- 1Department of Biological Oceanography, Leibniz Institute for Baltic Sea Research, Warnemünde (IOW), Rostock, Germany
- 2Department of Plant Physiology, Institute for Biosciences, University of Rostock, Rostock, Germany
Nodularia spumigena is a toxic, filamentous cyanobacterium capable of fixing atmospheric N2, which is often dominating cyanobacterial bloom events in the Baltic Sea and other brackish water systems worldwide. Increasing phosphate limitation has been considered as one environmental factor promoting cyanobacterial mass developments. In the present study, we analyzed the response of N. spumigena strain CCY9414 toward strong phosphate limitation. Growth of the strain was diminished under P-deplete conditions; however, filaments contained more polyphosphate under P-deplete compared to P-replete conditions. Using RNA-seq, gene expression was compared in N. spumigena CCY9414 after 7 and 14 days in P-deplete and P-replete conditions, respectively. After 7 days, 112 genes were significantly up-regulated in P-deplete filaments, among them was a high proportion of genes encoding proteins related to P-homeostasis such as transport systems for different P species. Many of these genes became also up-regulated after 14 days compared to 7 days in filaments grown under P-replete conditions, which was consistent with the almost complete consumption of dissolved P in these cultures after 14 days. In addition to genes directly related to P starvation, genes encoding proteins for bioactive compound synthesis, gas vesicles formation, or sugar catabolism were stimulated under P-deplete conditions. Collectively, our data describe an experimentally validated P-stimulon in N. spumigena CCY9414 and provide the indication that severe P limitation could indeed support bloom formation by this filamentous strain.
1. Introduction
Mass developments, so-called blooms, of toxic cyanobacteria occur worldwide in freshwater or coastal brackish water systems and are of increasing concern, as they negatively impact the use of water for drinking and/or for recreation purposes. Global warming and climate change scenarios are expected to increase the frequency of bloom events in the next years (Paerl and Huisman, 2008). Cyanobacterial blooms are usually dominated by toxic colony-forming Microcystis spp. strains in freshwater. Filamentous cyanobacteria capable of atmospheric N2 fixation in heterocysts are frequently occurring in brackish water blooms, where Nodularia spumigena is often dominating. This filamentous cyanobacterium can produce the potent hepatotoxin nodularin and occurs worldwide in coastal waters (Sivonen et al., 1989; Gehringer and Wannicke, 2014).
Key factors favoring growth and bloom formation of N2-fixing, filamentous cyanobacteria in the Baltic Sea include the availability of phosphorus (P) sources in combination with low to undetectable combined nitrogen concentrations (Sellner, 1997). The virtual absence of combined nitrogen sources after the diatom spring bloom promotes the dominance of N2-fixing cyanobacteria in the Baltic Sea during the summer. The increase in the cyanobacterial population then leads to a further decrease of available P sources triggering by so far unknown mechanisms a subsequent mass development of filamentous N2-fixing cyanobacterial strains. These nutrient relations are the prevailing conditions in summer when the upper water layer is thermally stratified. The gas vesicles of N. spumigena and other bloom-forming cyanobacteria provide buoyancy, leading to the formation of large surface scums in the absence of mixing. A correlation between biologically available dissolved inorganic and organic P forms and Nodularia spp. bloom formation is also evident in the Baltic Sea (Nausch et al., 2008; Vahtera et al., 2010). Moreover, increased expression of the gene cluster for nodularin synthesis during P-depletion has been reported (Jonasson et al., 2008), although the amount of nodularin did not change at different P conditions (Repka et al., 2001).
Molecular regulation of the acclimation to different P availability has been intensively studied in model cyanobacteria such as Synechocystis sp. PCC 6803. Basically, the sensing and acclimation to P limitation seems to be similar to that in Escherichia coli, i.e., a two-component system PhoB/PhoR (SphB/SphR) senses the P status and induces a defined P-regulon under P-limiting conditions as has been shown using DNA-microarray-based transcriptomics (e.g., Suzuki et al., 2004). As in many other bacteria, the regulon comprises transporters for inorganic phosphate, such as two ABC type transporters of different affinity for orthophosphate (from here on o-phosphate) import, Pst1 and Pst2 (Pitt et al., 2010). Furthermore, alkaline phosphatase, an exoenzyme known to be involved in the release of o-phosphate from organic phosphates is part of the Sph-regulon in Synechocystis sp. PCC 6803 (Suzuki et al., 2004), which is typically overexpressed under P limitation in many other cyanobacteria as well (e.g., Kelly et al., 2019; Yuan et al., 2019). A similar role of PhoB and PhoR in the regulation of P-limitation associated genes has been shown for marine Synechococcus sp. WH8102 (Tetu et al., 2009). Subsequent studies revealed that an additional regulator protein PtrA is also necessary for the coordinated expression of P-regulated genes in this marine cyanobacterium (Ostrowski et al., 2010).
Furthermore, cyanobacteria and microalgae can accumulate polyphosphate that can serve as storage for excess phosphate and/or energy (Sanz-Luque et al., 2020). In enterobacteria as in model cyanobacteria such as Synechocystis sp. PCC 6803 polyphosphate accumulation is induced when cells are shifted from P-deplete into P-replete conditions to store the surplus phosphate inside the cell (Voronkov and Sinetova, 2019). Consistently, a knock-out mutant in polyphosphate degradation cannot properly acclimate to P limitation (Hiyoshi et al., 2021). However, there are also hints that polyphosphate accumulation and mobilization is not always strictly related to the P status among different cyanobacteria (Li et al., 2019; Wan et al., 2019). It has been reported that polyphosphate is stored in filamentous strains under P-deplete conditions without its mobilization to sustain growth (e.g., Hagemann et al., 2019), which may serve as P storage for the next year’s generation. In contrast to model strains such as Synechocystis sp. PCC 6803, environmentally important filamentous strains such as Nodularia spp. seem to have a much greater capability to deal with different P availability. For example, in the genome of N. spumigena strain CCY9414 many genes for the acquisition of different inorganic and organic P-sources have been annotated (Voss et al., 2013). In addition to o-phosphate, transporters and associated enzymes for the utilization of phosphonates and phosphites are present in its genome. Recently, it has been shown that many bacteria including diverse cyanobacterial species in marine and brackish systems can produce and utilize phosphonates to sustain growth under different nutrient availability (e.g., Acker et al., 2022; Rabouille et al., 2022; Zhao et al., 2022).
The Baltic Sea isolate N. spumigena strain CCY9414 (from here on Nodularia CCY9414) represents an ecologically relevant model organism suitable to analyze the molecular response to P limitation and elucidate its role in the formation of recurring summer blooms. Therefore, we grew Nodularia CCY9414 in P-replete and P-deplete media and compared polyphosphate accumulation as well as gene expression changes to gain insights into the acquisition and utilization of P sources under conditions representative of bloom events. Our results showed that P-limited cultures of Nodularia CCY9414 accumulate more polyphosphate than P-replete filaments despite a harsh P starvation as indicated by the induction of many genes related to low P availability. In addition to genes for transporters involved in the uptake of inorganic and organic P sources, genes for bioactive compounds, gas vesicle formation, and sugar metabolism were strongly up-regulated under P limitation in Nodularia CCY9414.
2. Materials and methods
2.1. Strain and cultivation
Nodularia spumigena is a toxic, filamentous planktonic, heterocystous, gas-vacuolate cyanobacterium, which is representative of surface bloom-forming cyanobacteria in brackish waters. The strain N. spumigena CCY9414 was initially isolated from samples collected from surface water in the Bornholm Sea (Hayes and Barker, 1997; Stal et al., 2003; Voss et al., 2013). Before the experiment, Nodularia CCY9414 cells were pre-cultivated in P-replete medium for 10 days to induce exponential growth. The P-replete medium was composed as follows: 33% ASNIII and 67% BG11 medium mixture [as described by Hagemann et al. (2019)], with 0.02 g L–1 K2HPO4 in the modified ASNIII (Rippka et al., 1979) and 0.00078 mg L–1 K2HPO4 in BG11 medium modified with 20 mM TES buffer to pH 8 (Rippka et al., 1979). Omitting the inorganic N-source nitrate induced N2-fixing conditions. Furthermore, the final NaCl concentration of the growth medium was set to 10.3 g L–1, which is corresponding to the salt optimum of Nodularia CCY9414 (Möke et al., 2013). Sterile cell culture flasks (Greiner Bio-One GmbH, Frickenhausen, Germany) were used for both pre-cultivation and afterward for the experimental cultures. Each culture flask contained 10 mL of starting culture suspension and 90 mL of fresh medium and was manually mixed daily. Incubations were performed at a temperature of 19.5 – 20°C at a light/dark cycle of 16 h/8 h. During the light phase, cell suspensions were constantly exposed to 40 μmol photons m–2 s–1. Every 5 days the cells were transferred into fresh P-replete medium to prolong the exponential growth until the planned start of the experiment.
At the beginning of the experiment, all the pre-cultures were combined and mixed in a glass bottle for a total volume of 500 mL. Filaments were then separated from the medium by filtration through glass fiber filters (25 mm circle diameter; GE Healthcare, Chicago, IL, United States). Half of the biomass on the filters were washed off and inoculated into 1.2 L of fresh medium but without added inorganic phosphate (P-deplete), which was then divided into 12 culture flasks with 100 mL suspension each. These P-deplete cultures contained only traces of inorganic phosphate impurities from other chemicals in the medium and from the inoculation of the pre-experimental cultures. The final concentration of o-phosphate was less than 1 μM. The remaining half of the biomass was washed off and inoculated into 1.2 L of fresh P-replete medium equally divided in 12 culture flasks. Samples were taken by sacrificing three culture flasks of each P condition at the beginning of the experiment (d0), after 7 (d7) and 14 days (d14), and at the end of the experiment after 21 days (d21). The experiment was repeated two times independently.
2.2. Dry weight and polyphosphate extractions
For dry weight estimations, three 5 mL aliquots from each culture were filtered on pre-weighted glass fiber filters of 25 mm diameter (GE Healthcare, Chicago, IL, United States) and dried at 65°C overnight. The mean dry weight of these three filters was taken to obtain a representative estimate for each culture flask.
Cells for polyphosphate quantification were collected from 3 mL aliquots on polycarbonate filters of 0.22 μm pore size and 22.5 mm diameter (GE Healthcare, Chicago, IL, United States), placed on ice and stored at −80°C until further processing. Extraction and fluorometric quantification of polyphosphate was done as described by Martin and Van Mooy (2013). Briefly, reagents were obtained from the following providers: Proteinase K (BP1700) was obtained from Sigma-Aldrich Chemie GmbH (Schnelldorf, Germany); lysozyme (BP535), ADP (A2754), Ambion recombinant DNase (AM2235) and RNase cocktail (AM2286), and DAPI (4′,6-diamidino-2-phenylindole) were from Thermo Fischer Scientific (Langenselbold, Germany). Before the polyphosphate extraction, cells on filter were dried for 5 h at 35°C to estimate their dry weight to standardize polyphosphate content. Then, cells were scraped off the filters with a spatula and resuspended in 3 mL polyphosphate buffer (20 mM HEPES, 100 mM NaCl, 2 mM EDTA, and 2 mM MgCl2). The dissolved polyphosphate extraction required eight freeze-thaw cycles (freeze at −20°C for 2 h and 30 min, thaw at 35°C for 40 min) for lysis of the cells. Subsequent boiling step and enzymatic digestion were performed as described in the method of Martin and Van Mooy (2013). To stain the dissolved polyphosphates 5 μL of DAPI (1 mg mL–1) were added to each extract. The measurements were made on a Tecan Infinite 200 PRO multimode plate reader (Tecan Austria GmbH, Grödig, Austria) at an excitation wavelength of 415 nm and emission wavelength of 550 nm, with an integration time of 500 μs. Fluorescence emission spectra were acquired at 415 nm and emission from 230 nm to 850 nm in 1-nm increments and integrated for 20 μs. All bandwidths were 5 nm. The calibration curve was constructed from commercial polyphosphate with a chain length of 45 ± 5 residues (Sigma-Aldrich, S4379) in the range of 0.2–7.0 nmol polyphosphates.
2.3. Inorganic phosphate content
During the second iteration of the experiment, aliquots of 10 mL were collected from each culture flask to measure the level of the o-phosphate (PO43–) in the medium. Each aliquot was filtered through a glass fiber filter (25 mm circle diameter; GE Healthcare, Chicago, IL, United States), and o-phosphate and ammonium concentrations of the filtrate were measured colorimetrically according to Grasshoff et al. (2009) by means of a Seal Analytical QuAAtro constant flow analyzer (Seal Analytical GmbH, Norderstedt, Germany).
2.4. DAPI staining and epifluorescence microscopy
An aliquot of 1.5 mL was collected from each culture, fixed with 4% formaldehyde (v/v) and stored at −20°C until DAPI staining. After thawing, the filaments were harvested on polycarbonate membrane filters (0.88 μm pore size, 22.5 mm circle diameter, GE Healthcare, Chicago, IL, United States) and stained with 50 μl of DAPI solution (1 mg mL–1, Thermo Fischer Scientific, Langenselbold, Germany). Staining was performed in the dark at room temperature for 2 min. Then, the dye was filtered off, the filter was rinsed with ultra-pure water and dried in the dark at room temperature for 5 min. Polyphosphates in the stained filaments were visualized under a fluorescence microscope (Axioskop 2 mot PLUS, Carl Zeiss, Jena, Germany) with the specific DAPI filter set (excitation: BP 390/22, beam Splitter: FT 420, emission: 460/50). Polyphosphate chains longer than 15 P-subunits formed a complex with the dye that increased its fluorescence and shifted its absorption spectrum from 456 nm – when only DNA is visualized via blue fluorescence emission – to 526 nm via bright yellow fluorescence of the accumulated polyphosphate granules (Tijssen et al., 1985; Ohtomo et al., 2008; Diaz and Ingall, 2010).
2.5. RNA isolation
Filaments from aliquots of 30 mL of each culture were transferred to a 50 mL tube (Sarstedt, Nümbrecht, Germany) and immediately fixed for RNA extraction with 6 mL of a solution containing 95% (v/v) ethanol (molecular biology grade; Roth, Karlsruhe, Germany) and 5% (v/v) Roti-Aqua-Phenol (Roth, Karlsruhe, Germany). The fixed suspension was incubated in the dark at room temperature for 20 min to complete the collapse of gas vesicles before each sample was placed on ice. Then, cells were harvested by centrifugation at 12.000 g at 4°C for 8 min. The supernatant was quickly removed, and the collected pellets were flash frozen in liquid nitrogen until being processed for RNA isolation. The cell pellets were then suspended in 1 mL TRIzol reagent (Sigma-Aldrich, Steinheim am Albuch, Germany) and three spoons of acid-washed mixed glass beads of 425–600 μm (Sigma-Aldrich, Steinheim am Albuch, Germany) were added for bead beating for 3 × 30 s. Then, the suspension was heated at 65°C for 10 min and RNA was isolated as described by Steunou et al. (2006). RNA extracts were treated three times with two units of DNase I RNase-free (New England Biolabs, Frankfurt am Main, Germany) for 30 min as recommended by the manufacturer. DNase I was inactivated and removed with phenol/chloroform and total RNA was then precipitated with 3 M Na-Acetate pH 5.2 and 2.5 volumes of absolute Ethanol. RNA extracts were checked for DNA contamination by PCR using primers specific for the gene encoding Fe-superoxide dismutase subunit B (sodB forward: GACTCCTCTAAGGTGGGAATC; sodB reverse: CCCAGACATCCAAGGTTAAG) as was previously done by Kopf et al. (2015).
2.6. cDNA library preparation and sequence processing
Non-stranded rRNA-depleted libraries were prepared by the sequencing company LGC (LGC, Biosearch Technologies, Berlin, Germany) for 12 samples, collected at d7 and d14 for both P conditions from the second iteration of the experiment. Briefly, the total RNA was first depleted of rRNA using the Pan-Prokaryote riboPOOL kit (siTOOLs Biotech). The RNA was then converted into cDNA using the NEBNext RNA First Strand Synthesis and NEBNext RNA Second Strand Synthesis Modules (New England Biolabs). For the preparation of the indexed Illumina libraries the Encore Rapid DR Multiplex System 1–96 (NuGEN) was used. Libraries were amplified with 12 cycles and sequenced on Illumina NextSeq500/550.
Between 6.8 and 11.6 million single 75 bp reads were generated per sample. Adapter-clipped reads provided by the sequencing company were quality trimmed with BBDuk (Bushnell, 2014) using a sliding window approach with a window size of 4 bp and an average base quality of 15. Poly-G repeats longer than 10 bp were removed and reads shorter than 50 bp were discarded. Quality-trimmed reads were mapped against the reference genome of Nodularia CCY9414 (NCBI RefSeq accession: GCF_000340565.2) using the program bwa-mem (Li, 2013). Remaining hits to ribosomal RNA genes were excluded. Mapping results were further filtered to remove secondary and supplementary alignments, as well as alignments shorter than 50 bp and those with less than 95% sequence identity across the whole read to the reference. Read counts per gene were then calculated with featureCounts (Love et al., 2014) and converted to transcript percentages accounting for variable gene length. Operons were predicted with OperonMapper1 (Taboada et al., 2018). To perform a functional enrichment analysis based on the KEGG pathway hierarchy, the reference genome of Nodularia CCY9414 was re-annotated against KEGG (release January 2022) using diamond blastp version 2.0.14.152 (Buchfink et al., 2021) in sensitive mode, retaining hits with an e-value below 1e-5 and a blast score ratio of more than 0.4. KEGG ortholog (KO) annotations were assigned based on the best hit. Genes without a KO assignment in the blastp search were attributed a KO number according to kofamscan version 1.3.0 (Aramaki et al., 2019) at an e-value of 0.01. Mapping of genes to KEGG pathways was performed based on KO assignments, excluding pathways from overview and structural maps and those exclusive to viruses or eukaryotic organisms. The transcriptomic reads and processed feature counts are accessible on the GEO database2 with the following accession number: GSE213384.
2.7. Statistical data analysis
All the statistical data analyses and visualization were performed in R (R Core Team, 2021) using the additional packages tidyverse (Wickham et al., 2019), DESeq2 (Love et al., 2014), vegan (Oksanen et al., 2020), car (Fox and Weisberg, 2019), emmeans (Lenth, 2022), multcomp (Hothorn et al., 2008), multcompView (Graves et al., 2019), lmerTest (Kuznetsova et al., 2017), hrbrthemes (Rudis, 2020), and RColorBrewer (Neuwirth, 2014). Post-processing of the figures was performed using the software Inkscape (Inkscape Project, 2020). Further details about the bioinformatic sequence processing and statistical data analysis are available on http://doi.io-warnemuende.de/10.12754/misc-2022-0005.
Dry weight and polyphosphate concentrations were analyzed in a generalized linear mixed model to assess the effect of sampling time point and P conditions with experiment iteration as random factor. To meet the assumption of normality, polyphosphate concentrations were square-root transformed. Outlier observations with a Cook’s distance of more than 4 divided by sample size (Cook and Weisberg, 1984; Williams, 1987) were removed from the analysis. Post hoc tests, i.e., pairwise comparisons between experimental conditions and sampling time points, were implemented in emmeans (Lenth, 2022). Differential gene expression was assessed using DESeq2 (Love et al., 2014) between P-replete and P-deplete conditions at each sampling time point and between d7 and d14 in each P treatment. Genes were detected as differentially expressed at a Benjamini–Hochberg adjusted p-value of 0.1 and an absolute log2-fold change of at least 1. Functional enrichment analysis was conducted for each pathway using the proportion of genes per pathway of the total number of genes in the genome in a X2 goodness-of-fit analysis. The aim was to assess if the number of differentially expressed genes per pathway was higher than expected by chance. Significance was assessed at a family-wise error rate of 0.05 after Benjamini–Hochberg adjustment of p-values to account for multiple testing. Cases with expected frequencies below 1 were marked as potentially unreliable in the results.
3. Results
3.1. Physiological and biochemical characterization of P-limitation
All experiments were performed under conditions that are prevailing in the Baltic Sea during Nodularia spp. summer blooms. Dry weight (DW) measurements were performed for each culture in both conditions (P-deplete and P-replete) to evaluate growth throughout the two independent experiments. The increase in biomass was faster in P-replete than in P-deplete conditions (Figure 1A). However, even the cyanobacteria in the P-deplete medium were able to almost triple their biomass throughout the 3 weeks’ incubation time, whereas P-replete conditions permitted a fourfold increase.
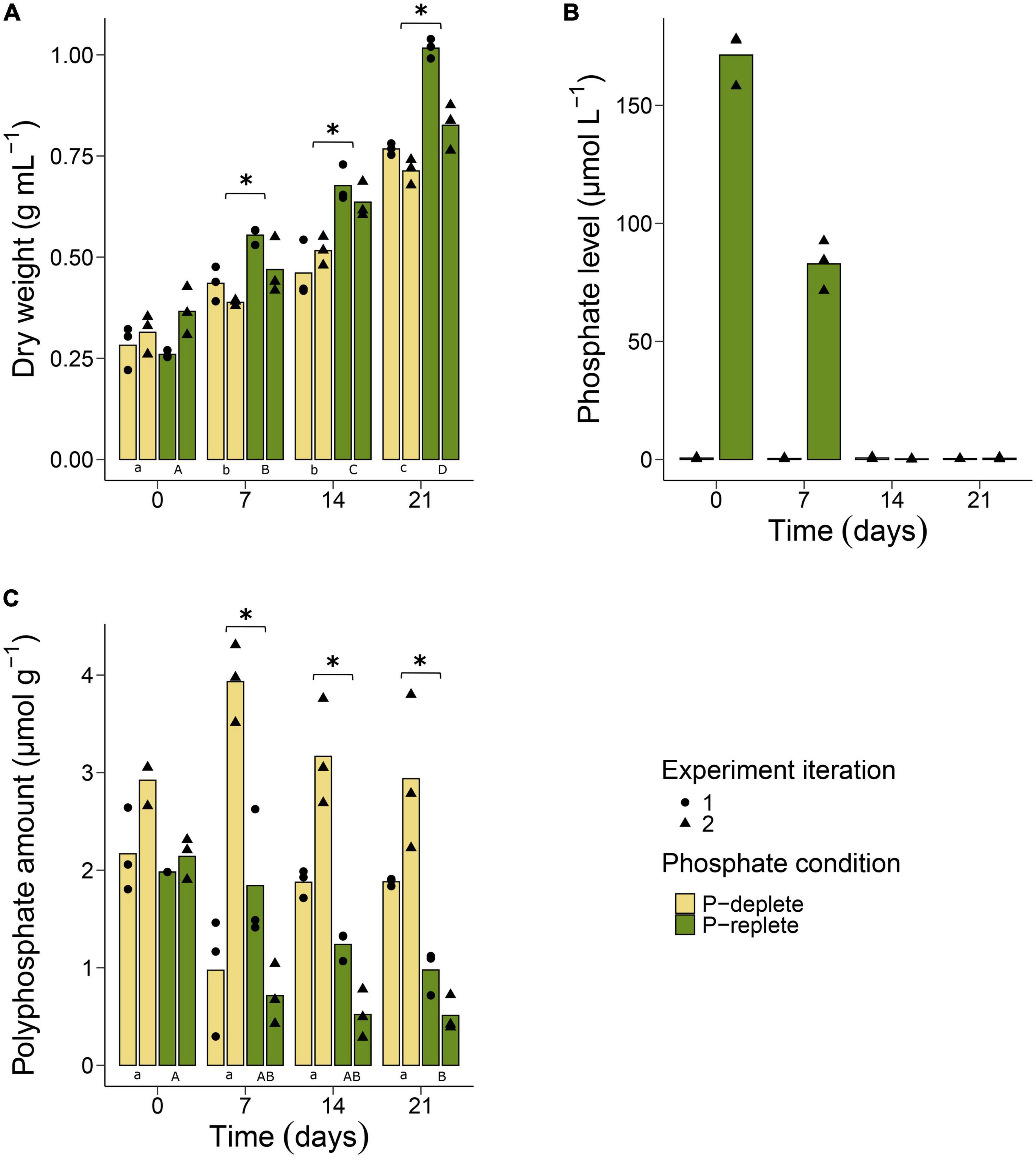
Figure 1. Physiological and biochemical assessment of Nodularia spumigena CCY9414 under P-replete (+P) and P-deplete (–P) conditions. Growth expressed as increase in dry weight (A). O-phosphate (PO43–) concentration in the cultivation media during the second experiment iteration (B). Accumulation of polyphosphates in cells of N. spumigena (C). Bars represent means of individual measurements shown as single points. Asterisks indicate significant differences between the P-replete and the P-deplete cultures at each sampling time point. Significant differences between sampling time points are indicated by different lower-case (P-deplete) and upper-case (P-replete) letters. The results of the statistical tests can be found in the Supplementary Tables 1, 2.
In addition, o-phosphate and ammonium concentrations were measured weekly. In media of P-deplete cultures, the amount of o-phosphate was always near the detection limit with values between 0.9 and 0.2 μM (Figure 1B). In the P-replete medium, o-phosphate concentrations were approximately 170 μM and dropped during the first 7 days of the experiment to about 50% of the initial amounts. Almost all o-phosphate was consumed by the cells in the P-replete conditions after 14 days, i.e., at that time point similarly low o-phosphate levels were detected in P-deplete and P-replete cultures. In contrast to o-phosphate, similar levels of ammonium, released from the N2-fixing filaments, were found in P-replete and P-deplete cultures during the entire experimental period. Its amount was always approximately 100 μM and did not differ significantly with time between the two experimental treatments (Supplementary Figure 1).
Polyphosphates were quantified in filaments at all sampling time points in P-replete and P-deplete cultures in two independent experiments (Figure 1C). At time point 0, cultures of both P conditions started with a relatively high internal polyphosphate amount of approximately 2.5 μmol g–1 DW. This amount continuously decreased in filaments grown under P-replete conditions, although the rate of decrease differed between the two experiment iterations. Especially in the second iteration of the experiment, the polyphosphate content decreased to less than 50% already after 7 days, when still substantial amounts of o-phosphate were available for the cells (Figure 1B). In contrast, filaments grown under P-deplete conditions did not significantly alter their internal polyphosphate pool during the 3-week time period, despite the observed divergent measurements between the experiment iterations at day 7 (Figure 1C). In addition to the chemical quantification of polyphosphate, we used DAPI-staining to obtain a qualitative picture of polyphosphate accumulation in different cell types of the Nodularia CCY9414 filaments. Stained filaments from P-replete and P-deplete cultures showed yellow inclusions at each time point in vegetative cells and in heterocysts (Supplementary Figure 2).
3.2. RNA-seq characterization of P-limitation
RNA was isolated and sequenced from Nodularia CCY9414 cells cultivated for 7 and 14 days (d7 and d14) under P-replete and P-deplete conditions, respectively, during the second experiment iteration. The RNA-seq approach enabled the differential expression analysis of 4752 non-ribosomal genes (Supplementary Data 1). Thereby, a larger number of genes showed increased compared to decreased expression in the three comparisons: day 7 P-replete relative to day 7 P-deplete conditions, day 14 P-replete relative to day 14 P-deplete conditions, and day 14 P-replete relative to day 7 P-replete conditions.
At day 7, 112 genes were up-regulated and 87 down-regulated comparing P-deplete and P-replete cultures, respectively, whereas these numbers increased after 14 days to 344 up-regulated and 231 down-regulated genes (Figure 2). Among them, 77 genes were commonly up-regulated in P-deplete cultures at the two time points and 40 remained down-regulated. Furthermore, the gene expression changes at day 14 compared to day 7 in P-replete cultures resembled the changes observed in P-deplete versus P-replete conditions when compared at the same sampling time point. Overall, 28 of the 88 up-regulated genes at day 14 in P-replete cultures were also up-regulated at day 7 in P-deplete Nodularia CCY9414 filaments (Figure 2). A similar relative overlap is also found between down-regulated genes in day 14 P-replete relative to day 7 P-deplete (each compared to day 7 P-replete) cultures. These relations indicate that during the long-term growth under initially P-replete conditions many P-limitation-induced genes became up-regulated, which is consistent with the complete consumption of o-phosphate between 7 and 14 days in these cultures (Figure 1B).
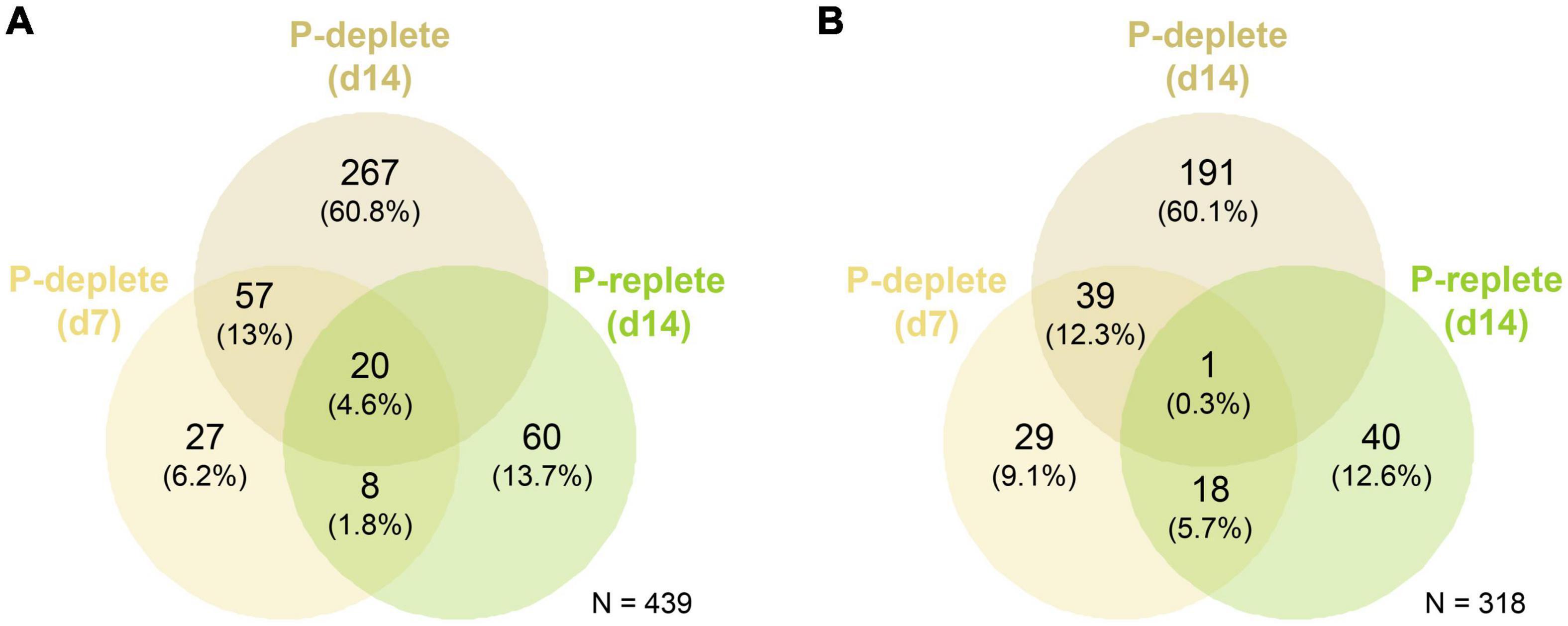
Figure 2. Venn diagrams for the global quantitative comparison of expression changes in Nodularia spumigena CCY9414 under P-replete (+P) or P-deplete (–P) conditions. Number of up-regulated (A) and down-regulated (B) genes under P starvation in the comparison of P-deplete and P-replete conditions at day 7, P-deplete and P-replete conditions at day 14, and between P-replete conditions at day 14 and day 7, respectively. The RNA-seq experiment was conducted with samples from the second experiment iteration.
Specific functional pathways were over-represented among the differentially expressed genes under specific P conditions (Figure 3). Especially at day 7, a disproportionally large number of genes up-regulated under P-deplete conditions were recruited from ABC transporters, among them 17 transporters for different P sources, and many genes for phosphonate and phosphinate metabolism. In addition, there were hints that P starvation had marked influence on the overall cell metabolism, e.g., many genes encoding regulatory proteins from two-component systems or enzymes involved in the metabolism of secondary metabolites as well as the oxidative pentose-phosphate (OPP) pathway were higher expressed, whereas genes for photosynthetic complexes or enzymes of the Calvin-Benson cycle appeared down-regulated at day 7 (Supplementary Data 1). After 14 days of growth without P, many more genes became stimulated. In addition to the previously mentioned P-associated genes, genes for proteins involved in energy metabolism, such as photosynthesis, oxidative phosphorylation and carbon fixation, were among the functional groups enriched in low P-stimulated genes after 14 days (Figure 2). Many genes encoding P limitation-related transport or regulatory proteins also became higher expressed when we compared the gene expression of P-replete cultures at day 14 to day 7, when all available o-phosphate had been consumed from the medium (Figure 1B).
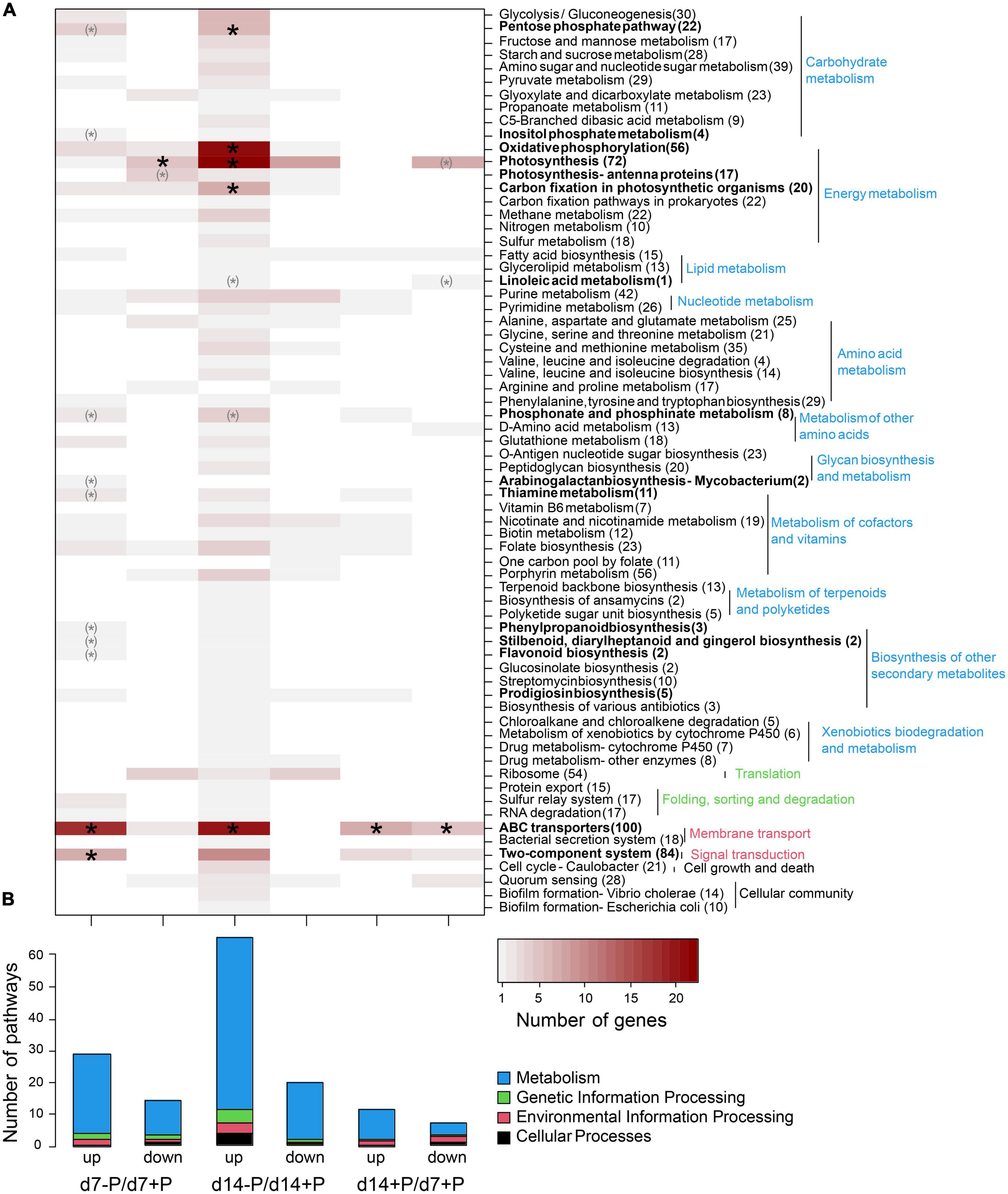
Figure 3. Assignment of differentially expressed genes to KEGG pathways. Up- and down-regulated genes are shown for the comparison of P-deplete and P-replete conditions at d7, P-deplete and P-replete conditions at d14, and between P-replete conditions at d14 and d7. (A) Number of differentially expressed genes per KEGG pathway. Asterisks indicate significant functional enrichment (X2 goodness-of-fit test, p < 0.05, parentheses indicate expected frequencies below 1). Number of genes per pathway in the genome is given after the pathway name. (B) Number of pathways containing differentially expressed genes. The RNA-seq experiment was conducted with samples from the second experiment iteration.
3.3. Defining the P-specific stimulon
In the initial genome annotation study, the authors provided a comprehensive overview of putatively P-associated genes in Nodularia CCY9414 (Voss et al., 2013). To confirm their involvement in the acclimation to P-limitation in our experimental set-up, the relative expression of all these genes was reviewed in the RNA-seq data (Supplementary Table 3 and Supplementary Data 1). This comparison permitted an experimentally supported annotation of the specific P-related stimulon in Nodularia CCY9414 (Table 1).
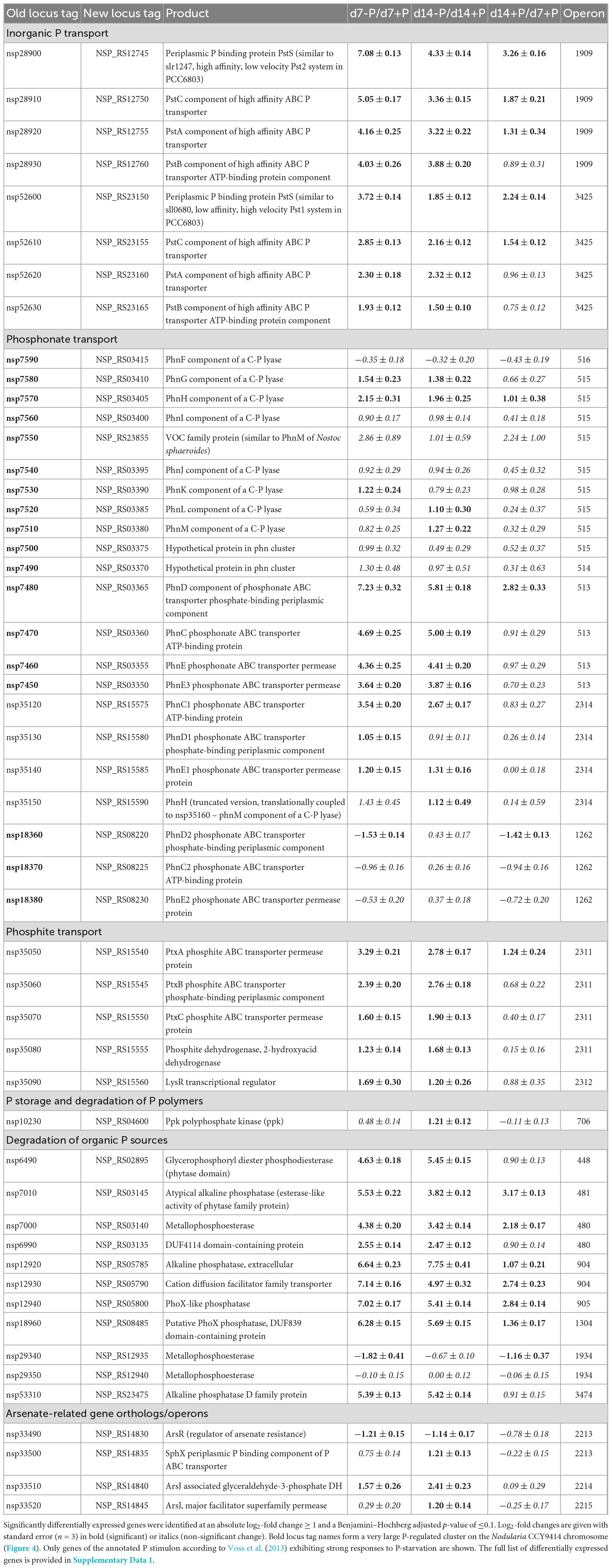
Table 1. P-related genes in Nodularia spumigena CCY9414 according to Voss et al. (2013) and their differential expression after 7 or 14 days of P-limitation as well as during growth for 14 days in P-replete medium.
Approximately half of the previously suggested P-associated genes (Voss et al., 2013) were among the differentially expressed genes identified in our experiment, comprising genes which exhibited the strongest responses including transport systems for different inorganic and organic P-sources (Supplementary Figure 3). Among them were two Pst systems (organized in operons 1909 and 3425) for o-phosphate that were concordantly stimulated but to a different extent. The operon 1909 that encodes for proteins similar to the high affinity but low velocity Pst2 system in Synechocystis sp. PCC 6803 (Pitt et al., 2010) is about three times higher expressed than the genes for the lower affinity but higher velocity Pst1 system. In addition, a large chromosomal region was found on which many P-regulated genes were situated. It comprises the genes nsp7450 to nsp7590 that encode at least two phosphonate transport systems in the operons 515 and 513, which were induced after 7 as well as 14 days of P-limitation (Table 1 and Figure 4). Interestingly, a third annotated phosphonate uptake system encoded by the operon 1262 was weakly down-regulated after 7-day growth under P-deplete conditions (Table 1). Moreover, the phosphite ABC transporter encoded by the operon 2311 was also stimulated after 7 and 14 days of P-limitation. In addition, many genes encoding alkaline or acid phosphatases, phytases and related proteins known to be able to release o-phosphate from different organic P-sources belonged to the most strongly induced genes. Among them, the alkaline phosphatase NSP_RS05785 was predicted with high confidence by TargetP3 to possess a transit peptide at the N-terminus, which indicates that this enzyme is likely released from the cell to acquire extracellular P (Table 1).
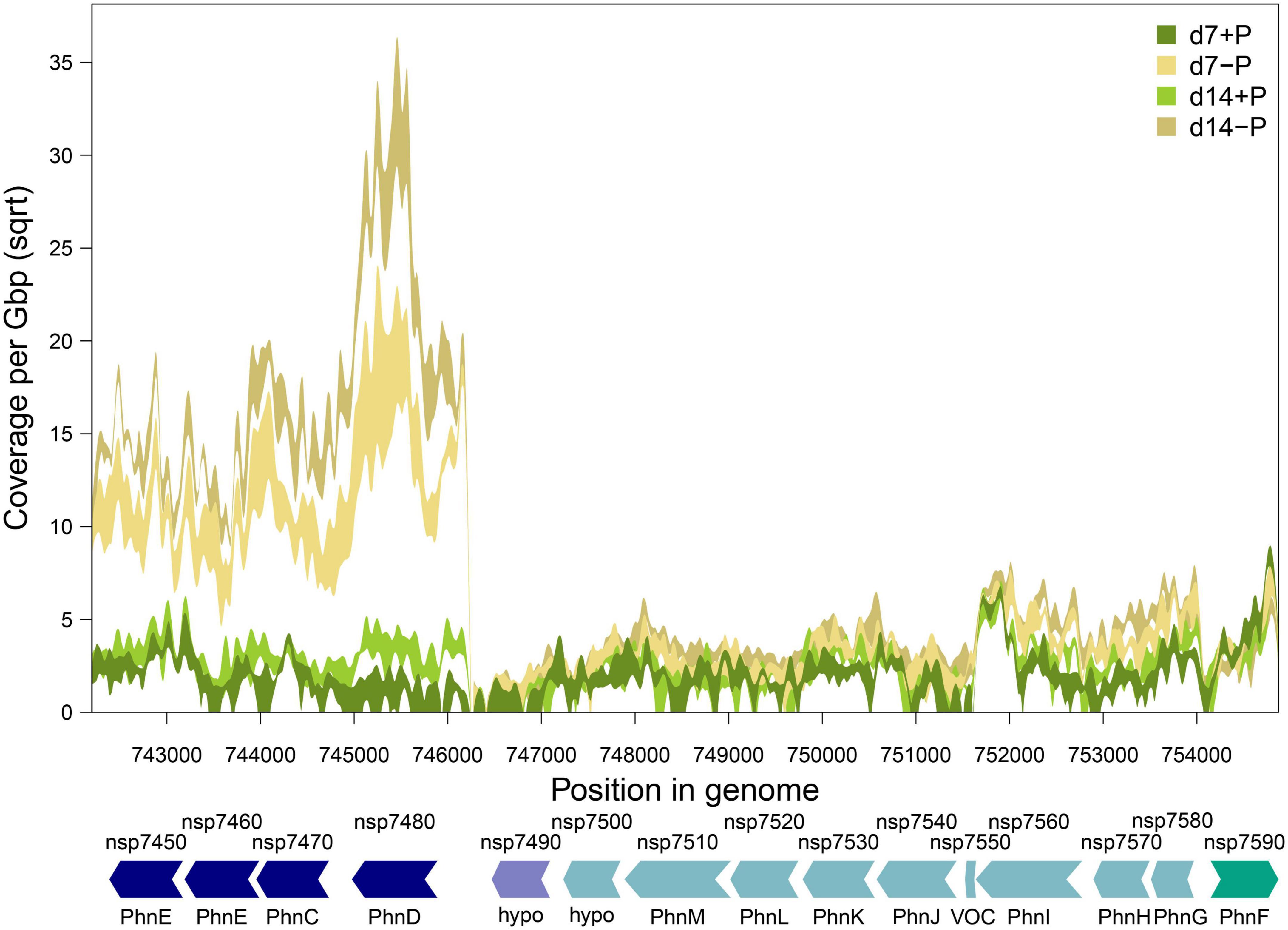
Figure 4. Gene expression of phosphate-regulated gene cluster nsp7450 to nsp7590 in N. spumigena CCY9414 under P-replete (+P) and P-deplete (–P) conditions. Shaded bands show minimum and maximum gene expression (normalized as square-root transformed per base pair coverage per Gbp sequencing effort) at each experimental condition. Arrows indicate gene arrangement and orientation and were colored according to operon assignment as provided in Table 1. Locus tags and gene names are according to Table 1 (hypo: hypothetical protein).
However, many genes that were previously suggested to be part of the P-stimulon (Voss et al., 2013) did not show pronounced phosphate-related gene expression changes, such as metallophosphoesterases, haloacid dehalogenase-like hydrolases and the majority of the arsenate-related genes with a few exceptions (Supplementary Table 3 and Supplementary Data 1). Furthermore, most genes annotated to be involved in polyphosphate synthesis and mobilization did not clearly change the expression upon P-limitation. Only the ppk gene, encoding the polyphosphate synthesizing kinase, became significantly higher expressed after 14 days of growth under P-deplete conditions, which was consistent with the largely stably maintained polyphosphate pool in these filaments (Figure 1C). Finally, despite the induction of many transporters for different P-sources, the expression of the related transcriptional regulators, i.e., the two-component regulatory system PhoB (SphR) and PhoR (SphS), were not significantly changed (Supplementary Table 3 and Supplementary Data 1). Only one LysR-type transcriptional regulator that is found downstream of the operon 2311 encoding the phosphite-specific ABC transporter showed significantly enhanced expression under P-limiting conditions (Table 1).
3.4. Further P-regulated genes
In addition to the above-mentioned genes encoding proteins that were likely directly involved in the acclimation to P limitation, many more genes appeared to be directly or indirectly P-regulated in Nodularia CCY9414. Amongst them, many genes encode hypothetical proteins with no functional annotation, i.e., at the time point day 7 this group comprised approximately 25% all up-regulated genes under P-deplete conditions (Supplementary Data 1). However, several functionally annotated and physiologically important genes were also strongly induced in P-starved cells (Table 2). One example is represented by three genes (new locus tags NSP_RS14115, NSP_RS14120, and NSP_RS14125) encoding proteins involved in the synthesis of a CTB family bacteriocin, which were induced after 7 and 14 days of growth in the P-deplete medium. The genome of Nodularia CCY9414 harbored a second cluster of genes (NSP_RS06270, NSP_RS06275, NSP_RS06280, NSP_RS06285, and NSP_RS06290) for bacteriocins, which was not differentially expressed under different P conditions (Supplementary Data 1). Furthermore, several genes associated with bloom formation of Nodularia CCY9414 appeared to be up-regulated under P starvation. Amongst them, several of the encoded proteins were related to N2 fixation, such as the Mo-dependent nitrogenase C-terminal domain-containing protein (NSP_RS06835), the nitrogen fixation protein NifX (NSP_RS18010), the nitrogenase-stabilizing/protective protein NifW (NSP_RS18025), which were higher expressed at day 7 in N-deplete filaments (Table 2). Additional bloom-related genes became induced after 14 days of growth under P-deplete conditions. Notably, a cluster of genes encoding different gas vesicle constituents, such as GvpA and GvpC (operon 1062: NSP_RS06890, NSP_RS06895, NSP_RS06900, as well as NSP_RS06910), or a non-ribosomal peptide synthetase operon 3225 (NSP_RS21715, NSP_RS21720, and NSP_RS21725) were among them. It further seemed that after 2 weeks of P starvation, the iron homeostasis in Nodularia CCY9414 was affected, because genes for iron uptake systems, such as the TonB receptor and the iron-siderophore ABC transporter substrate-binding protein (NSP_RS05344 and NSP_RS05350), as well as iron ABC transporter permeases (NSP_RS05360 and NSP_RS05365) became up-regulated (Table 2). Finally, the sugar-phosphate metabolism appeared to be altered due to strong P-limitation. Especially genes for enzymes involved in the OPP pathway, including the entrance enzyme glucose 6-phosphate dehydrogenase (zwf, NSP_RS22185), were found to be up-regulated after 7 and 14 days of P starvation.
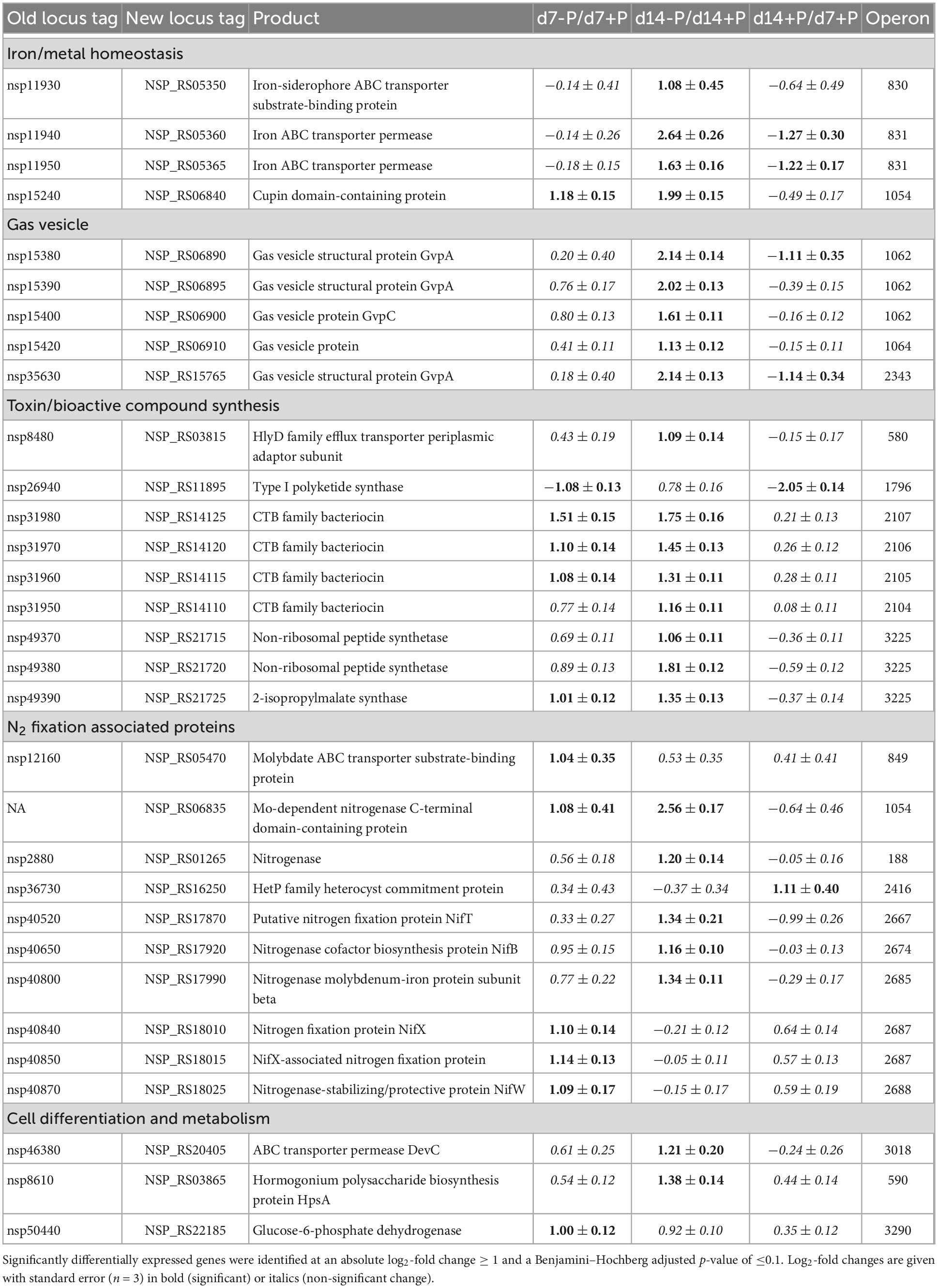
Table 2. Expression changes of genes encoding proteins indirectly involved in P acclimation of N. spumigena CCY9414 after 7 or 14 days of P-limitation as well as during growth for 14 days in P-replete medium.
4. Discussion
Acclimation to long-term, harsh P limitation was studied in the strain N. spumigena CCY9414, which was isolated from a cyanobacterial summer bloom in the Baltic Sea and can thus serve as model for bloom-forming Nodularia spp. under such conditions (Stal et al., 2003; Voss et al., 2013). In contrast to our previous study (Hagemann et al., 2019), Nodularia CCY9414 was pre-cultivated two times for 10 days in P-containing medium, which resulted in a good P-status of the starting cultures. This assumption is supported by the low expression of P-regulated genes in cells grown for 7 days in P-replete conditions and the relatively high polyphosphate content in Nodularia CCY9414 filaments at the beginning of the experiment (Figure 1C). Despite the significant slower growth of cells under P-deplete conditions, these cultures were still able to triple their biomass during our experimental period of 3 weeks (Figure 1A). The question arises, where the necessary P is coming from, because the stored polyphosphate pool remained almost stable. The amount of available o-phosphate in the P-deplete medium was close to 1 μM, which is comparable to the values measured in the Baltic Sea during summer bloom events (Nausch et al., 2008). However, it has been shown that the high affinity Pst2 system in the model strain Synechocystis sp. PCC 6803 has an o-phosphate affinity of 0.07 μM (Pitt et al., 2010). Genes encoding proteins with high similarity to this Pst2 were highly up-regulated in Nodularia CCY9414 filaments grown under P-deplete conditions, hence, it can be assumed that this ABC transporter is still able to acquire o-phosphate even in the presumably P-free medium. It is also known that acclimation to P limitation induces other P-saving strategies such as reduction in the copy number of cellular DNA or a decrease in the proportion of phospholipids in cyanobacteria (van Mooy et al., 2009; Zerulla et al., 2016).
It remains unknown why Nodularia CCY9414 is not consuming the stored polyphosphate after sudden transfer to P-deplete conditions at the beginning of the experiment. In contrast, when the cells gradually acclimated to P limitation in the P-replete medium after 14 days, then the polyphosphate pool was stepwise consumed. However, still a substantial amount of polyphosphate was kept in these cells after 3 weeks of incubation although the dissolved o-phosphate in the medium was utterly consumed (Figures 1B, C). Presumably, the immediate drop in P from normal availability to very low amounts transforms the cell into a state, in which the polyphosphate reserves are stabilized, whereas under slowly P-decreasing conditions it is at least partially consumed as reported from P-limited model organisms (Lawrence et al., 1998; Gómez-García et al., 2003). Interestingly, only few expression changes were observed for genes encoding proteins for polyphosphate synthesis or breakdown in Nodularia CCY9414 filaments under P-replete or P-deplete conditions (Supplementary Table 3). This finding suggests that polyphosphate accumulation is mostly regulated at post-transcriptional level in Nodularia CCY9414 and possibly other cyanobacteria, because related genes for polyphosphate accumulation are also not part of the P-regulon in Synechocystis sp. PCC 6803 (Suzuki et al., 2004). Comparatively high polyphosphate contents have been also reported from other filamentous cyanobacteria such as Trichodesmium spp. under low external P concentrations in the environment (Orchard et al., 2010; Martin et al., 2014). It can be speculated whether the stable polyphosphate pool at low external P contents is saved to support the development the next Nodularia spp. generation or akinete formation, or whether it is a strategy to keep all the available phosphate in non- or slow-growing cells to minimize the amount potentially available for competing microorganisms in the microbial community.
In the unicellular model Synechocystis sp. PCC 6803, P-induced genes could be assigned to a P-regulon, because almost all P-induced genes are characterized by a defined P-box (YTTAAYYW NNN YTTAAYYW NNN YTTAAYYW) upstream of their promoters that is targeted by the P-sensing two component system PhoB/PhoR (SphB/SphR) (Suzuki et al., 2004). Here, we used RNA-seq differential gene expression analysis to define the P-stimulon in Nodularia CCY9414. Our attempts to find a similar, conserved P-box in front of homologous genes/operons in Nodularia CCY9414 were not successful. This result may indicate that the more diverse and larger P-stimulon is regulated by different transcriptional factors. The P-regulon of Synechocystis sp. PCC 6803 mostly contains genes for o-phosphate uptake, alkaline phosphatase and regulatory proteins for their expression (Suzuki et al., 2004), whereas Nodularia CCY9414 has a much wider capacity to acclimate to low P conditions as an adaptation to its ecological niche. In addition to o-phosphate, transporters and enzymes for the utilization of alternative P-sources are not only encoded in the genome but mostly also induced in cells shifted into P-deplete medium, which supports the view that they contribute to the growth of Nodularia spp. in brackish waters under P-limiting conditions. These include proteins for the utilization of phosphonates and phosphites as well as different external organic P-sources. Similarly, many genes for the uptake and utilization of different inorganic or organic P-sources have been up-regulated in Trichodesmium spp. occurring in regions of the Atlantic Ocean with increasingly limiting P availability (Cerdan-Garcia et al., 2022). Phosphonate utilization has recently been verified to sustain growth of marine cyanobacteria under different nutrient availability (e.g., Acker et al., 2022; Rabouille et al., 2022; Zhao et al., 2022). Interestingly, not all annotated transport systems are induced under our P-limiting conditions, for example the operon 1262 encoding for one probable phosphonate uptake system is rather down-regulated after 7-day growth under P-deplete conditions (Table 1). In the future, it would be interesting to study the response of this and other operons/genes for the metabolism of alternative P-sources at varying amounts of phosphonates or related compounds in the growth medium.
In addition to up-regulation of genes for proteins directly involved in P acclimation, many annotated genes from different functional categories were co-regulated with the P-stimulon in Nodularia CCY9414. This group included several genes, which are assumed to be of importance in the context of bloom events. In addition to genes for proteins in N2-fixation and gas vesicle formation, genes for bioactive compound synthesis were found to be overexpressed in filaments from P-deplete cultures (Table 2). Nodularia CCY9414 has many operons that are potentially capable to produce different toxins such as nodularin and other bioactive compounds (Voss et al., 2013). Such gene clusters are especially widespread in cyanobacterial strains known to form blooms, where they can play different functions, such as defense against grazers, intra- or extracellular info-chemicals within the population, or as allelopathic signals between different microorganisms in the scum of the bloom (Dittmann et al., 2013; Guljamow et al., 2021). Among them, one operon for the synthesis of a CTB family bacteriocin was identified, which was induced after 7 and 14 days of growth in P-deplete medium. Bacteriocins are known allelopathic compounds that inhibit growth of other (cyano)bacteria in microbial consortia (Flores and Wolk, 1986; Aharonovich and Sher, 2016). Bacteriocin synthesis clusters have not only been identified in the genomes of bloom-forming filamentous cyanobacteria, but are also present in the genomes of picocyanobacteria with smaller genomes, highlighting their importance for the organism because they are kept despite substantial genome reduction (Paz-Yepes et al., 2013). Recently, it has been shown that such strains occur very frequently in the waters of different salinities including the Baltic Sea in summer (Cabello-Yeves et al., 2022). Since most bloom formation associated genes, such as nitrogenase-related, gas-vesicle, bacteriocin and iron-acquisition genes, became more highly expressed only after 14 days under P-limiting conditions, we conclude that long-term harsh P-starvation can indeed be regarded as a potential trigger for bloom formation in Nodularia CCY9414 and likely other cyanobacteria.
Data availability statement
The original contributions presented in this study are included in the article and the associated Supplementary material. Furthermore, the transcriptomic reads and processed feature counts are accessible from the GEO database (https://www.ncbi.nlm.nih.gov/geo/) with the following accession number: GSE213384. The scripts for bioinformatic sequence processing and statistical data analysis are available at http://doi.io-warnemuende.de/10.12754/misc-2022-0005. Data from all other measured parameters are available on PANGAEA (Santoro et al., 2022).
Author contributions
MH, MS, and ML designed the study. MS performed the cyanobacterial cultivations, poly-P estimation, and RNA extractions. CH analyzed the RNA-seq data. MH and ML supervised the experiments. MH, MS, and CH evaluated the data. MH and MS wrote the manuscript that was reviewed by all co-authors. All authors contributed to the article and approved the submitted version.
Funding
MS was funded by the Leibniz Science Campus Phosphorus Research Rostock in the funding line strategic networks of the Leibniz Association. Additional financial support by the University of Rostock and the IOW was acknowledged.
Acknowledgments
The strain Nodularia spumigena CCY9414 was kindly provided by Annick Wilmotte from the BCCM/ULC collection (University of Liège, Belgium). We thank Klaudia Michl (University of Rostock) for technical assistance in strain cultivation and sample preparation. We strongly acknowledge Christian Burmeister (Department of Biological Oceanography – IOW) for having performed the nutrient measurements.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1082763/full#supplementary-material
Footnotes
- ^ https://biocomputo.ibt.unam.mx/operon_mapper/
- ^ https://www.ncbi.nlm.nih.gov/geo/
- ^ https://services.healthtech.dtu.dk/service.php?TargetP-2.0
References
Acker, M., Hogle, S. L., Berube, P. M., Hackl, T., Coe, A., Stepanauskas, R., et al. (2022). Phosphonate production by marine microbes: exploring new sources and potential function. PNAS 119:e2113386119. doi: 10.1073/pnas.2113386119
Aharonovich, D., and Sher, D. (2016). Transcriptional response of prochlorococcus to co-culture with a marine alteromonas: differences between strains and the involvement of putative infochemicals. ISME J. 10, 2892–2906. doi: 10.1038/ismej.2016.70
Aramaki, T., Blanc-Mathieu, R., Endo, H., Ohkubo, K., Kanehisa, M., Goto, S., et al. (2019). KofamKOALA: KEGG ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 36, 2251–2252. doi: 10.1093/bioinformatics/btz859
Buchfink, B., Reuter, K., and Drost, H. G. (2021). Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 18, 366–368. doi: 10.1038/s41592-021-01101-x
Bushnell, B. (2014). BBMap: a fast, accurate, splice-aware aligner (No. LBNL-7065E). Berkeley, CA: Lawrence Berkeley National Lab.(LBNL).
Cabello-Yeves, P. J., Scanlan, D. J., Callieri, C., Picazo, A., Schallenberg, L., Huber, P., et al. (2022). α-cyanobacteria possessing form IA RuBisCO globally dominate aquatic habitats. ISME J. 22:1282. doi: 10.1038/s41396-022-01282-z
Cerdan-Garcia, E., Baylay, A., Polyviou, D., Woodward, E. M. S., Wrightson, L., Mahaffey, C., et al. (2022). Transcriptional responses of trichodesmium to natural inverse gradients of Fe and P availability. ISME J. 16, 1055–1064. doi: 10.1038/s41396-021-01151-1
Cook, R. D., and Weisberg, S. (1984). “Residuals and influence in regression,” in Applied regression, linear models, and related methods, ed. J. Fox (Sage Publications, Inc).
Diaz, J. M., and Ingall, E. D. (2010). Fluorometric quantification of natural inorganic polyphosphate. Environ. Sci. Technol. 44, 4665–4671. doi: 10.1021/es100191h
Dittmann, E., Fewer, D. P., and Neilan, B. A. (2013). Cyanobacterial toxins: biosynthetic routes and evolutionary roots. FEMS Microbiol. Rev. 37, 23–43. doi: 10.1111/j.1574-6976.2012.12000.x
Flores, E., and Wolk, C. P. (1986). Production, by filamentous, nitrogen-fixing cyanobacteria, of a bacteriocin and of other antibiotics that kill related strains. Arch. Microbiol. 145, 215–219. doi: 10.1007/BF00443648
Fox, J., and Weisberg, S. (2019). An R companion to applied regression, 3rd Edn. Thousand Oaks, CA: Sage.
Gehringer, M. M., and Wannicke, N. (2014). Climate change and regulation of hepatotoxin production in cyanobacteria. FEMS Microbiol. Ecol. 88, 1–25. doi: 10.1111/1574-6941.12291
Gómez-García, M. R., Losada, M., and Serrano, A. (2003). Concurrent transcriptional activation of ppa and ppx genes by phosphate deprivation in the cyanobacterium Synechocystis sp. strain PCC 6803. Biochem. Biophys. Res. Commun. 302, 601–609. doi: 10.1016/S0006-291X(03)00162-1
Grasshoff, K., Kremling, K., and Ehrhardt, M. (eds) (2009). Methods of seawater analysis. New York: John Wiley & Sons.
Graves, S., Piepho, H. P., Selzer, L., and Dorai-Raj, S. (2019). multcompView: visualizations of paired comparisons. R package version 0.1-8.
Guljamow, A., Barchewitz, T., Große, R., Timm, S., Hagemann, M., and Dittmann, E. (2021). Diel variations of extracellular microcystin influence the subcellular dynamics of rubisco in microcystis aeruginosa pcc 7806. Microorganisms 9:1265. doi: 10.3390/microorganisms9061265
Hagemann, M., Möke, F., Springer, A., Westermann, L., Frank, M., Wasmund, N., et al. (2019). Cyanobacterium Nodularia spumigena strain CCY9414 accumulates polyphosphate under long-term P-limiting conditions. Aquatic Microbial. Ecol. 82, 265–274. doi: 10.3354/ame01896
Hayes, P. K., and Barker, G. L. A. (1997). Genetic diversity within baltic sea populations of. nodularia (cyanobacteria). School Biol. Sci. Plant Agric. Sci. 923, 919–923.
Hiyoshi, T., Oyanagi, K., Niki, T., Fujiwara, S., and Sato, N. (2021). Requirement of the exopolyphosphatase gene for cellular acclimation to phosphorus starvation in a cyanobacterium, Synechocystis sp. PCC 6803. Biochem. Biophys. Res. Commun. 540, 16–21. doi: 10.1016/j.bbrc.2020.12.095
Hothorn, T., Bretz, F., and Westfall, P. (2008). Simultaneous inference in general parametric models. Biomet. J. J. Mathe. Methods Biosci. 50, 346–363.
Jonasson, S., Vintila, S., Sivonen, K., and El-Shehawy, R. (2008). Expression of the nodularin synthetase genes in the baltic sea bloom-former cyanobacterium nodularia spumigena strain AV1. FEMS Microbiol. Ecol. 65, 31–39. doi: 10.1111/j.1574-6941.2008.00499.x
Kelly, L. T., Ryan, K. G., and Wood, S. A. (2019). Differential strain response in alkaline phosphatase activity to available phosphorus in Microcoleus autumnalis. Harmful Algae 89:101664. doi: 10.1016/j.hal.2019.101664
Kopf, M., Möke, F., Bauwe, H., Hess, W. R., and Hagemann, M. (2015). Expression profiling of the bloom-forming cyanobacterium nodularia CCY9414 under light and oxidative stress conditions. ISME J. 9, 2139–2152. doi: 10.1038/ismej.2015.16
Kuznetsova, A., Brockhoff, P. B., and Christensen, R. H. B. (2017). lmerTest package: tests in llinear mixed effects models. J. Statist. Soft. 82, 1–26. doi: 10.18637/jss.v082.i13
Lawrence, B. A., Suarez, C., de Pina, A., Click, E., Kolodny, N. H., and Allen, M. M. (1998). Two internal pools of soluble polyphosphate in the cyanobacterium Synechocystis sp. strain PCC 6308: an in vivo 31P NMR spectroscopic study. Arch. Microbiol. 169, 195–200. doi: 10.1007/s002030050560
Lenth, R. V. (2022). emmeans: estimated marginal means, aka least-squares means. R package version 1.7.3.
Li, H. (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv [preprint]. arXiv:1303.3997.
Li, J., Plouchart, D., Zastepa, A., and Dittrich, M. (2019). Picoplankton accumulate and recycle polyphosphate to support high primary productivity in coastal Lake Ontario. Sci. Rep. 9, 1–10. doi: 10.1038/s41598-019-56042-5
Love, M., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. bioRxiv [Preprint]. doi: 10.1101/002832
Martin, P., and Van Mooy, B. A. (2013). Fluorometric quantification of polyphosphate in environmental plankton samples: extraction protocols, matrix effects, and nucleic acid interference. Appl. Environ. Microbiol. 79, 273–281.
Martin, P., Dyhrman, S. T., Lomas, M. W., Poulton, N. J., and van Mooy, B. A. S. (2014). Accumulation and enhanced cycling of polyphosphate by sargasso sea plankton in response to low phosphorus. Proc. Natl. Acad. Sci. U.S.A. 111, 8089–8094. doi: 10.1073/pnas.1321719111
Möke, F., Wasmund, N., Bauwe, H., and Hagemann, M. (2013). Salt acclimation of Nodularia spumigena CCY9414 - a cyanobacterium adapted to brackish water. Aquat. Microb. Ecol. 70, 207–214. doi: 10.3354/ame01656
Nausch, M., Nausch, G., Wasmund, N., and Nagel, K. (2008). Phosphorus pool variations and their relation to cyanobacteria development in the baltic sea: a three-year study. J. Mari. Syst. 71, 99–111. doi: 10.1016/j.jmarsys.2007.06.004
Ohtomo, R., Sekiguchi, Y., Kojima, T., and Saito, M. (2008). Different chain length specificity among three polyphosphate quantification methods. Analyt. Biochem. 383, 210–216. doi: 10.1016/j.ab.2008.08.002
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2020). vegan: community ecology package. R package version 2.5-7.
Orchard, E. D., Ammerman, J. W., Lomas, M. W., and Dyhrman, S. T. (2010). Dissolved inorganic and organic phosphorus uptake in Trichodesmium and the microbial community: the importance of phosphorus ester in the sargasso sea. Limnol. Oceanogr. 55, 1390–1399. doi: 10.4319/lo.2010.55.3.1390
Ostrowski, M., Mazard, S., Tetu, S. G., Phillippy, K., Johnson, A., Palenik, B., et al. (2010). PtrA is required for coordinate regulation of gene expression during phosphate stress in a marine synechococcus. ISME J. 4, 908–921. doi: 10.1038/ismej.2010.24
Paz-Yepes, J., Brahamsha, B., and Palenik, B. (2013). Role of a microcin-C-like biosynthetic gene cluster in allelopathic interactions in marine synechococcus. Proc. Natl. Acad. Sci. U.S.A. 110, 12030–12035. doi: 10.1073/pnas.1306260110
Pitt, F. D., Mazard, S., Humphreys, L., and Scanlan, D. J. (2010). Functional characterization of Synechocystis sp. strain PCC 6803 pst1 and pst2 gene clusters reveals a novel strategy for phosphate uptake in a freshwater cyanobacterium. J. Bacteriol. 192, 3512–3523. doi: 10.1128/JB.00258-10
Rabouille, S., Tournier, L., Duhamel, S., Claquin, P., Crispi, O., Talec, A., et al. (2022). Organic phosphorus scavenging supports efficient growth of diazotrophic cyanobacteria under phosphate depletion. Front. Microbiol. 13:848647. doi: 10.3389/fmicb.2022.848647
R Core Team (2021). R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing.
Repka, S., Mehtonen, J., Vaitomaa, J., Saari, L., and Sivonen, K. (2001). Effects of nutrients on growth and nodularin production of nodularia strain GR8b. Microb. Ecol. 42, 606–613. doi: 10.1007/s00248-001-0026-8
Rippka, R., Deruelles, J., and Waterbury, J. B. (1979). Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. General Microbiol. 111, 1–61. doi: 10.1099/00221287-111-1-1
Rudis, B. (2020). hrbrthemes: additional themes, theme components and utilities for ‘ggplot2’. R package version 0.8.0.
Santoro, M., Hassenrück, C., Labrenz, M., and Hagemann, M. (2022). Experimental phosphate limitation in Nodularia spumigena CCY9414. Pangaea 1594:9510787. doi: 10.1594/PANGAEA.951078
Sanz-Luque, E., Bhaya, D., and Grossman, A. R. (2020). Polyphosphate: a multifunctional metabolite in cyanobacteria and algae. Front Plant Sci. 11:1–21. doi: 10.3389/fpls.2020.00938
Sellner, K. G. (1997). Physiology, ecology, and toxic properties of marine cyanobacteria blooms. Limnol. Oceanogr. 42, 1089–1104. doi: 10.4319/lo.1997.42.5_part_2.1089
Sivonen, K., Kononen, K., Carmichael, W. W., Dahlem, A. M., Rinehart, K. L., Kiviranta, J., et al. (1989). Occurrence of the hepatotoxic cyanobacterium nodularia spumigena in the baltic sea and structure of the toxin. Appl. Environ. Microbiol. 55:8. doi: 10.1128/aem.55.8.1990-1995.1989
Stal, L. J., Albertano, P., Bergman, B., von Bröckel, K., Gallon, J. R., Hayes, P. K., et al. (2003). BASIC: baltic sea cyanobacteria. an investigation of the structure and dynamics of water blooms of cyanobacteria in the baltic sea - responses to a changing environment. Contin. Shelf Res. 23, 1695–1714. doi: 10.1016/j.csr.2003.06.001
Steunou, A. S., Bhaya, D., Bateson, M. M., Melendrez, M. C., Ward, D. M., Brecht, E., et al. (2006). In situ analysis of nitrogen fixation and metabolic switching in unicellular thermophilic cyanobacteria inhabiting hot spring microbial mats. Proc. Natl. Acad. Sci. 103, 2398–2403. doi: 10.1073/pnas.0507513103
Suzuki, S., Ferjani, A., Suzuki, I., and Murata, N. (2004). The SphS-SphR two component system is the exclusive sensor for the induction of gene expression in response to phosphate limitation in Synechocystis. J. Biol. Chem. 279, 13234–13240. doi: 10.1074/jbc.M313358200
Taboada, B., Estrada, K., Ciria, R., and Merino, E. (2018). Operon-mapper: a web server for precise operon identification in bacterial and archaeal genomes. Bioinformatics 34, 4118–4120. doi: 10.1093/bioinformatics/bty496
Tetu, S. G., Brahamsha, B., Johnson, D. A., Tai, V., Phillippy, K., Palenik, B., et al. (2009). Microarray analysis of phosphate regulation in the marine cyanobacterium synechococcus sp. WH8102. ISME J. 3, 835–849. doi: 10.1038/ismej.2009.31
Tijssen, J. P. F., van Steveninck, J., and de Bruijn, W. C. (1985). Cytochemical staining of a yeast polyphosphate fraction, localized outside the plasma membrane. Protoplasma 125, 124–128. doi: 10.1007/BF01297357
Vahtera, E., Autio, R., Kaartokallio, H., and Laamanen, M. (2010). Phosphate addition to phosphorus-deficient baltic sea plankton communities benefits nitrogen-fixing cyanobacteria. Aquat. Microb. Ecol. 60, 43–57. doi: 10.3354/ame01408
van Mooy, B. A. S., Fredricks, H. F., Pedler, B. E., Dyhrman, S. T., Karl, D. M., Koblížek, M., et al. (2009). Phytoplankton in the ocean use non-phosphorus lipids in response to phosphorus scarcity. Nature 458, 69–72. doi: 10.1038/nature07659
Voronkov, A., and Sinetova, M. (2019). Polyphosphate accumulation dynamics in a population of Synechocystis sp. PCC 6803 cells under phosphate overplus. Protoplasma 256, 1153–1164. doi: 10.1007/s00709-019-01374-2
Voss, B., Bolhuis, H., Fewer, D. P., Kopf, M., Möke, F., Haas, F., et al. (2013). Insights into the physiology and ecology of the brackish-water-adapted cyanobacterium Nodularia spumigena CCY9414 based on a genome-transcriptome analysis. PLoS One 8:60224. doi: 10.1371/journal.pone.0060224
Wan, L., Chen, X., Deng, Q., Yang, L., Li, X., Zhang, J., et al. (2019). Phosphorus strategy in bloom-forming cyanobacteria (dolichospermum and microcystis) and its role in their succession. Harmful Algae 84, 46–55. doi: 10.1016/j.hal.2019.02.007
Wickham, H., Averick, M., Bryan, J., Chang, W., McGowan, L. D. A., François, R., et al. (2019). Welcome to the tidyverse. J. Open Source Soft. 4:1686. doi: 10.21105/joss.01686
Williams, D. A. (1987). Generalized linear model diagnostics using the deviance and single case deletions. Appl. Statist. 36, 181–191.
Yuan, R., Li, J., Li, Y., Ren, L., Wang, S., and Kong, F. (2019). Formation mechanism of the Microcystis aeruginosa bloom in the water with low dissolved phosphorus. Mari. Pollut. Bull. 148, 194–201. doi: 10.1016/j.marpolbul.2019.07.074
Zerulla, K., Ludt, K., and Soppa, J. (2016). The ploidy level of Synechocystis sp. PCC 6803 is highly variable and is influenced by growth phase and by chemical and physical external parameters. Microbiology 162, 730–739. doi: 10.1099/mic.0.000264
Keywords: alkaline phosphatase, cyanobacterial bloom, diazotroph, polyphosphate, toxin, transcriptomics, transport
Citation: Santoro M, Hassenrück C, Labrenz M and Hagemann M (2023) Acclimation of Nodularia spumigena CCY9414 to inorganic phosphate limitation – Identification of the P-limitation stimulon via RNA-seq. Front. Microbiol. 13:1082763. doi: 10.3389/fmicb.2022.1082763
Received: 28 October 2022; Accepted: 05 December 2022;
Published: 04 January 2023.
Edited by:
Rehab El-Shehawy, Stockholm University, SwedenReviewed by:
Iris Maldener, University of Tübingen, GermanyOmer Murik, Shaare Zedek Medical Center, Israel
Copyright © 2023 Santoro, Hassenrück, Labrenz and Hagemann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Martin Hagemann, ✉ bWFydGluLmhhZ2VtYW5uQHVuaS1yb3N0b2NrLmRl
†These authors have contributed equally to this work
 Mariano Santoro
Mariano Santoro Christiane Hassenrück
Christiane Hassenrück Matthias Labrenz
Matthias Labrenz Martin Hagemann
Martin Hagemann