
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 21 December 2022
Sec. Evolutionary and Genomic Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1087212
This article is part of the Research Topic Insights on Fungal Diversity of Ascomycetes and Basidiomycetes: Taxonomy and Interaction with Their Host View all 24 articles
Sanguinoderma is distributed in tropical and subtropical areas as a member of Amauroderma s. lat., and the economic values of Sanguinoderma led to high attention in the taxonomic studies. Previously, 16 species have been developed into Sanguinoderma. In this study, the taxonomic system of Sanguinoderma was reconducted based on morphological and multi-gene phylogenetic analyses, especially making a distinction for Sanguinoderma rugosum complex. Morphological analysis was based on the notes of macro- and micro morphological observations. Multi-gene phylogenetic analyses were used maximum likelihood (ML) and Bayesian inference (BI) analyses inferred from combined dataset of ITS, nLSU, rpb2, tef1, mtSSU, and nSSU. Combined with morphological characters and phylogenetic evidence, the results demonstrated that S. rugosum complex consists of five taxa, in which Sanguinoderma leucomarginatum was described as a new species, and it is characterized by the orbicular pilei with white to buff margin when fresh and clavate apical cells of pileipellis with septa. In addition, Amauroderma preussii was transferred to Sanguinoderma as a new combination due to its blood-red color-changed pore surface; it is characterized by the funnel-shaped, greyish brown, and glabrous pilei with strongly incurved margin. Detailed descriptions and photographs of the two species were provided. With the extension of this study, 18 species were accepted in Sanguinoderma, and 12 species among them were distributed in China. A key to accepted species of Sanguinoderma was also provided.
Ganodermataceae is an important family of macrofungi according to its high economic and ecological values. Some species in this family, such as Ganoderma lingzhi, Ganoderma sinense, Ganoderma tsugae, Amauroderma rude, and Amauroderma rugosum, have been domesticated successfully in China and commonly used as traditional medicine for anti-cancer treatment, for lowering blood pressure, and for improving immunity (Wang et al., 1993; Dai et al., 2009; Cao et al., 2012; Chan et al., 2013; Jiao et al., 2013; Li et al., 2015; Zhao et al., 2015; Fung et al., 2017; Xiao et al., 2017; Zhang et al., 2019). As white-rot fungi, some species like G. australe, G. lingzhi, G. lucidum, and A. rugosum can secrete a series of carbohydrate hydrolase, peroxidase enzymes, and laccases to degrade the organic matters in forests, and this performance has been widely used as biofuel, for industrial applications and pollution abatement (Jong et al., 2017; Si et al., 2019, 2021; Wang et al., 2021). Besides, Ganoderma boninense, Ganoderma philippii, and A. rugosum as pathogenic species in Ganodermataceae can cause stem rot or root rot in forests leading to economic damage (Pilotti, 2005; Glen et al., 2009; Abubakar et al., 2022). To further understand how the economic and ecological values produced by Ganodermataceae species, genomics, transcriptomics, and proteomics were introduced by biologists to explore the mechanism of evolution, lignocellulose degradation, secondary metabolites biosynthesis, and plant-pathogenic (Chen et al., 2012; Kües et al., 2015; Zhu et al., 2015; Dhillon et al., 2021; Jiang et al., 2021; Lin et al., 2021; Liu et al., 2021; Sun et al., 2022a).
In view of the demand for health preservation and the utilization of biological resources, the mycologists were devoted to explore the potential species resources of Ganodermataceae. Since the first introduction of Ganodermataceae, the taxonomy and phylogeny studies of this family have been conducted over the past 100 years, and now the number of genera has increased from 2 to 14 (Murrill, 1905; Donk, 1948; Imazeki, 1952; Steyaert, 1972; Costa-Rezende et al., 2017, 2020; Sun et al., 2020, 2022b). Besides, the rise of species diversity is impressive but uneven. Ganoderma, as the biggest genus in this family, has expanded to 188 species based on credible morphological and phylogenetic evidence; however, the sum of species number of the other 13 genera is only half of that of Ganoderma (Ryvarden, 2020; Wu et al., 2020; Decock and Ryvarden, 2021; He et al., 2022; Sun et al., 2022b; Vinjusha and Kumar, 2022).
Sun et al. (2020) clarified the taxonomy and phylogeny of Amauroderma s. lat. in Ganodermataceae, in which Sanguinoderma was established with S. rude as type species, and five new species were presented based on the morphological and multi-gene phylogenetic evidence. The distinguished characters of Sanguinoderma are the dull pileal surface, the color of fresh pore surface changing to blood red when bruised, and the double-walled basidiospores with obvious spinules on endospore walls (Sun et al., 2020). The phylogenetic tree showed that Sanguinoderma rugosum was performed as two lineages with high support; yet, no morphological differences between them were observed. Sun et al. (2022b) evaluated 22 specimens with color-changed pore surfaces and described six new species of Sanguinoderma. Unfortunately, the differentiation in S. rugosum was ignored again due to the inappreciable differences. In fact, the variable morphological description of S. rugosum from different collections was proposed 40 years ago, for example, thin to thick and flexible to rigid pilei, dark brown to fuscous brown or black pileal surface with or without concentric zones in variable color, globose to subglobose basidiospores from 6.5 to 13 μm × 7 to 11 μm and so on Ryvarden and Johansen (1980), Corner (1983), Núñez and Ryvarden (2000). These differences indicated that the S. rugosum complex should be further excavated to solve the problem of subspecies differentiation.
During our investigations of Sanguinoderma, numerous specimens of S. rugosum complex were collected. The macro-/micro-morphological differences and phylogenetic relationships reflected their divergences indeed. Based on the morphological and phylogenetic analyses, five species were discovered in the S. rugosum complex, Sanguinoderma leucomarginatum was described as a new species, and another three species were identified as suspected new species due to their sterile basidiomata. In addition, Amauroderma preussii was transferred to Sanguinoderma as a new combination.
The studied specimens are deposited at the herbaria of the Institute of Microbiology, Beijing Forestry University (BJFC, Beijing, China), and the Institute of Microbiology, Chinese Academy of Sciences, China (HMAS). Macro-morphological descriptions of the taxa were based on field notes and herbarium specimens. Micro-morphological data were obtained from dried specimens and observed under a compound microscope following by Sun et al. (2022b) and Liu et al. (2022). Sections were studied at a magnification up to 1,000× using a Nikon Digital Sight DS-Fi2 microscope (Nikon Corporation, Tokyo, Japan) and quantified by the Image-Pro Plus 6.0 software (Media Cybernetics, Silver Spring, USA). Special color terms followed Petersen (1996). Morphological descriptions and abbreviations used in this study followed Cui et al. (2019) and Sun et al. (2022b).
The total genomic DNA was extracted from the dried specimens using CTAB rapid plant genome extraction kit-DN14 (Aidlab Biotechnologies Co., Ltd, Beijing, China) and a FH plant DNA kit II (Demeter Biotech Co., Ltd., Beijing, China). The detailed methods of DNA extraction and polymerase chain reaction (PCR) were according to the manufacturer's instructions with some modifications (Sun et al., 2020; Liu et al., 2022). The internal transcribed spacer regions (ITS) were amplified with primer pairs ITS5 and ITS4 (White et al., 1990). The large subunit of nuclear ribosomal RNA gene (nLSU) was amplified with primer pairs LR0R and LR7, and the primer LR5 was used sometimes as an alternative to LR7 (Vilgalys and Hester, 1990). The second subunit of RNA polymerase II (rpb2) was amplified with primer pairs fRPB2-5F and fRPB2-7CR (Liu et al., 1999). The translation elongation factor 1-α gene (tef1) was amplified with primer pairs EF1-983F and EF1-1567R (Rehner and Buckley, 2005). The small subunit mitochondrial rRNA gene (mtSSU) was amplified with primer pairs MS1 and MS2 (White et al., 1990). The small subunit nuclear ribosomal RNA gene (nSSU) was amplified with primer pairs PNS1 and NS41 (White et al., 1990).
The PCR volume contained 1 μl each primer, 1 μl extracted DNA, 12 μl ddH2O, and 15 μl 2 × EasyTaq PCR SuperMix (TransGen Biotech Co., Ltd., Beijing, China). The PCR cycling schedules for six-gene regions of ITS, nLsu, rpb2, tef1, nSSU, and mtSSU was followed by Sun et al. (2020, 2022b). The PCRs were performed on S1000™ Thermal Cycler (Bio-Rad Laboratories, California, USA), and the PCR products were purified and sequenced with the same primers at the Beijing Genomics Institute (BGI), China. All sequences used in this study were deposited at GenBank and are listed in Table 1.
The ITS, nLSU, rpb2, tef1, mtSSU, and nSSU sequences used in this study were combined into a dataset. Magoderna subresinosum was used as the outgroup, which is a sister clade with Sanguinoderma (Sun et al., 2022b). Phylogenetic analyses used in this study followed the approach of Cui et al. (2019). These sequences were aligned in online MAFFT v. 7 (Katoh et al., 2019; https://mafft.cbrc.jp/alignment/server/) and manually adjusted using BioEdit (Hall, 1999). Each alignment of ITS, nLSU, rpb2, tef1, mtSSU, and nSSU was catenated in Mesquite (Maddison and Maddison, 2017). The congruencies of six-gene loci were evaluated with the partition homogeneity test (PHT) (Farris et al., 1994) using PAUP v. 4.0b10 (Swofford, 2002) under 1,000 homogeneity replicates. The best-fit evolutionary model was calculated in MrModeltest v. 2.3 (Nylander, 2008) using hierarchical-likelihood ratio tests (hLRTs) and Akaike information criterion (AIC) strategies.
Based on the combined dataset, the maximum-likelihood (ML) analyses were conducted in RAxML-HPC v. 8.2.3 (Stamatakis, 2014). The best topology was obtained during 1 000 ML searches under the GTRGAMMA model, and 1,000 rapid bootstrap replicates were run with the GTRCAT model to assess the ML bootstrap values of the nodes. Bayesian inference analyses were calculated using MrBayes v. 3.1.2 (Ronquist and Huelsenbeck, 2003). The analyses were run with four Markov chains, starting trees for 12 M generations until the average standard deviation of split deviation frequency < 0.01, and sampled every 100 generations. The first 25% of the sampled trees were discarded as burn-in, and the remaining ones were used to reconstruct a majority rule consensus and calculate Bayesian posterior probability (BPP) of the clades.
All trees were visualized in FigTree v. 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/). The branches received ML bootstrap ≥ 70%, and Bayesian posterior probabilities ≥0.95 were regarded as credibly supported. The final alignments and the phylogenetic tree were deposited in TreeBASE (http://www.treebase.org), under accession ID: 29788 (http://purl.org/phylo/treebase/phylows/study/TB2:S29788).
In this study, 340 sequences of ITS, nLSU, rpb2, tef1, mtSSU, and nSSU were used to construct phylogenetic trees of Sanguinoderma, including 61 ITS sequences, 60 nLSU sequences, 44 rpb2 sequences, 56 tef1 sequences, 60 mtSSU sequences, and 59 nSSU sequences. The inferred sequences were obtained from 65 specimens representing 21 taxa in Sanguinoderma and Magoderna subresinosum as the outgroup. The combined six-gene (ITS+nLSU+rpb2+tef1+mtSSU+nSSU) sequence datasets had an aligned length of 5 017 total characters including gaps, of which 4 374 are constant, 207 are variable and parsimony-uninformative, and 436 are parsimony-informative.
The partition homogeneity test indicated all six different genes displayed a congruent phylogenetic signal (P = 1.00). The best-fit evolutionary models selected by MrModeltest v. 2.3 for each region of the six genes were K80+I (ITS1), K80 (5.8S), HKY+G (ITS2), GTR+I (nLSU), K80 (rpb2 introns), K80+I (rpb2 1st codon), GTR+I+G (rpb2 2nd codon), K80+G (tef1 introns), HKY+I (tef1 1st codon), SYM+I+G (tef1 2nd codon), GTR+G (tef1 3rd codon), HKY+I+G (mtSSU), and GTR (nSSU). These models were applied in Bayesian analyses for the combined dataset.
The average standard deviation of split frequencies in the Bayesian analyses reached 0.004273. The ML analyses resulted in a similar topology as Bayesian analyses, and only the ML topology with the calculated values is shown in Figure 1. The lineages presented in the phylogenetic tree were S. leucomarginatum as new species (98% ML, 0.98 BPP), S. preussii as new combination (96% ML, 1.00 BPP), S. bataanense (99% ML, 1.00 BPP), S. elmerianum (100% ML, 1.00 BPP), S. flavovirens, S. guangdongense (99% ML, 1.00 BPP), S. laceratum (92% ML, 1.00 BPP), S. longistipitum (98% ML, 1.00 BPP), S. infundibulare (96% ML, 1.00 BPP), S. melanocarpum (99% ML, 1.00 BPP), S. microporum (88% ML, 1.00 BPP), S. microsporum (92% ML, 1.00 BPP), S. perplexum (100% ML, 1.00 BPP), S. reniforme, S. rude (100% ML, 1.00 BPP), S. rugosum (93% ML, 1.00 BPP), S. sinuosum (88% ML, 1.00 BPP), S. tricolor (100% ML, 1.00 BPP), and three undetermined taxa: Sanguinoderma sp.1 (95% ML, 0.99 BPP), Sanguinoderma sp.2 (98% ML, 1.00 BPP), and Sanguinoderma sp.3 (100% ML, 0.97 BPP). Sanguinoderma rugosum complex comprised of S. rugosum, S. leucomarginatum, Sanguinoderma sp.1, Sanguinoderma sp.2, and Sanguinoderma sp.3, sharing the similar morphological characters.
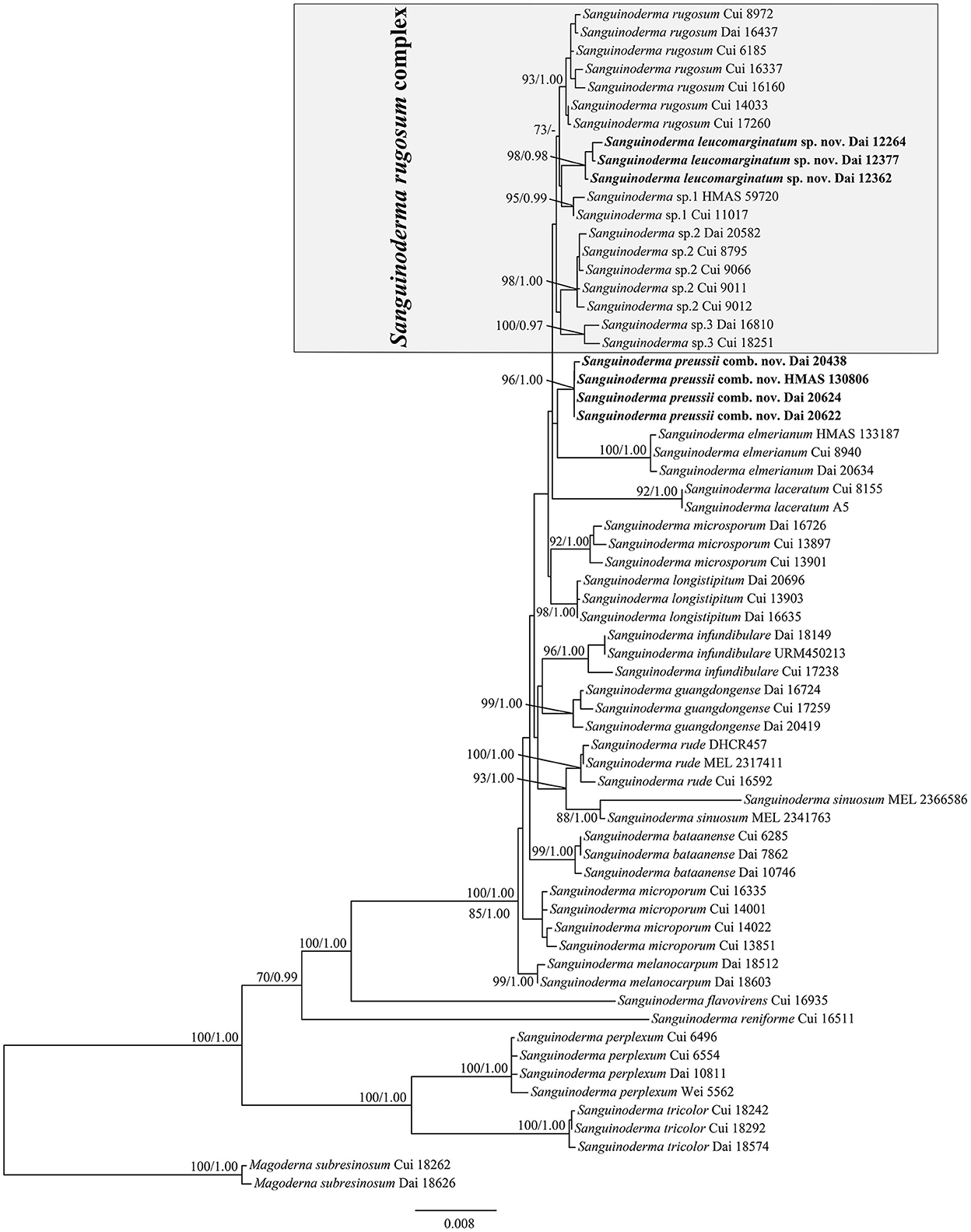
Figure 1. Maximum-likelihood (ML) analyses of Sanguinoderma based on the dataset of ITS+nLSU+rpb2+tef1+mtSSU+nSSU. Branches are labeled with maximum-likelihood bootstrap values equal to or higher than 70% and Bayesian posterior probability values equal to or higher than 0.95. New species or combinations are in bold.
Sanguinoderma leucomarginatum B. K. Cui and Y. F. Sun, sp. nov. (Figure 2)
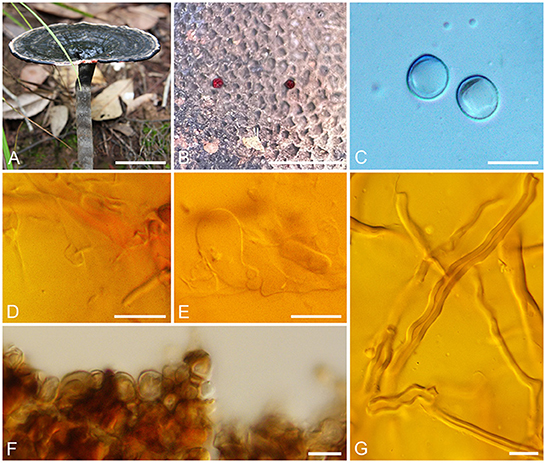
Figure 2. Basidiomata and microscopic structures of Sanguinoderma leucomarginatum. (A) Basidiomata. (B) Pores. (C) Basidiospores. (D) Clamp connections on generative hyphae. (E) Basidioles. (F) Pileipellis. (G) Skeletal hyphae. Scale bars: (A) = 2 cm, (B) = 1 mm, (C–G) = 10 μm.
MycoBank number: MB 846192
Diagnosis: Differs from other species in the genus by having near orbicular pilei with white to buff margin when fresh and clavate apical cells of pileipellis with septa.
Etymology: leucomarginatum (Lat.) refers to the white to buff margin of pilei.
Holotype: CHINA. Yunnan Province, Pu'er City, Laiyanghe Nature Reserve, on ground of forest, 9 June 2011, Yu-Cheng Dai, Dai 12377 (BJFC 010657).
Description: Basidiomata annual, laterally stipitate, hard corky to woody hard. Pilei solitary, near orbicular, up to 8 cm in diameter and 7-mm thick. Pileal surface fawn to vinaceous gray or near black, margin white to buff, dull, glabrous, with fuscous concentric zones or edges, and radial wrinkles near the margin; margin acute to obtuse, entire, slightly incurved and wavy when dry. Pore surface becoming blood red when bruised and then quickly darkening, pale mouse gray to ash-gray when dry; pores circular to angular, 5–6 per mm; dissepiments slightly thick, entire. Context cream to buff yellow, with dark melanoid lines, hard corky, up to 3-mm thick. Tubes light vinaceous gray to ash-gray, up to 3-mm long. Stipe clay buff to fawn, cylindrical and hollow, up to 8.5-cm long and 8 mm in diameter.
Hyphal system trimitic; generative hyphae with clamp connections, all hyphae IKI–, CB+; tissues darkening in KOH. Generative hyphae in context colorless, thin-walled, 3–6 μm in diameter; skeletal hyphae in context faint yellow, thick-walled with a wide to narrow lumen or sub-solid, arboriform and flexuous, 3–7 μm in diameter; binding hyphae in context faint yellow, sub-solid, branched and flexuous, up to 2 μm in diameter. Generative hyphae in tubes colorless, thin-walled, 3–6 μm in diameter; skeletal hyphae in tubes faint yellow, thick-walled with a wide to narrow lumen or sub-solid, arboriform and flexuous, 3–6 μm in diameter; binding hyphae in tubes faint yellow, sub-solid, branched, and flexuous, up to 2 μm in diameter. Pileipellis composed of clamped generative hyphae, thick-walled, apical cells clavate with septa, slightly inflated, yellow to reddish brown, about 40–70 × 4–7 μm, forming a regular palisade. Cystidia and cystidioles absent. Basidia barrel-shaped, colorless, thin-walled, 14–20 × 14–16 μm; basidioles in shape like the basidia, colorless, thin-walled, 12–23 × 6–15 μm. Basidiospores subglobose to broadly ellipsoid, pale yellow, IKI–, CB+, double-walled with slightly thick walls, exospore wall smooth, endospore wall with dense spinules (8.5–)8.8–10.1 × (7.4–)7.8–9 μm, L = 9.32 μm, W = 8.3 μm, Q = 1.12 (n = 60/1).
Additional specimens examined: CHINA. Yunnan Province, Pu'er City, Laiyanghe Nature Reserve, on ground of angiosperm forest, 9 June 2011, Yu-Cheng Dai, Dai 12264 (BJFC 010547), Dai 12390 (BJFC 010670); on root of Castanea, 9 June 2011, Yu-Cheng Dai, Dai 12362 (BJFC 010642); Jinghong City, Xishuangbanna Nature Reserve, on ground of forest, 7 June 2011, Yu-Cheng Dai, Dai 12324 (BJFC 010605).
Notes: Sanguinoderma leucomarginatum was described from Yunnan Province of Southwestern China. It is distinguished by its more or less orbicular pilei with white to buff margin when fresh and the clavate apical cells of pileipellis with septa. According to the previous studies, four species of Sanguinoderma had been reported from Yunnan Province, viz. S. elmerianum, S. guangdongense, S. laceratum, and S. longistipitum (Sun et al., 2020, 2022b). Compared to these species, S. leucomarginatum has the medially sized pores (5–6 per mm) with entire dissepiments, the stipe in medium length (up to 8.5 cm), and smaller basidiospores (8.8–10.1 × 7.8–9 μm). In the phylogenetic tree, S. leucomarginatum was presented as a distinct lineage with high support (Figure 1).
Sanguinoderma preussii (Henn.) B. K. Cui and Y. F. Sun, comb. nov. (Figure 3)
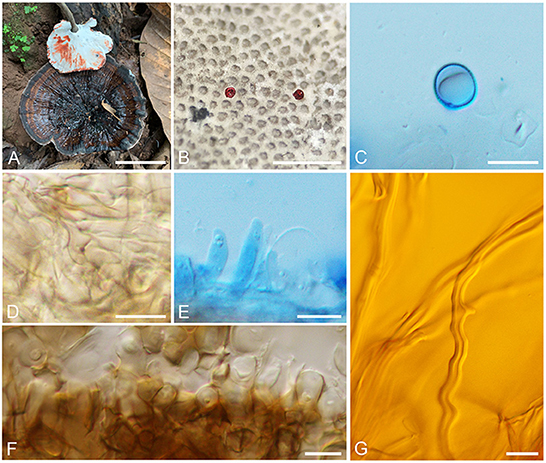
Figure 3. Basidiomata and microscopic structures of Sanguinoderma preussii. (A) Basidiomata. (B) Pores. (C) Basidiospores. (D) Clamp connections on generative hyphae. (E) Cystidioles. (F) Pileipellis. (G) Skeletal hyphae. Scale bars: (A) = 3 cm, (B) = 1 mm, (C–G) = 10 μm.
MycoBank number: MB 846193
Basionym: Ganoderma preussii Henn., Bot. Jb. 14(4): 342 (1891).
=Amauroderma preussii (Henn.) Steyaert, Persoonia 7(1): 107 (1972).
=Fomes preussii (Henn.) Sacc., Syll. fung. (Abellini) 11: 89 (1895).
=Scindalma preussii (Henn.) Kuntze, Revis. gen. pl. (Leipzig) 3(3): 519 (1898).
=Polyporus preussii (Henn.) Lloyd, Mycol. Writ. 3 (Syn. Stip. Polyporoids) (Cincinnati): 124 (1912).
=Ganoderma rubeolum Bres., Mycologia 17(2): 73 (1925).
=Ganoderma sikorae Bres., Annln K. K. naturh. Hofmus. Wien 26: 157 (1912).
=Polyporus salebrosus Lloyd, Mycol. Writ. (Cincinnati) 4(Letter 42): 14 (1912).
=Polyporus zambesianus Lloyd, Mycol. Writ. 3 (Syn. Stip. Polyporoids) (Cincinnati): 128 (1912).
=Polyporus rugosissimus Lloyd, Mycol. Writ. (Cincinnati) 4(Letter 48): 3 (1913).
=Ganoderma puberulum Pat., Bull. Soc. mycol. Fr. 30(3): 343 (1914).
=Fomes versicolor Bres., in Beeli, Bull. Jard. bot. État Brux. 8: 91 (1922).
Description: Basidiomata annual, centrally stipitate, hard corky to woody hard. Pilei solitary, funnel-shaped, up to 10.5 cm in diameter and 3-mm thick. Pileal surface grayish brown, dull, glabrous, with black and concentric zones and radial wrinkles; margin acute, entire, petaloid, strongly incurved, and wavy when dry. Pore surface becoming to blood red when bruised and then quickly darkening, white to cream when dry; pores circular to angular or irregular, 6–7 per mm; dissepiments medially thick, entire. Context buff yellow, with dark melanoid lines, hard corky, up to 1-mm thick. Tubes ash-gray, up to 2-mm long. Stipe grayish brown, cylindrical, and hollow, up to 11.5-cm long and 8 mm in diameter.
Hyphal system trimitic; generative hyphae with clamp connections, all hyphae IKI–, CB+; tissues are darkening in KOH. Generative hyphae in context colorless, thin-walled, 3–4 μm in diameter; skeletal hyphae in context pale yellow, thick-walled with a wide to narrow lumen or sub-solid, arboriform and flexuous, 3–7 μm in diameter; binding hyphae in context pale yellow, sub-solid, branched, and flexuous, up to 2 μm in diameter. Generative hyphae in tubes colorless, thin-walled, 4–5 μm in diameter; skeletal hyphae in tubes pale yellow, thick-walled with a wide to narrow lumen or sub-solid, arboriform and flexuous, 4–6 μm in diameter; binding hyphae in tubes pale yellow, sub-solid, branched and flexuous, up to 2 μm in diameter. Pileipellis composed of clamped generative hyphae, thick-walled to sub-solid, apical cells clavate, inflated, pale yellow to yellowish brown, about 45–65 × 5–8 μm, forming a regular palisade. Cystidia absent; cystidioles clavate and apices constricted, colorless, thin-walled, 12–24 × 2–4 μm. Basidia near orbicular to barrel-shaped, colorless, thin-walled, 15–23 × 11–12 μm; basidioles barrel-shaped to clavate, colorless, thin-walled, 16–22 × 7–15 μm. Basidiospores subglobose to broadly ellipsoid, pale yellow, IKI–, CB+, double-walled with slightly thick walls, exospore wall smooth, endospore wall with dense spinules, 9–10.5(−10.8) × 8–9(−9.5) μm, L = 9.54 μm, W = 8.46 μm, Q = 1.13 (n = 60/2).
Specimens examined: THAILAND. Chiang Rai, Mae Salong Nok, on ground of angiosperm forest, 22 July 2016, Yu-Cheng Dai, Dai 16646 (BJFC 022756); on ground of forest, 24 July 2016, Yu-Cheng Dai, Dai 16725 (BJFC 022832). CHINA. Yunnan Province, Pu'er City, Pu'er Forestry Park, on ground of forest, 17 August 2019, Yu-Cheng Dai, Dai 20438 (BJFC 032106), Dai 20456 (BJFC 032124), Dai 20467 (BJFC 032135), Dai 20468 (BJFC 032136); Mengla County, Shangyong Nature Reserve, on ground of forest, 20 August 2019, Yu-Cheng Dai, Dai 20622 (BJFC 032289), Dai 20624 (BJFC 032291); Bakaxiaozhai Nature Reserve, on ground, 5 August 2003, Tie-Zheng Wei, HMAS 130806.
Notes: Ganoderma preussii was described from Cameroon and temporarily transferred to Amauroderma in Steyaert (1972) by its dull pileal surface and double-walled basidiospores without truncated apex. Here, A. preussii was transferred to Sanguinoderma due to the color-changed pore surface when bruised. The specimens used in this study were collected from East Asia, and the morphological characters of basidiomata are mostly consistent with the original description of A. preussii (Steyaert, 1972). However, Steyaert (1972) mentioned that the hyphae of pileipellis extend externally free and anticlinal at the base, while the structural characters of pileipellis of specimens observed in this study are forming as a palisade, which are similar to most species in Amauroderma s. lat.
Sanguinoderma infundibulare is another species with funnel-shaped pilei in Sanguinoderma, and it can be characterized by the yellowish brown and tomentose pileal surface with uncurved margin and large basidiospores (10.2–12 × 9–10.2 μm; Sun et al., 2022b). Besides, S. preussii and S. infundibulare were supported as two distinct lineages in the phylogenetic tree (Figure 1).
In this study, the multi-gene phylogenetic analyses of Sanguinoderma were conducted based on the combined dataset of ITS+nLSU+rpb2+tef1+mtSSU+nSSU sequences. In the phylogenetic tree, 21 taxa of Sanguinoderma clustered together with high support (100% ML, 1.00 BPP; Figure 1), in which 16 species were shown as well-supported respective lineages in accordance with previous studies by Sun et al. (2020, 2022b).
Sun et al. (2020, 2022b) have improved the classification of Sanguinoderma and reported 16 species in the genus with detailed morphological and phylogenetic evidence, while the differentiation in phylogeny of Sanguinoderma rugosum was still not studied. The variable morphological characters observed from different collections (Ryvarden and Johansen, 1980; Corner, 1983; Núñez and Ryvarden, 2000) provided an auxiliary basis for this divergence. During this study, more than 80 specimens were collected from East Asia, which were identified as S. rugosum for the first time. These specimens can be divided into five groups roughly in the analysis tests, and more concise lineages were presented in this article with high support (Figure 1). We treated the five lineages as five different taxa of the S. rugosum complex, which are similar in morphology.
Sanguinoderma rugosum as the core species of this complex is easily confused in morphology with the other four taxa, except the deeply concentric furrows on pileal surface, clavate cystidioles, and lager basidiospores (9.5–11.6 × 8–9.5 μm). Sanguinoderma leucomarginatum was separated from other taxa of the S. rugosum complex according to its white to buff pileal margin with fuscous concentric zones or edges, cream to buff context, absent cystidioles, and smaller basidiospores (8.8–10.1 × 7.8–9 μm). The other morphological characters, such as the wrinkled pileal surface, pale mouse gray to ash-gray pore surface when dry, and 5–6 pores per mm, are indistinguishable from the other four taxa. The other three suspected new species were discovered in this study based on the morphological differences and independent phylogenetic relationships. However, the failure to observe the mature basidiospores in morphological studies was the biggest obstacle to clarify the taxonomic status of these species; these three suspected new species were treated as undescribed taxa due to the sterile specimens, even though the structure of pileipellis in Sanguinoderma sp.1, the thickness of pore dissepiments in Sanguinoderma sp.2, and the color of pore surface in Sanguinoderma sp.3 can distinguish them availably (Table 2). The problem of the sterility of specimens is still unavoidable in taxonomic studies.
Sanguinoderma preussii can be easily distinguished by the funnel-shaped and thin pilei with an incurved margin-like petals. Hapuarachchi et al. (2018) examined the specimens of S. preussii collected from Xiengkhouang Province in Laos and Hainan Province in China, but the recorded size of pores (2–4 per mm) is quite different from the observation in this study (6–7 per mm). The funnel-shaped pilei were also observed in Amauroderma wuzhishanense according to the description by Zhao and Zhang (1987), but the tubercles and broad radial wrinkles on pileal surface make A. wuzhishanense (= A. rugosum) different from the smooth pileal surface with lender radial wrinkles in S. preussii. The collections from East Asia enriched the distributions of S. preussii, and it implies that the species of Sanguinoderma may be widespread in Palaeotropics, such as S. rugosum and S. rude.
After the morphological and phylogenetic analyses, one new species called S. leucomarginatum was separated from S. rugosum complex. Besides, there are three suspected new species in Sanguindoerma rugosum complex without valid taxonomic status due to the sterile specimens. In addition, one new combination called S. preussii was transferred from Amauroderma. In summary, 18 species were accepted in Sanguinoderma around the world, in which 12 species were distributed in China; a key to accepted species of Sanguinoderma is provided. In further studies, more fertile specimens need to be collected to enrich the species diversity and clarify the taxonomic status of the suspected species.
(1) Pore dissepiments extremely thick………………………..2
(1) Pore dissepiments thin to distinctly thick…………………3
(2) Pileal surface pale yellowish brown, pore surface yellowish brown, context with dark melanoid lines……S. microporum
(2) Pileal surface rust brown to almost black, pore surface white to pale yellow, context without dark melanoid lines… …………………………………………………S. tricolor
(3) Pore dissepiments lacerate, tubes fascicular when dry…… ………………………………………………S. laceratum
(3) Pore dissepiments entire, tubes unchanged when dry…… …………………………………………………………4
(4) Pores less than or equal to 4 per mm……………………...5
(4) Pores more than 4 per mm………………………………7
(5) Pores sinuate; basidiospores more than 13.5 μm in length…………………………………………S. sinuosum
(5) Pores circular to irregular; basidiospores less than 13.5 μm in length…….……….……………………………………6
(6) Pore dissepiments thin; basidiospores globose to subglobose…………………………………..S. bataanense
(6) Pore dissepiments slightly thick; basidiospores subglobose to broadly ellipsoid……… …………………………..S. rude
(7) Basidiospores less than 6 μm in length…………… …………………………………………...S. microsporum
(7) Basidiospores more than 6 μm in length…………………8
(8) Pileal surface coal black; basidiospores slightly dextrinoid in Melzer's reagent…………………………S. melanocarpum
(8) Pileal surface brown to almost black; basidiospores IKI- in Melzer's reagent…………………………………………9
(9) Pilei funnel-shape………………………………………10
(9) Pilei flat…………….….….……………………………11
(10) Pileal margin uncurved; larger basidiospores (10.2–12 × 9–10.2 μm)…………………………………S. infundibulare
(10) Pileal margin strongly incurved; smaller basidiospores (9–10.5 × 8–9 μm)…………………………………S. preussii
(11) Basidiospores reniform…………….….………S. reniforme
(11) Basidiospores globose to subglobose or broadly ellipsoid…12
(12) Pore surface yellowish green when fresh………… ……………………………………………S. flavovirens
(12) Pore surface pale white to cream or pale grey……………13
(13) Cystidioles absent………………………………………14
(13) Cystidioles present……….….….………………………15
(14) Pileal margin white to buff; basidiospores less than 9 μm in width…………………………………S. leucomarginatum
(14) Pileal margin dark brown to nearly black; basidiospores more than 9 μm in width…………………………S. elmerianum
(15) Basidiomata sessile to subsessile; basidiospores more than or equal to 14 μm in length……………… ………………………………………………S. perplexum
(15) Basidiomata stipitate; basidiospores less than 14 μm in length………….….……………………………………16
(16) Pileal surface with shades of brown concentric zones and dense radial lines………………………...S. guangdongense
(16) Pileal surface with concentric furrows and radial wrinkles………………………………………………...17
(17) Basidiomata small, with lateral stipe; cystidioles fusiform…………………………………...S. longistipitum
(17) Basidiomata large, with central to lateral stipe; cystidioles clavate………………………….….….………S. rugosum.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
B-KC designed the research. B-KC and Y-FS prepared the samples and drafted the manuscript. Y-FS and Y-XF conducted the molecular experiments and analyzed the data. All authors have read and agreed to the published version of the manuscript.
The research was supported by the National Natural Science Foundation of China (nos. 31870008, U2003211, and 32270010), Beijing Forestry University Outstanding Young Talent Cultivation Project (no. 2019JQ03016), and scientific research startup project in School of Ecology and Nature Conservation, Beijing Forestry University (BH2022-04).
We express our gratitude to Prof. Yu-Cheng Dai (Beijing Forestry University, China) for his help during field collections.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abubakar, A., Ishak, M. Y., Bakar, A. A., and Uddin, M. K. (2022). Ganoderma boninense basal stem rot induced by climate change and its effect on oil palm. Environ. Sustain. 5, 289–303. doi: 10.1007/s42398-022-00244-7
Cao, Y., Wu, S. H., and Dai, Y. C. (2012). Species clarification of the prize medicinal Ganoderma mushroom “Lingzhi”. Fungal Divers. 56, 49–62. doi: 10.1007/s13225-012-0178-5
Chan, P. M., Kanagasabapathy, G., Tan, Y. S., Sabaratnam, V., and Kuppusamy, U. R. (2013). Amauroderma rugosum (Blume and T. Nees) Torrend: nutritional composition and antioxidant and potential anti-inflammatory properties. Evid Based Comp. Alternat. Med. 2013, 304713. doi: 10.1155/2013/304713
Chen, S. L., Xu, J., Liu, C., Zhu, Y. J., Nelson, D. R., Zhou, S. G., et al. (2012). Genome sequence of the model medicinal mushroom Ganoderma lucidum. Nat. Commun. 3, 913. doi: 10.1038/ncomms1923
Corner, E. J. H. (1983). Ad polyporaceae I. Amauroderma and Ganoderma. Beihefte zur Nova Hedwigia 75, 1–182.
Costa-Rezende, D. H., Robledo, G. L., Drechsler-Santos, E. R., Glen, M., Gates, G., de Madrignac Bonzi, B. R., et al. (2020). Taxonomy and phylogeny of polypores with ganodermatoid basidiospores (Ganodermataceae). Mycol. Prog. 19, 725–741. doi: 10.1007/s11557-020-01589-1
Costa-Rezende, D. H., Robledo, G. L., Góes-Neto, A., Reck, M. A., Crespo, E., and Drechsler-Santos, E. R. (2017). Morphological reassessment and molecular phylogenetic analyses of Amauroderma s. lat. raised new perspectives in the generic classification of the Ganodermataceae family. Persoonia 39, 254–269. doi: 10.3767/persoonia.2017.39.10
Cui, B. K., Li, H. J., Ji, X., Zhou, J. L., Song, J., Si, J., et al. (2019). Species diversity, taxonomy and phylogeny of Polyporaceae (Basidiomycota) in China. Fungal Divers. 97, 137–392. doi: 10.1007/s13225-019-00427-4
Dai, Y. C., Yang, Z. L., Cui, B. K., Yu, C. J., and Zhou, L. W. (2009). Species diversity and utilization of medicinal mushrooms and fungi in China. Int. J. Med. Mushrooms 11, 287–302. doi: 10.1615/IntJMedMushr.v11.i3.80
Decock, C., and Ryvarden, L. (2021). Aphyllophorales of Africa 48 some poroid species from São Tomé. Synopsis Fungorum 44, 14–18.
Dhillon, B., Hamelin, R. C., and Rollins, J. A. (2021). Transcriptional profile of oil palm pathogen, Ganoderma boninense, reveals activation of lignin degradation machinery and possible evasion of host immune response. BMC Genomics 22, 326. doi: 10.1186/s12864-021-07644-9
Donk, M. A. (1948). Notes on Malesian fungi. I. Bulletin du Jardin Botanique de Buitenzorg 17, 473–482.
Farris, J. S., Kallersjo, M., Kluge, A. G., and Bult, C. (1994). Testing significance of incongruence. Cladistics 10, 315–319.
Fung, S. Y., Tan, N. H., Kong, B. H., Lee, S., Tan, Y. S., and Sabaratnam, V. (2017). Acute toxicity study and the in vitro cytotoxicity of a black lingzhi medicinal mushroom, Amauroderma rugosum (Agaricomycetes) from Malaysia. Int. J. Med. Mushrooms 19, 1093–1099. doi: 10.1615/IntJMedMushrooms.2017024550
Glen, M., Bougher, N. L., Francis, A. A., Nigg, S. Q., Lee, S. S., Lrianto, R., et al. (2009). Ganoderma and Amauroderma species associated with root-rot disease of Acacia mangium plantation trees in Indonesia and Malaysia. Austral. Plant Pathol. 38, 345–356. doi: 10.1071/AP09008
Hall, T. A. (1999). Bioedit: a user-friendly biological sequence alignment editor and analyses program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41,95–98.
Hapuarachchi, K. K., Karunarathna, S. C., Phengsintham, P., Kakumyan, P., Hyde, K. D., and Wen, T. C. (2018). Amauroderma (Ganodermataceae, Polyporales) – bioactive compounds, beneficial properties and two new records from Laos. Asian J. Mycol. 1, 121–136. doi 10.5943/ajom/1/1/10
He, J., Han, X., Luo, Z. L., Li, E. X., Tang, S. M., Luo, H. M., et al. (2022). Species diversity of Ganoderma (Ganodermataceae, Polyporales) with three new species and a key to Ganoderma in Yunnan Province, China. Front. Microbiol. 13, 1035434. doi: 10.3389/fmicb.2022.1035434
Imazeki, R. (1952). A contribution to the fungus flora of Dutch New Guinea. Bull. Government Forest Exp. Station Meguro 57, 87–128.
Jiang, N., Hu, S., Peng, B., Li, Z. H., Yuan, X. H., Xiao, S. J., et al. (2021). Genome of Ganoderma species provides insights into the evolution, conifers substrate utilization, and terpene synthesis for Ganoderma tsugae. Front. Microbiol. 12, 724451. doi: 10.3389/fmicb.2021.724451
Jiao, C. W., Xie, Y. Z., Yang, X. L., Li, H. R., Li, X. M., Pan, H. H., et al. (2013). Anticancer activity of Amauroderma rude. PLoS ONE 8, e66504. doi: 10.1371/journal.pone.0066504
Jong, W. Y. L., Show, P. L., Ling, T. C., and Tan, Y. S. (2017). Recovery of lignin peroxidase from submerged liquid fermentation of Amauroderma rugosum (Blume and T. Nees) Torrend using polyethylene glycol/salt aqueous two-phase system. J. Biosci. Bioeng. 124, 91–98. doi: 10.1016/j.jbiosc.2017.02.008
Katoh, K., Rozewicki, J., and Yamada, K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinformatics 20, 1160–1166. doi: 10.1093/bib/bbx108
Kües, U., Nelson, D. R., Liu, C., Yu, G. J., Zhang, J. H., Li, J. Q., et al. (2015). Genome analysis of medicinal Ganoderma spp. with plant-pathogenic and saprotrophic life-styles. Phytochemistry 114, 18–37. doi: 10.1016/j.phytochem.2014.11.019
Li, M. J., and Yuan, H. S. (2015). Type studies on Amauroderma species described by J.D. Zhao et al. and the phylogeny of species in China. Mycotaxon 130, 79–89. doi: 10.5248/130.79
Li, X. M., Wu, Q. P., Xie, Y. Z., Ding, Y. R., Du, W. W., Sdiri, M., et al. (2015). Ergosterol purified from medicinal mushroom Amauroderma rude inhibits cancer growth in vitro and in vivo by up-regulating multiple tumor suppressors. Oncotarget 6, 17832–17846. doi: 10.18632/oncotarget.4026
Lin, W. P., Shi, Y. H., Jia, G. T., Sun, H. Y., Sun, T. Y., and Hou, D. H. (2021). Genome sequencing and annotation and phylogenomic analysis of the medicinal mushroom Amauroderma rugosum, a traditional medicinal species in the family Ganodermataceae. Mycologia 113, 268–277. doi: 10.1080/00275514.2020.1851135
Liu, S., Chen, Y. Y., Sun, Y. F., He, X. L., Song, C. G., Si, J., et al. (2022). Systematic classification and phylogenetic relationships of the brown-rot fungi within the Polyporales. Fungal Divers. doi: 10.1007/s13225-022-00511-2. [Epub ahead of print].
Liu, Y. C., Huang, L. H., Hu, H. P., Cai, M. J., Liang, X. W., Li, X. M., et al. (2021). Whole-genome assembly of Ganoderma leucocontextum (Ganodermataceae, Fungi) discovered from the Tibetan Plateau of China. G3 Genes Genomes 11, jkab337. doi: 10.1093/g3journal/jkab337
Liu, Y. J., Whelen, S., and Hall, B. D. (1999). Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 16, 1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092
Maddison, W. P., and Maddison, D. R. (2017). Mesquite: A Modular System for Evolutionary Analysis. Version 3.2. Available online at: http://mesquiteproject.org
Petersen, J. H. (1996). Farvekort. The Danish Mycological Society's colourchart. Foreningen til Svampekundskabens Fremme. Greve.
Pilotti, C. A. (2005). Stem rots of oil palm caused by Ganoderma boninense: pathogen biology and epidemiology. Mycopathologia 159, 129–137. doi: 10.1007/s11046-004-4435-3
Rehner, S. A., and Buckley, E. (2005). A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97, 84–98. doi: 10.1080/15572536.2006.11832842
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Ryvarden, L., and Johansen, I. (1980). A Preliminary Polypores Flora of East Africa. Oslo: Fungiflora.
Si, J., Meng, G., Wu, Y., Ma, H. F., Cui, B. K., and Dai, Y. C. (2019). Medium composition optimization, structural characterization, and antioxidant activity of exopolysaccharides from the medicinal mushroom Ganoderma lingzhi. Int. J. Biol. Macromol. 124, 1186–1196. doi: 10.1016/j.ijbiomac.2018.11.274
Si, J., Wu, Y., Ma, H. F., Cao, Y. J., Sun, Y. F., and Cui, B. K. (2021). Selection of a pH- and temperature-stable laccase from Ganoderma australe and its application for bioremediation of textile dyes. J. Environ. Manage. 299, 113619. doi: 10.1016/j.jenvman.2021.113619
Song, J., Xing, J. H., Decock, C., He, X. L., and Cui, B. K. (2016). Molecular phylogeny and morphology reveal a new species of Amauroderma (Basidiomycota) from China. Phytotaxa 260, 47–56. doi: 10.11646/phytataxa.260.1.5
Stamatakis, A. (2014). RAxML Version 8: a tool for phylogenetic analyses and post analyses of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Steyaert, R. L. (1972). Species of Ganoderma and related genera mainly of the Bogor and Leiden Herbaria. Persoonia 7, 55–118.
Sun, Y. F., Costa-Rezende, D. H., Xing, J. H., Zhou, J. L., Zhang, B., Gibertoni, T. B., et al. (2020). Multi-gene phylogeny and taxonomy of Amauroderma s. lat.(Ganodermataceae). Persoonia 44, 206–239. doi: 10.3767/persoonia.2020.44.08
Sun, Y. F., Lebreton, A., Xing, J. H., Fang, Y. X., Si, J., Morin, E., et al. (2022a). Phylogenomics and comparative genomics highlight specific genetic features in Ganoderma species. J. Fungi 8, 311. doi: 10.3390/jof8030311
Sun, Y. F., Xing, J. H., He, X. L., Wu, D. M., Song, C. G., Liu, S., et al. (2022b). Species diversity, systematic revision and molecular phylogeny of Ganodermataceae (Polyporales, Basidiomycota) with an emphasis on Chinese collections. Stud. Mycol. 101, 287–415. doi: 10.3114/sim.2022.101.05
Swofford, D. L. (2002). Paup*. Phylogenetic Analyses Using Parsimony (* and Other Methods) Version 4.0b10. Sunderland, MA: Sinauer Associates.
Vilgalys, R., and Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990
Vinjusha, N., and Kumar, T. K. A. (2022). Revision of Ganoderma species associated with stem rot of coconut palm. Mycologia 114, 157–174. doi: 10.1080/00275514.2021.1974724
Wang, G. Y., Zhang, J., Mizuno, T., Zhuang, C., Ito, H., Mayuzumi, H., et al. (1993). Antitumor active polysaccharides from the Chinese mushroom Songshan Lingzhi, the fruiting body of Ganoderma tsugae. Biosci. Biotechnol. Biochem. 57, 894–900. doi: 10.1271/bbb.57.894
Wang, H., Deng, W., Shen, M. H., Yan, G., Zhao, W., and Yang, Y. (2021). A laccase Gl-LAC-4 purified from white-rot fungus Ganoderma lucidum had a strong ability to degrade and detoxify the alkylphenol pollutants 4-n-octylphenol and 2-phenylphenol. J. Hazard. Mater. 408, 1–16. doi: 10.1016/j.jhazmat.2020.124775
White, T. J., Bruns, T., Lee, S., and Taylor, J. W. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols, A Guide to Methods and Applications, eds M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (San Diego, CA: Academic Press), 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1
Wu, S. H., Chern, C. L., Wei, C. L., Chen, Y. P., Akiba, M., and Hattori, T. (2020). Ganoderma bambusicola sp. nov. (Polyporales, Basidiomycota) from southern Asia. Phytotaxa 456, 75–85. doi: 10.11646/phytotaxa.456.1.5
Xiao, Z. T., Liu, M., and He, H. Q. (2017). Domestication and antioxidant activities of Amauroderma rugosum. Mycosystema 36, 358–366. doi: 10.13346/j.mycosystema.160029
Zhang, Y., Jiang, Y., Zhang, M., and Zhang, L. J. (2019). Ganoderma sinense polysaccharide: an adjunctive drug used for cancer treatment. Prog. Mol. Biol. Transl. Sci. 163, 165–177. doi: 10.1016/bs.pmbts.2019.02.008
Zhao, J. D., and Zhang, X. Q. (1987). Studies on the taxonomy of Ganodermataceae in China VIII. Acta Mycol. Sin. 5, 219–225.
Zhao, Z. Z., Yin, R. H., Chen, H. P., Feng, T., Li, Z. H., Dong, Z. J., et al. (2015). Two new triterpenoids from fruiting bodies of fungus Ganoderma lucidum. J. Asian Nat. Products Res. 17, 750–755. doi: 10.1080/10286020.2014.996139
Keywords: Ganodermataceae, macrofungi, morphology, multi-gene phylogeny, new taxa
Citation: Sun Y-F, Fang Y-X and Cui B-K (2022) Taxonomy and phylogeny of Sanguinoderma rugosum complex with descriptions of a new species and a new combination. Front. Microbiol. 13:1087212. doi: 10.3389/fmicb.2022.1087212
Received: 02 November 2022; Accepted: 24 November 2022;
Published: 21 December 2022.
Edited by:
Yong-Zhong Lu, Guizhou Institute of Technology, ChinaReviewed by:
ZongLong Luo, Dali University, ChinaCopyright © 2022 Sun, Fang and Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bao-Kai Cui, Y3VpYmFva2FpQGJqZnUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.