- Key Laboratory of Bio-Resource and Eco-Environment of Ministry of Education, College of Life Sciences, Sichuan University, Animal Disease Prevention and Food Safety Key Laboratory of Sichuan Province, Chengdu, China
Proteus mirabilis is an opportunistic pathogen frequently associated with nosocomial infection and food poisoning cases. Contamination of P. mirabilis in retail meat products may be important transmission routes for human infection with P. mirabilis. In this study a total of 89 P. mirabilis strains were isolated from 347 samples in 14 food markets in China and subjected to whole-genome sequencing. Phylogenetic analysis showed that all 89 strains were divided into 81 different clones (SNPs >5), indicating high genetic diversity of P. mirabilis in food markets. Antimicrobial susceptibility testing showed that 81 (91.01%) strains displayed multidrug resistance profiles. Seventy-three different resistance genes (or variants) were found, including various clinically important antimicrobial resistance genes aac(6′)-Ib-cr (77.53%), blaCTX-M (39.33%), fosA3 (30.34%), as well as multiresistance gene cfr (4.50%), tigecycline resistance gene cluster tmexCD3-toprJ1 (4.50%) and carbapenemase gene blaNDM-1 (1.12%). Diverse genetic elements including Tn7 transposon, plasmid, SXT/R391 integrative conjugative element were associated with the horizontal transfer of cfr. tmexCD3-toprJ1 and blaNDM-1 were located on ICEPmiChnJZ26 and Salmonella genomic island 1, respectively. Our study emphasized high contamination of P. mirabilis harbouring various clinically important antimicrobial resistance genes in retail meat and aquatic products, which might be an important issue in terms of food safety and human health.
Introduction
Food contamination caused by foodborne and opportunistic pathogenic bacteria that are leading causes of foodborne illnesses and deaths is a serious threat to global public health (Tropea, 2022). Proteus mirabilis is a species of Gram-negative bacteria belonging to genus Proteus of Enterobacterales (Janda and Abbott, 2021), and is widely distributed in the natural environment and intestines of humans and animals. P. mirabilis is one of the most frequently opportunistic pathogen causing nosocomial infection, especially causing urinary tract infection (Schaffer and Pearson, 2015). In addition, P. mirabilis infections can also cause bacteremia and the formation of urinary stones. It is worth noting that some food poisoning cases associated with P. mirabilis had been reported in China, as well as other countries (Gong et al., 2019). Given that P. mirabilis is a resident in the intestinal tract of chickens and swines, it can potentially be present in retail meat products due to fecal contamination at the slaughtering stage. Manipulation and consumption of retail meat products may be important transmission routes for human infection with P. mirabilis. Contamination of P. mirabilis in retail meat products has become an important issue for food safety and human health.
The wide use of antimicrobials for the treatment of bacterial infections in humans and food-producing animals has led to widespread dissemination of antimicrobial resistance genes. Emergence and spread of multidrug resistant (MDR) P. mirabilis isolates, including those producing extended-spectrum beta-lactamases (ESBLs), AmpC cephalosporinases and carbapenemases, raised concerns in the past few years (Girlich et al., 2020). P. mirabilis is intrinsically resistant to nitrofurantoin, polymyxin and tigecycline, which brings a serious challenge for the treatment of P. mirabilis infections. Many mobile genetic elements, including plasmids, integrons, insertion sequences and transposons (Partridge et al., 2018), as well as SXT/R391 integrative conjugative elements (ICEs) and genomic islands (such as Salmonella/Proteus genomic islands, SGI/PGI), are found to mediate horizontal transfer of various antimicrobial resistance genes in P. mirabilis (Cummins et al., 2020; He et al., 2021). For example, the New Delhi metallo-β-lactamase gene blaNDM-1 was sporadically found in P. mirabilis, and was located on plasmids (Bitar et al., 2020), PGI1 and SGI1 (Girlich et al., 2015; Yang et al., 2021). The multiresistance gene cfr that encodes an RNA methyltransferase conferring resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins and streptogramin A (Shen et al., 2013), as well as the plasmid-encoding RND efflux pump gene cluster tmexCD3-toprJ3 that reduces the sensitivity of multiple antimicrobials including tigecycline (Lv et al., 2020), were found to be located on SXT/R391 ICEs (Wang et al., 2021b; Song et al., 2022). These findings indicate that P. mirabilis is an important reservoir of clinically important antimicrobial resistance genes and mobile genetic elements that have the potential to spread to other pathogenic bacteria in Enterobacterales.
Traditionally, pulsed field gel electrophoresis was used for molecular typing, and there is no standard multilocus sequence typing for P. mirabilis. With the development of the next-generation sequencing (NGS) technology, phylogenetic analysis based on the single nucleotide polymorphisms (SNPs) in core genomes provides higher distinguishability for tracing the transmission of associated pathogens (Schurch et al., 2018). In China, food markets that sell retail meat (especially chicken and pork meat) and aquatic products are widely distributed in communities. However, the data of the contamination of P. mirabilis in those products is limited. The aims of the present study were to (i) investigate the prevalence of P. mirabilis in retail meat and aquatic products in food markets using NGS, (ii) detect antimicrobial resistance phenotypes and genotypes, and (iii) determine the mobile genetic elements associated with the clinically important antimicrobial resistance genes.
Results
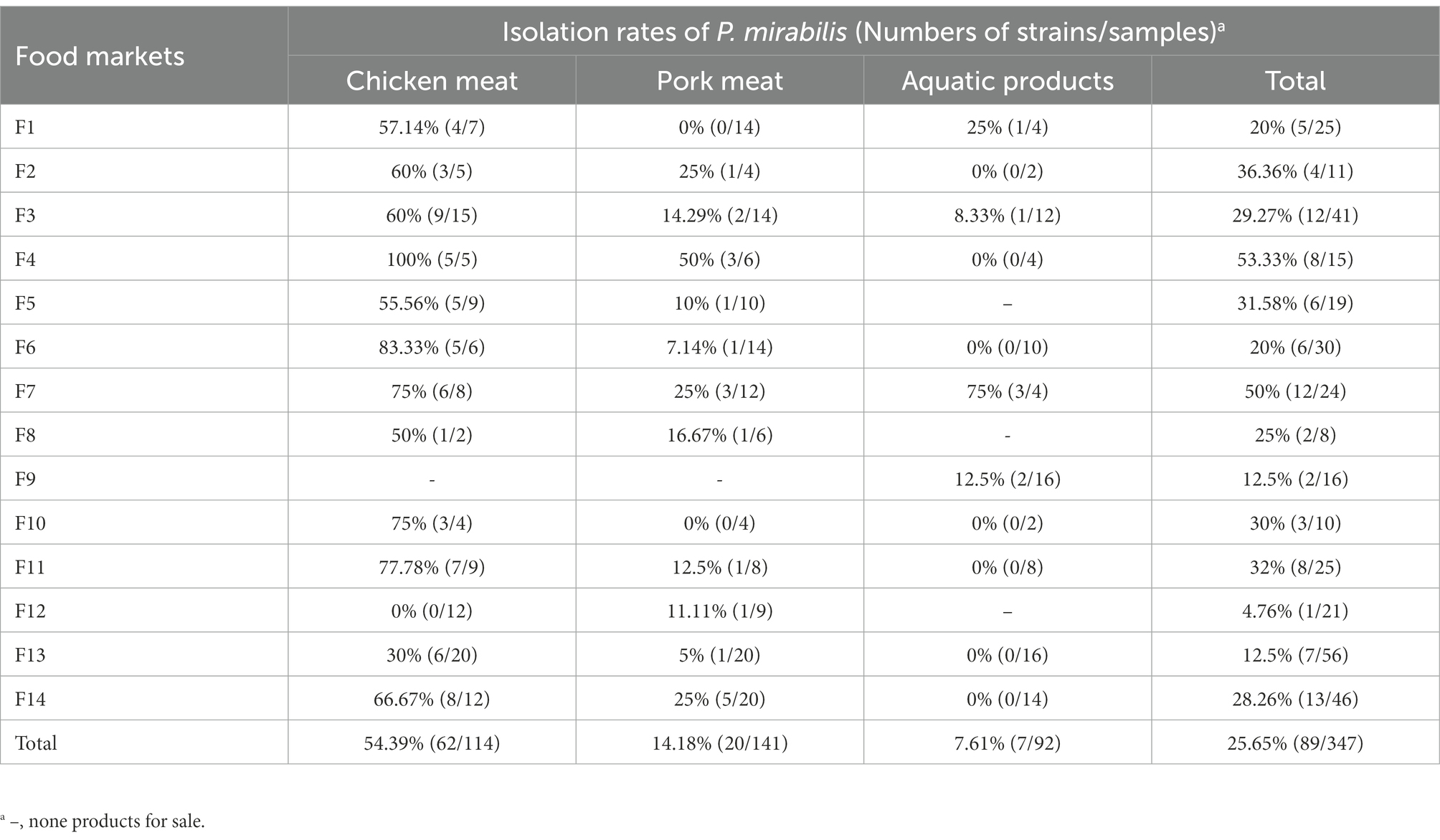
Prevalence of Proteus mirabilis in retail meat and aquatic products from food markets
A total of 89 P. mirabilis strains (62 from chicken meat, 20 from pork meat and 7 from aquatic products) were isolated from 347 samples, and the overall isolation rate of P. mirabilis was 25.65% (89/347). The chicken meat samples had the highest isolation rate, which reached 54.39% (62/114). The isolation rates of P. mirabilis in pork and aquatic products were low, which were 14.18% (20/141) and 7.61% (7/92), respectively. In 14 food markets, the isolation rates of P. mirabilis varied from 4.76% (1/21) to 53.33% (8/15) (Table 1). The detailed information of the 89 P. mirabilis strains reported in this study is listed in Supplementary Table S1.
Phylogenetic analysis of Proteus mirabilis strains
The mean of the draft genomes of 89 P. mirabilis strains was 4.00 Mb (from 3.79 to 4.32 Mb) in size with average GC content of 38.8%. The draft genomes consisted of an average of 95 (from 49 to 260) contigs above 200 bp (Supplementary Table S1). A total of 83,751 SNPs were identified in the genomes of the 89 P. mirabilis strains, and the phylogenetic tree based on the SNPs was conducted (Figure 1). All 89 strains were divided into 81 different clones (SNPs >5), indicating high genetic diversity of P. mirabilis in food markets. However, four clonal transmission events were also found: (i) four clonal strains JZ5, JZ35, JZ47 and JZ66 (0–1 SNPs) were isolated from pork meat samples in food market F14; (ii) two strains JZ6 and JZ37 (4 SNPs) that harboured blaDHA-1 came from chicken meat samples in F11; (iii) four strains (JZ4, JZ7, JZ53 and JZ70) were clonally related (0–1 SNPs), in which three from aquatic products in F7, but JZ70 from chicken meat in F5; and (iv) strains JZ26 and JZ117 (5 SNPs) respectively came from chicken meat in F14 and aquatic product in F1, both of which harboured tigecycline resistance gene cluster tmexCD3-toprJ1.
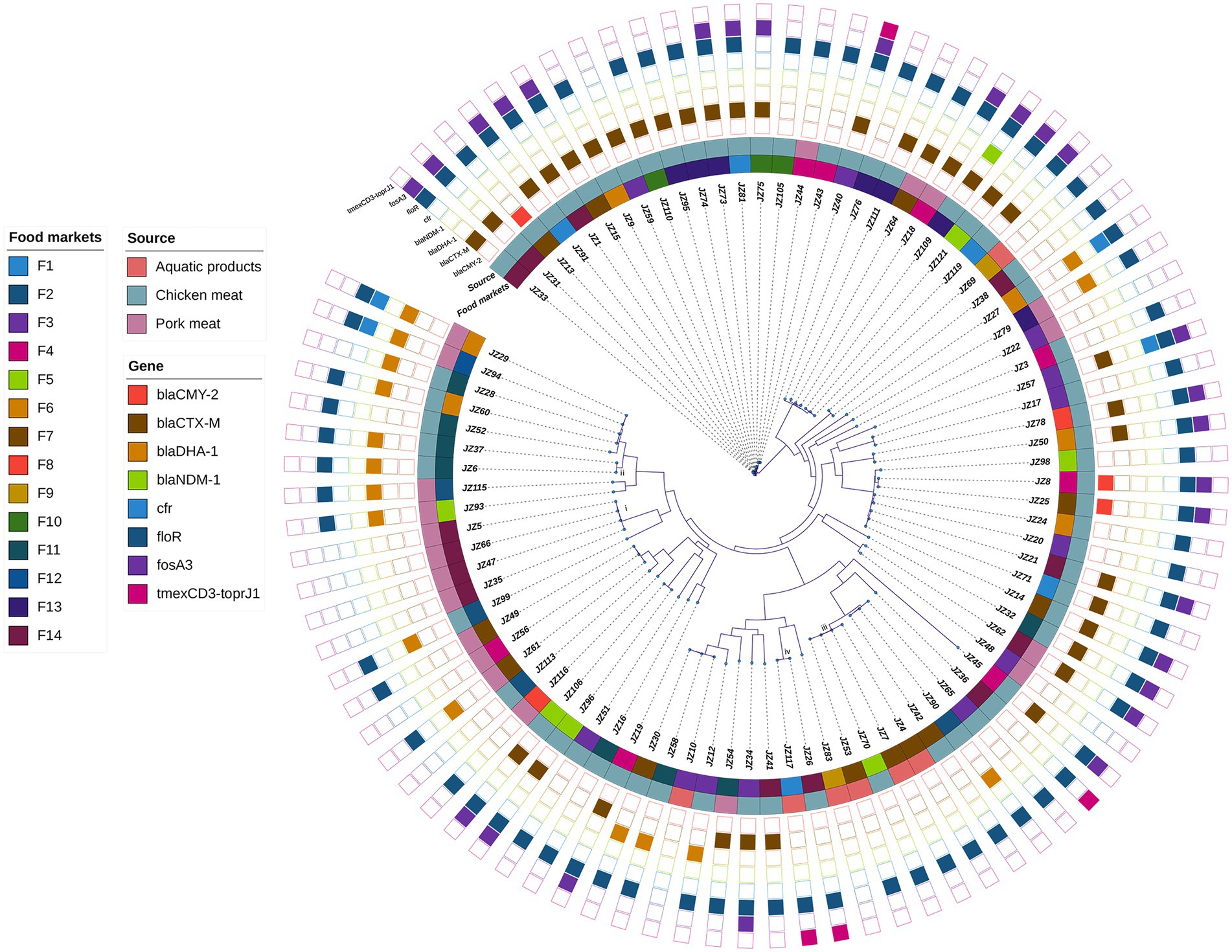
Figure 1. Phylogenetic analysis of 89 P. mirabilis strains. The food markets, sources and the presence of some clinically important antimicrobial resistance genes are indicated by different colours. i–iv represent four clonal transmission events (SNPs ≤5 among strains in the same clade).
Antimicrobial resistance profiles of Proteus mirabilis
Antimicrobial susceptibility testing showed that the P. mirabilis strains exhibited high resistance rate to SXT (94.38%), STR (93.26%), CHL (86.52%), FFC (83.15%) and AMP (83.15%), and low resistance rate to CAZ (16.85%), IPM (13.48%) and AMK (5.62%). The resistance percentage to other agents was as follows: NAL (79.78%), NOR (69.66%), GEN (49.44%), CRO (49.44%), CIP (42.70%), FOS (30.34%), FOX (26.97%) and AMC (22.47%). It’s worth noting that 81 (91.01%) strains displayed MDR profiles.
A total of 73 different resistance genes (or variants) were found in the 89 P. mirabilis strains (Supplementary Table S1). The prevalence of β-Lactam, quinolone, fosfomycin, aminoglycoside, chloramphenicol, sulfonamide, trimethoprim, tetracycline and lincosamide resistance genes were shown in Figure 2. Intrinsical resistance genes tet(J) (tetracycline) and cat (chloramphenicol) were detected in 89 (100%) and 87 (97.75%) strains, respectively. blaOXA-1 was the most prevalent β-lactamase genes, and found in 55 (61.80%) strains. Thirty-five strains carried ESBLs gene blaCTX-M, including 30 blaCTX-M-65, 3 blaCTX-M-14, 1 blaCTX-M-3 and 1 blaCTX-M-55. AmpC β-lactamases genes blaDHA-1 and blaCMY-2 were found in 16 and 3 strains, respectively. More importantly, one strain (JZ109) carried carbapenemase gene blaNDM-1. Quinolone resistance gene aac(6′)-Ib-cr was found in 69 (77.53%) strains. Nineteen strains carried plasmid-mediated quinolone resistance gene qnrD, including 16 qnrD1, 1 qnrD2 and 2 qnrD3. Fosfomycin resistance gene fosA3 was detected in 27 (30.34%) strains. The prevalence of other resistance genes above 60% was as follows: aminoglycoside resistance genes aph(4)-Ia (75.28%), aac(3)-IVa (74.16%), aadA1 (74.16%), aadA2 (73.03%) and aph(3′)-Ia (70.79%); chloramphenicol resistance genes floR (85.39%) and catB3 (62.92%); sulfonamide resistance genes sul1 (88.76%) and sul2 (88.76%). It’s worth noting that multiresistance gene cfr and tigecycline resistance gene cluster tmexCD3-toprJ1 were found in 4 and 4 strains, respectively.
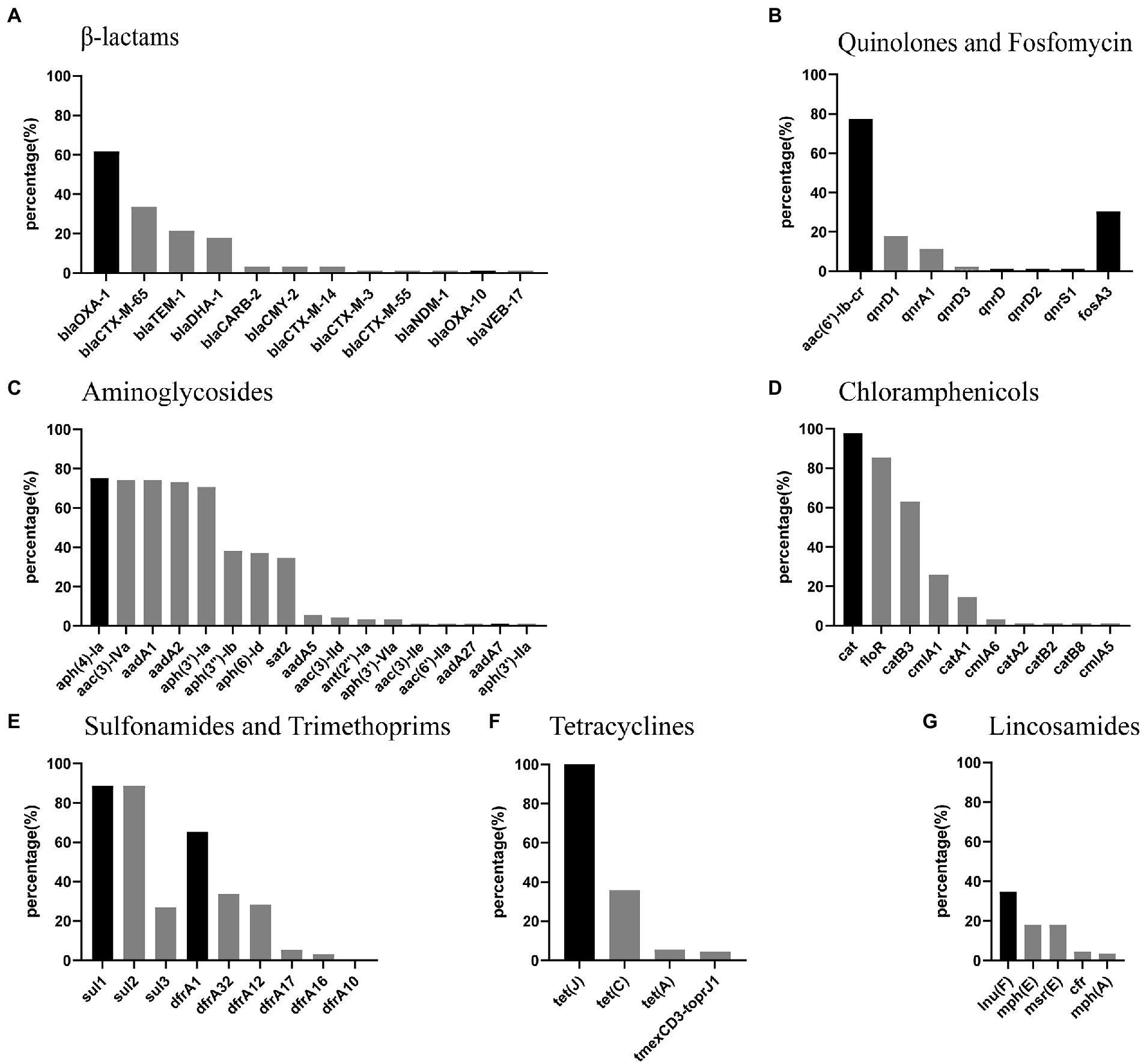
Figure 2. Percentage of antimicrobial resistance genes in P.mirabilis strains from food markets. (A-G) the most prevalent resistance genes for each class of antimicrobials are in deep black. Note: cfr is a multiresistance gene conferring resistance to lincosamides, phenicols, oxazolidinones, pleuromutilins, and streptogramin A.
Genetic environments and transferability of clinically important antimicrobial resistance genes
Genetic environments of clinically important antimicrobial resistance genes cfr, tmexCD3-toprJ1 and blaNDM-1 were characterized, which showed that those genes were located on diverse genetic elements including Tn7 transposon, plasmid, SXT/R391 ICEs and SGI1 (Figure 3).
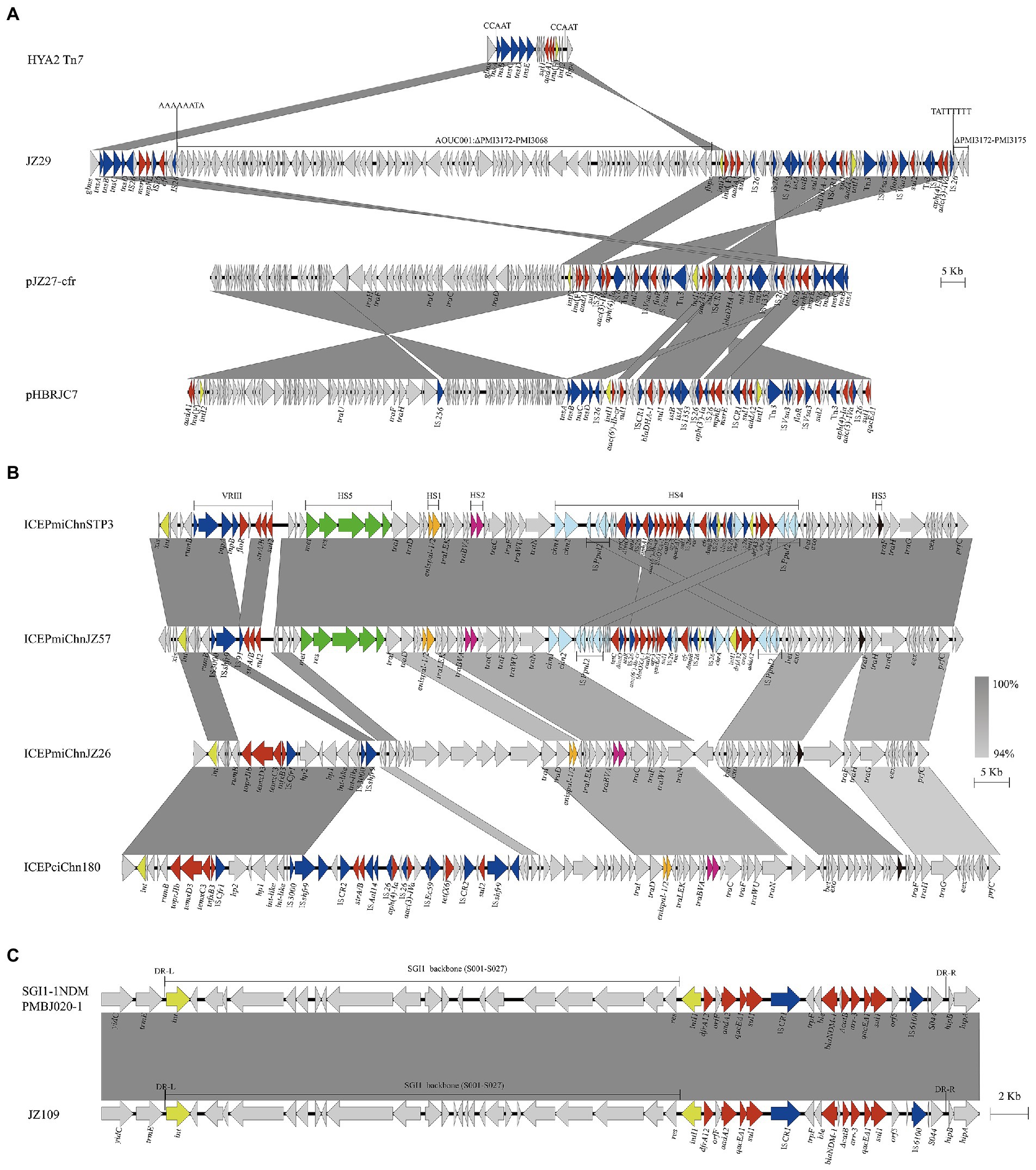
Figure 3. Genetic environments of clinically important antimicrobial resistance genes cfr, tmexCD3-toprJ1 and blaNDM-1. These genes were associated with Tn7 transposons in chromosome or plasmid (A), SXT/R391 ICEs (B), and SGI1 (C). Genes and ORFs are shown as arrows, and their orientations of transcription are indicated by the arrowheads. Shared regions of >99% nucleotide sequence identity are shaded in gray. Integrase genes, resistance genes and transposase genes are in yellow, red, and blue, respectively. HS1-HS5 and VRIII represent the hot spots 1–5 and variable region III in SXT/R391 ICE. DR-L and DR-R represent the 18-bp direct repeats at the ends of SGI1.
cfr gene was located on Tn7 in strains JZ29 and JZ94, pJZ27-cfr in strain JZ27 and ICEPmiChnJZ57 in strain JZ57. Strains JZ29 and JZ94 showed high genetic similarity (18 SNPs) and shared the same genetic environment for cfr. The cfr gene in four strains was flanked by insertion sequence IS26. The Tn7 region in JZ29 chromosome harboured various resistance genes including cfr and blaDHA-1, and was divided into two parts separated by a chromosomal DNA fragment of 112.54 kb (PMI3068-PMI3171 and truncated PMI3172; Figure 3A). We hypothesized that an initial single IS26 transposition event occurred in JZ29 chromosome in locus PMI3172 and has subsequently generated an inversion between two IS26 elements in opposite orientation, resulting in the right end of Tn7 and this 112.54 kb chromosomal DNA fragment being in inverse orientation. The cfr gene in strain JZ27 was also located on a Tn7 in plasmid pJZ27-cfr. pJZ27-cfr was 132,678 bp in size, and showed 100% nucleotide identity to plasmid pHBRJC7 (MK630213) recovered from P. mirabilis strain of broiler origin in China (Chen et al., 2022). Compared with pHBRJC7, pJZ27-cfr supplemented IS26-cfr-repUS18 region (Figure 3A). The ICEPmiChnJZ57 harbouring cfr was 110,965 bp is size, and showed 99.99% nucleotide identity to ICEPmiChnSTP3 (MT449450) in P. mirabilis strain from swine in China (He et al., 2021). ICEPmiChnJZ57 lost floR in variable region VRIII and aphA1 gene in hot spot HS4 compared with ICEPmiChnSTP3 (Figure 3B). Conjugation experiments indicated that pJZ27-cfr and ICEPmiChnJZ57 that harboured cfr gene could be conjugated into E. coli EC600, and ICEPmiChnJZ57 was chromosomally integrated into the 5′ end of the prfC gene.
tmexCD3-toprJ1 was located on a novel SXT/R391 ICE designated as ICEPmiChnJZ26 according to the SXT/R391 nomenclature (Burrus et al., 2006). ICEPmiChnJZ26 was 105,682 bp in size, and had 61% coverage to tmexCD3-toprJ1-harbouring ICEPciChn180 (CP073356) in Proteus terrae subsp. cibarius strain SDQ8C180-2 T from chicken in China (Wang et al., 2021a). tmexCD3-toprJ1 gene cluster was located in VRIII in both ICEPmiChnJZ26 and ICEPciChn180 associated with ISCfr1 (Figure 3B). Conjugative transfer of ICEPmiChnJZ26 failed despite three independent attempts to recover transconjugants carrying ICEPmiChnJZ26. However, the tmexCD3-toprJ1-harbouring ICE was detected in four strains, three of which were not clonally related, indicating the mobility of tmexCD3-toprJ1-harbouring ICE.
The blaNDM-1 in JZ109 was located on the chromosomal SGI1 (Figure 3C), which was 40,275 bp in size and showed 100% nucleotide identity (only one base change) to SGI1-1NDM that was firstly reported in clinical P. mirabilis strain PmBJ020-1 (CP065146) from China (Yang et al., 2021). Mobilization assays confirmed that SGI1-1NDM in JZ109 could be successfully transferred to E. coli and incorporated into the 3′ end of trmE with the help of IncC plasmid pR55.
Materials and methods
Sample collection and Proteus mirabilis identification
A total of 347 fresh samples (114 chicken meat, 141 pork meat and 92 aquatic products) were purchased from 14 food markets at different locations in Chengdu city, Sichuan province of China from December 2021 to January 2022. The numbers of market stalls selling retail meat and aquatic products were varied from 0 to 20. We collected only one sample from each market stall. The meat samples were stored in sterile bags at 4°C between sampling and further processing. The processing method of P. mirabilis identification was modified from the protocol reported previously (Wong et al., 2013). All culture mediums were purchased from Beijing Land Bridge Technology Co., Beijing, China. Briefly, 25 g of meat or aquatic product was placed in 100 ml of buffered peptone water (BPW) and incubated at 37°C for 24 h. Then, the 1 ml BPW-incubated culture was mixed with 9 ml of Rappaport-Vassiliadis Salmonella enrichment broth and incubated at 42°C for 24 h. One loopful of broth was streaked onto Mueller Hinton Agar and incubated at 37°C for 16 h to check for the motility of the isolates. The colonies with swarming growth phenotype were streaked on Shigella and Salmonella agar to obtain single clone. P. mirabilis isolates were confirmed by using the BD Phoenix™ 100 Automated Microbiology System (Becton Dickinson, United States). Only one isolate from each sample was selected for further study.
Genome sequencing, assembly, and bioinformatics analysis
Genomic DNA was extracted using a bacterial genomic DNA extraction kit (Tiangen, China). Whole genomes were sequenced using the Illumina HiSeq platform (150 bp paired-end reads with about 200-fold average coverage), and the clear data were assembled to draft genomes using the software SPAdes v3.15.4. Antimicrobial resistance genes were found using ResFinder 4.1.1 SNPs from genomes of the P. mirabilis strains were called and a phylogeny based on the concatenated alignment of the high-quality SNPs was inferred by using CSI Phylogeny 1.42 with parameters as defaults. A threshold of 5 SNPs among isolates was considered likely to have an clonal relationship (Dallman et al., 2015; Kanagarajah et al., 2018). The phylogenetic tree was modified in Itol.3
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed using the standard Kirby–Bauer disk diffusion method according to Clinical and Laboratory Standards Institutes (CLSI) guidelines (CLSI, 2020). Susceptibilities to 16 antimicrobial agents were tested including ampicillin (AMP), amoxicillin-clavulanate acid (AMC), cefotaxime (CTX), ceftazidime (CAZ), cefoxitin (FOX), imipenem (IPM), nalidixic acid (NAL), norfloxacin (NOR), ciprofloxacin (CIP), streptomycin (STR), gentamicin (GEN), amikacin (AMK), chloramphenicol (CHL), florfenicol (FFC), fosfomycin (FOS), and trimethoprim-sulfamethoxazole (SXT). Escherichia coli ATCC25922 was used as quality control. MDR was defined as resistance to at least one agent in three or more chemical classes of antimicrobials.
Genetic environments analysis of clinically important antimicrobial resistance genes
The genomes of the P. mirabilis strains harbouring cfr, tmexCD3-toprJ1 and blaNDM-1 were further sequenced using Nanopore MinION using Rapid Sequencing Kit. The complete genome sequences were assembled by Unicycler v0.5.0 using Nanopore sequencing data combined with Illumina sequencing data. Genetic environments of cfr, tmexCD3-toprJ1 and blaNDM-1 were analyzed using the BLAST program4 and Easyfig v2.2.2.5
Conjugation and mobilization assays
Conjugation experiments were performed by using P. mirabilis JZ27 (harbouring plasmid pJZ27-cfr), JZ26 (harbouring ICEPmiChnJZ26), and JZ57 (harbouring ICEPmiChnJZ57) as the donor strains and rifampin-resistant E. coli EC600 as the recipient strain with selection on nutrient agar plates containing 300 mg/l rifampin and 8 mg/l florfenicol (for cfr) or 2 mg/l tigecycline (for tmexCD3-toprJ1). The positive transconjugants were further determined by detection of antimicrobial resistance profile and the location of ICEPmiChnJZ26 and ICEPmiChnJZ57 in E. coli with primers LE1/LE4 and RE4/RE1 (McGrath et al., 2006). Mobilization assay for SGI1 harbouring blaNDM-1 was carried out as previously described (Siebor et al., 2016), using E. coli EC600 harboring an IncC plasmid pR55 as the recipient strain. The transconjugants (EC600 carrying SGI1) were further examined for the presence of the blaNDM-1 and the location of SGI1 in E. coli (Girlich et al., 2015; Siebor et al., 2016).
Nucleotide sequence accession numbers
The genomes of 89 P. mirabilis strains reported in this study have been deposited in National Center for Biotechnology Information and registered BioProject number PRJNA847328. The nucleotide sequences of ICEPmiChnJZ57, Tn7 in JZ29, pJZ27-cfr, SGI1 in JZ109 and ICEPmiChnJZ26 have been deposited in GenBank under accession numbers ON553412, ON553413, ON553414, ON553415 and ON630400, respectively.
Discussion
Food contamination caused by foodborne pathogenic bacteria, such as Salmonella and Campylobacter, has attracted worldwide attention. However, the prevalence data of the opportunistic pathogen in food products is limited. In this study we focused on P. mirabilis, a species frequently associated with nosocomial infection and food poisoning cases. By using NGS, we confirmed high contamination of P. mirabilis that harboured various clinically important antimicrobial resistance genes in retail meat and aquatic products.
Our study found that the isolation rate of P. mirabilis in retail meat products, especially in chicken meat (isolation rate, 54.39%), was higher than that in aquatic products. This might be due to the fact that the meat products were contaminated by P. mirabilis from intestinal tracts of chickens and swines at slaughter. A study from Hong Kong, China reported that 85% (50/58) raw chicken carcass samples were positive for P. mirabilis (Wong et al., 2013). Another study from Belgium also reported that P. mirabilis was isolated from 29 out of 80 broiler carcasses (36.25%) (Yu et al., 2021). These results indicated that meat products, especially chicken products, were seriously contaminated with P. mirabilis. More stringent biosecurity measures are needed in slaughtering, processing and retailing to control t0not only pathogenic bacteria but also opportunistic pathogens.
NGS technology is more and more widely used in tracing the transmission of pathogenic microorganisms. Molecular typing based on whole genome data has higher resolution than traditional PFGE, and can obtain genes information including antimicrobial resistance genes (Boolchandani et al., 2019), which has greater advantage as the cost of NGS decreases. However, genomic data of P. mirabilis from food sources is limited. In this study, we provided genomic data of 89 P. mirabilis strains, and the phylogenetic analysis showed that the strains had high genetic diversity, indicating that those strains had diverse origin. We also found that the clonal transmission could have occurred in different food markets and different meat products. Given that different meat products in different food markets in Chengdu city might come from the same wholesale meat market, It might be due to the presence of cross contamination of P. mirabilis in different meat products from the same wholesale market. Further studies can obtain more genomic data of P. mirabilis from animal, food and clinical sources to trace the spread of P. mirabilis in the food chain.
Antimicrobial resistance in P. mirabilis has become a serious issue for public health (Girlich et al., 2020). The ESBLs and fluoroquinolones resistance genes have been prevalent in P. mirabilis (Wong et al., 2013; Huang et al., 2015; Li et al., 2022). In this study, we found that 91.01% strains displayed MDR profiles. Fluoroquinolones and aminoglycosides resistance gene aac(6′)-Ib-cr, ESBLs blaCTX-M (especially variant blaCTX-M-65) and fosfomycin resistance gene fosA3 were prevalent in P. mirabilis in retail meat and aquatic products. It is worth noting P. mirabilis strains also harboured cfr, tmexCD3-toprJ1 and blaNDM-1 conferring resistance to the last-resort drugs oxazolidinones, tigecycline and carbapenems, respectively. Diverse genetic elements including Tn7 transposon, plasmid, SXT/R391 integrative conjugative elements and Salmonella genomic island 1, were associated with the horizontal transfer of those resistance genes, which would not only promote the spread of those resistance genes in P. mirabilis, but also might spread to Enterobacteriaceae pathogens including Salmonella.
Conclusion
Our study revealed high contamination of P. mirabilis harbouring various clinically important antimicrobial resistance genes in retail meat and aquatic products from food markets in China. The last-resort drugs resistance genes were associated with diverse mobile genetic elements, and had potential to spread to Enterobacteriaceae pathogens. More attention should be paid to monitor the presence of MDR opportunistic pathogen in retail meat products.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
W-QM, Y-YH, and LZ wrote the manuscript and collected the data. Other authors have given some help and suggestions during the experiment and the writing of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (grant nos. 32100147 and U21A20257), Natural Science Foundation of Sichuan Province (2022NSFSC0076) and the Sichuan Science and Technology Program (2021ZDZX0010).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1086800/full#supplementary-material
Footnotes
1. ^https://cge.cbs.dtu.dk/services/ResFinder/
2. ^https://cge.cbs.dtu.dk/services/CSIPhylogeny/
References
Bitar, I., Mattioni Marchetti, V., Mercato, A., Nucleo, E., Anesi, A., Bracco, S., et al. (2020). Complete genome and plasmids sequences of a clinical Proteus mirabilis isolate producing plasmid mediated NDM-1 from Italy. Microorganisms 8 :339. doi: 10.3390/microorganisms8030339
Boolchandani, M., Dsouza, A. W., and Dantas, G. (2019). Sequencing-based methods and resources to study antimicrobial resistance. Nat. Rev. Genet. 20Placeholder Text, 356–370. doi: 10.1038/s41576-019-0108-4
Burrus, V., Marrero, J., and Waldor, M. K. (2006). The current ICE age: biology and evolution of SXT-related integrating conjugative elements. Plasmid 55Placeholder Text, 173–183. doi: 10.1016/j.plasmid.2006.01.001
Chen, X., Lei, C. W., Liu, S. Y., Li, T. Y., Chen, Y., Wang, Y. T., et al. (2022). Characterisation of novel Tn7-derivatives and Tn7-like transposon found in Proteus mirabilis of food-producing animal origin in China. J. Glob. Antimicrob. Resist. 28, 233–237. doi: 10.1016/j.jgar.2022.01.012
Clinical and Laboratory Standards Institute(2020). Performance Standards for Antimicrobial Susceptibility Testing. 30th Edn. M100. Wayne, PA.
Cummins, M. L., Hamidian, M., and Djordjevic, S. P. (2020). Salmonella Genomic Island 1 is broadly disseminated within Gammaproteobacteriaceae. Microorganisms 8Placeholder Text,:161. doi: 10.3390/microorganisms8020161
Dallman, T. J., Byrne, L., Ashton, P. M., Cowley, L. A., Perry, N. T., Adak, G., et al. (2015). Whole-genome sequencing for national surveillance of Shiga toxinproducing Escherichia coli O157. Clin. Infect. Dis. 61Placeholder Text, 305–312. doi: 10.1093/cid/civ318
Girlich, D., Bonnin, R. A., Dortet, L., and Naas, T. (2020). Genetics of acquired antibiotic resistance genes in Proteus spp. Front. Microbiol. 11:256. doi: 10.3389/fmicb.2020.00256
Girlich, D., Dortet, L., Poirel, L., and Nordmann, P. (2015). Integration of the blaNDM-1 carbapenemase gene into Proteus genomic island 1 (PGI1-PmPEL) in a Proteus mirabilis clinical isolate. J. Antimicrob. Chemother. 70 Placeholder Text, 98–102. doi: 10.1093/jac/dku371
Gong, Z., Shi, X., Bai, F., He, X., Zhang, H., Li, Y., et al. (2019). Characterization of a novel Diarrheagenic strain of Proteus mirabilis associated with food poisoning in China. Front. Microbiol. 10:2810. doi: 10.3389/fmicb.2019.02810
He, J., Lei, C., Li, C., Wang, X., Cui, P., and Wang, H. (2021). Identification of a novel genomic resistance island PmGRI1-STP3 and an SXT/R391 integrative conjugative element in Proteus mirabilis of swine origin in China. J. Glob. Antimicrob. Resist. 25, 77–81. doi: 10.1016/j.jgar.2021.02.018
Huang, C. W., Chien, J. H., Peng, R. Y., Tsai, D. J., Li, M. H., Lee, H. M., et al. (2015). Molecular epidemiology of CTX-M-type extended-spectrum beta-lactamase-producing Proteus mirabilis isolates in Taiwan. Int. J. Antimicrob. Agents 45 Placeholder Text, 84–85. doi: 10.1016/j.ijantimicag.2014.09.004
Janda, J. M., and Abbott, S. L. (2021). The changing face of the family Enterobacteriaceae (order: "Enterobacterales"): new members, taxonomic issues, geographic expansion, and new diseases and disease syndromes. Clin. Microbiol. Rev. 34Placeholder Text, e00174–e00120. doi: 10.1128/CMR.00174-20
Kanagarajah, S., Waldram, A., Dolan, G., Jenkins, C., Ashton, P. M., Martin, C., et al. (2018). Whole genome sequencing reveals an outbreak of salmonella Enteritidis associated with reptile feeder mice in the United Kingdom, 2012-2015. Food Microbiol. 71, 32–38. doi: 10.1016/j.fm.2017.04.005
Li, Z., Peng, C., Zhang, G., Shen, Y., Zhang, Y., Liu, C., et al. (2022). Prevalence and characteristics of multidrug-resistant Proteus mirabilis from broiler farms in Shandong Province. China. Poult. Sci. 101 Placeholder Text:101710. doi: 10.1016/j.psj.2022.101710
Lv, L., Wan, M., Wang, C., Gao, X., Yang, Q., Partridge, S. R., et al. (2020). Emergence of a plasmid-encoded resistance-nodulation-division efflux pump conferring resistance to multiple drugs, including Tigecycline, in Klebsiella pneumoniae. MBio 11Placeholder Text, e02930–e02919. doi: 10.1128/mBio.02930-19
McGrath, B. M., O'Halloran, J. A., Piterina, A. V., and Pembroke, J. T. (2006). Molecular tools to detect the IncJ elements: a family of integrating, antibiotic resistant mobile genetic elements. J. Microbiol. Methods 66Placeholder Text, 32–42. doi: 10.1016/j.mimet.2005.10.004
Partridge, S. R., Kwong, S. M., Firth, N., and Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31Placeholder Text, e00088–e00017. doi: 10.1128/CMR.00088-17
Schaffer, J. N., and Pearson, M. M. (2015). Proteus mirabilis and urinary tract infections. Microbiol. Spectr. 3Placeholder Text. doi: 10.1128/microbiolspec.UTI-0017-2013
Schurch, A. C., Arredondo-Alonso, S., Willems, R. J. L., and Goering, R. V. (2018). Whole genome sequencing options for bacterial strain typing and epidemiologic analysis based on single nucleotide polymorphism versus gene-by-gene-based approaches. Clin. Microbiol. Infect. 24Placeholder Text, 350–354. doi: 10.1016/j.cmi.2017.12.016
Shen, J., Wang, Y., and Schwarz, S. (2013). Presence and dissemination of the multiresistance gene cfr in gram-positive and gram-negative bacteria. J. Antimicrob. Chemother. 68Placeholder Text, 1697–1706. doi: 10.1093/jac/dkt092
Siebor, E., de Curraize, C., Amoureux, L., and Neuwirth, C. (2016). Mobilization of the salmonella genomic island SGI1 and the Proteus genomic island PGI1 by the a/C2 plasmid carrying blaTEM-24 harboured by various clinical species of Enterobacteriaceae. J. Antimicrob. Chemother. 71Placeholder Text, 2167–2170. doi: 10.1093/jac/dkw151
Song, Z., Lei, C. W., Zuo, L., Li, C., Wang, Y. L., Tian, Y. M., et al. (2022). Whole genome sequence of Proteus mirabilis ChSC1905 strain harbouring a new SXT/R391-family ICE. J. Glob. Antimicrob. Resist. 30, 279–281. doi: 10.1016/j.jgar.2022.07.004
Tropea, A. (2022). Microbial contamination and public health: an overview. Int. J. Environ. Res. Public Health 19Placeholder Text,:7441. doi: 10.3390/ijerph19127441
Wang, C. Z., Gao, X., Lv, L. C., Cai, Z. P., Yang, J., and Liu, J. H. (2021a). Novel tigecycline resistance gene cluster tnfxB3-tmexCD3-toprJ1b in Proteus spp. and Pseudomonas aeruginosa, co-existing with tet(X6) on an SXT/R391 integrative and conjugative element. J. Antimicrob. Chemother. 76Placeholder Text, 3159–3167. doi: 10.1093/jac/dkab325
Wang, Q., Peng, K., Liu, Y., Xiao, X., Wang, Z., and Li, R. (2021b). Characterization of TMexCD3-TOprJ3, an RND-type efflux system conferring resistance to Tigecycline in Proteus mirabilis, and its associated integrative conjugative element. Antimicrob. Agents Chemother. 65Placeholder Text:e0271220. doi: 10.1128/AAC.02712-20
Wong, M. H., Wan, H. Y., and Chen, S. (2013). Characterization of multidrug-resistant Proteus mirabilis isolated from chicken carcasses. Foodborne Pathog. Dis. 10Placeholder Text, 177–181. doi: 10.1089/fpd.2012.1303
Yang, L., He, H., Chen, Q., Wang, K., Lin, Y., Li, P., et al. (2021). Nosocomial outbreak of Carbapenemase-producing Proteus mirabilis with two novel salmonella Genomic Island 1 variants carrying different blaNDM-1 gene copies in China. Front. Microbiol. 12:800938. doi: 10.3389/fmicb.2021.800938
Keywords: Proteus mirabilis, food markets, whole genome sequencing, multidrug resistance, NDM, mobile genetic elements
Citation: Ma W-Q, Han Y-Y, Zhou L, Peng W-Q, Mao L-Y, Yang X, Wang Q, Zhang T-J, Wang H-N and Lei C-W (2022) Contamination of Proteus mirabilis harbouring various clinically important antimicrobial resistance genes in retail meat and aquatic products from food markets in China. Front. Microbiol. 13:1086800. doi: 10.3389/fmicb.2022.1086800
Edited by:
Haijian Zhou, National Institute for Communicable Disease Control and Prevention, ChinaReviewed by:
Tao He, Jiangsu Academy of Agricultural Sciences, ChinaBasil Britto Xavier, University of Antwerp, Belgium
Valentine Usongo, Health Canada, Canada
Copyright © 2022 Ma, Han, Zhou, Peng, Mao, Yang, Wang, Zhang, Wang and Lei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chang-Wei Lei, leichangwei@126.com; leichangwei@scu.edu.cn
†These authors have contributed equally to this work
 Wan-Qing Ma†
Wan-Qing Ma† Ying-Yue Han
Ying-Yue Han Lin Zhou
Lin Zhou Wen-Qi Peng
Wen-Qi Peng Tie-Jun Zhang
Tie-Jun Zhang Hong-Ning Wang
Hong-Ning Wang Chang-Wei Lei
Chang-Wei Lei