- 1Laboratory of Molecular Chemistry, Materials and Catalysis, Faculty of Sciences and Technics, Sultan Moulay Slimane University, Beni-Mellal, Morocco
- 2Higher School of Technology Fkih Ben Salah, Sultan Moulay Slimane University, Fkih Ben Salah, Morocco
- 3Laboratory of Microbiology and Virology, Faculty of Medicine and Pharmacy, Cadi Ayyad University, Marrakesh, Morocco
- 4AgroBiosciences Program, Mohammed VI Polytechnic University (UM6P), Ben Guerir, Morocco
- 5Labelled Research Unit N°4 CNRST, Laboratory of Microbial Biotechnologies, Agrosciences and Environment (BioMAgE), Faculty of Sciences Semlalia, Cadi Ayyad University, Marrakesh, Morocco
- 6Institut de Chimie des Milieux et Matériaux de Poitiers (IC2MP–CNRS UMR 7285), Université de Poitiers, Poitiers, France
Actinomycetota in the phylum of bacteria has been explored extensively as a source of antibiotics and secondary metabolites. In addition to acting as plant growth-promoting agents, they also possess the potential to control various plant pathogens; however, there are limited studies that report the facultative predatory ability of Actinomycetota spp. Furthermore, the mechanisms that underline predation are poorly understood. We assessed the diversity of strategies employed by predatory bacteria to attack and subsequently induce the cell lysing of their prey. We revisited the diversity and abundance of secondary metabolite molecules linked to the different predation strategies by bacteria species. We analyzed the pros and cons of the distinctive predation mechanisms and explored their potential for the development of new biocontrol agents. The facultative predatory behaviors diverge from group attack “wolfpack,” cell-to-cell proximity “epibiotic,” periplasmic penetration, and endobiotic invasion to degrade host-cellular content. The epibiotic represents the dominant facultative mode of predation, irrespective of the habitat origins. The wolfpack is the second-used approach among the Actinomycetota harboring predatory traits. The secondary molecules as chemical weapons engaged in the respective attacks were reviewed. We finally explored the use of predatory Actinomycetota as a new cost-effective and sustainable biocontrol agent against plant pathogens.
1. Introduction
Cellular tropism also referred to as “cellular predation,” is a regular interspecific antagonistic that occurs in diverse living habitats. It also defines an act of a predatory organism to kill and devour a prey organism for its nutritional requirements (Pérez et al., 2016). Predation behavior expands from primitive prokaryotic microbes to highly evolved mammals in the animal kingdom (Sinclair et al., 2003; Ripple and Beschta, 2004). Among the microorganisms, a family member of the virus, bacteria have developed predatory behaviors that are well investigated under in vivo and ex vivo conditions (Curds, 1982; Gonzalez et al., 1990; Parry, 2004). Myxobacteria and Bdellovibrio are δ-Proteobacteria in the bacteria phyla with well-known bacteriophagic nature (Stolp and Starr, 1963; Casida, 1982; Berleman et al., 2006; Martins et al., 2022). Through predation attitude, Myxobacteria, and Bdellovibrio contribute to community structuring and carbon recycling in the soil food web systems and play an important ecosystem function are well-known keystone taxa (Bratanis et al., 2020; Cavallo et al., 2021; Mookherjee and Jurkevitch, 2022; Ogundero et al., 2022; Whitworth, 2022; Wu et al., 2022). Thus, bacteria species in the actinomycetes species are gram-positive and mostly studied for secondary metabolites production and were recently discovered to exert a facultative predatory role (Zeph and Casida, 1986; Hoshino et al., 2015; Ait Barka et al., 2016; Katz and Baltz, 2016; Baltz, 2019; Ouchari et al., 2019; Ibrahimi et al., 2020; Korichi et al., 2021). Studies deciphering or investigating the facultative predation role of Actinomycetota are gaining growing research attention (Kumbhar et al., 2014; Ibrahimi et al., 2019, 2020; Baig et al., 2021) owing to their lifestyles adjustment to adapt to complex limited resources (Ibrahimi et al., 2019, 2020) and importantly potential application to design bio-pesticides molecules/products alternatives to heavy toxic pesticides inorganic molecules (Palaniyandi et al., 2013). The approach is driven by increasing consumer demands for safe, healthy, and organically produced foods globally (Alvarez et al., 2017, Sathya et al., 2017; AbdElgawad et al., 2020). However, in the context of Actinomycetota species, the mechanisms that underline opportunistic predation behaviors are under-investigated (Ibrahimi et al., 2019, 2020).
Predation is a bacteria co-evolutional trait in the Actinomycetota, owing to adjustments in lifestyles among the species, possible geographical local adaptations, and habitat change (Jousset, 2012; Ibrahimi et al., 2020). For a long time, Actinomycetota were only viewed as competitive, rather than predatory organisms (Kumbhar and Watve, 2013; Kumbhar et al., 2014; Ibrahimi et al., 2020; Pérez et al., 2020). As a consequence, despite their widespread ecological importance in the environment, there are very few investigations on Actinomycetota predation (Bentley et al., 2002; Mawang et al., 2021; Boubekri et al., 2022; Santos-Aberturas and Vior, 2022). The first evidence of the Actinomycetota opportunistic predation behaviors is described in the Streptomyces and Agromyces genera (Waksman and Woodruff, 1941; Casida, 1980). The author examined the utilization of Micrococcus luteus as prey by Streptomyces species. As versatile-opportunistic Actinomycetota, Streptomyces is also a non-obligate epibiotic predator of various microorganisms, specifically, Staphylococcus aureus, Escherichia coli, Bacillus sp., Pseudomonas aeruginosa, and Klebsiella sp. (Kumbhar et al., 2014). In addition, It was reported in the literature that under in vivo conditions, Streptomyces isolates exhibited the predatory ability on various prey bacteria cells (Gram+, Gram−) and multidrug-resistant strains (Ibrahimi et al., 2020). The latest supportive evidence of good predatory behavior by Streptomyces against various types of prey is reported by Baig et al. (2021). However, studies examining the mechanisms that underline predator–prey relationships and the diversity or identity of small signal molecules are lacking (Ibrahimi et al., 2020).
Increasing plant diseases due to pathogenic microbes represent an important global constraint for agricultural production and economic losses (Collinge and Sarrocco, 2022). Current interventions are toward the use of synthetic heavy toxic molecules pesticides for crop protection that negatively poses acute risks to human health and the environment (Rani et al., 2021). It is imperative to find alternative solutions for a sustainable crop yield (Collinge and Sarrocco, 2022). The increasing knowledge and understanding of plant–microbe interactions, in particular the predatory capability of Actinomycetota to design as a biopesticide product to combat plant-associated pathogenic microbes (Shivlata and Satyanarayana, 2017). The objective of the present study was to:
a) critically review the facultative predatory mechanisms of bacteria and Actinomycetota
b) discuss the various small molecules synthesized from bacteria and Actinomycetota species during their opportunistic predatory lifestyle stage; and
c) explore the potential beneficial use of Actinomycetota synthesized small molecules during the predation stages in the development of biocontrol agents for plant disease suppression and protection.
2. Mechanisms of predation by bacteria and Actinomycetota species
Predation by bacteria has traditionally attracted lower attention than their multicellular development or their production of bioactive compounds (Furness et al., 2020). In recent years, many aspects of bacterial predation are starting to be explored through research (Table 1). Since the purpose of a predatory bacteria is to kill and digest its prey, it remains necessary to understand the hunting and killing behavior of the predator. Most authors propose to classify bacterial hunting strategies into four general categories: epibiotic, group attack, or wolfpack, periplasmic penetration, and endobiotic predation or direct invasion (Martin, 2002; Jurkevitch, 2007; Berleman and Kirby, 2009; Pasternak et al., 2014; Pérez et al., 2016). Epibiotic is a tactic that requires close cell-to-cell proximity (Shi et al., 1993). When the predation is extracellular, the predator attached to the prey from outside does not invade either the periplasm or the cytoplasm of the prey, degrades and assimilates host molecules through specialized structures, but without penetrating the prey (Martin, 2002; Pérez et al., 2016), consuming it from the exterior before dividing into the daughter cells via binary division (Koval et al., 2013; Figure 1). Ensifer adhaerens (Druga et al., 2011), Myxococcus xanthus (Nair, 2016; Thiery and Kaimer, 2020), and Streptomyces (Casida, 1988; Ibrahimi et al., 2020) are some examples of epibiotic strategy. The cell-to-cell contact between the epibiotic predator and its prey is crucial for the transfer of compounds between their cells (Castelle and Banfield, 2018; Yakimov et al., 2022). As a consequence of the limited literature on culture-based investigations, our current state of understanding of the behavior of epibiotic lifestyles is relatively poor (Bor et al., 2020; Batinovic et al., 2021; Yakimov et al., 2022). However, Cross et al. (2019) suggested that to clearly demonstrate cell-to-cell contact, conditions that could enhance predator abundance to the degree that allows detailed microscopic characterization must be evaluated.
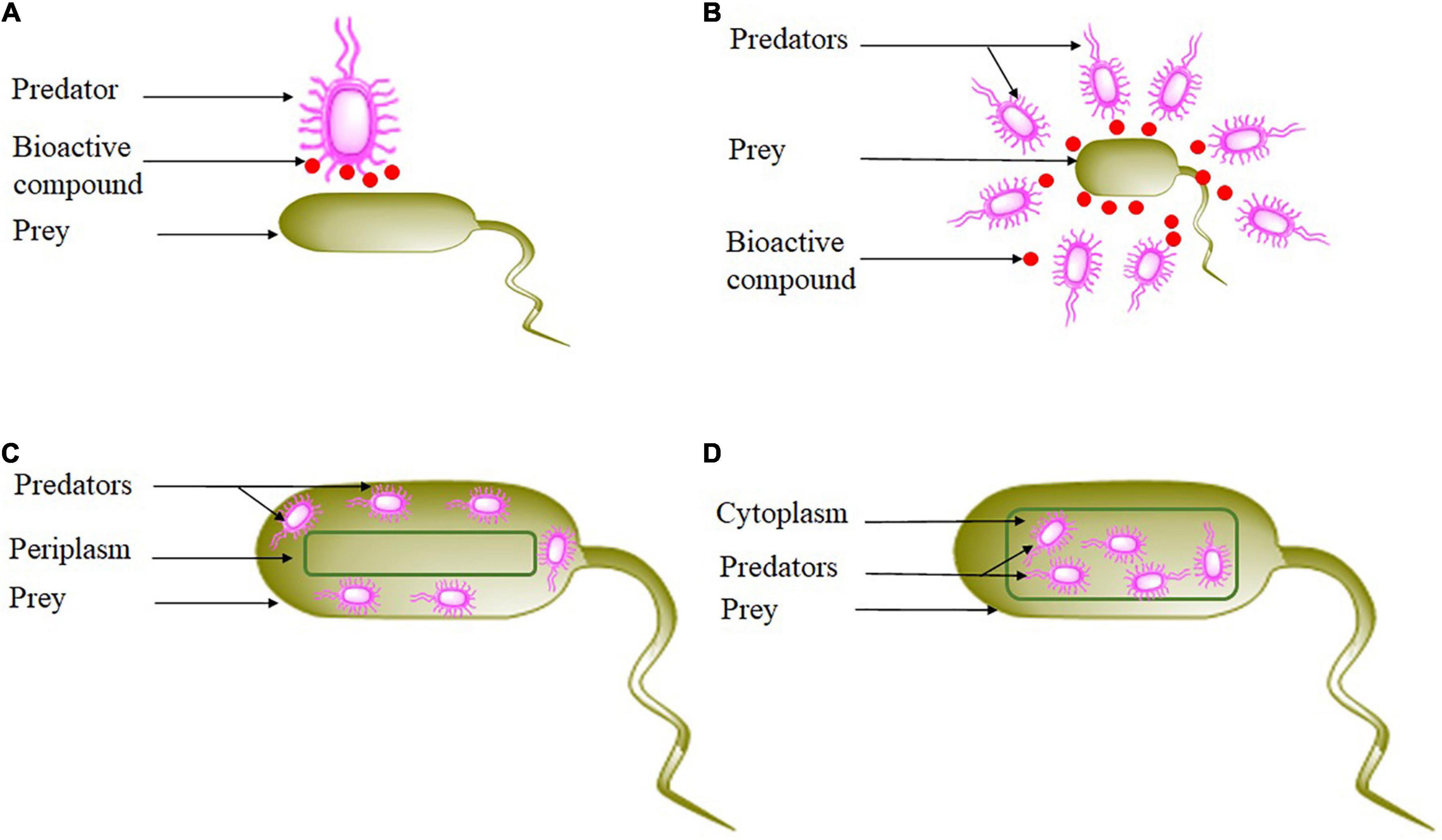
Figure 1. Bacterial hunting strategies. (A) Epibiotic strategy, (B) wolfpack strategy, (C) periplasmic strategy, and (D) endobiotic strategy.
The second strategy is wolfpack or group attack or group predation, predatory bacteria in this kind of predation work as a group (Marshall and Whitworth, 2019), they are assumed to hunt collectively to attack prey (Figure 1). They assemble and collectively secrete a diversity of diffusible compounds like hydrolytic enzymes and extracellular antibiotics that degrade and kill nearby bacteria. Wolfpack strategy lyse prey cells from the exterior through concerted action (Martin, 2002). Furthermore, the aim of the process of lysis is to produce small molecules that are easily assimilated by the predator (Xiao et al., 2011). Moreover, higher predatory cell densities suggest higher diffusible compounds (Keane and Berleman, 2016). The most important property of wolfpack is the lysed prey will be consumed by the predatory bacteria secreting and non-secreting (Mendes-Soares and Velicer, 2013). Lysobacter sp. (Hashizume et al., 2004; Li et al., 2008), Myxobacteria (McBride and Zusman, 1996; Thiery and Kaimer, 2020), and Herpetosiphon sp. (Nett et al., 2006) are examples of predatory bacteria using this strategy.
The third approach is when cells enter the prey periplasm (Figure 1). The predator invades and develops in the periplasm of gram-negative bacteria (Hobot et al., 1984; Ferguson, 1990; Ferguson et al., 1992). Predator produces hydrolytic enzymes that promote penetration and damage the prey cell wall (Lerner et al., 2012). In parallel, the infected prey is destroyed once respiration comes to a halt and the outer membrane is damaged (Thomashow and Rittenberg, 1978). The invading predator attaches to the prey’s cytoplasmic membrane and initiates growth using the cytoplasm of the prey as a nutrient supply. The predator grows like a polynucleotide filament, the length of which is dependent on the prey size (Kessel and Shilo, 1976). In the end, filaments septate into individual attack-phase cells that grow a flagellum, induce the formation of pores in the cell wall and burst into the external medium to engage in another cycle (Fenton et al., 2010). Therefore, the predator kills the prey by ingesting its cytoplasm. Bdellovibrionales (Jurkevitch and Davidov, 2006), Bdellovibrio bacteriovorus, Bacteriovorax marinus, Bacteriolyticum stolpii, and Peredibacter starrii, use periplasmic predation (Sockett and Lambert, 2004; Pasternak et al., 2014). However, the mechanism used by predators to release their intracellular contents to achieve their original cell cycle is poorly described (Laloux, 2020).
Moreover, some predatory bacteria can utilize more than one hunting strategy, like Bdellovibrio, which employs the periplasmic and epibiotic strategy; also, Myxobacteria can use both epibiotic and wolfpack strategies.
The last category of predation includes all predators that penetrate the host cytoplasm (Figure 1). This approach is also known as the invasion of cytoplasm or diacytotic strategy (Moulder, 1985). After the penetration, the predator grows and divides inside the cytoplasm. Daptobacter is the only bacteria that employ this strategy (Guerrero et al., 1986), but no other study has been reported about this group. Among predatory Actinomycetota, a few investigations have highlighted strategies used in predation because they have not received the level of attention of competitors (Ibrahimi, 2020). Pérez et al. (2016) suggest that bacteria secreting secondary metabolites, including Actinomycetota, can attack their prey in groups, which is supported by the fact that they are social prokaryotes that form and develop in multicellular structures. Correspondingly, Kumbhar and Watve (2013) reported that antibiotic producers such as Actinomycetota are unable to use an endobiotic strategy and direct contact with the prey is not required. Kumbhar et al. (2014) showed that Actinomycetota are non-obligate epibiotic predators of diverse prey such as S. aureus, E. coli, Bacillus spp., Pseudomonas aeruginosa, and Klebsiella spp. Recently, a study by Zeng et al. (2021) demonstrated that Streptomyces globisporus, a predatory Actinomycetota, preyed on Microcystis through an epibiotic mode of predation. Overall, among the phylum Actinomycetota, only two genera, Agromyces and Streptomyces, are known to have an epibiotic predatory behavior against other bacterial species (Casida, 1983; Arcamone et al., 2000; Ibrahimi et al., 2020). All this information allows us to believe that the predatory Actinomycetota can hunt their prey through wolfpack and epibiotic strategies. Therefore, our understanding of Actinomycetota predation is still very fragmentary, including gaps in their mechanisms of predation. Understanding these mechanisms by Actinomycetota species is required for better knowledge and understanding of their effect on prey structure to develop a new strategy to control plant disease and multidrug-resistant pathogens.
3. Identity and diversity of small molecules produced from bacteria and Actinomycetota during predation lifestyles
Predatory bacteria represent a diversified collection of prokaryotic organisms that have the ability to consume other bacteria (Jurkevitch, 2007). While some of these bacteria act as solitary hunters, others are known to hunt in groups in a wide mixture before they attack their prey (Martin, 2002). This predatory strategy generally implicates the production of lytic enzymes and small bioactive compounds as predatory weapons (Rosenberg and Varon, 1984; Reichenbach and Höfle, 1993; Berleman and Kirby, 2009), while genome sequencing programs of these microorganisms have revealed the presence of very broad and varied secondary metabolites (Kiss et al., 2011). Therefore, extraction and purification of antimicrobial molecules from predatory bacteria have yielded the discovery of numerous novel molecules, as illustrated by jahnellamides (Plaza et al., 2013), salimyxins (Felder et al., 2013), cystomanamides (Etzbach et al., 2014), and precorallopyronin (Schäberle et al., 2015). It is confirmed by Xiao et al. (2011) that this variety of secondary metabolites is supposed to be involved in the death of the prey. It has been found that a defect in the production of these substances significantly affects predatory activity. In this section, all research relative to the results of chemical studies of compounds produced by predatory bacteria will be reviewed and critically analyzed in Table 2.
Traditionally, Actinomycetota species are renowned for their excellent potential to produce secondary metabolites and antibiotics compounds (Hoshino et al., 2015; Ait Barka et al., 2016; Ouchari et al., 2019; Ibrahimi et al., 2020; Korichi et al., 2021). These molecules are produced to antagonize the growth of surrounding microbes (Ortiz-Ortiz et al., 2013). To date, up to 33 new secondary metabolites have been successfully isolated from 12 Actinomycetota through the co-culture (Hoshino et al., 2019). With their novel roles as predatory microbes, the diversity and identity of molecules produced during predation strategies should be considered an untapped source of biomolecules. A further co-culturing of predator and prey monitored for 15 days induced an increase in the total amount of methylated fatty acids biomarker of the predatory Actinomycetota responsible for the predation process (Ibrahimi et al., 2019). Therefore, secondary metabolites engaged in the facultative predation and antagonistic mechanism are likely to differ, from the Actinomycetota strain, the origin of habitat, or the ecology of isolated strains (McBride and Zusman, 1996; Jurkevitch, 2007; Octaviana, 2021).
Predatory Actinomycetota are mostly isolated from marine and soil environments (Kumbhar et al., 2014; Ibrahimi et al., 2020; Baig et al., 2021). They exhibit a wide range of predatory activities against diverse bacteria (Kumbhar et al., 2014; Ibrahimi et al., 2020; Baig et al., 2021). This is explained by the bioactive compound that they produce, which possess a range of antimicrobial activities. These molecules are used as a weapon by predatory Actinomycetota to kill their prey (Kumbhar and Watve, 2013). Recently, Baig et al. (2021) demonstrated a strong correlation between predation and enzyme inhibition, particularly trypsin and chymotrypsin inhibition, in which predatory Actinomycetota were found to release more enzymes in the presence of prey. On this basis, it has become apparent that Actinomycetota can exhibit the great potential to produce antibiotics and enzymes during predation. Also, it should be noted that, to date, no compounds produced by predatory Actinomycetota during predation behavior have been elucidated. Therefore, it will be interesting to conduct further studies to isolate new predatory Actinomycetota as well as the extraction and identification of their molecules involved in predation lifestyle.
4. Predatory Actinomycetota as an eco-friendly and promising tool in agricultural and environmental sustainability
4.1. Plant pathogens
Recent years have been marked by an expanding array of virulent infectious diseases caused by pests which are increasingly recognized as presenting a worldwide threat to food security (Olsen et al., 2011; Bosso et al., 2015; Hartmann, 2022). In addition, the extensive use of agrochemicals molecules has led to the development of bacterial resistance, causing significant risks to the environment and human health (Ebele Mbachu et al., 2022). Consequently, providing food for the world’s population without disrupting the environmental balance is becoming eminent (Pandit et al., 2022). It is highly recommended to provide sustainable solutions for agriculture (Jamiołkowska, 2020). Microbes are an alternative to agrochemical molecules like synthetic pesticides for controlling plant pathogens (Elnahal et al., 2022). Different microorganisms are used as biocontrol agents, such as bacteria, fungi, and Actinomycetota (Table 3). Direct antagonism and predation are the possible modes of action of biocontrol agents to eliminate plant parasites (Ebele Mbachu et al., 2022).
Predatory bacteria can be used as alternative applications in biological control (Olanya and Lakshman, 2015; Swain et al., 2017). Most of the Bacillus strains can control the plant pathogen as Fusarium fungi (Ongena and Jacques, 2008). Bdellovibrio also attacks a large variety of different plant pathogens (Baer et al., 2000; Jurkevitch et al., 2000; Dwidar et al., 2012). In addition, a very promising result in the control by Bdellovibrio of Pseudomonas glycinea blight of soybean was demonstrated by Scherff (1973). Recently, Ye et al. (2020) demonstrated the use of the Myxobacterium Corallococcus coralloides to control cucumber Fusarium wilt by migrating to the plant root and regulating the soil microbial community.
In the case of Actinomycetota, several investigations showed their successful ability to function as biocontrol agents against plant pathogens (Goudjal et al., 2014; Braga et al., 2016; Ebrahimi-Zarandi et al., 2022). The co-cultivation of Actinomycetota with other microorganisms generates several new secondary metabolites, which are not present during pure culture conditions (Scherlach and Hertweck, 2009; Sung et al., 2017; Wakefield et al., 2017; Shin et al., 2018; Vikeli et al., 2019; Yu M. et al., 2019). Indeed, the production of secondary metabolites in co-culture is enhanced by competitive or antagonistic interactions (Ibrahimi et al., 2020). For example, the co-cultivation of the Streptomyces coelicolor with the agricultural pathogen Aspergillus niger has activated the actinorhodin silent pathway in Actinomycetota (Wu et al., 2015).
Prior research suggests that Actinomycetota in co-culture can inhibit pathogens’ growth, decrease, and degrade toxins. For example, a recent investigation demonstrated that coculturing Streptomyces roseolus with the phytopathogen Aspergillus flavus could reduce the contamination generated by the mycotoxin aflatoxin B1, which is produced by A. flavus (Caceres et al., 2018). In addition, several Streptomyces strains showed the ability to inhibit Aspergillus flavus growth and decrease and degrade mycotoxin (Verheecke et al., 2015; Campos-Avelar et al., 2021). Another field that seems promising for the use of Actinomycetota in agriculture is their function as eco-friendly biofertilizers since they are involved in nutrient management, soil quality, decomposing of organic matter, enhancing plant growth promoting, recycling organic residues, and activating plant immune responses (Shivlata and Satyanarayana, 2017; Boubekri et al., 2022; Ebrahimi-Zarandi et al., 2022).
Using predatory Actinomycetota in co-culture may be a potential biocontrol agent and biofertilizer used in agriculture. Indeed, the plant can benefit on several different levels. It was reported in the literature that the co-cultivation of Streptomyces with the phytopathogenic fungus Sclerotinia sclerotiorum induces the deformation and fragmentation of the fungal mycelium through the production of hydrolytic enzymes and secondary metabolites (Liu et al., 2019). In parallel, the same study linked the promotion of plant growth through the solubilization of inorganic phosphate and the production of 1-aminocyclopropane-1-carboxylate deaminase and indole acetic acid by the Actinomycetota (Liu et al., 2019).
The predation of gram-negative and gram-positive bacteria by Actinomycetota may enhance their potential application for the biocontrol of foodborne and plant pathogens (Ibrahimi et al., 2019, 2020). Thus, predatory Actinomycetota can provide an ecologically sustainable solution for agricultural farming. This is explained by the fact that they will not increase the accumulation of antibiotics in the environment, which may generate antibiotic resistance (Pérez et al., 2020).
4.2. Cyanobacterial bloom
Cyanobacterial bloom can produce toxic molecules called cyanotoxins in freshwater (Saraf et al., 2018). The presence of such molecules can affect the functionality of ecosystems and water quality for recreation, drinking water, fisheries, and agriculture (O’Neil et al., 2012; Corbel et al., 2014). In the agriculture field, several publications have appeared in recent years documenting the bioaccumulation of cyanotoxins in plants used for human and animal food (Corbel et al., 2014; Machado et al., 2017; Ai et al., 2020; Melaram et al., 2022). Consequently, several physical, chemical, and biological strategies were deployed to control cyanobacterial bloom (Jia et al., 2018; Sun et al., 2018; Yu Y. et al., 2019; El Amrani Zerrifi et al., 2020). Although the physic-chemical techniques represent a high cost, the risk of contamination and toxicity to humans has limited their general use (Moreira et al., 2014). Whereas biological techniques involving microorganisms have attracted researchers for their promising eco-friendly tools and high potential (Yu Y. et al., 2019).
One of the most promising aspects is the use of microbes of predatory bacteria as a control agent of cyanobacterial blooms. However, despite some initial promising discoveries, this field has been almost completely ignored. The first predatory bacteria able to lyse various species of cyanobacteria was first reported in 1967 (Shilo, 1967). Since then, numerous strains of lytic gliding bacteria, mainly members of the Myxobacteria and Cytophaga groups, have been isolated (Daft and Stewart, 1971; Granhall and Berg, 1972; Gumbo et al., 2008) and lysed cyanobacteria cells by attachment and secretion of diffusible lytic substances. These bacteria produce a variety of different exoenzymes capable of hydrolyzing the cyanobacterial cell wall (Sudo and Dworkin, 1972; Gnosspelius, 1978). Subsequently, Rashidan and Bird (2001) isolated two strains of Cytophaga sp., with lytic activity on different cyanobacteria with a restricted host range. The same study showed that the lysis of cyanobacteria by predatory bacteria may be an important factor in their population dynamics in lakes and may contribute to the prevention or the sudden disappearance of cyanobacteria blooms and draw attention to the possibilities of using host-specific lytic bacteria in biological control of harmful cyanobacterial blooms (Rashidan and Bird, 2001).
Furthermore, Bdellovibrio and Myxococcus have received a lot of investigation as predators of cyanobacterial bloom (Bauer and Forchhammer, 2021). In contrast, the use of Actinomycetota as a predator is still broadly uncharacterized. However, recently Zeng et al. (2021) demonstrated the use of a predatory Actinomycetota to face harmful cyanobacterial algal blooms. The research demonstrated that Streptomyces globisporus could predate Microcystis aeruginosa via cell-to-cell contact with high algicidal activity (Zeng et al., 2021). The present findings confirm the use of predatory Actinomycetota as promising eco-friendly tools to combat harmful cyanobacteria blooms. Also, further investigations are highly recommended to assess the predation toward cyanobacteria. In addition, it is necessary to recognize the predatory Actinomycetota-cyanobacteria ratio due to their important role as a key to achieving effective lysis of cyanobacteria.
5. Future perspectives and conclusion
Nowadays, one of the most alarming world wild problems is the increase of plant diseases caused by pathogenic bacteria, which causes great economic, environmental, and human health damage. Therefore, finding alternative and sustainable solutions to confront these pathogens represent one of the biggest challenges. In this review, we suggest the use of predatory Actinomycetota as an effective biocontrol agent. Predation is an important cause of mortality and determines the structure and activity of microbial communities in both terrestrial and aquatic ecosystems, with a complex process involving several components such as prey finding, recognition, consumption, and digestion. The literature has concentrated largely on the presence of predatory Actinomycetota and their effective activity against a panel of pathogenic microorganisms but has not examined the mechanisms of the prey lysis. Indeed, a good knowledge of the predation mechanism used by predatory Actinomycetota is essential to improve the efficiency of the lysis of the prey cells and predation performance. The application of predatory Actinomycetota in large-scale systems and field experiments must be examined to determine if there will be ecological consequences. Furthermore, it is suggested that research evaluating predators’ efficiency should include molecular viability assays, such as EMAqPCR and EMA-Illumina, to determine the efficacy of treatment. Finally, during this review, we noticed that antibiotics produced during the process of predation by predatory Actinomycetota are demonstrated but have not yet been identified. Consequently, it seems advisable to discover such compounds. In the future, actinobacterial predation could be a new approach to control plant pathogen, cyanobacterial blooms; however, intensive research efforts are required to pursue this aim.
Author contributions
MI wrote the manuscript. YO and MJ refining and critical reading of the manuscript. LL, SL, and MH revised the manuscript. All authors contributed to the manuscript and approved the submitted version.
Funding
This study was financially supported by the French Ministries of Europe and Foreign Affairs (MEAE), French Ministries of Higher Education, Research and Innovation (MESRI), the Moroccan Ministry of Higher Education, Research and Managerial Training via the Partnership Hubert Curien (PHC) Toubkal n° 17/48–Campus France n° 36856WA, and the OCP Africa to Mohamed VI Polytechnic University to develop sustainable production in Africa (no. 01/OCP AFRICA/UM6P).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
AbdElgawad, H., Abuelsoud, W., Madany, M. M., Selim, S., Zinta, G., Mousa, A. S., et al. (2020). Actinomycetes enrich soil rhizosphere and improve seed quality as well as productivity of legumes by boosting nitrogen availability and metabolism. Biomolecules 10:1675. doi: 10.3390/biom10121675
Ai, Y., Lee, S., and Lee, J. (2020). Drinking water treatment residuals from cyanobacteria bloom-affected areas: Investigation of potential impact on agricultural land application. Sci. Total Environ. 706:135756. doi: 10.1016/j.scitotenv.2019.135756
Ait Barka, E. A., Vatsa, P., Sanchez, L., Gaveau-Vaillant, N., Jacquard, C., Klenk, H.-P., et al. (2016). Taxonomy, physiology, and natural products of actinobacteria. Microbiol. Mol. Biol. Rev. 80, 1–43. doi: 10.1128/MMBR.00019-15
Alvarez, A., Saez, J. M., Costa, J. S. D., Colin, V. L., Fuentes, M. S., Cuozzo, S. A., et al. (2017). Actinobacteria: Current research and perspectives for bioremediation of pesticides and heavy metals. Chemosphere 166, 41–62. doi: 10.1016/j.chemosphere.2016.09.070
Arcamone, F., Cassinelli, G., Fantini, G., Grein, A., Orezzi, P., Pol, C., et al. (2000). Adriamycin, 14-Hydroxydaunomycin, a new antitumor antibiotic from S. peucetius var. caesius. Biotechnol. Bioeng. 67, 704–713. doi: 10.1002/(SICI)1097-0290(20000320)67:6<704::AID-BIT8>3.0.CO;2-L
Baer, M. L., Ravel, J., Chun, J., Hill, R. T., and Williams, H. N. (2000). A proposal for the reclassification of Bdellovibrio stolpii and Bdellovibrio starrii into a new genus, Bacteriovorax gen. nov. as Bacteriovorax stolpii comb. nov. and Bacteriovorax starrii comb. nov., respectively. Int. J. Syst. Evol. Microbiol. 50, 219–224. doi: 10.1099/00207713-50-1-219
Baig, U., Dahanukar, N., Shintre, N., Holkar, K., Pund, A., Lele, U., et al. (2021). Phylogenetic diversity and activity screening of cultivable actinobacteria isolated from marine sponges and associated environments from the western coast of India. Access Microbiol. 3:000242. doi: 10.1099/acmi.0.000242
Baltz, R. H. (2019). Natural product drug discovery in the genomic era: Realities, conjectures, misconceptions, and opportunities. J. Ind. Microbiol. Biotechnol. 46, 281–299. doi: 10.1007/s10295-018-2115-4
Banning, E. C. (2010). Biology and potential biogeochemical impacts of novel predatory flavobacteria. DTIC Document, Massachusetts Institute of Technology: Cambridge, MA.
Batinovic, S., Rose, J. J., Ratcliffe, J., Seviour, R. J., and Petrovski, S. (2021). Cocultivation of an ultrasmall environmental parasitic bacterium with lytic ability against bacteria associated with wastewater foams. Nat. Microbiol. 6, 703–711. doi: 10.1038/s41564-021-00892-1
Bauer, A., and Forchhammer, K. (2021). Bacterial predation on cyanobacteria. Microb. Physiol. 31, 99–108. doi: 10.1159/000516427
Baumann, S., Herrmann, J., Raju, R., Steinmetz, H., Mohr, K. I., Hüttel, S., et al. (2014). Cystobactamids: Myxobacterial topoisomerase inhibitors exhibiting potent antibacterial activity. Angew. Chem. Int. Ed. 53, 14605–14609. doi: 10.1002/anie.201409964
Be’Er, A., Zhang, H. P., Florin, E.-L., Payne, S. M., Ben-Jacob, E., and Swinney, H. L. (2009). Deadly competition between sibling bacterial colonies. Proc. Natl. Acad. Sci. U.S.A. 106, 428–433. doi: 10.1073/pnas.0811816106
Bentley, S. D., Chater, K. F., Cerdeño-Tárraga, A.-M., Challis, G. L., Thomson, N. R., James, K. D., et al. (2002). Complete genome sequence of the model actinomycete Streptomyces coelicolor A3 (2). Nature 417:141.
Berleman, J. E., and Kirby, J. R. (2009). Deciphering the hunting strategy of a bacterial wolfpack. FEMS Microbiol. Rev. 33, 942–957. doi: 10.1111/j.1574-6976.2009.00185.x
Berleman, J. E., Chumley, T., Cheung, P., and Kirby, J. R. (2006). Rippling is a predatory behavior in Myxococcus xanthus. J. Bacteriol. 188, 5888–5895. doi: 10.1128/JB.00559-06
Bor, B., Collins, A. J., Murugkar, P. P., Balasubramanian, S., To, T. T., Hendrickson, E. L., et al. (2020). Insights obtained by culturing Saccharibacteria with their bacterial hosts. J. Dent. Res. 99, 685–694. doi: 10.1177/0022034520905792
Bosso, L., Scelza, R., Testa, A., Cristinzio, G., and Rao, M. A. (2015). Depletion of pentachlorophenol contamination in an agricultural soil treated with Byssochlamys nivea, Scopulariopsis brumptii and urban waste compost: A laboratory microcosm study. Water Air Soil Pollut. 226:183. doi: 10.1007/s11270-015-2436-0
Boubekri, K., Soumare, A., Mardad, I., Lyamlouli, K., Ouhdouch, Y., Hafidi, M., et al. (2022). Multifunctional role of actinobacteria in agricultural production sustainability: A review. Microbiol. Res. 261:127059. doi: 10.1016/j.micres.2022.127059
Braga, R. M., Dourado, M. N., and Araújo, W. L. (2016). Microbial interactions: Ecology in a molecular perspective. Braz. J. Microbiol. 47, 86–98. doi: 10.1016/j.bjm.2016.10.005
Bratanis, E., and Lood, R. (2019). A novel broad-spectrum elastase-like serine protease from the predatory bacterium Bdellovibrio bacteriovorus facilitates elucidation of site-specific IgA glycosylation pattern. Front. Microbiol. 10, 971. doi: 10.3389/fmicb.2019.00971
Bratanis, E., Andersson, T., Lood, R., and Bukowska-Faniband, E. (2020). Biotechnological potential of Bdellovibrio and like organisms and their secreted enzymes. Front. Microbiol. 11:662. doi: 10.3389/fmicb.2020.00662
Bratanis, E., Molina, H., Naegeli, A., Collin, M., and Lood, R. (2017). BspK, a serine protease from the predatory bacterium Bdellovibrio bacteriovorus with utility for analysis of therapeutic antibodies. Appl. Environ. Microbiol. 83, e03037-16. doi: 10.1128/AEM.03037-16
Bull, C. T., Shetty, K. G., and Subbarao, K. V. (2002). Interactions between myxobacteria, plant pathogenic fungi, and biocontrol agents. Plant Dis. 86, 889–896. doi: 10.1094/PDIS.2002.86.8.889
Caceres, I., Snini, S. P., Puel, O., and Mathieu, F. (2018). Streptomyces roseolus, a promising biocontrol agent against Aspergillus flavus, the main aflatoxin B1 producer. Toxins 10:442. doi: 10.3390/toxins10110442
Cain, C. C., Lee, D., Waldo, R. H., Henry, A. T., Casida, E. J., Wani, M. C., et al. (2003). Synergistic antimicrobial activity of metabolites produced by a nonobligate bacterial predator. Antimicrob. Agents Chemother. 47, 2113–2117. doi: 10.1128/AAC.47.7.2113-2117.2003
Campos-Avelar, I., Colas De La Noue, A., Durand, N., Cazals, G., Martinez, V., Strub, C., et al. (2021). Aspergillus flavus growth inhibition and aflatoxin B1 decontamination by Streptomyces isolates and their metabolites. Toxins 13:340. doi: 10.3390/toxins13050340
Cao, H., An, J., Zheng, W., and He, S. (2015). Vibrio cholerae pathogen from the freshwater-cultured whiteleg shrimp Penaeus vannamei and control with Bdellovibrio bacteriovorus. J. Invertebr. Pathol. 130, 13–20. doi: 10.1016/j.jip.2015.06.002
Cao, H., Wang, H., Yu, J., An, J., and Chen, J. (2019). Encapsulated Bdellovibrio powder as a potential bio-disinfectant against whiteleg shrimp-pathogenic vibrios. Microorganisms 7:244. doi: 10.3390/microorganisms7080244
Casida, L. E. (1980). Bacterial predators of Micrococcus luteus in soil. Appl. Environ. Microbiol. 39, 1035–1041. doi: 10.1128/aem.39.5.1035-1041.1980
Casida, L. E. (1983). Interaction of Agromyces ramosus with other bacteria in soil. Appl. Environ. Microbiol. 46, 881–888. doi: 10.1128/aem.46.4.881-888.1983
Casida, L. E. (1988). Minireview: Nonobligate bacterial predation of bacteria in soil. Microb. Ecol. 15, 1–8. doi: 10.1007/BF02012948
Casida, L. E. (1992). Competitive ability and survival in soil of Pseudomonas strain 679-2, a dominant, nonobligate bacterial predator of bacteria. Appl. Environ. Microbiol. 58, 32–37. doi: 10.1128/aem.58.1.32-37.1992
Casida, L. E. Jr. (1982). Ensifer adhaerens gen. nov., sp. nov.: A bacterial predator of bacteria in soil. Int. J. Syst. Evol. Microbiol. 32, 339–345. doi: 10.1099/00207713-32-3-339
Castelle, C. J., and Banfield, J. F. (2018). Major new microbial groups expand diversity and alter our understanding of the tree of life. Cell 172, 1181–1197. doi: 10.1016/j.cell.2018.02.016
Cavallo, F. M., Jordana, L., Friedrich, A. W., Glasner, C., and van Dijl, J. M. (2021). Bdellovibrio bacteriovorus: A potential ‘living antibiotic’ to control bacterial pathogens. Crit. Rev. Microbiol. 47, 630–646. doi: 10.1080/1040841X.2021.1908956
Chernyakovskaya, T. F., Dobrovol’skaya, T. G., and Bab’eva, I. P. (2004). The ability of saprotrophic bacteria isolated from natural habitats to lyse yeasts. Microbiology 73, 482–484. doi: 10.1023/B:MICI.0000036995.67735.df
Collinge, D. B., and Sarrocco, S. (2022). Transgenic approaches for plant disease control: Status and prospects 2021. Plant Pathol. 71, 207–225. doi: 10.1111/ppa.13443
Corbel, S., Mougin, C., and Bouaïcha, N. (2014). Cyanobacterial toxins: Modes of actions, fate in aquatic and soil ecosystems, phytotoxicity and bioaccumulation in agricultural crops. Chemosphere 96, 1–15. doi: 10.1016/j.chemosphere.2013.07.056
Cortina, N. S., Krug, D., Plaza, A., Revermann, O., and Müller, R. (2012). Myxoprincomide: A natural product from Myxococcus xanthus discovered by comprehensive analysis of the secondary metabolome. Angew. Chem. Int. Ed. 51, 811–816. doi: 10.1002/anie.201106305
Cotter, T. W., and Thomashow, M. F. (1992). A conjugation procedure for Bdellovibrio bacteriovorus and its use to identify DNA sequences that enhance the plaque-forming ability of a spontaneous host-independent mutant. J. Bacteriol. 174, 6011–6017. doi: 10.1128/jb.174.19.6011-6017.1992
Cross, K. L., Campbell, J. H., Balachandran, M., Campbell, A. G., Cooper, C. J., Griffen, A., et al. (2019). Targeted isolation and cultivation of uncultivated bacteria by reverse genomics. Nat. Biotechnol. 37, 1314–1321. doi: 10.1038/s41587-019-0260-6
Curds, C. R. (1982). The ecology and role of protozoa in aerobic sewage treatment processes. Annu. Rev. Microbiol. 36, 27–28. doi: 10.1146/annurev.mi.36.100182.000331
Daft, M. J., and Stewart, W. D. P. (1971). Bacterial pathogens of freshwater blue-green algae. New Phytol. 70, 819–829. doi: 10.1111/j.1469-8137.1971.tb02582.x
Dashiff, A., Junka, R. A., Libera, M., and Kadouri, D. E. (2011). Predation of human pathogens by the predatory bacteria Micavibrio aeruginosavorus and Bdellovibrio bacteriovorus. J. Appl. Microbiol. 110, 431–444. doi: 10.1111/j.1365-2672.2010.04900.x
Davidov, Y., Huchon, D., Koval, S. F., and Jurkevitch, E. (2006). A new α-proteobacterial clade of Bdellovibrio-like predators: Implications for the mitochondrial endosymbiotic theory. Environ. Microbiol. 8, 2179–2188. doi: 10.1111/j.1462-2920.2006.01101.x
Douillet, P. (2003). Strains of Bacillus for biological control of pathogenic fungi. Google Patents. Available online at: https://www.google.com/patents/US6589524 (accessed June 16, 2017).
Druga, B., Suteu, D., Rosca-Casian, O., Parvu, M., and Dragos, N. (2011). Two novel Alliin lyase (alliinase) genes from twisted-leaf garlic (Allium obliquum) and mountain garlic (Allium senescens ssp. montanum). Notulae Bot. Horti Agrobot. Cluj Napoca 39:293. doi: 10.15835/nbha3926355
Dwidar, M., Monnappa, A. K., and Mitchell, R. J. (2012). The dual probiotic and antibiotic nature of Bdellovibrio bacteriovorus. BMB Rep. 45, 71–78. doi: 10.5483/BMBRep.2012.45.2.71
Ebele Mbachu, A., Obianom, A. O., Ogbonna, U. S., Mbachu, N. A. and Abumchukwu Okoli, F. (2022). Mode of attack of microbiological control agents against plant pathogens for sustainable agriculture and food security. Asian J. Agric. Hortic. Res. 9, 1–16. doi: 10.9734/ajahr/2022/v9i130132
Ebrahimi-Zarandi, M., Saberi Riseh, R., and Tarkka, M. T. (2022). Actinobacteria as effective biocontrol agents against plant pathogens, an overview on their role in eliciting plant defense. Microorganisms 10:1739. doi: 10.3390/microorganisms10091739
El Amrani Zerrifi, S., El Khalloufi, F., Mugani, R., El Mahdi, R., Kasrati, A., Soulaimani, B., et al. (2020). Seaweed essential oils as a new source of bioactive compounds for cyanobacteria growth control: Innovative ecological biocontrol approach. Toxins 12:527. doi: 10.3390/toxins12080527
Elnahal, A. S., El-Saadony, M. T., Saad, A. M., Desoky, E. S. M., El-Tahan, A. M., Rady, M. M., et al. (2022). The use of microbial inoculants for biological control, plant growth promotion, and sustainable agriculture: A review. Eur. J. Plant Pathol. 162, 759–792. doi: 10.1007/s10658-021-02393-7
Etzbach, L., Plaza, A., Garcia, R., Baumann, S., and Müller, R. (2014). Cystomanamides: Structure and biosynthetic pathway of a family of glycosylated lipopeptides from myxobacteria. Org. Lett. 16, 2414–2417. doi: 10.1021/ol500779s
Ezzedine, J. A., Jacas, L., Desdevises, Y., and Jacquet, S. (2020). Bdellovibrio and like organisms in Lake Geneva: An unseen elephant in the room? Front. Microbiol. 11:98. doi: 10.3389/fmicb.2020.00098
Felder, S., Kehraus, S., Neu, E., Bierbaum, G., Schäberle, T. F., and König, G. M. (2013). Salimyxins and enhygrolides: Antibiotic, sponge-related metabolites from the obligate marine myxobacterium Enhygromyxa salina. ChemBioChem 14, 1363–1371. doi: 10.1002/cbic.201300268
Feng, S., Tan, C. H., Constancias, F., Kohli, G. S., Cohen, Y., and Rice, S. A. (2017). Predation by Bdellovibrio bacteriovorus significantly reduces viability and alters the microbial community composition of activated sludge flocs and granules. FEMS microbiol. Ecol. 93:fix020. doi: 10.1093/femsec/fix020
Fenton, A. K., Lambert, C., Wagstaff, P. C., and Sockett, R. E. (2010). Manipulating each MreB of Bdellovibrio bacteriovorus gives diverse morphological and predatory phenotypes. J. Bacteriol. 192, 1299–1311. doi: 10.1128/JB.01157-09
Ferguson, S. J. (1990). Periplasm underestimated. Trends Biochem. Sci. 15:377. doi: 10.1016/0968-0004(90)90234-3
Ferguson, S. J., Mohan, S., Dow, C., and Cole, J. A. (1992). The periplasm. New York, NY: Cambridge University Press.
Fratamico, P. M., and Cooke, P. H. (1996). Isolation of Bdellovibrios that prey on Escherichia coli O157: H7 and Salmonella species and application for removal of prey from stainless steel surfaces1. J. Food Safety 16, 161–173. doi: 10.1111/j.1745-4565.1996.tb00157.x
Fratamico, P. M., and Whiting, R. C. (1995). Ability of Bdellovibrio bacteriovorus 109J to lyse gram-negative food-borne pathogenic and spoilage bacteria. J. Food Prot. 58, 160–164. doi: 10.4315/0362-028X-58.2.160
Fujimoto, H., Kinoshita, T., Suzuki, H., and Umezawa, H. (1970). Studies on the mode of action of althiomycin. J. Antibiot. 23, 271–275. doi: 10.7164/antibiotics.23.271
Furness, E., Whitworth, D. E., and Zwarycz, A. (2020). “Predatory interactions between myxobacteria and their prey,” in The ecology of predation at the microscale, eds E. Jurkevitch and R. Mitchell (Cham: Springer), 1–36. doi: 10.1007/978-3-030-45599-6_1
Furusawa, G., Yoshikawa, T., Yasuda, A., and Sakata, T. (2003). Algicidal activity and gliding motility of Saprospira sp. SS98-5. Can. J. Microbiol. 49, 92–100. doi: 10.1139/w03-017
Germida, J. J., and Casida, L. E. (1983). Ensifer adhaerens predatory activity against other bacteria in soil, as monitored by indirect phage analysis. Appl. Environ. Microbiol. 45, 1380–1388. doi: 10.1128/aem.45.4.1380-1388.1983
Gerth, K., Irschik, H., Reichenbach, H., and Trowitzsch, W. (1982). The myxovirescins, a family of antibiotics from Myxococcus virescens (Myxobacterales). J. Antibiot. 35, 1454–1459. doi: 10.7164/antibiotics.35.1454
Gerth, K., Jansen, R., Reifenstahl, G., Höfle, G., Irschik, H., Kunze, B., et al. (1983). The myxalamids, new antibiotics from Myxococcus xanthus (Myxobacterales). J. Antibiot. 36, 1150–1156. doi: 10.7164/antibiotics.36.1150
Gnosspelius, G. (1978). Purification and properties of an extracellular protease from Myxococcus virescens. J. Bacteriol. 133, 17–25. doi: 10.1128/jb.133.1.17-25.1978
Gonzalez, J. M., Sherr, E. B., and Sherr, B. F. (1990). Size-selective grazing on bacteria by natural assemblages of estuarine flagellates and ciliates. Appl. Environ. Microbiol. 56, 583–589. doi: 10.1128/aem.56.3.583-589.1990
Goudjal, Y., Toumatia, O., Yekkour, A., Sabaou, N., Mathieu, F., and Zitouni, A. (2014). Biocontrol of Rhizoctonia solani damping-off and promotion of tomato plant growth by endophytic actinomycetes isolated from native plants of Algerian Sahara. Microbiol. Res. 169, 59–65. doi: 10.1016/j.micres.2013.06.014
Granhall, U., and Berg, B. (1972). Antimicrobial effects of Cellvibrio on blue-green algae. Arch. Microbiol. 84, 234–242. doi: 10.1007/BF00425201
Guerrero, R., Pedrós-Alió, C., Esteve, I., Mas, J., Chase, D., and Margulis, L. (1986). Predatory prokaryotes: Predation and primary consumption evolved in bacteria. Proc. Natl. Acad. Sci. U.S.A. 83, 2138–2142. doi: 10.1073/pnas.83.7.2138
Gumbo, J. R., Ross, G., and Cloete, T. E. (2010). The isolation and identification of predatory bacteria from a Microcystis algal bloom. Afr. J. Biotechnol. 9, 663–671. doi: 10.5897/AJB09.834
Gumbo, R. J., Ross, G., and Cloete, E. T. (2008). Biological control of Microcystis dominated harmful algal blooms. Afr. J. Biotechnol. 7, 4765–4773.
Hartmann, F. E. (2022). Using structural variants to understand the ecological and evolutionary dynamics of fungal plant pathogens. New Phytol. 234, 43–49. doi: 10.1111/nph.17907
Hashizume, H., Hirosawa, S., Sawa, R., Muraoka, Y., Ikeda, D., Naganawa, H., et al. (2004). Tripropeptins, novel antimicrobial agents produced by Lysobacter sp. J. Antibiot. 57, 52–58. doi: 10.7164/antibiotics.57.52
Hobley, L., Lerner, T. R., Williams, L. E., Lambert, C., Till, R., Milner, D. S., et al. (2012). Genome analysis of a simultaneously predatory and prey-independent, novel Bdellovibrio bacteriovorus from the River Tiber, supports in silico predictions of both ancient and recent lateral gene transfer from diverse bacteria. BMC Genomics 13:670. doi: 10.1186/1471-2164-13-670
Hobot, J. A., Carlemalm, E., Villiger, W., and Kellenberger, E. (1984). Periplasmic gel: New concept resulting from the reinvestigation of bacterial cell envelope ultrastructure by new methods. J. Bacteriol. 160, 143–152. doi: 10.1128/jb.160.1.143-152.1984
Hoshino, S., Onaka, H., and Abe, I. (2019). Activation of silent biosynthetic pathways and discovery of novel secondary metabolites in actinomycetes by co-culture with mycolic acid-containing bacteria. J. Ind. Microbiol. Biotechnol. 46, 363–374. doi: 10.1007/s10295-018-2100-y
Hoshino, S., Wakimoto, T., Onaka, H., and Abe, I. (2015). Chojalactones A–C, cytotoxic butanolides isolated from Streptomyces sp. Cultivated with mycolic acid containing bacterium. Org. Lett. 17, 1501–1504. doi: 10.1021/acs.orglett.5b00385
Ibrahimi, M. (2020). Extraction et caractérisation de nouveaux antibactériens produits par les actinobactéries prédatrices d’origine marine. Doctoral dissertation, Marrakech: Université Cadi Ayyad.
Ibrahimi, M., Korichi, W., Hafidi, M., Lemee, L., Ouhdouch, Y., and Loqman, S. (2020). Marine actinobacteria: Screening for predation leads to the discovery of potential new drugs against multidrug-resistant bacteria. Antibiotics 9:91. doi: 10.3390/antibiotics9020091
Ibrahimi, M., Korichi, W., Loqman, S., Hafidi, M., Ouhdouch, Y., and Lemee, L. (2019). Thermochemolysis–GC-MS as a tool for chemotaxonomy and predation monitoring of a predatory Actinobacteria against a multidrug resistant bacteria. J. Anal. Appl. Pyrolysis 145:104740. doi: 10.1016/j.jaap.2019.104740
Iebba, V., Totino, V., Santangelo, F., Gagliardi, A., Ciotoli, L., Virga, A., et al. (2014). Bdellovibrio bacteriovorus directly attacks Pseudomonas aeruginosa and Staphylococcus aureus Cystic fibrosis isolates. Front. Microbiol. 5:280. doi: 10.3389/fmicb.2014.00280
Irschik, H., Jansen, R., Höfle, G., Gerth, K., and Reichenbach, H. (1985). The corallopyronins, new inhibitors of bacterial RNA synthesis from myxobacteria. J. Antibiot. 38, 145–152. doi: 10.7164/antibiotics.38.145
Jafarian, N., Sepahi, A. A., Naghavi, N. S., Hosseini, F., and Nowroozi, J. (2020). Using autochthonous Bdellovibrio as a predatory bacterium for reduction of Gram-negative pathogenic bacteria in urban wastewater and reuse it. Iran. J. Microbiol. 12:556. doi: 10.18502/ijm.v12i6.5030
Jamiołkowska, A. (2020). Natural compounds as elicitors of plant resistance against diseases and new biocontrol strategies. Agronomy 10:173. doi: 10.3390/agronomy10020173
Jia, P., Zhou, Y., Zhang, X., Zhang, Y., and Dai, R. (2018). Cyanobacterium removal and control of algal organic matter (AOM) release by UV/H2O2 pre-oxidation enhanced Fe (II) coagulation. Water Res. 131, 122–130. doi: 10.1016/j.watres.2017.12.020
Jousset, A. (2012). Ecological and evolutive implications of bacterial defences against predators. Environ. Microbiol. 14, 1830–1843. doi: 10.1111/j.1462-2920.2011.02627.x
Jurkevitch, E. (2007). Predatory behaviors in bacteria-diversity and transitions. Microbe Am. Soc. Microbiol. 2:67. doi: 10.1128/microbe.2.67.1
Jurkevitch, E., and Davidov, Y. (2006). “Phylogenetic diversity and evolution of predatory prokaryotes,” in Predatory prokaryotes microbiology monographs, ed. E. Jurkevitch (Berlin: Springer), 11–56. doi: 10.1007/7171_052
Jurkevitch, E., and Ramati, B. (2000). Design and uses of Bdellovibrio 16S rRNA-targeted oligonucleotides. FEMS Microbiol. Lett. 184, 265–271. doi: 10.1111/j.1574-6968.2000.tb09025.x
Jurkevitch, E., Minz, D., Ramati, B., and Barel, G. (2000). Prey range characterization, ribotyping, and diversity of soil and rhizosphere Bdellovibrio spp. isolated on phytopathogenic bacteria. Appl. Environ. Microbiol. 66, 2365–2371. doi: 10.1128/AEM.66.6.2365-2371.2000
Kadouri, D. E., To, K., Shanks, R. M., and Doi, Y. (2013). Predatory bacteria: A potential ally against multidrug-resistant Gram-negative pathogens. PLoS One 8:e63397. doi: 10.1371/journal.pone.0063397
Kadouri, D., Venzon, N. C., and O’Toole, G. A. (2007). Vulnerability of pathogenic biofilms to Micavibrio aeruginosavorus. Appl. Environ. Microbiol. 73, 605–614. doi: 10.1128/AEM.01893-06
Katz, L., and Baltz, R. H. (2016). Natural product discovery: Past, present, and future. J. Ind. Microbiol. Biotechnol. 43, 155–176. doi: 10.1007/s10295-015-1723-5
Keane, R., and Berleman, J. (2016). The predatory life cycle of Myxococcus xanthus. Microbiology 162, 1–11. doi: 10.1099/mic.0.000208
Kessel, M., and Shilo, M. (1976). Relationship of Bdellovibrio elongation and fission to host cell size. J. Bacteriol. 128, 477–480. doi: 10.1128/jb.128.1.477-480.1976
Kiss, H., Nett, M., Domin, N., Martin, K., Maresca, J. A., Copeland, A., et al. (2011). Complete genome sequence of the filamentous gliding predatory bacterium Herpetosiphon aurantiacus type strain (114-95 T). Stand. Genomic Sci. 5:356. doi: 10.4056/sigs.2194987
Kohl, W., Irschik, H., Reichenbach, H., and Höfle, G. (1983). Antibiotika aus Gleitenden Bakterien, XVII. Myxopyronin A und B – zwei neue Antibiotika aus Myxococcus fulvus Stamm Mx f50. Liebigs Ann. Chem. 1983, 1656–1667. doi: 10.1002/jlac.198319831003
Korichi, W., Ibrahimi, M., Loqman, S., Ouhdouch, Y., Younes, K., and Lemée, L. (2021). Assessment of Actinobacteria use in the elimination of multidrug-resistant bacteria of Ibn Tofail hospital wastewater (Marrakesh, Morocco): A chemometric data analysis approach. Environ. Sci. Pollut. Res. 28, 26840–26848. doi: 10.1007/s11356-021-12445-4
Korp, J., Gurovic, M. S. V., and Nett, M. (2016). Antibiotics from predatory bacteria. Beilstein J. Organ. Chem. 12:594. doi: 10.3762/bjoc.12.58
Koval, S. F., Hynes, S. H., Flannagan, R. S., Pasternak, Z., Davidov, Y., and Jurkevitch, E. (2013). Bdellovibrio exovorus sp. nov., a novel predator of Caulobacter crescentus. Int. J. Syst. Evol. Microbiol. 63, 146–151. doi: 10.1099/ijs.0.039701-0
Kreutzer, M. F., Kage, H., and Nett, M. (2012). Structure and biosynthetic assembly of cupriachelin, a photoreactive siderophore from the bioplastic producer Cupriavidus necator H16. J. Am. Chem. Soc. 134, 5415–5422. doi: 10.1021/ja300620z
Krug, D., Zurek, G., Revermann, O., Vos, M., Velicer, G. J., and Müller, R. (2008). Discovering the hidden secondary metabolome of Myxococcus xanthus: A study of intraspecific diversity. Appl. Environ. Microbiol. 74, 3058–3068. doi: 10.1128/AEM.02863-07
Kumbhar, C., and Watve, M. (2013). Why antibiotics: A comparative evaluation of different hypotheses for the natural role of antibiotics and an evolutionary synthesis. Nat. Sci. 5, 26–40. doi: 10.4236/ns.2013.54A005
Kumbhar, C., Mudliar, P., Bhatia, L., Kshirsagar, A., and Watve, M. (2014). Widespread predatory abilities in the genus Streptomyces. Arch. Microbiol. 196, 235–248. doi: 10.1007/s00203-014-0961-7
Kunze, B., Bedorf, N., Kohl, W., Höfle, G., and Reichenbach, H. (1989). Myxochelin A, a new iron-chelating compound from Angiococcus disciformis (Myxobacterales). Production, isolation, physico-chemical and biological properties. J. Antibiot. 42, 14–17. doi: 10.7164/antibiotics.42.14
Kunze, B., Reichenbach, H., Augustiniak, H., and Höfle, G. (1982). Isolation and identification of althiomycin from Cystobacter fuscus (Myxobacterales). J. Antibiot. 35, 635–636. doi: 10.7164/antibiotics.35.635
Laloux, G. (2020). Shedding light on the cell biology of the predatory bacterium Bdellovibrio bacteriovorus. Front. Microbiol. 10:3136. doi: 10.3389/fmicb.2019.03136
Lerner, T. R., Lovering, A. L., Bui, N. K., Uchida, K., Aizawa, S.-I., Vollmer, W., et al. (2012). Specialized peptidoglycan hydrolases sculpt the intra-bacterial niche of predatory Bdellovibrio and increase population fitness. PLoS Pathog. 8:e1002524. doi: 10.1371/journal.ppat.1002524
Li, S., Jochum, C. C., Yu, F., Zaleta-Rivera, K., Du, L., Harris, S. D., et al. (2008). An antibiotic complex from Lysobacter enzymogenes strain C3: Antimicrobial activity and role in plant disease control. Phytopathology 98, 695–701. doi: 10.1094/PHYTO-98-6-0695
Liu, D., Yan, R., Fu, Y., Wang, X., Zhang, J., and Xiang, W. (2019). Antifungal, plant growth-promoting, and genomic properties of an endophytic actinobacterium Streptomyces sp. NEAU-S7GS2. Front. Microbiol. 10:2077. doi: 10.3389/fmicb.2019.02077
Machado, J., Campos, A., Vasconcelos, V., and Freitas, M. (2017). Effects of microcystin-LR and cylindrospermopsin on plant-soil systems: A review of their relevance for agricultural plant quality and public health. Environ. Res. 153, 191–204. doi: 10.1016/j.envres.2016.09.015
Makkar, N. S., and Casida, L. E. Jr. (1987). Cupriavidus necator gen. nov., sp. nov.; a nonobligate bacterial predator of bacteria in soil. Int. J. Syst. Evol. Microbiol. 37, 323–326. doi: 10.1099/00207713-37-4-323
Markelova, N. Y. (2010). Predacious bacteria, Bdellovibrio with potential for biocontrol. Int. J. Hygiene Environ. Health 213, 428–431. doi: 10.1016/j.ijheh.2010.08.004
Marshall, R. C., and Whitworth, D. E. (2019). Is “Wolf-Pack” predation by antimicrobial bacteria cooperative? Cell behaviour and predatory mechanisms indicate profound selfishness, even when working alongside kin. Bioessays 41:1800247. doi: 10.1002/bies.201800247
Martin, M. O. (2002). Predatory prokaryotes: An emerging research opportunity. J. Mol. Microbiol. Biotechnol. 4, 467–478.
Martins, S. J., Taerum, S. J., Triplett, L., Emerson, J. B., Zasada, I., de Toledo, B. F., et al. (2022). Predators of soil bacteria in plant and human health. Phytobiomes J. 6, 184–200. doi: 10.1094/PBIOMES-11-21-0073-RVW
Maurhofer, M., Reimmann, C., Schmidli-Sacherer, P., Heeb, S., Haas, D., and Défago, G. (1998). Salicylic acid biosynthetic genes expressed in Pseudomonas fluorescens strain P3 improve the induction of systemic resistance in tobacco against tobacco necrosis virus. Phytopathology 88, 678–684. doi: 10.1094/PHYTO.1998.88.7.678
Mawang, C. I., Azman, A. S., Fuad, A. S. M., and Ahamad, M. (2021). Actinobacteria: An eco-friendly and promising technology for the bioaugmentation of contaminants. Biotechnol. Rep. 32:e00679. doi: 10.1016/j.btre.2021.e00679
McBride, M. J., and Zusman, D. R. (1996). Behavioral analysis of single cells of Myxococcus xanthus in response to prey cells of Escherichia coli. FEMS Microbiol. Lett. 137, 227–231. doi: 10.1111/j.1574-6968.1996.tb08110.x
McBride, M. J., Xie, G., Martens, E. C., Lapidus, A., Henrissat, B., Rhodes, R. G., et al. (2009). Novel features of the polysaccharide-digesting gliding bacterium Flavobacterium johnsoniae as revealed by genome sequence analysis. Appl. Environ. Microbiol. 75, 6864–6875. doi: 10.1128/AEM.01495-09
Meiser, P., Bode, H. B., and Müller, R. (2006). The unique DKxanthene secondary metabolite family from the myxobacterium Myxococcus xanthus is required for developmental sporulation. Proc. Natl. Acad. Sci. U.S.A. 103, 19128–19133. doi: 10.1073/pnas.0606039103
Melaram, R., Newton, A. R., and Chafin, J. (2022). Microcystin contamination and toxicity: Implications for agriculture and public health. Toxins 14:350. doi: 10.3390/toxins14050350
Mendes-Soares, H., and Velicer, G. J. (2013). Decomposing predation: Testing for parameters that correlate with predatory performance by a social bacterium. Microbial Ecol. 65, 415–423. doi: 10.1007/s00248-012-0135-6
Miyanaga, S., Obata, T., Onaka, H., Fujita, T., Saito, N., Sakurai, H., et al. (2006). Absolute configuration and antitumor activity of myxochelin a produced by Nonomuraea pusilla TP-A0861†. J. Antibiot. 59, 698–703. doi: 10.1038/ja.2006.93
Monnappa, A. K., Dwidar, M., Seo, J. K., Hur, J.-H., and Mitchell, R. J. (2014). Bdellovibrio bacteriovorus inhibits Staphylococcus aureus biofilm formation and invasion into human epithelial cells. Sci. Rep. 4:3811. doi: 10.1038/srep03811
Mookherjee, A., and Jurkevitch, E. (2022). Interactions between Bdellovibrio and like organisms and bacteria in biofilms: Beyond predator–prey dynamics. Environ. Microbiol. 24, 998–1011. doi: 10.1111/1462-2920.15844
Moreira, C., Ramos, V., Azevedo, J., and Vasconcelos, V. (2014). Methods to detect cyanobacteria and their toxins in the environment. Appl. Microbiol. Biotechnol. 98, 8073–8082. doi: 10.1007/s00253-014-5951-9
Moulder, J. W. (1985). Comparative biology of intracellular parasitism. Microbiol. Rev. 49:298. doi: 10.1128/mr.49.3.298-337.1985
Nakano, C., Oshima, M., Kurashima, N., and Hoshino, T. (2015). Identification of a new diterpene biosynthetic gene cluster that produces O-Methylkolavelool in Herpetosiphon aurantiacus. ChemBioChem 16, 772–781. doi: 10.1002/cbic.201402652
Nett, M., Erol, Ö, Kehraus, S., Köck, M., Krick, A., Eguereva, E., et al. (2006). Siphonazole, an unusual metabolite from Herpetosiphon sp. Angew. Chemie Int. Ed. 45, 3863–3867. doi: 10.1002/anie.200504525
O’Neil, J. M., Davis, T. W., Burford, M. A., and Gobler, C. J. (2012). The rise of harmful cyanobacteria blooms: The potential roles of eutrophication and climate change. Harmful Algae 14, 313–334. doi: 10.1016/j.hal.2011.10.027
O’Sullivan, J., McCullough, J. E., Tymiak, A. A., Kirsch, D. R., Trejo, W. H., and Principe, P. A. (1988). Lysobactin, a novel antibacterial agent produced by Lysobacter sp. J. Antibiot. 41, 1740–1744. doi: 10.7164/antibiotics.41.1740
Octaviana, S. (2021). Exploring the diversity and antimicrobial potential of predatory bacteria from Indonesian mangroves. Ph.D. dissertation. Braunschweig: Universitätsbibliothek Braunschweig.
Ogundero, A., Vignola, M., Connelly, S., and Sloan, W. T. (2022). Validating flow cytometry as a method for quantifying Bdellovibrio predatory bacteria and its prey for microbial ecology. Microbiol. Spectrum 10:e01033-21. doi: 10.1128/spectrum.01033-21
Olanya, O. M., and Lakshman, D. K. (2015). Potential of predatory bacteria as biocontrol agents for foodborne and plant pathogens. J. Plant Pathol. 97, 405–417.
Olsen, L., Choffnes, E. R., Relman, D. A., and Pray, L. (2011). Fungal diseases: An emerging threat to human, animal and plant health. Workshop summary. Washington, DC: National Academies Press.
Ongena, M., and Jacques, P. (2008). Bacillus lipopeptides: Versatile weapons for plant disease biocontrol. Trends Microbiol. 16, 115–125. doi: 10.1016/j.tim.2007.12.009
Ortiz-Ortiz, L., Bojalil, L. F., and Yakoleff, V. (2013). Biological, biochemical, and biomedical aspects of actinomycetes. Amsterdam: Elsevier.
Ouchari, L., Boukeskasse, A., Bouizgarne, B., and Ouhdouch, Y. (2019). Antimicrobial potential of actinomycetes isolated from the unexplored hot Merzouga desert and their taxonomic diversity. Biol. Open 8:bio035410. doi: 10.1242/bio.035410
Palaniyandi, S. A., Yang, S. H., Zhang, L., and Suh, J. W. (2013). Effects of Actinobacteria on plant disease suppression and growth promotion. Appl. Microbiol. Biotechnol. 97, 9621–9636. doi: 10.1007/s00253-013-5206-1
Pandit, M. A., Kumar, J., Gulati, S., Bhandari, N., Mehta, P., Katyal, R., et al. (2022). Major biological control strategies for plant pathogens. Pathogens 11:273. doi: 10.3390/pathogens11020273
Parry, J. D. (2004). Protozoan grazing of freshwater biofilms. Adv. Appl. Microbiol. 54, 167–196. doi: 10.1016/S0065-2164(04)54007-8
Pasternak, Z., Njagi, M., Shani, Y., Chanyi, R., Rotem, O., Lurie-Weinberger, M. N., et al. (2014). In and out: An analysis of epibiotic vs periplasmic bacterial predators. ISME J. 8, 625–635. doi: 10.1038/ismej.2013.164
Pérez, J., Contreras-Moreno, F. J., Marcos-Torres, F. J., Moraleda-Muñoz, A., and Muñoz-Dorado, J. (2020). The antibiotic crisis: How bacterial predators can help. Comput. Struct. Biotechnol. J. 18, 2547–2555. doi: 10.1016/j.csbj.2020.09.010
Pérez, J., Moraleda-Muñoz, A., Marcos-Torres, F. J., and Muñoz-Dorado, J. (2016). Bacterial predation: 75 years and counting! Environ. Microbiol. 18, 766–779. doi: 10.1111/1462-2920.13171
Plaza, A., Viehrig, K., Garcia, R., and Müller, R. (2013). Jahnellamides, α-Keto-β-methionine-containing peptides from the terrestrial myxobacterium Jahnella sp.: Structure and biosynthesis. Org. Lett. 15, 5882–5885. doi: 10.1021/ol402967y
Quinn, G. R., and Skerman, V. B. (1980). Herpetosiphon—nature’s scavenger? Curr. Microbiol. 4, 57–62. doi: 10.1007/BF02602893
Rani, L., Thapa, K., Kanojia, N., Sharma, N., Singh, S., Grewal, A. S., et al. (2021). An extensive review on the consequences of chemical pesticides on human health and environment. J. Clean. Prod. 283:124657. doi: 10.1016/j.jclepro.2020.124657
Rashidan, K. K., and Bird, D. F. (2001). Role of predatory bacteria in the termination of a cyanobacterial bloom. Microb. Ecol. 41, 97–105. doi: 10.1007/s002480000074
Raza, W., Yang, W., and Shen, Q. R. (2008). Paenibacillus polymyxa: Antibiotics, hydrolytic enzymes and hazard assessment. J. Plant Pathol. 90, 419–430.
Reichenbach, H., and Höfle, G. (1993). Biologically active secondary metabolites from myxobacteria. Biotechnol. Adv. 11, 219–277. doi: 10.1016/0734-9750(93)90042-L
Richards, G. P., Fay, J. P., Dickens, K. A., Parent, M. A., Soroka, D. S., and Boyd, E. F. (2012). Predatory bacteria as natural modulators of Vibrio parahaemolyticus and Vibrio vulnificus in seawater and oysters. Appl. Environ. Microbiol. 78, 7455–7466. doi: 10.1128/AEM.01594-12
Richards, G. P., Watson, M. A., Boyd, E. F., Burkhardt, W., Lau, R., Uknalis, J., et al. (2013). Seasonal levels of the Vibrio predator Bacteriovorax in Atlantic, Pacific, and gulf coast seawater. Int. J. Microbiol. 2013:375371. doi: 10.1155/2013/375371
Ripple, W. J., and Beschta, R. L. (2004). Wolves and the ecology of fear: Can predation risk structure ecosystems? BioScience 54, 755–766. doi: 10.1641/0006-3568(2004)054[0755:WATEOF]2.0.CO;2
Rosenberg, E., and Varon, M. (1984). “Antibiotics and lytic enzymes,” in Myxobacteria, ed. E. Rosenberg (New York, NY: Springer), 109–125. doi: 10.1007/978-1-4613-8280-5_5
Ryan, R. P., Monchy, S., Cardinale, M., Taghavi, S., Crossman, L., Avison, M. B., et al. (2009). The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat. Rev. Microbiol. 7, 514–525. doi: 10.1038/nrmicro2163
Santos-Aberturas, J., and Vior, N. M. (2022). Beyond soil-dwelling actinobacteria: Fantastic antibiotics and where to find them. Antibiotics 11:195. doi: 10.3390/antibiotics11020195
Saraf, S. R., Frenkel, A., Harke, M. J., Jankowiak, J. G., Gobler, C. J., and McElroy, A. E. (2018). Effects of microcystis on development of early life stage Japanese medaka (Oryzias latipes): Comparative toxicity of natural blooms, cultured Microcystis and microcystin-LR. Aquat. Toxicol. 194, 18–26. doi: 10.1016/j.aquatox.2017.10.026
Sathya, A., Vijayabharathi, R., and Gopalakrishnan, S. (2017). Plant growth-promoting Actinobacteria: A new strategy for enhancing sustainable production and protection of grain legumes. 3 Biotech 7, 1–10. doi: 10.1007/s13205-017-0736-3
Saw, J. H., Yuryev, A., Kanbe, M., Hou, S., Young, A. G., Aizawa, S.-I., et al. (2012). Complete genome sequencing and analysis of Saprospira grandis str. Lewin, a predatory marine bacterium. Stand. Genomic Sci. 6:84.
Saxon, E. B., Jackson, R. W., Bhumbra, S., Smith, T., and Sockett, R. E. (2014). Bdellovibrio bacteriovorus HD100 guards against Pseudomonas tolaasii brown-blotch lesions on the surface of post-harvest Agaricus bisporus supermarket mushrooms. BMC Microbiol. 14:163. doi: 10.1186/1471-2180-14-163
Schäberle, T. F., Schmitz, A., Zocher, G., Schiefer, A., Kehraus, S., Neu, E., et al. (2015). Insights into structure–activity relationships of bacterial RNA polymerase inhibiting corallopyronin derivatives. J. Nat. Prod. 78, 2505–2509. doi: 10.1021/acs.jnatprod.5b00175
Scherff, R. H. (1973). Control of bacterial blight of soybean by Bdellovibrio bacteriovorus. Phytopathology 63, 400–402. doi: 10.1094/Phyto-63-400
Scherlach, K., and Hertweck, C. (2009). Triggering cryptic natural product biosynthesis in microorganisms. Org. Biomol. Chem. 7, 1753–1760. doi: 10.1039/b821578b
Schieferdecker, S., Domin, N., Hoffmeier, C., Bryant, D. A., Roth, M., and Nett, M. (2015). Structure and absolute configuration of Auriculamide, a natural product from the predatory bacterium Herpetosiphon aurantiacus. Eur. J. Org. Chem. 2015, 3057–3062. doi: 10.1002/ejoc.201500181
Schieferdecker, S., König, S., Weigel, C., Dahse, H.-M., Werz, O., and Nett, M. (2014). Structure and biosynthetic assembly of gulmirecins, macrolide antibiotics from the predatory bacterium Pyxidicoccus fallax. Chem. Eur. J. 20, 15933–15940. doi: 10.1002/chem.201404291
Schisler, D. A., Khan, N. I., Boehm, M. J., and Slininger, P. J. (2002). Greenhouse and field evaluation of biological control of Fusarium head blight on durum wheat. Plant Dis. 86, 1350–1356. doi: 10.1094/PDIS.2002.86.12.1350
Schwudke, D., Strauch, E., Krueger, M., and Appel, B. (2001). Taxonomic studies of predatory Bdellovibrios based on 16S rRNA analysis, ribotyping and the hit locus and characterization of isolates from the gut of animals. Syst. Appl. Microbiol. 24, 385–394. doi: 10.1078/0723-2020-00042
Seccareccia, I., Kost, C., and Nett, M. (2015). Quantitative analysis of Lysobacter predation. Appl. Environ. Microbiol. 81, 7098–7105. doi: 10.1128/AEM.01781-15
Shi, W., Köhler, T., and Zusman, D. R. (1993). Chemotaxis plays a role in the social behaviour of Myxococcus xanthus. Mol. Microbiol. 9, 601–611. doi: 10.1111/j.1365-2958.1993.tb01720.x
Shilo, M. (1967). Formation and mode of action of algal toxins. Bacteriol. Rev. 31, 180–193. doi: 10.1128/br.31.3.180-193.1967
Shin, D., Byun, W. S., Moon, K., Kwon, Y., Bae, M., Um, S., et al. (2018). Coculture of marine Streptomyces sp. with Bacillus sp. Produces a new piperazic acid-bearing cyclic peptide. Front. Chem. 6:498. doi: 10.3389/fchem.2018.00498
Shivlata, L., and Satyanarayana, T. (2017). “Actinobacteria in agricultural and environmental sustainability,” in Agro-environmental sustainability, eds J. Singh and G. Seneviratne (Cham: Springer), 173–218. doi: 10.1007/978-3-319-49724-2_9
Sinclair, A. R. E., Mduma, S., and Brashares, J. S. (2003). Patterns of predation in a diverse predator–prey system. Nature 425, 288–290. doi: 10.1038/nature01934
Sockett, R. E. (2009). Predatory lifestyle of Bdellovibrio bacteriovorus. Annu. Rev. Microbiol. 63, 523–539. doi: 10.1146/annurev.micro.091208.073346
Sockett, R. E., and Lambert, C. (2004). Bdellovibrio as therapeutic agents: A predatory renaissance? Nat. Rev. Microbiol. 2, 669–675. doi: 10.1038/nrmicro959
Stolp, H., and Starr, M. P. (1963). Bdellovibrio bacteriovorus gen. et sp. n., a predatory, ectoparasitic, and bacteriolytic microorganism. Antonie Van Leeuwenhoek 29, 217–248. doi: 10.1007/BF02046064
Sudo, S., and Dworkin, M. (1972). Bacteriolytic enzymes produced by Myxococcus xanthus. J. Bacteriol. 110, 236–245. doi: 10.1128/jb.110.1.236-245.1972
Sun, R., Sun, P., Zhang, J., Esquivel-Elizondo, S., and Wu, Y. (2018). Microorganisms-based methods for harmful algal blooms control: A review. Bioresour. Technol. 248, 12–20. doi: 10.1016/j.biortech.2017.07.175
Sung, A., Gromek, S., and Balunas, M. (2017). Upregulation and identification of antibiotic activity of a marine-derived Streptomyces sp. via co-cultures with human pathogens. Mar. Drugs 15:250. doi: 10.3390/md15080250
Swain, D. M., Yadav, S. K., Tyagi, I., Kumar, R., Kumar, R., Ghosh, S., et al. (2017). A prophage tail-like protein is deployed by Burkholderia bacteria to feed on fungi. Nat. Commun. 8, 1–9. doi: 10.1038/s41467-017-00529-0
Thiery, S., and Kaimer, C. (2020). The predation strategy of Myxococcus xanthus. Front. Microbiol. 11:2. doi: 10.3389/fmicb.2020.00002
Thomashow, M. F., and Rittenberg, S. C. (1978). Penicillin-induced formation of osmotically stable spheroplasts in nongrowing Bdellovibrio bacteriovorus. J. Bacteriol. 133, 1484–1491. doi: 10.1128/jb.133.3.1484-1491.1978
Trick, I., and Lingens, F. (1984). Characterization of Herpetosiphon spec. —A gliding filamentous bacterium from bulking sludge. Appl. Microbiol. Biotechnol. 19, 191–198. doi: 10.1007/BF00256453
Verheecke, C., Liboz, T., Anson, P., Zhu, Y., and Mathieu, F. (2015). Streptomyces–Aspergillus flavus interactions: Impact on aflatoxin B accumulation. Food Addit. Contam. Part A 32, 572–576. doi: 10.1080/19440049.2014.1003336
Vikeli, E., Widdick, D. A., Batey, S. F., Heine, D., Holmes, N. A., Bibb, M. J., et al. (2019). In situ activation and heterologous production of a cryptic lantibiotic from a plant-ant derived Saccharopolyspora species. Appl. Environ. Microbiol. 86:e01876-19. doi: 10.1128/AEM.01876-19
Wakefield, J., Hassan, H. M., Jaspars, M., Ebel, R., and Rateb, M. E. (2017). Dual induction of new microbial secondary metabolites by fungal bacterial co-cultivation. Front. Microbiol. 8:1284. doi: 10.3389/fmicb.2017.01284
Waksman, S. A., and Woodruff, H. B. (1941). Actinomyces antibioticus, a new soil organism antagonistic to pathogenic and non-pathogenic bacteria. J. Bacteriol. 42:231. doi: 10.1128/jb.42.2.231-249.1941
Wenzel, S. C., and Müller, R. (2009). Myxobacteria—‘microbial factories’ for the production of bioactive secondary metabolites. Mol. BioSyst. 5, 567–574. doi: 10.1039/b901287g
Whitworth, D. E. (2022). Myxobacteria: Physiology and regulation. Microorganisms 10:805. doi: 10.3390/microorganisms10040805
Wu, C., Zacchetti, B., Ram, A. F., Van Wezel, G. P., Claessen, D., and Hae Choi, Y. (2015). Expanding the chemical space for natural products by Aspergillus-Streptomyces co-cultivation and biotransformation. Sci. Rep. 5, 1–10. doi: 10.1038/srep10868
Wu, Z., Li, Y., Chen, H., Rao, J., and Sun, Q. (2022). Effects of straw mulching on predatory myxobacterial communities in different soil aggregates under wheat-corn rotation. Environ. Sci. Pollut. Res. 29, 29062–29074. doi: 10.1007/s11356-021-18350-0
Xiao, Y., Gerth, K., Müller, R., and Wall, D. (2012). Myxobacterium-produced antibiotic TA (myxovirescin) inhibits type II signal peptidase. Antimicrob. Agents Chemother. 56, 2014–2021. doi: 10.1128/AAC.06148-11
Xiao, Y., Wei, X., Ebright, R., and Wall, D. (2011). Antibiotic production by myxobacteria plays a role in predation. J. Bacteriol. 193, 4626–4633. doi: 10.1128/JB.05052-11
Yakimov, M. M., Merkel, A. Y., Gaisin, V. A., Pilhofer, M., Messina, E., Hallsworth, J. E., et al. (2022). Cultivation of a vampire: ‘Candidatus Absconditicoccus praedator’. Environ. Microbiol. 24, 30–49. doi: 10.1111/1462-2920.15823
Ye, X., Li, Z., Luo, X., Wang, W., Li, Y., Li, R., et al. (2020). A predatory myxobacterium controls cucumber Fusarium wilt by regulating the soil microbial community. Microbiome 8, 1–17. doi: 10.1186/s40168-020-00824-x
Yu, M., Li, Y., Banakar, S. P., Liu, L., Shao, C., Li, Z., et al. (2019). New metabolites from the co-culture of marine-derived actinomycete Streptomyces rochei MB037 and fungus Rhinocladiella similis 35. Front. Microbiol. 10:915. doi: 10.3389/fmicb.2019.00915
Yu, Y., Zeng, Y., Li, J., Yang, C., Zhang, X., Luo, F., et al. (2019). An algicidal Streptomyces amritsarensis strain against Microcystis aeruginosa strongly inhibits microcystin synthesis simultaneously. Sci. Total Environ. 650, 34–43. doi: 10.1016/j.scitotenv.2018.08.433
Zeng, Y., Wang, J., Yang, C., Ding, M., Hamilton, P. B., Zhang, X., et al. (2021). A Streptomyces globisporus strain kills Microcystis aeruginosa via cell-to-cell contact. Sci. Total Environ. 769:144489. doi: 10.1016/j.scitotenv.2020.144489
Keywords: bacteria, interactions, diversity, ecology, survival mechanism, prokaryotic predation
Citation: Ibrahimi M, Loqman S, Jemo M, Hafidi M, Lemee L and Ouhdouch Y (2023) The potential of facultative predatory Actinomycetota spp. and prospects in agricultural sustainability. Front. Microbiol. 13:1081815. doi: 10.3389/fmicb.2022.1081815
Received: 27 October 2022; Accepted: 28 December 2022;
Published: 25 January 2023.
Edited by:
Akram Sadeghi, Agricultural Biotechnology Research Institute of Iran, IranCopyright © 2023 Ibrahimi, Loqman, Jemo, Hafidi, Lemee and Ouhdouch. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yedir Ouhdouch,  eW91aGRvdWNoQGdtYWlsLmNvbQ==
eW91aGRvdWNoQGdtYWlsLmNvbQ==
 Manar Ibrahimi
Manar Ibrahimi Souad Loqman
Souad Loqman Martin Jemo
Martin Jemo Mohamed Hafidi
Mohamed Hafidi Laurent Lemee6
Laurent Lemee6 Yedir Ouhdouch
Yedir Ouhdouch