein recognition of human cell junction protein PALS1. Nat. Commun. 12:3433. doi: 10.1038/s41467-021-23533-x
Chang, C.-K., Sue, S.-C., Yu, T.-H., Hsieh, C.-M., Tsai, C.-K., Chiang, Y.-C., et al. (2005). The dimer interface of the SARS coronavirus nucleocapsid protein adapts a porcine respiratory and reproductive syndrome virus-like structure. FEBS Lett. 579, 5663–5668. doi: 10.1016/j.febslet.2005.09.038
Channappanavar, R., Lu, L., Xia, S., Du, L., Meyerholz, D. K., Perlman, S., et al. (2015). Protective effect of intranasal regimens containing peptidic middle east respiratory syndrome coronavirus fusion inhibitor against MERS-CoV infection. J. Infect. Dis. 212, 1894–1903. doi: 10.1093/infdis/jiv325
Cheng, Y. W., Chao, T. L., Li, C. L., Chiu, M. F., Kao, H. C., Wang, S. H., et al. (2020). Furin inhibitors block SARS-CoV-2 spike protein cleavage to suppress virus production and cytopathic effects. Cell Rep. 33:108254. doi: 10.1016/j.celrep.2020.108254
Chu, L. H., Chan, S. H., Tsai, S. N., Wang, Y., Cheng, C. H., Wong, K. B., et al. (2008). Fusion core structure of the severe acute respiratory syndrome coronavirus (SARS-CoV): in search of potent SARS-CoV entry inhibitors. J. Cell. Biochem. 104, 2335–2347. doi: 10.1002/jcb.21790
Chuck, C. P., Ke, Z. H., Chen, C., Wan, D. C. C., Chow, H. F., and Wong, K. B. (2014). Profiling of substrate-specificity and rational design of broad-spectrum peptidomimetic inhibitors for main proteases of coronaviruses. Hong Kong Med. J. 20, 22–25.
Conzelmann, C., Gilg, A., Groß, R., Schütz, D., Preising, N., Ständker, L., et al. (2020). An enzyme-based immunodetection assay to quantify SARS-CoV-2 infection. Antivir. Res. 181:104882. doi: 10.1016/j.antiviral.2020.104882
Edara, V.-V., Manning, K. E., Ellis, M., Lai, L., Moore, K. M., Foster, S. L., et al. (2022). mRNA-1273 and BNT162b2 mRNA vaccines have reduced neutralizing activity against the SARS-CoV-2 omicron variant. Cell Rep. Med. 3:100529. doi: 10.1016/j.xcrm.2022.100529
Efaz, F. M., Islam, S., Talukder, S. A., Akter, S., Tashrif, M. Z., Ali, M. A., et al. (2021). Repurposing fusion inhibitor peptide against SARS-CoV-2. J. Comput. Chem. 42, 2283–2293. doi: 10.1002/jcc.26758
Fraser, B. J., Beldar, S., Seitova, A., Hutchinson, A., Mannar, D., Li, Y., et al. (2022). Structure and activity of human TMPRSS2 protease implicated in SARS-CoV-2 activation. Nat. Chem. Biol. 18, 963–971. doi: 10.1038/s41589-022-01059-7
Fu, L., Ye, F., Feng, Y., Yu, F., Wang, Q., Wu, Y., et al. (2020). Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease. Nat. Commun. 11:4417. doi: 10.1038/s41467-020-18233-x
Gan, Y.-R., Huang, H., Huang, Y.-D., Rao, C.-M., Zhao, Y., Liu, J.-S., et al. (2006). Synthesis and activity of an octapeptide inhibitor designed for SARS coronavirus main proteinase. Peptides 27, 622–625. doi: 10.1016/j.peptides.2005.09.006
Gao, Y., Fang, H., Fang, L., Liu, D., Liu, J., Su, M., et al. (2018). The modification and design of antimicrobial peptide. Curr. Pharm. Des. 24, 904–910. doi: 10.2174/1381612824666180213130318
Ghosh, S. K., and Weinberg, A. (2021). Ramping up antimicrobial peptides against severe acute respiratory syndrome coronavirus-2. Front. Mol. Biosci. 8:620826. doi: 10.3389/fmolb.2021.620806
Gomes, C. P., Fernandes, D. E., Casimiro, F., Da Mata, G. F., Passos, M. T., Varela, P., et al. (2020). Cathepsin L in COVID-19: from pharmacological evidences to genetics. Front. Cell. Infect. Microbiol. 10:589505. doi: 10.3389/fcimb.2020.589505
Guo, Y., Wang, W., Sun, Y., Ma, C., Wang, X., Wang, X., et al. (2016). Crystal structure of the core region of hantavirus nucleocapsid protein reveals the mechanism for ribonucleoprotein complex formation. J. Virol. 90, 1048–1061. doi: 10.1128/JVI.02523-15
Han, D. P., Penn-Nicholson, A., and Cho, M. W. (2006). Identification of critical determinants on ACE2 for SARS-CoV entry and development of a potent entry inhibitor. Virology 350, 15–25. doi: 10.1016/j.virol.2006.01.029
Heydari, H., Golmohammadi, R., Mirnejad, R., Tebyanian, H., Fasihi-Ramandi, M., and Moosazadeh Moghaddam, M. (2021). Antiviral peptides against Coronaviridae family: a review. Peptides 139:170526. doi: 10.1016/j.peptides.2021.170526
Ho, T.-Y., Wu, S.-L., Chen, J.-C., Wei, Y.-C., Cheng, S.-E., Chang, Y.-H., et al. (2006). Design and biological activities of novel inhibitory peptides for SARS-CoV spike protein and angiotensin-converting enzyme 2 interaction. Antivir. Res. 69, 70–76. doi: 10.1016/j.antiviral.2005.10.005
Hodge, C. D., Rosenberg, D. J., Grob, P., Wilamowski, M., Joachimiak, A., Hura, G. L., et al. (2021). Rigid monoclonal antibodies improve detection of SARS-CoV-2 nucleocapsid protein. MAbs 13:1905978. doi: 10.1080/19420862.2021.1905978
Hoffmann, M., Kleine-Weber, H., and Pöhlmann, S. (2020a). A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell 78, 779–784.e5. doi: 10.1016/j.molcel.2020.04.022
Hoffmann, M., Kleine-Weber, H., Schroeder, S., Krüger, N., Herrler, T., Erichsen, S., et al. (2020b). SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cells 181, 271–280.e8. doi: 10.1016/j.cell.2020.02.052
Hoffmann, M., Krüger, N., Schulz, S., Cossmann, A., Rocha, C., Kempf, A., et al. (2022). The Omicron variant is highly resistant against antibody-mediated neutralization: Implications for control of the COVID-19 pandemic. Cells 185, 447–456.e411. doi: 10.1016/j.cell.2021.12.032
Hu, H., Li, L., Kao, R. Y., Kou, B., Wang, Z., Zhang, L., et al. (2005). Screening and identification of linear B-cell epitopes and entry-blocking peptide of severe acute respiratory syndrome (SARS)-associated coronavirus using synthetic overlapping peptide library. J. Comb. Chem. 7, 648–656. doi: 10.1021/cc0500607
Hu, Y., Ma, C., Szeto, T., Hurst, B., Tarbet, B., and Wang, J. (2020). Boceprevir, calpain inhibitors II and XII, and GC-376 have broad-spectrum antiviral activity against coronaviruses. ACS Infect. Dis. 7, 586–597. doi: 10.1021/acsinfecdis.0c00761
Huang, X., Li, M., Xu, Y., Zhang, J., Meng, X., An, X., et al. (2019). Novel gold nanorod-based HR1 peptide inhibitor for middle east respiratory syndrome coronavirus. ACS Appl. Mater. Interfaces 11, 19799–19807. doi: 10.1021/acsami.9b04240
Huang, C., Lokugamage, K. G., Rozovics, J. M., Narayanan, K., Semler, B. L., and Makino, S. (2011). SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog. 7:e1002433. doi: 10.1371/journal.ppat.1002433
Huang, L., Sexton, D. J., Skogerson, K., Devlin, M., Smith, R., Sanyal, I., et al. (2003). Novel peptide inhibitors of angiotensin-converting enzyme 2*. J. Biol. Chem. 278, 15532–15540. doi: 10.1074/jbc.M212934200
Huang, Y., Yang, C., Xu, X.-F., Xu, W., and Liu, S.-W. (2020). Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 41, 1141–1149. doi: 10.1038/s41401-020-0485-4
Iwata-Yoshikawa, N., Kakizaki, M., Shiwa-Sudo, N., Okura, T., Tahara, M., Fukushi, S., et al. (2022). Essential role of TMPRSS2 in SARS-CoV-2 infection in murine airways. Nat. Commun. 13:6100. doi: 10.1038/s41467-022-33911-8
Jaimes, J. A., André, N. M., Chappie, J. S., Millet, J. K., and Whittaker, G. R. (2020). Phylogenetic analysis and structural modeling of SARS-CoV-2 spike protein reveals an evolutionary distinct and proteolytically sensitive activation loop. J. Mol. Biol. 432, 3309–3325. doi: 10.1016/j.jmb.2020.04.009
Jaiswal, G., and Kumar, V. (2020). In-silico design of a potential inhibitor of SARS-CoV-2 S protein. PLoS One 15:e0240004. doi: 10.1371/journal.pone.0240004
Javorsky, A., Humbert, P. O., and Kvansakul, M. (2021). Structural basis of coronavirus E protein interactions with human PALS1 PDZ domain. Commun. Biol. 4:724. doi: 10.1038/s42003-021-02250-7
Kasuga, Y., Zhu, B., Jang, K.-J., and Yoo, J.-S. (2021). Innate immune sensing of coronavirus and viral evasion strategies. Exp. Mol. Med. 53, 723–736. doi: 10.1038/s12276-021-00602-1
Ke, M., Chen, Y., Wu, A., Sun, Y., Su, C., Wu, H., et al. (2012). Short peptides derived from the interaction domain of SARS coronavirus nonstructural protein nsp10 can suppress the 2′-O-methyltransferase activity of nsp10/nsp16 complex. Virus Res. 167, 322–328. doi: 10.1016/j.virusres.2012.05.017
Kirchdoerfer, R. N., and Ward, A. B. (2019). Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 10:2342. doi: 10.1038/s41467-019-10280-3
Konno, Y., Kimura, I., Uriu, K., Fukushi, M., Irie, T., Koyanagi, Y., et al. (2020). SARS-CoV-2 ORF3b is a potent interferon antagonist whose activity is increased by a naturally occurring elongation variant. Cell Rep. 32:108185. doi: 10.1016/j.celrep.2020.108185
Lei, X., Dong, X., Ma, R., Wang, W., Xiao, X., Tian, Z., et al. (2020). Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 11:3810. doi: 10.1038/s41467-020-17665-9
Li, J., Guo, M., Tian, X., Wang, X., Yang, X., Wu, P., et al. (2021). Virus-host Interactome and proteomic survey reveal potential virulence factors influencing SARS-CoV-2 pathogenesis. Medicine 2, 99–112.e7. doi: 10.1016/j.medj.2020.07.002
Li, J., Jia, H., Tian, M., Wu, N., Yang, X., Qi, J., et al. (2022). SARS-CoV-2 and emerging variants: unmasking structure, function, infection, and immune escape mechanisms. Front. Cell. Infect. Microbiol. 12:869832. doi: 10.3389/fcimb.2022.869832
Ling, R., Dai, Y., Huang, B., Huang, W., Yu, J., Lu, X., et al. (2020). In silico design of antiviral peptides targeting the spike protein of SARS-CoV-2. Peptides 130:170328. doi: 10.1016/j.peptides.2020.170328
Liu, C., Mendonça, L., Yang, Y., Gao, Y., Shen, C., Liu, J., et al. (2020). The architecture of inactivated SARS-CoV-2 with postfusion spikes revealed by Cryo-EM and Cryo-ET. Structure 28, 1218–1224.e4. doi: 10.1016/j.str.2020.10.001
Liu, S., Xiao, G., Chen, Y., He, Y., Niu, J., Escalante, C. R., et al. (2004). Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet 363, 938–947. doi: 10.1016/S0140-6736(04)15788-7
Lo, Y.-S., Lin, S.-Y., Wang, S.-M., Wang, C.-T., Chiu, Y.-L., Huang, T.-H., et al. (2013). Oligomerization of the carboxyl terminal domain of the human coronavirus 229E nucleocapsid protein. FEBS Lett. 587, 120–127. doi: 10.1016/j.febslet.2012.11.016
Lu, L., Liu, Q., Zhu, Y., Chan, K.-H., Qin, L., Li, Y., et al. (2014). Structure-based discovery of middle east respiratory syndrome coronavirus fusion inhibitor. Nat. Commun. 5:3067. doi: 10.1038/ncomms4067
Lu, X., Pan, J., Tao, J., and Guo, D. (2011). SARS-CoV nucleocapsid protein antagonizes IFN-β response by targeting initial step of IFN-β induction pathway, and its C-terminal region is critical for the antagonism. Virus Genes 42, 37–45. doi: 10.1007/s11262-010-0544-x
Ma, C., Sacco, M. D., Hurst, B., Townsend, J. A., Hu, Y., Szeto, T., et al. (2020). Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 30:678. doi: 10.1038/s41422-020-0356-z
Mahtarin, R., Islam, S., Islam, M. J., Ullah, M. O., Ali, M. A., and Halim, M. A. (2022). Structure and dynamics of membrane protein in SARS-CoV-2. J. Biomol. Struct. Dyn. 40, 4725–4738. doi: 10.1080/07391102.2020.1861983
Malone, B., Urakova, N., Snijder, E. J., and Campbell, E. A. (2022). Structures and functions of coronavirus replication–transcription complexes and their relevance for SARS-CoV-2 drug design. Nat. Rev. Mol. Cell Biol. 23, 21–39. doi: 10.1080/07391102.2020.1861983
Matsuyama, S., Nao, N., Shirato, K., Kawase, M., Saito, S., Takayama, I., et al. (2020). Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. U. S. A. 117, 7001–7003. doi: 10.1073/pnas.2002589117
Matsuyama, S., Shirato, K., Kawase, M., Terada, Y., Kawachi, K., Fukushi, S., et al. (2018). Middle east respiratory syndrome coronavirus spike protein is not activated directly by cellular furin during viral entry into target cells. J. Virol. 92, e00683–e00718. doi: 10.1128/JVI.00683-18
Mcbride, R., Van Zyl, M., and Fielding, B. C. (2014). The coronavirus nucleocapsid is a multifunctional protein. Viruses 6, 2991–3018. doi: 10.3390/v6082991
Miorin, L., Kehrer, T., Sanchez-Aparicio, M. T., Zhang, K., Cohen, P., Patel, R. S., et al. (2020). SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc. Natl. Acad. Sci. U. S. A. 117, 28344–28354. doi: 10.1073/pnas.2016650117
Nieto-Torres, J. L., Verdiá-Báguena, C., Jimenez-Guardeño, J. M., Regla-Nava, J. A., Castaño-Rodriguez, C., Fernandez-Delgado, R., et al. (2015). Severe acute respiratory syndrome coronavirus E protein transports calcium ions and activates the NLRP3 inflammasome. Virology 485, 330–339. doi: 10.1016/j.virol.2015.08.010
O'keefe, B. R., Giomarelli, B., Barnard, D. L., Shenoy, S. R., Chan, P. K., Mcmahon, J. B., et al. (2010). Broad-spectrum in vitro activity and in vivo efficacy of the antiviral protein griffithsin against emerging viruses of the family Coronaviridae. J. Virol. 84, 2511–2521. doi: 10.1128/JVI.02322-09
Peacock, T. P., Goldhill, D. H., Zhou, J., Baillon, L., Frise, R., Swann, O. C., et al. (2021). The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat. Microbiol. 6, 899–909. doi: 10.1038/s41564-021-00908-w
Rawson, T. M., Ming, D., Ahmad, R., Moore, L. S. P., and Holmes, A. H. (2020). Antimicrobial use, drug-resistant infections and COVID-19. Nat. Rev. Microbiol. 18, 409–410. doi: 10.1038/s41579-020-0395-y
Redondo, N., Zaldívar-López, S., Garrido, J. J., and Montoya, M. (2021). SARS-CoV-2 accessory proteins in viral pathogenesis: knowns and unknowns. Front. Immunol. 12:708264. doi: 10.3389/fimmu.2021.708264
Rockett, R., Basile, K., Maddocks, S., Fong, W., Agius, J. E., Johnson-Mackinnon, J., et al. (2022). Resistance mutations in SARS-CoV-2 Delta variant after Sotrovimab use. N. Engl. J. Med. 386, 1477–1479. doi: 10.1056/NEJMc2120219
Schoeman, D., and Fielding, B. C. (2019). Coronavirus envelope protein: current knowledge. Virol. J. 16:69. doi: 10.1186/s12985-019-1182-0
Schütz, D., Ruiz-Blanco, Y. B., Münch, J., Kirchhoff, F., Sanchez-Garcia, E., and Müller, J. A. (2020). Peptide and peptide-based inhibitors of SARS-CoV-2 entry. Adv. Drug Deliv. Rev. 167, 47–65. doi: 10.1016/j.addr.2020.11.007
Shah, J. N., Guo, G.-Q., Krishnan, A., Ramesh, M., Katari, N. K., Shahbaaz, M., et al. (2022). Peptides-based therapeutics: emerging potential therapeutic agents for COVID-19. Therapie 77, 319–328. doi: 10.1016/j.therap.2021.09.007
Shang, J., Han, N., Chen, Z., Peng, Y., Li, L., Zhou, H., et al. (2020). Compositional diversity and evolutionary pattern of coronavirus accessory proteins. Brief. Bioinform. 22, 1267–1278. doi: 10.1093/bib/bbaa262
Shapira, T., Monreal, I. A., Dion, S. P., Buchholz, D. W., Imbiakha, B., Olmstead, A. D., et al. (2022). A TMPRSS2 inhibitor acts as a pan-SARS-CoV-2 prophylactic and therapeutic. Nature 605, 340–348. doi: 10.1038/s41586-022-04661-w
Shen, L. W., Mao, H. J., Wu, Y. L., Tanaka, Y., and Zhang, W. (2017). TMPRSS2: a potential target for treatment of influenza virus and coronavirus infections. Biochimie 142, 1–10. doi: 10.1016/j.biochi.2017.07.016
Şimşek-Yavuz, S., and Komsuoğlu elikyurt, F. I. (2021). An update of anti-viral treatment of COVID-19. Turk. J. Med. Sci. 51, 3372–3390. doi: 10.3906/sag-2106-250
Snijder, E. J., and Limpens, R. (2020). A unifying structural and functional model of the coronavirus replication organelle: tracking down RNA synthesis. PLoS Biol. 18:e3000715. doi: 10.1371/journal.pbio.3000715
Struck, A.-W., Axmann, M., Pfefferle, S., Drosten, C., and Meyer, B. (2012). A hexapeptide of the receptor-binding domain of SARS corona virus spike protein blocks viral entry into host cells via the human receptor ACE2. Antivir. Res. 94, 288–296. doi: 10.1016/j.antiviral.2011.12.012
Su, C.-M., Wang, L., and Yoo, D. (2021). Activation of NF-κB and induction of proinflammatory cytokine expressions mediated by ORF7a protein of SARS-CoV-2. Sci. Rep. 11:13464. doi: 10.1038/s41598-021-92941-2
Sugrue, R. J., Brown, C., Brown, G., Aitken, J., and Mcl Rixon, H. W. (2001). Furin cleavage of the respiratory syncytial virus fusion protein is not a requirement for its transport to the surface of virus-infected cells. J. Gen. Virol. 82, 1375–1386. doi: 10.1099/0022-1317-82-6-1375
Sun, Y., Zhang, H., Shi, J., Zhang, Z., and Gong, R. (2017). Identification of a novel inhibitor against middle east respiratory syndrome coronavirus. Viruses 9:255. doi: 10.3390/v9090255
Tannock, G. A., Kim, H., and Xue, L. (2020). Why are vaccines against many human viral diseases still unavailable; an historic perspective? J. Med. Virol. 92, 129–138. doi: 10.1002/jmv.25593
Tavassoly, O., Safavi, F., and Tavassoly, I. (2020). Heparin-binding peptides as novel therapies to stop SARS-CoV-2 cellular entry and infection. Mol. Pharmacol. 98, 612–619. doi: 10.1124/molpharm.120.000098
Thoms, M., Buschauer, R., Ameismeier, M., Koepke, L., Denk, T., Hirschenberger, M., et al. (2020). Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science 369, 1249–1255. doi: 10.1126/science.abc8665
Ujike, M., Nishikawa, H., Otaka, A., Yamamoto, N., Yamamoto, N., Matsuoka, M., et al. (2008). Heptad repeat-derived peptides block protease-mediated direct entry from the cell surface of severe acute respiratory syndrome coronavirus but not entry via the endosomal pathway. J. Virol. 82, 588–592. doi: 10.1128/JVI.01697-07
Vagner, J., Qu, H., and Hruby, V. J. (2008). Peptidomimetics, a synthetic tool of drug discovery. Curr. Opin. Chem. Biol. 12, 292–296. doi: 10.1016/j.cbpa.2008.03.009
Volchkov, V. E., Feldmann, H., Volchkova, V. A., and Klenk, H.-D. (1998). Processing of the Ebola virus glycoprotein by the proprotein convertase furin. Proc. Natl. Acad. Sci. U. S. A. 95, 5762–5767. doi: 10.1073/pnas.95.10.5762
Walls, A. C., Park, Y.-J., Tortorici, M. A., Wall, A., Mcguire, A. T., and Veesler, D. (2020). Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cells 181, 281–292.e6. doi: 10.1016/j.cell.2020.02.058
Wang, C., Wang, S., Li, D., Chen, P., Han, S., Zhao, G., et al. (2021). Human cathelicidin inhibits SARS-CoV-2 infection: killing two birds with one stone. ACS Infect. Dis. 7, 1545–1554. doi: 10.1021/acsinfecdis.1c00096
Wang, C., Wang, S., Li, D., Wei, D. Q., Zhao, J., and Wang, J. (2020). Human intestinal defensin 5 inhibits SARS-CoV-2 invasion by cloaking ACE2. Gastroenterology 159, 1145–1147.e4. doi: 10.1053/j.gastro.2020.05.015
Wang, C., Xia, S., Zhang, P., Zhang, T., Wang, W., Tian, Y., et al. (2018). Discovery of hydrocarbon-stapled short α-helical peptides as promising middle east respiratory syndrome coronavirus (MERS-CoV) fusion inhibitors. J. Med. Chem. 61, 2018–2026. doi: 10.1021/acs.jmedchem.7b01732
Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C.-L., Abiona, O., et al. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263. doi: 10.1126/science.abb2507
Wu, F., Zhao, S., Yu, B., Chen, Y.-M., Wang, W., Song, Z.-G., et al. (2020). A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269. doi: 10.1038/s41586-020-2008-3
Xia, S., Lan, Q., Pu, J., Wang, C., Liu, Z., Xu, W., et al. (2019a). Potent MERS-CoV fusion inhibitory peptides identified from HR2 domain in spike protein of bat coronavirus HKU4. Viruses 11:56. doi: 10.3390/v11010056
Xia, S., Liu, M., Wang, C., Xu, W., Lan, Q., Feng, S., et al. (2020a). Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 30, 343–355. doi: 10.1038/s41422-020-0305-x
Xia, S., Yan, L., Xu, W., Agrawal, A. S., Algaissi, A., Tseng, C.-T. K., et al. (2019b). A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci. Adv. 5:eaav4580. doi: 10.1126/sciadv.aav4580
Xia, S., Zhu, Y., Liu, M., Lan, Q., Xu, W., Wu, Y., et al. (2020b). Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell. Mol. Immunol. 17, 765–767. doi: 10.1038/s41423-020-0374-2
Yan, L., Ge, J., Zheng, L., Zhang, Y., Gao, Y., Wang, T., et al. (2021). Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis. Cells 184, 184–193.e10. doi: 10.1016/j.cell.2020.11.016
Yan, W., Zheng, Y., Zeng, X., He, B., and Cheng, W. (2022). Structural biology of SARS-CoV-2: open the door for novel therapies. Signal Transduct. Target. Ther. 7:26. doi: 10.1038/s41392-022-00884-5
Yang, K., Wang, C., Kreutzberger, A. J. B., Ojha, R., Kuivanen, S., Couoh-Cardel, S., et al. (2022). Nanomolar inhibition of SARS-CoV-2 infection by an unmodified peptide targeting the prehairpin intermediate of the spike protein. Proc. Natl. Acad. Sci. U. S. A. 119:e2210990119. doi: 10.1073/pnas.2210990119
Yao, Y., Luo, Z., and Zhang, X. (2020). In silico evaluation of marine fish proteins as nutritional supplements for COVID-19 patients. Food Funct. 11, 5565–5572. doi: 10.1039/d0fo00530d
Yao, H., Song, Y., Chen, Y., Wu, N., Xu, J., Sun, C., et al. (2020). Molecular architecture of the SARS-CoV-2 virus. Cells 183, 730–738.e13. doi: 10.1016/j.cell.2020.09.018
Yuan, K., Yi, L., Chen, J., Qu, X., Qing, T., Rao, X., et al. (2004). Suppression of SARS-CoV entry by peptides corresponding to heptad regions on spike glycoprotein. Biochem. Biophys. Res. Commun. 319, 746–752. doi: 10.1016/j.bbrc.2004.05.046
Zhang, Q., Chen, X., Li, B., Lu, C., Yang, S., Long, J., et al. (2022). A database of anti-coronavirus peptides. Sci. Data 9:294. doi: 10.1038/s41597-022-01394-3
Zhao, H., To, K. K. W., Lam, H., Zhou, X., Chan, J. F.-W., Peng, Z., et al. (2021). Cross-linking peptide and repurposed drugs inhibit both entry pathways of SARS-CoV-2. Nat. Commun. 12:1517. doi: 10.1038/s41467-021-21825-w
Zhao, H., To, K. K. W., Sze, K.-H., Yung, T. T.-M., Bian, M., Lam, H., et al. (2020). A broad-spectrum virus-and host-targeting peptide against respiratory viruses including influenza virus and SARS-CoV-2. Nat. Commun. 11:4252. doi: 10.1038/s41467-020-17986-9
Zhao, M. M., Yang, W.-L., Yang, F.-Y., Zhang, L., Huang, W.-J., Hou, W., et al. (2021). Cathepsin L plays a key role in SARS-CoV-2 infection in humans and humanized mice and is a promising target for new drug development. Signal Transduct. Target. Ther. 6:134. doi: 10.1038/s41392-021-00558-8
Zhao, H., Zhou, J., Zhang, K., Chu, H., Liu, D., Poon, V. K.-M., et al. (2016). A novel peptide with potent and broad-spectrum antiviral activities against multiple respiratory viruses. Sci. Rep. 6:22008. doi: 10.1038/srep22008
Zheng, B.-J., Guan, Y., He, M.-L., Sun, H., Du, L., Zheng, Y., et al. (2005). Synthetic peptides outside the spike protein heptad repeat regions as potent inhibitors of sars-associated coronavirus. Antivir. Ther. 10, 393–403. doi: 10.1177/135965350501000301
Zhou, P., Yang, X.-L., Wang, X.-G., Hu, B., Zhang, L., Zhang, W., et al. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273. doi: 10.1038/s41586-020-2012-7
Zhu, Y., Yu, D., Yan, H., Chong, H., and He, Y. (2020). Design of potent membrane fusion inhibitors against SARS-CoV-2, an emerging coronavirus with high fusogenic activity. J. Virol. 94, e00635–e00640. doi: 10.1128/jvi.00635-20
Keywords: SARS-CoV-2, viral infection, spike protein, anti-viral peptides, host protease, host receptors
Citation: Tang M, Zhang X, Huang Y, Cheng W, Qu J, Gui S, Li L and Li S (2023) Peptide-based inhibitors hold great promise as the broad-spectrum agents against coronavirus. Front. Microbiol. 13:1093646. doi: 10.3389/fmicb.2022.1093646
Edited by:
Jian Shang, Zhengzhou University, ChinaReviewed by:
Jun Wang, Rutgers, The State University of New Jersey, United StatesCopyright © 2023 Tang, Zhang, Huang, Cheng, Qu, Gui, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuiqing Gui, ✉ Z3Vpc2h1aXFpbmdAMTYzLmNvbQ==; Liang Li, ✉ bGlsQHN1c3RlY2guZWR1LmNu; Shuo Li, ✉ U2h1b2xpQGVtYWlsLnN6dS5lZHUuY24=
†These authors share first authorship
 Mingxing Tang1,2,3†
Mingxing Tang1,2,3† Liang Li
Liang Li Shuo Li
Shuo Li